- Verspeeten Family Cancer Centre, London Health Sciences Centre, Lawson Health Research Institute, Victoria Hospital, London, ON, Canada
Introduction: The receptor for hyaluronan-mediated motility (RHAMM), a centrosomal protein expressing in multiple isoforms, is implicated in telomerase-independent aging. However, its involvement in telomerase regulation is unproven. This study aims to investigate whether RHAMM correlates with telomerase activity in mammalian cells.
Methods: Mouse embryonic fibroblasts expressing or lacking full-length RHAMM (RHAMMFL, amino acids 1–794) and the shorter isoform RHAMMΔ163 (amino acids 164–794), were explored to examine the effect of RHAMM isoforms on mRNA expression of telomerase reverse transcriptase (TERT) and selective shelterin proteins regulating telomere maintenance.
Results: The preliminary findings revealed that RHAMM regulated Tert expression based on its isoforms. RHAMMΔ163 enhanced Tert mRNA expression and promoted telomerase activity by stimulating sirtuin 1 (Sirt1), shelterin proteins Tpp1, and Pot1a and repressing the telomerase inhibitor Pinx1 levels. In contrast, RHAMMFL did not have significant effect on TERT expression and telomerase activity. Increasing Tert mRNA expression by blocking leucine zipper sequence with function-blocking RHAMM peptide NP-110 in a TERT-deficient mouse model of idiopathic pulmonary fibrosis, alongside suppressing Tpp1 and Pot1a expression in mouse embryonic fibroblasts using ERK1 inhibitor PD98059, highlights the importance of the HATABD domain (amino acids 718–751), which includes leucine zipper and ERK-binding sequences at the C-terminus of mouse RHAMM in regulating telomerase function. Increased telomerase activity raised Hmmr expression, suggesting a potential feedback loop between RHAMM and TERT expression.
Discussion: Taken together, this report provides the first evidence that RHAMMΔ163 regulates TERT and shelterin expression and telomerase activity in mammalian cells.
1 Introduction
Telomeres safeguard chromosome ends with the shelterin complex, which includes six proteins, including telomeric repeat binding factor 1 (TRF1 or TERF1), protection of telomeres protein 1 (POT1), and TPP1 (de Lange, 2005). Each cell cycle leads to telomere attrition due to incomplete synthesis of the G-rich leading strand and the C-rich lagging strand. Telomerase, comprising of telomerase reverse transcriptase (TERT) and an RNA template (TERC) in addition to dyskerin (Dkc1), extends the G-rich strand, while CST-Pola/primase maintains C-rich repeats through fill-in synthesis in coordination with shelterin (Greider and Blackburn, 1985; Cohen et al., 2007; Cai et al., 2024). Notably, telomerase activity and TERT expression in human placenta-derived mesenchymal stem cells are significantly affected when cultured on hyaluronan-coated tissue-culture plate (Wong et al., 2017). Hyaluronan, a key glycosaminoglycan in the extracellular matrix (ECM), plays critical roles in cell migration, proliferation, and differentiation through its receptors, CD44, and receptor for hyaluronan-mediated motility (RHAMM/HMMR) (Heldin et al., 2013), and binding proteins, transcription factors and growth factors (Basu et al., 2025). Although CD44 influences TERT expression (Chung et al., 2013), RHAMM can interact with TERC’s noncoding RNA (Terc-53) but not directly with TERC or TERT (Wu et al., 2025). Notably, neither of these interactions needed hyaluronan (Chung et al., 2013; Wu et al., 2025). While RHAMM is involved in telomerase-independent aging (Wu et al., 2025), its role in telomerase regulation remains unproven.
RHAMM is a unique nuclear protein that expresses negligibly in healthy tissue, lacks an N-terminal signal peptide for conventional export via Golgi/endoplasmic reticulum and is shuttled to the cell surface under stress where it interacts with hyaluronan and CD44 and facilitates cell motility, activating the MAPK/ERK1,2 signaling pathway (Maxwell et al., 2008; Tolg et al., 2006). It binds to actin filaments, microtubules, and centrosomes and helps in spindle formation (Assmann et al., 1999). RHAMM binds hyaluronan via hyaluronan binding domains, HABD1 (amino acids 719–729, mouse) and HABD2 (amino acids 741–750, mouse) in its C-terminus (Figure 2F). HABD1 contains ERK1-binding sequence: 718LKQKIKHVVK727 which induces interaction with CD44. HABD2 is critical for hyaluronan binding as it contains part of the leucine zipper sequence (728LKDENSQLKSEVSKL742) which stabilizes the helical hyaluronan binding sequences and is critical for RHAMM’s interaction with targeting protein for XKlp2 (TPX2), and aurora kinase A (AURKA) (Tolg et al., 2010). Combination of both the sequences results in ‘Hyaluronan-Tubulin-AURKA Binding Domain’ (HATABD) (amino acids 718–751, mouse). Notably, Terc-53 binds to Hmmr sequence (amino acids 596–794) which includes ‘HATABD’ domain (Wu et al., 2021).
RHAMM is expressed as multiple isoforms due to alternative splicing and the use of alternate start codons (Messam et al., 2021). In mice, in addition to the five natural isoforms of RHAMM, a truncated isoform RHAMMΔ163 (also referred to as RHAMMv4, amino acids 164–794) was variably detected in 3T3 cells using techniques such as 5′RACE, primer extension, and RT-PCR, corresponding to a protein size of 70–73 kDa (Zhang et al., 1998) (Figure 1A). This isoform is capable of transforming fibroblasts and exhibits oncogenic potential (Hall et al., 1995). Notably, high telomerase activity is common in cancers (Blasco, 2003). The protein expression of the full-length RHAMM (RHAMMFL, amino acids 1–794, MW: 95 kDa) and RHAMMΔ163 isoforms was confirmed in different cell lines using in vitro translation followed by Western Blot analysis (Telmer et al., unpublished). Of note, high telomerase activity is common in cancers (Blasco, 2003). The shorter isoform in mice, RHAMM X1 (MW: 87 kDa), binds more strongly to Terc-53 than RHAMMFL (Wu et al., 2025). Additionally, the isoform RHAMMΔ163 can enter the nucleus but RHAMMFL cannot (Tolg et al., 2010). This suggests that the shorter isoform of RHAMM behaves differently from the full-length RHAMM and has distinct functions. Taken together, it was hypothesized that RHAMMΔ163, unlike RHAMMFL, could be linked to TERT expression through the HATABD domain.
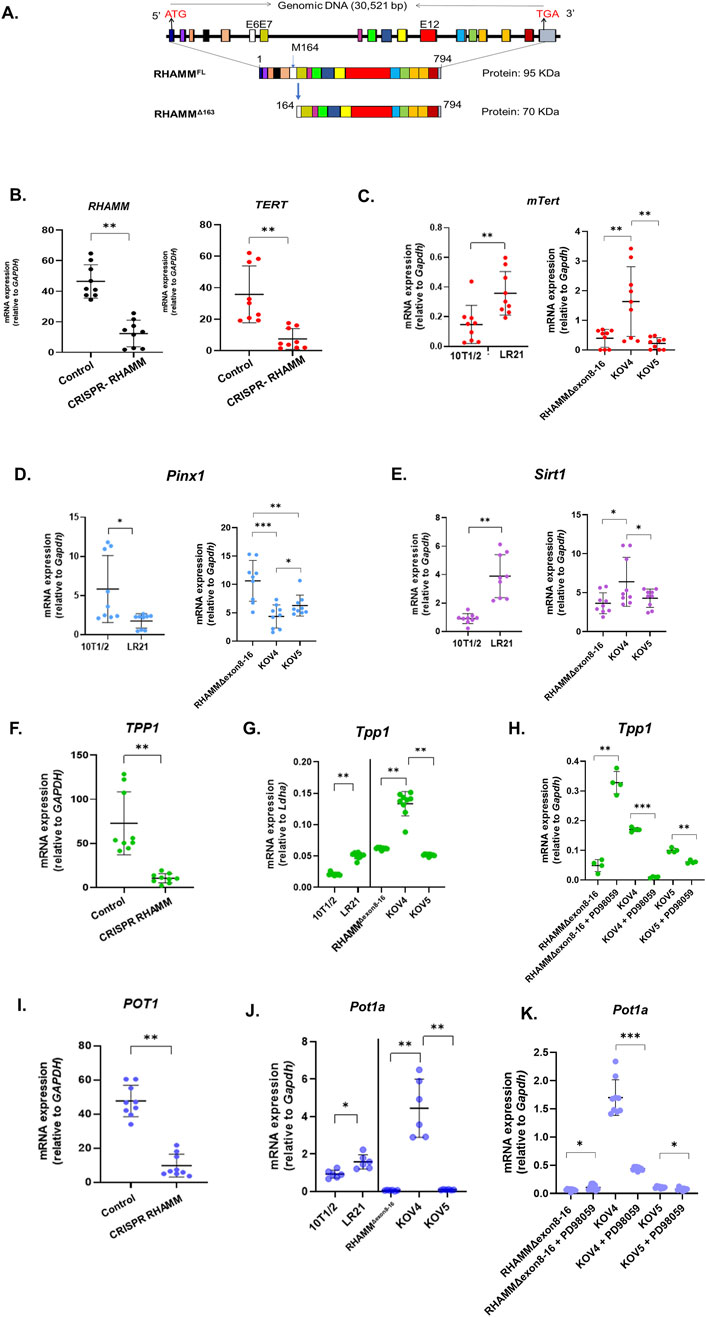
Figure 1. RHAMMΔ163 regulates telomerase-mediated telomere elongation. RHAMM is present in multiple isoforms, including RHAMMFL and RHAMMΔ163 in mice (A). HMMR was eliminated using CRISPR-Cas9 in MDA-MB-231 cells. RNA was isolated, and qRT-PCR was used to measure the mRNA expression of TERT, TPP1, and POT1, normalized to Gapdh. Data are means ± SD; n = 3 experiments; *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test (B,F,I). RNA was also isolated from 10T1/2, LR21, RHAMMΔexon8-16, KOV4, and KOV5 cell lines, with mRNA levels for mTert, Pinx1, Sirt1, Tpp1, and Pot1a measured similarly (C,D,E,G,J). For RHAMMΔexon8-16 (KO) and rescued lines (KOV4 and KOV5), treatment with PD98059 at 50 µM for 24 h was followed by qRT-PCR analysis to measure the mRNA expression of Tpp1, and Pot1a, normalized to Gapdh. Results are means ± SD; n = 2 experiments; *p < 0.05, **p < 0.01, ***p < 0.001 (H,K).
To test the hypothesis, seven cell lines were used. Commercially available 10T1/2, a mouse embryonic fibroblast cell line expressing endogenous RHAMMFL and a small amount of RHAMMΔ163 (Tolg et al., 2010), was modified in Dr. Eva Turley’s laboratory at Western University, Canada, to overexpress RHAMMΔ163, resulting in LR21 cells (Tolg et al., 2006). The other three murine cell lines were generated from mouse model in E. Turley’s research group. RHAMM consists of 18 exons, in which exon 16 contains the leucine zipper sequence and a microtubule-binding sequence facilitating binding to tubulin, essential for the integrity of the mitotic spindle and centrosome. A RHAMM−/− mouse model was developed by deleting exons 8–16 by homologous recombination, retaining exons 1–7 and 17–18, leading to a fusion between exon 7 and 17, causing a frameshift between exon 7 and the hyaluronan-binding region in exon 18 (Tolg et al., 2003). This deletion allows for the expression of a shorter 920 bp N-terminal mRNA but not the C-terminus and HATABD sequence. From the embryos of these mice, a RHAMMΔexon8-16 (KO) cell line was generated, which expresses non-oncogenic truncated N-terminal isoforms of RHAMM (RHAMMΔexon8-16), which promoted pancreatic cancer progression in partnership with heterozygous p53 knockout (Tolg et al., 2003; Lin et al., 2021). RHAMMFL and RHAMMΔ163 were rescued in the RHAMMΔexon8-16 (KO) cell line, labeled KOV5 and KOV4, respectively. Additionally, the human breast cancer MDA-MB-231 cell line, known for its high telomerase activity, was explored, along with RHAMM−/− MDA-MB-231 cells, to investigate the role of RHAMM in telomerase activity.
2 Materials and methods
2.1 Cell culture
The mouse embryonic fibroblast (MEF) cell lines: 10T1/2 (purchased from ATCC (Manassas, VA)), RHAMMΔ163-overexpressing (LR21) cells, RHAMMΔexon8-16 (KO), RHAMMΔexon8-16 rescued with RHAMMΔ163 (KOV4) and RHAMMΔexon8-16 rescued with RHAMMFL (KOV5) and the human breast cancer cell lines: MDA-MB-231 wild type and depleted of RHAMM using CRISPR/Cas9 were kind gifts from Dr. E. Turley, Western University, Canada and Dr. J. McCarthy, University of Minnesota. Because of different origin, the analysis was done by comparing 10T1/2 vs. LR21 and by comparing KO vs. KOV4 vs. KOV5. All the cell lines used in the study are telomerase positive. MDA-MB-231 cell line was used because telomerase activity is high in these human cells. The panel of murine cells were used to study the importance of N- and C- terminus of RHAMM and different isoforms of RHAMM. The cells were prepared and cultured to sub-confluency using Dulbecco’s modified Eagle’s medium (DMEM) (DMEM, 4.5 g/L Glucose; ThermoFisher, Cat # 21068028) containing 10% fetal bovine serum (FBS; Sigma Aldrich, cat #F0926), 4 μg/mL insulin, 8 μg/mL transferrin under the standard culture conditions of 37oC in a humidified 5% CO2 atmosphere as previously described (Tolg et al., 2003). The medium was changed every 2–3 days. The confluent cells were gently washed with sterile Phosphate Buffer Saline (PBS), pH 7.2–7.4, followed by harvested using TrypLE™ Express Enzyme (1X) (Gibco, Cat # 12563011).
2.2 Cell treatment
The 10T1/2 fibroblasts were cultured in a complete medium for 24 h, then switched to a serum-starved medium (SSM) with 1% FBS. Cycloastragenol (CAG; Cat # SML1448, Sigma-Aldrich, United States) was added at 3 µM for 6 h, while control cells received DMEM in SSM. After 6 h, removing the medium, the cells were washed twice with cold PBS for qRT-PCR analysis. RHAMMΔexon8-16 (KO), KOV4 and KOV5 were treated with PD98059 at 50 µM in SSM for 24 h before qRT-PCR analysis.
2.3 Telomeric Repeat Amplification Protocol
The TRAPeze Gel-based Telomerase Detection Kit (Cat #S7700, Millipore Sigma, United States) was used to perform the telomerase repeat amplification protocol (TRAP) assay according to the manufacturer’s instructions. Telomerase adds an AG sequence and telomeric repeats to the 3′end of substrate oligonucleotide (TS), followed by amplification using polymerase chain reaction (PCR) with unlabeled TS and reverse (RP) primers. This produces a ladder of products starting at 50 nucleotides in six-base increments. The PCR conditions were 94°C for 30 s, 59°C for 30 s, and 72°C for 1 min over 30 cycles. The products were analyzed on a 1% agarose gel, including controls for positive telomerase activity, PCR contamination, and a TSR-8 template as provided by the kit. Two independent experiments (n = 2) were conducted to validate the results.
2.4 Extracellular hyaluronan
The 10T1/2 fibroblasts were cultured in six-well plates with complete medium for 24 h, followed by serum starvation with 1% FBS for another 24 h. The conditioned medium was collected, and extracellular hyaluronan was quantified using the Hyaluronan Quantikine ELISA Kit (R&D, Cat # DHYAL0) according to the manufacturer’s instructions. In this assay, standards, controls, and samples were added to a microplate pre-coated with recombinant human (rh) aggrecan, allowing hyaluronan to bind to it. After washing to remove unbound substances, enzyme-linked rh aggrecan was added, followed by a substrate solution that developed color proportional to the bound hyaluronan. After stopping color development, the optical density was measured within 30 min at 450 nm. hyaluronan concentration was normalized to 1 μg of RNA extracted from cells, with data presented as means ± standard deviation (SD) from three replicates.
2.5 RHAMM functional assay
The significance of the hyaluronan binding and leucine zipper sequences was evaluated using a function-blocking RHAMM peptide, NP-110, in lung tissue from C57BL/6J mice. This tissue was sourced from a previous study (Wu et al., 2021) (kind gifts from Dr. E. Turley, Western University, Canada) in which the animal work was conducted by Stelic MC, Inc. in Tokyo, Japan, with institutional ethics approval and per the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan). In total, 36 six-week-old female C57BL/6J mice were randomly assigned to one of three groups: (i) the control group receiving 50 µL of 0.9% saline; (ii) the bleomycin (BLM) group receiving 50 µL of 1.5 mg/mL Bleomycin sulfate (Lot #15180, Nippon Kayaku, Japan) for 28 days; and (iii) BLM group receiving 3 mg/kg of NP-110 (sequence: 644KLKDENSQLKSEVSK) every fourth day (i.e., day 0, 4, 8, 12, 16, 20, 24) after BLM injections. Mice were subjected to sacrifice on day 28, and lungs were harvested and stored at −80 ± 5oC for future use.
2.6 RNA extraction and quantitative real time polymerase chain reaction (qRT-PCR)
RNA isolation and qRT-PCR were performed according to established protocols (Tolg et al., 2017). Briefly, TRIZOL reagent (Ambion, United States) was used to isolate total RNA from the cells and lung tissue following the manufacturer’s instructions. Complementary DNA (cDNA) synthesis was conducted using iScript Reverse Transcription Supermix (BioRad, United States). Primers were designed with Primer three software (version 0.4.0) and synthesized by Life Technologies, United States (Supplementary Table S2). PCR reactions were set up with 1 μg of cDNA using the SsoAdvanced Universal SYBR Green Supermix kit (BioRad, United States). The PCR cycling conditions included an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min. All samples were analyzed in triplicate. Expression analysis and calculation of relative changes in gene expression were carried out using Stratagene Mx3000Pro software and MS Excel. Mouse gene expression was normalized to the Gapdh, Ldha, and Snrpd3 while human gene expression was normalized to GAPDH and HPRT1.
2.7 In-silico analysis
The Protein sequences of RHAMM from different animals, shown in Figure 2I were sourced from the NCBI Protein database (https://www.ncbi.nlm.nih.gov/protein/). NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) checked the homology of the protein sequences of the ‘HATABD’ domain in RHAMM across the phylogenetic tree in the animal kingdom. Phylogenetic tree was drawn with iTOL (Letunic and Bork, 2024), and silhouettes are available on phylopic.org. We analyzed mRNA expression data of HMMR and hTERT using clinical datasets of ten different types of cancer retrieved from the TCGA database using cBioPortal (Cerami et al., 2012).
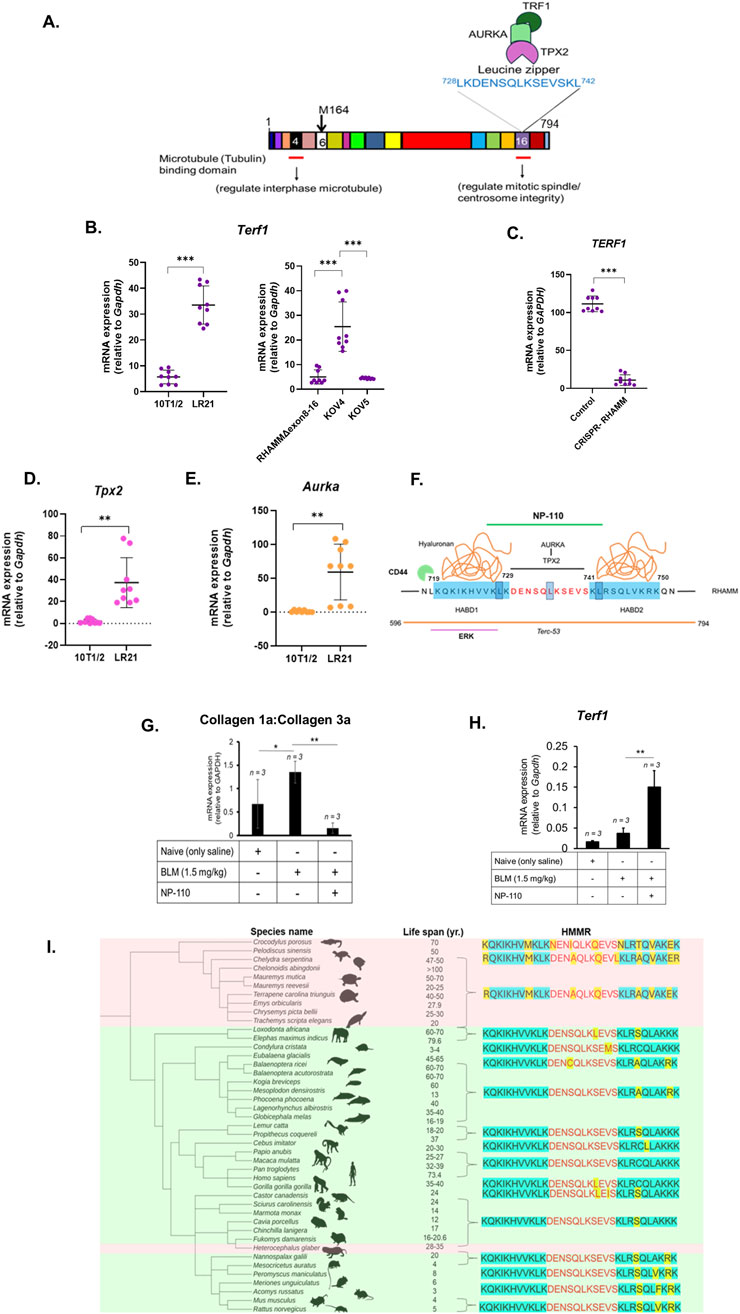
Figure 2. RHAMM regulates TERT expression via shelterin and ‘HATABD’ domain. A cartoon illustrates mouse RHAMM’s interaction with microtubules, TPX2, AURKA, and TRF1. The leucine zipper motif is highlighted (A). qRT-PCR was used to measure the mRNA expression of Terf1, Aurka, Tpx2, col1A and Col3A, normalized to Gapdh. Data are means ± SD; n = 3 experiments; *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test (B,D,E,G,H). HMMR was eliminated using CRISPR-Cas9 in MDA-MB-231 cells. RNA was isolated, and qRT-PCR was used to measure the mRNA expression of TERF1 normalized to Gapdh. Data are means ± SD; n = 3 experiments; *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test (C). Functional-blocking RHAMM peptide NP-110 (green line) inhibits hyaluronan (orange) binding (blue) and TPX2-AURKA binding domain with leucine zipper sequence (red) in RHAMM. The orange line denotes the Terc-53 binding domain, and the purple line indicates the ERK1-binding sequence (F). Importance of ‘HATABD’ domain of RHAMM in aging across the animal kingdom. Phylogenetic correlation between ectotherm (pink) and endotherm (green) vs. HMMR functional domains (HATABD) with reference to human HMMR sequence. Molecular changes are marked yellow (I).
2.8 Statistical analysis
The difference between experimental groups of two-sample unequal variance was analyzed using Student’s t-test, with *p < 0.05, **p < 0.01, ***p < 0.001 being considered significant. The experiments were performed at least two to three times independently and represented as bar diagrams or dot plots using GraphPad Prism, ver. 10.3.1 (URL: https://app.graphpad.com).
3 Results
3.1 RHAMMΔ163 enhances, but RHAMMFL does not affect TERT mRNA expression and telomerase activity
The mRNA expression levels of RHAMM and TERT are positively correlated (p < 0.05) across ten types of human cancer and cancer cell lines, obtained from the TCGA Pan-Cancer Atlas database (Supplementary Table S1). To investigate this correlation, RHAMM was eliminated in the MDA-MB-231 cancer cell line using CRISPR/Cas9 (kind gift from E. Turley), leading to a significant 3-fold decrease (p < 0.01) in TERT mRNA expression (Figure 1B). A TRAP assay revealed that RHAMM-depleted cells had lower telomerase activity than those expressing RHAMMΔ163 (Supplementary Figure S1A; original gel image shown in Supplementary Figure S3). Because RHAMM is expressed in multiple isoforms exhibiting different functions, further investigation revealed that RHAMMΔ163, significantly increased TERT mRNA levels (p < 0.01; Figure 1C). However, TERT mRNA level remained unchanged in RHAMMΔexon8-16 cells rescued with RHAMMFL or depleted of RHAMMFL and RHAMMΔ163, suggesting that RHAMMFL has no effect on TERT mRNA expression (Figure 1C). Furthermore, this result underscores that RHAMM C-terminus is crucial for TERT expression, but not the N-terminus. Similar patterns were observed for mRNA expression of dyskerin (Dkc1), which is critical for telomerase biogenesis (Cohen et al., 2007) (Supplementary Figure S1B). The TRAP assay confirmed that RHAMMΔexon8-16 cells expressing RHAMMΔ163 affected telomerase activity significantly compared to those with RHAMMΔexon8-16 cells lacking RHAMMΔ163 (Supplementary Figure S1A).
3.2 RHAMMΔ163 suppresses PINX1 expression and enhances TPP1-POT1 complex
Telomere length is controlled through multiple mechanisms involving telomerase regulators and shelterin proteins (de Lange, 2005). PINX1 directly binds to TERT and inhibits telomerase catalytic activity endogenously (Zhou and Lu, 2001). Conversely, the NAD-dependent deacetylase sirtuin 1 (SIRT1) interacts with telomeric repeats and positively regulates telomere elongation (Palacios et al., 2010). SIRT1 directly interacts with TERT and regulates its nuclear localization and stability (Lee et al., 2024). In contrast, its abrogation results in increased telomere erosion. SIRT1 acts upstream of TPP1 and interacts with it (Lee et al., 2024) to regulate telomere function and cellular senescence. A reduction in TPP1 levels leads to telomere attrition and cellular senescence associated with SIRT1 (Ahmad et al., 2017). TPP1 forms complex with POT1 which is essential for recruiting telomerase to telomeres (Sekne et al., 2022). TPP1 does not bind to telomeric DNA but interacts with the telomerase essential N-terminal (TEN) domain, which is critical for the TTAGGG repeat addition processivity of telomerase and its recruitment to telomeres, while POT1 flexibly binds both telomeric DNA and telomerase. Together, TPP1 and POT1 enhance telomerase processivity by minimizing DNA dissociation during repeat synthesis, while TPP1-POT1 depletion impairs processive DNA synthesis. Hence, the TPP1-POT1 complex is vital for telomerase processivity (Sekne et al., 2022). This complex protects telomeres (Kibe et al., 2010).
The qRT-PCR analysis showed that RHAMMΔ163 significantly reduced the negative regulator of TERT, Pinx1 mRNA levels while RHAMMΔexon8-16 depleted of C-terminus RHAMM and RHAMMΔexon8-16 rescued with RHAMMFL increased Pinx1 expression (Figure 1D). Pinx1 expression was higher in C-terminus RHAMM-depleted cells compared to RHAMMFL-expressing cells (p < 0.01). It could be possible that the N-terminus of RHAMM might enhance Pinx1 expression, while the C-terminus RHAMM represses it, which remains elusive and warrants further investigation. Thus, RHAMMΔ163 may potentially activate TERT expression and lengthen telomeres, while RHAMMFL is likely to inactivate TERT and shorten telomeres. Furthermore, RHAMMΔ163 increased the mRNA levels of Sirt1 (positive regulator of TERT) while neither RHAMMΔexon8-16 depleted of C-terminus RHAMM, nor RHAMMΔexon8-16 rescued with RHAMMFL had any effect (Figure 1E). This finding indicates that most likely, the N-terminus of RHAMM suppresses Sirt1 expression, while the C-terminus RHAMM elevates it. Eliminating RHAMM reduced TPP1 and POT1 mRNA expression in MDA-MB-231 (Figures 1F,I), indicating that RHAMM may regulate TPP1-POT1 complex. Further investigation revealed that RHAMMΔ163 increased the mRNA levels of Tpp1 and Pot1a, while RHAMMFL exerted no effect on these gene expression (Figure 1G). Taken together, these results show that RHAMMΔ163 may promote telomerase activity by increasing SIRT1 and TPP1 expression and suppressing PINX1.
To further understand how RHAMM controls Tpp1 and Pot1a expression, RHAMMΔexon8-16 (KO), RHAMMΔexon8-16 rescued with RHAMMΔ163 (KOV4) and RHAMMΔexon8-16 rescued with RHAMMFL (KOV5) were treated with PD98059 (50 µM), an MEK/ERK pathway inhibitor. As previously mentioned, RHAMM binds directly to ERK1 and indirectly to ERK2 and MEK through the sequence 718LKQKIKHVVK727 at the C-terminus of RHAMM (mouse) (Tolg et al., 2010). This binding is vital for microtubule dynamics and hyaluronan- and CD44-mediated cell motility. The results show that PD98059 significantly decreased Tpp1 (Figure 1H) and Pot1a mRNA expression (Figure 1K) in KOV4 and KOV5 cells, while RHAMMΔexon8-16 (KO) which lacks ERK1-binding domain (amino acids 718–727, mouse) promoted decreased Tpp1 and Pot1a levels. This result suggests that RHAMMΔ163 may regulate TPP1 and POT1 expression through the ERK-mediated signaling pathway, possibly via the ERK1-binding domain, which needs further validation. Notably, RHAMM, CD44, and ERK1,2 form a complex that is required for activating ERK1,2 (Tolg et al., 2006). qRT-PCR analysis revealed a significant increase in Cd44 mRNA expression in RHAMMΔexon8-16 rescued with RHAMMΔ163 (KOV4) compared to RHAMMΔexon8-16, with and without RHAMMFL (Supplementary Figure S2A).
3.3 RHAMMΔ163 enhances TRF1 mRNA expression
During mitosis, chromosome segregation relies on the organization of polymeric tubulin into bipolar spindles, with RHAMM decorating these spindles. As demonstrated in Figure 2A, the interaction of RHAMM with the cytoskeleton and centrosomes through TPX2 and AURKA is crucial for mitotic spindle assembly (Chen et al., 2014). Excessive AURKA upregulation disrupts normal mitosis, causing multinucleated cells and centrosome amplification. This is linked to TRF1, which stabilizes microtubule-kinetochore connections essential for chromosome segregation. AURKA phosphorylates TRF1, and excessive phosphorylation can lead to mitotic abnormalities (Ohishi et al., 2010). Notably, TRF1 is linked to TPP1 and POT1 via TIN2 and utilizes them to prevent ATR kinase during telomere replication and suppress sister telomere associations (Zimmermann et al., 2014).
RHAMMΔ163 increased Trf1 levels significantly (Figure 2B). In contrast, RHAMMΔexon8-16, with and without RHAMMFL did not affect Trf1 expression (Figure 2B). The elimination of RHAMM decreased TRF1 mRNA expression in MDA-MB-231 cells indicating that RHAMM positively regulates TRF1 (Figure 2C). Ectopic expression of RHAMMΔ163 elevated Tpx2 (Figure 2D) and Aurka mRNA levels (Figure 2E), provokes to hypothesize that RHAMMΔ163 may regulate TRF1 transcription through TPX2-AURKA-mediated pathways, which needs further investigation. Notably, TRF1 interacts with BubR1, Nek2, Mad1, and Mad2 in the mitotic spindle checkpoint, playing a role in spindle formation (Muñoz et al., 2009; Prime and Markie, 2005). It is not clear if RHAMM and TRF1 act together in mitotic spindle regulation. However, evaluation of unpublished microarray analysis of 10T1/2 and LR21 cells (Tolg et al., 2012) indicates that RHAMMΔ163 increases mRNA expression of Mad1l1, Mad2l1, and Nek2 (Table 1). This suggests that RHAMMΔ163 affects TRF1-partners in mitotic spindle checkpoint (Figure 2E). Furthermore, RHAMMΔ163 increases Rtel1 and Blm mRNA levels (Table 1). Notably TERF1 recruits these essential helicases, which resolve replication-associated issues and suppress the fragile-telomere phenotype. Additionally, RHAMMΔ163 represses the mRNA level of telomerase-associated protein 1 (Tep1) (Table 1). Taken together, this pilot study suggests that RHAMMΔ163 affects TERF1 expression, which may have paramount importance in telomere elongation and mitotic spindle regulation.
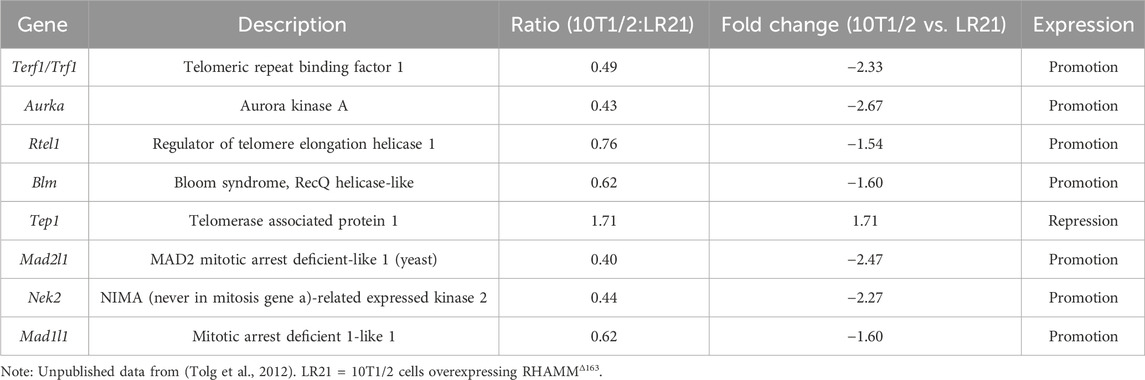
Table 1. Gene expression related to telomere function and mitotic spindle integrity in 10T1/2 mouse embryonic fibroblasts overexpressing RHAMMΔ1.63.
3.4 Importance of HATABD sequence of RHAMM for TERT expression
To investigate the importance of the ‘HATABD’ sequence of RHAMM on TERT mRNA expression, a mouse model with idiopathic pulmonary fibrosis (IPF) was chosen as IPF is characterized by TRF1 deletion or Tert deficiency, especially following a bleomycin (BLM) challenge (Povedano et al., 2015). HAS2 dysregulation in IPF (Li et al., 2016) and the effects of RHAMM antibodies and peptide mimetics in reducing macrophage recruitment and early fibrosis (Zaman et al., 2005) highlight the connection between RHAMM, hyaluronan, and IPF. In a prior study, the function-blocking RHAMM peptide, NP-110, which sterically disrupts RHAMM’s association with hyaluronan, TPX2, and AURKA (Figure 2F), was injected into BLM-treated mice, reducing fibrosis and increasing antifibrotic adipokines in skin (Wu et al., 2021). The current study analyzed the retained lung tissues from the previous study (a generous gift from Dr. E. Turley, Western University), revealing that NP-110 administration decreased the collagen 1a to collagen 3a ratio (Figure 2G) and elevated mTert mRNA expression almost three-fold (Figure 2H). These findings emphasize that blocking the interaction of RHAMM with hyaluronan, TPX2, tubulin, and AURKA via HATABD sequence may control IPF by regulating TERT expression.
The HATABD domain’s significance in regulating TERT expression by RHAMM prompted to screen the sequence across the animal kingdom in silico to understand if this sequence could be linked to longevity and telomere maintenance. The animals were selected based on their lifespan. The protein sequence homology of HATABD domain was examined in selected long-lived and short-lived species, using human RHAMM (HMMR) as a reference, with key amino acid changes highlighted in yellow (Figure 2I). The LKQKIKHVVK sequence in HABD1 is crucial for binding ERK1 (Tolg et al., 2010). Long-lived ectothermic reptiles showed two modifications (K636R and V644M), except crocodiles (demonstrating only V644M). These changes may influence RHAMM-ERK binding, although their connection to aging and telomere function is unclear. Stable HABD1 sequences were found in endothermic animals and long-lived naked mole rats. Additionally, the leucine zipper sequence (LKDENSQLKSEVSKL) contains three serine residues that were modified in long-lived ectothermic reptiles but not in short-lived species (Figure 2I). In-depth functional study is required to investigate the importance of these modifications on RHAMM interactions. Ectothermic tetrapods like crocodiles, turtles, and salamanders tend to age more slowly and have longer lifespans than birds and mammals (Reinke et al., 2022). This bioinformatic study prompts to hypothesize that amino acid substitutions in the ‘HATABD’ domain of RHAMM, under selection pressure, may relate to telomere maintenance and longevity, which warrants experimental validation.
3.5 Telomerase activity affects RHAMM mRNA expression
To examine the impact of telomerase activity and TERT expression on RHAMM, 10T1/2 fibroblasts were treated with telomerase activator, cycloastragenol (CAG), a triterpenoid saponin from Astragalus membranaceus (Yang et al., 2023). This treatment significantly increased mRNA levels of Hmmr and Tert (p < 0.05) (Figures 3A,B) and enhanced mRNA expression of Tpx2 and Aurka (Figures 3C,D). It enhanced Has2 mRNA expression (Figure 3E), resulting in a nearly four-fold increase in hyaluronan synthesis in treated cells (600 ng/mL) compared to untreated cells (150 ng/mL, p < 0.05) (Figure 3fF. These findings suggest that telomerase activation and TERT upregulation may significantly enhance Hmmr expression and indicate a potential feedback loop with TERT, as noted for CD44 (Chung et al., 2013).
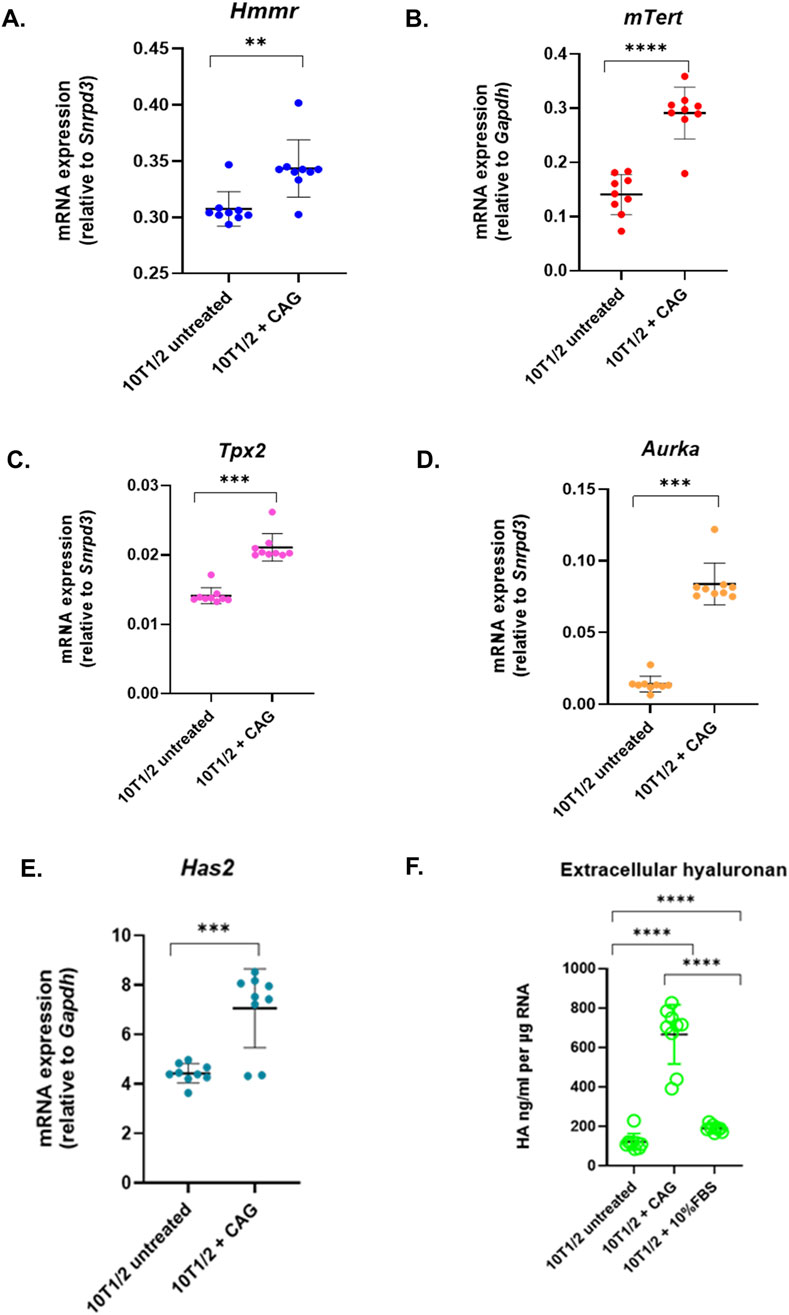
Figure 3. Telomerase activity affects RHAMM expression. Cycloastragenol (3 µM) was added to 10T1/2 fibroblasts for 6 h, followed by RNA isolation and qRT-PCR analysis to measure the mRNA expression of Hmmr, mTert, Tpx2, Aurka, and Has2, normalized to Gapdh or Snrpd3. Data are means ± SD; n = 3 experiments; *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test (A–E). The extracellular hyaluronan concentration was analyzed by hyaluronan-based ELISA assay. The hyaluronan level was normalized by total RNA concentration (F).
4 Discussion
This pilot study reveals, for the first time, that RHAMM regulates telomerase expression and activity, with different isoforms having distinct roles. The C-terminus of RHAMM is critical for TERT expression via the HATABD domain, which contains the leucine zipper (LKDENSQLKSEVSKL) and ERK1-binding sequences (LKQKIKHVVK). The study demonstrates that RHAMMΔ163 affects the expression of protein components of telomerase, i.e., TERT and dyskerin, as well as the expression of shelterin proteins TRF1, TPP1, POT1, and TRF2IP (Supplementary Figure S2B) and telomerase activity, supporting TERT-mediated telomere maintenance. Notably, POT1/TPP1 protects telomeres (Kibe et al., 2010) and regulates the synthesis of the C-rich lagging strand through their interaction with CST-Polα/primase (Cai et al., 2024). The possibility of RHAMMΔ163’s contribution to these processes cannot be ruled out. In contrast, RHAMMFL does not affect the mRNA expression of TERT and shelterin proteins. Based on previous findings (Messam et al., 2021), it is speculated that nuclear localization of RHAMMΔ163 but not RHAMMFL may play a significant role in triggering TERT mRNA expression. However, the possibility of a potential telomerase inhibitor domain (TID) at the N-terminus of RHAMM (amino acids 1–163, mouse) cannot be ruled out. Because the RHAMMΔexon8-16 isoform is non-oncogenic (Lin et al., 2021) and originated from the N-terminus, it is unlikely that it will affect TERT expression and have an additive effect on RHAMMΔ163. However, further investigation may be required to understand the role of RHAMMΔexon8-16 in telomerase regulation. An in vivo assay with the function-blocking RHAMM peptide NP-110 highlights the importance of RHAMM’s interaction with hyaluronan, tubulin, ERK1, and aurora kinase A via HATABD domain to regulate TERT expression. NP-110 peptide may offer a therapeutic approach to modulate telomerase function and control idiopathic pulmonary fibrosis which needs further validation in larger cohorts of mice. A limitation of the study is that it exclusively investigated mRNA expression through qRT-PCR analysis, without addressing protein analysis. Furthermore, it is unclear whether telomerase affects RHAMM expression through TERT activity or an off-target mechanism. Additional experiments are necessary to address this question. However, this study, along with Wu et al. (2021), identifies RHAMM as a key telomere-associated protein involved in both telomerase-dependent and independent aging, suggesting it as a promising biomarker for aging.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study used the retained lung tissues obtained from a previous study (Wu et al., 2021). This is a kind gift from Dr. E. Turley, Western University. Therefore, no ethics approval is required for this study. However, it should be noted that in the original study (Wu et al., 2021), the animal experiments were conducted with institutional ethics approval and compliant with the Act of Welfare and Management of Animals, Standards Relating to the Care and Management of Animals and Relief of Pain (Ministry of the Environment, Act No. 105 and No. 88), and the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KB: Conceptualization, Visualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The author thanks Dr. Eva Turley, Western University, Canada, for providing essential resources, including cell lines, murine lung tissues, and reagents to conduct this study in her laboratory, and critical inputs to improve the manuscript. The author acknowledges Dr. Len Luyt, Western University, Canada, for providing the RHAMM peptide used in this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fragi.2025.1604051/full#supplementary-material
References
Ahmad, T., Sundar, I. K., Tormos, A. M., Lerner, C. A., Gerloff, J., Yao, H., et al. (2017). Shelterin telomere protection protein 1 reduction causes telomere attrition and cellular senescence via sirtuin 1 deacetylase in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 56 (1), 38–49. doi:10.1165/rcmb.2016-0198OC
Assmann, V., Jenkinson, D., Marshall, J. F., and Hart, I. R. (1999). The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J. Cell Sci. 112 (Pt 22), 3943–3954. doi:10.1242/jcs.112.22.3943
Basu, K., Li, Y., Parnigoni, A., Vasilaki, E., Kolliopoulos, C., and Heldin, P. (2025). p53 and ΔNp63 transcriptional programs in coordination with TGF-β1 and RAS activation regulate HAS2 and HAS3 in epithelial cells. Proteoglycan Res. 3, e70015. doi:10.1002/pgr2.70015
Blasco, M. A. (2003). Telomeres and cancer: a tale with many endings. Curr. Opin. Genet. Dev. 13 (1), 70–76. doi:10.1016/s0959-437x(02)00011-4
Cai, S. W., Takai, H., Zaug, A. J., Dilgen, T. C., Cech, T. R., Walz, T., et al. (2024). POT1 recruits and regulates CST-Polα/primase at human telomeres. Cell 187 (14), 3638–3651.e18. doi:10.1016/j.cell.2024.05.002
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.CD-12-0095
Chen, H., Mohan, P., Jiang, J., Nemirovsky, O., He, D., Fleisch, M. C., et al. (2014). Spatial regulation of Aurora A activity during mitotic spindle assembly requires RHAMM to correctly localize TPX2. Cell Cycle 13 (14), 2248–2261. doi:10.4161/cc.29270
Chung, S. S., Aroh, C., and Vadgama, J. V. (2013). Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PLoS One 8 (12), e83971. doi:10.1371/journal.pone.0083971
Cohen, S. B., Graham, M. E., Lovrecz, G. O., Bache, N., Robinson, P. J., and Reddel, R. R. (2007). Protein composition of catalytically active human telomerase from immortal cells. Science 315 (5820), 1850–1853. doi:10.1126/science.1138596
de Lange, T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19 (18), 2100–2110. doi:10.1101/gad.1346005
Greider, C. W., and Blackburn, E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43 (2 Pt 1), 405–413. doi:10.1016/0092-8674(85)90170-9
Hall, C. L., Yang, B., Yang, X., Zhang, S., Turley, M., Samuel, S., et al. (1995). Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell 82 (1), 19–26. doi:10.1016/0092-8674(95)90048-9
Heldin, P., Basu, K., Olofsson, B., Porsch, H., Kozlova, I., and Kahata, K. (2013). Deregulation of hyaluronan synthesis, degradation and binding promotes breast cancer. J. Biochem. 154 (5), 395–408. doi:10.1093/jb/mvt085
Kibe, T., Osawa, G. A., Keegan, C. E., and de Lange, T. (2010). Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol. Cell Biol. 30 (4), 1059–1066. doi:10.1128/MCB.01498-09
Kishi, S., Wulf, G., Nakamura, M., and Lu, K. P. (2001). Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is downregulated in human breast tumors. Oncogene 20 (12), 1497–1508. doi:10.1038/sj.onc.1204229
Lee, S. E., Lee, S. B., Roh, J. I., Kim, K. P., Lee, J. H., and Lee, H. W. (2024). SIRT1 regulates the localization and stability of telomerase protein by direct interaction. Biochem. Biophys. Res. Commun. 720, 150098. doi:10.1016/j.bbrc.2024.150098
Letunic, I., and Bork, P. (2024). Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52 (W1), W78–W82. doi:10.1093/nar/gkae268
Li, Y., Liang, J., Yang, T., Monterrosa Mena, J., Huan, C., Xie, T., et al. (2016). Hyaluronan synthase 2 regulates fibroblast senescence in pulmonary fibrosis. Matrix Biol. 55, 35–48. doi:10.1016/j.matbio.2016.03.004
Lin, A., Feng, J., Chen, X., Wang, D., Wong, M., Zhang, G., et al. (2021). High levels of truncated RHAMM cooperate with dysfunctional p53 to accelerate the progression of pancreatic cancer. Cancer Lett. 514, 79–89. doi:10.1016/j.canlet.2021.05.011
Maxwell, C. A., McCarthy, J., and Turley, E. (2008). Cell-surface and mitotic-spindle RHAMM: moonlighting or dual oncogenic functions? J. Cell Sci. 121 (Pt 7), 925–932. doi:10.1242/jcs.022038
Messam, B. J., Tolg, C., McCarthy, J. B., Nelson, A. C., and Turley, E. A. (2021). RHAMM is a multifunctional protein that regulates cancer progression. Int. J. Mol. Sci. 22 (19), 10313. doi:10.3390/ijms221910313
Muñoz, P., Blanco, R., de Carcer, G., Schoeftner, S., Benetti, R., Flores, J. M., et al. (2009). TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell Biol. 29 (6), 1608–1625. doi:10.1128/MCB.01339-08
Ohishi, T., Hirota, T., Tsuruo, T., and Seimiya, H. (2010). TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 70 (5), 2041–2052. doi:10.1158/0008-5472.CAN-09-2008
Palacios, J. A., Herranz, D., De Bonis, M. L., Velasco, S., Serrano, M., and Blasco, M. A. (2010). SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 191 (7), 1299–1313. doi:10.1083/jcb.201005160
Povedano, J. M., Martinez, P., Flores, J. M., Mulero, F., and Blasco, M. A. (2015). Mice with pulmonary fibrosis driven by telomere dysfunction. Cell Rep. 12 (2), 286–299. doi:10.1016/j.celrep.2015.06.028
Prime, G., and Markie, D. (2005). The telomere repeat binding protein Trf1 interacts with the spindle checkpoint protein Mad1 and Nek2 mitotic kinase. Cell Cycle 4 (1), 121–124. doi:10.4161/cc.4.1.1351
Reinke, B. A., Cayuela, H., Janzen, F. J., Lemaître, J. F., Gaillard, J. M., Lawing, A. M., et al. (2022). Diverse aging rates in ectothermic tetrapods provide insights for the evolution of aging and longevity. Science 376 (6600), 1459–1466. doi:10.1126/science.abm0151
Sekne, Z., Ghanim, G. E., van Roon, A. M., and Nguyen, T. H. D. (2022). Structural basis of human telomerase recruitment by TPP1-POT1. Science 375 (6585), 1173–1176. doi:10.1126/science.abn6840
Sohr, S., and Engeland, K. (2008). RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53. Cell Cycle 7 (21), 3448–3460. Epub 2008 Nov 17. doi:10.4161/cc.7.21.7014
Tolg, C., Hamilton, S. R., Morningstar, L., Zhang, J., Zhang, S., Esguerra, K. V., et al. (2010). RHAMM promotes interphase microtubule instability and mitotic spindle integrity through MEK1/ERK1/2 activity. J. Biol. Chem. 285 (34), 26461–26474. doi:10.1074/jbc.M110.121491
Tolg, C., Hamilton, S. R., Nakrieko, K. A., Kooshesh, F., Walton, P., McCarthy, J. B., et al. (2006). Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J. Cell Biol. 175 (6), 1017–1028. doi:10.1083/jcb.200511027
Tolg, C., Hamilton, S. R., Zalinska, E., McCulloch, L., Amin, R., Akentieva, N., et al. (2012). A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am. J. Pathol. 181 (4), 1250–1270. doi:10.1016/j.ajpath.2012.06.036
Tolg, C., Poon, R., Fodde, R., Turley, E. A., and Alman, B. A. (2003). Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor). Oncogene 22 (44), 6873–6882. doi:10.1038/sj.onc.1206811
Tolg, C., Yuan, H., Flynn, S. M., Basu, K., Ma, J., Tse, K. C. K., et al. (2017). Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biol. 63, 117–132. doi:10.1016/j.matbio.2017.02.003
Wong, T. Y., Chang, C. H., Yu, C. H., and Huang, L. L. H. (2017). Hyaluronan keeps mesenchymal stem cells quiescent and maintains the differentiation potential over time. Aging Cell 16 (3), 451–460. doi:10.1111/acel.12567
Wu, K. Y., Kim, S., Liu, V. M., Sabino, A., Minkhorst, K., Yazdani, A., et al. (2021). Function-blocking RHAMM peptides attenuate fibrosis and promote antifibrotic adipokines in a bleomycin-induced murine model of systemic sclerosis. J. Invest Dermatol 141 (6), 1482–1492.e4. doi:10.1016/j.jid.2019.11.032
Wu, S., Cai, Y., Zhang, L., Li, X., Liu, X., Zhou, G., et al. (2025). Noncoding RNA Terc-53 and hyaluronan receptor Hmmr regulate aging in mice. Protein Cell 16 (1), 28–48. doi:10.1093/procel/pwae023
Yang, H., Ou, C. C., Feldman, R. I., Nicosia, S. V., Kruk, P. A., and Cheng, J. Q. (2004). Aurora-A kinase regulates telomerase activity through c-Myc in human ovarian and breast epithelial cells. Cancer Res. 64 (2), 463–467. doi:10.1158/0008-5472.can-03-2907
Yang, M. H., Hwang, S. T., Um, J. Y., and Ahn, K. S. (2023). Cycloastragenol exerts protective effects against UVB irradiation in human dermal fibroblasts and HaCaT keratinocytes. J. Dermatol Sci. 111 (2), 60–67. doi:10.1016/j.jdermsci.2023.07.001
Yonekawa, T., Yang, S., and Counter, C. M. (2012). PinX1 localizes to telomeres and stabilizes TRF1 at mitosis. Mol. Cell Biol. 32 (8), 1387–1395. doi:10.1128/MCB.05641-11
Yoo, J. E., Oh, B. K., and Park, Y. N. (2009). Human PinX1 mediates TRF1 accumulation in nucleolus and enhances TRF1 binding to telomeres. J. Mol. Biol. 388 (5), 928–940. doi:10.1016/j.jmb.2009.02.051
Zaman, A., Cui, Z., Foley, J. P., Zhao, H., Grimm, P. C., Delisser, H. M., et al. (2005). Expression and role of the hyaluronan receptor RHAMM in inflammation after bleomycin injury. Am. J. Respir. Cell Mol. Biol. 33 (5), 447–454. doi:10.1165/rcmb.2004-0333OC
Zhang, S., Chang, M. C., Zylka, D., Turley, S., Harrison, R., and Turley, E. A. (1998). The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J. Biol. Chem. 273 (18), 11342–11348. doi:10.1074/jbc.273.18.11342
Zhou, X. Z., and Lu, K. P. (2001). The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107 (3), 347–359. doi:10.1016/s0092-8674(01)00538-4
Keywords: HMMR, TERT, TRF1, telomerase, shelterin, hyaluronan, aging
Citation: Basu K (2025) The truncated isoform of the receptor for hyaluronan-mediated motility (RHAMMΔ163) modulates shelterin and telomerase reverse transcriptase transcription affecting telomerase activity. Front. Aging 6:1604051. doi: 10.3389/fragi.2025.1604051
Received: 01 April 2025; Accepted: 10 June 2025;
Published: 30 June 2025.
Edited by:
Karl Miller, Sanford Burnham Prebys Medical Discovery Institute, United StatesReviewed by:
Seung-Hwa Woo, Mayo Clinic, United StatesBeatrice Toia, Salk Institute for Biological Studies, United States
Copyright © 2025 Basu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaustuv Basu, a2F1c3R1dmJhc3U3QGdtYWlsLmNvbQ==
 Kaustuv Basu
Kaustuv Basu