Abstract
Crop improvement programmes began with traditional breeding practices since the inception of agriculture. Farmers and plant breeders continue to use these strategies for crop improvement due to their broad application in modifying crop genetic compositions. Nonetheless, conventional breeding has significant downsides in regard to effort and time. Crop productivity seems to be hitting a plateau as a consequence of environmental issues and the scarcity of agricultural land. Therefore, continuous pursuit of advancement in crop improvement is essential. Recent technical innovations have resulted in a revolutionary shift in the pattern of breeding methods, leaning further towards molecular approaches. Among the promising approaches, marker-assisted selection, QTL mapping, omics-assisted breeding, genome-wide association studies and genome editing have lately gained prominence. Several governments have progressively relaxed their restrictions relating to genome editing. The present review highlights the evolutionary and revolutionary approaches that have been utilized for crop improvement in a bid to produce climate-resilient crops observing the consequence of climate change. Additionally, it will contribute to the comprehension of plant breeding succession so far. Investing in advanced sequencing technologies and bioinformatics will deepen our understanding of genetic variations and their functional implications, contributing to breakthroughs in crop improvement and biodiversity conservation.
1 Introduction
Agriculture has undergone transformation significantly over the past century as a result of scientific expansion, shifting societal mores and transitions in the political, economic and social environment. Plant breeding is considered to be a co-evolutionary process between human civilization and food sources (Breseghello and Coelho, 2013). From the Green Revolution of the mid-20th century, which aimed to increase crop yields through the use of high-yield varieties to the more recent Genome Revolution, which is leveraging the latest in genomic and biotechnological tools to create crops that are more resilient to environmental instability, the field of plant breeding has been constantly evolving. Over the years, plant breeders and farmers have long been exploiting natural variations to develop and improve crop cultivars (Sadia et al., 2021). However, overexploitation of any practice facilitates disturbances in the balanced ecosystem. For instance, the green revolution in rice in the 1960s led to the invention of the rice cultivar, IR8 (Hargrove and Cabanilla, 1979; Khush, 1999). The widespread adoption of this particular rice variety averted the issue of food shortage across the Globe. However, several socio-ecological concerns were observed with the widespread use of the variety IR8. Several studies have highlighted that IR8 requires a high amount of fertilizer input, which in the long run would cause adverse effects on the environment (Cassman, 1999). In addition, the large-scale utilization of this rice cultivar narrowed down the genetic base, which could result in being more susceptible to certain pests or diseases (Sanchez et al., 2013; Shakiba and Eizenga, 2014). Henceforth, it is undeniable that the excessive crop production generally has a detrimental effect on the environment. But it is also crucial to keep in mind that the scope and character of these effects might be changed based on advanced techniques and technological improvements. It makes it substantially more crucial to carefully consider the benefits and drawbacks of approaches often used in crop development. To address the issues of genetic vulnerability, the application of different advanced molecular breeding tools is highly essential in order to create variability.
Among the existing breeding tools, some can create novel variability (Rothan and Causse, 2007; Karthika et al., 2020; Awan et al., 2021; Schleif et al., 2021; Suprasanna et al., 2021) while others can exploit already existing variability (Begna, 2021). The main drawback of conventional breeding is the generation time required for screening potential genotypes over the years in different environmental conditions (Lasley et al., 1994; Prohens, 2011). For instance, mutation as an ultimate source of variation consequently provides better opportunities for novel genetic variability (Holme et al., 2019; Ahmar et al., 2020). However, as a genetic improvement technique, induced mutagenesis has long been an ineffectual approach to obtaining new alleles of genes. This is mostly due to the necessity to generate and evaluate massive populations of presumed mutants, the prevalence of chimaeras, and the recessive character of mutations (Mba, 2013). With the advancement of biotechnology, via the transgenic breeding approach, successful desirable gene introduction into the host organism has now been possible (Visarada et al., 2009). However, the environmental and ethical issues have also imposed problems related to the release of transgenic varieties (Dale et al., 2002; Snow et al., 2005). Numerous reports from different committees have provided clear evidence that contradicts the notion that transgenic crops had a detrimental impact on the environment. (Genetically Engineered Crops: Experiences and Prospects, 2016). But again, disagreements among experts have imposed issues with the global dissemination of transgenic crops. To create desirable variability in our crops, a more sophisticated and time saving technology is indeed required (Figure 1). The rise of molecular approaches such as marker-assisted breeding (MAB), genome wide association mapping (GWAS), targeting induced local lesions (TILLING), and others has rendered it simpler to evaluate potential genotypes. Breeding schemes such as speed breeding (SB) and rapid generation advancement (RGA) were developed to further expedite generation advancement.
FIGURE 1
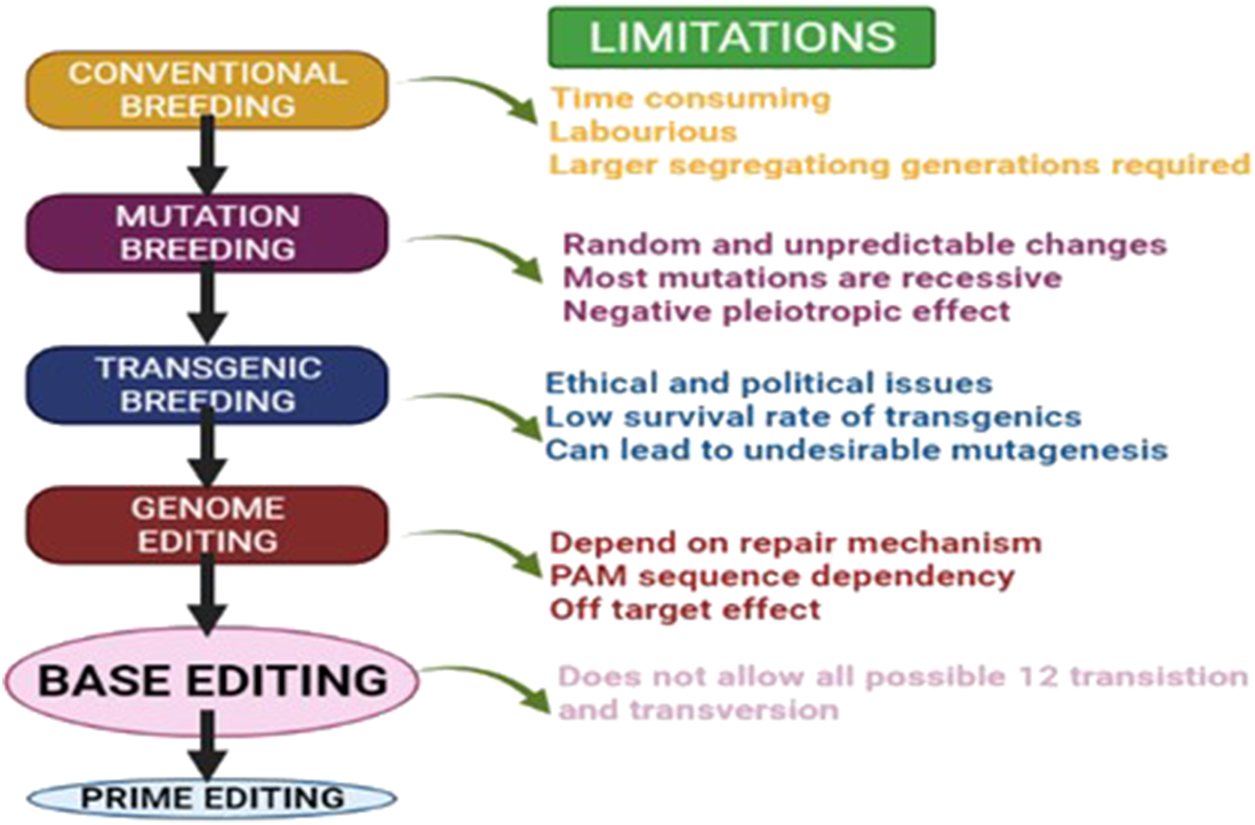
Evolution of major breeding techniques and their limitations.
Along with the technological progression, the DNA/RNA repair mechanisms viz., homologous direct repair (HDR) and non-homologous end joining (NHEJ), genome editing techniques were uncovered to create double-strand breaks. Over the years, genome editing techniques have evolved, from mega nucleases (MegNs) to zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Recently, the genome editing technique; clustered regularly interspaced short palindromic repeats (CRISPR) made a scientific breakthrough for agricultural applications. ZFNs and TALENs were regarded as first-generation (Razzaq et al., 2019) while CRISPR is considered as a second-generation editing tool and further revolutionized precision for genome editing (Jinek et al., 2012; Doudna and Charpentier, 2014). For instance, speed breeding-CRISPR is one of the techniques that can be implemented to hasten the generation cycle and development of new cultivars (Samantara et al., 2022). The primary objective of this review is to discuss the overall aspects starting from the conventional breeding approaches to the modern breeding tools (classified in different phases) considering the ease of use and hastening the entire breeding programme in order to generate novel desirable crop cultivars. This review also highlighted previously applied tools, their drawbacks, mitigating strategies and evolving technologies in the field of plant breeding. Furthermore, the discussion here aims to investigate how plant breeders are utilizing the most recent scientific tools and methods to cultivate crops that are better equipped to survive the difficulties encountered by an evolving environment and a growing population.
2 Phase I: unleashing nature’s potential: conventional plant breeding
Plant breeding is critical in addressing our population’s rising food demand along with upgrading nutrition (Borlaug, 1983; Huang et al., 2002; Bouis and Saltzman, 2017). In addition, it has given us the ability to develop and improve existing cultivars that are better adapted to climate change and new disease strains. It has been established that the domestication of food and animal crops progresses together with human civilization (Gupta, 2004). Historically, plant breeding has evolved from conventional to genomics-assisted breeding. Conventional breeding entails selection through phenotypic evaluation. Breeders use crossing strategies to combine favorable attributes (reside in the genetic background) from various yet related plants to a new variety (Acquaah, 2015). The screening of certain traits would largely be from a large set of populations. However, this type of breeding process is laborious and time-consuming.
Mutation breeding contributed significantly to crop breeding by creating evolutionary divergence (Yu et al., 1991; Kharkwal et al., 2004; Oladosu et al., 2016). This has been successfully used across various crops such as maize, barley, rice, lettuce, and other crops (Stadler, 1928a; 1928b; Sawada et al., 2016; Yamatani et al., 2018). As compared with conventional hybridization, mutation breeding has a shorter time frame for varietal development (Saima Mir et al., 2021). Breakthroughs in breeding have led to the discovery of novel technologies for eliciting variation in target crops. Mutation breeding is still utilized in conventional modern breeding today, despite the more sophisticated procedures given by contemporary biotechnology (Bado et al., 2013). Succeeding this method are other advanced techniques such as Eco-TILLING, plant tissue culture, and genetic engineering (Figure 2).
FIGURE 2
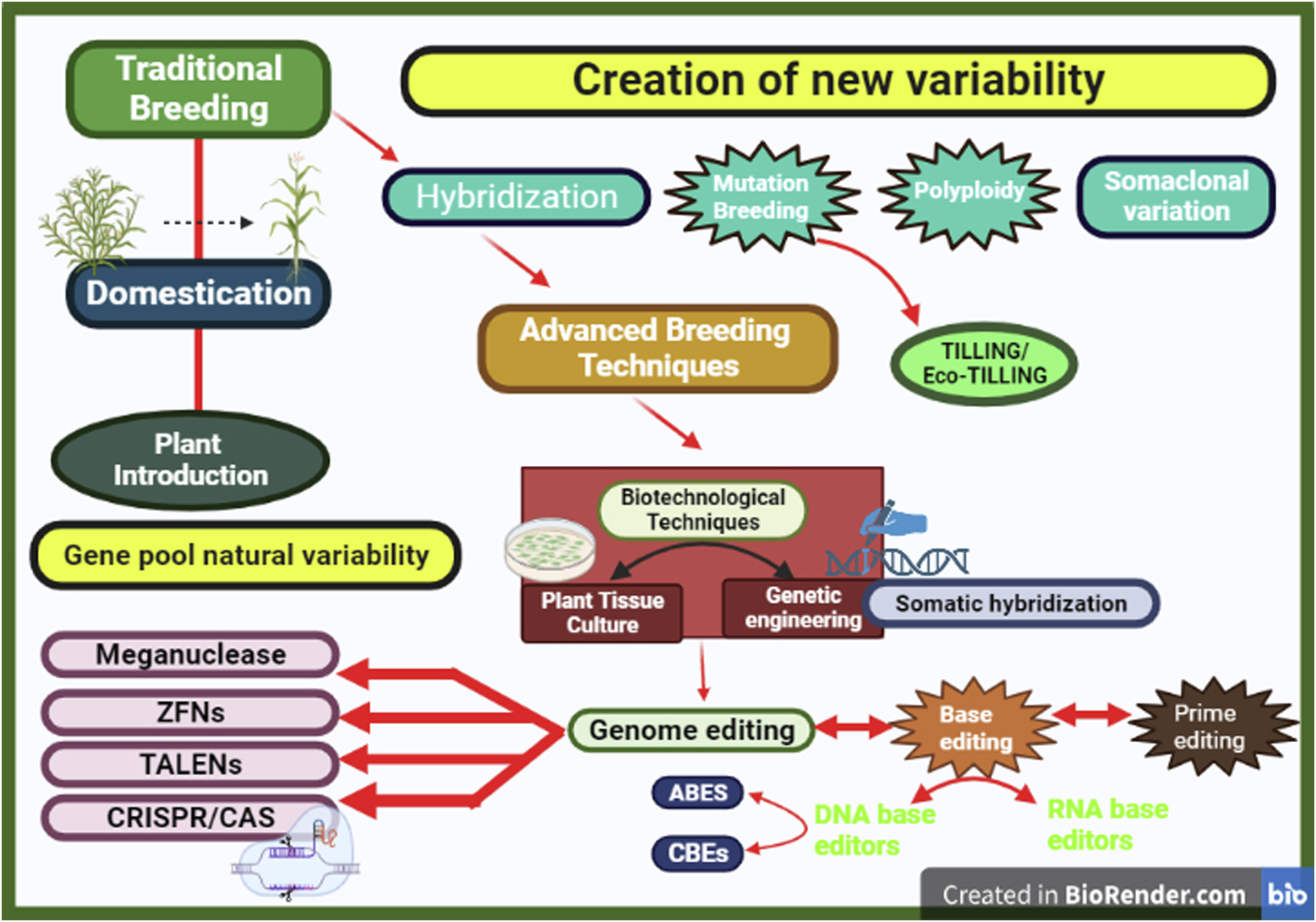
Breeding techniques across eras: tracing advances from tradition to innovation.
2.1 Crop domestication
Domestication is a process of coevolution where wild species were brought under human cultivation through artificial selection which led to the differentiation and development of new species (Fuller et al., 2014; Schaal, 2019; Chen et. al., 2021a). The transformation occurred many times independently around the world during the Neolithic period (Childe, 1949), where hunter-gatherer tribes were settled as agricultural societies (Diamond, 2002; Diamond and Bellwood, 2003) and manipulated the rise of modern civilization (Chen et. al., 2021a). The spontaneous mutation had a significant role in modifying the attributes of domesticated plants. Significant changes in necessities, such as land for agriculture, food demand, and the use of agricultural technologies, came from the coevolution of civilization and the human race. As a result, the necessary crops were eventually expanded from their restricted growing zone to be produced in a newer, undiscovered location. As a result, wild plant species were cultivated and favored plants were chosen based on their requirements. Although many initiatives are underway to broaden the genetic background of prominent crops by introgression of genes from their wild relatives (Sadiki et al., 2007). Yet there is a considerable gap slot in the knowledge of forbears, origin and domestication time frames for different crops (Meyer et al., 2012). To get a fuller insight into these unanswered concerns regarding domestication, genomics and related disciplines (transcriptomics and epigenomics) have resulted in new findings (Barrera-Redondo et al., 2020). This demonstrates a bridge between traditional and modern breeding approaches. One of the aspects of domestication is de-domestication, also known as feralization, which is a fascinating phenomenon in which there is a deliberate establishment of a population of domesticated crops in the wild (Figure 3). De-domestication research has helped in better grasping the complexities of crop evolution, as well as the genetic novelties of de-domesticates that are useful in current crop breeding (Wu et al., 2021).
FIGURE 3
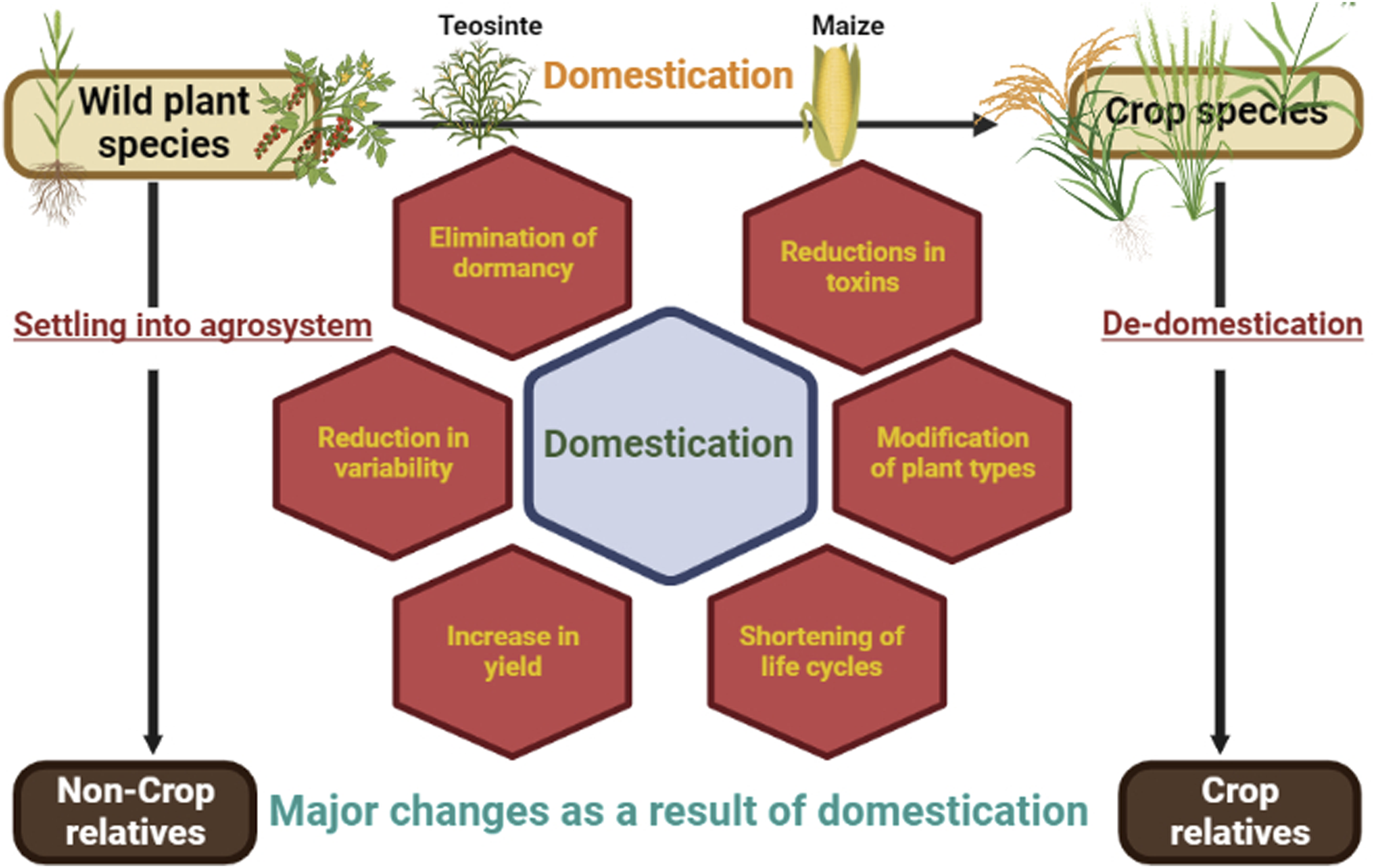
Domestication and de-domestication: transformations in plant traits.
Humans have domesticated different plants in need of food, fodder, fibre and tools throughout the past 12,000 years, which has had an influence on human culture as well as germplasm under domesticated species. Domestication is significant since it makes plant species more accessible to agricultural development programmes. During the phase of domestication, there is the role of artificial as well as natural forces in selection which leads to major changes (domestication syndrome) (Hammer, 1984) (Figure 3). This includes for instance, 1) loss of dormancy [in Chenopodium, the outer layer of the testa is accountable for the black appearance of wild seeds which is diminished or nonexistent in domesticates, resulting in pale-colored seeds (Wilson, 1981; Bruno, 2006)] 2) reduced toxins (wild potato tubers contain amounts of bitter glycoalkaloids that may be hazardous to humans (Johns and Alonso, 1990)] 3) increase in size (Pericarps and placentas of near-isogenic tomato fruits with small vs. big fruits have cells of similar size, but the giant fruits have more cells (Cong et al., 2002)]. Therefore, domestication is regarded as the most crucial and centric method of plant breeding (Table 1) since all other breeding methods become relevant only after it has been domesticated with success. Many of the crops under current cultivation have been domesticated from ancient times. It alleviates a significant number of problems posed by intensive agriculture. A better comprehension of the development of adaptation among crop species might lead to novel ideas for developing new varieties/species that can address present and potential environmental challenges sustainably (Purugganan, 2019).
TABLE 1
| Crops | Traits improved | Target genes | Remarks | References |
|---|---|---|---|---|
| Domestication | ||||
| Rice | Plant architecture | Prostrate growth 1 (PROG1) | Upright plant growth | Jin et al. (2008) |
| Tan et al. (2008) | ||||
| Number of grains per panicle | Frizzy panicle (FZP gene | Boosted number of secondary branches | Huang et al. (2018) | |
| Seed shattering | SHA1 (a single dominant gene Shattering 1)/SH4 (Shattering 4) and QTL present in chromosome 1(qSH1) controls seed shattering | Reduced shattering of seeds | Li et al. (2006) | |
| Konishi et al. (2006) | ||||
| Lin et al. (2007) | ||||
| Sweeney and McCouch (2007) | ||||
| Vaughan et al. (2008) | ||||
| Seed dormancy | Seed dormancy 4 on chromosome 7 and OsG on chromosome 4 | Genes governing seed dormancy | Sugimoto et al. (2010) | |
| Wang et al. (2018a) | ||||
| Maize | Apical dominance | tb1gene (Teosinte branched1) | Regulating the inflorescence architecture | Doebley et al. (1997) |
| Cubas et al. (1999) | ||||
| Increase in ear | zfl2 (Zeafloricaula leafy2) | Increase in ear rank number | Bomblies and Doebley (2006) | |
| Sexual conversion | tru1 gene (tassels replace upper ears1) | Conversion of a tassel in teosinte to an ear | Doebley et al. (1995) | |
| Dong et al. (2017) | ||||
| Chen et al. (2019) | ||||
| Selection and hybridization | ||||
| Maize | Inflorescence | Remosa 1 (ra1) gene | Ear and tassel are suppressed | Vollbrecht et al. (2005) |
| Sigmon and Vollbrecht (2010) | ||||
| Xu et al. (2017) | ||||
| Chen et al. (2019) | ||||
| Hybrid crop breeding/cross breeding | ||||
| Rice | Yield increases | Cross Breeding | Witcombe et al. (2013) | |
| Increased spikelet number per panicle | Panigrahi et al. (2019) | |||
| Wheat | Grain yield | Basnet et al. (2019) | ||
| Mutation breeding | ||||
| Wheat | Dwarf height of plant | Rht-B1b & Rht-D1b | Single base pair mutation | Peng et al. (1999) |
| Ellis et al. (2002) | ||||
| Rust resistance | Lr34 | SNPs | Dakouri et al. (2010) | |
| Krattinger et al. (2009) | ||||
| Chauhan et al. (2015) | ||||
| Rice | Increased 1000-grain weight | GW2 gene | Loss of function mutation | Song et al. (2007) |
| Higher nitrogen use efficiency | NRT | NRT1.1B (SNP) | Hu et al. (2015) | |
| Li et al. (2018a) | ||||
| Disease resistance (bacterial blight) | Os8N3 | Loss of function mutation | Kim et al. (2019) | |
Application of different classical techniques in crop improvement.
2.2 Crop introduction
The plant has been introduced when it has been brought by humans across a vast geographic border (Richardson et al., 2000). Richardson et al. (2000) differentiated between introduction and invasion, proposing invasion as being spontaneous and without human intervention [earlier supported by ecologists (Clement and Foster, 1994)]. It is one of the easiest methods to use naturally existing variability from one area to be introduced in an area of absence. Plant variety to be introduced in new areas can be used directly as an elite commercial variety without any alteration in its genotype, referred as primary introduction. On the contrary, for secondary introduction, the cultivar is reinforced in selection and hybridization programmes and modified with some new characters. A classic example of introduction is the dwarf wheat variety Sonora-64 and Lerma Rojo as a primary introduction and Kalyan Sona and Sonalika as a secondary introduction (Allard, 1999). Although it is a beneficial method in adding variation to the gene pool in particular areas, some instances in history which led to the introduction of pests, weeds and fungal diseases along with crops unintentionally (Knezevic, 2017).
2.3 Crop selection
The differential rate of reproduction of different genotypes is referred to as selection (Lennox and Wilson, 1994). This can be performed in the presence of variability. However, the effectiveness of selection is mainly dependent on the presence of variability since selection cannot create new variations and heritability of the trait (Gregory, 2009). Evidence for selection can be seen from ancient times when farmers used this in agriculture. Selection in crop varieties has been reported by agriculturists, viz., Vans Mons in Belgium, Andrew Knight in England and Cooper in the United States. Publications from 1843 show the evidence for selection practiced in the Isle of Jersey. Individual plant selection was practiced by Patrick Shireff in wheat and oats at the same time (Sneep, 1966). Hallet also reported individual plant selection in wheat, oats and barley in the year 1857. Vilmorin came up with the individual plant selection with progeny testing in the later years (Vilmorin, 1930). Artificial selection represents a sophisticated way of establishing the underlying genetic variation and hence the evolvability of certain features (Table 1). It imparts a known intensity and direction of selection to specific morphological characters (Gregory, 2009).
2.4 Crop hybridisation
The genetic base of our current crop population has been narrowed down, hence the need to create variation. The process of hybridization aims to create variation in the population (Anderson and Stebbins, 1954). In layman’s terms, this refers to a method of crossing between individuals of different plant populations that differ from each other in their heritable characters (Harrison, 1990). The process involves crossing two distinct genotypes, resulting in a Filial 1 (F1) hybrid. Within the F1 generation, genes segregate and recombine, leading to the formation of new gene combinations and thereby generating genetic variability. Different methods can be used; however, most continued inbreeding is done in the lines taken from the source population which is coupled with selection. Hybridization allows exploiting of heterosis (Timberlake, 2013). However, several factors may affect hybridization frequency such as reproductive barriers and incompatibilities between populations and taxon-specific differences.
2.5 Mutation breeding
Sudden heritable change in the characteristics of the organism that has not been acquired by genetic recombination is referred to as mutation (Van Harten, 1998). The term mutation breeding (“Mutationszuchtung”) was first coined by Freisleben and Lein (Figure 4) in the year 1944 which refers to the development of mutant lines to improve crops (Freisleben and Lein, 1944). Crop improvement can only be achieved when enough variation is present for the trait to help the breeder to exercise his breeding skills. However, because of the overexploitation of commercial varieties, the genetic base has narrowed down. Even if the desired variation is present, it is present in wild relatives/old landraces which makes breeding tedious and time taking to retrieve this variation. Mutation breeding provides an opportunity without extensive upgrading crossing and selection (Shu et al., 2012; Rani et al., 2016). Induced mutagenesis serves as one of the most productive strategies for creating evolutionary divergence as well as identifying critical regulatory genes for commercially significant features for crop improvement (Mehta and Basha, 2018; Chaudhary et al., 2019a). Forward genetics as well as reverse genetics both played crucial roles in achieving significant improvement for various economic traits (Aklilu, 2021). The former involves the identification of an induced or random mutant gene that is responsible for a particular phenotype, while in the latter the function of a gene is not known (Jankowicz-Cieslak and Till, 2015).
FIGURE 4
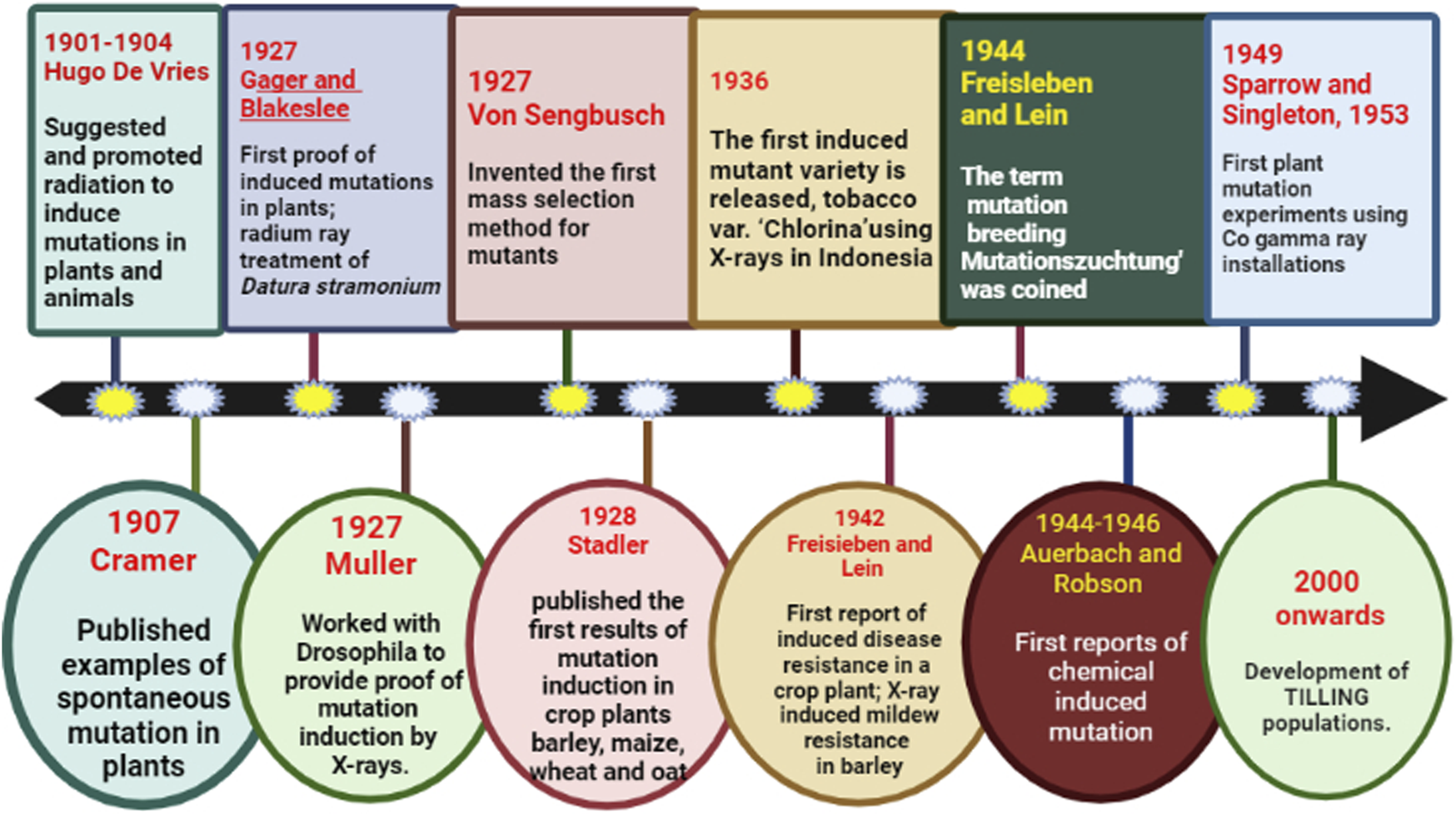
Timeline of milestones in mutation breeding.
Gene inactivation has been implemented extensively to figure out the role of the unknown gene in different crops. To inactivate endogenous genes, T-DNA or transposon is injected into the gene to modify and tag it (Munoz-Lopez and Garcia-Perez, 2010; Kyndt et al., 2015). Insertional mutagenesis in crops is commonly used in crop improvement, particularly for creating disease-resistant varieties. The inserted DNA often contains genes that confer resistance to specific pathogens, making the plant more resistant to disease (Gachomo et al., 2003; Webb et al., 2006). However, insertional mutagenesis can also have unintended consequences, such as altering the expression of important genes and leading to unexpected phenotypic changes in the plant (Miki et al., 2009; Schnell et al., 2015). In addition, these techniques require effective plant transformation, which is only accessible in a few crops, prominently maize and rice (Kolesnik et al., 2004). This led to the discovery of a new approach, “TILLING,” which is now preferred over other methods of reverse genetics.
Although physical and chemical-induced mutagenesis have expedited crop improvement (Mba et al., 2010; Mostafa, 2011; Kozgar et al., 2012), these approaches are still less popular due to random mutation and its expensive nature. In contrast, innovations in genome editing techniques have reimagined the ability to accurately manufacture a specific modification in the genome.
3 Phase II: molecular and advanced plant breeding technology
3.1 Marker-assisted selection
In today’s world, molecular markers provide significant potential in terms of systems, creative and enhanced genetic mapping methodologies. In 1986, the first authentic restriction fragment length polymorphism (RFLP) map in an agricultural crop (tomato) was generated (Bernatzky and Tanksley, 1986). The advantages of MAS included assessing plants at the beginning of the growing season, screening several qualities that would ordinarily be epistatic with one another, deterministically removing linkage drag, and fast retrieving the genotype of a recurring parent (Tanksley et al., 1989). Even the technological constraints of early RFLPs seemed to be overcome as newer and simpler molecular marker systems like RAPDs (Williams et al., 1990) AFLPs (Vos et al., 1995), and microsatellites (Akkaya et al., 1992) were developed. Within a few years, high-density DNA marker maps for nearly every key crop species had been created (O’Brien, 1993), each one promising the use of strategic MAS to supplement, if not replace, traditional plant breeding strategies. This allows plant breeders to make more informed decisions about which plants to use in their breeding programs, leading to faster and more effective improvement of crops (Garrett et al., 2017; Boopathi, 2020). MAS can also be used to reduce the threat of inadvertently integrating undesirable characteristics into the genome (Bhatnagar-Mathur et al., 2015). Soon after, QTL mapping rose to prominence, facilitating the precise mapping of genetic loci influencing complex characteristics (Phillips and Vasil, 2003). Before the emergence of molecular markers, the concept of promptly identifying the loci driving polygenic characteristics appeared to be a utopian dream.
3.2 QTL mapping
Quantitative trait locus (QTL) mapping is a strong technique for the investigation of complex attributes in plants. It is the way of discovering particular locations in the genome that are accountable for variation in a certain characteristic of relevance (Mather, 1938). QTL mapping involves several steps, including the identification of phenotypic variation in a population of individuals, the genotyping of those individuals, and the statistical analysis of the genotype-phenotype data to identify QTLs (Dhingani et al., 2015). The first step in QTL mapping is to generate a mapping population, which can be done by crossing two parental lines with contrasting phenotypes or by selecting individuals from a natural population with phenotypic variation.
Once the mapping population is generated, the phenotypic data is collected, and the individuals are genotyped using molecular markers, such as SNPs or SSRs (Park et al., 2021). The markers are used to construct a genetic map of the genome, which provides a framework for the location of the QTLs. The next step is to perform a statistical analysis of the genotype-phenotype data to identify QTLs. The most commonly used method for QTL mapping is linkage analysis, which compares the phenotypic data with the molecular markers to identify regions of the genome that are associated with the phenotypic variation (Akond et al., 2019). Once the QTLs are identified, further fine-mapping can be done to narrow down the chromosomal region relevant to the trait variation (Raihan et al., 2016b). QTL mapping has been used effectively in various crops, including rice, maize, and tomato, and has revealed insightful information into the genetic basis of complex characteristics including yield, biotic and abiotic stress resistance (Bai et al., 2018; Yadav et al., 2019; Saleem et al., 2020; Goering et al., 2021). In addition, QTL mapping has also been used to identify candidate genes for the targeted improvement of crops (Luo et al., 2019b; Guo et al., 2019; Kumar et al., 2020). In conclusion, QTL mapping provides valuable information for plant breeding and improvement, as well as for understanding the genetic basis of phenotypic variation.
The acronym MAGIC stands for “Multiparent Advanced Generation Inter-Cross,” and it refers to the process of establishing a population by crossing many distinct founder parents across several generations in order to harness the genetic diversity contained in the founding lines (Samantara et al., 2021). Magic populations provide higher genetic variety and improved possibilities for identifying the genetic basis of complex characteristics by mixing many founding parents and allowing for recombination over several generations (Huang et al., 2015). Magic populations are exceedingly helpful for quantitative trait mapping and genetic research. The increasing genetic diversity and the enormous number of recombination events in these populations improve the ability to discover and map quantitative trait loci (QTL) associated with complex characteristics (Huynh et al., 2018; Diaz et al., 2020).
3.3 Association mapping (AM)
The idea of linkage disequilibrium (LD) underpins association mapping, which is the non-random association of alleles (gene versions) at distinct loci (positions) on a chromosome (Joiret et al., 2019). In the context of association mapping, LD refers to the fact that some genetic variations tend to occur together more frequently than expected by chance. Association mapping plays a crucial role in crop improvement by identifying the genetic variation underlying traits of interest in crops (Zhu et al., 2008). For example, association mapping studies have identified genes that regulate grain size and weight, which are important components of yield in cereal crops (Cockram et al., 2010; Neumann et al., 2011; Sukumaran et al., 2012; Alipour and Darvishzadeh, 2019; Alqudah et al., 2020) The goal of association mapping is to identify the specific genetic variations that are most likely to be associated with a particular phenotype, such as height, weight, or susceptibility to a disease or abiotic stress (Poland et al., 2011; Sehgal et al., 2015; Hanson et al., 2018). The approach is further expanded to minor millets, offering a food security perspective (Babu et al., 2014; Gupta et al., 2014; Puranik et al., 2020). The method can be used to identify novel alleles and genes that can be used to improve crops through conventional breeding methods or genetic engineering.
Additionally, association mapping enables researchers to identify the relationships between the genetic makeup of crops and the environment, allowing for the development of more sustainable and resilient crop varieties. This information can be used to design breeding programs that are better suited to the specific environmental conditions faced by farmers. Some of the main limitations of association mapping (Korte and Farlow, 2013; Gupta et al., 2019a) include false positive results (AM can sometimes produce false positive results, which means the association between a particular genetic variation and a trait may be coincidental, rather than causal, also may be due to confounding factors, such as population structure, or to multiple testing errors), low statistical power (AM studies often have low statistical power, that they are not always able to detect the true associations between genetic variations and traits, Missing data (AM requires high-quality genotyping data, which is not always available and missing data can result in reduced statistical power or can lead to the exclusion of important individuals from the analysis), Complex trait analysis (these are often influenced by multiple genetic variations, some of which may be rare or have small effects). AM is not well suited for analyzing these traits, as it can only identify common genetic variations that have moderate to large effects, Cost and time: large numbers of individuals are required to be genotyped and phenotype.
Integrating association mapping with a secondary mapping population is a powerful strategy to validate and refine the findings of association mapping. Secondary mapping populations have been essential in enhancing crop breeding efforts in a variety of crops. Secondary mapping populations, for example, have proved useful in mapping and dissecting complex characteristics. Researchers found quantitative trait loci (QTLs) linked with these qualities by crossing divergent parental lines and analyzing the resultant segregating populations, allowing for marker-assisted selection and the generation of superior varieties. Consistent and overlapping associations provide confidence in the validity of the markers and associations. Further fine mapping and candidate gene analysis can be conducted to narrow down the genomic regions and identify specific genes associated with the trait. Secondary mapping populations have been utilized in crops such as rice, maize, wheat, tomato, cotton and soybean to map and confirm QTLs connected to characteristics such as disease resistance, yield potential, fruit quality features, and abiotic stress tolerance (Wissuwa et al., 2002; Niu et al., 2017; Chen et al., 2018).
The primary goal of NAM (Nested Association Mapping) is to combine the advantages of linkage mapping with association mapping. It involves creating a population of recombinant inbred lines (RILs) by crossing a common parent with multiple diverse founder parents. It plays a pivotal role in crop breeding by combining the benefits of linkage and association mapping, enabling trait dissection, facilitating marker-assisted selection, broadening the genetic base, and serving as a resource for functional genomics. These contributions enhance our understanding of complex traits, accelerate breeding efforts, and drive the development of improved crop varieties with desirable agronomic and quality traits. Researchers may locate sections of the genome related to certain qualities of interest by phenotyping the NAM population and genotyping it using high-density markers (Yu et al., 2008; Gage et al., 2020).
3.4 TILLING (targeting induced local lesions in genome)
TILLING is a non-transgenic high-throughput method (Henikoff et al., 2004). It involves the creation of chemically induced point mutations in the genome with PCR-based screening of the hetero duplex formed for crop improvement (McCallum et al., 2000a). The word “targeting” refers to focusing on genes of interest, usually taken as 1,500 bp in a single reaction. This technique overcomes the limitations associated with other tools of reverse genetics like insertional mutagenesis, RNA interference, and transposons (McCallum et al., 2000b). TILLING can also be used to study naturally present single nucleotide polymorphism in the genes of interest as a variant named Eco-TILLING (Gilchrist et al., 2006; Nieto et al., 2007; Ibarra et al., 2017). TILLING technique was first discovered by McCallum in the late 1990s who worked on Arabidopsis to characterize the two chromo methylase (CMT 2) gene functions. The goal of TILLING is to introduce specific changes into the DNA of crops to improve their traits, such as resistance to pests and diseases, increased yield, and improved nutritional content (Wang et al., 2012; Raihan et al., 2016a; Acevedo-Garcia et al., 2017) (Table 2). The process involves chemically inducing errors in the DNA and then screening for plants with the desired mutations. TILLING can be used in conjunction with CRISPR/Cas9 to deliver precise modifications to the genome and achieve particular breeding goals.
TABLE 2
| Crops | Traits improved | Target genes | Remarks | References |
|---|---|---|---|---|
| QTL Mapping/Marker assisted selection | ||||
| Rice | Blast resistance | Pi9, Pi2 | Blast resistance is imparted by the hybrid of Hui 316 (restorer line) and Pi9, Pi2, respectively | Tian et al. (2019) |
| Wheat | Drought tolerance | QTL found on chromosome 2A | QTLs for cell membrane stability, water content, and photosynthesis | Malik and Malik (2015) |
| Maize | Earliness and yield | QTL | Chromosomes 5, 8, and 10 have QTLs | Bouchez et al. (2002) |
| European corn borer and Mediterranean corn borer | 42 SIR MQTL | The two chromosomes with the highest SIR MQTL are 2 and 5. Fibre and hydroxycinnamate are cross-linked to prevent mechanical harm from insects | Badji et al. (2018) | |
| Maize rough dwarf disease (MRDD) | QTL qMrdd | QMrdd8 from X178 is introduced into top germplasm using a conventional technique and MAS. | Xu et al. (2020) | |
| Association Mapping | ||||
| Wheat | Karnal bunt resistance | 13,098 SNPs | Population size of 339 | Gupta et al. (2019b) |
| Brassica | Improving yield | 74 significant QTNs detected | important loci associated with seed per silique and thousand-seed weight across the chromosomes of rapeseed by QTL and GWAS studies | Khan et al. (2019) |
| Rice | Abiotic stress | NAC42 acts as a transcription factor that regulates the expression of the nitrate transporter gene | NUE-related agronomic traits | Tang et al. (2019) |
| Maize | Abiotic stress | Identified candidate genes associated with phosphorus deficiency tolerance | Metabolites under low Pi | Luo et al. (2019a) |
| TILLING Approach | ||||
| Wheat | Powdery mildew resistance | TaMlo | Partial loss-of-function | Acevedo-Garcia et al. (2017) |
| Wheat (Triticum turgidum) | Increased in amount of amylose | SBEIIa | The amalgamation of two non-sense mutations leads in a high amylose phenotype | Sestili et al. (2015) |
| Rice | Salt tolerance | OsAKT1, OsHKT6, OsNSCC2, OsHAK11 and OsSOS1 | Variation in membrane transport genes (expression levels and protein structures) | Hwang et al. (2016) |
| Barley | Changed starch phenotype | MY1, GBSSI, LDA, SSI and SSIIa | 29 novel alleles were discovered in five genes linked to starch metabolism that are active in the endosperm during grain filling | Sparla et al. (2014) |
| Groundnut | Drought tolerance | PLD | Phospholipase D expression increase | Guo et al. (2015) |
| Accelerated Plant Breeding Approach (Speed breeding) | ||||
| Rice | Achieved 4–5 generations in a year (Salt tolerance) | Speed Breeding | Rana et al. (2019) | |
| Collard et al. (2017) | ||||
| Wheat | Achieved 4–6 generations in a year | Mukade (1974) | ||
| Watson et al. (2018) | ||||
| Oat | Achieved 7 generations in a year | Liu et al. (2016) | ||
| Chickpea | Achieved 4–6 generations in a year | O’connor et al. (2013) | ||
| Brassica napus | Achieved 5 generations in a year | Watson et al. (2018) | ||
Application of advanced breeding techniques in crop improvement.
4 Accelerated plant breeding techniques
Traditional breeding is a time cumbersome process that includes different important phases. Three to 7 years are required for crossing and inbreeding to develop homozygous stable lines. Thereafter, four to 5 years of testing and selection for traits like quality, pest and disease resistance and yield are performed. Lastly, one to 3 years are required before release; in the multiplication of seeds. However, the period was reduced to half with the introduction of methods like shuttle breeding (Rajaram et al., 2002; Ortiz et al., 2007), which allowed the screening of 2 generations/year instead of one generation (Figure 5). Another technology that rapidly produced inbred lines without self-pollination cycles is doubled haploid technology (Ren et al., 2017; Wu et al., 2020). This method decreased the time for inbred making from 7 to 2 years. Nevertheless, there are several limitations to using this technology. For instance, there is no scope for early generation selection, lack of recombination and its effectiveness varies across crosses. While considering the larger scale of breeding programs, doubled haploid breeding turns out to be expensive (Chaudhary et al., 2019b; Chaikam and Prasanna, 2020).
FIGURE 5
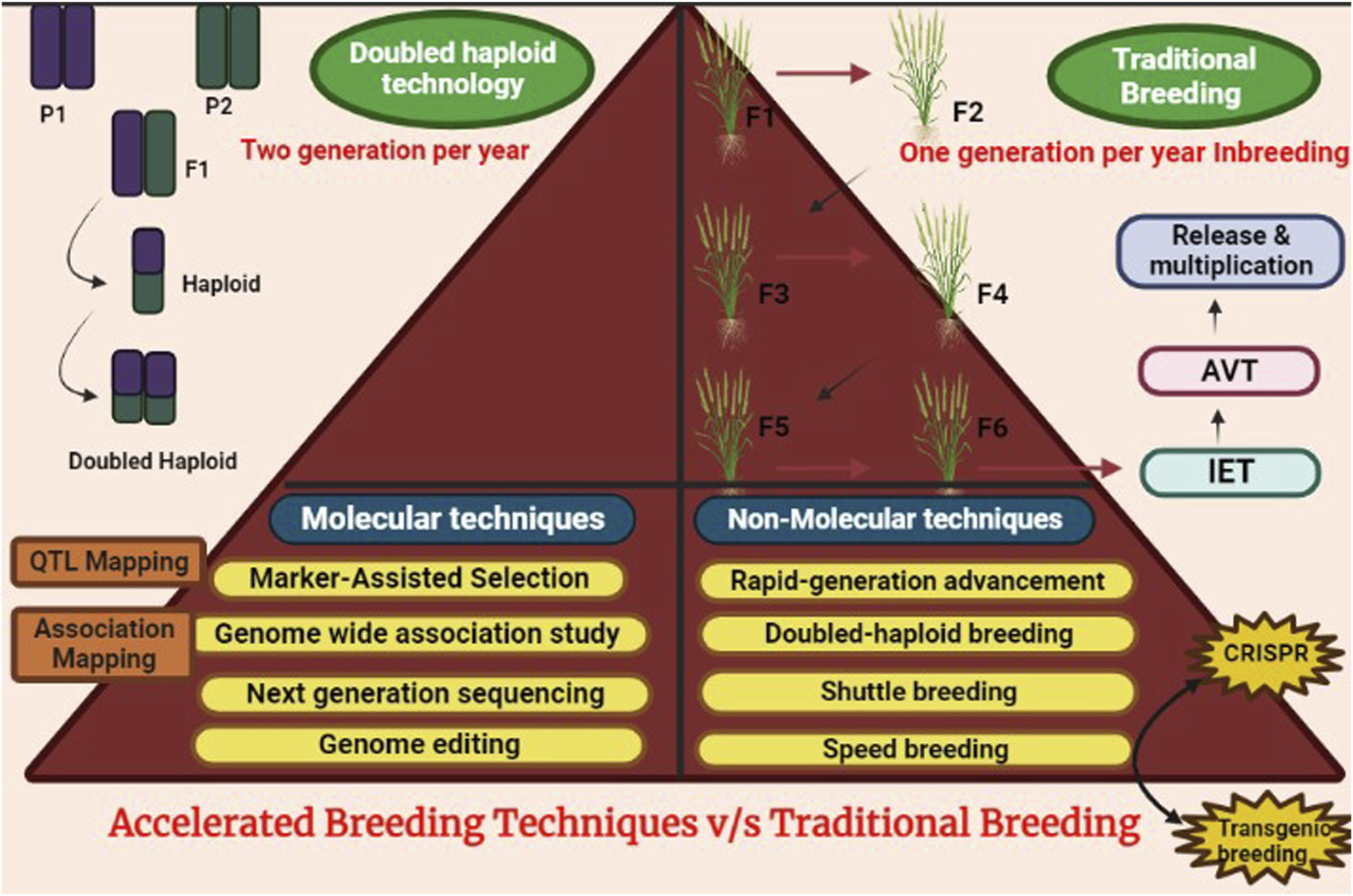
Comparison of accelerated breeding methods v/s traditional methods.
Speed breeding innovation was stimulated by NASA, aiming to cultivate wheat in space. This technique utilizes specialized greenhouse facilities and carefully controlled lighting regimes to provide optimal growth conditions, allowing for the rapid acceleration of the breeding process. Usually, 22 h of light phase at around 22°C and 2 h of the dark period at 17°C are given to promote early flowering (Watson et al., 2018). For crops such as Triticum, Hordeum and Cicer spp., speed breeding can accomplish up to six cycles per year, resulting in faster selection and development of new cultivars with improved traits such as yield, disease resistance, and tolerance to environmental stress (Ghosh et al., 2018; Samineni et al., 2020). Although for crops like canola 4 generation/year can be achieved, which is still commendable, while working for pod-shattering phenotyping (Watson et al., 2018). Therefore, speed breeding has huge scope when it comes to doubling genetic gain and accelerated transfer of new alleles into adapted material through rapid backcrossing. Tremendous research is still going on in developing protocols for different crops like pepper, cassava, amaranthus, etc. (Stetter et al., 2016; Souza et al., 2018; Borovsky et al., 2020) (Table 2). DS Faraday was the first wheat variety produced via speed breeding that had excellent protein milling quality and resistance to PHS. It was released in partnership with DOW Agrosciences. The technique involves repeated cycles of selection for grain dormancy and backcrossing (Schwager, 2017). One of the key advancements in speed breeding’s future is its integration with CRISPR technology and the enhancement of transgenic techniques. Speed breeding will be closely linked with the revolutionary gene-editing tool CRISPR, allowing for more precise and efficient modifications in plant genomes (Murovec et al., 2018; Bao et al., 2020).
5 Phase III: genome editing technologies
This is a form of genetic manipulation in which DNA is incorporated to, or removed/substituted from a living organism’s genome for any desirable trait expression. In contrast to genetic engineering, which inserts the target gene into the host organism at random, genome editing targets the introduction to certain predefined locations. A gene knockout occurs when a frameshift mutation occurs in a gene, resulting in the cell no longer expressing any functional protein. ZFNs, TALENS and CRISPR can be used for gene knockouts. A gene knockdown is when a gene expression is reduced but not eliminated. This is usually accomplished by degrading or inhibiting the gene’s mRNA transcript from being translated.
5.1 Exploitation of natural DNA repair system in the host organism (through ZFNs, TALENs, and CRISPR)
Using appropriate genome editing technology techniques, double-stranded breaks may be generated. These DNA breaks stimulate the cellular DNA repair processes, allowing site-specific genomic changes to be introduced more easily (Rouet et al., 1994; Choulika et al., 1995). Artemis’ unique nuclease actions result in the formation of INDELs, rendering non-homologous end-joining repair systems inappropriate for precise alterations (Chang and Lieber, 2016). In situations where Homology-Directed Repair (HDR) is involved, a process that relies on a matching pair of chromosomes, the preservation of sequence information during the repair process is exceptionally high, exhibiting either minimal loss or no loss at all (known as the conservative type). At the site of the DSB, a required gene from the donor DNA strand is inserted using the homologous chromosome. Mammalian cells were formerly presumed to repair potentially lethal chromosomal double-strand breaks (DSBs) in part by non-homologous processes. Yet, it was later discovered that DSBs can increase homologous recombination by three or four orders of magnitude, suggesting that homology-directed repair is possible (Liang et al., 1998). As a consequence, the DNA-repair mechanism might be employed to insert the requisite genetic material, allowing for high-precision genome editing of a target cell. This type of genome editing can be employed to insert new genes or wipe out existing ones (Mali et al., 2013). Genome editing technologies can be categorized into four major groups: 1) Meganucleases; 2) ZFNs; 4) TALENs; 4) CRISPR/Cas9 system. However, the CRISPR/Cas9 system predominates as the preeminent and extensively employed methodology for genome editing.
The term “mega” is used to describe a massive recognition site. Because they are endonucleases, this location usually only appears once in each genome (Gallagher et al., 2014). The LAGLIDADG family contains the most well-known meganucleases proteins. LAGLIDADG proteins have one of two main functions 1) function as RNA maturase and 2) cleaving the exon-exon junction sequence where their intron is located, earning them the nickname “homing endonuclease.” This approach has a significant benefit in terms of safety since it is less harmful to cells than other naturally occurring restriction enzymes. Nevertheless, this procedure is both expensive and time-consuming.
Construction of the first chimeric restriction endonucleases gene by linking the finger domain to the non-specific cleavage domain Fok 1(Flavobacterium okeanokoites) (Kim et al., 1996). Zinc fingers use a mix of cysteine and histidine residues to coordinate zinc ions. Each domain’s-helix (also known as the “recognition helix”) may create sequence-specific interactions with DNA bases. There were two distinct realms, one DNA-binding domain (zinc finger motifs), which is made up of a chain of two-finger modules and recognizes three nucleotide sequences of DNA (one amino acid), and another DNA-cleaving domain, which is made up of Fok I nuclease domain. This method is described to have rapid disruption and integration into any genomic loci. Also, it can create gene knockouts in multiple cell lines. As compared to mega nucleases, this can have an off-target effect, and construction is complex as it needs two zinc finger motifs and a nuclease to create double-strand breaks.
Transcription Activator-Like Effector Nucleases (TALEN), like zinc finger nuclease is chimera that includes TALEs and Fok 1 endonuclease. The ability of TALENs to bind to DNA and promote the expression of their target genes by mimicking eukaryotic transcription factors is used as a DNA binding domain, in conjunction with the cleaving domain Fok 1, which causes double-stranded breaks. As compared to the Mega nucleases and zinc finger nuclease (ZFNs), Transcription Activator-Like Effector Nucleases TALEN is considered to have a simpler design and a higher specificity. However, two caveats were recognized in this technique 1) challenging to use in viral systems due to large protein size and 2) repetitive sequences may induce undesirable recombination events within the TALEN array.
The acronym CRISPR for Clustered Regularly Interspaced Short Palindromic Repeats was first time used in 2002 by Jansen et al. (2002). The word repeats refer to palindromic sequences which are interspaced by unique sequences called spacer. These spacers are molecular records in bacteria that are part of the virus genome which has earlier attacked the bacteria. This system works like an antigen/antibody system. The spacer is formed from protospacer sequences present in the virus which gets into the bacterial genome in the form of unique spacer sequences. Therefore, CRISPR is referred to as an adaptive immunity system. This system consists of RNA molecules and Cas enzymes that work together to identify and cut specific DNA sequences.
CRISPR-associated protein (Cas9) is an RNA-guided endonuclease that uses a single-guide RNA to cleave DNA at specific target sites. CRISPR/Cas9-based genome editing relies on creating a double-strand break in the DNA and then utilizing the cell’s natural DNA repair mechanisms. Within the native CRISPR/Cas9 system, the mature crRNA and transactivating crRNA come together to form a complex called tracrRNA: crRNA (Zhan et al., 2014). This complex serves as a guide for Cas9, directing it to the desired location on the DNA. While Cas9 is widely known, there are other forms of Cas proteins with distinct properties and functions. For example, Cas1 and Cas2 are involved in the adaptation phase of CRISPR systems, while Cas3 plays a role in the destruction of foreign DNA. Cas12 (Cpf1) is another variant that cleaves DNA with staggered ends, and Cas13 proteins target and cleave RNA molecules instead of DNA. Scientists have adapted this system for use in a wide range of organisms to precisely and efficiently edit their genomes. The ability to edit genes with unprecedented precision and ease has opened up many new possibilities for basic research and applied biotechnology.
CRISPR/Cas technology has significant potential for crop improvement by enabling precise and targeted genetic modifications in crops (Table 3). This technology has several advantages over traditional breeding methods, including speed, precision, and accuracy. With CRISPR/Cas, scientists can target specific genes in crop plants and make precise modifications, such as creating mutations or introducing new traits, without the need for introducing foreign DNA. This system consists of RNA molecules and Cas enzymes that work together to identify and cut specific DNA sequences. Scientists have adapted this system for use in a wide range of organisms, including plants, to precisely and efficiently edit their genomes. The ability to edit genes with unprecedented precision and ease has opened up many new possibilities for basic research and applied biotechnology. One application of CRISPR/Cas in crop improvement is to enhance the nutritional value of crops (Zhu et al., 2019). For example, researchers have used CRISPR/Cas to increase the iron content in rice, which is a significant dietary source of iron for many people. While CRISPR/Cas technology holds tremendous potential for genome editing, there are also some limitations and challenges that need to be addressed (Chen et al., 2021b; Yumlu and Stumm, 2021). This involves off-target effects (sometimes cut DNA at unintended locations, leading to unintended mutations), mosaicism, challenges in delivering methods, etc. Despite these challenges, researchers and companies around the world are working to overcome these limitations and leverage the potential of CRISPR/Cas technology for a wide range of applications.
TABLE 3
| Target gene | Trait improved | Remarks | References |
|---|---|---|---|
| Rice | |||
| (Hpt) Hygromycin phosphotransferase | ZFNs | Cantos et al. (2014) | |
| Resistance to bacterial blight | TALENs | Li et al. (2012) | |
| Fragrant rice | Shan et al. (2015) | ||
| Resistance to disease & tolerant to abiotic stresses | CRISPR/CAS9 | Xie and Yang (2013) | |
| Increase resistance to blast | Liu et al. (2012) | ||
| Cold resistance resistant | Shen et al. (2017) | ||
| Tiller spreading | Miao et al. (2013) | ||
| Increase in grain number, grain size with thick erect panicles | Li et al. (2016a) | ||
| High amylose content | Sun et al. (2017) | ||
| Production of haploid plants | Yao et al. (2018) | ||
| Resistance to rice root-knot nematode | CRISPR/CAS9 | Huang et al. (2023) | |
| Reduce Cd accumulation | Chen et al. (2023) | ||
| Improving fragrance efficiency | Imran et al. (2023) | ||
| Agronomic traits and starch composition | Zheng et al. (2023) | ||
| Increase Photosynthesis | Caddell et al. (2023) | ||
| Broad-spectrum disease resistance | Liu et al. (2023) | ||
| Herbicide resistance | CRISPR/CAS9 | Sun et al. (2016) | |
| Endo et al. (2016) | |||
| Butt et al. (2017) | |||
| Li et al. (2016b) | |||
| Shimatani et al. (2017) | |||
| Nutritional quality improvement | CBEs | Li et al. (2017a) | |
| Enhance nitrogen use efficiency | Lu and Zhu (2017) | ||
| Regulate senescence and death | Zong et al. (2017) | ||
| Resistance to blast | Ren et al. (2018) | ||
| Defence response | Ren et al. (2018) | ||
| Pathogen-responsive gene | ABEs | Yan et al. (2018) | |
| Della protein for plant height | Hua et al. (2018) | ||
| Regulation of architecture of plant and grain yield | Hua et al. (2018) | ||
| Amylose synthesis | Hao et al. (2019) | ||
| Defence response | Hao et al. (2019) | ||
| Wheat | |||
| TaMLO | Powdery mildew disease resistance | Shan et al. (2014) | |
| TaDREB2 | Dehydration responsive element | Shan et al. (2014) | |
| TaERF3 | Ethylene responsive factor | Shan et al. (2014) | |
| TaGW2 | Negative regulator of grain traits | Wang et al. (2018b) | |
| EDR1 | Resistance to powdery mildew | Zhang et al. (2017) | |
| TaSPL13 | Improve multiple agronomic traits | Gupta et al. (2023) | |
| SPO11-1 | Fertility and synapsis | Hyde et al. (2023) | |
| Tamyb10 | Pre-harvest sprouting-resistant red wheat | Zhu et al. (2023) | |
| Ppd-1 | Spike architecture | Errum et al. (2023) | |
| Lipid metabolism | CBEs | Zong et al. (2017) | |
| Panicle length and grain weight | ABEs | Li et al. (2018b) | |
| Maize | |||
| ZmIPK1 | Responsible for herbicide tolerance and reduction of phytate content | ZFNs | Shukla et al. (2009) |
| ZmGL2 | Reduced epicuticular wax in leaves | Char et al. (2015) | |
| ZmMTL | Production of haploids | TALENs | Kelliher et al. (2017) |
| ARGOS8 | Expressed well under drought stress with increase in grain yield | Shi et al. (2017) | |
| ZmIPK1A, ZmIPK and ZmMRP4 | Phytic acid synthesis | Liang et al. (2014) | |
| PSY1 | Phytoene synthase | Zhu et al. (2016) | |
| Zmzb7 | Knockout of gene resulted in albino plant | Feng et al. (2016) | |
| ZmTMS5 | Thermosensitive genic male-sterile | Li et al. (2017b) | |
| Wx1 | High amylopectin content | Pioneer (2016) | |
| ALS | Herbicide resistance | Svitashev et al. (2015) | |
| ARGOS8 | Drought stress tolerance | Shi et al. (2017) | |
| ipdC | Promote maize growth | Figueredo et al. (2023) | |
| 36 genes potentially involved in leaf growth | 10% increase in leaf size | Impens et al. (2023) | |
| pipeline BREEDIT | Improve complex traits such as yield and drought tolerance | Lorenzo et al. (2023) | |
| First time multiplex gene editing | CRISPR/Cas9 (tRNA-RNAprocessing system) | Qi et al. (2016) | |
| Chromosomal segregation | CBEs | Zong et al. (2017) | |
| Tomato | |||
| Production of purple tomatoes | TALENs | Cermak et al. (2015) | |
| Powdery mildew resistance | CRISPR/Cas9 | Nekrasov et al. (2017) | |
| Bacterial speck resistance | Ortigosa et al. (2019) | ||
| Tomato domestication | Li et al. (2018c) | ||
| Earlier harvest time | Soyk et al. (2017) | ||
| Parthenocarpy | Klap et al. (2017) | ||
| Repression of fruit ripening | Ito et al. (2015) | ||
| Prevents tomato fruit ripening | Yang et al. (2017b) | ||
| Increase shelf life | Yu et al. (2017) | ||
| Leaf shape variations and seedless fruits | Ueta et al. (2017) | ||
| Drought tolerance | Wang et al. (2017) | ||
| PSY1, MYB12, and SGR1 | Fruit colour-related genes | Yang et al. (2023) | |
| SlATG5 | Resistance to Botrytis cinerea | Li et al. (2023) | |
| SlHyPRP1 | Multi-stress tolerance | Tran et al. (2023) | |
| SlDYT1 and SlGSTAA | Male Sterility | Zhou et al. (2023) | |
| Herbicide resistance | CBEs | Veillet et al. (2019) | |
| SlRIN | Tomato fruit ripening | ABEs | Niu et al. (2023) |
| Soyabean | |||
| High oleic acid contents | TALENs | Haun et al. (2014) | |
| High oleic & low linoleic contents | Demorest et al. (2016) | ||
| Herbicide resistance | CRISPR/Cas9 | Li et al. (2015) | |
| Disease resistance against Phytophthora sojae | Fang and Tyler (2016) | ||
| Flowering time | Cai et al. (2018) | ||
| Carotenoid biosynthesis | Du et al. (2016) | ||
| Potato | |||
| Minimizing reducing sugars | TALENs | Clasen et al. (2016) | |
| Herbicide resistance | CRISPR/Cas9 | Butler et al. (2016) | |
| High amylopectin content | Andersson et al. (2017) | ||
| Herbicide resistance | Veillet et al. (2019) | ||
| VInv | Quality of potato tubers | Sattar et al. (2023) | |
| VInv and AS1 | Reduced Browning | Ly et al. (2023) | |
| Sugarcane | |||
| Improved cell wall composition | TALENs | Jung and Altpeter (2016) | |
| Improved efficiency of saccharification | Kannan et al. (2018) | ||
| Arabidopsis | |||
| MIR169a | Drought tolerance | CRISPR/Cas9 | Zhao et al. (2016) |
| Turnip mosaic virus (TuMV) resistance | CRISPR/Cas9 | Pyott et al. (2016) | |
| Increased stomatal closure against abscisic acid | CRISPR/Cas9 | Osakabe et al. (2016) | |
| High-light acclimation and photomorphogenesis | CRISPR/Cas9 | Atanasov et al. (2023) | |
Application of genome editing in crop improvement.
5.2 Base editing
After the discovery of CRISPR/Cas9 system, another mechanism called base editing has also become a popular tool for genome editing. Base editing is revolutionizing crop improvement by allowing alteration (Gaudelli et al., 2017) of the genome by making specific changes to the DNA base pairs (Komor et al., 2016; Porto et al., 2020). This technology can be used to change one DNA letter into another, which can then lead to new traits or characteristics appearing in the plants (Henikoff and Comai, 2003). Base editors can make precise, single-nucleotide changes to the genome without inducing double-stranded breaks (Rees and Liu, 2018; Molla and Yang, 2019). This makes base editing an attractive alternative to traditional CRISPR/Cas9 methods, which often result in unwanted indels. Base editors are made up of two components: a CRISPR protein that can be configured to target a particular position in the genome and an enzyme that can chemically alter the DNA base at that position. The most often used base editors are cytosine base editors (CBEs) and adenine base editors (ABEs), which are used to change a C-G base pair to a T-A base pair or an A-T base pair to a G-C base pair, respectively. CBEs use a modified version of the CRISPR protein Cas9 that has been fused to a cytidine deaminase enzyme (Li et al., 2018a). The deaminase enzyme converts the C base to a U base, and then the DNA repair machinery in the cell converts the U to a T, resulting in a C to T base change (Zhang et al., 2020). ABEs, on the other hand, employ a separate enzyme known as an adenine deaminase to convert an A base to an inosine base, which the DNA repair machinery interprets as a G base, resulting in an A to G base conversion (Kang et al., 2018; Hua et al., 2020).
The benefits of base editing include its precision, efficiency and lack of off-target effects (Lee et al., 2020). Base editing has already had a positive impact on crop production, and it is predicted to play a more significant role in the future as more genes are discovered that can be edited with this technique (Eid et al., 2018). Crop improvement by base editing is a technique that entails amending DNA to enhance its attributes (Bharat et al., 2020). This can be done by either mutating existing genes or adding new ones. Base editing is different from traditional methods of genetic modification, as it does not require the use of foreign DNA. This overcomes regulatory issues (Jones, 2015). There are several ways in which base editing can be used to improve crops. One way is by increasing resistance to biotic and abiotic stress (Zhang et al., 2018a; Sood et al., 2022). This can be done by altering the genes that encode proteins that are targets of disease-causing organisms. Another way is by improving the nutritional value of crops. This can be done by modifying the genes that control the production of vitamins and minerals (Kumar et al., 2021).
5.3 Prime editing: unlocking the precision of genetic rewriting
It is a breakthrough advanced technique over existing CRISPR/Cas9 tools. Prime editing is complementary to base editing to correct small mutations including indels (Anzalone et al., 2019; Kantor et al., 2020). CRISPR/CAS9 as stated above causes double-stranded breaks. However, it was seen in a few studies that double-stranded breaks are not safe (Carusillo and Mussolino, 2020; Ochoa-Sanchez et al., 2021). It can cause mutagenic activity, a complex mix of undesirable products or may cause translocation of DNA. These breaks can trigger P53 activity which can induce cell death (Shen and Li, 2022). On the other hand, base editing is limited to four possible transition mutations (C to T, A to G, T to C, and G to A) and is still prone to off-target effects. Base editing has not proved to be useful in the case of mutations like insertions and deletions. Therefore, to overcome these obstacles, prime editing was developed (Marzec and Hensel, 2020).
Prime editors have three major components referred as pegRNA, CAS9 H840A nickase fused with M-MLV reverse transcriptase and single guide RNA. Guide RNA is lengthened including mutant target recognition sequences and correction sequences which in combination are referred as pegRNA. The CAS enzyme domains are also modified like in base editing such that only one strand is cut. Finally, instead of a base editing domain, a reverse transcriptase activity domain uses the corrected sequence of the peg RNA as a template to synthesize the corresponding stretch of the DNA strand. The newly synthesized stretch of DNA will then bind to the untampered original DNA along with creating a flap that the stretch of new DNA is supposed to replace. The flap will be cut out since it is an unnatural DNA structure. Although a new segment of DNA contains the correct DNA sequence, the other segment still does not. This mismatch will be corrected by a triggered natural repair mechanism (Marzec et al., 2020; Molla et al., 2021; Hillary and Ceasar, 2022). Its major inference lies in the precise gene editing with reduced off-target mutations and wider scope of applications. This technique can bring revolution by allowing for precise alterations to specific genes which will eventually increase yield by providing tolerance to biotic and abiotic stress. (Gao, 2021; Lu et al., 2022).
5.4 Achievements of genome editing in plant breeding
Genome editing in plant breeding has achieved several desirable outcomes (Table 2). Most significant is breeding for resistance in staple crops like rice and wheat by modifying the genes responsible for susceptibility to various diseases (Wang et al., 2014; Zhou et al., 2015; Wang et al., 2018b). Additionally, genome editing can modify genes that control yield-related traits (Ainley et al., 2013; Zhang et al., 2018a; Zong et al., 2018). For instance, researchers have altered a gene in rice that controls the plant’s sensitivity to nitrogen using CRISPR/Cas9, resulting in plants that yield more with less fertilizer. The nutritional content of crops may be improved through genome editing (Connorton et al., 2017). For instance, by altering the genes that produce beta-carotene in rice, scientists were able to produce a type of grain known as “golden rice” that had greater quantities of vitamin A (Busch and Schneeberger, 2019). Another important application is for adaptation to environmental stress. This may lead to crops that are more resilient to the difficulties posed by a changing climate (Zhang et al., 2014).
5.5 Unlocking nature’s blueprint: target genes for precision genome editing
CRISPR techniques have revolutionized genome editing and have made it possible to target any gene of interest with a high degree of precision. Researchers can modify genes involved in various traits, such as disease resistance, quality attributes, yield, and many other desired characteristics. For instance, TaDREB2 is a transcription factor that plays a crucial role in drought stress response in wheat. It is involved in regulating the expression of stress-responsive genes that help the plant cope with water deficit conditions. Additionally, TaERF3 is another transcription factor that is associated with abiotic stress responses, including drought, heat and salinity stresses in wheat (Kim et al., 2018). In maize, the mLG1, UB2 and UB3 genes have been utilized for the development of a haploid-inducer mediated genome editing system (Wang et al., 2019a). The mLG1 gene is a maternally expressed gene that plays a role in inhibiting embryo development when it is paternally inherited. The UB2 gene is a ubiquitin-conjugating enzyme that is involved in protein degradation and regulation. The UB3 gene is a ubiquitin ligase that plays a role in the degradation of proteins through the ubiquitin-proteasome system. In tomato crops, one of the target genes for improving quality traits is ACS2 (Ito et al., 2021). The purpose of editing this gene is to regulate the ripening process and extend the shelf life of tomatoes (Peng et al., 2022). For herbicide tolerance, the target gene is EPSPS (5-enolpyruvylshikimate-3-phosphate synthase). By editing this gene, scientists aim to confer resistance to the widely used herbicide glyphosate (Achary et al., 2020; Wang et al., 2021a; Li et al., 2021). CRISPR technology provides a powerful tool for gene editing (Table 3), but there are still challenges to overcome, such as off-target effects and ensuring precise and accurate edits. However, ongoing research and advancements in CRISPR techniques continue to improve the precision and efficiency of gene editing, making it a promising technology for various applications.
6 Phase IV: beyond and next-generation smart crops
These are sophisticated agricultural technologies and approaches that increase crop productivity, efficiency, and sustainability. This includes various techniques like Haplotype crop breeding, genomic selection, use of secondary population, artificial intelligence (AI), etc.
6.1 Genomic selection
Genomic selection utilizes genomic information to predict the performance of plants and select the best individuals for breeding. Genomic selection has been used in a variety of crops to advance breeding programmes and increase crop improvement efficiency (Benavente and Giménez, 2021). Genomic selection, for example, has been used in cereal breeding to improve yield, drought tolerance, disease resistance, and nutritional quality (Haile et al., 2020; Simmons et al., 2021). Breeders can find maize lines with high genomic estimated breeding values (GEBVs) for these characteristics by utilizing genomic information and marker-assisted selection, accelerating the production of better varieties (Crossa et al., 2017). GS method facilitates the early selection of individuals with favorable genomic profiles, resulting in the production of high-performing wheat cultivars (Sinha et al., 2021). By harnessing the power of genomics, breeders can make more accurate selections, enhance breeding efficiency, and develop improved crop varieties with desired traits.
6.2 Haplotype-based breeding
It is a cutting-edge technology that entails discovering and choosing precise allele combinations within certain genomic areas known as haplotypes to obtain desired crop attributes (Bhat et al., 2021). For instance, in wheat the method may be used to boost disease resistance against rusts and Fusarium head blight, as well as attributes such as drought tolerance and yield potential (Athiyannan et al., 2022; Alemu et al., 2023). Advances in genomic technologies, such as high-throughput genotyping and genome sequencing, have facilitated the identification and characterization of haplotypes in rice. This has opened up new opportunities for breeders to accelerate the development of improved rice varieties by incorporating favorable haplotypes into their breeding programs. It helps in developing blast-resistant, bacterial blight-resistant, and submergence-tolerant cultivars while also increasing grain quality traits (Tanweer et al., 2015; Verma et al., 2023). Few studies have highlighted the use of haplotype analysis to understand germplasm diversity in maize breeding programs. Haplotype-based approaches provide advantages over single-marker-based methods in assessing population structure and capturing additional information compared to individual SNPs (Coffman et al., 2020). Improved features in maize include drought tolerance, insect resistance, and nitrogen usage efficiency (Simmons et al., 2021). Similarly, haplotype-based breeding may be used to improve disease resistance, qualitative traits, and stress tolerance in diverse crops such as tomato, potato, cotton, and barley. Various studies have discussed the advantages of haplotype-based breeding, such as its ability to capture the combined effects of multiple genetic variants and its potential to increase the accuracy of trait selection.
6.3 Omics-based breeding
Omics-based plant breeding is the use of high-throughput technology and data-driven methodologies for investigating and altering plant genetic and molecular features for crop development. Omics-based approaches have found successful applications in various crops, and one notable example is in the field of rice breeding (Dai et al., 2022; Zaghum et al., 2022). Genomics has proven critical in decoding the rice genome sequence and discovering genes linked to crucial agronomic features (Peng et al., 2020). Transcriptomic studies on rice have shown gene expression patterns at various developmental stages and stress responses. Understanding the activities of individual proteins involved in grain quality and stress tolerance processes has been improved by proteomics (Choi, 2019). Metabolomics has aided in the discovery of compounds associated with nutritional characteristics and stress responses in rice. Overall, in crop breeding programmes, these omics-based techniques have aided in the identification of candidate genes, molecular markers, and important pathways linked with desirable features (Alotaibi et al., 2021; Cao et al., 2022; Shen et al., 2022).
7 Convergence of genome-assisted breeding and genome editing
The confluence of genome-assisted breeding and genome editing entails merging genomic information with precise gene editing methods to boost agricultural yield. Genome-assisted breeding assists in finding significant genes linked with desired qualities, while genome editing enables precise gene alteration. By combining these techniques, breeders can minimize the time and resources necessary for the production of superior cultivars with increased attributes, as well as enable the transfer of beneficial genetic variants from wild relatives. This confluence provides a tremendous tool for expediting crop development efforts and tackling agricultural concerns.
8 Empowering food security with resilient orphan crops through diverse breeding approaches
A range of orphan crop species exhibit regional significance and possess stress resilient traits in order to thrive extreme climatic conditions owing to their relevant genes and stress combating mechanisms. However, these species lack global trade and substantial recognition from a research standpoint. Hence, research initiatives attempt to exploit their potential to improve major crops, address nutritional challenges and enhance food system sustainability (Dawson et al., 2019; Yaqoob et al., 2023). Lemmon et al. (2018) elucidated the viability of different breeding approaches and underscored the significance of allocating resources to study and acquire further knowledge concerning genomes, genes, and cellular mechanisms underlying plant characteristics. Several conventional as well as advanced breeding techniques have been employed to enhance the desirable traits of orphan crops yet.
Historically, the genetic enhancement of orphan crops were primarily restricted to conventional breeding practices, employing pedigree-based selection methods to enhance desired traits such as larger seed size, increased yield, ease of propagation, reduced seed dispersal, etc. The emphasis was predominantly placed on improving the domestication process of these crops (Kamenya, et al., 2021). For orphan legumes (pea, lupin) mutation breeding and interspecific introgression have been successful in generating genetic diversity leading to favorable traits like stress tolerance, high yield, etc., (Chongtham et al., 2022). A robust repertoire of molecular markers serves as a crucial asset in the breeding endeavors of all crop species; however, it is often deficient in broad array of orphan crops. Among the techniques employed for molecular marker development in orphan crops, Diversity Arrays Technology has emerged as a highly significant method. The advent of this technology ushered in a paradigm shift in the genetic profiling and establishment of genetic linkages in several crops (predominantly pigeon pea and cassava) that were previously deemed as orphaned, a span of approximately 20 years ago (Kamenya, et al., 2021). However, with the sequencing of more than 35 orphan crops, rapid SNP discovery has become possible (finger millet, Bambara groundnut, lupin, etc.). Certain researchers have pinpointed specific genes or quantitative trait loci (QTLs) associated with adaptive traits in orphan legume species. These genetic elements possess the potential to be utilized in crop enhancement efforts to confer stress tolerance onto other cultivated crops (Chongtham et al., 2022). Although research on speed breeding in orphan crops is still limited, there are a few examples where it has been explored. For instance, chickpea, peanut and amaranth speed breeding protocol has been developed (Chiurugwi et al., 2019). Speed breeding could be used to shorten the breeding cycle of fonio and facilitate the development of improved varieties with increased yield, disease resistance, and drought tolerance (Ibrahim Bio Yerima and Achigan-Dako, 2021). Some initiatives have been there for implementing TILLING in various orphan crops like pearl millet, teff, cassava, mung bean, chickpea, banana, etc. (Esfeld et al., 2013). Genome editing has been exploited in orphan crops like sorghum (modulating flowering time), foxtail millet (male sterility), chickpea (draught tolerance), etc. (Venezia and Creasey Krainer, 2021).
These days, with the advent of genome editing techniques, these underutilized crops are undergone targeted improvements, leading to advancements in their characteristics and overall performance (Table 4). Enhancing essential nutrients such as iron, zinc, a range of vitamins can have significant impacts on improving the nutritional quality of diets that heavily rely on these crops (Tadele, 2018). A study showed that in the future, the CRISPR/Cas9 tool holds potential for application in the Ca transporter genes of finger millet. The docking study proposed that EcCBL4 has a strong binding affinity with EcCIPK24 and might play a significant role in the accumulation of Ca in seeds (Chinchole et al., 2017). Classical domesticated genes possess characteristics that make them ideal candidates for Cas base editing. They are well characterized, exhibit simple genetic architecture, and typically have a monogenetic nature -(Rasheed et al., 2021). Studies have shown the role of TALENs to target a gene involved in the lignin biosynthesis pathway in pearl millet. By knocking out this gene, they achieved improved resistance against downy mildew, a significant foliar disease in pearl millet (Maurya et al., 2022). Cassava yield is significantly affected by disease-causing pathogens nearly around 50% of total yield loss attributed to the African cassava mosaic virus (ACMV) and cassava brown streak disease (CBSD). Hence, targeted mutation using Cas9/gRNA, have been developed to address such challenges. Orphan crop genomes were annotated through whole-genome sequencing and their transcriptomes were generated. In the annotation process, transcriptomes from the same species were preferred, as seen in examples like the african eggplant, wild mustard, tef, etc. (Cannarozzi et al., 2014; Bhardwaj et al., 2015; Chen et al., 2021c). However, in some cases, closely associated or model crop transcriptomes were used, as in finger millet, which utilized data from maize. RNA sequencing (RNA-seq) became the method of choice for generating specific crop transcriptomes (Ozsolak and Milos, 2011). Microarrays were the preferred method before NGS technology for transcriptome analysis in various orphan crops to uncover expression profiles associated with abiotic stress resilience. Among the crops studied were buckwheat, tef (Golisz et al., 2008), white lupine (Zhu et al., 2010), etc. Another study detected 2,416 DEGs in quinoa (Chenopodium quinoa) during salt stress profiling (Ranasinghe et al., 2019). Additionally, transcription analysis in jute-mallow helped identify genes related to drought stress response (Yang et al., 2017a).
TABLE 4
| Crop | Target gene | Trait improved | Remarks | References |
|---|---|---|---|---|
| Sorghum | Alpha-Kafirin gene family | Increase digestibility and protein quality | CRISPR/Cas | Li et al. (2018d) |
| FLOWERING TIME (FT); Gibberellin 2-oxidase 5 (Ga2ox5) | Flowering time | Char et al. (2020) | ||
| Finger millet | Bhlh57 | Salinity resistance | Babitha et al. (2015) | |
| Foxtail millet | Phytochrome C (PHYC) | Photoperiodic flowering | Yang et al. (2020) | |
| DROOPY LEAF1 (DPY1) | Plant architecture | Zhao et al. (2020) | ||
| SiMTL | Haploid embryo induction | Cheng et al. (2021) | ||
| Sweet potato | Granule-bound starch synthase I (GBSSI) | Availability of more digestible sugars | Wang et al. (2019b) | |
| Starch branching enzyme II (SBEII) | ||||
| Cassava | Protein targeting to starch 1 (PTST1) | Increase in digestible sugars | Bull et al. (2018) | |
| EPSPS | Glyphosate tolerance | Hummel et al. (2018) | ||
| Pigeon Pea | CcFT8 | Florigen producing gene | Tribhuvan et al. (2020) | |
| Quinoa | WUSCHEL, BABY BOOM and LEAFY COTYLEDON1 | Improve the transformation efficiency | Wang et al. (2021b) | |
| Lettuce | GDP-L-galactose phosphorylase 1 (GGP1) | Increase in the vitamin C | Zhang et al. (2018b) | |
| GDP-L-galactose phosphorylase 2 (GGP2) | ||||
| Yam | Phytoene Desaturase (PDS) | Carotenoid biosynthesis | Syombua et al. (2020) | |
| ERF (ethylene-responsive factor) | Resistance to anthracnose | Ntui et al. (2021) | ||
| Eggplant | Polyphenol oxidase (PPO) | Decreased browning | Maioli et al. (2020) | |
| Bambara groundnut | KUP | Abiotic stress tolerance | Sharma et al. (2022) | |
| Fonio | DeSh1-9A | Reduced seed shattering | Abrouk et al. (2020) | |
| Teff | SEMIDWARF-1 (SD-1) | Semi-dwarfism and lodging resistance | Beyene et al. (2022) | |
| OsSPL14, OsmiR397 | Panicle branching trait | Numan et al. (2021) | ||
| Watermelon | Acetolactate synthase (ALS) | Herbicide-resistant | Tian et al. (2018) |
Potential role of genome editing in orphan crops.
9 Conclusion and future thrust
The trend in plant breeding has evolved significantly over the years, shifting from conventional breeding to more advanced molecular breeding, and this shift is likely to continue in the future with novel biotechnology tools. Traditional breeding relies on the genetic diversity of the parent plants and necessitates maturing plants and waiting for the next-generation to be produced. While breakthroughs in biotechnology have substantially improved breeding precision and speed, a shortage of facilities and financial resources keeps conventional breeding still in demand. Many breeders now use a combination of conventional and molecular breeding techniques to produce crops with improved traits. This has led to the development of new crop varieties that are more resilient to disease, pests, and environmental stresses, with better yields and improved nutritional quality. In summary, the future thrust of plant breeding will involve developing crop varieties that are resilient to climate change, provide nutritional security, are sustainable, use precision agriculture techniques, utilize gene editing technologies, leverage genomics and big data, and involve collaborative research efforts.
Statements
Author contributions
Conceptualization, RC, AP, and PP; writing—original draft preparation, RC, AP, KS, SM, SN, AM, and ID; writing—review and editing, KS, SN, SM, KS, ID, and AM; supervision, PP and ID. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abrouk M. Ahmed H. I. Cubry P. Simoníková D. Cauet S. Pailles Y. et al (2020). Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat. Commun.11 (1), 4488–4513. 10.1038/s41467-020-18329-4
2
Acevedo-Garcia J. Spencer D. Thieron H. Reinstädler A. Hammond-Kosack K. Phillips A. L. et al (2017). mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J.15 (3), 367–378. 10.1111/pbi.12631
3
Achary V. M. M. Sheri V. Manna M. Panditi V. Borphukan B. Ram B. et al (2020). Overexpression of improved EPSPS gene results in field level glyphosate tolerance and higher grain yield in rice. Plant Biotechnol. J.18 (12), 2504–2519. 10.1111/pbi.13428
4
Acquaah G. (2015). “Conventional plant breeding principles and techniques,” in Advances in plant breeding strategies: Breeding, biotechnology and molecular tools. Editors Al-KhayriJ.JainS.JohnsonD. (Cham: Springer), 115–158. 10.1007/978-3-319-22521-0_5
5
Ahmar S. Gill R. A. Jung K. H. Faheem A. Qasim M. U. Mubeen M. et al (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci.21 (7), 2590. 10.3390/ijms21072590
6
Ainley W. M. Sastry-Dent L. Welter M. E. Murray M. G. Zeitler B. Amora R. et al (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J.11 (9), 1126–1134. 10.1111/pbi.12107
7
Akkaya M. S. Bhagwatt A. A. Cregan P. B. (1992). Length polymorphisms of simple sequence repeat DNA in soybean. Genetics132, 1131–1139. 10.1093/genetics/132.4.1131
8
Aklilu E. (2021). Review on forward and reverse genetics in plant breeding. All Life14 (1), 127–135. 10.1080/26895293.2021.1888810
9
Akond Z. Alam M. J. Hasan M. N. Uddin M. S. Alam M. Mollah M. N. H. (2019). A comparison on some interval mapping approaches for QTL detection. Bioinformation15 (2), 90–94. 10.6026/97320630015090
10
Alemu A. Batista L. Singh P. K. Ceplitis A. Chawade A. (2023). Haplotype-tagged SNPs improve genomic prediction accuracy for Fusarium head blight resistance and yield-related traits in wheat. Theor. Appl. Genet.136 (4), 92. 10.1007/s00122-023-04352-8
11
Alipour H. Darvishzadeh R. (2019). Association mapping of quantitative traits in molecular cereal breeding. Cereal Res.9 (3), 271–298. 10.22124/CR.2019.14333.1518
12
Allard R. W. (1999). Principles of plant breeding. John Wiley & Sons.
13
Alotaibi F. Alharbi S. Alotaibi M. Al Mosallam M. Motawei M. Alrajhi A. (2021). Wheat omics: classical breeding to new breeding technologies. Saudi J. Biol. Sci.28 (2), 1433–1444. 10.1016/j.sjbs.2020.11.083
14
Alqudah A. M. Sallam A. Baenziger P. S. Borner A. (2020). Gwas: fast-forwarding gene identification and characterization in temperate cereals: lessons from barley–a review. J. Adv. Res.22, 119–135. 10.1016/j.jare.2019.10.013
15
Anderson E. Stebbins G. L. Jr (1954). Hybridization as an evolutionary stimulus. Evolution8, 378–388. 10.2307/2405784
16
Andersson M. Turesson H. Nicolia A. Fält A. S. Samuelsson M. Hofvander P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep.36, 117–128. 10.1007/s00299-016-2062-3
17
Anzalone A. V. Randolph P. B. Davis J. R. Sousa A. A. Koblan L. W. Levy J. M. et al (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature576 (7785), 149–157. 10.1038/s41586-019-1711-4
18
Atanasov V. Schumacher J. Muino J. M. Larasati C. Wang L. Kaufmann K. et al (2023). Arabidopsis thaliana BBX14 is a target of GLK1 and involved in high-light acclimation, photomorphogenesis and GUN-type retrograde signaling. BioRxiv, 2023–2103. 10.1101/2023.03.03.530939
19
Athiyannan N. Zhang P. McIntosh R. Chakraborty S. Hewitt T. Bhatt D. et al (2022). Haplotype variants of the stripe rust resistance gene Yr28 in Aegilops tauschii. Theor. Appl. Genet.135 (12), 4327–4336. 10.1007/s00122-022-04221-w
20
Awan F. S. Sadia B. Altaf J. Habib M. Hameed K. Hussain S. (2021). “Genetic variability through induced mutation,” in Genetic variation (IntechOpen).
21
Babitha K. C. Vemanna R. S. Nataraja K. N. Udayakumar M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS One10, e0137098. 10.1371/JOURNAL.PONE.0137098
22
Babu B. K. Dinesh P. Agrawal P. K. Sood S. Chandrashekara C. Bhatt J. C. et al (2014). Comparative genomics and association mapping approaches for blast resistant genes in finger millet using SSRs. PloS one9 (6), e99182. 10.1371/journal.pone.0099182
23
Badji A. Otim M. Machida L. Odong T. Kwemoi D. B. Okii D. et al (2018). Maize combined insect resistance genomic regions and their co-localization with cell wall constituents revealed by tissue-specific QTL meta-analyses. Front. Plant Sci.9, 895. 10.3389/fpls.2018.00895
24
Bado S. Kozak K. Sekander H. Alhajaj N. Ghanim A. Forster B. P. et al (2013). “Resurgence of X-rays in mutation breeding,” in Plant genetics and breeding technologies, plant diseases and resistance mechanisms, Proceedings, 18–20 February 2013, Vienna. Editor AustriaM-M. (Pianoro: International Proceedings Division), 13–16.
25
Bai Y. Kissoudis C. Yan Z. Visser R. G. van der Linden G. (2018). Plant behaviour under combined stress: tomato responses to combined salinity and pathogen stress. Plant J.93 (4), 781–793. 10.1111/tpj.13800
26
Bao A. Zhang C. Huang Y. Chen H. Zhou X. Cao D. (2020). Genome editing technology and application in soybean improvement. Oil Crop Sci.5 (1), 31–40. 10.1016/j.ocsci.2020.03.001
27
Barrera-Redondo J. Pinero D. Eguiarte L. E. (2020). Genomic, transcriptomic and epigenomic tools to study the domestication of plants and animals: A field guide for beginners. Front. Genet.11, 742. 10.3389/fgene.2020.00742
28
Basnet B. R. Crossa J. Dreisigacker S. Perez-Rodriguez P. Manes Y. Singh R. P. et al (2019). Hybrid wheat prediction using genomic, pedigree, and environmental covariables interaction models. Plant Genome12 (1), 180051. 10.3835/plantgenome2018.07.0051
29
Begna T. (2021). Role and economic importance of crop genetic diversity in food security. J. Agric. Sc. Food Technol.7 (1), 164–169. 10.17352/2455-815X.00010
30
Benavente E. Giménez E. (2021). Modern approaches for the genetic improvement of rice, wheat and maize for abiotic constraints-related traits: a comparative overview. Agronomy11 (2), 376. 10.3390/agronomy11020376
31
Bernatzky R. Tanksley S. D. (1986). Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics112, 887–898. 10.1093/genetics/112.4.887
32
Beyene G. Chauhan R. D. Villmer J. Husic N. Wang N. Gebre E. et al (2022). CRISPR/Cas9-mediated tetra-allelic mutation of the ‘Green Revolution’ semidwarf-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef). Plant Biotechnol. J.20, 1716–1729. 10.1111/pbi.13842
33
Bharat S. S. Li S. Li J. Yan L. Xia L. (2020). Base editing in plants: current status and challenges. Crop J.8 (3), 384–395. 10.1016/j.cj.2019.10.002
34
Bhardwaj A. R. Joshi G. Kukreja B. Malik V. Arora P. Pandey R. et al (2015). Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol.15 (1), 9–15. 10.1186/s12870-014-0405-1
35
Bhat J. A. Yu D. Bohra A. Ganie S. A. Varshney R. K. (2021). Features and applications of haplotypes in crop breeding. Commun. Biol.4 (1), 1266. 10.1038/s42003-021-02782-y
36
Bhatnagar-Mathur P. Sunkara S. Bhatnagar-Panwar M. Waliyar F. Sharma K. K. (2015). Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci.234, 119–132. 10.1016/j.plantsci.2015.02.009
37
Bomblies K. Doebley J. F. (2006). Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics172, 519–531. 10.1534/genetics.105.048595
38
Boopathi N. M. (2020). “Marker-assisted selection (MAS),” in Genetic mapping and marker assisted selection (Singapore: Springer), 343–388.
39
Borlaug N. E. (1983). Contributions of conventional plant breeding to food production. Science219 (4585), 689–693. 10.1126/science.219.4585.689
40
Borovsky Y. Mohan V. Shabtai S. Paran I. (2020). CaFT-LIKE is a flowering promoter in pepper and functions as florigen in tomato. Plant Sci.301, 110678. 10.1016/j.plantsci.2020.110678
41
Bouchez A. Hospital F. Causse M. Gallais A. Charcosset A. (2002). Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics162 (4), 1945–1959. 10.1093/genetics/162.4.1945
42
Bouis H. E. Saltzman A. (2017). Improving nutrition through biofortification: a review of evidence from harvest plus, 2003 through 2016. Glob. Food Sec.12, 49–58. 10.1016/j.gfs.2017.01.009
43
Breseghello F. Coelho A. S. G. (2013). Traditional and modern plant breeding methods with examples in rice (Oryza sativa L). J. Agric. Food Chem.61 (35), 8277–8286. 10.1021/jf305531j
44
Bruno M. C. (2006). A morphological approach to documenting the domestication of Chenopodium in the Andes. Documenting Domest. New Genet. Archaeol. paradigms, 32–45.
45
Bull S. E. Seung D. Chanez C. Mehta D. Kuon J.-E. Truernit E. et al (2018). Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv.4, eaat6086. 10.1126/sciadv.aat6086
46
Busch W. Schneeberger K. (2019). Precision breeding for a better world. Nat. Biotechnol.37 (3), 243–254. 10.1038/s41587-019-0047-0
47
Butler N. M. Baltes N. J. Voytas D. F. Douches D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum L) using sequence-specific nucleases. Front. Plant Sci.7, 1045. 10.3389/fpls.2016.01045
48
Butt H. Eid A. Ali Z. Atia M. A. Mokhtar M. M. Hassan N. et al (2017). Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front. Plant Sci.8, 1441. 10.3389/fpls.2017.01441
49
Caddell D. Langenfeld N. J. Eckels M. J. Zhen S. Klaras R. Mishra L. et al (2023). Photosynthesis in rice is increased by CRISPR/Cas9-mediated transformation of two truncated light-harvesting antenna. Front. Plant Sci.14, 1050483. 10.3389/fpls.2023.1050483
50
Cai Y. Chen L. Liu X. Guo C. Sun S. Wu C. et al (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soyabean. Plant Biotechnol. J.16 (1), 176–185. 10.1111/pbi.12758
51
Cannarozzi G. Plaza-Wüthrich S. Esfeld K. Larti S. Wilson Y. S. Girma D. et al (2014). Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genom15 (1), 581–621. 10.1186/1471-2164-15-581
52
Cantos C. Francisco P. Trijatmiko K. R. Slamet-Loedin I. Chadha-Mohanty P. K. (2014). Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci.5, 302. 10.3389/fpls.2014.00302
53
Cao P. Zhao Y. Wu F. Xin D. Liu C. Wu X. et al (2022). Multi-omics techniques for soybean molecular breeding. Int. J. Mol. Sci.23 (9), 4994. 10.3390/ijms23094994
54
Carusillo A. Mussolino C. (2020). DNA damage: from threat to treatment. Cells9 (7), 1665. 10.3390/cells9071665
55
Cassman K. G. (1999). Ecological intensification of cereal production systems: yield potential, soil quality, and precision agriculture. Proc. Natl. Acad. Sci.96 (11), 5952–5959. 10.1073/pnas.96.11.5952
56
Cermak T. Baltes N. J. Cegan R. Zhang Y. Voytas D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol.16, 232–315. 10.1186/s13059-015-0796-9
57
Chaikam V. Prasanna B. M. (2020). “Doubled haploid technology for rapid and efficient maize breeding,” in Accelerated plant breeding (Cham: Springer International Publishing), Vol. 1, 257–292. Cereal Crops. 10.1007/978-3-030-41866-3_11
58
Chang H. H. Lieber M. R. (2016). Structure-specific nuclease activities of artemis and the artemis: DNA-PKcs complex. Nucleic Acids Res.44 (11), 4991–4997. 10.1093/nar/gkw456
59
Char S. N. Unger-Wallace E. Frame B. Briggs S. A. Main M. Spalding M. H. et al (2015). Heritable site-specific mutagenesis using TALEN s in maize. Plant Biotechnol. J.13 (7), 1002–1010. 10.1111/pbi.12344
60
Char S. N. Wei J. Mu Q. Li X. Zhang Z. J. Yu J. et al (2020). An agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol. J.18, 319–321. 10.1111/pbi.13229
61
Chaudhary H. K. Sharma P. Manoj N. V. Singh K. (2019b). New frontiers in chromosome elimination-mediated doubled haploidy breeding: focus on speed breeding in bread and durum wheat. Indian J. Genet. Plant Breed.79 (1), 254–263. 10.31742/IJGPB.79S.1.16
62
Chaudhary J. Deshmukh R. Sonah H. (2019a). Mutagenesis approaches and their role in crop improvement. Plants8 (11), 467. 10.3390/plants8110467
63
Chauhan H. Boni R. Bucher R. Kuhn B. Buchmann G. Sucher J. et al (2015). The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J.84 (1), 202–215. 10.1111/tpj.13001
64
Chen F. Pruett-Miller S. M. Huang Y. (2021b). Gaps and opportunities of CRISPR/Cas9 in vivo gene editing. J. Mol. Cell Biol.13 (1), 3–15.
65
Chen H. Ye R. Liang Y. Zhang S. Liu X. Sun C. et al (2023). Generation of low-cadmium rice germplasms via knockout of OsLCD using CRISPR/Cas9. J. Environ. Sci.126, 138–152. 10.1016/j.jes.2022.05.047
66
Chen L. M. Li X. W. He T. J. Li P. J. Liu Y. Zhou S. X. et al (2021c). Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics113 (4), 2108–2121. 10.1016/j.ygeno.2021.05.002
67
Chen Q. Li W. Tan L. Tian F. (2021a). Harnessing knowledge from maize and rice domestication for new crop breeding. Mol. Plant.14, 9–26. 10.1016/j.molp.2020.12.006
68
Chen Q. Yang C. J. York A. M. Xue W. Daskalska L. L. DeValk C. A. et al (2019). TeoNAM: a nested association mapping population for domestication and agronomic trait analysis in maize. Genetics213 (3), 1065–1078. 10.1534/genetics.119.302594
69
Chen Y. Liu G. Ma H. Song Z. Zhang C. Zhang J. et al (2018). Identification of introgressed alleles conferring high fiber quality derived from Gossypium barbadense L. in secondary mapping populations of. G. hirsutum L. Front. Plant Sci.9, 1023. 10.3389/fpls.2018.01023
70
Cheng Z. Sun Y. Yang S. Zhi H. Yin T. Ma X. et al (2021). Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J.19 (6), 1089–1091. 10.1111/pbi.13584
71
Childe V. G. (1949). The origin of Neolithic culture in northern Europe. Antiquity23 (91), 129–135. 10.1017/S0003598X00020184
72
Chinchole M. Pathak R. K. Singh U. M. Kumar A. (2017). Molecular characterization of EcCIPK 24 gene of finger millet (Eleusine coracana) for investigating its regulatory role in calcium transport. 3 Biotech.7, 267–310. 10.1007/s13205-017-0874-7
73
Chiurugwi T. Kemp S. Powell W. Hickey L. T. (2019). Speed breeding orphan crops. Theor. Appl. Genet.132, 607–616. 10.1007/s00122-018-3202-7
74
Choi H. K. (2019). Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding. Genes Genom41, 133–146. 10.1007/s13258-018-0751-8
75
Chongtham S. K. Devi E. L. Samantara K. Yasin J. K. Wani S. H. Mukherjee S. et al (2022). Orphan legumes: harnessing their potential for food, nutritional and health security through genetic approaches. Planta256 (2), 24. 10.1007/s00425-022-03923-1
76
Choulika A. Perrin A. Dujon B. Nicolas J. F. (1995). Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol.15 (4), 1968–1973. 10.1128/MCB.15.4.1968
77
Clasen B. M. Stoddard T. J. Luo S. Demorest Z. L. Li J. Cedrone F. et al (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J.14 (1), 169–176. 10.1111/pbi.12370
78
Clement E. J. Foster M. C. (1994). Alien plants of the British isles. London: Botanical Society of the British Isles.
79
Cockram J. White J. Zuluaga D. L. Smith D. Comadran J. Macaulay M. et al (2010). Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc. Natl. Acad. Sci.107 (50), 21611–21616. 10.1073/pnas.1010179107
80
Coffman S. M. Hufford M. B. Andorf C. M. Lubberstedt T. (2020). Haplotype structure in commercial maize breeding programs in relation to key founder lines. Theor. Appl. Genet.133, 547–561. 10.1007/s00122-019-03486-y
81
Collard B. C. Beredo J. C. Lenaerts B. Mendoza R. Santelices R. Lopena V. et al (2017). Revisiting rice breeding methods–evaluating the use of rapid generation advance (RGA) for routine rice breeding. Plant Prod. Sci.20 (4), 337–352. 10.1080/1343943X.2017.1391705
82
Cong B. Liu J. Tanksley S. D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci.99 (21), 13606–13611. 10.1073/pnas.172520999
83
Connorton J. M. Jones E. R. Rodriguez-Ramiro I. Fairweather-Tait S. Uauy C. Balk J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol.174 (4), 2434–2444. 10.1104/pp.17.00672
84
Crossa J. Perez-Rodríguez P. Cuevas J. Montesinos-Lopez O. Jarquín D. De Los Campos G. et al (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci.22 (11), 961–975. 10.1016/j.tplants.2017.08.011
85
Cubas P. Lauter N. Doebley J. Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J.18, 215–222. 10.1046/j.1365-313X.1999.00444.x
86
Dai L. Li P. Li Q. Leng Y. Zeng D. Qian Q. (2022). Integrated multi-omics perspective to strengthen the understanding of salt tolerance in rice. Int. J. Mol. Sci.23 (9), 5236. 10.3390/ijms23095236
87
Dakouri A. McCallum B. D. Walichnowski A. Z. Cloutier S. (2010). Fine-mapping of the leaf rust Lr34 locus in Triticum aestivum (L) and characterization of large germplasm collections support the ABC transporter as essential for gene function. Theor. Appl. Genet.121, 373–384. 10.1007/s00122-010-1316-7
88
Dale P. J. Clarke B. Fontes E. M. (2002). Potential for the environmental impact of transgenic crops. Nat. Biotechnol.20 (6), 567–574. 10.1038/nbt0602-567
89
Dawson I. K. Powell W. Hendre P. Bancic J. Hickey J. M. Kindt R. et al (2019). The role of genetics in mainstreaming the production of new and orphan crops to diversify food systems and support human nutrition. New Phytol.224 (1), 37–54. 10.1111/nph.15895
90
Demorest Z. L. Coffman A. Baltes N. J. Stoddard T. J. Clasen B. M. Luo S. et al (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol.16, 225–228. 10.1186/s12870-016-0906-1
91
Dhingani R. M. Umrania V. V. Tomar R. S. Parakhia M. V. Golakiya B. (2015). Introduction to QTL mapping in plants. Ann. Plant Sci.4 (4), 1072–1079.
92
Diamond J. Bellwood P. (2003). Farmers and their languages: the first expansions. Science300 (5619), 597–603. 10.1126/science.1078208
93
Diamond J. (2002). Evolution, consequences and future of plant and animal domestication. Nature418 (6898), 700–707. 10.1038/nature01019
94
Diaz S. Ariza-Suarez D. Izquierdo P. Lobaton J. D. de la Hoz J. F. Acevedo F. et al (2020). Genetic mapping for agronomic traits in a MAGIC population of common bean (Phaseolus vulgaris L) under drought conditions. BMC Genom21 (1), 799–820. 10.1186/s12864-020-07213-6
95
Doebley J. Stec A. Gustus C. (1995). Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics141, 333–346. 10.1093/genetics/141.1.333
96
Doebley J. Stec A. Hubbard L. (1997). The evolution of apical dominance in maize. Nature386 (6624), 485–488. 10.1038/386485a0
97
Dong Z. Li W. Unger-Wallace E. Yang J. Vollbrecht E. Chuck G. (2017). Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci.114, E8656–E8664. 10.1073/pnas.1714960114
98
Doudna J. A. Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science346 (6213), 1258096. 10.1126/science.1258096
99
Du H. Zeng X. Zhao M. Cui X. Wang Q. Yang H. et al (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol.217, 90–97. 10.1016/j.jbiotec.2015.11.005
100
Eid A. Alshareef S. Mahfouz M. M. (2018). CRISPR base editors: genome editing without double-stranded breaks. Biochem. J.475 (11), 1955–1964. 10.1042/BCJ20170793
101
Ellis M. Spielmeyer W. Gale K. Rebetzke J. Richards A. (2002). Perfect" markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor. Appl. Genet.105, 1038–1042. 10.1007/s00122-002-1048-4
102
Endo M. Mikami M. Toki S. (2016). Biallelic gene targeting in rice. Plant physiol.170 (2), 667–677. 10.1104/pp.15.01663
103
Errum A. Rehman N. Uzair M. Inam S. Ali G. M. Khan M. R. (2023). CRISPR/Cas9 editing of wheat Ppd-1 gene homoeologs alters spike architecture and grain morphometric traits. Funct. Integr. Genom.23 (1), 66. 10.1007/s10142-023-00989-2
104
Esfeld K. Uauy C. Tadele Z. (2013). “Application of TILLING for orphan crop improvement,” in Biotechnology of neglected and underutilized crops. Editors JainS.Dutta GuptaS. (Dordrecht: Springer). 10.1007/978-94-007-5500-0_6
105
Fang Y. Tyler B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol.17 (1), 127–139. 10.1111/mpp.12318
106
Feng C. Yuan J. Wang R. Liu Y. Birchler J. A. Han F. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics43 (1), 37–43. 10.1016/j.jgg.2015.10.002
107
Figueredo E. F. da Cruz T. A. de Almeida J. R. Batista B. D. Marcon J. de Andrade P. A. M. et al (2023). The key role of indole-3-acetic acid biosynthesis by Bacillus thuringiensis RZ2MS9 in promoting maize growth revealed by the ipdC gene knockout mediated by the CRISPR-Cas9 system. Microbiol. Res.266, 127218. 10.1016/j.micres.2022.127218
108
Freisleben R. Lein A. (1944). Moglichkeiten und praktische Durchfuhrung der Mutationszuchtung. Kuhn-Arhiv60, 211–222.
109
Fuller D. Q. Denham T. Arroyo-Kalin M. Lucas L. Stevens C. J. Qin L. et al (2014). Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc. Natl. Acad. Sci.111 (17), 6147–6152. 10.1073/pnas.1308937110
110
Gachomo E. W. Shonukan O. O. Kotchoni S. O. (2003). The molecular initiation and subsequent acquisition of disease resistance in plants. Afr. J. Biotechnol.2 (2), 26–32. 10.5897/AJB2003.000-1005
111
Gage J. L. Monier B. Giri A. Buckler E. S. (2020). Ten years of the maize nested association mapping population: impact, limitations, and future directions. Plant Cell32 (7), 2083–2093. 10.1105/tpc.19.00951
112
Gallagher R. R. Li Z. Lewis A. O. Isaacs F. J. (2014). Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat. Protoc.9 (10), 2301–2316. 10.1038/nprot.2014.082
113
Gao C. (2021). Genome engineering for crop improvement and future agriculture. Cell184 (6), 1621–1635. 10.1016/j.cell.2021.01.005
114
Garrett K. A. Andersen K. F. Asche F. Bowden R. L. Forbes G. A. Kulakow P. A. et al (2017). Resistance genes in global crop breeding networks. Phytopathology107 (10), 1268–1278. 10.1094/PHYTO-03-17-0082-FI
115
Gaudelli N. M. Komor A. C. Rees H. A. Packer M. S. Badran A. H. Bryson D. I. et al (2017). Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature551 (7681), 464–471. 10.1038/nature24644
116
Genetically Engineered Crops: experiences and Prospects- New Report (2016). News release. Available at: https://www.nationalacademies.org/news/2016/05/genetically-engineered-crops-experiences-and prospects-new-report.
117
Ghosh S. Watson A. Gonzalez-Navarro O. E. Ramirez-Gonzalez R. H. Yanes L. Mendoza-Suárez M. et al (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc.13 (12), 2944–2963. 10.1038/s41596-018-0072-z
118
Gilchrist E. J. Haughn G. W. Ying C. C. Otto S. P. Zhuang J. U. N. Cheung D. et al (2006). Use of Ecotilling as an efficient SNP discovery tool to survey genetic variation in wild populations of Populus trichocarpa. Mol. Ecol.15 (5), 1367–1378. 10.1111/j.1365-294X.2006.02885.x
119
Goering R. Larsen S. Tan J. Whelan J. Makarevitch I. (2021). QTL mapping of seedling tolerance to exposure to low temperature in the maize IBM RIL population. Plos one16 (7), e0254437. 10.1371/journal.pone.0254437
120
Golisz A. Sugano M. Fujii Y. (2008). Microarray expression profiling of Arabidopsis thaliana L. in response to allelochemicals identified in buckwheat. J. Exp. Bot.59 (11), 3099–3109. 10.1093/jxb/ern168
121
Gregory T. R. (2009). Understanding natural selection: essential concepts and common misconceptions. Evol. Educ. Outreach2 (2), 156–175. 10.1007/s12052-009-0128-1
122
Guo T. Yang J. Li D. Sun K. Luo L. Xiao W. et al (2019). Integrating GWAS, QTL, mapping and RNA-seq to identify candidate genes for seed vigor in rice (Oryza sativa L). Mol. Breed.39, 87–16. 10.1007/s11032-019-0993-4
123
Guo Y. Abernathy B. Zeng Y. Ozias-Akins P. (2015). TILLING by sequencing to identify induced mutations in stress resistance genes of peanut (Arachis hypogaea). BMC Genom16 (1), 157–213. 10.1186/s12864-015-1348-0
124
Gupta A. Hua L. Zhang Z. Yang B. Li W. (2023). CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol. J.21 (3), 536–548. 10.1111/pbi.13969
125
Gupta A. K. (2004). Origin of agriculture and domestication of plants and animals linked to early Holocene climate amelioration. Curr. Sci., 54–59. Available at: https://www.jstor.org/stable/24107979.
126
Gupta P. K. Kulwal P. L. Jaiswal V. (2019a). Association mapping in plants in the post-GWAS genomics era. Adv. Genet.104, 75–154. 10.1016/bs.adgen.2018.12.001
127
Gupta S. Kumari K. Muthamilarasan M. Parida S. K. Prasad M. (2014). Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep.33, 881–893. 10.1007/s00299-014-1564-0
128
Gupta V. He X. Kumar N. Fuentes-Davila G. Sharma R. K. Dreisigacker S. et al (2019b). Genome wide association study of Karnal bunt resistance in a wheat germplasm collection from Afghanistan. Int. J. Mol. Sci.20 (13), 3124. 10.3390/ijms20133124
129
Haile J. K. N’Diaye A. Sari E. Walkowiak S. Rutkoski J. E. Kutcher H. R. et al (2020). Potential of genomic selection and integrating “omics” data for disease evaluation in wheat. Crop Breed. Genet. Genomics4 (3). 10.20900/cbgg20200016
130
Hammer K. (1984). Das domestikationssyndrom. Die Kult.32, 11–34. 10.1007/bf02098682
131
Hanson A. A. Lorenz A. J. Hesler L. S. Bhusal S. J. Bansal R. Michel A. P. et al (2018). Genome-wide association mapping of host-plant resistance to soybean aphid. Plant Genome11 (3), 180011. 10.3835/plantgenome2018.02.0011
132
Hao L. I. Ruiying Q. I. N. Xiaoshuang L. I. U. Shengxiang L. I. A. O. Rongfang X. U. Jianbo Y. A. N. G. et al (2019). CRISPR/Cas9-mediated adenine base editing in rice genome. Rice Sci.26 (2), 125–128. 10.1016/j.rsci.2018.07.002
133
Hargrove T. R. Cabanilla V. L. (1979). The impact of semidwarf varieties on Asian rice-breeding programs. Bioscience29 (12), 731–735. 10.2307/1307667
134
Harrison R. G. (1990). Hybrid zones: windows on evolutionary process. Oxf. Surv. Evol. Biol.7, 69–128.
135
Haun W. Coffman A. Clasen B. M. Demorest Z. L. Lowy A. Ray E. et al (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J.12 (7), 934–940. 10.1111/pbi.12201
136
Henikoff S. Comai L. (2003). Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol.54 (1), 375–401. 10.1146/annurev.arplant.54.031902.135009
137
Henikoff S. Till B. J. Comai L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol.135, 630–636. 10.1104/pp.104.041061
138
Hillary V. E. Ceasar S. A. (2022). Prime editing in plants and mammalian cells: mechanism, achievements, limitations, and future prospects. BioEssays44 (9), 2200032. 10.1002/bies.202200032
139
Holme I. B. Gregersen P. L. Brinch-Pedersen H. (2019). Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Front. Plant Sci.10, 1468. 10.3389/fpls.2019.01468
140
Hu B. Wang W. Ou S. Tang J. Li H. Che R. et al (2015). Variation in NRT1 1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet.47 (7), 834–838. 10.1038/ng.3337
141
Hua K. Tao X. Liang W. Zhang Z. Gou R. Zhu J. K. (2020). Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J.18 (3), 770–778. 10.1111/pbi.13244
142
Hua K. Tao X. Yuan F. Wang D. Zhu J. K. (2018). Precise A· T to G· C base editing in the rice genome. Mol. Plant11 (4), 627–630. 10.1016/j.molp.2018.02.007
143
Huang B. E. Verbyla K. L. Verbyla A. P. Raghavan C. Singh V. K. Gaur P. et al (2015). MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet.128, 999–1017. 10.1007/s00122-015-2506-0
144
Huang J. Pray C. Rozelle S. (2002). Enhancing the crops to feed the poor. Nature418 (6898), 678–684. 10.1038/nature01015
145
Huang Q. Lin B. Cao Y. Zhang Y. Song H. Huang C. et al (2023). CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 in rice confers enhanced resistance to rice root-knot nematode. Front. Plant Sci.14, 1134653. 10.3389/fpls.2023.1134653
146
Huang Y. Zhao S. Fu Y. Sun H. Ma X. Tan L. et al (2018). Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J.96 (4), 716–733. 10.1111/tpj.14062
147
Hummel A. W. Chauhan R. D. Cermak T. Mutka A. M. Vijayaraghavan A. Boyher A. et al (2018). Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J.16 (7), 1275–1282. 10.1111/pbi.12868
148
Huynh B. L. Ehlers J. D. Huang B. E. Munoz-Amatriaín M. Lonardi S. Santos J. R. et al (2018). A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L Walp). Plant J.93 (6), 1129–1142. 10.1111/tpj.13827
149
Hwang J. E. Jang D. S. Lee K. J. Ahn J. W. Kim S. H. Kang S. Y. et al (2016). Identification of gamma ray irradiation-induced mutations in membrane transport genes in a rice population by TILLING. Genes Genet. Syst.91 (5), 245–256. 10.1266/ggs.15-00052
150
Hyde L. Osman K. Winfield M. Sanchez-Moran E. Higgins J. D. Henderson I. R. et al (2023). Identification, characterization, and rescue of CRISPR/Cas9 generated wheat SPO11-1 mutants. Plant Biotechnol. J.21 (2), 405–418. 10.1111/pbi.13961
151
Ibarra A. C. M. Galvez H. F. Gardoce R. R. De Chavez J. A. M. (2017). Eco-TILLING for natural single nucleotide polymorphism towards genomics-assisted mutation breeding in pineapple (Ananas comosus (L) Merr). Philipp. J. Crop Sci.42, 99–100. Available at: https://www.ukdr.uplb.edu.ph/journal-articles/5579.
152
Ibrahim Bio Yerima A. R. Achigan-Dako E. G. (2021). A review of the orphan small grain cereals improvement with a comprehensive plan for genomics-assisted breeding of fonio millet in West Africa. Plant Breed.140 (4), 561–574. 10.1111/pbr.12930
153
Impens L. Lorenzo C. D. Vandeputte W. Wytynck P. Debray K. Haeghebaert J. et al (2023). Combining multiplex gene editing and doubled haploid technology in maize. New Phytol.239 (4), 1521–1532. 10.1111/nph.19021
154
Imran M. Shafiq S. Tang X. (2023). CRISPR-Cas9-mediated editing of BADH2 gene triggered fragrance revolution in rice. Physiol. Plant175, e13871. 10.1111/ppl.13871
155
Ito Y. Nakamura N. Kotake-Nara E. (2021). Semi-dominant effects of a novel ripening inhibitor (rin) locus allele on tomato fruit ripening. Plos one16 (4), e0249575. 10.1371/journal.pone.0249575
156
Ito Y. Nishizawa-Yokoi A. Endo M. Mikami M. Toki S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun.467 (1), 76–82. 10.1016/j.bbrc.2015.09.117
157
Jankowicz-Cieslak J. Till B. J. (2015). “Forward and reverse genetics in crop breeding,” in Advances in plant breeding strategies: Breeding, biotechnology and molecular tools. (Switzerland: Springer), 215–224. 10.1007/978-3-319-22521-0_8
158
Jansen R. Embden J. D. V. Gaastra W. Schouls L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol.43 (6), 1565–1575. 10.1046/j.1365-2958.2002.02839.x
159
Jin J. Huang W. Gao J. P. Yang J. Shi M. Zhu M. Z. et al (2008). Genetic control of rice plant architecture under domestication. Nat. Genet.40, 1365–1369. 10.1038/ng.247
160
Jinek M. Chylinski K. Fonfara I. Hauer M. Doudna J. A. Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science337 (6096), 816–821. 10.1126/science.1225829
161
Johns T. Alonso J. G. (1990). Glycoalkaloid change during the domestication of the potato, solanum section petota. Euphytica50, 203–210. 10.1007/BF00023646
162
Joiret M. Mahachie John J. M. Gusareva E. S. Van Steen K. (2019). Confounding of linkage disequilibrium patterns in large scale DNA based gene-gene interaction studies. BioData Min.12, 11–23. 10.1186/s13040-019-0199-7
163
Jones H. D. (2015). Regulatory uncertainty over genome editing. Nat. Plants1 (1), 14011–14013. 10.1038/nplants.2014.11
164
Jung J. H. Altpeter F. (2016). TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol.92, 131–142. 10.1007/s11103-016-0499-y
165
Kamenya S. N. Mikwa E. O. Song B. Odeny D. A. (2021). Genetics and breeding for climate change in Orphan crops. Theor. Appl. Genet.134, 1787–1815. 10.1007/s00122-020-037551
166
Kang B. C. Yun J. Y. Kim S. T. Shin Y. Ryu J. Choi M. et al (2018). Precision genome engineering through adenine base editing in plants. Nat. Plants4 (7), 427–431. 10.1038/s41477-018-0178-x
167
Kannan B. Jung J. H. Moxley G. W. Lee S. M. Altpeter F. (2018). TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J.16 (4), 856–866. 10.1111/pbi.12833
168
Kantor A. McClements M. E. MacLaren R. E. (2020). CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci.21 (17), 6240. 10.3390/ijms21176240
169
Karthika V. Babitha K. C. Kiranmai K. Shankar A. G. Vemanna R. S. Udayakumar M. (2020). Involvement of DNA mismatch repair systems to create genetic diversity in plants for speed breeding programs. Plant Physiol. Rep.25 (2), 185–199. 10.1007/s40502-020-00521-9
170
Kelliher T. Starr D. Richbourg L. Chintamanani S. Delzer B. Nuccio M. L. et al (2017). MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature542 (7639), 105–109. 10.1038/nature20827
171
Khan S. U. Yangmiao J. Liu S. Zhang K. Khan M. H. U. Zhai Y. et al (2019). Genome-wide association studies in the genetic dissection of ovule number, seed number, and seed weight in Brassica napus L. Ind. Crop. Prod.142, 111877. 10.1016/j.indcrop.2019.111877
172
Kharkwal M. C. Pandey R. N. Pawar S. E. (2004). Mutation breeding for crop improvement. Plant Breed. Mendelian Mol. approaches, 601–645. 10.1007/978-94-007-1040-5_26
173
Khush G. S. (1999). Green revolution: preparing for the 21st century. Genome42 (4), 646–655. 10.1139/g99-044
174
Kim D. Alptekin B. Budak H. (2018). CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom.18, 31–41. 10.1007/s10142-017-0572-x
175
Kim Y. A. Moon H. Park C. J. (2019). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Xanthomonas oryzae pv. Oryzae. Rice12 (1), 67–13. 10.1186/s12284-019-0325-7
176
Kim Y. G. Cha J. Chandrasegaran S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci.93 (3), 1156–1160. 10.1073/pnas.93.3.1156
177
Klap C. Yeshayahou E. Bolger A. M. Arazi T. Gupta S. K. Shabtai S. et al (2017). Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J.15 (5), 634–647. 10.1111/pbi.12662
178
Knezevic S. Z. (2017). Invasive plant species, Editor(s): Brian thomas, brian G murray, denis J murphy, encyclopedia of applied plant sciences (Second Edition), Academic Press, 300–303.
179
Kolesnik T. Szeverenyi I. Bachmann D. Kumar C. S. Jiang S. Ramamoorthy R. et al (2004). Establishing an efficient Ac/ds tagging system in rice: large-scale analysis of ds flanking sequences. Plant J.37 (2), 301–314. 10.1046/j.1365-313X.2003.01948.x
180
Komor A. C. Kim Y. B. Packer M. S. Zuris J. A. Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature533 (7603), 420–424. 10.1038/nature17946
181
Konishi S. Izawa T. Lin S. Y. Ebana K. Fukuta Y. Sasaki T. et al (2006). An SNP caused loss of seed shattering during rice domestication. Science312 (5778), 1392–1396. 10.1126/science.1126410
182
Korte A. Farlow A. (2013). The advantages and limitations of trait analysis with GWAS: a review. Plant methods9 (1), 29–9. 10.1186/1746-4811-9-29
183
Kozgar M. I. Khan S. Wani M. R. (2012). Variability and correlations studies for total iron and manganese contents of chickpea (Cicer arietinum L) high yielding mutants. Am. J. Food Technol.7 (7), 437–444. 10.3923/ajft.2012.437.444
184
Krattinger S. G. Lagudah E. S. Spielmeyer W. Singh R. P. Huerta-Espino J. McFadden H. et al (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Sci. (New York, N.Y.)323 (5919), 1360–1363. 10.1126/science.1166453
185
Kumar A. Anju T. Kumar S. Chhapekar S. S. Sreedharan S. Singh S. et al (2021). Integrating omics and gene editing tools for rapid improvement of traditional food plants for diversified and sustainable food security. Int. J. Mol. Sci.22 (15), 8093. 10.3390/ijms22158093
186
Kumar R. Janila P. Vishwakarma M. K. Khan A. W. Manohar S. S. Gangurde S. S. et al (2020). Whole-genome resequencing-based QTL-seq identified candidate genes and molecular markers for fresh seed dormancy in groundnut. Plant Biotechnol. J.18 (4), 992–1003. 10.1111/pbi.13266
187
Kyndt T. Quispe D. Zhai H. Jarret R. Ghislain M. Liu Q. et al (2015). The genome of cultivated sweet potato contains agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc. Natl. Acad. Sci.112 (18), 5844–5849. 10.1073/pnas.1419685112
188
Lasley B. L. Loskutoff N. M. Anderson G. B. (1994). The limitation of conventional breeding programs and the need and promise of assisted reproduction in nondomestic species. Theriogenology41 (1), 119–132. 10.1016/S0093-691X(05)80057-3
189
Lee S. Ding N. Sun Y. Yuan T. Li J. Yuan Q. et al (2020). Single C-to-T substitution using engineered APOBEC3G-nCas9 base editors with minimum genome-and transcriptome-wide off-target effects. Sci. Adv.6 (29), eaba1773. 10.1126/sciadv.aba1773
190
Lemmon Z. H. Reem N. T. Dalrymple J. Soyk S. Swartwood K. E. Rodriguez-Leal D. et al (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants4 (10), 766–770. 10.1038/s41477-018-0259-x
191
Lennox J. G. Wilson B. E. (1994). Natural selection and the struggle for existence. Stud. Hist. Philos. Sci. Part A25 (1), 65–80. 10.1016/0039-3681(94)90020-5
192
Li A. Jia S. Yobi A. Ge Z. Sato S. J. Zhang C. et al (2018d). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol.177, 1425–1438. 10.1104/pp.18.00200
193
Li C. Zhou A. Sang T. (2006). Rice domestication by reducing shattering. Science311 (5769), 1936–1939. 10.1126/science.1123604
194
Li J. Meng X. Zong Y. Chen K. Zhang H. Liu J. et al (2016a). Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. plants2 (10), 16139–16146. 10.1038/nplants.2016.139
195
Li J. Sun Y. Du J. Zhao Y. Xia L. (2017a). Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant10 (3), 526–529. 10.1016/j.molp.2016.12.001
196
Li J. Zhang X. Sun Y. Zhang J. Du W. Guo X. et al (2018a). Efficient allelic replacement in rice by gene editing: a case study of the NRT1 1B gene. J. Integr. Plant Biol.60 (7), 536–540. 10.1111/jipb.12650
197
Li J. Zhang H. Si X. Tian Y. Chen K. Liu J. et al (2017b). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics44 (9), 465–468. 10.1016/j.jgg.2017.02.002
198
Li M. Li X. Zhou Z. Wu P. Fang M. Pan X. et al (2016b). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci.7, 377. 10.3389/fpls.2016.00377
199
Li T. Liu B. Spalding M. H. Weeks D. P. Yang B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol.30 (5), 390–392. 10.1038/nbt.2199
200
Li T. Yang X. Yu Y. Si X. Zhai X. Zhang H. et al (2018c). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol.36 (12), 1160–1163. 10.1038/nbt.4273
201
Li X. Wang Y. Liu Y. Yang B. Wang X. Wei J. et al (2018b). Base editing with a Cpf1–cytidine deaminase fusion. Nat. Biotechnol.36 (4), 324–327. 10.1038/nbt.4102
202
Li Y. Shu P. Xiang L. Sheng J. Shen L. (2023). CRISPR/Cas9-Mediated SlATG5 mutagenesis reduces the resistance of tomato fruit to Botrytis cinerea. Foods12 (14), 2750. 10.3390/foods12142750
203
Li Z. Li X. Cui H. Zhao G. Zhai D. Chen J. (2021). Vegetative and fecundity fitness benefit found in a glyphosate-resistant Eleusine indica population caused by 5-enolpyruvylshikimate-3-phosphate synthase overexpression. Front. Plant Sci.12, 776990. 10.3389/fpls.2021.776990
204
Li Z. Liu Z. B. Xing A. Moon B. P. Koellhoffer J. P. Huang L. et al (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol.169 (2), 960–970. 10.1104/pp.15.00783
205
Liang F. Han M. Romanienko P. J. Jasin M. (1998). Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci.95 (9), 5172–5177. 10.1073/pnas.95.9.5172
206
Liang Z. Zhang K. Chen K. Gao C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics41 (2), 63–68. 10.1016/j.jgg.2013.12.001
207
Lin Z. Griffith M. E. Li X. Zhu Z. Tan L. Fu Y. et al (2007). Origin of seed shattering in rice (Oryza sativa L). Planta226, 11–20. 10.1007/s00425-006-0460-4
208
Liu D. Chen X. Liu J. Ye J. Guo Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot.63 (10), 3899–3911. 10.1093/jxb/ers079
209
Liu H. Zwer P. Wang H. Liu C. Lu Z. Wang Y. et al (2016). A fast generation cycling system for oat and triticale breeding. Plant Breed.135 (5), 574–579. 10.1111/pbr.12408
210
Liu X. Yu Y. Yao W. Yin Z. Wang Y. Huang Z. et al (2023). CRISPR/Cas9-mediated simultaneous mutation of three salicylic acid 5-hydroxylase (OsS5H) genes confers broad-spectrum disease resistance in rice. Plant Biotechnol. J.10.1111/pbi.14099
211
Lorenzo C. D. Debray K. Herwegh D. Develtere W. Impens L. Schaumont D. et al (2023). Breedit: A multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell35 (1), 218–238. 10.1093/plcell/koac243
212
Lu C. Kuang J. Shao T. Xie S. Li M. Zhu L. et al (2022). Prime editing: an all-rounder for genome editing. Int. J. Mol. Sci.23 (17), 9862. 10.3390/ijms23179862
213
Lu Y. Zhu J. K. (2017). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant10 (3), 523–525. 10.1016/j.molp.2016.11.013
214
Luo B. Ma P. Nie Z. Zhang X. He X. Ding X. et al (2019a). Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J.97 (5), 947–969. 10.1111/tpj.14160
215
Luo H. Pandey M. K. Khan A. W. Guo J. Wu B. Cai Y. et al (2019b). Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L). Plant Biotechnol. J.17 (7), 1248–1260. 10.1111/pbi.13050
216
Ly D. N. P. Iqbal S. Fosu-Nyarko J. Milroy S. Jones M. G. (2023). Multiplex CRISPR-cas9 gene-editing can deliver potato cultivars with reduced browning and acrylamide. Plants12 (2), 379. 10.3390/plants12020379
217
Maioli A. Gianoglio S. Moglia A. Acquadro A. Valentino D. Milani A. M. et al (2020). Simultaneous CRISPR/Cas9 editing of three PPO genes reduces fruit flesh browning in Solanum melongena L. Front. Plant Sci.11, 607161. 10.3389/fpls.2020.607161
218
Mali P. Yang L. Esvelt K. M. Aach J. Guell M. DiCarlo J. E. et al (2013). RNA-guided human genome engineering via Cas9. Science339 (6121), 823–826. 10.1126/science.1232033
219
Malik S. Malik T. A. (2015). Genetic mapping of potential QTLs associated with drought tolerance in wheat. J. Anim. Plant Sci.25 (4), 1032–1040.
220
Marzec M. Brąszewska-Zalewska A. Hensel G. (2020). Prime editing: A new way for genome editing. Trends Cell Biol.30 (4), 257–259. 10.1016/j.tcb.2020.01.004
221
Marzec M. Hensel G. (2020). Prime editing: game changer for modifying plant genomes. Trends Plant Sci.25 (8), 722–724. 10.1016/j.tplants.2020.05.008
222
Mather K. (1938). The measurement of linkage in heredity. The measurement of linkage in heredity.
223
Maurya J. Singh R. K. Sett S. Prasad M. (2022). “Genomic tools and proteomic determinants for abiotic stress tolerance in pearl millet (Pennisetum glaucum) and foxtail millet (Setaria italica L),” in Omics approach to manage abiotic stress in cereals (Singapore: Springer Nature Singapore), 531–557.
224
Mba C. Afza R. Bado S. Jain S. M. (2010). Induced mutagenesis in plants using physical and chemical agents. Plant Cell Cult. Essent. Methods20, 111–130.
225
Mba C. (2013). Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy3 (1), 200–231. 10.3390/agronomy3010200
226
McCallum C. M. Comai L. Greene E. A. Henikoff S. (2000a). Targeted screening for induced mutations. Nat. Biotechnol.18 (4), 455–457. 10.1038/74542
227
McCallum C. M. Comai L. Greene E. A. Henikoff S. (2000b). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant physiol.123 (2), 439–442. 10.1104/pp.123.2.439
228
Mehta A. K. Basha M. H. (2018). “Application of mutation breeding in plants,” in Advanced molecular plant breeding: Meeting the challenge of food security (CRC Press), 139–234.
229
Meyer R. S. DuVal A. E. Jensen H. R. (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol.196 (1), 29–48. 10.1111/j.1469-8137.2012.04253.x
230
Miao J. Guo D. Zhang J. Huang Q. Qin G. Zhang X. et al (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res.23 (10), 1233–1236. 10.1038/cr.2013.123
231
Miki B. Abdeen A. Manabe Y. MacDonald P. (2009). Selectable marker genes and unintended changes to the plant transcriptome. Plant Biotechnol. J.7 (3), 211–218. 10.1111/j.1467-7652.2009.00400.x
232
Molla K. A. Sretenovic S. Bansal K. C. Qi Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants7 (9), 1166–1187. 10.1038/s41477-021-00991-1
233
Molla K. A. Yang Y. (2019). CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol.37 (10), 1121–1142. 10.1016/j.tibtech.2019.03.008
234
Mostafa G. G. (2011). Effect of sodium azide on the growth and variability induction in helianthus annuus L. Int. J. Plant Breed. Genet.5, 76–85. 10.3923/ijpbg.2011.76.85
235
Mukade K. (1974). New procedures for accelerating generation advancement in wheat breeding. JARQ8, 1–5.
236
Munoz-Lopez M. Garcia-Perez J. L. (2010). DNA transposons: nature and applications in genomics. Curr. Genomics11 (2), 115–128. 10.2174/138920210790886871
237
Murovec J. Gucek K. Bohanec B. Avbelj M. Jerala R. (2018). DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci.9, 1594. 10.3389/fpls.2018.01594
238
Nekrasov V. Wang C. Win J. Lanz C. Weigel D. Kamoun S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep.7 (1), 482–486. 10.1038/s41598-017-00578-x
239
Neumann K. Kobiljski B. Dencic S. Varshney R. K. Borner A. (2011). Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L). Mol. Breed.27, 37–58. 10.1007/s11032-010-9411-7
240
Nieto C. Piron F. Dalmais M. Marco C. F. Moriones E. Gomez-Guillamón M. L. et al (2007). EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol.7 (1), 34–39. 10.1186/1471-2229-7-34
241
Niu J. Guo N. Sun J. Li L. Cao Y. Li S. et al (2017). Fine mapping of a resistance gene RpsHN that controls Phytophthora sojae using recombinant inbred lines and secondary populations. Front. Plant Sci.8, 538. 10.3389/fpls.2017.00538
242
Niu Q. Wu S. Xie H. Wu Q. Liu P. Xu Y. et al (2023). Efficient A· T to G· C base conversions in dicots using adenine base editors expressed under the tomato EF1α promoter. Plant Biotechnol. J.21 (1), 5–7. 10.1111/pbi.13736
243
Ntui V. O. Uyoh E. A. Ita E. E. Markson A. A. A. Tripathi J. N. Okon N. I. et al (2021). Strategies to combat the problem of yam anthracnose disease: status and prospects. Mol. Plant Pathol.22 (10), 1302–1314. 10.1111/mpp.13107
244
Numan M. Khan A. L. Asaf S. Salehin M. Beyene G. Tadele Z. et al (2021). From traditional breeding to genome editing for boosting productivity of the ancient grain tef [Eragrostis tef (Zucc) Trotter]. Plants10, 628. 10.3390/plants10040628
245
O'Brien S. J. (1993). Genetic maps: Locus maps of complex genomes. Cold Spring Harbor Laboratory Press.
246
O'Connor D. J. Wright G. C. Dieters M. J. George D. L. Hunter M. N. Tatnell J. R. et al (2013). Development and application of speed breeding technologies in a commercial peanut breeding program. Peanut Sci.40 (2), 107–114. 10.3146/PS12-12.1
247
Ochoa-Sanchez A. Perez-Sanchez G. Torres-Ledesma A. M. Valdez J. P. R. Rinaldi G. Moguel B. B. et al (2021). Prime Editing, a novel genome-editing tool that may surpass conventional CRISPR-Cas9. Re Gen. Open1 (1), 75–82. 10.1089/regen.2021.0016
248
Oladosu Y. Rafii M. Y. Abdullah N. Hussin G. Ramli A. Rahim H. A. et al (2016). Principle and application of plant mutagenesis in crop improvement: a review. Biotechnol. Biotechnol. Equip.30 (1), 1–16. 10.1080/13102818.2015.1087333
249
Ortigosa A. Gimenez-Ibanez S. Leonhardt N. Solano R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotechnol. J.17 (3), 665–673. 10.1111/pbi.13006
250
Ortiz R. Trethowan R. Ferrara G. O. Iwanaga M. Dodds J. H. Crouch J. H. et al (2007). High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica157, 365–384. 10.1007/s10681-007-9375-9
251
Osakabe Y. Watanabe T. Sugano S. S. Ueta R. Ishihara R. Shinozaki K. et al (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep.6 (1), 26685. 10.1038/srep26685
252
Ozsolak F. Milos P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet.12 (2), 87–98. 10.1038/nrg2934
253
Panigrahi R. Kariali E. Panda B. B. Lafarge T. Mohapatra P. K. (2019). Controlling the trade-off between spikelet number and grain filling: the hierarchy of starch synthesis in spikelets of rice panicle in relation to hormone dynamics. Funct. Plant Biol.46 (6), 507–523. 10.1071/FP18153
254
Park S. H. Scheffler J. A. Ray J. D. Scheffler B. E. (2021). Identification of simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) that are associated with the nectariless trait of Gossypium hirsutum L. Euphytica217, 1–4. 10.3348/kjr.2020.1429
255
Peng H. Wang K. Chen Z. Cao Y. Gao Q. Li Y. et al (2020). MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res.48, D1085–D1092. 10.1093/nar/gkz921
256
Peng J. Richards D. E. Hartley N. M. Murphy G. P. Devos K. M. Flintham J. E. et al (1999). ‘Green revolution’genes encode mutant gibberellin response modulators. Nature400 (6741), 256–261. 10.1038/22307
257
Peng Z. Liu G. Li H. Wang Y. Gao H. Jemrić T. et al (2022). Molecular and genetic events determining the softening of fleshy fruits: A comprehensive review. Int. J. Mol. Sci.23 (20), 12482. 10.3390/ijms232012482
258
Phillips R. L. Vasil I. K. (2003). Phillips, R.L. and Vasil, I.K. DNA-based markers in plants 2nd edn. Ann. Bot.91 (6), 749–750. 10.1093/aob/mcg078
259
Pioneer (2016). DuPont announces intentions to commercialize first CRISPR-cas product. Press Release.
260
Porto E. M. Komor A. C. Slaymaker I. M. Yeo G. W. (2020). Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov.19 (12), 839–859. 10.1038/s41573-020-0084-6
261
Poland J. A. Bradbury P. J. Buckler E. S. Nelson R. J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci.108 (17), 6893–6898. 10.1073/pnas.1010894108
262
Prohens J. (2011). Plant breeding: a success story to be continued thanks to the advances in genomics. Front. Plant Sci.2, 51. 10.3389/fpls.2011.00051
263
Puranik S. Sahu P. P. Beynon S. Srivastava R. K. Sehgal D. Ojulong H. et al (2020). Genome-wide association mapping and comparative genomics identifies genomic regions governing grain nutritional traits in finger millet (Eleusine coracana L. Gaertn. . . Plants, People, Planet2 (6), 649–662. 10.1002/ppp3.10120
264
Purugganan M. D. (2019). Evolutionary insights into the nature of plant domestication. Curr. Biol.29 (14), R705–R714. 10.1016/j.cub.2019.05.053
265
Pyott D. E. Sheehan E. Molnar A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol.17, 1276–1288. 10.1111/mpp.12417
266
Qi W. Zhu T. Tian Z. Li C. Zhang W. Song R. (2016). High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol.16, 58–8. 10.1186/s12896-016-0289-2
267
Raihan A. Nahiyan A. S. M. Rahman A. Rahman L. (2016a). Molecular characterization of selected M5 lines of rice after TILLING for salinity tolerance using 20 SSR primers. Br. Biotechnol. J.12 (4), 1–11. 10.9734/BBJ/2016/23868
268
Raihan M. S. Liu J. Huang J. Guo H. Pan Q. Yan J. (2016b). Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58× SK maize population. Theor. Appl. Genet.129, 1465–1477. 10.1007/s00122-016-2717-z
269
Rajaram S. Borlaug N. E. Van Ginkel M. (2002). CIMMYT international wheat breeding. Bread wheat improvement and production. Rome: FAO, 103–117.
270
Rana M. M. Takamatsu T. Baslam M. Kaneko K. Itoh K. Harada N. et al (2019). Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci.20 (10), 2585. 10.3390/ijms20102585
271
Ranasinghe R. A. S. N. Maduwanthi S. D. T. Marapana R. A. U. J. (2019). Nutritional and health benefits of jackfruit (Artocarpus heterophyllus lam): a review. Int. J. Food Sci.2019, 4327183. 10.1155/2019/4327183
272
Rani R. Yadav P. Barbadikar K. M. Baliyan N. Malhotra E. V. Singh B. K. et al (2016). CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol. Lett.38, 1991–2006. 10.1007/s10529-016-2195-z
273
Rasheed A. Gill R. A. Hassan M. U. Mahmood A. Qari S. Zaman Q. U. et al (2021). A critical review: recent advancements in the use of CRISPR/Cas9 technology to enhance crops and alleviate global food crises. Curr. Issues Mol. Biol.43 (3), 1950–1976. 10.3390/cimb43030135
274
Razzaq A. Saleem F. Kanwal M. Mustafa G. Yousaf S. Imran Arshad H. M. et al (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci.20 (16), 4045. 10.3390/ijms20164045
275
Rees H. A. Liu D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Genetics19 (12), 770–788. 10.1038/s41576-018-0059-1
276
Ren B. Yan F. Kuang Y. Li N. Zhang D. Zhou X. et al (2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-Guided hyperactive hAID mutant. Mol. Plant11 (4), 623–626. 10.1016/j.molp.2018.01.005
277
Ren J. Wu P. Trampe B. Tian X. Lubberstedt T. Chen S. (2017). Novel technologies in doubled haploid line development. Plant Biotechnol. J.15 (11), 1361–1370. 10.1111/pbi.12805
278
Richardson D. M. Pysek P. Rejmanek M. Barbour M. G. Panetta F. D. West C. J. (2000). Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib.6 (2), 93–107. 10.1046/j.1472-4642.2000.00083.x
279
Rothan C. Causse M. (2007). “Natural and artificially induced genetic variability in crop and model plant species for plant systems biology,”. Plant systems biology. Editors BaginskyS.FernieA. R. (Experientia Supplementum), 97, 21–53. 10.1007/978-3-7643-7439-6_2
280
Rouet P. Smih F. Jasin M. (1994). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol.14 (12), 8096–8106. 10.1128/mcb.14.12.8096
281
Sadia B. Awan F. S. Saleem F. Altaf J. Umar A. B. Nadeem M. et al (2021). “Exploring plant genetic variations with morphometric and molecular markers,” in Genetic variation (Intech Open).
282
Sadiki M. Jarvis D. I. Rijal D. Bajracharya J. Hue N. N. Camacho-Villa T. C. et al (2007). “Variety names: an entry point to crop genetic diversity and distribution in agroecosystems?,”. Managing biodiversity in agricultural ecosystems. Editors JarvisD. I.PadochC.CooperC. D. (New York, NY, USA: Columbia University Press), 34, 76.
283
Saima Mir A. Maria M. Muhammad S. Mahboob Ali S. (2021). “Potential of mutation breeding to sustain food security,” in Genetic variation. Editors Trindade MaiaR.de Araujo CamposM. (Intech Open). 10.5772/intechopen.94087
284
Saleem S. Bari A. Abid B. Tahir ul Qamar M. Atif R. M. Khan M. S. (2020). QTL mapping for abiotic stresses in cereals. Environ. Clim. plant Veg. growth, 229–251. 10.1007/978-3-030-49732-3_10
285
Samantara K. Bohra A. Mohapatra S. R. Prihatini R. Asibe F. Singh L. et al (2022). Breeding more crops in less time: a perspective on speed breeding. Biology11 (2), 275. 10.3390/biology11020275
286
Samantara K. Reyes V. P. Agarwal N. Mohapatra S. R. Jena K. K. (2021). Advances and trends on the utilization of multi parent advanced generation intercross (MAGIC) for crop improvement. Euphytica217, 189. 10.1007/s10681-021-02925-6
287
Samineni S. Sen M. Sajja S. B. Gaur P. M. (2020). Rapid generation advance (RGA) in chickpea to produce up to seven generations per year and enable speed breeding. Crop J.8 (1), 164–169. 10.1016/j.cj.2019.08.003
288
Sanchez P. L. Wing R. A. Brar D. S. (2013). The wild relative of rice: Genomes and genomics. Plant genetics and genomics: Crops and models. New York: Springer, 9–25. 10.1007/978-1-4614-7903-1_2
289
Sattar T. Yaseen S. Waheed U. Farid H. N. Khan Z. (2023). CRISPR/Cas9-mediated inhibition of vacuolar invertase (VInv) gene in potato: CRISPR in potato. Pak. J. Biochem. Biotechnol.4 (1), 23–34. 10.52700/pjbb.v4i1.155
290
Sawada R. Hirano T. Iimure K. Abe T. Ozaki Y. (2016). Low polyphenol oxidase mutant induced by 12C6+ ion beam irradiation to protoplasts of lettuce (Lactuca sativa L). Hort. Res.15 (4), 347–353. 10.2503/hrj.15.347
291
Schaal B. (2019). Plants and people: our shared history and future. Plants, People, Planet1 (1), 14–19. 10.1002/ppp3.12
292
Schleif N. Kaeppler S. M. Kaeppler H. F. (2021). Generating novel plant genetic variation via genome editing to escape the breeding lottery. Vitro Cell. Dev. Biol.57, 627–644. 10.1007/s11627-021-10213-0
293
Schnell J. Steele M. Bean J. Neuspiel M. Girard C. Dormann N. et al (2015). A comparative analysis of insertional effects in genetically engineered plants: considerations for pre-market assessments. Transgenic Res.24, 1–17. 10.1007/s11248-014-9843-7
294
Schwager R. (2017). The land. Fairfax Media.
295
Sehgal D. Skot L. Singh R. Srivastava R. K. Das S. P. Taunk J. et al (2015). Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS One10 (5), e0122165. 10.1371/journal.pone.0122165
296
Sestili F. Palombieri S. Botticella E. Mantovani P. Bovina R. Lafiandra D. (2015). TILLING mutants of durum wheat result in a high amylose phenotype and provide information on alternative splicing mechanisms. Plant Sci.233, 127–133. 10.1016/j.plantsci.2015.01.009
297
Shakiba E. Eizenga G. C. (2014). Unraveling the secrets of rice wild species. UK: Rice-Germplasm, Genetics and Improvement. InTech, 1–58. 10.5772/58393
298
Shan Q. Wang Y. Li J. Gao C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc.9, 2395–2410. 10.1038/nprot.2014.157
299
Shan Q. Zhang Y. Chen K. Zhang K. Gao C. (2015). Creation of fragrant rice by targeted knockout of the Os BADH 2 gene using TALEN technology. Plant Biotechnol. J.13 (6), 791–800. 10.1111/pbi.12312
300
Sharma K. K. Palakolanu S. R. Bhattacharya J. Shankhapal A. R. Bhatnagar-Mathur P. (2022). CRISPR for accelerating genetic gains in under-utilized crops of the drylands: progress and prospects. Front. Genet.13, 999207. 10.3389/fgene.2022.999207
301
Shen C. Que Z. Xia Y. Tang N. Li D. He R. et al (2017). Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol.60, 539–547. 10.1007/s12374-016-0400-1
302
Shen H. Li Z. (2022). DNA double-strand break repairs and their application in plant DNA integration. Genes13 (2), 322. 10.3390/genes13020322
303
Shen Y. Zhou G. Liang C. Tian Z. (2022). Omics-based interdisciplinarity is accelerating plant breeding. Curr. Opin. Plant Biol.66, 102167. 10.1016/j.pbi.2021.102167
304
Shi J. Gao H. Wang H. Lafitte H. R. Archibald R. L. Yang M. et al (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J.15 (2), 207–216. 10.1111/pbi.12603
305
Shimatani Z. Kashojiya S. Takayama M. Terada R. Arazoe T. Ishii H. et al (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol.35 (5), 441–443. 10.1038/nbt.3833
306
Shu Q. Y. Forster B. P. Nakagawa H. (2012). “Principles and applications of plant mutation breeding,” in Plant mutation breeding and biotechnology (Wallingford UK: CABI), 301–325. 10.1079/9781780640853.0301
307
Shukla V. K. Doyon Y. Miller J. C. DeKelver R. C. Moehle E. A. Worden S. E. et al (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature459 (7245), 437–441. 10.1038/nature07992
308
Sigmon B. Vollbrecht E. (2010). Evidence of selection at the ramosa1 locus during maize domestication. Mol. Ecol.19, 1296–1311. 10.1111/j.1365-294X.2010.04562.x
309
Simmons C. R. Lafitte H. R. Reimann K. S. Brugiere N. Roesler K. Albertsen M. C. et al (2021). Successes and insights of an industry biotech program to enhance maize agronomic traits. Plant Sci.307, 110899. 10.1016/j.plantsci.2021.110899
310
Sinha P. Singh V. K. Bohra A. Kumar A. Reif J. C. Varshney R. K. (2021). Genomics and breeding innovations for enhancing genetic gain for climate resilience and nutrition traits. Theor. Appl. Genet.134 (6), 1829–1843. 10.1007/s00122-021-03847-6
311
Sneep J. (1966). Some facts about plant breeding before the discovery of mendelism. Euphytica15 (2), 135–140. 10.1007/BF00022315
312
Snow A. A. Andow D. A. Gepts P. Hallerman E. M. Power A. Tiedje J. M. et al (2005). Genetically engineered organisms and the environment: current status and recommendations. Ecol. Appl.15 (2), 377–404. 10.1890/04-0539
313
Song X. J. Huang W. Shi M. Zhu M. Z. Lin H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet.39 (5), 623–630. 10.1038/ng2014
314
Sood S. Kumar A. Sundaresha S. Bhardwaj V. (2022). “Genome editing prospects to develop disease/pest-resistant potato varieties,” in Sustainable management of potato pests and diseases (Singapore: Springer Singapore), 413–434. 10.1007/978-981-16-7695-6_17
315
Souza L. S. Diniz R. P. de Jesus Neves R. Alves A. A. C. de Oliveira E. J. (2018). Grafting as a strategy to increase flowering of cassava. Sci. Hortic.240, 544–551. 10.1016/j.scienta.2018.06.070
316
Soyk S. Müller N. A. Park S. J. Schmalenbach I. Jiang K. Hayama R. et al (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet.49 (1), 162–168. 10.1038/ng.3733
317
Sparla F. Falini G. Botticella E. Pirone C. Talame V. Bovina R. et al (2014). New starch phenotypes produced by TILLING in barley. PloS one9 (10), e107779. 10.1371/journal.pone.0107779
318
Stadler L. J. (1928a). Genetic effects of X-rays in maize. Proc. Natl. Acad. Sci.14 (1), 69–75. 10.1073/pnas.14.1.69
319
Stadler L. J. (1928b). Mutations in barley induced by X-rays and radium. Science68 (1756), 186–187. 10.1126/science.68.1756.186
320
Stetter M. G. Zeitler L. Steinhaus A. Kroener K. Biljecki M. Schmid K. J. (2016). Crossing methods and cultivation conditions for rapid production of segregating populations in three grain amaranth species. Front. Plant Sci.7, 816. 10.3389/fpls.2016.00816
321
Sugimoto K. Takeuchi Y. Ebana K. Miyao A. Hirochika H. Hara N. et al (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci.107 (13), 5792–5797. 10.1073/pnas.0911965107
322
Sukumaran S. Xiang W. Bean S. R. Pedersen J. F. Kresovich S. Tuinstra M. R. et al (2012). Association mapping for grain quality in a diverse sorghum collection. Plant Genome5 (3), plantgenome2012.07.0016. 10.3835/plantgenome2012.07.0016
323
Sun Y. Jiao G. Liu Z. Zhang X. Li J. Guo X. et al (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci.8, 298. 10.3389/fpls.2017.00298
324
Sun Y. Zhang X. Wu C. He Y. Ma Y. Hou H. et al (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant9 (4), 628–631. 10.1016/j.molp.2016.01.001
325
Suprasanna P. Saddhe A. Ghuge S. A. Ingle K. P. (2021). New and novel genetic tools for improving crops. CABI Rev.2021. 10.1079/PAVSNNR202116028
326
Svitashev S. Young J. K. Schwartz C. Gao H. Falco S. C. Cigan A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol.169 (2), 931–945. 10.1104/pp.15.00793
327
Sweeney M. McCouch S. (2007). The complex history of the domestication of rice. Ann. Bot.100 (5), 951–957. 10.1093/aob/mcm128
328
Syombua E. D. Zhang Z. Tripathi J. N. Ntui V. O. Kang M. George O. O. et al (2020). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea spp). Plant Biotechnol. J.19, 645–647. 10.1111/pbi.13515
329
Tian S. Jiang L. Cui X. Zhang J. Guo S. Li M. et al (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep.37 (9), 1353–1356. 10.1007/s00299-018-2299-0
330
Tadele Z. (2018). African orphan crops under abiotic stresses: challenges and opportunities. Scientifica2018, 1451894. 10.1155/2018/1451894
331
Tan L. Li X. Liu F. Sun X. Li C. Zhu Z. et al (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet.40 (11), 1360–1364. 10.1038/ng.197
332
Tang W. Ye J. Yao X. Zhao P. Xuan W. Tian Y. et al (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun.10 (1), 5279. 10.1038/s41467-019-13187-1
333
Tanksley S. D. Young N. D. Paterson A. H. Bonierbale M. W. (1989). RFLP mapping in plant breeding: new tools for an old science. Bio/technology7 (3), 257–264. 10.1038/nbt0389-257
334
Tanweer F. A. Rafii M. Y. Sijam K. Rahim H. A. Ahmed F. Latif M. A. (2015). Current advance methods for the identification of blast resistance genes in rice. C. R. Biol.338 (5), 321–334. 10.1016/j.crvi.2015.03.001
335
Tian D. Guo X. Zhang Z. Wang M. Wang F. (2019). Improving blast resistance of the rice restorer line, Hui 316, by introducing Pi9 or Pi2 with marker-assisted selection. Biotechnol. Biotechnol. Equip.33 (1), 1195–1203. 10.1080/13102818.2019.1649095
336
Timberlake W. E. (2013). Heterosis. Brenner’s Encycl. Genet., 451–453. 10.1016/b978-0-12-374984-0.00705-1
337
Tran M. T. Son G. H. Song Y. J. Nguyen N. T. Park S. Thach T. V. et al (2023). CRISPR-Cas9-based precise engineering of SlHyPRP1 protein towards multi-stress tolerance in tomato. Front. Plant Sci.14, 1186932. 10.3389/fpls.2023.1186932
338
Tribhuvan K. U. Das A. Srivastava H. Kumar K. Durgesh K. Mithra S. V. et al (2020). Identification and characterization of PEBP family genes reveal CcFT8 a probable candidate for photoperiod insensitivity in C. cajan. 3 Biotech.10, 194. 10.1007/s13205-020-02180-x
339
Ueta R. Abe C. Watanabe T. Sugano S. S. Ishihara R. Ezura H. et al (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep.7 (1), 507. 10.1038/s41598-017-00501-4
340
Van Harten A. M. (1998). Mutation breeding: Theory and practical applications. Cambridge: Cambridge University Press, 137–158.
341
Vaughan D. A. Lu B. R. Tomooka N. (2008). The evolving story of rice evolution. Plant Sci.174 (4), 394–408. 10.1016/j.plantsci.2008.01.016
342
Veillet F. Perrot L. Chauvin L. Kermarrec M. P. Guyon-Debast A. Chauvin J. E. et al (2019). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci.20 (2), 402. 10.3390/ijms20020402
343
Venezia M. Creasey Krainer K. M. (2021). Current advancements and limitations of gene editing in orphan crops. Front. Plant Sci.12, 742932. 10.3389/fpls.2021.742932
344
Verma H. Theunuo S. Devi E. L. Sarma R. N. (2023). Abiotic and biotic stress tolerance in rice: recent advances in molecular breeding approaches. QTL Mapp. Crop Improv., 219–234. 10.1016/B978-0-323-85243-2.00003-9
345
Vilmorin R. D. (1930). Crop plants, methods of selection and creation of new varieties. Bull. Soc. Enc. Ind. Nat. Paris.129, 779–792.
346
Visarada K. B. R. S. Meena K. Aruna C. Srujana S. Saikishore N. Seetharama N. (2009). Transgenic breeding: perspectives and prospects. Crop Sci.49 (5), 1555–1563. 10.2135/cropsci2008.10.0581
347
Vollbrecht E. Springer P. S. Goh L. Buckler IV E. S. Martienssen R. (2005). Architecture of floral branch systems in maize and related grasses. Nature436 (7054), 1119–1126. 10.1038/nature03892
348
Vos P. Hogers R. Bleeker M. Reijans M. Lee T. V. D. Hornes M. et al (1995). Aflp: a new technique for DNA fingerprinting. Nucleic Acids Res.23 (21), 4407–4414. 10.1093/nar/23.21.4407
349
Wang B. Zhu L. Zhao B. Zhao Y. Xie Y. Zheng Z. et al (2019a). Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant12 (4), 597–602. 10.1016/j.molp.2019.03.006
350
Wang H. Wu Y. Zhang Y. Yang J. Fan W. Zhang H. et al (2019b). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea batatas) for the improvement of starch quality. IJMS20, 4702. 10.3390/ijms20194702
351
Wang L. Chen L. Li R. Zhao R. Yang M. Sheng J. et al (2017). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem.65 (39), 8674–8682. 10.1021/acs.jafc.7b02745
352
Wang M. Li W. Fang C. Xu F. Liu Y. Wang Z. et al (2018a). Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat. Genet.50 (10), 1435–1441. 10.1038/s41588-018-0229-2
353
Wang T. L. Uauy C. Robson F. Till B. (2012). TILLING in extremis. Plant Biotechnol. J.10 (7), 761–772. 10.1111/j.1467-7652.2012.00708.x
354
Wang W. Pan Q. He F. Akhunova A. Chao S. Trick H. et al (2018b). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J.1 (1), 65–74. 10.1089/crispr.2017.0010
355
Wang Y. Cheng X. Shan Q. Zhang Y. Liu J. Gao C. et al (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol.32 (9), 947–951. 10.1038/nbt.2969
356
Wang Y. Zhang Y. Dai C. Ma J. Zhou Y. Jaikishun S. et al (2021b). The establishment of two efficient transformation systems in quinoa. Res. Sq.10.21203/rs.3.rs-364280/v1
357
Wang Z. Wan L. Xin Q. Zhang X. Song Y. Wang P. et al (2021a). Optimizing glyphosate tolerance in rapeseed by CRISPR/Cas9-based geminiviral donor DNA replicon system with Csy4-based single-guide RNA processing. J. Exp. Bot.72 (13), 4796–4808. 10.1093/jxb/erab167
358
Watson A. Ghosh S. Williams M. J. Cuddy W. S. Simmonds J. Rey M. D. et al (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants4 (1), 23–29. 10.1038/s41477-017-0083-8
359
Webb C. A. Szabo L. J. Bakkeren G. Garry C. Staples R. C. Eversmeyer M. et al (2006). Transient expression and insertional mutagenesis of Puccinia triticina using biolistics. Funct. Integr. Genomics6, 250–260. 10.1007/s10142-005-0009-9
360
Williams J. G. Kubelik A. R. Livak K. J. Rafalski J. A. Tingey S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res.18 (22), 6531–6535. 10.1093/nar/18.22.6531
361
Wilson H. D. (1981). Domesticated Chenopodium of the Ozark bluff dwellers. Econ. Bot.35, 233–239. 10.1007/BF02858690
362
Wissuwa M. Wegner J. Ae N. Yano M. (2002). Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet.105, 890–897. 10.1007/s00122-002-1051-9
363
Witcombe J. R. Gyawali S. Subedi M. Virk D. S. Joshi K. D. (2013). Plant breeding can be made more efficient by having fewer, better crosses. BMC Plant Biol.13, 22–12. 10.1186/1471-2229-13-22
364
Wu D. Lao S. Fan L. (2021). De-domestication: an extension of crop evolution. Trends Plant Sci.26 (6), 560–574. 10.1016/j.tplants.2021.02.003
365
Wu M. F. Goldshmidt A. Ovadya D. Larue H. (2020). I am all ears: maximize maize doubled haploid success by promoting axillary branch elongation. Plant Direct4 (5), e00226. 10.1002/pld3.226
366
Xie K. Yang Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant6 (6), 1975–1983. Available at: https://doi.org/10.1093/mp/sst119.
367
Xu G. Wang X. Huang C. Xu D. Li D. Tian J. et al (2017). Complex genetic architecture underlies maize tassel domestication. New Phytol.214 (2), 852–864. 10.1111/nph.14400
368
Xu Z. Hua J. Wang F. Cheng Z. Meng Q. Chen Y. et al (2020). Marker-assisted selection of qMrdd8 to improve maize resistance to rough dwarf disease. Breed. Sci.70 (2), 183–192. 10.1270/jsbbs.19110
369
Yadav S. Sandhu N. Singh V. K. Catolos M. Kumar A. (2019). Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance. Sci. Rep.9 (1), 14326–14412. 10.1038/s41598-019-50880-z
370
Yamatani H. Kohzuma K. Nakano M. Takami T. Kato Y. Hayashi Y. et al (2018). Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J. Exp. Bot.69 (5), 1027–1035. 10.1093/jxb/erx468
371
Yan F. Kuang Y. Ren B. Wang J. Zhang D. Lin H. et al (2018). Highly efficient A· T to G· C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant11 (4), 631–634. 10.1016/j.molp.2018.02.008
372
Yang M. M. Wang J. Dong L. Kong D. J. Teng Y. Liu P. et al (2017a). Lack of association of C3 gene with uveitis: additional insights into the genetic profile of uveitis regarding complement pathway genes. Sci. Rep.7 (1), 879. 10.1038/s41598-017-00833-1
373
Yang T. Ali M. Lin L. Li P. He H. Zhu Q. et al (2023). Recoloring tomato fruit by CRISPR/Cas9-mediated multiplex gene editing. Hortic. Res.10 (1), uhac214. 10.1093/hr/uhac214
374
Yang Y. Zhu G. Li R. Yan S. Fu D. Zhu B. et al (2017b). The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol.175 (4), 1690–1702. 10.1104/pp.17.01265
375
Yang Z. Zhang H. Li X. Shen H. Gao J. Hou S. et al (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants6, 1167–1178. 10.1038/s41477-020-0747-7
376
Yao L. Zhang Y. Liu C. Liu Y. Wang Y. Liang D. et al (2018). OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants4 (8), 530–533. 10.1038/s41477-018-0193-y
377
Yaqoob H. Tariq A. Bhat B. A. Bhat K. A. Nehvi I. B. Raza A. et al (2023). Integrating genomics and genome editing for orphan crop improvement: a bridge between orphan crops and modern agriculture system. Gm. Crops Food14 (1), 1–20. 10.1080/21645698.2022.2146952
378
Yu J. Holland J. B. McMullen M. D. Buckler E. S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics178 (1), 539–551. 10.1534/genetics.107.074245
379
Yu Q. H. Wang B. Li N. Tang Y. Yang S. Yang T. et al (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep.7 (1), 11874. 10.1038/s41598-017-12262-1
380
Yu Z. Deng J. He J. Huo Y. Wu Y. Wang X. et al (1991). Mutation breeding by ion implantation. Nucl. Instrum. Methods Phys. Res. B Beam Interact. Mat. At.59, 705–708. 10.1016/0168-583X(91)95307-Y
381
Yumlu S. Stumm J. (2021). CRISPR/Cas-mediated genome editing in mammalian cells: limitations, challenges, and perspectives. Biochem. Soc. Trans.49 (3), 1435–1448.
382
Zaghum M. J. Ali K. Teng S. (2022). Integrated genetic and omics approaches for the regulation of nutritional activities in rice (Oryza sativa L). Agriculture12 (11), 1757. 10.3390/agriculture12111757
383
Zhang H. Si X. Ji X. Fan R. Liu J. Chen K. et al (2018a). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol.36 (9), 894–898. 10.1038/nbt.4202
384
Zhang H. Zhang J. Wei P. Zhang B. Gou F. Feng Z. et al (2014). The CRISPR/C as9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J.12 (6), 797–807. 10.1111/pbi.12200
385
Zhang X. Chen L. Zhu B. Wang L. Chen C. Hong M. et al (2020). Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol.22 (6), 740–750. 10.1038/s41556-020-0518-8
386
Zhang Y. Bai Y. Wu G. Zou S. Chen Y. Gao C. et al (2017). Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J.91 (4), 714–724. 10.1111/tpj.13599
387
Zhang Y. Li D. Zhang D. Zhao X. Cao X. Dong L. et al (2018b). Analysis of the functions of Ta GW 2 homoeologs in wheat grain weight and protein content traits. Plant J.94 (5), 857–866. 10.1111/tpj.13903
388
Zhao M. Tang S. Zhang H. He M. Liu J. Zhi H. et al (2020). DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc. Natl. Acad. Sci.117 (35), 21766–21774. 10.1073/pnas.2002278117
389
Zhao Y. Zhang C. Liu W. Gao W. Liu C. Song G. et al (2016). An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep.6 (1), 23890–23911. 10.1038/srep23890
390
Zheng X. Zhang S. Liang Y. Zhang R. Liu L. Qin P. et al (2023). Loss-function mutants of OsCKX gene family based on CRISPR-Cas systems revealed their diversified roles in rice. Plant Genome16 (2), e20283. 10.1002/tpg2.20283
391
Zhou J. Peng Z. Long J. Sosso D. Liu B. O. Eom J. S. et al (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J.82 (4), 632–643. 10.1111/tpj.12838
392
Zhou M. Deng L. Yuan G. Zhao W. Ma M. Sun C. et al (2023). A CRISPR-cas9-derived male sterility system for tomato breeding. Agronomy13 (7), 1785. 10.3390/agronomy13071785
393
Zhu C. Gore M. Buckler E. S. Yu J. (2008). Status and prospects of association mapping in plants. Plant Genome1 (1), plantgenome2008.02.0089. 10.3835/plantgenome2008.02.0089
394
Zhu J. Song N. Sun S. Yang W. Zhao H. Song W. et al (2016). Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-cas9. J. Genet. Genomics43 (1), 25–36. 10.1016/j.jgg.2015.10.006
395
Zhu R. Luo Y. Bai Y. (2019). Improving nutritional quality of crops through genome editing. Crit. Rev. Food Sci. Nutr.59 (1), 17–30. 10.1080/10408398.2018.1437040
396
Zhu Y. Lin Y. Fan Y. Wang Y. Li P. Xiong J. et al (2023). CRISPR/Cas9-mediated restoration of Tamyb10 to create pre-harvest sprouting-resistant red wheat. Plant Biotechnol. J.21 (4), 665–667. 10.1111/pbi.13981
397
Zhu Y. Y. Zeng H. Q. Dong C. X. Yin X. M. Shen Q. R. Yang Z. M. (2010). microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L). Plant Sci.178 (1), 23–29. 10.1016/j.plantsci.2009.09.011
398
Zong Y. Song Q. Li C. Jin S. Zhang D. Wang Y. et al (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol.36 (10), 950–953. 10.1038/nbt.4261
399
Zong Y. Wang Y. Li C. Zhang R. Chen K. Ran Y. et al (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol.35 (5), 438–440. 10.1038/nbt.3811
Summary
Keywords
climate change, genome editing, genome revolution, green revolution, marker-assisted selection, omics-assisted breeding, QTL mapping
Citation
Chawla R, Poonia A, Samantara K, Mohapatra SR, Naik SB, Ashwath MN, Djalovic IG and Prasad PVV (2023) Green revolution to genome revolution: driving better resilient crops against environmental instability. Front. Genet. 14:1204585. doi: 10.3389/fgene.2023.1204585
Received
12 April 2023
Accepted
11 August 2023
Published
31 August 2023
Volume
14 - 2023
Edited by
Vandana Jaiswal, Institute of Himalayan Bioresource Technology (CSIR), India
Reviewed by
Kalyani Makarand Barbadikar, Indian Institute of Rice Research (ICAR), India
Stuart Smyth, University of Saskatchewan, Canada
Updates
Copyright
© 2023 Chawla, Poonia, Samantara, Mohapatra, Naik, Ashwath, Djalovic and Prasad.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sourav Ranjan Mohapatra, mohapatrasourav23@gmai.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.