- 1Division of Forest, Nature and Landscape, Department of Earth and Environmental Sciences, KU Leuven, Geel, Belgium
- 2Plant Sciences Unit, Flanders Research Institute for Agriculture, Fisheries and Food (ILVO), Merelbeke, Belgium
- 3Department of Biochemistry and Microbiology, Ghent University, Ghent, Belgium
- 4Division of Crop Biotechnics, Department of Biosystems, KU Leuven, Geel, Belgium
- 5Centre for Environmental Sciences, Environmental Biology, UHasselt, Diepenbeek, Belgium
Introduction: Plant growth and health are significantly influenced by the composition, activity, and diversity of the rhizosphere microbiome. Developing strategies to modify the rhizosphere microbiome to foster beneficial interactions with plants therefore is a promising avenue for sustainable plant production. One such strategy involves the addition of artificial root exudate compounds. This study evaluates the effects of artificial root exudates on the growth and performance of Chrysanthemum ‘Merlino’ and its rhizosphere microbiome.
Methods: Horticultural substrates, both peatand compost-based, were supplemented with two concentrations of artificial root exudates containing fructose, glucose, sucrose, succinic acid, malic acid, arginine, serine, and cysteine. Twelve different treatments were tested in total. Plant physiological parameters, including root development, plant growth, and photosynthetic performance, were monitored to assess plant growth and stress responses. Microbial diversity shifts were analyzed using amplicon metabarcoding of the 16S rRNA gene (bacteria) and ITS region (fungi), alongside assessments of enzyme activity (alkaline phosphatase and urease) and microbial metabolic diversity.
Results: Our findings revealed significant impacts on microbial communities in the rhizosphere. Addition of artificial root exudates caused shifts in the bacterial and fungal community composition in both peatand compost-based substrates. The relative abundance of certain fungi increased in treatments with artificial root exudates, particularly those capable of metabolizing hexose or pentose sugars. That lead to a decrease in overall fungal diversity. Although bacterial diversity was not affected, the addition of artificial root exudates enhanced their metabolic diversity. Moreover, the application of artificial root exudates increased the activities of alkaline phosphatase and urease enzymes. Anticipated positive effects on plant growth were not observed: high concentrations of artificial root exudates (three weekly applications of 250 mg C g−1 substrate) resulted in reduced root development across all four horticultural substrates. Additionally, the highest concentration of artificial root exudates appeared to induce plant stress in peat-based substrates, as evidenced by lower dry mass of the plants and reduced Fv/Fm and PIabs.
Discussion: While artificial root exudates significantly alter the rhizosphere microbiome, they do not necessarily promote plant growth and may, depending on the composition and concentration, induce stress and inhibit root development in horticultural substrates.
1 Introduction
The rhizosphere harbors a great diversity of microorganisms, including bacteria, archaea, protozoa, and fungi, and represents a niche for interactions between the plant and microorganisms (Ali et al., 2017; Kour et al., 2019; Ryan et al., 2009). Plants have developed complex interactions with the rhizosphere microbiome, that plays an important role in plant growth and development, productivity, nutrition, disease susceptibility and overall plant performance (Ali et al., 2017; Dessaux et al., 2016; Kour et al., 2019; Mendes et al., 2013; Qu et al., 2020). In mineral soils, a high microbial richness, biomass, and activity have been associated with positive effects on plant growth and health. Several of these functions are conducted by rhizosphere microorganisms including nitrogen-fixing bacteria, mycorrhizal fungi, plant-growth promoting rhizobacteria (PGPR) and fungi (PGPF) (Berendsen et al., 2012). These organisms conduct a range of mechanisms involving the improvement of nutrient uptake by the plant, the production of various phytohormones, and the enhancement of tolerance to biotic and abiotic stresses (Ali et al., 2017; Kour et al., 2019; Qu et al., 2020).
Various strategies have been proposed to modify the rhizosphere microbiome to obtain beneficial interactions between the plant and the microbial community and optimize plant growth and health (Chaparro et al., 2012; Dessaux et al., 2016; Mueller and Sachs, 2015; Pascale et al., 2020; Ryan et al., 2009). First, organic amendments such as composts, have been shown to influence microbial communities of the rhizosphere (Brussaard et al., 2007; de Brito et al., 1995; Neher et al., 2022; Stewart-Wade, 2020; Viti et al., 2010). Composts have physiochemical and (bio)chemical characteristics that make them suitable for horticultural substrates (Bustamante et al., 2008; Hernandez-Apaolaza et al., 2005; Herrera et al., 2008; Vandecasteele et al., 2021). Previous research has shown that green composts are suitable peat replacers for Chrysanthemum (Delcour et al., unpublished data; Hummel et al., 2001; Papafotiou and Vagena, 2012). Moreover, green compost has been shown to have high microbial biomass and high relative abundances of beneficial microorganisms such as Pseudomonas spp., Bacillus spp., and Paenibacillus spp (Pot et al., 2022a).
A second strategy to manipulate the rhizosphere microbiome is the addition of artificial root exudate compounds. Plants actively or passively secrete a wide range of compounds, known as root exudates, into the rhizosphere, which is a major source of organic carbon released by plant roots. Root exudates mainly consist of carbon-containing substances, including low-molecular-weight compounds, such as amino acids and sugars, and high-molecular-weight exudates, such as proteins and polysaccharides. In addition, organic acids, phenolic compounds, flavonoids, terpenoids and alkaloids can be categorized as root exudates (Badri and Vivanco, 2009; Bais et al., 2006; Haichar et al., 2014). Root exudates play an important role in mediating the interactions between plants and rhizosphere microorganisms (Bais et al., 2006; Haichar et al., 2014; Philippot et al., 2013; Singh et al., 2004). Root exudates can act as chemical attractants and repellants to shape the rhizosphere microbiome and are a key factor in increasing microbial abundance and activity in the rhizosphere (Berendsen et al., 2012; Quiza et al., 2015; Walker et al., 2003). The composition and function of the rhizosphere microbial community are, in this way, actively influenced to ultimately shape a rhizosphere microbiome to be beneficial for the plant The remark needs to be made however, that the release of carbon rich root exudates can also attract pathogens, potentially negative for the plant (Dubey and Sharma, 2019; Quiza et al., 2015).
Artificially adding root exudate solutions to the rhizosphere may therefore be a promising approach to modify the rhizosphere microbiome to benefit plant growth and health. Artificial root exudates have the potential to alter microbial community composition and metabolic fingerprint and to increasing microbial richness, biomass, and activity in soils (Alam et al., 2014; Brussaard et al., 2014; Kolton et al., 2017; Mendes et al., 2011; Neher et al., 2022; Shen et al., 2016; Wagg et al., 2011; Weidner et al., 2015). For example, it can change the production of alkaline phosphatase by microorganisms, an important enzyme involved in phosphate solubilization, increasing the availability of phosphate, and is predominantly produced by bacteria (Amri et al., 2022; Wan et al., 2020). In addition, urease produced by microorganisms is involved in the degradation of urea, generating available N for plant and microbial growth (Cordero et al., 2019). Whether or not the inclusion of artificial root exudates results in a positive effect on plant growth or health has, to our knowledge, not been studied yet. In addition, effects of artificial root exudates on the microbiome were only studied in mineral soils, not in horticultural substrates. It is therefore unclear how the horticultural sector could benefit from strategies to influence the root microbiome, i.e. by combining composts and artificial root exudates, to ultimately contribute to sustainable solutions for plant growth and health.
In this study, the effect of artificial root exudates in different horticultural substrates on plant growth and performance of Chrysanthemum ‘Merlino’ was studied in relation to the rhizosphere microbiome. Chrysanthemum is an ornamental plant belonging to the Asteraceae family and holds significant economic value worldwide, ranking second only to rose in the cut flower trade. In addition to its use as a cut flower, Chrysanthemum is widely cultivated for gardening, ornamental landscaping, and as a potted plant. Furthermore, the species finds applications in various industries, including medicine, food, and beverages.
We hypothesize that artificial root exudates will change the composition and activity of the rhizosphere microbiome in horticultural media and that these effects benefit plant growth and performance. Moreover, as composts are richer in microorganisms than peat-based horticultural media, we expect a synergistic effect of artificial root exudates on the rhizosphere microbiome and plant growth in compost-based media.
To study this, Chrysanthemum ‘Merlino’ plants were grown in four substrates, either with or without compost in which artificial root exudates were added in two different compositions. Plant growth promotion was measured by focusing on plant physiological parameters. In addition, we studied changes in the microbial community composition making use of 16S rRNA (bacteria) and ITS (fungi) metabarcoding for taxonomical changes and BioLog Ecoplates to study metabolic profiles.
2 Materials and methods
2.1 Container trial with Chrysanthemum ‘Merlino’
A container trial with Chrysanthemum ‘Merlino’ (Asteraceae, Asterales) was set up in which the growth and performance of the plants were evaluated. An overview of the experiment can be found in Figure 1.
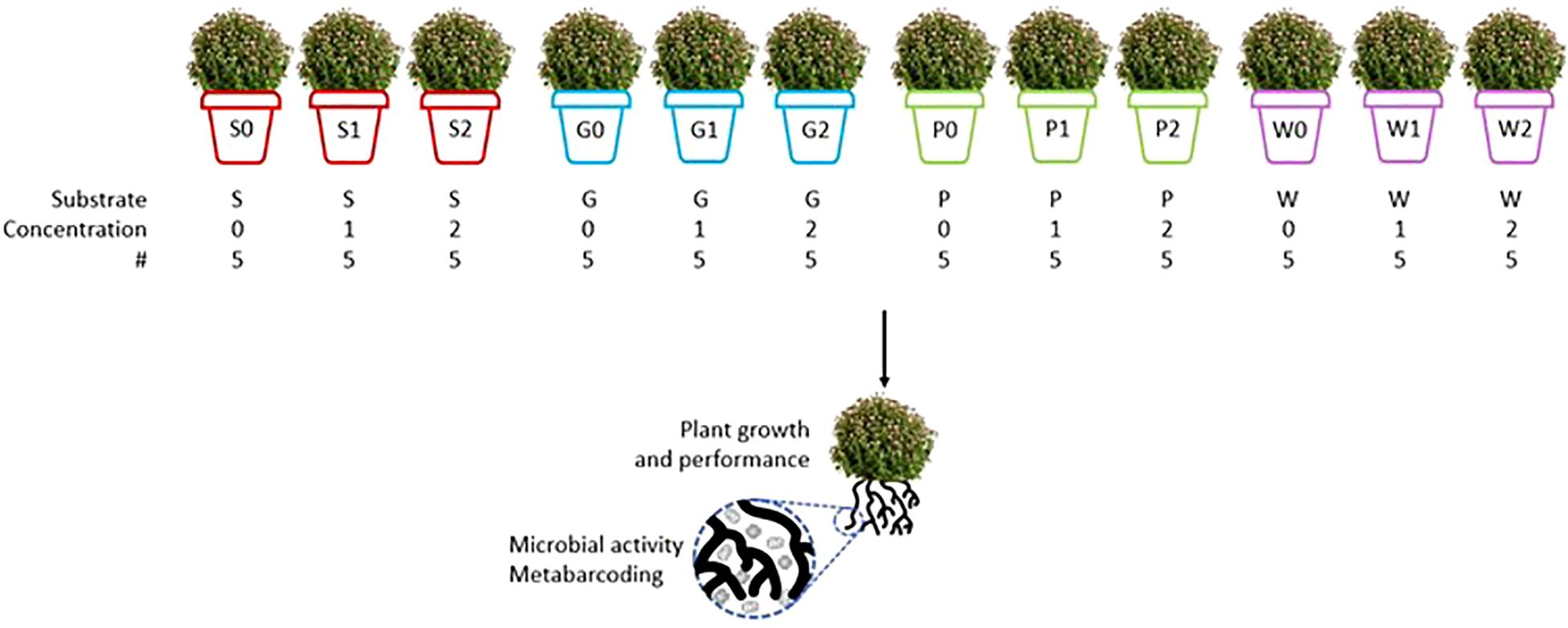
Figure 1. Set-up of the container trial with Chrysanthemum ‘Merlino’. Four substrates were used: one 100 vol% peat-based substrate (S) and three different compost-based substrates with 40 vol% peat and 60 vol% green compost (G, P, and W). In addition, three concentrations of artificial root exudates were used: 0 μg C g−1 substrate and 0 μg N g−1 substrate (demineralized water; 0), 100 μg C g−1 substrate and 6 μg N g−1 substrate (1) and 250 μg C g−1 substrate and 15 μg N g−1 substrate (2). Five biological replicates were planted for each treatment, resulting in 60 plants. (Icons from thenounproject.com; created by Webtechops LLP, Vectors Point, and Guillhem from the Noun project).
Four different horticultural substrates were used. Three green composts (G, P, and W) from different installations, i.e., Limburg.net (Belgium), IOK (Belgium), and IGEAN (Belgium), were mixed with peat in a 60:40 vol% ratio. Although these green composts were sourced from different waste management installations within the same region, variations in local waste streams and composting techniques likely resulted in distinct compost characteristics. All three composts have a VLACO quality label, indicating they comply with the legal requirements to be used as a fertilizer or soil improver. A standard 100 vol% peat-based substrate (Agaris, Belgium) was used as a reference (S). To account for differences in nitrogen level between the different horticultural substrates, fertilizers were mixed in the four different substrates based on their initial NO3-N and NH4-N content (Supplementary Table S1), which was determined using a Skalar San ++ Continuous Flow Analyzer (Skalar, Breda, The Netherlands). The N content of each substrate was equated to 618 mg N L−1 with Osmocote Exact Standard 3-4 M (16-9-12 (NPK) + 2 MgO + TE; ICL Speciality Fertilizers, Belgium). P, Ca, Mg, K, and Na were extracted from the fertilized substrates (1:5 v/v) in 0.5 M ammonium acetate buffered with 96% acetic acid at pH 4.65, and the K, Mg, Ca, and P concentration in the extract was measured by ICP-OES. The pH-H2O was measured in a 1:5 v/v water extract according to EN13037 (CEN, 2011) (Supplementary Table S2).
The fertilized substrates were put in pots (600 g substrate/pot). Subsequently, a rooted cutting of Chrysanthemum ‘Merlino’ was planted in each pot (obtained from Dataflor, Beselare, Belgium). In total, 60 plants were planted. The plants were arranged in a growth chamber in a completely random design (21°C, 12 h light – 12 h dark, RH 70%) and grown for 13 weeks. The substrates were kept moist during the experiment by adding water by hand three times a week. Each plant was given the same amount of water each time they were watered. No additional fertilizer was applied during the experiment.
Starting from the third week of the experiment, artificial root exudates were applied to the plants three times a week. A stock solution with a concentration of 2.5 mg C mL−1 and 150 mg N mL−1 (C/N of 16.67) was prepared according to Griffiths et al. (1998) (Table 1). This solution was based on a range of common root exudate compounds. Three different concentrations of artificial root exudates were added to the plants: 0 mg C g−1 substrate and 0 mg N g−1 substrate (demineralized water; 0), 100 mg C g−1 substrate and 6 mg N g−1 substrate (1), and 250 mg C g−1 substrate and 15 mg N g−1 substrate (2). For concentration 1, each pot received 2.34 g C and 0.14 g N throughout the experiment. For concentration 2, each pot received 5.85 g C and 0.35 g N throughout the experiment. The combination of four substrates and three concentrations of artificial root exudates resulted in 12 treatments (S0, S1, S2, G0, G1, G2, P0, P1, P2, W0, W1, W2).
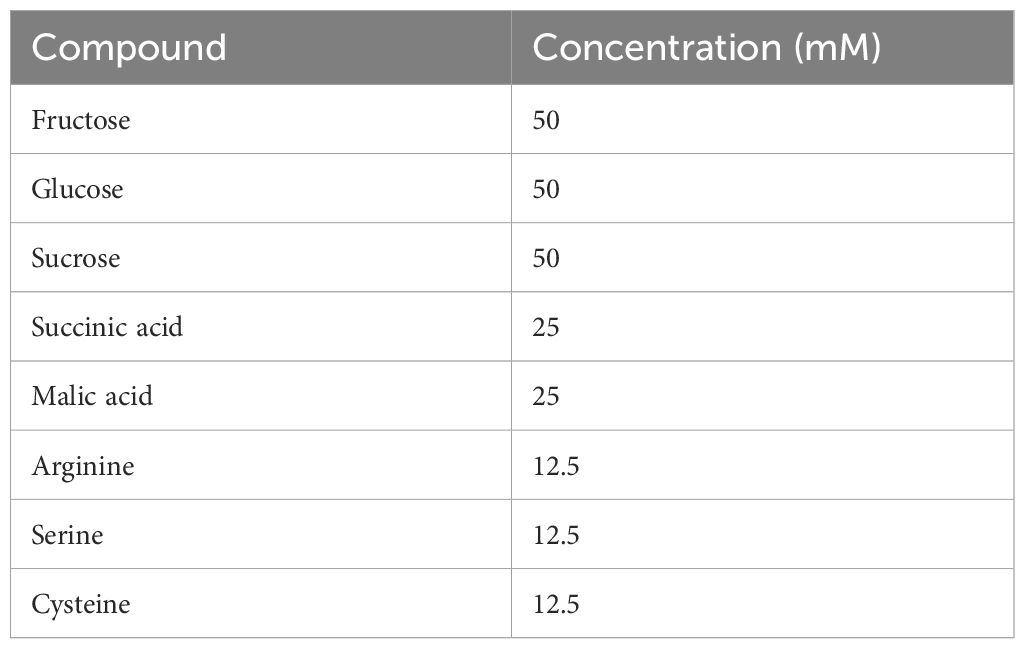
Table 1. Composition of the stock solution of artificial root exudates based on Griffiths et al. (1998).
2.2 Measurements of plant growth and performance
Plant height was measured weekly with a ruler. At the end of the experiment, plant height and diameter were manually measured with a ruler. Root development was scored using a scoring system based on the root system visible at the surface of the substrate (Supplementary Figure S1). A 1–3 score was given based on the number of visible lateral roots, with 1, few lateral roots; 2, a moderate number of lateral roots; and 3, a large number of lateral roots, which are all over the substrate surface. Above ground fresh and dry mass was determined at the end of the experiment. By cutting, aboveground plant material was separated from the roots and weighed. The plant material was then dried at 70°C for 4 days and weighed again.
Photosynthetic performance was monitored weekly and at the end of the experiment using nondestructive chlorophyll a fluorescence measurements. Chlorophyll a fluorescence measurements were carried out using a Handy PEA fluorometer (Plant Efficiency Analyser, Hansatech, King’s Lynn, United Kingdom) and were taken on young, fully developed leaves. Measurements were performed using a saturating pulse of 3,500 μmol m–2 s–1, pulse duration of 1 s, and fixed gain (1.1 ×). Before the measurements, leaves were allowed to dark adapt for 15 min using light-withholding clips (Hansatech). The light level of the saturating pulse and the minimal dark period were experimentally determined beforehand to obtain true values for the chlorophyll a fluorescence parameters. Chlorophyll fluorescence parameters describing the maximum quantum efficiency of PSII (Fv/Fm) and performance index (PIabs) were calculated.
At the end of the experiment, the quality of the plants was evaluated using a scoring system used in the horticultural sector to determine the commercial value of a plant (PCS Ornamental Plant Research). It is based on an assessment of the overall look of the plants (Supplementary Figure S2). A 1–3 score was given, with 1 = low quality; 2 = moderate quality; and 3 = high quality.
Relative water content of the leaves was determined according to Ceusters et al. (2008) at the end of the experiment by sampling punches of young, fully developed leaves of each plant and calculated with the formula: (fresh weight − dry weight)/(turgid weight − dry weight). Leaf punches were weighed after sampling to determine fresh weight. Turgid weight was determined by floating the leaf punches in plastic tubes filled with demineralized water for 6 h in darkness at 4°C. Dry weight was determined by putting the leaf punches in an oven at 70°C for 4 days.
To determine leaf chlorophyll and carotenoid contents, leaves were sampled from each plant (about 1 g) and chopped into small pieces. Plant pigments were extracted by immersing the leaf material in N, N-dimethylformamide (DMFA) at room temperature for 72 h in darkness. The supernatant was used to determine the absorbance at 647 nm (A647), 664 nm (A664), and 470 nm (A470) (Genesys 10S UV-VIS, Thermo Fisher Scientific, United States). These data were used to calculate the content of chlorophyll a (Ca), chlorophyll b (Cb), total chlorophyll (C), and total carotenoids using the empirical formulas: Ca = 11.65 A664 – 2.69 A647; Cb = 20.81 A647 – 4.53 A664; C = Ca + Cb; carotenoids = (1000 A470 – 0.89 Ca – 52.02 Cb)/245 (Moran and Porath, 1980; Porra et al., 1989; Wellburn, 1994).
2.3 Rhizosphere sampling
The rhizosphere of all samples was sampled three days after the last application of artificial root exudates as described in detail in Lundberg et al. (2012). In this process, soil loosely adhering to the roots was manually dislodged, and the roots were placed within a 50 mL tube along with 25 mL of phosphate buffer. Through vortexing, the rhizosphere soil was liberated from the roots. The resulting mixture was subsequently passed through a 100 µm nylon mesh cell strainer into a fresh 50 mL tube to remove plant components and large sediments. To create a pellet (weighing 250 mg) representing the rhizosphere sample, the filtered solution underwent centrifugation at 3,000 g for 15 minutes. Samples for determination of enzyme activities and community-level physiological profiling (Biolog EcoPlates) were stored for two weeks at 4°C awaiting further analysis, and samples for metabarcoding were stored at −20°C.
2.4 Determination of enzyme activities
The activity of urease, alkaline phosphatase, and dehydrogenase was determined in all rhizosphere samples, according to Shen et al. (2006).
For urease activity, 1 g of the rhizosphere samples was added to 5 mL of phosphate buffer (0.1 mol L−1, pH 6.7) and 0.5 mL of toluene. After 15 minutes, 5 mL of 10% urea was added. The mixture was incubated at 37°C for 24 h. After incubation, 10 mL of 1 mol L−1 1 KCl was added to the mixture. The mixture was shaken at 200 rpm for 10 minutes and centrifuged at 17,000 g for 10 minutes. Subsequently, 1 mL of supernatant was mixed with 8 mL distilled water, 0.5 mL of 25% (w/w) potassium sodium tartrate, 0.3 mL of 1 mol L−1 NaOH, and 0.1 mL of Nessler reagent. After 10 minutes, the absorbance at 460 nm was measured. A control without urea was used for each sample. Urease activity was expressed as g NH3 produced L−1 substrate.
For alkaline phosphatase activity, 1 g of the rhizosphere samples was added to 5 mL of 0.5% disodium phenyl phosphate and 0.2 mL of toluene. This solution was incubated at 37°C for 2 h and centrifuged at 17,000 g for 10 min. Subsequently, 1 mL of supernatant was diluted to 5 mL with distilled water, 4 mL of borate buffer (0.05 mol L−1, pH 10), 0.5 mL of 2% (w/w) 4-amino antipyrine, and 0.5 mL of 8% (w/w) potassium ferricyanide were added. The absorbance at 510 nm was measured. Phosphatase activity was expressed as g P released L−1 substrate.
For dehydrogenase activity, 1 g of the rhizosphere samples was added to 0.1 g of CaCO3, 2 mL of 1% 2,3,5-triphenyl tetrazolium (TTC), and 1.5 mL of distilled water. The solution was incubated at 30°C for 24 h. After incubation, the oxidized TTC was extracted by shaking at 200 rpm at room temperature with 7 mL of acetone for 30 min and centrifuging at 17,000 g for 10 min. The absorbance of the supernatant was measured (485 nm). Dehydrogenase activity was expressed as g TTC oxidized L−1 substrate.
2.5 Community-level physiological profiling using Biolog EcoPlates
Each rhizosphere sample was analyzed using Biolog EcoPlates (Biolog, Inc., CA, USA) as described in Pot et al. (2022a). The metabolic activity, functional diversity, and Rsi values were calculated as described in Pot et al. (2022a).
2.6 DNA extraction and 16S rRNA gene and ITS2 region metabarcoding
DNA was extracted from each rhizosphere sample (250 mg) using the DNeasy Powersoil Pro Kit (QIAGEN, Germantown, MD, USA), according to the manufacturer’s instructions. DNA was stored at −20°C until metabarcoding. Metabarcoding on the V3–V4 fragment of the 16S rRNA gene and the rDNA-ITS2 region was done as described in (Pot et al., 2022a). Sequencing of the reads was done through Illumina MiSeq v3 technology, 2x 300bp by Admera Health (United States). Reads are available for download at the NCBI SRA under project number PRJNA913777. Sequences were processed as described in detail in (Pot et al., 2022a).
2.7 Statistical analysis
To determine the effect of substrate and concentration of the artificial root exudates on plant growth, diameter, fresh and dry plant mass, fluorescence parameters, relative water content, chlorophyll, and carotenoid content, general linear models 20with substrate and concentration of the artificial root exudate solution as main effects and including all interaction terms, were used. Assumptions were checked by looking into the diagnostic plots of the models. Generalized linear models were used to test differences in plant quality and root development, which included substrate and concentration of the artificial root exudates as the main effects and the interaction between them. Assumptions of linearity, homogeneity of variances, and normality were checked before analysis. Pairwise comparisons were made using contrasts between least square means and were corrected for multiple testing by Tukey’s HSD.
General linear models were used to determine differences in enzyme activity, including substrate and concentration of the artificial root exudate solution as main effects and their interaction. Assumptions of linearity, homogeneity of variances, and normality were checked before analysis. Pairwise comparisons were made using contrasts between least square means.
A general linear model, including substrate and concentration of the artificial root exudate solution as main effects and their interaction, was used to determine differences in metabolic activity and functional diversity between the treatments. Assumptions of linearity, homogeneity of variances, and normality were checked before analysis. Pairwise comparisons were made using contrasts between least square means and were corrected for multiple testing by Tukey’s HSD. Differences in functional community composition determined with Biolog EcoPlates were visualized using PCA on the Rsi values. The effect of substrate and concentration of the artificial root exudate solution on the functional community composition was determined using PERMANOVA analysis.
For the metabarcoding data, a sequence table was constructed for bacteria and fungi, respectively. All analyses were done for each of these sequence tables. First, ASVs with less than two counts per million in at least five samples were removed to exclude low abundant reads. Second, the Shannon diversity index was calculated using the vegan package (version 2.5.7) in R (version 4.0.4) (Oksanen et al., 2022) to determine alpha diversity. A general linear model including substrate and concentration as main effects and their interaction was used to find significant differences in diversity between the treatments. Assumptions of linearity, homogeneity of variances, and normality were checked before analysis. Pairwise comparisons were studied using contrasts between least square means. Third, beta diversity was studied. Absolute ASV counts were transformed to relative abundances, and a dissimilarity matrix (based on the Bray-Curtis dissimilarity index) was calculated. The homogeneity of the variances was checked for this dissimilarity matrix using the betadisper function. The condition of homogeneity of variances was fulfilled in all substrates, except for substrates P and W (P = 0.002 for both substrates) in the fungal dataset, indicating sample heterogeneity. The effect of substrate and concentration on the community composition was tested using a PERMANOVA analysis of the dissimilarity matrix, making use of the adonis function in R. PCoA was used on the dissimilarity matrix to visualize the observed differences. Fourth, relative abundances of the most abundant genera in each treatment were studied. Fifth, the effect of substrate and concentration on relative abundances was tested using the edgeR package (version 3.32.1) (Robinson et al., 2010), as described in Pot et al. (2021). P-values less than 0.05 were considered significant. Briefly, differential abundance in the soil microbiome was assessed for the effect of different substrates (S, G, P and W) and the different concentrations (0, 1, 2) of root exudates added. The hypotheses of interest were tested by building a specific contrast, comparing the estimated mean differences in between root exudates treatments and substrate types. The data were normalized with the trimmed mean of M values (TMM) that corrects the effective library size of the count tables. A generalized linear model with negative binomial distribution (nbGLM) was applied to the counts for each ASV with substrate and root exudate concentration as two main effects. The significance of the ASV was inferred with a quasi-likelihood F-test with a Benjamin-Hochberg False Discovery Rate [FDR] of 5% to adjust for multiple corrections.
3 Results
3.1 A high concentrations of artificial root exudates results in lower root development
No significant effect of artificial root exudates on fresh plant mass was found within the different substrates, whereas for substrate S, concentration 0 showed significantly higher dry plant mass than concentration 2 (P = 0.04). In addition, aboveground plant growth, diameter, and quality were not altered by the presence of artificial root exudates in the different substrates. However, concentration 2 of artificial root exudates resulted in significantly lower root development than concentration 0 (P = 0.04). (Figure 2). No significant effects of substrate or concentration were found on the relative water content of the plants or total chlorophyll and carotenoid concentrations.
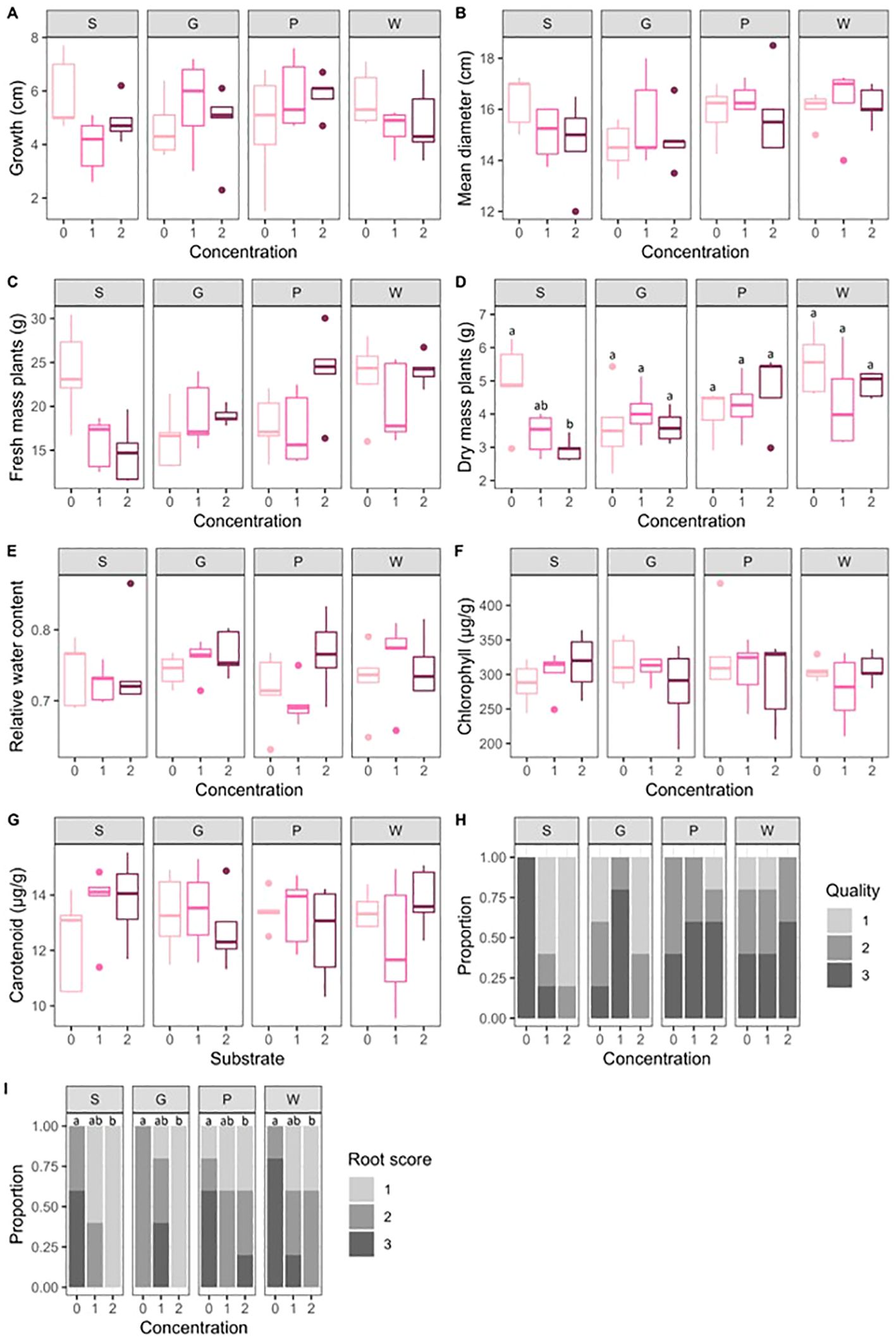
Figure 2. (A) Boxplots (n = 5) of aboveground plant growth (cm) (A), plant diameter (cm) (B), fresh mass of plants (g) (C), dry mass of plants (g) (D), relative water content of the plants (E), total chlorophyll concentrations (mg/g) (F), carotenoid concentrations (mg/g) (G), and bar plots (n=5) of proportions of quality scores (1: low quality; 2: moderate quality; and 3: good quality) (H), proportions of root scores (1: few lateral roots; 2: a moderate number of lateral roots; and 3: a large number of lateral roots all over the substrate surface) (I) in the different treatments. Letters indicate significant differences between the different concentrations of artificial root exudate solutions (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 µg C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate) within the peat-based substrate (S) and the three different compost-based substrates (G, P, and W) (general linear model, contrasts between least square means; P < 0.05).
No significant effect of artificial root exudates on Fv/Fm was found over time in substrates G, W, and P. From week 6 onwards, a significant difference in Fv/Fm was found between concentration 0 and concentration 2 (P < 0.001) and between concentration 1 and concentration 2 in substrate S (P < 0.001), with concentration 2 showing a significantly lower Fv/Fm (Figure 3A). No significant effect of artificial root exudates on PIabs was found over time in substrates G, P, and W. From week 5 onwards, a significant difference in PIabs was found between concentration 0 and concentration 2 in substrate S (P < 0.001), with concentration 2 showing a significant lower PIabs. Moreover, between weeks 5 and 7, a significant difference in PIabs was found between concentration 1 and concentration 2 in substrate S (P = 0.005), with concentration 2 showing significantly lower PIabs (Figure 3B).
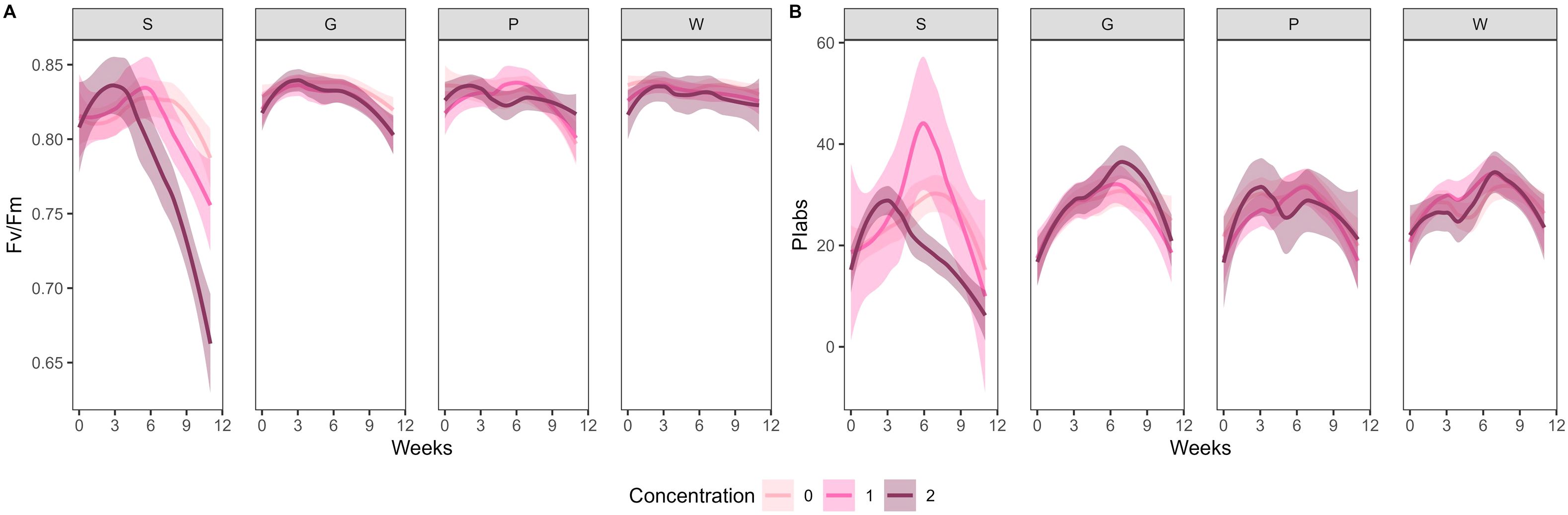
Figure 3. Fv/Fm (A) and PIabs (B) in the different treatments over 12 weeks. 95% confidence intervals are shown as shaded areas. Colors indicate the different concentrations (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 µg C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate) in the different substrates (S = peat-based substrate; G, P, and W = compost-based substrates).
3.2 Artificial root exudates alter the bacterial and fungal community in the rhizosphere of Chrysanthemum in peat- and compost-based substrates
On average 48,856 ± 2,535 reads per sample for the bacterial dataset and 65,461 ± 4,062 reads per sample for the fungal dataset were obtained, which resulted in the identification of 13,992 bacterial and 5,925 fungal ASVs. Artificial root exudates caused a shift in the bacterial and fungal community composition of peat-based and compost-based substrates (Figure 4). Based on the first axis, explaining 22.8% of the total variance for the bacteria and 45,1% for the fungi, we could see that there is a separate clustering based on the addition of the different concentrations of the artificial root exudates. Therefore, these were more determining than the different substrates, which clustered according to the second axes on the PCoA plot and explained 18,7% and 14.2% of the variance for the bacteria and fungi accordingly (Figure 4).
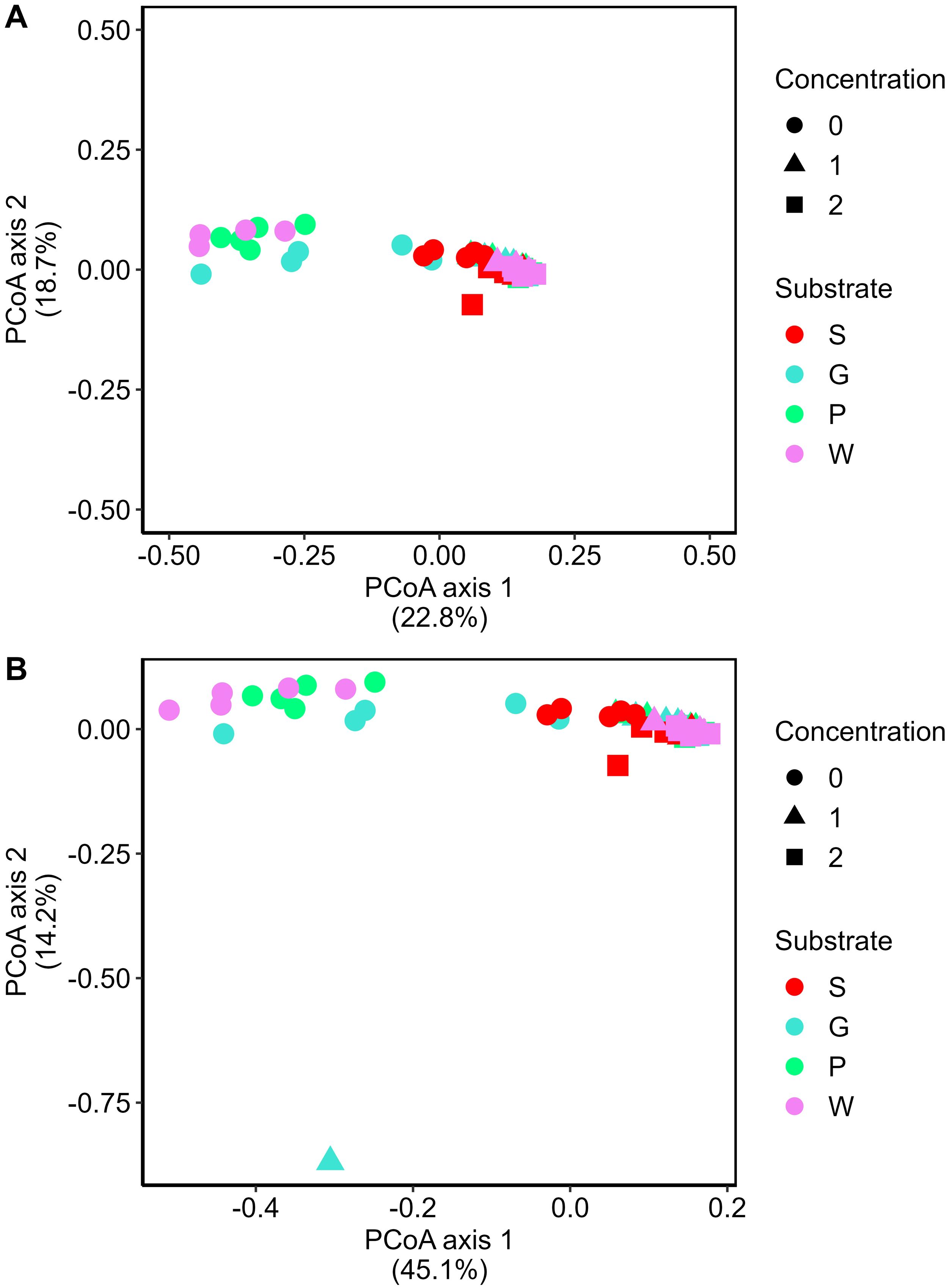
Figure 4. Shifts in bacterial and fungal community composition between the different treatments. Principal Coordinate Analysis (PCoA) profile of pairwise community dissimilarity (Bray-Curtis) indices of bacterial (16S V3-V4 rRNA gene) (A) and fungal (ITS2 region) (B) sequencing data. Colors indicate the four types of substrates (S = peat-based substrate; G, P, and W = compost-based substrates), whereas shapes show the three concentrations of artificial root exudates (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 µg C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate).
The interaction between the substrate and the concentration of artificial root exudates was shown to be significant (PERMANOVA; P = 0.001 and P = 0.001) for bacteria and fungi. Within each substrate, a significant effect of concentration was observed (bacteria: P = 0.001 for all four substrates, and fungi: P = 0.001 for S, G, and P, and P = 0.002 for W). Overall, the bacterial and fungal community composition of the peat-based substrate showed a clear shift compared to the three compost-based substrates, which were more equal concerning bacterial and fungal community composition. Moreover, artificial root exudates seemed to have the largest effect on the compost-based substrates, especially for the fungal community. The shift between the lowest concentration (concentration 0) and concentrations 1 and 2 was notably larger than between concentrations 1 and 2.
To study the organisms that drive these different community profiles, the microbial community composition between the different treatments was studied in more detail by investigating, (1) the composition of the most abundant genera and (2) differential abundances.
For the compost-based substrates (G, P, and W), the top ten most abundant bacterial and fungal genera were similar between concentrations 1 and 2 of artificial root exudates, while the top ten most abundant bacterial and fungal genera in concentration 0 showed more differences (Supplementary Figures S3 and Supplementary Figure S4). For the peat-based substrate (S), the top ten most abundant bacterial and fungal genera were similar between the three concentrations of artificial root exudates.
In addition, the differential abundances of bacterial and fungal phyla, families, and genera were studied (Supplementary Table S3) by difference in concentration. For bacteria and fungi, the number of differentially abundant taxa changed more between concentration 0 and concentration 1 or 2 compared to the number of differentially abundant taxa between concentration 1 and concentration 2, and this for all four substrates. This indicates that the addition of artificial root exudates had a higher effect than a change in its concentration. For bacteria, the largest number of differentially abundant genera for the compost-based substrates G, P, and W was seen between concentration 0 and concentration 2, while the largest number of differentially abundant genera for the peat-based substrate S was observed between concentration 0 and 1. The number of differentially abundant taxa between concentration 0 and concentration 1 and between concentrations 0 and 2 were more similar for fungi.
The differentially abundant bacterial and fungal genera (relative abundance > 0.01) between the different concentrations of artificial root exudates within each substrate are shown in more detail in Tables 2, 3, respectively. One bacterial genus, Taibaiella, was significantly more abundant in concentration 2 compared with concentration 0 for all four substrates. The bacterial genera Arachidococcus, Nakamurella, Pseudarthrobacter, and TM7a were significantly more present in concentrations 1 and 2 compared to concentration 0; whereas Panearthrobacter was more abundant in concentration 1 compared to concentration 0 in all three compost-based substrates. Moreover, the genera, Microbacterium, Pseudaminobacter, and Rhodanobacter, were significantly more abundant in concentration 2 compared to concentration 0 in all three compost-based substrates. For fungi, Clitopilus was significantly less present in concentration 1 and 2 compared with concentration 0 in all four substrates, while two fungal genera, Solicoccozyma and Saitozyma, were significantly more present in concentration 1 and concentration 2, respectively, compared with concentration 0 in all four substrates. The fungal genus, Coprinellus, was significantly less abundant in concentration 1 compared with concentration 0 in all three compost-based substrates. In addition, the fungal genus Candida was significantly more abundant in concentration 2 compared with concentration 0 in the three compost-based substrates. No genera commonly known to include plant-growth-promoting species were found to be increased in relative abundance in response to artificial root exudates in the four horticultural substrates.
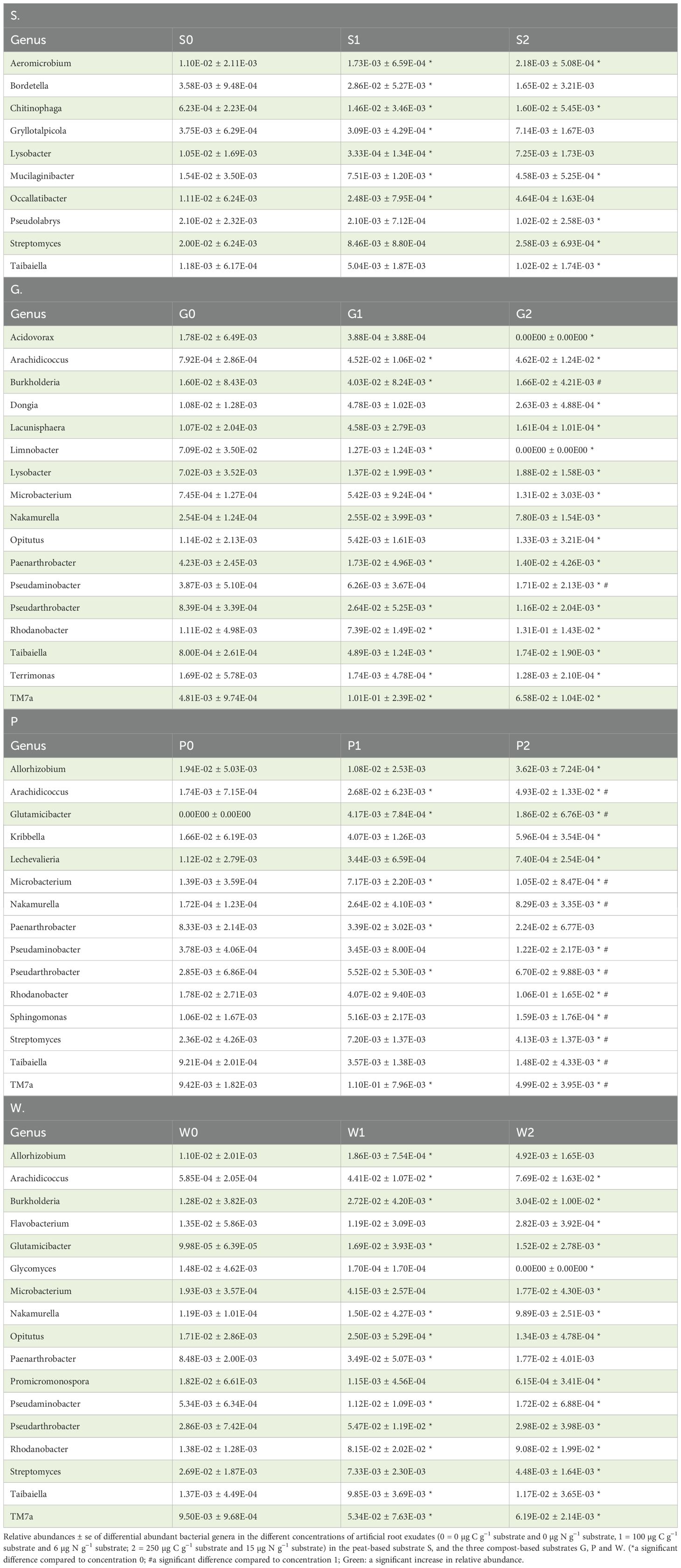
Table 2. Differential abundant bacterial genera in the different concentrations of artificial root exudates in each substrate (n=5).
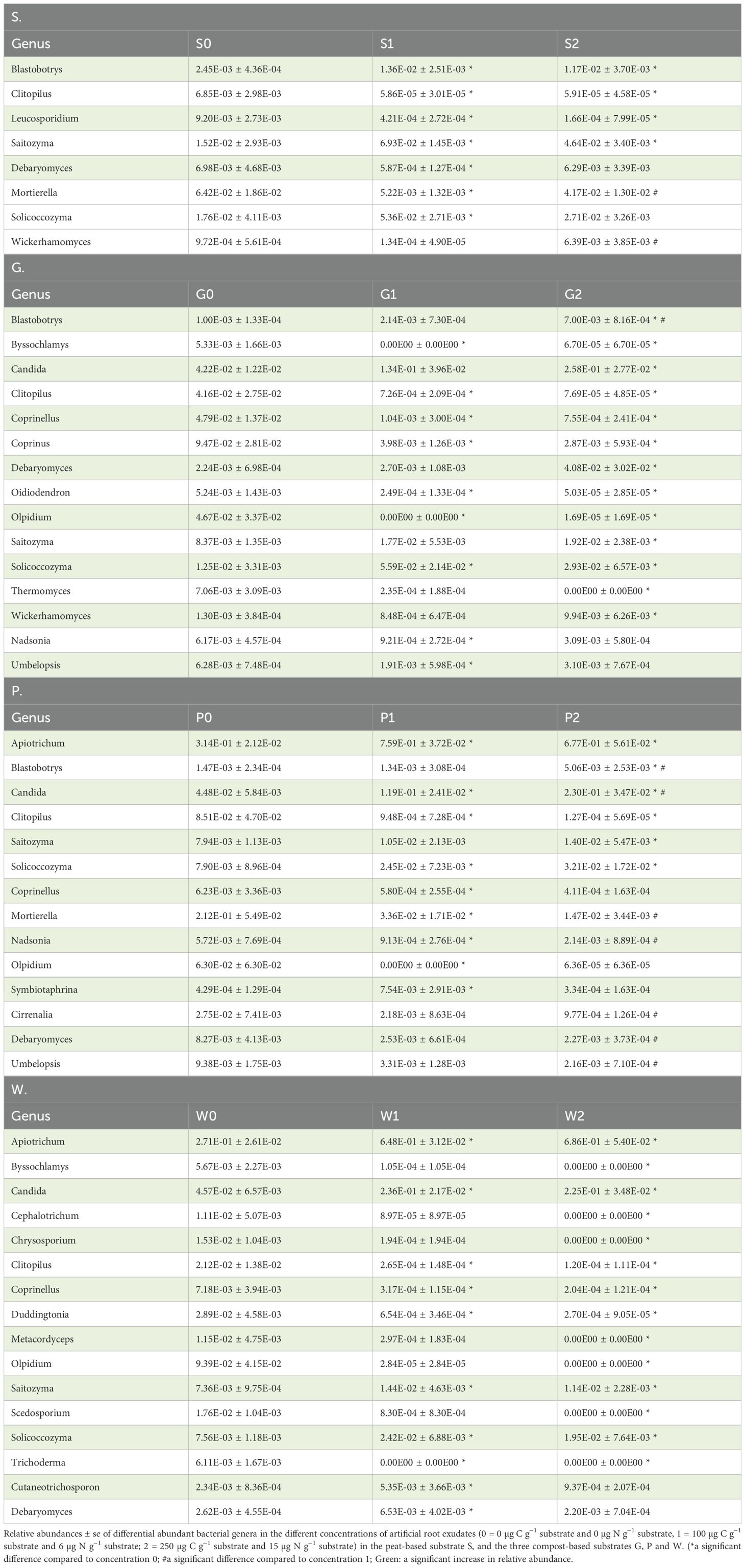
Table 3. Differential abundant fungal genera in the different concentrations of artificial root exudates in each substrate.
At last, bacterial and fungal diversity was compared between the different concentrations of artificial root exudates in each substrate. No significant effects of artificial root exudates were found for bacterial diversity in the different horticultural substrates. For fungal diversity, concentrations 1 and 2 showed a significantly lower fungal diversity than concentration 0 (P < 0.001 for both concentrations in the four horticultural substrates) (Figure 5).
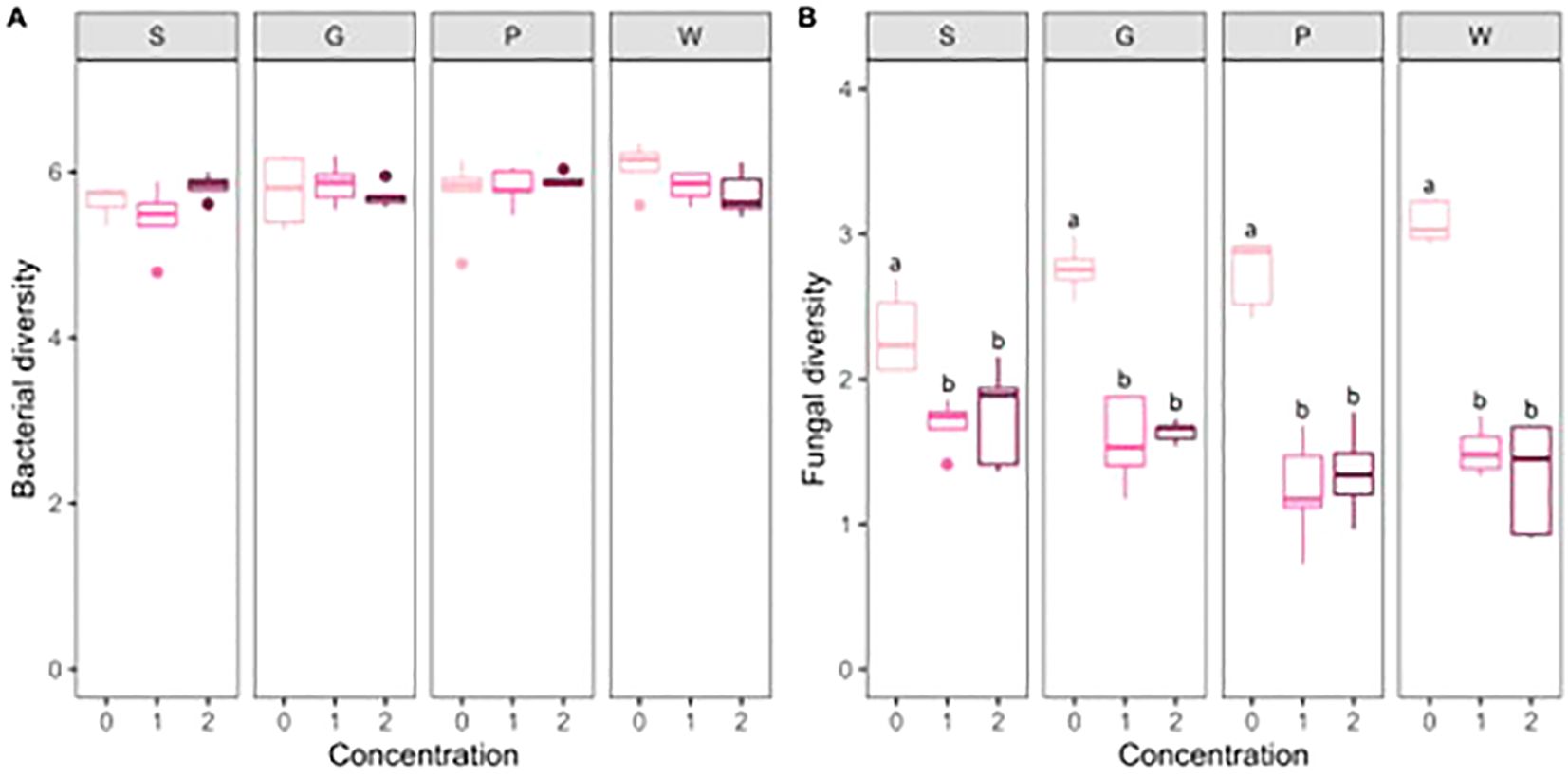
Figure 5. Boxplots (n = 5) of the bacterial (A) and fungal (B) diversity (n = 5) (calculated as the Shannon Diversity Index) for the different treatments. Letters indicate significant differences between concentrations (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 µg C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate) within the peat-based substrate (S) and the compost-based substrates (G, P, W) (linear model, contrasts between least square means; P < 0.05).
3.3 Artificial root exudates increase metabolic diversity and activity of alkaline phosphatase and urease
Multiple mechanisms have been suggested by which root exudates may reshape the microbiome s studied by the Biolog Ecoplates between treatments, which is also reflected in the PCA plot (Supplementary Figure S5). On the other hand, the concentration of the artificial root exudates significantly affected metabolic activity and functional diversity. Concentration 2 resulted in a significantly higher average well color development (AWCD) compared to concentration 1 (P = 0.02) and a significantly higher functional diversity compared to concentration 0 (P = 0.01) in all horticultural substrates. Moreover, concentration 2 showed a significantly higher AWCD of carboxylic acids (P = 0.02) and polymers (P = 0.02) than concentration 1 in all substrates (Figure 6). This indicates that, although the overall physiological profiles of the microbial communities remained stable, their metabolic activity and functional diversity were significantly affected by the concentration of the artificial root exudates.
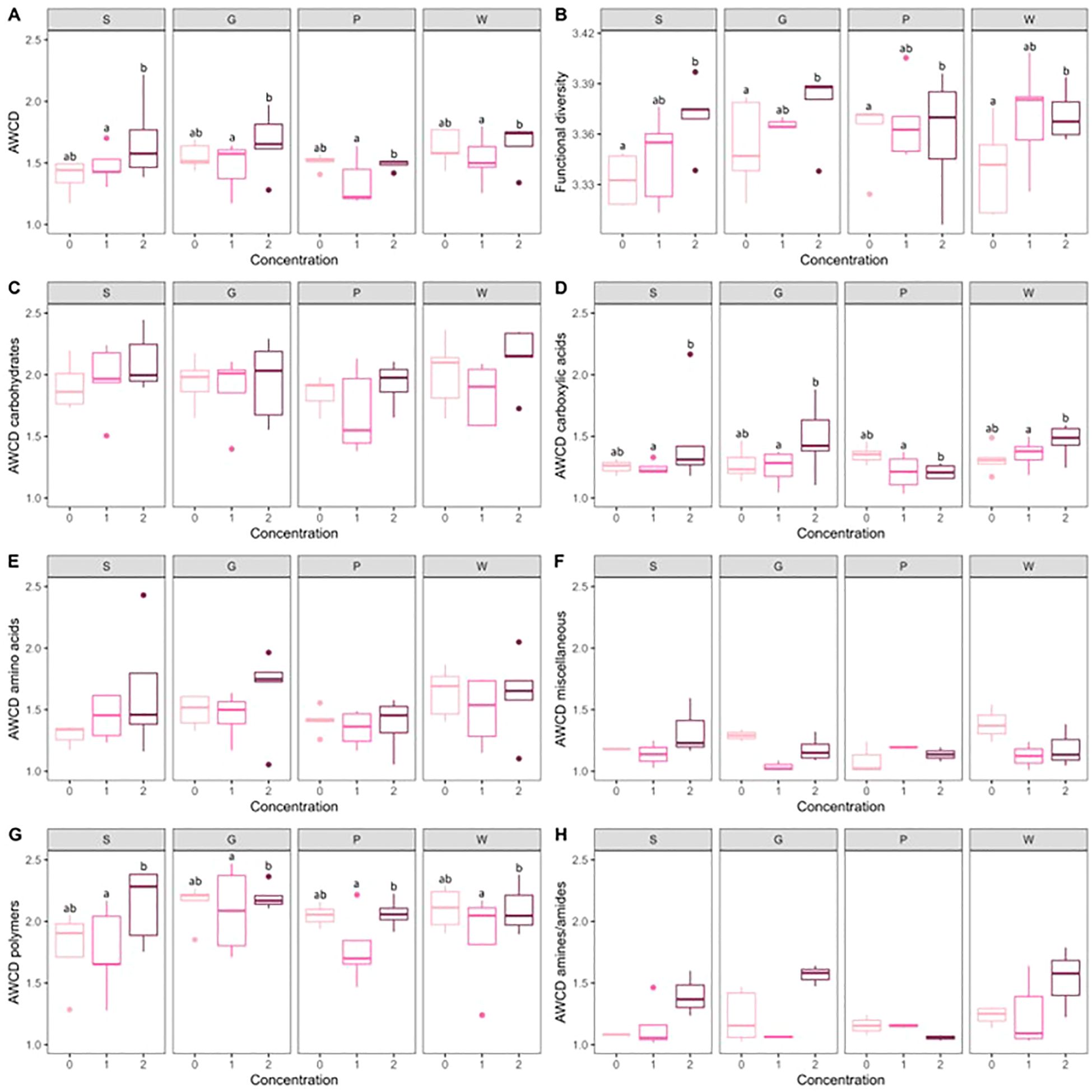
Figure 6. Metabolic characteristics of the different treatments obtained with community-level physiological profiling using Biolog EcoPlates. Boxplots (n = 5) of metabolic activity, expressed as average well color development (AWCD) (A), functional diversity calculated as the Shannon Diversity Index (B), AWCD of carbohydrates (C), AWCD of carboxylic acids (D), AWCD of amino acids (E), AWCD of miscellaneous C-courses (F), AWCD of polymers (G) and AWCD of amines/amides (H). Letters indicate significant differences between concentrations of artificial root exudates (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 gµ C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate) within the peat-based substrate (S) and the three compost-based substrates (G, P, and W) (linear model, contrasts between least square means; P < 0.05).
The substrate and concentration of artificial root exudates had no significant effects on the dehydrogenase activity. The concentration of artificial root exudates had a significant effect on the activity of alkaline phosphatase and urease, with concentration 2 showing a significantly higher alkaline phosphatase (P <no><</no> 0.001) and urease activity (P = 0.01) than concentration 0 in all substrates (Figure 7).
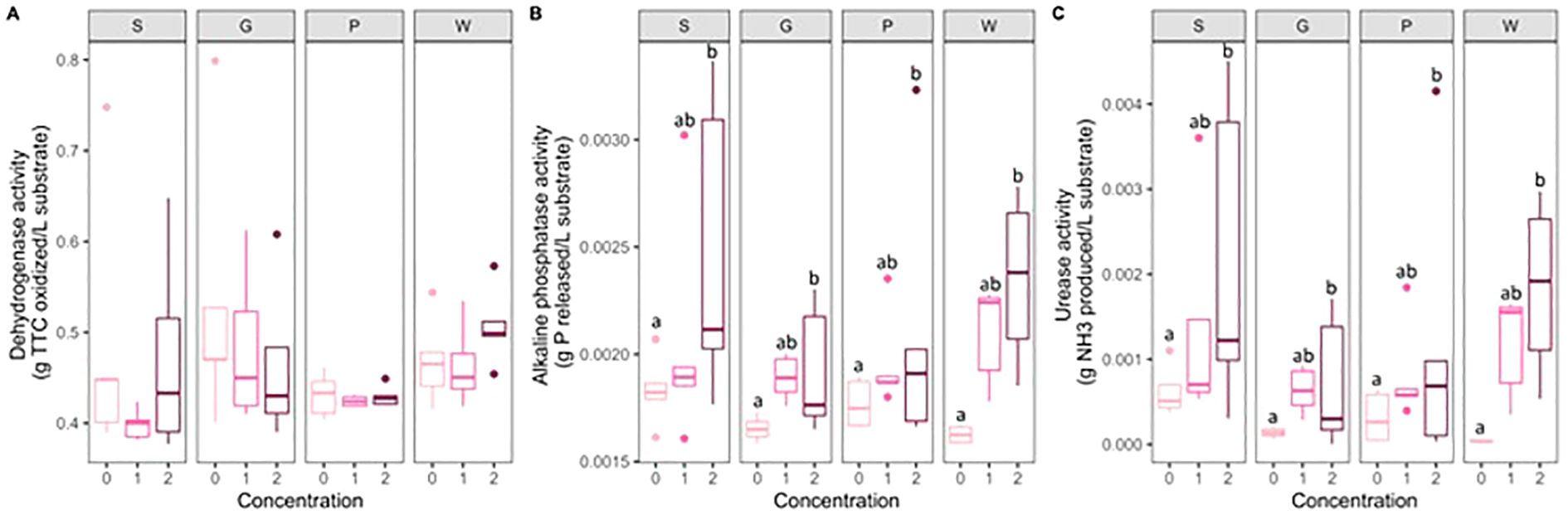
Figure 7. Boxplots (n = 5) of dehydrogenase activity (g TTC oxidized/L substrate) (A), alkaline phosphatase activity (g P released/L substrate) (B), and urease activity (g NH3 produced/L substrate) (C). Letters indicate significant differences between the concentrations of artificial root exudate solutions (0 = 0 µg C g−1 substrate and 0 µg N g−1 substrate, 1 = 100 µg C g−1 substrate and 6 µg N g−1 substrate; 2 = 250 µg C g−1 substrate and 15 µg N g−1 substrate) within the peat-based substrate (S) and the three compost-based substrates (G, P, and W) (linear model, contrasts between least square means; P < 0.05).
4 Discussion
In this study, we investigated the effect of two concentrations of artificial root exudates (100 µg C g−1 substrate and 250 µg C g−1 substrate) in one peat-based and three compost-based horticultural substrates on plant growth and performance of Chrysanthemum ‘Merlino’ in relation to the rhizosphere microbiome.
Our study only partially confirmed our initial hypothesis. While we successfully demonstrated that artificial root exudates can significantly alter the rhizosphere microbiome, the anticipated benefits to plant growth and performance were not realized. In fact, our findings revealed a more complex interaction between artificial root exudates, the rhizosphere microbiome, and plant health. As hypothesized, the artificial root exudates did indeed cause substantial shifts in both bacterial and fungal community compositions across peat-based and compost-based substrates. We observed significant changes in microbial community structure, with the concentration of artificial root exudates being a more determining factor than the substrate type. The metabolic diversity of bacteria increased, and enzyme activities were enhanced, suggesting active microbial responses to the artificial exudates. However, contrary to our expectations, these microbiome changes did not translate into improved plant growth or performance. Instead, higher concentrations of artificial root exudates led to reduced root development and, in peat-based substrates, signs of plant stress. The anticipated synergistic effect in compost-based media was not observed, challenging our initial hypothesis about the potential of artificial root exudates to promote plant growth.
Our results show that artificial root exudates can shift the bacterial and fungal community composition of peat-based and compost-based substrates. Our results corroborate the effects previously evidenced in mineral soils: it confirms the role of exudates as one of the main drivers for the rhizosphere microbiome (Walker et al., 2003) and furthermore evidences that mimicking this effect artificially (Steinauer et al., 2016) holds potential for the horticultural sector. For mineral soils, Baudoin et al. (2003) showed that daily inputs of artificial root exudates at 100 µg C g−1 dry soil for 14 days resulted in restructuring the bacterial communities in bulk and rhizosphere soils. Kozdrój and van Elsas (2000) showed that artificial root exudates, based on the composition of root exudates of maize, affected the structure of bacterial communities in mineral soils polluted with heavy metals. Li et al (2021) demonstrated that artificial root exudates, consisting of sugars, amino acids, and organic acids, affected the bacterial community composition of mineral soils affected by mining. Similar to the studies executed in soil, our study showed artificial root exudates mainly affects fungal diversity (Waldrop et al, 2006). On the other hand, Vieira et al. (2020) showed that different bacteria are associated with specific root exudates in soils, especially amino acids and organic acids (Vieira et al., 2020). Other studies also indicate that different artificial root exudates compounds influence specific taxa in soil (Eilers et al., 2010; Hugoni et al., 2018; Shi et al., 2011) and that organic acids have a greater impact on microbial community composition than carbohydrates (Chen et al., 2019; Shi et al., 2011). However, as our setup only included a single blend of different compounds, we could not differentiate which component has had the largest influence on certain taxa. Furthermore, this study utilized artificial root exudate compounds that were based on root exudate compounds commonly found in various plants. However, it is important to note that the composition of root exudates is highly plant-specific, yet very little information is available on the root exudate composition of most plants, including Chrysanthemum. Moreover, the artificial root exudate mix used in this study lacked signaling compounds naturally found in root exudates, such as phenols, terpenes, alkaloids, and flavonoids, which play key roles in shaping microbial communities. The absence of these compounds may have limited the ability of the artificial mix to fully mimic the complex interactions occurring in the rhizosphere. Future determination of which root exudate compounds may attract beneficial microorganisms may allow us to steer the microbial community composition in a more targeted way. Moreover, a qualitative analysis of the root exudates of different plant species may form the basis to produce a more effective, plant-specific artificial root exudate solution (Dundek et al., 2014).
Multiple mechanisms have been suggested by which root exudates may reshape the microbiome, including providing a substrate or nutrient source for microbial growth, eliciting chemotactic responses, facilitating root colonization, and limiting microbial growth by acting as antimicrobial compounds (Rolfe et al., 2019; Vieira et al., 2020), which is depending on the life strategies of the microbes however (Ho et al., 2017). In this study, we found a higher abundance of several fungi able to grow on hexose or pentose sugars such as glucose and fructose, such Saitozyma, Candida, and Solicoccozyma (Aliyu et al., 2021; Filippucci et al., 2016; Misra et al., 2012) in treatments with artificial root exudates, which is in line with a mechanism based on food availability. The decrease in fungal diversity in response to the addition of artificial root exudates may therefore be due to a shift toward a more specialized fungal community. This is likely driven by the proliferation of fungi capable of efficiently metabolizing carbon-rich substrates, leading to a competitive exclusion of less adapted taxa (Chigineva et al., 2009; de Graaff et al., 2010). For bacteria, such associations could not be observed. This could potentially be explained by differences in microbial reproduction rates. Bacteria generally have faster generation times compared to fungi, which may allow their populations to return to baseline levels more quickly after the cessation of artificial root exudate applications. Since rhizosphere sampling occurred three days after the last treatment, transient bacterial shifts may have diminished, whereas fungal communities, which are often more resilient but slower to respond, still exhibited measurable differences.
Artificial root exudates increased the metabolic diversity of bacteria. Li et al. (2015) also showed that amendments with organic carbon in the soil increased metabolic diversity. The increased metabolic diversity may be attributed to the promotion of microorganisms suited to metabolize carbon and nitrogen-rich substrates (Li et al., 2015). In this study, bacteria do not appear to exhibit the same level of specialization as fungi under these conditions. This may be due to their more generalist nature, with many bacterial species able to metabolize a wide range of substrates. As a result, bacterial communities may not undergo the same competitive exclusion dynamics observed in fungi. Higher metabolic diversity in microbial communities has been linked to disease suppression and plant growth promotion (Brussaard et al., 2007; Kolton et al., 2017; Neher et al., 2022). Artificial root exudates did not increase metabolic activity, which may be attributed to the complexity of the organic matter, which may not be readily available for bacteria (Baudoin et al., 2003).
In line with Steinauer et al. (2016); Shi et al. (2011) and Vaidya et al. (2022) in mineral soils, our results corroborate that exudates can alter the activity of extracellular enzymes produced by microorganisms. We observed an increase in alkaline phosphatase and urease activity was observed in response to the addition of artificial root exudates. Both alkaline phosphatase and urease production has been shown to be beneficial for plant and microbial growth (Cordero et al., 2019). Furthermore, high metabolic diversity and enzyme activity in the rhizosphere has been linked to disease suppression in mineral soils (Mendes et al., 2013; Neher et al., 2022). In further research, it would therefore be interesting to study the effect of artificial root exudates in the presence of root-borne pathogens.
Despite changes in the microbial community composition and the positive effects of artificial root exudates on metabolic diversity and enzyme activity, no positive effects on plant growth were observed in our study. Notably, higher concentrations of these exudates (administered at a rate of three weekly doses of 250 µg C per gram of substrate) led to significantly reduced root development in compost-based and peat-based horticultural substrates. The increased activity of urease and alkaline phosphatase resulting from the introduction of artificial root exudates might have contributed to the observed decline in root development, as root growth and proliferation can be influenced by the availability of soil nutrients: elevated nitrate levels can decrease primary root elongation, while increased nitrate and phosphate availability can hamper lateral root elongation (López-Bucio et al., 2003). Another important consideration is that the substrates that were used in this experiment may not have contained microbial species that can effectively interact with Chrysanthemum. Plant–microbe interactions are often highly selective. If the microbial communities present in these horticultural substrates were not well adapted to Chrysanthemum, this could explain why no synergistic effects on plant growth were observed despite changes in microbiome composition.
In peat-based substrates, the highest concentration of artificial root exudates (three weekly 250 µg C g−1 substrate) caused plant stress, which was reflected in reduced Fv/Fm and PIabs values. Both of these chlorophyll fluorescence derived values measure the photosynthetic performance of plants and have been related with various types of plant stress (Ceusters et al., 2019; Maxwell and Johnson, 2000). Consequently, the above ground dry mass of these plants was also significantly lower. In compost-based media, no such effect was observed. Unraveling the mechanism behind this loss in the efficiency of the photosynthetic light reactions and growth in the peat-based substrates is not straightforward. The increased enzymatic activity described above could suppress plant-microbe symbiosis and increase plant–microbe competition, yet no significant effects of artificial root exudates were found in the plants’ relative water content or chlorophyll concentrations. The latter may be related to the suppressed growth of the stressed plants but for now cannot be confirmed. The cause for the lack of plant stress in response to artificial root exudates in compost-based substrates also can only be speculated at this point, but the larger shift observed in the rhizosphere microbiome in response to artificial root exudates in compost-based substrates may provide a clue: research has shown that the flexibility of the microbiome in response to changes in the environment can reduce plant stress (Liu et al., 2020; Voolstra and Ziegler, 2020). Further research is therefore needed to study plant stress in response to artificial root exudates in peat-based substrates. Monitoring changes in the chemical properties of horticultural substrates and nutrient and water availability in response to artificial root exudate additions in further research could shed light on possible stress causes.
Furthermore, it is worth mentioning that in this study, we worked with an high fertilization in each horticultural substrate. Substrate fertilization regimes have been shown to influence plant-microbiome interactions, with differing outcomes for plant health (Pot et al., 2022b). Further research therefore is needed to evaluate if responses of plants to artificial root exudates differ according to different fertilization levels. Also, effects of dose of the artificial root exudates merit further consideration: this study used three-weekly artificial root exudates inputs of 100 µg C g−1 and 250 µg C g−1. Natural inputs of C to the soil by root exudates are difficult to determine but are often estimated at around 100 µg C g−1 day−1 (Baudoin et al., 2003; Griffiths et al., 1998). Other studies showed changes in the microbial communities in response to application rates of artificial root exudates between 100 µg C g−1 day−1 and 2500 µg C g−1 day−1 (Baudoin et al., 2003; Griffiths et al., 1998; Shi et al., 2011). In this study, we showed that three-weekly applications of 100 µg C g−1 and 250 µg C g−1 were able to affect the microbial community. Further research should focus on determining if optimal concentrations of artificial root exudates can be determined to promote plant growth.
While our experiment was conducted under favorable growth parameters, it’s crucial to consider the effect of the change in the rhizosphere microbiome caused by artificial root exudates under suboptimal or stress-inducing conditions. Composts are known for their pathogen-suppressive capabilities (Neher et al., 2022). Such protective effects could become particularly relevant when plants are challenged by disease or environmental stressors. Furthermore, the composition of the microbial community can drastically influence plant responses to stresses like salinity or drought (Allison and Martiny, 2008; Philippot et al., 2021). Although we did not observe significant growth promotion with artificial root exudates, the observed shifts in microbial community structure suggest a potential for modulating plant–microbe interactions in ways that could enhance stress tolerance. Responses noted here might also be attributed to the use of compost instead of mere the root exudates. We acknowledge that compost addition can enrich soil with beneficial microbes, although it must be said that those microbes are not necessarily PGPRs. In addition, we need to acknowledge that effects that occur can also be attributed to changes in nutrient composition in the growing media and that to ascertain the role of the microbial community, this should be followed up in future experiments. Future research should therefore explore how these microbial community changes manifest under various stress conditions, investigating whether the observed compositional shifts could provide plants with enhanced resilience or adaptive capabilities that are not apparent under optimal growth conditions.
5 Conclusion
This study investigated for the first time the effects of artificial root exudates on the rhizosphere microbiome and plant growth in horticultural substrates. As root exudates play a vital role in mediating plant–microbe interactions, we anticipated that introducing artificial root exudates could reshape the rhizosphere microbiome to enhance plant health and growth. However, our observations revealed significant shifts in microbial composition and increased enzyme activity without the expected positive effects on plant growth. Higher concentrations of artificial root exudates led to negative impacts on root development, particularly in compost-based and peat-based substrates. In addition, high concentrations of artificial root exudates led to plant stress in peat-based substrates. The unexpected findings regarding the negative effects of artificial root exudates on plant growth underscore the complexity of the interactions between the microbiome and plant growth and open avenues for further investigations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA913777.
Author contributions
SP: Conceptualization, Data curation, Writing – original draft. CDT: Data curation, Formal Analysis, Methodology, Writing – review & editing. JC: Funding acquisition, Project administration, Supervision, Writing – review & editing. JD: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. KV: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Flanders Innovation and Entrepreneurship grant HBC.2017.0815 and Research Foundation Flanders grant 12S9418N and fits in the scope of COST action CA22158 MicropBiomes of which Caroline De Tender is member and work group leader. The performed studies are in frame with research networking of COST Action MiCropBiomes CA22158.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fhort.2025.1562929/full#supplementary-material
References
Alam M. Z., Braun G., Norrie J., Mark Hodges D. (2014). Ascophyllum extract application can promote plant growth and root yield in carrot associated with increased root-zone soil microbial activity. Can. J. Plant Sci. 94, 337–348. doi: 10.4141/cjps2013-135
Ali M. A., Naveed M., Mustafa A., Abbas A. (2017). “The good, the bad, and the ugly of rhizosphere microbiome,”. Eds. Kumar V., Kumar M., Sharma S., Prasad R. Probiotics and plant health. (Singapore: Springer), 253–290. doi: 10.1007/978-981-10-3473-2_11
Aliyu H., Gorte O., Neumann A., Ochsenreither K. (2021). Global transcriptome profile of the oleaginous yeast Saitozyma podzolica DSM 27192 cultivated in glucose and xylose. J. Fungi 7, 9. doi: 10.3390/jof7090758
Allison S. D., Martiny J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sciences 105(Supplement 1), 11512–11519. doi: 10.1073/pnas.0801925105
Amri M. F., Husen E., Tjahjoleksono A., Wahyudi A. T. (2022). Alkaline phosphatase activity of plant growth-promoting Actinomycetes and their genetic diversity based on the phoD gene. HAYATI J. Biosci. 29, 360–369. doi: 10.4308/hjb.29.3.360-369
Badri D. V., Vivanco J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Bais H. P., Weir T. L., Perry L. G., Gilroy S., Vivanco J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Baudoin E., Benizri E., Guckert A. (2003). Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 35, 1183–1192. doi: 10.1016/S0038-0717(03)00179-2
Berendsen R. L., Pieterse C. M. J., Bakker P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Brussaard L., de Ruiter P. C., Brown G. G. (2007). Soil biodiversity for agricultural sustainability. Agriculture Ecosystems Environ. 121, 233–244. doi: 10.1016/j.agee.2006.12.013
Brussaard L., de Ruiter P. C., Brown G. G. (2014). Soil biodiversity for agricultural sustainability. Agriculture Ecosyst. Environ. 121, 233–244.
Bustamante M., Paredes C., Moral R., Agulló E., Murcia P., Abad M. (2008). Composts from distillery wastes as peat substitutes for transplant production. Resour. Conserv. Recycl. 52 (5), 792–799. doi: 10.1016/j.resconrec.2007.11.005
CEN (2011). EN 13037: Soil improvers and growing media. Determination of pH. Technical Committee CEN/TC 223 (European Committee For Standardization (CEN).
Ceusters J., Londers E., Verdoodt V., Ceusters N., De Proft M. (2008). Seasonal impact on physiological leaf damage risk of aechmea hybrid under greenhouse conditions. Scientia Hortic. 118 (3), 242–245.
Ceusters N., Valcke R., Frans M., Claes J. E., Van den Ende W., Ceusters J. (2019). Performance index and PSII connectivity under drought and contrasting light regimes in the CAM orchid Phalaenopsis. Front. Plant Sci. 10, 1012. doi: 10.3389/fpls.2019.01012
Chaparro J., Sheflin A., Manter D., Vivanco J. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chen S., Waghmode T. R., Sun R., Kuramae E. E., Hu C., Liu B. (2019). Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7, 136. doi: 10.1186/s40168-019-0750-2
Chigineva N. I., Aleksandrova A. V., Tiunov A. V. (2009). The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates. Appl. Soil Ecol. 42, 264–270. doi: 10.1016/j.apsoil.2009.05.001
Cordero I., Snell H., Bardgett R. D. (2019). High throughput method for measuring urease activity in soil. Soil Biol. Biochem. 134, 72–77. doi: 10.1016/j.soilbio.2019.03.014
de Brito A. M., Gagne S., Antoun H. (1995). Effect of compost on rhizosphere microflora of the tomato and on the incidence of plant growth-promoting rhizobacteria. Appl. Environ. Microbiol. 61, 194–199. doi: 10.1128/aem.61.1.194-199.1995
de Graaff M.-A., Classen A. T., Castro H. F., SChadt C. W. (2010). Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol. 188, 1055–1064. doi: 10.1111/j.1469-8137.2010.03427.x
Dessaux Y., Grandclément C., Faure D. (2016). Engineering the rhizosphere. Special Issue: Unravelling Secrets Rhizosphere 21, 266–278. doi: 10.1016/j.tplants.2016.01.002
Dubey S., Sharma ,. S. (2019). “Rhizospheric microbiome engineering as a sustainable tool in agriculture: Approaches and challenges,” in Microbial diversity in ecosystem sustainability and biotechnological applications: Volume 2. Soil & agroecosystems. Eds. Satyanarayana T., Das S. K., Johri B. N. (Singapore: Springer), 257–272. doi: 10.1007/978-981-13-8487-5_11
Dundek P., Holík L., Rohlík T., Hromádko L., Vranová V., Rejšek K., et al. (2014). Methods of plant root exudates analysis: A review. Acta Universitatis Agriculturae Silviculturae Mendelianae Brunensis 59, 241–246. doi: 10.11118/actaun201159030241
Eilers K. G., Lauber C. L., Knight R., Fierer N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903. doi: 10.1016/j.soilbio.2010.02.003
Filippucci S., Tasselli G., Scardua A., Di Mauro S., Cramarossa M. R., Perini D., et al. (2016). Study of Holtermanniella wattica, Leucosporidium creatinivorum, Naganishia adeliensis, Solicoccozyma aeria, and Solicoccozyma terricola for their lipogenic aptitude from different carbon sources. Biotechnol. Biofuels 9, 259. doi: 10.1186/s13068-016-0672-1
Griffiths B. S., Ritz K., Ebblewhite N., Dobson G. (1998). Soil microbial community structure: Effects of substrate loading rates. Soil Biol. Biochem. 31, 145–153. doi: 10.1016/S0038-0717(98)00117-5
Haichar F., Santaella C., Heulin T., Achouak W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Hernandez-Apaolaza L., Gasco A. M., Gasco J. M., Guerrero F. (2005). Reuse of waste materials as growing media for ornamental plants. Bioresource Technol. 96, 125–131. doi: 10.1016/j.biortech.2004.02.028
Herrera F., Castillo J. E., Chica A. F., Lopez Bellido L. (2008). Use of municipal solid waste compost (MSWC) as a growing medium in the nursery production of tomato plants. Bioresource Technol. 99, 287–296. doi: 10.1016/j.biortech.2006.12.042
Ho A., Ijaz U. Z., Janssens T. K. S., Ruijs R., Yoon Kim S., de Boer W., et al. (2017). Effects of bio-based residue amendments on greenhouse gas emission from agricultural soil are stronger than effects of soil type with different microbial community composition. Global Change Biol. Bioenergy 9, 1707–1720.
Hugoni M., Luis P., Guyonnet J., F. el Z. (2018). Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 28, 451–463. doi: 10.1007/s00572-018-0857-5
Hummel R. L., Johnson C. R., Riley R., Smith S. (2001). Yard trimmings compost as a growing medium component and nutrient source for chrysanthemum and fuchsia production©. (Washington: Department of horticulture and landscape), 51.
Kolton M., Graber E. R., Tsehansky L., Elad Y., Cytryn E. (2017). Biochar-stimulated plant performance is strongly linked to microbial diversity and metabolic potential in the rhizosphere. New Phytol. 213, 1393–1404. doi: 10.1111/nph.14253
Kour D., Rana K. L., Yadav N., Yadav A. N., Kumar A., Meena V. S., et al. (2019). “Rhizospheric microbiomes: Biodiversity, mechanisms of plant growth promotion, and biotechnological applications for sustainable agriculture,” in Plant growth promoting rhizobacteria for agricultural sustainability: From theory to practices. Eds. Kumar A., Meena V. S. (Singapore: Springer), 19–65. doi: 10.1007/978-981-13-7553-8_2
Kozdrój J., van Elsas J. D. (2000). Response of the bacterial community to root exudates in soil polluted with heavy metals assessed by molecular and cultural approaches. Soil Biol. Biochem. 32, 1405–1417. doi: 10.1016/S0038-0717(00)00058-4
Li J., Li Y., Yang X., Zhang J., Lin Z., Zhao B. (2015). Microbial community structure and functional metabolic diversity are associated with organic carbon availability in an agricultural soil. J. Integr. Agric. 14, 2500–2511. doi: 10.1016/S2095-3119(15)61229-1
Li Y., Yuan L., Xue S., Liu B., Jin G. (2021). Artificial root exudates excite bacterial nitrogen fixation in the subsurface of mine soils. Appl. Soil Ecol. 157, 103774. doi: 10.1016/j.apsoil.2020.103774
Liu H., Brettell L. E., Qiu Z., Singh B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
López-Bucio J., Cruz-Ramırez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. doi: 10.1016/S1369-5266(03)00035-9
Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Maxwell K., Johnson G. N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51, 659–668. doi: 10.1093/jxb/51.345.659
Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Misra S., Raghuwanshi S., Gupta P., Dutt K., Saxena R. K. (2012). Fermentation behavior of osmophilic yeast Candida tropicalis isolated from the nectar of Hibiscus rosa sinensis flowers for xylitol production. Antonie Van Leeuwenhoek 101, 393–402. doi: 10.1007/s10482-011-9646-2
Moran R., Porath D. (1980). Chlorophyll determination in intact tissues using N,N-dimethylformamide. Plant Physiol. 65, 478–479. doi: 10.1104/pp.65.3.478
Mueller U. G., Sachs J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Neher D. A., Hoitink H. A., Biala J., Rynk R., Black G. (2022). “Chapter 17—Compost use for plant disease suppression,” in The composting handbook. Ed. Rynk R. (New York: Academic Press), 847–878. doi: 10.1016/B978-0-323-85602-7.00015-7
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2022). vegan: Community ecology package (R package version 2.5-7. 2020).
Papafotiou M., Vagena A. (2012). Cotton gin trash compost in the substrate reduces the daminozide spray dose needed to produce compact potted chrysanthemum. Scientia Hortic. 143, 102–108. doi: 10.1016/j.scienta.2012.06.004
Pascale A., Proietti S., Pantelides I. S., Stringlis I. A. (2020). Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01741
Philippot L., Bru D., Saby N. P. A., Ranjard L. (2021). Bacteria-archaea interactions in soil: Cooperation, syntrophy, and niche partitioning. Microbiol. Mol. Biol. Rev. 85, e00026–e00020. doi: 10.1128/MMBR.00026-20
Philippot L., Raaijmakers J., Lemanceau P., Putten W. (2013). Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Porra R. J., Thompson W. A., Kriedemann P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA) - Bioenergetics 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Pot S., De Tender C., Ommeslag S., Delcour I., Ceusters J., Gorrens E., et al. (2021). Understanding the shift in the microbiome of composts that are optimized for a better fit-for-purpose in growing media. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.643679
Pot S., De Tender C., Ommeslag S., Delcour I., Ceusters J., Vandecasteele B., et al. (2022a). Elucidating the microbiome of the sustainable peat replacers composts and nature management residues. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.983855
Pot S., De Tender C., Ommeslag S., Delcour I., Ceusters J., Vandecasteele B., et al. (2022b). Suppression of Phytophthora on Chamaecyparis in sustainable horticultural substrates depends on fertilization and is linked to the rhizobiome. Phytobiomes J. 6, PBIOMES–05-22-0029-R. doi: 10.1094/PBIOMES-05-22-0029-R
Qu Q., Zhang Z., Peijnenburg W. J. G. M., Liu W., Lu T., Hu B., et al. (2020). Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 68, 5024–5038. doi: 10.1021/acs.jafc.0c00073
Quiza L., St-Arnaud M., Yergeau E. (2015). Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front. Plant Sci. 6507. doi: 10.3389/fpls.2015.00507
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rolfe S. A., Griffiths J., Ton J. (2019). Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 49, 73–82. doi: 10.1016/j.mib.2019.10.003
Ryan P. R., Dessaux Y., Thomashow L. S., Weller D. M. (2009). Rhizosphere engineering and management for sustainable agriculture. Plant Soil 321, 363–383. doi: 10.1007/s11104-009-0001-6
Shen R. F., Cai H., Gong W. (2006). Transgenic Bt cotton has no apparent effect on enzymatic activities or functional diversity of microbial communities in rhizosphere soil. Plant Soil 285, 149–159. doi: 10.1007/s11104-006-9000-z
Shen Y., Chen Y., Li S. (2016). Microbial functional diversity, biomass and activity as affected by soil surface mulching in a semiarid farmland. PloS One 11, e0159144. doi: 10.1371/journal.pone.0159144
Shi S., Richardson A. E., O’Callaghan M., DeAngelis K. M., Jones E. E., Stewart A., et al. (2011). Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 77, 600–610. doi: 10.1111/j.1574-6941.2011.01150.x
Singh B. K., Millard P., Whiteley A. S., Murrell J. C. (2004). Unravelling rhizosphere–microbial interactions: Opportunities and limitations. Trends Microbiol. 12, 386–393. doi: 10.1016/j.tim.2004.06.008
Steinauer K., Chatzinotas A., Eisenhauer N. (2016). Root exudate cocktails: The link between plant diversity and soil microorganisms? Ecol. Evol. 6, 7387–7396. doi: 10.1002/ece3.2454
Stewart-Wade S. M. (2020). Efficacy of organic amendments used in containerized plant production: Part 1 – Compost-based amendments. Scientia Hortic. 266, 108856. doi: 10.1016/j.scienta.2019.108856
Vaidya B. P., Hagmann D. F., Haramuniz J., Krumins J. A., Goodey N. M. (2022). Artificial root exudates restore microbial functioning in a metal contaminated, barren, inactive soil. Environ. Pollution 312, 120007. doi: 10.1016/j.envpol.2022.120007
Vandecasteele B., Pot S., Maenhout K., Delcour I., Vancampenhout K., Debode J. (2021). Acidification of composts versus woody management residues: Optimizing biological and chemical characteristics for a better fit in growing media. J. Environ. Manage. 277, 111444. doi: 10.1016/j.jenvman.2020.111444
Vieira S., Sikorski J., Dietz S., Herz K., Schrumpf M., Bruelheide H., et al. (2020). Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 14, 2. doi: 10.1038/s41396-019-0543-4
Viti C., Tatti E., Decorosi F., Lista E., Rea E., Tullio M., et al. (2010). Compost effect on plant growth-promoting rhizobacteria and mycorrhizal fungi population in maize cultivations. Compost Sci. Utilization 18, 273–281. doi: 10.1080/1065657X.2010.10736966
Voolstra C. R., Ziegler M. (2020). Adapting with microbial help: Microbiome flexibility facilitates rapid responses to environmental change. BioEssays 42, 2000004. doi: 10.1002/bies.202000004
Wagg C., Jansa J., Schmid B., van der Heijden M. G. A. (2011). Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol. Lett. 14, 1001–1009. doi: 10.1111/j.1461-0248.2011.01666.x
Waldrop M. P., Zak D. R., Blackwood C. B., Curtis C. D., Tilman D. (2006). Resource availability controls fungal diversity across a plant diversity gradient. Ecol. Lett. 9, 1127–1135. doi: 10.1111/j.1461-0248.2006.00965.x
Walker T. S., Bais H. P., Grotewold E., Vivanco J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Wan W., Wang Y., Tan J., Qin Y., Zuo W., Wu H., et al. (2020). Alkaline phosphatase-harboring bacterial community and multiple enzyme activity contribute to phosphorus transformation during vegetabl e waste and chicken manure composting. Bioresource Technol. 297, 122406. doi: 10.1016/j.biortech.2019.122406
Weidner S., Koller R., Latz E., Kowalchuk G., Bonkowski M., Scheu S., et al. (2015). Bacterial diversity amplifies nutrient-based plant–soil feedbacks. Funct. Ecol. 29, 1341–1349. doi: 10.1111/1365-2435.12445
Keywords: horticultural substrates, root exudates, Chrysanthemum, rhizosphere microbiome, horticultural plants
Citation: Pot S, De Tender C, Ceusters J, Debode J and Vancampenhout K (2025) Unraveling microbial shifts and plant growth responses in Chrysanthemum ‘Merlino’ due to artificial root exudates in horticultural substrates. Front. Hortic. 4:1562929. doi: 10.3389/fhort.2025.1562929
Received: 18 January 2025; Accepted: 31 March 2025;
Published: 28 April 2025.
Edited by:
Chris Blok, Wageningen University and Research, NetherlandsReviewed by:
Annelein Meisner, Wageningen University and Research, NetherlandsGabriele Bellotti, Catholic University of the Sacred Heart, Italy
Aditi Satpute, Texas A&M University Kingsville, United States
Copyright © 2025 Pot, De Tender, Ceusters, Debode and Vancampenhout. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caroline De Tender, Y2Fyb2xpbmUuZGV0ZW5kZXJAdWdlbnQuYmU=
 Steffi Pot1
Steffi Pot1 Caroline De Tender
Caroline De Tender Johan Ceusters
Johan Ceusters Jane Debode
Jane Debode Karen Vancampenhout
Karen Vancampenhout