- 1Microbial Engineering Laboratory, Microbial Research Institute of Liaoning Province, Chaoyang, Liaoning, China
- 2College of Chemistry and Environmental Engineering, China University of Mining and Technology, Beijing, China
Introduction: This study aimed to evaluate the ability of Pseudomonas E sp017968885 to mobilize insoluble phosphate under different phosphorus (P) fertilizer application rates, as well as its impacts on soil properties and pepper yield—providing insights for optimizing P fertilizer application strategies in sustainable agriculture.
Methods: A pot experiment was conducted, consisting of six treatments: five inoculated groups (P1+H50 to P5+H50) treated with varying P fertilizer rates (1.17~1.96 g/pot) and one non-inoculated group (Control) as the reference. Key soil and crop indicators, along with microbial gene expression, were measured to assess the strain’s effects.
Results: At the fruit setting stage, the content of Olsen-P (available phosphorus) in the P1+H50 to P3+H50 treatments increased by 66.36~102.08% compared with the Control, with a significant difference (P<0.05). The P2+H50 treatment significantly enhanced the activity of soil acid phosphatase (S-ACP) by 16.79% relative to the Control (P<0.05). Pepper P uptake in the inoculated treatments increased by 32.67~84.06% (P<0.05), which was correlated with a 23.98~65.80% increase in yield compared with the Control (P<0.05). The expression of phosphate-solubilizing ability-related genes (e.g., pqq, gdh) supported the observed changes in P-related physicochemical indicators. Additionally, Pseudomonas E sp017968885 exhibited genetic potential in secondary metabolite synthesis, protein secretion, and siderophore production—providing a molecular basis for its advantages in environmental adaptation and ecological competition.
Discussion: These findings confirm that Pseudomonas E sp017968885 is a promising biofertilizer. It can reduce external P input while improving crop productivity by enhancing nutrient cycling, which is of great significance for promoting sustainable agricultural development.
1 Introduction
Chili pepper (Capsicum annuum L.) is a significant economic crop and condiment, occupying a crucial position in China's agricultural production and food industry (Deng et al., 2024). China has a long history of chili pepper cultivation, with a wide geographic distribution (Ao et al., 2024). A core planting pattern has been established in the Southwest, Central China, and North China regions. Provinces such as Guizhou, Sichuan, Hunan, Hubei, and Henan form the main production bases (Zou et al., 2020). In terms of consumer markets, as of 2021, the consumer base for chili peppers in China has exceeded 500 million people (Zou et al., 2025). With the increase in residents' income levels and the diversification of dietary structures, the application of chili peppers in food processing, catering services, and health products continues to expand (Azlan et al., 2022). China is not only the world's largest producer and consumer of chili peppers but also a significant exporter. According to statistical data from 2022, the total trade value of chili peppers in China reached 2.11 billion US dollars, with export value increasing by 11.6% year-on-year to 1.71 billion US dollars, demonstrating strong international market competitiveness (Akram et al., 2017) (http://www.e658.cn/jck/).
However, the phosphorus requirements of chili peppers during cultivation and the current status of phosphorus fertilization have not received adequate attention. Phosphorus is a key nutrient for the growth and development of chili peppers, especially during the flowering and fruiting stages, where sufficient phosphorus supply is critical for yield formation and quality improvement (Cui et al., 2023). Research indicates that for every 1,000 kg of chili pepper fruit produced, approximately 0.5-0.8 kg of phosphorus is absorbed (Liang et al., 2024). Nevertheless, the phosphorus status in the soils of major chili pepper production areas in China varies significantly, with some soils having low available phosphorus content that fails to meet the growth requirements of chili peppers (Lu and Tian, 2017). There are also pronounced issues in phosphorus fertilizer management within chili pepper cultivation systems. Survey data show that in some regions, the application rate of phosphorus fertilizer in chili pepper cultivation reaches 2–3 times the theoretical crop requirement, yet the phosphorus fertilizer utilization rate is generally below 20% (Bindraban et al., 2020). Because of exogenous P is often rapidly immobilized in soils, limiting its availability for plant uptake (Dejene et al., 2023). This excessive fertilization not only leads to resource wastage but may also trigger environmental risks such as soil compaction and heavy metal accumulation (Richardson et al., 2011; Wei et al., 2020). Therefore, optimizing phosphorus fertilizer management strategies and improving phosphorus fertilizer utilization efficiency have become key scientific issues urgently needing resolution for the sustainable production of chili peppers.
Certain non-agricultural plants have demonstrated the ability to thrive in low-phosphorus soils, leading to research on reducing phosphorus requirements in crops through breeding. Plants with low phosphorus levels can achieve this by either decreasing inorganic phosphorus concentration in the vacuole or by enhancing the transfer of phosphorus from inactive to active tissues (Warner et al., 2001). However, diminishing phosphorus input to essential compounds like ribosomal RNA and nucleotides may impede protein synthesis and growth rates (Perry, 2007; Matzek and Vitousek, 2009; Rose et al., 2010). This raises concerns about the feasibility of decreasing phosphorus concentrations in plants through breeding. Other studies have concentrated on enhancing phosphorus utilization by crops. Crops that utilize phosphorus more efficiently in the plant produce more dry matter per unit of phosphorus absorbed, thereby reducing the phosphorus needed per unit of yield. Nonetheless, the phosphorus harvest index was found to be closely linked with the grain harvest index (Batten, 1992; Batten et al., 1999; Zhang et al., 2018), indicating that exploiting genetic variation in this trait could be counterproductive. Further research is needed on the efficiency of phosphorus transport in plants and the genetic mechanisms behind phosphorus transport to seeds.
Soil Phosphorus-Solubilizing Microorganisms (PSM) are capable of converting phosphorus into a more plant-accessible form in low-available phosphorus agricultural soils (Sharma et al., 2013; Zheng et al., 2017). These microorganisms play a significant role in phosphorus cycling by affecting adsorption-desorption, solution-precipitation, and mineralization-immobilization processes (Rfaki et al., 2019). Research has shown that soil PSM release various organic acids, such as oxalic acid, citric acid and gluconic acid, that bind to phosphorus via hydroxyl and carboxyl chelated cations (Al, Fe and Ca), making phosphorus more easily absorbed by plants (Xu et al., 2019). Additionally, soil PSM facilitate organic phosphate mineralization through the production of hydrolase enzymes, particularly phytase and phosphatase, which catalyze the hydrolysis of phosphate ester bonds (Delgado et al., 2002). These mechanisms can occur simultaneously within the same microorganism strain. The presence of organic anions, such as humic acid and citric acid, may hinder stable calcium-phosphorus formation, thereby increasing phosphorus availability in the soil (Alori et al., 2017). Studies have shown that phosphatase enzymes produced by phosphorus solubilizing bacteria are essential for phosphorus dissolution (Wang et al., 2009). Furthermore, research demonstrated that Penicillium oxalate C2 has a high capacity for dissolving calcium phosphate and calcium phytate, with malic acid and tartaric acid potentially playing a key role in phosphorus dissolution (Gupta et al., 2011).
Although the field efficacy of PSM is subject to ongoing discussion (Kalayu, 2019), several greenhouse and field studies show that soil PSM can enhance phosphorus uptake in plants, leading to increased yields in various vegetable and cereal crops. In stevia plants, for example, stem length, root length, leaf dry weight, stem dry weight, and total stem biomass all showed significant increases after phosphate-solublizing bacteria (PSB) inoculation, with yield increasing by 291% (Han et al., 2006). Furthermore, PSB inoculation has been found to enhance phosphorus and potassium availability in soil, improve nitrogen, phosphorus, and potassium absorption in both roots and shoots, and promote the growth of pepper and cucumber plants (Wang et al., 2015). In watermelon and peanut rhizospheres, significant increases in root length, tip number, and root volume were observed after inoculation (Baccouri et al., 2019). Penicillium, known for its high efficiency, broad spectrum, and timely effects, not only improves phosphorus utilization but also secretes hormones and proteins that promote plant growth and enhance resistance (Hamdali et al., 2008).
The role of phospholytic microorganisms in promoting soil phosphorus release has been extensively validated. PSB can activate insoluble phosphorus in the soil through the secretion of organic acids and extracellular enzymes (Pan and Cai, 2023; Babu et al., 2025). This process ultimately leads to improved plant yields, highlighting the significant application potential and economic value of PSB in agricultural production (Sharma et al., 2013). Currently, most phosphate-solubilizing bacteria strains demonstrate phosphate solubilization capacities ranging from approximately 100 to 500 mg/L in NBRIP medium (National Botanical Research Institute's Phosphate medium) (Wan et al., 2020; Chen et al., 2024). However, it should be noted that in vitro phosphate solubilizing capacity does not always correlate with in planta performance. This discrepancy may arise from factors such as rhizosphere incompetence (e.g., failure to colonize the root zone), competition with indigenous soil microorganisms, and differences in soil conditions (e.g., the TCP (tricalcium phosphate) used in NBRIP medium may not reflect the phosphorus speciation and dissolution dynamics in acidic fields) (De Zutter et al., 2022; Rawat et al., 2021). Over 30 genera of PSB have been reported, including Bacillus, Paenibacillus, Pseudomonas, Rhizobium, Burkholderia, and Microbacterium, and their growth-promoting effects on plants have been validated in specific soil and crop systems, though such effects may vary under different environmental conditions (Ma and Diao, 2018; Xiong and Zhao, 2024).
However, despite the fact that the impact of chemical fertilizers on grain yield per unit area has dwindled to nearly zero, their contribution rate to China's overall grain production remains at 40%-50% (Liao et al., 2023). Therefore, alternative strategies must be explored to enhance food production. Combining fertilizers with PSB could be a more effective approach to boost phosphorus absorption and increase crop yield. PSB activate soil phosphorus through biological pathways, which can not only reduce phosphorus fertilizer application costs and increase crop yields but also mitigate environmental pollution and resource waste, thereby achieving both economic and ecological benefits (Aswitha et al., 2024). This study aims to investigate the effects of combined application of varying phosphorus fertilizer rates with PSB strain Pseudomonas E sp017968885 on the activation of soil insoluble phosphorus and chili pepper growth. The research provides a feasible approach for "phosphorus reduction and efficiency enhancement" in chili cultivation while offering a scientific basis for integrating phosphorus fertilizers with PSB in fertilization practices. This work holds significant importance for ensuring the sustainable development of chili industry and safeguarding farmer income.
2 Materials and methods
2.1 Materials
2.1.1 Test pepper
The pepper seedlings used in the study were 18 days old and were sourced from Xuhua Planting, Production, and Marketing Cooperative in Gongyingzi Township, Kazuo County. These seedlings belonged to the Sao Paulo variety.
2.1.2 Test strain and preparation of the experimental bacterial suspension
The Pseudomonas E sp017968885 used in this study was isolated and purified from the rhizosphere soil of Picris japonica Thunb. It is currently preserved in the Laboratory of Agricultural Microbiology at the Microbial Research Institute of Liaoning Province. The preservation medium (PVK) was formulated as follows:10g of glucose, 0.5g of (NH4)2SO4, 0.3g of NaCl, 0.3g of KCl, 0.3g of MgSO4·7 H2O, 10g of Ca3(PO4)2, 0.03g of FeSO4·7 H2O, 0.03g of MnSO4·H2O, 18g of agar powde, and 1000mL of distilled water, with the adjusted to pH 7.0-7.5.
The experimental bacterial suspension was prepared through the following steps:
1. The strain Pseudomonas E sp017968885 was inoculated into PVK liquid medium.
2. The inoculated medium was cultured at 30°Cwith a shaking speed of 160 rpm for 72 hours.
3. After cultivation, the bacterial culture was centrifuged to collect the bacterial biomass.
4. The collected biomass was resuspended in liquid medium to a final concentration of 2×109 CFU/mL.
The nitrogen and phosphorus contents in the bacterial suspension were 39.7µg/L and 0.78µg/L, respectively, indicating that its direct fertilization effect was negligible.
2.1.3 Test soil
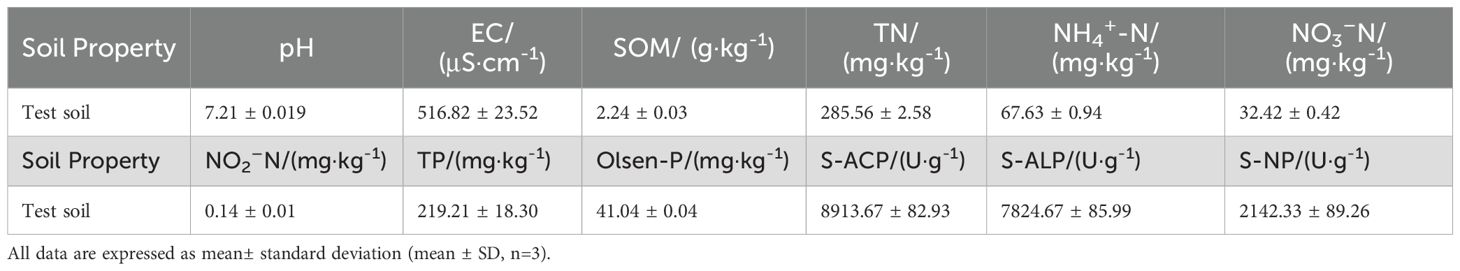
The experiment was conducted in May 2023 at the Microbial Research Institute of Liaoning Province. Soil samples were collected from the vegetable field of Pinai Chi Village, Chaoyang County. The soil was air-dried naturally after collection, and residual roots and gravel were removed. Table 1 presents the physical and chemical properties of the soil. The test basin used was a 3-gallon flowerpot with an inner diameter of 24cm.
2.2 Methods
2.2.1 Experimental design
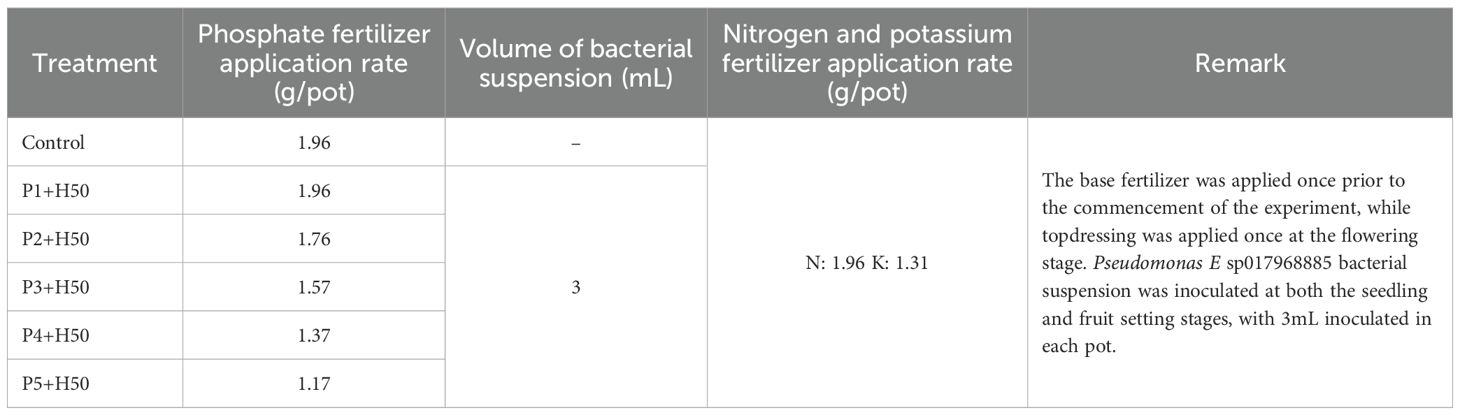
A pot design was utilized in this study, with six different treatments established, The specific contents of each processing are shown in the following Table 2. The phosphorus, nitrogen, and potassium fertilizers utilized in each treatment were calcium phosphate (P2O5 45.8%), urea (N 46%), and potassium sulfate (K2O 34.3%), respectively.
2.2.2 Sample collection
The pot experiment spanned 111 days (from May 17, 2023, to September 5, 2023). Soil samples in the root zone were collected at different growth stages: 21st day (seedling stage), 57th day (flowering stage), 82nd day (fruit setting stage), and 111th day (post-cultivation), the various indicators of the soil before cultivation (that is, the data in Table 1) will also be involved in the analysis, referred to in this paper as “pre-cultivation”. At each stage, three basins were destructively sampled. Soil samples from the root zone were air-dried, sieved through a 2 mm mesh, and analyzed to determine soil physicochemical indices. Pay individual attention to the four pots of chili peppers and pick them in real time according to their ripening conditions. The picked peppers were heated at 105°C for 30 minutes, followed by drying at 80°C until a constant weight was achieved, and all dry matter was accumulated and preserved. After the cultivation was completed, destructive sampling was conducted on four pots of chili peppers, the plants were divided into fruit and stalk+leaf components. The drying process was consistent with that described previously. The weight of dried fruits was accumulated into the early-stage fruit weight for calculating chili pepper yield (Formula 1). Additionally, data on total aboveground biomass—encompassing fruits, stalks, and leaves—were used to determine plant phosphorus mass fraction (Formula 2), phosphorus uptake (Formula 3), and phosphate fertilizer utilization efficiency (Formula 4).
2.2.3 Soil physicochemical index determination
Soil organic matter (SOM) was determined via dry combustion in a laboratory muffle furnace at 550°C ± 20°C for 3 hours, with subsequent weighing. The pH value and electrical conductivity (EC) of the soil samples were measured using a pH meter and conductivity meter, respectively, with a soil-water ratio of 5:1(Xu et al., 2024). The contents of ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3–N) and nitrite nitrogen (NO2–N) in the soil were calculated using the 2 mol·L-1 KCl leaching distillation method and the phenol disulfonic acid colorimetric method, respectively. Total nitrogen (TN) content in the soil was determined using the Kjeldahl method (Li et al., 2025). Total phosphorus (TP) content in the soil was determined using the Ascorbic acid reduction method, while the available phosphorus (Olsen-P) content was determined using a colorimetric method with a 0.5 mol·L-1 NaHCO3 solution (Eduah et al., 2024; Yang et al., 2024; Bao, 2021). Soil acid phosphatase (S-ACP), soil alkaline phosphatase (S-ALP), soil neutral phosphatase (S-NP) were determined by soil phosphatase activity assay kit, purchased from Sangon Biotech (Shanghai) Co., Ltd.
Formula 1: Yield per unit area (kg/ha) = Total output (kg) / area (ha).
Formula 2: Plant phosphorus mass fraction: w(P)= ×100 (Tao et al., 2022).
(P)―Mass fraction of total P in plants %.
―The mass concentration of P in the test solution µg/L.
v―Measure liquid volume L.
ts―Partition multiple.
m―Dried sample mass g.
Note: All the fruits of the four pots of chili peppers and the dry matter of the plants after cultivation were uniformly mixed and measured to calculate the phosphorus mass fraction of the above-ground parts of the plants. According to the formula 2, the phosphorus mass fraction of the chili pepper plants in this experiment throughout the growth process was 0.108%.
Formula 3: Phosphorus absorption of pepper (g)= w(P)× plant dry weight (g).
Formula 4: Phosphate fertilizer utilization efficiency % =The amount of phosphorus absorbed by plants and fruits (P) / the amount of phosphorus fertilizer used (Ca3(PO4)2)×100.
2.3.4 Gene prediction and annotation
Genomic DNA extraction and genome sequencing:
A single colony selected from Luria-Bertani medium (LB medium) was inoculated into liquid Luria-Bertani medium and cultured at 30°C with shaking at 170 rpm for 24 hours. The bacterial cells were harvested by centrifugation at 10,000 rpm for 10 minutes. The cetyltrimethylammonium bromide (CTAB) method was employed to extract the genomic DNA of strain Pseudomonas E sp017968885 (Murray and Thompson, 1980), followed by purification using the Wizard Genomic DNA Purification Kit (Vazyme Biotech Co., Ltd, Nanjing, China). The whole genome of Pseudomonas E sp017968885 was sequenced by Shanghai Majorbio Biopharm Technology Co., Ltd (Shanghai, China) utilizing two different sequencing techniques: Illumina HiSeq and the PacBio RSII system (Menlo Park, CA, USA). Subsequently, SPAdes was utilized to assemble the reads into scaffolds.
Homology comparisons were conducted using the BLAST online tool with model strain sequences obtained from the NCBI database. The sequences of the model strains were downloaded for analysis. A phylogenetic tree of the strains was constructed using the adjacency method implemented in MEGA 6.0 software. Additionally, the genomic circular map of the sample was generated using Circos software. The Gene Ontology (GO) database (http://geneontology.org/), the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/), and the COG database (https://www.ncbi.nlm.nih.gov/research/cog/) were utilized, along with the non-redundant protein sequence database (NR) from NCBI (ftp://ftp.ncbi.nlm.nih.gov/blast/db/), to provide functional annotation for the genes of strain Pseudomonas E sp017968885. The antiSMASH website (http://antismash.secondarymetabolites.org/) was employed for the rapid identification and analysis of secondary metabolite synthesis gene clusters.
2.3.5 Statistical analysis
Data processing and calculations were performed using Microsoft Excel 2021. Statistical analyses were conducted in SPSS 22.0, with one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test for multiple comparisons. Graphs were generated using GraphPad Prism.
3 Results
3.1 Effect of Pseudomonas E sp017968885 on yield and activation rate of phosphate fertilizer in pepper
The results presented in Table 3 demonstrate that the yield of pepper significantly increased with the application of Pseudomonas E sp017968885. Specifically, the yield of treatments increased by 23.98%, 31.06%, 53.47%, 65.80%, and 61.58% respectively (P<0.05), compared to the control. Furthermore, after the application of Pseudomonas E sp017968885, phosphorus absorption in each treatment showed a significant increase of 23.98%, 61.58%, 65.8%, 53.47%, and 31.06% (P<0.05). The utilization efficiency of phosphorus fertilizer also saw improvements, with rates of 23.98%, 79.94%, 1.06 times, 1.19 times, and 1.19times for the respective treatments (P<0.05).
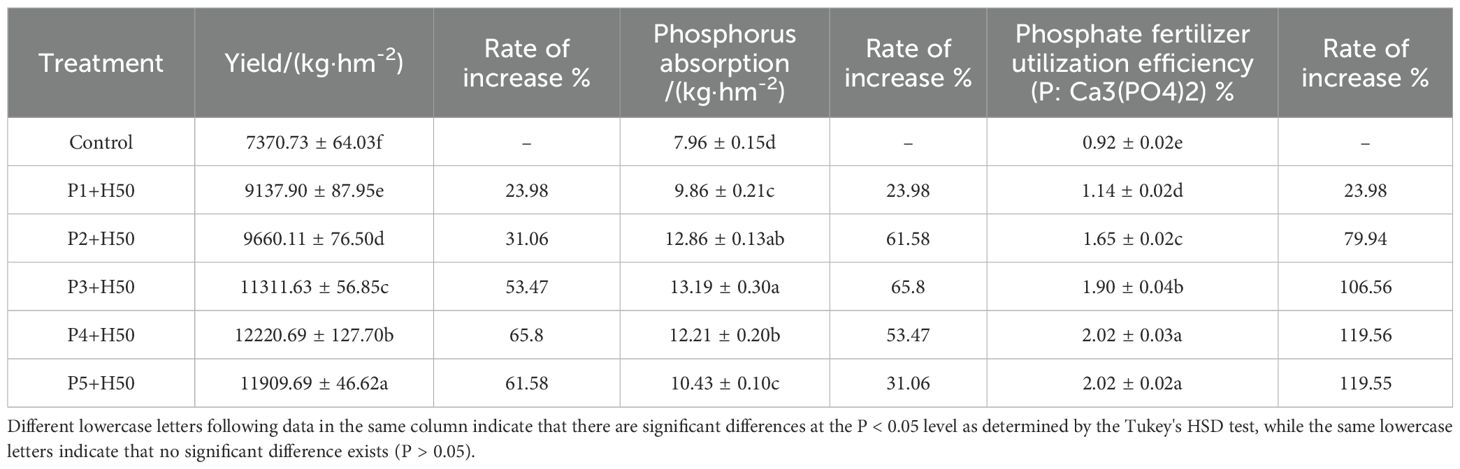
Table 3. Effects of Pseudomonas E sp017968885 on pepper yield and absorption and utilization of phosphate fertilizer.
3.2 Effect of Pseudomonas E sp017968885 on soil physicochemical index at different growth stages
With the exception of the P5+H50, the EC of all treatments generally followed a pattern of initial increase followed by decrease as the growth stage progressed (Figure 1a). At seeding stage, the EC of P2+H50 and P3+H50 treatments showed a significant decrease of 26.65% and 26.07%, respectively compared to the control (P<0.05). In the flowering stage, there was no significant difference in EC among all treatments (P>0.05). By fruit setting stage, P4+H50 treatments showed significant reductions of 41.92% compared to control (P<0.05). Finally, at post-cultivation, the EC of all treatments decreased to a range of 271.03μS·cm-1 to 349.53μS·cm-1, significantly lower than that at pre-cultivation (516.83μS·cm-1) (P<0.05).
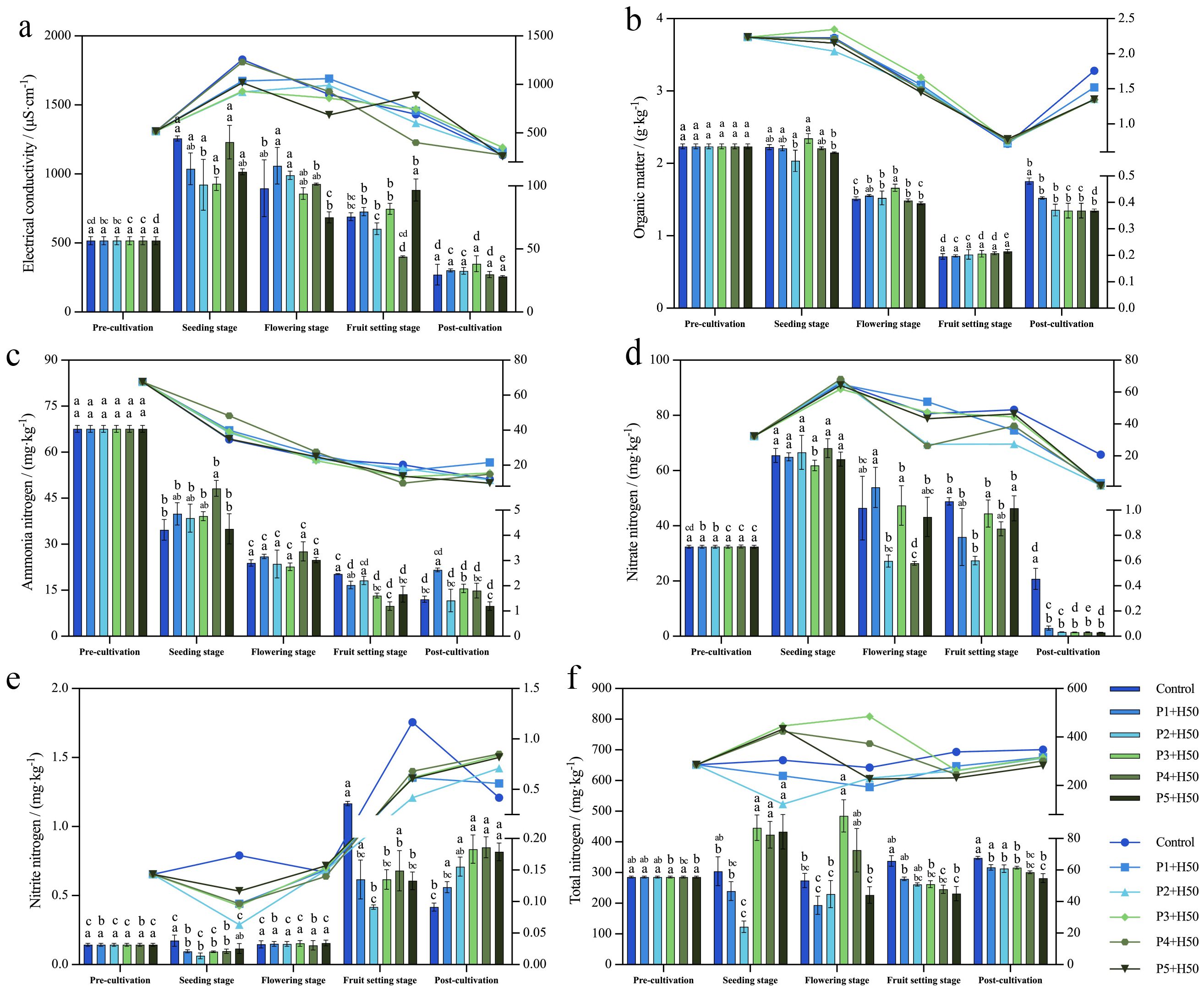
Figure 1. Changes of soil physicochemical indexes with growth stage. The figure depicts two distinct representations of a dataset. On the left side, there is a column chart scale, while the right side features a line chart scale. The letters at the bottom of the columns indicate the difference comparison results between various treatments at the same growth stage, whereas the letters at the top represent the difference comparison results between different growth stages of the same treatment. If the lowercase letters are the same, it signifies no significant difference (P>0.05), whereas different lowercase letters indicate a significant difference (P<0.05).
SOM content initially decreased as the growth stages progressed, reaching its lowest point at fruit setting stage before gradually increasing (Figure 1b). Specifically, the fluctuation in SOM content between pre-cultivation and seeding stage was minimal, whereas substantial fluctuations were observed from seeding stage to fruit setting stage across treatments, with significant decreases ranging from 63.73% to 68.09% (P<0.05). It indicates that the flowering stage and fruit setting stage may be the critical periods for organic matter consumption, and therefore the content is the lowest throughout the growth period. Following fruit setting stage, SOM content in each treatment started to rise, reaching values of 1.76g·kg-1, 1.52g·kg-1, 1.36g·kg-1, 1.35g·kg-1, 1.35g·kg-1, and 1.35g·kg-1 at post-cultivation, respectively. At post-cultivation, the SOM content of treatments was notably lower than that of control by 13.64%, 22.73%, 23.30%, 22.30%, and 22.30% (P<0.05), indicating that the application of Pseudomonas E sp017968885 facilitated SOM conversion and utilization by capsicum plants, ultimately leading to reduced soil organic matter stocks after pepper cultivation.
The analysis of NH4+-N indicated a continuous decline among all treatments as the growth stage progressed, reaching its minimum at post-cultivation (Figure 1c). In comparison to the initial 67.63mg·kg-1 before cultivation, the NH4+-N content in post-cultivation treatment saw a significant reduction by 82.17%, 68.03%, 82.74%, 76.95%, 77.95%, and 85.47% (P<0.05), respectively. These findings suggest a substantial utilization of NH4+-N during pepper growth. Notably, at the fruit setting stage, NH4+-N contents in the P3+H50 to P5+H50 treatments were significantly lower than that in the control group (P<0.05), indicating that peppers in these treatments absorbed and utilized more NH4+-N compared to the control. These results suggest that inoculation with Pseudomonas E sp017968885 significantly enhances nitrogen utilization during the fruit setting period.
The analysis of NO3–N show that, the NO3–N content initially increased and then decreased in all treatments as the plants grew (Figure 1d). The seeding stage had the highest NO3–N content among all growth stages. In contrast, there was notable fluctuation in NO3–N content among treatments at flowering stage and fruit setting stage, but it became similar by post-cultivation. Notably, at post-cultivation, there was no significant difference in NO3–N content between P1+H50 to P5+H50 treatments, but there was a substantial reduction compared to control (P<0.05), indicating that the application of Pseudomonas E sp017968885 promoted nitrate nitrogen utilization by pepper plants and reduced soil NO3–N content by post-cultivation.
Compared with other growth periods, the NO2–N content in each treatment was lower during the seedling stage and flowering stage (Figure 1e). Specifically, in seeding stage, the NO2–N content of P1+H50~P4+H50 treatments was significantly lower than that of control by 41.18%, 64.71%, 47.06%, and 41.18% (P<0.05). Following flowering stage, the NO2–N content increased. In fruit setting stage, the NO2–N content of all treatments was significantly lower than that of control treatment by 47%, 64.10%, 47%, 41.88%, and 57% (P<0.05), indicating that Pseudomonas E sp017968885 application enhanced the utilization of NO2–N and its conversion to other nitrogen forms during the late growth stages of pepper plants. By post-cultivation, the NO2–N content in all treatments was significantly higher than in control, suggesting that Pseudomonas E sp017968885 application reduced the utilization and transformation of nitrite nitrogen in pepper plants toward the end of their growth cycle.
The results regarding changes in TN content indicate that the TN content in the control and P1+H50 treatments fluctuated to a lesser extent during the growth period (Figure 1f). Moreover, TN content in P1+H50 consistently remained lower than that in control, and at the fruit setting stage and post-cultivation, it was significantly lower than the control by 17.26% and 8.93% (P<0.05), indicating the significant effects of Pseudomonas E sp017968885 on soil nitrogen transformation, absorption by pepper plants and nitrogen utilization. At the fruit setting stage and post-cultivation, TN content in all treatments was significantly lower than that in control, with reductions ranging from 17.27%~31.57% and 8.93%~19.06% (P<0.05). These results suggest that the application of Pseudomonas E sp017968885 enhanced the conversion and utilization of TN during pepper plant growth, leading to a decrease in soil TN content in the later stages of pepper growth.
3.3 Effects of Pseudomonas E sp017968885 on soil phosphorus related indexes of pepper at different growth stages
Results indicated that, with the exception of P3+H50, P4+H50, and P5+H50 treatments, TP content in all treatments exhibited a notable fluctuation pattern throughout the growth stages, showing an initial increase, followed by a decrease, another increase, and a subsequent decrease, that is, two peaks appeared at the seeding stage and fruit setting stage respectively (Figure 2a). In P3+H50 and P5+H50, TP content gradually increased as the plants grew, peaking at fruit setting stage before declining. The TP content in post-cultivation was lower than that of control, and treatments P4+H50 experienced significant decreases of 17.47% (P<0.05). This suggests that the application of Pseudomonas E sp017968885 facilitated the conversion and utilization of TP from pepper plants, leading to a decrease in soil TP content after cultivation.
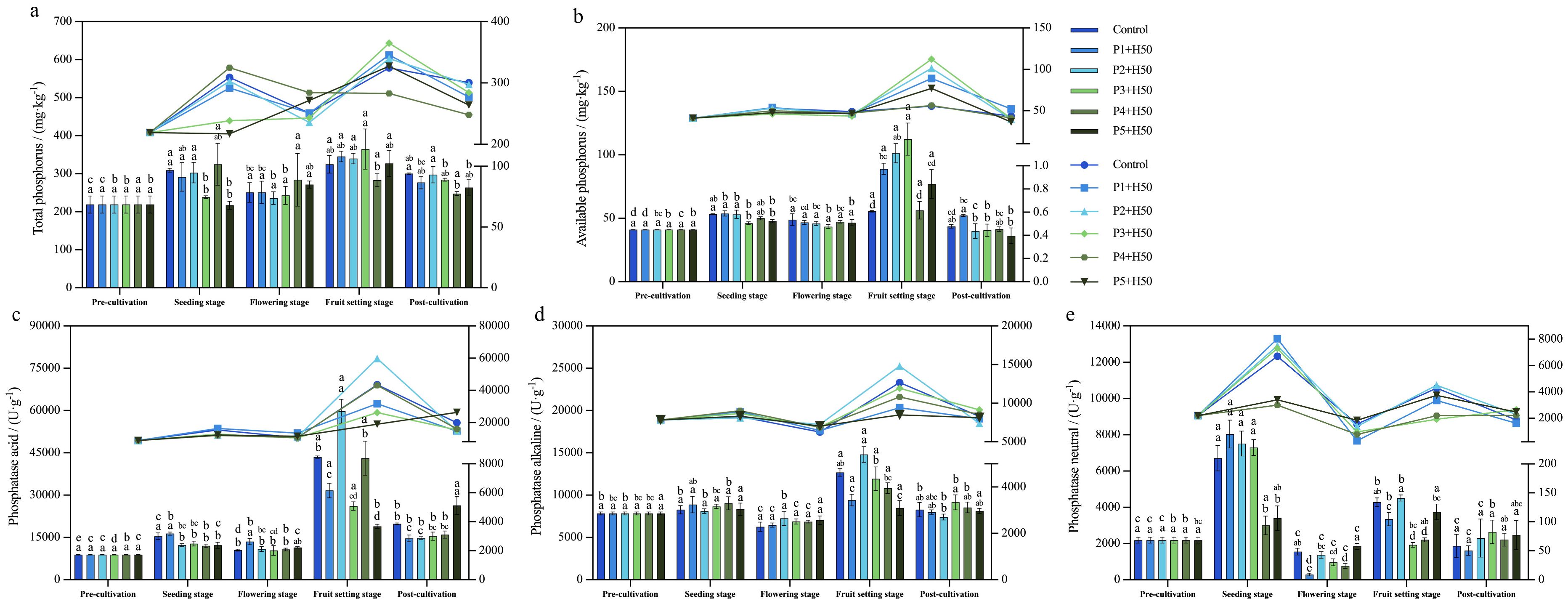
Figure 2. Changes of phosphorus related soil physicochemical indexes with growth stage. Different lowercase letters indicate significant differences at the P < 0.05 level as tested by Tukey's HSD, while the same lowercase letters indicate no significant difference (P > 0.05).
The results revealed that Olsen-P content remained relatively stable in stages seeding and flowering (Figure 2b). However, in fruit setting stage, Olsen-P content significantly increased across all treatments compared to flowering stage, with notable differences observed among treatments. Specifically, in stage fruit setting stage, the Olsen-P content for treatments of P1+H50, P2+H50, and P3+H50 was significantly higher than that of control by 66.36%, 78.32%, 102.08% (P<0.05) respectively. This suggests that the application of Pseudomonas E sp017968885 facilitated the conversion of soil phosphorus into available forms, thereby enhancing the available phosphorus content during the fruit setting stage of pepper plants. Subsequently, Olsen-P content decreased in all treatments post stage fruit setting. At the post-cultivation stage, no significant difference in available phosphorus content was observed among the various treatments (P>0.05). However, compared with the fruit-setting stage, the available phosphorus content in all treatments decreased significantly (P<0.05). Specifically, the P1+H50, P2+H50, and P3+H50 treatments exhibited significant reductions of 41.36%, 60.55%, and 63.87%, respectively (P<0.05). These results indicate that under the treatment with Pseudomonas E sp017968885, the P1+H50, P2+H50, and P3+H50 treatments exerted favorable phosphorus activation effects throughout the growth cycle of pepper plants.
The findings showed that the activities of both S-ACP and S-ALP exhibited a similar trend across growth stages: they remained relatively stable until the flowering stage, increased after flowering, reached a peak, and then declined to initial levels by the post-cultivation stage (Figures 2c, d). Among them, the differences among the treatments during the fruit setting period were greater than those in other periods, and during the fruit setting period, the S-ACP and S-ALP treatments of P2+H50 had the highest activities. S-NP activity exhibited fluctuations throughout the growth process, ultimately returning to baseline levels by post-cultivation. Notably, Olsen-P, S-ACP, and S-ALP exhibited a similar trend across growth stages, suggesting that available phosphorus is likely to be significantly positively correlated with phosphatase activity. This conclusion is consistent with the results of the correlation analysis of soil physicochemical indices provided below (Figure 3).
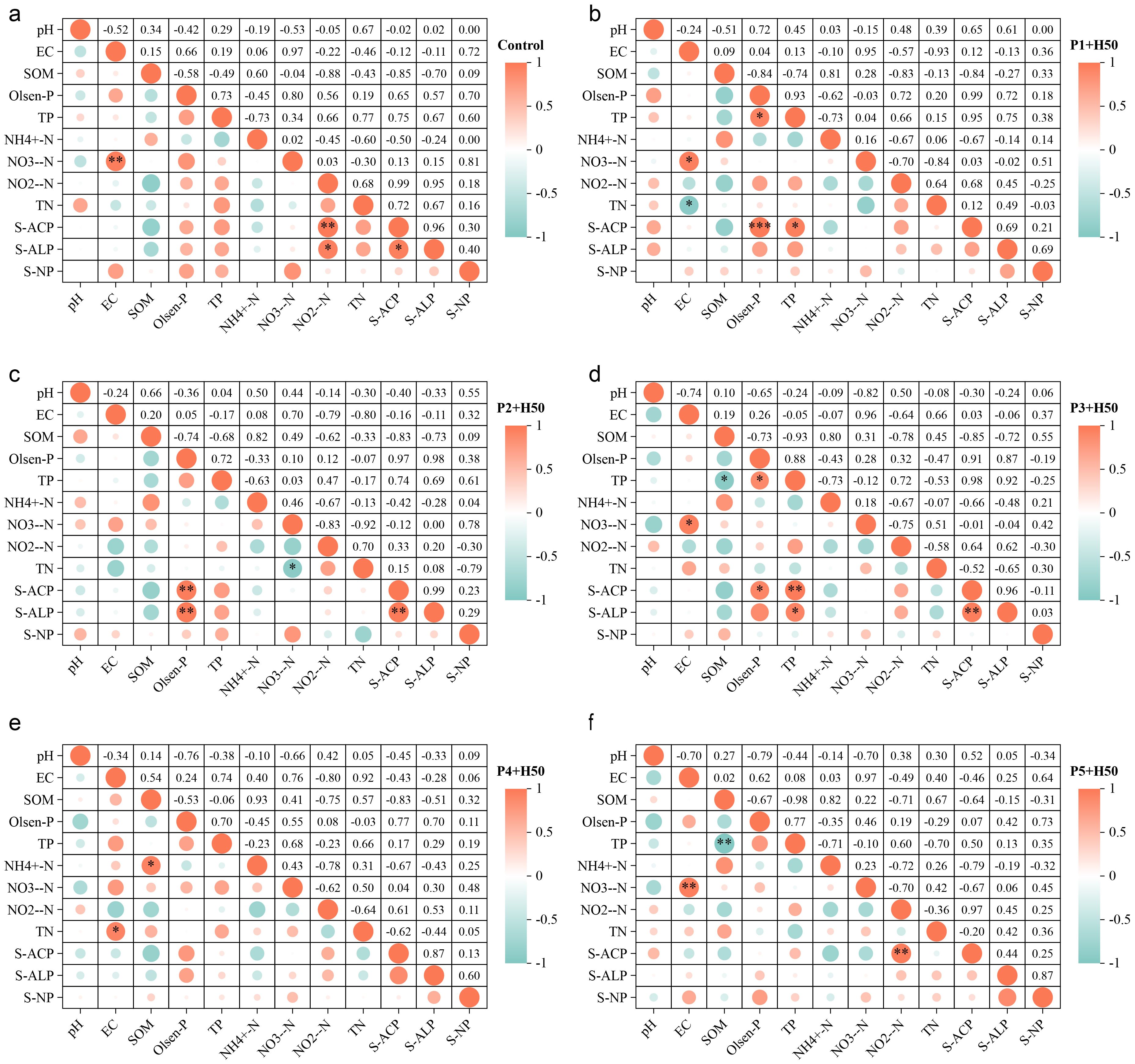
Figure 3. Correlation analysis of soil physicochemical indexes under different treatments. “*” means P<0.05, “**” means P<0.01, and “***” means P<0.001.
3.4 Correlation analysis of soil physicochemical indexes under different treatments
Figure 3 shows the results of correlation analysis of soil physicochemical indexes under each treatment. As shown in Figure 3a, the activity of S-ACP and S-ALP were significantly positively correlated with NO2–N (P<0.05), and the EC was significantly positively correlated with NO3–N (P<0.01). Figure 3b shows that S-ACP is significantly positively correlated with Olsen-P (P<0.001) and TP (P<0.05), and EC is significantly positively correlated with TN and NO3–N (P<0.05). Figure 3c shows that both S-ACP and S-ALP activities are significantly positively correlated with Olsen-P (P<0.01). Figure 3d shows that S-ACP activity is significantly positively correlated with Olsen-P (P<0.05) and TP (P<0.01). S-ALP activity was positively correlated with TP (P<0.05). Figure 3e shows that there is a significant positive correlation between EC and TN (P<0.05), and a significant positive correlation between SOM and NH4+-N (P<0.05). Figure 3f shows that there is a significant positive correlation between S-ACP activity and NO3–N (P<0.01). There was a significant positive correlation between EC and NO2–N (P<0.01). Overall, S-ACP and S-ALP underwent a process from "N-correlated" to "P-correlated" and then back to "N-correlated" (Figure 4). This might be related to the dosage of phosphorus fertilizer. That is, under the "reasonable" dosage of phosphorus fertilizer, the inoculation of Pseudomonas E sp017968885 has a more effective phosphorus activation function, and the high availability phosphorus content of P1+H50 to P3+H50 treatment during the fruit setting period supports this conclusion.
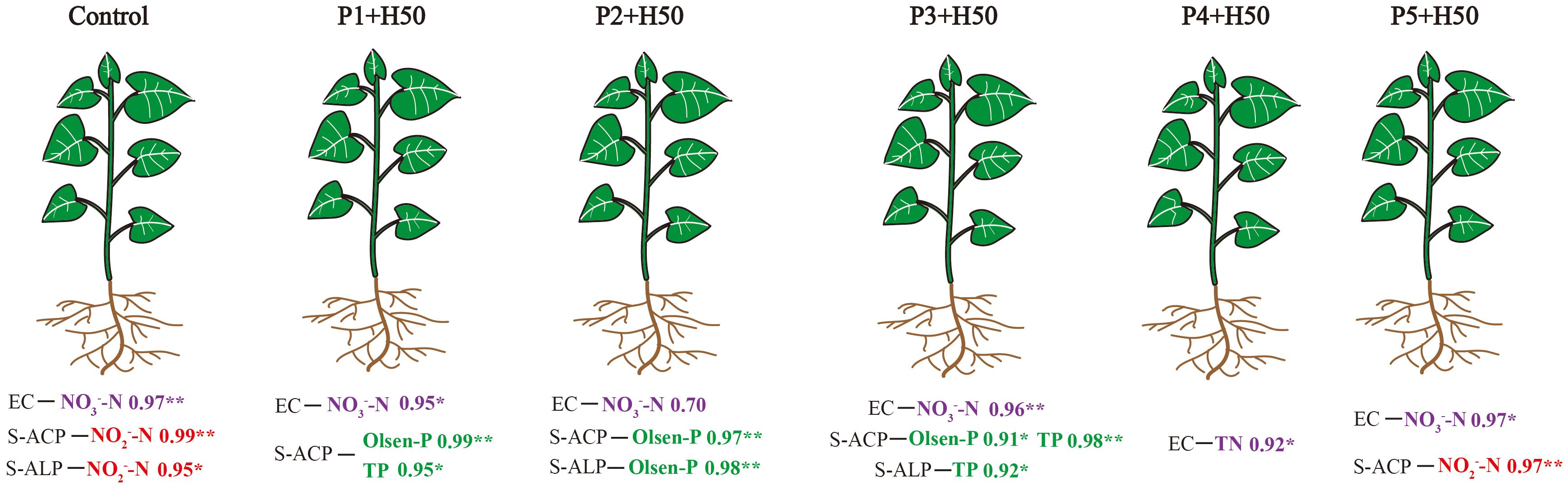
Figure 4. Schematic diagram of the correlation of physical and chemical indicators of each treatment.
3.5 Genome chatacteristics of Pseudomonas E sp017968885
The genomic DNA of Pseudomonas E sp017968885 was used for amplifying the 16S rRNA fragments. The Pseudomonas E sp017968885 whole genome has a total length of 6,715,831 bp. A total of 5,892 genes were predicted, with a combined gene length of 15,352,435 bp, accounting for 88.34% of the total genome length. Statistical analysis revealed that the average coding gene length is 1,006.89 bp (Table 4). A 9,272 bp fragment was obtained and submitted to NCBI for phylogenetic tree construction. The 16S rRNA sequence exhibited the highest resemblance with Pseudomonas E sp017968885(GCF 017968885.1 rRNA4) (Figure 5a). The circular genomic map of Pseudomonas E sp017968885 is depicted in Figure 5b. The outermost circle of the map represents the genome size scale. The second and third circles display the coding sequences (CDSs) on the positive and negative strands, respectively, with different colors indicating the functional classification of CDSs according to the Clusters of Orthologous Groups (COG). The fourth circle illustrates the locations of rRNA and tRNA genes. The fifth circle represents the GC content, where outward red peaks indicate regions with GC content higher than the average genomic GC content (61.13%), with the height of the peaks reflecting the magnitude of deviation from the average. Inward blue peaks indicate regions with GC content lower than the average genomic GC content, with the height of the peaks similarly reflecting the magnitude of deviation. The innermost circle displays the GC-Skew values, which provide insights into the strand-specific distribution of guanine (G) and cytosine (C) bases.
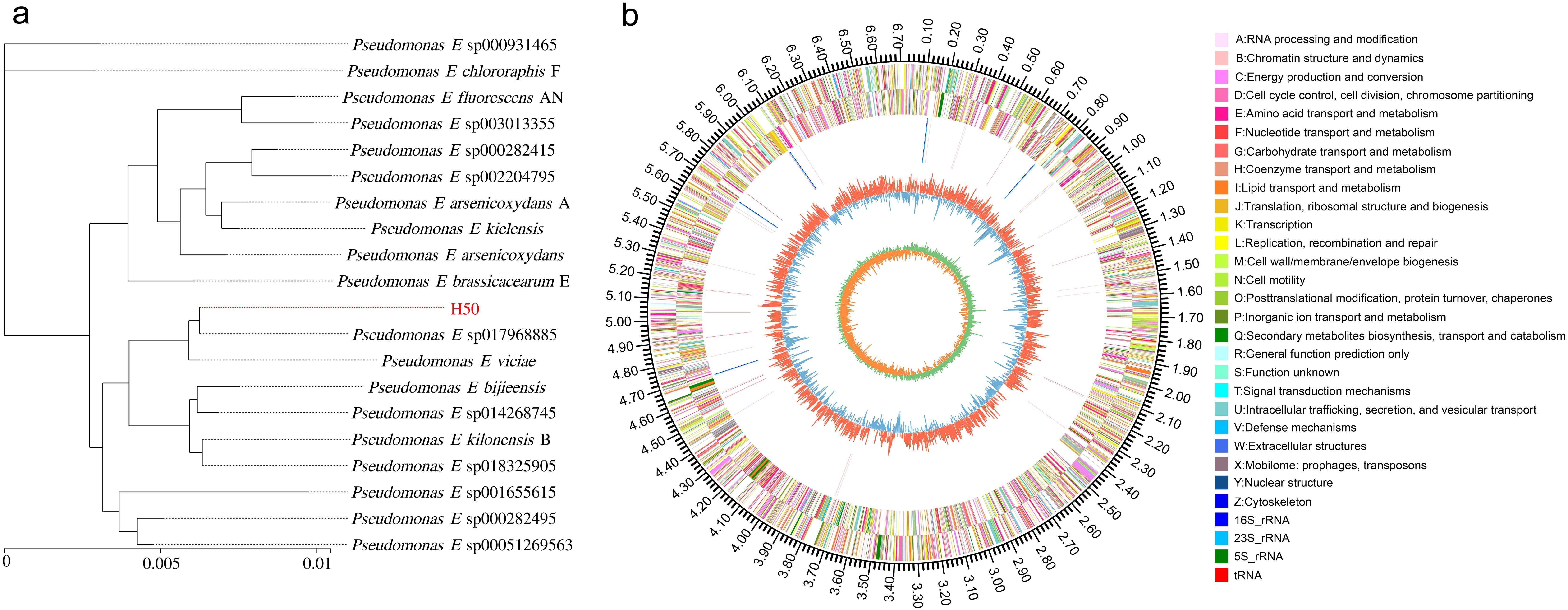
Figure 5. (a) The phylogenetic tree of strain H50 based on 16 S rRNA gene sequence analysis using the MEGA 6.0 software’s neighbour-joining algorithm; (b) Circular genomic map of H50. The genome circle map was drawn by Circos, and the corresponding information from the outer circle to the inner circle was: Genome size, forward CDS, reverse CDS, rRNA and tRNA, GC content, and GC skew.
The genome function of strain Pseudomonas E sp017968885 was predicted to further explore its characteristics. Through annotation, the genome provided genetic information pertaining to the species, as well as valuable insights into the study of organic metabolites. A total of 5892 genes were predicted, with a gene/genome ratio of 88.34%, encompassing 65 tRNAs, 16 rRNAs, and 108 sRNAs. Regarding functional annotation, 5868, 4443, 5165, 4878, 2712, and 4498 genes were annotated by the NR, Swiss-Prot, Pfam, GO, COG, and KEGG databases, respectively. As depicted in Figure 6a, among the six KEGG pathway classifications, the category of Metabolism harbored the greatest number of genes (3508), followed by Environmental Information Processing (688) and Cellular Processes (536), which exhibited a comparable quantity of genes. Within the level 2 classification of KEGG pathways annotated for the metabolism of Pseudomonas E sp017968885, the primary functions of this strain were identified as Global and overview maps, Amino acid metabolism, Signal transduction, and Carbohydrate metabolism, containing 1385, 426, 366, and 352 genes, respectively. Furthermore, several Cellular Processes were discerned, including Cellular community-prokaryotes (305) and Cell motility (145). As depicted in Figure 6b, a total of 2712 genes (accounting for 46.03%) were annotated into the GO database, which encompassed three main categories: biological process (comprising 1484 genes), cellular component (including 1265 genes), and molecular function (encompassing 2219 genes). In addition, 4878 genes (representing 82.79% of the total) from Pseudomonas E sp017968885 were classified into four categories according to the COG database. Notably, 611 genes were identified as being associated with amino acid transport and metabolism, as shown in Figure 6c.
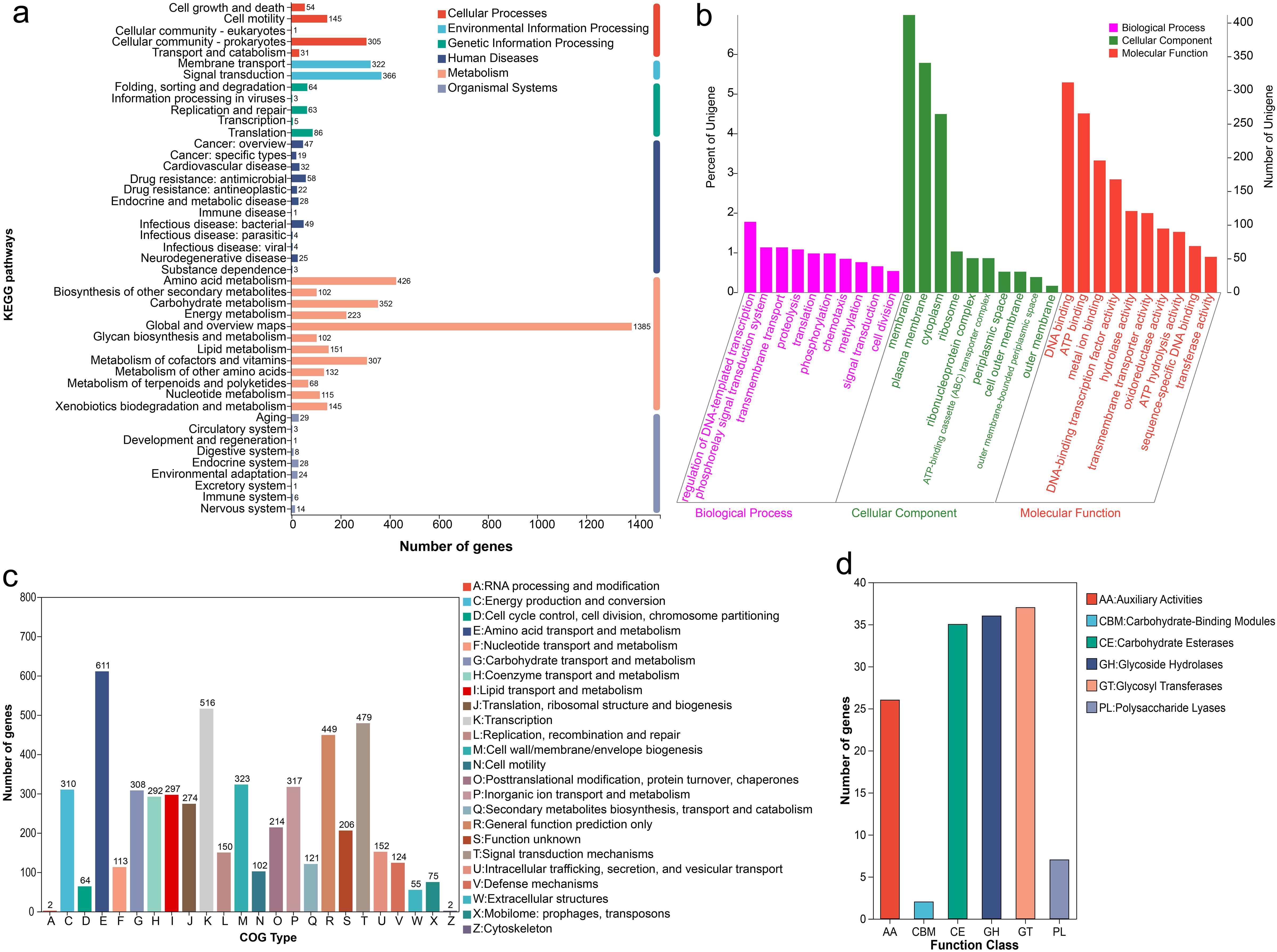
Figure 6. Genome features of H50. (a) KEGG classification; (b) GO classification; (c) COG classification; (d) CAZy classification.
The CAZy annotations were analyzed to identify the genes involved in the degradation of inorganic phosphate using the bCAN carbohydrate-active enzymes (CAZy) annotation algorithm. A total of 143 genes were identified from the CAZy families. As depicted in Figure 6d, about 36 glycoside hydrolases (GHs), 35 Carbohydrate Esteraseswere (CEs), 37 Glycosyl Transferases (GTs), and 26 Auxiliary Activities (AAs) were found in the strain Pseudomonas E sp017968885.
Based on the analysis of biosynthetic gene clusters in the Pseudomonas E sp017968885 genome, as shown in Supplementary Figure S, a total of 12 classes of gene clusters were identified. Among these, cluster 2 contains a complete set of genes related to the protein secretion system (gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, and gspM ). Clusters 5, 9, and 10 all harbor large fragments of the bacC gene, while clusters 9 and 10 additionally contain the tycC and ituB genes, respectively, all of which are closely associated with antibiotic biosynthesis. Regarding genes involved in siderophore synthesis, clusters 1 and 10 both contain the dhbF gene, whereas cluster 9 contains the entF gene. Notably, cluster 12 was also found to include the pqq and gdh genes, which are related to gluconic acid metabolism. These findings demonstrate that the Pseudomonas E sp017968885 genome possesses a rich potential for the synthesis of secondary metabolites, particularly exhibiting significant features in antibiotic and siderophore biosynthesis.
4 Discussion
Microorganisms play an important role in soil nutrient cycling, and the application of microorganisms or their preparations can improve plant growth and yield. The same is true of phosphate solublizing bacteria. For instance, Dai et al. reported that Bacillus Y8 can decompose organophosphorus compounds and dissolve calcium phosphate (Ca3(PO4)2), and its use as a microbial fertilizer can reduce the amount of chemical fertilizer required without compromising yield (Dai et al., 2010). Similarly, Tang et al. demonstrated that PSB X-P18 significantly increased the yield of Chinese cabbage in phosphorus-deficient environments (Tang et al., 2020). The application of PSB has also been shown to promote the growth of various crops, including wheat (Kumar et al., 2014), rice (Aarab et al., 2017), tomato (Nassal et al., 2017), mung bean (Biswas et al., 2018), rape (Valetti et al., 2018), and soybean (Ku et al., 2018). In the present study, the application of Pseudomonas E sp017968885 in combination with fertilizer was found to enhance the yield of capsicum. Notably, a reduction in the amount of phosphorus fertilizer further increased the yield. These findings are consistent with previous research, suggesting that lower doses of phosphorus fertilizer are more conducive to the activity of PSB when applied in conjunction with these microorganisms. This is supported by the study of Kumar et al., which showed that low-dose conventional fertilizer combined with microbial fertilizer can significantly improve corn yield (Kumar et al., 2014). Similarly, Nassal et al. found that Pseudomonas sp. RU47 enhanced phosphorus supply and growth of tomato plants under limited phosphorus conditions (Nassal et al., 2017).
Soil salinity exerts a profound influence on agricultural productivity, as excessive salinity hinders seed germination and diminishes crop yield (El-Ramady et al., 2024). The electrical conductivity of the soil saturated leaching solution is a crucial factor in determining soil salinity (Sun and Selim, 2019). Pseudomonas E sp017968885 has been found to significantly reduce the electrical conductivity of the soil. This indicates that Pseudomonas E sp017968885 may have potential applications in improving soil quality and increasing agricultural productivity. SOM constitutes a vital source of organic nutrients for plants and a key determinant of soil fertility and quality (Yang et al., 2024). The mineralization of SOM primarily originates from fertilization inputs and the natural decomposition of indigenous soil organic components (Malone et al., 2023). Notably, the application of Pseudomonas E sp017968885 led to a significant reduction in SOM content following pepper cultivation, even in the absence of supplementary fertilization that could potentially alter SOM dynamics. This suggests that Pseudomonas E sp017968885 facilitated the transformation of soil organic matter, enhancing its utilization efficiency by pepper plants.
PSM play a crucial role in enhancing phosphorus uptake by plants through activation and fixation mechanisms. Activation involves the secretion of organic acids and phosphatases to break down insoluble phosphorus, carbon and phosphorus exchange with arbuscular mycorrhizal fungi, nitrogen fixation, and plant hormone secretion. Fixation refers to the conversion of soil available phosphorus into biomass phosphorus, which is subsequently released for plant absorption when needed (Fu et al., 2022; Dietrich et al., 2023). The results of the correlation analysis showed that the significant positive correlations among the elements all occurred between phosphatase and N/P elements (Figure 4). Compared with the control, phosphatases in treatments P1+H50 to P3+H50 shifted from "N-correlated" to "P-correlated" patterns (Figure 4). PSB is an important producer of phosphatase (Pan and Cai, 2023). The application of Pseudomonas E sp017968885 promotes the secretion of phosphatase, hydrolyzing organic phosphorus compounds (e.g., nucleic acids, phospholipids, and phytic acid) in the soil into phosphate ester bonds and releasing inorganic phosphorus (Manzoor et al., 2022). PSB also solubilize organic acids and synergistically dissolve phosphorus through "acidification-chelation-ion exchange" (Chen et al., 2016). Citric acid chelates Ca²+, directly destroying the structure of Ca3(PO4)2 (Barrow et al., 2017), with 1mg of citric acid dissolving 0.8 mg of phosphorus (Roy et al., 2018). Oxalic acid combines with Fe3+, releasing PO43- from FePO4 and promoting the dissolution of iron phosphate (FePO4) (Panhwar et al., 2013). That is, during the fruit setting stage, Olsen-P contents in P1+H50, P2+H50, and P3+H50 treatments were significantly higher than the control (Figure 2b), and acidic phosphatase activities in P2+H50 were notably elevated (Figure 2c), forming a "P-correlated" pattern. However, P5+H50 reverted to an "N-correlated" pattern (Figure 4), likely because phosphatase (a protein) synthesis requires N sources. When NO3–N and NH4+-N in the soil are more abundant than PO43-, microorganisms can preferentially utilize inorganic nitrogen synthase proteins (e.g., alkaline/acidic phosphatase), thereby increasing the secretion of phosphatase and forming a cascade reaction of "abundant nitrogen → enhanced phosphorus hunger response → increased phosphatase secretion" (Santos-Beneit, 2015; Medici et al., 2019), i.e., "nitrogen correlation". This is supported by NO3–N ratio in control/P5+H50 exceeding control/P1+H50 to control/P3+H50 (Figure 2b), while Olsen-P ratio in control/P1+H50 to control/P3+H50 surpassed control/P5+H50 (Figures 1d, e). Simultaneously, this change of recovery also indicates that within the reasonable range of phosphate fertilizer dosage (P3+H50, P4+H50), the phosphate activation effect of Pseudomonas E sp017968885 is better. As a highly active cation, NO3–N concentration influences solution conductivity via ionic and environmental ion synergistic effects, rendering EC values significantly correlated with NO3–N (Figure 4). This relationship is used in agriculture for N fertilizer assessment, with EC >4.0 mS/cm indicating reduced N application to avoid ionic toxicity (Baker, 2008; Curti et al., 2014).
Currently, the molecular mechanisms underlying microbial solubilization of insoluble phosphates are widely recognized in academia to primarily include the secretion of organic acids and the hydrolytic action of phosphatases by microorganisms (Lei et al., 2025). Research on phosphate-solubilizing mechanisms is often closely associated with the exploration of phosphate-solubilizing genes (Mayer et al., 2025). Among these, pyrroloquinoline quinone (PQQ), as a coenzyme of glucose dehydrogenase (GDH), plays a crucial role in the conversion of glucose to gluconic acid (Chen et al., 2024). In 1987, Goldstein et al. cloned the pqq gene from Erwinia herbicola and transformed it into Escherichia coli, thereby conferring the ability to solubilize inorganic phosphate to the latter (Goldstein and Liu, 1987). This study not only validated the phosphate-solubilizing function of the pqq gene but also observed a significant increase in the production of gluconic acid (GA) as a metabolic byproduct (Goldstein and Liu, 1987). Subsequently, Kim et al. cloned a series of pqq genes from Enterobacter intermedium, a gluconic acid-producing bacterium, further confirming that the synthesis of gluconic acid depends on the assistance of the pqq factor (Kim et al., 1998). In 1990, Cleton-Jansen et al. cloned and analyzed the gdh gene encoding quinoprotein in E. coli, demonstrating that GDH catalyzes the conversion of glucose to gluconic acid with the assistance of PQQ (Cleton-Jansen et al., 1900). After years of research, Goldstein et al. proposed in 1999 that the phosphate-solubilizing ability of Gram-negative bacteria is primarily attributed to the solubilizing effect of gluconic acid, and the synthesis of gluconic acid requires the joint participation of both gdh and pqq genes, with neither being dispensable (Goldstein et al., 1999). The absence of either factor significantly impairs the ability of microorganisms to solubilize inorganic phosphate. In this investigation, the analysis of secondary metabolite biosynthetic gene clusters in strain Pseudomonas E sp017968885 demonstrated that the pqqC, pqqD, and pqqE genes are positioned within cluster 12, whereas the gdh gene is located in cluster 10 (Supplementary Figure S. Cluster 10, Cluster 12). This finding is consistent with the theoretical premise that both pqq and gdh are essential. Additionally, a set of genes related to protein secretion (gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, and gspM) was identified in the Pseudomonas E sp017968885 genome (Supplementary Figure S. Cluster 2). These genes encode components of the type II secretion system (T2SS), a system widely present in Gram-negative bacteria responsible for transporting proteins from the cytoplasm to the extracellular environment (Konstantin et al., 2013). The Pseudomonas E sp017968885 genome also contains large fragments of antibiotic biosynthesis genes (bacC, tycC, and ituB), which encode nonribosomal peptide synthetases or related modifying enzymes involved in the biosynthesis of antibiotics, thereby aiding the microorganism in gaining a competitive advantage in ecological niches (Kim et al., 2024). Furthermore, siderophore synthesis-related genes dhbF (Supplementary Figure S. Cluster 1, Cluster 10) and entF (Supplementary Figure S. Cluster 9) were detected in the Pseudomonas E sp017968885 genome. These genes encode nonribosomal peptide synthetases or related modifying enzymes, facilitating the synthesis of efficient siderophores, which play a crucial role in iron acquisition, environmental adaptation, and pathogenicity of microorganisms (Kumar et al., 2025). In summary, strain Pseudomonas E sp017968885 not only exhibits phosphate-solubilizing potential but also demonstrates significant genetic potential in secondary metabolite synthesis, protein secretion, and siderophore production, providing a molecular basis for its advantages in environmental adaptation and ecological competition. This genome characteristics of Pseudomonas E sp017968885 also support the observed changes in physicochemical indicators.
5 Conclusion
The application of Pseudomonas E sp017968885 has exhibited pronounced advantages in ameliorating soil health, enhancing nutrient utilization efficiency, and optimizing phosphorus management during pepper cultivation. Specifically, Pseudomonas E sp017968885 promoted the transformation and utilization of SOM and nitrogen forms (NH4+-N, NO3–N, TN). In addition, this strain stimulates the activity of phosphorus-related enzymes (S-ACP) through the expression of genes related to phosphate-solubilizing ability, thereby promoting the rhizosphere phosphorus cycle, improving the bioavailability and absorption of phosphorus (Olsen-P, TP), and further increasing the yield. In conclusion, the reasonable application of phosphorus fertilizer combined with Pseudomonas E sp017968885 represents an effective strategy to enhance phosphorus utilization efficiency and facilitate yield formation during pepper growth. Future experimental investigations will entail high-performance liquid chromatography (HPLC) analysis to characterize the types and concentrations of organic acids secreted by Pseudomonas E sp017968885, thereby further validating its application efficacy. Concurrently, field trials will be extended to translate these laboratory findings into practical agricultural applications.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
QH: Data curation, Methodology, Writing – original draft, Writing – review & editing, Formal Analysis. HW: Conceptualization, Writing – review & editing. MY: Methodology, Investigation, Supervision, Writing – review & editing. LS: Investigation, Software, Methodology, Writing – review & editing. JF: Formal Analysis, Project administration, Writing – review & editing. YM: Funding acquisition, Validation, Writing – review & editing. FD: Resources, Visualization, Writing – review & editing. LG: Funding acquisition, Project administration, Writing – review & editing. ZW: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The following two projects: Key Technology Development for Soil Health, Biodiversity Conservation and Productivity Enhancement in Black Soil Regions (2024JH2/102500010); The Vegetable Technology Special Mission Team in Beipiao City, Liaoning Province (2024JH5/10400072) covered all the expenses related to this experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2025.1628083/full#supplementary-material
References
Aarab S., Ollero F. J., Megias M., Laglaoui A., Bakkali M., and Arakrak A. (2017). Simultaneous p-solubilizing and biocontrol activity of rhizobacteria isolated from rice rhizosphere soil. Probiotics Agroecosyst., 207–215. doi: 10.1007/978-981-10-4059-7_11
Akram M., Hussain S., Hamid A., Majeed S., Chaudary S. A., Shah Z. A., et al. (2017). Interactive effect of phosphorus and potassium on growth, yield, quality and seed production of chili (Capsicum annuum L.). J. Hortic. 4, 192. doi: 10.4172/2376-0354.1000192
Alori E. T., Glick B. R., and Babalola O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00971
Ao N., Zou H., Li J., Shao H., Kageyama K., and Feng W. (2024). First report of pythium aphanidermatum and pythium myriotylum causing root rot on chili pepper (Capsicum annuum L.) in Guizhou, China. Crop Prot. 181, 106704. doi: 10.1016/j.cropro.2024.106704
Aswitha K., Malarvizhi P., Chitdeshwari T., Lakshmanan A., and Kalarani M. K. (2024). Bioactivation of legacy phosphorus in calcareous soil and its impact on maize yield. Agric. Sci. Dig. 44, 47–52. doi: 10.18805/ag.D-5644
Azlan A., Sultana S., Chan S., and Razman M. (2022). Antioxidant, anti-obesity, nutritional and other beneficial effects of different chili pepper: a review. Molecules. 27, 898. doi: 10.3390/molecules27030898
Babu P. M., Panda N., Nayak R. K., Sethi D., Biswal S., Mishra M. K., et al. (2025). Isolation, characterization and screening. BMC Plant Biol. 25, 362. doi: 10.1186/s12870-025-06385-1
Baccouri O., Boukerb A. M., Farhat L. B., Boukerb A. M., Zébré A., Zimmermann K., et al. (2019). Probiotic potential and safety evaluation of enterococcus faecalis OB14 and OB15, isolated from Traditional Tunisian Testouri Cheese and Rigouta, using physiological and genomic analysis. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00881
Baker N. R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59
Bao S. D. (2021). Soil and agricuitural chemistry analysis. 3rd ed. (Chaoyang, Beijing: China Agricultural Press).
Barrow N. J., Debnath A., and Sen A. (2017). Mechanisms by which citric acid increases phosphate availability. Plant Soil. 423, 1–12. doi: 10.1007/s11104-017-3490-8
Batten G. D. (1992). A review of phosphorus efficiency in wheat. Plant Soil. 146, 163–168. doi: 10.1007/BF00012009
Batten G. D., Fettell N. A., Mead J. A., and Khan M. A. (1999). Effect of sowing date on the uptake and utilisation of phosphorus by wheat (cv. Osprey) grown in central New South Wales. Aus. J. Exp. Agric. 39, 161–170. doi: 10.1071/EA97104
Bindraban P. S., Dimkpa C. O., and Pandey R. (2020). Exploring phosphorus fertilizers and fertilization strategies for improved human and environmental health. Bio. Fertil. Soils. 56, 299–317. doi: 10.1007/s00374-019-01430-2
Biswas J. K., Banerjee A., Rai M., Naidu R., Biswas B., Vithanage M., et al. (2018). Potential application of selected metal resistant phosphate solubilizing bacteria isolated from the gut of earthworm (Metaphire posthuma) in plant growth promotion. Geoderma. 330, 117–124. doi: 10.1016/j.geoderma.2018.05.034
Chen W., Yang F., Zhang L., and Wang J. (2016). Organic acid secretion and phosphate solubilizing efficiency of pseudomonas sp. PSB12: effects of phosphorus forms and carbon sources. Geomicrobiol. J. 33, 870–877. doi: 10.1080/01490451.2015.1123329
Chen X., Zhao Y., Huang S., Peñuelas J., Sardans J., Wang L., et al. (2024). Genome-based identification of phosphate-solubilizing capacities of soil bacterial isolates. AMB Express. 85, 14. doi: 10.1186/s13568-024-01745-w
Cleton-Jansen A. M., Goosen N., Fayet O., and van P. (1900). Cloning, mapping, and sequencing of the gene encoding escherichia coli quinoprotein glucose dehydrogenase. J. Appl. Bacteriol. 172, 6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990
Cui Y., Li S., Wang Y., Sun K., Li H., and Zhang W. (2023). Effects of phosphorus application rates and methods on the yield, phosphorus uptake and utilization of pepper. J. Plant Nutr. Fertil. 29, 2322–2331. doi: 10.11674/zwyf.2023205
Curti R. N., de la Vega A. J., Andrade A. J., Bramardi S. J., and Bertero H. D. (2014). Multi-environmental evaluation for grain yield and its physiological determinants of quinoa genotypes across Northwest Argentina. Field Crops Res. 166, 46–57. doi: 10.1016/j.fcr.2014.06.011
Dai S., He Y., Shen W., Zhong W., and Pen Y. (2010). Mutagenesis of a phosphate dissolving bacterial strain by UV and its application to rice cultivation in red soil. Ecol. Enviro. Sci. 19, 1646–1652. doi: 10.1080/00949651003724790
Dejene M., Abera G., and Desalegn T. (2023). The effect of phosphorus fertilizer sources and lime on acidic soil properties of mollic rhodic nitisol in Welmera District, Central Ethiopia. Appl. Environ. Soil Sci. 2023, 7002816. doi: 10.1155/2023/7002816
Delgado A., Madrid A., Kassem S., Andreu L., and del Campillo M. C. (2002). Phosphorus fertilizer recovery from calcareous soils amended with humic and fulvic acids. Plant Soil. 245, 277–286. doi: 10.1023/A:1020445710584
Deng C., Zhong Q., Shao D., Ren Y., Li Q., Wen Q., et al. (2024). Potential suitable habitats of chili pepper in China under climate change. Plants. 13, 1027. doi: 10.3390/plants13071027
De Zutter N., Ameye M., Vermeir P., Verwaeren J., De Gelder L., and Audenaert K. (2022). Innovative rhizosphere-based enrichment under P-limitation selects for bacterial isolates with high-performance P-solubilizing traits. Microbiol. Spectr. 10, e02052–e02022. doi: 10.1128/spectrum.02052-22
Dietrich P., Ferlian O., Huang Y., Luo S., Quosh J., and Eisenhauer N. (2023). Tree diversity effects on productivity depend on mycorrhizae and life strategies in a temperate forest experiment. Ecology. 104, e3896. doi: 10.1002/ecy.3896
Eduah J. O., Arthur A., Dogbatse J. A., Amoako-Attah I., and Afful E. A. (2024). Ecological effects of soil physicochemical properties and copper speciation on the microbial properties associated with land use management in cacao production. Environ. Technol. Innov. 33, 103538. doi: 10.1016/j.eti.2024.103538
El-Ramady H., Prokisch J., Mansour H., Bayoumi Y. A., Shalaby T. A., Veres S., et al. (2024). Review of crop response to soil salinity stress: possible approaches from leaching to nano-management. Soil Syst. 8, 11. doi: 10.3390/soilsystems8010011
Fu W., Chen B., Rillig M. C., Jansa J., Ma W., Xu C., et al. (2022). Community response of arbuscular mycorrhizal fungi to extreme drought in a cold-temperate grassland. New Phytol. 234, 2003–2017. doi: 10.1111/nph.17692
Goldstein A. H., Braverman K., and Osorio N. (1999). Evidence for mutualism between a plant growing in a phosphate-limited desert environment and a mineral phosphate solubilizing (MPS) rhizobacterium. FEMS Microbiol. Ecol. 30, 295–300. doi: 10.1111/j.1574-6941.1999.tb00657.x
Goldstein A. H. and Liu S. T. (1987). Molecular cloning and regulation of a mineral phosphate solubilizing gene from erwinia herbicola. Biotechnol. 5, 72–74. doi: 10.1038/nbt0187-72
Gupta M., Bisht S., Singh B., Gulati A., and Tewari R. (2011). Enhanced biomass and steviol glycosides in Stevia rebaudiana treated with phosphate-solubilizing bacteria and rock phosphate. Plant Growth Regul. 65, 449–457. doi: 10.1007/s10725-011-9615-9
Hamdali H., Bouizgarne B., Hafidi M., Lebrihi A., Virolle M. J., and Ouhdouch Y. (2008). Screening for rock phosphate solubilizing actinomycetes from moroccan phosphate mines. Appl. Soil Ecol. 38, 12–19. doi: 10.1016/j.apsoil.2007.08.007
Han H. S., Supanjani, and Lee K. D. (2006). Effect of co-inoculation with phosphate and potassium solubilizing bacteria on mineral uptake and growth of pepper and cucumber. Plant Soil Environ. 52, 130–136. doi: 10.17221/3356-PSE
Kalayu G. (2019). Phosphate solubilizing microorganisms: promising approach as biofertilizers. Int. J. Agronomy. 7, 4917256. doi: 10.1155/2019/4917256
Kim M. S., Jeong D., Jang J., Jang J., and Choi S. (2024). Mining biosynthetic gene clusters in Paenibacillus genomes to discover novel antibiotics. BMC Microbiol. 24, 226. doi: 10.1186/s12866-024-03375-5
Kim K. Y., Jordan D., and Krishinan H. G. (1998). Expression of genes from ranella apuatilis that are necessary for mineral phosphate solubilization in escherichia coli. FEMS Microbiol. Letters. 159, 121–127. doi: 10.1111/j.1574-6968.1998.tb12850.x
Konstantin V. K., Maria S., and Wim G. J. H. (2013). The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351. doi: 10.1038/nrmicro2762
Ku Y., Xu G., Tian X., Xie H., Yang X., Cao C., et al. (2018). Root colonization and growth promotion of soybean, wheat and Chinese cabbage by Bacillus cereus YL6. PloS One 13, e0200181. doi: 10.1371/journal.pone.0210035
Kumar S., Bauddh K., Barman S. C., and Singh R. P. (2014). Amendments of microbial biofertilizers and organic substances reduces requirement of urea and DAP with enhanced nutrient availability and productivity of wheat (Triticum aestivum L.). Ecol. Eng. 71, 432–437. doi: 10.1016/j.ecoleng.2014.07.007
Kumar R., Singh A., and Srivastava A. (2025). Xenosiderophores: bridging the gap in microbial iron acquisition strategies. World J. Microbiol. Biotechnol. 41, 69. doi: 10.1007/s11274-025-04287-w
Lei Y., Kuai Y., Guo M., Zhang H., Yuan Y., and Hong H. (2025). Phosphate-solubilizing microorganisms for soil health and ecosystem sustainability: a forty-year scientometric analysis, (1984-2024). . Front. Microbiol. 16. doi: 10.3389/fmicb.2025.1546852
Li Y., Long Y., Liu T., Zhang D., He M., Xie Q., et al. (2025). The role of soil microorganisms and physicochemical properties in determining the germinate of invasive Solidago canadensis L. Plant Soil. 507, 897–914. doi: 10.1007/s11104-024-06776-7
Liang C., Xing D., He J., Tu D., and Wang Y. (2024). Correlation Analysis of soil nutrients and quality index in pepper planting areas. Agronomy. 14, 2752. doi: 10.3390/agronomy14122752
Liao G., Wang Y., Yu H., He P., Lin Z., Dai T., et al. (2023). Nutrient use efficiency has decreased in southwest China since 2009 with increasing risk of nutrient excess. Commu. Earth Environ. 4, 388. doi: 10.1038/s43247-023-01036-5
Lu C. and Tian H. (2017). Global nitrogen and phosphorus fertilizer use for agriculture production in the past half century: shifted hot spots and nutrient imbalance. Earth Syst. Sci. 9, 181–192. doi: 10.5194/essd-9-181-2017
Ma K. and Diao G. (2018). Research on the contribution rate of fertilizer to grain yield in China. J. Plant Nutr. Fertil. 24, 1113–1120. doi: 10.11674/zwyf.17375
Malone Z., Berhe A. A., and Ryals R. (2023). Impacts of organic matter amendments on urban soil carbon and soil quality: A meta-analysis. J. Clean. Prod. 419, 138148. doi: 10.1016/j.jclepro.2023.138148
Manzoor A., Dippold M. A., Loeppmann S., and Blagodatskaya E. (2022). Two-phase conceptual framework of phosphatase activity and phosphorus bioavailability. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.935829
Matzek V. and Vitousek P. M. (2009). N:P stoichiometry and protein:RNA ratios in vascular plants: an evaluation of the growth-rate hypothesis. Ecol. Lett. 12, 765–771. doi: 10.1111/j.1461-0248.2009.01310.x
Mayer C., Abanto M., Urrutia C., Jerez-Quezada C., Barra P. J., and Abantoand M. (2025). Genomic Insights into Phosphorus Solubilization of Pseudomonas extremaustralis. Microorganisms. 13, 911. doi: 10.1111/brv.12779
Medici A., Szponarski W., Dangeville P., Safi A., Dissanayake I. M., Saenchai C., et al. (2019). Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell. 31, 656. doi: 10.1105/tpc.18.00656
Murray M. G. and Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nassal D., Spohn M., Eltlbany N., Jacquiod S., Smalla K., Marhan S., et al. (2017). Effects of phosphorus-mobilizing bacteria on tomato growth and soil microbial activity. Plant Soil. 427, 17–37. doi: 10.1007/s11104-017-3528-y
Pan L. and Cai B. Y. (2023). Phosphate-solubilizing bacteria: advances in their physiology, molecular mechanisms and microbial community effects. Microorganisms. 11, 2904. doi: 10.3390/microorganisms11122904
Panhwar Q. A., Jusop S., Naher U. A., Othman R., and Razi M. I. (2013). Application of potential phosphate-solubilizing bacteria and organic acids on phosphate solubilization from phosphate rock in aerobic rice. Sci. World J. 272409, 1–10. doi: 10.1155/2013/272409
Perry R. P. (2007). Balanced production of ribosomal proteins. Gene. 401, 1–32007. doi: 10.1016/j.gene.2007.07.007
Rawat P., Das S., Shankhdhar D., and Shankhdhar S. C. (2021). Phosphate-solubilizing microorganisms: mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 21, 49–68. doi: 10.1007/s42729-020-00342-7
Rfaki A., Zennouhi O., Aliyat F. Z., Nassiri L., and Ibijbijen J. (2019). Isolation, Selection and characterization of root-associated rock phosphate solubilizing bacteria in moroccan wheat (triticum aestivum L.). Geomicrobiol. J. 37, 230–241. doi: 10.1080/01490451.2019.1694106
Richardson A. E., Lynch J. P., Ryan P. R., Delhaize E., Smith F. A., Smith S. E., et al. (2011). Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 349, 121–156. doi: 10.1007/s11104-011-0950-4
Rose T. J., Pariasca-Tanaka J., Rose M. T., Fukuta Y., and Wissuwa M. (2010). Genotypic variation in grain phosphorus concentration, and opportunities to improve P-use efficiency in rice. Field Crops Res. 119, 154–160. doi: 10.1016/j.fcr.2010.07.004
Roy T., Biswas D. R., Datta S. C., Sarkar A., and Biswas S. (2018). Citric acid loaded nano clay polymer composite for solubilization of Indian rock phosphates: a step towards sustainable and phosphorus secure future. Arch. Agron. Soil Sci. 64, 1564–1581. doi: 10.1080/03650340.2018.1444275
Santos-Beneit F. (2015). The pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00402
Sharma S. B., Sayyed R. Z., Trivedi M. H., and Gobi T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus. 2, 587. doi: 10.1186/2193-1801-2-587
Sun W. and Selim M. H. (2019). Transport and retention of Molybdenum(VI) on iron oxide-coated sand: A modified multi reaction model. Appl. Geochemistry. 108, (104387). doi: 10.2136/sssaj2018.05.0189
Tang M., Li W., Song T., and Xie J. (2020). Screening of a highly efficient phosphate-solubilizing bacterium and validation of its phosphate-solubilizing effect. Biotechnol. Bull. 36, 102–109. doi: 10.13560/j.cnki.biotech.bull.1985.2019-0969
Tao S., He Z., Zhang M., Chen Y., Dai M., Zhang H., et al. (2022). Effects of organic (biogas slurry) substitution and reduced fertilization on crop yield and fertilizer utilization under wheat jade rotation mode. J. Sichuan Agric. Univ. 40, 714–720. doi: 10.16036/j.issn.1000-2650.202112064
Valetti L., Iriarte L., and Fabra A. (2018). Growth promotion of rapeseed (Brassica napus) associated with the inoculation of phosphate solubilizing bacteria. Appl. Soil Ecol. 132, 1–10. doi: 10.1016/j.apsoil.2018.08.017
Wan W., Qin Y., Wu H., Zuo W., He H., and Tan J. (2020). Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capability and their potential for lead immobilization in soil. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00752
Wang J. L., Gao X. R., Lu J., and An L. J. (2009). Phosphate-solubilizing mechanism of C2' and its actual phosphate-solubilizing effect in soil. Chin. J. Soil Sci. 40, 771–775. doi: 10.19336/j.cnki.trtb.2009.04.012
Wang Z. G., Hu Y. L., Xu W. H., Hu Y., Liu S., Zhang Y., et al. (2015). Separation and identification of a phosphorus solubilizing bacteria and its promoting effect on watermelon root. Acta Agric. Zhejiangensis. 27, 798–803. doi: 10.3969/j.issn.1004-1524.2015.05.16
Warner J. R., Vilardell J., and Sohn J. H. (2001). Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quan. Biol. 66, 567–574. doi: 10.1101/sqb
Wei K., Chen Z., Jiang N., Zhang Y., Feng J., Tian J., et al. (2020). Effects of mineral phosphorus fertilizer reduction and maize straw incorporation on soil phosphorus availability, acid phosphatase activity, and maize grain yield in northeast China. Arch. Agron. Soil Sci. 67, 66–78. doi: 10.1080/03650340.2020.1714031
Xiong C. and Zhao X. (2024). Impacts of chemical fertilizer reduction on grain yield: A case study of China. PloS One 19, e0298600. doi: 10.1371/journal.pone.0298600
Xu J. C., Huang L. M., Chen C., Wang J., and Long X. X. (2019). Effective lead immobilization by phosphate rock solubilization mediated by phosphate rock amendment and phosphate solubilizing bacteria. Chemosphere. 237, 124540. doi: 10.1016/j.chemosphere.2019.124540
Yang Y., Wang H., Li C., Liu H., Fang X., Wu M., et al. (2024). Identification of the soil physicochemical and bacterial indicators for soil organic carbon and nitrogen transformation under the wheat straw returning. PloS One 19, e0299054. doi: 10.1371/journal.pone.0299054
Yang X., Zhang K., Chang T., Shaghaleh H., Qi Z., Zhang J., et al. (2024). Interactive effects of microbial fertilizer and soil salinity on the hydraulic properties of salt-affected soil. Plants (Basel). 13, 473. doi: 10.3390/plants13040473
Zhang L., Feng G., and Declerck S. (2018). Signal beyond nutrient, fructose, exuded by an arbuscular mycorrhizal fungus triggers phytate mineralization by a phosphate solubilizing bacterium. Int. Soc. Microb. Ecol. J. 12, 2339–2351. doi: 10.1038/s41396-018-0171-4
Zheng B. X., Hao X. L., Ding K., Zhou G. W., Chen Q. L., Zhang J. B., et al. (2017). Long-term nitrogen fertilization decreased the abundance of inorganic phosphate solubilizing bacteria in an alkaline soil. Sci. Rep. 7, 42284. doi: 10.1038/srep42284
Zou X., Ma Y., Dai X., Li X., and Yang S. (2020). Spread and industry development of pepper in China. Acta Hortic. Sin. 47 , 1715–1726. doi: 10.16420/j.issn.0513-353x.2020-0103
Keywords: pepper, phosphate solubilizing bacteria, soil physicochemical index, soil phosphatase, phosphorus fertilizer efficiency
Citation: Hu Q, Wu H, Yu M, Song L, Feng J, Ma Y, Ding F, Guo L and Wang Z (2025) Optimizing the application strategy of phosphorus fertilizer by Pseudomonas E sp017968885 to increase phosphorus yield of pepper. Front. Agron. 7:1628083. doi: 10.3389/fagro.2025.1628083
Received: 13 May 2025; Accepted: 07 October 2025;
Published: 27 October 2025.
Edited by:
Muhammad Yahya Khan, University of Agriculture, Faisalabad, PakistanReviewed by:
Amit Anil Shahane, Central Agricultural University, IndiaAdnan Akhter, University of the Punjab, Pakistan
Copyright © 2025 Hu, Wu, Yu, Song, Feng, Ma, Ding, Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingling Guo, TG53c3cyMDEzQDE2My5jb20=
 Qinqin Hu
Qinqin Hu Hongyan Wu1
Hongyan Wu1