Abstract
Introduction:
As long-lived top predators inhabiting the Arctic and subarctic, belugas are under threat of anthropogenic stressors including climate change, pollution, noise, and habitat degradation, which in turn can negatively affect their health and viability. There is currently a need for health indicators that can be easily collected and used to assess and monitor the response to stressors in whales. Comparative transcriptomics using skin tissue can be used to provide understanding of organismal responses to stressors at the cellular level.
Methods:
For this study, intra- and inter-population comparisons were performed using the skin transcriptomes obtained from Bristol Bay (BB) belugas sampled in spring and late summer, and Eastern Chukchi Sea (ECS) belugas sampled in early summer in Alaska to investigate significantly differentially expressed genes over 2-fold change (padj<0.05).
Results:
Both principal component and hierarchical clustering analysis showed separate clustering of ECS whales, with further clustering of BB whales based on season. Intra-population comparisons carried out between different sexes and age groups did not result in any significant changes. However, the samples collected in spring versus summer within BB stock resulted in 541 significantly regulated genes, with significant activation (z-score≥|2|) predictions in pathways related with extracellular matrix organization, collagen biosynthesis and degradation, wound healing and cytokine signaling, potentially suggesting epidermal changes occurring in preparation for the seasonal molt in BB whales. The inter-population comparisons performed separately for BB-Spring versus ECS and BB-Summer versus ECS resulted in 574 and 938 significantly regulated genes, respectively. The significantly enriched canonical pathways common to both comparisons suggest increased cell survival and host defense responses along with increased cellular maintenance and growth in BB whales, and increased inflammation in ECS whales.
Discussion:
These changes observed could potentially be due to differences in molting, bias in hunting preferences and/or differences in environmental conditions during the time of sampling. Findings from this study suggest comparative skin transcriptomics as a useful tool towards understanding biologically relevant gene expression differences at different temporal and spatial scales among beluga stocks with potential to inform and contribute to conservation and recovery of endangered beluga stocks.
1 Introduction
As an ice-associated species mainly inhabiting Arctic and subarctic regions, belugas are increasingly exposed to multiple stressors potentially exacerbated by changes in sea ice conditions and ocean temperature (i.e., climate change). Even though the adaptive capacity of belugas to respond to these changes is currently unknown (Moore and Huntington, 2008; Gulland et al., 2022), some of the factors potentially affecting their distribution, behavior, reproduction and health, directly or indirectly, include contaminants, anthropogenic noise (e.g., offshore energy development, commercial shipping), changing prey populations, increased predation pressures, and emerging pathogens (Burek et al., 2008; Simmonds and Eliott, 2009; Gulland et al., 2022). There is a growing body of evidence about the effects of some of these stressors on the neuroendocrine and immune systems, reproduction, metabolism, as well as cellular DNA integrity and gene expression (Kight and Swaddle, 2011). Belugas are also an integral part of Alaska Native communities and are traditionally hunted for subsistence (Breton-Honeyman et al., 2021). As such, belugas are not only vitally important for meeting nutritional needs of many communities in Alaska, but they are also crucial for their medicinal, spiritual, economic and social network. Therefore, any changes in their distribution and health will in turn have negative consequences for the indigenous communities that depend on them.
There are six confirmed genetically distinct beluga stocks in Alaska (O’Corry-Crowe et al., 1997; O’Corry-Crowe, 2018; O’Corry-Crowe et al., 2021). Among these, the northerly located Eastern Chukchi Sea (ECS) stock is currently stable with a negatively biased estimate of 6,456 – 16,598 individuals observed during the months of July and August between 2012-2017 (Givens et al., 2020). Inhabiting both shallow nearshore and deepwater offshore habitats, the ECS whales mostly consume bottom invertebrates including shrimp along with some cod (Quakenbush et al., 2015). This stock is migratory, performing long-range seasonal migrations from oceanic wintering areas in the Bering Sea to ice-free estuaries in Chukchi Sea and western Beaufort Sea in late spring and early summer, and offshore habitats in the Beaufort Sea during summer, returning to the Bering Sea in the fall (Frost and Lowry, 1990; Suydam et al., 2005). The ECS whales are thought to perform these long-range migrations for several reasons, including their need for warmer water temperatures and appropriate substrate for rubbing off old skin for their seasonal summer molt which occurs during late June to late July (St. Aubin et al., 1990; Frost et al., 1993; Boily, 1995), and food sources, particularly in the Beaufort Sea and northeastern Chukchi Sea. The southern Bristol Bay (BB) stock remains in the bay the entire year, showing only small seasonal shifts in distribution (Citta et al., 2016), with an estimated average population size of ~2,040 individuals (Citta et al., 2019). Typically inhabiting shallow estuarine bays with strong tidal influences, BB whales mostly consume Pacific salmon as prey (Quakenbush et al., 2015). When salmon return to the bay in summer, BB whales are found at the river entrances, and in coastal areas, moving into deeper water when ice begins to form within BB in the autumn and early winter (Citta et al., 2016).
Cetacean skin has evolved for adaptation to life in the aquatic environment acting as a metabolically active barrier without the presence of a fully cornified layer of epidermis as observed in terrestrial mammals (Ehrlich et al., 2019; Espregueira Themudo et al., 2020; Menon et al., 2022). Cetacean epidermal skin is composed of lipokeratinocytes which are responsible for the production of keratin and lipid droplets, enhancing the capability to act as a physical barrier within a hypertonic environment, also providing buoyancy and an energy reserve (Elias et al., 1987; Elias, 1988; Mouton and Botha, 2012)). It is also an active immune organ providing essential protection from injury and infection, composed of structural proteins and inflammatory mediators (Zabka and Romano, 2003; Wang et al., 2021; Menon et al., 2022). Moreover, skin is a rich source of expressed genes with diverse functions in several physiological processes such as inflammatory, immune and stress responses (Ierardi et al., 2009; Van Dolah et al., 2015; Neely et al., 2018; Unal et al., 2018). Skin is also an abundant source of contaminants for cetaceans due to direct exposure or bioaccumulation, and skin gene expression profiling has successfully been utilized in cetaceans to assess long-term environmental and anthropogenic influences including contaminant exposure and toxicological responses, general physiological responses to stressors, immunosuppression, and seasonal differences (Trego et al., 2019a, b; Menon et al., 2022; Buckman et al., 2011; Fossi and Marsili, 2011; Panti et al., 2011; Van Dolah et al., 2015; Lunardi et al., 2016; Neely et al., 2018). Moreover, skin more likely represents chronic health status and/or seasonal changes as compared to blood, since the acute changes are almost immediately reflected in blood but not in skin (Unal et al., 2018; Menon et al., 2022), potentially due to the slow rates of epidermal cell turnover estimated to be between 45 – 75 days in cetaceans (Hicks et al., 1985; St. Aubin et al., 1990; Bechshoft et al., 2020). Due to these reasons and easy accessibility, cetacean skin is an ideal organ to study and to monitor the impact of anthropogenic stressors and environmental change (Menon et al., 2022), and can be obtained as a part of health-assessment studies and subsistence hunts, or through remote biopsy darts.
The seasonal molt in belugas occurs through shedding of the outer layer of degenerative epidermal skin cells to maintain skin health which appears to be facilitated by relatively warm and brackish waters in estuaries and lagoons in summer months (St. Aubin et al., 1990; Frost et al., 1993; Boily, 1995). Due to their continuous exposure to subfreezing temperatures in their wintering grounds, belugas conserve body heat by diverting blood flow away from the skin, which in turn inhibits continuous skin regeneration (St. Aubin et al., 1990; Pitman et al., 2020). Their movement to warmer waters is thought to facilitate this process as warmer temperatures can help with skin turnover by resuming the blood supply without losing heat (Boily, 1995; Pitman et al., 2020).
Functional genomics is frequently used in conservation biology to provide an understanding of organismal responses to environmental stressors at the cellular level. Gene expression studies have previously been carried out for dolphins (Panti et al., 2011; Van Dolah et al., 2015; Lunardi et al., 2016; Neely et al., 2018; Trego et al., 2019a), sperm whales (Wang et al., 2021) and belugas (Unal et al., 2018; Simond et al., 2019) by using skin samples to investigate transcriptomic resources and responses to various factors. However, to date, high-throughput skin transcriptome studies from belugas have not been reported, providing us with a unique opportunity to identify the physiological changes occurring in these populations. Transcriptome analysis through RNA sequencing allows the entire transcriptome to be surveyed in a quantitative manner (Wang et al., 2009) and has the potential to identify genes and pathways associated with immune/endocrine dysfunction, toxic exposure, reproductive disorders, and issues related to malnutrition and metabolism or other stressors, such as the warming environment.
The major goals of this study are to investigate population level gene expression differences in signaling pathways, biological functions and potential disease associated gene markers between the BB and ECS beluga stocks by carrying out differential gene expression analysis of transcriptomes. Not only will this study represent the first skin transcriptome analysis of belugas, but it will also provide an invaluable source of information for identification of new markers to monitor important biological activity essential for health and the stress response. In the long term, the data generated may potentially be utilized for comparative analysis with other beluga stocks in order to help inform management decisions for conservation success, especially for those stocks that are at risk.
2 Methods
2.1 Sample collection
For this study, epidermal skin samples of up to 5mm thickness collected from 24 BB belugas (14 males, 10 females), and 10 ECS belugas (7 males, 3 females) were utilized (Table 1). The BB skin samples were collected during spring (May, 6 samples) and late summer (August-September, 18 samples) in 2008, 2012, 2013 and 2014 during wild live-capture release beluga health assessment studies, coordinated by the National Marine Fisheries Service (NMFS), Alaska Department of Fish and Game, and Alaska SeaLife Center (Norman et al., 2012; Goertz et al., 2019) (NMFS Scientific Research Permits #782-1719 and #14245). These health assessments included capture, physical examination, morphological measurements, biological sampling, and satellite tagging followed by release in less than 2 hours (Goertz et al., 2019). ECS skin samples were collected in late June to early July 2010, 2012, 2014 and 2017 from 7 subsistence-hunted belugas (NMFS Permit #17350 by the Department of Wildlife Management, North Slope Borough, Utqiaġvik, AK), and 3 live-captured released belugas in association with satellite tagging and health assessment and preserved in 5-10 volumes of RNAlater® solution. All samples were then kept frozen at -80°C until further processing.
Table 1
| Animal ID | Stock origin | Sample group | Sex | Stage | Season | Date |
|---|---|---|---|---|---|---|
| BBN0208 | Bristol Bay, AK | Live-capture | F | Adult | Spring | 05/17/2008 |
| BBN0308 | Bristol Bay, AK | Live-capture | F | Adult | Spring | 05/18/2008 |
| BBN1008 | Bristol Bay, AK | Live-capture | F | Subadult | Spring | 05/21/2008 |
| DLBB16-02 | Bristol Bay, AK | Live-capture | F | Subadult | Spring | 05/13/2016 |
| DLBB16-07 | Bristol Bay, AK | Live-capture | M | Subadult | Spring | 05/16/2016 |
| DLBB16-09 | Bristol Bay, AK | Live-capture | M | Adult | Spring | 05/17/2016 |
| BBN1208 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 09/19/2008 |
| BBN1408 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 09/20/2008 |
| BBN1508 | Bristol Bay, AK | Live-capture | F | Subadult | Late Summer | 09/21/2008 |
| BBN1708 | Bristol Bay, AK | Live-capture | F | Adult | Late Summer | 09/21/2008 |
| BBN1808 | Bristol Bay, AK | Live-capture | M | Subadult | Late Summer | 09/24/2008 |
| DLBB12-04 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 09/08/2012 |
| DLBB12-06 | Bristol Bay, AK | Live-capture | F | Adult | Late Summer | 09/10/2012 |
| DLBB12-07 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 09/12/2012 |
| DLBB13-02 | Bristol Bay, AK | Live-capture | F | Adult | Late Summer | 08/24/2013 |
| DLBB13-09 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/302013 |
| DLBB13-10 | Bristol Bay, AK | Live-capture | F | Subadult | Late Summer | 08/302013 |
| DLBB14-01 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/25/2014 |
| DLBB14-02 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/25/2014 |
| DLBB14-03 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/26/2014 |
| DLBB14-07 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/29/2014 |
| DLBB14-08 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 08/31/2014 |
| DLBB14-09 | Bristol Bay, AK | Live-capture | F | Adult | Late Summer | 08/31/2014 |
| DLBB14-10 | Bristol Bay, AK | Live-capture | M | Adult | Late Summer | 09/03/2014 |
| LDL10-13 | E. Chukchi Sea, AK | Subs. hunt | M | Adult | Early Summer | 06/30/2010 |
| LDL10-14 | E. Chukchi Sea, AK | Subs. hunt | M | Adult | Early Summer | 06/30/2010 |
| LDL10-17 | E. Chukchi Sea, AK | Subs. hunt | F | Adult | Early Summer | 06/30/2010 |
| LDL14-13 | E. Chukchi Sea, AK | Subs. hunt | M | Subadult | Early Summer | 07/05/2014 |
| LDL14-14 | E. Chukchi Sea, AK | Subs. hunt | M | Adult | Early Summer | 07/05/2014 |
| LDL14-16 | E. Chukchi Sea, AK | Subs. hunt | F | Subadult | Early Summer | 07/05/2014 |
| LDL17-3 | E. Chukchi Sea, AK | Subs. hunt | M | Adult | Early Summer | 06/29/2017 |
| LC14303 | E. Chukchi Sea, AK | Live-capture | M | Adult | Early Summer | 06/29/2017 |
| LC14304 | E. Chukchi Sea, AK | Live-capture | M | Subadult | Early Summer | 06/29/2017 |
| LC108772 | E. Chukchi Sea, AK | Live-capture | F | Subadult | Early Summer | 07/09/2012 |
Skin samples utilized for the current study from wild live-captured released (Live-capture, LC) belugas from Bristol Bay (BBN, DLBB) and Eastern Chukchi Sea (LC), and from subsistence-hunted (Subs. hunt) belugas from Eastern Chukchi Sea (LDL).
F, Females; M, Males.
2.2 RNA extraction
The skin samples were processed utilizing a two-step tissue disruption protocol with a sterile mortar and pestle filled with liquid nitrogen, followed by homogenization in PureZOL™ RNA isolation reagent (Bio-Rad, Hercules, CA) and an Omni Tissue Homogenizer (Omni International, Inc., Kennesaw, GA) as previously described (Unal et al., 2018). Sterile hard tissue probes were used for homogenization, and both the mortar/pestle and the probes were changed in between samples to prevent cross-contamination. Total RNA extractions were then performed using Aurum™ Total RNA Fatty and Fibrous Kit (Bio-Rad, Hercules, CA) following the manufacturer’s protocol.
2.3 RNA sequencing and mapping
Total RNA samples were quantified, and the samples were submitted for RNA Sequencing at the University of Connecticut, Center for Genome Innovation. Quality control analyses for RNA samples were carried out using the Agilent TapeStation Automated Electrophoresis System (Agilent, Santa Clara, CA). Following strand-specific library generation using the Illumina TruSeq® stranded mRNA library preparation kit (Illumina, Inc., San Diego, CA), RNA sequencing was carried out using Illumina NovaSeq S4 instrument to generate paired-end reads for differential expression analysis.
Raw reads were trimmed with Trimmomatic (version 0.39) (Bolger et al., 2014), with a quality threshold of 25 and length threshold of 45. The quality of the reads was accessed using FastQC and MultiQC (Andrews, 2010; Ewels et al., 2016). A taxonomic classification analysis was then carried out to identify the sequences that are of non-mammalian origin using Kraken2 (Wood et al., 2019), utilizing a database built from the reference sequence libraries of bacteria, viruses, and archaea. The small percentage (<3%) of contaminating reads were filtered out. The remaining reads were mapped against the previously published Delphinapterus leucas genome (Jones et al., 2017) (NCBI Genome Assembly No: ASM228892v3) using HISAT2 (version 2.1.0) (Kim et al., 2015). The resulting SAM files were converted into BAM format using samtools (version 1.9) (Li et al., 2009), and gene expression was estimated by generating a count matrix with htseq-count (Anders et al., 2015).
2.4 Differential gene expression analysis and data visualization
For initial data visualization, Principal Component Analysis (PCA) was carried out based on the variance stabilized transformation of the count data for 500 genes that showed the largest variability across all the samples. The differential expression of genes between conditions were evaluated using DESeq2 (Love et al., 2014) installed within RStudio (Rstudio PBC, 2021.09.1). Sex and age group (adults or subadults based on observations) were included as covariates in the DESeq2 analysis to account for any potential effects. Shrunken log2 fold changes were utilized for DESeq2 to reduce the noise due to the genes with low counts and high dispersion values (Zhu et al., 2019).
Intra-population variability was assessed by carrying out differential gene expression analysis of different sexes (females versus males) and life stages (subadults versus adults) for both stocks. Differential gene expression analysis was also carried out between live-captured and hunted whales from ECS stock, and between different seasons (spring versus summer) in BB stock. Sea surface temperature (SST) data for BB was obtained from seatemperature.info website. Correlation and linear regression analysis were carried out for the top differentially expressed genes for seasonal comparison. Comparisons between ECS and BB stocks were then performed separately for spring and summer BB samples. The analysis was carried out with a false discovery rate-controlled (Benjamini and Hochberg, 1995) significance value (padj<0.05), and log2 fold-change cutoff value (log2FC ≥ |1|). Genes which did not have any counts in at least half of the samples for each comparison were removed from the analysis based on published recommendations (Deyneko et al., 2022).
For each comparison, a Bland-Altman plot (MA plot) was generated to display the magnitude of change as log2FC versus the mean expression using the normalized counts in order to visualize the gene expression differences between two stocks. A volcano plot was generated to show the statistical significance (-log10(padj)) versus the magnitude of change (log2FC). Heatmap analysis was carried out using regularized log transformation of counts and the shrunken log2 fold changes of the top 200 differentially regulated genes in order to visualize the hierarchical clustering of the data for samples and genes. The top significantly (padj<0.05) regulated genes among the pairwise comparisons were then inspected to see if their normalized counts seem consistent with expectations using the plotCounts function in RStudio, and were reported along with their fold change values, significance and putative functions.
2.5 Pathway analysis
Canonical pathway analysis was carried out using the QIAGEN Ingenuity Pathway Analysis (IPA, Version 84978992) platform incorporating the algorithms developed for QIAGEN IPA (Krämer et al., 2013). QIAGEN IPA software utilizes extensive and manually curated content maintained in the Ingenuity Knowledge Base to investigate the activation status of the pathways based on the differentially expressed genes between comparisons (Krämer et al., 2013). The index of cell signaling and metabolic canonical pathways are defined in Pathways Knowledge Library of the Ingenuity Knowledge Base which are utilized by QIAGEN IPA software (https://geneglobe.qiagen.com/us/knowledge/pathways). The pathways related to cancer and cardiovascular signaling were excluded from the analysis due to irrelevance with skin tissue. Human orthologs of the gene names were used as gene identifiers. Upregulated and downregulated genes were analyzed separately and genes that were significantly associated with a canonical pathway were identified. A right-tailed Fisher’s Exact Test was used to calculate a Benjamini-Hochberg corrected p-value (FDR) determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone with FDR<0.05 cutoff for significant associations. Among the significantly enriched pathways, only those with an activation z-score value of ≥ |2| were reported. The z-score ≥2 was defined as the threshold of significant activation, whereas z-score ≤−2 was defined as the threshold of significant inhibition. Intra-population pathway analysis was performed between two different seasons for BB samples only (BB-Spring versus BB-Summer). Seasonal data was not available for the ECS population. Inter-population analyses were then performed by utilizing separate comparisons of BB-Spring versus ECS and BB-Summer versus ECS. The canonical pathways showing significant differences common to both comparisons were then identified using the comparison analysis heatmap functionality of IPA. This comparison analysis enables visualization of the paired canonical pathways common to both comparisons as a heatmap. The canonical pathways that are predicted to be activated or inhibited in the same way in both comparisons were reported as a means to minimize the effect of differences between spring and summer seasons on gene expression.
3 Results
3.1 Transcriptome generation and data visualization
RNA sequencing was carried out for a total of 34 beluga skin samples obtained from two different stocks (BB and ECS), including samples collected from two different seasons within the BB stock generating an average of 20 million reads per sample. The taxonomic classification analysis (Kraken2) resulted in an average of 2.9% of the reads being represented by microorganisms. These reads were then removed from the downstream analysis. Following trimming for sequencing adapters and filtering of low-quality reads, the samples had an average of 12.2 million reads with a mean read size of 140bp and per base quality (Phred) scores of over 36 out of 40. Upon mapping of these reads to the beluga genome, the average alignment rate was 82.32%, resulting in a total of 12,350 genes available for differential expression analysis (Supplementary Table S1) out of a total of 15,002 genes identified in beluga skin.
Principal Component Analysis (PCA) of the top 500 genes that displayed the largest variability in gene expression resulted in separate clustering of the two stocks with the first principal component (PC1) explaining 60% and the second principal component (PC2) explaining 11% of the variance observed (Figure 1). Within the BB stock, the samples collected from spring and summer seasons also displayed separation (Figure 1). Live-captured ECS belugas clustered with the hunted ECS belugas, except for one individual (LC108778-12) which did not cluster with either group displaying a very different gene expression profile than the rest of the individuals. This individual, which might potentially have a different health status, was then removed from the dataset for the downstream analysis.
Figure 1
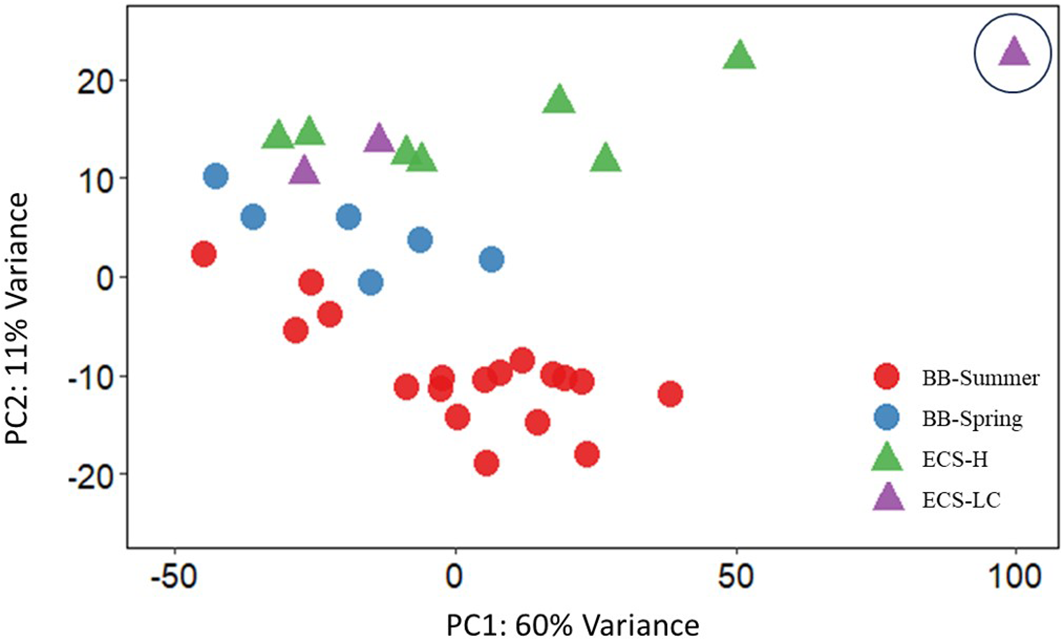
Principal Component Analysis (PCA) of top 500 genes that display the largest variability among the two beluga stocks for the first principal component (PC1) explaining 60% of the variance, and the second principal component (PC2) explaining 11% of the variance. Bristol Bay live-captured whales sampled during September are shown in red circles (BB-Summer) and those sampled during May are shown in blue circles (BB-Spring). Eastern Chukchi Sea subsistence-hunted whales (ECS-H) are shown in green triangles and Eastern Chukchi Sea live-captured whales (ECS–LC) are shown in purple triangles. The single individual circled indicates the outlier sample (LC108778-12) which was removed from the differential expression analysis.
Hierarchical cluster (heatmap) analysis of the top 200 genes that are significantly differentially expressed (FDR<0.05, log2FC ≥1) in ECS vs BB beluga stocks displayed distinct clustering of the individuals consistent with their stock assignments (Figure 2). Clustering by individuals (i.e. columns) displayed two main clusters representing each stock (ECS or BB), with further sub-clustering of the BB stock based on season. Clustering by rows (i.e. genes) displayed two main clusters of genes. While the top gene cluster showed upregulation in ECS whales, the bottom cluster showed downregulation when compared to BB belugas (Figure 2).
Figure 2
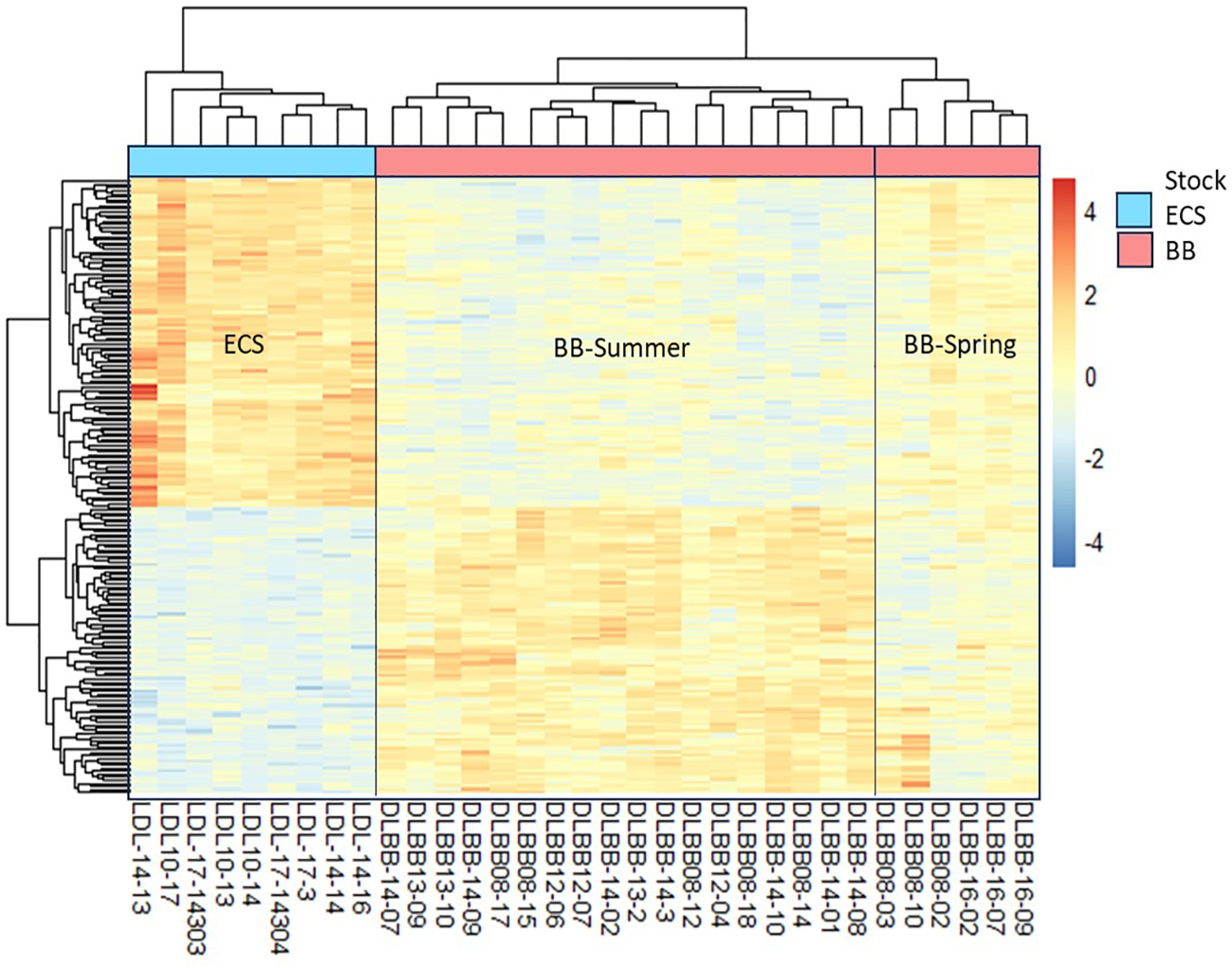
Hierarchical clustering (heatmap) analysis of the top 200 significantly differentially expressed genes between BB and ECS beluga stocks based on -log(FDR) values of shrunken log2 fold-changes. BB samples showed clustering based on the two seasons (BB-Summer and BB-Spring). The expression data is scaled by row, and clustered by rows and columns. The colors in the heatmap represent the gene expression level in terms of log2 fold change from the mean. Mean expression across the whole dataset is set to 0 indicated by yellow in the color scale. Red indicates upregulation and blue indicates downregulation in ECS vs BB stocks.
3.2 Differential gene expression and pathway analyses
3.2.1 Intra-population variability
Differential expression analysis performed between females versus males, subadults versus adults, or hunted versus live-captured ECS whales did not result in any significant gene expression differences. Year was not included in the analyses because of small sample sizes. However, differential expression analysis performed between the samples collected in spring (May) versus summer (September) seasons within the BB stock resulted in 3.6% of the entire transcriptome exhibiting significant changes between spring and summer, resulting in a total of 541 significantly (padj<0.05) regulated genes with a log2FC range from -3.78 to 3.42 (Figure 3). Out of these, 391 genes were successfully mapped with 75% of the mapped genes (293 total) showing higher expression in summer, and 25% of the genes (98 total) showing higher expression in spring (Supplementary Table S2).
Figure 3
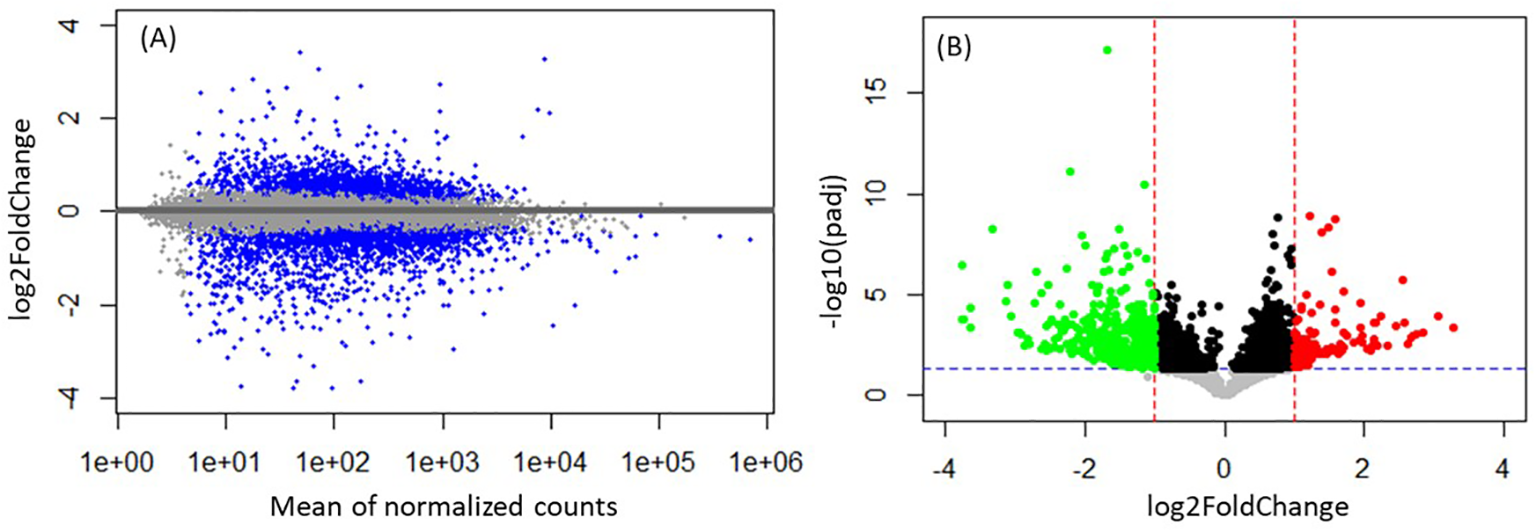
Differential expression analysis overview for seasonal (spring versus summer) comparison within the BB stock (A) Bland-Altman (MA plot) displaying shrunken log2 fold change (y-axis) versus mean of normalized counts (x-axis). The horizontal (x = 0) line represents the mean expression value, and the gray dots represent the genes that did not pass the significance criteria. The blue dots above the line represent upregulation in spring vs summer, and the blue dots below the line represent downregulation in spring vs summer (i.e. upregulation in summer vs spring) (B) Volcano plot displaying the statistical significance (-log10(padj), y-axis) versus magnitude of change (log2FoldChange, x-axis). The horizontal dashed line represents the significance cutoff value of padj = 0.05, and the vertical dashed lines represent log2 fold change cutoff value of |1|. Black and gray dots represent the genes that did not pass the significance criteria. Significantly downregulated genes are shown in green on the left (log2FoldChange ≤ -1), and significantly upregulated genes are shown in red on the right (log2FoldChange ≥ 1).
The most significantly upregulated genes in spring vs summer were mostly related with mitotic cell cycle, signal transduction, and cellular growth, proliferation and development. The most significantly (padj = 9.12E-10) upregulated gene in spring vs late summer within the BB stock was tubulin beta 2A class IIa (TUBB2A) which takes part in cytoskeleton organization and the mitotic cell cycle (Table 2; Figure 4A). The additional genes that were consistently upregulated in spring samples included those that take part in mitotic cell cycle (CENPT), cellular proliferation and skin aging (LMNB1), mitochondrial maintenance (MTFR2), mitochondrial respiration (PHB1), and oxidative stress response (GPX2) (Table 2; Figure 4A).
Table 2
| Gene ID | Gene Name | Log2FC | padj | Function |
|---|---|---|---|---|
| Upregulated in spring (May) | ||||
| TUBB2A | tubulin beta 2A class IIa | 1.466 | 9.12E-10 | Cytoskeleton organization, microtubule formation, mitotic cell cycle |
| GPX2 | glutathione peroxidase 2 | 1.377 | 1.81E-09 | Antioxidant defense, oxidative damage/stress response |
| CENPT | centromere protein T | 1.448 | 3.19E-06 | Centromere assembly, kinetochore formation, chromosome segregation |
| LMNB1 | lamin B1 | 1.135 | 8.89E-06 | Intracellular structural support, DNA replication, gene expression |
| MTFR2 | mitochondrial fission regulator 2 | 1.337 | 2.62E-05 | Mitochondrial maintenance, aging |
| RRM2 | ribonucleotide reductase regulatory subunit M2 | 1.874 | 3.26E-05 | DNA synthesis and repair, cell cycle regulation |
| PHB1 | prohibitin 1 | 1.068 | 4.83E-05 | Maintenance of mitochondrial respiration, cell signaling, glucose homeostasis |
| Upregulated in late summer (September) | ||||
| TECTA | tectorin alpha | -1.644 | 3.24E-17 | Auditory receptor cell organization, mechanotransduction, cell-matrix adhesion |
| GUCY2D | guanylate cyclase 2D | -2.187 | 7.04E-12 | Phototransduction, photoreceptor functioning in light/dark cycle, response to stimulus |
| GRB14 | growth factor receptor bound protein 14 | -1.205 | 7.92E-12 | Inhibition of insulin receptor signaling skin health and metabolism |
| RORC | Retinoid acid related orphan receptor C | -1.597 | 8.05E-12 | Circadian rhythm signaling, inflammation, autoimmune function |
| NPM2 | nucleophosmin/nucleoplasmin 2 | -2.070 | 3.62E-09 | Expressed in melanocytes in pigmented skin |
| SMIM2 | small integral membrane protein 2 | -3.246 | 4.04E-09 | Barrier function, phospholipid metabolism, maintenance of skin integrity |
| MTMR4 | myotubularin related protein 4 | -1.421 | 1.45E-08 | Macrophage phagocytosis, pathogen removal |
| OBSCN | obscurin, cytoskeletal calmodulin | -1.410 | 2.33E-08 | Calcium regulation, organization of myofibrils, cell adhesion |
| RIMS3 | regulating synaptic membrane exocytosis 3 | -1.140 | 2.85E-08 | Regulation of synaptic function, neurotransmitter release |
Top significantly (padj < 0.05) differentially regulated genes in spring vs late summer within the Bristol Bay stock.
Log2 fold-change values (Log2FC), adjusted significance value (padj) and putative gene functions in skin are indicated.
Figure 4
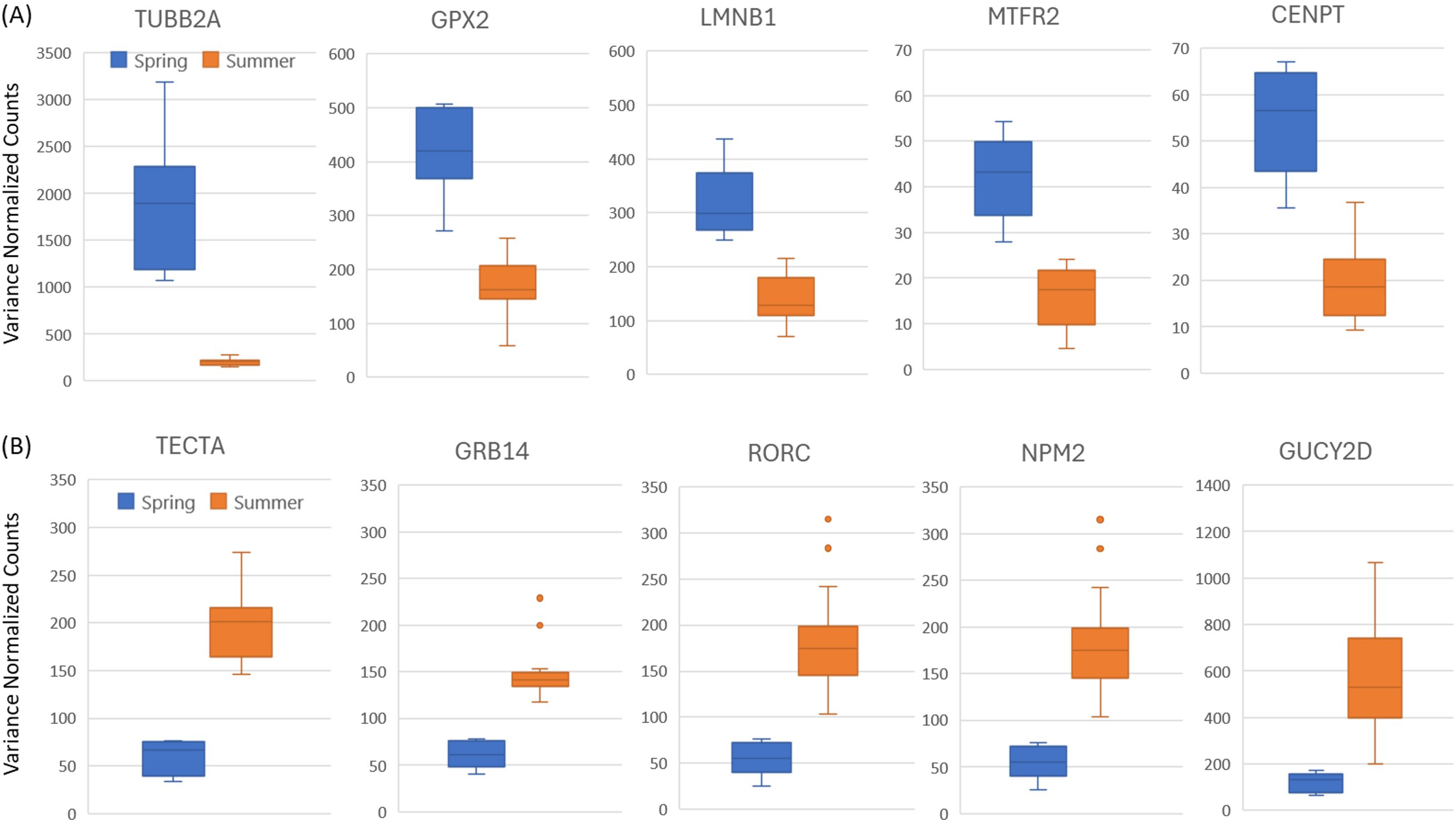
Variance normalized counts for the top five significant genes (with lowest padj values) that are differentially expressed in spring vs summer samples collected from the BB stock. (A) Genes with higher expression in spring (B) Genes with higher expression in summer. Blue color represents spring samples, and orange color represents summer samples.
The most significantly upregulated genes in summer vs spring were mostly related with skin structural maintenance, sensory perception and circadian rhythm. The most significantly (padj =3.24E-17) upregulated gene in late summer vs spring was tectorin alpha (TECTA), an extracellular matrix constituent important in sound perception and mechanotransduction. Additional genes that were consistently upregulated in summer included those that take part in sensory perception and circadian rhythm (GUCY2D, RORC), skin homeostasis, skin barrier function and phospholipid metabolism (GRB14, NPM2, SMIM2, MTMR4), and calcium regulation and neurotransmitter release (OBSCN, RIMS3) (Table 2; Figure 4B).
Pathway analysis resulted in a total of 24 canonical pathways that were predicted to be significantly activated using the upregulated genes in spring versus late summer within BB population (Table 3; Figure 5). These pathways were mostly represented in the general categories of extracellular matrix organization involving collagen metabolism, cellular stress and injury, wound healing, mitotic cell cycle and other intracellular signaling, cytokine signaling, and metabolism of proteins with 4 to 11 genes involved per pathway (Figure 5; Table 3). The canonical pathway that was the most significantly enriched was “Collagen Biosynthesis and Modifying Enzymes” with a z-score of 3.32 (FDR=1.58E-12). This pathway also had the greatest number of genes involved (11 genes) (Table 3). The upregulated genes in the summer samples, however, did not result in any significant pathway enrichment based on the cutoff criteria.
Table 3
| Ingenuity Canonical Pathways | FDR | Ratio | z-score | Gene ID |
|---|---|---|---|---|
| Extracellular Matrix Organization | ||||
| Collagen Biosynthesis and Modifying Enzymes | 1.58E-12 | 0.17 | 3.32 | ADAMTS2, COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1, P4HA1, SERPINH1 |
| Assembly of Collagen Fibrils and other Multimeric Structures | 8.51E-10 | 0.15 | 3.00 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1, LOXL2 |
| Collagen Chain Trimerization | 1.29E-09 | 0.19 | 2.83 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1 |
| Extracellular Matrix Organization | 2.45E-09 | 0.09 | 3.16 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, SPARC, TGFB3, VCAN |
| Collagen Degradation | 1.23E-08 | 0.13 | 2.83 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1 |
| Integrin Cell Surface Interactions | 1.58E-07 | 0.09 | 2.83 | COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1, FBN1 |
| Syndecan Interactions | 1.05E-04 | 0.15 | 2.00 | COL1A1, COL1A2, COL3A1, COL5A1 |
| Elastic Fibre Formation | 6.03E-04 | 0.09 | 2.00 | ELN, FBN1, LOXL2, TGFB3 |
| Cellular Stress and Injury | ||||
| GP6 Signaling Pathway | 2.57E-06 | 0.06 | 2.83 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1 |
| Wound Healing Signaling Pathway | 2.69E-05 | 0.04 | 3.00 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1, TGFB3 |
| Cell Cycle | ||||
| Mitotic Metaphase and Anaphase | 2.04E-05 | 0.04 | 3.00 | BIRC5, CDCA5, CENPT, FBXO5, LMNB1, PTTG1, TUBB2A, TUBB6, ZWINT |
| Mitotic Prometaphase | 5.62E-05 | 0.04 | 2.83 | BIRC5, CDCA5, CENPT, NCAPG, SMC2, TUBB2A, TUBB6, ZWINT |
| Cytokine Signaling | ||||
| IL-17A Signaling in Fibroblasts | 4.27E-05 | 0.07 | 2.45 | COL11A2, COL1A1, COL1A2, COL3A1, P4HA1, SERPINH1 |
| IL-4 Signaling | 1.10E-03 | 0.02 | 3.16 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1, COL6A1, COL6A3, COL8A1, CREB3L1, TGFB3 |
| Signal Transduction | ||||
| Signaling by MET | 3.98E-04 | 0.06 | 2.24 | COL11A2, COL1A1, COL1A2, COL3A1, COL5A1 |
| Signaling by PDGF | 1.48E-03 | 0.07 | 2.00 | COL3A1, COL5A1, COL6A1, COL6A3 |
| RHO GTPases Activate Formins | 3.89E-03 | 0.04 | 2.24 | BIRC5, CENPT, TUBB2A, TUBB6, ZWINT |
| Metabolism of Proteins | ||||
| O-linked Glycosylation | 1.48E-03 | 0.05 | 2.24 | ADAMTS2, ADAMTS8, ADAMTSL2, GALNT16, SPON1 |
| Regulation of Insulin-like Growth Factor (IGF) Transport and Uptake by IGFBPs | 2.34E-03 | 0.04 | 2.24 | FBN1, IGF1, IGFBP5, RCN1, VCAN |
| Post-translational Protein Phosphorylation | 1.20E-02 | 0.04 | 2.00 | FBN1, IGFBP5, RCN1, VCAN |
| Vesicle Mediated Transport | ||||
| Binding and Uptake of Ligands by Scavenger Receptors | 1.20E-02 | 0.04 | 2.00 | COL1A1, COL1A2, COL3A1, SPARC |
| Organismal Growth and Development | ||||
| Activin Inhibin Signaling Pathway | 1.95E-02 | 0.02 | 2.24 | COL11A2, COL1A1, COL1A2, COL3A1, TGFB3 |
| Intracellular and Second Messenger Signaling | ||||
| NAD Signaling Pathway | 2.95E-02 | 0.03 | 2.00 | H1-4, IGF1, SLC29A1, SLC7A5 |
| Cellular Growth, Proliferation and Development | ||||
| Ribonucleotide Reductase Signaling Pathway | 4.17E-02 | 0.02 | 2.00 | BIRC5, CREB3L1, E2F7, RRM2 |
The canonical pathways that are significantly enriched (FDR < 0.05) among upregulated genes in spring (May) versus summer (September) within Bristol Bay stock (see Figure 5).
The pathways are listed under prospective pathway categories shown in bold in each section. All the enriched pathways were predicted to be activated in BB-Spring whales (z-score ≥ 2). For each pathway, multiple test-controlled significance of overlap (FDR), overlap ratio statistics (Ratio), activity predictions (z-score), and the genes that are involved (Gene ID) are listed (see Supplementary Table S1 for gene names).
Figure 5
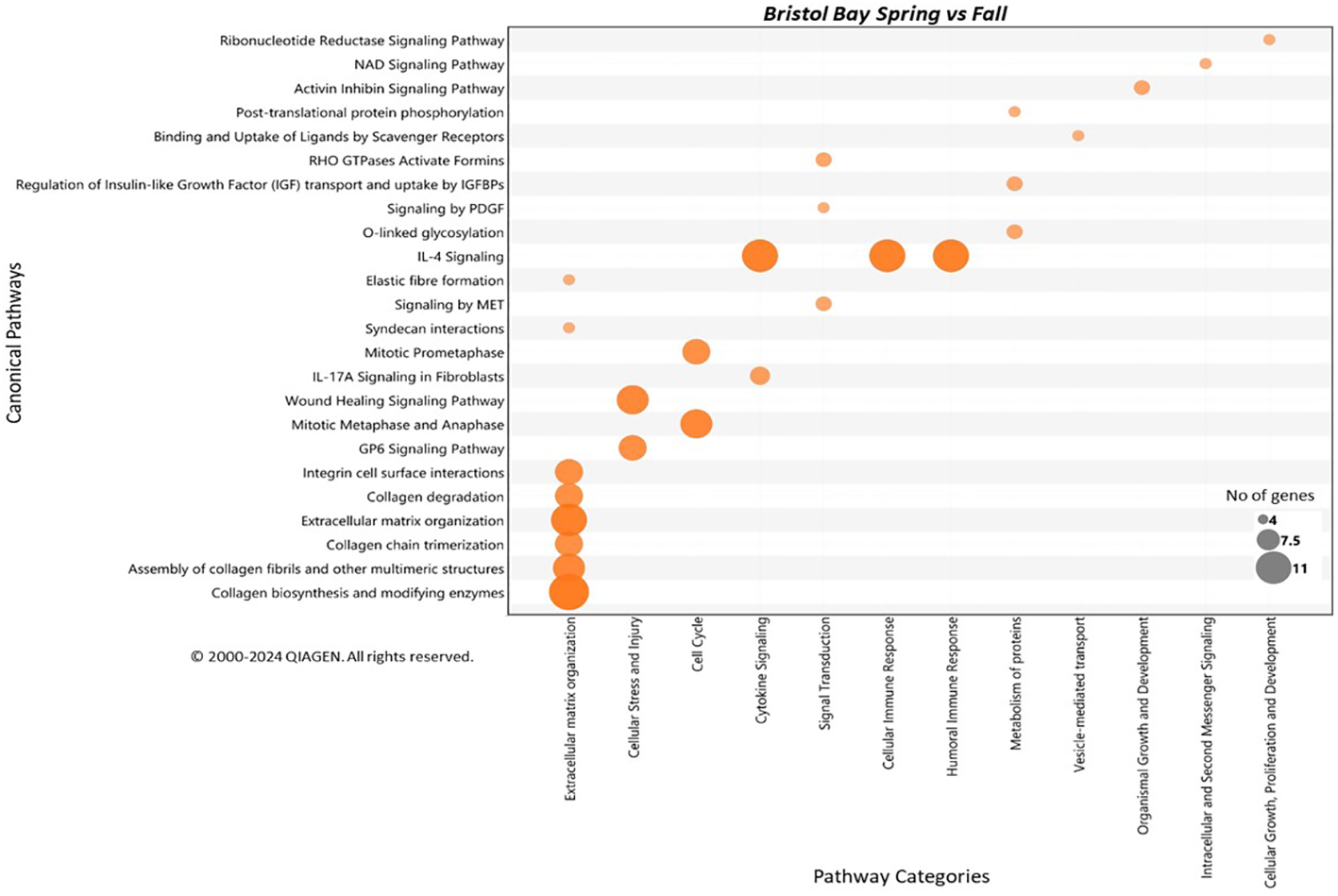
Canonical pathways that are significantly enriched (FDR < 0.05) among the upregulated genes in BB-Spring versus BB-Summer comparison. All the enriched pathways were also predicted to be activated in BB-Spring whales (z-score ≥ 2). The enriched canonical pathway names are shown on the y-axis, and the pathway categories are shown on the x-axis. The size of the circles corresponds to the number of significantly upregulated genes that overlap the pathway, as indicated in the legend. The color intensity represents the degree of z-score activity predictions with darker colors indicating larger z-scores and higher activation. The pathways are ordered based on their ascending significance values, with those at the bottom showing the most significant predictions.
3.2.2 Inter-population variability
Differential expression analysis performed between BB-Spring versus ECS samples yielded in 3.8% of the transcriptome (574 total) exhibiting significant changes between them (padj<0.05, log2FC of -4.01 to 4.93) (Figures 6A, B). Out of these, 489 were mapped, with 77.5% of the mapped genes (379 total) showing higher expression in BB-Spring, and 22.5% (110 total) showing higher expression in ECS (Supplementary Table S3). Differential expression analysis performed between BB-Summer vs ECS samples yielded in 6.2% of the transcriptome (938 total) being significantly regulated (padj<0.05, log2FC of -3.96 to 7.82) (Figures 6C, D). Out of these, 702 genes were mapped with 79.2% of the mapped genes (556 total) showing higher expression in BB-Summer samples, and the remaining 20.8% (146 total) showing higher expression in ECS samples (Supplementary Table S4).
Figure 6
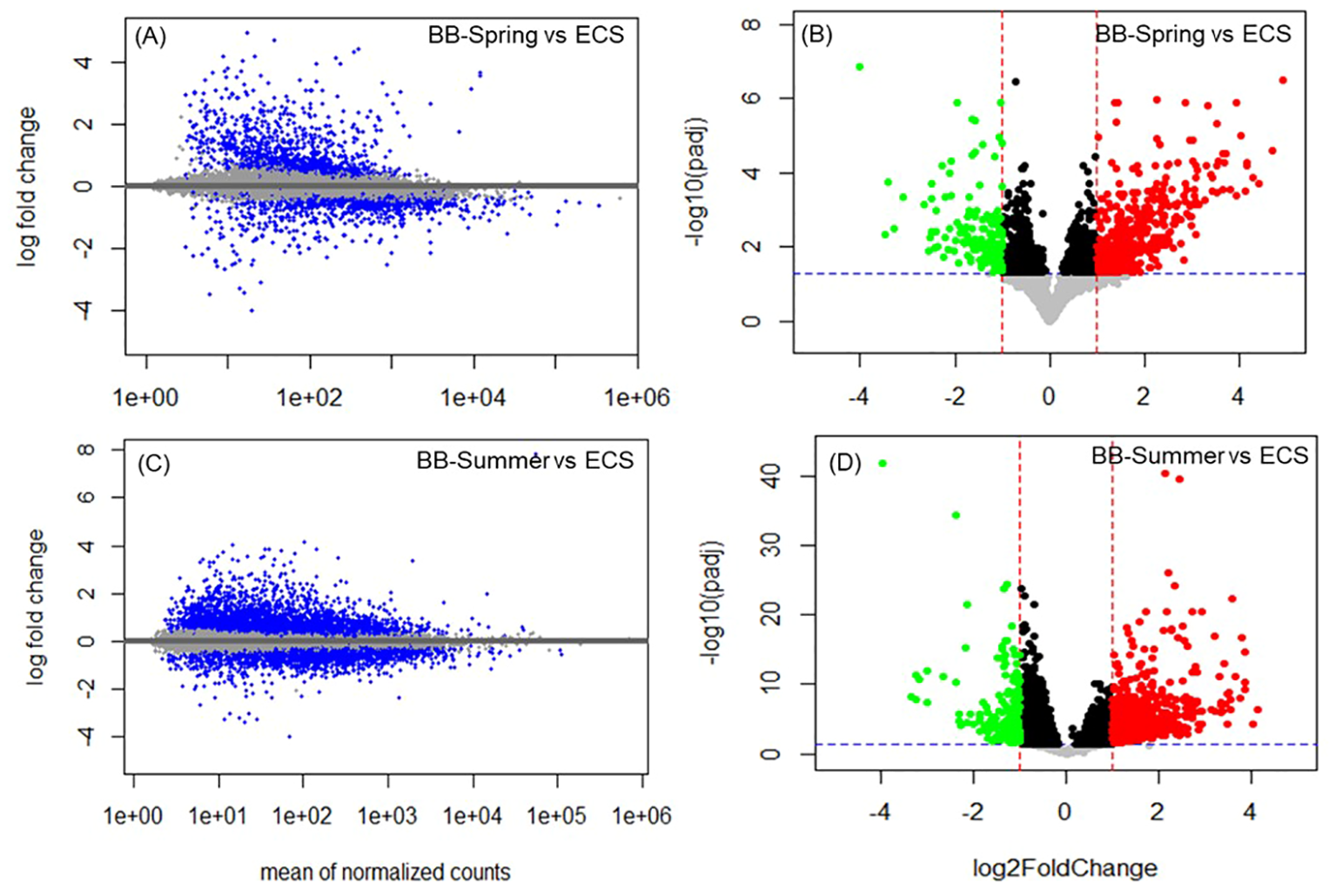
Differential expression analysis overview for population level comparisons of Bristol Bay (BB) and Eastern Chukchi Sea (ECS) belugas. Relative gene expression values for BB-Spring versus ECS comparison are shown on the top panels, and those of BB-Summer versus ECS comparison are shown on the bottom panels. (A, C) Bland-Altman (MA) plot displaying shrunken log2 fold change (y-axis) versus mean of normalized counts (x-axis). The horizontal (x = 0) line represents the mean expression value, and the gray dots represent the genes that did not pass the significance criteria. The blue dots above the line represent upregulation in spring vs summer, and the blue dots below the line represent downregulation in spring vs summer (i.e. upregulation in summer vs spring). (B, D) Volcano plots displaying the statistical significance (-log10(padj), y-axis) versus magnitude of change (log2FoldChange, x-axis). The horizontal dashed line represents the significance cutoff value of padj = 0.05, and the vertical dashed lines represent log2 fold change cutoff value of |1|. Black and gray dots represent the genes that did not pass the significance criteria. Significantly downregulated genes are shown in green on the left (log2FoldChange ≤ -1), and significantly upregulated genes are shown in red on the right (log2FoldChange ≥ 1).
The canonical pathway comparison of the core analysis results that are common to both BB-Spring and BB-Summer samples relative to ECS showed significant activity predictions for a total of 13 pathways (Figure 7; Table 4). While 12 of these pathways were significantly activated in BB with z-scores ranging from 2.45 to 5.20, only one pathway was significantly activated in ECS with a z-score of -2.45 with 1-acylglycerol-3-phosphate-O-acyltransferase 2 (AGPAT2) being the single shared gene in both comparisons (Table 4). The number of genes associated with these pathways ranged between 5 and 30, with the highest numbers observed for cellular immune response related “S100 Family Signaling Pathway” which also had the highest z-score (Table 4). The activity z-scores were higher, and significance values were much lower for the BB-Spring vs ECS than BB-Summer vs ECS comparison, especially for extracellular matrix organization related pathways.
Figure 7
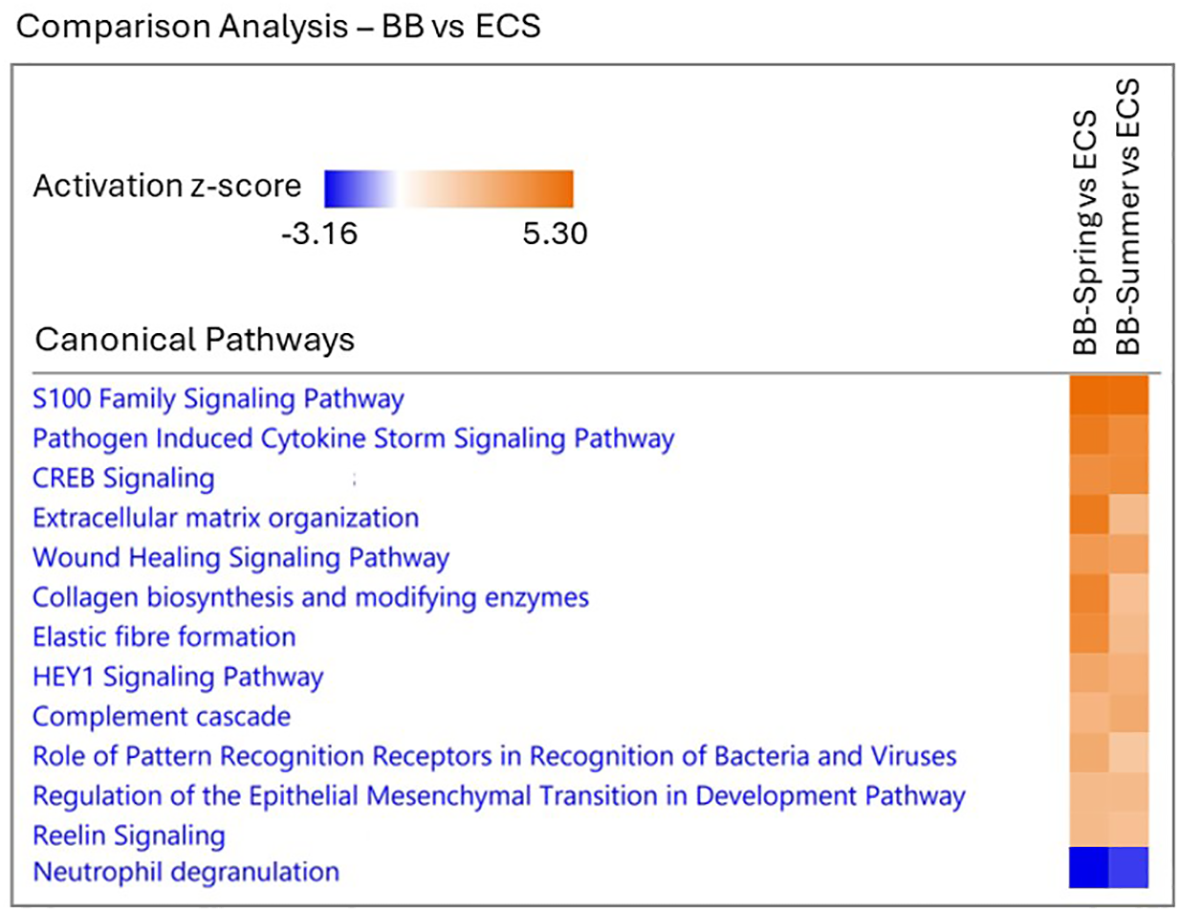
Heatmap of significantly (p < 0.05) enriched canonical pathways common to both comparisons between Bristol Bay (BB) and Eastern Chukchi Sea (ECS) belugas. The significantly enriched canonical pathways are listed for BB-Spring versus ECS (the first column of heatmap) and BB-Summer versus ECS (the second column of heatmap). The color scale represents the pathway activation z-score range across the comparisons. Orange color represents activation in BB and blue color represents inhibition in BB (i.e. activation in ECS). The darker the colors are, the higher the activation or inhibition in that pathway.
Table 4
| Ingenuity Canonical Pathways | BB-Spring vs ECS | BB-Summer vs ECS | Common Genes | Pathway Function in Skin | ||||
|---|---|---|---|---|---|---|---|---|
| p-value | z-score | #genes | p-value | z-score | #genes | |||
| Activated in BB | ||||||||
| S100 Family Signaling | 1.34E-04 | 5.20 | 27 | 4.16E-03 | 5.11 | 30 | WNT11, SMAD9, ESR1, PTGER2, GPR173 | Pro-inflammatory function, cytokine production, cell proliferation and differentiation, calcium homeostasis, antimicrobial activity |
| Pathogen Induced Cytokine Storm Signaling | 1.83E-07 | 4.69 | 22 | 6.54E-03 | 4.12 | 17 | COL12A1, COL21A1, CXCL14, CXCL9, IL21R, TLR5 | Activation of immune cells, inflammatory response |
| CREB Signaling | 9.41E-03 | 4.00 | 18 | 4.48E-02 | 4.15 | 21 | GPR173, PTGER2 | Regulation of gene expression, response to UV radiation, wound healing, inflammatory response |
| Extracellular Matrix Organization | 2.75E-18 | 4.69 | 22 | 4.15E-02 | 2.45 | 6 | DCN, FMOD, TNXB | Structural support, wound healing, barrier function, tissue homeostasis |
| Wound Healing Signaling | 7.69E-07 | 3.64 | 17 | 3.42E-02 | 3.32 | 11 | COL12A1, COL21A1 | Re-epithelialization, angiogenesis, collagen production, remodeling |
| Collagen Biosynthesis and Modifying Enzymes | 7.05E-19 | 4.36 | 19 | 2.04E-02 | 2.24 | 5 | COL12A1, COL21A1, PCOLCE2 | Maintenance of skin integrity, skin strength and elasticity functions |
| Elastic Fibre Formation | 1.53E-19 | 4.12 | 17 | 5.79E-04 | 2.45 | 6 | BMP4, FBN3, LOX, LTBP2 | Maintenance of skin integrity, skin elasticity and recoil functions |
| HEY1 Signaling | 3.15E-04 | 3.16 | 10 | 3.68E-02 | 2.83 | 8 | BMP4, PRKAB2, SMAD9 | Regulation of cell differentiation, wound healing, angiogenesis in skin |
| Complement Cascade | 5.81E-03 | 2.65 | 7 | 3.97E-03 | 3.00 | 9 | C2, CFH, SERPING1, C1QA, C1QB, C1QC | Clearance of pathogens and damaged cells, inflammatory response, tissue repair |
| Role of Pattern Recognition Receptors of Bacteria and Viruses | 4.27E-05 | 3.00 | 11 | 2.83E-02 | 2.00 | 8 | TLR5, C1QA, C1QB, C1QC | Antimicrobial response, inflammatory response, skin barrier protection, cytokine production, wound healing |
| Regulation of the Epithelial Mesenchymal Transition in Development | 2.97E-03 | 2.45 | 6 | 4.43E-03 | 2.45 | 7 | WNT11, LOX | Wound healing; skin development |
| Reelin Signaling | 4.76E-04 | 2.45 | 9 | 5.85E-03 | 2.24 | 9 | APOE, MAPK8IP2, MAPK8IP3 | Maintaining fluid balance and immune function; regulation of lymphatic vessel formation |
| Activated in ECS | ||||||||
| Neutrophil Degranulation | 9.40E-04 | -2.45 | 6 | 2.68E-02 | -3.16 | 10 | AGPAT2 | Increased inflammation and infection, production of reactive oxygen species |
Significantly activated canonical pathways (p-value < 0.05, z-score ≥ 2) common to both BB-Spring and BB-Summer versus ECS comparison (see Figure 7).
For each pathway, significance of overlap (p-value), activity predictions (z-score), the total number of genes that are involved in each pathway (#genes), the genes that are common to both comparisons, and putative pathway functions in skin tissue are listed (see Supplementary Table S1 for gene names).
The most significantly activated pathways in BB whales included “S100 Family Signaling Pathway” involving 27-30 differentially regulated genes that broadly took part in cellular immune and inflammatory response and cell proliferation (Table 4). The other immune response related pathways that were activated in BB whales were “Pathogen Induced Cytokine Storm Signaling”, “Complement Cascade” and “Role of Pattern Recognition Receptors in Bacteria and Viruses” taking part in antimicrobial and inflammatory responses, and skin barrier protection. The remaining activated pathways in BB whales were mostly related with skin structural maintenance, cell differentiation and growth, and tissue repair including “Extracellular Matrix Organization” “Collagen Biosynthesis and Modifying Enzymes”, “Elastic Fibre Formation” and “Wound Healing Signaling” (Table 4). The only significantly activated pathway in ECS whales was “Neutrophil Degranulation”, taking part in inflammation and infection involving 6-10 genes (Table 4).
4 Discussion
Skin is a relatively accessible and a very informative tissue matrix for investigation and health monitoring of marine mammals with potential to measure multiple parameters (i.e. hormones/proteins, contaminants, lipids, gene expression, age, sex) to maximize the information gained. Skin samples obtained from beluga whales utilized in this study proved to be a robust source of gene expression, in agreement with the previous studies (Ierardi et al., 2009; Lunardi et al., 2016; Neely et al., 2018; Unal et al., 2018). Skin closely reflects the characteristics of the environment the animal is living in, its physiological status (e.g., molting), or life stage with potential to identify population-level differences in their health and physiology.
In this study, differential expression analyses of skin transcriptomes generated from the two beluga stocks in Alaska (ECS and BB) displayed a clear separation based on PCA and heatmap analyses for both seasonal and population-level variability. The differences observed between the two stocks were consistent with the previous finding utilizing differentially regulated target genes obtained from the same individuals showing similar separation (Unal et al., 2018). However, there was also a large amount of variability among individuals within each stock, potentially due to differences in life history parameters such as age, interannual changes, phenotype and physiology, or due to differences in tissue handling such as time passed until tissue preservation. Additionally, the differences in sample sizes between the two stocks and biased sampling based on hunter’s preferences might have affected the results.
The inclusion of post-mortem skin samples following subsistence-hunts, particularly with the ECS animals being chased for hours before being harvested, can potentially raise some concerns. Even though preliminary analysis indicated that there were no significant gene expression differences between live-captured (n=3) versus subsistence-hunted (n=7) whales from the ECS stock, the results might change with the inclusion of more samples. However, this finding may not be surprising as both the harvested and live-captured whales were chased the same way until they were driven to a lagoon for the hunt or tagging. Furthermore, it has previously been shown that the effect of hormonal response to an acute stressor in cetacean skin could not be detected before 46 days (Bechshoft et al., 2020). These findings all together seem to indicate the usefulness of post-mortem skin collection to assess skin gene expression provided that the animal is considerably fresh which occurs in cold water conditions such as the Arctic.
4.1 Variability within stocks: seasonal effects
The lack of gene expression differences based on sex observed in this study is consistent with findings from Van Dolah et al. (2015), but not with Trego et al. (2019b). However, the differences likely resulted from variable cut-off criteria for significant expression. Even though Trego et al. (2019b) reports the presence of subtle but significant differences based on sex, only six genes had a minimum of 2-fold difference in expression levels between males and females, four of which were non-coding RNAs. Even though the current study did not find any significant differences between sexes in epidermal skin transcriptome in belugas, it remains to be determined if gene expression in other layers of skin might be affected by sex. Additionally, even though no significant changes were observed between the two age categories (adult and sub-adult) in this analysis, these categories are too broad to accurately reflect differences during maturation.
Significant seasonal variability observed among the beluga skin samples collected from the BB stock was consistent with other studies using cetacean skin (Van Dolah et al., 2015; Trego et al., 2019b). This study identified 3.6% of the transcriptome (541 genes) differentially expressed between spring and summer in beluga skin using the cutoff criteria of minimum log2FC of 1 (corresponding to fold change of 2) and padj<0.05. Previously reported values for a similar comparison in dolphin skin was 1.5% using the cutoff criteria of minimum fold change of 1.5 and padj<0.01 (Van Dolah et al., 2015). This reported value was significantly lower than the proportion of the differentially expressed genes identified in this study for belugas using the same criteria (5,4%). The proportion of the significantly expressed genes potentially reflects the presence of a greater response to seasonal changes between these two species. The thickness and structure of cetacean epidermis, as well as epidermal proliferation rates, vary considerably among species based on their aquatic adaptations to different ocean environments (Menon et al., 2022 and references therein). As an Arctic species, belugas are routinely exposed to a wide range of temperature and salinity changes during the process of migration associated with seasonal epidermal molt in spring/summer months. The molting process in cetaceans involves dramatic changes in skin involving proteolysis and inflammation in the earlier stages followed by protein synthesis and tissue regeneration, somewhat similar to wound healing (Keith, 2021; Su et al., 2022). On the other hand, as a temperate water species, dolphins are known to carry out continuous epidermal regeneration (Hicks et al., 1985). The more dramatic gene expression changes between spring and fall seasons observed in belugas versus dolphins could potentially be indicative of the molting process in belugas, in addition to other environmental or species-specific differences.
The effect of sea surface temperature on cetacean skin transcriptome has been previously documented for dolphins with 4279 genes (23.7% of the transcriptome) significantly differentially expressed with respect to mean sea surface temperature based on their geographic location (Trego et al., 2019b). The mean sea surface temperature in BB varied from 38.3°C in May to 51.9°C in September during the sampling period (https://seatemperature.info). Even though the temperature data was not included in this analysis, the seasonal gene expression changes observed could potentially be influenced by this exogenous variation, in addition to other oceanographic parameters. Additional analysis incorporating a variety of exogenous parameters may help tease apart the source of this seasonal variation.
4.1.1 Upregulation in spring
The upregulation and the pathway activation observed in spring samples were indicative of active re-organization of skin structural constituents in aging epidermal skin, along with increased energy production and inflammation, consistent with the initial stages of a seasonal molt.
Among the first group of genes with higher expression in spring, TUBB2A was the most significantly regulated gene, having a major role in microtubule formation for maintenance of cellular architecture and function in a wide range of cellular processes, including those vital for skin health and development. In the skin, it may also play a role in sensory nerve endings or other neuronal components (Uhlén et al., 2015). Centromere protein T (CENPT) also plays a fundamental role in cell division for accurate chromosome segregation during mitosis (Uhlén et al., 2015). Lamin B1 (LMNB1) is crucial in maintaining the structural integrity of the cell nucleus, and its expression levels impact cellular senescence and aging in skin tissue (Dreesen et al., 2013). The activation of these genes in spring is therefore indicative of increased cellular regeneration and cytoskeletal rearrangements (Hodge and Ridley, 2016).
The other group of genes that show higher expression in spring were mostly related with increased energy production through mitochondrial regulation. For example, MTFR2 is a mitochondrial fission regulator essential for maintaining mitochondrial health in aged keratinocytes in humans by breaking them apart (Sreedhar et al., 2020). Since mitochondrial dynamics is mainly manipulated by the balance between mitochondrial fission and fusion, increased expression in MTFR2 indicates elevated mitochondrial fragmentation. PHB1 is another gene with mitochondrial maintenance function by regulating mitochondrial respiration, and RRM2 is a part of the ribonucleotide reductase enzyme that takes part in DNA synthesis and repair showing increased expression associated with increased DNA damage response (Uhlén et al., 2015). The activation of these genes is potentially indicative of increased cellular energy demands to support drastic changes in the cellular architecture as expected from a molting skin.
4.1.2 Upregulation in summer
Even though the most consistently upregulated genes in summer samples (i.e. downregulated in spring) did not result in significant pathway enrichment based on the cutoff criteria, the genes with significantly higher expression in summer were mostly related with skin homeostasis, skin barrier, cellular immune system, circadian rhythm and sensory functions. Interestingly, the gene that showed the most significant increase in summer samples was tectorin alpha (TECTA), which has been well studied for its role in tectorial membrane that is critical for normal hearing in humans (Kim et al., 2019). Likewise, another sensory perception gene, retinal guanylate cyclase 2D (GUCY2D), also showed increased expression in summer. GUCY2D is mainly known for its role in visual phototransduction in humans, however, is also known to encode for an olfactory sensory protein in mice and rats (Fulle, 1995). Therefore, the function of the protein coded by GUCY2D gene varies across species and is currently unknown for cetaceans. Both GUCY2D and TECTA protein expression have been detected in human skin tissue (Uhlén et al., 2015). In addition, retinoic acid related orphan receptor gamma (RORC) gene is known to be expressed in all skin cell types, including epidermal keratinocytes and melanocytes. RORC plays a crucial role in circadian rhythms regulating several clock genes, while taking part in modulation of lipid/glucose metabolism (Fan et al., 2018). The increased expression of these sensory genes observed in summer samples in belugas might be indicative of increased levels of sensory stimulation, in addition to metabolic and/or immune challenges in their environment. Cetacean skin is a part of a complex somatosensory system allowing the perception of external stimuli unique to their marine environment through a variety of neuroanatomical innervations, however, the functions of this system have not yet been fully clarified (Eldridge et al., 2022; De Vreese et al., 2023). The cetacean skin has been shown to have exceptionally dense low threshold mechanosensory system innervation (Eldridge et al., 2022). It has also previously been suggested that skin receptors of toothed whales are extremely sensitive to vibrations, and dolphins can detect changes in hydrodynamic and hydrostatic pressure through their skin, including low frequency sound (Ridgway and Carder, 1990; Simmonds et al., 2004). However, the exact functions of these sensory genes in beluga skin warrants further investigation.
The genes related to the immune system that were activated in summer samples included myotubularin MTMR4 which regulates macrophage phagocytosis for immune control of infection and selective pathogen clearance of invading microorganisms and apoptotic bodies (Sheffield et al., 2019). In addition to its other roles, RORC is also an important regulator of the immune response as it is expressed in natural killer T cells in skin (Fan et al., 2018). Nucleophosmin NPM2 is shown to be uniquely expressed in pigment producing melanocytes in humans (Reemann et al., 2014), and pigmented parts of the beluga skin is known to contain large and well-developed melanocytes with abundant melanosomes (Menon et al., 2022). The genes related with metabolism included growth factor receptor protein GRB14, which is known to modulate insulin signaling which is crucial for maintaining glucose homeostasis, influencing cellular responses related to growth and metabolism. Increased GRB14 expression is shown to result in inhibition of insulin signaling and increased lipid storage in humans (Sun et al., 2022). SMIM2 is also predicted to be an integral component of the cell membrane, act as a receptor capable of mediating phospholipid uptake (Conrad et al., 2017). Overall, the results suggest an activation in immune response, growth and lipid/glucose metabolism in summer samples when compared to spring.
4.2 Variability between stocks
The skin samples included in this study were a subset of samples analyzed within the framework of another study reporting significantly lower cytokine gene expression in ECS whales in comparison to BB whales using real-time PCR (Unal et al., 2018), which corresponds to a higher expression in BB belugas. Results of our current study agree with the former study displaying increased expression in a number of cytokine genes in BB belugas. Cytokines are regularly produced by keratinocytes and other skin resident cells to maintain the barrier function of skin through cell proliferation and differentiation modulated by a complex network of signaling molecules (Hänel et al., 2013). The higher cytokine transcription observed in BB whale skin could in part be indicative of differences in skin condition and barrier function in between these two stocks potentially as a result of additional seasonal differences and/or molting. Moreover, the majority of the enriched canonical pathways predicted to be activated in the BB stock in relation to the ECS stock were related with cellular immune response, and neurotransmitter & other nervous system signaling. These results could be indicative of metabolically active skin in BB whales with increased exposure to pathogens, or higher level of immune system functioning in BB whales as explained in detail below.
Some of the differentially expressed genes identified were common to multiple canonical pathways indicating involvement in several biological processes. In skin, S100 proteins participate in innate and adaptive immune responses, and tissue development & repair (Halawi et al., 2014). Among the top regulated genes in this pathway highly conserved WNT11 plays an important role in fibrogenesis in skin including the last stages of cutaneous wound healing involving tissue remodeling and extracellular matrix maturation (Bukowska et al., 2021). Additionally, several G-protein coupled receptors including the prostaglandin receptor PTGDR2 are known to have regulatory roles in skin homeostasis controlling epithelial cell renewal and keratinocyte proliferation (Pedro et al., 2020), and the membrane-bound matrix metalloproteinase MMP24 is known to be involved in tissue remodeling (Verma and Hansch, 2007). The upregulation of these genes in BB whales might be indicative of increased tissue regeneration as well as increased immune functions. On the other hand, the only common gene that was upregulated in ECS versus both seasons of BB stock was AGPAT2 which is known to be involved in triglyceride synthesis and adipogenesis playing a crucial role in synthesis of phospholipids and development of adipocytes as the cells that store lipids for energy (Gale et al., 2006; Agarwal, 2012). The increased expression of AGPAT2 in ECS whales might be indicative of increased fat storage in these whales.
Among other significantly enriched pathways “Complement Cascade” and “Role of Pattern Recognition Receptors in Bacteria and Viruses”, which were predicted to be activated in BB whales, provide evidence of the active immune system in this stock. Even though the BB stock is considered to be stable, the population growth has slowed down or ceased based on the most recent estimates (Citta et al., 2019; Lowry et al., 2019). In addition, some concerns have been raised about increases in infectious disease outbreaks as a result of warming oceans and increased ship traffic (Gulland et al., 2022). Prevalence of alphaherpesvirus has also been reported in a high proportion of animals in this stock causing skin lesions (Nielsen et al., 2017). More recently, prevalence of marine-origin Brucella sp. exposure has also been reported in both BB and ECS stocks which has the potential to cause skin lesions among other clinical symptoms (Thompson et al., 2022). All these factors might have potentially contributed to the differences observed in these two stocks. Expression of immune response-related genes in both stocks may suggest that many animals are exposed to and/or experiencing skin infection. Therefore, measuring expression of immune markers in different populations could be a way to assess exposure to pathogens and animals’ immune capacity in responding to those pathogens.
The differences in gene expression between beluga stocks might also have been influenced by different levels of contaminant exposure (i.e. POP/PFAS) between the populations. Belugas, being top predators, are known to bioaccumulate contaminants which can potentially have adverse effects on the development of immune, nervous and reproductive systems as well as causing cancer (Wilson et al., 2005; Lair et al., 2016). Cook Inlet is located by the increasingly industrialized waters of Anchorage as Alaska’s most populated and fastest-growing city. Being geographically close to BB, Cook Inlet belugas generally have higher levels of contaminants compared to Bristol Bay belugas. Recent studies have shown that Cook Inlet belugas are exposed to various pollutants, including persistent organic pollutants (POPs), per- and polyfluoroalkyl substances (PFAS), and mercury (Burek-Huntington et al., 2022). Even though blubber is known to serve as storage organ for most of the listed contaminants, an unfortunate consequence of utilization of blubber as an energy source is the mobilization of lipophilic contaminants (i.e., POPs) closely associated with these lipid reserves into the blood and surrounding internal tissues (Yordy et al., 2010; Hoguet et al., 2013). Moreover, associations of POPs in blubber with thyroid- and steroid-related gene expression in skin has previously been identified in St Lawrence belugas (Simond et al., 2019). Gene expression in skin has also been previously investigated for the role of vitamin D3 pathway in innate immunity including proliferation, differentiation, apoptosis, immunomodulation, stress response and antimicrobicity, similar to what has been reported in humans and other terrestrial mammals (Ellis et al., 2009; Mancia, 2018 and the references therein). These studies provide evidence for the utilization of gene expression in skin for assessing the health of cetaceans.
Even though it is not possible to identify the exact causes for the differences observed between ECS and BB belugas, the increased expression of genes associated with the functions of cellular movement, cellular growth and proliferation and immune system in BB whales could potentially be indicative of the presence of increased number of pathogens or differences in environmental conditions, and/or increased exposure to these changes due to a thinner skin. A decrease in skin thickness in fall has previously been documented in belugas based on observations of Inuit hunters. This traditional ecological knowledge indicated obvious physiological changes occurring in beluga skin during annual molt, reporting that the outer layer of dorsal body surface is thick and yellow in spring migration, of intermediate thickness in summer when molting occurs, and thin and white in fall after the molt is completed (St. Aubin et al., 1990). ECS belugas were sampled in late June to early July when they were just arriving at the inlet in Point Lay, AK most likely still going through their seasonal molt with dead epidermal skin creating a barrier with the newly regenerating skin underneath. On the other hand, BB belugas were sampled in August to September, when they had already completed their molt two or three months ago. It is also likely that different environmental conditions might have resulted in different cutaneous structure of the skin between these populations. Cutaneous lesions in cetaceans including belugas have previously been suggested as indicators of ecosystem status (Mouton and Botha, 2012), however, an in-depth investigation of differences in structural components and lesions is needed before drawing a solid conclusion.
4.3 Limitations
Major limitations of this study include small sample sizes especially for ECS stock, biased sampling of the hunted whales as the hunters tend to primarily focus on larger (and potentially older) animals, and the potential effects of interannual and seasonal changes. Even though two age categories (adult and sub-adult) were accounted for in this analysis, they are too broad to accurately reflect differences during maturation. The results might have been affected not only by the small sample sizes and age differences, but also by the time period of chasing and harvesting or tagging of the ECS belugas by hunters, and the differences in molting status between the two stocks. Inclusion of additional samples from both populations representing different age and size classes, and inclusion of samples collected from different times of the year from the same population to investigate seasonal differences will be necessary to characterize gene expression changes more comprehensively in these stocks.
Another limitation for this study is the low availability of live-captured skin samples from ECS stock, therefore, the limitation of including skin samples collected post-mortem from subsistence-hunted whales for ECS stock is recognized. Even though it appears that the gene expression changes in skin due to death within this time period is unlikely, it is not a definitive conclusion without experimental validation. Additionally, the results of the pathway analyses should be interpreted with caution as these are not causations but rather predictions based on statistically significant gene expression differences between the two stocks. Moreover, pathway databases are known to be mostly human/rodent-biased and many genes have multiple, different functions, some of which may not be well-represented in pathway databases (e.g., see comment about AGPAT2). Even though most differences observed can potentially be explained by differences in molting, this is also not a definitive conclusion as the molt status for the majority of the whales had not been determined. The differences between these two stocks could also be explained by differences in environmental parameters between these two regions, exposure to contaminants and/or pathogens, prey preferences, resulting in differences in physiology and metabolism represented in skin morphology.
Even with potential limitations, the comparative skin transcriptomics technique utilized here provides physiological data on differences in skin function between these two stocks. Importantly, this methodology can be incorporated in future analyses with larger datasets involving the endangered Cook Inlet stock to reveal the key gene activity differences, and to provide health data to inform the conservation and recovery efforts of Cook Inlet belugas.
5 Conclusions
This study compared transcriptomic profiles of ECS versus BB beluga stocks in Alaska investigating canonical pathways and diseases & functions that showed significant differences based on their gene expression profiles. In total, BB belugas were predicted to have higher cellular immune response along with higher skin structural maintenance functions. On the other hand, ECS belugas were predicted to have higher levels of inflammation most likely due to the differences in molting status. Top differentially regulated genes associated with these pathways and functions that showed the most consistent differences were identified for future development of hypotheses and studies. The utilization of in-depth functional analysis of skin transcriptomes as shown in this study has the potential to provide valuable insights on the physiology of beluga whales, especially if combined with better controlled samples (i.e. samples from individuals of known age, sex, and molt status) along with other types of analysis that can readily be applied to skin samples such as proteomics, metabolomics, and lipidomics, and other biological information such as age, reproductive status, diet and contaminant levels. Overall, the methodology and data analysis steps outlined in this study have the potential to identify the biologically relevant gene expression differences within or between beluga populations in the wild, which can also be applied to other marine mammal species to infer population level differences, and/or to understand gene expression changes at the individual level in relation to phenotypic differences. Moreover, if meaningful differences are found to persist when the aforementioned sample biases can be controlled, then the results can potentially be utilized to inform management decisions to enhance conservation efforts for recovery of the beluga stocks at risk.
Statements
Data availability statement
The original contributions presented in the study are included in the article and the Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by all applicable international, national, and/or institutional guidelines for the care and use of animals. Sampling of belugas in Bristol Bay, AK, was conducted under NMFS Scientific Research Permit #782-1719 (for 2008) and #14245 (for 2012–2014), and IACUC #AFSC/NWFC 2012-1 (for 2012–2014). Sampling of the Eastern Chukchi Sea whales was conducted under NMFS Scientific Research Permit #17350 issued to the Department of Wildlife Management, North Slope Borough, AK. The research samples admitted at Mystic Aquarium between 2008 and 2012 were under NOAA/NMFS Permit #42-1908; samples admitted from 2013 and 2014 were under permit #17298. Live-capture methods were approved by ADF&G Animal Care and Use Committee protocols #2012-20, #2013-20, #2014-03, and #2017-27. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
EU: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. VS: Formal analysis, Methodology, Software, Visualization, Writing – review & editing, Data curation. RS: Investigation, Resources, Writing – review & editing. CG: Investigation, Resources, Writing – review & editing. TR: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the North Pacific Research Board (NPRB Award No: NA21NMF4720289, Project No: 2209) and National Oceanic and Atmospheric Administration (NOAA Contract No: 1305M320PNFFS0401).
Acknowledgments
The authors thank the Department of Wildlife Management, North Slope Borough, Utqiaġvik AK, the Point Lay field team and the community of Point Lay, AK. The authors also thank the Bristol Bay field team along with the Bristol Bay Marine Mammal Council and Bristol Bay Native Association. The Alaska Beluga Whale Committee encouraged health studies on belugas. This constitutes scientific contribution #360 from the Sea Research Foundation, Inc.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1282210/full#supplementary-material
References
1
Agarwal A. K. (2012). Lysophospholipid acyltransferases: 1-acylglycerol-3-phosphate O-acyltransferases. From discovery to disease. Curr. Opin. Lipidol.23, 290–302. doi: 10.1097/MOL.0b013e328354fcf4
2
Anders S. Pyl P. T. Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169. doi: 10.1093/bioinformatics/btu638
3
Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed September 20, 2024).
4
Bechshoft T. Wright A. J. Styrishave B. Houser D. (2020). Measuring and validating concentrations of steroid hormones in the skin of bottlenose dolphins (Tursiops truncatus). Conserv. Physiol.8, coaa32. doi: 10.1093/conphys/coaa032 (accessed September 19, 2024).
5
Benjamini Y. Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B Stat. Method.57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
6
Boily P. (1995). Theoretical heat flux in water and habitat selection of phocid seals and beluga whales during the annual molt. J. Theor. Biol.172, 233–244. doi: 10.1006/jtbi.1995.0020
7
Bolger A. M. Lohse M. Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120. doi: 10.1093/bioinformatics/btu170
8
Breton-Honeyman K. Huntington H. P. Basterfield M. Campbell K. Dicker J. Gray T. et al . (2021). Beluga whale stewardship and collaborative research practices among Indigenous peoples in the Arctic. Polar Res.40, 5522. doi: 10.33265/polar.v40.5522
9
Buckman A. H. Veldhoen N. Ellis G. Ford J. K. Helbing C. C. Ross P. S. (2011). PCB-associated changes in mRNA expression in killer whales (Orcinus orca) from the NE Pacific Ocean. Environ. Sci. Technol.45, 10194–101202. doi: 10.1021/es201541j
10
Bukowska J. Walendzik K. Kopcewicz M. Cierniak P. Gawronska-Kozak B. (2021). Wnt signaling and the transcription factor Foxn1 contribute to cutaneous wound repair in mice. Connect. Tissue Res.62, 238–248. doi: 10.1080/03008207.2019.1688314
11
Burek K. A. Gulland F. M. D. O’Hara T. M. (2008). Effects of climate change in Arctic marine mammal health. Ecol. Appl.18, S126–S134. doi: 10.1890/06-0553.1
12
Burek-Huntington K. Shelden K. Goetz K. Mahoney B. Vos D. Reiner J. et al . (2022). Life History, contaminant and histopathologic assessment of beluga whales, Delphinapterus leucas, harvested for subsistence in Cook Inlet, Alaska 1989-2005 (NOAA Tech. Memo). U.S. Dep. Commer. Available at: https://tsapps.nist.gov/publication/get_pdf.cfm?pub_id=935389. NMFS-AFSC-440.
13
Citta J. J. Frost K. J. Quakenbush L. (2019). Aerial surveys of Bristol Bay beluga whales, Delphinapterus leucas, in 2016. Mar. Fish. Rev.81, 98–104. Available at: https://spo.nmfs.noaa.gov/sites/default/files/pdf-content/mfr813-45_0.pdf.
14
Citta J. J. Quakenbush L. T. Frost K. J. Lowry L. Hobbs R. C. Aderman H. (2016). Movements of beluga whales (Delphinapterus leucas) in Bristol Bay, Alaska. Mar. Mam. Sci.32, 1272–1298. doi: 10.1111/mms.12337
15
Conrad K. S. Cheng T. W. Ysselstein D. Heybrock S. Hoth L. R. Chrunyk B. A. et al . (2017). Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies. Nat. Commun.8, 1908. doi: 10.1038/s41467-017-02044-8
16
De Vreese S. Orekhova K. Morell M. Gerussi T. Graïc J. (2023). Neuroanatomy of the cetacean sensory systems. Animals14, 66. doi: 10.3390/ani14010066
17
Deyneko I. V. Mustafaev O. N. Tyurin A. A. Zhukova K. V. Varzari A. Goldenkova-Pavlova I. (2022). Modeling and cleaning RNA-seq data significantly improve detection of differentially expressed genes. BMC Bioinform.23, 488. doi: 10.1186/s12859-022-05023-z
18
Dreesen O. Chojnowski A. Ong P. F. Zhao T. Y. Common J. E. Lunny D. et al . (2013). Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol.200 (5), 605–617. doi: 10.1083/jcb.201206121
19
Ehrlich F. Fischer H. Langbein L. Praetzel-Wunder S. Ebner B. Figlak K. et al . (2019). Differential evolution of the epidermal keratin cytoskeleton in terrestrial and aquatic mammals. Mol. Biol. Evol.36, 328–340. doi: 10.1093/molbev/msy214
20
Eldridge S. A. Mortazavi F. Rice F. L. Ketten D. R. Wiley D. N. Lyman E. et al . (2022). Specializations of somatosensory innervation in the skin of humpback whales (Megaptera novaeangliae). Anat. Rec.305, 514–534. doi: 10.1002/ar.24856
21
Elias P. M. (1988). Structure and function of the stratum corneum permeability barrier. Drug Dev. Res.13, 97–105. doi: 10.1002/ddr.430130203
22
Elias P. M. Menon G. K. Grayson S. Brown B. E. Rehfeld S. J. (1987). Avian sebokeratocytes and marine mammal lipokeratinocytes: structural, lipid biochemical, and functional considerations. Am. J. Anat.180, 161–177. doi: 10.1002/aja.1001800206
23
Ellis B. C. Gattoni-Celli S. Mancia A. Kindy M. S. (2009). The vitamin D3 transcriptomic response in skin cells derived from the Atlantic bottlenose dolphin. Dev. Comp. Immunol.33, 901–912. doi: 10.1016/j.dci.2009.02.008
24
Espregueira Themudo G. Alves L. Q. MaChado A. M. Lopes-Marques M. da Fonseca R. R. Fonseca M. et al . (2020). Losing genes: the evolutionary remodeling of cetacea skin. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.592375
25
Ewels P. Magnusson M. Lundin S. Käller M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics32, 3047–3048. doi: 10.1093/bioinformatics/btw354
26
Fan J. Lv Z. Yang G. Liao T. T. Xu J. Wu F. et al . (2018). Retinoic acid receptor-related orphan receptors: Critical roles in tumorigenesis. Front. Immunol.31. doi: 10.3389/fimmu.2018.01187
27
Fossi M. C. Marsili L. (2011). “Multi-Trial Ecotoxicological Diagnostic Tool in Cetacean Skin Biopsies,” in Skin Biopsy – Perspectives, U. Khopkar (Rajeka, Croatia: InTech), 317–336. doi: 10.5772/19947
28
Frost K. Lowry L. (1990). Distribution, abundance, and movements of beluga whales, Dephinapterus leucas, in coastal waters of western Alaska. Can. J. Fish. Aquat. Sci.224, 39–57. Available at: https://waves-vagues.dfo-mpo.gc.ca/library-bibliotheque/119072.pdf.
29
Frost K. Lowry L. Carroll G. (1993). Beluga whale and spotted seal use of a coastal lagoon system in the Northeastern Chukchi Sea. Arctic. 46, 8–16. doi: 10.14430/arctic1316
30
Fulle H. J. (1995). A receptor guanylyl cyclase expressed specifically in olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A.92, 3571–3575. doi: 10.1073/pnas.92.8.3571
31
Gale S. E. Frolov A. Han X. Bickel P. E. Cao L. Bowcock A. et al . (2006). A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem.281, 11082–11089. doi: 10.1074/jbc.M509612200
32
Givens G. Ferguson M. Clarke J. Willoughby A. Brower A. Suydam R. (2020). Abundance of the eastern Chukchi sea stock of beluga whales, 2012–17. Arctic. 73, 485–497. doi: 10.14430/arctic71592
33
Goertz C. Burek-Huntington K. Royer K. Quakenbush L. Clauss T. Hobbs R. et al . (2019). Comparing progesterone in blubber and serum to assess pregnancy in wild beluga whales (Delphinapterus leucas). Conserv. Physiol.7, coz071. doi: 10.1093/conphys/coz071
34
Gulland F. M. D. Baker J. D. Howe M. LaBrecque E. Leach L. Moore S. E. et al . (2022). A review of climate change effects on marine mammals in United States waters: Past predictions, observed impacts, current research and conservation imperatives. Clim. Change Ecol.3, 100054. doi: 10.1016/j.ecochg.2022.100054
35
Halawi A. Abbas O. Mahalingam M. (2014). S100 proteins and the skin: a review. J. Eur. Acad. Dermatol. Venereol.28, 405–414. doi: 10.1111/jdv.12237
36
Hänel K. H. Cornelissen C. Lüscher B. Baron J. M. (2013). Cytokines and the skin barrier. Int. J. Mol. Sci.14, 6720–6745. doi: 10.3390/ijms14046720
37
Hicks B. D. St. Aubin D. J. Geraci J. R. Brown W. R. (1985). Epidermal growth in the bottlenose dolphin, tursiops truncatus. J. Invest. Dermatol.85, 60–63. doi: 10.1111/1523-1747.ep12275348
38
Hodge R. G. Ridley A. J. (2016). Regulating Rho GTPases and their regulators. Nature17, 496–510. doi: 10.1038/nrm.2016.67
39
Hoguet J. Keller J. Reiner J. Kucklick J. Bryan C. Moors A. et al . (2013). Spatial and temporal trends of persistent organic pollutants and mercury in beluga whales (Delphinapterus leucas) from Alaska. Sci. Total Environ.449, 285–294. doi: 10.1016/j.scitotenv.2013.01.072
40
Ierardi J. L. Mancia A. McMillan J. Lundqvist M. L. Romano T. A. Wise J. P. et al . (2009). Sampling the skin transcriptome of the North Atlantic Right Whale. Comp. Biochem. Physiol. Part D. Genomics Proteomics4, 154–158. doi: 10.1016/j.cbd.2009.01.004
41
Jones S. J. M. Taylor G. A. Chan S. Warren R. L. Hammond S. A. Bilobram S. et al . (2017). The genome of the beluga whale (Delphinapterus leucas). Genes8, 378. doi: 10.3390/genes8120378
42
Keith A. (2021). Molecular Responses to Catastrophic Molting in a Wild Marine Mammal. Master’s Thesis. Stockton (CA), University of the Pacific.
43
Kight C. R. Swaddle J. P. (2011). How and why environmental noise impacts animals: An integrative, mechanistic review. Ecol. Lett.14, 1052–1061. doi: 10.1111/j.1461-0248.2011.01664.x
44
Kim D. Kim J. A. Park J. Niazi A. Almishaal A. Park S. (2019). The release of surface-anchored α-tectorin, an apical extracellular matrix protein, mediates tectorial membrane organization. Sci. Adv.5, eaay6300. doi: 10.1126/sciadv.aay6300
45
Kim D. Langmead B. Salzberg S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods12, 357–360. doi: 10.1038/nmeth.3317
46
Krämer A. Green J. Pollard J. Jr. Tugendreich S. (2013). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics30, 523–530. doi: 10.1093/bioinformatics/btt703
47
Lair S. Measures L. N. Martineau D. (2016). Pathologic findings and trends in mortality in the beluga (Delphinapterus leucas) population of the St Lawrence estuary, Quebec, Canada, from 1983 to 2012. Vet. Pathol.53, 22–36. doi: 10.1177/0300985815604726
48
Li H. Handsaker B. Wysoker A. Fennell T. Ruan J. Homer N. et al . (2009). The sequence alignment/map format and SAMtools. Bioinformatics25, 2978–2079. doi: 10.1093/bioinformatics/btp352
49
Love M. I. Huber W. Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550. doi: 10.1186/s13059-014-0550-8
50
Lowry L. F. Citta J. J. O’Corry-Crowe G. Quakenbush L. T. Frost K. J. Suydam R. et al . (2019). Distribution, abundance, harvest, and status of western Alaska beluga whale, delphinapterus leucas, stocks. Mar. Fish. Rev.81, 54–71. doi: 10.7755/MFR.81.3–4.2
51
Lunardi D. Abelli L. Panti C. Marsili L. Fossi M. C. Mancia A. (2016). Transcriptomic analysis of bottlenose dolphin (Tursiops truncatus) skin biopsies to assess the effects of emerging contaminants. Mar. Environ. Res.114, 74–79. doi: 10.1016/j.marenvres.2016.01.002
52
Mancia A. (2018). “New Technologies for Monitoring Marine Mammal Health,” in Marine Mammal Ecotoxicology. Eds. FossiM. C.PantiC. (Academic Press), Marine Mammal Ecotoxicology291–320. doi: 10.1016/B978-0-12-812144-3.00011-5
53
Menon G. K. Elias P. M. Wakefield J. S. Crumrine D. (2022). Cetacean epidermal specialization: A review. Anat. Histol. Embryol.51, 563–575. doi: 10.1111/ahe.12829
54
Moore S. E. Huntington H. P. (2008). Arctic marine mammals and climate change: Impacts and resilience. Ecol. Appl.18, S157–S165. doi: 10.1890/06-0571.1
55
Mouton M. Botha A. (2012). “Cutaneous Lesions in Cetaceans: An Indicator of Ecosystem Status?,” in New Approaches to the Study of Marine Mammals. Eds. RomeroA.KeithE. O. (IntechOpen, Rijeka). doi: 10.5772/54432
56
Neely M. G. Morey J. S. Anderson P. Balmer B. C. Ylitalo G. M. Zolman E. S. et al . (2018). Skin Transcriptomes of common bottlenose dolphins (Tursiops truncatus) from the northern Gulf of Mexico and southeastern U.S. Atlantic coasts.38, 45–58. doi: 10.1016/j.margen.2017.08.002
57
Nielsen O. Burek-Huntington K. Loseto L. L. Morell M. Romero C. H. (2017). Alphaherpesvirus: isolation, identification, partial characterisation, associated pathologic findings, and epidemiology in beluga whales (Delphinapterus leucas) in Alaska and Arctic Canada. Artic Sci.4, 338–357. doi: 10.1139/as-2017-0043
58
Norman S. A. Goertz C. E. Burek K. A. Quakenbush L. T. Cornick L. A. Romano T. A. et al . (2012). Seasonal hematology and serum chemistry of wild beluga whales (Delphinapterus leucas) in Bristol Bay, Alaska, USA. J. Wildl. Dis.48, 21–32. doi: 10.7589/0090-3558-48.1.21
59
O’Corry-Crowe G. M. (2018). “Beluga Whale: Delphinapterus leucas,” in Encyclopedia of Marine Mammals, 3rd ed., vol. 93-96 . Eds. WürsigB.ThewissenJ. G. M.KovacsK. M. (Academic Press). doi: 10.1016/B978-0-12-804327-1.00065-0
60
O’Corry-Crowe G. Ferrer T. Citta J. Suydam R. Quakenbush L. Burns J. et al . (2021). Genetic history and stock identity of beluga whales in Kotzebue Sound. Polar Res.40, 7623. doi: 10.33265/polar.v40.7623
61
O’Corry-Crowe G. M. Suydam R. S. Rosenberg A. Frost K. J. Dizon A. E. (1997). Phylogeography, population structure and dispersal patterns of the beluga whale Delphinapterus leucas in the western Nearctic revealed by mitochondrial DNA. Mol. Ecol.6, 955–970. doi: 10.1046/j.1365-294X.1997.00267.x
62
Panti C. Spinsanti G. Marsili L. Casini S. Frati F. Fossi M. C. (2011). Ecotoxicological diagnosis of striped dolphin (Stenella coeruleoalba) from the Mediterranean basin by skin biopsy and gene expression approach. Ecotoxicology20, 1791–1800. doi: 10.1007/s10646-011-0713-2
63
Pedro M. P. Lund K. Iglesias-Bartolome R. (2020). The landscape of GPCR signaling in the regulation of epidermal stem cell fate and skin homeostasis. Stem Cells38, 1520–1531. doi: 10.1002/stem.3273
64
Pitman R. L. Durban J. W. Joyce T. Fearnbach H. Panigada S. Lauriano G. (2020). Skin in the game: Epidermal molt as a driver of long-distance migration in whales. Mar. Mam. Sci.36, 565–594. doi: 10.1111/mms.12661
65
Quakenbush L. Suydam R. Bryan A. L. Lowry L. Frost K. Mahoney B. (2015). Diet of beluga whales, Delphinapterus leucas, in Alaska from stomach contents, March-November. Mar. Fish. Rev.77, 70–84. doi: 10.7755/MFR.77.1.7
66
Reemann P. Reimann E. Ilmjärv S. Porosaar O. Silm H. Jaks V. et al . (2014). Melanocytes in the skin–comparative whole transcriptome analysis of main skin cell types. PloS One9, e0173792. doi: 10.1371/journal.pone.0115717
67
Ridgway S. H. Carder D. A. (1990). “Tactile Sensitivity, Somatosensory Responses, Skin Vibrations, and the Skin Surface Ridges of the Bottle-Nose Dolphin, Tursiops Truncatus,” in Sensory Abilities of Cetaceans, vol. 196 . Eds. ThomasJ. A.KasteleinR. A. (Springer, Boston, MA). doi: 10.1007/978-1-4899-0858-2_9
68
Sheffield D. A. Jepsen M. R. Feeney S. J. Bertucci M. C. Sriratana A. Naughtin M. J. et al . (2019). The myotubularin MTMR4 regulates phagosomal phosphatidylinositol 3-phosphate turnover and phagocytosis. J. Biol. Chem.294, 16684–16697. doi: 10.1074/jbc.RA119.009133
69
Simmonds M. Dolman S. Weilgart L. C. S. Whale and Dolphin (2004). Oceans of Noise: A WDCS Science Report (Whale and Dolphin Conservation UK, WDC Publications and Reports). Available at: https://uk.whales.org/whales-dolphins/wdc-publications-and-reports/.
70
Simmonds M. P. Eliott W. J. (2009). Climate change and cetaceans: concerns and recent developments. J. Mar. Biol. Assoc. U. K89, 203–210. doi: 10.1017/S0025315408003196
71
Simond A. E. Houde M. Lesage V. Michaud R. Zbinden D. Verreault J. (2019). Associations between organohalogen exposure and thyroid- and steroid-related gene responses in St. Lawrence Estuary belugas and minke whales. Mar. pollut. Bull.145, 174–184. doi: 10.1016/j.marpolbul.2019.05.029
72
Sreedhar A. Aguilera-Aguirre L. Singh K. K. (2020). Mitochondria in skin health, aging, and disease. Cell Death Dis.11, 444. doi: 10.1038/s41419-020-2649-z
73
St. Aubin D. J. Smith T. G. Geraci J. R. (1990). Seasonal epidermal molt in beluga whales, Delphinapterus leucas. Can. J. Zool.68, 359–367. doi: 10.1139/z90-051
74
Su C. Hughes M. W. Liu T. Chuong C. Wang H. Yang W. (2022). Defining Wound Healing Progression in Cetacean Skin: Characteristics of Full-Thickness Wound Healing in Fraser’s dolphins (Lagenodelphis hosei). Anim. (Basel)12, 537. doi: 10.3390/ani12050537
75
Sun C. Förster F. Gutsmann B. Moulla Y. Stroh C. Dietrich A. et al . (2022). Metabolic effects of the waist-to-hip ratio associated locus GRB14/COBLL1 are related to GRB14 expression in adipose tissue. Int. J. Mol. Sci.23, 8558. doi: 10.3390/ijms23158558
76
Suydam R. Lowry L. Frost K. (2005). Distribution and movements of beluga whales from the eastern Chukchi sea stock during summer and early autumn. OCS Study MMS 2005-035 Final Report, 48. Available at: https://www.north-slope.org/wp-content/uploads/2022/03/Suydam_et_al_beluga_tracks_CMI_Final_Report_2005-1-1.pdf.
77
Thompson L. A. Goertz C. E. C. Quakenbush L. T. Burek Huntington K. Suydam R. S. Stimmelmayr R. et al . (2022). Serological detection of marine origin brucella exposure in two Alaska beluga stocks. Anim. (Basel)12, 1932. doi: 10.3390/ani12151932
78
Trego M. L. Hoh E. Whitehead A. Kellar N. M. Lauf M. Datuin D. O. et al . (2019a). Contaminant exposure linked to cellular and endocrine biomarkers in Southern California bottlenose dolphins. Environ. Sci. Technol.53, 3811–3822. doi: 10.1021/acs.est.8b06487
79
Trego M. L. Whitehead A. Kellar N. M. Lauf M. Lewison R. L. (2019b). Tracking transcriptomic responses to endogenous and exogenous variation in cetaceans in the Southern California Bight. Conserv. Physiol.7, coz018. doi: 10.1093/conphys/coz018
80
Uhlén M. Fagerberg L. Hallström B. M. Lindskog C. Oksvold P. Mardinoglu A. et al . (2015). Tissue-based map of the human proteome. Science347, 1260419. doi: 10.1126/science.1260419
81
Unal E. Goertz C. E. C. Hobbs R. C. Suydam R. Romano T. (2018). Investigation of molecular biomarkers as potential indicators of health in wild belugas (Delphinapterus leucas). Mar. Biol.165, 165–182. doi: 10.1007/s00227-018-3439-3
82
Van Dolah F. M. Neely M. G. McGeorge L. E. Balmer B. C. Ylitalo G. M. Zolman E. S. et al . (2015). Seasonal variation in the skin transcriptome of common bottlenose dolphins (Tursiops truncatus) from the northern Gulf of Mexico. PloS One10, e0130934. doi: 10.1371/journal.pone.0130934
83
Verma R. P. Hansch C. (2007). Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorg. Med. Chem.15, 2223–2268. doi: 10.1016/j.bmc.2007.01.011
84
Wang Z. Gerstein M. F. Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet.10, 57–63. doi: 10.1038/nrg2484
85
Wang D. Li Y. Aierken R. Kang Q. Wang X. Zeng Q. et al . (2021). Integrated full-length transcriptome and RNA-seq to identify immune system genes from the skin of sperm whale (Physeter macrocephalus). Genes.12, 233. doi: 10.3390/genes12020233
86
Wilson J.Y. Cooke Suzy R. Moore Michael J. Daniel M. Igor M. Metner Donald A. et al . (2005). Systemic effects of arctic pollutants in beluga whales indicated by CYP1A1 expression. Environ. Health Perspect.113, 1594–1599. doi: 10.1289/ehp.7664
87
Wood D. E. Lu J. Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biol.20, 257. doi: 10.1186/s13059-019-1891-0
88
Yordy J. E. Wells R. S. Balmer B. C. Schwacke L. H. Rowles T. K. Kucklick J. R. (2010). Partitioning of persistent organic pollutants between blubber and blood of wild bottlenose dolphins: implications for biomonitoring and health. Environ. Sci. Technol.44, 4789–4795. doi: 10.1021/es1004158
89
Zabka T. S. Romano T. A. (2003). Distribution of MHC II (+) cells in skin of the Atlantic bottlenose dolphin (Tursiops truncatus): An initial investigation of dolphin dendritic cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol.273A, 636–647. doi: 10.1002/ar.a.10077
90
Zhu A. Ibrahim J. G. Love M. I. (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics35, 2084–2092. doi: 10.1093/bioinformatics/bty895
Summary
Keywords
beluga, skin, transcriptome, gene expression, Bristol Bay, Eastern Chukchi Sea, whale, cetacean
Citation
Unal E, Singh V, Suydam R, Goertz CEC and Romano TA (2024) Comparative skin transcriptome analysis as a potential tool to investigate intra- and inter-population differences in belugas. Front. Mar. Sci. 11:1282210. doi: 10.3389/fmars.2024.1282210
Received
23 August 2023
Accepted
08 November 2024
Published
29 November 2024
Volume
11 - 2024
Edited by
Mason Weinrich, Center for Coastal Studies, United States
Reviewed by
Shannon Barber-Meyer, Pacific Whale Foundation, United States
Jane Khudyakov, The University of the Pacific, United States
Updates
Copyright
© 2024 Unal, Singh, Suydam, Goertz and Romano.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ebru Unal, eunal@mysticaquarium.org
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.