- 1Department of Chemistry, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
- 2Department of Biology, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
- 3Department of Medical Microbiology and Immunology, College of Medicine and Health Sciences, United Arab Emirates University, Al Ain, United Arab Emirates
- 4Zayed Center for Health Sciences, United Arab Emirates University, Al Ain, United Arab Emirates
- 5The Big Data Analytics Center, United Arab Emirates University, Al Ain, United Arab Emirates
Vibrio is a genus of gram-negative, rod-shaped, motile bacteria commonly found in saltwater. One species in particular, Vibrio gazogenes PB1, sourced from an estuarine environment, is known to produce the secondary metabolite, prodigiosin. This high-value compound has potential uses as an antibiotic, a fungicide, and an anti-cancer agent. To further explore its metabolic and genetic features for biotechnological purposes, the complete genome sequence of V. gazogenes PB1 was determined by Illumina and Pacbio sequencing. Two chromosomes were assembled with a mean coverage of 293x. Chromosome 1 is 3.5 Mbp in size with 45.3% GC content and chromosome 2 is 1.2 Mbp in size with 45.1% GC content. The entire genome harbours 4178 genes, of which 3988 are protein-coding and 114 are RNA-coding. A total of 55 virulence-related genes, 38 antimicrobial resistance genes, 48 transposase sequences, 2 intact prophage regions, and 10 genomic islands were present within the genome. Six genes associated with the degradation of cellulose and starch were also identified within the genome. Four of them were strongly up-regulated, as confirmed by RT-qPCR, thus providing strong evidence for their involvement in starch and cellulose degradation. Quite importantly, we demonstrate for the first time that starch and cellulose is associated with the synthesis of prodigiosin in a native prodigiosin-producing bacterium. The prodigiosin titres obtained in the presence of cellulose were on par with glucose as the carbon source which lends further support in the use of V. gazogenes PB1 as a biotechnological host for prodigiosin production.
1 Introduction
In 1854, the Italian anatomist, Filippo Pacini, isolated bacteria from patients with cholera. He described them as ‘vibrions’, a term used to describe motile, worm-like organism (Lippi and Gotuzzo, 2014). This term eventually evolved into the bacterial genus now known as Vibrio. Members of this genus are gram negative, non-sporulating, curve-shaped, facultative anaerobes with flagella. They are widely distributed within the aquatic environment, of which several are pathogenic (Osunla and Okoh, 2017).
Vibrio cholerae, the causative agent of cholera, was the first Vibrio species to be sequenced (Heidelberg et al., 2000). The genomes of other pathogenic species such as V. vulnificus, V. parahaemolyticus, V. alginolyticus, V. harveyi and V. campbelli have also been sequenced (Chen et al., 2003; Makino et al., 2003; Liu et al., 2015; Wang et al., 2015b; Colston et al., 2019). Analyses of Vibrio genomes have revealed that these species typically possess two chromosomes, each with their own distinct and independent origin of replication (Duigou et al., 2006; Rasmussen et al., 2007). The large chromosome is approximately 3 Mb, while the small chromosome can vary from 0.8 to 2.4 Mb (Okada et al., 2005). The presence of two chromosomes within the bacterial kingdom is highly unusual and has been the subject of much debate (Heidelberg et al., 2000; Xu et al., 2003; Okada et al., 2005; Kirkup et al., 2010). Based on insights from functional genomics, the general view is that the genes on chromosome 1 perform the housekeeping roles, while the genes on chromosome 2 confer evolutionary benefit for adapting to different ecological niches.
Vibrio species are receiving a great deal of attention on the biotechnological front. V. natriegens, for example, has recently been employed as a novel host platform for protein expression (Xu et al., 2021). The sequence of its genome was reported in 2013 (Wang et al., 2013). This Vibrio species possesses a rapid rate of metabolism, which on an industrial scale would greatly reduce the production costs for recombinant proteins (Long et al., 2017). The use of V. natriegens can sometimes lead to better protein expression levels compared to traditional bacterial hosts such as E. coli (Xu et al., 2021). Moreover, cell-free systems for the in vitro synthesis of proteins have been developed using the protein-synthesising machinery from V. natriegens (Des Soye et al., 2018; Failmezger et al., 2018; Wiegand et al., 2018). On a more ambitious note, recombinant DNA techniques for V. natriegens have paved the way for metabolic engineering approaches (Dalia et al., 2017; Long et al., 2017). Several studies have already demonstrated the production of 1,3-propanediol (Zhang et al., 2021), melanin (Wang et al., 2020), β-carotene (Ellis et al., 2019), violacein (Ellis et al., 2019), and even selenium nanoparticles (Fernández-Llamosas et al., 2017).
The natural metabolic traits of Vibrio species such as V. fluvialis and V. alginolyticus can also be used in a bioremedial capacity for the removal of heavy metals and diesel oil (Saranya et al., 2017; Li et al., 2020; Purwanti et al., 2020). There is also strong evidence to suggest their involvement in the degradation of plastics (Raghul et al., 2013). The diverse metabolic pathways of Vibrio species can also be applied for the production of several secondary metabolites such as andrimids, (Buijs et al., 2020), aqabamycin (Al-Zereini et al., 2010) and vibrindole A (Bell et al., 1994), all of which have potential clinical uses (Mansson et al., 2011). One particular species, V. gazogenes PB1 (ATCC: 29988, KCTC: 12695), originally isolated from an estuary, is capable of producing the secondary metabolite, prodigiosin (Harwood, 1978). This red pigment possesses antibiotic (Gohil et al., 2020), fungicidal (John Jimtha et al., 2017) and anti-cancer properties (Pérez-Tomás and Viñas, 2010). Recently, we had shown that up to 231 mg/L of prodigiosin can be produced in V. gazogenes PB1 (Vijay et al., 2022a; Vijay et al., 2022b). Although V. gazogenes PB1 was discovered almost five decades ago, the genome sequence of this organism still remains unknown (Harwood, 1978). In this study, we present the complete genome sequence and genetic annotation of V. gazogenes PB1. Furthermore, we demonstrate for the first time its biotechnological versatility in utilising cellulose and starch for the production of prodigiosin, reveal the genes most likely responsible for this useful biotechnological trait, and report on the titres that can be obtained in the presence of these carbon sources.
2 Materials and methods
2.1 Genome extraction from V. gazogenes PB1 and its analysis
V. gazogenes PB1 (KCTC: 12695, ATCC: 29988) was obtained from the Korean Collection for Type Cultures (KCTC). For genome extraction, V. gazogenes PB1 was cultivated as described in an earlier study (Vijay et al., 2022b). The genomic DNA was shipped, in a freeze-dried form, at room temperature for genome sequencing. Whole-genome sequencing and analysis was performed as described previously by Vijay et al., 2022b. For genomic analysis, systems and subsystems were identified using Pathosystems Resource Integration Center (PATRIC) (Davis et al., 2020) (https://patricbrc.org) in conjunction with Rapid Annotation Subsystem Technology (RAST) (Brettin et al., 2015). The genome map was generated using the Circular Genome Viewer tool of the Bacterial and Viral Bioinformatics Resource Center (BV-BRC; https://www.bv-brc.org) (Olson et al., 2023). REBASE (Roberts et al., 2015) (http://rebase.neb.com) was searched to establish the identity of candidate restriction-modification enzymes. Insertion sequences present in the genome were assessed using genome annotations and ISESCAN 1.7.2.3 (Xie and Tang, 2017). PHASTER (Arndt et al., 2016) (https://phaster.ca) was used to identify prophage sequences in the genome. Genomic islands were assessed using IslandPath-DIMOB 1.0.6 (Bertelli and Brinkman, 2018).
2.2 Comparative genomics
The genome of V. gazogenes PB1 was compared with various members of the Vibrio genus as described within the text. Progressive MAUVE (Darling et al., 2010) was used to align the genomes and to identify locally collinear blocks (LCB). Average nucleotide identity (ANI) was computed using the OrthoANIu algorithm (Yoon et al., 2017) (https://www.ezbiocloud.net/tools/ani) and the average amino acid identity (AAI) was computed using EzAAI 1.2.1 (Kim et al., 2021). The percentage of conserved proteins (POCP) was determined based on the method suggested by Qin et al., 2014. Homologous proteins in V. gazogenes PB1 required for cellulose and starch degradation were identified by performing a BLAST analysis of the proteins encoded by V. gazogenes PB1 using protein sequences from other Vibrio species obtained from UniProt (https://www.uniprot.org).
2.3 RT-qPCR analysis
RNA was isolated from the bacterial pellets using TRIzol Reagent (Invitrogen, Waltham, MA, USA) protocols. A total of 2 µg RNA was DNase-treated and reverse transcribed to cDNA using Promega A3500 kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Approximately 50ng of cDNA was amplified using power SYBR® Green PCR master mix (Applied Biosystems, Waltham, MA, USA) in a 40-cycle reaction carried out in a QuantStudioTM 7 Flex System (Applied Biosystems, Waltham, MA, USA). Melt curve analysis was performed. The 16S gene was used as the endogenous control and for normalisation of the expression levels of the candidate genes. RT-qPCR experiments were performed in duplicates and repeated three times for two pairs of primer sets.
2.4 Identification of CRISPR arrays and cas genes
CRISPR loci were predicted using CRISPRCasFinder (Couvin et al., 2018) (https://crisprcas.i2bc.paris-saclay.fr) with default parameters. CRISPR arrays with evidence level 1 or 2, which are indicative of false-positive or spurious arrays, were not considered for further analysis. All predicted loci were manually checked, and arrays located in the coding regions were discarded. To identify potential targets of CRISPR spacers, NCBI BLASTN searches of these sequences were performed. CRISPRTarget (http://bioanalysis.otago.ac.nz/CRISPRTarget/crispr_analysis.html) was used to identify potential targets of CRISPR spacers (Biswas et al., 2013). BLASTN search of the spacer sequences was performed against the GenBank phage and Refseq plasmid databases to identify matches with at least 90% identity and 90% query coverage. Genomic comparisons and gene arrangements were generated using Easyfig 2.2.5 (Sullivan et al., 2011).
2.5 In vivo production and quantification of prodigiosin
For evaluating prodigiosin production in the presence of various carbon sources, cultivation was carried out in minimal media as described previously (Vijay et al., 2022b). For the quantification of prodigiosin, a 10 μl cell culture was added to 190 μl of acidified ethanol in a microplate well and mixed. The absorbance was recorded at 535nm, as described by Vijay et al., 2022b. Cell culture density was determined by measuring the scattering of light at 650nm. Viability of cells were evaluated by streaking out on growth on LB medium/agar plates supplemented with 3% (w/v).
2.6 Statistical analysis
All quantitative data represent the means ± SD of at least 3 independent culture experiments. Any significant differences between sample treatments were determined using a one-way ANOVA test followed by post-hoc analyses to identify the sample treatments which showed statistically significantly differences. A p value of less than 0.05 was considered significant.
3 Results
3.1 Genome sequencing, assembly and annotation
The genomic DNA of V. gazogenes PB1 was used to generate Illumina and PacBio sequencing libraries. After quality control and filtering of raw reads, de novo genome assembly of V. gazogenes PB1 was performed using Unicycler version 0.4.9 by combining PacBio and Illumina sequencing data. This resulted in two circular contigs (Table S1). Chromosome 1 is 3.5 Mbp in size with 45.3% GC content (NCBI Accession: CP092587.1/NZ_CP092587.1), and chromosome 2 is 1.2 Mbp in size with 45.1% GC content (NCBI Accession: CP092588.1/NZ_CP092588.1). Based on NCBI PGAP annotations, the genome encodes 4178 genes, of which 3988 are protein-coding, and 76 are pseudogenes. The genome also has 114 RNA-encoding genes, which includes 87 tRNAs, 22 rRNAs and 5 non-coding RNAs (Figure 1). Assessment of the genome using BUSCO version 5.2.2 based on the vibrionales_odb10 dataset indicated the presence of 1378 out of a core of 1445 Vibrionales genes (95.4%); 1275 were single-copy and 3 were duplicated BUSCOs. Annotated genes were grouped into subsystems in PATRIC using RAST. The most abundant subsystems were metabolism (92), protein processing (44), energy (32), stress response, defence and virulence (36), membrane transport (22), DNA processing (21), and cellular processes (18) (Figure S1).
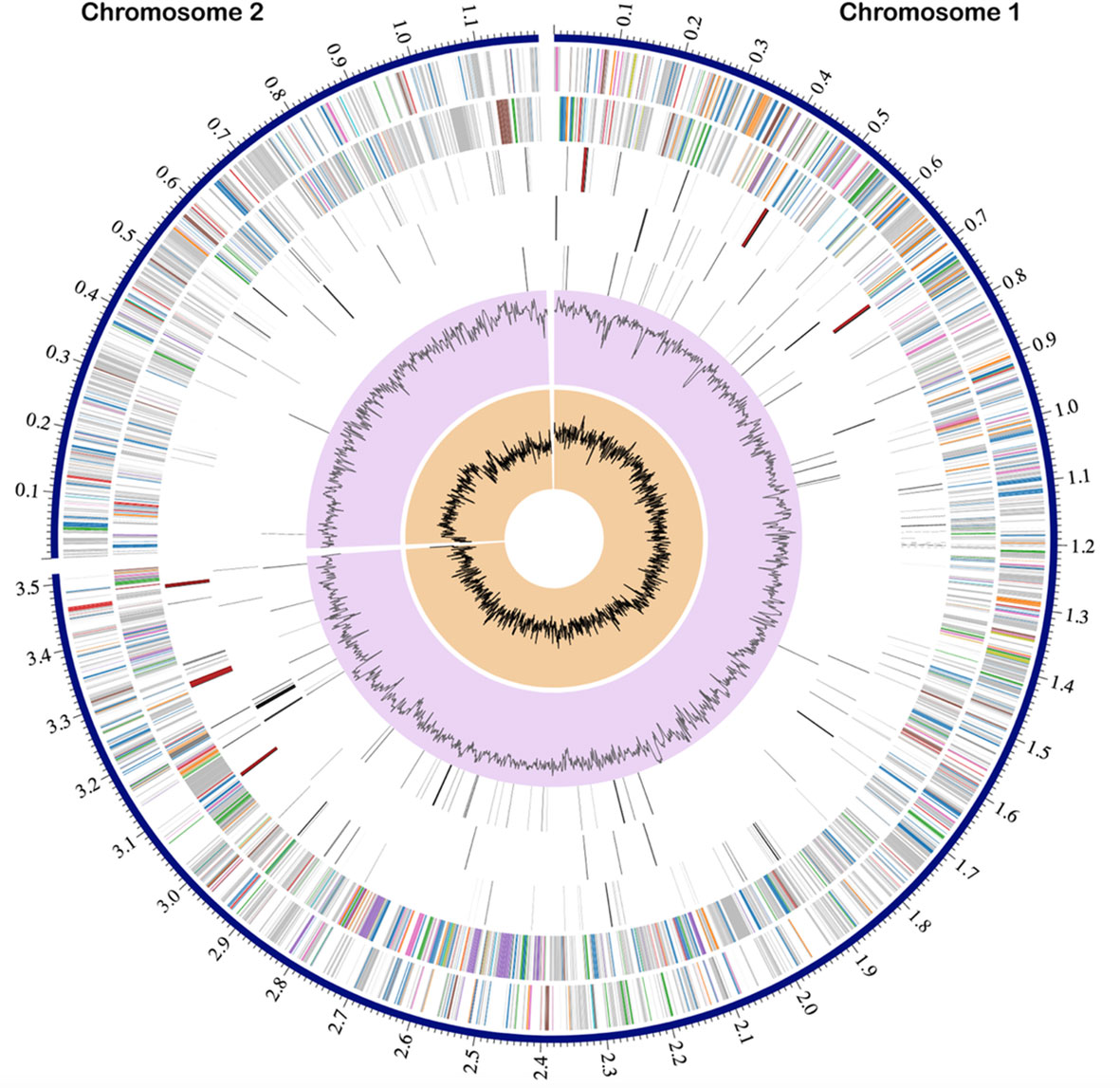
Figure 1 Circular genomic map of V. gazogenes PB1. From outside to inside the tracks represent the genome position in Mbp, the two chromosomes of sizes 3.5 Mbp and 1.2 Mbp, coding sequences on the forward strand, coding sequences on the reverse strand, non-coding features including tRNA and rRNA, antimicrobial resistance genes, virulence factor genes, GC content and GC skew.
3.2 Presence of genes associated with virulence
To evaluate its potential for pathogenicity, the genome of V. gazogenes PB1 was analysed in PATRIC for virulence factors. A total of 55 genes were identified with virulence potential (Table S2). Based on PATRIC, these were related to adherence, secretion, regulation, motility, and metabolism and included dksA, fur, ompR, rpoE, rpoS, phoU, fleN and flgP. Genes relating to quorum sensing (luxS), a type VI secretion system (T6SS) (hcp2, vipA, vipB) antimicrobial resistance (catB, qnrB), and efflux systems (emrAB-tolC, macA/macB/tolC) were also encoded in the genome (Table S3). An inspection of the genome for potential restriction endonucleases (REase) identified the annotated CDS “MvaI/BcnI restriction endonuclease family protein” which is a putative Type II restriction enzyme with potential specificity for the motif CCWGG. Though methylation (M) and DNA sequence-recognition (S) subunits of a putative methyltransferase (MTase) were identified on chromosome 1, a required restriction enzyme was not identified to complete a Type I REase (Loenen et al., 2014). Based on analysis by ISESCAN and the genome annotations, 48 transposases were identified and annotated including IS110, IS200/IS605, IS256, IS3, IS30, IS630, ISNCY family transposases as well as tnpA, DDE-type integrase/transposase/recombinase and Mu transposase C-terminal domain-containing protein. PHASTER identified four prophage regions in the V. gazogenes PB1 genome, of which two regions are intact, one region is incomplete, and one region is questionable (Table S4). The two intact regions have proteins with the highest number of hits from Vibrio phage VP882 (NCBI RefSeq accession: NC_009016) located on chromosome 1, and Vibrio_12B12 (NCBI RefSeq accession: NC_021070) located on chromosome 2. Genomic islands (GIs), which are indicative of horizontal gene transfer, were assessed using IslandPath-DIMOB. Six GIs were located on chromosome 1, and four on chromosome 2 (Table S5).
3.3 Comparison with V. gazogenes ATCC 43942
The most similar genome was that of Vibrio genus ATCC 43942 with average nucleotide and average amino acid identities of 94.22% and 96.66%, respectively (Table S6). A comparison of the two proteomes in PATRIC showed that around 6% of the proteins shared 100% sequence identity, while 83.6% shared 90-99.99% identity (Figure S2). The percentage of conserved proteins (POCP) between the two genomes was computed to be 85.46%. Chromosome 1 of V. gazogenes PB1 is 3.52 Mbp and is comparable to the 3.47 Mbp chromosome 1 of V. gazogenes ATCC 43942. Alignment of chromosome 1 of V. gazogenes PB1 and V. gazogenes ATCC 43942 using progressiveMauve indicated that the genome orientation is largely preserved apart from two shorts regions of lengths which have been swapped and inverted (Figure S2). Chromosome 2 of V. gazogenes PB1 is 1.2 Mbp while chromosome 2 of V. gazogenes ATCC 43942 is 1.3 Mbp long. The genomic rearrangement in the two species includes the inversion of a 400 kbp region in chromosome 2 (Figure S3). The region absent from V. gazogenes PB1 was found to encode for several transposases, toxins (toxin HipA, zeta toxin), endonuclease and several hypothetical genes (Table S7).
3.4 Presence of CRISPR-Cas systems
The genome of V. gazogenes PB1 were found to harbour 3 CRISPR-Cas operons and 4 CRISPR arrays without cas genes. Three CRISPR arrays were identified on chromosome 1, while chromosome 2 has 3 CRISPR-Cas operons and one CRISPR-array without cas genes. Based on CRISPR-Cas classification, all the CRISPR-Cas operons identified belong to Class 1 CRISPR system. Chromosome 2 has cas3, cas8, cse2, cas7, cas5e, cas6, cas1 and cas2 genes followed by the CRISPR array. Therefore, it belongs to the type I-E CRISPR-Cas system (Figure 2A). Type III-B and type III-D CRISPR system for RNA degradation were also identified on chromosome 2 (Table S8). Genomic comparisons, of the homologous regions between V. gazogenes PB1 and V. gazogenes ATCC 43942, and gene arrangements were constructed using Easyfig (Figures 2A, B). The Type III-B system contains the cmr6, cmr5, cmr4, cmr3, cmr2, cas10, csm3, cas1 and cas2 genes followed by the CRISPR array while the type III-D CRISPR system has the cas10, csm3, csm3, cas1 and cas2 genes with the CRISPR array located upstream of cas10 (Figures 2B, C). In the type I-E CRISPR system, an additional gene between the repeats, which encodes a hypothetical protein, was annotated between the CRISPR array and cas3 gene (Figure 2A). In type III-B system of V. gazogenes PB1, the presence of a reverse transcriptase (RT) domain in cas1 proteins was also observed (Figure 2B).
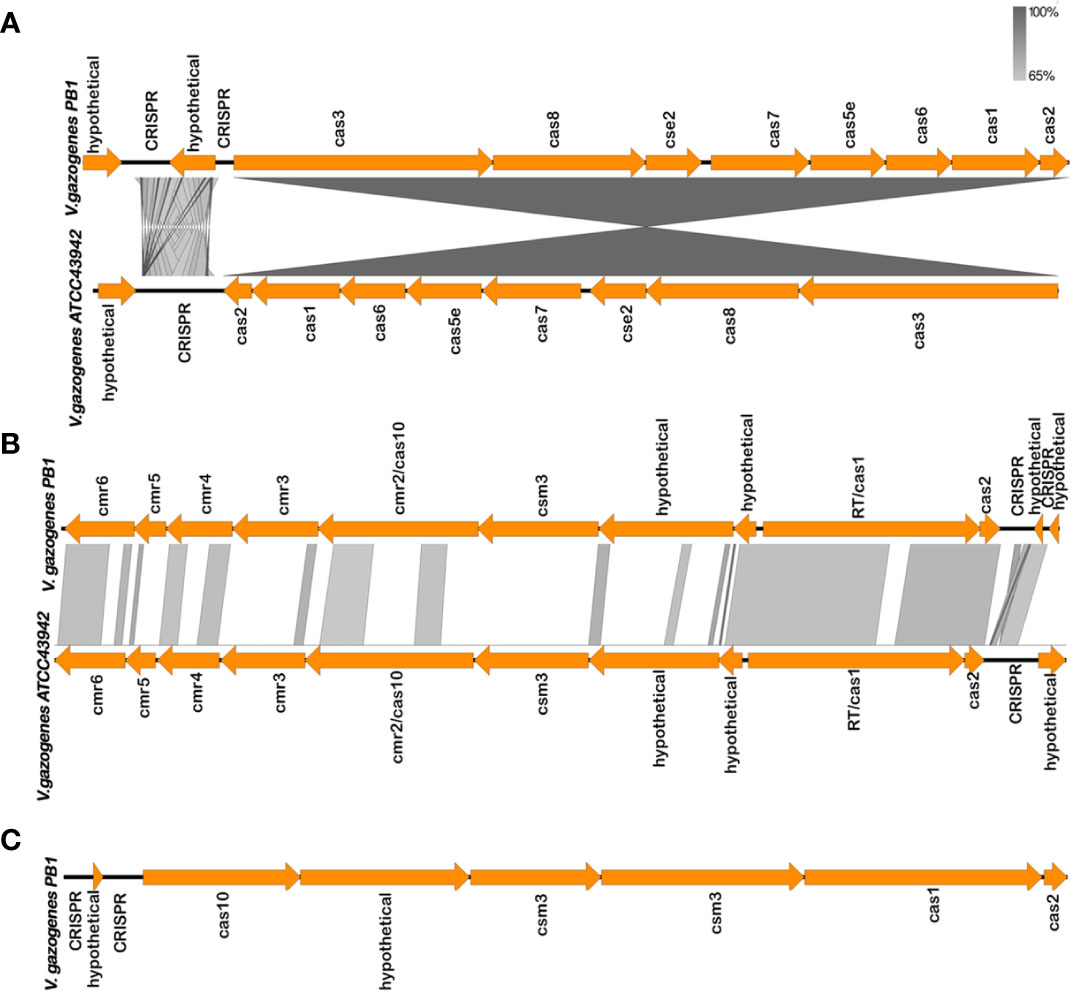
Figure 2 CRISPR-Cas systems in V. gazogenes PB1 and comparison of gene arrangement with V. gazogenes ATCC 43942 constructed using Easyfig. (A) CRISPR-Cas type I-E systems of V. gazogenes PB1. (B) CRISPR-Cas type III-B systems of V. gazogenes PB1. (C) CRISPR-Cas type III-D of V. gazogenes PB1. Grayscale bars represent nucleotide homology.
BLASTN analysis identified spacer sequences which were identical to phage and plasmid sequences, and are potential targets of the CRISPR-Cas immune systems. CRISPR arrays in V. gazogenes PB1 contained 64 spacers and two of these spacer sequences were identical to sequences of Vibrio phage vB VpaM MAR, and V. parahaemolyticus strain 20130629002S01 plasmid pVPGX2 (Table S9). The canonical type I-E CRISPR-Cas system was identified with 12 spacers, and the type III-B and III-D were identified with 6 and 8 spacers respectively.
3.5 Identification of genes associated with the degradation of starch and cellulose
Several genes encoding for enzymes capable of breaking down the polysaccharides cellulose and starch were identified within the V. gazogenes PB1 genome (Figure 3A). These genes were annotated under different protein names by the NCBI’s PGAP annotation pipeline: endoglucanase as a ‘glycoside hydrolase family 9’ protein (NCBI Accession: USP15540.1), exoglucanase as a ‘glycoside hydrolase family 6’ protein (NCBI Accession: USP14760.1), β-glucosidase as a ‘β-glucosidase’ (NCBI Accession: USP15545.1), α-amylase as a ‘starch-binding protein’ (NCBI Accession: USP15696.1) and α-glucosidase as an ‘α-glucosidase’ (NCBI Accession: USP13629.1). The presence of these genes suggested that V. gazogenes PB1 is capable of degrading starch and cellulose to its constituent glucose monomers. To confirm if this was the case, V. gazogenes PB1 cells were streaked out on plates containing NaCl/minimal media/agar solid media supplemented with 1% (w/v) of starch and cellulose (Figure 3B). Additional plates containing glucose and another without a supplemented carbon source were included as controls (Figure 3B). The appearance of V. gazogenes PB1 cells on plates containing cellulose and starch confirmed that V. gazogenes PB1 was capable of metabolising cellulose and starch for growth. Additionally, RT-qPCR was performed to evaluate the transcript levels of the genes identified for starch and cellulose degradation based on the PGAP annotation pipeline (Figure 3C, Table S10). The significant differences (as much as 50-fold) between the transcript levels of the genes encoding for the α-amylase (USP15696.1), α-glucosidase (USP13629.1) and the exoglucanase (USP14760.1) genes under the different experimental conditions indicated that these genes are most likely involved in starch and cellulose metabolism (Figures 3C, S4; Table S11). Contrary to our expectation, the transcript levels of the isoamylase (USP16020.1) and β-glucosidase (USP15545.1) genes were not upregulated suggesting that these genes have no role to play in cellulose or starch metabolism (Figures 3C, S4). The endo-1,4-β-glucanase gene (USP15540.1) could not be amplified whatsoever. Given that the RT-qPCR protocol was carried out with two primer sets, the failure to amplify this particular transcript could have been due to its high susceptibility to degradation in vivo or the presence of RNA secondary structures that inhibited reverse transcriptase enzyme activity.
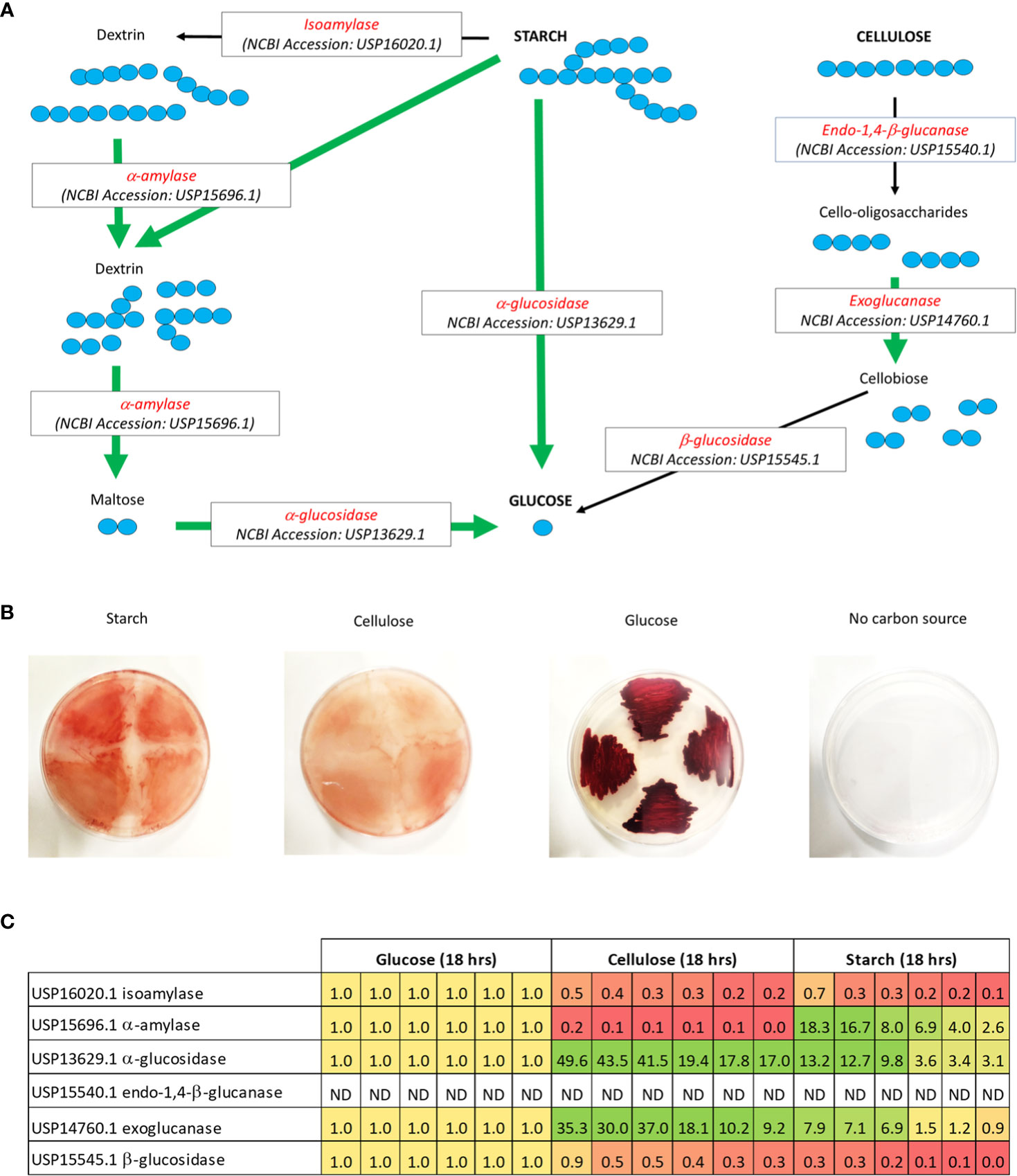
Figure 3 Possible metabolic routes of degradation for starch and cellulose in V. gazogenes PB1. (A) Analysis of the V. gazogenes PB1 genome revealed several genes that were capable of degrading starch and cellulose. For starch degradation, which consists of amylopectin (branched-chain) and amylose (straight-chain), these included: i) isoamylase for cleavage of the α-1,6-glycosidic bonds of amylopectin and dextrins to release straight-chain polysaccharides, ii) α-amylase for cleavage of the α-1,4-glycosidic bonds of starch to release dextrins and the disaccharide maltose, and iii) α-glucosidase for cleavage of the α-1,4-glycosidic bonds of the terminal non-reducing α-D-glucosyl residues of maltose or oligosaccharides to the glucose monomer. For cellulose degradation, these included: i) endo-1,4-b-glucanase for cleavage of the β-1,4-glycosidic bonds of cellulose to release cello-oligosaccharides, ii) exoglucanase for cleavage of the β-1,4-glycosidic bonds of cellulose or cello-oligosaccharides to release the disaccharide, cellobiose, and iii) β-glucosidase for cleavage of the β-1,4-glycosidic bonds of the terminal non-reducing β-D-glucosyl residue of cellobioase or oligosaccharides to release glucose. The green arrows highlight the genes that are up-regulated according to RT-qPCR analysis (refer to next part). (B) Growth of V. gazogenes PB1 in the presence of starch and cellulose. Cells were streaked out on solid minimal media plates supplemented with 2% (w/v) NaCl and different carbon sources. Plates were sealed with parafilm and incubated at 29°C for up to 14 days. Growth of V. gazogenes PB1 was monitored with and without the supplementation of starch, cellulose and glucose. (C) Heat-map of the transcript levels of genes associated with starch and cellulose degradation. Based on RT-qPCR analysis, several gene-products involved in starch and cellulose degradation were found to be up-regulated in the presence of starch and/or cellulose. The number in each box (sample replicate) signifies the fold increase (green) or decrease (red) of the transcript levels of the specific genes relative to transcript levels (orange) observed under glucose condition. ND, no detection of amplified signal.
3.6 Utilisation of starch and cellulose is associated with the production of prodigiosin
We noted that the growth of V. gazogenes PB1 in the presence of starch and cellulose was associated with the production of the secondary metabolite, prodigiosin (Figure 3C). A previous study had reported on the growth of PB1 in the presence of starch and carboxymethylcellulose though not for cellulose (Harwood, 1978). Moreover, the quantitative levels of prodigiosin in the presence of these carbon sources were not reported. Therefore, we set up cultivation conditions to monitor the levels of prodigiosin in the presence of starch and cellulose. We used laboratory-grade (>99% purity) starch and cellulose prepared in minimal media to firmly establish whether starch and/or cellulose can serve as the sole carbon source for prodigiosin production in V. gazogenes PB1 (Figure 4). For the first 2 days of cultivation, prodigiosin titres reached as high as 50mg/Litre. Under the same conditions and with prolonged incubation, a similar level could be attained in the presence of 1% (w/v) cellulose. An increase in the starting cellulose content to 2% (w/v) resulted in two-fold higher prodigiosin titres. In contrast, the titres in the presence of 1% (w/v) starch, however, reached approximately 30 mg/liter indicating that starch may be the less effective carbon source for prodigiosin production compared to cellulose.
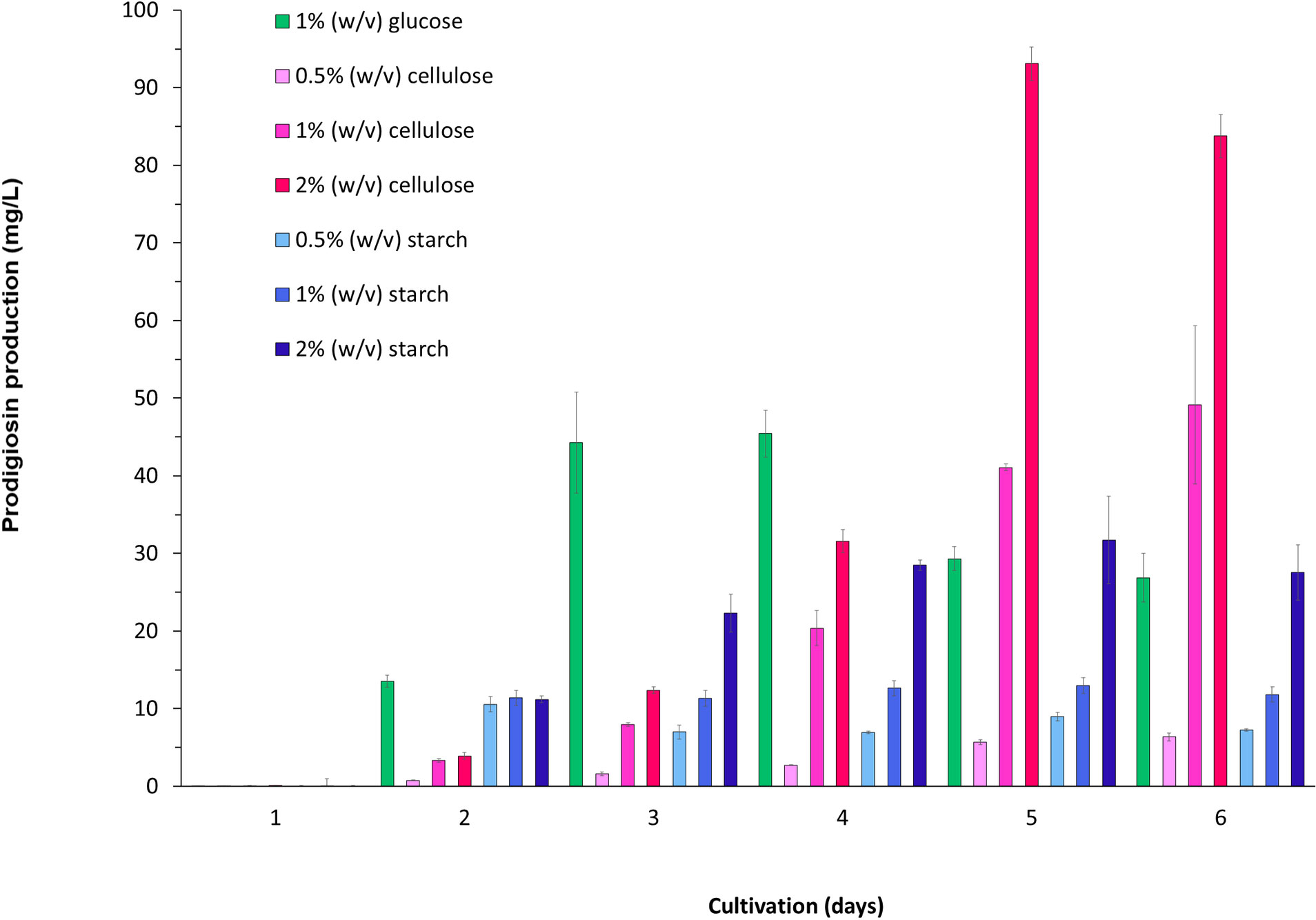
Figure 4 Starch- and cellulose-associated production of prodigiosin in V. gazogenes PB1. Prodigiosin production was monitored over a 6-day period by recording the absorbance at 535nm.
Discussion
In this study, we reveal the full genome sequence of V. gazogenes PB1, previously known as Beneckea gazogenes, This species was assigned by Harwood (Harwood, 1978) to the Vibrio genus after a careful reassessment of its morphological, biochemical and genetic characteristics. Its genome size is similar in size to other members of the Vibrio genus such as V. cholera, V. parahaemolyticus. V. gazogenes PB1 harbours two chromosomes: a large one with a size of 3.5 Mbp and a small one with a size of 1.2 Mbp. The genome encodes 4178 genes, of which 3988 are protein-coding and 114 RNA-coding genes. Type I and type III CRISPR systems present within this species affords it some level of protection against DNA- or RNA-based phages. A recent study (Watson et al., 2019) had shown that the presence of a natural CRISPR Cas system present within the gram-negative plant bacterium, Pectobacterium atrosepticum, can help to provide population-level immunity. Thus, bacterial hosts that harbour natural immune system may be beneficial in reducing phage contamination during scale-ups. For research and development purposes, organisms that can be safely used and handled are particularly advantageous. Given its low-risk, class 1 status and based on our analysis for virulence factors, V. gazogenes PB1 would be considered safe for experimental research and scale-up applications.
Earlier we had demonstrated that V. gazogenes PB1 is capable of tolerating high concentrations of NaCl (Vijay et al., 2022b). For biotechnological scale-ups, this is a useful trait for two reasons. Firstly, it avoids the strict requirement for desalinated water which is a costly process (Mohammadi et al., 2020). Secondly, high-salt media would hinder the growth of microbial contaminants during a scale-up (Skinner and Leathers, 2004). Marine bacteria achieve this high-salt tolerance, via the intracellular synthesis of compatible solutes (Gregory and Boyd, 2021). These small soluble metabolites are accumulated within the cell in order to maintain the cell’s turgor pressure within a high-salt environment. Like many bacterial species, V. gazogenes PB1 is capable of synthesising the well-known compatible solutes, ectoine and glycine betaine. As an alternative mechanism, salt tolerance could also be achieved with the intracellular uptake of compatible solutes, via the activities of solute carriers and transporters, which is a more energetically favourable process (Gregory and Boyd, 2021). In V. gazogenes PB1, several ABC transporters and solute carriers were identified, which permits a range of compatible solutes such as ectoine, glycine betaine, and DMSP among others, to be taken up from the external environment.
Starch and cellulose are abundant, naturally occurring polysaccharides found in the cell wall of algae and plants. The degradation of cellulose to glucose could potentially occur via the enzyme activities of endo-1,4-β-glucanase, exoglucanase and β-glucosidase, while for starch this could occur via the activities of an isoamylase, α-amylase, and α-glucosidase. The up-regulation of α-amylase and α-glucosidase gene in the presence of starch strongly indicates the involvement of these genes in the degradation of starch, while the up-regulation of the exoglucanase gene suggests its involvement for the degradation of cellulose. Thus, our data provides empirical evidence that these genes are correctly annotated and in accordance with their assigned functions. The failure to observe the up-regulation of the β-glucosidase gene and the isoamylase gene would lead us to conclude either that these gene products act on other polysaccharides or that their levels of expression are specifically down-regulated during starch or cellulose degradation as a possible transcriptional mechanism for regulating carbohydrate synthesis. Currently, very little is known about the transcriptional regulation of the cellulose or starch degradation in bacteria and is an area of research that has only just recently emerged due to the affordability of DNA sequencing and the development of RNA-seq (Xu et al., 2013). At least at the post-translational level, cellulose-degrading enzymes such as β-glucosidase are known to become inhibited in the presence of glucose, as well as other intermediates of cellulose breakdown (Andrić et al., 2010; Salgado et al., 2018).
The most exciting and novel observation observed in this study is that cultivation in the presence of either starch or cellulose is associated with the production of prodigiosin. This high-value compound possesses a wide range of biological properties including as an antibiotic (Lapenda et al., 2015), anti-cancer agent (Montaner et al., 2000), algicide (Zhang et al., 2016) and anti-viral (Suryawanshi et al., 2020). The earlier study by Harwood, 1978 used hydrolysed plant material as the source of carbon for the cultivation of V. gazoegenes PB1. Since crude plant hydrolysates will contain a variety of carbon-based materials, this earlier experimental approach would not have conclusively demonstrated that cellulose can act as the sole carbon source for the growth of V. gazogenes PB1. In our experimental approach, we used minimal media supplemented with laboratory-grade starch and cellulose. Thus, we can conclude that starch and cellulose can serve as sole carbon sources not just for growth but also for prodigiosin production. Furthermore, we show that with respect to titres, cellulose is as effective as glucose for prodigiosin production in V. gazogenes PB1. The recalcitrance of starch and cellulose, as well as the requirement of additional enzyme activities to degrade starch and cellulose would explain the slower rates of prodigiosin synthesis. This bottle-neck could be alleviated with the co-cultivation of microbes evolved for polysaccharide degradation, as demonstrated in a previous study (Wang et al., 2015a). Quite importantly, this is the first report to demonstrate the production of prodigiosin in V. gazogenes PB1 using either starch and cellulose as carbon sources. This biotechnologically relevant finding paves the way for the use of V. gazogenes PB1 as a bacterial host for the production of prodigiosin using plant- or algae- derived feedstocks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MKA conceptualised the study. BB and RV assembled and annotated the genome. BB, RV, and MKA analysed the sequencing data. DV carried out the genomic extraction and purification. DV, AAA, HMA, and SOA carried out the in vivo cultivation experiments. BB and DV presented the in vivo cultivation results. DV and MKA analysed the in vivo cultivation data. PPP performed the qPCR. PPP, GK, and MKA analysed the qPCR data. MKA and RV drafted the manuscript. BB, DV, PPP, GK, RV, and MKA edited the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the United Arab Emirates University (UAEU) via the UAEU Program for Advanced Research (fund codes: 12S011 and 12S006) awarded to MKA and RV, as well as the 2022 Sustainable Development Goals Program (grant code: G00003981) awarded to MKA, 2023 SURE Plus Program (grant code: G00004393) awarded to MKA and the Zayed Center-based grant (fund code: 12R001) awarded to GK.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1028319/full#supplementary-material
References
Al-Zereini W., Fotso Fondja Yao C., Laatsch H., Anke H. (2010). Aqabamycins a-G: novel nitro maleimides from a marine Vibrio species. I. taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo). 63, 297–301. doi: 10.1038/JA.2010.34
Andrić P., Meyer A. S., Jensen P. A., Dam-Johansen K. (2010). Reactor design for minimizing product inhibition during enzymatic lignocellulose hydrolysis: I. Significance and mechanism of cellobiose and glucose inhibition on cellulolytic enzymes. Biotechnol. Adv. 28, 308–324. doi: 10.1016/J.BIOTECHADV.2010.01.003
Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/NAR/GKW387
Bell R., Carmeli S., Sar N. (1994). Vibrindole a, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod. 57, 1587–1590. doi: 10.1021/NP50113A022
Bertelli C., Brinkman F. S. L. (2018). Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 34, 2161–2167. doi: 10.1093/BIOINFORMATICS/BTY095
Biswas A., Gagnon J. N., Brouns S. J. J., Fineran P. C., Brown C. M. (2013). CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 10, 817–827. doi: 10.4161/RNA.24046
Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/SREP08365
Buijs Y., Isbrandt T., Zhang S.-D., Larsen T. O., Gram L. (2020). The antibiotic andrimid produced by Vibrio coralliilyticus increases expression of biosynthetic gene clusters and antibiotic production in Photobacterium galatheae. Front. Microbiol. 0. doi: 10.3389/FMICB.2020.622055
Chen C. Y., Wu K. M., Chang Y. C., Chang C. H., Tsai H. C., Liao T. L., et al. (2003). Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13, 2577. doi: 10.1101/GR.1295503
Colston S. M., Judson Hervey W., Horne W. C., Haygood M. G., Petersen B. D., van J. C., et al. (2019). Complete genome sequence of Vibrio campbellii DS40M4. Microbiol. Resour. Announc. 8, e01187-18. doi: 10.1128/MRA.01187-18
Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Néron B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/NAR/GKY425
Dalia T. N., Hayes C. A., Stolyar S., Marx C. J., McKinlay J. B., Dalia A. B. (2017). Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth. Biol. 6, 1650–1655. doi: 10.1021/ACSSYNBIO.7B00116
Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/JOURNAL.PONE.0011147
Davis J. J., Wattam A. R., Aziz R. K., Brettin T., Butler R., Butler R. M., et al. (2020). The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612. doi: 10.1093/NAR/GKZ943
Des Soye B. J., Davidson S. R., Weinstock M. T., Gibson D. G., Jewett M. C. (2018). Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 7, 2245–2255. doi: 10.1021/ACSSYNBIO.8B00252
Duigou S., Knudsen K. G., Skovgaard O., Egan E. S., Løbner-Olesen A., Waldor M. K. (2006). Independent control of replication initiation of the two vibrio cholerae chromosomes by DnaA and RctB. J. Bacteriol. 188, 6419–6424. doi: 10.1128/JB.00565-06
Ellis G. A., Tschirhart T., Spangler J., Walper S. A., Medintz I. L., Vora G. J. (2019). Exploiting the feedstock flexibility of the emergent synthetic biology chassis Vibrio natriegens for engineered natural product production. Mar. Drugs 17, 679. doi: 10.3390/MD17120679
Failmezger J., Scholz S., Blombach B., Siemann-Herzberg M. (2018). Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.01146
Fernández-Llamosas H., Castro L., Blázquez M. L., Díaz E., Carmona M. (2017). Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 7 (1), 1–9. doi: 10.1038/s41598-017-16252-1
Gohil N., Bhattacharjee G., Singh V. (2020). Synergistic bactericidal profiling of prodigiosin extracted from Serratia marcescens in combination with antibiotics against pathogenic bacteria. Microb. Pathog. 149, 104508. doi: 10.1016/J.MICPATH.2020.104508
Gregory G. J., Boyd E. F. (2021). Stressed out: bacterial response to high salinity using compatible solute biosynthesis and uptake systems, lessons from Vibrionaceae. Comput. Struct. Biotechnol. J. 19, 1014–1027. doi: 10.1016/J.CSBJ.2021.01.030
Harwood C. S. (1978). Beneckea gazogenes sp. nov., a red, facultatively anaerobic, marine bacterium. Curr. Microbiol. 1, 233–238. doi: 10.1007/BF02602849
Heidelberg J. F., Elsen J. A., Nelson W. C., Clayton R. A., Gwinn M. L., Dodson R. J., et al. (2000). DNA Sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
John Jimtha C., Jishma P., Sreelekha S., Chithra S., Radhakrishnan E. (2017). Antifungal properties of prodigiosin producing rhizospheric Serratia sp. Rhizosphere 3, 105–108. doi: 10.1016/j.rhisph.2017.02.003
Kim D., Park S., Chun J. (2021). Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 59, 476–480. doi: 10.1007/S12275-021-1154-0
Kirkup B. C., Chang L., Chang S., Gevers D., Polz M. F. (2010). Vibrio chromosomes share common history. BMC Microbiol. 10, 1–13. doi: 10.1186/1471-2180-10-137
Lapenda J. C., Silva P. A., Vicalvi M. C., Sena K. X., Nascrimento S. C. (2015). Antimicrobial activity of prodigiosin isolated from Serratia marcescens UFPEDA 398. World J. Microbiol. Biotechnol. 31 (2), 399–406. doi: 10.1007/s11274-014-1793-y
Li J., Zhou X., Zhang Y., Zhong F., Lin C., McCormick P. J., et al. (2020). Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci. Bull. 66, 661. doi: 10.1016/j.scib.2020.10.018
Lippi D., Gotuzzo E. (2014). The greatest steps towards the discovery of Vibrio cholerae. Clin. Microbiol. Infect. 20, 191–195. doi: 10.1111/1469-0691.12390
Liu X. F., Cao Y., Zhang H. L., Chen Y. J., Hu C. J. (2015). Complete genome sequence of Vibrio alginolyticus ATCC 17749T. Genome Announc. 3, 1500–1514. doi: 10.1128/GENOMEA.01500-14
Loenen W. A. M., Dryden D. T. F., Raleigh E. A., Wilson G. G. (2014). Type I restriction enzymes and their relatives. Nucleic Acids Res. 42, 20–44. doi: 10.1093/NAR/GKT847
Long C. P., Gonzalez J. E., Cipolla R. M., Antoniewicz M. R. (2017). Metabolism of the fast-growing bacterium Vibrio natriegens elucidated by 13C metabolic flux analysis. Metab. Eng. 44, 191–197. doi: 10.1016/J.YMBEN.2017.10.008
Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet (London England) 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Mansson M., Gram L., Larsen T. O. (2011). Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 9, 1440. doi: 10.3390/MD9091440
Mohammadi F., Sahraei-Ardakani M., Al-Abdullah Y., Heydt G. T. (2020). Cost-benefit analysis of desalination: a power market opportunity. Electr. Power Components Syst. 48, 1091–1101. doi: 10.1080/15325008.2020.1829188
Montaner B., Navarro S., Piqué M., Vilaseca M., Martinell M., Giralt E., et al. (2000). Prodigiosin from the supernatant of Serratia marcescens induces apoptosis in haematopoietic cancer cell lines. Br. J. Pharmacol. 131 (3), 585–593. doi: 10.1038/sj.bjp.0703614
Okada K., Iida T., Kita-Tsukamoto K., Honda T. (2005). Vibrios commonly possess two chromosomes. J. Bacteriol. 187, 752–757. doi: 10.1128/JB.187.2.752-757.2005
Olson R. D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J. J., et al. (2023). Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689. doi: 10.1093/NAR/GKAC1003
Osunla C. A., Okoh A. I. (2017). Vibrio pathogens: a public health concern in rural water resources in sub-Saharan Africa. Int. J. Environ. Res. Public Heal. 14, 1188. doi: 10.3390/IJERPH14101188
Pérez-Tomás R., Viñas M. (2010). New insights on the antitumoral properties of prodiginines. Curr. Med. Chem. 17, 2222–2231. doi: 10.2174/092986710791331103
Purwanti I. F., Obenu A., Tangahu B. V., Kurniawan S. B., Imron M. F., Abdullah S. R. S. (2020). Bioaugmentation of Vibrio alginolyticus in phytoremediation of aluminium-contaminated soil using Scirpus grossus and Thypa angustifolia. Heliyon 6, e05004. doi: 10.1016/J.HELIYON.2020.E05004
Qin Q. L., Xie B., Zhang X. Y., Chen X. L., Zhou B. C., Zhou J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Raghul S. S., Bhat S. G., Chandrasekaran M., Francis V., Thachil E. T. (2013). Biodegradation of polyvinyl alcohol-low linear density polyethylene-blended plastic film by consortium of marine benthic Vibrios. Int. J. Environ. Sci. Technol. 11, 1827–1834. doi: 10.1007/S13762-013-0335-8
Rasmussen T., Jensen R., Ole S. (2007). The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 26, 3124–3131. doi: 10.1038/sj.emboj.7601747
Roberts R. J., Vincze T., Posfai J., Macelis D. (2015). REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299. doi: 10.1093/NAR/GKU1046
Salgado J. C. S., Meleiro L. P., Carli S., Ward R. J. (2018). Glucose tolerant and glucose stimulated β-glucosidases – a review. Bioresour. Technol. 267, 704–713. doi: 10.1016/J.BIORTECH.2018.07.137
Saranya K., Sundaramanickam A., Shekhar S., Swaminathan S., Balasubramanian T. (2017). Bioremediation of mercury by Vibrio fluvialis screened from industrial effluents. BioMed. Res. Int. 2017, 6509648. doi: 10.1155/2017/6509648
Skinner K. A., Leathers T. D. (2004). Bacterial contaminants of fuel ethanol production. J. Ind. Microbiol. Biotechnol. 31, 401–408. doi: 10.1007/S10295-004-0159-0
Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/BIOINFORMATICS/BTR039
Suryawanshi R. K., Koujah L., Patil C. D., Ames J. M., Agelidis A., Yadavalli T., et al. (2020). Bacterial pigment prodigiosin demonstrates a unique antiherpesvirus activity that is mediated through inhibition of prosurvival signal transducers. J. Virol. 94, 00251-20. doi: 10.1128/JVI.00251-20
Vijay D., Alshamsi N. S., Moussa Z., Akhtar M. K. (2022a). Extraction of the anticancer and antimicrobial agent, prodigiosin, from Vibrio gazogenes PB1 and its identification by 1D and 2D NMR. Molecules 27, 6030. doi: 10.3390/MOLECULES27186030
Vijay D., Baby B., Alhayer M. S., Vijayan R., Akhtar M. K. (2022b). Native production of prodigiosin in the estuarine bacterium, Vibrio gazogenes PB1, and identification of the associated pig genes. Front. Mar. Sci. 0. doi: 10.3389/FMARS.2022.940888
Wang Z., Cao G., Zheng J., Fu D., Song J., Zhang J., et al. (2015a). Developing a mesophilic co-culture for direct conversion of cellulose to butanol in consolidated bioprocess. Biotechnol. Biofuels 8, 84. doi: 10.1186/S13068-015-0266-3
Wang Z., Hervey W. J., Kim S., Lin B., Vora G. J. (2015b). Complete genome sequence of the bioluminescent marine bacterium Vibrio harveyi ATCC 33843 (392 [MAV]). Genome Announc. 3, 1493–1507. doi: 10.1128/GENOMEA.01493-14
Wang Z., Lin B., Hervey W. J., Vora G. J. (2013). Draft genome sequence of the fast-growing marine bacterium Vibrio natriegens strain ATCC 14048. Genome Announc. 1, e00589-13. doi: 10.1128/GENOMEA.00589-13
Wang Z., Tschirhart T., Schultzhaus Z., Kelly E. E., Chen A., Oh E., et al. (2020). Melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis: characterization and application. Appl. Environ. Microbiol. 86, e02749-19. doi: 10.1128/AEM.02749-19
Watson B. N. J., Vercoe R. B., Salmond G. P. C., Westra E. R., Staals R. H. J., Fineran P. C. (2019). Type I-f CRISPR-cas resistance against virulent phages results in abortive infection and provides population-level immunity. Nat. Commun. 10, 5526. doi: 10.1038/S41467-019-13445-2
Wiegand D., Lee H. H., Ostrov N., Church G. M. (2018). Establishing a cell-free Vibrio natriegens expression system. ACS Synth. Biol. 7, 2475–2479. doi: 10.1021/ACSSYNBIO.8B00222
Xie Z., Tang H. (2017). ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33, 3340–3347. doi: 10.1093/BIOINFORMATICS/BTX433
Xu J., Dong F., Wu M., Tao R., Yang J., Wu M., et al. (2021). Vibrio natriegens as a pET-compatible expression host complementary to Escherichia coli. Front. Microbiol. 0. doi: 10.3389/FMICB.2021.627181
Xu Q., Dziejman M., Mekalanos J. J. (2003). Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 1286–1291. doi: 10.1073/PNAS.0337479100
Xu C., Huang R., Teng L., Wang D., Hemme C. L., Borovok I., et al. (2013). Structure and regulation of the cellulose degradome in Clostridium cellulolyticum. Biotechnol. Biofuels 6, 1–15. doi: 10.1186/1754-6834-6-73/FIGURES/5
Yoon S. H., Ha S., Lim J., Kwon S., Chun J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/S10482-017-0844-4
Zhang Y., Li Z., Liu Y., Cen X., Liu D., Chen Z. (2021). Systems metabolic engineering of Vibrio natriegens for the production of 1,3-propanediol. Metab. Eng. 65, 52–65. doi: 10.1016/J.YMBEN.2021.03.008
Keywords: high-value compound, polysaccharide, CRISPR, marine, virulence, degradation
Citation: Baby B, Vijay D, Philip PS, Alnuaimi AA, Almansoori HM, Areidat SO, Khan G, Vijayan R and Akhtar MK (2023) Complete genome sequence of Vibrio gazogenes PB1: an estuarine bacterium capable of producing prodigiosin from starch or cellulose. Front. Mar. Sci. 10:1028319. doi: 10.3389/fmars.2023.1028319
Received: 25 August 2022; Accepted: 16 June 2023;
Published: 14 July 2023.
Edited by:
Lory Z. Santiago-Vazquez, University of Houston–Clear Lake, United StatesReviewed by:
Xuewei Yang, Shenzhen University, ChinaJimmy H. Saw, George Washington University, United States
Copyright © 2023 Baby, Vijay, Philip, Alnuaimi, Almansoori, Areidat, Khan, Vijayan and Akhtar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjit Vijayan, cmFuaml0LnZAdWFldS5hYy5hZQ==; M. Kalim Akhtar, bWsuYWtodGFyQHVhZXUuYWMuYWU=
 Bincy Baby
Bincy Baby Dhanya Vijay
Dhanya Vijay Pretty S. Philip3
Pretty S. Philip3 Gulfaraz Khan
Gulfaraz Khan Ranjit Vijayan
Ranjit Vijayan M. Kalim Akhtar
M. Kalim Akhtar