- 1Molecular Biology Research Center and Center for Medical Genetics, School of Life Sciences, Central South University, Changsha, China
- 2School of Medical Laboratory, Shao Yang University, Shaoyang, China
- 3Molecular Science and Biomedicine Laboratory, College of Biology, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha, China
- 4State Key Laboratory for Chemo/Biosensing and Chemometrics, College of Biology, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan Univers ity, Changsha, China
- 5Department of Histology and Embryology, Xiangya School of Medicine, Central South University, Changsha, China
- 6Department of Oncology, The Second Xiangya Hospital of Central South University, Central South University, Changsha, China
Non–small-cell lung cancer (NSCLC) is divided into three major histological types, namely, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large-cell lung carcinoma (LCLC). We previously identified that 4.1N/EPB41L1 acts as a tumor suppressor and is reduced in NSCLC patients. In the current study, we explored the underlying epigenetic mechanisms of 4.1N/EPB41L1 reduction in NSCLC. The 4.1N/EPB41L1 gene promoter region was highly methylated in LUAD and LUSC patients. LUAD patients with higher methylation level in the 4.1N/EPB41L1 gene promoter (TSS1500, cg13399773 or TSS200, cg20993403) had a shorter overall survival time (Log-rank p = 0.02 HR = 1.509 or Log-rank p = 0.016 HR = 1.509), whereas LUSC patients with higher methylation level in the 4.1N/EPB41L1 gene promoter (TSS1500 cg13399773, TSS1500 cg07030373 or TSS200 cg20993403) had a longer overall survival time (Log-rank p = 0.045 HR = 0.5709, Log-rank p = 0.018 HR = 0.68 or Log-rank p = 0.014 HR = 0.639, respectively). High methylation of the 4.1N/EPB41L1 gene promoter appeared to be a relatively early event in LUAD and LUSC. DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine restored the 4.1N/EPB41L1 expression at both the mRNA and protein levels. MiR-454-3p was abnormally highly expressed in NSCLC and directly targeted 4.1N/EPB41L1 mRNA. MiR-454-3p expression was significantly correlated with 4.1N/EPB41L1 expression in NSCLC patients (r = −0.63, p < 0.0001). Therefore, we concluded that promoter hypermethylation of the 4.1N/EPB41L1 gene and abnormally high expressed miR-454-3p work at different regulation levels but in concert to restrict 4.1N/EPB41L1 expression in NSCLC. Taken together, this work contributes to elucidate the underlying epigenetic disruptions of 4.1N/EPB41L1 deficiency in NSCLC.
Introduction
Lung cancer is the most commonly diagnosed cancer and the most lethal cause of cancer mortality worldwide (Bray et al., 2018). Non–small-cell lung cancer (NSCLC), consisting of lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large-cell lung carcinoma (LCLC) (Rodriguez-Canales et al., 2016), represents major types of lung cancers (80–85%) (D'Addario et al., 2010). Owing to a lack of obvious early symptoms and early-stage diagnosis, most patients with NSCLC are diagnosed in the advanced clinical stage—that is—III or IV (Norouzi and Hardy, 2021). Despite recent advances in NSCLC treatment, less than 15% of the patients eventually survived (Quintanal-Villalonga and Molina-Pinelo, 2019; Norouzi and Hardy, 2021).
Gene promoter methylation and miRNA dysregulation are typical markers of cancer epigenetics (Nebbioso et al., 2018). Gene promoter methylation most commonly occurs at the CpG islands and regulates the gene expression at the transcriptional level (Yang et al., 2014; Zhou et al., 2017; Arechederra et al., 2018). 5–10% of CpG islands in the promoter of genes have been identified as cancer-specifically methylated, which should not be methylated in normal cells (Heller et al., 2013; Olbromski et al., 2020). The methylations of certain genes are of clinical relevance for patients with NSCLC (Heller et al., 2013). MiRNAs are endogenous small non-coding RNAs, which directly bind to the 3′-untranslated regions (3′UTRs) of target mRNAs to regulate the gene expression at the posttranscriptional level. NSCLC patients have widespread dysregulation of miRNA expression (Du et al., 2018; Uddin and Chakraborty, 2018). It has been well-documented that 4.1 family members 4.1N/EPB41L1 and its homologs (4.1B/EPB41L3, 4.1G/EPB41L2, and 4.1R/EPB41) are lost in various cancers (Yang et al., 2021). However, epigenetic silencing of 4.1 family members in cancers is still largely unknown. Loss of 4.1B/EPB41L3 is the only case that has been linked to high promoter methylation in cancers (Kikuchi et al., 2005; Zhang et al., 2012). No miRNAs have been found to regulate 4.1 family members.
Our previous studies suggested that 4.1N/EPB41L1 is abnormally low expressed and exerts anticancer effects in NSCLC (Wang et al., 2016; Yang et al., 2016; Yang et al., 2021). In the current study, for the first time, we focus on identifying the underlying epigenetic disruptions of 4.1N/EPB41L1 deficiency in NSCLC. We report that promoter hypermethylation and aberrant miR-454-3p expression regulate 4.1N/EPB41L1 expression at transcriptional and posttranscriptional levels, respectively, but work in concert to restrict its expression in NSCLC.
Materials and Methods
Antibodies
Rabbit anti-4.1N antibody was purchased from ATLAS (Bromma, Sweden). Rabbit anti-GAPDH antibody was purchased from Santa Cruz (Santa Cruz Biotechnology, United States).
Cell Culture
MRC5, 95C, and 95D cells were grown in DMEM medium (Gibco, United States) and supplemented with 10% fetal bovine serum (Gibco, United States). H460 and A549 cells were grown in RPMI 1640 medium (Gibco, United States) and supplemented with 10% fetal bovine serum. All the cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
NSCLC Tissue Samples
Tumor tissues and tumor-adjacent tissues were obtained from the Second Xiangya Hospital of Central South University (Changsha, China). The tissue samples were subjected to qPCR experiments after approval by the Ethics Committee of the Second Xiangya Hospital. Informed consent was obtained from all participating subjects.
Methylation-Based Analysis
The MethSurv tool (https://biit.cs.ut.ee/methsurv/) (Modhukur et al., 2018) was used to perform the assessment of methylation-based analysis for the 4.1N/EPB41L1 gene in LUAD and LUSC. The raw data for LUAD and LUSC could be downloaded from the website (https://biit.cs.ut.ee/methsurv/).
Cell Transfection and Western Blot
MiR-454-3p and the control mimics were purchased from RiboBio (Guangzhou, China) and transfected according to our previously published protocol (Li et al., 2016). Western blot was also performed according to our previous protocol (Yang et al., 2016).
Targeted Bisulfite Sequencing
The cells were sent to Biomarker Acegene Corporation, Shenzhen, China for targeted bisulfite sequencing (TBS-seq) analysis. 4.1N/EPB41L1 promoter methylation was assessed according to the previously published method (Gao et al., 2014a; Gao et al., 2014b; Gao et al., 2015; Pan et al., 2018). Methylation levels are defined as the fraction of read counts of ‘C’ in the total read counts of both ‘C’ and ‘T’ for each covered C site. On the basis of such read fraction, methylated cytosine was called using a binomial distribution as in the method described by Lister et al. (2009), whereby a probability mass function is calculated for each methylation context (CpG). Two-tailed Fisherʼs exact test was used to identify cytosines that are differentially methylated between two samples or groups. Only those CGs covered by at least 200 reads in at least one sample were considered for testing.
5-Aza-2′-deoxycytidine(5-Aza-CdR)treatment
5-Aza-CdR (Merck, Germany) was diluted in PBS. The cells were seeded in a 6-well plate and treated with 0, 1, or 10 μM 5-Aza-CdR for 48 h. 5-Aza-CdR was replaced every 24 h.
RNA Extraction and qPCR
Total RNA was isolated using the RNeasy kit (QIAGEN, United States). cDNA was synthesized using the RevertAid H Minus First-Strand cDNA Synthesis Kit (Thermo Scientific, United States). Stem-loop RT primers (RiboBio, China) were used in reverse transcription for miR-454-3p. qPCR was performed using the One-Step qRT-PCR SYBR® Green Kit (Vazyme Biotech, China). The sequences of primers targeting 4.1N/EPB41L1 and miR-454-3p were used as described earlier (Wang et al., 2010) and designed by Vazyme Biotech (Nanjing, China). U6 small nuclear RNA was used as an internal control for miR-454-3p analysis.
Dual-Luciferase Reporter Gene Assay
The 3′UTR target sites of 4.1N/EPB41L1 mRNA were amplified by PCR with genomic DNA from MRC5 cells. The PCR product was cloned in the psiCHECK2 vector (Promega, United States) to construct the wild-type plasmid (psiCHECK2-4.1N-wt). The corresponding mutant psiCHECK2-4.1N-mut was constructed by in vitro site-directed mutagenesis (Mut ExpressMultiS Fast Mutagenesis Kit, Vazyme Biotech, China). Bidirectional sequencing was applied to confirm the correct sequence of the two constructs. For the dual-luciferase reporter gene assay, A549 and H460 cells were cultured in a 24-well plate for 24 h and transfected with psiCHECK2-4.1N-wt or psiCHECK2-4.1N-mut plasmids and miR-454-3p mimics or miR-negative-control using the RiboFECT™CP transfection kit (Ribo Biotechnology, China) and Lipofectamine 2000 (Invitrogen, United States). 48 hours after the transfection, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States) was used to measure the luciferase activity according to the manufacturer’s protocol.
Statistics
All the experiments were performed in triplicate, and statistical analyses were conducted using GraphPad Prism 5.0. The data were presented as the mean ± standard deviation (SD). Student’s t-tests were used to calculate the results. A p-value < 0.05 was considered significant statistically. DNA methylation values were represented as beta values (range from 0 to 1). Any beta value equal to or greater than 0.6 was considered fully methylated. Any beta value equal to or less than 0.2 was considered to be fully unmethylated. Beta values between 0.2 and 0.6 were considered to be partially methylated. Differential methylation for individual CpG loci was assessed by comparing the beta values. The patients were classified into high-methylation and low-methylation levels based on maxstat (Modhukur et al., 2018). Cox proportional hazards models were used to perform the survival analysis based on methylation levels of the CpG sites. The methylation levels and overall survival time were used as explanatory variables and response variables, respectively, to perform overall survival analysis.
Results
Hypermethylation of the 4.1N/EPB41L1 Gene in NSCLC
Aberrant hypermethylations in the promoter region of genes are considered a major reason for gene silencing in cancer (Lamy et al., 2001). The CpG island methylation prediction using the CpGPNP program (http://forensicdna.kr/cpgpnp/) showed four CpG islands in the 4.1N/EPB41L1 gene promoter (2,000 bp upstream to 1,000 bp downstream of the transcription start site, Figure 1A). NSCLC predominantly encompasses the LUAD (40% prevalence) and LUSC subtypes (25% prevalence). The MethSurv tool (https://biit.cs.ut.ee/methsurv/) (Modhukur et al., 2018) was used to perform the assessment of methylation-based analysis for the 4.1N/EPB41L1 gene promoter region (TSS200 and TSS1500) in LUAD and LUSC. The heat map showed that high methylation of the 4.1N/EPB41L1 gene was prevalent in both LUAD (Figure 1B) and LUSC (Figure 1C). Because the MethSurv tool (https://biit.cs.ut.ee/methsurv/) lacks LCLC data, we investigated the methylation of the 4.1N/EPB41L1 gene in LCLC cells (95C, 95D, and H460) and normal lung fibroblast cells (MRC5). TBS-seq results showed that promoter methylation of 4.1N/EPB41L1 was significantly higher in LCLC cells (95C, 95D, and H460) than in normal lung cells (MRC5) (p < 0.001) (Figures 1D,E). To further validate the role of methylation in 4.1N/EPB41L1 gene repression, we treated the LCLC cells (95C and H460) and LUAD cells (A549) with DNA methyltransferase inhibitor 5-Aza-CdR. After demethylation treatment, the 4.1N/EPB41L1 gene was restored both at mRNA (Figures 2A–C) and protein levels (Figures 2D–F). Taken together, these results indicated that 4.1N/EPB41L1 gene methylation is a cause of decreased 4.1N/EPB41L1 expression in NSCLC patients.
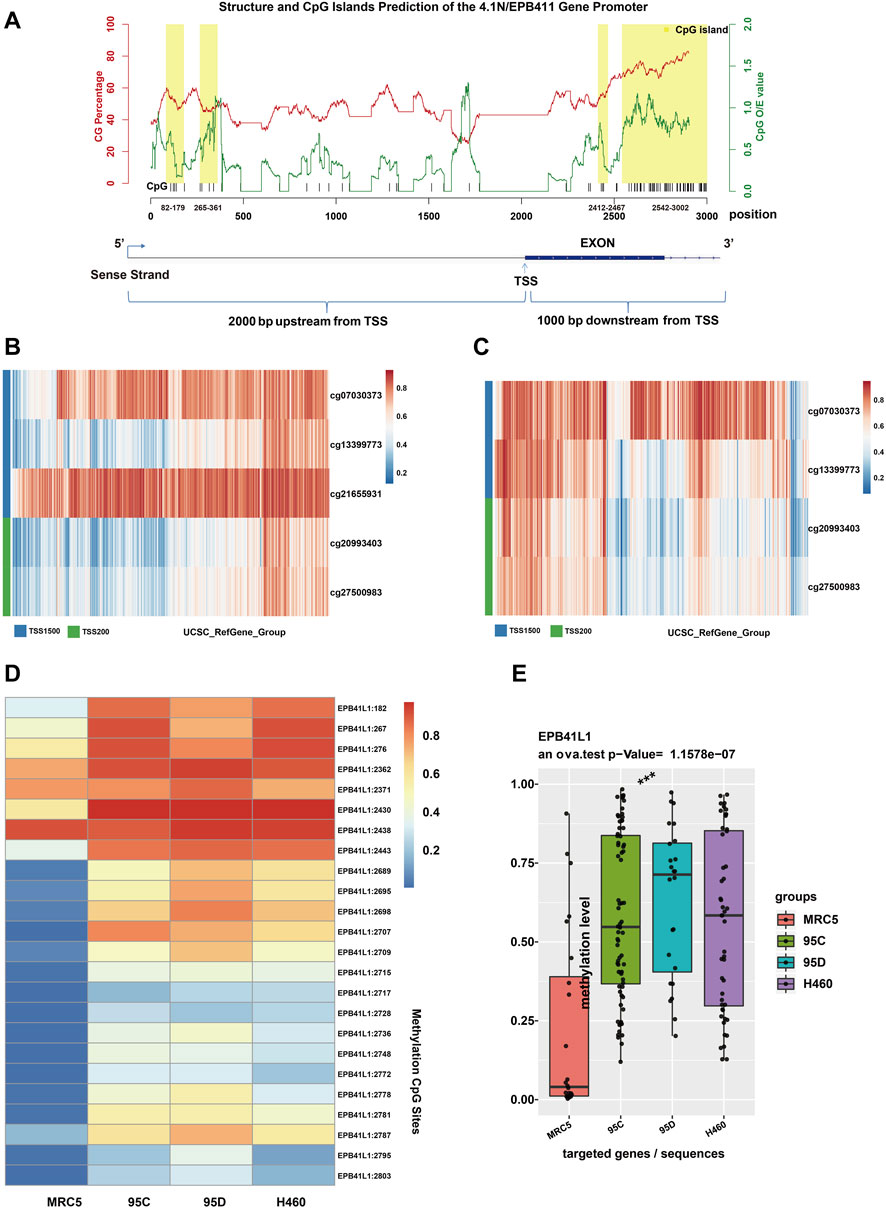
FIGURE 1. Promoter methylation of the 4.1N/EPB41L1 gene in NSCLC. (A) In the upper panel, CpG island prediction of the 4.1N/EPB41L1 gene promoter was integrated using the CpGPNP program. The red line represented GC content, the green line represents CpG O/E value, and the yellow box represented four predicted locations of CpG island (range from 82 to 179, 265–361, 2412–2467, and 2542–3002, sequence length >100 bp, GC content >50%, O/E value >0.6). The lower panel represented the structure of the 4.1N/EPB41L1 gene promoter (range from TSS -2000 to TSS 1000). Coordinate values of abscissas in the lower and upper panels were correlated. TSS -2000 in the lower panel corresponded to 0 in the upper panel. The TSS site in the lower panel corresponded to 2000 in the upper panel. TSS 1000 in the lower panel corresponded to 3000 in the upper panel. (B,C) Heatmap depicting the CpG methylation level of the 4.1N/EPB41L1 gene promoter in LUAD and LUSC patients. Rows and columns represented the CpGs and the patients, respectively. (D) Heatmap depicting the methylation levels of the 4.1N/EPB41L1 gene promoter in normal lung fibroblast cells MRC5 and LCLC cells (95C, 95D, and H460). The row label was the methylation site. The number of row labels corresponded to the coordinate value of abscissa in the upper panel of Panel 1A. Methylation levels were represented as beta values and shown as a continuous variable from blue to red. Any beta value equal to or greater than 0.6 was considered fully methylated. Any beta value equal to or less than 0.2 was considered to be fully unmethylated. Beta values between 0.2 and 0.6 were considered to be partially methylated. (E) Boxplots indicating the methylation differences between normal lung fibroblast cells MRC5 and LCLC cells (95C, 95D, and H460). Median methylation levels (show by a thick black line). ***p < 0.001.
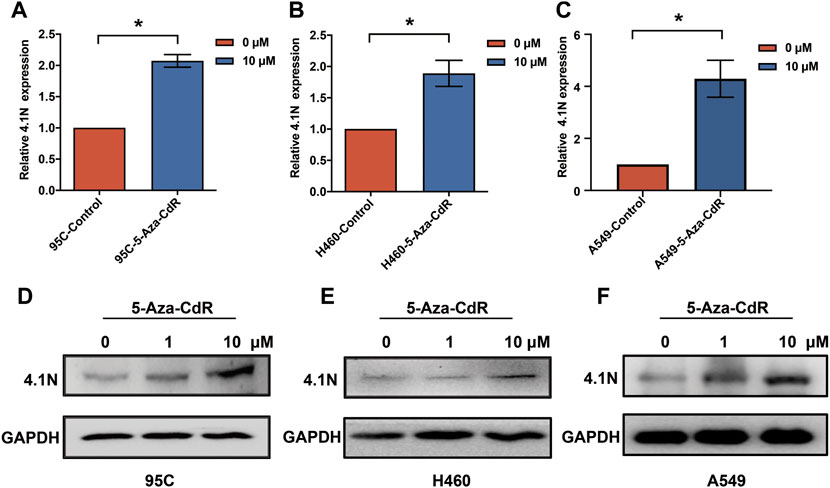
FIGURE 2. 4.1N/EPB41L1 was restored by methyltransferase inhibitor 5-Aza-CdR in NSCLC cells. (A–C) qPCR was performed to evaluate expressions of 4.1N/EPB41L1 mRNA in 95C, H460, and A549 cells after 0 μM or 1 μM 5-Aza-CdR treatments for 48 h (D–F) 95C, H460, and A549 cells were treated with 0, 1 or 10 μM 5-Aza-CdR for 48 h, and then the protein was detected by Western blot. The data were presented as the mean ± SD, *p < 0.05.
Prognostic Relevance of 4.1N/EPB41L1 Hypermethylation in LUAD and LUSC
In most cases, evaluating DNA methylation signature in the promoter region is highly desirable and sensitive for cancer diagnosis and prognosis. The Kaplan–Meier survival curve showed that the higher methylation levels in the promoter (TSS1500, cg13399773 or TSS200, cg20993403) of the 4.1N/EPB41L1 gene were significantly associated with a shorter overall survival time (Log-rank p = 0.02, HR = 1.509 or Log-rank p = 0.016, HR = 1.509, respectively) for LUAD patients (Figures 3A,C). Median methylation levels of the two CpG sites (cg13399773 and cg20993403) were high (beta>0.5) at stage I and did not essentially change in tumors of more advanced stages (Figures 3B,D). Unlike the LUAD, the Kaplan–Meier survival curve showed that the higher methylation levels in the promoter (TSS1500 cg13399773, TSS1500 cg07030373, or TSS200 cg20993403) of the 4.1N/EPB41L1 gene were significantly associated with a shorter overall survival time (Log-rank p = 0.045 HR = 0.5709, Log-rank p = 0.018 HR = 0.68 or Log-rank p = 0.014 HR = 0.639 respectively) for LUSC patients (Figures 4A,C,E). Median methylation levels of the three CpG sites (TSS1500 cg13399773, TSS1500 cg07030373, or TSS200 cg20993403) were high (beta>0.5) from stage I to IV and tended to decline with tumor progression (Figures 4B,D,F). We did not further explore the contrasting prognostic relevance of 4.1N/EPB41L1 hypermethylation between LUAD and LUSC. However, methylation of the 4.1N/EPB41L1 gene might be an important mechanism for tumor formation in LUAD and LUSC because high 4.1N/EPB41L1 gene methylations were observed at stage I. Because the MethSurv tool (https://biit.cs.ut.ee/methsurv/) lacks LCLC data, the prognostic relevance of 4.1N/EPB41L1 hypermethylation in LCLC was unknown.
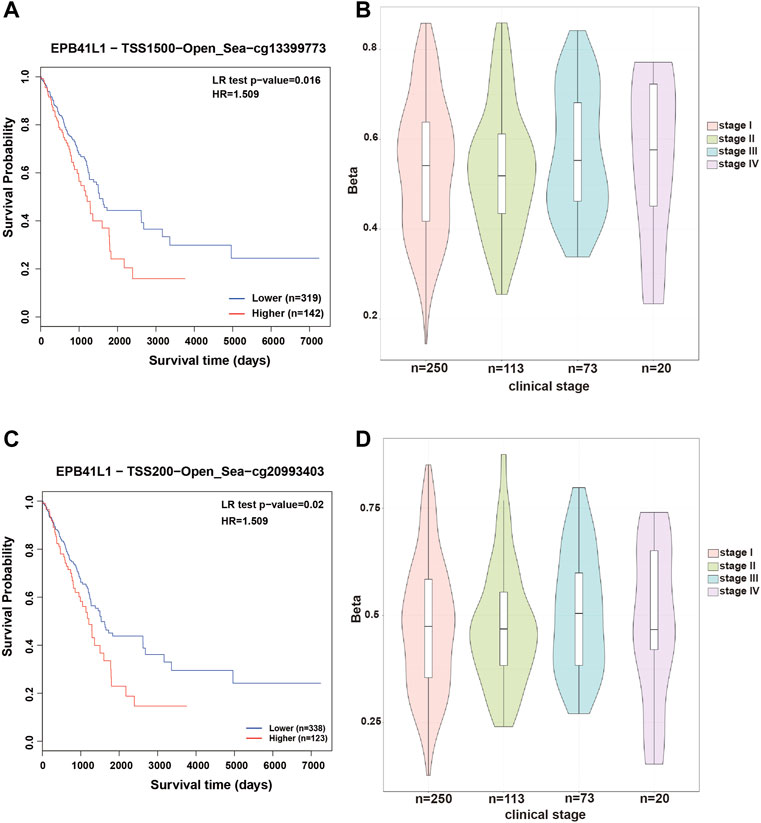
FIGURE 3. Methylation levels and prognosis of the 4.1N/EPB41L1 promoter in patients with LUAD. (A) Kaplan–Meier curves for cg13399773-4.1N showing survival in lower (beta <0.611, shown in blue) and higher methylation groups (beta > 0.611, shown in red) dichotomized by the maxstat method. (C) Kaplan–Meier curves for cg20993403-4.1N showing survival in lower (beta <0.59, shown in blue) and higher methylation groups (beta > 0.59, shown in red) dichotomized by the maxstat method. (B,D) Violin plots showing the methylation levels of cg13399773-4.1N and cg20993403-4.1N among stage I–IV LUAD patients. Median methylation levels (show by a thick black line) and interquartile range were summarized by the boxplot within each violin plot. HR: hazard ratio; LR: log-likelihood ratio; LUAD: lung adenocarcinoma. Methylation levels were represented as beta values. Any beta value equal to or greater than 0.6 was considered fully methylated. Any beta value equal to or less than 0.2 was considered to be fully unmethylated. Beta values between 0.2 and 0.6 were considered to be partially methylated.
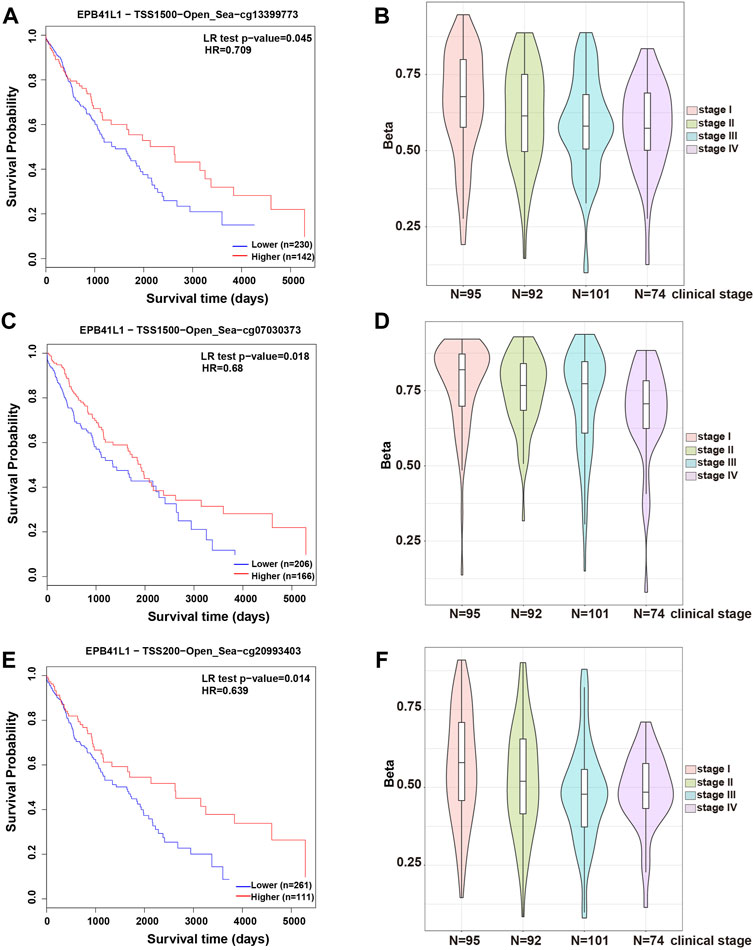
FIGURE 4. Methylation levels and prognosis of the 4.1N/EPB41L1 promoter in patients with LUSC. (A) Kaplan–Meier curves for cg13399773-4.1N showing survival in lower (beta <0.675, shown in blue) and higher methylation groups (beta > 0.675, shown in red) dichotomized by the maxstat method. (C) Kaplan–Meier curves for cg07030373-4.1N showing survival in lower (beta <0.796, shown in blue) and higher methylation groups (beta > 0.796, shown in red) dichotomized by the maxstat method. (E) Kaplan–Meier curves for cg20993403-4.1N showing survival in lower (beta <0.611, shown in blue) and higher methylation groups (beta > 0.611, shown in red) dichotomized by the maxstat method. (B,D,F) Violin plots showing the methylation levels of cg13399773-4.1N, cg07030373-4.1N, and cg20993403-4.1N among stage I–IV LUAD patients. Median methylation levels (show by a thick black line) and interquartile range were summarized by the boxplot within each violin plot. HR: hazard ratio; LR: log-likelihood ratio; LUSC: lung squamous cell carcinoma. Methylation levels were represented as beta values. Any beta value equal to or greater than 0.6 was considered fully methylated. Any beta value equal to or less than 0.2 was considered to be fully unmethylated. Beta values between 0.2 and 0.6 were considered to be partially methylated.
4.1N/EPB41L1 mRNA is a Direct Target of miR-454-3p
We previously described that the 95D cells had remarkably lower 4.1N/EPB41L1 expression than the 95C cells (Yang et al., 2016). Unexpectedly, although the methylation level of the 95D promoter was higher than that of 95C, there was no significant difference between the two homologous NSCLC subclones 95C/95D (Figure 5A). Over half of all protein-encoding genes are regulated by miRNAs (Turchinovich et al., 2012). Aberrantly high-expressed oncomiRNA silencing cancer suppressing genes are frequently found in NSCLC. Therefore, we investigated the potential miRNAs regulating 4.1N/EPB41L1 expression. The miRNA-target gene database TargetScan (miRBase:www.mirbase.org) was used to predict the potential miRNAs regulating 4.1N/EPB41L1, and miR-454-3p was suggested as a potential miRNA (Figure 5B). qPCR results showed that the miR-454-3p was expressed significantly lower in the 95D cells than in 95C cells (Figure 5C). After overexpressing the miR-454-3p in A549 cells (Figure 5D), the expression level of protein 4.1N/EPB41L1 was downregulated (Figure 5E).
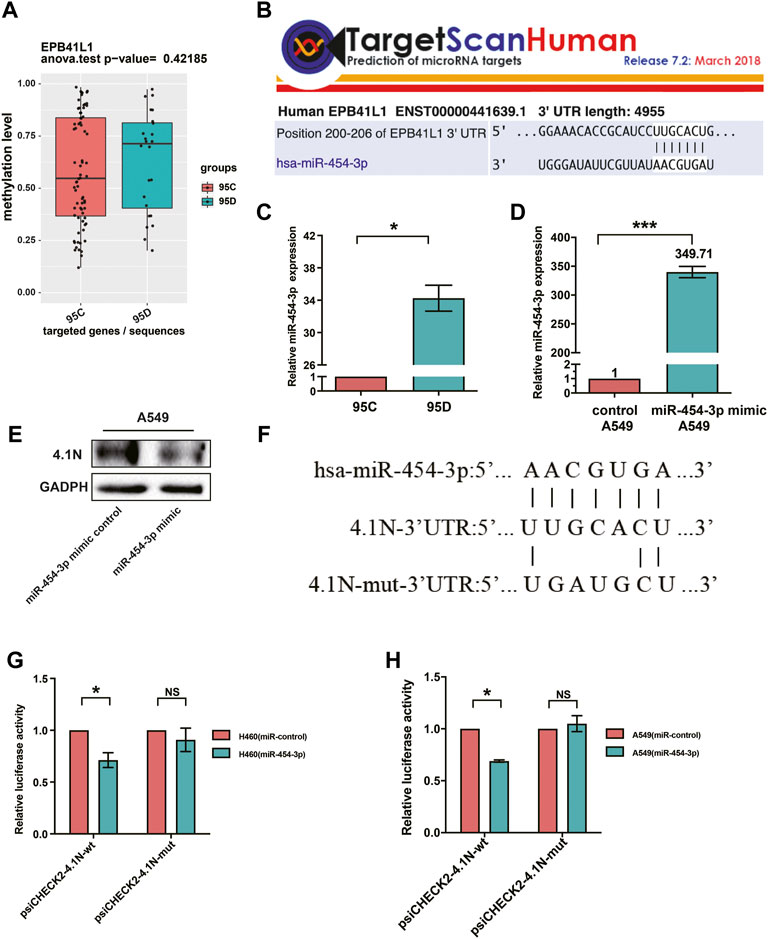
FIGURE 5. MiR-454-3p directly bound to 3′UTR of 4.1N/EPB41L1 mRNA. (A) There was no significant difference in methylation levels of the 4.1N/EPB41L1 gene promoter between the two homologous NSCLC subclones 95C and 95D. (B) MiR-454-3p was a predicted binding partner of 4.1N/EPB41L1. (C,D) After transfecting A549 cells with miR-454-3p or negative control mimics, the expression levels of miR-454-3p and protein 4.1N/EPB41L1 were detected using qPCR and Western blot, respectively. (E) miR-454-3p expression was significantly lower in 95C cells than 95D cells. (F) Schematic representation of the predicted binding sites of miR-454-3p and 4.1N/EPB41L1 mRNA and the mutated sequences in potential binding sites. (G,H) A549 or H460 cells were co-transfected with miR-454-3p or control mimics and wild-type (psiCHECK2-4.1N-wt) or mutant-type (psiCHECK2-4.1N-mut) plasmids. 48 h later, dual-luciferase activity was measured. The data were presented as the mean ± SD. *p < 0.05 GADPH and U6 were used as loading control of 4.1N/EPB41L1 mRNA and miR-454-3p, respectively.
To further examine if 4.1N/EPB41L1 was a target gene of miR-454-3p, the dual-luciferase activity assay was applied in A549 and H460 cells. The predicted binding sites of miR-454-3p and 4.1N/EPB41L1 mRNA and mutant sequences containing four mutated nucleotides are shown in Figure 5F. MiR-454-3p significantly suppressed the luciferase activity, and this suppressive effect was abolished by the mutation in the miR-454-3p-binding region of the 4.1N/EPB41L1 mRNA 3′UTR in H460 and A549 cells (Figures 5G,H). The abovementioned results signified that abnormally highly expressed miR-454-3p is another epigenetic cause of 4.1N/EPB41L1 decreasing in NSCLC patients.
Abnormally High-Expressed miR-454-3p Decreases 4.1N/EPB41L1 in NSCLC
TCGA data were used to extract RNA transcript levels of the miR-454-3p. We found that the miR-454-3p was significantly higher in the LUAD (Figure 6A) and LUSC (Figure 6C) tissues in comparison to the adjacent tissues, whereas the 4.1N/EPB41L1 mRNA was significantly lower in LUAD (Figure 6B) and LUSC (Figure 6D) tissues than the corresponding adjacent tissues. Then, we used qPCR to measure the miR-454-3p and 4.1N/EPB41L1 mRNA expressions in 37 NSCLC tissues and 31 tumor-adjacent tissues. We found that the expression patterns are consistent with TCGA data (Figures 6E,F). Moreover, the association between miR-454-3p and 4.1N/EPB41L1 mRNA was validated by Spearman’s coefficient analysis. The MiR-454-3p expression level showed a significantly negative correlation with 4.1N/EPB41L1 mRNA (r = −0.63, p < 0.0001, Figure 6G). Expression patterns of miR-454-3p and 4.1N/EPB41L1 mRNA showed that abnormally high expression of miR-454-3p negatively regulates 4.1N/EPB41L1 in NSCLC.
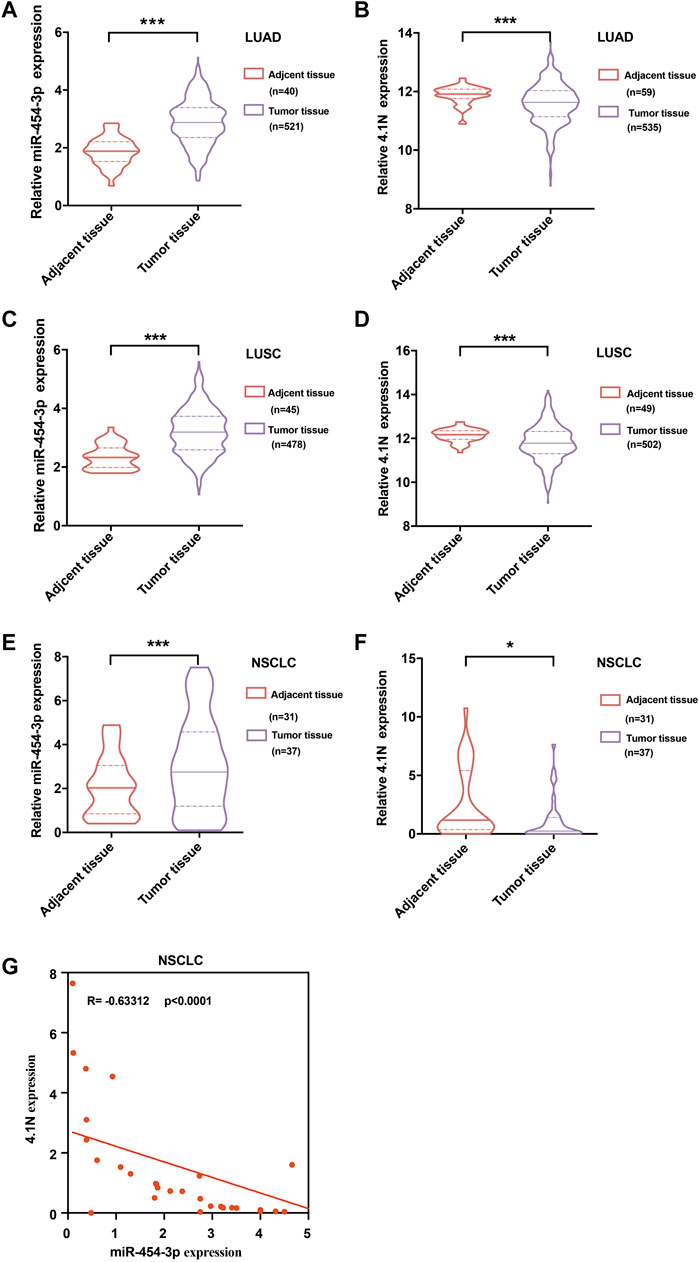
FIGURE 6. Validation of expression patterns of miR-454-3p and 4.1N/EPB41L1 mRNA in NSCLC. Relative expressions of miR-454-3p and 4.1N/EPB41L1 mRNA in LUAD. (A,B) and LUSC (C,D) were analyzed using the data from the TCGA dataset. (E-F) Relative expression of miR-454-3p and 4.1N/EPB41L1 mRNA in NSCLC and adjacent tissues was measured by qPCR. GADPH and U6 were used as loading control of 4.1N/EPB41L1 mRNA and miR-454-3p, respectively. (G) Correlation between miR-454-3p and 4.1N/EPB41L1 mRNA was determined using Spearman’s coefficient analysis. The data were presented as the mean ± SD. *p < 0.05 and ***p < 0.001.
Discussions
DNA methylation in the promoter region results in gene repression, which is one of the most well-defined epigenetic hallmarks. We previously revealed that downregulated 4.1N/EPB41L1 exerts antitumor effects by activating the classical Wnt pathway and C-MYC expression in NSCLC (Wang et al., 2016; Yang et al., 2016; Yang et al., 2021). The Wnt pathway disruption driven by methylation of promoter regions plays a key driving role in the high CpG island methylated phenotype LUAD subtype. This subtype is also significantly correlated with the overexpressed MYC gene (Cancer Genome Atlas Research, 2014; Duruisseaux and Esteller, 2018). 4.1N/EPB41L1 has many similar biological characteristics to its homologous 4.1B/EPB41L3. High promoter methylation of the 4.1B/EPB41L3 gene–induced gene-silencing frequently occurs in NSCLC (Kikuchi et al., 2005; Zhang et al., 2012), breast cancer (Heller et al., 2007), renal clear cell carcinoma (Yamada et al., 2006), and prostate cancer (Schulz et al., 2007; Schulz et al., 2010). Thus, we decided to explore the underlying epigenetic disruptions of 4.1N/EPB41L1 deficiency in NSCLC. Similarly, we found that high 4.1N/EPB41L1 gene methylation was prevalent in LUAD and LUSC. 4.1B/EPB41L3 gene methylation is a potential indicator for poor prognosis in NSCLC patients, especially in LUAD patients (Kikuchi et al., 2005). We found that a higher methylation level of 4.1N/EPB41L1 gene CpG sites (cg13399773 and cg20993403) was potential predictive markers of shorter overall survival in LUAD patients. It is acceptable that patients with higher promoter methylation within the 4.1N/EPB41L1 gene have a shorter overall survival time because the methylation inhibits tumor suppressor 4.1N/EPB41L1 expression at the transcriptional level. However, it is a different matter for LUSC, which remains to be elucidated in the future. The 5-year survival rate is less than 15% for NSCLC patients, but the rate can increase to 63% with the early stage of initial diagnosis (van Rens et al., 2000; Geng et al., 2017), thus demonstrating the value of the early diagnosis of NSCLC. 4.1B/EPB41L3 gene promoter methylation is regarded as an early event in renal clear cell carcinoma (Yamada et al., 2006). In this study, high 4.1N/EPB41L1 gene methylation was also observed to be a relatively early event in LUAD and LUSC patients, indicating its valuable role in tumorigenesis and potential as an early detection marker.
qPCR results of our sample set and the TCGA database together showed that the miR-454-3p was upregulated in the NSCLC tumor tissue, which contrasted with the results from another independent study (Jin et al., 2019). Interestingly, tumor expression of miR-454-5p in NSCLC is reported as upregulated (Zhu et al., 2016), but another report suggests its expression to be downregulated (Liu et al., 2018). It is not rare when various clinical specimens are evaluated; the expression levels of some miRNAs are different. Because of the complexity of NSCLC development, for these miRNAs, the specimen integration of different NSCLC stages and histological types could impede their expressions together (Zhong et al., 2021). In NSCLC, our study showed aberrantly high-expressed miR-454-3p directly bound to 4.1N/EPB41L1 mRNA 3′UTR and led to the depression of tumor suppressor 4.1N/EPB41L1 at the posttranscriptional level, uncovering the known miRNAs regulating 4.1N/EPB41L1. It has been reported that the miR-454-3p also acts as oncomiRNA in oral squamous cell carcinoma (Shi et al., 2021), cervical cancer (Guo et al., 2018; Shukla et al., 2019; Song et al., 2019; Li et al., 2021), liver cancer (Li et al., 2019), breast cancer (Ren et al., 2019; Wang et al., 2020), and colon cancer (Li et al., 2018). Moreover, an integrative bioinformatics analysis indicates that miR-454-3p is a biomarker for diagnosing some cancers, including the LUAD (Botling et al., 2013), which needs further confirmation studies in the future.
Epigenetic alterations have been demonstrated to be highly orchestrated from the initiation step to therapy resistance step in lung cancer (Quintanal-Villalonga and Molina-Pinelo, 2019). Although many research studies have revealed the vital anticancer roles of 4.1 family members, the knowledge of the underlying epigenetic mechanism behind their loss in cancers is nearly empty. This study showed that promoter methylation and miR-454-3p were implicated in the expression deregulation of the 4.1N/EPB41L1 at transcriptional and posttranscriptional levels, respectively. The epigenetic abnormalities are reversible; the application of upregulating 4.1N/EPB41L1 by targeting DNA methylation and miR-454-3p may represent a promising therapy for NSCLC treatment.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: To investigate the 4.1N/EPB41L1 gene methylation and its relation to NSCLC patient prognosis, TCGA methylation data (https://cancergenome.nih.gov/) of the array features 450k CpG sites covering the 4.1N gene promoter region (TSS200 and TSS1500) were analyzed. The MethSurv tool (https://biit.cs.ut.ee/methsurv/) was used to perform the assessment of methylation-based analysis for the 4.1N/EPB41L1 gene in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). TCGA data for LUAD and LUSC were used to extract RNA transcript levels of the miR-454-3p.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Xiangya Hospital, Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
QY, LZ, HL, and JL contributed the central idea, performed the experiments, analyzed the data, and wrote the initial draft of the manuscript. The remaining authors discussed the results and finalized this manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos 81770107 and 82003286); the Natural Science Foundation of Hunan Province (Grant No. 2020JJ4560); Scientific Research Foundation of Hunan Provincial Education Department (Grant No. 20B528); Guidance Science and Technology Program of Shaoyang City (Grant No. 2019ZD07); the Fellowship of China Postdoctoral Science Foundation (Grant Nos 2020M672474 and 2021T140195); and the Changsha Municipal Natural Science Foundation (Grant Number kq20140421); Postgraduate Innovation and Research Project of Central South University (Grant No. 2021zzts0569).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
3′UTRs, 3′-untranslated regions; 5-Aza-CdR, 5-Aza-2′-deoxycytidine; HR, hazard ratio; LCLC, large-cell lung carcinoma; LR, log-likelihood ratio; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC, non–small-cell lung cancer; SD, standard deviation; TBS-seq, targeted bisulfite sequencing; TSS, transcription start site.
References
Arechederra, M., Daian, F., Yim, A., Bazai, S. K., Richelme, S., Dono, R., et al. (2018). Hypermethylation of Gene Body CpG Islands Predicts High Dosage of Functional Oncogenes in Liver Cancer. Nat. Commun. 9 (1), 3164. doi:10.1038/s41467-018-05550-5
Botling, J., Edlund, K., Lohr, M., Hellwig, B., Holmberg, L., Lambe, M., et al. (2013). Biomarker Discovery in Non-small Cell Lung Cancer: Integrating Gene Expression Profiling, Meta-Analysis, and Tissue Microarray Validation. Clin. Cancer Res. 19 (1), 194–204. doi:10.1158/1078-0432.ccr-12-1139
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Cancer Genome Atlas Research (2014). Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature 511 (7511), 543–550. doi:10.1038/nature13385
D'Addario, G., Früh, M., Reck, M., Baumann, P., Klepetko, W., and Felip, E. (2010). Metastatic Non-small-cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 21 (Suppl. 5), v116–v119. doi:10.1093/annonc/mdq189
Du, X., Zhang, J., Wang, J., Lin, X., and Ding, F. (2018). Role of miRNA in Lung Cancer-Potential Biomarkers and Therapies. Cpd 23 (39), 5997–6010. doi:10.2174/138161282366617071415011
Duruisseaux, M., and Esteller, M. (2018). Lung Cancer Epigenetics: From Knowledge to Applications. Seminars cancer Biol. 51, 116–128. doi:10.1016/j.semcancer.2017.09.005
Gao, F., Liang, H., Lu, H., Wang, J., Xia, M., Yuan, Z., et al. (2015). Global Analysis of DNA Methylation in Hepatocellular Carcinoma by a Liquid Hybridization Capture-Based Bisulfite Sequencing Approach. Clin. Epigenet 7, 86. doi:10.1186/s13148-015-0121-1
Gao, F., Xia, Y., Wang, J., Lin, Z., Ou, Y., Liu, X., et al. (2014). Integrated Analyses of DNA Methylation and Hydroxymethylation Reveal Tumor Suppressive Roles of ECM1, ATF5, and EOMESin Human Hepatocellular Carcinoma. Genome Biol. 15 (12), 533. doi:10.1186/s13059-014-0533-9
Gao, F., Zhang, J., Jiang, P., Gong, D., Wang, J.-W., Xia, Y., et al. (2014). Marked Methylation Changes in Intestinal Genes during the Perinatal Period of Preterm Neonates. BMC Genomics 2615, 716. doi:10.1186/1471-2164-15-716
Geng, X., Pu, W., Tan, Y., Lu, Z., Wang, A., Tan, L., et al. (2017). Quantitative Assessment of the Diagnostic Role of FHIT Promoter Methylation in Non-small Cell Lung Cancer. Oncotarget 8 (4), 6845–6856. doi:10.18632/oncotarget.14256
Guo, Y., Tao, M., and Jiang, M. (2018). MicroRNA-454-3p Inhibits Cervical Cancer Cell Invasion and Migration by Targeting C-Met. Exp. Ther. Med. 15 (3), 2301–2306. doi:10.3892/etm.2018.5714
Heller, G., Babinsky, V. N., Ziegler, B., Weinzierl, M., Noll, C., Altenberger, C., et al. (2013). Genome-wide CpG Island Methylation Analyses in Non-small Cell Lung Cancer Patients. Carcinog. Mar. 34 (3), 513–521. doi:10.1093/carcin/bgs363
Heller, G., Geradts, J., Ziegler, B., Newsham, I., Filipits, M., Markis-Ritzinger, E.-M., et al. (2007). Downregulation of TSLC1 and DAL-1 Expression Occurs Frequently in Breast Cancer. Breast Cancer Res. Treat. 103 (3), 283–291. doi:10.1007/s10549-006-9377-7
Jin, C., Lin, T., and Shan, L. (2019). Downregulation of Calbindin 1 by miR-454-3p Suppresses Cell Proliferation in Nonsmall Cell Lung Cancer In Vitro. Cancer Biotherapy Radiopharm. 34 (2), 119–127. doi:10.1089/cbr.2018.2598
Kikuchi, S., Yamada, D., Fukami, T., Masuda, M., Sakurai-Yageta, M., Williams, Y. N., et al. (2005). Promoter Methylation of DAL-1/4.1B Predicts Poor Prognosis in Non-small Cell Lung Cancer. Clin. Cancer Res. 11 (8), 2954–2961. doi:10.1158/1078-0432.ccr-04-2206
Lamy, A., Metayer, J., Thiberville, L., Frebourg, T., and Sesboue, R. (2001). Re: Promoter Methylation and Silencing of the Retinoic Acid Receptor- Gene in Lung Carcinomas. JNCI J. Natl. Cancer Inst. 93 (1), 66–67. doi:10.1093/jnci/93.1.66
Li, H., Ouyang, R., Wang, Z., Zhou, W., Chen, H., Jiang, Y., et al. (2016). MiR-150 Promotes Cellular Metastasis in Non-small Cell Lung Cancer by Targeting FOXO4. Sci. Rep. 6, 39001. doi:10.1038/srep39001
Li, P., Wang, J., Zhi, L., and Cai, F. (2021). Linc00887 Suppresses Tumorigenesis of Cervical Cancer through Regulating the miR-454-3p/FRMD6-Hippo axis. Cancer Cell Int. 21 (1), 33. doi:10.1186/s12935-020-01730-w
Li, W., Feng, Y., Ma, Z., and Lu, L. (2018). Expression of miR-454-3p and its Effect on Proliferation, Invasion and Metastasis of Colon Cancer. Nan Fang. Yi Ke Da Xue Xue Bao 38 (12), 1421–1426. doi:10.12122/j.issn.1673-4254.2018.12.04
Li, Y., Jiao, Y., Fu, Z., Luo, Z., Su, J., and Li, Y. (2019). High miR-454-3p Expression Predicts Poor Prognosis in Hepatocellular Carcinoma. Cmar Vol. 11, 2795–2802. doi:10.2147/cmar.s196655
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA Methylomes at Base Resolution Show Widespread Epigenomic Differences. Nature 462 (7271), 315–322. doi:10.1038/nature08514
Liu, S., Ge, X., Su, L., Zhang, A., and Mou, X. (2018). MicroRNA-454 Inhibits Nonsmall Cell Lung Cancer Cells Growth and Metastasis via Targeting Signal Transducer and Activator of Transcription-3. Mol. Med. Rep. 17 (3), 3979–3986. doi:10.3892/mmr.2017.8350
Modhukur, V., Iljasenko, T., Metsalu, T., Lokk, K., Laisk-Podar, T., and Vilo, J. (2018). MethSurv: a Web Tool to Perform Multivariable Survival Analysis Using DNA Methylation Data. Epigenomics 10 (3), 277–288. doi:10.2217/epi-2017-0118
Nebbioso, A., Tambaro, F. P., Dell’Aversana, C., and Altucci, L. (2018). Cancer Epigenetics: Moving Forward. PLoS Genet. 14 (6), e1007362. doi:10.1371/journal.pgen.1007362
Norouzi, M., and Hardy, P. (2021). Clinical Applications of Nanomedicines in Lung Cancer Treatment. Acta Biomater. 121, 134–142. doi:10.1016/j.actbio.2020.12.009
Olbromski, M., Podhorska-Okołów, M., and Dzięgiel, P. (2020). Role of SOX Protein Groups F and H in Lung Cancer Progression. Cancers (Basel) 12 (11), 13235. doi:10.3390/cancers12113235
Pan, X., Gong, D., Nguyen, D. N., Zhang, X., Hu, Q., Lu, H., et al. (2018). Early Microbial Colonization Affects DNA Methylation of Genes Related to Intestinal Immunity and Metabolism in Preterm Pigs. DNA Res. Int. J. rapid Publ. Rep. Genes Genomes 25 (3), 287–296. doi:10.1093/dnares/dsy001
Quintanal-Villalonga, Á., and Molina-Pinelo, S. (2019). Epigenetics of Lung Cancer: a Translational Perspective. Cell Oncol. 42 (6), 739–756. doi:10.1007/s13402-019-00465-9
Ren, L., Chen, H., Song, J., Chen, X., Lin, C., Zhang, X., et al. (2019). MiR-454-3p-Mediated Wnt/β-Catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics 9 (2), 449–465. doi:10.7150/thno.29055
Rodriguez-Canales, J., Parra-Cuentas, E., and Wistuba, (2016). Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat. Res. 170, 25–46. doi:10.1007/978-3-319-40389-2_2
Schulz, W. A., Alexa, A., Jung, V., Hader, C., Hoffmann, M. J., Yamanaka, M., et al. (2007). Factor Interaction Analysis for Chromosome 8 and DNA Methylation Alterations Highlights Innate Immune Response Suppression and Cytoskeletal Changes in Prostate Cancer. Mol. Cancer 6, 14. doi:10.1186/1476-4598-6-14
Schulz, W. A., Ingenwerth, M., Djuidje, C. E., Hader, C., Rahnenführer, J., and Engers, R. (2010). Changes in Cortical Cytoskeletal and Extracellular Matrix Gene Expression in Prostate Cancer Are Related to Oncogenic ERG Deregulation. BMC Cancer 10, 505. doi:10.1186/1471-2407-10-505
Shi, D., Li, H., Zhang, J., and Li, Y. (2021). CircGDI2 Regulates the Proliferation, Migration, Invasion and Apoptosis of OSCC via miR-454-3p/FOXF2 Axis. Cancer. Manag. Res. Vol. 13, 1371–1382. doi:10.2147/cmar.s277096
Shukla, V., Varghese, V. K., Kabekkodu, S. P., Mallya, S., Chakrabarty, S., Jayaram, P., et al. (2019). Enumeration of Deregulated miRNAs in Liquid and Tissue Biopsies of Cervical Cancer. Gynecol. Oncol. 155 (1), 135–143. doi:10.1016/j.ygyno.2019.08.012
Song, Y., Guo, Q., Gao, S., and Hua, K. (2019). miR-454-3p Promotes Proliferation and Induces Apoptosis in Human Cervical Cancer Cells by Targeting TRIM3. Biochem. biophysical Res. Commun. 516 (3), 872–879. doi:10.1016/j.bbrc.2019.06.126
Turchinovich, A., Weiz, L., and Burwinkel, B. (2012). Extracellular miRNAs: the Mystery of Their Origin and Function. Trends Biochem. Sci. 37 (11), 460–465. doi:10.1016/j.tibs.2012.08.00
Uddin, A., and Chakraborty, S. (2018). Role of miRNAs in Lung Cancer. J. Cell Physiol. 20, 1–10. doi:10.1002/jcp.26607
van Rens, M. T. M., van den Bosch, J. M. M., Brutel de la Rivière, A., and Elbers, H. R. J. (2000). Prognostic Assessment of 2,361 Patients Who Underwent Pulmonary Resection for Non-small Cell Lung Cancer, Stage I, II, and IIIA. Chest 117 (2), 374–379. doi:10.1378/chest.117.2.374
Wang, H., Liu, C., Debnath, G., Baines, A. J., Conboy, J. G., Mohandas, N., et al. (2010). Comprehensive Characterization of Expression Patterns of Protein 4.1 Family Members in Mouse Adrenal Gland: Implications for Functions. Histochem Cell Biol. 134 (4), 411–420. doi:10.1007/s00418-010-0749-z
Wang, L., He, M., Fu, L., and Jin, Y. (2020). Exosomal Release of microRNA-454 by Breast Cancer Cells Sustains Biological Properties of Cancer Stem Cells via the PRRT2/Wnt axis in Ovarian Cancer. Life Sci. 257, 118024. doi:10.1016/j.lfs.2020.118024
Wang, Z., Ma, B., Li, H., Xiao, X., Zhou, W., Liu, F., et al. (2016). Protein 4.1N Acts as a Potential Tumor Suppressor Linking PP1 to JNK-C-Jun Pathway Regulation in NSCLC. Oncotarget 7 (1), 509–523. doi:10.18632/oncotarget.6312
Yamada, D., Kikuchi, S., Williams, Y. N., Sakurai-Yageta, M., Masuda, M., Maruyama, T., et al. (2006). Promoter Hypermethylation of the Potential Tumor suppressorDAL-1/4.1Bgene in Renal Clear Cell Carcinoma. Int. J. Cancer 118 (4), 916–923. doi:10.1002/ijc.21450
Yang, Q., Liu, J., and Wang, Z. (2021). 4.1N-Mediated Interactions and Functions in Nerve System and Cancer. Front. Mol. Biosci. 8, 711302. doi:10.3389/fmolb.2021.711302
Yang, Q., Zhu, M., Wang, Z., Li, H., Zhou, W., Xiao, X., et al. (2016). 4.1N Is Involved in a Flotillin-1/β-catenin/Wnt Pathway and Suppresses Cell Proliferation and Migration in Non-small Cell Lung Cancer Cell Lines. Tumor Biol. 37 (9), 12713–12723. doi:10.1007/s13277-016-5146-3
Yang, X., Han, H., De Carvalho, D. D., Lay, F. D., Jones, P. A., and Liang, G. (2014). Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 26 (4), 577–590. doi:10.1016/j.ccr.2014.07.028
Zhang, Y., Xu, R., Li, G., Xie, X., Long, J., and Wang, H. (2012). Loss of Expression of the Differentially Expressed in Adenocarcinoma of the Lung (DAL-1) Protein Is Associated with Metastasis of Non-small Cell Lung Carcinoma Cells. Tumor Biol. 33 (6), 1915–1925. doi:10.1007/s13277-012-0452-x
Zhong, S., Golpon, H., Zardo, P., and Borlak, J. (2021). miRNAs in Lung Cancer. A Systematic Review Identifies Predictive and Prognostic miRNA Candidates for Precision Medicine in Lung Cancer. Transl. Res. 230, 164–196. doi:10.1016/j.trsl.2020.11.012
Zhou, S., Shen, Y., Zheng, M., Wang, L., Che, R., Hu, W., et al. (2017). DNA Methylation of METTL7A Gene Body Regulates its Transcriptional Level in Thyroid Cancer. Oncotarget 8 (21), 34652–34660. doi:10.18632/oncotarget.16147
Keywords: 4.1N/EPB41L1, lung adenocarcinoma (LUAD), methylation, miR-454-3P, non–small-cell lung cancer (NSCLC), lung squamous cell carcinoma (LUSC)
Citation: Yang Q, Zhu L, Ye M, Zhang B, Zhan P, Li H, Zou W and Liu J (2022) Tumor Suppressor 4.1N/EPB41L1 is Epigenetic Silenced by Promoter Methylation and MiR-454-3p in NSCLC. Front. Genet. 13:805960. doi: 10.3389/fgene.2022.805960
Received: 31 October 2021; Accepted: 08 April 2022;
Published: 20 June 2022.
Edited by:
Biaoru Li, Augusta University, United StatesCopyright © 2022 Yang, Zhu, Ye, Zhang, Zhan, Li, Zou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, amluZ2xpdWNzdUBob3RtYWlsLmNvbQ==; Wen Zou, em91d2VuMjl3QGNzdS5lZHUuY24=; Hui Li, bGlodWlzY2llbmNlQDE2My5jb20=
†These authors contributed equally to this work
 Qin Yang
Qin Yang Lin Zhu1†
Lin Zhu1† Bin Zhang
Bin Zhang Wen Zou
Wen Zou Jing Liu
Jing Liu