- 1School of Agriculture and Food Systems, West Virginia University, Morgantown, WV, United States
- 2West Virginia University Extension, Morgantown, WV, United States
This study aimed to investigate breed-specific differences in the liver transcriptomic response to lipopolysaccharide (LPS) in St. Croix (STC) and Suffolk (SUF) sheep. A total of 18 sheep (9 STC and 9 SUF) were administered LPS (E. coli O111), at a dosage of 2.5 mg/kg via I.V. After euthanasia, liver samples were collected at three time points: HR0 (n = 3 per breed; no LPS), HR2 (n = 3 per breed; 2 hours after LPS), and HR6 (n = 3 per breed; 6 hours after LPS). RNA sequencing and pathway enrichment analysis were conducted to assess differentially expressed genes (DEG; FDR ≤ 0.05) and associated biological processes. At baseline (HR0), 141 DEGs were identified between STC and SUF. SUF sheep upregulated pathways related to immune readiness, including defense responses and innate immune responses. At HR2, STC had 1,719 DEGs with top upregulated genes including SELE and CCL20. Alternatively, SUF had 2,742 DEGs at HR2, with similar immune response genes expressed but at lower magnitudes. At HR6, STC had a significant shift in gene expression, with 5,568 DEGs. Notably, STC shifted from immunity-related pathways to metabolic processes. In contrast, SUF had 5,862 DEGs at HR6, continuing to express pathways related to cytokine signaling and cellular responses. Overall, STC exhibited a more intense initial response to LPS, followed by a shift towards increased differential expression of genes associated with metabolism rather than immunity. SUF, however, maintained heightened immune activity even after 6 hours, suggesting a less effective resolution of the LPS-induced challenge.
Introduction
Previous studies have demonstrated a connection between parasite resistance and disease resistance (Colditz et al., 1996; Miller and Horohov, 2006). However, the relationship between these traits and immune competency remains poorly understood. Bentley et al. (2023) examined the association between parasite resistance and antibody production in Katahdin, a hair sheep breed. The study found that sheep with low post-weaning fecal egg counts (PWFEC) exhibited a greater response to vaccination compared to sheep with high PWFEC. Similarly, Muñoz-Guzmán et al. (2006) reported that parasite-resistant Barbados Blackbelly sheep showed higher antibody production and significantly lower fecal egg counts (FEC) compared to parasite-susceptible Columbia sheep. These findings highlight the interplay between parasite resistance, vaccination responses, and immune competency.
Parasitic infections are a major challenge for sheep production worldwide. The overuse of anthelmintics has led to widespread parasite resistance, necessitating alternative approaches to parasite control (Taylor, 2012). One promising strategy is the selection of sheep breeds with enhanced parasite resistance (Sayers and Sweeney, 2005). Understanding the genetic and physiological basis of breed-specific immune responses is essential for developing effective breeding programs and maintaining healthy and efficient animals.
Breed-specific differences in parasite resistance have been documented, notably between St. Croix (STC), a hair sheep breed, and Suffolk (SUF), a wool sheep breed (Alba-Hurtado and Muñoz-Guzmán, 2012). STC sheep demonstrate superior resistance to gastrointestinal nematodes, particularly Haemonchus contortus, compared to SUF sheep. Previous studies have shown that STC sheep mount a robust T-helper type 2 (Th2) immune response to parasitic infections, while SUF sheep exhibit a less specific, proinflammatory response (Jacobs et al., 2020, 2016; MacKINNON et al., 2010). Investigating breed-specific responses in parasite-resistant and susceptible sheep enhances our understanding of mechanisms underlying immune competency.
The liver plays a crucial role in clearing lipopolysaccharide (LPS) and modulating systemic inflammatory response (Shimada et al., 2006). Lipopolysaccharide, a component of gram-negative bacterial cell walls, serves as a valuable model for studying innate immune responses (Aksel and Akyüz, 2021). It activates pattern recognition receptors, particularly Toll-like receptors, triggering the release of proinflammatory cytokines (Lynn and Golenbock, 1992). Our recent liver metabolomics study revealed differences in how STC and SUF sheep process LPS, with STC showing a more effective resolution of the inflammatory challenge (Johnson et al., 2024).
The application of multi-omics techniques allows for a more comprehensive understanding of breed-specific responses at both metabolic and transcriptomic levels. Transcriptomics, in particular, is a powerful tool for elucidating gene expression profiles and identifying molecular mechanisms underlying immune responses. RNA sequencing (RNA-seq) enables the discovery of novel genes and pathways involved in disease resistance (Salleh et al., 2017). For example, Jacobs et al., 2020 used RNA-seq analysis of ovine peripheral blood mononuclear cells (PBMC) exposed to Haemonchus contortus. The study identified key differences in the early immune responses of STC and SUF sheep, showing that STC sheep quickly upregulate proinflammatory genes and receptors, enabling rapid parasite expulsion. In contrast, SUF sheep lack the specific responses required for effective parasite clearance.
The objective of this study was to investigate LPS-induced changes in the liver transcriptome of STC and SUF sheep. By comparing the gene expression profiles of these breeds in response to an inflammatory challenge, we aimed to uncover the molecular basis of their divergent immune capabilities.
Materials and methods
Animals
All research procedures were approved by the Institutional Animal Care and Use Committee of West Virginia University (protocol number # 2303064503). A total of 18 sheep were used, including 9 STC wethers (average body weight 66.8 ± 8.2 kg, 2.5 years old) and 9 SUF sheep (7 wethers and 2 ewes; average body weight 83.0 ± 8.7 kg, 2.5 years old). The sheep were housed at the West Virginia University Agronomy Farm. All sheep were maintained under the same environmental and management conditions. Complete details of procedures have been previously described (Johnson et al., 2024). Briefly, a licensed veterinarian evaluated the health status of all sheep 24 h before the start of the experiment (May, 2023), and only animals deemed healthy were included. Health assessment was based on rectal temperature and the absence of observable clinical signs of illness or disease. Lambs were vaccinated at 28 d of age with Bovilis Covexin 8 (Merck & Co. Inc., Rahway, NJ), followed by an annual booster. In addition, sheep received a rabies vaccination (Boehringer Ingelheim, Duluth, GA) at 1 yr of age. All vaccinations were administered several months prior to the start of the study. Live weights were recorded to determine the appropriate dosage of lipopolysaccharide (LPS; E. coli O111, Sigma-Aldrich, St. Louis, MO), which was administered via I.V at a dose of 2.5 mg/kg (Graham et al., 2018; Hadfield et al., 2018). The animals were euthanized using a captive bolt gun at three designated time points: HR0 (n = 3 per breed; no LPS administration), HR2 (n = 3 per breed; 2 hours post-LPS administration), and HR6 (n = 3 per breed; 6 hours post- LPS administration). All STC sheep in the study were wethers. For SUF, the HR0 group consisted of three wethers, while the HR2 and HR6 groups each included two wethers and one ewe. Following euthanasia, sheep were exsanguinated, and after confirmation of death, liver and spleen tissues were collected.
Sample collection, RNA extraction and sequencing
The liver from each animal was removed and weighed. A pair of biopsy punches from the right lobe were collected and placed in cryogenic tubes and stored in liquid nitrogen until the time of analysis. RNA was isolated from the eighteen liver samples, using the phenol-chloroform method. Approximately 0.5 g of liver tissue was homogenized in 1 mL Trizol™ reagent (Thermo Fisher Scientific, Hampton, NH) and then incubated for 5 minutes at room temperature. Following incubation, 0.27 ml of chloroform (Sima-Aldrich, St. Louis, MO) was added, samples were vortexed briefly and then incubated for 5 minutes at room temperature. The samples were then centrifuged at 2,000 x g for 15 minutes at 4°C. The aqueous layer was transferred to a new 1.5 mL microcentrifuge tube and 0.65 ml of 100% isopropanol (Vedco Inc, St. Joseph, MO) was added, tubes vortexed briefly, and incubated at room temperature for 5 minutes. Samples were then centrifuged at 2,000 x g for 10 minutes at 4°C, to precipitate RNA. The isopropanol was poured off and 1 mL cold 75% ethanol (Pharmco, Brookfield, CT) was added followed by centrifugation at 2,000 x g for 2 minutes at 4°C. Ethanol was removed, and remaining pellet was left to air dry then reconstituted with RNAse-free water. Purity and concentration were determined and then sent to the West Virginia University Genomics Core facility for RNA sequencing. All samples had A260:A280 ratios between 1.8 and 2.0 and their average RNA integrity number was approximately 8.0.
Messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads for library construction. The RNA was fragmented, followed by synthesis of the first cDNA strand using random hexamer primers, and subsequently the second cDNA strand. The library was prepared through a series of steps, including end repair, A-tailing, adapter ligation, size selection, amplification, and purification. Library quality was assessed using Qubit and real-time PCR for quantification, and a bioanalyzer for detecting size distribution. The qualified libraries were then sequenced on a dual-indexed 2 × 50 paired-end run on an Illumina NextSeq2000 equipped with a P3 flow.
Sequence data and statistical analyses
Analysis of the raw data was done using CLC Genomics Workbench 23.0 (Qiagen CLC Genomics Workbench). Briefly, the raw data were filtered by removing low-quality reads (Qscore ≤ 5), reads containing adapter sequences, and reads with more than 10% N base calls. The resulting clean reads were mapped to the ovine reference genome (Oar_v4.0) using STAR: Ultrafast Universal RNA-Seq Aligner v.2.5.2b (Dobin et al., 2013). Unique gene counts were calculated using FeatureCounts from the Subread package v.1.5.2, and these counts were used for downstream differential expression analysis. The differential gene expression statistical package in CLC, based on General Linear Model with a negative binomial distribution algorithm, was applied to compare gene expression between STC and SUF at HR0 (before the LPS challenge). This was also applied to compare gene expression changes following the LPS challenge (at 2 and 6 hours) with those at HR0 for both STC and SUF sheep. Differentially expressed genes (DEGs) were identified based on a False Discovery Rate (FDR) of ≤ 0.05. Gene ontology terms and pathway analysis of DEGs were performed using a web-based SR-plot platform (Tang et al., 2023). Significantly enriched pathways were cataloged using FDR ≤ 0.05.
Results
Transcriptome profile prior to LPS challenge
A total of 141 DEGs were identified between STC and SUF sheep before LPS administration (FDR ≤ 0.05; Supplementary Table 1a). Eighty-five of these were upregulated, while 56 were downregulated in SUF compared to STC (Supplementary Table 1a). Table 1 lists the 10 most upregulated DEGs including PWWP3B, SLITRK4, ESRRB, RAD51AP1, MEI1 and the 10 most downregulated DEGs including S100B, SV2C, GADL1, CD22, and MYOM2. Results of the pathway enrichment analysis of all the DEGs revealed several biological processes associated with immunological response including defense response, response to virus, immune effector process and innate immune response were enriched in SUF, compared to STC with respective enrichment scores of 3.92, 3.7, 3.4 and 3.1 (Figure 1; Supplementary Table 1b).
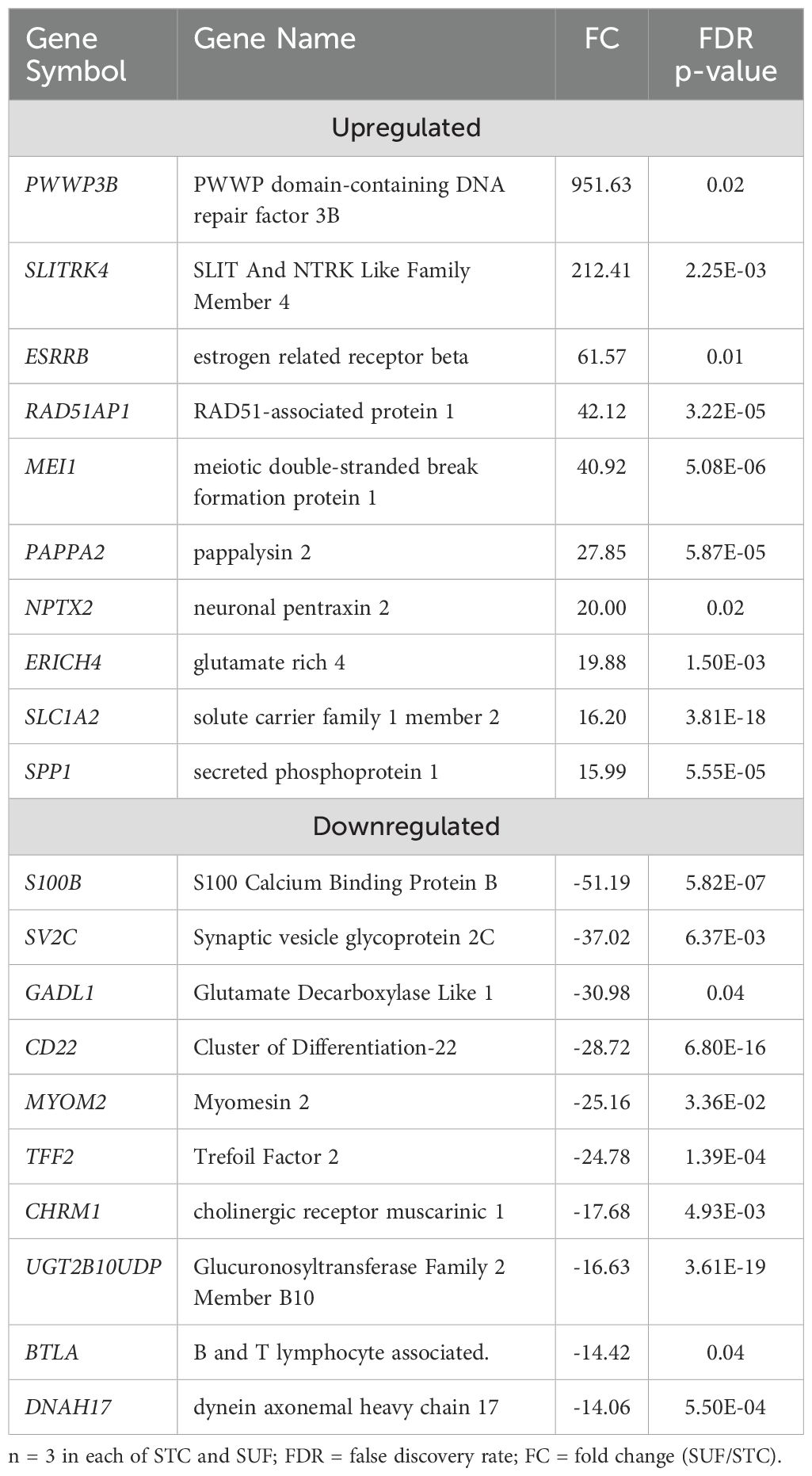
Table 1. Top 10 upregulated and downregulated differentially expressed genes in SUF compared to STC at HR0.
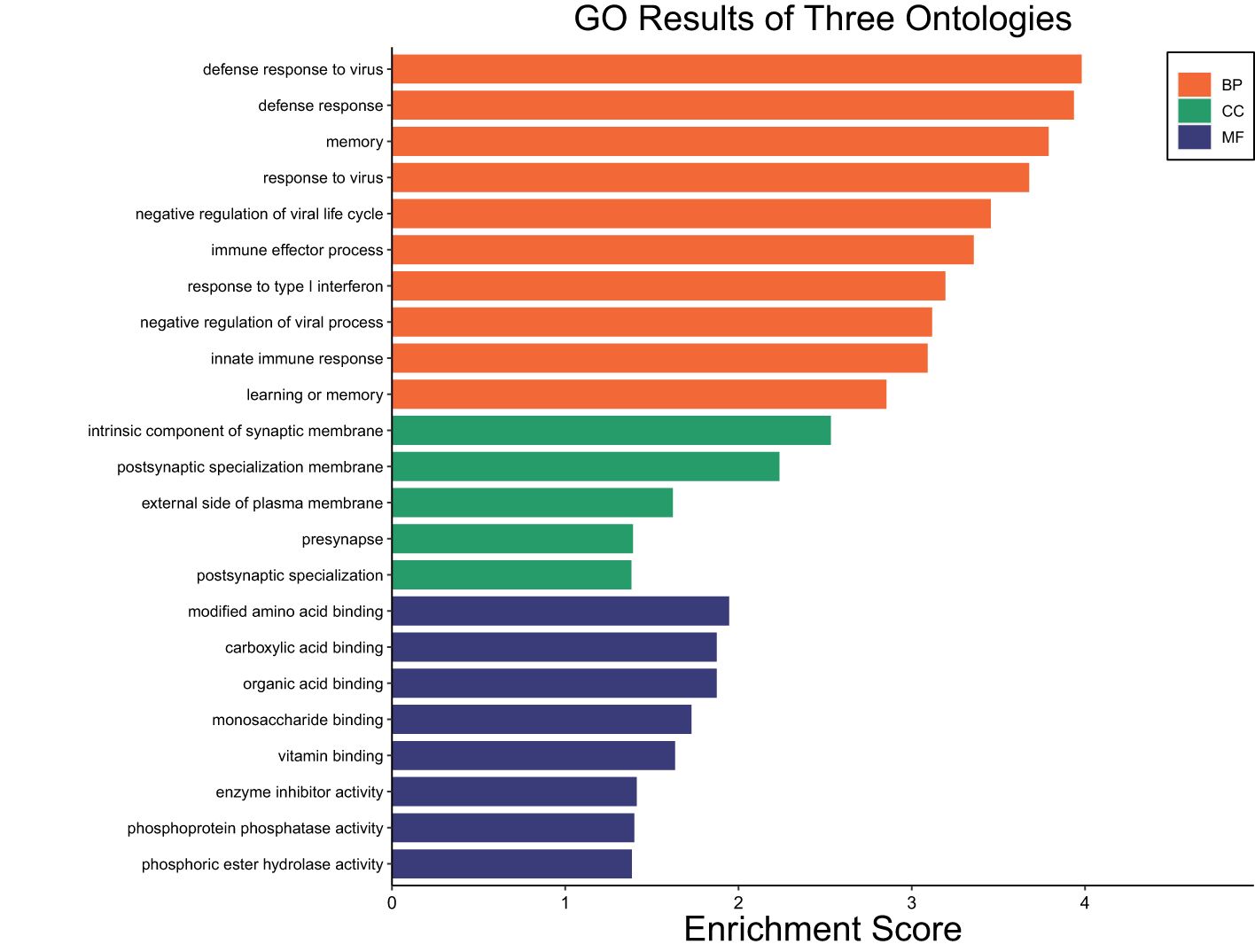
Figure 1. Gene ontology showing enriched biological processes, molecular functions, and cellular components in SUF compared to STC before LPS administration.
Transcriptomic changes in STC sheep following LPS challenge
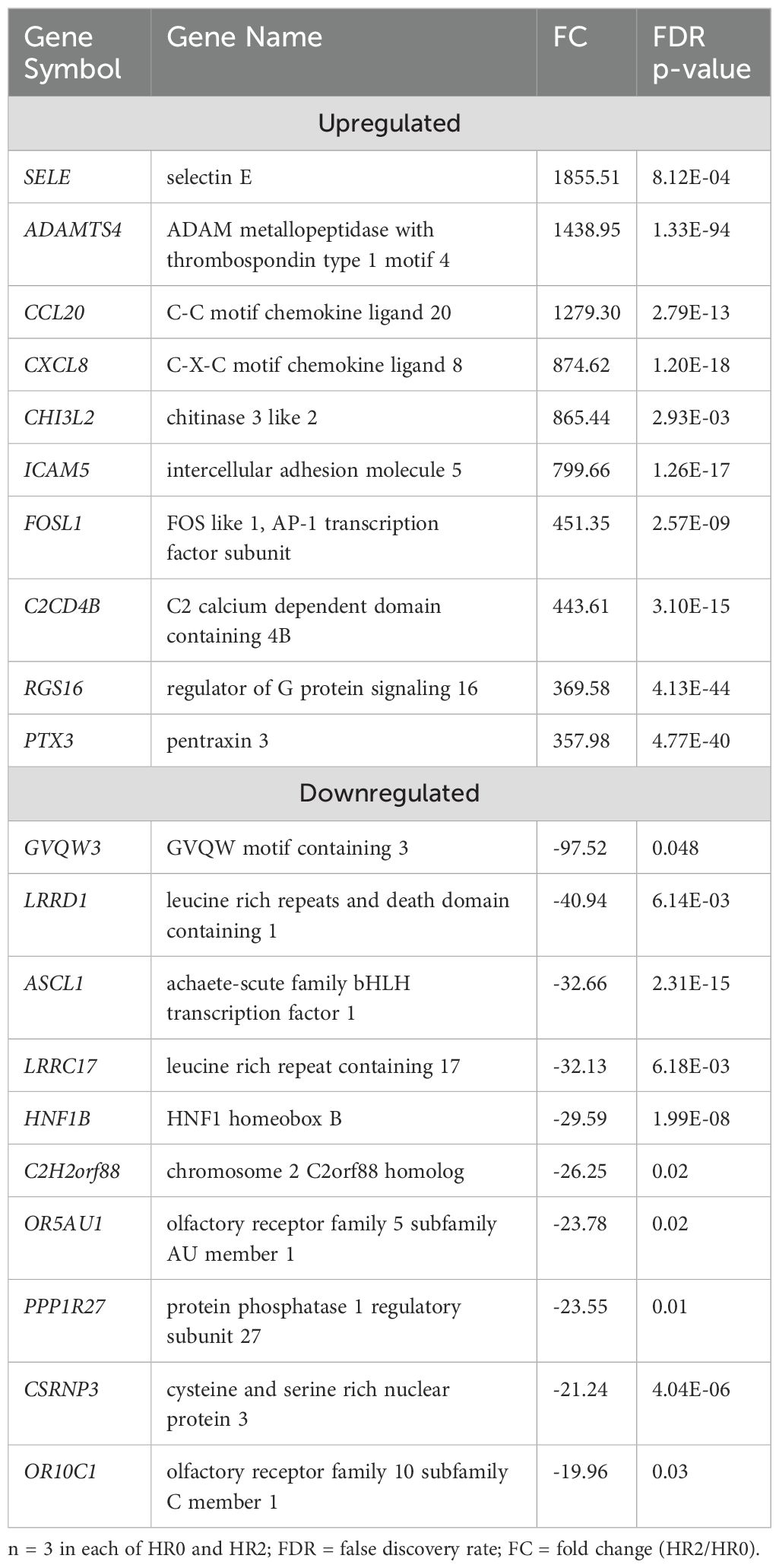
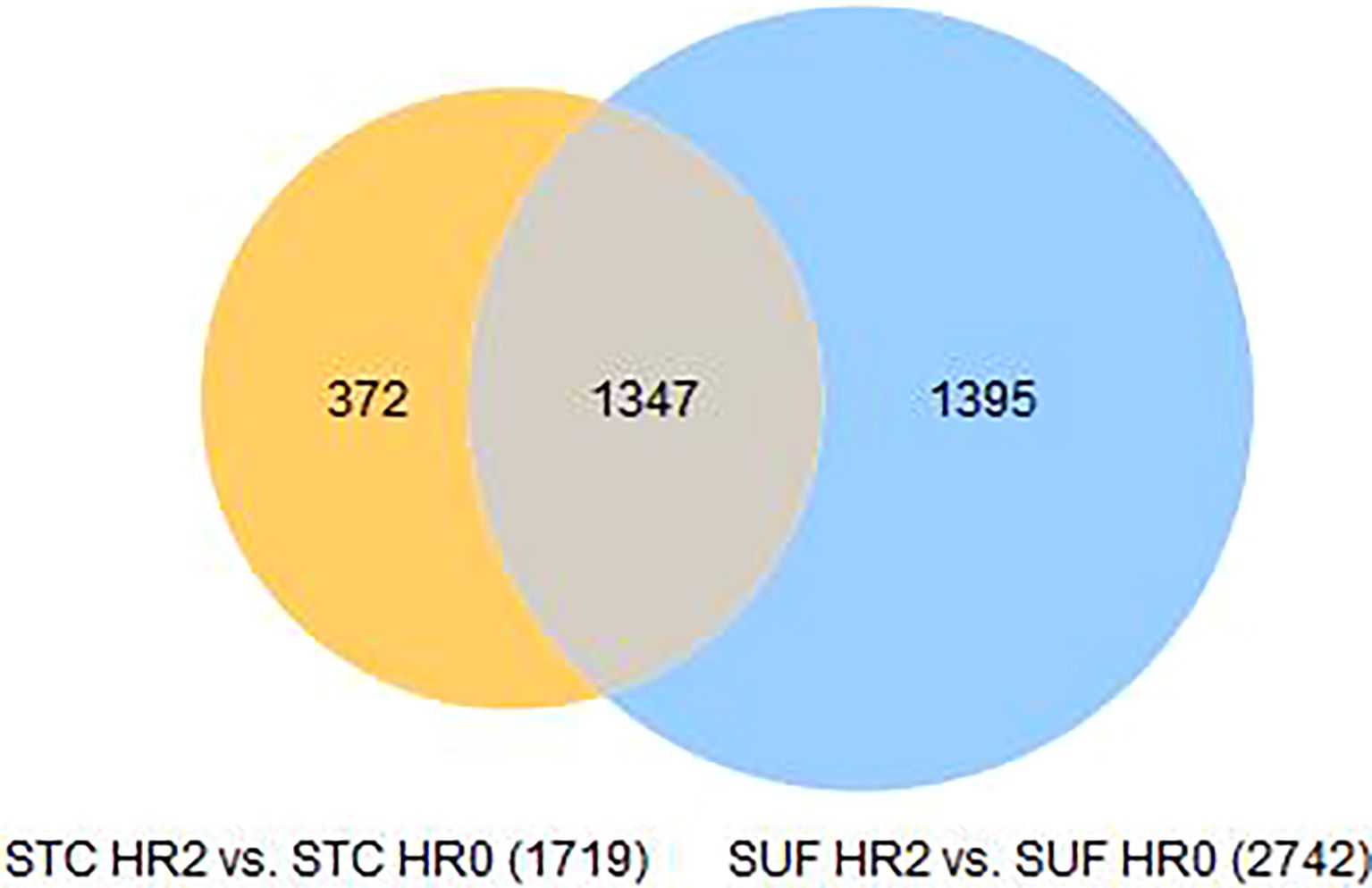
The transcriptome profiles of STC sheep after 2 or 6 hours post-LPS challenge were each compared to HR0 (before LPS challenge). A total of 1719 DEGs were identified in STC at HR2 compared to HR0 (FDR ≤ 0.05; Supplementary Table 2a). Of which, 658 were downregulated and 1061 were upregulated (Supplementary Table 2a). Table 2 lists the 10 most upregulated DEGs including CCL20, CXCL8, and CHI3L2 and the 10 most downregulated DEGs including GVQW3, IGIP, and TOR4A. Results of pathway enrichment analysis revealed that the DEGs were associated with several biological processes including immune response, cellular response to organic substances, defense response, and response to cytokine with respective enrichment scores of 9.4, 11.7, 11.9 and 14.9 (Figure 2: Supplementary Table 2b).
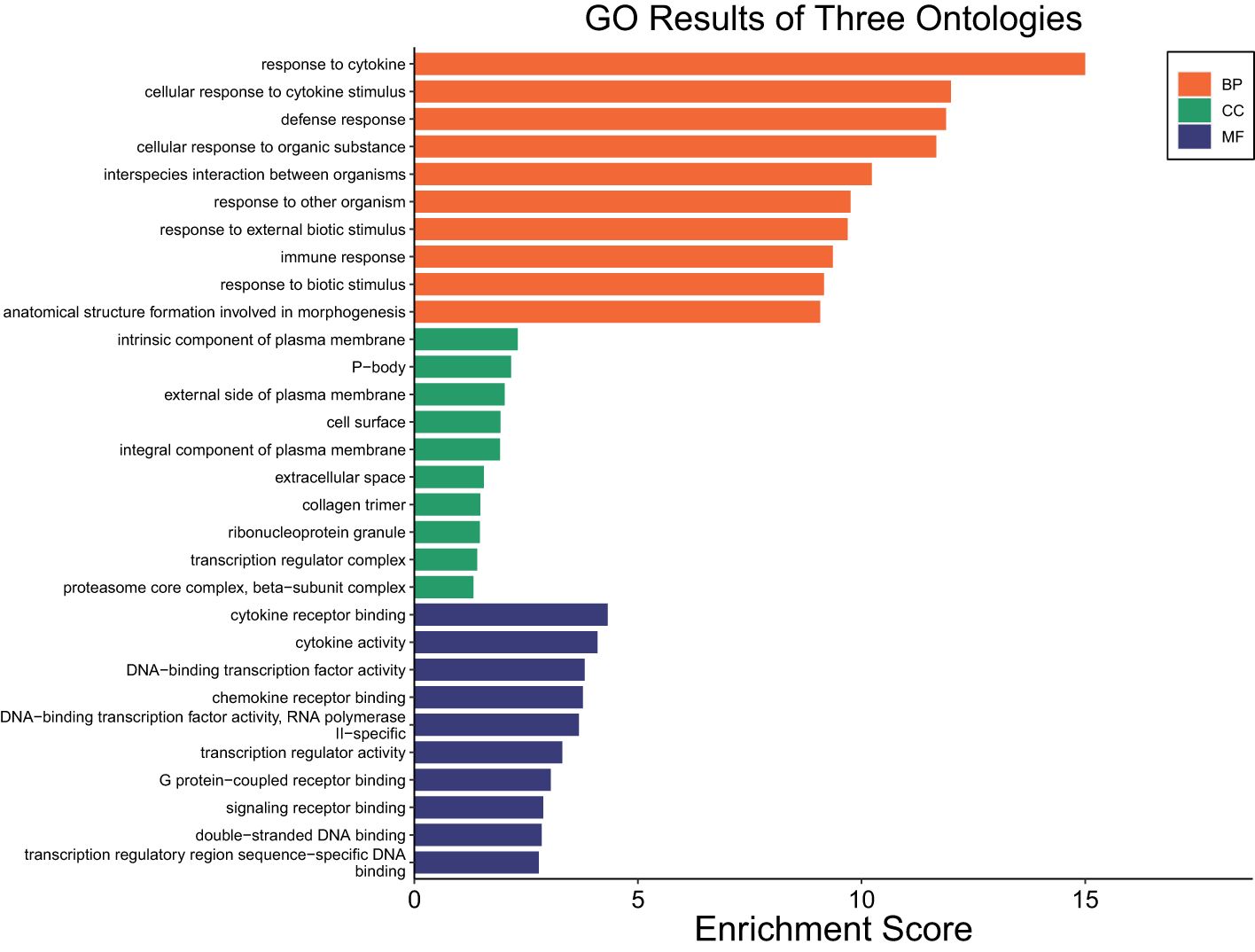
Figure 2. Gene ontology showing enriched biological processes, molecular functions, and cellular components in STC at HR2 relative to HR0.
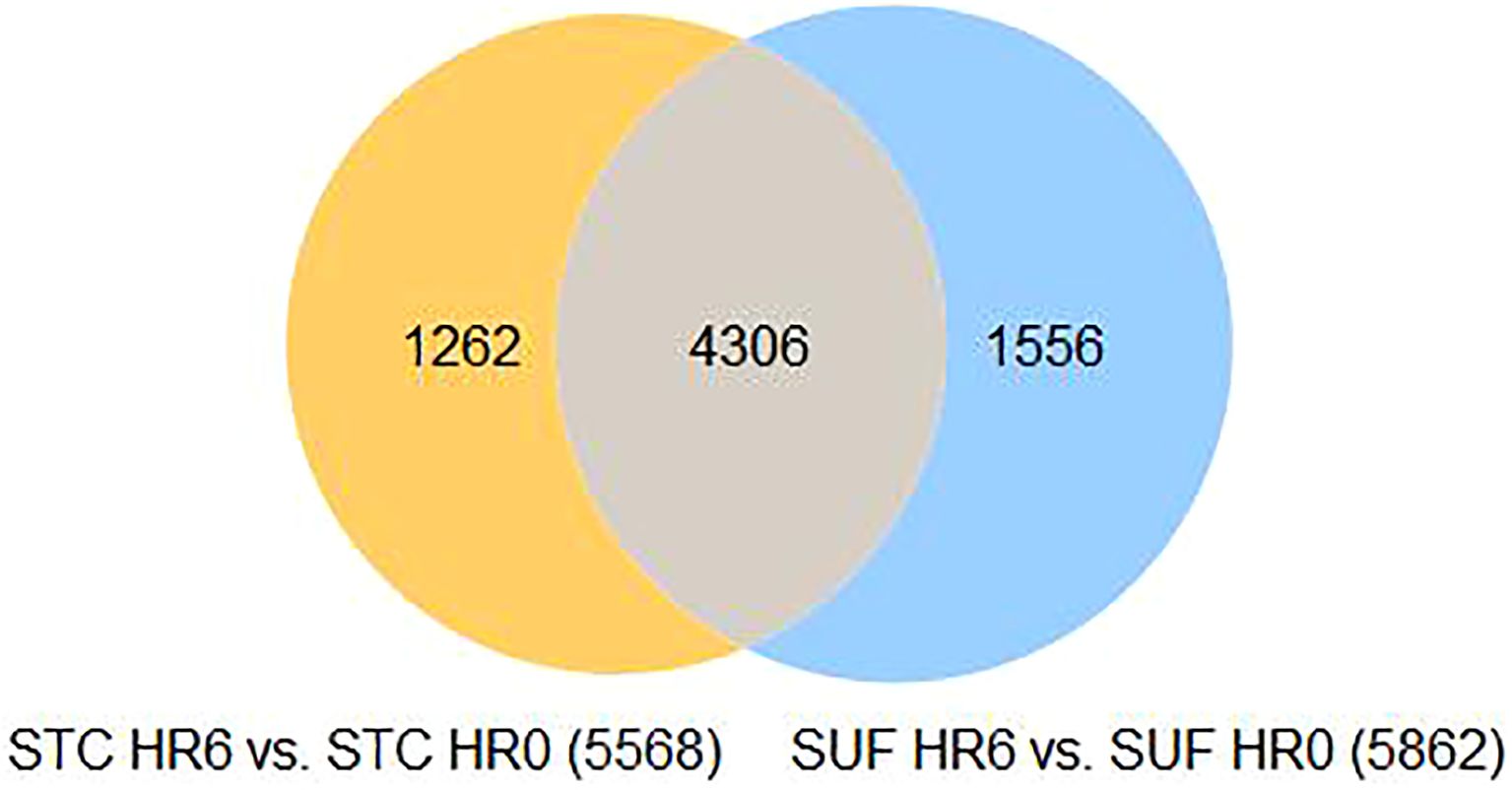
At HR 6, a total of 5,568 DEGs were identified in STC (FDR ≤ 0.05; Supplementary Table 3a). Of these, 3114 were downregulated and 2454 were upregulated (Supplementary Table 3a).
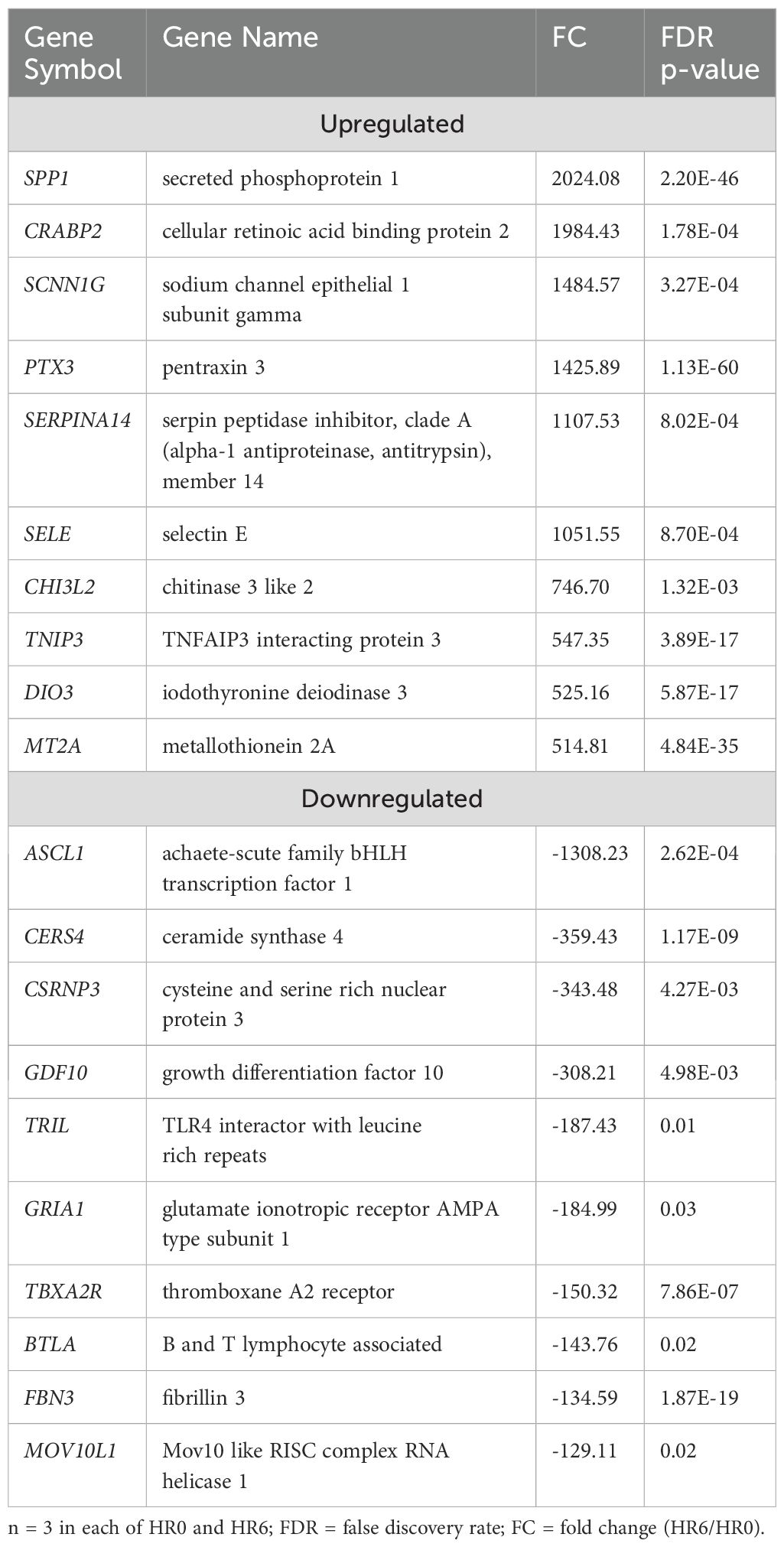
Table 3 lists the top 10 most upregulated DEGs including SPP1, CRABP2, SCNN1G, and PTX3 and the 10 most downregulated DEGs including ASCL1, CERS4, CSRNP3, GDF10, and TRIL. Results of the pathway enrichment analysis showed that these DEGs were associated with metabolic processes such as organic acid metabolic process, oxoacid metabolic process, carboxylic acid metabolic process, and small molecule biosynthetic process with respective enrichment scores of 8.5, 8.1, 7.8, and 7.1 (Figure 3: Supplementary Table 3b). Notably, no immunity-related pathways were enriched at HR 6.

Figure 3. Gene ontology showing enriched biological processes, molecular functions, and cellular components in STC at HR6 relative to HR0.
Transcriptomic changes in SUF sheep following LPS challenge
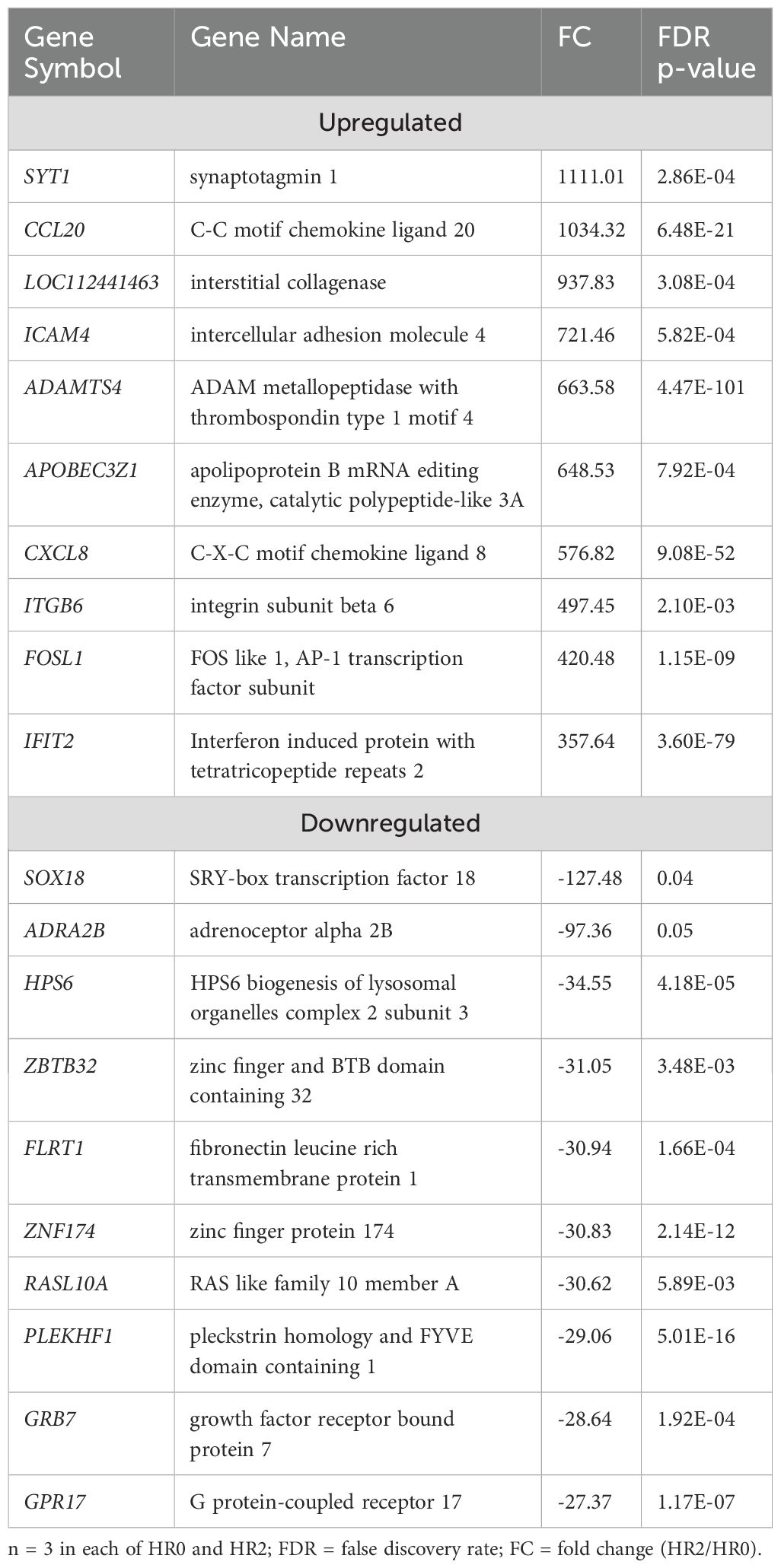
The transcriptome profiles of SUF at HR2 or HR6 were each compared to HR0 (before LPS challenge). A total of 2,742 DEGs were identified in SUF at HR2 compared to HR0 (FDR ≤ 0.05; Supplementary Table 4a). Of these, 1,175 DEGs were downregulated and 1,567 were upregulated (Supplementary Table 4a). Table 4 depicts the top 10 most upregulated DEGs such as SYT1, CCL20, ICAM4, and ADAMTS4 and the 10 most downregulated DEGs such as TCEAL8, SOX18, ADRA2B, HPS6, and OR5G1BP. Results of pathway enrichment analysis revealed that the DEGs were associated with several biological processes including immune response, defense response, interspecies interaction between organisms and response to cytokines with respective enrichment scores of 9, 9.4, 7.1 and 8.4 (Figure 4: Supplementary Table 4b).
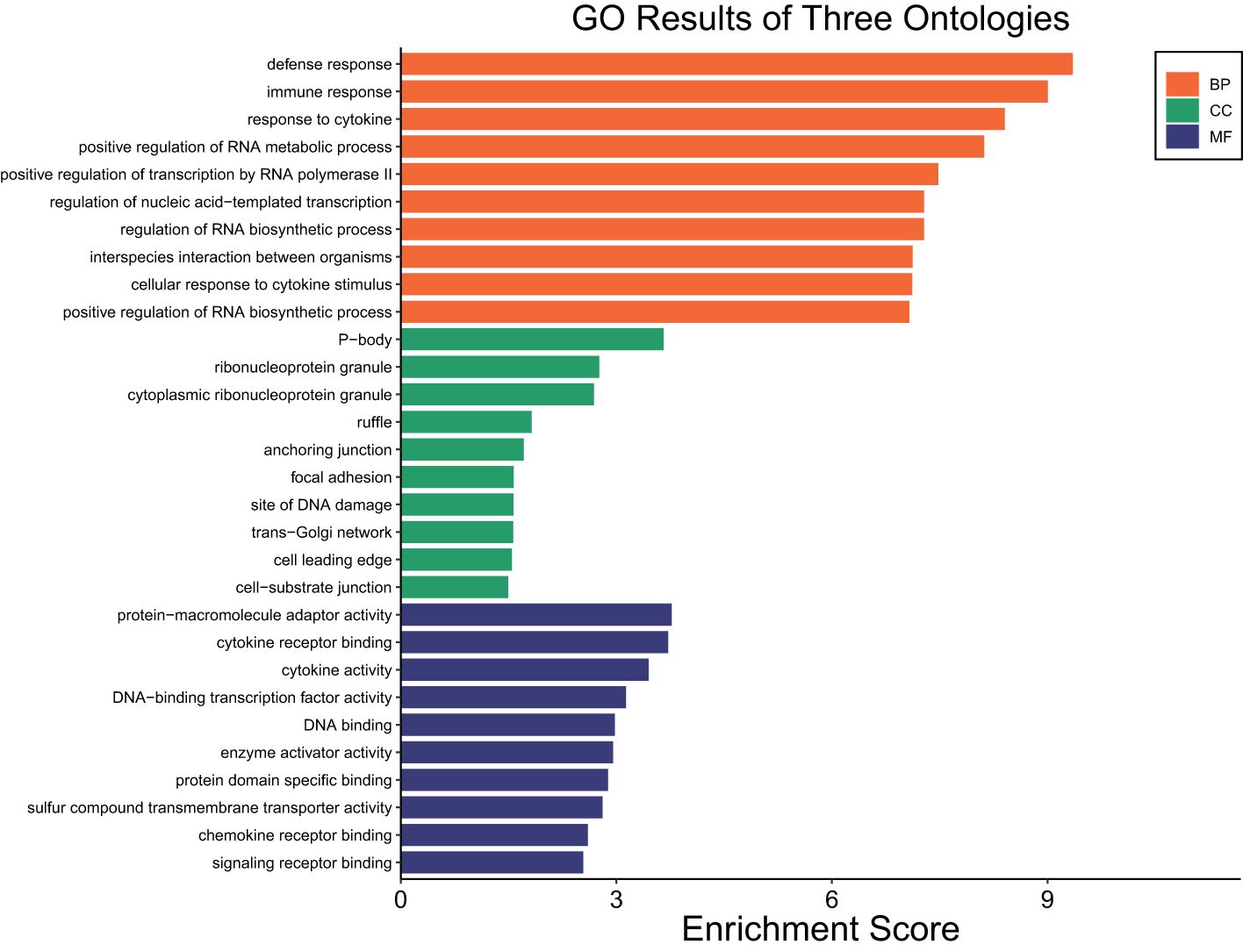
Figure 4. Gene ontology showing enriched biological processes, molecular functions, and cellular components in SUF at HR2 relative to HR0.
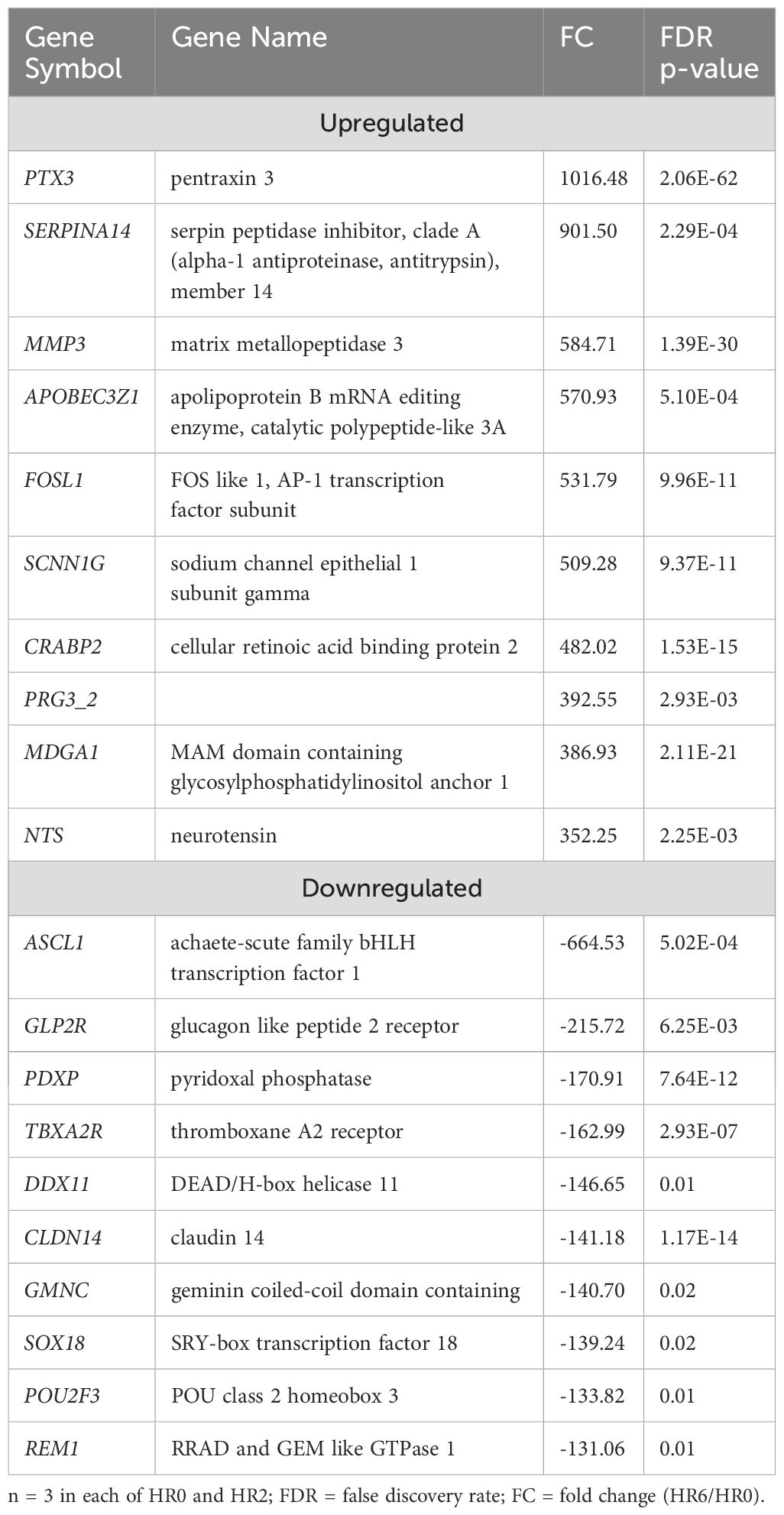
A total of 5,862 DEGs were identified at HR 6 in SUF (FDR ≤ 0.05; Supplementary Table 5a). Of which, 3,089 DEGs were downregulated and 2,773 were upregulated (Supplementary Table 5a). Table 5 lists the 10 most upregulated DEGs such as PTX3, SERPINA14, and MMP3 and the 10 most downregulated DEGs such as ASCL1, GLP2R, PDXP, and DDX1. Pathway enrichment analysis revealed that the DEGs were associated with several biological processes including cellular response to cytokine stimulus, response to cytokine, cellular response to organic substance, response to molecule of bacterial origin, and response to lipopolysaccharide with respective enrichment scores of 4.4, 5.9, 4.5, 4.9 and 5 (Figure 5: Supplementary Table 5b). The comparison of the number of DEGs between STC and SUF at HR2 relative to HR0 as well as HR6 relative to HR0 are shown in Figures 6 and 7, respectively.
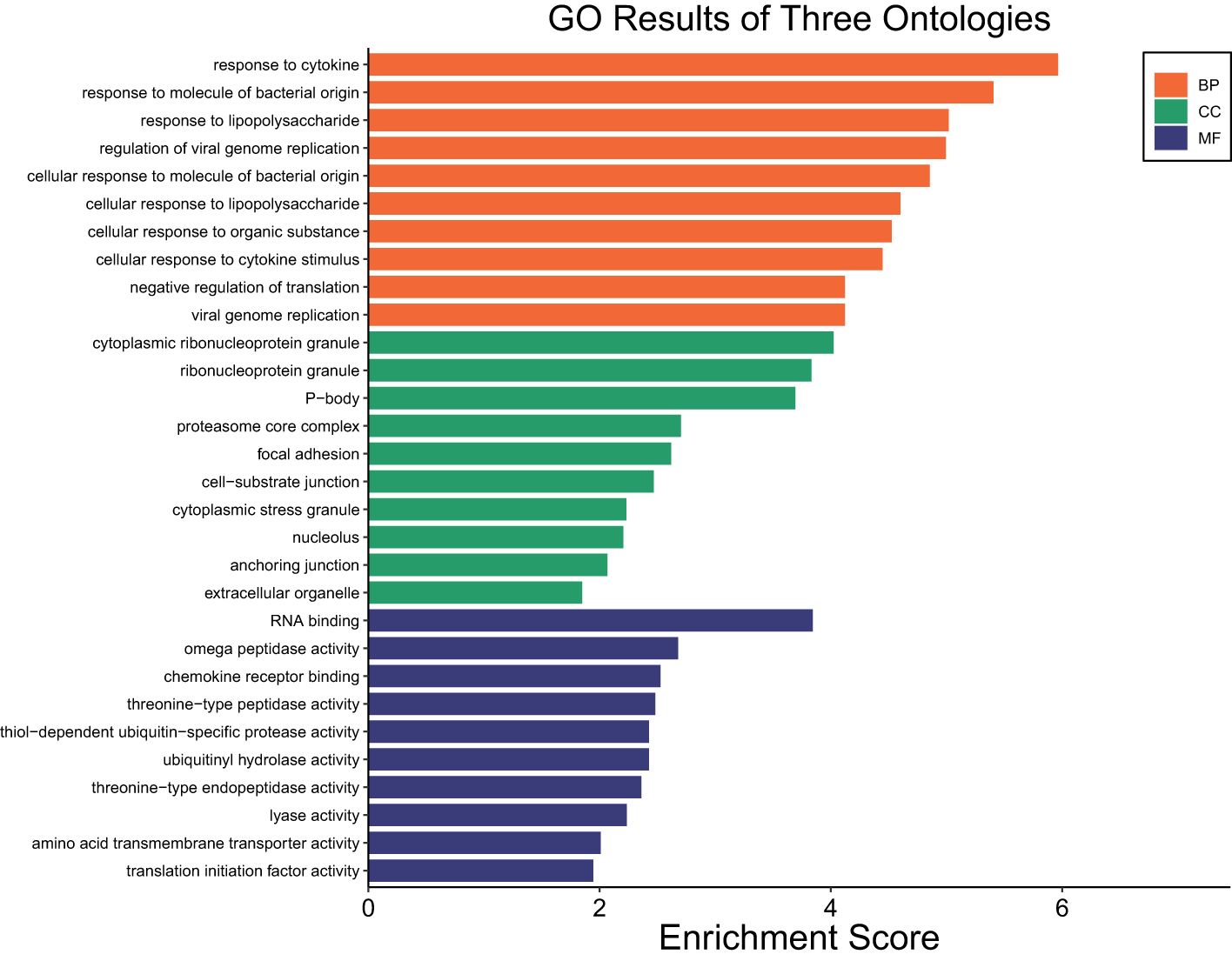
Figure 5. Gene ontology showing enriched biological processes, molecular functions, and cellular components in SUF at HR6 relative to HR0.
Discussion
Our previous study indicated that STC sheep, although less metabolically active than SUF under steady-state conditions, exhibit a greater capacity to mitigate LPS-induced oxidative stress and display a shorter duration of sickness behavior following LPS administration (Johnson et al., 2024). Suffolk sheep demonstrated greater changes in body temperature and higher sheep grimace scores (SGS) in response to LPS compared with STC (Johnson et al., 2024). Similarly, Bentley et al. (2025) reported that SUF exhibited more immediate and prolonged behavioral responses to LPS, as reflected in SGS, compared with STC. These findings underscore breed-specific differences in physiological responses to LPS. Additionally, no significant differences were observed between ewes and wethers in response to LPS (Nikpour, 2017), suggesting that the observed differences in this study are attributable to breed rather than sex.
At HR0, in the steady state (pre-challenge), SUF sheep showed enrichment of several pathways compared to STC, including defense response, immune effector processes, innate immune response, and response to viral infections (FDR ≤ 0.05). The DEGs shared across these pathways included interferon alpha-inducible protein 6 (IFI6), interferon-stimulated gene 15 (ISG15), MX dynamin-like GTPase 1 and 2 (MX1, MX2), radical S-adenosyl methionine domain-containing 2 (RSAD2), and 2’-5’-oligoadenylate synthetase 2 (OAS2) (see Supplementary Table 1B). These genes are interferon-stimulated genes (ISGs) that enhance interferon (IFN) signaling as part of the innate immune response to viral, bacterial, and parasitic infections (Schneider et al., 2014). Among the ISGs, ISG15 has been shown to regulate immune pathways involving NFκB, IFN, and JNK, as well as trigger IFN-γ production (dos Santos and Mansur, 2017). The regulation of NFκB is crucial for the production and release of proinflammatory cytokines such as TNF-α and IL-6 (Kuzmich et al., 2017).
The previously reported higher metabolic activity in SUF (Johnson et al., 2024) may explain the differential expression and enrichment of immunity-related pathways in SUF during steady-state conditions. This suggests that SUF sheep expend significant energy to maintain heightened immune readiness in the absence of active immunological challenges. This strategy may reflect a trade-off between growth and immune regulation, as Suffolk sheep are known for their favorable growth rates and carcass traits (Leymaster, 1991), However, selection for these production traits may have led to a metabolic burden that compromises their immune adaptability. Consistent with this, Yang et al. (2021) observed that native Kazakh sheep, despite their smaller size and inferior production traits, demonstrated superior immune adaptability to harsh environments compared to SUF sheep. Yang et al. also reported that SUF sheep exhibited upregulation of immunity-related genes, including those involved in hematopoietic cell lineage and natural killer cell-mediated cytotoxicity, under steady-state conditions. These findings align with our results, suggesting that SUF sheep maintain a heightened inflammatory state even in the absence of immunological challenge.
Divergence in responses of STC and SUF to Haemonchus contortus infection has been previously observed (Jacobs et al., 2020). Jacobs and colleagues reported that STC upregulated a greater number of genes, particularly those involved in inflammation, compared to SUF when challenged with H. contortus. In the current study, STC also upregulated immune response related gene at a greater magnitude to LPS than SUF. Both breeds responded to LPS, with functional pathways related to defense responses, immune responses, and cytokine signaling enriched at HR2 relative to HR0 in both groups. However, STC displayed greater pathway enrichment, with a score exceeding 14, compared to an enrichment score of approximately 9 in SUF. This suggests a more intense response in STC, likely due to broader gene activation following LPS challenge. A similar pattern was observed in PBMCs from STC and SUF exposed to H. contortus larval antigen, where STC activated the same set of genes as SUF but at higher magnitudes (Jacobs et al., 2020). Likewise (Middleton et al., 2024), found that in response to LPS, STC PBMC had significant upregulation of proinflammatory genes such as TLR4, IFNγ, NFκB, TNFα compared to SUF. These findings confirmed that STC mount a stronger early immune response not only to parasite but also to LPS.
In the same context, RNA-seq of liver tissue in this study allowed for not just immune cell interaction in response to LPS but also endothelial and non-immune cell interactions. Lipopolysaccharide triggers a cascade of immune events, including NFκB activation through tolllike receptors (TLRs), release of proinflammatory cytokines such as IL-1β, IL-8, and TNF-α via MYD88-dependent pathways and CXCL10, IFN-β, and CCL5 via MYD88-independent pathways (Page et al., 2022). Recognition of LPS through TLRs requires LPS binding protein (LBP), a crucial mediator of this response (Akira et al., 2006). Interestingly, LBP was not differentially expressed in SUF at HR2, raising the question of whether SUF rely on alternative pathways to regulate downstream proinflammatory gene expression or exhibits a diminished immune response. This may also suggest that LBP may be a function of endothelial cells responding to a TLR ligand. LBP is known to enhance LPS-induced responses, such as TNF-α production, which effects systemic response (Schumann, 1992). Bannerman et al. (2003) observed increased LBP concentrations in bovine milk and blood following LPS challenge, which may explain the more intense immune response observed in STC compared to SUF, further supporting the stronger response of STC to LPS challenge.
Inflammatory response involves intercellular adhesion molecules (ICAM) and selectins, cellular influx and apoptosis (Duffield, 2002). Adhesion molecules like SELE and ICAM5 facilitate cell migration to site of infection while PTX3 aids in inflammation regulation and tissue remodeling as well as phagocytosis (Hogg and Clive Landis, 1993; Daigo et al., 2016). While FOSL1 is a transcription factor, it has been also identified as an IFN-type I regulator that helps to prevent overactivation of immune response (Cai et al., 2017). The pattern of inflammatory gene expression in STC sheep by HR2 reveals an advantage not observed in SUF sheep. Both breeds upregulated CCL20 and CXCL8, two chemokines involved in proinflammatory responses (see Supplementary Tables 2A, 4A). CXCL8, also known as interleukin-8 (IL-8), is induced by TNF-α and promotes neutrophil migration (Fitzgerald et al., 2007). Similarly, CCL20 is induced by IL-17 and contributes to the Th17 inflammatory response, involving neutrophil infiltration (Singh et al., 2008). These findings emphasize the proinflammatory nature of responses of both STC and SUF to LPS. Notably, STC also had significant enrichment of pathways related to apoptotic processes (see Supplementary Table 2B), while SUF’s unique expression profile focused on transcriptional regulation (Supplementary Table 4B). Apoptosis plays a critical role in controlling immune overreactions and removing damaged cells, helping maintain homeostasis (Redza-Dutordoir and Averill-Bates, 2016). This apoptotic activity complements the efficient innate immune response seen in STC.
By HR6, the differences between the breeds became more pronounced. STC had a shift towards pathways related to metabolic processes, suggesting that they had begun resolving the LPS challenge. In contrast, SUF continued to express pathways associated with cytokine response and cellular response to organic substances, indicating an ongoing immune response to LPS insult.
Additionally, presence of PTX3 in SUF at HR6 suggest they may be shifting towards repair of LPS-induced damages, which is seen as early as HR2 in STC. One of the most enriched pathways in STC at HR 6 was the organic acid metabolic process, which is important for stimulating ruminal microbial fermentation (Martin, 1998). Organic acids such as acetate, propionate and butyrate are substantial energy sources in ruminants (Bergman, 1990). More specifically, propionate is a major precursor of glucose, via gluconeogenesis, a significant metabolic currency in ruminants, facilitating health and growth performance (Young, 1977). This finding suggests that STC may have redirected metabolic energy towards reestablishing homeostasis. In SUF, similar pathways to those enriched at HR2 remained active at HR6, though the enrichment score was lower, dropping from < 9.5 to < 6. This reduction suggests that SUF were still actively combating LPS but at a diminished intensity. The prolonged response in SUF raises questions about the duration and effectiveness of their immune response, potentially contributing to their increased disease susceptibility. Further exploration of the underlying mechanisms driving the observed immune response differences between STC and SUF sheep would strengthen the study. While our current analysis focuses on differential gene expression patterns, follow-up functional validation studies, such as gene knockdown or overexpression experiments, could provide direct evidence of the roles of the observed DEGs, such as LBP and PTX3, in immune regulation of STC and SUF.
Conclusion
The present study highlights the differential hepatic responses of STC and SUF to LPS administration. STC demonstrated a more robust and efficient immune response, as evidenced by higher pathway enrichment scores at HR2 and shift in gene type from immune response at HR2 to more metabolic activity-related genes by HR6. In contrast, SUF exhibited an initial heightened metabolic activity prior to LPS exposure, which may have led to the expression of interferon-stimulated genes but also contributed to a prolonged inflammatory response and slower recovery. This study represents a foundational step in understanding gene expression responses to LPS challenges in this context, laying the groundwork for future investigations that can build upon these findings with expanded datasets.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be here: https://www.ncbi.nlm.nih.gov/, SRA PRJNA1196424.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of West Virginia University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SJ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. KB: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Visualization, Writing – review & editing. SB: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. IO: Data curation, Formal Analysis, Investigation, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research study was funded by West Virginia University Animal Health hatch project #923.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2025.1565253/full#supplementary-material
References
Akira S., Uematsu S., and Takeuchi O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Aksel E. G. and Akyüz B. (2021). Effect of LPS and LTA stimulation on the expression of TLR-pathway genes in PBMCs of Akkaraman lambs in vivo. Trop. Anim. Health Prod. 53, 65. doi: 10.1007/s11250-020-02491-4
Alba-Hurtado F. and Muñoz-Guzmán M. A. (2012). Immune responses associated with resistance to haemonchosis in sheep. BioMed. Res. Int. 2013, e162158. doi: 10.1155/2013/162158
Bannerman D. D., Paape M. J., Hare W. R., and Sohn E. J. (2003). Increased levels of LPSBinding protein in bovine blood and milk following bacterial lipopolysaccharide challenge. J. Dairy Sci. 86, 3128–3137. doi: 10.3168/jds.S0022-0302(03)73914-9
Bentley K. L., Greiner S., Kelley E., Ogunade I., and Bowdridge S. A. (2025). Evaluating differential lipopolysaccharide-induced behavioral, immune and plasma metabolome responses of St. Croix and Suffolk sheep. Small Rumin. Res. 245. doi: 10.1016/j.smallrumres.2025.107461
Bentley K. L., Weaver A. R., Wright D. L., Greiner S. P., and Bowdridge S. A. (2023). Postweaning fecal egg count estimated breeding value is associated with greater antibody production after clostridial vaccination in Katahdin lambs. Small Rumin. Res. 229, 107128. doi: 10.1016/j.smallrumres.2023.107128
Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Cai B., Wu J., Yu X., Su X., and Wang R.-F. (2017). FOSL1 inhibits type I interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. mBio 8. doi: 10.1128/mbio.02161-16
Colditz I. G., Watson D. L., Gray G. D., and Eady S. J. (1996). Some relationships between age, immune responsiveness and resistance to parasites in ruminants. Int. J. Parasitol. 26, 869–877. doi: 10.1016/S0020-7519(96)80058-0
Daigo K., Inforzato A., Barajon I., Garlanda C., Bottazzi B., Meri S., et al. (2016). Pentraxins in the activation and regulation of innate immunity. Immunol. Rev. 274, 202–217. doi: 10.1111/imr.12476
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
dos Santos P. F. and Mansur D. S. (2017). Beyond ISGlylation: functions of free intracellular and extracellular ISG15. J. Interferon Cytokine Res. 37, 246–253. doi: 10.1089/jir.2016.0103
Duffield J. S. (2002). The inflammatory macrophage: a story of Jekyll and Hyde. Clin. Sci. 104, 27–38. doi: 10.1042/cs1040027
Fitzgerald D. C., Meade K. G., McEvoy A. N., Lillis L., Murphy E. P., MacHugh D. E., et al. (2007). Tumour necrosis factor-α (TNF-α) increases nuclear factor κB (NFκB) activity in and interleukin-8 (IL-8) release from bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 116, 59–68. doi: 10.1016/j.vetimm.2006.12.008
Graham M. R., Bowdridge E. C., Bowdridge S. A., Holásková I., Elsasser T. H., and Dailey R. A. (2018). Effect of lipopolysaccharide-induced immune responses on pregnancy loss in ewes. Open J. Anim. Sci. 8, 421–431. doi: 10.4236/ojas.2018.84031
Hadfield J. M., Bowdridge E. C., Holásková I., Elsasser T. H., and Dailey R. A. (2018). Breed-specific differences in the immune response to lipopolysaccharide in ewes. J. Anim. Sci. 96, 4220–4228. doi: 10.1093/jas/sky288
Hogg N. and Clive Landis R. (1993). Adhesion molecules in cell interactions. Curr. Opin. Immunol. 5, 383–390. doi: 10.1016/0952-7915(93)90057-Y
Jacobs J. R., Middleton D., Greiner S. P., and Bowdridge S. A. (2020). RNA-Sequencing of ovine PBMC after exposure to Haemonchus contortus larval antigen. Parasite Immunol. 42, e12697. doi: 10.1111/pim.12697
Jacobs J. R., Sommers K. N., Zajac A. M., Notter D. R., and Bowdridge S. A. (2016). Early IL-4 gene expression in abomasum is associated with resistance to Haemonchus contortus in hair and wool sheep breeds. Parasite Immunol. 38, 333–339. doi: 10.1111/pim.12321
Johnson S. R., Bentley K., Bowdridge S., and Ogunade I. M. (2024). Lipopolysaccharideinduced alterations in the liver metabolome of St. Croix and Suffolk sheep. Front. Anim. Sci. 5. doi: 10.3389/fanim.2024.1407533
Kuzmich N. N., Sivak K. V., Chubarev V. N., Porozov Y. B., Savateeva-Lyubimova T. N., and Peri F. (2017). TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines 5, 34. doi: 10.3390/vaccines5040034
Leymaster K. A. (1991). Straightbred comparison of a composite population and the Suffolk breed for performance traits of sheep. J. Anim. Sci. 69, 993–999. doi: 10.2527/1991.693993x
Lynn W. A. and Golenbock D. T. (1992). Lipopolysaccharide antagonists. Immunol. Today 13, 271–276. doi: 10.1016/0167-5699(92)90009-V
MacKINNON K. M., Zajac A. M., Kooyman F. N. J., and Notter D. R. (2010). Differences in immune parameters are associated with resistance to Haemonchus contortus in Caribbean hair sheep. Parasite Immunol. 32, 484–493. doi: 10.1111/j.1365-3024.2010.01211.x
Martin S. A. (1998). Manipulation of ruminal fermentation with organic acids: a review. J. Anim. Sci. 76, 3123–3132. doi: 10.2527/1998.76123123x
Middleton D., Hanlon K., Greiner S. P., and Bowdridge S. A. (2024). Variants of NLRP3 protein in haemonchus contortus infected sheep: impact on immune cell responsiveness to LPS in vitro. Parasite Immunol. 46, e13054. doi: 10.1111/pim.13054
Miller J. E. and Horohov D. W. (2006). Immunological aspects of nematode parasite control in sheep1. J. Anim. Sci. 84, E124–E132. doi: 10.2527/2006.8413_supplE124x
Muñoz-Guzmán M. A., Cuéllar-Ordaz J. A., Valdivia-Anda A. G., Buendía-Jiménez J. A., and Alba-Hurtado F. (2006). Correlation of parasitological and immunological parameters in sheep with high and low resistance to haemonchosis. Can. J. Anim. Sci. 86, 363–371. doi: 10.4141/A06-010
Nikpour H. (2017). Influence of Sex and Blood Collection Method on Innate Immune Response following Lipopolysaccharide Administration in Sheep [M.S.]. United States -- West Virginia: West Virginia University, United States. Available online at: https://www.proquest.com/docview/1901441407/abstract/9C5AC071520A4A98PQ/1.
Page M. J., Kell D. B., and Pretorius E. (2022). The role of lipopolysaccharide-induced cell signalling in chronic inflammation. Chronic Stress 6, 24705470221076390. doi: 10.1177/24705470221076390
Qiagen CLC Genomics Workbench. Available online at: https://digitalinsights.qiagen.com (Accessed December 20, 2024).
Redza-Dutordoir M. and Averill-Bates D. A. (2016). Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta BBA - Mol. Cell Res. 1863, 2977–2992. doi: 10.1016/j.bbamcr.2016.09.012
Salleh M. S., Mazzoni G., Höglund J. K., Olijhoek D. W., Lund P., Løvendahl P., et al. (2017). RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high- and low-residual feed intake in Nordic dairy cattle. BMC Genomics 18, 258. doi: 10.1186/s12864-017-3622-9
Sayers G. and Sweeney T. (2005). Gastrointestinal nematode infection in sheep – a review of the alternatives to anthelmintics in parasite control. Anim. Health Res. Rev. 6, 159–171. doi: 10.1079/AHR2005108
Schneider W. M., Chevillotte M. D., and Rice C. M. (2014). Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schumann R. R. (1992). Function of lipopolysaccharide (LPS)-binding protein (LBP) and CD14, the receptor for LPS/LBP complexes: a short review. Res. Immunol. 143, 11–15. doi: 10.1016/0923-2494(92)80074-U
Shimada H., Hasegawa N., Koh H., Tasaka S., Shimizu M., Yamada W., et al. (2006). Effects of initial passage of endotoxin through the liver on the extent of acute lung injury in A rat model. Shock 26, 311. doi: 10.1097/01.shk.0000224960.17274.6f
Singh S. P., Zhang H. H., Foley J. F., Hedrick M. N., and Farber J. M. (2008). Human T cells that are able to produce IL-17 express the chemokine receptor CCR61. J. Immunol. 180, 214–221. doi: 10.4049/jimmunol.180.1.214
Tang D., Cheng M., Huang X., Zhang G., Zeng L., Zhang G., et al. (2023). SRplot: A free online platform for data visualization and graphing. PLOS ONE 18, e0294236. doi: 10.1371/journal.pone.0294236
Taylor M. A. (2012). Emerging parasitic diseases of sheep. Vet. Parasitol. 189, 2–7. doi: 10.1016/j.vetpar.2012.03.027
Yang H., Yang Y.-L., Li G.-Q., Yu Q., and Yang J. (2021). Identifications of immuneresponsive genes for adaptative traits by comparative transcriptome analysis of spleen tissue from Kazakh and Suffolk sheep. Sci. Rep. 11, 3157. doi: 10.1038/s41598-02182878-x
Young J. W. (1977). Gluconeogenesis in cattle: significance and methodology1. J. Dairy Sci. 60, 1–15. doi: 10.3168/jds.S0022-0302(77)83821-6
References
Akira S., Uematsu S., and Takeuchi O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Aksel E. G. and Akyüz B. (2021). Effect of LPS and LTA stimulation on the expression of TLR-pathway genes in PBMCs of Akkaraman lambs in vivo. Trop. Anim. Health Prod. 53, 65. doi: 10.1007/s11250-020-02491-4
Alba-Hurtado F. and Muñoz-Guzmán M. A. (2012). Immune responses associated with resistance to haemonchosis in sheep. BioMed. Res. Int. 2013, e162158. doi: 10.1155/2013/162158
Bannerman D. D., Paape M. J., Hare W. R., and Sohn E. J. (2003). Increased levels of LPSBinding protein in bovine blood and milk following bacterial lipopolysaccharide challenge. J. Dairy Sci. 86, 3128–3137. doi: 10.3168/jds.S0022-0302(03)73914-9
Bentley K. L., Greiner S., Kelley E., Ogunade I., and Bowdridge S. A. (2025). Evaluating differential lipopolysaccharide-induced behavioral, immune and plasma metabolome responses of St. Croix and Suffolk sheep. Small Rumin. Res. 245. doi: 10.1016/j.smallrumres.2025.107461
Bentley K. L., Weaver A. R., Wright D. L., Greiner S. P., and Bowdridge S. A. (2023). Postweaning fecal egg count estimated breeding value is associated with greater antibody production after clostridial vaccination in Katahdin lambs. Small Rumin. Res. 229, 107128. doi: 10.1016/j.smallrumres.2023.107128
Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Cai B., Wu J., Yu X., Su X., and Wang R.-F. (2017). FOSL1 inhibits type I interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. mBio 8. doi: 10.1128/mbio.02161-16
Colditz I. G., Watson D. L., Gray G. D., and Eady S. J. (1996). Some relationships between age, immune responsiveness and resistance to parasites in ruminants. Int. J. Parasitol. 26, 869–877. doi: 10.1016/S0020-7519(96)80058-0
Daigo K., Inforzato A., Barajon I., Garlanda C., Bottazzi B., Meri S., et al. (2016). Pentraxins in the activation and regulation of innate immunity. Immunol. Rev. 274, 202–217. doi: 10.1111/imr.12476
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
dos Santos P. F. and Mansur D. S. (2017). Beyond ISGlylation: functions of free intracellular and extracellular ISG15. J. Interferon Cytokine Res. 37, 246–253. doi: 10.1089/jir.2016.0103
Duffield J. S. (2002). The inflammatory macrophage: a story of Jekyll and Hyde. Clin. Sci. 104, 27–38. doi: 10.1042/cs1040027
Fitzgerald D. C., Meade K. G., McEvoy A. N., Lillis L., Murphy E. P., MacHugh D. E., et al. (2007). Tumour necrosis factor-α (TNF-α) increases nuclear factor κB (NFκB) activity in and interleukin-8 (IL-8) release from bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 116, 59–68. doi: 10.1016/j.vetimm.2006.12.008
Graham M. R., Bowdridge E. C., Bowdridge S. A., Holásková I., Elsasser T. H., and Dailey R. A. (2018). Effect of lipopolysaccharide-induced immune responses on pregnancy loss in ewes. Open J. Anim. Sci. 8, 421–431. doi: 10.4236/ojas.2018.84031
Hadfield J. M., Bowdridge E. C., Holásková I., Elsasser T. H., and Dailey R. A. (2018). Breed-specific differences in the immune response to lipopolysaccharide in ewes. J. Anim. Sci. 96, 4220–4228. doi: 10.1093/jas/sky288
Hogg N. and Clive Landis R. (1993). Adhesion molecules in cell interactions. Curr. Opin. Immunol. 5, 383–390. doi: 10.1016/0952-7915(93)90057-Y
Jacobs J. R., Middleton D., Greiner S. P., and Bowdridge S. A. (2020). RNA-Sequencing of ovine PBMC after exposure to Haemonchus contortus larval antigen. Parasite Immunol. 42, e12697. doi: 10.1111/pim.12697
Jacobs J. R., Sommers K. N., Zajac A. M., Notter D. R., and Bowdridge S. A. (2016). Early IL-4 gene expression in abomasum is associated with resistance to Haemonchus contortus in hair and wool sheep breeds. Parasite Immunol. 38, 333–339. doi: 10.1111/pim.12321
Johnson S. R., Bentley K., Bowdridge S., and Ogunade I. M. (2024). Lipopolysaccharideinduced alterations in the liver metabolome of St. Croix and Suffolk sheep. Front. Anim. Sci. 5. doi: 10.3389/fanim.2024.1407533
Kuzmich N. N., Sivak K. V., Chubarev V. N., Porozov Y. B., Savateeva-Lyubimova T. N., and Peri F. (2017). TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines 5, 34. doi: 10.3390/vaccines5040034
Leymaster K. A. (1991). Straightbred comparison of a composite population and the Suffolk breed for performance traits of sheep. J. Anim. Sci. 69, 993–999. doi: 10.2527/1991.693993x
Lynn W. A. and Golenbock D. T. (1992). Lipopolysaccharide antagonists. Immunol. Today 13, 271–276. doi: 10.1016/0167-5699(92)90009-V
MacKINNON K. M., Zajac A. M., Kooyman F. N. J., and Notter D. R. (2010). Differences in immune parameters are associated with resistance to Haemonchus contortus in Caribbean hair sheep. Parasite Immunol. 32, 484–493. doi: 10.1111/j.1365-3024.2010.01211.x
Martin S. A. (1998). Manipulation of ruminal fermentation with organic acids: a review. J. Anim. Sci. 76, 3123–3132. doi: 10.2527/1998.76123123x
Middleton D., Hanlon K., Greiner S. P., and Bowdridge S. A. (2024). Variants of NLRP3 protein in haemonchus contortus infected sheep: impact on immune cell responsiveness to LPS in vitro. Parasite Immunol. 46, e13054. doi: 10.1111/pim.13054
Miller J. E. and Horohov D. W. (2006). Immunological aspects of nematode parasite control in sheep1. J. Anim. Sci. 84, E124–E132. doi: 10.2527/2006.8413_supplE124x
Muñoz-Guzmán M. A., Cuéllar-Ordaz J. A., Valdivia-Anda A. G., Buendía-Jiménez J. A., and Alba-Hurtado F. (2006). Correlation of parasitological and immunological parameters in sheep with high and low resistance to haemonchosis. Can. J. Anim. Sci. 86, 363–371. doi: 10.4141/A06-010
Nikpour H. (2017). Influence of Sex and Blood Collection Method on Innate Immune Response following Lipopolysaccharide Administration in Sheep [M.S.]. United States -- West Virginia: West Virginia University, United States. Available online at: https://www.proquest.com/docview/1901441407/abstract/9C5AC071520A4A98PQ/1.
Page M. J., Kell D. B., and Pretorius E. (2022). The role of lipopolysaccharide-induced cell signalling in chronic inflammation. Chronic Stress 6, 24705470221076390. doi: 10.1177/24705470221076390
Qiagen CLC Genomics Workbench. Available online at: https://digitalinsights.qiagen.com (Accessed December 20, 2024).
Redza-Dutordoir M. and Averill-Bates D. A. (2016). Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta BBA - Mol. Cell Res. 1863, 2977–2992. doi: 10.1016/j.bbamcr.2016.09.012
Salleh M. S., Mazzoni G., Höglund J. K., Olijhoek D. W., Lund P., Løvendahl P., et al. (2017). RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high- and low-residual feed intake in Nordic dairy cattle. BMC Genomics 18, 258. doi: 10.1186/s12864-017-3622-9
Sayers G. and Sweeney T. (2005). Gastrointestinal nematode infection in sheep – a review of the alternatives to anthelmintics in parasite control. Anim. Health Res. Rev. 6, 159–171. doi: 10.1079/AHR2005108
Schneider W. M., Chevillotte M. D., and Rice C. M. (2014). Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schumann R. R. (1992). Function of lipopolysaccharide (LPS)-binding protein (LBP) and CD14, the receptor for LPS/LBP complexes: a short review. Res. Immunol. 143, 11–15. doi: 10.1016/0923-2494(92)80074-U
Shimada H., Hasegawa N., Koh H., Tasaka S., Shimizu M., Yamada W., et al. (2006). Effects of initial passage of endotoxin through the liver on the extent of acute lung injury in A rat model. Shock 26, 311. doi: 10.1097/01.shk.0000224960.17274.6f
Singh S. P., Zhang H. H., Foley J. F., Hedrick M. N., and Farber J. M. (2008). Human T cells that are able to produce IL-17 express the chemokine receptor CCR61. J. Immunol. 180, 214–221. doi: 10.4049/jimmunol.180.1.214
Tang D., Cheng M., Huang X., Zhang G., Zeng L., Zhang G., et al. (2023). SRplot: A free online platform for data visualization and graphing. PLOS ONE 18, e0294236. doi: 10.1371/journal.pone.0294236
Taylor M. A. (2012). Emerging parasitic diseases of sheep. Vet. Parasitol. 189, 2–7. doi: 10.1016/j.vetpar.2012.03.027
Yang H., Yang Y.-L., Li G.-Q., Yu Q., and Yang J. (2021). Identifications of immuneresponsive genes for adaptative traits by comparative transcriptome analysis of spleen tissue from Kazakh and Suffolk sheep. Sci. Rep. 11, 3157. doi: 10.1038/s41598-02182878-x
Keywords: transcriptome, LPS, inflammatory, DEGs, response
Citation: Johnson SR, Bentley KL, Bowdridge SA and Ogunade IM (2025) Lipopolysaccharide-induced changes in the hepatic transcriptome revealed breed-specific response in St. Croix and Suffolk sheep. Front. Anim. Sci. 6:1565253. doi: 10.3389/fanim.2025.1565253
Received: 22 January 2025; Accepted: 13 June 2025;
Published: 08 July 2025.
Edited by:
Mohan Mondal, Eastern Regional Station, IndiaReviewed by:
Gerardo Ramírez-Rico, National Autonomous University of Mexico, MexicoHua Yang, Xinjiang Academy of Agricultural and Reclamation Sciences (XAARS), China
Esma Gamze Aksel, Erciyes University, Türkiye
Copyright © 2025 Johnson, Bentley, Bowdridge and Ogunade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibukun M. Ogunade, SWJ1a3VuLm9ndW5hZGVAbWFpbC53dnUuZWR1; Scott A. Bowdridge, c2NvdHQuYm93ZHJpZGdlQG1haWwud3Z1LmVkdQ==
†Present address: Kelsey L. Bentley, Animal Sciences and Industry Department, Kansas State University, Manhattan, KS, United States
 Samanthia R. Johnson
Samanthia R. Johnson Kelsey L. Bentley
Kelsey L. Bentley Scott A. Bowdridge
Scott A. Bowdridge Ibukun M. Ogunade
Ibukun M. Ogunade