- Department of Biology, Mount Royal University, Calgary, AB, Canada
Of the about 1850 species of Hymenoptera for which chromosome counts are known, only just over 200 of these are bees (Apoidea). Haploid numbers (n) range from 3-28, which probably does represent the true range of chromosome numbers in this superfamily. The modal number is 17, with another peak at n=9, representing a clade of meliponid bees which has been well studied. Although much is known about the chromosomes of bees there is still much to learn about overall trends in haploid number and chromosome organization. We are still lacking this information for many important families of bees. The only andrenid bee karyotyped, Andrena togashii has the low n of 3, so we certainly need to know which other species in this family have low chromosome numbers to see if this is an exception and to further test the Minimum Interaction Theory (MIT) of Imai and colleagues which predicts the evolutionary increase in chromosome number. In general, an overall increase from low numbers (n=3-8) to the higher numbers found in the Apidae, Colletidae, Halictidae, and Megachilidae (modal numbers 17, 16, 16, 16, respectively) does appear to be followed. However, within groups this is not always the case; the Meliponid clade with n=9 being an example. The potential adaptive value of chromosome number per se is of great interest. I propose a hypothesis to account for the high (n=25) chromosome number found in the social parasitic bumble bee subgenus Psithyrus. More sophisticated techniques beyond chromosome counting and karyotyping using C-banding, will yield much more detailed information about chromosomal rearrangements as shown by the work on the neotropical meliponid bees by the Brazilian cytogeneticists, and when these are applied to other taxa of bees will undoubtedly reveal features of great interest. Genomic approaches are starting to identify chromosomal rearrangements such as inversions and this holds much potential to explore their adaptive significance.
1 Introduction
There is a vast amount known about the chromosomes of animals (White, 1977), and the study of chromosomes has been central to the discipline of genetics since its early days. Similarly, chromosome rearrangements within species are of evolutionary significance, and have also been of importance for revealing phylogenetic patterns among different species (White, 1977; Wellenreuther and Bernatchez, 2018). Indeed, interest in the former has also now undergone resurgence in the era of genomics (Wellenreuther and Bernatchez, 2018). It is a striking fact that species, even of one taxonomic group, can vary dramatically in chromosome number; ants (see below) are a good example of this. However, almost paradoxically, although chromosome numbers can vary by almost an order of magnitude [for example the Lycaenoidea butterflies have a range of haploid chromosome number from 10 to a high of 223 (White, 1977)] there is no obvious visible effect on the phenotypes; all the butterflies look like butterflies in gross morphology. In some ways this is not really surprising as differences in chromosome number just represent different ways of packaging the total genetic material – the genome. Nevertheless, it would be naïve to think that there is no importance to this variation, and at the very least there are likely to be limits to the effectiveness of meiosis and mitosis with very large numbers of chromosomes. The events of chromosome replication and meiosis are obviously critical as well known from classical genetics. Even a mutation at a single base (not even a base pair) can be transmitted to the offspring as a half-chromatid mutation and can potentially result in a very large phenotypic effect such as phenotypic mosaics and even gynandromorphs in the Hymenoptera (Owen, 2023). This is the apparent paradox; large scale changes in chromosome number may have no obvious phenotypic effect on an organism but a single base change can have a drastic effect. Chromosomal variation simply cannot be without significance and thus still needs to be understood. As Imai et al. (2001) remark, “…karyotypes might be important as an isolating mechanism in speciation and have their own evolutionary trends independent of genetic evolution (King, 1993)”. However the exact role of chromosomes and chromosomal changes in speciation remains unclear and is still subject to considerable debate (King, 1993; Coyne and Orr, 2004).
In this review I will summarize some of what is known about chromosome variation in bees, and discuss some possible causes and adaptive consequences of this variation. I will start by putting bee chromosome numbers in the context of chromosome variation the Hymenoptera as a whole.
2 Hymenopteran chromosomes
The most obvious, and dramatic, difference in chromosome number in the Hymenoptera is that between males and females (Figure 1) which arises because of the genetic system of haplodiploidy, whereby females arise from fertilized eggs and males from unfertilized eggs. Thus, males have just one haploid (n) set of chromosomes while the females have the full diploid complement of 2n. This represents the successive breaking of constraints that allows development to proceed by mitosis from the unfertilized egg (Gallis and van Alphen, 2020). In many species of Hymenoptera there is a genic system of sex-determination underlying this; the single-locus complementary sex-determination (sl-CSD) proposed by Whiting (1933) to explain sex-determination in Habrobracon. Sex is determined a single locus with multiple alleles x1, x2, x3,…,xk(where k varies from 12–20 in many natural populations); any heterozygote (e.g. x1x3) is female, haploids are normal males while homozygotes are diploid males. In any finite population diploid males are expected to occur regularly at frequency of 5-10% (Owen and Packer, 1994). Diploid males are inviable in some species, while in others are viable and fertile (Garofalo and Kerr, 1975). Karyotypes have been obtained of diploid males for a number of species of bees, for example honeybees and bumble bees (Hoshiba, 1984b; Hoshiba et al., 1995). That these dramatic ploidy differences between males, females and diploid males results in perfectly viable adults suggests that the Hymenoptera may be able to tolerate changes in chromosome number rather well. Also, somatic polyploidy is well known and widespread in the Hymenoptera (Crozier, 1975) but will not be discussed in this review.

Figure 1. Bombus (Thoracobombus) fervidus male (left) and female (right) with haploid number N = 18, and diploid number 2N = 36 respectively.
2.1 Chromosome nomenclature
The conventional system of classifying and naming chromosomes follows that of Levan et al. (1964) which is based on the ratio of length between the long and short arms. Thus, we have metacentric (m), submetacentric (sm), subtelocentric (st), and acrocentric (a) chromosomes, and this is widely used for most plant and animal taxa. In contrast a rather more detailed, and complex system of naming chromosomes has been used by Imai and coworkers and has been applied particularly to the chromosomes of the Hymenoptera (Hoshiba and Imai, 1993; Imai, 1978). It is worth going into this in some detail as it forms the basis of the minimum interaction theory to account for chromosomal evolution in the Hymenoptera (Imai et al., 1986; Hoshiba and Imai, 1993). This theory is interesting and important because it attempts an adaptive explanation for changes in chromosome number and of the amount and distribution of heterochromatin observed.
The system of chromosome morphology, called the TAM system by Imai (1978, 1991) classifies chromosomes into three basic categories, telocentric (T) acrocentric (A) and metacentric () chromosomes (Hoshiba and Imai, 1993). The A and M chromosomes are defined by two features; (i) chromosome arms are euchromatic or heterochromatic, and (ii) ratio of the width (i.e., length) of short (WS) and long arms (WL) as follows:
A, heterochromatic short arm, euchromatic long arm, and WS < WL
M, both arms are euchromatic, and WS = WL
Modified types of the A chromosomes are Ae, AM (pseudoacrocentric), and Ah which are defined as
Ae, both arms are euchromatic, and WS < WL
AM, either short or long arms heterochromatic and WS = WL
Ah, both arms are heterochromatic, and WS < WL
Heterochromatic regions are located by the C-banding technique (see below), and C-band(s) located at the pericentromeric, interstitial or, terminal regions of chromosome arms are represented as AC, Ai, Ae, MC, Mj, Mt, etc. The terms “A group” and “M group” chromosomes are used by Hoshiba and Imai (1993), and include respectively A, AM, AMc, AMc, etc., and M, Mt, Mc, Mi, etc. Some examples of the A and M group chromosomes with C-banding found in bees and wasps show in Figure 2, which is modified from Hoshiba and Imai (1993). It is the cyclic transition between these types of chromosomes that is the essence of the minimum interaction theory (Imai, 1978). Imai (1978) proposed that centric fission of an M chromosome → T + T (two telocentric chromosomes), this is followed by tandem growth of heterochromatin, thus T → A, and finally pericentric inversion reverting A → M (Imai, 1978). Thus, the overall trend is the evolution of higher chromosome numbers and numbers of chromosome arms within lineages (Imai, 1978). Telocentric chromosomes although rare in nature, do occur (Marks, 1957; White, 1977); for example, are common in some birds (Takagi and Sasaki, 1974). Telocentric chromosomes (or telosomes) are thought to be unstable as Koo et al. (2015) state “they arise through misdivision of centromeres in normal chromosomes, and their cytological stability depends on the structure of their kinetochores. The instability of telosomes may be attributed to the relative centromere size and the degree of completeness of their kinetochore”. Thus, in their analysis of chromosomes evolution in the Hymenoptera Hoshiba and Imai (1993) treat the T chromosome as only a theoretical construct and it is used only for theoretical discussions of chromosome evolution, and is regarded as a member of A chromosome group (Hoshiba and Imai, 1993). In this system the T chromosomes transform into acrocentrics through increase in heterochromatin (T → A, see above).

Figure 2. A selection of some of the main “A” group and “ ” group chromosomes found in bees and wasps showing C banding. Modified and simplified from Hoshiba and Imai (1993).
2.2 Theories of chromosomal evolution
From the study of the diversity of mammalian chromosome numbers three different mechanisms or hypotheses emerged to account for this karyotype evolution (Imai and Crozier, 1980; Menezes et al., 2014). The hypotheses are general enough to be applicable to any group of animals:
1. The fusion hypothesis (White, 1977) assumes that chromosome numbers have decreased by centric fusion from an ancestral high number of acrocentric chromosomes.
2. The fission hypothesis is the opposite, assuming ancestral chromosome numbers to be low and subsequently increasing by centric fissions and pericentric inversions (Todd, 1970; Imai and Crozier, 1980). Centric fission of chromosomes was long thought to be unlikely given the nature of the centromere (Imai et al., 2001). However, more recent molecular genetics has revised this idea, (Imai et al., 2001) and there is now direct evidence of fission in hymenopteran chromosomes (Rousselet et al., 2000).
3. The modal hypothesis (Matthey, 1973) assumes that ancestral chromosome numbers are intermediate and subsequently have increased or decreased in lineages through fission and fusion.
As an alternative to all of the above, as already mentioned, Imai et al. (1986) have proposed a modified form of the fission hypothesis, which is:
4. The minimum interaction theory (MIT) to account for chromosomal evolution in ants, bees and wasps (Imai et al., 1986; Hoshiba and Imai, 1993). As succinctly put by Hoshiba and Imai (1993) under the minimum interaction hypothesis “…chromosome evolution proceeds toward minimizing chromosomal interactions which induce deleterious chromosomal mutations such as reciprocal translocation, and predicts that increasing chromosome number by centric fission is one of the adaptive solutions”.
Here I am following Imai et al. (2001) by describing “chromosome evolution’’ as a general term covering three distinct concepts: (i) morphological alteration of individual chromosomes, (ii) evolution of individual karyotypes, and (iii) evolution of mass-karyotypes, which refers to evolution of karyotype at the generic, familial or ordinal level Imai et al. (2001).
Keeping these hypotheses in mind, I will briefly summarize some trends in chromosomal evolution as seen in some of the other major groups of Hymenoptera, and then turn to the bees in detail.
2.3 Hymenopteran chromosomes: variation in number
Chromosome counts for 1846 species of Hymenoptera are given in Table 1. Chromosome numbers vary widely in the Hymenoptera, from an n = 1 to a high of n = 60 (both ants)! and their numbers, karyotypes and evolution have been studied intensively in many groups and have been reviewed elsewhere (e.g. Crozier, 1975; Gokhman, 2023; Lorite and Palomeque, 2010; Ross et al., 2015). However, in spite of this variation, haploid numbers of the vast majority of Hymenoptera lie within the range typical for animals as a whole which range from 6-20, and which presumably represents the limits of the spindle apparatus to function with either large or small numbers of chromosomes (White, 1977). Interestingly Hymenoptera do exhibit some of the lowest chromosome numbers in the animal kingdom. The lowest possible chromosome number, n = 1, in the Australian ant Myrmeca croslandi (Crossland and Crozier, 1986) is rivaled only by one species of nematode (White, 1977), nonetheless the low haploid number of three (n=3) is found in some bees, parasitoids and ants (Table 1). On the other hand, the highest n of 60 in the South American ant Dinoponera lucida (Mariano et al., 2008) is much less than the highest chromosome numbers recorded in animals, the record being an n = 217–223 in the butterfly Lysandra atlantica (White, 1977).
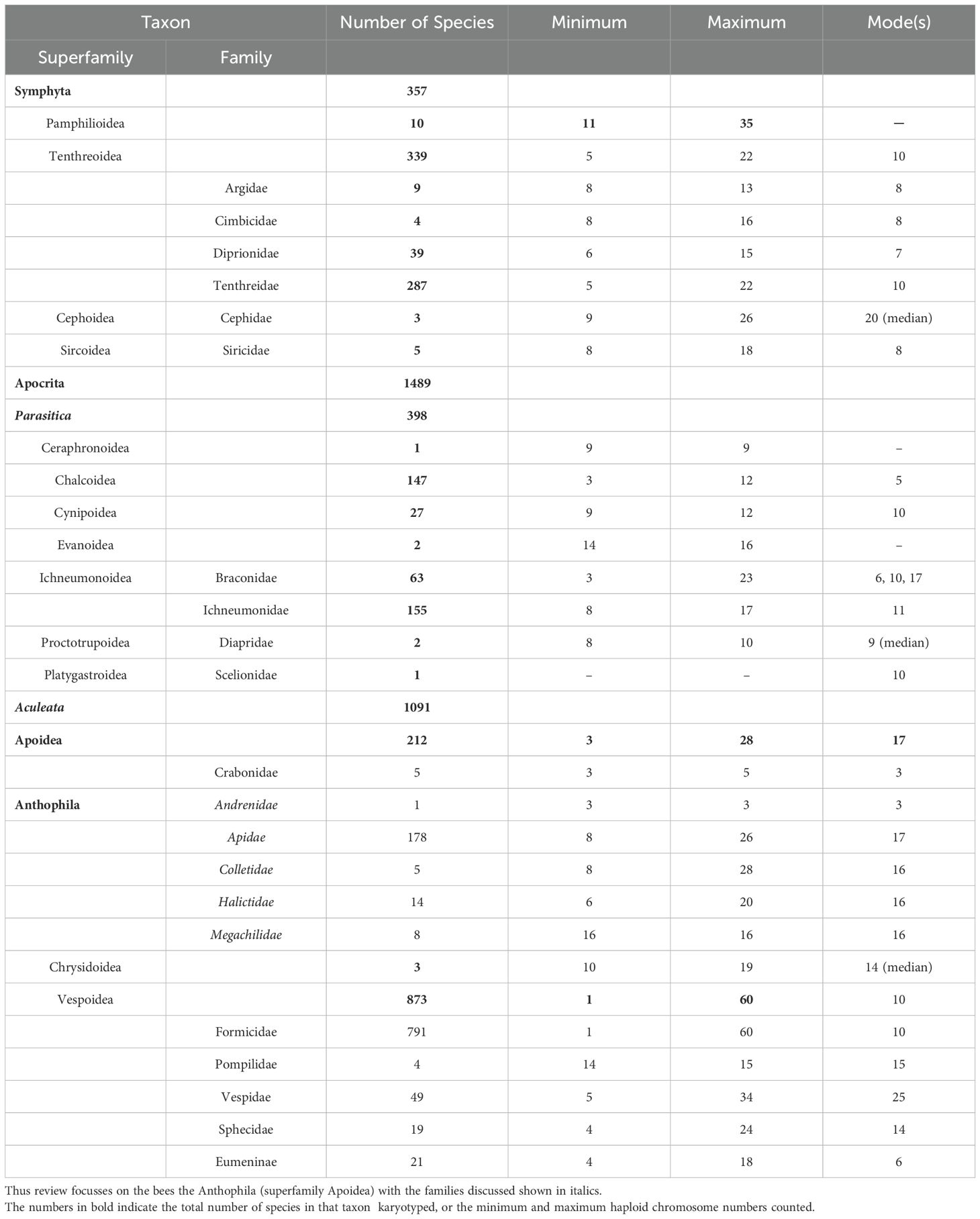
Table 1. A summary of chromosome numbers in the Hymenoptera. Haploid numbers (n) for 1846 species of Hymenoptera are given.
Polyploidy (of the germ line, as distinct from somatic polyploidy) has been suggested as a cause of major trends in hymenopteran chromosomal evolution (reviewed by Crozier, 1975) and in certain groups (for example bees, Kerr and Silveira, 1972). However, is little evidence for this except in some specific cases and other types of chromosomal rearrangements can account for the observed karyotypic evolution in the Hymenoptera (Crozier, 1975). An interesting exception is the gall wasp Diplolepis eglanteriae (Sanderson, 1988). Three other species in this genus have an n=18, whereas as D. eglanteriae has n=27, and triploidy is strikingly seen by the presence of three distinctively large chromosomes (Figure 3, in Sanderson, 1988). Similarly, Naito and Inomata (2006) identified a triploid thelytokous sawfly, Pachyprotasis youngiae. Thirty chromosomes were seen at mitotic metaphase and the karyotype clearly consists of three sets of n = 10 chromosomes (Figure 4, Naito and Inomata, 2006). Given that most other Japanese species of Pachyprotasis have a haploid number of 10, triploidy of P. youngiae is strongly implied (Naito and Inomata, 2006).
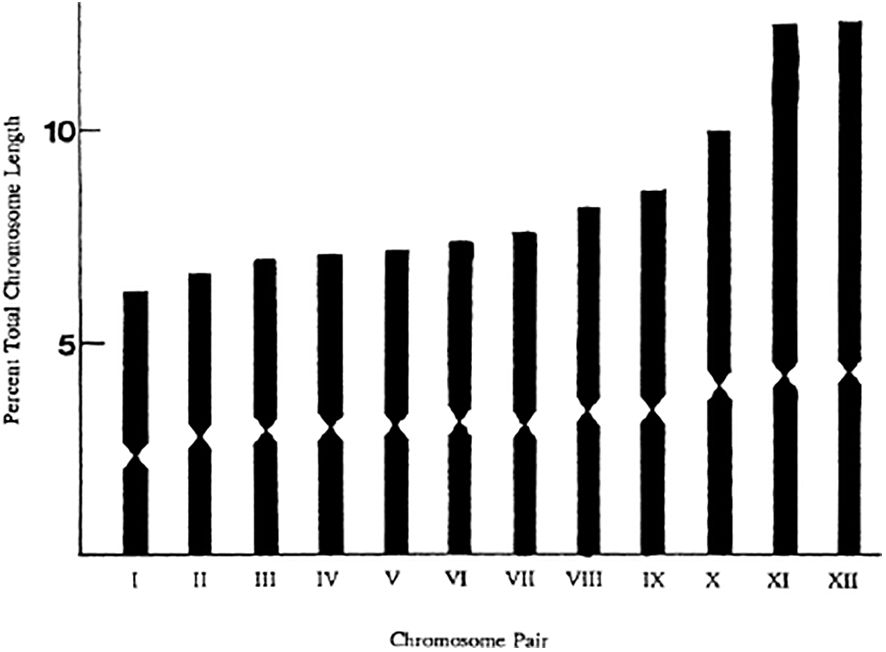
Figure 3. Ideogram of B. (Pyrobombus) perplexus. Mean percent chromosome length based on a sample of 14 nuclei, with a mean haploid total complement length of 24.1μm. From: Owen et al. (1995).
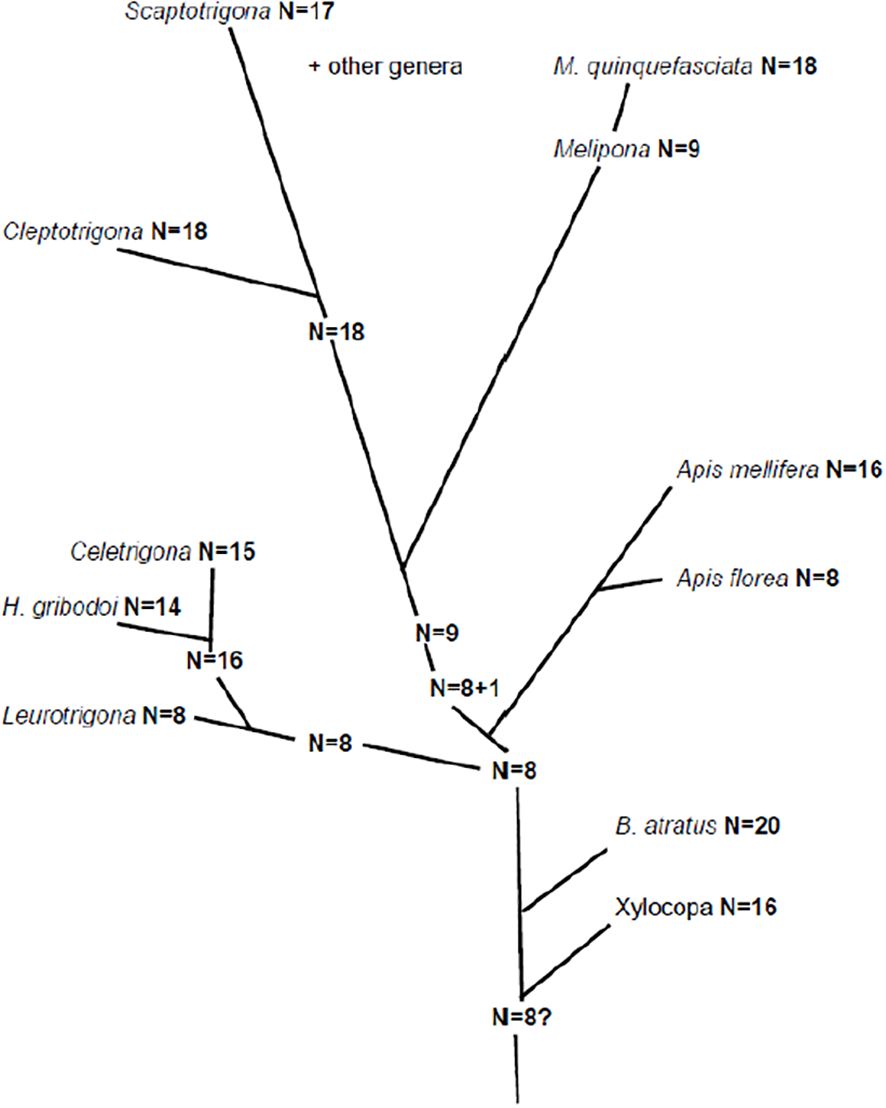
Figure 4. The polyploid hypothesis of Kerr and Silveira (1972).
In terms of total DNA content, it is worth noting that there is no obvious relationship between C-value and chromosome number in the Hymenoptera (Ardila-Garcia et al., 2010). This is the so-called C-value paradox (Thomas, 1971), for which there appears to be no one satisfactory explanation (Lakhotia, 2023). The haploid genome sizes (picograms, pg) for 131 species of Hymenoptera range from 0.1 pg in a braconid wasp to 1.14 pg in the yellow mud dauber Sceliphron caementarium (Ardila-Garcia et al., 2010). Ardila-Garcia et al. (2010) give additional genome size data for 89 species of Hymenoptera, which have a mean genome size of 0.38 pg ± 0.20 SD. For the 18 species of bees (Anthophila) they examined, taking the data from their Table 1, gives a mean of 0.57 pg ± 0.18 SD. The values ranged from 0.24 pg in Apis mellifera to 0.90 pg in the halicted Augochloropsis metallica. Thus, there appears to be no clear connection between chromosome number and genome size in the Hymenoptera as a whole, or the bees in particular.
2.4 Symphyta
Only a small fraction (about 360/7170 ≈ 5%, Table 1) of the described species in this suborder have been studied cytogenetically and for many families chromosome numbers are unknown (Westendorff, 2006). Likewise modal chromosome numbers cannot be confidently given for all families which have been studied (Westendorff, 2006). However, some families, such as the large family Tenthreidae are relatively well studied (Westendorff et al., 1999; Kuznetsova et al., 2001), but few species have been karyotyped in the Argidae, Cimbicidae and Cephidae (Westendorff and Taeger, 2002) Overall, haploid (n) chromosome numbers in the Symphyta vary from 5-35 (Table 1) (Westendorff, 2006), with some genera showing great variation both in number and morphology of chromosomes. For example, Cephalcia (Pamphiliidae) n varies from 23-35, and chromosome morphology is also highly diverse with metacentrics predominating in some species and acrocentrics in others (Westendorff, 2006). Nevertheless, given the phylogeny of the suborder (Vilhelmsen, 2006) and its position as a basal group of the Hymenoptera, it is reasonable to conclude that low n values of 7–8 is ancestral to the group, and as Hoshiba and Imai (1993) and Gokhman and Quicke (1995) suggest the ancestral chromosome number of the Hymenoptera and also the Apocrita was low; such as n = 8 or less. Thus, in the Symphyta and Apocrita chromosome evolution has, to a large extent, proceeded by fission, a major tenet of the minimum interaction theory of Imai et al. (1986). Interestingly there is direct evidence of chromosomal fission in sawflies as Rousselet et al. (2000) suggested that the origin of Neodiprion abietis with n = 8 was due to fission from a karyotype with n = 7 (typical for the Diprionidae), and that the break point of the chromosome fission was located close to an rRNA gene cluster.
2.5 Parasitica
Gokhman (2006) reviewed the trends in chromosome evolution of parasitic Hymenoptera. Haploid chromosome numbers of parasitic wasps (Parasitica + Chrysoidea) for the approximately 400 species examined range from 3-23 (Gokhman, 2006, 2009, 2023) (Table 1). He considers chromosome numbers (n) of 14–17 with a symmetrical karyotype to be ancestral. A symmetrical karyotype is one containing metacentric chromosomes of a similar size, in contrast to an asymmetrical karyotype which has predominantly acrocentric and telocentric chromosomes (intrachromosomal asymmetry) and highly variable chromosome sizes (interchromosomal asymmetry) (White, 1977; Peruzzi and Eroğlu, 2013).
The two major trends have been independent reduction in chromosome number in the major lineages and increasing asymmetry of the karyotype. Reductions to n ≤ 10–11 occurred in some groups of the Ichneumonoidea, and independently in the common ancestor of the Proctotrupoidea, Ceraphronoida, Cynipodea and Chalcidoidea (Gokhman, 2006).
2.6 Vespoidea: Formicidae
Ants (Formicidae) are the most variable chromosomally of all Hymenoptera (Table 1), and a great deal is known about their karyotypic evolution, as reviewed by Lorite and Palomeque (2010). Robersonian centric fusions and fissions, and also inversions and translocations appear to be the most important mechanisms for karyotypic evolution in ants whereas polyploidy and aneuploidy probably have a minor role (Lorite and Palomeque, 2010). For example, Santos et al. (2012) found large numbers of acrocentic chromosomes in four closely related Dinoponera ants, D. australis (n = 57), D. gigantea, (n = 41), D. lucida, (n = 59/60), D. quadriceps (n = 46) suggesting that these arose from centric fissions of metacentric chromosomes. Imai et al. (1986) have proposed the minimum-interaction theory (MIT) to account for ant chromosomal evolution, and under which chromosome numbers tend to increase, but obviously to an upper limit (Lorite and Palomeque, 2010; Cardoso and Cristiano, 2021).
2.7 Vespidae and Sphecidae
The haploid chromosome numbers of the wasps (Vespidae) vary widely from 5-34 (Table 1). Hoshiba and Imai (1993) concluded that the pattern of chromosome evolution in both of these groups conformed with the MIT starting with an ancestral low n of around three, and then proceeding in a characteristic zig-zag pattern through the karyograph and evolving high numbers. However, a more recent study by Menezes et al. (2014) on the Epiponini (swarm-founding Polistine wasps) concluded that in this group a high chromosome number of n = 33 was ancestral and that a gradual reduction in chromosome number had occurred. For example, chromosome numbers had decreased to an n = 16 in Polybia, and even within this genus a similar reduction in n had occurred within each subgenus (Menezes et al., 2014). These trends contradict the MIT of Imai et al. (1986) (Menezes et al., 2021). Menezes et al. (2014) used the computer program chromEvol v1.3 of Mayrose et al. (2010) to evaluate the direction and type of chromosome change. The program uses a probabilistic approach to estimate the most likely chromosome changes (polyploidy, gain or loss of chromosomes by fission or fusion) that have occurred when as set of chromosomes is mapped onto a known phylogeny (Mayrose et al., 2010). Menezes et al. (2014) found that a process of constant gain and loss, and no duplication (polyploidy) gave the best fit to the data and indicated the ancestral n of 33.
Twenty one species from 10 genera of the subfamily Eumeninae have been karyotyped with n values ranging from 5-18 (Tavares and Teixeira, 2021). Tavares and Teixeira (2021, 2022, 2023) have examined in detail chromosomal evolution in various solitary wasps. Notable among these is the variation they observed in Ancistrocerus flavomarginatus (Tavares and Teixeira, 2023). In the karyotype they found two larger chromosome pairs, which were almost entirely heterochromatic, and many subtelocentric chromosomes with heterochromatic short arms. They concluded that the latter resulted from chromosomal fissions (Tavares and Teixeira, 2023).
3 Chromosome numbers and karyotypes in bees
Here I will examine in some detail trends in evolution of chromosome numbers in the major lineages of bees, which are a monophyletic group – the Anthophila (Sann et al., 2018) - of the superfamily Apoidea (Table 1). Also included in the Apoidea is the Crabronidae which are a polyphyletic group of wasps (Sann et al., 2018) which will not be considered here.
I will first briefly describe some of the advances in cytological techniques that have led to an improved understanding of bee chromosomes. Conceptually it may seem that counting the chromosome number of a species is one of the easiest tasks. However, what may not be realized is that definite chromosome counts of even some of the most common and well-known species on Earth, such as Homo sapiens, and some Apis species have only been relatively recently been determined! It wasn’t until 1956 that the chromosome number of humans was definitely established as 46 by Tjio and Levan (1956). This was possible by using the squash technique rather than the old method of sectioning of testicular tissue embedded in paraffin (O’Connor, 2008).
Petrunkewitsch (1910) (quoted in Milne, 1986) in 1901 correctly determined the chromosome number of A. mellifera when he observed 32 very small chromosomes in oogonia (Milne, 1986). This was subsequently confirmed with better techniques and differences in chromosome size were observed (Sanderson and Hall, 1948). However, it wasn’t until 1977 that Fahrenhorst was able to lay to rest “the contradictory reports in the existing literature” regarding chromosomes numbers in the other Apis species (Fahrenhorst, 1977) and state unequivocally (in the English abstract) “The haploid chromosome number is 16 and this was found uniformly in Apis mellifera, A. cerana, A. dorsata and A. florea. As such the reported number of 8 haploid chromosomes in the last two species mentioned above should be rejected. It is doubtless, that in all the species of Apis, the diploid set of female germline cells consists of 32 chromosomes, as this was established for A. mellifera” (Fahrenhorst, 1977).
3.1 Cytological techniques
As with humans (O’Connor, 2008), sectioning of tissue embedded in paraffin was previously used for hymenopteran chromosomes counts; for example, an incorrect (Owen et al., 1995) n = 12 for Bombus fervidus was obtained by Whelden (1954). Although reliable chromosome counts have been obtained with sectioning (e.g. Deodikar et al., 1959), generally this is an unreliable technique (Crozier, 1975). Squash methods using fresh brain, testis, or ovary tissue yield much more better chromosome preparations (Figure 5) and have been successfully used to count bee chromosomes (e.g. Owen, 1983; Owen et al., 1995). Following dissection and fixation, the tissue can be stained using either the Feulgen technique or aceto-carmine method (Darlington and LaCour, 1976; Owen, 1983; Owen et al., 1995). Slides are prepared by teasing out a small piece of tissue into a drop of 50% acetic acid and then squashing under a cover slip (Figures 1, 5). Although the results are acceptable, a simpler air-drying technique that yields superior preparations was developed by Imai et al. (1977) and is widely used (Figure 6). In brief, the tissue to be examined is, after treatment in a colchicine-hypotonic and Fixative (I) solution, macerated on a slide under a dissecting microscope to separate out single cells and clumps of cells, then Fixative II solution is added which is then drained off the slide (Imai et al., 1977). The slide is left to dry for one day (Barcia, 2007) and then stained with Giemsa solution, and after brief washing the slide is again left to dry; it is now permanent and needs no coverslip (Imai et al., 1977). Furthermore, it also gives C-bands spontaneously (Imai et al., 1977). C-bands occur where Giemsa stains intensely, and identify regions of constitutive heterochromatin on the chromosome; however, it is not clear whether this results from DNA denaturation-renaturation (Gokhman, 2009; Kumar et al., 2021) or staining of heterochromatin-specific proteins (Gokhman, 2009). C-banding has been widely used on hymenopteran chromosomes, particularly by Imai and coworkers on bees and wasps (Hoshiba, 1984a, b; Hoshiba and Imai, 1993; Imai, 1991). The classic Giemsa stain, perfected by Gustav Giemsa in 1904, and developed originally to identify malarial plasmodia in the blood, was successful due to its stability and because of its ability to stain chromatin deeply (Fleischer, 2004; Barcia, 2007).

Figure 5. Squash preparations of (a) Bombus (Pyrobombus) ephipiatus (male brain tissue), n = 18, and (b) B. (Pyrobombus) impatiens (testes) n = 18. Scale bar = 1μm. From Owen (1983).

Figure 6. Left: Chromosomes of Lasioglossum (Evylaeus) cooleyi from the testes of a male pink-eyed pupa showing the haploid chromosome number, n = 18. From: Packer and Owen (1989); Right: Bombus appositus (male), n = 16 from Owen et al. (1995). Both preparations were made using the air-drying technique of Imai et al. (1977) followed by Giemsa staining.
There are numerous other staining techniques in addition to C-banding that give chromosome bands (e.g. Q, G, R, T) or identify certain regions on the chromosome (Kumar et al., 2021), one of these is NOR banding. Nucleolus organizer regions (NOR) of metaphase chromosomes can be visualized by silver staining techniques, either Ag-As or Ag (Kumar et al., 2021). Maffei et al. (2001) localized NORs in the bees Euglossa sp., Melipona marginate, Plebia sp., and the parasitic wasp Mellitobia australica, with AgNOR. Similarly, Rocha et al. (2002) used NOR banding on ten Brazilian species of Melipona. Similarly, Beye and Moritz (1993) used DNA probes specific to Drosophila melanogaster, coding for 28S and 18S rRNA, and found that these hybridized in situ to distinct regions of two chromosomes of the honeybee, identifying NORs.
More recently the development of single-stranded DNA probes that anneal to complementary DNA (Bishop, 2010) has allowed very specific regions of chromosomes to be identified. Of these Fluorescence In Situ Hybridization (FISH) has been used extensively for the Hymenoptera (Gokhman, 2009), with ribosomal DNA and microsatellite-satellite sequences commonly mapped (Cunha et al., 2023). For example, Cunha et al. (2023) used probes for four microsatellites to help identify possible Robertson fusion events in Neotropical meliponid bees (see below).
3.2 Variation in size and number of chromosomes
There is great interspecific variation in the number and the size of chromosomes within a taxonomic group, (White, 1977) which are generally inversely correlated because if we view this as a resulting from “…merely in packaging, if we regard chromosomes as packages of genetic material.” (Hsu and Mead, 1969). Gokhman (2009) points out this applies equally well to the Hymenoptera. Hymenopteran chromosomes at metaphase have an average length of 3-5 μm, but range in size from 0.5-17 μm (Gokhman, 2009). A dramatic example of size variation is seen in the ant subfamily Myrmeciinae where the Dinosaur Ant, Nothomyrmecia macrops, with n=47 has chromosomes of sizes 1-4 μm (Imai et al., 1988, 1990) whereas Myrmecia croslandi (Crossland and Crozier, 1986) a most unusual species in the same subfamily, has a single pair of chromosomes (the minimum possible haploid number, n=1) of length 17 μm (Gokhman, 2009). However, although the total genome size may be about the same in species with very different chromosome numbers and sizes there is no doubt that karyotypes are of adaptive significance and not just differences in “packaging” (White, 1977). Having an n of 1 may be risky since any chromosomal change such as a deletion however small could be lethal, whereas individuals with higher chromosome numbers might be able to tolerate this. Indeed M. crosslandi is known from only a few locations and is one of a set of sibling species with various chromosome numbers (Crossland and Crozier, 1986; Taylor, 1991). Ants themselves exhibit the total range of chromosome number as found in the Hymenoptera as a whole (Imai et al., 1990; Cardoso and Cristiano, 2021) from the Australian M. croslandi with n=1 to the Brazilian giant ant Dinoponera lucida with n=60 (Mariano et al., 2008). Bees show a smaller range of variation with the lowest found in the andrenid Andrena togashii (n=3) and the highest (n=28) of colletid Hylaeus sp. 2, both from Japan (Hoshiba and Imai, 1993).
The range of bee haploid chromosome numbers is shown in Figure 7 with data taken from the Bee Chromosome Database (www.bees.ufop.br) established by Cunha et al. (2021) which
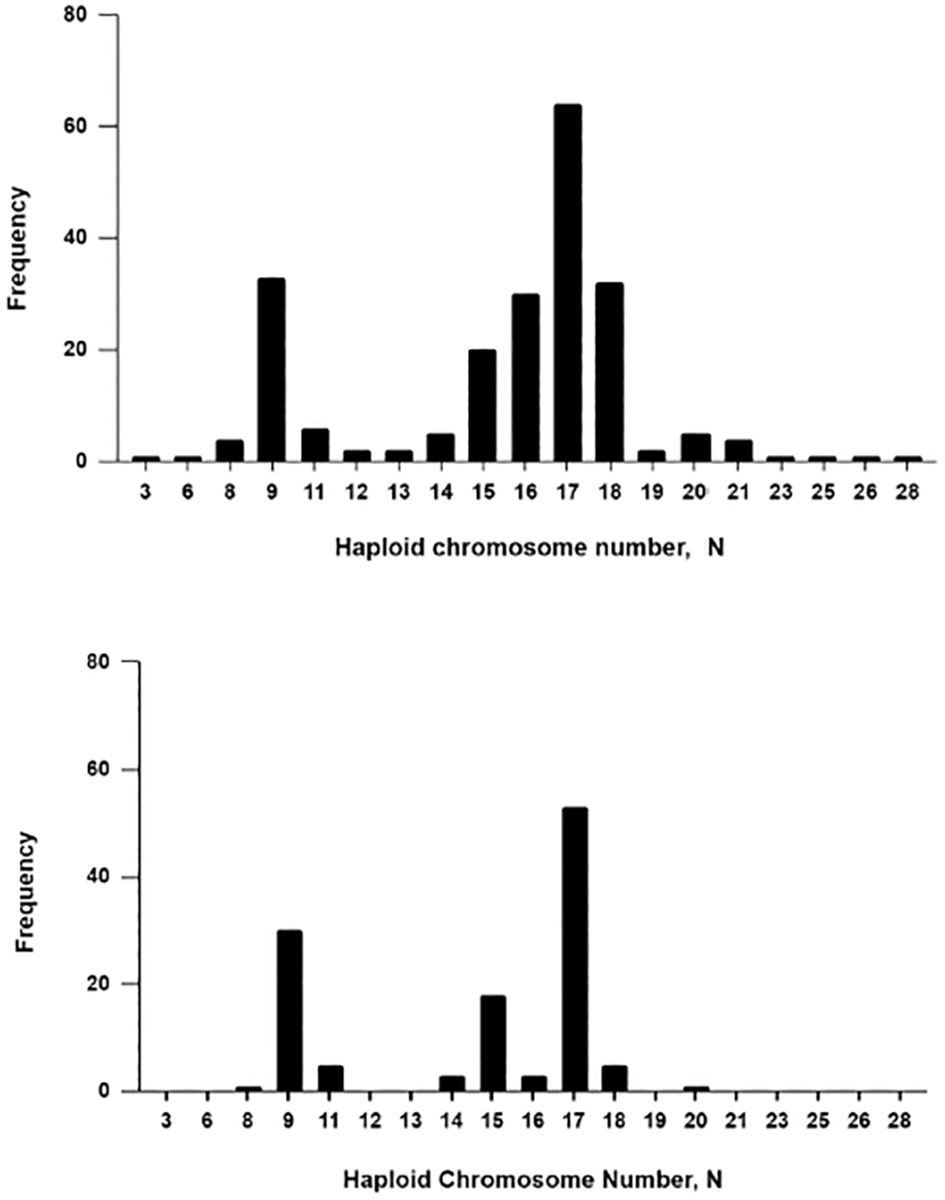
Figure 7. Top: Haploid chromosome number distribution for 215 species of Apoidea. Bottom: Haploid chromosome numbers in the 119 species of Meliponini. Data taken from: Cunha et al. (2021) The Bee Chromosome Database) https://doi.org/10.1007/s13592-020-00838-2.
“… is an online resource to gather information on chromosome number and nuclear genome size on bee species from all over the world. Considering the importance of cytogenetic studies for taxonomy, phylogeny, genetics, systematics, conservation, and evolution, the main goal of this database is to outline what has been done in the field of bee cytogenetics over the last century.”
This is a very valuable initiative and it is to be hoped that as new studies are published that the authors will upload their results to the database.
Although relatively few chromosome counts are available for bees (Apoidea) - only 215 species out of a total of 1846 hymenopterans (Table 1) and over 20,000 species of bees - it is possible that most of the entire range of chromosome numbers has been discovered (Figure 7). However, the frequency distribution may not be completely representative, since over half of the counts (119) are of meliponid species which have a distinct clade with an n=9 (Cunha et al., 2023) as seen in Figure 7. It would appear that the modal haploid number for the Apoidea as a whole is 17 and the second mode of 9 is exaggerated given the number of meliponid species that have been studied; but this is not to say that this is not of significance nor of importance. Higher chromosome numbers may be found, since ants have a maximum n value of 60 (Table 1) numbers this high could occur in bees also.
In addition to the standard chromosome complement of a species there may be supernumerary or B chromosomes in some (or many) individuals in a population, which as defined by White (1977) are “ones additional to the normal karyotype and not homologous, or only partly homologous to members of the regular set”. They are usually heterochromatic and there can be geographical variation among populations (White, 1977). B chromosomes are found in some Hymenoptera; various species of ants, a sphecid wasp (Gokhman, 2009) and the meliponid bees Melipona quinquefasciata, Tetragonisca fiebrigi (Cunha et al., 2023), Partamona cupira and P. helleri (Costa et al., 1992; Brito et al., 1997).
Returning to the question of variation in chromosome size, it is clear that the cellular mechanisms, such as spindle formation, etc, and the processes of mitosis and meiosis, are intimately related to the number and morphology of the chromosomes (White, 1977). There are also genetical implications, for example it is well known that recombination rates increase with increasing chromosome number and not just total map length (White, 1977; Sherman, 1979; Templeton, 1979). There will be genetic consequences of chromosomal fusions as this inevitable results in some loss of genes (White, 1977). Within taxonomic groups there is variation of total chromosome complement length as well as variation in size of chromosomes in some species. For example, the bumble bee Bombus pennsylvanicus shows little variation in individual chromosome length while B. fervidus shows somewhat more (Figure 8). The mean haploid total complement lengths (TCL) being 24.00 μm and 23.3 μm respectively (Owen et al., 1995). Owen et al. (1995) did find significant, but not great, differences in TCL among different bumble bee species (see also Table 2).
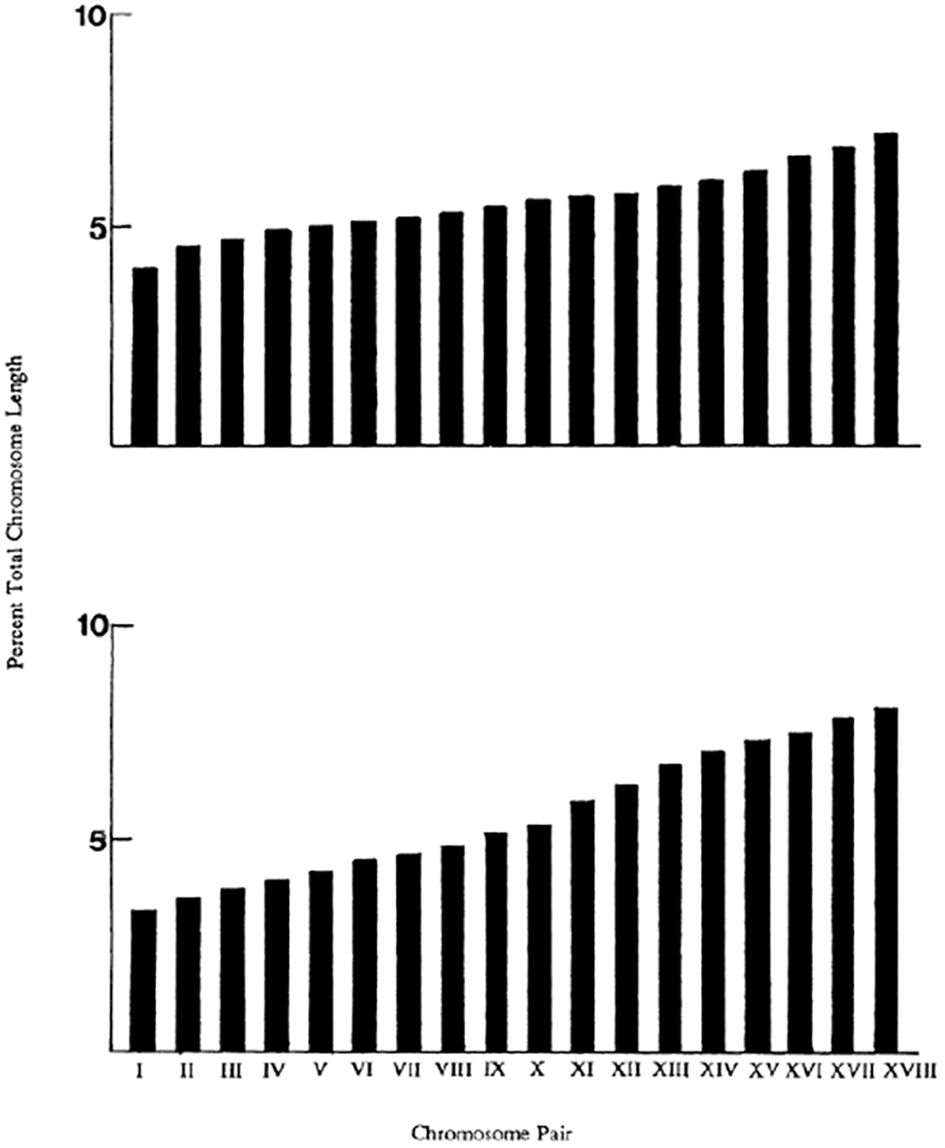
Figure 8. Representative ideograms of bumble bee chromosomes. Top, B. pennsylvanicus. Mean percent chromosome length based on a sample of 26 nuclei, with a mean haploid total complement length (TCL) of 24.00 μm. Bottom, B. fervidus. Based on 23 nuclei, with mean haploid TCL of 23.3 μm. From: Owen et al. (1995).

Table 2. Chromosome lengths in bumble bee (Bombus spp.) arranged and numbered according to subgenus (Williams et al., 2008).
3.3 Chromosome evolution in bees (Apoidea); overall trends
An early attempt to describe and understand chromosome evolution in bees was made by Kerr and Silveira (1972) and they advanced the idea that this had proceeded by various rounds of polyploidy (Figure 4). In their own words:
“The chromosome numbers published for all bees … suggest that polyploidy originated independently at least 5 times: (1) From an ancestor of Augochloropsis sparsilis n=8, to Pseudoaugoclloropsis graminea n=16; (2) from an ancestor of Leurotrigona muelleri n=8 to Frieseomelitta (3 species) n=15; (3) from an ancestral Trigonini n=9 to Plebeia (6 species) n=18; (4) from an ancestor of Melipona quadrifasciata, Melipona arginate and other species n=9 to Melipona quinquefasciata n =18; (5) from an ancestor of Apis florea n = 8, to Apis cerana and Apis mellifera n = 16.”
At the time they were writing this was plausible given the data available at that time, however even soon after it was published it was regarded skeptically (Crozier, 1975) and is no longer accepted. There is no evidence of widespread polyploidy, and where it has rarely been found it is quite obviously unusual, as already discussed earlier with the triploid gall wasp (Sanderson, 1988) and sawfly (Naito and Inomata, 2006). Also, some of the chromosome numbers were incorrect; as we have seen it was not until 1977 that it was established that all Apis species had an n=16 (Fahrenhorst, 1977), also Kerr’s (1972) count of 16 for Melipona quinquefasciata was incorrect; it is in fact just 8 (Cunha et al., 2023).
Treating the bees as the superfamily Apoidea and the major groups as families (Table 1) rather than as subfamilies as done by Cunha et al. (2021), we can map chromosome numbers on a phylogeny (Bossert et al., 2019), as shown in Figure 9. This gives the modal haploid chromosome numbers for the major families of bees with the Apidae split into the five recognized tribes (Bossert et al., 2019).
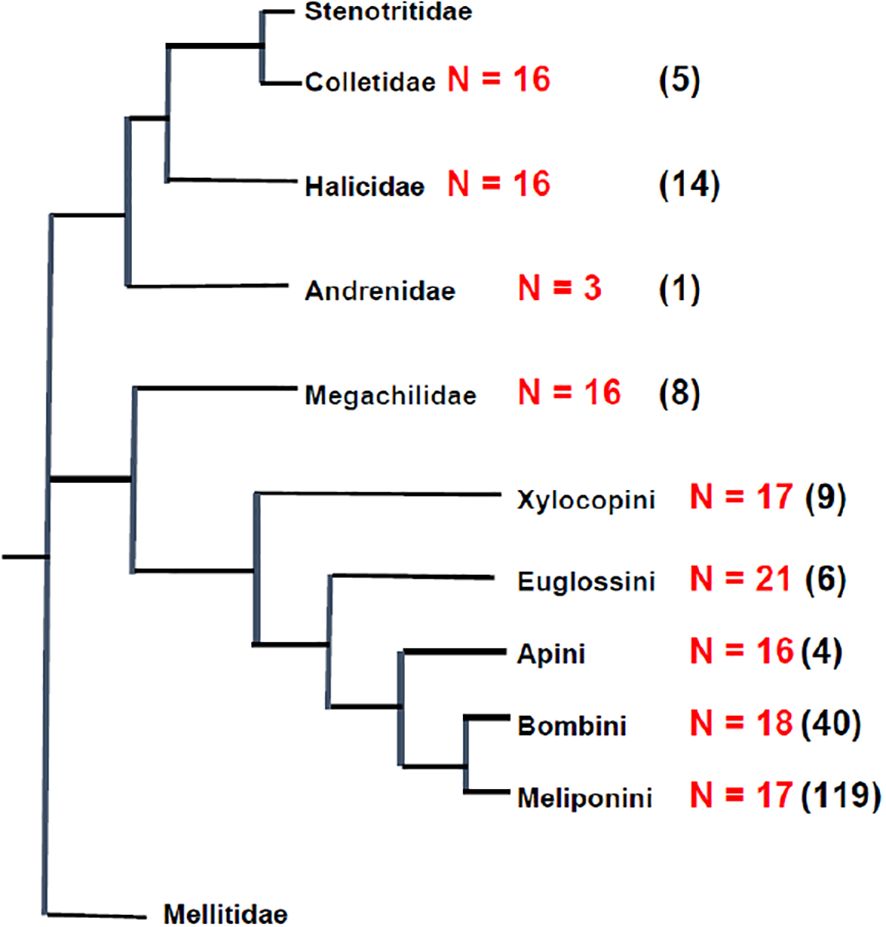
Figure 9. Phylogeny of the major lineages of the bees, superfamily Apoidea, as given in Table 1, with the modal haploid chromosome number of each tribe where known, and number of species on which this is based in parentheses. Phylogeny redrawn and simplified from Bossert et al. (2019).
As mentioned earlier, Hoshiba and Imai (1993) and Gokhman and Quicke (1995) suggest that the ancestral chromosome number of the Apocrita was low, n = 8 or less. Thus, according to this hypothesis, chromosome evolution in the Apoidea has proceeded by fission to generate the higher chromosome numbers in the families Halictidae, Megachilidae and the Apidae (Figure 9) corresponding to the general predictions of the minimum interaction theory of Imai et al. (1986). Reduction in haploid number would have occurred in the Andrenidae. Although the overall trends of the MIT seem to be met it does not necessarily mean that within groups chromosome evolution always follow the MIT, and this is particularly true with the meliponid bees (Cunha et al., 2023).
3.4 Karyotypic variation in the major taxa of bees
Relatively few species (28 total) have been karyotyped in the families Andrenidae (1), Colletidae (5), Halictidae (14), and Megachilidae (8). The only andrenid bee kayyotyped, Andrena togashii with n=3, just happens to have a particularly low chromosome number. It clearly is of importance to obtain more chromosome counts for the Andrenidae. However, it must be noted that haploid numbers of two other Andrena species are given by Goodpasture, (19741, unpublished doctoral dissertation) but which are used by Ross et al. (2015) and given in their Supplementary table of data. These are A. duboisi with n=3, and an Andrena sp. with n=10 (Goodpasture, 1974). These are not included in the Bee Chromosome Database (Cunha et al., 2021) and Cunha et al. (2021) do point out that only one andrenid bee (A. togashii) has been karyotyped. Since these observations were not published in the primary literature that is good reason to exclude them, nevertheless this does suggest a low n in this group but still with some variation. Also, it is important to recognize that Goodpasture (1974) published high quality chromosome preparations of five species of eumenid wasps so there is no reason to doubt his results for the Andrena species. The two of the three chromosomes of A. togashii are relatively long with lengths (when measured from Figure 6J of Hoshiba and Imai (1993) using their 5 μm as reference) of about 8.9 μm and 6.7 μm, the other being 3.3 μm (Hoshiba and Imai, 1993). In contrast all the 28 chromosomes of Hylaeus sp. 2 are of shorter length being about 3.3 μm (Figure 6I of Hoshiba and Imai, 1993). This does illustrate the inverse relationship between size and number of chromosomes (Gokhman, 2009).
The five species of the Colletidae show the range of haploid numbers from 8 to the highest in the bees of 28, with a mode of 16. Again, is unlikely to be a coincidence that one of the five species examined happened to have the highest n of the bees, so one might expect other species to also have high haploid numbers. Since Hylaeus is a large genus with 47 subgenera and over 650 described species globally (Michener, 2007; Almeida and Danforth, 2009), much chromosome variation is likely. It would be particularly interesting to look at the 60 Hawaiian species since they form a single clade, although the arrival of the common ancestor and the subsequent rapid adaptive radiation only occurred about 0.4–0.7 MYBP (Magnacca and Danforth, 2006). It would also be very informative to karyotype bees in the closely allied genus of Chilean bees, Xeromelissa (Almeida and Danforth, 2009), some of which, such as X. rozeni, have extremely long tongues as a result of extreme ecological adaptation (Miklasevskaja and Packer, 2015).
The 14 Halictidae karyotyped have a mode of n =16, but range of 6-20 (Figures 6, 9). A count of 21 for the former Nomia nevadensis angelesia (Cockerell, 1910), now a subspecies of Dieunomia nevadensis, was reported by Goodpasture (1974a) and is not included by Cunha et al. (2021) in the bee chromosome database and is not included “officially” here.
Chromosome counts have been made for eight leafcutter bees (Megachilidae) all of which have n=16. However, another 12 species were also karyotyped by Goodpasture (1974a) and of these all had n=16 except for one with n=15 and another with n=17.
Turning now to the five tribes of the Apidae. The modal haploid chromosome number of each tribe where known, and number of species on which this is based is shown in Figure 9.
3.4.1 Xylocopini
The nine Xylocopini species karyotyped do not show great variation in chromosome number. Small carpenter bees, Ceratina: n=17 (5 spp.), n=14 (1 spp.); large Carpenter bees, Xylocopa: n=16 (1spp.), n=17 (1 spp.) and Exoneura robusta n=13 (Bousjein et al., 2019).
3.4.2 Euglossini
Orchid bees are essential pollinators in the neotropics with hundreds of species, often very abundant and many of which are endangered (Roubik et al., 2021). Although allozyme and microsatellite variation is quite well known in these bees (e.g. López-Uribe et al., 2007; Soro et al., 2017; Souza et al., 2007, 2010) only six species have been examined cytogenetically. Of these four have n=21, one n=15, Eufriesea violacea (Gomes et al., 1998) and the other, Eg. hyacinthine n=20 (Eltz et al., 1997). Intriguingly, Fernandes et al. (2013) found high heterochromatin content in Euglossa carolina with euchromatin only at the chromosome ends, while Eg. townsendi had low heterochromatin content throughout, both species had n = 21. Gomes et al. (1998) found a similar distribution of chromatin in Eu. violacea, in which the long arm of 13 of the chromosome pairs consisted of constitutive heterochromatin, whereas in two pairs the end of the long arm was more euchromatic. Fernandes et al. (2013) suggest that these high and low degrees of heterochromatization represent a differnt mechanism of chromosome evolution in these solitary bees than in other Hymenoptera and which is not consistent with the MIT of Imai et al. (1986).
3.4.3 Apini
The tribe Apini consists of a single genus Apis comprised of three recognized clades or subgenera; Apis – the cavity-nesting species (A. mellifera, A. cerana, A. koschevnikovi, A. nigrocincta A. nulensis); Megapis – the Giant bees (A. dorsata, A. laboriosa), and Micrapis – the Dwarf bees (A. florea, A. andreniformis), thus nine species are formally distinguished (Gupta, 2014; Shanas et al., 2022). However, Shanas et al. (2022) recently described a new species, Apis karinjodian, endemic to the Western Ghats Mountain range (a biodiversity hotspot and UNESCO World Heritage Site) which runs north to south in southwestern India. Shanas et al. (2022) used both morphometrics and mitochondrial COI and COII sequence data to identify this as a new species. Their analysis also suggested that the specific status of A. indica Fabricius, 1798 be restored (Shanas et al., 2022) as it is currently treated as synonym of A. cerana (Radloff et al., 2010). Eleven therefore, would be the total number of distinct honeybee species. There are numerous subspecies or races of most species also identified on the basis of morphology, behavior, ecology and genomic sequencies (Ruttner, 1986; Le Conte and Navajas, 2008; Gupta, 2014; Carr, 2023). Interestingly there is still considerable debate over the origins of A. mellifera; either out of Africa, out of Asia, from the Middle East to Europe (Gupta, 2014), or a sole European origin (Carr, 2023).
Much is known about honeybee genetics from the pioneering work of Sladen (1913) on the Mendelian inheritance of body colour to the current genome sequencing. For example, complete mitochondrial DNA sequences are known for 21 of the 25 subspecies (Le Conte and Navajas, 2008) of A. mellifera alone (Carr, 2023). Although much cytogenetic work has been done on honeybees (e.g. Deodikar et al., 1959; Fahrenhorst, 1977; Hoshiba and Kusanagi, 1978; Milne, 1986; Stanimirovic et al., 2005) it is surprising that actual chromosome counts have only been made for four species: A. mellifera, A. dorsata, A. cerana and A. florea, and all have n=16 (Figure 9). Although sectioning techniques are not ideal, Deodikar et al. (1959) did achieve good preparations with both diploid (worker destined) and haploid eggs and larvae using this method with A. indica. Their photographs clearly show 16 and 32 chromosomes for haploid and diploid eggs respectively (Deodikar et al., 1959). Brito and Oldroyd (2010) refined the technique for preparing karyotypes from eggs and found that three-day old eggs yielded cells in metaphase with clearly seen chromosomes.
As mentioned earlier, Fahrenhorst (1977) demonstrated that all four Apis species all had the same haploid number of chromosomes. He used testes of white eyed drone pupae and a maceration and evaporation method which gave excellent results with the sister chromatids clearly visible in his preparations (Fahrenhorst, 1977). Hoshiba and Kusanagi (1978) using male and female gonadal tissue and drone head ganglia, provided a more detailed analysis and classified the A. mellifera chromosomes as consisting of 8 metacentric (m) and 8 submetacentric (sm) pairs with lengths of 1.3-4.3 μm. Later, Hoshiba (1984a, b) using tissue (testes) from young larvae of haploid and diploid males and C- and G-banding refined the classification to 4 m and 12 sm pairs, and also found that each chromosome had a unique banding pattern. Some comparison in banding pattern of chromosomes between honeybee species was done by Hoshiba and Imai (1993) where they found different c-banding patterns in A. cerana japonica and A. mellifera ligustica in at least six of their chromosomes as shown in their Figures 3F, G.
Stanimirovic et al. (1999a, b, 2005) in their extremely detailed studies of chromosomal variation in A. mellifera carnica, found differences in length of some chromosomes and in G-banding patterns among different ecotypes in Serbia. They studied three populations corresponding to three ecotypes2; Banat (B), Timok (T) and Syenichko – Peshterski (S-P), distributed roughly north to south in present day Serbia (Figure 1 in Stanimirovic et al., 2005). Stanimirovic et al. (2005) consider that “…honey bees of each ecotype investigated … are adapted to specific microclimatic and floristic conditions of the region they inhabit.” That these represent semi-isolated populations is very likely since Stanimirovic et al. (2005) sampled small apiaries at least 7 km distant from any others, and which had been established for at least 50 years. Moreover, traditional honeybee keeping practices had been followed and requeening of colonies was strictly natural (Stanimirovic et al., 2005). Stanimirovic et al. (1999a) observed significant differences in the relative chromosome and arm lengths between bees from the B and S-P ecotypes; chromosomes 12, 2, 3, 1 and 6 being longer in the S-P ecotype, while chromosomes 15, 14 and 11 were longer in the B ecotype (Stanimirovic et al., 1999a). G-bands of chromosomes 2, 4, 11 and 13 showed different patterns in T and B ecotypes, and for the T and S–P ecotypes, there were differences for chromosomes 1, 12, 15 and 16 Stanimirovic et al. (1999b, 2005). Overall, the B and S-P ecotypes showed the largest differences in G-band number and distribution for chromosomes 1, 2, 4, 11, 12, 13, 15 and 16 Stanimirovic et al. (2005). All these differences are clearly illustrated in Figures 7 and 8 of Stanimirovic et al. (2005). Muñoz et al. (2012) followed this up with mitochondrial DNA analysis to look for other evidence of genetic differentiation among the ecotypes and for any indication of hybridization between the subspecies A. mellifera carnica and A. m. macedonica. Both of these belong to the East Mediterranean or carnica (C-branch) as defined morphometrically (Ruttner, 1986). Muñoz et al. (2012) analyzed the tRNAleu-cox2 gene and identified seven mt haplotypes of the C-branch present, including two new ones; C2o and C2p restricted to the regions B and S-P respectively. Only the C2d haplotype was present throughout the country at frequencies 0.615, 0.500, 0.400 and 0.750 in the regions B, T, S-P and SE (Southeast3) respectively (Muñoz et al., 2012). Since C2d is found in A. m. macedonica in Greece and in countries neighbouring Serbia, this suggests introgression from A. m. macedonica into A. m. carnica (Muñoz et al., 2012). Similarly, C1a, present in A. m. carnica (region T), implies introgression from A. m. ligustica (Muñoz et al., 2012). Comparisons with surrounding honeybee populations suggest a hybrid situation between A. m. carnica and A. m. macedonica and also introgression from A. m. ligustica.
Inversions in some populations of honeybees have been detected, not through classical cytogenetics but by genomics. Wallberg et al. (2017) and Christmas et al. (2019) found genomic regions on chromosomes 7 and 9 that differed between highland and lowland populations of the honeybee A. mellifera in Kenya in East Africa. These were 573kb and 1639kb in length, and dated at ages 3.2MYBP and 1.28MYBP for chromosomes 7 and 9 respectively (Wallberg et al., 2017). These blocks are interpreted as inversions and presumably maintained by balancing selection in each region as each retain genes in a complex coadapted to the differing environmental conditions in the cooler highlands and warmer lowlands (Wellenreuther and Bernatchez, 2018). Also, one of the genomic regions contains octopamine receptor genes which may regulate differences in foraging behavior between the bees in the two habitats (Wallberg et al., 2017). Since different highland populations share the same inversions (Wallberg et al., 2017; Christmas et al. (2019), they may have had the same initial origin and then as Westram et al. (2022) suggest, have “travelled” across areas where they are maladaptive; a process consistent with some theoretical models (Westram et al., 2022).
3.4.4 Bombini
Of the 289 currently recognized species of bumble bees (Williams et al., 2022), chromosome numbers of 40 have been reported (Kerr and Silveira, 1972; Garófalo, 1973; Owen, 1983; Hoshiba and Imai, 1993; Hoshiba et al., 1995; Owen et al., 1995; Ayabe et al., 2004; Chauhan et al., 2015). Ideograms and c-banding patterns have determined for some species (Hoshiba and Imai, 1993; Owen et al., 1995; Chauhan et al., 2015). Given that the modal number of 18 is most likely to be the ancestral chromosome number for Bombus, it is not surprising that there is relatively little variation among subgenera (Figure 10), with the exception of Psithyrus, Figures 10, 11), which will be discussed in more detail shortly. Haploid numbers range from n =12 in B. Pyrobombus perplexus (Figure 3), and B. hypnorum Koch et al. (2024) to n =26 in B. (Psithyrus) citrinus with a mode of 18 (26 species, Owen et al., 1995).

Figure 10. The simplified subgenera of bumble bees (Bombus) as proposed by Williams et al. (2008) with the modal haploid chromosome numbers shown for each subgenus, and number of species on which this is based in parentheses.
Figure 11. Bombus (Psithyrus) ashtoni male, N = 25. Photograph and preparation by A. Wilkes, previouly unpublished.
Given that the divergence of the Bombini from the Meliponini is estimated to be about 34 MYBP (Hines, 2008) the n of 18 found in most species of bumble bees may represent a stable chromosome number that is optimal for this taxon. However, there is some interesting variation within and among subgenera (Figure 10). The subgenus Thoracobombus shows variation at the higher end of the scale, with n values from 17-23. The higher values being 20 in B. atratus and B. morio (Kerr and Silveira, 1972) and B. deuteronymus with n=23 (Hoshiba and Imai, 1993). These higher chromosome numbers must have resulted from fission of some chromosomes. Fusion of chromosomes likely resulted in the n of 16 for the two Subterraneobombus species appositus and borealis (Owen et al., 1995) and certainly for the exceptionally low n in B. perplexus and B. hypnorum (Figure 3, Table 2).
Chromosome length has been measured directly in some species (Owen et al., 1995; Chauhan et al., 2015), and in others chromosome size in megabases (Mb, where 1 Mb = 1 million bases) has been assessed using genomic techniques (Sun et al., 2021), or through linkage analysis for B. terrestris (Gadau et al., 2001). These different estimates are given in Table 2.
Where possible I have shown the equivalents using the conversion factor derived from the data in Gadau et al. (2001) on the genome of B. terrestris. They determined the physical size of haploid genome to be 274Mb, and the total minimum recombination size to be 1073 cM, thus 1 cM is about equal to 255 kb. Similarly, using the rough estimate that 1 Mb ≈ 1 cM (true for the human genome) we can convert the chromosome sizes given by Sun et al. (2021) to cMs and total complement lengths in μm. It is important to realize that these conversion factors appear inconsistent because those based on linkage analysis depend on the underlying recombination rate of that particular genome, so as Gadau et al. (2001) point out in the honeybee 1 cM = 50 kb due to the five times higher recombination rate in Apis as compared to Bombus. However, Stolle et al. (2011) provided a second generation linkage map for B. terrestris and estimated the size of the genome to be 433 Mb, and the total corrected map length to be 2047 cM, giving 1 cM ≈ 210 kb. Stolle et al. (2011) do quote another (unpublished) estimate of genome assembly size of 250 Mb from the Baylor College of Medicine Human Genome Sequencing Center, thus I have used the earlier estimate of Gadau et al. (2001). For B. impatiens, we do have an actual measurement of mean total complement length of 25.25 μm from Owen et al. (1995), and a genome assembly size of 242.00 Mb from Koch et al. (2024). These two independent estimates allow us to check the accuracy of the conversions. Converting the Mb back to μm gives a length of 21.22 μm (Table 2) which is reasonably close to 25.25 μm. Thus, looking at Table 2, we can see that even with these approximations, there is considerable consistency among these measurements and estimates of bumble bee chromosome sizes and total complement lengths. The latter ranges from about 21 to 30 μm, total chromosome size is about 240 Mb, and total map length about 1000 cM. The only anomaly, apart from the revised linkage map estimate of Stolle et al. (2011), is the observation by Chauhan et al. (2015) of the shorter chromosome complement length (15 μm) in male B. haemorroidalis as compared to that in females of 29 μm. This is difficult to understand. Although total chromosome lengths are reasonably consistent among species there is some variation of chromosome size within species (Figures 8, 3).
Relatively little has been done on other types of chromosome variation in bumble bees. B. deuteronymus (n=23) has one pair that has polymorphic chromosomes (Hoshiba and Imai, 1993). The recent molecular work of Sun et al. (2021), in addition to confirming the basic 18 chromosome complement of the genus (with the exception of the subgenus Psithyrus) estimated some chromosomal rearrangement (inversion) rates. These ranged from 0.0016 to 0075 inversions/Mb/My, which as they point out are much lower than rates in various Diptera, thus in this regard bumble bee chromosome evolution is relatively slow (Sun et al., 2021).
The high haploid number of the species in the subgenus Psithyrus, as compared to the rest of the Bombus is of considerable interest. It certainly has occurred by fission of some of the chromosomes. As Owen et al. (1995) point out the total complement length of B. citrinus at 25.51 μm is no longer than that of other species (Table 2). Moreover, Sun et al. (2021) have shown by genomic analysis, that the 25 chromosomes of B. turneri have arisen from fission, fusion and conservation of ancestral chromosomes.
The subgenus Psithyrus (originally classified as a separate genus of bumble bees), is a very well defined monophyletic group on the basis of many characters (Plowright and Stephen, 1973; Williams, 1985; Pamilo et al., 1987; Sun et al., 2021) and is comprised of obligate socially parasitic bees. The queens infiltrate eusocial Bombus colonies, and kill the host queen, or cohabit with the host queen and use chemical mimicry to “blend-in” and not be recognized by the host workers (Fisher, 1985; Martin et al., 2010; Lhomme and Hines, 2019; Dozier et al., 2023). Three of the 27 Psithyrus species have been karyotyped; B. (P) ashtoni n = 25 (Owen, 1983), B. (P) turneri n = 25 (Sun et al., 2021), B. (P) citrinus n = 26 (Owen et al., 1995). Note that B. (P) ashtoni may be conspecific with B. (P) bohemicus, similarly B. fernaldae and B. flavidus are also probably conspecific (Cameron et al., 2007; Hines, 2008) which would reduce the number of species to 25. When I initially observed (Owen, 1983) the B. (P) ashtoni chromosome number I regarded it as a possible outlier, however the finding of equally high n’s in the two other species suggests that an n = 25 is possibly ancestral in this subgenus. This is likely since all three species are quite well separated phylogenetically within this subgenus (Cameron et al., 2007). The following hypotheses could account for the high n of Psithyrus: (i) the common ancestor of the subgenera Thoracobombus and Psithyrus (Figure 10) could have had an n of 25, which has subsequently been reduced in Thoracobombus but retained in Psithyrus, (ii) the common ancestor of Psithyrus could have had an n of 25 which has been retained in some species but lost in others, (iii) the common ancestor of Psithyrus could have had an n of 25 which has been retained in all species. Clearly it is essential to determine haploid numbers of other Psithyrus species to help distinguish among these possibilities. For hypotheses (i) and (iii) if the n of 25 has been retained, then there are at least two obvious sub-hypotheses; either (a) there has not yet been enough time for subsequent fusion of chromosomes to occur as expected under the minimum interaction theory (Imai et al., 1986), which assumes fission-fusion cycles, or just fusion as other theories assume (King, 1993). Since the subgenus diverged quite recently, about 9 MYBP compared to other bumble bee subgenera (Hines, 2008) this is plausible. Alternatively (b) there may a selective advantage for this higher n which actively maintains it, and this is what I will now explore.
As is well known, Darwin (1859) saw the existence of sterile workers in social insect colonies as a very real problem for his idea of natural selection4, and he solved this by invoking the idea of family or colony-level selection, which although relevant in other contexts (Owen, 1986) is not really a satisfactory solution for the evolution of eusociality and altruism. However, other later authors, even R.A. Fisher (1930) pursued similar models (see Owen, 2014), even relatively recently (Nowak et al., 2010). The problem with this group-selection type approach is that the model itself implicitly assumes the outcome. Hamilton (1964) solved this conundrum with his formalization of the concept of inclusive fitness and also suggested that haplodiploidy was a major driver social evolution in the Hymenoptera since the ¾ degree of relatedness among workers would lower the cost/benefit ratio required for an altruistic allele to spread. Subsequently this “kin-selection” has become the dominant paradigm for the evolution of eusociality (Wilson, 1971; Nowak et al., 2010; Ratnieks et al., 2011; Owen, 2014). Early on Sherman (1979) pointed out that although the average relatedness among workers was ¾, the actual fraction of alleles shared, those identical by descent (IBD) will vary due to Mendelian segregation and the rules of meiosis. Thus, if workers can recognize and preferentially assist the siblings to which they are most similar, this will reduce the reproductive success of their mother (Sherman, 1979). However, as Sherman (1979) also argued, if the same number of gene loci, including ones with recognition alleles, were distributed over a large number of chromosomes then the actual number of alleles IBD would approach the average. Thus, he predicted that eusocial species should have higher chromosome numbers (2n) than their solitary counterparts, and tested this by comparing eusocial and solitary Hymenoptera, and did find a significant effect of n on eusociality (Sherman, 1979). However, since then the association has broken down and there appears to be no association per se between chromosome number and sociality, although eusocial taxa may show increased recombination rates (Kent and Zayed, 2013; Ross et al., 2015).
However, we can turn this around and can use this reasoning to postulate how high chromosome numbers may be of selective advantage to the social parasite bees, Psithyrus. Templeton (1979) extended Sherman’s (1979) reasoning to account for chromosomes of different lengths and derived the variance in the coefficient of kinship among full sisters in haplodiploid species as:
where N = the haploid chromosome number, lj = the map length (Morgans) of chromosome j, L = the total map length per genome summed over all chromosomes. If it is assumed that the chromosomes are of equal length (lj = L/N for all j) then Equation 1 reduces to,
which approaches zero as N increases (Templeton, 1979). This is a generalization of Sherman’s (1979) demonstration that the variance in relatedness among siblings decreases as chromosome numbers increase. Although the equations apply specifically to the coefficient of kinship, since this is measure of alleles IBD, it will apply to all genes. This means that as chromosome number and length increases offspring will become more and more uniform. Cuticular hydrocarbons provide Psithyrus species with the means of chemical mimicry with which to evade chemical recognition by the host species (Martin et al., 2010; Kather and Martin, 2015). If they are produced by many genes [(although not polygenic inheritance sensu quantitative genetics (Owen, 1989)] we can expect these loci to be spread out over the chromosomes so the young queen offspring of the successful Psithyrus queen will be more uniform in cuticular hydrocarbon composition the higher the chromosome number. Therefore, my hypothesis is that it will of selective advantage to a Psithyrus queen who has successfully invaded a Bombus nest to have female offspring with a chemical profile similar to hers, as their success is also more likely. The higher chromosome number will help to achieve this by reducing the variance of genes IBD, and so a larger proportion of the female offspring will have a more advantageous profile than if n was lower. Of course, the original queen will mate with a male who will probably carrying genes for a different profile (unless there is also some assortative mating as well) and the female offspring carry only half of the queen’s genes. Monoandry is the usual condition in bumble bees (Payne et al., 2003; Owen and Whidde, 2013). However, even given this, the slight, but definite selective advantage my be enough to help maintain this high chromosome number found in this subgenus. To quantify this, I have plotted Equation 2 for various n (Figure 12) assuming that the chromosomes are of equal length, and total map length constant at 1073 cM, typical of that found in bumble bees (Table 2). Increasing the number of chromosomes from 18 to 25 decreases the variance among progeny considerably, from 0.001804 to 0.001521, or 15.7%.
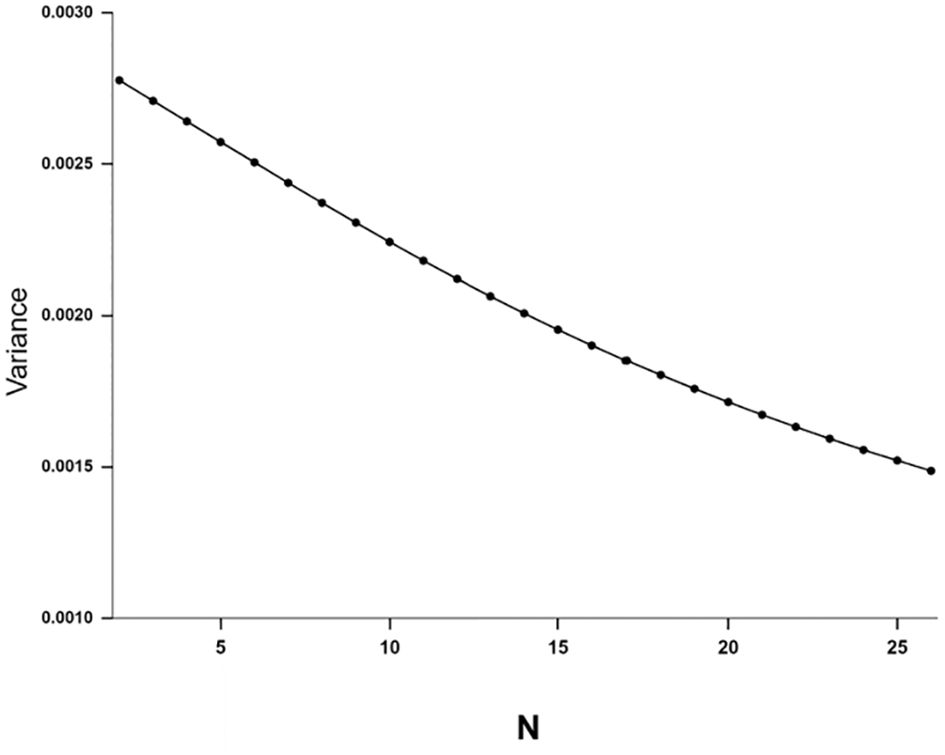
Figure 12. The variance in the coefficient of kinship among full sisters in haplodiploid species plotted using the equation where N = the haploid chromosome number, L = the total map length per genome summed over all chromosomes, assuming that the chromosomes are of equal length. The total map length was kept constant at 1073 cM. See text for more details.
Given that Psithyrus species, as are all parasites, are under intense selective pressure to evade their host defenses (Lhomme and Hines, 2019; Dozier et al., 2023) as part of the co-evolutionary “arms-race” between host and parasite (Wurdack et al., 2015) this additional selective advantage may help to retain the high n in thus subgenus. The hypothesis predicts that Psithyrus offspring in a colony will be very uniform in their cuticular hydrocarbon profile, more so than their hosts. The hypothesis can be disproved if this is not the case, also, as pointed out earlier, if not all species in this subgenus retain the high n of 25 then this will be a disproof of the hypothesis. There still has to be variation among Psithyrus females to keep up with the constant selection for increased discrimination on the part of the host species, and there is variation in Psithyrus cuticular hydrocarbon composition (see Figure 5 in Martin et al., 2010), so presumably there must be an optimal balance between variation and uniformity, which possibly occurs at this number of chromosomes. I do admit that this hypothesis is somewhat tenuous, but it is an attempt to explain the high chromosome number found in this subgenus, which is considerably greater than those of other bumble bee subgenera, and is unlikely to be coincidental.
3.4.5 Meliponini
Extensive work has been done on the chromosomes of the stingless bees (Rocha et al., 2002; Costa et al., 1992; Tavares et al., 2017, 2021; Pereira et al., 2021; Cunha et al., 2023) and at least 119 species have been karyotyped (Figure 7). Most of the species are from South America (Brazil) with a few Afrotopical and Indo-Malayan/Australasia species examined (Tavares et al., 2017). Although only 32 of the 54 extant genera of Meliponini have been karyotyped and chromosome numbers vary from n=8-20 (Tavares et al., 2017). Three main clades occur with n = 9, 15 and 17 (Tavares et al., 2017; Cunha et al., 2023) as shown in Figure 13.
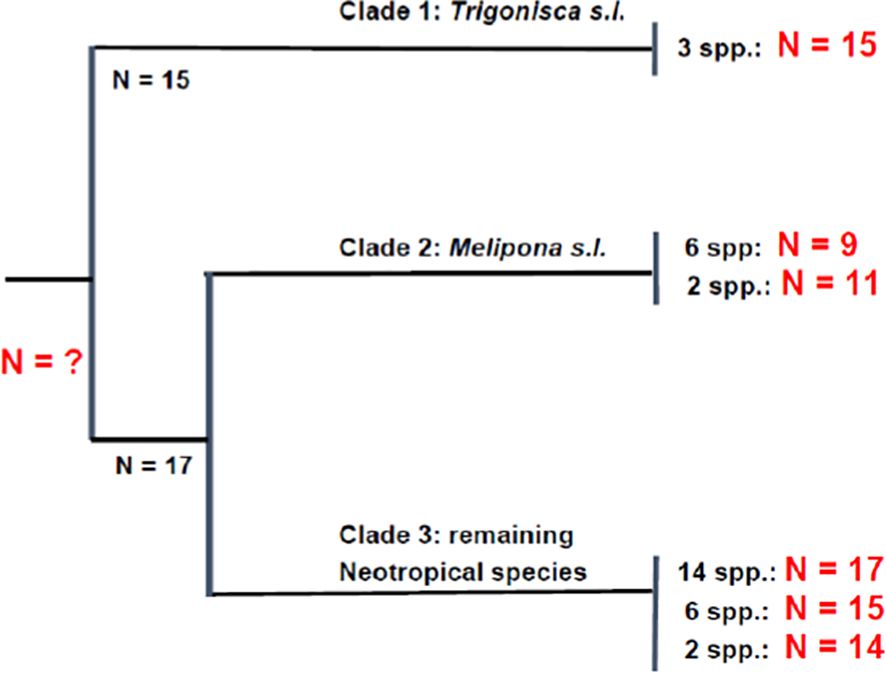
Figure 13. Haploid chromosome numbers in three clades of neotropical stingless bees (Meliponini) as identified by Cunha et al. (2023) using chromosomal mapping of 18S rDNA and five microsatellite loci. Modified and simplified from Cunha et al. (2023).
It is the number of meliponid species with n = 9 that accounts for the bimodal distribution of haploid numbers in the bees as a whole (Figure 7). Assuming that the ancestral chromosome number of the Meliponini was relatively high (n =17-18) as for the other Apidae (Figure 9) then clearly reduction by fusion has occurred to yield the chromosome numbers in clade 2 (Cunha et al. (2023). Tavares et al. (2017) discuss chromosome evolution in stingless bees as it relates to the Minimum Interaction Theory (MIT). They point out that for some taxa it applies quite well, but not for others. For example, Pompolo and Campos (1995) found that it explained quite well the karyotypic difference between two Leurotrigona species, whereas it does not satisfactorily account for chromosome evolution in some species of Melipona (Rocha and Pompolo, 1998).
Under MIT ancestral chromosome numbers should be low, but then would increase by a series of fissions, followed by accumulation of heterochromatin in one chromosome arm (Hoshiba and Imai, 1993; Rocha et al., 2002). In most genera of Meliponini, this is seen, but not in Melipona because of its lower chromosome numbers (n = 9, 11, Figure 13) and the position of the heterochromatin in some species (Rocha et al., 2002).
Similarly, it is difficult to account for the clade with n = 9 purely on the basis of the MIT, and Tavares et al. (2017) conclude that chromosome evolution in the stingless bees cannot simply be explained by a single process.
The Meliponini are notable for various types of chromosomal variation (Tavares et al., 2017). For example, 12 different B chromosomes occur in Partamona helleri, with geographical variation between populations (Martins et al., 2009). Tavares et al. (2021) found geographical karyotypic variation in Trigona spinioes in Brazil. Although chromosome number of 2n = 34 was constant there was variation in chromosome type (i.e. metacentric vs submetacentric, etc.), and in the localization of rDNA clusters and of a repetitive DNA sequence (Tavares et al., 2021). Meliponid bees have been studied using sophisticated techniques such as FISH, and chromosomal mapping (Figure 13) of 18S rDNA and microsatellites (Cunha et al., 2023).
4 The minimum interaction hypothesis as applied to bees
The Minimum Interaction Theory or MIT, proposed and elaborated by Imai et al. (1977, 1986, 2001) postulates that karyotypes evolve to minimize deleterious interactions between chromosomes. This occurs in cycles; centric fissions first increase the number of chromosomes, which reduces their size and the interactions between them. Subsequently there will be an increase in heterochromatin in one of the chromosomal arms to restore the stability of the telomeres. Initially the karyotypes of a group consist of a small number of large chromosomes which would evolve to give a larger number of smaller acrocentric chromosomes resulting from fissions. In many groups of bees the general trends predicted by the MIT appear to hold, increasing from a low chromosome number (n=2-8) as seen in the Halictidae and Andreniidae (Figure 9) and to higher numbers of 17-18 (Hoshiba and Imai, 1993). However, this trend certainly does not hold in all clades, for example in the stingless bees (Tavares et al., 2017). Nevertheless, the MIT is a very helpful theoretical framework with which to view chromosomal evolution in the Hymenoptera.
The karyograph method, devised by Imai et al. (1977, 1986) shows these changes visually, and allows actual changes to be plotted. Chromosomes are constrained within the two border lines KA and KM where KA are karyotypes having only acrocentric chromosomes and KM are those with only metacentric chromosomes. Summarizing from Imai et al. (2001), a haploid karyotype, K is defined as K = aA + mM with “a’’ numbers of A-chromosomes and “m’’ numbers of M-chromosomes. The haploid chromosome number (n) is n = a + m. Since the number of euchromatin arms in each A- and M-chromosome is, respectively, one and two, the haploid arm number (AN, the total arm number in K) is AN = a + 2m. Karyotypes having only A- or M-chromosomes are denoted, respectively, as acrocentric karyotypes (KA) and metacentric karyotypes (KM) as special cases.
Actual chromosome evolution is plotted on the karyograph, as done by Hoshiba and Imai (1993) by following a series of steps: (1) classify A and group chromosomes and arrange these by frequency, (2) list the chromosomal rearrangements showing morphological alterations between A and groups (see Figure 2), i.e centric fissions, fusions, inversions, etc., (3) list the various transitions that can occur, e.g. –(fis)→t-(C+)→A, (4) reconstruct chromosomal networks (not shown here, Figure 12 in Hoshiba and Imai, 1993), (5) rank each chromosomal alteration by frequency of the chromosome types involved. The results can then be plotted on the karyograph (Figure 13, in Hoshiba and Imai, 1993). This is a complicated method and requires detailed C-banding of chromosomes from many species, but does give a convincing pattern of chromosome evolution for many taxa of Hymenoptera, for example ants (Lorite and Palomeque, 2010) and many wasps (Hoshiba and Imai, 1993) and bees.
Imai et al. (2001) have modeled chromosome evolution under the MIT. They view chromosome evolution as a stochastic process and used Monte Carlo methods to simulate mass-karyotype evolution, and were able to generate theoretical karyographs similar to those derived from empirical data (Hoshiba and Imai, 1993). The MIT, although not necessarily applicable for all taxa is a very useful theoretical framework, but nevertheless does put selective limits on the extent of chromosomal rearrangements.
5 Conclusions
Although much is known about the chromosomes of bees there is still much to learn about overall trends in haploid number and chromosome organization. In this review I have focused on largely on chromosome number – the most basic aspect of all, but we are still lacking this information for many important families of bees. Only 28 species in total have been karyotyped for the families Andrenidae, Colletidae, Halictidae, and Megachilidae. The only andrenid bee karyotyped, A. togashii has the low n of 3, so we certainly need to know which other species in these families have low chromosome numbers to see if this is an exception and to further test the MIT prediction of the evolutionary increase in chromosome number. The potential adaptive value of chromosome number per se is of great interest. I propose a hypothesis to account for the high (n=25) chromosome number found in the social parasitic bumble bee subgenus Psithyrus. Straightforward counts of additional species would help resolve this question. More sophisticated techniques beyond chromosome counting and karyotyping using C-banding, yields much more detailed information about chromosomal rearrangements as shown by the work on the neotropical meliponid bees by the Brazilian cytogeneticist (Rocha et al., 2002; Costa et al., 2004; Tavares et al., 2017, 2021; Pereira et al., 2021; Cunha et al., 2023). When these techniques are applied to other taxa of bees they will undoubtedly reveal features of great interest. Genomic approaches are starting to identify chromosomal rearrangements such as inversions and this holds much potential to explore their adaptive significance.
Author contributions
RO: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
I thank two reviewers for helpful suggestions.
In memoriam
The late Dr. Klaus Rothfels to a great extent inspired my interest in cytogenetics and introduced me to various techniques of chromosome preparation.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ Goodpasture, C. (1974). Cytological data and classification of the Hymenoptera. University of California, Davis. Unpublished Ph.D.
- ^ The concept of an ecotype was introduced by Turesson (1922) and a revised version by Le Moan et al. (2016) who define ecotypes as “…populations of the same species which have evolved heritable physiological, morphological, behavioral or life history differences that are closely associated with environmental variation”.
- ^ another region added in this study.
- ^ In fact, he saw the saw the evolution of different morphological castes within the same colony as the real difficulty (Ratnieks et al., 2011).
References
Almeida E. A. B. and Danforth B. N. (2009). Phylogeny of colletid bees (Hymenoptera: Colletidae) inferred from four nuclear genes. Mol. Phylogenet. Evol. 50, 290–309. doi: 10.1016/j.ympev.2008.09.028
Ardila-Garcia A. M., Umphrey G. J., and Gregory T. R. (2010). An expansion of the genome size dataset for the insect order Hymenoptera, with a first test of parasitism and eusociality as possible constraints. Insect Mol. Biol. 19, 337–346. doi: 10.1111/j.1365-2583.2010.00992.x
Ayabe T., Hoshiba H., and Ono M. (2004). Cytological evidence for triploid males and females in the bumblebee, Bombus terrestris. Chromosome Res. 12, 215–223. doi: 10.1023/B:CHRO.0000021880.83639.4b
Barcia J. J. (2007). The Giemsa stain: its history and applications. Int. J. Surg. Pathol. 15, 292–296. doi: 10.1177/1066896907302239
Beye M. and Moritz R. F. A. (1993). In situ hybridization of rDNA on chromosomes of the honeybee, Apis mellifera L. Experientia 49, 337–338. doi: 10.1007/BF01923416
Bishop R. (2010). Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. BioscienceHorizons 3, 85–95. doi: 10.1093/biohorizons/hzq009
Bossert S., Murray E. A., Almeida E. A., Brady S. G., Blaimer B. B., and Danforth B. N. B. (2019). Combining transcriptomes and ultraconserved elements to illuminate the phylogeny of Apidae. Mol. Phylogenet. Evol. 130, 121–131. doi: 10.1016/j.ympev.2018.10.012
Bousjein N. S., Zahed M. A., and Oryan S. (2019). The first contribution to karyotyping of the Australian allodapine bee Exoneura robusta. J. Apicultural Res. 59, 1–3. doi: 10.1080/00218839.2019.1637225
Brito R. M., Costa M. A., and Pompolo S. G. (1997). Characterization and distribution of supernumerary chromosomes in 23 colonies of Partamona helleri (Hymenoptera, Apidae, Meliponinae). Braz. J. Genet. 20, 185–188.
Brito R. M. and Oldroyd B. P. (2010). A scientific note on a simple method for karyotyping honey bee (Apis mellifera) eggs. Apidologie 41, 178–180. doi: 10.1051/apido/2009058
Cameron S. A., Hines H. M., and Williams P. H. (2007). A comprehensive phylogeny of the bumble bees (Bombus). Biol. J. Linn. Soc. 91, 161–188. doi: 10.1111/j.1095-8312.2007.00784.x
Cardoso D. C. and Cristiano M. P. (2021). Karyotype diversity, mode, and tempo of the chromosomal evolution of attina (Formicidae: myrmicinae: attini): is there an upper limit to chromosome number? Insects. 12, 1084. doi: 10.3390/insects12121084
Carr S. M. (2023). Multiple mitogenomes indicate things fall apart with out of Africa or Asia hypotheses for the phylogeographic evolution of honey bees (Apis mellifera). Sci. Rep. 13, 9386. doi: 10.1038/s41598-023-35937-4
Chauhan A., Rana B. S., and Kumari. S. (2015). Karyological studies on bumble bee, Bombus haemorrhoidalis Smith in Himachal Pradesh. Int. J. Farm Sci. 5, 129–139.
Christmas M. J., Wallberg A., Bunikis I., Olsson A., Wallerman O., and Webster M. T. (2019). Chromosomal inversions associated with environmental adaptation in honeybees. Mol. Ecol. 28, 1358–1374. doi: 10.1111/mec.2019.28.issue-6
Costa M. A., Pompolo S. G., and Campos L. A. O. (1992). Supernumerary chromosomes in Partamona cupira (Hymenoptera, Apidae, Meliponinae). Rev. Bras Genet. 15, 801–806.
Costa K. F., Brito R. M., and Miyazawa C. S. (2004). Karyotypic description of four species of Trigona (Jurine, 1807) (Hymenoptera, Apidae, Meliponini) from the State of Mato Grosso. Brazil Genet Mol Biol. 27, 187–190.
Crossland M. W. J. and Crozier R. H. (1986). Myrmecia pilosula, an ant with only one pair of chromosomes. Science 231, 1278.
Crowley L. and Sivell O. (2023). The genome sequence of the Tree Bumblebee, Bombus hypnorum (Linnaeus 1758). Wellcome Open Res. 8, 21. doi: 10.12688/wellcomeopenres
Crozier R. H. (1975). Animal Cytogenetics, Vol. 3. Insecta. 7. Hymenoptera (Berlin: Gebruder Borntraeger), 95pp.
Cunha M. S., Cardoso D. C., Cristiano M. P., Campos L. A. O., and Lopes D. M. (2021). The bee chromosome database (Hymenoptera: apidae). Apidologie 52, 493–502. doi: 10.1007/s13592-020-00838-2
Cunha M. S., Garcia M. V. B., Campos L. A. O., and Lopes D. M. (2023). Cytotaxonomy and karyotype evolution in Neotropical Meliponini (Hymenoptera: Apidae) inferred by chromosomal mapping of 18S rDNA and five microsatellites. J. Apicultural Res. 63, 208–218. doi: 10.1080/00218839.2023.2179228
Darlington C. D. and LaCour L. F. (1976). The Handling of Chromosomes. 6th ed (London: Allen & Unwin Ltd), 201pp.
Darwin C. R. (1859). On the Origin of Species by means of Natural Selection, or the preservation of favoured races in the struggle for life (London: John Murray).
Deodikar G. B., Thakar C. V., and Shah P. N. (1959). Cyto-genetic studies in Indian honey-bees. Proc. Indian Acad. Sci. 49, 194–206. doi: 10.1007/BF03051656
Dozier A. E., Straw E. A., and Stanley D. A. (2023). Systematic review of cuckoo bumblebee research reveals data gaps and understudied species. Ecol. Entomology 48, 636–649. doi: 10.1111/een.13260
Eltz T., Schmid M., and Roubik D. W. (1997). Haploid karyotypes of two species of orchid bees (Hymenoptera: apidae, euglossini). J. Kansas Entomological Soc. 70, 142–144.
Fahrenhorst H. (1977). Nachweis ubereinstimmender Chromosomen-Zahlen (n = 16) bei allen 4 Apis-Arten. Apidologie 8, 89–100. doi: 10.1051/apido:19770107
Fernandes A., Werneck H. A., Pompolo S. G., and Lopes D. M. (2013). Evidence of separate karyotype evolutionary pathway in Euglossa orchid bees by cytogenetic analyses. An. Acad. Bras. Cien.c 85, 937–944. doi: 10.1590/S0001-37652013005000050
Fisher R. M. (1985). Evolution and host specificity: dichotomous invasion success of Psithyrus citrinus (Hymenoptera: Apidae), a bumblebee social parasite in colonies of its two hosts. Can. J. Zool 63, 977–981. doi: 10.1139/z85-144
Fleischer B. (2004). Editorial: 100 years ago: Giemsa’s solution for staining of plasmodia. Trop. Med. Int. Health 9, 755–756. doi: 10.1111/j.1365-3156.2004.01278.x
Gadau J., Gerloff C., Krüger N., Chan H., Schmid-Hempel P., Wille A., et al. (2001). A linkage analysis of sex determination in Bombus terrestris (L.) (Hymenoptera: Apidae). Heredity 87, 234–242. doi: 10.1046/j.1365-2540.2001.00919.x
Gallis F. and van Alphen J. M. (2020). Parthenogenesis and developmental constraints. Evol. Dev. 22, 205–217. doi: 10.1111/ede.12324
Garófalo C. A. (1973). Occurrence of diploid drones in a neotropical bumblebee. Experientia 29, 726–727. doi: 10.1007/BF01944800
Garofalo C. A. and Kerr W. E. (1975). Sex determination in bees. I. Balance between femaleness and maleness genes in Bombus atratus Franklin (Hymenoptera: Apidae). Genetica 45, 203–209.
Gokhman V. E. (2006). Karyotypes of parasitic Hymenoptera: diversity, evolution and taxonomic significance. Insect Sci. 13, 237–241. doi: 10.1111/j.1744-7917.2006.00089.x
Gokhman V. E. (2023). Chromosome study of the Hymenoptera: History, current State, perspectives. Biol. Bull. Rev. 13, 247–257. doi: 10.1134/S2079086423030040
Gokhman V. E. and Quicke D. L. J. (1995). The last twenty years of parasitic hymenoptera karyology: an update and phylogenetic implications. J. Hymenoptera Res. 4, 41–63.
Gomes L. F., Brito R. M., Pompolo S. G., Campos L. A. O., and Peruquetti R. C. (1998). Karyotype and C-and G-banding patterns of Eufriesea violacea (Hymenoptera, Apidae, Euglossinae). Hereditas 128, 73–76. doi: 10.1111/j.1601-5223.1998.00073.x
Goodpasture C. (1974). Karyology and taxonomy of some species of Eumenid wasps (Hymenoptera: Eumenidae). J. Kansas Entomological Soc. 47, 364–372.
Gupta R. K. (2014). “Taxonomy and distribution of different honeybee species,” in Beekeeping for Poverty Alleviation and Livelihood Security. Ed. Gupta R. K., et al (Springer Science+Business Media, Dordrecht), 63–103.
Hamilton W. D. (1964). The genetical evolution of social behavior I and II. J. Theor. Biol. 7, 1–52. doi: 10.1016/0022-5193(64)90038-4
Heraghty S. D., Sutton J. M., Pimsler M. L., Fierst J. L., Strange J. P., and Lozier J. D. (2020). De Novo genome assemblies for three North American bumble bee species: Bombus bifarius, Bombus vancouverensis, and Bombus vosnesenskii. G3 (Bethesda) 10, 2585–2592. doi: 10.1534/g3.120.401437
Hines H. M. (2008). Historical biogeography, divergence times, and diversification patterns of bumble bees (Hymenoptera: Apidae: Bombus). Systematic Biol. 57, 58–75. doi: 10.1080/10635150801898912
Hoshiba H. (1984a). Karyotype and banding analyses on haploid males of the honey bee (Apis mellifera). Proc. Japan Acad. Ser. B 60, 122–124. doi: 10.2183/pjab.60.122
Hoshiba H. (1984b). The C-banding analysis of the diploid male and female honeybee (Apis mellifera). Proc. Japan Acad. Ser. B 60, 238–240. doi: 10.2183/pjab.60.238
Hoshiba H., Duchateau M. J., and Velthuis H. H. W. (1995). Diploid males in the bumble bee Bombus terrestris (Hymenoptera) karyotype analyses of diploid females, diploid males and haploid males. Jpn J. Entomol 63, 203–207.
Hoshiba H. and Imai H. (1993). Chromosome evolution of bees and wasps (Hymenoptera, Apocrita) on the basis of C-banding pattern analyses. Japanese J. Entomology 61, 465–492.
Hoshiba H. and Kusanagi A. (1978). Karyological study of honeybee. J. Apicultural Res. 17, 105–109. doi: 10.1080/00218839.1978.11099913
Hsu T. C. and Mead R. A. (1969). “Mechanisms of chromosomal change in mammalian speciation,” in Comparative Mammalian Cytogenetics. Ed. Benirschke K. (Springer-Verlag, New York), 8–17.
Imai H. T. (1978). On the origin of telocentric chromosomes in mammals. J. Theor. Biol. 71, 619–637. doi: 10.1016/0022-5193(78)90328-4
Imai H. T. (1991). Mutability of constitutive heterochromatin (C-bands) during eukaryotic chromosomal evolution and their cytological meaning. Jpn. J. Genet. 66, 635–661. doi: 10.1266/jjg.66.635
Imai H. T. and Crozier R. T. (1980). Quantitative analysis of directionality in mammalian karyotype evolution. Am. Nat. 16, 537–569. doi: 10.1086/283646
Imai H. T., Crozier R. T., and Taylor R. W. (1977). Karyotype evolution in Australian ants. Chromosoma 59, 341–393. doi: 10.1007/BF00327974
Imai H. T., Maruyama T., Gojobori Y. I., and Crozier R. H. (1986). Theoretical bases for karyotype evolution. 1. The minimum interaction hypothesis. Am. Nat. 128, 900–920. doi: 10.1086/284612
Imai H. T., Satta, and Takahata (2001). Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J. Theor. Biol. 210, 475–497. doi: 10.1006/jtbi.2001.2327
Imai H. T., Taylor R. W., and Crosland M. W. J. (1988). Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Jpn. J. Genet. 63, 159–185. doi: 10.1266/jjg.63.159
Imai H. T., Taylor R. W., Kubota M., Ogata K., and Wada M. Y. (1990). Notes on the remarkable karyology of the primitive ant Nothomyrmecia macrops, and of the related genus Myrmecia (Hymenoptera: Formicidae). Psyche 97, 133–140. doi: 10.1155/psyc.v97.3-4
Kather R. and Martin S. J. (2015). Evolution of cuticular hydrocarbons in the hymenoptera: a meta-Analysis. J. Chem. Ecol. 41, 871–883. doi: 10.1007/s10886-015-0631-5
Kent C. F. and Zayed A. (2013). Evolution of recombination and genome structure in eusocial insects. Communicative Integr. Biol. 6, 2. doi: 10.4161/cib.22919
Kerr W. E. (1972). Number of chromosomes in some species of bees. J. Kansas Entomol. Soc 45, 111–122.
Kerr W. E. and Silveira Z. V. D. (1972). Karyotypic evolution of bees and corresponding taxonomic implications. Evolution 26, 197–202. doi: 10.1111/j.1558-5646.1972.tb00187.x
King M. (1993). Species Evolution. The Role of Chromosome Change (New York: Cambridge University Press).
Koch J. B. U., Sim S. B., Scheffler B., Lozier J. D., and Geib S. M. (2024). Chromosome-scale genome assembly of the hunt bumble bee, Bombus huntii Greene 1860 a species of agricultural interest. G3 14, jkae160. doi: 10.1093/g3journal/jkae160
Koo D.-H., Sehgal S. K., Friebe B., and Gill B. S. (2015). Structure and stability of telocentric chromosomes in wheat. PloS One 10, e0137747. doi: 10.1371/journal.pone.0137747
Kumar S., Kiso A., and Kithan N. A. (2021). “Chromosome Banding and Mechanism of Chromosome Aberrations” in Cytogenetics-classical and molecular strategies for analysing heredity material. Eds. Larramendy M. L. and Soloeski S. (London: IntechOpen), 45–60.
Kuznetsova V. G., Westendorff M., and Nokkala S. (2001). Patterns of chromosome banding in the sawfly family Tenthredinidae (Hymenoptera, Symphyta). Caryologia 54, 227–233. doi: 10.1080/00087114.2001.10589230
Lakhotia S. C. (2023). C-value paradox: Genesis in misconception that natural selection follows anthropocentric parameters of ‘economy’ and ‘optimum’. BBA Adv. 4, 100107. doi: 10.1016/j.bbadva.2023.100107
Le Conte Y. and Navajas M. (2008). Climate change: Impact on honey bee populations and diseases. Rev. Sci. Tech. Off. Int. Epiz. 27, 499–510.
Le Moan A., Gagnaire P.-A., and Bonhomme F. (2016). Parallel genetic divergence among coastal–marine ecotype pairs of European anchovy explained by differential introgression after secondary contact. Mol. Ecol. 25, 3187–3202. doi: 10.1111/mec.2016.25.issue-13
Levan A., Fredga K., and Sandberg A. A. (1964). Nomenclature for centromeric position on chromosomes. Hereditas 52, 201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x
Lhomme P. and Hines H. M. (2019). Ecology and evolution of cuckoo bumble bees. Ann. Entomological Soc. America 112, 122–140. doi: 10.1093/aesa/say031
López-Uribe M. M., Almanza M. T., and Ordoñez M. (2007). Diploid male frequencies in Colombian populations of euglossine bees. Biotropica 39, 660–662. doi: 10.1111/j.1744-7429.2007.00287.x
Lorite P. and Palomeque T. (2010). Karyotype evolution in ants (Hymenoptera: Formicidae), with a review of the known ant chromosome numbers. Myrmecol News 13, 89–102.
Maffei E. M. D., Pompolo S. G., Silva-Junior J. C., Caixeiro A. P. A., Rocha M. P., and Dergam J. A. (2001). Silver staining of nucleolar organizer regions (NOR) in some species of Hymenoptera (bees and parasitic wasp) and Coleoptera (lady-beetle). Cytobios 104, 119–125.
Magnacca K. N. and Danforth B. N. (2006). Evolution and biogeography of native Hawaiian Hylaeus bees (Hymenoptera: Colletidae). Cladistics 22, 393–411. doi: 10.1111/j.1096-0031.2006.00119.x
Mariano C., dos S. F., Pompolo S. D. G., Barros L. A. C., Mariano-Neto E., Campiolo S., et al. (2008). A biogeographical study of the threatened ant Dinoponera lucida Emery (Hymenoptera: formicidae: ponerinae) using a cytogenetic approach. Insect Conserv. Divers. 1, 161–168. doi: 10.1111/j.1752-4598.2008.00022.x
Martin S. J., Carruthers J. M., Williams P. H., and Drijfhout F. P. (2010). Host specific social parasites (Psithyrus) indicate chemical recognition system in bumblebees. J. Chem. Ecol. 36, 855–863. doi: 10.1007/s10886-010-9805-3
Martins C. C. C., Duarte O. M. P., Waldschmidt A. M., Alves R. M. O., and Costa M. A. (2009). New occurrence of B chromosomes in Partamona helleri (Friese 1900) (Hymenoptera, Meliponini). Gen. Mol. Biol. 32, 782–785. doi: 10.1590/S1415-47572009005000065
Matthey R. (1973). “The chromosome formulae of eutherian mammals,” in Cytotaxonomy and vertebrate evolution. Eds. Chiarelli A. B. and Capanna E. (Academic Press, London), 531–616.
Mayrose I., Backer M. S., and Otto S. P. (2010). Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst. Biol. 59, 132–144. doi: 10.1093/sysbio/syp083
Menezes R. S. T., Cabral-de-Mello D. C., Milani D., Bardella V. B., and Almeida E. A. B. (2021). The relevance of chromosome fissions for major ribosomal DNA dispersion in hymenopteran insects. J. Evol. Biol. 34, 1466–1476. doi: 10.1111/jeb.13909
Menezes R. S. T., Carvalho A. F., Correia J. P. S. O., Silva T. S., Somavilla A., Costa M. A., et al. (2014). Evolutionary trends in the chromosome numbers of swarm-founding social wasps. Insect. Soc 61, 385–393. doi: 10.1007/s00040-014-0365-3
Michener C. (2007). The Bees of the World (2nd Ed.) (Baltimore: The Johns Hopkins University Press).
Miklasevskaja M. and Packer L. (2015). Fluctuating asymmetry in an extreme morphological adaptation in the Chilean bee Xeromelissa rozeni (Hymenoptera: Colletidae). Can. J. Zoology. 93, 833–840. doi: 10.1139/cjz-2015-0073
Milne C. P. (1986). “Cytology and cytogenetics,” in Bee Genetics and Breeding. Ed. Rinderer T. E. (Academic Press Inc., Orlando), 205–233.
Muñoz L., Stevanovic J., Stanimirovic L., and de la Rúa P. (2012). Genetic variation of Apis mellifera from Serbia inferred from mitochondrial analysis. J. Apicultural Sci. 56, 59–69. doi: 10.2478/v10289-012-0007-9
Naito T. and Inomata R. (2006). “A new triploid thelytokous species of the genus Pachyprotasis Hartig 1837 (Hymenoptera: tenthredinidae) from Japan and Korea,” in Recent Sawfly Research: Synthesis and Prospects. Eds. Blank S. M., Schmidt S., and Taeger A. (Goecke & Evers, Keltern), 279–283.
Nowak M. A., Tarnita C., and Wilson E. O. (2010). The evolution of eusociality. Nature 466, 1057–1062. doi: 10.1038/nature09205
Owen R. E. (1983). Chromosome numbers of 15 North American bumble bee species (Hymenoptera, Apidae, Bombini). Can. J. Genet. Cytology 25, 26–29. doi: 10.1139/g83-004
Owen R. E. (1986). Colony-level selection in the social insects: Single locus additive and nonadditive models. Theor. Population Biol. 29, 198–234. doi: 10.1016/0040-5809(86)90009-2
Owen R. E. (1989). Differential size variation of male and female bumble bees (Hymenoptera, Apidae, Bombus). J. Heredity 80, 39–43. doi: 10.1093/oxfordjournals.jhered.a110786
Owen R. E. (2014). R.A. Fisher and social insects: The Fisher-Darwin model of the evolution of eusociality. Biol. Theory 9, 347–356. doi: 10.1007/s13752-014-0168-9
Owen R. E. (2023). Half-chromatid mutation as a possible cause of mosaic males and females in Hymenoptera and rare fertile male tortoiseshell cats. Genome 66, 295–304. doi: 10.1139/gen-2023-0006
Owen R. E. and Packer L. (1994). Estimation of the proportion of diploid males in populations of Hymenoptera. Heredity 72, 219–227. doi: 10.1038/hdy.1994.31
Owen R. E., Richards K. W., and Wilkes A. (1995). Chromosome numbers and karyotypic variation in bumble bees (Hymenoptera: Apidae; Bombini). J. Kansas Entomological Soc. 68, 290–302.
Owen R. E. and Whidden T. L. (2013). Monandry and polyandry in three species of North American bumble bees (Hymenoptera: Apidae, Bombus) determined using microsatellite DNA markers. Can. J. Zoology 91, 523–528. doi: 10.1139/cjz-2012-0288
Packer L. and Owen R. E. (1989). Notes on the biology of Lasioglossum (Evylaevs) cooleyi (Crawford), An eusocial Halictine bee (Hymenoptera: Halicttdae). Can. Entomologist 121, 431–438. doi: 10.4039/Ent121431-6
Pamilo P., Pekkarinen A., and Vario S.-L. (1987). Clustering of bumble bee subgenera based on interspecific genetic relationships (Hymenoptera, Apidae: Bombus and Psithyrus). Ann. Zool. Fenn. 24, 19–27.
Payne C. M., Laverty T. M., and Lachance M. A. (2003). The frequency of multiple paternity in bumble bee (Bombus) colonies based on microsatellite DNA at the B10 locus. Insectes Soc 50, 375–378. doi: 10.1007/s00040-003-0692-2
Pereira J. A., Travenzoli N. m., Oliveira M. P., Werneck H. A., Salomão T. M. F., and Lopes D. M. (2021). Molecular cytogenetics in the study of repetitive sequences helping to understand the evolution of heterochromatin in Melipona (Hymenoptera, Meliponini). Genetica 149, 55–62.
Peruzzi L. and Eroğlu H. E. (2013). Karyotype asymmetry: again, how to measure and what to measure? Comp. Cytogenetics 7, 1–9. doi: 10.3897/compcytogen.v7i1.4431
Petrunkewitsch A. (1910). Die Richtunskorper und ihr Schicksal im befruchteten und unbefruchteten Bienenei. ZooL Hahrb. Abt. Anat. u. Ont. 14, 573–608.
Plowright R. C. and Stephen W. P. (1973). A numerical taxonomic analysis of the evolutionary relationship of Bombus and P.sirhyus (Apidae: Hymenoptera). Can. Entomol. 105, 733–743. doi: 10.4039/Ent105733-5
Pompolo S. G. and Campos L. A. O. (1995). Karyotypes of two species of stingless bees, Leurotrigona muelleri and Leurotrigona pusilla (Hymenoptera, Meliponinae). Rev. Bras Genet. 18, 181–184.
Radloff S. E., Hepburn C., Hepburn H. R., Fuchs S., Hadisoesilo S., Tan K., et al. (2010). Population structure and classification of Apis cerana. Apidologie 41, 589–601. doi: 10.1051/apido/2010008
Ratnieks F. L. W., Foster K. R., and Wenseleers T. (2011). Darwin’s special difficulty: the evolution of “neuter insects” and current theory. Behav. Ecol. Sociobiol 65, 481–492. doi: 10.1007/s00265-010-1124-8
Rocha M. P. and Pompolo S. G. (1998). Karyotypes and heterochromatin variation (C-bands) in Melipona species (Hymenoptera, Apidae, Meliponinae). Gen. Mol. Biol. 21, 41–45. doi: 10.1590/S1415-47571998000100008
Rocha M. P., Pompolo S. G., Dergam J. A., Fernandes A., and Campos L. A. O. (2002). DNA characterization and karyotypic evolution in the bee genus Melipona (Hymenoptera, Meliponini). Hereditas 136, 19–27. doi: 10.1034/j.1601-5223.2002.1360104.x
Ross L., Blackmon H., Lorite P., Gokhman V. E., and Hardy N. B. (2015). Recombination, chromosome number and eusociality in the Hymenoptera. J. Evol. Biol. 28, 105–116. doi: 10.1111/jeb.12543
Roubik D. W., Basset Y., Lopez Y., Bobadilla R., Perez, and Ramírez S. J. A. (2021). Long-term, (1979–2019) dynamics of protected orchid bees in Panama. Conserv. Sci. Pract. 3, e543. doi: 10.1111/csp2.543
Rousselet J., Monti L., Auger-Rozenberg M.-A., Parker J. S., and Lemeunier F. (2000). Chromosome fission associated with growth of ribosomal DNA in Neodiprion abietis (Hymenoptera: Diprionidae). Proc. R. Soc. London Ser. B Biol. Sci. 267, 1819–1823. doi: 10.1098/rspb.2000.1216
Ruttner F. (1986). “Geographical variation and classification,” in Bee Genetics and Breeding. Ed. Rinderer T. E. (Academic Press Inc., Orlando), 23–56.
Sanderson A. R. (1988). Cytological investigation of parthenogenesis in gall wasps (Cynipidae, Hymenoptera). Genetica 77, 189–216. doi: 10.1007/BF00122389
Sanderson A. R. and Hall (1948). The cytology of the honeybee - Apis mellifica L. Nature. 162, 34–35. doi: 10.1038/162034a0
Sann M., Niehuis O., Peters R. S., Mayer C., Kozlov A., Podsiadlowski L., et al. (2018). Phylogenomic analysis of Apoidea sheds new light on the sister group of bees. BMC Evol. Biol. 18, 71. doi: 10.1186/s12862-018-1155-8
Santos I. S., Delabie J. H. C., Silva J. G., Costa M. A., Barros L. A. C., and Pompolo S.G. (2012). Karyotypic differentiation among four Dinoponera (Formicidae: Ponerinae) species. Florida Entomologist. 95, 737–742. doi: 10.1653/024.095.0324
Shanas S., Anju K. G., and Mashhoor K. (2022). Identity of cavity nesting honey bees of the Indian subcontinent with description of a new species (Hymenoptera: Apidae: Apinae: Apini: Apis). Entomon 47, 197–220. doi: 10.33307/entomon.v47i3.755
Sherman P. (1979). Insect chromosome numbers and eusociality. Am. Nat. 113, 925–935. doi: 10.1086/283445
Sladen F. W. L. (1913). Queen-rearing in England: With notes on a scent-producing organ in the worker-bee; and how pollen is collected by the honey-bee and bumble-bee. 2nd. ed (London: Madgwick Houlston & Co.), 86pp.
Soro A., Javier J., Quezada-Euan G., Theodorou P., Moritz R. F. A., and Paxton R. J. (2017). The population genetics of two orchid bees suggests high dispersal, low diploid male production and only an effect of island isolation in lowering genetic diversity. Conserv Genet. 18, 607–619. doi: 10.1007/s10592-016-0912-8
Souza R. O., Cervini M., Del Lama A., and Paxton R. J. (2007). Microsatellite loci for euglossine bees (Hymenoptera: Apidae). Mol. Ecol. Resour 7, 1352–1356. doi: 10.1111/j.1471-8286.2007.01878.x
Souza R. O., Del Lama M. A., Cervini M., Mortari N., Eltz T., Zimmermann Y., et al. (2010). Conservation genetics of neotropical pollinators revisited: microsatellite analysis suggests that diploid males are rare in orchid bees. Evolution 64, 3318–3326. doi: 10.1111/j.1558-5646.2010.01052.x
Stanimirovic Z., Popeskovic D., and Pejovic D. (1999a). Biodiversity of the honeybee Apis mellifera, Linne, (1758), from the Yugoslav regions: I – The biometric variability of the chromosomes of the Banat and Syenichko–Peshterski ecotypes. Acta Vet. (Beograd) 49, 199–206.
Stanimirovic Z., Stevanovic J., and Andjelkovic M. (2005). Chromosomal diversity in Apis mellifera carnica from Serbia. Apidologie 36, 31–42. doi: 10.1051/apido:2004067
Stanimirovic Z., Vucinic M., and Stevanovic J. (1999b). Biodiversity of the honeybee Apis mellifera, Linne, (1758), from some Yugoslav regions: II – Ultrastructural chromosomal differences between Banat and Syenichko–Peshterski honeybee ecotype. Acta Vet. (Beograd) 49, 207–214.
Stolle E., Wilfert L., Schmid-Hempel R., Kube M., Reinhardt R., FA Moritz R. F. A., et al. (2011). A second generation genetic map of the bumblebee Bombus terrestris (Linnaeus 1758) reveals slow genome and chromosome evolution in the Apidae. BMC Genomics 12, 48. doi: 10.1186/1471-2164-12-48
Sun C., Huang J., Wang Y., Zhao X., Su L., Thomas G. W. C., et al. (2021). Genus-wide characterization of bumblebee genomes provides insights into their evolution and variation in ecological and behavioral traits. Mol. Biol. Evolution. 38, 486–501. doi: 10.1093/molbev/msaa240
Takagi N. and Sasaki M. (1974). A phylogenetic study of bird karyotypes. Chromosoma 46, 91–120. doi: 10.1007/BF00332341
Tavares M. G., Ferreira R.d., Travenzoli N. M., and Lopes D. M. (2021). Karyotypic variation in the stingless bee Trigona spinipes (Hymenoptera: Apidae: Meliponini) from different geographical regions of Brazil. Apidologie 52, 1358–1367. doi: 10.1007/s13592-021-00906-1
Tavares M. G., Lopes D. M., and Campos L. A. O. (2017). An overview of cytogenetics of the tribe Meliponini (Hymenoptera: Apidae). Genetica 145, 241–258. doi: 10.1007/s10709-017-9961-2
Tavares M. G. and Teixeira G. A. (2021). Comparative cytogenetic analysis of three eumeninae species (Hymenoptera, vespidae). Cytogenet. Genome Res. 61, 203–212. doi: 10.1159/000515082
Tavares M. G. and Teixeira G. A. (2022). Classic and molecular cytogenetic analysis unveils different chromosome rearrangements shaping the karyotype of Monobia angulosa Saussure 1852 (Hymenoptera: vespidae: eumeninae). Biol. J. Linn. Soc. 136, 145–154. doi: 10.1093/biolinnean/blac011
Tavares M. G. and Teixeira G. A. (2023). Cytogenetic characterization of solitary wasp Ancistrocerus flavomarginatus (Brèthes 1906) (Hymenoptera, Vespidae) with insights into the chromosomal evolution in the genus. Genome. 66, 62–67. doi: 10.1139/gen-2022-0095
Taylor R. W. (1991). Myrmecia croslandi sp. n., a karyologically remarkable new Australian jack-jumper ant (Hymenoptera: Formicidae: Myrmeciinae). J. Aust. Ent. Soc 30, 288.
Templeton A. R. (1979). Chromosome number, quantitative genetics, and eusociality. Am. Nat. 113, 937–941. doi: 10.1086/283446
Thomas C. A. J. (1971). The genetic organization of chromosomes. Annu. Rev. Genet. 5, 237–256. doi: 10.1146/annurev.ge.05.120171.001321
Tjio J. H. and Levan A. (1956). The chromosome number of man. Hereditas 42, 1–6. doi: 10.1111/j.1601-5223.1956.tb03010.x
Todd N. B. (1970). Karyotypic fissioning and canid phylogeny. J. Theor. Biol. 26, 445–480. doi: 10.1016/0022-5193(70)90096-2
Turesson G. (1922). The species and variety as ecological units. Hereditas 3, 100–113. doi: 10.1111/j.1601-5223.1922.tb02727.x
Vilhelmsen L. (2006). “Developments in ‘Symphytan’ phylogenetics in the 20th Century: Towards a consensus,” in Recent Sawfly Research: Synthesis and Prospects. Eds. Blank S. M., Schmidt S., and Taeger A. (Goecke & Evers, Keltern), 31–38.
Wallberg A., SchoÈning C., Webster M. T., and Hasselmann M. (2017). Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. PloS Genet. 13, e1006792. doi: 10.1371/journal.pgen.1006792
Wellenreuther M. and Bernatchez L. (2018). Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427–440. doi: 10.1016/j.tree.2018.04.002
Westendorff M. (2006). “Chromosomes of sawflies (Hymenoptera: Symphyta) – a survey including new data,” in Recent Sawfly Research: Synthesis and Prospects. Eds. Blank S. M., Schmidt S., and Taeger A. (Goecke & Evers, Keltern), 39–46.
Westendorff M., Kuznetsova V., Taeger A., and Blank S. (1999). Karyotype diversity in the sawfly family tenthredinidae (Symphyta, hymenoptera): new data and review. Cytologia. 64, 401–409. doi: 10.1508/cytologia.64.401
Westendorff M. and Taeger A. (2002). New data on chromosomes of sawflies in the families Argidae, Cimbicidae and Cephidae (Hymenoptera, Symphyta). Beitr. Ent. 52, 347–352. doi: 10.21248/contrib.entomol.52.2.347-352
Westram A. M., Faria R., Johannesson K., Butlin R., and Barton N. (2022). Inversions and parallel evolution. Phil.Trans. R. Soc B 377, 20210203. doi: 10.1098/rstb.2021.0203
Whelden R. M. (1954). Notes on the bumble-bee (Bombus fervidus Fabricius) and its chromosomes. J.N.Y. Entomol. Soc. 32, 91–97.
White M. J. D. (1977). Animal Cytology and Evolution. 3rd. ed (Cambridge: Cambridge University Press).
Whiting P. W. (1933). Selective fertilization and sex-determination in Hymenoptera. Science 78, 537–553. doi: 10.1126/science.78.2032.537.b
Williams P. H. (1985). A preliminary cladistic investigation of relationships among the bumble bees (Hymenoptera, Apidae). Systematic Entomology 10, 239–255. doi: 10.1111/j.1365-3113.1985.tb00529.x
Williams P. H., Cameron S. A., Hines H. M., Cederberg B., and Rasmont P. (2008). A simplified subgeneric classification of the bumblebees (genus Bombus). Apidologie 39, 46–74. doi: 10.1051/apido:2007052
Williams P. H., Françoso E., Martinet B., Orr M. C., Ren Z., Santos Júnior J. E., et al. (2022). When did bumblebees reach South America? Unexpectedly old montane species may be explained by Mexican stopover (Hymenoptera: Apidae). Systematics Biodiversity 20, 1–24. doi: 10.1080/14772000.2022.2092229
Keywords: Hymenoptera, bees, Apidae, chromosomes, karyotypes, inversions
Citation: Owen RE (2025) Chromosome evolution in bees. Front. Bee Sci. 3:1395037. doi: 10.3389/frbee.2025.1395037
Received: 02 March 2024; Accepted: 17 April 2025;
Published: 26 May 2025.
Edited by:
David De Jong, University of Sao Paulo, BrazilReviewed by:
Sean Brady, Smithsonian National Museum of Natural History (SI), United StatesTiago Mauricio Francoy, University of São Paulo, Brazil
Thiago S. Depintor, University of São Paulo, Brazil
Copyright © 2025 Owen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin E. Owen, cm93ZW5AbXRyb3lhbC5jYQ==
 Robin E. Owen
Robin E. Owen