- 1Division of Animal and Nutritional Science, West Virginia University, Morgantown, WV, United States
- 2Agricultural Research Station, Fort Valley State University, Fort Valley, GA, United States
- 3Department of Animal Sciences, North Carolina Agricultural and Technical State University, Greensboro, NC, United States
We evaluated the mRNA expression of genes involved in hepatic fatty acid, amino acid, and mitochondrial energy metabolism in crossbred beef steers with divergent low and high residual feed intake (RFI). Low-RFI beef steers (n = 8; RFI = - 1.93 kg/d) and high-RFI beef steers (n = 8; RFI = + 2.01kg/d) were selected from a group of 56 growing crossbred beef steers (average BW = 261 ± 18.5 kg) fed a high-forage total mixed ration after a 49-d performance testing period. At the end of the 49-d performance testing period, liver biopsies were collected from the low-RFI and high-RFI beef steers for RNA extraction and cDNA synthesis. The mRNA expression of 84 genes each related to fatty acid metabolism, amino acid metabolism, and mitochondrial energy metabolism were analyzed using pathway-focused PCR-based arrays. The mRNA expression of 8 genes (CRAT, SLC27A5, SLC27A2, ACSBG2, ACADL, ACADSB, ACAA1, and ACAA2) involved fatty acid transport and β-oxidation were upregulated (FC ≥ 2.0, FDR ≤ 0.05) in low-RFI, compared to high-RFI steers. Among those involved in amino acid metabolism, hepatic mRNA expression of a gene encoding for aminoadipate aminotransferase, an enzyme related to lysine degradation, was downregulated (FC = -5.45, FDR = 0.01) in low-RFI steers, whereas those of methionine adenosyltransferase I and aspartate aminotransferase 2, which both link amino acid and lipid metabolism, were upregulated (FC ≥ 2.0, FDR ≤ 0.05). Two mitochondrial energy metabolism genes (UQCRC1 and ATP5G1) involved in ATP synthesis via oxidative phosphorylation were upregulated (FC ≥ 2.0, FDR ≤ 0.05) in low-RFI beef steers, compared to high-RFI beef steers. The results of this study demonstrated that low-RFI beef steers exhibit upregulation of molecular mechanisms related to fatty acid transport, fatty acid β-oxidation, and mitochondrial ATP synthesis, which suggest that low-RFI beef steers have enhanced metabolic capacity to maximize capture of energy and nutrients from feeds consumed.
Introduction
Due to high feed costs associated with animal production, residual feed intake (RFI), calculated as the difference between actual and expected dry matter intake required for maintenance and growth in animals (Koch et al., 1963), is of great economic importance. Compared to beef cattle with high (or positive) RFI, those with low (or negative) RFI are more feed efficient because they consume less feed than expected while maintaining similar growth performance.
Despite the great economic importance of RFI, the underlying biological mechanisms controlling this trait in beef cattle are still not well understood. Recent studies have suggested that low-RFI beef cattle possess several mechanisms that enable them to maximize capture of energy and nutrients from feed consumed (Elolimy et al., 2019). One of the most important organs for metabolic process is the liver, which regulates whole-body energy metabolism and acts as the main site of nutrient and energy metabolism that are essential for growth and productivity of animals (Bauchart et al., 1996; Baldwin et al., 2004). Studies have shown that fatty acids (including acetate and long-chain fatty acids) and acetyl-CoA produced from catabolism of fatty acids and amino acids are the primary carbon sources oxidized in the liver to provide energy for the body in ruminants (Drackley et al., 2001). Indeed, work from other researchers using liver transcriptomics (Alexandre et al., 2015; Mukiibi et al., 2018) have demonstrated that several biologically relevant pathways, such as lipid and amino acid metabolism, are associated with RFI in beef cattle, given their roles in energy production and tissue protein synthesis.
Furthermore, the major energy-generating organelles in body tissues, including the liver, are the mitochondria which are known to help the body cells adapt to conditions of nutrient deficiency and excess (Bottje, 2019). Thus, mitochondria are considered the metabolic hub for the regulation of hepatic metabolism of nutrients such as lipids and proteins (Saraste, 1999). Due to the role of mitochondria in energy metabolism, several studies have demonstrated that variation in RFI could be associated with differences in mitochondrial energy metabolism (Rolfe and Brand, 1997; Kolath et al., 2006; Lancaster et al., 2014). For instance, Lancaster et al., 2014 revealed that low-RFI steers had greater hepatic mitochondrial rate than high-RFI beef steers. In a similar study, Ramos and Kerley (2013) observed increased activities of some proteins of respiratory complexes in muscle of low-RFI beef steers when compared with high-RFI beef steers. In poultry, enhanced expression of mitochondrial proteins in breast muscle was observed in high feed-efficient broilers, compared to low feed-efficient ones (Bottje et al., 2017). Due to the fact that hepatic metabolism is controlled in large part by transcriptional regulation of several genes/enzymes which catalyze key nutrient and energy metabolic reactions, it was necessary to characterize expression of nutrient and energy metabolism pathway-focused genes in low- and high-RFI beef steers. Therefore, the objective of the present study was to analyze the mRNA expression of genes involved in hepatic fatty acid, amino acid, and mitochondrial energy metabolism in crossbred growing beef steers with low- or high-RFI fed a high forage diet using pathway-focused PCR-based arrays to give more insight into the biological mechanisms associated with RFI divergence. We hypothesized that differences in the expression of some nutrient or/and energy metabolism-related genes would be associated with divergence in RFI in beef steers.
Materials and Methods
Animals, Feeding, RFI Determination
The Institutional Animal Care and Use Committees of West Virginia University (protocol number 1608003693) approved the research procedures used in this study. A group of 56 crossbred growing beef steers (average BW = 261 ± 18.5 kg; age = 305 + 27 d) were obtained from a single source (raised together since birth), had been previously dewormed, vaccinated, and pre-conditioned for at least 60 d before arrival at West Virginia University farm. The beef steers were fed a high-forage total mixed ration formulated to achieve gains of about 0.9 kg per day (Table 1), which is a typical backgrounding diet fed in West Virginia. The beef steers were kept in five dry lot pens (10 – 12 steers per pen) each equipped with three GrowSafe 8000 intake nodes (GrowSafe Systems Ltd., Airdrie, Alberta, Canada) to measure individual feed intake and In-Pen Weighing Positions (IPW, Vytelle LLC), positioned at a water trough in each pen to measure BW of individual animals several times daily (Wells et al., 2021). The use of IPW to measure BW has enabled the measurement of feed efficiency with sufficient accuracy with a test period of 49 d (MacNeil et al., 2021; Wells et al., 2021). Steers were identified with a passive, half-duplex, transponder ear tag (Allflex USA Inc., Dallas–Fort Worth, TX) before entry into the test facility. The steers were first allowed to adjust to the facilities and diet for 15 days before the start of the trial. After the adjustment period, individual feed intake was measured over 49 days. The IPW Positions measured the partial BW by weighing the front end of an animal every second the animals stayed on the scale while drinking. Details on the use and accuracy of IPW positions have been published in previous studies (MacNeil et al., 2021; Wells et al., 2021). Approximately 617 ± 92 daily BW data points (after filtering outliers) per animal were generated in this study and were regressed on time using simple linear regression to calculate beginning BW, mid-test BW, and average daily gain (ADG). Values of steer’s ADG and mid-test metabolic BW (mid-test BW0.75) were regressed against individual average daily intake (in dry matter basis; 40 days of valid intake data were used) and RFI was calculated as the residual or the difference between the predicted value of the regression and the actual measured value based on the following equation: Y = β0 + β1X1 + β2X2 + ϵ, where Y is the observed DMI (kg/d), β0 is the regression intercept, β1 and β2 are the partial regression coefficients, X1 is the mid-test metabolic BW (kg), X2 is the ADG (kg/d), and ϵ indicates the RFI (kg/d; Durunna et al., 2011). At the end of the RFI testing period, all animals were ranked by RFI coefficients. Based on the RFI coefficients, the most-efficient with the lowest RFI (low-RFI; n = 8) and the least-efficient with the highest RFI (high-RFI; n = 8) beef steers were selected.
Liver Biopsy Collection, RNA Extraction, and Gene Expression
Liver biopsies were collected from low- and high-RFI beef steers by needle biopsy under local anesthesia. After cutting the skin, liver tissue was extracted using a 14-gauge biopsy needle (Tru-Core-II Automatic Biopsy Instrument: Angiotech, Lausanne, Switzerland). Approximately 1 g of liver tissue samples obtained by one puncture were immediately stored in RNAprotect tissue tubes (Cat No: 76163; Qiagen, Germantown, MD), which contain RNAprotect tissue reagent that immediately stabilizes RNA in tissue samples to preserve the gene expression profile, and immediately stored at -80°C until analyzed. Total RNA was isolated with RNeasy Micro Kit (Cat No: 74004; Qiagen) following the manufacturer’s protocol. Total RNA concentration (>100 ng/ul) was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and RNA integrity was verified using Agilent 2100 bioanalyzer (Agilent Technologies; Santa Clara, CA). All the RNA samples had integrity number > 8.0.
The RNA samples were then used to synthesize complementary DNA (cDNA) using RT2 First Strand Kit (Cat. No. 330401; Qiagen) following the manufacturer’s instructions.
The mRNA expression of 84 genes each related to fatty acid metabolism, amino acid metabolism, and mitochondrial energy metabolism were analyzed using the RT² Profiler PCR Array Cow Fatty Acid Metabolism (PABT-007ZA; Qiagen), RT² Profiler PCR Array Human Amino Acid Metabolism I (PAHS-129ZA; Qiagen), and RT² Profiler PCR Array Cow Mitochondrial Energy Metabolism (PABT-008ZA; Qiagen), respectively following the manufacturer’s instructions. Each array consisted 84 metabolism-related genes, five housekeeping genes (actin, glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine phosphoribosyltransferase 1, TATA box binding protein, and tyrosine 3-monooxygenase), one genomic DNA control to detect gDNA contamination, three reverse transcription controls, and three positive PCR controls. Real-time PCR was performed using a QuantStudio 5 Real-Time PCR System (Applied Biosystems, Foster City, CA). The PCR cycling conditions were as follows: 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s and 60°C for 1 min.
Statistical Analysis
Differences in average RFI values, ADG and DMI between low- and high-RFI groups were determined by student’s t-test. All mRNA expression data were analyzed using the Qiagen web-based platform, GeneGlobe (https://geneglobe.qiagen.com). The comparative cycle threshold (Ct) method was used for relative quantification of the gene expression (Pfaffl, 2001). Delta-delta-Ct (ΔΔCt) method with normalization of the raw data using the geometric mean of the 5 housekeeping genes was used to calculate the differences in mRNA expression of the genes between low- and high-RFI beef steers (Pfaffl, 2001). The PCR Arrays used have an average amplification efficiency of 99% with a 95% CI from 90 – 110%, which enable them to accurately analyze multiple genes simultaneously utilizing the ΔΔCT method. The mRNA expression of genes with absolute fold change (FC) ≥ 2.0 having false discovery rate-adjusted P-values [FDR; Benjamini and Hochberg, 1995)] ≤ 0.05 were considered to be differentially expressed.
Results
Table 2 shows the results of the growth performance of the low- and high-RFI beef steers. The average RFI values of low- and high-RFI steers were -1.93 and 2.01 kg/d, respectively (P = 0.01). The initial BW, final BW, and ADG were similar between the two groups (P > 0.05); however, high-RFI steers had greater (P = 0.01) DMI and tended to have lower (P = 0.09) gain:feed ratio than low-RFI steers.
The mRNA expression of the 84 genes involved in hepatic fatty acid metabolism are shown in Table S1. Out of the 84 genes analyzed, the mRNA expression of 8 genes encoding several enzymes such as acetyl-CoA transferases, acyl-CoA dehydrogenases, acyl-CoA synthetases, and fatty acid transport were upregulated (FC ≥ 2.0, FDR ≤ 0.05) in low-RFI, compared to high-RFI steers (Table 3).
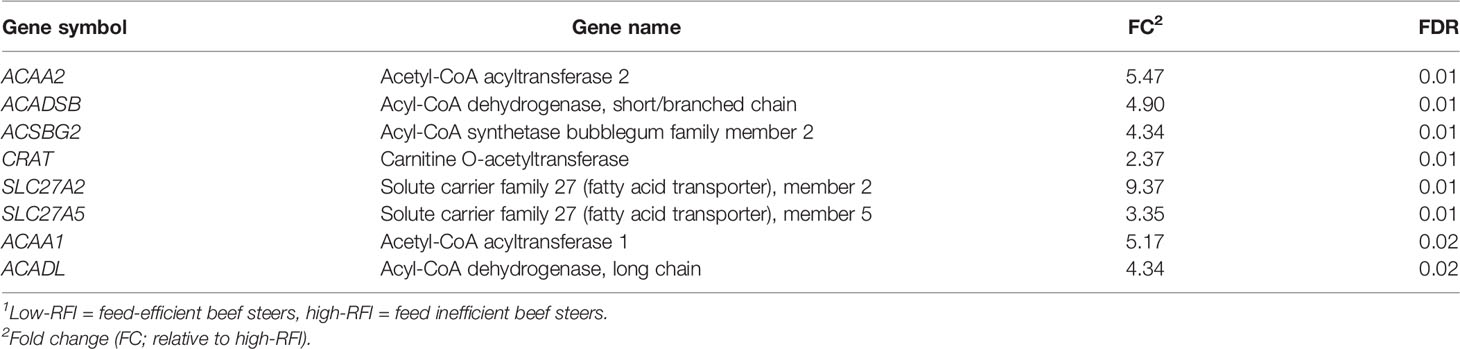
Table 3 Fold change in hepatic fatty acid metabolism gene expression in low compared with high-RFI beef steers1.
The mRNA expression of the 84 genes involved in hepatic amino acid metabolism are shown in Table S2. The mRNA expression of only 3 genes encoding aminoadipate aminotransferase (FC = -5.45, FDR = 0.01), methionine adenosyltransferase I (FC = 2.07, FDR = 0.03), and aspartate aminotransferase 2 (FC = 3.96, FDR = 0.05) were differentially expressed (Table 4).

Table 4 Fold change in hepatic amino acid metabolism gene expression in low compared with high-RFI beef steers1.
The mRNA expression of the 84 genes involved in mitochondrial energy metabolism are shown in Table S3. The mRNA expression of 2 genes (UQCRC1 and ATP5G1) encoding ubiquinol-cytochrome C reductase core protein 1 and ATP synthase, respectively were upregulated (FC > 2; FDR < 0.05) in low-RFI beef steers, compared to high-RFI beef steers (Table 5). No other mitochondrial energy metabolism genes were differentially expressed.
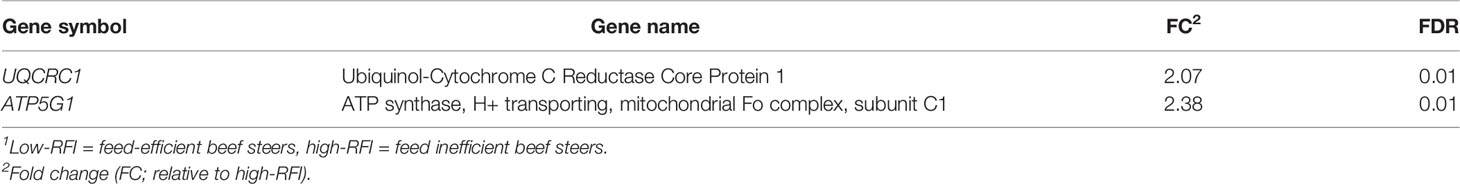
Table 5 Fold change in hepatic mitochondrial energy metabolism gene expression in low compared with high-RFI beef steers1.
Discussion
Hepatic mRNA Expression of Fatty Acid Metabolism Genes
The mRNA expression of genes involved in fatty acid transport (CRAT, SLC27A5, and SLC27A2) were upregulated in low-RFI beef steers. Fatty acids (in the form of fatty acyl-CoA) must be transported across the mitochondrial outer and inner membranes for β-oxidation to occur (Kerner and Hoppel, 2000). Carnitine O-acetyltransferase is a member of the carnitine acyltransferase family which promotes the translocation of long-chain fatty acids across the mitochondrial membrane (Kerner and Hoppel, 2000). Both SLC27A2 and SLC27A4 genes are members of the solute carrier family 27 that encode fatty acid transport protein 2 and 4, respectively (Schaffer and Lodish, 1994; Krammer et al., 2011). Fatty acid transport proteins have been proposed to function as both direct transporters of long chain fatty acids (LCFA) as well as enzymes that activate LCFA via conjugation with Co-enzyme A, a reaction catalyzed by Acyl-CoA synthetases (Anderson and Stahl, 2013). Thus, upregulation of SLC27A2 and SLC27A4 in low-RFI steers partly explained the greater mRNA expression of gene ACSBG2 encoding acyl-CoA synthetase in low-RFI beef steers, compared to high-RFI beef steers. Acyl-CoA synthetase catalyzes the activation of free fatty acids to their CoA thioesters before they can participate in any cellular metabolic pathways such as oxidation, elongation or unsaturation, protein acylation, and conversion into phospholipids (Steinberg et al., 2000). ACSBG2 belongs to a family of genes encoding enzymes capable of activating long chain fatty acids which are known to originate primarily from the diet (Steinberg et al., 2000).
The first and rate-limiting step in fatty acid β-oxidation in mitochondria is catalyzed by acyl-CoA dehydrogenases (ACADs; Ghisla and Thorpe, 2004). The mRNA expression of two genes, ACADL and ACADSB, encoding acyl-CoA dehydrogenases were upregulated in low-RFI steers. ACADL and ACADSB catalyze the first step in mitochondrial β-oxidation of long and short-chain fatty acids, respectively (Ghisla and Thorpe, 2004). Acetyl-CoA transferases are a group of enzymes involved in β-oxidation pathway of fatty acid degradation and various fatty acid biosynthetic pathways (Wanders et al., 2001). Acetyl-CoA acyltransferase 1 and 2 (ACAA1 and ACAA2), both of which were upregulated in low-RFI steers, are key regulators of fatty acid β-oxidation in peroxisomes and mitochondria, respectively (Wanders et al., 2001; Li, 2008). Acetyl-CoA acyltransferase 1 catalyzes the splitting of 3-ketoacyl-CoA to acetyl-CoA and acyl-CoA, which are both involved in fatty acid elongation and degradation in peroxisomes, while ACAA2 catalyzes the last step of mitochondrial β-oxidation of fatty acids (Wanders et al., 2001; Li, 2008). Collectively, upregulated expression of these aforementioned genes suggests enhanced mitochondrial and peroxisomal fatty acid β-oxidation in low-RFI steers.
The major function of hepatic β-oxidation of fatty acids is energy production (Kunau et al., 1995). Fatty acid oxidation can generate 2.5 times more energy than metabolism of carbohydrate via oxidative phosphorylation. Metabolism of fatty acids is a major source of energy for the skeletal muscle and β-oxidation of fatty acids in the liver produces ketone bodies that serve as essential energy source for extra-hepatic organs. These results agree with several hepatic transcriptomic studies that demonstrated lipid oxidation and transport as the most significant pathway associated with RFI divergence in beef cattle (Alexandre et al., 2015; Mukiibi et al., 2018). For instance, Mukiibi et al., 2018 analyzed the hepatic transcriptome of three breeds of beef cattle fed a grain-based finishing diet and revealed that expression of genes related to lipid synthesis and accumulation were observed to be downregulated in the liver tissues of low-RFI animals, which was consistent across the three beef breeds. In a similar study, the gene, ACACB, encoding fatty acid synthase, an enzyme that promotes lipid synthesis, was observed to be downregulated in low-RFI beef steers fed a grain-based finishing diet (Alexandre et al., 2015). In dairy cattle, Salleh et al., 2018 reported upregulation of co-expressed genes involved in lipid and cholesterol biosynthesis in liver of high-RFI cows fed a high-forage diet, compared to low-RFI cows. Previous studies in pigs have also demonstrated that altered hepatic lipid metabolism is closely related to feed efficiency (Lkhagvadorj et al., 2010; Zhao et al., 2016). Taken together, increased hepatic expression of these fatty acid metabolism-related genes in low-RFI beef steers, relative to high-RFI beef steers, suggests an improved efficiency of energy utilization, thus allowing for a similar level of growth performance despite lower DMI.
Hepatic mRNA Expression of Amino Acid Metabolism Genes
Hepatic mRNA expression of a gene, AADAT, encoding for aminoadipate aminotransferase was downregulated in low-RFI steers. Aminoadipate aminotransferase is a protein involved in a metabolic pathway that synthesizes kynurenine and glutaric acid, products of tryptophan and lysine metabolism, respectively (Sauer et al., 2011). Although most previous studies have focused on its role in kynurenine biosynthesis, the function of AADAT in hepatic lysine degradation to aminoadipate has been well described in several studies (Higashino et al., 1971; Goh et al., 2002). In fact, the enzyme has been reported to have a higher catalytic efficiency for biosynthesis of aminoadipate than for kynurenine (Han et al., 2008). Downregulation of the activity of aminoadipate aminotransferase in the liver tissue of low-RFI steers suggests increased availability of lysine for tissue protein synthesis in low-RFI steers. This result probably explains the reduced plasma concentrations of 5-aminopentanoic acid and 2-aminohexanedioic acid (aminoadipic acid), and increased plasma concentration of chloro-lysine in low-RFI beef steers observed in our companion paper (Taiwo et al., 2021). In agreement with our results, a recent study in sheep demonstrated reduced serum concentration of aminoadipic acid and increased serum concentration of lysine in low-RFI sheep, relative to high-RFI sheep (Goldansaz et al., 2020).
The mRNA expression of a gene, GOT2, encoding for mitochondrial aspartate aminotransferase isoenzyme 2 was upregulated in the liver tissue of low-RFI steers. Mitochondrial aspartate aminotransferase is involved in several metabolic processes; the enzyme links amino acid metabolism to carbohydrate metabolism by catalyzing the reaction of L-aspartate and α-ketoglutarate to form oxaloacetate and L-glutamate, which both fuel the tricarboxylic acid cycle for ATP synthesis (Jiang et al., 2016). Mitochondrial aspartate aminotransferase also has high affinity for LCFA and is known to facilitate cellular transport of both saturated and unsaturated LCFA, a key step in energy-generating mitochondrial beta-oxidation of fatty acids (Roepstorff et al., 2004), which is in line with upregulation of fatty acid metabolism genes observed in this study.
Methionine adenosyltransferase is an enzyme specific to the liver and is primarily involved in the conversion of methionine to S-adenosyl-methionine, a biological methyl donor (Avila et al., 2002; Mato et al., 2002). S-adenosylmethionine is the key methyl donor for the synthesis of several compounds (Pérez-Mato et al., 1999), including phosphatidylcholine that is required for export of very-low-density lipoproteins from the liver to adipose and muscle tissues where they can either be hydrolyzed to provide fatty acids as substrates for ATP-generating fatty acid oxidation or stored as fat when energy is not needed (Avila et al., 2002; Alves-Bezerra and Cohen, 2017). Methionine is also a precursor for synthesis of succinyl-CoA, homocysteine, cysteine, choline, creatine, methylarginine, and carnitine which are directly or indirectly involved in lipid metabolism; carnitine, in particular, is essential for the transfer of LCFA across the inner mitochondrial membrane (Roe and Ding, 2001). Increased mRNA expression of MAT1A, a gene encoding methionine adenosyltransferase in low-RFI steers suggests increased conversion of methionine to s-adenosyl-methionine in the liver. In our companion paper (Taiwo et al., 2021), we applied a chemical group-based metabolomics technique to identify blood metabolic signatures associated with RFI and observed lower plasma concentration of methionine in low-RFI beef steers when compared with high-RFI beef steers, which is in line with the result of the current study. It is important to note that the PCR array panel used was designed for human mRNA and its cross-reaction with the specific bovine mRNA was not validated in this study. Therefore, these results should be interpreted with caution.
Hepatic mRNA Expression of Mitochondrial Energy Metabolism Genes
Ubiquinol-cytochrome c reductase core protein 1 is a sub-unit of mitochondrial respiratory complex III and is known to play an essential role in regulating mitochondrial function, although its exact function is not yet determined (Yi et al., 2020). Mitochondrial respiratory complex III is one of the series of multi-subunit protein complexes of the electron transport chain (ETC), a key player in mitochondrial energy production (Lehninger et al., 1993). The ETC generates a potential difference across the mitochondrial membrane, by pumping protons from the mitochondrial matrix to the intermembrane space, which is used to power ATP synthesis (Osellame et al., 2012). Casal et al., 2018 determined the mRNA expression of some select genes involved in mitochondrial respiratory chain in the liver of Hereford steers fed high-forage diet and observed upregulation of some genes, including UQCRC1 in low-RFI steers. Similar results were observed in beef steers fed a grain-based diet. For instance, Lancaster et al., 2014 observed greater ADP-stimulated respiration rates (state 3 respiration rates) in liver of low-RFI beef steers than their high-RFI counterparts. Kolath et al. (2006) reported greater ADP-stimulated respiration rates in muscle of low-RFI beef steers compared to steers with high-RFI. In another study, Ramos and Kerley, 2013 reported greater quantity of mitochondrial complex proteins in beef cattle with low-RFI, relative to high-RFI cattle. In contrast to our results, other studies that utilized total RNA sequencing or microarray, reported no differentially expressed genes related to electron transport chains between beef cattle with divergent RFI (Alexandre et al., 2015; Tizioto et al., 2015; Zarek et al., 2017), which may be due to several reasons, including difference in methodology because transcriptomics analysis is not as specific and targeted as qPCR-based analysis.
The gene, ATP5G1, encodes a subunit of the mitochondrial ATP synthase and it is an important component of complex V of the oxidative phosphorylation chain (He et al., 2017). Mitochondrial ATP synthase catalyzes ATP synthesis during oxidative phosphorylation using the energy provided by the proton electrochemical gradient to meet cellular energy needs (Keogh et al., 2015; He et al., 2017). In rats, low feed intake has been demonstrated to result in increased expression of muscle and hepatic gene transcripts involved in ATP production to compensate for the lower caloric intake per unit weight (Gredilla et al., 2001; Sreekumar et al., 2002). This suggests that increased expression of ATP5G1 is probably an adaptive response of low-RFI steers to low DMI that may result in increased ATP production. Taken together, upregulation of UQCRC1 and ATP5G1 supports the notion that low-RFI beef steers can synthesize ATP more efficiently than high-RFI beef steers because mitochondrial oxidative phosphorylation provides over 90% of the energy needed for mammalian metabolism (Bermejo-Nogales et al., 2015).
It is important to note that the potential impact of these genes on liver function was not fully evaluated in this study as it was not determined if these mRNA changes are translated into proteins or how they affect important metabolic changes that could influence feed efficiency. In addition, the validation of the use of the reference genes and amplification efficiency of the PCR array was based on the information provided by the manufacturer. However, the results of this study provide evidence to support the concept that divergence in RFI is associated with changes in mRNA expression of genes involved in lipid metabolism, amino acid metabolism, and mitochondrial ATP production in the liver of crossbred beef steers.
Conclusion
This study revealed differential hepatic mRNA expression of multiple genes involved in amino acid, fatty acid, and mitochondrial energy metabolism in growing beef steers with divergently low- or high-RFI. Future studies are needed to determine how these mRNA changes are translated into proteins or how they affect important metabolic changes that could influence measures of feed efficiency in beef cattle.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The research procedures were approved by the Institutional Animal Care and Use Committees of West Virginia University (protocol number 1608003693).
Author Contributions
IO and MW designed the experiment. GT, IO, and MI conducted the experiment and analyzed the data. IO, GT, AP-C, and ZE-R drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by West Virginia University Experimental Station (scientific article number 3436) in support of U.S. Department of Agriculture hatch multi-state regional project W-3010.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2022.828591/full#supplementary-material
References
Alexandre P. A., Kogelman L. J., Santana M. H., Passarelli D., Pulz L. H., Fantinato-Neto P., et al. (2015). Liver Transcriptomic Networks Reveal Main Biological Processes Associated With Feed Efficiency in Beef Cattle. BMC Genomics 16, 1–13. doi: 10.1186/s12864-015-2292-8
Alves-Bezerra M., Cohen D. E. (2017). Triglyceride Metabolism in the Liver. Comp. Physiol. 8, 1–8. doi: 10.1002/cphy.c170012
Anderson C. M., Stahl A. (2013). SLC27 Fatty Acid Transport Proteins. Mol. Aspects. Med. 34, 516–528. doi: 10.1016/j.mam.2012.07.010
Avila M. A., Garcıía-Trevijano E. R., Martıínez-Chantar M. L., Latasa M. U., Pérez-Mato I., Martınez-Cruz L. A., et al. (2002). S-Adenosylmethionine Revisited: Its Essential Role in the Regulation of Liver Function. Alcohol 27, 163–167. doi: 10.1016/S0741-8329(02)00228-8
Baldwin R. L., McLeod K. R., Capuco A. V. (2004). Visceral Tissue Growth and Proliferation During the Bovine Lactation Cycle. J. Dairy. Sci. 87, 2977–2986. doi: 10.3168/jds.S0022-0302(04)73429-3
Bauchart D., Gruffat D., Durand D. (1996). Lipid Absorption and Hepatic Metabolism in Ruminants. P. Nutr. Soc. 55, 39–47. doi: 10.1079/PNS19960010
Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bermejo-Nogales A., Calduch-Giner J. A., Pérez-Sánchez J. (2015). Unraveling the Molecular Signatures of Oxidative Phosphorylation to Cope With the Nutritionally Changing Metabolic Capabilities of Liver and Muscle Tissues in Farmed Fish. PloS One 10, e0122889. doi: 10.1371/journal.pone.0122889
Bottje W. G. (2019). BOARD INVITED REVIEW: Oxidative Stress and Efficiency: The Tightrope Act of Mitochondria in Health and Disease. J. Anim. Sci. 97, 3169 3179. doi: 10.1093/jas/skz219
Bottje W. G., Lassiter K., Dridi S., Hudson N., Kong B. W. (2017). Enhanced Expression of Proteins Involved in Energy Production and Transfer in Breast Muscle of Pedigree Male Broilers Exhibiting High Feed Efficiency. Poult. Sci. 96, 2454–2458. doi: 10.3382/ps/pew453
Casal A., Garcia-Roche M., Navajas E. A., Cassina A., Carriquiry M. (2018). Hepatic Mitochondrial Function in Hereford Steers With Divergent Residual Feed Intake Phenotypes. J. Anim. Sci. 96, 4431–4443. doi: 10.1093/jas/sky285
Drackley J. K., Overton T. R., Douglas G. N. (2001). Adaptations of Glucose and Long-Chain Fatty Acid Metabolism in Liver of Dairy Cows During the Periparturient Period. J. Dairy. Sci. 84, 100–112. doi: 10.3168/jds.S0022-0302(01)70204-4
Durunna O. N., Mujibi F. D. N., Goonewardene L., Okine E. K., Wang Z., Moore S. S. (2011). Feed Efficiency Differences and Re-Ranking in Beef Steers Fed Grower and Finisher Diets. J. Anim. Sci. 89, 158–167. doi: 10.2527/jas.2009–2514
Elolimy A. A., Abdel-Hamied E., Hu L., McCann J. C., Shike D. W., Loor J. J. (2019). RAPID COMMUNICATION: Residual Feed Intake in Beef Cattle is Associated With Differences in Protein Turnover and Nutrient Transporters in Ruminal Epithelium. J. Anim. Sci. 97, 2181–2187. doi: 10.1093/jas/skz080
Ghisla S., Thorpe C. (2004). Acyl-CoA Dehydrogenases. A Mechanistic Overview. Eur. J. Biochem. 271, 494–508. doi: 10.1046/j.1432-1033.2003.03946.x
Goh D. L., Patel A., Thomas G. H., Salomons G. S., Schor D. S., Jakobs C., et al. (2002). Characterization of the Human Gene Encoding α-Aminoadipate Aminotransferase (AADAT). Mol. Gen. Met. 76, 172–180. doi: 10.1016/S1096-7192(02)00037-9
Goldansaz S. A., Markus S., Berjanskii M. (2020). Candidate Serum Metabolite Biomarkers of Residual Feed Intake and Carcass Merit in Sheep. J. Anim. Sci. 98, skaa298. doi: 10.1093/jas/skaa298
Gredilla R., Barja G., López-Torres M. (2001). Effect of Short-Term Caloric Restriction on H2O2 Production and Oxidative DNA Damage in Rat Liver Mitochondria and Location of the Free Radical Source. J. Bioenerg. Biomembr. 33, 279–287. doi: 10.1023/A:1010603206190
Han Q., Cai T., Tagle D. A., Robinson H., Li J. (2008). Substrate Specificity and Structure of Human Aminoadipate Aminotransferase/Kynurenine Aminotransferase II. Biosci. Rep. 28, 205–215. doi: 10.1042/BSR20080085
He J., Ford H. C., Carroll J., Ding S., Fearnley I. M., Walker J. E. (2017). Persistence of the Mitochondrial Permeability Transition in the Absence of Subunit C of Human ATP Synthase. Proc. Natl. Acad. Sci. United. States America 114, 3409–3414. doi: 10.1073/pnas.1702357114
Higashino K., Fujioka M., Yamamura Y. (1971). The Conversion of L-Lysine to Saccharopine and α-Aminoadipate in Mouse. Arch. Biochem. Biophy. 142, 606–614. doi: 10.1016/0003-9861(71)90525-X
Jiang X., Wang J., Chang H., Zhou Y. (2016). Recombinant Expression, Purification and Crystallographic Studies of the Mature Form of Human Mitochondrial Aspartate Aminotransferase. Bio. Trends. doi: 10.5582/bst.2015.01150. 2015-01150.
Keogh K., Waters S. M., Kelly A. K., Kenny D. A. (2015). Feed Restriction and Subsequent Realimentation in Holstein Friesian Bulls: I. Effect on Animal Performance; Muscle, Fat, and Linear Body Measurements; and Slaughter Characteristics1. J. Anim. Sci. 93, 3578–3589. doi: 10.2527/jas.2014-8470
Kerner J., Hoppel C. (2000). Fatty Acid Import Into Mitochondria. Biochim. Biophys. Acta 1486, 1–17. doi: 10.1016/s1388-1981(00)00044-5. 26.
Koch R. M., Swiger L. A., Chambers D., Gregory K. E. (1963). Efficiency of Feed Use in Beef Cattle. J. Anim. Sci. 22, 486–494. doi: 10.2527/jas1963.222486x
Kolath W. H., Kerley M. S., Golden J. W., Keisler D. H. (2006). The Relationship Between Mitochondrial Function and Residual Feed Intake in Angus Steers. J. Anim. Sci. 84, 861–865. doi: 10.2527/2006.844861x
Krammer J., Digel M., Ehehalt F., Stremmel W., Füllekrug J., Ehehalt R. (2011). Overexpression of CD36 and Acyl-CoA Synthetases FATP2, FATP4 and ACSL1 Increases Fatty Acid Uptake in Human Hepatoma Cells. Inter. J. Med. Sci. 8, 599–614. doi: 10.7150/ijms.8.599
Kunau WH, Dommes V, Schulz H (1995). Beta-Oxidation of Fatty Acids in Mitochondria, Peroxisomes, and Bacteria: A Century Of continued Progress. Prog Lipid Res 34, 267–342. doi: 10.7150/ijms.8.599
Lancaster P. A., Carstens G. E., Michal J. J., Brennan K. M., Johnson K. A., Davis M. E. (2014). Relationships Between Residual Feed Intake and Hepatic Mitochondrial Function in Growing Beef Cattle. J. Anim. Sci. 92, 3134–3141. doi: 10.2527/jas.2013-7409
Lehninger A. L., Nelson D. L., Cox M. M. (1993). Principles of Biochemistry. 2nd ed (New York: Worth Publishers).
Li W. J. (2008). Screening and High-Spirited Regulation of Functional Genes of Chicken Related Fat Metabolism (Beijing: Chinese Academy of Agricultural Sciences).
Lkhagvadorj S., Qu L., Cai W., Couture O. P., Barb C. R., Hausman G. J., et al. (2010). Gene Expression Profiling of the Shortterm Adaptive Response to Acute Caloric Restriction in Liver and Adipose Tissues of Pigs Differing in Feed Efficiency. Am. J. Phys. Regul. Integr. Comp. Phys. 298 (2), R494–R507. doi: 10.1152/ajpregu.00632.2009
MacNeil M. D., Berry D. P., Clark S. A., Crowley J. J., Scholtz M. M. (2021). Evaluation of Partial Body Weight for Predicting Body Weight and Average Daily Gain in Growing Beef Cattle. Trans. Anim. Sci. 5, txab126. doi: 10.1093/tas/txab126
Mato J. M., Corrales F. J., Lu S. C., Avila M. A. (2002). S-Adenosylmethionine: A Control Switch That Regulates Liver Function. FASEB J. 16, 15–26. doi: 10.1096/fj.01-0401rev
Mukiibi R., Vinsky M., Keogh K. A., Fitzsimmons C., Stothard P., Waters S. M., et al. (2018). Transcriptome Analyses Reveal Reduced Hepatic Lipid Synthesis and Accumulation in More Feed Efficient Beef Cattle. Sci. Rep. 8, 7303. doi: 10.1038/s41598-018-25605-3
Osellame L. D., Blacker T. S., Duchen M. R. (2012). Cellular and Molecular Mechanisms of Mitochondrial Function. Best Practice and Research. Clin. Endocrinol. Metab. 26, 711–723. doi: 10.1016/j.beem.2012.05.003
Pérez-Mato I., Castro C., Ruiz F. A., Corrales F. J., Mato J. M. (1999). Methionine Adenosyltransferase S-Nitrosylation is Regulated by the Basic and Acidic Amino Acids Surrounding the Target Thiol. J. Biol. Chem. 274, 17075–17079. doi: 10.1074/jbc.274.24.17075
Pfaffl M. W. (2001). A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 29, e45. doi: 10.1093/nar/29.9.e45
Ramos M. H., Kerley M. S. (2013). Mitochondrial Complex I Protein Differs Among Residual Feed Intake Phenotype in Beef Cattle. J. Anim. Sci. 91, 3299–3304. doi: 10.2527/jas.2012-5589
Roe C., Ding J. (2001). “Mitochondrial Fatty Acid Oxidation Disorders,” in The Metabolic and Molecular Bases of Inherited Disease. Eds. Scriver C., Beaudet A., Sly W., Valle D. (New York: McGraw-Hill), 2297–2326.
Roepstorff C., Helge J. W., Vistisen B., Kiens B. (2004). Studies of Plasma Membrane Fatty Acid-Binding Protein and Other Lipid-Binding Proteins in Human Skeletal Muscle. Proc. Nutt. Soc. 63, 239–244. doi: 10.1079/PNS2004332
Rolfe D. F., Brand M. D. (1997). The Physiological Significance of Mitochondrial Proton Leak in Animal Cells and Tissues. Biosci. Rep. 17, 9–16. doi: 10.1023/A:102732701595
Salleh S. M., Mazzoni G., Løvendahl P., Kadarmideen N. (2018). Gene Co-Expression Networks From RNA Sequencing of Dairy Cattle Identifies Genes and Pathways Affecting Feed Efficiency. BMC Bioinf. 19, 513. doi: 10.1186/s12859-018-2553-z
Saraste M. (1999). Oxidative Phosphorylation at the Fin De Siècle. Science 283, 1488–1493. doi: 10.1126/science.283.5407.1488
Sauer S. W., Opp S., Hoffmann G. F., Koeller D. M., Okun J. G., Kölker S. (2011). Therapeutic Modulation of Cerebral L-Lysine Metabolism in a Mouse Model for Glutaric Aciduria Type I. Brain 134, 157–170. doi: 10.1093/brain/awq269
Schaffer J. E., Lodish H. F. (1994). Expression Cloning and Characterization of a Novel Adipocyte Long Chain Fatty Acid Transport Protein. Cell 79, 427–436. doi: 10.1016/0092-8674(94)90252-6
Sreekumar R., Unnikrishnan J., Fu A., Nygren J., Short K. R., Schimke J., et al. (2002). Effects of Caloric Restriction on Mitochondrial Function and Gene Transcripts in Rat Muscle. Am. J. Physiology-Endocrinol. Metabol. 283, 38–43. doi: 10.1152/ajpendo.00387.2001
Steinberg S. J., Morgenthaler J., Heinzer A. K., Smith K. D., Watkins P. A. (2000). Very Long-Chain Acyl-CoA Synthetases. Human "Bubblegum" Represents a New Family of Proteins Capable of Activating Very Long-Chain Fatty Acids. J. Biol. Chem. 275 (45), 35162–35169. doi: 10.1074/jbc.M006403200
Taiwo G. A., Idowu M., Collins S., Sidney T., Wilson M., Pech Cervantes A. A., et al. (2021). Chemical Group-Based Metabolome Analysis Identifies Candidate Plasma Biomarkers Associated With Residual Feed Intake in Beef Steers. Front. Anim. Sci. 2, 783314. doi: 10.3389/fanim.2021.783314
Tizioto P. C., Coutinho L. L., Decker J. E., Schnabel R. D., Rosa K. O. P., Oliveira S., et al. (2015). Global Liver Gene Expression Differences in Nelore Steers With Divergent Residual Feed Intake Phenotypes. BMC Genomics 16, 242. doi: 10.1186/s12864-015-1464-x
Wanders R. J., Vreken P., Ferdinandusse S., Jansen G. A., Waterham H. R., Van Roermund C. W. (2001). Peroxisomal Fatty Acid Alpha- and Beta-Oxidation in Humans: Enzymology, Peroxisomal Metabolite Transporters and Peroxisomal Diseases. Biochem. Soc Trans. 29, 250–267. doi: 10.1042/bst0290250
Wells R. S., Interrante S. M., Sakkuma S. S., Walker R. S., Butler T. J. (2021). Accuracy of the VYTELLE SENSE In-Pen Weighing Positions. Appl. Anim. Sci. 37, 626–634. doi: 10.15232/aas.2021-02183
Yi T., Wu X., Li H. (2020). Ubiquinol-Cytochrome C Reductase Core Protein 1 Overexpression Protects H9c2 Cardiac Cells Against Mimic Ischemia/Reperfusion Injury Through PI3K/Akt/GSK-3β Pathway. Bioch. Biophys. Res. Comm. 529 (4), 904–909. doi: 10.1016/j.bbrc.2020.06.089
Zarek C. M., Lindholm-Perry A. K., Kuehn L. A., Freetly H. C. (2017). Differential Expression of Genes Related to Gain and Intake in the Liver of Beef Cattle. BMC Res. Notes 10, 1. doi: 10.1186/s13104-016-2345-3
Keywords: ATP, beta oxidation, feed efficiency, mitochondria, methionine
Citation: Taiwo G, Idowu MD, Wilson M, Pech-Cervantes A, Estrada-Reyes ZM and Ogunade IM (2022) Residual Feed Intake in Beef Cattle Is Associated With Differences in Hepatic mRNA Expression of Fatty Acid, Amino Acid, and Mitochondrial Energy Metabolism Genes. Front. Anim. Sci. 3:828591. doi: 10.3389/fanim.2022.828591
Received: 03 December 2021; Accepted: 27 May 2022;
Published: 24 June 2022.
Edited by:
Katie Wood, University of Guelph, CanadaReviewed by:
Dave Seymour, Trouw Nutrition R&D, NetherlandsAllison M. Meyer, University of Missouri, United States
Copyright © 2022 Taiwo, Idowu, Wilson, Pech-Cervantes, Estrada-Reyes and Ogunade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibukun M. Ogunade, SWJ1a3VuLm9ndW5hZGVAbWFpbC53dnUuZWR1
 Godstime Taiwo1
Godstime Taiwo1 Modoluwamu D. Idowu
Modoluwamu D. Idowu Andres Pech-Cervantes
Andres Pech-Cervantes Ibukun M. Ogunade
Ibukun M. Ogunade