- 1Key Laboratory of Livestock and Forage Resources Utilization Around Tarim, College of Animal Science and Technology, Tarim University, Alar, Xinjiang, China
- 2Key Laboratory of Tarim Animal Husbandry Science and Technology, College of Animal Science and Technology, Tarim University, Alar, Xinjiang, China
- 3Animal Husbandry and Veterinary Department of Tashkurgan Tajik Autonomous County, Kashi, China
- 4State Key Laboratory Incubation Base for Conservation and Utilization of Bio-Resource in Tarim Basin, College of Life Sciences and Technology, Tarim University, Alar, Xinjiang, China
Introduction: Follicle-stimulating hormone (FSH) plays a crucial role in regulating ovarian follicular development in sheep. This study investigated the regulatory mechanism of FSH in periostin (POSTN) expression in ovine granulosa cells (GCs).
Methods: Immunofluorescence staining revealed POSTN expression throughout the cytoplasm and nucleus of the GCs. The treatment of GCs with 10 ng/mL FSH significantly increased POSTN mRNA and protein levels in a time-dependent manner, peaking at 24 h. Knockdown of POSTN using siRNA severely impaired GCs viability, which could not be rescued by FSH, indicating the essential role of POSTN in GCs function. Cloning and analysis of the POSTN promoter identified putative binding sites for serum response factor (SRF) and JunD transcription factors within the core promoter region (-387 to +1). Targeted mutagenesis of the SRF site significantly reduced POSTN transcriptional activity, whereas JunD mutation had no effect. Chromatin immunoprecipitation assays confirmed SRF enrichment at the POSTN promoter following FSH stimulation.
Results and Discussion: These findings indicate that FSH regulates POSTN expression in ovine GCs through the transcription factor SRF, potentially by activating signaling cascades such as cAMP/PKA and MAPK/ERK to promote SRF phosphorylation and activation, a mechanism that remains to be further explored. This study provides new insights into the molecular mechanisms of folliculogenesis and offers a potential target for enhancing reproductive efficiency in sheep.
1 Introduction
GCs are fundamental cells responsible for ovum maturation in ovines. They play a crucial role in ovarian selection, dominance, maturation, and ovulation. However, ovarian development is frequently accompanied by apoptosis in 99% of the GCs cases. Therefore, the most direct way to obtain a mature ovum is to maintain the normal morphological structure and physiological function of GCs. Several studies have confirmed that FSH promotes normal development of mammalian GCs (Orisaka et al., 2021). It has been demonstrated that the addition of 10 ng/mL FSH promotes the proliferation of ovine follicular GCs (Li et al., 2022; Liu et al., 2024; Xu et al., 2025). Suocheng et al. found that FSH mediates the FSH receptor (FSHR) to reduce the rate of apoptosis in GCs and increase the rate of oocyte maturation in ovines (Suocheng et al., 2017). In a study of GCs in rats, it was found that FSH action on the receptor activates signal transduction, relying primarily on cAMP synthesis and PKA activation (Chen et al., 2008). FSH also inhibits the PI3K and ERK pathways, promotes the expression of TSP1, and inhibits the proliferation of ovarian GCs (Jiapeng et al., 2023). FSH promotes the biochemical functions of GCs by activating relevant downstream signaling pathways within GCs and increasing the transcription levels of specific relevant genes. However, the specific regulatory mechanisms involved require further elucidation.
POSTN is a protein secreted by stromal cells with multiple biological functions and is a cell adhesion protein first cloned and characterized by Takeshita from mouse osteoblasts MC3r13-E1 (Takeshita et al., 1993). POSTN is involved in angiogenesis, promotion of cell proliferation, inhibition of apoptosis, and extracellular matrix remodeling (Ratajczak and Dziegiel, 2015). The current body of research on POSTN focuses on its role in cancer and tumors. These findings indicated that POSTN facilitates the adhesion and migration of ovarian epithelial cancer cells by binding to αvβ3 and αvβ5 integrins. Furthermore, overexpression of POSTN in ovarian cancer cells significantly enhances the migration and invasion abilities of ovarian cancer cells (Yu et al., 2021). Research has shown that POSTN mRNA expression is upregulated in ovarian tumor tissues and is involved in the ECM-mediated cell adhesion signaling pathway, which in turn is regulated by estrogen (Ismail et al., 2000; Syed et al., 2005; Takayama and Kudo, 2012). POSTN also mediates angiogenesis by increasing the VEGF receptor expression in endothelial cells, which may be associated with luteal ovarian angiogenesis (Ruan et al., 2009). POSTN expression is upregulated in porcine GCs cultured in vitro and may be involved in regulating cell proliferation in vitro. Increased expression of this gene has been observed in many different tissues such as the placenta, heart valves, and ligaments, where tissue repair and angiogenesis occur (Kulus et al., 2020). These studies suggest that POSTN may be involved in post-ovulatory-related life processes such as corpus luteum formation and embryo implantation. A recent study demonstrated that POSTN overexpression in GCs significantly enhances cell proliferation and inhibits apoptosis. In GCs overexpressing POSTN, phosphorylation levels of focal adhesions and autophagy are elevated, thereby affecting GCs proliferation and follicular development (Abudureyimu et al., 2024).
We hypothesize that POSTN expression in ovine GCs is mediated by FSH through certain transcription factors. Therefore, in the present study, FSH induction of POSTN expression in granulosa cells and the transcription factors involved in this process were investigated. This study will serve to elucidate the mechanism by which transcription factors are regulated in FSH-induced POSTN expression, spurring further research in this field.
2 Materials and methods
All experimental protocols involving ovines strictly followed the relevant guidelines set by the Science and Technology Ethics Committee of Tarim University (Approval ID: TUEC2023-060).
2.1 GCs isolation and primary culture in ovine follicles
Ovaries from Altai sheep were collected at a slaughterhouse. Immediately after collection, they were placed in pre-warmed (37°C) sterile PBS (100 IU/mL penicillin and 50 mg/mL streptomycin) and kept in a thermos for transport back to the laboratory. Granulosa cells were isolated from follicles larger than 2 mm in diameter, washed twice with PBS, and then seeded in DMEM/F12 (Gibco) for 24 h. After incubation, the cells were collected by centrifugation, rinsed twice with PBS, and resuspended in DMEM/F12 supplemented with 10% FBS (ExCell). Once cultures reached ≥80% confluency, cells were cryopreserved in liquid nitrogen.
2.2 GCs resuscitation and cultivation
Frozen GCs were thawed for 30 s in a 37°C water bath, and cryotube exteriors were disinfected with 70% ethanol. GCs were transferred to RNase-free tubes and resuspended in 1 mL pre-warmed medium. The cells were centrifuged at 1,400 rpm for 5 min and resuspended in fresh medium twice. The cell suspension was seeded in culture flasks containing 8 mL medium and incubated at 37°C in 5% CO2. After 24 h, the medium was replaced, and the cells were collected at 80% confluence.
2.3 Immunofluorescence detection of POSTN expression in GCs
When GCs on coverslips reached 60–70% confluence, they were washed three times for 5 min with pre-warmed PBS. The cells were fixed in 4% paraformaldehyde at room temperature for 30 min and washed three times with PBS. The cells were permeabilized with 0.2% Triton X-100 for 10 min, washed three times with PBS, blocked in PBST with 0.3% bovine serum albumin (BSA) for 30 min at room temperature, and incubated overnight at 4°C with POSTN primary antibody (1:500, abcom). The next day, the cells were washed three times with PBS, incubated with POSTN secondary antibody (1:500, abcom) at room temperature for 1 h in the dark, and washed three times with PBS. Nuclei were stained with DAPI, washed three times with PBS, blocked with goat serum, and imaged using a fluorescence microscope.
2.4 GCs RNA extraction and reverse transcription
Total RNA was extracted from GCs using TRIzol reagent, according to the manufacturer’s protocol, and dissolved in RNase-free water. RNA purity and concentration were assessed by UV spectrophotometry. cDNA was synthesized from total RNA by reverse transcription. Reverse transcription was carried out in a 25 μL reaction mixture containing 2 μg RNA, 50 µM random primers, 1 mM dNTPs, 5× RT buffer, 100 IU RNase inhibitor, and 10 U M−MLV reverse transcriptase. Reactions were incubated at 37°C for 1 h, inactivated at 95°C, and cDNA was stored at −80°C.
2.5 qRT-PCR
POSTN expression was quantified by qRT-PCR. The primers used were: 5’-TACCTTCAAAGAAATCCCCAT-3’ (upstream) and 5’-GGTGAAACGGTAACTGAAG-3’ (downstream). The 20 µL reaction contained 10 µL of 2× LightCycler DNA Master SYBR Green I, 0.5 µL of each primer (10 µM), 2 µL of cDNA, and 7 µL ddH2O. Cycling began at 95°C for 5 min, followed by 35 cycles at 95°C for 5 s, 60°C for 20 s, and 72°C for 15 s, with fluorescence recorded during the extension phase. Melt curve analysis was performed from 70°C to 95°C. β-Actin served as an internal control, and the reactions were run in triplicate.
2.6 Western blot
Protein were extracted with RIPA buffer, quantified by BCA assay, mixed with 5× SDS-PAGE loading buffer (4:1), and denatured at 95°C for 5 min. SDS-PAGE was performed on 5% stacking and 12% resolving gels with 20 μg of protein per well at 100 V for 90–120 min. Proteins were transferred to PVDF membranes at 300 mA for 40–60 min, and the transfer efficiency was verified by Coomassie Brilliant Blue staining. PVDF membranes were blocked with 5% skimmed milk in TBST at room temperature for 30 min. Mouse anti-POSTN mAbs were incubated overnight at 4°C on a shaker. On the second day, the PVDF membranes were washed three times with TBST for 10 min each. PVDF membranes were incubated with horseradish peroxidase (HRP)-goat anti-mouse IgG for 1 h, detected in enhanced chemiluminescence (ECL) liquid, and imaged using a chemiluminescence imager.
2.7 POSTN siRNA chemical synthesis
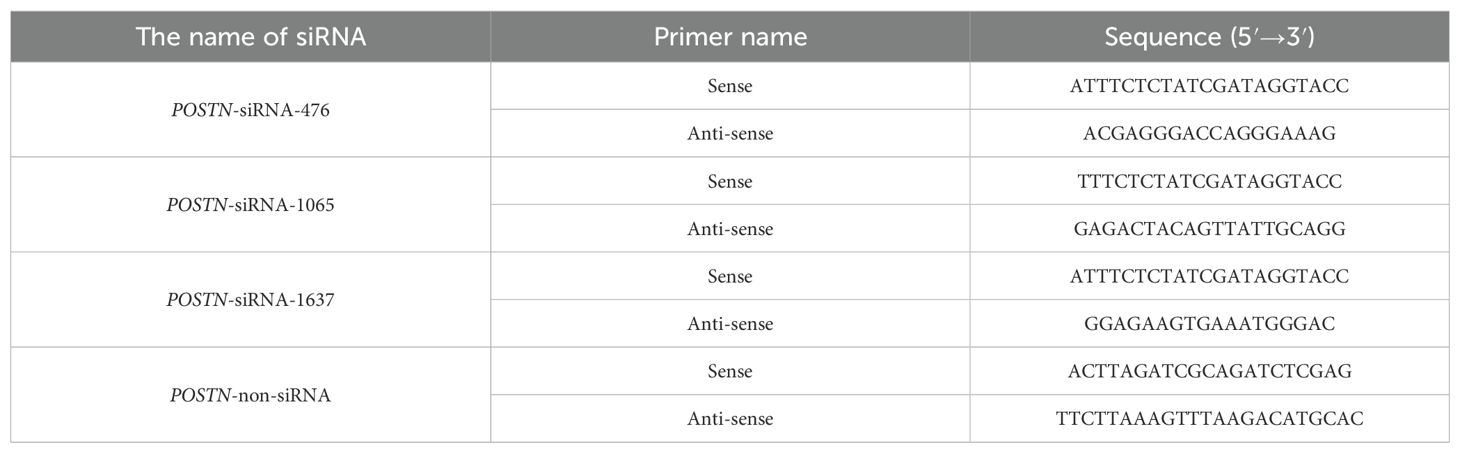
siRNA fragments targeting the ovine POSTN gene were designed and the sequences are listed in Table 1. A non-homologous siRNA fragment was used as a negative control. All siRNA sequences were synthesized by RuiBo Company (Guangzhou, China).
2.8 POSTN siRNA cell transfection and CCK-8 proliferation assay
To assess GCs transfection efficiency, POSTN-siRNA was labeled with FAM, and after transfection the percentage of FAM-positive cells was quantified by fluorescence microscopy. GCs were collected 24 h before transfection, digested, counted, and seeded in 1.5 mL of complete medium without antibiotics. At ~50% confluence, 3 µL of POSTN-siRNA was diluted in 250 µL of Opti-MEM serum-free medium and incubated at room temperature for 5 min. Similarly, 1 µL of Lipofectamine™ 3000 was diluted in 1 mL of Opti-MEM serum-free medium and incubated at room temperature for 5 min. Finally, the two samples were mixed and incubated for 5 minutes. For transfection, cells in 12-well plates were replaced with 1 mL of Opti-MEM serum-free medium, and after 6 h, the cells were switched to serum-containing medium. Transfected GCs were harvested 24 h after siRNA delivery (defined as “day 0”), then trypsinized, counted, and reseeded into 96-well plates at 2×103 cells per well in 100 µL of complete DMEM/F12 (n=6 wells per condition). For each siRNA type (POSTN-siRNA or NC-siRNA), two treatment groups were established: basic medium (no FSH) and FSH-supplemented medium (10 ng/mL recombinant ovine FSH, Sigma F2293), added at seeding on day 0. At each time point (days 0 through 6, at 24 h intervals), 10 µL of CCK-8 solution (10% v/v, Dojindo) was added to each well. The plates were gently shaken to mix, then incubated for 2 h at 37°C in 5% CO2. Absorbance at 450 nm was measured using a microplate reader. Wells containing medium plus CCK-8, but no cells, served as blanks for background subtraction.
2.9 Ovine POSTN promoter cloning and bioinformatics analysis
The ovine POSTN sequence was retrieved from the UCSC database (http://genome.ucsc.edu/). Primers were designed using Primer 5.0 (F1:5’-TGAACCAGATTTAAAGAAC-3’, R1:5’-TGTGAGCCAAGATCTTGTC-3’, 2,197 bp). DNA from ovine GCs was used to clone the 5’-UTR of POSTN. Transcription start sites were determined by comparing the cloned POSTN mRNA sequences. The POSTN promoter was analyzed using online transcription factor prediction tools (Gene−Regulation.com) and JASPAR (http://jaspar.genereg.net/). The relative profile score threshold was set at 85%.
2.10 Cloning of the 5’ deletion fragments of ovine POSTN promoter and vector construction
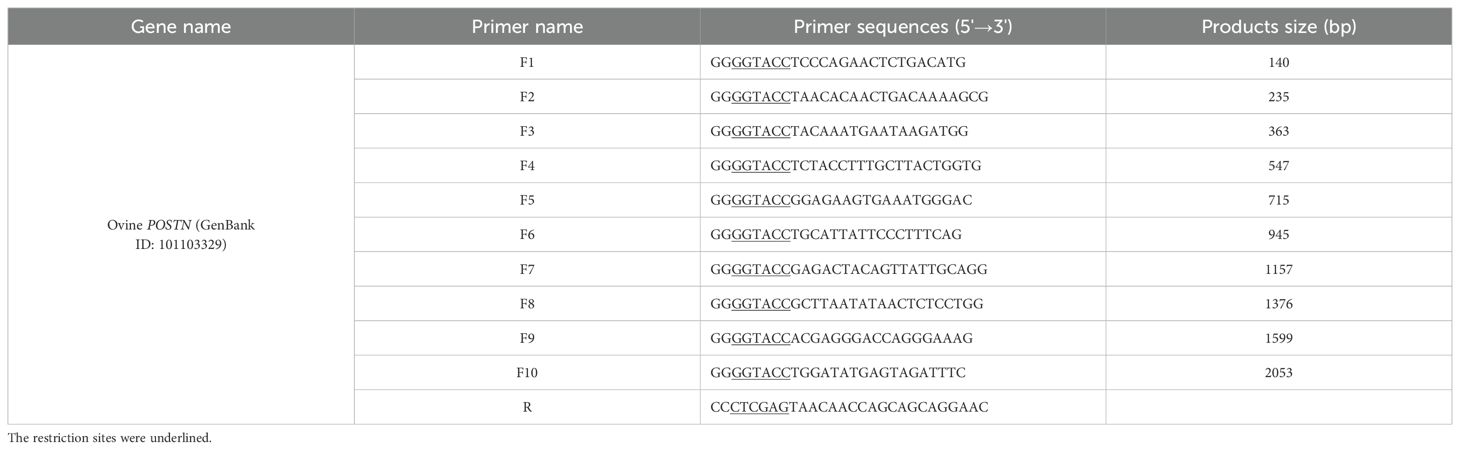
Based on section 2.9 promoter analysis, ten primer pairs were designed using a constant downstream primer and variable upstream primers (Table 2). The POSTN promoter fragments of varying lengths were named by size: POSTN-140, POSTN-235, POSTN-363, POSTN-547, POSTN-715, POSTN-945, POSTN-1157, POSTN-1376, POSTN-1599, and POSTN-2053. The fragments and pGL3-Basic vector were digested with KpnI and XhoI using primer-added restriction sites in a 100 µL reaction containing 10 µL Buffer M, 5 µL each enzyme, 45 µL DNA, and ddH2O to 100 µL. The linearized vector and digested fragments were purified, respectively. Ligation was performed using ClonExpress II Kit with 100 ng each of carrier DNA and PCR product, 4 µL CE II Buffer, 2 µL Express® II, and ddH2O to 20 µL. After 37°C incubation and ice cooling, 10 µL product was transformed into dH5α competent cells. Positive clones were verified by PCR, restriction digestion and sequencing. Constructs were designated POSTN-140 to POSTN-2053, reflecting promoter boundaries from +21/+160 to -1893/+160.
2.11 Transfection of GCs and analysis of luciferase activity
Since primary GCs have low transfection efficiency, 293T cells were used to validate the pGL3 plasmid construction. One day before transfection, the 293T cells were seeded in 12×well plates at 2×105cells/well to reach 70–80% confluence. The next morning, the medium was replaced with antigen-free DMEM and the cells were incubated for 1 h at 37°C with 5% CO2. pGL3 constructs and pRL-TK were dissolved at a 10:1 ratio in 50 μL Opti-MEM, and GCs were transfected with Lip3000. pGL3-basic and pRL-TK served as controls. Transfection was performed according to the Lip3000 protocol and repeated 3–4 times per group. After 4–6 h, the medium was replaced and GCs were harvested for subsequent experiments. After transfection, 100 μL of the culture supernatant was mixed with 100 μL LARII. A Dual-Luciferase Reporter system was used to measure the POSTN promoter activity across different fragment lengths. Subsequently, 100 μL of STOP&GLO reagent was added to each well and luciferase activity was measured.
2.12 Mutation analysis of POSTN promoter transcription factor binding sites
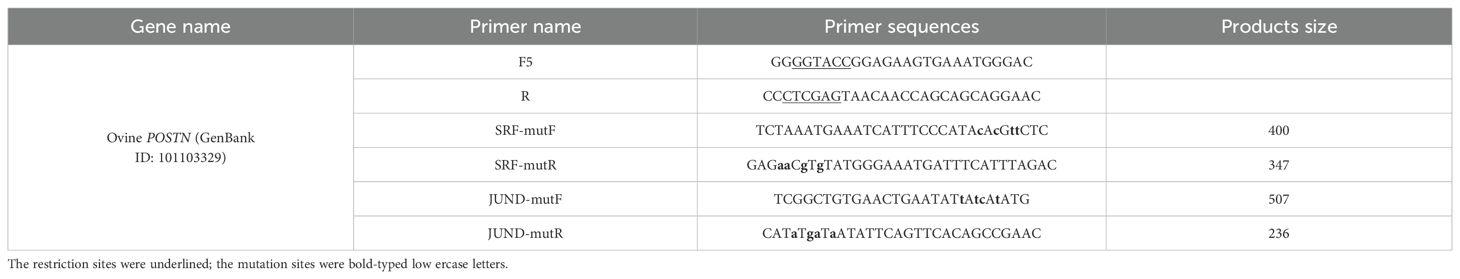
Transcription factor-binding sites within the POSTN-547 (-387/+160) region were predicted using NCBI BLAST and JASPAR. JunD and SRF sites with high scores were selected, and primers for these mutations were designed (Table 3). Overlapping PCR on the POSTN-547 (-387/+160) vector yielded mutant fragments. POSTN-715 plasmids were used to amplify fragments 1 and 2 with primers F5+SRF-mutR and F5+SRF-mutF, and fragments were purified and combined to amplify fragment 3 using primers F5+R, producing the mutated sequence. PCR was performed with annealing temperatures of ≥60 °C for 25 cycles and high-fidelity Taq. The mutant sequences were ligated into the pGL3 vector, followed by transient transfection and luciferase analysis.
2.13 Chromatin immunoprecipitation
Ovine GCs were seeded at 2×105 cells/well in 12-well plates. The cells were treated with 0.1% DMSO (control) and 10 ng/mL FSH for 24 h and then cross-linked with 1% formaldehyde for 10 min. Glycine (10×, 1.1 mL) was then added at room temperature and quenched on ice for 5 min. The dishes were washed with PBS (PMSF), and the cells were scraped into PBS and centrifuged at 12,000 rpm at 4°C for 2 min. The pellets were lysed in 200 µL of SDS buffer (PMSF) on ice for 10 min and sonicated (10 s on/20 s off) for 10 min. Sonicates were incubated with 5 M NaCl (8 µL) at 65°C for 4 h, centrifuged, and diluted in ChIP buffer (containing PMSF). Aliquots were incubated with Protein A/G agarose + salmon sperm DNA and antibodies (anti-SRF or IgG control) and washed sequentially with low-salt, high-salt, LiCl, and TE buffers. DNA was eluted with SDS/NaHCO3 buffer, reversed cross-linked, treated with EDTA/Tris/Proteinase K, purified, and resuspended in ddH2O. The eluted DNA was used for PCR with ChIP primers (F: TCTGAAAGGTATGGATTAC, R: TACTTCCTGACTCATTTC, 120 bp).
2.14 Statistical analysis
Data were analyzed with SPSS 26.0 and presented as mean ± SD (n=3). Normality was assessed using the Shapiro–Wilk test. Two-group comparisons were performed using the t-test, ≥3 groups were compared using one-way ANOVA with Duncan’s post-hoc test. p< 0.05 was considered significant.
3 Results
3.1 Localization and expression of POSTN in GCs
Protein expression in GCs was assessed by immunofluorescence staining, revealing that POSTN signals were distributed throughout the cytoplasm and nucleus of ovine GCs (Figure 1).
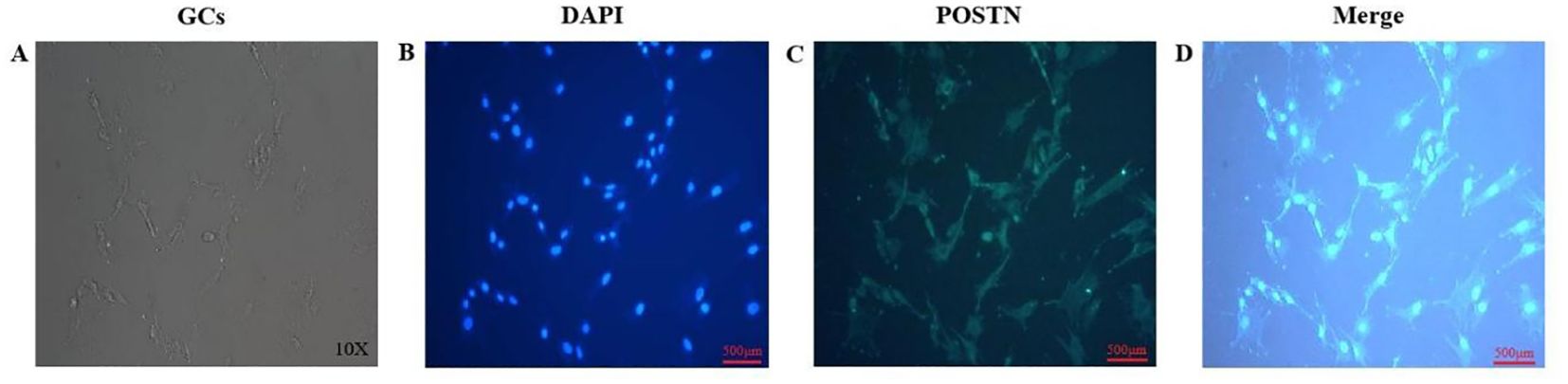
Figure 1. The distribution and expression of POSTN in ovine GCs. GCs were cultured in complete medium (A). Nuclei were re-stained with DAPI (B). POSTN antibody was used for immunofluorescence staining of GCs (C). Merged image combining DAPI-stained nuclei (blue) and POSTN immunofluorescence (green) (D). Scale bar: 500 µm (B-D).
3.2 Effect of FSH on the expression of POSTN at different times in GCs
POSTN mRNA expression was detected in ovine GCs treated with/without 10 ng/mL FSH at different times (0, 4, 8, 12, 16, 20, 22, 24, and 26 h) by qRT-PCR. Results (Figure 2A) showed POSTN mRNA expression in FSH-treated GCs was significantly higher than untreated GCs after 8 h. POSTN mRNA expression increased significantly after 4 h of FSH treatment. At 24 h, POSTN expression reached its maximum (p<0.05), then decreased at 26 h (p<0.05). Western blotting determined FSH’s effect on POSTN protein expression in GCs at different intervals. Results showed POSTN protein expression increased significantly after 4 h of FSH treatment (p<0.05) (Figure 2B). POSTN protein expression peaked at 24 h (p<0.05), followed by a significant decrease at 26 h (p<0.05). Treatment with 10 ng/mL FSH for 24 h significantly increased POSTN gene and protein expression in follicular GCs (p<0.05), suggesting FSH regulates POSTN expression in ovarian follicular GCs.
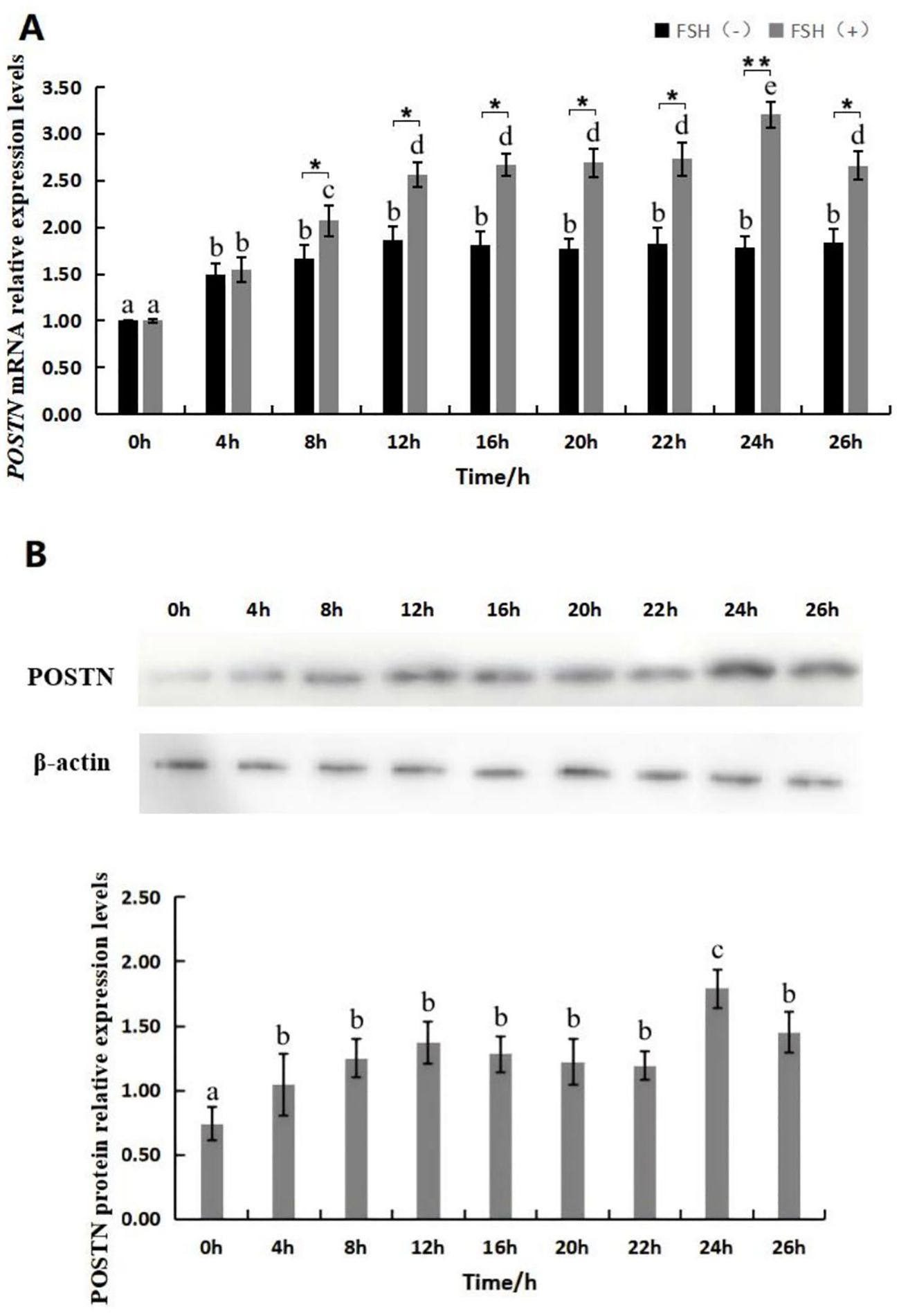
Figure 2. Effect of FSH on the expression of POSTN gene and protein in GCs. Expression levels of POSTN mRNA at various time points following treatment with 10 ng/mL FSH (with no FSH added as the control group) (A), different lowercase letters denote significant differences among time points within the same treatment group (p<0.05), and asterisks denote significant differences between the FSH-treated and control groups at the same time point (*p<0.05, **p<0.01). Protein expression levels of POSTN at various time points following treatment with 10 ng/mL FSH (B). Different letters indicate significant differences (p<0.05). n=3 independent experiments (mean ± SD).
3.3 POSTN-siRNA interference efficiency assay
The efficiency of siRNA interference was tested after GC transfection with POSTN-siRNA. 100 nM siRNA-NC, POSTN-siRNA-476, POSTN-siRNA-1065, and POSTN-siRNA-1637 were transfected into GCs for 6 h, then the medium was replaced for 24 h. RNA and proteins from GCs were extracted, and POSTN mRNA and protein expression were detected by qRT-PCR and western blotting. The results are shown in Figure 3. POSTN-siRNA was labeled with FAM, and the results showed that approximately 50% of GCs grew normally (Figure 3A). All three POSTN-siRNA fragments significantly inhibited POSTN expression compared to control siRNA, with POSTN-siRNA-476 showing stronger inhibition than POSTN-siRNA-1065 and POSTN-siRNA-1637 (Figure 3B). Western blot results matched qRT-PCR findings: all POSTN-siRNA fragments significantly reduced POSTN protein expression compared to NC-siRNA, with POSTN-siRNA-476 showing the strongest effect (p<0.05) (Figure 3C).
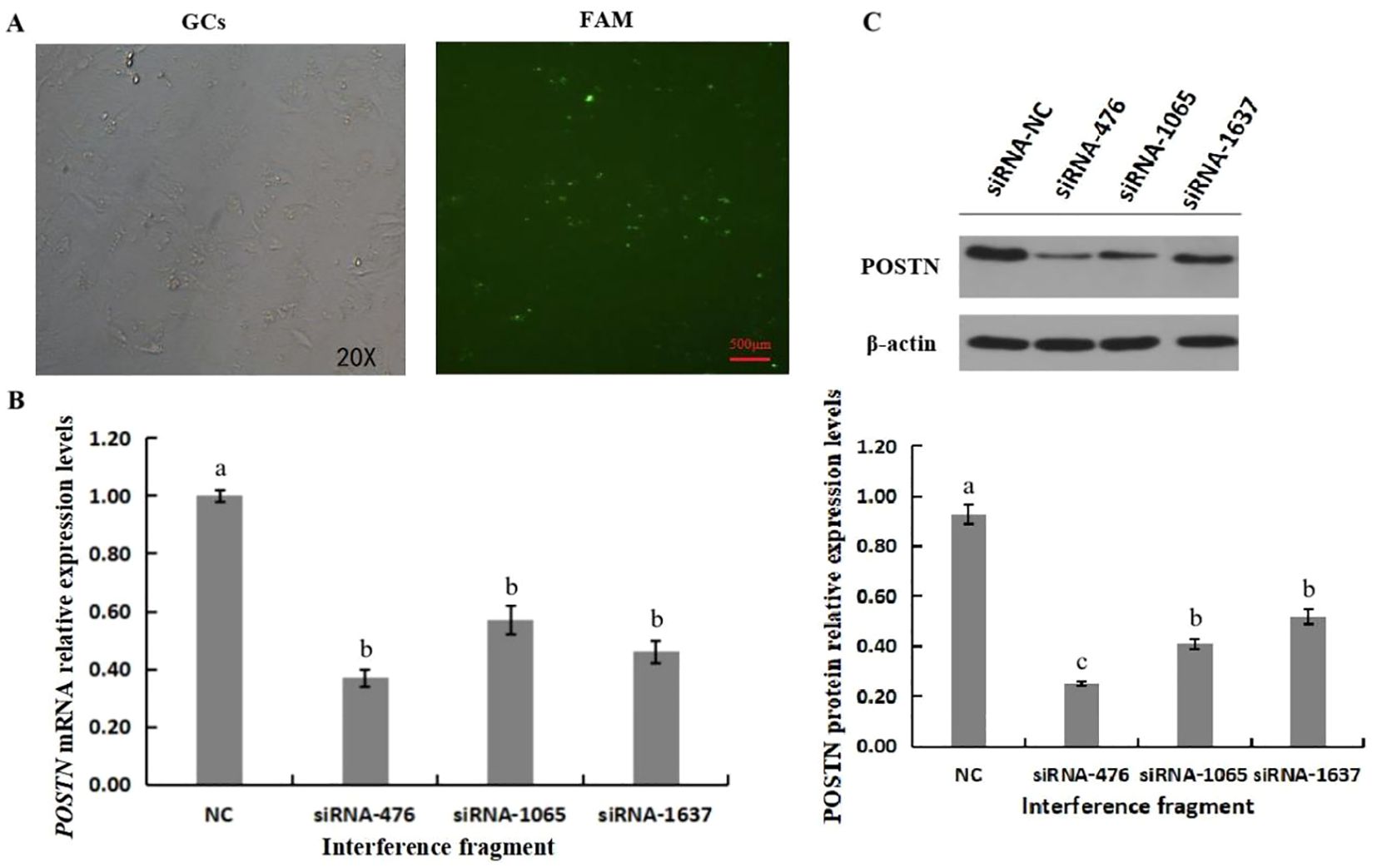
Figure 3. Interference Effect of POSTN-siRNA. FAM marker transfection efficiency of POSTN-siRNA (A). Effect of POSTN siRNA on the expression level of POSTN mRNA (B). Effect of siRNA on POSTN protein levels (C). Bars with no common superscripts are significantly different (p<0.05). n=3 independent experiments (mean ±SD).
3.4 The effect of POSTN-siRNA transfection on the activity of GCs in vitro
The results were shown in Figure 4, compared with the NC siRNA control group, the proliferation of GCs transfected with POSTN-siRNA gradually decreased over time. A significant difference was observed on day 2, with a marked reduction in GCs activity. These results indicate that interference with POSTN severely affected GCs proliferation. Furthermore, addition of 10 ng/mL FSH did not counteract the inhibitory effect of POSTN-siRNA on GCs activity. This result suggested that POSTN may be a key factor influencing GCs activity, and the subsequent interference duration for POSTN-siRNA was determined to be 48 h.
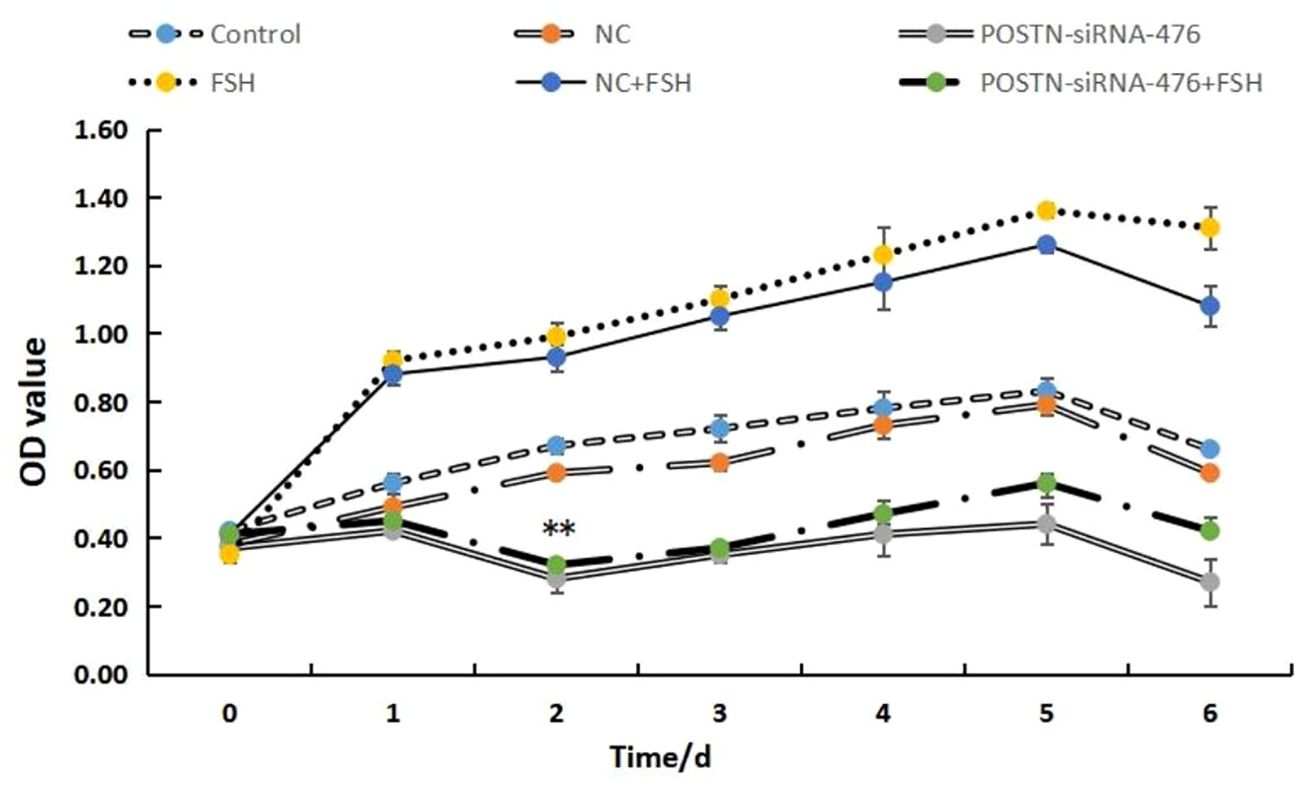
Figure 4. Effect of POSTN-siRNA on the activity of GCs in vitro. NC: negative control. Control: Blank control. POSTN-siRNA: transfected POSTN-siRNA. FSH: After 6 h of basal medium culture, 10 ng/mL FSH was added to the fresh culture medium of the GCs. NC + FSH: 6 h after transfection with NC, 10 ng/mL FSH was added to the replaced GCs medium. POSTN-siRNA + FSH:6 h after transfection of POSTN-siRNA, add 10 ng/mL to the replaced GCs medium FSH. ** indicates extremely significant difference (p<0.01).
3.5 POSTN promoter cloning and analysis
Analysis of the POSTN promoter using NCBI BLAST, TFSEARCH, and JASPAR software revealed transcription factor-binding sites, including JUND and SRF (Figure 5).

Figure 5. Putative transcription factor binding sites (boxed sequences) were predicted by a JASPAR program.
3.6 Analysis of POSTN promoter activity
The promoter activities of POSTN were analyzed and are shown in Figure 6A. The result showed that the activity of plasmid pPOSTN-547(-387/+160) was significantly higher than that of pGL3-basic(p<0.05). The activity of plasmid pPOSTN-715(-555/+160) was slightly higher than that of pPOSTN-547(-387/+160), but there was no statistically significant difference (p>0.05). The activity of plasmid pPOSTN-945(-785/+160) was higher than pPOSTN-715(-555/+160) but no significant different with that of plasmid pPOSTN-1157(-997/+160), pPOSTN-1376(-1216/+160), pPOSTN-1599(-11439/+160) and pPOSTN-2053(11893/+160) (p>0.05). All these data suggested that there was a core positive regulatory element in the region within -387 to -203 bp.
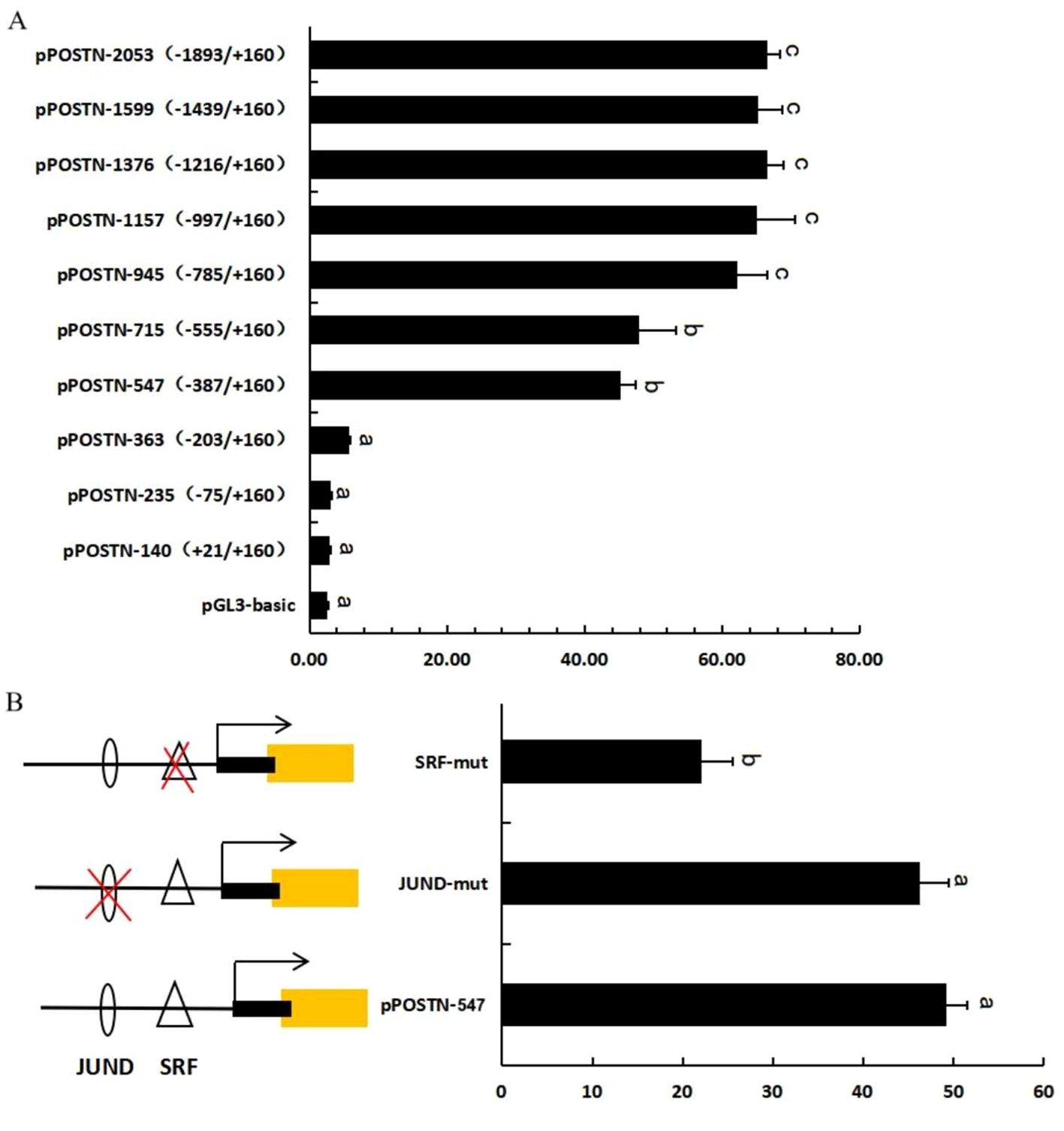
Figure 6. POSTN promoter activity and site-directed mutagenesis.Activity of the ovine POSTN promoter in 293T cells (A). The effect of site-directed mutagenesis on POSTN promoter activity in ovine GCs (B). Bars with different letters are significantly different (p<0.05). The X-axis represents relative luciferase activity.
Analysis of the POSTN core promoter region (-387 to -203 bp) using TESEARCH and JASPAR software revealed one SRF and one JUND-binding site. Base mutations in transcription factor-binding sites in the pPOSTN-547 vector were identified by overlapping PCR and constructing mutant plasmids. The mutant plasmids were transfected into 293T cells and detected using a dual luciferase assay (Figure 6B). Results showed mutations in the SRF-binding site at -222 to -210 bp significantly reduced POSTN transcriptional activity, while mutations in the JUND site did not significantly affect POSTN activity. This indicates SRF may be the core transcription factor of POSTN.
3.7 ChIP identification of transcription factor binding
To validate the binding of SRF and POSTN promoter, ChIP assay was conducted. After cross-linking genomic DNA with protein, the fragment was fragmented to 300–600 bp (Figure 7A) for ChIP reaction. ChIP-purified samples served as templates for PCR amplification. If the PCR sample showed a band, the DNA fragment could bind to the protein corresponding to the antibody. Results in Figure 7B demonstrated that SRF antibody precipitated the POSTN promoter fragment containing the SRF binding site in GCs. The anti-SRF group showed a deep band after 24 h of FSH treatment, which increased SRF enrichment in the promoter region of POSTN. We verified that SRF binds directly to the predicted SRF-binding site in the -222 to -210 region of POSTN promoter to regulate POSTN expression.
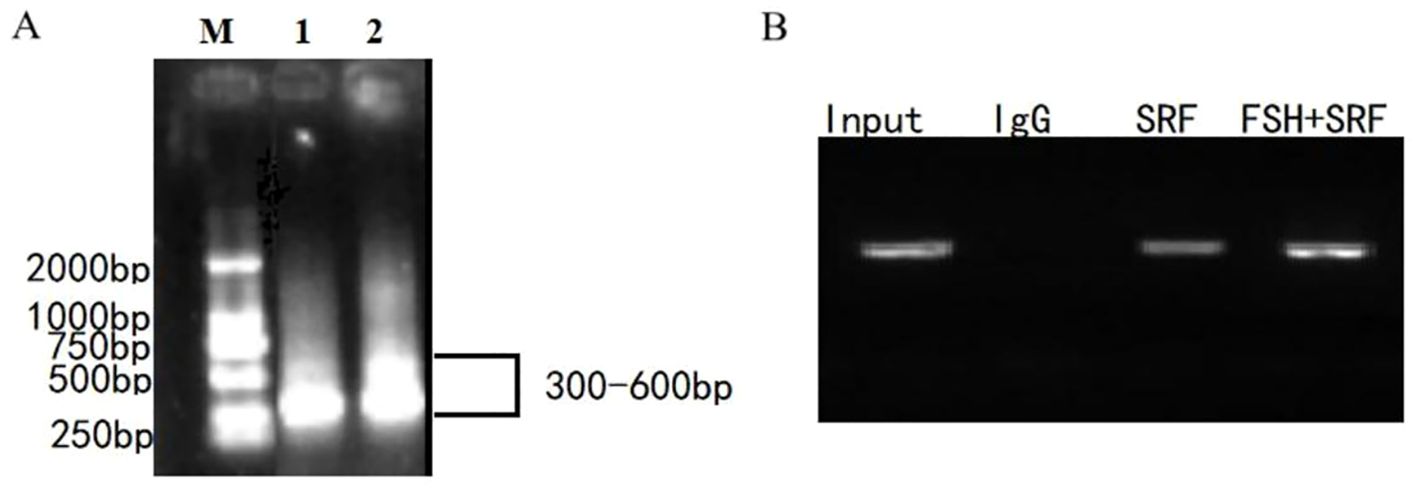
Figure 7. SRF binding to the POSTN promoter.Fragments of 300-600 bp were generated after Chromatin in GCs was sheared with sonication (A). ChIP analysis of SRF binding to the POSTN promoter region in ovine GCs (B). DNA was isolated from the ChIP precipitation complex and used as the PCR template. The PCR products were detected using a 2% agarose gel. POSTN DNA precipitated with the SRF antibody indicated the degree of enrichment of the SRF protein. IgG was used as the negative control.
4 Discussion
Follicular development in mammals depends on the intricate interplay between endocrine hormones and intrafollicular factors. It has been demonstrated that FSH is an important signaling molecule involved in follicular GCs growth. It forms a complex regulatory network with follicular development-related target genes that work together to maintain follicular life activities including cell proliferation, differentiation, and programmed cell death (Asselin et al., 2000; Regan et al., 2015; Rybska et al., 2018; Yilong et al., 2020). In recent years, the genes involved in cell proliferation and apoptosis have attracted considerable attention. POSTN is highly expressed in various tissues (periosteum, teeth, lungs, heart, uterus, and blood vessels) and activates FAK and PI3K/Akt via extracellular matrix (ECM) receptors to regulate cellular functions. It mediates cell recruitment, migration, adhesion, proliferation, and differentiation (Dong et al., 2017; Li et al., 2021; Jia et al., 2021). At different doses and durations, POSTN promoted cellular senescence and ECM metabolism in a dose- and time-dependent manner (Zhu et al., 2024). Intramuscular GnRH injection for 24 h increased POSTN mRNA levels and increased the dominant follicle count in bovine follicles (Lussier et al., 2017). These results implied that POSTN is involved in follicular development and ovulation.
In this study, we found that FSH treatment significantly increased the POSTN mRNA and protein levels in ovine GCs. POSTN expression increased in a time-dependent manner, peaking 24 h post-treatment. These results indicated that POSTN acts as a key downstream mediator of GCs. POSTN knockdown via siRNA significantly inhibited GCs viability, which could not be rescued by FSH, indicating that POSTN was indispensable for GCs function. To explore the transcriptional regulation of POSTN, we cloned its promoter and identified two putative binding sites, SRF and JunD. Luciferase assays showed that SRF binding significantly enhanced POSTN promoter activity, whereas JunD mutation had no effect. ChIP assays confirmed SRF enrichment at the POSTN promoter after FSH stimulation. These transcription factors may regulate POSTN expression through different signaling pathways.
SRF is a serum response factor and the main member of the MADS-box (MCM1, AGAMOUS, DEFICIENS, and SRF) family of transcription factors in animals (Wardle, 2019). SRF and MEF2 regulate downstream initiation and expression by specifically recognizing and binding to their corresponding cis-acting DNA elements (Morita and Hayashi, 2023). Both SRF and MEF2 transcription factors contain MADS-box functional domains and are homologous proteins, and their DNA binding sites are rich in A/T sequences, however, at the same time, there are obvious differences between the two protein structures and their DNA binding sites. SRF can correctly recognize and bind to the functional CArG box, which helps activate downstream target molecules for transcription (Onuh and Qiu, 2021). SRF also regulates cardiac homeostasis, maintains skeletal muscle function, and is involved in the regulation of cancer, digestive disorders, and neurological disorders (Luo et al., 2020). It has been shown that SRF expression is associated with the regulation of NLRP 3 inflammatory vesicles, autophagy, and necrosis in vascular endothelial cells through siRNA-mediated gene silencing. SRF expression controls the expression of cytoskeletal proteins and its expression increases with age (Chiu et al., 2024). Regulatory signaling in the SRF response occurs mainly through mitogen-activated protein kinase (MAPK) or RhoA GTPase pathways (Miano, 2010). SRF is a ubiquitously expressed transcriptional regulator that binds to the promoter regions of target molecules and directly regulates the activity of early genes, thus participating in cellular processes, such as apoptosis, cell proliferation, and differentiation (Kong et al., 2019). Mutations or abnormalities in SRF may cause the death of an organism. Knockdown during the mouse embryonic stage failed to form proto-gut embryos and mesoderm, resulting in death. It can be seen that SRF plays a critical regulatory role during embryonic development (Schwartz et al., 2014). Upstream of SRF, FSH signaling via its G-protein-coupled receptor raises intracellular cAMP and activates PKA, while concurrently engaging the MEK/ERK and p38/MK2 cascades. These pathways not only phosphorylate downstream effectors but also enhance SRF recruitment to Serum Response Elements (such as the Egr-1 promoter), thereby driving target gene transcription (Heidenreich et al., 1999; Russell et al., 2003). Our study suggests that SRF acts as a direct transcriptional activator of POSTN, mediating FSH’s effect of FSH on GCs. JunD family of proteins includes multifunctional activator protein-1 (AP-1) (Diaz-Cañestro et al., 2019). AP-1 transcription factors are dimerization complexes composed of members of three families of DNA-binding proteins: Jun (c-Jun, JunB, v-Jun, and JunD), Fos (Fra-1, Fra-2, c-Fos, and FosB), and ATF/CREB (ATF1-4, ATF-6, β-ATF, and ATFx) (Ruiz et al., 2021). JunD is specifically involved in oxidative stress, cell differentiation, transcriptional repression, and the activation of proliferative genes. The absence of JunD leads to the abnormal activation of the genetic program, resulting in cell death (Maślikowski et al., 2016). JunD inhibits the proliferation of mouse embryonic fibroblasts (MEFs). Although JunD is commonly implicated in oxidative stress responses and transcriptional repression, it appears dispensable for POSTN promoter activation in this context. In ovine GCs, other AP-1 family members likely compensate for JunD loss to sustain transcriptional activity, whereas JunD itself may predominantly regulate stress- or differentiation-related targets, contributing minimally to POSTN gene activation.
Overall, this study elucidated a novel FSH–POSTN regulatory axis in ovine GCs, offering new insights into folliculogenesis and potential targets for reproductive regulation. However, the intricate and time-consuming process of follicular development involving a multitude of pivotal regulators within the follicle remains poorly understood. Further investigation is required to elucidate the relationship between the intrafollicular regulatory factors.
5 Conclusion
FSH (10 ng/mL) markedly upregulates POSTN expression in ovine follicular GCs. The POSTN core promoter resides between -387 and -203 bp, encompassing SRF and JunD binding motifs. Among these, SRF emerged as the dominant transcriptional regulator of POSTN, and FSH enhances POSTN promoter activity primarily through SRF.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Science and Technology Ethics Committee of Tarim University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LP: Formal analysis, Writing – original draft, Investigation. WW: Data curation, Investigation, Writing – original draft. PG: Writing – original draft, Visualization, Data curation. XD: Resources, Writing – original draft. XX: Software, Writing – review & editing, Methodology, Funding acquisition. CL: Supervision, Conceptualization, Writing – review & editing, Methodology, Funding acquisition, Project administration, Validation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research grant is for study on the selection of sheep reproductive performance by transcriptome methylation and application of timed insemination technology (2023A02011-2-3) (CL, E-mail addresses: Z3VpbHQzNjlAMTYzLmNvbQ==), and the use of PE system to achieve precision editing in the sheep genome (TDZKBS202417) (XX, E-mail addresses: MTI0MTE0MTY5N0BxcS5jb20=).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2025.1637526/full#supplementary-material
References
Abudureyimu G., Wu Y., Wang L., Hao G., Chen Y., Yu J., et al. (2024). POSTN promotes granulosa cell proliferation in sheep follicles through focal adhesion. Gene Rep. 35, 18–90. doi: 10.1016/j.genrep.2024.101890
Asselin E., Xiao C. W., Wang Y. F., and Tsang B. K. (2000). Mammalian follicular development and atresia: role of apoptosis. Biol. Signals Recept 9, 87–95. doi: 10.1159/000014627
Chen Y. J., Lee M. T., Yao H. C., Hsiao P. W., Ke F. C., and Hwang J. J. (2008). Crucial role of estrogen receptor-α Interaction with transcription coregulators in follicle-stimulating hormone and transforming growth factor β1 up-regulation of steroidogenesis in rat ovarian granulosa cells. Endocrinology 149, 4658–4668. doi: 10.1210/en.2008-0063
Chiu H., Chou C., Lee K., Shih C., Huang T., and Sung L. (2024). Nattokinase attenuates endothelial inflammation through the activation of SRF and THBS1. Int. J. Biol. macromolecules 268, 131779. doi: 10.1016/j.ijbiomac.2024.131779
Diaz-Cañestro C., Reiner M. F., Bonetti N. R., Liberale L., Merlini M., Wüst P., et al. (2019). AP-1 (Activated protein-1) transcription factor junD regulates ischemia/reperfusion brain damage via IL-1β (Interleukin-1β). Stroke 50, 469–477. doi: 10.1161/STROKEAHA.118.023739
Dong Y., Zhi S. S., Shi J. Q., Qun L., and Guo L. W. (2017). Role and underlying mechanisms of the interstitial protein periostin in the diagnosis and treatment of Malignant tumors (Review). Oncol. Lett. 1, 5099–5106. doi: 10.3892/ol.2017.6866
Heidenreich O., Neininger A., Schratt G., Zinck R., Cahill M. A., Engel K., et al. (1999). MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J. Biol. Chem. 274, 14434. doi: 10.1074/jbc.274.20.14434
Ismail R. S., Baldwin R. L., Fang J., Browning D., Karlan B. Y., Gasson J. C., et al. (2000). Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 60, 6744–6749.
Jia Y. Y., Yu Y., and Li H. J. (2021). POSTN promotes proliferation and epithelial-mesenchymal transition in renal cell carcinoma through ILK/AKT/mTOR pathway. J. Cancer 12, 4183–4195. doi: 10.7150/jca.51253
Jiapeng L., Chunjie L., Liqin W., Ying C., Xiaolin L., Yangsheng W., et al. (2023). Expression analysis of POSTN gene in ovine follicles. Can. J. Anim. Sci. 103, 66–72. doi: 10.1139/cjas-2021-0036
Kong M., Chen X., Lv F., Ren H., Fan Z., Qin H., et al. (2019). Serum response factor (SRF) promotes ROS generation and hepatic stellate cell activation by epigenetically stimulating NCF1/2 transcription. Redox Biol. 26, :101302. doi: 10.1016/j.redox.2019.101302
Kulus M., Kranc W., Sujka-Kordowska P., Mozdziak P., Jankowski M., Konwerska A., et al. (2020). The processes of cellular growth, aging, and programmed cell death are involved in lifespan of ovarian granulosa cells during short-term IVC – Study based on animal model. Theriogenology 148, 76–88. doi: 10.1016/j.theriogenology.2020.02.044
Li C., Cheng D., Xu P., Nie H., Zhang T., and Pang X. (2021). POSTN promotes the proliferation of spermatogonial cells by activating the wnt/β-catenin signaling pathway. Reprod. Sci. 28, 2906–2915. doi: 10.1007/s43032-021-00596-1
Li X., Lin J., Chen Y., Wang L., Han B., Jia B., et al. (2022). FSH promotes the proliferation of sheep granulosa cells by inhibiting the expression of TSP1. Anim. Biotechnol. 33, 260–272. doi: 10.1080/10495398.2020.1789868
Liu Z., Dai L., Sun T., Liu Y., Bao Y., Gu M., et al. (2024). Massively parallel CRISPR-cas9 knockout screening in sheep granulosa cells for FSH response genes. Animals 14, 898. doi: 10.3390/ani14060898
Luo J., Jin F. Q., Yin M., and Jin Z. G. (2020). Regulation of srf protein stability by an autophagy-dependent pathway. Biochem. Biophys. Res. Commun. 521, 279–284. doi: 10.1016/j.bbrc.2019.09.104
Lussier J. G., Diouf M. N., Lévesque V., Sirois J., and Ndiaye K. (2017). Gene expression profiling of upregulated mRNAs in granulosa cells of bovine ovulatory follicles following stimulation with hCG. Reprod. Biol. Endocrinol. 15, 88. doi: 10.1186/s12958-017-0306-x
Maślikowski B. M., Wang L., Wu Y., Fielding B., and Bédard P. A. (2016). JunD/AP-1 antagonizes the induction of DAPK1 to promote the survival of v-src-transformed cells. J. Virol. 91, e01925–e01916. doi: 10.1128/JVI.01925-16
Miano J. M. (2010). Role of serum response factor in the pathogenesis of disease. Lab. investigation; J. Tech. Methods Pathol. 90, 1274–1284. doi: 10.1038/labinvest.2010.104
Morita T. and Hayashi K. (2023). Actin-related protein 5 suppresses the cooperative activation of cardiac gene transcription by myocardin and MEF2. FEBS Open Bio 13, 363–379. doi: 10.1002/2211-5463.13549
Onuh J. O. and Qiu H. (2021). Serum response factor-cofactor interactions and their implications in disease. FEBS J. 288, 3120–3134. doi: 10.1111/febs.15544
Orisaka M., Miyazaki Y., Shirafuji A., Tamamura C., Tsuyoshi H., Tsang B. K., et al. (2021). The role of pituitary gonadotropins and intraovarian regulators in follicle development: A mini-review. Reprod. Med. Biol. 20, 169–175. doi: 10.1002/rmb2.12371
Ratajczak K. and Dziegiel P. (2015). The role of periostin in neoplastic processes. Folia histochemica cytobiologica 53, 120–132. doi: 10.5603/FHC.a2015.0014
Regan S. L., McFarlane J. R., O’Shea T., Andronicos N., Arfuso F., Dharmarajan A., et al. (2015). Flow cytometric analysis of FSHR, BMRR1B, LHR and apoptosis in granulosa cells and ovulation rate in merino sheep. Reprod. (Cambridge England) 150, 151–163. doi: 10.1530/REP-14-0581
Ruan K., Bao S., and Ouyang G. (2009). The multifaceted role of periostin in tumorigenesis. Cell. Mol. Life sciences: CMLS 66, 2219–2230. doi: 10.1007/s00018-009-0013-7
Ruiz E. J., Lan L., Diefenbacher M. E., Riising E. M., Da Costa C., Chakraborty A., et al. (2021). JunD, not c-Jun, is the AP-1 transcription factor required for Ras-induced lung cancer. JCI Insight 6, e124985. doi: 10.1172/jci.insight.124985
Russell D. L., Doyle K. M., Gonzales-Robayna I., Pipaon C., and Richards J. S. (2003). Egr-1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3’,5’-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1. Mol. Endocrinol. (Baltimore Md.) 17, 520–533. doi: 10.1210/me.2002-0066
Rybska M., Knap S., Jankowski M., Ješeta M., Bukowska D., Antosik P., et al. (2018). Characteristic of factors influencing the proper course of folliculogenesis in mammals. Med. J. Cell Biol. 6, 33–38. doi: 10.2478/acb-2018-0006
Schwartz B., Marks M., Wittler L., Werber M., Währisch S., Nordheim A., et al. (2014). SRF is essential for mesodermal cell migration during elongation of the embryonic body axis. Mech. Dev. 133, 23–35. doi: 10.1016/j.mod.2014.07.001
Suocheng W., Zhuandi G., Li S., Haoqin L., Luju L., and Yingying D. (2017). Maturation rates of oocytes and levels of FSHR, LHR and GnRHR of COCs response to FSH concentrations in IVM media for sheep. J. Appl. Biomedicine 15, 180–186. doi: 10.1016/j.jab.2017.01.001
Syed V., Zhang X., Lau K. M., Cheng R., Mukherjee K., and Ho S. M. (2005). Profiling estrogen-regulated gene expression changes in normal and Malignant human ovarian surface epithelial cells. Oncogene 24, 8128–8143. doi: 10.1038/sj.onc.1208959
Takayama I. and Kudo A. (2012). Periostin in dental science. Japanese Dental Sci. Rev. 48, 92–98. doi: 10.1016/j.jdsr.2012.02.001
Takeshita S., Kikuno R., Tezuka K., and Amann E. (1993). Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem. J. 294, 271–278. doi: 10.1042/bj2940271
Wardle F. C. (2019). Master control: transcriptional regulation of mammalian Myod. J. Muscle Res. Cell Motil. 40, 211–226. doi: 10.1007/s10974-019-09538-6
Xu X., Wang R., Pei L., Wang Q., and Liu C. (2025). Glucose transport by follicle-stimulating hormone is mediated through the akt/FOXO1 pathway in ovine granulosa cells. Veterinary Med. Sci. 11, e70294. doi: 10.1002/vms3.70294
Yilong Y., Yefen X., Suolang S., Yuheng W., Yun C., Qiangba Y., et al. (2020). BMP15 miR-31 FSHR axis regulates yak ovarian granulosa cell proliferation and P4 secretion. Arch. Clin. Obstetrics Gynecology Res. 1, 1–8. doi: 10.33425/2768-0304.1004
Yu Y., Tan C. M., and Jia Y. Y. (2021). Research status and the prospect of POSTN in various tumors. Neoplasma 68, 673–682. doi: 10.4149/neo_2021_210223N239
Zhu D., Chen S., Sheng P., Wang Z., Li Y., and Kang X. (2024). POSTN promotes nucleus pulposus cell senescence and extracellular matrix metabolism via activing Wnt/β-catenin and NF-κB signal pathway in intervertebral disc degeneration. Cell. Signalling, 121, 111277. doi: 10.1016/j.cellsig.2024.111277
Keywords: GCS, Postn, FSH, SRF, folliculogenesis, ovis
Citation: Pei L, Wang W, Guo P, Duan X, Xu X and Liu C (2025) FSH-mediated regulation of POSTN expression in ovine granulosa cells via SRF transcription factor. Front. Anim. Sci. 6:1637526. doi: 10.3389/fanim.2025.1637526
Received: 29 May 2025; Accepted: 18 July 2025;
Published: 20 August 2025.
Edited by:
Shaobin Li, Gansu Agricultural University, ChinaCopyright © 2025 Pei, Wang, Guo, Duan, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Xu, MTI0MTE0MTY5N0BxcS5jb20=; Chunjie Liu, Z3VpbHQzNjlAMTYzLmNvbQ==
 Linlin Pei
Linlin Pei Wenhao Wang1,2
Wenhao Wang1,2 Peilin Guo
Peilin Guo Xin Xu
Xin Xu Chunjie Liu
Chunjie Liu