- 1State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 2State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, China
- 3Genomic & Phenomic Data Center, Chengdu 23Mofang Biotechnology Co., Ltd, Chengdu, China
- 4Department of Biobank, Chengdu 23Mofang Biotechnology Co., Ltd, Chengdu, China
- 5Department of Laboratory Medicine/Research Center of Clinical Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, China
- 6Department of Dermatology, Rare Disease Center, West China Hospital, Sichuan University, Chengdu, China
- 7Lung Cancer Center, West China Hospital, Sichuan University, Chengdu, China
- 8State Key Laboratory of Biotherapy, Department of Thoracic Oncology, Cancer Center, West China Hospital, Sichuan University, Chengdu, China
Background: The aesthetic facial traits are closely related to life quality and strongly influenced by genetic factors, but the genetic predispositions in the Chinese population remain poorly understood.
Methods: A genome-wide association studies (GWAS) and subsequent validations were performed in 26,806 Chinese on five facial traits: widow’s peak, unibrow, double eyelid, earlobe attachment, and freckles. Functional annotation was performed based on the expression quantitative trait loci (eQTL) variants, genome-wide polygenic scores (GPSs) were developed to represent the combined polygenic effects, and single nucleotide polymorphism (SNP) heritability was presented to evaluate the contributions of the variants.
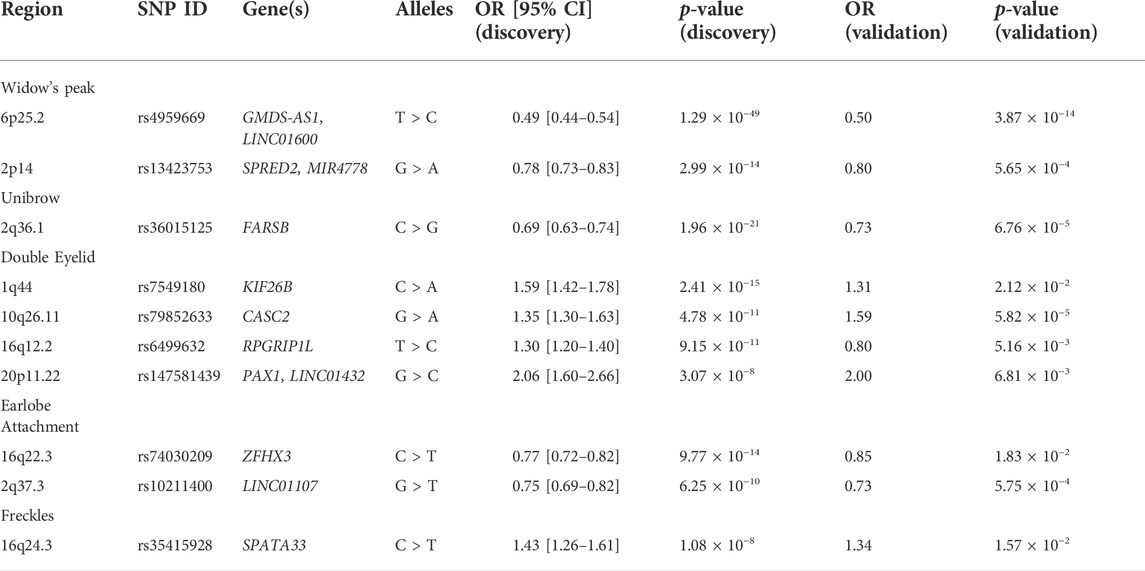
Results: In total, 21 genetic associations were identified, of which ten were novel: GMDS-AS1 (rs4959669, p = 1.29 × 10−49) and SPRED2 (rs13423753, p = 2.99 × 10−14) for widow’s peak, a previously unreported trait; FARSB (rs36015125, p = 1.96 × 10−21) for unibrow; KIF26B (rs7549180, p = 2.41 × 10−15), CASC2 (rs79852633, p = 4.78 × 10−11), RPGRIP1L (rs6499632, p = 9.15 × 10−11), and PAX1 (rs147581439, p = 3.07 × 10−8) for double eyelid; ZFHX3 (rs74030209, p = 9.77 × 10−14) and LINC01107 (rs10211400, p = 6.25 × 10−10) for earlobe attachment; and SPATA33 (rs35415928, p = 1.08 × 10−8) for freckles. Functionally, seven identified SNPs tag the missense variants and six may function as eQTLs. The combined polygenic effect of the associations was represented by GPSs and contributions of the variants were evaluated using SNP heritability.
Conclusion: These identifications may facilitate a better understanding of the genetic basis of features in the Chinese population and hopefully inspire further genetic research on facial development.
1 Introduction
Facial features exhibit a higher degree of variability than other physical features, thus making human faces unique and recognizable. Appearance variations impact quality of life, in most cases, from the perspective of aesthetics. Although sometimes acquired over the lifespan due to external factors, the variation is closely connected with the inherited complexity of facial morphogenesis (Weinberg et al., 2013; Cole et al., 2017). The correlation has been extensively researched in genetic studies and experimental animal models (Weinberg et al., 2019), and a thorough understanding of the genetic basis of specific facial traits provides insights into, for instance, the mechanisms of facial morphogenesis as well as biometrics and forensic science (Kayser and De Knijff, 2011; Claes, 2014; Sturm and Duffy, 2018).
Despite evidence accumulated to illustrate the association between facial traits and genetic variants (Liu et al., 2012; Huang et al., 2021), a considerable fraction remains to be discovered. Our study aimed at five aesthetic facial features: widow’s peak, unibrow, double eyelid, earlobe attachment, and freckles. To date, the correlated genetic factors involved in some of these traits have been studied. For instance, unibrow has been reported with associations in 2q36 near the PAX3 gene (Adhikari et al., 2016). Regarding eyelid trait that has a pronounced level of variation in East Asians, genome-wide association studies (GWASs) have revealed HOXD-MTX2 to be relevant to eyelid curvature in Koreans (Seongwon et al., 2018) and EMX2 associated with eyelid folding in Japanese (Chihiro et al., 2018). Meanwhile, a large-scale multiethnic GWAS revealed multiple loci associated with earlobe attachment harboring several candidate genes (e.g. MRPS22, EDAR, and PAX9) (Shaffer et al., 2017). Moreover, several variants of pigmentary genes, such as BNC2, IRF4, and MC1R, have been identified by recent studies, especially in Caucasians (Maarten et al., 2001; Eriksson et al., 2010; Jacobs et al., 2015; Kim et al., 2022), while only a few studies have been performed in Asians, mainly in Japanese and Korean population (Chihiro et al., 2018; Shido et al., 2019; Shin et al., 2021). In spite of the previous findings, the genetic background of these facial traits remains far from fully understood, especially in the Chinese population. Besides, although long been understood to have a genetic basis, genetic predispositions to widow’s peak, an important aesthetic trait, have not been reported.
Therefore, we performed a large-scale GWAS on the Chinese population to gain insights into genetic variants contributing to the five aesthetic facial traits (widow’s peak, unibrow, double eyelid, earlobe attachment, and freckles). Functional annotation of the genome-wide significant single nucleotide polymorphisms (SNPs) was performed based on the expression quantitative trait loci (eQTL) variants and genome-wide polygenic scores (GPSs) were subsequently developed to represent the combined polygenic effects in these five traits (Supplementary Figure S1). In total, 21 associations were identified and ten of them were novel (Figure 1). Specifically, to our knowledge, this is the first genetic report of widow’s peak, identifying GMDS-AS1 (rs4959669, p = 1.29 × 10−49) and SPRED2 (rs13423753, p = 2.99 × 10−14) as genome-wide significant associations in the Chinese population. The other novel associations included FARSB (rs36015125, p = 1.96 × 10−21) for unibrow; KIF26B (rs7549180, p = 2.41 × 10−15), CASC2 (rs79852633, p = 4.78 × 10−11), RPGRIP1L (rs6499632, p = 9.15 × 10−11), and PAX1 (rs147581439, p = 3.07 × 10−8) for double eyelid; ZFHX3 (rs74030209, p = 9.77 × 10−14) and LINC01107 (rs10211400, p = 6.25 × 10−10) for earlobe attachment; and SPATA33 (rs35415928, p = 1.08 × 10−8) for freckles. This study was expected to facilitate a better understanding of the genetic basis of the facial features and inspire further research on the biological functions of the relevant genes.
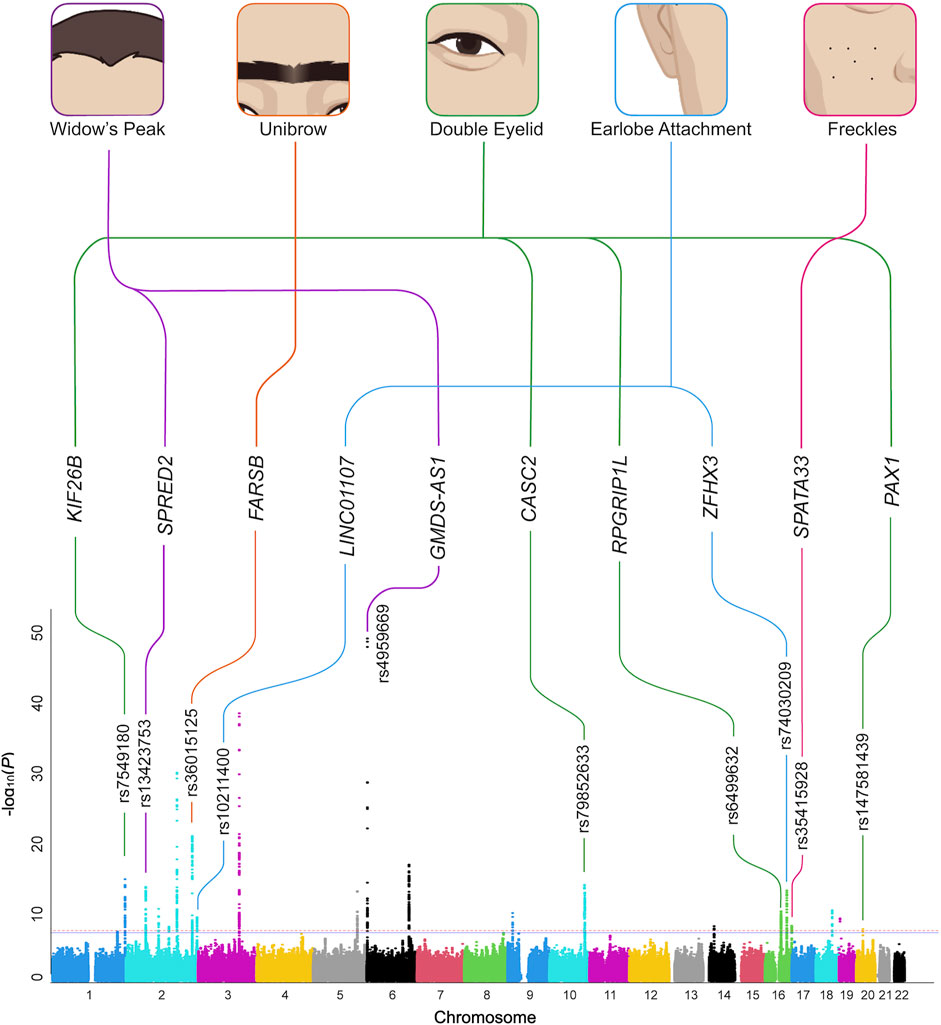
FIGURE 1. Overview of the GWAS results. The five aesthetic facial traits studied in the Chinese study sample (top) are connected with the candidate genes identified in regions with novel genome-wide significant associations. The GWAS results of the five traits were summarized on a single composite Manhattan plot (bottom). The rs ID of the SNP with the smallest p-value at the top of each association peak is given. GWAS, genome-wide association study. SNP, single nucleotide polymorphism.
2 Materials and methods
2.1 Subject, sample, and phenotypes
Subjects were voluntarily enrolled in the study and filled out the questionnaires designed by West China Hospital, Sichuan University. Questionnaires soliciting trait information including “widow’s peak,” “unibrow,” “double eyelid,” “earlobe attachment,” and “freckles,” were collected (Supplementary Methods). Subsequently, phenotypical data were filtered and merged on the grounds of the verification questions to ensure the authenticity and accuracy of the preprocessed data. The study was approved by the local ethics committee [West China Hospital, Sichuan University, approval no. 2017(241)] and all participants signed an electronic informed consent form. Methods were performed following the relevant guidelines and regulations.
2.2 DNA extraction and genotyping
Each participant donated 2 ml of their saliva into a sample tube which was later sent to the laboratory to extract DNA, and DNA quality was determined by examining the OD260/OD280 ratio and integrity in agarose gels. Due to the long time span of the project, samples were randomly genotyped with one of the three highly correlated versions of chip arrays – Mofang v1.0, Mofang v2.0, and Mofang v2.1, which were all Affymetrix Axiom Precision Medicine Research Array (PMRA)-based high-throughput SNP chip arrays (Affymetrix, Santa Clara, CA, United States).
2.3 Quality controls
To control the genotyping quality, QCs were performed at both the individual and SNP levels: 1) SNP with genotype call rate (CR) below 0.98, 2) individual CR below 0.98, 3) gender inconsistencies, 4) number of alleles >2, 5) minor allele frequency (MAF) below 0.01 (Supplementary Table S1), 6) deviation from Hardy–Weinberg equilibrium (p-value < 1 × 10−6), 7) outliers ±3 SD from the samples’ heterozygosity rate, 8) individuals with cryptic relatedness, 9) outliers from multidimensional scaling (MDS) analysis (Xu et al., 2015a; Xu et al., 2015b; Qian et al., 2019; Zhang et al., 2019; Hao et al., 2021; Hertz et al., 2021) (Supplementary Methods; Supplementary Figure S1).
2.4 Genome-wide association study
For each of the five traits, 80% of the samples were randomly selected to perform GWASs as the discovery set, and the rest 20% were used for validation (Supplementary Table S2). Additional QC was further performed before the association analyses: inclusion of SNPs with CR 0.98 and MAF ≥0.01, removal of heterozygosity outliers, removal of individuals with cryptic relatedness and population structure outliers. The genotype frequency between cases and controls was compared with sex, age, and five top principal components (PCs) as covariates, by the logistic regression model using PLINK v1.90b5.4 (Supplementary Methods) (Chang et al., 2015).
2.5 Functional annotation
The genome-wide SNPs were subjected to HaploReg database (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) (Ward and Kellis, 2012), WashU EpiGenome Browser (http://epigenomegateway.wustl.edu/browser/) (Zhou et al., 2015) and Genotype-Tissue Expression (GTEx) dataset (https://gtexportal.org/home/) (GTEx Consortium, 2015) for functional annotation.
2.6 Construction of the GPSs
28 candidate GPSs based on a pruning and thresholding (P-T) method were derived for each trait using the GWAS discovery set and discovery GWAS summary statistics from the previous step. The best scores, defined by the maximal area under curve (AUC), were applied to the validation set with 20% samples to generate a polygenic score for each individual. The individuals were binned into 20 groups according to the GPS quantile and the prevalence of each trait (Supplementary Methods) (Khera et al., 2018).
2.7 Estimation of SNP heritability
To evaluate the contribution of the variants to heritability, SNP heritability of the five traits was estimated based on the GWAS summary statistics using linkage disequilibrium score regression analysis (LDSC) (Bulik-Sullivan et al., 2015).
3 Results
In this study, a total of 26,806 Chinese volunteers who passed QC were enrolled to investigate the associations between genetic variants and the five facial traits: widow’s peak (N = 11,946), unibrow (N = 7,254), double eyelid (N = 7,473), earlobe attachment (N = 9,977), and freckles (N = 8,251). All volunteers had phenotype information of one or more traits. The discovery and validation sets for each trait were randomly drawn respectively (Supplementary Table S2; Supplementary Figure S1). GWAS was performed for each trait, showing no obvious inflation (Supplementary Figure S2). Associations of genome-wide significant signals in the discovery sets with a predisposition to each trait were estimated in the respective validation sets to verify their reliability (Table 1; Supplementary Table S3).
3.1 Association analyses of the five facial traits
3.1.1 Widow’s peak
Although widow’s peak is regarded as a genetic heritable phenotypic pattern (Rassman et al., 2013; Kyriakou et al., 2021), genetic study of this trait is still lacking. In the present study, three loci reached genome-wide significance (p-value < 5 × 10−8) in the discovery cohort, including the strongest signals at 6p25.2 downstream GMDS-AS1 (top SNP: rs4959669, p = 1.29 × 10−49), followed by 2p14 downstream SPRED2 (top SNP: rs13423753, p = 3.0 × 10−14) and 2q22.3 downstream ARHGAP15 (top SNP: rs4662351, p = 1.42 × 10−8) (Table 1; Figure 2A, Figures 3A,B). However, associations could only be reproduced for rs4959669 and rs13423753, but not for rs4662351 (Supplementary Table S3).
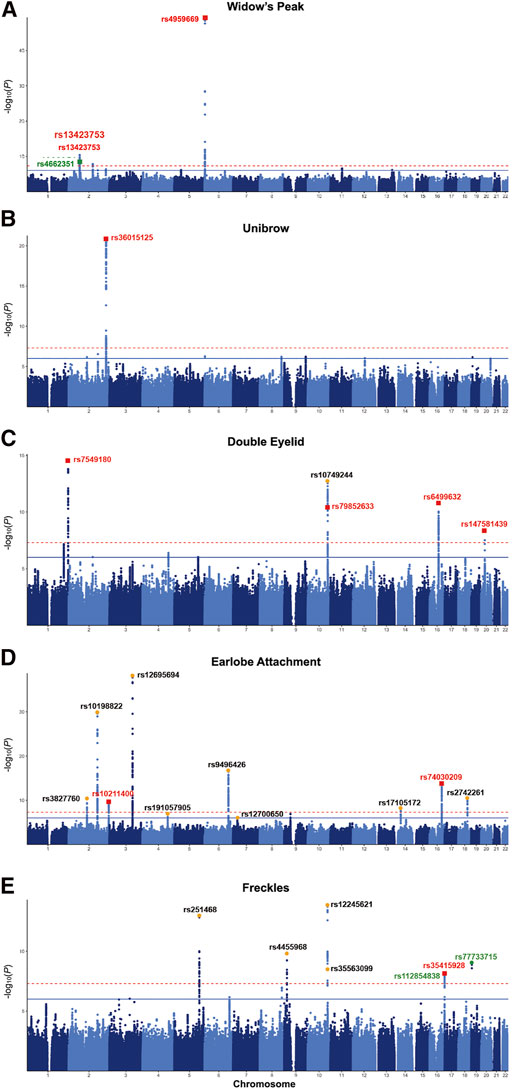
FIGURE 2. Manhattan plots of the discovery GWAS. Manhattan plot for (A) widow’s peak; (B) unibrow; (C) double eyelid; (D) earlobe attachment; (E) freckles. Bonferroni corrected threshold and candidate threshold correspond to 7.30 and 5.30, respectively, with regard to −log10 (P). Previously unreported SNPs are marked RED, previously reported SNPs are dotted ORANGE, and the SNPs failing validation are marked GREEN. GWAS, genome-wide association study. SNP, single nucleotide polymorphism.
FIGURE 3. Regional association plots for eight regions with novel SNPs showing genome-wide significant associations with the five facial traits. Two novel associations for widow’s peak and novel associations with the smallest p-values for unibow, double, eyelid, earlobe attachment, and freckles are shown (A) and (B), Regional association plot for (A) 6p25.2 and (B) 2p14 with novel SNP showing genome-wide significant association with widow’s peak; (C) regional association plot for 2q36.1 showing association with unibrow; (D) regional association plot for 1q44 showing association with double eyelid; (E) regional association plot for 16q22.3 showing association with earlobe attachment; (F) regional association plot for 16q24.3 showing association with freckles. SNP, single nucleotide polymorphism.
3.1.2 Unibrow
As for unibrow, the only previously reported significant association signal, rs2218065 at 2q36.1 (Adhikari et al., 2016), could not be validated in our cohort (p = 3.11 × 10−3). Instead, we identified a locus 300–500 kb downstream of rs2218065 with the top signal at FARSB rs36015125 (p = 1.96 × 10−21) (Figure 2B, Figure 3C) and rs36015125 was not in linkage disequilibrium (LD) with rs2218065 (r2<0.2).
3.1.3 Double eyelid
Concerning double eyelid, five genetic loci were genome-wide significant and validated in the validation set, four of which have not been reported to our knowledge (Figure 2C). The loci at 10q26.11 replicated signals reported in Japanese women (Chihiro et al., 2018) with the top hit at rs10749244 near EMX2 and RAB11FIP2 (p = 1.96 × 10−13), in high LD with the reported variant rs1415425 (r2 = 0.97) (Supplementary Figure S3D). A novel locus ∼500 kb downstream of the reported one, overlapping the long noncoding RNA (lncRNA) gene CASC2 (top SNP: rs79852633, p = 4.78 × 10−11) exhibited an independent association (Supplementary Figure S3A). The other three unreported genome-wide significant loci overlapped KIF26B (1q44, top SNP: rs7549180, p = 5.75 × 10−39), RPGRIP1L (16q12.2, top SNP: rs6499632, p = 9.2 × 10−11), and PAX1/LINC01432 (20p11.22, top SNP: rs147581439, p = 3.07 × 10−8), respectively (Figure 3D; Supplementary Figure S3).
3.1.4 Earlobe attachment
For earlobe attachment, eight loci reached genome-wide significance in the discovery stage and were validated in a validation set (Figure 2D; Supplementary Table S3). Among them, two loci were novel to our knowledge, including a series of variants at 16q22.3 (top SNP: rs74030209, p = 9.8 × 10−14) and 2q37.3 (top SNP: rs10211400, p = 6.3 × 10−10) (Table 1; Figure 3E; Supplementary Figure S4A). The other six have been either previously reported or in LD with the reported SNPs in other ethnic populations (Supplementary Table S3; Supplementary Figure S4).
3.1.5 Freckles
In pursuit of genetic associations with freckles, seven genome-wide significant loci were identified in the discovery stage, while only five passed validation (Figure 2E; Supplementary Table S3), among which only one has not been previously reported (16q24.3, SPATA33, top SNP: rs35415928, p = 1.1 × 10−8) (Table 1; Figure 3F). The other significant loci that passed validation overlapped HSPA12A (10q25.3), PPARGC1B (5q32), BNC2 (9p22), and EMX2/RAB11FIP2 (10q26.11) (Supplementary Figure S5).
3.2 The possible impact of the genome-wide significant associations
Functionally, the variants identified by GWAS may impact the corresponding phenotype by either altering the amino acids or regulating the expression of their nearby genes (Moriyama et al., 2016; Zhu et al., 2018; Tam et al., 2019; Moriyama et al., 2021). In this study, almost all the genome-wide significant variants are located in noncoding regions, except for rs3827760, a missense mutation point of the EDAR gene (Supplementary Table S3).
Meanwhile, seven variants are in LD (r2 ≥ 0.2) with the coding variants based on LD calculations using 1,000 Genome Project data according to the HaploReg database (Supplementary Table S4) (Ward and Kellis, 2012). Therefore, we consider that the significant variants might mainly function by affecting gene expressions. Altogether, six of the genome-wide significant SNPs have GTEx eQTL associations (p < 1 × 10−4) in skin tissue and cultured fibroblasts that are of potential relevance to the five facial traits (Supplementary Table S5) (GTEx Consortium, 2015). Some of the SNPs show associations with only one gene. For instance, the previously unreported double eyelid-associated SNP rs6499632 is in LD with a missense variant of RPGRIP1L (r2 = 0.26) and is a strong eQTL for RP11-36I17.2 expression in cultured fibroblasts (Supplementary Table S4; Supplementary Figure S6). Meanwhile, some SNPs may have a tissue-specific eQTL association with multiple genes. The novel freckles-associated variant rs35415928 in SPATA33 serves as a strong eQTL for several genes in both skin tissue (sun-exposed and no-sun-exposed) and fibroblasts, showing the strongest association with DBNDD1 (Supplementary Figure S7), a gene involved in tanning ability (Nan et al., 2009) and squamous cell carcinoma (Asgari et al., 2016). Unibrow-associated FARSB rs36015125 is an eQTL for RP11-16P6.1, SGPP2, and FARSB expression in skin tissues (sun-exposed and no-sun-exposed) and cultured fibroblasts (Supplementary Figure S6), but information regarding these genes’ function in unibrow or hair appearance is unavailable.
3.3 Genome-wide polygenic score analysis
GPS was constructed to manifest the genomic polygenic effect. For each trait, we derived 28 GPS predictors based on a P-T method from the discovery GWAS summary statistics and selected one best predictor defined by the maximal AUC in the discovery set (Supplementary Table S6). Taking widow’s peak as an example, the AUCs of the predictors ranged from 0.563 to 0.598 and reached the maximum when p = 1 × 10−5 and r2 = 0.6 (Supplementary Table S6). Afterward, polygenic scores were generated in the validation set. Across the population, GPS was distributed with the empirical risk of the traits, showing a generally rising trend from 0.322 in the lowest quantile to 0.566 in the highest quantile (Figure 4A). Odds ratios (ORs) based on the quantile were given (Supplementary Table S7). Likewise, GPSs with the best performance were selected in the other four traits, and phenotype prevalence according to GPS was generated (Figures 4B–E). AUC reached maximum values of 0.594, 0.665, 0.657, and 0.625 in unibrow (p = 1 × 10−5, r2 = 0.4), double eyelid (p = 1 × 10−4, r2 = 0.2 or 0.8), earlobe attachment (p = 1 × 10−5, r2 = 0.4), and freckles (p = 1 × 10−7, r2 = 0.4), respectively (Supplementary Table S6).
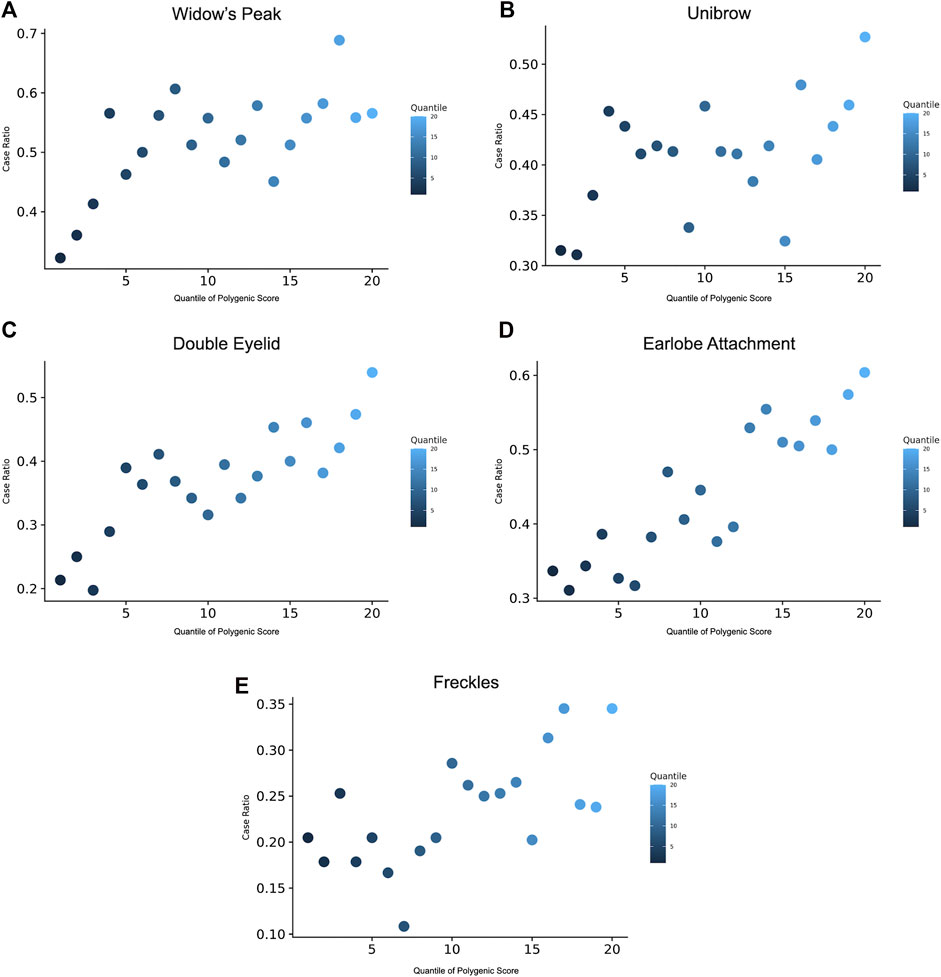
FIGURE 4. Prevalence of the traits according to the GPS quantile. 20 groups of the validation were derived based on the percentile of the GPS. Prevalence of phenotype displayed for the risk of (A) widow’s peak, (B) unibrow, (C) double eyelid, (D) earlobe attachment, and (E) freckles, within each quantile. GPS, genome-wide polygenic score.
3.4 SNP heritability of each trait
SNP heritability (h2) using LDSC for the five traits was presented. The highest h2 was seen for double eyelid (h2 = 0.4487, standard error [SE] = 0.0765), and the h2 for widow’s peak, unibrow, earlobe attachment, and freckles were estimated to be 0.3046 (SE = 0.0591), 0.433 (SE = 0.0881), 0.2443 (SE = 0.0765), and 0.1431 (SE = 0.0733), respectively.
4 Discussion
It is important to understand the complicated genetic background of the facial traits since it may facilitate further understanding of the basic mechanism of facial development. It becomes even more significant upon the notion that, while some facial traits only represent nonsyndromic conditions, some can be clinical manifestations of certain syndromes.
To our knowledge, we are the first to provide public GWAS results on widow’s peak, presenting two associated loci. The strongest signal was at 6p25.2 (rs4959669), near RNA genes GMDS-AS1 and LINC01600, but further studies are needed to verify how the genes and their variants contribute to hairline morphology. The other association (rs13423753) occurred at 2p14 near SPRED2. Of potential relevance, some other members of the SPRED family have been reported to activate MAPK cascade (Nonami et al., 2004) which is implicated in hair follicle cell development (Yoon et al., 2011). Moreover, widow’s peak sometimes manifests as a symptom of certain syndromes, such as Donnai-Barrow syndrome (Longoni et al., 1993; Khalifa et al., 2015), Waardenburg syndrome type 1 (WS1), and Aarskog syndrome (Pingault et al., 2010), but variants related to these syndromes did not reach genome-wide significance in our study, probably due to the rare syndromic incidence and the difference among populations. It is important to take into consideration rare genetic-based disorders and diseases when discussing variant and phenotype association. Noteworthily, 23andMe Co. attempted to identify significant variants for unibrow and widow’s peak. According to the regional plot released on their website (https://medical.23andme.com/), associated loci for unibrow existed on several chromosomes, while significant variants for widow’s peak were located at 2q and 6p. Since detailed information from 23andMe Co.’s research is restrained, associations in our present GWAS provide novel insights for the public.
Unibrow is also related to attractiveness in many cultures. As far as we know, the only published associations for unibrow were in 2q36 with the lead SNP of rs2218065 near the PAX3 gene, which was not validated in the present study (Adhikari et al., 2016). PAX3 is a key transcription factor that guides the normal development of neural crest derivatives (Sang et al., 2012), and its mutations have been shown to cause WS1, 85% of which has manifestations including unibrow (Pingault et al., 2010). Of potential relevance, the PAX3 locus has previously been shown to control the location of “nasion,” the point at the middle of two eyebrows (Liu et al., 2012; Paternoster et al., 2012). In the present study, unlinked significant signals occurred near PAX3 at 2q36.1, overlapping with the FARSB gene, a member of the ARS class IIc subfamily (Rodova et al., 1999). Other ARS members such as KARS, CARS, and TARS have been associated with hair phenotype (Santos-Cortez et al., 2013; Theil et al., 2019), suggesting a potential connection between FARSB and hair/brow development.
Interestingly, although East Asians are genetically closely-related, the present-day populations from different countries may have distinct genetic makeup (Wang et al., 2018), as seen in the pursuit of eyelid-associated variants. Among the five double eyelid-associated variants identified in our research, only rs10749244 at 10q26.11 is in high LD with previously reported rs1415425 (r2 = 0.97) in Japanese (Chihiro et al., 2018). Among the four novel signals, the strongest association was observed for KIF26B rs7549180. In light of findings of Kif26b in the development of face (Marikawa et al., 2004), our results may suggest a regulatory role of KIF26B in the development of facial structure and concomitant upper eyelid differences. Functionally, RPGRIP1L rs6499632 serves as a strong GTEx eQTL for RPGRIP1L in cultured fibroblasts. The gene has been suggested to be involved in mechanisms such as craniofacial development, patterning of the limbs, and formation of the left-right axis (Delous et al., 2007). Another novel association (top SNP: rs147581439) overlapped with PAX1, a member of the PAX transcription factor family that plays a critical role during fetal development. Specifically, PAX1 functions in pattern formation during embryogenesis (Wallin et al., 1994), and a missense mutation in PAX1 has been shown to cause autosomal recessive Oto-Facio-Cervical syndrome, a disorder characterized by markedly skeletal and facial abnormalities (Pohl et al., 2013).
Regarding earlobe attachment, we identified two novel associations. One (rs74030209) is an intron point of ZFHX3, in LD with mutation points of the gene (Supplementary Table S4). ZFHX3 is of potential relevance to ear development since it is involved in myogenic control by modulating myoblast differentiation (Berry et al., 2001), lack of which has been found to influence organogenesis in the inner ear phenotype (Rot et al., 2017). The other (rs10211400) is at the noncoding RNA (ncRNA) LINC01107. As some ncRNAs are correlated with nearby gene expression (Cabili et al., 2011; Guil and Esteller, 2012), the variant has a chance to be related to the regulation of TWIST2 314 kb downstream. Mutations of TWIST2 have been associated with ectodermal dysplasia, such as Ablepharon-Macrostomia syndrome and Barber-Say syndrome (Marchegiani et al., 2015) whose manifestations include dysmorphic ears. Further studies are still needed to verify the conjecture. Besides these two unreported variants, our results of the attached earlobe mostly replicated the previous findings in diverse cohorts (Dutta and Ganguly, 1965; Adhikari et al., 2015; Shaffer et al., 2017). For instance, the strongest association was seen for the intergenic SNP rs12695694 near MRPS22, showing a strong GTEx eQTL association with MRPS22 expression in cultured fibroblasts. Mutations of MRPS22 have been previously implicated in earlobe size in Latin Americans and in lobe attachment in multiple cohorts (Adhikari et al., 2015; Shaffer et al., 2017), and relatively, a homozygous mutation in MRPS22 has been reported to lead to oxidative phosphorylation system deficiency, which may manifest as dysmorphic features including low implanted posteriorly rotated ears (Smits et al., 2011). Meanwhile, the previously reported EDAR exonic variant rs3827760 (Bryk et al., 2008; Mou et al., 2008) also reached genome-wide significance in the present study. EDAR is involved in the prenatal development of ectoderm (Mikkola, 2009), and its deficiency has been suggested to result in abnormally shaped ears in mice (Adhikari et al., 2015).
Despite that pigmentary traits could be induced by extrinsic factors such as sun exposure, genetic predisposition has been suggested among different populations (Jacobs et al., 2015; Crawford et al., 2017; Chihiro et al., 2018; Adhikari et al., 2019; Shin et al., 2021). This may be because pigmentation is mainly contributed by a complicated process of melanin synthesis which is tightly associated with multiple genetic variants, and response after sun exposure is also genetically controlled (Nan et al., 2009; Shido et al., 2019). The freckles-associated variants identified in the present study were highly consistent with findings from Japanese and Korean cohorts (Chihiro et al., 2018; Shin et al., 2021), presumably due to the shared genetic backgrounds of East Asian populations. The only novel variant was SPATA33 rs35415928. SPATA33 has long been associated with facial pigmentation (Jacobs et al., 2015), cutaneous squamous cell carcinoma (Asgari et al., 2016), and melanoma (Fang et al., 2019). Closely downstream of the associations also lies the well-defined freckles-associated gene MC1R. (Maarten et al., 2001; Sulem et al., 2008; Eriksson et al., 2010). Among the identified associations, BNC2 has been identified in Europeans (Jacobs et al., 2015). The top signal within BNC2 (rs4455968) is in high LD with rs16935073 (r2 = 0.94) and rs10816035 (r2 = 0.83) that have been associated with pigmentary traits or tanning ability in Koreans (Shin et al., 2021) and Japanese (Chihiro et al., 2018; Shido et al., 2019). Another significant association existed in 10q25.3, led by HSPA12A rs12245621 which is in LD with the reported variant rs12259842 (r2 = 0.76) (Chihiro et al., 2018). HSPA12A is affiliated to HSP70 family whose members (HSP70 and HSP47) are expressed in the dermis and epidermis following laser irradiation, which has been related to pigmentation (Sajjadi et al., 2013). Interestingly, the nearby RAB11FIP2 has been proved to facilitate melanin exocytosis from melanocytes and filopodia-mediated melanin transfer (Beaumont et al., 2011; Tarafder et al., 2014), and SNP rs35563099 192 kb upstream of RAB11FIP2 also reached a genome-wide significance. To be noted, rs77733715 that has been associated with ease of tanning and darker skin color in UK BioBank samples (Sturm and Duffy, 2018) reached genome-wide significance in the discovery set but failed validation in our cohort. rs77733715 lies near the pigmentary gene MFSD12 (Crawford et al., 2017; Adhikari et al., 2019; Hédan et al., 2019; Tanaka et al., 2019) and is in LD with the well-documented pigmentation-associated missense variant MFSD12 rs2240751 (r2 = 0.56) in the Korean and Latin American populations (Adhikari et al., 2019; Shin et al., 2021).
The study also has room for improvement. First, the individuals were recruited based on their self-reported traits instead of professional assessment. As the traits included are easily distinguished aesthetic traits and illustrations were added for each question, we consider the reports reliable. Second, functional annotation suggested that some of the variants may be associated with phenotype by impacting the expression level or coding sequence of the nearby genes, but the functions of these variants should be further determined and experimentally evaluated. Moreover, SNP heritability was presented for each trait, but since it can only include contributions from causal variants tagged by the measured SNPs, it is lower than total narrow-sense heritability, such as estimation from twin or family studies. For instance, an adult twin study on the relative contribution of genetic and environmental effects on the expression of nevi and freckles suggested that additive genetic effects explained 91% of the variance in freckle counts (Bataille et al., 2000). Future studies on narrow-sense heritability could be adopted to understand the genetic contribution of the five aesthetic facial traits.
5 Conclusion
This GWAS of five aesthetic facial traits in a large Chinese cohort of 26,806 uncovered ten novel genetic associations. Specifically, this is the first study, to our knowledge, to report genetic predispositions to widow’s peak. The identified variants indicated both important similarities and differences among different ethnic groups. Hopefully, the findings would facilitate an understanding of the genetic basis of facial traits and, more importantly, facial development.
Data availability statement
The GWAS summary statistics can be found at the publicly available at http://www.biosino.org/node/project/detail/OEP002975.
Ethics statement
The studies involving human participants were reviewed and approved by West China Hospital, Sichuan University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
PW, XS, QM, HX, DB, and YZ contributed to the conceptualization; XS and QM contributed to the methodology; XS, HM, MC, and YZ performed the formal analysis; PW, QM, and MC performed the investigation; XS, HM, MC took charge in the resources; MC, SZ, and YW contributed to the data curation; PW prepared the original draft; PW, XS, and MC prepared the figures; DB and YZ supervised the study; All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
The study was supported by the National Key Research and Development Program of China (2021YFA1301203), grants from National Natural Science Foundation of China (No. 81973408, 81903735, 82002569, and 82071146), and 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYYC20003 and ZYJC18004).
Conflict of interest
Xinghan Sun and Hao Mi are employees of Chengdu 23Mofang Biotechnology. The remaining authors declare no competing interests.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.967684/full#supplementary-material
References
Adhikari, K., Reales, G., Smith, A. J. P., Konka, E., Palmen, J., Quinto-Sanchez, M., et al. (2015). A genome-wide association study identifies multiple loci for variation in human ear morphology. Nat. Commun. 6, 7500. doi:10.1038/ncomms8500
Adhikari, K., Fontanil, T., Cal, S., Mendoza-Revilla, J., Fuentes-Guajardo, M., Chacón-Duque, J.-C., et al. (2016). A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat. Commun. 7 (1), 10815. doi:10.1038/ncomms10815
Adhikari, K., Mendoza-Revilla, J., Sohail, A., Fuentes-Guajardo, M., Lampert, J., Chacón-Duque, J. C., et al. (2019). A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat. Commun. 10 (1), 358. doi:10.1038/s41467-018-08147-0
Asgari, M. M., Wang, W., Ioannidis, N. M., Itnyre, J., Hoffmann, T., Jorgenson, E., et al. (2016). Identification of susceptibility loci for cutaneous squamous cell carcinoma. J. Investigative Dermatol. 136 (5), 930–937. doi:10.1016/j.jid.2016.01.013
Bataille, V., Snieder, H., MacGregor, A. J., Sasieni, P., and Spector, T. D. (2000). Genetics of risk factors for melanoma: an adult twin study of nevi and freckles. J. Natl. Cancer Inst. 92 (6), 457–463. doi:10.1093/jnci/92.6.457
Beaumont, K. A., Hamilton, N. A., Moores, M. T., Brown, D. L., Ohbayashi, N., Cairncross, O., et al. (2011). The recycling endosome protein Rab17 regulates melanocytic filopodia formation and melanosome trafficking. Traffic 12 (5), 627–643. doi:10.1111/j.1600-0854.2011.01172.x
Berry, F. B., Miura, Y., Mihara, K., Kaspar, P., Sakata, N., Hashimoto-Tamaoki, T., et al. (2001). Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 276 (27), 25057–25065. doi:10.1074/jbc.M010378200
Bryk, J., Hardouin, E., Pugach, I., Hughes, D., Strotmann, R., Stoneking, M., et al. (2008). Positive selection in East Asians for an EDAR allele that enhances NF-κB activation. PloS one 3 (5), e2209. doi:10.1371/journal.pone.0002209
Bulik-Sullivan, B. K., Loh, P. R., Loh, P.-R., Finucane, H. K., Ripke, S., Yang, J., et al. (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47 (3), 291–295. doi:10.1038/ng.3211
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25 (18), 1915–1927. doi:10.1101/gad.17446611
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci 4, 7. doi:10.1186/s13742-015-0047-8
Chihiro, E., Johnson, T. A., Ryoko, M., Kazuyuki, N., Shigeo, K., Masanori, A., et al. (2018). Genome-wide association study in Japanese females identifies fifteen novel skin-related trait associations. Sci. Rep. 8 (1), 8974. doi:10.1038/s41598-018-27145-2
Cole, J. B., Manyama, M., Larson, J. R., Liberton, D. K., Ferrara, T. M., Riccardi, S. L., et al. (2017). Human facial shape and size heritability and genetic correlations. Genetics 205 (2), 967–978. doi:10.1534/genetics.116.193185
Crawford, N. G., Kelly, D. E., Hansen, M. E. B., Beltrame, M. H., Fan, S., Bowman, S. L., et al. (2017). Loci associated with skin pigmentation identified in African populations. Science 358 (6365), eaan8433. doi:10.1126/science.aan8433
Claes, P. (2014). Modeling 3D facial shape from DNA. PLoS Genet. 10 (3), e1004224. doi:10.1371/journal.pgen.1004224
Delous, M., Baala, L., Salomon, R., Laclef, C., Vierkotten, J., Tory, K., et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39 (7), 875–881. doi:10.1038/ng2039
Dutta, P., and Ganguly, P. (1965). Further observations on ear lobe attachment. Hum. Hered. 15, 77–86. doi:10.1159/000151894
Eriksson, N., Macpherson, J. M., Tung, J. Y., Hon, L. S., Naughton, B., Saxonov, S., et al. (2010). Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 6 (6), e1000993. doi:10.1371/journal.pgen.1000993
Fang, S., Lu, J., Zhou, X., Wang, Y., Ross, M. I., Gershenwald, J. E., et al. (2019). Functional annotation of melanoma risk loci identifies novel susceptibility genes. Carcinogenesis 41 (4), 452–457. doi:10.1093/carcin/bgz173
GTEx Consortium (2015). Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348 (6235), 648–660. doi:10.1126/science.1262110
Guil, S., and Esteller, M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19 (11), 1068–1075. doi:10.1038/nsmb.2428
Hao, Q., Cao, M., Zhang, C., Yin, D., Wang, Y., Ye, Y., et al. (2021). Age-related differences of genetic susceptibility to patients with acute lymphoblastic leukemia. Aging 13 (9), 12456–12465. doi:10.18632/aging.202903
Hédan, B., Cadieu, E., Botherel, N., Dufaure de Citres, C., Letko, A., Rimbault, M., et al. (2019). Identification of a missense variant in MFSD12 involved in dilution of phaeomelanin leading to white or cream coat color in dogs. Genes 10 (5), 386. doi:10.3390/genes10050386
Hertz, D. L., Douglas, J. A., Kidwell, K. M., Gersch, C. L., Desta, Z., Storniolo, A.-M., et al. (2021). Genome-wide association study of letrozole plasma concentrations identifies non-exonic variants that may affect CYP2A6 metabolic activity. Pharmacogenet Genomics 31 (5), 116–123. doi:10.1097/fpc.0000000000000429
Huang, Y., Li, D., Qiao, L., Liu, Y., Peng, Q., Wu, S., et al. (2021). A genome-wide association study of facial morphology identifies novel genetic loci in Han Chinese. J. Genet. Genomics 48 (3), 198–207. doi:10.1016/j.jgg.2020.10.004
Jacobs, L. C., Hamer, M. A., Gunn, D. A., Deelen, J., Lall, J. S., van Heemst, D., et al. (2015). A genome-wide association study identifies the skin color genes IRF4, MC1R, ASIP, and BNC2 influencing facial pigmented spots. J. Investigative Dermatol. 135 (7), 1735–1742. doi:10.1038/jid.2015.62
Kayser, M., and De Knijff, P. (2011). Improving human forensics through advances in genetics, genomics and molecular biology. Nat. Rev. Genet. 12 (3), 179–192. doi:10.1038/nrg2952
Khalifa, O., Al-Sahlawi, Z., Imtiaz, F., Ramzan, K., Allam, R., Al-Mostafa, A., et al. (2015). Variable expression pattern in Donnai-Barrow syndrome: report of two novel LRP2 mutations and review of the literature. Eur. J. Med. Genet. 58 (5), 293–299. doi:10.1016/j.ejmg.2014.12.008
Khera, A. V., Chaffin, M., Aragam, K. G., Haas, M. E., Roselli, C., Choi, S. H., et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50 (9), 1219–1224. doi:10.1038/s41588-018-0183-z
Kim, Y., Yin, J., Huang, H., Jorgenson, E., Choquet, H., and Asgari, M. M. (2022). Genome-wide association study of actinic keratosis identifies new susceptibility loci implicated in pigmentation and immune regulation pathways. Commun. Biol. 5 (1), 386. doi:10.1038/s42003-022-03301-3
Kyriakou, G., Glentis, A., and Papanikolaou, S. (2021). Widow's peak: a usually overlooked, yet significant morphogenetic trait. J. Dtsch. Dermatol. Ges. 19 (9), 1271–1275. doi:10.1111/ddg.14502
Liu, F., van der Lijn, F., Schurmann, C., Zhu, G., Chakravarty, M. M., Hysi, P. G., et al. (2012). A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 8 (9), e1002932. doi:10.1371/journal.pgen.1002932
Longoni, M., Kantarci, S., Donnai, D., and Pober, B. R. (1993). “Donnai-barrow syndrome,” in GeneReviews(®). Editors M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. Stephenset al. (Seattle, WA: University of Washington). Copyright © 1993-2020, GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.
Maarten, B., ter Huurne, J., Nelleke, G., Wilma, B., Rudi, W., Vermeer, B.-J., et al. (2001). The melanocortin-1-receptor gene is the major freckle gene. Hum. Mol. Genet. 10 (16), 1701. doi:10.1093/hmg/10.16.1701
Marchegiani, S., Davis, T., Tessadori, F., van Haaften, G., Brancati, F., Hoischen, A., et al. (2015). Recurrent mutations in the basic domain of TWIST2 cause Ablepharon macrostomia and barber-say syndromes. Am. J. Hum. Genet. 97 (1), 99–110. doi:10.1016/j.ajhg.2015.05.017
Marikawa, Y., Fujita, T. C., and Alarcón, V. B. (2004). An enhancer-trap LacZ transgene reveals a distinct expression pattern of Kinesin family 26B in mouse embryos. Dev. Genes Evol. 214 (2), 64–71. doi:10.1007/s00427-003-0377-x
Mikkola, M. L. (2009). Molecular aspects of hypohidrotic ectodermal dysplasia. Am. J. Med. Genet. 149a (9), 2031–2036. doi:10.1002/ajmg.a.32855
Moriyama, T., Nishii, R., Perez-Andreu, V., Yang, W., Klussmann, F. A., Zhao, X., et al. (2016). NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 48 (4), 367–373. doi:10.1038/ng.3508
Moriyama, T., Yang, W., Smith, C., Pui, C.-H., Evans, W. E., Relling, M. V., et al. (2021). Comprehensive characterization of pharmacogenetic variants in TPMT and NUDT15 in children with acute lymphoblastic leukemia. Pharmacogenet Genomics 32, 60–66. doi:10.1097/fpc.0000000000000453
Mou, C., Thomason, H. A., Willan, P. M., Clowes, C., Harris, W. E., Drew, C. F., et al. (2008). Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum. Mutat. 29 (12), 1405–1411. doi:10.1002/humu.20795
Nan, H., Kraft, P., Qureshi, A. A., Guo, Q., Chen, C., Hankinson, S. E., et al. (2009). Genome-wide association study of tanning phenotype in a population of European ancestry. J. Investigative Dermatol. 129 (9), 2250–2257. doi:10.1038/jid.2009.62
Nonami, A., Kato, R., Taniguchi, K., Yoshiga, D., Taketomi, T., Fukuyama, S., et al. (2004). Spred-1 negatively regulates interleukin-3-mediated ERK/mitogen-activated protein (MAP) kinase activation in hematopoietic cells. J. Biol. Chem. 279 (50), 52543–52551. doi:10.1074/jbc.M405189200
Paternoster, L., Zhurov, A. I., Toma, A. M., Kemp, J. P., Pourcain, B. S., Timpson, N. J., et al. (2012). Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am. J. Hum. Genet. 90, 478. doi:10.1016/j.ajhg.2011.12.021
Pingault, V., Ente, D., Dastot-Le Moal, F., Goossens, M., Marlin, S., and Bondurand, N. (2010). Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 31 (4), 391–406. doi:10.1002/humu.21211
Pohl, E., Aykut, A., Beleggia, F., Karaca, E., Durmaz, B., Keupp, K., et al. (2013). A hypofunctional PAX1 mutation causes autosomal recessively inherited otofaciocervical syndrome. Hum. Genet. 132 (11), 1311–1320. doi:10.1007/s00439-013-1337-9
Qian, M., Xu, H., Perez-Andreu, V., Roberts, K. G., Zhang, H., Yang, W., et al. (2019). Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in Hispanics. Blood 133 (7), 724–729. doi:10.1182/blood-2018-07-862946
Rassman, W. R., Pak, J. P., and Kim, J. (2013). Phenotype of normal hairline maturation. Facial Plastic Surg. Clin. N. Am. 21 (3), 317–324. doi:10.1016/j.fsc.2013.04.001
Rodova, M., Ankilova, V., and Safro, M. G. (1999). Human phenylalanyl-tRNA synthetase: cloning, characterization of the deduced amino acid sequences in terms of the structural domains and coordinately regulated expression of the α and β subunits in chronic myeloid leukemia cells. Biochem. Biophys. Res. Commun. 255 (3), 765–773. doi:10.1006/bbrc.1999.0141
Rot, I., Baguma-Nibasheka, M., Costain, W. J., Hong, P., Tafra, R., Mardesic-Brakus, S., et al. (2017). Role of skeletal muscle in ear development. Histol. Histopathol. 32 (10), 987–1000. doi:10.14670/hh-11-886
Sajjadi, A. Y., Mitra, K., and Grace, M. (2013). Expression of heat shock proteins 70 and 47 in tissues following short-pulse laser irradiation: assessment of thermal damage and healing. Med. Eng. Phys. 35 (10), 1406–1414. doi:10.1016/j.medengphy.2013.03.011
Lee, S. H., Goddard, M. E., Wray, N. R., and Visscher, P. M. (2012). A better coefficient of determination for genetic profile Analysis. Genet. Epidemiol. 36 (3), 214–224. doi:10.1002/gepi.21614
Santos-Cortez, R. L. P., Lee, K., Azeem, Z., Antonellis, P. J., Pollock, L. M., Khan, S., et al. (2013). Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am. J. Hum. Genet. 93 (1), 132–140. doi:10.1016/j.ajhg.2013.05.018
Seongwon, C., Eun, L. J., Yeon, P. A., Jun-Hyeong, D., Woo, L. S., Chol, S., et al. (2018). Identification of five novel genetic loci related to facial morphology by genome-wide association studies. Bmc Genomics 19 (1), 481. doi:10.1186/s12864-018-4865-9
Shaffer, L. J., Lee, M. K., Roosenboom, J., Orlova, E., Adhikari, K., Gallo, C., et al. (2017). Multiethnic GWAS reveals polygenic architecture of earlobe attachment. Am. J. Hum. Genet. 101, 913. doi:10.1016/j.ajhg.2017.10.001
Shido, K., Kojima, K., Yamasaki, K., Hozawa, A., Tamiya, G., Ogishima, S., et al. (2019). Susceptibility loci for tanning ability in the Japanese population identified by a genome-wide association study from the tohoku medical megabank project cohort study. J. Investigative Dermatol. 139 (7), 1605–1608. doi:10.1016/j.jid.2019.01.015
Shin, J.-G., Leem, S., Kim, B., Kim, Y., Lee, S.-G., Song, H. J., et al. (2021). GWAS analysis of 17,019 Korean women identifies the variants associated with facial pigmented spots. J. Investigative Dermatol. 141 (3), 555–562. doi:10.1016/j.jid.2020.08.007
Smits, P., Saada, A., Wortmann, S. B., Heister, A. J., Brink, M., Pfundt, R., et al. (2011). Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 19 (4), 394–399. doi:10.1038/ejhg.2010.214
Sturm, R. A., and Duffy, D. L. (2018). Toward the full spectrum of genes for human skin colour. Pigment. Cell Melanoma Res. 31, 457. doi:10.1111/pcmr.12691
Sulem, P., Gudbjartsson, D. F., Stacey, S. N., Helgason, A., Rafnar, T., Jakobsdottir, M., et al. (2008). Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 40 (7), 835–837. doi:10.1038/ng.160
Tam, V., Patel, N., Turcotte, M., Bossé, Y., Paré, G., and Meyre, D. (2019). Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20 (8), 467–484. doi:10.1038/s41576-019-0127-1
Tanaka, J., Leeb, T., Rushton, J., Famula, T. R., Mack, M., Jagannathan, V., et al. (2019). Frameshift variant in MFSD12 explains the mushroom coat color dilution in shetland ponies. Genes 10 (10), 826. doi:10.3390/genes10100826
Tarafder, A. K., Bolasco, G., Correia, M. S., Pereira, F. J. C., Iannone, L., Hume, A. N., et al. (2014). Rab11b mediates melanin transfer between donor melanocytes and acceptor keratinocytes via coupled exo/endocytosis. J. Investigative Dermatology 134 (4), 1056–1066. doi:10.1038/jid.2013.432
Theil, A. F., Botta, E., Raams, A., Smith, D. E. C., Mendes, M. I., Caligiuri, G., et al. (2019). Bi-Allelic TARS mutations are associated with brittle hair phenotype. Am. J. Hum. Genet. 105 (2), 434–440. doi:10.1016/j.ajhg.2019.06.017
Wallin, J., Wilting, J., Koseki, H., Fritsch, R., Christ, B., and Balling, R. (1994). The role of Pax-1 in axial skeleton development. Development 120 (5), 1109–1121. doi:10.1242/dev.120.5.1109
Wang, Y., Lu, D., Chung, Y.-J., and Xu, S. (2018). Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas 155 (1), 19. doi:10.1186/s41065-018-0057-5
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40 (Database issue), D930–D934. doi:10.1093/nar/gkr917
Weinberg, S. M., Parsons, T. E., Marazita, M. L., and Maher, B. S. (2013). Heritability of face shape in twins: A preliminary study using 3D stereophotogrammetry and geometric morphometrics. Dent. 3000 1, 14. doi:10.5195/d3000.2013.14
Weinberg, S. M., Roosenboom, J., Shaffer, J. R., Shriver, M. D., Wysocka, J., and Claes, P. (2019). Hunting for genes that shape human faces: Initial successes and challenges for the future. Orthod. Craniofac Res. 22 Suppl 1(Suppl 1), 207–212. doi:10.1111/ocr.12268
Xu, H., Robinson, G. W., Huang, J., Lim, J. Y.-S., Zhang, H., Bass, J. K., et al. (2015a). Common variants in ACYP2 influence susceptibility to cisplatin-induced hearing loss. Nat. Genet. 47 (3), 263–266. doi:10.1038/ng.3217
Xu, H., Zhang, H., Yang, W., Yadav, R., Morrison, A. C., Qian, M., et al. (2015b). Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat. Commun. 6, 7553. doi:10.1038/ncomms8553
Yoon, S.-Y., Kim, K.-T., Jo, S. J., Cho, A.-R., Jeon, S.-I., Choi, H.-D., et al. (2011). Induction of hair growth by insulin-like growth factor-1 in 1,763 MHz radiofrequency-irradiated hair follicle cells. PLOS ONE 6 (12), e28474. doi:10.1371/journal.pone.0028474
Zhang, S. Y., Zhou, X. Y., Zhou, X. L., Zhang, Y., Deng, Y., Liao, F., et al. (2019). Subtype specific inherited predisposition to pemphigus in the Chinese population. Br. J. Dermatol. 180 (4), 828–835. doi:10.1111/bjd.17191
Zhou, X., Li, D., Zhang, B., Lowdon, R. F., Rockweiler, N. B., Sears, R. L., et al. (2015). Epigenomic annotation of genetic variants using the roadmap epigenome browser. Nat. Biotechnol. 33 (4), 345–346. doi:10.1038/nbt.3158
Zhu, Y., Yin, D., Su, Y., Xia, X., Moriyama, T., Nishii, R., et al. (2018). Combination of common and novel rare NUDT15 variants improves predictive sensitivity of thiopurine-induced leukopenia in children with acute lymphoblastic leukemia. Haematologica 103 (7), e293–e295. doi:10.3324/haematol.2018.187658
Keywords: facial trait, aesthetics, genome-wide association study, genome-wide polygenic score, widow’s peak
Citation: Wang P, Sun X, Miao Q, Mi H, Cao M, Zhao S, Wang Y, Shu Y, Li W, Xu H, Bai D and Zhang Y (2022) Novel genetic associations with five aesthetic facial traits: A genome-wide association study in the Chinese population. Front. Genet. 13:967684. doi: 10.3389/fgene.2022.967684
Received: 13 June 2022; Accepted: 27 June 2022;
Published: 12 August 2022.
Edited by:
Guanglin He, Nanyang Technological University, SingaporeReviewed by:
Lan Zhang, Southwest Jiaotong University, ChinaHaiyang Yu, Tianjin University of Traditional Chinese Medicine, China
Copyright © 2022 Wang, Sun, Miao, Mi, Cao, Zhao, Wang, Shu, Li, Xu, Bai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ding Bai, YmFpZGluZ0BzY3UuZWR1LmNu; Yan Zhang, emhhbmcueWFuQHNjdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Peiqi Wang
Peiqi Wang Xinghan Sun
Xinghan Sun Qiang Miao5†
Qiang Miao5† Minyuan Cao
Minyuan Cao Yang Shu
Yang Shu Heng Xu
Heng Xu Ding Bai
Ding Bai Yan Zhang
Yan Zhang