- 1Center for Integrated Fungal Research, Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC, United States
- 2Department of Plant Pathology, The Ohio State University, Wooster, OH, United States
- 3Department of Plant and Environmental Sciences, Clemson University, Coastal Research and Education Center, Charleston, SC, United States
Over the past two decades, significant changes in the population structure of Pseudoperonospora cubensis have been reported worldwide. These changes have been associated with, among other things, severe epidemics of cucurbit downy mildew that are now much more destructive particularly on cucumber, than has previously been reported. Host specificity has complicated disease control as host resistance and fungicides that were previously effective in controlling the disease have become less effective. In response to this resurgence, significant research efforts have been made to better understand disease epidemiology, pathogen biology and host resistance, to generate information to improve disease management. Oospores have been reported under natural field settings in the United States, however, uncertainty remains regarding their role as a source of inoculum for initial disease outbreaks in northern latitudes that experience hard frost. Further, recent work indicates that the initial source of inoculum in the continental United States is southern Florida and along the edge of the Gulf of Mexico. Network analysis of disease outbreaks has identified key locations in the eastern United States that could be critical for disease monitoring in an effort to limit epidemic spread during the growing season. Lineage-specific biosurveillance of P. cubensis using spore traps complements existing disease monitoring efforts and is providing opportunities for precision management by determining cucurbit crops at risk of infection during the season. This review summarizes the substantial progress that has been made in understanding the biology of P. cubensis, disease epidemiology and control, which could inform better the management of cucurbit downy mildew.
1 Introduction
Cucurbit downy mildew (CDM) is a foliar disease caused by the oomycete Pseudoperonospora cubensis [(Berkeley and M. A. Curtis) Rostovzev, 1903)]. This foliar disease of cucurbits is widespread and has devastating impacts on cucurbit production. When poorly managed, the disease has been reported to cause significant yield losses annually in the United States, China, Israel, throughout Europe, and in other parts of the world (Mirzwa-Mróz et al., 2024; Lebeda and Cohen, 2011; Thomas, 1996). The disease has been reported in a range geographical environments, from temperate, semi-arid to the tropics, and in different production systems of open fields, managed tunnels, and greenhouses (Savory et al., 2011; Naegele et al., 2016; Spring et al., 2018; Egel et al., 2022). Cucurbit downy mildew affects the foliar portions of the plant, resulting in significant losses in fruit quality and yield among members of the Cucurbitaceae family that include cucumber (Cucumis sativa), cantaloupe (Cucumis melo), watermelon (Citrullus lanatus), pumpkin (Cucurbita maxima, C. pepo), winter and summer squash (Cucurbita moschata, C. maxima, C. pepo) (Savory et al., 2011; Palti and Cohen, 1980).
The resurgence of cucurbit downy mildew in the United States in 2004 was described as a unique phenomenon characterized by devastating epidemics, rapid spread, and failure of fungicide management programs, complete crop loss in several regions, and abandonment of cucumber fields prior to harvest (Holmes et al., 2015). This resurgence dramatically altered the production of cucurbits and disease management systems at multiple scales (Ojiambo et al., 2015). For nearly two decades, growers in the eastern United States have had to apply fungicides prophylactically at regular intervals to protect their crops from cucurbit downy mildew. Prior to the resurgence of the disease in the United States in 2004 and 2005, host resistance was the most effective disease management tool in cucumber production. While cucurbit downy mildew still occurred in the United States, management of the disease on cucumber crops did not require fungicide applications, and other cucurbit hosts such as squashes, were produced with minimal use of fungicides (Holmes et al., 2015). This article builds on previous reviews (Cohen et al., 2015; Holmes et al., 2015; Ojiambo et al., 2015; Lebeda and Cohen, 2011; Savory et al., 2011) and highlights some recent research on the biology of P. cubensis, disease epidemiology and control. The goal of this review is to focus on how the knowledge gained thereof has been applied to improve disease management. Where appropriate, we also point out how this recent knowledge provides opportunities for more improved management of cucurbit downy mildew.
2 Cucurbit downy mildew symptoms and signs
Under favorable environmental conditions and in the presence of a susceptible host tissue, P. cubensis can infect different cucurbit host types, and the reaction results in salient symptoms after infection. However, disease symptoms differ greatly among cucurbit host species (Figure 1). Typically, symptoms of cucurbit downy mildew are characterized by lesion development and form, color, and size of lesions, which varies depending on the cucurbit host species, cultivar, infected part of the host, and environmental conditions (Savory et al., 2011). Generally, symptom development in CDM-infected cucumber plants is characterized by the formation of angular, chlorotic lesions confined by leaf veins on the upper leaf surface (Figure 1A), with hyaline sporangiophores bearing lemon-shaped sporangia on the underside of the leaves. Under favorable weather conditions, the disease progresses rapidly, leading to coalescence of chlorotic lesions and subsequent necrosis, ultimately resulting in complete leaf curling and collapse.
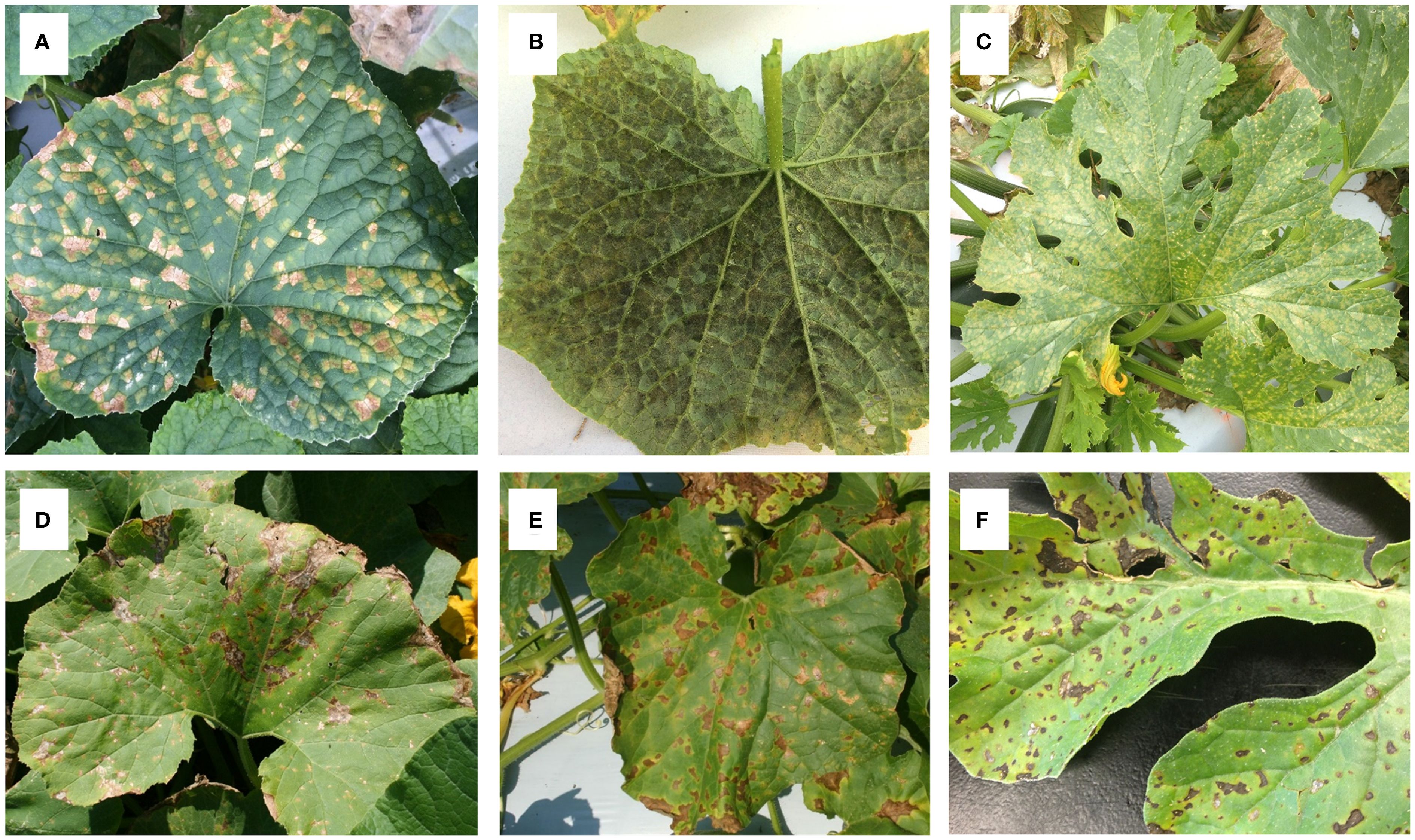
Figure 1. Symptoms of cucurbit downy mildew caused by Pseudoperonospora cubensis in species of Cucurbitaceae. (A) Chlorotic angular lesions on the adaxial side of cucumber leaf; (B) Chlorotic lesions covered with sporangia on the lower side of cucumber leaf; (C) Chlorotic spots on the upper side of summer squash; (D) Necrotic lesions on ‘Hubbard’ squash (Cucurbita maxima); (E) leaf spot symptoms on ‘Rocio’ honeydew; (F) Necrotic lesions on watermelon.
In other cucurbit species such as cantaloupe, watermelon, and squash, leaf spots may start as pale green then enlarge and turn brown (Figures 1C-F). While cucumber lesions are often confined by leaf veins and appear angular in shape, lesions on cantaloupe appear round to irregular in shape. Lesions on watermelon are circular, of varying size, and not restricted by leaf veins (Lebeda and Schwinn, 1994). As the disease progresses, lesions expand and coalesce, leading to affected cucumber, cantaloupe, and watermelon fields to look brown and “crispy”. Sporulation on the underside of the leaf is characterized by gray to deep violet-colored growth on the lower surface of leaves (Figure 1B), consisting of sporangia and sporangiophores of P. cubensis. Dense patches of sporangia are a key diagnostic feature of the disease. Sporangia are more prominent under high humidity conditions, particularly in the morning. Typically, sporangia production is very limited on watermelon compared to all other cultivated cucurbit host types.
3 Life and disease cycle
3.1 Asexual reproduction
Asexual spores are the main infective propagules of P. cubensis (Lebeda and Cohen, 2011). The role of the asexual phase (sporangia production and subsequent tissue infection) of P. cubensis in the epidemiology of cucurbit downy mildew has been well described (Ojiambo and Holmes, 2011). Below we highlight the most salient features of the disease cycle and where appropriate, link specific stages to the disease initiation, progress and spread in the field.
3.1.1 Primary inoculum
In the United States, the survival of P. cubensis depends on the presence of cucurbit hosts in the regions where production of cucurbits occurs year-round due to favorable warm winter conditions such as South Florida, the Caribbean, the Rio Grande Valley of Texas, Gulf of Mexico, and greenhouse production (Holmes et al., 2015; Nandwani et al., 2014; Nusbaum, 1944, 1945, 1948; Ojiambo and Holmes, 2011; Thomas, 1996). The primary source of inoculum is airborne sporangia, which are dispersed by wind from infected regions to fields with susceptible hosts in new locations (Ojiambo and Holmes, 2011; Savory et al., 2011).
3.1.2 Infection, colonization and sporulation
During spring when cucurbit hosts are present in northern latitudes, long distance, aerially dispersed sporangia land on host leaf surfaces where they undergo indirect germination in the presence of free moisture (Cohen and Eyal, 1977; Lebeda and Cohen, 2011). The pathogen forms hyaline, coenocytic, intercellular mycelium (Figure 2A) that colonizes the mesophyll and palisade tissues of its host. During asexual reproduction, sporangiophores (Figure 2B) emerge from stomata on the lower leaf surface, bearing dichotomously branched, hyaline structures that terminate in gray-violet, lemon-shaped sporangia (Figures 2C, E) that are characteristic signs of the pathogen (Choi et al., 2005; Palti, 1975). Sporangia are ovoid to ellipsoid, measuring approximately 17 - 39 µm in length and 11 - 24 µm in width (Choi et al., 2005; Runge et al., 2011). Sexual reproduction results in the formation of thick walled oospores (Figure 2D) that serve as resting structures for the pathogen (see details below).
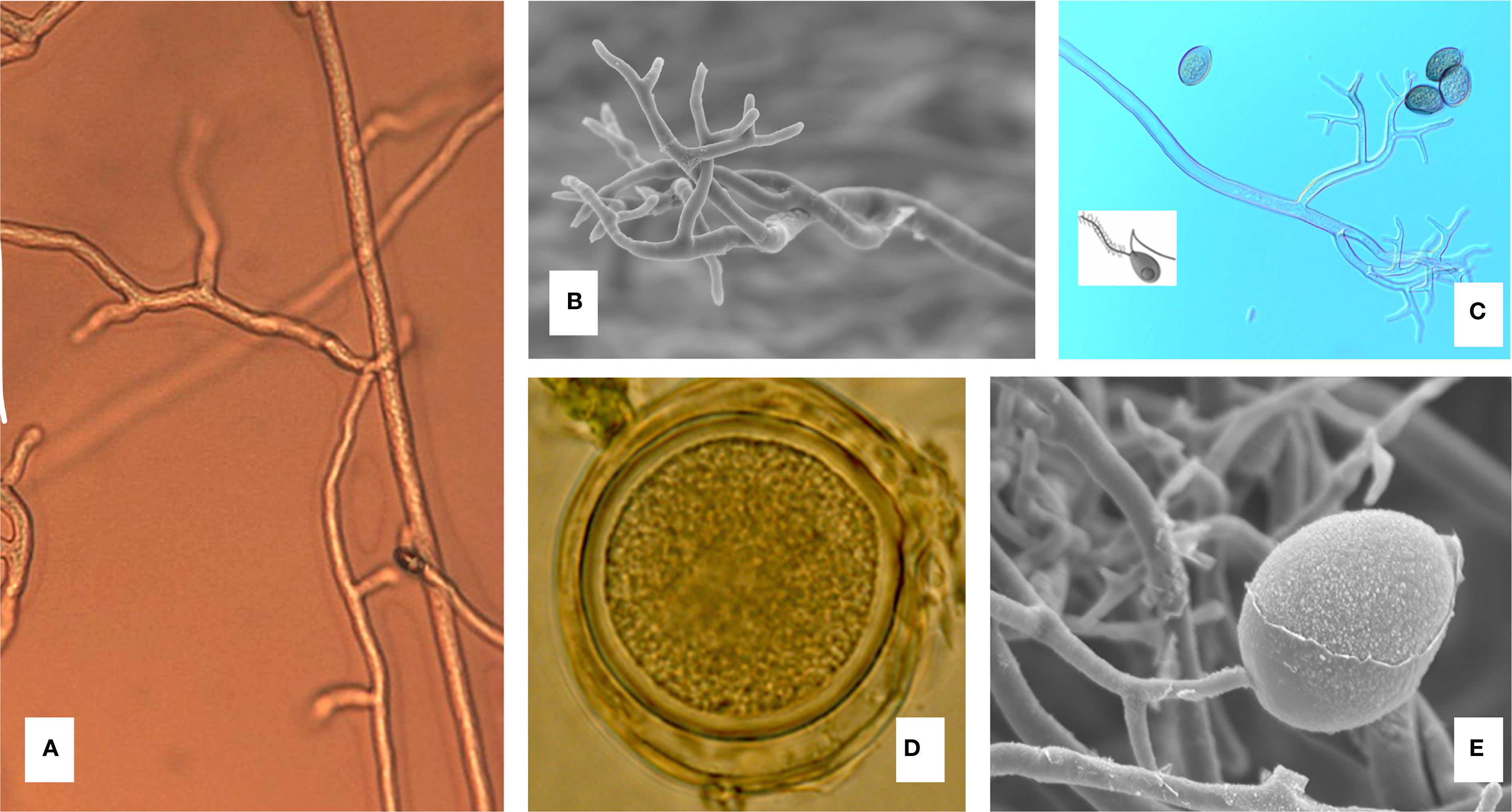
Figure 2. Morphology of Pseudoperonospora cubensis. (A) Coenocytic mycelium. (B) Sporangiophores. (C) Detached lemon shaped sporangium and biflagellate zoospores released by zoosporangium (inset). (D) Thick cell wall sexual and resting structure (oospore) viewed at ×1000 total magnification (×10 eyepiece and ×100 oil immersion objective). (E) Scanning electron micrograph of a sporangium attached to a sporangiophore at ×2000 magnification (Photo credit: Gerald J. Holmes, NC State University).
Under favorable conditions characterized by free moisture (irrigation water, rain, or dew) and an optimum temperature ranging from 15 to 22°C (Lebeda et al., 2011), sporangia (Figure 3A) deposited on leaves undergo cytoplasmic cleavage to release 5 to 15 biflagellate zoospores (Figure 3B) that swim chemotactically toward host stomata where they encyst (Cohen, 1981). After a germ tube emerges from a cystospore, an appressorium penetrates host tissue via a stoma (Lebeda and Cohen, 2011). Infection hyphae grow into the host intercellular space and colonize mesophyll tissue (Figure 3C). The pathogen establishes web-like structures known as haustoria, which play a crucial role in nutrient absorption and the delivery of effector proteins that suppress host defense mechanisms (Lange et al., 1989; Savory et al., 2011; Hahn and Mendgen, 2001; Whisson et al., 2007). Depending on environmental conditions, inoculum load, and host genotype (resistance/susceptibility), the incubation period from penetration and colonization until visible symptoms (Figure 3E) appear ranges from 4 to 12 days (Lebeda and Cohen, 2011). After 5 to 7 days post-infection, asexual reproduction is characterized by the emergence of branched sporangiophores bearing lemon-shaped sporangia at the tips (Figure 3D). These sporangia are easily dislodged from sporangiophores by air currents, diurnal changes in relative humidity, or rain splash. They serve as either primary or secondary inoculum locally or some distance away from the source as they are wind-borne and can be dispersed over long distances (Cohen et al., 2015; Lange et al., 1989; Granke and Hausbeck, 2011; Ojiambo and Holmes, 2011).
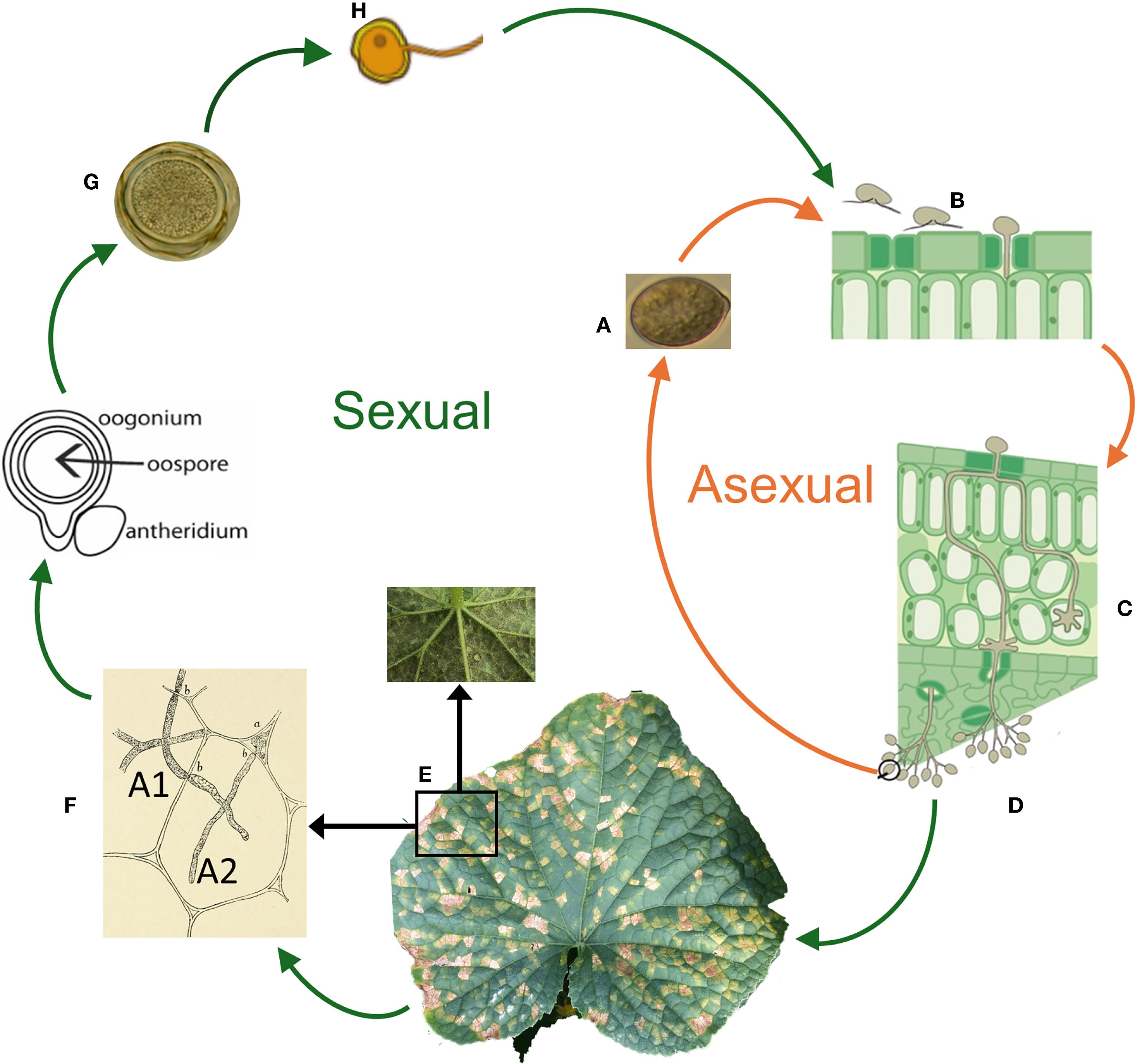
Figure 3. Life cycle of Pseudoperonospora cubensis. Asexual reproductive stage involves the production of sporangia, while sexual reproductive cycle involving mating of strains of opposite mating type, A1 and A2. (A) Aerially dispersed, lemon shaped, melanized, grey-purple sporangia land on leaf surface and germinate by cytoplasmic cleavage in free moisture to produce biflagellate motile zoospores; (B) Motile zoospores swim and encyst in stomata, then penetrate leaf surface via germ tube; (C) Coenocytic hyphae colonize the mesophyll layer, then establish clavate-branched haustoria within plant cells for nutrient absorption and host-pathogen interaction; (D) Optimum environmental conditions triggers sporulation where sporangiophores bearing lemon-shaped sporangia at the tips emerge through each stomata. Dislodged sporangia from sporangiophores may result in local and long distance dispersal by wind currents that carry them to their next host; (E) Chlorotic, angular lesions bound by leaf veins (cucumber) are classical symptoms of P. cubensis infection that are visible on the adaxial leaf surface. Visible grey-purple sporulation on the lower leaf surface are signs of P. cubensis (inset); (F) Sexual reproduction occurs in the presence of heterothallic A1 and A2 mating types resulting in formation of oogonium and antheridium and formation of an oospore (G); (H) Germination, infectivity and the role of the sexual stage in the disease cycle is not well established.
3.1.3 Disease progress and secondary infections
Sporangia are dispersed by air currents, initiating secondary cycles of infection (Ojiambo and Holmes, 2011; Ojiambo et al., 2011). The polycyclic nature of cucurbit downy mildew allows for multiple infection cycles per season, leading to a rapid increase in disease under conducive conditions. Environmental factors such as rain, wind, and anthropogenic activities contribute to dispersal of sporangia, while prolonged leaf wetness (≥ 6 h) facilitates new infections (Cohen and Eyal, 1977). Within a growing season, successive generations of P. cubensis sporangia produced and aerially disseminated over long distances from the southern-most states (Florida and Texas) to northern states (e.g., Massachusetts, Wisconsin) in the continental United States (Ojiambo and Holmes, 2011).
3.2 Sexual reproduction and overwintering
In oomycetes, mating systems vary considerably even within same species due to various factors. Pseudoperonospora cubensis exhibits heterothallism, where the sexual process occurs only when mycelia of opposite, compatible mating types interact, leading to the formation of oospores (Cohen and Rubin, 2012). Oomycete species such as Sclerospora sorghi, Plasmopara halstedii, and Pseudoperonospora humuli are homothallic, while Pseudoperonospora cubensis, Phytophthora infestans, Plasmopara viticola, and Bremia lactucae are heterothallic (Gent et al., 2017; Spring et al., 2018; Kitner et al., 2021). Laboratory studies conducted in Israel and the United States have confirmed that P. cubensis is heterothallic with both A1 and A2 mating types (Figure 3F) coexisting under natural field conditions. When sporangia of opposite mating types are co-inoculated on detached cucumber or cantaloupe leaves and incubated at 15°C or 21°C, hyaline to golden yellow, spherical, thick-walled oospores (Figures 3G, H) develop within the mesophyll in 6 – 11 days (Thomas et al., 2017a). Oospore production depends on the isolate pairing, host type, and environmental conditions. Reports of oospore production in P. cubensis in the field date back to Russia (Rostovzev, 1903), Japan (Hiura and Kawada, 1933), North China (Chen et al., 1959), and later under greenhouse conditions in China (Zhang et al., 2012). Oospores are resting structures and are mechanism that generates genetic diversity within the pathogen population allowing the pathogen to survive unfavorable environmental conditions (McDonald and Linde, 2002).
Formation of oospores in P. cubensis is considered rare, and reports of oospore formation under field conditions have previously been limited to Asia and Europe (Cohen, 2015). Recently, oospores were reported in leaves of naturally infected Cucumis spp. (Figures 4A, D) with typical disease symptoms late in the season in the United States (Kikway et al., 2022a). These oospores are characterized by two thick layers with an outer smooth surface and an intact cytoplasm (Figures 4B, C). Typically, oospores are spherical and hyaline to golden yellow or light brown (Cohen and Rubin, 2012; Cohen et al., 2015; Thomas et al., 2017a; Kikway et al., 2022a; Rani et al., 2022). Viability and infectivity of oospores has been established in P. cubensis (Cohen and Rubin, 2012; Kikway et al., 2022a). Oospore viability was determined based on the integrity of the cytoplasm. When exposed to osmotic stress, viable oospores show a contracted (i.e., plasmolyzed) cytoplasm that is detached from the wall due to loss of cell wall membrane integrity (Kikway et al., 2022a) when treated with a hypertonic solution, while the cytoplasm of non-viable oospores remains intact with no loss of differential permeability of the cellular membrane.
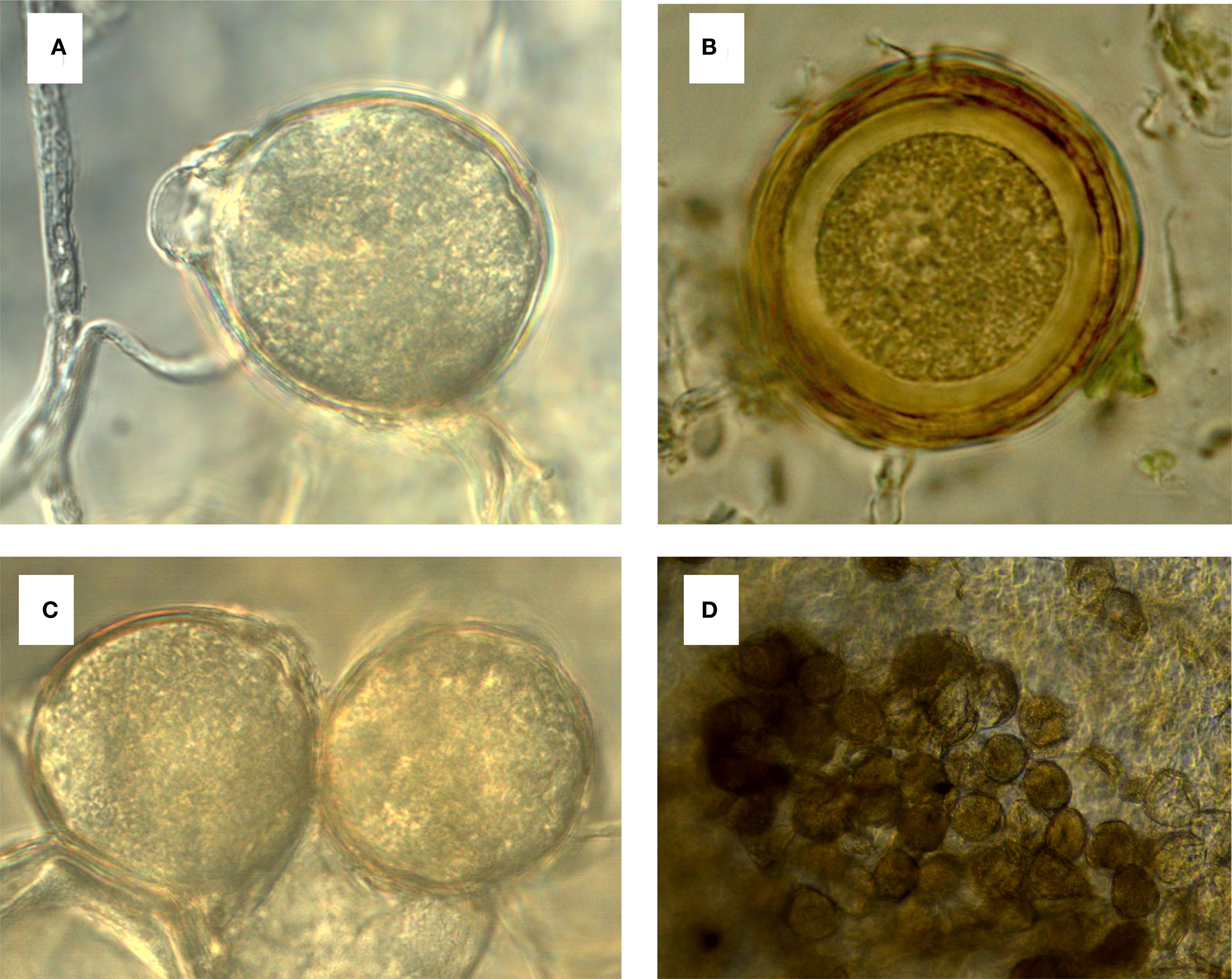
Figure 4. Oospores of Pseudoperonospora cubensis observed in clarified tissue of naturally infected leaves of cucumber cultivar ‘Straight Eight’. (A) Spherical hyaline to golden yellow intact oospore (×1000 magnification); (B) A close-up of the oospore showing the typical two thick layers and intact cytoplasm (×1000 magnification); (C) A pair of spherical hyaline to golden yellow oospores (×1000 magnification); (D) A mass of oospores embedded in a clarified leaf tissue (×400 magnification).
Oospores enable the pathogen to persist between seasons in soil, plant debris, fruits, and seeds of infected hosts, potentially acting as primary inoculum in regions where host plants cannot survive harsh winter conditions (Lebeda and Cohen, 2011; Zhang et al., 2012). Given their resilience, oospores are of significant concern for disease management, as they can persist in the environment for extended periods, often exceeding 10 months, allowing the pathogen to persist in the absence of living host plants (Zhang et al., 2012; Rani et al., 2022; Kikway et al., 2022a). Over a six-month winter exposure in outdoor conditions, the viability of P. cubensis oospores declined from 68% in November to about 20% by April-May, the following year, a period that coincides with the start of cucurbit planting in North Carolina. While the recovered oospores were considered viable and potentially infective, they remained dormant and failed to germinate and induce infection when inoculated on cucumber or cantaloupe leaves (Kikway et al., 2022a). Instances of successful germination and infectivity of oospores of P. cubensis are still very rare, with the frequency of oosporic infection being less than 0.2% (Cohen and Rubin, 2012). However, oosporic inoculum appears to be epidemiologically important for cucurbit downy mildew in China (Zhang et al., 2012), where inoculation of cucumber seedlings in the greenhouse with oospore suspension resulted in infection. Due to the low frequency of infections due to oospores as reported in Israel, and the unsuccessful germination of oospores and resultant infection reported in the United States, further studies are needed to better understand factors that influence germination of P. cubensis oospores, the potential role of oospores as an initial source of primary inoculum as well as their impact on the disease epidemiology and management.
4 Ecology, epidemiology and annual disease epidemics
The disease and life cycles of P. cubensis, along with symptom expression and the dynamics of annual epidemics, are greatly influenced by environmental and ecological conditions. Understanding these factors is thus essential for predicting disease outbreaks and effective implementation of integrated management strategies. In this review, we focus on the key ecological factors that greatly impact the epidemiology of P. cubensis and management of the disease.
4.1 Source of primary inoculum and dispersal
The primary source of inoculum and means of dispersal for P. cubensis is its sporangia that serve as the main infective propagules of the pathogen. In the United States, P. cubensis overwinters in the form of sporangia or mycelia on wild hosts in nature or open production fields in subtropical and tropical regions such as Southern Florida, the Caribbean, Texas, and Mexico (Bains and Jhooty, 1976; Ojiambo et al., 2015; Savory et al., 2011; Wallace et al., 2015). Sporulation of P. cubensis has also been reported on wild hosts like Momordica balsamina, M. charantia, and Cucurbita foetidissima that were collected from natural fields. This suggests that these wild hosts may act as a reservoir for the pathogen during the season and between growing seasons (Wallace et al., 2014, 2015, 2020). Wind is the primary driver of sporangia dispersal over long distances. Sporangia are dispersed by wind currents from southern regions to northern latitudes in the United States (Neufeld et al., 2013; Savory et al., 2011), where disease occurrence follows a regular annual pattern of spread along the eastern seaboard region due to south to north dispersal of sporangia (Ojiambo et al., 2015). However, up to 5% of disease outbreaks in the northern tier has not been associated with the predicted pattern of long-distance dispersal. These atypical outbreaks could be caused by alternative sources of primary inoculum, such as overwintering inoculum in the year-round production of cucurbits in greenhouses in areas surrounding Michigan, and dispersal of sporangia from cucurbit producing regions in Canada from greenhouse systems (Ojiambo et al., 2011, 2015). Additionally, P. cubensis has been demonstrated to be seed-borne and seed-transmission via hyphae and sporangia in the seed coat or embryonic tissue (such as cotyledon or embryo axis) of cucumber, melon, squash, and pumpkin, can provide possible pathways for early-season inoculum introduction (Cohen et al., 2014; Spring et al., 2018).
In the Great Lakes, mid-Atlantic and northern tier regions, cucumbers are produced year-around as protected winter crops in greenhouses. Greenhouses can be important reservoirs of P. cubensis pathogen as a ‘green-bridge’ for over-wintering, and thus protected cultures of cucurbit plants were highly suspected as sources of downy mildew primary inoculum and a source of early epidemics for field-grown crops grown early in spring and summer (Lebeda and Cohen, 2011; Naegele et al., 2016; Ojiambo et al., 2015; Savory et al., 2011). Indeed, there has been speculation and circumstantial evidence that inoculum sources outside of Florida may be important for the disease in more northern latitudes in the United States (Ojiambo et al., 2015; Cohen et al., 2015). However, a recent study did not find any evidence of an annually occurring alternate source of P. cubensis in northern latitudes but rather found support for an alternate inoculum source on the western edge of the Gulf of Mexico (Ojwang’ et al., 2021). Thus, inoculum sources in the southern United States outside of Florida may be responsible for disease outbreaks in more northern latitudes as had been previously reported by Ojiambo and Holmes (2011). In addition, Naegele et al. (2016) found genetic differentiation between isolates of P. cubensis from field-grown cucumbers in Michigan and various field-grown cucurbits in Ontario, although greenhouse-grown cucumbers from Ontario were not sampled in that study.
Studies on sexual reproduction in P. cubensis around the world have shown the pathogen is capable of overwintering via oospores in the greenhouse or in production fields (Cohen et al., 2015; Lebeda and Cohen, 2011; Kikway et al., 2022a; Rani et al., 2022; Zhang et al., 2012). In the United States, P. cubensis oospores were found in the 2022 growing season in naturally infected cucurbit research plots in North Carolina and South Carolina (Kikway et al., 2022a). However, the role of P. cubensis oospores as primary inoculum for subsequent disease epidemics is still not well established, although oospore inoculum appears to be epidemiologically important for cucurbit downy mildew in other regions (Zhang et al., 2012).
4.2 Temperature and humidity
Environmental conditions such as temperature and humidity favor infection, colonization, reproduction, and the release, as well as dissemination of inoculum of P. cubensis (Cohen and Eyal, 1977; Cohen and Rotem, 1971; Sun et al., 2017). Production of sporangia depends on diurnal temperature, relative humidity and light. The temperatures for sporangial germination and infection range between 5 - 25°C with an optimum infection temperature of about 19°C (Lebeda and Cohen, 2011; Sun et al., 2017). Temperatures lower than 5°C will require a longer period of leaf wetness for successful infection (Cohen and Eyal, 1977). Dry and hot temperatures >35°C may suppress disease development by limiting sporulation (Sun et al., 2017). However, high relative humidity (i.e., > 90%) and frequent rainfall or dew promote sporulation, infection and pathogen dispersal. Sporulation of P. cubensis can occur at temperatures ranging from 5 to 30°C, with the reported optimum temperature range being between 15 - 25°C (Lebeda and Cohen, 2011; Sun et al., 2017). Rainfall increases humidity and duration of leaf wetness which favor host infection. According to Lebeda and Cohen (2011), the lifespan for sporangia after dislodging from the sporangiophore does not exceed 48 h and a minimal wetting period of 2 h is required for sporangia to infect leaves. Dew and guttation droplets on the host leaves can promote sporangia germination (Lebeda and Cohen, 2011). While light promotes sporulation, a minimum of 6 hours of darkness is required for sporangia to undergo differentiation (Cohen and Eyal, 1977). Hygroscopic twisting of sporangiophores that allows sporangia to dislodge and be released into air currents is favored by declining relative humidity late in the morning (Granke and Hausbeck, 2011; Lange et al., 1989). Peak concentration of sporangia in the atmosphere has been reported to coincide with this time period regardless of the level of disease severity (Neufeld et al., 2013). Sporangia are sensitive to ultraviolet light and exposure to sunlight determines the survival during aerial dispersal and the ability to germinate and cause disease thereafter upon landing on a host following a rain event. Solar radiation of 8.9 MJ m−2 for 4 – 5 h reduced sporangial germination by 50%, and lengthening of the exposure period to 9 h reduced germination by 80% (Kanetis et al., 2010). On sunny days, the effective cumulative solar radiation dose to inactivate 95% of the sporangia is about 29.5 MJ/m2 (Kanetis et al., 2010).
5 Diversity, mating types, pathotypes and races
Pseudoperonospora cubensis is a highly diverse pathogen with several pathotypes documented in the United States, Israel, Czech Republic, and Kazakhstan (Lebeda et al., 2013, 2024; Rsaliyev et al., 2018; Thomas et al., 2017b). This pathogen is highly aggressive and has demonstrated a high degree of host adaptation that has enabled it to increase its host range by infecting a wide range of cultivated and wild plants of the Cucurbitaceae family. Studies conducted in Israel, China, and the United States have shown the existence of A1 and A2 mating types (Cohen and Rubin, 2012; Thomas et al., 2017a) with a distinct association of mating type with specific cucurbit host types.
In their study on P. cubensis host specialization, Thomas et al. (2017a, b) found that the A1 mating type was primarily associated with Cucumis spp. (cucumber and melon) while the A2 mating type was primarily associated with Cucurbita spp. (squash, pumpkin, or butternut squash). Wallace et al. (2020) and Shirley et al. (2024) conducted population analyses revealing that cucurbit host species play a key role in shaping pathogen populations in North Carolina and Florida, respectively. Their findings indicate that lineage I/A2 mating type isolates preferentially infect cultivated Cucurbita pepo, C. maxima, and Citrullus lanatus and wild Momordica charantia and M. balsamina. In contrast, lineage II/A1 mating type isolates show preference for cultivated species such as Cucumis sativus and C. melo and Lagenaria siceraria. Lineage I also was found on slicing cucumber in Florida (Shirley et al., 2024), which indicates that Lineage I isolates have a broader host range than Lineage II isolates. As a confirmation of this association between mating type and host, all 106 isolates of P. cubensis collected from Cucumis melo in the eastern United States displayed an inverse relationship between lineage and mating type, with 88 isolates in lineage II/mating type A1 and the remaining 18 isolates in lineage I/mating type A2 (Toporek and Keinath, 2021). Out of nine locations sampled in two years, Charleston, South Carolina, United States, was the only location where both mating types were recovered. This observation matches the finding of oospores by Kikway et al. (2022a) in field-grown cantaloupe at this location.
The pathogenic variability of P. cubensis populations has been investigated in several countries around the globe and the pathogen has a complex virulence structure (Thomas et al., 1987, 2017b; Cohen, 2015; Lebeda et al., 2013, 2018). The virulence level is much higher in the Czech Republic (Lebeda et al., 2013) compared to the United States (Thomas et al., 2017b). For example, the mean virulence factors per isolate in the United States population was 5.05 (Thomas et al., 2017b) compared to 9.0 in the European population (Lebeda et al., 2013). Pathotypes are also associated with the mating type of the pathogen. For example, pathotypes 1 and 3 in the United States population are associated with A1, while pathotypes 4, 5 and 6 isolates are of the A2 mating type (Thomas et al., 2017b). A similar observation was reported for the European population (Kitner et al., 2015; Lebeda et al., 2014). Further, European pathotypes (Lebeda et al., 2006, 2011, 2013) differ significantly from the five pathotypes of P. cubensis previously detected in Japan, Israel, and the United States (Thomas et al., 1987; Cohen et al., 2003). These differences are likely due to the limited diversity of virulent isolates affecting C. lanatus in the United States, where infection and sporangia production is very limited on C. lanatus. Upon infection, oospores produce recombinant offspring isolates, and such offspring may differ from their parents in host range and fungicide sensitivity (Cohen and Rubin, 2012). These offspring may also have enhanced virulence towards resistant host genotypes as has been the case with P. infestans in tomato and potato (Klarfeld et al., 2009). Thus, an integrated approach involving a combination of timely application of fungicides, rotation of fungicides with different modes of action, and introduction of cultivars with new resistance genes is required for effective management of cucurbit downy mildew.
Until recently, pathotypes were used to differentiate and characterize virulence of P. cubensis (Lebeda et al., 2024). However, recent study conducted in the Czech Republic introduced the use of races for virulence differentiation. In that study, Lebeda et al. (2024) identified pathotypes and races as two key concepts of virulence differentiation in P. cubensis. Pathotypes were defined by the interactions between pathogen isolates and a range of host genotypes representing different species or genera within the host plant family, Cucurbitaceae. In contrast, races were characterized by their interaction with genotypes within host species, such as cultivars, breeding lines, or accession (Lebeda et al., 2024). Clarification of races of P. cubensis could help resolve the inconsistent genetic patterns of CDM that has been observed in cucumber and melon (Ding et al., 2024). Further, clarification of races can lead to identification of downy mildew resistance genes and could help breeding efforts in cucurbit crops as reported for cucumber (Chen et al., 2020).
6 Disease control and management
Management of plant diseases involves three key strategies namely; cultural, biological and chemical control (Adeniyi et al., 2021). However, while different management strategies exist, chemical control remains the most reliable management method for cucurbit downy mildew due to its high efficacy relative to other methods.
6.1 Cultural practices
6.1.1 Early planting
The annual northward movement of P. cubensis sporangia in the eastern United States allows growers in states north of Florida and Texas, the opportunity to plant and potentially harvest cucurbits, particularly early maturing cucumber and summer squash, before downy mildew affects the crop (Ojiambo and Holmes, 2011; Ojiambo et al., 2011). The benefits of early planting were demonstrated on cucumber planted on 24 March, 15 April, and 12 May 2021 in Charleston, South Carolina, United States. Disease symptoms were detected on 25 May, which was after the first three harvests of the early planted crop and just before the first harvest for the middle planting date (Keinath et al., 2021). At early planting, marketable yield and disease severity (relative area under the disease progress curve) did not differ (P > 0.05) between fungicide-treated and non-treated plots. At the late planting, marketable yield was greater and disease severity was lower in fungicide-treated plots than non-treated plots, as expected. Planting early and not spraying the crop for CDM is potentially risky, as sporangia of P. cubensis were detected consistently before planting in multiple locations and years in Michigan (Granke et al., 2014). Early planting can be combined with fungicide application at a reduced frequency to sustainably manage CDM on cucumber and other cucurbits. Early planting is particularly important for organic production, as effective CDM biofungicides are limited (Marine et al., 2016; Uebbing et al., 2024).
6.1.2 Resistant varieties
P. cubensis exhibits considerable variability in pathogenicity within Cucurbitaceae family, with significant spatio-temporal fluctuations driven by pathotype and race specificity, which affect not only the effectiveness of resistance breeding but also the stability of resistance over time (Lebeda et al., 2016a, 2016b, 2017, 2019). Advances in molecular breeding, such as marker-assisted selection (MAS), genomic selection (GS), and CRISPR/Cas9, have accelerated the development of resistant cultivars (Mirzwa-Mróz et al., 2024). Further, a large number of quantitative trait loci (QTLs) associated with CDM resistance have been identified, including dm1.1, dm2.1, dm4.1, dm4.1.2, dm4.1.3, dmG2.1 and dmG7.1 (Berg et al., 2020; Win et al., 2017). Among the identified QTLs that has been tested, dm1.1 has the most substantial effect on disease resistance (Yoshioka et al., 2014). Further, a combination of bulked segregant analysis (BSA) and next-generation sequencing (NGS) has identified five major QTLs (dm2.2, dm4.1, dm5.1, dm5.2, and dm6.1), with dm2.2 showing the most significant effect (Win et al., 2017). The integration of molecular breeding techniques with traditional breeding approaches is now providing opportunities to accelerate the development of cucumber varieties that are resistant to CDM.
For decades, CDM was managed with minimal applications of fungicides, especially on cucumber. However, disease resurgence around the globe due to new strains that overcame previously resistant genes in cucumber and other host types, leading to a substantial increase in fungicide use. In the United States, pickling and slicing cucumber varieties with partial resistance can be included in disease management programs (Brzozowski et al., 2016; Call et al., 2013; Keinath, 2019). To avoid disease damage and yield loss, it is advisable to select cucumber varieties with resistance to downy mildew (Kemble et al., 2025). Compared to susceptible host types, a partially resistant host types hinder infection, colonization, or reproduction. This characteristic is manifested as a slower rate of disease development on partially resistant hosts than on susceptible hosts (Kikway et al., 2022b). Host tolerance allows production of marketable fruit even after downy mildew infections. Selection of host type can be important in disease management. Summer squash, zucchini, and acorn squash (horticultural types of Cucurbita pepo) have been found not only to tolerate but also to reduce the rate of disease development as compared to cucumber (Kikway et al., 2022b; Call et al., 2013). The inclusion of moderately resistant or tolerant host cultivars in the management program is important. Such cultivars do not need to be sprayed as often with high-risk single site fungicides, and protectant fungicides might be sufficient to prevent yield loss (Holmes et al., 2006). Further, the number of sprays per season is reduced and this may slow down the potential of P. cubensis to develop resistance to currently effective chemistries.
6.2 Disease forecasting and biosurveillance
Globally, plant diseases pose a significant threat to food security and food safety. Thus, the need for timely disease management is paramount to curb yield losses and fulfil the ever-increasing food demand (Fenu and Malloci, 2021). While the risk for disease outbreaks varies based on environment, time and space, decisions on disease management techniques that utilize preventative measures depend heavily on early detection of disease and effective forecasting. Forecasting aims to predict the probability of disease outbreak, giving growers a decision platform for effective management of disease in near-real time. Prediction of disease outbreaks or an increase in intensity and the severity of disease epidemics may allow decision making on an appropriate and effective management strategy (Charaya et al., 2021; Keinath et al., 2007).
For cucurbit downy mildew, annual epidemics in the continental United States are associated with long distance dispersal of inoculum from overwintering sources in the south (Ojiambo et al., 2009). Thus, early detection of inoculum arrival from overwintering regions is key to effective management of cucurbit downy mildew. The timing of the first fungicide spray is key in limiting the northward advance of disease epidemics during the growing season (Ojiambo et al., 2015). To this end, an integrated aerobiological modeling system (Neufeld et al., 2018a) was developed to predict the risk of disease occurrence and to facilitate timely use of fungicides for disease management as part of the CDM ipmPIPE program (Ojiambo et al., 2011). The forecasting system combined information on known inoculum sources (from sentinel and non-sentinel plots), long-distance atmospheric spore transport and spore deposition. The system was designed to provide cucurbit growers with valuable information that alerted them to increased risks of disease outbreaks and thereby help to improve the timing of the initial fungicide spray. The potential benefits of the forecasting system and making timely fungicide applications include not only a reduction of disease epidemics but also lower input costs, reduced negative impacts of fungicide use on the environment, and reduced risk for resistance development in the pathogen population. In a 2021 survey of CDM ipmPIPE users, over 84% of grower and industry respondents used website resources to make fungicide application decisions with an estimated median savings of $6,500 to $10,000 (Gugino et al., 2022). Like with many IPM related tools, the CDM ipmPIPE has faced, in the recent past, financial challenges and the ability to continue providing alerts to growers due to a reduction in disease monitoring via the sentinel plot network in the eastern United States, and a lack of funding to maintain web resources and computer programmers who consolidate and package model outputs and disease risk information in a form that stakeholders can easily utilize to make decisions on fungicide application.
Based on Granke and Hausbeck (2011), it is recommended that a timely fungicide application should be initiated when airborne sporangia are detected in the production regions and the environmental conditions favor cucurbit downy mildew. A network of spore traps, scouting of cucurbit fields and monitoring environmental conditions such as temperature and relative humidity are important sources of timely inputs for effective forecasting (Bello et al., 2022). While the latter is useful, relating sporangia availability to actual disease onset in subsequent days continues to be a challenge. Following the initial spray, growers typically apply subsequent sprays prophylactically on a calendar-based approach regardless of the disease risk during the growing season. While such an approach can maximize the effectiveness of disease control, it negates the benefits of forecasting. Models to guide fungicide applications based on disease risk during the season have been developed (Neufeld et al., 2017, 2018b) to maximize the effectiveness of fungicide applications made during the season.
Isolates of P. cubensis exhibit host-specialization, with lineage II isolates preferentially infecting cucumber and cantaloupe, while lineage I isolates primarily infecting squash, pumpkin, watermelon and wild cucurbit hosts (Thomas et al., 2017b; Wallace et al., 2020). Genomic evidence suggests that P. cubensis isolates display host adaptation based on their respective lineages and as such, mitochondrial genomic markers have been developed to detect and quantify P. cubensis sporangia (Rahman et al., 2017, 2021; Mandal et al., 2025). Deployment of spore sampling systems and analysis of samplers using these lineage-specific markers can complement the CDM ipmPIPE and further improve disease prediction during the season (Rahman et al., 2021). Further, combining information on the inoculum level with the risk of infection can also facilitate fungicide applications only to at-risk crops and thus reduce potential economic losses and prolong the efficacy of currently effective fungicides against cucurbit downy mildew.
6.3 Chemical control and fungicide resistance
Fungicide application affects specific components of the P. cubensis life cycle by reducing the reproduction efficiency and the proportion of pathogen inoculum available for infection, increasing the latent period, which is the time from infection to spore production, and reducing the infectious period by reducing the rate of sporulation (van den Bosch et al., 2015a; Hobbelen et al., 2011a, b). Chemical control of cucurbit downy mildew relies heavily on frequent fungicide applications, often applied on a weekly or biweekly basis (Holmes et al., 2015; D’Arcangelo et al., 2021). In the United States, the annual cost of fungicide applications for cucurbit downy mildew control exceeds $100 million (Holmes et al., 2015). Similar fungicide expenses can be expected in Europe, where the disease is the most destructive on cucumber in Central and Northern Europe leading to losses of up to 80% (Lebeda and Schwinn, 1994; Lebeda and Urban, 2004). The disease still remains a persistent threat in the eastern United States, where high humidity and moderate temperatures favor for disease development (Keinath et al., 2019). The ongoing challenge of fungicide resistance underscores the need for an integrated approach to CDM management that combines host resistance, cultural practices, disease monitoring and forecasting for effective disease management (Lebeda and Cohen, 2012; Ojiambo et al., 2015; Urban and Lebeda, 2006).
Management of CDM requires multiple timely fungicide applications on a 7-day schedule to limit the severity of the disease to an economic threshold. In 2022, a majority of the cucumber (85%), pumpkin (71%), and watermelon (62%) acreage in the United States was treated with fungicides (Table 1) (U.S. Department of Agriculture, National Agricultural Statistics Service, 2023a, 2025). Of the 32 fungicide active ingredients applied in 2020, 15 (47%) are effective against downy mildew, if all copper formulations are counted as one active ingredient (U.S. Department of Agriculture, National Agricultural Statistics Service, 2021). Fungicides to control cucurbit downy mildew are applied in rotations of two or three products to conform to fungicide residue tolerance limits on harvested fruit set by the U.S. Environmental Protection Agency and to conform to fungicide label restrictions on the amount and frequency of use to reduce the risk of fungicide resistance.
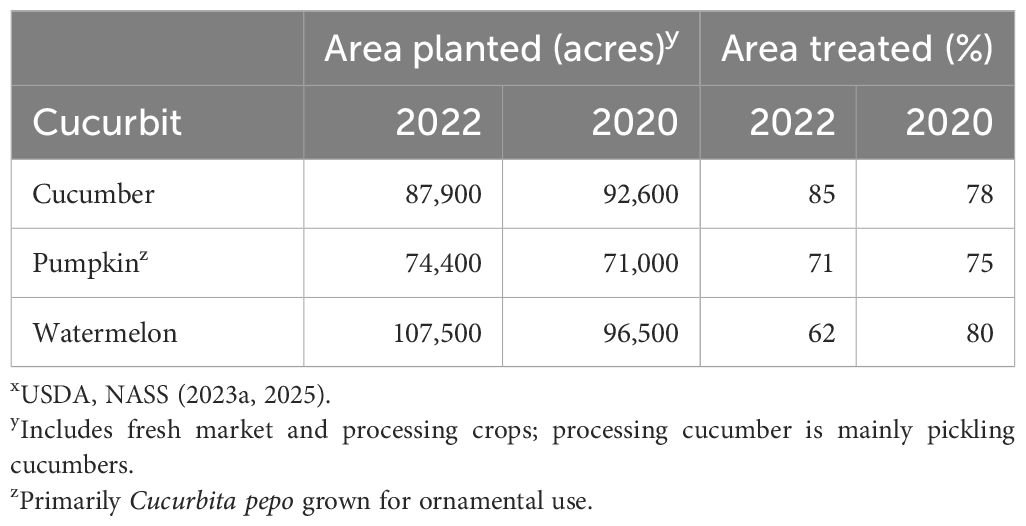
Table 1. Acreage and area treated with fungicides for major cucurbits produced in the United Statesx.
Over the past 10 years, reduced efficacy of several fungicides has been observed in the eastern United States in the field or confirmed in laboratory bioassays (D’Arcangelo et al., 2021; Goldenhar and Hausbeck, 2019; Jones et al., 2021; Keinath, 2023; Keinath et al., 2019; Keinath and Silva, 2022; Thomas et al., 2018). Reduced sensitivity of P. cubensis to fungicides in FRAC groups 4 (mefenoxam), 11 (quinone outside inhibitors, QoIs), 28 (propamocarb), 40 (carboxylic acid amides, CAAs), and 43 (fluopicolide), has been reported in the United States. While the mechanisms of resistance to mefenoxam, fluopicolide, and propamocarb are not yet fully understood, resistances to QoI and CAA fungicides are associated with specific single nucleotide polymorphisms in the cytochrome b (cytb) and cellulose synthase 3 (CesA3) genes, respectively. In most Peronosporales, including P. cubensis and Plasmopara viticola, QoI resistance involves a G143A mutation in the cytb gene, where glycine is replaced by alanine at position 143 (Gisi et al., 2002; Sharma et al., 2025). CAA fungicides inhibit cellulose biosynthesis by targeting the CesA3 gene. Resistance-conferring mutations in CesA3 include substitutions at position 1105 from glycine to alanine, serine, valine, or tryptophan (G1105A/S/V/W), and at position 1109 from valine to leucine or methionine (V1109L/M) (Blum et al., 2011; D'Arcangelo et al., 2023).
Due to these reports, growers have focused more and more on a few highly effective fungicides, notably cyazofamid (Ranman) and oxathiapiprolin products (Orondis) (Toporek and Keinath, 2022; U.S. Department of Agriculture, National Agricultural Statistics Service, 2021, 2023b). Oxathiapiprolin, an oxysterol binding protein homologue inhibitor with FRAC Code 49, is classified as having a medium to high risk of resistance (FRAC, 2022). Cyazofamid, a quinone-inside inhibitor with FRAC Code 21, also has a medium to high risk of resistance. In the US, oxathiapiprolin is marketed only as a pre-mix with mandipropamid (Orondis Ultra) or chlorothalonil (Orondis Opti), which are registered for application on all cucurbits; however, US and Canadian vegetable pathologists have discouraged use of Orondis Ultra, because lineage 2 of P. cubensis is resistant to mandipropamid (Goldenhar and Hausbeck, 2019; Jones et al., 2021; Keinath et al., 2019; Olaya et al., 2009).
Reduced efficacy of oxathiapiprolin was detected in Charleston, SC, United States, in the fall 2020, spring 2021, and fall 2021 populations of P. cubensis. Linear regression estimated the EC50 values to be 5.6, 4.5, and 13.9 mg/L oxathiapiprolin a.i. for lineage II isolates from cucumber (Keinath, 2022). Lineage I isolates from butternut squash and watermelon were more sensitive than lineage II isolates from cucumber but significantly less sensitive than baseline isolates (Cohen, 2015). Reduced efficacy of oxathiapiprolin applied without a mixing partner was also observed on cucumber in research plots in Georgia in fall 2020 (Dutta, 2021). In addition, a gene associated with insensitivity to oxathiapiprolin was detected recently in isolates of P. cubensis collected in North Carolina (Prieto Torres and Quesada-Ocampo, 2024).
Fungicide use data collected by the USDA National Agricultural Statistics Service (NASS) for the 2020 and 2022 growing seasons were analyzed to compare reported use patterns on cucumber (slicing and pickling combined), pumpkin (primarily Cucurbita pepo), and watermelon, the three largest acreage cucurbits grown in the United States (Table 1). Percentage area treated was statistically greater on cucumber (P < 0.001) and pumpkin (P < 0.05) than on watermelon for four of five fungicides used to manage cucurbit downy mildew: cyazofamid, oxathiapiprolin, propamocarb, and chlorothalonil (Figure 5). The percentage of area treated corresponds to the relative susceptibility and disease occurrence on these three crops (Marine et al., 2016; Keinath, 2022). Mandipropamid had a unique use pattern, as percentage area treated was greater on pumpkin than on cucumber and watermelon. Chlorothalonil was applied to the largest area (>35%) on all crops (mean of 54%, range of 37 to 68%). Ignoring chlorothalonil, mean cucumber percentage area treated with cyazofamid and oxathiapiprolin was greater than area of pumpkin and watermelon treated with any fungicide. Use of fluopicolide, ametoctradin, and cymoxanil was ≤5% on all crops (data not shown). On cucumber, the percentage of area treated with chlorothalonil, cyazofamid, and oxathiapiprolin was greater than area treated with propamocarb and mandipropamid (P < 0.0001). On pumpkin and watermelon, percentage area treated with chlorothalonil was higher than for the other four fungicides, and percentage area treated with propamocarb was lower than for the other four fungicides. On watermelon, percentage area treated with oxathiapiprolin was greater (P = 0.0300) than mandipropamid, which suggests that Orondis Opti is used more frequently than Orondis Ultra. This use pattern may be due to overlapping acreages of cucumber and watermelon on the same farms. Thus, cyazofamid and oxathiapiprolin are the primary fungicides used on cucumber, a host for lineage II, and cyazofamid, oxathiapiprolin, and mandipropamid are the primary fungicides used to control lineage I that affects pumpkin and watermelon. Thus, reduced sensitivity of P. cubensis lineage II cannot be attributed to inappropriate use of oxathiapiprolin plus mandipropamid on cucumber.
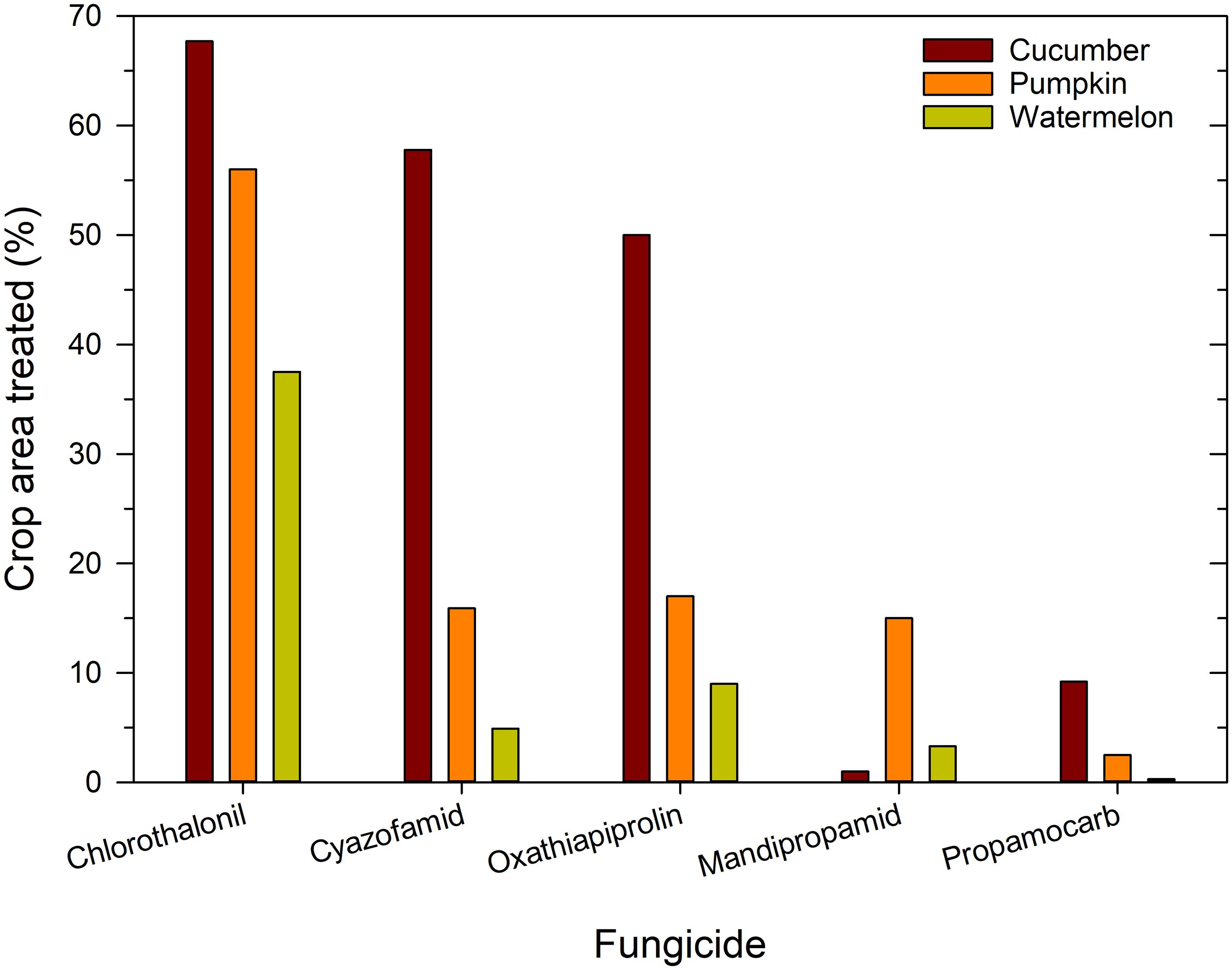
Figure 5. Proportion of United State cucurbit acreage treated with five fungicides that are currently effective against cucurbit downy mildew.
The concentration of chlorothalonil in Orondis Opti is 0.81 to 1.16 kg/ha a.i., depending on application rate, whereas the concentration of chlorothalonil in Bravo Weatherstik is greater, 1.68 kg/ha a.i. Apparently, the lower concentration of chlorothalonil in Orondis Opti was not sufficient to suppress isolates with the oxathiapiprolin resistance mutation, which then were able to reproduce, spread, and establish in the population. In several fungicide evaluations, chlorothalonil applied season long as the sole fungicide was more effective than the non-treated control but often less effective than oomycete-specific fungicides (D’Arcangelo et al., 2021; Uebbing et al., 2024). When chlorothalonil was applied biweekly, which would simulate use as a rotation partner with an ineffective fungicide, chlorothalonil did not suppress cucurbit downy mildew (Keinath and Silva, 2022). Some fungicide recommendations for disease management call for tank mixing additional chlorothalonil with Orondis Opti to bring the concentration of chlorothalonil up to the concentration in chlorothalonil products applied alone (Hausbeck and Mongeon, 2024).
The discovery of host adaptation by P. cubensis lineages allows for more precise determination of fungicide resistance occurrence. Although previously it was assumed that resistance to mefenoxam occurred throughout the P. cubensis population, D'Arcangelo et al. (2023) found that resistant alleles were much more common among lineage II isolates (68.5%) than in lineage I (9.3% of isolates). Similarly, Keinath (2023) reported lack of efficacy (relative to the nontreated control) for mandipropamid against lineage II, while propamocarb was ineffective against lineage I isolates. Lineage-specific fungicide resistance and host adaptation of lineages of P. cubensis offers the option to tailor fungicide recommendations to specific cucurbit crops. For example, mandipropamid can be used on squash and watermelon to reduce reliance on cyazofamid.
In Europe, where fungicide resistance monitoring is mandatory and carried out by fungicide registrants, Plasmopara viticola appears to exhibit negative cross-resistance between azoxystrobin and cyazofamid in that all isolates resistant to azoxystrobin are sensitive to cyazofamid (Torriani and Sierotzki, 2023). The widespread resistance (>70%) to azoxystrobin of isolates of P. cubensis in both lineages may explain why resistance to cyazofamid has not been detected despite frequent use of cyazofamid, particularly on cucumber (D'Arcangelo et al., 2023; U.S. Department of Agriculture, National Agricultural Statistics Service, 2023b).
6.4 Management of fungicide resistance
Fungicide resistance occurs when the frequency of resistant isolates in a population increases relatively faster than the frequency of sensitive isolates (van den Bosch et al., 2015a). Fungicides not only affect growth rates of pathogen populations but also the life cycle components of the pathogen (e.g., infection, latent period and sporulation) in a sensitive population (van den Bosch et al., 2015a; van den Berg et al., 2016). Insensitive populations are not affected by fungicide active ingredients and therefore have opportunities for increased growth rates compared to those of sensitive strains. The increase in the resistant phenotype depends on the existence of resistance gene(s) in the pathogen population, the nature of fungicide mode of action, application dose, and fungicide exposure time (Staub and Sozzi, 1983; van den Bosch et al., 2015b; Lucas et al., 2015; Toquin et al., 2007). While it is generally considered that poor disease control with fungicide applications may arise due to development of resistance in pathogens, resistance is not the only cause of reduced fungicide efficacy (Thind, 2022). Other factors such as fungicide application practices, environmental conditions, and plant characteristics may lead to failure in chemical disease control. Ineffective fungicide applications can be attributed to inadequate dose, improper application timing, poor spray coverage, wrong choice of fungicide, and incompatible tank mixture partner (Thind, 2022).
The CDM pathogen has a short life cycle, high spore production rate, ability to rapidly disperse, and extraordinary genetic diversity. Thus, the Fungicide Resistance Action Committee (FRAC) classifies it as a high risk pathogen for developing resistance to various fungicides. For instance, resistance in P. cubensis populations has been reported in fungicides corresponding to FRAC groups 4, 11, 28, 40, and 43 (Gisi and Sierotzki, 2008; Ishii and Hollomon, 2015; Thomas et al., 2018). In addition, reduced efficacy of field rates of cymoxanil (FRAC Code 27) have been reported (Keinath, 2023). The level of resistance to mefenoxam in the oomycete, Phytophthora capsici, is determined by the number of resistant alleles present in oospores resulting from crosses of parental isolates with different sensitivities, with 0, 1, or 2 resistant alleles present in sensitive, intermediately sensitive, and insensitive isolates, respectively (Lamour and Hausbeck, 2000). Since P. cubensis is diploid, it is possible that the same genetic control of resistance may operate in in the cucurbit downy mildew pathogen. However, since oospores of P. cubensis are constitutively dormant and germination of oosporic inoculum is still very rare (Cohen and Rubin, 2012; Kikway et al., 2022a), the inheritance of fungicide resistance in progeny isolates has not been demonstrated.
Components of an effective fungicide resistance management strategy include; (i) reliable disease monitoring and forecasting, (ii) adhering to legal application doses, (iii) restricting the number and frequency of sprays per season, (iv) the combination of different modes of action as fungicide mixtures, (v) alternating applications with different modes of action, (vi) the avoidance of curative or eradicant use of fungicides, and (viii) use of non-chemical control methods such as resistant crop cultivars, biocontrol agents and cultural control to reduce selection pressure for resistant isolates (Gisi and Sierotzki, 2008; Yin et al., 2023; Torriani and Sierotzki, 2023). For example, to reduce selection pressure for resistance to oxathiapiprolin and cyazofamid in P. cubensis populations of lineage I and II isolates, a third effective fungicide with a FRAC Code other than 43 or 21 should be added to the all-too-common rotation of oxathiapiprolin and cyazofamid.
6.5 Biological control
In conventional production of cucurbits, synthetic fungicides are an important management tool for disease management. The use of chemical pesticides together with moderately resistant cultivars has been the main strategy to manage oomycete diseases. However, frequent fungicide application is costly, and can lead to the development of resistant pathogen populations and can also negatively impact the environment (Sun et al., 2022; Zhang et al., 2019; Taboadela-Hernanz et al., 2025). Thus, there has been increasing interest in developing fungicide alternatives such as biological control agents (BCAs) that have reduced risk of resistance development in target pathogens and are environment friendly (Sun et al., 2022; Taboadela-Hernanz et al., 2025). BCAs suppress plant pathogens through several modes of action such as, antibiosis, competition for nutrients, hyperparasitism, and enhancement of plant immunity, that often acting in combination (Taboadela-Hernanz et al., 2025). Several BCAs belonging to bacterial genera such as Bacillus, Paenibacillus, Enterobacter, Streptomyces, Pseudomonas, Derxia, and Aneurinibacillus, as well as a few fungal genera including Trichoderma, Pestalotiopsis, and Fusarium have demonstrated effective biocontrol activity against diseases caused oomycetes, primarily in experimental greenhouse conditions (Sun et al., 2022; Taboadela-Hernanz et al., 2025). In the United States, more than 200 biopesticide active ingredients have been registered (Marine et al., 2016; Chandler et al., 2011). Currently, Serenade® ASO (1.34% Bacillus subtilis QST 713), Serenade Opti (26.2% Bacillus subtilis QST 713), and Sonata® (1.38% Bacillus pumilus strain QST 2808) are approved microbial biopesticides commercially available for managing CDM disease (Taboadela-Hernanz et al., 2025). The Organic Materials Review Institute (OMRI) lists biorational products for organic production. In organic agriculture, naturally derived products with low selection for resistance such as copper, sulfur, and biorationals (i.e., microbial biopesticides or biochemical biopesticides) are recommended. Compared to synthetic fungicides, biological products are generally more variable in their efficacy and less effective in managing diseases in the field (Uebbing et al., 2024). For example, Marine et al. (2016), reported that in organic production, rotation programs of copper products with biopesticides were moderately effective against cucurbit downy mildew. Other studies, however, showed that repeated applications of copper result in elevated levels in the soil and thus may negatively impact soil invertebrates, alter soil fungal communities, and lead to phytotoxicity to the crop (Van Zwieten et al., 2004; Adrees et al., 2015). In other studies, biopesticides did not provide consistent suppression of CDM on cucumber and were largely ineffective compared to conventional synthetic fungicides (Uebbing et al., 2024). However, biopesticides may be more effective when applied to tolerant hosts or cultivars. For example, copper and other biopesticides were more effective in controlling the disease on Cucurbita spp. than Cucumis spp. (Marine et al., 2016). This highlights the benefits of integrating host resistance with biological control strategies in CDM control. Some of the resistance management strategies for conventional cucurbit production include alternation or tank mixing of at-risk products with microbial biopesticides or biochemical biopesticides (Marine et al., 2016).
7 Future directions
Recent research advances in cucurbit downy mildew are providing opportunities to improve disease control and maximize cucurbit yield. Fungicide stewardship will continue to be an important element in disease control to mitigate the emergence of fungicide resistance in P. cubensis, a pathogen that has a high evolutionary potential. While significant efforts have been made in disease monitoring with the recent research in lineage-specific biosurveillance, a real-time disease monitoring network is required to allow growers to track yearly epidemic development and, when necessary, implement disease management in real-time. Advances in artificial intelligence and machine learning coupled with the connectivity of network products based on the Internet of Things (IoT) are fueling the application of precision tools in crop production (Antony et al., 2020; Vidya Madhuri et al., 2025). The IoT smart technologies should be explored to establish how they can assist growers and researchers in making decisions to improve CDM management practices. Further, there is a need to explore other emerging technologies such as nanotechnology to improve the efficacy, accuracy and targeting of cucurbit downy mildew fungicides using nanopesticides (Atanda et al., 2025). Nanopesticides utilize nanomaterials as carriers for fungicide active ingredients. Fungicide application via nanomaterials improves bioavailability of active ingredients thereby increasing efficacy. The latter can reduce opportunities of exposing the pathogen to sub-lethal doses that can encourage the development of fungicide resistance, which is a major problem with P. cubensis. Nanopesticides also have the added advantage of limiting environmental exposure although the high production costs still make their use in agriculture a low-margin industry (Younis et al., 2021). Host disease resistance offers a great potential in developing long-term strategies to sustainable manage CDM. However, this will require traditional breeding methods to work in combination with modern breeding techniques to exploit the potential of molecular mechanisms that confer CDM resistance to develop varieties with durable disease resistance.
Author contributions
IK: Formal analysis, Data curation, Writing – original draft. AK: Formal analysis, Writing – review & editing, Data curation. PO: Writing – review & editing, Formal Analysis, Supervision, Conceptualization, Funding acquisition.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeniyi D. O., Kunwar D., Dongo L. N., Animasaun D. A., and Aravind T. (2021). “New-generation fungicides for sustainable production and disease suppression,” in Emerging Trends in Plant Pathology. Eds. Singh K. P., Jahagirdar S., and Sarma B. K. (Springer Nature, Singapore, Pte Ltd), 249–256.
Adrees M., Ali S., Rizwan M., Ibrahim M., Abbas F., Farid M., et al. (2015). The effect of excess copper on growth and physiology of important food crops: a review. Environ. Sci. pollut. Res. 22, 8148–8162. doi: 10.1007/s11356-015-4496-5
Antony A. P., Leith K., Jolley C., Lu J., and Sweeney D. J. (2020). A review of practice and implementation of the internet of things (IoT) for smallholder agriculture. Sustainability 12, 3750. doi: 10.3390/su12093750
Atanda S. A., Shaibu R. O., and Agunbiade F. O. (2025). Nanoparticles in agriculture: balancing food security and environmental sustainability. Discov. Agric. 3, 26. doi: 10.1007/s44279-025-00159-x
Bains S. S. and Jhooty J. S. (1976). Over wintering of Pseudoperonospora cubensis causing downy mildew of muskmelon. Indian Phytopathol. 29, 213–214.
Bello J. C., Higgins D. S., Sakalidis M. L., Quesada-Ocampo L. M., Martin F., and Hausbeck M. K. (2022). Clade-specific monitoring of airborne Pseudoperonospora spp. sporangia using mitochondrial DNA markers for disease management of cucurbit downy mildew. Phytopathology 112, 2110–2125. doi: 10.1094/PHYTO-12-21-0500-R
Berg J. A., Hermans F. W. K., Beenders F., Lou L., Vriezen W. H., Visser R. G. F., et al. (2020). Analysis of QTL DM4.1 for downy mildew resistance in cucumber reveals multiple subQTL: A novel RLK as candidate gene for the most important subQTL. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.569876
Blum M., Waldner M., Olaya G., Cohen Y., Gisi U., and Sierotzki H. (2011). Resistance mechanism to carboxylic acid amide fungicides in the cucurbit downy mildew pathogen. Pseudoperonospora cubensis. Pest Manage. Sci. 67, 1211–1214. doi: 10.1002/ps.2238
Brzozowski L., Holdsworth W. L., and Mazourek M. (2016). ‘DMR-NY401’: A new downy mildew-resistant slicing cucumber. HortScience 51, 1294–1296. doi: 10.21273/HORTSCI10857-16
Call A. D., Wehner T. C., Holmes G. J., and Ojiambo P. S. (2013). Effects of host plant resistance and fungicides on severity of cucumber downy mildew. HortScience 48, 53–59. doi: 10.21273/HORTSCI.48.1.53
Chandler D., Bailey A. S., Tatchell G. M., Davidson G., Greaves J., and Grant W. P. (2011). The development, regulation and use of biopesticides for integrated pest management. Philos. Trans. R. Soc Lond. B Biol. Sci. 366, 1987–1998. doi: 10.1098/rstb.2010.0390
Charaya M. U., Upadhyay A., Bhati H. P., and Kumar A. (2021). “Plant disease forecasting: Past practices to emerging technologies,” in : Plant Disease: Management Strategies. Ed. Nehra S. (Jodhpur, India: Agrobios Research), 1–30.
Chen C. P., Sung C.-C., and Ho C.-C. (1959). A brief report of the discovery of oospores of downy mildew of cucumbers (Pseudoperonospora cubensis (B. and C.) Rost.). Zhibing Zhishi 3, 144–145.
Chen T., Katz D., Ben Naim Y., Hammer R., Ben Daniel B. H., Rubin A. E., et al. (2020). Isolate-dependent inheritance of resistance against Pseudoperonospora cubensis in cucumber. Agronomy 10, 1086. doi: 10.3390/agronomy10081086
Choi Y. J., Hong S. B., and Shin H. D. (2005). A re-consideration of Pseudoperonospora cubensis and P. humuli based on molecular and morphological data. Mycol. Res. 109, 841–848. doi: 10.1017/S0953756205002534
Cohen Y. (1981). “Downy mildew of cucurbits,” in The Downy Mildews. Ed. Spencer D. M. (Academic Press, London), 341–354.
Cohen Y. (2015). The novel oomycide oxathiapiprolin inhibits all stages in the asexual life cycle of Pseudoperonospora cubensis-causal agent of cucurbit downy mildew. PloS One 10, e0140015. doi: 10.1371/journal.pone.0140015
Cohen Y. and Eyal H. (1977). Growth and differentiation of sporangia and sporangiophores of Pseudoperonospora cubensis on cucumber cotyledons under various combinations of light and temperature. Physiol. Plant Pathol. 10, 93–103. doi: 10.1016/0048-4059
Cohen Y., Meron I., Mor N., and Zuriel S. (2003). A new pathotype of Pseudoperonospora cubensis causing downy mildew in cucurbits in Israel. Phytoparasitica 31, 458–466. doi: 10.1007/BF02979739
Cohen Y. and Rotem J. (1971). Field and growth chamber approach to epidemiology of Pseudoperonospora cubensis on cucumbers. Phytopathology 61, 736–737. doi: 10.1094/Phyto-61-736
Cohen Y. and Rubin A. E. (2012). Mating type and sexual reproduction of Pseudoperonospora cubensis, the downy mildew agent of cucurbits. Eur. J. Plant Pathol. 132, 577–592. doi: 10.1007/s10658-011-9902-3
Cohen Y., Rubin A. E., Galperin M., Ploch S., Runge F., and Thines M. (2014). Seed transmission of Pseudoperonospora cubensis. PloS One 9, e109766. doi: 10.1371/journal.pone.0109766
Cohen Y., Van den Langenberg K. M., Wehner T. C., Ojiambo P. S., Hausbeck M., Quesada-Ocampo L. M., et al. (2015). Resurgence of Pseudoperonospora cubensis: The causal agent of cucurbit downy mildew. Phytopathology 105, 998–1012. doi: 10.1094/PHYTO-11-14-0334-FI
D'Arcangelo K. N., Wallace E. C., Miles T. D., and Quesada-Ocampo L. M. (2023). Carboxylic acid amide but not quinone outside inhibitor fungicide resistance mutations show clade-specific occurrence in Pseudoperonospora cubensis causing downy mildew in commercial and wild cucurbits. Phytopathology 113, 80–89. doi: 10.1094/PHYTO-05-22-0166-R
D’Arcangelo K. N., Adams M. L., Kerns J. P., and Quesada-Ocampo L. M. (2021). Assessment of fungicide product applications and program approaches for control of downy mildew on pickling cucumber in North Carolina. Crop Prot. 140, 105412. doi: 10.1016/j.cropro.2020.105412
Ding Z., Zhu Z. C., Shi Y. N., Li Y. H., Meng X. B., and Cui H. N. (2024). Molecular genetic basis of resistance to downy mildew in cucumber and melon. J. Plant Pathol. 106, 499–506. doi: 10.1007/s42161-024-01602-6
Dutta B. (2021). Evaluation of individual fungicides for downy mildew control in cucumber in Tift County, Georgi. Plant Dis. Manage. Rep. 15, V021.
Egel D. S., Adkins S. T., Wintermantel W. M., Keinath A. P., D’Arcangelo K. N., Parada-Rojas C. H., et al. (2022). “Diseases of Cucumbers, Melons, Pumpkins, Squash, and Watermelons,” in Handbook of Vegetable and Herb Diseases. Eds. Elmer W. H., McGrath M., and McGovern R. J. (Springer International Publishing, Cham), 1–105.
Fenu G. and Malloci F. M. (2021). Forecasting plant and crop disease: an explorative study on current algorithms. Big Data Cogn. Comput. 5, 2. doi: 10.3390/bdcc5010002
FRAC (2022). Fungicide Action Resistance Committee Code List 2022: Fungal control agents sorted by cross resistance pattern and mode of action (including coding for FRAC Groups on product labels). Available online at: https://mssoy.org/sites/default/files/documents/frac-code-list-2022_1.pdf (Accessed September 8, 2025).
Gent D. H., Cohen Y., and Runge F. (2017). Homothallism in Pseudoperonospora humuli. Plant Pathol. 66, 1508–1516. doi: 10.1111/ppa.12689
Gisi U. and Sierotzki H. (2008). Fungicide modes of action and resistance in downy mildews. Eur. J. Plant Pathol. 122, 157–167. doi: 10.1007/s10658-008-9290-5
Gisi U., Sierotzki H., Cook A., and McCaffery A. (2002). Mechanisms influencing the evolution of resistance to Qo inhibitor fungicides. Pest Manage. Sci. 58, 859–867. doi: 10.1002/ps.565
Goldenhar K. E. and Hausbeck M. K. (2019). Fungicides for control of downy mildew on pickling cucumber in Michigan. Plant Health Prog. 20, 165–169. doi: 10.1094/PHP-04-19-0025-RS
Granke L. L. and Hausbeck M. K. (2011). Dynamics of Pseudoperonospora cubensis sporangia in commercial cucurbit fields in Michigan. Plant Dis. 95, 1392–1400. doi: 10.1094/PDIS-11-10-0799
Granke L. L., Morrice J. J., and Hausbeck M. K. (2014). Relationships between airborne Pseudoperonospora cubensis sporangia, environmental conditions, and cucumber downy mildew severity. Plant Dis. 98, 674–681. doi: 10.1094/PDIS-05-13-0567-RE
Gugino B. K., Britton W., Keinath A. P., McGrath M. T., Melanson R. A., Miller S. A., et al. (2022). Cucurbit downy mildew ipmPIPE: A valued resource for information dissemination and in-season disease management decisions for diverse stakeholders. (Abstr.). Phytopathology 112, S3.163.
Hahn M. and Mendgen K. (2001). Signal and nutrient exchange at biotrophic plant–fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. doi: 10.1016/S1369-5266(00)00180-1
Hausbeck M. and Mongeon E. (2024). Downy mildew confirmed on cucumbers in eight Michigan counties. MSU Extension Vegetables. Available online at: https://www.canr.msu.edu/news/downy-mildew-confirmed-on-cucumbers-in-eight-michigan-counties (Accessed September 8, 2025).
Hiura M. and Kawada S. (1933). On the overwintering of Peronoplasmopara cubensis. Jpn. J. Bot. 6, 507–513.
Hobbelen P. H. F., Paveley N. D., Fraaije B. A., Lucas J. A., and van den Bosch F. (2011b). Derivation and testing of a model to predict selection for fungicide resistance. Plant Pathol. 60, 304–313. doi: 10.1111/j.1365-3059.2010.02380.x
Hobbelen P. H. F., Paveley N. D., and van den Bosch F. (2011a). Delaying selection for fungicide insensitivity by mixing fungicides at a low and high risk of resistance development: A modeling analysis. Phytopathology 101, 1224–1233. doi: 10.1094/PHYTO-10-10-0290
Holmes G. J., Adams M. L., and Colucci S. J. (2006). Evaluation of fungicides for control of downy mildew of winter squas. Fungic. Nematic. Tests 61, V110.
Holmes G. J., Ojiambo P. S., Hausbeck M. K., Quesada-Ocampo L., and Keinath A. P. (2015). Resurgence of cucurbit downy mildew in the United States: A watershed event for research and extension. Plant Dis. 99, 428–441. doi: 10.1094/PDIS-09-14-0990-FE
Ishii H. and Hollomon D. W. (2015). Fungicide Resistance in Plant Pathogens (Tokyo, Japan: Springer). doi: 10.1007/978-4-431-55642-8
Jones J. G., Everts K. L., McGrath M. T., and Gugino B. K. (2021). Efficacy of fungicides for Pseudoperonospora cubensis determined using bioassays over multiple years in the mid-Atlantic and northeastern United States. Plant Health Prog. 22, 355–361. doi: 10.1094/PHP-10-20-0086-FI
Kanetis L., Holmes G. J., and Ojiambo P. S. (2010). Survival of Pseudoperonospora cubensis sporangia exposed to solar radiation. Plant Pathol. 59, 313–323. doi: 10.1111/j.1365-3059.2009.02211.x
Keinath A. P. (2019). Integrated management of downy mildew on slicing cucumber with fungicides and host resistance but not trellising. Plant Dis. 103, 2592–2598. doi: 10.1094/PDIS-02-19-0323-RE
Keinath A. P. (2022). Reduced sensitivity of Pseudoperonospora cubensis clades 1 and 2 to oxathiapiprolin in South Carolina. Plant Health Prog. 23, 256–259. doi: 10.1094/PHP-12-21-0148-SC
Keinath A. P. (2023). Congruent and differential responses of Pseudoperonospora cubensis clades 1 and 2 to downy mildew fungicides. Plant Health Prog. 24, 405–410. doi: 10.1094/PHP-01-23-0007-SC
Keinath A. P., Holmes G. J., Everts K. L., Egel D. S., and Langston D. B. (2007). Evaluation of combinations of chlorothalonil with azoxystrobin, harpin, and disease forecasting for control of downy mildew and gummy stem blight on melon. Crop Prot. 26, 83–88. doi: 10.1016/j.cropro.2006.04.004
Keinath A. P., Miller S. A., and Smart C. D. (2019). Response of Pseudoperonospora cubensis to preventative fungicide applications varies by state and year. Plant Health Prog. 20, 142–146. doi: 10.1094/PHP-04-19-0028-RS
Keinath A. P. and Silva F. (2022). Economic impacts of reduced fungicide efficacy against downy mildew on slicing cucumber. Crop Prot. 155, 105934. doi: 10.1016/j.cropro.2022.105934
Keinath A. P., Silva F. D., Du Bose. V. B., and Zardus. S. H. (2021). Evaluation of seeding dates and fungicide application to manage downy mildew on slicing cucumber 2021. Plant Dis. Manage. Rep. 16, V073.
Kemble J. M., Albornoz K., Bertucci M. B., Bilbo T. R., Jennings K. M., Meadows I. M., et al. (2025). Southeast U.S. 2025 Vegetable Crop Handbook. Available online at: http://vegcrophandbook.com/ (Accessed September 8, 2025).
Kikway I., Keinath A. P., and Ojiambo P. S. (2022a). Field occurrence and overwintering of oospores of Pseudoperonospora cubensis in the southeastern United States. Phytopathology 112, 1946–1955. doi: 10.1094/PHYTO-11-21-0467-R
Kikway I., Keinath A. P., and Ojiambo P. S. (2022b). Temporal dynamics and severity of cucurbit downy mildew epidemics as affected by chemical control and cucurbit host type. Plant Dis. 106, 1009–1019. doi: 10.1094/PDIS-09-21-1992-RE
Kitner M., Lebeda A., Sharma R., Runge F., Dvořák P., Tahir A., et al. (2015). Coincidence of virulence shifts and population genetic changes of Pseudoperonospora cubensis in the Czech Republic. Plant Pathol. 64, 1461–1470. doi: 10.1111/ppa.12370
Kitner M., Runge F., Lebeda A., Vaculná L., Sedláková B., and Thines M. (2021). Pseudoperonospora humuli might be an introduced species in Central Europe with low genetic diversity but high distribution potential. Eur. J. Plant Pathol. 159, 903–915. doi: 10.1007/s10658-021-02214-x
Klarfeld S., Rubin A., and Cohen Y. (2009). Pathogenic fitness of oosporic progeny isolates of Phytophthora infestans on late-blight-resistant tomato lines. Plant Dis. 93, 947–953. doi: 10.1094/pdis-93-9-0947
Lamour K. H. and Hausbeck M. K. (2000). Mefenoxam insensitivity and the sexual stage of Phytophthora capsici in Michigan cucurbit fields. Phytopathology 90, 396–400. doi: 10.1094/PHYTO.2000.90.4.396
Lange L., Eden U., and Olson L. W. (1989). Zoosporogenesis in Pseudoperonospora cubensis, the causal agent of cucurbit downy mildew. Nord. J. Bot. 8, 497–504. doi: 10.1111/j.1756-1051.1989.tb00527.x
Lebeda A. and Cohen Y. (2011). Cucurbit downy mildew (Pseudoperonospora cubensis) Biology, ecology, epidemiology, host-pathogen interaction, and control. Eur. J. Plant Pathol. 129, 157–192. doi: 10.1007/s10658-010-9658-1
Lebeda A. and Cohen Y. (2012). “Fungicide resistance in Pseudoperonospora cubensis, the causal pathogen of cucurbit downy mildew,” in Fungicide Resistance in Crop Protection: Risk and Management. Ed. Thind T. S. (CABI, Wallingford, UK), 44–63.
Lebeda A., Kitner M., Sedláková B., Sharma R., Runge F., and Thines M. (2014). “Biological and molecular evidences about changes in the host range and virulence of Pseudoperonospora cubensis populations in the Czech Republic,” in Proceedings Cucurbitaceae 2014. Eds. Havey M., Weng Y., Day B., and Grumet R. (Michigan State University and University of Wisconsin-Madison, American Society for Horticultural Science, Alexandria, VA), 24–27.
Lebeda A., Křístková E., Roháčková J., Sedláková B., Widrlechner M. P., and Paris H. S. (2016b). Race-specific resistance of Cucurbita germplasm to Pseudoperonospora cubensis. Euphytica 212, 145–156. doi: 10.1007/s10681-016-1783-2
Lebeda A., Křístková E., and Sedláková B. (2024). Pathotypes and races of Pseudoperonospora cubensis: Two concepts of virulence differentiation. Plant Pathol. 73, 2537–2547. doi: 10.1111/ppa.13993
Lebeda A., Křístková E., Sedláková B., Kitner M., and Widrlechner M. P. (2019). Status of research, breeding and protection of cucurbits in relation to cucurbit downy mildew: their limits and perspectives. Acta Hortic. 1242, 421–426. doi: 10.17660/ActaHortic.2019.1242.60
Lebeda A., Křístková E., Sedláková B., McCreight J. D., and Kosman E. (2018). Virulence variation of cucurbit powdery mildews in the Czech Republic–population approach. Eur. J. Plant Pathol. 152, 309–326. doi: 10.1007/s10658-018-1476-x
Lebeda A., Křístková E., Sedláková B., Štěpánková J., and Widrlechner M. P. (2017). Race-specificity in interactions between Cucumis melo germplasm and Pseudoperonospora cubensis. Acta Hortic. 1151, 203–210. doi: 10.17660/ActaHortic.2017.1151.32
Lebeda A., Křístková E., Štěpánková J., Sedláková B., and Widrlechner M. P. (2016a). Response of Cucumis melo accessions to isolates of Pseudoperonospora cubensis with different levels of virulence. Sci. Hortic. 200, 45–54. doi: 10.1016/j.scienta.2015.09.028
Lebeda A., Pavelková J., Sedláková B., and Urban J. (2013). Structure and temporal shifts in virulence of Pseudoperonospora cubensis populations in the Czech Republic. Plant Pathol. 62, 336–345. doi: 10.1111/j.1365-3059.2012.02642.x
Lebeda A., Pavelková J., Urban J., and Sedláková B. (2011). Distribution, host range and disease severity of Pseudoperonospora cubensis on cucurbits in the Czech Republic. J. Phytopathol. 159, 589–596. doi: 10.1111/j.1439-0434.2011.01811.x
Lebeda A. and Schwinn F. J. (1994). The downy mildews–an overview of recent research progress. J. Plant Dis. Prot. 101, 225–254.
Lebeda A. and Urban J. (2004). “Disease impact and pathogenicity variation in Czech populations of Pseudoperonospora cubensis,” in Progress in Cucurbit Genetics and Breeding Research. Proceedings of Cucurbitaceae 2004, 8th EUCARPIA Meeting on Cucurbit Genetics and Breeding. Eds. Lebeda A. and Paris H. S. (Palacký University, Olomouc. Czech Republic), 267–273.
Lebeda A., Widrlechner M. P., and Urban J. (2006). “Individual and population aspects of interactions between cucurbits and Pseudoperonospora cubensis: pathotypes and races,” in Proceedings of Cucurbitaceae. Ed. Holmes G. J. (Universal, Raleigh), 453–467.
Lucas J. A., Hawkins N. J., and Fraaije B. A. (2015). Chapter two-the evolution of fungicide resistance. Adv. Appl. Microbiol. 90, 29–92. doi: 10.1016/bs.aambs.2014.09.001
Mandal M. K., Turechek W. W., Ikerd J. L., Wallace E., Quesada-Ocampo L., and Kousik C. S. (2025). Temporal dynamics of two genetically diverse downy mildew mitochondrial clades on cucumber cultivars in South Carolina and Florida. Plant Health Progr. 26, 120–129. doi: 10.1094/PHP-01-24-0003-RS
Marine S. C., Newark M. J., Korir R. C., and Everts K. L. (2016). Evaluation of rotational biopesticide programs for disease management in organic cucurbit production. Plant Dis. 100, 2226–2233. doi: 10.1094/PDIS-02-16-0252-RE
McDonald B. A. and Linde C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
Mirzwa-Mróz E., Zieniuk B., Yin Z., and Pawełkowicz M. (2024). Genetic insights and molecular breeding approaches for downy mildew resistance in cucumber (Cucumis sativus L.): Current progress and future prospects. Int. J. Mol. Sci. 25, 12726. doi: 10.3390/ijms252312726
Naegele R. P., Quesada-Ocampo L. M., Kurjan J. D., Saude C., and Hausbeck M. K. (2016). Regional and temporal population structure of Pseudoperonospora cubensis in Michigan and Ontario. Phytopathology 106, 372–379. doi: 10.1094/PHYTO-02-15-0043-R
Nandwani D., Williamson J. R., Crossman S., and Forbes V. (2014). Cucumber cultivar study in the U.S. Virgin Islands. Proc. Carib. Food Crops Soc. 50, 157–161.
Neufeld K. N., Isard S. A., and Ojiambo P. S. (2013). Relationship between disease severity and escape of Pseudoperonospora cubensis sporangia from a cucumber canopy during downy mildew epidemics. Plant Pathol. 62, 1366–1377. doi: 10.1111/ppa.12040
Neufeld K. N., Keinath A. P., Gugino B. K., McGrath M. T., Sikora E. J., Miller S. A., et al. (2018a). Predicting the risk of cucurbit downy mildew in the eastern United States using an integrated aerobiological model. Int. J. Biometeorol. 62, 655–668. doi: 10.1007/s00484-017-1474-2
Neufeld K. N., Keinath A. P., and Ojiambo P. S. (2017). A model to predict the risk of infection of cucumber by Pseudoperonospora cubensis. Microbial Risk Anal. 6, 21–30. doi: 10.1016/j.mran.2017.05.001
Neufeld K. N., Keinath A. P., and Ojiambo P. S. (2018b). Evaluation of a model for predicting the risk of squash and cantaloupe by Pseudoperonospora cubensis. Plant Dis. 102, 855–862. doi: 10.1094/PDIS-07-17-1046-RE
Nusbaum C. J. (1944). The seasonal spread and development of cucurbit downy mildew in the Atlantic coastal states. Plant Dis. Rep. 28, 82–85.
Nusbaum C. J. (1945). The seasonal spread and development of cucurbit downy mildew in Atlantic coastal states in 1944. Plant Dis. Rep. 29, 141–143.
Nusbaum C. J. (1948). A summary of cucurbit downy mildew reports from Atlantic coastal states in 1947. Plant Dis. Rep. 32, 44–48.
Ojiambo P. S., Gent D. H., Quesada-Ocampo L. M., Hausbeck M. K., and Holmes G. J. (2015). Epidemiology and population biology of Pseudoperonospora cubensis: A model system for management of downy mildews. Annu. Rev. Phytopathol. 53, 223–246. doi: 10.1146/annurev-phyto-080614-120048
Ojiambo P. S. and Holmes G. J. (2011). Spatiotemporal spread of cucurbit downy mildew in the eastern United States. Phytopathology 101, 451–461. doi: 10.1094/PHYTO-09-10-0240
Ojiambo P. S., Holmes G. J., Britton W., Keever T., Adams M. L., Babadoost M., et al. (2011). Cucurbit downy mildew ipmPIPE: A next generation web-based interactive tool for disease management and extension outreach. Plant Health Prog. 12, 1–14. doi: 10.1094/PHP-2011-0411-01-RV
Ojiambo P. S., Kanetis L., and Holmes G. (2009). Forecasting long distance movement of Pseudoperonospora cubensis and the Cucurbit ipmPIPE. Phytopathology 99, S171.
Ojwang’ A. M. E., Ruiz T., Bhattacharyya S., Chatterjee S., Ojiambo P. S., and Gent D. H. (2021). A general framework for spatio-temporal modeling of epidemics with multiple epicenters: application to an aerially dispersed plant pathogen. Front. Appl. Math. Stat. 7. doi: 10.3389/fams.2021.721352
Olaya G., Kuhn P., Hert A., Holmes G., and Colucci S. (2009). Fungicide resistance in cucurbit downy mildew. Phytopathology 99, S171.
Palti J. (1975). Pseudoperonospora cubensis (Berk and M. A. Curtis) rost. CMI Descriptions Pathogenic Fungi Bacteria 457, 1–2.
Palti J. and Cohen Y. (1980). Downy mildew of cucurbits (Pseudoperonospora cubensis): the fungus and its hosts, distribution, epidemiology, and control. Phytoparasitica 8, 109–147. doi: 10.1007/BF02994506
Prieto Torres M. and Quesada-Ocampo L. M. (2024). Oxathiapiprolin fungicide resistance mutation occurrence over time in Pseudoperonospora cubensis samples from North Carolina. (Abstr.). Phytopathology 114, S1.70.
Rahman A., Miles T. D., Martin F. N., and Quesada-Ocampo L. M. (2017). Molecular approaches for biosurveillance of the cucurbit downy mildew pathogen, Pseudoperonospora cubensis. Can. J. Plant Pathol. 39, 282–296. doi: 10.1080/07060661.2017.1357661
Rahman A., Standish J. R., D’arcangelo K. N., and Quesada-Ocampo L. M. (2021). Clade-specific biosurveillance of Pseudoperonospora cubensis using spore traps for precision disease management of cucurbit downy mildew. Phytopathology 111, 312–320. doi: 10.1094/PHYTO-06-20-0231-R
Rani R., Negi P., Sharma S., and Jain S. (2022). Occurrence of oosporic stage of Pseudoperonospora cubensis on cucumber in Punjab, India: a first report. Crop Prot. 155, 105939. doi: 10.1016/j.cropro.2022.105939
Rostovzev S. J. (1903). Beiträge zur Kenntnis der Peronosporeen. Flora oder Botanische Zeitung 92, 405–433.
Rsaliyev A. S., Amirkhanova N. T., Rametov N. M., Pahratdinova Z. U., Ojiambo P. S., and Lebeda A. (2018). Pseudoperonospora cubensis virulence and pathotype structure in Kazakhstan. Plant Pathol. 67, 1924–1935. doi: 10.1111/ppa.12890
Runge F., Choi Y.-J., and Thines M. (2011). Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur. J. Plant Pathol. 129, 135–146. doi: 10.1007/s10658-010-9714-x
Savory E. A., Granke L. L., Quesada-ocampo L. M., Varbanova M., Hausbeck M. K., and Day B. (2011). The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol. Plant Pathol. 12, 217–226. doi: 10.1111/j.1364-3703.2010.00670.x
Sharma N., Heger L., Combs D. B., Smith W. M., Holland L., Brannen P., et al. (2025). Prevalence of mutations associated with QoIs, QiIs, QioSIs, and CAA fungicide resistance within Plasmopara viticola in North America and a tool to detect CAA-resistant isolates. Phytopathology 115, 495–506. doi: 10.1094/PHYTO-08-24-0257-R
Shirley A. M., Vallad G. E., Quesada-Ocampo L., Dufault N., and Raid R. (2024). Effect of cucurbit host, production region, and season on the population structure of Pseudoperonospora cubensis in Florida. Plant Dis. 108, 442–450. doi: 10.1094/PDIS-12-22-2939-RE
Spring O., Gomez-Zeledon J., Hadziabdic D., Trigiano R. N., Thines M., and Lebeda A. (2018). Biological characteristics and assessment of virulence diversity in pathosystems of economically important biotrophic oomycetes. Crit. Rev. Plant Sci. 37, 439–495. doi: 10.1080/07352689.2018.1530848
Staub T. and Sozzi D. (1983). “Recent practical experiences with fungicide resistance,” in 10th International Congress of Plant Protection: Plant Protection for Human Welfare (Brighton, England), 591–598. doi: 10.1146/annurev.py.29.090191.002225
Sun S., Lian S., Feng S., Dong X., Wang C., Li B., et al. (2017). Effects of temperature and moisture on sporulation and infection by Pseudoperonospora cubensis. Plant Dis. 101, 562–567. doi: 10.1094/PDIS-09-16-1232-RE
Sun Z., Yu S., Hu Y., and Wen Y. (2022). Biological control of the cucumber downy mildew pathogen Pseudoperonospora cubensis. Horticulturae 8, 410. doi: 10.3390/horticulturae8050410
Taboadela-Hernanz J., Hieno A., and Shimizu M. (2025). Toward effective biocontrol of oomycete plant pathogens: traits and modes of action of biocontrol agents, and their screening approaches. Rev. Agric. Sci. 13, 32–51. doi: 10.7831/ras.13.1_32
Thind T. S. (2022). New insights into fungicide resistance: a growing challenge in crop protection. Indian Phytopathol. 75, 927–939. doi: 10.1007/s42360-022-00550-4
Thomas C. E. (1996). “Downy mildew,” in Compendium of Cucurbit Diseases. Eds. Zitter T. A., Hopkins D. L., and Thomas C. E. (APS Press, St. Paul, MN), 25–27.
Thomas A., Carbone I., Cohen Y., and Ojiambo P. S. (2017a). Occurrence and distribution of mating types of Pseudoperonospora cubensis in the United States. Phytopathology 107, 313–321. doi: 10.1094/PHYTO-06-16-0236-R
Thomas A., Carbone I., Lebeda A., and Ojiambo P. S. (2017b). Virulence structure within populations of Pseudoperonospora cubensis in the United States. Phytopathology 107, 777–785. doi: 10.1094/PHYTO-07-16-0277-R
Thomas C. E., Inaba T., and Cohen Y. (1987). Physiological specialization in Pseudoperonospora cubensis. Phytopathology 77, 1621–1624. doi: 10.1094/Phyto-77-1621
Thomas A., Neufeld K. N., Seebold K. W., Braun C. A., Schwarz M. R., and Ojiambo P. S. (2018). Resistance to fluopicolide and propamocarb and baseline sensitivity to ethaboxam among isolates of Pseudoperonospora cubensis from the eastern United States. Plant Dis. 102, 1619–1626. doi: 10.1094/PDIS-10-17-1673-RE
Toporek S. M. and Keinath A. P. (2021). Clade and mating type distribution and population structure of Pseudoperonospora cubensis on Cucumis melo in the eastern United States. (Abstr.). Phytopathology 111, S2.35.
Toporek S. M. and Keinath A. P. (2022). Efficacy of fungicides used to manage downy mildew in cucumber, assessed with multiple meta-analysis techniques. Phytopathology 112, 1651–1658. doi: 10.1094/PHYTO-10-21-0432-R
Toquin V., Barja F., Sirven C., and Bffa R. (2007). “Fluopicolide, a new anti-oomycetes fungicide with a new mode of action inducing perturbation of a spectrin-like protein,” in Modern Crop Protection Compounds. Eds. Krämer W. and Schirmer U. (Wiley-VCH Verlag GmbH & Co. KgaA, Weinheim, Germany), 675–682. doi: 10.1002/9783527619580.ch19
Torriani S. F. F. and Sierotzki H. (2023). “Quinone outside inhibitor fungicide resistance: selection patterns and the current situation,” in Understanding and Minimizing Fungicide Resistance. Ed. Lopez-Ruiz F. J. (Burleigh Dodds Series in Agricultural Science Number 132. Burleigh Dodds Science Publishing, London, UK), 273–297.
Uebbing M. R., Hayden Z. D., and Hausbeck M. K. (2024). Conventional and biopesticide fungicides for cucurbit downy mildew control on cucumber in Michigan. Plant Health Prog. 25, 9–18. doi: 10.1094/PHP-03-23-0024-RS
Urban J. and Lebeda A. (2006). Fungicide resistance in cucurbit downy mildew–methodological, biological and population aspects. Ann. Appl. Biol. 149, 63–75. doi: 10.1111/j.1744-7348.2006.00070.x
U.S. Department of Agriculture, National Agricultural Statistics Service (2021). 2020 Vegetable Chemical Use, Pre-Defined Queries: Environmental, Data and Statistics. Available online at: https://www.nass.usda.gov/Data_and_Statistics/Pre-efined_Queries/2020_Vegetables/index.php (Accessed September 8, 2025).
U.S. Department of Agriculture, National Agricultural Statistics Service (2023a). Vegetables 2022 Summary. USDA Economics, Statistics and Market Information System. Available online at: https://usda.library.cornell.edu/concern/publications/02870v86p?locale=en (Accessed September 8, 2025).
U.S. Department of Agriculture, National Agricultural Statistics Service (2023b). 2022 Vegetable Chemical Use, Pre-Defined Queries: Environmental, Data and Statistics. Available online at: https://www.nass.usda.gov/Data_and_Statistics/Pre-efined_Queries/2022_Vegetables/index.php (Accessed September 8, 2025).
U.S. Department of Agriculture, National Agricultural Statistics Service (2025). Vegetables 2024 Summary. USDA Economics, Statistics and Market Information System. Available online at: https://usda.library.cornell.edu/concern/publications/02870v86p?locale=en (Accessed September 8, 2025).
van den Berg F., Paveley N. D., and van den Bosch F. (2016). Dose and number of applications that maximize fungicide effective life exemplified by Zymoseptoria tritici on wheat–a model analysis. Plant Pathol. 65, 1380–1389. doi: 10.1111/ppa.12558
van den Bosch F., Fraaije B., Oliver R., van den Berg F., and Paveley N. (2015a). “The use of mathematical models to guide fungicide resistance management decisions,” in Fungicide Resistance in Plant Pathogens. Eds. Ishii H. and Hollomon D. W. (Springer, Tokyo), 49–62.
van den Bosch F., Paveley N., Fraaije B., van den Berg F., and Oliver R. (2015b). “Evidence-based resistance management: a review of existing evidence,” in Fungicide Resistance in Plant Pathogens. Eds. Ishii H. and Hollomon D. W. (Springer, Tokyo), 63–76.
Van Zwieten L., Rust J., Kingston T., Merrington G., and Morris S. (2004). Influence of copper fungicide residues on occurrence of earthworms in avocado orchard soils. Sci. Total Environ. 329, 29–41. doi: 10.1016/j.scitotenv.2004.02.014
Vidya Madhuri E., Rupali J. S., Sharan S. P., Pooja Sai N., Sujatha G. S., Singh D. P., et al. (2025). Transforming pest management with artificial intelligence technologies: the future of crop protection. J. Crop Health 77, 48. doi: 10.1007/s10343-025-01109-9
Wallace E., Adams M., Ivors K., Ojiambo P. S., and Quesada-Ocampo L. M. (2014). First report of Pseudoperonospora cubensis causing downy mildew on Momordica balsamina and M. charantia in North Carolina. Plant Dis. 98, 1279–1279. doi: 10.1094/PDIS-03-14-0305-PDN
Wallace E., Adams M., and Quesada-Ocampo L. M. (2015). First report of downy mildew on buffalo gourd (Cucurbita foetidissima) caused by Pseudoperonospora cubensis in North Carolina. Plant Dis. 99, 1861–1861. doi: 10.1094/PDIS-03-15-0348-PDN
Wallace E. C., D’Arcangelo K. N., and Quesada-Ocampo L. M. (2020). Population analyses reveal two host-adapted clades of Pseudoperonospora cubensis, the causal agent of cucurbit downy mildew, on commercial and wild cucurbits. Phytopathology 110, 1578–1587. doi: 10.1094/PHYTO-01-20-0009-R
Whisson S. C., Boevink P. C., Moleleki L., Avrova A. O., Morales J. G., Gilroy E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. doi: 10.1038/nature06203
Win K. T., Vegas J., Zhang C., Song K., and Lee S. (2017). QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor. Appl. Genet. 130, 199–211. doi: 10.1007/s00122-016-2806-z
Yin Y. N., Miao J., Shao W., Liu X., Zhao Y., and Ma Z. (2023). Fungicide resistance: Progress in understanding mechanism, monitoring and management. Phytopathology 113, 707–718. doi: 10.1094/PHYTO-10-22-0370-KD
Yoshioka Y., Sakata Y., Sugiyama M., and Fukino N. (2014). Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 198, 265–276. doi: 10.1007/s10681-014-1102-8
Younis S. A., Kim K. H., Shaheen S. M., Antoniadis V., Tsang Y. F., Rinklebe J., et al. (2021). Advancements of nanotechnologies in crop promotion and soil fertility: benefits, life cycle assessment, and legislation policies. Renew. Sust. Energ. Rev. 152, 111686. doi: 10.1016/j.rser.2021.111686
Zhang Y. J., Pu Z. J., Qin Z. W., Zhou X. Y., Liu D., Dai L. T., et al. (2012). A study on the overwintering of cucumber downy mildew oospores in China. J. Phytopathol. 160, 469–474. doi: 10.1111/j.1439-0434.2012.01926.x
Keywords: biosurveillance, cucurbit downy mildew, disease control, disease monitoring, fungicides, inoculum source, network analysis
Citation: Kikway I, Keinath AP and Ojiambo PS (2025) A comprehensive review of Pseudoperonospora cubensis: biology, epidemiology, and disease management. Front. Hortic. 4:1636345. doi: 10.3389/fhort.2025.1636345
Received: 27 May 2025; Accepted: 26 August 2025;
Published: 19 September 2025.
Edited by:
Thomas M. Chappell, Texas A and M University, United StatesReviewed by:
Sandeep Jain, Punjab Agricultural University, IndiaAles Lebeda, Palacký University in Olomouc, Czechia
Thomas Isakeit, Texas A&M University San Antonio, United States
Copyright © 2025 Kikway, Keinath and Ojiambo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter S. Ojiambo, cG9qaWFtYkBuY3N1LmVkdQ==
 Isaack Kikway1,2
Isaack Kikway1,2 Peter S. Ojiambo
Peter S. Ojiambo