- College of Animal Science and Technology, Yangzhou University, Yangzhou, China
With the increasing global demand for dairy products, the improvement of cow performance has become the core topic of the development of dairy farming industry, and the synergy of genetics and nutrition will become one of the keys to solve this problem. Based on the latest research progress and integration of existing studies, we systematically reviewed the effects of genetic and nutritional factors (as well as their synergistic interactions) on dairy cow performance. Genetic factors can improve key performance indicators such as milk yield, milk protein percentage and milk fat percentage by regulating specific genes and their key pathways. At the same time, nutritional intervention for dairy cows, such as optimizing diet structure and adding relevant functional additives, can also significantly improve the key performance indicators of dairy cows. The synergy of the two factors, through the bridge mediated by the rumen microbial community, can improve the genetic potential of production traits with genetic factors as the dominant factor and nutritional factors as the auxiliary factor to maximize the expression of genetic potential, or break through the limitation of a single factor. However, there is a lack of in-depth analysis and research on the underlying molecular mechanisms of this key pathway. The vast majority of the related studies that have been conducted only focus on the changing patterns of apparent traits through comparative experiments. As a result, an accurate linkage model between genetic and nutritional factors could not be established, limiting the development of practical solutions for sustainable and efficient animal husbandry. Future research can focus on more in-depth analysis using multi-omics technology, especially on the deeper molecular mechanism of the nutrition-genetic-rumen microbial-performance pathway, so as to promote a closer linkage between genetic and nutritional factors and provide a feasible method for the continuous and efficient development of animal husbandry.
1 Introduction
In the past few decades, the dairy industry has made remarkable achievements, and some breeds of dairy cows have greatly increased their yields (Brito et al., 2021). Some of these gains were due to selection and genetic improvement, and the rest were due to advances in nutrition and management (Baumgard et al., 2017). In the aspect of genetics, the macroscopic manipulation of genetic breeding and the microscopic analysis of molecular mechanisms all confirm that human beings are strengthening the improvement of production performance step by step through this key factor. The breeding of hybrid Holstein dairy cows and the analysis of mTOR and its pathways that regulate milk protein and fat content (Burgos et al., 2010) provided experimental support for this viewpoint. With the development of genomic technology (Risch and Merikangas, 1996; Klein et al., 2005), the use of genetic factors to improve the performance of dairy cattle will be more systematic and refined, the excavation of genetic factors affecting the performance of cattle will be more in-depth, and the genetic improvement of dairy cattle will therefore move towards a new era.
In terms of nutrition, the feeding method for dairy cows has evolved from the early extensive grazing to the partial addition of concentrate feed to roughage after the recognition of the importance of nutrition to the production performance of dairy cows, and then to total mixed ration (TMR) (McCoy et al., 1966) and the addition of some nutrients in the feed (Anil et al., 1993; Oliveira et al., 2017). Human beings have increasingly delved into the regulation of dairy cow production performance through nutritional factors, from macro nutrient supply to a precise intervention system based on digestion and metabolism regulation. The development of metabolomics (Nicholson et al., 1999) has not only accelerated this process but also significantly improved the relevant production performance of dairy cows.
It can be seen from this that both genetic and nutritional factors have significant regulatory and influential roles on the production performance of dairy cows. However, the regulation of a single factor has obvious limitations. These limitations are specifically manifested as follows: adequate nutritional supply is an indispensable condition for the full expression of genetic potential, while the lack of a favorable genetic basis severely restricts the upper limit of the final effect that nutritional intervention can achieve. For example, under the same nutritional conditions, the genomic estimated breeding value (GEBV) of 305-day milk yield of genotyped samples can differ by up to 800 kg (Oliveira et al., 2023). Therefore, when genetic and nutritional factors can form a synergy, it is expected to overcome these inherent constraints of single-factor regulation, thereby opening up more effective paths for the regulation of production performance.
To achieve the coordinated regulation of dairy cow production performance by genetic and nutritional factors, a bridge that links the regulatory relationship between the two is needed. As a unique symbiotic microbial community in ruminants, rumen microorganisms in dairy cows can be regulated by nutritional factors (Henderson et al., 2015; Belanche et al., 2020), are closely related to individual genes (Weimer et al., 2010), and can have a certain impact on the production performance of dairy cows (Li et al., 2019). Therefore, this bridge of gene-microbe-nutrition may provide a new focus for breaking through the existing production bottlenecks.
2 Effects of genetic factors on performance of dairy cows
2.1 Evolution of genetically-driven breeding techniques to improve performance in dairy cattle
The production performance of dairy cows is significantly influenced by genetic factors. For instance, although Holstein cows produce more milk than Jersey cows, their milk quality indicators such as fat content are lower than those of Jersey cows (Palladino et al., 2010). People have long recognized the impact of these genetic factors on the production performance of dairy cows. As early as the late 19th century, Danish farmers proposed that “the ability of a cow to produce more or less milk fat from the feed it consumes depends on genetics” and verified this on their own farms (BONNIER, 1936). The United States Department of Agriculture established a dedicated dairy department as early as 1895, exploring the impact of genetic effects on milk production through various breeding strategies and proving through experiments that genetic factors could be utilized through breeding to enhance the production capacity of dairy cows (Hodgson, 1956). By the early 20th century, it was observed in production practice that there might be a connection between a cow’s milk production and that of its mother, and research (Edwards, 1932) confirmed the relative accuracy of comparing mother and daughter. However, this comparison method ignored the changes in genetic trends over time and the influence of the environment on production performance data, leading to significant deviations in genetic prediction values (Weigel et al., 2017). With the innovation of genetic evaluation techniques, the methods of dairy cow breeding have also advanced. In the mid-20th century, the contemporary (herdmate) comparison method was proposed (Robertson and Rendel, 1954). By grouping the cattle according to their common physiological characteristics and lifestyle, it was found that this method could take into account the specific management and environmental conditions at the time of phenotypic expression (Robertson et al., 1956), marking another step forward in understanding the impact of genetic factors on the production performance of dairy cows. In the following years, the demand for more precise selection of dairy cows with higher production performance based on genetic factors has grown increasingly. With the development of methods such as animal model best linear unbiased prediction (BLUP) (Henderson, 1953), whole-genome selection (Nejati-Javaremi et al., 1997; Meuwissen et al., 2001), and genomic prediction through machine learning methods (Long et al., 2007), the selection of high-production-performance dairy cows based on genetic factors has become increasingly accurate and convenient for humans.
The content above can be clearly and concisely presented by Figure 1.
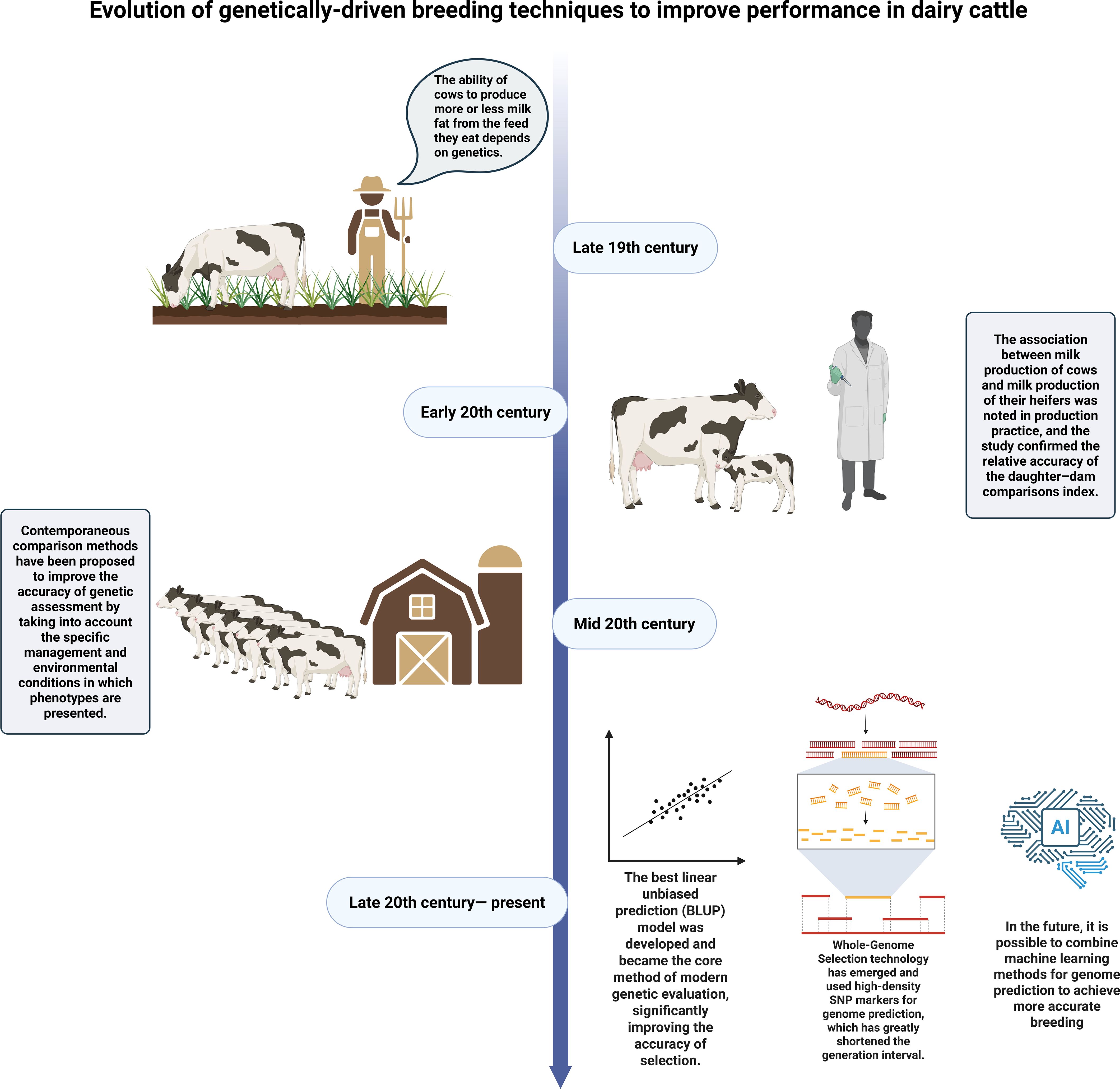
Figure 1. Evolution of genetically-driven breeding techniques to improve performance in dairy cattle.
2.2 Genetic factors affect key indicators of dairy cow performance through the regulation of specific genes and their involved pathways
Recent experiments have shown that the Csp6I polymorphism of the FGF-2 gene has a significant impact on milk production indicators such as peak milk yield (PMY), lactation milk yield (LMY), milking time (MT), and 305-day milk yield. Moreover, without affecting the reproductive performance of Holstein cows, the a allele and AA genotype of the FGF-2/Csp6I gene can significantly increase milk production (Kibar and Aytekin, 2025). Besides directly regulating milk production traits, genetic factors can also indirectly regulate lactation performance by influencing host-microbe interactions. Experimental results have indicated that chr25:111,177 (5s_rRNA) may promote the ecological niche advantage of Veillonella parabutyrica in the rumen microbiota through the action of lysozyme GH24, altering the rumen microbiota composition and thereby affecting nutrient absorption and milk component synthesis, which in turn influences lactation performance (Zhang et al., 2024). To identify the genomic regions that regulate lactation in dairy cows, one can compare the genomic differences between lactating and non-lactating groups to initially determine the genomic regions that may regulate lactation. For instance, the study (Si et al., 2025) identified a unique selection signature in the genomic region containing the casein genes (CSN1S1, CSN2, and CSN1S2) by exploring the genomic differences between dairy water buffaloes and non-dairy water buffaloes. These genes were later found to be significantly associated with lactation traits in mammals (Bonfatti et al., 2010, 2012). With the continuous progress of genomic analysis, genes related to lactation traits are constantly being discovered and analyzed, and new candidate genes are being sought. Recently, Teng et al. used the genotypes of 1,059 Holstein cows from the 1,000 Bull Genomes Project as a reference panel to impute the chip data of 6,470 Chinese Holstein cows to whole-genome sequence data. Then, based on a random regression test-day model, they conducted a longitudinal genome-wide association analysis of milk production traits using the imputed sequence data and identified 130 QTL regions related to milk production traits such as milk yield and estimated the 95% confidence intervals for each QTL region. Among these confidence intervals, not only genes such as DGAT1, HSF1, MGST1, GHR, ABCG2, ADCK5, and CSN1S1 that have been reported in various literature to be related to milk production traits in dairy cows were found, but also candidate genes such as CCSER1, CUX2, SNTB1, RGS7, OSR2, and STK3 that show good potential were discovered (Teng et al., 2023). The key genes and pathways obtained from these experiments have demonstrated that genetic factors can influence the lactation performance of dairy cows through their regulatory roles. These key genes and pathways will serve as important anchors for future breeding or improvement of high-lactation dairy cows.
Apart from increasing milk yield, improving the efficiency and quality of milk protein synthesis is also an ideal goal for the dairy industry, which remains a continuous challenge (Bionaz et al., 2012). Currently, many genes related to milk protein synthesis have been reported, among which STAT5 and mTOR genes are particularly crucial and important. Studies have found that the expression of milk protein genes in dairy cows’ mammary glands is mainly regulated by prolactin through the STAT5A pathway, and the STAT5A gene may be a marker gene for dairy cow production performance. Mutations in the STAT5A gene can cause changes in milk yield and composition (Brym et al., 2004). Research has shown that amino acids promote milk protein synthesis in vitro when mammary alveoli are cultured under different nutritional and hormonal conditions, leading to the conclusion that the mTOR gene mediates the regulation of milk protein synthesis through nutrients and hormones (Burgos et al., 2010). Regarding milk protein quality, in 2003, Brophy et al. introduced gene copies encoding bovine β- and κ-casein (CSN2 and CSN3, respectively) into fibroblast cells from adult cows. After nuclear transfer using four independent donor cell lines, transgenic calves expressing additional copies of β- and κ-casein were obtained (Brophy et al., 2003). Subsequent detailed analysis of the milk produced by these transgenic calves revealed that the expression of these transgenes led to changes in individual milk proteins and micelle size, but did not affect the total protein concentration (Laible et al., 2016). Finally, to identify which genes affect milk protein synthesis, it is possible to compare the expression profiles of high and low milk protein rate dairy cow mammary tissues. To test whether lactation affects the expression of individual genes, it is also necessary to compare the common differentially expressed genes (DEGs) in mammary tissues during peak lactation and non-lactation periods. After integrated analysis, key genes affecting milk protein synthesis can be proposed (Li et al., 2016). Similar to the lactation traits, the discovery of these key genes not only demonstrates that genetic factors can regulate and influence the milk protein-related traits of dairy cows through specific genes, but also paves the way for future breeding of high-milk-protein dairy cows and the improvement of milk protein yield and quality.
In addition to the quantity and quality of milk proteins, the indicators of milk quality also include the amount and rate of milk fat, as well as the content of fatty acids. These indicators together describe the quantity and quality of milk fat. Among them, the synthesis of milk fat in the mammary gland is regulated by numerous genes and the pathways they participate in. For instance, Gu et al. identified miRNAs in cattle by cloning small RNAs from adipose tissue and mammary glands, and concluded that miRNAs affect milk fat metabolism and biosynthesis (Gu et al., 2007). On this basis, Wang et al. clarified the inhibitory effect of miR-34b on milk fat synthesis in bovine mammary epithelial cells and demonstrated that miR-34b inhibits milk fat synthesis in bovine mammary epithelial cells by reducing phosphorylation levels and modulating the Akt/mTOR signaling pathway (Wang et al., 2021). In contrast, a team studying buffalo mammary epithelial cells (BuMEC) found that liver X receptor (LXR) can enhance lipid synthesis in bovine and caprine mammary epithelial cells by mediating the transcription of SREBP1 gene (Zhang et al., 2021). Milk, as a common drink, not only has extremely high nutritional value but also has good preventive and health care functions for many diseases (Pereira, 2014; Stergiadis et al., 2019), and these functions are closely related to the composition and content of fatty acids in milk (Jiao et al., 2020), thus making it a key indicator of milk quality. Research has shown that RNA interference of EEF1D in bovine mammary epithelial cells leads to abnormal formation of lipid droplets, and EEF1D gene can regulate the content of triglycerides in bovine mammary epithelial cells through insulin (PI3K-Akt), AMPK and PPAR pathways. Additionally, Chen et al. found that circRNA (circ09863) can promote the synthesis of triglycerides (Triacylglyceride, TAG) in bovine mammary epithelial cells and up-regulate the expression levels of C16:1 and C18:1 (Chen et al., 2020). All these studies indicate that genetic factors regulate milk quality through specific genes and the pathways they participate in.
Except for the milk quantity and quality traits that are highly related to genetic factors as mentioned above, the body size and growth rate of dairy cows are also regulated by genetic factors. Body size and growth rate have always been important selection indicators in livestock breeding, although they are more crucial in the breeding of meat cattle. However, they cannot be ignored in dairy cattle breeding, especially in comprehensive breeding strategies where they need to be balanced and taken into account. Bai et al. innovatively used the Ancestral Recombination Graph (ARG) analysis method to precisely identify the key mutations that drive the selection sweep of the LCORL and STC2 loci in beef cattle breeds, which can significantly enhance the body size and growth rate of beef cattle (Bai et al., 2025). Coincidentally, Rodrigues’s team strictly selected 24 Charolais crossbred cattle from 344 individuals and divided them into two groups based on the LCORL-NCAPG haplotype: 12 carrying the double-copy “high growth” haplotype (QQ) and 12 carrying the ancestral haplotype (qq). Longissimus dorsi muscle samples were collected from these cattle at 300 days of age for RNA sequencing. The study found that the expression of LCORL was significantly increased in QQ individuals, while the expression of key fat synthesis genes FASN (fatty acid synthase) and LEP (leptin) was downregulated by 1.2 and 3.6 times, respectively. When the results were extended to the entire population of 344 Charolais crossbred cattle, 50K chip data analysis showed that QQ haplotype carriers had significantly higher birth weight (BW), yearling weight (YW), and eye muscle area (REA), while backfat thickness (BF) and marbling score (MS) were reduced. This confirmed the consistent effect of this haplotype in promoting growth and reducing fat at the population level (Rodrigues et al., 2025). Although these studies were conducted on beef cattle breeds such as Simmental and Charolais in Central Europe, the discovery of genes directly related to body size and growth traits can provide important references for the genetic mechanism of body size scoring in dairy cattle. This key mutation can be further validated through targeted experimental studies to determine its occurrence in dairy cows and its potential effects on body conformation and growth performance.
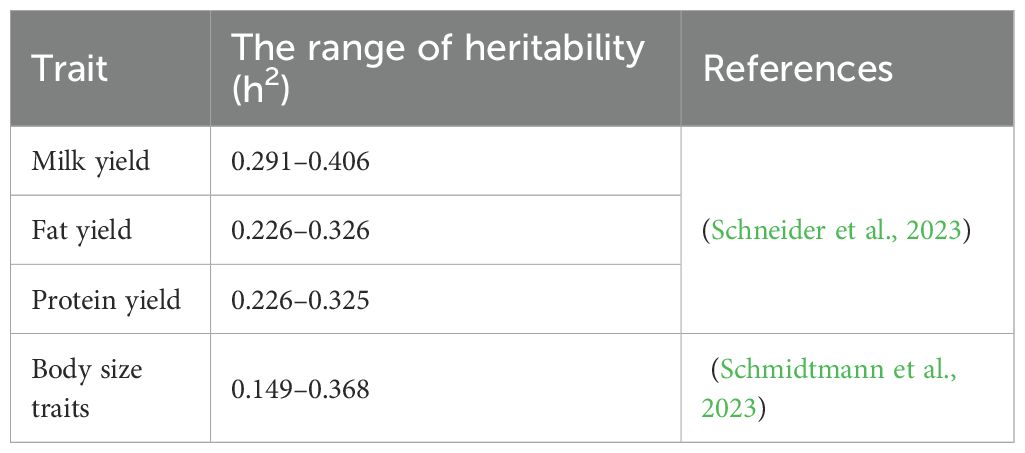
The response degree of the above-mentioned traits to genetic regulation varies, and this difference is essentially determined by the heritability of the traits. Therefore, this paper integrates relevant literature data and summarizes the estimated heritability values of the above-mentioned main traits in Table 1.
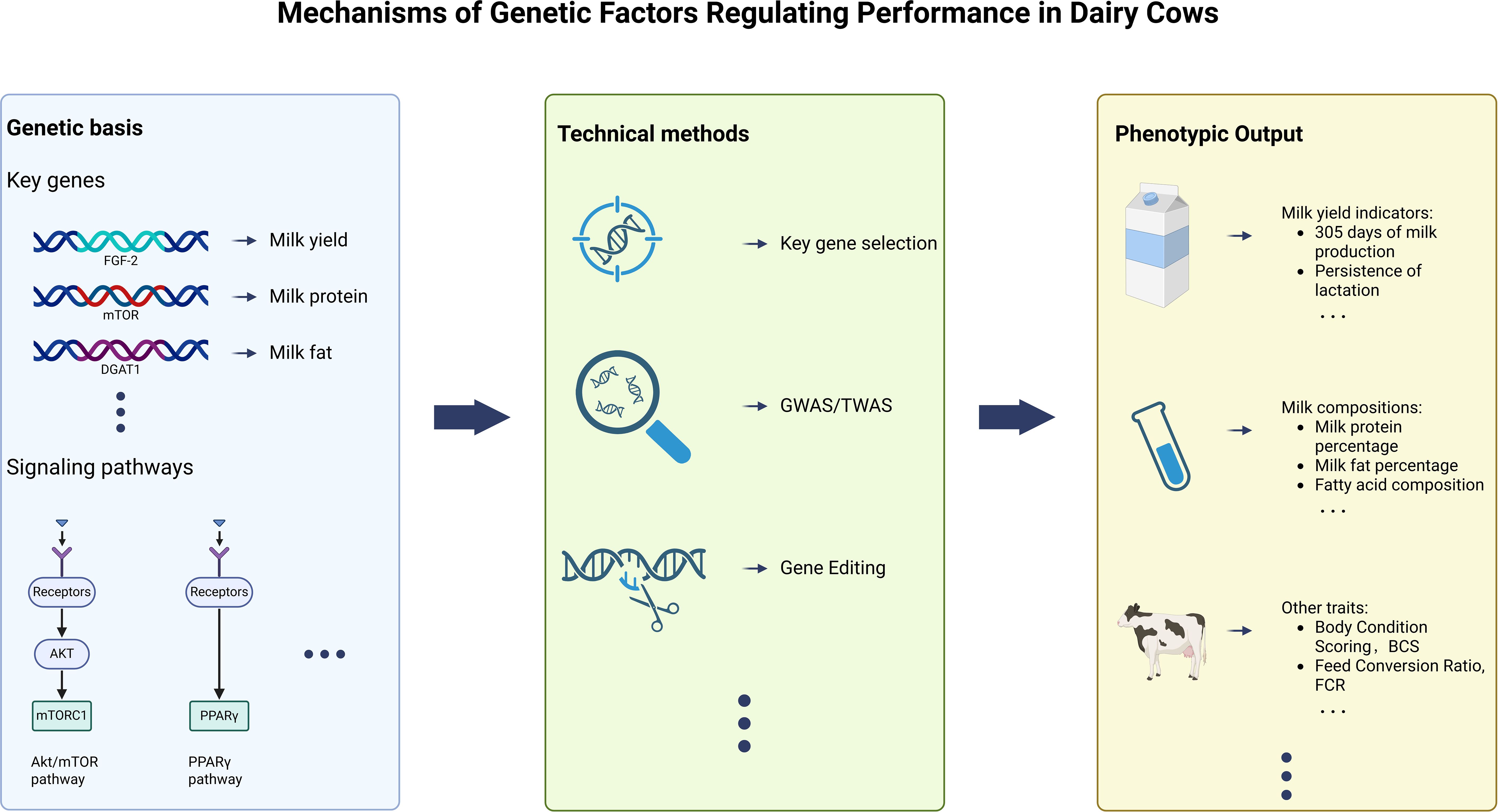
The process by which the aforementioned genetic factors influence the key indicators of dairy cow production performance through specific genes and their involved pathways can be visually presented in Figure 2.
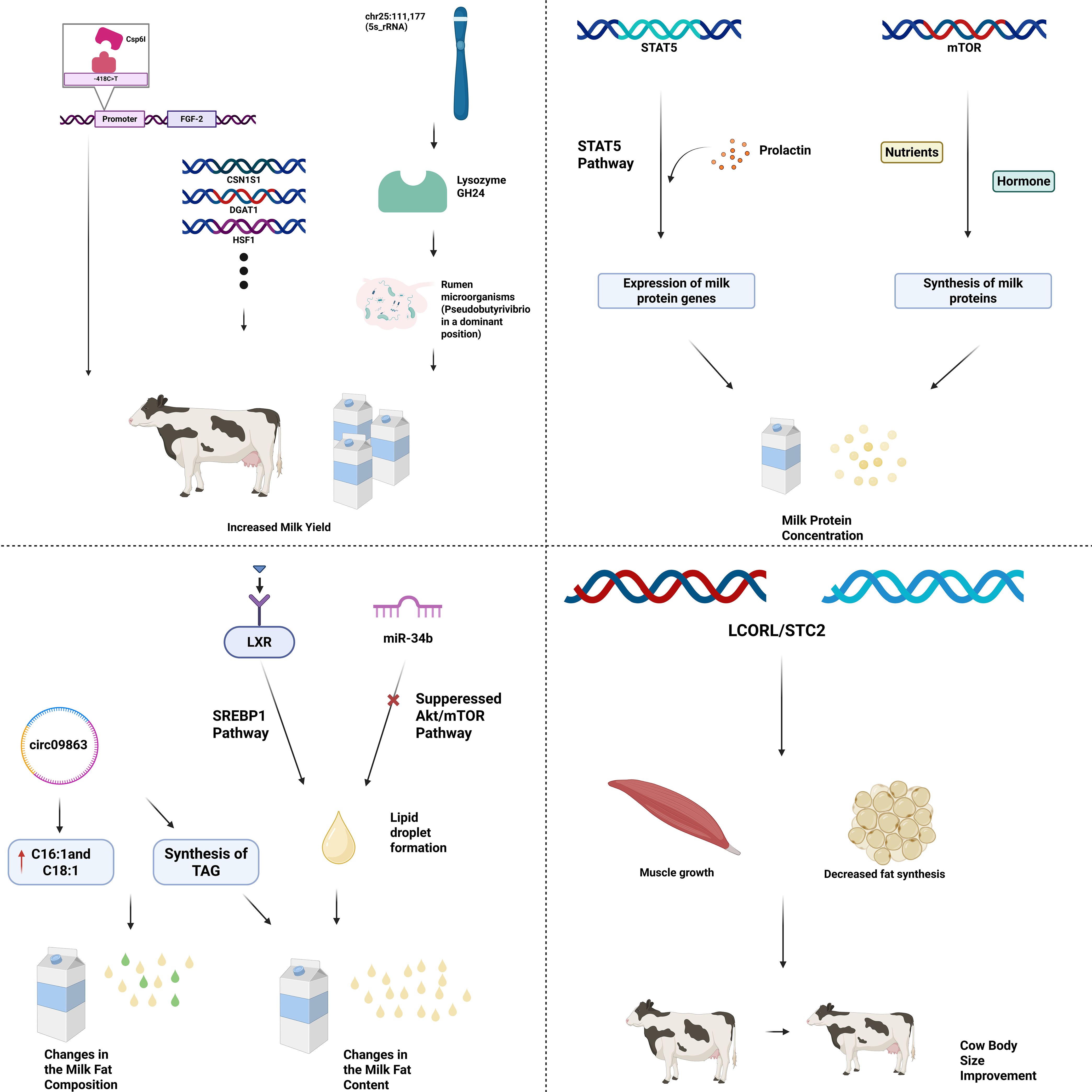
Figure 2. Genetic factors affect key indicators of dairy cow performance through the regulation of specific genes and their involved pathways.
2.3 Further analysis of genetic factors in dairy cow performance
With the development of genomics technology, genetic breeding has become increasingly systematic and precise. From Mendel’s early observations of the dominant and recessive traits of pea offspring to infer the transmission pattern of genetic factors, to chromosome karyotype analysis, metabolic pathway research by analyzing metabolic products, and then to genetic marker and linkage analysis as well as candidate gene association studies, human research on genetic variation has gradually deepened. However, traditional methods have significant limitations, including but not limited to the inability to directly analyze the entire genome sequence, the need to infer gene function through phenotypes, low throughput, and longtime consumption. In 1996, Neil Risch (Risch and Merikangas, 1996) proposed to break away from the constraints of candidate genes and directly scan the association between genome-wide variations and phenotypes. This idea became the theoretical foundation of GWAS(Genome-wide association study), an analytical method used for interpreting the association between genomic variations and biological phenotypes. Based on this, Klein et al. first applied GWAS to human diseases in 2005, opening a new era in the study of complex genetic mechanisms. Thus, despite its many limitations, including genetic loss, complex traits, limited functional understanding, and sample size requirements (Walsh et al., 2023), it has been widely used in the identification of genetic variations in human and livestock populations, and has identified thousands of genetic variations related to complex traits (Buniello et al., 2019; Hu et al., 2019). Since the genetic markers obtained by GWAS are often located in non-coding regions, identifying candidate genes for complex traits becomes relatively more difficult. To understand the molecular regulatory mechanisms of important economic traits in cattle and improve the genetic progress of artificial selection, Liu et al. used 7,180 public transcriptome data to systematically construct the Cattle Genotype-Tissue Expression atlas (CattleGTEx). Using this atlas, the team linked complex traits with gene expression through transcriptome-wide association analysis (TWAS), thereby systematically locating candidate genes regulating 43 important economic traits in cattle. Among them, the team verified the major gene DGAT1 affecting milk protein content, milk fat percentage and other milk production traits in dairy cows, and further revealed that the expression of this gene in the liver is regulated by genetic variations, thereby affecting the milk production performance of dairy cows (Liu et al., 2022a). This research can be regarded as a further application of GWAS in the new era, and has a milestone role in exploring genetic factors affecting the production performance of cattle and improving the accuracy of genomic selection.
At the end of this section, Figure 3 can simply summarize in graphical form the mechanism by which genetic factors regulate the production performance of dairy cows.
3 Effects of nutritional factors on performance of dairy cows
3.1 History of using nutritional intervention to regulate dairy cow performance
The development of dairy cow nutrition regulation technology is closely related to the transformation of the livestock production model. In the very early days of traditional dairy farming, there were almost no human efforts to improve the production performance of dairy cows through nutritional conditions. Extensive grazing was the dominant feature of early dairy cow rearing, and the nutrients required for the maintenance of life, lactation, and reproduction of dairy cows mainly relied on the intake of forage, that is, roughage. As time moved into the 20th century, before the 1960s, most dairy herds in the United States could be kept in stalls or tie stalls. People gradually realized that dairy cows might need more nutrients during the production process, so they chose to supplement with concentrated feed. According to the report, this type of feed was usually directly added to the roughage at that time (Schingoethe, 2017). The initial guidelines for the addition of concentrated feed were proposed in the 1930s (Huffman, 1939) and were refined in subsequent research. From the early 1960s, with the popularization of milking parlors and the increasing scale of dairy herds, more concentrated feed feeding systems were developed (Coppock et al., 1981). In 1966, the Total Mixed Ration (TMR) technology emerged (McCoy et al., 1966), with a primary advantage that every mouthful of food consumed by dairy cows is a uniformly mixed and as comprehensive as possible ration, preventing selective feeding caused by uneven mixing in traditional rations, and reducing the occurrence of indigestion and anorexia (Hernandez-Urdaneta et al., 1976). Most importantly, both research experiments and production practices in large dairy farms have proven that the use of TMR can achieve high yields (Coppock, 1977). Experiments have shown that dairy cows fed TMR have a higher efficiency in converting metabolic energy into milk and a higher milk yield (Holter et al., 1977). Therefore, TMR can be regarded as a key step in human intervention in the regulation of dairy cow production performance through nutritional conditions.
Another type of feed - silage - is also widely used in dairy farming. This may be because silage can minimize the loss of nutrients from harvest to storage, and silage is usually more convenient for farms to mix and handle than dry feed (Mahanna and Chase, 2003), which can better improve production efficiency and reduce production costs. The application of these feeds indicates that humans have recognized the important role of nutritional factors in the regulation of dairy cow production performance and have continuously improved some of the most basic feed types in order to enhance the production performance of dairy cows.
Apart from applying different types of feed, people often add specific components to the feed in production to regulate the nutrients consumed by dairy cows, thereby controlling the production performance of dairy cows by regulating the nutrients. For example, Oliveira et al. treated a batch of silage with homofermentative lactic acid bacteria and facultative heterofermentative lactic acid bacteria as inoculants. The results showed that inoculating lactic acid bacteria increased the milk yield of dairy cows (Oliveira et al., 2017). Another study showed that dairy cows fed silage inoculated with lactic acid bacteria had higher total rumen volatile fatty acid (VFA) content than those fed untreated silage, and the ratio of volatile fatty acids did not change (Mohammed et al., 2012). In addition, lactic acid is the main fermentation end product in silage (Webster, 1992), indicating that lactic acid, as one of the nutritional factors, can increase the milk yield of dairy cows. In terms of application in feed, acetate is also widely used. It can inhibit the intake of hay or silage by lactating dairy cows in a nearly linear manner (Anil et al., 1993). This may be because acetate (such as sodium acetate) can affect the regulation of intake by increasing the osmotic pressure of rumen contents (Forbes et al., 1992). Thus, from extensive grazing to precise regulation of nutritional factors in feeding, human understanding and regulation of nutrients in feed have gradually helped us further regulate and improve the production performance of dairy cows.
3.2 Nutritional factors affect key indicators of dairy cow performance through digestion and metabolism
Milk production is one of the important indicators of dairy cow performance. Cueva et al. found that a genetically engineered corn can enhance the endogenous α-amylase activity in the endosperm, and experiments have shown that this type of corn, when used as silage corn feed, can increase milk production in dairy cows (Cueva et al., 2021). Because the amylase activity of this corn can improve the digestion rate of starch in the rumen, some teams have speculated whether increasing the content of starch substances in the dry matter of feed would also increase milk production in dairy cows. Later research showed that increasing the content of starch substances in feed from 25% to 30% of dry matter (DM) would reduce the neutral detergent fiber digestibility (NDFD), but still increase milk production in dairy cows (Krogstad and Bradford, 2023). Besides improving the feed source, adding some additional additives to the feed is also commonly applied in production. For example, supplementing rumen-protected choline (RPC) in the diets of dairy cows during the perinatal period has been shown to increase milk production (Arshad et al., 2020). This may result from improved postpartum dry matter intake (DMI) (Holdorf et al., 2023), and a review has suggested that RPC can regulate lactation by altering liver energy and lipid metabolism (McFadden et al., 2020), thereby explaining its effect on dairy cow performance. Besides this, a team explored the effect of adding isoacid (ISO) to roughage on the lactation performance of Holstein dairy cows and determined that the supplementation of ISO indeed increased the digestibility of nutrients and improved milk production in dairy cows. The mechanism behind ISO’s ability to increase milk production in dairy cows may be that ISO can significantly promote the growth of cellulolytic bacteria (Bryant and Doetsch, 1955), and the accelerated growth of cellulolytic bacteria will greatly increase the activity of cellulase in the rumen, promoting the digestion of fiber, thereby enabling the host animal to obtain more nutrients and improving the production performance of the animal (Dai et al., 2015; Wang et al., 2019). Whether it is the improvement from the feed source or the addition of additives to the feed, these operations carried out by humans to increase milk production in dairy cows strongly demonstrate that nutritional factors can influence the key indicators of dairy cow production performance through the regulation of metabolism and digestion.
The content of milk protein is also one of the indicators that can be used to evaluate the production performance of dairy cows, and this indicator can be regulated by controlling nutritional factors artificially. Many studies have shown that adding butyric acid to the feed of dairy cows can improve the digestibility of nutrients (Huhtanen et al., 1998; Fukumori et al., 2020). Experiments have also found that during the period when the amount of butyric acid infused into the rumen of dairy cows changes from 0 to 600 grams per day, the milk protein content shows a positive linear response (Huhtanen et al., 1993), and it means that under the nutritional conditions of butyric acid infusion, there is a positive regulatory effect on the milk protein content. Although some studies have found that adding a certain amount of butyric acid to the basic diet has no effect on the milk protein content of dairy cows (Urrutia et al., 2019), this may be due to the different administration methods and doses of butyric acid, because other experiments have found an increase in milk protein content with a lower supplementation dose (Zhang et al., 2023). The reason why butyric acid can regulate the content of milk protein may be that the supplementation of butyric acid promotes gastrointestinal function and improves the digestibility of nutrients. Studies have shown that rumen infusion or dietary addition of butyric acid or butyrate can stimulate the growth of rumen epithelium, increase the length of rumen papillae (Mentschel et al., 2001), accelerate the blood flow rate of rumen epithelium (Rémond et al., 1993), increase the absorption of volatile fatty acids (VFA) (Storm et al., 2011), and stimulate the secretion of digestive enzymes (Guilloteau et al., 2010). Through these improvements in digestive capabilities, nutritional factors can regulate the content of milk protein. It is worth noting that although the nutritional intervention strategies targeting milk proteins have a relatively clear mechanism, there are still controversies regarding the dose effect in practical applications. Specifically, the effects of different doses of butyric acid addition on the milk protein rate of dairy cows are inconsistent, and this uncertainty to some extent restricts the formulation of nutritional standards. Therefore, subsequent research is necessary to clarify the optimal additive amount through systematic experiments, thereby providing a basis for formulating scientific and efficient nutritional plans.
The fat content and fatty acid composition in milk are also key parameters for evaluating milk quality. In the early postpartum period, dairy cows have limited dry matter intake (DMI) and microbial amino acid synthesis, leading to insufficient supply of essential amino acids, including methionine, which may reduce the supply of phosphatidylcholine and thereby affect lipid metabolism (Lima et al., 2024). Therefore, under conditions where choline supply may be limited in perinatal cows, adding rumen-protected choline (RPC) to the transition diet may help improve lactation performance (Humer et al., 2019). Based on this, Lima et al. conducted experiments to determine whether adding RPC before or around calving could improve the metabolic status and lactation performance of dairy cows, and concluded that cows fed RPC before and after calving could increase milk fat content. On the contrary, another experiment provided a negative proof of the importance of nutritional factors in regulating milk fat percentage. The team selected 16 Holstein×Normande crossbred cows and divided them into a control diet group and a restricted diet group. After nine weeks of feeding, the restricted diet group had a lower milk fat yield compared to the control diet group (Dessauge et al., 2011). This experiment provided negative evidence that nutritional factors are important in regulating milk fat percentage. The composition of fatty acids has a significant impact on the nutritional value of milk. Reducing saturated medium-chain fatty acids while increasing long-chain polyunsaturated fatty acids can enhance the nutritional value of milk. The simplest way to change the fat composition of milk may be to supplement unsaturated fats to dairy cows (Chilliard et al., 2007). Studies have shown that after dietary fat supplementation, the increase in C18 flow in the duodenum leads to a linear decrease in the production rate of C4 to C16, while the production rate of C18 in milk increases quadratically (Glasser et al., 2008), achieving a change in the composition of fatty acids in milk. This may be due to the increased substrate competition of C18 for the esterification of short-chain fatty acids caused by fatty acid supplementation, and a decrease in dry matter intake (DMI) (Allen, 2000) may also be a contributing factor. In summary, nutritional factors can significantly regulate milk fat percentage and fatty acid composition through digestive metabolism.
Body size is also an important factor related to the production performance of dairy cows, and this factor is also regulated and influenced by nutritional factors. For example, high milk feeding can promote the growth performance of dairy calves and reduce their non-nutritive behaviors (Silper et al., 2014). The nutritional requirements for the growth of calves mainly lie in net energy. For instance, a 50-kilogram calf needs to consume 0.78 kilograms of dry matter (about 5.0 to 6.5 liters of liquid feed) per day to achieve an average daily weight gain of 600 grams (van Niekerk et al., 2021). Similar to dry matter, the crude protein content in starter feed also plays a regulatory role in average daily weight gain. Kazemi-Bonchenari et al. pointed out that compared with 18% crude protein content, 23% crude protein content in starter feed increased the average daily weight gain of pre-weaned calves (Kazemi-Bonchenari et al., 2022). Thus, it can be seen that nutritional factors also have a significant regulatory effect on body size.
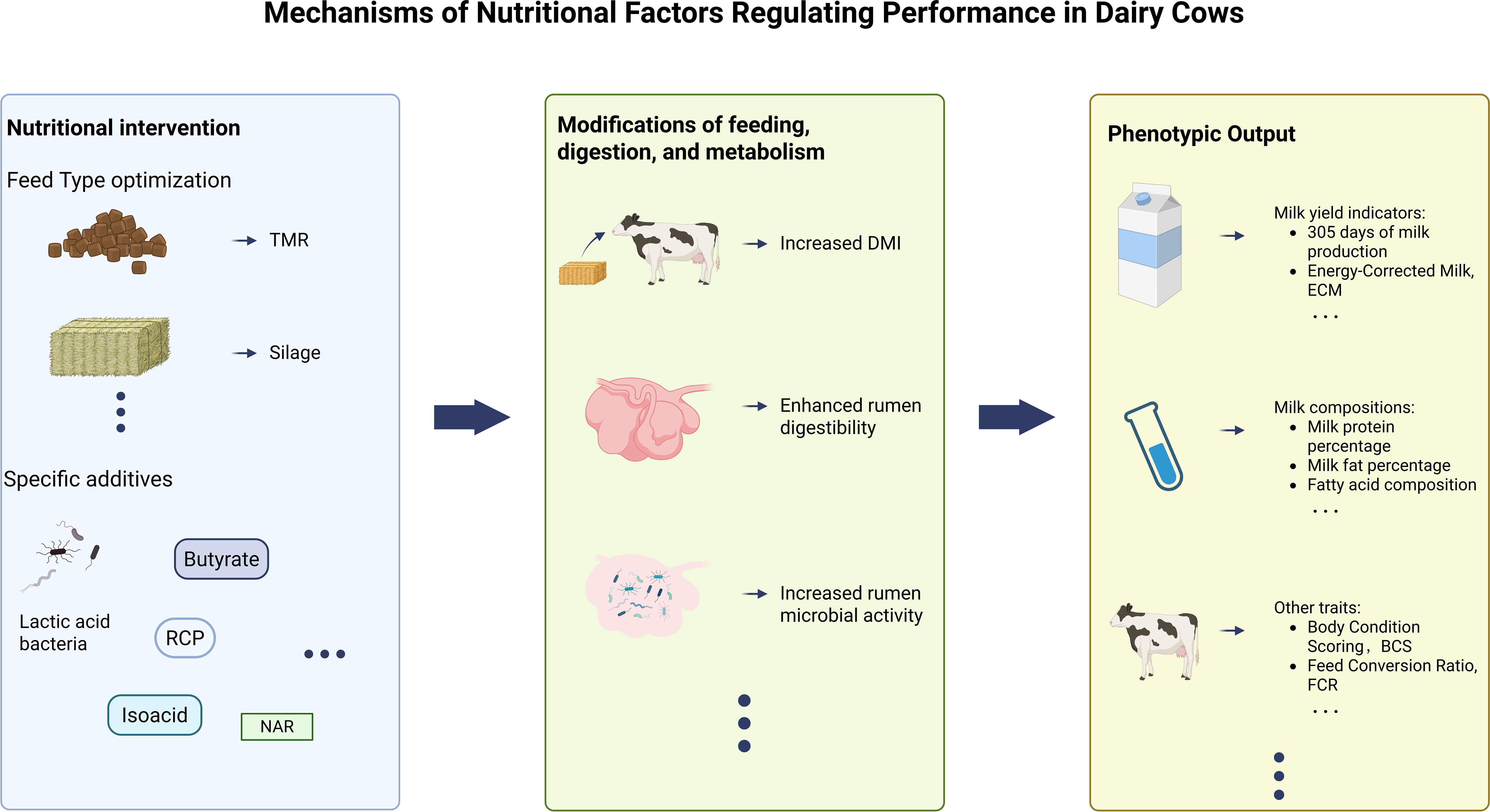
Similar to the schematic diagram of genetic factor regulation (Figure 2), the key indicators for evaluating the production performance of dairy cows through the digestion and metabolism regulation of nutritional factors can also be intuitively presented through Figure 4.
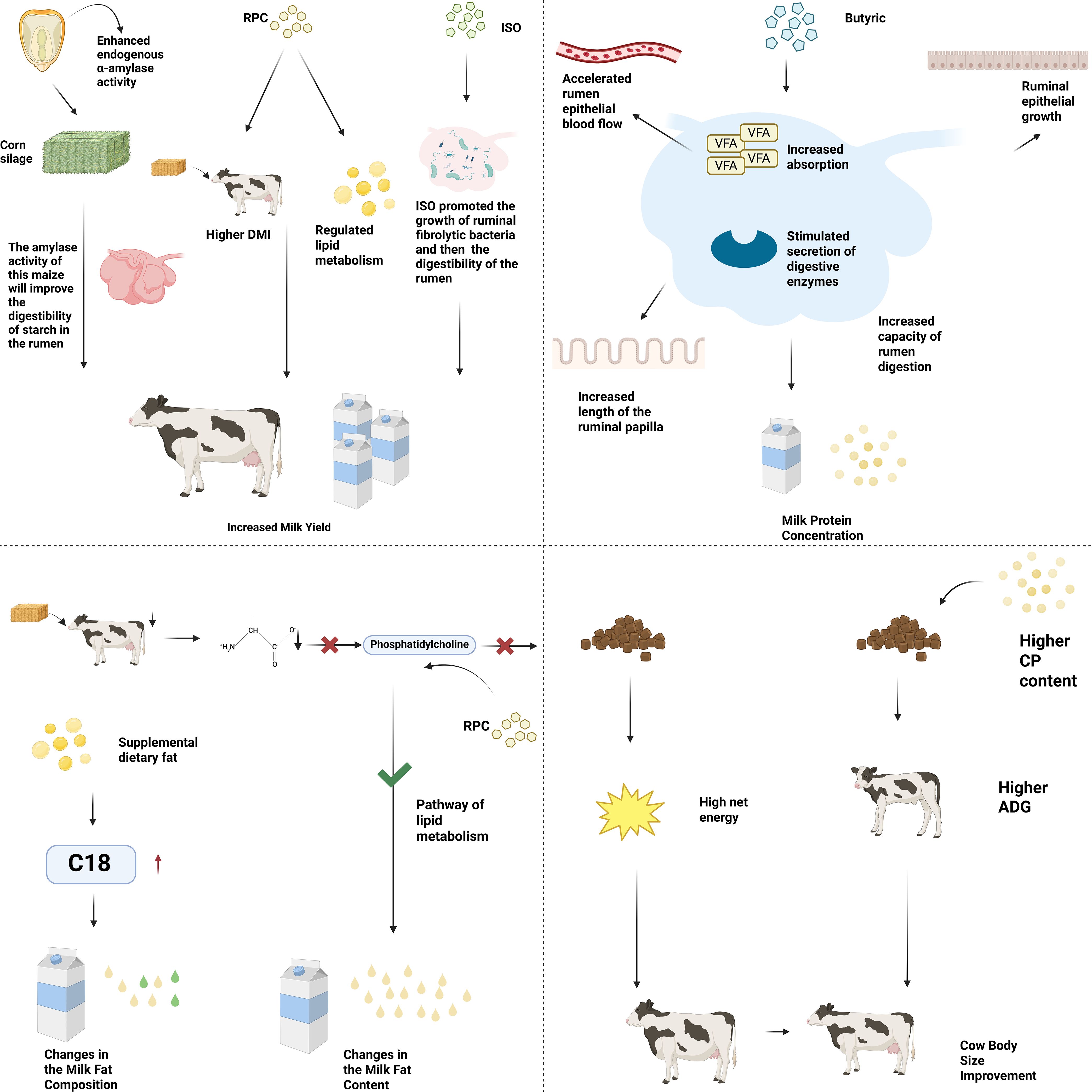
Figure 4. Nutritional factors affect key indicators of dairy cow performance through digestion and metabolism.
3.3 Further analysis of nutritional factors in dairy cow performance
The regulation of dairy cow production performance by nutritional factors has become increasingly refined and precise along with the rise in production demands and technological advancements. The control of the addition amount of conventional nutritional factors and the verification of their effects are still limited to production experience and individual-level comparisons of yields before and after addition, lacking more detailed research on the underlying principles. Metabolomics was first proposed by Nicholson et al. (1999). By using metabolomics technology, the metabolic products of dairy cows after consuming feed supplemented with new nutrients can be qualitatively and quantitatively analyzed, allowing for a deeper understanding of the regulatory role of certain nutritional factors on dairy cow production performance. For instance, in the previous study where significant differences in rumen bacterial communities were found among early lactating dairy cows with different feed intakes (Li et al., 2020), Huang et al. (2025) further explored through metabolomics that the most differentially expressed COG and KEGG pathways in the rumen microbiome of high feed intake cows were mainly concentrated in carbohydrate metabolism and protein biosynthesis, explaining the reason why high feed intake cows can produce more energy. Furthermore, a research team observed an increase in milk production when adding naringin dietary supplements. Through lipidomics and proteomics analysis, it was found that naringin directly affected lipid metabolic pathways, reducing sphingolipids in adipose tissue and increasing glycerophospholipids. These lipid changes may help alleviate inflammation and oxidative stress. Immune responses can increase energy demands by up to 55% (Burdick Sanchez et al., 2021), and changes in adipose tissue immune activity may help save energy in this regard, thereby leading to an increase in milk production (Li et al., 2024). The combined application of multi-omics technologies such as metabolomics and lipidomics will further advance the work of regulating dairy cow production performance through nutritional factors towards a more refined and precise direction.
Similarly, at the end of this section, Figure 5 can still simply summarize in graphical form the mechanism by which nutritional factors regulate the production performance of dairy cows.
4 Genetic and nutritional factors synergetically regulate dairy cow performance in different proportions
4.1 Genetic and nutritional factors synergize to regulate the performance of dairy cows by using rumen microorganisms as a bridge
The rumen and rumen microbiota, as the most crucial part of the unique digestive system of ruminants, can produce volatile fatty acids (VFA) by decomposing plant fibers through rumen microbiota to meet the majority of energy demands of ruminants (Bergman, 1990). They can also serve as a bridge for the coordinated regulation of dairy cow performance by genetic and nutritional factors. In terms of nutritional factors, they have a significant impact on the structure and function of the gastrointestinal microbiota of cattle (Henderson et al., 2015; Belanche et al., 2020). The production performance of dairy cows can be controlled by adjusting the nutritional conditions to regulate the structure and function of the rumen microbial community. For instance, compared with the diet based on alfalfa hay, the diet based on corn straw can significantly reduce the protein content of milk by altering the rumen microbiota (Sun et al., 2020). Meanwhile, adding extra non-fiber carbohydrates to the diet based on corn straw can increase the number of certain bacterial groups in the rumen microbiota and the ability to synthesize amino acids, thereby enhancing the milk production efficiency of dairy cows (Wei et al., 2021). Regarding genetic factors, since the rumen microbiome is closely related to the genome of individual cattle [after artificially exchanging the rumen contents of dairy cows with completely different rumen microbiota, the rumen microbiome remained similar after two months (Weimer et al., 2010)]. Abbas et al. (2020) found that Prevotella is associated with multiple loci on chromosomes 2, 6, 9, 19, 23, and 27 of cattle, further establishing the link between rumen microorganisms and the host cattle. Beyond the association with the host cows themselves, some experiments have also revealed that certain rumen microbiota are related to the production characteristics of dairy cows, such as feed efficiency (Li et al., 2019). Upon further exploration, a core rumen microbiome was identified that is phylogenetically linked and exhibits a preserved hierarchical structure. A subset of 39 members within this core forms hubs in the co-occurrence network. These hubs bridge the microbial community structure with host genetics and phenotypes (such as the feed efficiency mentioned above). Consequently, by leveraging machine learning algorithms, these phenotypes can be predicted from the core microbiome (Wallace et al., 2019), thereby enabling macroscopic regulation of dairy cow production performance. In conclusion, genetic and nutritional factors can work together to regulate rumen microbiota and thereby influence the performance of dairy cows.
4.2 Genetic and nutritional factors exhibit distinct proportions to the synergistic regulation of dairy cow production performance
The progress achieved through genetic selection requires reasonable measures in nutrition and overall management to enable dairy cows to fully realize their production potential. Similarly, the increase in production brought about by the development of basic dairy cow biological technologies also requires a good genetic foundation (Baumgard et al., 2017). Although both genetic and nutritional factors influence the production performance of dairy cows, when regulating certain production performances, genetic and nutritional factors have different degrees of emphasis on the regulation of that production performance. For instance, by statistically comparing the average breeding values of Holstein cows born in 1980 with those of selected birth years, it was found that the cumulative genetic gain in the production performance of Holstein cows from 1980 to 2000, genetic factors accounted for more than 55% in milk yield and milk protein rate (Shook, 2006). This might be because the long-term cumulative effect of genetic advantages across generations makes them have a higher proportion in the regulation of production performance traits with generally higher heritability compared to nutritional factors. Compared to the dominant genetic factors in the regulation of dairy cow production performance, nutritional factors are more in an auxiliary position to complement the deficiencies of genetic factors. For example, the additional demands resulting from genetic gains will be met by maintaining a balanced intake of basic roughage and concentrate while appropriately increasing the intake of concentrate (Shook, 2006). This does not imply the unimportance of nutritional factors; on the contrary, without the supplementation of nutritional factors, the gains brought by genetic factors cannot be reflected in the actual production products, highlighting their crucial supporting role in regulating the production performance of dairy cows. In summary, genetic factors play a dominant role in regulating the production performance of dairy cows, building the upper limit of the potential that dairy cows may have in production performance through the cumulative effect of generations, while the satisfaction of nutritional factors is a necessary condition for dairy cows to reach this upper limit. Both are important but have different emphases, and they need to work together to better regulate the production performance of dairy cows.
At the end of this section, Figure 6 briefly summarizes and illustrates how genetic and nutritional factors, in different proportions, synergistically regulate the production performance of dairy cows through rumen microorganisms.
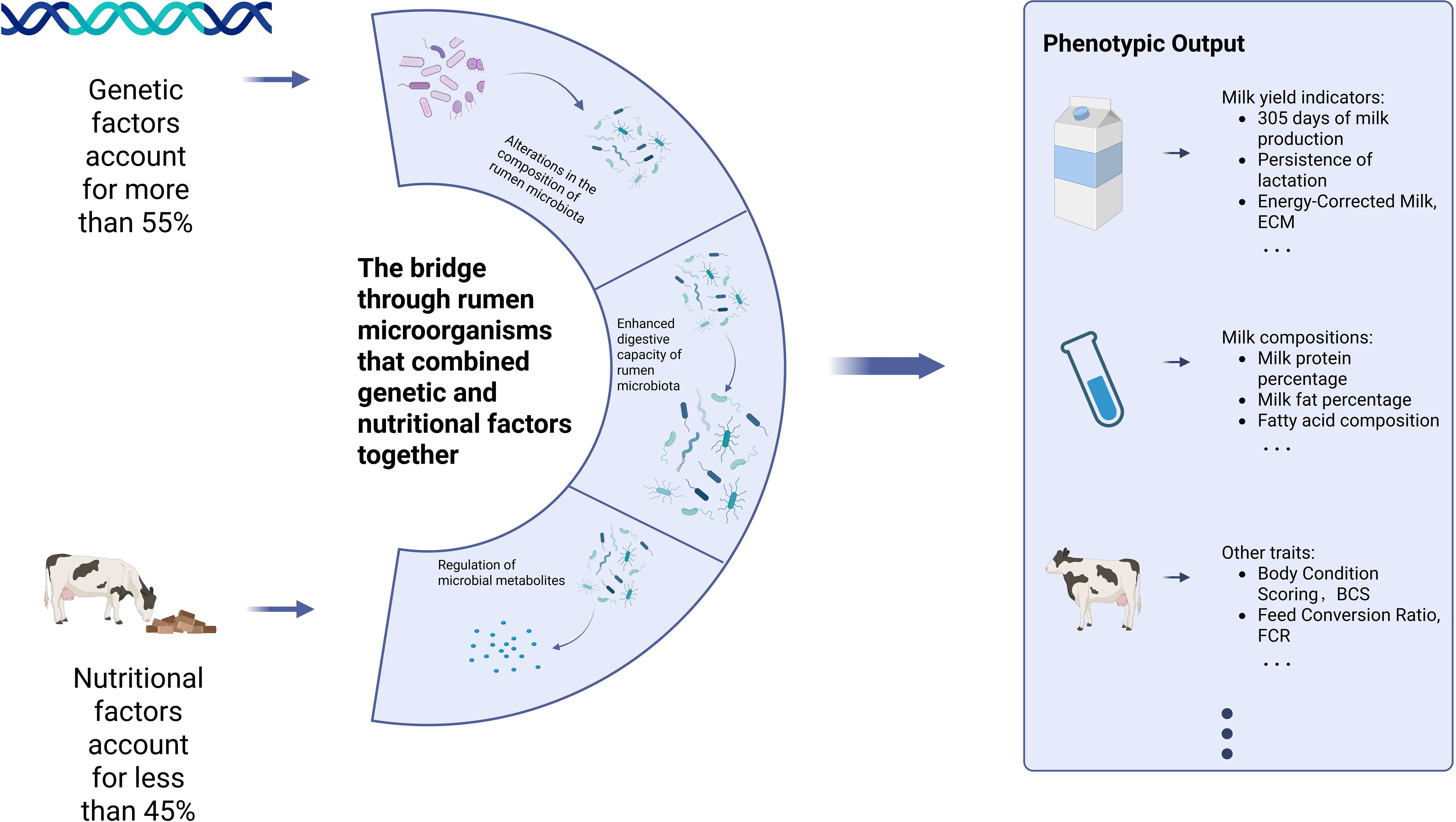
Figure 6. Genetic and nutritional factors synergetically regulate dairy cow performance in different proportions.
5 Summary and prospect
This article systematically reviews the impact of genetics, nutrition, and their synergy on the production performance of dairy cows. It also reveals their complementary roles in regulating milk yield, milk composition, and other production traits. Genetic factors establish the upper limit of the production potential of dairy cows through the regulation of genes and their pathways, while nutritional factors provide the material basis for the actual expression of genetic potential through digestive and metabolic functions. Specifically, genetic technologies such as whole-genome selection and molecular marker-assisted breeding have significantly enhanced the genetic progress of key production traits, while nutritional strategies such as total mixed ration (TMR) technology and functional additives have converted genetic potential into actual production benefits by regulating nutrient utilization efficiency and microbial metabolic networks. Finally, the synergy of genetics and nutrition, with rumen microorganisms as the bridge, can also form unique interaction patterns, transcending the bottlenecks of individual factors. Notably, the weights of the synergy of genetics and nutrition are trait-specific. Traits with higher heritability will rely more on genetic improvement, while indicators more strongly regulated by the environment are more likely to be optimized through nutritional intervention. Therefore, the methods for regulating specific production performance indicators using genetic and nutritional factors are not fixed but require the formulation of specific strategies based on the different heritabilities of the indicators.
Most of the current related research focuses on observable traits such as meat quality and feed conversion rate (Liu et al., 2022b), without exploring the underlying molecular mechanisms that cause these patterns. Therefore, future research should focus more on a deeper analysis of the mechanisms of genetic and nutritional synergy, and then summarize the rules of the coordinated action of multi-level biological processes. Furthermore, it should extend from the synergy of genetics and nutrition to the coordinated regulation of other factors that may affect the production performance of dairy cows. It is also necessary to combine actual production situations and establish different rules for different production scenarios to avoid production losses caused by ignoring other practical factors. Finally, it is worth noting that the damage to the environment caused by the cattle industry is an objective fact, and cattle are also one of the main factors causing annual methane emissions (Scholtz et al., 2011). Therefore, finding a balance between improving the production efficiency of cattle and reducing the environmental pollution caused by cattle will also be one of the key points of future development. At present, the feasible pathways involve enhancing the production per constant unit input, carrying out hybridization to a certain extent, and implementing selection and improvement within the breed for a particular trait. Based on the three aforementioned pathways and incorporating the article’s discussion on the synergy between genetic and nutritional factors mediated by rumen microorganisms, potential strategies may involve identifying feed efficiency-associated genotypes through genetic screening in order to enhance the host’s capacity for nutritional conversion, implementing precision nutrition strategies to optimize dietary formulations and regulate rumen microbial fermentation dynamics and leveraging hybridization advantages to promote host-microbiome symbiotic adaptation, thereby fostering a microbial ecosystem characterized by reduced methane emissions. These methods will cost - effectively and durably reduce the carbon footprint of beef cattle, thereby achieving a balance between industrial development and environmental protection (Scholtz et al., 2011).
Author contributions
DQ: Conceptualization, Writing – review & editing, Writing – original draft. RG: Writing – review & editing. DJ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This project was funded by the Innovation and Entrepreneurship Training Program for College Students (202411117095Z) and the National Natural Science Foundation of China (32172690), Jiangsu Higher Education Key Program (21KJA230002).
Acknowledgments
Thanks to the College of Animal Science and Technology of Yangzhou University, China, for its guidance and assistance in this review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas W., Howard J. T., Paz H. A., Hales K. E., Wells J. E., Kuehn L. A., et al. (2020). Influence of host genetics in shaping the rumen bacterial community in beef cattle. Sci. Rep. 10, 15101. doi: 10.1038/s41598-020-72011-9
Allen M. S. (2000). Effects of diet on short-term regulation of feed intake by lactating dairy cattle. J. Dairy Sci. 83, 1598–1624. doi: 10.3168/jds.S0022-0302(00)75030-2
Anil M. H., Mbanya J. N., Symonds H. W., and Forbes J. M. (1993). Responses in the voluntary intake of hay or silage by lactating cows to intraruminal infusions of sodium acetate or sodium propionate, the tonicity of rumen fluid or rumen distension. Br. J. Nutr. 69, 699–712. doi: 10.1079/bjn19930071
Arshad U., Zenobi M. G., Staples C. R., and Santos J. E. P. (2020). Meta-analysis of the effects of supplemental rumen-protected choline during the transition period on performance and health of parous dairy cows. J. Dairy Sci. 103, 282–300. doi: 10.3168/jds.2019-16842
Bai F., Cai Y., Qi M., Liang C., Pan L., Liu Y., et al. (2025). LCORL and STC2 variants increase body size and growth rate in cattle and other animals. Genomics Proteomics Bioinf. 23 (3). doi: 10.1093/gpbjnl/qzaf025
Baumgard L. H., Collier R. J., and Bauman D. E. (2017). A 100-Year Review: Regulation of nutrient partitioning to support lactation. J. Dairy Sci. 100, 10353–10366. doi: 10.3168/jds.2017-13242
Belanche A., Patra A. K., Morgavi D. P., Suen G., Newbold C. J., and Yáñez-Ruiz D. R. (2020). Editorial: gut microbiome modulation in ruminants: enhancing advantages and minimizing drawbacks. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.622002
Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Bionaz M., Periasamy K., Rodriguez-Zas S. L., Everts R. E., Lewin H. A., Hurley W. L., et al. (2012). Old and new stories: revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PloS One 7. doi: 10.1371/journal.pone.0033268
Bonfatti V., Di Martino G., CecChinato A., Vicario D., and Carnier P. (2010). Effects of β-κ-casein (CSN2-CSN3) haplotypes and β-lactoglobulin (BLG) genotypes on milk production traits and detailed protein composition of individual milk of Simmental cows. J. Dairy Sci. 93, 3797–3808. doi: 10.3168/jds.2009-2778
Bonfatti V., Giantin M., Gervaso M., Coletta A., Dacasto M., and Carnier P. (2012). Effect of CSN1S1-CSN3 (αs1-κ-casein) composite genotype on milk production traits and milk coagulation properties in Mediterranean water buffalo. J. Dairy Sci. 95, 3435–3443. doi: 10.3168/jds.2011-4901
BONNIER G. (1936). Progeny tests of dairy sires. Hereditas 22, 145–166. doi: 10.1111/j.1601-5223.1936.tb02645.x
Brito L. F., Bedere N., Douhard F., Oliveira H. R., Arnal M., Peñagaricano F., et al. (2021). Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 15, 100292. doi: 10.1016/j.animal.2021.100292
Brophy B., Smolenski G., Wheeler T., Wells D., L’Huillier P., and Laible G. (2003). Cloned transgenic cattle produce milk with higher levels of β-casein and κ-casein. Nat. Biotechnol. 21, 157–162. doi: 10.1038/nbt783
Bryant M. P. and Doetsch R. N. (1955). Factors necessary for the growth of bacteroides succinogenes in the volatile acid fraction of rumen fluid. J. Dairy Sci. 38, 340–350. doi: 10.3168/jds.S0022-0302(55)94984-5
Brym P., Kaminski S., and Rusc A. (2004). New SSCP polymorphism within bovine STAT5A gene and its associations with milk performance traits in Black-and-White and Jersey cattle. J. Appl. Genet. 45, 445–452.
Buniello A., MacArthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012. doi: 10.1093/nar/gky1120
Burdick Sanchez N. C., Broadway P. R., and Carroll J. A. (2021). Influence of yeast products on modulating metabolism and immunity in cattle and swine. Animals 11, 371. doi: 10.3390/ani11020371
Burgos S. A., Dai M., and Cant J. P. (2010). Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J. Dairy Sci. 93, 153–161. doi: 10.3168/jds.2009-2444
Chen Z., Zhou J., Wang M., Liu J., Zhang L., Loor J. J., et al. (2020). Circ09863 regulates unsaturated fatty acid metabolism by adsorbing miR-27a-3p in bovine mammary epithelial cells. J. Agric. Food Chem. 68, 8589–8601. doi: 10.1021/acs.jafc.0c03917
Chilliard Y., Glasser F., Ferlay A., Bernard L., Rouel J., and Doreau M. (2007). Diet, rumen biohydrogenation and nutritional quality of cow and goat milk fat. Eur. J. Lipid Sci. Technol. 109, 828–855. doi: 10.1002/ejlt.200700080
Coppock C. E. (1977). Feeding methods and grouping systems. J. Dairy Sci. 60, 1327–1336. doi: 10.3168/jds.S0022-0302(77)84030-7
Coppock C. E., Bath D. L., and Harris B. (1981). From feeding to feeding systems. J. Dairy Sci. 64, 1230–1249. doi: 10.3168/jds.S0022-0302(81)82698-7
Cueva S. F., Stefenoni H., Melgar A., Räisänen S. E., Lage C. F. A., Wasson D. E., et al. (2021). Lactational performance, rumen fermentation, and enteric methane emission of dairy cows fed an amylase-enabled corn silage. J. Dairy Sci. 104, 9827–9841. doi: 10.3168/jds.2021-20251
Dai X., Tian Y., Li J., Su X., Wang X., Zhao S., et al. (2015). Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 81, 1375–1386. doi: 10.1128/AEM.03682-14
Dessauge F., Lollivier V., Ponchon B., Bruckmaier R., Finot L., Wiart S., et al. (2011). Effects of nutrient restriction on mammary cell activity and hormonal statement in lactating dairy cows. J. Dairy Sci. 93, 4623–4635. doi: 10.3168/jds.2010-4012
Edwards J. (1932). The progeny test as a method of evaluating the dairy sire. J. Agric. Sci. 22, 811–837. doi: 10.1017/S0021859600054617
Forbes J. M., Mbanya J. N., and Anil M. H. (1992). Effects of intraruminal infusions of sodium acetate and sodium chloride on silage intake by lactating cows. Appetite 19, 293–301. doi: 10.1016/0195-6663(92)90169-7
Fukumori R., Oba M., Izumi K., Otsuka M., Suzuki K., Gondaira S., et al. (2020). Effects of butyrate supplementation on blood glucagon-like peptide-2 concentration and gastrointestinal functions of lactating dairy cows fed diets differing in starch content. J. Dairy Sci. 103, 3656–3667. doi: 10.3168/jds.2019-17677
Glasser F., Ferlay A., Doreau M., Schmidely P., Sauvant D., and Chilliard Y. (2008). Long-chain fatty acid metabolism in dairy cows: a meta-analysis of milk fatty acid yield in relation to duodenal flows and de novo synthesis. J. Dairy Sci. 91, 2771–2785. doi: 10.3168/jds.2007-0383
Gu Z., Eleswarapu S., and Jiang H. (2007). Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 581, 981–988. doi: 10.1016/j.febslet.2007.01.081
Guilloteau P., Savary G., Jaguelin-Peyrault Y., Romé V., Le Normand L., and Zabielski R. (2010). Dietary sodium butyrate supplementation increases digestibility and pancreatic secretion in young milk-fed calves. J. Dairy Sci. 93, 5842–5850. doi: 10.3168/jds.2009-2751
Henderson C. R. (1953). Estimation of variance and covariance components. Biometrics 9, 226–252. doi: 10.2307/3001853
Henderson G., Cox F., Ganesh S., Jonker A., Young W., Abecia L., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 14567. doi: 10.1038/srep14567
Hernandez-Urdaneta A., Coppock C. E., McDowell R. E., Gianola D., and Smith N. E. (1976). Changes in forage-concentrate ratio of complete feeds for dairy cows. J. Dairy Sci. 59, 695–707. doi: 10.3168/jds.S0022-0302(76)84260-9
Hodgson R. E. (1956). Dairy production research in the United States department of agriculture. J. Dairy Sci. 39, 674–682. doi: 10.3168/jds.S0022-0302(56)91188-2
Holdorf H. T., Kendall S. J., Ruh K. E., Caputo M. J., Combs G. J., Henisz S. J., et al. (2023). Increasing the prepartum dose of rumen-protected choline: Effects on milk production and metabolism in high-producing Holstein dairy cows. J. Dairy Sci. 106, 5988–6004. doi: 10.3168/jds.2022-22905
Holter J. B., Urban W. E., Hayes H. H., and Davis H. A. (1977). Utilization of diet components fed blended or separately to lactating cows1, 2. J. Dairy Sci. 60, 1288–1293. doi: 10.3168/jds.S0022-0302(77)84024-1
Hu Z.-L., Park C. A., and Reecy J. M. (2019). Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 47, D701–D710. doi: 10.1093/nar/gky1084
Huang S., Shang Y., Wang L., Wang S., Zheng G., Wang D., et al. (2025). Variations of rumen metagenome and metabolome in dairy cows with different feed intake levels during the postpartum period. Anim. Nutriomics 2, e4. doi: 10.1017/anr.2024.29
Huffman C. F. (1939). Roughage quality and quantity in the dairy ration, A review. J. Dairy Sci. 22, 889–980. doi: 10.3168/jds.S0022-0302(39)92948-6
Huhtanen P. J., Blauwiekel R., and Saastamoinen I. (1998). Effects of intraruminal infusions of propionate and butyrate with two different protein supplements on milk production and blood metabolites in dairy cows receiving grass silage-based diet. J. Sci. Food Agric. 77, 213–222. doi: 10.1002/(SICI)1097-0010(199806)77:2<213::AID-JSFA28>3.0.CO;2-6
Huhtanen P., Miettinen H., and Ylinen M. (1993). Effect of increasing ruminal butyrate on milk yield and blood constituents in dairy cows fed a grass silage-based diet. J. Dairy Sci. 76, 1114–1124. doi: 10.3168/jds.S0022-0302(93)77440-8
Humer E., Bruggeman G., and Zebeli Q. (2019). A meta-analysis on the impact of the supplementation of rumen-protected choline on the metabolic health and performance of dairy cattle. Animals 9, 566. doi: 10.3390/ani9080566
Jiao P., Yuan Y., Zhang M., Sun Y., Wei C., Xie X., et al. (2020). PRL/microRNA-183/IRS1 pathway regulates milk fat metabolism in cow mammary epithelial cells. Genes 11, 196. doi: 10.3390/genes11020196
Kazemi-Bonchenari M., Khanaki H., Jafari A., Eghbali M., Poorhamdollah M., and Ghaffari M. H. (2022). Milk feeding level and starter protein content: Effects on growth performance, blood metabolites, and urinary purine derivatives of Holstein dairy calves. J. Dairy Sci. 105, 1115–1130. doi: 10.3168/jds.2021-21208
Kibar M. and Aytekin B. (2025). Strong associations between the FGF-2 gene and productivity traits of Holstein-Friesian dairy cattle. Gene 940, 149027. doi: 10.1016/j.gene.2024.149027
Klein R. J., Zeiss C., Chew E. Y., Tsai J. Y., Sackler R. S., Haynes C., et al. (2005). Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389. doi: 10.1126/science.1109557
Krogstad K. C. and Bradford B. J. (2023). The effects of feeding α-amylase-enhanced corn silage with different dietary starch concentrations to lactating dairy cows on milk production, nutrient digestibility, and blood metabolites. J. Dairy Sci. 106, 4666–4681. doi: 10.3168/jds.2022-23030
Laible G., Smolenski G., Wheeler T., and Brophy B. (2016). Increased gene dosage for β- and κ-casein in transgenic cattle improves milk composition through complex effects. Sci. Rep. 6. doi: 10.1038/srep37607
Li L., Bai S., Zhao H., Tan J., Wang Y., Zhang A., et al. (2024). Dietary supplementation with naringin improves systemic metabolic status and alleviates oxidative stress in transition cows via modulating adipose tissue function: A lipid perspective. Antioxidants 13, 638. doi: 10.3390/antiox13060638
Li C., Cai W., Zhou C., Yin H., Zhang Z., Loor J. J., et al. (2016). RNA-Seq reveals 10 novel promising candidate genes affecting milk protein concentration in the Chinese Holstein population. Sci. Rep. 6. doi: 10.1038/srep26813
Li Y. Q., Xi Y. M., Wang Z. D., Zeng H. F., and Han Z. (2020). Combined signature of rumen microbiome and metabolome in dairy cows with different feed intake levels. J. Anim. Sci. 98. doi: 10.1093/jas/skaa070
Li F., Zhang H., Fu W., Shi W., Wang C., Lei Y., et al. (2019). Lift-off flames of propane under a variety of co-flow conditions. Chem. Eng. Process. - Process Intensification 136, 92–100. doi: 10.1016/j.cep.2018.12.013
Lima F. S., MF S. F., Greco L. F., and Santos J. E. P. (2024). Rumen-protected choline improves metabolism and lactation performance in dairy cows. Animals 14. doi: 10.3390/ani14071016
Liu S., Gao Y., Canela-Xandri O., Wang S., Yu Y., Cai W., et al. (2022a). A multi-tissue atlas of regulatory variants in cattle. Nat. Genet. 54, 1438–143+. doi: 10.1038/s41588-022-01153-5
Liu X., Tang Y., Wu J., Liu J.-X., and Sun H.-Z. (2022b). Feedomics provides bidirectional omics strategies between genetics and nutrition for improved production in cattle. Anim. Nutr. 9, 314–319. doi: 10.1016/j.aninu.2022.03.002
Long N., Gianola D., Rosa G. J. M., Weigel K. A., and Avendano S. (2007). Machine learning classification procedure for selecting SNPs in genomic selection:: application to early mortality in broilers. J. Anim. Breed. Genet. 124, 377–389. doi: 10.1111/j.1439-0388.2007.00694.x
Mahanna B. and Chase L. E. (2003). Practical applications and solutions to silage problems. Silage Science and Technology. 42, 855–895. doi: 10.2134/agronmonogr42.c19
McCoy G. C., Thurmon H. S., Olson H. H., and Reed A. (1966). Complete feed rations for lactating dairy cows1, 2. J. Dairy Sci. 49, 1058–1063. doi: 10.3168/jds.S0022-0302(66)88017-7
McFadden J. W., Girard C. L., Tao S., Zhou Z., Bernard J. K., Duplessis M., et al. (2020). Symposium review: One-carbon metabolism and methyl donor nutrition in the dairy cow. J. Dairy Sci. 103, 5668–5683. doi: 10.3168/jds.2019-17319
Mentschel J., Leiser R., Mülling C., Pfarrer C., and Claus R. (2001). Butyric acid stimulates rumen mucosa development in the calf mainly by a reduction of apoptosis. Arch. Tierernahr 55, 85–102. doi: 10.1080/17450390109386185
Meuwissen T. H., Hayes B. J., and Goddard M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi: 10.1093/genetics/157.4.1819
Mohammed R., Stevenson D. M., Beauchemin K. A., Muck R. E., and Weimer P. J. (2012). Changes in ruminal bacterial community composition following feeding of alfalfa ensiled with a lactic acid bacterial inoculant1. J. Dairy Sci. 95, 328–339. doi: 10.3168/jds.2011-4492
Nejati-Javaremi A., Smith C., and Gibson J. P. (1997). Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 7), 1738. doi: 10.2527/1997.7571738x
Nicholson J. K., Lindon J. C., and Holmes E. (1999). ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189. doi: 10.1080/004982599238047
Oliveira C. S., Camargo L. S. A., da Silva M., Saraiva N. Z., Quintão C. C., and MaChado M. A. (2023). Embryo biopsies for genomic selection in tropical dairy cattle. Anim. Reprod. 20, e20230064. doi: 10.1590/1984-3143-ar2023-0064
Oliveira A. S., Weinberg Z. G., Ogunade I. M., Cervantes A. A. P., Arriola K. G., Jiang Y., et al. (2017). Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J. Dairy Sci. 100, 4587–4603. doi: 10.3168/jds.2016-11815
Palladino R. A., Buckley F., Prendiville R., Murphy J. J., Callan J., and Kenny D. A. (2010). A comparison between Holstein-Friesian and Jersey dairy cows and their F1 hybrid on milk fatty acid composition under grazing conditions. J. Dairy Sci. 93, 2176–2184. doi: 10.3168/jds.2009-2453
Pereira P. C. (2014). Milk nutritional composition and its role in human health. Nutrition 30, 619–627. doi: 10.1016/j.nut.2013.10.011
Rémond D., Chaise J. P., Delval E., and Poncet C. (1993). Net transfer of urea and ammonia across the ruminal wall of sheep. J. Anim. Sci. 71, 2785–2792. doi: 10.2527/1993.71102785x
Risch N. and Merikangas K. (1996). The future of genetic studies of complex human diseases. Science 273, 1516–1517. doi: 10.1126/science.273.5281.1516
Robertson A. and Rendel J. M. (1954). The performance of heifers got by artificial insemination. J. Agric. Sci. 44, 184–192. doi: 10.1017/S002185960004627X
Robertson A., Stewart A., and Ashton E. D. (1956). The progeny assessment of dairy sires for milk: the use of contemporary comparisons. Proc. Br. Soc. Anim. Production 1956, 43–50. doi: 10.1017/S0369852100001688
Rodrigues F. M., Majeres L. E., Dilger A. C., McCann J. C., Cassady C. J., Shike D. W., et al. (2025). Characterizing differences in the muscle transcriptome between cattle with alternative LCORL-NCAPG haplotypes. BMC Genomics 26. doi: 10.1186/s12864-025-11665-z
Schingoethe D. J. (2017). A 100-Year Review: Total mixed ration feeding of dairy cows. J. Dairy Sci. 100, 10143–10150. doi: 10.3168/jds.2017-12967
Schmidtmann C., Segelke D., Bennewitz J., Tetens J., and Thaller G. (2023). Genetic analysis of production traits and body size measurements and their relationships with metabolic diseases in German Holstein cattle. J. Dairy Sci. 106, 421–438. doi: 10.3168/jds.2022-22363
Schneider H., Heise J., Tetens J., Thaller G., Wellmann R., and Bennewitz J. (2023). Genomic dominance variance analysis of health and milk production traits in German Holstein cattle. J. Anim. Breed. Genet. 140, 390–399. doi: 10.1111/jbg.12765
Scholtz M., Steyn Y., van Marle-Koster E., and Theron H. E. (2011). Improved production efficiency in cattle to reduce their carbon footprint for beef production. South Afr. J. Anim. Sci. 42, 450–453. doi: 10.4314/sajas.v42i5.1
Shook G. E. (2006). Major advances in determining appropriate selection goals. J. Dairy Sci. 89, 1349–1361. doi: 10.3168/jds.S0022-0302(06)72202-0
Si J., Dai D., Gorkhali N. A., Wang M., Wang S., Sapkota S., et al. (2025). Complete genomic landscape reveals hidden evolutionary history and selection signature in Asian water buffaloes (Bubalus bubalis). Advanced Sci. 12. doi: 10.1002/advs.202407615
Silper B. F., Lana A. M., Carvalho A. U., Ferreira C. S., Franzoni A. P., Lima J. A., et al. (2014). Effects of milk replacer feeding strategies on performance, ruminal development, and metabolism of dairy calves. J. Dairy Sci. 97, 1016–1025. doi: 10.3168/jds.2013-7201
Stergiadis S., Norskov N. P., Purup S., Givens I., and Lee M. R. F. (2019). Comparative nutrient profiling of retail goat and cow milk. Nutrients 11, 2282. doi: 10.3390/nu11102282
Storm A. C., Hanigan M. D., and Kristensen N. B. (2011). Effects of ruminal ammonia and butyrate concentrations on reticuloruminal epithelial blood flow and volatile fatty acid absorption kinetics under washed reticulorumen conditions in lactating dairy cows. J. Dairy Sci. 94, 3980–3994. doi: 10.3168/jds.2010-4091
Sun H. Z., Zhou M., Wang O., Chen Y., Liu J. X., and Guan L. L. (2020). Multi-omics reveals functional genomic and metabolic mechanisms of milk production and quality in dairy cows. Bioinformatics 36, 2530–2537. doi: 10.1093/bioinformatics/btz951
Teng J., Wang D., Zhao C., Zhang X., Chen Z., Liu J., et al. (2023). Longitudinal genome-wide association studies of milk production traits in Holstein cattle using whole-genome sequence data imputed from medium-density chip data. J. Dairy Sci. 106, 2535–2550. doi: 10.3168/jds.2022-22277
Urrutia N., Bomberger R., Matamoros C., and Harvatine K. J. (2019). Effect of dietary supplementation of sodium acetate and calcium butyrate on milk fat synthesis in lactating dairy cows. J. Dairy Sci. 102, 5172–5181. doi: 10.3168/jds.2018-16024
van Niekerk J. K., Middeldorp M., Guan L. L., and Steele M. A. (2021). Preweaning to postweaning rumen papillae structural growth, ruminal fermentation characteristics, and acute-phase proteins in calves. J. Dairy Sci. 104, 3632–3645. doi: 10.3168/jds.2020-19003
Wallace R. J., Sasson G., Garnsworthy P. C., Tapio I., Gregson E., Bani P., et al. (2019). A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 5, eaav8391. doi: 10.1126/sciadv.aav8391
Walsh R., Jurgens S. J., Erdmann J., and Bezzina C. R. (2023). Genome-wide association studies of cardiovascular disease. Physiol. Rev. 103, 2039–2055. doi: 10.1152/physrev.00024.2022
Wang C., Liu Q., Guo G., Huo W. J., Zhang Y. L., Pei C. X., et al. (2019). Effects of rumen-protected folic acid and branched-chain volatile fatty acids supplementation on lactation performance, ruminal fermentation, nutrient digestion and blood metabolites in dairy cows. Anim. Feed Sci. Technol. 247, 157–165. doi: 10.1016/j.anifeedsci.2018.11.015
Wang Y., Wang X., Wang M., Zhang L., Zan L., and Yang W. (2021). Bta-miR-34b controls milk fat biosynthesis via the Akt/mTOR signaling pathway by targeting RAI14 in bovine mammary epithelial cells. J. Anim. Sci. Biotechnol. 12. doi: 10.1186/s40104-021-00598-8
Webster J. (1992). The Biochemistry of Silage (Second Edition). By McDonald P., Henderson A. R., and Heron S. J. E.. Marlow, Bucks, UK: Chalcombe Publications, (1991). Exp. Agric. 28, 125–125. doi: 10.1017/S0014479700023115
Wei Z., Xie X., Xue M., Valencak T. G., Liu J., and Sun H. (2021). The effects of non-fiber carbohydrate content and forage type on rumen microbiome of dairy cows. Animals 11, 3519. doi: 10.3390/ani11123519
Weigel K. A., VanRaden P. M., Norman H. D., and Grosu H. (2017). A 100-Year Review: Methods and impact of genetic selection in dairy cattle—From daughter–dam comparisons to deep learning algorithms. J. Dairy Sci. 100, 10234–10250. doi: 10.3168/jds.2017-12954
Weimer P. J., Stevenson D. M., Mantovani H. C., and Man S. L. C. (2010). Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents1. J. Dairy Sci. 93, 5902–5912. doi: 10.3168/jds.2010-3500
Zhang J., Bu L., Liu Y., Huo W., Xia C., Pei C., et al. (2023). Dietary supplementation of sodium butyrate enhances lactation performance by promoting nutrient digestion and mammary gland development in dairy cows. Anim. Nutr. 15, 137–148. doi: 10.1016/j.aninu.2023.08.008
Zhang Y., Fan X., Qiu L., Zhu W., Huang L., and Miao Y. (2021). Liver X receptor α promotes milk fat synthesis in buffalo mammary epithelial cells by regulating the expression of FASN. J. Dairy Sci. 104, 12980–12993. doi: 10.3168/jds.2021-20596
Zhang C., Liu H., Jiang X., Zhang Z., Hou X., Wang Y., et al. (2024). An integrated microbiome- and metabolome-genome-wide association study reveals the role of heritable ruminal microbial carbohydrate metabolism in lactation performance in Holstein dairy cows. Microbiome 12. doi: 10.1186/s40168-024-01937-3
Keywords: dairy cattle, performance, genetic effect, nutritional intervention, synergy
Citation: Qin D, Gao R and Ji D (2025) Synergy between genetics and nutrition: a systematic review of factors affecting performance in dairy cows. Front. Anim. Sci. 6:1653052. doi: 10.3389/fanim.2025.1653052
Received: 24 June 2025; Accepted: 15 September 2025;
Published: 25 September 2025.
Edited by:
Juliana Petrini, Clinica do Leite Ltda, BrazilReviewed by:
Ozden Cobanoglu, Bursa Uludağ University, TürkiyeZixin Liu, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Qin, Gao and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dejun Ji, MDAyMDQxQHl6dS5lZHUuY24=
 Danqing Qin
Danqing Qin Rui Gao
Rui Gao