Abstract
Fucoidanases are hydrolytic enzymes that degrade fucoidan to a lower molecular weight while retaining the side substituent groups of the polymer. Fucoidanases are produced by marine organisms: bacteria, fungi, algae, molluscs, echinoderms. Fucoidanases are rare and little studied enzymes. There is currently no information on the structural organization of fucoidanases, the size of active centers, their secondary and tertiary structures. This review summarizes the data on fucoidanase sources and factors influencing fucoidanase activity. It was found that that such factors include medium pH, temperature, and the presence of metal ions. The principles of classification of fucoidanases were analyzed. Fucoidanase was found to have high biological activity. Fucoidanases are known to hydrolyze fucoidan to oligosaccharides that have anti-inflammatory, antiangiogenic, anticancer, antiviral, prebiotic, and anticoagulant properties. Thus, research into sources, isolation methods, the effect of fucoidanase on fucoidan, and its enzymatic activity is promising, and can be used to build the body’s resistance to adverse environmental factors (difficult working conditions, stress, and overwork), as well as restore and stimulate the immune response.
1 Introduction
In nature, algae are an ancient form of existing for photosynthetic organisms that belong to the plant kingdom and make up its largest group. Along with that, they constitute a considerable part of human diet in many parts of the globe, mainly in Southeast Asian countries. Since prehistoric times, seaweeds have been widely used as an alternative medicine for different purposes. Besides, they are a source of polysaccharides with potentially active biological compounds. Brown algae fucoidan, is the most widely studied type of polysaccharide so far (Ale and Meyer, 2013; Ermakova et al., 2015; Biancarosa et al., 2018; Quitério et al., 2021). However, alginate is the most used polysaccharide in the industry. In general, the brown algae Ectocarpales and Laminariales can evolve polysaccharides with a structure consisting of 1 → 3-α-L-Fucp residues. Fucales is the main chain of fucoidan within the brown alga; it consists of linked 1 → 3 and 1 → 4 residues of α-L-fucoidan (Kusaykin et al., 2008; Holtkamp et al., 2009; Usov and Bilan, 2009; Hentati et al., 2020; Shiau et al., 2022). It (fucoidan) has a higher degree of polymerization. Therefore, depolymerization is required for fucoidan before medical or downstream applications. Enzymes that degrade polysaccharides have practical application in science, i.e. in structural studies, bioactivity studies, and drug manufacturing industries (Kusaykin et al., 2006; Drira et al., 2021).
Fucoidan is exposed to hydrolyze of some chemicals such as acids and bases, but enzymatic hydrolysis by fucoidanase is believed to be the main functional process involved in various biological activities towards sulfated polysaccharides. Till date, fucoidanase enzyme has only been described in a few or limited studies. It might be due to several factors, including a small number of a quantitative method for determining fucoidanase hydrolyzing activity, as well as the study’s use of structurally uncharacterized or partially purified fucoidan as a substrate (Hentati et al., 2020; Drira et al., 2021).
Fucoidanases are a collective name for the enzymes involved in the hydrolysis of polysaccharides and fucoidan. This group includes fucoidanase (EC 3.2.1.44, α-L- fucosidase, poly(1,2-α-L-fucoside-4- sulfate) glycanohydrolase), an endoenzyme that catalyzes the hydrolysis of the (1→2)-α-L-fucoidan bond by retaining sulfate in fucoidan, while alpha-L-fucosidase (EC 3.2.1.51) contain L-fucose and α(1→ 3)-, α(1→ 4)-, α(1→ 2)-O-fucosyl bond. Various microbial fucoidanases promoted the degradation of algal fucoidan from different sources, resulting in a variety of degradation patterns and product structures. Enzymatic hydrolysis of fucoidan is generally considered to be an advanced sustainable green technology because it does not allow to hydrolyze fucoidan in chemically (Sørensen et al., 2007; Dobrinčić et al., 2020; Mikkelsen et al., 2021).
Such studies need to consider the structural diversity of fucoidan. In the case of source, bacteria that can hydrolyze fucoidan molecules has been under investigation since 1959 (Yaphe and Morgan, 1959) when Fucoidanase was discovered in the Haliotis sp. (Thanassi and Nakada, 1967; Zayed and Ulber, 2020). In addition to the marine bacteria (Bakunina et al., 2000; Descamps et al., 2006; Silchenko et al., 2013; Trang et al., 2022) and marine invertebrates (Kitamura et al., 1992; Daniel et al., 1999; Kusaykin et al., 2003; Bilan et al., 2005), fucoidanase was also found in some listed fungi (Rodríguez-Jasso et al., 2010; Cui et al., 2018). Though, fucoidanase obtained from such agents, is comparatively far less common that the other enzymes like glycosidases produced from cellulases, amylases, laminarinases. Fucoidanases (formerly known as α-L-fucosidases) can be extracted as an intracellular as well as an extra-cellular metabolite. The information concerning the fermentative power of fucoidanases i.e. site of dissociation of glycoside bond in fucoidan, the coalition in their catalytic action, can rarely be found. Even though the sequence of genes affecting the structure of fucoidanase has been reported, and the specific amino acid structure of fucoidanases isolated from Mariniflexile fucanivorans SW5 (Silchenko et al., 2013) and Alteromonas sp. SN-1009, however, endofucosidase does not yet exist in the commercial form. Since, fucoidanases are less studied enzymes therefore, actual mode of action is still unclear (Birolli et al., 2019).
Hence, the present article has been designed to highlight the fucoidans, generated by specifically brown seaweed, along with to discuss new information or data on the fucoidanase enzyme hydrolyzing fucoidan, as well as to determine its properties and biological activity for possible downstream applications.
2 Sources of fucoidanase and its enzymatic activity
To date, enzymes that decompose fucoidan are produced by a wide range of living organisms (marine bacteria, seaweed, sea urchins). The fucoidanase enzyme was produced by marine bacteria, seaweeds, and sea urchins as a result of the formation of fucoidan during their vital activity, which is cleaved by fucoidanase to ensure the metabolism of these organisms. List of organisms producing fucoidannase are presented in Table 1.
Table 1
| Organisms | Type of organism | Source |
|---|---|---|
| Vibrio sp. N-5 | marine bacteria | (Furukawa et al., 1992; Bilan et al., 2006; Usoltseva et al., 2019; Choi et al., 2022; Lin et al., 2022) |
| Fucobacter marina SI-0098 | marine bacteria | (Wang et al., 2008) |
| Pseudoalteromonas citrea КММ 3296 | marine bacteria | (Bakunina et al., 2002) |
| Pseudoalteromonas citrea КММ 3297 | marine bacteria | (Bakunina et al., 2002 |
| Pseudoalteromonas citrea КММ 3298 | marine bacteria | (Bakunina et al., 2002 |
| Fucobacter marina SA-0082 | marine bacteria | (Wang et al., 2008) |
| Fucophilus fucoidanolyticus SI-1234 | marine bacteria | (Bakunina et al., 2002; Urvantseva et al., 2006) |
| Mesonia algae KMM 3909 | marine bacteria | (Urvantseva et al., 2006) |
| Maribacter sp. КММ 6211 | marine bacteria | (Urvantseva et al., 2006) |
| Gramella sp. KMM 6054 | marine bacteria | (Ohshiro et al., 2012) |
| Alteromonas sp. SN-1009 | marine bacteria | (Yarza et al., 2014) |
| Luteolibacter sp. H18 | marine bacteria | (Nagao et al., 2018) |
| Mariniflexile fucanivorans SW5 | marine bacteria | (Silchenko et al., 2013) |
| Sphingomonas paucimobilis PF-1 | marine bacteria | (Silchenko et al., 2013) |
| Formosa KM3553 | marine algae | (Silchenko et al., 2013) |
| Fusarium sp. LD8 | marine fungi | (Qianqian et al., 2011) |
| Pseudomonas atlantica | marine bacteria | (Vuillemin et al., 2020) |
| Pseudomonas carrageenovora, | marine bacteria | (Vuillemin et al., 2020) |
| Formosa | marine algae | (Vuillemin et al., 2020) |
| Streptomyces sp. | marine bacteria | (Manivasagan and Oh, 2015) |
| Pseudoalteromonas sp. SB 1493 | marine bacteria | (Lee et al., 2012) |
| Sphingomonas paucimobilis PF-1 | marine bacteria | (Kim et al., 2015) |
| Dendryphiella arenaria TM94 | marine fungi | (Bilan et al., 2002; Bilan et al., 2004; Bilan et al., 2006; Rodríguez-Jasso et al., 2010; Wu et al., 2011; Ahmad et al., 2022; Choi et al., 2022; Iliou et al., 2022), (Foley et al., 2011), (Wu et al., 2011) |
| Lambis sp. | marine mollusk | (Silchenko et al., 2014) |
| Littorina kurila | marine mollusk | (Kusaykin et al., 2003; Bilan et al., 2005; Yang et al., 2021b) |
| Haliotus sp. | marine mollusk | (Thanassi and Nakada, 1967) |
| Mizuhopecten yessoensis (Patinopecten yessoensis) | marine mollusk | (Kitamura et al., 1992) |
| Vasticardium flavum | marine mollusk | (Khanh et al., 2019b) |
| Echinodermata | echinoderms | (Chevolot et al., 2001; Larsson et al., 2003; Bilan et al., 2004; Bilan et al., 2005; Yang et al., 2021b; Abdel-Latif et al., 2022; Ahmad et al., 2022) |
| Strongylocentrotus nudus | Sea urchins | (Sasaki et al., 1996) |
Fucoidanase-producing organisms.
To date, there are several studies have highlighted about the isolation and characterization of fucoidanase including various possible methods for isolating and purifying fucoidanase. The general protocols followed for isolation and purification of some fucoidanases producing organisms from different marine sources are summarized in the Table 2.
Table 2
| Organism | Source | Isolated from | Species | Concentration Method | Used chromatography column | Total Protein (mg) | Total Activity (U) | Mw (kDa) | Purification (-Fold) | Specific activity (U/mg) | Yield (%) d.w. | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Busan coast (Lat 35°09′ N; Long129°07′ E), South Korea | Marine sediment | Streptomyces sp. | (NH4)2SO4 precipitation | Sephadex G-100 column | 16.74 | 12.81 | ND (Not Determined) | 3.1 | (Manivasagan and Oh, 2015) | ||
| Aomori, Japan | Coastal seawater | Strain SN-1009 | (NH4)2SO4 precipitation | DEAE-Cellulofine A-800, DEAE-Sepharose FF, Sephacryl S-200, Phenyl-Sepharose CL-4B, and DEAE-Cellulofine A-800 | 4.32 | 11,800 (mU) | 100 | 3500 | 2,730 (mU/mg) | 12 | (Sakai et al., 2004) | |
| Sewha Beach, Jeju Island, Korea | Seawater | Strain PF-1 | (NH4)2SO4 precipitation | Bio-Gel P-4 column | 4.4 | 84.08 | 60-130 | 112.8 | 0.019 (U·mg−1) | 13.6 | (Kim et al., 2008) | |
| PIBOC* Marine Microorganisms Collection | Luteolibacter algae H18 | (NH4)2SO4 precipitation | Resource Q column | ND (Not Determined) | 112 | 25 | ND (Not Determined) | - | (Nagao et al., 2018) | |||
| PIBOC* Marine Microorganisms Collection | Brown algae Fucusevanescens | Formosa algae strain KMM 3553T | Ultrafiltration | DEAE-MacroPrep column, Sephacryl S-200 column | ND (Not Determined) | 20.7 | 33.4 | ND (Not Determined) | 9.4 | (Silchenko et al., 2013) | ||
| Nha Trang Bay, Vietnam | Formosa haliotis | Ethanol precipitation | Ion-exchange | ND (Not Determined) | 71 | ND (Not Determined) | 0.98 | (Vuillemin et al., 2020) | ||||
| NA (Not available) |
Sea sand | Vibrio sp. N-5 | – | – | ND (Not Determined) | – | (Furukawa et al., 1992) | |||||
| Fungi | Baltic Sea in Germany | Sand | Dendryphiellaarenaria TM94 | Acetone precipitation | Sephadex G-100 | 210.05 | 67.07 (IU) | 180 | 26.67 | 0.32 (IU·mL−1) | 17.69 | (Wu et al., 2011) |
| North Sea in German | Sand | Fusarium sp. LD8 | Acetone precipitation | Sephadex G-100 | 120.43 | 30.64 (IU) | 64 | 22.7 | 0.25 (IU·mg−1) | 23.9 | (Qianqian et al., 2011) | |
| Mollusk | NA (Not available) |
Digestive glands | Lambis sp. | (NH4)2SO4 precipitation | Phenyl-sepharose, Sephacryl S-100, DEAE-MacroPrep, CM-MacroPrep, TSK 2000 | 0.11 | 0.28 | 50 | 99.0 | 2.571 | 0.75 | (Silchenko et al., 2014) |
| Shoji Store Co. (Hokkaido) | Hepatopancreas | Patinopectenyessoensis | (NH4)2SO4 precipitation | DEAE-Toyopearl 650 M, Isoelectric focusing, Sephacryl S-30 | 6.2 | 24.2 | 84 | 14 | 3.9 | 27 | (Kitamura et al., 1992) | |
| Sea urchin | NA (Not available) |
Digestive tract | Strongylocentrotusnudus | (NH4)2SO4 precipitation | SP-Toyopearl 650, CM -Sepharose FF, Sephacryl S-200 | 5.6 | 864 | 130 | 307 | 154 | 1.6 | (Sasaki et al., 1996) |
Isolation and purification of some fucoidanase-producing organisms from various sources.
*- G.B. Elyakov Pacific Institute of Bioorganic Chemistry of the Far-Eastern Branch of the Russian Academy of Sciences.
So far, different marine ecosystems such as Busan coast at South Korea (Manivasagan and Oh, 2015); Aomori at Japan (Sakai et al., 2004); Sewha Beach, Jeju Island at Korea (Kim et al., 2008); Baltic Sea and North Sea at Germany (Qianqian et al., 2011; Wu et al., 2011) have been studied with the aim to determine isolation of fucoidanases producing by marine organisms. Mentioned bacteria and fungi producing fucoidanase enzymes have been isolated from marine sediment (Manivasagan and Oh, 2015), seawater (Sakai et al., 2004), brown algae F. evanescens L. (Silchenko et al., 2013), sea sand (Furukawa et al., 1992; Qianqian et al., 2011; Wu et al., 2011), and mollusks and sea urchin producing fucoidanase enzymes have been isolated from digestive glands (Silchenko et al., 2014), hepatopancreas (Kitamura et al., 1992), and digestive tract (Sasaki et al., 1996).
2.1 Fucoidanase from marine bacteria
In another finding, a marine isolated such as Fucophilus fucoidanolyticus L. has been reported to synthesize numerous types of fucoidan-degrading enzymes in that way they were proficient in utilizing fucoidans of various structures. Additionally, intracellular fucoidanase also demonstrated the ability to hydrolyze 1,3- α-glycosidic bonds and to exhibit deacetylase activity on acetylated fucoidans produced by Cladosiphon okamuranus Tokida (Sakai et al., 2004).
Fucoidanase was preliminarily isolated from several marine microbial florae namely, Vibrio sp., Alteromonas sp., Pseudoalteromonas sp., and the members of Flavobacteriaceae (Furukawa et al., 1992; Shin-Ichi et al., 1992; Bakunina et al., 2000; Descamps et al., 2006). Subsequently, 25 epiphytic bacterial cultures isolated from brown algae, as well as more than 53 sea cucumber strains were screened and showed a significant result in fucoidanase production. The highest rate of fucoidanase activity was noted in Cytophaga and Alteromonas/Pseudoalteromonas species (Bilan et al., 2005; Yang et al., 2021a).
Furthermore, the novel marine bacterium Fucanobacter lyticus is an effective indicator of extracellular and intracellular fucoidanase causing degradation of fucoidan isolated from brown algae belonging to the genera Fucales and Chordariales and found to be effective for hydrolysis of fucoidan. Thus, Chordariales demonstrated the specificity of endo-1,3-α-fucoidanase (Sakai et al., 2004).
For example, fucoidanase from marine bacteria strain SN-1009 was purified using DEAE-Cellulofine A-800, DEAE-Sepharose FF, Sephacryl S-200, Phenyl-Sepharose CL-4B, and DEAE-Cellulofine A-800 column chromatography (Sakai et al., 2004). The purified enzyme showed specific activity of 2730 (mU/mg), and its output was 12%; the enzyme had molecular weight of 100 kDa (Sakai et al., 2004; Silchenko et al., 2014). Marine mollusk Lambis sp. produced fucoidanase using a five-step procedure in this case fuconoidase was isolated from Phenyl-sepharose, Sephacryl S-100, DEAE-MacroPrep, and CM-MacroPrep, TSK 2000 after concentration with ammonium sulfate precipitate. The enzyme demonstrated specific activity of 2.571 U/mg, and the purification efficiency was 99 times higher (Silchenko et al., 2014).
Bioactive fucoidan oligosaccharides obtained by the fucoidanase action are one of the promising components for biomedical applications. The high biological activity of fucoidans depends on the degree of their sulfation. In the study (Trang et al., 2022), scientists identified a new fucoidanase Fhf2 isolated from the genome of the marine bacterium Formosa haliotis. Thus, Fhf2 was found to be similar to endo-α(1,4)-fucoidanases (EC 3.2.1.212) of the glycoside hydrolase class.
2.2 Fucoidanase from seaweed
According to the study (Tran et al., 2022b), endofucoidanases such as endo-α-1,3-L-fucanase EC 3.2.1.211 and endo-α-1,4-L-fucanase EC 3.2.1.212 catalyze the depolymerization of fucoidans. Quantitive evaluation of endofucoidanase activity is of exceptional importance for characterizing the endofucoidanase kinetics and for comparing the effects of different endofucoidanases on various types of fucoidans. The study used the Fourier Transform Infrared Spectroscopy (FTIR) along with the sensitive factor analysis to quantify endofucoidanase. The results coincide with the data obtained by monitoring microbial enzymes FcnAΔ229, FFA2 and Fhf1Δ470 on fucoidan substrates. These substrates were isolated from the algae Fucus evanescens L. and Fucus vesiculosus L. The results of the studies showed that the peak of the spectra of fucoidans during fermentation fell in the range of 1220–1260 cm-1; however, their profiles on different substrates differentiated. Spectral peaks in the range 1220–1260 cm-1¹ correspond to the absorption spectra of sulfated fucosyls. In this range of the spectra, sulfate-ether bonds and stretching vibrations of CO-groups were absorbed. One unit of endofucoidanase activity or Uf is the amount of enzymes affecting the change in FTIR-PARAFAC by 0.01 in 498 seconds of reaction for 20g/l of pure fucoidan isolated from F. evanescens L. at 42°C, acidity 7.4, for solutions NaCl (100 mM) and CaCl2 (10 mM). The proposed quantitative analysis of endofucoidanase reveals new possibilities for studying endofucoidanases (Tran et al., 2022b).
Endo-α(1,4)-fucoidanase with molecular weight of 46 kDa was found to be more stable than the Fhf2 enzyme. Fhf2 was used for fermentation of seaweed fucoidans including F. evanescens L., F. vesiculosus L., Sargassum mcclurei Setchell, and Sargassum polycystum C.Agardh. The highest enzymatic activity of Fhf2 was shown in relation to fucoidan from F. evanescens L. The enzyme Fhf2 was active at 20–45°C and pH 6–9. In addition, the activity of this enzyme solely depended on the calcium content. NMR analysis showed that Fhf2 hydrolyzes α(1,4) bonds between sulfated L-fucosyl residues and releases oligosaccharides with a large amount of 2,4-disulfated fucose residues. Thus, it can be concluded that the Fhf2 fucoidanase is promising for the isolation of highly sulfated oligosaccharides, which are planned to be used to study the biological activity of fucoidan (Trang et al., 2022).
In the study (Silchenko et al., 2017b), it was established that it was possible to clone the gene encoding fucoidanase FFA2 in strains KMM 3553 of seaweed Formosa and in the bacterium Escherichia coli. Recombinant fucoidanase FFA2 was purified and its biochemical properties were studied. It was found that FFA2 has amino acid sequence identical to the one of fucoidanase FcnA isolated from M. fucanivorans. The molecular weight of the gene FFA2 was 101.2 kDa (918 amino acid residues). Analysis of this sequence showed that the studied fucoidanase FFA2 belongs to the class of glycoside hydrolases GH107 (CAZy). Substrate specificity was studied in details by using fucoidans from brown algae and synthetic fucooligosaccharides. Degradation of (1→4)-α-glycosidic bonds in the fucoidan of the brown alga F. evanescens L. FFA2 catalyzed within the fragment of fucoidan structure (→3) -α - L -Fuc p 2S-(1→4) -α - L -Fuc p 2S -(1→) n, but if the fragment structure was (→3) - α - L -Fuc p 2S,4S-(1→4) -α- L-Fuc p 2S-(1→) n, FFA2 enzymatic activity was absent.
2.3 Enzymatic activity of fucoidanases
Use of fucoidanase helps to assess the exact molecular structure responsible for the protective effect of biological activity. In addition, fucoidan-cleaving enzymes may help elucidate the various structures of fucoidan, paving the way for industrial applications of these enzymes.
To date, the activity of enzymes in relation to fucoidans has been little studied. The study (Vickers et al., 2018) presented an analysis of three enzymes from the class of glycoside hydrolases 107 (GH107): MfFcnA, P5AFcnA, and P19DFcnA, isolated from the culture liquid of bacteria of the genus Psychromonas. These enzymes were shown to be substrate-specific and active against fucoidans. Studies established that GH107 family enzymes share a common structure and catalytic mechanism with the GH29 α-l-fucosidase enzyme, the fact was proved by X-ray crystallography and NMR analysis. However, GH107 enzymes used the histidine side chain as an acid-base catalyst in the retention mechanism. Interpretation of the structure of the enzymes showed that these classes of enzymes differ in the architecture of the active sites for different substructures of fucoidan. According to the study (Vickers et al., 2018), these results illuminate the mechanism of molecular biological based processing of fucoidans.
3 Classification of fucoidanases
Due to the complex structure of fucoidans, which are heteropolysaccharides, fucoidanases are difficult to be classified correctly. Fragmentary data on the structures of fucoidans lead to an incorrect classification of fucoidanases. For example, fucoidan isolated from F. vesiculosus L. was determined incorrectly as a unit with structure 1→2-α-L-fucan. This definition led to the incorrect assignment of fucoidanases to EC 3.2.1.44. Subsequently, the structure of the enzyme was refined and assigned to α-1→3- and α-1→4 bond residues of fucose. However, there is a lack of knowledge for a complete and correct classification of fucoidanases. In addition, it is noted that enzymes that hydrolyze any glycosidic bonds in fucanes (polysaccharides from fucose residues) are called fucanases rather than fucoidanases (Kusaykin et al., 2016).
Comparison of amino acid sequences has led to an alternative classification of fucoidanases based on hydrolyzed bond types. The classification by carbohydrate active enzymes or CAZy (www.cazy.org) is called homologous. According to CAZy, glycoside hydrolase (GH) enzymes are divided into 133 classes (GH classes). Peptides with unknown properties can be found in each class, if they are similar in amino acid structure and sequence, they can also be classified by CAZy. CAZy can be used to predict the catalytic and molecular properties of fucoidanases (Gebler et al., 1992) and the structural geometry of cleavable glycosidic bonds (Henrissat et al., 1995). The CAZy system includes 133 GH classes united into 14 families of closely related classes. Due to the presence of sites that belong to several classes, some GHs are multifunctional enzymes. By applying this classification, fucoidanases can be assigned to class 107 GH. Fucoidanase FcnA isolated from the marine bacterium M. fucanivorans strain SW5T is one of the first members of this class. A comparative analysis of the secondary structure elements of the fucoidanase FcnA and other known fucoidanases subsequently made it possible to state that this entire class contains data on four sequences: FcnA (CAI47003.1) from M. fucanivorans SW5T, Fda1 (AAO00508.1) and Fda2 (AAO00509.1) from Alteromonas sp. SN-1009 and gene SVI_0379 (BAJ00350.1) from Shewanella violacea DSS12T (Kusaykin et al., 2016).
4 Factors influencing fucoidanase activity
4.1 pH influence
In Table 2 we can observe that fucoidanase of marine bacterium origin is most active in the neutral or slightly alkaline pH medium. Furthermore, fucoidanase isolated from the seaweed strain Formosa KMM3553 (Silchenko et al., 2013) showed supreme activity in an extensive range of acidity (pH: 6.5 to 9.1). Invertebrate enzymes usually have an optimal acidic pH, apart from the enzymes contained in the liver-pancreas of the marine mollusk Littorina kurila (Kusaykin et al., 2003). In marine bacterium, Formosa algae strain KMM 3553, two forms of fucoidanase were found in this organism, with acidic pH (about 5.5) and basic pH (about 8) being optimal (Table 3). Fucoidanase isolated from mollusca Haliotus sp. (Thanassi and Nakada, 1967), Lambis sp. (Silchenko et al., 2014), Vasticardium flavum (Khanh et al., 2019a), and echinus Strongylocentrotus nudus (Sasaki et al., 1996), showed supreme activity in the pH range of 3.5–5.5 (Table 2). There are only a few publications so far on marine fungal fucoidanase. Nowadays, scientists studied and characterized only fucoidanases isolated from Dendryphiella arenaria TM94 (Wu et al., 2011) and Fusarium sp. LD8 (Qianqian et al., 2011). It shows that fucoidanase of marine fungi origin showed supreme activity at pH 6.0 (Table 3).
Table 3
| Fucoidanases source | Species | pH Opt | Temp Opt | pH stability | Temp stability (°C) | Inhibited by | Activated by | Km (mg ml−1) |
Vmax (mg·mL−1 ·min−1) |
Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Luteolibacter algae H18 | 7.3 | 40 | stable at pH 5.5-8.5 after incubation at 30°C for 30 min | retained more than 90% of its activity after incubation at 40° C for 30 min but enzyme activity was lost above 50°C | Co2+, Cu2+, Mn2+, Zn2+ | Ba2+, Fe2+, Ca2+, Al3+ | – | – | (Nagao et al., 2018) |
| Formosa haliotis | 8 | 37-40 | – | – | – | Ca2+, Mn2+, Zn2+ or Ni2+ | – | – | (Vuillemin et al., 2020) | |
| Pseudoalteromonassp strain SB 1493 | 8 | 50 | – | – | – | – | – | – | (Lee et al., 2012) | |
| Formosa algae strain KMM 3553 | 6.5 to 9.1 | – | – | Activity of fucoidanase was considerably reduced after prolonged (about 60 min) incubation of the enzyme solution at 45°C. The enzyme was completely inactivated after 40 min of incubation at 55° C | Cu2+ and Zn2+ | Mg2+, Ca2+ and Ba2+ | – | – | (Silchenko et al., 2013) | |
| Vibrio sp. N-5 | 6 | 40 | stable between pH 5 and 8 | Completely inactivated over 70°C | – | – | – | – | (Furukawa et al., 1992) | |
| Formosa haliotis | 8 | 37 | – | Inactive at temperatures of 55°C and above. | – | – | – | – | (Vuillemin et al., 2020) | |
| Fungi | Dendryphiellaarenaria TM94 | 6 | 50 | 3.0 and 8.0, an activity loss of about 50% occurred after 4 and 6 h, respectively | At 30°C the activity of fucoidanase decreased to 12.5%, while at 80°C to 18.75%. Fifty percent inactivation of the fucoidanase activity occurred at 56°C for 1 h, and total fucoidanase inactivation could be achieved at 100°C for 30 min. | – | – | 6.56 | 6.55 | (Wu et al., 2011) |
| Fusarium sp. (LD8) | 6 | 60 | Displayed stability at pH 6.0, whereas at pH 5.0 and 8.0, an activity loss of about 50% occurred after 6 h incubation at room temperature (25°C), respectively. | half inactivation was 50°C | – | – | 8.9 | 2.02 | (Qianqian et al., 2011) | |
| Mollusk | Lambis sp. | 4.9 | 37 | – | half-inactivation time was 20 min at 54 °C | Hg2+, Zn2+, Cu2+ | Mg2+, Ba2+, Ca2+ | 1.3 | – | (Silchenko et al., 2014) |
| Littorina kurila | 5.4 and 8.5 | – | – | – | – | – | – | – | (Kusaykin et al., 2003) | |
| Haliotis sp. | 5.4 | 38 | 2-10 | At 50°C about 85% of the activity remained. purified enzyme apparently resisted denaturation until the temperature was above 50 |
Hg++, Ag+ | – | – | – | (Thanassi and Nakada, 1967) | |
| Vasticardium flavum | 3-4 | – | – | Activity was greatly reduced after 60 min of the denaturation at 45 °C. | Cu2+, Sn2+, Fe2+, Al3+ | Ca2+, Ba2+, Co2+, Mg2+ | – | – | (Khanh et al., 2019a) | |
| Sea urchin | Strongylocent rotusnudus | 3 | 45 | 2.0 to 5.0 | below 50°C | – | – | – | – | (Sasaki et al., 1996) |
Physico-chemical properties of fucoidanase produced from various sources.
Fucoidanase from the marine fungus Dentryphiella renaria TM94 is stable at pH 6–7; but when pH is 3.0 and 8.0, there is a loss of activity by 50% after 4 and 6 hours, respectively, and complete inactivation of fucoidanase is achieved at 100°C in 30 minutes (Wu et al., 2011). The most responsive to temperature changes appeared to be marine fungus Fusarium sp. LD8, as they loss half of their activity in about 1 hour at 50°C; although, and the enzyme had stable character at pH 6.0 (Qianqian et al., 2011) (Table 3).
4.2 Temperature influence
For marine invertebrate fucoidanase, the optimal temperature range is 38-45°C (Silchenko et al., 2014) (Kusaykin et al., 2003) (Thanassi and Nakada, 1967; Sasaki et al., 1996; Khanh et al., 2019a), and for marine bacteria and fungi it is 37-50°C (Furukawa et al., 1992; Lee et al., 2012; Silchenko et al., 2013; Nagao et al., 2018; Vuillemin et al., 2020) and 50-60°C (Qianqian et al., 2011; Wu et al., 2011). Marine bacterium Vibrio sp. N-5 produces fucoidanase that is completely inactivated at temperatures above 70°C (Furukawa et al., 1992); while marine bacterium F. haliotis gives fucoidanase inactivated at temperatures above 55°C (Vuillemin et al., 2020). These data indicates that fucoidanase can be characterized as active and stable in different medium with a varied range of pH; it is typical of fucoidanases isolated from the major part of marine sources as bacteria and invertebrate organisms (Silchenko et al., 2013). Besides, we found that this fucoidanase is functional even at high temperatures. Meanwhile, elevated temperature at industrial treatment and processing are considered prominent as this let reducing microbial contamination in large scale industrial reactions of prolonged durations (Ahmed et al., 2009; Garuba et al., 2020).
4.3 Influence of metal ions
Dependence of fucoidanase activity on impact of inhibitors and activators has not been thoroughly studied. Several publications devoted to the effect of metal ions on the fucoidanase activity. They include studies on divalent metal cations’ impact on the enzymatic activity of fucoidanases of different origin: isolated from marine bacterium Vibrio sp. N-5 (Furukawa et al., 1992) and from marine mollusk Haliotis sp. (Thanassi and Nakada, 1967). The research showed that these enzymes do not depend on metals. Though, cations of some metals can affect fucoidanase activity; but they are inhibited by Ag+ and ions of divalent metals (Hg2+, Fe2+, and Mn2+). Ions of Cu2+ and Pb2+ produce no effect on the activity of these enzymes, while ions of Co2+ and Mg2+ slightly activate the enzymes (Thanassi and Nakada, 1967; Furukawa et al., 1992). The activity of fucoidanase from Vasticardium flavum is enhanced by Ca2+, Ba2+, Co2+, and Mg2+ cations, but inhibited by Cu2+, Sn2+, Fe2+, and Al3+ cations (Khanh et al., 2019a). In one study (Nagao et al., 2018), the activity of the enzyme was intense by divalent cations such as Ba2+, Fe2+, and Ca2+, while inhibited by Co2+, Cu2+, Mn2+, and Zn2+ (Nagao et al., 2018) and Formosa KMM3553 fucoidanase requires metal ions for its activity (Silchenko et al., 2013) (Table 3). These facts propose that metal ions to have influence on either substrate binding or catalytic activity.
In consequences, fucoidanase enzyme activity primly depends on type of organisms and its physicochemical parameters. Hence, right identification of organism for the fucoidanase enzyme production is a need-based approach.
5 Biological activity of fucoidanases
Significant interest in health-promoting properties of polysaccharide bio-macromolecules of marine origin, particularly acidic polysaccharides, has been sparked by developments particularly in biopharmaceutical and genetic engineering techniques. Fucoidan is one of these and is thought to be promising (Wang et al., 2019). The structure of these polysaccharides is so irregular that it is difficult to study them chemically and analyze their structure in detail (Sergeevich and Olegovna, 2015). To address this issue, specific enzymes capable to hydrolyze complex fucoidan molecules are required.
Fucoidanase catalyzes the O-glycosidic bond hydrolysis in the main chain of the fucoidan molecule, while exhibiting biological activity. Moreover, fucoidanase is a rare and little-studied enzyme. To date, there is no information or very scanty information on the complete structure of fucoidanase, active sites, substrate binding sites, secondary and tertiary structures. The mechanism of fucoidanase action has not yet been completely understood. Moreover, due to their heterogeneity, fucoidans are still largely underutilized as a base of pharmaceuticals despite having a wide range of biological activity and being non-toxic when taken orally (Kusaykin et al., 2016). Therefore, one possible solution to address such issue is to define the sequence of amino acid in fucoidanase and obtain enough recombinant enzymes to study the enzyme’s properties and spatial structure (Tran et al., 2022a).
There was determined the sequence of amino acid in fucoidanase isolated from the marine bacterium Formosa algae. They were obtained by recording the nucleotide sequence of the F. algae genome and homologs of gene products contained in the genome of algae were found (Sergeevich and Olegovna, 2015). As a result, it can be concluded that, the algae genome contains two genes encoding fucoidanase (FFA1 and FFA2) and the review of the amino acid sequences of FFA1 and FFA2 by multiple alignment showed 57% identity. The amino acid sequence of FcnA is 67% and 57% identical for FFA1 and FFA2, respectively.
Cadherin-like domains were found in bacterial enzymes that accelerate decomposition of various compounds (proteins, carbohydrates, nucleic acids) and lectins that bind to anionic polysaccharides (Abdian et al., 2013). It should be noted that the functions of these domains described in the literature are common for binding to one or another biopolymer using calcium ions (calcium dependent binding).
The gene construct encodes full-length fucoidanases (FFA1 and FFA2) and their truncated derivatives without the C-terminal domain of FFA1-SD and FFA2-SD, and without the cadherin-like repeating domain of FFA2-KD and FFA1-KD, containing nucleotide sequences (Sergeevich and Olegovna, 2015).
The full-length recombinant fucoidanase and all its truncated derivatives catalyze the breakdown of fucoidan. The fucoidanase activity of recombinant and native enzymes is manifested only if there are ions of certain divalent metals. The presence of fucoidanase activity in recombinant FFA1-SD fucoidanase and truncated FFA2-SD derivatives suggests that the C-terminal domain may not be involved in catalysis. So, it could be stated that, fucoidanase FFA1-KD and FFA2-KD also have the ability to hydrolyze fucoidan, but to a much lesser extent. This modification of the protein can reduce its ability to bind to substrate molecules. If the cadherin-like domain plays a stabilizing role, then similar results can be achieved by decreasing the molecule stability (Abdian et al., 2013; Silchenko et al., 2013).
Nonetheless, both the enzymes rapidly hydrolyze fucoidan. The fucoidan chain alternates with residues of sulfated fucose that are linked to α-1 → 3 and α-1 → 4. The obtained data shows that both fucoidanases are specific for splitting of the α-1 → 4-glycosidic bond within residues of sulfated fucose (Tran et al., 2022a).
At present, three principal mechanisms of fucoidan modifying enzymes have been identified and listed below: a-L-fucosidase acts as a catalyst for the degradation of the a-L-fucosyl linkages at the nonreducing ends, thereby liberating fucose units from the major backbone of fucose in fucoidans (Figure 1).
Figure 1
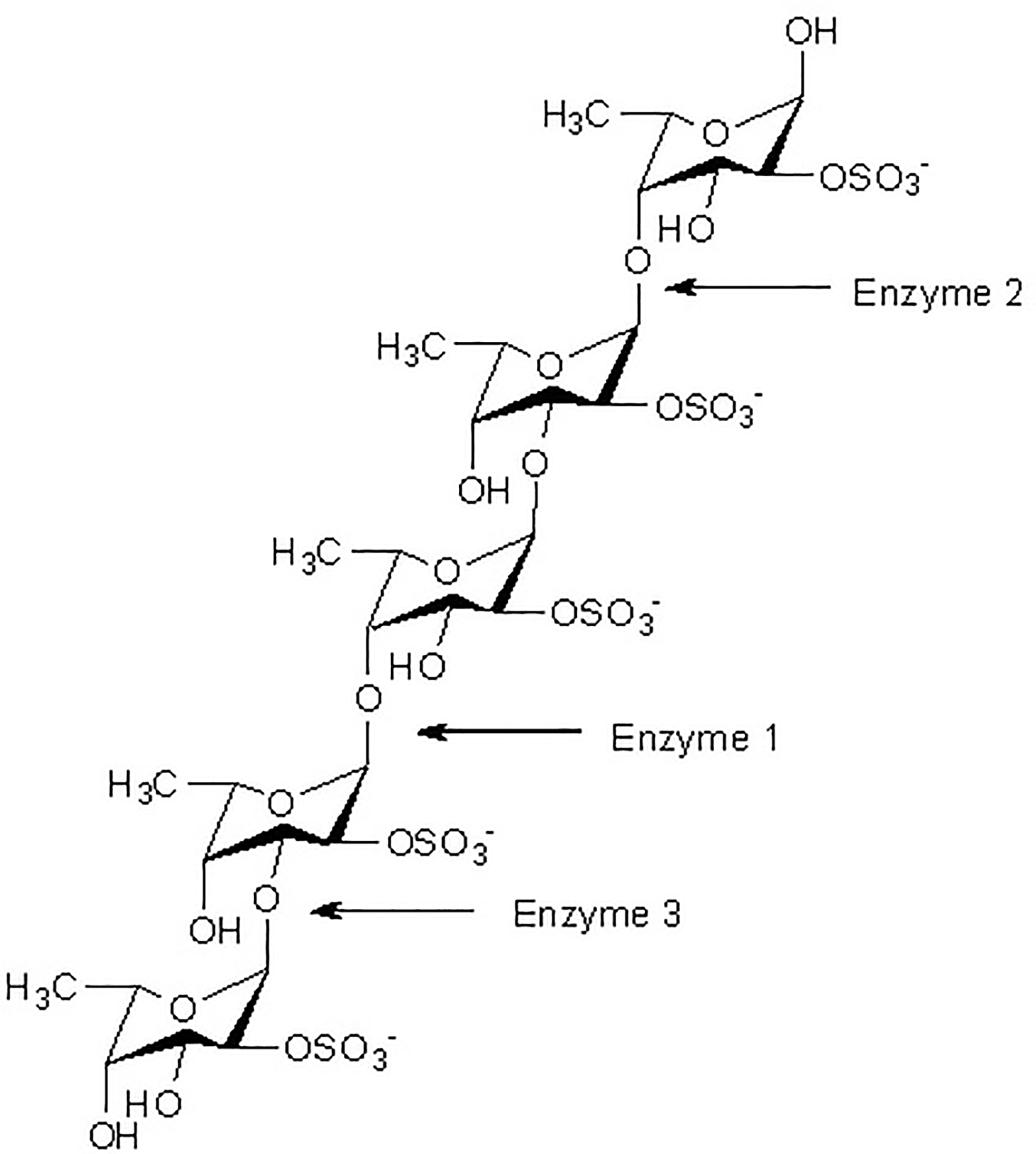
Fucoidanase assisted degradation pattern of fucoidan. Enzymatic attack patterns of α-L-fucoidan endohydrolase and fucosidase, respectively: For simplicity, we have distinguished the cleavage patterns as type 1 for (1→4)-α-L-fucoidan endohydrolase and type 2 for (1→3)-α-L-fucoidan endohydrolase; and the exo-action, EC 3.2.1.51, as type 3. (Since the EC 3.2.1.44 fucoidanase activity only designates catalysis of the endo-hydrolysis of (1→2)-α-L-fucosides, the enzymes type 1 and type 2 α-L-fucoidan endohydrolases are in principle not EC categorized.) (Ale and Meyer, 2013).
While the other type of fucoidanase causes random endo-type hydrolysis of glycosidic linkages (both α (1→3)-, and α (1→4)-) found in the fucose (Figure 1).
The study (Silchenko et al., 2017a) observed the anticancer activity of fucoidan from S. horneri var. densum C.Agardh, described its structure and established the process of its transformation under the action of fucoidanases. The gene encoding fucoidanase from the marine bacterium F. algae was identified and cloned; subsequently FFA1 was generated in Escherichia coli. The product of gene FFA1 had a molecular weight of 111 kDa. Amino acid sequence analysis indicated that the FFA1 fucoidanase belongs to the class of glycoside hydrolases GH107 (CAZy). The resulting recombinant fucoidanase FFA1 was used to obtain fucooligosaccharides. NMR spectroscopy was used to determine the structure of 5 sulfated oligosaccharides with a degree of polymerization of 4–10. It was shown that the fucoidan extracted from S. horneri var. densum C.Agardh was an almost pure fucane (Silchenko et al., 2017a).
6 Conclusion
The marine organisms serve as producers of bioactive compounds in a large amount and diversity presenting elements and compounds of different chemical classes (Barzkar et al., 2017; Barzkar et al., 2018; Jahromi and Barzkar, 2018a; Jahromi and Barzkar, 2018b; Barzkar, 2020; Barzkar and Sohail, 2020; Barzkar et al., 2021a; Barzkar et al., 2021b; Barzkar et al., 2021c; Barzkar et al., 2022a; Barzkar et al., 2022b). Meanwhile, seaweeds play the fundamental role in oxygen producing in the aquatic media, being prolific sources of bioactive agents (Barzkar et al., 2019). Seaweed polysaccharides such as brown algae fucoidan are of value and it is a negatively charged hygroscopic molecule that is chemically saturated with fucose and sulfated polysaccharides, mainly from the extracellular matrix of brown seaweeds. Fucoidan enzymes are a group of hydrolases that hydrolyze complex fucoidan polymers to produce low molecular weight fucoidan.
To date, only the basic principles of the construction of fucoidan molecules have been established. Chemical studies of these polysaccharides are difficult due to the extreme irregularity of their structure, and therefore a detailed analysis of the structure presents significant difficulties. Therefore, a reliable correlation between the chemical structure and the biological action of fucoidans has not yet been elicited. To solve these problems, it is important to use enzymes with established specificity and mechanism of action that can hydrolyze complex fucoidan molecules. A detailed analysis of the structure of small fragments of fucoidans will allow establishing the chemical structure of this polysaccharide. The use of fucoidan fragments of various structures will help to determine the relationship between certain structural elements and their biological action.
Enzymes that use fucoidans as a substrate are called fucoidanases or fucanases. These enzymes act as a hydrolysis catalyst for O-glycosidic bonds within the main chain of fucoidan molecules (EC 3.2.1.44, GH107 (CAZy database)). Fucoidanases are rare and poorly studied enzymes. To date, there is no information on the structural organization of fucoidanases, sizes of active centers, substrate-binding sites, secondary and tertiary structures. Fucoidanases presented a particularly not studied mechanism of action. Such a small number of publications on the structure of fucoidanases and their catalytic properties is due to the low content of these enzymes in organisms used as sources. One solution is to determine the sequences of amino acids inside fucoidanases and obtain recombinant enzymes in sufficient quantities to study specific properties and spatial structure of enzymes (Bakunina et al., 2000).
Fucoidans fermented by fucoidanase can be absorbed into the blood in a small amount and have a direct effect on the circulatory system (Tokita et al., 2010; Nagamine et al., 2014). Nagamine et al. (Nagamine et al., 2014) picturized that low molecular weight fucoidan oligo- and polysaccharides supplied with food are cleaved into small soluble fragments of about 3 kDa and secreted into the extracellular space. Further, these fragments can bind to receptors on the membranes of target cells, mediating effects similar to those during parenteral administration of high molecular weight polysaccharides (Nagamine et al., 2014).
Thus, both direct and indirect biological effects can be expected from fermented fucoidans (Trang et al., 2022). As in the case when fucoidan with low molecular weight fermented by fucoidanase causes a prebiotic effect that can be considered as a direct effect, while the anti-inflammatory effect can be considered as indirect and implemented through an improvement in the composition of the intestinal microflora, an increase in the proportion of symbiotic, and a decrease in the proportion of facultative microflora that has an anti-inflammatory effect (Luthuli et al., 2019; Wang et al., 2019). Fucoidanases can be engaged in the process of sulfated fucooligosaccharides’ production (in biotechnology), thus it has potential applications as biologically active additives and drugs.So, the collection of information on this subject indeed need-based attempt or research, and there is an ample scope to explore such enzyme or bioactive compounds for many beneficial purposes, including health, biomedical, bioengineering and life-science research. Hence, the future research strategies may be adopted or designed by the researchers with such latest updated information mentioned in this review article.
Statements
Author contributions
Conceptualization and writing original draft, NB. Editing, QL, STJ, VR and RD. Review and funding acquisition, SS, OB All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the financial support of the Ministry of Science and Higher Education of the Russian Federation as part of the implementation of the Presidential Grant 075-15- 2022-393 (MK-484.2022.1.4).
Acknowledgments
Special thanks go to the KMUTNB Postdoctoral Fellowship Program (KMUTNB-Post-66-07) for the financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abdel-Latif H. M. Dawood M. A. Alagawany M. Faggio C. Nowosad J. Kucharczyk D. (2022). Health benefits and potential applications of fucoidan (FCD) extracted from brown seaweeds in aquaculture: an updated review. Fish Shellfish Immunol.122, 115–130. doi: 10.1016/j.fsi.2022.01.039
2
Abdian P. L. Caramelo J. J. Ausmees N. Zorreguieta A. (2013). RapA2 is a calcium-binding lectin composed of two highly conserved cadherin-like domains that specifically recognize rhizobium leguminosarum acidic exopolysaccharides. J. Biol. Chem.288, 2893–2904. doi: 10.1074/jbc.M112.411769
3
Ahmad T. Ishaq M. Eapen M. Singh N. Park A. Karpiniec S. et al . (2022). Fucoidan as an inhibitor of pro-inflammatory cytokines: potential candidate for treating inflammatory-related conditions. FASEB J.36, L7837. doi: 10.1096/fasebj.2022.36.S1.L7837
4
Ahmed S. Riaz S. Jamil A. (2009). Molecular cloning of fungal xylanases: an overview. Appl. Microbiol. Biotechnol.84, 19–35. doi: 10.1007/s00253-009-2079-4
5
Ale M. T. Meyer A. S. (2013). Fucoidans from brown seaweeds: an update on structures, extraction techniques and use of enzymes as tools for structural elucidation. Rsc Advances.3, 8131–8141. doi: 10.1039/C3RA23373A
6
Bakunina I. Y. Nedashkovskaya O. Alekseeva S. Ivanova E. Romanenko L. Gorshkova N. et al . (2002). Degradation of fucoidan by the marine proteobacterium pseudoalteromonas citrea. Microbiology71, 41–47. doi: 10.1023/A:1017994131769
7
Bakunina I. I. Shevchenko L. Nedashkovskaia O. Shevchenko N. Alekseeva S. Mikhaĭlov V. et al . (2000). Screening of marine bacteria for fucoidan hydrolases. Mikrobiologiia69, 370–376. doi: 10.1007/BF02756738
8
Barzkar N. (2020). Marine microbial alkaline protease: an efficient and essential tool for various industrial applications. Int. J. Biol. Macromolecules.161, 1216–1229. doi: 10.1016/j.ijbiomac.2020.06.072
9
Barzkar N. Attaran Fariman G. Taheri A. (2017). Proximate composition and mineral contents in the body wall of two species of sea cucumber from Oman Sea. Environ. Sci. pollut. Res.24, 18907–18911. doi: 10.1007/s11356-017-9379-5
10
Barzkar N. Homaei A. Hemmati R. Patel S. (2018). Thermostable marine microbial proteases for industrial applications: scopes and risks. Extremophiles22, 335–346. doi: 10.1007/s00792-018-1009-8
11
Barzkar N. Jahromi S. T. Poorsaheli H. B. Vianello F. (2019). Metabolites from marine microorganisms, micro, and macroalgae: immense scope for pharmacology. Mar. Drugs17, 464. doi: 10.3390/md17080464
12
Barzkar N. Jahromi S. T. Vianello F. (2022a). Marine microbial fibrinolytic enzymes: an overview of source, production, biochemical properties and thrombolytic activity. Mar. Drugs20, 46. doi: 10.3390/md20010046
13
Barzkar N. Khan Z. Jahromi S. T. Pourmozaffar S. Gozari M. Nahavandi R. (2021a). A critical review on marine serine protease and its inhibitors: a new wave of drugs? Int. J. Biol. Macromolecules.170, 674–687. doi: 10.1016/j.ijbiomac.2020.12.134
14
Barzkar N. Sheng R. Sohail M. Jahromi S. T. Babich O. Sukhikh S. et al . (2022b). Alginate lyases from marine bacteria: an enzyme ocean for sustainable future. Molecules27, 3375. doi: 10.3390/molecules27113375
15
Barzkar N. Sohail M. (2020). An overview on marine cellulolytic enzymes and their potential applications. Appl. Microbiol. Biotechnol.104, 6873–6892. doi: 10.1007/s00253-020-10692-y
16
Barzkar N. Sohail M. Tamadoni Jahromi S. Gozari M. Poormozaffar S. Nahavandi R. et al . (2021b). Marine bacterial esterases: emerging biocatalysts for industrial applications. Appl. Biochem. Biotechnol.193, 1187–1214. doi: 10.1007/s12010-020-03483-8
17
Barzkar N. Sohail M. Tamadoni Jahromi S. Nahavandi R. Khodadadi M. (2021c). Marine microbial l-glutaminase: from pharmaceutical to food industry. Appl. Microbiol. Biotechnol.105, 4453–4466. doi: 10.1007/s00253-021-11356-1
18
Biancarosa I. Belghit I. Bruckner C. G. Liland N. S. Waagbø R. Amlund H. et al . (2018). Chemical characterization of 21 species of marine macroalgae common in Norwegian waters: benefits of and limitations to their potential use in food and feed. J. Sci. Food Agricult.98, 2035–2042. doi: 10.1002/jsfa.8798
19
Bilan M. I. Grachev A. A. Shashkov A. S. Nifantiev N. E. Usov A. I. (2006). Structure of a fucoidan from the brown seaweed fucus serratus l. Carbohydr. Res.341, 238–245. doi: 10.1016/j.carres.2005.11.009
20
Bilan M. I. Grachev A. A. Ustuzhanina N. E. Shashkov A. S. Nifantiev N. E. Usov A. I. (2002). Structure of a fucoidan from the brown seaweed fucus evanescens c. Ag. Carbohydr. Res.337, 719–730. doi: 10.1016/S0008-6215(02)00053-8
21
Bilan M. I. Grachev A. A. Ustuzhanina N. E. Shashkov A. S. Nifantiev N. E. Usov A. I. (2004). A highly regular fraction of a fucoidan from the brown seaweed fucus distichus l. Carbohydr. Res.339, 511–517. doi: 10.1016/j.carres.2003.10.028
22
Bilan M. Kusaykin M. Grachev A. Tsvetkova E. Zvyagintseva T. Nifantiev N. et al . (2005). Effect of enzyme preparation from the marine mollusk littorina kurila on fucoidan from the brown alga fucus distichus. Biochem. (Moscow).70, 1321–1326. doi: 10.1007/s10541-005-0264-3
23
Birolli W. G. Lima R. N. Porto A. L. (2019). Applications of marine-derived microorganisms and their enzymes in biocatalysis and biotransformation, the underexplored potentials. Front. Microbiol.10, 1453. doi: 10.3389/fmicb.2019.01453
24
Chevolot L. Mulloy B. Ratiskol J. Foucault A. Colliec-Jouault S. (2001). A disaccharide repeat unit is the major structure in fucoidans from two species of brown algae. Carbohydr. Res.330, 529–535. doi: 10.1016/S0008-6215(00)00314-1
25
Choi S. Jeon S. A. Heo B. Y. Kang J.-G. Jung Y. Duong P. T. T. et al . (2022). Gene set enrichment analysis reveals that fucoidan induces type I IFN pathways in BMDC. Nutrients14, 2242. doi: 10.3390/nu14112242
26
Cui K. Tai W. Shan X. Hao J. Li G. Yu G. (2018). Structural characterization and anti-thrombotic properties of fucoidan from nemacystus decipiens. Int. J. Biol. macromolecules.120, 1817–1822. doi: 10.1016/j.ijbiomac.2018.09.079
27
Daniel R. Berteau O. Jozefonvicz J. Goasdoue N. (1999). Degradation of algal (Ascophyllum nodosum) fucoidan by an enzymatic activity contained in digestive glands of the marine mollusc pecten maximus. Carbohydr. Res.322, 291–297. doi: 10.1016/S0008-6215(99)00223-2
28
Descamps V. Colin S. Lahaye M. Jam M. Richard C. Potin P. et al . (2006). Isolation and culture of a marine bacterium degrading the sulfated fucans from marine brown algae. Mar. Biotechnol.8, 27–39. doi: 10.1007/s10126-005-5107-0
29
Dobrinčić A. Balbino S. Zorić Z. Pedisić S. Bursać Kovačević D. Elez Garofulić I. et al . (2020). Advanced technologies for the extraction of marine brown algal polysaccharides. Mar. Drugs18, 168. doi: 10.3390/md18030168
30
Drira M. Hentati F. Babich O. Sukhikh S. Larina V. Sharifian S. et al . (2021). Bioactive carbohydrate polymers–between myth and reality. Molecules26, 7068. doi: 10.3390/molecules26237068
31
Ermakova S. Kusaykin M. Trincone A. Tatiana Z. (2015). Are multifunctional marine polysaccharides a myth or reality? Front. Chem.3, 39. doi: 10.3389/fchem.2015.00039
32
Foley S. A. Szegezdi E. Mulloy B. Samali A. Tuohy M. G. (2011). An unfractionated fucoidan from ascophyllum nodosum: extraction, characterization, and apoptotic effects in vitro. J. Natural prod.74, 1851–1861. doi: 10.1021/np200124m
33
Furukawa S.-I. Fujikawa T. Koga D. Ide A. (1992). Production of fucoidan-degrading enzymes, fucoidanase, and fucoidan sulfatase by vibrio sp. n-5. Nippon Suisan Gakkaish.58, 1499–1503. doi: 10.2331/suisan.58.1499
34
Garuba E. O. Adeleye P. A. Onilude A. A. (2020). Purification and properties of thermostable fucoidanase produced by recently isolated terrestrial aspergillus flavus FS018: Characteristics of fucoidanase extracted from aspergillus flavus FS018. Trends Pept. Protein Sci5 (e3), 1–7. doi: 10.22037/tpps.v5i0.30913
35
Gebler J. Gilkes N. Claeyssens M. Wilson D. B. Beguin P. Wakarchuk W. et al . (1992). Stereoselective hydrolysis catalyzed by related beta-1, 4-glucanases and beta-1, 4-xylanases. J. Biol. Chem.267, 12559–12561. doi: 10.1016/S0021-9258(18)42313-7
36
Henrissat B. Callebaut I. Fabreg S. Lehn P. Mornon J.-P. Davies G. (1995). Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci.92, 7090–7094. doi: 10.1073/pnas.92.15.7090
37
Hentati F. Tounsi L. Djomdi D. Pierre G. Delattre C. Ursu A. V. et al . (2020). Bioactive polysaccharides from seaweeds. Molecules25, 3152. doi: 10.3390/molecules25143152
38
Holtkamp A. D. Kelly S. Ulber R. Lang S. (2009). Fucoidans and fucoidanases–focus on techniques for molecular structure elucidation and modification of marine polysaccharides. Appl. Microbiol. Biotechnol.82, 1–11. doi: 10.1007/s00253-008-1790-x
39
Iliou K. Kikionis S. Ioannou E. Roussis V. (2022). Marine biopolymers as bioactive functional ingredients of electrospun nanofibrous scaffolds for biomedical applications. Mar. Drugs20, 314. doi: 10.3390/md20050314
40
Jahromi S. T. Barzkar N. (2018a). Future direction in marine bacterial agarases for industrial applications. Appl. Microbiol. Biotechnol.102, 6847–6863. doi: 10.1007/s00253-018-9156-5
41
Jahromi S. T. Barzkar N. (2018b). Marine bacterial chitinase as sources of energy, eco-friendly agent, and industrial biocatalyst. Int. J. Biol. macromolecules.120, 2147–2154. doi: 10.1016/j.ijbiomac.2018.09.083
42
Khanh H. H. N. Dieu V. Thinh T. Pt S. (2019a). Catalytic conditions of fucoidanase from vasticardium flavum. Vietnam J. Sci. Technol.57, 28–37. doi: 10.15625/2525-2518/57/1/12249
43
Khanh H. H. N. Trang V. T. D. Thinh P. D. San P. T. (2019b). Catalytic conditions of fucoidan degrading enzymes from vasticardium flavum. Vietnam J. Sci. Technol.57, 28–28. doi: 10.15625/2525-2518/57/1/12249
44
Kim W.-J. Kim S.-M. Lee Y.-H. Kim H.-G. Kim H.-K. Moon S.-H. et al . (2008). Isolation and characterization of marine bacterial strain degrading fucoidan from Korean undaria pinnatifida sporophylls. J. Microbiol. Biotechnol.18, 616–623.
45
Kim W. J. Park J. W. Park J. K. Choi D. J. Park Y. I. (2015). Purification and characterization of a fucoidanase (FNase s) from a marine bacterium sphingomonas paucimobilis PF-1. Mar. Drugs13, 4398–4417. doi: 10.3390/md13074398
46
Kitamura K. Matsuo M. Tsuneo Y. (1992). Enzymic degradation of fucoidan by fucoidanase from the hepatopancreas of patinopecten yessoensis. Biosci. biotechnol. Biochem.56, 490–494. doi: 10.1271/bbb.56.490
47
Kusaykin M. Bakunina I. Sova V. Ermakova S. Kuznetsova T. Besednova N. et al . (2008). Structure, biological activity, and enzymatic transformation of fucoidans from the brown seaweeds. Biotechnol. Journal: Healthcare Nutr. Technol.3, 904–915. doi: 10.1002/biot.200700054
48
Kusaykin M. Burtseva Y. V. Svetasheva T. Sova V. Zvyagintseva T. (2003). Distribution of O-glycosylhydrolases in marine invertebrates. enzymes of the marine mollusk littorina kurila that catalyze fucoidan transformation. Biochem. (Moscow).68, 317–324. doi: 10.1023/A:1023058301392
49
Kusaykin M. Chizhov A. Grachev A. Alekseeva S. Bakunina I. Y. Nedashkovskaya O. et al . (2006). A comparative study of specificity of fucoidanases from marine microorganisms and invertebrates. J. Appl. Phycol.18, 369–373. doi: 10.1007/s10811-006-9042-x
50
Kusaykin M. I. Silchenko A. S. Zakharenko A. M. Zvyagintseva T. N. (2016). Fucoidanases. Glycobiology26, 3–12. doi: 10.1093/glycob/cwv072
51
Larsson A. M. Andersson R. Ståhlberg J. Kenne L. Jones T. A. (2003). Dextranase from penicillium minioluteum: reaction course, crystal structure, and product complex. Structure11, 1111–1121. doi: 10.1016/S0969-2126(03)00147-3
52
Lee Y.-R. Lim J. M. Kim K.-Y. Mun S.-B. Kwak I. Sohn J. H. (2012). Isolation and characteristics of fucoidan degrading bacterium from marine. J. Life Sci.22, 1724–1728. doi: 10.5352/JLS.2012.22.12.1724
53
Lin J. Jiao G. Kermanshahi-Pour A. (2022). Algal polysaccharides-based hydrogels: extraction, synthesis, characterization, and applications. Mar. Drugs20, 306. doi: 10.3390/md20050306
54
Luthuli S. Wu S. Cheng Y. Zheng X. Wu M. Tong H. (2019). Therapeutic effects of fucoidan: a review on recent studies. Mar. Drugs17, 487. doi: 10.3390/md17090487
55
Manivasagan P. Oh J. (2015). Production of a novel fucoidanase for the green synthesis of gold nanoparticles by streptomyces sp. and its cytotoxic effect on HeLa cells. Mar. Drugs13, 6818–6837. doi: 10.3390/md13116818
56
Mikkelsen M. D. Cao H. T. T. Roret T. Rhein-Knudsen N. Holck J. Tran V. T. T. et al . (2021). A novel thermostable prokaryotic fucoidan active sulfatase PsFucS1 with an unusual quaternary hexameric structure. Sci. Rep.11, 1–12. doi: 10.1038/s41598-021-98588-3
57
Nagamine T. Nakazato K. Tomioka S. Iha M. Nakajima K. (2014). Intestinal absorption of fucoidan extracted from the brown seaweed, cladosiphon okamuranus. Mar. Drugs13, 48–64. doi: 10.3390/md13010048
58
Nagao T. Arai Y. Yamaoka M. Komatsu F. Yagi H. Suzuki H. et al . (2018). Identification and characterization of the fucoidanase gene from luteolibacter algae H18. J. biosci. bioeng.126, 567–572. doi: 10.1016/j.jbiosc.2018.05.016
59
Ohshiro T. Harada N. Kobayashi Y. Miki Y. Kawamoto H. (2012). Microbial fucoidan degradation by luteolibacter algae H18 with deacetylation. Biosci. biotechnol. Biochem.76 (3), 620–623. doi: 10.1271/bbb.110911
60
Qianqian W. Shuang M. Hourong X. Min Z. Jingmin C. (2011). Purification and the secondary structure of fucoidanase from fusarium sp. LD8. Evidence-Based Complement. Altern. Med.2011, 196190. doi: 10.1155/2011/196190
61
Quitério E. Soares C. Ferraz R. Delerue-Matos C. Grosso C. (2021). Marine health-promoting compounds: recent trends for their characterization and human applications. Foods10, 3100. doi: 10.3390/foods10123100
62
Rodríguez-Jasso R. M. Mussatto S. I. Pastrana L. Aguilar C. N. Teixeira J. A. (2010). Fucoidan-degrading fungal strains: screening, morphometric evaluation, and influence of medium composition. Appl. Biochem. Biotechnol.162, 2177–2188. doi: 10.1007/s12010-010-8992-2
63
Sørensen H. R. Pedersen S. Jørgensen C. T. Meyer A. S. (2007). Enzymatic hydrolysis of wheat arabinoxylan by a recombinant “minimal” enzyme cocktail containing β-xylosidase and novel endo-1, 4-β-xylanase and α-l-arabinofuranosidase activities. Biotechnol. Progress.23, 100–107. doi: 10.1021/bp0601701
64
Sakai T. Kawai T. Kato I. (2004). Isolation and characterization of a fucoidan-degrading marine bacterial strain and its fucoidanase. Mar. Biotechnol.6, 335–346. doi: 10.1007/s10126-003-0033-5
65
Sasaki K. Sakai T. Kojima K. Nakayama S. Nakanishi Y. Kato I. (1996). Partial purification and characterization of an enzyme releasing 2-sulfo-α-l-fucopyranose from 2-sulfo-α-l-fucopyranosyl-(1→ 2) pyridylaminated fucose from a sea urchin, strongylocentrotus nudus. Biosci. biotechnol. Biochem.60, 666–668. doi: 10.1271/bbb.60.666
66
Sergeevich S. A. Olegovna Z. A. (2015). Characterization of the catalytic properties of two novel recombinant fucoidanases from the marine bacterium Formosa algae. Eurasian Union Sci.8, 154–157.
67
Shiau J.-P. Chuang Y.-T. Yang K.-H. Chang F.-R. Sheu J.-H. Hou M.-F. et al . (2022). Brown algae-derived fucoidan exerts oxidative stress-dependent antiproliferation on oral cancer cells. Antioxidants11, 841. doi: 10.3390/antiox11050841
68
Shin-Ichi F. Tatsuo F. Daizo K. Akio I. D. E. (1992). Purification and some properties of exo-type fucoidanases from vibrio sp. n-5. Biosci. Biotechnol. Biochem.56 (11), 1829–1834. doi: 10.1271/bbb.56.1829
69
Silchenko A. S. Kusaykin M. I. Kurilenko V. V. Zakharenko A. M. Isakov V. V. Zaporozhets T. S. et al . (2013). Hydrolysis of fucoidan by fucoidanase isolated from the marine bacterium, Formosa algae. Mar. Drugs11, 2413–2430. doi: 10.3390/md11072413
70
Silchenko A. S. Kusaykin M. I. Zakharenko A. M. Menshova R. V. Khanh H. H. N. Dmitrenok P. S. et al . (2014). Endo-1, 4-fucoidanase from Vietnamese marine mollusk lambis sp. which producing sulphated fucooligosaccharides. J. Mol. Catalysis B: Enzymatic.102, 154–160. doi: 10.1016/j.molcatb.2014.02.007
71
Silchenko A. S. Rasin A. B. Kusaykin M. I. Kalinovsky A. I. Miansong Z. Changheng L. et al . (2017a). Structure, enzymatic transformation, anticancer activity of fucoidan and sulphated fucooligosaccharides from sargassum horneri. Carbohydr. polymers.175, 654–660. doi: 10.1016/j.carbpol.2017.08.043
72
Silchenko A. S. Ustyuzhanina N. E. Kusaykin M. I. Krylov V. B. Shashkov A. S. Dmitrenok A. S. et al . (2017b). Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiology27, 254–263. doi: 10.1093/glycob/cww138
73
Thanassi N. M. Nakada H. I. (1967). Enzymic degradation of fucoidan by enzymes from the hepatopancreas of abalone, haliotus species. Arch. Biochem. Biophys.118, 172–177. doi: 10.1016/0003-9861(67)90294-9
74
Tokita Y. Nakajima K. Mochida H. Iha M. Nagamine T. (2010). Development of a fucoidan-specific antibody and measurement of fucoidan in serum and urine by sandwich ELISA. Biosci. biotechnol. Biochem.74 (2), 350–357. doi: 10.1271/bbb.90705
75
Tran V. H. N. Nguyen T. T. Meier S. Holck J. Cao H. T. T. Van T. T. T. et al . (2022a). The endo-α (1, 3)-fucoidanase Mef2 releases uniquely branched oligosaccharides from saccharina latissima fucoidans. Mar. Drugs20, 305. doi: 10.3390/md20050305
76
Tran V. H. N. Perna V. Mikkelsen M. D. Nguyen T. T. Trang V. T. D. Baum A. et al . (2022b). A new FTIR assay for quantitative measurement of endo-fucoidanase activity. Enzyme Microbial. Technol.158, 110035. doi: 10.1016/j.enzmictec.2022.110035
77
Trang V. T. D. Mikkelsen M. D. Vuillemin M. Meier S. Cao H. T. T. Muschiol J. et al . (2022). The endo-α (1, 4) specific fucoidanase Fhf2 from Formosa haliotis releases highly sulfated fucoidan oligosaccharides. Front. Plant sci.13, 823668. doi: 10.3389/fpls.2022.823668
78
Urvantseva A. Bakunina I. Y. Nedashkovskaya O. Kim S. Zvyagintseva T. (2006). Distribution of intracellular fucoidan hydrolases among marine bacteria of the family flavobacteriaceae. Appl. Biochem. Microbiol.42, 484–491. doi: 10.1134/S0003683806050073
79
Usoltseva R. V. Shevchenko N. M. Malyarenko O. S. Anastyuk S. D. Kasprik A. E. Zvyagintsev N. V. et al . (2019). Fucoidans from brown algae laminaria longipes and saccharina cichorioides: structural characteristics, anticancer and radiosensitizing activity in vitro. Carbohydr. polymers.221, 157–165. doi: 10.1016/j.carbpol.2019.05.079
80
Usov A. I. Bilan M. I. (2009). Fucoidans–sulfated polysaccharides of brown algae. Russian Chem. Rev.78, 785. doi: 10.1070/RC2009v078n08ABEH004063
81
Vickers C. Liu F. Abe K. Salama-Alber O. Jenkins M. Springate C. M. et al . (2018). Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-l-fucosidases from GH29. J. Biol. Chem.293, 18296–18308. doi: 10.1074/jbc.RA118.005134
82
Vuillemin M. Silchenko A. S. Cao H. T. T. Kokoulin M. S. Trang V. T. D. Holck J. et al . (2020). Functional characterization of a new GH107 endo-α-(1, 4)-fucoidanase from the marine bacterium Formosa haliotis. Mar. Drugs18, 562. doi: 10.3390/md18110562
83
Wang Y. Xing M. Cao Q. Ji A. Liang H. Song S. (2019). Biological activities of fucoidan and the factors mediating its therapeutic effects: a review of recent studies. Mar. Drugs17, 183. doi: 10.3390/md17030183
84
Wang J. Zhang Q. Zhang Z. Li Z. (2008). Antioxidant activity of sulfated polysaccharide fractions extracted from laminaria japonica. Int. J. Biol. macromolecules.42, 127–132. doi: 10.1016/j.ijbiomac.2007.10.003
85
Wu Q. Zhang M. Wu K. Liu B. Cai J. Pan R. (2011). Purification and characteristics of fucoidanase obtained from dendryphiella arenaria TM94. J. Appl. phycol.23, 197–203. doi: 10.1007/s10811-010-9588-5
86
Yang J. Yang X. Pan W. Wang M. Lu Y. Zhang J. et al . (2021a). Fucoidan-supplemented diet potentiates immune checkpoint blockage by enhancing antitumor immunity. Front. Cell Dev. Biol.9, 2198. doi: 10.3389/fcell.2021.733246
87
Yang J. Yang X. Pan W. Wang M. Lu Y. Zhang J. et al . (2021b). Fucoidan-supplemented diet potentiates immune checkpoint blockage by enhancing antitumor immunity. Front. Cell Dev. Biol.9, 733246. doi: 10.3389/fcell.2021.733246
88
Yaphe W. Morgan K. (1959). Enzymic hydrolysis of fucoidin by pseudomonas atlantica and pseudomonas carrageenovora. Nature183, 761–762. doi: 10.1038/183761b0
89
Yarza P. Yilmaz P. Pruesse E. Glöckner F. O. Ludwig W. Schleifer K.-H. et al . (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol.12, 635–645. doi: 10.1038/nrmicro3330
90
Zayed A. Ulber R. (2020). Fucoidans: downstream processes and recent applications. Mar. Drugs18, 170. doi: 10.3390/md18030170
Summary
Keywords
brown seaweed, fucoidan, fucoidanase, biological activity, hydrolysis, polysaccharides, fermentation mechanism
Citation
Barzkar N, Rungsardthong V, Tamadoni Jahromi S, Laraib Q, Das R, Babich O and Sukhikh S (2023) A recent update on fucoidonase: source, Isolation methods and its enzymatic activity. Front. Mar. Sci. 10:1129982. doi: 10.3389/fmars.2023.1129982
Received
22 December 2022
Accepted
20 April 2023
Published
16 May 2023
Volume
10 - 2023
Edited by
Sachin Kumar, Sardar Swaran Singh National Institute of Renewable Energy, India
Reviewed by
Diana Pacheco, University of Coimbra, Portugal; Sasi Nayar, South Australian Research and Development Institute, Australia
Updates
Copyright
© 2023 Barzkar, Rungsardthong, Tamadoni Jahromi, Laraib, Das, Babich and Sukhikh.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noora Barzkar, noora.barzkar@gmail.com
This article was submitted to Marine Biotechnology and Bioproducts, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.