Human-induced environmental changes have been linked directly with loss of biodiversity. Coral reefs, which have been severely impacted by anthropogenic activities over the last few decades, exemplify this global problem and provide an opportunity to develop research addressing key knowledge gaps through “omics”-based approaches. While many stressors, e.g., global warming, ocean acidification, overfishing, and coastal development have been identified, there is an urgent need to understand how corals function at a basic level in order to conceive strategies for mitigating future reef loss. In this regard, availability of fully sequenced genomes has been immensely valuable in providing answers to questions of organismal biology. Given that corals are metaorganisms comprised of the coral animal host, its intracellular photosynthetic algae, and associated microbiota (i.e., bacteria, archaea, fungi, viruses), these efforts must focus on entire coral holobionts. The Reef Future Genomics 2020 (ReFuGe 2020) Consortium has formed to sequence hologenomes of 10 coral species representing different physiological or functional groups to provide foundation data for coral reef adaptation research that is freely available to the research community.
Overview
Scleractinian or stony corals are a foundation species of reef ecosystems, as their carbonate skeletons provide the structural habitat complexity necessary to maintain millions of vertebrate and invertebrate marine organisms (Reaka-Kudla, 1997). Over 500 million people and billion-dollar industries, including fisheries and tourism, depend on healthy reef ecosystems (Cesar, 2002). Increases in atmospheric CO2 concentrations place reef-building corals at risk due to potential rises in water temperatures and ocean acidity. While research focusing on adaptability and response of corals to “future ocean” conditions is steadily increasing (Kleypas et al., 2006; Hoegh-Guldberg et al., 2007a; Császár et al., 2010; Hofmann et al., 2010; Iguchi et al., 2011; Voolstra et al., 2011; Sawall et al., 2015), these studies typically focus on ecosystem scale consequences of environmental change or study the effect on distinct coral compartments.
Relatively few studies target the entire coral holobiont (i.e., the coral metaorganism consisting of the coral animal host, its intracellular algae, and other microbiota) as the functional unit, potentially missing important interactions among members of the holobiont association that contribute to stress tolerance. However, a growing number of studies are now emphasizing the validity of the metaorganism concept (McFall-Ngai et al., 2013), with functional studies elucidating these fundamental interactions also in aquatic and marine organisms (Bolnick et al., 2014; Franchini et al., 2014; McFall-Ngai, 2014). These studies commonly focus on bacterial-host interactions and demonstrate how bacteria fundamentally alter host organism development, ecology, and evolution (Bosch and McFall-Ngai, 2011). An exception is the study of coral-algal symbioses. Symbiodinium sp., the photosynthetic algal symbiont of reef-building corals was formally described in 1962 (Freudenthal, 1962), and the partnership between the coral animal host and its algal symbiont has long been recognized to provide the foundation of coral reef ecosystems. Whereas the algal symbionts provide energy to their hosts in the form of photosynthates, the coral animal provides a sheltered, light-rich environment and inorganic nutrients (Muscatine and Cernichiari, 1969; Falkowski et al., 1984). Currently, there is an urgent requirement for understanding coral resilience and their capacity to adapt in a holobiont framework to provide threshold values for coral reef stewardship (sensu Steffen et al., 2015). The long history of research detailing the intricacies of the coral-algal symbiosis and more recent studies on bacterial interactions can now provide the building blocks to understand coral metaorganism function using the new suite of genomic-based approaches.
Although we focus here primarily on coral species from Australia's Great Barrier Reef, our considerations are likely to apply to all coral reefs. We use the term “resilience” in reference to the capacity of an ecosystem or organism to absorb and recover from a perturbation (e.g., toxic agricultural runoffs or temperature spikes) and “adaptability” in reference to the capacity for a lineage or species to adjust to (rapidly) changing circumstances (e.g., a higher average ambient temperature), including potential changes to the composition of the ecosystem. In this context, resilience and adaptation can result in the ecosystem maintaining critical functions and services.
Rather than defining a set of “critical” species to investigate, we define a set of signature genera that represent different physiological or functional coral groups that together form the core of the ecosystem. We acknowledge that coral reefs are highly complex systems in which mechanisms of resilience may not be obvious (e.g., Bellwood et al., 2006), but we also believe that modern genomic approaches may provide equally important insight to the ability of corals to respond to climate change (e.g., Shinzato et al., 2011). We provide an overview of how contemporary genomic tools can be used to provide information pertinent to better understanding how corals function at a basic level, which we expect to be helpful in informing management and conservation of coral reefs. Specifically, we highlight how such tools can be used to explore genetic diversity within and among these species, the genetic basis of relevant traits and their patterns of inheritance, and the dynamic changes that occur during acclimation and adaptation.
This article is structured into four parts: (1) strategic considerations of the composition of complex ecosystems and selection of core functional coral groups; (2) insights from pioneering studies on adaptation in Drosophila, stickleback fish and other species groups that might inform similar research on corals; (3) description of coral ecosystems and the threats arising from environmental change; and (4) a case study on genomic approaches to understand adaptability of corals.
Strategic Considerations and Approach
Environmental change may intensify selective pressures acting on organisms. Although our overall concern is the resilience of a given ecosystem as a whole, it is more practical to consider the responses of individual component species, especially those that play key roles in the ecosystem. Understanding which species can or cannot cope will provide a baseline (i.e., which species might be expected to persist) and constrain models of ecosystem function that depend on the sum of individual responses and the synergies among them. “Foundation species”, those that provide habitat for others and simultaneously facilitate higher diversity, are increasingly recognized as significant factors at the ecosystem level (Bruno et al., 2003). The heritable effects of genes in these foundation species extend to higher levels, contributing to ecosystem phenotypes that other species may rely on (Whitham et al., 2006). In the example at hand, the coral is a metaorganism consisting of the coral animal, photosynthetic dinoflagellate symbionts, and species-specific assemblages of microorganisms, collectively referred to as the coral holobiont (Rohwer et al., 2002). Population and quantitative genetic analyses of the coral holobiont allow us to elucidate heritable components of ecosystem phenotypes and provide an evolutionary framework in which to understand the effect of climate change on organisms that in turn affect ecosystem processes. In this regard, it is important to incorporate knowledge about metaorganism function from ecological and evolutionary studies, since an important aspect of coral resilience might lie in understanding how the coral-algae-microbe symbioses co-evolved and how this in turn connects genomes and phenotypes (Bosch and McFall-Ngai, 2011). In other words, an increased understanding of the evolutionary forces that shape metaorganisms and their hologenomes will help to assist and mitigate detrimental effects of environmental change on corals and their reef ecosystems, and support efforts of building coral resilience through assisted evolution (van Oppen et al., 2015).
The primary question is how to identify the species that are likely to be most informative for assessing resilience and adaptability of the ecosystem in question, and thus should be prioritized for study. We posit that there exist certain core coral genera that reflect distinct physiological groups, which should become the initial focus, but will be expanded as the ability to gather data advances. Given the above, the criteria for selection should be (i) genera or species that occur (or have very close relatives) in equivalent ecosystems around the world, and are thus generally typical and representative of that type of ecosystem; and (ii) among these, species that are amenable to experimental assessment and manipulation in enclosed environments (i.e., aquaria). Given the ongoing advance of sequencing technology, selecting species with relatively small genomes (for efficiency of assembly and analysis) is not a primary criterion, but might be considered downstream in the selection process. Compromises will have to be made, and known and unknown blind spots accepted, if we are to make a start and not be inhibited by the scope of the problem and our current ignorance.
For the experimental component, we propose a three-pronged approach: (i) identification of genomic, epigenomic, and transcriptomic variation across natural clines of the environmental factor of interest; (ii) experimental assessment of the ability of variants from each end of the clines to withstand a controlled progression to the other extreme and determine how time-sensitive and reversible this response may be; and (iii) genome-scale sequencing to examine the genomic, epigenomic, and transcriptomic changes that may have occurred as part of an adaptive response and how they correspond to the variation that is observed naturally. In making these suggestions, we note the enormous recent increase in DNA-sequencing capacity, and the corresponding reduction in cost, that make large-scale surveys increasingly feasible. We suggest that any strategy must be tractable, yet scalable to benefit from ever-improving sequencing technologies. Further, although there are many sequencing projects underway, the concerted work of a group of researchers will facilitate coordination, and thus helps to streamline and channel sequencing efforts. For instance, the Global Invertebrate Genomics Alliance (GIGA) was formed to coordinate current efforts generating and analyzing invertebrate non-arthropod genomic data (GIGA Community of Scientists, 2014). This becomes especially important in conducting down-stream comparative analyses and providing a data infrastructure that allows easy access and retrieval of genomic baseline data.
The Genetics of Adaptation: Insights from the Fruit Fly Drosophila and Other Model Systems
Studies on various other species groups including the fruit fly Drosophila, the rockcress Arabidopsis, stickleback fish Gasterosteus, and other model organisms have been particularly informative in understanding the heritability of different traits and the response of different species to environmental change and selection at the genetic and genomic levels. The Drosophila work was initiated in D. melanogaster and has focused on environmental gradients, particularly latitudinal climatic gradients (see Schmidt et al., 2005; Hoffmann and Weeks, 2007). Genetic analyses along gradients have been used to identify genes and genetic polymorphisms that appear involved in evolutionary adaptation to different climates (e.g., Schmidt et al., 2005; Paaby et al., 2010; Telonis-Scott et al., 2011). Some of these polymorphisms are shifting in response to recent climate change (Umina et al., 2005). Regions of the genome that have diverged between the ends of climatic gradients have been identified (González et al., 2010; Fabian et al., 2012), and transcriptomes from gradient ends have also been compared (Chen et al., 2012). Environmental gradients have been used to link adaptation to genetic polymorphisms in many other species; notable examples include polymorphisms affecting salinity in Gasterosteus (Barrett et al., 2008) or flowering time in Arabidopsis (Caicedo et al., 2004).
Although great progress has been made in understanding the genetic basis of adaptive variation in Drosophila and other organisms, it remains difficult to make direct connections between genetic polymorphisms and traits associated with climate adaptation (Chung et al., 2014). The effects of candidate genetic polymorphisms identified from different types of experiments and/or clinal analysis need to be tested on randomized genetic backgrounds to assess their impact on traits that vary along gradients (Lee et al., 2011). This is facilitated by tools available in model organisms, including D. melanogaster, which allow the expression of specific genes to be modified, and effects of polymorphisms to be assessed in controlled backgrounds. A combination of approaches has allowed the adaptive significance of particular genetic changes to be identified: for instance, expression of the ebony gene affects body pigmentation (Pool and Aquadro, 2007; Telonis-Scott et al., 2011), which in turn may influence fitness under different climatic conditions (Parkash et al., 2008) and shows clinal patterns (Telonis-Scott et al., 2011); a set of polymorphisms in the regulatory region influences expression of this gene (Takahashi and Takano-Shimizu, 2011).
The genus Drosophila itself and related genera contain an enormous range of species adapted to different climatic regions. It is now possible to map traits that influence climate adaptation onto Drosophila phylogeny to develop a detailed understanding of limits to climate adaptation, controlling for environmental effects by rearing the species in a common environment (Strachan et al., 2011; Kellermann et al., 2012). Clear links have been established between traits and species distributions including resistance to desiccation, cold, and heat. There is strong phylogenetic signature in these traits, and clades can be identified that seem to lack evolutionary potential because all the related species show a similar stress response (e.g., desiccation resistance in D. birchii and other species from wet tropical environments (Hoffmann et al., 2003; Kellermann et al., 2009). To understand why trait evolution might be limited within particular species and clades, candidate genes and genetic processes are being mapped onto phylogenies (Reis et al., 2011). Within Drosophila phylogenies, evolutionary responses to environmental factors appear related to gene duplication and gene loss within particular gene families (Zhong et al., 2013). De-novo evolution of genes seems to be a rich source for adaptive evolution in Drosophila lineages (Chen et al., 2010). While immediate responses to selection under climate change may depend on standing genetic variation, genomic changes might nevertheless contribute to adaptive changes across a few decades (Izutsu et al., 2012). Genetic adaptation will also depend on other factors like patterns of gene flow that can be maladaptive and limit local adaptation (Magiafoglou and Hoffmann, 2003) and/or maternal effects that can result in the transmission of stress resistance from mothers to offspring (Jenkins and Hoffmann, 1994).
Coral Reefs and Environmental Change
Phylogenetic constraints outlined in species of Drosophila are likely to occur in many other species groups, particularly for upper thermal limits (Araújo et al., 2013). This is of particular significance as most corals live close to their thermal tolerance limits, and hence may be severely affected by global warming and increasing sea surface temperatures (SST). To better understand the consequences of environmental change for coral reefs, a closer look at the ecology of coral organisms is needed.
Over the past century, coral reefs have been in global decline—more than 50% of extant reefs are severely degraded and most of the remainder are under serious threat (Hoegh-Guldberg, 1999). The causes of reef loss are varied and complex, but a major current concern is that global changes in climate may drive increasingly more frequent and widespread coral bleaching events and thus exacerbate reef loss (Hoegh-Guldberg, 1999). Reefs flourished in pre-industrial times, when the pH of the ocean was around 8.1, but the advent of the industrial period has led to increasing atmospheric CO2, and the oceans have become both warmer and more acidic. Average SSTs in the tropical oceans have increased by around 0.6°C over the past century, and much greater changes are forecast to occur in the twenty-first century if present rates of emission continue (Bindoff et al., 2007). Mass coral bleaching events, resulting in widespread mortality, are a consequence of thermal anomalies that are likely to increase in frequency and extent as a function of higher ocean temperatures (Hughes et al., 2003). This effect may be compounded by the decreases in pH that occur as atmospheric CO2 equilibrates with seawater, as calcification becomes increasingly more energetically costly under more acidic conditions (Anthony et al., 2008).
Although corals may be able to adapt to a changing world, the main concern is that they may be unable to do so rapidly enough to keep pace with current rates of change, which are orders of magnitude more rapid than occurred during ice age transitions (Hoegh-Guldberg et al., 2007b). Corals have relatively long generation times (10–100 years), and thus evolve on far longer timescales than, for example, fruit flies. Factors that can potentially mitigate the slow evolutionary rate include high levels of genetic diversity and the large effective population sizes in at least some species. Moreover, the coral holobiont as a metaorganism may evolve much more rapidly than can the coral animal because the symbiotic dinoflagellates and microorganisms associated with corals are themselves highly variable and have short generation times, provided that there is enough flexibility in these associations. Of note, the contribution of phenotypic plasticity in corals, i.e., the development of different phenotypes from a single genotype depending on the environment, might be an important contributor to the response to environmental change, and hence, coral resilience (Todd, 2008). While light and water movement are recognized important variables in determining coral shape and form, it was recently shown that seawater acidification can also cause a shift in coral skeleton morphology, in concert with and probably to evade the consequences of a decrease in coral calcification rates (Tambutté et al., 2015). As such, plastic responses in corals (including all holobiont compartments) might in themselves be adaptive and improve fitness, i.e., growth, reproduction, or survival (Gotthard and Nylin, 1995). Availability of large-scale gene expression data will make it possible to study plasticity from a molecular perspective, e.g., determining how many and which genes vary in expression between different phenotypes of a plastic trait, thereby helping to uncover the cellular mechanism(s) involved (Aubin-Horth and Renn, 2009).
Genetic diversity within a location may also provide scope for selection for change in local thermal tolerance. For instance, recent work on Acropora hyacinthus has shown that within a single site, local acclimatization and adaptation contribute equally to thermal tolerance (Palumbi et al., 2014). Importantly, this was reflected in patterns of gene expression, providing a direction for further studies. However, our understanding of genetic diversity within coral populations is generally at an early stage, leaving a large series of unanswered questions including (i) which species will be most sensitive to the predicted change in sea surface temperature and ocean acidity; (ii) where resistant genotypes might be found for potential use in captive breeding, translocation, or assisted migration programs, and for DNA banks; (iii) what is the natural level of gene flow among populations within a reef system, e.g., along the Great Barrier Reef; (iv) the expected rate of adaptive response, if any, in different species; and (v) how differences in the microbial communities associated with a coral affect its capacity for phenotypic response, and how heritable that might be. We can begin to address these questions within an initial set of signature genera as further outlined below.
Case Study: Genomic Approaches to Assess Adaptability of Corals to Climate Change
Building on insights into the genomic bases of adaptation in model organisms including Drosophila, genome science has the potential to rapidly advance our understanding of the adaptive capacity of reef-building corals (Stapley et al., 2010; Shinzato et al., 2011). While we acknowledge the inherent differences between fruit flies and corals, development and testing of hypotheses in model organisms can act as a springboard to inform coral biology (Baumgarten et al., 2015). Given the urgency of the problem and decreasing costs of sequencing technologies, the authors of this paper have formed the Reef Future Genomics 2020 Consortium (ReFuGe 2020), within which our “Sea-quence” project has identified a framework of molecular datasets that we anticipate will provide novel insights into the adaptive landscape of reef-building corals, their dinoflagellate symbionts, and the associated microbial communities (http://refuge2020.com/).
For the Sea-quence project, we have initially selected 10 species of tropical corals from a diverse set of genera for deep genomic and transcriptomic sequencing of all coral holobiont compartments, i.e., de-novo genome and transcriptome sequencing of the coral host, associated Symbiodinium types, and microbial (including viral) metagenomes and metatranscriptomes for each selected coral species. We included corals that are broadly distributed across environmental gradients, physiologically diverse, and easily identified in the field. While it is impossible to cover the full diversity of the order Scleractinia, we selected species within genera that represent different physiological groups, e.g., thermally susceptible as well as tolerant genera (e.g., Porites and Acropora) (Marshall and Baird, 2000). We also considered the mode of reproduction, extent of existing resources and datasets, ease of collection and cultivation, and diversity and transmission mode of the symbiont. The 10 species are broadly distributed across the Indo-Pacific and occupy a wide range of thermal environments, making it likely that the results will be broadly applicable (Table 1). We anticipate the initial species selection will be expanded in the course of the work of the ReFuGe 2020 Consortium. A current overview is available on the ReFuGe 2020 webpage (http://refuge2020.com/our-research/research-progress). At the time of writing of this manuscript, genomic DNA from several coral holobionts has been sequenced (data are accessible at https://ccgapps.com.au/bpa-metadata/gbr/).
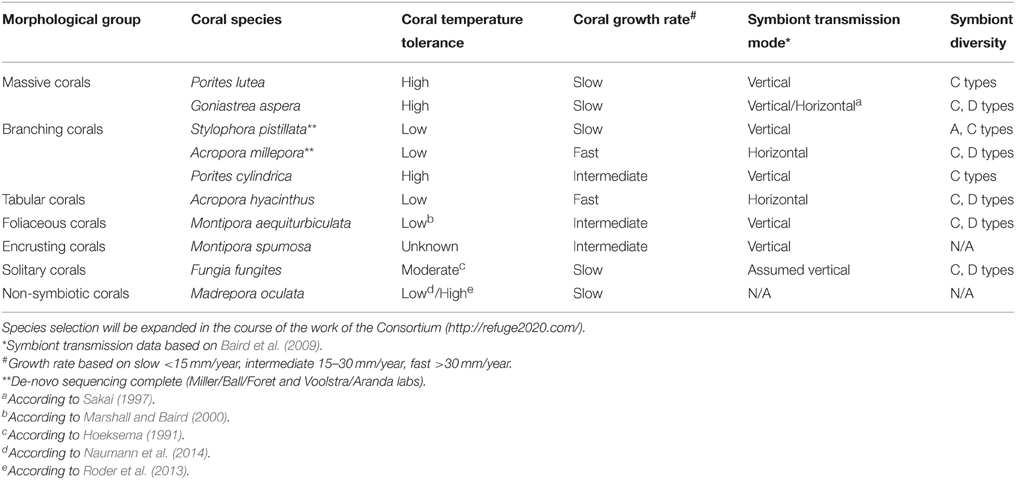
Table 1. Selection of 10 initial coral species and organismal attributes targeted for hologenome sequencing in the ReFuGe 2020 Consortium.
Availability of de-novo reference genome sequences of 10 coral genomes and associated microorganisms will be instrumental in providing answers to basic and specific questions of coral biology. For instance, the provision of annotated holobiont gene sets will allow the elucidation of the metabolic, immunological, and physiological capacities encoded in coral metaorganisms as well as the determination of a core set of orthologous genes that allows deciphering the mechanistic underpinnings of coral biology. The availability of reference genomic resources is invaluable for informing downstream experimental approaches, such as transcriptional, protein, and epigenetic profiling. As described above for Drosophila, we aim to explore genomic, epigenomic, and transcriptomic variation along latitudinal gradients to uncover acclimation and adaptation potential of selected genera. For instance, along the 1000-km north-south gradient of the Great Barrier Reef water temperatures differ by about 2°C. Along this transect, experiments have revealed that corals from the southern Great Barrier Reef are much more susceptible to thermal stress than are those from the northern region (Ulstrup et al., 2006; Cooper et al., 2011). Complementing such data with analyses of selected target or similar species from “extreme” environments (“extreme phenotype sequencing”), e.g., the Red Sea (Arif et al., 2014; Sawall et al., 2014) or Arabian Gulf (Hume et al., 2013, 2015), provides the opportunity to determine how standing genetic variation, phenotypic plasticity, population size, and genomic architecture translate into adaptability of coral organisms and resilience of reef ecosystems. In combination with manipulative experiments, genetic variability can potentially be partitioned into that relating to tolerance to water temperature vs. the many other environmental factors that co-vary with it. Understanding which elements are associated with “climate change tolerant” coral species will present opportunities for management strategies aiming to preserve the resilience of coral reef regions. Generation of holobiont genome reference data is not an end (Richards, 2015), but rather the foundational necessity for research targeting resilience and adaptability of coral holobionts. As such, it substantiates, rather than replaces, other efforts of coral reef research.
Conclusions
Here we have outlined a genomics framework focusing on signature coral genera for which hologenomes will be generated to better understand resilience and adaptability of coral holobionts and by extension reef ecosystems. Under the “Sea-quence” project the ReFuGe 2020 Consortium generates and analyzes a set of genomic reference data to interrogate the adaptive potential of different functional groups. Our approach leverages the enormous scope and capacity of sequencing technology to query genetic diversity and to profile real-time genomic responses to environmental stress. On this road we have adopted lessons from adaptation research in Drosophila and other species. Our approach is an important first step to generate foundation data for coral reef adaptation research, freely available to the research community, to make better predictions about which species may be expected to persist, under what conditions, and why.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anthony, K. R. N., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U.S.A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Araújo, M. B., Ferri-Yáñez, F., Bozinovic, F., Marquet, P. A., Valladares, F., and Chown, S. L. (2013). Heat freezes niche evolution. Ecol. Lett. 16, 1206–1219. doi: 10.1111/ele.12155
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Aubin-Horth, N., and Renn, S. C. (2009). Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 18, 3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x
Baird, A. H., Guest, J. R., and Willis, B. L. (2009). Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evol. Syst. 40, 551–571. doi: 10.1146/annurev.ecolsys.110308.120220
Barrett, R. D. H., Rogers, S. M., and Schluter, D. (2008). Natural selection on a major armor gene in threespine stickleback. Science 322, 255–257. doi: 10.1126/science.1159978
Baumgarten, S., Simakov, O., Esherick, L. Y., Liew, Y. J., Lehnert, E. M., Michell, C. T., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.1513318112. [Epub ahead of print].
Bellwood, D. R., Hughes, T. P., and Hoey, A. S. (2006). Sleeping functional group drives coral-reef recovery. Curr. Biol. 16, 2434–2439. doi: 10.1016/j.cub.2006.10.030
Bindoff, N., Willebrand, J., Artale, V., Cazenave, A., Gregory, J., Gulev, S., et al. (2007). “Observations: oceanic climate change and sea level,” in The Physical Science Basis. Working Group I Contribution to the Intergovernmental Panel on Climate Change Fourth Assessment Report, eds S. Solomon, D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor, and H. L. Miller (Cambridge, UK; New York, NY: Cambridge University Press), 385–432.
Bolnick, D. I., Snowberg, L. K., Hirsch, P. E., Lauber, C. L., Knight, R., Caporaso, J. G., et al. (2014). Individuals' diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 17, 979–987. doi: 10.1111/ele.12301
Bosch, T. C., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology (Jena.) 114, 185–190. doi: 10.1016/j.zool.2011.04.001
Bruno, J. F., Stachowicz, J. J., and Bertness, M. D. (2003). Inclusion of facilitation into ecological theory. Trends Ecol. Evol. (Amst.) 18, 119–125. doi: 10.1016/S0169-5347(02)00045-9
Caicedo, A. L., Stinchcombe, J. R., Olsen, K. M., Schmitt, J., and Purugganan, M. D. (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. U.S.A. 101, 15670–15675. doi: 10.1073/pnas.0406232101
Cesar, H. (2002). “The biodiversity benefits of coral reef ecosystems: values and markets,” in Working Party on Global and Structural Policies Working Group on Economic Aspects of Biodiversity (Paris: OECD).
Chen, S. D., Zhang, Y. E., and Long, M. Y. (2010). New genes in drosophila quickly become essential. Science 330, 1682–1685. doi: 10.1126/science.1196380
Chen, Y., Lee, S. F., Blanc, E., Reuter, C., Wertheim, B., Martinez-Diaz, P., et al. (2012). Genome-wide transcription analysis of clinal genetic variation in drosophila. PLoS ONE 7:e34620. doi: 10.1371/journal.pone.0034620
Chung, H., Loehlin, D. W., Dufour, H. D., Vaccarro, K., Millar, J. G., and Carroll, S. B. (2014). A single gene affects both ecological divergence and mate choice in drosophila. Science 343, 1148–1151. doi: 10.1126/science.1249998
Cooper, T. F., Berkelmans, R., Ulstrup, K. E., Weeks, S., Radford, B., Jones, A. M., et al. (2011). Environmental factors controlling the distribution of symbiodinium harboured by the coral Acropora millepora on the Great Barrier Reef. PLoS ONE 6:e25536. doi: 10.1371/journal.pone.0025536
Császár, N. B. M., Ralph, P. J., Frankham, R., Berkelmans, R., and van Oppen, M. J. H. (2010). Estimating the potential for adaptation of corals to climate warming. PLoS ONE 5:e9751. doi: 10.1371/journal.pone.0009751
Fabian, D. K., Kapun, M., Nolte, V., Kofler, R., Schmidt, P. S., Schlötterer, C., et al. (2012). Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21, 4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and Porter, J. W. (1984). Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709. doi: 10.2307/1309663
Franchini, P., Fruciano, C., Frickey, T., Jones, J. C., and Meyer, A. (2014). The gut microbial community of midas cichlid fish in repeatedly evolved limnetic-benthic species Pairs. PLoS ONE 9:e95027. doi: 10.1371/journal.pone.0095027
Freudenthal, H. D. (1962). Symbiodinium gen. nov. and Symbiodinium microadriaticum sp. nov., a Zooxanthella: Taxonomy, Life Cycle, and Morphology*. J. Protozool. 9, 45–52. doi: 10.1111/j.1550-7408.1962.tb02579.x
GIGA Community of Scientists. (2014). The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. J. Hered. 105, 1–18. doi: 10.1093/jhered/est084
González, J., Karasov, T. L., Messer, P. W., and Petrov, D. A. (2010). Genome-wide patterns of adaptation to temperate environments associated with transposable elements in drosophila. PLoS Genet. 6:e1000905. doi: 10.1371/journal.pgen.1000905
Gotthard, K., and Nylin, S. (1995). Adaptive plasticity and plasticity as an adaptation: a selective review of plasticity in animal morphology and life history. Oikos 74, 3–17. doi: 10.2307/3545669
Hoegh-Guldberg, O. (1999). Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007b). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Hoegh-Guldberg, O., Mumby, P., Hooten, A., Steneck, R., Greenfield, P., Gomez, E., et al. (2007a). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Hoeksema, B. W. (1991). Control of bleaching in mushroom coral populations (Scleractinia: Fungiidae) in the Java Sea: stress tolerance and interference by life history strategy. Mar. Ecol. Prog. Ser. 74, 225–237. doi: 10.3354/meps074225
Hoffmann, A. A., Hallas, R. J., Dean, J. A., and Schiffer, M. (2003). Low potential for climatic stress adaptation in a rainforest Drosophila species. Science 301, 100–102. doi: 10.1126/science.1084296
Hoffmann, A. A., and Weeks, A. R. (2007). Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129, 133–147. doi: 10.1007/s10709-006-9010-z
Hofmann, G. E., Barry, J. P., Edmunds, P. J., Gates, R. D., Hutchins, D. A., Klinger, T., et al. (2010). The effect of ocean acidification on calcifying organisms in marine ecosystems: an organism-to-ecosystem perspective. Annu. Rev. Ecol. Evol. Syst. 41, 127–147. doi: 10.1146/annurev.ecolsys.110308.120227
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Hume, B. C. C., D'Angelo, C., Smith, E. G., Stevens, J. R., Burt, J., and Wiedenmann, J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world's hottest sea, the Persian/Arabian Gulf. Sci. Rep. 5:8562. doi: 10.1038/srep08562
Hume, B., D'Angelo, C., Burt, J., Baker, A. C., Riegl, B., and Wiedenmann, J. (2013). Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar. Pollut. Bull. 72, 313–322. doi: 10.1016/j.marpolbul.2012.11.032
Iguchi, A., Shinzato, C., Forêt, S., and Miller, D. J. (2011). Identification of fast-evolving genes in the Scleractinian Coral Acropora Using Comparative EST Analysis. PLoS ONE 6:e20140. doi: 10.1371/journal.pone.0020140
Izutsu, M., Zhou, J., Sugiyama, Y., Nishimura, O., Aizu, T., Toyoda, A., et al. (2012). Genome Features of “Dark-Fly,” a drosophila line reared long-term in a dark environment. PLoS ONE 7:e33288. doi: 10.1371/journal.pone.0033288
Jenkins, N. L., and Hoffmann, A. A. (1994). Genetic and maternal variation for heat-resistance in Drosophila from the field. Genetics 137, 783–789.
Kellermann, V., Loeschcke, V., Hoffmann, A. A., Kristensen, T. N., Fløjgaard, C., David, J., et al. (2012). Phylogenetic constraints in key functional traits behind species' climate niches: patterns of desiccation and cold resistance across 95 Drosophila species. Evolution 66, 3377–3389. doi: 10.1111/j.1558-5646.2012.01685.x
Kellermann, V., Van Heerwaarden, B., Sgro, C. M., and Hoffmann, A. A. (2009). Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science 325, 1244–1246. doi: 10.1126/science.1175443
Kleypas, J., Feely, R., Fabry, V., Langdon, C., Sabine, C., and Robbins, L. (2006). “Impacts of ocean acidification on coral reefs and other marine calcifiers: a guide for future research,” Report of a Workshop Held 18–20 April 2005 (St. Petersburg, FL,: NSF, NOAA, The US Geological Survey).
Lee, S. F., Chen, Y., Varan, A. K., Wee, C. W., Rako, L., Axford, J. K., et al. (2011). Molecular basis of adaptive shift in body size in Drosophila melanogaster: functional and sequence analyses of the Dca Gene. Mol. Biol. Evol. 28, 2393–2402. doi: 10.1093/molbev/msr064
Magiafoglou, A., and Hoffmann, A. (2003). Thermal adaptation in Drosophila serrata under conditions linked to its southern border: unexpected patterns from laboratory selection suggest limited evolutionary potential. J. Genet. 82, 179–189. doi: 10.1007/BF02715817
Marshall, P., and Baird, H. (2000). Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19, 155–163. doi: 10.1007/s003380000086
McFall-Ngai, M. (2014). Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 12:e1001783. doi: 10.1371/journal.pbio.1001783
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Muscatine, L., and Cernichiari, E. (1969). Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol. Bull. 137, 506–523. doi: 10.2307/1540172
Naumann, M. S., Orejas, C., and Ferrier-Pagès, C. (2014). Species-specific physiological response by the cold-water corals Lophelia pertusa and Madrepora oculata to variations within their natural temperature range. Deep Sea Res. Part II Top. Stud. Oceanogr. 99, 36–41. doi: 10.1016/j.dsr2.2013.05.025
Paaby, A. B., Blacket, M. J., Hoffmann, A. A., and Schmidt, P. S. (2010). Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 19, 760–774. doi: 10.1111/j.1365-294X.2009.04508.x
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., and Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898. doi: 10.1126/science.1251336
Parkash, R., Sharma, V., and Kalra, B. (2008). Climatic adaptations of body melanisation in Drosophila melanogaster from Western Himalayas. Fly 2, 111–117. doi: 10.4161/fly.6351
Pool, J. E., and Aquadro, C. F. (2007). The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol. Ecol. 16, 2844–2851. doi: 10.1111/j.1365-294X.2007.03324.x
Reaka-Kudla, M. L. (1997). “The global biodiversity of coral reefs: A comparison with rainforests,” in Biodiversity II: Understanding and Protecting Our Natural Resources, eds M. L. Reaka-Kudla, D. E. Wilson, and O. E. Wilson (Washington, DC: Joseph Henry/National Academy Press), 83–103.
Reis, M., Vieira, C. P., Morales-Hojas, R., Aguiar, B., Rocha, H., Schlotterer, C., et al. (2011). A comparative study of the short term cold resistance response in distantly related Drosophila species: the role of regucalcin and frost. PLoS ONE 6:e25520. doi: 10.1371/journal.pone.0025520
Richards, S. (2015). It's more than stamp collecting: how genome sequencing can unify biological research. Trends Genet. 31, 411–421. doi: 10.1016/j.tig.2015.04.007
Roder, C., Berumen, M. L., Bouwmeester, J., Papathanassiou, E., Al-Suwailem, A., and Voolstra, C. R. (2013). First biological measurements of deep-sea corals from the Red Sea. Nat. Sci. Rep. 3:2802. doi: 10.1038/srep02802
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Sakai, K. (1997). Gametogenesis, spawning, and planula brooding by the reef coral Goniastrea aspera (Scleractinia) in Okinawa, Japan. Oceanogr. Lit. Rev. 44, 1149. doi: 10.3354/meps151067
Sawall, Y., Al-Sofyani, A., Banguera-Hinestroza, E., and Voolstra, C. R. (2014). Spatio-temporal analyses of symbiodinium physiology of the coral Pocillopora verrucosa along large-scale nutrient and temperature gradients in the Red Sea. PLoS ONE 9:e103179. doi: 10.1371/journal.pone.0103179
Sawall, Y., Al-Sofyani, A., Hohn, S., Banguera-Hinestroza, E., Voolstra, C. R., and Wahl, M. (2015). Extensive phenotypic plasticity of a Red Sea coral over a strong latitudinal temperature gradient suggests limited acclimatization potential to warming. Sci. Rep. 5:8940. doi: 10.1038/srep08940
Schmidt, P. S., Paaby, A. B., and Heschel, M. S. (2005). Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution 59, 2616–2625. doi: 10.1111/j.0014-3820.2005.tb00974.x
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Steffen, W., Richardson, K., Rockstroem, J., Cornell, S. E., Fetzer, I., Bennett, E. M., et al. (2015). Planetary boundaries: Guiding human development on a changing planet. Science 347:6223. doi: 10.1126/science.1259855
Stapley, J., Reger, J., Feulner, P. G., Smadja, C., Galindo, J., Ekblom, R., et al. (2010). Adaptation genomics: the next generation. Trends Ecol. Evol. (Amst.) 25, 705–712. doi: 10.1016/j.tree.2010.09.002
Strachan, L. A., Tarnowski-Garner, H. E., Marshall, K. E., and Sinclair, B. J. (2011). The evolution of cold tolerance in Drosophila Larvae. Physiol. Biochem. Zool. 84, 43–53. doi: 10.1086/657147
Takahashi, A., and Takano-Shimizu, T. (2011). Divergent enhancer haplotype of ebony on inversion In(3R)Payne associated with pigmentation variation in a tropical population of Drosophila melanogaster. Mol. Ecol. 20, 4277–4287. doi: 10.1111/j.1365-294X.2011.05260.x
Tambutté, E., Venn, A. A., Holcomb, M., Segonds, N., Techer, N., Zoccola, D., et al. (2015). Morphological plasticity of the coral skeleton under CO2-driven seawater acidification. Nat. Commun. 6:7368. doi: 10.1038/ncomms8368
Telonis-Scott, M., Hoffmann, A. A., and Sgró, C. M. (2011). The molecular genetics of clinal variation: a case study of ebony and thoracic trident pigmentation in Drosophila melanogaster from eastern Australia. Mol. Ecol. 20, 2100–2110. doi: 10.1111/j.1365-294X.2011.05089.x
Todd, P. A. (2008). Morphological plasticity in scleractinian corals. Biol. Rev. 83, 315–337. doi: 10.1111/j.1469-185X.2008.00045.x
Ulstrup, K. E., Berkelmans, R., Ralph, P. J., and Oppen, M. J. H. V. (2006). Variation in bleaching sensitivity of two coral species across a latitudinal gradient on the Great Barrier Reef: the role of zooxanthellae. Mar. Ecol. Prog. Ser. 314, 135–148. doi: 10.3354/meps314135
Umina, P. A., Weeks, A. R., Kearney, M. R., Mckechnie, S. W., and Hoffmann, A. A. (2005). A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693. doi: 10.1126/science.1109523
van Oppen, M. J. H., Oliver, J. K., Putnam, H. M., and Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Voolstra, C. R., Sunagawa, S., Matz, M. V., Bayer, T., Aranda, M., Buschiazzo, E., et al. (2011). Rapid evolution of coral proteins responsible for interaction with the environment. PLoS ONE 6:e20392. doi: 10.1371/journal.pone.0020392
Whitham, T. G., Bailey, J. K., Schweitzer, J. A., Shuster, S. M., Bangert, R. K., Leroy, C. J., et al. (2006). A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523. doi: 10.1038/nrg1877
Zhong, Y., Jia, Y. X., Gao, Y., Tian, D. C., Yang, S. H., and Zhang, X. H. (2013). Functional requirements driving the gene duplication in 12 Drosophila species. BMC Genomics 14:12. doi: 10.1186/1471-2164-14-555
Appendix
*Christian R. Voolstra, Division of Biological and Environmental Science and Engineering, Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
David J. Miller, ARC Centre of Excellence for Coral Reef Studies, and Comparative Genomics Centre, James Cook University, Townsville, QLD, Australia
Mark A. Ragan, ARC Centre of Excellence in Bioinformatics, and Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
Ary A. Hoffmann, Departments of Genetics and Zoology, Bio21 Institute, The University of Melbourne, Melbourne, VIC, Australia
Ove Hoegh-Guldberg, Global Change Institute, The University of Queensland, Brisbane, QLD, Australia
David G. Bourne, Australian Institute of Marine Science, Centre for Marine Microbiology and Genetics, Townsville, QLD, Australia
Eldon E. Ball, Division of Evolution, Ecology and Genetics, The Australian National University, Canberra, ACT, Australia
Hua Ying, Division of Evolution, Ecology and Genetics, The Australian National University, Canberra, ACT, Australia
Sylvain Forêt, Division of Evolution, Ecology and Genetics, The Australian National University, Canberra, ACT, Australia
Shunichi Takahashi, Division of Environmental Photobiology, National Institute for Basic Biology, Okazaki, Japan
Karen D. Weynberg, Australian Institute of Marine Science, Centre for Marine Microbiology and Genetics, Townsville, QLD, Australia
Madeleine J. H. van Oppen, ARC Centre of Excellence for Coral Reef Studies, and Australian Institute of Marine Science, Centre for Marine Microbiology and Genetics, Townsville, QLD, Australia; School of BioSciences, The University of Melbourne, Parkville, VIC, Australia
Kathleen Morrow, Department of Molecular, Cellular, and Biomedical Sciences, University of New Hampshire, Durham, NH, USA
Cheong Xin Chan, ARC Centre of Excellence in Bioinformatics, and Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
Nedeljka Rosic, School of Biological Sciences, The University of Queensland, Brisbane, QLD, Australia
William Leggat, ARC Centre of Excellence for Coral Reef Studies, and School of Pharmacy and Molecular Sciences, James Cook University, Townsville, QLD, Australia
Susanne Sprungala, ARC Centre of Excellence for Coral Reef Studies, and School of Pharmacy and Molecular Sciences, James Cook University, Townsville, QLD, Australia
Michael Imelfort, Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD, Australia
Gene W. Tyson, Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD, Australia
Karin S. Kassahn, Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia, Present address: Karin S. Kassahn, SA Pathology, Women's and Children's Hospital, North Adelaide, Australia
Petra B. Lundgren, Great Barrier Reef Foundation, Brisbane, QLD, Australia
Roger J. Beeden, Great Barrier Reef Marine Park Authority, Townsville, QLD, Australia
Timothy Ravasi, KAUST Environmental Epigenetics Program, Division of Biological and Environmental Science and Engineering, and Division of Applied Mathematics and Computer Science, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
Michael L. Berumen, Division of Biological and Environmental Science and Engineering, Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
Eva Abal, Great Barrier Reef Foundation, Brisbane, QLD, Australia
Theresa Fyffe, Great Barrier Reef Foundation, Brisbane, QLD, Australia
Keywords: coral reef ecosystem, global environmental change, adaptation, resilience, Great Barrier Reef, Red Sea, holobiont, metaorganism
Citation: ReFuGe 2020 Consortium (2015) The ReFuGe 2020 Consortium—using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front. Mar. Sci. 2:68. doi: 10.3389/fmars.2015.00068
Received: 17 June 2015; Accepted: 28 August 2015;
Published: 15 September 2015.
Edited by:
Fengping Wang, Shanghai Jiao Tong University, ChinaReviewed by:
Carmelo Fruciano, University of Konstanz, GermanyEmre Keskin, Ankara University, Turkey
Copyright © 2015 ReFuGe 2020 Consortium. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian R. Voolstra, Division of Biological and Environmental Science and Engineering, Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal 23955, Saudi Arabia,Y2hyaXN0aWFuLnZvb2xzdHJhQGthdXN0LmVkdS5zYQ==
†Authors' names and addresses are listed in the Appendix.
 ReFuGe 2020 Consortium†
ReFuGe 2020 Consortium†