- Department of Plant Pathology, Dr. Yashwant Singh Parmar University of Horticulture and Forestry, Solan, Himachal Pradesh, India
Apple is most important fruit crop in Himachal Pradesh, contributing substantially to the state’s economy. However, soilborne diseases have emerged as a major concern affecting nursery-raised apples. Trichoderma species produce chitinase, an enzyme that degrades chitin, a major component of the fungal cell wall. This study aimed to optimize the growth parameters for chitinase production, extraction, purification, and characterization and to assess the antifungal potential against soilborne pathogens of apple. A total of 14 isolates of Trichoderma spp. produced chitinases in a colloidal chitin agar (CCA) medium to varying extents. The optimal incubation period, pH, substrate concentration, and incubation temperature were 7 days, 5, 1%, and 30°C, respectively, while the thermal and pH stability ranged from 30°C to 50°C and from 4 to 6, respectively. Chitinases were purified from Trichoderma atroviride UHFTA005 and UHFTA006 and from Trichoderma virens UHFTV017 with a molecular mass of 40 kDa. The chitinase from T. atroviride UHFTA005 at 0.60 μl inhibited the in vitro growth of Dematophora necatrix (92.22%) and Sclerotium rolfsii (91.11%). In a further in vivo evaluation of the chitinases, T. atroviride UHFTA005 was found to be more effective against white root rot and seedling blight of apple, with disease control of 86.67% and 73.33%, respectively, and with 86.67% white root rot disease control in nursery field conditions suggesting its strong potential as a biocontrol agent in nursery field conditions.
1 Introduction
Apple (Malus × domestica) is a deciduous tree in the Rosaceae family. It is one of the world’s most well-known fruits, with an annual production of more than 80 million tonnes (http://faostat.fao.org). It is also one of the most profitable fruit types in terms of foreign exchange. Long-term continuous cropping can disrupt the microbial community, reducing the number of beneficial microbes while increasing the number of soilborne diseases (Chadfield et al., 2022). Apple production is under threat due to global diseases, particularly those produced by soilborne pathogens due to their persistence in the soil and the limited effectiveness of chemical treatments. Climate change hampers management by shifting the soil temperature and moisture levels, promoting pathogen survival and infection.
Rosellinia necatrix (Pliego et al., 2012), Phytopythium cactorum (Zhou et al., 2022), Sclerotium rolfsii (Corazza et al., 1999), Armillaria mellea (Marsh, 1952), Helicobasidium mompa (Lee et al., 2022), and Macrophomina phaseolina, among others, cause major economic losses in agriculture production (Shafique et al., 2016; Surendra et al., 2025). Preventive techniques against soilborne pathogens are needed. The detection of soilborne pathogens is difficult at the early stage; as a result, once infected, plants could not survive (Singh et al., 2019). Traditionally, agrochemicals have been used for their management, but continuous and long-term use has led to soil and water pollution and microbial imbalance, which could result in the emergence of new pathogenic variants (Zhou et al., 2024). The overuse of fungicides has resulted in resistance to some diseases (Jamar et al., 2010).
A more economical and sustainable option to reduce crop losses is “biological control,” in which beneficial microbes are used for disease control (Velivelli et al., 2014). Trichoderma spp., Bacillus spp., Pseudomonas spp., and Streptomyces spp. are among the most promising biological control agents for plant disease management (Mukhopadhyay and Kumar, 2020). Apple rhizosphere microorganisms with biocontrol ability can be used against soilborne pathogens while maintaining microbial balance (Velivelli et al., 2014; Zang et al., 2021). Trichoderma is widespread, grows quickly, and is easily identified by its enormous number of green conidia. Trichoderma, a greenish filamentous fungus, was first reported by Persoon (1794). The morphological characteristics of Trichoderma include the presence of hyphae, conidiophores, and phialides (Tulasne and Tulasne, 1865). It is classified under the order Hypocreales, the family Hypocreaceae, and the genus Trichoderma. This genus includes saprophytic fungi that are widespread, mycoparasitic, and are beneficial to improving the soil microbial diversity (Mukherjee et al., 2013; Ribeiro dos Santos and Lima dos Santos, 2025).
Trichoderma exhibits different mechanisms of action, e.g., competition, hyperparasitism, direct antagonism, secondary metabolite production, and cell wall lysis through the production of different enzymes, including chitinase, glucanase, proteases, and cellulases (Wonglom et al., 2020). Trichoderma exhibits mycoparasitism on many pathogens, such as Pythium, Phytophthora, Sclerotium, and Dematophora, among others (Shaw et al., 2016). Mycoparasitism is caused by Trichoderma-produced secondary metabolites and cell wall disintegrating enzymes (CWDEs). Trichoderma species coil around the hyphae of the target fungal pathogens and enter the cell wall by producing CWDEs (Gajera et al., 2012). CWDEs target the pathogen’s cell wall components (β-1,3-glucan and chitin) and utilize them as nutrients, promoting Trichoderma growth (Viterbo et al., 2002).
Trichoderma species, in addition to controlling plant diseases, improve plant heath, increase plant growth, provide better nutrient utilization, and increase plant resistance. Among all the enzymes, chitinase is of upmost importance as most of the fungal plant pathogenic species inherit chitin in their cell wall as an imperative constituent (Baek et al., 1999; Ulhoa and Peberdy, 1991). Furthermore, Trichoderma species produce enzymes that target certain plant diseases (Bastakoti et al., 2017) and have the ability to hydrolyze proteins, cellulose, hemicellulose, and chitin, which directly limit plant diseases (Sharma et al., 2019).
Chitin is a polymer of N-acetylglucosamine (NAG), which is the second most abundant polymer after cellulose and is considered as a major component of the fungal cell wall (Haran et al., 1996). The cell wall is mainly composed of Chytridiomycota, Ascomycota, Basidiomycota, and the exoskeleton of insects (Flach et al., 1992). It strengthens the cell wall of fungi and the exoskeleton of insects (Elieh-Ali-Komi and Hamblin, 2016). Chitinase hydrolyzes the β-1,4-glycosidic bond between the NAG residues of chitin (Kitamura and Kamei, 2003). The molecular weights of chitinases range from 20 to 90 kDa. The Trichoderma spp. chitinases belong to family 18 of glycosyl hydrolase and are further classified into class III and class V. There are other microorganisms that show chitinolytic activity, including Bacillus spp., Serratia spp., and Streptomyces spp (Dahiya et al., 2006). As chitin is not easily soluble in water, it is chemically altered to create colloidal chitin, which can more easily form a homogeneous distribution on agar medium due to its small particle size (Lingappa and Lockwood, 1962). Therefore, it is one of the effective methods for the prevention and control of soilborne pathogens that can effectively resist chemical pesticides.
In this study, Trichoderma spp. were isolated from the rhizospheric region of apple for the extraction of chitinase. The present study also focused on the optimization of the growth parameters for chitinase production, extraction, purification, and characterization and on the assessment of the antifungal potential against soilborne pathogens of apple.
2 Materials and methods
2.1 Study area
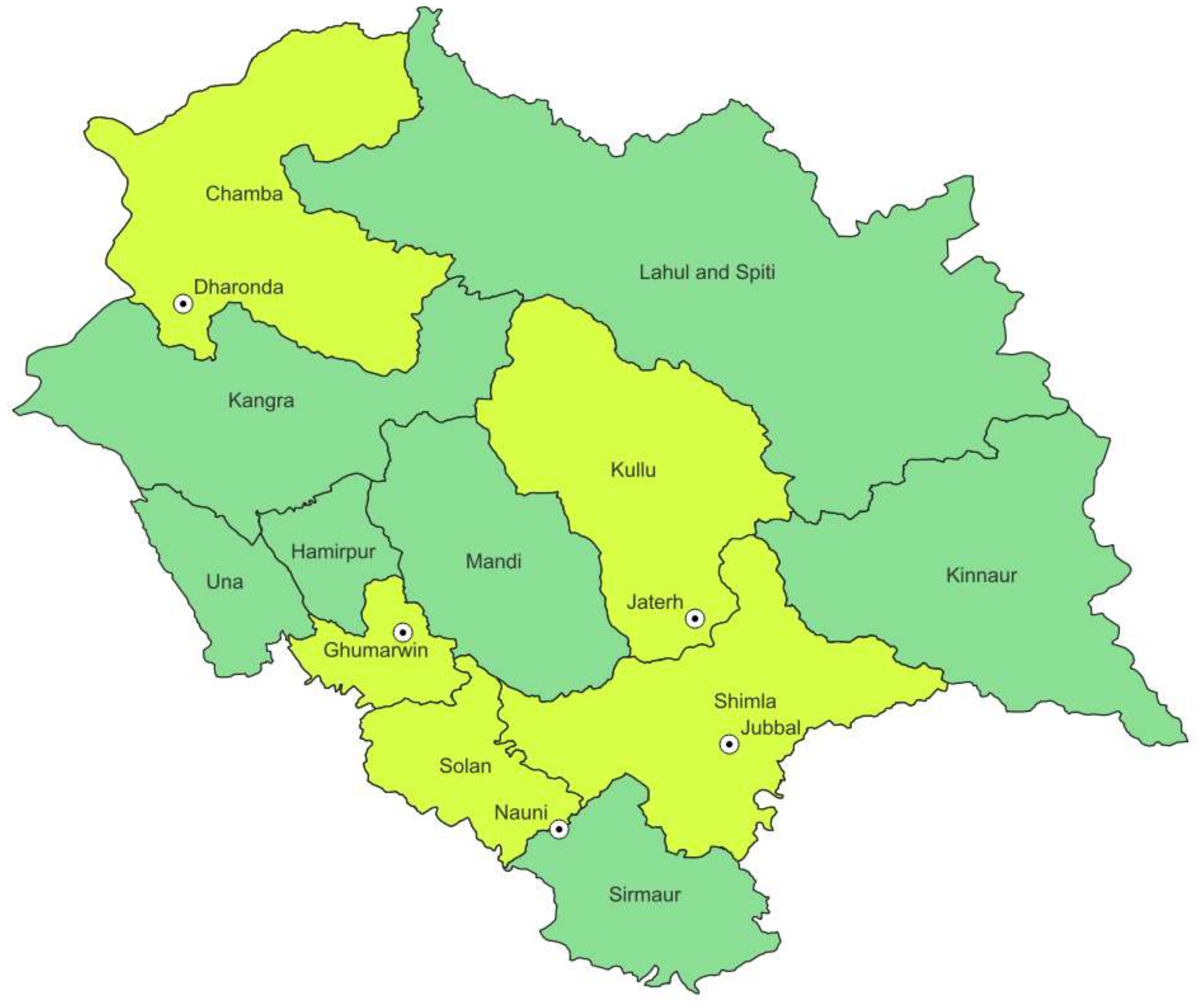
A survey was conducted across various prominent apple-growing areas in Himachal Pradesh, in particular Bilaspur, Mandi, Kullu, Solan, and Shimla (Figure 1). At each selected site, four sub-sites were selected in orchards/nurseries. Soil samples were collected from the rhizospheric area of apple at 10- to 15-cm soil depth into polythene bags (Singh et al., 2003).
2.2 Trichoderma
2.2.1 Isolation of resident Trichoderma species
For isolation, the serial dilution technique was followed at 105 dilution for each sample. Of each sample, 100 μl was pipetted out into a plate with potato dextrose agar (PDA) medium and incubated at 30°C for 7 days. Isolate UHFTB002 was procured from the Department of Plant Pathology, Dr Yashwant Singh Parmar University of Horticulture and Forestry Nauni, Solan, Himachal Pradesh.
2.2.2 Isolation of soilborne pathogens of apple
Dematophora necatrix was isolated from the roots of the affected seedlings/plants following standard procedure, while S. rolfsii was procured from the Department of Plant Pathology, Dr. Yashwant Singh Parmar University of Horticulture and Forestry Nauni, Solan, Himachal Pradesh.
2.3 Genomic DNA extraction and sequencing
Genomic DNA was extracted using the CTAB method (Sambrook and Fritsch, Maniatis, 1989). PCR amplification was carried out in a thermal cycler. ITS1 (5′ TCC GTA GGT GAA CCT TGC GG 3′) and ITS4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) primers were used during PCR amplification. The PCR conditions were as follows: 95°C for 5 min (initial denaturation), 95°C for 1 min (denaturation), 60°C for 1 min (annealing), 72°C for 1 min (extension), and 72°C for 10 min (final extension) with 35 cycles. The PCR product was electrophoresed at 1% agarose gel for 1 h at 100 V and visualized under Gel Doc. The 600-bp PCR product was sent to Eurofins Genomics India Pvt. Ltd., Bengaluru, Karnataka. Sequenced data were analyzed with BLAST (Basic Local Alignment Search Tool; www.ncbi.nlm.nih.gov).
2.4 Phylogenetic analysis
For phylogenetic analysis, the downloaded sequences of Trichoderma spp. were aligned using ClustalW with the neighborhood joining method (Thompson et al., 1994) and were edited using BioEdit. Alignment was trimmed using MEGA 11 software (Tamura et al., 2013).
2.5 Medium and culture conditions
CCA medium (in grams per liter): MgSo4·7H20, 0.3; (NH4)2·SO4, 3.0; KH2PO4, 2.0; citric acid, 1.0; agar powder, 15; Tween-80, 200 μl; colloidal chitin, 4.5; and bromocresol purple, 0.15. pH was set to 4.7.
PDA medium (in grams per liter): potato extract, 4; dextrose, 20; and agar, 20. The final pH was adjusted to 5.6. This was used for preservation of the fungal cultures.
2.6 Preparation of colloidal chitin
Colloidal chitin was prepared from chitin powder. Of the chitin powder, 10 g was treated with 40–50 ml (37%) of HCl. In a 1,000-ml beaker, HCl was slowly added into the chitin powder, with continuous stirring until becoming a slurry. The chitin–HCl mixture was then kept in a refrigerator (4°C) for 1 h incubation. The mixture was treated with 50% (pre-chilled) ethanol and kept in the refrigerator overnight to facilitate better precipitation of colloidal chitin. This mixture was filtered through Whatman no. 1 filter and washed with 1–2 L of pre-chilled distilled water. The pH was set between 6 and 7 by adding NaOH/HCl and was kept in shade after filtration for drying. The ground powder was kept at 4°C for further use.
2.7 Preparation of colloidal chitin agar medium for chitinase-producing isolates
The modified protocol of Gomez Ramirez et al. (2004) was used to prepare the CCA mixture, which was then autoclaved at 121°C for 20 min. The sterilized medium was then poured into Petri plates. After solidification of the medium, different isolates of Trichoderma were kept at the center of the plate to examine the chitinase-producing isolates with three replications each and incubated at 30°C for 3 days.
2.8 Optimization of the growth parameters on chitinase activity and stability
The effects of different growth parameters, such as the incubation period, pH, substrate concentration, and incubation temperature, on chitinase activity were examined. The effect of pH was assessed between 3 and 8 at 1-unit intervals with NaOH/HCl. The effects of five temperature values ranging from 20°C to 40°C with a variation of 5°C and five concentrations of colloidal chitin, i.e., 0.5%, 1%, 1.5%, 2%, and 2.5%, were evaluated for 9 days. The enzyme activity was measured (Miller, 1959).
2.9 Enzyme activity assay
The enzyme activity assay was performed according to the method of Miller (1959). The reaction mixture [1 ml crude enzyme solution + 1 ml colloidal chitin (1%) + 1 ml of 0.1 M citrate buffer (pH 6.0)] was incubated in a water bath at 37°C for 30 min. The reaction was stopped by adding 2 ml of a dinitrosalicylic acid (DNS) reagent and incubated in a water bath for 5 min at 60°C for color development. The reaction mixture without crude enzyme was used as blank. The optical density (OD) value was measured at 540 nm using the standard curve of N-acetyl glucosamine unit (100–600 ppm). One unit (U) of chitinase activity was defined as the amount of enzyme that released micromoles of NAG per minute per milliliter, while specific activity (SA) represents the micromoles of NAG released per minute per milligram of protein.
2.10 Protein estimation using the Bradford method
The concentration of protein was estimated according to Bradford (1976) using Coomassie Brilliant Blue G-250. The OD value was measured at 595 nm using the standard curve of bovine serum albumin (BSA).
2.11 Enzyme extraction
Colloidal chitin broth medium with a pH of 5 was prepared in 500-ml flasks, and four to five pieces of 5 mm size were inoculated in each flask for the different isolates of Trichoderma and were incubated at 30°C for 7 days. After 7 days, the flasks were kept on a rotary shaker for 1 day and the broth centrifuged at 10,000 rpm for 20 min. The supernatant was collected and then stored in a refrigerator for purification. The supernatant was used as a crude enzyme.
2.12 Partial purification
Purification of the crude enzyme was performed with the ammonium sulfate precipitation method (Xu et al., 2014). Ammonium sulfate precipitation was conducted using 0%–45% concentration of ammonium sulfate under 4°C. The precipitates were collected by centrifugation at 10,000 rpm for 20 min and the supernatant was discarded. The pellet was dissolved in citrate buffer (pH 6). Ammonium sulfate traces were removed through dialysis. Sephadex G-100 beads were dissolved in distilled water (10 g of Sephadex G-100 was added into 500 ml of distilled water), which was then poured over the top of the silica and washed with a large amount of citrate buffer for the smooth downward flow of protein. The dialyzed fraction was suspended in a Sephadex G-100 chromatographic column, and the fraction showing maximum chitinase activity was pooled (Laemmli, 1970).
2.13 Sodium dodecyl sulfate polyacrylamide gel electrophoresis
The molecular weight of the purified protein was determined using SDS-PAGE. The purified fraction was submitted into 12% SDS-PAGE (Laemmli, 1970), and 40 μg of the sample was loaded and electrophoresis carried out with 150 V of current for approximately 1–2 h. The gel was carefully removed and then placed into a staining solution (Coomassie Brilliant Blue G-250) (Candiano et al., 2004). Thereafter, the staining solution was poured off and the gel was kept in a destaining solution for 3–4 h with continuous shaking for band visualization.
2.14 Statistical analysis
Statistical analysis was performed with R software (R Development Core Team, 2008). Experiments were conducted using a completely randomized design (CRD) in triplicates and analyzed using one-way ANOVA. Qualitative analysis of chitinase was performed using Duncan’s multiple range test (DMRT). A p-value <0.05 was considered statistically significant.
3 Results and discussion
3.1 Phylogenetic analysis of isolates of Trichoderma species
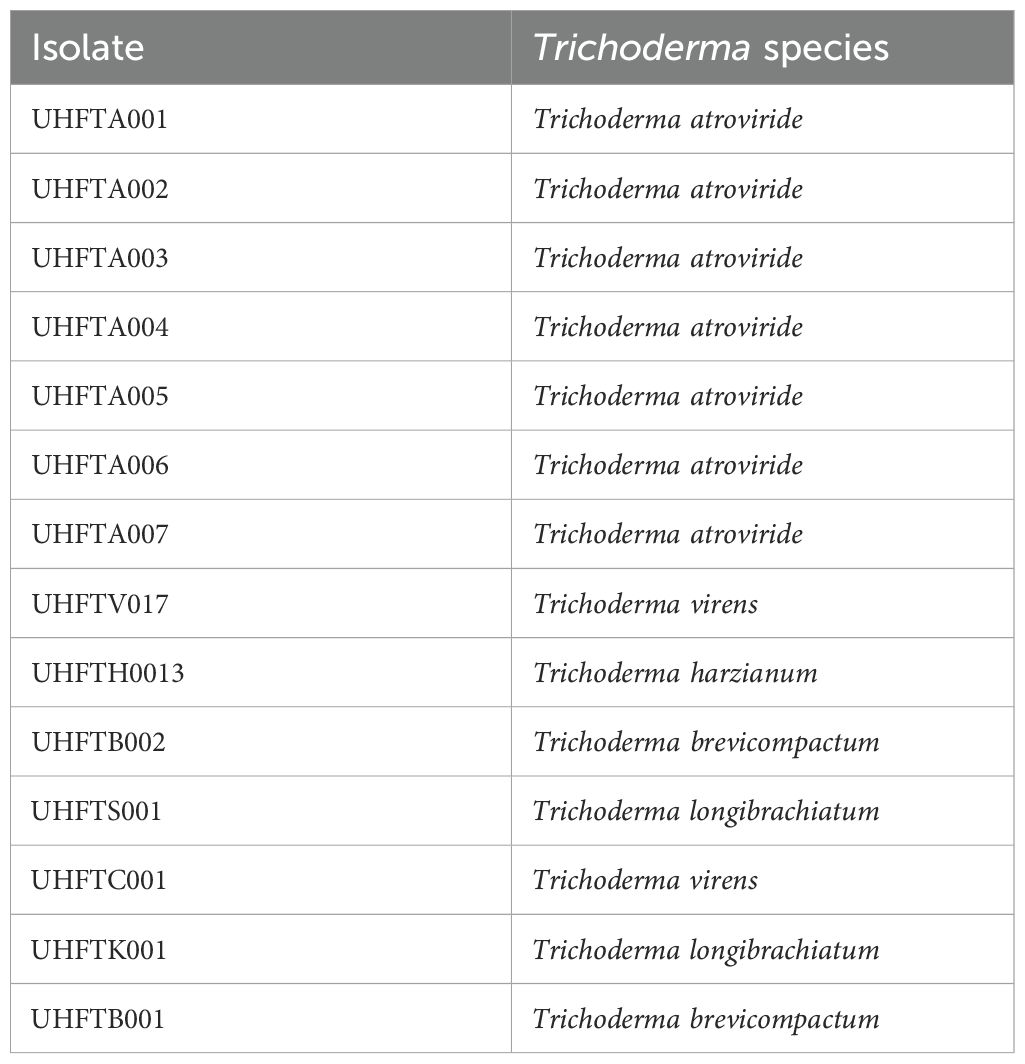
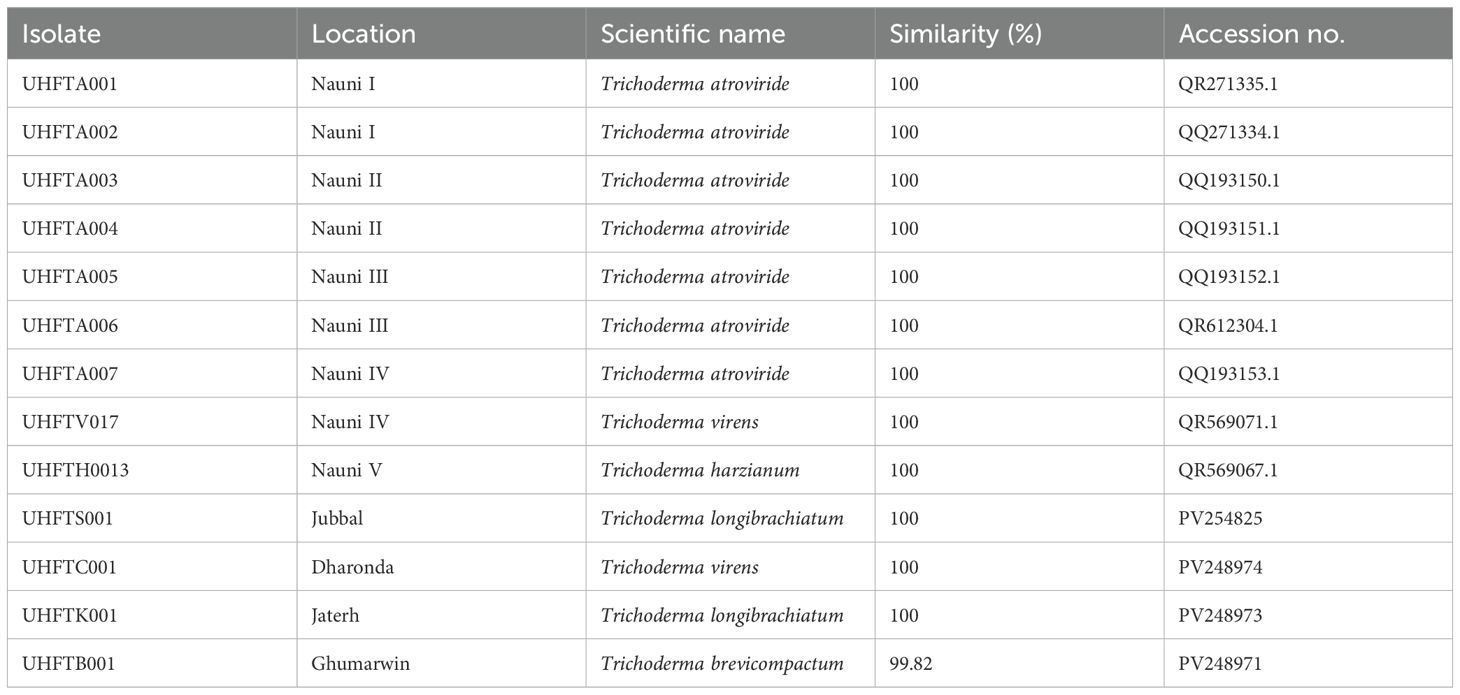
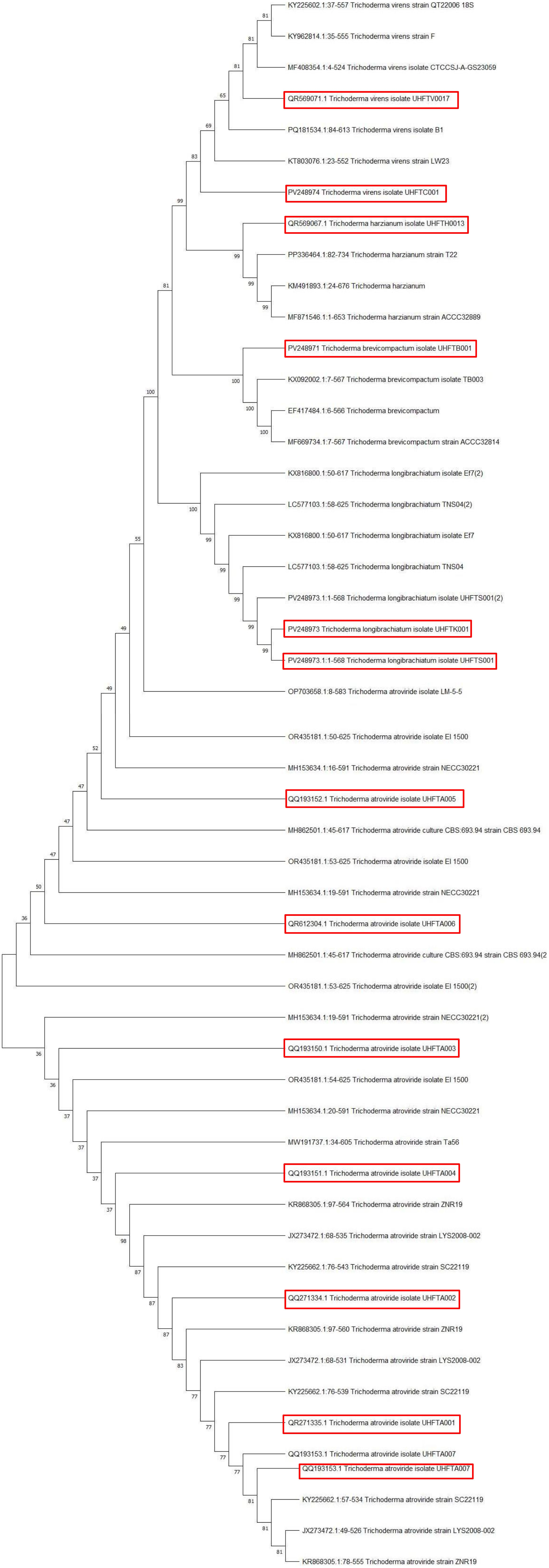
Different isolates of Trichoderma species were isolated and used for phylogenetic analysis (Table 1). The phylogenetic tree showed that Trichoderma virens UHFTV017 (QR569071.1) shared close affinity with the T. virens isolate CTCCSJ-A-CS23059 with 81% bootstrap support, T. virens UHFTC001 (PV248974) shared close affinity with the T. virens isolate LW23 with 69% bootstrap support, Trichoderma harzianum UHFTH0013 (QR569067.1) shared close affinity with the T. harzianum isolate T22 with 99% bootstrap support, Trichoderma brevicompactum UHFTB001 (PV248971) shared close affinity with the T. brevicompactum isolate TB003 with 100% bootstrap support, Trichoderma longibrachiatum UHFTS001 (PV254825) and T. longibrachiatum UHFTK001 (PV248973) shared close affinity with the T. longibrachiatum isolate Ef72 with 100% bootstrap support, Trichoderma atroviride UHFTA005 (QQ193152.1) shared close affinity with the T. atroviride isolate LM-5-5 with 100% bootstrap support, T. atroviride UHFTA006 (QR612304.1) shared close affinity with the T. atroviride isolate CBS693.94 with 52% bootstrap support, T. atroviride UHFTA003 (QQ193150.1) shared close affinity with the T. atroviride isolate CBS693.94 with 50% bootstrap support and the T. atroviride isolate CBS693.94 with 52% bootstrap support, T. atroviride UHFTA004 (QQ193151.1) shared close affinity with the T. atroviride isolate Ta56 with 37% bootstrap support, T. atroviride UHFTA002 (QQ271334.1) shared close affinity with the T. atroviride isolate ZNR19 with 83% bootstrap support, T. atroviride UHFTA001 (QR271335.1) shared close affinity with the T. atroviride isolate ZNR19 with 98% bootstrap support, and T. atroviride UHFTA007 (QQ193153.1) shared close affinity with the T. atroviride isolate ZNR19 with 81% bootstrap support (Table 2; Figures 2, 3).
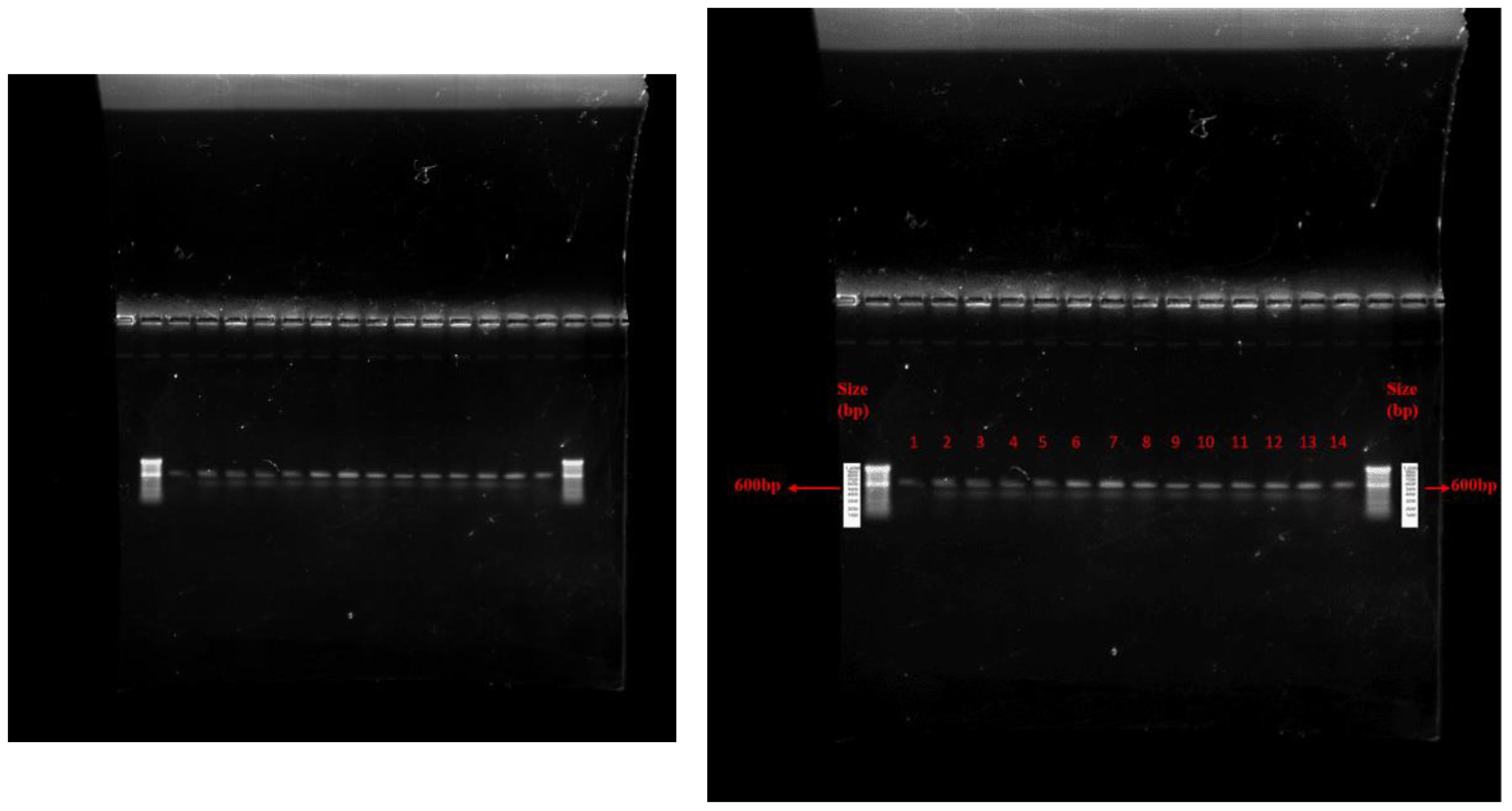
Figure 2. Agarose gel electrophoresis of PCR amplicon from fungal DNA. Where, 1= Trichoderma atroviride UHFTA001, 2= Trichoderma atroviride UHFTA002, 3= Trichoderma atroviride UHFTA003, 4= Trichoderma atroviride UHFTA004, 5= Trichoderma atroviride UHFTA005, 6= Trichoderma atroviride UHFTA006, 7= Trichoderma atroviride UHFTA007, 8= Trichoderma virens UHFTV017, 9= Trichoderma harzianum UHFTH0013, 10= Trichoderma longibrachiatum UHFTS001, 11= Trichoderma virens UHFTC001, 12= Trichoderma longibrachiatum UHFTK001, 13= Trichoderma brevicompactum UHFTB001, 14= Dematophora necatrix.
Garcia-Nunez et al. (2017) identified 10 isolates of Trichoderma with ITS-1 and ITS-4 of 600 bp amplicon size. Hermosa et al. (2000) identified several biocontrol isolates of Trichoderma of 500–600 bp amplicon size using ITS-1 and ITS-4 primers.
3.2 Qualitative screening of chitinase-producing strains
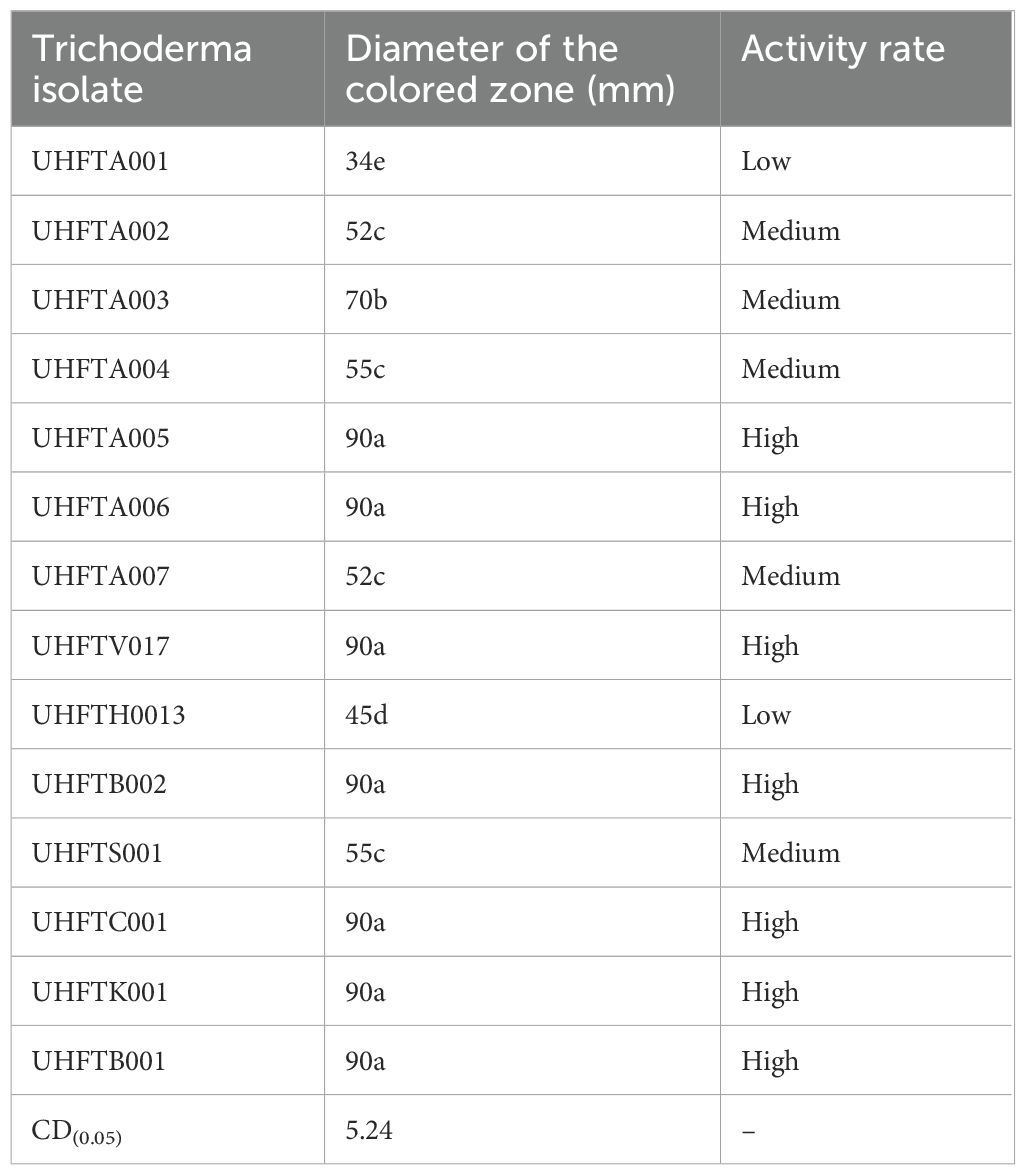
The results of the qualitative screening for chitinase production showed that all isolates of Trichoderma spp. showed chitinolytic activity at varying extents. UHFTA005, UHFTA006, UHFTV017, UHFTB002, UHFTC001, UHFTK001, and UHFTB001 showed maximum chitinase activity; isolates UHFTA002, UHFTA003, UHFTA004, and UHFTA007 showed medium chitinase activity; and isolates UHFTA001 and UHFTH0013 showed minimum chitinase activity (Table 3; Figure 4).
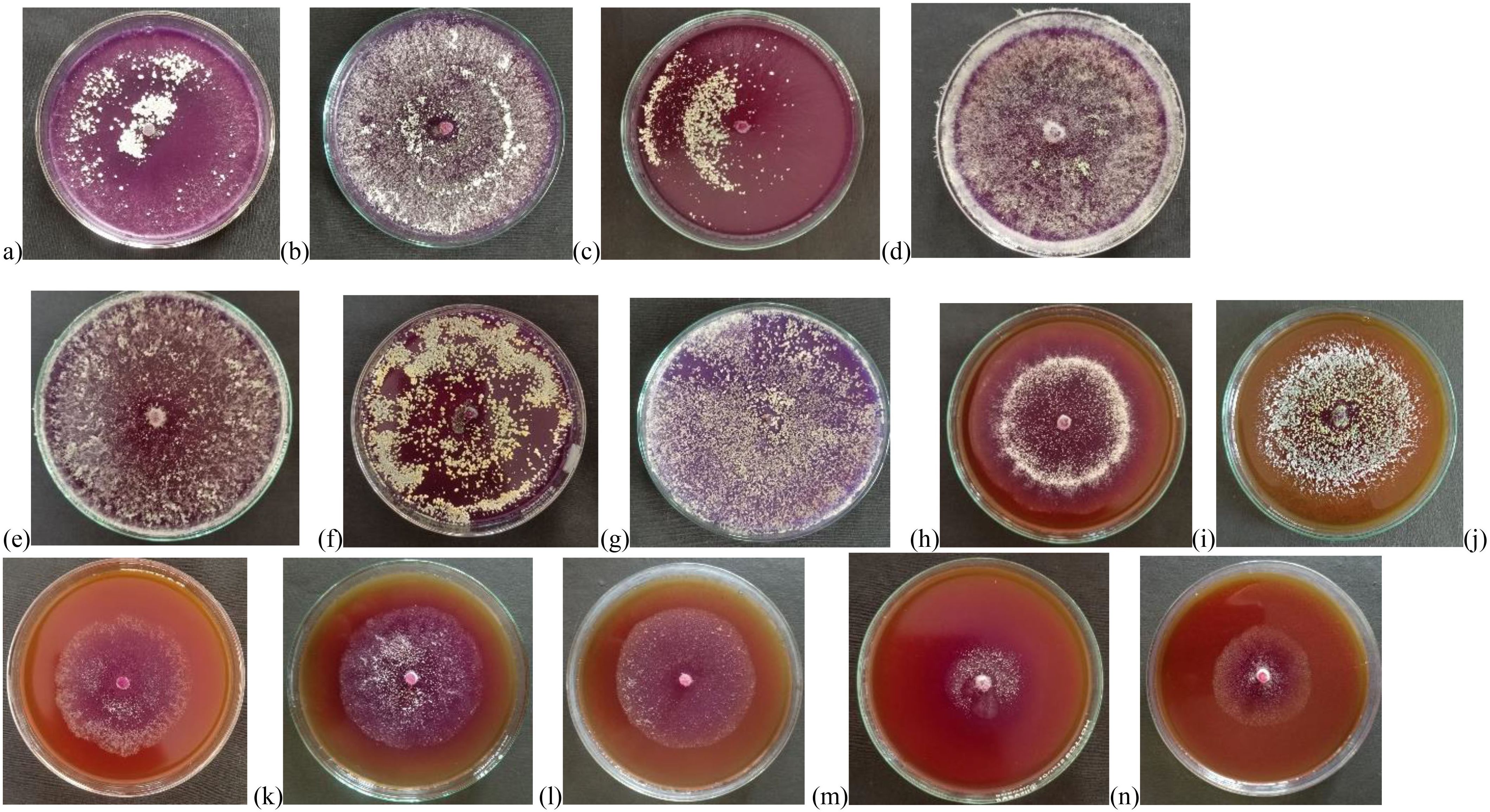
Figure 4. Qualitative analysis of chitinases producing isolates of Trichoderma spp. Where, (a) UHFTA005, (b) UHFTA006, (c) UHFTV017, (d) UHFTB002, (e) UHFTB001, (f) UHFTC001, (g) UHFTK001 (High chitinolytic activity), (h) UHFTA002, (i) UHFTA003, (j) UHFTA004, (k) UHFTA007, (l) UHFTS001 (Medium chitinolytic activity), (m) UHFTA001, (n) UHFTH0013 (Low chitinolytic activity).
Trichoderma species produce the hydrolytic enzyme chitinase that is key to the hydrolysis of chitin, which is present in the cell wall of many phytopathogenic fungi. In the present study, the production of a hydrolytic zone around the colony indicated the breakdown of chitin by chitinases. The bromocresol purple dye resulted in a change in the pH toward alkalinity by changing the yellow color of the dye to purple. Different isolates of Trichoderma species showed significant differences in zone size, which is due to the varying capacities of the isolates to produce chitinase for the hydrolysis of chitin. Agrawal and Kotasthane (2012) reported that, out of 61 isolates of Trichoderma spp., 17 showed highest, 8 medium, and 12 low chitinase activity. Gonzalez et al. (2023) observed that T. harzianum, the Trichoderma asperellum isolate 12.2, and the T. asperellum isolate BP60 produced chitinase in a CCA medium by changing the color from orange to purple.
High activity (H), 76–100; moderate activity (M), 51–75; low activity (L), 25–50. Means in each column followed by the same lowercase letter(s) did not differ at CD(0.05) according to Duncan’s multiple range test.
CD(0.05), critical difference at 0.05 significance
High chitinolytic activity
Medium chitinolytic activity
Low chitinolytic activity
3.3 Optimization of the incubation period for chitinase production
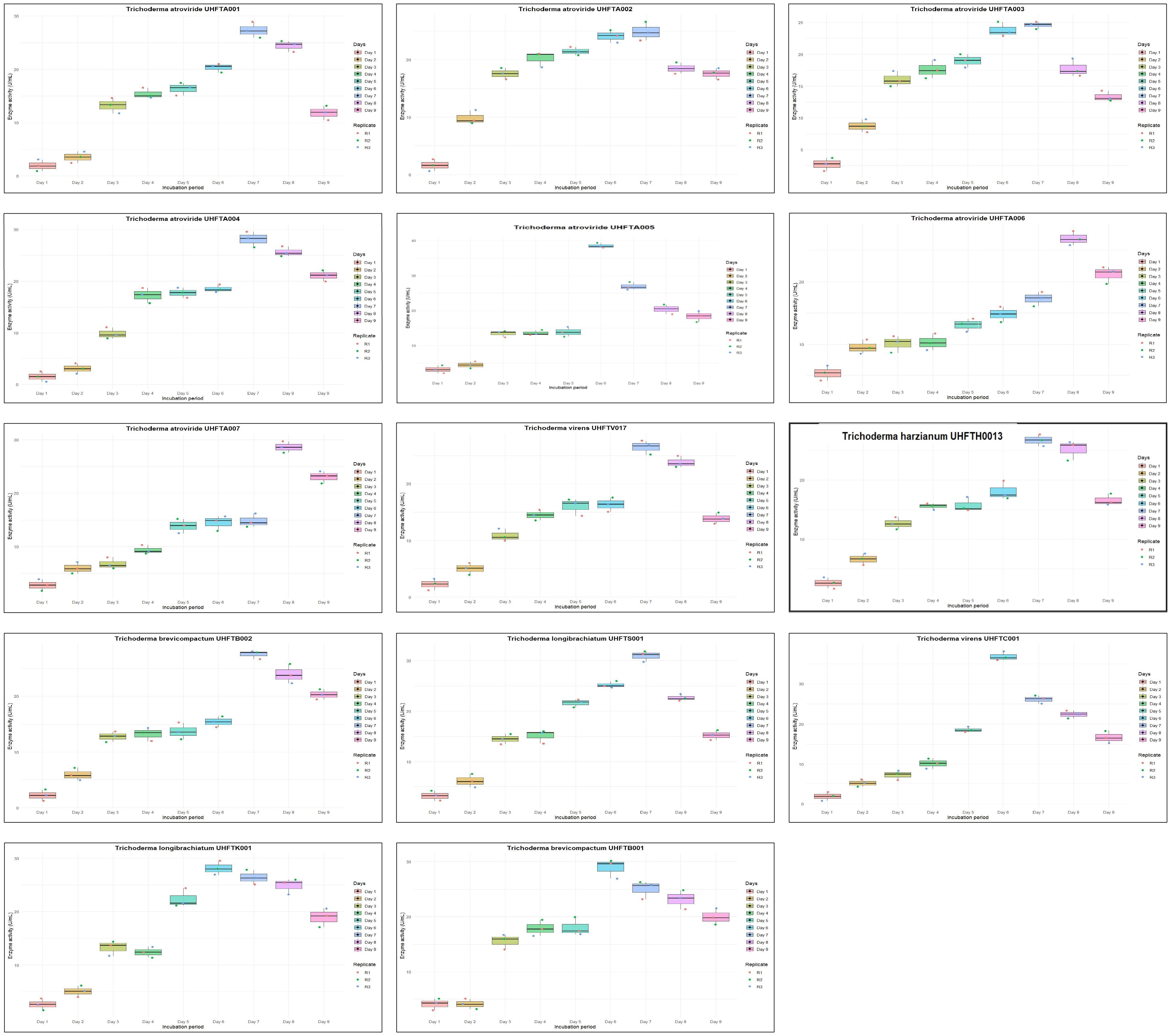
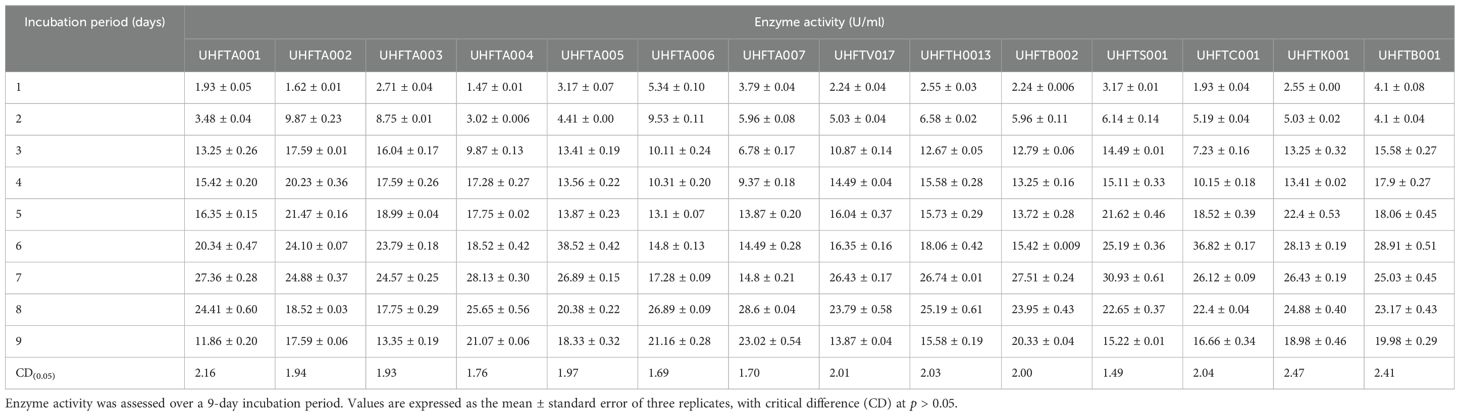
Different isolates of Trichoderma species were evaluated for chitinase production under in vitro conditions, and chitinase activity was monitored regularly for 9 days at 30°C. Table 4 illustrates the relationship between enzyme activity and incubation period, which revealed that the highest chitinase activity of the T. atroviride isolates UHFTA001, UHFTA002, UHFTA003, and UHFTA004; T. harzianum UHFTH0013; T. virens UHFTV017; T. brevicompactum UHFTB002; and T. longibrachiatum UHFTS001 peaked on day 7 of incubation, with values of 27.36, 24.88, 24.57, 28.13, 26.74, 26.43, 27.51, and 30.93 U/ml, respectively. However, in the case of T. atroviride UHFTA005, T. virens UHFTC001, T. longibrachiatum UHFTK001, and T. brevicompactum UHFTB001, the highest chitinase activity was achieved on day 6 of incubation, with activity values of 38.52, 36.82, 28.13, and 28.91 U/ml, respectively. In most of the isolates of Trichoderma species, the chitinase activity started increasing from the second day of incubation and continued its lag phase from the second day onward until day 7, where the chitinase activity reached its peak. With the increase in incubation period, the chitinase activity also increased, but declined beyond day 7 of incubation (Figure 5).
Optimization of the incubation period is a crucial factor for increasing the production of chitinase as it directly influences the secretion of enzymes and their metabolic activity. By optimizing the depletion of nutrients, instability of the enzymes and metabolic changes occur. Below the optimal incubation period, the activity decreases due to the slow binding of the substrate to the active site of the enzyme. At optimal incubation, the activity reaches its peak due to the presence of sufficient nutrients and the more efficient substrate binding to the enzyme’s active site. Other researchers have also optimized the incubation period for chitinase production across different microorganisms. El-Katatny et al. (2000) observed the maximum production of chitinase on day 7 of incubation in T. harzianum, while Gueye et al. (2020) observed a progressive increase of the chitinase activity of T. asperellum between 3 and 5 days, with the highest activity observed on day 5 of incubation with an enzyme activity of 2.81 U/ml. Brzezinska and Jankiewicz (2012) reported maximum chitinase production of the Aspergillus niger isolate LOCK 62 on day 6 of incubation. These findings provide evidence for the role of the incubation period in maximizing the enzymatic activity, with many microorganisms showing a peak in activity around days 6 and 7 of incubation.
3.4 Optimization of the substrate concentration for chitinase production
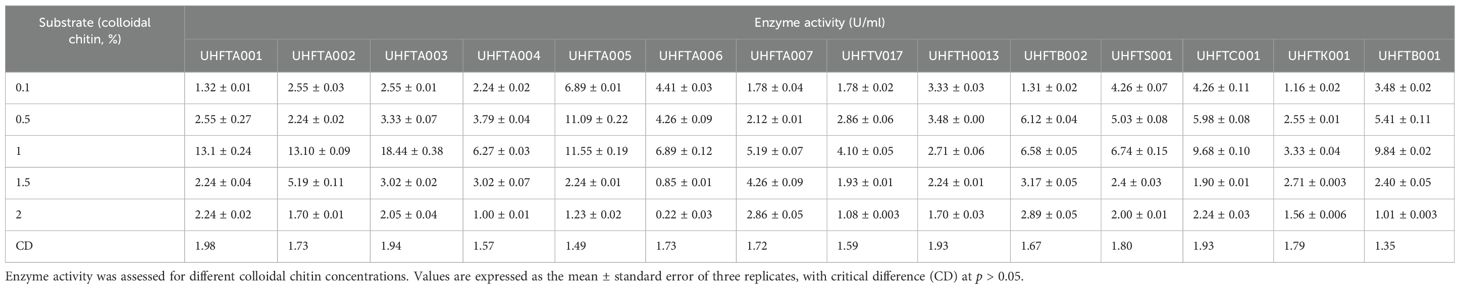
The effect of the concentration of colloidal chitin (0.1%–2%) on enzyme production was examined to elucidate the best substrate concentration under controlled conditions of 30°C temperature and pH 5 over a period of 7 days. Table 5 reveals that the highest chitinase activity in UHFTA001 (13.10 U/ml), UHFTA002 (13.10 U/ml), UHFTA003 (18.44 U/ml), UHFTA004 (6.27 U/ml), UHFTA005 (11.55 U/ml), UHFTA006 (6.89 U/ml), UHFTA007 (5.19 U/ml), UHFTV017 (4.10 U/ml), UHFTB002 (6.58 U/ml), UHFTS001 (6.74 U/ml), UHFTC001 (9.68 U/ml), UHFTK001 (3.33 U/ml), and UHFTB001 (9.84 U/ml) was observed at 1% concentration of colloidal chitin, indicating optimal chitinase production at this concentration, except for UHFTH0013 (3.48 U/ml) that had the highest activity at 0.5% concentration of colloidal chitin (Figure 6).
Initially, more active sites of the enzymes are available for binding. Therefore, with the increase in substrate concentration, the reaction rate also increases due to more substrates binding to the active sites of the enzymes, leading to the release of more products. However, at a certain point, a further increase in the substrate concentration does not increase the reaction rate due to the saturation of all active sites of the enzymes, which prevents new substrates from binding, and due to the loss of enzyme activity. Optimal concentration of the substrate is a crucial factor for maximizing the production of chitinase and for increasing its biological efficacy against the chitin-exhibited phytopathogens. El-Katatny et al. (2000) concluded that the chitinase production of T. harzianum increased with an increase in the concentration of chitin up to 1%. These results are also in accordance with those of Elad et al. (1982), who concluded that the chitinase activity of T. harzianum increased up to a concentration of 1%.
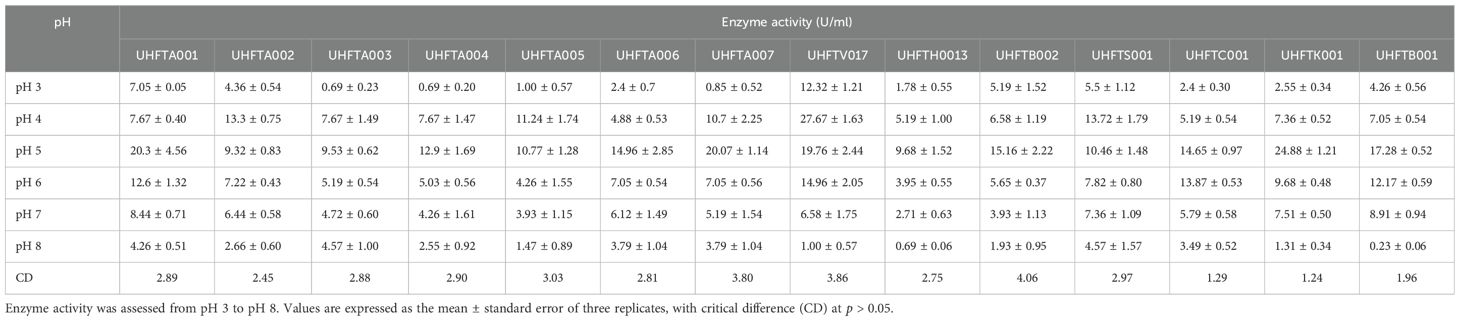
3.5 Optimization of the pH for chitinase production
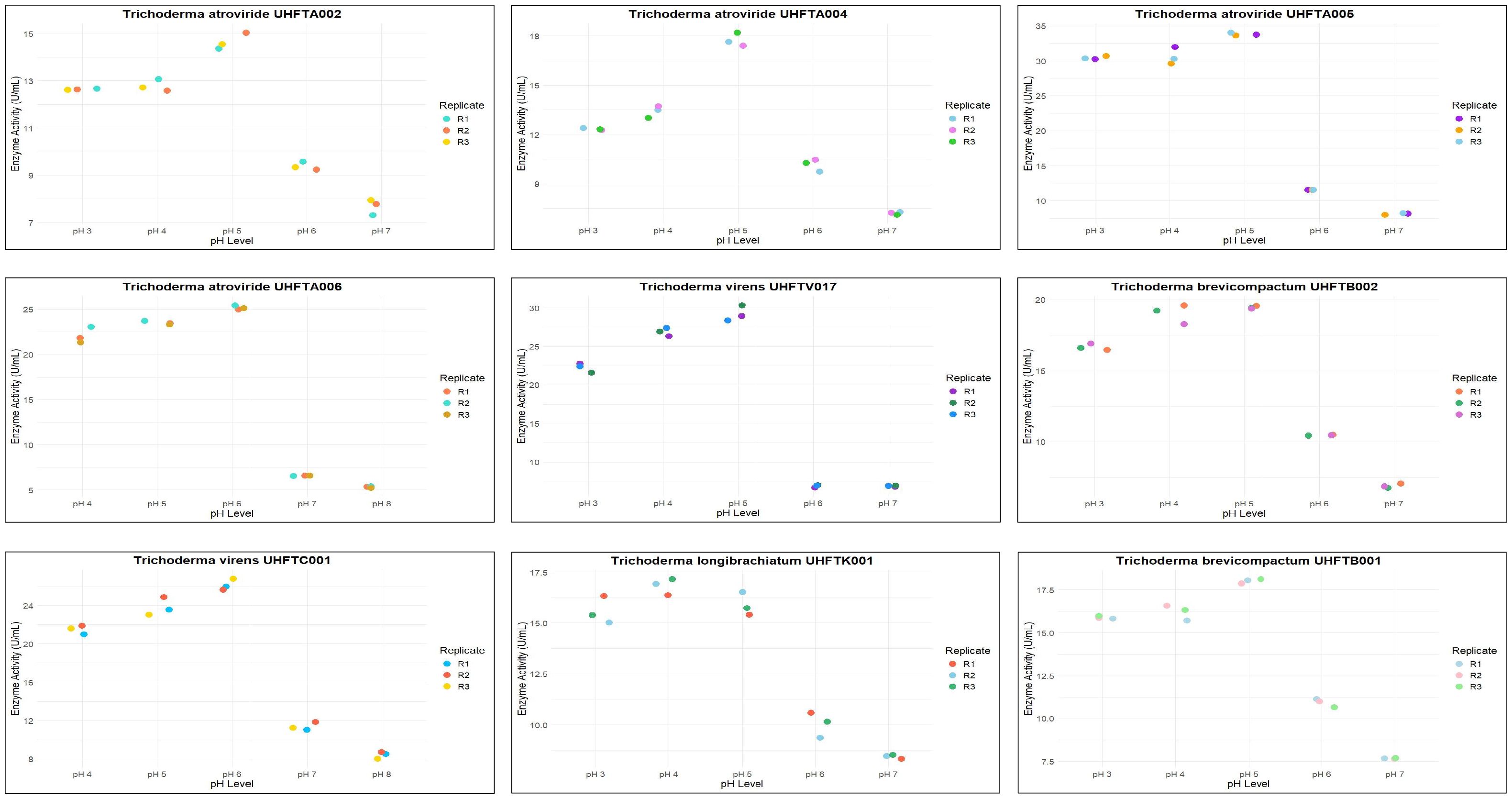
All isolates of Trichoderma species were examined for the optimization of the pH to maximize chitinase production. The enzyme activity was estimated between pH 3 and pH 8 using colloidal chitin broth medium at 30°C, incubated for 7 days. As depicted in Table 6, there was a clear relationship between the pH of the medium and the activity of chitinase. The maximum production of chitinase of UHFTA001, UHFTA003, UHFTA004, UHFTA006, UHFTA007, UHFTH0013, UHFTB002, UHFTC001, UHFTK001, and UHFTB001 was recorded at pH 5, with enzymatic activity of 20.38, 9.53, 12.94, 14.96, 20.07, 9.68, 15.16, 14.65, 24.88, and 17.28 U/ml, respectively. On the other hand, the maximum production of chitinase of UHFTA002, UHFTA005, UHFTV017, and UHFTS001 was recorded at pH 4, with enzymatic activity of 13.35, 11.24, 27.67, and 13.72 U/ml, respectively (Figure 7).
Chitinase production was at its peak in the acidic medium, which declined under an alkaline environment, clearly suggesting the importance of the pH of the medium for maximum production of enzyme. During the exponential phase, the production of chitinase was the highest, enabling a more efficient breakdown of chitin. However, beyond this phase, the production entered a death phase, which was due to the exhaustion of all active sites of the enzyme. The denaturation of chitinase beyond the optimal pH reduced the enzyme’s efficiency. Therefore, maintaining an optimum pH is critical for its effective application in the management of targeted pathogens. Ekundayo et al. (2016) reported an optimum pH of 5 for the maximum chitinolytic activity of Trichoderma viride, while Sharma et al. (2021) reported the maximum production of chitinase by T. harzianum at pH 5.5.
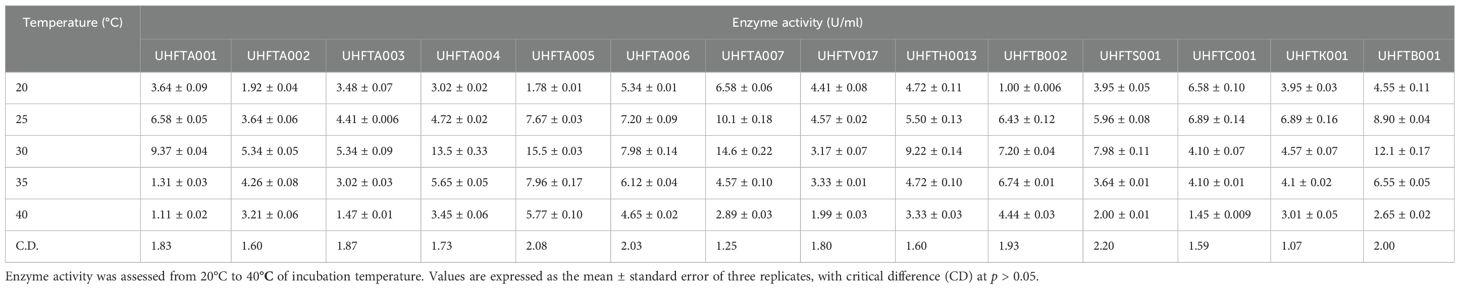
3.6 Optimization of the temperature for chitinase production
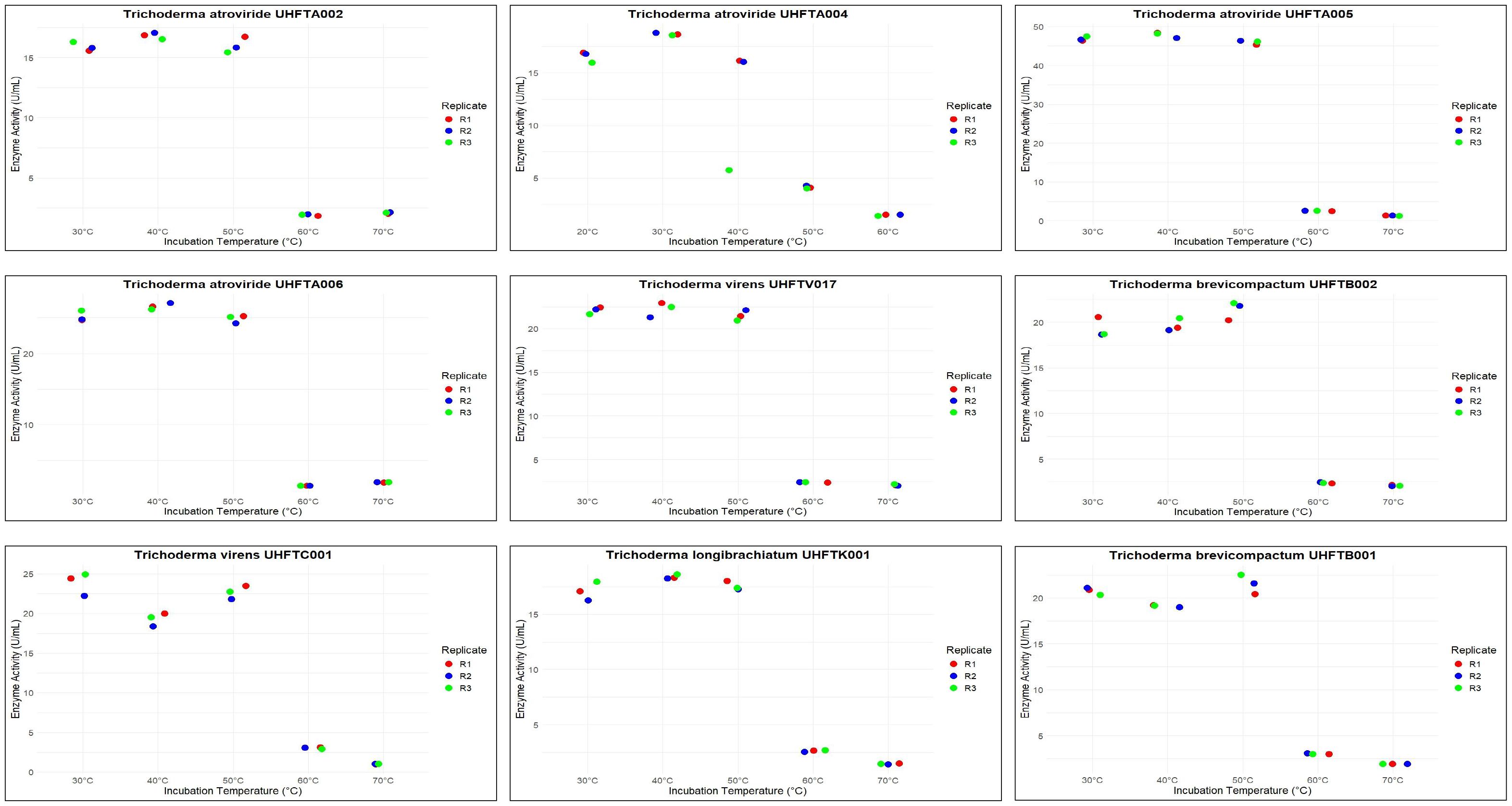
To maximize the production of chitinase, an optimal temperature is a crucial factor. Different isolates of Trichoderma spp. were assessed for the optimization of the temperature between 20°C and 40°C. As shown in Table 7, the optimum temperature for chitinase production was at 30°C for most of the examined isolates of Trichoderma spp., including UHFTA001 (9.37 U/ml), UHFTA002 (5.34 U/ml), UHFTA003 (5.34 U/ml), UHFTA004 (13.56 U/ml), UHFTA005 (15.58 U/ml), UHFTA006 (7.98 U/ml), UHFTA007 (14.65 U/ml), UHFTH0013 (9.22 U/ml), UHFTS001 (7.98 U/ml), and UHFTB001 (12.12 U/ml). However, in UHFTV017 (4.57 U/ml) and UHFTB002 (7.20 U/ml), maximum chitinase activity was achieved when incubated at 25°C and 35°C in UHFTK001 (6.89 U/ml) and UHFTC001 (6.89 U/ml) (Figure 8).
The active site of an enzyme is a specific region where a substrate binds. When the temperature is below optimal for chitinase production by Trichoderma spp., the enzymatic activity is low due to the slower binding of the substrate; when the temperature is higher than optimal, denaturation of the enzyme occurs due to the distortion of its active site. Optimal temperature maximizes the enzyme activity due to the perfect shape of the active site, allowing enzyme to bind to the substrate in its maximum efficiency. Mohiddin et al. (2021) reported that T. harzianum showed the highest average chitinase activity at 30°C, except for the T. harzianum isolate BT3, which exhibited maximum production at 25°C. Mukhammadiev et al. (2020) reported that the highest activity (3.2 ± 0.08 U/ml) of T. viride was observed at 30°C compared with the other temperatures tested, and the enzyme synthesis declined at temperatures over 38°C. El-Katatny et al. (2000) reported that the optimum temperature for the production of chitinase by T. harzianum was around 30°C, with practically no growth being observed beyond 40°C.
3.7 Extraction, purification, and quantification of chitinase from Trichoderma species
Colloidal chitin broth medium was used for the extraction of chitinases from different isolates of Trichoderma spp. The supernatant collected from the medium was used as a crude enzyme extract, which was further purified with ammonium sulfate precipitation. The crude enzyme extract was suspended in 0%–45% ammonium sulfate, and the resulting precipitates were resuspended in citrate buffer and loaded into a Sephadex G-100 column for purification.
3.7.1 Sodium dodecyl sulfate polyacrylamide gel electrophoresis
SDS-PAGE was performed to determine the molecular weight and the purity of the enzyme. The partially purified protein from the ammonium sulfate precipitation was evaluated on 12% acrylamide gel. The presence of clear bands confirmed the purity of chitinase from the different species of Trichoderma. A protein molecular weight marker (10–180 kDa size) was used to determine the molecular weight of the protein. It was found that the molecular weight of the purified chitinases from the different species of Trichoderma, e.g., T. atroviride UHFTA006, T. virens UHFTC001, T. brevicompactum UHFTB001, T. virens UHFTV017, T. atroviride UHFTA002, T. longibrachiatum UHFTK001, T. atroviride UHFTA005, T. atroviride UHFTA004, and T. brevicompactum UHFTB002, was 40 kDa, which lies between 35 and 42 kDa of the molecular marker (Figures 9).
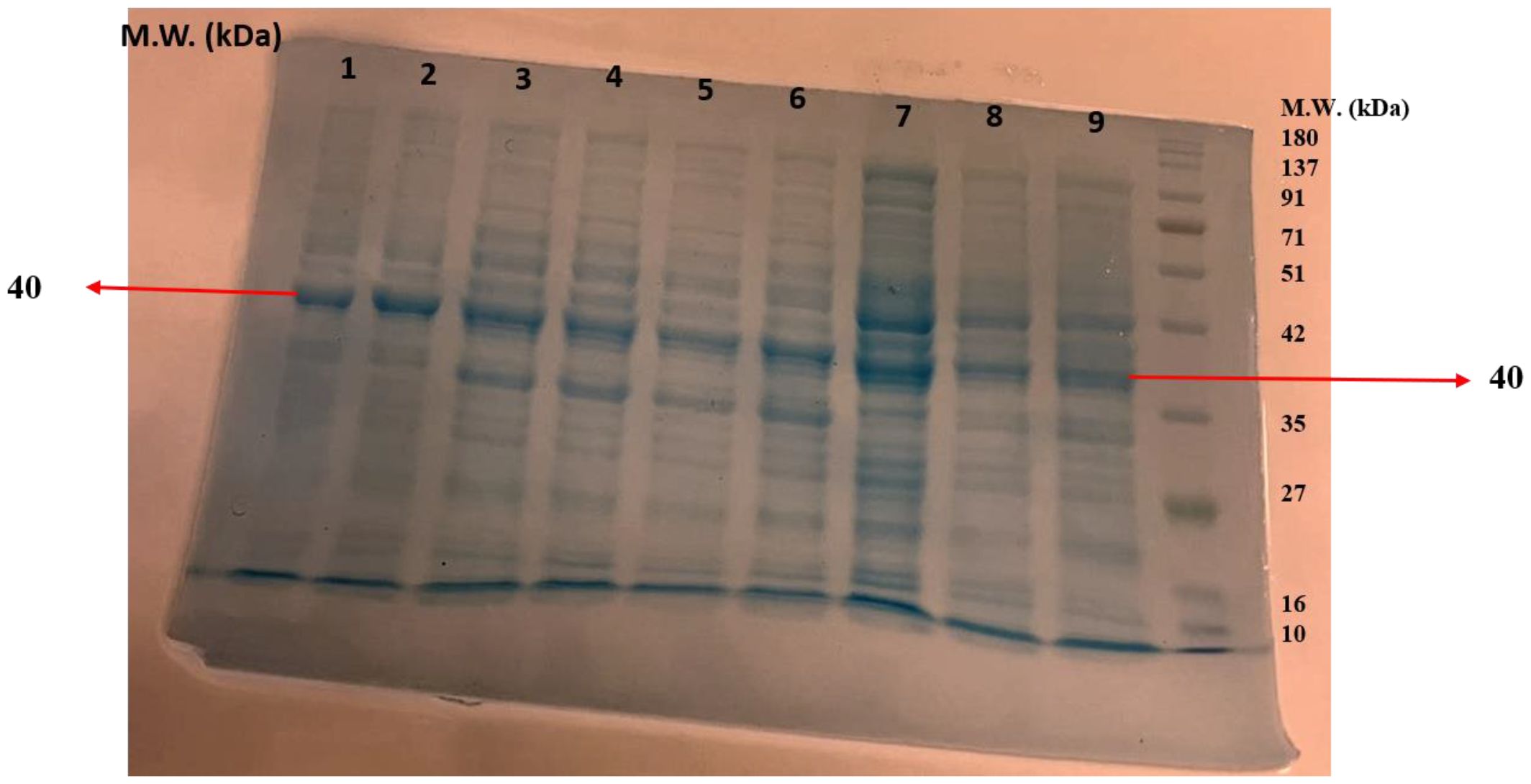
Figure 9. Molecular weight of chitinase (1: T. atroviride UHFTA006, 2: T. virens UHFTC001, 3: T. brevicompactum UHFTB001, 4: T. virens UHFTV017, 5: T. atroviride UHFTA002, 6: T. longibrachiatum UHFTK001, 7: T. atroviride UHFTA005, 8: T. atroviride UHFTA004, 9: T. brevicompactum UHFTB002).
Purification is essential to remove contaminants and other associated proteins. The higher the purity of the enzyme, the higher the activity, stability, and efficiency against targeted fungal phytopathogens. A higher purity is also beneficial in the bioformulation of biofungicides to target specific pathogens. The molecular weight of an enzyme affects its stability in various growth conditions, its solubility, and its ability to bind to the substrate. The higher the molecular weight, the higher the efficiency of binding, whereas a lower molecular weight results in a fast diffusion rate. Harighi et al. (2007) recovered a 42-kDa chitinase (Chit42) from the supernatants of T. atroviride, and Kulkarni et al. (2010) also reported that the Trichoderma isolates Tv-10, Tv-21, Tv-23, and Th-13 exhibited 36- and 45-kDa bands in crude protein estimation using SDS-PAGE. Jenifer et al. (2014) also estimated a molecular mass of 46 kDa using SDS-PAGE.
3.7.2 Quantification of the purified chitinases from the different isolates of Trichoderma species
Analysis of the data presented in Table 8 highlighted that the highest specific activity of the purified chitinases was achieved in T. atroviride UHFTA005 (43.32 U/mg), followed by T. atroviride UHFTA006, with 38.20 U/mg specific activity. In contrast, the lowest specific activity was observed in the purified chitinase obtained from T. atroviride UHFTA001 (7.00 U/mg), followed by T. harzianum UHFTH0013 (7.10 U/mg), with 3.50 mg/ml protein concentration.
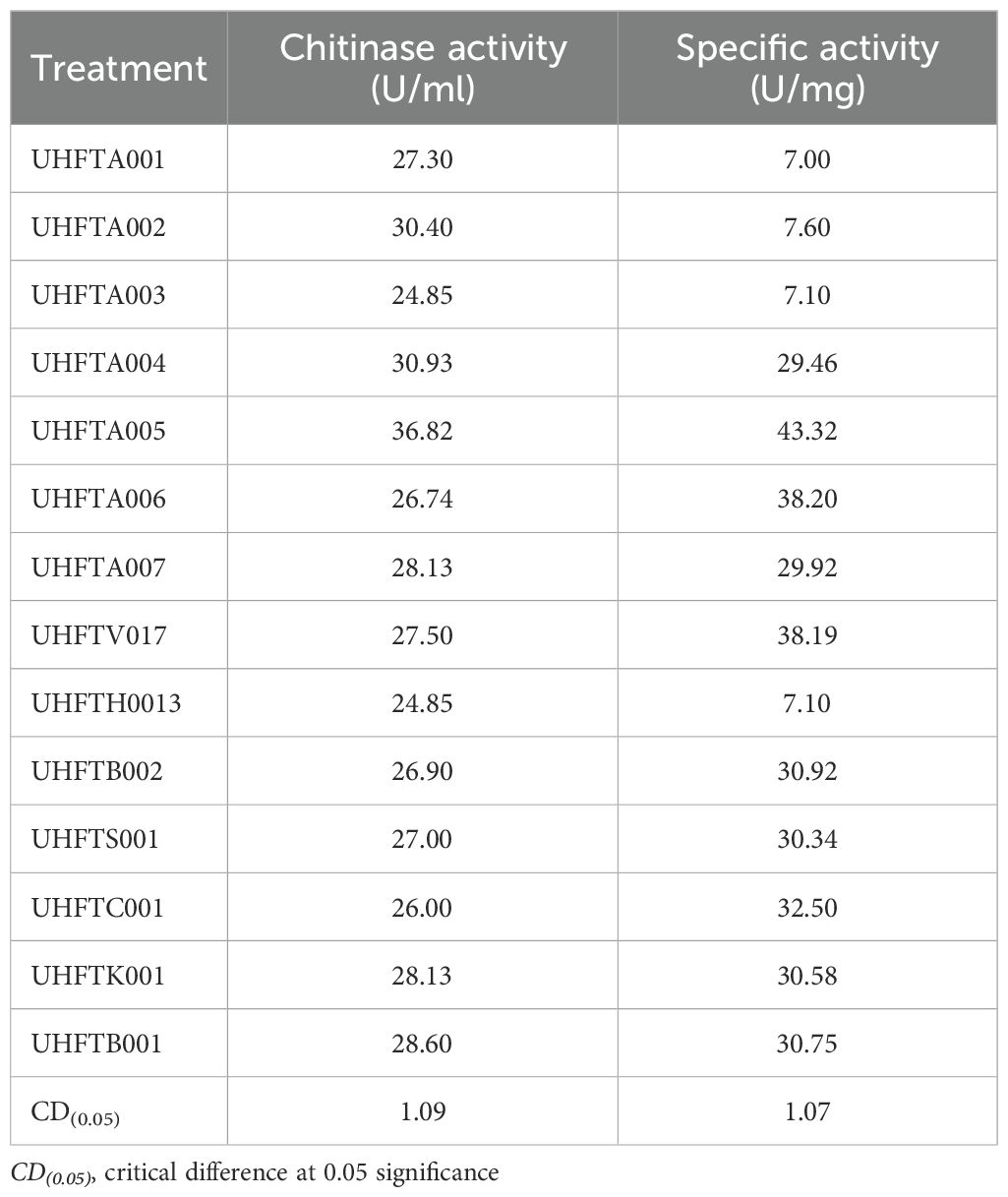
Table 8. Quantification of the purified chitinase from the different isolates of Trichoderma species.
Specific activity is a key indicator in determining the purity and functionality of an enzyme. The higher the specific activity, the lower the non-enzymatic protein. A high specific activity value determines the higher purity of the protein, indicating that the total protein contains more active chitinase. This is a key parameter for the characterization of an enzyme, and it also represents the efficiency of the total protein present. Sayed et al. (2019) reported that, among all the tested species, Trichoderma asperelloides exhibited the highest chitinase activity (1.736 U/ml), with the highest total protein content (9.861 mg) and specific activity (0.176 U/mg). Similarly, Karaca and Eltem (2024) found that the chitinase enzyme activity of Trichoderma spp. ranged from 0.01 to 0.40 U/ml, with specific activity from 0.15 to 30.66 U/mg.
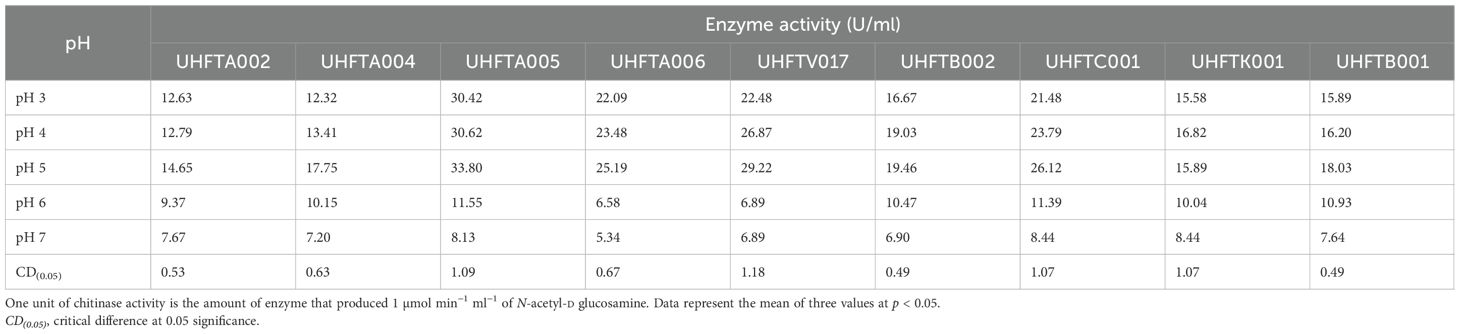
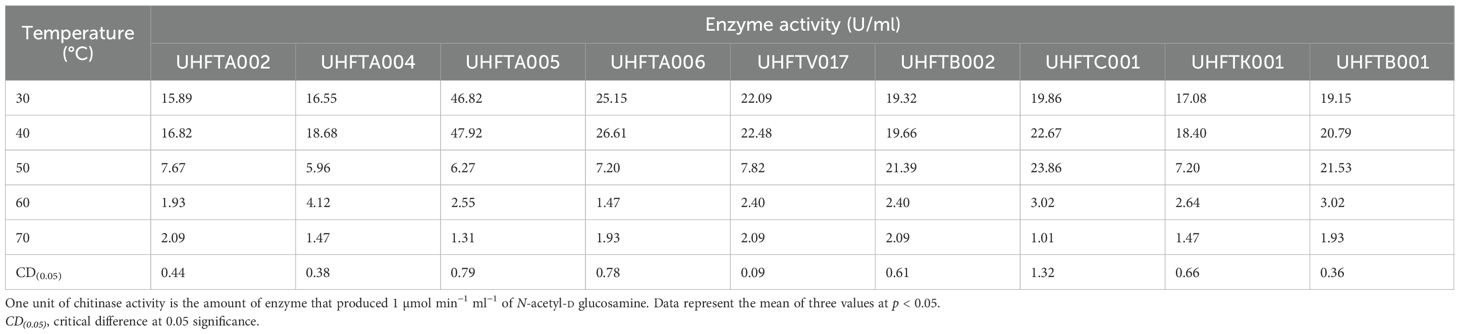
3.8 Effects of pH and temperature on the stability of chitinase
Different isolates of Trichoderma spp. were selected on the basis of the highest specific activity of the purified chitinase and were assessed for stability at various pH levels (pH 3–7) and incubation temperatures (from 30°C to 70°C). The results demonstrated that all of the examined isolates of Trichoderma spp., including UHFTA002, UHFTA004, UHFTA005, UHFTA006, UHFTV017, UHFTB002, UHFTC001, UHFTK001, and UHFTB001, maintained high stability between a pH range of 3–7 and temperature of 30–50°C. A drastic fall in activity was observed beyond pH 5 and beyond 50°C. The stability of the chitinases from all isolates of Trichoderma spp. declined drastically, and a significant inactivation of the enzyme was observed at 70°C (Tables 9, 10 and Figures 10, 11).
The active site of an enzyme is highly sensitive to pH change, which affects the binding of the substrate and its catalytic function. A high pH can lead to denaturation due to the disruption of hydrogen bonding. Acidic pH can reduce the binding of the substrate to the active site of the enzyme due to the unfolding of protein. Microorganisms produce enzymes that must remain active in various environmental and soil conditions; therefore, it is important to determine the stability of the particular enzyme produced by a particular microorganism. In various bioformulations such as biopesticides and biofungicides, the activity of chitinase is crucial for maintaining its stability at a broad pH range.
The incubation temperature is an important factor that influences the stability of an enzyme, as high temperatures can disrupt the structure of the protein, leading to its denaturation. This suggests that the chitinases from the different species of Trichoderma are well adapted for functioning at medium to slightly high temperatures, i.e., 30–50°C, making them suitable for use in the biological control of various phytopathogens in the different agroclimatic zones of Himachal Pradesh. Jenifer et al. (2014) concluded that the chitinase from T. viride remained stable at pH 3–6 and up to 50°C. Ekundayo et al. (2016) also concluded that the optimal temperature for the chitinolytic activity of T. viride was 50°C and that it was stable at temperatures of 40–50°C and pH 6–7.
3.9 In vitro antifungal activity of chitinase against Dematophora necatrix
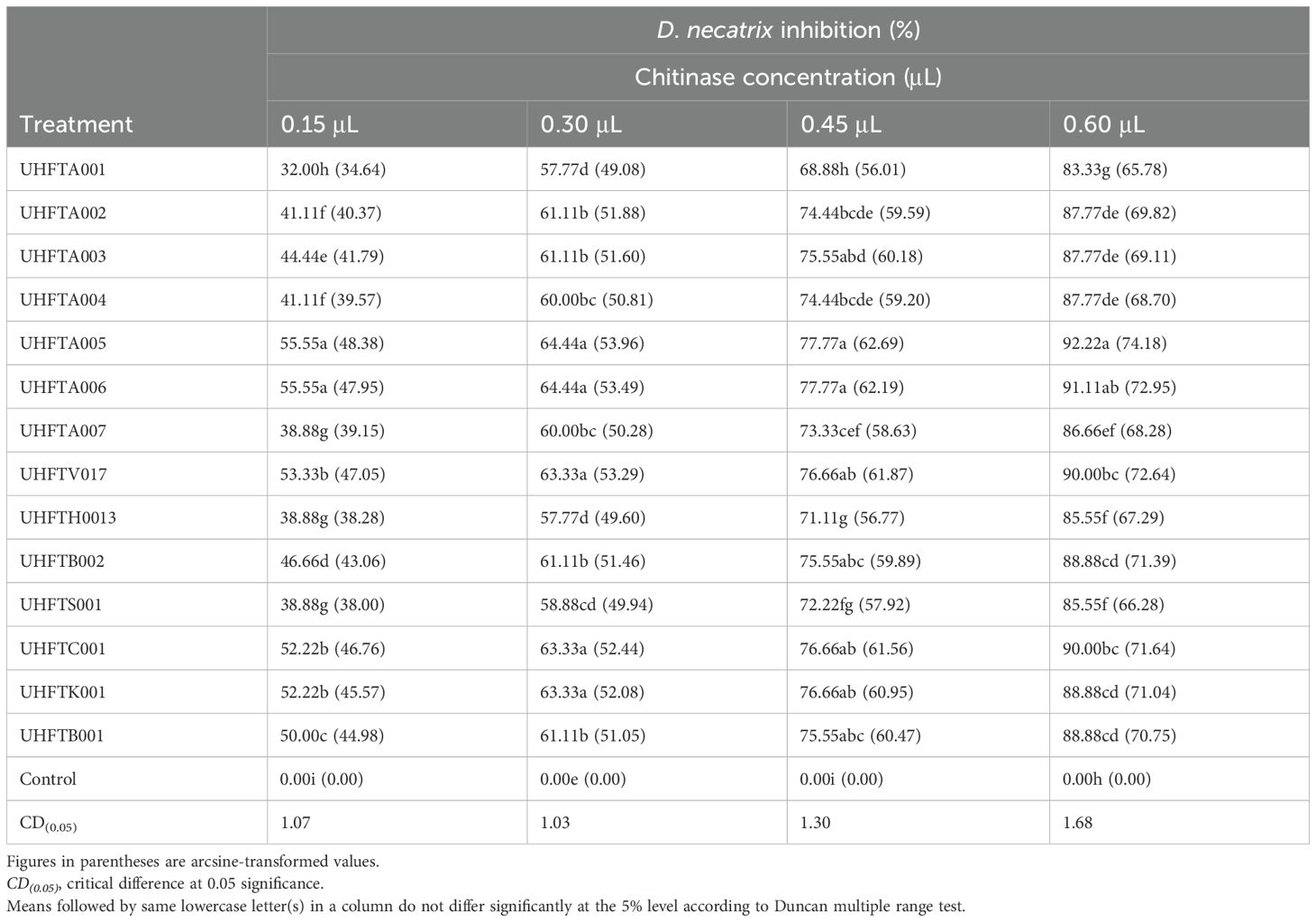
Different concentrations of the purified chitinases of 14 isolates of Trichoderma spp. were evaluated for their antifungal activity against D. necatrix. From Table 11, it is evident that, with the increase in the concentration of chitinase, the inhibition of D. necatrix also increased, with the highest inhibition observed in T. atroviride UHFTA005 at all concentrations, i.e., 0.15, 0.30, 0.45, and 0.60 μl with 55.55%, 64.44%, 77.77%, and 92.22% inhibition, respectively. However, minimum inhibition at 0.15 μl concentration was recorded in T. atroviride UHFTA001 (32.00%), and at 0.30 μl concentration, it was observed in T. atroviride UHFTA001 (57.77%). At 0.45 and 0.60 μl chitinase concentrations, minimum percent inhibition was recorded in T. atroviride UHFTA001, i.e., 68.88% and 83.33%, respectively, when compared with the control. All treatments were statistically significant.
3.10 In vitro antifungal activity of chitinase against Sclerotium rolfsii
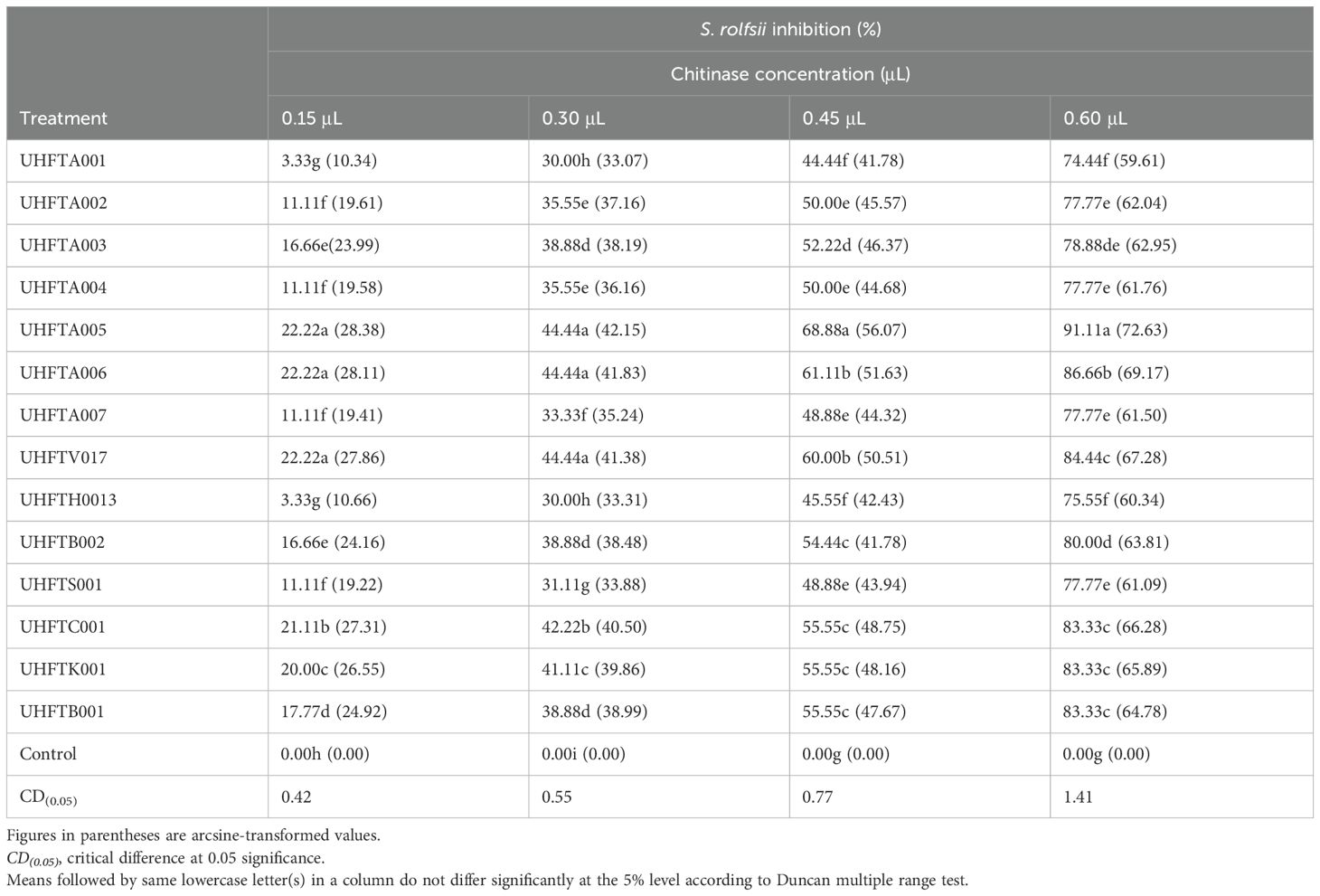
The purified chitinases from the different isolates of Trichoderma spp. were assessed for their antifungal activity against S. rolfsii. As depicted in Table 12, the highest inhibition at 0.15 μl was recorded in T. atroviride UHFTA005 (22.22%), which was statistically at par with that in T. atroviride UHFTA006 (22.22%) and T. virens UHFTV017 (22.22%). Similarly, maximum inhibition at 0.30 μl was recorded in T. atroviride UHFTA005 (44.44%), which was statistically at par with that in T. atroviride UHFTA006 (44.44%) and T. virens UHFTV017 (44.44%). However, at 0.45 and 0.60 μl concentrations, maximum inhibition was observed in T. atroviride UHFTA005, with 68.88% and 91.11%, respectively. Minimum inhibition at 0.15 μl concentration was observed in T. atroviride UHFTA001 (3.33%) when compared with the control. All treatments were statistically significant.
In the biological control of soilborne pathogens, namely, D. necatrix and S. rolfsii, through Trichoderma spp., chitinase plays a major role by degrading the chitin present in the cell wall of these fungi, thereby hydrolyzing chitin in the fungal cell wall, leading to the weakening of the cell structure, lysis of the cell, and, ultimately, death of the fungus. The degradation of chitin by chitinase also disrupts the integrity of fungal hyphae, making it more susceptible to antagonism by other microbes. In addition to suppressing the growth of fungal pathogens, chitinase plays an important role in the growth promotion of plants. Sharma et al. (2021) highlighted the potential inhibition of D. necatrix through chitinase from Trichoderma spp., while Gonzalez et al. (2023) highlighted that chitinase significantly inhibited the growth of Fusarium oxysporum.
3.11 Antifungal activity of chitinase against white root rot in pot culture conditions
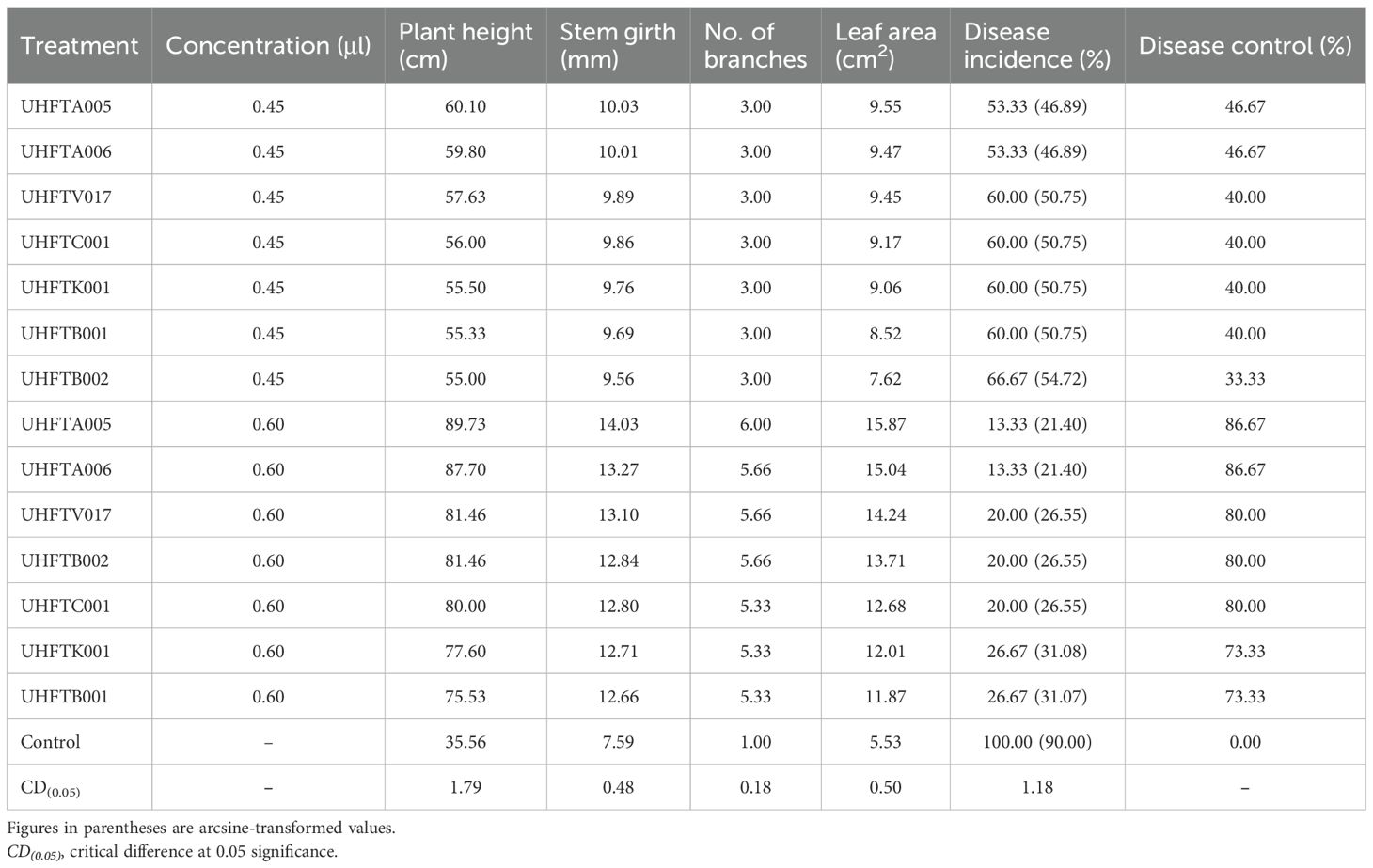
From Table 13, it can be inferred that the chitinases from the different isolates of Trichoderma spp. not only reduced the incidence of white root rot but also promoted the health of plants in terms of plant height, stem girth, number of branches, and leaf area. Data pertaining to the effectiveness of Trichoderma spp. in Table 13 revealed that minimum disease incidence (13.33%) was recorded in seedlings treated with 0.60 μl of T. atroviride UHFTA005 and T. atroviride UHFTA006, with 86.67% disease control compared with the control, thereby depicting maximum plant height, i.e., 89.73 and 87.70 cm, respectively. Similarly, maximum stem girth, leaf area, and number of branches were also recorded in both these treatments, i.e., 14.03 mm stem girth, 6.00 number of branches, and 15.87 cm2 leaf area in seedlings treated with T. atroviride UHFTA005, followed by T. atroviride UHFTA006 with 13.27 mm stem girth, 5.66 average number of branches, and 15.04 cm2 leaf area. However, minimum plant height, stem girth, number of branches, and leaf area were recorded in seedlings treated with 0.45 μl chitinase of T. brevicompactum UHFTB002, with respective values of 55.00 cm, 9.56 mm, 3.00, and 7.62 cm2. All treatments were statistically significant.
Hood et al. (2023) demonstrated that T. harzianum β-1,4-glucosidase binds to and degrades the key structural polysaccharides of M. phaseolina—including α-1,3-glucan, β-1,3-glucan, β-1,3/1,4-glucan, and chitin—suggesting its role in the reduction of pathogen incidence by weakening the integrity of the fungal cell wall. Our research validated similar results, as the Trichoderma isolates producing chitinases significantly reduced the incidence of white root rot and enhanced plant health, highlighting chitinase as a crucial enzyme in the suppression of fungal pathogens. These findings underline the critical enzymatic involvement of Trichoderma in biocontrol.
3.12 Antifungal activity of chitinase against seedling blight in pot culture conditions
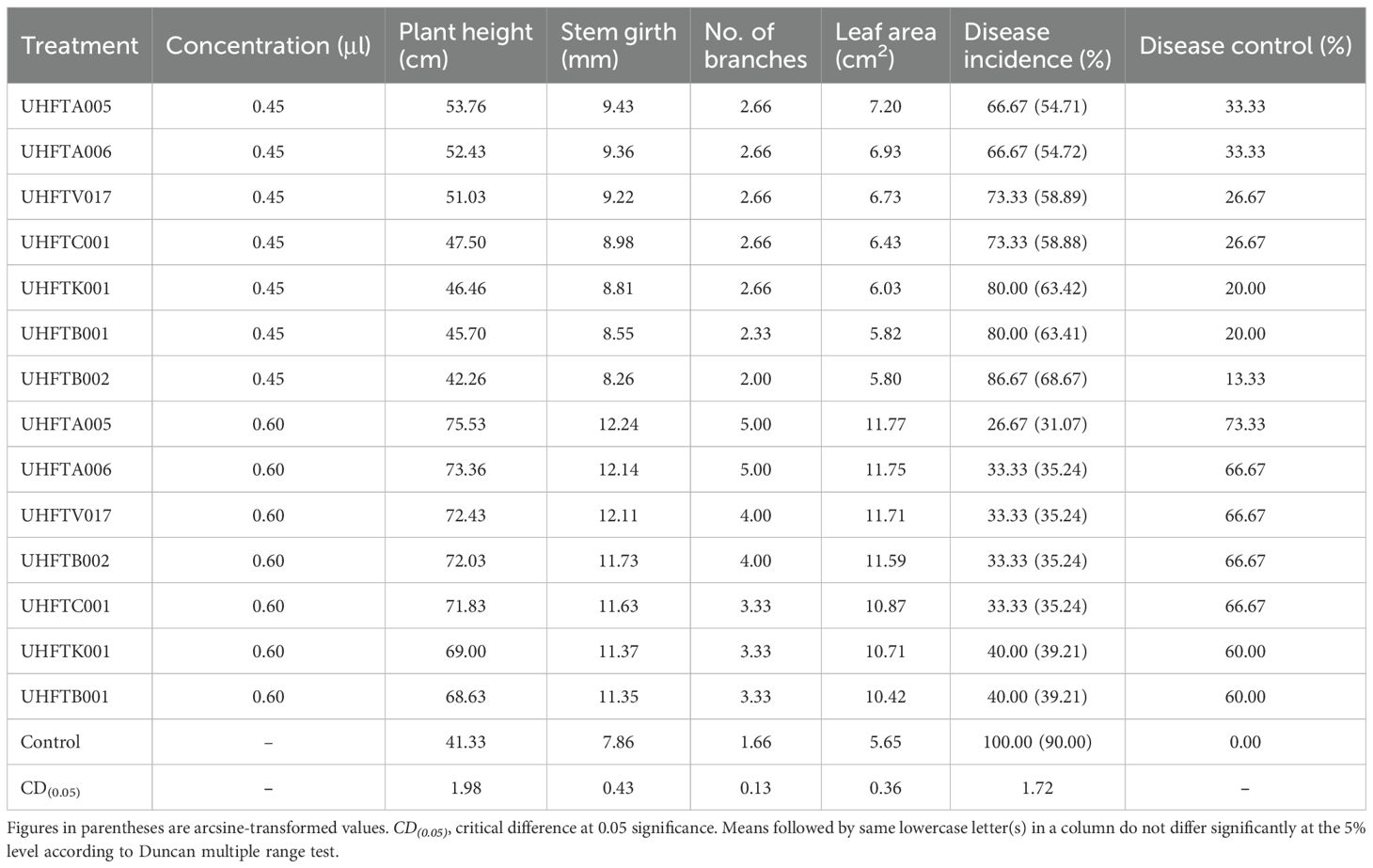
From Table 14, it can be inferred that the chitinases from the different isolates of Trichoderma spp. not only reduced the incidence of seedling blight but also promoted the health of plants in terms of plant height, stem girth, no of branches, and leaf area. Minimum disease incidence (26.67%) was recorded in seedlings treated with 0.60 μl of chitinase from T. atroviride UHFTA005, with 73.33% disease control, followed by T. atroviride UHFTA006, T. virens UHFTV017, T. brevicompactum UHFTB002, and T. virens UHFTC001, with 66.67% disease control in each case. Maximum disease incidence was recorded (86.67%) when seedlings were treated with 0.45 μl chitinase from T. brevicompactum UHFTB002. Data also revealed that maximum plant height (75.53 cm), stem girth (12.24 mm), number of branches (5.00), and leaf area (11.77 cm2) were recorded in seedlings treated with 0.60 μl chitinase from T. atroviride UHFTA005, followed by seedlings treated with 0.60 μl chitinase from T. atroviride UHFTA006, with 73.36 cm plant height, 12.14 mm stem girth, 5.00 average number of branches, and 11.75 cm2 leaf area. However, minimum plant height, stem girth, number of branches, and leaf area were recorded in seedlings treated with 0.45 μl chitinase from T. brevicompactum UHFTB002, with values of 42.26 cm, 8.26 mm, 2.00, and 5.80 cm2, respectively. All treatments were statistically significant.
The findings of Ribeiro dos Santos and Lima dos Santos (2025) underscored the multifaceted role of Trichoderma species in agriculture, emphasizing their capacity to colonize the rhizosphere, promote plant growth, and produce enzymes and secondary metabolites that contribute to pathogen suppression.
4 Conclusions
According to the results of this study, purified chitinase from the isolates of Trichoderma spp. can be successfully used not only to control soilborne pathogens of apple but also to promote plant health. The experimental results indicated that all of the isolates of Trichoderma spp. produced chitinases, but to varying extents. However, maximum production was observed when incubated for 7 days at pH 5 with 1% concentration of colloidal chitin at 30°C, whereas the enzyme was stable between pH 3 and 7 and temperature of 30–50°C. The isolates that showed the highest specific activity exhibited a molecular weight of 40 kDa. The chitinases from UHFTA005 and UHFTA006 exhibited the highest inhibition and disease control in vitro and in pot culture conditions. From all of the above, it can be concluded that the purified chitinases from isolates UHFTA005 and UHFTA006 can be effectively used against soilborne pathogens of apple.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The authors declare that there is no fabrication of data and does not contain any human and animal study.
Author contributions
A: Methodology, Writing – review & editing, Writing – original draft, Formal Analysis, Data curation, Software, Conceptualization. SS: Supervision, Formal Analysis, Writing – review & editing, Methodology, Validation, Investigation, Data curation, Conceptualization. BG: Writing – review & editing, Conceptualization, Methodology, Formal Analysis, Visualization, Resources. NR: Resources, Writing – review & editing, Validation, Methodology, Supervision. AS: Formal Analysis, Software, Supervision, Data curation, Writing – review & editing. PV: Validation, Writing – review & editing, Visualization, Resources.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors gratefully acknowledge the support and facilities provided by Dr. Yashwant Singh Parmar University of Horticulture and Forestry, Nauni, Solan (HP), India. We extend our sincere thanks to the Department of Plant Pathology and the Department of Basic Sciences (Laboratory of Biochemistry) for their technical support, and encouragement throughout the course of this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Correction note
This article has been corrected with minor changes. These changes do not impact the scientific content of the article.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agrawal T. and Kotasthane A. S. (2012). Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. Springer Plus. 1, 73. doi: 10.1186/2193-1801-1-73, PMID: 23526575
Baek J. M., Howell C. R., and Kenerley C. M. (1999). The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr. Genet. 35, 41–50. doi: 10.1007/s002940050431, PMID: 10022948
Bastakoti S., Belbase S., Manandhar S., and Arjyal C. (2017). Trichoderma species as biocontrol agent against soil borne fungal pathogens. Nepal J. Biotechnol. 5, 39–45. doi: 10.3126/njb.v5i1.18492
Bradford M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Analyt. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3, PMID: 942051
Brzezinska M. S. and Jankiewicz U. (2012). Production of antifungal chitinase by Aspergillus Niger LOCK 62 and its potential role in the biological control. Curr. Microbiol. 65, 666–672. doi: 10.1007/s00284-012-0208-2, PMID: 22922773
Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., et al. (2004). Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 25, 1327–1333. doi: 10.1002/elps.200305844, PMID: 15174055
Chadfield V. G., Hartley S. E., and Redeker K. R. (2022). Associational resistance through intercropping reduces yield losses to soil-borne pests and diseases. New. Phytol. 235, 2393–2405. doi: 10.1111/nph.18302, PMID: 35678712
Corazza L., Belisario A., and Forti E. (1999). First report of root and coliar rot by Sclerotium rolfsii on apple trees in Italy. Plant Dis. 125, 111–119. doi: 10.1094/PDIS.1999.83.7.695B, PMID: 30845628
Dahiya N., Tewari R., and Hoondal G. S. (2006). Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71, 773–782. doi: 10.1007/s00253-005-0183-7, PMID: 16249876
Ekundayo E., Ekundayo F., and Bamidele F. (2016). Production, partial purification and optimization of a chitinase produced from Trichoderma viride, an isolate of maize cob. Mycosphere. 7, 786–793. doi: 10.5943/mycosphere/7/6/9
Elad Y., Chet I., and Henis Y. (1982). Degradation of plant pathogenic fungi by Trichoderma harzianum. Can. J. Microbiol. 28, 719–772. doi: 10.1139/m82-110
Elieh-Ali-Komi D. and Hamblin M. R. (2016). Chitin and chitosan: production and application of versatile biomedical nanomaterials. Int. J. Adv. Res. 4, 411–427., PMID: 27819009
El-Katatny M. H., Somitsch W., Robra K. H., El-Katatny M. S., and Gubitz G. M. (2000). Production of chitinase and beta-1,3-glucanase by T. harzianum. Food Technol. Biotechnol. 38, 173–180.
Flach J., Pilet P. E., and Jolles P. (1992). What’s new in chitinases research. Experientia. 48, 701–716. doi: 10.1007/BF02124285, PMID: 1516675
Gajera H. P., Vakharia D. N., and Vakharia D. N. (2012). Antagonistic and genetic diversity assessment of Trichoderma isolates against Sclerotium rolfsii causing stem rot in groundnut. J. Plant Pathol. 28, 1–9. doi: 10.5423/PPJ.NT.06.2011.0111
Garcia-Nunez H. G., Martínez-Campos A. R., Hermosa-Prieto M. R., Monte-Vázquez E., Aguilar-Ortigoza C. J., and González-Esquivel C. E. (2017). Morphological and molecular characterization of native isolates of Trichoderma and its potential biocontrol against Phytophthora infestans. Rev. Mex. Fitopatol. 35, 58–79. doi: 10.18781/R.MEX.FIT.1605-4>
Gomez Ramirez M., Rojas Avelizapa L. I., Rojas Avelizapa N. G., and Cruz Camarillo R. (2004). Colloidal chitin stained with Remazol Brilliant Blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J. Microbiological Methods 56, 213–219., PMID: 14744450
Gonzalez M. F., Galarza L., Valdez L. L., and Quizhpe G. M. (2023). Antifungal activity of metabolites from Trichoderma spp. against Fusarium oxysporum. Revista. Bionatura. 8, 1–7.
Gueye N., Kumar G., Ndiaye M., Sall-Sy D., Arama M., Ndiaye F., et al. (2020). Factors affecting the chitinase activity of Trichoderma asperellum isolated from agriculture field soils. Int. J. Biol. Biotechnol. 8, 41–44. doi: 10.7324/JABB.2020.80207
Haran S., Schickler H., and Chet I. (1996). Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology. 142, 2321–2331. doi: 10.1099/00221287-142-9-2321
Harighi M. J., Zamani M. R., and Motallebi M. (2007). Evaluation of antifungal activity of purified chitinase 42 from Trichoderma atroviride PTCC5220. Biotechnology. 6, 28–33.
Hermosa R., Grondona I., Iturriaga E., Díaz E., Castro C., Monte E., et al. (2000). Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl. Environ. Microbiol. 66, 1890–1898. doi: 10.1128/AEM.66.5.1890-1898.2000, PMID: 10788356
Hood M. H. M., Tengku H. T. A., Wahab R. A., Huyop F. Z., Kaya Y., and Abdul H. (2023). Molecular interactions of Trichoderma β-1,4-glucosidase with mycelial cell wall components of M. phaseolina. J. Biomolecul. Struct. Dynamic. 41, 2831–2847. doi: 10.1080/07391102.2022.2039772, PMID: 35174777
Jamar L., Lateur M., and Van Huylenbroeck J. (2010). Reduced fungicide inputs in organic apple production using resistant cultivars and microbiological control. Crop Protection. 29, 535–544. doi: 10.1016/j.cropro.2009.12.016
Jenifer S., Jeyasree J., Laveena D. K., and Manikandan K. (2014). Purification and characterization of chitinase from Trichoderma viride (N9) and its antifungal activity against phytopathogenic fungi. World. J. Pharm. Pharm. Sci. 3, 1604–1611.
Karaca K. and Eltem R. (2024). Investigating plant growth promoting properties of trichoderma species for sustainable agriculture. Authorea.doi: 10.22541/au.172499496.61669454/v1
Kitamura E. and Kamei Y. (2003). Molecular cloning, sequencing and expression of the gene encoding a novel chitinase from marine bacterium, Pseudomonas sp. PE2, and its domain structure. App. Microbiol. Biotechnol., 61., PMID: 12655456
Kulkarni S., Ramanujam B., Rabindra R. J., Nagesh M., and Roa N. R. (2010). Isolation, identification and documentation of efficient chitinase enzyme production ability strains of Trichoderma. J. Plant Dis. 5, 198–202.
Laemmli U. K. (1970). Cleavage of structural proteins during assembly of the head of bacteriophage. Nature. 227, 680–685. doi: 10.1038/227680a0, PMID: 5432063
Lee S. H., Shin H., Lee H. I., and Lee S. (2022). The control efficacy of sodium hypochlorite against violet root rot caused by Helicobasidium mompa in apple. Plant Pathol. J. 38, 513–521. doi: 10.5423/PPJ.OA.04.2022.0057, PMID: 36221923
Lingappa Y. and Lockwood J. L. (1962). Chitin media for selective isolation and culture of actinomycetes. Phytopathology. 52, 317–323.
Marsh R. W. (1952). Field observations on the spread of Armillaria mellea in apple orchards and in a blackcurrant plantation. Trans. Br. Mycol. Soc 35, 201–207. doi: 10.1016/S0007-1536(52)80049-X
Miller G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analyt. Chem. 31, 426–429. doi: 10.1021/ac60147a030
Mohiddin F. A., Padder S. A., Bhat A. H., Ahanger M. A., Shikari A. B., Wani S. H., et al. (2021). Phylogeny and optimization of Trichoderma harzianum for chitinase production: evaluation of their antifungal behaviour against the prominent soil borne phyto-pathogens of temperate India. Microorganisms. 9, 1–20. doi: 10.3390/microorganisms9091962, PMID: 34576858
Mukhammadiev R., Mukhammadiev R., Skvortsov E., Cheremisin A., Zavriev S., Gerner A., et al. (2020). Chitinase production by Trichoderma viride in submerged state fermentation. Earth. Environ. Sci. 578, 1–8. doi: 10.1088/1755-1315/578/1/012009
Mukherjee P. K., Horwitz B. A., Herrera-Estrella A., Schmoll M., and Kenerley C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353, PMID: 23915132
Mukhopadhyay R. and Kumar D. (2020). Trichoderma: a beneficial antifungal agent and insights into its mechanism of biocontrol potential. Egypt. J. Biol. Pest. Control. 30, 1–8. doi: 10.1186/s41938-020-00333-x
Pliego C., López-Herrera C., Ramos C., and Cazorla F. M. (2012). Developing tools to unravel the biological secrets of Rosellinia necatrix, an emergent threat to woody crops. Mol. Plant Pathol. 13, 226–239. doi: 10.1111/j.1364-3703.2011.00753.x, PMID: 22014332
R Development Core Team (2008). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Ribeiro dos Santos U. and Lima dos Santos J. (2025). Lessons from the field: Trichoderma in agriculture and human health. Can. J. Microbiol. 71, 1–15. doi: 10.1139/cjm-2024-0227, PMID: 40227123
Sambrook J. E. F. and Fritsch, Maniatis T. (1989). Molecular cloning: a laboratory manual (Cold Spring Harbor, New York: Cold Spring Harbor Press).
Sayed M. A., Tahany, Ragab A. A., and Abdellatif A. M. (2019). Biocontrol of root-knot nematode Meloidogyne incognita by chitinolytic Trichoderma spp. Egypt. J. Agronematol. 18, 30–47. doi: 10.21608/ejaj.2019.52842
Shafique S., Bajwa R., Shafique S., and Naz S. (2016). Fungal pathogens as a major cause of plant diseases. Mycopath. 14, 85–94.
Sharma R., Katoch M., and Sharma M. (2021). Phylogeny and optimization of Trichoderma harzianum for chitinase production: evaluation of their antifungal behaviour against the prominent soil-borne phyto-pathogens of temperate India. Microorganisms. 9 1962., PMID: 34576858
Sharma S., Kour D., Rana K. L., Dhiman A., Thakur S., Thakur P., et al. (2019). “Trichoderma: biodiversity, ecological significances, and industrial applications,” in Recent advancement in white biotechnology through fungi: volume 1: diversity and enzymes perspectives, 85–120. doi: 10.1007/978-3-030-10480-13
Shaw S., Le Cocq K., Paszkiewicz K., Moore K., Winsbury R., and de Torres Zabala M. (2016). Transcriptional reprogramming underpins enhanced plant growth promotion by the biocontrol fungus Trichoderma hamatum GD12 during antagonistic interactions with Sclerotinia sclerotiorum in soil. Mol. Plant Pathol. 17, 1425–1441. doi: 10.1111/mpp.12429, PMID: 27187266
Singh J., Silva K. J. P., Fuchs M., and Khan A. (2019). Potential role of weather, soil and plant microbial communities in rapid decline of apple trees. PLoS. One 14, e0213293. doi: 10.1371/journal.pone.0213293, PMID: 30840713
Singh B. K., Walker A., Morgan J. A. W., and Wright D. J. (2003). Effect of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Appl. Environ. Microbiol. 69, 5198–5206. doi: 10.1128/AEM.69.9.5198-5206.2003, PMID: 12957902
Surendra P., Rao D. M., Ramesh K., and Anitha S. (2025). Efficacy and economic viability of biopolymer-based Trichoderma for managing safflower soil-borne diseases. J. Crop Health 77, 45. doi: 10.1007/s10343-024-01073-w
Tamura K., Stecher G., Filipski A., and Kumar S. (2013). MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197, PMID: 24132122
Thompson J., Higgins D. G., and Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acid Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673, PMID: 7984417
Tulasne L. R. and Tulasne C. (1865). “Selecta fungorum carpologia,” in Imperiali typographeo excudebatur(Jussu Paris).
Ulhoa C. J. and Peberdy J. F. (1991). Regulation of chitinase synthesis in Trichoderma harzianum. J. Gen. Microbiol. 137, 2163–2169. doi: 10.1099/00221287-137-9-2163, PMID: 1748872
Velivelli S. L. S., De Vos P., Kromann P., Declerck S., and Prestwich B. D. (2014). Biological control agents: from field to market, problems, and challenges. Trends. Biotechnol. 32, 493–496. doi: 10.1016/j.tibtech.2014.07.002, PMID: 25246168
Viterbo A., Ramot O., Chernin L., and Chet I. (2002). Significance of lytic enzymes from Trichoderma spp. in the biocontrol of fungal plant pathogens. Antonie van Leeuwenhoek. 81, 549–556. doi: 10.1023/A:1020552226195, PMID: 12448750
Wonglom P., Ito S., and Sunpapao A. (2020). Volatile organic compounds emitted from endophytic fungus Trichoderma asperellum T1 mediate antifungal activity, defense response and promote plant growth in lettuce (Lactuca sativa). Fungal Ecol. 43, 100867. doi: 10.1016/j.funeco.2019.100867
Xu L., Wang D., Lu L., Jin L., Lui J., Song D., et al. (2014). Purification, cloning, characterization and n-glycosylation analysis of a novel β-fructosidase from Aspergillus oryzae FS4 synthesizing levan- and neolevan-type fructiooligosaccharides. Pone. 9, 1–20., PMID: 25501957
Zang L. S., Wang S., Zhang F., and Desneux N. (2021). Biological control with Trichogramma in China: history, present status, and perspectives. Annu. Rev. Entomol. 66, 463–484. doi: 10.1146/annurev-ento-060120-091620, PMID: 32976724
Zhou W., Li M., and Achal V. (2024). A comprehensive review on environmental and human health impacts of chemical pesticide usage. Emerg. Contamin., 100410. doi: 10.1016/j.emcon.2024.100410
Keywords: biological control, chitin, chitinase, soil borne, Trichoderma
Citation: Akanksha, Sharma SK, Gupta BK, Rana N, Sharma A and Verma P (2025) Biocontrol potential of Trichoderma-derived chitinase: optimization, purification, and antifungal activity against soilborne pathogens of apple. Front. Fungal Biol. 6:1618728. doi: 10.3389/ffunb.2025.1618728
Received: 26 April 2025; Accepted: 16 June 2025;
Published: 24 July 2025; Corrected: 25 July 2025.
Edited by:
Benjamin A. Horwitz, Technion Israel Institute of Technology, IsraelReviewed by:
Laith Khalil Tawfeeq Al-Ani, Universiti Sains Malaysia, MalaysiaYilmaz Kaya, Ondokuz Mayıs University, Türkiye
Copyright © 2025 Akanksha, Sharma, Gupta, Rana, Sharma and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satish K. Sharma, c2F0aXNoc2hhcm1hMjAyNkBnbWFpbC5jb20=
 Akanksha
Akanksha Satish K. Sharma
Satish K. Sharma Bhupesh K. Gupta
Bhupesh K. Gupta Neerja Rana
Neerja Rana Anju Sharma
Anju Sharma Pramod Verma
Pramod Verma