- Institute of Protein Biochemistry, National Research Council, Naples, Italy
The enormous potential in diversity of the marine life is still not fully exploited due to the difficulty in culturing many of the microorganisms under laboratory conditions. In this mini-review we underlined the importance of using an omics technique, such as metagenomics, to access the uncultured majority of microbial communities. We report examples of several hydrolytic enzymes and natural products isolated by functional sequenced-based and function- screening strategies assisted by new high-throughput DNA sequencing technology and recent bioinformatics tools. This article ends with an overview of the potential future perspectives of the metagenomics in bioprospecting novel biocatalysts and bioactive compounds from marine sources.
Marine Environment
Marine environment is the largest aquatic ecosystem on the planet and it is estimated to be one of the most important sources of biodiversity in the world (Zhao, 2011; Felczykowska et al., 2012). Marine ecosystems have peculiar characteristics that result from the unique combination of several physical factors. These habitats allow the growth of a multitude of organisms, such as bacteria, algae, sponges, fungi, and fishes, which are able to face with these harsh conditions. In particular, certain microorganisms are able to live in the cold sea of Arctic and Antarctic regions, and these species growth under high pressure and low temperatures, at different pH and salinity, or in seas characterized by high level of pollutions (Norway, Red Sea). For their particular features, many of these microorganisms are used in a wide range of biotechnological applications, providing novel bioactive compounds (Faulkner, 2001) and biocatalysts for modern industries (Kennedy et al., 2011; De Pascale et al., 2012).
However, most of the marine biodiversity is still unexplored, because of the difficulties in reproducing marine microenvironment in laboratory. In fact, it has been estimated that less than 0.1% of all microbes in the oceans today has been discovered so far (Simon and Daniel, 2009) and therefore it is clear we have explored just a minimal part of the vast potential of the ocean in terms of natural product discovery.
For these reasons most of the marine bioma remained so far uncultured, therefore the marine treasure still remains in the abysses.
Metagenomics, with its culture-independent principle, offers novel opportunities for studying marine biodiversity and its biotechnological application.
Herein, we analyzed the state of the art of marine metagenomics, focusing on the discovery and application of novel enzymatic biocatalysts and bioactive compounds as well as the novel technological improvement in the field.
Bioprospecting
Marine bioprospecting is aimed to search novel organisms or genes. Sediments and biota sampling is generally carried out in less explored regions, such as at extreme depths, on the high seas or on the deep seabed, specifically around sub-marine trenches, cold seeps, and hydrothermal vents. Bioprospecting in the oceans has been potentiated by the integration of high-throughput DNA sequencing methods to evaluate marine biodiversity. Such genomic data can provide a useful starting point to identify new enzymes involved in the biosynthesis of secondary metabolites (Arrieta et al., 2010; Abida et al., 2013). Genomics and biotechnology are examples of modern approaches to expand our knowledge of the processes influencing the diversity of life in the oceans (Huete-Perez and Quezada, 2013).
Metagenomic Approach
Culture-Independent Method
Many organisms require special growth parameters (physical and chemical) that are hard or even impossible to reproduce in the laboratory (Pace, 2009). In addition the interdependence with other organisms in Nature is a crucial point to be considered when culturing microorganisms under unnatural conditions. This is the reason why more than 70 bacterial phyla have no cultured representatives.
Although the cultivation success rate certainly can be improved (Tyson and Banfield, 2005), there are recent approaches that microbiologists are employing to convert currently unculturable bacteria into cultured isolates in laboratory, as the capillary-based system of cells culturing based on porous hollow-fiber membranes (Stewart, 2012). To explore this vast source of genetic diversity, omics techniques are currently used.
Metagenomics is an extremely powerful omics technique. It refers togenetic analysis of microorganisms by direct extraction method and cloning of DNA recovered from mixed combination of organisms collected from Nature (Handelsman, 2004). Metagenomics may be focused on gene clusters or genes encoding enzymes and on the discovery of biocatalysts for synthesis and production of secondary metabolites like bioactive compounds (Wong, 2010). Three categories of environments are often considered: (1) highly diverse environments (e.g., soil and seawater), (2) naturally or artificially enriched environments for the target gene/biocatalyst, or (3) extreme environments.
Screening, Sequencing, and Data Analysis of Metagenomic Libraries
Screening of metagenomic libraries can be divided in sequence-based screening and function-based screening; in the sequence-based screening PCR amplifications are used to identify target genes from conserved regions of known genes, while the function-based screening is often carried out using robotic systems looking for a well-defined phenotype.
The screening may be often considered a drawback for a successful metagenomic approach due to the sensitivity of agar plate-based screening and faint signals. Furthermore, the expression of the target genes may be considered the major bottleneck due to the choice of an adequate expression vector and a suitable recombinant host.
However, over the last years some of these issues have been solved thank to the continuous progresses in robotic machinery and sequencing technologies.
A crucial connection from the screening to the data analysis is represented by the development of new sequencing systems. This new platform concept consists on sequencing of a dense array of amplified DNA fragments through iterative cycles of enzymatic manipulation and imaging-based data collection (Shendure and Ji, 2008). More recent technologies, the so-called “third generation sequencing technologies,” involve sequencing of individual molecules, the single-cell sequencing (Xu et al., 2009). Third generation sequencing technologies, based on fluorescence detection, have already been launched (Xu et al., 2009; Metzker, 2010).
One example is the Next-generation Sequencing Simulator for Metagenomics (NeSSM), which is a fast simulation system for high-throughput metagenome sequencing (Jia et al., 2013). The large number of metagenomic information obtained by the sequencing platforms must be processed and suitable data-analysis tools are required. Bio-informatics software tools have been developed in order to manage enormous datasets (reads- or contigs-). Examples are the metagenome analyzer MEGAN, a recently software tool able to analyze the taxonomic content of large metagenomic datasets of short DNA fragments obtained through 454 sequencing (Huson et al., 2007). Another recent ultrafast program, named Kraken, is a highly accurate program to assign taxonomic labels to metagenomic DNA sequences (Wood and Salzberg, 2014) and the newest Meta-QC-Chain provides an useful and high-performance QC (quality control) tool for metagenomic data (Zhou et al., 2014).
Marine Products
Enzymes
Oceanic microorganisms have to face extreme environmental conditions such as low temperature, high salinity, and extreme pressures and they have evolved special metabolites to survive and proliferate during the evolution. Thus, the general life conditions are reflected into the enzymes that potentially may endow of unique properties.
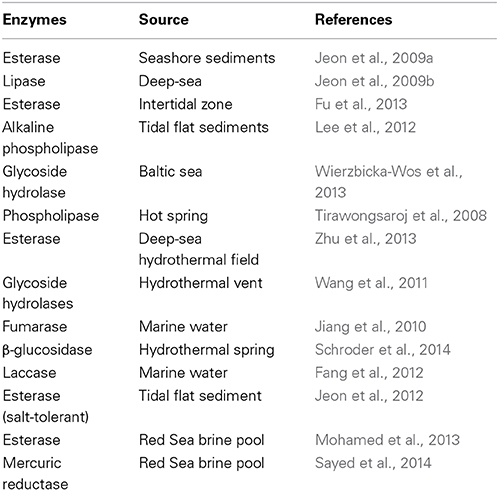
Metagenomics resulted in being very effective in the discovery of novel extremozymes, isolated from extreme marine environments (Table 1). Cold-adapted enzymes represent a class of extremozymes and compared to their mesophilic or thermophilic homologs, can be up to 10 times more active at low and moderate temperatures (Cipolla et al., 2012; Karan et al., 2012).
These enzymes are already being used in many biotechnological applications providing economic benefits and energy savings. As a result of their high activity at mild temperatures or fast heat-inactivation, a lower concentration of the enzyme is required to reach a given activity reducing the costs of enzyme preparation. Also, they can minimize undesirable chemical reactions that can occur at higher temperatures (Cipolla et al., 2012). These properties are of particular relevance for the food and feed industry to avoid spoilage and change in nutritional value and flavor of the original heat-sensitive substrates and products (Cavicchioli et al., 2011; Florczak et al., 2013).
It has been demonstrated that cold-adapted enzymes possess peculiar structural features that confer them a flexible configuration. Comparative genome analyzes suggested that this typical features of psychrophilic enzymes are most probably due to a combination of changes in the overall amino-acid composition, by which psychrophilic proteins lose their rigidity and gain increased structural flexibility enhancing catalytic function at low temperatures (Liszka et al., 2012; De Pascale et al., 2012).
Lipase and esterase are prominent industrial enzymes, being employed in the food, laundry, textile, pulp and paper industries, production of biodiesel, and in the synthesis of fine chemicals. Furthermore, they are very easy to detect from a functional agar screening by using synthetic substrates (Jeon et al., 2009a,b).
A metagenomic library screening of an Arctic intertidal zone allowed the isolation of a novel cold active esterase called Est97 (Fu et al., 2013). The recombinant enzyme demonstrated to retain almost the 60% of relative activity at 20°C and a very low thermostability, suggesting its utilization in cold biotransformation. Metagenomic techniques also allow the isolation of other classes of enzymes, in fact, a phospholipase A with lipase activity (after called MPlaG) is the first obtained from a metagenomic library from tidal flat sediments on the Korean west coast. It shown a maximum activity at 25°C and also presented specific catalytic properties against olive oil and phosphatidylcholine, which means that MPlaG is a lipid-preferred phospholipase (Lee et al., 2012).
A monomeric cold-active glycoside hydrolase family 1 enzyme, named BglMKg, was identified from metagenomic library of Baltic Sea water sample. This enzyme is characterized by a wide range of enzymatic activities including β-galactosidase, β-fucosidase, and β-glucosidase activities and, it demonstrated to be stable below 30°C, in the range from pH 6.0 to 8.0. The results of the kinetic studies revealed that BglMKg more efficiently hydrolyzed β-glucosidase substrates than β-galactosidase (Wierzbicka-Wos et al., 2013). The β-galactosidases are mainly implied in dairy industry because they are able to specifically hydrolyze the lactose into galactose and glucose.
Apart from the cold-adapted enzymes, the oceans host many other classes of microorganism (and therefore of enzymes) in specific and extreme ecological niches. Thermophilic and hyperthermophilic microorganisms, halophiles, and barophiles possess biocatalysts with relevant biotechnological applications (Bruins et al., 2001), showing stability and catalytic efficiency in the presence of high temperature, high salt concentration, and high pressure.
Novel thermostable biocatalysts have also been isolated through metagenomic approach. Several classes of enzymes have been identified and characterized: lipolytic enzymes (Tirawongsaroj et al., 2008; Zhu et al., 2013), glycoside hydrolases (Wang et al., 2011), fumarase (Jiang et al., 2010), and β-glucosidase (Schroder et al., 2014). Interesting, salt-tolerant enzymes have also been discovered. Fang and coworkers discovered a novel bacterial laccase, with alkaline activity, whose activity is enhanced by chloride addiction (Fang et al., 2012). A group of Korean researcher was able to identify salt-tolerant esterases belonged to a new subfamily, through metagenomics (Jeon et al., 2012).
Bioprospecting in polluted and contaminated seas can lead to the isolation of enzymes that also display tolerance for high concentration of heavy metals. Mohamed et al. (2013) isolated a novel esterase from a metagenomic library from a Red Sea brine pool. This esterase combined a thermophilic activity with high resistant to several heavy metals, making this enzymes appealing for applications in bioremediation (Mohamed et al., 2013). Similarly, a novel mercuric reductase was, again, discovered from samples collected from Red Sea. This enzyme showed enhanced catalytic activity in presence of high temperature, high salt, and heavy metals concentrations (Sayed et al., 2014).
Bioactive Compounds
Compounds from marine environments
The oceans may be considered a vast “container” of natural products that could be exploited in medicine. Marine macro/micro organisms, during the evolution, acquired the capability to produce secondary metabolites with unique biological activity (Imhoff et al., 2011). These compounds have found a wide range of applications as antibacterial (Teasdale et al., 2009; Plaza et al., 2010), antifungal (Nishimura et al., 2010), antimalarial, antiprotozoa (Dos Santos et al., 2011), and antiviral (Cheng et al., 2010), as well as being active in diseases related to the cardiovascular, immune, and nervous systems (Asolkar et al., 2009; Sakurada et al., 2010; Mayer et al., 2013). Metagenomics revealed to be a very powerful tool also for the exploitation of bioactive compounds from marine bacterial communities, since it is extremely hard to isolate and cultivate symbiotic bacteria of marine macroorganisms, e.g., sponges that has been recently indicated as promising source of novel compounds, in particular as anticancer, by a large body of literature (Schirmer et al., 2005; Kennedy et al., 2007).
Compounds from sequence-based screening
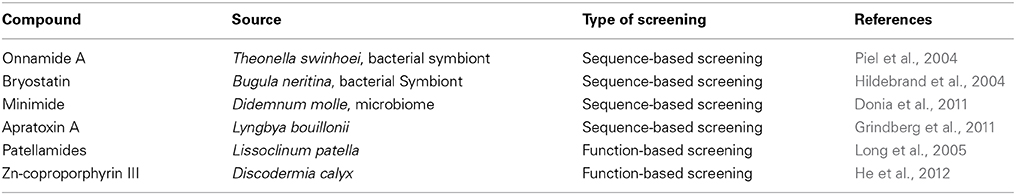
The continuous progress in sequencing technology (e.g., pyrosequencing), the bioinformatic tools and the acquired information on bacterial gene clusters that produce natural compounds, such as the Non-Ribosomial Peptide Synthases (NRPS), and the Polyketide synthases (PKSs) (Fischbach and Walsh, 2006; Fieseler et al., 2007; Hochmuth and Piel, 2009) contributed in making the sequence-based screening a valid approach for the drug-discovery of novel bioactive compounds (Table 2). A first success of this approach was in 2002 by using beetles (Piel, 2002), and it gave the input to perform metagenomic on the marine sponges. The first work employing this strategy on sponges dates back 2004 and were performed again by Piel et al. (2004). They isolated and identified several putative PKS clusters from a highly complex metagenome of the marine sponge Theonella swinhoei. The total DNA was extracted, cloned in cosmids and the library was screened by using appropriate PCR primers. With this strategy, the authors isolated the PKS and NRPS clusters responsible for the production of onnamides and theopederins, a group of polyketides with clear antitumoral activity. The isolated genes resulted belonging to bacterial symbionts, due to the absence of introns and the presence of Shine-Dalgarno sequences. Between 2004 and 2007 the Haygood research group was able to identify, through metagenomic approach, the putative bryostatin PKS gene cluster. In a previous work (Davidson et al., 2001), the researchers identified a 300 bp fragments of a β-ketoacyl-synthase (KSa) involved in the production of bryostatin, that was used as a probe for the screening of a metagenomic library enriched with DNA of Candidatus Endobugula sertula, a bacterial symbiont of the marine bryozoans Bugula neritina. This screening led to the identification of a 65 kb cluster responsible for the bryostatin production (Hildebrand et al., 2004; Sudek et al., 2007).
Donia and co-workers showed the validity of metagenomics also for ribosomal peptides. In their work (Donia et al., 2011), they described the isolation and identification of a novel cyanobactin peptide that was called “minimide” from environmental DNA extracted from dotting colonies of Didemnum molle, an ubiquitous ascidian that inhabits diverse tropical marine habitats. The biosynthetic pathway was isolated by PCR screening, identified, cloned and the optimization of the recombinant expression in E. coli was FINALLY performed.
The recent advances of the genomics contributed to provide novel tools for developing of new metagenomic strategies. The single cell-genomics helped to reduce the metagenomic complexity. A single cell can now be isolated from complex microbial mixtures and the genome amplified for sequencing or PCR screening (Kvist et al., 2007). This approach, combined with metagenomic screening, led the isolation apr gene cluster that proved to be responsible for the biosynthesis of the antitumor natural product apratoxin A (Grindberg et al., 2011).
Compounds from function-based screening
The principal advantage of the function-based screening is that it does not require information regarding the biosynthetic origin of the compounds.
The functional approach has proven its validity thank to the work of Long and collaborators in 2005 (Table 2). In this study, a metagenomic BAC library was obtained from bacterial DNA extracted from the ascidian Lissoclinum patella. The library was then screened searching for clones producing patellamides compounds. The authors identified the recombinant-producing clones and performed the optimization of the heterologous expression in E. coli (Long et al., 2005). In a similar way, a Japanese group in 2012 developed a rapid and efficient functional screening for the detection of natural compounds. The 250.000-fosmid library, prepared using microbiome DNA from marine sponge Discodermia calyx, was rapidly screened on agar plates using a color selection to identify red E. coli clones that indicated the production of phorphyrin. These procedures led the isolation two positive clones that were then cultured in large scale and analyzed. The red pigments were then isolated and structurally elucidated (He et al., 2012). Despite these two successes, the functional screenings displayed several drawbacks, among the others the poor expression of interesting genes in heterologous host strains. However, this problem was addressed by the use of more suitable bacterial strains belonging to Streptomyces, Pseudomonas, and Bacillus genera (Ekkers et al., 2012) or by setting up of specific heterologous expression systems in E. coli (Yuzawa et al., 2012). Other liability of this strategy regards the few number of positive clones obtained in a screening and the missing of an easy and quick screening techniques for analyzing a huge amount of samples. However, there is an incessant development of new screening strategies (Ballestriero et al., 2010; Owen et al., 2012) as well as new approaches to improve the effectiveness of screenings (Penesyan et al., 2013), that will make functional screening more feasible in the future.
Conclusion
In this paper, we highlighted the importance of metagenomics for marine bioprospecting. Metagenomics allows the study and the biotechnological implementation of the marine biodiversity, proved by the huge number of novel biocatalysts and compounds discovered in the last 10 years. It is expected that metagenomics will acquire further interest, despite improvements of culturing techniques. Our suggestion is validated by the rapid advancement in the methods and tools, especially in sequencing and bioinformatic analysis.
As evidence of the metagenomics strength and validity, currently new interdisciplinary projects, aiming at the marine ecosystem access from a biotechnology point of view, have been funded in the EU FP7. Many of these projects are focused on the identification of new marine microbial strains from extreme environments to discovery novel products in the following industry sectors: health, personal care and nutrition. So far, the interest in the exploitation of the marine environment is still growing, as demonstrated by the large number of EU calls recently launched over the EU Horizon 2020 Framework.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the EU FP7-KBBE 2012-2016 project PharmaSea: Increasing Value and Flow in the Marine Biodiscovery Pipeline, grant N° 312184.
References
Abida, H., Ruchaud, S., Rios, L., Humeau, A., Probert, I., De Vargas, C., et al. (2013). Bioprospecting marine plankton. Mar. Drugs 11, 4594–4611. doi: 10.3390/md11114594
Arrieta, J. M., Arnaud-Haond, S., and Duarte, C. M. (2010). What lies underneath: conserving the oceans' genetic resources. Proc. Natl. Acad. Sci. U.S.A. 107, 18318–18324. doi: 10.1073/pnas.0911897107
Asolkar, R. N., Freel, K. C., Jensen, P. R., Fenical, W., Kondratyuk, T. P., Park, E. J., et al. (2009). Arenamides A-C, cytotoxic NFkappaB inhibitors from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 72, 396–402. doi: 10.1021/np800617a
Ballestriero, F., Thomas, T., Burke, C., Egan, S., and Kjelleberg, S. (2010). Identification of compounds with bioactivity against the nematode Caenorhabditis elegans by a screen based on the functional genomics of the marine bacterium Pseudoalteromonas tunicata D2. Appl. Environ. Microbiol. 76, 5710–5717. doi: 10.1128/AEM.00695-10
Bruins, M. E., Janssen, A. E., and Boom, R. M. (2001). Thermozymes and their applications: a review of recent literature and patents. Appl. Biochem. Biotechnol. 90, 155–186. doi: 10.1385/ABAB:90:2:155
Cavicchioli, R., Charlton, T., Ertan, H., Mohd Omar, S., Siddiqui, K. S., and Williams, T. J. (2011). Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 4, 449–460. doi: 10.1111/j.1751-7915.2011.00258.x
Cheng, S. Y., Hsue, C. S., Hwang, G. H., Tsai, L. C., and Pei, S. C. (2010). Hourly oral misoprostol administration for terminating midtrimester pregnancies: a pilot study. Taiwan. J. Obstet. Gynecol. 49, 438–441. doi: 10.1016/S1028-4559(10)60095-2
Cipolla, A., Delbrassine, F., Da Lage, J. L., and Feller, G. (2012). Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie 94, 1943–1950. doi: 10.1016/j.biochi.2012.05.013
Davidson, S. K., Allen, S. W., Lim, G. E., Anderson, C. M., and Haygood, M. G. (2001). Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 67, 4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001
De Pascale, D., De Santi, C., Fu, J., and Landfald, B. (2012). The microbial diversity of Polar environments is a fertile ground for bioprospecting. Mar. Genomics 8, 15–22. doi: 10.1016/j.margen.2012.04.004
Donia, M. S., Ruffner, D. E., Cao, S., and Schmidt, E. W. (2011). Accessing the hidden majority of marine natural products through metagenomics. Chembiochem 12, 1230–1236. doi: 10.1002/cbic.201000780
Dos Santos, A. O., Britta, E. A., Bianco, E. M., Ueda-Nakamura, T., Filho, B. P., Pereira, R. C., et al. (2011). 4-Acetoxydolastane diterpene from the Brazilian brown alga Canistrocarpus cervicornis as antileishmanial agent. Mar. Drugs 9, 2369–2383. doi: 10.3390/md9112369
Ekkers, D. M., Cretoiu, M. S., Kielak, A. M., and Elsas, J. D. (2012). The great screen anomaly–a new frontier in product discovery through functional metagenomics. Appl. Microbiol. Biotechnol. 93, 1005–1020. doi: 10.1007/s00253-011-3804-3
Fang, Z. M., Li, T. L., Chang, F., Zhou, P., Fang, W., Hong, Y. Z., et al. (2012). A new marine bacterial laccase with chloride-enhancing, alkaline-dependent activity and dye decolorization ability. Bioresour. Technol. 111, 36–41. doi: 10.1016/j.biortech.2012.01.172
Felczykowska, A., Bloch, S. K., Nejman-Falenczyk, B., and Baranska, S. (2012). Metagenomic approach in the investigation of new bioactive compounds in the marine environment. Acta Biochim. Pol. 59, 501–505.
Fieseler, L., Hentschel, U., Grozdanov, L., Schirmer, A., Wen, G., Platzer, M., et al. (2007). Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl. Environ. Microbiol. 73, 2144–2155. doi: 10.1128/AEM.02260-06
Fischbach, M. A., and Walsh, C. T. (2006). Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496. doi: 10.1021/cr0503097
Florczak, T., Daroch, M., Wilkinson, M. C., Bialkowska, A., Bates, A. D., Turkiewicz, M., et al. (2013). Purification, characterisation and expression in Saccharomyces cerevisiae of LipG7 an enantioselective, cold-adapted lipase from the Antarctic filamentous fungus Geomyces sp. P7 with unusual thermostability characteristics. Enzyme Microb. Technol. 53, 18–24. doi: 10.1016/j.enzmictec.2013.03.021
Fu, J., Leiros, H. K., De Pascale, D., Johnson, K. A., Blencke, H. M., and Landfald, B. (2013). Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 97, 3965–3978. doi: 10.1007/s00253-012-4276-9
Grindberg, R. V., Ishoey, T., Brinza, D., Esquenazi, E., Coates, R. C., Liu, W. T., et al. (2011). Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE 6:e18565. doi: 10.1371/journal.pone.0018565
Handelsman, J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. doi: 10.1128/MMBR.68.4.669-685.2004
He, R., Wakimoto, T., Takeshige, Y., Egami, Y., Kenmoku, H., Ito, T., et al. (2012). Porphyrins from a metagenomic library of the marine sponge Discodermia calyx. Mol. Biosyst. 8, 2334–2338. doi: 10.1039/c2mb25169h
Hildebrand, M., Waggoner, L. E., Liu, H., Sudek, S., Allen, S., Anderson, C., et al. (2004). bryA: an unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem. Biol. 11, 1543–1552. doi: 10.1016/j.chembiol.2004.08.018
Hochmuth, T., and Piel, J. (2009). Polyketide synthases of bacterial symbionts in sponges–evolution-based applications in natural products research. Phytochemistry 70, 1841–1849. doi: 10.1016/j.phytochem.2009.04.010
Huete-Perez, J. A., and Quezada, F. (2013). Genomic approaches in marine biodiversity and aquaculture. Biol. Res. 46, 353–361. doi: 10.4067/S0716-97602013000400007
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Imhoff, J. F., Labes, A., and Wiese, J. (2011). Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol. Adv. 29, 468–482. doi: 10.1016/j.biotechadv.2011.03.001
Jeon, J. H., Kim, J. T., Kang, S. G., Lee, J. H., and Kim, S. J. (2009a). Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar. Biotechnol. 11, 307–316. doi: 10.1007/s10126-008-9145-2
Jeon, J. H., Kim, J. T., Kim, Y. J., Kim, H. K., Lee, H. S., Kang, S. G., et al. (2009b). Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 81, 865–874. doi: 10.1007/s00253-008-1656-2
Jeon, J. H., Lee, H. S., Kim, J. T., Kim, S. J., Choi, S. H., Kang, S. G., et al. (2012). Identification of a new subfamily of salt-tolerant esterases from a metagenomic library of tidal flat sediment. Appl. Microbiol. Biotechnol. 93, 623–631. doi: 10.1007/s00253-011-3433-x
Jia, B., Xuan, L., Cai, K., Hu, Z., Ma, L., and Wei, C. (2013). NeSSM: a next-generation sequencing simulator for metagenomics. PLoS ONE 8:e75448. doi: 10.1371/journal.pone.0075448
Jiang, C., Wu, L. L., Zhao, G. C., Shen, P. H., Jin, K., Hao, Z. Y., et al. (2010). Identification and characterization of a novel fumarase gene by metagenome expression cloning from marine microorganisms. Microb. Cell Fact. 9, 91. doi: 10.1186/1475-2859-9-91
Karan, R., Capes, M. D., and Dassarma, S. (2012). Function and biotechnology of extremophilic enzymes in low water activity. Aquat. Biosyst. 8, 4. doi: 10.1186/2046-9063-8-4
Kennedy, J., Marchesi, J. R., and Dobson, A. D. (2007). Metagenomic approaches to exploit the biotechnological potential of the microbial consortia of marine sponges. Appl. Microbiol. Biotechnol. 75, 11–20. doi: 10.1007/s00253-007-0875-2
Kennedy, J., O'Leary, N. D., Kiran, G. S., Morrissey, J. P., O'Gara, F., Selvin, J., et al. (2011). Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 111, 787–799. doi: 10.1111/j.1365-2672.2011.05106.x
Kvist, T., Ahring, B. K., Lasken, R. S., and Westermann, P. (2007). Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl. Microbiol. Biotechnol. 74, 926–935. doi: 10.1007/s00253-006-0725-7
Lee, M. H., Oh, K. H., Kang, C. H., Kim, J. H., Oh, T. K., Ryu, C. M., et al. (2012). Novel metagenome-derived, cold-adapted alkaline phospholipase with superior lipase activity as an intermediate between phospholipase and lipase. Appl. Environ. Microbiol. 78, 4959–4966. doi: 10.1128/AEM.00260-12
Liszka, M. J., Clark, M. E., Schneider, E., and Clark, D. S. (2012). Nature versus nurture: developing enzymes that function under extreme conditions. Annu. Rev. Chem. Biomol. Eng. 3, 77–102. doi: 10.1146/annurev-chembioeng-061010-114239
Long, P. F., Dunlap, W. C., Battershill, C. N., and Jaspars, M. (2005). Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem 6, 1760–1765. doi: 10.1002/cbic.200500210
Mayer, A. M., Rodriguez, A. D., Taglialatela-Scafati, O., and Fusetani, N. (2013). Marine pharmacology in 2009-2011: marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 11, 2510–2573. doi: 10.3390/md11072510
Metzker, M. L. (2010). Sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Mohamed, Y. M., Ghazy, M. A., Sayed, A., Ouf, A., El-Dorry, H., and Siam, R. (2013). Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea brine pool. Sci. Rep. 3:3358. doi: 10.1038/srep03358
Nishimura, S., Arita, Y., Honda, M., Iwamoto, K., Matsuyama, A., Shirai, A., et al. (2010). Marine antifungal theonellamides target 3beta-hydroxysterol to activate Rho1 signaling. Nat. Chem. Biol. 6, 519–526. doi: 10.1038/nchembio.387
Owen, J. G., Robins, K. J., Parachin, N. S., and Ackerley, D. F. (2012). A functional screen for recovery of 4'-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ. Microbiol. 14, 1198–1209. doi: 10.1111/j.1462-2920.2012.02699.x
Pace, N. R. (2009). Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73, 565–576. doi: 10.1128/MMBR.00033-09
Penesyan, A., Ballestriero, F., Daim, M., Kjelleberg, S., Thomas, T., and Egan, S. (2013). Assessing the effectiveness of functional genetic screens for the identification of bioactive metabolites. Mar. Drugs 11, 40–49. doi: 10.3390/md11010040
Piel, J. (2002). A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. U.S.A. 99, 14002–14007. doi: 10.1073/pnas.222481399
Piel, J., Hui, D., Wen, G., Butzke, D., Platzer, M., Fusetani, N., et al. (2004). Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. U.S.A. 101, 16222–16227. doi: 10.1073/pnas.0405976101
Plaza, A., Keffer, J. L., Lloyd, J. R., Colin, P. L., and Bewley, C. A. (2010). Paltolides A–C, anabaenopeptin-type peptides from the palau sponge Theonella swinhoei. J. Nat. Prod. 73, 485–488. doi: 10.1021/np900728x
Sakurada, T., Gill, M. B., Frausto, S., Copits, B., Noguchi, K., Shimamoto, K., et al. (2010). Novel N-methylated 8-oxoisoguanines from Pacific sponges with diverse neuroactivities. J. Med. Chem. 53, 6089–6099. doi: 10.1021/jm100490m
Sayed, A., Ghazy, M. A., Ferreira, A. J., Setubal, J. C., Chambergo, F. S., Ouf, A., et al. (2014). A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red Sea. J. Biol. Chem. 289, 1675–1687. doi: 10.1074/jbc.M113.493429
Schirmer, A., Gadkari, R., Reeves, C. D., Ibrahim, F., Delong, E. F., and Hutchinson, C. R. (2005). Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71, 4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005
Schroder, C., Elleuche, S., Blank, S., and Antranikian, G. (2014). Characterization of a heat-active archaeal beta-glucosidase from a hydrothermal spring metagenome. Enzyme Microb. Technol. 57, 48–54. doi: 10.1016/j.enzmictec.2014.01.010
Shendure, J., and Ji, H. (2008). Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145. doi: 10.1038/nbt1486
Simon, C., and Daniel, R. (2009). Achievements and new knowledge unraveled by metagenomic approaches. Appl. Microbiol. Biotechnol. 85, 265–276. doi: 10.1007/s00253-009-2233-z
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
Sudek, S., Lopanik, N. B., Waggoner, L. E., Hildebrand, M., Anderson, C., Liu, H., et al. (2007). Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula,” the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70, 67–74. doi: 10.1021/np060361d
Teasdale, M. E., Liu, J., Wallace, J., Akhlaghi, F., and Rowley, D. C. (2009). Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl. Environ. Microbiol. 75, 567–572. doi: 10.1128/AEM.00632-08
Tirawongsaroj, P., Sriprang, R., Harnpicharnchai, P., Thongaram, T., Champreda, V., Tanapongpipat, S., et al. (2008). Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 133, 42–49. doi: 10.1016/j.jbiotec.2007.08.046
Tyson, G. W., and Banfield, J. F. (2005). Cultivating the uncultivated: a community genomics perspective. Trends Microbiol. 13, 411–415. doi: 10.1016/j.tim.2005.07.003
Wang, H., Gong, Y., Xie, W., Xiao, W., Wang, J., Zheng, Y., et al. (2011). Identification and characterization of a novel thermostable gh-57 gene from metagenomic fosmid library of the Juan de Fuca Ridge hydrothemal vent. Appl. Biochem. Biotechnol. 164, 1323–1338. doi: 10.1007/s12010-011-9215-1
Wierzbicka-Wos, A., Bartasun, P., Cieslinski, H., and Kur, J. (2013). Cloning and characterization of a novel cold-active glycoside hydrolase family 1 enzyme with beta-glucosidase, beta-fucosidase and beta-galactosidase activities. BMC Biotechnol. 13:22. doi: 10.1186/1472-6750-13-22
Wong, D. (2010). “Applications of metagenomics for industrial bioproducts,” in Metagenomics: Theory, Methods and Applications. 1st Edn. ed D. Marco (Norwich, UK: Horizon Scientific Press), 141–158.
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46. doi: 10.1186/gb-2014-15-3-r46
Xu, M., Fujita, D., and Hanagata, N. (2009). Perspectives and challenges of emerging single-molecule DNA sequencing technologies. Small 5, 2638–2649. doi: 10.1002/smll.200900976
Yuzawa, S., Kim, W., Katz, L., and Keasling, J. D. (2012). Heterologous production of polyketides by modular type I polyketide synthases in Escherichia coli. Curr. Opin. Biotechnol. 23, 727–735. doi: 10.1016/j.copbio.2011.12.029
Zhao, X. Q. (2011). Genome-based studies of marine microorganisms to maximize the diversity of natural products discovery for medical treatments. Evid. Based Complement. Alternat. Med. 2011:384572. doi: 10.1155/2011/384572
Zhou, Q., Su, X., Jing, G., and Ning, K. (2014). Meta-QC-Chain: comprehensive and fast quality control method for metagenomic data. Genomics Proteomics Bioinformatics 12, 52–56. doi: 10.1016/j.gpb.2014.01.002
Keywords: marine environments, metagenomics, biocatalysts, bioactive compounds, biotechnological applications
Citation: Barone R, De Santi C, Palma Esposito F, Tedesco P, Galati F, Visone M, Di Scala A and De Pascale D (2014) Marine metagenomics, a valuable tool for enzymes and bioactive compounds discovery. Front. Mar. Sci. 1:38. doi: 10.3389/fmars.2014.00038
Received: 28 April 2014; Accepted: 20 August 2014;
Published online: 04 September 2014.
Edited by:
Conxita Avila, Universitat de Barcelona, SpainReviewed by:
Susanna López-Legentil, University of North Carolina Wilmington, USAAna Riesgo, University of Barcelona, Spain
Copyright © 2014 Barone, De Santi, Palma Esposito, Tedesco, Galati, Visone, Di Scala and De Pascale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donatella De Pascale, Institute of Protein Biochemistry, National Research Council, Via P. Castellino, 111, Naples, I-80131, Italy e-mail:ZC5kZXBhc2NhbGVAaWJwLmNuci5pdA==
†These authors have contributed equally to this work.
 Rosalba Barone†
Rosalba Barone† Pietro Tedesco
Pietro Tedesco Donatella De Pascale
Donatella De Pascale