- 1Department of Marine Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2The Marine Science Institute and the Department of Ecology Evolution and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA, United States
- 3Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, United States
- 4Department of Earth, Ocean, and Atmospheric Sciences, University of British Columbia, Vancouver, BC, Canada
- 5Department of Marine and Coastal Sciences, Rutgers, the State University of New Jersey, New Brunswick, NJ, United States
- 6Department of Ocean Sciences, University of California, Santa Cruz, Santa Cruz, CA, United States
- 7Chemistry Department, Humboldt State University, Arcata, CA, United States
- 8Department of Oceanography and Coastal Sciences, College of the Coast and Environment, Louisiana State University, Baton Rouge, LA, United States
Changes in iron (Fe) bioavailability influence diatom physiology and community composition, and thus have a profound impact on primary productivity and ecosystem dynamics. Iron limitation of diatom growth rates has been demonstrated in both oceanic and coastal waters of the Northeast Pacific Ocean and is predicted to become more pervasive in future oceans. However, it is unclear how the strategies utilized by phytoplankton to cope with low Fe bioavailability and resupply differ across these ocean provinces. We investigated the response of diatom communities to variable Fe conditions through incubation experiments performed in the Fe mosaic of the California Upwelling Zone and along a natural Fe gradient in the Northeast Pacific Ocean. Through coupling gene expression of two dominant diatom taxa (Pseudo-nitzschia and Thalassiosira) with biological rate process measurements, we provide an in-depth examination of the physiological and molecular responses associated with varying Fe status. Following Fe enrichment, oceanic diatoms showed distinct differential expression of gene products involved in nitrogen assimilation, photosynthetic carbon fixation, and vitamin production compared to diatoms from low-Fe coastal sites, possibly driven by the chronic nature of Fe stress at the oceanic site. Genes of interest involved in Fe and N metabolism additionally exhibited divergent expression patterns between the two diatom taxa investigated, demonstrating that diverse diatoms may invoke alternative strategies when dealing with identical changes in their environment. We report here several mechanisms used distinctly by coastal or oceanic diatom communities as well as numerous taxa-specific strategies for coping with Fe stress and rearranging nutrient metabolism following Fe enrichment.
Introduction
Phytoplankton growth is limited by iron (Fe) availability in ~30–40% of the ocean (Moore et al., 2001, 2004). The subarctic Northeast (NE) Pacific Ocean is one of the most well-characterized of these high-nutrient, low chlorophyll (HNLC) regions. Productivity in the NE Pacific Ocean remains low as a result of low Fe concentrations regardless of sufficient nitrate () levels and is typically dominated by small cells such as the cyanobacterium Synechococcus and eukaryotic picophytoplankton (Varela and Harrison, 1999). In this region, Fe is supplied to surface waters mainly through atmospheric deposition of dust from arid continental regions and volcanic emissions, with Fe inputs from continental margin sediments fueling winter phytoplankton blooms when atmospheric deposition is low (Lam et al., 2006; Lam and Bishop, 2008). A gradient in surface nutrient concentrations is observed from this oceanic region eastwards toward the continent; bioavailable Fe increases and supports higher phytoplankton biomass while concentrations in the upper mixed layer decrease to limiting levels on the continental shelf (Taylor and Haigh, 1996; Harris et al., 2009; Ribalet et al., 2010).
Iron-limited growth of phytoplankton may also occur in coastal waters, notably in regions of the California Upwelling Zone (CUZ; Hutchins et al., 1998; Bruland et al., 2001). These regions of the CUZ are characterized by high concentrations of upwelled macronutrients, but relatively low dissolved Fe (dFe) such that phytoplankton blooms ultimately become Fe-stressed. Low Fe levels result from the lack of Fe inputs from rivers and narrow continental shelves that prevent mixing of upwelled water with Fe derived from Fe-rich shelf sediments (Johnson et al., 1999; Bruland et al., 2001) and consequently, the primary Fe source in the CUZ is winter river sediment deposition (Hutchins et al., 2002; Chase et al., 2005).
Phytoplankton that subsist in Fe-limited environments are equipped with strategies to sustain growth during periods of physiological Fe stress and to rapidly respond to sudden increases in bioavailable Fe. Strategies employed by phytoplankton include replacement of Fe-containing proteins with Fe-independent ones to decrease cellular Fe requirements (La Roche et al., 1996; Peers and Price, 2006; Allen et al., 2008; Lommer et al., 2012), increasing Fe uptake rates through induction of high affinity Fe uptake systems (Maldonado and Price, 2001; Morrissey et al., 2015) and using Fe storage through specialized proteins or vacuoles (Marchetti et al., 2009; Nuester et al., 2012). In some diatom laboratory isolates and natural communities, these low-Fe strategies are rapidly reversed when Fe concentrations increase (Kustka et al., 2007; Lommer et al., 2012), whereas in others these strategies are permanent adaptations (Lommer et al., 2010; Marchetti et al., 2012). Phytoplankton species from low-Fe oceanic environments generally have lower growth requirements for cellular Fe than species from higher Fe coastal waters, largely linked to differences in Fe-containing photosynthetic proteins and complexes (Sunda and Huntsman, 1995; Strzepek and Harrison, 2004; Peers and Price, 2006; Behrenfeld and Milligan, 2013). While we have an understanding of how a few phytoplankton species alter their nutrient metabolism in response to chronic Fe limitation from laboratory experiments, how the nutrient strategies invoked by intermittently Fe-limited coastal taxa might differ from those used by species residing in chronically Fe-limited regions of the open ocean has not been directly compared.
A large amount of genetic diversity exists among diatom taxa, possibly due to differences in environmental pressures at the time of evolutionary emergence (Sims et al., 2006; Armbrust, 2009; Rabosky and Sorhannus, 2009). A genomic comparison between the evolutionarily older centric Thalassiosira pseudonana and the more recently evolved pennate Phaeodactylum tricornutum demonstrates the two diatoms share only 57% of their genes with each other, suggesting a tremendous amount of genomic diversity exists between members of these two diatom lineages (Bowler et al., 2008). Furthermore it is often observed that pennate diatoms, especially those belonging to the genus Pseudo-nitzschia, tend to dominate large Fe-induced blooms in HNLC waters (de Baar et al., 2005; Marchetti et al., 2012). These observations may suggest that the pennate diatoms have evolved distinct strategies for optimizing their potential for rapid growth when transitioning from low to relatively high Fe conditions, resulting in a competitive advantage over older lineages of diatoms as well as other types of phytoplankton.
To better understand whether major diatom genera from coastal and oceanic regions differ in their gene expression responses to changes in Fe availability, a comparative analysis across distinct nutrient regimes was performed through a combination of metatranscriptomic and physiological approaches. Microcosm incubation experiments were conducted at geographically diverse sites with different Fe regimes, macronutrient concentrations, and phytoplankton community compositions—at an Fe-limited oceanic site and a coastal site in the subarctic NE Pacific Ocean, and at three biogeochemically distinct sites within the Fe mosaic of the coastal CUZ. For our study, we focused on the changes in gene expression patterns between two dominant taxa across all sites—the pennate diatom Pseudo-nitzschia and the centric diatom Thalassiosira. These two taxa were classified by the Tara Oceans circumnavigation expedition to be two of the eight most abundant diatom genera in the global ocean (Malviya et al., 2016). Given the large amount of genetic and physiological variation observed between major diatom groups (Bowler et al., 2008; Marchetti et al., 2009; Sutak et al., 2012; Alexander et al., 2015), differences in molecular responses to changing Fe availabilities across the NE Pacific Ocean and CUZ were anticipated.
Materials and Methods
Experimental Design
Incubation experiments were conducted on two separate cruises: within regions of the CUZ during July 3–26th 2014 onboard the R/V Melville and along the Line-P transect of the subarctic NE Pacific Ocean during June 7–23rd 2015 onboard the Canadian Coast Guard Ship (CCGS) John P. Tully (Figure 1). The incubated phytoplankton community response was assessed using a combination of physiological measurements and metatranscriptomics to examine the effects of Fe status on diatom physiology and gene expression. Each experiment included three treatments: (1) a 5 nmol L−1 FeCl3 addition (Fe), (2) a 200 nmol L−1 desferroxamine B (DFB) addition, and (3) an unamended control (Ctl), each sampled at two time points.
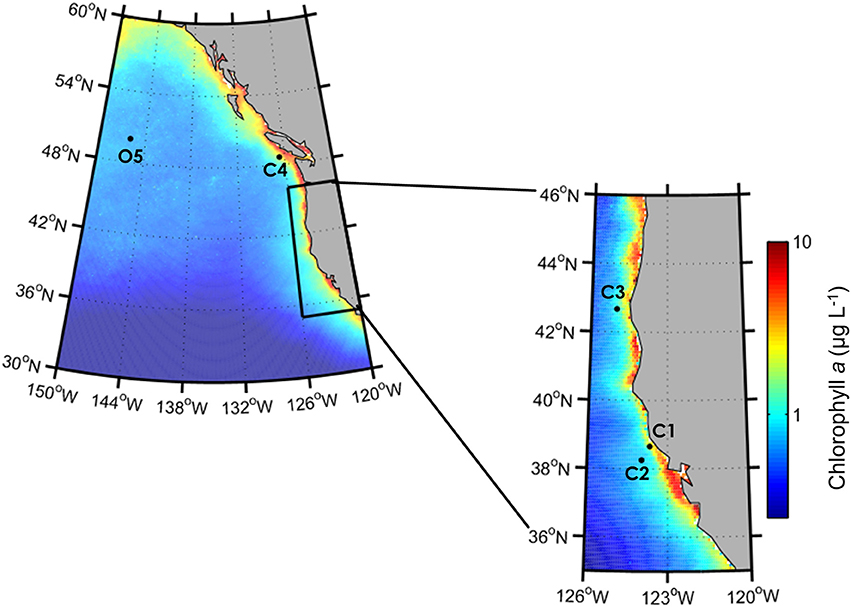
Figure 1. Locations of incubation experiments in the California Upwelling Zone (C1, C2, C3) and along Line P (C4, O5) in the Northeast Pacific Ocean. Color bar indicates climatological-averaged chlorophyll a concentrations (μg L−1) from SeaWiFS (1997-2010).
During the CUZ cruise, three incubation experiments were performed at separate locations corresponding to distinct Fe and macronutrient regimes (Supplementary Table 1), including sites of high dFe, macronutrients, and phytoplankton biomass (C1: 38°39.30 N, 123°39.87 W), relatively low dFe, high macronutrients and high phytoplankton biomass (C2: 38°15.31 N, 123°57.98 W), and low dFe with high macronutrients and low phytoplankton biomass (C3: 42°40.00 N, 125°0.00 W) (Figure 1). Near-surface seawater was collected from a depth of ~15 m using a trace-metal clean sampling system consisting of a tow-fish sampler attached to Kevlar™ line, PFA Teflon tubing, and a Teflon dual-diaphragm pump that pumped seawater directly into a positive pressure trace-metal clean bubble. The seawater was placed in a large 200 L acid-cleaned HDPE drum for homogenization before being distributed into 10 L flexible acid-cleaned polyethylene cubitainers (Hedwin Corporation). Cleaning protocols for the cubitainers included successive soaks in 1.2 mol L−1 hydrochloric acid (reagent grade) for 3 days, 1.2 mol L−1 hydrochloric acid (trace metal grade) for 1 week and 0.1 mol L−1 acetic acid (trace-metal grade) until use. Prior to filling the cubitainers with seawater, the dilute acetic acid was removed and the cubitainers were rinsed thoroughly three times with ambient seawater from the collection site. The primary objective of these experiments was to elucidate the responses of target diatom genera and the phytoplankton community to variable Fe conditions. Therefore, sites were targeted that would ensure adequate macronutrient concentrations to support phytoplankton growth. However, at C2, 15 μmol L−1 of Si(OH)4 was added to all cubitainers to support growth of diatoms due to the initially low Si(OH)4 concentration (<4.7 μmol L−1).
During the Line-P cruise, incubation experiments were conducted at the low coastal station P4 (48°39 N, 126°40 W; referred to as C4 in this analysis) and at the chronically Fe-limited, HNLC oceanic station P26, also known as Ocean Station Papa (OSP, 50°00 N, 145°00 W; Harrison, 2002; referred to as O5). Seawater was collected at depths corresponding to ~30% of incident irradiance (8–12 m) at both stations using a trace-metal clean sampling system consisting of a Teflon air bellows pump and PTFE lined Kevlar™ tubing attached to a Kevlar™ line. The seawater was pumped directly into 10 L acid-cleaned polyethylene cubitainers placed within an on-deck trace-metal clean positive pressure flowhood. At site C4, 10 μmol L−1 of was added to all cubitainers to support growth of diatoms due to the initially low concentration (<1.5 μmol L−1).
At the start of the experiments, ambient seawater was filtered for all initial measurements (T0). For each incubation experiment, cubitainers were filled to serve as a control (Ctl) or amended with FeCl3 or DFB just prior to dawn. All cubitainers were placed in on-deck Plexiglass incubators with flow-through seawater to maintain near-ambient surface water temperatures. Incubators were covered with neutral density screening to achieve ~30% of the incident irradiance (Supplementary Figure 1). Following 24–96 h of incubation (Supplementary Table 1; depending on the measured macronutrient drawdown) the cubitainers for a specific time point were removed from the incubators and filtered immediately. The goal for each time point was to achieve measureable drawdowns in macronutrients that would infer stimulation of phytoplankton growth without complete macronutrient depletion. However, for some experiments and time points, depletion of or other macronutrients occurred. All filtrations were conducted at dawn. Subsamples for dissolved and particulate nutrients, size-fractionated uptake rates of dissolved inorganic carbon (DIC) and , size-fractionated chlorophyll a, Fv/Fm, and RNA were collected at T0 and from each cubitainer according to the protocols described below.
Nutrient Concentrations, Uptake Rates, and Biogenic Silica Concentrations
For CUZ experiments, dissolved nitrate and nitrite ( + ), phosphate (), and silicic acid (H4SiO4) concentrations were measured onboard using a Lachat Quick Chem 8000 Flow Injection Analysis system (Parsons et al., 1984) with detection limits of 0.05 μM for + , 0.03 μM for , and 0.2 μM for H4SiO4 (Bruland et al., 2008). Particles were removed by filtration through a Whatman GF/F filter (25 mm). Reference standards for nutrients in seawater were run for quality control. During Line-P sampling, ~15 mL of seawater was filtered through a Whatman GF/F filter into acid-rinsed polypropylene tubes and frozen at −20°C in aluminum blocks until onshore analysis. Shortly following the cruise, the dissolved + , , and H4SiO4 concentrations were determined using an Astoria nutrient analyzer (Barwell-Clarke and Whitney, 1996). Nutrient detection limits were 0.2 μM for + , 0.02 μM for , and 0.5 μM for H4SiO4 (Frank Whitney and Mark Belton [IOS], pers. comm.).
For biogenic silica (bSi) measurements, 335 mL (CUZ) or 250 mL (Line P) of seawater was filtered onto polycarbonate filters (1.2 μm pore size for CUZ and 0.6 μm pore size for Line-P, 25 mm), digested with NaOH in Teflon tubes, and measured with the colorimetric ammonium molybdate method (Krause et al., 2013).
Size-fractionated particulate nitrogen (PN), particulate carbon (PC), and uptake rates were obtained by adding 15N-NaNO3 to 618 mL subsample of experimental seawater placed within clear polycarbonate bottles. The concentration of added was no more than 10% of ambient level within CUZ incubations, and was 1 μmol L−1 within Line-P incubations (corresponding to levels of 68% at T0 and 10% within -amended incubations at C4, and ~10% at O5). DIC uptake within Line-P incubations was measured by additionally spiking subsamples with 120 μmol L−1 NaH13CO3. Bottles were incubated in the same flow-through Plexiglass incubators where cubitainers were kept. Following 8 h of incubation, seawater samples were filtered in series through a polycarbonate filter (5 μm pore size, 47 mm) via gravity filtration, and then through a pre-combusted (450°C for 5 h) GF/F filter by gentle vacuum (<100 mg Hg). Particulates collected on the 5 μm polycarbonate filter were then rinsed onto a separate pre-combusted GF/F filter using an artificial saline solution. Filters were stored at −20°C until onshore analysis. In the laboratory, filters were heated to 60°C for 24 h and pelletized in tin capsules (Elemental Microanalysis) in preparation for analysis of the atom % 15N, atom % 13C (for Line-P), particulate nitrogen (PN), and particulate carbon (PC) using an elemental analyzer paired with an isotope ratio mass spectrometer (EA-IRMS). Biomass-normalized uptake rates (PN-VNO3) and DIC uptake rates (PC-VDIC) for the Line-P experiments were obtained by dividing the measured and DIC biological uptake rates by PN and PC concentrations, respectively.
To quantify VDIC in CUZ incubations, incorporation of 14C was determined using a protocol adapted from Taylor et al. (2013). Briefly, 60 mL of seawater from each cubitainer was distributed into acid-cleaned light and dark polycarbonate bottles. In each bottle, 1.2 μCi of NaH14CO3 was added. Bottles were incubated in the same flow-through Plexiglass incubators where cubitainers were kept for 6.5–8 h. Following incubation, samples were filtered through stacked 47 mm polycarbonate filters (5 and 1 μm) separated with a mesh spacer during filtration. Filters were vacuum dried, placed in 7 mL scintillation vials containing 0.5 mL of 6 M HCl and permitted to degas for 24 h. Disintegrations per minute (DPM) retained on the filters were measured using a Beckman Coulter LS 6500 scintillation counter. Reported values are light bottle DPMs minus dark bottle DPMs. To obtain VDIC, DIC uptake rates were normalized to PC concentrations obtained as part of the uptake measurements within each incubation and size fraction.
Dissolved Iron Concentrations
Seawater samples for Fe analysis within the CUZ were acidified at sea with the equivalent of 4 mL 6 N quartz-distilled HCl per L of seawater (pH ~ 1.7) and stored in acid-cleaned LDPE bottles for at least 2 months prior to analysis. Samples were analyzed using an adaption of Biller and Bruland (2012) as described in Parker et al. (2016). Briefly, this method involves preconcentrating the Fe from buffered (pH 6.0) seawater on Nobias-chelate PA1 resin and eluting with 1 N quartz-distilled HNO3. The eluent was analyzed with a Thermo-Element high resolution XR ICP-MS in counting mode. Line-P dissolved Fe samples were stored in acid-cleaned LDPE bottles, acidified post-cruise with Optima-grade HCl (1 mL 12 N HCl per L of seawater), and allowed to sit for >3 months. Dissolved Fe was measured via ICP-MS by P. Morton at Florida State University following resin preconcentration using the protocol of Milne et al. (2010).
Chlorophyll a
Four hundred milliliters of seawater was gravity-filtered through a polycarbonate filter (5 μm pore size, 47 mm diameter) followed by vacuum filtration through a GF/F filter (0.7 μm nominal pore size, 25 mm diameter) using a series filter cascade for size fractionation. Filters were frozen at −80°C until analysis. Chlorophyll a extraction was performed using 90% acetone at −20°C for 24 h and the extracted Chl a was quantified by fluorometry with a Turner Designs 10-AU fluorometer using the acidification method (Parsons et al., 1984).
Domoic Acid
Approximately 250 mL of seawater from each CUZ site was filtered through GF/F filters (25 mm) via vacuum pressure (<100 mm Hg) and the filters were frozen at −80°C. The filters were extracted with 20% methanol (MeOH) in water. The mixture was sonicated in an ice bath for 2 min at 30–40 W with a Sonicator 3000, followed by centrifugation (10 min, 1,399 × g). The supernatant was collected and passed through a 0.22 μm syringe filter. Samples were stored at −20°C until analysis. Concentrations with a detection limit of 0.01 μg L−1 were obtained using an enzyme-linked immunosorbent assay (Abraxis, Warminster, PA, USA) following the manufacturer's protocol, including running each sample in duplicate at several dilutions. Final concentrations (pg DA mL extract−1) were calculated using the manufacturer supplied analysis spreadsheet.
Photophysiology
The maximum photochemical yield of PSII (Fv/Fm) was measured by fast repetition rate fluorometry (FRRF) using a custom-built fluorescence-induction and relaxation system (Kolber et al., 1998; Gorbunov and Falkowski, 2004). Before each measurement, a 5 mL subsample of seawater from each cubitainer was acclimated to low light for 20 min. A saturating pulse (20,000 μmol photons m−2 s−1) of blue light (450 nm) was applied to dark-acclimated cells for a duration of 100–200 μs. Measurements were obtained using the single-turnover flash (STF) setting with the average of 50 iterations for the CUZ experiments, and a single iteration for the Line-P experiments. Data were blank corrected using 0.2 μm filtered seawater.
RNA Extraction and Bioinformatic Analysis
Phytoplankton in seawater samples were filtered onto 0.8 μm Pall Supor filters (142 mm) via peristaltic pumping, immediately flash frozen in liquid nitrogen and stored at −80°C until extraction onshore. The filters were briefly thawed on ice before being extracted individually using the ToTALLY RNA Kit (Ambion). The extraction procedure followed manufacturer protocols with the modified first step of glass bead addition and vortexing to facilitate disruption of cells. Removal of DNA was performed with one round of DNAse I (Ambion) incubation. For the Line P experiments, due to low yields in treatments, RNA from the triplicate cubitainers was pooled prior to sequencing. Within CUZ experiments all triplicate incubation samples were sequenced separately. At the oceanic site O5, RNA yields were too low to successfully sequence metatranscriptomes at the T1 timepoint, and consequently, transcriptomic analyses were performed using the T0, T2 Fe, and T2 Ctl treatments. Metatranscriptomic library preparation was performed with the Illumina TruSeq Stranded mRNA Library Preparation Kit and HiSeq v4 reagents. Samples were barcoded and run across three lanes of Illumina HiSeq 2000 (125 bp, paired-end) yielding on average 23 million paired-end reads per sample (Supplementary Table 2). The RNA-seq data reported here has been deposited in the National Center for Biotechnology (NCBI) sequence read archive (SRA) under the BioProject accession no. PRJNA320398 and PRJNA388329.
Raw reads were trimmed for quality bases and removal of adapters using Trimmomatic v0.32 (paired-end mode, adaptive quality trim with 40 bp target length, and strictness of 0.6, minimum length of 36 bp; Bolger et al., 2014). Trimmed paired reads were merged into single reads with BBMerge v8.0. For each site, the resulting merged pairs and non-overlapping paired-end reads were assembled using ABySS v1.5.2 with a multi-kmer approach (Birol et al., 2009). The different k-mer assemblies were merged to remove redundant contigs using Trans-ABySS v1.5.3 (Robertson et al., 2010). Read counts were obtained by mapping raw reads to assembled contigs with Bowtie2 v2.2.6 (end-to-end alignment; Langmead and Salzberg, 2012). Alignments were filtered by mapping quality score (MAPQ) of 10 or higher as determined by SAMtools v1.2 (Li et al., 2009). Taxonomic and functional annotations were assigned based on sequence homology to reference databases via BLASTx v2.3.0 with an e-value cutoff of 10−3 (Altschul et al., 1990). Functional annotations were assigned according to the top hit using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Release 75), while taxonomic assignments were obtained according to the top hit using MarineRefII (Laboratory of Mary Ann Moran, University of Georgia), a custom-made database comprised of protein sequences of marine prokaryotes and eukaryotes including all sequenced transcriptomes from Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (Keeling et al., 2014). Taxonomic information was obtained from NCBI's Taxonomy Database (each isolate in MarineRefII is assigned a NCBI taxonomic ID). The information from NCBI was manually curated to ensure proper assignment and use of common phytoplankton taxonomic ranks. For our analysis, we have grouped diatom-associated sequences at the genus level. Therefore, the patterns in gene expression observed could be driven by one dominant species or many equally distributed species belonging to a genus within each site.
All diatom-assigned counts were summed to both the genus taxonomic rank and KEGG Orthology (KO) functional annotation level. For genes of interest without a KO assignments but with an annotated gene definition (i.e., ISIPs and rhodopsin), raw counts corresponding to KEGG gene definitions were summed. EdgeR v3.12.0 was used to calculate Pseudo-nitzschia- or Thalassiosira-specific normalized fold change and counts-per-million (CPM) from pairwise comparisons using the exactTest (Robinson and Smyth, 2008; Robinson and Oshlack, 2010; Robinson et al., 2010; Klingenberg and Meinicke, 2017). Significance (p < 0.05) was calculated with edgeR's estimate of tagwise dispersions across all samples within CUZ sites. Heatmaps were produced with the R package pheatmap v1.0.8, and dendrograms created using Euclidean distance and hierarchical clustering. Assembled contigs, read counts, and functional annotations of contigs are available at marchettilab.web.unc.edu/data.
In order to directly compare transcript abundance across locations for principal component analyses (PCA), the assemblies for all sites were merged with Trans-ABySS. The removal of redundant contigs was verified with GenomeTools v.1.5.1. Counts were obtained by aligning raw reads against this merged metatranscriptome using Salmon v.0.7.3-beta. Normalized counts were then obtained with edgeR v3.12.0. PCA biplots were created using log-transformed normalized counts for genes of interest with ggbiplot v.0.5.
Phylogenetic Analysis of Environmental Sequences
Environmental Pseudo-nitzschia and Thalassiosira contigs functionally annotated as RubisCO (RBCL), rhodopsin (RHO), or superoxide dismutase (SOD) and containing a large number of mapped reads were compared to diatom reference sequences for phylogenetic characterization. Diatom sequences used in reference alignments were obtained through a sequence homology search using BLASTx v2.2.28 with Pseudo-nitzschia RBCL, RHO, and SOD against the database MMETSP using an E-value cutoff of 10−5 (Altschul et al., 1990). Sequences were aligned using MUSCLE within Geneious v5.6.4 software (Edgar, 2004).
Results
Nutrient Regimes of Experimental Sites
CUZ site C1 (Figure 1) was characterized by high macronutrient and dFe concentrations in the mixed layer supporting a high biomass, nutrient-replete phytoplankton community. The community was dominated by phytoplankton cells in the >5 μm chlorophyll a (chl a) size fraction, constituting 88% of the total chl a concentration (Figure 2B; Supplementary Table 1). Macronutrient concentrations were rapidly consumed during the first 24 h of incubation (T1), with near complete depletion of the (≤1 μmol L−1 remaining by 48 h [T2]; Figure 2A). The initially Fe-replete phytoplankton community (dFe: 3.57 nmol L−1) was mostly unaffected by the additions of Fe or DFB as demonstrated through relatively constant Fv/Fm, phytoplankton biomass, particulate nitrogen (PN)-specific nitrate uptake rates (VNO3, or nitrate assimilation rates), and particulate carbon (PC)-specific dissolved inorganic carbon uptake rates (VDIC, or carbon assimilation rates) across treatments at each time point (Figures 2B–E). Furthermore, the NO3:Fe ratio of the initial (T0) seawater (3.8 μmol:nmol. Supplementary Table 1) was substantially below the predicted threshold ratio for eventual Fe stress of 12 μmol:nmol for phytoplankton in this region as proposed by King and Barbeau (2007), albeit this ratio is subject to variation as a function of phytoplankton iron demands (Bruland et al., 2001), suggesting this phytoplankton community was not likely to be driven into Fe limitation prior to complete utilization. However, indications of molecular-level responses to Fe and DFB additions were observed; 74 genes were differentially expressed (p < 0.05) in Pseudo-nitzschia between the Fe and DFB treatments (Supplementary Figure 2A). Fe-stress bioindicator genes (FLDA, PETE, and ISIP2A; Whitney et al., 2011; Morrissey et al., 2015; Graff van Creveld et al., 2016) increased in expression following the addition of DFB relative to the added Fe treatment, suggesting the onset of Fe stress following the addition of DFB by the end of the first time point.
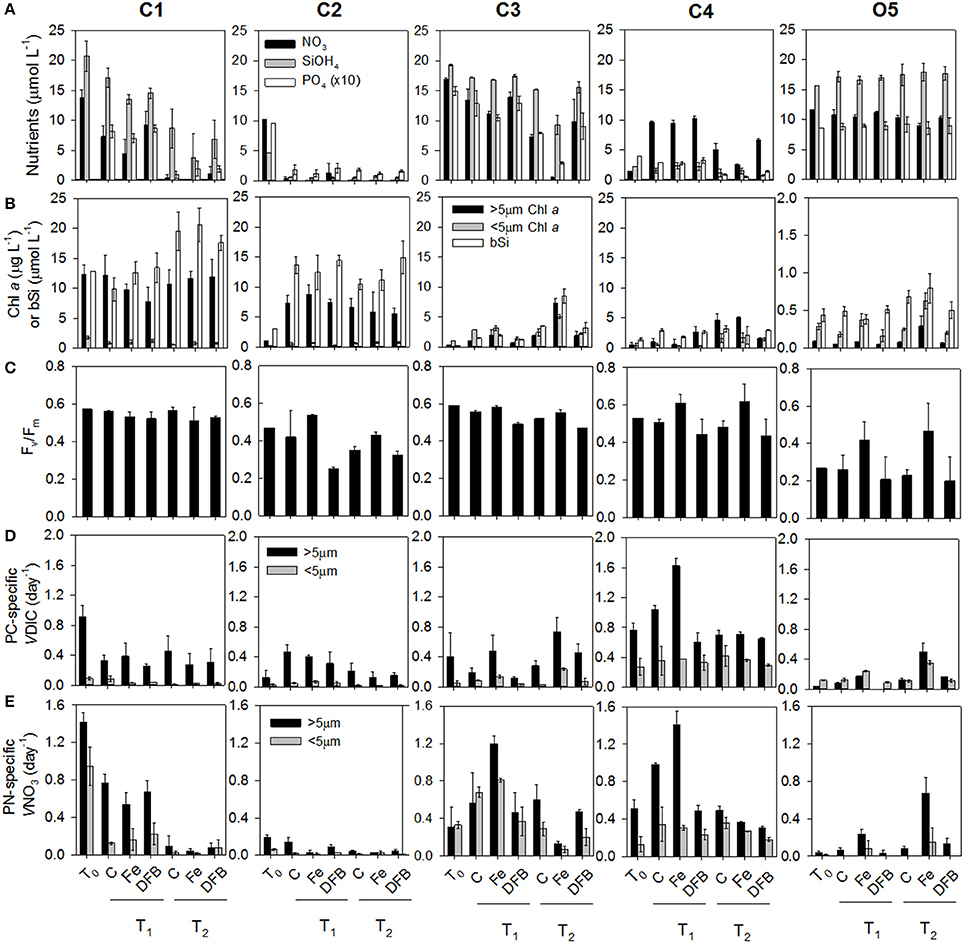
Figure 2. Dissolved macronutrient concentrations: nitrate (), silicic acid (SiOH4), and ortho-phosphate () (A), Size-fractionated chlorophyll a (μg L−1) within the large (>5 μm) and small (<5 μm) size fractions or biogenic silica (μmol L−1) (B), Maximum photochemical yield of PSII (Fv/Fm) (C), Particulate carbon (PC)-specific dissolved inorganic carbon (DIC) uptake rates [VDIC (day−1)] within the large and small size fractions (D), and particulate nitrogen (PN)-specific nitrate uptake rates [VNO3 (day−1)] within the large and small size fractions (E) in each treatment and sample time points across sites (see Supplementary Table 1). Where present, error bars represent the standard deviation associated with the mean of triplicate incubations.
CUZ site C2 was located in close geographical proximity to C1 (Figure 1), yet exhibited different mixed layer properties in relation to phytoplankton biomass, silicic acid (Si[OH]4) and dFe concentrations (0.44 nmol L−1). Nitrate and ortho-phosphate () concentrations were similarly high (10.3 and 0.96 μmol L−1, respectively) as found at site C1, although Si(OH)4 levels were appreciably lower (4.7 μmol L−1) and possibly growth-limiting to certain diatoms (Nelson et al., 1996). Therefore, incubations were amended with 15 μmol L−1 Si(OH)4 to support potential diatom growth with added Fe (Brzezinski, 1985). Although the chl a concentration in the >5 μm size fraction was initially <1 μg L−1 and biogenic silica (bSi) concentrations were <3 μmol L−1, by 48 h (T1) the >5 μm chl a fraction reached 5–8 μg L−1, and bSi increased to 10–15 μmol L−1 in all treatments, accompanied by appreciable decreases in , , and Si(OH)4 concentrations (Figures 2A,B). Since this community quickly depleted concentrations during the experimental period, this site presented an opportunity to couple the physiological indicators of stress with N-related transport and assimilation genes observed to be elevated in NO3-starved laboratory diatom cultures (Hildebrand, 2005; Song and Ward, 2007; Bender et al., 2014; Rogato et al., 2015). Apart from Fv/Fm reaching relatively low values in the DFB treatments, indications of Fe stress in bulk physiological measurements across treatments were absent (Figure 2C). However, the initial seawater NO3:Fe ratio of 23.4 μmol:nmol suggests this community may have been driven into Fe limitation provided sufficient Si(OH)4 was present. Additionally, a total of 414 Pseudo-nitzschia–associated genes were differentially expressed (p < 0.05) by T1 between the Fe and DFB treatments (Supplementary Figure 2). This greater number of differentially expressed genes in Pseudo-nitzschia when compared to C1 suggests the C2 diatom community in the DFB treatment experienced a higher degree of Fe stress during the incubation period. The initially low dissolved Si(OH)4:NO3 ratio at this site furthermore implies a possible increase in the Si:N ratios of Fe-stressed diatoms (Hutchins and Bruland, 1998; Marchetti and Cassar, 2009; Brzezinski et al., 2015). Interestingly, concentrations of domoic acid (DA), a neurotoxin produced by Pseudo-nitzschia, was 90 pg mL−1 in initial seawater (T0) and exceeded 3,000 pg mL−1 in the control treatment by T1 (Supplementary Figure 3). This increase in DA concentration may be linked to both the increase in Pseudo-nitzschia abundance and depletion of Si(OH)4 resulting in Si-limited cells which has been shown to greatly enhance DA production (Pan et al., 1996).
Site C3 (Figure 1) contained the lowest dFe concentrations (0.31 nmol L−1) among the CUZ sites along with high macronutrient concentrations [17 μmol L−1 , 19 μmol L−1 Si(OH)4, and 1.5 μmol L−1 ; Figure 2A]. The corresponding NO3:Fe ratio of the initial seawater was ~54.9 μmol:nmol (Supplementary Table 1). Following incubation, the chl a, bSi, PN-specific VNO3, and PC-specific VDIC were all higher in the Fe-amended treatment relative to the unamended control by T1 (Figures 2B,D,E). By 72 h, was completely drawn down within the Fe treatment (T2). Despite the pronounced influence of Fe enrichment on bulk parameters, Fv/Fm values were only slightly higher in the Fe treatment than the control, but they were substantially higher than in the DFB treatment (Figure 2C). This is likely a reflection of the different phytoplankton composition at this location compared to site C2, which did not show indications of an Fe-addition response on the measured bulk parameters, but did demonstrate elevated Fv/Fm values in the added Fe treatment. Site C3 represented the only phytoplankton community in the CUZ that displayed a definite physiological response to Fe addition relative to the control treatment (Supplementary Table 1). The Fe-induced molecular response in diatoms was demonstrated by the differential expression of 458 genes in Pseudo-nitzschia and 1,223 genes in Thalassiosira between the Fe and DFB treatments (Supplementary Figure 2C), and 365 genes in Pseudo-nitzschia and 837 genes in Thalassiosira between the Fe and Ctl treatments (p < 0.05).
Coastal site C4 was located at station P4 of the Line-P transect in the subarctic NE Pacific Ocean (Figure 1). Initial mixed-layer seawater properties were characterized by low concentrations of macronutrients and dFe, which supported a low phytoplankton biomass. Nitrate concentrations were initially 1.5 μmol L−1 (Figure 2A). To facilitate a potential phytoplankton growth response to added Fe, 10 μmol L−1 of was added to each treatment. Si(OH)4 concentrations were also initially low (2.2 μmol L−1) and incubation concentrations dropped to <2 μmol L−1 in most treatments by the second time point (T2; Figure 2A). These low concentrations restricted biomass accumulation as bSi (Figure 2B) and it is likely that the resulting diatom community experienced Si(OH)4 limitation by the end of the incubation period. Despite its relatively close proximity to land and relatively high dFe concentration (0.64 nmol L−1), there was a pronounced response to Fe addition at C4 as demonstrated through higher Fv/Fm, PN-specific VNO3, and PC-specific VDIC in the Fe treatment compared to values in the unamended control by T1 (Figures 2D,E; Supplementary Table 1). The NO3:Fe ratio following artificial addition was 18.8 μmol:nmol, sufficiently high to cause Fe stress with phytoplankton growth following an increase in phytoplankton biomass.
Oceanic site O5 was located at Ocean Station Papa (OSP), station P26 of the Line-P transect (Figure 1). This site demonstrated characteristically high macronutrients and low dFe (0.05 nmol L−1), resulting in the highest NO3:Fe ratio observed across all experimental sites (234 μmol:nmol; Supplementary Table 1). Phytoplankton biomass was initially low, consistent with historical observations from this well-characterized Fe-limited region (Figure 2A; Supplementary Table 1; Boyd and Harrison, 1999). In contrast to most of the coastal sites, the majority of the phytoplankton biomass was dominated by picophytoplankton and other small cells (<5 μm) initially and throughout the incubation period (Supplementary Table 1; Figure 2B). Biogenic Si concentrations only increased after 96 h with similar responses in controls and Fe treatments (Figure 2B). Both large and small chl a size fractions, Fv/Fm, PN-specific VNO3, and PC-specific VDIC were higher in the Fe treatment than in the unamended control (Ctl), confirming that the phytoplankton community in the initial seawater and in all incubation treatments without added Fe were experiencing Fe limitation (Figures 2B–E).
Community Composition across Sites
Metatranscriptomic assembly of sequence data and subsequent taxonomic annotation yielded the relative transcript proportions of phytoplankton functional groups (Figure 3). The CUZ site C1 was predominantly comprised of diatom transcripts at T0; however, there was a 26% decrease in diatom transcripts in both the Fe and DFB treatments by T1, accompanied by genus-level shifts within the diatoms. CUZ site C3 contained a phytoplankton community transcript pool almost equally represented by diatoms, prasinophytes, haptophytes, and dinoflagellates with little change in community composition among treatments following incubation. The coastal subarctic Pacific site C4 yielded an initial phytoplankton community transcript pool dominated by dinoflagellate-assigned sequences (24%), although these sequences decreased by ~10% in the Fe treatment, concurrent with a 9% increase in diatom transcripts. At the oceanic site O5, there were initially equal proportions of prasinophyte (22%) and haptophyte (23%) transcripts, with little representation by diatoms (4%). However, diatom-assigned transcripts constituted 9% of the community transcript pool by T2 in the Fe addition treatment. Pseudo-nitzschia and Thalassiosira were among the top five diatom genera at all sites examined based on relative transcript abundance (Figure 3). These two genera together constituted between 9 and 53% of the transcript proportions in the initial diatom communities, and 25–58% of the Fe-enriched diatom communities.
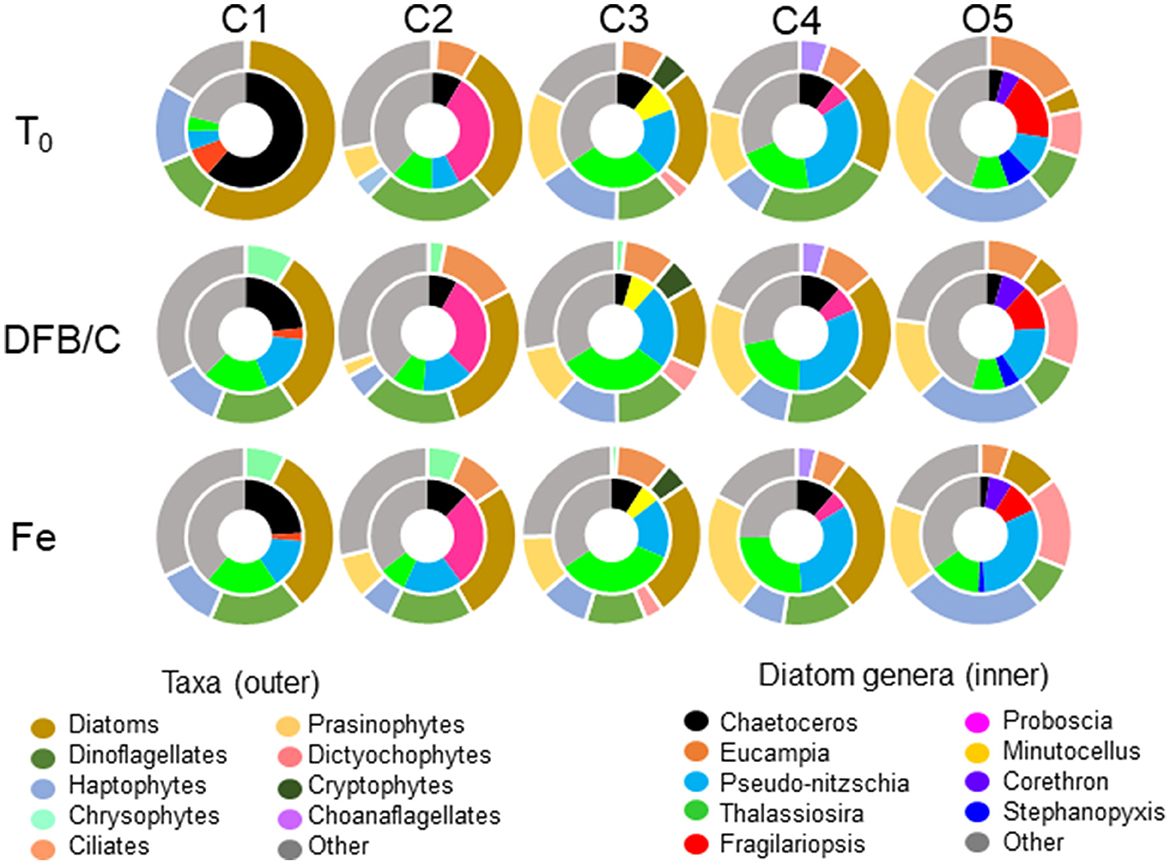
Figure 3. The average transcript proportions of phytoplankton taxa (outer charts) and diatom genera (inner charts) from initial seawater (T0) and during the first time point (T1; see Supplementary Table 1) within the Fe addition (Fe) and DFB addition (DFB) treatments at each site. Note that for site O5, the T2 control (Ctl) treatment is provided as the Fe-limited comparison.
Gene Expression Responses to Fe Status across Sites
Gene expression responses among sites were compared using Euclidian distance similarity analyses between Fe and DFB treatments (Fe/DFB, Fe/Ctl for O5) within the diatom genera Pseudo-nitzschia and Thalassiosira (Figure 4). Expression responses within coastal sites clustered together, while the oceanic site O5 displayed distinctly different patterns in both taxa. At site O5, 83 out of 1,334 KEGG Orthology genes (KOs) in Pseudo-nitzschia demonstrated >16-fold higher expression in the added Fe treatment than in the Fe-limited control treatment (Figure 4, Supplemental Table 4). By comparison, 155 out of 1,241 KOs in Thalassiosira showed >16-fold higher expression in the added Fe treatment compared to the low Fe control treatment. The most highly differentially expressed genes in oceanic Pseudo-nitzschia following Fe enrichment were ferritin (FTN, 290-fold), a metal transporter (CNNM, 32-fold), a putative bicarbonate (HCO3) transporter (ICTB, 133-fold), and an NADPH-dependent glutamate synthase (GLT; 146-fold). In oceanic Thalassiosira, highly differentially expressed genes included ferredoxin-dependent sulfite reductase (Fd-SIR, 74-fold) and ferredoxin-dependent glutamate synthase (Fd-GLT; 416-fold). Fe addition induced both genera to increase the expression of several genes involved in photosynthesis by >16-fold exclusively at this location. Both taxa overexpressed gene products involved in vitamin biosynthesis, including the Fe-dependent vitamin B7 synthesis protein biotin synthase (BIOB), which increased expression in the Fe enriched treatment by 84- and 49-fold in Pseudo-nitzschia and Thalassiosira, respectively. Furthermore, Pseudo-nitzschia increased expression of the vitamin B1 (thiamine) biosynthetic gene THIC (by 179-fold) and vitamin B6 (pyridoxine) biosynthetic genes pyridoxine kinase (PDXK; by 74-fold) and pyridoxine 4-dehydrogenase (PLDH; by 152-fold) following Fe enrichment at the oceanic site.
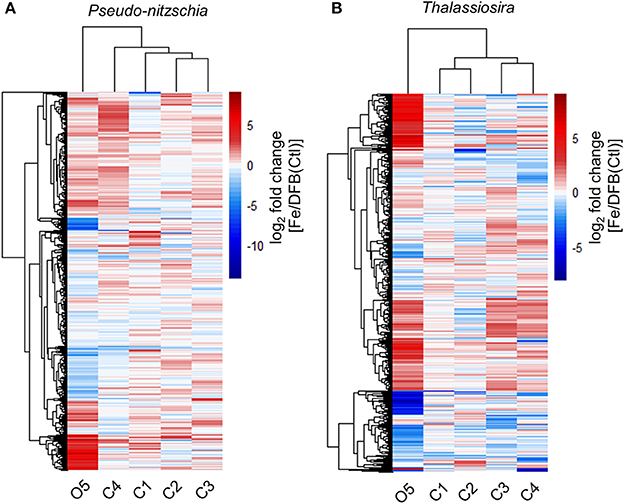
Figure 4. Differential expression response of shared KEGG Orthologs (KOs) between the Fe and DFB treatments at T1 in the diatom genera Pseudo-nitzschia (A) and Thalassiosira (B). Heatmap represents the log2 fold change in gene expression within the Fe addition treatment relative to the DFB addition treatment at each site. For site O5, the T2 control (Ctl) treatment is used as the Fe-limited comparison. Only KOs with transcript abundances >5 log2 CPM are included. Dendrograms reflect similarity in expression responses among sites (columns) or KOs (rows).
A number of genes demonstrated higher expression in the Fe-limited control treatment at O5. Forty-eight out of 1,334 genes in Pseudo-nitzschia and 77 out of 1,241 genes in Thalassiosira showed >16-fold higher expression in the Ctl treatment than in the added Fe treatment, patterns that were not found in diatoms from the examined coastal sites (Figure 4). In Thalassiosira, these genes encode proteins such as the copper (Cu)/zinc (Zn) superoxide dismutase (Cu-Zn SOD), an enzyme that removes toxic superoxide radicals by dismuting them into molecular oxygen and hydrogen peroxide, and a divalent metal transporter belonging to the ZIP family (ZIP7) (Marchetti and Maldonado, 2016). In both taxa, ribulose-1,5-bisphosphate carboxylase oxygenase (RubisCO; large subunit; RBCL), which catalyzes C-fixation in the Calvin cycle, had ≥24-fold higher expression in the Ctl treatment at O5.
Influence of Fe Availability on Fe Metabolism
The expression of genes involved in cellular growth and function, including N and C assimilation, vitamin synthesis, Fe-related metabolism, and trace metal acquisition, were compared in the dominant diatom genera Pseudo-nitzschia and Thalassiosira between the Fe and DFB/Ctl treatments (Figure 5). Genes encoding proteins involved in metal transport were detected at all locations, with expression patterns varying depending on site and taxa. Pseudo-nitzscha increased expression of the Fe transporter ABC.FEV.S by >2-fold under Fe enrichment at all locations where incubated communities showed a physiological Fe effect (C3, C4, O5; Supplementary Table 1). Transcripts for another Fe uptake protein, the high affinity iron permease FTR, were generally more abundant in the DFB/Ctl treatments in Thalassiosira, although the gene was more highly expressed following Fe enrichment in Pseudo-nitzschia at sites C2, C3, and O5 (Figure 5). The putative metal transporter CNNM was 32-fold more highly expressed following Fe enrichment in Pseudo-nitzschia at the oceanic site, but was not detected in Thalassiosira. Conversely, the non-specific metal transporter ZIP7 was 21-fold more highly expressed under Fe-limiting conditions in oceanic Pseudo-nitzschia and similarly not detected in oceanic Thalassiosira. Transcripts for Fe starvation induced proteins (ISIPs), including the recently-identified Fe acquisition protein ISIP2A that binds Fe at the cell surface and is thought to be involved in intracellular Fe transport (Morrissey et al., 2015), were highly abundant in Fe-stressed treatments (e.g., DFB and/or Ctl depending on the site) across all sites and in both taxa (Figure 5). Although their specific functions in diatoms are unclear, other ISIPs were markedly abundant and differentially expressed in the DFB/Ctl treatments, with ISIP1 one of the most differentially expressed genes between Fe-replete and Fe-limited treatments at each experimental site and in both taxa (Supplementary Figure 2).
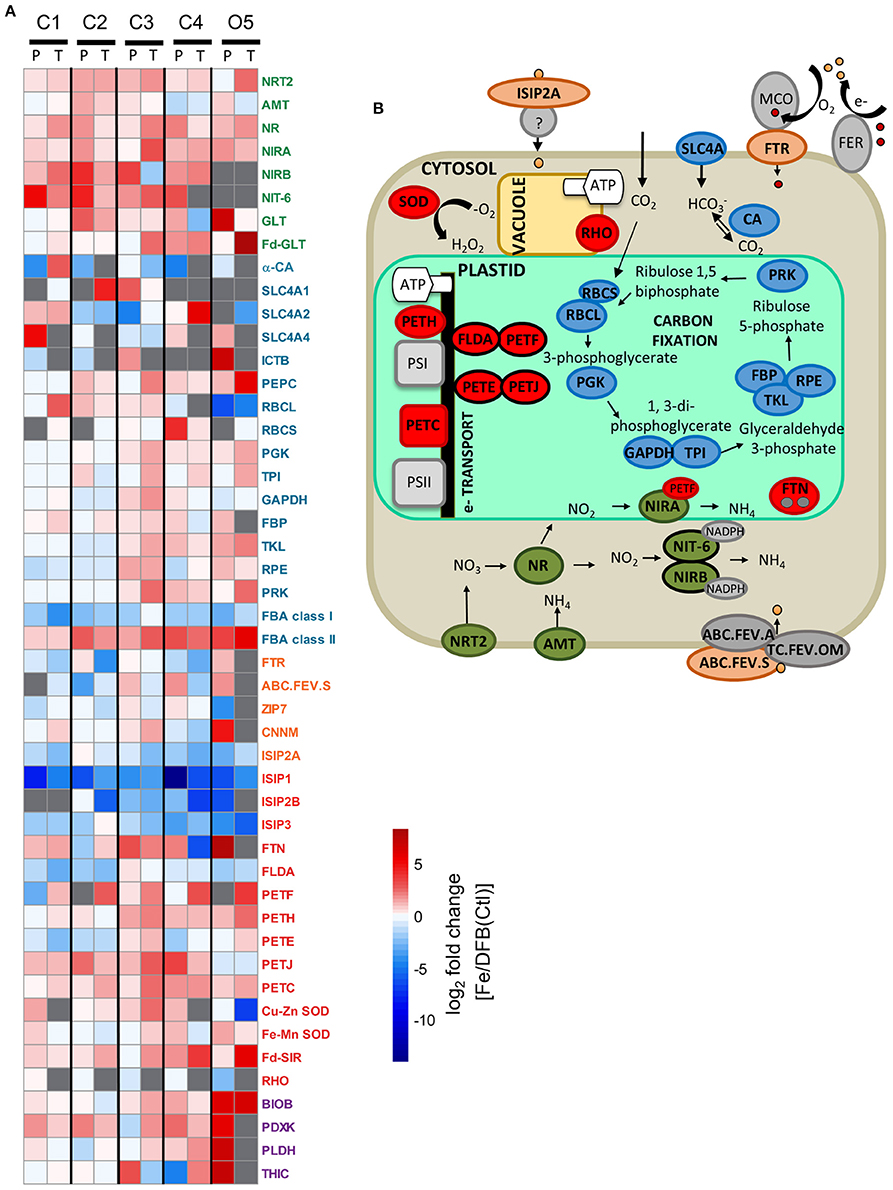
Figure 5. Differential expression responses of select genes involved in nitrogen (N; green), carbon (C; blue), metal transport (orange), iron (Fe; red), and vitamin (purple)-related processes between the T1 Fe and DFB/Ctl treatments within the diatom genera Pseudo-nitzschia (P) and Thalassiosira (T) (A). Heatmap represents the log2 fold change of gene expression within the Fe addition treatment relative to the DFB treatment at each site. For site O5, the T2 control (Ctl) treatment is used as the Fe-limited comparison. Gray boxes indicate transcripts were not detected in either treatment. White boxes signify no change in expression between treatments. A schematic representation of select N, C, Fe, metal transport, and vitamin-related processes within a diatom cell, color-coded by genes of interest included in (A) is provided (B). Adjacent proteins with black borders indicate similar cellular functions (e.g., FLDA, PETF). Gene abbreviations are NRT2, nitrate transporter; AMT, ammonium transporter; URTA, urea transporter; NR, nitrate reductase; NIRA, ferredoxin-nitrite reductase; NIRB, nitrite reductase; NIT-6, nitrite reductase; GLT, glutamate synthase; Fd-GLT, ferredoxin-glutamate synthase; α-CA, carbonic anhydrase (α family); SLC4A, solute carrier family (bicarbonate transporters); ICTB, putative bicarbonate transporter; PEPC, phosphoenolpyruvate carboxylase; RBCL, RubisCO large subunit; RBCS, RubisCO small subunit; PGK, phosphoglycerate kinase; TPI, triseophosphate isomerase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; FBP, fructose-1,2-bisphosphatase I; TKL, transketolase; RPE, ribulose-phosphate 3-epimerase; PRK, phosphoribulokinase; FBA class I, fructose bisphosphate aldolase (class I); FBA class II, fructose bisphosphate aldolase (classII); FTR, high affinity iron permease; ABC.FEV.S: iron complex transport system substrate-binding protein; ZIP7, zinc transporter 7; CNNM, metal transporter; ISIP2A, iron starvation induced protein 2A; ISIP1, iron starvation induced protein 1; ISIP2B, iron starvation induced protein 2B; ISIP3, iron starvation induced protein 3; FTN, ferritin; FLDA, flavodoxin I; PETF: ferredoxin; PETH, ferredoxin-NADP+ reductase; PETE, plastocyanin; PETJ, cytochrome c6; PETC, cytochrome b6f complex; Cu-Zn SOD, superoxide dismutase containing Cu and Zn as cofactors; Fe-Mn SOD, superoxide dismutase containing Fe or Mn as cofactor; Fd-SIR, ferredoxin-sulfite reductase; RHO, rhodopsin (note the localization of RHO within the vacuole membrane is speculative); BIOB, biotin synthase; PDXK, pyridoxal kinase; PLDH, pyridoxal 4-dehydrogenase; THIC, phosphomethylpyrimidine synthase.
Other Fe-related metabolic processes similarly varied depending on both site and taxa. Differences in expression patterns between taxa were generally greater for these Fe-related genes than in the N- and C-related genes investigated (Figure 5). At most sites, transcripts for the Fe storage protein ferritin (FTN) were higher in the Fe addition treatments than in the DFB/Ctl treatments. However, at two sites (C2 and C4), FTN transcripts were more abundant in the DFB treatment compared to the Fe addition treatment for one of the two genera (e.g., at site C2, 3.5-fold higher in Pseudo-nitzschia, p = 1 × 10−3 and at site C4, 90-fold higher in Thalassiosira). SODs were additionally differentially expressed, but they showed different expression patterns depending on the enzymes' metal cofactor(s) and the diatom genus. Cu-Zn SOD, which contains both Cu and Zn at its active site, showed a >100-fold higher expression in Thalassiosira in the Fe-limited control than in the added Fe treatment at the Fe-limited site O5. In contrast, in the same Fe-limited control treatment at this location, Pseudo-nitzschia demonstrated 2-fold higher expression of Fe-Mn SOD, which contains either Fe or manganese (Mn) as its metal cofactor. Based on the presence of Mn-coordinating amino acids at sites G-77 and Q-146 of the most highly expressed Fe-Mn SOD contigs, this Pseudo-nitzschia SOD was determined to specifically utilize Mn as its metal cofactor (Crowley et al., 2000; Groussman et al., 2015) (Supplementary Figure 4C).
Transcriptional responses of genes encoding Fe-dependent proteins and their functional replacements in photosynthetic electron transport were examined in both diatom genera (Figure 5). Transcripts for the Fe-independent protein flavodoxin (FLDA), which functionally replaces the Fe-protein ferredoxin (PETF) in photosynthetic electron transport, were generally more abundant in the DFB/Ctl treatments than in the Fe treatments in both genera (Figure 5). Conversely, transcripts of PETF were >2-fold higher in the high-Fe treatment only in Thalassiosira and across all sites. In Pseudo-nitzschia, PETF transcripts were either constitutively expressed (C3 and C4), more highly expressed in the DFB treatment (C1), or not present (C2 and O5) (Figure 5). Transcripts of cytochrome c6 (PETJ) and its functional non-Fe replacement, the copper-protein plastocyanin, also showed differences in gene expression. PETJ transcripts were more abundant in the high Fe treatment at all sites and in both genera, except O5, where it was slightly more abundant in the Fe-limited control treatment (Figure 5). By contrast, transcripts for plastocyanin (PETE) displayed inconsistent expression trends in response to Fe status across sites, being relatively more abundant following Fe enrichment in both genera at C3 (1.4-fold in Pseudo-nitzschia; 1.9-fold in Thalassiosira, p = 5 × 10−4) and at the initially Fe-limited oceanic site, O5 (1.4-fold in Thalassiosira; Figure 5). At all other locations PETE transcripts were either more abundant under DFB conditions or not detected.
Transcripts for the proton-pumping protein rhodopsin (RHO) furthermore demonstrated differences in expression patterns among genera. This protein can supplement Fe-intensive photosynthesis in the light-driven production of membrane proton gradients and ATP in some diatoms (Marchetti et al., 2015). Rhodopsin was not detected in Thalassiosira at any location while its expression increased in Pseudo-nitzschia by >2-fold in the DFB/Ctl treatments relative to the Fe treatment at the two lowest dFe sites [C3 (p = 0.01) and O5; Figure 5; Supplementary Table 1]. At the other sites RHO expression was constitutive. These rhodopsin contigs were structurally similar to diatom rhodopsins identified within the MMETSP database (≥55% similarity; Supplementary Figure 4B).
Relationships among Fe-related transcript abundance, experimental site and treatment were determined using Principal Components Analysis (PCA) individually for each diatom genus. Principle components P1 and P2 explained 54% of the variation in transcript abundance in Pseudo-nitzschia and 76% in Thalassiosira (Figure 6C). In Pseudo-nitzschia, transcripts for the photosynthetic genes ferredoxin-NADP+ reductase (PETH), PETJ, a cytochrome b6/f complex protein (PETC), FTN, and Cu-Zn SOD were in higher relative abundance within Fe addition treatments while RHO, ISIPs, FLDA, PETE, and FTR were generally more abundant in the Ctl and/or DFB treatments, as the principle component P1 separated these samples based on Fe treatment. In Thalassiosira, a similar response was observed, although RHO was not detected, and PETF, which was sporadically found and not abundant in Pseudo-nitzschia, strongly co-varied with the other genes highly expressed in the treatments where Fe was added (Figure 6C).
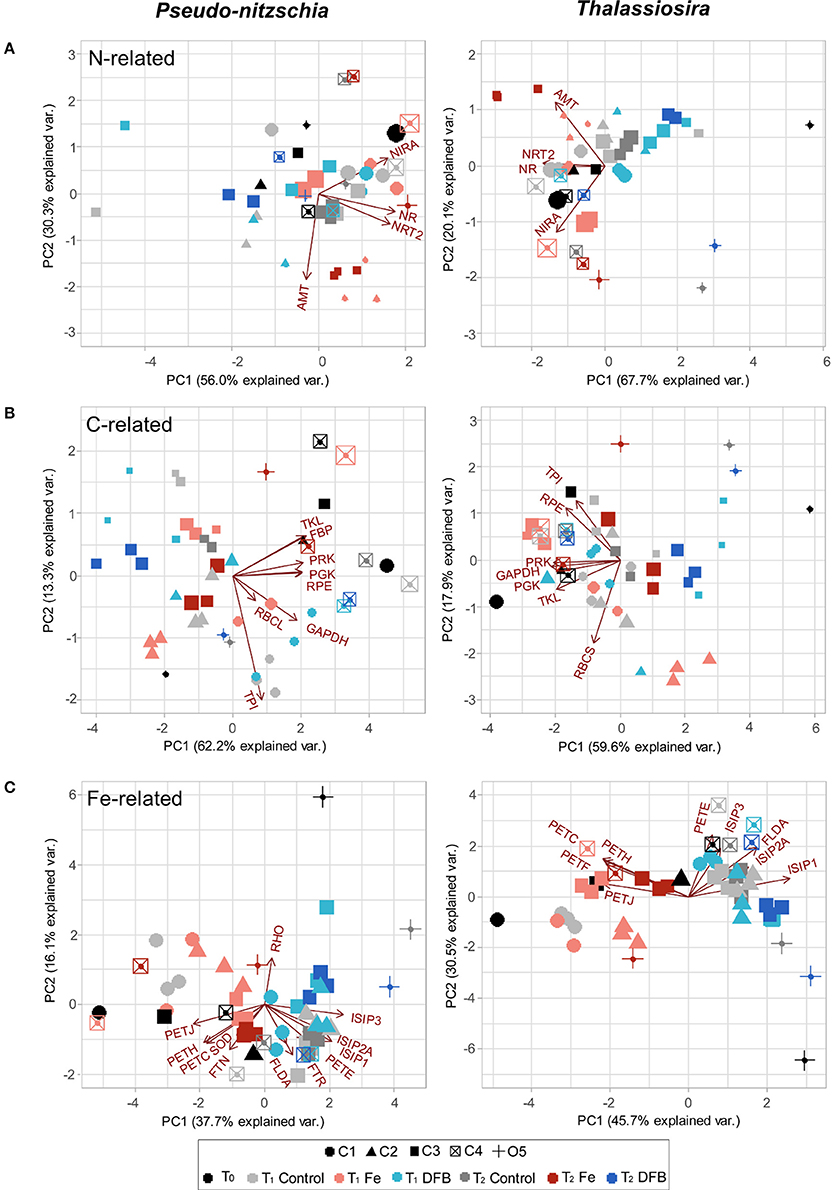
Figure 6. PCA bi-plots depicting the relationship between treatment (color), site (shape), and biomass-normalized N and C rates for transcript abundances of genes involved in N, C, and Fe-related processes in Pseudo-nitzschia (left) and Thalassiosira (right). Size of points scales with increasing PN-specific VNO3 (day−1) from 0.04 to 1.63 day−1 (A), and with increasing PC-specific VDIC (day−1) from 0.01 to 1.50 day−1 (B). For the Fe-related genes transcript abundance PCA bi-plot, sizes remain constant across samples (C). See Figure 5 for the list of gene abbreviations.
Influence of Fe Availability on N Metabolism
Genes involved in N transport and metabolism were investigated to assess the influence of varying Fe status on N assimilation. Transcripts for genes encoding nitrate (NRT2) and ammonium (AMT) transporters were detected at all locations, with NRT2 increasing in expression by >2-fold in response to Fe addition relative to the DFB/Ctl treatment at the majority of sites in both taxa, while AMT expression varied depending on site (Figure 5). For instance, C4 was the only location with a >2-fold increase in AMT expression in the DFB treatment in both Pseudo-nitzschia and Thalassiosira. Transcripts corresponding to genes encoding components of assimilation, including nitrate (NR) and nitrite reductases (NIRA, NIRB, NIT-6) were generally more abundant in the treatments with added Fe, although NIRA and NIRB displayed opposite expression patterns in Pseudo-nitzschia and Thalassiosira at site C3 (Figure 5). Furthermore Pseudo-nitzschia increased gene expression of one group of nitrite reductases [NIRB and NIT-6, which use NADPH as the reductant (Brown et al., 2009)] by 11- and 3.6-fold, respectively, following added Fe while Thalassiosira conversely increased NIRB expression by 3.7-fold in the DFB treatment (Figure 5). In addition, Thalassiosira increased gene expression of another form of nitrite reductase (NIRA, which uses ferredoxin/flavodoxin as reductant; Brown et al., 2009) by 8-fold (p = 3 × 10−22) following Fe enrichment while Pseudo-nitzschia constitutively expressed NIRA at this location. Noticeably, transcripts for the genes encoding NIRB and NIT-6 were present in at least one of the two diatom taxa examined at all sites except the oceanic site, O5.
The relationships among transcript abundance for N uptake and assimilation-related genes, experimental sites, treatments and PN-specific VNO3 measurements within the >5 μm size fraction of the phytoplankton community were examined via PCA bi-plots. Principle components P1 and P2 explained 86% of the variation in N-related transcript abundance in Pseudo-nitzschia and 88% in Thalassiosira (Figure 6A). Sites generally contained high transcript abundances of NRT2 and NR in the added Fe treatment, with the two genes strongly co-varying with one another in both Pseudo-nitzschia and Thalassiosira. Furthermore, the Fe addition treatments at two sites that experienced depletion following incubation, C2 and C3, clustered together and contained the highest AMT transcript abundance at T1 and T2, respectively. Phytoplankton communities within these incubation treatments concomitantly displayed low PN-specific VNO3 (0.03–0.13 day−1; Figure 6A). The highest PN-specific VNO3 were observed in the added Fe treatment at site C4 at T1 and at C1 within the initial (T0) phytoplankton community (1.4 day−1), which coincided with high abundances of NIRA transcripts in both genera at these locations.
Influence of Fe Availability on C Metabolism
To further gain insight into how variable Fe status influences macronutrient resource utilization and regional biogeochemistry, genes involved in C transport and fixation were examined among sites and between diatom genera. Transcripts corresponding to a carbonic anhydrase belonging to the α-family (α-CA), involved in the carbon concentrating mechanism (CCM) within photosynthetic eukaryotes (Reinfelder, 2011), were either constitutively expressed, not detected, or more highly expressed in the DFB treatment at all locations apart from C1, where expression was 7-fold higher following Fe addition in Thalassiosira (Figure 5).
Members of the solute carrier (SLC) family of bicarbonate transporters (SLC4A-1, -2, and -4), which import bicarbonate ions from the environment also thought to be involved in the CCM (Nakajima et al., 2013), were detected intermittently among sites, though in low transcript abundance (Figure 5). These genes share sequence homology with the P. tricornutum genes PtSLC4-1, -2, and -4 (BLASTP; E <2 × 10−69) and displayed inconsistent patterns of gene expression with each another, with no clear relationship to carbon assimilation rates. Another putative bicarbonate transporter (ICTB) was detected intermittently across sites and solely in Pseudo-nitzschia, where it was notably more highly expressed by 128-fold following Fe addition at O5. Conversely in Thalassiosira, the gene encoding phosphoenolpyruvate carboxylase (PEPC), which is part of a C4-CCM in some species of this genus (Reinfelder, 2011), was more highly expressed by 73-fold following Fe addition at O5.
Gene expression of RubisCO (RBCL) was higher by >24-fold in the Fe-limited control treatment in both genera at site O5 while at other sites the gene was either constitutively expressed, increased expression in the added Fe treatment, or not detected (Figure 5). In addition, other genes involved in the Calvin Cycle, including phosphoglycerate kinase (PGK), transketolase (TKL), ribulose-phosphate 3-epimerase (RPE), and phosphoribulokinase (PRK), generally increased in expression following Fe addition compared to the DFB/Ctl treatment at one or more of the three sites experiencing some degree of Fe limitation (C3, C4, and O5; Supplementary Table 1; Figure 5). At the CUZ sites C1 and C2, transcripts for these genes were either not differentially expressed or were more abundant in the DFB treatment within both diatom genera. Fructose-bisphosphate aldolases (FBA), involved in the Calvin Cycle, glycolysis, and gluconeogenesis, demonstrated strong Fe-dependent transcriptional patterns regardless of site and taxa (Figure 5). Transcripts corresponding to class II FBA, likely a metal-dependent aldolase, increased by 1.5 to 69-fold in the added Fe treatment as compared to DFB treatments with the largest fold change attributed to Pseudo-nitzschia from O5. Class II FBA has been previously demonstrated to be abundant under high-Fe conditions in diatoms and is hypothesized to contain Fe2+ as a metal cofactor (Horecker et al., 1972; Allen et al., 2012; Lommer et al., 2012). Transcripts corresponding to class I FBA, the metal-independent version of class II FBA, conversely increased by 1.3 to 16-fold in DFB compared to Fe treatments.
The relationships in transcript abundance among C fixation-related genes, experimental sites, incubated treatments and PC-specific VDIC measurements were assessed using PCA bi-plots. Principle components P1 and P2 together explained 80% of the variation in C-related transcript abundance in Pseudo-nitzschia and 78% in Thalassiosira (Figure 6B). Site C4 contained some of the highest PC-specific VDIC measurements within the >5 μm size fraction (0.65–1.6 day−1), and coincided with the highest transcript abundances of PGK, PRK, FBP, TKL, RPE, and GAPDH in Pseudo-nitzschia (Figure 6B). Conversely, Fe-limited treatments from C3 and O5 had the lowest transcript abundances of these genes in both Pseudo-nitzschia and Thalassiosira, with principle component P1 separating these samples from other sites and treatments (Figure 6C). Fe-limited sites C3 and O5 phytoplankton communities additionally displayed some of the lowest PC-specific VDIC observed (0.11–0.17 day−1).
Discussion
Prior to this study, our understanding of the strategies utilized by phytoplankton to cope with low Fe bioavailability and resupply across different coastal and oceanic regions was limited. Furthermore, whether diverse diatom genera from identical environments would respond similarly when exposed to changes in Fe availability was unresolved. The gene expression patterns presented here demonstrate that the cosmopolitan diatom genera Pseudo-nitzschia and Thalassiosira rely on diverse sets of strategies to handle Fe stress, and that oceanic diatoms from both groups are highly responsive to changes in Fe availability with a greater degree of differentially expressed genes involved in nitrate assimilation, carbon fixation, and vitamin production compared to their coastal counterparts.
Iron-Related Gene Expression Responses across Sites
Differences in gene expression patterns in response to Fe status were observed between the coastal (C1-C4) and oceanic sites (O5) examined in this study. This included the >16-fold higher expression of genes in the added Fe treatment relative to the Fe-limited control encoding proteins involved in B7 synthesis (BIOB) in both taxa, and B1 (THIC) and B6 (PDXK, PDLH) synthesis in Pseudo-nitzschia. These increases are consistent with previous field observations demonstrating that Fe enrichment of previously Fe-limited oceanic diatom communities stimulates B-vitamin transcript production (Cohen et al., 2017). Genes encoding an Fe storage protein (ferritin [FTN]) and components of amino acid metabolism (glutamate synthase [GLT] in Pseudo-nitzschia; ferredoxin-dependent glutamate synthase [Fd-GLT] in Thalassiosira) were similarly more highly expressed by >16-fold following Fe addition exclusively at site O5. Conversely, in the Fe-limited control, we observed the >16-fold higher expression of genes encoding the proteins Cu-Zn superoxide dismutase (Cu-Zn SOD) and RubisCO (RBCL), in either one or both taxa investigated. These distinct transcriptomic patterns of genes involved in diverse metabolic processes reflect differences in environmental factors selecting for diatom growth between the chronically Fe-limited open ocean and sporadically Fe-limited coastal regions.
In contrast, many photosynthetic genes were highly expressed following Fe addition regardless of location. A subset of these genes displayed distinct expression responses depending on whether the incubated communities experienced Fe limitation of growth rate (e.g., C3 and O5) or only Fe stress (C4; Supplementary Table 1). One such gene encodes the putative Fe transporter ABC.FEV.S, in which expression increased following Fe addition in Pseudo-nitzschia only at sites C3, C4, and O5. Additional genes include flavodoxin (FLDA) and plastocyanin (PETE), in which transcripts were generally more abundant in the DFB or Fe-limited Ctl treatments, consistent with flavodoxin's role as an Fe-independent photosynthetic electron carrier and plastocyanin's role as a Cu-dependent replacement for Fe-dependent cytochrome c6. At the Fe-stressed CUZ site C3 however, FLDA was either constitutively expressed or slightly more abundant after Fe addition, depending on the diatom genus. Plastocyanin (PETE) transcripts were similarly more abundant after Fe addition in both diatom genera at C3 and in Thalassiosira at O5. This pattern suggests coastal diatoms from higher-Fe systems tend to temporarily replace Fe-dependent photosynthetic proteins with Fe-independent ones, while certain diatoms in chronically Fe-limited environments may rely exclusively on the Fe-free alternatives (Marchetti et al., 2012).
Fe-Related Gene Expression Responses between Diatom Taxa
Pseudo-nitzschia and Thalassiosira demonstrated several distinct responses to changes in Fe status despite co-existing under identical environmental conditions. Ferredoxin (PETF), ferredoxin-dependent glutamate synthase (Fd-GLT), and ferredoxin-dependent sulfite reductase (Fd-SIR) transcripts were more abundant in Thalassiosira at oceanic site O5 following Fe addition with these responses absent in Pseudo-nitzschia. In contrast, ferredoxin-related transcripts in oceanic Pseudo-nitzschia were constitutively expressed or not detected. These patterns may suggest oceanic Thalassiosira strongly utilizes ferredoxin and ferredoxin-dependent proteins following Fe addition while Pseudo-nitzchia relies on Fe-independent machinery. Site O5 was additionally the only location in which Thalassiosira increased gene expression of Cu-Zn SOD under Fe-limitation. This pattern was not evident in oceanic Pseudo-nitzschia, where gene expression of this protein was constitutive, or by either genus at coastal sites, suggesting that the oceanic Thalassiosira species have distinctly evolved to rely on this Cu- and Zn-containing enzyme as the preferred superoxide dismutase in their Fe-limited environment. Pseudo-nitzschia conversely increased expression of Mn SOD following Fe addition, likely as a result of iron-induced increases in photosynthetic rates and photosynthetic production of superoxide radicals (Asada, 2006). These patterns highlight differences in preferred metal cofactors as a function of Fe status and transcriptional tendencies between the two taxa.
Transcripts corresponding to rhodopsin (RHO) increased in abundance within Pseudo-nitzschia in the DFB/Ctl treatments at the two sites experiencing pronounced Fe limitation (C3 and O5), but were not identified in Thalassiosira at any location. This is consistent with rhodopsin being undetected in sequenced Thalassiosira spp. transcriptomes (Marchetti et al., 2015) and supports the notion that Pseudo-nitzschia may have a competitive advantage over non-rhodopsin containing taxa, allowing for an Fe-independent alternative to photosynthesis for ATP generation during times of Fe stress. Ferritin (FTN) gene expression patterns furthermore diverged between the two taxa at the coastal sites C4 (Line-P) and C2 (CUZ). This supports laboratory findings suggesting FTN may exhibit different expression patterns among diverse phytoplankton (Marchetti et al., 2009; Botebol et al., 2015), even between taxa residing in the same location. Lastly, ABC.FEV.S, encoding a membrane Fe transport system protein, displayed divergent expression patterns between the examined genera with only Pseudo-nitzschia increasing ABC.FEV.S expression after Fe addition in all incubations exhibiting signs of iron limitation (C3, C4, and O5).
Taken together, these patterns in gene expression demonstrate that members of the pennate diatom genus Pseudo-nitzschia and the centric diatom genus Thalassiosira restructure their functional metabolisms in response to changes in Fe availability in distinct manners, possibly allowing both species to co-exist in the same environment. Both taxa are equipped with strategies to sustain growth under chronic Fe limitation in the open ocean, as supported by their equal transcript abundance during initial sampling. Following pulse Fe additions however, oceanic Pseudo-nitzschia relies in part on the strategies discussed above to gain a competitive advantage over Thalassiosira and quickly dominates the phytoplankton community. It remains unclear however which combination of environmental factors in the NE Pacific Ocean would select for the preferential growth of Thalassiosira over Pseudo-nitzschia. We conclude that substantial differences in molecular responses to changes in Fe status are observed across taxonomic groups, and patterns in gene expression should not be assumed universal across diverse taxa or environments.
Nitrogen-Related Gene Expression as a Function of Fe Status
The majority of N transport and assimilation genes investigated increased in expression following Fe addition in both Pseudo-nitzschia and Thalassiosira. Several site- and taxa-specific patterns were identified, with some trends also possibly explained by each site's initial concentration. For example, most gene copies encoding the transporter, NRT2, have been demonstrated in laboratory cultures to increase in expression in NO3-stressed diatoms (Bender et al., 2014; Rogato et al., 2015), and transcripts corresponding to this gene were some of the most abundant in both Pseudo-nitzschia and Thalassiosira at C2—the CUZ site where concentrations were depleted in all incubations by the first sampling time point. This gene also showed expression trends that correlated with Fe status; NRT2 transcripts were more abundant after Fe addition at all locations, regardless of initial concentrations. Based on these observations, NRT2 in diatoms also appears to be linked to Fe status and follows the expression of other N-related genes involved in Fe-dependent assimilation, including those encoding nitrate reductase (NR) and nitrite reductase (NIRA; Marchetti et al., 2012).
Diatoms were perhaps relying on NH4 in place of as a source of N based on gene expression patterns at several CUZ sites. Fe-enriched treatments at C2 contained the lowest after 48 h of incubation (0.06 μmol L−1), and the genes encoding the ammonium transporters AMTs concomitantly increased in expression in the Fe relative to DFB treatment (Figure 6). Furthermore at C3, Fe-enriched communities entered stress by the end of the incubation period, and AMT expression simultaneously increased in both Pseudo-nitzschia and Thalassiosira. This negative relationship between concentrations and AMT transcript abundance in natural diatom assemblages is consistent with those in laboratory Pseudo-nitzschia multiseries and Fragilariopsis cylindrus cultures (Bender et al., 2014; Rogato et al., 2015), and is reported here as one of the first observations of this relationship in natural phytoplankton communities.
High AMT transcript abundance at some of these locations may also represent NH4 rather than being preferred as an N source by Fe-stressed diatoms conserving their cellular Fe supply, as assimilation depends on various Fe-dependent processes (Milligan and Harrison, 2000). This is supported by the increased expression of AMT transcripts in both Pseudo-nitzschia and Thalassiosira from the Fe-stressed coastal Line-P incubations at C4. Pseudo-nitzschia from the Fe-limited site O5 also exhibited this pattern whereas Pseudo-nitzschia from C3 and Thalassiosira from both C3 and O5 did not, suggesting other environmental parameters aside from Fe status are influencing whether diatoms utilize NH4- or NO3-specific N uptake pathways.
Similar to our Fe-related gene expression results, several N-related genes demonstrated divergent expression responses between Pseudo-nitzschia and Thalassiosira. Expression of the reductase genes, NIRA and NIRB, displayed opposite patterns between the two genera at the CUZ site where Fe-stress occurred in incubations (C3), with Pseudo-nitzschia highly expressing the gene encoding non-ferredoxin-utilizing reductase (NIRB) following Fe addition, and Thalassiosira highly expressing the gene encoding the ferredoxin-utilizing nitrite reductase (NIRA). Furthermore at site O5, Pseudo-nitzschia increased expression of AMT and NADPH-dependent glutamate synthase (GLT) following Fe addition while Thalassiosira increased expression of NRT2 and ferredoxin-dependent glutamate synthase (Fd-GLT). These transcriptomic patterns may suggest Pseudo-nitzschia continues to rely on the non-Fe requiring metabolic pathways for assimilating N once Fe becomes available (AMT, NIRB, GLT), whereas Thalassiosira shifts over to Fe-dependent ones (NRT2, NIRA, Fd-GLT) upon Fe resupply.
These expression patterns furthermore support that substantial variations exist between the two diatom taxa in terms of N acquisition and assimilation strategies following changes in Fe supply. Both Pseudo-nitzschia and Thalassiosira are equipped with distinct strategies to compete under a variety of Fe and N conditions, and this may contribute to how multiple diatom species relying upon the same limiting resources in identical environments co-exist (i.e., paradox of the plankton; Hutchinson, 1961). These patterns are consistent with previous reports of resource partitioning among diatoms based on N and phosphate utilization (Alexander et al., 2015). Varying environmental pressure likely maintain populations of diverse diatom genera in the open ocean, with certain species outcompeting others depending on specific sets of external factors, including both macro- and micronutrients (Godhe and Rynearson, 2017).
Carbon-Related Gene Expression Responses as a Function of Fe Status
Genes encoding proteins involved in C uptake and assimilation were surveyed in order to determine the influence of Fe addition or stress on C metabolism. We observed site-specific expression patterns of the diatom RubisCO large subunit protein (RBCL), where gene expression was substantially elevated at site O5 in the Fe-limited control treatment relative to the Fe addition response in both diatom genera. A sequence analysis of RubisCO contigs obtained across experimental sites demonstrates that O5 protein sequences are structurally less similar to known Pseudo-nitzschia and Thalassiosira RubisCO protein sequences within the MMETSP database than those at the four coastal sites (Supplementary Figure 4A; Supplementary Table 3). This distinction in both protein structure and transcriptional expression may indicate a distinct adaptation and utilization of RubisCO in the oceanic diatoms than in those from high-Fe coastal waters. Phylogenetically diverse diatom species have been demonstrated to vary in their RubisCO enzyme kinetics in laboratory cultures, with their RubisCO content inversely linked to the strength of their carbon concentrating mechanism (CCM; Young et al., 2016). The CCM increases CO2 concentrations in chloroplast stroma in the vicinity of RubisCO and is fueled by the energy (ATP) generated from the Fe-intensive process of photosynthesis (Reinfelder, 2011; Young et al., 2016). We hypothesize that chronically Fe-limited oceanic diatoms are ATP-limited by the scarcity of Fe needed to support photosynthesis, and instead increase their RubisCO protein content to maintain high rates of carbon fixation rather than allocate scarce energy resources to the CCM. Furthermore, the genes encoding a putative bicarbonate transporter (ICBT) and a C4-CCM component (PEPC; Reinfelder et al., 2000; Sage, 2004; Reinfelder, 2011) were highly expressed following Fe addition in Pseudo-nitzschia and Thalassiosira, respectively, exclusively at O5. This supports that diatoms may be capable of shuffling energy pools into either the CCM or RubisCO production depending on Fe bioavailability. Interestingly, in laboratory-based proteomic analyses with cultures of the coastal diatom T. pseudonana, RubisCO was similarly more highly expressed under Fe limitation, while PEPC protein levels were higher under Fe-replete conditions (Nunn et al., 2013). Consistent with our hypothesis, Hopkinson et al. (2010) attributed increases in biomass following CO2-enrichment of an Fe-limited phytoplankton community in the HNLC Northeast Pacific Ocean to downregulation of the CCM in order to conserve iron and photosynthetically-produced energy. Laboratory-based RubisCO kinetic work with cultured diatom isolates is needed to confirm whether diatoms from HNLC regions minimize their photosynthetic demand for Fe by synthesizing more RubisCO enzymes rather than allocating scarce energy resources into the CCM.
Other C fixation-related gene expression patterns were largely consistent with C assimilation rates, and generally varied as a function of both Fe status and ocean province. The genes PGK, TKL, RPE, and PRK did not exhibit site-specific expression patterns similar to RBCL, and instead increased in expression following Fe enrichment at sites where Fe addition increased C assimilation rates (C3, C4, and O5). Increased expression of these genes is expected with Fe stimulation of C-fixation and growth. These expression patterns are in agreement with laboratory cultures of the diatom P. tricornutum, which increased expression of genes involved in C fixation during the light portion of their diel cycle, when DIC is being taken up to support photosynthesis (Chauton et al., 2013).
Conclusion
Gene expression characterization coupled with biological rate processes across geographically diverse communities suggests regional and taxa-specific strategies are utilized by diatoms when rapidly responding to variations in environment. Our analysis demonstrates that chronically Fe-limited oceanic diatoms will restructure Fe, N, and C metabolism in a distinctive manner following Fe addition when compared to the response of coastal diatom communities that receive inherently more variable Fe inputs. Pseudo-nitzschia and Thalassiosira, two cosmopolitan diatom taxa found at all locations investigated, at times demonstrated divergent transcriptomic responses to changes in Fe status in terms of photosynthetic processes and N metabolism, even under identical environmental conditions.
Potential limitations to our approach include gene expression analyses being conducted on specific diatom genera while the physiological rate process measurements correspond to bulk phytoplankton communities. We therefore assumed the physiological characteristics to be representative of all phytoplankton members present. Furthermore, the metatranscriptomic approach used here consisted of analyzing cumulative expression responses of pooled gene copies; however, distinct gene copies have been shown to vary in their transcriptional response to environmental conditions within a single organism (Bender et al., 2014; Levitan et al., 2015; Rogato et al., 2015). In order to gain further resolution, we recommend laboratory-based studies be performed investigating the direct relationships between nutrient uptake rates and expression of specific gene copies encoding proteins involved in nutrient assimilation in distinct members from each of the genera Pseudo-nitzschia and Thalassiosira.
The findings presented here support the notion that a tremendous degree of genetic diversity is contained within the diatom lineage, and this may strongly influence the abundance and distribution of phytoplankton communities. Since Fe bioavailability to phytoplankton is predicted to change with increasing temperature and acidification of surface seawater (Shi et al., 2010; Sunda, 2010; Capone and Hutchins, 2013; Hutchins and Boyd, 2016), these findings will aid in predicting the consequences of changing ocean conditions on phytoplankton productivity and community growth dynamics.
Author Contributions
AM, BT, and KB designed the study; NC, KE, BT, and AM performed the incubation experiments; NC conducted the metatranscriptomic and physiological analysis; RL provided bioinformatic support; FK and KT obtained photophysiological measurements onboard the R/V Melville; MB and HM quantified biogenic silica; MM provided primary productivity measurements; CT and BT quantified trace metals; WS contributed to iron metabolism interpretations; SB quantified domoic acid; NC and AM wrote the manuscript. All authors contributed to intellectual content and approved the final manuscript.
Funding
Research was funded through National Science Foundation grants OCE-1334935 to AM, OCE-1334632 to BT, OCE-1334387 to MB, OCE-1333929 to KT, and OCE-1259776 to KB, as well as through Discovery NSERC grant 261521-13 to MM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank J. Roach (UNC), S. Haines (UNC), W. Gong (UNC), M. Kanke (UNC), S. Davies (UNC), and M. Love (UNC) for sequencing analysis advice and guidance. P. Morton (FSU) analyzed the dissolved Fe samples from the Line P cruise (C4 and O5). Dissolved nutrients were analyzed by T. Coale (SIO) onboard the R/V Melville (C1-C3), and by M. Belton (IOS) onboard the CCGS J.P. Tully (C4 & O5). Z. Li (Duke) provided climatological remote sensing images in Figure 1. We are grateful to the scientists and crew of the CCGS J.P. Tully (Line-P cruise 2015-09) and the R/V Melville (cruise 1405) for their support and assistance at sea. RNA-Seq data was processed using UNC's Research Computing clusters. Finally, we would like to thank Carolyn Duckham for her help collecting and processing samples for DIC uptake rates at stations C1–C3.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2017.00360/full#supplementary-material
References
Alexander, H., Jenkins, B. D., Rynearson, T. A., and Dyhrman, S. T. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. U.S.A. 112, E2182–E2190. doi: 10.1073/pnas.1421993112
Allen, A. E., LaRoche, J., Maheswari, U., Lommer, M., Schauer, N., Lopez, P. J., et al. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443. doi: 10.1073/pnas.0711370105
Allen, A. E., Moustafa, A., Montsant, A., Eckert, A., Kroth, P. G., and Bowler, C. (2012). Evolution and functional diversification of fructose bisphosphate aldolase genes in photosynthetic marine diatoms. Mol. Biol. Evol. 29, 367–379. doi: 10.1093/molbev/msr223
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Armbrust, E. V. (2009). The life of diatoms in the world' s oceans. Nature 459, 185–192. doi: 10.1038/nature08057
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/pp.106.082040
Barwell-Clarke, J., and Whitney, F. (1996). Institute of Ocean Sciences Nutrient Methods and Analysis. Canadian Technical Report of Hydrography and Ocean Sciences no. 182. Available online at: http://linep.waterproperties.ca/2012-13/documents/Barwell-Clarke_Whitney_1996.pdf
Behrenfeld, M. J., and Milligan, A. J. (2013). Photophysiological expressions of iron stress in phytoplankton. Annu. Rev. Mar. Sci. 5, 217–246. doi: 10.1146/annurev-marine-121211-172356.
Bender, S., Durkin, C., Berthiaume, C., Morales, R., and Armbrust, E. V. (2014). Transcriptional responses of three model diatoms to nitrate limitation of growth. Front. Mar. Sci. 1:3. doi: 10.3389/fmars.2014.00003
Biller, D. V., and Bruland, K. W. (2012). Analysis of Mn, Fe, Co, Ni, Cu, Zn, Cd, and Pb in seawater using the Nobias-chelate PA1 resin and magnetic sector inductively coupled plasma mass spectrometry (ICP-MS). Mar. Chem. 130–131, 12–20. doi: 10.1016/j.marchem.2011.12.001
Birol, I., Jackman, S. D., Nielsen, C. B., Qian, J. Q., Varhol, R., Stazyk, G., et al. (2009). De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877. doi: 10.1093/bioinformatics/btp367
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Botebol, H., Lesuisse, E., Sutak, R., Six, C., Lozano, J.-C., Schatt, P., et al. (2015). Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 112, 1–6. doi: 10.1073/pnas.1506074112
Boyd, P., and Harrison, P. J. (1999). Phytoplankton dynamics in the NE subarctic Pacific. Deep-Sea Res. II 46, 2405–2432. doi: 10.1016/S0967-0645(99)00069-7
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Brown, K. L., Twing, K. I., and Robertson, D. L. (2009). Unraveling the regulation of nitrogen assimilation in the marine diatom Thalassiosira pseudonana (Bacillariophyceae): diurnal variations in transcript levels for five genes involved in nitrogen assimilation. J. Phycol. 45, 413–426. doi: 10.1111/j.1529-8817.2009.00648.x
Bruland, K. W., Lohan, M. C., Aguilar-Islas, A. M., Smith, G. J., Sohst, B., and Baptista, A. (2008). Factors influencing the chemistry of the near-field Columbia River plume: Nitrate, silicic acid, dissolved Fe, and dissolved Mn. J. Geophys. Res. Ocean. 113:C00B02. doi: 10.1029/2007JC004702
Bruland, K. W., Rue, E. L., and Smith, G. J. (2001). Iron and macronutrients in California coastal upwelling regimes: implications for diatom blooms. Limnol. Oceanogr. 46, 1661–1674. doi: 10.4319/lo.2001.46.7.1661
Brzezinski, M. A. (1985). The Si:C:N ratio of marine diatoms: interspecific variability and the effect of some environmental variables. J. Phycol. 21, 347–357. doi: 10.1111/j.0022-3646.1985.00347.x
Brzezinski, M. A., Krause, J. W., Bundy, R. M., Barbeau, K. A., Franks, P., Goericke, R., et al. (2015). Enhanced silica ballasting from iron stress sustains carbon export in a frontal zone within the California Current. J. Geophys. Res. Ocean. 120, 4654–4669. doi: 10.1002/2015JC010829
Capone, D. G., and Hutchins, D. A. (2013). Microbial biogeochemistry of coastal upwelling regimes in a changing ocean. Nat. Geosci. 6, 711–717. doi: 10.1038/ngeo1916
Chase, Z., Hales, B., Cowles, T., Schwartz, R., and van Geen, A. (2005). Distribution and variability of iron input to Oregon coastal waters during the upwelling season. J. Geophys. Res. C Ocean. 110, 1–14. doi: 10.1029/2004JC002590
Chauton, M. S., Winge, P., Brembu, T., Vadstein, O., and Bones, A. M. (2013). Gene regulation of carbon fixation, storage, and utilization in the diatom phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161, 1034–1048. doi: 10.1104/pp.112.206177
Cohen, N. R., Ellis, K. A., Burns, W. G., Lampe, R. H., Schuback, N., Johnson, Z., et al. (2017). Iron and vitamin interactions in marine diatom isolates and natural assemblages of the Northeast Pacific Ocean. Limnol. Oceanogr. 62, 2076–2096. doi: 10.1002/lno.10552
Crowley, J. D., Traynor, D. A., and Weatherburn, D. C. (2000). Enzymes and proteins containing manganese: an overview. Met. Ions Biol. Syst. 37, 209–278.
de Baar, H. J. W., Boyd, P. W., Coale, K. H., Landry, M. R., Tsuda, A., Assmy, P., et al. (2005). Synthesis of iron fertilization experiments: from the iron age in the age of enlightenment. J. Geophys. Res. C Ocean. 110, 1–24. doi: 10.1029/2004JC002601
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Godhe, A., and Rynearson, T. (2017). The role of intraspecific variation in the ecological and evolutionary success of diatoms in changing environments. Philos. Trans. R. Soc. B Biol. Sci. 372:20160399. doi: 10.1098/rstb.2016.0399
Gorbunov, M. Y., and Falkowski, P. (2004). “Fluorescence Induction and Relaxation (FIRe) technique and instrumentation for monitoring photosynthetic processes and primary production in aquatic ecosystems,” in 13th International Congress of Photosynthesis, Vol. 2, eds A. van der Est and D. Bruce (Montreal, QC: Allen Press), 1029–1031.
Graff van Creveld, S., Rosenwasser, S., Levin, Y., and Vardi, A. (2016). Chronic iron limitation confers transient resistance to oxidative stress in marine diatoms. Plant Physiol. 172, 968–979. doi: 10.1104/pp.16.00840
Groussman, R. D., Parker, M. S., and Armbrust, E. V. (2015). Diversity and evolutionary history of iron metabolism genes in diatoms. PLoS ONE 10:e0129081. doi: 10.1371/journal.pone.0129081
Harris, S. L., Varela, D. E., Whitney, F. W., and Harrison, P. J. (2009). Nutrient and phytoplankton dynamics off the west coast of Vancouver Island during the 1997/98 ENSO event. Deep Sea Res. Part II Top. Stud. Oceanogr. 56, 2487–2502. doi: 10.1016/j.dsr2.2009.02.009
Harrison, P. J. (2002). Station papa time series: insights into ecosystem dynamics. J. Oceanogr. 58, 259–264. doi: 10.1023/A:1015857624562
Hildebrand, M. (2005). Cloning and functional characterization of ammonium transcporters fromt he marine diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 41, 105–113. doi: 10.1111/j.1529-8817.2005.04108.x
Hopkinson, B. M., Xu, Y., Shi, D., McGinn, P. J., and Morel, F. M. M. (2010). The effect of CO2 on the photosynthetic physiology of phytoplankton in the Gulf of Alaska. Limnol. Oceanogr. 55, 2011–2024. doi: 10.4319/lo.2010.55.5.2011
Horecker, B. L., Tsolas, O., and Lai, C.-Y. (1972). “6 Aldolases,” in The Enzymes, ed P. D. Boyer (San Diego, CA: Academic Press), 213–258.
Hutchins, D. A., and Boyd, P. W. (2016). Marine phytoplankton and the changing ocean iron cycle. Nat. Clim. Change 6, 1072–1079. doi: 10.1038/nclimate3147
Hutchins, D. A., and Bruland, K. W. (1998). Iron-limited diatom growth and Si:N uptake ratios in a coastal upwelling regime. Nature 393, 561–564. doi: 10.1038/31203
Hutchins, D. A., DiTullio, G. R., Zhang, Y., and Bruland, K. W. (1998). An iron limitation mosaic in the California upwelling regime. Limnol. Oceanogr. 43, 1037–1054. doi: 10.4319/lo.1998.43.6.1037
Hutchins, D. A., Hare, C. E., Weaver, R. S., Zhang, Y., Firme, G. F., DiTullio, G. R., et al. (2002). Phytoplankton iron limitation in the humboldt current and peru upwelling. Limnol. Oceanogr. 47, 997–1011. doi: 10.4319/lo.2002.47.4.0997
Johnson, K. S., Chavez, F. P., and Friederich, G. E. (1999). Continental-shelf sediment as a primary source of iron for coastal phytoplankton. Nature 398, 697–700. doi: 10.1038/19511
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
King, A. L., and Barbeau, K. (2007). Evidence for phytoplankton iron limitation in the southern California Current System. Mar. Ecol. Prog. Ser. 342, 91–103. doi: 10.3354/meps342091
Klingenberg, H., and Meinicke, P. (2017). How to normalize metatranscriptomic count data for differential expression analysis. bioRxiv. doi: 10.1101/134650
Kolber, Z. S., Prášil, O., and Falkowski, P. G. (1998). Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: defining methodology and experimental protocols. Biochim. Biophys. Acta 1367, 88–106. doi: 10.1016/S0005-2728(98)00135-2
Krause, J. W., Brzezinski, M. A., Villareal, T. A., and Wilson, C. (2013). Biogenic silica cycling during summer phytoplankton blooms in the North Pacific subtropical gyre. Deep Sea Res. Part I Oceanogr. Res. Pap. 71, 49–60. doi: 10.1016/j.dsr.2012.09.002
Kustka, A. B., Allen, A. E., and Morel, F. M. M. (2007). Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J. Phycol. 43, 715–729. doi: 10.1111/j.1529-8817.2007.00359.x
La Roche, J., Boyd, P. W., McKay, R. M. L., and Geider, R. J. (1996). Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382, 802–805. doi: 10.1038/382802a0
Lam, P. J., and Bishop, J. K. B. (2008). The continental margin is a key source of iron to the HNLC North Pacific Ocean. Geophys. Res. Lett. 35, 1–5. doi: 10.1029/2008GL033294
Lam, P. J., Bishop, J. K. B., Henning, C. C., Marcus, M. A., Waychunas, G. A., and Fung, I. Y. (2006). Wintertime phytoplankton bloom in the subarctic Pacific supported by continental margin iron. Global Biogeochem. Cycles 20, 1–12. doi: 10.1029/2005GB002557
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Levitan, O., Dinamarca, J., Zelzion, E., Lun, D. S., Guerra, L. T., Kim, M. K., et al. (2015). Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc. Natl. Acad. Sci. U.S.A. 112, 412–417. doi: 10.1073/pnas.1419818112
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lommer, M., Roy, A.-S., Schilhabel, M., Schreiber, S., Rosenstiel, P., and LaRoche, J. (2010). Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11:718. doi: 10.1186/1471-2164-11-718
Lommer, M., Specht, M., Roy, A.-S., Kraemer, L., Andreson, R., Gutowska, M. A., et al. (2012). Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 13:R66. doi: 10.1186/gb-2012-13-7-r66
Maldonado, M. T., and Price, N. M. (2001). Reduction and transport of organically bound iron by Thalassiosira oceanica (Bacillariophyceae). J. Phycol. 37, 298–310. doi: 10.1046/j.1529-8817.2001.037002298.x
Malviya, S., Scalco, E., Audic, S., Vincent, F., Veluchamy, A., Poulain, J., et al. (2016). Insights into global diatom distribution and diversity in the world's ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525. doi: 10.1073/pnas.1509523113
Marchetti, A., and Cassar, N. (2009). Diatom elemental and morphological changes in response to iron limitation: a brief review with potential paleoceanographic applications. Geobiology 7, 419–431. doi: 10.1111/j.1472-4669.2009.00207.x
Marchetti, A., Catlett, D., Hopkinson, B. M., Ellis, K., and Cassar, N. (2015). Marine diatom proteorhodopsins and their potential role in coping with low iron availability. ISME J. 9, 2745–2748. doi: 10.1038/ismej.2015.74
Marchetti, A., and Maldonado, M. T. (2016). “Iron,” in The Physiology of Microalgae, eds M. A. Borowitzka, J. Beardall, and J. A. Raven (Cham: Springer International Publishing), 233–279.
Marchetti, A., Parker, M. S., Moccia, L. P., Lin, E. O., Arrieta, A. L., Ribalet, F., et al. (2009). Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470. doi: 10.1038/nature07539
Marchetti, A., Schruth, D. M., Durkin, C. A., Parker, M. S., Kodner, R. B., Berthiaume, C. T., et al. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U.S.A. 109, E317–E325. doi: 10.1073/pnas.1118408109
Milligan, A. J., and Harrison, P. J. (2000). Effects of non-steady-state iron limitation on nitrogen assimilatory enzymes in the marine diatom Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 36, 78–86. doi: 10.1046/j.1529-8817.2000.99013.x
Milne, A., Landing, W., Bizimis, M., and Morton, P. (2010). Determination of Mn, Fe, Co, Ni, Cu, Zn, Cd and Pb in seawater using high resolution magnetic sector inductively coupled mass spectrometry (HR-ICP-MS). Anal. Chim. Acta 665, 200–207. doi: 10.1016/j.aca.2010.03.027
Moore, J. K., Doney, S. C., Glover, D. M., and Fung, I. Y. (2001). Iron cycling and nutrient-limitation patterns in surface waters of the World Ocean. Deep. Res. II 49, 463–507. doi: 10.1016/S0967-0645(01)00109-6
Moore, J. K., Doney, S. C., and Lindsay, K. (2004). Upper ocean ecosystem dynamics and iron cycling in a global three-dimensional model. Global Biogeochem. Cycles 18:GB4028. doi: 10.1029/2004GB002220
Morrissey, J., Sutak, R., Paz-Yepes, J., Tanaka, A., Moustafa, A., Veluchamy, A., et al. (2015). A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Curr. Biol. 25, 364–371. doi: 10.1016/j.cub.2014.12.004
Nakajima, K., Tanaka, A., and Matsuda, Y. (2013). SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl. Acad. Sci. U.S.A. 110, 1767–1772. doi: 10.1073/pnas.1216234110
Nelson, D. M., DeMaster, D. J., Dunbar, R. B., and Smith, W. O. (1996). Cycling of organic carbon and biogenic silica in the Southern Ocean: estimates of water-column and sedimentary fluxes on the Ross Sea continental shelf. J. Geophys. Res. 101, 18519. doi: 10.1029/96JC01573
Nuester, J., Vogt, S., and Twining, B. S. (2012). Localization of iron within centric diatoms of the genus Thalassiosira. J. Phycol. 48, 626–634. doi: 10.1111/j.1529-8817.2012.01165.x
Nunn, B. L., Faux, J. F., Hippmann, A. A., Maldonado, M. T., Harvey, H. R., Goodlett, D. R., et al. (2013). Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation. PLoS ONE 8:e75653. doi: 10.1371/journal.pone.0075653
Pan, Y., Subba Roa, D. V., Mann, K. H., Brown, R. G., and Pocklington, R. (1996). Effects of silicate limitation on production of domoic acid, a neurotoxin, by the diatom Pseudo- nitzschia m ultiseries. I. Batch culture studies. Mar. Ecol. Prog. Ser. 131, 225–233. doi: 10.3354/meps131235
Parker, C. E., Brown, M. T., and Bruland, K. W. (2016). Scandium in the open ocean: a comparison with other group 3 trivalent metals. Geophys. Res. Lett. 43, 2758–2764. doi: 10.1002/2016GL067827
Parsons, T. R., Maita, Y., and Lalli, C. M. (1984). A Manual of Chemical and Biological Methods for Seawater Analysis. Oxford; New York, NY: Pergamon Press.
Peers, G., and Price, N. M. (2006). Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 441, 341–344. doi: 10.1038/nature04630
Rabosky, D. L., and Sorhannus, U. (2009). Diversity dynamics of marine planktonic diatoms across the Cenozoic. Nature 457, 183–186. doi: 10.1038/nature07435
Reinfelder, J. R. (2011). Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Annu. Rev. Mar. Sci. 3, 291–317. doi: 10.1146/annurev-marine-120709-142720
Reinfelder, J. R., Kraepiel, A. M. L., and Morel, F. M. M. (2000). Unicellular C4 photosynthesis in a marine diatom. Nature 407, 996–999. doi: 10.1038/35039612
Ribalet, F., Marchetti, A., Hubbard, K. A., Brown, K., Durkin, C. A., Morales, R., et al. (2010). Unveiling a phytoplankton hotspot at a narrow boundary between coastal and offshore waters. Proc. Natl. Acad. Sci. U.S.A. 107, 16571–16576. doi: 10.1073/pnas.1005638107/-/DCSupplemental
Robertson, G., Schein, J., Chiu, R., Corbett, R., Field, M., Jackman, S. D., et al. (2010). De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912. doi: 10.1038/nmeth.1517
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Robinson, M. D., and Smyth, G. K. (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332. doi: 10.1093/biostatistics/kxm030
Rogato, A., Amato, A., Iudicone, D., Chiurazzi, M., Ferrante, M. I., and d'Alcalà, M. R. (2015). The diatom molecular toolkit to handle nitrogen uptake. Mar. Genomics 24 (Pt 1), 95–108. doi: 10.1016/j.margen.2015.05.018
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. doi: 10.1111/j.1469-8137.2004.00974.x
Shi, D., Xu, Y., Hopkinson, B. M., and Morel, F. M. M. (2010). Effect of ocean acidification on iron availability to marine phytoplankton. Science 327, 676–679. doi: 10.1126/science.1183517
Sims, P. A., Mann, D. G., and Medlin, L. K. (2006). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. doi: 10.2216/05-22.1
Song, B., and Ward, B. B. (2007). Molecular cloning and characterization of high-affinity nitrate transporters in marine phytoplankton. J. Phycol. 43, 542–552. doi: 10.1111/j.1529-8817.2007.00352.x
Strzepek, R. F., and Harrison, P. J. (2004). Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. doi: 10.1038/nature02954
Sunda, W. G., and Huntsman, S. A. (1995). Iron uptake and growth limitation in oceanic and coastal phytoplankton. Mar. Chem. 50, 189–206. doi: 10.1016/0304-4203(95)00035-P
Sutak, R., Botebol, H., Blaiseau, P.-L., Léger, T., Bouget, F.-Y., Camadro, J.-M., et al. (2012). A comparative study of iron uptake mechanisms in marine microalgae: Iron binding at the cell surface is a critical step. Plant Physiol. 160, 2271–2284. doi: 10.1104/pp.112.204156.
Taylor, F. J. R., and Haigh, R. (1996). Spatial and temporal distributions of microplankton during the summers of 1992–1993 in Barkley Sound, British Columbia, with emphasis on harmful species. Can. J. Fish. Aquat. Sci. 53, 2310–2322. doi: 10.1139/f96-181
Taylor, R. L., Semeniuk, D. M., Payne, C. D., Zhou, J., Tremblay, J.-É., Cullen, J. T., et al. (2013). Colimitation by light, nitrate, and iron in the Beaufort Sea in late summer. J. Geophys. Res. Ocean. 118, 3260–3277. doi: 10.1002/jgrc.20244
Varela, D. E., and Harrison, P. J. (1999). Effect of ammonium on nitrate utilization by Emiliania huxleyi, a coccolithophore from the oceanic northeastern Pacific. Mar. Ecol. Prog. Ser. 186, 67–74.
Whitney, L. P., Lins, J. J., Hughes, M. P., Wells, M. L., Chappell, P. D., and Jenkins, B. D. (2011). Characterization of putative iron responsive genes as species-specific indicators of iron stress in thalassiosiroid diatoms. Front. Microbiol. 2:234. doi: 10.3389/fmicb.2011.00234
Keywords: diatoms, Thalassiosira, Pseudo-nitzschia, iron, metatranscriptomics, California Upwelling Zone, Northeast Pacific Ocean
Citation: Cohen NR, Ellis KA, Lampe RH, McNair H, Twining BS, Maldonado MT, Brzezinski MA, Kuzminov FI, Thamatrakoln K, Till CP, Bruland KW, Sunda WG, Bargu S and Marchetti A (2017) Diatom Transcriptional and Physiological Responses to Changes in Iron Bioavailability across Ocean Provinces. Front. Mar. Sci. 4:360. doi: 10.3389/fmars.2017.00360
Received: 13 August 2017; Accepted: 26 October 2017;
Published: 14 November 2017.
Edited by:
Kristian Spilling, Finnish Environment Institute (SYKE), FinlandReviewed by:
Ulisses Miranda Azeiteiro, University of Aveiro, PortugalPenelope Ann Ajani, University of Technology Sydney, Australia
Copyright © 2017 Cohen, Ellis, Lampe, McNair, Twining, Maldonado, Brzezinski, Kuzminov, Thamatrakoln, Till, Bruland, Sunda, Bargu and Marchetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrian Marchetti, YW1hcmNoZXR0aUB1bmMuZWR1
 Natalie R. Cohen
Natalie R. Cohen Kelsey A. Ellis1
Kelsey A. Ellis1 Robert H. Lampe
Robert H. Lampe Heather McNair
Heather McNair Benjamin S. Twining
Benjamin S. Twining Maria T. Maldonado
Maria T. Maldonado Fedor I. Kuzminov
Fedor I. Kuzminov Kimberlee Thamatrakoln
Kimberlee Thamatrakoln Claire P. Till
Claire P. Till Kenneth W. Bruland
Kenneth W. Bruland William G. Sunda
William G. Sunda Sibel Bargu
Sibel Bargu Adrian Marchetti
Adrian Marchetti