- National Key Laboratory of Pig Genetic Improvement and Germplasm Innovation, Jiangxi Agricultural University, Nanchang, Jiangxi, China
Skeletal muscle fiber traits are fundamental to meat production and the meat quality of agricultural animals. The rich genetic resources and diverse phenotypic expression of muscle traits in agricultural animal species provide invaluable materials for investigating the genetic and molecular regulatory mechanisms underlying myofiber development and characteristics, optimizing breeding strategies, and developing models for human muscle-related diseases. This review presents an integrative perspective on the genetic and epigenetic regulation of skeletal muscle fiber development, incorporating evolutionary, genomic, epigenomic, and multi-omics insights. We focus on genetic architecture and causative or candidate genes for muscle fiber traits, as revealed by genome-wide association studies (GWAS) and selective sweep signatures, underscoring their adaptive significance and potential for selective breeding. The role of epigenetic mechanisms, such as DNA methylation, histone modifications, and non-coding RNAs, in linking genetic variation and phenotypic expression is also discussed. By synthesizing multi-omic data, we provide a comprehensive understanding of the molecular networks driving muscle fiber growth and differentiation. This review aims to consolidate current knowledge and offer actionable insights to advance research, breeding strategies, and applications in agricultural and biomedical fields.
1 Introduction
Despite ongoing efforts, hunger and food insecurity persist, with billions of individuals lacking access to nutritious, safe, and adequate food (UNICEF, 2024). With the global population projected to reach 9.7 billion by 2050 (UN 2015), the demand for food is anticipated to surge. Agricultural animals are pivotal in global food production (Davis and White, 2020), with meat serving as a crucial dietary protein source. By 2033, poultry meat is expected to constitute 43% of protein intake from all meat sources, trailed by pork, beef, and mutton. Moreover, meat is a vital provider of essential nutrients, including vitamins and minerals (Gehring, 2023). According to the Organization for Economic Cooperation and Development (Oecd, 2024)/Food and Agriculture Organization of the United Nations (FAO) (Oecd, 2024), it is projected that by the year 2033, there will be a global increase in the consumption of poultry, pork, beef, and sheep meat by 16%, 8%, 11%, and 16%, respectively. Furthermore, per capita meat consumption is anticipated to rise by 2%. This escalating demand, combined with heightened consumer expectations regarding meat quality, underscores the imperative to enhance the production efficiency of agricultural animals.
Skeletal muscle, which constitutes approximately 40% of an animal’s body weight (Erimbetov et al., 2019), is pivotal to meat production and performs essential functions in metabolism, movement, and energy storage. The development of skeletal muscle is governed by intricate interactions between genetic factors, such as regulatory genes and non-coding RNA, and environmental influences. These interactions determine critical muscle fiber traits, including number, diameter, and type, directly affecting meat yield and quality. Through natural selection and selective breeding, agricultural animals have acquired distinct characteristics that set them apart from humans and conventional model organisms. Their genetic diversity and economic importance make them valuable subjects for muscle development research. Notably, swine have emerged as prominent models for human medical research due to their genetic and physiological resemblances with humans, thereby propelling advancements in the domains of genetics, developmental biology, disease mechanisms, and xenotransplantation (Prather, 2013; Fang et al., 2024; Meyerholz et al., 2024; Peterson et al., 2024). Therefore, a comprehensive understanding of the genetic and regulatory mechanisms underlying skeletal muscle growth is imperative for enhancing meat production, optimizing breeding strategies, and contributing to advancements in medical science.
Recent advances in molecular biology techniques and high-throughput sequencing technologies have significantly expanded our knowledge of the genetic and epigenetic frameworks underlying muscle fiber traits in agricultural animals. This review provides a comprehensive overview of how genetic and epigenetic factors contribute to skeletal muscle growth and myofiber characteristics in agricultural animals from evolutionary, genomic, epigenomic, and multi-omics perspectives.
2 Muscle morphological diversity in agricultural animals and its impact on meat productivity and quality
Agricultural animals exhibit substantial genetic diversity, shaped by natural evolution and intensive artificial selection (Andersson, 2001; Koshkina et al., 2023). This diversity is vital for expressing economically significant and adaptive traits, as well as understanding molecular mechanisms behind muscle growth and development (Zhong et al., 2020). Unlike genetically uniform model organisms like mice, livestock and poultry display significant variation within and across breeds (Jiang et al., 2019; Jiang et al., 2018). For example, Chinese local pig breeds exhibit superior traits such as high fertility and excellent meat quality (Huang et al., 2020), while poultry and cattle breeds reflect diverse genetic and phenotypic outcomes from domestication and targeted breeding (Makarova et al., 2019; Wang et al., 2020; Bennett et al., 2008; García-Ruiz et al., 2016). In cattle, small effective population sizes due to rigorous selection enhance the detection of recessive deleterious mutations, which are less prevalent than in natural populations due to purification mechanisms (Kemper et al., 2012; Eyre-Walker, 2010). This genetic diversity not only underpins agricultural advancements but also offers valuable insights for human biomedical research.
Given the challenges associated with direct human disease studies, including the limitations in available samples, environmental constraints, and ethical considerations, the use of animal models is imperative for elucidating disease mechanisms. Agricultural animals like pigs, cows, and sheep surpass traditional models like mice due to their closer physiological and genetic alignment with humans (Muroya, 2022; Petersen, 2023). Pigs, in particular, are considered to be significant candidates for xenotransplantation, vaccine development, and modeling human development, owing to their anatomical and genomic similarities (Abdoli et al., 2018; Lunney et al., 2021). For instance, porcine models of Duchenne muscular dystrophy (DMD) replicate human clinical features more effectively than mice, with studies demonstrating gene restoration and phenotypic parallels (Perleberg et al., 2018; Yu et al., 2016). These advantages highlight the critical role of agricultural animals in biomedical research.
Beyond their biomedical value, the genetic and phenotypic traits of agricultural animals are pivotal for enhancing meat production and quality. Myofiber characteristics, including their number, size, type, and metabolism, play a fundamental role in determining meat yield and quality (Lefaucheur, 2010; Joo et al., 2013). A comprehensive understanding of these traits is imperative for the optimization of breeding and management practices, thereby ensuring the fulfillment of consumer demand for meat products of superior quality (Godfray et al., 2018; Kopler et al., 2023).
Myofiber traits vary due to factors like breed, sex, physiological stage, nutrition, and environment. For instance, Chinese local pig breeds show higher levels of type I, IIa, and IIx myofibers compared to foreign breeds (Kahane et al., 2001), while Turkish sheep demonstrate a link between fiber diameter and tenderness (Şirin et al., 2017). Dairy cows possess more oxidized fibers than beef cattle, reflecting metabolic differences (Schreurs et al., 2008), and ducks have higher myofiber counts but smaller fiber sizes than chickens (Kim et al., 2008). Generally, pigs and poultry favor fast-twitch fibers for rapid growth, whereas cattle and sheep rely on slow-twitch fibers for endurance, showcasing species-specific adaptations.
These muscle fiber traits directly impact meat quality and production efficiency, influencing attributes like color, pH, and tenderness (Ismail and Joo, 2017). Type I fibers’ myoglobin content affects color and flavor (Buckingham and Rigby, 2014; Matarneh et al., 2021; Mashima et al., 2019), while glycolytic fibers correlate with shear force (Kim et al., 2016; Park et al., 2024). Genetic analyses reveal strong links between fiber density and quality indicators such as eye muscle area and color (Larzul et al., 1997; Choi and Kim, 2009). Myofiber number drives postnatal growth and feed efficiency (Wigmore and Stickland, 1983), yet selection for traits like increased muscle mass can reduce fiber counts, risking quality declines (Felício et al., 2013). Balancing fiber number and size is thus essential for optimal meat production (Callaghan et al., 2020; Li et al., 2015). In general, a higher number of muscle fibers combined with an appropriately sized muscle fiber cross-sectional area can improve the final lean meat productivity and ensure normal meat quality, while increasing the number and area ratio of type I and IIA muscle fibers and reducing the number and area ratio of type IIB muscle fibers to achieve a balance between meat quality and yield. A deeper grasp of the genetic and biological underpinnings of these traits can steer breeding programs toward improved meat quality and efficiency.
3 Genetic signals of selection in muscle mass and fiber traits
Agricultural animals, subjected to prolonged natural selection and artificial breeding, have exhibited marked adaptive alterations in morphology, physiology, and behavior. These phenotypic modifications represent the genome’s response to environmental challenges and human intervention, offering a distinct research opportunity to explore how genetic variation influences phenotypic diversity. With recent advancements in genomics technology, scientists have been able to pinpoint genetic selection signals linked to key economic traits, facilitating a deeper understanding of the molecular underpinnings of muscle growth and development.
Leif Andersson et al. (Rubin et al., 2010) employed massively parallel sequencing to discern advantageous alleles and candidate mutations that have significantly influenced chicken domestication. Notably, a selective sweep in broiler chickens aligns with genes including IGF1 and TBC1D1, which are implicated in growth, appetite, and metabolic regulation. In addition, Tan et al. (2024) found that the SOX6 gene has a key regulatory role in broiler breast muscle traits and adaptation to artificial selection. These insights elucidate the genetic underpinnings of rapid muscle growth in contemporary broiler breeds. Zhou et al. (2018) pinpointed two mutations in Pekin duck genes linked to white feathers, augmented body size, and enhanced feed-conversion efficiency through a comparative genomic analysis across diverse duck breeds. One mutation in the MITF gene correlates with white feather coloration, while another distinct mutation might induce sustained expression of the IGF2BP1 gene postnatally in Pekin ducks, thereby augmenting meat yield. Skeletal muscle consists of multiple heterogeneous cell populations whose interactions are critical for the maintenance of muscle homeostasis. Xu et al. (2023) analyzed neonatal skeletal muscle cell populations from wild boar, Duroc, and Laiwu pigs using single-cell RNA sequencing (scRNA-seq) to investigate the effects of artificial selection on muscle cell profiles. It was found that compared to wild boar, domestic pigs had significantly more fiber adipose-forming progenitor (FAP) cells and fewer myoblasts, while the proliferation rate of myogenic progenitor cells was higher, suggesting a greater muscle growth potential in domestic pigs. In addition, cross-species comparisons identified 186 shared active transcription factors, as well as some (e.g., CREB3L1, TGIF1, NFIC, and CEBPZ) gene regulatory networks that have been shown for the first time to have a potential role in muscle development. These results provide new perspectives for understanding the effects of artificial selection on the spectrum of muscle-resident cells. Furthermore, Liu et al. (2024) unveiled epigenetic mechanisms that account for disparities in muscle growth between eastern and western pig breeds. Their analyses suggest that artificial selection impacts DNA methylation and gene regulation, subsequently affecting muscle growth and meat quality. The research highlighted pivotal genes such as GHSR and BDH1 in modulating skeletal muscle development and meat production. Specifically, the GHSR gene fosters myoblast proliferation but impedes their differentiation and fusion. A C>T mutation in its intronic region exerts an allelic genetic effect of 4.22 kg on carcass weight at 240 days of age. Concurrently, an insertion acts as a potential enhancer regulating the BDH1 gene in eastern pigs, causing variances in myofiber proliferation and differentiation across breeds. BDH1 is a crucial rate-limiting enzyme in ketone metabolism and ATP synthesis. The insertion-triggered elevated expression of BDH1 may be instrumental in myofiber proliferation and differentiation.
Research into genomic selection signals in agricultural animals has unveiled numerous critical genes and mutations associated with muscle growth and development. These insights not only elucidate the role of genetic variations in shaping phenotypic diversity but also offer a scientific foundation for enhancing agricultural animal breeds and boosting the efficiency of meat production.
4 Genetic architecture of myofiber traits
Heritability, a significant concept in animal breeding, quantifies the proportion of phenotypic variation in a trait attributable to genetic factors (Visscher et al., 2008). Concerning myofiber traits in agricultural animals, heritability estimation serves as a critical metric for evaluating the potential of these traits for genetic selection. A comprehensive understanding of myofiber trait heritability can inform the development of more scientifically rigorous and effective breeding strategies, thereby enhancing meat yield and quality. In recent years, advancements in quantitative genetics and genomic technologies have led to increasingly precise estimates of myofiber trait heritability, thereby providing crucial reference points for genetic improvement programs.
Heritability estimates for muscle fiber traits demonstrate considerable variation across different agricultural animal species. For instance, a study (Li H. et al., 2024) on Gushi chickens reported low heritability for both leg muscle fiber density and diameter (ranging from 0 to 0.2), with moderate heritability observed for intramuscular fat (0.35). On the other hand, heritability estimates for fiber traits in Kashmiri Merino sheep varied between 0.189 and 0.300 (Ahmad et al., 2022), suggesting some potential for genetic selection within these traits. In the case of Large White pigs (Larzul et al., 1997), myofiber traits exhibited moderate to high heritabilities, such as a heritability of 0.46 for type I fiber percentage and up to 0.58 for type IIB fiber percentage.
Further studies showed that myofiber traits also have large genetic variation across breeds and populations. Yan et al. (2024) conducted an analysis of the muscle fiber types and meat quality traits in 318 pigs from four distinct Shanxia Changhei pig populations. They discovered that the heritability range for meat quality traits spanned from 0.06 (pH at 24 h) to 0.47 (shearing force), while the heritability for muscle fiber types varied between 0.04 and 0.4. Notably, the highest heritability was observed for total fiber density (0.40), whereas the lowest was for the percentage of type IIA fibers (0.04). These findings align with those of previous studies (Lee et al., 2022), which reported heritability values ranging from 0.13 to 0.55. Furthermore, Huang et al. (Huang et al., 2022) determined that the heritability of muscle fiber number is approximately 0.3–0.54, considered moderate, and noted a coefficient of variation as high as 18.64% for the total number of muscle fibers. In a study involving the White Duroc × Erhualian F2 resource population, Li W. et al. (2009) observed a substantial 3.6-fold difference between the minimum and maximum values of the total number of muscle fibers. These estimates imply that pigs and broilers, with moderate to high heritability, are prime candidates for selection to improve myofiber traits, potentially enhancing meat quality and yield efficiently. Conversely, the lower heritability in sheep suggests that selection may yield slower progress, requiring alternative strategies like crossbreeding to capitalize on hybrid vigor. The variability across studies underscores how species, population structure, and genetic background shape trait inheritance, necessitating customized breeding approaches.
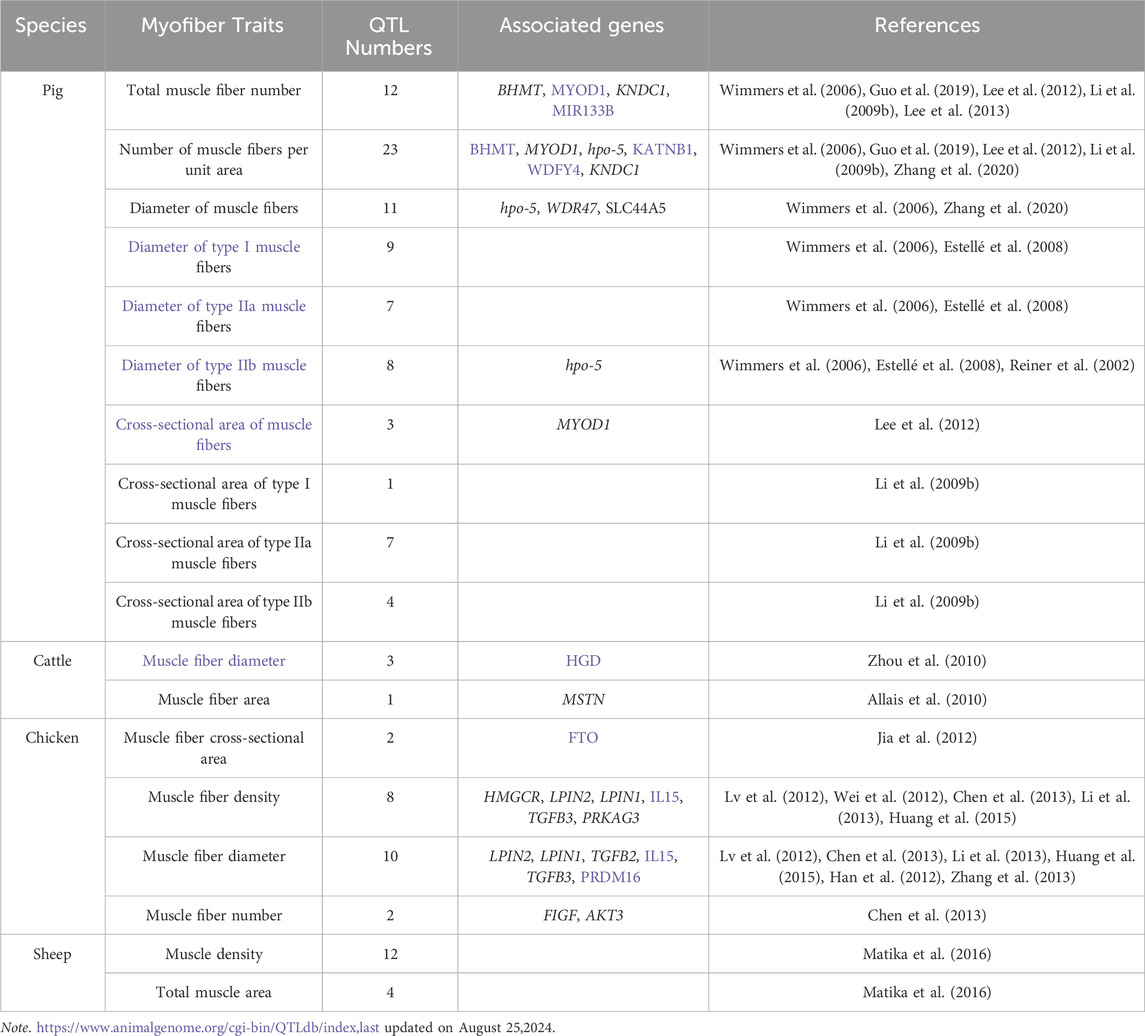
The majority of myofiber traits in agricultural animals are controlled by multiple quantitative trait loci (QTL). There have been, and continue to be, successful attempts to localize QTL affecting myofiber traits (Wimmers et al., 2006; Guo et al., 2019). Table 1 provides a summary of the QTLs that impact the muscle fiber traits of pigs, cattle, sheep, and chickens as listed in the Animal QTLdb database. To date, there have been 12 documented QTLs that affect the total number of muscle fibers in pigs (https://www.animalgenome.org/cgi-bin/QTLdb/index, last updated on August 25, 2024). Further, we analyzed the biological pathway enrichment of these QTL-associated genes in Table 1 and found that they were significantly enriched in pathways related to muscle growth and development, as shown in Figure 1. In addition, Huang et al. (2022) conducted a genome-wide association study (GWAS) for nine myofiber traits using whole-genome sequence data from a heterogeneous population of eight breed crosses. Sixty-seven quantitative trait loci (QTL) for these traits were revealed, several loci were found to be significantly associated with the myofiber number phenotype, and some key candidate genes were identified. These results have facilitated the process of resolving the genetics of myofiber traits. However, despite the success of GWAS in identifying multiple loci, the heritability explained by these loci is usually lower than the total heritability estimate, a phenomenon known as “missing heritability” (Eichler et al., 2010; Trerotola et al., 2015). This discrepancy may stem from undetected factors such as epistatic interactions, rare variants, or epigenetic modifications. Current genomic tools, while powerful, struggle to capture these complexities, limiting the resolution of trait genetics. Addressing this requires advanced approaches like whole-genome sequencing, multi-omics integration, and the utilization of extensive, diverse datasets to elucidate the comprehensive genetic underpinnings of myofiber traits.
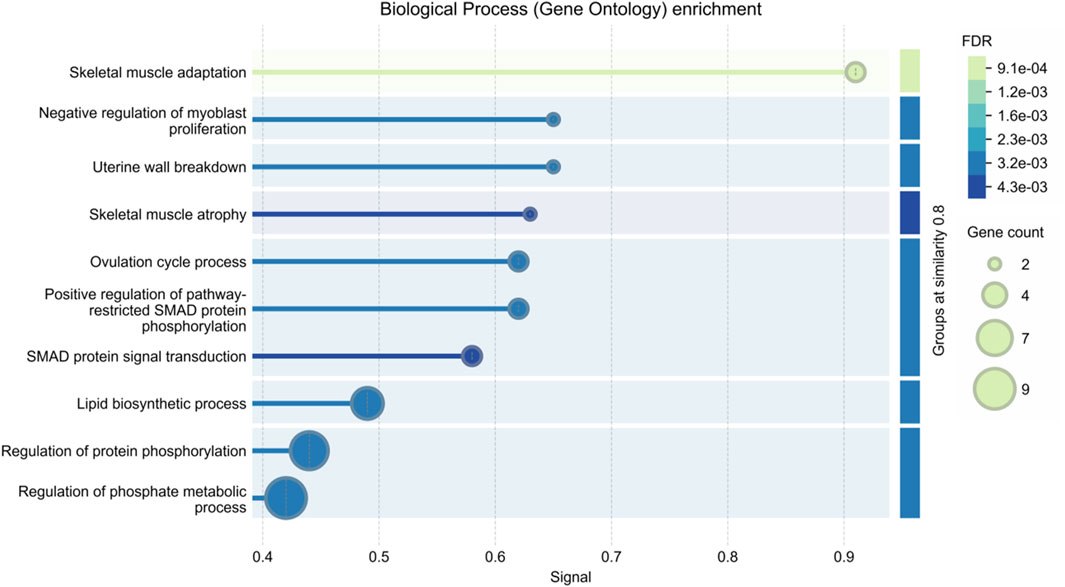
Figure 1. Enrichment analysis of biological pathways in quantitative trait loci (QTL)-associated genes for muscle fiber traits in agricultural animals.
Overall, muscle fiber traits in agricultural animals show different levels of estimated heritability, which highlights their potential application value in future genetic improvement programs, especially in improving meat quality and yield. However, the effective application of these traits in breeding practices requires a deep understanding of the species-specific genetic structure, as heritability and its trait expression can vary significantly among different livestock species. In addition, it is crucial to use advanced genomic technologies to develop breeding strategies tailored to these genetic differences to optimize meat production sustainably and efficiently.
5 The molecular regulatory network underlying skeletal muscle fiber development and phenotypes
Muscle fibers, the fundamental units of muscle, determine muscle mass through their number and size. The development of skeletal muscle fibers is a complex, multi-step process that is tightly coordinated. This process can be divided into three stages: primary myogenesis, occurring during the embryonic period, where primary muscle fibers are generated; secondary myogenesis, taking place during the fetal period, marked by the formation of secondary muscle fibers around the primary fibers; and the maturation of muscle fibers post-birth. The total number of muscle fibers in an individual is primarily determined by the primary fibers, with secondary fibers forming additional muscle fibers based on the framework established by the primary ones (Gros et al., 2004). Figure 2 provides a summary of the myogenesis time for major agricultural animals such as pigs, cattle, sheep, and chickens. The two waves of muscle fiber formation during pregnancy dictate the overall number of muscle fibers (Wigmore and Stickland, 1983; Stickland and Handel, 1986), significantly influencing the growth rate and degree of postnatal skeletal muscle development (Dwyer et al., 1993). It is widely accepted that the total number of muscle fibers is fixed at birth, with the postnatal process primarily involving muscle fiber hypertrophy and maturation (Grefte et al., 2007). However, this view has been challenged by Bérard et al. (2011), who found that the total fiber number (TFN) in pig muscles is not entirely fixed at birth. Instead, they observed a postnatal increase in TFN, potentially attributed to the elongation of existing muscle fibers and the formation of tertiary fibers, primarily occurring within 3 weeks after birth. Similarly, a study (Li et al., 2015) using the Cre-loxP system to track muscle fiber formation in various skeletal muscles of mice during development revealed that the TFN in the longissimus dorsi, gastrocnemius, and rectus femoris muscles is established before birth. In contrast, the anterior tibial muscle and extensor digitorum longus exhibited different developmental patterns, with their fiber numbers continuing to increase during the first week post-birth before stabilizing. These findings suggest that the developmental dynamics of muscle fibers may vary significantly across different muscles. Given that the development and growth of skeletal muscle determine the yield and quality of meat, it is crucial to systematically understand and study the regulatory mechanisms of skeletal muscle development in agricultural animals.
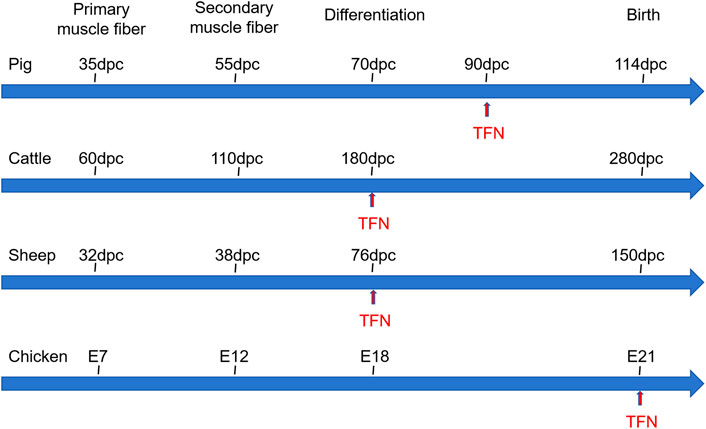
Figure 2. The skeletal muscle developmental processes of pig, cattle, sheep, and chicken. Dpc: day postconception, E: Embryonic days, TFN: total fiber number.
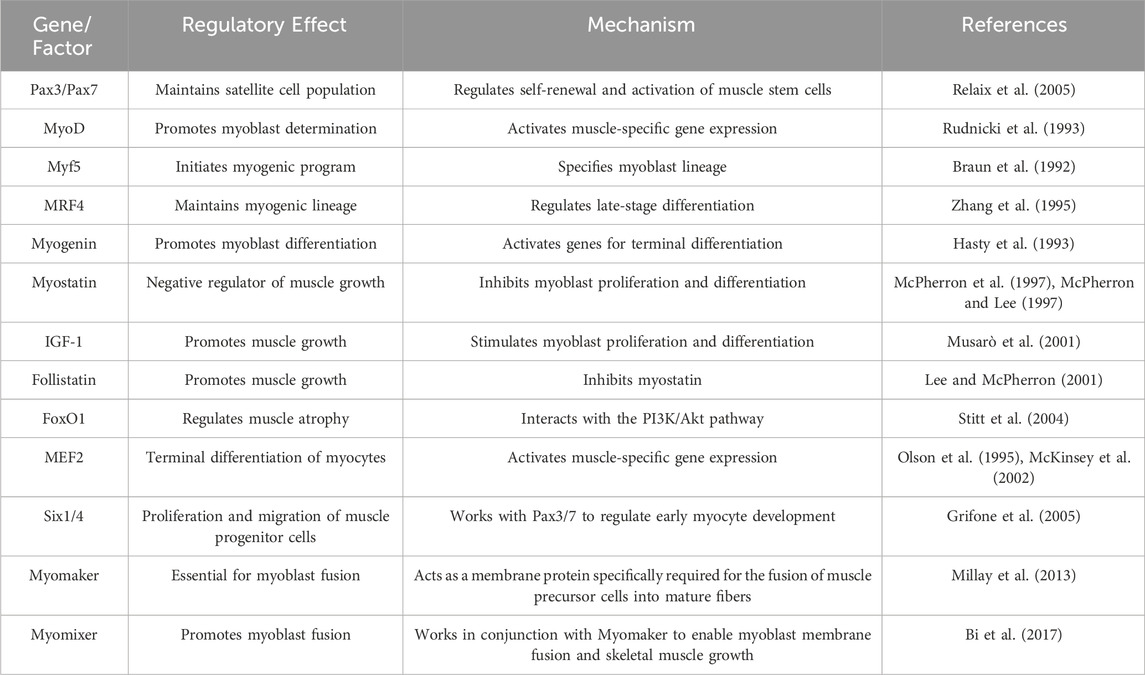
The process of myogenesis is governed by a complex network of gene expression regulation, primarily facilitated through the meticulous control of intercellular signals and specific gene expression. This intricate developmental process involves multiple elements, with genetic factors playing a pivotal role. These are regulated by numerous genes, signaling pathways (Schiaffino et al., 2007), and transcription factors such as Myogenic Regulatory Factors (MRFs) (Zanou and Gailly, 2013) and Muscle Enhancer Factor 2 (MEF2) (Rullman et al., 2018), among others. Table 2 provides a summary of key regulatory factors involved in muscle fiber development. A multitude of genes contribute to the growth and development of skeletal muscle, interacting synergistically to form a complex regulatory network that ensures the stability of skeletal muscle growth and development (Braun and Gautel, 2011; Perry and Rudnick, 2000).
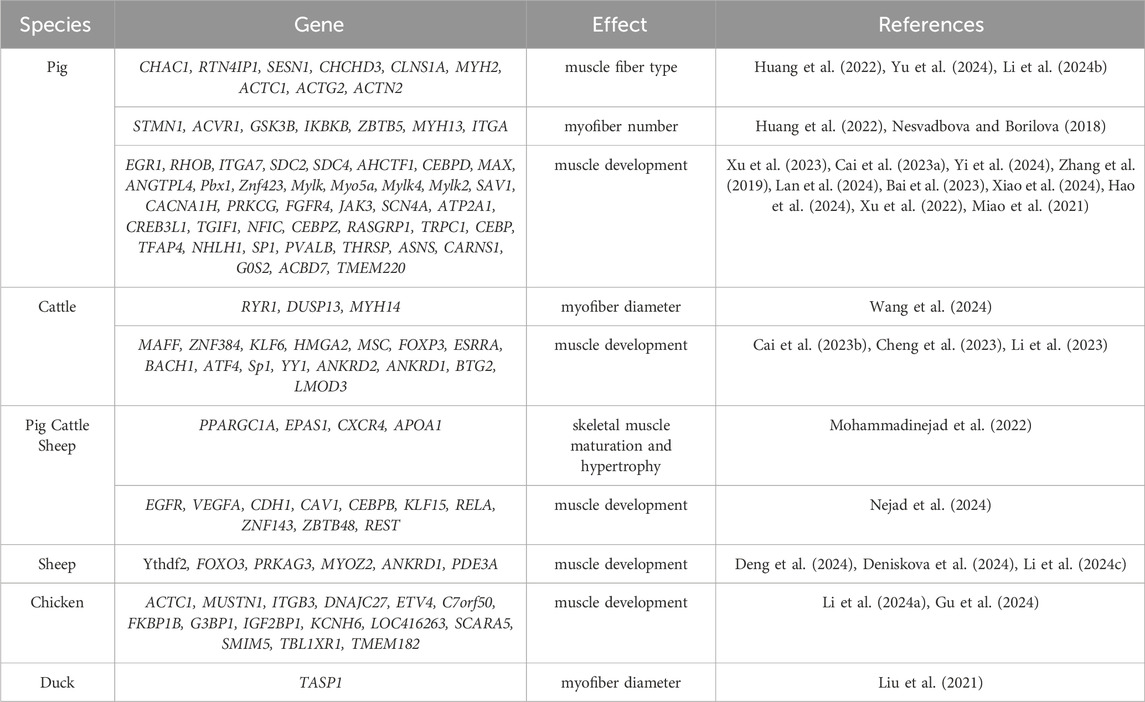
In recent years, candidate genes regulating myofiber development have been continuously identified with the rapid advancement of sequencing technology and the booming development of multi-omics technology. These studies have revealed the important effects of myofiber size and number on muscle mass and yield and provided new perspectives on the molecular regulatory mechanisms of myofiber development.
The diameter of myofibers directly affects muscle mass. Wang et al. (2024) compared gene expression differences between oxidative and glycolytic muscles in bulls by transcriptome sequencing and identified 23 candidate genes (e.g., RYR1, DUSP13, and MYH14) that are related to myofiber diameter. Functional studies of these genes can help to reveal the molecular basis of the differences in fiber size between oxidative and glycolytic muscles. Yu et al. (2024) further constructed a transcriptome profile of skeletal muscle from 27 developmental stages from embryo to adulthood in long white pigs and identified 11 candidate genes (e.g., CHAC1, RTN4IP1, and SESN1), which may play a key role in the myofiber type switching and muscle volume expansion.
An increase in myofiber number is essential to enhance muscle production. A study (Nesvadbova and Borilova, 2018) in Landrace pigs identified several genes (e.g., STMN1, ACVR1, GSK3B, IKBKB, and ITGA) associated with increased myofiber number and regulation of late-stage muscle production. In mice, Myogenin, Klf5, and Tead4 were found to synergistically activate the transcription of genes in developing muscle (Dos Santos et al., 2023), whereas, in mature muscle fibers, the high expression of the transcription factor Maf revealed its important regulatory role in myofiber maturation. Maf was identified as a key factor in myofiber maturation by ChIP-seq experiments and analysis of Maf knockout mice.
Gene editing technology has shown considerable promise in furthering our comprehension of gene function and regulatory mechanisms. Chen et al. (2024) constructed for the first time “double muscle” sheep with double gene editing of MSTN and FGF5 and revealed the molecular mechanism by which double gene editing regulates myofiber proliferation through activating the MEK-ERK pathway. The study showed that the activation of FOSL1 could promote cell proliferation by accelerating the cell cycle transition from the G0/G1 phase to the S phase and inhibit the differentiation of skeletal muscle satellite cells by suppressing the expression of Myod1. In addition, MSTN and FGF5 editing further mediated myofiber proliferation by inhibiting CaMKII-dependent myotube fusion. This study not only revealed the complex regulatory network of myofiber development but also bred a new breed of sheep that combines high meat yield, low fat deposition, and high-quality fine wool.
Recent advances in the molecular regulation of muscle fiber development from the above studies provide important opportunities for enhancing muscle growth and meat production in agricultural animals. The identification of species-specific regulatory mechanisms highlights the importance of studying these processes directly in agricultural animals rather than relying solely on model organism findings.
To systematically summarize the findings in recent years, we have compiled a selection of new candidate genes related to muscle development in agricultural animals (Table 3) and mapped the interaction network of candidate genes related to muscle development in pigs and cattle (Figure 3). For example, YY1 is a transcription factor involved in the regulation of a variety of biological processes, including skeletal muscle development and differentiation (Chen et al., 2019). PPARGC1A is a key transcriptional co-activator that plays an important role in the regulation of energy metabolism and myofiber type transformation in skeletal muscle (Ma et al., 2022). The collaborative impact of these two genes may play an important role in myofiber differentiation, type composition, and muscle regeneration, which may provide some theoretical basis and practical guidance for further studies in the future.
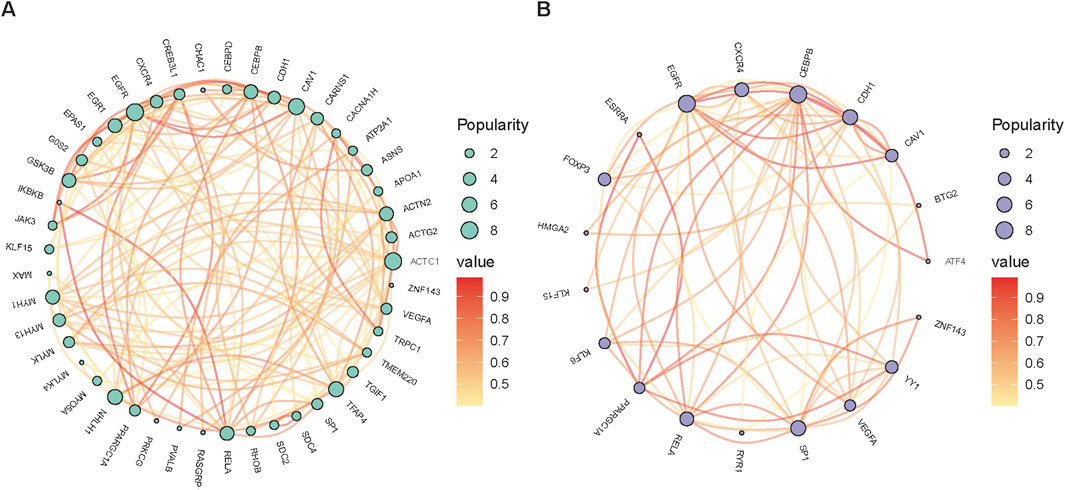
Figure 3. The network of candidate genes linked to muscle development in farm animals, specifically pigs (A) and cattle (B).
6 Epigenetic regulation of myogenesis
In recent years, the rapid development of epigenetics has provided significant breakthroughs in the study of muscle development and its regulatory mechanisms in agricultural animals. Epigenetic regulation affects gene expression by not altering the DNA sequence, and the main mechanisms include DNA methylation, histone modification, and non-coding RNAs, which play a key role in the growth and development of muscle fibers (Gao et al., 2017; Cao et al., 2018; Shi et al., 2015).
DNA methylation is one of the core mechanisms of epigenetic regulation, which modulates gene expression through adding methyl to the cytosine of DNA (usually occurs at CpG islands) (Baik et al., 2014; Park et al., 2018). The hypomethylated state is usually associated with gene transcriptional activity, whereas hypermethylation leads to gene silencing or expression inhibition (Moore et al., 2013; Besselink et al., 2023). For instance, differential methylation of the cofilin-1 gene was confirmed to be closely related to the changes in myofiber formation, which has a direct impact on livestock meat quality (Sun et al., 2022). Moreover, studies (Carrió et al., 2016; Laker and Ryall, 2016) showed that myogenic stem cells exhibited an increase in DNA methylation during the proliferation stage, while the differentiation stage was accompanied by demethylation of specific gene regions, such as the enhancer region, which can activate the expression of key myogenic genes such as MyoD and Myf5 (Carrio et al., 2015; Brunk et al., 1996).
Histone modification is another important means of epigenetic regulation (Zhao et al., 2007; Mikkelsen et al., 2007), which dynamically adjusts the open state of chromatin and gene expression activity through acetylation, methylation, ubiquitination, and other modifications of histone tails (Zhao and Shilatifard, 2019; Roma-Rodrigues et al., 2020). For example, H3K27me3 and H3K4me3, as key epigenetic markers in muscle development (Tan et al., 2021; Yang Y. et al., 2021), are respectively related to gene silencing and activation; histone acetyltransferase (HAT) (He et al., 2021; Yucel et al., 2019) and histone deacetylase (HDAC) (Cohen et al., 2015) play a key role in regulating muscle stem cell activation and myofiber type determination. Research has found that Taf1 regulates Pax3 protein through monoubiquitination (Boutet et al., 2010), thereby affecting the differentiation of skeletal muscle progenitor cells. The dynamic changes of these modifications determine the fate of skeletal muscle cells and the final phenotype of muscle development (Wang and Ibeagha-Awemu, 2021).
Non-coding RNAs, including miRNAs, lncRNAs, and circRNAs, play a pivotal role in muscle growth and development in agricultural animals (Huang et al., 2021; Cesana et al., 2011; Ouyang et al., 2018; Luo et al., 2013). The advent of high-throughput sequencing technology has facilitated the identification of numerous non-coding RNAs as key regulators of myofiber development. MiRNAs, for example, inhibit the expression of target genes by binding to their mRNA, thereby affecting the proliferation and differentiation of myoblasts and satellite cells. A precise study has revealed a strong correlation between the eQTL mutation site of miR-4331-5p and pig muscle fiber types (Zhong et al., 2023). LncRNAs not only influence gene expression by regulating miRNA or directly acting on gene promoters but can also encode functional short peptides to regulate muscle development. For instance (Dou et al., 2020), MyHC-IIA/X-AS, a lncRNA specifically expressed in skeletal muscle, regulates miR-130b through a competitive endogenous RNA (ceRNA) mechanism to maintain fast-twitch muscle fiber phenotypes. CircRNAs, owing to their stability and abundance, occupy a significant position in skeletal muscle (Yang Z. et al., 2021). For instance, circMEF2As has been demonstrated in studies in chickens and mice to promote satellite cell differentiation and skeletal muscle formation via the ceRNA mechanism (Shen et al., 2023). Figure 4 provides a summary of the key non-coding RNAs identified in recent years that regulate muscle development in agricultural animals, offering new molecular targets for livestock muscle trait improvement.
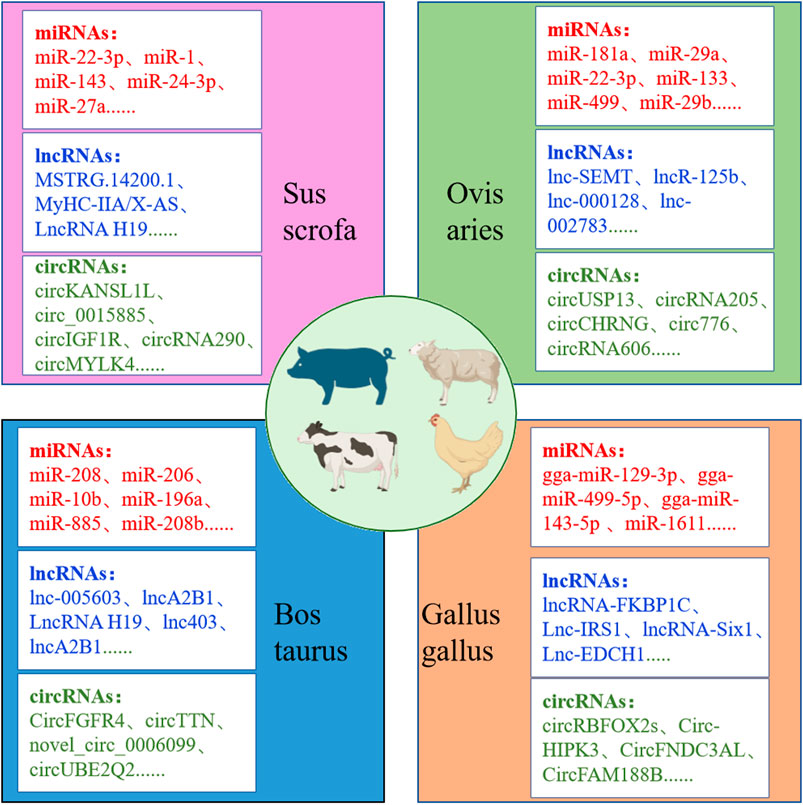
Figure 4. Non-coding RNAs affecting growth and development of muscle fibers in agricultural animals.
Intricate interactions frequently occur between various epigenetic mechanisms. For instance, non-coding RNAs can affect chromatin states by regulating DNA methylation enzymes or histone modification enzymes, and the synergy between DNA methylation and histone modification further refines the gene expression regulatory network. During muscle development, epigenetic mechanisms dynamically regulate gene expression to adapt to the developmental needs of different stages. For example, the synergy between DNA methylation and chromatin accessibility (ATAC-seq) regulates the expression of genes related to muscle hypertrophy and fiber transition in the growth process of Hu sheep skeletal muscle (Cao et al., 2023). Furthermore, research indicates that some epigenetic markers not only play a role in individual development but may also be affected by environmental factors such as parental diet or environmental exposure, thereby affecting the muscle development of offspring through transgenerational transmission mechanisms (González-Recio et al., 2015).
Advanced technologies have accelerated progress in this field. Single-cell multi-omics enables high-resolution analysis of epigenetic heterogeneity within muscle tissues, precisely tracing developmental changes (Chen et al., 2018). Integrating spatiotemporal omics data (Fomchenko et al., 2020) reveals unique epigenetic characteristics of different muscle cell populations, providing new optimization strategies.
The epigenetic insights outlined above offer significant practical applications for agricultural animal breeding and production systems. Epigenetic markers could serve as powerful selection tools in breeding programs, enabling the identification of animals predisposed to superior muscle development and meat quality traits before phenotypic expression. For example, specific DNA methylation patterns in the cofilin-1 gene could be used as early biomarkers for selecting animals with desirable myofiber characteristics. Nutritional intervention strategies targeting epigenetic mechanisms represent another promising application. Feed formulations could be designed to influence histone modifications and DNA methylation patterns during critical developmental windows, enhancing muscle growth efficiency and quality. Additionally, the identified non-coding RNAs could provide targets for novel biotechnological interventions in livestock improvement. RNA-based therapeutics or gene editing approaches targeting key regulatory non-coding RNAs might enable precise modulation of muscle fiber types to meet specific market demands for meat texture and quality. For the meat processing industry, understanding epigenetic profiles could improve processing protocols by predicting meat quality characteristics and enabling customized handling of carcasses based on their epigenetic signatures. This would reduce waste and increase value across the production chain.
In conclusion, epigenetic research is incrementally unveiling the intricate regulatory mechanisms underlying muscle development in agricultural animals, providing both theoretical frameworks and practical applications for precision breeding and meat quality optimization. As technology advances, this field will continue to drive unprecedented progress in muscle development research in agricultural animals.
7 Summary and future perspectives
The regulatory mechanisms of muscle fiber growth and development in agricultural animals represent critical research areas with significant implications for meat quality, yield, and economic outcomes. This review has systematically summarized heritability estimates of muscle fiber traits in agricultural animals, their effects on meat quality and yield, and the molecular and epigenetic regulatory mechanisms governing these traits.
Despite considerable progress in recent years, a substantial gap persists between muscle fiber research in agricultural animals compared to humans or model organisms. A notable deficiency is the absence of comprehensive, publicly accessible biobanks for agricultural animals, limiting analyses of relationships among phenotypes, genotypes, and environmental factors. This hinders a deeper understanding of the genetic principles underlying muscle fiber development. Additionally, while CRISPR-Cas9 technology has revolutionized gene function research, the development of CRISPR interference systems (CRISPRi) and other high-throughput screening technologies remains inadequate for agricultural animals, complicating the efficient identification of key regulatory genes and functional modules. Furthermore, cross-species comparative research on species-specific gene regulatory networks is lacking, necessitating unified databases and standardized analysis frameworks to enhance data integration.
We propose the establishment of standardized biobanks for agricultural animals as the most critical initiative for advancing muscle fiber research. This priority recommendation addresses a fundamental infrastructure gap and would catalyze progress across multiple research fronts. Launch a multi-institutional pilot biobank, focusing on economically important species such as pigs and chickens. The project will standardize the collection of genomic, transcriptomic, and phenotypic data from different breeds. Seek funding from agricultural research institutions, industry partners (e.g., meat production companies), and international partners (e.g., FAO partners). Ultimately, it creates an open-access database modeled after the human biobank, providing standardized data formats, detailed metadata, and an intuitive interface for researchers worldwide.
In summary, the exploration of muscle fiber growth and development regulation in agricultural animals is experiencing significant advancement. Bridging gaps in data and technology will catalyze the integration of emerging technologies and interdisciplinary collaborations, offering new insights into regulatory mechanisms. This will enhance meat production efficiency while contributing to global food security and sustainable development objectives.
Author contributions
XC: Writing – original draft, Writing – review and editing. JM: Funding acquisition, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by funding from the Biological Breeding-National Science and Technology Major Project (2023ZD0404501) and the National Natural Science Foundation of China (32272855).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdoli, R., Zamani, P., and Ghasemi, M. (2018). Genetic similarities and phylogenetic analysis of human and farm animal species based on mitogenomic nucleotide sequences. Meta Gene 15, 23–26. doi:10.1016/j.mgene.2017.10.004
Ahmad, S. F., Rather, M. A., Shanaz, S., Khan, N. N., and Ganai, N. A. (2022). Multivariate animal model-based analysis of fiber traits in kashmir Merino sheep. J. Nat. Fibers 19 (14), 7948–7955. doi:10.1080/15440478.2021.1958425
Allais, S., Levéziel, H., Payet-Duprat, N., Hocquette, J.-F., Lepetit, J., Rousset, S., et al. (2010). The two mutations, Q204X and nt821, of the myostatin gene affect carcass and meat quality in young heterozygous bulls of French beef breeds. J. animal Sci. 88 (2), 446–454. doi:10.2527/jas.2009-2385
Andersson, L. (2001). Genetic dissection of phenotypic diversity in farm animals. Nat. Rev. Genet. 2 (2), 130–138. doi:10.1038/35052563
Bai, J., Lin, Y., Zhang, J., Chen, Z., Wang, Y., Li, M., et al. (2023). Profiling of chromatin accessibility in pigs across multiple tissues and developmental stages. Int. J. Mol. Sci. 24 (13), 11076. doi:10.3390/ijms241311076
Baik, M., Vu, T., Piao, M., and Kang, H. (2014). Association of DNA methylation levels with tissue-specific expression of adipogenic and lipogenic genes in longissimus dorsi muscle of Korean cattle. Asian-Australasian J. animal Sci. 27 (10), 1493–1498. doi:10.5713/ajas.2014.14283
Bennett, G., Thallman, R., Snelling, W., and Kuehn, L. (2008). Experimental selection for calving ease and postnatal growth in seven cattle populations. II. Phenotypic differences. J. animal Sci. 86 (9), 2103–2114. doi:10.2527/jas.2007-0768
Bérard, J., Kalbe, C., Lösel, D., Tuchscherer, A., and Rehfeldt, C. (2011). Potential sources of early-postnatal increase in myofibre number in pig skeletal muscle. Histochem. cell Biol. 136, 217–225. doi:10.1007/s00418-011-0833-z
Besselink, N., Keijer, J., Vermeulen, C., Boymans, S., de Ridder, J., van Hoeck, A., et al. (2023). The genome-wide mutational consequences of DNA hypomethylation. Sci. Rep. 13 (1), 6874. doi:10.1038/s41598-023-33932-3
Bi, P., Ramirez-Martinez, A., Li, H., Cannavino, J., McAnally, J. R., Shelton, J. M., et al. (2017). Control of muscle formation by the fusogenic micropeptide myomixer. Science 356 (6335), 323–327. doi:10.1126/science.aam9361
Boutet, S. C., Biressi, S., Iori, K., Natu, V., and Rando, T. A. (2010). Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol. cell 40 (5), 749–761. doi:10.1016/j.molcel.2010.09.029
Braun, T., and Gautel, M. (2011). Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12 (6), 349–361. doi:10.1038/nrm3118
Braun, T., Rudnicki, M. A., Arnold, H.-H., and Jaenisch, R. (1992). Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell 71 (3), 369–382. doi:10.1016/0092-8674(92)90507-9
Brunk, B. P., Goldhamer, D. J., and Emerson, Jr C. P. (1996). Regulated demethylation of themyoddistal enhancer during skeletal myogenesis. Dev. Biol. 177 (2), 490–503. doi:10.1006/dbio.1996.0180
Buckingham, M., and Rigby, P. W. (2014). Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. cell 28 (3), 225–238. doi:10.1016/j.devcel.2013.12.020
Cai, C., Wan, P., Wang, H., Cai, X., Wang, J., Chai, Z., et al. (2023b). Transcriptional and open chromatin analysis of bovine skeletal muscle development by single-cell sequencing. Cell Prolif. 56 (9), e13430. doi:10.1111/cpr.13430
Cai, S., Hu, B., Wang, X., Liu, T., Lin, Z., Tong, X., et al. (2023a). Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig. BMC Biol. 21 (1), 19. doi:10.1186/s12915-023-01519-z
Callaghan, M., Rodgers, R., and Perry, V. (2020). Supplementation of rangeland primiparous Bos indicus x Bos taurus beef heifers during lactation. 1. Effects on dam milk production and liveweight, bull calf growth, live carcass characteristics and metabolic hormone concentrations. Theriogenology 152, 69–82. doi:10.1016/j.theriogenology.2020.04.030
Cao, Y., Ai, Y., Zhang, X., Zhang, J., Long, X., Zhu, Y., et al. (2023). Genome-wide epigenetic dynamics during postnatal skeletal muscle growth in Hu sheep. Commun. Biol. 6 (1), 1077. doi:10.1038/s42003-023-05439-0
Cao, Y., You, S., Yao, Y., Liu, Z.-J., Hazi, W., Li, C.-Y., et al. (2018). Expression profiles of circular RNAs in sheep skeletal muscle. Asian-Australasian J. animal Sci. 31 (10), 1550–1557. doi:10.5713/ajas.17.0563
Carrio, E., Díez-Villanueva, A., Lois, S., Mallona, I., Cases, I., Forn, M., et al. (2015). Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage. Stem Cells 33 (6), 2025–2036. doi:10.1002/stem.1998
Carrió, E., Magli, A., Muñoz, M., Peinado, M. A., Perlingeiro, R., and Suelves, M. (2016). Muscle cell identity requires Pax7-mediated lineage-specific DNA demethylation. BMC Biol. 14, 30–15. doi:10.1186/s12915-016-0250-9
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 (2), 358–369. doi:10.1016/j.cell.2011.09.028
Chen, F., Zhou, J., Li, Y., Zhao, Y., Yuan, J., Cao, Y., et al. (2019). YY1 regulates skeletal muscle regeneration through controlling metabolic reprogramming of satellite cells. Embo J. 38 (10), e99727. doi:10.15252/embj.201899727
Chen, M.-M., Zhao, Y., Yu, K., Xu, X.-L., Zhang, X.-S., Zhang, J.-L., et al. (2024). A MSTNDel73C mutation with FGF5 knockout sheep by CRISPR/Cas9 promotes skeletal muscle myofiber hyperplasia. Elife 12, RP86827. doi:10.7554/eLife.86827
Chen, S., An, J., Lian, L., Qu, L., Zheng, J., Xu, G., et al. (2013). Polymorphisms in AKT3, FIGF, PRKAG3, and TGF-β genes are associated with myofiber characteristics in chickens. Poult. Sci. 92 (2), 325–330. doi:10.3382/ps.2012-02766
Chen, X., Miragaia, R. J., Natarajan, K. N., and Teichmann, S. A. (2018). A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 9 (1), 5345. doi:10.1038/s41467-018-07771-0
Cheng, J., Cao, X., Wang, X., Wang, J., Yue, B., Sun, W., et al. (2023). Dynamic chromatin architectures provide insights into the genetics of cattle myogenesis. J. Animal Sci. Biotechnol. 14 (1), 59. doi:10.1186/s40104-023-00855-y
Choi, Y., and Kim, B. (2009). Muscle fiber characteristics, myofibrillar protein isoforms, and meat quality. Livest. Sci. 122 (2-3), 105–118. doi:10.1016/j.livsci.2008.08.015
Cohen, T. J., Choi, M.-C., Kapur, M., Lira, V. A., Yan, Z., and Yao, T.-P. (2015). HDAC4 regulates muscle fiber type-specific gene expression programs. Mol. cells 38 (4), 343–348. doi:10.14348/molcells.2015.2278
Davis, T., and White, R. (2020). Breeding animals to feed people: the many roles of animal reproduction in ensuring global food security. Theriogenology 150, 27–33. doi:10.1016/j.theriogenology.2020.01.041
Deng, K., Su, Y., Liu, Z., Hu, S., Ren, C., Wei, W., et al. (2024). Ythdf2 facilitates precursor miR-378/miR-378-5p maturation to support myogenic differentiation. Cell. Mol. Life Sci. 81 (1), 445–520. doi:10.1007/s00018-024-05456-0
Deniskova, T., Koshkina, O., Petrov, S., Sermyagin, A., and Zinovieva, N. (2024). Identification of candidate genes associated with growth and development of sheep from a crossbred population using genome-wide association studies. Agric. Sci. Euro-North-East 25 (2), 236–250. doi:10.30766/2072-9081.2024.25.2.236-250
Dos Santos, M., Shah, A. M., Zhang, Y., Bezprozvannaya, S., Chen, K., Xu, L., et al. (2023). Opposing gene regulatory programs governing myofiber development and maturation revealed at single nucleus resolution. Nat. Commun. 14 (1), 4333. doi:10.1038/s41467-023-40073-8
Dou, M., Yao, Y., Ma, L., Wang, X., Yang, G., Li, X., et al. (2020). The long noncoding RNA MyHC IIA/X-AS contributes to skeletal muscle myogenesis and maintains the fast fiber phenotype. J. Biol. Chem. 295 (15), 4937–4949. doi:10.1074/jbc.RA119.010498
Dwyer, C., Fletcher, J., and Stickland, N. (1993). Muscle cellularity and postnatal growth in the pig. J. Animal Sci. 71 (12), 3339–3343. doi:10.2527/1993.71123339x
Eichler, E. E., Flint, J., Gibson, G., Kong, A., Leal, S. M., Moore, J. H., et al. (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11 (6), 446–450. doi:10.1038/nrg2809
Erimbetov, K., Obvintseva, O., Fedorova, A., Zemlyanoy, R., and Solovieva, A. (2019). Phenotypic regulation of animal skeletal muscle protein metabolism. Ukrainian J. Ecol. 9 (4), 651–656. doi:10.15421/2019_804
Estellé, J., Gil, F., Vazquez, J., Latorre, R., Ramirez, G., Barragan, M., et al. (2008). A quantitative trait locus genome scan for porcine muscle fiber traits reveals overdominance and epistasis. J. animal Sci. 86 (12), 3290–3299. doi:10.2527/jas.2008-1034
Eyre-Walker, A. (2010). Evolution in health and medicine Sackler colloquium: genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. 107 (Suppl. l_1), 1752–1756. doi:10.1073/pnas.0906182107
Fang, M., Yang, Y.-G., and Hu, Z. (2024). Current status and challenges of pig-to-human organ xenotransplantation. Sci. China Life Sci. 67 (4), 829–831. doi:10.1007/s11427-023-2500-9
Felício, A. M., Gaya, L. G., Ferraz, J. B. S., Moncau, C. T., Mattos, E. C., Santos, N. P., et al. (2013). Heritability and genetic correlation estimates for performance, meat quality and quantitative skeletal muscle fiber traits in broiler. Livest. Sci. 157 (1), 81–87. doi:10.1016/j.livsci.2013.08.005
Fomchenko, K. M., Walsh, E. M., Yang, X., Verma, R. X., Lin, B. L., Nieuwenhuis, T. O., et al. (2020). Spatial proteomic approach to characterize skeletal muscle myofibers. J. proteome Res. 20 (1), 888–894. doi:10.1021/acs.jproteome.0c00673
Gao, P., Guo, X., Du, M., Cao, G., Yang, Q., Pu, Z., et al. (2017). LncRNA profiling of skeletal muscles in Large White pigs and Mashen pigs during development. J. animal Sci. 95 (10), 4239–4250. doi:10.2527/jas2016.1297
García-Ruiz, A., Cole, J. B., VanRaden, P. M., Wiggans, G. R., Ruiz-López, F. J., and Van Tassell, C. P. (2016). Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. 113 (28), E3995–E4004. doi:10.1073/pnas.1519061113
Godfray, H. C. J., Aveyard, P., Garnett, T., Hall, J. W., Key, T. J., Lorimer, J., et al. (2018). Meat consumption, health, and the environment. Science. 361 (6399), eaam5324. doi:10.1126/science.aam5324
González-Recio, O., Toro, M. A., and Bach, A. (2015). Past, present, and future of epigenetics applied to livestock breeding. Front. Genet. 6, 305. doi:10.3389/fgene.2015.00305
Grefte, S., Kuijpers-Jagtman, A. M., Torensma, R., and Von den Hoff, J. W. (2007). Skeletal muscle development and regeneration. Stem cells Dev. 16 (5), 857–868. doi:10.1089/scd.2007.0058
Grifone, R., Demignon, J., Houbron, C., Souil, E., Niro, C., Seller, M. J., et al. (2005). Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. 132, 2235, 2249. doi:10.1242/dev.01773
Gros, J., Scaal, M., and Marcelle, C. (2004). A two-step mechanism for myotome formation in chick. Dev. cell 6 (6), 875–882. doi:10.1016/j.devcel.2004.05.006
Gu, S., Huang, Q., Jie, Y., Sun, C., Wen, C., and Yang, N. (2024). Transcriptomic and epigenomic landscapes of muscle growth during the postnatal period of broilers. J. Animal Sci. Biotechnol. 15 (1), 91. doi:10.1186/s40104-024-01049-w
Guo, T., Gao, J., Yang, B., Yan, G., Xiao, S., Zhang, Z., et al. (2019). A whole genome sequence association study of muscle fiber traits in a White Duroc× Erhualian F2 resource population. Asian-Australasian J. animal Sci. 33 (5), 704–711. doi:10.5713/ajas.18.0767
Han, R., Wei, Y., Kang, X., Chen, H., Sun, G., Li, G., et al. (2012). Novel SNPs in the PRDM16 gene and their associations with performance traits in chickens. Mol. Biol. Rep. 39, 3153–3160. doi:10.1007/s11033-011-1081-y
Hao, X., Fu, Y., Li, S., Nie, J., Zhang, B., and Zhang, H. (2024). Porcine transient receptor potential channel 1 (TRPC1) regulates muscle growth via the Wnt/β-catenin and Wnt/Ca2+ pathways. Int. J. Biol. Macromol. 265, 130855. doi:10.1016/j.ijbiomac.2024.130855
Hasty, P., Bradley, A., Morris, J. H., Edmondson, D. G., Venuti, J. M., Olson, E. N., et al. (1993). Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364 (6437), 501–506. doi:10.1038/364501a0
He, R., Dantas, A., and Riabowol, K. (2021). Histone acetyltransferases and stem cell identity. Cancers 13 (10), 2407. doi:10.3390/cancers13102407
Huang, C., Ge, F., Ma, X., Dai, R., Dingkao, R., Zhaxi, Z., et al. (2021). Comprehensive analysis of mRNA, lncRNA, circRNA, and miRNA expression profiles and their ceRNA networks in the longissimus dorsi muscle of cattle-yak and yak. Front. Genet. 12, 772557. doi:10.3389/fgene.2021.772557
Huang, Y., Cai, L., Duan, Y., Zeng, Q., He, M., Wu, Z., et al. (2022). Whole-genome sequence-based association analyses on an eight-breed crossed heterogeneous stock of pigs reveal the genetic basis of skeletal muscle fiber characteristics. Meat Sci. 194, 108974. doi:10.1016/j.meatsci.2022.108974
Huang, Y., Zhang, C., Zhang, W., Zhang, P., Kang, X., and Chen, W. (2015). Variation in the chicken LPIN2 gene and association with performance traits. Br. Poult. Sci. 56 (2), 175–183. doi:10.1080/00071668.2015.1008994
Huang, Y., Zhou, L., Zhang, J., Liu, X., Zhang, Y., Cai, L., et al. (2020). A large-scale comparison of meat quality and intramuscular fatty acid composition among three Chinese indigenous pig breeds. Meat Sci. 168, 108182. doi:10.1016/j.meatsci.2020.108182
Ismail, I., and Joo, S.-T. (2017). Poultry meat quality in relation to muscle growth and muscle fiber characteristics. Korean J. food Sci. animal Resour. 37 (6), 873–883. doi:10.5851/kosfa.2017.37.6.87
Jia, X., Nie, Q., Lamont, S., and Zhang, X. (2012). Variation in sequence and expression of the avian FTO, and association with glucose metabolism, body weight, fatness and body composition in chickens. Int. J. Obes. 36 (8), 1054–1061. doi:10.1038/ijo.2011.221
Jiang, J., Cole, J. B., Da, Y., VanRaden, P. M., and Ma, L. (2018). Fast Bayesian fine-mapping of 35 production, reproduction and body conformation traits with imputed sequences of 27K Holstein bulls. BioRxiv, 428227. doi:10.1101/428227
Jiang, J., Cole, J. B., Freebern, E., Da, Y., VanRaden, P. M., and Ma, L. (2019). Functional annotation and Bayesian fine-mapping reveals candidate genes for important agronomic traits in Holstein bulls. Commun. Biol. 2 (1), 212. doi:10.1038/s42003-019-0454-y
Joo, S., Kim, G., Hwang, Y., and Ryu, Y. (2013). Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 95 (4), 828–836. doi:10.1016/j.meatsci.2013.04.044
Kahane, N., Cinnamon, Y., Bachelet, I., and Kalcheim, C. (2001). The third wave of myotome colonization by mitotically competent progenitors: regulating the balance between differentiation and proliferation during muscle development. Development 128 (12), 2187–2198. doi:10.1242/dev.128.12.2187
Kemper, K. E., Visscher, P. M., and Goddard, M. E. (2012). Genetic architecture of body size in mammals. Genome Biol. 13, 244–313. doi:10.1186/gb4016
G. Kim, J. Jeong, S. Moon, Y. Hwang, G. Park, and S. Joo (2008). “Effects of muscle fibre type on meat characteristics of chicken and duck breast muscle,” 54th international congress of meat science and technology (54th ICoMST).
Kim, G.-D., Yang, H.-S., and Jeong, J.-Y. (2016). Comparison of characteristics of myosin heavy chain-based fiber and meat quality among four bovine skeletal muscles. Korean J. Food Sci. Animal Resour. 36 (6), 819–828. doi:10.5851/kosfa.2016.36.6.819
Kopler, I., Marchaim, U., Tikász, I. E., Opaliński, S., Kokin, E., Mallinger, K., et al. (2023). Farmers’ perspectives of the benefits and risks in precision livestock farming in the EU pig and poultry sectors. Animals 13 (18), 2868. doi:10.3390/ani13182868
Koshkina, O., Deniskova, T., Dotsev, A., Kunz, E., Selionova, M., Medugorac, I., et al. (2023). Phylogenetic analysis of Russian native sheep breeds based on mtDNA sequences. Genes 14 (9), 1701. doi:10.3390/genes14091701
Laker, R. C., and Ryall, J. G. (2016). DNA methylation in skeletal muscle stem cell specification, proliferation, and differentiation. Stem cells Int. 2016 (1), 5725927. doi:10.1155/2016/5725927
Lan, Y., Yan, D., Li, X., Zhou, C., Bai, Y., and Dong, X. (2024). Muscle growth differences in Lijiang pigs revealed by ATAC-seq multi-omics. Front. Veterinary Sci. 11, 1431248. doi:10.3389/fvets.2024.1431248
Larzul, C., Lefaucheur, L., Ecolan, P., Gogue, J., Talmant, A., Sellier, P., et al. (1997). Phenotypic and genetic parameters for longissimus muscle fiber characteristics in relation to growth, carcass, and meat quality traits in large white pigs. J. animal Sci. 75 (12), 3126–3137. doi:10.2527/1997.75123126x
Lee, E., Kim, J., Lim, K., Ryu, Y., Jeon, W., and Hong, K. (2012). Effects of variation in porcine MYOD1 gene on muscle fiber characteristics, lean meat production, and meat quality traits. Meat Sci. 92 (1), 36–43. doi:10.1016/j.meatsci.2012.03.018
Lee, J. S., Kim, J. M., Lim, K. S., Hong, J. S., Hong, K. C., and Lee, Y. S. (2013). Effects of polymorphisms in the porcine microRNA MIR206/MIR133B cluster on muscle fiber and meat quality traits. Anim. Genet. 44 (1), 101–106. doi:10.1111/j.1365-2052.2012.02362.x
Lee, S.-H., Kim, S., and Kim, J.-M. (2022). Genetic correlation between biopsied and post-mortem muscle fibre characteristics and meat quality traits in swine. Meat Sci. 186, 108735. doi:10.1016/j.meatsci.2022.108735
Lee, S.-J., and McPherron, A. C. (2001). Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. 98 (16), 9306–9311. doi:10.1073/pnas.151270098
Lefaucheur, L. (2010). A second look into fibre typing–relation to meat quality. Meat Sci. 84 (2), 257–270. doi:10.1016/j.meatsci.2009.05.004
Li, C., Wang, Y., Sun, X., Yang, J., Ren, Y., Jia, J., et al. (2024b). Identification of different myofiber types in pigs muscles and construction of regulatory networks. BMC genomics 25 (1), 400. doi:10.1186/s12864-024-10271-9
Li, H., Li, S., Zhang, H., Gu, J., Dai, Y., Wu, R., et al. (2024a). Integrated GWAS and transcriptome analysis reveals key genes associated with muscle fibre and fat traits in Gushi chicken. Br. Poult. Sci. 66, 31–41. doi:10.1080/00071668.2024.2400685
Li, J., Chen, Z., Bai, Y., Wei, Y., Guo, D., Liu, Z., et al. (2023). Integration of ATAC-seq and RNA-seq analysis to identify key genes in the longissimus dorsi muscle development of the tianzhu white yak. Int. J. Mol. Sci. 25 (1), 158. doi:10.3390/ijms25010158
Li, M., Zhou, X., Chen, Y., Nie, Y., Huang, H., Chen, H., et al. (2015). Not all the number of skeletal muscle fibers is determined prenatally. BMC Dev. Biol. 15, 42–46. doi:10.1186/s12861-015-0091-8
Li, S., Chen, W., Kang, X., Han, R., Sun, G., and Huang, Y. (2013). Distinct tissue expression profiles of chicken Lpin1-α/β isoforms and the effect of the variation on muscle fiber traits. Gene 515 (2), 281–290. doi:10.1016/j.gene.2012.11.075
Li, T., Jin, M., Wang, H., Zhang, W., Yuan, Z., and Wei, C. (2024c). Whole-genome scanning for selection signatures reveals candidate genes associated with growth and tail length in sheep. Animals 14 (5), 687. doi:10.3390/ani14050687
Li, W., Ren, J., Zhu, W., Guo, B., Yang, B., Liu, L., et al. (2009a). Mapping QTL for porcine muscle fibre traits in a White Duroc x Erhualian F(2) resource population. J. Animal Breed. Genet. 126 (6), 468–474. doi:10.1111/j.1439-0388.2009.00805.x
Li, W. B., Ren, J., Zhu, W. C., Guo, B. L., Yang, B., Liu, L. T., et al. (2009b). Mapping QTL for porcine muscle fibre traits in a White Duroc x Erhualian F(2) resource population. J. Anim. Breed. Genet. 126 (6), 468–474. doi:10.1111/j.1439-0388.2009.00805.x
Liu, D., Fan, W., Xu, Y., Yu, S., Liu, W., Guo, Z., et al. (2021). Genome-wide association studies demonstrate that TASP1 contributes to increased muscle fiber diameter. Heredity 126 (6), 991–999. doi:10.1038/s41437-021-00425-w
Liu, L., Yi, G., Yao, Y., Liu, Y., Li, J., Yang, Y., et al. (2024). Multiomics analysis reveals signatures of selection and loci associated with complex traits in pigs. iMeta 3, e250. doi:10.1002/imt2.250
Lunney, J. K., Van Goor, A., Walker, K. E., Hailstock, T., Franklin, J., and Dai, C. (2021). Importance of the pig as a human biomedical model. Sci. Transl. Med. 13 (621), eabd5758. doi:10.1126/scitranslmed.abd5758
Luo, W., Nie, Q., and Zhang, X. (2013). MicroRNAs involved in skeletal muscle differentiation. J. Genet. genomics 40 (3), 107–116. doi:10.1016/j.jgg.2013.02.002
Lv, S.-J., Su, L., Li, H., Han, R.-L., Sun, G.-R., and Kang, X.-T. (2012). Polymorphisms of the interleukin-15 gene and their associations with fatness and muscle fiber traits in chickens. J. Appl. Genet. 53, 443–448. doi:10.1007/s13353-012-0111-3
Ma, M., Cai, B., Kong, S., Zhou, Z., Zhang, J., Zhang, X., et al. (2022). PPARGC1A is a moderator of skeletal muscle development regulated by miR-193b-3p. Int. J. Mol. Sci. 23 (17), 9575. doi:10.3390/ijms23179575
Makarova, A., Mitrofanova, O., Vakhrameev, A., and Dementeva, N. (2019). Molecular-genetic bases of plumage coloring in chicken. Vavilov J. Genet. Breed. 23 (3), 343–354. doi:10.18699/vj19.499
Mashima, D., Oka, Y., Gotoh, T., Tomonaga, S., Sawano, S., Nakamura, M., et al. (2019). Correlation between skeletal muscle fiber type and free amino acid levels in Japanese Black steers. Animal Sci. J. 90 (4), 604–609. doi:10.1111/asj.13185
Matarneh, S. K., Silva, S. L., and Gerrard, D. E. (2021). New insights in muscle biology that alter meat quality. Annu. Rev. animal Biosci. 9 (1), 355–377. doi:10.1146/annurev-animal-021419-083902
Matika, O., Riggio, V., Anselme-Moizan, M., Law, A. S., Pong-Wong, R., Archibald, A. L., et al. (2016). Genome-wide association reveals QTL for growth, bone and in vivo carcass traits as assessed by computed tomography in Scottish Blackface lambs. Genet. Sel. Evol. 48, 11–15. doi:10.1186/s12711-016-0191-3
McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27 (1), 40–47. doi:10.1016/s0968-0004(01)02031-x
McPherron, A. C., Lawler, A. M., and Lee, S.-J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387 (6628), 83–90. doi:10.1038/387083a0
McPherron, A. C., and Lee, S.-J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. 94 (23), 12457–12461. doi:10.1073/pnas.94.23.12457
Meyerholz, D. K., Burrough, E. R., Kirchhof, N., Anderson, D. J., and Helke, K. L. (2024). Swine models in translational research and medicine. Vet. Pathol. 61 (4), 512–523. doi:10.1177/03009858231222235
Miao, W., Ma, Z., Tang, Z., Yu, L., Liu, S., Huang, T., et al. (2021). Integrative ATAC-seq and RNA-seq analysis of the longissimus muscle of luchuan and Duroc pigs. Front. Nutr. 8, 742672. doi:10.3389/fnut.2021.742672
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 (7153), 553–560. doi:10.1038/nature06008
Millay, D. P., O'Rourke, J. R., Sutherland, L. B., Bezprozvannaya, S., Shelton, J. M., Bassel-Duby, R., et al. (2013). Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499 (7458), 301–305. doi:10.1038/nature12343
Mohammadinejad, F., Mohammadabadi, M., Roudbari, Z., and Sadkowski, T. (2022). Identification of key genes and biological pathways associated with skeletal muscle maturation and hypertrophy in Bos taurus, Ovis aries, and Sus scrofa. Animals 12 (24), 3471. doi:10.3390/ani12243471
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 (1), 23–38. doi:10.1038/npp.2012.112
Muroya, S. (2022). - Invited Review - postmortem skeletal muscle metabolism of farm animals approached with metabolomics. Anim. Biosci. 36 (2), 374–384. doi:10.5713/ab.22.0370
Musarò, A., McCullagh, K., Paul, A., Houghton, L., Dobrowolny, G., Molinaro, M., et al. (2001). Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 27 (2), 195–200. doi:10.1038/84839
Nejad, F. M., Mohammadabadi, M., Roudbari, Z., Gorji, A. E., and Sadkowski, T. (2024). Network visualization of genes involved in skeletal muscle myogenesis in livestock animals. BMC genomics 25 (1), 294. doi:10.1186/s12864-024-10196-3
Nesvadbova, M., and Borilova, G. (2018). Molecular regulation of skeletal muscle tissue formation and development. Veterinární medicína. 63 (11), 489–499. doi:10.17221/7/2018-vetmed
Olson, E. N., Perry, M., and Schulz, R. A. (1995). Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev. Biol. 172 (1), 2–14. doi:10.1006/dbio.1995.0002
Ouyang, H., Chen, X., Wang, Z., Yu, J., Jia, X., Li, Z., et al. (2018). Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 25 (1), 71–86. doi:10.1093/dnares/dsx039
Park, J., Moon, S. S., Song, S., Cheng, H., Im, C., Du, L., et al. (2024). Comparative review of muscle fiber characteristics between porcine skeletal muscles. J. Animal Sci. Technol. 66 (2), 251–265. doi:10.5187/jast.2023.e126
Park, S. J., Beak, S.-H., Kim, S. Y., Jeong, I. H., Piao, M. Y., Kang, H. J., et al. (2018). Genetic, management, and nutritional factors affecting intramuscular fat deposition in beef cattle—a review. Asian-Australasian J. animal Sci. 31 (7), 1043–1061. doi:10.5713/ajas.18.0310
Perleberg, C., Kind, A., and Schnieke, A. (2018). Genetically engineered pigs as models for human disease. Dis. models & Mech. 11 (1), dmm030783. doi:10.1242/dmm.030783
Perry, R., and Rudnick, M. A. (2000). Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 5 (3), D750–D767. doi:10.2741/perry
Petersen, B. (2023). Editorial: genetic engineering in farm animals. Front. Genet. 14, 1155201. doi:10.3389/fgene.2023.1155201
Peterson, L., Yacoub, M. H., Ayares, D., Yamada, K., Eisenson, D., Griffith, B. P., et al. (2024). Physiological basis for xenotransplantation from genetically modified pigs to humans. Physiol. Rev. 104 (3), 1409–1459. doi:10.1152/physrev.00041.2023
Prather, R. S. (2013). Pig genomics for biomedicine. Nat. Biotechnol. 31 (2), 122–124. doi:10.1038/nbt.2490
Reiner, G., Heinricy, L., Müller, E., Geldermann, H., and Dzapo, V. (2002). Indications of associations of the porcine FOS proto-oncogene with skeletal muscle fibre traits. Anim. Genet. 33 (1), 49–55. doi:10.1046/j.1365-2052.2002.00805.x
Relaix, F., Rocancourt, D., Mansouri, A., and Buckingham, M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435 (7044), 948–953. doi:10.1038/nature03594
Roma-Rodrigues, C., Rivas-García, L., Baptista, P. V., and Fernandes, A. R. (2020). Gene therapy in cancer treatment: why go nano? Pharmaceutics 12 (3), 233. doi:10.3390/pharmaceutics12030233
Rubin, C.-J., Zody, M. C., Eriksson, J., Meadows, J. R., Sherwood, E., Webster, M. T., et al. (2010). Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464 (7288), 587–591. doi:10.1038/nature08832
Rudnicki, M. A., Schnegelsberg, P. N., Stead, R. H., Braun, T., Arnold, H.-H., and Jaenisch, R. (1993). MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75 (7), 1351–1359. doi:10.1016/0092-8674(93)90621-v
Rullman, E., Fernandez-Gonzalo, R., Mekjavić, I. B., Gustafsson, T., and Eiken, O. (2018). MEF2 as upstream regulator of the transcriptome signature in human skeletal muscle during unloading. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 315 (4), R799–R809. doi:10.1152/ajpregu.00452.2017
Schiaffino, S., Sandri, M., and Murgia, M. (2007). Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology 22 (4), 269–278. doi:10.1152/physiol.00009.2007
Schreurs, N., Garcia, F., Jurie, C., Agabriel, J., Micol, D., Bauchart, D., et al. (2008). Meta-analysis of the effect of animal maturity on muscle characteristics in different muscles, breeds, and sexes of cattle. J. animal Sci. 86 (11), 2872–2887. doi:10.2527/jas.2008-0882
Shen, X., Zhao, X., He, H., Zhao, J., Wei, Y., Chen, Y., et al. (2023). Evolutionary conserved circular MEF2A RNAs regulate myogenic differentiation and skeletal muscle development. PLoS Genet. 19 (9), e1010923. doi:10.1371/journal.pgen.1010923
Shi, L., Zhou, B., Li, P., Schinckel, A. P., Liang, T., Wang, H., et al. (2015). MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell. Signal. 27 (9), 1895–1904. doi:10.1016/j.cellsig.2015.05.001
Şirin, E., Aksoy, Y., Uğurlu, M., Cicek, Ü., Önenc, A., Ulutaş, Z., et al. (2017). The relationship between muscle fiber characteristics and some meat quality parameters in Turkish native sheep breeds. Small Ruminant Res. 150, 46–51. doi:10.1016/j.smallrumres.2017.03.012
Stickland, N., and Handel, S. (1986). The numbers and types of muscle fibres in large and small breeds of pigs. J. Anat. 147, 181–189.
Stitt, T. N., Drujan, D., Clarke, B. A., Panaro, F., Timofeyva, Y., Kline, W. O., et al. (2004). The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. cell 14 (3), 395–403. doi:10.1016/s1097-2765(04)00211-4
Sun, Y., Ma, Y., Zhao, T., Li, M., Mao, Y., and Yang, Z. (2022). Epigenetic regulation mechanisms of the cofilin-1 gene in the development and differentiation of bovine primary myoblasts. Genes 13 (5), 723. doi:10.3390/genes13050723
Tan, B., Wang, S., Wang, S., Zeng, J., Hong, L., Li, Z., et al. (2021). Genome-wide analysis of H3K27me3 in porcine embryonic muscle development. Front. Cell Dev. Biol. 9, 739321. doi:10.3389/fcell.2021.739321
Tan, X., Liu, R., Zhao, D., He, Z., Li, W., Zheng, M., et al. (2024). Large-scale genomic and transcriptomic analyses elucidate the genetic basis of high meat yield in chickens. J. Adv. Res. 55, 1–16. doi:10.1016/j.jare.2023.02.016
Trerotola, M., Relli, V., Simeone, P., and Alberti, S. (2015). Epigenetic inheritance and the missing heritability. Hum. genomics 9, 17–12. doi:10.1186/s40246-015-0041-3
UNICEF (2024). The state of food security and nutrition in the world 2024: financing to end hunger, food insecurity and malnutrition in all its forms.
Visscher, P. M., Hill, W. G., and Wray, N. R. (2008). Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet. 9 (4), 255–266. doi:10.1038/nrg2322
Wang, M., and Ibeagha-Awemu, E. M. (2021). Impacts of epigenetic processes on the health and productivity of livestock. Front. Genet. 11, 613636. doi:10.3389/fgene.2020.613636
Wang, M.-S., Thakur, M., Peng, M.-S., Jiang, Y., Frantz, L. A. F., Li, M., et al. (2020). 863 genomes reveal the origin and domestication of chicken. Cell Res. 30 (8), 693–701. doi:10.1038/s41422-020-0349-y
Wang, W., Zhang, T., Du, L., Li, K., Zhang, L., Li, H., et al. (2024). Transcriptomic analysis reveals diverse expression patterns underlying the fiber diameter of oxidative and glycolytic skeletal muscles in steers. Meat Sci. 207, 109350. doi:10.1016/j.meatsci.2023.109350
Wei, Y., Zhu, S., Zhang, S., Han, R., Tian, Y., Sun, G., et al. (2012). Two novel SNPs of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene associated with growth and meat quality traits in the chicken. Genet. Mol. Res. 11 (4), 4765–4774. doi:10.4238/2012.November.12.10
Wigmore, P., and Stickland, N. (1983). Muscle development in large and small pig fetuses. J. Anat. 137 (Pt 2), 235–245.
Wimmers, K., Fiedler, I., Hardge, T., Murani, E., Schellander, K., and Ponsuksili, S. (2006). QTL for microstructural and biophysical muscle properties and body composition in pigs. BMC Genet. 7, 15–14. doi:10.1186/1471-2156-7-15
Xiao, L., Qiao, J., Huang, Y., Tan, B., Hong, L., Li, Z., et al. (2024). RASGRP1 targeted by H3K27me3 regulates myoblast proliferation and differentiation in mice and pigs: RASGRP1 regulates myoblast proliferation and differentiation. Acta Biochimica Biophysica Sinica 56 (3), 452–461. doi:10.3724/abbs.2024011
Xu, D., Wan, B., Qiu, K., Wang, Y., Zhang, X., Jiao, N., et al. (2023). Single-cell RNA-sequencing provides insight into skeletal muscle evolution during the selection of muscle characteristics. Adv. Sci. 10 (35), 2305080. doi:10.1002/advs.202305080
Xu, Z., Wu, J., Zhou, J., Zhang, Y., Qiao, M., Sun, H., et al. (2022). Integration of ATAC-seq and RNA-seq analysis identifies key genes affecting intramuscular fat content in pigs. Front. Nutr. 9, 1016956. doi:10.3389/fnut.2022.1016956
Yan, M., Li, L., Huang, Y., Tang, X., Shu, Y., Cui, D., et al. (2024). Investigation on muscle fiber types and meat quality and estimation of their heritability and correlation coefficients with each other in four pig populations. Animal Sci. J. 95 (1), e13915. doi:10.1111/asj.13915
Yang, Y., Fan, X., Yan, J., Chen, M., Zhu, M., Tang, Y., et al. (2021a). A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic acids Res. 49 (3), 1313–1329. doi:10.1093/nar/gkaa1203
Yang, Z., He, T., and Chen, Q. (2021b). The roles of CircRNAs in regulating muscle development of livestock animals. Front. Cell Dev. Biol. 9, 619329. doi:10.3389/fcell.2021.619329
Yi, X.-D., Yu, H., Li, R., Zhao, T.-T., He, Z.-Z., and Pang, W.-j. (2024). Single-cell transcriptional profiling of porcine muscle satellite cells and myoblasts during myogenesis. Agric. Commun. 2 (1), 100026. doi:10.1016/j.agrcom.2024.100026
Yu, H.-H., Zhao, H., Qing, Y.-B., Pan, W.-R., Jia, B.-Y., Zhao, H.-Y., et al. (2016). Porcine zygote injection with Cas9/sgRNA results in DMD-modified pig with muscle dystrophy. Int. J. Mol. Sci. 17 (10), 1668. doi:10.3390/ijms17101668
Yu, M., Feng, Y., Yan, J., Zhang, X., Tian, Z., Wang, T., et al. (2024). Transcriptomic regulatory analysis of skeletal muscle development in landrace pigs. Gene 915, 148407. doi:10.1016/j.gene.2024.148407
Yucel, N., Wang, Y. X., Mai, T., Porpiglia, E., Lund, P. J., Markov, G., et al. (2019). Glucose metabolism drives histone acetylation landscape transitions that dictate muscle stem cell function. Cell Rep. 27 (13), 3939–3955. doi:10.1016/j.celrep.2019.05.092
Zanou, N., and Gailly, P. (2013). Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell. Mol. Life Sci. 70, 4117–4130. doi:10.1007/s00018-013-1330-4
Zhang, L., Guo, Y., Wang, L., Liu, X., Yan, H., Gao, H., et al. (2020). Genomic variants associated with the number and diameter of muscle fibers in pigs as revealed by a genome-wide association study. animal 14 (3), 475–481. doi:10.1017/S1751731119002374
Zhang, S., Li, S., Chen, W., Lu, W., and Huang, Y. (2013). A single-nucleotide polymorphism in the 3′ untranslated region of the LPIN1 gene and association analysis with performance traits in chicken. Br. Poult. Sci. 54 (3), 312–318. doi:10.1080/00071668.2013.791383
Zhang, W., Behringer, R. R., and Olson, E. N. (1995). Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes & Dev. 9 (11), 1388–1399. doi:10.1101/gad.9.11.1388
Zhang, X., Cai, S., Chen, L., Yuan, R., Nie, Y., Ding, S., et al. (2019). Integrated miRNA–mRNA transcriptomic analysis reveals epigenetic-mediated embryonic muscle growth differences between Wuzhishan and Landrace pigs. J. Animal Sci. 97 (5), 1967–1978. doi:10.1093/jas/skz091
Zhao, X. D., Han, X., Chew, J. L., Liu, J., Chiu, K. P., Choo, A., et al. (2007). Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell stem cell 1 (3), 286–298. doi:10.1016/j.stem.2007.08.004
Zhao, Z., and Shilatifard, A. (2019). Epigenetic modifications of histones in cancer. Genome Biol. 20, 245–316. doi:10.1186/s13059-019-1870-5
Zhong, L., Zheng, M., Huang, Y., Jiang, T., Yang, B., Huang, L., et al. (2023). An atlas of expression quantitative trait loci of microRNAs in longissimus muscle of eight-way crossbred pigs. J. Genet. Genomics 50 (6), 398–409. doi:10.1016/j.jgg.2023.02.007
Zhong, Y., Tang, Z., Huang, L., Wang, D., and Lu, Z. (2020). Genetic diversity of Procambarus clarkii populations based on mitochondrial DNA and microsatellite markers in different areas of Guangxi, China. Mitochondrial DNA Part A 31 (2), 48–56. doi:10.1080/24701394.2020.1721484
Zhou, G., Dudgeon, C., Li, M., Cao, Y., Zhang, L., and Jin, H. (2010). Molecular cloning of the HGD gene and association of SNPs with meat quality traits in Chinese red cattle. Mol. Biol. Rep. 37, 603–611. doi:10.1007/s11033-009-9860-4
Keywords: agricultural animal, myofiber development, meat quality, regulation mechanism, epigenetic
Citation: Chang X and Ma J (2025) Integrative genetic and epigenetic control of skeletal muscle fiber traits in agricultural animals. Front. Genet. 16:1566553. doi: 10.3389/fgene.2025.1566553
Received: 25 January 2025; Accepted: 21 April 2025;
Published: 02 May 2025.
Edited by:
Demin Cai, Yangzhou University, ChinaReviewed by:
Jianning He, Qingdao Agricultural University, ChinaZhuying Wei, University of Michigan, United States
Copyright © 2025 Chang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junwu Ma, bWFfanVud3VAaG90bWFpbC5jb20=
 Xiaolong Chang
Xiaolong Chang Junwu Ma
Junwu Ma