- 1College of Agronomy and Life Sciences, Zhaotong University, Zhaotong, China
- 2College of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
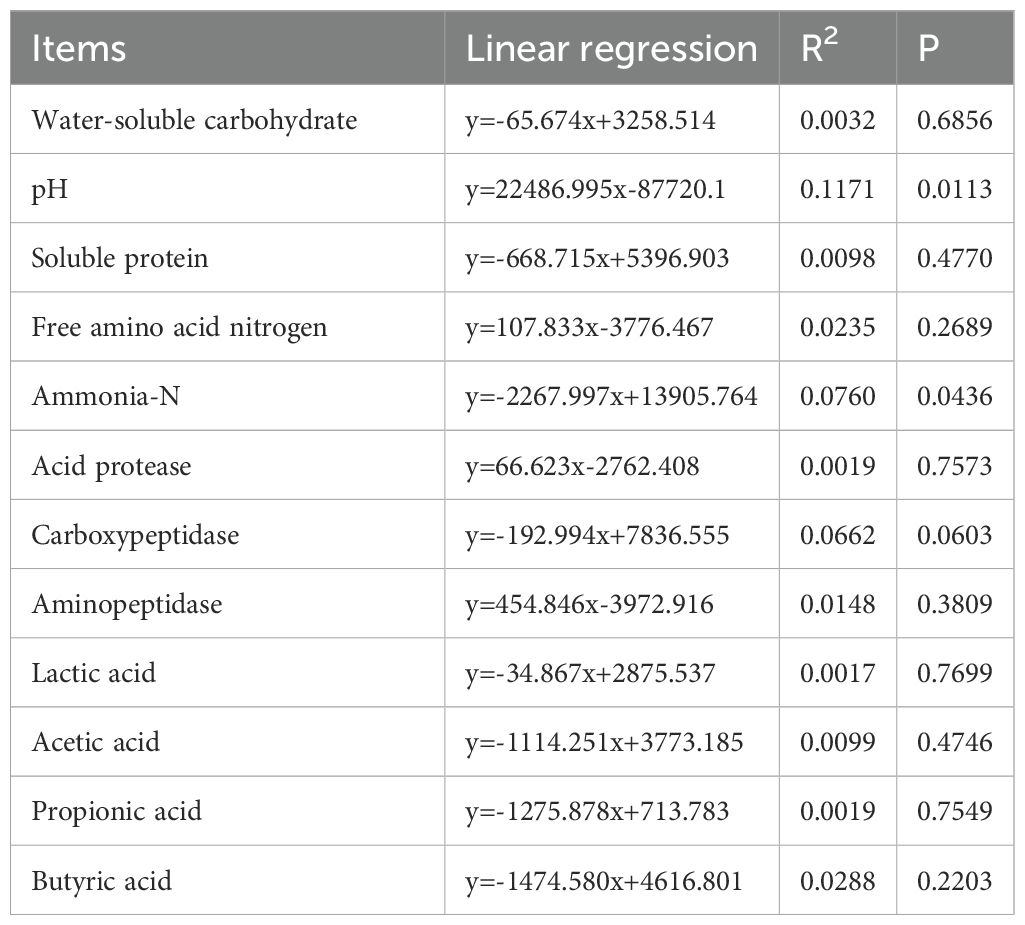
Leaf spot is a widespread disease that poses a severe challenge to the forage industry. Lactic acid bacteria (LAB) additives inhibit the growth of harmful bacteria through mechanisms such as competitive exclusion and the production of antimicrobial substances, thereby improving the fermentation environment. The aim of this study was to explore the effects of LAB on the regulation of nitrogen compounds and related bacterial communities in silage infected with leaf spot disease. The experiments included two forage types (Italian ryegrass [IR] and oat), three additives (deionized water [control, CK], Lactobacillus plantarum YM3, and Lactobacillus rhamnosus HT1), and four ensiling periods (0, 15, 30, and 60 d). Ammonia-N concentration and LAB numbers increased with increasing ensiling time. The ammonia-N concentration of IR was significantly lower than that of oat (P < 0.05). The aminopeptidase activity of CK silage (10.5 units h-1 g-1 dry matter [DM]) was higher than that of YM3 (7.19 units h-1 g-1 DM) and HT1 silages (7.07 units h-1 g-1 DM) (P < 0.05). The YM3 silage (94.5 g kg-1 DM) had a higher lactic acid concentration than that of CK (69.5 g kg-1 DM) and HT1 silages (83.3 g kg-1 DM) (P < 0.05). Among the 12 environmental factors analyzed, pH (R2 = 0.1171, P = 0.0113) and ammonia-N (R2 = 0.076, P = 0.0436) had the greatest influence on bacterial relative abundance. The normalized shuffle test index demonstrated a significant impact on microbial stability, with differences observed under varying treatment conditions (IR > oat, CK > HT1 > YM3, and ES60 > ES15 > ES30 (P < 0.05)). In conclusion, oat silage exhibited better fermentation quality and bacterial stability than that of IR. Extending the ensiling time significantly improved the overall fermentation quality of silage. Furthermore, the application of LAB additives, particularly YM3, significantly enhanced the fermentation quality and bacterial stability of silage from forage infected with leaf spot disease.
Introduction
Leaf spot is a widespread disease that poses a substantial challenge to the forage industry. It leads to a significant reduction in forage yield and severely impairs the nutritional value and quality of forage (Johnson et al., 2023; Lan et al., 2024; Qian et al., 2024). Infections by Leptosphaeria avenaria and Parastagonospora nodorum reduce forage quality, compromising its nutritional value (Qian et al., 2024). Agronomic management measures such as rational fertilization (Grewal and Williams, 2002) and scientific pesticide application (Atri et al., 2022) have been implemented to prevent and control leaf spot with some success. However, forage producers still lack a deep understanding of how to further enhance the utilization potential of these diseased forages, improve production outcomes, and evaluate the practical effectiveness of these improvement measures when they fail to promptly treat the forages affected by leaf spot. Recent studies have highlighted the pivotal role of lactic acid bacteria (LAB) additives in forage silage (Wang et al., 2021; Zhou et al., 2024; Zong et al., 2023). These additives inhibit the growth of harmful bacteria through mechanisms such as competitive exclusion and the production of antimicrobial substances, thereby improving the fermentation environment, reducing protein degradation, and enhancing the nutritional value of the silage (Zhang et al., 2023). For example, LAB inhibit the quorum sensing signaling of Pseudomonas aeruginosa, reducing the mRNA expression levels of the lasI and rhlI genes, thereby decreasing the synthesis of N-acyl homoserine lactone and ultimately suppressing its virulence (Rana et al., 2020). However, under conditions where forage plants are infected with diseases like leaf spot, the specific mechanisms and effects of LAB additives on silage quality remain poorly understood. Therefore, exploring the potential application and effects of LAB additives in the silage process of forage infected with leaf spot diseases is crucial for ensuring feed safety and enhancing the efficiency of animal husbandry production.
During silage fermentation, protein degradation is primarily driven by plant and microbial proteases, resulting in the production of a large amount of non-protein nitrogen compounds, including peptide nitrogen, free amino acid nitrogen (FAA-N), and ammonia-N (Huang et al., 2023; Xu et al., 2021, 2023). Excessive protein degradation in silage reduces nitrogen utilization by animals, resulting in economic losses. As the pH of silage decreases, some peptidases are inactivated, and some microorganisms metabolize and produce proteases that further promote protein degradation (Fijałkowska et al., 2015). Factors like pH (Li et al., 2018) and temperature (Liu et al., 2016) directly influence protein degradation in silage, and microorganisms such as Pantoea (Jiang et al., 2023), Weissella (Zhang et al., 2023), and Pseudomonas (Li et al., 2024) have a substantial effect on silage pH (Der Bedrosian and Kung, 2019). However, the impact of microbial metabolites on silage protease activity remains unclear.
Previous studies have primarily focused on the silage fermentation characteristics of healthy forage, while this study systematically reveals the fermentation dynamics and quality changes of leaf spot disease-infected forage during the ensiling process, filling the gap in research on diseased silage. Additionally, this study provides an in-depth analysis of the regulatory mechanisms of LAB additives on the microbial community structure and stability of diseased silage, clarifying the impact of key environmental factors on microbial metabolism and nitrogen compound degradation. Therefore, the results of this study not only contribute to improving the utilization efficiency of diseased forage but also provide a theoretical basis for the optimization and regulation of nitrogen compounds in diseased silage, deepen the understanding of the dynamic succession mechanisms of microbial communities, and offer scientific support for enhancing the quality and sustainable use of diseased silage. In this study, we aimed to investigate the regulatory mechanisms of LAB additives on nitrogen compounds and related bacterial communities during the ensiling process of oat (Avena sativa) and Italian ryegrass (Lolium multiflorum, IR) infected with leaf spot disease. The specific objectives include: 1) investigating the inhibitory effect of LAB additives on nitrogen compounds and related bacterial communities during ensiling; and 2) analyzing the impact of LAB additives on the fermentation quality of silage, with a focus on inhibiting protein degradation.
Materials and methods
Experimental site
The experimental site was set up at the experimental field of Zhaotong University (Zhaoyang District, Zhaotong City, Yunnan Province; 27°36′N, 103°74′E; 1985 m altitude). No herbicides have been applied at the site. According to data from the Zhaotong Meteorological Bureau, the average annual temperature over the past 20 years was 12.3°C and the annual total rainfall was 682 mm. Every year, the locally grown forage in this area is affected by various leaf diseases, with leaf spot disease being particularly prevalent.
Raw materials and silage preparation
Forage with more than 30% leaf spot was selected for ensiling. The forage was harvested during the late flowering and early milk stages for IR and oat, respectively. Fresh matter (FM) was transported to the laboratory, chopped into 20–30 mm lengths, and thoroughly mixed. Subsequently, the materials were inoculated with Lactobacillus plantarum YM3 and Lactobacillus rhamnosus HT1 at a concentration of 1.0 × 105 cfu g-1 FM. The control (CK) was sprayed with an equal amount of deionized water. Each treatment (including raw materials) was prepared in triplicate. All samples were placed in bags (polyethylene bag, 300 by 400 by 0.2 mm, Mingkang Packing, Zhongshan, China), which were placed in a laboratory cabinet (22°C, dark environment) for 0, 15, 30, and 60 days (ES0d, ES15d, ES30d, and ES60d groups, respectively). The two LAB strains were isolated from the forage processing laboratory of South China Agricultural University and have been previously used in studies to improve the fermentation quality of silage (Liu et al., 2018b; Xie et al., 2012). Next, 300 g of the treated materials were placed into polyethylene bags (300 mm × 400 mm × 0.2 mm, Mingkang Packing Co. Ltd., Zhongshan, China), which were degassed and sealed using a vacuum sealing machine (Sinbo Vacuum Sealer, Hong Tai Home Electrical Appliance Co., Ltd., Hong Kong, China).
Chemical and microbial analysis
The LAB, aerobic bacteria (AB), yeasts and molds were counted on de Man, Rogosa, and Sharpe agar, nutrient agar and potato dextrose agar, respectively (Tang et al., 2023). The LAB were cultured under anaerobic conditions at 37°C for 2 d. AB, yeasts, and molds were cultured under aerobic conditions at 37°C for 3 d. The dry matter (DM) concentration was determined by drying samples at 70°C for 48 h in an oven with forced air circulation, followed by grinding the dried material to pass a 1.0 mm screen using a knife mill (Blast drying oven 9003A, Shandong Jinan Haineng Instrument Co., Ltd., China Co., Ltd). Crude ash concentration was measured by incinerating samples in a muffle furnace at 550°C for 5 h. Buffering capacity was assessed using acid-base titration (Playne and McDonald, 1966). Water-soluble carbohydrate (WSC) concentration was determined using the sulfuric acid-anthrone method (use sucrose to make a standard curve) (Murphy, 1958).
Furthermore, the ammonia-N concentration was measured using phenol method after determining the pH of the silage filtrate (Broderick and Kang, 1980). The pH of the filtrate (10 g of FM added to 90 mL distilled water) was determined using a pH meter (LE438 pH meter, Mettler Toledo, Shanghai, China). The soluble protein concentration was determined using the Coomassie brilliant blue method (Bradford, 1976). The supernatant was used to determine the FAA-N concentration (Broderick and Kang, 1980).
Moreover, acid protease, aminopeptidase, and carboxypeptidase activities were analyzed according to the method recommended by Guo et al. (2007). After bags were opened, 20 g subsample of each silo was homogenized with 100 mL of 0.1 M sodium phosphate buffer (pre-cooling, pH 6.0 with 5 mM hyposulfite) for 1 min. The homogenates were filtered through 4 layers of cheesecloth and centrifuged at 8,000 × g for 10 min at 4°C. The supernatant samples were stored at −80°C for analysis of protease activities. Acid proteinase activity was measured by the methods of Peterson and Huffaker (1975). Aminopeptidase and carboxypeptidase activities were determined according to the method of Feller et al. (1977). The concentrations of organic acids (lactic acid, acetic acid, propionic acid, and butyric acid) were determined by high-performance liquid chromatography using a Sodex RS Pak KC-811 column (Showa Denko K.K., Kawasaki, Japan), a diode array detector set at 210 nm (serial presence detect-20A, Shimadzu Co., Ltd, Kyoto, Japan), and an eluent of 3 mmol L-1 HClO4 (flow rate: 1.0 mL min-1; temperature: 60°C).
Bacterial community analysis
Total microbial community DNA was extracted using an EZNAPlant DNA kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. DNA was extracted using 1% agarose gel electrophoresis, and DNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer. To address the common issue of sequence similarity between plant endophytic bacteria and chloroplast or mitochondrial rRNA coding sequences, a nested polymerase chain reaction (PCR) assay was used (Guo et al., 2017). The V5–V7 regions of the 16S rRNA genes of the strains were amplified with primers 779F (5′AACMGGATTAGATACCCKG-3′) and 1193R (5′-ACGTCATCCCCACCTTCC-3’) using the thermocycler PCR system (GeneAmp 9700, ABI, USA). PCR products from each same sample were mixed and resolved on a 2% agarose gel. The recovered DNA fragments were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions, and their concentrations were quantified using a Quantus Fluorometer (Axygen Biosciences, Union City, CA, USA). Library construction of the purified PCR products was performed using a NEXTFLEX Rapid DNA-Sequencing Kit. Sequencing was performed using Illumina’s PE300 platform. Quality control and sequence assembly were performed using Fastp software (OriginPro®2021b, OriginLab Corp., WA, USA).
Statistical analysis
Analysis of variance was used to assess the effects of ensiling time, crop type, and additives on fermentation quality, microbial community, and protein degradation. The mean values of ensiling time, crop type, and additives were calculated for fermentation quality, microbial community, and protein degradation, respectively using SPSS software (version 17.0 for Windows; SPSS Inc., Chicago, IL, USA). The statistical model used was (Li et al., 2018): Y=μ + Tj + ϵij, where Y represents the response variable, μ is the overall mean, Tj represents the effect of treatment, and ϵij is the residual error.
Analyses of fermentation quality, microbial community, and protein degradation across different ensiling times, crop types, and additives were performed using the principal component analysis (PCA) function (OriginPro®2021b software, OriginLab Corp., WA, USA). A normalized shuffle test (NST) index was used to evaluate the stability of microbial communities.
Results
Characteristics of raw materials
The chemical composition and microbial population of the FM before ensiling are listed in Table 1. The DM concentration was 321 g kg-1 (IR) and 293 g kg-1 (oat) (P < 0.05). However, the neutral detergent fiber and acid detergent fiber concentrations of oat were higher than those of IR (P < 0.05). The carboxypeptidase activity in IR (26.4 units h-1 g-1 DM) was lower than that of oat (10.2 units h-1 g-1 DM) (P < 0.05), while the aminopeptidase activity was higher (12.4 vs. 19.1 units h-1 g-1 DM) (P < 0.05).
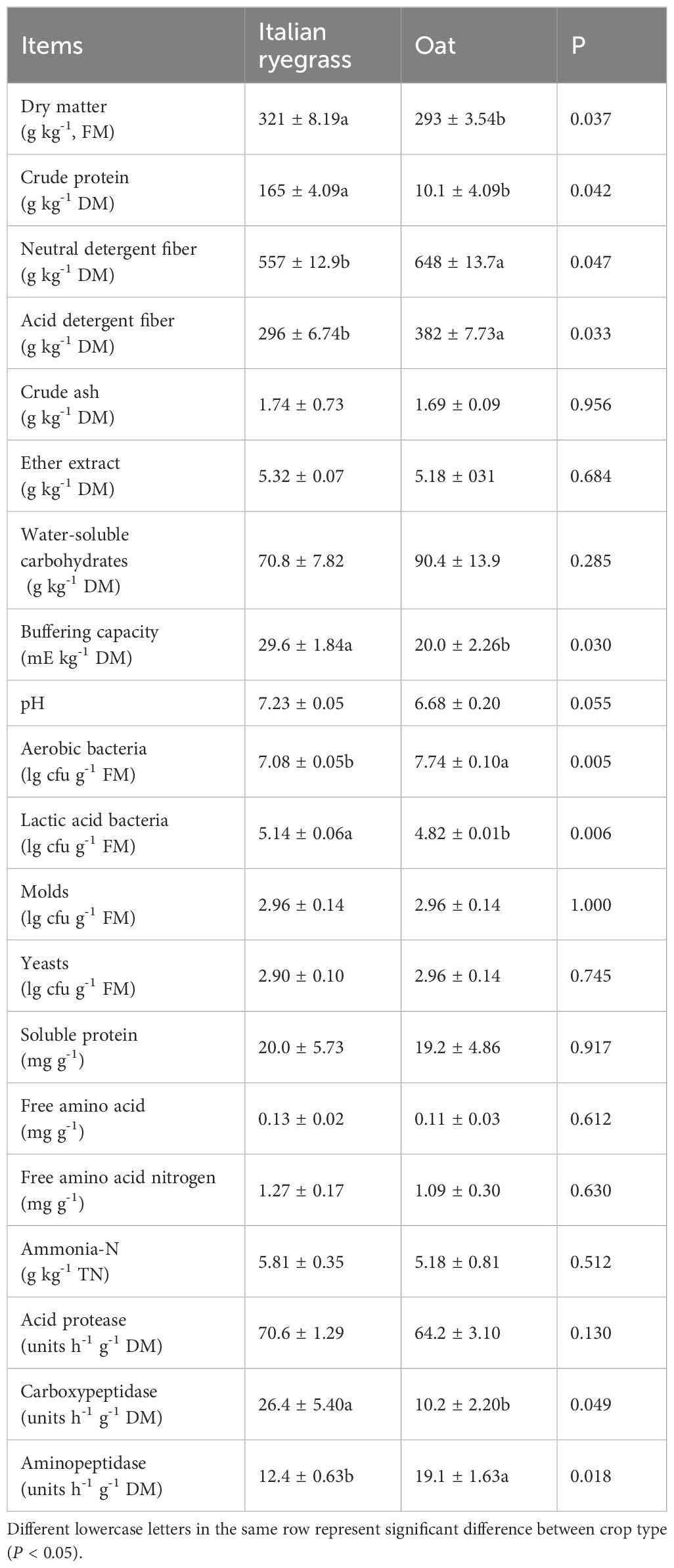
Table 1. Chemical compositions, buffering capacity, epiphytic microbes and protease activities of Italian ryegrass and oat.
Fermentation quality, nitrogen compounds, and protease activities of silage
As shown in Table 2, WSC and ammonia-N concentrations were affected by two sources of interactive variation: ensiling time × crop type and ensiling time × additives. Ammonia-N concentration and LAB numbers generally increased with increasing ensiling time. The soluble protein (7.25 mg g-1) and ammonia-N (6.28 g kg-1 TN) concentrations in IR were lower than those in oat (8.73 mg g-1 and 5.34 g kg-1 TN, respectively) (P < 0.05). The YM3 silage had a higher FAA-N concentration (4.48 mg kg-1) than the CK (1.96 mg kg-1) and HT1 (2.41 mg kg-1) silages (P < 0.05, Table 2).
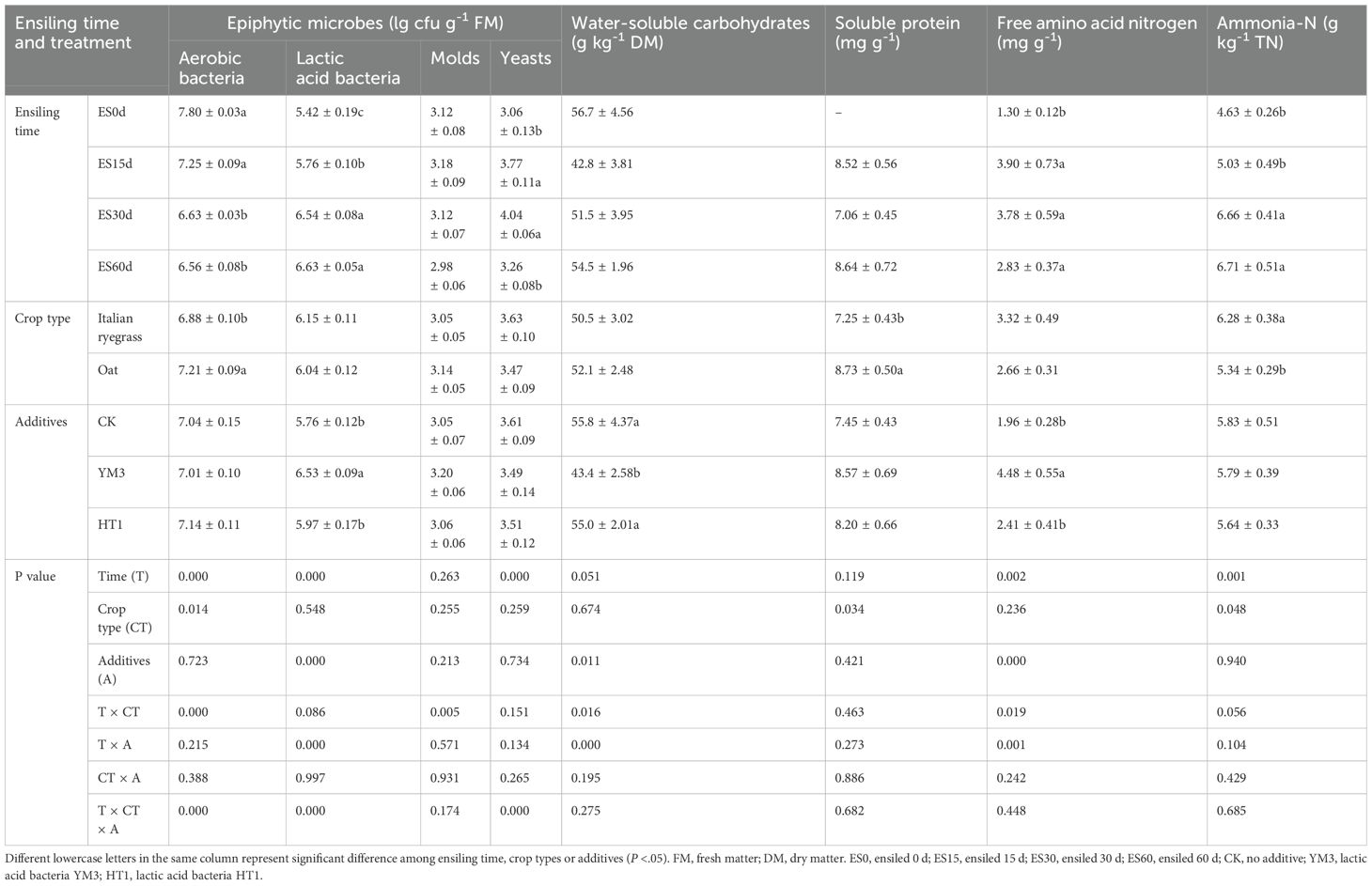
Table 2. Epiphytic microbes and nitrogen compounds of different additives, crop type and ensiling time treatments.
As shown in Table 3, The activities of aminopeptidase (47.5 units h-1 g-1 DM) and carboxypeptidase (11.4 units h -1 g-1 DM) in the ES15d group were significantly higher than those in the ES0d group (20.5 and 6.77 units h-1 g-1 DM, respectively) (P < 0.05). The pH decreased over time (P < 0.05), indicating progressive acidification. Carboxypeptidase and aminopeptidase activity were higher in IR (41.7 and 9.89 h-1 g-1 DM) than in oat (30.7 and 6.92 h-1 g-1 DM, P < 0.05), suggesting greater protein degradation in Italian ryegrass. The aminopeptidase activity of CK silage (10.5 units h-1 g-1 DM) was higher than that of YM3 (7.19 units h-1 g-1 DM) and HT1 silages (7.07 units h-1 g-1 DM) (P < 0.05). The YM3 (94.5 g kg-1 DM) and HT1 (83.3 g kg-1 DM) silages had a higher lactic acid concentration than that of CK (69.5 g kg-1 DM) and HT1 silages (P < 0.05). Ensiling time, crop type, additives, and their interaction had no significant effects on the propionic and butyric acids concentrations of the silage (P > 0.05, Table 3).
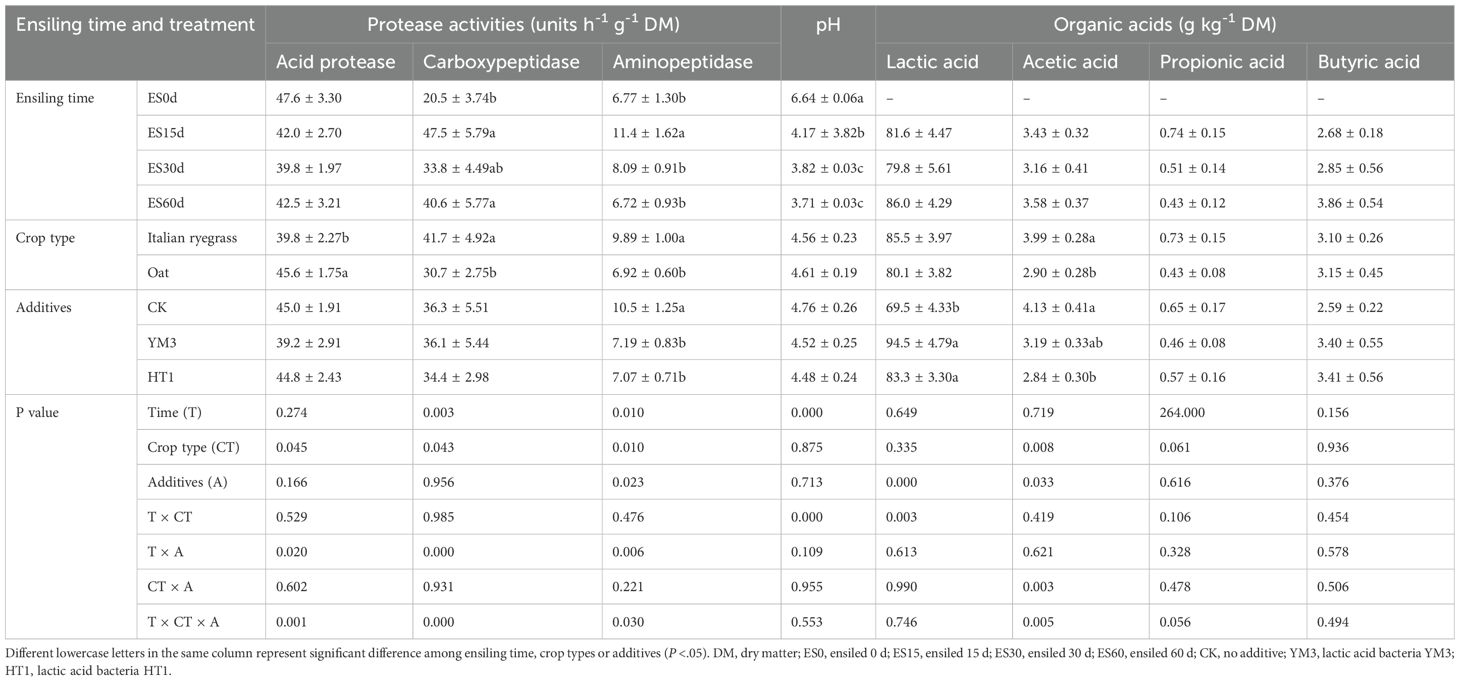
Table 3. Protease activities and organic acids of different additives, crop type and ensiling time treatments.
Bacterial community and influencing factors of silage
At the genus level, the relative abundances of Lactobacillus and Pseudomonas were predominant and remained consistent across different crop types and additives (Figure 1). During ensiling, the overlap of operational taxonomic units (OTUs) in different crop types and additives was high. A total of 36, 43, and 85 OTUs were found in the ES15d, ES30d, and ES60d silage sample groups, respectively. Furthermore, 73, 82, and 76 OTUs were found in the CK, YM3, and HT1 silages, respectively (Supplementary Figure S1). To explore the variation in bacterial community composition, a scatter plot was generated and subjected to PCA, revealing that the relative abundance of bacteria varied with ensiling duration, crop type, and additives (Figure 2). The first two principal components (PC1 and PC2) accounted for a cumulative variance of 73.8%. Notably, the IR silage samples did not differentiate from oat silage samples, and the CK samples did not distinctly differentiate from YM3 and HT1 silages. However, a significant differentiation was observed within the 95% confidence intervals between ES60 and both ES30 and ES15 samples (Figure 2).
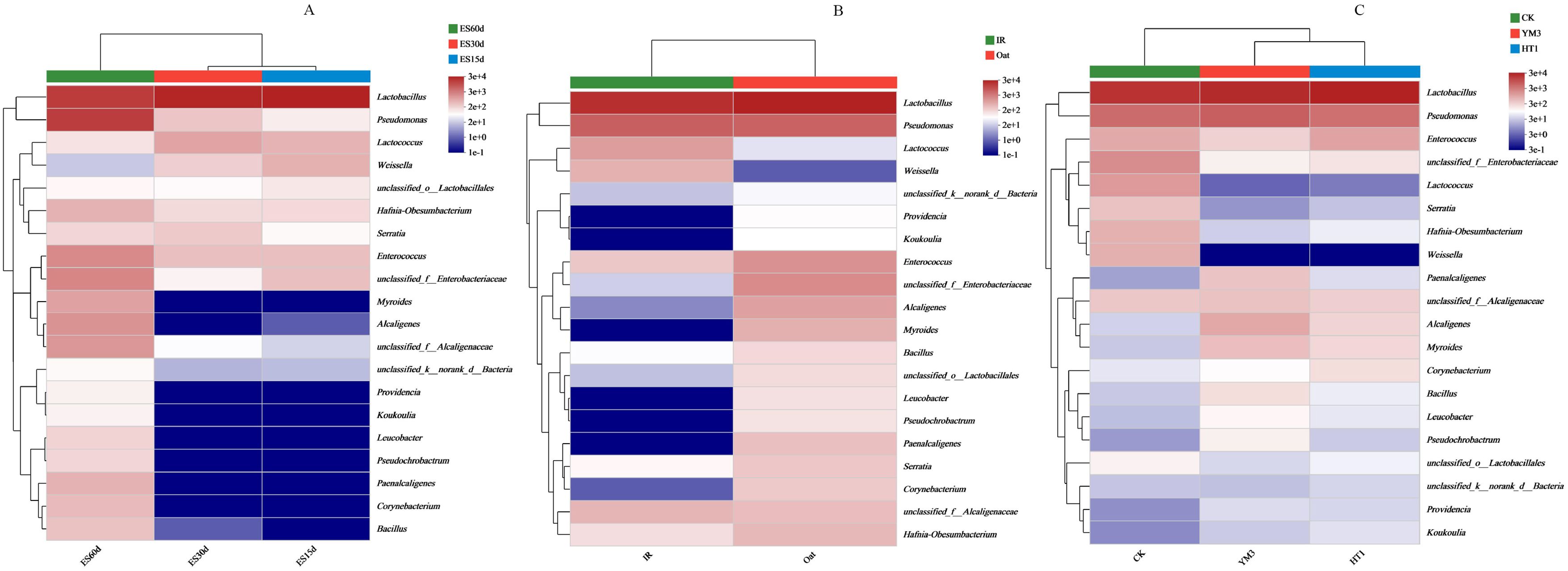
Figure 1. Community heatmap of (20 most abundant) silage bacterial genus under different ensiling time (A), crop type (B) or additives (C). ES15d, ensiled 15 d; ES30d, ensiled 30 d; ES60d, ensiled 60 d; CK, no additive; YM3, lactic acid bacteria YM3; HT1, lactic acid bacteria HT1; IR, Italian ryegrass.
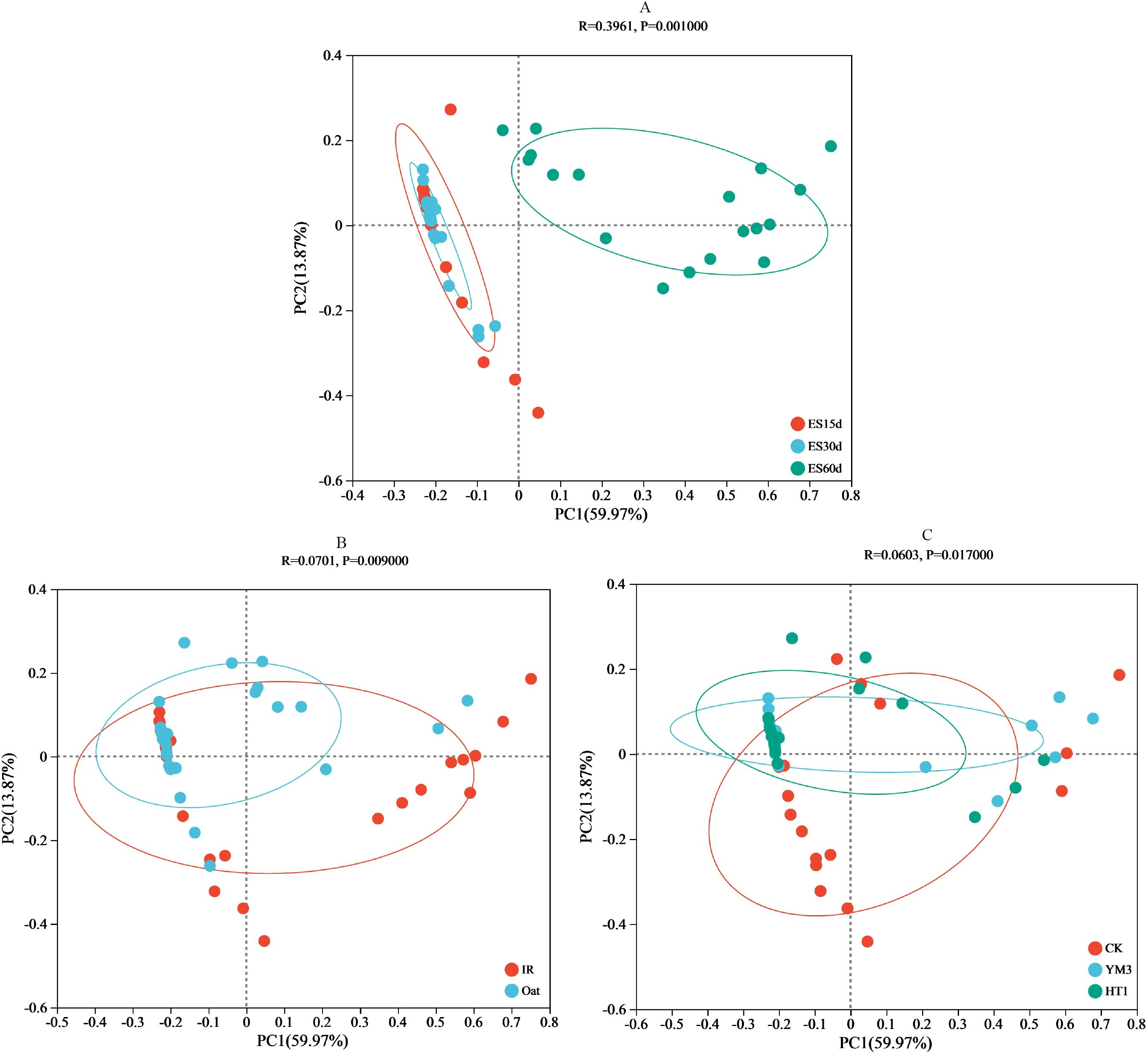
Figure 2. Principal coordinate analysis plot of silage microbial composition at different ensiling time (A), crop type (B) or additives (C). Note: The first axis accounts for 59.97% of the total variance and the second for 13.87%. The original attributes, with their vectors intersecting at (0, 0), are also inserted. The length of each attribute vector is proportional to its contribution to the principal component axis. The ellipse indicates 95% confidence. ES15d, ensiled 15 d; ES30d, ensiled 30 d; ES60d, ensiled 60 d; CK, no additive; YM3, lactic acid bacteria YM3; HT1, lactic acid bacteria HT1; IR, Italian ryegrass.
Environmental factors, such as pH and acid protease, carboxypeptidase, and aminopeptidase activities, significantly influenced the microbial communities in the silage (Figure 3). Butyric acid concentration was positively correlated with the relative abundances of Myroides, Alcaligenes, Pseudochrobactrum, Leucobacter, and Providencia (P < 0.01). Additionally, the relative abundance of Weissella was positively correlated with pH and carboxypeptidase and aminopeptidase activities, as well as acetic and propionic acid concentrations (P < 0.05). Acid protease activity was positively associated with the relative abundances of Paenalcaligenes, Pseudochrobactrum, Leucobacter, and Bacillus (P < 0.05).
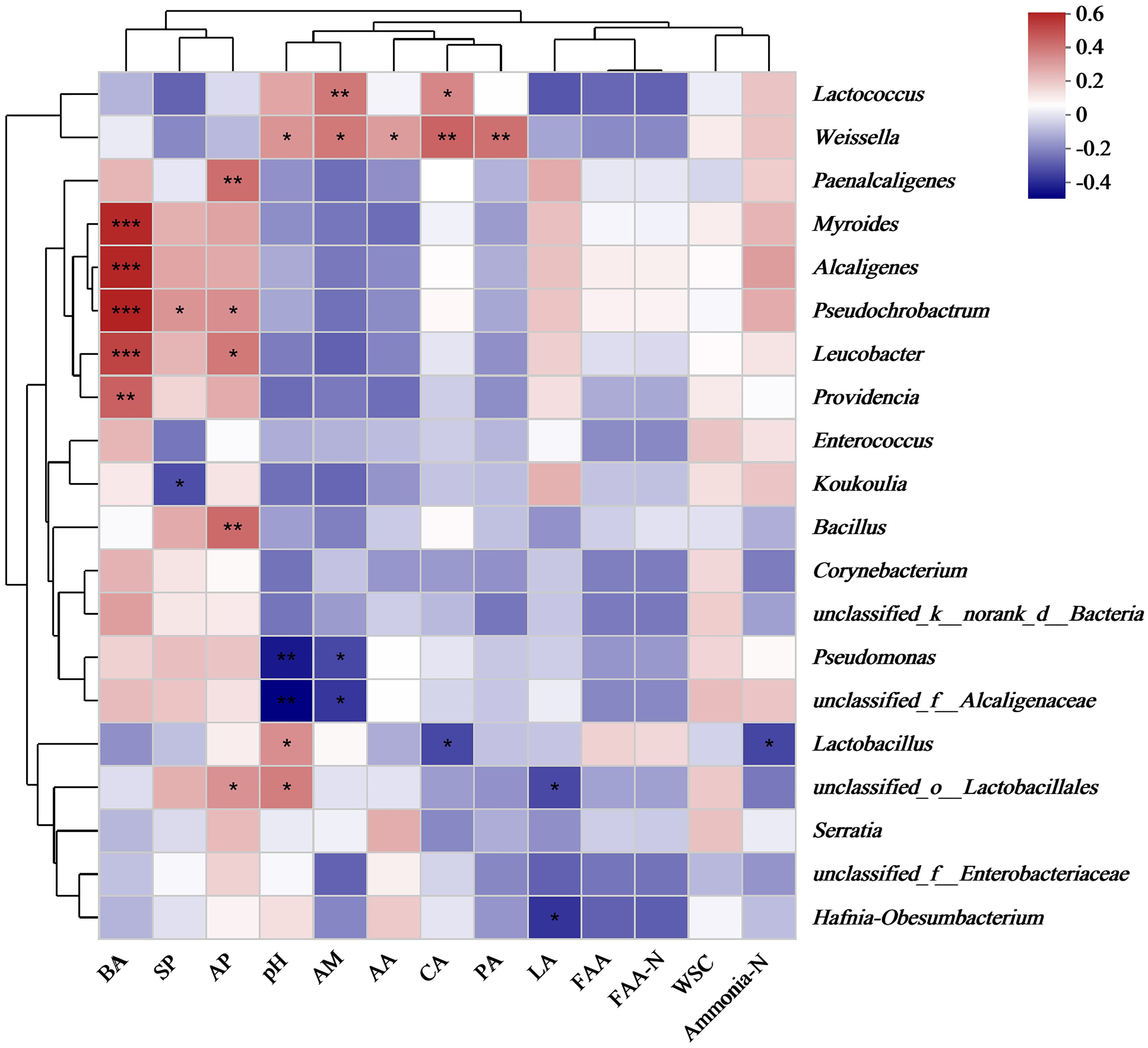
Figure 3. Relationship between chemical properties and bacterial communities of silage. Note: The X-and Y-axes are environmental factors and species, respectively, and correlation R and P values are obtained by calculation. R values are presented in different colors in the figure, P values are marked with * if P < 0.05, and the legend on the right side shows the color intervals of different R values; asterisks indicate significant differences at P < 0.05 (*), P < 0.01 (**), and P < 0.01 (***), respectively. WSC, water-soluble carbohydrate; SP, soluble protein; FAA, free amino acid; FAA-N, free amino acid nitrogen; AP, acid protease; CA, carboxypeptidase; AM, aminopeptidase; LA, lactic acid; AA, acetic acid; PA, propionic acid; BA, butyric acid.
Upon examining the influence of bacterial diversity on environmental factors under various treatments, IR was positively correlated with most environmental factors (Figure 4A), while CK was negatively correlated with most factors (Figure 4B). Among the 12 environmental factors analyzed, pH (R2 = 0.1171, P = 0.0113) and ammonia-N (R2 = 0.076, P = 0.0436) had the greatest influence on bacterial relative abundance, while other factors had minimal impact (R2 < 0.07, P > 0.05) (Table 4). Moreover, the NST index demonstrated a significant impact on microbial stability, revealing under different treatment conditions that: IR > oat, CK > HT1 > YM3, and ES60 > ES15 > ES30 (P < 0.001) (Figure 5).
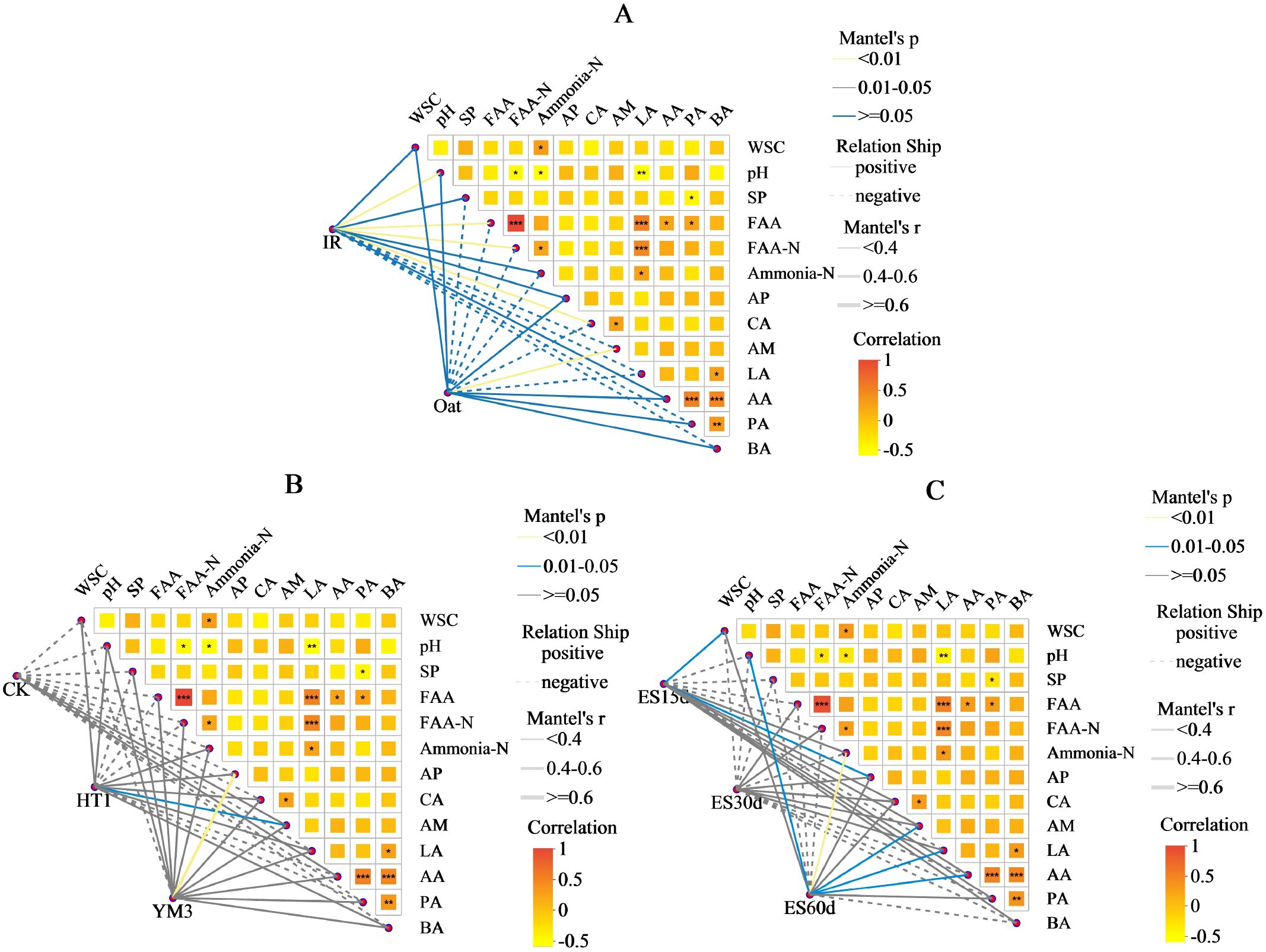
Figure 4. Mantel test heatmap of the effect of among crop type (A), additives (B), and ensiling time (C) or environmental factors on bacteria relative abundance. (A-C) the lines represent the correlation among bacterial communities and environmental factors, while the heatmap represents the correlation among environmental factors. In the heatmap, different colors represent positive or negative correlations, and color depth represents the magnitude of positive or negative correlations. * P<0.05, ** P<0.01, *** P <0.001.
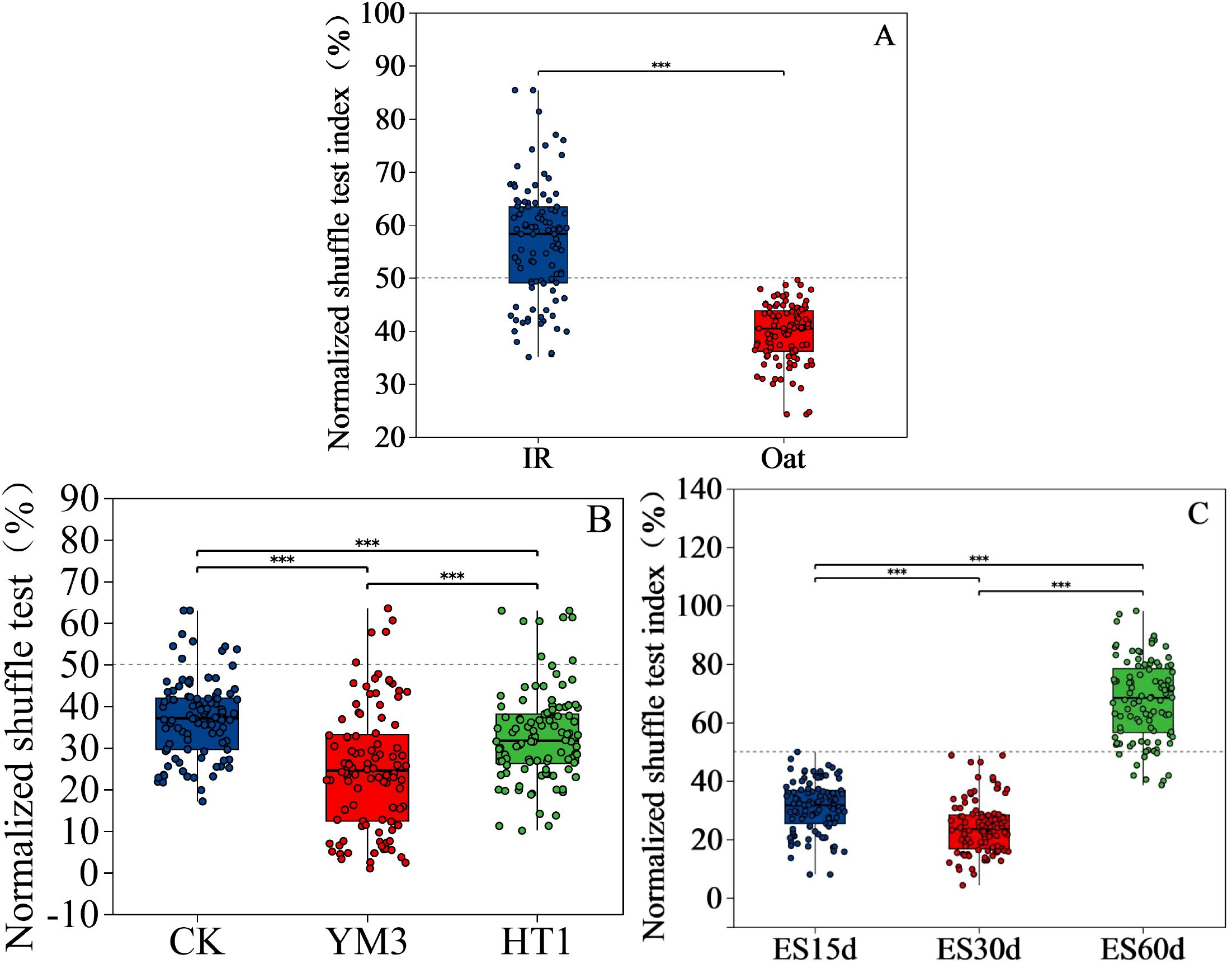
Figure 5. Significance and normalized shuffle test index of bacteria relative abundance analyses of different ensiling time (A), crop type (B), and additives (C). Y-axis indicates the normalized shuffle test index, X-axis indicates the mean relative abundance in different subgroups of the species, and different coloured columns indicate different subgroups; P-values are shown on the rightmost side. The dotted lines were thresholds for deterministic and random divisions, demonstrating significance markers for between-group analysis of variance. *** P <0.001. NST, normalized shuffle test index.
Discussion
Effects of ensiling time, crop type, and additives on the fermentation quality, nitrogen compounds, and protease activities of silage
The WSC concentration and epiphytic LAB numbers in the forage are crucial factors influencing silage fermentation quality. Optimal silage production requires a WSC concentration of 60–70 g kg-1 DM and LAB numbers of at least 105 lo cfu g-1 FM (Liu et al., 2018b; Wilkinson, 2003). Despite the widespread occurrence of leaf spot disease in IR and oat used in this study, the higher WSC concentration and abundant LAB significantly improved the fermentation quality of silage (Tables 1 and 3). The reason was that WSC and free amino acids provided nutrients for the proliferation of LAB, and as time passed, the proportion of LAB increased accordingly. This effect was particularly evident during the silage process, maintaining the same trend despite the influence of different crop types and additives. This study highlights that even under disease pressure, the overall quality of silage can still be effectively improved by regulating key biochemical factors. Moreover, the LAB additives were particularly effective in improving silage quality and inhibiting protease activity, with the most pronounced effect observed after 60 days of ensiling (Table 3). In general, with the prolongation of ensiling time, LAB metabolize and produce large amounts of lactic acid, which gradually inhibited the growth of other microorganisms (especially not acid-intolerant microorganisms) (Liu et al., 2023). In this study, ES60d silage had the highest LAB numbers and lactic acid concentrations (Table 2 and 3). Lactobacillus spp. were the dominant microorganisms (Figure 1), consistent with findings from previous studies (Wang et al., 2019; Zhang et al., 2023). The proteolytic activity in silage involves plant proteases hydrolyzing true proteins into peptide nitrogen and FAA-N, which are further broken down into ammonia-N. This process contributed to the increased ammonia-N concentration observed in ES60d silage. In general, non-protein nitrogen in plants mainly consists of ammonia-N, amines, amides, and free amino acids (Fijałkowska et al., 2015). LAB, which require high levels of free amino acids and peptides, rely on the hydrolysis of proteins into amino acids to thrive and acidify the silage (Kim et al., 2021). This need for amino acids can intensify protein degradation, leading to higher ammonia-N concentrations.
Typically, peptides and free amino acids, which are the hydrolysis products of true proteins, can be deaminated by microbial activity to produce ammonia-N; however, plant proteases also contribute to ammonia-N production. Therefore, both were able to respond to the relative activity of protein hydrolysis and deamidation (Yuan et al., 2017). In this study, ammonia-N concentrations were generally lower than FAA-N concentrations across all treatments and treatment conditions, along with reduced butyric acid concentrations. This suggests that plant proteases were the primary source of ammonia-N. The buffering capacity of IR silage, which was higher than that of oat, resulted in a slower rate of pH decline during ensiling. This slower decline allowed plant proteases to hydrolyze true proteins into FAA-N. Thus, deamination was more active during the pre-silage period. This conclusion was supported by the changes in aminopeptidase activity at different ensiling times (Table 3). Previous studies have shown that protease activity is closely related to pH, with aminopeptidase activity being higher at pH 6.6–7.1 during ensiling, while decreasing as pH decreases (Li et al., 2018; Xu et al., 2023). Moreover, acid protease activity (with azocasein as substrate) was higher at pH 4.5 (Hashmi et al., 2022; Shan et al., 2022). Therefore, acidic protease activity was lower than aminopeptidase activity in the present study (Table 3).
Many studies have shown that a rapid decrease in pH not only promotes lactic acid production but also inhibits protein degradation (Li et al., 2018; Zong et al., 2023). In this study, CK silage showed higher aminopeptidase activity and lower lactic acid concentration than those of YM3 and HT1 silages. In generally, lactic acid is produced in large quantities as LAB colonize the silage, leading to a gradual decrease in pH (Zhao et al., 2024a). However, a low pH can also inhibit microbial activity, including LAB number and protease activity, in the later stages of ensiling (Zhou et al., 2024). Thus, there were no differences in protease activity, organic acid, or nitrogenous compound concentrations between ES30d and ES60d silage. Conversely, the growth of most molds is inhibited under anaerobic and low pH conditions, whereas some yeasts can thrive in these environments (Liu et al., 2018a). In this study, yeast numbers initially increased and then decreased as ensiling time increased, which might be related to strain-specific characteristics; unfortunately, this study did not analyze the fungal species and diversity.
Effects of ensiling time, crop type, and additives on the bacterial community of silage
Lactobacillus, Lactococcus, and Enterococcus spp. are ideal functional bacteria for silage fermentation and have been widely used to improve silage quality (Okoye et al., 2023; Zhao et al., 2024b). In this study, the community relative abundance of Lactobacillus was higher during ensiling (Figure 4), which aligns with previous findings (Wang et al., 2019; Yan et al., 2019). Lactococcus in silage was gradually replaced by the more acid-tolerant Lactobacillus as the ensiling time increased and lactic acid accumulated because suitable DM and WSC concentrations and LAB population of the raw material promotes the rapid fermentation of lactic acid. Therefore, during ensiling (Figure 6), Lactobacillus (relative abundance of 90.9%, 92.5%, and 34.4% in the ES15d, ES30d, and ES60d treatments, respectively; P < 0.001), Pseudomonas (0.37%, 0.86%, and 45.1% in the ES15d, ES30d, and ES60d treatments, respectively; P < 0.001), Enterococcus (1.33%, 1.18%, and 2.87% in the ES15d, ES30d, and ES60d treatments, respectively; P < 0.001), and Lactococcus (2.22%, 2.37%, and 0.56% in the ES15d, ES30d, and ES60d treatments, respectively; P < 0.001) were the four most differentially abundant genera. Pseudomonas produces biogenic amines, not favored in silage (Dunière et al., 2013; Jiang et al., 2020). Pseudomonas abundance was negatively correlated with pH and aminopeptidase activity in silage (Figure 2), indicating its minimal involvement in protein degradation in this study. The LAB Weissella grows well in mildly acidic environments and can metabolize WSC to produce a mixture of lactic and acetic acids, with the end product being mostly lactate (Graf et al., 2016). Notably, Weissella had a higher relative abundance in treatments related to ensiling time and crop type. Moreover, Weissella abundance was positively correlated with pH, carboxypeptidase and aminopeptidase activities, and propionic and acetic acid concentrations. The gradual decrease in the relative abundance of Weissella with prolonged ensiling time suggests its role in protein degradation during the pre-silage period, which is consistent previous findings (Cai et al., 1998; Jiang et al., 2023). Therefore, acid protease, carboxypeptidase, and aminopeptidase activities were affected by two sources of interactive variation: ensiling time × additives and ensiling time × crop type × additives. It was worth noting that despite the impact of leaf spot disease on the forage, after 60 d of silage, beneficial bacteria, represented by lactic acid bacteria, still effectively inhibited a large number of pathogens and microorganisms prone to participating in protein degradation in this study.
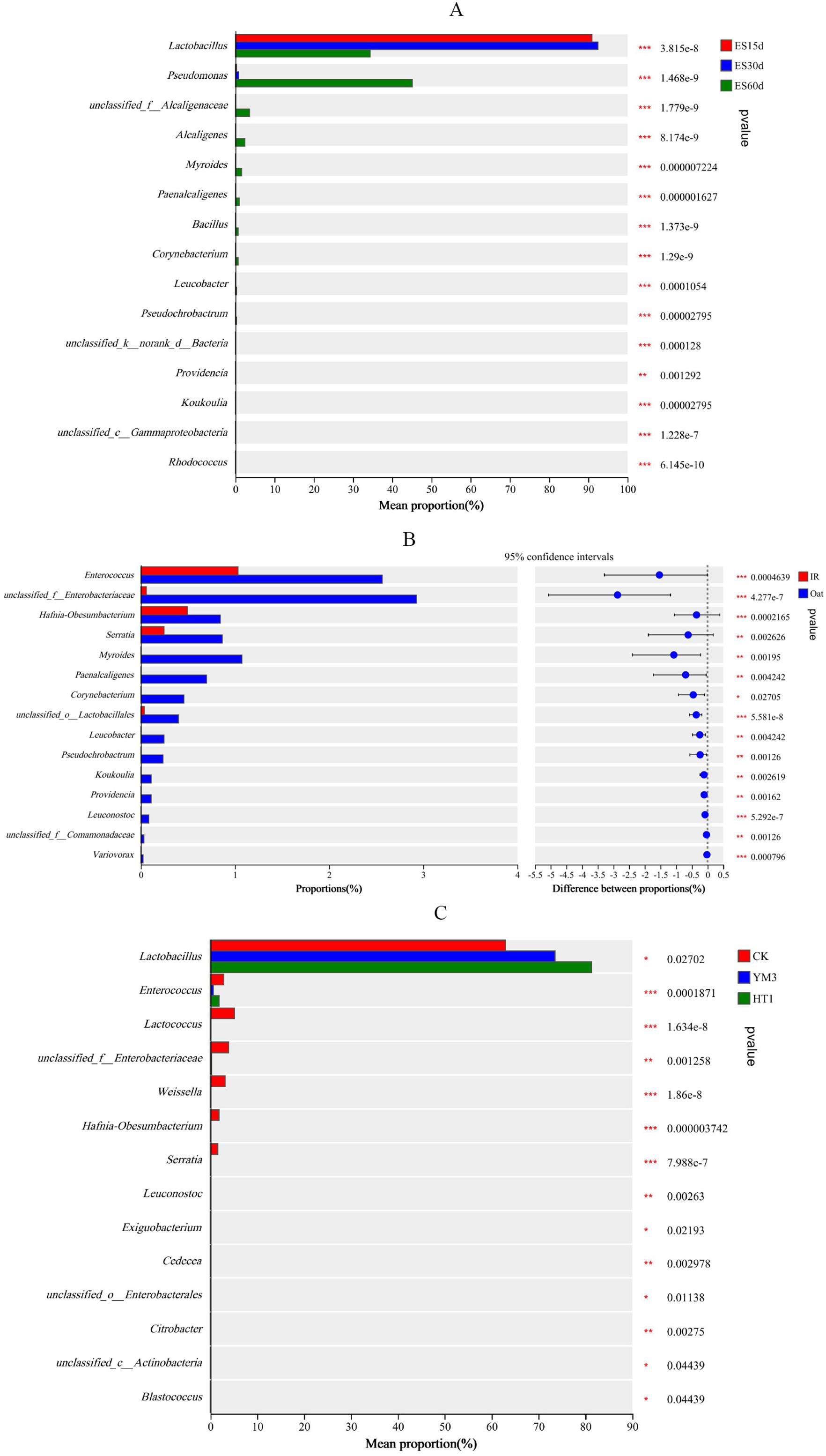
Figure 6. Microbiological differential analysis of different crop type (A), additives (B) ensiling time (C). The vertical axis on the left half of the graph shows differentially abundant species, and the horizontal axis shows mean species abundance. On the right half, the horizontal axis shows the difference in abundance between groups, and the color of the dots indicates the group with higher abundance. The error bars of the dots indicate fluctuations in the 95% confidence interval of the difference. The vertical axis indicates the significance of the difference between corresponding species groups (i.e., the magnitude of the pvalue).
Silage quality mainly depends on the competition between LAB and spoilage microorganisms, and variations in microbial communities partly explain the differences in silage quality (Ni et al., 2018; Zhang et al., 2023). In this study, the relative abundance of Enterococcus (67.6% and 77.7% in the IR and oat treatments, respectively; P < 0.001), Enterobacteriaceae (0.06% and 2.93% in the IR and oat treatments, respectively; P < 0.001), Hafnia-Obesumbacterium (0.50% and 0.85% in the IR and oat treatments, respectively; P < 0.001), and Serratia (0.25% and 0.87% in the IR and oat treatments, respectively; P < 0.01) (Figure 6) varied greatly between the two crops. However, the relative abundance of Lactococcus was lower in all treatments, which was related to the characteristics of the raw material and the weaker acid tolerance of Lactococcus (He et al., 2020). LAB act as key microorganism to improve silage fermentation quality, and the number of LAB in IR was significantly higher than that in oat before fermentation, which provided conditions for the rapid accumulation of lactic acid and the growth of acid-tolerant microorganisms. This was reflected in the relative abundance of Bacillus, which typically increases during the aerobic spoilage phase of silage (Li et al., 2021; Okoye et al., 2023). Bacillus secretes acid proteases that promote protein degradation, contributing to the poor nutritional quality of the silage (Figure 3). Notably, LAB additives had less of an effect on the numbers and species of silage microbes than the CK, which was inconsistent with most studies (Ma et al., 2022; Zhang et al., 2023). This discrepancy may be attributed to the high fermentation potential of the raw material used in this study, even in the presence of severe leaf spot disease. However, Lactobacillus was more abundant in the HT1 and YM3 treatment groups than in the CK group (Figure 6). This indicates that the LAB additives further improved the silage fermentation quality of forage with leaf spot disease, which was mainly due to the rapid decline in pH (Table 4). Although the OTU level was higher in oat, the NST index of oat was lower than that of IR (P < 0.001). This suggests a more intense competition among bacteria and a greater environmental complexity in IR. In contrast, the impact of oat on environmental factors was generally negative, which enhanced the stability of the bacterial network structure (Fan et al., 2018). Similarly, CK also increased the instability and complexity of the silage environment, which was confirmed by its highest NST index. This indicates that despite the improvement in microbial stability of silage with leaf spot disease by adding LAB, the silage environment with leaf spot disease remained more complex and unstable (Figure 5).
Conclusion
In this study, we comprehensively investigated the effects of different treatments, including ensiling time and LAB additives, on the fermentation quality, nitrogen compound content, and protease activity of forage infected with leaf spot disease. We elucidated the mechanisms by which LAB additives regulate harmful bacteria and nitrogen compound conversion during ensiling. Additionally, we found that oat silage exhibited better fermentation quality and bacterial stability than that of IR. The extension of ensiling time significantly improved the overall fermentation quality of silage. Furthermore, the application of LAB additives, particularly YM3, significantly enhanced the fermentation quality and bacterial stability of silage from forage infected with leaf spot disease. In conclusion, these findings fill the knowledge gap in the field and provides a solid theoretical foundation and practical guidance for the scientific application of LAB additives in animal husbandry. By continuously optimizing the ensiling process and selecting appropriate LAB additives, the negative impact of leaf spot disease on silage quality can be effectively mitigated, leading to reduced production costs and significant economic benefits in the livestock industry.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://resolve.pid21.cn/13913.11.micro.data.sra.NMDC40041972, NMDCR1693659043309.
Author contributions
XZ: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. YM: Investigation, Writing – original draft, Writing – review & editing. LX: Conceptualization, Data curation, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Yunnan Fundamental Research Projects (202301AU070035), Xingzhao Talent Support Program (2023), and Special Basic Cooperative Research Programs of Yunnan Provincial Undergraduate Universities’ Association (202301BA070001-120).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2025.1577383/full#supplementary-material
Supplementary Figure 1 | Venn diagram of shared and unique operational taxonomic units of silage bacterial communities at different different ensiling time (A), crop type (B) or additives (C). ES15d, ensiled 15 d; ES30d, ensiled 30 d; ES60d, ensiled 60 d; CK, no additive; YM3, lactic acid bacteria YM3; HT1, lactic acid bacteria HT1; IR, Italian ryegrass.
Abbreviations
AB, aerobic bacteria; CK, control; DM, dry matter; FM, fresh matter; HT1, Lactobacillus rhamnosus HT1; IR, Italian ryegrass; LAB, lactic acid bacteria; NST, normalized shuffle test; OUT, operational taxonomic units; PCR, polymerase chain reaction; WSC, Water-soluble carbohydrate; YM3, Lactobacillus plantarum YM3.
References
Atri A., Banyal D. K., Bhardwaj N. R., and Roy A. K. (2022). Exploring the integrated use of fungicides, bio-control agent and biopesticide for management of foliar diseases (anthracnose, grey leaf spot and zonate leaf spot) of sorghum. Int. J. Pest Management 2, 1–12. doi: 10.1080/09670874.2022.2039799
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of proteindye binding. Analytical Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Broderick G. A. and Kang J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Cai Y., Benno Y., Ogawa M., Ohmomo S., Kumai S., and Nakase T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1089/oli.1.1998.8.351
Der Bedrosian M. C. and Kung L. Jr. (2019). The effect of various doses of an exogenous acid protease on the fermentation and nutritive value of corn silage. J. Dairy Sci. 102, 10925–10933. doi: 10.3168/jds.2019-16436
Dunière L., Sindou J., Chaucheyras-Durand F., Chevallier I., and Thévenot-Sergentet D. (2013). Silage processing and strategies to prevent persistence of undesirable microorganisms. Anim. Feed Sci. Technol. 182, 1–15. doi: 10.1016/j.anifeedsci.2013.04.006
Fan K., Weisenhorn P., Gilbert J. A., Shi Y., Bai Y., and Chu H. (2018). Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol. Biochem. 121, 185–192. doi: 10.1016/j.soilbio.2018.03.017
Feller U. K., Soong T. S. T., and Hageman R. H. (1977). Leaf proteolytic activities and senescence during grain development of field-grown corn (Zea mays L.). Plant Physiol. 59, 290–294. doi: 10.1104/pp.59.2.290
Fijałkowska M., Pysera B., Lipiński K., and Strusińska D. (2015). Changes of nitrogen compounds during ensiling of high protein herbages–a review. Ann. Anim. Sci. 15, 289–305. doi: 10.1515/aoas-2015-0008
Graf K., Ulrich A., Idler C., and Klocke M. (2016). Bacterial community dynamics during ensiling of perennial ryegrass at two compaction levels monitored by terminal restriction fragment length polymorphism. J. Appl. Microbiol. 120, 1479–1491. doi: 10.1111/jam.13114
Grewal H. S. and Williams R. (2002). Influence of potassium fertilization on leaf to stem ratio, nodulation, herbage yield, leaf drop, and common leaf spot disease of alfalfa. J. Plant Nutrition 25, 781–795. doi: 10.1081/pln-120002959
Guo M., Wu F., Hao G., Qi Q., Li R., Li N., et al. (2017). Bacillus subtilis improves immunity and disease resistance in rabbits. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00354
Guo X. S., Zhou H., Yu Z., and Zhang Y. (2007). Changes in the distribution of nitrogen and plant enzymatic activity during ensilage of lucerne treated with different additives. Grass Forage sci. 62, 35–43. doi: 10.1111/j.1365-2494.2007.00559.x
Hashmi S., Iqbal S., Ahmed I., and Janjua H. A. (2022). Production, optimization, and partial purification of alkali-thermotolerant proteases from newly isolated Bacillus subtilis S1 and Bacillus amyloliquefaciens KSM12. Processes 10, 1050. doi: 10.3390/pr10061050
He L., Lv H., Xing Y., Wang C., You X., Chen X., et al. (2020). The nutrients in Moringa oleifera leaf contribute to the improvement of stylo and alfalfa silage: Fermentation, nutrition and bacterial community. Biores. Technol. 301, 122733. doi: 10.1016/j.biortech.2020.122733
Huang R., Zhang F., Wang X., and Ma C. (2023). Effect of condensed tannins on nitrogen distribution and metabolome after aerobic exposure of sainfoin silage. Fermentation 9, 739. doi: 10.3390/fermentation9080739
Jiang D., Li B., Zheng M., Niu D., Zuo S., and Xu C. (2020). Effects of Pediococcus pentosaceus on fermentation, aerobic stability and microbial communities during ensiling and aerobic spoilage of total mixed ration silage containing alfalfa (Medicago sativa L.). Grassland Sci. 66, 215–224. doi: 10.1111/grs.12272
Jiang H., Wang H., Bao B., Qu H., Wang J., Sun L., et al. (2023). Effect of compound additives on nutritional composition, fermentation quality, and bacterial community of high-moisture alfalfa silage. Fermentation 9, 453. doi: 10.3390/fermentation9050453
Johnson E. T., Dowd P. F., Ramirez J. L., and Behle R. W. (2023). Potential biocontrol agents of corn tar spot disease isolated from overwintered Phyllachora maydis stromata. Microorganisms 11, 1550. doi: 10.3390/microorganisms11061550
Kim D., Dong Lee K., and Choon Choi K. (2021). Role of LAB in silage fermentation: Effect on nutritional quality and organic acid production—An overview. AIMS Agric. Food 6, 216–234. doi: 10.3934/agrfood.2021014
Lan Y., Zhou W., Duan T., Li Y., Matthew C., and Nan Z. (2024). Alfalfa spring black stem and leaf spot disease caused by Phoma medicaginis: epidemic occurrence and impacts. Microorganisms 12, 1279. doi: 10.3390/microorganisms12071279
Li T., Mann R., Kaur J., Spangenberg G., and Sawbridge T. (2021). Transcriptome analyses of barley roots inoculated with novel Paenibacillus sp. and Erwinia gerundensis strains reveal beneficial early-stage plant–bacteria interactions. . Plants 10, 1802. doi: 10.3390/plants10091802
Li X., Tian J., Zhang Q., Jiang Y., Wu Z., and Yu Z. (2018). Effects of mixing red clover with alfalfa at different ratios on dynamics of proteolysis and protease activities during ensiling. J. Dairy Sci. 101, 8954–8964. doi: 10.3168/jds.2018-14763
Li M., Zi X., Sun R., Ou W., Chen S., Hou G., et al. (2024). Co-ensiling whole-plant cassava with corn stalk for excellent silage production: fermentation characteristics, bacterial community, function profile, and microbial ecological network features. Agronomy 14, 501. doi: 10.3390/agronomy14030501
Liu Q. H., Dong Z. H., and Shao T. (2018b). Effect of additives on fatty acid profile of high moisture alfalfa silage during ensiling and after exposure to air. Anim. Feed Sci. Technol. 236, 29–38. doi: 10.1016/j.anifeedsci.2017.11.022
Liu H., Li X., Hu J., Zhao J., Xu G., Dong D., et al. (2023). Fermentation quality and aerobic stability evaluation of rice straw silage with different ensiling densities. Fermentation 10, 20. doi: 10.3390/fermentation10010020
Liu Q., Lindow S. E., and Zhang J. (2018a). Lactobacillus parafarraginis ZH1 producing anti-yeast substances to improve the aerobic stability of silage. Anim. Sci. J. 89, 1302–1309. doi: 10.1111/asj.13063
Liu Q. H., Shao T., and Bai Y. F. (2016). The effect of fibrolytic enzyme, Lactobacillus plantarum and two food antioxidants on the fermentation quality, alpha-tocopherol and beta-carotene of high moisture napier grass silage ensiled at different temperatures. Anim. Feed Sci. Technol. 221, 1–11. doi: 10.1016/j.anifeedsci.2016.08.020
Ma Z. Y., Ungerfeld E., Ouyang Z., Zhou X. L., Han X. F., Zeng Y. Q., et al. (2022). Effect of Lactobacillus plantarum inoculation on chemical composition, fermentation, and bacterial community composition of ensiled sweet corn whole plant or stover. Fermentaion 8, 24. doi: 10.3390/fermentation8010024
Murphy R. (1958). A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food Agriculture 9, 714–717. doi: 10.1002/jsfa.2740091104
Ni K., Zhao J., Zhu B., Su R., Pan Y., Ma J., et al. (2018). Assessing the fermentation quality and microbial community of the mixed silage of forage soybean with crop corn or sorghum. Biores. Technol. 265, 563–567. doi: 10.1016/j.biortech.2018.05.097
Okoye C. O., Wu Y., Wang Y., Gao L., Li X., and Jiang J. (2023). Fermentation profile, aerobic stability, and microbial community dynamics of corn straw ensiled with Lactobacillus buchneri PC-C1 and Lactobacillus plantarum PC1-1. Microbiological Res. 270, 127329. doi: 10.1016/j.micres.2023.127329
Peterson L. W. and Huffaker R. C. (1975). Loss of ribulose 1, 5-diphosphate carboxylase and increase in proteolytic activity during senescence of detached primary barley leaves. Plant Physiol. 55, 1009–1015. doi: 10.1104/pp.55.6.1009
Playne M. J. and McDonald P. (1966). The buffering constituents of herbage and of silage. J. Sci. Food Agriculture 17, 264–268. doi: 10.1002/jsfa.2740170609
Qian Y., Jin Y., Han X., Malik K., Li C., and Yu B. (2024). Effects of grazing and leaf spot disease on the structure and diversity of phyllosphere microbiome communities in Leymus chinensis. Plants 13, 2128. doi: 10.3390/plants13152128
Rana S., Bhawal S., Kumari A., Kapila S., and Kapila R. (2020). pH-dependent inhibition of AHL-mediated quorum sensing by cell-free supernatant of lactic acid bacteria in Pseudomonas aeruginosa PAO1. Microbial. Pathogenesis 142, 104105. doi: 10.1016/j.micpath.2020.104105
Shan P., Ho C.-T., Zhang L., Gao X., Lin H., Xu T., et al. (2022). Degradation mechanism of soybean protein B3 subunit catalyzed by prolyl endopeptidase from aspergillus Niger during soy sauce fermentation. J. Agric. Food Chem. 70, 5869–5878. doi: 10.1021/acs.jafc.2c01796
Tang G., Fan Y., Li X., Tian R., Tang R., Xu L., et al. (2023). Effects of leaf properties on the counts of microbes on the leaf surfaces of wheat, rye and triticale. FEMS Microbiol. Ecol. 99, 1–10. doi: 10.1093/femsec/fiad024
Wang Y., He L., Xing Y., Zheng Y., Zhou W., Pian R., et al. (2019). Dynamics of bacterial community and fermentation quality during ensiling of wilted and unwilted moringa oleifera leaf silage with or without lactic acid bacterial inoculants. mSphere 4, e00341–e00319. doi: 10.1128/mSphere.00341-19
Wang X., Liu H., Xie Y., Zhang Y., Lin Y., Zheng Y., et al. (2021). Effect of sucrose and lactic acid bacteria additives on fermentation quality, chemical composition and protein fractions of two typical woody forage silages. Agriculture 11, 256. doi: 10.3390/agriculture11030256
Wilkinson J. M. (2003). A laboratory evaluation of comfrey (Symphytum officinale L.) as a forage crop for ensilage. Anim. Feed Sci. Technol. 104, 227–233. doi: 10.1016/s0377-8401(02)00293-6
Xie Z. L., Zhang T. F., Chen X. Z., Li G. D., and Zhang J. G. (2012). Effects of maturity stages on the nutritive composition and silage quality of whole crop wheat. Asian-Australasian J. Anim. Sci. 25, 1374–1380. doi: 10.5713/ajas.2012.12084
Xu L., Hu Y., Li X., Yin X., Tang G., and Zhang J. (2021). Effects of wheat-legume cultures on the fermentation quality and protein degradation of silage. Grassland Sci. 68, 13–22. doi: 10.1111/grs.12329
Xu L., Tang G., Wu D., and Zhang J. (2023). Effects of sowing methods on nitrogen compounds and protease activities of whole-crop wheat silage. Grassland Res. 2, 46–55. doi: 10.1002/glr2.12041
Yan Y., Li X., Guan H., Huang L., Ma X., Peng Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Biores. Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
Yuan X., Wen A., Desta S. T., Dong Z., and Shao T. (2017). Effects of four short-chain fatty acids or salts on the dynamics of nitrogen transformations and intrinsic protease activity of alfalfa silage. J. Sci. Food Agriculture 97, 2759–2766. doi: 10.1002/jsfa.8103
Zhang X., Zhao S., Wang Y., Yang F., Wang Y., Fan X., et al. (2023). The effect of Lactiplantibacillus plantarum ZZU203, cellulase-producing Bacillus methylotrophicus, and their combinations on alfalfa silage quality and bacterial community. Fermentation 9, 278. doi: 10.3390/fermentation9030287
Zhao M., Bao J., Wang Z., Sun P., Liu J., Yan Y., et al. (2024a). Utilisation of Lactiplantibacillus plantarum and propionic acid to improve silage quality of amaranth before and after wilting: fermentation quality, microbial communities, and their metabolic pathway. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1415290
Zhao X., Sun Y., Chang Z., Yao B., Han Z., Wang T., et al. (2024b). Innovative lactic acid production techniques driving advances in silage fermentation. Fermentation 10, 533. doi: 10.3390/fermentation10100533
Zhou H., Jia S., Gao Y., Li X., Lin Y., Yang F., et al. (2024). Characterization of phyllosphere endophytic lactic acid bacteria reveals a potential novel route to enhance silage fermentation quality. Commun. Biol. 7, 117. doi: 10.1038/s42003-024-05816-3
Keywords: lactic acid bacteria, leaf spot disease, microbial community, protein degradation, silage fermentation quality
Citation: Zhang X, Meng Y and Xu L (2025) Lactic acid bacteria additives effectively regulate nitrogen compounds and related bacterial communities in silage infected with leaf spot disease. Front. Anim. Sci. 6:1577383. doi: 10.3389/fanim.2025.1577383
Received: 19 February 2025; Accepted: 25 April 2025;
Published: 19 May 2025.
Edited by:
Francisco José Fernández Perrino, Universidad Autónoma Metropolitana, MexicoReviewed by:
Sanjay Kumar Singh Patel, Hemwati Nandan Bahuguna Garhwal University, IndiaAlejandro Azaola-Espinosa, Autonomous Metropolitan University, Mexico
Copyright © 2025 Zhang, Meng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyan Meng, MTA1NjAzNzc1MUBxcS5jb20=; Liuxing Xu, MzMxNDA1NzE5QHFxLmNvbQ==
 Xiaolong Zhang1
Xiaolong Zhang1 Liuxing Xu
Liuxing Xu