- 1Second College of Clinical Medicine, Shanxi Medical University, Taiyuan, Shanxi, China
- 2The First College of Clinical Medicine, Shanxi Medical University, Taiyuan, Shanxi, China
- 3Shanxi Medical University, Taiyuan, Shanxi, China
- 4Department of Cardiology, The Second Hospital of Shanxi Medical University, Taiyuan, China
- 5School of Medicine, Shanxi Medical University, Taiyuan, China
A Correction on
Non-coding RNAs in heart failure: epigenetic regulatory mechanisms and therapeutic potential
by Ren Y, Zhao B, Lv L, Yang J, Nan X, Li B and Yang B (2025). Front. Genet. 16:1677797. doi: 10.3389/fgene.2025.1677797
There was a mistake in Figure 4 as published. In Figure 4, the word “faiure” was incorrectly spelled. The correct spelling is “failure.” The corrected Figure 4 appears below.

Figure 4. Differential ncRNA Expression in HF Types. This figure outlines the characteristic ncRNAs signatures across in patients with different types of HF. The pathogenesis and diagnoses depicted are all related to heart failure. The potential biomarkers mentioned require validation through large-scale, multi-center clinical trials. This does not imply that these biomarkers are without impact on the disease pathogenesis. (A) In HFrEF, miR-375 and lncRNA GDE1-1:1 are downregulated, while miR-208, miR-423-5p, exo-miR-92b-5p, lncRNA SRA1, lncRNA HEAT2, and circRNA DEPCS are upregulated. These molecules are potential biomarkers for HFrEF diagnosis. (B) In HFpEF, miR-19b-3p is downregulated, while miR-222/221, lncRNA TUG1, lncRNA MHRT, and circRNA HECW2 are upregulated. These molecules serve as potential biomarkers for HFpEF, with lncRNA XIST and circRNA HECW2 linked to profibrotic and inflammatory pathways. The figure is created in https://BioRender.com.
There was a mistake in Figure 5 as published. In Figure 5, the words “faiure” and “diabete” were incorrectly spelled. The correct spellings are “failure” and “diabetes,” respectively. The corrected Figure 5 appears below.
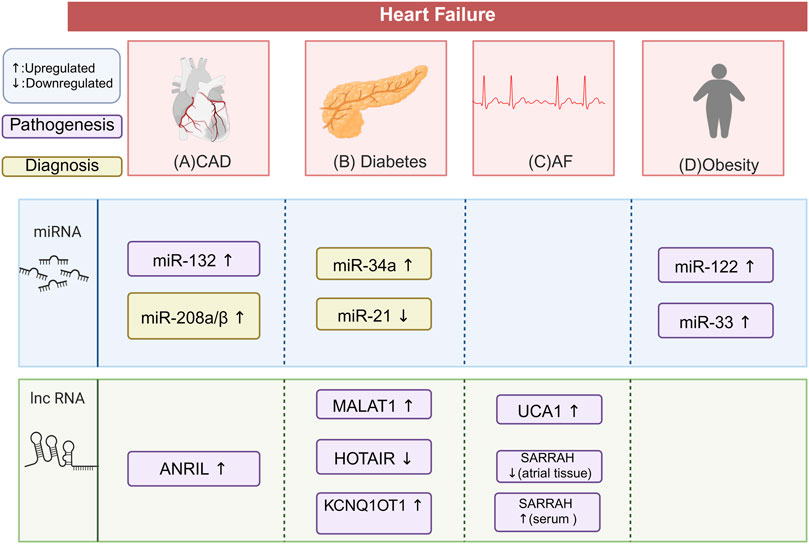
Figure 5. Differential ncRNA Expression in HF Comorbidities and Complications. This figure illustrates the characteristic ncRNA signatures in patients with HF and its comorbidities and complication. The potential biomarkers highlighted need to be validated through large-scale, multi-center clinical trials. This does not suggest that these biomarkers are irrelevant to the disease process. (A) In HF patients with CAD, miR-132 and lncRNA ANRIL are upregulated, contributing to the pathogenesis of CAD. miR-208a/β is also upregulated, helping diagnose myocardial injury alongside troponin. (B) In HF patients with diabetes, miR-34a is upregulated and miR-21 is downregulated, both serving as biomarkers for HFrEF. lncRNA MALAT1 is upregulated, increasing myocardial apoptosis and fibrosis. LncRNA KCNQ1OT1 is also upregulated, promoting cardiomyocyte pyroptosis and fibrosis. lncRNA HOTAIR is downregulated, promoting cardiac oxidative stress. (C) In HF patients with AF, lncRNA UCA1 is upregulated, promoting myocardial hypertrophy. lncRNA SARRAH is downregulated in atrial tissue but upregulated in serum, linked to resistance to oxidative stress and ischemic injury. (D) In HF patients with obesity, miR-33 and miR-122 are both upregulate. miR-122 promotes cardiac hypertrophy and fibrosis. miR-33 Participates in cardiovascular remodeling. The figure is created in https://BioRender.com.
The original article has been updated.
Generative AI statement
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: heart failure, non-coding RNA, epigenomics, microRNAs, long non-coding RNA, circular RNA, therapeutics
Citation: Ren Y, Zhao B, Lv L, Yang J, Nan X, Li B and Yang B (2025) Correction: Non-coding RNAs in heart failure: epigenetic regulatory mechanisms and therapeutic potential. Front. Genet. 16:1746339. doi: 10.3389/fgene.2025.1746339
Received: 14 November 2025; Accepted: 17 November 2025;
Published: 24 November 2025.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2025 Ren, Zhao, Lv, Yang, Nan, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao Li, bGliYW94eXNAMTYzLmNvbQ==; Bin Yang, eWFuZ2J4eXNAMTYzLmNvbQ==
 Yubo Ren
Yubo Ren Bomeng Zhao
Bomeng Zhao Luo Lv
Luo Lv Jingyuan Yang
Jingyuan Yang Xiangting Nan3
Xiangting Nan3 Bin Yang
Bin Yang