- Department of Plant Biology, Rutgers University, New Brunswick, NJ, United States
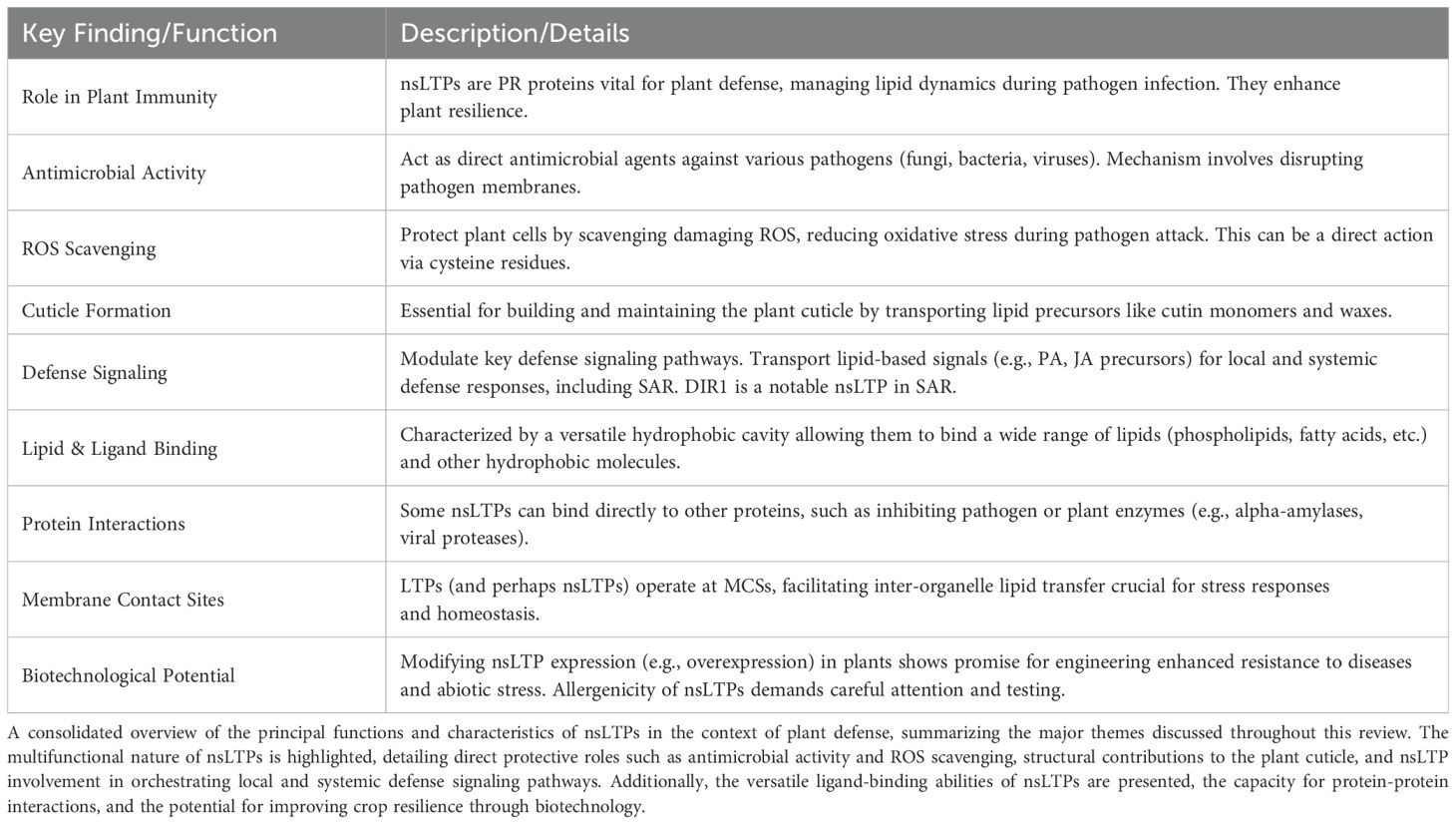
Non-specific lipid transfer proteins (nsLTPs) are vital and versatile components of plant cellular systems. They are characterized by a conserved eight-cysteine motif and are increasingly recognized for their dual roles in direct defense and stress modulation. nsLTPs serve critical structural and signaling functions in plant immunity. In contrast, other lipid transfer proteins, which lack the conserved cysteine motif, are primarily localized at membrane contact sites, specialized inter-organelle junctions that act as central hubs for lipid trafficking and signaling. This review explores the diverse roles of nsLTPs from structural, functional, and evolutionary perspectives, and examines current classification methodologies for the plant nsLTP superfamily. Functionally, nsLTPs contribute to the formation of protective barriers by transporting cutin monomers and other lipids, while also possessing lipid-specific antimicrobial properties that disrupt pathogen membranes. They support redox balance by scavenging reactive oxygen species, thereby minimizing oxidative stress. Additionally, nsLTPs are involved in defense signaling by transporting lipid-derived molecules essential to systemic acquired resistance. Their structural adaptability enables binding to a wide range of lipid species, underpinning their involvement in cuticle integrity, immune responses, and abiotic stress tolerance. These attributes position nsLTPs as promising targets for engineering durable, broad-spectrum disease resistance in crops. However, significant knowledge gaps remain regarding their structure-function relationships, lipid transport mechanisms, and roles in defense signaling and pathogen resistance. Addressing these challenges through advanced molecular and genetic tools could unlock the potential of nsLTPs to enhance crop resilience and contribute significantly to global food security.
1 Introduction
Lipid-transfer proteins (LTPs) mediate non-vesicular lipid transport, ensuring the optimal distribution of various lipids essential for cellular function, metabolism, and signaling (Chiapparino et al., 2016; Lev, 2010; Reinisch and Prinz, 2021; Neuman et al., 2022). LTPs were initially identified in the 1970s as small proteins that facilitate phospholipid transfer between membranes in vitro. This mode of transport is crucial for the proper distribution of lipids across the various membranes within a cell and between organelles (Lahiri et al., 2015; Wong et al., 2019, 2017). Initial LTP studies aimed to elucidate how these proteins contribute to fundamental biological processes related to lipid transport and metabolism. In vitro studies showed that LTPs could transfer lipids between lipid vesicles (Wirtz and Zilversmit, 1968; McMurray and Dawson, 1969; Wirtz, 1974). Molecular and structural biology research in yeast and in mammalian systems has revealed details of LTP transport that helps better understand sporulation and mitochondrial stress (Gao and Yang, 2018; Neiman, 2024; Shiiba et al., 2025). Plant research has advanced by investigating how LTPs use non-vesicular transport to control lipid homeostasis and signaling (Wang et al., 2012; Hurlock et al., 2014; de Oliveira Carvalho and Gomes, 2007).
Plants utilize a complex network of defense mechanisms to combat pathogen attack (Dodds et al., 2024; Shah and Chaturvedi, 2009; Staskawicz, 2001). Central to this defense system are lipid-mediated processes, with lipids serving as structural components, signaling molecules, and precursors for defense compounds (Kuźniak and Gajewska, 2024). Plant pathogens have evolved to manipulate host plants to promote successful infection and disease development (Shah and Chaturvedi, 2009; Christensen and Kolomiets, 2011). In response, plants have co-evolved intricate defense mechanisms to counteract pathogens, highlighting the ongoing evolutionary arms race between plants and their pathogens (Jones and Dangl, 2006). The role of LTPs has evolved from being perceived merely as lipid shuttles to key players in plant immunity (Kader, 1996; Hamaï and Drin, 2024; Cavaco et al., 2021).
Non-specific lipid transfer proteins (nsLTPs) are a subset of LTPs classified as pathogenesis-related proteins (PR-14), characterized by their conserved eight-cysteine motif (8CM) (José-Estanyol et al., 2004; Van Loon et al., 2006) which is largely confined to plant proteins, as searches have not revealed the complete 8CM outside the plant kingdom (Edstam et al., 2011). nsLTP-like proteins have been identified in Acinetobacter baumannii and Paenibacillus sp. and were considered possible cases of horizontal gene transfer (HGT) (Santos-Silva et al., 2023). nsLTPs are classified as pathogenesis-related (PR) proteins due to their induced expression in response to pathogens, abiotic factors such as cold/drought/heat, and stress-inducing chemicals like H2O2 (Sels et al., 2008; Santos-Silva et al., 2023; Van Loon and Van Strien, 1999; Liu et al., 2015; Kader, 1996).
Historically, nsLTPs were linked to cuticular barrier formation, seed development, and responses to abiotic stresses such as drought and salinity due to their ability to bind diverse lipid molecules, influencing cell wall modifications and membrane stabilization (Thoma et al., 1993; Cavaco et al., 2021; Christensen and Kolomiets, 2011; de Oliveira Carvalho and Gomes, 2007). More recently, their role in plant defense has gained prominence. Multiple nsLTPs have been found to exhibit antimicrobial activity by disrupting pathogen membranes and are integral to signaling pathways that activate immune responses, including the recognition of pathogen-associated molecular patterns (PAMPs) and inducing the accumulation of defensive compounds such as phytoalexins, lignin, and callose (Gonçalves et al., 2024c; Missaoui et al., 2022; Edstam et al., 2011).
Research continues to uncover novel defense functions of nsLTPs. Recently, a direct link to effector triggered immunity (ETI) was discovered in rice. The Xanthomonas oryzae pv. oryzae (Xoo) avirulence protein TalAE73PXO61 was found to trigger ETI by activating the nsLTP OsLTPL23. Xoo is a causal agent of rice bacterial blight (BB), a major rice disease. The TalAE73PXO61 effector, a transcription activator-like effector (TALE), binds to an effector binding element (EBE) located in the promoter region of the OsLTPL23 gene in the rice. OsLTPL23 expression was linked to ROS levels, nitrate uptake, and SA homeostasis. This represents the first documented instance of a bacterial effector protein directly targeting a plant nsLTP to trigger ETI (Jia et al., 2025). Another novel function of nsLTPs was found recently when a cowpea (Vigna unguiculata) nsLTP (LTP1) was found to interact with the cowpea mosaic virus (CPMV)-encoded cysteine protease 24KPro, interfering with viral replication and boosting resistance to the virus (Ji et al., 2024).
nsLTPs have also been found to contribute to systemic acquired resistance (SAR) by mediating the transport of lipid-derived signaling molecules, orchestrating whole-plant defense strategies (Carella et al., 2017; Champigny et al., 2013; David et al., 2021; Lascombe et al., 2008). Advances in genomics and proteomics have uncovered extensive nsLTP family diversity, suggesting specialized functions and an evolutionary arms race with pathogens (Edstam et al., 2011; Jang et al., 2008; Amador et al., 2021). Their conserved structural features, such as disulfide bonds linking cysteine residues, afford stability or resistance to degradation/denaturation under stress, underscoring their defensive roles. nsLTPs are crucial plant defense proteins of significant interest in agricultural biotechnology (Cheng et al., 2004; Iqbal et al., 2023; Cammue et al., 1995; Gao et al., 2022; García-Olmedo et al., 1995). Understanding the importance of nsLTPs, both historically and with modern bioinformatic tools, enhances our knowledge of plant immunity and facilitates engineering disease-resistant crops by manipulating nsLTP expression.
2 Structure of the review
This review examines the current understanding of nsLTPs in plant disease resistance, exploring their structural features, mechanisms of action, and involvement in defense pathways and stress homeostasis. The structure and classification of nsLTPs are presented, with an emphasis on the highly conserved eight-cysteine motif (8CM) that is critical for lipid binding. This review distinguishes between nsLTPs and other LTPs; classical LTPs are more established in their role in lipid remodeling, while the unique characteristics and functions of nsLTPs in plant defense are still being elucidated.
We examine the multifunctional roles of nsLTPs in plant defense, including antimicrobial activity, direct and indirect involvement in reactive oxygen species (ROS) scavenging, the ability to bind to fungal chitin, and participation in SAR. In this context, it’s important to highlight the emerging role of membrane contact sites (MCS) in lipid transfer. MCS are crucial for maintaining cellular homeostasis and potentially play distinct roles in plant-pathogen interactions (Michaud and Jouhet, 2019; Paul and Tiwari, 2023; Yuen et al., 2021; Lahiri et al., 2015). While LTPs, distinct from nsLTPs, are known to be associated with MCS and function in lipid exchange between organelles, the precise role of LTPs at MCS in response to biotic stress requires further investigation.
Challenges and opportunities in harnessing nsLTPs for crop protection are considered, such as addressing allergenicity and the necessity for tissue-specific targeting strategies. The functional dichotomy of nsLTPs, where certain isoforms enhance susceptibility to specific fungal pathogens while others confer resistance, necessitates mechanistic investigations to elucidate the underlying molecular determinants. This contrasting behavior underscores the importance of distinguishing nsLTP function from that of other LTPs, especially in the context of lipid dynamics and localization within the cell, including at MCS.
Key knowledge gaps remain. The mechanisms by which pathogens potentially counteract plant LTPs, including whether they employ virulence effectors that target these host proteins to compromise plant immunity, are not well understood. It is also unclear why structurally similar nsLTPs exhibit disparate functions or variable allergenic potential, and the precise mechanisms determining lipid binding specificity require further clarification. A more comprehensive understanding of nsLTPs’ involvement in specific processes like ROS scavenging is needed. We also recognize the need to better understand the role of LTPs in lipid remodeling, particularly at MCS, during disease, and their integration with other defense pathways.
This review offers insights into enhancing plant immunity with nsLTPs. A central theme is the importance of understanding how specific nsLTPs uniquely contribute to plant defense, and how their mechanisms and functions diverge from the established roles of LTPs, especially those at MCS, in lipid remodeling. We explore optimal strategies for deploying nsLTPs to engineer robust and sustainable pathogen resistance in crops. Understanding the complex interplay between lipids and plant-pathogen interactions, particularly the distinct roles of LTPs and nsLTPs, presents significant opportunities for advancing agricultural biotechnology.
3 Non-specific nature of nsLTPs
Plant nsLTPs are termed “non-specific” because they can interact with a diverse array of lipid molecules rather than a single, specific lipid species (Kader, 1996; Fleury et al., 2019; Santos-Silva et al., 2023; Amador et al., 2021). nsLTPs bind a wide variety of lipids including phospholipids, glycolipids, and fatty acids (Li et al., 2021b; Scheurer and Schülke, 2018; McLaughlin et al., 2021). This versatility in lipid binding underlies their multifaceted roles in plant defense, from maintaining cellular membrane integrity to facilitating long-distance immune signaling (Goyal and Mattoo, 2014). This class of small proteins contribute to both biotic and abiotic stress tolerance, participating in processes like membrane stabilization, cell wall organization, cuticle synthesis, and signal transduction (de Oliveira Carvalho and Gomes, 2007; Salminen et al., 2016; Goyal and Mattoo, 2014). However, “non-specificity” does not equate to a complete lack of selectivity. While nsLTPs can bind various lipids, they exhibit preferences and varying affinities, dictated by the structural properties of both the lipid molecules and the hydrophobic binding cavity within the nsLTP (Aldakhil et al., 2023; Madni et al., 2020; Han et al., 2001; Gonçalves et al., 2024a). This broad binding capability enables nsLTPs to engage in diverse physiological processes such as cuticle formation by transporting lipid-derived monomers, membrane stabilization and organization by modulating membrane lipid composition and fluidity, signal transduction as signal transducers in plant-pathogen interactions, responses to biotic and abiotic stresses including defense against pathogens, and plant growth and development, including roles in embryogenesis, reproduction, and germination (de Oliveira Carvalho and Gomes, 2007; Fleury et al., 2019; Santos-Silva et al., 2023; Amador et al., 2021).
Lipid overlay assays, employing lipids immobilized on hydrophobic membranes, are effective in revealing the diverse lipid affinities of nsLTPs (Dowler et al., 2002). For example, lipid-protein interaction assays can identify these affinities by incubating purified nsLTPs with commercially available membrane lipid strips which are available from companies like Echelon Biosciences (Catalog P-6002) (Shirey et al., 2017). A total of fifteen lipids are present on the strip, consisting of three important phosphoinositides and twelve other biologically significant lipids, including cardiolipin, cholesterol, and sphingomyelin. A western blot-like approach is taken to visualize the protein bound to the strip. For example, AtLTP4.4 binds a number of different lipids and with different affinities, the three strongest being phosphatidic acid (PA), phosphatidylinositol-4-phosphate (Ptdlns(4)P), Phosphatidylinositol (3,4,5)-trisphosphate (PtdLns(3,4,5)P3) (McLaughlin et al., 2021). A similar study working with a wheat nsLTP, TaMs1, which plays a role in pollen development, used lipid strips to show that orthologous proteins from rice (OsLTPg29) and maize (ZmLTPg11) are able to bind PA and several phosphoinositides (Li et al., 2021b). The authors were able to confirm and differentiate the roles of the orthologous rice and maize nsLTPs in pollen development in Poaceae using complementation of the male sterility phenotype of the wheat tams1 mutant with the wildtype (OsLTPg29 or ZmLTPg11) genes.
4 Structure and classification of nsLTPs
4.1 Type I vs Type II cavity structures
Initial classifications of nsLTPs were based on molecular mass and are categorized into two main types: Type I nsLTPs, which are roughly 9 kDa, and Type II nsLTPs, at around 7 kDa (Kader, 1996). nsLTPs are defined by a conserved eight-cysteine motif (8CM), crucial for their lipid-binding function (Madni et al., 2020; Wang et al., 2012; José-Estanyol et al., 2004). This 8CM, typically arranged as C-Xn-C-XnC-C-Xn-C-Xn-C-Xn-C-C (where ‘C’ is cysteine and ‘Xn’ is a variable number of amino acids), forms a hydrophobic cavity lined by hydrophobic amino acid side chains.
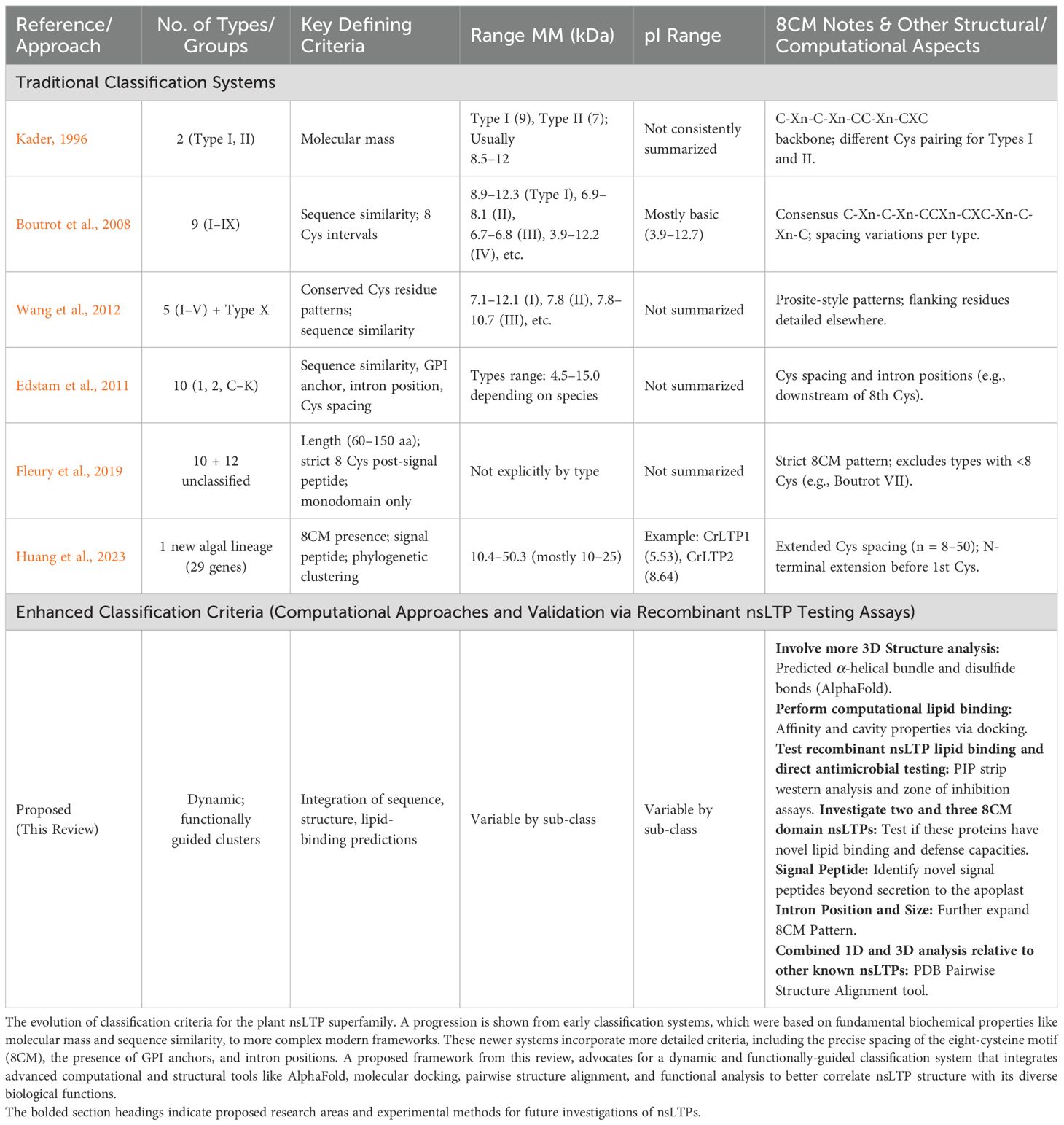
Classifications were later expanded upon by several bioinformatics labs (Boutrot et al., 2008; Santos-Silva et al., 2023; Wang et al., 2012; Amador et al., 2021; Fleury et al., 2019). A side-by-side comparison of the key criteria used in these evolving classification systems, from Kader (1996) to Huang et al. (2023), is provided in Supplementary Table S1. Boutrot et al. (2008) proposed nine types (I-IX) based on sequence similarity and cysteine spacing, with subsequent additions including groups X and XI (Liu et al., 2010). The major classification differences between Type I and Type II nsLTPs are shown in Table 1.
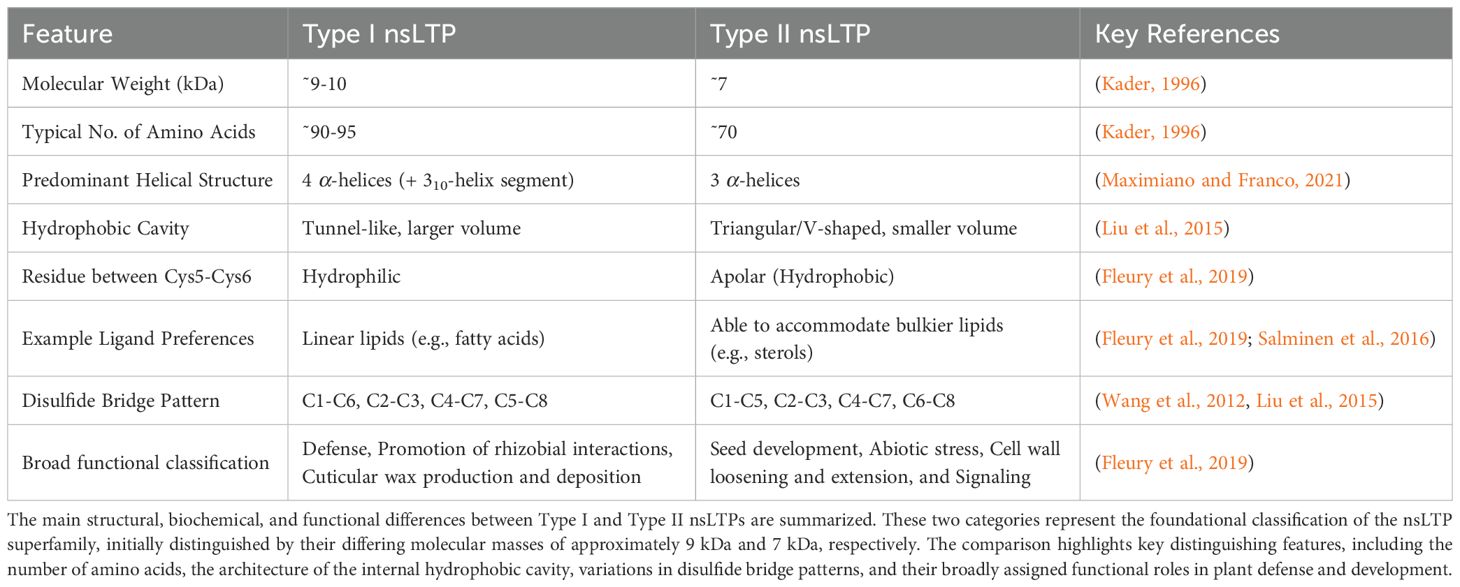
Table 1. Comparative structural and functional characteristics of plant nsLTP subtypes (Type I vs. Type II).
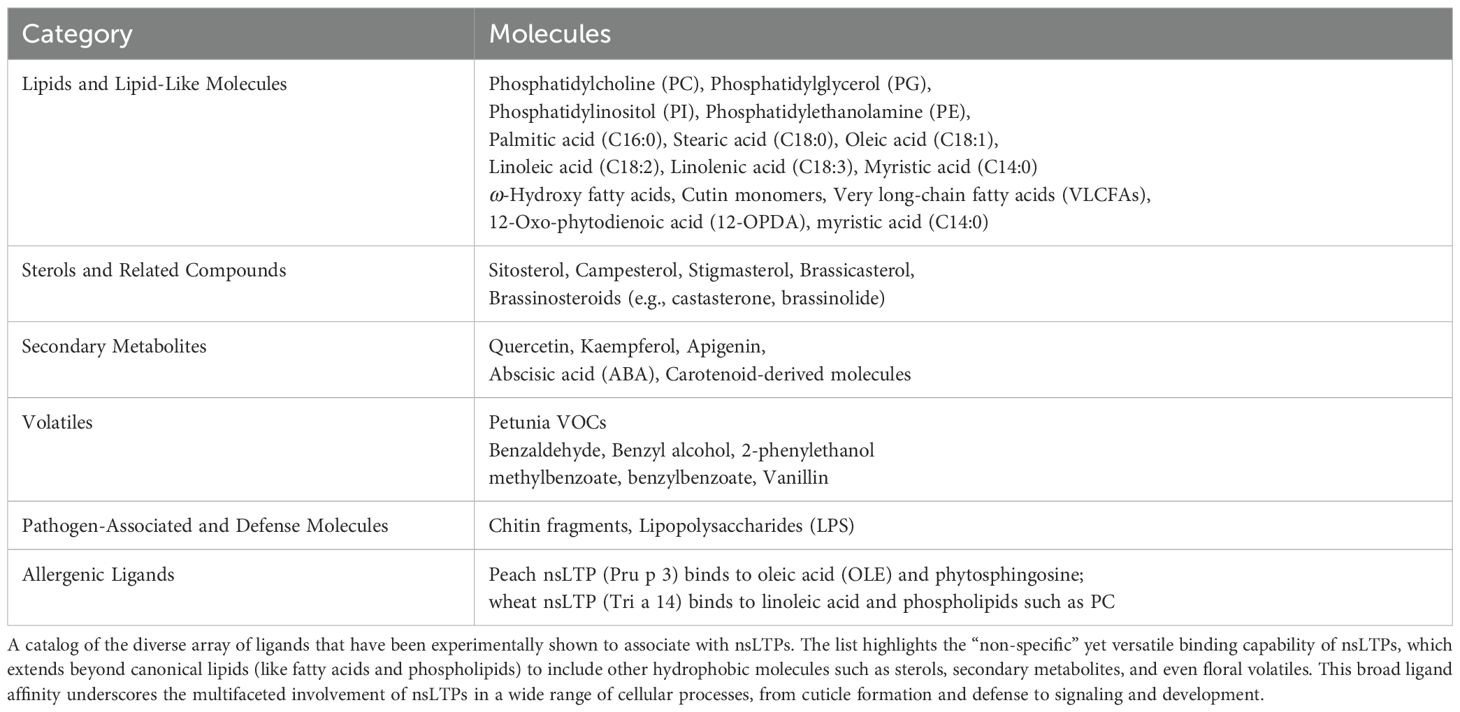
nsLTPs have been found to accommodate a diverse group of lipids such as fatty acids, phospholipids, and sterols (Salminen et al., 2016; Fleury et al., 2019). nsLTPs have also been found to bind to other hydrophobic molecules. This ability to bind a variety of lipids in addition to other diverse ligands further emphasizes the versatility of nsLTPs and their involvement in a wide range of cellular processes as highlighted in Table 2.
4.2 Disulfide bridge patterns
Type I nsLTPs are characterized by a conserved disulfide bond pattern, typically C1-C6, C2-C3, C4-C7, C5-C8 (Wang et al., 2012; Liu et al., 2015). This arrangement underpins a single, elongated, tunnel-like hydrophobic cavity that is highly conducive to binding and transporting a single lipid molecule. Proteindocking simulations indicate that ligands often lack a preferred orientation within these cavities, with hydrophobic interactions strongly dominating the protein-ligand interface (Fleury et al., 2019).
In contrast, Type II nsLTPs exhibit a different set of disulfide linkages, commonly C1-C5, C2-C3, C4-C7, C6-C8 (Liu et al., 2015). This altered bonding leads to a markedly different internal architecture, often featuring two adjacent hydrophobic cavities or a more triangular conformation (Liu et al., 2015). These variations suggest that Type II nsLTPs may accommodate different types or multiple ligands, or bind through distinct mechanisms. Molecular dynamics (MD) simulations have revealed the importance of specific amino acid residues for binding particular fatty acids, such as myristic acid and oleic acid in Ajwain [(Trachyspermum ammi)] nsLTP1 (Nazeer et al., 2019). Furthermore, subtle sequence differences, particularly in loop regions like H1–H2 and H1, influence ligand binding modes, as observed between barley HvLTP1.1 and maize ZmLTP1.6 (Salminen et al., 2016). The orientation of the ligand cavity entrance differs between the two folds. In Type I fold, it’s along an axis perpendicular to the C-terminal loop, while in Type II fold, it’s approximately parallel to it (Fleury et al., 2019).
The eight cysteines form four disulfide bridges, stabilizing the compact, α-helical rich fold and maintaining cavity integrity, facilitating lipid encapsulation and transfer. The N-terminal signal peptides often direct nsLTPs to the extracellular apoplast via the secretory pathway, or other compartments such as the endoplasmic reticulum, mitochondria, chloroplast, or vacuoles, depending on the type of nsLTP and associated specific signal peptide (Levesque-Tremblay et al., 2009; Nishimura et al., 2008; Li et al., 2024; Santos-Silva et al., 2023).
4.3 Expanded classification systems for plant nsLTPs
The nsLTP superfamily has been organized into at least eleven types based on a combination of features, including molecular mass, sequence homology, the spacing of cysteines within the 8CM domains, posttranslational modifications, and subcellular localization (Edstam et al., 2011; Boutrot et al., 2008; Santos-Silva et al., 2023) as shown in Supplementary Table S2. These classifications align with clear structural distinctions described above. While all nsLTPs share a conserved right-handed superhelix fold, a fundamental structural dichotomy exists between the “Type I fold” and the “Type II fold.” This divergence is driven by differences in amino acid sequence and disulfide bond patterns, which result in significant variations in their 3D architecture, internal cavities, and ligand-binding characteristics (Fleury et al., 2019).
4.4 Bioinformatic and structural tools to study nsLTPs
The classification and understanding of nsLTPs are being revolutionized by advanced computational tools. The European Bioinformatics Institute’s InterPro database (Blum et al., 2024) is an invaluable bioinformatics resource, unifying protein classification by integrating predictive signatures from various member databases like Pfam, SMART, PROSITE, CATH-Gene3D, and SUPERFAMILY. This approach mitigates redundancy and provides crucial insights into sequence conservation and predicted features. For instance, InterPro’s largest nsLTP family, “Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain” (IPR016140), comprises over 43,000 proteins, 362 domain architectures, and over 2,000 taxons. It also integrates data from 75 PDB solved structures and over 27,000 AlphaFold 3 predicted structures, partly based on PROSITE DOC (PDOC00516) and the PLANT LTP signature (PS00597): [LIVM]-[PA]-x(2)-C-x(1,2)-[LIVM]-x(1,2)-[LIVMST]-x-[LIVMFY]x(1,2)-[LIVMF]-[STRD]-x(3)-[DN]-C-x(2)-[LIVM].
Beyond broad databases, custom scripting, particularly in Python within the Anaconda environment (Anaconda, 2016), offers unparalleled flexibility for detailed sequence analysis. This enables researchers to precisely identify and categorize disulfide bond linkages (e.g., C1-C6, C2-C3, C4-C7, C5-C8 for Type I versus C1-C5, C2-C3, C4-C7, C6-C8 for Type II nsLTPs) in a flexible and scalable manner in addition to looking for novel patterns. For instance, by extending the 8CM spacing from n = 8–30 to n = 8–50, a new class of nsLTPs was identified in algae (Huang et al., 2023). Algal nsLTPs have divergent 8CM spacing and binding pocket residue property differences compared to land plants.
The use of the RapGreen tool developed by the de Lamotte research group was used to organize the nsLTPs superfamily into an interactive phylogenetic tree which and included phylogenetic, structural, and both Plant Ontology (PO) and Gene Ontology (GO) information (Fleury et al., 2019; Dufayard et al., 2021). The RapGreen phylogenetic analysis of 797 nsLTPs can be explored using the following website https://phylogeny.southgreen.fr/treedisplay/index.php?data=msdmind. The seven broad nsLTP classes in this phylogenetic tree range include Type I (409 nsLTPs) and Type II (118 nsLTPs) and range to Type VIII nsLTP. This type of analysis can be applied to new nsLTPs which are identified in protein databases and with advanced deep and machine learning tools (e.g., UniProt, RCSB Protein Data Bank (RCSB PDB), and AlphaFold). Frequently Aligned Symbol Tree (FAST) and Structural Trace Analysis (STD), revealed additional information about amino acid residues that might confer functional specificity in defense but require additional study to connect with protein function (Fleury et al., 2019).
The advent of AlphaFold for highly accurate 3D protein structure prediction (Jumper et al., 2021) has provided an unparalleled dataset of predicted structures, complementing laborious experimental methods. Tools like the RCSB PDB’s Pairwise Structure Alignment tool (Bittrich et al., 2024), utilizing algorithms such as TM-align (Zhang and Skolnick, 2005), efficiently compare predicted and experimentally determined structures. This allows for robust identification of structural homologs, revealing subtle variations in folds, loop regions, and cavity architectures that refine existing classifications and can identify novel structural subgroups (Gonçalves et al., 2024a; Morales-Quintana et al., 2024). As an example of this, we downloaded the 12,880 proteins (in FASTA format) classified in the Plant non-specific lipid-transfer protein (IPR000528) group from InterPro and removed the 981 redundant sequences using FASTA file manipulation tool seqkit2 (Shen et al., 2024). A python script (McLaughlin, 2025) was used to identify 8CM motifs revealing 11,861 individual proteins with one 8CM domain, 35 proteins with two 8CM domains (UniProt format: A0A067JWZ8, A0A0D3HHF7, A0A251VQ94, A0A498ILS8, A0A498J7V5, A0A4D6NFG6, A0A4S8IBN1, A0A4Y1RRI6, A0A5B6W1L1, A0A6P5FSB9, A0A7J6HF70, A0A7J6HFY0, A0A7J7KVB4, A0A7J7MH43, A0A803MFK2, A0A834TCD7, A0A835KDA7, A0A835KME6, A0A8J5YXI7, A0A8J5ZEW1, A0A9D3VEA1, A0A9D5CJ42, A0A9E7F1U6, A0A9E7F2M4, A0A9J5YS56, A0A9Q0F1H8, A0AA38W3R9, A0AA88R1U2, A0AAD5D2I7, A0AAD6QXP8, A0AAD6WBZ6, A0AAP0LW68, A0AAQ3STU6, A0AAV7H7B0, and A0ABD3GH35), two proteins with three 8CM domains (A0AAD8S0M5, A0A6N2MXE4), and one protein with four 8CM domains (A0A0D3HQM8). Multi 8CM domain proteins have been detected before within the nsLTP family (Edstam et al., 2011). In that work, four proteins (MpLTPg2 from Marchantia polymorpha and PpLTPg1, PpLTPg5, PpLTPj5 from Physcomitrella patens) were found to have two 8CMs, while one protein (PpLTPg7 from P. patens) was detected with three 8CMs.
A bioinformatic analysis of the UniProt database using a custom Python script identified two novel multidomain nsLTPs in African wild rice (Oryza barthii). These proteins, A0A0D3HHF7 and A0A0D3HQM8, were found to possess two and four eight-cysteine motif (8CM) domains, respectively. AlphaFold3 structures are available for these novel proteins. For comparison, the AlphaFold3 structures for a single 8CM domain (A0A0D3GPP4) (A,E) and a GPI-anchored, single 8CM domain (A0A0D3FEW3) (B,F), a two 8CM domain (A0A0D3HHF7) (C,G), and a four 8CM domain protein (A0A0D3HQM8) (D,H), all from Oryza barthii, are shown in Figures 1A–D, which represent AlphaFold3 predicted structures with predicted Local Distance Difference Test (pLDDT) scores. The pLDDT score is a per-residue confidence metric that indicates how closely a predicted protein structure is expected to match its experimentally determined 3D structure. Figures 1E–H represent the same structures but highlighting a domain defined by The Encyclopedia of Domains (TED). The TED domain represents a systematically identified and classified protein domain within the AlphaFold Protein Structure Database (Lau et al., 2024). To confirm lipid binding activity of these novel proteins, an assay using recombinant versions of these proteins challenged against PIP (phosphoinositides) strips could be used to identify potential protein-lipid interactions (Shirey et al., 2017). The nsLTP classification systems are summarized in Table 3.
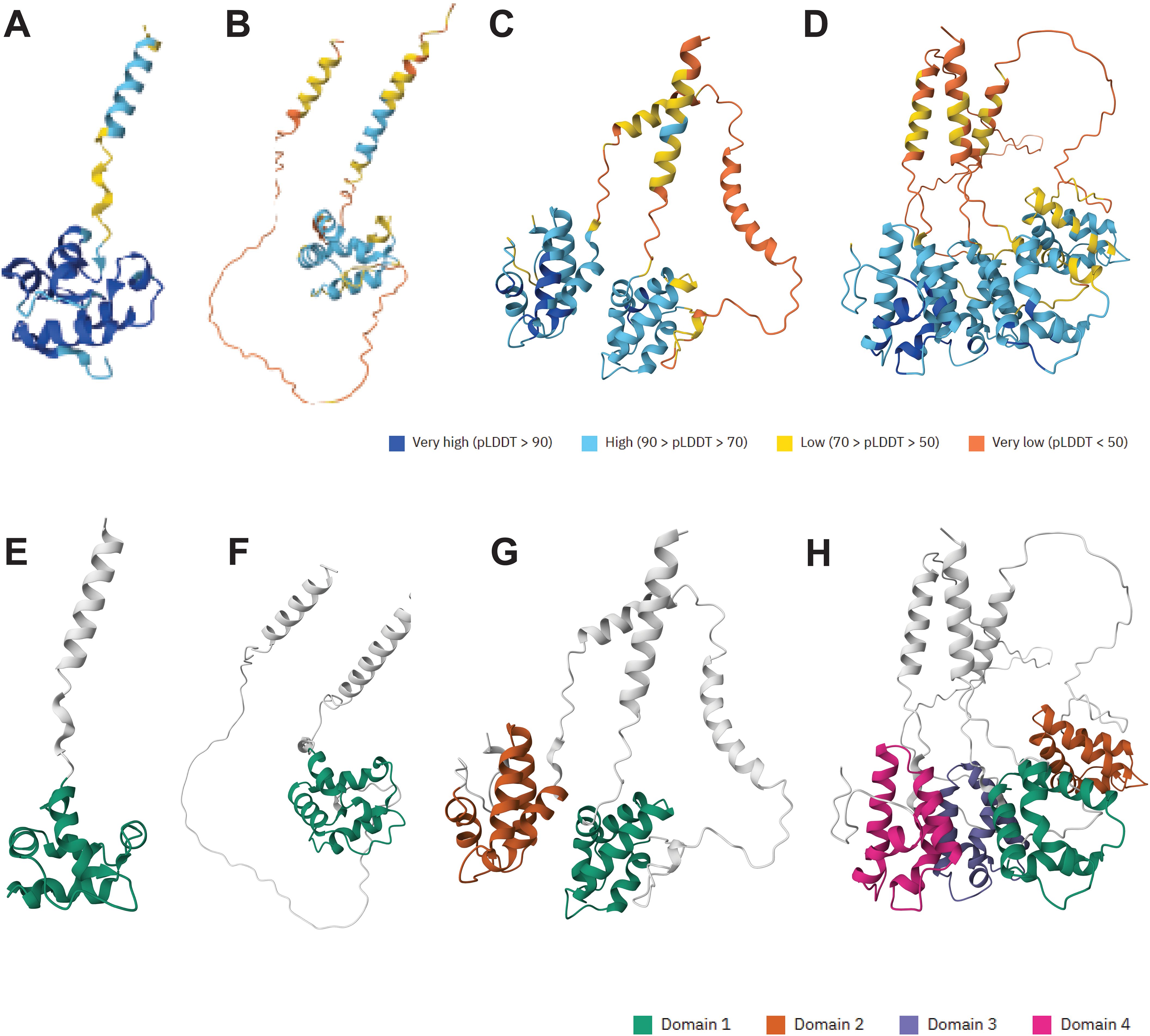
Figure 1. The structural diversity and novel multi-domain architecture of nsLTPs from African wild rice (Oryza barthii), is illustrated based on AlphaFold3 predictions. The top row (A-D) shows the 3D structures colored by their predicted Local Distance Difference Test (pLDDT) confidence scores, while the bottom row (E-H) highlights the functional eight-cysteine motif (8CM) domains as classified by The Encyclopedia of Domains (TED). The panels display a progression from a typical single 8CM domain nsLTP [(A, E); A0A0D3GPP4] and a GPI-anchored variant [(B, F); A0A0D3FEW3], to the novel discovery of an nsLTP with two 8CM domains [(C, G); A0A0D3HHF7] and a particularly novel protein possessing four 8CM domains [(D, H); A0A0D3HQM8]. The identification of these multi-domain proteins significantly expands the known structural variety within the nsLTP superfamily.
Furthermore, the integration of advanced molecular docking programs and MD simulations (Paggi et al., 2024; Neubergerová and Pleskot, 2024) allows for atomic-level modeling of nsLTP-ligand interactions. These simulations quantify interaction energies, identify critical binding residues, and explore ligand dynamics (Fleury et al., 2019; Nazeer et al., 2019; Salminen et al., 2016). This capability is invaluable for correlating structural variations with specific ligand preferences, adding a functional dimension to structural classification. In essence, the synergistic application of comprehensive bioinformatics databases (e.g., InterPro), flexible Perl/Python scripting, AlphaFold’s structural predictions, advanced structural comparison tools (e.g., TM-align), and sophisticated molecular docking/MD simulations enables a more granular and functionally relevant classification of nsLTPs. Future nsLTP classifications can take advantage of these tools and the accumulated knowledge built up from sequence and structural information that is available in UniProt and AlphaFold databases. The PDB Pairwise Structure Alignment tool, for instance, enables the simultaneous alignment of up to 10 protein 3D structures (Bittrich et al., 2024). This provides an excellent method to compare to Type I, Type II, GPI-anchored nsLTPs, and other nsLTPs in a single tool providing alignment quality structure-based scores, such as Root-Mean-Square-Deviation (RMSD, measured in Angstroms)˚ and TM-score (ranging from 0.00 to 1.00), alongside sequence-based metrics like percent identity and the number of aligned residues. This multidisciplinary computational approach will accelerate the discovery and characterization of novel subfamilies, providing a more complete picture of their diverse roles in plant physiology and defense.
4.5 Relationship between nsLTP structure and defense function
The most complete discussion of the relationship between nsLTP structure and plant defense is given by the de Lamotte laboratory (Fleury et al., 2019; Dufayard et al., 2021). They utilized a comprehensive approach combining phylogenetic and structural information to classify nsLTPs and investigate their involvement in defense mechanisms. To understand nsLTP function and variability across the superfamily, researchers analyzed a large dataset of 797 nsLTP protein sequences, which included both experimental 3D structures, including X-ray crystallography, NMR spectroscopy, cryo-electron microscopy data, and computer modeling. Type I nsLTPs formed a well-supported monophyletic group and predominated, making up over half of their dataset (417 out of 797 sequences). Type II nsLTPs were the second most abundant, with 126 sequences. Using the structural information available with these sequences, the study then classified the nsLTPs into two distinct structural categories: the Type I fold and the Type II fold. As expected, phylogenetic Type I nsLTPs were consistently found to have the Type I fold.
The Type I family is the predominant group for defense-related nsLTPs, encompassing 28 proteins that have been functionally classified as defense-related. In contrast, the Type II group contained only 3 classified defense-related nsLTPs, including the known protein AtDIR1 (Q8W453). This skewed distribution strongly suggests that Type I nsLTPs play a more significant role in plant defense (Fleury et al., 2019). Further experimental validation is required to refine the functional classification of Type I and Type II nsLTPs, many of which currently lack specific GO annotations.
4.6 Evolutionary aspects of nsLTPs
Investigation of the evolutionary history of nsLTPs can provide valuable insights into their functional diversity and role in plant adaptation to different environments and pathogens. nsLTPs are found in all land plants and recently in green algae (Edstam et al., 2011; Edqvist et al., 2018; Huang et al., 2023). The diversity of nsLTP subfamilies is more limited in non-seed plants compared to seed plants, suggesting that new nsLTPs may have evolved during land plant evolution (Edstam et al., 2011). Phylogenetic analyses indicate that nsLTPs have undergone both tandem and segmental duplications, contributing to their functional diversity (Edstam et al., 2011; Wei et al., 2025). The adoption of novel nsLTP types likely assisted plants in adjusting to the harsh new environments and disease pressures.
Differences in nsLTPs between plant species and within families have been revealed via structural analysis and bioinformatic comparisons. For example, a survey of nsLTPs in rice and Arabidopsis thaliana revealed 52 rice nsLTPs and 49 Arabidopsis nsLTPs (Boutrot et al., 2008). The authors employed comparative genomics, using the identified rice nsLTPs as a basis, to identify 156 putative nsLTPs in wheat. The nsLTP gene family in maize (Zea mays) includes 65 genes, which can be divided into six types (1, 2, C, D, G, and a unique type X), each with distinct expression patterns and functions (Fang et al., 2023). Similarly, in Brassica rapa, 63 nsLTP genes were identified and grouped into nine types (I, II, III, IV, V, VI, VIII, IX, and XI), with specific roles in defense, reproduction, and stress responses (Li et al., 2014). In barley, 70 nsLTPs were classified, based on phylogeny, protein characteristics and gene structures, and placed in groups 1, 2, C, D, and G (Zhang et al., 2019). This diversity highlights the extensive expansion and functional specialization of the nsLTP family across the plant kingdom. Supplementary Table S2 provides a comprehensive overview of the number, types, and key characteristics of nsLTPs identified in representative species, from green algae and bryophytes to angiosperms like rice and A. thaliana. Research into barley nsLTPs has been extensive due to the connections with the brewing industry as these proteins significantly influence key beer quality attributes such as foam stability, head retention, haze formation, and flavor stability (antioxidant capacity) during storage (Cai et al., 2019; Stanislava, 2007; Wu et al., 2011; Douliez et al., 2001).
The adaptation and evolution of nsLTPs in response to different pathogens or environments are driven by selective pressures, including those from pathogens. For instance, nsLTPs involved in pathogen defense have evolved to recognize and respond to specific PAMPs, impacting the plant’s immune response. A good example of this was shown by the work of Situ et al. (2024). The oomycete Peronophythora litchii secretes a pectin acetylesterase (PlPAE5), which was found to destabilize the litchi (Litchi chinensis) lipid transport protein (LcLTP1), reducing salicylic acid (SA) production and promoting infection. This interaction indicates that pathogens target LTPs to disrupt key plant defense signaling pathways. The interaction between PlPAE5 and LcLTP1 exemplifies a counter-defense strategy, where the pathogen interferes with the plant’s defense mechanism.
Additionally, nsLTPs have adapted to various environmental stresses, such as drought and salinity, by modulating lipid composition and signaling pathways (Xiao et al., 2023; Fang et al., 2023; Santos-Silva et al., 2023; Riahi et al., 2021). With their varied structures and evolutionary adaptations, nsLTPs play critical roles in plant defense, development, and stress responses. Investigating their molecular mechanisms and evolutionary history reveal valuable insights into their functions and potential applications in agricultural biotechnology.
5 Lipid transfer by nsLTPs
5.1 Mechanism of lipid binding and transfer
As detailed in the nsLTP structure section, the highly conserved 8CM, which forms four disulfide bridges, stabilizes nsLTPs. This stabilization is crucial for maintaining the integrity of the hydrophobic cavity, a feature that allows these proteins to bind and encapsulate lipid molecules (Wang et al., 2012). The lipid-binding process begins when lipid molecules interact with the hydrophobic cavity of nsLTPs. The internal cavity, lined with hydrophobic amino acid residues, creates an environment conducive to the stable binding of lipid molecules. Key amino acid residues involved in lipid binding include hydrophobic residues such as leucine, isoleucine, valine, and phenylalanine, which interact with the lipid tails (Madni et al., 2020). Mutational and modeling approaches have been used to define key residues for both protein stability and antimicrobial properties (Fleury et al., 2019; Gonçalves et al., 2024a).
5.2 Influence of hydrophobic cavity structure on lipid specificity
The structure of the hydrophobic cavity significantly influences lipid specificity. The cavity’s size, shape, and flexibility determine which lipid molecules can be accommodated (Reinisch and Prinz, 2021; Missaoui et al., 2022; Sawano et al., 2008). For example, the hydrophobic cavity of nsLTPs can adjust its volume to accommodate various ligands, ranging from C10 to C18 fatty acids (Han et al., 2001; Liu et al., 2015). This structural plasticity allows nsLTPs to bind a wide array of lipid molecules, including multiple lipids simultaneously, contributing to their non-specific binding nature (Wang et al., 2012; Sodano et al., 1997; Charvolin et al., 1999; Wong et al., 2019; Douliez et al., 2001). For example, Figure 2 presents a Pymol rendering of Solanum melongena nsLTP determined by X-ray crystallography which demonstrates the presence of two lauric acid molecules sequestered within the protein’s hydrophobic cavity (Madni et al., 2023).
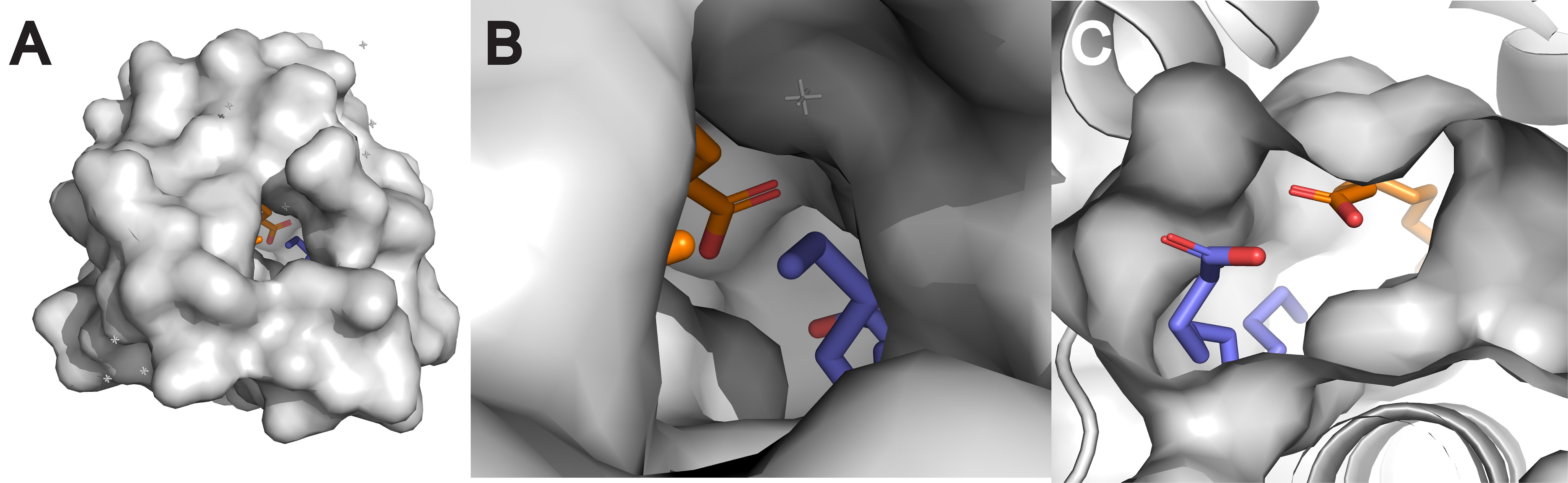
Figure 2. Surface structures of nsLTP from eggplant (Solanum melongena) (PDB ID: 7W9A) (Madni et al., 2023) visualized with PyMOL 3.1. The visualizations show the overall surface of the protein (A), a magnified view into its hydrophobic cavity (B), and a cross-section revealing two distinct lauric acid molecules sequestered within (C). This ability to bind multiple lipids at once is a key demonstration of the structural plasticity that underlies the “non-specific” binding nature of nsLTPs.
The hydrophobic cavity’s ability to shield the hydrophobic tails of lipid molecules during transfer is essential for preventing the exposure of these tails to the aqueous cellular environment, thereby facilitating efficient lipid transport (Avula et al., 2021). The binding sites of nsLTPs exhibit such flexibility that they have even been shown to accommodate volatiles in Petunia hybrida (Liao et al., 2023). This research provides mechanistic insights into how hydrophobic compounds, including volatiles, cross the hydrophilic cell wall. Recent advancements have unveiled the molecular details of lipid transport by yeast LTP VPS13, providing crucial insights into its function (Adlakha et al., 2022; Melia and Reinisch, 2022; Park et al., 2016; Neiman, 2024). This work revealed that VPS13 forms a bridge containing a hydrophobic channel at membrane contact sites to facilitate bulk glycerolipid transport between organelles. This bridge-conduit model is highly relevant for plants, where proteins like ATG2 likely employ a similar mechanism to mediate the large-scale lipid flux required for processes such as autophagy-related membrane biogenesis and constitutive inter-organelle lipid homeostasis (Osawa et al., 2019; Valverde et al., 2019; McEwan and Ryan, 2022). These lessons from model organisms such as yeast provide important clues into lipid transport mechanisms of actions in plants.
5.3 Energetics and thermodynamics of lipid transfer
nsLTPs facilitate non-vesicular lipid transfer, which is distinct from the energy-dependent vesicular transport (Wong et al., 2017). The energetics of lipid transfer by nsLTPs involve both enthalpic and entropic contributions (Wong et al., 2019). The binding of lipid molecules to nsLTPs is primarily driven by hydrophobic interactions, which release water molecules from the hydrophobic cavity, resulting in a favorable entropic gain (Mouritsen, 2013). nsLTPs and LTPs function by reducing the energy barrier for lipids to leave bilayers, as the lipid is transferred into the hydrophobic cavity of the protein rather than into the aqueous phase (Reinisch and Prinz, 2021). The formation of disulfide bridges within the 8CM also contributes to the stability of the lipid-protein complex, providing an enthalpic contribution to the binding process (Maldonado et al., 2002). In general, metabolic energy is not required for LTPs to transfer lipids. LTPs facilitate lipid transfer by lowering the energy needed for a lipid to detach from a membrane. This occurs because the lipid enters the protein’s hydrophobic cavity rather than the less favorable aqueous environment (Reinisch and Prinz, 2021). Active lipid transport, particularly against a concentration gradient, necessitates energy. ATP-binding cassette (ABC) transporters are often involved in these processes, using ATP hydrolysis to power the movement of lipids (Christensen and Kolomiets, 2011; Roston et al., 2012; Sakuragi and Nagata, 2023; Norris et al., 2024).
6 nsLTPs and plant resistance
Plant resistance to pathogens relies on a multi-layered defense strategy that includes physical barriers, direct antimicrobial action, and sophisticated stress response mechanisms. nsLTPs and, more broadly, LTPs, play significant and interconnected roles in all these aspects of plant immunity (García-Olmedo et al., 1995; Gao et al., 2022; Carella et al., 2017).
The first line of defense is often the plant cuticle, a hydrophobic layer composed primarily of cutin and waxes that prevent pathogen penetration and reduce water loss (Arya et al., 2021; Zhao et al., 2020). Crucially, nsLTPs and LTPs are essential for the formation and maintenance of this barrier. They transport the precursors of cutin and waxes from their sites of synthesis within the endoplasmic reticulum (ER) to the plant surface (Jacq et al., 2017; DeBono et al., 2009; Wang et al., 2022). Specific examples include Arabidopsis thaliana LTPG1 and LTPG2, which transport wax precursors (DeBono et al., 2009).
Beyond their structural role in cuticle formation, nsLTPs exhibit direct antimicrobial activity against a wide range of pathogens, including fungi, bacteria, and viruses (Cammue et al., 1995; Christensen and Kolomiets, 2011; Kirubakaran et al., 2008; Avula et al., 2021). This activity stems from the inherent ability of nsLTPs to bind and transport lipids. By doing so, nsLTPs can bind to and disrupt pathogen membranes, limiting pathogen growth and spread (Diz et al., 2006; Regente et al., 2005; Chen et al., 2024). The mechanisms of antimicrobial action are diverse, including membrane permeabilization, pore formation, ion leakage, inhibition of pathogen enzymes, and disruption of crucial metabolic pathways (Melnikova et al., 2022). Notably, the antimicrobial activity of nsLTPs often shows specificity toward particular pathogens and not self, suggesting that they may recognize and target specific lipid molecules present in pathogen membranes (Terras et al., 1992; Maximiano and Franco, 2021; Cammue et al., 1995).
In addition to direct antimicrobial effects of nsLTPs, plants must balance ROS production and scavenging during pathogen attack. While ROS are produced as part of the plant’s defense response, excessive accumulation can lead to oxidative damage. nsLTPs have been found to play a role in responding to ROS stress in plants. One feature has been the discovery that nsLTPs contribute to ROS scavenging, thereby maintaining cellular redox balance under stress. This would tend to disfavor biotrophs such as the powdery mildew causing fungi (Ascomycetes), the Basidiomycetes which cause fungal rusts, and Oomycetes causing downy mildew diseases (Mapuranga et al., 2022) but favor resistance to fungi such as Botrytis cinerea and Sclerotinia sclerotiorum which show hemibiotrophic or necrotrophic lifestyles (Barna et al., 2012). Some nsLTPs, such as Arabidopsis thaliana AtLTP4.4 was shown to enhance resistance to the hemibiotrophic fungal pathogen Fusarium graminearum (F.g.), possess antioxidant activity and directly scavenge ROS (McLaughlin et al., 2015, 2021). In Brassica napus, LTP-II was found to function as a ROS scavenger and antioxidant in guard cells, as evidenced by ltp-II mutant plants showing higher ROS and lower free thiols after flg22 treatment. Furthermore, treating ltp-II mutants with the ROS scavenger catalase restored stomatal aperture differences, confirming the role of LTP-II in mitigating oxidative stress (Balmant et al., 2021). nsLTPs have also been found to negatively regulate resistance by impacting the abundance of H2O2 and reducing the hypersensitive response (HR) to fungal infection. Virus-induced gene silencing (VIGS) of wheat ortholog of the Arabidopsis DEFECTIVE IN INDUCED RESISTANCE 1 (DIR1) gene, TaDIR1–2 significantly increased H2O2, SA, and resistance to the biotrophic stripe rust pathogen, Puccinia striiformis f. sp. tritici (Pst) (Ahmed et al., 2017). Other nsLTPs were found to contribute indirectly by regulating the expression of antioxidant enzymes or by transporting lipids that are involved in ROS detoxification (Song et al., 2023; Xu et al., 2018; Zhu et al., 2023; Li et al., 2024; Hsouna et al., 2021).
Finally, nsLTPs are integral to plant defense signaling pathways. Plant hormones, including abscisic acid (ABA), SA, ethylene, and methyl jasmonate (MeJA), have demonstrated involvement in regulating the expression of nsLTP genes (Missaoui et al., 2022). The ability of nsLTPs to influence the synthesis, transport, and signaling of phytohormones such as SA, JA, and ethylene—central regulators of plant defense—can establish a feedback loop that modulates the overall defense response (Chen et al., 2017). Specifically, nsLTPs facilitate the transport of lipid-based secondary messengers, such as phosphatidic acid (PA) and lysophosphatidylcholine (LPC), which are known to activate downstream defense responses (Lee et al., 1997). Furthermore, nsLTPs are key players in SAR, transporting lipid-derived signals in the phloem to uninfected tissues, effectively priming systemic defenses. DIR1 is a well-studied example of a protein that facilitates long-distance signaling within the SAR pathway (Cameron et al., 2016; Lascombe et al., 2008). nsLTPs are pivotal in activating SAR (Carella et al., 2017; Lascombe et al., 2008; Champigny et al., 2013), functioning as carriers for lipid-based signaling molecules, including precursors of JA and SA. While DIR1’s precise cargo and mechanism remain under investigation (Maldonado et al., 2002), this signaling, along with nsLTP activity, contributes to defense-related gene expression (Blein et al., 2002) (potentially through interaction with elicitin receptors (Buhot et al., 2001) and modulation of JA and SA pathways. Furthermore, nsLTPs contribute to the reinforcement of plant membranes during SAR. The SAR-inducing activity of signals such as azelaic acid (AzA), dehydroabietinal (DA), and glycerol-3-phosphate (G3P) are dependent on functional DIR1 (Duan et al., 2024). Collectively, these functions highlight the crucial involvement of nsLTPs in multiple aspects of the plant immune system.
6.1 The role of lipids and nsLTPs in plant immunity
Mechanistically, nsLTPs orchestrate plant immunity through a multifaceted strategy. nsLTPs have been shown to play a role in immune responses against fungi, bacteria, and viruses (Wang et al., 2004; Molina et al., 1993; Ji et al., 2024; Zhu et al., 2023; Zhao et al., 2020; Sun et al., 2008). This includes direct antimicrobial actions, such as disrupting pathogen membranes (Missaoui et al., 2022), a process that may involve permeabilization, pore formation (Regente et al., 2005), and interaction with specific lipids like PA (Madni et al., 2020). They also directly inhibit pathogen growth by interfering with crucial metabolic pathways (McLaughlin et al., 2021; Molina et al., 1993; Jiang et al., 2013; Schmitt et al., 2018), including inhibition of glycosidases (Resende et al., 2023). A nsLTP, Ca-LTP1 isolated from Capsicum annuum is able to inhibit α-amylase in vitro (Bessiatti Fava Oliveira et al., 2025; Kirubakaran et al., 2008), and a nsLTP isolated from Ginkgo biloba can function to inhibit the aspartic acid proteinase, pepsin and the cysteine proteinase papain (Sawano et al., 2008). Finally, nsLTPs integrate with other defense pathways, collaborating with ROS signaling and phytohormone signaling. Their antioxidant properties (Aldakhil et al., 2023; Balmant et al., 2021) are integral to maintaining redox homeostasis and preventing excessive ROS-mediated damage.
Lipids are essential for plant immunity, serving diverse roles beyond their structural function in membranes. They form protective barriers such as the cuticle, composed of cutin and waxes, which prevent pathogen entry and water loss, serving as a first line of defense against pathogens (Zhao et al., 2020; Jacq et al., 2017). Additionally, membrane lipids, including phospholipids and sphingolipids, maintain cellular integrity and are crucial for proper cellular function and immune responses (Dodds et al., 2024; Pretorius et al., 2021; Situ et al., 2024; Sarowar et al., 2009; Guo et al., 2022). Lipids also act as signaling molecules, with phosphatidic acid (PA) and diacylglycerol (DAG) being key examples. PA is a lipid second messenger involved in various stress responses, including the activation of defense-related genes and the production of ROS (Yao and Xue, 2018). DAG plays a role in protein kinase activation and downstream defense signaling pathways (Kalachova et al., 2022). The transport of these signaling lipids by LTPs is crucial for modulating defense responses and intercellular communication. Lipids also serve as precursors for antimicrobial compounds, such as oxylipins and phytoalexins, which combat pathogen infection (Cavaco et al., 2021; Kachroo and Kachroo, 2009). Membrane lipids, such as phospholipids and sphingolipids, are essential for maintaining membrane fluidity and integrity, which are critical for proper cellular function and immune responses. Additionally, certain lipids act as precursors for defense compounds, such as oxylipins and phytoalexins, which are antimicrobial compounds produced by plants to combat pathogen infection (Pretorius et al., 2021; Blée, 1998).
nsLTPs directly support these diverse roles, functioning as important components of plant defense against pathogens (Gao et al., 2022; Kader, 1996; Shah and Chaturvedi, 2009). A survey of the literature highlights the complexity of lipid-mediated plant immunity but also the opportunity to better understand plant-pathogen interactions. Advances in understanding pathogen recognition, signaling pathways, pathogenesis-related proteins, and lipid homeostasis, particularly in relation to nsLTPs, offer a framework for designing bioengineered crops with enhanced resistance to pathogens (Dodds et al., 2024).
nsLTPs are also recognized to play critical roles in the symbiotic relationships between plants and microorganisms, a prime example being the role of nsLTPs in legume–rhizobia symbioses (Zhou et al., 2019; Gao et al., 2022; Gasser et al., 2023; Wei et al., 2022; Chen et al., 2023). In Medicago truncatula (Barrel Medic), the nsLTP MtN5 displays a dual role by acting as an antimicrobial agent for plant defense while also being essential for promoting symbiotic root nodulation with Sinorhizobium meliloti (Pii et al., 2009). The expression level of the MtN5 gene directly dictates the quantity of root nodules, as silencing the gene reduced nodule formation by 50% while overexpressing it resulted in a threefold increase in nodule formation. A nodule-specific nsLTP in Chinese Milk Vetch (Astragalus sinicus), AsE246, was found to play an important role in the symbiosis with nitrogen-fixing bacterium Mesorhizobium huakuii (Lei et al., 2014). Similar to the findings for MtN5 in M. truncatula, the expression level of AsE246 was shown to be a key determinant of symbiotic efficacy. Overexpression of AsE246 promoted an increase in nodule formation while silencing the gene resulted in a suite of symbiotic defects, including reduced nodulation, lower lipid content within nodules, impaired nitrogen fixation, and aberrant symbiosome development. Later, AsE246, was found to directly bind the high temperature protein G (HtpG) from M. huakuii (Zhou et al., 2019), impacting the lipid profile of the root nodules. Gasser et al. (2023) provides a recent review of nsLTPs in nitrogen-fixing symbiosis. Interestingly, the role of nsLTPs in common bean (Phaseolus vulgaris) and soybean (Glycine max) nodule development is not clear. RT-qPCR expression analysis of roots inoculated with rhizobia has shown the upregulation of PvLTPs (PvLTPd.4, PvLTPd.6, PvLTPd.10, and PvLTPg.11) and GmLTPs GmLTPd.1 and GmLTP1.1 but functional studies are needed (Fonseca-García et al., 2021).
The lipid biology underlying plant defense mechanisms is complex and dynamic. During pathogen challenge, plants rapidly remodel their membrane composition, produce signaling lipids, and mobilize antimicrobial lipid compounds (Pretorius et al., 2021). nsLTPs orchestrate many of these responses by ensuring the appropriate trafficking of lipid molecules to their required locations for membrane repair, signal transmission, or direct pathogen confrontation (Santos-Silva et al., 2023; Boutrot et al., 2008; Blein et al., 2002; Buhot et al., 2001; Cheng et al., 2004; de Oliveira Carvalho and Gomes, 2007; Edqvist et al., 2018).
6.2 Antimicrobial activity of nsLTPs
Some nsLTPs exhibit direct antimicrobial activity, functioning as antimicrobial peptides (AMPs) that target and disrupt pathogen membranes (Gao et al., 2022; Santos-Silva et al., 2023; Amador et al., 2021). Molina et al. (1993) provided an early study on the antifungal nature of nsLTPs. nsLTPs purified from barley (Hordeum vulgare) (Cw18 and Cw21) and maize (Zea mays) leaves (Cw41) showed activity against phytopathogenic bacteria and fungi. In vitro assays using a recombinant nsLTP Ltp 3F1 cloned from wheat (Sumai 3), was shown to have broad antifungal properties, including Aspergillus species, Candida species, Fusarium species, Rhizoctonia solani, Pyricularia oryzae, Alternaria species, Botrytis cinerea, Pythium debaryanum, Phytophthora infestans, and Magnaporthe poae (Kirubakaran et al., 2008).
The rice LTP110 protein was shown to directly inhibit fungal spores of P. oryzae (Ge et al., 2002). Using site-directed mutagenesis, the authors showed that the Cys50–Cys89 disulfide bridge was not essential for antimicrobial activity (Ge et al., 2003). Enhanced resistance to Alternaria solani and B. cinerea was observed in transgenic Arabidopsis plants that expressed the TdLTP4 gene (Safi et al., 2015). Further research found that recombinant TdLTP4 protein had a broad antimicrobial spectrum and was effective against bacteria and fungi, including S. aureus, L. monocytogenes, F. oxysporum, F. g. (Hsouna et al., 2021).
McLaughlin et al. (2021) showed that recombinant AtLTP4.4 expressed in Pichia pastoris was able to inhibit F. g. in zone of inhibition assays, possibly by disrupting membranes based on its ability to bind and transfer lipids, leading to alterations in membrane permeability and integrity. Antifungal activity was also noted for the recombinant Brassica rapa BrLTP2.1 protein when tested against F. oxysporum and P. syringae pv. tomato. Site-directed mutagenesis of select cysteine residues, for example, Cys69 of BrLTP2.1 impacted the antifungal nature of the protein (Schmitt et al., 2018). Recently, a potato (S. tuberosum nsLTP, SpLTPa was shown to bind to and disrupt the plasma membrane (PM) of the oomycete pathogen P. infestans but not the PM of the potato cells (Chen et al., 2024). The ability of plant LTPs, including Coffea canephora Cc-LTP1 and Cc-LTP2, to permeabilize the membranes of Candida albicans, a fungal pathogen associated with nosocomial infections, was also observed (Zottich et al., 2011; Bard et al., 2016).
Do nsLTPs specifically interact with fungal membranes? There are multiple lines of evidence that there are direct interactions. The Ha-AP10 nsLTP from common sunflower (Helianthus annuus) was the first nsLTP shown to permeabilize fungal cell membranes, specifically inhibiting F. solani spore germination (Regente and De La Canal, 2000). Ha-AP10 was subsequently shown, using fluorescent probes and a liposome leakage assay, to interact directly with phospholipids and this produces the fungicidal effect (Regente et al., 2005). Using chitin affinity chromatography, chitin-binding nsLTPs were isolated from Capsicum chinense were identified and characterized (Gonçalves et al., 2024b). These proteins were found to have antifungal activity against Candida and Fusarium (Gonçalves et al., 2024a, c). Chen et al. (2024) found that PI(3,5)P2 and PI(3)P phosphoinositides competitively inhibit the binding of StLTPa to fungal plasma membranes, diminishing the inhibitory effect of StLTPa. Madni et al. (2020) also demonstrated that a nsLTP isolated from eggplant (Solanum melongena) can disrupt fungal membranes. They used the SYTOX Green uptake assay, which measures membrane integrity, to show this disruption (characterized as “bleaching” or increased porosity) via the dye entering the fungal cells upon exposure to the nsLTP.
6.3 Role of nsLTPs in cuticle development and integrity
The plant cuticle is composed of a complex mixture of lipids, including cutin and waxes, which create a hydrophobic layer that prevents water loss and protects against pathogen entry. The lipids on the plant surface consist of cutin and waxes (very-long-chain fatty acids (VLCFAs) and their derivatives) (Serrano et al., 2014). Cutin consists of a polymeric network formed by C16 and C18 fatty acids, cross-linked through ester bonds. nsLTPs transport cutin monomers and wax to the plant surface for cuticle assembly and deposition (Jacq et al., 2017; Zhao et al., 2020; Missaoui et al., 2022; Nakamura et al., 2016). nsLTPs facilitate the transport and deposition of lipid molecules essential for cuticle formation by binding and transferring various lipids, such as fatty acids, phospholipids, and glycolipids, from their synthesis sites in the endoplasmic reticulum to epidermal cells. This process ensures a continuous supply of lipids necessary for cuticle development and maintenance. Some evidence of this has come from overexpression studies in plants. For instance, Arabidopsis plants engineered to overexpress an nsLTP from saltwater cress (Thellungiella salsuginea), TsnsLTP4, showed an increase in epicuticular wax deposition (Sun et al., 2015). Likewise, a nsLTP in tomato, SlLTPG3, was found to play a crucial role in transporting cuticular wax and cutin to the tomato fruit surface, contributing to enhanced cuticle thickness and reduced permeability. Overexpression of SlLTPG3 resulted in increased cuticular wax and cutin accumulation, leading to delayed fruit softening and an extended shelf life in tomatoes compared to wild-type plants (Wang et al., 2022).
Another notable example is Arabidopsis LTPG (Type G nsLTP), which is a glycosylphosphatidylinositol-anchored lipid transfer protein identified as crucial for exporting lipids to the plant surface. It plays a significant role in transporting cutin and wax precursors, thereby contributing to the proper formation and integrity of the cuticle. Research by DeBono et al. (2009) highlighted the importance of Arabidopsis LTPG in maintaining cuticle integrity and function. nsLTPs contribute to cuticle development through their hydrophobic cavity, which allows for stable binding and transport of lipid molecules, as well as targeted delivery facilitated by signal peptide sequences that guide these proteins to specific subcellular locations. Disrupting the nsLTP GPI-anchored 1 (LTPG1) gene in Arabidopsis altered the plant’s cuticular lipid composition and ultrastructure, negatively impacting its immunity as shown by increased susceptibility to fungal infection (Alternaria brassicicola) (Lee et al., 2009).
Additionally, nsLTPs play a role in the plant’s defense mechanisms against abiotic stresses such as drought and salinity by reducing water loss and protecting against desiccation (Nakamura et al., 2016). Their antimicrobial properties further reinforce the cuticle’s barrier function against pathogen attack (Kuźniak and Gajewska, 2024). In summary, nsLTPs, including Arabidopsis LTPG, are integral to the development and maintenance of the plant cuticle, enhancing the plant’s ability to withstand environmental stresses and pathogen invasion through the continuous transport of lipid molecules required for cuticle formation.
The broad classes of nsLTP1 and nsLTP2 are known to participate in the transport of phospholipids and glycolipids, contributing to the maintenance of membrane integrity and the formation of lipid-based defense barriers (Aldakhil et al., 2023; Gangadhar et al., 2016; Gasser et al., 2023; Jacq et al., 2017; Schmitt et al., 2018). The integrity of the cuticle is directly linked to plant resistance as a compromised cuticle significantly increases susceptibility to both pathogen invasion and dehydration. Studies have revealed that an nsLTP from Arabidopsis plays a structural role in maintaining the adhesion between the hydrophobic cuticle and the hydrophilic cell wall (Jacq et al., 2017; Hairat et al., 2018). During fungal infection, fungal cutinases release cutin which can activate defense responses (Serrano et al., 2014; Arya et al., 2021). In rice, the application of cutin monomer 16-hydroxypalmitic acid (HPA) induces OsLTP5 expression along with other defense genes (Kim et al., 2008), indicating that plants respond to cuticle damage.
6.4 Protein binding by nsLTPs: beyond lipid transport
While nsLTPs are traditionally associated with their ability to bind and transport lipids, emerging evidence underscores their versatile role in protein-protein interactions. This protein-binding capability extends beyond simple lipid binding and exchange, offering a direct mechanism for nsLTPs to modulate cellular processes and interfere with pathogen virulence.
A prominent example of nsLTP protein binding involves their interaction with alpha-amylases. Studies have demonstrated that nsLTPs can bind to and inhibit the enzymatic activity of these hydrolytic enzymes (da Silva et al., 2018b; Bessiatti Fava Oliveira et al., 2025; Zottich et al., 2011). This suggests a resistance mechanism against insect alpha-amylases, potentially disrupting their function and thereby inhibiting carbohydrate digestion, which would serve as a potent plant defense mechanism against herbivory (da Silva et al., 2018b; Saxena et al., 2023). Subsequent research using C. canephora Cc-LTP1 and a recombinant Vigna unguiculata Vu-LTP1 confirmed that nsLTPs cause potent inhibition of insect intestinal alpha-amylases, leading to reduced larval development in the cowpea weevil, Callosobruchus maculatus and supporting the role of these nsLTPs as an active defense mechanism against insect attack by disrupting their digestive processes (da Silva et al., 2018a).
The ability of nsLTPs to modulate carbohydrate metabolism could also play a crucial role in plant stress responses to other threats, such as fungal or bacterial challenge, by influencing the availability of energy and building blocks needed for defense or recovery. A recent study found a potential link between nsLTPs, rice germination rates, and carbohydrate levels in seeds. CRISPR knockouts of OsLTPL23 negatively impacted rice germination rates and resulted in significantly lower starch levels and higher soluble sugar levels in the edited seeds, the authors hypothesizing that OsLTPL23 may have alpha-amylase inhibitor activity (Li et al., 2023). Further research is needed to understand the exact role of alpha-amylase inhibition by nsLTPs in plants and if there is a role in disease resistance, specifically how this might affect the availability of sugars crucial for both plant defense and pathogen growth.
More recently, the ability of nsLTPs to directly interact with and modulate pathogen-derived proteins has garnered significant attention. Although a mechanism was not identified, antiviral and antiproliferative activities have been demonstrated for a nsLTP derived from bunch-flowered daffodil (Narcissus tazetta) (Ooi et al., 2008). However, more recent work has shown how nsLTPs exert antiviral action. For instance, the cowpea Vigna unguiculata LTP1 has been shown to bind to and inhibit the proteolytic activity of the cysteine protease encoded by the CPMV (Ji et al., 2024). This interaction directly interferes with a key viral protein necessary for replication and spread, showcasing a novel antiviral defense mechanism mediated by nsLTPs. The implications of this protein-binding capacity extends beyond viral defense. It is plausible that nsLTPs can interact with other pathogen-derived proteins, such as bacterial or fungal effectors, thereby disrupting their virulence functions (Guo et al., 2022). Exploring the diversity of pathogen proteins targeted by nsLTPs is crucial for understanding the full scope of their contribution to plant immunity. Cataloging the contents of nsLTPs in plants using proteomic and lipomics would help better understand the scope of nsLTP involvement in plant disease response.
6.5 Lipid dynamics and signaling in plant immunity: the orchestrating role of nsLTPs
Lipids are dynamic participants in plant immunity, functioning beyond structural roles to generate potent signaling molecules during plant-pathogen interactions (Li et al., 2024). The lipid landscape itself is a dynamic battleground where plants modify lipids for defense, while pathogens exploit them for survival. Upon perception of microbial threats, plants rapidly remodel their lipidome, producing signaling lipids such as phosphatidic acid (PA), lysophosphatidic acid (LPA), oxylipins like jasmonic acid (JA) and 12oxo-phytodienoic acid (OPDA), and sphingolipids such as ceramides (Kuźniak and Gajewska, 2024; Pretorius et al., 2021). These lipid mediators activate diverse pathways, including ROS production, calcium signaling via phosphoinositides, and programmed cell death, collectively shaping robust defense responses. Sphingolipids, like ceramides, induce programmed cell death (PCD), and glycosyl inositolphosphoryl ceramides (GIPCs) contribute to membrane structure and are known to be bound to nsLTPs (Berkey et al., 2012; Gonzalez-Klein et al., 2021). Phosphoinositides (PIs), such as PI4P and PI(4,5)P2, serve as precursors for second messengers like IP3, regulating calcium signaling (Boss and Im, 2012). Additionally, fatty acid derived signals like N-acylethanolamines (NAEs) modulate stress responses (Chapman, 2004; Blancaflor et al., 2014; Hu et al., 2023). These lipid mediators initiate complex signaling cascades, ultimately leading to defense gene expression.
nsLTPs emerge as central regulators of these lipid-mediated defenses, contributing to antimicrobial lipid delivery, membrane reinforcement, lipid signal transduction (e.g., PA, LPC), and long-distance signaling (e.g., DIR1 in SAR) (Santos-Silva et al., 2023; Amador et al., 2021; Chen et al., 2017; Salminen et al., 2016). They facilitate the targeted movement of signaling lipids, reinforce membrane barriers, and orchestrate long-distance immune communication, notably through SAR (Chen et al., 2017; Finkina et al., 2016; Li et al., 2024). The Arabidopsis DIR1 protein exemplifies this function, mediating the movement of lipid-based systemic signals (Carella et al., 2017; Champigny et al., 2013; Lascombe et al., 2008). Beyond mere transport, nsLTPs actively amplify signaling cascades by mobilizing lipid messengers, including PA and lysophosphatidylcholine (LPC). By controlling the spatial and temporal distribution of these lipids, nsLTPs integrate localized and systemic immune responses.
nsLTPs operate within a broader lipid signaling network through coordinated interactions with lipid kinases, phosphatases, and membrane transporters. By delivering lipid substrates to diacylglycerol kinases and phosphoinositide kinases, nsLTPs potentiate lipid signal amplification, while their collaboration with lipid phosphatases ensures signal resolution (Lev, 2010). In parallel, they complement energy-dependent export mediated by ATP-binding cassette (ABC) transporters, particularly during defense-induced membrane remodeling (Roston et al., 2012). At membrane contact sites (MCS), LTPs contribute to lipid exchange between organelles, linking lipid homeostasis with stress responses (Lahiri et al., 2015; Michaud and Jouhet, 2019; Prinz et al., 2020; Zhang et al., 2022). Thus, nsLTPs and LTPs serve not merely as passive shuttles but as dynamic integrators of lipid signaling, tightly coordinating immune activation and cellular resilience.
6.6 Mechanisms of action of nsLTPs in plant defense
nsLTPs play diverse roles in plant defense, as illustrated by the following specific examples. A. thaliana DIR1, a nsLTP, is crucial for SAR by facilitating long-distance transport of lipid-based signals (Champigny et al., 2013). Medicago sativa MsLTP1 exhibits broad-spectrum antimicrobial activity, potentially via a pore-forming mechanism in pathogen membranes (Barashkova et al., 2023). AtLTP4.4 enhances resistance to Fusarium head blight through antifungal and antioxidant activities (McLaughlin et al., 2021). Nicotiana benthamiana NbLTP1 boosts immunity against tobacco mosaic virus (TMV) by upregulating SA biosynthesis and downstream signaling components like NPR1 (Zhu et al., 2023).
nsLTPs interact with pattern recognition receptors (PRRs), triggering pattern-triggered immunity (PTI). They are also involved in activating mitogen-activated protein kinase (MAPK) pathways, crucial for defense signaling, partly by transporting lipid-based secondary messengers like phosphatidic acid (PA). Furthermore, nsLTPs integrate with hormonal signaling (SA, JA, ET), enhancing SA biosynthesis and signaling and modulating JA and ET pathways.
The expression of nsLTP genes is regulated by biotic and abiotic stress factors. PAMP recognition by PRRs initiates signaling cascades that activate defense-related genes, including nsLTPs. Key transcription factors (WRKY, MYB, NAC) bind to specific cis-regulatory elements (e.g., W-box, MYB-box) in nsLTP gene promoters. Post-translational modifications, such as glycosylation and phosphorylation, are also important. Glycosylation can enhance nsLTP stability, activity, and localization, while phosphorylation can alter conformation, affecting interactions and activity.
The nsLTPs often contain N-terminal signal peptides and are directed to specific cellular locations like the extracellular apoplast (via the secretory pathway), endoplasmic reticulum, the chloroplast, or vacuoles (Boutrot et al., 2008; Liu et al., 2010; Wang et al., 2012; Nishimura et al., 2008; Chiu et al., 2020). The presence of numerous nsLTP genes raises questions about functional redundancy. While some nsLTPs show overlapping functions in lipid binding and transport, others have specialized roles in cuticle formation, pathogen defense, or reproduction, potentially providing robust defense. Some nsLTPs exhibit direct antimicrobial activity, while others are crucial for SAR by facilitating long-distance transport of lipid signals or maintaining cuticle integrity. The redox-sensitive nsLTP, LTP-II, was shown to be important for guard cell closure in response to the bacterial protein flg22 (Balmant et al., 2021).
SAR is a long-lasting, broad-spectrum immune response triggered by an initial localized pathogen attack. Following local infection, mobile signals are generated and transported to distal tissues, priming them for enhanced defense upon subsequent pathogen exposure. Lipids, lipid-derived molecules, and lipid-associated proteins, particularly nsLTPs, are key mediators of this systemic immune communication.
Among nsLTPs, Arabidopsis DIR1 is a well-characterized example essential for SAR establishment (Champigny et al., 2013; David et al., 2021; Lascombe et al., 2008; Carella et al., 2017). DIR1 facilitates the movement of lipid-based signals through the plant vascular system. Loss of DIR1 function impairs systemic defenses despite normal local responses. Structural and biochemical studies suggest DIR1 carries lipid molecules, likely glycerolipids or phospholipid-derived messengers, crucial for priming distal tissues. While the exact cargo is still under investigation, candidates include lipid derivatives like azelaic acid (AzA) and dehydroabietinal (DA) (Champigny et al., 2013; Lascombe et al., 2008; Carella et al., 2017; David et al., 2021).
Beyond DIR1, other lipid-associated proteins contribute to SAR. AZI1 (Azelaic Acid Induced 1), an LTP-like protein from the hybrid proline-rich protein (HyPRP) family, is implicated in amplifying SAR signaling, potentially by facilitating azelaic acid mobilization (Priya Reddy and Oelmüller, 2024; Gao et al., 2022). AZI1 functions in concert with DIR1, suggesting cooperative action of multiple lipid carriers for robust signal fidelity. The transport of lipid signals by nsLTPs likely occurs through the apoplast and phloem, enabling rapid, energy-efficient dissemination of immune signals.
At the molecular level, SAR involves transcriptional reprogramming, including systemic upregulation of PR genes and increased SA biosynthesis (Vidhyasekaran, 2015). nsLTPs likely interface with these pathways by delivering lipid signals that trigger SA accumulation and by reinforcing membrane and cell wall integrity, enhancing overall stress resilience. Thus, through lipid transport and signal integration, nsLTPs act as key orchestrators of SAR, linking local pathogen recognition to global plant-wide immune readiness.
6.7 nsLTPs: versatile proteins with ROS scavenging potential
ROS are highly reactive molecules produced as byproducts of normal cellular metabolism. While ROS are essential signaling molecules in various physiological processes, including defense, excessive accumulation of ROS leads to oxidative stress, causing damage to lipids, proteins, and DNA (Mittler, 2017; Shetty et al., 2008; Wang et al., 2024). nsLTPs mitigate oxidative stress during pathogen attack through direct ROS scavenging or by indirectly upregulating antioxidant genes (McLaughlin et al., 2021, 2015; Yang et al., 2023; Gangadhar et al., 2016; Xu et al., 2018; Wang et al., 2014; Balmant et al., 2021; Hsouna et al., 2021; Safi et al., 2015). In Nicotiana benthamiana, overexpression of the type-I nsLTP, NbLTP1, activated genes related to ROS scavenging and enhanced resistance to TMV (Zhu et al., 2023). Under thermal stress, tobacco plants overexpressing NtLTPI.38 exhibited a significant upregulation of genes encoding antioxidant enzymes and thermal stress-related proteins (Song et al., 2023). Similarly, in NtLTP4 overexpression lines, several important ROS-scavenging enzyme encoding genes, such as SOD, APX, CAT, and GST, dramatically increased (Xu et al., 2018). Overexpression of NtLTP25 significantly increased the enzyme activities of CAT, GST, APX, and SOD, as well as the transcription levels of their encoding gene (Li et al., 2024).
The ROS scavenging activity of nsLTPs has profound implications for plant immunity. By neutralizing ROS, nsLTPs mitigate oxidative damage, protecting cellular components and ensuring the proper function of essential cellular processes. Furthermore, because ROS also act as signaling molecules, nsLTPs can fine-tune ROS levels, influencing the activation of defense pathways and preventing excessive, damaging oxidative stress. Ultimately, by scavenging ROS, nsLTPs enhance the overall stress tolerance of plants, allowing them to better cope with adverse environmental conditions, including pathogen attack. Understanding the mechanisms of their ROS scavenging activity provides valuable insights for improving plant resistance to both biotic and abiotic stresses.
nsLTPs contribute significantly to the plant’s antioxidant defense system. Several mechanisms underlie their ROS scavenging activity. First, the cysteine-rich nature of nsLTPs provides abundant free thiol groups (Wu et al., 2011). These thiol groups can, theoretically, directly interact with and neutralize ROS, such as H2O2 and superoxide radicals . Second, nsLTPs impact the degree of lipid peroxidation, a chain reaction initiated by ROS that severely damages cellular membranes (Priya Reddy and Oelmüller, 2024; Levesque-Tremblay et al., 2009). For instance, overexpression of NtLTP25 in tobacco significantly reduced the degree of lipid peroxidation, as indicated by a reduction in malondialdehyde (MDA) levels in the leaves (Li et al., 2024). By stabilizing lipid membranes, nsLTPs may prevent the propagation of lipid peroxidation and protect cellular integrity (Song et al., 2023).
Several examples illustrate the ROS-scavenging capabilities of nsLTPs. Barley LTP1 exhibits strong antioxidant activity, effectively scavenging ROS and protecting cells from oxidative damage (Cai et al., 2019; Stanislava, 2007; Wu et al., 2011). CmnsLTP6.9, from Chinese chestnut, regulates ROS scavenging and remodels lipid profiles, contributing to stress tolerance (Xiao et al., 2023). Similarly, tobacco NtLTPI.38 displays antioxidant capacity, suggesting its involvement in ROS detoxification (Yang et al., 2023). To better understand how the sugar beet responses to abiotic stress, quantitative redox proteomics (iodoTMTRAQ) was used to identify and quantify redox posttranslational modifications (PTMs). Several proteins were identified to be chemically reduced during salt stress, including a nsLTP (A0A0K9RNM7), a novel discovery (Li et al., 2021a). Another example is the LTP-II, which was found to be redox-responsive in response to flg22 during stomatal closure and played a role in plant resistance to Pseudomonas syringe pv. tomato DC3000 (Balmant et al., 2021).
How might the cysteines in nsLTPs function to scavenge or otherwise impact ROS accumulation and thereby provide some measure of resistance against oxidative stress? Molecular details from the study of the serine protease inhibitor (serpin) superfamily protein maspin in mouse mammary cells may provide useful information (Mahajan et al., 2013). Maspin is rich in redox-sensitive cysteine residues, containing a total of eight, similar to that found in nsLTPs. It was shown that structurally exposed cysteine thiols were oxidized in the presence of sulfenic acid and that the protein binds to glutathione S-transferase (GST). Another research group, following up on that work, showed that oxidized maspin increases GST, which may lead to the inhibition of oxidative stress-induced ROS generation (Yin et al., 2005). These compelling studies provide a powerful framework for investigating nsLTPs in plants and their potential to modulate ROS. A critical question emerges: does oxidized nsLTP, analogous to maspin, bind to GST in plants, thereby directly influencing its activity and the plant’s capacity to manage oxidative stress? If so, that might help explain why glutathione levels were found to be higher in transgenic Arabidopsis overexpressing AtLTP4.4 and why ROS levels were significantly lower upon exposure to oxidizing agents such as trichothecenes and H2O2 (McLaughlin et al., 2015). Other nsLTPs may contribute indirectly to ROS accumulation by regulating the expression of antioxidant enzymes or by transporting lipids that are involved in ROS detoxification (Song et al., 2023; Xu et al., 2018; Zhu et al., 2023; Li et al., 2024; Hsouna et al., 2021).
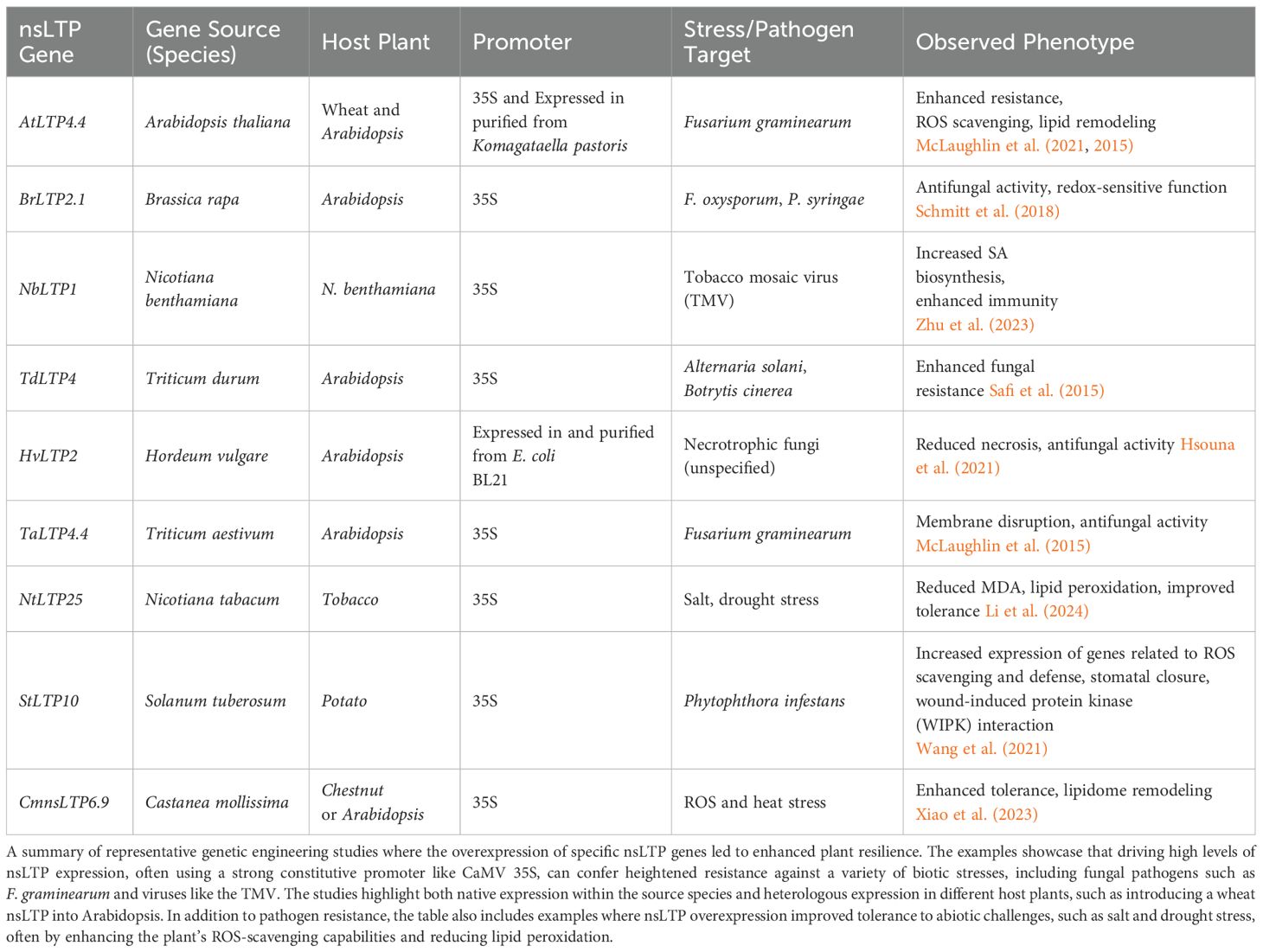
7 Genetic engineering of nsLTPs for enhanced plant disease resistance
Genetic engineering offers a powerful strategy for leveraging the inherent disease-protective capabilities of nsLTPs to enhance plant resistance against a broad spectrum of pathogens. Enhanced disease resistance in transgenic plants has been achieved by strategically modifying non-specific lipid transfer proteins (nsLTPs). These modifications include modulating expression levels, altering functional properties, and controlling spatiotemporal expression patterns. Heterologous expression has proved useful in the study and application of nsLTPs. For example, expression of the nsLTP gene LJAMP2 from motherwort in transgenic poplar trees significantly enhanced their resistance to the fungal pathogens Alternaria alternata and Colletotrichum gloeosporioides (Jia et al., 2010). Overexpression of Triticum durum TdLTP2 in A. thaliana enhanced resistance against fungal pathogens Aspergillus niger, F. graminearum, B. cinerea, and A. solani. Transgenic Arabidopsis expressing barley LTP2 showed reduced necrotic effects caused by infection by Pseudomonas (Molina et al., 1993). Transgenic Arabidopsis overexpressing pepper CALTP1 showed resistance to P. syringae and Botrytis cinerea (Diz et al., 2011). Expression of NbLTP1 using a 35S promoter in tobacco enhanced resistance to tobacco mosaic virus (TMV) (Zhu et al., 2023).
Several key approaches can be pursued to enhance disease resistance with nsLTPs. Overexpression of native nsLTP genes, often driven by strong constitutive promoters (e.g., CaMV 35S, maize ubiquitin) or pathogen-inducible promoters, can significantly increase resistance. Two nsLTP genes, nsLTP4.4 and nsLTP4.5 were identified from a screen of an activation tagged A. thaliana population for resistance to trichothecin, a type B trichothecene in the same class as deoxynivalenol (DON). Overexpression of nsLTP4.4 provided resistance to Tcin in Arabidopsis (McLaughlin et al., 2015). Overexpression of the tomato SlLTPG1 gene enhanced resistance to Botrytis cinerea (Liu et al., 2024). Introduction of heterologous nsLTP genes from other plant species or even non-plant organisms can confer novel defense capabilities. An example is the introduction of the Arabidopsis AtLTP4.4 gene into wheat, which enhanced resistance to F.g (McLaughlin et al., 2021). nsLTP overexpression studies cited in this review are shown in Table 4. Creation of chimeric nsLTP variants, combining functional domains or structural modules from different nsLTPs, offers the potential to generate proteins with enhanced or broadened antimicrobial properties. Rational design and protein engineering techniques are crucial for optimizing these chimeric nsLTPs. Targeted expression and subcellular localization using tissue-specific promoters and subcellular targeting signals allows for precise control over where and when nsLTPs are active, minimizing potential off-target effects and maximizing their efficacy. Finally, genome editing technologies, particularly CRISPRCas9, enable precise modification of endogenous nsLTP genes, potentially enhancing their expression or function without the introduction of foreign DNA.
nsLTP overexpression has also been observed to improve abiotic stress resistance. Overexpression of native nsLTPs driven the CaMV 35S promoter in tobacco has increased salt and drought stress resistance (Xu et al., 2018). Overexpressing the Chinese chestnut (Castanea mollissima) protein CmnsLTP6.9L in Arabidopsis enhanced tolerance to both osmotic and drought stress (Xiao et al., 2023). Increased reactive oxygen species (ROS)-scavenging enzyme activity (SOD and POD) was recorded. Similarly, Xu et al. (2018) overexpressed the native NtLTP4 in N. tabacum to reveal enhanced resistance to salt and drought stresses.
Despite the promise, several challenges and limitations must be addressed. Allergenicity is of significant concern, as nsLTPs are known allergens (e.g., peach Pru p 3 and wheat Tri a 14) (Egger et al., 2010; Gonzalez-Klein et al., 2021; Ruano-Zaragoza et al., 2021; Safi et al., 2019). The low molecular mass and high thermal and proteolytic stability of nsLTPs enable this class of proteins to maintain their tertiary structure and provide a means for the proteins to reach the immune system in a biologically intact form, thereby eliciting an immunoglobulin E (IgE)-mediated hypersensitivity reaction in sensitized individuals. Efforts have been made to generate hypoallergenic nsLTPs in the context of immunotherapy (Gómez-Casado et al., 2013) but these modifications would need to be tested for their impact on nsLTP function (Scheurer and Schülke, 2018). Rigorous allergenicity assessments are essential to ensure the safety of engineered crops for human consumption (Hoffmann-Sommergruber, 2002; Singh and Bhalla, 2008; Salcedo et al., 2007). Pathogen evasion is another critical consideration; pathogens may evolve mechanisms to overcome nsLTP-mediated resistance, necessitating continuous monitoring and development of new strategies.
Deployment strategies can utilize either traditional transgenic approaches by introducing nsLTP genes from other species, or CRISPR-based genome editing, allowing for precise modifications of the endogenous genes. CRISPR offers advantages in terms of precision and potentially fewer regulatory hurdles. To maximize effectiveness and durability, nsLTP-based strategies can be combined with other control methods. Combining nsLTPs with other AMPs, such as defensins or thionins, can broaden the spectrum of antimicrobial activity. Combining nsLTPs with genes that enhance membrane repair and homeostasis (e.g, AtCHL for damaged chloroplasts; Levesque-Tremblay et al., 2009) can improve the plant’s ability to maintain cellular integrity during pathogen attack. Integrating nsLTPs into broader integrated pest management (IPM) strategies, which include cultural practices, biological control, and judicious use of chemical treatments, offers a holistic approach that reduces reliance on any single method.
8 LTPs and membrane contact sites
Membrane contact sites (MCSs) are dynamic junctions where organelles connect through protein tethers, serving as critical hubs for the non-vesicular transfer of lipids, ions like calcium, and small molecules between organelles (Scorrano et al., 2019; Michaud and Jouhet, 2019). These specialized zones facilitate efficient molecular transfer by allowing organelles to closely appose without fusing, forming essential microdomains (Lahiri et al., 2015). This inter-organelle communication is vital for maintaining cellular homeostasis (Michaud and Jouhet, 2019; Wang et al., 2023; Man et al., 2024). While extensively studied in yeast and mammalian systems for their roles in calcium signaling, mitochondrial dynamics, and lipid metabolism (Michaud and Jouhet, 2019; Banday et al., 2022), the understanding of MCS function in plants remains limited, representing a significant knowledge gap.
LTPs are key players at MCSs, binding lipids within hydrophobic pockets and facilitating their transfer across the aqueous cytosol between membranes (Michaud and Jouhet, 2019). The close proximity of organelle membranes at MCSs significantly shortens the diffusion distance for LTPs, thereby accelerating lipid transfer (Zhang et al., 2022). LTPs are highly conserved and classified by lipid-binding specificity, including sphingolipid-, sterol-, and phospholipid-transfer proteins. Notable examples include oxysterol-binding protein (OSBP) and its related proteins (ORPs), which mediate sterol and phosphoinositide transfer, and ceramide transfer protein (CERT), involved in ceramide transfer between the ER and Golgi (Renna et al., 2024; Nakatsu and Kawasaki, 2021; Kumagai and Hanada, 2019). In plants, LTPs at MCSs contribute significantly to both lipid homeostasis and defense mechanisms by facilitating the transfer of various lipids, including phospholipids and glycolipids (Michaud and Jouhet, 2019).
In plants, MCSs are crucial for efficient lipid trafficking and signaling, supporting cellular homeostasis and mediating responses to both biotic and abiotic stresses. The presence of plant-specific organelles, such as plastids, introduces unique MCS functionalities linked to photosynthesis, nutrient assimilation, and stress responses (Yao et al., 2023). During pathogen attack, MCSs are hypothesized to be critical for the rapid exchange of signaling molecules that trigger defense responses (Michaud and Jouhet, 2019; Paul and Tiwari, 2023). For instance, they likely mediate the transfer of lipids like PA and phosphatidylinositol phosphates (PIPs), which are critical in plant defense. However, the precise molecular mechanisms and specific proteins involved in these plant MCS-mediated processes is not well understood. Recently, LaBrant (2025) identified chloroplast MCS proteins using a transient expression screen in N. benthamiana. The Arabidopsis nsLTP GPI-anchored 20 (LTPG20, At3g22620) was found to localize to chloroplasts in this study. Additional research will be necessary to verify LTPG20 functions in chloroplast lipid trafficking at MCSs but this type of research highlights efforts to catalog nsLTPs and LTPs at plant MCSs.
Mechanisms of stress resistance involving MCSs are also being discovered in mammalian systems. Under stress conditions, mammalian (HeLa) mitochondria can generate oxidized lipids, which are efficiently transferred to the ER via LTPs localized at mitochondria-ER MCSs (Sassano et al., 2022; Shiiba et al., 2025). These contact sites facilitate the non-vesicular transfer of peroxidized lipids, enabling rapid responses to oxidative stress (Shiiba et al., 2025). Similar LTP mechanisms may operate in plants during pathogen-induced stress; for example, certain virulence factors, such as trichothecene mycotoxins, are known to induce mitochondrial stress (Bin-Umer et al., 2014). It remains to be determined if damaged plant mitochondria utilize a similar mechanism to manage oxidized lipids during biotic stress. This is an emerging aspect of plant biology with likely important roles in plant responses to pathogens.
9 Future research directions
Understanding the dynamic interplay of nsLTPs in lipid transport and signaling offers valuable insights into their application in developing disease-resistant crops. Biotechnological strategies that leverage the functional capabilities of nsLTPs hold significant potential to enhance plant defense mechanisms. Moreover, emerging research reveals that nsLTPs serve roles far beyond immunity, including oxidative stress mitigation and long-distance signaling, positioning them as key molecular players in the broader landscape of cellular lipid metabolism.
To unlock the full potential of nsLTPs in plant defense and enable their effective deployment in agricultural biotechnology, several strategic research directions must be pursued. Central to this is the detailed elucidation of their structural and functional properties. High-resolution techniques such as X-ray crystallography and cryo-electron microscopy are essential to visualize nsLTPs in complex with diverse lipid ligands, thereby advancing our understanding of ligand specificity and binding mechanisms. Biochemical and biophysical characterization, using methods such as isothermal titration calorimetry (ITC) or surface plasmon resonance (SPR), can yield critical data on lipid binding affinities and kinetics. In parallel, site-directed mutagenesis can identify key residues involved in lipid binding, transfer, and potential antimicrobial activity, guiding efforts in protein engineering and synthetic biology.
A comprehensive understanding lipid dynamics and trafficking is also vital. Mass spectrometry-based lipidomics can profile shifts in lipid composition during pathogen attack, illuminating how specific lipids are mobilized and potentially interact with nsLTPs. This approach can also delineate how pathogen-derived effectors disrupt lipid signaling networks or alter membrane composition to suppress host defenses. Integrating lipidomics with functional genomics and proteomics will further clarify the downstream consequences of pathogen-mediated lipid manipulation. Furthermore, utilizing lipidomics in organelle trafficking studies may reveal the nuanced roles nsLTPs play in inter-organelle lipid exchange, particularly under stress conditions.
Further investigation into the antifungal and antiviral properties of nsLTPs, their crosstalk with other defense signaling pathways (e.g., salicylic acid, jasmonic acid, ethylene), and the evolutionary strategies pathogens employ to circumvent nsLTP-mediated immunity are also critical areas of future inquiry. Mechanistic studies are needed to determine how nsLTPs exert antifungal and antiviral effects, identify targets, and evaluate the spectrum of their protective activity. Understanding how pathogen effectors directly target or evade nsLTP function can inform the design of more durable, resistance-conferring interventions.
Several promising avenues exist for biotechnological applications. The rational design of engineered nsLTP variants guided by structural and functional insights offers significant promise. Variants with enhanced lipid-binding affinity, broader antimicrobial spectra, or increased biochemical stability can be tailored for targeted expression in specific tissues (e.g., epidermis, vascular tissue) or subcellular compartments (e.g., apoplast, chloroplast). Such targeted deployment can optimize defense responses while minimizing potential fitness trade-offs. Combinatorial strategies that integrate nsLTP-based interventions with traditional resistance genes or biocontrol agents may yield robust, multi-layered plant protection systems.
In addition, the role of nsLTPs in membrane repair and oxidative stress management warrants focused investigation, particularly in relation to chloroplast integrity under pathogen-induced damage. Studies examining nsLTP interactions with specific chloroplast lipids and antioxidant pathways may uncover novel mechanisms of stress resilience. These integrated efforts will be instrumental in developing crops with superior adaptability and disease resistance. Rigorous field validation and a thorough evaluation of biosafety and regulatory considerations will be essential before widespread adoption of nsLTP-engineered crops. If successful, such advances will contribute meaningfully to agricultural sustainability and global food security.
10 Conclusion
nsLTPs are emerging as pivotal regulators of plant defense, orchestrating lipid transport, membrane remodeling, and signaling cascades in response to pathogenic threats. Their multifunctional nature, which encompasses antimicrobial activity, ROS scavenging, and modulation of systemic defense pathways, which are summarized in Table 5, underscores their vital role in maintaining cellular homeostasis during biotic stress.
Equally important is their function in sustaining lipid homeostasis. nsLTPs facilitate membrane integrity, organelle crosstalk particularly at MCS, and dynamic lipid signaling, all of which are essential for robust immune responses. By shuttling lipids between compartments, they ensure both structural stability and signaling precision.
Genetic manipulation of nsLTP expression has demonstrated promise in conferring broad-spectrum resistance across a range of crop species. Exploring the interplay between nsLTPs and lipid signaling in plant disease contexts presents a compelling avenue for next-generation crop improvement strategies.
A multidisciplinary approach, which integrates lipidomics, advanced imaging, systems biology, and plant pathology will be critical to fully exploit the potential of nsLTPs. This integrated framework can inform the design of novel, sustainable crop protection strategies that are both scientifically rigorous and commercially viable. Continued innovation and research are imperative to establish nsLTPs as central components of modern agricultural biotechnology and to address the growing global demand for resilient, high-yield crops.
Author contributions
JM: Conceptualization, Funding acquisition, Investigation, Writing – original draft. NT: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by a grant from the U.S. Department of Agriculture Agricultural Research Service (number 59-0206-2-134) to JM in cooperation with the United States Wheat and Barley Scab Initiative. Additional funding came from the Rutgers Center for Turfgrass Science.
Acknowledgments
We thank members of the Plant Biology Department for discussions about the role of nsLTPs in plant defense in model organisms, turf, and cereal crops.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2025.1640465/full#supplementary-material
Supplementary Table 1 | Chronological comparison of the major classification systems developed for the plant nsLTP superfamily, summarizing the key literature from (Kader, 1996) to Huang et al. (2023). This timeline illustrates the evolution of the defining criteria used to categorize these proteins, beginning with early systems based on fundamental physical properties like molecular mass. The table tracks the progression to more sophisticated, multifaceted approaches that incorporate sequence homology, the precise spacing of the 8CM, and genomic features such as intron positioning. It also documents the recent expansion of the nsLTP family to include novel lineages discovered in algae, which was made possible by adapting broader bioinformatic search parameters. This comprehensive overview serves as a foundational reference for understanding the various nsLTP types and the scientific rationale behind their classification as discussed throughout this review.
Supplementary Table 2 | A detailed overview of the nsLTP superfamily across the plant kingdom, organized by major evolutionary lineages. The table provides a species-specific look at the number of nsLTPs identified, the major types present, and their key structural and functional characteristics. Important evolutionary patterns are highlighted, such as the existence of specific nsLTP types (like Type D) in early land plants like bryophytes, the general absence of Types I and II in those early lineages, and the recent discovery of a distinct nsLTP lineage in green algae that challenges previous assumptions about their origins. This comparative summary illustrates the diversification of the nsLTP family as plants adapted to new environments and pathogens.
References
Adlakha J., Hong Z., Li P., and Reinisch K. M. (2022). Structural and biochemical insights into lipid transport by VPS13 proteins. J. Cell Biol. 221, e202202030. doi: 10.1083/jcb.202202030
Ahmed S. M., Liu P., Xue Q., Ji C., Qi T., Guo J., et al. (2017). TaDIR1-2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici. Front. Plant Sci. 8, 521. doi: 10.3389/fpls.2017.00521
Aldakhil T., Alshammari S. O., Siraj B., El-Aarag B., Zarina S., Salehi D., et al. (2023). The structural characterization and bioactivity assessment of nonspecific lipid transfer protein 1 (nsLTP1) from caraway (Carum carvi) seeds. BMC Complement. Med. Ther. 23, 254. doi: 10.1186/s12906-023-04083-9
Amador V. C., Santos-Silva C., Vilela L. M. B., Oliveira-Lima M., de Santana Rêgo M., Roldan-Filho R. S., et al. (2021). Lipid transfer proteins (LTPs)—structure, diversity and roles beyond antimicrobial activity. Antibiotics 10, 1281. doi: 10.3390/antibiotics10111281
Anaconda (2016). Anaconda Software Distribution. Available online at: https://anaconda.com (Accessed July 15, 2025).
Arya G. C., Sarkar S., Manasherova E., Aharoni A., and Cohen H. (2021). The plant cuticle: an ancient guardian barrier set against long-standing rivals. Front. Plant Sci. 12, 663165. doi: 10.3389/fpls.2021.663165
Avula K., Singh B., Kumar P. V., and Syed G. H. (2021). Role of lipid transfer proteins (LTPs) in the viral life cycle. Front. Microbiol. 12, 673509. doi: 10.3389/fmicb.2021.673509
Balmant K. M., Lawrence S. R. II, Duong B. V., Zhu F., Zhu N., Nicklay J., et al. (2021). Guard cell redox proteomics reveals a role of lipid transfer protein in plant defense. J. Proteomics 242, 104247. doi: 10.1016/j.jprot.2021.104247
Banday Z. Z., Cecchini N. M., Speed D. J., Scott A. T., Parent C., Hu C. T., et al. (2022). Friend or foe: hybrid proline-rich proteins determine how plants respond to beneficial and pathogenic microbes. Plant Physiol. 190, 860–881. doi: 10.1093/plphys/kiac263
Barashkova A. S., Smirnov A. N., Zorina E. S., and Rogozhin E. A. (2023). Diversity of cationic antimicrobial peptides in black cumin (Nigella sativa l.) seeds. Int. J. Mol. Sci. 24, 8066. doi: 10.3390/ijms24098066
Bard G., Zottich U., Souza T., Ribeiro S., Dias G., Pireda S., et al. (2016). Purification, biochemical characterization, and antimicrobial activity of a new lipid transfer protein from Coffea canephora seeds. Genet. Mol. Res. 15, 1–16. doi: 10.4238/gmr15048859
Barna B., Fodor J., Harrach B., Pogány M., and Király Z. (2012). The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 59, 37–43. doi: 10.1016/j.plaphy.2012.01.014
Berkey R., Bendigeri D., and Xiao S. (2012). Sphingolipids and plant defense/disease: the “death” connection and beyond. Front. Plant Sci. 3, 68. doi: 10.3389/fpls.2012.00068
Bessiatti Fava Oliveira A. P., Resende L. M., da Silva M. S., de Azevedo dos Santos L., Carvalho A. O., Chaves R. P., et al. (2025). Lipid transfer proteins (LTPs) partially purified from Capsicum chinense jacq. seeds: Antifungal properties and α-amylase inhibitory activity. Protein J. 44, 1–12. doi: 10.1007/s10930-025-10256-x
Bin-Umer M. A., McLaughlin J. E., Butterly M. S., McCormick S., and Tumer N. E. (2014). Elimination of damaged mitochondria through mitophagy reduces mitochondrial oxidative stress and increases tolerance to trichothecenes. Proc. Natl. Acad. Sci. 111, 11798–11803. doi: 10.1073/pnas.1403145111
Bittrich S., Segura J., Duarte J. M., Burley S. K., and Rose Y. (2024). RCSB protein data bank: exploring protein 3D similarities via comprehensive structural alignments. Bioinformatics 40, btae370. doi: 10.1093/bioinformatics/btae370
Blancaflor E. B., Kilaru A., Keereetaweep J., Khan B. R., Faure L., and Chapman K. D. (2014). N-Acylethanolamines: lipid metabolites with functions in plant growth and development. Plant J. 79, 568–583. doi: 10.1111/tpj.12427
Blée E. (1998). Phytooxylipins and plant defense reactions. Prog. Lipid Res. 37, 33–72. doi: 10.1016/S0163-7827(98)00004-6
Blein J.-P., Coutos-Thevenot P., Marion D., and Ponchet M. (2002). From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 7, 293–296. doi: 10.1016/S1360-1385(02)02284-7
Blum M., Andreeva A., Florentino L., Chuguransky S., Grego T., Hobbs E., et al. (2024). Interpro: the protein sequence classification resource in 2025. Nucleic Acids Res. 53, D444–D456. doi: 10.1093/nar/gkae1082
Boss W. F. and Im Y. J. (2012). Phosphoinositide signaling. Annu. Rev. Plant Biol. 63, 409–429. doi: 10.1146/annurev-arplant-042110-103840
Boutrot F., Chantret N., and Gautier M.-F. (2008). Genome-wide analysis of the rice and arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsltp genes by EST data mining. BMC Genomics 9, 1–19. doi: 10.1186/1471-2164-9-86
Buhot N., Douliez J.-P., Jacquemard A., Marion D., Tran V., Maume B., et al. (2001). A lipid transfer protein binds to a receptor involved in the control of plant defence responses. FEBS Lett. 509, 27–30. doi: 10.1016/S0014-5793(01)03116-7
Cai L., Brennan C. S., Yang H., Li W., and Zhao H. (2019). Evolution of oxidative and structural characteristics of proteins, especially lipid transfer protein 1 (LTP1) in beer during forced-ageing. Int. J. Food Sci. Technol. 54, 3166–3174. doi: 10.1111/ijfs.14250
Cameron R. K., Carella P., Isaacs M., Champigny M., Merl-Pham J., Dey S., et al. (2016). Using DIR1 to investigate long-distance signal movement during systemic acquired resistance. Can. J. Plant Pathol. 38, 19–24. doi: 10.1080/07060661.2016.1147497
Cammue B. P. A., Thevissen K., Hendriks M., Eggermont K., Goderis I. J., Proost P., et al. (1995). A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 109, 445–455. doi: 10.1104/pp.109.2.445
Carella P., Kempthorne C. J., Wilson D. C., Isaacs M., and Cameron R. K. (2017). Exploring the role of DIR1, DIR1-like and other lipid transfer proteins during systemic immunity in Arabidopsis. Physiol. Mol. Plant Pathol. 97, 49–57. doi: 10.1016/j.pmpp.2016.12.005
Cavaco A. R., Matos A. R., and Figueiredo A. (2021). Speaking the language of lipids: the crosstalk between plants and pathogens in defence and disease. Cell. Mol. Life Sci. 78, 4399–4415. doi: 10.1007/s00018-021-03791-0
Champigny M. J., Isaacs M., Carella P., Faubert J., Fobert P. R., and Cameron R. K. (2013). Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 4, 230. doi: 10.3389/fpls.2013.00230
Chapman K. D. (2004). Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog. Lipid Res. 43, 302–327. doi: 10.1016/j.plipres.2004.03.002
Charvolin D., Douliez J.-P., Marion D., Cohen-Addad C., and Pebay-Peyroula E. (1999). The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 a˚ resolution. Eur. J. Biochem. 264, 562–568. doi: 10.1046/j.1432-1327.1999.00667.x
Chen X., Feng J., Li Z., Feng H., Song C., Cai L., et al. (2024). Lipid transfer protein StLTPa enhances potato disease resistance against different pathogens by binding and disturbing the integrity of pathogens plasma membrane. Plant Biotechnol. J. 22, 1913–1925. doi: 10.1111/pbi.14310
Chen D., Li D., Li Z., Song Y., Li Q., Wang L., et al. (2023). Legume nodulation and nitrogen fixation require interaction of DnaJ-like protein and lipid transfer protein. Plant Physiol. 193, 2164–2179. doi: 10.1093/plphys/kiad437
Chen Y., Ma J., Zhang X., Yang Y., Zhou D., Yu Q., et al. (2017). A novel non-specific lipid transfer protein gene from sugarcane (NsLTPs), obviously responded to abiotic stresses and signaling molecules of SA and MeJA. Sugar Tech. 19, 17–25. doi: 10.1007/s12355-016-0431-4
Cheng C.-S., Samuel D., Liu Y.-J., Shyu J.-C., Lai S.-M., Lin K.-F., et al. (2004). Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry 43, 13628–13636. doi: 10.1021/bi048873j
Chiapparino A., Maeda K., Turei D., Saez-Rodriguez J., and Gavin A.-C. (2016). The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog. Lipid Res. 61, 30–39. doi: 10.1016/j.plipres.2015.10.004
Chiu L.-Y., Chen I.-H., Hsu Y.-H., and Tsai C.-H. (2020). The lipid transfer protein 1 from Nicotiana benthamiana assists bamboo mosaic virus accumulation. Viruses 12, 1361. doi: 10.3390/v12121361
Christensen S. A. and Kolomiets M. V. (2011). The lipid language of plant–fungal interactions. Fungal Genet. Biol. 48, 4–14. doi: 10.1016/j.fgb.2010.05.005
da Silva F. C. V., do Nascimento V. V., Fernandes K. V., MaChado O. L. T., da Silva Pereira L., Gomes V. M., et al. (2018a). Recombinant production and α-amylase inhibitory activity of the lipid transfer protein from Vigna unguiculata (l. walp.) seeds. Process Biochem. 65, 205–212. doi: 10.1016/j.procbio.2017.10.018
da Silva F. C. V., do Nascimento V. V., MaChado O. L. T., Pereira L., d. S., Gomes V. M., et al. (2018b). Insight into the α-amylase inhibitory activity of plant lipid transfer proteins. J. Chem. Inf. Model. 58, 2294–2304. doi: 10.1021/acs.jcim.8b00540
David L., Kang J., Nicklay J., Dufresne C., and Chen S. (2021). Identification of DIR1-dependant cellular responses in guard cell systemic acquired resistance. Front. Mol. Biosci. 8, 746523. doi: 10.3389/fmolb.2021.746523
DeBono A., Yeats T. H., Rose J. K., Bird D., Jetter R., Kunst L., et al. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21, 1230–1238. doi: 10.1105/tpc.108.064451
de Oliveira Carvalho A. and Gomes V. M. (2007). Role of plant lipid transfer proteins in plant cell physiology- A concise review. Peptides 28, 1144–1153. doi: 10.1016/j.peptides.2007.03.004
Diz M. S., Carvalho A. O., Ribeiro S. F., Da Cunha M., Beltramini L., Rodrigues R., et al. (2011). Characterisation, immunolocalisation and antifungal activity of a lipid transfer protein from chili pepper (Capsicum annuum) seeds with novel α-amylase inhibitory properties. Physiol. Plant. 142, 233–246. doi: 10.1111/j.1399-3054.2011.01464.x
Diz M. S., Carvalho A. O., Rodrigues R., Neves-Ferreira A. G. C., Da Cunha M., Alves E. W., et al. (2006). Antimicrobial peptides from chilli pepper seeds causes yeast plasma membrane permeabilization and inhibits the acidification of the medium by yeast cells. Biochim. Biophys. Acta (BBA) General Subj. 1760, 1323–1332. doi: 10.1016/j.bbagen.2006.04.010
Dodds P. N., Chen J., and Outram M. A. (2024). Pathogen perception and signaling in plant immunity. Plant Cell 36, 1465–1481. doi: 10.1093/plcell/koae020
Douliez J.-P., Jégou S., Pato C., Mollé D., Tran V., and Marion D. (2001). Binding of two monoacylated lipid monomers by the barley lipid transfer protein, LTP1, as viewed by fluorescence, isothermal titration calorimetry and molecular modelling. Eur. J. Biochem. 268, 384–388. doi: 10.1046/j.1432-1033.2001.01889.x
Dowler S., Kular G., and Alessi D. R. (2002). Protein lipid overlay assay. Science’s STKE 2002, pl6–pl6. doi: 10.1126/stke.2002.129.pl6
Duan M., Bao L., Eman M., Han D., Zhang Y., Zheng B., et al. (2024). The ectopic expression of the MpDIR1(t) gene enhances the response of plants from Arabidopsis thaliana to biotic stress by regulating the defense genes and antioxidant flavonoids. Plants 13, 2692. doi: 10.3390/plants13192692
Dufayard J.-F., Bocs S., Guignon V., Lariviere D., Louis A., Oubda N., et al. (2021). RapGreen, an interactive software and web package to explore and analyze phylogenetic trees. NAR Genomics Bioinf. 3, lqab088. doi: 10.1093/nargab/lqab088
Edqvist J., Blomqvist K., Nieuwland J., and Salminen T. A. (2018). Plant lipid transfer proteins: are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 59, 1374–1382. doi: 10.1194/jlr.R083139
Edstam M. M., Viitanen L., Salminen T. A., and Edqvist J. (2011). Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 4, 947–964. doi: 10.1093/mp/ssr019
Egger M., Hauser M., Mari A., Ferreira F., and Gadermaier G. (2010). The role of lipid transfer proteins in allergic diseases. Curr. Allergy Asthma Rep. 10, 326–335. doi: 10.1007/s11882-010-0128-9
Fang C., Wu S., Li Z., Pan S., Wu Y., An X., et al. (2023). A systematic investigation of lipid transfer proteins involved in male fertility and other biological processes in maize. Int. J. Mol. Sci. 24, 1660. doi: 10.3390/ijms24021660
Finkina E., Melnikova D., Bogdanov I., and Ovchinnikova T. (2016). Lipid transfer proteins as components of the plant innate immune system: structure, functions, and applications. Acta Naturae 8, 47–61. doi: 10.32607/20758251-2016-8-2-47-61
Fleury C., Gracy J., Gautier M.-F., Pons J.-L., Dufayard J.-F., Labesse G., et al. (2019). Comprehensive classification of the plant non-specific lipid transfer protein superfamily towards its sequence–structure– function analysis. PeerJ 7, e7504. doi: 10.7717/peerj.7504
Fonseca-García C., Solis-Miranda J., Pacheco R., and Quinto C. (2021). Non-specific lipid transfer proteins in legumes and their participation during root-nodule symbiosis. Front. Agron. 3, 660100. doi: 10.3389/fagro.2021.660100
Gangadhar B. H., Sajeesh K., Venkatesh J., Baskar V., Abhinandan K., Yu J. W., et al. (2016). Enhanced tolerance of transgenic potato plants over-expressing non-specific lipid transfer protein-1 (StnsLTP1) against multiple abiotic stresses. Front. Plant Sci. 7, 1228. doi: 10.3389/fpls.2016.01228
Gao H., Ma K., Ji G., Pan L., and Zhou Q. (2022). Lipid transfer proteins involved in plant-pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 23, 1815–1829. doi: 10.1111/mpp.13264
Gao M. and Yang H. (2018). VPS13: A lipid transfer protein making contacts at multiple cellular locations. J. Cell Biol. 217, 3322–3324. doi: 10.1083/jcb.201808151
García-Olmedo F., Molina A., Segura A., and Moreno M. (1995). The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol. 3, 72–74. doi: 10.1016/S0966-842X(00)88879-4
Gasser M., Keller J., Fournier P., Pujic P., Normand P., and Boubakri H. (2023). Identification and evolution of nsLTPs in the root nodule nitrogen fixation clade and molecular response of Frankia to AgLTP24. Sci. Rep. 13, 16020. doi: 10.1038/s41598-023-41117-1
Ge X.-C., Chen J.-C., Lin Y., Sun C.-R., and Cao K.-M. (2002). Expression, purification and function of rice nonspecific lipid transfer protein. Sheng wu hua xue yu Sheng wu wu li xue bao Acta Biochim. Biophys. Sin. 34, 83–87.
Ge X., Chen J., Sun C., and Cao K. (2003). Preliminary study on the structural basis of the antifungal activity of a rice lipid transfer protein. Protein Eng. 16, 387–390. doi: 10.1093/protein/gzg055
Gómez-Casado C., Garrido-Arandia M., Gamboa P., Blanca-López N., Canto G., Varela J., et al. (2013). Allergenic characterization of new mutant forms of Pru p 3 as new immunotherapy vaccines. J. Immunol. Res. 2013, 385615. doi: 10.1155/2013/385615
Gonçalves G. R., da Silva M. S., Dos Santos L. A., Guimaraes T. Z. A., Taveira G. B., Almeida F. A., et al. (2024a). Structural and functional characterization of new lipid transfer proteins with chitin-binding properties: Insights from protein structure prediction, molecular docking, and antifungal activity. Biochemistry 63, 1824–1836. doi: 10.1021/acs.biochem.4c00124
Gonçalves G. R., de Azevedo dos Santos L., da Silva M. S., Taveira G. B., da Silva T. M., Almeida F. A., et al. (2024b). Purification, structural characterization, and anticandidal activity of a chitinbinding peptide with high similarity to hevein and endochitinase isolated from pepper seeds. Curr. Microbiol. 81, 319. doi: 10.1007/s00284-024-03839-x
Gonçalves G. R., Silva M. S., dos Santos L. A., Resende L. M., Taveira G. B., Guimaraes T. Z. A., et al. (2024c). Chitin-binding peptides from capsicum annuum with antifungal activity and low toxicity to mammalian cells and Galleria mellonella larvae. Pept. Sci. 116, e24338. doi: 10.1002/pep2.24338
Gonzalez-Klein Z., Cuevas-Zuviria B., Wangorsch A., Hernandez-Ramirez G., Pazos-Castro D., Oeo-Santos C., et al. (2021). The key to the allergenicity of lipid transfer protein (LTP) ligands: A structural characterization. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1866, 158928. doi: 10.1016/j.bbalip.2021.158928
Goyal R. K. and Mattoo A. K. (2014). Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 228, 135–149. doi: 10.1016/j.plantsci.2014.05.012
Guo Z.-H., Lung S.-C., Hamdan M. F., and Chye M.-L. (2022). Interactions between plant lipid-binding proteins and their ligands. Prog. Lipid Res. 86, 101156. doi: 10.1016/j.plipres.2022.101156
Hairat S., Baranwal V. K., and Khurana P. (2018). Identification of Triticum aestivum nsLTPs and functional validation of two members in development and stress mitigation roles. Plant Physiol. Biochem. 130, 418–430. doi: 10.1016/j.plaphy.2018.07.030
Hamaï A. and Drin G. (2024). Specificity of lipid transfer proteins: An in vitro story. Biochimie. 227, 85–110. doi: 10.1016/j.biochi.2024.09.007
Han G. W., Lee J. Y., Song H. K., Chang C., Min K., Moon J., et al. (2001). Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution x-ray crystallography. J. Mol. Biol. 308, 263–278. doi: 10.1006/jmbi.2001.4559
Hoffmann-Sommergruber K. (2002). Pathogenesis-related (PR)-proteins identified as allergens. Biochem. Soc. Trans. 30, 930–935. doi: 10.1042/bst0300930
Hsouna A. B., Saad R. B., Dhifi W., Mnif W., and Brini F. (2021). Novel non-specific lipid-transfer protein (TdLTP4) isolated from durum wheat: Antimicrobial activities and anti-inflammatory properties in lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophages. Microb. Pathogen. 154, 104869. doi: 10.1016/j.micpath.2021.104869
Hu Z., Shi J., Feng S., Wu X., Shao S., and Shi K. (2023). Plant N-acylethanolamines play a crucial role in defense and its variation in response to elevated CO2 and temperature in tomato. Hortic. Res. 10, uhac242. doi: 10.1093/hr/uhac242
Huang M.-D., Wu C.-W., Chou H.-Y., Cheng S.-Y., and Chang H.-Y. (2023). The revealing of a novel lipid transfer protein lineage in green algae. BMC Plant Biol. 23, 21. doi: 10.1186/s12870-023-04040-1
Hurlock A. K., Roston R. L., Wang K., and Benning C. (2014). Lipid trafficking in plant cells. Traffic 15, 915–932. doi: 10.1111/tra.12187
Iqbal A., Khan R. S., Shah D. A., Hussain S. A., Abdalla A. N., Wadood A., et al. (2023). Lipid transfer proteins: structure, classification and prospects of genetic engineering for improved disease resistance in plants. Plant Cell Tissue Organ Culture (PCTOC) 153, 3–17. doi: 10.1007/s11240-023-02445-2
Jacq A., Pernot C., Martinez Y., Domergue F., Payré B., Jamet E., et al. (2017). The arabidopsis lipid transfer protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Sci. 8, 263. doi: 10.3389/fpls.2017.00263
Jang C. S., Yim W. C., Moon J.-C., Jung J. H., Lee T. G., Lim S. D., et al. (2008). Evolution of non-specific lipid transfer protein (nsLTP) genes in the Poaceae family: their duplication and diversity. Mol. Genet. Genomics 279, 481–497. doi: 10.1007/s00438-008-0327-4
Ji J., Du S., Wang K., Qi Z., Zhang C., Wang R., et al. (2024). Cowpea lipid transfer protein 1 regulates plant defense by inhibiting the cysteine protease of cowpea mosaic virus. Proc. Natl. Acad. Sci. 121, e2403424121. doi: 10.1073/pnas.2403424121
Jia Z., Gou J., Sun Y., Yuan L., Tang Q., Yang X., et al. (2010). Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP-like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiol. 30, 1599–1605. doi: 10.1093/treephys/tpq093
Jia Y., Li C., Qin J., Xiong M., Gou B., Zhai W., et al. (2025). Xoo effector TalAE73-targeted OsLTPL23 mediates bacterial blight resistance in rice. J. Agric. Food Chem. 73, 6567–6579. doi: 10.1021/acs.jafc.4c12956
Jiang Y., Fu X., Wen M., Wang F., Tang Q., Tian Q., et al. (2013). Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Physiol. Mol. Plant Pathol. 82, 81–87. doi: 10.1016/j.pmpp.2012.11.001
Jones J. D. and Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
José-Estanyol M., Gomis-Rüth F. X., and Puigdomènech P. (2004). The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 42, 355–365. doi: 10.1016/j.plaphy.2004.03.009
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kachroo A. and Kachroo P. (2009). Fatty acid–derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Kader J.-C. (1996). Lipid-transfer proteins in plants. Annu. Rev. Plant Biol. 47, 627–654. doi: 10.1146/annurev.arplant.47.1.627
Kalachova T., Škrabálková E., Pateyron S., Soubigou-Taconnat L., Djafi N., Collin S., et al. (2022). DIACYLGLYCEROL KINASE 5 participates in flagellin-induced signaling in arabidopsis. Plant Physiol. 190, 1978–1996. doi: 10.1093/plphys/kiac354
Kim T. H., Park J. H., Kim M. C., and Cho S. H. (2008). Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. J. Plant Physiol. 165, 345–349. doi: 10.1016/j.jplph.2007.06.004
Kirubakaran S. I., Begum S. M., Ulaganathan K., and Sakthivel N. (2008). Characterization of a new antifungal lipid transfer protein from wheat. Plant Physiol. Biochem. 46, 918–927. doi: 10.1016/j.plaphy.2008.05.007
Kumagai K. and Hanada K. (2019). Structure, functions and regulation of CERT, a lipid-transfer protein for the delivery of ceramide at the ER-Golgi membrane contact sites. FEBS Lett. 593, 2366–2377. doi: 10.1002/1873-3468.13511
Kuźniak E. and Gajewska E. (2024). Lipids and lipid-mediated signaling in plant–pathogen interactions. Int. J. Mol. Sci. 25, 7255.
LaBrant E. W. (2025). Screening for chloroplast inner envelope-thylakoid membrane contact site proteins. Ph.D. thesis. Lincoln, Nebraska: The University of Nebraska-Lincoln.
Lahiri S., Toulmay A., and Prinz W. A. (2015). Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 33, 82–87. doi: 10.1016/j.ceb.2014.12.004
Lascombe M.-B., Bakan B., Buhot N., Marion D., Blein J.-P., Larue V., et al. (2008). The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci. 17, 1522–1530. doi: 10.1110/ps.035972.108
Lau A. M., Bordin N., Kandathil S. M., Sillitoe I., Waman V. P., Wells J., et al. (2024). Exploring structural diversity across the protein universe with The Encyclopedia of Domains. Science 386, eadq4946. doi: 10.1126/science.adq4946
Lee S. B., Go Y. S., Bae H.-J., Park J. H., Cho S. H., Cho H. J., et al. (2009). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 150, 42–54. doi: 10.1104/pp.109.137745
Lee S., Suh S., Kim S., Crain R. C., Kwak J. M., Nam H.-G., et al. (1997). Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 12, 547–556. doi: 10.1046/j.1365-313X.1997.00547.x
Lei L., Chen L., Shi X., Li Y., Wang J., Chen D., et al. (2014). A nodule-specific lipid transfer protein AsE246 participates in transport of plant-synthesized lipids to symbiosome membrane and is essential for nodule organogenesis in Chinese milk vetch. Plant Physiol. 164, 1045–1058. doi: 10.1104/pp.113.232637
Lev S. (2010). Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11, 739–750. doi: 10.1038/nrm2971
Levesque-Tremblay G., Havaux M., and Ouellet F. (2009). The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects arabidopsis against oxidative stress. Plant J. 60, 691–702. doi: 10.1111/j.1365-313X.2009.03991.x
Li J., Gao G., Xu K., Chen B., Yan G., Li F., et al. (2014). Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PloS One 9, e84556. doi: 10.1371/journal.pone.0084556
Li H., Liu Y., Gao W., Zhu J., Zhang H., Wang Z., et al. (2024). Genome-wide characterization of small secreted peptides in Nicotiana tabacum and functional assessment of NtLTP25 in plant immunity. Physiol. Plant. 176, e14436. doi: 10.1111/ppl.14436
Li J., Wang Z., Chang Z., He H., Tang X., Ma L., et al. (2021b). A functional characterization of TaMs1 orthologs in Poaceae plants. Crop J. 9, 1291–1300. doi: 10.1016/j.cj.2020.12.002
Li J., Wang K., Ji M., Zhang T., Yang C., Liu H., et al. (2021a). Cys-SH based quantitative redox proteomics of salt induced response in sugar beet monosomic addition line M14. Botanical Stud. 62, 1–18. doi: 10.1186/s40529-021-00320-x
Li Q., Zhai W., Wei J., and Jia Y. (2023). Rice lipid transfer protein, OsLTPL23, controls seed germination by regulating starch-sugar conversion and ABA homeostasis. Front. Genet. 14, 1111318. doi: 10.3389/fgene.2023.1111318
Liao P., Maoz I., Shih M.-L., Lee J. H., Huang X.-Q., Morgan J. A., et al. (2023). Emission of floral volatiles is facilitated by cell-wall non-specific lipid transfer proteins. Nat. Commun. 14, 330. doi: 10.1038/s41467-023-36027-9
Liu W., Huang D., Liu K., Hu S., Yu J., Gao G., et al. (2010). Discovery, identification and comparative analysis of non-specific lipid transfer protein (nsLtp) family in Solanaceae. Genom. Proteomics Bioinf. 8, 229–237. doi: 10.1016/S1672-0229(10)60024-1
Liu F., Zhang X., Lu C., Zeng X., Li Y., Fu D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. doi: 10.1093/jxb/erv313
Liu J., Zhu J., Yang R., Su C., Wang Z., Meng J., et al. (2024). SlLTPg1, a tomato lipid transfer protein, positively regulates in response to biotic stresses. Int. J. Biol. Macromol. 279, 135219. doi: 10.1016/j.ijbiomac.2024.135219
Madni Z. K., Kumar A., Kumar U., Jaiswal D., and Salunke D. M. (2023). Dynamics of lipid displacement inside the hydrophobic cavity of a nonspecific lipid transfer protein from Solanum melongena. J. Biomol. Struct. Dyn. 41, 5839–5849. doi: 10.2210/pdb00007w9a/pdb
Madni Z. K., Tripathi S. K., and Salunke D. M. (2020). Structural insights into the lipid transfer mechanism of a non-specific lipid transfer protein. Plant J. 102, 340–352. doi: 10.1111/tpj.14627
Mahajan N., Shi H. Y., Lukas T. J., and Zhang M. (2013). Tumor-suppressive maspin functions as a reactive oxygen species scavenger: importance of cysteine residues. J. Biol. Chem. 288, 11611–11620. doi: 10.1074/jbc.M112.410852
Maldonado A. M., Doerner P., Dixon R. A., Lamb C. J., and Cameron R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in arabidopsis. Nature 419, 399–403. doi: 10.1038/nature00962
Man Y., Zhang Y., Chen L., Zhou J., Bu Y., Zhang X., et al. (2024). The VAMP-associated protein VAP27–1 plays a crucial role in plant resistance to ER stress by modulating ER-PM contact architecture in Arabidopsis. Plant Commun. 5, f100929. doi: 10.1016/j.xplc.2024.100929
Mapuranga J., Zhang N., Zhang L., Chang J., and Yang W. (2022). Infection strategies and pathogenicity of biotrophic plant fungal pathogens. Front. Microbiol. 13, 799396. doi: 10.3389/fmicb.2022.799396
Maximiano M. R. and Franco O. L. (2021). Biotechnological applications of versatile plant lipid transfer proteins (LTPs). Peptides 140, 170531. doi: 10.1016/j.peptides.2021.170531
McEwan D. G. and Ryan K. M. (2022). ATG2 and VPS13 proteins: molecular highways transporting lipids to drive membrane expansion and organelle communication. FEBS J. 289, 7113–7127. doi: 10.1111/febs.16280
McLaughlin J. E. (2025). 2 8CM: Python code to identify two or more 8CM motifs in nsLTPs. Available online at: https://github.com/mclaughj23/nsLTP-search-tool-ofFASTA-files-Proteins-containing-eight-cysteine-motifs-8CM-/tree/main.
McLaughlin J. E., Bin-Umer M. A., Widiez T., Finn D., McCormick S., and Tumer N. E. (2015). A lipid transfer protein increases the glutathione content and enhances Arabidopsis resistance to a trichothecene mycotoxin. PloS One 10, e0130204. doi: 10.1371/journal.pone.0130204
McLaughlin J. E., Darwish N. I., Garcia-Sanchez J., Tyagi N., Trick H. N., McCormick S., et al. (2021). A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology 111, 671–683. doi: 10.1094/PHYTO-04-20-0153-R
McMurray W. and Dawson R. (1969). Phospholipid exchange reactions within the liver cell. Biochem. J. 112, 91–108. doi: 10.1042/bj1120091
Melia T. J. and Reinisch K. M. (2022). A possible role for VPS13-family proteins in bulk lipid transfer, membrane expansion and organelle biogenesis. J. Cell Sci. 135, jcs259357. doi: 10.1242/jcs.259357
Melnikova D. N., Finkina E. I., Bogdanov I. V., Tagaev A. A., and Ovchinnikova T. V. (2022). Features and possible applications of plant lipid-binding and transfer proteins. Membranes 13, 2. doi: 10.3390/membranes13010002
Michaud M. and Jouhet J. (2019). Lipid trafficking at membrane contact sites during plant development and stress response. Front. Plant Sci. 10, 2. doi: 10.3389/fpls.2019.00002
Missaoui K., Gonzalez-Klein Z., Pazos-Castro D., Hernandez-Ramirez G., Garrido-Arandia M., Brini F., et al. (2022). Plant non-specific lipid transfer proteins: An overview. Plant Physiol. Biochem. 171, 115–127. doi: 10.1016/j.plaphy.2021.12.026
Molina A., Segura A., and Garcıa-Olmedo F. (1993). Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 316, 119–122. doi: 10.1016/0014-5793(93)81198-9
Morales-Quintana L., Rabert C., Mendez-Yanez A., and Ramos P. (2024). Transcriptional and structural analysis of non-specific lipid transfer proteins modulated by fungal endophytes in Antarctic plants under drought. Physiol. Plant. 176, e14359.
Mouritsen O. G. (2013). “Thermodynamics of lipid interactions,” in Encyclopedia of Biophysics (Berlin, Heidelberg: Springer), 2606–2613.
Nakamura Y. and Li-Beisson Y..eds., (2016). Lipids in Plant and Algae Development (Cham, Switzerland: Springer International Publishing).
Nakatsu F. and Kawasaki A. (2021). Functions of oxysterol-binding proteins at membrane contact sites and their control by phosphoinositide metabolism. Front. Cell Dev. Biol. 9, 664788. doi: 10.3389/fcell.2021.664788
Nazeer M., Waheed H., Saeed M., Ali S. Y., Choudhary M. I., Ul-Haq Z., et al. (2019). Purification and characterization of a nonspecific lipid transfer protein 1 (nsLTP1) from ajwain (Trachyspermum ammi) seeds. Sci. Rep. 9, 4148. doi: 10.1038/s41598-019-40574-x
Neiman A. M. (2024). Membrane and organelle rearrangement during ascospore formation in budding yeast. Microbiol. Mol. Biol. Rev. 88, e00013–e00024. doi: 10.1128/mmbr.00013-24
Neubergerová M. and Pleskot R. (2024). Plant protein–lipid interfaces studied by molecular dynamics simulations. J. Exp. Bot. 75, 5237–5250.
Neuman S. D., Levine T. P., and Bashirullah A. (2022). A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol. 32, 962–974. doi: 10.1016/j.tcb.2022.03.011
Nishimura S., Tatano S., Gomi K., Ohtani K., Fukumoto T., and Akimitsu K. (2008). Chloroplastlocalized nonspecific lipid transfer protein with anti-fungal activity from rough lemon. Physiol. Mol. Plant Pathol. 72, 134–140. doi: 10.1016/j.pmpp.2008.07.003
Norris A. C., Mansueto A. J., Jimenez M., Yazlovitskaya E. M., Jain B. K., and Graham T. R. (2024). Flipping the script: Advances in understanding how and why P4-ATPases flip lipid across membranes. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 187, 119700. doi: 10.1016/j.bbamcr.2024.119700
Ooi L. S., Tian L., Su M., Ho W.-S., Sun S. S., Chung H.-Y., et al. (2008). Isolation, characterization, molecular cloning and modeling of a new lipid transfer protein with antiviral and antiproliferative activities from Narcissus tazetta. Peptides 29, 2101–2109. doi: 10.1016/j.peptides.2008.08.020
Osawa T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., et al. (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288. doi: 10.1038/s41594-019-0203-4
Paggi J. M., Pandit A., and Dror R. O. (2024). The art and science of molecular docking. Annu. Rev. Biochem. 93, 389–410. doi: 10.1146/annurev-biochem-030222-120000
Park J.-S., Thorsness M. K., Policastro R., McGoldrick L. L., Hollingsworth N. M., Thorsness P. E., et al. (2016). Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 27, 2435–2449. doi: 10.1091/mbc.e16-02-0112
Paul P. and Tiwari B. (2023). Organelles are miscommunicating: Membrane contact sites getting hijacked by pathogens. Virulence 14, 2265095. doi: 10.1080/21505594.2023.2265095
Pii Y., Astegno A., Peroni E., Zaccardelli M., Pandolfini T., and Crimi M. (2009). The Medicago truncatula N5 gene encoding a root-specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 22, 1577–1587. doi: 10.1094/MPMI-22-12-1577
Pretorius C. J., Zeiss D. R., and Dubery I. A. (2021). The presence of oxygenated lipids in plant defense in response to biotic stress: A metabolomics appraisal. Plant Signaling Behav. 16, 1989215. doi: 10.1080/15592324.2021.1989215
Prinz W. A., Toulmay A., and Balla T. (2020). The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7–24. doi: 10.1038/s41580-019-0180-9
Priya Reddy Y. and Oelmüller R. (2024). Lipid peroxidation and stress-induced signalling molecules in systemic resistance mediated by azelaic acid/AZELAIC ACID INDUCED1: signal initiation and propagation. Physiol. Mol. Biol. Plants 30, 305–316. doi: 10.1007/s12298-024-01420-1
Regente M. C. and De La Canal L. (2000). Purification, characterization and antifungal properties of a lipid-transfer protein from sunflower (Helianthus annuus) seeds. Physiol. Plant. 110, 158–163. doi: 10.1034/j.1399-3054.2000.110203.x
Regente M., Giudici A., Villalaín J., and de la Canal L. (2005). The cytotoxic properties of a plant lipid transfer protein involve membrane permeabilization of target cells. Lett. Appl. Microbiol. 40, 183–189. doi: 10.1111/j.1472-765X.2004.01647.x
Reinisch K. M. and Prinz W. A. (2021). Mechanisms of nonvesicular lipid transport. J. Cell Biol. 220, e202012058. doi: 10.1083/jcb.202012058
Renna L., Stefano G., Puggioni M. P., Kim S.-J., Lavell A., Froehlich J. E., et al. (2024). ER-associated VAP27–1 and VAP27–3 proteins functionally link the lipid-binding ORP2A at the ER-chloroplast contact sites. Nat. Commun. 15, 6008. doi: 10.1038/s41467-024-50425-7
Resende L. M., de Oliveira Mello E., de Lima Aguieiras M. C., Nagano C. S., Chaves R. P., Taveira G. B., et al. (2023). Inhibition of serine protease, α-amylase and growth of phytopathogenic fungi by antimicrobial peptides from Capsicum chinense fruits. Probiotics Antimicrob. Proteins 15, 502–515. doi: 10.1007/s12602-021-09865-6
Riahi M., Mostajeran A., and Miroliaei M. (2021). Azospirillum brasilense can modulate salt stress in Triticum aestivum via MN052803-LTP regulation and phosphatidylcholines content. Russian J. Plant Physiol. 68, 890–900. doi: 10.1134/S1021443721050150
Roston R. L., Gao J., Murcha M. W., Whelan J., and Benning C. (2012). TGD1,-2, and-3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J. Biol. Chem. 287, 21406–21415. doi: 10.1074/jbc.M112.370213
Ruano-Zaragoza M., Somoza M. L., Jiménez-Rodriguez T. W., Soriano-Gomis V., González-Delgado P., Esteban-Rodriguez A., et al. (2021). Lipid transfer protein sensitization: risk of anaphylaxis and molecular sensitization profile in Pru p 3-sensitized patients. Int. Arch. Allergy Immunol. 182, 425–432. doi: 10.1159/000511977
Safi H., Saibi W., Alaoui M. M., Hmyene A., Masmoudi K., Hanin M., et al. (2015). A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 89, 64–75. doi: 10.1016/j.plaphy.2015.02.008
Safi H., Wangorsch A., Lidholm J., Brini F., Spiric J., Rihs H.-P., et al. (2019). Identification and molecular characterization of allergenic non-specific lipid-transfer protein from durum wheat (Triticum turgidum). Clin. Exp. Allergy 49, 120–129. doi: 10.1111/cea.13271
Sakuragi T. and Nagata S. (2023). Regulation of phospholipid distribution in the lipid bilayer by flippases and scramblases. Nat. Rev. Mol. Cell Biol. 24, 576–596. doi: 10.1038/s41580-023-00604-z
Salcedo G., Sánchez-Monge R., Barber D., and Diaz-Perales A. (2007). Plant non-specific lipid transfer proteins: An interface between plant defence and human allergy. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1771, 781–791. doi: 10.1016/j.bbalip.2007.01.001
Salminen T. A., Blomqvist K., and Edqvist J. (2016). Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244, 971–997. doi: 10.1007/s00425-016-2585-4
Santos-Silva C. A., Ferreira-Neto J. R. C., Amador V. C., Bezerra-Neto J. P., Vilela L. M. B., Binneck E., et al. (2023). From gene to transcript and peptide: A deep overview on non-specific lipid transfer proteins (nsLTPs). Antibiotics 12, 939. doi: 10.3390/antibiotics12050939
Sarowar S., Kim Y. J., Kim K. D., Hwang B. K., Ok S. H., and Shin J. S. (2009). Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 28, 419–427. doi: 10.1007/s00299-008-0653-3
Sassano M. L., Felipe-Abrio B., and Agostinis P. (2022). ER-mitochondria contact sites; a multifaceted factory for Ca2+ signaling and lipid transport. Front. Cell Dev. Biol. 10, 988014. doi: 10.3389/fcell.2022.988014
Sawano Y., Hatano K.-I., Miyakawa T., Komagata H., Miyauchi Y., Yamazaki H., et al. (2008). Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol. 146, 1909–1919. doi: 10.1104/pp.107.111500
Saxena H., Negi H., Keshan R., Chitkara P., Kumar S., Chakraborty A., et al. (2023). A comprehensive investigation of lipid-transfer proteins from Cicer arietinum disentangles their role in plant defense against Helicoverpa armigera-infestation. Front. Genet. 14, 1195554. doi: 10.3389/fgene.2023.1195554
Scheurer S. and Schülke S. (2018). Interaction of non-specific lipid-transfer proteins with plant-derived lipids and its impact on allergic sensitization. Front. Immunol. 9, 1389. doi: 10.3389/fimmu.2018.01389
Schmitt A. J., Sathoff A. E., Holl C., Bauer B., Samac D. A., and Carter C. J. (2018). The major nectar protein of Brassica rapa is a non-specific lipid transfer protein, BrLTP2. 1, with strong antifungal activity. J. Exp. Bot. 69, 5587–5597. doi: 10.1093/jxb/ery319
Scorrano L., De Matteis M. A., Emr S., Giordano F., Hajnóczky G., Kornmann B., et al. (2019). Coming together to define membrane contact sites. Nat. Commun. 10, 1287. doi: 10.1038/s41467-019-09253-3
Sels J., Mathys J., De Coninck B. M., Cammue B. P., and De Bolle M. F. (2008). Plant pathogenesisrelated (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Serrano M., Coluccia F., Torres M., L’Haridon F., and Métraux J.-P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5, 274. doi: 10.3389/fpls.2014.00274
Shah J. and Chaturvedi R. (2009). Lipid signals in plant-pathogen interactions. Annu. Plant Rev. 34, 292–333. doi: 10.1002/9781444301441
Shen W., Sipos B., and Zhao L. (2024). SeqKit2: A Swiss army knife for sequence and alignment processing. Imeta 3, e191. doi: 10.1002/imt2.191
Shetty N. P., Jørgensen H. J. L., Jensen J. D., Collinge D. B., and Shetty H. S. (2008). Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280. doi: 10.1007/s10658-008-9302-5
Shiiba I., Ito N., Oshio H., Ishikawa Y., Nagao T., Shimura H., et al. (2025). ER-mitochondria contacts mediate lipid radical transfer via RMDN3/PTPIP51 phosphorylation to reduce mitochondrial oxidative stress. Nat. Commun. 16, 1508. doi: 10.1038/s41467-025-56666-4
Shirey C. M., Scott J. L., and Stahelin R. V. (2017). Notes and tips for improving quality of lipid-protein overlay assays. Anal. Biochem. 516, 9–12. doi: 10.1016/j.ab.2016.10.009
Singh M. B. and Bhalla P. L. (2008). Genetic engineering for removing food allergens from plants. Trends Plant Sci. 13, 257–260. doi: 10.1016/j.tplants.2008.04.004
Situ J., Song Y., Feng D., Wan L., Li W., Ning Y., et al. (2024). Oomycete pathogen pectin acetylesterase targets host lipid transfer protein to reduce salicylic acid signaling. Plant Physiol. 194, 1779–1793. doi: 10.1093/plphys/kiad638
Sodano P., Caille A., Sy D., de Person G., Marion D., and Ptak M. (1997). 1H NMR and fluorescence studies of the complexation of dmpg by wheat non-specific lipid transfer protein. Global fold of the complex. FEBS Lett. 416, 130–134. doi: 10.1016/S0014-5793(97)01185-X
Song H., Yao P., Zhang S., Jia H., Yang Y., and Liu L. (2023). A non-specific lipid transfer protein, NtLTPI.38, positively mediates heat tolerance by regulating photosynthetic ability and antioxidant capacity in tobacco. Plant Physiol. Biochem. 200, 107791. doi: 10.1016/j.plaphy.2023.107791
Stanislava G. (2007). Barley grain non-specific lipid-transfer proteins (ns-LTPs) in beer production and quality. J. Institute Brewing 113, 310–324. doi: 10.1002/j.2050-0416.2007.tb00291.x
Staskawicz B. J. (2001). Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 125, 73–76. doi: 10.1104/pp.125.1.73
Sun J.-Y., Gaudet D. A., Lu Z.-X., Frick M., Puchalski B., and Laroche A. (2008). Characterization and antifungal properties of wheat nonspecific lipid transfer proteins. Mol. Plant-Microbe Interact. 21, 346–360. doi: 10.1094/MPMI-21-3-0346
Sun W., Li Y., Zhao Y., and Zhang H. (2015). The TsnsLTP4, a nonspecific lipid transfer protein involved in wax deposition and stress tolerance. Plant Mol. Biol. Rep. 33, 962–974. doi: 10.1007/s11105-014-0798-x
Terras F. R., Goderis I. J., Van Leuven F., Vanderleyden J., Cammue B. P., and Broekaert W. F. (1992). In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant Physiol. 100, 1055–1058. doi: 10.1104/pp.100.2.1055
Thoma S., Kaneko Y., and Somerville C. (1993). A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 3, 427–436. doi: 10.1046/j.1365-313X.1993.t01-25-00999.x
Valverde D. P., Yu S., Boggavarapu V., Kumar N., Lees J. A., Walz T., et al. (2019). ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798. doi: 10.1083/jcb.201811139
Van Loon L. C., Rep M., and Pieterse C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Van Loon L. C. and Van Strien E. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Vidhyasekaran P. (2015). Salicylic acid signaling in plant innate immunity. Plant Hormone Signaling Syst. Plant Innate Immun. 2, 27–122. doi: 10.1007/978-94-017-9285-1
Wang P., Duckney P., Gao E., Hussey P. J., Kriechbaumer V., Li C., et al. (2023). Keep in contact: multiple roles of endoplasmic reticulum-membrane contact sites and the organelle interaction network in plants. New Phytol. 238, 482–499. doi: 10.1111/nph.18745
Wang C., Gao H., Chu Z., Ji C., Xu Y., Cao W., et al. (2021). A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato. Mol. Plant Pathol. 22, 48–63. doi: 10.1111/mpp.13007
Wang Y., He Y., Zhang M., Li J., Xu X., Shi X., et al. (2022). Slltpg3, a non-specific lipid transfer protein, acts on the cuticle synthetic pathway to delay water loss and softening of tomato fruit. Postharvest Biol. Technol. 188, 111899. doi: 10.1016/j.postharvbio.2022.111899
Wang N.-J., Lee C.-C., Cheng C.-S., Lo W.-C., Yang Y.-F., Chen M.-N., et al. (2012). Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genom. 13, 1–9. doi: 10.1186/1471-2164-13-S1-S9
Wang R., Li J., and Liang Y. (2024). Role of ROS signaling in the plant defense against vascular pathogens. Curr. Opin. Plant Biol. 81, 102617. doi: 10.1016/j.pbi.2024.102617
Wang S. Y., Wu J. H., Ng T., Ye X. Y., and Rao P. F. (2004). A non-specific lipid transfer protein with antifungal and antibacterial activities from the mung bean. Peptides 25, 1235–1242. doi: 10.1016/j.peptides.2004.06.004
Wang F., Zang X.-S., Kabir M. R., Liu K.-L., Liu Z.-S., Ni Z.-F., et al. (2014). A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis. Gene 550, 18–26. doi: 10.1016/j.gene.2014.08.007
Wei F., Liu Y., Zhou D., Zhao W., Chen Z., Chen D., et al. (2022). Transcriptomic identification of a unique set of nodule-specific cysteine-rich peptides expressed in the nitrogen-fixing root nodule of Astragalus sinicus. Mol. Plant-Microbe Interact. 35, 893–905. doi: 10.1094/MPMI-03-22-0054-R
Wei H., Lu Z., Xue C., Jiang H., Xu X., Liu G., et al. (2025). Comprehensive analysis of the LTPG gene family in willow: Identification, expression profiling, and stress response. Int. J. Biol. Macromol. 295, 139600. doi: 10.1016/j.ijbiomac.2025.139600
Wirtz K. (1974). Transfer of phospholipids between membranes. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 344, 95–117. doi: 10.1016/0304-4157(74)90001-X
Wirtz K. and Zilversmit D. (1968). Exchange of phospholipids between liver mitochondria and microsomes in vitro. J. Biol. Chem. 243, 3596–3602. doi: 10.1016/S0021-9258(19)34182-1
Wong L. H., Čopič A., and Levine T. P. (2017). Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 42, 516–530. doi: 10.1016/j.tibs.2017.05.001
Wong L. H., Gatta A. T., and Levine T. P. (2019). Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20, 85–101. doi: 10.1038/s41580-018-0071-5
Wu M. J., Clarke F. M., Rogers P. J., Young P., Sales N., O’Doherty P. J., et al. (2011). Identification of a protein with antioxidant activity that is important for the protection against beer ageing. Int. J. Mol. Sci. 12, 6089–6103. doi: 10.3390/ijms12096089
Xiao Y., Xiao C., He X., Yang X., Tong Z., Wang Z., et al. (2023). A novel non-specific lipid transfer protein gene, CmnsLTP6. 9, enhanced osmotic and drought tolerance by regulating ROS scavenging and remodeling lipid profiles in chinese chestnut (Castanea mollissima Blume). Plants 12, 3916. doi: 10.3390/plants12223916
Xu Y., Zheng X., Song Y., Zhu L., Yu Z., Gan L., et al. (2018). NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 8, 8873. doi: 10.1038/s41598-018-27274-8
Yang Y., Song H., Yao P., Zhang S., Jia H., and Ye X. (2023). NtLTPI. 38, a plasma membrane-localized protein, mediates lipid metabolism and salt tolerance in Nicotiana tabacum. Int. J. Biol. Macromol. 242, 125007. doi: 10.1016/j.ijbiomac.2023.125007
Yao H.-Y., Lu Y.-Q., Yang X.-L., Wang X.-Q., Luo Z., Lin D.-L., et al. (2023). Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development. Proc. Natl. Acad. Sci. 120, e2221637120. doi: 10.1073/pnas.2221637120
Yao H.-Y. and Xue H.-W. (2018). Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 60, 851–863. doi: 10.1111/jipb.12655
Yin S., Li X., Meng Y., Finley R. L., Sakr W., Yang H., et al. (2005). Tumor-suppressive maspin regulates cell response to oxidative stress by direct interaction with glutathione s-transferase. J. Biol. Chem. 280, 34985–34996. doi: 10.1074/jbc.M503522200
Yuen E. L. H., Savage Z., Adamkova V., Vuolo C., Zhou Y., Tumtas Y., et al. (2021). Membrane contact sites between chloroplasts and pathogen interface underpin plant focal immune responses. bioRxiv, 2021–2010. doi: 10.1101/2021.10.08.463641
Zhang Y., Ge J., Bian X., and Kumar A. (2022). Quantitative models of lipid transfer and membrane contact formation. Contact 5, 25152564221096024. doi: 10.1177/25152564221096024
Zhang M., Kim Y., Zong J., Lin H., Dievart A., Li H., et al. (2019). Genome-wide analysis of the barley non-specific lipid transfer protein gene family. Crop J. 7, 65–76. doi: 10.1016/j.cj.2018.07.009
Zhang Y. and Skolnick J. (2005). TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309. doi: 10.1093/nar/gki524
Zhao Z., Yang X., Lü S., Fan J., Opiyo S., Yang P., et al. (2020). Deciphering the novel role of AtMIN7 in cuticle formation and defense against the bacterial pathogen infection. Int. J. Mol. Sci. 21, 5547. doi: 10.3390/ijms21155547
Zhou D., Li Y., Wang X., Xie F., Chen D., Ma B., et al. (2019). Mesorhizobium huakuii HtpG interaction with nsLTP AsE246 is required for symbiotic nitrogen fixation. Plant Physiol. 180, 509–528. doi: 10.1104/pp.18.00336
Zhu F., Cao M.-Y., Zhu P.-X., Zhang Q.-P., and Lam H.-M. (2023). Non-specific LIPID TRANSFER PROTEIN 1 enhances immunity against tobacco mosaic virus in Nicotiana benthamiana. J. Exp. Bot. 74, 5236–5254. doi: 10.1093/jxb/erad202
Zottich U., Da Cunha M., Carvalho A. O., Dias G. B., Silva N. C., Santos I. S., et al. (2011). Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with α-amylase inhibitor properties. Biochim. Biophys. Acta (BBA) General Subj. 1810, 375–383. doi: 10.1016/j.bbagen.2010.12.002
Keywords: non-specific lipid transfer proteins (nsLTPs), lipid biology, plant disease resistance, plant immunity, antimicrobial peptides (AMPs), genetic engineering, lipid signaling, lipidomics
Citation: McLaughlin JE and Tumer NE (2025) Roles of non-specific lipid transfer proteins in plant defense: structural and functional perspectives. Front. Fungal Biol. 6:1640465. doi: 10.3389/ffunb.2025.1640465
Received: 03 June 2025; Accepted: 18 August 2025;
Published: 16 September 2025.
Edited by:
Sergio Casas-Flores, Instituto Potosino de Investigación Científica y Tecnológica (IPICYT), MexicoReviewed by:
Mitzuko Dautt-Castro, National Council of Science and Technology (CONACYT), MexicoJorge Verdín, CONACYT Centro de Investigación y Asistencia en Tecnología y Diseño del Estado de Jalisco (CIATEJ), Mexico
Copyright © 2025 McLaughlin and Tumer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John E. McLaughlin, bWNsYXVnaGpAc2Vicy5ydXRnZXJzLmVkdQ==
 John E. McLaughlin
John E. McLaughlin Nilgun E. Tumer
Nilgun E. Tumer