- Department of Chemistry, Clemson University, Clemson, SC, United States
Cellular damage and death caused by oxidative stress by reactive oxygen species play an important role in disease development. Testing the ability of hydrophobic compounds to prevent radical-generated oxidative stress typically involves radical scavenging assays; however, these oxidative stress assays often do not accurately reflect biological outcomes. We present an in vitro assay that quantifiably evaluates the ability of hydrophobic compounds to prevent DNA damage, a biological endpoint that is linked to disease development. This gel electrophoresis assay enables evaluation of a wide range of hydrophobic compounds for metal-mediated hydroxyl radical damage prevention by using high-ethanol concentrations in electrophoretic conditions, and the effects of these high-ethanol conditions on iron- and copper-mediated DNA damage are established. This assay was used to compare the effects of metal-mediated DNA damage and its prevention by polyphenols, bipyridine, and selone antioxidant compounds as well as the radical scavenger edaravone. We also demonstrated that the well-studied glutathione peroxidase mimic ebselen and a group of ebselen derivatives prevent copper-mediated DNA damage with IC50 values in the 280–450 µM range. These same compounds do not inhibit iron-mediated DNA damage under similar conditions. This DNA-damage assay allows determination of antioxidant properties for hydrophobic antioxidants and drugs using a model system based on metal-mediated oxidative DNA damage, a primary cause of cell death.
1 Introduction
Oxidative stress is the increased production or decreased elimination of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), superoxide (O2•-) and hydroxyl radical (•OH). ROS control physiological responses such as inflammation, changes in gene expression, apoptosis, and cell proliferation (Alfadda and Sallam, 2012). Increased ROS levels also play an important role in disease development, including atherosclerosis, neurodegenerative diseases, cancer, and aging (Alfadda and Sallam, 2012; Collin, 2019; Yang and Lian, 2020). Halliwell (2000) and Halliwell and Aruoma (1991) emphasized that DNA is a primary target for cellular ROS damage, making oxidative DNA damage an important biomarker for oxidative stress.
Cellular dioxygen (O2) is converted to H2O2 and O2•- by direct oxidation of flavoproteins, and H2O2 is also produced by superoxide disproportionation (Fridovich, 1989) and cellular respiration (Duthie, 1999). Redox-active transition metals, particularly iron and copper, also play an essential role in ROS production (Angelé-Martínez, Goodman and Brumaghim, 2014). Iron and copper react with hydrogen peroxide to produce •OH (Dixon and Stockwell, 2014; Linder, 2012) (Equation 1) that can oxidize DNA bases or abstract a hydrogen atom from deoxyribose (Park and Imlay, 2003) to break the DNA backbone, in addition to oxidizing cellular lipids, proteins, and small molecules (Dixon and Stockwell, 2014). Oxidized Fe3+ or Cu2+ can be reduced by nicotinamide adenine dinucleotide (NADH) (Imlay and Linn, 1988) or ascorbic acid (B. Halliwell and Aruoma, 1991) making cellular •OH generation catalytic. In fact, iron-mediated DNA damage by •OH is the primary mechanism for cell death in both prokaryotes and eukaryotes (B. Halliwell and Aruoma, 1991; Imlay and Linn, 1988) highlighting the biological importance of metal-mediated radical generation.
For decades, measuring the antioxidant abilities of dietary antioxidants in a variety of foods has been an active research area. Recently, many drugs have also been designed to have antioxidant properties, including edaravone (Figure 1A) for treating ALS (Homma, S., Sato, and J., 2019). For many of these drugs, their hydrophobicity allows them to more readily cross the blood-brain-barrier as well as cellular and nuclear membranes. Thus, it is of significant interest to establish the ability of these more hydrophobic compounds to prevent DNA damage.
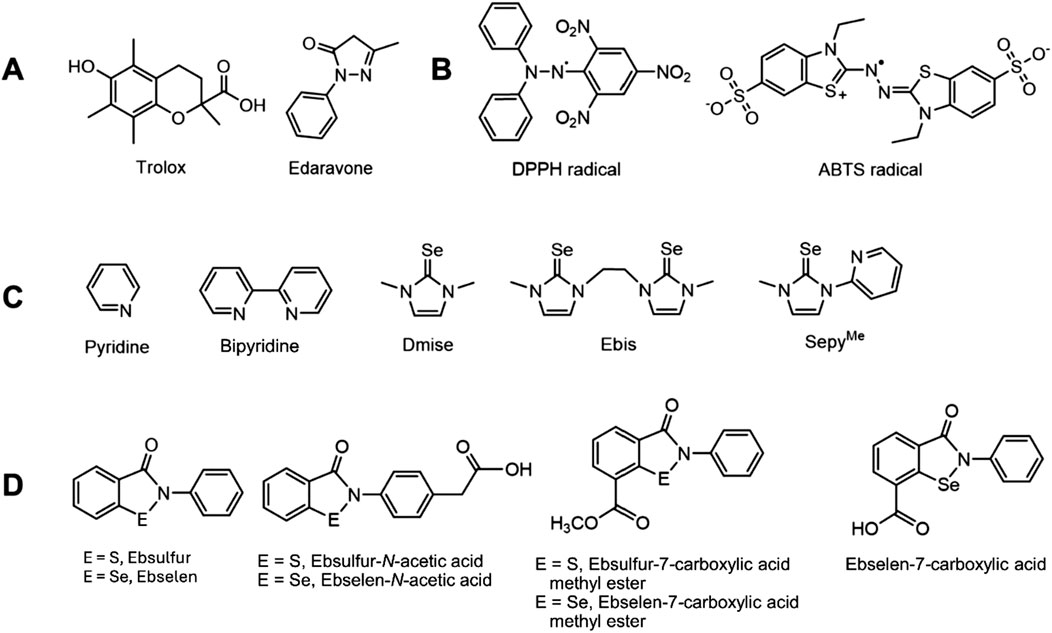
Figure 1. Structures of (A) Trolox and edaravone, known radical scavengers; (B) DPPH and ABTS radicals commonly used in antioxidant assays; (C) various selones and pyridine derivatives; and (D) ebselen (E = Se) and ebsulfur (E = S) derivatives.
Duthie (1999) described the ability of antioxidants to prevent oxidation by three mechanisms: 1) coordination to transition metals to prevent radical formation, 2) decreasing localized O2•- concentrations to reduce oxidation reactions, and 3) scavenging radicals that can abstract H• from molecules. Common in vitro assays for screening antioxidant activity primarily focus on radical scavenging ability. Radicals in these assays include 2,2′-diphenyl-1-picryl-hydrazyl (DPPH) and 2,2′azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS; Figure 1B) (Schaich, Tian and Xie, 2015). DPPH and ABTS form stable organic radicals with very different properties compared to small, oxygen-based ROS. In addition, DPPH and ABTS scavenging assays are typically performed in organic solvents (Pérez-Jiménez and Saura-Calixto, 2006).
Separate assays are also performed to evaluate the ability of antioxidants to reduce Fe3+ or Cu2+, including the ferric reduction antioxidant power (FRAP) assay (Benzie and Devaki, 2018) or the cupric reducing antioxidant capacity (CUPRAC) assay (Özyürek et al., 2011). Whereas CUPRAC assays are performed at pH 7, FRAP assays are conducted at a non-biological pH of 3.6. Collectively, these measures used to determine antioxidant radical inhibition typically only examine a single potential mechanism of action and do not reflect biological endpoints such as DNA damage (Frankel and Meyer, 2000).
The inadequacy of such antioxidant assays to mirror biological endpoints is highlighted by the failure of the oxygen radical absorbance capacity (ORAC) assay that measured scavenging of singlet oxygen, peroxynitrite, hydroxyl radical, and superoxide radical (Cao, Alessio and Cutler, 1993) using standard protocols adaptable for both aqueous and organic solvents (Cao, Verdon, Wu, Wang and Prior, 1995). From 2007 to 2012, the United States Department of Agriculture (USDA) published tables of ORAC values to enable comparison of the antioxidant activity of various foods and food additives (Bank and Schauss, 2004; Singh and Singh, 2008). Due to the lack of correlations between ORAC results and observed biological effects, as well as incorrect methodology, the USDA entirely discontinued the use of this assay and deleted the ORAC database in 2012 (Schaich et al., 2015). No other standard assay or method of evaluating and comparing antioxidant activity has been put into place since 2012.
To increase the biological relevance of antioxidant determinations, cell survival assays are also used to establish and compare antioxidant ability (López-Alarcón, 2013). These assays are very relevant because they explore antioxidant behavior in living systems, but they have drawbacks for screening antioxidant efficacy. The use of a wide variety of cell lines for these assays limits the ability to compare results, and toxicity issues prevent the screening of many compounds (Singh and Singh, 2008). All these radical scavenging, metal reduction, and cellular assays generate different scales of antioxidant activity because they are based on different underlying mechanisms for antioxidant activity, and many use different conditions for the same assay, further complicating antioxidant comparisons.
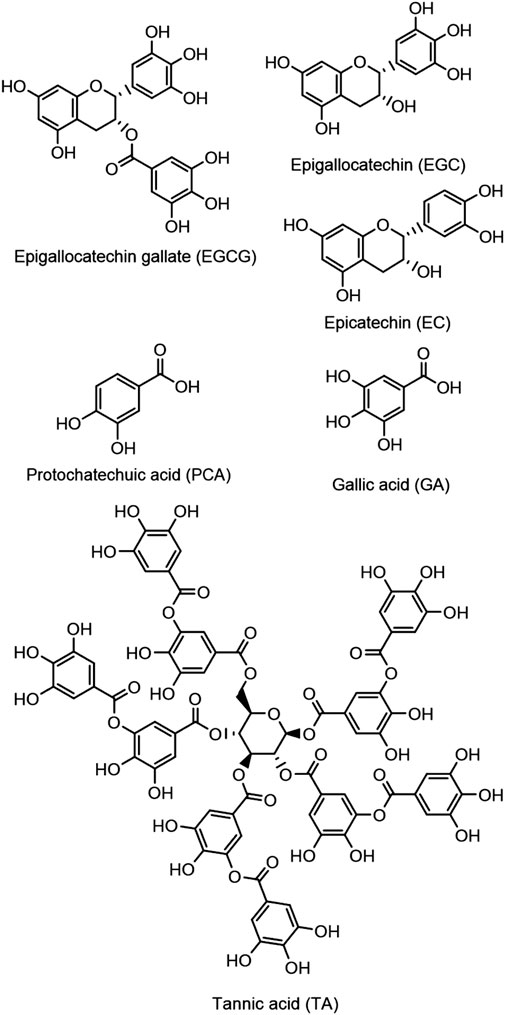
Our hydrophobic DNA damage assay overcomes many limitations of these antioxidant assays by using an in vitro assay with DNA damage as an endpoint. We use ethanol to dissolve hydrophobic antioxidants in these DNA electrophoresis studies and report the effects of high ethanol in gel electrophoresis studies. Adding ethanol allows dissolution of hydrophobic compounds, and these electrophoretic methods provide a quantitative method to directly compare antioxidant efficacy for polyphenol antioxidants (Figure 2) as well as a variety of water-insoluble antioxidant compounds that act through radical inhibition and/or metal binding mechanisms, including edaverone, ebselen, and ebselen derivatives (Figures 1A,C,D).
2 Materials and methods
2.1 Materials
Water was deionized (diH2O) using a Nano Pure DIamond Ultrapure H2O system (Barnstead International). 3-(N-Morpholino) propanesulfonic acid (MOPS; Sigma), 2-(N-morpholino) ethanesulfonic acid (MES; BDH), NaCl (99.999% Alfa Aesar), CuSO4 (Fisher), FeSO4 (Acros), H2O2 (30% w/v, Fisher), DMPO (Cayman Chemicals), ascorbic acid (Alfa Aesar), DPPH (Alfa Aesar), edaravone (Acros), Trolox (Acros), methanol (Sigma-Aldrich), pyridine (Alfa Aesar), bipyridine (Chem-Implex), gallic acid (Acros), epicatechin (Sigma), tannic acid (Sigma Aldrich), protocatechuic acid (Frontier Scientific), epigallocatechin (TCI), epigallocatechin gallate (Enzo), agarose (Sigma Life Science), Chelex (Sigma), and ebselen (Acros) were used as received.
2.2 EPR spectroscopy to measure radical generation
To prepare EPR samples, Cu(SO4)2∙3H2O (300 µM), ascorbic acid (375 µM), and H2O2 (2.5 mM), and indicated ethanol concentrations were added to an aqueous solution of MOPS buffer (pH 7, 10 mM) containing DMPO (30 mM) as a spin trap. Iron-containing samples were prepared with FeSO4 (300 µM), H2O2 (2.5 mM) and DMPO (30 mM) with indicated ethanol concentrations in MES buffer (pH 6, 10 mM). All samples were diluted with the appropriate buffer solution to achieve a final volume of 500 µL. EPR spectra were measured on a Bruker EMX spectrometer at room temperature in a quartz flat cell. Spectra centered at 3,431.24 G were acquired with a sweep width of 100 G. The modulation amplitude was 2.00 G with time and conversion constants of 81.92 s, and microwave power and frequency were 1.99 mW and 9.756 GHz, respectively. EPR spectra can be found in Supplementary Figures S1–S6 in the Supplementary Material.
2.3 Plasmid DNA transfection, amplification, and purification
Plasmid DNA (pBSSK) was purified from DH1 E. coli competent cells using a Zyppy™ Plasmid Miniprep Kit (400 count, Zymo Research). Tris-EDTA buffer (pH 8.0) was used to elute the plasmid DNA from the spin columns. Plasmid was dialyzed against 130 mM NaCl for 24 h at 4 °C to ensure all Tris-EDTA buffer and metal contaminates were removed, and plasmid concentration was determined by UV-vis spectroscopy at a wavelength of 260 nm. Absorbance ratios of A260/A230 ≥ 2.0 and A260/A280 ≥ 1.8 were determined for DNA used in all experiments. Plasmid purity was determined through digestion of plasmid (0.1 pmol) with Sac 1 and KpnI restriction endonucleases at 37 °C for 90 min. Digested plasmid was compared to undigested plasmid and a 1 kb molecular weight marker using gel electrophoresis.
2.4 DNA gel electrophoresis studies
DNA electrophoresis samples were prepared in acid-washed (1 M HCl for 1 h) microcentrifuge tubes that were triple-rinsed with DI water and dried. MOPS (10 mM, pH 7.0 for copper studies) or MES (10 mM, pH 6 for iron studies to prevent precipitation) buffer was treated with Chelex resin prior to preparing the samples. For low-ethanol gels, ethanol was added to achieve a final concentration of 10 mM, and the antioxidant compound was added in the indicated concentrations in buffer solution. For high-ethanol gel samples (10% v/v, 1.7 M), the antioxidant compound to be tested was dissolved in cold absolute ethanol and added to the buffer solution to achieve the indicated final concentrations. Then CuSO4∙5H2O and ascorbate (1.25× the copper concentration, to reduce Cu2+ to Cu+) were added at the indicated concentrations to the antioxidant solution and the samples were allowed to stand for 5 min at room temperature. Plasmid (pBSSK, 0.1 pmol in 130 mmol NaCl) was then added and samples again were allowed to stand for 5 min at room temperature. H2O2 (50 µM) was added to start hydroxyl radical generation, and samples were allowed to stand at room temperature for 30 min. EDTA (50 µM) was added after 30 min to stop radical generation by chelating the metal ions. For the Fe2+ DNA damage experiments, the indicated concentrations of freshly prepared FeSO4∙7H2O and MES (10 mM, pH 6.0) buffer were used to prevent iron precipitation, and no ascorbate was added. For control samples without antioxidant, metal, and/or H2O2, additional buffer was added to make up the missing volume. All sample concentrations are reported as final concentrations in a 10 µL volume, and final samples were mixed with 2 µM of 6× loading dye (1.59 mL diH2O, 2.08 mL 1% xylene cyanol FF solution, 2.08 mL 0.5% bromophenol blue solution, and 3.75 mL of 80% glycerol).
Samples were loaded into a 1% agarose gel in a TAE running buffer (50×), and damaged (nicked) and undamaged (supercoiled) plasmid was separated by horizontal gel electrophoresis (140 V for 60 min). Gels were stained using ethidium bromide and imaged using UV light for comparison to published results using this staining method. It is possible to use safer alternatives to ethidium bromide, such as SYBR dyes (Bourzac, LaVine and Rice, 2003; Dragan et al., 2012), for DNA visualization.
The amounts of nicked (damaged) and supercoiled (undamaged) DNA were analyzed using a gel imager and UViProMW software (Jencons Scientific Inc.). The intensity of the supercoiled plasmid band was multiplied by 1.24, due to its lower binding affinity of ethidium bromide compared to nicked plasmid (Hertzberg and Dervan, 1982; Lloyd, Haidle and Robberson, 1978). Intensities of the nicked and supercoiled bands were normalized for each lane so that % nicked + % supercoiled = 100%. All nicked band intensities were corrected for residual nicked DNA in the DNA-only control lanes prior to calculation. Results were obtained in triplicate for all experiments, and standard deviations are represented as error bars. The plots of percent DNA damage versus log concentration of copper or iron were fit to a variable-slope sigmoidal dose-response curve using SigmaPlot (v. 11.0, Systat Software, Inc.).
2.5 IC50 value determination
Plots of percent inhibition of DNA damage versus log concentration of the indicated compound were fit to a variable-slope sigmoidal dose-response curve using SigmaPlot, version 11 (Systat Software, Inc.), and IC50 values with standard deviations were determined from the best-fit dose-response curves. Statistical significance was determined by calculating p values at 95% confidence (p < 0.05 indicates significance) (Perkowski and Perkowski, 2007). Data from all DNA damage assays are provided in Supplementary Tables S1–S33.
2.6 DPPH assay studies
DPPH solutions (1.2 mg in 30 mL methanol, 100 µM) were prepared fresh before each experiment: 0.5 mL of antioxidant sample in methanol at various concentrations were combined with 1 mL of DPPH solution (100 µM) with a final concentration of 67 µM DPPH in methanol. Antioxidant concentration ranges tested varied by DPPH scavenging ability and appropriate ranges were selected to obtain IC50 plots. The samples were incubated for 30 min in the dark, and spectra were taken at 515 nm on a Thermo Electron Corporation BioMate3 UV-visible spectrometer. Percentages of DPPH scavenging were calculated using the equation %DPPH scavenged = ((A-A0)/(AT-A0))*100, where A is the absorbance of the incubated sample and DPPH, A0 is the absorbance of DPPH in methanol, and AT is the absorbance of DPPH incubated with 50 µM Trolox. DPPH assay results and antioxidant concentration ranges can be found in Supplementary Tables S34–S46; Supplementary Figures S19–S30 in the Supplementary Material.
3 Results and discussion
3.1 DNA gel electrophoresis with hydrophobic compounds
Our hydrophobic DNA gel electrophoresis assays determine the ability of compounds to prevent copper- or iron-mediated plasmid DNA damage by hydroxyl radical (Equation 1) at pH 7 for copper and pH 6 for iron (to prevent precipitation). To generate damaging hydroxyl radical, Cu+ or Fe2+ and hydrogen peroxide were used in low micromolar concentrations (6 and 15 μM, respectively). Using gel electrophoresis, supercoiled (undamaged) and nicked (damaged) plasmid DNA was separated, allowing a quantitative analysis of antioxidant activity.
To enable DNA damage prevention studies with hydrophobic compounds, ethanol was added to the DNA-damaging reactions. Ethanol was selected as the organic solvent because double-stranded DNA is most stable in low-dielectric solvents such as ethanol (Nakano and Sugimoto, 2016). Hydrophobic antioxidant compounds were dissolved in 100% ethanol and 1 µL of this stock solution was added to the aqueous reagents (9 µL), resulting in a 10% (v/v; 1.7 M) final ethanol concentration.
Ethanol is not commonly added to plasmid DNA gel-electrophoresis studies, and we report the effects of ethanol addition in these experiments. Few reports of ethanol or methanol addition in these types of assays exist (Battin, Zimmerman, Ramoutar, Quarles and Brumaghim, 2011; Briviba, Roussyn, Sharov and Sies, 1996; Henle et al., 1999; Roussyn, Briviba, Masumoto and Sies, 1996) and only one focused on antioxidant prevention of DNA damage. Sies et al. (Roussyn et al., 1996) attempted to evaluate peroxynitrite-induced DNA damage prevention by the hydrophobic antioxidant ebselen in aqueous methanolic solutions, but analysis of the results was problematic because radical scavenging by methanol was not accounted for, only one concentration (50 µM) of ebselen was tested, and necessary controls were missing.
3.2 Effects of increased ethanol concentration on DNA damage
Ethanol is a known radical scavenger, and Linn et al. (Henle et al., 1999; Linn, 2015) studied the effect of ethanol on DNA damage caused by iron and hydrogen peroxide (Equation 1). In the presence of Fe2+ and H2O2, they identified modes of DNA damage by •OH: Mode I damage occurs at low H2O2 concentrations, is caused by Fe2+ loosely bound to DNA, and is moderately reduced by ethanol scavenging; Mode II damage occurs at higher H2O2 concentrations, results from Fe2+ coordinated to DNA bases, and is resistant to ethanol scavenging. The ethanyl radical, like other alkyl radicals, can lead directly or indirectly to the production of DNA-derived radicals, DNA single-strand breaks, and 8-alkylguanine adducts (Nakao, Fonseca and Augusto, 2002).
A previous report from this laboratory (Perron et al., 2010) tested the antioxidant activity of water-soluble compounds using similar plasmid DNA damage assays with a lower concentration of ethanol (10 mM) added to mimic naturally occurring organic compounds in cells that act as radical scavengers. When we tested our hydrophobic antioxidant assay with higher-ethanol conditions (10% v/v; 1.7 M), this 1700-fold increase in ethanol showed no effect on copper-mediated DNA damage with the same Cu+ (6 µM) and H2O2 (50 μM; pH 7) conditions: 93 ± 4% and 94 ± 4% DNA damage at 10 mM and 1.7 M ethanol, respectively.
In contrast, prevention of iron-mediated DNA damage is dependent upon ethanol concentration. Under low-ethanol conditions (10 mM), 2 µM Fe2+ in the presence of H2O2 (50 µM) results in 92 ± 3% DNA damage, but under high-ethanol conditions (1.7 M), no DNA damage is observed at this Fe2+ concentration. Increasing DNA damage with increasing Fe2+ concentration is observed under both ethanol concentrations (Figure 3), but more Fe2+ is required to damage the same percentage of DNA under high-ethanol conditions. This concentration effect of ethanol on DNA damage has not been previously determined.
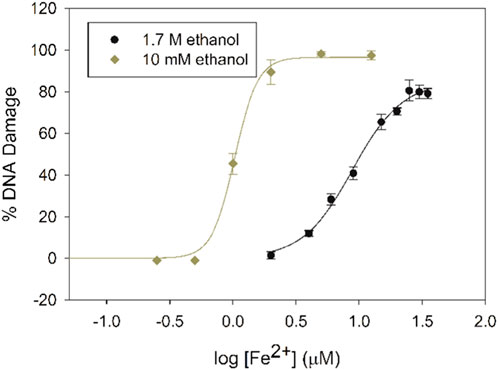
Figure 3. Dose-response curves for iron-mediated DNA damage with 10 mM (diamonds) and 1.7 M ethanol (circles) with increasing Fe2+ concentrations in the presence of H2O2 (50 μM).
A concentration of 15 µM Fe2+ was chosen for our gel electrophoresis studies under high-ethanol conditions, since the percentage of DNA damage (∼90%) at this concentration of iron was similar to the DNA damage percentage under low-ethanol conditions with 2 µM Fe2+. This higher Fe2+ concentration is equal to the labile Fe2+ pools in E. coli (15–30 µM) (Park and Imlay, 2003; Woodmansee and Imlay, 2002) and in mammalian cells (30–210 µM) (Jhurry, Chakrabarti, McCormick, Holmes-Hampton and Lindahl, 2012).
Differences between ethanol effects on copper- and iron-mediated DNA damage is likely due to these metals producing different damaging species. Fe2+ transfers an electron directly to H2O2 to produce •OH (Floyd and Lewis, 1983; B. Halliwell and Gutteridge, 1992), as has been thoroughly established in vitro and in cells (Burkitt and Mason, 1991). In contrast, the precise DNA-damaging species produced by Cu+ is a topic of current debate. Although many studies propose direct •OH generation from the reaction of Cu+ with H2O2 (Aruoma OI et al., 1991), other studies suggest at least initial formation of a separate reactive species (Yamamoto and Kawanishi, 1989), such as inner sphere complexation of copper to H2O2. It is clear, however, that compared to Fe2+ in the presence of H2O2, Cu+ forms a more stable oxidant that is much less susceptible to ethanol radical scavenging (Stoewe and Prütz, 1987).
3.3 Electron paramagnetic spectroscopy studies
EPR experiments of these reaction mixtures were conducted in the presence of the spin trap 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) were conducted to further explore the influence of ethanol concentration on copper- and iron-generated radical species. Both Cu+ (300 µM) and Fe2+ (300 µM) react with H2O2 (2.5 mM) to generate 1:2:2:1 four-line EPR signals typical of the DMPO-OH adduct, although signals were more intense for the iron system. High-ethanol concentrations (1.7 M) inhibit formation of the DMPO-OH adduct (Taniguchi and Madden, 2000). When the ethanol concentration is reduced, DMPO-OH resonances appear (Figure 4A), indicating that high-ethanol concentrations inhibit •OH generation.
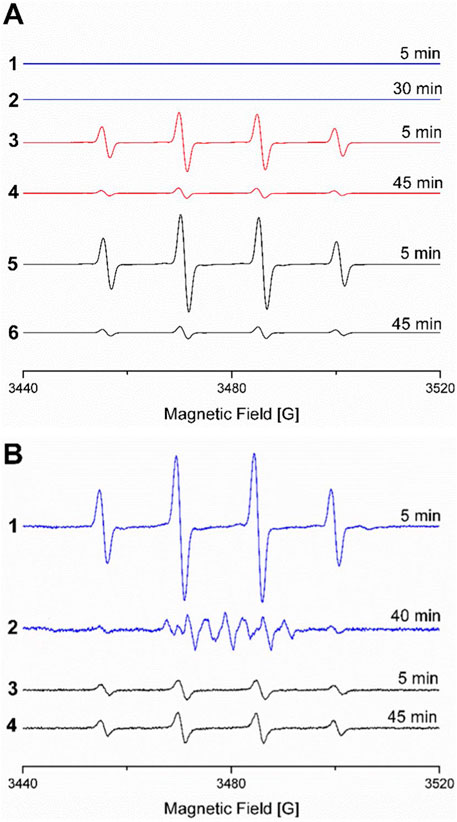
Figure 4. (A) EPR spectra of Fe2+, H2O2, and DMPO in MES buffer (10 mM, pH 6) with 1.7 M ethanol after 1) 5 min and 2) 30 min; with 425 mM ethanol after 3) 5 min and 4) 45 min; and without ethanol after 5) 5 min and 6) 45 min (due to signal overload, spectra without ethanol were collected with lower receiver gain of 103 versus 105 for all other spectra). (B) EPR spectra of Cu2+ ascorbate, H2O2, and DMPO in MOPS buffer (10 mM, pH 7) with 1.7 M ethanol after 1) 5 min and 2) 40 min and without ethanol after 3) 5 min and 4) 45 min. EPR spectra under all conditions followed for up to 115 min are provided in Supplementary Figures S1-S6.
3.4 Effects of ethanol on antioxidant DNA damage prevention assays
To evaluate the impact of high-ethanol concentration on antioxidant prevention of iron-mediated DNA damage, a set of six polyphenols (Figure 2) were tested for their ability to prevent DNA damage under high- and low-ethanol conditions. Polyphenol antioxidant activity has been extensively studied, and previous research from this laboratory (Perron and Brumaghim, 2009; Perron, Hodges, Jenkins and Brumaghim, 2008) established that polyphenol compounds prevent iron-mediated DNA damage under low-ethanol conditions by coordinating iron, making them ideal for a comparison study. These DNA electrophoresis assays were conducted with Fe2+ (2 and 15 μM for low-ethanol and high-ethanol conditions, respectively) plus H2O2 (50 μM) and either 10 mM or 1.7 M ethanol. Increasing concentrations of polyphenol antioxidants were added to these basic reaction conditions.
Tannic acid (TA) in the presence of H2O2 did not damage DNA (Figure 5, lane 3), but Fe2+/H2O2 caused over 90% damage in the presence of 10 mM ethanol (Figure 5, lane 4). The same percentage of DNA damage occurred with Fe2+/H2O2 in the presence of 1.7 M ethanol (Figure 5, lane 5). Adding increasing concentrations of TA up to 20 µM prevented this iron-mediated DNA damage (Figure 5, lanes 6–12).
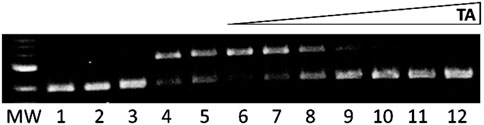
Figure 5. Gel electrophoresis images showing tannic acid (TA) prevention of iron-mediated DNA damage. MW: 1 kb molecular weight marker; lane 1: plasmid DNA (p); lane 2: p + H2O2; lane 3: p + 20 μM TA + H2O2 + 1.7 M ethanol; lane 4: p + Fe2+ (2 µM) + H2O2 + ethanol (10 mM); lane 5: p + Fe2+ (15 µM) + H2O2 + ethanol (1.7 M); lanes 6-12: Fe2+ (15 µM) + H2O2 + ethanol (1.7 M) + TA (0.1, 1, 2.5, 5, 7.5, 10, and 20 μM, respectively).
The plasmid DNA band intensities were quantified, and the resulting data were fit with a dose-response curve to determine the TA concentration required to inhibit 50% DNA damage (IC50 value; Figure 6). IC50 value for TA prevention of iron-mediated DNA damage with 1.7 M ethanol is 2.27 ± 0.01 μM, 7.6 times higher than the IC50 value of 0.3 ± 0.1 μM determined under low-ethanol conditions (Table 1). This increase in IC50 value with increased ethanol concentration is expected, since iron concentrations are 7.5 times higher in the high-ethanol than in the low-ethanol assay conditions (15 and 2 µM Fe2+, respectively) due to the susceptibility of iron-generated •OH to ethanol scavenging. This trend of higher IC50 values for iron-mediated DNA damage prevention under high-ethanol conditions is observed for all the tested polyphenols (Table 1).
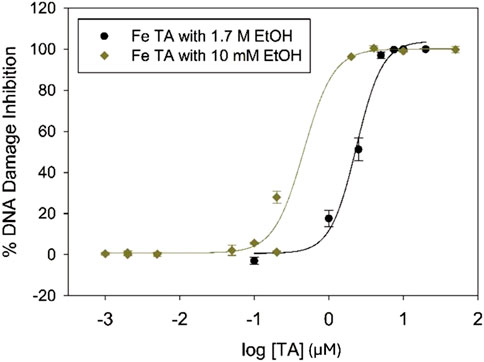
Figure 6. IC50 plots for prevention of iron-mediated DNA damage in the presence of 10 mM and 1.7 M ethanol for tannic acid.
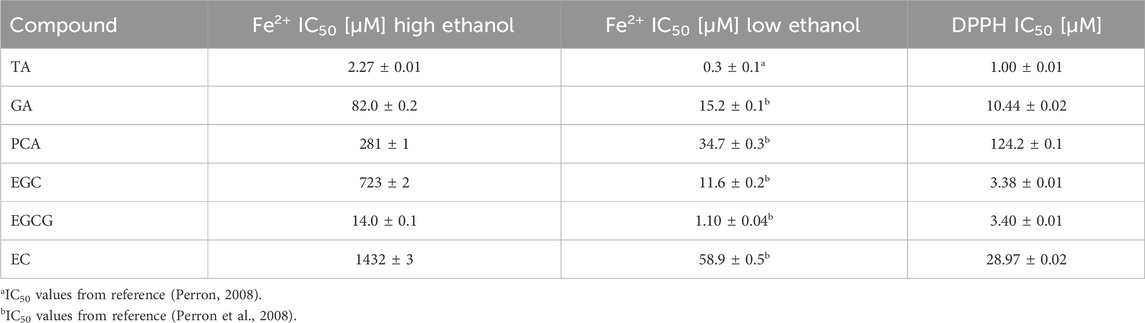
Table 1. IC50 values for polyphenol prevention of iron-mediated DNA damage under high (1.7 M) and low (10 mM) ethanol conditions and polyphenol scavenging of DPPH.
To investigate the correlation between polyphenol IC50 values from plasmid DNA assays under low- and high-ethanol conditions, the log IC50 values were plotted together (Figure 7). The graph is linear, and can be fit with a best-fit line with an R2 value of 0.87. Since polyphenols exhibit both antioxidant and prooxidant behavior in DNA damage prevention assays with copper under low-ethanol conditions (Perron, García, Pinzón, Chaur and Brumaghim, 2011) similar correlations with polyphenol effects on copper-mediated DNA damage IC50 values under high- and low-ethanol conditions were not explored.
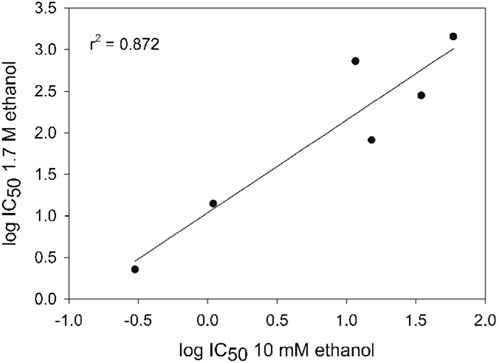
Figure 7. Correlation of polyphenol IC50 values for prevention of DNA damage in the presence of high and low ethanol concentrations (R2 value for linear fit is 0.87). Error bars are within the data symbols.
3.5 Comparing the ability of metal binding and radical scavenging on DNA damage
Since studies with polyphenols reinforced the importance of metal interaction for the observed DNA damage prevention, we chose five additional compounds that systematically differ in their metal-coordination properties to test and compare the effects of metal chelation on DNA damage under high-and low-ethanol conditions. N,N′-dimethylimidazole selone (dmise) is an imidazole selone well-studied for its ability to coordinate copper and iron through selenium (Kimani et al., 2015; Stadelman, Kimani, Bayse, McMillen and Brumaghim, 2016), and ethyl-bis(imidazole) selone (ebis) is a bidentate ligand, binding metals through both selenium atoms (Stadelman et al., 2016). Pyridine and 2,2′-bipyridine (bipy) are extremely well-studied nitrogen-containing ligands for iron and copper (Figure 1C; Fábián, 1989). The selenium and nitrogen metal-binding motifs are combined in (2-mercapto-1-methylimidazolyl) pyridine selone (sepyMe), a bidentate ligand that can coordinate through the selone Se and the pyridine N atoms. Pyridine and bipy are borderline bases and are expected to coordinate more strongly to borderline Fe2+ than to soft Cu+. Selones, one the other hand, are soft bases and therefore bind more strongly to Cu+ than Fe2+. In addition, bidentate ligands, such as bipy and ebis, should coordinate more strongly to metals than their monodentate analogs. If metal coordination plays a significant role for the DNA damage prevention abilities of these compounds, these trends should be reflected in their IC50 values.
Increasing the ethanol concentration increases copper-mediated DNA damage prevention IC50 value of dmise from ∼240 to 312.8 ± 0.7 µM (Table 2). For iron-mediated DNA damage prevention, the dmise IC50 value increases from 3.68 ± 0.01 to 658 ± 2 μM, respectively, a 179-fold difference (Table 2). For ebis, IC50 values for copper-mediated DNA damage decrease somewhat with increasing ethanol concentration: 8.20 ± 0.02 and 13.09 ± 0.03 µM, respectively, for high- and low-ethanol conditions. Similar to dmise, the ebis IC50 value for iron-mediated DNA damage prevention increases more dramatically with increasing ethanol concentrations (IC50 values of 3.2 ± 0.9 and 140.5 ± 0.3 µM, respectively).
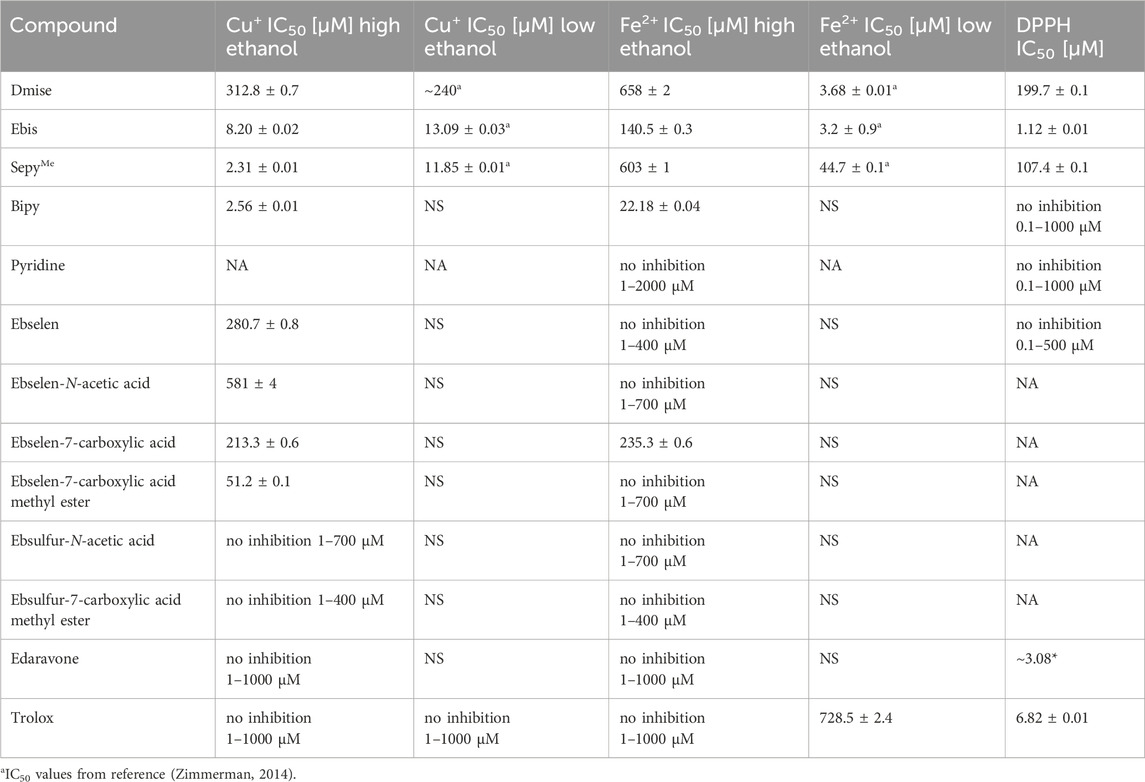
Table 2. IC50 values for metal-mediated DNA damage prevention and DPPH scavenging of selones and ebselen and ebsulfur derivatives (NS = IC50 value not obtained due to solubility issues, NA = IC50 value not tested).
The increase in IC50 values for iron-compared to copper-mediated DNA damage prevention is consistent with the expected trend of favored selenium-copper over selenium-iron coordination. In addition, IC50 values for the bidentate ebis compared to the monodentate dmise are 38- and 18-fold higher for prevention of copper-mediated DNA damage under high and low ethanol concentrations, respectively, and 5-fold higher for prevention of iron-mediated DNA damage under high ethanol concentrations.
The same denticity trends hold true for monodentate and bidentate nitrogen donor ligands: pyridine does not exhibit any iron-mediated DNA damage prevention (1 – 2000 µM), but bipy has IC50 values of 2.56 ± 0.01 and of 22.18 ± 0.04 µM for copper- and iron-mediated DNA damage prevention under high-ethanol conditions, respectively (no low-ethanol IC50 values could be obtained due to the poor water solubility of bipy). For the mixed-donor, bidentate sepyMe, IC50 values are 44.7 ± 0.1 and 603 ± 1 µM for iron-mediated DNA damage prevention under low- and high-ethanol conditions, respectively. The IC50 value for sepyMe is smaller than that determined for dmise, indicating that addition of the pyridine substituent increases iron-mediated DNA-damage prevention, consistent with metal-binding being a factor in these high-ethanol DNA damage assays. This differs from the copper-mediated DNA damage prevention for sepyMe, 11.85 ± 0.01 and 2.31 ± 0.01 for low and high ethanol, respectively, a 5-fold decease upon increasing the ethanol concentration.
In addition to well-established metal binding ligands, the well-known radical scavengers edaravone and Trolox (Figure 1A) were tested to establish the importance of radical scavenging in this assay. Edaravone, a substituted 2-pyrazolin-5-one, was approved by the Federal Drug Administration (FDA) in 2017 as the first drug for the treatment of amyotrophic lateral sclerosis (ALS) (Homma et al., 2019). The exact mechanism of action of edaravone in the treatment of ALS is unknown, but it is thought to be a radical scavenger in vivo, and edaravone is soluble in ethanol, but poorly soluble in water (Cruz, 2018). In contrast, Trolox (Figure 1A), is a fairly water-soluble vitamin E derivative that is also a well-studied radical scavenger (Alberto, Russo, Grand and Galano, 2013; Oliveira, Geraldo and Bento, 2014). It is commonly used as a standard against which antioxidant activity is compared in ROS scavenging assays, expressed as Trolox equivalents (TE) (Huang et al., 2005). Neither edaravone nor Trolox coordinate metals, so we compared results of our DNA gel electrophoresis assays under high-ethanol conditions to those of the DPPH radical scavenging assay to determine the effects of radical scavenging abilities in our assay.
Edaravone does not prevent copper- or iron-mediated DNA damage but scavenges DPPH with an IC50 value of 3.08 µM (Table 2), consistent with reported results (Tokumaru et al., 2018; Wang and Zhang, 2003). Trolox also does not prevent copper- or iron-mediated DNA damage under high-ethanol conditions, but it does have a very high IC50 value of 728 µM for iron-mediated DNA damage prevention under low-ethanol conditions. These results highlight a shortcoming with the use of Trolox as a gold standard for antioxidant assays: comparing every antioxidant to a compound that primarily scavenges radicals could result in the neglect of other potential antioxidant mechanisms such as metal chelation.
3.6 Examining DNA damage prevention abilities for hydrophobic ebselen derivatives
Hydrophobic antioxidants have significant potential, especially for neuropharmaceuticals, where biodistribution of drugs is limited by the blood-brain barrier (BBB) that prevents transit of >98% of small molecules. Water-soluble drugs can be structurally modified to become lipid-soluble drugs that can cross the BBB, but issues arise with in vitro screening since current models vary greatly in cost, technical demands, and intended applications (Pardridge, 2007).
The hydrophobic antioxidant drug ebselen is another example of how an assay to evaluate hydrophobic compounds for their abilities to prevent DNA damage could benefit development of more potent antioxidant drugs. Ebselen (Figure 1D) was developed in the early 1980s by Sies et al. (Piętka-Ottlik et al., 2008) as a glutathione peroxidase (GPx) mimic to prevent oxidative damage by hydrogen peroxide, and these GPx-mimic measurements were performed in organic solvents due to ebselen’s poor water solubility (Azad and Tomar, 2014; Zade, Panda, Tripathi, Singh and Wolmershäuser, 2004).
Ebselen also prevents oxidative stress in cultured cells as well as in Se-deficient mice, an effect independent of endogenous GPx expression (Steinbrenner and Brigelius-Floh, 2015). In the 1980s and 1990s, ebselen was examined in clinical trials for treatment of brain ischemia during stroke, and pproved in Japan for this purpose (Yamaguchi et al., 1998). In the U.S., ebselen has failed in clinical trials for treatment of asthma, atherosclerosis, cerebral infarction, myocardial ischemia, peptic ulcer, rheumatic disorder, and moderate and severe COVID-19 due to insufficient efficacy compared to placebo and concerns regarding its toxicity (Kil et al., 2017; Sies and Parnham, 2020). Ebselen is currently in Phase II clinical trials for hearing loss and tinnitus and in phase I/II trials for Meniere’s disease, tobramycin-induced ototoxicity, chemotherapy-induced hearing loss, and as a treatment for bipolar disorder (Azad and Tomar, 2014; Kil et al., 2017; Parnham, 1990).
Despite its setbacks in clinical trials, ebselen continues to be the standard for measuring small-molecule GPx-like activity, and it is a well-established ROS scavenger (Fujisawa and Kadoma, 2005; Maiorino, Roveri, Coassin and Ursini, 1988; Masumoto and Sies, 1996; Sies and Masumoto, 1997), although its biological mechanisms are not firmly established. One major issue preventing examination of the antioxidant activity of ebselen and ebselen derivatives is very limited water solubility (up to 13.6 μg/mL; 50 µM). Our high-ethanol DNA damage prevention assay is capable of testing DNA damage prevention for hydrophobic compounds such as ebselen with water solubilities of as little as 25 μM, depending on their antioxidant potencies.
Sies et al. (Roussyn et al., 1996) tested the ability of ebselen to inhibit peroxynitrite-induced DNA damage in a 1% methanol system (0.2 M). Since methanol scavenges radicals similarly to ethanol, the DNA-damage control lane exhibited only 25% damage in this study. Although it was reported that 50 µM ebselen inhibits 43% of DNA damage caused by peroxynitrite (100 µM) (Roussyn et al., 1996), this translates to prevention of damage to only 14% of the DNA. Due to this small difference in DNA damage inhibition, it is not clear that the observed inhibition is significantly different from the control. Thus, it was worthwhile to retest ebselen activity in our hydrophobic assay for DNA damage.
Under high-ethanol conditions, ebselen and H2O2 alone do not damage DNA (Figure 8A, lane 3), but Cu+ and H2O2 cause 86 ± 4% DNA damage, similar to damage observed for the same Cu+ and H2O2 concentrations under low-ethanol conditions (85 ± 2%; Figure 8A, lanes 5 and 4, respectively; ascorbate is added to reduce Cu2+ to Cu+ in situ). In the presence of Cu+ and H2O2, increasing ebselen concentrations up to 400 µM (Figure 8A, lanes 6–13) prevent copper-mediated DNA damage. At the same concentrations, ebselen prevents no iron-mediated DNA damage. Even under these more hydrophobic conditions, the upper concentration range for these DNA damage assays is limited by ebselen’s solubility in aqueous ethanolic solution.

Figure 8. Gel electrophoresis images showing ebselen prevention of (A) copper- and (B) iron-mediated DNA damage. MW: 1 kb molecular weight marker; lane 1: plasmid DNA (p); lane 2: p + H2O2 and (A) lane 3: p + 400 µM ebselen + H2O2 + 1.7 M ethanol; lane 4: p + Cu2+ (6 µM) + ascorbate (7.5 µM) + H2O2 + ethanol (10 mM); lane 5: p + Cu2+ (6 µM) + ascorbate (7.5 µM) + H2O2 + ethanol (1.7 M); lanes 6-12: Cu2+ (6 µM) + ascorbate (7.5 µM) + H2O2 + ethanol (1.7 M) + ebselen (1, 10, 50, 100, 200, 300, and 400 μM, respectively). (B) lane 3: p + 400 µM ebselen + H2O2 + 1.7 M ethanol; lane 4: p + Fe2+ (2 µM) + H2O2 + ethanol (10 mM); lane 5: p + Fe2+ (15 µM) + H2O2 + ethanol (1.7 M); lanes 6-12: Fe2+ (15 µM) + H2O2 + ethanol (1.7 M) + ebselen (1, 10, 50, 100, 200, 300, and 400 μM, respectively).
These ebselen gel data were fit with a dose-response curve (Figure 9), and the IC50 value for ebselen prevention of copper-mediated DNA damage was found to be 280.7 ± 0.8 μM. Due to ebselen’s low water solubility, our method is the first to determine an IC50 value for its ability to prevent copper-mediated DNA damage. Under these conditions, ebselen shows only limited ability to prevent metal-mediated DNA damage in this assay.
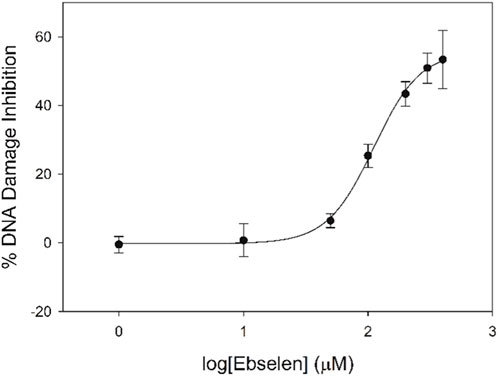
Figure 9. Dose-response curve for ebselen prevention of copper-mediated DNA damage under high-ethanol conditions.
The ability to test hydrophobic compounds such as ebselen for their ability to prevent metal-mediated DNA damage opens up a new area of antioxidant investigations, including development of ebselen derivatives that may act through metal binding. Using our high-ethanol DNA damage assay, we tested a series of five new ebselen derivatives (Gordhan et al., 2016) that had structural features that might enhance metal coordination (Figure 1D). Similar to ebselen, most of the tested ebselen derivatives more effectively prevent copper-mediated DNA damage than iron-mediated damage, with only ebselen-7-carboxylic acid able to prevent iron-mediated DNA damage (IC50 of 235.3 ± 0.6 µM; Table 2). Ebsulfur-N-acetic acid and ebsulfur-N-carboxylic acid methyl ester prevent neither copper- nor iron-mediated damage. As expected, the selenium-containing ebselen derivatives are more effective than their sulfur analogs, highlighting the importance of selenium for antioxidant activity of these ebselen derivatives.
Adding a carboxylate group to the nitrogen atom in the scaffold in ebselen-N-acetic acid decreases its ability to prevent copper-mediated DNA damage relative to ebselen, perhaps due to the acetate substituent creating a potentially competing, bidentate metal binding site with the keto group on the other side of the ring from Se. Addition of a carboxylate group near the Se, but without creating a potentially competing metal binding site, results in a lower IC50 value for copper-mediated DNA damage prevention for ebselen-7-carboxylic acid compared to ebselen (213.3 ± 0.6 and 280.7 ± 0.8 μM, respectively). The methyl ester of ebselen-7-carboxylic acid is by far the most effective at preventing copper-mediated DNA damage, with an IC50 value of 51.2 ± 0.1 μM, an over-four-fold increase in activity compared to ebselen-7-carboxylic acid.
Ebselen-7-carboxylic acid (Figure 1D) is also the only ebselen derivative that prevents iron-mediated DNA damage, likely due to the potentially bidentate carboxylate oxygen and selenium binding site. Blocking this carboxylate oxygen with a methyl group as in ebselen-7-carboxylic acid methyl ester prevents all activity, supporting this theory. Ebselen and its derivatives more effectively prevent copper-over iron-mediated DNA damage, a result that likely arises because of the soft selenium more strongly interacting with the soft Cu+ than the borderline Fe2+, although adding a hard oxygen donor near the selenium site with the potential for bidentate binding makes ebselen-7-carboxylic acid nearly equivalent in its ability to prevent copper- and iron-mediated DNA damage.
3.7 Implications of a DNA gel electrophoresis method for hydrophobic compounds
Our DNA damage prevention assay permits assessment of hydrophobic antioxidants, including edaravone, ebselen, and ebselen derivatives, that cannot otherwise be investigated using other DNA-based methods. Since we are examining DNA damage prevention directly, this assay avoids the need to extend hydrophobic radical scavenging results to the more complex system of DNA damage prevention. Typical DNA damage assays have been limited to examination of very water-soluble compounds, so for hydrophobic compounds, results of hydrophobic radical assays such as DPPH and ABTS have been correlated with biological outcomes such as DNA damage prevention without testing the compounds directly with DNA. We present the first gel electrophoresis assay that allows direct evaluation of copper- and iron-mediated DNA damage for hydrophobic compounds. Our electrophoresis and EPR results show that iron-mediated DNA damage is much more susceptible to radical scavenging by increased ethanol concentrations compared to copper-mediated DNA damage, illustrating the differences between the reactive species generated by these two metals.
Metal-antioxidant interactions play a major role in this hydrophobic gel electrophoresis assay, as highlighted by the trends observed for polyphenol prevention of iron-mediated DNA damage, since iron binding is an established antioxidant mechanism for these compounds. The importance of metal coordination was also demonstrated by examining the IC50 value trends of a group of nitrogen- and selenium-containing ligands for prevention of copper- and iron-mediated DNA damage. Edaravone and Trolox, radical scavengers that do not coordinate metals, show no activity in this assay. Although edaravone is used to treat ALS that has been associated with elevated copper concentrations, our results suggest that edaravone’s mechanism of action likely does not involve copper coordination.
This assay allowed DNA damage prevention testing for ebselen and ebselen derivatives, compounds with limited water solubility. Ebselen prevents only copper-mediated DNA damage, but addition of a carboxylate group to form a potential metal chelating site allows ebselen-7-carboxylic acid to prevent both copper- and iron-mediated DNA damage. This more hydrophobic DNA damage prevention assay is a significant step forward that will allow screening of hydrophobic compounds for their metal-binding antioxidant activity, an antioxidant mechanism that is not examined by common radical-scavenging methods. By testing a variety of potential antioxidants using this hydrophobic DNA damage assay, we demonstrated that it can be used to evaluate and compare new classes of compounds, give insight into mechanisms for antioxidant behavior, and aid in the development of hydrophobic antioxidants and drugs that cross the blood-brain barrier for treatment of Alzheimer’s, Parkinson’s, and other diseases caused by oxidative stress.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
AG: Writing – original draft, Investigation, Formal Analysis, Writing – review and editing, Methodology. NP: Writing – review and editing, Formal Analysis, Investigation. HA: Writing – review and editing, Investigation, Formal Analysis. DW: Resources, Writing – review and editing, Supervision. JB: Formal Analysis, Methodology, Project administration, Conceptualization, Supervision, Writing – review and editing, Resources, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors declare that financial support was received for the research and/or publication of this article through National Science Foundation grants CHE 1213912, CHE 1807709, and CHE 2203847.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchbi.2025.1677610/full#supplementary-material
References
Alberto, M. E., Russo, N., Grand, A., and Galano, A. (2013). A physicochemical examination of the free radical scavenging activity of trolox: mechanism, kinetics and influence of the environment. Phys. Chem. Chem. Phys. 15 (13), 4642–4650. doi:10.1039/C3CP43319F
Alfadda, A. A., and Sallam, R. M. (2012). Reactive oxygen species in health and disease. J. Biomed. Biotechnol. 2012, 1–14. doi:10.1155/2012/936486
Angelé-Martínez, C., Goodman, C., and Brumaghim, J. (2014). Metal-mediated DNA damage and cell death: mechanisms, detection methods, and cellular consequences. Metallomics 6 (8), 1358–1381. doi:10.1039/c4mt00057a
Aruoma O, H. B., Gajewski, E., and Dizdaroglu, M. (1991). Copper-ion-dependent damage to the bases in DNA in the presence of hydrogen peroxide. Biochem. J. 273 (3), 601–604. doi:10.1042/bj2730601
Azad, G. K., and Tomar, R. S. (2014). Ebselen, a promising antioxidant drug: mechanisms of action and targets of biological pathways. Mol. Biol. Rep. 41 (8), 4865–4879. doi:10.1007/s11033-014-3417-x
Battin, E. E., Zimmerman, M. T., Ramoutar, R. R., Quarles, C. E., and Brumaghim, J. L. (2011). Preventing metal-mediated oxidative DNA damage with selenium compounds. Metallomics 3 (5), 503–512. doi:10.1039/C0MT00063A
Benzie, I. F. F., and Devaki, M. (2018). “The ferric reducing/antioxidant power (FRAP) assay for non-enzymatic antioxidant capacity: concepts, procedures, limitations and applications,” in Measurement of antioxidant activity and capacity. Recent trends and applications. Editors R. Apak, E. Çapanoğlu, and F. Shahidi (Hoboken, NJ, USA: Wiley), 77–106.
Bourzac, K. M., LaVine, L. J., and Rice, M. S. (2003). Analysis of DAPI and SYBR green I as alternatives to ethidium bromide for nucleic acid staining in agarose gel electrophoresis. J. Chem. Educ. 80, 1292–1296. doi:10.1021/ed080p1292
Briviba, K., Roussyn, I., Sharov, V. S., and Sies, H. (1996). Attenuation of oxidation and nitration reactions of peroxynitrite by selenomethionine, selenocystine and ebselen. Biochem. J. 319 (1), 13–15. doi:10.1042/bj3190013
Burkitt, M. J., and Mason, R. P. (1991). Direct evidence for in vivo hydroxyl-radical generation in experimental iron overload: an ESR spin-trapping investigation. Proc. Natl. Acad. Sci. U. S. A. 88 (19), 8440–8444. doi:10.1073/pnas.88.19.8440
Cao, G., Alessio, H. M., and Cutler, R. G. (1993). Oxygen-radical absorbance capacity assay for antioxidants. Free Radic. Biol. Med. 14 (3), 303–311. doi:10.1016/0891-5849(93)90027-R
Cao, G., Verdon, C. P., Wu, A. H., Wang, H., and Prior, R. L. (1995). Automated assay of oxygen radical absorbance capacity with the COBAS FARA II. Clin. Chem. 41 (12), 1738–1744. doi:10.1093/clinchem/41.12.1738
Collin, F. (2019). Chemical basis of reactive oxygen species reactivity and involvement in neurodegenerative diseases. Int. J. Mol. Sci. 20, 2407. doi:10.3390/ijms20102407
Cruz, M. P. (2018). Edaravone (radicava): a novel neuroprotective agent for the treatment of amyotrophic lateral sclerosis. P T 43 (1), 25–28.
Dixon, S. J., and Stockwell, B. R. (2014). The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 10 (1), 9–17. doi:10.1038/nchembio.1416
Dragan, A. I., Pavlovic, R., McGivney, J. B., Casas-Finet, J. R., Bishop, E. S., Strouse, R. J., et al. (2012). SYBR green I: fluorescence properties and interaction with DNA. J. Fluoresc. 22 (4), 1189–1199. doi:10.1007/s10895-012-1059-8
Duthie, G. G. (1999). Determination of activity of antioxidants in human subjects. Proc. Nutr. Soc. 58 (4), 1015–1024. doi:10.1017/s0029665199001330
Fabian, I. (1989). Hydrolytic reactions of copper(II) bipyridine complexes. Inorg. Chem. 28 (20), 3805–3807. doi:10.1021/ic00319a011
Floyd, R. A., and Lewis, C. A. (1983). Hydroxyl free radical formation from hydrogen peroxide by ferrous iron-nucleotide complexes. Biochemistry 22 (11), 2645–2649. doi:10.1021/bi00280a008
Frankel, E. N., and Meyer, A. S. (2000). The problems of using one-dimensional methods to evaluate multifunctional food and biological antioxidants. J. Sci. Food Agric. 80 (13), 1925–1941. doi:10.1002/1097-0010(200010)80:13<1925::AID-JSFA714>3.0.CO;2-4
Fridovich, I. (1989). Superoxide dismutases. J. Biol. Chem. 264 (14), 7761–7764. doi:10.1016/S0021-9258(18)83102-7
Fujisawa, S., and Kadoma, Y. (2005). Kinetic studies of the radical-scavenging activity of ebselen, a seleno-organic compound. Anticancer Res. 25 (6B), 3989–3994.
Gordhan, H. M., Patrick, S. L., Swasy, M. I., Hackler, A. L., Anayee, M., Golden, J. E., et al. (2017). Evaluation of substituted ebselen derivatives as potential trypanocidal agents. Bioorg. and Med. Chem. Lett. 27 (3), 537–541. doi:10.1016/j.bmcl.2016.12.021
Halliwell, B. (2000). Why and how should we measure oxidative DNA damage in nutritional studies? How far have we come? Am. J. Clin. Nutr. 72 (5), 1082–1087. doi:10.1093/ajcn/72.5.1082
Halliwell, B., and Aruoma, O. I. (1991). DNA damage by oxygen-derived species its mechanism and measurement in mammalian systems. FEBS Lett. 281 (1-2), 9–19. doi:10.1016/0014-5793(91)80347-6
Halliwell, B., and Gutteridge, J. M. C. (1992). Biologically relevant metal ion-dependent hydroxyl radical generation an update. FEBS Lett. 307 (1), 108–112. doi:10.1016/0014-5793(92)80911-y
Henle, E. S., Han, Z., Tang, N., Rai, P., Luo, Y., and Linn, S. (1999). Sequence-specific DNA cleavage by Fe2+-mediated Fenton reactions has possible biological implications. J. Biol. Chem. 274 (2), 962–971. doi:10.1074/jbc.274.2.962
Hertzberg, R. P., and Dervan, P. B. (1982). Cleavage of double helical DNA by methidium-propyl-EDTA-iron(II). J. Am. Chem. Soc. 104 (1), 313–315. doi:10.1021/ja00365a069
Homma, T. S. K., Kobayashi, S., Sato, H., and Fujii, J. (2019). Edaravone, a free radical scavenger, protects against ferroptotic cell death in vitro. Exp. Cell Res. 384 (1), 111592. doi:10.1016/j.yexcr.2019.111592
Huang, D., Ou, B., and Prior, R. L. (2005). The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 53 (6), 1841–1856. doi:10.1021/jf030723c
Imlay, J. A., and Linn, S. (1988). DNA damage and oxygen radical toxicity. Science 240 (4857), 1302–1309. doi:10.1126/science.3287616
Jhurry, N. D., Chakrabarti, M., McCormick, S. P., Holmes-Hampton, G. P., and Lindahl, P. A. (2012). Biophysical investigation of the ironome of human Jurkat cells and mitochondria. Biochemistry 51 (26), 5276–5284. doi:10.1021/bi300382d
Kil, J., Lobarinas, E., Spankovich, C., Griffiths, S. K., Antonelli, P. J., Lynch, E. D., et al. (2017). Safety and efficacy of ebselen for the prevention of noise-induced hearing loss: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 390 (10098), 969–979. doi:10.1016/S0140-6736(17)31791-9
Kimani, M. M., Watts, D., Graham, L. A., Rabinovich, D., Yap, G. P. A., and Brumaghim, J. L. (2015). Dinuclear copper(I) complexes with N-heterocyclic thione and selone ligands: synthesis, characterization, and electrochemical studies. Dalton Trans. 44 (37), 16313–16324. doi:10.1039/C5DT02232K
Linder, M. C. (2012). The relationship of copper to DNA damage and damage prevention in humans. Mutat. Research/Fundamental Mol. Mech. Mutagen. 733 (1-2), 83–91. doi:10.1016/j.mrfmmm.2012.03.010
Linn, S. (2015). Radicals in Berkeley? J. Biol. Chem. 290 (14), 8748–8757. doi:10.1074/jbc.X115.644989
Lloyd, R. S., Haidle, C. W., and Robberson, D. L. (1978). Bleomycin-specific fragmentation of double-stranded DNA. Biochemistry 17 (10), 1890–1896. doi:10.1021/bi00603a014
López-Alarcón, C., and Denicola, A. (2013). Evaluating the antioxidant capacity of natural products: a review on chemical and cellular-based assays. Anal. Chim. Acta 763, 1–10. doi:10.1016/j.aca.2012.11.051
Maiorino, M., Roveri, A., Coassin, M., and Ursini, F. (1988). Kinetic mechanism and substrate specificity of glutathione peroxidase activity of ebselen (PZ51). Biochem. Pharmacol. 37 (11), 2267–2271. doi:10.1016/0006-2952(88)90591-6
Masumoto, H., and Sies, H. (1996). The reaction of ebselen with peroxynitrite. Chem. Res. Toxicol. 9 (1), 262–267. doi:10.1021/tx950115u
Nakano, S.-I., and Sugimoto, N. (2016). The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents. Biophys. Rev. 8, 11–23. doi:10.1007/s12551-015-0188-0
Nakao, L. S., Fonseca, E., and Augusto, O. (2002). Detection of C8-(1-hydroxyethyl)guanine in liver RNA and DNA from control and ethanol-treated rats. Chem. Res. Toxicol. 15 (10), 1248–1253. doi:10.1021/tx0255166
Oliveira, R., Geraldo, D., and Bento, F. (2014). Radical scavenging activity of antioxidants evaluated by means of electrogenerated HO radical. Talanta 129, 320–327. doi:10.1016/j.talanta.2014.05.047
Özyürek, M., Güçlü, K., Tütem, E., Başkan, K. S., Erçağ, E., Esin Çelik, S., et al. (2011). A comprehensive review of CUPRAC methodology. Anal. Methods 3, 2439. doi:10.1039/C1AY05320E
Pardridge, W. M. (2007). Blood-brain barrier delivery. Drug Discov. Today 12 (1-2), 54–61. doi:10.1016/j.drudis.2006.10.013
Park, S., and Imlay, J. A. (2003). High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 185 (6), 1942–1950. doi:10.1128/JB.185.6.1942-1950.2003
Parnham, M. J. (1990). Biological activities and clinical potential of ebselen. Adv. Exp. Med. Biol. 264, 193–197. doi:10.1007/978-1-4684-5730-8_31
Pérez-Jiménez, J., and Saura-Calixto, F. (2006). Effect of solvent and certain food constituents on different antioxidant capacity assays. Food Res. Int. 39 (7), 791–800. doi:10.1016/j.foodres.2006.02.003
D. A. Perkowski, and M. Perkowski (2007). Data and probability connections (New Jersey, USA: Pearson Prentice Hall), 318–337.
Perron, N. R. (2008). Effects of polyphenol compounds on Iron- and copper-mediated DNA damage mechanism and predictive models. Clemson University. Available online at: https://open.clemson.edu/all_dissertations/256/.
Perron, N. R., and Brumaghim, J. L. (2009). A review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell biochem. Biophys. 53 (2), 75–100. doi:10.1007/s12013-009-9043-x
Perron, N. R., Hodges, J. N., Jenkins, M., and Brumaghim, J. L. (2008). Predicting how polyphenol antioxidants prevent DNA damage by binding to iron. Inorg. Chem. 47 (14), 6153–6161. doi:10.1021/ic7022727
Perron, N. R., Wang, H. C., Deguire, S. N., Jenkins, M., Lawson, M., and Brumaghim, J. L. (2010). Kinetics of iron oxidation upon polyphenol binding. Dalton Trans. 39, 9982–9987. doi:10.1039/C0DT00752H
Perron, N. R., García, C. R., Pinzón, J. R., Chaur, M. N., and Brumaghim, J. L. (2011). Antioxidant and prooxidant effects of polyphenol compounds on copper-mediated DNA damage. J. Inorg. Biochem. 105 (5), 745–753. doi:10.1016/j.jinorgbio.2011.02.009
Piętka-Ottlik, M., Wójtowicz-Młochowska, H., Kołodziejczyk, K., Piasecki, E., and Młochowski, J. (2008). New organoselenium compounds active against pathogenic bacteria, fungi and viruses. Chem. Pharm. Bull. 56 (10), 1423–1427. doi:10.1248/cpb.56.1423
Roussyn, I., Briviba, K., Masumoto, H., and Sies, H. (1996). Selenium-containing compounds protect DNA from single-strand breaks caused by peroxynitrite. Archives Biochem. Biophysics 330 (1), 216–218. doi:10.1006/abbi.1996.0245
Schaich, K. M., Tian, X., and Xie, J. (2015). Reprint of hurdles and pitfalls in measuring antioxidant efficacy: a critical evaluation of ABTS, DPPH, and ORAC assays. J. Funct. Foods 18 (B), 782–796. doi:10.1016/j.jff.2015.05.024
Sies, H., and Masumoto, H. (1997). Ebselen as a glutathione peroxidase mimic and as a scavenger of peroxynitrite. Adv. Pharmacol. 38, 229–246. doi:10.1016/s1054-3589(08)60986-2
Sies, H., and Parnham, M. J. (2020). Potential therapeutic use of ebselen for COVID-19 and other respiratory viral infections. Free Radic. Biol. Med. 156, 107–112. doi:10.1016/j.freeradbiomed.2020.06.032
Singh, S., and Singh, R. P. (2008). In vitro methods of assay of antioxidants: an overview. Food Rev. Int. 24 (4), 392–415. doi:10.1080/87559120802304269
Stadelman, B. S., Kimani, M. M., Bayse, C. A., McMillen, C. D., and Brumaghim, J. L. (2016). Synthesis, characterization, DFT calculations, and electrochemical comparison of novel iron(II) complexes with thione and selone ligands. Dalton Trans. 45 (11), 4697–4711. doi:10.1039/c5dt03384e
Steinbrenner, H., and Brigelius-Floh, R. (2015). Das essenzielle Spurenelement Selen. Selenbedarf in Gesundheit und Krankheit. Aktuel Ernahrungsmed 40 (6), 368–378. doi:10.1055/s-0035-1552774
Stoewe, R., and Prütz, W. A. (1987). Copper-catalyzed DNA damage by ascorbate and hydrogen peroxide: Kinetics and yield. Free Radic. Biol. Med. 3 (2), 97–105. doi:10.1016/s0891-5849(87)80003-5
Taniguchi, H., and Madden, K. P. (2000). DMPO-alkyl radical spin trapping: an in situ radiolysis steady-state ESR study. Radiat. Res. 153 (4), 447–453. doi:10.1667/0033-7587(2000)153[0447:darsta]2.0.co;2
Tokumaru, O., Shuto, Y., Ogata, K., Kamibayashi, M., Bacal, K., Takei, H., et al. (2018). Dose-dependency of multiple free radical-scavenging activity of edaravone. J. Surg. Res. 228, 147–153. doi:10.1016/j.jss.2018.03.020
Wang, L.-F., and Zhang, H.-Y. (2003). A theoretical investigation on DPPH radical-scavenging mechanism of edaravone. Bioorg. and Med. Chem. Lett. 13 (21), 3789–3792. doi:10.1016/j.bmcl.2003.07.016
Woodmansee, A. N., and Imlay, J. A. (2002). Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods Enzym. 349, 3–9. doi:10.1016/S0076-6879(02)49316-0
Yamaguchi, T., Sano, K., Takakura, K., Saito, I., Shinohara, Y., Asano, T., et al. (1998). Ebselen in acute ischemic stroke: a placebo-controlled, double-blind clinical trial. Ebselen study group. Stroke 29 (1), 12–17. doi:10.1161/01.str.29.1.12
Yamamoto, K., and Kawanishi, S. (1989). Hydroxyl free radical is not the main active species in site-specific DNA damage induced by copper (II) ion and hydrogen peroxide. J. Biol. Chem. 264 (26), 15435–15440. doi:10.1016/S0021-9258(19)84847-0
Yang, S., and Lian, G. (2020). ROS and diseases: role in metabolism and energy supply. Mol. Cell. Biochem. 467, 1–12. doi:10.1007/s11010-019-03667-9
Zade, S. S., Panda, S., Tripathi, S. K., Singh, H. B., and Wolmershäuser, G. (2004). Convenient synthesis, characterization and GPx-Like catalytic activity of novel ebselen derivatives. Eur. J. Org. Chem. 2004 (18), 3857–3864. doi:10.1002/ejoc.200400326
Zimmerman, M. T. (2014). Determining DNA damage prevention mechanisms for multifunctional selenium and sulfur antioxidants and the DNA-damaging capabilities of clotrimazole and pseudophedrine-derived metal complexes. Clemson University. Available online at: https://open.clemson.edu/all_dissertations/1759/.
Keywords: ebselen, ebsulfur, metal-mediated DNA damage, edaravone, trolox, selones
Citation: Gaertner AAE, Perron NR, Anderholm HMG, Whitehead DC and Brumaghim JL (2025) Quantifying antioxidant activity of hydrophobic compounds using metal-mediated DNA damage. Front. Chem. Biol. 4:1677610. doi: 10.3389/fchbi.2025.1677610
Received: 01 August 2025; Accepted: 06 October 2025;
Published: 17 October 2025.
Edited by:
Debbie C. Crans, Colorado State University, United StatesReviewed by:
Aviva Levina, The University of Sydney, AustraliaChristina Banti, University of Ioannina, Greece
Copyright © 2025 Gaertner, Perron, Anderholm, Whitehead and Brumaghim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia L. Brumaghim, YnJ1bWFnaEBjbGVtc29uLmVkdQ==
 Andrea A. E. Gaertner
Andrea A. E. Gaertner Heeren M. G. Anderholm
Heeren M. G. Anderholm Julia L. Brumaghim
Julia L. Brumaghim