- 1College of Forestry, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Forest Protection of Sichuan Education Department, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of National Forestry & Grassland Administration on Forest Resources Conservation and Ecological Safety in the Upper Reaches of the Yangtze River, Sichuan Agricultural University, Chengdu, China
Introduction: The rust disease caused by the fungus Coleosporium zanthoxyli in Zanthoxylum armatum leads to significant leaf damage. However, the genetic regulatory mechanisms underlying disease resistance in this plant remain unclear.
Methods: Through transcriptome analysis and bioinformatics screening, the candidate gene CCoAOMT was identified as potentially involved in rust resistance. Genetic engineering techniques were employed to construct recombinant plasmids for both overexpression and RNAi mediated of CCoAOMT. Transient transformation and stable transformation methods successfully generated CCoAOMT-RNAi and CCoAOMT-overexpression transgenic plants and calli, respectively.
Results: qRT-PCR analysis revealed that the expression levels of CCoAOMT and six related genes were downregulated in RNAi-silenced plants but upregulated in overexpression lines. Following inoculation with fungal spore suspensions, CCoAOMT-RNAi plants exhibited significantly higher disease incidence and severity indices compared to wild-type controls, whereas overexpression plants showed the opposite trend with reduced susceptibility. Furthermore, total lignin and flavonoid contents were markedly decreased in RNAi lines and increased in overexpression lines relative to wild-type plants. Subcellular localization assays using GFP fusion constructs confirmed dual localization of CCoAOMT protein in both the cytoplasm and nucleus.
Conclusion: These findings collectively demonstrate that CCoAOMT enhances rust resistance in Z. armatum by regulating lignin and flavonoid biosynthesis pathways.
1 Introduction
Z. armatum, a member of the Rutaceae family, is valued as a multipurpose plant providing spices, essential oils, and traditional Chinese medicinal materials. Recognized as a “medicinal-food homologous” species, it has been recorded in Pharmacopoeia of the People’s Republic of China for nearly two millennia (Zhang and Ye, 2010; National Pharmacopoeia Commission, 2020). Among the Zanthoxylum genus, Z. armatum (commonly known as green pepper) and Z. bungeanum represent two economically significant species. Cultivated across China with a planting area exceeding 20 million mu, Z. armatum demonstrates remarkable ecological adaptability while generating an annual economic value surpassing USD 2 billion (Feng et al., 2020; Feng et al., 2021). As China’s predominant woody spice crop, Z. armatum is recognized for its critical contributions to ecosystem stabilization, rural economic development through specialty agriculture, post-poverty-alleviation sustainability, and culinary innovation. Sichuan Province, recognized as both the geographical origin and primary production zone for premium peppercorns in China, serves as the epicenter of Z. armatum cultivation, with production spanning 144 counties across 5.7 million mu (≈380,000 hectares). Annual yields exceed 100,000 metric tons of dried Z. armatum, generating economic returns valued at USD 1.1 billion (Ye, 2020). However, with the rapid development of Z. armatum industry, the pests and diseases of Z. armatum have increased year by year (Wu et al., 2023). One of the most dangerous of these is rust, which is caused by Coleosporium zanthoxyli (Zheng and Zhu, 2012; Yuan et al., 2024). This obligate biotrophic fungus exhibits broad host specificity, with individual Coleosporium species demonstrating polyphagous behavior while frequently occurring as multispecies Spore morphology on infected hosts. The genus comprises 239 documented species and formae speciales globally (You, 2012; Xi et al., 2018). C. zanthoxyli infection triggers premature defoliation, photosynthetic impairment, and fruit quality deterioration, causing yield losses up to 60% that jeopardize the economic viability of Z. armatum cultivation systems. While recent investigations have elucidated aspects of C. zanthoxyli epidemiology and control strategies (Yue, 2010; Mao et al., 2014; Tang et al., 2015; Chen et al., 2022; Wei et al., 2023; Yang, 2023), critical knowledge gaps persist regarding the molecular determinants of Z. armatum rust resistance, particularly the genetic regulation of host-pathogen interactions.
Plant diseases compromise crop productivity, product quality, and commercial viability. The development of disease-resistant cultivars has been recognized as an environmentally sustainable alternative to synthetic agrochemicals while enhancing phytopathological resilience in perennial woody species. Recent advances in molecular biology, transgenic breeding, and plant tissue culture techniques have enabled precision utilization of resistance (R) genes for cultivar development, offering a cost-effective strategy for integrated disease management. This molecular breeding paradigm holds particular promise for addressing persistent challenges in Z. armatum cultivation systems, where conventional control methods frequently prove inadequate against rapidly evolving rust pathogens (Aktar-uz-zaman et al., 2017; Li et al., 2020; Luo et al., 2022).
Stress activates the plant phenylpropaniod metabolic pathway and regulates the expression of related genes, which can accumulate secondary metabolites, slow down membrane oxidation and scavenge reactive oxygen species (Gharibi et al., 2019; Sharma et al., 2019; Wang et al., 2021). Flavonoids, a predominant class of plant secondary metabolites, are abundantly synthesized in fruits, seeds, leaves, and flowers. These phytochemicals regulate critical physiological processes, including auxin polar transport, pollen germination, and pollen tube elongation, while functioning as multifunctional defense compounds against biotic and abiotic stressors (Wang et al., 2020; Wang et al., 2022; Zhuang et al., 2023). Lignin is a complex compound formed by the synthesis of lignin monomers via the phenylacetone metabolic pathway followed by transport and polymerisation (He et al., 2023; Zhu et al., 2023). The phenylalanine generated by the upstream glycolysis system and the shikimate pathway is the starting point for the phenylpropanoid biosynthesis pathway in higher plants. It serves as the initial substrate for the reaction and supplies a precursor for the synthesis of the products of the subsequent pathway. It then undergoes a sequence of enzymatic reactions to generate intermediate products, lignin and flavonoids, such as cinnamic acid, ferulic acid, naringenin, and caffeic acid (Gho et al., 2020). Three core enzymes govern the general phenylpropanoid pathway: phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL). Under the catalytic activity of hydroxycinnamoyl transferase (HCT), coumaroyl-CoA is directed into the lignin biosynthesis pathway. The production of lignin monomers involves sequential biochemical reactions: continuous hydroxylation of aromatic rings mediated by HCT, caffeoyl shikimate esterase (CSE), coumarate 3-hydroxylase (C3H), and ferulate 5-hydroxylase (F5H), followed by O-methylation via ferulate O-methyltransferase (COMT) and caffeoyl-CoA O-methyltransferase (CCoAOMT), culminating in the conversion of hydroxyl groups to methoxyl groups. Subsequent reduction reactions are catalyzed by cinnamoyl-CoA reductase (CCR) and coniferyl alcohol dehydrogenase (CAD) (Cao et al., 2023; Cao et al., 2025). These reactions yield the three primary lignin monomers—p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol—which are synthesized in the cytoplasm and subsequently transported to the cell wall. Finally, lignin is deposited onto the secondary cell wall through the polymerization of 4-hydroxyphenyl (H-type), guaiacyl (G-type), and syringyl (S-type) lignin subunits (Alejandro et al., 2012). In the flavonoid biosynthesis pathway, p-coumaroyl-CoA is channeled into flavonoid-specific pathways by chalcone synthase (CHS). Subsequent enzymatic modifications, including hydroxylation by flavanone 3-hydroxylase (F3H), isomerization by chalcone isomerase (CHI), and oxidation by flavonol synthase (FLS), ultimately yield diverse flavonoid compounds. Dynamic regulation of phenylpropanoid pathway enzymes constitutes a central mechanism underlying plant stress adaptation throughout ontogeny (Wu et al., 2022). Dynamic regulation of phenylpropanoid pathway enzymes constitutes a central mechanism underlying plant stress adaptation throughout ontogeny.
Caffeoyl CoA O-methyltransferase, a S-adenosylmethionine (SAM) utilizing O-methyltransferase, catalyzes the methylation of caffeoyl-CoA at the 3-hydroxyl position of its aromatic ring, generating feruloyl-CoA. This metabolic intermediate is subsequently converted into lignin monomers through coordinated actions of CCR and COMT (Walker et al., 2016). As a rate-limiting enzyme in monolignol biosynthesis, CCoAOMT critically regulates lignin deposition. CCoAOMT expression is typically suppressed via antisense or RNA interference strategies to modulate lignification levels. Antisense-mediated CCoAOMT suppression reduces total lignin content by 8%–15% in Nicotiana tabacum (Parvathi et al., 2001), 12% in Populus trichocarpa (Shi et al., 2010), and 8%–44% in Medicago sativa (Nakashima et al., 2008) compared to wild-type counterparts. Conversely, CCoAOMT overexpression consistently enhances lignification, with transgenic Arabidopsis thaliana and Populus hybrids exhibiting 26% and 20% increases in lignin content, respectively (Li et al., 2023). Despite this well-characterized role in model species, CCoAOMT remains understudied in Z. armatum, with its functional significance in rust resistance mechanisms remaining wholly uncharacterized.
In this study, transcriptome analysis was employed to identify rust resistance-associated candidate genes in Z. armatum. RNAi and overexpression approaches were subsequently implemented to suppress or elevate CCoAOMT expression, enabling functional validation of this key resistance gene and mechanistic exploration of Z. armatum molecular defenses against rust pathogenesis. These findings establish a molecular framework for resistance gene-assisted breeding programs in Z. armatum and inform the design of targeted, sustainable rust management protocols.
2 Results
2.1 Gene sequence analysis and protein structure prediction
The NCBI BLAST sequence alignment tool was employed to analyze the target sequence against the nucleotide database. Alignment results revealed that the coding sequence (CDS) of ZaCCoAOMT exhibited 79.58% nucleotide identity with Pistacia vera (XM_031400822.1). Protein sequence analysis demonstrated that the CCoAOMT-encoded protein shared 75.80% amino acid similarity with Citrus sinensis (XP_052287466.1) within the Rutaceae family. Based on these alignment results, orthologous sequences encoding conserved methyltransferase domains were selected across phylogenetically diverse taxa. A phylogenetic tree was reconstructed using the Neighbor-Joining algorithm in MEGA7 (v7.0.26) with 1000 bootstrap replicates (Figure 1a). Conserved protein motifs were subsequently predicted using MEME (v5.5.3) under default parameters (Figure 1c). Phylogenetic analysis demonstrated strong conservation of ZaCCoAOMT within the Rutaceae family, with MEME identifying 17 conserved motifs (E-values < 1e-10). Phylogenetic analysis revealed that ZaCCoAOMT clusters within the same clade as Citrus clementina and Citrus sinensis (Rutaceae), demonstrating its closest evolutionary affinity to members of this family. Multiple sequence alignment of 11 CCoAOMT homologs was performed using DNAMAN 8.0, focusing on conserved methyltransferase domains. The Z. armatum CCoAOMT exhibited 85.98% pairwise identity with Rutaceae CCoAOMT homologs, confirming strong sequence conservation (Figure 1). Bioinformatic characterization using ExPASy ProtParam predicted a 246-residue polypeptide with a theoretical pI of 4.96 and molecular mass of 27.7 kDa. The protein contained 37 acidic residues (Asp + Glu) and 26 basic residues (Arg + Lys), consistent with its predicted pI. Hydropathy analysis via ProtScale yielded a grand average of hydropathy (GRAVY) index of -0.169, confirming the hydrophilic nature of the protein. Signal peptide analysis conducted with SignalP-5.0 indicated negligible secretion potential (probability score: 0.0008), suggesting cytoplasmic localization. Transmembrane topology prediction using TMHMM 2.0 revealed no α-helical transmembrane helices, supporting its classification as a soluble protein. The CCoAOMT gene comprises a 741 bp coding sequence (CDS) encoding a 246-amino acid open reading frame (ORF). Secondary structure prediction via SOPMA algorithm revealed a composition of 40.24% α-helices, 18.29% extended strands, 8.94% β-sheets, and 32.52% random coils (Appendix A.2), dominated by helical and coil conformations. Homology modeling of the CCoAOMT tertiary structure was performed using SWISS-MODEL, with template identification based on structural similarity (Figure 1d). The top structural homolog identified was Sorghum bicolor caffeoyl-CoA O-methyltransferase (PDB: 5KVA.A), sharing 59.91% sequence identity with CCoAOMT residues 52–278 and containing conserved SAM-binding motifs. The model exhibited a GMQE score of 0.85, reflecting high confidence in the predicted SAM-dependent methyltransferase fold. The tertiary structure model displayed characteristic α-helix-rich regions, consistent with canonical SAM-dependent methyltransferase architectures. These, computational analyses confirmed evolutionary conservation, of, CCoAOMT methyltransferase function across Rutaceae, species, supporting its putative role in lignin-related phenylpropanoid metabolism.
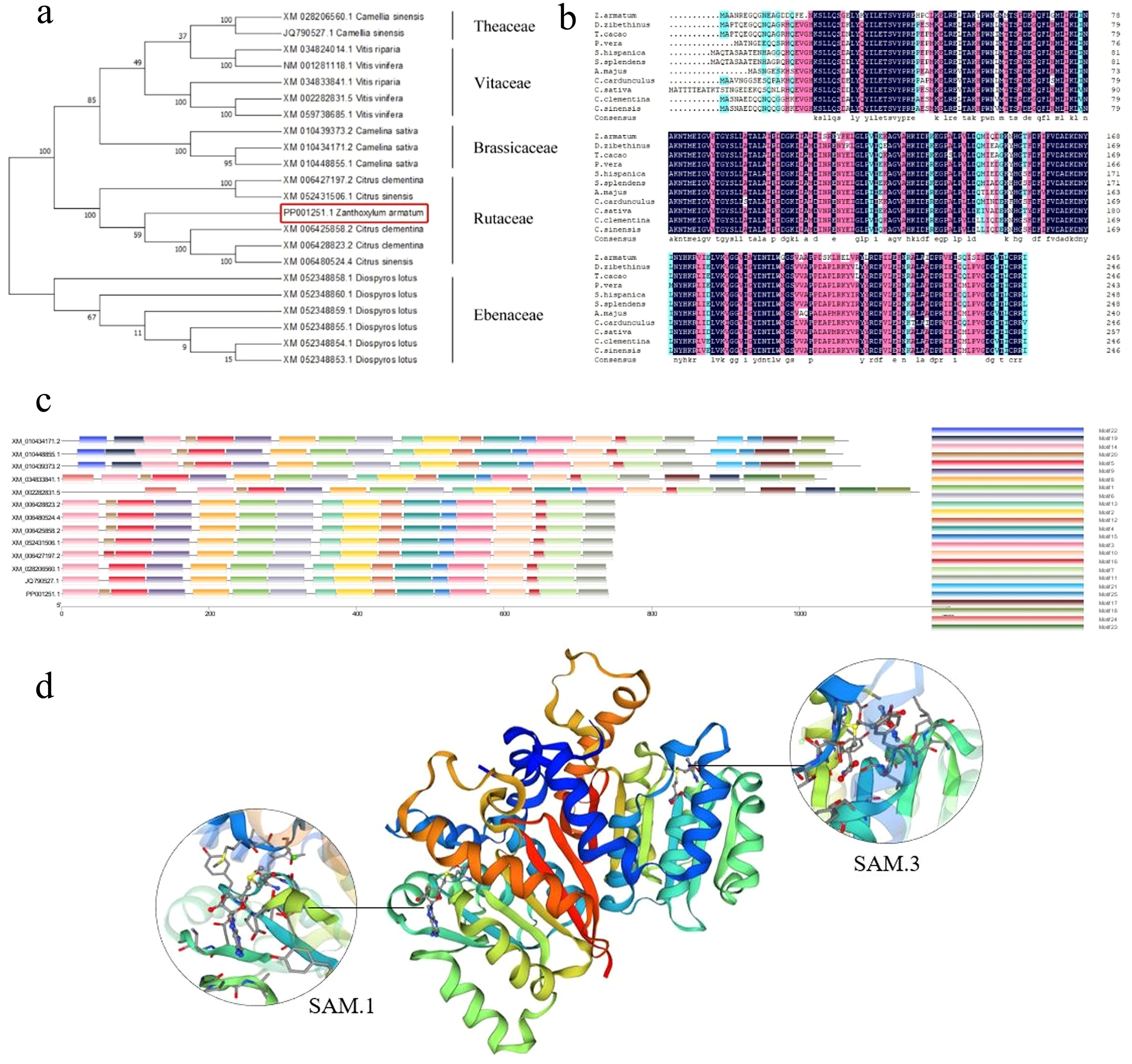
Figure 1. Bioinformatics analysis of the ZaCCoAOMT gene. (a) Phylogenetic tree of the ZaCCoAOMT gene constructed based on the adjacency algorithm,the red box represents the target gene; (b) Protein multiple sequence alignment; (c) Conservative structural domain prediction of ZaCCoAOMT; (d) Homologous modeling of the tertiary structure, SAM.1 and SAM.3 are two predicted functional domains of S-adenosylmethionine, respectively.
2.2 Tissue-specific expression analysis of ZaCCoAOMT gene
Relative expression analysis of ZaCCoAOMT transcript abundance revealed tissue-specific expression, with the highest level detected in leaves, low levels observed in stems, and almost no expression in roots (Figure 2). These findings suggest that ZaCCoAOMT is predominantly accumulated in Z. armatum foliar tissues, where it likely modulates phenylpropanoid-derived metabolite biosynthesis, contributing to biochemical defenses against phytopathogens.
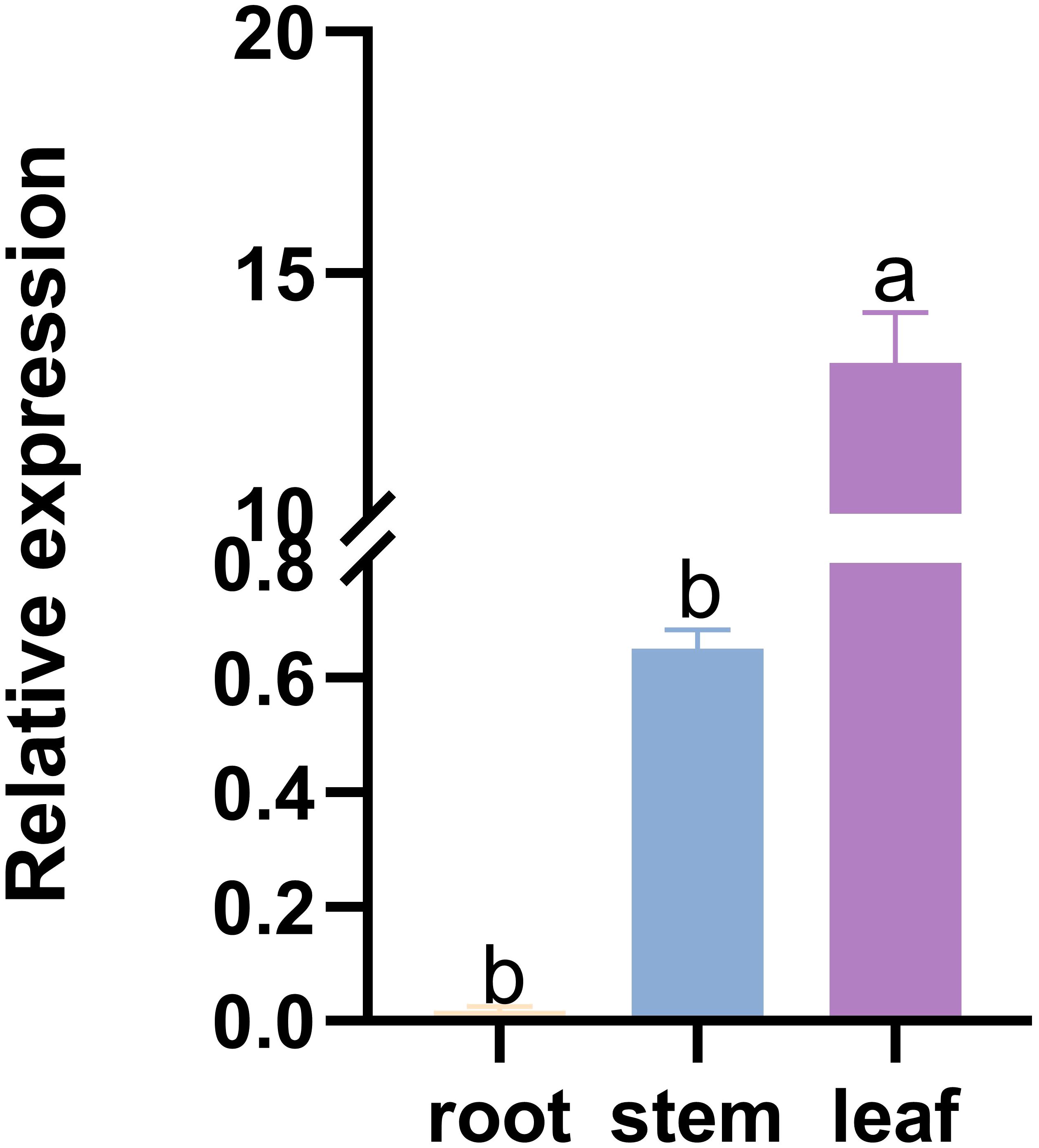
Figure 2. Tissue-specific expression analysis of ZaCCoAOMT gene. The abscissa represents the plant tissue site, the ordinate represents the relative expression of ZaCCoAOMT, and the different lowercase letters indicate that there are significant differences between different combinations (P < 0.05).
2.3 Construction of the ZaCCoAOMT-RNAi plasmid
Z. armatum cDNA served as the template for amplifying the ZaCCoAOMT fragment, which included flanking sequences homologous to the pCAMBIA1301-35SN vector. Amplification products were resolved on 1% agarose gels. The ZaCCoAOMT coding sequence (741 bp) was flanked by 18 bp homology arms overlapping the KpnI-digested pCAMBIA1301-35SN vector, with an additional 6 bp restriction site for directional cloning. This design yielded predicted insertion fragments of 765 bp. Electropherogram of the 765 bp fragments aligned with the 750 bp region of the 2 kb Marker, confirming expected sizes (Appendix A.2.5b). Purified fragments exhibited sharp electrophoretic bands without nonspecific products, meeting quality standards for downstream homologous recombination. The 769 bp intronic spacer was engineered with 26 bp terminal homology to the target gene for recombination. The final 795 bp intron-containing construct migrated at the 750 bp position relative to the 2 kb marker (Appendix A.2.5d). KpnI-linearized pCAMBIA1301-35SN was recombined with sense and antisense ZaCCoAOMT fragments and the intronic spacer (1:3 molar ratio) via homologous recombination, followed by transformation into E. coli DH5α competent cells. To prevent hairpin structure formation during antisense strand synthesis, vector-specific primers spanning the insertion sites were designed to confirm bidirectional cassette assembly. Colony PCR and restriction digestion confirmed successful tripartite recombination in E. coli (Appendix A.2.5e). Transformed colonies were selected on LB agar plates after 48 h incubation at 28°C. The validated recombinant plasmid was chemically transformed into A. tumefaciens GV3101. PCR amplification confirmed successful Agrobacterium transformation.
2.4 Construction of the ZaCCoAOMT-overexpression plasmid
The cDNA of Z. armatum was used as a template to amplify the ZaCCoAOMT overexpression gene fragment, and the BG Plant-Express MCS vector was constructed for high expression of the disease resistance candidate gene. Positive recombinant clones were identified using primers yz-F and yz-R (Appendix A.1) following transformation into E. coli. The verified recombinant plasmid was subsequently introduced into A. tumefaciens, followed by selection of a single colony for expansion in liquid culture to generate ZaCCoAOMT-overexpression Agrobacterium strains (Appendix A.2.6).
2.5 Agrobacterium-mediated transformation
In this study, gene function verification was performed using two complementary approaches. First, calli were induced from Z. armatum leaves using the Purohit method (Purohit et al., 2020), followed by co-culture with Agrobacterium infection solution. After co-culture, calli were transferred to screening media to promote stable genetic transformation through regeneration of new tissue. Second, Agrobacterium-mediated transient expression (Xia et al., 2023) was employed for in planta transformation. Calli cultures are shown in Figure 3. PCR screening of silencing and overexpression lines was conducted using primer pairs hyg501-F/R & hyg359-F/R and kan270-F/R & kan300-F/R (Appendix A.1). Transformants with confirmed silencing or overexpression were identified using the same primer sets. Electrophoretic analysis revealed distinct bands at 359 bp and 501 bp in silenced calli and transformants, whereas overexpression lines exhibited bands approximately 270 bp and 300 bp, respectively (Figures 3b, c, e, f). All screened calli and transformants tested positive, confirming their suitability for subsequent experiments.
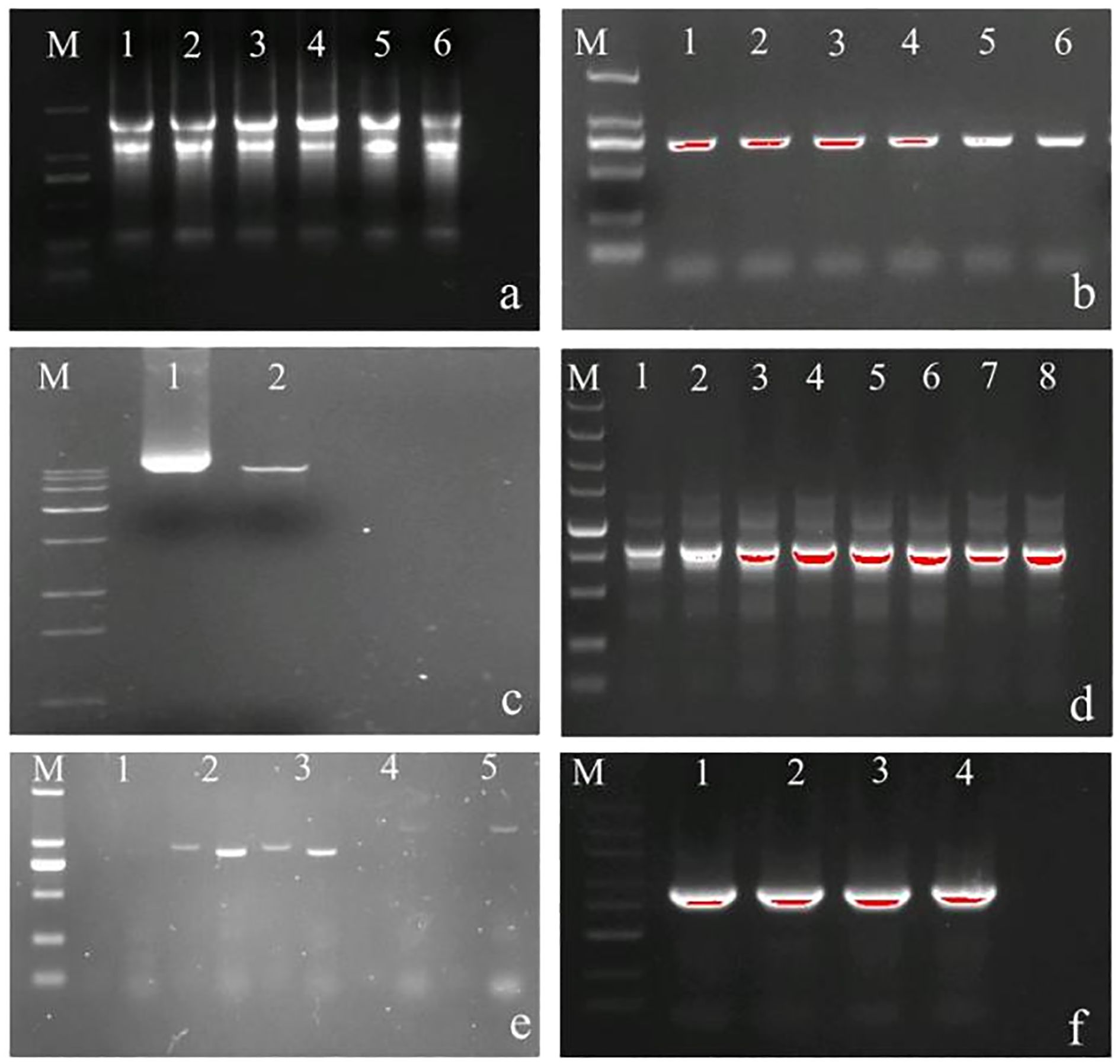
Figure 3. Detection of positive ZaCCoAOMT-RNAi and ZaCCoAOMT-overexpression transformants and calli. (a) Calli induction and proliferation were performed under four experimental conditions: positive-control, negative-control, silencing-treatment, and overexpression-treatment groups (arranged top to bottom). (b) Silencing efficacy in transgenic calli was confirmed through qPCR analysis. (c) Overexpression efficacy in transgenic calli was confirmed through qPCR analysis. (d) Transient transformation via Agrobacterium-mediated delivery included silencing-treatment, overexpression-treatment, positive-control, and negative-control groups (left to right). (e) Detection of positive transient silencing transformants; (f) Detection of positive transient overexpression transformants. In the figure, M represents marker, P represents positive control, N represents negative control, and the number represents repetition.
2.6 Expression analysis of ZaCCoAOMT
2.6.1 Expression of ZaCCoAOMT in stable calli
Total RNA was extracted from calli of wild-type, ZaCCoAOMT-RNAi groups, and ZaCCoAOMT-overexpression plants, followed by reverse transcription into cDNA and subsequent qPCR analysis of target genes. As shown in Figure 4, ZaCCoAOMT expression in RNAi-treated calli exhibited the lowest levels, with an 8.35-fold downregulation relative to actin compared to the wild type, whereas overexpression resulted in a 7.36-fold upregulation. These findings demonstrate that ZaCCoAOMT expression in Z. armatum decreased following the transfer of sense and antisense fragments of CCoAOMT. Conversely, the introduction of the CCoAOMT coding sequence (CDS) significantly enhanced its expression.
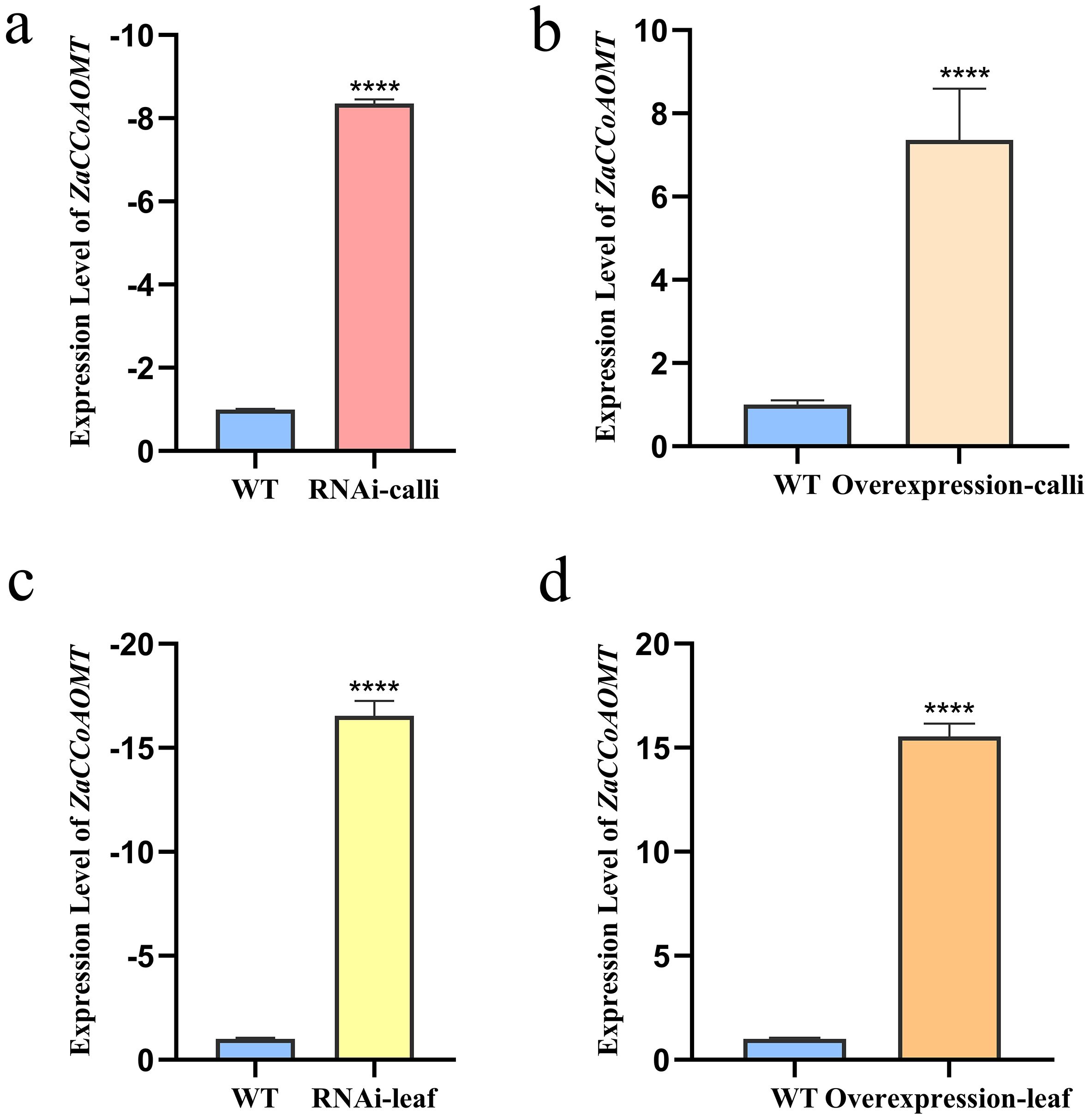
Figure 4. Relative expression level of ZaCCoAOMT. (a) Expression level of ZaCCoAOMT-RNAi in calli; (b) Expression level of ZaCCoAOMT-overexpression in calli; (c) Expression level of ZaCCoAOMT-RNAi in transformants; (d) Expression level of ZaCCoAOMT-overexpression in transformants. "* * * *" indicates a highly significant difference between the control group and the treatment group, p<0.001.
2.6.2 Expression of ZaCCoAOMT in transient transformants
The results of qPCR analysis showed that compared to the wild type, ZaCCoAOMT expression in the ZaCCoAOMT-RNAi transformants were significantly reduced, exhibiting a 16.55-fold downregulation relative to actin. Conversely, ZaCCoAOMT-overexpression transformants displayed a 15.54-fold upregulation (Figure 4). These observations align with the target gene detection results in positive calli following Agrobacterium coculture, demonstrating that interference and overexpression of the target gene can be achieved through in vitro synthesis of recombinant vectors.
2.7 Detection of disease resistance level of positive transformants
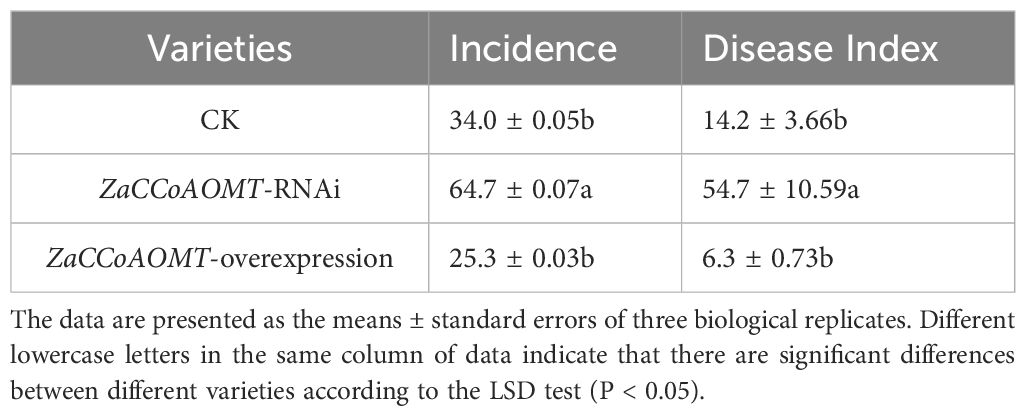
The wild-type plants served as the control, and a spore suspension (106 spores/mL) was prepared using freshly harvested urediospores. The control, ZaCCoAOMT-RNAi, and ZaCCoAOMT-overexpression groups were inoculated with the spore suspension. Disease incidence and severity index were assessed 20 days post-inoculation (Table 1). Notably, the ZaCCoAOMT-overexpression groups exhibited the lowest disease incidence (25.3 ± 0.03) and severity index (6.3 ± 0.73), demonstrating that overexpression of the disease resistance candidate gene significantly reduced susceptibility in Z. armatum compared to the control. In contrast, the ZaCCoAOMT-RNAi groups displayed the highest values (64.7 ± 0.07 incidence; 54.7 ± 10.59 severity index), confirming that RNAi-mediated suppression of the candidate gene markedly increased disease progression relative to the control.
Twenty days post-inoculation, symptom severity between the control and experimental groups exhibited visually distinct and statistically significant differences (Figure 5). In the control group, five sporulation foci developed on the abaxial leaf surface following spore suspension inoculation, with limited radial expansion. In contrast, the ZaCCoAOMT-overexpression group displayed only three sporulation foci, exhibiting markedly attenuated symptom severity compared to the control. Conversely, ZaCCoAOMT-RNAi plants manifested the most severe pathology, characterized by nine sporulation foci and extensive peripheral spread.

Figure 5. Symptoms of different treatment groups after spore suspension inoculation. (a) Symptom development on the adaxial leaf surface of control plants following inoculation with a spore suspension. (b) Symptom development on the abaxial leaf surface of control plants following inoculation with a spore suspension. (c) Symptom development on the adaxial leaf surface of ZaCCoAOMT-overexpression plants following inoculation with a spore suspension. (d) Symptom development on the abaxial leaf surface of ZaCCoAOMT-overexpression plants following inoculation with a spore suspension. (e) Symptom development on the adaxial leaf surface of ZaCCoAOMT-RNAi plants following inoculation with a spore suspension. (f) Symptom development on the abaxial leaf surface of ZaCCoAOMT-RNAi plants following inoculation with a spore suspension.
2.8 Analysis of total lignin and total flavonoid content
2.8.1 Analysis of total lignin content
The lignin content in leaves of wild-type, ZaCCoAOMT-overexpression, and ZaCCoAOMT-RNAi plants was quantified using the acetyl bromide method (Table 2). Notably, ZaCCoAOMT-overexpression Z. armatum exhibited the highest total lignin content (26.5 mg/g), representing a 15% increase compared to wild-type plants. Conversely, RNAi-mediated suppression of ZaCCoAOMT resulted in the lowest lignin accumulation (11.7 mg/g), corresponding to a 49% reduction relative to the wild type. Statistical analysis confirmed that lignin levels in ZaCCoAOMT-RNAi transgenic plants differed significantly from those in wild-type plants, whereas no significant difference was observed between ZaCCoAOMT-overexpression and wild-type lines.
2.8.2 Analysis of total flavonoid content
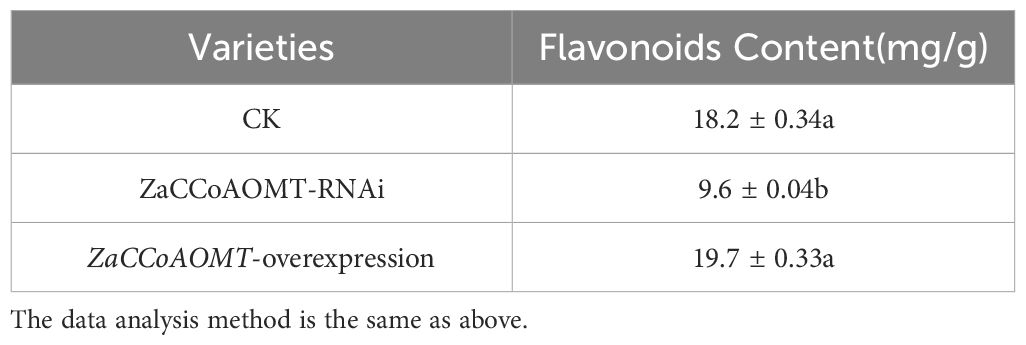
The flavonoid content in leaves of wild-type, ZaCCoAOMT-overexpression, and ZaCCoAOMT-RNAi plants was quantified via UV-Vis spectrophotometry (Table 3). ZaCCoAOMT-overexpression lines exhibited the highest flavonoid accumulation (19.7 mg/g), representing an 8% increase compared to wild-type plants. Conversely, RNAi-mediated silencing of ZaCCoAOMT resulted in the lowest total flavonoid content (9.6 mg/g), corresponding to a 47% reduction relative to the wild type. Statistical analysis revealed a significant difference in flavonoid levels between RNAi-silenced transformants and wild-type controls, whereas no significant variation was observed between overexpression lines and the control group.
2.8.3 Correlation analysis
Pearson correlation analysis was conducted to evaluate relationships between ZaCCoAOMT expression levels and disease incidence, disease severity index, lignin content, and flavonoid content, as well as pairwise interactions among these variables (Table 4). A significant negative correlation was observed between ZaCCoAOMT expression and both disease incidence and severity index, whereas positive correlations were detected with lignin and flavonoid accumulation. In RNAi-silenced transformants, lignin and flavonoid levels exhibited significant negative correlations with disease parameters. In contrast, no such correlations were observed in ZaCCoAOMT-overexpression plants. Notably, lignin and flavonoid content showed no significant interdependence in silenced transformants. These findings collectively demonstrate that ZaCCoAOMT modulates lignin and flavonoid biosynthesis pathways, which are integral to Z. armatum defense mechanisms against rust pathogens.
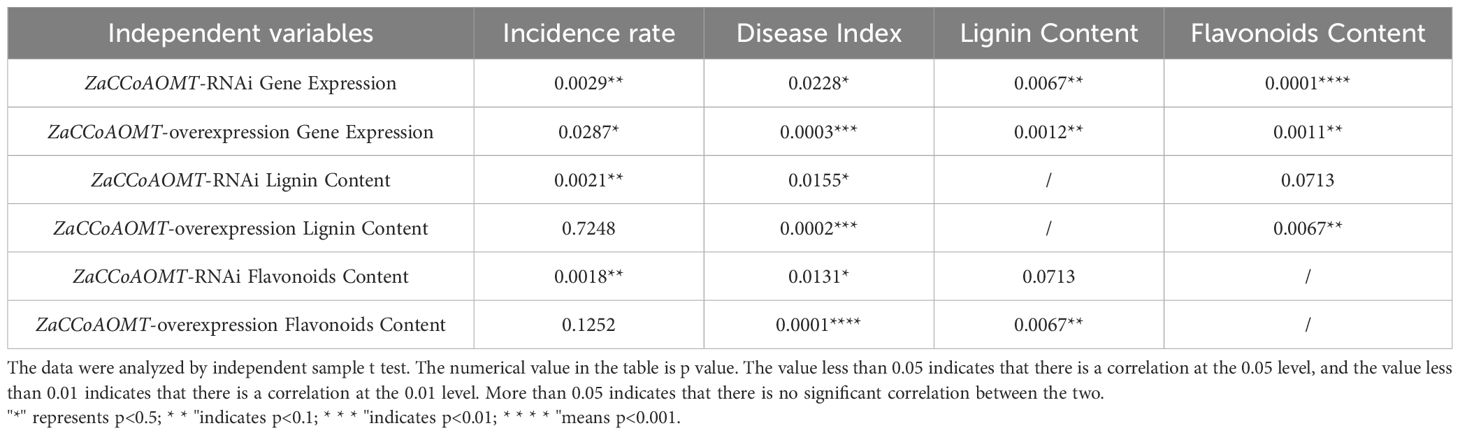
Table 4. Correlation analysis of gene expression, incidence, disease index, lignin content and total flavonoid content.
2.9 Gene expression analysis of transformants
2.9.1 Gene expression analysis in stable transformation
The STRING v11 protein-protein interaction database was utilized to analyze CCoAOMT in Z. armatum. Based on transcriptome annotations and prior qPCR validation of gene expression, six interacting proteins—HCT, F3H, F3M, 4CL, C3H, and C4H—were identified. To validate transcriptional expression levels of these proteins, primers (Appendix A.1) were designed using Primer 5, followed by qPCR quantification (Figure 6). In ZaCCoAOMT-RNAi transformants, transcriptional downregulation was observed in all six genes relative to wild-type plants. Notably, HCT, F3H, F3M, 4CL, and C4H exhibited significant downregulation, whereas C3H displayed a comparatively reduced but statistically significant decrease in expression. In contrast, ZaCCoAOMT-overexpression lines exhibited distinct transcriptional profiles, characterized by significant upregulation of HCT and F3H relative to wild-type plants, whereas F3M and C3H expression remained statistically unchanged. Notably, 4CL and C4H were markedly downregulated in overexpression lines, potentially attributable to lignin and flavonoid accumulation post-overexpression. This metabolic shift may activate a negative feedback mechanism, suppressing specific gene expression to limit further metabolite biosynthesis. Collectively, these findings demonstrate that ZaCCoAOMT orchestrates transcriptional networks governing phenylpropanoid biosynthesis—a critical pathway underpinning rust resistance in Z. armatum.
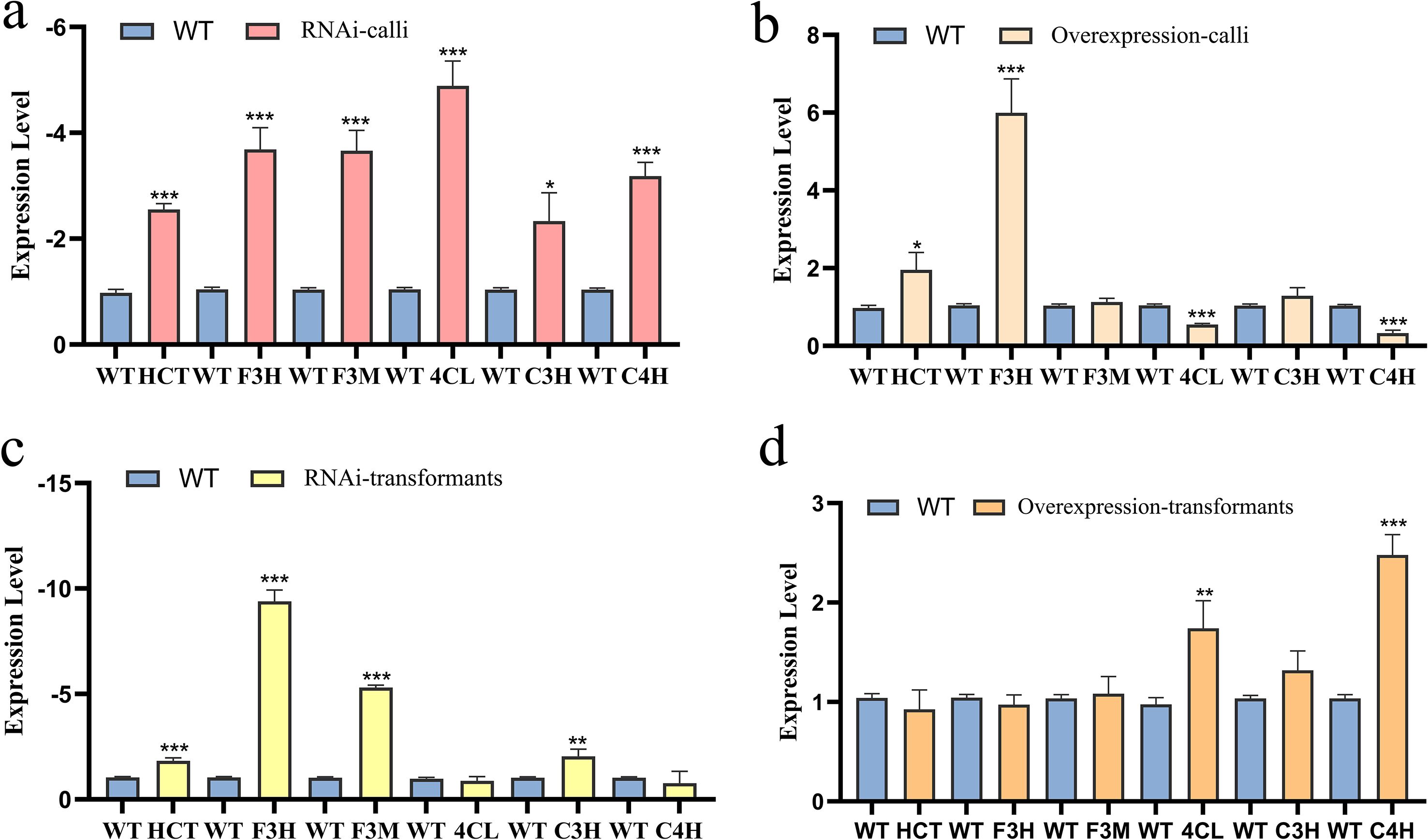
Figure 6. The gene expression levels of related proteins. (a) Expression levels of related genes in ZaCCoAOMT-RNAi calli; (b) Expression levels of related genes in ZaCCoAOMT-overexpresssion calli; (c) Expression levels of related genes in ZaCCoAOMT-RNAi transformants; (d) Expression levels of related genes in ZaCCoAOMT-overexpression transformants. In the figure, WT represents the relative expression levels of relevant genes in the wild type, and different colored bars represent the treatment groups. “*” indicates p<0.05, “* *” indicates p<0.01, and “* * *” indicates p<0.001.
2.9.2 Gene expression analysis of transient expression transformants
Changes in the transcriptional expression levels of six associated proteins following Agrobacterium-mediated transient transformation in Z. armatum are presented in Figure 6. In ZaCCoAOMT-RNAi transformants, HCT, F3H, F3M, and C3H were significantly downregulated relative to wild-type plants, whereas no significant differences in 4CL or C4H expression were observed. Conversely, in ZaCCoAOMT-overexpression lines, 4CL and C4H exhibited significant upregulation compared to wild-type controls, with no detectable changes in the remaining four genes. Comparative analysis of callus tissue and stable transformants revealed that transcriptional responses among the six genes diverged following silencing or overexpression treatments. These disparities could stem from tissue-specific heterogeneity, subtle variations in regulatory efficiency between experimental systems, or additional layers of transcriptional regulation that necessitate further investigation.
2.10 Subcellular localization of ZaCCoAOMT
The complete coding sequence of ZaCCoAOMT and homologous arms of the green fluorescent protein (GFP) vector were amplified, followed by verification and purification of the target fragments. The pSuper1300-GFP vector was linearized via double digestion with KpnI and XbaI, and the target fragment was ligated into the linearized vector. The ligation product was introduced into E. coli DH5α competent cells, and individual colonies were screened for insert size verification. Plasmids were extracted from liquid cultures and subsequently transformed into Agrobacterium competent cells. The resulting fusion construct was designated ZaCCoAOMT-GFP. Following 2–3 days of incubation on LB agar plates, a single colony was isolated and validated. A 15 bp homologous arm and 6 bp restriction site were engineered into the target fragment, yielding a theoretical product size of 770 bp. Agarose gel electrophoresis revealed bands within the 750–1000 bp range, with a prominent band near 750 bp (Appendix A.2.7), confirming successful transfer of the target fragment into Agrobacterium and its suitability for subsequent experiments.
The confirmed Agrobacterium tumefaciens strain carrying the target construct was cultured to logarithmic phase and infiltrated into the abaxial epidermis of Nicotiana benthamiana leaves grown for approximately one month. Following a 72 h incubation in a plant growth chamber at 25°C, cellular fluorescence was visualized and captured using laser-scanning confocal microscopy. The pSuper1300-GFP positive control exhibited cytoplasmic GFP accumulation with a diffuse localization pattern, while nuclear, and plasma membrane markers displayed expected compartmentalization. The ZaCCoAOMT-GFP fusion protein showed partial colocalization with nuclear markers but no overlap with plasma membrane signals, suggesting dual localization in both the nucleus and cytoplasm (Figure 7).
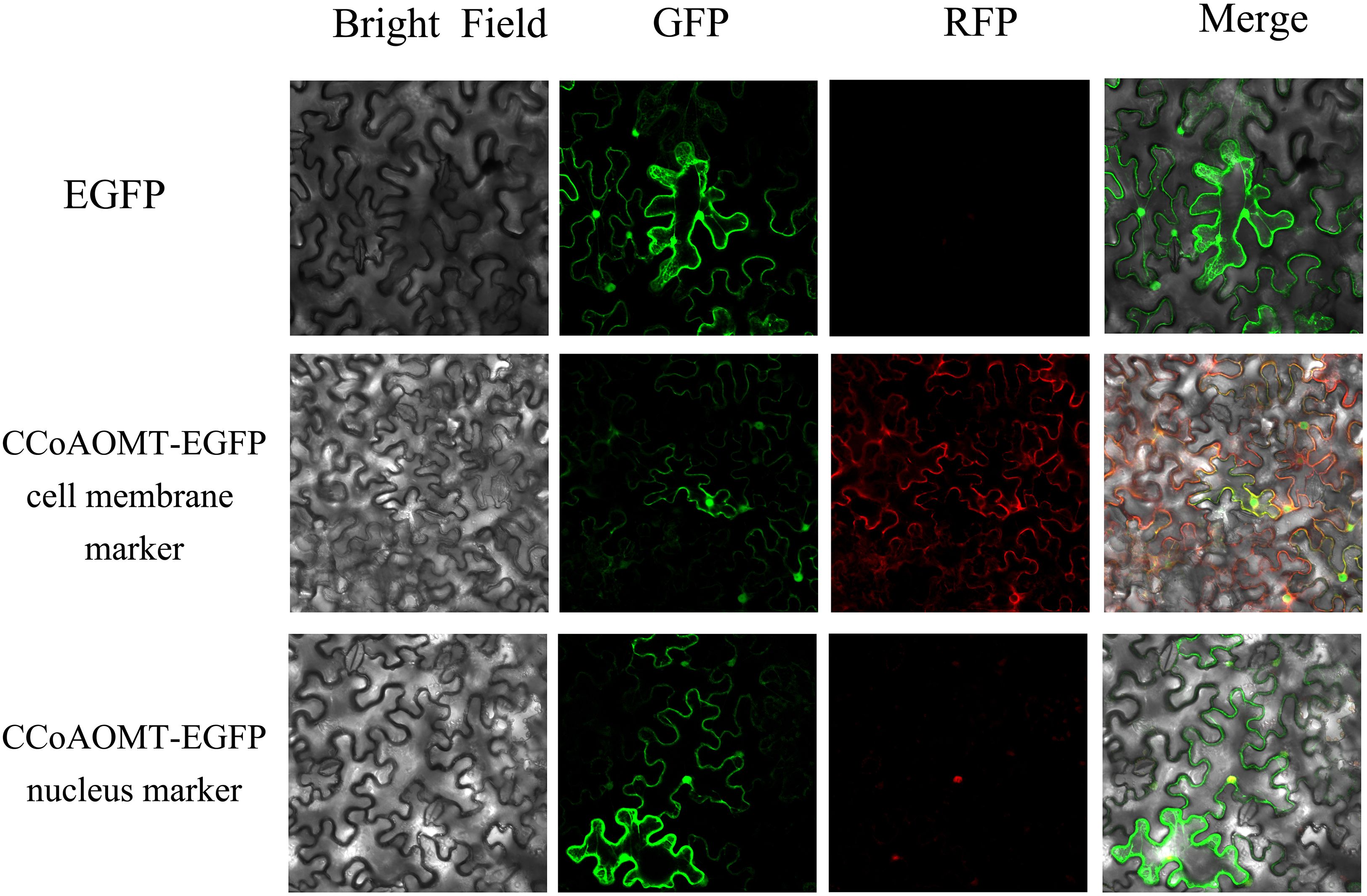
Figure 7. Subcellular localization of ZaCCoAOMT. The first row depicts the localization of green fluorescent protein (GFP) in bright field, green fluorescence, red fluorescence, and merged channels. The second row illustrates the subcellular distribution of the ZaCCoAOMT-GFP fusion protein and plasma membrane markers across bright field, green fluorescence, red fluorescence, and merged channels. The third row demonstrates the co-localization of the ZaCCoAOMT-GFP fusion protein with nuclear markers in bright field, green fluorescence, red fluorescence, and merged channels.
3 Discussions
Plant O-methyltransferases (OMTs) are classified into two categories based on protein size, substrate specificity, and Mg2+ dependence. Class I OMTs, designated as caffeoyl-CoA O-methyltransferases (CCoAOMTs), require Mg2+ for catalytic activity and exhibit a molecular mass range of 26–29 kDa. The cloned CCoAOMT sequence in Z. armatum encodes a 27.9 kDa protein, closely aligned with typical Class I plant OMTs in both molecular mass and structural features, consistent with reports in liverworts (Xu et al., 2022). Structural analysis revealed that the cloned CCoAOMT contains a conserved S-adenosylmethionine (SAM)-binding domain, indicating its functional capacity as an O-methyltransferase.This enzyme catalyzes methyl group transfer from SAM to caffeoyl-CoA substrates, a mechanism previously documented in Eriobotrya japonica (Liu et al., 2015), Camellia sinensis (Lin et al., 2019), and Sorghum bicolor (Walker et al., 2016). The structural determinants of CCoAOMT and its mechanistic role in rust resistance remain poorly characterized. To address this knowledge gap, the CCoAOMT ortholog from Z. armatum was cloned and functionally characterized. Mechanistic studies demonstrated that CCoAOMT modulates rust resistance in Z. armatum through dual pathways: (1) regulating lignin biosynthesis via monolignol methylation, thereby altering cell wall composition and structural integrity; (2) enhancing flavonoid stability and protein-binding capacity, collectively inhibiting progression of Z. armatum infections.
RNA interference (RNAi), an evolutionarily conserved sequence-specific regulatory mechanism in eukaryotes, mediates gene silencing at transcriptional or post-transcriptional levels (Bologna et al., 2018). Sequence-specific hairpin RNA production necessitates assembly of recombinant plasmid constructs containing inverted repeats. When regulated by constitutive promoters, inserted inverted repeat sequences generate long double-stranded hairpin RNAs that are efficiently processed by Dicer enzymes into siRNAs, enabling targeted gene suppression (Dang et al., 2011; Zhao and Guo, 2022).dsRNA recombinant constructs and overexpression vectors were developed, enabling successful establishment of both Agrobacterium-mediated transient transformation in Z. armatum and stable transgenic calli. Quantitative PCR analysis revealed substantial CCoAOMT suppression in RNAi lines and marked overexpression in transgenic systems compared to wild-type controls. These findings demonstrate the operational siRNA-mediated RNAi pathway in Z. armatum, where exogenous dsRNA introduction via recombinant constructs enables sequence-specific gene silencing.This approach establishes both methodological framework and mechanistic foundation for functional genomics studies in Zanthoxylum species, paralleling conserved RNAi mechanisms characterized in Arabidopsis thaliana (Dare et al., 2013) and Arnebia euchroma (Wang et al., 2005).
Caffeoyl-CoA O-methyltransferase contributes to plant cell wall fortification during induced disease resistance responses, with the cell wall serving as the primary structural barrier against pathogen penetration, thereby establishing ZaCCoAOMT critical role in phytopathogen defense (Xie et al., 2014). ZaCCoAOMT-RNAi transformants exhibited significantly higher infection rates (64.7%) compared to wild-type controls (34%), as quantified through standardized pathogen challenge assays. Conversely, ZaCCoAOMT-overexpression transgenic lines demonstrated markedly reduced infection incidence (25.3%) relative to control cohorts. Quantitative disease assessments revealed significantly elevated disease indices in silenced lines compared to control specimens. This functional validation aligns with conserved resistance mechanisms reported across plant taxa, confirming CCoAOMT essential role in mediating Z. armatum resistance against C. zanthoxyli induced rust pathogenesis (Schmitt et al., 1991).
Quantitative spectrophotometric assays revealed that RNAi-mediated suppression of CCoAOMT in Z. armatum resulted in significant reductions in both lignin (49% decrease) and total flavonoids (47% decrease), whereas ectopic overexpression elevated these metabolites by 15% and 8%, respectively, relative to wild-type controls. These findings demonstrate CCoAOMT dual regulatory role in lignin and flavonoid biosynthesis, corroborating prior functional studies (Li et al., 2013). A strong inverse correlation was observed between lignin accumulation and pathogen susceptibility metrics, with RNAi lines showing elevated disease indices (54.7 ± 10.59) and overexpression lines exhibiting enhanced resistance (disease index: 6.3 ± 0.73). Experimental data demonstrated a direct correlation between ZaCCoAOMT activity and rust resistance in Z. armatum. Elevation or reduction in lignin and total flavonoid content corresponded to proportional changes in the incidence rate and disease index of transformants. These opposing phenotypic trends established ZaCCoAOMT as a key regulator of a dual-layered defense strategy against C. zanthoxyli infection. Upon activation of the disease resistance response, this gene initiates cell wall fortification via lignin biosynthesis, whereby the plant cell wall serves as the primary barrier against pathogen invasion, thereby creating structural impediments to fungal penetration. Furthermore, ZaCCoAOMT activates chemical defenses by enhancing the accumulation of antifungal metabolites. These metabolites, either induced or pre-formed during pathogen invasion, function as phytotoxins that inhibit fungal hyphal elongation, thereby protecting plants from pathogenic colonization. The pivotal role of ZaCCoAOMT in, coordinating these defense mechanisms underscores its potential, as a molecular target for developing rust-resistant Z. armatum cultivars.
Lignin biosynthesis is mediated by a multi-enzyme consortium, including C3H, HCT, CCoAOMT, CCR, CAD, F5H, and COMT, working in conjunction with the core phenylpropanoid pathway enzymes PAL, C4H, and 4CL (Xu et al., 2011). For instance, transcriptional downregulation of C4H was observed to reduce mRNA abundance of PAL and 4CL, with PAL exhibiting more pronounced suppression (42% reduction) compared to C4H (28% reduction). Transcriptional abundance of key phenylpropanoid genes CCoAOMT and CAD decreased by 19–29% relative to wild-type controls (Kumar et al., 2016). RNAi-mediated suppression of CCoAOMT and CAD induced systemic transcriptional perturbations across the phenylpropanoid network, including downstream effects on 4CL, C4H, F3H, HCT, F3M, and C3H. Consequently, coordinated downregulation of lignin biosynthetic regulators impaired flux through the phenylpropanoid pathway, disrupting hydroxylation and reduction reactions essential for metabolite biosynthesis. This metabolic bottleneck reduced lignin deposition by 49% and flavonoid accumulation by 47% in silenced lines, whereas transgenic overexpression elevated these compounds by 15% and 8%, respectively. Therefore, after the expression of ZaCCoAOMT gene related to lignin synthesis was interfered, other genes in the phenylpropanoid metabolic pathway such as 4CL, C4H, F3H, HCT, F3M, and C3H genes were affected by the down-regulation of ZaCCoAOMT gene, resulting in the formation of, phenylpropanoid compounds. This impairs catalytic activity,, hydroxylation, and reduction processes critical to phenylpropanoid compound synthesis, thereby reducing lignin and flavonoid content.Despite the absence of significant differences in lignin and total flavonoid levels between ZaCCoAOMT-overexpression strains and controls, a paradoxical increase in rust resistance was observed, suggesting that resistance mechanisms extend beyond quantitative phenylpropanoid accumulation. Potential explanations for this phenomenon include: (i) localized metabolite deposition at infection sites. Inoculation with C. zanthoxyli occurs primarily in leaves, where ZaCCoAOMT expression is most pronounced. Lignin and flavonoid measurements were conducted in leaf tissues. Thus, ZaCCoAOMT overexpression may induce localized changes in secondary metabolite content within leaf tissues, conferring partial resistance to C. zanthoxyli; (ii) structural modifications of lignin polymers or flavonoid derivatives may enhance antifungal activity. These metabolites may function as defense compounds, either induced or pre-formed during infection, which inhibit fungal hyphal elongation to limit pathogenic colonization; (iii) activation of JA/SA-mediated defense signaling pathways. Following inoculation with C. zanthoxyli, JA/SA-mediated signaling pathways are potentially activated, synergizing with secondary metabolites to confer rust resistance in Z. armatum. These hypotheses necessitate validation through spatial metabolomics, histological staining, and transcriptomic profiling, and we intend to investigate these mechanisms in subsequent studies.
To elucidate CCoAOMT regulatory function in lignin and flavonoid biosynthesis, stable transgenic systems were generated through Agrobacterium-mediated transformation of Z. armatum calli, yielding distinct overexpression and RNAi-silenced lines. Quantitative analysis revealed systemic downregulation of lignin/flavonoid biosynthetic genes in silenced calli, whereas overexpression lines exhibited marked upregulation, mirroring transcriptional patterns observed in transient transformation assays. These coordinated transcriptional responses collectively demonstrate CCoAOMT pivotal role in modulating phenylpropanoid pathway activity across transformation platforms.
Prior to experimental validation, the subcellular localization of the CCoAOMT protein was predicted using bioinformatic tools, which indicated that the protein was most likely localized in the cytoplasm and nucleus at low probability. However, previous studies have documented variability in the subcellular localization of this gene across plant species (Widiez et al., 2011; Lv et al., 2018; Ma et al., 2021; Guo et al., 2023). To confirm this prediction experimentally, the complete coding sequence (CDS) of CCoAOMT was ligated into the pSuper 1300 plasmid fused with GFP for transient expression. Subcellular localization analysis demonstrated that CCoAOMT localized to both the nucleus and cytoplasm, consistent with the predicted subcellular distribution. Furthermore, analysis using TMHMM confirmed the absence of transmembrane domains, excluding its localization to the cell membrane and further corroborating the reliability of the experimental findings.
Z. armatum rust disease represents the most prevalent and severe foliar disease affecting this species. Current control strategies predominantly rely on chemical pesticides, which are associated with issues such as pathogen resistance and environmental contamination, underscoring the need for sustainable alternatives. Consequently, investigations into the resistance mechanisms of Z. armatum against the rust fungus C. zanthoxyli have been initiated. Exogenous application of 0.4 mg/mL salicylic acid (SA) was found to enhance the activities of CAT, POD, and PAL, elevate H2O2, lignin, and endogenous SA levels, and upregulate the SA signaling pathway-associated gene ZaPR1, thereby bolstering rust resistance in Z. armatum (Ren et al., 2024). Integrated transcriptomic and metabolomic analyses have suggested that lipid-derived plant hormone DAMs, particularly methyl jasmonate (MeJA), may activate rust resistance in Z. armatum by modulating phenylpropanoid and flavonoid biosynthesis pathways in leaf tissues (Han et al., 2023). Additionally, a strain of Ramularia coleosporii exhibiting potent parasitic activity against C. zanthoxyli was isolated and screened from Z. armatum urediospores. Formulated as a wettable powder, the strain demonstrated growth-inhibitory effects on C. zanthoxyli in both pot and field trials (Yuan, 2024). In this study, ZaCCoAOMT is proposed to function as a disease resistance gene, conferring rust tolerance in Z. armatum. Thus, elucidating plant resistance regulatory mechanisms, identifying robust rust-resistant genes in Z. armatum, and deciphering molecular resistance pathways are essential for achieving targeted rust resistance breeding. These findings hold significant implications for guiding future disease-resistant breeding programs in Z. armatum and establishing a scientific foundation for improved rust disease management.
4 Materials and methods
4.1 Materials
Two-year-old Z. armatum plants averaging 40 cm in height were selected from a cultivated Z. armatum garden in Hongya County, Sichuan Province (geographic coordinates: 33°59’N, 106°39’E). The plants were transferred to a greenhouse located in the Fifth Teaching Building of Sichuan Agricultural University, Wenjiang District, Chengdu. Growth conditions were maintained at 25–28°C, 70–80% relative humidity, and a 16-hour photoperiod followed by 8 hours of darkness. Urediospores from Z. armatum plants were aseptically collected using sterile surgical blades. Following one week of air-drying, the spores were separately stored at -20°C and -80°C in the Forest Protection Laboratory.
4.2 Methods
4.2.1 Extraction of total RNA and synthesis of cDNA from Z. armatum
Total RNA was isolated from young leaves of Z. armatum using the EASYspin Plus Rapid Plant RNA Extraction Kit. RNA integrity and concentration were assessed through 1% agarose gel electrophoresis and an ultra-micro spectrophotometer. Subsequently, cDNA synthesis was performed via reverse transcription of the RNA template using the Evo M-MLV Plus One-Step Kit. The synthesized cDNA was stored at –20°C for subsequent analyses.
4.2.2 Cloning of candidate genes
Based on the transcriptome-derived candidate gene sequence, primers for amplifying the full-length coding sequence (CDS) of CCoAOMT were designed using Primer 5.0 software, with the primer pair designated as CCoAOMT-F/R (Appendix A.1). The target sequence was amplified using Z. armatum cDNA as a template, followed by ligation of the purified PCR product into the pEASY®-T5 Zero Cloning Vector via TA cloning. Recombinant plasmids were confirmed through sequencing, after which the validated plasmid DNA was stored at –20°C. The corresponding bacterial glycerol stock was preserved at –80°C for long-term archival.
4.2.3 Gene sequence analysis and protein structure prediction
The coding sequence of the CCoAOMT gene was translated into amino acid sequences using standard bioinformatic tools. Corresponding CCoAOMT protein sequences from over 20 representative species, including members of the families Theaceae, Rutaceae, and Vitaceae, were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/). A phylogenetic tree of CCoAOMT homologs was constructed using MEGA 7.0 software with the neighbor-joining method. Additionally, multiple sequence alignment of the CCoAOMT amino acid sequences across species was performed using DNAMAN 8.0 software to identify conserved domains and functional residues.
The CCoAOMT protein sequence was retrieved, and its physicochemical properties were analyzed using multiple bioinformatics tools. Key parameters, including the isoelectric point (pI), instability index, grand average of hydropathicity (GRAVY), and molecular weight, were determined using the ExPASy ProtParam tool (http://web.expasy.org/protparam/). Conserved motifs within the protein were identified using the MEME Suite (https://meme-suite.org/meme/tools/meme). Signal peptide prediction was performed with SignalP-3.0 (https://services.healthtech.dtu.dk/service/SignalP-3.0), while secondary structure analysis was conducted using the SOPMA tool via the NPS@ server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). Finally, the tertiary structure of CCoAOMT was modeled using SWISS-MODEL (http://swissmodel.expasy.org/), with structural validation based on global quality estimation scores.
4.2.4 Tissue-specific expression analysis of CCoAOMT gene
To investigate the tissue-specific expression profile of CCoAOMT, root (R), stem (S), and leaf (L) tissues were collected from two-year-old Z. armatum plants, with three biological replicates per tissue type (nine samples total). Total RNA was isolated from the tissues, and cDNA was synthesized for downstream analysis. Gene-specific primers (qCCoAOMT-F/R; Appendix A.1) were designed using Primer 5.0 software and commercially synthesized for quantitative PCR (qPCR). The relative expression of the CCoAOMT gene across tissues was quantified via qPCR, with β-actin employed as the internal control gene. All reactions were performed in triplicate using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) with SYBR® Green Pro Taq HS Premix, and relative expression levels were calculated using the 2−ΔΔCT method (Zhou et al., 2023). Statistical significance (P < 0.05) between tissue types was determined through Duncan’s multiple range test, and graphical representations of the data were generated using GraphPad Prism 5.0.
4.2.5 Construction and transformation of the CCoAOMT-RNAi expression vector and CCoAOMT-overexpression vector
Gene-specific primers (T-ZX-F/R, T-FX-F/R, T-PDK-F/R, and BGT-F/R) were designed using SnapGene software. Amplification of each fragment was performed following the manufacturer’s protocol for the 2× TransTaq HiFi PCR SuperMix (TransGen Biotech). PCR products were subjected to agarose gel electrophoresis to confirm amplification success, followed by size-specific purification of the forward, reverse, and overexpression fragments for CCoAOMT RNA interference and overexpression constructs. The intermediate vector pHANNIBAL was linearized using XbaI restriction enzyme, and the linearized vector was purified post-digestion. The interference intron fragment was amplified using the same protocol as the forward and reverse fragments. The plant binary vectors pCAMBIA1301-35SN and BG Plant-Express MCS were digested with KpnI and BamHI, respectively, followed by purification of the digested vectors. Homologous recombination was performed using the Uniclone One-Step Seamless Cloning Kit, with the CCoAOMT forward, reverse, and intron fragments assembled into the linearized pHANNIBAL vector at a molar ratio of 1:1–1:10 (vector:insert). The target fragment was similarly ligated into the BG Plant-Express MCS vector. Recombinant plasmids were transformed into chemically competent E. coli DH5α cells, and colony PCR screening was conducted using primers 01yz-F/R and yz-F/R (Appendix A.1) to verify positive clones for both pCAMBIA1301-35SN and BG Plant-Express MCS constructs. Validated RNAi and overexpression vectors were preserved in sterile 25% glycerol and stored at –80°C for long-term use.
4.2.6 Agrobacterium-mediated transformation and identification of positive calli and positive plants
The RNAi recombinant product and overexpression recombinant product were transformed into DH5α, and a monoclonal expansion culture was selected. The plasmid was extracted, and the recombinant plasmid was transferred to Agrobacterium competent GV3101 by the liquid nitrogen freeze-thaw method. A single colony was selected to verify the positive Agrobacterium, and the correct positive Agrobacterium liquid culture was verified. The positive Agrobacterium solution was shaken to OD600 = 0.8, and centrifuged at 5000 rpm/min for 10 min, after which the supernatant was discarded. The cells were resuspended in MS (MS + 30 g/L sucrose, pH=5.7-5.9; acetosyringone was added before resuspension, and the final concentration was 100 μM) (Zhao et al., 2021).
Twelve Z. armatum plants exhibiting uniform growth were selected for transformation. Following the protocol described by Yang (Zhou et al., 2022), an A. tumefaciens suspension was applied to the adaxial and abaxial surfaces of the leaves via foliar spray. Leaf tissues were harvested three days post-infiltration, and genomic DNA was isolated using a standardized extraction protocol. To confirm successful transformation, PCR amplification of the hygromycin resistance gene (hptII) was performed using primer pairs hyg501-F/R and hyg359-F/R (Appendix A.1). Similarly, the kanamycin resistance gene (kan) was amplified with primers kan270-F/R and kan300-F/R to screen putative transgenic leaves. The infection protocol was repeated at three-day intervals for a total duration of one month to ensure sustained transformation efficiency.
Calli induction in Z. armatum was performed according to the protocol established by Yang et al (Yang, 2023), and calli exhibiting vigorous growth were selected for subsequent transformation. Calli from ZaCCoAOMT-RNAi, ZaCCoAOMT-overexpression, empty vector control, and sterile water negative control groups were immersed in an A. tumefaciens suspension for 10 minutes. Post-infection, calli were blotted dry on sterile filter paper and rinsed five times with sterile water to remove residual bacterial cells. The calli were then transferred to Murashige and Skoog (MS) medium supplemented with 1.5 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), 0.5 mg/L 6-benzylaminopurine (6-BA), 0.75 mg/L naphthaleneacetic acid (NAA), and 0.4 g/L acid-hydrolyzed casein, followed by incubation in darkness for 3 days. After co-cultivation, calli were rinsed five times with sterile water containing 500 mg/L cefotaxime and 250 mg/L timentin to eliminate residual Agrobacterium, blotted dry, and transferred to selection medium (MS basal salts, 1.5 mg/L 2,4-D, 0.5 mg/L 6-BA, 0.75 mg/L NAA, 0.4 g/L acid-hydrolyzed casein, 50 μg/L kanamycin, and 25 μg/L timentin) for secondary induction under dark conditions.
4.2.7 Relative expression level analysis of ZaCCoAOMT in transformants by qPCR
cDNA was isolated from wild-type, ZaCCoAOMT-RNAi, and ZaCCoAOMT-overexpression lines using the methodology described in preceding sections. Gene-specific primers (qCCoAOMT-F/R; Appendix A.1) targeting the RNAi fragment were designed using Primer 3.0 software, and qPCR amplification was performed using SYBR Green Pro Taq HS Premix (TransGen Biotech), with β-actin serving as the internal control. RNA extracted from wild-type calli and calli transformed with ZaCCoAOMT-RNAi or ZaCCoAOMT-overexpression constructs was reverse-transcribed into cDNA, which was subsequently used as the template for qPCR with the aforementioned primers. All reactions were conducted in triplicate, and gene expression levels were quantified using the 2−ΔΔCT method, with mean values calculated for statistical analysis.
4.2.8 Pathogen inoculation and disease resistance detection
Leaves of Z. armatum exhibiting rust infection symptoms were collected from field-grown plants, wrapped in sterile filter paper, and transported to the laboratory. Urediospores were carefully dislodged from infected leaf surfaces using a sterilized surgical blade, collected into sterile 1.5 mL microcentrifuge tubes, and dehydrated in a desiccator containing silica gel for 7 days. Dehydrated spores were stored at –20°C until further use. Prior to inoculation, urediospores were retrieved from storage and surface-sterilized by suspending them in sterile distilled water. A spore suspension was prepared by evenly distributing spores onto 1.3% (w/v) water agar medium and incubating at 22°C for 24 hours to hydrate and activate germination. Activated spores were washed twice via gentle centrifugation (500 × g, 10 min) and resuspended in sterile water to a final concentration of 106 spores/mL. For inoculation, leaf surfaces were pre-treated with 0.1% (v/v) Triton X-20 detergent solution to enhance spore adhesion. The spore suspension was uniformly sprayed onto adaxial and abaxial leaf surfaces using an atomizer. Inoculated plants were maintained at 100% relative humidity for 12 hours in a growth chamber (25°C, 16/8 h light/dark cycle). Disease incidence and severity indices were quantified 20 days post-inoculation according to the standardized protocol described by Zhang et al (Luo et al., 2022).
Disease incidence (DI) was calculated using the following formula (Equation 1):
where I represents the incidence rate, D represents the number of leaves exhibiting rust symptoms, and T denotes the total number of leaves assessed.
Disease severity was classified into five grades based on the percentage of leaf area covered by uredinia (or teliospore sori):
a. Grade 0: No visible uredinia/teliospores.
b. Grade 1: Uredinia/teliospores occupying ≤10% of the leaf area.
c. Grade 2: 11–33% of the leaf area affected.
d. Grade 3: 34–50% of the leaf area affected.
e. Grade 4: >50% of the leaf area affected.
The disease severity index (DSI) was calculated as follows (Equation 2):
4.2.9 Total lignin content and total flavonoid content
Lignin content in ZaCCoAOMT-RNAi, ZaCCoAOMT-overexpression, and wild-type Z. armatum leaves was quantified via the acetyl bromide method following the protocol described by Moreira-Vilar et al (Wang et al., 2013). Briefly, leaf tissues were freeze-dried at 80°C until a constant weight was achieved, homogenized using liquid nitrogen, and sieved through a 40-mesh screen. A 2 mg aliquot of the resulting powder was processed according to the manufacturer’s instructions provided with the GraceBio Lignin Content Assay Kit (Grace Biotechnology Co., Ltd.). Absorbance measurements were performed at 280 nm using a UV-Vis spectrophotometer (Model UV-2600, Shimadzu), and lignin concentration was calculated using the formula (Equation 3):
where W is the sample weight; and 4 is the dilution multiple of the supernatant when detected in the cuvette.
In an alkaline nitrite solution, flavonoids form a red complex with aluminum ions, which exhibits a characteristic absorption peak at 510 nm. Based on this principle, the total flavonoid content in plant samples was quantified using the NaNO2-Al(NO3)3-NaOH chromogenic method (Cheng et al., 2024). Samples were heat-inactivated at 105°C for 3 minutes, dried at 60°C to a constant weight, ground into powder, and sieved through a 40-mesh sieve to obtain homogenized dry material. A 0.03 g aliquot of the dried sample was mixed with 1.5 mL of 60% ethanol, incubated at 60°C for 2 h with shaking, and centrifuged to collect the supernatant for analysis. The supernatant was subsequently centrifuged and assayed in accordance with the manufacturer’s protocol for the designated kit (Equation 4).
where W is the sample weight; V is the volume of the extract; and D is the dilution factor.
4.2.10 Analysis of genes related to phenylpropanoid metabolic pathway
The ZaCCoAOMT gene was analyzed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins), a functional protein interaction database, and integrated with annotated transcriptomic data from Z. armatum. Six interacting proteins were identified through qPCR data analysis of previously identified candidate genes. These included 4-coumarate-CoA ligase, hydroxycinnamoyltransferase, flavanone 3-hydroxylase, flavanone 3-monooxygenase, coumarate 3-hydroxylase, and cinnamate 4-hydroxylase. qPCR primers (Appendix A.1) for the six interacting proteins were designed using Primer-BLAST software, with wild-type Z. armatum cDNA (control) and the β-actin housekeeping gene as an internal reference. Amplification and analysis were performed via real-time fluorescence quantification, as outlined in Section 4.2.7.
4.2.11 Subcellular localization of the ZaCCoAOMT gene
The primers ZaCCoAOMT-EGFP-F/R (Appendix A.1) were designed according to the coding sequence (CDS) of ZaCCoAOMT. The ZaCCoAOMT coding sequence, flanked by homologous arms, was amplified via PCR using Z. armatum cDNA as a template, and the resulting PCR products were purified. KpnI and XbaI restriction enzymes were selected to linearize the green fluorescent protein vector Super1300. The purified target fragment was ligated into the linearized Super1300 vector and subsequently transformed into DH5α competent cells. Positive transformants were confirmed by PCR amplification using the 00yz-F/R primers and validated by Sanger sequencing. Plasmid DNA was extracted from verified monoclonal bacterial cultures and introduced into A. tumefaciens GV3101 competent cells via chemical transformation. N. benthamiana leaves were infiltrated with recombinant Agrobacterium suspension, and tissue sections were prepared from infiltrated regions 48–72 hours post-infiltration. Fluorescence was visualized in leaf tissues using laser scanning confocal microscopy.
4.2.12 Statistical analysis
Gene expression levels of target genes, disease incidence, disease index, total lignin content, and total flavonoid content were visualized using GraphPad Prism software and analyzed for correlations via Pearson coefficient calculations. Statistical significance was evaluated using SPSS software (IBM) through appropriate parametric tests.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XX: Conceptualization, Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. YH: Formal Analysis, Methodology, Writing – review & editing. YML: Formal Analysis, Methodology, Writing – review & editing. TZ: Conceptualization, Supervision, Writing – review & editing. SjL: Methodology, Resources, Writing – review & editing. YL: Resources, Writing – review & editing. SyL: Methodology, Writing – review & editing. CY: Methodology, Software, Writing – review & editing. SH: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the General Project of Sichuan Provincial Department of Science and Technology (24NSFSC0799).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. AI was used to polish the manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2025.1604811/full#supplementary-material
References
Aktar-uz-zaman M. D., Tuhina-khatun M., Hanafi M. M., and Sahebi M. (2017). Genetic analysis of rust resistance genes in global wheat cultivars: an overview. Biotechnol. Biotechnol. Equip. 31, 431–445. doi: 10.1080/13102818.2017.1304180
Alejandro S., Lee Y., Tohge T., Sudre D., Osorio S., Park J., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22, 1207–1212. doi: 10.1016/j.cub.2012.04.064, PMID: 22704988
Bologna N. G., Iselin R., Abriata L. A., Sarazin A., Pumplin N., Jay F., et al. (2018). Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell 69, 709–719. doi: 10.1016/j.molcel.2018.01.007, PMID: 29398448
Cao Y., Chen Y., Zhang L., and Cai Y. (2023). Two monolignoid biosynthetic genes 4-coumarate: coenzyme A ligase (4CL) and p-coumaric acid 3-hdroxylase (C3H) involved in lignin accumulation in pear fruits. Physiol. Mol. Biol. Plants 29, 791–798. doi: 10.1007/s12298-023-01329-1, PMID: 37520811
Cao Y., Feng X., Ding B., Huo H., Abdullah M., Hong J., et al. (2025). Gap-free genome assemblies of two Pyrus bretschneideri cultivars and GWAS analyses identify a CCCH zinc finger protein as a key regulator of stone cell formation in pear fruit. Plant Commun. 6, 3. doi: 10.1016/j.xplc.2024.101238, PMID: 40071379
Chen L., Ren J., Xiang P., Mou H., Li L., Qiao X., et al. (2022). Optimisation of prevention and control technologies for pepper rust in Chongqing. South. Agric. 16, 71–74. doi: 10.19415/j.cnki.1673-890x.2022.05.018
Cheng Y., Ding M., Wang S., and Fan Z. (2024). Preparation of liposomes of Vaccinium myrtillus leaf flavonoids and their effects on HepG2 cells. Fine Chem. Industry 2024, 1–15. doi: 10.13550/j.jxhg.20230866
Dang Y., Yang Q., Xue Z., and Liu Y. (2011). RNA interference in fungi: pathways, functions, and applications. Eukaryotic Cell 10, 1148–1155. doi: 10.1128/EC.05109-11, PMID: 21724934
Dare A. P., Tomes S., Jones M., Mcghie T.K., Stevenson D.E., Johnson R.A., et al. (2013). Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus× domestica). Plant J. 74, 398–410. doi: 10.1111/tpj.12140, PMID: 23398045
Feng S., Liu Z., Cheng J., Li Z., Tian L., Liu M., et al. (2021). Zanthoxylum-specific whole genome duplication and recent activity of transposable elements in the highly repetitive paleotetraploid Z. bungeanum genome. Horticulture Res. 8(205). doi: 10.1038/s41438-021-00665-1, PMID: 34480029
Feng S., Liu Z., Hu Y., Tian J., Yang T., Wei A., et al. (2020). Genomic analysis reveals the genetic diversity, population structure, evolutionary history and relationships of Chinese pepper. Horticulture Res. 2020, 7. doi: 10.1038/s41438-020-00376-z, PMID: 33082965
Gharibi S., Tabatabaei B.E.S., Saeidi G., Talebi M., and Matkowski A. (2019). The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech. f. Phytochemistry 162, 90–98. doi: 10.1016/j.phytochem.2019.03.004, PMID: 30875522
Gho Y., Kim S., and Jung K. (2020). Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice. Genes Genomics 42, 67–76. doi: 10.1007/s13258-019-00879-7, PMID: 31736007
Guo Z., Si X.g., Jiao L., and Liu D. (2023). Cloning and structural analysis of soybean caffeoyl coenzyme A-O-methyltransferase (CCoAOMT) gene. Fujian J. Agric. 38, 616–623. doi: 10.19303/j.issn.1008-0384.2023.05.013
Han S., Xu X., Yuan H., Li S., Lin T., Liu Y., et al. (2023). Integrated transcriptome and metabolome analysis reveals the molecular mechanism of rust resistance in resistant(Youkang) and susceptive(Tengjiao) zanthoxylun armatun cultivars. Int. J. Mol. Sci. 24(19), 14761. doi: 10.3390/ijms241914761, PMID: 37834210
He Y., Hao Q., Chen P., Qin Y., Peng M., Yao S., et al. (2023). Cloning of PmMYB6 in Pinus massoniana and an analysis of its function. Int. J. Mol. Sci. 24, 13766. doi: 10.3390/ijms241813766, PMID: 37762069
Kumar R., Vashisth D., Misra A., Akhtar M.Q., Jalil S.U., Shanker K., et al. (2016). RNAi down-regulation of cinnamate-4-hydroxylase increases artemisinin biosynthesis in Artemisia annua. Sci. Rep. 6, 26458. doi: 10.1038/srep26458, PMID: 27220407
Li J., Ji X., Mao R., Cao F., and Li M.. (2023). Increasing lignin accumulation in arabidopsis and poplar by overexpressing a CCoAOMT gene from the dove tree (Davidia involucrata baill.). J. Plant Growth Regul. 42, 4095–4105. doi: 10.1007/s00344-022-10872-2
Li W., Deng Y., Ning Y., He Z., and Wang G. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603. doi: 10.1146/annurev-arplant-010720-022215, PMID: 32197052
Li X., Chen W., Zhao Y., Xiang Y., Jiang H., Zhu S., et al. (2013). Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet. Mol. Biol. 36, 540–546. doi: 10.1590/S1415-47572013005000039, PMID: 24385858
Lin H., Luo Y., Chen S., Yu S., Zeng Z., Liu Z., et al. (2019). Cloning of tea tree CCoAOMT gene and structural analysis of promoter. Mol. Plant Breed. 17, 1804–1813. doi: 10.13271/j.mpb.017.001804
Liu Y., Zou D., Wu B., Lin D., Zhang Z., Wu J., et al. (2015). Cloning and expression analysis of a CCoAOMT homolog in loquat fruit in response to low-temperature storage. Postharvest Biol. Technol. 105, 45–50. doi: 10.1016/j.postharvbio.2015.03.008
Luo F., Yan P., Xie L., Li S., Zhu T., Han S., et al. (2022). Molecular Mechanisms of Phenylpropane-Synthesis-Related Genes Regulating the Shoot Blight Resistance of Bambusa pervariabilis × Dendrocalamopsis grandis. Int. J. Mol. Sci. 23, 6760. doi: 10.3390/ijms23126760, PMID: 35743217
Lv X., Wang Y., and Jin Y. (2018). Cloning, expression and bioinformatics analysis of BvM14-CCoAOMT gene in sugar beet. J. Natural Sci. Heilongjiang Univ. 35, 317–323. doi: 10.13482/j.issn1001-7011.2018.05.113
Ma Q., Yan Q., Zhang Z., Wu F., and Zhang J. (2021). Identification, evolution and expression analysis of the CCoAOMT gene family in alfalfa. J. Grass Industry 30, 144–156. doi: 10.11686/cyxb2020429
Mao C., Zheng L., Zhu T., and Du X. (2014). Research on the incidence pattern and pharmaceutical control of vine pepper rust. Sichuan Forestry Sci. Technol. 35, 1–8. doi: 10.16779/j.cnki.1003-5508.2014.06.001
Nakashima J., Chen F., Jackson L., Shadle G., and Dixon R.A. (2008). Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol. 179, 738–750. doi: 10.1111/j.1469-8137.2008.02502.x, PMID: 18547377
National Pharmacopoeia Commission (2020). Pharmacopoeia of the People’s Republic of China - A Part (Haidian District, Beijing, China: China Pharmaceutical Science and Technology Press).
Parvathi K., Chen F., Guo D., Blount J.W., and Dixon R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25, 193–202. doi: 10.1111/j.1365-313X.2001.00956.x, PMID: 11169195
Purohit S., Joshi K., Rawat V., Bhatt I.D., and Nandi S.K. (2020). Efficient plant regeneration through callus in Zanthoxylum armatum DC: an endangered medicinal plant of the Indian Himalayan region. Plant Biosystems-An Int. J. Dealing all Aspects Plant Biol. 154, 288–294. doi: 10.1080/11263504.2019.1610107
Ren Y., Liu J., He L., Jiang J., Gong T., Zhou Q., et al. (2024). Effect of Salicylic Acid Treatment on Disease Resistance to Coleosporium zanthoxyli in Chinese Pepper(Zanthoxylun armatun). Plant Dis. 108. doi: 10.1094/PDIS-09-23-1880-RE, PMID: 37858967
Schmitt D., Pakusch A., and Matern U. (1991). Molecular cloning, induction and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J. Biol. Chem. 266, 17416–17423. doi: 10.1016/S0021-9258(19)47389-4, PMID: 1894629
Sharma A., Shahzad B., Rehman A., Bhardwaj R., Landi M., Zheng B., et al. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24, 2452. doi: 10.3390/molecules24132452, PMID: 31277395
Shi R., Sun Y., Li Q., Heber S., Sederoff R., Chiang V.L., et al. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51, 144–163. doi: 10.1093/pcp/pcp175, PMID: 19996151
Tang Y., Cao Z., and Wang J. (2015). Research on the occurrence pattern and chemical control of pepper rust. J. Northwest Forestry Coll. 30, 150–153. doi: 10.3969/j.issn.1001-7461
Walker A. M., Sattler S. A., Regner M., Jones J.P., Ralph J., Vermerris W., et al. (2016). The structure and catalytic mechanism of Sorghum bicolor caffeoyl-CoA O-methyltransferase. Plant Physiol. 172, 78–92. doi: 10.1104/pp.16.00845, PMID: 27457122
Wang B., Wu C., Wang G., He J., and Zhu S. (2021). Transcriptomic analysis reveals a role of phenylpropanoid pathway in the enhancement of chilling tolerance by pre-storage cold acclimation in cucumber fruit. Scientia Hortic. 288, 110282. doi: 10.1016/j.scienta.2021.110282
Wang J., Cao D., and Zhang Y. (2013). Determination of lignin in cottonseed hulls by acetyl bromide method. Textile J. 34, 12–16. doi: 10.13475/j.fzxb.2013.09.011
Wang L., Chen M., Lam P., Dini-Andreote F., Dai L., Wei Z., et al. (2022). Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 10, 233. doi: 10.1186/s40168-022-01420-x, PMID: 36527160
Wang L., Lam L., Liu A., Zhu Chen F. M., and Liu H. (2020). Flavonoids are indispensable for complete male fertility in rice. J. Exp. Bot. 71, 4715–4728. doi: 10.1093/jxb/eraa204, PMID: 32386058
Wang T., Iyer L. M., Pancholy R., Shi X., and Hall T.C. (2005). Assessment of penetrance and expressivity of RNAi-mediated silencing of the Arabidopsis phytoene desaturase gene. New Phytol. 167, 751–760. doi: 10.1111/j.1469-8137.2005.01454.x, PMID: 16101912
Wei Z., Guo J., Wang M., Li D., He C., Chen Q., et al. (2023). Hazards of pepper rust disease and comprehensive prevention and control measures. Sichuan Agric. Sci. Technol. 2023, 56–58. doi: 10.3969/j.issn.1004-1028
Widiez T., Hartman T. G., Dudai N., Yan Q., Lawton M., Havkin-Frenkel D., et al. (2011). Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol. Biol. 76, 475–488. doi: 10.1007/s11103-011-9772-2, PMID: 21629984
Wu Y., Luo H., Wang L., Zhu J., Yang X., Liu J., et al. (2023). Major pests and diseases of pepper in Guizhou and their coping strategies. Anhui Agric. Bull. 29, 96–101. doi: 10.3969/j.issn.1007-7731
Wu Y., Zhang C., Huang Z., Lyu L., Li W., Wu W., et al. (2022). Integrative analysis of the metabolome and transcriptome provides insights into the mechanisms of flavonoid biosynthesis in blackberry. Food Res. Int. 153, 110948. doi: 10.1016/j.foodres.2022.110948, PMID: 35227472
Xi G., Zhao N., Zhao J., and Zhao G. (2018). Analysis on the identification of a new disease of Dendrobium rosenbergii pepper sheath rust. J. Southwest Forestry Univ. (Natural Science) 38, 202–205. doi: 10.11929/j.issn.2095-1914.2018.02.033
Xia H., Lin Z., He Z., Guo Y., Liu X., Deng H., et al. (2023). AcMADS32 positively regulates carotenoid biosynthesis in kiwifruit by activating AcBCH1/2 expression. Int. J. Biol. Macromolecules 242, 124928. doi: 10.1016/j.ijbiomac, PMID: 37224896
Xie T., Liu Y., Wang S., Liu T., Kang L., Guo L., et al. (2014). Overexpression and RNAi vectors built for key secondary metabolic pathway genes PAL, HMGR, PGT of Arnebia euchroma. China J. Chin. Materia Med. 39, 4538–4543. doi: 10.4268/cjcmm20142312, PMID: 25911797
Xu B., Escamilla Treviño L. L., Sathitsuksanoh N., Shen Z., Shen H., Zhang Y., et al. (2011). Silencing of 4-coumarate: coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 192, 611–625. doi: 10.1111/j.1469-8137.2011.03830.x, PMID: 21790609
Xu R., Ni R., Gao S., Fu J., Xiong R., Zhu T., et al. (2022). Molecular cloning and characterization of two distinct caffeoyl CoA O-methyltransferases (CCoAOMTs) from the liverwort Marchantia paleacea. Plant Sci. 314, 111102. doi: 10.1016/j.plantsci.2021.111102, PMID: 34895539
Yang J. (2023). Identification and control of major disease pathogens of pepper in Guizhou Vol. 2023 (Guizhou University). doi: 10.27047/d.cnki.ggudu.2023.003312
Yang B. (2023). Mechanism of lignin synthesis regulation by VcMYB4a transcription factor in lingonberry and screening of interacting proteins Vol. 2023 (Guiyang City, Guizhou Province: Jilin University). doi: 10.27162/d.cnki.gjlin.2022.002912
Ye M. (2020). Development status and prospect of Sichuan pepper industry. China Rural Sci. Technol. 2020, 70–73. doi: 10.3969/j.issn.1005-9768
You C. (2012). Taxonomy and molecular phylogeny of the Chinese sheath rust fungus Vol. 2012 (Haidian District, Beijing: Beijing Forestry University).
Yuan H. (2024). Establishment of a Molecular Detection System for CHINA Pepper Rust and Development Application of Biocontrol Agents Vol. 2024 (Chengdu City, Sichuan Province: Sichuan Agricultural University). doi: 10.27345/d.cnki.gsnyu.2024.000735
Yuan H., Fang H., Xu X., et al. (2024). Isolation, identification and optimisation of spore-producing conditions of the pathogenic heavy parasitic bacteria of bamboo leaf pepper rust. J. Sichuan Agric. Univ. 42, 85–93. doi: 10.16036/j.issn.1000-2650.202308300
Yue X. (2010). Studies on the inherent resistance of pepper to Sphingomonas sp Vol. 2010 (Xianyang City, Shaanxi Province: Northwest Agriculture and Forestry University).
Zhang H. and Ye M. (2010). Taxonomic status and current status of compositional studies on green peppercorns. Northern Horticulture. 14), 199–203.
Zhao J. and Guo H. (2022). RNA silencing: From discovery and elucidation to application and perspectives. J. Integr. Plant Biol. 64, 476–498. doi: 10.1111/jipb.13213, PMID: 34964265
Zhao F., Wang J., Gong W., Fan J., and Xi W. (2021). Progress in tissue culture of pepper plants. World Forestry Res. 34, 32–37. doi: 10.13348/j.cnki.sjlyyj.2020.0112.y
Zheng L. and Zhu T. (2012). Research on the pathogenic bacteria of vine pepper rust. Sichuan Forestry Sci. Technol. 33, 28–31. doi: 10.3969/j.issn.1003-5508
Zhou R., Dong Y., Liu X., Feng S., Wang C., Ma X., et al. (2022). JrWRKY21 interacts with JrPTI5L to activate the expression of JrPR5L for resistance to Colletotrichum gloeosporioides in walnut. Plant J. 111, 1152–1166. doi: 10.1111/tpj.15883, PMID: 35765867
Zhou L., Zhu T., Han S., Li S., Liu Y., Lin T., et al. (2023). Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis. Int. J. Mol. Sci. 24, 4879. doi: 10.3390/ijms24054879, PMID: 36902308
Zhu P., Zhong Y., Luo L., Shen J., Sun J., Li L., et al. (2023). The MPK6-LTF1L1 module regulates lignin biosynthesis in rice through a distinct mechanism from Populus LTF1. Plant Sci. 2023, 111890. doi: 10.1016/j.plantsci.2023.111890, PMID: 37813192
Keywords: Z. armatum, resistance genes, CCoAOMT, RNAi, overexpression
Citation: Xu X, Hu Y, Lai Y, Zhu T, Li S, Liu Y, Li S, Yang C and Han S (2025) CCoAOMT positively regulates the biosynthesis of secondary metabolites in Zanthoxylum armatum. Front. Agron. 7:1604811. doi: 10.3389/fagro.2025.1604811
Received: 02 April 2025; Accepted: 13 June 2025;
Published: 28 July 2025.
Edited by:
Aziz Aziz, Université de Reims Champagne-Ardenne, FranceReviewed by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaPeitao Lü, Fujian Agriculture and Forestry University, China
Copyright © 2025 Xu, Hu, Lai, Zhu, Li, Liu, Li, Yang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shan Han, MTM3MjJAc2ljYXUuZWR1LmNu
 Xiu Xu
Xiu Xu Yingxia Hu1
Yingxia Hu1 Shujiang Li
Shujiang Li Chunlin Yang
Chunlin Yang