- Department of Bone and Joint Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Osteoarthritis (OA) is a complicated disease with both hereditary and environmental causes. Despite an increase in reports of possible OA risk loci, it has become clear that genetics is not the sole cause of osteoarthritis. Epigenetics, which can be triggered by environmental influences and result in transcriptional alterations, may have a role in OA pathogenesis. The majority of recent research on the epigenetics of OA has been focused on DNA methylation, histone modification, and non-coding RNAs. However, this study will explore epigenetic regulation in OA at the present stage. How genetics, environmental variables, and epigenetics interact will be researched, shedding light for future studies. Their possible interaction and control processes open up new avenues for the development of innovative osteoarthritis treatment and diagnostic techniques.
Introduction
Osteoarthritis (OA) is a widespread joint disease that affects around 15% of the world’s population (Glyn-Jones et al., 2015). OA was once thought to be a degenerative illness arose from chronic wear-and-tear and mechanical stress. However, the emerging paradigm now sees it as a complex process involving interactions between a wide range of internal and environmental factors (Figure 1). The genetic component of the disease is complex. Current evidence supports the theory of a polygenic inheritance, as published studies have reported a variety of OA-related risk loci (Styrkarsdottir et al., 2018; Zengini et al., 2018; Styrkarsdottir et al., 2019; Tachmazidou et al., 2019). However, studies have shown that more than 80% of the disease-related variants are located in non-coding regions of the genome (Maurano et al., 2012). As a result, it has been proposed that changes in gene expression, rather than changes in the genetic code sequence, are more likely to influence OA development. Indeed, epigenetics, a crucial method of gene expression regulation, has been implicated in the start and progression of OA in recent research (Ramos and Meulenbelt, 2017).
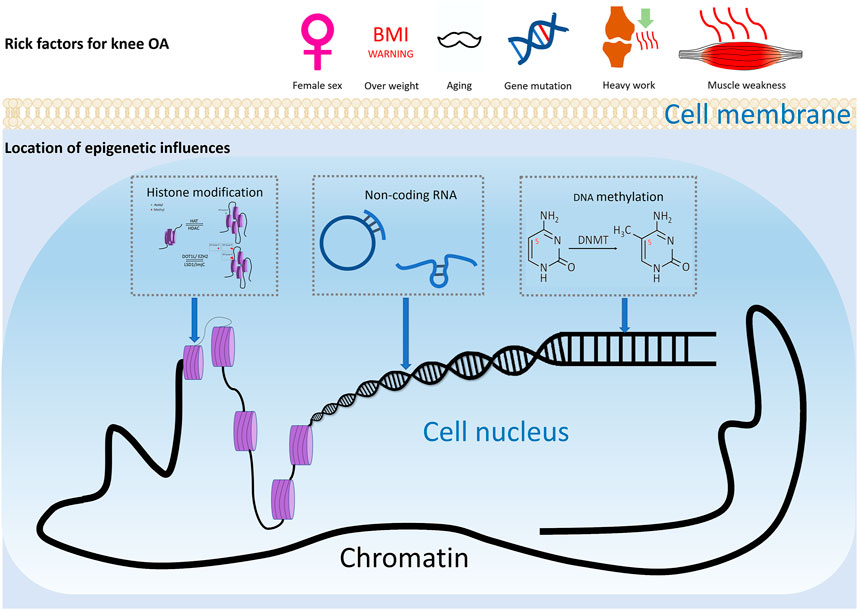
FIGURE 1. Risk factors of knee OA. Risk factors for knee osteoarthritis includes ageing, gender, injury, and joint overloading, etc. Epigenetics may play a considerable role in how these environmental factors lead to altered gene expression and ultimately pathophysiologic manifestations such as cartilage damage and subchondral osteosclerosis.
Epigenetics is an essential gene-environment interaction process. Epigenetic modification, unlike genomic modifications, is more versatile and reversible. Regulations of epigenetics vary by cell type and gene. Epigenetic phenomena including DNA methylation, genomic imprinting, maternal effects, post-translational modifications of histones, RNA (controlled by non-coding regulatory RNAs), and epigenetic chromatin remodeling (three-dimensional structure of chromatin) have been well studied. (Simon and Jeffries, 2017). With the help of modern testing techniques, epigenetic studies of knee osteoarthritis are now possible to reveal how the external environment affects changes in somatic cells and tissues. Many studies have been conducted on the epigenetics of osteoarthritis, although they have primarily focused on DNA methylation, histone modification, and miRNA (Ramos and Meulenbelt, 2017). This section will review the epigenetic regulation and reciprocity based on previous findings, trying to seek the connection, investigate how genetics and epigenetics interact, and steer future research toward identifying biologically significant alterations and gaining a better understanding of the mechanisms involved.
DNA Methylation
The most fully studied methylation is 5-methylcytosine (5 mC) (Miranda-Duarte, 2018). CpG dinucleotides are distributed unevenly throughout the human genome, with the bulk of CpG islands found in the promoter and exon regions of genes 300 to 3000bp in length. As a result, the methylation level of CpGs can be dynamically controlled by DNMTs and TETs to regulate transcription (Figure 2) (Rice et al., 2018).
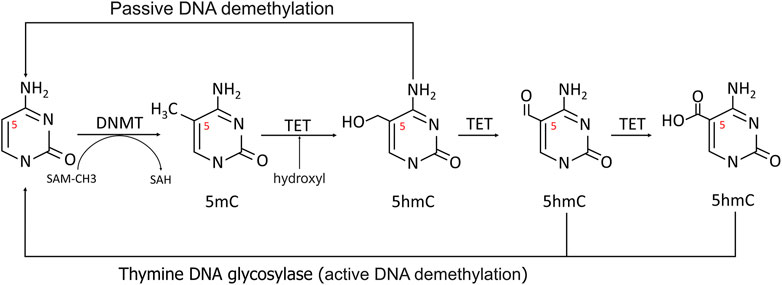
FIGURE 2. Methylation and demethylation of DNA. In methylation process, the 5′carbon of cytosine was methylated to 5-MC with S-adenosine methionine (SAM) as the methyl donor. During active demethylation, the methyl group of 5 mC can be modified by the TETs-mediated addition of hydroxyl groups to generate 5-hydroxymethylcytosine (5hmC), which can be subsequently processed in demethylation process.
Recent research into global and gene-specific methylation has proved a clear link between methylation and the development of OA. Helmtrud I. et al. were the first to study individual methylation loci. They investigated the amount of methylation of proximal promoter regions of aggrecanases1 (ADAMTS 4) and matrix metalloproteinase (MMPs) 2 (gelatinase A), MMP3 (stromelysin 1), MMP9 (gelatinase B), and MMP13 (collagenase 3) (Roach et al., 2005). Despite the fact that the demethylation and sensitivity to demethylation of these four enzymes differed significantly, the degree of demethylation of at least one CpG site in each enzyme in OA was statistically higher than in the control group.
Additional DNA methylation locations encompassing numerous OA-related chemicals or pathways have been discovered in subsequent cartilage tissue studies, and these genes can be classified into four main categories. The first category addresses the extracellular matrix’s homeostasis, which includes COL2A1, COL9A1, COL10A1, ACAN, MMP2, MMP3, MMP9, MMP13, ADAMTS-4 (Pöschl et al., 2005; Roach et al., 2005; Dickhut et al., 2009; Imagawa et al., 2014). Inflammation-related molecules, such as IL1b, IL8, IL32, TGF-β2, IL1RN, and WNT11, fall under the second category (Hashimoto et al., 2009; Moazedi-Fuerst et al., 2014; Takahashi et al., 2015). SOX4, SOX9, RUNX2, and SOD2 are members of the third group, which are involved in cartilage maintenance (Ezura et al., 2009; Kim et al., 2013). Growth factors such as BMP7, SOST, and GDF5 fall within the fourth category (Loeser et al., 2009; Reynard et al., 2011; Papathanasiou et al., 2015). DNA methylation is known to vary greatly between tissues and stages of illness. Such studies only identify methylation sites that may be related to disease due to sampling issues. Still, these findings add to the current understanding of OA epigenetics.
CpG Methylation Patterns Correlated With OA-Associated SNPs
Direct SNP correlations or gene-environment interactions may be mediated by these methylation quantitative trait loci (meQTLs). Eilis H. et al. investigated DNA methylation at 850,000 locations across the genome using a MethylationEPIC BeadChip (850K chip) in samples from the Understanding Society UK Household Longitudinal Study (UKHLS) (n = 1,111) (Hannon et al., 2018). They discovered 548 significant DNA methylation quantitative trait loci correlations between 2,907,234 genetic variations and 93,268 DNA methylation sites, resulting in 548 significant DNA methylation quantitative trait loci. They offered two key concepts: 1) mQTL-associated SNP mutations were more common within 500 kb of the DNA methylation site (designated as cis), and the cis SNP variation had a much more considerable influence on DNA methylation; 2) proximal methylation sites shared mQTL-associated SNP mutations. Differential methylation region refers to the methylation region regulated by such SNPs (DMR). These results explained the association between SNP changes and surrounding DMR in the proximal regions of several genes correlated with OA risk in osteoarthritis (RUNX2, PLEC, ALDH1A2, GDF5, MGP, COLgalt2, and COL11A2) (Figure 3) (Reynard et al., 2014; Shepherd et al., 2018; Rice et al., 2019a; Shepherd et al., 2019). Further mQTL research revealed 24 CpGs linked to genes including COLgalt2, COL11A2, RAPH1, FGFR3 and WWP2. Importantly, WWP2 encloses miRNA 140, which have been shown to be critical in cartilage maintenance, as miRNA 140 have severe cartilage defects (Duan et al., 2020). These findings demonstrate that DNA methylation can influence gene and non-coding RNA expression, as well as gene transcription and translation (Table 1).
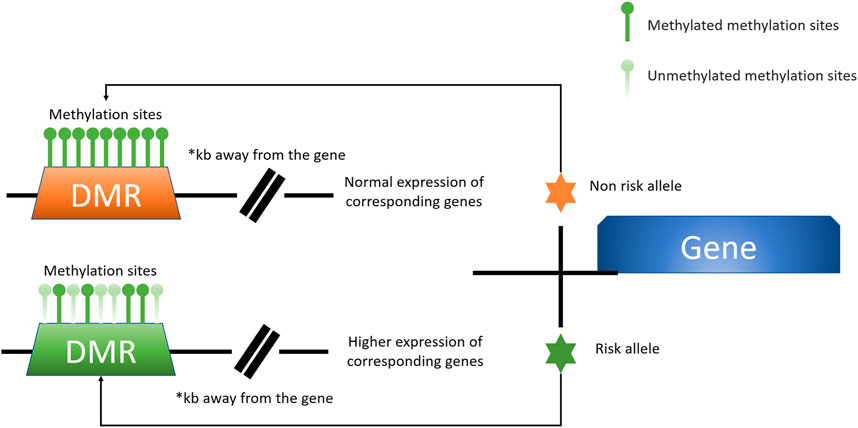
FIGURE 3. A model for the presence of altered levels of DMR methylation due to specific risk alleles is shown here. Certain OA-associated SNP mutations may lead to alterations in the degree of DMR methylation at a distance from the associated causative gene, which affects the expression of the associated gene. This unique mode of action is better studied and defined in the locus of chromosome 6p21.1 labelled rs10948172, with the associated gene RUNX2 (Zhang G. et al., 2020).
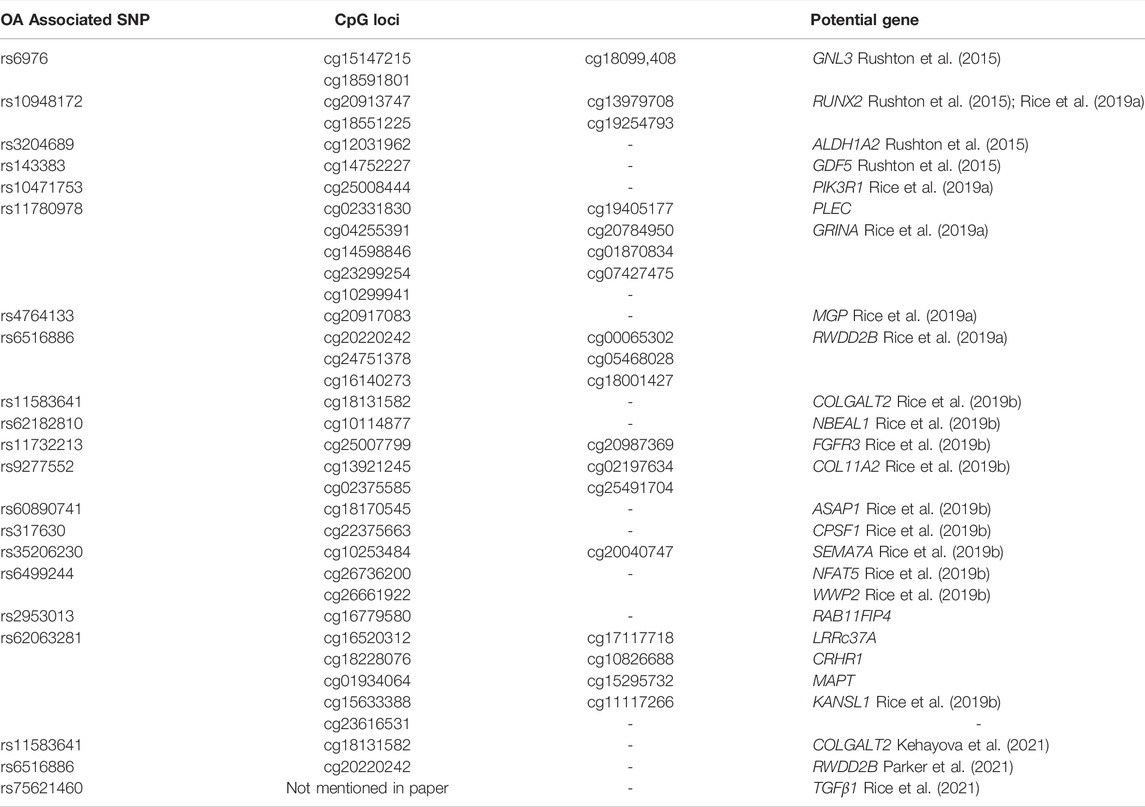
TABLE 1. Genes with a differentially methylated region potentially related to single nucleotide polymorphism
Risk Factors and DNA Methylation in OA
Animal models have provided light on the relationship between recognized OA risk factors like injury, aging and altered methylation patterns in the formation of OA. Destabilization of the medial meniscus, for example, can lead to OA development and altered 5mc and 5hmc patterns in 12-week-old male mice (Singh et al., 2019). Furthermore, tamoxifen-induced articular chondrocyte specific deletion of DNMT3b (AGCCREERT2; DNMT3bfl/fl), the de novo methyltransferase, can result in OA-like phenotypes in knee joints; whereas DNMT3b overexpression in articular chondrocytes can delay OA development after meniscus ligament injury (Shen et al., 2017).
Other OA risk factors, such as obesity, appear to be mediated by DNA methylation. Obesity has long been assumed to cause OA due to increased mechanical loading and wear and tear. Recent evidence shows that even in non-weight-bearing joints, such as hands, obesity still seems to be a significant risk factor. Leptin, a cytokine-like peptide hormone secreted by white adipose tissue, has long been stipulated to connect obesity with OA. Because obese patients generally have a high level of leptin in their bodies, their blood level is regulated significantly by body fat content. This is most likely owing to the obese patient’s altered methylation pattern in the leptin (lep) promoter region. According to a recent study, patients after bariatric surgery have lower serum leptin levels and a different methylation profile in the lep and lepr gene promoter regions. These findings back up previous studies in rat models, which indicated that a high-energy diet could alter lep methylation patterns (Milagro et al., 2009). While inhibiting leptin signaling in ob−/− and DB−/− mice models resulted in obesity, it did not affect the occurrence of knee OA (Griffin et al., 2014). The suppression of lep methylation by 5′-Aza-2-deoxycytidine (AZA) increased MMP13 activity in patient chondrocytes in vitro. MMP-13 was elevated after leptin’s epigenetic reactivation, and small interfering RNA against lep inhibited it directly (Iliopoulos et al., 2007). Overall, existing evidence suggests that leptin methylation could link obesity and OA development.
The Role of DNA Methyltransferase in OA
Members of the DNMTs family include DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3l. The main function of DNMT2 is to catalyze the methylation of RNA, while DNMT3l does not have catalytic activity. CpG methylation is determined by three DNA methyltransferases: DNMT1, DNMT3A, and DNMT3B. DNMT1 is responsible for maintaining methylation during DNA replication and damage repair. DNMT3A and DNMT3B play major role in de novo methylation (Gao L. et al., 2020). A previous matched case-control study found a connection between DNMT polymorphisms and primary knee OA. Under a co-dominant and dominant paradigm, the CT haplotype of DNMT1 polymorphisms was related to a lower risk of OA. In contrast, the CC genotype of rs2424913 of DNMT3b was associated with an increased risk (Miranda-Duarte et al., 2020).
Differential expression of DNMT1 in human chondrocytes collected from different areas in articular chondrocytes can be triggered by IL-1, a pro-inflammatory cytokine. Notably, deep and superficial zone chondrocytes, instead of transition zone chondrocytes, enhanced DNMT1 protein expression and activity in response to IL-1 (Akhtar and Haqqi, 2013). The demethylation of DMNT1 by AZA can greatly increase the expression of pro-inflammatory or matrix carbolic proteins such as COX2, MMP9, and MMP13. Only DNMT3b was expressed in the mature normal cartilage of the knee joint and TMJ, but not DNMT3a or DNMT1. Additionally, and the occurrence of OA in both joints would lead to the decrease of DNMT3b expression (Shen et al., 2017). Overexpression of DNMT3b also appears to slow the onset of osteoarthritis caused by trauma or chemicals (Shen et al., 2017; Zhou et al., 2019).
Histone Modification
The most in-depth study on OA’s overall accessible chromatin landscape change was performed by comparing cartilage from both the outer region of the lateral tibial plateau and the inner region of the medial tibial plateau from the same patient. The accessible chromatin landscapes of injured and undamaged tissues display strikingly distinct chromatin signatures, notwithstanding patient-to-patient differences (Liu et al., 2018). Further analysis revealed that enhancers account for the majority of differentially accessible peaks, including enhancers from BMPR1b, WNT5a, and FGFR2, all of which are known to play a role in OA-related cellular activity. Histone modification is shown to regulate some essential chondrocyte regulator genes, such as SOX9, as well as functioning in conjunction with epigenetic regulators to affect downstream genes (Kim et al., 2013). SOX11 and WNT5b, two OA-related genes, had differing accessible peaks in their promoter regions.
Histone Acetylation in OA
Histone acetylation is a dynamic process governed by two distinct enzyme families: HATs and HDACs. HDACs can modify nonhistone proteins to affect a variety of cellular processes in addition to their effects on chromatin structure (Figure 4).
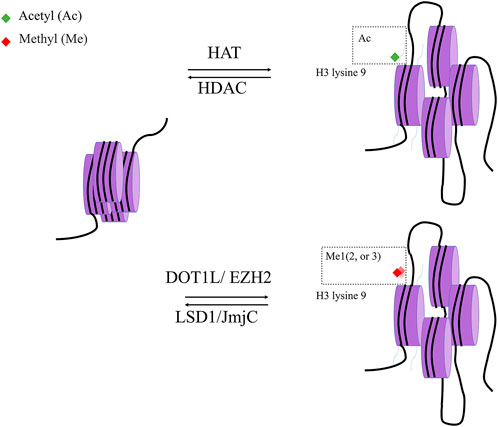
FIGURE 4. histone modification. Histone acetylase (HAT) and Histone deacetylase (HDAC) are two classes of enzymes responsible for the structural modification of chromosomes and regulation of gene expression. In general, acetylation of histones facilitates the dissociation of DNA from histone octamers and the relaxation of nucleosome structure, thus allowing various transcription factors and co-transcription factors to bind specifically to DNA binding sites and activate gene transcription. In contrast, deacetylation of histones exerts the opposite effect with Histone acetylase.
Currently, most research on HDACs’ functions in OA pathogenesis shows their interactions with non-histone proteins, which change target gene transcription. HDAC1 and HDAC2 were detected in large amounts in human chondrocytes and synovial cells from OA patients. They may alter the expression of cartilage-specific genes such as COL9A1, COL11A1, COMP, AGGRECAN, and Dermatopontin, likely via interaction with SNAIL protein (Hong et al., 2009). HDAC4, which is not indicated in normal knee cartilage, was highly expressed in OA cartilage in other research employing knee cartilage specimens from OA patients and normal donors. modulate JNK and ERK1/2 activation to mediate IL1b-induced matrix catabolic protein synthesis (Wang et al., 2018). The HDAC9-PIASy-RNF4 axis promotes chondrocyte hypertrophy by regulating the sumo and ubiquitination of Nkx3.2/Bapx1, which is degraded by the proteasome, according to studies on HDAC9 in osteoarthritis (Choi et al., 2016).
A few studies on the regulation of mesenchymal stem cell destiny discloses that the epigenetic role of HDAC may also be involved in the development of OA. In rate bone marrow stromal cells, HDAC8 can inhibit osteogenesis via two pathways: it can inhibit histone H3 lysine 9 acetylation, which reduces the osteogentic protein RUNX2, Osterix, Osteopotin and ALP. It can also associate with RUNX2 to repress its transcriptional activity (Lee et al., 2019). Human MSCs increased global histone acetylation following in situ stiffening in a hydrogel environment by reducing HDAC activity. Histone acetylation and Lamin A/C, the mechanosensor signaling protein, were considerably low in subchondral bone isolated from OA patients, while HDAC activity was significantly higher (Lee et al., 2019).
Sirtuins (SIRTs) are histone and protein deacetylases dependent on nicotinamide dinucleotide (NAD+). SIRTs and SIRT-dependent epigenetic control have been widely reported to play vital roles in DNA repair, inflammation, and aging-related illnesses (Lee et al., 2019). Chondrocyte hypertrophy and proteoglycan production were reduced in SIRT6-deficient mice from the mesenchyme (Ailixiding et al., 2015). Sirtuin 1 (SIRT1), like SOX9, is essential for maintaining cartilage homeostasis (Tsuda et al., 2003). Boosted SIRT1 expression increased the expression of cartilage ECM genes such as COL2A1, COL9A1, AGGRECAN and COMP. SIRT1 has been found to connect with SOX9 and p300, GN5, both of which are involved in nucleosome acetylation in the COL2 promoter region. SIRT3 is mostly found in mitochondria, and its deacetylation activity regulates mitochondrial function, regeneration, and kinetics (Ansari et al., 2017). These activities are hypothesized to maintain REDOX equilibrium in cell metabolism, preventing oxidative stress (Bause and Haigis, 2013).
Studies concentrating on deacetylase/acetylases still have significant limitations because HATs/HDACs can modify transcription factors as well as signaling molecules, thus affecting the expression of other distant genes (Zhao et al., 2008). HDAC inhibitors, for example, can stimulate the acetylation of other proteins in several physiological pathways in addition to increasing histone acetylation. The non-targeted protein deacetylase inhibitors Zolinza (Vorinostat) and Istodax (Romidepsin) have been approved for the treatment of cutaneous T-cell lymphoma. Histone modification medicines for osteoarthritis, on the other hand, have not yet been developed.
Histone Methylation
Typically, histone methylation is more stable than histone acetylation. Histone methylation is performed by histone methylation transferase (HMT) on lysine and arginine residues. Common locations of methylation include Lys 4, 9, 27, 36, 79 and Arg 2, 17, 26 on H3, Lys 20 and Arg 3 on H4, and Lys 4, 9, 27, 36, 79 and Arg 2, 17, 26 on H3 and H4. Studies have shown that arginine methylation on histone is a relatively dynamic marker. Arginine methylation is associated with gene activation, whereas arginine methylation loss in H3 and H4 is linked to gene silence (Greer and Shi, 2012). In contrast, lysine methylation appears to be a reasonably persistent marker for gene expression regulation.
Similar to histone acetylation, changes in histone methylation are frequently related to altered gene expression and signaling pathways in chondrocytes. For instance, H3k79 methylation was reduced in OA and RA patients (He et al., 2017), while H3K9 methylation was decreased in the temporomandibular joints of elderly mice (Ukita et al., 2020). DOT1L, an enzyme involved in histone methylation of Lys79 of H3 (H3K79), was a cartilage homeostasis regulator (Monteagudo et al., 2017). Dot1l polymorphism was found to be associated with hip joint cartilage thickness and OA risk in research involving 6,532 people (Castaño Betancourt et al., 2012). According to an in vitro study, DOT1L can methylate H3K79 of the LEFL and TCF1 genes, hence reducing WNT pathway activation and causing chondrocyte hypertrophy (Monteagudo et al., 2017). EZH2 is another histone methyltransferase discovered to modulate the WNT pathway. EZH2, which is elevated in OA articular chondrocytes, can increase trimethylation of the SFRP1 promoter and is a WNT inhibitor, leading to hyperactivation of the WNT/β-catenin signaling pathway (Chen L. et al., 2016). Overexpression of EZH2 increases the expression of MMP13, ADAMTS5, and COLX, via methylation of miR-138 promoter, resulting in cartilage breakdown (Chen L. et al., 2016). In addition to histone methylation, the demethylation process is implicated in OA as well. Jumonji domain-containing 3 (JMJD3), HEK27Me3 demethylase, was found at increased level in cartilage from the tibial plateau of the OA knee. Its inhibition in vitro by GSK4, significantly increases the expression of OA-related genes MMP13 and PTGS2.
Non-coding RNA
microRNA (miRNA) and OA
Mature miRNAs bind to complementary messenger RNA (mRNA) sequences of target genes via RNA-induced silencing complexes (RISCs) or block gene expression directly. Interactions between miRNAs and highly complementary targets lead to mRNA degradation, while incomplete interactions between miRNAs and target transcripts usually lead to translation suppression. In 2008, Dimitrios et al. found that in the chondrocytes of arthroplasty patients, miR-483, miR-22, miR-377, miR-103, miR-16, miR-223, miR-2b, miR-23b, and miR509 were elevated, and miR-29a, miR-140, miR-25, miR-337, miR-210, miR-26a, and miR-373 were decreased. The genetic signatures of these 17 miRNAs clearly distinguish OA chondrocytes from normal chondrocytes, and 17 miRNA-protein pairs that may be involved in the progression of osteoarthritis were revealed by matching microRNAs and proteomics. In this study, miR-22 and miR-103 expression were favorably connected with BMI, but miR-25, and miR-337, while miR-29a expression was inversely correlated with BMI (Iliopoulos et al., 2008). Since then, numerous research has investigated the connection between miRNA expression and effector genes in OA, including inflammation, aging, transcription factors, apoptosis, autophagy, and other pathogenic events in the evolution of the illness (Zhang et al., 2015; Wang et al., 2016; D'Adamo et al., 2016; Miyaki et al., 2010). MiR-140 is a well-known miRNA associated with osteoarthritis that can be found in peripheral blood circulation or synovial fluid during the progression of OA (Miyaki et al., 2010). In articular cartilage of OA patients, the extracellular matrix is actively remodeled under inflammatory conditions, altering the local biomechanical properties of chondrocytes and accelerating the course of OA, attesting that MiR-146a has a significant function in OA inflammatory induction (Yamasaki et al., 2009). Other previously unmentioned miRNAs associated with ECM degradation include miR-137, miR-449, miR221, miR-30a, miR-19b-3p, miR-634, miR-29, miR-107, miR-497-5p, miR-26a, miR-101 (Akbari Dilmaghnai et al., 2021). Certain miRNAs, such as miR-378 in synovial fluids and let-7e in blood, might change their expression level as the disease progresses (Beyer et al., 2015; Li et al., 2016). The intra-articular injection of certain microRNAs has the ability to reverse disease progression, which will revolutionize the treatment of osteoarthritis.
lncRNA and OA
Unlike microRNAs that rely mostly on RNA sequence complementary pairing to suppress target genes, lncRNAs operate in a significantly more sophisticated manner. lncRNAs have a unique role in a variety of gene expression regulation mechanisms, including epigenetic regulation, transcriptional regulation, and post-transcriptional regulation (Hoolwerff et al., 2020; Wang Z. et al., 2021; Statello et al., 2021).
Current research has revealed the way of axial regulation of the highly expressed lncRNA in OA tissue to the appropriate miRNA, which subsequently influences the OA target genes. HOTTIP, for instance, was engaged in the proliferation and death of OA chondrocytes via the miR-663a/FRK axis (He et al., 2021). Other lncRNA and miRNA interaction modes are more unique. Through spatial conformation, the secondary structure created by lncRNA can exert a sponge-like adsorption effect on miRNA, alter the actual contact concentration of miRNA, and the production of inflammatory genes or transcription factors in OA tissue (Chen et al., 2020a; Liu et al., 2020a; Zhang Y. et al., 2020; Zhou et al., 2021; Fan et al., 2021; Jiang et al., 2021).
Combination of Epigenetic Factors
In the pathological process of osteoarthritis, a multitude of epigenetic variables may influence the expression of OA-related genes. For instance, during articular chondrocyte death, DNMT3b facilitated the downregulation of miR-29 by increasing its promoter methylation. This, in turn, led to the overexpression of PTHLH, a process strikingly similar to that observed in tumor disorders (Dou et al., 2020). Numerous studies have identified a connection between HDACs and miRNA cooperation patterns of OA, including HDAC3, HDAC4, HDAC7, and HDAC8 (Chen et al., 2016b; Chen et al., 2016a; Mao et al., 2018; Meng et al., 2018; Zhang et al., 2019). For instance, miRNA-381 targets the 3′UTR of HDAC4, which in turn leads to increase in acetylation of H3, RUNX2 and MMP13, and ultimately result in chondrocyte hypertrophy (Chen et al., 2016a). In Table 2, a selection of the reasonably well-studied regulatory mechanisms of these models is listed.
Conclusion
As a form of non-coding genetic information, epigenetics plays distinct regulatory roles in disease onset and progression. Epigenetics has made great strides in oncology research, and drugs targeting DNA methylation and histone deacetylases have also emerged. Numerous studies in the field of OA research have demonstrated that epigenetics regulations produce interconnected and dynamic disease progression changes. OA-related signaling pathways, transcription factors, inflammatory factors, extracellular matrix (ECM) proteins, and other variables traditionally associated with OA are all sensitive to epigenetic control to varying degrees. The combination of these genetic and epigenetic variables provides more insight into the pathophysiology of osteoarthritis. Epigenetic control can link a range of genetic and environmental factors, which could provide a holistic understanding of the etiology of osteoarthritis (OA) and guide treatment.
At present, the epigenetics of osteoarthritis focuses primarily on chondrocytes, with only two published works addressing the epigenetic manifestations of subchondral bone (Jeffries et al., 2016; Zhang et al., 2016). Zhang Y et al. discovered that not only did the subchondral bone share 111 differential methylated probes (DMPs) and 41 differential methylated genes (DMGs) with chondrocytes, but they also proposed a novel hypothesis that the subchondral compartment epigenetic changes take precedence over cartilage in the development of osteoarthritis by comparing the tibia zoning analysis (Zhang et al., 2016). Due to the differential expression of epigenetics in various tissues, subchondral bone, synovium, ligaments, and other joint tissues besides chondrocytes may contradict experimental results.
For future approaches to epigenetics in the realm of osteoarthritis, it would be prudent to investigate refined single-cell research. In cartilage tissue isolated from OA samples with variable degrees of injury and degeneration, there exist chondrocytes, macrophages, fibroblasts, and numerous other types of tissue. This heterogeneity can compromise the accuracy of current knowledge of gene function and epigenetic regulation. In the case of osteoarthritic cartilage, single-cell investigations provide more precise subtyping of diseased tissues. Embryonic cell research has demonstrated the validity of single-cell methylation investigations (Lorthongpanich et al., 2013). Using EpiTOF, histone modification investigations on a single cell are now also feasible. Adopting lanthanide metal isotopes labeled antibodies and mass spectrometry, this method allows epigenetic landscape profiling at single-cell precision (Cheung et al., 2018). In the future, these techniques could be deployed in osteoarthritis research, and this refinement will bring a fresh viewpoint to the field of osteoarthritis research. Large samples and extensive genetic and epigenetic analysis, along with prospective studies of pertinent patient histories and imaging presentations, will eventually provide solid evidence to guide the tertiary prevention and individualized and precise therapy of osteoarthritis.
Author Contributions
ZC and TL contributed equally to the writing of the article and shared the title of co-first author. TL and YW share the corresponding author.
Funding
This study was funded by Natural Science Foundation of China (no. 81501855).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Special thanks go to other Department of Joint Surgery colleagues for their insightful comments on this study and Qianxia Huang for English language editing.
References
Ailixiding, M., Aibibula, Z., Iwata, M., Piao, J., Hara, Y., Koga, D., et al. (2015). Pivotal Role of Sirt6 in the Crosstalk Among Ageing, Metabolic Syndrome and Osteoarthritis. Biochem. Biophysical Res. Commun. 466 (3), 319–326. doi:10.1016/j.bbrc.2015.09.019
Akbari Dilmaghnai, N., Shoorei, H., Sharifi, G., Mohaqiq, M., Majidpoor, J., Dinger, M. E., et al. (2021). Non-coding RNAs Modulate Function of Extracellular Matrix Proteins. Biomed. Pharmacother. 136, 111240. doi:10.1016/j.biopha.2021.111240
Akhtar, N., and Haqqi, T. M. (2013). In Human Chondrocytes DNMT-1 Plays a Key Role in the Expression of Several Genes Associated with the Pathogenesis of Osteoarthritis (OA). Osteoarthr. Cartil. 21 (Suppl. l), S35–S36. doi:10.1016/j.joca.2013.02.094
Ansari, A., Rahman, M. S., Saha, S. K., Saikot, F. K., Deep, A., and Kim, K.-H. (2017). Function of the SIRT3 Mitochondrial Deacetylase in Cellular Physiology, Cancer, and Neurodegenerative Disease. Aging Cell 16 (1), 4–16. doi:10.1111/acel.12538
Bause, A. S., and Haigis, M. C. (2013). SIRT3 Regulation of Mitochondrial Oxidative Stress. Exp. Gerontol. 48 (7), 634–639. doi:10.1016/j.exger.2012.08.007
Beyer, C., Zampetaki, A., Lin, N.-Y., Kleyer, A., Perricone, C., Iagnocco, A., et al. (2015). Signature of Circulating microRNAs in Osteoarthritis. Ann. Rheum. Dis. 74 (3), e18. doi:10.1136/annrheumdis-2013-204698
Castaño Betancourt, M. C., Cailotto, F., Kerkhof, H. J., Cornelis, F. M. F., Doherty, S. A., Hart, D. J., et al. (2012). Genome-wide Association and Functional Studies Identify the DOT1L Gene to Be Involved in Cartilage Thickness and Hip Osteoarthritis. Proc. Natl. Acad. Sci. U.S.A. 109 (21), 8218–8223. doi:10.1073/pnas.1119899109
Chen, K., Fang, H., and Xu, N. (2020). LncRNA LOXL1-AS1 Is Transcriptionally Activated by JUND and Contributes to Osteoarthritis Progression via Targeting the miR-423-5p/KDM5C axis. Life Sci. 258, 118095. doi:10.1016/j.lfs.2020.118095
Chen, L., Wu, Y., Wu, Y., Wang, Y., Sun, L., and Li, F. (2016). The Inhibition of EZH2 Ameliorates Osteoarthritis Development through the Wnt/β-Catenin Pathway. Sci. Rep. 6, 29176. doi:10.1038/srep29176
Chen, W., Sheng, P., Huang, Z., Meng, F., Kang, Y., Huang, G., et al. (2016a). MicroRNA-381 Regulates Chondrocyte Hypertrophy by Inhibiting Histone Deacetylase 4 Expression. Int. J. Mol. Sci. 17 (9), 1377. doi:10.3390/ijms17091377
Chen, W., Chen, L., Zhang, Z., Meng, F., Huang, G., Sheng, P., et al. (2016b). MicroRNA-455-3p Modulates Cartilage Development and Degeneration through Modification of Histone H3 Acetylation. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1863 (12), 2881–2891. doi:10.1016/j.bbamcr.2016.09.010
Chen, Y., Guo, H., Li, L., Bao, D., Gao, F., Li, Q., et al. (2020a). Long Non-coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. Med. Sci. Monit. 26, e923868. doi:10.12659/MSM.923868
Chen, Y., Zhang, L., Li, E., Zhang, G., Hou, Y., Yuan, W., et al. (2020b). Long-chain Non-coding RNA HOTAIR Promotes the Progression of Osteoarthritis via Sponging miR-20b/PTEN axis. Life Sci. 253, 117685. doi:10.1016/j.lfs.2020.117685
Cheung, P., Vallania, F., Warsinske, H. C., Donato, M., Schaffert, S., Chang, S. E., et al. (2018). Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 173 (6), 1385–1397. doi:10.1016/j.cell.2018.03.079
Choi, H.-J., Kwon, S., and Kim, D.-W. (2016). A Post-translational Modification Cascade Employing HDAC9-PIASy-RNF4 axis Regulates Chondrocyte Hypertrophy by Modulating Nkx3.2 Protein Stability. Cell. Signal. 28 (9), 1336–1348. doi:10.1016/j.cellsig.2016.06.006
D'Adamo, S., Alvarez-Garcia, O., Muramatsu, Y., Flamigni, F., and Lotz, M. K. (2016). MicroRNA-155 Suppresses Autophagy in Chondrocytes by Modulating Expression of Autophagy Proteins. Osteoarthr. Cartil. 24 (6), 1082–1091. doi:10.1016/j.joca.2016.01.005
Dickhut, A., Pelttari, K., Janicki, P., Wagner, W., Eckstein, V., Egermann, M., et al. (2009). Calcification or Dedifferentiation: Requirement to Lock Mesenchymal Stem Cells in a Desired Differentiation Stage. J. Cell. Physiol. 219 (1), 219–226. doi:10.1002/jcp.21673
Dou, P., He, Y., Yu, B., and Duan, J. (2020). Downregulation of microRNA-29b by DNMT3B Decelerates Chondrocyte Apoptosis and the Progression of Osteoarthritis via PTHLH/CDK4/RUNX2 axis. Aging (Albany NY) 13, 7676–7690. doi:10.18632/aging.103778
Duan, L., Liang, Y., Xu, X., Xiao, Y., and Wang, D. (2020). Recent Progress on the Role of miR-140 in Cartilage Matrix Remodelling and its Implications for Osteoarthritis Treatment. Arthritis Res. Ther. 22 (1), 194. doi:10.1186/s13075-020-02290-0
Ezura, Y., Sekiya, I., Koga, H., Muneta, T., and Noda, M. (2009). Methylation Status of CpG Islands in the Promoter Regions of Signature Genes during Chondrogenesis of Human Synovium-Derived Mesenchymal Stem Cells. Arthritis Rheum. 60 (5), 1416–1426. doi:10.1002/art.24472
Fan, H., Ding, L., and Yang, Y. (2021). lncRNA SNHG16 Promotes the Occurrence of Osteoarthritis by Sponging miR-373-3p. Mol. Med. Rep. 23 (2). doi:10.3892/mmr.2020.11756
Feng, L., Yang, Z. M., Li, Y. C., Wang, H. X., Lo, J. H. T., Zhang, X. T., et al. (2021). Linc-ROR Promotes Mesenchymal Stem Cells Chondrogenesis and Cartilage Formation via Regulating SOX9 Expression. Osteoarthr. Cartil. 29 (4), 568–578. doi:10.1016/j.joca.2020.12.020
Fu, Q., Zhu, J., Wang, B., Wu, J., Li, H., Han, Y., et al. (2021). LINC02288 Promotes Chondrocyte Apoptosis and Inflammation through miR-374a-3p Targeting RTN3. J. Gene Med. 23, e3314. doi:10.1002/jgm.3314
Gao, L., Emperle, M., Guo, Y., Grimm, S. A., Ren, W., Adam, S., et al. (2020). Comprehensive Structure-Function Characterization of DNMT3B and DNMT3A Reveals Distinctive De Novo DNA Methylation Mechanisms. Nat. Commun. 11 (1), 3355. doi:10.1038/s41467-020-17109-4
Gao, S. T., Yu, Y. M., Wan, L. P., Liu, Z. M., and Lin, J. X. (2020). LncRNA GAS5 Induces Chondrocyte Apoptosis by Down-Regulating miR-137. Eur. Rev. Med. Pharmacol. Sci. 24 (21), 10984–10991. doi:10.26355/eurrev_202011_23582
Glyn-Jones, S., Palmer, A. J. R., Agricola, R., Price, A. J., Vincent, T. L., Weinans, H., et al. (2015). Osteoarthritis. Lancet 386 (9991), 376–387. doi:10.1016/s0140-6736(14)60802-3
Greer, E. L., and Shi, Y. (2012). Histone Methylation: a Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 13 (5), 343–357. doi:10.1038/nrg3173
Griffin, T. M., Huebner, J. L., Kraus, V. B., and Guilak, F. (2014). Extreme Obesity Due to Impaired Leptin Signaling in Mice Does Not Cause Knee Osteoarthritis. Arthritis Rheum. 60 (10), 2935–2944. doi:10.1002/art.24854
Hannon, E., Gorrie-Stone, T. J., Smart, M. C., Burrage, J., Hughes, A., Bao, Y., et al. (2018). Leveraging DNA-Methylation Quantitative-Trait Loci to Characterize the Relationship between Methylomic Variation, Gene Expression, and Complex Traits. Am. J. Hum. Genet. 103 (5), 654–665. doi:10.1016/j.ajhg.2018.09.007
Hashimoto, K., Oreffo, R. O. C., Gibson, M. B., Goldring, M. B., and Roach, H. I. (2009). DNA Demethylation at Specific CpG Sites in theIL1Bpromoter in Response to Inflammatory Cytokines in Human Articular Chondrocytes. Arthritis Rheum. 60 (11), 3303–3313. doi:10.1002/art.24882
He, D., Liu, J., Hai, Y., Zhu, Q., Shen, Y., Guo, S., et al. (2017). Increased DOT1L in Synovial Biopsies of Patients with OA and RA. Clin. Rheumatol. 37 (5), 1327–1332. doi:10.1007/s10067-017-3941-x
He, X., Gao, K., Lu, S., and Wu, R. (2021). LncRNA HOTTIP Leads to Osteoarthritis Progression via Regulating miR-663a/Fyn-Related Kinase axis. BMC Musculoskelet. Disord. 22 (1), 67. doi:10.1186/s12891-020-03861-7
Hong, S., Derfoul, A., Pereira-Mouries, L., and Hall, D. J. (2009). A Novel Domain in Histone Deacetylase 1 and 2 Mediates Repression of Cartilage‐specific Genes in Human Chondrocytes. FASEB J. 23 (10), 3539–3552. doi:10.1096/fj.09-133215
Hoolwerff, M., Metselaar, P. I., Tuerlings, M., Suchiman, H. E. D., Lakenberg, N., Ramos, Y. F. M., et al. (2020). Elucidating Epigenetic Regulation by Identifying Functional Cis ‐Acting Long Noncoding RNAs and Their Targets in Osteoarthritic Articular Cartilage. Arthritis Rheumatol. 72 (11), 1845–1854. doi:10.1002/art.41396
Iliopoulos, D., Malizos, K. N., Oikonomou, P., and Tsezou, A. (2008). Integrative microRNA and Proteomic Approaches Identify Novel Osteoarthritis Genes and Their Collaborative Metabolic and Inflammatory Networks. PLoS One 3 (11), e3740. doi:10.1371/journal.pone.0003740
Iliopoulos, D., Malizos, K. N., and Tsezou, A. (2007). Epigenetic Regulation of Leptin Affects MMP-13 Expression in Osteoarthritic Chondrocytes: Possible Molecular Target for Osteoarthritis Therapeutic Intervention. Ann. Rheumatic Dis. 66 (12), 1616–1621. doi:10.1136/ard.2007.069377
Imagawa, K., de Andrés, M. C., Hashimoto, K., Itoi, E., Otero, M., Roach, H. I., et al. (2014). Association of Reduced Type IX Collagen Gene Expression in Human Osteoarthritic Chondrocytes with Epigenetic Silencing by DNA Hypermethylation. Arthritis & Rheumatology 66 (11), 3040–3051. doi:10.1002/art.38774
Jeffries, M. A., Donica, M., Baker, L. W., Stevenson, M. E., Annan, A. C., Beth Humphrey, M., et al. (2016). Genome-Wide DNA Methylation Study Identifies Significant Epigenomic Changes in Osteoarthritic Subchondral Bone and Similarity to Overlying Cartilage. Arthritis & Rheumatology 68 (6), 1403–1414. doi:10.1002/art.39555
Jiang, H., Pang, H., Wu, P., Cao, Z., Li, Z., and Yang, X. (2021). LncRNA SNHG5 Promotes Chondrocyte Proliferation and Inhibits Apoptosis in Osteoarthritis by Regulating miR-10a-5p/H3F3B axis. Connect. Tissue Res. 62 (6), 605–614. doi:10.1080/03008207.2020.1825701
Kehayova, Y. S., Watson, E., Wilkinson, J. M., Loughlin, J., and Rice, S. J. (2021). Genetic and Epigenetic Interplay Regulates COLGALT2, Contributing to Osteoarthritis Genetic Risk. Arthritis Rheumatol. doi:10.1002/art.41738
Kim, K.-I., Park, Y.-S., and Im, G.-I. (2013). Changes in the Epigenetic Status of the SOX-9 promoter in Human Osteoarthritic Cartilage. J. Bone Min. Res. 28 (5), 1050–1060. doi:10.1002/jbmr.1843
Lee, S.-H., Lee, J.-H., Lee, H.-Y., and Min, K.-J. (2019). Sirtuin Signaling in Cellular Senescence and Aging. BMB Rep. 52 (1), 24–34. doi:10.5483/bmbrep.2019.52.1.290
Li, Y.-H., Tavallaee, G., Tokar, T., Nakamura, A., Sundararajan, K., Weston, A., et al. (2016). Identification of Synovial Fluid microRNA Signature in Knee Osteoarthritis: Differentiating Early- and Late-Stage Knee Osteoarthritis. Osteoarthr. Cartil. 24 (9), 1577–1586. doi:10.1016/j.joca.2016.04.019
Lian, L. P., and Xi, X. Y. (2020). Long Non-coding RNA XIST Protects Chondrocytes ATDC5 and CHON-001 from IL-1β-induced Injury via Regulating miR-653-5p/SIRT1 axis. J. Biol. Regul. Homeost. Agents 34 (2), 379–391. doi:10.23812/19-549-A-65
Liu, Y., Chang, J.-C., Hon, C.-C., Fukui, N., Tanaka, N., Zhang, Z., et al. (2018). Chromatin Accessibility Landscape of Articular Knee Cartilage Reveals Aberrant Enhancer Regulation in Osteoarthritis. Sci. Rep. 8 (1), 15499. doi:10.1038/s41598-018-33779-z
Liu, Y., Liu, K., Tang, C., Shi, Z., Jing, K., and Zheng, J. (2020b). Long Non-coding RNA XIST Contributes to Osteoarthritis Progression via miR-149-5p/DNMT3A axis. Biomed. Pharmacother. 128, 110349. doi:10.1016/j.biopha.2020.110349
Liu, Y., Yang, Y., Ding, L., Jia, Y., and Ji, Y. (2020a). LncRNA MIR4435-2HG Inhibits the Progression of Osteoarthritis through miR-510-3p Sponging. Exp. Ther. Med. 20 (2), 1693–1701. doi:10.3892/etm.2020.8841
Loeser, R. F., Im, H.-J., Richardson, B., Lu, Q., and Chubinskaya, S. (2009). Methylation of the OP-1 Promoter: Potential Role in the Age-Related Decline in OP-1 Expression in Cartilage. Osteoarthr. Cartil. 17 (4), 513–517. doi:10.1016/j.joca.2008.08.003
Lorthongpanich, C., Cheow, L. F., Balu, S., Quake, S. R., Knowles, B. B., Burkholder, W. F., et al. (2013). Single-cell DNA-Methylation Analysis Reveals Epigenetic Chimerism in Preimplantation Embryos. Science 341 (6150), 1110–1112. doi:10.1126/science.1240617
Lu, J. F., Qi, L. G., Zhu, X. B., and Shen, Y. X. (2020). LncRNA RMRP Knockdown Promotes Proliferation and Inhibits Apoptosis in Osteoarthritis Chondrocytes by miR-206/CDK9 axis. Pharmazie 75 (10), 500–504. doi:10.1691/ph.2020.0591
Mao, G., Hu, S., Zhang, Z., Wu, P., Zhao, X., Lin, R., et al. (2018). Exosomal miR-95-5p Regulates Chondrogenesis and Cartilage Degradation via Histone Deacetylase 2/8. J. Cell Mol. Med. 22 (11), 5354–5366. doi:10.1111/jcmm.13808
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 337 (6099), 1190–1195. doi:10.1126/science.1222794
Meng, F., Li, Z., Zhang, Z., Yang, Z., Kang, Y., Zhao, X., et al. (2018). MicroRNA-193b-3p Regulates Chondrogenesis and Chondrocyte Metabolism by Targeting HDAC3. Theranostics 8 (10), 2862–2883. doi:10.7150/thno.23547
Meng, Y., Qiu, S., Sun, L., and Zuo, J. (2020). Knockdown of Exosomemediated lncPVT1 Alleviates Lipopolysaccharideinduced Osteoarthritis Progression by Mediating the HMGB1/TLR4/NFkappaB Pathway via miR935p. Mol. Med. Rep. 22 (6), 5313–5325. doi:10.3892/mmr.2020.11594
Milagro, F. I., Campión, J., García-Díaz, D. F., Goyenechea, E., Paternain, L., and Martínez, J. A. (2009). High Fat Diet-Induced Obesity Modifies the Methylation Pattern of Leptin Promoter in Rats. J. Physiol. Biochem. 65 (1), 1–9. doi:10.1007/bf03165964
Miranda-Duarte, A., Borgonio-Cuadra, V. M., González-Huerta, N. C., Rojas-Toledo, E. X., Ahumada-Pérez, J. F., Sosa-Arellano, M., et al. (2020). DNA Methyltransferase Genes Polymorphisms Are Associated with Primary Knee Osteoarthritis: a Matched Case-Control Study. Rheumatol. Int. 40 (4), 573–581. doi:10.1007/s00296-019-04474-7
Miranda-Duarte, A. (2018). DNA Methylation in Osteoarthritis: Current Status and Therapeutic Implications. Open Rheumatol. J. 12, 37–49. doi:10.2174/1874312901812010037
Miyaki, S., Sato, T., Inoue, A., Otsuki, S., Ito, Y., Yokoyama, S., et al. (2010). MicroRNA-140 Plays Dual Roles in Both Cartilage Development and Homeostasis. Genes Dev. 24 (11), 1173–1185. doi:10.1101/gad.1915510
Moazedi-Fuerst, F. C., Hofner, M., Gruber, G., Weinhaeusel, A., Stradner, M. H., Angerer, H., et al. (2014). Epigenetic Differences in Human Cartilage between Mild and Severe OA. J. Orthop. Res. 32 (12), 1636–1645. doi:10.1002/jor.22722
Monteagudo, S., Cornelis, F. M. F., Aznar-Lopez, C., Yibmantasiri, P., Guns, L.-A., Carmeliet, P., et al. (2017). DOT1L Safeguards Cartilage Homeostasis and Protects against Osteoarthritis. Nat. Commun. 8, 15889. doi:10.1038/ncomms15889
Papathanasiou, I., Kostopoulou, F., Malizos, K. N., and Tsezou, A. (2015). DNA Methylation Regulates Sclerostin (SOST) Expression in Osteoarthritic Chondrocytes by Bone Morphogenetic Protein 2 (BMP-2) Induced Changes in Smads Binding Affinity to the CpG Region of SOST Promoter. Arthritis Res. Ther. 17, 160. doi:10.1186/s13075-015-0674-6
Parker, E., Hofer, I. M. J., Rice, S. J., Earl, L., Anjum, S. A., Deehan, D. J., et al. (2021). Multi‐Tissue Epigenetic and Gene Expression Analysis Combined with Epigenome Modulation Identifies RWDD2B as a Target of Osteoarthritis Susceptibility. Arthritis Rheumatol. 73 (1), 100–109. doi:10.1002/art.41473
Pöschl, E., Fidler, A., Schmidt, B., Kallipolitou, A., Schmid, E., and Aigner, T. (2005). DNA Methylation Is Not Likely to Be Responsible for Aggrecan Down Regulation in Aged or Osteoarthritic Cartilage. Ann. Rheum. Dis. 64 (3), 477–480. doi:10.1136/ard.2004.022509
Ramos, Y. F. M., and Meulenbelt, I. (2017). The Role of Epigenetics in Osteoarthritis: Current Perspective. Curr. Opin. Rheumatol. 29 (1), 119–129. doi:10.1097/bor.0000000000000355
Reynard, L. N., Bui, C., Canty-Laird, E. G., Young, D. A., and Loughlin, J. (2011). Expression of the Osteoarthritis-Associated Gene GDF5 Is Modulated Epigenetically by DNA Methylation. Hum. Mol. Genet. 20 (17), 3450–3460. doi:10.1093/hmg/ddr253
Reynard, L. N., Bui, C., Syddall, C. M., and Loughlin, J. (2014). CpG Methylation Regulates Allelic Expression of GDF5 by Modulating Binding of SP1 and SP3 Repressor Proteins to the Osteoarthritis Susceptibility SNP Rs143383. Hum. Genet. 133 (8), 1059–1073. doi:10.1007/s00439-014-1447-z
Rice, S. J., Roberts, J. B., Tselepi, M., Brumwell, A., Falk, J., Steven, C., et al. (2021). Genetic and Epigenetic Factors Fine-Tune TGFB1 Expression within the Osteoarthritic Articular Joint. Arthritis Rheumatol. 73 (10), 1866–1877. doi:10.1002/art.41736
Rice, S. J., Aubourg, G., Sorial, A. K., Almarza, D., Tselepi, M., Deehan, D. J., et al. (2018). Identification of a Novel, Methylation-dependent, RUNX2 Regulatory Region Associated with Osteoarthritis Risk. Hum. Mol. Genet. 27 (19), 3464–3474. doi:10.1093/hmg/ddy257
Rice, S. J., Cheung, K., Reynard, L. N., and Loughlin, J. (2019b). Discovery and Analysis of Methylation Quantitative Trait Loci (mQTLs) Mapping to Novel Osteoarthritis Genetic Risk Signals. Osteoarthr. Cartil. 27 (10), 1545–1556. doi:10.1016/j.joca.2019.05.017
Rice, S. J., Tselepi, M., Sorial, A. K., Aubourg, G., Shepherd, C., Almarza, D., et al. (2019a). Prioritization of PLEC and GRINA as Osteoarthritis Risk Genes through the Identification and Characterization of Novel Methylation Quantitative Trait Loci. Arthritis Rheumatol. 71 (8), 1285–1296. doi:10.1002/art.40849
Roach, H. I., Yamada, N., Cheung, K. S. C., Tilley, S., Clarke, N. M. P., Oreffo, R. O. C., et al. (2005). Association between the Abnormal Expression of Matrix-Degrading Enzymes by Human Osteoarthritic Chondrocytes and Demethylation of Specific CpG Sites in the Promoter Regions. Arthritis Rheum. 52 (10), 3110–3124. doi:10.1002/art.21300
Rushton, M. D., Reynard, L. N., Young, D. A., Shepherd, C., Aubourg, G., Gee, F., et al. (2015). Methylation Quantitative Trait Locus Analysis of Osteoarthritis Links Epigenetics with Genetic Risk. Hum. Mol. Genet. 24 (25), 7432–7444. doi:10.1093/hmg/ddv433
Shen, J., Wang, C., Li, D., Xu, T., Myers, J., Ashton, J. M., et al. (2017). DNA Methyltransferase 3b Regulates Articular Cartilage Homeostasis by Altering Metabolism. JCI Insight 2 (12). doi:10.1172/jci.insight.93612
Shepherd, C., Reese, A. E., Reynard, L. N., and Loughlin, J. (2019). Expression Analysis of the Osteoarthritis Genetic Susceptibility Mapping to the Matrix Gla Protein Gene MGP. Arthritis Res. Ther. 21 (1), 149. doi:10.1186/s13075-019-1934-7
Shepherd, C., Zhu, D., Skelton, A. J., Combe, J., Threadgold, H., Zhu, L., et al. (2018). Functional Characterization of the Osteoarthritis Genetic Risk Residing at ALDH1A2 Identifies Rs12915901 as a Key Target Variant. Arthritis Rheumatol. 70 (10), 1577–1587. doi:10.1002/art.40545
Simon, T. C., and Jeffries, M. A. (2017). The Epigenomic Landscape in Osteoarthritis. Curr. Rheumatol. Rep. 19 (6), 30. doi:10.1007/s11926-017-0661-9
Singh, P., Lessard, S. G., Mukherjee, P., Carballo, C. B., Rodeo, S. A., and Otero, M. (2019). The Progression of Post-traumatic Osteoarthritis in the Murine DMM Model Is Marked by Distinctive Epigenomic Patterns Associated with Transcriptomic Changes in Articular Cartilage. Osteoarthr. Cartil. 27, S282. doi:10.1016/j.joca.2019.02.664
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Styrkarsdottir, U., Stefansson, O. A., Gunnarsdottir, K., Thorleifsson, G., Lund, S. H., Stefansdottir, L., et al. (2019). GWAS of Bone Size Yields Twelve Loci that Also Affect Height, BMD, Osteoarthritis or Fractures. Nat. Commun. 10, 2054. doi:10.1038/s41467-019-09860-0
Styrkarsdottir, U., Lund, S. H., Thorleifsson, G., Zink, F., Stefansson, O. A., Sigurdsson, J. K., et al. (2018). Meta-analysis of Icelandic and UK Data Sets Identifies Missense Variants in SMO, IL11, COL11A1 and 13 More New Loci Associated with Osteoarthritis. Nat. Genet. 50 (12), 1681–1687. doi:10.1038/s41588-018-0247-0
Tachmazidou, I., Hatzikotoulas, K., Hatzikotoulas, K., Southam, L., Esparza-Gordillo, J., Haberland, V., et al. (2019). Identification of New Therapeutic Targets for Osteoarthritis through Genome-wide Analyses of UK Biobank Data. Nat. Genet. 51 (2), 230–236. doi:10.1038/s41588-018-0327-1
Takahashi, A., de Andrés, M. C., Hashimoto, K., Itoi, E., and Oreffo, R. O. C. (2015). Epigenetic Regulation of Interleukin-8, an Inflammatory Chemokine, in Osteoarthritis. Osteoarthr. Cartil. 23 (11), 1946–1954. doi:10.1016/j.joca.2015.02.168
Tan, F., Wang, D., and Yuan, Z. (2020). The Fibroblast-like Synoviocyte Derived Exosomal Long Non-coding RNA H19 Alleviates Osteoarthritis Progression through the miR-106b-5p/TIMP2 Axis. Inflammation 43 (4), 1498–1509. doi:10.1007/s10753-020-01227-8
Tsuda, M., Takahashi, S., Takahashi, Y., and Asahara, H. (2003). Transcriptional Co-activators CREB-Binding Protein and P300 Regulate Chondrocyte-specific Gene Expression via Association with Sox9. J. Biol. Chem. 278 (29), 27224–27229. doi:10.1074/jbc.m303471200
Ukita, M., Matsushita, K., Tamura, M., and Yamaguchi, T. (2020). Histone H3K9 Methylation Is Involved in Temporomandibular Joint Osteoarthritis. Int. J. Mol. Med. 45 (2), 607–614. doi:10.3892/ijmm.2019.4446
Wang, B., Li, J., and Tian, F. (2021). Downregulation of lncRNA SNHG14 Attenuates Osteoarthritis by Inhibiting FSTL-1 Mediated NLRP3 and TLR4/NF-Κb Pathway through miR-124-3p. Life Sci. 270, 119143. doi:10.1016/j.lfs.2021.119143
Wang, C.-L., Zuo, B., Li, D., Zhu, J.-F., Xiao, F., Zhang, X.-L., et al. (2020). The Long Noncoding RNA H19 Attenuates Force-Driven Cartilage Degeneration via miR-483-5p/Dusp5. Biochem. Biophysical Res. Commun. 529 (2), 210–217. doi:10.1016/j.bbrc.2020.05.180
Wang, J., Chen, L., Jin, S., Lin, J., Zheng, H., Zhang, H., et al. (2016). MiR-98 Promotes Chondrocyte Apoptosis by Decreasing Bcl-2 Expression in a Rat Model of Osteoarthritis. Acta Biochim. Biophys. Sin. 48 (10), 923–929. doi:10.1093/abbs/gmw084
Wang, P., Mao, Z., Pan, Q., Lu, R., Huang, X., Shang, X., et al. (2018). Histone Deacetylase-4 and Histone Deacetylase-8 Regulate Interleukin-1β-Induced Cartilage Catabolic Degradation through MAPK/JNK and ERK Pathways. Int. J. Mol. Med. 41 (4), 2117–2127. doi:10.3892/ijmm.2018.3410
Wang, Z., Ni, S., Zhang, H., Fan, Y., Xia, L., and Li, N. (2021). Silencing SGK1 Alleviates Osteoarthritis through Epigenetic Regulation of CREB1 and ABCA1 Expression. Life Sci. 268, 118733. doi:10.1016/j.lfs.2020.118733
Xiao, P., Zhu, X., Sun, J., Zhang, Y., Qiu, W., Li, J., et al. (2021). LncRNA NEAT1 Regulates Chondrocyte Proliferation and Apoptosis via Targeting miR-543/PLA2G4A axis. Hum. Cell 34 (1), 60–75. doi:10.1007/s13577-020-00433-8
Xu, J., Pei, Y., Lu, J., Liang, X., Li, Y., Wang, J., et al. (2021). LncRNA SNHG7 Alleviates IL-1β-induced Osteoarthritis by Inhibiting miR-214-5p-Mediated PPARGC1B Signaling Pathways. Int. Immunopharmacol. 90, 107150. doi:10.1016/j.intimp.2020.107150
Yamasaki, K., Nakasa, T., Miyaki, S., Ishikawa, M., Deie, M., Adachi, N., et al. (2009). Expression of MicroRNA-146a in Osteoarthritis Cartilage. Arthritis Rheum. 60 (4), 1035–1041. doi:10.1002/art.24404
Zengini, E., Hatzikotoulas, K., Tachmazidou, I., Steinberg, J., Hartwig, F. P., Southam, L., et al. (2018). Genome-wide Analyses Using UK Biobank Data Provide Insights into the Genetic Architecture of Osteoarthritis. Nat. Genet. 50 (4), 549–558. doi:10.1038/s41588-018-0079-y
Zhang, C., Zhang, Z., Chang, Z., Mao, G., Hu, S., Zeng, A., et al. (2019). miR‐193b‐5p Regulates Chondrocytes Metabolism by Directly Targeting Histone Deacetylase 7 in Interleukin‐1β‐induced Osteoarthritis. J Cell. Biochem. 120 (8), 12775–12784. doi:10.1002/jcb.28545
Zhang, D., Cao, X., Li, J., and Zhao, G. (2015). MiR-210 Inhibits NF-Κb Signaling Pathway by Targeting DR6 in Osteoarthritis. Sci. Rep. 5, 12775. doi:10.1038/srep12775
Zhang, G., Zhang, Q., Zhu, J., Tang, J., and Nie, M. (2020). LncRNA ARFRP1 Knockdown Inhibits LPS-Induced the Injury of Chondrocytes by Regulation of NF-Κb Pathway through Modulating miR-15a-5p/TLR4 axis. Life Sci. 261, 118429. doi:10.1016/j.lfs.2020.118429
Zhang, H., Li, J., Shao, W., and Shen, N. (2020). LncRNA SNHG9 Is Downregulated in Osteoarthritis and Inhibits Chondrocyte Apoptosis by Downregulating miR-34a through Methylation. BMC Musculoskelet. Disord. 21 (1), 511. doi:10.1186/s12891-020-03497-7
Zhang, P., Sun, J., Liang, C., Gu, B., Xu, Y., Lu, H., et al. (2020). lncRNA IGHCγ1 Acts as a ceRNA to Regulate Macrophage Inflammation via the miR-6891-3p/TLR4 Axis in Osteoarthritis. Mediat. Inflamm. 2020, 9743037. doi:10.1155/2020/9743037
Zhang, X., Huang, C.-r., Pan, S., Pang, Y., Chen, Y.-s., Zha, G.-c., et al. (2020). Long Non-coding RNA SNHG15 Is a Competing Endogenous RNA of miR-141-3p that Prevents Osteoarthritis Progression by Upregulating BCL2L13 Expression. Int. Immunopharmacol. 83, 106425. doi:10.1016/j.intimp.2020.106425
Zhang, Y., Dong, Q., and Sun, X. (2020). Positive Feedback Loop LINC00511/miR-150-5p/SP1 Modulates Chondrocyte Apoptosis and Proliferation in Osteoarthritis. DNA Cell Biol. 39 (9), 1506–1512. doi:10.1089/dna.2020.5718
Zhang, Y., Fukui, N., Yahata, M., Katsuragawa, Y., Tashiro, T., Ikegawa, S., et al. (2016). Identification of DNA Methylation Changes Associated with Disease Progression in Subchondral Bone with Site-Matched Cartilage in Knee Osteoarthritis. Sci. Rep. 6, 34460. doi:10.1038/srep34460
Zhao, W., Kruse, J.-P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008). Negative Regulation of the Deacetylase SIRT1 by DBC1. Nature 451 (7178), 587–590. doi:10.1038/nature06515
Zhou, L., Gu, M., Ma, X., Wen, L., Zhang, B., Lin, Y., et al. (2021). Long Non-coding RNA PCAT-1 Regulates Apoptosis of Chondrocytes in Osteoarthritis by Sponging miR-27b-3p. J. Bone Min. Metab. 39 (2), 139–147. doi:10.1007/s00774-020-01128-8
Zhou, Y., Chen, M., O'Keefe, R. J., Shen, J., Li, Z., Zhou, J., et al. (2019). Epigenetic and Therapeutic Implications of Dnmt3b in Temporomandibular Joint Osteoarthritis. Am. J. Transl. Res. 11 (3), 1736–1747.
Keywords: epignetics, knee osteoarthritis, DNA methylation, histone modification, noncoding RNA
Citation: Cai Z, Long T, Zhao Y, Lin R and Wang Y (2022) Epigenetic Regulation in Knee Osteoarthritis. Front. Genet. 13:942982. doi: 10.3389/fgene.2022.942982
Received: 13 May 2022; Accepted: 20 June 2022;
Published: 08 July 2022.
Edited by:
Heng Sun, University of Macau, ChinaReviewed by:
Korosh Morshedi, Kashan University of Medical Sciences, IranJianjie Li, Xuzhou Medical University, China
Copyright © 2022 Cai, Long, Zhao, Lin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teng Long, bG9uZ3RlbmdAcmVuamkuY29t; You Wang, eW91d2FuZ0ByZW5qaS5jb20=
†These authors have contributed equally to this work and share first authorship
 Zhengyu Cai
Zhengyu Cai Teng Long
Teng Long Yaochao Zhao
Yaochao Zhao Ruixin Lin
Ruixin Lin You Wang
You Wang