- 1Lab of Animal Disease Prevention & Control and Animal Model, Hunan Provincial Key Laboratory of Protein Engineering in Animal Vaccines, College of Veterinary Medicine, Hunan Agricultural University (HUNAU), Changsha, China
- 2North Guangdong Collaborative Innovation and Development Center of Pig Farming and Disease Control, Shaoguan University, Shaoguan, China
The emergence and widespread of porcine circovirus-associated diseases (PCVADs), mainly caused by porcine circovirus type 2 (PCV2), threatens the Chinese swine industry. In this study, to investigate the recent prevalence of PCV2 in northern Guangdong Province of China, 573 tissue samples from 132 pig farms were collected during 2016–2021 and analyzed via PCR. Overall, 51.38% (297/573, 95%CI 47.74–55.92) samples were tested PCV2 positive. The detection rate of PCV2 was significantly lower in samples collected before 2016-2018 than after the outbreak of African Swine Fever (2019-2021), being 59.85% (158/264, 95%CI 53.94–65.76) and 41.47% (141/340, 95%CI 36.43–46.71), respectively. On the other end, the genetic characteristics of 26 PCV2 strains were further analyzed. These PCV2 strains belonged to three genotypes, including PCV2a, PCV2b, and PCV2d. Specifically, the predominant genotype prevalent during two periods (2016–2018 and 2019–2021) wasPCV2b (81.82%, 9/11) and PCV2d (80.0%, 12/15), respectively. The results above illustrated the high prevalence and the genetic evolution feature of PCV2 in Guangdong Province in recent years.
Introduction
Porcine circoviruses (PCVs) are small, circular, single-stranded DNA viruses belonging to the genus of Circovirus of the family Circoviridae (1). Currently, four genotypes of PCVs have been identified, termed porcine circovirus type 1 (PCV1), PCV2, PCV3, and PCV4 (2). Since the first identification of PCV2 in Canada in the 1990s, this pathogen has been considered the primary causative agent of porcine circovirus-associated diseases (PCVADs) (3, 4). PCVADs are mainly characterized by postweaning multisystemic wasting syndromes (PMWS), such as respiratory distress in piglets, slow growth in fattening pigs, reproductive failures in sows, etc., which threatens the development of pig industry worldwide (3, 4).
The PCV2 genome is 1,766–1,768 nucleotide (nt) in length, which mainly comprises two open reading frames (ORFs). The ORF1 encodes the replication-related proteins (Rep and Rep'), while the ORF2 encodes the capsid protein (Cap), which induces the neutralization antibodies production and viral entry (3, 5). According to the genomic characteristics of PCV2 strains, they are divided into eight subtypes (PCV2a-PCV2h) (6). PCV2 strains prevalent before 2008 belonged to three genotypes (PCV2a-PCV2c). The PCV2d was identified in 2010, and then prevalent in pig populations worldwide since 2012 (4). In recent years, other novel PCV2 genotypes (PCV2e-PCV2h) have been identified (5). Other than these, owing to the high sequence similarity among different PCV2 genotypes, the novel recombinant strains generated from different PCV2 genotype strains were also documented (7, 8).
A number of studies have investigated the epidemiology and genetic features of PCV2 in certain regions of China (9–11), which confirmed the rapid evolution of PCV2, and the genotype shift from PCV2b to PCV2d in China. The prevalence and genetic characteristics of PCV2 in Guangdong Province have been documented in previous studies (12, 13). However, the corresponding information for recent years is still missing, particularly after the outbreak of African Swine Fever (ASF). In this study, polymerase chain reaction (PCR) was performed to investigate the epidemiological characteristics of PCV2 in Guangdong Province from 2016 to 2021. Moreover, the complete genomes of 26 PCV2 strains from different periods were sequenced and analyzed.
Materials and methods
Sample collection
From April 2016 to September 2021, 573 tissue samples (lung, tonsil, and lymph node) were collected from 132 pig farms across the northern Guangdong Province (Shaoguan, Qingyuan, Heyuan, Zhaoqing, Guangzhou, and Meizhou cities). Most diseased pigs from these farms showed clinical symptoms characterized by PMWS and/or porcine dermatitis and nephropathy syndrome (PDNS) or reproductive failures. Tissue samples with detailed information including collection sites, collection dates, and clinical symptoms were sent to Shaoguan University for further processing.
DNA extraction and PCV2 detection
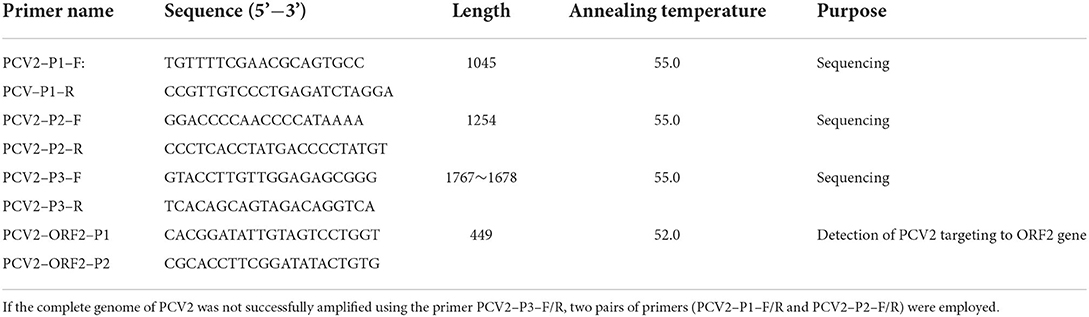
Viral DNA genomes were extracted from the tissue sample using commercial kits (GDSBio, Guangdong, China) according to the manufacturer's instructions. Subsequently, the presence of PCV2 nucleic acid was detected by PCR with primers (PCV2-ORF2-P1 and PCV2-ORF2-P2) as described previously (14). The PCR products were analyzed in 1% agarose gel electrophoresis, in which the samples with the expected DNA bands, approximately 450 bp, were noted as PCV2-positive samples.
PCV2 complete genome sequencing
According to the collected regions and years of the positive samples, 26 PCV2-positive samples were selected to amplify the complete genome of the virus with three pairs of primers (Table 1). Each PCR reaction (50 μl) contained 25 μl 2 × Taq Master mix (Takara Biotechnology, Dalian, China), 12.0 μl of each primer (10 pmol), 3.0 μl DNA template, and 20.0 μl sterilized water. The PCR amplification was performed with the following steps: 95°C for 5 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60/90 s; followed by 72°C for 7 min. The PCR products were purified, cloned into the pUCm-T vector, and sequenced by Sangon Biotech Co. Ltd (Shanghai, China). The sequences of these novel PCV2 strains were submitted to the GenBank (ON361010-ON361035).
Bioinformatics analyses
The complete genomes of 20 reference PCV2 strains (including PCV2a, PCV2b, PCV2c, PCV2d, PCV2e, and PCV2h) were downloaded from the GenBank database (Supplementary Table S1). The nucleotide sequences and their corresponding amino acid sequences (Cap and Rep) variations of 26 novel PCV2 strains and reference strains were analyzed via the Lasergene DNAStar software. Phylogenetic trees were generated based on the complete genome and ORF2 gene sequences using the neighbor-joining method in the MEGA 7.0 software (Kimura 2-parameter model, 1,000 bootstrap replications).
Data analyses
The statistical significance of the detection rates of PCV2 in pigs among different groups was analyzed using the chi-square test in the SPSS 21.0 software (SPSS Inc., Chicago, IL, USA), in which, P-values < 0.05 were taken as statistically significant. Meanwhile, the minimum infection rate (MIR) with 95% CI was determined via the SPSS 21.0 software.
Results
The epidemiology of PCV2 in northern Guangdong Province from 2016 to 2021
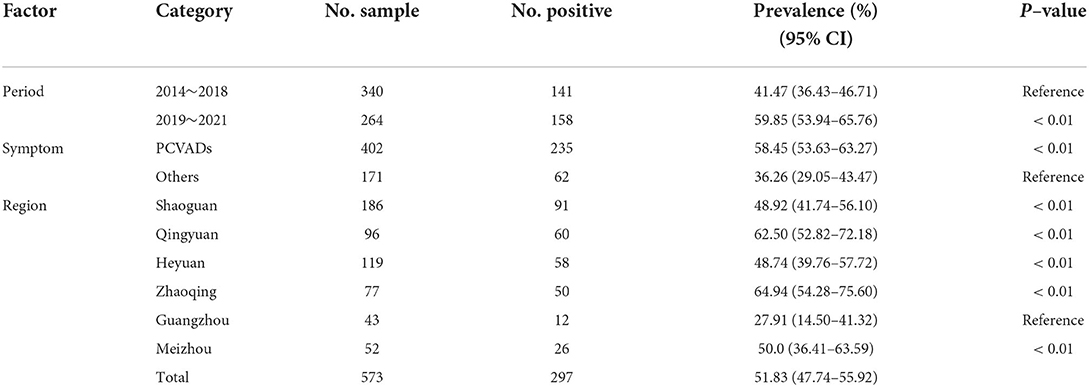
In this study, a total of 573 tissue samples from 132 pig farms were collected from northern Guangdong Province for the PCV2 nucleic acids test by conventional PCR. The results showed that 98 out of 132 (74.24%) investigated pig farms were tested PCV2-positive. Overall, 297 tissue samples (51.83%, 95%CI 47.74–55.92) were PCV2-positive, with the positive rates of PCV2 in different regions varying from 27.91–64.94% (Table 2). Moreover, The detection rate of PCV2 was significantly lower in samples collected before 2016–2018 than after the outbreak of African Swine Fever (2019–2021), being 59.85% (158/264, 95%CI 53.94–65.76) and 41.47% (141/340, 95%CI 36.43–46.71), respectively (Table 2). Additionally, the positive rate of PCV2 among pigs with PCVADs (58.45%, 95%CI 53.63–63.27) was significantly higher than in pigs without PCVADs (36.26%, 95%CI 29.05–43.47) (Table 2).
Genome sequence analysis
To further investigate the genetic features of recent PCV2 strains prevalent in Guangdong Province, 26 PCV2 strains were randomly selected among PCV2-positive samples from different regions, whose complete genome sequences were amplified, sequenced, and analyzed (Table 3). The complete genomes of all 26 novel PCV2 strains were 1,764–1,767 nt in length. Particularly, the length of the ORF1 gene encoding the Rep and ORF2 gene encoding the Cap was 945 nt and 702–705 nt, respectively. Pairwise-sequence comparisons among 26 novel isolates ranged from 95.9–99.9% (complete genome), 97.0–99.7% (ORF1), and 94.0–100.0% (ORF2) at the nt level, respectively, and 94.6–99.4% (Rep protein) and 98.6–100.0% (Cap protein) at amino acid (aa) level, respectively.
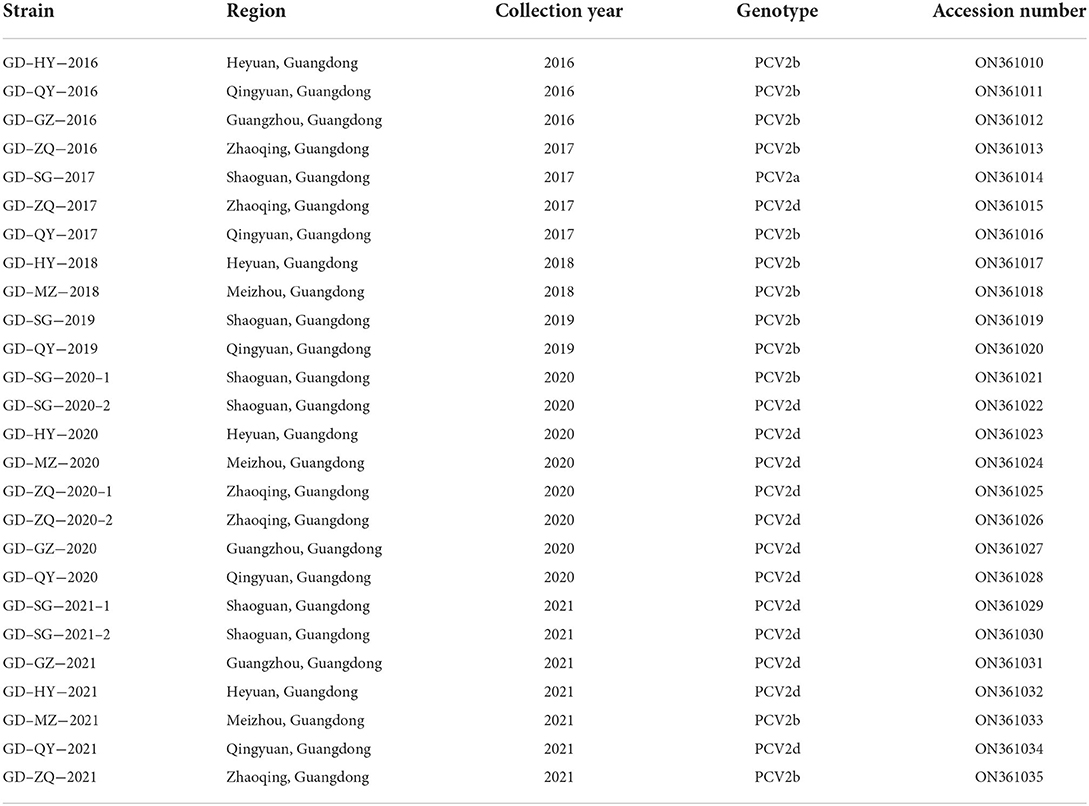
Table 3. Detail information of PCV2 strains obtained in this study, including strain name, collection year, isolation region, genotype, and GenBank accession numbers.
Phylogenetic analysis
Phylogenetic trees were generated according to the complete genome and ORF2 sequences of the 26 PCV2 strains in this study and 20 reference strains from GenBank. The results showed that the 26 Guangdong strains were divided into three sub-genotypes (PCV2a, PCV2b, and PCV2d) (Figures 1A,B), 12 of 26 (46.15%) isolates belonged to the genotype 2b; and half of all isolates belonged to the genotype 2d; only one isolate, GD-SG-2017, was clustered with PCV2a strains. Remarkably, the PCV2a isolate (GD-SG-2017) and 9 of 12 PCV2b isolates were collected between 2016 and 2018, whereas 92.31% (12/13) PCV2d strains were isolated during 2019-2021.
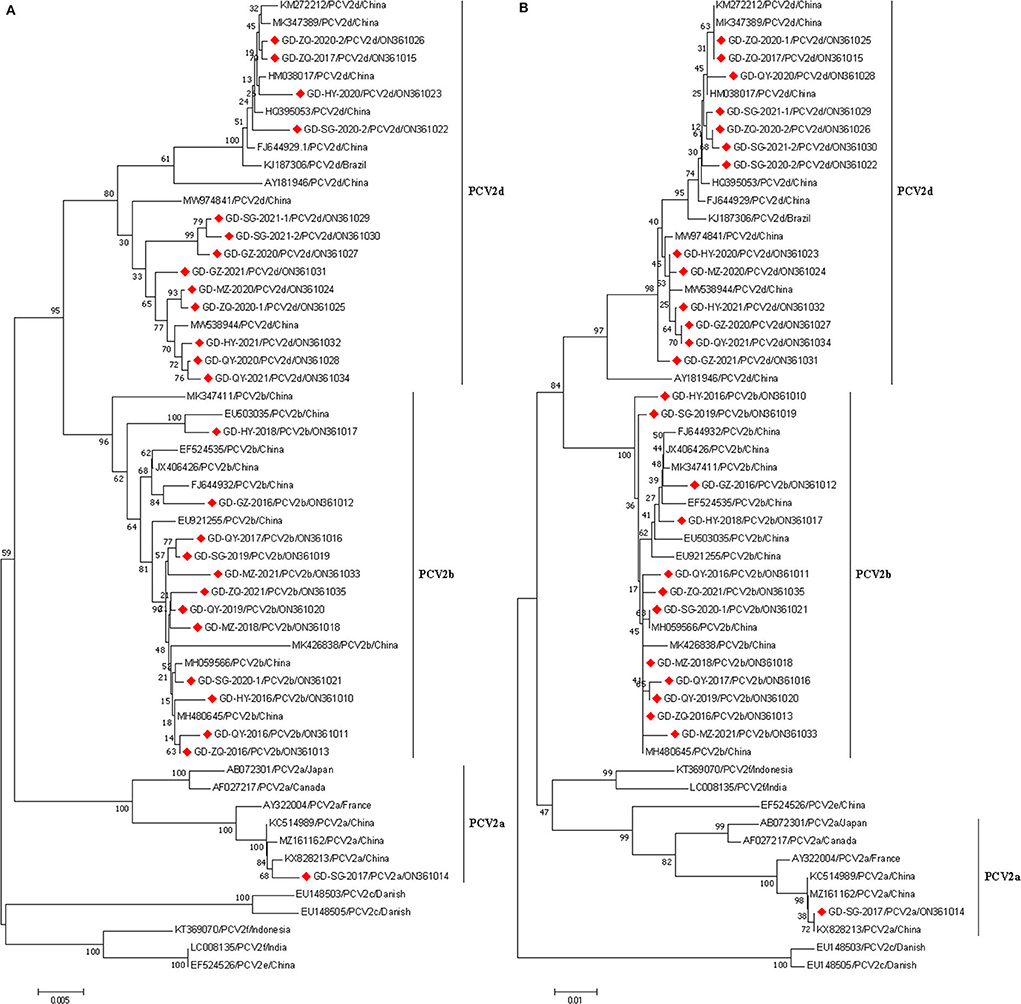
Figure 1. Phylogenetic analysis based on the complete genome (A) and ORF2 gene (B) sequences of 26 PCV2 strains obtained in this study and other reference strains. phylogenetic trees were constructed using the neighbor-joining method with 1,000 bootstrap replicates in MEGA7.0 software. Red squares represented PCV2 isolates obtained in this study.
Analysis of Cap amino acid sequences
To investigate the sequence characteristics of the Cap aa sequences obtained in this study, the aa sequence alignment was performed between the 26 PCV2 strains and the 20 reference PCV2 strains. The results showed that the lengths of Cap sequences of 26 PCV2 strains were 233 aa (12 PCV2b isolate and GD-GZ-2021) or 234 aa (12 PCV2d isolates and GD-SG-2017). As shown in Supplementary Table S2, a total of 8 and 4 aa substitutions were found in Cap among 12 PCV2b and 13 PCV2d isolates, respectively. To be more specific, these substitutions were located at sites 25 (R → H), 59 (K → R), 63 (R → K),84 (G → E), 131 (T → P), 151 (T → P), 173 (Y → H), and 190 (T → A) in the Cap of PCV2b isolates; and 30 (V → L), 169 (G → R), 206 (I → K), and 233 (N → K) in the Cap of PCV2d isolates, respectively. Remarkably, a series of unique aa substitutions in the Cap, for instance, the substitutions at sites 25, 59, and 63 were found in GD-GZ-2016 isolate; and site 151 in GD-QY-2016 isolate; and 59, 63, 84, 173, and 190 in GD-HY-2018 isolate, respectively.
Discussion
PCV2 has been prevalent in pig populations in China for many years. The disease (PCVAD) caused by PCV2 is considered a major factor threatening the pig industry (11). Vaccine pressure, viral evolution, natural selection, and international pig transportation contribute to the rapid evolution rate of PCV2 (15). In recent years, two genotype shifts of PCV2 have been observed in China. PCV2a was the predominant prevalent genotype before 2003, which has been gradually replaced by PCV2b from 2003 to 2010. Owing to the high prevalence of PCV2 and the widely applied PCV2 vaccines since 2010, PCV2d is an emerging genotype prevalent in China (4, 16). Moreover, the co-prevalence of multiple PCV2 genotypes was often observed in the same region, even on the same pig farm (4, 7, 14, 17). Thus, an investigation of the prevalence and genetic characteristics of PCV2 will provide scientific evidence for the prevention and control of PCV2.
In this study, 573 samples from pigs were collected to investigate the epidemiological characteristics of PCV2 in the northern Guangdong Province of China. Several features were summarized: 1) high detection rate of PCV2 (51.83%, 297/573) was observed in nearly 75% of the investigated pig farms. The PCV2-positive rate among these samples in northern Guangdong Province was similar to other areas in China, such as Henan (62.4%, 73/117) (18), Shanghai (57.78%, 115/199) (4), and Yunnan (60.93%, 170/279) (11), but higher than that in Shandong Province (36.98%, 490/1325) (19); taken together, these bodies of evidence showed PCV2 high prevalence in China; 2) the positive rate of PCV2 among specimens collected before (2014–2018) was lower than that after (2019–2021) the outbreak of ASF. Owing to the prevalence of ASF in China (from 2019 to 2021), PCV2-positive pigs were introduced into pig farms to keep the breeding scale. Nevertheless, some pig farmers mainly focused on ASF prevention, but neglected PCV2 prevention, which resulted in the high prevalence of PCV2 from 2019 to 2021 in northern Guangdong province; 3) the PCV2 detection rate among pigs with PCVADs was higher than those without PCVADs. To better prevent the PCV2 spread, the PCV2 positive pigs without PCVADs should not be neglected.
Currently, PCV2 strains have been divided into eight genotypes (PCV2a–h) based on their genomic characteristics, in which, multiple genotypes were prevalent in China (4, 16). More recently, PCV2d has been considered a novel emerging major genotype prevalent in Chinese pig populations (15, 20). Among 26 PCV2 strains obtained in this study, only three genotypes were identified, PCV2a, PCV2b, and PCV2d, the proportions of which were 3.85% (1/26), 46.15% (12/26), and 50.0% (13/26), respectively, suggesting that PCV2b and PCV2d, rather than PCV2a, predominated in these investigated regions. According to the findings from other studies, more than 30% of PCV2 strains prevalent in China before 2018 belonged to the PCV2d genotype (16, 18), particularly, PCV2d has become the major dominant genotype circulating in Shandong Province from 2015 to 2018 (19). However, the results in this study showed that the predominant genotypes of PCV2 during 2014–2018 and 2019–2021 were PCV2b and PCV2d, the proportions of which were 81.82% (9/11) and 80.0% (12/15) in northern Guangdong Province, respectively. Guangdong Province is a major pig breeding area with millions of pigs transferred out, whereas very few pigs were introduced into this province before 2018. These factors resulted in the relatively stable PCV2b genotype in these areas. However, after the outbreak of ASF in China, a large number of pigs were introduced into Guangdong Province, which might lead to the PCV2 genotype shifting from PCV2b to PCV2d.
The PCV2 Cap is the sole structural protein that is responsible for a series of biological processes, such as virus entry into host cells, replication, and activation of host immune responses (21, 22). Therefore, the aa mutations in Cap may determine viral biological characteristics (23). In this study, a series of aa substitutions were found in the Cap protein (Supplementary Table S2), such as the sites of 30 (V → L) (n = 6), and 59 (K → R) (n = 2), and 63 (R → K) (n = 2). It has been confirmed that the aa residues 1–41 of Cap participate in viral nuclear localization (24), while the aa residues 47–63 are crucial for PCV2 epitope recognition (25). Further experiments will be performed to investigate the effects of these aa substitutions on PCV2 infection.
In conclusion, this study revealed the epidemiology and genetic characteristics of PCV2 in recent years in northern Guangdong Province of China and indicated the severe prevalence of PCV2 in these regions. Moreover, this study confirmed the prevalence of multiple PCV2 genotypes in Guangdong Province from 2016 to 2021, while, currently the PCV2d has become the predominant genotype. These findings highlight the importance to investigate the genetic features of currently prevalent PCV2 strains, and to develop novel vaccines to control PCV2d.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Conceptualization: ZD. Methodology: WN, ST, JW, and HH. Software: WN and ST. Formal analysis and investigation: WN, ST, JW, GP, and HH. Resources and original draft writing: WN. Review and revised and funding acquisition: WN and ZD. All authors participated in the article editing and approved the final manuscript.
Funding
This work was supported by Natural Science Foundation of Hunan Province (2021JJ30316) and the Project of Swine Innovation Team in Guangdong Modern Agricultural Research System (2021KJ126).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.932612/full#supplementary-material
References
1. Tischer I, Gelderblom H, Vettermann W, Koch MAA. Very small porcine virus with circular single-stranded DNA. Nature. (1982) 295:64–6. doi: 10.1038/295064a0
2. Ge M, Ren J, Xie YL, Zhao D, Fan FC, Song XQ, et al. Prevalence and genetic analysis of Porcine circovirus 3 in China from 2019 to 2020. Front Vet Sci. (2021) 8:773912. doi: 10.3389/fvets.2021.773912
3. Yu WT, Sun YA, He Q, Sun CY, Dong T, Zhang LH, et al. Mitochondrial localization signal of porcine circovirus type 2 capsid protein plays a critical role in Cap-induced apoptosis. Vet Sci. (2021) 8:272. doi: 10.3390/vetsci8110272
4. Kang L, Wahaab A, Shi K, Mustafa BE, Zhang Y, Zhang JJ, et al. Molecular epidemic characteristics and genetic evolution of porcine circovirus type 2 (PCV2) in swine herds of Shanghai, China. Viruses. (2022) 14:289. doi: 10.3390/v14020289
5. Li N, Liu J, Qi JL, Hao F, Xu L, Guo KK, et al. Genetic diversity and prevalence of porcine circovirus type 2 in China during 2000-2019. Front Vet Sci. (2021) 8:788172. doi: 10.3389/fvets.2021.788172
6. Franzo G, Segalés J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE. (2018) 13:e0208585. doi: 10.1371/journal.pone.0208585
7. Qu TL, Li RC, Yan MJ, Luo BY, Yang TT, Yu XL, et al. High prevalence of PCV2d in Hunan Province, China: a retrospective analysis of samples collected from 2006 to 2016. Arch Virol. (2018) 163:1897–906. doi: 10.1007/s00705-018-3823-9
8. Wen LB, He KW. Genomic rearrangement and recombination of porcine circovirus type 2 and porcine circovirus-like virus P1 in China. Front Vet Sci. (2021) 8:736366. doi: 10.3389/fvets.2021.736366
9. Li LM, Yuan WZ, Guo HY, Ma ZJ, Song QY, Wang XB, et al. Prevalence and genetic variation of porcine circovirus type 2 in Hebei, China from 2004 to 2014. Gene. (2016) 586:222–7. doi: 10.1016/j.gene.2016.04.014
10. Xu T, Zhang YH, Tian RB, Hou CY, Li XS, Zheng LL, et al. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in central China. Infect Genet Evol. (2021) 94:105016. doi: 10.1016/j.meegid.2021.105016
11. Lv NC, Zhu L, Li WG, Li ZL, Qian QS, Zhang TY, et al. Molecular epidemiology and genetic variation analyses of porcine circovirus type 2 isolated from Yunnan Province in China from 2016-2019. BMC Vet Res. (2020) 16:96. doi: 10.1186/s12917-020-02304-8
12. Wei CY, Zhang MZ, Chen Y, Xie JX, Huang Z, Zhu WJ, et al. Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect Genet Evol. (2013) 17:87–92. doi: 10.1016/j.meegid.2013.03.041
13. Zhai SL, Chen SN, Liu W, Li XP, Deng SF, Wen XH, et al. Molecular detection and genome characterization of porcine circovirus type 2 in rats captured on commercial swine farms. Arch Virol. (2016) 161:3237–44. doi: 10.1007/s00705-016-3004-7
14. Hu Y, Zhan Y, Wang DD, Xie XH, Liu TB, Liu W, et al. Evidence of natural co-infection with PCV2b subtypes in vivo. Arch Virol. (2017) 162:2015–20. doi: 10.1007/s00705-017-3303-7
15. Xu PL, Zhao Y, Zheng HH, Tian RB, Han HY, Chen HY, et al. Analysis of genetic variation of porcine circovirus type 2 within pig populations in central China. Arch Virol. (2019) 164:1445–51. doi: 10.1007/s00705-019-04205-0
16. Wang HJ, Gu JY, Xing G, Qiu XH, An ST, Wang YX, et al. Genetic diversity of porcine circovirus type 2 in China between 1999-2017. Transbound Emerg Dis. (2019) 66:599–605. doi: 10.1111/tbed.13040
17. Ouyang T, Zhang XW, Liu XH, Ren LZ. Co-infection of swine with porcine circovirus type 2 and other swine viruses. Viruses. (2019) 11:185. doi: 10.3390/v11020185
18. Zheng GM, Lu QX, Wang FY, Xing GX, Feng H, Jin QY, et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet Res. (2020) 16:6. doi: 10.1186/s12917-019-2193-1
19. Ma ZC, Liu MD, Liu ZH, Meng FL, Wang HY, Cao LL, et al. Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong Province in China from 2015 to 2018. BMC Vet Res. (2021) 17:17. doi: 10.1186/s12917-020-02718-4
20. Hou Z, Wang H, Feng Y, Song M, Li Q, Genetic LJ, et al. Variation and phylogenetic analysis of Porcine circovirus type 2 in China from 2016 to 2018. Acta Virol. (2019) 63:459–68. doi: 10.4149/av_2019_413
21. Huang Y, Chen XH, Long YZ, Yang L, Song WB, Liu JJ, et al. Epidemiological analysis from 2018 to 2020 in China and prevention strategy of porcine circovirus type 2. Front Vet Sci. (2021) 8:753297. doi: 10.3389/fvets.2021.753297
22. Zhan Y, Yu WT, Cai X, Lei XN, Lei HY, Wang AB, et al. The carboxyl terminus of the porcine circovirus type 2 capsid protein is critical to virus-like particle assembly, cell entry, and propagation. J Virol. (2020) 94:e00042–e00020. doi: 10.1128/JVI.00042-20
23. Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS, et al. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol. (2000) 81:2281–7. doi: 10.1099/0022-1317-81-9-2281
24. Hou Q, Hou SH, Chen Q, Jia H, Xin T, Jiang YT, et al. Nuclear localization signal regulates porcine circovirus type 2 capsid protein nuclear export through phosphorylation. Virus Res. (2018) 246:12–22. doi: 10.1016/j.virusres.2017.12.012
Keywords: porcine circovirus type 2, epidemiology, complete genome, genetic characteristics, Guangdong Province
Citation: Nan W, Wu J, Hu H, Peng G, Tan S and Deng Z (2022) Prevalence and genetic diversity of porcine circovirus type 2 in northern Guangdong Province during 2016–2021. Front. Vet. Sci. 9:932612. doi: 10.3389/fvets.2022.932612
Received: 30 April 2022; Accepted: 15 July 2022;
Published: 10 August 2022.
Edited by:
Zhou Mo, Heilongjiang University, ChinaReviewed by:
Fei Gao, Shanghai Veterinary Research Institute (CAAS), ChinaKayode Olayinka Afolabi, Anchor University Lagos, Nigeria
Copyright © 2022 Nan, Wu, Hu, Peng, Tan and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhibang Deng, emJhbmdkQGh1bmF1LmVkdS5jbg==
 Wenjin Nan
Wenjin Nan Jingbo Wu2
Jingbo Wu2