- 1Molecular Medicine Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Student Research Committee, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran
- 4Department of Molecular Genetics, School of Biological Sciences, Tarbiat Modares University, Tehran, Iran
- 5Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 6Department of Social Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
- 7Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 8Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
LncRNAs act as part of non-coding RNAs at high levels of complex and stimulatory configurations in basic molecular mechanisms. Their extensive regulatory activity in the CNS continues on a small scale, from the functions of synapses to large-scale neurodevelopment and cognitive functions, aging, and can be seen in both health and disease situations. One of the vast consequences of the pathological role of dysregulated lncRNAs in the CNS due to their role in a network of regulatory pathways can be manifested in Alzheimer's as a neurodegenerative disease. The disease is characterized by two main hallmarks: amyloid plaques due to the accumulation of β-amyloid components and neurofibrillary tangles (NFT) resulting from the accumulation of phosphorylated tau. Numerous studies in humans, animal models, and various cell lines have revealed the role of lncRNAs in the pathogenesis of Alzheimer's disease. This scoping review was performed with a six-step strategy and based on the Prisma guideline by systematically searching the publications of seven databases. Out of 1,591 records, 69 articles were utterly aligned with the specified inclusion criteria and were summarized in the relevant table. Most of the studies were devoted to BACE1-AS, NEAT1, MALAT1, and SNHG1 lncRNAs, respectively, and about one-third of the studies investigated a unique lncRNA. About 56% of the studies reported up-regulation, and 7% reported down-regulation of lncRNAs expressions. Overall, this study was conducted to investigate the association between lncRNAs and Alzheimer's disease to make a reputable source for further studies and find more molecular therapeutic goals for this disease.
Introduction
Alzheimer's disease (AD) is a disease known for its clinical symptoms, including gradual memory loss and language problems and cognitive impairments such as the inability to solve problems and spatial cognition and difficulty changing to mood (Cacace et al., 2016; Zhang and Wang, 2021). Accumulation of dense and insoluble beta-amyloid (Aβ) fragments outside and around neurons and neurofibrillary tangles (NFTs) resulted from the accumulation of hyper-phosphorylated Tau proteins inside cells are neuropathological symptoms of AD (Tiraboschi et al., 2004; McKhann et al., 2011). These lesions lead to neuronal degeneration, loss of synapses, and reduced neurotransmitter transport (Graham et al., 2017). AD dementia may affect 13.8 million Americans aged 65 and up by the middle of the century (Alzheimer's Association, 2020), and causes 50–75% of dementias (Association As, 2019). In terms of the time of onset, the disease is divided into two forms of early-onset AD (EOAD) and late-onset AD (LOAD). EOAD is diagnosed in patients under the age of 65, with a more significant genetic influence being reported for this form. LOAD accounts for 90% of cases seen in patients over 65 (Dursun et al., 2008; Wingo et al., 2012; Cacace et al., 2016). The genetic and etiological dimensions of the disease, focus on several specific genes, including amyloid precursor protein (APP), presenilin-1 (PSEN1), and presenilin-2 (PSEN2). Highly influential mutations in these genes can increase the susceptibility to AD, particularly the EOAD (Atri, 2019). Meanwhile, we should not forget the effect of non-coding RNAs in the pathogenesis of the disease.
Long non-coding RNAs (lncRNAs) are part of non-coding RNAs with sizes between 200 nucleotides and several kbs and high tissue specificity. They have fundamental role in regulation of gene expression (Zhou et al., 2021). According to the Annotations of the FANTOM5 project, about 28,000 lncRNA genes have been identified so far (Hon et al., 2017). Like mRNAs, lncRNAs are capped and polyadenylated and undergo the splicing process (Derrien et al., 2012). At the molecular level, lncRNAs play an essential role in transcription, translation, and regulation of gene expression, and chromatin remodeling and genomic imprinting (Statello et al., 2021), and at the biological level, they are one of the significant factors in the regulation of proliferation (Ponting et al., 2009), survival (Shen et al., 2015), and differentiation (Cesana et al., 2011). These non-coding RNAs are also involved in the pathogenesis and progression of AD due to their structural diversity and important biochemical properties (Idda et al., 2018).
Of the thousands of lncRNAs encoded in organs, around 40% of these lncRNAs are specifically expressed in brain tissue (Briggs et al., 2015). Many studies have shown an association between their expression dysregulation and many neurodegenerative diseases, including AD (Ni et al., 2017; Lyu et al., 2019). Studies performed on the 3xTg-AD model mice brain show that the expression of hundreds of lncRNAs is significantly changed compared with the control group (Zhou and Xu, 2015). Transcriptome analysis studies on human post-mortem brain tissues show changes in the expression levels of several lncRNAs in AD patients (Cao et al., 2019). Overall, both animal and human studies confirm the potential effect of lncRNAs on AD. To date, many studies have been done on AD and the physiopathology of the disease. On the other hand, more and more attention has been paid to lncRNAs, their structure, and their effect on AD development, progression, or treatment. In the present study, our focus has been on conducting a systematic scoping review of all clinical studies to summarize these studies and strengthen the link between the effect of lncRNAs on AD.
Methods
The General Framework for Review
The strategy for writing this article is based on the method proposed by Arksey and O'Malley (2005). This strategy was later improved by Levac et al. (2010) and Colquhoun et al. (2014). In this review, five steps of the 6-step framework are followed, which include:
1. Identifying the research question.
2. Search strategy.
3. Study selection.
4. Charting the data.
5. Collating, summarizing, and reporting the results.
Consultation is the optional sixth step and is not included in this article. The Preferred Reporting Items for Systematic Reviews and Meta-Analysis Extension for Scoping Reviews (PRISMA-ScR) Checklist is used to consider and observe two essential factors of clarity transparency in writing the article (Tricco et al., 2018).
Identifying the Research Question
Our article was guided by the following questions in order to study, review, and discuss all original studies on lncRNAs in AD:
• What studies have been done on lncRNAs in AD?
• What are the results and findings of these studies?
Search Strategy
Seven databases were searched for access to the publications: Pubmed, Scopus, Cochrane, Google Scholar, Embase, Web of Science, and ProQuest. The search did not apply a filter restricting the date, language, subject, or publication type. Review publications were also revised to reduce the possibility of missing related articles. “Alzheimer Disease” and “RNA, long non-coding” keywords were medical subject heading (MeSH) used in search strategy in PubMed and Embase database. The last search was conducted on APR 19, 2021. The references were managed using EndNote X8.1.
Study Selection
Studies of AD concerning lncRNAs in humans, cell lines, and animal model studies were screened from publications obtained during the search process. All publication types were assessed, including journal articles, conference presentations, Erratum, conference abstracts, and reports. The screening was done in two stages by two reviewers (MRA, MH) separately. At this stage, the titles and abstracts of the articles were examined according to Table 1. The article's full text was reviewed, and irrelevant articles were deleted, and the articles remained utterly consistent with the research questions. Any contradiction in agreement with the opinion of the third person was resolved.
Charting the Data
After reaching the final articles that fulfill the research questions, we developed the data-charting. Study variables were created using the following headings: author's name, year of publication, country, type of study, human samples, animal models, cell lines, lncRNAs, methods, major findings, and references. Two reviewers (MRA, MH) separately extracted data from articles based on charts.
Collating, Summarizing, and Reporting the Results
Quantitative and qualitative analysis was accomplished on the findings from the publications represented in tables and charts. A descriptive numerical summary of the extent, nature, and distribution of the studies was reviewed in the quantitative analysis section, and the presented data affirmation on the broader context suggested by Levac et al. (2010), conducted in a narrative review.
Results
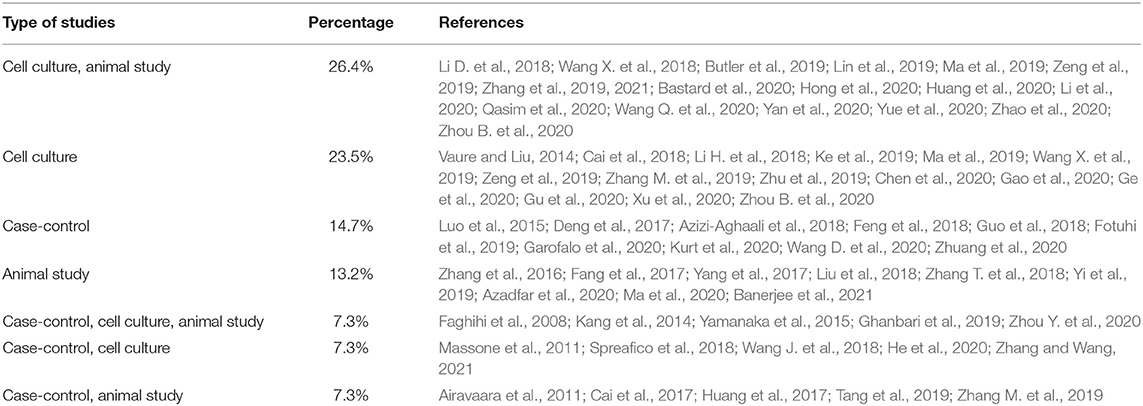
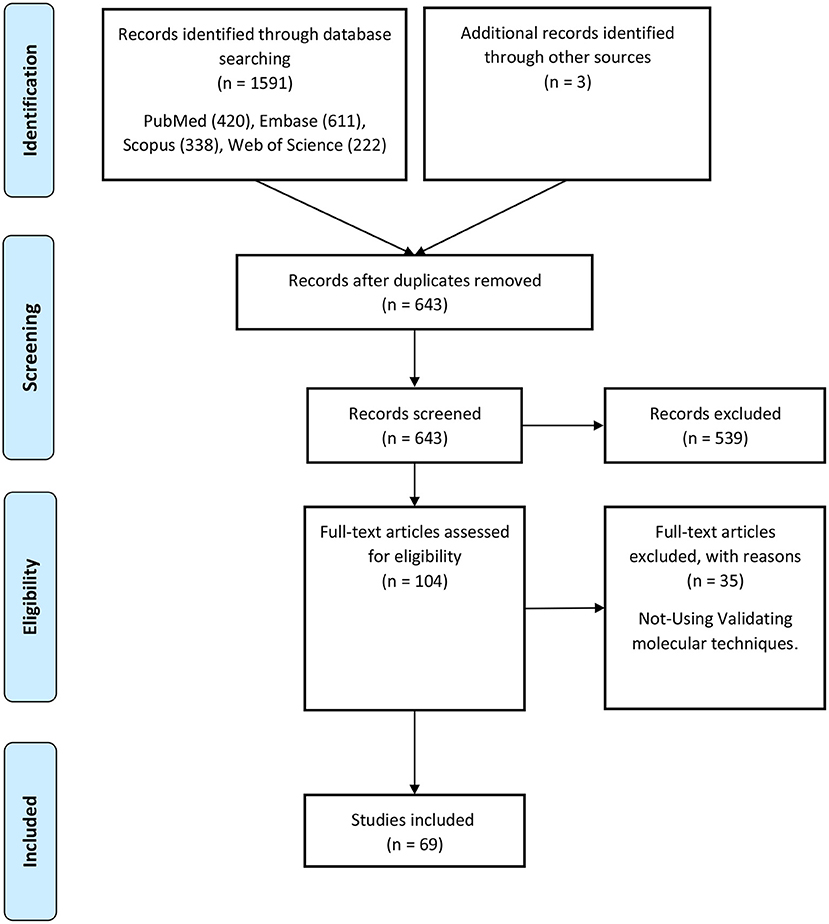
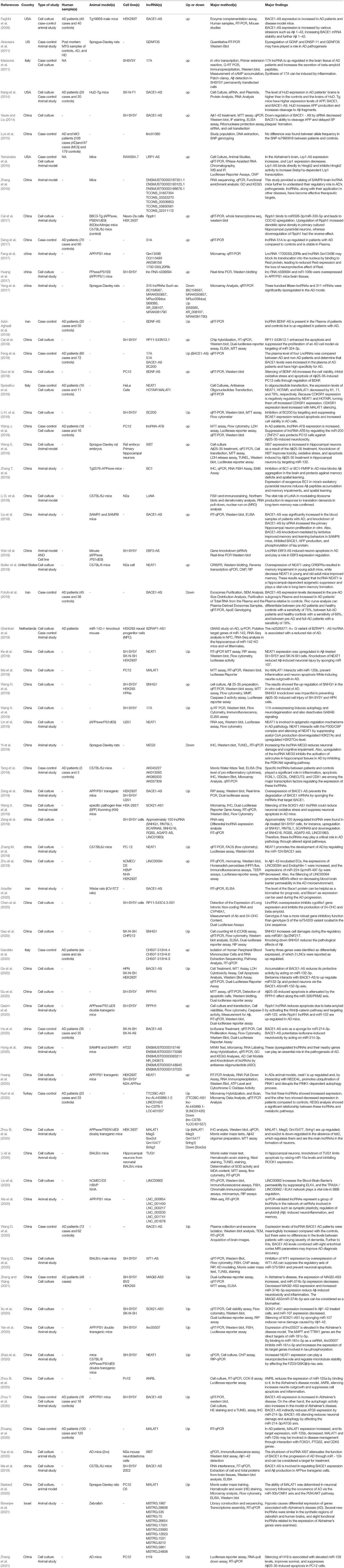
A keyword search in seven databases yielded 1,591 records. In the meantime, three records were identified from other sources and added to the total number of articles. A total of 951 duplicate records were identified and deleted by Endnote software, and the total number of articles reached 643. After reviewing the titles and abstracts of the articles, 104 articles based on the research question were selected. Since it is impossible to select the exact desired studies from the abstract and the title alone, and the full text of the articles needs to be inspected, at this stage, by reviewing the full text of 104 articles, 69 articles were eligible to be included in Table 2 for the charting data stage. The process of selecting eligible articles and studies is described in detail in Figure 1. Eligible studies have been published from 2008 to 2021. Table 2 was designed to rank studies from top to bottom for faster access to article division based on the frequency of studies. Based on the mentioned number, 744 samples of AD patients and 771 healthy controls were included in these studies. In most cases, the sex of patients and controls is not mentioned. Mice were used as the model in 36 animal studies, and zebrafish was used in one study. There are 16 different cell lines used in these studies, including SH-SY5Y in 16 studies (Massone et al., 2011; Vaure and Liu, 2014; Huang et al., 2017, 2020; Cai et al., 2018; Li H. et al., 2018; Ke et al., 2019; Ma et al., 2019; Wang X. et al., 2019; Zeng et al., 2019; Zhang M. et al., 2019; Chen et al., 2020; Qasim et al., 2020; Wang Q. et al., 2020; Xu et al., 2020; Yan et al., 2020; Zhao et al., 2020; Zhou Y. et al., 2020; Zhang and Wang, 2021), HEK293 in 12 studies (Faghihi et al., 2008; Cai et al., 2017; Ghanbari et al., 2019; Ke et al., 2019; Zeng et al., 2019; Zhang et al., 2019; Zhu et al., 2019; Ge et al., 2020; Huang et al., 2020; Zhou B. et al., 2020; Zhang and Wang, 2021), PC12 in seven studies (Guo et al., 2018; Wang J. et al., 2018; Ma et al., 2019; Zhao et al., 2019; Bastard et al., 2020; Zhou B. et al., 2020; Zhang et al., 2021), SK-N-SH in 5 studies (Ke et al., 2019; Gao et al., 2020; Ge et al., 2020; He et al., 2020; Xu et al., 2020), N2A in five studies (Cai et al., 2017; Li D. et al., 2018; Butler et al., 2019; Huang et al., 2020; Yue et al., 2020), U251 in two studies (Lin et al., 2019; Zeng et al., 2019), Human peripheral neurons (HPNs) in two studies (Zeng et al., 2019; Ge et al., 2020), SK-N-F1 (Kang et al., 2014), RAW264.7 (Yamanaka et al., 2015), Hela (Spreafico et al., 2018), CHP212 (Gao et al., 2020), SK-N-AS (He et al., 2020), HT22 (Hong et al., 2020), BV2 (Zhang and Wang, 2021), and 20E2 (Ma et al., 2019) cell lines each were used in one study. The number and frequency of lncRNAs are shown in Figure 2. The following is a schematic view of the contribution of LncRNAs in studies and a comparison chart of up-regulated LncRNAs compared to down-regulated ones. Due to the large volume of methods and tests performed in these studies, only the major methods are mentioned. The distribution of studies is limited to only seven countries, in which China with 54 studies has a significant share (Vaure and Liu, 2014; Luo et al., 2015; Zhang et al., 2016, 2021; Cai et al., 2017, 2018; Deng et al., 2017; Fang et al., 2017; Huang et al., 2017, 2020; Yang et al., 2017; Feng et al., 2018; Guo et al., 2018; Li D. et al., 2018; Li H. et al., 2018; Liu et al., 2018; Wang J. et al., 2018; Wang X. et al., 2018, 2019; Zhang T. et al., 2018; Ke et al., 2019; Lin et al., 2019; Ma et al., 2019, 2020; Tang et al., 2019; Yi et al., 2019; Zeng et al., 2019; Zhang M. et al., 2019; Zhu et al., 2019; Bastard et al., 2020; Chen et al., 2020; Gao et al., 2020; Ge et al., 2020; He et al., 2020; Hong et al., 2020; Li et al., 2020; Qasim et al., 2020; Wang D. et al., 2020; Wang Q. et al., 2020; Xu et al., 2020; Yan et al., 2020; Yue et al., 2020; Zhao et al., 2020; Zhou B. et al., 2020; Zhou Y. et al., 2020; Zhuang et al., 2020; Zhang and Wang, 2021), followed by the United States with five studies (Faghihi et al., 2008; Airavaara et al., 2011; Kang et al., 2014; Yamanaka et al., 2015; Butler et al., 2019), Iran (Azizi-Aghaali et al., 2018; Fotuhi et al., 2019; Azadfar et al., 2020), and Italy (Massone et al., 2011; Spreafico et al., 2018; Garofalo et al., 2020) with three studies, and the Netherlands (Ghanbari et al., 2019) and Turkey (Kurt et al., 2020) and Israel (Banerjee et al., 2021) each with one study.
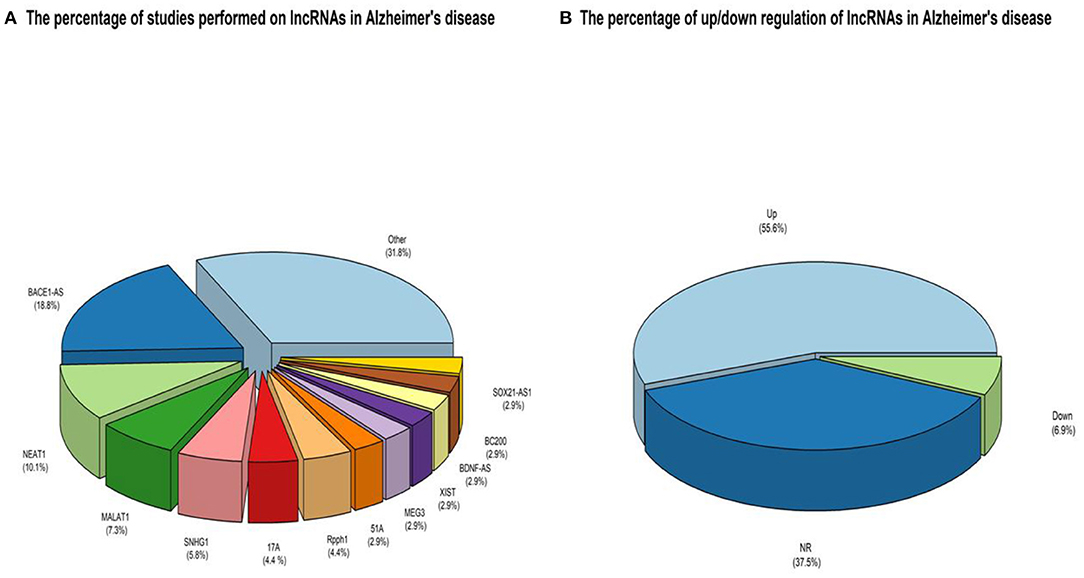
Figure 2. (A) An overview of the LncRNAs proportion is considered in the qualified studies. The other part is about lncRNAs that have only been studied once that can be accessed by referring to Table 3. (B) An overview of the ratio of up-regulated LncRNAs compared to down-regulated ones. NR also reflects non-reported studies of regulation levels.
The Perspective of Up-Regulated lncRNAs in AD
BACE1-AS
Aβ plays an essential role in AD. The cleavage of APP causes the production of Aβ by β-secretase 1 (BACE1) and γ-secretase. Compared to normal individuals, BACE1 levels are increased in AD patients. Hence, increased BACE1 expression plays a critical role in AD (Zeng et al., 2019). Increasing the expression of some lncRNAs such as BACE1-AS induces BACE1 expression. BACE1-AS, as an antisense RNA, can positively regulate BACE1 mRNA and protein expression in vivo and in vitro (Faghihi et al., 2008; Zhang et al., 2018). BACE1-AS plays a crucial role in BACE1 stability through RNA duplex formation and can positively regulate BACE1 and protein expression (Zeng et al., 2019). The cortex of patients with AD showed significantly higher levels of HuD, and an increase in APP, BACE1, BACE1AS, and Aβ compared to the cortical tissue of healthy individuals (Kang et al., 2014).
Additionally, up-regulation of BACE1-AS leads to the prevention of the binding of miRNA to BACE1. The knockdown of BACE1-AS leads to an increase in the level of miRNAs, a reduction in the level of BACE1 expression (Zeng et al., 2019). Zhang et al. reported that BACE1-AS was significantly increased in the blood samples of patients with AD, and knockdown of BACE1-AS by siRNA increased the primary hippocampal neuron proliferation in vitro. BACE1-AS knockdown improved memory and learning behaviors in SAMP8 mice, inhibited BACE1, APP production, and tau protein phosphorylation in hippocampi (Zhang et al., 2018). In the Plasma of AD patients and SK-N-SH and SK-N-AS cells treated with Aβ and isoflurane, the BACE1-AS was upregulated, while miR-214-3p was downregulated. Additionally, miR-214-3p improved cognitive status in mouse models by preventing autophagy and reducing apoptosis via suppressing Atg12 expression (He et al., 2020). Therefore, BACE1-AS can play a critical role in the monitoring and management of AD.
NEAT1
LncRNA nuclear enriched abundant transcript 1 (NEAT1) is highly evolutionarily conserved between humans and rodents, especially in the 5' region of the transcript (Hutchinson et al., 2007). Increased NEAT1 is associated with several cognitive and neurodegenerative disorders such as AD, schizophrenia, Huntington's, and Parkinson's. Studies in human and rodent samples have shown that NEAT1 may play an important role in neuroplasticity (Butler et al., 2019). NEAT1 is also involved in epigenetic regulation mechanisms in AD pathology (Lin et al., 2019). Wang et al. Found that NEAT1 interacts with the P300/CBP complex, and silencing of NEAT1 by suppressing acetyl-CoA production downregulated H3K27Ac and upregulated H3K27Cro level (Lin et al., 2019). Anderson et al. reported that NEAT1 is epigenetically involved in hippocampus-dependent, long-term memory formation, and knockdown of Neat1 resulted in extensive changes in gene expression and histone H3 lysine-9 dimethylations (H3K9me2) disturbances in the hippocampus of aged rodents (Butler et al., 2019). Huang et al. showed that NEAT1 is upregulated in the APP/PS1 transgenic mice and regulated the interaction between PINK1 and NEDD4L. Upregulation of NEAT1 induces ubiquitination and degradation of PINK1, leading to autophagy signaling, increased amyloid accumulation, and decreased cognition (Huang et al., 2020). In Aβ-treated SH-SY5Y and SK-N-SH cells, NEAT1 was increased, and its decrease inhibited Aβ-induced by reducing survival and p-Tau levels and promoting apoptosis. Also, NEAT1 acted as decoy and sponge of miR-107. miR-107 abundance was decreased in Aβ-treated cells (Ke et al., 2019). Hence, NEAT1 could provide new therapeutic approaches for AD.
SNHG1
Small nucleolar RNA host gene 1 (SNHG1) is increased in various diseases and plays an oncogenic role in cancer (Gao et al., 2020). Silencing of SNHG1 promoted neuronal autophagy and prevented cell death in Parkinson's disease. Also, knockdown of SNHG1 has effectively prevented Aβ (25-35)-induced cell injury of SH-SY5Y and HPN cells (Wang H. et al., 2019). Gao et al. reported that SNGH1 could induce ZFN217 expression to modulate Aβ-induced cell injury by sponging miR-361-3p (Gao et al., 2020). Additionally, a study has shown that several lncRNAs and miRNAs, including SNHG1, are dysregulated in aged 2 × Tg-AD mice, and SNHG1 was targeted by Tet2 (Zhou B. et al., 2020).
17A
GABAB receptors (GABABR) are activated potassium channels and inhibit adenylate cyclase via G proteins. GABABR is a heterodimer of G protein-coupled receptors consisting of two subunits (GABABR1 and GABABR2). The GPR51 gene encodes GABABR2. LncRNA 17A strictly controls alternative splicing of GPR51. RNA polymerase III transcribes 17A from intron 3 of GPR51 (Massone et al., 2011; Luo and Chen, 2016). One study reported that a lack of 17A leads to inhibition of apoptosis, migration, and increased autophagy (Wang X. et al., 2019). Massone et al. showed that the expression of 17A was increased in the cerebral tissue of AD patients and demonstrated that its expression in neuroblastoma cells increased Aβ secretion in response to inflammatory stimuli. So, 17A may be a potential target for treating AD (Massone et al., 2011).
51A
SORL1 gene encodes SORLA protein, a receptor for apolipoprotein E, associated with AD (Motoi et al., 1999; Ciarlo et al., 2013). SORLA controls APP trafficking and processing and restricts Aβ peptide production. Allelic variants of the SORL1 gene are associated with AD disease, and the function of this gene is reduced in AD (Willnow and Andersen, 2013). 51A lncRNA is antisense of the SORL1 gene and is frequently increased in AD patients' cerebral cortices (Ciarlo et al., 2013). Ciarlo et al. reported that expression of 51A alters SORL1 splicing and shifts from the canonical long protein variant A to an alternatively spliced protein form. This process reduces the synthesis of variant A of SORL1, and with impaired APP processing, it leads to increased Aβ formation (Ciarlo et al., 2013). 51A expression has also been increased in the AD brain and in vitro models (Feng et al., 2018). However, the plasma level of lncRNA 51A did not show a significant difference between AD patients and healthy controls (Feng et al., 2018). This evidence suggests that 51A by reducing SORLA levels may be involved in AD progression, but more studies are needed in the future.
XIST
The lncRNA X inactive specific transcript (XIST) is involved in developing many malignant tumors (Yue et al., 2020). XIST can act as an oncogenic lncRNA and induce growth in pancreatic and bladder tumors by interacting and inhibiting miR-124 (Liang et al., 2017; Xiong et al., 2017). It has been reported that miR-124 regulates the expression of BACE1 and is decreased in the AD tissues, implying that XIST might play a critical role in AD. Yue et al. Treatment of Na2 cells with H2O2 has created an AD model in vitro. Silencing of XIST has reduced the effect of H2O2 on miR-124, BACE1, and Aβ1–42 expression in N2a cells (Yue et al., 2020). A study in primary cultured rat hippocampal neurons showed that knockdown of XIST reduced Aβ25-35-induced neurotoxicity, apoptosis, and oxidative stress through upregulation of miR-132 (Wang X. et al., 2018). Therefore, XIST may be a new potential target therapy for AD (Yue et al., 2020).
RPPH1
Ribonuclease P RNA component H1 (RPPH1) is part of the RNase P ribonucleoprotein RNA complex and converts precursor tRNA into mature tRNA by cleavage. RPPH1 enhances cdc45 expression levels and induces dendritic spine formation by binding to miR-330-5p (Cai et al., 2017). Gu et al. showed that the levels of rpph1 and miR-122 are increased in AD mice, and rpph1 by binding to miR-122 leads to the activation of the Wnt/β-catenin and Aβ-induced neuronal apoptosis in SH-SY5Y cells (Qasim et al., 2020). Also, Aβ25-35-induced apoptosis and ER stress in SH-SY5Y cells could be reduced by RPPH1. RPPH1 targets miR-326; thereby, the inhibitory effect of miR-326 on Pyruvate kinase M2 (PKM2) is removed. PKM2 regulates cell death and apoptosis by modulating glycolysis metabolism. Therefore, RPPH1 could be involved in AD (Gu et al., 2020).
TUG1
Taurine Upregulated Gene 1 (TUG1) encodes a new lncRNA that is 6.7 kb in length and is located on chromosome 22q12. At first, the essential role in retinal development and the formation of photoreceptors was identified (Lin et al., 2016). It was later found that TUG1 promotes apoptosis by sponging miR-9 and up-regulation of BCL2L11 under ischemia. Up-regulation of TUG1 is associated with the pathogenesis of Huntington's disease, which is a neurodegenerative disease (Chen et al., 2017). Li et al. Reported that knockdown of TUG1 inhibits the apoptosis of hippocampal neurons in AD by upregulating miR-15a and downregulating ROCK1 expression. Therefore, it may serve as a new therapeutic target in AD (Li et al., 2020).
LoNA
Long nucleolus-specific lncRNA (LoNA) reduces rRNA production by reducing nucleolin (NCL) transcription and decreases rRNA 2′-O-methylation by reducing active fibrillarin (FBL). The 5' portion of LoNA has NCL binding site, and the 3' portion of LoNA has a snoRNA for binding to FBL (Decatur and Fournier, 2002; Li D. et al., 2018). Decreased LoNA leads to increased rRNA and ribosome levels and increased translation. Also, the transport of ribosomes to synapses is enhanced, leading to increasing AMPA/NMDA receptors, synapse flexibility, and ultimately enhancing long-term memory. Knockdown of LoNA, in addition to increasing long-term memory in WT mice, improved memory function in APP/PS1 transgenic mice (Li D. et al., 2018).
SOX21-AS1
The Wnt signaling pathway is involved in the proliferation, differentiation, and survival of neuronal cells (Kishimoto et al., 2008). Traces of this pathway have also been identified in carcinogenesis and neurodegenerative disorders such as AD (Inestrosa et al., 2007). SOX21-AS1 is increased in AD patients. Silencing of SOX21-AS1 in AD mice could reduce neuronal oxidative stress and inhibit apoptosis in neuronal cells by upregulation of FZD3/5 and activating the Wnt pathway. Frizzled 3/5 (FZD3/5) are two receptors required for the Wnt signaling pathway, which play a role in developing the central nervous system, including synaptogenesis and structural plasticity (Zhang et al., 2019). Therefore, future studies can assess SOX21-AS1 as a new target for AD treatment.
BC-200
LncRNA BC-200 encodes from Brain Cytoplasmic RNA 1 (BCYRN1) gene by RNA pol III. The BC-200 is a translational regulator that targets the eukaryotic initiation factor 4A (eIF4A), maintaining long-term synaptic plasticity. BC-200 levels in the brains of AD patients are increased (Li H. et al., 2018). However, in 2007, a study reported a decrease in its expression (Mus et al., 2007). This discrepancy may be due to differences in brain regions and the severity of the disease. In a study of post-mortem specimens in the control group, BC-200 levels were reduced. However, in AD brains, compared with normal brains, BC-200 levels were significantly up-regulated (Ahmadi et al., 2020). Li et al. showed that the expression BC-200 and BACE1 are increased in Aβ1-42 induced AD cell model. They also reported that inhibition of BC-200 by targeting and suppressing BCAE1 expression reduced apoptosis and increased cell viability in AD cells. So BC-200 could provide new insights into AD gene therapy (Li H. et al., 2018).
BDNF-AS
BDNF is involved in neurogenesis and synaptic plasticity, and its decrease in the brain led to damage to memory and learning. Levels of BDNF are decreased in patients with advanced and mild AD (Azizi-Aghaali et al., 2018). LncRNA BDNF-AS is an antisense transcript to BDNF and could negatively regulate BDNF (Guo et al., 2018). Real-time PCR data showed a significant increase in BDNF-AS levels in the plasma of patients compared to controls (Azizi-Aghaali et al., 2018). Guo et al. reported that in Aβ25-35-induced PC12 cells, BDNF-AS is increased, but BDNF is decreased. These expression changes promote apoptosis and reduce cell viability. Additionally, silencing of BDNF-AS increases the cell viability and inhibits oxidative stress and apoptosis of Aβ25-35-induced PC12 cells through upregulation of BDNF (Guo et al., 2018).
ANRIL
Lnc-antisense non-coding RNA in the INK4 locus (lnc-ANRIL) is located on chromosome 9 and regulates neuronal functions and inflammation. A study in diabetic rats revealed that silencing of this lncRNA inhibited the NF-κB signaling pathway and subsequently improved memory and reduced apoptosis of hippocampal neurons (Wen et al., 2018). Inflammation is involved in the pathogenesis of AD, and lnc-ANRIL can regulate inflammation and cytokine expression through association with the NF-κB or other inflammatory pathways such as the BRCC3 signaling pathway. Zhou et al. reported that ANRIL silencing increases neurite outgrowth, suppresses cell apoptosis and inflammation by binding to miR-125a in the Pc12 cell line. Therefore, ANRIL may be a potential therapeutic target for AD (Zhou B. et al., 2020).
LncRNA-ATB
Dysregulation of the lncRNA activated by transforming growth factor-β (lncRNA ATB) involves various pathological processes, such as colorectal cancer and pancreatic cancer (Yue et al., 2016). There are limited reports on the role of lncRNA ATB in neurodegenerative diseases such as AD. The role of lncRNA ATB in Aβ25-35-induced PC12 cell injury has been investigated. The results showed that in AD patients, lncRNA ATB expression is increased. In P12 cells, lncRNA ATB negatively regulates the expression of miR-200, and miR-200 can negatively regulate ZNF217. Thus, suppression of lncRNA ATB reduced Aβ25-35-induced PC12 cell injury by regulating the miR-200/ZNF217 axis (Wang J. et al., 2018).
The Perspective of Down-Regulated lncRNAs in AD
MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is abundantly expressed in neurons. MALAT1 is involved in synaptic density (Wu et al., 2013), Schwann cell proliferation and migration, and in initiating regenerative responses after peripheral nerve injury. Also, MALAT1 has the potential to protect neurons and modify anti-inflammatory effects, and it possibly plays a protective role in AD pathology. Ma et al. reported that MALAT1 boosts neurite outgrowth and prevents neuron apoptosis and inflammation in AD through interaction with miR-125b (Ma et al., 2019). Also, MALAT1 can act as a sponge for miR-30b and increase CNR1 expression, which stimulates PI3K and AKT phosphorylation and ultimately could improve neuronal recovery following AD in animal and cell models (Bastard et al., 2020). These studies suggested the critical role of MALAT1 in neuronal loss and inflammation.
LncRNA MEG3
The PI3K/Akt pathway plays an essential role in protecting neurons and inhibiting apoptosis by increasing SOD expression. This pathway appears to be vital in AD because it is related to hyper-phosphorylated tau protein (Matsuda et al., 2018). Maternally expressed gene 3 (MEG3) lncRNA is involved in PI3K/Akt pathway. Yi et al. reported that MEG3 expression is decreased in the tissues of AD rats. Also, upregulation of MEG3 led to improved cognitive impairment, reduced neuronal damage, reduced Aβ positive expression, and inhibits activation of astrocytes in hippocampus tissues in AD rats via inactivation of the PI3K/Akt signaling pathway (Yi et al., 2019). Therefore, increased expression of MEG3 may lead to an improvement in AD.
WT1-AS
Another notable lncRNA associated with AD is WT1-AS. Wang et al. reported low expression of WT1-AS in cell models induced by Aβ25-35. Overexpression of WT1-AS through inhibition of WT1 expression can suppress miR-375 expression and promote SIX4 expression, thus preventing neuronal apoptosis and oxidative stress injury (Wang Q. et al., 2020).
Other lncRNAs
LRP1-AS is another lncRNA that is dysregulated in AD. LRP1 locus produces both LRP1 mRNA and a spliced LRP1-AS of the LRP1 gene. LRP1 plays a role in the systemic clearance of AD amyloid-beta (Aβ), and LRP1 expression levels are critical for AD progression. Yamanaka et al. reported that in the AD brain, Lrp1-AS expression is increased and negatively regulates the expression of Lrp1. Lrp1-AS directly binds to Hmgb2 and inhibits Hmgb2 activity to increase Srebp1a-dependent Lrp1 transcription (Yamanaka et al., 2015). In a recent study, MAGI2-AS3 is significantly increased in AD patients and act as a sponge and negative regulator for miR-374b-5p (Zhang and Wang, 2021). Also, they reported that decreased MAGI2-AS3 expression and increased miR-374b-5p expression reduce Aβ-induced neurotoxicity and inflammation. The results of luciferase activity provide evidence for the interaction of miR-374b-5p with BACE1 (Zhang and Wang, 2021). Therefore, the MAGI2-AS3/miR-374b-5p axis can be considered as a biomarker for AD (Zhang and Wang, 2021). Linc00507 was significantly increased in AD mice and AD-like SH-SY5Y cells. linc00507 through binding to miR-181c-5p regulates expression of microtubule-associated Tau protein (MAPT) and microtubule tau tubulin kinase (TTBK1). Also, linc00507 can mediate tau protein hyperphosphorylation by activating the P25/P35/GSK3β signaling pathway through MAPT/TTBK1 regulation (Yan et al., 2020). It has recently been reported that RP11-543N12.1 enhanced apoptosis and suppresses an AD cell model's proliferation via targeting miR324-3p. Thus, it is suggested that RP11-543N12.1 and miR-324-3p may serve as practical biomarkers and therapeutic targets for AD in the future (Cai et al., 2018). Additional studies have been conducted on the role of other lncRNAs in AD, such as LINC00094, RP11-543C4.3-001, GDNFOS, n336694 (Airavaara et al., 2011; Huang et al., 2017; Zhu et al., 2019; Chen et al., 2020) (Table 3).
Discussion
Expression Regulation, the Most Crucial Step in the Process of LncRNA Function
The precise expression of LncRNAs is vital because their expression is low compared to the genes encoding proteins and they are much less expressed than them. The specificity of tissue expression and low expression means that the expression of LncRNAs must be highly regulated (Hansen et al., 2011). Remarkably, as much as the genes encoding proteins are sensitive to developmental conditions and environmental stress changes, these changes affect LncRNAs (Cawley et al., 2004; Yang et al., 2013). On the other hand, because LncRNAs themselves are involved in regulating the expression of other genes, small changes in their expression can manifest as a significant milestone in the expression regulating of other genes, disrupting the co-expression network between LncRNAs and mRNAs (Lim et al., 2019). There have not been many studies on the mechanisms of regulation of lncRNA expression. However, a few can be mentioned, including chromatin state, which can be extensively altered by DNA methylation and histone modification. Promoter hyper-methylation in the MEG3 lncRNA gene causes downregulation of expression, which is increased by interfering with DNA methyltransferase activity (Braconi et al., 2011). In addition, methylated cytosines are found in critical functional regions of LncRNAs such as XIST and HOTAIR and show their effect on the function of LncRNAs (Amort et al., 2013).
The effects of histone acetylation on the chromatin state, which prevents the formation of its super-condensing structure and facilitates the expression of surrounding (lncRNA)genes, can also be mentioned as LncRNAs expression regulation mechanisms (Chen and Pikaard, 1997). The high sensitivity of regulating the expression of LncRNAs, tissue specificity, and their essential and indispensable roles in regulating the expression of other genes predispose them in the case of dysregulation to the pathogenesis of various diseases, including neurodegenerative disorders, in particular, AD. Among these, GWAS studies identify the potential of several LncRNAs in the pathogenesis of AD by examining many polymorphisms. One study discovered eight variants in lncRNA genes that had never been studied before in AD. These polymorphisms can result in changes in lncRNA secondary structures, resulting in the loss or increase of microRNA binding sites (miRNAs) and downstream pathway regulation (Kretzschmar et al., 2021). According to the present study, the significant contribution of dysregulated LncRNAs in AD is assigned to Bace1-AS, NEAT1, MALAT1, SNHG1, 17A, and Rpph1 LncRNAs, respectively. Among the reported dysregulation of lncRNA expressions, the significant share of these dysregulations with 56% is assigned to the up-regulated, and 6.9% of the total cases reported are down-regulated, and 37.5% of the studies They also did not report up or down-regulation of LncRNAs. Interestingly, the scales in these dysregulations of expressions in AD weigh heavily toward up-regulation, and down-regulated ones are meaningfully less.
LncRNA Authority in Transcription Regulation
One of the essential functional areas of LncRNAs is gene expression regulation. LncRNAs affect gene expression through various molecular mechanisms. Some lncRNAs can function simultaneously through several of these mechanisms, so these mechanisms cannot be considered in isolation. LncRNAs can act as guides, signals, decoys, and scaffolds (Wang and Chang, 2011). As a guide, lncRNAs can bind to proteins such as chromatin-modifying enzymes and direct them to their specific target and mediate epigenetic modification. In this mechanism, lncRNAs can change the pattern of gene expression in cis or trans. LncRNAs such as ANRIL, XIST, HOTAIR, and KCNQ1OT1 can serve as chromatin modifier enzymes to reprogram epigenetic status (Bhat et al., 2016). LncRNAs can also act as molecular signals to change chromatin structure and recruit transcriptional proteins to the target gene to enhance gene expression (Wang and Chang, 2011; Bhat et al., 2016). Functional flexibility in the structure of LncRNAs as a decoy mechanism provides the ability to act as “molecular sinks” for RNA-binding proteins (RBPs), including transcription factors, regulatory factors, and chromatin modifiers and these groups of lncRNAs are likely to be negative regulators. Also, in this mechanism, lncRNAs sponge miRNAs in a ceRNA network and prevent them from binding to the target RNA (Wang and Chang, 2011). miRNAs bind to the 3'UTR sequences or the coding sequences in mRNA molecules, reducing mRNA stability and the abundance of target proteins (Baek et al., 2008; Bartel, 2009). Scaffolds as a nest for connecting several effective partners and transporting them simultaneously to one place can be considered one of the capabilities of LncRNAs in the transcription process. These molecular companions can activate or suppress transcription (Wang and Chang, 2011; Bhat et al., 2016). The following is a schematic of the four regulatory mechanisms in the transcription regulation process (Figure 3). Because lncRNAs are involved in various human diseases which AD can be considered one of the main ones, knowing the mechanism of action and their characteristics facilitate their application in targeted diagnostics, monitoring progression, and treatment (Bhat et al., 2016). The following section provides a comprehensive overview of up and down-regulated LncRNAs.
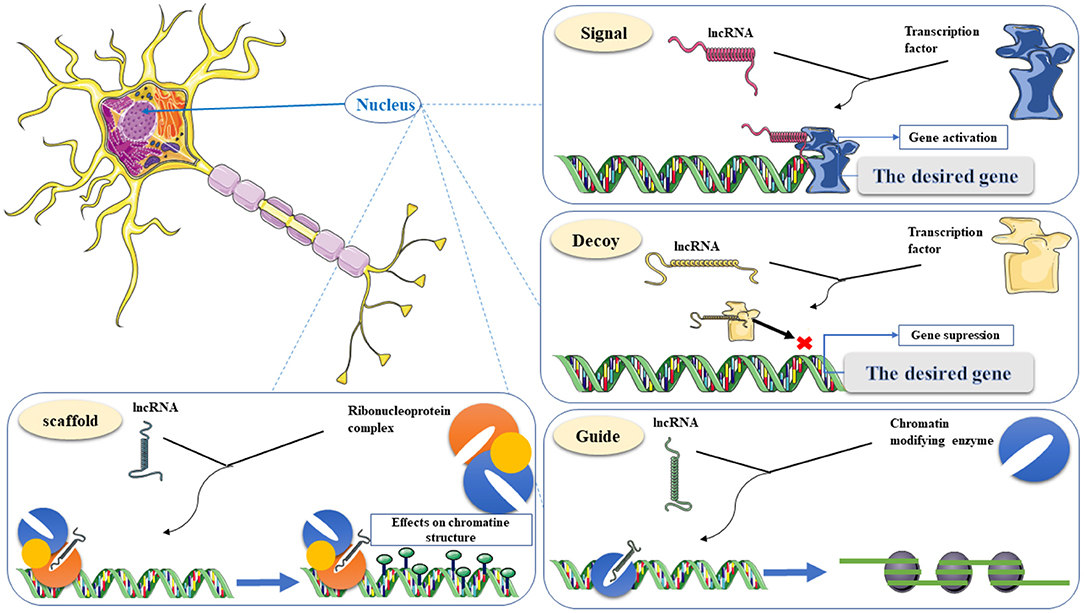
Figure 3. Classification of lncRNA functions in gene transcriptional regulations, including Signal, Decoy, Guide, and Scaffold. Signal, recruiting transcriptional proteins to the target gene to enhance gene expression. Decoy, a “molecular sink” for RNA-binding proteins (RBPs). Guide, binding to chromatin-modifying enzymes and direct to the target for epigenetic modification. Scaffold, a nest for connecting several effective partners and transporting them simultaneously to one place. This graphical figure was created using the vector image bank of Servier Medical Art (http://smart.servier.com).
Conclusion
In addition to regulating the expression of other genes, LncRNAs play critical regulatory roles by interacting with miRNAs in the ceRNA network. Tissue expression specificity is another factor that makes LncRNAs more sensitive. Low expression of LncRNAs compared to other genes and their essential role in vital cell mechanisms causes the slightest change or dysregulation in the expression of LncRNAs as a disorder, especially neurodegenerative diseases. Among these, AD can be considered the most important member of this group of diseases that LncRNAs also play an important role in its etiology due to the tissue expression specificity of 40% of them related to the brain. So far, many studies have examined the expression of LncRNAs in AD. In this review, we tried to provide a comprehensive summary of studies that have used validated molecular methods and provide an overview of the role of LncRNAs in the pathogenesis of this disease. The same project could be carried out in other neurodegenerative diseases, such as Parkinson's or ALS, and the role of LncRNAs in it can be discussed. On the other hand, further studies on the existing pathways for each of the mentioned LncRNAs have sound potential. Finally, it is decent to mention that there were some limitations to our study. First, we can mention the searching process. During it, all efforts were focused on selecting the keywords to cover the studies on the subject entirely. On the other hand, during screening studies, a study may be lost. It should also be noted that there were several studies that, despite much effort, could not provide their full text (Yang et al., 2018).
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MT, MR, and SG-F wrote the draft and revised it. MA, MH, SK, HS, MM, and MK designed the tables and figures and collected the data. All the authors read and approved submitted version.
Funding
The research protocol was approved and supported by the Molecular Medicine Research Center, Tabriz University of Medical Sciences (grant number: 67060).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadi, S., Zobeiri, M., and Bradburn, S. (2020). Molecular mechanisms underlying actions of certain long noncoding RNAs in Alzheimer's disease. Metabolic Brain Dis. 35, 681–693. doi: 10.1007/s11011-020-00564-9
Airavaara, M., Pletnikova, O., Doyle, M. E., Zhang, Y. E., Troncoso, J. C., and Liu, Q.-R. (2011). Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 286. 45093–45102. doi: 10.1074/jbc.M111.310250
Alzheimer's Association (2020). Alzheimer's disease facts and figures. Alzheimer's Dement. 16, 391–460. doi: 10.1002/alz.12068
Amort, T., Soulière, M. F., Wille, A., Jia, X. Y., Fiegl, H., Wörle, H., et al. (2013). Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 10, 1003–1008. doi: 10.4161/rna.24454
Arksey, H., and O'Malley, L. (2005). Scoping studies: towards a methodological framework. Int. J. Soc. Res. Methodol. 8, 19–32. doi: 10.1080/1364557032000119616
Association As (2019). Alzheimer's disease facts and figures. Alzheimer's Dement. 15, 321–87. doi: 10.1016/j.jalz.2019.01.010
Atri, A. (2019). The Alzheimer's disease clinical spectrum: diagnosis and management. Medical Clin. North America 103, 263–293. doi: 10.1016/j.mcna.2018.10.009
Azadfar, P., Noormohammadi, Z., Noroozian, M., Eidi, A., and Mortazavi, P. (2020). Effect of memantine on expression of Bace1-as and Bace1 genes in STZ-induced Alzheimeric rats. Mol. Biol. Rep. 47, 5737–5745. doi: 10.1007/s11033-020-05629-7
Azizi-Aghaali, R., Khalaj-Kondori, M., Zeinalzadeh, N., Hoseinpour Feizi, M. A., Farhoudi, M., and Talebi, M. (2018). Comparison between the plasma levels of long noncoding RNA BDNF-as in patients with Alzheimer's disease and healthy subjects. J. Babol Univ. Medical Sci. 20, 24–29. doi: 10.18869/ACADPUB.JBUMS.20.4.24
Baek, D., Villén, J., Shin, C., Camargo, F. D., Gygi, S. P., and Bartel, D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64–71. doi: 10.1038/nature07242
Banerjee, B., Koner, D., Karasik, D., and Saha, N. (2021). Genome-wide identification of novel long non-coding RNAs and their possible roles in hypoxic zebrafish brain. Genomics 113, 29–43. doi: 10.1016/j.ygeno.2020.11.023
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bastard, P., Rosen, L. B., Zhang, Q., Michailidis, E., Hoffmann, H. H., Zhang, Y., et al. (2020). Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370:abd4585. doi: 10.1126/science.abd4585
Bhat, S. A., Ahmad, S. M., Mumtaz, P. T., Malik, A. A., Dar, M. A., Urwat, U., et al. (2016). Long non-coding RNAs: mechanism of action and functional utility. Non-coding RNA Res. 1, 43–50. doi: 10.1016/j.ncrna.2016.11.002
Braconi, C., Kogure, T., Valeri, N., Huang, N., Nuovo, G., Costinean, S., et al. (2011). microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30, 4750–4756. doi: 10.1038/onc.2011.193
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Butler, A. A., Johnston, D. R., Kaur, S., and Lubin, F. D. (2019). Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 12:aaw9277. doi: 10.1126/scisignal.aaw9277
Cacace, R., Sleegers, K., and Van Broeckhoven, C. (2016). Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimer's Dement. J. Alzheimer's Assoc. 12, 733–748. doi: 10.1016/j.jalz.2016.01.012
Cai, M., Wang, Y. W., Xu, S. H., Qiao, S., Shu, Q. F., Du, J. Z., et al. (2018). Regulatory effects of the long non-coding RNA RP11-543N12.1 and microRNA-324-3p axis on the neuronal apoptosis induced by the inflammatory reactions of microglia. Int. J. Mol. Med. 42, 1741–1755. doi: 10.3892/ijmm.2018.3736
Cai, Y., Sun, Z., Jia, H., Luo, H., Ye, X., Wu, Q., et al. (2017). Rpph1 upregulates CDC42 expression and promotes hippocampal neuron dendritic spine formation by competing with miR-330-5p. Front. Mol. Neurosci. 10:27. doi: 10.3389/fnmol.2017.00027
Cao, M., Li, H., Zhao, J., Cui, J., and Hu, G. (2019). Identification of age- and gender-associated long noncoding RNAs in the human brain with Alzheimer's disease. Neurobiol. Aging 81, 116–126. doi: 10.1016/j.neurobiolaging.2019.05.023
Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509. doi: 10.1016/S0092-8674(04)00127-8
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chen, S., Wang, M., Yang, H., Mao, L., He, Q., Jin, H., et al. (2017). LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem. Biophys. Res. Commun. 485, 167–173. doi: 10.1016/j.bbrc.2017.02.043
Chen, Y., Li, H. Y., Zeng, F., Chen, L., Zhou, F. Y., Peng, Z. Y., et al. (2020). LincRNA plays a role in the effect of CYP46A1 polymorphism in Alzheimer's disease – related pathology. Front. Aging Neurosci. 11:381. doi: 10.3389/fnagi.2019.00381
Chen, Z. J., and Pikaard, C. S. (1997). Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136. doi: 10.1101/gad.11.16.2124
Ciarlo, E., Massone, S., Penna, I., Nizzari, M., Gigoni, A., Dieci, G., et al. (2013). An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis. Models Mechanisms 6, 424–433. doi: 10.1242/dmm.009761
Colquhoun, H. L., Levac, D., O'Brien, K. K., Straus, S., Tricco, A. C., Perrier, L., et al. (2014). Scoping reviews: time for clarity in definition, methods, and reporting. J. Clin. Epidemiol. 67, 1291–1294. doi: 10.1016/j.jclinepi.2014.03.013
Decatur, W. A., and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351. doi: 10.1016/S0968-0004(02)02109-6
Deng, Y., Xiao, L., Li, W., Tian, M., Feng, X., Feng, H., et al. (2017). Plasma long noncoding RNA 51A as a stable biomarker of Alzheimer's disease. Int. J. Clin. Exp. Pathol. 10, 4694–4699.
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Dursun, E., Gezen-Ak, D., Eker, E., Ertan, T., Engin, F., Hanagasi, H., et al. (2008). Presenilin-1 gene intronic polymorphism and late-onset Alzheimer's disease. J. Geriatr. Psychiatry Neurol. 21, 268–273. doi: 10.1177/0891988708324941
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Fang, M., Zhang, P., Zhao, Y., and Liu, X. (2017). Bioinformatics and co-expression network analysis of differentially expressed lncRNAs and mRNAs in hippocampus of APP/PS1 transgenic mice with Alzheimer disease. Am. J. Transl. Res. 9, 1381–1391.
Feng, L., Liao, Y. T., He, J. C., Xie, C. L., Chen, S. Y., Fan, H. H., et al. (2018). Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 18:1008. doi: 10.1186/s12883-017-1008-x
Fotuhi, S. N., Khalaj-Kondori, M., Hoseinpour Feizi, M. A., and Talebi, M. (2019). Long non-coding RNA BACE1-AS may serve as an Alzheimer's disease blood-based biomarker. J. Mol. Neurosci. 69, 351–359. doi: 10.1007/s12031-019-01364-2
Gao, Y., Zhang, N., Lv, C., Li, N., Li, X., and Li, W. (2020). LncRNA SNHG1 knockdown alleviates amyloid-β-induced neuronal injury by regulating ZNF217 via sponging miR-361-3p in Alzheimer's disease. J. Alzheimer's Dis. 77, 85–98. doi: 10.3233/JAD-191303
Garofalo, M., Pandini, C., Bordoni, M., Pansarasa, O., Rey, F., Costa, A., et al. (2020). Alzheimer's, parkinson's disease and amyotrophic lateral sclerosis gene expression patterns divergence reveals different grade of RNA metabolism involvement. Int. J. Mol. Sci. 21, 1–16. doi: 10.3390/ijms21249500
Ge, Y., Song, X., Liu, J., Liu, C., and Xu, C. (2020). The combined therapy of berberine treatment with lncRNA BACE1-AS depletion attenuates Aβ25–35 induced neuronal injury through regulating the expression of miR-132-3p in neuronal cells. Neurochem. Res. 45, 741–751. doi: 10.1007/s11064-019-02947-6
Ghanbari, M., Munshi, S. T., Ma, B., Lendemeijer, B., Bansal, S., Adams, H. H., et al. (2019). A functional variant in the miR-142 promoter modulating its expression and conferring risk of Alzheimer disease. Hum. Mutat. 40, 2131–2145. doi: 10.1002/humu.23872
Graham, W. V., Bonito-Oliva, A., and Sakmar, T. P. (2017). Update on Alzheimer's disease therapy and prevention strategies. Ann. Rev. Med. 68, 413–430. doi: 10.1146/annurev-med-042915-103753
Gu, R., Liu, R., Wang, L., Tang, M., Li, S. R., and Hu, X. (2020). LncRNA RPPH1 attenuates Aβ25-35-induced endoplasmic reticulum stress and apoptosis in SH-SY5Y cells via miR-326/PKM2. Int. J. Neurosci. 2020:1746307. doi: 10.1080/00207454.2020.1746307
Guo, C. C., Jiao, C. H., and Gao, Z. M. (2018). Silencing of LncRNA BDNF-AS attenuates Aβ(25-35)-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol. Res. 40, 795–804. doi: 10.1080/01616412.2018.1480921
Hansen, T. B., Wiklund, E. D., Bramsen, J. B., Villadsen, S. B., Statham, A. L., Clark, S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. doi: 10.1038/emboj.2011.359
He, W., Chi, S., Jin, X., Lu, J., Zheng, W., Yan, J., et al. (2020). Long non-coding RNA BACE1-AS modulates isoflurane-induced neurotoxicity to Alzheimer's disease through sponging miR-214-3p. Neurochem. Res. 45, 2324–2335. doi: 10.1007/s11064-020-03091-2
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5' ends. Nature 543, 199–204. doi: 10.1038/nature21374
Hong, H., Mo, Y., Li, D., Xu, Z., Liao, Y., Yin, P., et al. (2020). Aberrant expression profiles of lncRNAs and their associated nearby coding genes in the hippocampus of the SAMP8 mouse model with AD. Mol. Therapy Nucl. Acids 20, 140–54. doi: 10.1016/j.omtn.2020.02.008
Huang, W. Z., Li, Z. Y., Zhao, L. D., and Zhao, W. (2017). Simvastatin ameliorate memory deficits and inflammation in clinical and mouse model of Alzheimer's disease via modulating the expression of miR-106b. Biomed. Pharmacother. 92, 46–57. doi: 10.1016/j.biopha.2017.05.060
Huang, Z., Zhao, J., Wang, W., Zhou, J., and Zhang, J. (2020). Depletion of LncRNA NEAT1 rescues mitochondrial dysfunction through NEDD4L-dependent PINK1 degradation in animal models of Alzheimer's disease. Front. Cell. Neurosci. 14:28. doi: 10.3389/fncel.2020.00028
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 8, 1–16. doi: 10.1186/1471-2164-8-39
Idda, M. L., Munk, R., Abdelmohsen, K., and Gorospe, M. (2018). Noncoding RNAs in Alzheimer's disease. Wiley Interdiscipl. Rev. RNA 9:1463. doi: 10.1002/wrna.1463
Inestrosa, N. C., Varela-Nallar, L., Grabowski, C. P., and Colombres, M. (2007). Synaptotoxicity in Alzheimer's disease: the Wnt signaling pathway as a molecular target. IUBMB Life 59, 316–321. doi: 10.1080/15216540701242490
Kang, M. J., Abdelmohsen, K., Hutchison, E. R., Mitchell, S. J., Grammatikakis, I., Guo, R., et al. (2014). HuD regulates coding and noncoding RNA to induce APP → Aβ processing. Cell Rep. 7, 1401–1409. doi: 10.1016/j.celrep.2014.04.050
Ke, S., Yang, Z. H., Yang, F., Wang, X. M., Tan, J., and Liao, B. (2019). Long noncoding RNA NEAT1 aggravates a beta-induced neuronal damage by targeting miR-107 in Alzheimer's disease. Yonsei Med. J. 60, 640–650. doi: 10.3349/ymj.2019.60.7.640
Kishimoto, M., Ujike, H., Okahisa, Y., Kotaka, T., Takaki, M., Kodama, M., et al. (2008). The Frizzled 3 gene is associated with methamphetamine psychosis in the Japanese population. Behav. Brain Funct. 4, 1–7. doi: 10.1186/1744-9081-4-37
Kretzschmar, G. C., Alencar, N. M., da Silva, S. S. L., Sulzbach, C. D., Meissner, C. G., Petzl-Erler, M. L., et al. (2021). GWAS-Top polymorphisms associated with late-onset Alzheimer disease in Brazil: pointing out possible new culprits among non-coding RNAs. Front. Mol. Biosci. 8:632314. doi: 10.3389/fmolb.2021.632314
Kurt, S., Tomatir, A. G., Tokgun, P. E., and Oncel, C. (2020). Altered expression of long non-coding RNAs in peripheral blood mononuclear cells of patients with Alzheimer's disease. Mol. Neurobiol. 57, 5352–5361. doi: 10.1007/s12035-020-02106-x
Levac, D., Colquhoun, H., and O'Brien, K. K. (2010). Scoping studies: advancing the methodology. Implement. Sci. 5:69. doi: 10.1186/1748-5908-5-69
Li, D., Zhang, J., Wang, M., Li, X., Gong, H., Tang, H., et al. (2018). Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 9:1726. doi: 10.1038/s41467-018-04072-4
Li, H., Zheng, L., Jiang, A., Mo, Y., and Gong, Q. (2018). Identification of the biological affection of long noncoding RNA BC200 in Alzheimer's disease. NeuroReport 29, 1061–1067. doi: 10.1097/WNR.0000000000001057
Li, X., Wang, S. W., Li, X. L., Yu, F. Y., and Cong, H. M. (2020). Knockdown of long non-coding RNA TUG1 depresses apoptosis of hippocampal neurons in Alzheimer's disease by elevating microRNA-15a and repressing ROCK1 expression. Inflamm. Res. 69, 897–910. doi: 10.1007/s00011-020-01364-8
Liang, S., Gong, X., Zhang, G., Huang, G., Lu, Y., and Li, Y. (2017). The lncRNA XIST interacts with miR-140/miR-124/iASPP axis to promote pancreatic carcinoma growth. Oncotarget 8:113701. doi: 10.18632/oncotarget.22555
Lim, L. J., Wong, S. Y. S., Huang, F., Lim, S., Chong, S. S., Ooi, L. L., et al. (2019). Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 79, 5131–5139. doi: 10.1158/0008-5472.CAN-19-0255
Lin, L., Li, X., Pan, C., Lin, W., Shao, R., Liu, Y., et al. (2019). ATXN2L upregulated by epidermal growth factor promotes gastric cancer cell invasiveness and oxaliplatin resistance. Cell Death Dis. 10:2. doi: 10.1038/s41419-019-1362-2
Lin, P.-C., Huang, H.-D., Chang, C.-C., Chang, Y.-S., Yen, J.-C., Lee, C.-C., et al. (2016). Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer 16, 1–10. doi: 10.1186/s12885-016-2569-6
Liu, M., Liu, K., Zhang, L., Cai, J., Yao, H., Bai, Y., et al. (2018). Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 8248–8256. doi: 10.26355/eurrev_201812_16519
Liu, Q., Zhu, L., Liu, X., Zheng, J., Liu, Y., Ruan, X., et al. (2020). TRA2A-induced upregulation of LINC00662 regulates blood-brain barrier permeability by affecting ELK4 mRNA stability in Alzheimer's microenvironment. RNA Biol. 17, 1293–1308. doi: 10.1080/15476286.2020.1756055
Luo, Q., and Chen, Y. (2016). Long noncoding RNAs and Alzheimer's disease. Clin. Intervent. Aging 11:867. doi: 10.2147/CIA.S107037
Luo, X., Zhu, J., Cheng, Z., Zhang, F., Zhang, G., Yuan, J., et al. (2015). Lack of association of a genetic variant in the long intergenic noncoding RNA (linc01080) with Alzheimer's disease and amnestic mild cognitive impairment in Han Chinese. Int. J. Neurosci. 125, 419–423. doi: 10.3109/00207454.2014.944616
Lyu, Y., Bai, L., and Qin, C. (2019). Long noncoding RNAs in neurodevelopment and Parkinson's disease. Anim. Models Exp. Med. 2, 239–251. doi: 10.1002/ame2.12093
Ma, N., Tie, C., Yu, B., Zhang, W., and Wan, J. (2020). Identifying lncRNA-miRNA-mRNA networks to investigate Alzheimer's disease pathogenesis and therapy strategy. Aging 12, 2897–2920. doi: 10.18632/aging.102785
Ma, P., Li, Y., Zhang, W., Fang, F., Sun, J., Liu, M., et al. (2019). Long non-coding RNA MALAT1 inhibits neuron apoptosis and neuroinflammation while stimulates neurite outgrowth and its correlation with MiR-125b mediates PTGS2, CDK5 and FOXQ1 in Alzheimer's disease. Curr. Alzheimer Res. 16, 596–612. doi: 10.2174/1567205016666190725130134
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., et al. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317. doi: 10.1016/j.nbd.2010.09.019
Matsuda, S., Nakagawa, Y., Tsuji, A., Kitagishi, Y., Nakanishi, A., and Murai, T. (2018). Implications of PI3K/AKT/PTEN signaling on superoxide dismutases expression and in the pathogenesis of Alzheimer's disease. Diseases 6:28. doi: 10.3390/diseases6020028
McKhann, G. M., Knopman, D. S., Chertkow, H., Hyman, B. T., Jack, C. R. Jr., Kawas, C. H., et al. (2011). The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. J. Alzheimer's Assoc. 7, 263–269. doi: 10.1016/j.jalz.2011.03.005
Motoi, Y., Aizawa, T., Haga, S., Nakamura, S., Namba, Y., and Ikeda, K. (1999). Neuronal localization of a novel mosaic apolipoprotein E receptor, LR11, in rat and human brain. Brain Res. 833, 209–215. doi: 10.1016/S0006-8993(99)01542-5
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl. Acad. Sci U. S. A. 104:10679. doi: 10.1073/pnas.0701532104
Ni, Y., Huang, H., Chen, Y., Cao, M., Zhou, H., and Zhang, Y. (2017). Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson's disease. Cell. Mol. Neurobiol. 37, 329–338. doi: 10.1007/s10571-016-0373-0
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Qasim, S. S. B., Al-Otaibi, D., Al-Jasser, R., Gul, S. S., and Zafar, M. S. (2020). An evidence-based update on the molecular mechanisms underlying periodontal diseases. Int. J. Mol. Sci. 21:3829. doi: 10.3390/ijms21113829
Shen, J., Siegel, A. B., Remotti, H., Wang, Q., Shen, Y., and Santella, R. M. (2015). Exploration of deregulated long non-coding RNAs in association with hepatocarcinogenesis and survival. Cancers 7, 1847–1862. doi: 10.3390/cancers7030865
Spreafico, M., Grillo, B., Rusconi, F., Battaglioli, E., and Venturin, M. (2018). Multiple layers of CDK5R1 regulation in Alzheimer's disease implicate long non-coding RNAs. Int. J. Mol. Sci. 19:72022. doi: 10.3390/ijms19072022
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
Tang, L., Liu, L., Li, G., Jiang, P., Wang, Y., and Li, J. (2019). Expression profiles of long noncoding RNAs in intranasal LPS-mediated Alzheimer's disease model in mice. BioMed Res. Int. 2019:9642589. doi: 10.1155/2019/9642589
Tiraboschi, P., Hansen, L. A., Thal, L. J., and Corey-Bloom, J. (2004). The importance of neuritic plaques and tangles to the development and evolution of AD. Neurology 62, 1984–1989. doi: 10.1212/01.WNL.0000129697.01779.0A
Tricco, A. C., Lillie, E., Zarin, W., O'Brien, K. K., Colquhoun, H., Levac, D., et al. (2018). PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 169, 467–473. doi: 10.7326/M18-0850
Vaure, C., and Liu, Y. (2014). A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5:316. doi: 10.3389/fimmu.2014.00316
Wang, D., Wang, P., Bian, X., Xu, S., Zhou, Q., Zhang, Y., et al. (2020). Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer's disease. Mol. Med. Rep. 22, 227–238. doi: 10.3892/mmr.2020.11118
Wang, H., Lu, B., and Chen, J. (2019). Knockdown of lncRNA SNHG1 attenuated Aβ(25-35)-inudced neuronal injury via regulating KREMEN1 by acting as a ceRNA of miR-137 in neuronal cells. Biochem. Biophys. Res. Commun. 518, 438–444. doi: 10.1016/j.bbrc.2019.08.033
Wang, J., Zhou, T., Wang, T., and Wang, B. (2018). Suppression of lncRNA-ATB prevents amyloid-β-induced neurotoxicity in PC12 cells via regulating miR-200/ZNF217 axis. Biomed. Pharmacother. 108, 707–715. doi: 10.1016/j.biopha.2018.08.155
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, Q., Ge, X., Zhang, J., and Chen, L. (2020). Effect of LncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer's disease via inhibition of the miR-375/SIX4 axis. Aging 12:104079. doi: 10.18632/aging.104079
Wang, X., Wang, C., Geng, C., and Zhao, K. (2018). LncRNA XIST knockdown attenuates Aβ(25-35)-induced toxicity, oxidative stress, and apoptosis in primary cultured rat hippocampal neurons by targeting miR-132. Int. J. Clin. Exp. Pathol. 11, 3915–3924.
Wang, X., Zhang, M., and Liu, H. (2019). LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer's disease. Biosci. Biotechnol. Biochem. 83, 609–621. doi: 10.1080/09168451.2018.1562874
Wen, X., Han, X. R., Wang, Y. J., Wang, S., Shen, M., Zhang, Z. F., et al. (2018). Down-regulated long non-coding RNA ANRIL restores the learning and memory abilities and rescues hippocampal pyramidal neurons from apoptosis in streptozotocin-induced diabetic rats via the NF-κB signaling pathway. J. Cell. Biochem. 119, 5821–5833. doi: 10.1002/jcb.26769
Willnow, T. E., and Andersen, O. M. (2013). Sorting receptor SORLA–a trafficking path to avoid Alzheimer disease. J. Cell Sci. 126, 2751–2760. doi: 10.1242/jcs.125393
Wingo, T. S., Lah, J. J., Levey, A. I., and Cutler, D. J. (2012). Autosomal recessive causes likely in early-onset Alzheimer disease. Archiv. Neurol. 69, 59–64. doi: 10.1001/archneurol.2011.221
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bullet. 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Xiong, Y., Wang, L., Li, Y., Chen, M., He, W., and Qi, L. (2017). The long non-coding RNA XIST interacted with MiR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR). Cell. Physiol. Biochem. 43, 405–418. doi: 10.1159/000480419
Xu, W., Li, K., Fan, Q., Zong, B., and Han, L. (2020). Knockdown of long non-coding RNA SOX21-AS1 attenuates amyloid-β-induced neuronal damage by sponging miR-107. Biosci. Rep. 40:BSR20194295. doi: 10.1042/BSR20194295
Yamanaka, Y., Faghihi, M. A., Magistri, M., Alvarez-Garcia, O., Lotz, M., and Wahlestedt, C. (2015). Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 11, 967–976. doi: 10.1016/j.celrep.2015.04.011
Yan, Y., Yan, H., Teng, Y., Wang, Q., Yang, P., Zhang, L., et al. (2020). Long non-coding RNA 00507/miRNA-181c-5p/TTBK1/MAPT axis regulates tau hyperphosphorylation in Alzheimer's disease. J. Gene Med. 22:3268. doi: 10.1002/jgm.3268
Yang, B., Xia, Z.-A., Zhong, B., Xiong, X., Sheng, C., Wang, Y., et al. (2017). Distinct hippocampal expression profiles of long non-coding RNAs in an Alzheimer's disease model. Mol. Neurobiol. 54, 4833–4846. doi: 10.1007/s12035-016-0038-5
Yang, F., Huo, X. S., Yuan, S. X., Zhang, L., Zhou, W. P., Wang, F., et al. (2013). Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 49, 1083–1096. doi: 10.1016/j.molcel.2013.01.010
Yang, W., Zhang, S., Li, B., and Zhang, Y. (2018). MALAT1 inhibits proliferation and promotes apoptosis of SH-SY5Y cells induced by Aβ(25-35) via blocking PI3K/mTOR/GSK3β pathway. Chin. J. Cell. Mol. Immunol. 34, 434–441.
Yi, J., Chen, B., Yao, X., Lei, Y., Ou, F., and Huang, F. (2019). Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer's disease through inactivating the PI3K/Akt signaling pathway. J. Cell. Biochem. 120, 18053–18065. doi: 10.1002/jcb.29108
Yue, B., Qiu, S., Zhao, S., Liu, C., Zhang, D., Yu, F., et al. (2016). LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J. Gastroenterol. Hepatol. 31, 595–603. doi: 10.1111/jgh.13206
Yue, D., Guanqun, G., Jingxin, L., Sen, S., Shuang, L., Yan, S., et al. (2020). Silencing of long noncoding RNA XIST attenuated Alzheimer's disease-related BACE1 alteration through miR-124. Cell Biol. Int. 44, 630–636. doi: 10.1002/cbin.11263
Zeng, T., Ni, H. T., Yu, Y., Zhang, M. K., Wu, M. J., Wang, Q. L., et al. (2019). BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 98, 87–96. doi: 10.1016/j.jchemneu.2019.04.001
Zhang, J., and Wang, R. (2021). Deregulated lncRNA MAGI2-AS3 in Alzheimer's disease attenuates amyloid-β induced neurotoxicity and neuroinflammation by sponging miR-374b-5p. Exp. Gerontol. 144:111180. doi: 10.1016/j.exger.2020.111180
Zhang, L., Fang, Y., Cheng, X., Lian, Y. J., and Xu, H. L. (2019). Silencing of long noncoding RNA SOX21-AS1 relieves neuronal oxidative stress injury in mice with Alzheimer's disease by upregulating FZD3/5 via the Wnt signaling pathway. Mol. Neurobiol. 56, 3522–3537. doi: 10.1007/s12035-018-1299-y
Zhang, M., Zhang, Y. Q., Wei, X. Z., Lee, C., Huo, D. S., Wang, H., et al. (2019). Differentially expressed long-chain noncoding RNAs in human neuroblastoma cell line (SH-SY5Y): Alzheimer's disease cell model. J. Toxicol. Environ. Health A Curr. Issues 2019:1687183. doi: 10.1080/15287394.2019.1687183
Zhang, S., Qin, C., Cao, G., Xin, W., Feng, C., and Zhang, W. (2016). Systematic analysis of long noncoding RNAs in the senescence-accelerated mouse prone 8 brain using RNA sequencing. Mol. Ther. Nucl. Acids 5:e343–e. doi: 10.1038/mtna.2016.57
Zhang, T., Pang, P., Fang, Z., Guo, Y., Li, H., Li, X., et al. (2018). Expression of BC1 impairs spatial learning and memory in Alzheimer's disease via APP translation. Mol. Neurobiol. 55, 6007–6020. doi: 10.1007/s12035-017-0820-z
Zhang, W., Zhao, H., Wu, Q., Xu, W., and Xia, M. (2018). Knockdown of BACE1-AS by siRNA improves memory and learning behaviors in Alzheimer's disease animal model. Experimental and therapeutic medicine. 16, 2080–2086. doi: 10.3892/etm.2018.6359
Zhang, Y. Y., Bao, H. L., Dong, L. X., Liu, Y., Zhang, G. W., and An, F. M. (2021). Silenced lncRNA H19 and up-regulated microRNA-129 accelerates viability and restrains apoptosis of PC12 cells induced by Aβ(25-35) in a cellular model of Alzheimer's disease. Cell Cycle 20, 112–125. doi: 10.1080/15384101.2020.1863681
Zhao, M. Y., Wang, G. Q., Wang, N. N., Yu, Q. Y., Liu, R. L., and Shi, W. Q. (2019). The long-non-coding RNA NEAT1 is a novel target for Alzheimer's disease progression via miR-124/BACE1 axis. Neurol. Res. 41, 489–497. doi: 10.1080/01616412.2018.1548747
Zhao, Y., Wang, Z., Mao, Y., Li, B., Zhu, Y., Zhang, S., et al. (2020). NEAT1 regulates microtubule stabilization via FZD3/GSK3β/P-tau pathway in SH-SY5Y cells and APP/PS1 mice. Aging 12, 23233–23250. doi: 10.18632/aging.104098
Zhou, B., Li, L., Qiu, X., Wu, J., Xu, L., and Shao, W. (2020). Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer's disease. Mol. Med. Rep. 22, 1489–1497. doi: 10.3892/mmr.2020.11203
Zhou, Q., Zhang, M. M., Liu, M., Tan, Z. G., Qin, Q. L., and Jiang, Y. G. (2021). LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson's disease progression. Aging 13, 4115–4137. doi: 10.18632/aging.202378
Zhou, X., and Xu, J. (2015). Identification of Alzheimer's disease-associated long noncoding RNAs. Neurobiol. Aging 36, 2925–2931. doi: 10.1016/j.neurobiolaging.2015.07.015
Zhou, Y., Ge, Y., Liu, Q., Li, Y. X., Chao, X., Guan, J. J., et al. (2020). LncRNA BACE1-AS promotes autophagy-mediated neuronal damage through the miR-214-3p/ATG5 signalling axis in Alzheimer's disease. Neuroscience 10:28. doi: 10.1016/j.neuroscience.2020.10.028
Zhu, L., Lin, M., Ma, J., Liu, W., Gao, L., Wei, S., et al. (2019). The role of LINC00094/miR-224-5p (miR-497-5p)/Endophilin-1 axis in Memantine mediated protective effects on blood-brain barrier in AD microenvironment. J. Cell. Mol. Med. 23, 3280–3292. doi: 10.1111/jcmm.14214
Keywords: Alzheimer's disease, lncRNAs, β-amyloid, NFT, BACE1-AS, NEAT1, MALAT1, SNHG1
Citation: Asadi MR, Hassani M, Kiani S, Sabaie H, Moslehian MS, Kazemi M, Ghafouri-Fard S, Taheri M and Rezazadeh M (2021) The Perspective of Dysregulated LncRNAs in Alzheimer's Disease: A Systematic Scoping Review. Front. Aging Neurosci. 13:709568. doi: 10.3389/fnagi.2021.709568
Received: 14 May 2021; Accepted: 12 August 2021;
Published: 21 September 2021.
Edited by:
Robert Petersen, Central Michigan University, United StatesReviewed by:
Nobuyuki Kimura, National Center for Geriatrics and Gerontology (NCGG), JapanSafikur Rahman, Babasaheb Bhimrao Ambedkar Bihar University, India
Copyright © 2021 Asadi, Hassani, Kiani, Sabaie, Moslehian, Kazemi, Ghafouri-Fard, Taheri and Rezazadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Maryam Rezazadeh, cmV6YXphZGVobUB0YnptZWQuYWMuaXI=
†These authors have contributed equally to this work
 Mohammad Reza Asadi
Mohammad Reza Asadi Mehdi Hassani
Mehdi Hassani Shiva Kiani4
Shiva Kiani4 Hani Sabaie
Hani Sabaie Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Mohammad Taheri
Mohammad Taheri