- Genomics Division, Department of Agricultural Biotechnology, National Institute of Agricultural Sciences, Rural Development Administration, Wanju, Republic of Korea
Introduction
Rice cultivation in South Korea has a rich and extensive historical background, dating back to the Bronze Age (Jeong et al., 2021). Throughout history, rice has experienced various transformations in its status, from being a luxury food item to being used as animal feed owing to overproduction and reduced consumption. Recently, rice consumption in South Korea has declined owing to cultural changes and governmental policies. Consequently, the South Korean government is actively promoting rice consumption and focusing on rice exports to meet the global demand and supply chain requirements. The aroma of rice varies around the world, and South Korean rice varieties are primarily glossy, soft, and sticky rice, compared to other parts of the world. Additionally, factors such as enhancing the population-specific nutrients in rice, similar to the golden rice project (Wu et al., 2021), and reducing inputs for rice cultivation (Adu et al., 2022), which benefits the environment, the project such as Green Super Rice (Yu et al., 2022), are essential. Therefore, it is crucial to develop new rice varieties that cater to consumer preferences, combine traditional colored rice varieties, and enhance their nutritional value. Furthermore, the current scenario of rice overproduction is not stable, as Korean rice varieties have faced significant production losses due to temperature fluctuations in the past, such as a 17% loss in 1971 and 20% loss in 2003, as well as an 80% loss due to cold temperatures in 1980 (Jeong, et al., 2021). In light of the potential threat posed by current climate fluctuations to South Korea’s rice self-sufficiency, proactive measures must be taken to ensure continued rice production. Given the consequences of global warming and population expansion, which have resulted in a scarcity of rice worldwide, it is imperative to adopt proactive measures to promote sustainable rice production. To alleviate the strain on global food supply, the South Korean government is exploring the cultivation of non-japonica rice for export, which could also enable South Korea to establish itself as a formidable player in the global rice trade (Jeong, et al., 2021).
Considering rice breeding and its associated challenges, South Korea has been dedicated to extensive research and varietal improvements since the 1970s. This dedication began with the development of the high-yield cultivar “Tong-il,” which served as the foundation for the South Korean Green Revolution and allowed the country to achieve self-sufficiency (Kim et al., 2014). Over time, the emphasis has shifted from high-yield components to grain quality and stress resistance, resulting in the diversification of rice varieties. In total, 206 Korean rice varieties have been studied, among which a significant proportion has been categorized as good for consumption (Cho et al., 2009). It is noteworthy that the genetic diversity within these varieties is somewhat limited, with a large percentage originating from a few Korean-bred and Japanese stocks, which may pose challenges for future breeding owing to the potential reduction in hybrid vigor. In terms of disease resistance, a study of Korean rice varieties revealed the presence of major blast resistance genes, with a significant number of varieties containing genes that have origins in both Korean and Japanese japonica rice genotypes. The presence of these resistance genes contributes to the overall resistance of Korean rice to blast disease caused by Magnaporthe oryzae (Cho, et al., 2009). Although the presence of resistance gene pools ensures blast resistance, further breeding programs may be needed to address the genetic diversity issue to ensure the sustainable development of rice varieties in Korea (Cho, et al., 2009).
Despite significant progress in exploring the genetics of the rice genome using the available rice germplasm globally, a comprehensive analysis of 3,010 rice germplasms from diverse regions worldwide has been conducted as part of this endeavor (Wang et al., 2018). Additionally, contemporary rice breeding programs primarily incorporate genome sequencing to acquire detailed genetic knowledge and establish precision breeding to achieve desired outcomes (Paul, 2020; Wing et al., 2018). Furthermore, the era of genomics has facilitated researchers to pan-genome, which involves incorporating various cultivars and varieties to better comprehend genetics and biological functions associated with specific or multiple traits (Schreiber et al., 2024). Recently, the pan-genome of rice has shed light on the subpopulation structure of Asian rice varieties compared to the wild type (Zhou et al., 2023). The collective effort of the International Rice Genome Sequencing Project (IRGSP) consortium has resulted in a high-quality reference genome for rice Oryza sativa subsp. japonica cv Nipponbare (IRGSP v.1.0) (Kawahara et al., 2013), which offers comprehensive genomic information for O. sativa, a model species for monocotyledonous plants. This reference and its annotations are distributed with various additional assessments through primary rice databases such as RAP-db (https://rapdb.dna.affrc.go.jp), RGAP (http://rice.uga.edu), and Gramene (https://www.gramene.org). Additionally, insights into the rice pan-genome datasets can be obtained through RPAN (https://cgm.sjtu.edu.cn/3kricedb/index.php) and Rice RC (http://ricerc.sicau.edu.cn/RiceRC). Furthermore, the comprehensive catalogue of rice genes is organized at Rice Gene Index (RGI), and its variants are organized in databases such as Rice SNP-Seek, RPAN, and RiceVarMap (Yu et al., 2023). However, nationwide germplasm databases are the most valuable assets for researchers focusing on domestic varieties that are being improved. To facilitate this for Korean rice researchers, genome re-sequencing of the Korean rice population was conducted to identify effective breeding signatures for the green super rice strategy. This strategy aims to improve the Korean breeding efficiency for various associated issues. Despite the inclusion of 35 rice varieties from South Korea in the 3 K rice genome-resequencing project, additional elite lines were incorporated into the genome-sequencing process. The K-Rice database provides a comprehensive catalog of the elite Korean rice population and wild genomes, which will enable researchers to better understand the genetic variants present in these genomes and to identify breeding signatures primarily from Korean rice populations.
Value of the data
The dataset presented in this study constitutes a useful and informative resource for understanding the genetic diversity of the Korean rice population. This dataset may prove to be a valuable asset for rice breeders and researchers, enabling them to conduct research on Korean rice varieties and develop new varieties that can address the challenges posed by the ongoing global warming crisis and the impending population increase.
Materials and methods
Collection of rice germplasm and phenotypic data
A total of 105 rice germplasms (85 elite cultivars and 20 wild accessions, detailed in Supplementary Table 1) were procured from the National Institute of Crop and Food Science (NICS), RDA, Korea. Along with the germplasm, NICS provided data for 15 phenotypic traits that they had previously investigated for these lines. These traits include protein content, milling recovery ratio, grain filling ratio, taste evaluation, head rice ratio, grain number, height, yield, panicle length, panicle number, grain length/width ratio, 1000-grain weight of brown rice, heading ecotype, blast resistance, and RSV (rice stripe virus) resistance. This phenotypic data is accessible within the K-RICE database for each corresponding germplasm.
DNA sequencing and variant calling
Total DNA was isolated from the samples individually according to standard sequencing protocols. DNA was prepared using a TruSeq Nano DNA Prep Kit for Illumina sequencing. Each isolated DNA sample was sequenced using Novaseq6000 (Illumina), which is a short-read sequencing technique. The experiment was performed by Macrogen, an authorized service provider in South Korea. Illumina paired-end sequences were subjected to quality and adapter trimming using BBDuk v28.26. The processed reads were mapped to the O. sativa subsp. japonica cv Nipponbare (IRGSP v1.0) reference genome (Kawahara, et al., 2013) using Bowtie2 v.2.2.5(Langmead and Salzberg, 2012), and variant calling was performed with the Haplotype caller in the Genome Analysis Toolkit (GATK v4.2.0.0) (McKenna et al., 2010). SNPs were selected using GATK parameters, that is, a normalized quality score ≥2 and mapping quality ≥40. The SNPs were annotated using SnpEff v.4.2 (Cingolani et al., 2012).
Establish database framework
The entire database of webpages was encoded using Java, and the database was accessed via the URL (http://nabic.rda.go.kr/post_jbrowse.do?data=K_RICE). The database was designed to facilitate effective exploration of the variant region using the genome JBrowse for all 100 VCF file tracks, along with the reference genome, which includes the transcripts, gene, and exon regions of the genes. The respective VCF files were also available for downloading (Figure 1).
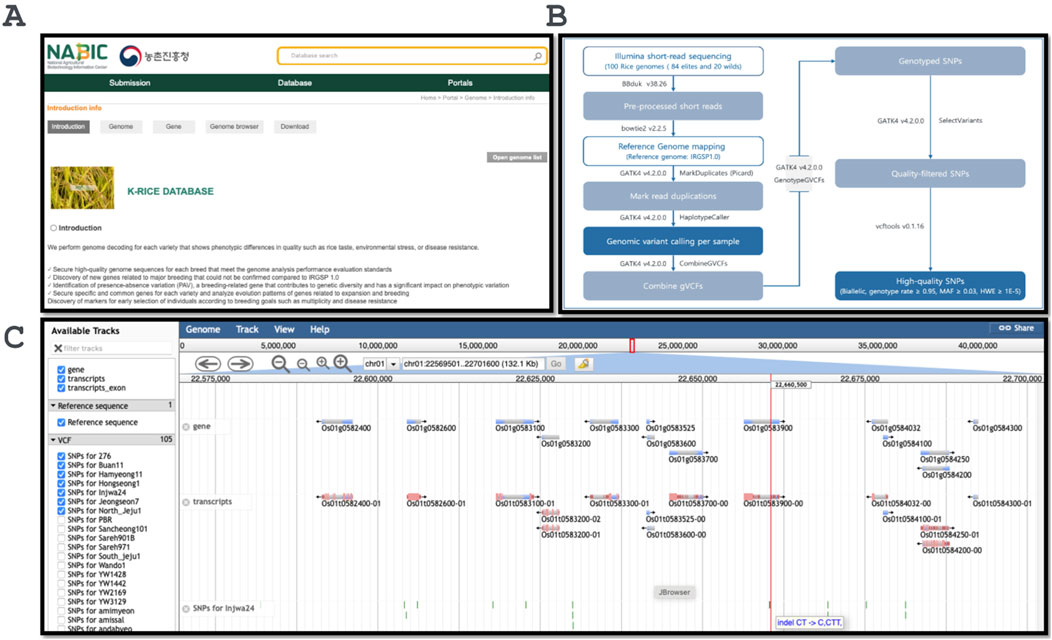
Figure 1. The design and output example of the database. (A) Index page of the K-Rice database; (B) The variants calling pipeline overview; (C) Genome browser with all 100 germplasm variant datasets.
Preliminary analysis report
The overall quantity of sequence data generated from 105 rice germplasm samples (including 85 elite and 20 wild samples) using whole-genome short-read sequencing. Each sample generated approximately 11.9 GB of raw reads, which became 11.6 GB after processing. Of the processed reads, 98.2% were successfully mapped to the rice genome, with the mapped reads covering the genome being approximately 26.6-fold. The dataset also provided 15 phenotypic values, such as protein content, milling recovery ratio, grain filling ratio, taste evaluation, rice head ratio, grain number, height, yield, panicle length, panicle number, grain length/width ratio, 1,000 grain weight of brown rice, heading ecotype, blast resistance, RSV (rice stripe virus) resistance, to evaluate individual trait vigor (Supplementary Table 1). Moreover, the genomes were categorized into three primary groups based on their resistance to blast, striped leaf blight, and flowering time, as illustrated in Table 1, and shown to represent the coverage of the Korean germplasm.
Recognizing the importance of nationwide germplasm variation in rice, it is crucial for researchers to conduct extensive research and contribute to the diversification of rice varieties. The Korean National Agricultural Biotechnology Information Center, which formerly maintained the Korean rice germplasm, has organized the genetic variant data into the K-Rice database, which is now accessible to the public.
Data availability statement
The complete sequences generated in this study were deposited in NCBI Project accession no. PRJNA1180626.
Author contributions
J-GK: Data curation, Writing – original draft. G-HP: Resources, Writing – review and editing. JK: Validation, Writing – review and editing. JJ: Resources, Writing – review and editing. T-HL: Conceptualization, Methodology, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the “National Institute of Agricultural Sciences” (PJ01721001) of the Rural Development Administration of the Republic of Korea. The authors extend their gratitude to the Research and Development Center of Insilicogen Inc. for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1544652/full#supplementary-material
References
Adu, B. G., Argete, A. Y. S., Egawa, S., Nagano, A. J., Shimizu, A., Ohmori, Y., et al. (2022). A koshihikari X Oryza rufipogon introgression line with a high capacity to take up nitrogen to maintain growth and panicle development under low nitrogen conditions. Plant Cell Physiol. 63, 1215–1229. doi:10.1093/pcp/pcac097
Cho, Y.-C., Suh, J.-P., Jeung, J.-U., Roh, J.-H., Yang, C.-I., Oh, M.-K., et al. (2009). “Resistance genes and their effects to blast in Korean rice varieties (Oryza sativa L.). In G-L wang, valent B, edtiors,” in Proceedings of the advances in genetics, genomics and control of rice blast disease (Netherlands: Springer).
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin) 6, 80–92. doi:10.4161/fly.19695
Jeong, O. Y., Park, H.-S., Baek, M.-K., Kim, W.-J., Lee, G.-M., Lee, C.-M., et al. (2021). Review of rice in Korea: current status, future prospects, and comparisons with rice in other countries. J. Crop Sci. Biotechnol. 24, 1–11. doi:10.1007/s12892-020-00053-6
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. doi:10.1186/1939-8433-6-4
Kim, B., Kim, D.-G., Lee, G., Seo, J., Choi, I.-Y., Choi, B.-S., et al. (2014). Defining the genome structure of “Tongil” rice, an important cultivar in the Korean “Green Revolution”. Rice 7, 22. doi:10.1186/s12284-014-0022-5
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. Sep. 20, 1297–1303. doi:10.1101/gr.107524.110
Paul, A. (2020). “Sequencing the rice genome: gateway to agricultural development,” in Rice research for quality improvement: genomics and genetic engineering: volume 1: breeding techniques and abiotic stress tolerance (Singapore: Springer Singapore), 109–157.
Schreiber, M., Jayakodi, M., Stein, N., and Mascher, M. (2024). Plant pangenomes for crop improvement, biodiversity and evolution. Nat. Rev. Genet. 25, 563–577. doi:10.1038/s41576-024-00691-4
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi:10.1038/s41586-018-0063-9
Wing, R. A., Purugganan, M. D., and Zhang, Q. (2018). The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 19, 505–517. doi:10.1038/s41576-018-0024-z
Wu, F., Wesseler, J., Zilberman, D., Russell, R. M., Chen, C., and Dubock, A. C. (2021). Opinion: allow golden rice to save lives. Proc. Natl. Acad. Sci. U. S. A. Dec 21, e2120901118. doi:10.1073/pnas.2120901118
Yu, S., Ali, J., Zhou, S., Ren, G., Xie, H., Xu, J., et al. (2022). From Green Super Rice to green agriculture: reaping the promise of functional genomics research. Mol. Plant 15, 9–26. doi:10.1016/j.molp.2021.12.001
Yu, Z., Chen, Y., Zhou, Y., Zhang, Y., Li, M., Ouyang, Y., et al. (2023). Rice Gene Index: a comprehensive pan-genome database for comparative and functional genomics of Asian rice. Mol. Plant 16, 798–801. doi:10.1016/j.molp.2023.03.012
Keywords: rice, database, elite lines, Korean rice population, Oryza sativa
Citation: Kim J-G, Park G-H, Kim J, Jeong J and Lee T-H (2025) K-rice: a comprehensive database of Korean rice germplasm variants. Front. Genet. 16:1544652. doi: 10.3389/fgene.2025.1544652
Received: 13 December 2024; Accepted: 22 May 2025;
Published: 05 June 2025.
Edited by:
Marcos Vinicius Bohrer Monteiro Siqueira, Minas Gerais State University, BrazilReviewed by:
Tian Qing Zheng, Chinese Academy of Agricultural Sciences, ChinaKelvin Dodzi Aloryi, University of Florida, United States
Copyright © 2025 Kim, Park, Kim, Jeong and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tae-Ho Lee, dGhsZWUwQGtvcmVhLmty
 Jeong-Gu Kim
Jeong-Gu Kim Gyu-Hwang Park
Gyu-Hwang Park Tae-Ho Lee
Tae-Ho Lee