- 1Department of Molecular and Computational Biology, University of Southern California, Los Angeles, CA, United States
- 2US Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 3Genome Sequencing Center, HudsonAlpha Institute for Biotechnology, Huntsville, AL, United States
- 4Arizona Genomics Institute, School of Plant Sciences, University of Arizona, Tucson, AZ, United States
- 5US Department of Agriculture, Agricultural Research Service (USDA-ARS), Ithaca, NY, United States
- 6Section On Plant Breeding and Genetics, School of Integrative Plant Sciences, Cornell University, Ithaca, NY, United States
- 7Department of Plant and Microbial Biology, University of California Berkeley, Berkeley, CA, United States
- 8Department of Ecology and Evolutionary Biology, University of Connecticut, Stamford, CT, United States
- 9Applied Ocean Physics and Engineering Department, Woods Hole Oceanographic Institution, Woods Hole, MA, United States
Increasing the genomic resources of emerging aquaculture crop targets can expedite breeding processes as seen in molecular breeding advances in agriculture. High quality annotated reference genomes are essential to implement this relatively new molecular breeding scheme and benefit research areas such as population genetics, gene discovery, and gene mechanics by providing a tool for standard comparison. The brown macroalga Saccharina latissima (sugar kelp) is an ecologically and economically important kelp that is found in both the northern Pacific and Atlantic Oceans. Cultivation of Saccharina latissima for human consumption has increased significantly this century in both North America and Europe, and its single blade morphology allows for dense seeding practices used in the cultivation of its Asian sister species, Saccharina japonica. While Saccharina latissima has potential as a human food crop, insufficient information from genetic resources has limited molecular breeding in sugar kelp aquaculture. We present scaffolded and annotated Saccharina latissima nuclear and organelle genomes from a female gametophyte collected from Black Ledge, Groton, Connecticut. This Saccharina latissima genome compares well with other published kelp genomes and contains 218 scaffolds with a scaffold N50 of 1.35 Mb, a GC content of 49.84%, and 25,012 predicted genes. We also validated this genome by comparing the synteny and completeness of this Saccharina latissima genome to other kelp genomes. Our team has successfully performed initial genomic selection trials with sugar kelp using a draft version of this genome. This Saccharina latissima genome expands the genetic toolkit for the economically and ecologically important sugar kelp and will be a fundamental resource for future foundational science, breeding, and conservation efforts.
1 Introduction
The demand for sustainable farming resources is increasing due to the combination of rising global temperatures (Abbass et al., 2022), increasing population levels (Pimentel, 1991), and decreasing amounts of arable land (Thaler et al., 2021). Increasing global food production can help raise the carrying capacity of Earth despite these environmental challenges escalating in the 21st century (Hopfenberg, 2003). One potential sustainable resource solution is to increase the amount of biomass produced in the ocean by deploying open ocean kelp farms, as kelp grows quickly and requires no arable land, freshwater, herbicides, or fertilizers (Kim et al., 2019a). Kelp are haplodiplontic brown macroalgae (North, 1987; Diehl et al., 2024) that provide vital ecosystem services, including habitat creation and primary production (Eger et al., 2023), that sustain some of the ocean’s most diverse communities (Dayton, 1985; Steneck et al., 2002; Wernberg et al., 2018). The emergence of the kelp lineage (Laminariales) is estimated to have occurred over 80 million years ago (Choi et al., 2024), followed by rapid radiation that gave rise to a globally distributed lineage of morphologically diverse, complex macroalgae (Silberfeld et al., 2010; Starko et al., 2019; Bringloe et al., 2020).
While kelps and other seaweeds have been consumed by humans since the Mesolithic era (Buckley et al., 2023), kelp aquaculture development lagged compared to terrestrial agriculture until the 20th century (Hwang et al., 2019). Large scale kelp farming initially started in Japan, Korea, and China in the 1950s–1970s (Tseng and Fei, 1987), and Asia currently accounts for 97% of the $6 billion global kelp market (FAO, 2023; 2024). The predominant kelp species farmed are Saccharina japonica ($4.6 billion) and Undaria pinnatifida ($1.9 billion) (Cai et al., 2021), with kelp aquaculture directly supporting a range of industries, from food to pharmaceuticals (Kim et al., 2017). As the industry expanded, kelp breeding programs were formed to address low quality seed, increasing biomass, disease resistance, and trait consistency using phenotypic selection (Hwang et al., 2019; Hu et al., 2023). In agriculture, emerging genetic resources, such as reference genomes and sequence information for breeding panels of plants, have accelerated genomics guided breeding programs to develop more productive and resilient cultivars (Heffner et al., 2009). Genomics can also accelerate kelp breeding programs (Budhlakoti et al., 2022; Song et al., 2023), if the proper genetic resources, such as reference genomes and breeding populations, are developed (DeWeese and Osborne, 2021).
The haplodiplontic life cycle of kelp is ideal for genomics-based breeding, as haploid gametophytes can be vegetatively propagated in culture (Dring and Lüning, 1975; Huang et al., 2022; 2023). These gametophyte cultures can then serve as kelp breeding germplasm, with scalable production of monoclonal gametophyte cultures producing ample material for crossing or sequencing experiments (Hoffmann and Santelices, 1991). Sequencing data can then be aligned to reference genomes, producing variant information that can be used with phenotypic data to produce genomic selection models (Moose and Mumm, 2008). These models produce genome estimated breeding values (GEBVs) (Meuwissen et al., 2001), which can then be used to predict the phenotypes, such as biomass, of potential crosses in the sequenced germplasm (Desta and Ortiz, 2014; Huang et al., 2022). Sequenced germplasm collections, along with the vegetative propagation of haploid gametophytes, compose an incredibly powerful tool for breeding programs for kelp (The Aquaculture Genomics, Genetics and Breeding Workshop et al., 2017; Wade et al., 2020).
The brown macroalga Saccharina latissima (sugar kelp, Laminariales) (Lane et al., 2006) is an ecologically and economically significant species found in the Pacific, Atlantic, and Arctic Oceans (Diehl et al., 2024). S. latissima is a sister species to the commercially cultivated Japanese sugar kelp S. japonica, with a similar morphology of an undivided single blade (Redmond et al., 2014), ideal for kelp farming (Peteiro and Freire, 2011). S. latissima is farmed in both northern Europe and North America, and is the most farmed kelp in the United States, accounting for a predominant share of the current >$300 million US kelp industry (Kim et al., 2019a; Heidkamp et al., 2022; Brayden and Coleman, 2023; Stekoll et al., 2024). Kelp mariculture is a rapidly developing sector in the United States and demands for fundamental research into cultivable kelp species have engendered significant investment (e.g., ARPA-E MARINER programs) in projects to increase the productivity of kelp farms (Li et al., 2022).
While population genetic studies of sugar kelp in both the United States and Europe have begun to provide some of the resources necessary for breeding programs (Breton et al., 2018; Mao et al., 2020; Huang et al., 2022), an annotated reference genome is foundational for genomic selection technologies. In fact, early results from a S. latissima genomic selection breeding program based on the reference genome described here produced cultivars that doubled biomass yield compared with non-selected kelps (Huang et al., 2023). The recently published European S. latissima genome (Denoeud et al., 2024) represents an important milestone in research and development for the sugar kelp aquaculture industry in Europe and for expanded brown algal genetic analyses through the Phaeoexplorer genome database. In the United States, a reference for North American S. latissima provides opportunities to further refine breeding models on local sugar kelp populations for the expanding kelp industry (Brakel et al., 2021), as well as to aid in kelp forest conservation and restoration efforts (Coleman and Glasby, 2024; Bemmels et al., 2025). We present the scaffolded and annotated nuclear and organelle genome assemblies of North American sugar kelp (S. latissima), key genomic resources for population genetic studies, conservation, and modern genomic breeding of sugar kelp in the United States.
2 Materials and methods
2.1 Sample collection and nucleotide extraction
Reproductive sorus tissue from a wild population of S. latissima sporophytes was sampled from Black Ledge, Groton, Connecticut, US (41°31′N, 72°07′W, 26 June 2014). Following induced sporulation, individual gametophytes were isolated to establish monoclonal gametophyte cultures in a laboratory setting, as outlined by Redmond et al. (2014) and Alsuwaiyan et al. (2019). One female S. latissima gametophyte (var. SL-CT1-FG3) was selected for long-read sequencing for reference genome assembly and cultured in Erlenmeyer flasks under red light with a 12:12 h light:dark photoperiod at 10°C to inhibit reproduction and promote growth and mitotic division (Redmond et al., 2014; Augyte et al., 2018) for genomic DNA extraction. Augyte et al. (2018) at the Marine Biotechnology Laboratory at the University of Connecticut extracted DNA from a 24 mg (fresh) female gametophyte culture of S. latissima (var. SL-CT1-FG3) using a modified protocol of the NucleoSpin Plant II Maxi Kit (Macherey-Nagel, Düren, Germany; cat # 740609). Gametophyte biomass was spun down in 1.5-mL tubes in an Eppendorf 5424 microcentrifuge (21,000 rcf, 2 min). Sealed tubes of gametophyte material were frozen in liquid nitrogen for 20 s before being ground with a plastic pestle for 30 s. Ground samples were extracted with CTAB buffer using repeated wash steps (Doyle and Doyle, 1990; Augyte et al., 2018).
We collected samples from Saccharina latissima sporophytes cultivated and harvested by University of Connecticut and GreenWave from a farm site in Branford, Connecticut, USA (41°15′13″N, 72°46′5″W, May 2020). Sporophytes were replicates of a single cross from S. latissima gametophyte collections (Augyte et al., 2018; Mao et al., 2020): a female gametophyte (var. LIS-F1-3) originating from Southern New England, and a male gametophyte (var. SL-CT1-MG2) cultured from the spore release that also generated SL-CT1-FG3. We flash-froze the samples in liquid nitrogen immediately after collection and subsequently stored at −80°C for less than 1 month before being sent on dry ice to Cornell University for RNA extraction. Sporophyte samples were ground in liquid nitrogen and RNA was extracted using the Quick-RNA Microprep Kit (Zymo Research, Irvine, CA, USA; cat #R1052), yielding 8.31 µg total RNA (average 1.66 µg/sample). An additional RNA extraction was performed at the HudsonAlpha Institute (Huntsville, Alabama, USA) from SL-CT1-FG3 gametophyte culture using the RNeasy Micro Kit (Qiagen Inc., Valencia, CA, USA; cat # 74004).
2.2 DNA and RNA sequencing
Extracted DNA from the S. latissima female gametophyte (var. SL-CT1-FG3) was sent to the HudsonAlpha Institute for whole genome sequencing using a whole genome shotgun sequencing strategy and standard sequencing protocols. Sequencing reads were collected using two platforms: Illumina reads were sequenced using the Illumina NovaSeq 6000 platform, and the PacBio reads were sequenced using the Sequel II platform. One 400bp insert 2 × 250 Illumina fragment library (66.03x) was sequenced along with one 2 × 150 Dovetail Hi-C library (145.33x) (Supplementary Table S1). Prior to use, the Illumina fragment reads were screened for phix contamination. Reads composed of >95% simple sequence were removed. Illumina reads <50bp after trimming for adapter and quality (q < 20) were removed. The final Illumina read set consisted of 282,564,006 reads for a total of 66.03x of high-quality bases. PacBio sequencing yielded 111.14 Gb of total raw sequence, representing 180.56x genomic coverage (Supplementary Table S2).
RNA extracted from five S. latissima sporophyte samples was sent to the HudsonAlpha Institute for library preparation and sequencing using standard protocols. All RNA libraries were prepared using TruSeq Stranded mRNA Library Prep (96 samples) (Illumina, San Diego, CA, USA; cat # 20020595) and indexed with IDT for Illumina TruSeq RNA UD Indexes v2 (96 indexes) (Illumina, San Diego, CA, USA; cat # 20040871) according to manufacturer instructions. cDNA was sequenced on the Illumina NovaSeq 6000 platform and generated a total of 287 million reads, with an average of 96 million reads per sample.
2.3 Nuclear genome assembly and decontamination
The initial assembly version 0 was generated by assembling 11,430,834 PacBio CCS reads of the female S. latissima gametophyte SL-CT1-FG3 using hifiasm v0.7 (Cheng et al., 2021) and subsequently polished using RACON v1.4 (Vaser et al., 2017). This produced an initial assembly (SL-CT1-FG3 v0) consisting of 4,854 contigs, with a contig N50 of 863.2 Kb, and a total assembly size of 925.8 Mb (Supplementary Table S3).
Hi-C sequencing of SL-CT1-FG3 yielded 626,664,456 2 × 150 Hi-C Illumina reads, an estimated 145.33x coverage. The reads were aligned to the SL-CT1-FG3 v0 assembly using BWA-MEM v0.7.17 (Li, 2013). Paired-end reads were mapped independently (as single-ends) due to the nature of the Hi-C pair, which captures conformation via proximity-ligated fragments. A small fraction of single-end mapped reads will contain a ligation junction, an indicator that they are chimeric because they do not originate from a contiguous piece of DNA. In these cases, only the 5′-side was retained, as the 3′-end generally originates from the same contiguous DNA as the 5′-side of the mated read. The resulting single end alignments were combined into a BAM file containing the paired, chimera-filtered Hi-C read alignments. The 3D-DNA (Dudchenko et al., 2017) suite of internal tools was used to generate a contact map using the resultant BAM file, and the contact map was visualized using Juicebox (Durand et al., 2016). Chromosome-scale scaffolding was attempted using 3D-DNA, but high levels of contamination in the v0 genome initially prevented meaningful scaffolding.
The assembled contigs were screened against bacterial proteins, organelle sequences, and the NCBI non-redundant protein sequence database (NR) (Sayers et al., 2024) and removed if found to be a contaminant according to standard practice (e.g., (Bennetzen et al., 2012; Motamayor et al., 2013; Bartholomé et al., 2015). Contigs were classified into bins depending on sequence content. Contamination was identified using BLASTn (Camacho et al., 2009) against the NCBI non-redundant nucleotide database (NT) (Sayers et al., 2024) and BLASTx using a set of known microbial proteins. Additional contigs were classified in the version 1 release as contaminants (2,863 contigs, 283.9 Mb), chloroplast (181 contigs, 11.0 Mb), prokaryote (14 contigs, 10.6 Mb), redundant (>95% masked with 24mers that occur more than 2 times in all contigs) (233 contigs, 6.2 Mb), repetitive (>95% masked with 24mers that occur more than 4 times in contigs greater than the contig N50) (24 contigs, 1.8 Mb), and unanchored rDNA (Ding et al., 2022) (3 contigs, 165.1 Kb).
2.4 Hi-C scaffolding and polishing
With contaminant contigs removed, we attempted to scaffold contigs together using the contact information from 3D-DNA (Dudchenko et al., 2017) and scaffold graph construction, as described by Ghurye et al. (2019). In brief, a graph was formed between all contigs in which graph edge weights, w(u, v), between any two contigs u and v were computed by dividing the number of counts, N(u, v), by the total number of cut sites, C, in both u and v: (Ghurye et al., 2019)
We then computed a normalized Best Buddy Weight, BBW(u, v), as the weight, w(u, v), divided by the maximal weight of any edge incident upon contigs u or v, excluding the (u, v) edge itself (Ghurye et al., 2019). All BBW(u, v) values >1 were retained, and a reverse Dijkstra’s algorithm (highest weight graph) (Dijkstra, 1959) was then utilized to generate the contig order and orientation for the join file. Using this algorithm, a total of 333 joins were made to form an additional 218 scaffolded contig sets. Each join was padded with an unsized gap of 10,000 Ns.
SNP and INDEL errors in the consensus were corrected with 282,564,006 Illumina fragment 2 × 250 reads (66.03x coverage) by aligning the reads using BWA-MEM (Li, 2013) and identifying SNPs and INDELs with GATK3 UnifiedGenotyper (Van der Auwera and O’Connor, 2020). A total of 502 SNPs and 20,723 INDELs were corrected in the release. The final version 1 release contained 612.21 Mb of sequence, consisting of 218 scaffolds and 1,513 contigs, with a contig N50 of 971.7 Kb (Supplementary Table S4).
2.5 Gene annotation
The JGI Annotation Pipeline (Grigoriev et al., 2006) was used to annotate the S. latissima nuclear genome assembly. The pipeline automatedly predicts, filters, and functionally annotates gene models as described by Grigoriev et al. (2006). Briefly: repeats in the genome assembly were masked with RepeatMasker (Smit et al., 2004), RepBase (Jurka et al., 2005), and RepeatScout (Price et al., 2005). The masked assembly was then used to predict protein-coding gene models with ab initio, homology, and transcriptomic modeling methods. Ab initio gene predictions were performed with Fgenesh (Salamov and Solovyev, 2000) and GeneMark (Ter-Hovhannisyan et al., 2008). Homology was assessed by BLASTx (Camacho et al., 2009) of the assembly against NCBI NR (Sayers et al., 2024). Resulting alignments were used to seed Fgenesh+ (Salamov and Solovyev, 2000) and Genewise (Birney et al., 2004) for homology-based gene prediction. A transcriptome assembly was generated for S. latissima with Illumina RNAseq reads with Trinity v2.11.0 (Grabherr et al., 2011), and RNA reads were mapped back to the genome assembly with HISAT2 (Kim et al., 2019b). With these inputs, the transcriptome-based programs Fgenesh (Salamov and Solovyev, 2000) and combest (Zhou et al., 2015) were used to generate gene models. A total of 41,561 gene models predicted across all methods (all models) were filtered based on protein homology and transcriptome evidence to a single representative model at each genomic locus (filtered models), generating a set of 24,790 gene models; for a detailed description of JGI model filtration, refer to Grigoriev et al. (2006). Protein sequences predicted from the set of filtered gene models were functionally annotated. Proteins were classified using SignalP v3 (Nielsen et al., 1997) for signal sequences, TMHMM (Melén et al., 2003) for transmembrane domains, and InterproScan (Quevillon et al., 2005) for functional domains. BLASTp (Camacho et al., 2009) alignments of proteins against NCBI NR (Sayers et al., 2024), Swiss-Prot (UniProt Consortium, 2012), KEGG (Kanehisa, 2004), and KOG (Koonin et al., 2004) databases to further informed functional interpretation of predicted proteins. The version 1 S. latissima genome assembly, associated annotations, and metadata are hosted on the JGI PhycoCosm (Grigoriev et al., 2021) comparative algal genome portal (https://phycocosm.jgi.doe.gov/SlaSLCT1FG3_1).
2.6 Comparative genomic analyses
Gene content was scored for the analyzed brown macroalgal genomes using BUSCO v5.7.1 (Manni et al., 2021) in genome mode with gene predictor set to Augustus v3.5.0 (Stanke et al., 2008) against the ortholog databases for Eukaryota (eukaryota_odb10) and Stramenopiles (stramenopiles_odb10). QUAST-LG v5.2.0 (Mikheenko et al., 2018) was used to compute relevant assembly quality metrics for each. Synteny of our v1 S. latissima assembly to related species was investigated by aligning to published genomes of S. japonica, Macrocystis pyrifera, U. pinnatifida, and Ectocarpus sp. using the Progressive Cactus v2.6.7 pipeline (Armstrong et al., 2020). The phylogenetic tree used to seed the Cactus alignment was pruned using tidytree v0.4.6 (Yu, 2022) and treeio v1.30.0 (Wang et al., 2020) from the Starko et al. (2019) kelp phylogeny constructed from plastid, mitochondrial, and ribosomal genes.
Blocks of synteny were extracted from the five-species hierarchical whole genome alignment (HAL) format (Hickey et al., 2013) into the BLAT-defined PSL format (Kent, 2002) using halSynteny v2.2 (Krasheninnikova et al., 2020). We applied a method put forward by Nosil et al. (2023) to establish one-to-one homology between chromosomes of related species using synteny blocks derived from pairwise Cactus alignments. To appropriately map our S. latissima scaffolds onto longer reference chromosomes, we modified the method to a many-to-one approach, allowing multiple S. latissima scaffolds to align to a single chromosome. For each alignment of a S. latissima scaffold to a reference species chromosome, lengths of syntenic blocks (“matches” in PSL format (Kent, 2002)) were summed. Best scaffold-chromosome pairs were identified with respect to each S. latissima scaffold, hereafter referred to as a “maximal syntenic match”, by calculating the maximum summed exact match per scaffold amongst the aligned chromosomes. FASTA genome assembly files for each of the five species, and a whole genome alignment file converted from HAL to MAF using hal2maf v2.2 (Hickey et al., 2013), were given as input to Ragout v2.3 (Kolmogorov et al., 2018), a reference-assisted scaffolding tool used to improve assembly contiguity.
Heatmaps representing synteny between reference chromosomes of each species versus S. latissima scaffolds were generated using ggplot2 v3.5.1 (Wickham, 2016) in R v4.4.0 (R Core Team, 2024). Ordering of our genome v1 assembly scaffolds onto synteny-constructed pseudochromosomes was rendered into genetic map representation using ggplot2 v3.5.1 (Wickham, 2016) in R v4.4.0 (R Core Team, 2024). Scripts used to perform these analyses and generate figures are hosted in a public GitHub repository (https://github.com/kdews/s-latissima-genome).
2.7 NCBI decontamination
The v1 assembly was screened for vector using the NCBI UniVec database (https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec) using the standard command blastall -p blastn -d Saccharina_latissima.mainGenome.fasta -i UniVec -q −5 -G 3 -E 3 -F ″m D” -e 700 -Y 1.75e12 -m 8 (Camacho et al., 2009). A total of 43 vector hits were identified with≥95% identity and≥31bp in length. Contaminants were identified using the contaminant screener FCS-GX (Astashyn et al., 2024). A total of 90 contaminated regions were identified (63 TRIM, 21 FIX, 6 EXCLUDE). All identified vector and contaminant regions were removed from the sequence. If a vector/contaminant region fell within a scaffold, then the region was replaced with the same number of N’s. If the vector/contaminant region fell on the front/end of a scaffold, the bases were eliminated, and the annotation GFF file was translated appropriately. These changes resulted in a net loss of 53 scaffolds, 4,498,393 bp, and a total of 209 annotated genes.
2.8 Organelle genome assemblies
Organelle genomes from the same female S. latissima gametophyte (SL-CT1-FG3) were also assembled using the PacBio reads generated for the nuclear genome assembly. Reference organelle genomes for S. latissima have been published (Wang et al., 2016; Fan et al., 2020b), but these genomes do not have corresponding nuclear genomes from the same individual. Gene transfers between organelle and nuclear genomes are more easily identified when using genomes sourced from the same individual (Cui et al., 2021). To identify organelle reads, raw PacBio reads were aligned to the S. latissima mitochondrial (Wang et al., 2016) and chloroplast (Fan et al., 2020b) genomes using minimap2 standard settings (Li, 2018). Read IDs that aligned to the organelle genomes were extracted using samtools (Li et al., 2009), and then corresponding PacBio reads that aligned to the respective organelle genomes were subset using seqtk subseq (Li, 2012). Organelle genomes were then assembled using the assembler flye v2.9.2-b1786 (Kolmogorov et al., 2019), with the--pacbio-raw flag and genome size estimates based on previously published sugar kelp chloroplast (Fan et al., 2020b) and mitochondrial (Wang et al., 2016) genomes. We used GeSeq2 (Tillich et al., 2017) to annotate and compare our sugar kelp organelle genomes versus available published reference S. latissima organelle genomes (Wang et al., 2016; Fan et al., 2020b), using genome annotations of U. pinnatifida mitochondria (Li et al., 2015) and chloroplast (Zhang Y. et al., 2016) as outgroups. A collection of scripts used in this analysis have been placed in a public GitHub repository (https://github.com/kdews/s-latissima-organelles).
3 Results
3.1 Nuclear genome assembly
3.1.1 General statistics
Long-read sequencing of an individual S. latissima gametophyte (var. SL-CT1-FG3) yielded 111 Gb of PacBio HiFi reads, estimated to represent ∼180x genomic coverage. Our initial de novo assembly (v0) was 925.8 Mb and consisted of 4,854 contigs (Supplementary Table S3). Following contaminant filtering, a total of 3,341 contigs (283.9 Mb) were removed. Hi-C scaffolding joined 333 contigs from the v0 genome into 218 scaffolds in the v1 genome. The final S. latissima genome (v1) described here contains a total of 218 scaffolds and 1,513 contigs, with a genome size of 615.5 Mb (0.5% gaps) and scaffold N50 of 1.35 Mb (Figure 1; Table 1). Gene annotation with the JGI annotation pipeline (Grigoriev et al., 2006) yielded 24,790 filtered gene models and 25,012 functionally annotated protein sequences. All annotations are available on the JGI PhycoCosm portal (Grigoriev et al., 2021) for our S. latissima v1 genome (https://phycocosm.jgi.doe.gov/SlaSLCT1FG3_1).
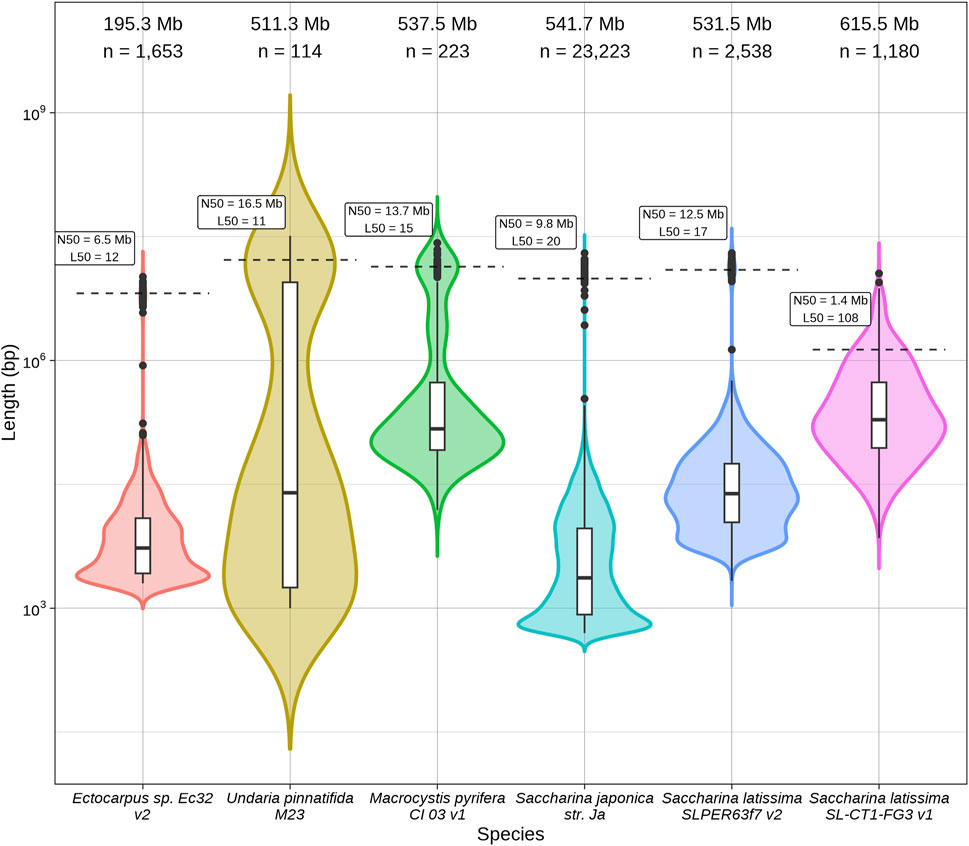
Figure 1. Log10-scaled size distribution for six brown algae genome assembly contigs and scaffolds: Ectocarpus sp. Ec32 v2 (Cormier et al., 2017), Undaria pinnatifida M23 (Shan et al., 2020), Macrocystis pyrifera CI_03 v1 (Diesel et al., 2023), Saccharina japonica str. Ja (Fan et al., 2020a), European Saccharina latissima SLPER63f7 v2 (Denoeud et al., 2024) and North American Saccharina latissima SL-CT1-FG3 v1 (this publication).
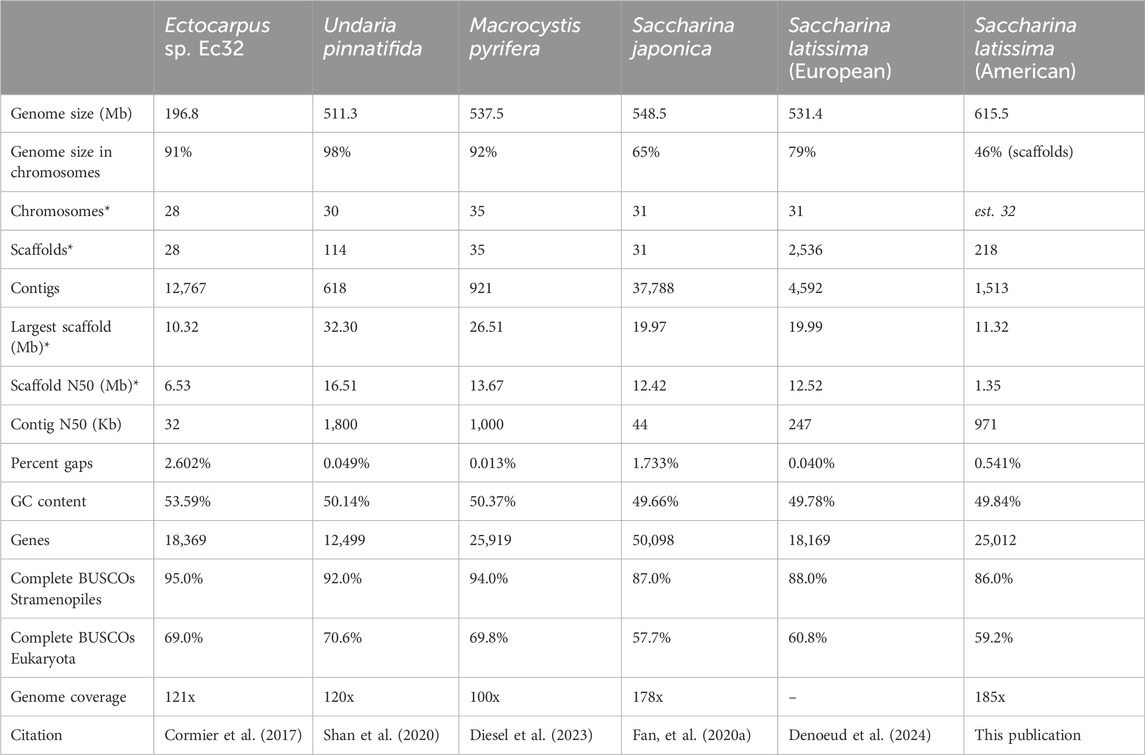
Table 1. Comparison of Saccharina latissima nuclear genome assembly statistics to those of related brown macroalgal species. Number of genes reported reflects conservatively curated gene models reported for each specific genome assembly. *Excludes artificial chromosomes.
To evaluate the quality and completeness of the North American S. latissima genome assembly reported here, we compared to the genomes of four related brown algae: the model brown alga Ectocarpus sp. (Cormier et al., 2017), giant kelp M. pyrifera (Diesel et al., 2023), and the widely cultivated kelps wakame U. pinnatifida (Shan et al., 2020) and Japanese sugar kelp S. japonica (Fan et al., 2020a) (Figure 1; Table 1). Following the recent publication of the Phaeoexplorer brown algal genome database (Denoeud et al., 2024), the European S. latissima nuclear assembly was also included for comparison on summary statistics (Figure 1; Table 1). Three early genome size predictions for S. latissima used standard methods of staining and flow cytometry to estimate a genome size of 588–720 Mb (Phillips et al., 2011). These genome size estimates agree with our v1 assembly size of 615.5 Mb. The sequenced length and GC content of this S. latissima genome assembly is generally comparable to the other two Saccharina genomes; our North American S. latissima assembly contains ∼84 Mb more sequence than the European assembly (Table 1).
3.1.2 Conserved gene content
Gene content was evaluated with BUSCO, which evaluates a given assembly against the set of single-copy, highly conserved orthologous genes predicted to be present in a specific clade (Manni et al., 2021). Detection of conserved gene orthologs in de novo assemblies infers genome completeness, especially when comparing single-copy orthologs amongst species within a monophyletic clade (Waterhouse et al., 2011). We benchmarked the six compared genomes against two relevant clades: Eukaryota, which provides a general metric for comparison across all conserved Eukaryota genes, and Stramenopiles genes, the monophyletic clade containing brown algae. The percentage of complete BUSCOs (comprising single copy and duplicate orthologs) detected in an assembly can be used as a proxy for genome completeness and to detect artificial duplications resulting from de novo assembly. Benchmarked against Eukaryota, our North American S. latissima assembly (59.2% complete BUSCOs) scores closely to S. japonica (57.7%) and the European S. latissima assembly (60.8%); similarly, against Stramenopiles this assembly scored 86%, compared to 88% in S. japonica and the European S. latissima (Table 1; Figure 2). Of the 100 BUSCOs in Stramenopiles, our genome contains 86 complete BUSCOs (86%), with 85 single copy and 1 duplicated, and the remainder fragmented (3) and missing (11) (Figure 2). Incomplete BUSCOs in the S. latissima genome could be attributed to lower contiguity, which would be further exacerbated by the Laminariales gene structure that often features long intronic regions. Long genes split between unassembled regions could be fragmented beyond the threshold of local alignment detection.
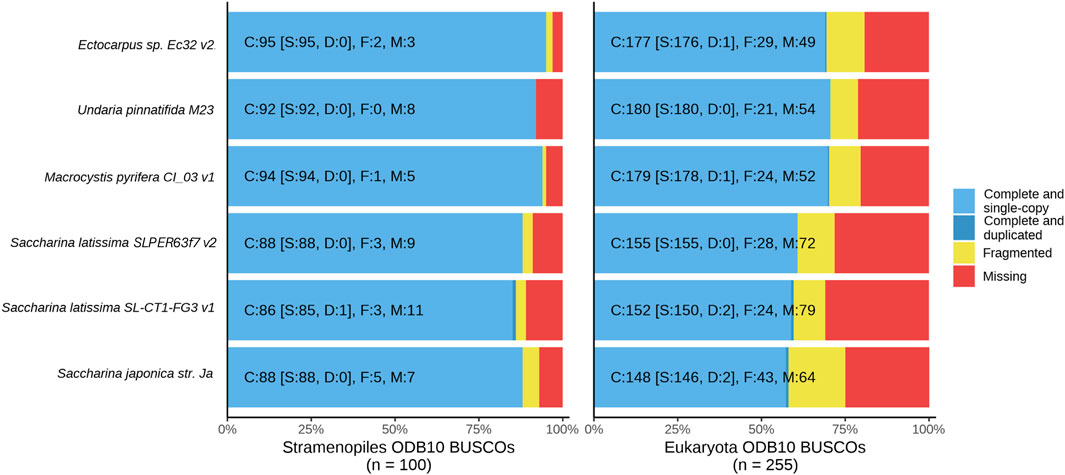
Figure 2. BUSCO scoring using the Stramenopiles and Eukaryota ortholog databases (odb10) shows relative counts of complete (blue), fragmented (yellow), and missing (red) orthologs in each compared brown algae assembly, Ectocarpus sp. Ec32 v2 (Cormier et al., 2017), Undaria pinnatifida M23 (Shan et al., 2020), Macrocystis pyrifera CI_03 v1 (Diesel et al., 2023), Saccharina japonica str. Ja (Fan et al., 2020a), European Saccharina latissima SLPER63f7 v2 (Denoeud et al., 2024) and North American Saccharina latissima SL-CT1-FG3 v1 (this publication).
3.2 Synteny analysis
3.2.1 Interspecies whole genome alignment
To capture canonical genomic rearrangements between related species, we performed hierarchal whole multi-genome alignments (Armstrong et al., 2020) of our S. latissima assembly against the genomes of four related brown macroalgal species, which formed the basis of our homology analysis. Sequence homology to at least one of the compared genomes was detected in 99% of our assembly across 1123 scaffolds and contigs (613.38 Mb). On average, exact matches spanned 15% of each scaffold in S. latissima. A core set of 858 scaffolds and contigs (525.43 Mb) in the S. latissima genome aligned the genome assemblies of each of the four compared brown algal species (Figure 3A).

Figure 3. (A) Venn diagram highlights intersects between sets of homologous Saccharina latissima v1 assembly scaffolds that uniquely mapped to the genomes of four related species: Ectocarpus sp. Ec32, Saccharina japonica, Undaria pinnatifida, and Macrocystis pyrifera. (B) Heatmap shows the maximal syntenic match of 1,110 S. latissima scaffolds and contigs to chromosomes of the same four related brown algal species.
As expected, S. latissima had the highest total exact matches to S. japonica of all species, both genome-wide (181.89 Mb) and averaged per chromosome (5.68 Mb) (Supplementary Table S5). Overall, summed exact matches between our v1 S. latissima assembly and each compared assembly increases with respective species relatedness to S. latissima, a trend that holds both genome-wide and per chromosome (Supplementary Figure S1B; Supplementary Table S5). Linear regression of S. latissima scaffold lengths versus their respective homologous reference chromosome lengths yielded slopes that closely correspond to genome size ratios, with the smallest reference, Ectocarpus sp. Ec32, reflecting a 3x shorter genome with a slope of 3.06 (Supplementary Figure S1A).
3.2.2 Genome contiguity and chromosome number estimation
Despite high genomic sequencing coverage (185x) with long PacBio reads, as well as Hi-C sequencing to 145x coverage, our reported S. latissima v1 genome assembly could not be scaffolded to the level of chromosomes based on sequencing and contact information alone. Our v1 assembly contains 218 scaffolds, and the scaffold N50 of our v1 S. latissima assembly (1.35 Mb) is an order of magnitude smaller than similarly sized brown macroalgal genomes (Table 1). Chromosome number estimates in brown macroalgae are complicated due to sex-specific polyteny and population-specific ploidy variation in sugar kelp meristem sporophyte tissue (Müller et al., 2016; Goecke et al., 2022). Chromosome number predictions in Saccharina species vary depending upon which method is used: flow cytometry has estimated a genome ranging from 588 to 720 Mb with 62 chromosomes (Phillips et al., 2011), while microscopy has estimated 31 chromosomes (Liu et al., 2012; Liu et al., 2022). Recent brown macroalgal genome assemblies have predicted 28–34 chromosomes (Cormier et al., 2017; Fan et al., 2020a; Shan et al., 2020; Diesel et al., 2023). The European S. latissima genome (Denoeud et al., 2024) was able to scaffold 2,085 of 4,592 contigs into 31 pseudochromosomes using genetic linkage mapping in its v2 release (February 2025), which aligns with microscopy estimates for chromosome count in Saccharina.
We applied the results of our hierarchal whole-genome alignments of S. latissima to Ectocarpus sp. Ec32, S. japonica, U. pinnatifida, and M. pyrifera to identify chromosomes homologous to our v1 scaffolds. For each scaffold in the S. latissima v1 genome assembly that aligned, we calculated a maximal syntenic match to a single chromosome in each related genome (see Methods 3.6). Accounting for the sub-chromosomal length of our scaffolds, we assigned maximal syntenic matches with respect to S. latissima, with each scaffold matching only one chromosome, while chromosomes could be assigned multiple homologous S. latissima scaffolds (one-chromosome-to-many-scaffolds). With this schema, we visualized S. latissima scaffold alignments along the length of chromosomes in each of the four aligned genomes (Figure 3B).
We estimated chromosome number in S. latissima by leveraging syntenic information to order our assembly scaffolds into pseudochromosomes. We repeatedly re-scaffolded our v1 assembly under varying parameters (Kolmogorov et al., 2018) into 32–40 pseudochromosomes that incorporated 92–464 scaffolds and contigs (Supplementary Figures S2, 3), reflecting 40%–55% of genome sequence. Restricting our re-scaffolding to the 155 longest S. latissima scaffolds and allowing for chimeric assembly yielded 32 pseudochromosomes (Supplementary Figures S2D, S3D), the closest result to the predicted 31 chromosomes for Saccharina. This 32-pseudochromosome assembly was then used to map gene orthology between the genomes of S. latissima and U. pinnatifida (Shan et al., 2020) (Figure 4B).
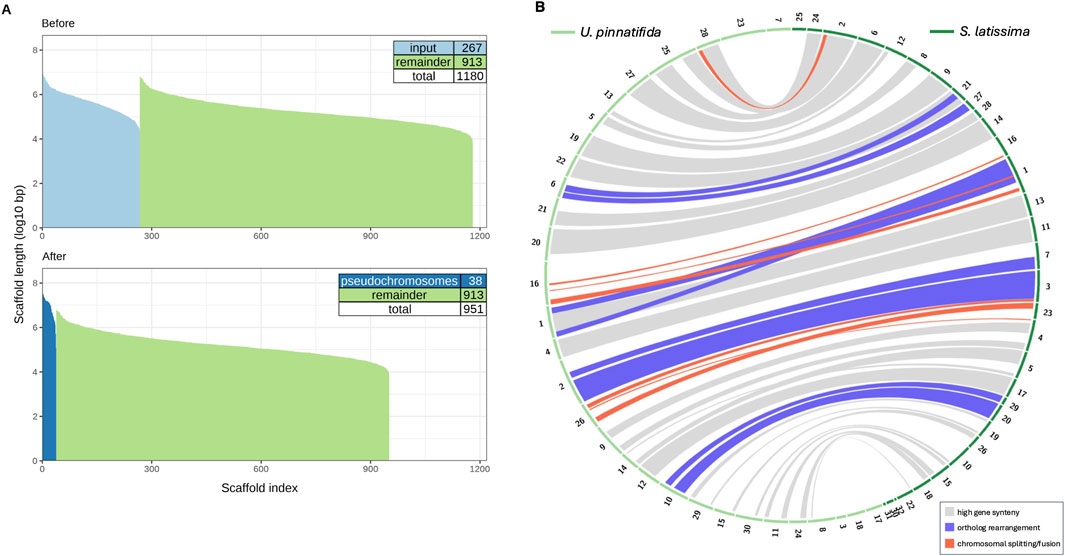
Figure 4. (A) Size distribution (log10 bp scale) of our v1 Saccharina latissima assembly before and after synteny-based re-scaffolding show 267 scaffolds (light blue) incorporated into 38 psuedochromosomes (dark blue). (B) Gene orthology between 32 S. latissima psuedochromosomes (dark green) and 38 Undaria pinnatifida chromosomes and contigs (light green) was mapped using 3 Mb windows containing 10 single-copy orthologs, with band colors denoting highest synteny (gray), ortholog rearrangement (red) and chromosomal splitting or fusion (purple).
3.3 Organelle genomes
Our assembled S. latissima chloroplast genome (130,613 bp) is almost the same length as the published chloroplast genome (130,619 bp) (Fan et al., 2020b) (Table 2; Supplementary Figure S4; Supplementary Figure S6B). Our assembled S. latissima mitochondrial genome is 37,510 bp and contains 39 genes. It is slightly smaller than the previously published mitochondrial genome (37,659 bp) (Wang et al., 2016), but contains an additional tRNA annotation (Table 2; Supplementary Figure S5; Supplementary Figure S6A). For each of the new organelle genomes reported here, a consensus sequence was generated through multiple separate assemblies with flye (Kolmogorov et al., 2019), a long-read assembler especially robust to sequencing errors and specialized to resolve repetitive regions. Long reads allowed for resolution of inverted repeat (IR) regions in the chloroplast genome that typically span ∼5.5 kb in brown macroalgae (Rana et al., 2019). The average sequencing read length (8,389 bp) also aided with assembly, as each read covers ∼22% and ∼6% of mitochondria and chloroplast genome lengths, respectively.
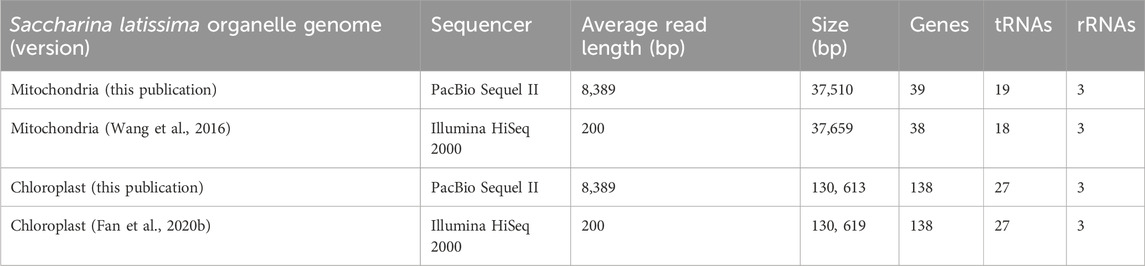
Table 2. Comparison of Saccharina latissima organelle genome versions on assembly and gene annotation statistics.
4 Discussion
4.1 Enhancing sugar kelp breeding with genomic tools
The decrease in sequencing costs has led to an increase in genome assemblies for non-model species including brown algae, which until recently have lacked genomic resources. Nuclear genomes are now becoming available for some Phaeophyta species, e.g., Ectocarpus sp. (Cock et al., 2010; Cormier et al., 2017), S. japonica (Ye et al., 2015; Liu et al., 2019; Fan et al., 2020a), U. pinnatifida (Shan et al., 2020; Graf et al., 2021), and S. latissima (Denoeud et al., 2024; this publication). This annotated and scaffolded genome of North American sugar kelp (S. latissima) represents a major step forward in kelp research and development in the United States. Our assembly provides a reference for future research into population genetics and gene expression in sugar kelp populations along the coast of the northeastern US and supports both marine conservation efforts and the nascent US kelp industry.
Many tools used in breeding rely on the availability of high-quality genomes and rigorous gene annotation. Demands on genome assembly coverage, accuracy, and annotation have increased as selective breeding methods have progressed (Varshney et al., 2014; International Wheat Genome Sequencing Consortium et al., 2018; Bayer et al., 2020). With sparse maps of genetic markers, marker-assisted selection (MAS) achieved modest improvements over phenotypic selection in the prediction of breeding values for crops and livestock (Dekkers and Hospital, 2002). Modern genomic selection (GS) models depend on high-density genotypic data to generate genomic estimated breeding values (GEBVs) (Meuwissen et al., 2001) that take genome-wide variation into account (Calus and Veerkamp, 2007; Heffner et al., 2009; Christensen and Lund, 2010). To accurately predict the performance of genetic crosses with these methods, chromosome rearrangements between them must be known (Nuzhdin et al., 2012). Additionally, genome engineering (e.g., CRISPR/Cas9 gene editing) relies on the construction of accurate genome assemblies for cultivated species to mitigate potential off-target genome modification (Guo et al., 2023). Here, we have assembled a genome for S. latissima of sufficient quality to assess genetic variation through a variety of markers (e.g., SNPs, INDELs, structural variants) useful for the progression of intraspecific breeding programs (Li et al., 2022), as well as possible crossbreeding with closely related species such as S. japonica (Zhang J. et al., 2016; 2018).
4.2 Improved organelle genome resources for sugar kelp
In addition to nuclear genomes of brown algae, publications of plastid and mitochondria genome assemblies are also mounting (Oudot-Le Secq et al., 2006; Chen et al., 2019; Rana et al., 2021). While genomics has greatly increased breeding efficiency in plants, animals, and kelp, most of these breeding programs rely strictly on nuclear genomes for establishing markers for selective breeding. However, integrating organelle genomes into breeding models may increase breeding efficiency (Kersten et al., 2016). By optimizing cytonuclear interactions, breeding efficiency for sugar kelp in the future can be accelerated (Colombo, 2019). As nuclear and organelle genomes can differ between individuals in the same species, pairing nuclear and cytoplasmic genomes from a single genotype can provide a reference for future breeding experiments. For S. latissima, short-read organelle genome assemblies have become available over the past decade (Wang et al., 2016; Fan et al., 2020b; Rana et al., 2021). Our use of long-read sequencing increases our confidence in the nuclear and organelle S. latissima genome assemblies described here to serve as a foundation for future sugar kelp scientific research and breeding.
4.3 Advancements in brown algal genome assembly and comparative genomics
The whole-genome hierarchal alignments conducted in this study produced syntenic data applicable to both our genome improvement and comparative genomic analyses, and future research into brown algae evolution, phylogenetics, and genomic breeding. High molecular weight DNA extraction is particularly challenging for macroalgal species (Snirc et al., 2010; Greco et al., 2014), which can pose a significant barrier to generating chromosome-level assemblies. In general, genome quality and completeness are assessed through gene annotation and evaluation of sequence contiguity. From a structural perspective, genome contiguity (i.e., N50, gaps/N content) informs the degree to which assembly and scaffolding methods have succeeded in reconstructing chromosomes from whole-genome shotgun sequencing.
In brown macroalgae, Ectocarpus sp. Ec32 v2 (Cormier et al., 2017), U. pinnatifida (Shan et al., 2020), and M. pyrifera (Diesel et al., 2023) represent the most highly scaffolded genome assemblies, with greater than 90% of sequence contained in chromosomal scaffolds. In contrast, although the S. japonica assembly (Fan et al., 2020a) has been mapped into chromosomes, almost 35% of the genome is not scaffolded. A similar pattern exists in the European S. latissima v2 assembly (Denoeud et al., 2024), in which >20% of the genome was not incorporated into pseudochromosomes. Our North American S. latissima v1 genome assembly contains 218 scaffolds that incorporate ∼45% of the genome by length. These results are despite high (>100x) genomic coverage with long reads and Hi-C-assisted assembly methods for all genomes compared in this study (Table 1). Though not conclusive, the relative difficulty of de novo chromosomal reconstruction in Saccharina genome assemblies could indicate physical traits or genome architecture specific to this genus that impede efforts to sequence and scaffold DNA, for example, elevated concentrations of polysaccharides and/or phenols or higher genomic repeat content in the genus Saccharina as compared to other kelps. To overcome obstacles to chromosome-level de novo assembly, we demonstrate the utility of syntenic information to estimate chromosome number and assist in scaffold placement along chromosomes in S. latissima.
Importantly, our comparative genomics analysis has identified syntenic regions and homologous chromosomes between five species of brown algae. Construction of the brown algal phylogeny has historically been complicated and contested, both in morphological and genetic studies, owing to a sparse brown algal fossil record, rapid diversification, and convergent evolution (Bolton, 2010; Silberfeld et al., 2010; Starko et al., 2019; Bringloe et al., 2020; Choi et al., 2024; Denoeud et al., 2024). Building on syntenic blocks we identified among brown algal genomes, phylogenomic analyses that evaluate whole genomes and investigate chromosome homology and evolution have the potential to resolve outstanding problems of placement within this clade radiation (Steenwyk and King, 2024). Comparative genomic analyses like those presented here help correlate chromosomes between species for more robust evolutionary analysis and genomic breeding prediction that leverage the increasing amount of high-quality, functionally annotated brown macroalgal genome assemblies.
5 Future directions
This scaffolded and annotated sugar kelp (S. latissima) genome is a fundamental resource for subsequent basic science research, genomic breeding, and conservation in sugar kelp. Already, this genome has served as the backbone of recent applied genomic advances of a sugar kelp selective breeding project, including a genomic selection model targeting higher yield (Umanzor et al., 2021; Li et al., 2022; Huang et al., 2023). Despite several years of work on the nuclear genome assembly for S. latissima, the reference can and should be further improved with the goal of a chromosome-level genome. As we have demonstrated, this and other high-quality brown macroalgae genomes allow for comparative genomics studies to examine the evolution of chromosome structure and gene content across brown algae. To investigate sugar kelp genetic diversity and evolution across important biogeographic regions such as Alaska, Gulf of Maine, and Europe, future work should look to assemble a sugar kelp pan-genome representing global genetic variation across known sugar kelp populations.
The importance of genomics to kelp domestication projects has been highlighted by other industry leaders (Goecke et al., 2022; Hu et al., 2023). Since sugar kelp is the most farmed kelp in the United States and Europe, publication of an annotated reference genome for North American sugar kelp alongside the existing European sugar kelp genome (Denoeud et al., 2024) will advance breeding opportunities for the kelp farming industry worldwide. The knowledge presented here is crucial to genomic selection for superior traits, such as high yield, disease resistance, and stress tolerance, to bring improved sugar kelp cultivars to United States markets.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA1156187 (https://www.ncbi.nlm.nih.gov) and SlaSLCT1FG3_1 (https://phycocosm.jgi.doe.gov/SlaSLCT1FG3_1).
Author contributions
KD: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. GM: Conceptualization, Formal Analysis, Methodology, Visualization, Writing – original draft, Writing – review and editing. SC: Data curation, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft. AL: Data curation, Formal Analysis, Methodology, Resources, Writing – original draft. JJ: Data curation, Formal Analysis, Methodology, Writing – original draft. MW: Data curation, Writing – original draft. CP: Data curation, Writing – original draft. JT: Data curation, Writing – original draft. JG: Data curation, Writing – original draft. J-LJ: Conceptualization, Investigation, Supervision, Writing – review and editing. IG: Data curation, Formal Analysis, Resources, Supervision, Writing – review and editing. JS: Data curation, Formal Analysis, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review and editing. CY: Conceptualization, Funding acquisition, Supervision, Writing – review and editing. SN: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review and editing. SL: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project was supported by the U.S. Department of Energy (DOE) Advanced Research Projects Agency–Energy (ARPA-E) program Macroalgae Research Inspiring Novel Energy Resources (MARINER) awards DE-AR0000914 and DE-AR0000915.
Acknowledgments
We would like to thank University of Connecticut researchers Simona Augyte, Yaoguang Li, Schery Umanzor, and Michael Marty-Rivera for establishing and culturing the Saccharina latissima germplasm for sequencing. We would also like to thank University of Southern California researchers José Diesel, Maxim Kovalev and Jordan Chancellor for their contributions to figure design and validation.
A portion of these data were produced by the U.S. Department of Energy Joint Genome Institute in collaboration with the user community. The work conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1494480/full#supplementary-material
References
Abbass, K., Qasim, M. Z., Song, H., Murshed, M., Mahmood, H., and Younis, I. (2022). A review of the global climate change impacts, adaptation, and sustainable mitigation measures. Environ. Sci. Pollut. Res. 29 (28), 42539–42559. doi:10.1007/s11356-022-19718-6
Alsuwaiyan, N. A., Mohring, M. B., Cambridge, M., Coleman, M. A., Kendrick, G. A., and Wernberg, T. (2019). A review of protocols for the experimental release of kelp (Laminariales) zoospores. Ecol. Evol. 9 (14), 8387–8398. doi:10.1002/ece3.5389
Armstrong, J., Hickey, G., Diekhans, M., Fiddes, I. T., Novak, A. M., Deran, A., et al. (2020). Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587 (7833), 246–251. doi:10.1038/s41586-020-2871-y
Astashyn, A., Tvedte, E. S., Sweeney, D., Sapojnikov, V., Bouk, N., Joukov, V., et al. (2024). Rapid and sensitive detection of genome contamination at scale with FCS-GX. Genome Biol. 25 (1), 60. doi:10.1186/s13059-024-03198-7
Augyte, S., Lewis, L., Lin, S., Neefus, C. D., and Yarish, C. (2018). Speciation in the exposed intertidal zone: the case of Saccharina angustissima comb. nov. and stat. nov. (Laminariales, Phaeophyceae). Phycologia 57 (1), 100–112. doi:10.2216/17-40.1
Bartholomé, J., Mandrou, E., Mabiala, A., Jenkins, J., Nabihoudine, I., Klopp, C., et al. (2015). High-resolution genetic maps of Eucalyptus improve Eucalyptus grandis genome assembly. New Phytol. 206 (4), 1283–1296. doi:10.1111/nph.13150
Bayer, P. E., Golicz, A. A., Scheben, A., Batley, J., and Edwards, D. (2020). Plant pan-genomes are the new reference. Nat. Plants 6 (8), 914–920. doi:10.1038/s41477-020-0733-0
Bemmels, J. B., Starko, S., Weigel, B. L., Hirabayashi, K., Pinch, A., Elphinstone, C., et al. (2025). Population genomics reveals strong impacts of genetic drift without purging and guides conservation of bull and giant kelp. Curr. Biol. 35 (3), 688–698.e8. doi:10.1016/j.cub.2024.12.025
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30 (6), 555–561. doi:10.1038/nbt.2196
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and genomewise. Genome Res. 14 (5), 988–995. doi:10.1101/gr.1865504
Bolton, J. J. (2010). The biogeography of kelps (Laminariales, Phaeophyceae): a global analysis with new insights from recent advances in molecular phylogenetics. Helgol. Mar. Res. 64 (4), 263–279. doi:10.1007/s10152-010-0211-6
Brakel, J., Sibonga, R. C., Dumilag, R. V., Montalescot, V., Campbell, I., Cottier-Cook, E. J., et al. (2021). Exploring, harnessing and conserving marine genetic resources towards a sustainable seaweed aquaculture. Plants People Planet 3 (4), 337–349. doi:10.1002/ppp3.10190
Brayden, C., and Coleman, S. (2023). Maine seaweed benchmarking report. Available online at: https://maineaqua.org/wp-content/uploads/2023/08/Maine-Seaweed-Benchmarking-Report.pdf.
Breton, T. S., Nettleton, J. C., O’Connell, B., and Bertocci, M. (2018). Fine-scale population genetic structure of sugar kelp, Saccharina latissima (Laminariales, Phaeophyceae), in eastern Maine, USA. Phycologia 57 (1), 32–40. doi:10.2216/17-72.1
Bringloe, T. T., Starko, S., Wade, R. M., Vieira, C., Kawai, H., Clerck, O. D., et al. (2020). Phylogeny and evolution of the Brown algae. Crit. Rev. Plant Sci. 39 (4), 281–321. doi:10.1080/07352689.2020.1787679
Buckley, S., Hardy, K., Hallgren, F., Kubiak-Martens, L., Miliauskienė, Ž., Sheridan, A., et al. (2023). Human consumption of seaweed and freshwater aquatic plants in ancient Europe. Nat. Commun. 14 (1), 6192. doi:10.1038/s41467-023-41671-2
Budhlakoti, N., Kushwaha, A. K., Rai, A., Chaturvedi, K. K., Kumar, A., Pradhan, A. K., et al. (2022). Genomic selection: a tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 13, 832153. doi:10.3389/fgene.2022.832153
Cai, J., Lovatelli, A., Aguilar-Manjarrez, J., Cornish, L., Dabbadie, L., et al. (2021). “Seaweeds and microalgae: an overview for unlocking their potential in global aquaculture development,”, 1229. Rome: FAO. doi:10.4060/cb5670en
Calus, M. P. L., and Veerkamp, R. F. (2007). Accuracy of breeding values when using and ignoring the polygenic effect in genomic breeding value estimation with a marker density of one SNP per cM. J. Animal Breed. Genet. 124 (6), 362–368. doi:10.1111/J.1439-0388.2007.00691.X
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinforma. 10 (1), 421. doi:10.1186/1471-2105-10-421
Chen, J., Zang, Y., Shang, S., and Tang, X. (2019). The complete mitochondrial genome of the brown alga Macrocystis integrifolia (Laminariales, Phaeophyceae). Mitochondrial DNA Part B 4 (1), 635–636. doi:10.1080/23802359.2018.1495114
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H., and Li, H. (2021). Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18 (2), 170–175. doi:10.1038/s41592-020-01056-5
Choi, S.-W., Graf, L., Choi, J. W., Jo, J., Boo, G. H., Kawai, H., et al. (2024). Ordovician origin and subsequent diversification of the brown algae. Curr. Biol. 34 (4), 740–754.e4. doi:10.1016/j.cub.2023.12.069
Christensen, O. F., and Lund, M. S. (2010). Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 42 (1), 2. doi:10.1186/1297-9686-42-2
Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465 (7298), 617–621. doi:10.1038/nature09016
Coleman, M. A., and Glasby, T. M. (2024). Breakthrough innovation will be essential for scaling up marine macrophyte restoration and achieving targets. J. Phycol. 60 (4), 781–784. doi:10.1111/jpy.13484
Colombo, N. (2019). Taking advantage of organelle genomes in plant breeding: an integrated approach. J. Basic Appl. Genet. 30 (1), 35–51. doi:10.35407/bag.2019.01.05
Cormier, A., Avia, K., Sterck, L., Derrien, T., Wucher, V., Andres, G., et al. (2017). Re-annotation, improved large-scale assembly and establishment of a catalogue of noncoding loci for the genome of the model brown alga Ectocarpus. New Phytol. 214 (1), 219–232. doi:10.1111/nph.14321
Cui, H., Ding, Z., Zhu, Q., Wu, Y., Qiu, B., and Gao, P. (2021). Comparative analysis of nuclear, chloroplast, and mitochondrial genomes of watermelon and melon provides evidence of gene transfer. Sci. Rep. 11 (1), 1595. doi:10.1038/s41598-020-80149-9
Dayton, P. K. (1985). Ecology of kelp communities. Annu. Rev. Ecol. Syst. 16 (1), 215–245. doi:10.1146/annurev.es.16.110185.001243
Dekkers, J. C. M., and Hospital, F. (2002). The use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 3 (1), 22–32. doi:10.1038/nrg701
Denoeud, F., Godfroy, O., Cruaud, C., Heesch, S., Nehr, Z., Tadrent, N., et al. (2024). Evolutionary genomics of the emergence of brown algae as key components of coastal ecosystems. Cell 187 (24), 6943–6965.e39. doi:10.1016/j.cell.2024.10.049
Desta, Z. A., and Ortiz, R. (2014). Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19 (9), 592–601. doi:10.1016/j.tplants.2014.05.006
DeWeese, K. J., and Osborne, M. G. (2021). Understanding the metabolome and metagenome as extended phenotypes: the next frontier in macroalgae domestication and improvement. J. World Aquac. Soc. 52 (5), 1009–1030. doi:10.1111/jwas.12782
Diehl, N., Li, H., Scheschonk, L., Burgunter-Delamare, B., Niedzwiedz, S., Forbord, S., et al. (2024). The sugar kelp Saccharina latissima I: recent advances in a changing climate. Ann. Bot. 133 (1), 183–212. doi:10.1093/aob/mcad173
Diesel, J., Molano, G., Montecinos, G. J., DeWeese, K., Calhoun, S., Kuo, A., et al. (2023). A scaffolded and annotated reference genome of giant kelp (Macrocystis pyrifera). BMC Genomics 24 (1), 543. doi:10.1186/s12864-023-09658-x
Dijkstra, E. W. (1959). A note on two problems in connexion with graphs. Numer. Math. 1 (1), 269–271. doi:10.1007/BF01386390
Ding, Q., Li, R., Ren, X., Chan, L., Ho, V. W. S., Xie, D., et al. (2022). Genomic architecture of 5S rDNA cluster and its variations within and between species. BMC Genomics 23 (1), 238. doi:10.1186/s12864-022-08476-x
Doyle, J. J., and Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus 12 (1), 13–15.
Dring, M. J., and Lüning, K. (1975). A photoperiodic response mediated by blue light in the brown alga Scytosiphon lomentaria. Planta 125 (1), 25–32. doi:10.1007/BF00388870
Dudchenko, O., Batra, S. S., Omer, A. D., Nyquist, S. K., Hoeger, M., Durand, N. C., et al. (2017). De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356 (6333), 92–95. doi:10.1126/science.aal3327
Durand, N. C., Shamim, M. S., Machol, I., Rao, S. S. P., Huntley, M. H., Lander, E. S., et al. (2016). Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3 (1), 95–98. doi:10.1016/j.cels.2016.07.002
Eger, A. M., Marzinelli, E. M., Beas-Luna, R., Blain, C. O., Blamey, L. K., Byrnes, J. E. K., et al. (2023). The value of ecosystem services in global marine kelp forests. Nat. Commun. 14 (1), 1894. doi:10.1038/s41467-023-37385-0
Fan, X., Han, W., Teng, L., Jiang, P., Zhang, X., Xu, D., et al. (2020a). Single-base methylome profiling of the giant kelp Saccharina japonica reveals significant differences in DNA methylation to microalgae and plants. New Phytol. 225 (1), 234–249. doi:10.1111/nph.16125
Fan, X., Xie, W., Wang, Y., Xu, D., Zhang, X., and Ye, N. (2020b). The complete chloroplast genome of Saccharina latissima. Mitochondrial DNA Part B 5 (3), 3481–3482. doi:10.1080/23802359.2020.1825999
FAO (2023). “Statistical Query panel - global aquaculture production (1984-2022),” in FAO fisheries and aquaculture division. Rome: FAO. Available online at: https://www.fao.org/fishery/statistics-query/en/aquaculture/aquaculture_value.
FAO (2024). The state of world fisheries and aquaculture 2024 – blue transformation in action. Rome: FAO. doi:10.4060/cd0683en
Ghurye, J., Rhie, A., Walenz, B. P., Schmitt, A., Selvaraj, S., Pop, M., et al. (2019). Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLOS Comput. Biol. 15 (8), e1007273. doi:10.1371/journal.pcbi.1007273
Goecke, F., Gómez Garreta, A., Martín–Martín, R., Rull Lluch, J., Skjermo, J., and Ergon, Å. (2022). Nuclear DNA content variation in different life cycle stages of sugar kelp, Saccharina latissima. Mar. Biotechnol. 24 (4), 706–721. doi:10.1007/s10126-022-10137-9
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Graf, L., Shin, Y., Yang, J. H., Choi, J. W., Hwang, I. K., Nelson, W., et al. (2021). A genome-wide investigation of the effect of farming and human-mediated introduction on the ubiquitous seaweed Undaria pinnatifida. Nat. Ecol. and Evol. 5 (3), 360–368. doi:10.1038/s41559-020-01378-9
Greco, M., Sáez, C. A., Brown, M. T., and Bitonti, M. B. (2014). A simple and effective method for high quality Co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model Brown alga. PLOS ONE 9 (5), e96470. doi:10.1371/journal.pone.0096470
Grigoriev, I. V., Hayes, R. D., Calhoun, S., Kamel, B., Wang, A., Ahrendt, S., et al. (2021). PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res. 49 (D1), D1004–D1011. doi:10.1093/nar/gkaa898
Grigoriev, I. V., Martinez, D. A., and Salamov, A. A. (2006). “Fungal genomic annotation,” in Applied mycology and Biotechnology (Elsevier), 123–142. doi:10.1016/S1874-5334(06)80008-0
Guo, C., Ma, X., Gao, F., and Guo, Y. (2023). Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 11, 1143157. doi:10.3389/fbioe.2023.1143157
Heffner, E. L., Sorrells, M. E., and Jannink, J. (2009). Genomic selection for crop improvement. Crop Sci. 49 (1), 1–12. doi:10.2135/cropsci2008.08.0512
Heidkamp, C. P., Krak, L. V., Kelly, M. M. R., and Yarish, C. (2022). Geographical considerations for capturing value in the U.S. sugar kelp (Saccharina latissima) industry. Mar. Policy 144, 105221. doi:10.1016/j.marpol.2022.105221
Hickey, G., Paten, B., Earl, D., Zerbino, D., and Haussler, D. (2013). HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29 (10), 1341–1342. doi:10.1093/bioinformatics/btt128
Hoffmann, A. J., and Santelices, B. (1991). Banks of algal microscopic forms: hypotheses on their functioning and comparisons with seed banks. Mar. Ecol. Prog. Ser. 79 (1/2), 185–194. doi:10.3354/meps079185
Hopfenberg, R. (2003). Human carrying capacity is determined by food availability. Popul. Environ. 25 (2), 109–117. doi:10.1023/b:poen.0000015560.69479.c1
Hu, Z., Shan, T., Zhang, Q., Liu, F., Jueterbock, A., Wang, G., et al. (2023). Kelp breeding in China: challenges and opportunities for solutions. Rev. Aquac. 16, 855–871. doi:10.1111/raq.12871
Huang, M., Robbins, K. R., Li, Y., Umanzor, S., Marty-Rivera, M., Bailey, D., et al. (2023). Genomic selection in algae with biphasic lifecycles: a Saccharina latissima (sugar kelp) case study. Front. Mar. Sci. 10. doi:10.3389/fmars.2023.1040979
Huang, M., Robbins, K. R., Li, Y., Umanzor, S., Marty-Rivera, M., Bailey, D., et al. (2022). Simulation of sugar kelp (Saccharina latissima) breeding guided by practices to accelerate genetic gains G3 (Bethesda) 12 (3). jkac003. doi:10.1093/g3journal/jkac003R. Wisser (ed.).
Hwang, E. K., Yotsukura, N., Pang, S. J., Su, L., and Shan, T. F. (2019). Seaweed breeding programs and progress in eastern Asian countries. Phycologia 58 (5), 484–495. doi:10.1080/00318884.2019.1639436
International Wheat Genome Sequencing Consortium (IWGSC) Appels, R., Eversole, K., Stein, N., Feuillet, C., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361 (eaar7191), eaar7191. doi:10.1126/science.aar7191
Jurka, J., Kapitonov, V. V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110 (1–4), 462–467. doi:10.1159/000084979
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 (90001), 277D–D280. doi:10.1093/nar/gkh063
Kent, W. J. (2002). BLAT —the BLAST-like alignment tool. Genome Res. 12 (4), 656–664. doi:10.1101/gr.229202
Kersten, B., Faivre Rampant, P., Mader, M., Le Paslier, M.-C., Bounon, R., Berard, A., et al. (2016). Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding A. Dhingra. PLOS ONE 11 (1), e0147209. doi:10.1371/journal.pone.0147209
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019a). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi:10.1038/s41587-019-0201-4
Kim, J., Stekoll, M., and Yarish, C. (2019b). Opportunities, challenges and future directions of open-water seaweed aquaculture in the United States. Phycologia 58 (5), 446–461. doi:10.1080/00318884.2019.1625611
Kim, J. K., Yarish, C., Hwang, E. K., Park, M., and Kim, Y. (2017). Seaweed aquaculture: cultivation technologies, challenges and its ecosystem services. Algae 32 (1), 1–13. doi:10.4490/algae.2017.32.3.3
Kolmogorov, M., Armstrong, J., Raney, B. J., Streeter, I., Dunn, M., Yang, F., et al. (2018). Chromosome assembly of large and complex genomes using multiple references. Genome Res. 28 (11), 1720–1732. doi:10.1101/gr.236273.118
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37 (5), 540–546. doi:10.1038/s41587-019-0072-8
Koonin, E. V., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Krylov, D. M., Makarova, K. S., et al. (2004). A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5 (2), R7. doi:10.1186/gb-2004-5-2-r7
Krasheninnikova, K., Diekhans, M., Armstrong, J., Dievskii, A., Paten, B., and O’Brien, S. (2020). halSynteny: a fast, easy-to-use conserved synteny block construction method for multiple whole-genome alignments. GigaScience 9 (6), giaa047. doi:10.1093/gigascience/giaa047
Lane, C. E., Mayes, C., Druehl, L. D., and Saunders, G. W. (2006). A multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic re-organization. J. Phycol. 42 (2), 493–512. doi:10.1111/j.1529-8817.2006.00204.x
Li, H. (2012). seqtk: toolkit for processing sequences in FASTA/Q formats. Available online at: https://github.com/lh3/seqtk.
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. doi:10.48550/arXiv.1303.3997
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34 (18), 3094–3100. doi:10.1093/bioinformatics/bty191
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Li, T.-Y., Qu, J.-Q., Feng, Y.-J., Liu, C., Chi, S., and Liu, T. (2015). Complete mitochondrial genome of Undaria pinnatifida (alariaceae, Laminariales, phaeophyceae). Mitochondrial DNA 26 (6), 953–954. doi:10.3109/19401736.2013.865172
Li, Y., Umanzor, S., Ng, C., Huang, M., Marty-Rivera, M., Bailey, D., et al. (2022). Skinny kelp (Saccharina angustissima) provides valuable genetics for the biomass improvement of farmed sugar kelp (Saccharina latissima). J. Appl. Phycol. 34, 2551–2563. doi:10.1007/s10811-022-02811-1
Liu, T., Wang, X., Wang, G., Jia, S., Liu, G., Shan, G., et al. (2019). Evolution of complex thallus alga: genome sequencing of Saccharina japonica. Front. Genet. 10 (MAY), 378. doi:10.3389/fgene.2019.00378
Liu, Y., Bi, Y., Gu, J., Li, L., and Zhou, Z. (2012). Localization of a female-specific marker on the chromosomes of the Brown seaweed Saccharina japonica using fluorescence in situ hybridization S. PLoS ONE 7 (11), e48784. doi:10.1371/journal.pone.0048784
Liu, Y., Bi, Y.-H., and Zhou, Z.-G. (2022). Dual-color fluorescence in situ hybridization with combinatorial labeling probes enables a detailed karyotype analysis of Saccharina japonica. Algal Res. 62, 102636. doi:10.1016/j.algal.2022.102636
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38 (10), 4647–4654. doi:10.1093/molbev/msab199
Mao, X., Augyte, S., Huang, M., Hare, M. P., Bailey, D., Umanzor, S., et al. (2020). Population genetics of sugar kelp throughout the northeastern United States using genome-wide markers. Front. Mar. Sci. 7 (694), 1–13. doi:10.3389/fmars.2020.00694
Melén, K., Krogh, A., and Von Heijne, G. (2003). Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327 (3), 735–744. doi:10.1016/S0022-2836(03)00182-7
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157 (4), 1819–1829. doi:10.1093/genetics/157.4.1819
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34 (13), i142–i150. doi:10.1093/bioinformatics/bty266
Moose, S. P., and Mumm, R. H. (2008). Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 147 (3), 969–977. doi:10.1104/PP.108.118232
Motamayor, J. C., Mockaitis, K., Schmutz, J., Haiminen, N., III, D. L., Cornejo, O., et al. (2013). The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 14 (6), r53. doi:10.1186/gb-2013-14-6-r53
Müller, D. G., Maier, I., Marie, D., and Westermeier, R. (2016). Nuclear DNA level and life cycle of kelps: evidence for sex-specific polyteny in Macrocystis (Laminariales, Phaeophyceae). J. Phycol. 52 (2), 157–160. doi:10.1111/jpy.12380
Nielsen, H., Engelbrecht, J., Brunak, S., and Von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. Des. Sel. 10 (1), 1–6. doi:10.1093/protein/10.1.1
North, W. J. (1987). “Biology of the Macrocystis resource in North America,” in Case study of seven commercial seaweed resources. Editors M. Doty, J. Caddy, and B. Santelices (Rome: FAO Fisheries Technical Paper), 265–311. Available online at: https://www.fao.org/4/x5819e/x5819e00.htm.
Nosil, P., Soria-Carrasco, V., Villoutreix, R., De-la-Mora, M., de Carvalho, C. F., Parchman, T., et al. (2023). Complex evolutionary processes maintain an ancient chromosomal inversion. Proc. Natl. Acad. Sci. 120 (25), e2300673120. doi:10.1073/pnas.2300673120
Nuzhdin, S. V., Friesen, M. L., and McIntyre, L. M. (2012). Genotype–phenotype mapping in a post-GWAS world. Trends Genet. 28 (9), 421–426. doi:10.1016/j.tig.2012.06.003
Oudot-Le Secq, M.-P., Goër, S. L., Stam, W. T., and Olsen, J. L. (2006). Complete mitochondrial genomes of the three brown algae (Heterokonta: phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr. Genet. 49 (1), 47–58. doi:10.1007/s00294-005-0031-4
Peteiro, C., and Freire, Ó. (2011). Offshore cultivation methods affects blade features of the edible seaweed Saccharina latissima in a bay of Galicia, Northwest Spain. Russ. J. Mar. Biol. 37 (4), 319–323. doi:10.1134/S1063074011040110
Phillips, N., Kapraun, D. F., Gómez Garreta, A., Ribera Siguan, M. A., Rull, J. L., Salvador Soler, N., et al. (2011). Estimates of nuclear DNA content in 98 species of brown algae (Phaeophyta). AoB PLANTS 2011, plr001. doi:10.1093/aobpla/plr001
Pimentel, D. (1991). Global warming, population growth, and natural resources for food production. Soc. and Nat. Resour. 4 (4), 347–363. doi:10.1080/08941929109380766
Price, A. L., Jones, N. C., and Pevzner, P. A. (2005). De novo identification of repeat families in large genomes. Bioinformatics 21 (Suppl. 1), i351–i358. doi:10.1093/bioinformatics/bti1018
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33 (Web Server), W116–W120. doi:10.1093/nar/gki442
Rana, S., Valentin, K., Bartsch, I., and Glöckner, G. (2019). Loss of a chloroplast encoded function could influence species range in kelp. Ecol. Evol. 9 (15), 8759–8770. doi:10.1002/ece3.5428
Rana, S., Valentin, K., Riehl, J., Blanfuné, A., Reynes, L., Thibaut, T., et al. (2021). Analysis of organellar genomes in brown algae reveals an independent introduction of similar foreign sequences into the mitochondrial genome. Genomics 113 (2), 646–654. doi:10.1016/j.ygeno.2021.01.003
R Core Team (2024). R: a language and environment for statistical computing. Available online at: https://www.R-project.org.
Redmond, S., Green, L., Yarish, C., Kim, J., and Neefus, C. (2014). New England seaweed culture handbook. Available online at: https://digitalcommons.lib.uconn.edu/seagrant_weedcult/1.
Salamov, A. A., and Solovyev, V. V. (2000). Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10 (4), 516–522. doi:10.1101/gr.10.4.516
Sayers, E. W., Beck, J., Bolton, E. E., Brister, J. R., Chan, J., Comeau, D. C., et al. (2024). Database resources of the national center for Biotechnology information. Nucleic Acids Res. 52 (D1), D33–D43. doi:10.1093/nar/gkad1044
Shan, T., Yuan, J., Su, L., Li, J., Leng, X., Zhang, Y., et al. (2020). First genome of the Brown alga Undaria pinnatifida: chromosome-level assembly using PacBio and Hi-C technologies. Front. Genet. 11, 140. doi:10.3389/fgene.2020.00140
Silberfeld, T., Leigh, J. W., Verbruggen, H., Cruaud, C., De Reviers, B., and Rousseau, F. (2010). A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Mol. Phylogenetics Evol. 56 (2), 659–674. doi:10.1016/j.ympev.2010.04.020
Smit, A. F. A., Hubley, R., and Green, P. (2004). RepeatMasker open-3.0. Available online at: https://www.repeatmasker.org/.
Snirc, A., Silberfeld, T., Bonnet, J., Tillier, A., Tuffet, S., and Sun, J.-S. (2010). Optimization of Dna extraction from Brown algae (phaeophyceae) based on a commercial Kit. J. Phycol. 46 (3), 616–621. doi:10.1111/j.1529-8817.2010.00817.x
Song, H., Dong, T., Yan, X., Wang, W., Tian, Z., Sun, A., et al. (2023). Genomic selection and its research progress in aquaculture breeding. Rev. Aquac. 15 (1), 274–291. doi:10.1111/raq.12716
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24 (5), 637–644. doi:10.1093/bioinformatics/btn013
Starko, S., Soto Gomez, M., Darby, H., Demes, K. W., Kawai, H., Yotsukura, N., et al. (2019). A comprehensive kelp phylogeny sheds light on the evolution of an ecosystem. Mol. Phylogenetics Evol. 136, 138–150. doi:10.1016/j.ympev.2019.04.012
Steenwyk, J. L., and King, N. (2024). The promise and pitfalls of synteny in phylogenomics. PLOS Biol. 22 (5), e3002632. doi:10.1371/journal.pbio.3002632
Stekoll, M., Pryor, A., Meyer, A., Kite-Powell, H. L., Bailey, D., Barbery, K., et al. (2024). Optimizing seaweed biomass production - a two kelp solution. J. Appl. Phycol. 36, 2757–2767. doi:10.1007/s10811-024-03296-w
Steneck, R. S., Graham, M. H., Bourque, B. J., Corbett, D., Erlandson, J. M., Estes, J. A., et al. (2002). Kelp forest ecosystems: biodiversity, stability, resilience and future. Environ. Conserv. 29 (4), 436–459. doi:10.1017/S0376892902000322
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O., and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18 (12), 1979–1990. doi:10.1101/gr.081612.108
Thaler, E. A., Larsen, I. J., and Yu, Q. (2021). The extent of soil loss across the US Corn Belt. Proc. Natl. Acad. Sci. 118 (8), e1922375118. doi:10.1073/pnas.1922375118
The Aquaculture Genomics, Genetics and Breeding Workshop Abdelrahman, H., ElHady, M., Alcivar-Warren, A., Allen, S., Al-Tobasei, R., et al. (2017). Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. BMC Genomics 18 (1), 191. doi:10.1186/s12864-017-3557-1
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45 (W1), W6–W11. doi:10.1093/nar/gkx391
Tseng, C. K., and Fei, X. G. (1987). “Macroalgal commercialization in the orient,” in Twelfth international seaweed symposium. Developments in hydrobiology. 14 August 1987. Editors M. A. Ragan, and C. J. Bird (Dordrecht: Springer), 167–172. doi:10.1007/978-94-009-4057-4_23
Umanzor, S., Li, Y., Bailey, D., Augyte, S., Huang, M., Marty-Rivera, M., et al. (2021). Comparative analysis of morphometric traits of farmed sugar kelp and skinny kelp, Saccharina spp., strains from the Northwest Atlantic. J. World Aquac. Soc. 52 (5), 1059–1068. doi:10.1111/jwas.12783
UniProt Consortium (2012). Update on activities at the universal protein resource (UniProt) in 2013. Nucleic Acids Res. 41 (D1), D43–D47. doi:10.1093/nar/gks1068
Van der Auwera, G. A., and O’Connor, B. D. (2020). Genomics in the cloud: using docker, GATK, and WDL in terra. 1st edition. Sebastopol: O’Reilly Media, Inc. Available online at: https://learning.oreilly.com/library/view/genomics-in-the/9781491975183/.
Varshney, R. K., Terauchi, R., and McCouch, S. R. (2014). Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12 (6), e1001883. doi:10.1371/journal.pbio.1001883
Vaser, R., Sović, I., Nagarajan, N., and Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27 (5), 737–746. doi:10.1101/gr.214270.116
Wade, R., Augyte, S., Harden, M., Nuzhdin, S., Yarish, C., and Alberto, F. (2020). Macroalgal germplasm banking for conservation, food security, and industry. PLOS Biol. 18 (2), e3000641. doi:10.1371/journal.pbio.3000641
Wang, L.-G., Lam, T.T.-Y., Xu, S., Dai, Z., Zhou, L., Feng, T., et al. (2020). Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 37 (2), 599–603. doi:10.1093/molbev/msz240
Wang, S., Fan, X., Guan, Z., Xu, D., Zhang, X., Wang, D., et al. (2016). Sequencing of complete mitochondrial genome of Saccharina latissima ye-C14. Mitochondrial DNA Part A 27 (6), 4037–4038. doi:10.3109/19401736.2014.1003832
Waterhouse, R. M., Zdobnov, E. M., and Kriventseva, E. V. (2011). Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol. Evol. 3 (1), 75–86. doi:10.1093/gbe/evq083
Wernberg, T., Coleman, M. A., Bennett, S., Thomsen, M. S., Tuya, F., and Kelaher, B. P. (2018). Genetic diversity and kelp forest vulnerability to climatic stress. Sci. Rep. 8 (1), 1851. doi:10.1038/s41598-018-20009-9
Wickham, H. (2016). “ggplot2: elegant graphics for data analysis,”. Use R. 2nd edition (New York: Springer-Verlag). doi:10.1007/978-3-319-24277-4
Ye, N., Zhang, X., Miao, M., Fan, X., Zheng, Y., Xu, D., et al. (2015). Saccharina genomes provide novel insight into kelp biology. Nat. Commun. 6 (1), 6986. doi:10.1038/ncomms7986
Yu, G. (2022). Data integration, manipulation and visualization of phylogenetic trees. 1st edition. Boca Raton: Chapman and Hall/CRC. doi:10.1201/9781003279242
Zhang, J., Liu, T., Bian, D., Zhang, L., Li, X., Liu, D., et al. (2016a). Breeding and genetic stability evaluation of the new Saccharina variety “Ailunwan” with high yield. J. Appl. Phycol. 28 (6), 3413–3421. doi:10.1007/s10811-016-0810-y
Zhang, J., Liu, T., Feng, R., Liu, C., Jin, Y., Jin, Z., et al. (2018). Breeding of the new Saccharina variety “Sanhai” with high-yield. Aquaculture 485, 59–65. doi:10.1016/j.aquaculture.2017.11.015
Zhang, Y., Guo, Y.-M., Li, T.-J., Chen, C.-H., Shen, K.-N., and Hsiao, C.-D. (2016b). The complete chloroplast genome of Wakame (Undaria pinnatifida), an important economic macroalga of the family Alariaceae. Mitochondrial DNA Part B 1 (1), 25–26. doi:10.1080/23802359.2015.1137791
Keywords: Saccharina latissima, sugar kelp, genome, comparative genomics, genomic breeding, brown macroalga, kelp aquaculture
Citation: DeWeese K, Molano G, Calhoun S, Lipzen A, Jenkins J, Williams M, Plott C, Talag J, Grimwood J, Jannink J-L, Grigoriev IV, Schmutz J, Yarish C, Nuzhdin S and Lindell S (2025) Scaffolded and annotated nuclear and organelle genomes of the North American brown alga Saccharina latissima. Front. Genet. 16:1494480. doi: 10.3389/fgene.2025.1494480
Received: 10 September 2024; Accepted: 23 April 2025;
Published: 14 May 2025.
Edited by:
Jianping Wang, University of Florida, United StatesReviewed by:
Josué Barrera-Redondo, Unidad Irapuato (CINVESTAV), MexicoLeonardo Alfredo Ornella, Cubiqfoods SL, Spain
Copyright © 2025 DeWeese, Molano, Calhoun, Lipzen, Jenkins, Williams, Plott, Talag, Grimwood, Jannink, Grigoriev, Schmutz, Yarish, Nuzhdin and Lindell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Scott Lindell, c2xpbmRlbGxAd2hvaS5lZHU=
 Kelly DeWeese
Kelly DeWeese Gary Molano
Gary Molano Sara Calhoun
Sara Calhoun Anna Lipzen
Anna Lipzen Jerry Jenkins
Jerry Jenkins Melissa Williams3
Melissa Williams3 Jane Grimwood
Jane Grimwood Jean-Luc Jannink
Jean-Luc Jannink Igor V. Grigoriev
Igor V. Grigoriev Jeremy Schmutz
Jeremy Schmutz Charles Yarish
Charles Yarish Sergey Nuzhdin
Sergey Nuzhdin Scott Lindell
Scott Lindell