- 1Department of Medical Laboratory, Affiliated Hospital of Jining Medical University, Shandong Key Laboratory of Multi-disciplinary Molecular Diagnosis Precision Medicine, Jining, Shandong, China
- 2Department of Medical Laboratory, Disease Prevention and Control Center of Jining Rencheng District, Jining, Shandong, China
- 3Medical Laboratory of Jining Medical University, Lin He’s Academician Workstation of New Medicine and Clinical Translation in Jining Medical University, Jining Medical University, Jining, Shandong, China
Introduction: Individuals with balanced chromosomal rearrangements are at an increased risk for infertility, recurrent miscarriages, and the birth of infants with congenital malformations. Traditional cytogenetic techniques are limited by their low resolution, whereas optical genome mapping offers enhanced capabilities for detecting chromosomal rearrangements and determining genomic localization and orientation. This study sought to evaluate the efficacy of optical genome mapping in identifying complex balanced chromosomal rearrangements that may contribute to fertility challenges.
Case presentation: A 21-year-old Asian female patient with a history of recurrent abortions was included in the study. Peripheral blood samples were collected for high-resolution karyotyping, chromosomal microarray analysis, and optical genome mapping. The high-resolution karyotype analysis identified complex chromosomal abnormalities. Optical genome mapping has revealed additional cryptic chromosomal aberrations, such as ins (2; 12) (p16.1; q12q12), inv (6) (q21q21), and inv (12) (q12q12), offering a novel perspective on this case. Notably, the disrupted genes, including CRIM1, MUC19, and PRDM1, have not been classified as pathogenic by existing databases.
Conclusion: This study underscores the capability of optical genome mapping to deliver comprehensive and precise information. It is anticipated that optical genome mapping will emerge as a valuable cytogenetic tool within clinical genetic methodologies, providing new references and insights for clinical practice in the future.
Introduction
Complex chromosomal rearrangements (CCRs) play a critical role in various human genetic disorders by disrupting protein-coding genes and cis-regulatory elements. These rearrangements encompass a range of distinct structural variations on the same allele within a single mutational signature event (Collins et al., 2020). Balanced reciprocal rearrangements (BCRs) occur due to the random de novo breakage and subsequent rejoining of two or more chromosomes. The incidence rate of BCRs is approximately 7% among infertile couples, a figure that is significantly elevated compared to the general human population (0.14%) (Kasikova et al., 2012). Most carriers of BCRs exhibit normal phenotypes, as the majority of breakpoints (BPs) occur in intergenic or noncoding regions, where the disruption has minimal interference with gene expression (Carvalho and Lupski, 2016). However, these individuals face an increased risk of recurrent miscarriages and the birth of newborns with chromosomal imbalances (Chow et al., 2020). Research indicates that the likelihood of identifying normal or balanced blastocysts in patients with CCRs is less than 6%, while the probability of achieving pregnancy is below 4%. These factors may contribute to a diminished number of transplantable embryos (L. Hu et al., 2018). This may manifest as implantation failure, miscarriage of established pregnancies, or the birth of infants with chromosomal syndromes, which poses significant concerns for affected couples (Wilch and Morton, 2018). In a study involving 300 couples experiencing recurrent miscarriages, chromosomal abnormalities were identified in 26 cases, representing 8.7% of the sample. Among these, structural anomalies being the most prevalent at 57.7%, followed by numerical chromosomal aberrations at 42.3% (Yildirim et al., 2019). Genetic analyses of BCRs associated with a normal phenotype hold significant clinical implications for fertility counseling and the prevention of genetic disorders in offspring.
Routine cytogenetic analyses are essential for identifying genetic biomarkers relevant to clinical diagnosis, however, each testing method presents specific limitations. Karyotyping, recognized as a fundamental first-line approach, is capable of identifying both numerical and structural chromosomal aberrations. Nonetheless, it is characterized by certain drawbacks, including a time-intensive process, limited microscopic resolution (averaging between 5 and 10 Mb), and a relatively low overall diagnostic yield (Hofherr et al., 2011; Dremsek et al., 2021; MacLeod and Drexler, 2013). Copy number variation sequencing (CNV-seq) and chromosomal microarray analysis (CMA) are capable of identifying variations as small as a few kilobases (Kb). However, these methods have limitations in detecting lower-proportion mosaicism and BCRs (Miller et al., 2010; Waggoner et al., 2018). Fluorescence in situ hybridization (FISH) serves as a viable method for confirming suspected cryptic BCRs. However, its application is constrained by challenges in determining the precise localization beforehand and the complexities involved in assigning specific fluorescent probes. Consequently, FISH is not appropriate for comprehensive gene detection (Cui et al., 2016). Whole genome sequencing (WGS) facilitates the identification of structural rearrangements and nucleotide variations within a single experimental framework, generating significant interest in its potential as a primary diagnostic tool (Lionel et al., 2017). However, its implementation is hindered by challenges such as limited read length, high costs, and reduced sensitivity in repetitive genomic regions (Savara et al., 2021). Phasing multiple rearrangement breakpoints in complex B Cell receptor structures using WGS can be a formidable task (Dong et al., 2017). Consequently, there is a pressing need for a rapid and reliable method to identify BCRs during clinical testing.
Optical genome mapping (OGM) represents a sophisticated, high-throughput cytogenomic methodology that utilizes imaging techniques to analyze exceptionally long linear single DNA molecules. OGM offers enhanced sensitivity and specificity in the detection of various structural variants (SVs), encompassing aneuploidies, deletions, duplications, inversions, insertions, translocations, and the absence of heterozygosity (AOH) (Mantere et al., 2021). The capabilities of preamplification-free techniques and high-resolution analysis are enhanced through the fluorescent labeling of specific sequence motifs (Sanders et al., 2019; Chan Saki et al., 2018). OGM offers significant advantages, including improved turnaround times and cost-effective generation of 300–500 × genome-wide coverage. Recent studies have demonstrated the efficacy of OGM in identifying a broad spectrum of genetic disorders associated with cancers (Neveling et al., 2021; Zhang et al., 2023a; Sahajpal et al., 2021). The finding indicates that OGM demonstrated complete concordance with cytogenetic assays in detecting genomic chromosomal aberrations across 85 individuals (Mantere et al., 2021). Furthermore, OGM has been effectively employed to elucidate the relationship between genes and phenotypes based on breakpoint locations. Additionally, OGM possesses the capability to identify intricate genetic abnormalities and facilitates the characterization of breakpoints (Wang et al., 2020; Chan EvaKF. et al., 2018; Jain et al., 2016).
Significant challenges have thus far constrained the objective assessment of BCRs. We uncovered a complex BCRs by OGM analysis, which was discribed as 46,XX,ins (6; 2) (q21; p16.1p22.2) ins (2; 12) (p16.1; q12q12) inv (6) (q21q21) t (6; 12) (q21; q12) inv (12) (q12q12), which has not been previously reported. This study aims to investigate the potential of OGM in detecting complex BCRs in the absence of various clinical indications, offering valuable references and insights for future clinical diagnoses.
Case description
A 21-year-old Asian woman participated in this study. She was diagnosed with pregnancy at the Affiliated Hospital of Jining Medical University. Eclampsia assessment revealed a high risk, with a Placental Growth Factor Multiple Of Median of 0.27. Additionally, mid-trimester Down syndrome screening indicated a heightened risk of open neural tube malformation, with a Multiple Of Median of 2.407. At 19 weeks of gestation, the ultrasound findings indicated the presence of multiple fetal developmental anomalies, including a reduced size of the cerebellar vermis and a cystic echo observed in the cerebellar bulbous cisterna. The results of the auxiliary examinations were normal.
The family opted for labor induction following an abnormal fetal ultrasound examination. CNV-seq (Copy Number Variation Sequencing) genetic analysis of the aborted tissue revealed a 22.5 Mb duplication in the p22.3p16.1 segment of chromosome 2 [seg (GRch38) dup (2) (p22.3p16.1) Chr2:g36,500,001-59,000,001dup], which is classified as pathogenic According to the ACMG guidelines (Riggs, E. R., et al. (2019). Genet Med. doi:10.1038/s 41436-019-0686-8. Relevant databases such as ClinGen, DECIPHER, OMIM, ClinVar and PubMed and others if required suggest that the identified duplicated fragment is linked to various clinical manifestations, including intellectual disability, growth retardation, neonatal hypotonia, syndactyly, gastroesophageal reflux, hydrocephalus, anal atresia, and short stature. Given the significant size of the fragment, the couple was advised to collect anticoagulant peripheral blood to undergo high-resolution karyotyping analysis and CMA. The subject exhibited normal phenotypes, and her CMA results indicated no imbalanced structural variations according to the American College of Medical Genetics and Genomics (ACMG) guidelines. Consequently, we hypothesized that she was a carrier of BCRs. To validate this hypothesis, we employed high-resolution karyotyping according to the International System for Human Cytogenetic Nomenclature (ISCN) 2020 and OGM based on the GRCh38 human reference genome. The subject’s husband’s karyotype was characterized as 46,XY (Figure 1A), with normal CMA results (Figure 1B).
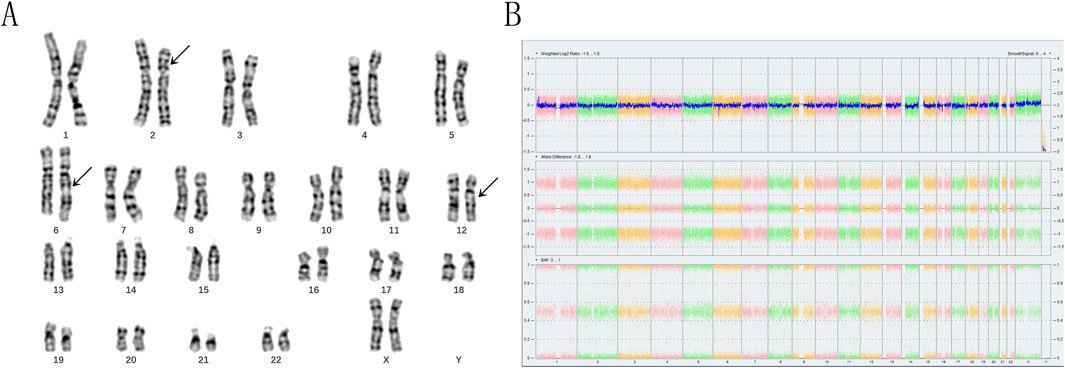
Figure 1. The karyotyping and chromosomal microarray analyses of the subject revealed significant findings. (A) The arrows indicate the presence of abnormal chromosomes. The karyotyping analysis demonstrated the insertion of the p16p22 segment from chromosome 2 into the q21 region of chromosome 6, as well as a translocation between the q21 segment of chromosome 6 and the q13 segment of chromosome 12. This was characterized as 46,XX,ins (6; 2) (q21; p16p22) t (6; 12) (q21; q13). (B) In contrast, the chromosomal microarray analysis yielded normal results.
Results
The karyotyping results confirmed that the subject was indeed a carrier of BCRs, characterized as 46,XX, with a specific rearrangement involving chromosomes 2, 6, and 12. Specifically, the p16p22 segment of chromosome two was inserted into the q21 segment of chromosome 6, accompanied by a mutual translocation between the q21 segment of chromosome six and the q13 segment of chromosome 12. This rearrangement was described as 46,XX,ins (6; 2) (q21; p16p22) t (6; 12) (q21; q13) (Figure 2A).
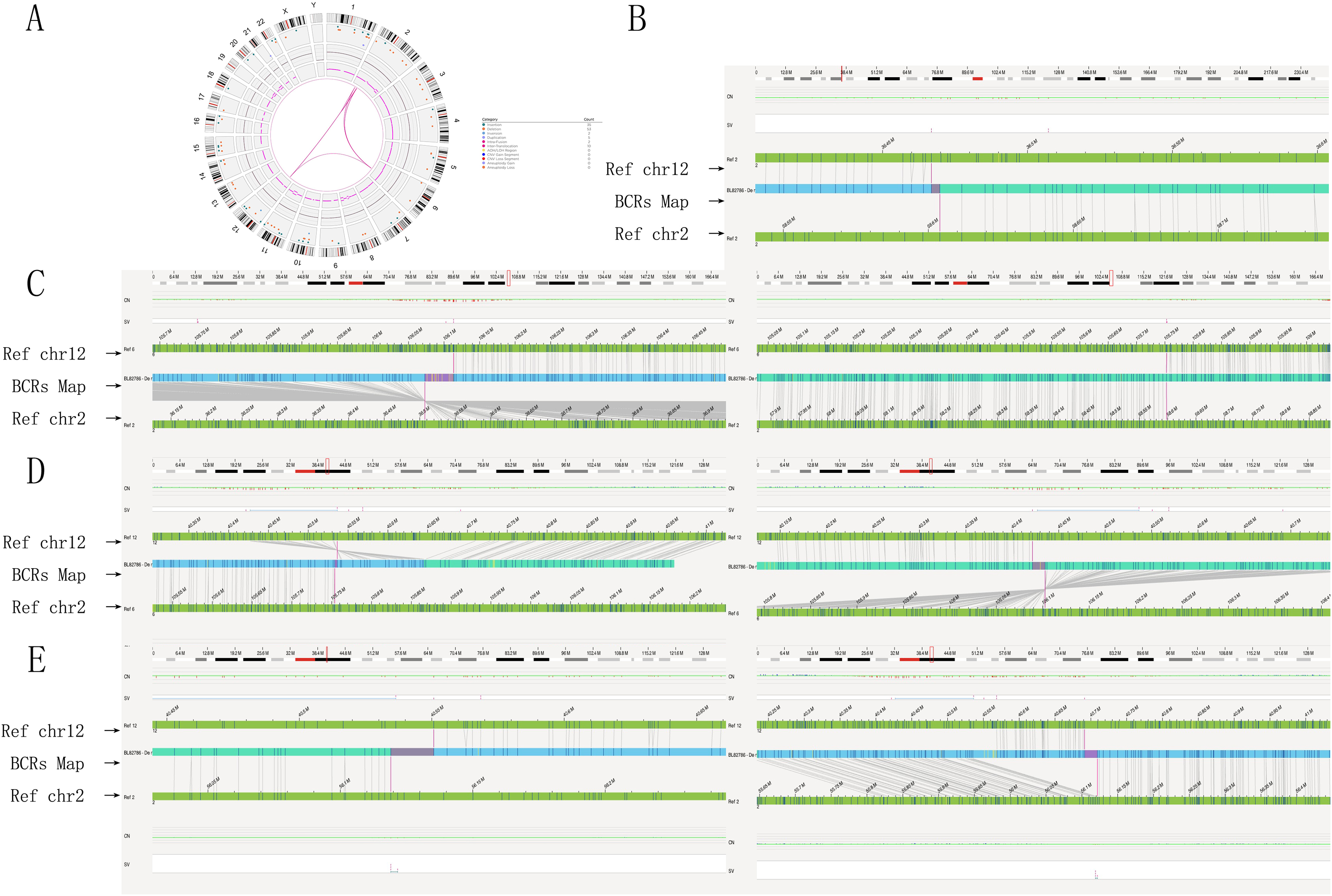
Figure 2. Genome-wide visualization of optical genome mapping reveals significant chromosomal structural rearrangements. (A) The Circos plot integrates all identified chromosomal alterations, with the default view organized from the outer to the inner circle: cytoband locations numbered 1 to 24 (1–22, XX). Color-coded interstitial structural variations are highlighted at specific locations, alongside observed copy number changes for each chromosome or region, illustrating translocations, insertions, and inversions represented as lines. Notably, the Circos plot indicates connections between chromosomes 2, 6, and 12 at genomic loci, depicted with red lines. (B) The genome map view illustrates the inversion of chromosome 2p16.1p22.2. (C) The genome map view details the breakpoint junction of an insertion, ins (6; 2) (q21; p16.1p22.2). (D) The genome map view presents the breakpoint junction of a translocation, t (6; 12) (q21; q12). (E) The genome map view highlights the breakpoint junction of another insertion, ins (2; 12) (p16.1; q12q12). Abbreviations used include INS for insertion, DEL for deletion, INV for inversion, DUP for duplication, and TRA for translocation.
OGM identified a reciprocal translocation between chromosomes 6q21 and 12q12, as well as the insertion of a 2p segment into 6q21, which closely aligned with the findings from karyotyping analysis (Figures 2C,D). Furthermore, the insertion segment of chromosome two was refined from p16p22 to p16.1p22.2 (Figure 2B). OGM also provided detailed information regarding additional fragments, including fus (2; 2) (p22.2; p16.1) involving a 22.1 Mb segment of chromosome 2 (BP: 36,466,829-58,603,981) (Figure 2B), ins (2; 12) (p16.1; q12q12) with a 141 Kb segment of chromosome 12 (BP: 40,550,771-40,691,368) and chromosome 2 (BP: 56,119,045-56,121,630) (Figure 2E), inv (6) (q21q21) with a 359 Kb segment of chromosome 6 (BP: 105,753,796-106,102,619) (Figure 2D), and inv (12) (q12q12) with a 115 Kb segment of chromosome 12 (BP: 40,550,771-40,691,368) (Figure 2D). Additionally, the orientations of the rearranged chromosomes were determined: the p22.2p16.1 segment of chromosome two was reversed and inserted into the q21 segment of chromosome 6, while the q12q12 segment of chromosome 12 was positively inserted into the p16.1 segment of chromosome 2. The refinement of breakpoints and the orientation of segments significantly contributed to the elucidation of the chromosome rearrangement pattern. The findings from the OGM analysis were characterized by the chromosomal alterations 46,XX,ins (6; 2) (q21; p16.1p22.2) ins (2; 12) (p16.1; q12q12)inv (6) (q21q21) t (6; 12) (q21; q12)inv (12) (q12q12). Detailed breakpoint information is shown in Table 1.
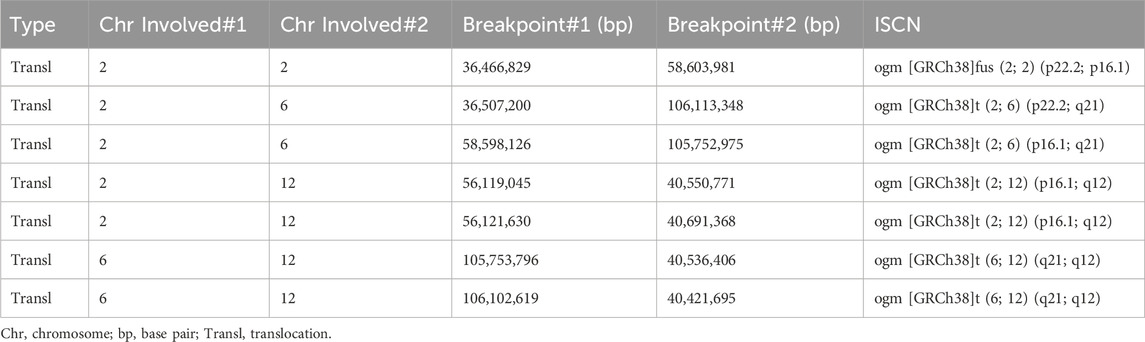
Table 1. Detailed characterisations of the rearrangements on chromosomes 2, 6, and 12 by Bionano Access Software.
Discussion
CCRs are known to encompass both balanced and imbalanced categories, which can lead to a range of phenotypic outcomes. Additionally, BCRs may result in congenital anomalies in the absence of identifiable causative agents, indicating potential disruptions in gene function and long-range regulatory interactions (Sanders et al., 2019; Sinnerbrink et al., 2013). While the majority of BCRs carriers do not exhibit clear clinical phenotypes, there is a significant increase in the risk of spontaneous abortion, infertility, or the birth of abnormal offspring (Liao et al., 2014), the refinement of cryptic BCRs presents a significant challenge.
In this study, high-resolution karyotyping indicated two breakpoints (2p16, 2p22) on chromosome 2, one breakpoint (6q21) on chromosome 6, and one breakpoint (12q13) on chromosome 12. Balanced rearrangements may not necessarily correlate with abnormal phenotypes; however, certain reverse transfers can alter the orientation of repetitive DNA, leading to the formation of inverted loops during meiosis, which results in a 50% probability of producing imbalanced gametes, characterized by partial duplications and deletions. Consequently, there is an elevated risk of genetic diseases in the offspring of carriers with inversions (Watson et al., 2014; Antonacci et al., 2010).
The precise region and orientation of rearrangements in BPs are crucial for elucidating the genetic underpinnings of diseases. However, existing molecular technologies are inadequate for accurately determining the location and orientation of abnormal segments that disrupt or fuse genes. OGM demonstrates the ability to detect significant types of chromosomal aberrations, facilitating the identification of genes located within breakpoint intervals, which can enhance the clinical interpretation of genetic disorders, particularly concerning intragenic rearrangements in prenatal diagnosis. Furthermore, OGM achieves 100% concordance with karyotype analysis for all aberrations involving non-centromeric breakpoints (Sahajpal et al., 2021; Mantere et al., 2021; Wang et al., 2020).
In our study, The findings of OGM, which were challenging to distinguish through karyotyping, provide new insights into this case. Additionally, the orientations of the rearranged chromosomes were determined: the p22.2p16.1 segment of chromosome two was reversed and inserted into the q21 segment of chromosome 6, while the q12q12 segment of chromosome 12 was positively inserted into the p16.1 segment of chromosome 2. The refinement of breakpoints and the orientation of segments significantly contributed to the elucidation of the chromosome rearrangement pattern. Collectively, our results suggest that OGM demonstrates superior potential and enhanced resolution in identifying specific breakpoints (Table 1) and the orientations of abnormal chromosomal segments, aligning with the observations made by Mantere T. et al. (Mantere et al., 2021). According to the GRCh38 assembly, the CRIM1 gene was disrupted in the region of chromosome 2 (BP: 36,466,829-36,507,200), the MUC19 gene was disrupted in the region of chromosome 12 (BP: 40,550,771-40,421,695), and the PRDM1 gene was disrupted in the region of chromosome 6 (BP: 105,752,975-106,102,619). However, no variants with established pathogenic significance for the genes have been reported. Based on the OGM analysis, we have simulated a schematic diagram of the rearrangement occurring in this case (Figure 3). OGM represents a cutting-edge methodology focused on the manipulation of megabase-length DNA molecules to develop a “fluorescent barcode DNA map.” This innovative approach integrates microfluidics, high-resolution microscopy, and automated image analysis, facilitating high-throughput whole-genome imaging and de novo assembly (Sahajpal et al., 2021). The data analysis of OGM employs two advanced pipelines that demonstrate heightened sensitivity for detecting large segments based on coverage depth, as well as small CNVs through the comparative analysis of assembly maps against reference maps. OGM offers significant advantages in identifying previously unrecognized clinically relevant SVs and accurately determining the location and orientation of BPs (P. Hu et al., 2024; Zhang et al., 2023b). The study conducted by Mantere et al. (2021) demonstrated that OGM achieved a remarkable 100% consistency with standard tests in identifying known chromosomal aberrations across 85 blood or cultured cell samples. The study demonstrated a 100% concordance between OGM and karyotype analysis and/or CMA, indicating the efficacy of OGM in identifying genomic alterations in prenatal samples (Goumy, 2023).
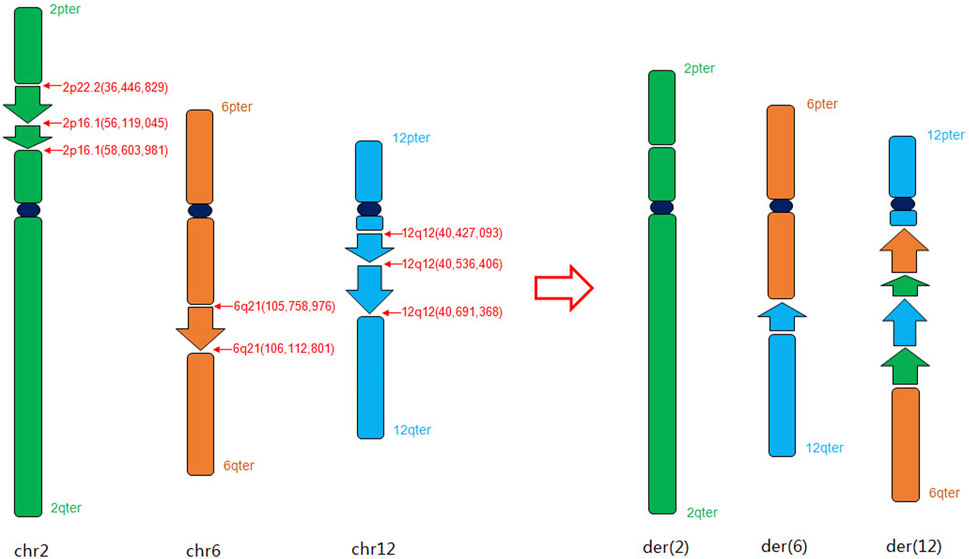
Figure 3. Schematic diagram of the complex chromosome rearrangement pattern in chromosomes 2, 6 and 12 based on OGM analysis.
OGM was originally primarily used in the diagnosis of hematological diseases (Levy et al., 2022; Nilius-Eliliwi et al., 2023) as well as germline SVs of individual study participants (Barseghyan et al., 2018; Du et al., 2018) and rare diseases (Shieh et al., 2021). Subsequently it was applied to the prenatal and postpartum diagnosis (Sahajpal et al., 2021; P; Hu et al., 2024; Zhang et al., 2023a; Goumy, 2023). Researches (Zhang et al., 2023a; Mantere et al., 2021) indicate that OGM is recognized as a precise and thorough technique for identifying cryptic balanced translocations. This capability is instrumental in the genetic diagnosis of patients experiencing unexplained recurrent abortion or infertility. Furthermore, OGM has the potential to forecast disruptions in genes caused by breakpoints, which may lead to the formation of fusion genes (Halgren et al., 2018). The destruction of known pathogenic genes may result in structural abnormalities that can subsequently give rise to clinical syndromes, attributable to inadequate haploid function of the associated genes (Lee et al., 2020; Schluth-Bolard et al., 2019). The study (Wang et al., 2020) identified breakpoint regions in nine subfertile patients with various balanced reciprocal translocations using OGM. It was determined that four disrupted genes, specifically NUP155, FNVDC3A, DPY19L1, and BAI3, are associated with male infertility. Additionally, three genes are currently no definitive pathogenic reports, which may account for the normal phenotype observed in the individual. To date, there have been no documented cases involving ins (6; 2), inv (6) (q21q21), or inv (12) (q12q12). Over the past 3 decades, only ten cases of t (6; 12) have been reported, with the involved sites differing and potentially associated with other chromosomal abnormalities (Vandeweyer et al., 2012; Bernheim et al., 2007; Bianco et al., 2011; Coccé et al., 2016), all of which exhibited a variety of clinical manifestations, while other instances may remain unreported due to the absence of identifiable clinical phenotypic abnormalities.
However, OGM presents certain limitations. Primarily, the absence of a comprehensive human reference genome, coupled with the presence of excessively long repeat sequences in the centromeric regions, hinders OGM’s ability to accurately detect balanced structural variations, breakpoints, and fusion sites within structural heterochromatin areas, including centromeres and the short arms of telomeric chromosomes (Qu et al., 2023). The successful completion of telomere-to-telomere genome assembly and the subsequent gap filling of the human reference genome offer promising prospects for overcoming the limitations associated with OGM (Gershman et al., 2022). A recent study indicates that the integration of sequencing techniques and OGM could facilitate the comprehensive assembly of the human X chromosome from telomere to telomere (Miga et al., 2020). Secondly, it is essential to provide specific technical support to ensure the accuracy of the BPs identified by OGM at the single nucleotide level. Furthermore, the relationship between the newly identified variations and the corresponding phenotypes requires additional verification.
Conclusion
The sensitivity of OGM in this study surpasses that of other established molecular technologies for detecting large-fragment translocations and small-fragment CNVs inversions. Furthermore, OGM offers specific breakpoint information for complex structural aberrations and can predict the potential destruction of breakpoint genes, thereby serving as a valuable reference for disease diagnosis, prevention, and assisted reproduction in patients with structural variations. Nonetheless, the high costs and the requirement for more sophisticated testing techniques and analytical processes currently limit its widespread application in clinical practice. Looking ahead, OGM is anticipated to become a widely adopted tool in clinical settings.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Institutional Medical Ethics Committee of Affiliated Hospital of Jining Medical University (No. 2024-03-C019). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
XH: Data curation, Funding acquisition, Methodology, Supervision, Validation, Writing – original draft, Writing – review and editing. JG: Software, Validation, Writing – review and editing. HS: Investigation, Resources, Visualization, Writing – review and editing. JY: Data curation, Methodology, Project administration, Writing – review and editing. HC: Formal Analysis, Supervision, Validation, Writing – review and editing. TL: Data curation, Project administration, Visualization, Writing – review and editing. HD: Formal Analysis, Investigation, Methodology, Project administration, Supervision, Writing – review and editing. MK: Conceptualization, Data curation, Formal Analysis, Methodology, Project administration, Supervision, Visualization, Writing – review and editing. YT: Conceptualization, Formal Analysis, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review and editing. LJ: Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Research Fund for Jining City Key Research and Development Plan Project (Nos 2022YXNS013 and 2023YXNS178).
Acknowledgments
We thank all participants. We certainly are thankful to Beijing Beikang Medical Laboratory for providing the process and results of OGM for this study. XH and LJ were involved in the study design, data analysis, and manuscript drafting. YT and JY edited and critically reviewed the manuscript. HC, JG, HS, and TL collected clinical data and provided assistance for the manuscript. HD and MK revised the manuscript and provided data analysis. All authors contributed to the final manuscript and approved the submitted version. We were also grateful to the doctors for their assistance in collecting samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antonacci, F., Kidd, J. M., Marques-Bonet, T., Brian, T., Ventura, M., Girirajan, S., et al. (2010). A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat. Genet. 42 (9), 745–750. doi:10.1038/ng.643
Barseghyan, H., Délot, E. C., and Vilain, E. (2018). New technologies to uncover the molecular basis of disorders of sex development. Mol. Cell. Endocrinol. 468, 60–69. doi:10.1016/j.mce.2018.04.003
Bernheim, A, Toujani, S, Guillaud-Bataille, M, Richon, C., Waxin, H., Dessen, P., et al. (2007). Intragenic breakpoints localized by array CGH in a t(2;6) familial translocation. Cytogenet. Genome. Res. 119 (3-4), 185–190.
Bianco, B., Christofolini, D., Gava, M., Mafra, F., Moraes, E., and Barbosa, C. (2011) Severe oligospermia associated with a unique balanced reciprocal translocation t(6;12)(q23;q24.3): male infertility related to t(6;12). Andrologia. 43 (2), 145–148.
Carvalho, C. M., and Lupski, J. R. (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17 (4), 224–238. doi:10.1038/nrg.2015.25
Chan, E. K. F., Cameron, D. L., Petersen, D. C., Lyons, R. J., Baldi, B. F., Papenfuss, A. T., et al. (2018a). Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 28 (5), 726–738. doi:10.1101/gr.227975.117
Chan, S., Lam, E., Saghbini, M., Bocklandt, S., Hastie, A., Cao, H., et al. (2018b). “Structural variation detection and analysis using Bionano optical mapping,” in Copy number variants: methods and protocols, derek M. Bickhart. New York, NY: Springer, 193–203. doi:10.1007/978-1-4939-8666-8_16
Chow, J. F. C., Cheng, H. H. Y., Lau, E. Y. L., Yeung, W. S. B., and Ng, E. H. Y. (2020). Distinguishing between carrier and noncarrier embryos with the use of long-read sequencing in preimplantation genetic testing for reciprocal translocations. Genomics 112 (1), 494–500. doi:10.1016/j.ygeno.2019.04.001
Coccé, M. C., Mardin, B. R., Bens, S., Stütz, A. M., Lubieniecki, F., Vater, I., et al. (2016) Identification of ZCCHC8 as fusion partner of ROS1 in a case of congenital glioblastoma multiforme with a t(6;12)(q21;q24.3). Genes Chromosomes Cancer. 55 (9), 677–687.
Collins, R. L., Brand, H., Karczewski, K. J., Zhao, X., Alföldi, J., Francioli, L. C., et al. (2020). A structural variation reference for medical and population genetics. Nature 581 (7809), 444–451. doi:10.1038/s41586-020-2287-8
Cui, C., Shu, W., and Li, P. (2016). Fluorescence in situ hybridization: cell-based genetic diagnostic and research applications. Front. Cell. Dev. Biol. 4, 89. doi:10.3389/fcell.2016.00089
Dong, Z., Wang, H., Chen, H., Jiang, H., Yuan, J., Yang, Z., et al. (2017). Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 Genomes Project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Official J. Am. Coll. Med. Genet. 20 (7), 697–707. doi:10.1038/gim.2017.170
Dremsek, P., Schwarz, T., Weil, B., Malashka, A., Franco, L., and Jürgen, N. (2021). Optical genome mapping in routine human genetic diagnostics—its advantages and limitations. Genes. 12 (12), 1958. doi:10.3390/genes12121958
Du, C., Mark, D., Wappenschmidt, B., Böckmann, B., Pabst, B., Chan, S., et al. (2018). A tandem duplication of BRCA1 exons 1–19 through DHX8 exon 2 in four families with hereditary breast and ovarian cancer syndrome. Breast Cancer Res. Treat. 172 (3), 561–569. doi:10.1007/s10549-018-4957-x
Gershman, A., Sauria, M. E. G., Guitart, X., Vollger, M. R., Hook, P. W., Hoyt, S. J., et al. (2022). Epigenetic patterns in a complete human genome. Sci. (New York, N.Y.) 376 (6588), eabj5089. doi:10.1126/science.abj5089
Goumy, C., Guy Ouedraogo, Z., Soler, G., Eymard-Pierre, E., Laurichesse, H., Delabaere, A., et al. (2023). Optical genome mapping for prenatal diagnosis: a prospective study. Int. J. Clin. Chem. 551, 117594. doi:10.1016/j.cca.2023.117594
Halgren, C., Nielsen, N. M., Nazaryan-Petersen, L., Silahtaroglu, A., Collins, R. L., Lowther, C., et al. (2018). Risks and recommendations in prenatally detected de novo balanced chromosomal rearrangements from assessment of long-term outcomes. Am. J. Hum. Genet. 102 (6), 1090–1103. doi:10.1016/j.ajhg.2018.04.005
Hofherr, S. E., Wiktor, A. E., Kipp, B. R., Brian Dawson, D., and Van Dyke, D. L. (2011). Clinical diagnostic testing for the cytogenetic and molecular causes of male infertility: the mayo clinic experience. J. Assisted Reproduction Genet. 28 (11), 1091–1098. doi:10.1007/s10815-011-9633-6
Hu, L., Wei, Y., Luo, K., Xie, P., Gong, F., Xiong, Bo, et al. (2018). Clinical outcomes in carriers of complex chromosomal rearrangements: a retrospective analysis of comprehensive chromosome screening results in seven cases. Fertil. Steril. 109 (3), 486–492. doi:10.1016/j.fertnstert.2017.11.021
Hu, P., Xu, Y., Zhang, Q., Zhou, R., Ji, X., Wang, Y., et al. (2024). Prenatal diagnosis of chromosomal abnormalities using optical genome mapping vs chromosomal microarray. Am. J. Obstetrics Gynecol. 230 (5), e82–e83. doi:10.1016/j.ajog.2023.12.012
Jain, A., Sheats, J., Reifenberger, J. G., Cao, H., and Dorfman, K. D. (2016). Modeling the relaxation of internal DNA segments during genome mapping in nanochannels. Biomicrofluidics 10 (5), 054117. doi:10.1063/1.4964927
Kasikova, K., Vozdova, M., Prinosilova, P., Gaillyova, R., Hanakova, M., and Jiri, R. (2012). Sperm meiotic segregation, aneuploidy and high risk of delivering an affected offspring in carriers of non-robertsonian translocation t(13;15). J. Assisted Reproduction Genet. 29 (7), 693–698. doi:10.1007/s10815-012-9767-1
Lee, K.-S., Choi, M., Kwon, D.-W., Kim, D., Choi, J.-M., Kim, A.-K., et al. (2020). A novel de novo heterozygous DYRK1A mutation causes complete loss of DYRK1A function and developmental delay. Sci. Rep. 10, 9849. doi:10.1038/s41598-020-66750-y
Levy, B., Baughn, L. B., Akkari, Y., Chartrand, S., LaBarge, B., Claxton, D., et al. (2022). Optical genome mapping in acute myeloid leukemia: a multicenter evaluation. Blood Adv. 7 (7), 1297–1307. doi:10.1182/bloodadvances.2022007583
Liao, Y., Wang, L., Zhang, D., and Liu, C. (2014). Identification of a balanced complex chromosomal rearrangement involving chromosomes 3, 18 and 21 with recurrent abortion: case report. Mol. Cytogenet. 7, 39. doi:10.1186/1755-8166-7-39
Lionel, A. C., Costain, G., Monfared, N., Walker, S., Reuter, M. S., Mohsen Hosseini, S., et al. (2017). Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 20 (4), 435–443. doi:10.1038/gim.2017.119
MacLeod, R. A. F., and Drexler, H. G. (2013). “Classical and molecular cytogenetic analysis. Basic cell culture protocols 946:39–60,” in Methods in molecular biology. Totowa, NJ: Humana Press. doi:10.1007/978-1-62703-128-8_4
Mantere, T., Neveling, K., Pebrel-Richard, C., Benoist, M., van der Zande, G., Kater-Baats, E., et al. (2021). Optical genome mapping enables constitutional chromosomal aberration detection. Am. J. Hum. Genet. 108 (8), 1409–1422. doi:10.1016/j.ajhg.2021.05.012
Miga, K. H., Koren, S., Rhie, A., Vollger, M. R., Gershman, A., Bzikadze, A., et al. (2020). Telomere-to-Telomere assembly of a complete human X chromosome. Nature 585 (7823), 79–84. doi:10.1038/s41586-020-2547-7
Miller, D. T., Adam, M. P., Aradhya, S., Biesecker, L. G., Brothman, A. R., Carter, N. P., et al. (2010). Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 86 (5), 749–764. doi:10.1016/j.ajhg.2010.04.006
Neveling, K., Mantere, T., Vermeulen, S., Oorsprong, M., van Beek, R., Kater-Baats, E., et al. (2021). Next-generation cytogenetics: comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am. J. Hum. Genet. 108 (8), 1423–1435. doi:10.1016/j.ajhg.2021.06.001
Nilius-Eliliwi, V., Gerding, W. M., Schroers, R., Nguyen, H. P., and Vangala, D. B. (2023). Optical genome mapping for cytogenetic diagnostics in AML. Cancers 15 (6), 1684. doi:10.3390/cancers15061684
Qu, J., Li, S., and Yu, D. (2023). Detection of complex chromosome rearrangements using optical genome mapping. Gene 884, 147688. doi:10.1016/j.gene.2023.147688
Riggs, E. R., Andersen, E. F., Cherry, A. M., Kantarci, S., Kearney, H., Patel, A., et al. (2019). Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Official J. Am. Coll. Med. Genet. 22 (2), 245–257. doi:10.1038/s41436-019-0686-8
Sahajpal, N. S., Barseghyan, H., Kolhe, R., Hastie, A., and Alka, C. (2021). Optical genome mapping as a next-generation cytogenomic tool for detection of structural and copy number variations for prenatal genomic analyses. Genes. 12 (3), 398. doi:10.3390/genes12030398
Sanders, A. D., Meiers, S., Ghareghani, M., Porubsky, D., Jeong, H., Alexandra, M., et al. (2019). Single-cell analysis of structural variations and complex rearrangements with tri-channel-processing. Nat. Biotechnol. 38 (3), 343–354. doi:10.1038/s41587-019-0366-x
Savara, J., Novosád, T., Gajdoš, P., and Kriegová, E. (2021). Comparison of structural variants detected by optical mapping with long-read next-generation sequencing. Bioinformatics 37 (20), 3398–3404. doi:10.1093/bioinformatics/btab359
Schluth-Bolard, C., Diguet, F., Chatron, N., Rollat-Farnier, P.-A., Bardel, C., Afenjar, A., et al. (2019). Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 56 (8), 526–535. doi:10.1136/jmedgenet-2018-105778
Shieh, J. T., Penon-Portmann, M., Wong, K. H. Y., Levy-Sakin, M., Verghese, M., Slavotinek, A., et al. (2021). Application of full-genome analysis to diagnose rare monogenic disorders. NPJ Genomic Med. 6, 77. doi:10.1038/s41525-021-00241-5
Sinnerbrink, I. B., Sherwen, A., Meiser, B., Halliday, J., Amor, D. J., Waters, E., et al. (2013). Long-term health and development of children diagnosed prenatally with a de novo apparently balanced chromosomal rearrangement. Prenat. Diagn. 33 (9), 831–838. doi:10.1002/pd.4131
Vandeweyer, G., Van der Aa, N., Ceulemans, B., van Bon, B. W., Rooms, L., and Kooy, R. F. (2012). A de novo balanced t(2;6)(p15;p22.3) in a patient with West Syndrome disrupts a lnc-RNA. Epilepsy Res. 99 (3), 346–349.
Waggoner, D., Wain, K. E., Dubuc, A. M., Conlin, L., Hickey, S. E., Lamb, A. N., et al. (2018). Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Official J. Am. Coll. Med. Genet. 20 (10), 1105–1113. doi:10.1038/s41436-018-0040-6
Wang, H., Jia, Z., Mao, A., Xu, B., Wang, S., Wang, Li, et al. (2020). Analysis of balanced reciprocal translocations in patients with subfertility using single-molecule optical mapping. J. Assisted Reproduction Genet. 37 (3), 509–516. doi:10.1007/s10815-020-01702-z
Watson, C. T., Marques-Bonet, T., Sharp, A. J., and Mefford, H. C. (2014). The genetics of microdeletion and microduplication syndromes: an update. Annu. Rev. Genomics Hum. Genet. 15, 215–244. doi:10.1146/annurev-genom-091212-153408
Wilch, E. S., and Morton, C. C. (2018). “Historical and clinical perspectives on chromosome translocation. 1044,” in Advances in experimental medicine and biology. Singapore: Springer Singapore. doi:10.1007/978-981-13-0593-1
Yildirim, M. E., Karakus, S., Kucuk Kurtulgan, H., Baser, B., and Sezgin, I. (2019). The type and prevalence of chromosomal abnormalities in couples with recurrent first trimester abortions: a Turkish retrospective study. J. Gynecol. Obstetrics Hum. Reproduction 48 (7), 521–525. doi:10.1016/j.jogoh.2019.05.014
Zhang, Q., Wang, Y., Xu, Y., Zhou, R., Huang, M., Qiao, F., et al. (2023a). Optical genome mapping for detection of chromosomal aberrations in prenatal diagnosis. Acta Obstetricia Gynecol. Scand. 102 (8), 1053–1062. doi:10.1111/aogs.14613
Keywords: optical genome mapping, high-resolution karyotyping analysis, chromosomal microarray analysis, balanced reciprocal rearrangements, complex chromosomal rearrangements
Citation: Hu X, Guo J, Sang H, Yan J, Chang H, Liu T, Dong H, Kong M, Tian Y and Jiang L (2025) Case Report: Optical genome mapping enables identification of complex balanced chromosomal rearrangements. Front. Genet. 16:1555485. doi: 10.3389/fgene.2025.1555485
Received: 04 January 2025; Accepted: 29 April 2025;
Published: 22 May 2025.
Edited by:
Terje Raudsepp, Texas A&M University, United StatesReviewed by:
Denilce Lopes, Universidade Federal de Viçosa, BrazilRoberto Valli, University of Insubria, Italy
Copyright © 2025 Hu, Guo, Sang, Yan, Chang, Liu, Dong, Kong, Tian and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjun Tian, dHlqai0yMDBAMTYzLmNvbQ==; Liqing Jiang, anlqaWFuZ2xpcWluZ0AxNjMuY29t
 Xiaohang Hu
Xiaohang Hu Jing Guo2
Jing Guo2 Ting Liu
Ting Liu Min Kong
Min Kong Yanjun Tian
Yanjun Tian Liqing Jiang
Liqing Jiang