- Department of Chemical Sciences, University of Naples Federico II, Complesso Universitario di Monte Sant’Angelo, Napoli, Italy
Most protein structures deposited in the Protein Data Bank have been determined at 100 K by collecting X-ray diffraction data while holding crystals under a liquid nitrogen flux. Recently, the growing awareness that low-temperature diffraction data collection is accompanied by artifacts and by a loss of physiologically relevant information has driven crystallographers to collect X-ray diffraction data at room temperature or at body temperature (37 °C). Here, the results obtained from recent structural determinations of metal-protein adducts at body temperature are briefly discussed.
Structural determinations at non-cryogenic temperatures
X-ray crystallography is the technique that has been used to determine most of the protein structures reported in the Protein Data Bank (PDB) (Berman, 2000). Although it is currently facing strong competition from cryo-electron microscopy (cryoEM) (Cheng, 2018), X-ray crystallography remains the technique that currently allows for the most precise data on atomic coordinates. More than 90% of the protein crystal structures reported in the PDB and solved by X-ray crystallography were determined at 100 K (Haas, 2020). The success of low-temperature crystallography is mainly due to the fact that crystals of biological macromolecules are stable at low temperatures and can be easily preserved, even for months, and subsequently transported in liquid nitrogen, with a low damage risk. In cryocooled crystals, conformational disorder is reduced. This is generally associated with a more regular repetition of motifs which leads to cleaner/better diffraction. Furthermore, cryocooled crystals can be exposed to X-ray radiation for longer time, as the production and diffusion of free radicals at low temperature is slower (Garman, 1999). Under these conditions, however, it is possible to obtain structures that contain artifacts (Fraser et al., 2011; Keedy et al., 2014; Thorne, 2023).
The structural models that have been refined using diffraction data collected at 100 K generally exhibit a single conformation of the polypeptide chain, with, at most alternative conformations of specific residue side chains. However, it is well known that the polypeptide chain in many protein structures is quite flexible, adopting multiple conformations that are associated with the predominant one (Fraser et al., 2009; 2011; Lang et al., 2010; 2014; Keedy et al., 2014; 2015). These conformations can also play important roles from a functional point of view: they can be crucial for the molecular recognition of small ligands or other proteins, for catalysis and/or allosteric regulation of enzymes. Knowing the structure of these minor conformations can be crucial for understanding these important molecular mechanisms. Furthermore, regions with well-ordered electron density at cryogenic temperature may become disordered at physiological temperature and vice versa. This can happen both due to temperature effects and as a result of crystal volume variations and, consequently, of crystal packing changes. To collect data at low temperatures, biological macromolecule crystals have to be placed in a solution that contains a cryoprotectant (Jang et al., 2022). The presence of the cryoprotectant can alter the conformation of specific regions of the macromolecule structure; moreover, during the cryoprotectant soaking process, protein crystals may undergo osmotic shock and become disordered or damaged.
It is well known that temperature significantly limits the atomic motions and therefore the structure of molecules, including biological macromolecules. It would be desirable, or at least of interest, to determine the structure of proteins at the living temperature of the organism from which they originate, or at least at a temperature that may be of physiological interest, also to gather information about the protein flexibility, which plays a crucial role in enzymatic catalysis and allosteric communications. In this area, in recent years, many efforts have been made to collect diffraction data at non-cryogenic temperatures (Thompson, 2023). Methods and suggestions for collecting diffraction data at room temperature and analyzing these data have been recently reviewed (Fischer, 2021). During the data collection at non-cryogenic temperature, fragile protein crystals have to be handled with greater care and in a way that prevents dehydration. At non-cryogenic temperatures, it is necessary to limit the exposure of protein crystals to X-rays because the diffusion of free radicals in these crystals can lead to numerous damages, such as the reduction of the oxidation state of the metal centers, the decarboxylation of glutamic and aspartic acid residues, and the breaking of disulfide bridges, which could significantly alter the protein structure (Vergara et al., 2018). However, this type of data collection has the advantage of producing cleaner diffraction patterns with a better signal-to-noise ratio since the diffraction spots cannot be obscured by ice rings.
Examples of protein metalation studies at body temperature
Although the effects of temperature on the structure of proteins have been studied in many works (Tilton et al., 1992; Dutta et al., 2022; Mehra and Kepp, 2022) and it is well known that temperature influences the actual conformation of proteins and the molecular recognition mechanisms (Busi et al., 2021; Skaist Mehlman et al., 2023), an analysis of the structures deposited in the PDB shows that there are only a few dozen protein structures determined at human body temperature, 37 °C, and even fewer structures of metal-protein adducts have been determined at 37 °C. The structure determination of adducts formed upon reaction of proteins with metal complexes at body temperature is of significant interest, because metallodrugs interact with proteins in the organism at this temperature. In this regard, it is important to recall that beyond protein conformation, the temperature can also affect the coordination number and geometry of metal complexes. In solution, oxidovanadium (IV) complexes of bidentate L ligands have the V center that can adopt different coordination numbers and geometries with equilibrium depending on the temperature. For example, when L = deferiprone (dhp) the square pyramidal VIVO(dhp)2 species exists at 298 K, while cis-VIVO(dhp)2(H2O) and trans-VIVO(dhp)2(H2O) species are also present at 120 K (Sanna et al., 2012; 2021). Other examples of structural changes in solution are given by Ni(II) and Cu(II) complexes (Anachini et al., 1977; Willett. et al., 1974). Examples of changes in the coordination numbers of metals have been also observed in the solid state. Zhang et al. reported a temperature-induced single-crystal-to-single-crystal (SCSC) transformation of [Ag6Cl(atz)4]OH·6H2O (Hatz = 3-amino-1,2,4-triazole), with a change in the Ag coordination number (Zhang et al., 2005). Along the same line, Hu and Englert showed that SCSC transformation of [ZnCl2(μ-bipy)]n (bipy = 4,4′-bipyridine) is associated with a variation in the Zn coordination number at temperatures >360 K and <130 K (Hu and Englert, 2005). Similarly, Xie et al. reported that UO2(C18H20N2O4@CB6)2Br2 with a pseudorotaxane motif C6BPCA@CB6 (C6BPCA = 1,1′-(hexane-1,6-diyl)bis(4-(carbonyl)pyridin-1-ium), CB6 = cucurbit [6]uril) as the organic linker, transforms from a 7-coordinated uranium (VI) to a 6-coordinated uranium (VI) form upon cooling and heating in the 170–320 K range (Xie et al., 2017). Moreover, Bernini et al. showed that Yb coordination in [Yb(C4H4O4)1.5] changes when it is heated above 130 °C, returning to its initial form when back at room temperature (Bernini et al., 2009).
Obviously, these changes could influence the interaction with proteins and thus it is not surprising that the binding of metal complexes to proteins is significantly influenced by temperature (Ferraro et al., 2016). Inductively Coupled Plasma Mass Spectrometry (ICP-MS) analyses conducted at different temperatures to detect the amount of Pt bound to the model protein hen egg white lysozyme (HEWL) when treated with cisplatin have shown that protein metalation by the Pt compound increases when temperature rises (Ferraro et al., 2016). Recent studies have also highlighted that the conformational changes induced by the temperature increase in the TRPM4 protein prevent the recognition of decavanadate ([VV10O28]6-), a molecule which alters TRPM4 voltage dependence (Hu et al., 2024).
Along this line Fukuda and Inoue observed a difference in substrate binding modes between cryogenic and high-temperature copper nitrite reductase structures (Fukuda and Inoue, 2015).
Jacobs, Helliwell and Brink determined the structure of the adduct that HEWL forms with the Re compound fac-[Et4N]2 [Re(CO)3(Br)3] in the presence of imidazole (Jacobs et al., 2024a). This structure, denoted as HEWL-Re-Im, has been compared with those of a series of adducts formed at room temperature and solved using data collected at cryogenic temperature (Jacobs et al., 2024b). The results of this comparison revealed that the Re binding sites, where Re-containing fragments covalently bind the protein, are retained at 37 °C with minor modifications of protein residue side chains (see, for example, Re binding sites close to the side chains of His15 and Asp101 in the structures at 100 K and at 37 °C reported in Figure 1). Lower occupancy or absence of Re-containing fragments are observed at 37 °C in the case of non-covalent binding sites.
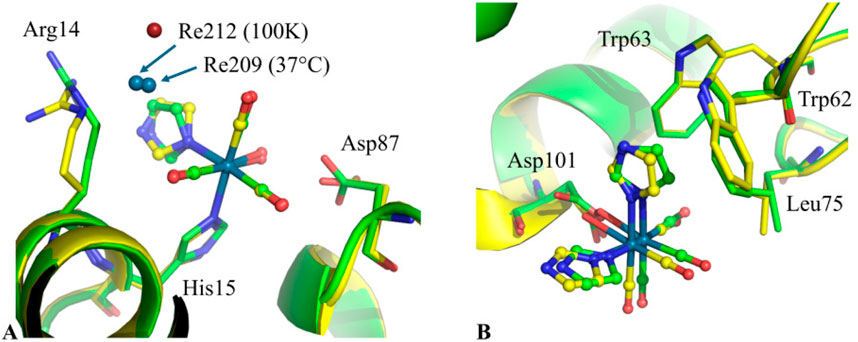
Figure 1. Superimposition of the Re binding sites close to the side chains of (A) His15 and (B) Asp101 in the structures of HEWL-Re-Im at cryogenic temperature (PDB code 8QCU, green) and at 37 °C (PDB code 9GHX, yellow). Additional non-covalent Re atom binding site close to His15 is also shown (Re212 and Re209 in the structures at 100K and at 37 °C, respectively). The superimpositions show minor displacements of metal-containing fragments and HEWL residues involved in their recognition in the structures of HEWL-Re-Im at 37 °C and at 100 K.
In my group, we also recently determined the structure of the adduct formed upon reaction of dirhodium tetraacetate ([Rh2 (ac)4], ac = acetate) with HEWL at 37 °C (Tito et al., 2025b), and those formed upon reaction of the same protein with [VIVO(acac)2], acac = acetylacetonato (Tito et al., 2025a), and with the dioxovanadium(V) complex of malic acid, Cs2 [VV2O4(mal)2]·2H2O, mal = malato (Paolillo et al., 2025). These experiments were conducted by growing protein crystals at 37 °C and treating these crystals at 37 °C with the metal complex. The results of the structural analyses performed using X-ray diffraction data collected at 37 °C were compared with those previously obtained in cryogenic conditions on crystals grown and exposed to metal complexes at 20 °C (Loreto et al., 2021; Tito et al., 2024; 2025b; Paolillo et al., 2025).
Data collected on HEWL crystals treated with [Rh2 (ac)4] show a substantial invariance of the Rh binding sites at different temperatures, with a significant difference in metal occupancy, which is higher at body temperature (Loreto et al., 2021; Tito et al., 2025b). Although this result was obtained on crystals of different sizes and may be influenced by factors not easily controllable during the experiment, such as the radial diffusion of the metal complex in the solvent channels within the crystal and possible small differences in the radiation damage induced by X-rays on the crystals collected at different temperatures, it is consistent with the ICP-MS results obtained in solution by treating the protein with cisplatin (Ferraro et al., 2016) and with the data discussed by Jacobs, Helliwell and Brink (Jacobs et al., 2024a). Even in the comparison of the structures obtained by reacting HEWL with [VIVO(acac)2] at different temperatures, shown in Figure 2, only small differences were observed in the protein binding of the species derived from the transformation of the V complex (Ferraro et al., 2023; Tito et al., 2024; 2025a). In particular, a [V20O51(H2O)]n–ion and a [VV7VIV8O33(H2O)]+ cation covalently bind HEWL surface at 37 °C (Figure 2). These polyoxidovanadates are found also in the 100 K structures of HEWL treated with [VIVO(acac)2], but at cryogenic temperature they were observed in two different crystals (Ferraro et al., 2023; Tito et al., 2024; 2025a).
![Two molecular structures labeled A and B demonstrate protein-ligand interactions. Both structures feature helices and strands in purple and yellow, with amino acids such as Lys1, Ser86, and Asn19 labeled. The ligands, depicted as polyoxometalate clusters, are engaged with the protein structures. Based on their chemical formulas, cluster A is labeled as \([V_{20}O_{51}(OH_2)]^n\) and cluster B as \([V_{15}O_{33}(OH_2)]^+\).](https://www.frontiersin.org/files/Articles/1670177/fchbi-04-1670177-HTML/image_m/fchbi-04-1670177-g002.jpg)
Figure 2. Superimposition of the (A) [V20O51(H2O)]n–ion binding sites close to the side chains of Asp87 and Asp87* and (B) [VV7VIV8O33(H2O)]+ cation binding sites close to Asp18 and Gly71*, in the structures at cryogenic temperature (PDB code 9EX0, in violet, and PDB code 9EX2, in light blue) and at 37 °C (PDB code 9I8L, yellow). The superimpositions show high similarity in polyoxidovanadate binding sites in the structures at 37 °C and at 100 K. Asterisk (*) indicates residues from symmetry-related molecules.
Different results were obtained by treating HEWL with Cs2 [VV2O4(mal)2]·2H2O (Paolillo et al., 2025). Crystallographic data, corroborated by NMR measurements, reveal that protein metalation (Merlino, 2021) appears to be negatively affected by temperature in this case: in cryogenic structures, HEWL binds and stabilizes [VIVO]2+ [VV2O5(mal)]2– [VV10O28]6-, and its derivative [VV10O26]2–, which can be considered a [VV10O28]6- ion with two oxygen atoms replaced by oxygens of the side chains of Glu residues, while at body temperature only the binding of [VIVO]2+ is observed. These molecules are formed by the speciation of Cs2[VV2O4(mal)2]·2H2O. Interestingly, the structure at 100 K shows covalent and non-covalent binding of the decavanadate ion to the protein (Figures 3A,B). Since the HEWL structure in the presence of [VV2O4(mal)2] at 37 °C and at 100 K is very similar in the decavanadate binding site (Figure 3C), a different flexibility of protein residues in this region has to be invoked to explain the inability of HEWL to bind [V10O28]6-/[VV10O26]2– at body temperature.
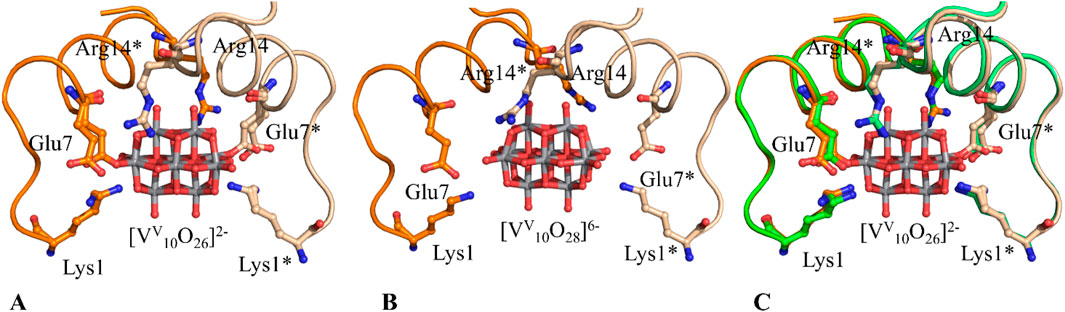
Figure 3. Covalent (A) and non-covalent (B) binding of decavanadate to HEWL in the 100 K structures of the adduct formed upon reaction of the protein with Cs2 [VV2O4(mal)2]·2H2O (PDB codes 9RBG and 9RBT, respectively) (C) Superimposition of the decavanadate binding sites close to the side chains of Glu7 and Glu7* in the structures at cryogenic temperature, reported in panel A (PDB code 9RBG, in orange), and at 37 °C (PDB code 9RBV, in green). The superimpositions show high similarity in the decavanadate binding sites in the structures at 37 °C and at 100 K. Asterisk (*) indicates residues from symmetry mates, that are shown with slightly softer colors.
Conclusion
Temperature is a key variable that governs the coordination number and geometry of metal complex, protein conformation and flexibility and can significantly alter ligand-protein recognition processes (Yeh et al., 2023; Hu et al., 2024).
Due to the traditional use of diffraction data collection at cryogenic temperatures, only a few structures of proteins have been solved at body temperature and just four X-ray structures of metal-protein adducts have been obtained at 37 °C. This is probably due to the difficulty to grow crystals at body temperature and to collect X-ray diffraction data at this temperature suitable for structural analyses. The experiments carried out up to now on metal/protein adducts show conflicting results: temperature can both enhance and hamper the protein metalation process. At the moment, it is not possible to predict the effects that temperature changes can have on protein metalation process, as protein metalation at body temperature critically depends likely on the nature of the metal complexes and the physico-chemical characteristics of the protein involved in the molecular recognition process. This suggests that further studies on the binding of metal complexes to proteins at different temperatures are needed. In this respect, it should be noted that although new methods that enable the collection of high-quality diffraction data from single crystals at different temperatures have been described (Doukov et al., 2020), it would be helpful to develop protocols and to standardize methods of growing and transporting protein crystals at 37 °C. Furthermore, to allow diffraction data collection at 37 °C, it is necessary to implement new technologies that facilitate the conduction of experiments at this temperature.
Future research could also focus on the stability and dynamics of metal-protein adducts at body temperature, since these data could provide us with new, useful information on the possible transport and release of metallodrugs by proteins under conditions close to physiological ones.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
AM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. AM thanks MIUR PRIN2022 (project code: 2022JMFC3X, “Protein Metalation by Anticancer Metal-based Drugs“) for financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer EG declared a past co-authorship with the author AM at the time of review.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anichini, A., Fabbrizzi, L., Paoletti, P., and Clay, R. M. (1977). Does a macrocyclic effect exist in the blue-to-yellow conversion of nickel(II) complexes with tetra-aza ligands? Inorganica Chim. Acta 24, L21–L23. doi:10.1016/S0020-1693(00)93812-X
Berman, H. M. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. doi:10.1093/nar/28.1.235
Bernini, M. C., Gándara, F., Iglesias, M., Snejko, N., Gutiérrez-Puebla, E., Brusau, E. V., et al. (2009). Reversible breaking and forming of metal–ligand coordination bonds: temperature-Triggered single-crystal to single-crystal transformation in a metal–organic framework. Chem. A Eur. J. 15, 4896–4905. doi:10.1002/chem.200802385
Busi, B., Yarava, J. R., Bertarello, A., Freymond, F., Adamski, W., Maurin, D., et al. (2021). Similarities and differences among protein dynamics studied by variable temperature nuclear magnetic resonance relaxation. J. Phys. Chem. B 125, 2212–2221. doi:10.1021/acs.jpcb.0c10188
Cheng, Y. (2018). Single-particle cryo-EM—How did it get here and where will it go. Science 361, 876–880. doi:10.1126/science.aat4346
Doukov, T., Herschlag, D., and Yabukarski, F. (2020). Instrumentation and experimental procedures for robust collection of X-ray diffraction data from protein crystals across physiological temperatures. J. Appl. Crystallogr. 53, 1493–1501. doi:10.1107/S1600576720013503
Dutta, P., Roy, P., and Sengupta, N. (2022). Effects of external perturbations on protein systems: a microscopic view. ACS Omega 7, 44556–44572. doi:10.1021/acsomega.2c06199
Ferraro, G., Pica, A., Russo Krauss, I., Pane, F., Amoresano, A., and Merlino, A. (2016). Effect of temperature on the interaction of cisplatin with the model protein hen egg white lysozyme. J. Biol. Inorg. Chem. 21, 433–442. doi:10.1007/s00775-016-1352-0
Ferraro, G., Tito, G., Sciortino, G., Garribba, E., and Merlino, A. (2023). Stabilization and binding of [V4 O12]4- and unprecedented [V20O54(NO3)]n- to lysozyme upon loss of ligands and oxidation of the potential drug VIV O(acetylacetonato)2. Angew. Chem. Int. Ed. 62 (50), e202310655. doi:10.1002/anie.202310655
Fischer, M. (2021). Macromolecular room temperature crystallography. Quart. Rev. Biophys. 54, e1. doi:10.1017/s0033583520000128
Fraser, J. S., Clarkson, M. W., Degnan, S. C., Erion, R., Kern, D., and Alber, T. (2009). Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673. doi:10.1038/nature08615
Fraser, J. S., Van Den Bedem, H., Samelson, A. J., Lang, P. T., Holton, J. M., Echols, N., et al. (2011). Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 108, 16247–16252. doi:10.1073/pnas.1111325108
Fukuda, Y., and Inoue, T. (2015). High-temperature and high-resolution crystallography of thermostable copper nitrite reductase. Chem. Commun. 51, 6532–6535. doi:10.1039/C4CC09553G
Garman, E. (1999). Cool data: quantity AND quality. Acta Crystallogr. D. Biol. Crystallogr. 55, 1641–1653. doi:10.1107/s0907444999008653
Haas, D. J. (2020). The early history of cryo-cooling for macromolecular crystallography. IUCrJ 7, 148–157. doi:10.1107/s2052252519016993
Hu, C., and Englert, U. (2005). Crystal-to-Crystal transformation from a chain polymer to a two-dimensional network at low temperatures. Angew. Chem. Int. Ed. 44, 2281–2283. doi:10.1002/anie.200462100
Hu, J., Park, S. J., Walter, T., Orozco, I. J., O‘Dea, G., Ye, X., et al. (2024). Physiological temperature drives TRPM4 ligand recognition and gating. Nature 630, 509–515. doi:10.1038/s41586-024-07436-7
Jacobs, F. J. F., Helliwell, J. R., and Brink, A. (2024a). Body temperature protein X-ray crystallography at 37 °C: a rhenium protein complex seeking a physiological condition structure. Chem. Commun. 60, 14030–14033. doi:10.1039/d4cc04245j
Jacobs, F. J. F., Helliwell, J. R., and Brink, A. (2024b). Time-series analysis of rhenium(I) organometallic covalent binding to a model protein for drug development. IUCrJ 11, 359–373. doi:10.1107/s2052252524002598
Jang, K., Kim, H. G., Hlaing, S. H. S., Kang, M., Choe, H.-W., and Kim, Y. J. (2022). A short review on cryoprotectants for 3D protein structure analysis. Crystals 12, 138. doi:10.3390/cryst12020138
Keedy, D. A., van den Bedem, H., Sivak, D. A., Petsko, G. A., Ringe, D., Wilson, M. A., et al. (2014). Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR. Structure 22, 899–910. doi:10.1016/j.str.2014.04.016
Keedy, D. A., Kenner, L. R., Warkentin, M., Woldeyes, R. A., Hopkins, J. B., Thompson, M. C., et al. (2015). Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography. eLife 4, e07574. doi:10.7554/elife.07574
Lang, P. T., Ng, H., Fraser, J. S., Corn, J. E., Echols, N., Sales, M., et al. (2010). Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 19, 1420–1431. doi:10.1002/pro.423
Lang, P. T., Holton, J. M., Fraser, J. S., and Alber, T. (2014). Protein structural ensembles are revealed by redefining X-ray electron density noise. Proc. Natl. Acad. Sci. U.S.A. 111, 237–242. doi:10.1073/pnas.1302823110
Loreto, D., Ferraro, G., and Merlino, A. (2021). Unusual structural features in the adduct of dirhodium tetraacetate with lysozyme. Int. J. Mol. Sci. 22, 1496. doi:10.3390/ijms22031496
Mehra, R., and Kepp, K. P. (2022). Structure and mutations of SARS-CoV-2 spike protein: a focused overview. ACS Infect. Dis. 8, 29–58. doi:10.1021/acsinfecdis.1c00433
Merlino, A. (2021). Recent advances in protein metalation: structural studies. Chem. Commun. 57, 1295–1307. doi:10.1039/d0cc08053e
Paolillo, M., Ferraro, G., Gumerova, N. I., Pisanu, F., Garribba, E., Rompel, A., et al. (2025). Speciation and structural transformation of a VV–malate complex in the absence and in the presence of a protein: from a dinuclear species to decavanadate. Inorg. Chem. Front. doi:10.1039/d5qi01384d
Sanna, D., Ugone, V., Micera, G., and Garribba, E. (2012). Temperature and solvent structure dependence of VO2+ complexes of pyridine-N-oxide derivatives and their interaction with human serum transferrin. Dalton Trans. 41, 7304–7318. doi:10.1039/c2dt12503j
Sanna, D., Lubinu, G., Ugone, V., and Garribba, E. (2021). Influence of temperature on the equilibria of oxidovanadium(IV) complexes in solution. Dalton Trans. 50, 16326–16335. doi:10.1039/d1dt02680a
Skaist Mehlman, T., Biel, J. T., Azeem, S. M., Nelson, E. R., Hossain, S., Dunnett, L., et al. (2023). Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B. eLife 12, e84632. doi:10.7554/elife.84632
Thompson, M. C. (2023). “Combining temperature perturbations with X-ray crystallography to study dynamic macromolecules: a thorough discussion of experimental methods,” in Methods in enzymology (Elsevier), 255–305. doi:10.1016/bs.mie.2023.07.008
Thorne, R. E. (2023). Determining biomolecular structures near room temperature using X-ray crystallography: concepts, methods and future optimization. Acta Crystallogr. D. Struct. Biol. 79, 78–94. doi:10.1107/s2059798322011652
Tilton, R. F., Dewan, J. C., and Petsko, G. A. (1992). Effects of temperature on protein structure and dynamics: x-ray crystallographic studies of the protein ribonuclease-A at nine different temperatures from 98 to 320K. Biochemistry 31, 2469–2481. doi:10.1021/bi00124a006
Tito, G., Ferraro, G., Pisanu, F., Garribba, E., and Merlino, A. (2024). Non-Covalent and covalent binding of new mixed-valence cage-like polyoxidovanadate clusters to lysozyme. Angew. Chem. Int. Ed. 63, e202406669. doi:10.1002/anie.202406669
Tito, G., Ferraro, G., Garribba, E., and Merlino, A. (2025a). Formation of mixed-valence Cage-Like polyoxidovanadates at 37 °C upon reaction of VIVO(acetylacetonato)2 with lysozyme. Chem. A Eur. J. 31, e202500488. doi:10.1002/chem.202500488
Tito, G., Ferraro, G., and Merlino, A. (2025b). Dirhodium tetraacetate binding to lysozyme at body temperature. Int. J. Mol. Sci. 26, 6582. doi:10.3390/ijms26146582
Vergara, A., Caterino, M., and Merlino, A. (2018). Raman-markers of X-ray radiation damage of proteins. Int. J. Biol. Macromol. 111, 1194–1205. doi:10.1016/j.ijbiomac.2018.01.135
Willett, R. D., Haugen, J. A., Lebsack, J., and Morrey, J. (1974). Thermochromism in copper(II) chlorides. Coordination geometry changes in tetrachlorocuprate(2-)anions. Inorg. Chem. 13, 2510–2513. doi:10.1021/ic50140a040
Xie, Z.-N., Mei, L., Wu, Q.-Y., Hu, K. q., Xia, L.-S., Chai, Z.-F., et al. (2017). Temperature-induced reversible single-crystal-to-single-crystal isomerisation of uranyl polyrotaxanes: an exquisite case of coordination variability of the uranyl center. Dalton Trans. 46, 7392–7396. doi:10.1039/C7DT01034F
Yeh, F., Jara-Oseguera, A., and Aldrich, R. W. (2023). Implications of a temperature-dependent heat capacity for temperature-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 120, e2301528120. doi:10.1073/pnas.2301528120
Keywords: protein metalation, body temperature, X-ray crystallography, metal-protein adducts, physiological temperature (37 °C)
Citation: Merlino A (2025) X-ray structures of metal-protein adducts at body temperature: concepts, examples and perspectives. Front. Chem. Biol. 4:1670177. doi: 10.3389/fchbi.2025.1670177
Received: 21 July 2025; Accepted: 12 September 2025;
Published: 29 September 2025.
Edited by:
Debbie C. Crans, Colorado State University, United StatesReviewed by:
Eugenio Garribba, University of Sassari, ItalyCraig C. McLauchlan, Illinois State University, United States
Copyright © 2025 Merlino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonello Merlino, YW50b25lbGxvLm1lcmxpbm9AdW5pbmEuaXQ=
 Antonello Merlino
Antonello Merlino