- Amity Institute of Molecular Medicine and Stem Cell Research (AIMMSCR), Amity University, Noida, Uttar Pradesh, India
The voltage gated calcium channels (Cav1 and Cav2) and sodium channels are modulated by calmodulin (CaM) via IQ motifs. But Cav3 (aka T-type) channels lack IQ motif and therefore, they transiently interact with CaM via the gating brake (GB) regions of T-type channels. However, the structural basis of the interactions remains unclear. This study employs molecular dynamics (MD) simulations to investigate the complete binding process of GB peptides (GB3.1 and GB3.2) with CaM at an atomic level, starting from a non-interacting state to a fully formed complex. We provide a detailed analysis of the binding trajectories, identifying how the GB peptides dynamically explore and engage their binding interfaces on CaM. Our results reveal that GB3.1 induces significant conformational rearrangement in CaM, bending its central helix by ∼90° and forming a compact structure. In contrast, GB3.2 binding does not induce such changes, and CaM remains in an extended conformation. Both peptides interact primarily with CaM’s N-lobe. The MM-PBSA analysis yielded negative binding energies indicating a spontaneous and favourable complex formation.
Introduction
T-type ion channels represent a class of voltage-gated ion channels that play a crucial role in regulating cellular excitability (Chemin et al., 2002). In contrast to high voltage activated (HVA) calcium channels (Cav1 and Cav2), the T-type channels (Cav3) exhibit distinctive biophysical properties marked by low activation thresholds and transient openings (Catterall, 2011a; Senatore et al., 2014; Simms and Zamponi, 2014). Their involvement in diverse physiological processes, such as neuronal firing (Chemin et al., 2002; Cain and Snutch, 2010; Shao et al., 2021), pacemaker activity in the heart (Ferron et al., 2002; Mesirca et al., 2015), hormone secretion (Barghouth et al., 2022), sensory processing in the retina and auditory system (Hong et al., 2014; Lundt et al., 2019), muscle contraction and pain perception (Marger et al., 2011; Hong et al., 2014; Korczeniewska et al., 2022), gene expression and cell proliferation (Santoni et al., 2012; Antal and Martin-Caraballo, 2019; Yabuki et al., 2021), and even learning and memory formation (Benkert et al., 2019; Fukunaga et al., 2019; Joksimovic et al., 2023), underscores their significance in maintaining cellular homeostasis.
Additionally, T-type channels are implicated in neuropathic pain, where heightened activity in sensory neurons influences pain signalling and perception (Kerckhove et al., 2014; Bourinet et al., 2016; Harding and Zamponi, 2022). In the cardiovascular system, dysfunctions in T-type channel regulation have been implicated in cardiac arrhythmias, disrupting the normal rhythm of the heart (Clozel et al., 1999; Shah et al., 2022). Furthermore, some studies suggest their diverse roles in oncogenesis. In prostate cancer, Cav3.2, a subtype of T-type VGCCs, is notably overexpressed at both mRNA and protein levels, influencing cellular morphology, mitogenic factor secretion, and neurite outgrowths (Mariot et al., 2002; Gackière et al., 2008; Weaver et al., 2015b; 2015a; Hall et al., 2018). Breast cancer studies reveal that inhibiting T-type Ca2+ channel function reduces the proliferation of MCF-7 cells (Taylor et al., 2008; Lu et al., 2020). Ovarian cancer cells also demonstrate decreased proliferation upon T-type channel inhibition (Li et al., 2011; Dziegielewska et al., 2016).
T-type ion channels are classified into three main types based on their subunit composition. First, Cav3.1 (α1G) encoded by the CACNA1G gene contributes to the generation of low-threshold calcium (Ca2+) spikes. Second, Cav3.2 (α1H) is encoded by the CACNA1H gene and contributes to the initiation of burst firing in neurons. Third, Cav3.3 (α1I) subtype is encoded by the CACNA1I gene and shares similarities with Cav3.1 and Cav3.2 in terms of its functional properties (Cribbs et al., 1998; Lee et al., 1999; Perez-Reyes, 2006; Szymanowicz et al., 2024). T-type ion channels consist of pore-forming α1 subunits with multiple transmembrane domains that span the cellular membrane. A distinctive feature is the “gating brake (GB)” located in the I-II linker region, playing a pivotal role in the channels’ low-voltage activation characteristics (Perez-Reyes, 2010).
Calmodulin (CaM), a universal Ca2+ sensor found ubiquitously in eukaryotes, plays a crucial role in mediating various calcium-dependent signalling pathways. With the ability to bind more than 300 targets, CaM regulates Ca2+ influx through mechanisms such as calcium-dependent inactivation (CDI) and calcium-dependent facilitation (CDF) of neuronal Ca2+ channels (Lee et al., 2000; Han et al., 2010). The highly conserved IQ motif [FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY], crucial for CDI, is found in the C-lobe of HVA Ca2+ channels. CaM interacts with these channels through their IQ motifs and point mutations in these motifs abolish CDI properties (Blaich et al., 2012). Further, CaM binds four Ca2+ ions via EF-hand motifs, and a mutant with altered motifs abolishes CDI in ion channels (Fallon et al., 2005; Gardill et al., 2019). Thus, all HVA channels are modulated by intracellular Ca2+ ions.
The intracellular I–II loop in HVA channels plays a major role in channel function including interaction with auxiliary β subunit, influencing channel trafficking, and potentially modulating Ca2+ influx (Buraei and Yang, 2010; Catterall, 2011b; Zamponi and Currie, 2012). In T-type channels, the equivalent region hosts the GB formed a helix-loop-helix structure. The second helix of the GB establishes important contacts with the gating machinery, thereby stabilizing a closed state of T-channels, and that this interaction is disrupted by depolarization, allowing the S6 segments to spread open and Ca2+ ions to flow into the cell (Baumgart et al., 2008; Monteil et al., 2015). Since T-type ion channels lack IQ motifs, rendering them unable to interact with CaM and, consequently, having no impact on CDI or CDF (Ben-Johny et al., 2014; 2015a). However, it was found that rise in submembrane Ca2+ concentration induces a large decrease in T-type current amplitude (Cazade et al., 2017a). Further, T-type channels availability is inhibited by the intracellular Ca2+ ions (Chemin et al., 2019). However, in contrast with HVA channels, the Ca2+-dependent modulation of Cav3 channels are independent of CaM (Zamponi, 2016; Cazade et al., 2017b; Chemin et al., 2019). Another study showed that T-type channels associate with CaM at the GB’s helix two in the I-II linker despite lacking IQ motifs (Chemin et al., 2017). Therefore, it seems that CaM regulates the Cav3 channels in a more “tonic” manner, whereas the “dynamic” regulation of Cav3 observed requires phosphorylation (Cazade et al., 2017b; Chemin et al., 2019). The high-nanomolar affinity between the GB and CaM highlights the unique regulatory potential of T-type channels (Chemin et al., 2017). Recently, several cryo-EM structures of T-type channels are reported, but all of them are without CaM (Zhao et al., 2019; He et al., 2022; Huang et al., 2024). Therefore, the intricate structure of the complex formed by CaM and T-type ion channels remains elusive. This study employs molecular dynamics (MD) simulations to elucidate the dynamic interplay between CaM and T-type ion channels, aiming to understand their association and regulatory mechanisms comprehensively.
Material and methods
CaM-GB3.1 and CaM-GB3.2 model generation
To decipher the molecular interactions between CaM and T-type ion channels, we established a complex system as follows: GB peptides from Cav3.1 and Cav3.2, denoted as GB3.1 and GB3.2, respectively, were selected and modelled. The peptide GB3.1 comprises the sequence (433YEELLKYLVYILRKAARRLAQVSRA457), while GB3.2 comprises the sequence (456CYEELLKYVGHIFRKVKRRSLRLYARW482) based on earlier study (Chemin et al., 2017). These peptides fragments are α helical in structure. CaM exists mainly in extended form in the absence of any binding partner. Therefore, the extended form of the CaM structure (1CLL) was chosen to monitor the whole complex formation process (Babu et al., 1988; Chattopadhyaya et al., 1992). In our initial models, GB peptides (GB3.1/GB3.2) were placed randomly ∼25 Å away from CaM (Figure 1A), ensuring that CaM and GB peptides were initially in non-interacting position. This system was subjected to MD. simulation. The rationale behind this setup was to allow the two components (peptide and CaM) to move freely, encounter each other and form complex mimicking the natural situation as in aqueous solution. The MD simulations were subsequently conducted for CaM/GB3.1 and CaM/GB3.2 system under similar conditions as described below.
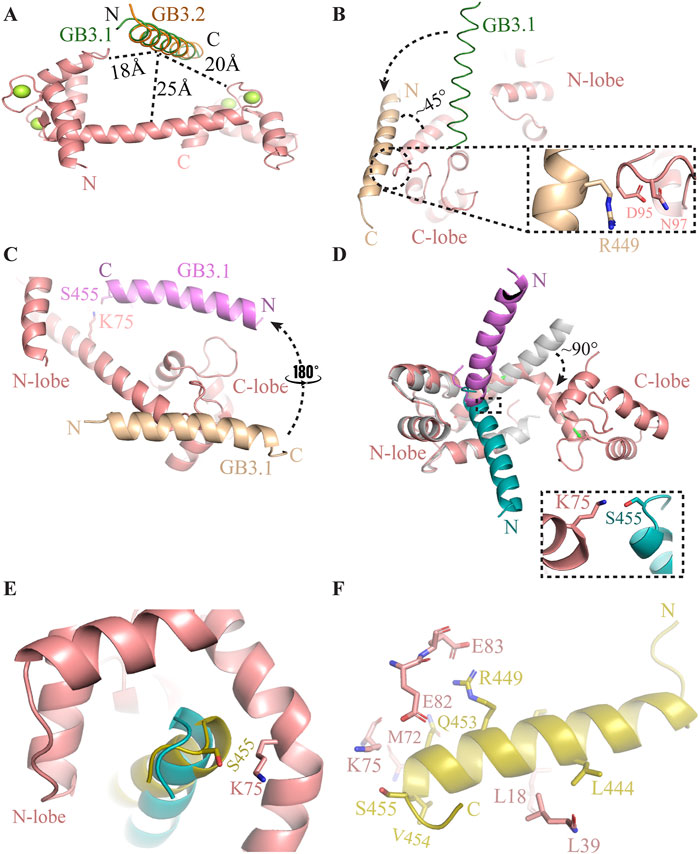
Figure 1. Process of complex formation between CaM and GB3.1 peptide to GB3.2 peptide. (A) Initial position of GB3.1 (green) and GB3.2 (orange) with respect to CaM. (B) On the onset of simulation, the GB3.1 (orange) has moved towards C-lobe from its original position (green). (C) Subsequently, GB3.1 (magenta) has made 180° transverse rotation and aligned parallelly to the central helix of CaM. (D) After that GB3.1 (cyan) had made 90° lateral rotation and become entrapped between two lobes of CaM. The central helix of CaM has made ∼90° bend to adopt compact conformation. The C-lobe in extended conformation (grey) has been deleted for clarity purpose. (E) Final position of GB3.1 (yellow) on compact ellipsoidal CaM. (F) The interaction between CaM and GB3.1 are shown in stick representation.
MD simulation
The simulations were conducted using Gromacs 2020.6 employing the CHARMM36 force field (Abraham et al., 2015). Simulation parameters were generated through the CHARMM-GUI web interface (Jo et al., 2008; Brooks et al., 2009; Lee et al., 2016). Protein molecules were placed in a cubic box, maintaining a 10 Å distance from the boundaries, and solvent water, modelled with the tip3p model, surrounded the protein molecules (Mark and Nilsson, 2001). Four Ca2+ ions were consistently present in the EF hand motifs of CaM throughout the simulations.
To maintain the ionic strength of solution, 0.15M NaCl was added, and simulations were performed under cubic periodic boundary conditions. Energy minimization procedures, involving a gradual reduction of side chain and backbone restraints, were executed over 250 picoseconds (ps). Subsequently, water equilibration was performed under NVT conditions for 125 ps, followed by an additional 125 ps under NPT ensembles at 303 K. The production run was conducted at 303 K in the NPT ensemble for 1.5 microseconds (µs) and 1.0 µs for CaM/GB3.1 and CaM/GB3.2 complexes, respectively. The time step was two femtoseconds (fs) and trajectories were saved every 10 ps. Temperature control utilized velocity scaling, while bond lengths were constrained using the LINCS algorithm. Pressure was regulated by isotropic coupling employing the Parrinello-Rahman barostat. Van der Waals and Particle Mesh Ewald electrostatic interactions (PME) within a 1.2 nm range were computed using a Verlet scheme, with van der Waals interactions switched above 1.0 nm. All MD simulations production runs were executed on the HPC facility at the Indian Institute of Technology Delhi, India (https://supercomputing.iitd.ac.in/).
Trajectory analysis
The analysis of MD results was performed using Gromacs 2020.6 package utilities. The MD trajectories were concatenated using the trjcat utility, and multiple chains of protein molecules were clustered with the trjconv utility. This clustering option accounted for periodic boundary conditions to maintain the structure integrity broken by the periodic boundary. The representative final PDB trajectory was utilized for image preparation in the manuscript. The angle between helices were calculated using the “anglebetweenhelices” script downloaded from the PyMOL Script Library. The simulation trajectory is available upon genuine request.
Molecular mechanics-poisson Boltzmann surface area (MM-PBSA)
The binding energies were computed using the MM-PBSA method (Kollman et al., 2000). To perform these calculations, the g_mmpbsa program within GROMACS was utilized, employing trajectories generated from molecular dynamics simulations (Kumari et al., 2014). The MM-PBSA method entails the assessment of three main energy components: the potential energy in vacuum, the polar solvation energy, and the non-polar solvation energy (Genheden and Ryde, 2015). Specifically, a 10 ns trajectory from the GB3.1/CaM (1460–1470 ns) and GB3.2/CaM (900–910 ns) simulation was selected for MM-PBSA analysis. These snapshot trajectories were selected after the CaM and GB peptides has associated as seen in RMSD profile (Figure 2A). Frames were extracted every 10 ps, resulting in a total of 1000 frames. The binding energy were computed as per the instructions of developers.
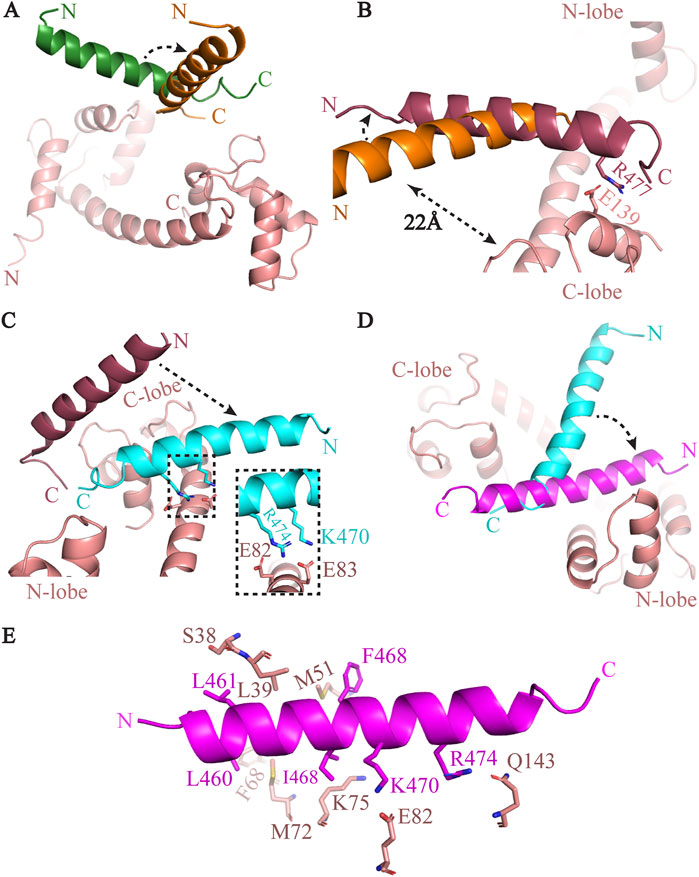
Figure 2. The complex formation process of between CaM and GB3.2 peptide. (A) The GB3.2 peptide from initial position (green) made 90° turn and aligned parallel to the central helix (shown in orange). (B) The GB3.2 has further rotated anticlockwise. (C) The GB3.2 has moved down and formed contact with CaM. The interacting residues are shown in inset. (D) The peptide has further rotated and shifted towards the N-lobe of CaM. (E) The interaction between CM and GB3.2 are shown in stick representation.
Principal component analysis (PCA)
PCA analysis was performed in Bio3D 2.2 (Grant et al., 2006; Skjaerven et al., 2014) under R environment (https://www.r-project.org/) using simulation trajectories. PCA is a multivariate technique that reduces data to two or three dimensions by maximizing the variance and analyzing inter-conformer relationships. PCA extracts biologically relevant movements of protein domains from MD simulation trajectory (Yang et al., 2009; Salsbury, 2010).
Structure predictions and comparison
The structure predictions of the CaM and GB peptide complexes were conducted using the AlphaFold2 (Jumper et al., 2021) program accessed via ChimeraX (Meng et al., 2023). AlphaFold2 requires fewer computational resources, and the accuracy of its predicted structures is nearly identical to experimentally solved structures. With the CaM and the peptides sequences the program was run with default settings. Additionally, the I-TASSER online program was also utilized for predicting the complex structures using default parameters (Zhou et al., 2022).
The hydropathicity, optimized matching hydrophobicity, and polarity profiles of the IQ motif peptides and GB peptides were calculated using ProtScale (Gasteiger et al., 2005), a bioinformatics tool available on the ExPASy Server (https://web.expasy.org/protscale/).
Results
The interaction between CaM and T-type ion channels via GB is transient, making experimental structure determination challenging. We employed a novel approach to elucidate the transient interactions between CaM and GB peptides. Our method integrates information from structurally homologous complexes with MD simulations to resolve the interactions within these elusive complexes. We chose peptides from GB3.1 and GB3.2, representing the GB regions of Cav3.1 and Cav3.2, respectively. The extended structure of CaM is obtained from PDB, and the GB peptide structure is predicted to be α-helical (Figure 1A). In the initial configuration, GB3.1 and GB3.2 peptides were positioned approximately 25 Å, 18 Å, and 20 Å away from the central helix, N-lobe, and C-lobe, respectively. The CaM structure was in the extended form. This arrangement ensured that initially, CaM and the peptides did not interact (Figure 1A). Subsequently, MD simulations were conducted for these systems, revealing that the peptides dynamically moved towards CaM and eventually occupied the natural binding site on CaM, as detailed below.
CaM-GB3.1 complex formation
Initially, the peptide was positioned perpendicularly to the central helix and in the middle of the two lobes of CaM (Figure 1A). At the onset of simulation, the peptide started moving towards the CaM. First, the peptide transitioned towards the C-lobe and aligned with central helix at ∼45°. The C-terminal of GB3.1 was close to the C-lobe of CaM. The Arg449 of GB3.1 formed very first contact with EF motif residues Asp95 and Asn97 at C-lobe of CaM (Figure 1B). However, this contact lost as the simulation progressed. After this transient initial contact, interestingly, the GB3.1 peptide took ∼180° turn and aligned parallelly to the central helix. Due to this rotation, the N-terminal of the peptide was close to the C-lobe of CaM and vice versa (Figure 1C). The interaction between residues Lys75 (CaM) and Ser455 (GB3.1) played a crucial role in facilitating this transient positioning of the GB3.1 peptide (Figures 1C,D). After that, the N-terminal of the GB3.1 peptide made a 90° rotation losing its N-terminal interactions with CaM C-lobe, while the C-terminal remain fixed (Figure 1D). Therefore, the interaction between Lys75 (CaM) and Ser455 (GB3.1) also remained unbroken (Figure 1D). This rotation was in the vertical direction to the central helix (Figure 1D). This was final position of C-terminal region of the GB3.1 on CaM as no further movement was observed in this region (Figures 1D,E).
After the peptide has occupied its natural position on CaM, the central helix has undergone a huge conformational change. A sharp kink was observed between residues Asp78-Ser81 which displaced the relative position of two lobes. To compare the bending of the central helix, we superimposed the simulated structure onto extended CaM structure on N-lobe. The central helix made a ∼90° turn at Thr79 converting the CaM into compact ellipsoidal conformation (Figure 1D). The two lobes came close to each other, and peptide was further moved away from C-lobe to N-lobe. Gradually the two lobes came further close to each other and formed contact with peptide (Figure 1E). The peptide also slipped in further. The N-terminal of the peptide moved towards the N-lobe of CaM, establishing robust hydrophobic interactions. The central helix curved, and the two lobes enveloped the GB3.1 peptide (Figure 1E). The interacting residues are shown in Figure 1F. The C-terminal of GB3.1 peptide is deeply buried at hydrophobic pocket and has made extensive contacts with both N- and C-lobe of CaM (Figure 1F).
CaM-GB3.2 complex formation
CaM/GB3.2 simulation system, we systematically monitored the trajectory of the GB3.2 peptide. The initial position of GB3.2 was same as GB3.1 (Figure 1A). The complex between CaM and GB3.2 peptide was formed within the 200 ns of simulation. Further, simulation didn’t result in significant movement. Therefore, the simulation was stopped at 1 µs (Figure 3A). First the peptide moved towards C-lobe of CaM and made a 90° rotation while pivoted at C-terminal. However, the peptide was still far from CaM to establish any contact with it (Figure 2A). Then the peptide further moved towards C-lobe and oriented perpendicular to the central helix of CaM. The Arg477 (GB3.2) and Glu139 (CaM) formed very first contact. These contacts were responsible for holding the GB3.2 peptide at this orientation. The rest of peptide was above the central helix at distance of ∼22Å (Figure 2B). As the simulation further progressed the peptide went down towards the central helix, and it formed additional contact with it. The CaM residues Glu82 and Glu83 formed contact with Lys470 and Arg474, respectively (Figure 2C). After simulation further progressed, the peptide didn’t go further down but it rotated laterally towards N-lobe of CaM, pivoted at C-terminal in the same plan. That was final position of GB3.2 on CaM. Thus, mirroring the behaviour observed with GB3.1, the GB3.2 peptide engaged with CaM at its N-terminal region. However, in contrast to CaM/GB3.1 complex, the CaM was in extended conformation. Further, the peptide GB3.2 made cross contact with both N-and C-terminal. The N-terminal of peptide was close to N-lobe and C-terminal was close to C-lobe (Figure 2D).
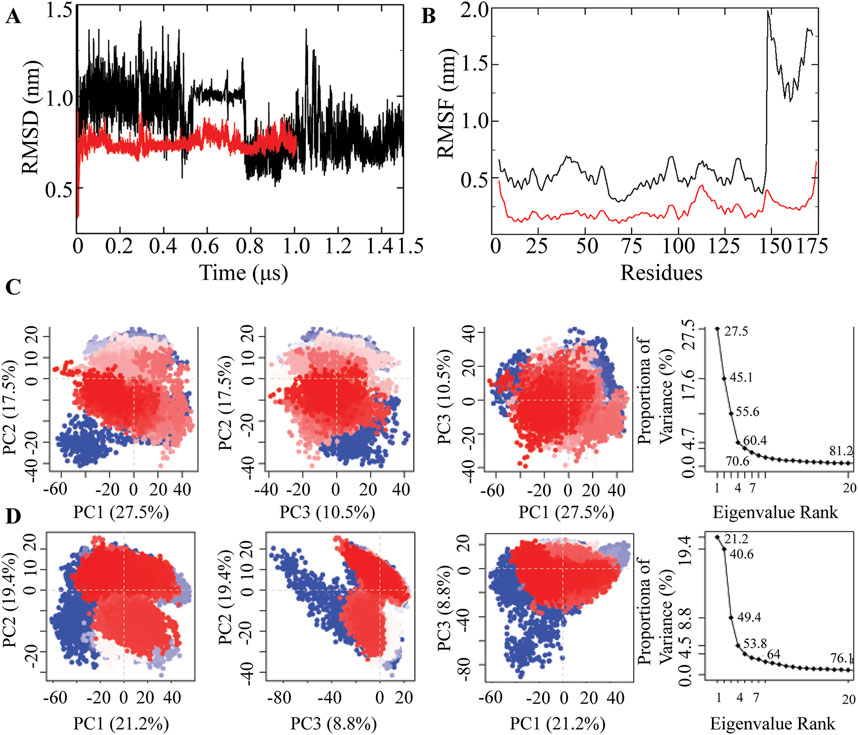
Figure 3. MD simulation of CaM/GB3.1 and CaM/GB3.2 complexes. (A) RMSD profile of CaM/GB3.1 (black) and CaM/GB3.2 (red). (B) RMSF profile of CaM/GB3.1 (black) and CaM/GB3.2 (red). The residues from 1 to 147 are from CaM and the residues from 148 to 175 are from peptides. (C) A scree plot was plotted showing the proportion of variance against its eigenvalue rank. The first three PC vectors are plotted to monitor the residual displacement in CaM/GB3.1 complex. (D) A similar analysis was carried out for CaM/GB3.2 complex. The 2D plot illustrates the ensemble distribution changes across different conformations during the simulation. Each point on the plot corresponds to a specific conformation at a given time. The colour gradient, ranging from blue to white to red, highlights significant periodic transitions between conformational states throughout the simulation. Blue points signify unstable conformations, white points represent intermediate states, and red points indicate stable conformations that persist during the trajectory.
The central helix residue Lys75 established contact with Ile467 of the GB3.2 peptide, while Glu82 of CaM interacted with Lys470 of GB3.2. Additionally, Met72 formed contacts with Val464, and Phe68 interacted with Leu460 of GB3.2. The N-terminal of the GB3.2 peptide exhibited strong hydrophobic interactions with the N-lobe of CaM (Figure 2E). Specific interactions included Ser38/Leu461, Leu39/Val464, and Met51/Phe468 (CaM/GB3.2) at the N-lobe, while at the C-lobe, Gln143 engaged in an electrostatic interaction with Arg474. Notably, previous studies also demonstrated a higher affinity of the GB3.2 peptide for the N-lobe of CaM (Chemin et al., 2017).
Root mean square deviation (RMSD) and root mean square fluctuation (RMSF)
The complexes’ dynamic behaviour was evaluated through time-dependent root mean square deviation (RMSD) of all protein atoms with the original complex as a reference. RMSD values were then plotted over time. Additionally, the dynamic behaviour at the residue level was assessed using Root Mean Square Fluctuation (RMSF) calculations and plotted against the residue numbers.
In CaM/GB3.1 complex, the observed RMSD fluctuated between 0.7 and 1.0 nm. Initially, the RMSD was approximately 1.0 nm, gradually decreasing to 0.7 nm as the simulation progressed and the complex stabilized (Figure 3A). The slightly higher RMSD was primarily attributed to the movement of peptide GB3.1 (Figure 1). CaM residues (residues 1-147) displayed RMSF within the range of 0.23–0.60 nm, while GB3.1 (residues 148-172) exhibited RMSF between 1.2 and 2.0 nm. The higher RMSD for peptide GB3.1 can be attributed to significant conformational changes during its complexation with CaM.
CaM/GB3.2 complex formed within very short period. Therefore, GB3.1, GB3.2 did not undergo significant conformational changes, leading to a consistent overall RMSD. The RMSD of the CaM/GB3.2 complex remained approximately 0.7 nm throughout the simulations. Therefore, simulation was concluded at 1000 ns as the RMSD has attained stable plateau. Residue-level fluctuations for both CaM and the peptide were within the range of 0.2–0.25 nm. Therefore, this complex was more stable than CaM/GB3.1 (Figures 3A,B).
Principal component analysis (PCA)
To gain deeper insight into the large-scale collective motions, we performed PCA for analysing the dominant protein conformational patterns in a principal component (PCs). We investigated the conformational behaviour of the Cα atoms of the proteins by projecting them along the direction of the first three eigenvectors (PC1, PC2, and PC3). These first three PCs were extracted from simulation trajectories in the form of cluster groups. PCA filters global and local motions in a simulation trajectory, systematically reducing dimensions to highlight essential protein dynamic motions. The analysis, based on Cα position covariance, reveals significant spatial-scale motions in the study.
In CaM/GB3.1 complex PCA analysis, the first five principal components (PCs) collectively explain approximately 70% of the total variance, with subsequent individual component contributions dropping below 5% (Figure 3C). Each dot on the plots represents a conformation of the proteins, red indicating the stable conformation. The first principal component corresponds to the motion of the CaM N-lobe and linker, while the second PC illustrates the movement of the peptide GB3.1. Additionally, the second PC is prominently influenced by the linker region (Figure 3C).
Similarly, for the CaM/GB3.2 complex, the initial five principal components (PCs) collectively explain approximately 64% of the total variance, with subsequent individual component contributions falling below 5% (Figure 3D). The first principal component primarily signifies the motion of the peptide in its quest to locate the binding site on CaM, and it is also influenced by the linker of CaM. The second principal component is predominantly influenced by the N-lobe residues engaged in the interaction with GB3.2, as elaborated earlier.
MM-PBSA
The GB peptides bind to apo-CaM with significantly lower affinity. Upon binding Ca2+ ions at its EF-hand motifs, CaM undergoes conformational changes that expose large hydrophobic surfaces essential for interactions with target proteins. GB peptides associate with intact Ca2+-bound CaM with high nanomolar affinity, whereas their interactions with the isolated N- or C-lobes of CaM are considerably weaker. Furthermore, mutations in the EF-hand motifs that disrupt Ca2+ binding markedly reduce the affinity of CaM for GB peptides.
MM-PBSA algorithm is widely used to compute the binding energies of protein-ligand (Boonyasuppayakorn et al., 2020; Sanachai et al., 2020) and protein-protein complexes (Lees et al., 2017; Isa et al., 2019). The MM-PBSA calculations are done using short MD simulation trajectories. The MM-PBSA analysis unveiled negative binding energies (Table 1) for the CaM/GB3.1 and CaM/GB3.2 complexes, indicating a spontaneous and favourable complex formation. The binding energies were found to be higher for CaM/GB3.2 complex compared to that of CaM/GB3.1 complex, suggesting a stronger affinity between CaM and GB3.2 peptide. This further supports spontaneous complex formation between CaM and GB3.2 as discussed above. The sequence differences between GB3.1 and 3.2 peptides contribute significantly differences in binding affinity.
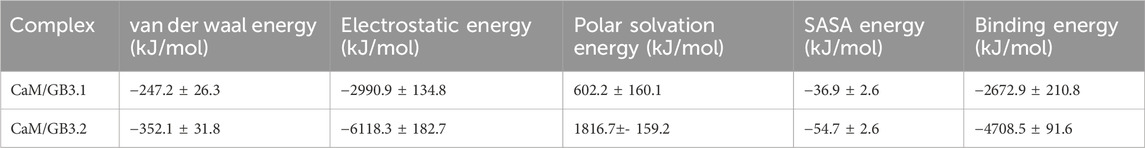
Table 1. The binding energy between CaM and GB3.1/GB3.2 peptides were calculated using MM-PBSA method.
Structural comparison
We compared the CaM/GB3.1 and CaM/GB3.2 complexes by superimposing CaM structures (Figure 4A). During the complex formation, the N- and C-lobe’s intramolecular interactions remains unaltered. In both complexes, N- and C-lobes are similar and superimposable. The only notable conformation changes are seen in the loop that connects the two EF-hand motifs in each lobe (Figure 4A). The EF-hand motifs coordinate the Ca2+ ions on each lobe throughout the simulations. However, the central helix has undergone a huge conformation change. The CaM in GB3.1 complex is in closed conformation whereas it is in open conformation in CaM/GB3.2 complex. The central helix in GB3.1 complex bends at Arg74 whereas no such bending was observed in GB3.2 complex. The GB3.1 and GB3.2 occupied similar position on CaM (Figure 4A). Both peptides formed extensive contact with N-lobe of CaM. Therefore, interactions with GB3.1 and GB3.2 peptides with CaM mainly affect the linker region of CaM.
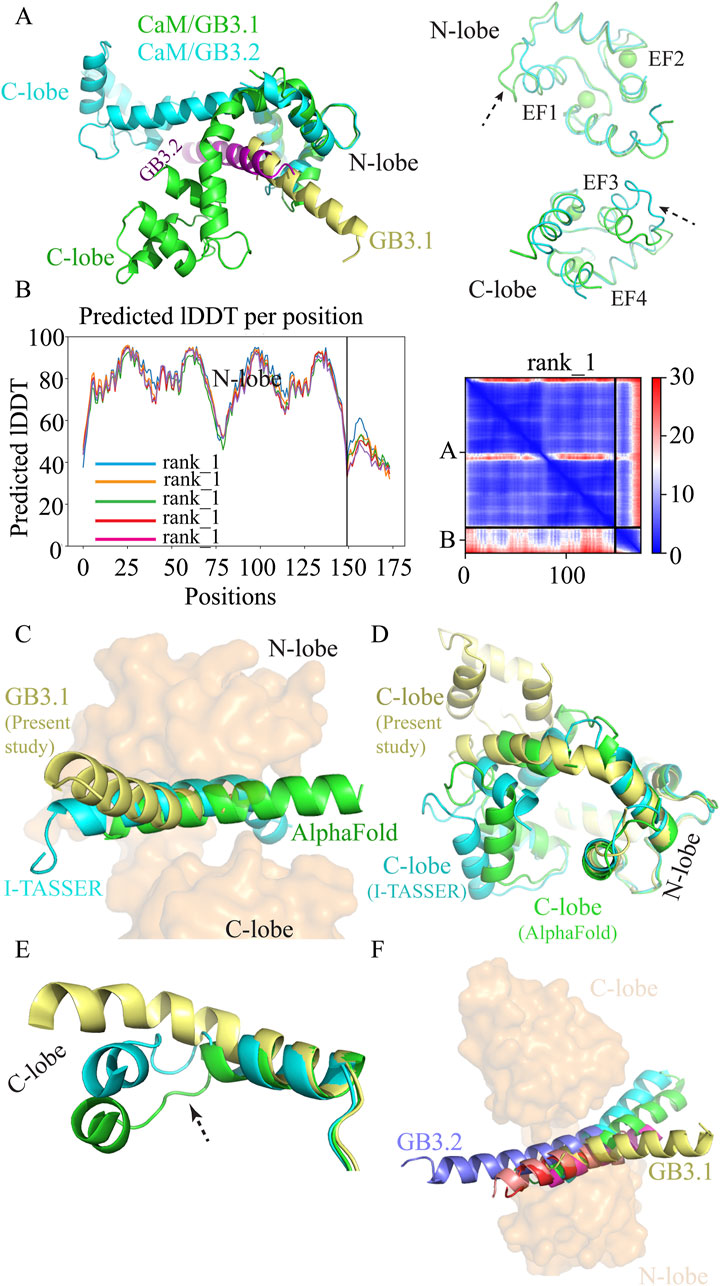
Figure 4. Structural comparison. (A) Superimposition of CaM/GB3.1 and CaM/GB3.2 on N-lobe of CaM. (B) [Left panel] Predicted local distance difference test (LDDT) score per position for the five CaM/GB3.1 complex models generated by alphafold2. The residue position is plotted against the predicted LDDT. The pLDDT values above 90 indicate very high accuracy equivalent to experimentally solved structures, pLDDT values range 70–90 indicate a high accuracy, and values between 50 and 70 indicate a lower accuracy. [Right Panel] Prediction aligned error (PAE) score for models ranked 1. PAE score displays the calculated error of the predicted distance for each pair of residues. (C) Superimposition of predicted CaM/GB3.1 structures. CaM is shown as surface (orange) whereas GB3.1 from I-TASSER (cyan), Alphafold 2 (green), and current study (yellow) are shown as cartoon. (D) Comparison of CaM conformation from I-TASSER (cyan), Alphafold 2 (green), and current study (yellow). (E) In predicted structures CaM central helix unwind that facilitates the bending of the helix whereas in MD simulated structure such unwinding didn’t take place. (F) Superimposition of various ion channels IQ motif with CaM/GB3.1. 2BE6 (green), 2VAY (cyan), 3BXL (magenta), 3DVJ (red), and 3DVM (salmon).
Structure prediction
We generated complex structure of CaM and GB3.1 peptide using AlphaFold2 and I-TASSER (Yang and Zhang, 2015; Jumper et al., 2021; Meng et al., 2023). The deep-learning Alphafold2 is a promising tool for structure predictions close to experimental accuracy. The predicted Local Distance Difference Test (pLDDT) scores indicated a high degree of confidence predicted structures (Figure 4B). However, it is important to note that pLDDT scores utilize the information available in the training data, which may not perfectly reflect specific biological contexts (Mariani et al., 2013). In the predicted structures, the peptides occupy the same site and orientation as in MD simulation (Figure 4C). Superimposition of various CaM structures reveals that the central α-helix undergoes a characteristic bend at residue Arg75, resulting in a compact conformation. In all such compact structures, the segment spanning Ala74 to Asp79 exhibits unwinding and variable bending near Arg75, indicating a high degree of flexibility in this region. This conformational variability contributes to functional adaptability of CaM in binding diverse target proteins. Due to this intrinsic flexibility, the Arg75 region is challenging to predict accurately using AlphaFold2, as reflected by the relatively low pLDDT score (∼50) observed in this study (Figure 4B).
Similarly, I-TASSER predicted structures were also in good agreement with MD simulation structures (Figure 4C). Thus, the MD simulation structure is in good agreement with the predicted structures.
CaM conformation
CaM exists in solution either in a compact ellipsoidal or in extended form. In all identified complexes of Ca2+-bound CaM with voltage-gated calcium channel (VGCC) IQ domains, CaM adopts a compact ellipsoidal conformation. However, across these CaM structures, the two lobes occupy distinct positions, resulting in non-superimposable overall structures (Yaduvanshi et al., 2021; Yaduvanshi and Kumar, 2024). The central helix of CaM is seen to bend to varying extents in reported CaM/VGCC IQ complexes, creating different relative orientations of the lobes. In apo and extended Ca2+-CaM, the two lobes assume trans conformations, while in compact ellipsoidal CaM structures, they adopt cis conformations.
We conducted a comparison between CaM structures predicted by AlphaFold2 and I-TASSER with the structure obtained from MD simulations. We aligned them based on the N-lobe. Up to residue Arg75, all structures aligned closely, but the C-lobe occupied different spatial positions, as expected (Figure 4D). In each of the three models, the central helix exhibited different degree of bending, positioning the two lobes in a compact ellipsoidal conformation that wraps around the peptide (Figure 4C). Interestingly, while the predicted models from AlphaFold2 and I-TASSER showed the two lobes in a fully closed arrangement, the CaM in MD simulated CaM/GB3.1 complex displayed a slightly less pronounced bend in the central helix, allowing for a partially open configuration (Figure 4D). A noteworthy observation in the predicted structures was the unwinding and increased flexibility of the K76-E83 segment in the central helix, potentially facilitating the bending that brings the two lobes into a compact ellipsoidal form. Conversely, in the MD simulation structure, the K76-E83 segment retained its α-helical structure, suggesting a more stable conformation for this region in the absence of external bending forces (Figure 4E).
Comparison with other VGCCs
Then we compared the CaM/GB peptide simulated complexes from this study with experimentally solved CaM/IQ motif complexes. The following CaM/IQ motifs complexes were chosen for comparison: Cav1.2 IQ (2BE6), Cav1.1 IQ (2VAY), Cav2.3 IQ (3BXL), Cav2.2 IQ (3DVJ), and Cav2.1 IQ (3DVM). In all these structures CaM is in ellipsoidal compact conformations and IQ motifs are trapped between two lobes (Figure 4F). The central helix bends to varying degree as discussed in our previous study (Yaduvanshi et al., 2021). Upon superimposition on N-lobe, the GB3.1 peptide occupied the same site as IQ motifs from Cav1 and Cav2 channels (Figure 4F). Thus, validating the binding of GB3.1 and GB3.2 with CaM in present study. The GB3.2 peptide was in the same plane as IQ motif whereas N-terminal of GB3.1 was slightly above the plane. The hydrophobic interactions between CaM and the C-terminal of GB3.1, which contains more hydrophobic residues is responsible for this orientation. Importantly, despite starting both GB3.1 and GB3.2 peptides from identical initial positions relative to CaM, the peptides followed distinct binding pathways and adopted different final conformations. This divergence in binding trajectories, despite the same starting setup, suggests that the observed interactions are not artifacts of the initial configuration and supports the reproducibility and robustness of the findings. Additionally, the simulations were performed using a 2fs time step, which is standard and sufficiently small to ensure numerical stability and accurate integration of atomic motions over microsecond timescales.
Discussion
In this paper, we attempted to understand the CaM-mediated regulation of T-type ion channels using MD simulations. MD simulation is a computer-based simulation method for analysing the physical movements of atoms and molecules by numerically solving Newton’s equations of motion (Hollingsworth & Dror, 2018). MD simulation mimics what atoms do in real life. Proteins or enzyme’s biological functions result by transient interactions with other proteins or ligands. Though experimental techniques like NMR, X-ray crystallography or Cryo-EM are very powerful in elucidating the structural details of protein-protein complexes. However, week affinity complex or transient protein complexes are difficult to analyse by experimental techniques because of low affinity, experiment often end up in one partner only. Therefore, trapping the transiently interacting protein complexes are highly challenging for structural or functional studies (Qin and Gronenborn, 2014; Omranian et al., 2022). Linking the two transiently interacting proteins at gene level via a suitable flexible linker is often used to study such complexes (Chichili et al., 2013; 2016; Kumar et al., 2013; Reddy Chichili et al., 2013). However, the linker may hamper the movement of the proteins and thereby their natural interactions. The present study relies on integration of existing structural information of homologous proteins into molecular dynamics simulation. The homologous structure will guide the initial orientation of molecules followed by atomic-level MD simulations. By this approach, we can monitor the progress of complex formation. How the two proteins associate and dissociate reversibly during a single trajectory. Further, mechanistic details like intermediate and transition states along the association pathway could be observed. Therefore, this technique allows characterization of structure and dynamics of protein–protein complexes. In this work, instead of docking the CaM and GB peptide followed by simulation, we simulated the CaM and peptide in the same system and allowed them to interact and form stable complex.
The calcium (Cav1 and Cav2) and sodium (Nav1) ion channel consist of IQ motif [FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY] through which they interact with CaM. We compared GB sequence from T-type with IQ motifs from various ion channels (Figure 5). The first amino acid of IQ motif is commonly Ile (I), but it can Phe (F), Leu (L), Val (V) as well, however, the second amino acid is invariably Gln (Q). In case of T-type GB peptides, first amino acid is I in all three channels. However, the second amino acid is L in GB3.1 and GB3.3 and F in GB3.2 (Figure 5A). Therefore, GB peptides consist of hydrophobic amino acids in contrast to polar glutamine in bona fide IQ motifs. However, the overall hydrophobicity of GB peptides is quite similar to the IQ motif peptides (Figure 5B). Further, IQ motif sequences consist of one or more bulky strong hydrophobic amino acid which anchors the peptide into the CaM. All 3 GB peptides consist of Tyr in this position. Charged amino acids are present in GB and IQ motifs in similar fashion (Figure 5). Further, our simulation suggested that GB peptides interact with CaM in similar way as IQ motifs. Thus, we hypothesized that the earliest Cav3 T-type channels possessed an IQ motif like other channels, but during evolutionary divergence, this motif transformed into a GB, giving these channels their unique identity as T-type. However, while the IQ motifs in Cav1 and Cav2 channels are located at the C-terminal, the GB sequence in Cav3 channels resides at the N-terminal. This positional divergence may have arisen through an evolutionary event involving DNA translocation. Mutations in the GB are associated with various disorders such as epilepsy or autism spectrum disorder (Chen et al., 2003; Splawski et al., 2006; Liang et al., 2007). Their interaction with CaM has been shown earlier in vitro (Chemin et al., 2017). Another study also has shown co-immunoprecipitation of CaM and rat Cav3.3 splice variant (Lee et al., 2017). Therefore, it supports the universality of CaM association with all T-type channels in the same manner as it associates with Cav1, Cav2, and Nav1 sodium channels (Sarhan et al., 2012; Ben-Johny et al., 2015b). However, further experimental structural verification is required to support this view.
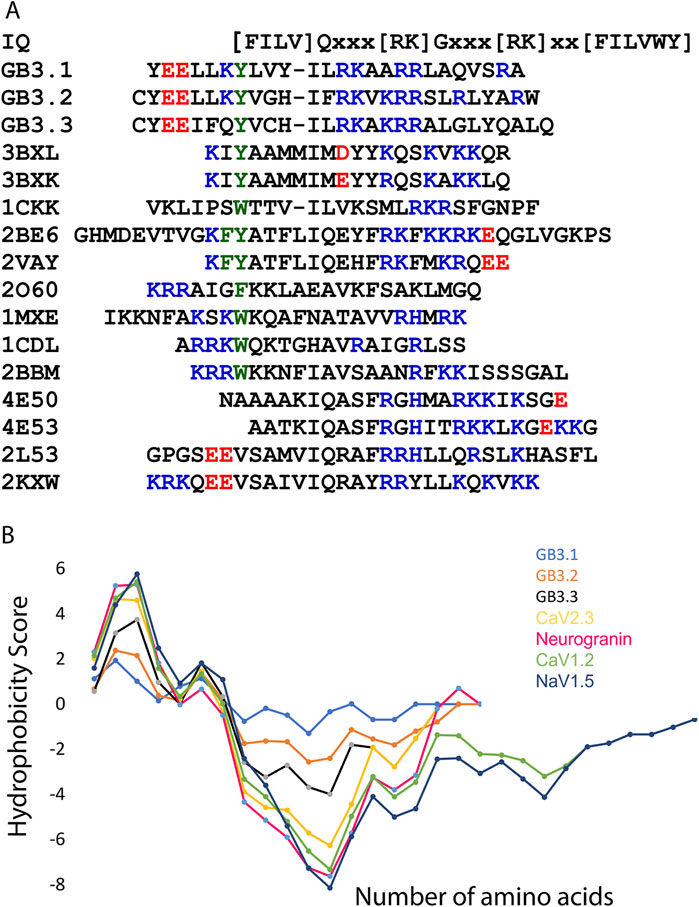
Figure 5. IQ motifs and GB peptide sequence analysis. (A) Sequence alignment of various IQ motifs and grating brake peptide from T-type ion channels. Positive and negative charged residues are shown in blue and red, respectively. The bulky hydrophobic residue is shown in green. This residue anchors the peptide into CaM. This sequence was fetched from PDB, and code is shown. (B) The hydrophobic plot of various IQ motifs and GB peptides from T-type ion channels. This plot was generated using ProtScale from Expasy server.
The results of this study reveal the distinct interactions and structural conformations between CaM and GB peptides from Cav3.1 and Cav3.2. The MD simulations demonstrate the dynamic and stepwise association of these peptides, with initial interactions at the C-lobe progressing to a stable binding state involving both lobes of CaM. In both type of complex formation, the peptides first transiently associate with C-lobe of CaM (Figures 1B, 2B). The hydrophobic interactions are the driving force for the initial interaction. Subsequently, the GB peptides orient themselves on CaM that further improves the structural complementarity and binding affinity to CaM. The GB3.1 makes ∼180° turn whereas GB3.2 goes down from its initial position (Figures 1C, 2C). Once the peptide is close to the CaM, then it rotates laterally and occupies its position on CaM. Notably, the pronounced bending of the central helix observed in the CaM/GB3.1 complex versus the extended configuration in the CaM/GB3.2 complex suggests differential modes of regulation for these channels. These findings align with prior experimental data on CaM interactions with other ion channel motifs, underscoring the evolutionary divergence of GB from canonical IQ motifs (Figure 4F). The spontaneous and favourable binding energies, validated by MM-PBSA, reinforce the physiological relevance of these interactions. While earlier studies established that CaM interacts with the GB of T-type calcium channels, our study provides, for the first time, a dynamic and atomistic view of how these complex forms over time. Through long-timescale MD simulations, we reveal distinct binding pathways and conformational transitions of CaM during complex formation with GB3.1 and GB3.2 peptides. Overall, this study provides structural and mechanistic insights into the regulation of T-type ion channels by CaM, opening avenues for targeted therapeutic interventions in disorders such as epilepsy and cardiac arrhythmias. Although this study was supported by various in silico methods, further experimental validation is necessary.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SY: Formal Analysis, Methodology, Writing – review and editing. VK: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We would like to acknowledge the Ramalingaswami Fellowship (BT/RLF/Re-entry/64/2017), Department of Biotechnology, Govt of India (VK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
Acknowledgments
MD simulations were performed on the IIT Delhi HPC facility (https://supercomputing.iitd.ac.in/). The authors thank Dr Manish Agarwal, IIT Delhi for his support in running MD simulation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. doi:10.1016/j.softx.2015.06.001
Antal, L., and Martin-Caraballo, M. (2019). T-Type calcium channels in cancer. Cancers (Basel) 11, 134. doi:10.3390/CANCERS11020134
Babu, Y. S., Bugg, C. E., and Cook, W. J. (1988). Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 204, 191–204. doi:10.1016/0022-2836(88)90608-0
Barghouth, M., Ye, Y., Karagiannopoulos, A., Ma, Y., Cowan, E., Wu, R., et al. (2022). The T-type calcium channel CaV3.2 regulates insulin secretion in the pancreatic β-cell. Cell Calcium 108, 102669. doi:10.1016/J.CECA.2022.102669
Baumgart, J. P., Vitko, I., Bidaud, I., Kondratskyi, A., Lory, P., and Perez-Reyes, E. (2008). I-II loop structural determinants in the gating and surface expression of low voltage-activated calcium channels. PLoS One 3, e2976. doi:10.1371/JOURNAL.PONE.0002976
Ben-Johny, M., Dick, I. E., Sang, L., Limpitikul, B., Kang, P. W., Niu, J., et al. (2015a). Towards a unified theory of calmodulin regulation (calmodulation) of voltage-gated calcium and sodium channels. Curr. Mol. Pharmacol. 8, 188–205.
Ben-Johny, M., Dick, I. E., Sang, L., Limpitikul, B., Kang, P. W., Niu, J., et al. (2015b). Towards a unified theory of calmodulin regulation (calmodulation) of voltage-gated calcium and sodium channels. Curr. Mol. Pharmacol. 8, 188–205. doi:10.2174/1874467208666150507110359
Ben-Johny, M., Yang, P. S., Niu, J., Yang, W., Joshi-Mukherjee, R., and Yue, D. T. (2014). Conservation of Ca2+/calmodulin regulation across Na and Ca2+ channels. Cell 157, 1657–1670. doi:10.1016/J.CELL.2014.04.035
Benkert, J., Hess, S., Roy, S., Beccano-Kelly, D., Wiederspohn, N., Duda, J., et al. (2019). Cav2.3 channels contribute to dopaminergic neuron loss in a model of Parkinson’s disease. Nat. Commun. 10 (1), 5094. doi:10.1038/s41467-019-12834-x
Blaich, A., Pahlavan, S., Tian, Q., Oberhofer, M., Poomvanicha, M., Lenhardt, P., et al. (2012). Mutation of the calmodulin binding motif IQ of the L-type Cav1.2 Ca2+ channel to EQ induces dilated cardiomyopathy and death. J. Biol. Chem. 287, 22616–22625. doi:10.1074/JBC.M112.357921
Boonyasuppayakorn, S., Saelee, T., Visitchanakun, P., Leelahavanichkul, A., Hengphasatporn, K., Shigeta, Y., et al. (2020). Dibromopinocembrin and dibromopinostrobin are potential anti-dengue leads with mild animal toxicity. Molecules 25, 4154. doi:10.3390/MOLECULES25184154
Bourinet, E., Francois, A., and Laffray, S. (2016). T-type calcium channels in neuropathic pain. Pain 157 (Suppl. 1), S15–S22. doi:10.1097/J.PAIN.0000000000000469
Brooks, B. R., Brooks, C. L., Mackerell, A. D., Nilsson, L., Petrella, R. J., Roux, B., et al. (2009). CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614. doi:10.1002/JCC.21287
Buraei, Z., and Yang, J. (2010). The β subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506. doi:10.1152/PHYSREV.00057.2009
Cain, S. M., and Snutch, T. P. (2010). Contributions of T-type calcium channel isoforms to neuronal firing. Channels 4, 475–482. doi:10.4161/CHAN.4.6.14106
Catterall, W. A. (2011a). Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947. doi:10.1101/cshperspect.a003947
Catterall, W. A. (2011b). Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947. doi:10.1101/CSHPERSPECT.A003947
Cazade, M., Bidaud, I., Lory, P., and Chemin, J. (2017a). Activity-dependent regulation of T-type calcium channels by submembrane calcium ions. Elife 6, e22331.
Cazade, M., Bidaud, I., Lory, P., and Chemin, J. (2017b). Activity-dependent regulation of T-type calcium channels by submembrane calcium ions. Elife 6, e22331. doi:10.7554/ELIFE.22331
Chattopadhyaya, R., Meador, W. E., Means, A. R., and Quiocho, F. A. (1992). Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 228, 1177–1192. doi:10.1016/0022-2836(92)90324-D
Chemin, J., Monteil, A., Perez-Reyes, E., Bourinet, E., Nargeot, J., and Lory, P. (2002). Specific contribution of human T-type calcium channel isotypes (alpha(1G), alpha(1H) and alpha(1I)) to neuronal excitability. J. Physiol. 540, 3–14. doi:10.1113/JPHYSIOL.2001.013269
Chemin, J., Stamenic, T. T., Cazade, M., Llinares, J., Blesneac, I., Todorovic, S. M., et al. (2019). A novel phospho-modulatory mechanism contributes to the calcium-dependent regulation of T-type Ca2+ channels. Sci. Rep. 9, 15642. doi:10.1038/S41598-019-52194-6
Chemin, J., Taiakina, V., Monteil, A., Piazza, M., Guan, W., Stephens, R. F., et al. (2017). Calmodulin regulates Cav3 T-type channels at their gating brake. J. Biol. Chem. 292, 20010–20031. doi:10.1074/jbc.M117.807925
Chen, Y., Lu, J., Pan, H., Zhang, Y., Wu, H., Xu, K., et al. (2003). Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann. Neurology 54, 239–243. doi:10.1002/ANA.10607
Chichili, V. P. R., Kumar, V., and Sivaraman, J. (2013). A method to trap transient and weak interacting protein complexes for structural studies. Intrinsically Disord. Proteins 1, e25464. doi:10.4161/idp.25464
Clozel, J. P., Ertel, E. A., and Ertel, S. I. (1999). Voltage-gated T-type Ca2+ channels and heart failure. Proc. Assoc. Am. Physicians 111, 429–437. doi:10.1111/PAA.1999.111.5.429
Cribbs, L. L., Lee, J. H., Yang, J., Satin, J., Zhang, Y., Daud, A., et al. (1998). Cloning and characterization of α1H from human heart, a member of the T-type Ca2+ channel gene family. Circulation Res. 83, 103–109. doi:10.1161/01.RES.83.1.103
Dziegielewska, B., Casarez, E. V., Yang, W. Z., Gray, L. S., Dziegielewski, J., and Slack-Davis, J. K. (2016). T-type Ca2+ channel inhibition sensitizes ovarian cancer to carboplatin. Mol. Cancer Ther. 15, 460–470. doi:10.1158/1535-7163.MCT-15-0456
Fallon, J. L., Halling, D. B., Hamilton, S. L., and Quiocho, F. A. (2005). Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Cav1.2 calcium channel. Structure 13, 1881–1886. doi:10.1016/J.STR.2005.09.021
Ferron, L., Capuano, V., Deroubaix, E., Coulombe, A., and Renaud, J. F. (2002). Functional and molecular characterization of a T-type Ca2 + channel during fetal and postnatal rat heart development. J. Mol. Cell. Cardiol. 34, 533–546. doi:10.1006/jmcc.2002.1535
Fukunaga, K., Izumi, H., Yabuki, Y., Shinoda, Y., Shioda, N., and Han, F. (2019). Alzheimer’s disease therapeutic candidate SAK3 is an enhancer of T-type calcium channels. J. Pharmacol. Sci. 139, 51–58. doi:10.1016/J.JPHS.2018.11.014
Gackière, F., Bidaux, G., Delcourt, P., Van Coppenolle, F., Katsogiannou, M., Dewailly, E., et al. (2008). CaV3.2 T-type calcium channels are involved in calcium-dependent secretion of neuroendocrine prostate cancer cells. J. Biol. Chem. 283, 10162–10173. doi:10.1074/jbc.M707159200
Gardill, B. R., Rivera-Acevedo, R. E., Tung, C. C., and Van Petegem, F. (2019). Crystal structures of Ca2+–calmodulin bound to NaV C-terminal regions suggest role for EF-hand domain in binding and inactivation. Proc. Natl. Acad. Sci. U. S. A. 116, 10763–10772. doi:10.1073/pnas.1818618116
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein analysis tools on the ExPASy server 571 571 from: the proteomics protocols handbook protein identification and analysis tools on the ExPASy server.
Genheden, S., and Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461. doi:10.1517/17460441.2015.1032936
Grant, B. J., Rodrigues, A. P. C., ElSawy, K. M., McCammon, J. A., and Caves, L. S. D. (2006). Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696. doi:10.1093/BIOINFORMATICS/BTL461
Hall, M., Todd, B., Allen, E. D., Nguyen, N., Kwon, Y.-J., Nguyen, V., et al. (2018). Androgen receptor signaling regulates T-type Ca2+ channel expression and neuroendocrine differentiation in prostate cancer cells. Am. J. Cancer Res. 8, 732–747.
Han, D. Y., Minobe, E., Wang, W. Y., Guo, F., Xu, J. J., Hao, L. Y., et al. (2010). Calmodulin- and Ca2+-dependent facilitation and inactivation of the Cav1.2 Ca2+ channels in Guinea-pig ventricular myocytes. J. Pharmacol. Sci. 112, 310–319. doi:10.1254/JPHS.09282FP
Harding, E. K., and Zamponi, G. W. (2022). Central and peripheral contributions of T-type calcium channels in pain. Mol. Brain 15, 39. doi:10.1186/S13041-022-00923-W
He, L., Yu, Z., Geng, Z., Huang, Z., Zhang, C., Dong, Y., et al. (2022). Structure, gating, and pharmacology of human CaV3.3 channel. Nat. Commun. 13, 2084–2089. doi:10.1038/s41467-022-29728-0
Hong, S. Z., Kim, H. R., and Fiorillo, C. D. (2014). T-type calcium channels promote predictive homeostasis of input-output relations in thalamocortical neurons of lateral geniculate nucleus. Front. Comput. Neurosci. 8, 98. doi:10.3389/fncom.2014.00098
Huang, J., Fan, X., Jin, X., Lyu, C., Guo, Q., Liu, T., et al. (2024). Structural basis for human Cav3.2 inhibition by selective antagonists. Cell Res. 34, 440–450. doi:10.1038/s41422-024-00959-8
Isa, D. M., Chin, S. P., Chong, W. L., Zain, S. M., Rahman, N. A., and Lee, V. S. (2019). Dynamics and binding interactions of peptide inhibitors of dengue virus entry. J. Biol. Phys. 45, 63–76. doi:10.1007/S10867-018-9515-6
Jo, S., Kim, T., Iyer, V. G., and Im, W. (2008). CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865. doi:10.1002/jcc.20945
Joksimovic, S. M., Ghodsi, S. M., Heinsbroek, J. A., Orfila, J. E., Busquet, N., Tesic, V., et al. (2023). CaV3.1 T-type calcium channels are important for spatial memory processing in the dorsal subiculum. Neuropharmacology 226, 109400. doi:10.1016/J.NEUROPHARM.2022.109400
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kerckhove, N., Mallet, C., François, A., Boudes, M., Chemin, J., Voets, T., et al. (2014). Ca(v)3.2 calcium channels: the key protagonist in the supraspinal effect of paracetamol. Pain 155, 764–772. doi:10.1016/J.PAIN.2014.01.015
Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., et al. (2000). Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897. doi:10.1021/AR000033J
Korczeniewska, O. A., Kohli, D., Benoliel, R., Baddireddy, S. M., and Eliav, E. (2022). Pathophysiology of post-traumatic trigeminal neuropathic pain. Biomolecules 12, 1753. doi:10.3390/BIOM12121753
Kumar, V., Chichili, V. P. R., Zhong, L., Tang, X., Velazquez-Campoy, A., Sheu, F. S., et al. (2013). Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin. Sci. Rep. 3, 1392. doi:10.1038/srep01392
Kumari, R., Kumar, R., and Lynn, A. (2014). g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model 54, 1951–1962. doi:10.1021/ci500020m
Lee, A., Scheuer, T., and Catterall, W. A. (2000). Ca2+/Calmodulin-Dependent facilitation and inactivation of P/Q-Type Ca2+ channels. J. Neurosci. 20, 6830–6838. doi:10.1523/JNEUROSCI.20-18-06830.2000
Lee, J., Cheng, X., Swails, J. M., Yeom, M. S., Eastman, P. K., Lemkul, J. A., et al. (2016). CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413. doi:10.1021/acs.jctc.5b00935
Lee, J. H., Daud, A. N., Cribbs, L. L., Lacerda, A. E., Pereverzev, A., Klöckner, U., et al. (1999). Cloning and expression of a novel member of the low voltage-activated T-type calcium channel family. J. Neurosci. 19, 1912–1921. doi:10.1523/jneurosci.19-06-01912.1999
Lee, N., Jeong, S., Kim, K. C., Kim, J. A., Park, J. Y., Kang, H. W., et al. (2017). Ca2+ regulation of Cav3.3 T-type Ca2+ channel is mediated by calmodulin. Mol. Pharmacol. 92, 347–357. doi:10.1124/MOL.117.108530
Lees, W. D., Stejskal, L., Moss, D. S., and Shepherd, A. J. (2017). Investigating substitutions in antibody-antigen complexes using molecular dynamics: a case study with broad-spectrum, influenza A antibodies. Front. Immunol. 8, 143. doi:10.3389/FIMMU.2017.00143
Li, W., Zhang, S. L., Wang, N., Zhang, B. B., and Li, M. (2011). Blockade of T-type Ca2+channels inhibits human ovarian cancer cell proliferation. Cancer Investig. 29, 339–346. doi:10.3109/07357907.2011.568565
Liang, J., Zhang, Y., Chen, Y., Wang, J., Pan, H., Wu, H., et al. (2007). Common polymorphisms in the CACNA1H gene associated with childhood absence epilepsy in Chinese Han population. Ann. Hum. Genet. 71, 325–335. doi:10.1111/J.1469-1809.2006.00332.X
Lu, C., Ma, Z., Cheng, X., Wu, H., Tuo, B., Liu, X., et al. (2020). Pathological role of ion channels and transporters in the development and progression of triple-negative breast cancer. Cancer Cell Int. 20, 377. doi:10.1186/S12935-020-01464-9
Lundt, A., Seidel, R., Soós, J., Henseler, C., Müller, R., Bakki, M., et al. (2019). Cav3.2 T-Type calcium channels are physiologically mandatory for the auditory system. Neuroscience 409, 81–100. doi:10.1016/J.NEUROSCIENCE.2019.04.024
Marger, F., Gelot, A., Alloui, A., Matricon, J., Ferrer, J. F. S., Barrère, C., et al. (2011). T-type calcium channels contribute to colonic hypersensitivity in a rat model of irritable bowel syndrome. Proc. Natl. Acad. Sci. U. S. A. 108, 11268–11273. doi:10.1073/pnas.1100869108
Mariani, V., Biasini, M., Barbato, A., and Schwede, T. (2013). lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728. doi:10.1093/BIOINFORMATICS/BTT473
Mariot, P., Vanoverberghe, K., Lalevée, N., Rossier, M. F., and Prevarskaya, N. (2002). Overexpression of an α1H (Cav3.2) T-type calcium channel during neuroendocrine differentiation of human prostate cancer cells. J. Biol. Chem. 277, 10824–10833. doi:10.1074/jbc.M108754200
Mark, P., and Nilsson, L. (2001). Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960. doi:10.1021/JP003020W
Meng, E. C., Goddard, T. D., Pettersen, E. F., Couch, G. S., Pearson, Z. J., Morris, J. H., et al. (2023). UCSF ChimeraX: tools for structure building and analysis. Protein Sci. a Publ. Protein Soc. 32, e4792. doi:10.1002/PRO.4792
Mesirca, P., Torrente, A. G., and Mangoni, M. E. (2015). Functional role of voltage gated Ca2+ channels in heart automaticity. Front. Physiol. 6, 19. doi:10.3389/FPHYS.2015.00019
Monteil, A., Chausson, P., Boutourlinsky, K., Mezghrani, A., Huc-Brandt, S., Blesneac, I., et al. (2015). Inhibition of Cav3.2 T-type calcium channels by its intracellular I-II loop. J. Biol. Chem. 290, 16168–16176. doi:10.1074/jbc.M114.634261
Omranian, S., Nikoloski, Z., and Grimm, D. G. (2022). Computational identification of protein complexes from network interactions: present state, challenges, and the way forward. Comput. Struct. Biotechnol. J. 20, 2699–2712. doi:10.1016/J.CSBJ.2022.05.049
Perez-Reyes, E. (2006). Molecular characterization of T-type calcium channels. Cell Calcium 40, 89–96. doi:10.1016/J.CECA.2006.04.012
Perez-Reyes, E. (2010). Characterization of the gating brake in the I–II loop of CaV3 T-type calcium channels. Channels 4, 453–458. doi:10.4161/CHAN.4.6.12889
Qin, J., and Gronenborn, A. M. (2014). Weak protein complexes: challenging to study but essential for life. FEBS J. 281, 1948–1949. doi:10.1111/FEBS.12744
Reddy Chichili, V. P., Kumar, V., and Sivaraman, J. (2013). Linkers in the structural biology of protein-protein interactions. Protein Sci. 22, 153–167. doi:10.1002/pro.2206
Reddy Chichili, V. P., Kumar, V., and Sivaraman, J. (2016). Application of linker technique to trap transiently interacting protein complexes for structural studies. J. Biol. Methods 3, e34. doi:10.14440/jbm.2016.81
Salsbury, F. R. (2010). Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr. Opin. Pharmacol. 10, 738–744. doi:10.1016/J.COPH.2010.09.016
Sanachai, K., Mahalapbutr, P., Choowongkomon, K., Poo-Arporn, R. P., Wolschann, P., and Rungrotmongkol, T. (2020). Insights into the binding recognition and susceptibility of tofacitinib toward janus kinases. ACS Omega 5, 369–377. doi:10.1021/ACSOMEGA.9B02800
Santoni, G., Santoni, M., and Nabissi, M. (2012). Functional role of T-type calcium channels in tumour growth and progression: prospective in cancer therapy. Br. J. Pharmacol. 166, 1244–1246. doi:10.1111/J.1476-5381.2012.01908.X
Sarhan, M. F., Tung, C. C., Van Petegem, F., and Ahern, C. A. (2012). Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. U. S. A. 109, 3558–3563. doi:10.1073/pnas.1114748109
Senatore, A., Guan, W., and Spafford, J. D. (2014). Cav3 T-type channels: regulators for gating, membrane expression, and cation selectivity. Pflügers Archiv - Eur. J. Physiology 466, 645–660. doi:10.1007/s00424-014-1449-7
Shah, K., Seeley, S., Schulz, C., Fisher, J., and Gururaja Rao, S. (2022). Calcium channels in the heart: disease states and drugs. Cells 11, 943. doi:10.3390/CELLS11060943
Shao, J., Liu, Y., Gao, D., Tu, J., and Yang, F. (2021). Neural burst firing and its roles in mental and neurological disorders. Front. Cell Neurosci. 15, 741292. doi:10.3389/FNCEL.2021.741292
Simms, B. A., and Zamponi, G. W. (2014). Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron 82, 24–45. doi:10.1016/j.neuron.2014.03.016
Skjaerven, L., Yao, X. Q., Scarabelli, G., and Grant, B. J. (2014). Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinforma. 15, 1–11. doi:10.1186/S12859-014-0399-6/TABLES/1
Splawski, I., Yoo, D. S., Stotz, S. C., Cherry, A., Clapham, D. E., and Keating, M. T. (2006). CACNA1H mutations in autism spectrum disorders. J. Biol. Chem. 281, 22085–22091. doi:10.1074/JBC.M603316200
Szymanowicz, O., Drużdż, A., Słowikowski, B., Pawlak, S., Potocka, E., Goutor, U., et al. (2024). A review of the CACNA gene family: its role in neurological disorders. Diseases 12, 90–12. doi:10.3390/DISEASES12050090
Taylor, J. T., Huang, L., Pottle, J. E., Liu, K., Yang, Y., Zeng, X., et al. (2008). Selective blockade of T-type Ca 2+ channels suppresses human breast cancer cell proliferation. Cancer Lett. 267, 116–124. doi:10.1016/j.canlet.2008.03.032
Weaver, E. M., Zamora, F. J., Hearne, J. L., and Martin-Caraballo, M. (2015a). Posttranscriptional regulation of T-type Ca2+ channel expression by interleukin-6 in prostate cancer cells. Cytokine 76, 309–320. doi:10.1016/j.cyto.2015.07.004
Weaver, E. M., Zamora, F. J., Puplampu-Dove, Y. A., Kiessu, E., Hearne, J. L., and Martin-Caraballo, M. (2015b). Regulation of T-type calcium channel expression by sodium butyrate in prostate cancer cells. Eur. J. Pharmacol. 749, 20–31. doi:10.1016/j.ejphar.2014.12.021
Yabuki, Y., Matsuo, K., Yu, M., Xu, J., Sakimura, K., Shioda, N., et al. (2021). Cav3.1 t-type calcium channel is critical for cell proliferation and survival in newly generated cells of the adult hippocampus. Acta Physiol. 232, e13613. doi:10.1111/APHA.13613
Yaduvanshi, S., Ero, R., and Kumar, V. (2021). The mechanism of complex formation between calmodulin and voltage gated calcium channels revealed by molecular dynamics. PLoS One 16, e0258112. doi:10.1371/JOURNAL.PONE.0258112
Yaduvanshi, S., and Kumar, V. (2024). Fungal alkaloid malbrancheamide reorients the lipid binding domain of GRK5. J. Biomol. Struct. and Dyn., 1–12. doi:10.1080/07391102.2024.2333987
Yang, J., and Zhang, Y. (2015). I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181. doi:10.1093/nar/gkv342
Yang, L. W., Eyal, E., Bahar, I., and Kitao, A. (2009). Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics. Bioinformatics 25, 606–614. doi:10.1093/bioinformatics/btp023
Zamponi, G. W. (2016). Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat. Rev. Drug Discov. 15, 19–34. doi:10.1038/NRD.2015.5
Zamponi, G. W., and Currie, K. P. M. (2013). Regulation of CaV2 calcium channels by G protein coupled receptors. Biochimica Biophysica Acta (BBA) - Biomembr. 1828, 1629–1643. doi:10.1016/J.BBAMEM.2012.10.004
Zhao, Y., Huang, G., Wu, Q., Wu, K., Li, R., Lei, J., et al. (2019). Cryo-EM structures of apo and antagonist-bound human Cav3.1. Nature 576, 492–497. doi:10.1038/s41586-019-1801-3
Keywords: calmodulin, gating brake, t-type ion channels, Cav3.1, Cav3.2, MD simulations
Citation: Yaduvanshi S and Kumar V (2025) Interaction studies between calmodulin and gating brake peptides from T-type channels. Front. Biophys. 3:1569091. doi: 10.3389/frbis.2025.1569091
Received: 31 January 2025; Accepted: 12 May 2025;
Published: 23 May 2025.
Edited by:
Oscar Brenes, University of Costa Rica, Costa RicaReviewed by:
William J. Zamora, University of Costa Rica, Costa RicaSeyedsajad Moazzeni, Rutgers, The State University of New Jersey, United States
Copyright © 2025 Yaduvanshi and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veerendra Kumar, dmt1bWFyMzRAYW1pdHkuZWR1
 Shivani Yaduvanshi
Shivani Yaduvanshi Veerendra Kumar
Veerendra Kumar