Abstract
As rate-limiting enzymes of β-oxidation of fatty acids in mitochondria, the carnitine palmitoyltransferase (CPT) played an important role in regulating energy homeostasis of aquatic animals. However, there was very little research on β-oxidation of fatty acids in crustaceans. In the present study, the full-length cDNA sequences of cpt-1a, cpt-1b and cpt-2 were isolated from the hepatopancreas of Scylla paramamosain, and contained 4206, 5303 and 3486 bp respectively. Sequence analysis showed that the CPT-1A, CPT-1B and CPT-2 encoded proteins with 777, 775 and 672 amino acids respectively, and only the CPT-1A possessed a transmembrane region. In addition, both the CPT-1B and CPT-2 contained conservative functional domains like N-terminal domain and acyltransferases choActase 2, while the CPT-1A lacked. The results of phylogenetic tree indicated that the CPT-1A, CPT-1B and CPT-2 of S. paramamosain gathered together with their corresponding orthologues from crustaceans. The tissue distribution exhibited that the cpt-1a was highly expressed in hepatopancreas, followed by muscle, eyestalk and cranial ganglia, and the muscle, eyestalk and heart were main expressed tissues of cpt-1b. Furthermore, the high expression levels of cpt-2 were mainly detected in hepatopancreas, muscle and heart. The transcriptional levels of cpt-1a, cpt-1b and cpt-2 were significantly up-regulated under chronic low salinity stress. Besides, at the acute low salinity stress condition, the expression levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas were dramatically increased in 14‰ and 4‰ salinity groups at the 6h and 48h, while the transcriptional levels of cpt-1a, cpt-1b and cpt-2 in muscle were signally up-regulated in 14‰ and 4‰ salinity groups at the 12h and 24h, showing an alternate response pattern. Similarly, the present study found that fasting could markedly increase the expression levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas and muscle, especially cpt-1a in hepatopancreas as well as cpt-1a and cpt-1b in muscle. The results above indicated that the cpt-1a, cpt-1b and cpt-2 played a crucial part in providing energy for coping with fasting and salinity stress. These results would contribute to enhancing the knowledge of cpt phylogenetic evolution and their roles in energy metabolism of crustaceans.
1 Introduction
Carnitine palmitoyltransferase (CPT) is a rate-limiting enzyme for β-oxidation of long-chain fatty acids in mitochondria including two subtypes CPT-1 and CPT-2, which plays an important role in maintaining energy balance of body (Bartlett and Eaton, 2004). The enzymes involved in β-oxidation of fatty acids are existed in the mitochondrial matrix, while the activated long-chain acyl-coenzyme A (CoA) in cytoplasm cannot pass freely through the mitochondrial inner membrane (Xu et al., 2017; Wang et al., 2021). The CPT-1 and CPT-2 are located in the inner and outer membrane of mitochondria respectively, and mutual collaboration participate in transport of long-chain fatty acids in mitochondria (Kerner and Hoppel, 2000). The CPT-1 can catalyze acyl groups of long-chain acyl-CoA and carnitine to form acyl carnitine, and the complex above further passed, and then the complex above passes through the inner membrane into the matrix via the carnitine-acylcarnitine translocase (De Paula et al., 2023). Ultimately, the acyl groups are separated from the hydroxyl group of carnitine to become long-chain acyl-CoA and carnitine under the catalysis of CPT-2 (Akieda et al., 2024). After entering the mitochondrial matrix, acyl CoA becomes the substrate of β-oxidation.
Currently, the study of cpt paid more attention to the cpt-1, and studies have confirmed that there were three subtypes of cpt-1 in mammals including cpt-1a (Liver type), cpt-1b (Muscle type) and cpt-1c (Brain type) (Price et al., 2002; Lopes-Marques et al., 2015). There were significant differences in the expression of these three genes in different tissues. The cpt-1a was mainly distributed in liver, kidney and colon, the cpt-1b was highly expressed in skeletal muscle, heart, and testicles (McGarry and Brown et al., 1997), while the cpt-1c was only observed in neurons (Price et al., 2002). By contrast, cpt-1 in teleosts only existed two subtypes, named cpt-1a and cpt-1b, which were found in Monopterus albus (Chao et al., 2024), Takifugu obscurus (Liu Q. et al., 2020), Pelteobagrus fulvidraco (Zheng et al., 2013), Larimichthys crocea (Wang et al., 2019), Ctenopharyngodon idellus (Shi et al., 2017), Oreochromis niloticus (Bayır et al., 2020), Sparus aurata (Boukouvala et al., 2010) and Oncorhynchus mykiss (Gutières et al., 2003). Besides, studies in C. idellus (Shi et al., 2017), O. niloticus (Bayır et al., 2020) and P. fulvidraco (Zheng et al., 2013) observed that their cpt-1a subtypes possessed multiple isoforms, which may be the results of the multiplication of fish genomes. The cpt-1 of most fish was primarily high expression in tissues with high-energy requirements like liver, heart and muscle (Zheng et al., 2013; Shi et al., 2017; Wang et al., 2019; Bayır et al., 2020; Chao et al., 2024). Compared with the cpt-1, the cpt-2 did not observe the presence of subtypes and there are few studies on cpt-2 in teleosts, which was merely covered in C. idellus (Shi et al., 2017) and M. albus (Chao et al., 2024). In addition, the structure and function of cpt-1 and cpt-2 were still poorly understood in crustaceans and only reported in Eriocheir sinensis (Liu et al., 2018).
β-oxidation of fatty acids is one of the most important lipolysis reactions in animals, which is precisely regulated. The currently known regulatory mechanisms about β-oxidation of fatty acids in teleosts were mainly involved in hormone, nuclear receptor and microRNA (Dreyer et al., 1992; Esau et al., 2006; Wu et al., 2016; Ning et al., 2019). The hormone regulation primarily included insulin and glucagon, the insulin was negatively correlated with β-oxidation of fatty acids, but glucagon showed positive correlation. Peroxisome proliferation-activated receptor (PPARs) is one of the main nuclear receptors controlling lipid catabolism. Studies have found that PPARs existed three subtypes named PPARα, the PPARβ and PPARγ, and the PPARα and PPARβ subtypes played a regulatory role in promoting β-oxidation of fatty acids, while the PPARγ focused on regulating the process in β-oxidation of fatty acids (Dreyer et al., 1992; Berger and Moller, 2002). In addition, the β-oxidation of fatty acids in fish was affected by many factors including physiology (growth stage and nutriture), environment (water temperature, salinity and pollutants) and diets (lipid sources, lipid levels, carnitine, choline, betaine and vitamins) (Ning et al., 2019). In comparison, influence factors and regulatory mechanisms about β-oxidation of fatty acids in crustaceans were still unclear.
The mud crab (Scylla paramamosain) is an economically important crustacean. Due to large size, fast growth and special flavor, the mud crab has been widely cultured in southern China and many Indo-Pacific countries, and the production of mud crab in China was reported as 154,661 tons in 2022 (China Fishery Statistical Yearbook, 2023). Improving the energy supply of lipid decomposition can help to exert the “protein-saving effect” of lipid as well as alleviate the problems of growth inhibition, decline in stress resistance and impaired food quality caused by abnormal accumulation of lipid, thus the study of lipid catabolism in aquatic animals has gradually become a hot spot in the current research of aquatic animal physiology. The β-oxidation of fatty acids was considered as the most important pathway of lipid catabolism in animals. However, the studies of β-oxidation of fatty acids in crustaceans were still in infancy. Therefore, in the present study, we investigated the molecular characterization of three rate-limiting enzymes of fatty acids β-oxidation (cpt-1a, cpt-1b and cpt-2) in mud crab and their roles in respond to fasting and ambient salinity stress. These results will contribute to providing new insights into cpt phylogenetic evolution and increasing the knowledge of cpt roles in lipid catabolism of crustaceans.
2 Materials and methods
2.1 Salinity and starvation stress experiments
The acute low salinity stress experiment was performed in the farming system of Ningde Normal University. Healthy mud crabs procured from a local crab farm in Sandu bay (Ningde, Fujian, China), were temporarily cultured for acclimatization before processing. Three salinity levels were set in the present study containing one control group (24‰ salinity) and two groups (14‰ and 4‰ salinity). Seventy-two vibrant mud crabs (average weight: 68.66 ± 1.38 g) were selected and randomly allocated into twelve polypropylene buckets (Zhongkehai, Qingdao, China). There were three treatments, each with four replicates, and each replicate with six crabs. During the experiment, the mud crabs were fed with a local bivalve mollusk (Sinonovacula constrzcta), and seawater was exchanged twice daily. The hepatopancreas, muscle and gill were obtained at 0, 6, 12, 24, 48 and 96 h after the salinity stress treatment, and the samples were immediately frozen in liquid nitrogen and kept in −80°C for further RNA extraction. In addition, the chronic low salinity stress experiment contained four experimental groups (22‰, 17‰, 12‰ and 7‰ salinity) and one control group (27‰ salinity), and the starvation stress experiment included starvation group (experimental group) and feeding group (control group). The crab size, design, experimental condition, and sampling of the chronic low salinity stress and starvation stress experiments have been presented in our previous study (Lin et al., 2023).
2.2 Gene cloning
As the center of lipid metabolism, the fresh hepatopancreas of mud crab was applied for isolating the total RNA by using TRNzol Universal Reagent (Tiangen, Beijing, China). After assessing integrity and purity of the total RNA, the high quality samples were reverse transcribed into first-strand cDNA by using SMART RACE cDNA Amplification kit (Clontech, USA) according to manufacturer’s protocol. The obtained cDNA was stored at -20°C for further processing. Partial cpt-1a, cpt-1b and cpt-2 cDNA sequences of mud crab were obtained from transcriptome data. Based on the partial sequences above, primers were designed for obtaining 5’ and 3’ untranslated region (UTR) sequence information of the cpt-1a, cpt-1b and cpt-2 cDNA. The primes used for cloning have been shown in Table 1. Combining the touch-down PCR (first round PCR) and nested PCR (second round PCR) strategies, 5’ and 3’ UTR of cpt-1a, cpt-1b and cpt-2 cDNA were obtained by 5’ and 3’ rapid amplification of cDNA ends (RACE) methods respectively. The amplification program and reaction system of touch-down PCR and nested PCR have been given in our previous study (Lin et al., 2023). The target fragments were separated with 1% agarose gel and purified with a TIANgel MIDI Purification Kit (Tiangen, China). The purified target fragments were further connected to pMD19-T vector (Takara, China) and sequenced by a commercial company (Tsingke, China).
Table 1
| Primer | Sequence (5’-3’) | Objective |
|---|---|---|
| Oligo- | AAGCAGTGGTATCAACGCAGAGTACXXXXX | First-Strand cDNA Synthesis |
| UPM (long) | CTAATACGACTCACTATAGGGCA AGCAGTGGTATCAACGCAGAGT |
RACE-PCR |
| UPM (short) | CTAATACGACTCACTATAGGGC | RACE-PCR |
| NUP | AAGCAGTGGTATCAACGCAGAGT | RACE-PCR |
| β-actin F | GCCCTTCCTCACGCTATCCT | qRT-PCR |
| β-actin R | GCGGCAGTGGTCATCTCCT | qRT-PCR |
| 18S rRNA F | TACCGATTGAATGATTTAGTGAGG | qRT-PCR |
| 18S rRNA R | CTACGGAAACCTTGTTACGACTT | qRT-PCR |
| M13F | CGCCAGGGTTTTCCCAGTCACGAC | PCR screening |
| M13R | AGCGGATAACAATTTCACACAGGA | PCR screening |
| For cpt-1a clone and qRT-PCR | ||
| cpt-1a 3-1 | GGCCGGTGTCTGATGAGGGATACG | 3’RACE |
| cpt-1a 3-2 | GTCCCAAGCCTGTGCTAGTTCCCG | 3’RACE |
| cpt-1a 5-1 | GCTTGAAGGACTTGGGTGGAGGCA | 5’RACE |
| cpt-1a 5-2 | CGCTGGCAGGAGTACAGGGAAGGCT | 5’RACE |
| Q-cpt-1a F | CCAGGCAGGTATCAAGAGCAGC | qRT-PCR |
| Q-cpt-1a R | GAACTCGCACAGTGGAGAAGAGG | qRT-PCR |
| For cpt-1b clone and qRT-PCR | ||
| cpt-1b 3-1 | TGATCCATTACCATGTGGCAGGAAGGC | 3’RACE |
| cpt-1b 3-2 | GGAAGCCAGCTTGTGAAGAGAACCATG | 3’RACE |
| cpt-1b 5-1 | CAGTTGCGTGGGATCATCAGGGCTAT | 5’RACE |
| cpt-1b 5-2 | CATGTGTCACATTGGCTGCCCGAG | 5’RACE |
| Q-cpt-1b F | AATGTGCAATGCGGTCTGTTTC | qRT-PCR |
| Q-cpt-1b R | ACACTTTGGGCTGGTTCCTCTAA | qRT-PCR |
| For cpt-2 clone and qRT-PCR | ||
| cpt-2 3-1 | TGGCCAGCTTGATGTCACACCTGTG | 3’RACE |
| cpt-2 3-2 | AGATGGTCATGGGGGAGCAGTGTGA | 3’RACE |
| cpt-2 5-1 | GAACTCCAGCCTGCGGACAAGTTTTG | 5’RACE |
| cpt-2 5-2 | GCTTTGGGATGCGAGTGGAGTTGAA | 5’RACE |
| Q-cpt-2 F | ACAAGGCAAATAAACACACCAGC | qRT-PCR |
| Q-cpt-2 R | ATGAACCTCAGAGACGACACCAA | qRT-PCR |
Names and sequences of primers used in the present study.
X, undisclosed base in the proprietary SMARTer oligo sequence.
2.3 Sequence and phylogenetic analysis
The identity and similarity of sequences were analyzed by using BLAST at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The open reading frame position was predicted by using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Transmembrane structure was forecasted by making use of TMHMM 2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). ExPASy ProtParam tool (https://web.expasy.org/protparam/) was applied to calculate the proteic molecular mass and isoelectric point. Proteic secondary structure and structural domain were analyzed by using NPS Server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) and InterPro (https://www.ebi.ac.uk/interpro/), respectively. SignalP5.0 Server (https://services.healthtech.dtu.dk/services/SignalP-5.0/) was chosen to predict signal peptide. SWISS-MODEL (https://swissmodel.expasy.org/) was used to evaluate tertiary protein structure. Multiple alignments were performed using DNAMAN software. The phylogenetic tree construction was built using the software MEGA version 7.0 with confidence in the resulting phylogenetic tree branch topology measured by bootstrapping through 10000 iterations.
2.4 Quantitative real-time PCR
For tissue distribution analysis, six mud crabs (body weight: 49.50 ± 1.18 g) were purchased from a local crab farm in Sandu bay (Ningde, Fujian, China). After being anesthetized with ice, the crabs above were dissected for obtaining tissues of hepatopancreas, cranial ganglia, thoracic ganglia, eyestalk, gill, intestine, muscle and heart. Quantitative real-time PCR was applied to determine the expression levels of cpt-1a, cpt-1b and cpt-2 in different tissues above as well as expression levels in respond to salinity and starvation stress. The TRNzol Universal Reagent (Tiangen, China) was used to extract the total RNA of samples. Then, 1 μg high-quality total RNA of each sample was applied to synthesize cDNA template with PrimeScript® RT reagent Kit with gDNA Eraser (Takara, Dalian, China), and further diluted into working fluid by adding 4 times ultra-pure water. The quantitative real-time PCR was performed in QuantStudio 3 (Thermo Fisher, USA). The procedure of quantitative real-time PCR contained a hold stage (95°C for 30 s), PCR stage (40 cycles of 95°C for 10 s and 60°C for 30 s) and melt curve stage (95°C for 15 s, 60°C for 60 s and 95°C for 15 s). The reaction system of quantitative real-time PCR executed in a total volume of 20 μL including 10 μL 2 × ChamQ Universal SYBR qPCR Master Mix (Q711-02/03, Vazyme Biotech Co., Ltd., Nanjing, China), 1.0 μL of the diluted cDNA template, 0.4 μL of each primer (10μM) and 8.2 μL of sterile distilled H2O. The 18S rRNA and β-actin were selected as reference genes. The amplification efficiency of primers was investigated by five different thinned cDNA samples and counted with formula E = 10(−1/Slope) – 1. The amplification efficiency of primers used in the presented study was between 90% and 110%. The relative mRNA levels of target genes were calculated by 2-ΔΔCt method (Livak and Schmittgen, 2001). The primes involved in quantitative real-time PCR have been presented in Table 1.
2.5 Statistical analysis
SPSS 20.0 software was applied to perform all statistical analysis in the present study, and P values less than 0.05 was deemed to be statistically significant. The results were given as means ± standard error. Independent-sample T-test was used to determine the significant differences between the feeding group and starvation group. Besides, one-way analysis of variance (ANOVA) followed by Duncan’s multiple comparison test was chosen to identify the significant differences of acute and chronic salinity stress experiments as well as tissue distribution after determining the normality and homogeneity.
3 Results
3.1 Sequence analysis
The full-length cDNA sequences of cpt-1a (GenBank accession number PP259098), cpt-1b (GenBank accession number PP259102) and cpt-2 (GenBank accession number PP259103) are 4206, 5303 and 3486 bp, respectively. Concretely, the cpt-1a consists of a 196 bp of 5’- UTR, 1676 bp of 3’-UTR with a poly (A) tail and 2334 bp open reading frame encoded a protein with 777 amino acids (Supplementary Figure 1). The cpt-1b is composed of a 117 bp of 5’-UTR, 2858 bp of 3’-UTR with a poly (A) tail and 2328 bp open reading frame encoded a protein with 775 amino acids (Supplementary Figure 2). In addition, the cpt-2 includes a 252 bp of 5’-UTR, 1215 bp of 3’-UTR with a poly (A) tail and 2019 bp open reading frame encoded a protein with 672 amino acids (Supplementary Figure 3). The molecular weights of CPT-1A, CPT-1B and CPT-2 proteins are 89.36, 88.16 and 75.37 kDa, respectively. The theoretical isoelectric point of CPT-1A protein is 8.86, the CPT-1B is 8.75 and the CPT-2 is 8.24.
The NPS Server prediction showed that α-helix, extended chain, random curl and β-corner together constitute the secondary structures of CPT-1A, CPT-1B and CPT-2 proteins, in which α-helix structure accounts for the largest proportion, 49.42% (CPT-1A), 48.90% (CPT-1B) and 46.28% (CPT-2), respectively. The predicted three-dimensional structures of CPT-1A, CPT-1B and CPT-2 proteins have been given in Figure 1. The SignalP 5.0 analysis showed that there are no signal peptides among the CPT-1A, CPT-1B and CPT-2. The TMHMM 2.0 predicted that the CPT-1A protein only contains a transmembrane structure (WLANLVGSVGLSFVFFVILM, 99~118 region) (Supplementary Figure 1), while the CPT-1B and CPT-2 have no transmembrane structures. In addition, the function domain prediction displayed that both the CPT-1B and CPT-2 possess typical characteristics of CPT proteins including the N-terminal domain (CPT-1B, 1~46 region; CPT-2, 48~149 region) and acyltransferases choActase 2 (CPT-1B, 455~482 region; CPT-2, 368~395 region), while the CPT-1A has no the structures of N-terminal domain and acyltransferases choActase (Figure 2).
Figure 1

Three-dimensional structure prediction of carnitine palmitoyltransferase proteins in Scylla paramamosain.
Figure 2

Multiple alignments of the carnitine palmitoyltransferase 1 [CPT-1; (A)] and CPT-2 (B) amino acid sequences from Scylla paramamosain with other species. The threshold for similarity shading was set at 50%. Identical residues are shaded black. The similarity of amino acid residues more than 75% and 50% are shaded in pink and cyan, respectively. The conserved acyltransferases ChoActase domain is marked with boxes, and the predicted N-terminal region is underlined. The sequences used for comparison are as follows: Homo sapiens CPT-1A (EAW74722), Mus musculus CPT-1A (EDL32931), Monopterus albus CPT-1A (WHB10902), H. sapiens CPT-1B (BAD96894), M. musculus CPT-1B (EDL04340), M. albus CPT-1B (WHB10903), Eriocheir sinensis CPT-1B (AXY87731), H. sapiens CPT-2 (EAX06753), M. musculus CPT-2 (EDL30761), Danio rerio CPT-2 (NP_001007448), Larimichthys crocea CPT-2 (TMS20131), M. albus CPT-2 (WHB10904), E. sinensis CPT-2 (AXY87732).
3.2 Phylogenetic analysis
The results of phylogenetic tree exhibited that the CPT-1A and CPT-1B of S. paramamosain gathered together with their corresponding orthologues from crustaceans and separated from mollusks and vertebrates (mammals, birds, reptiles, amphibians and fish) (Figure 3). Concretely, the S. paramamosain CPT-1A first grouped into a branch with Portunus trituberculatus CPT-1A, and then gathered with E. sinensis CPT-1A into a bigger branch. Subsequently, the CPT-1A from S. paramamosain, P. trituberculatus and E. sinensis further clustered together with other crustaceans including Cherax quadricarinatus, Procambarus clarkii, Penaeus japonicas, Penaeus monodon and Penaeus vannamei. Similarly, the S. paramamosain CPT-1B first gathered with P. trituberculatus CPT-1B, and then grouped with E. sinensis CPT-1B. The branch above was further clustered with Homarus americanus CPT-1, and finally grouped with mollusks (like Crassostrea gigas, Crassostrea virginica, Mytilus galloprovincialis and Pecten maximus) into a bigger branch. In addition, the S. paramamosain CPT-2 gathered with the CPT-2 from crustacean group and isolated from mammals, birds, reptiles, amphibians, fish and mollusks (Figure 4). Specifically, the S. paramamosain CPT-2 clustered most closely with P. trituberculatus CPT-2, and then further gathered together with CPT-2 of E. sinensis and Chionoecetes opilio, forming a bigger branch, finally grouped with other CPT-2 from other crustaceans.
Figure 3
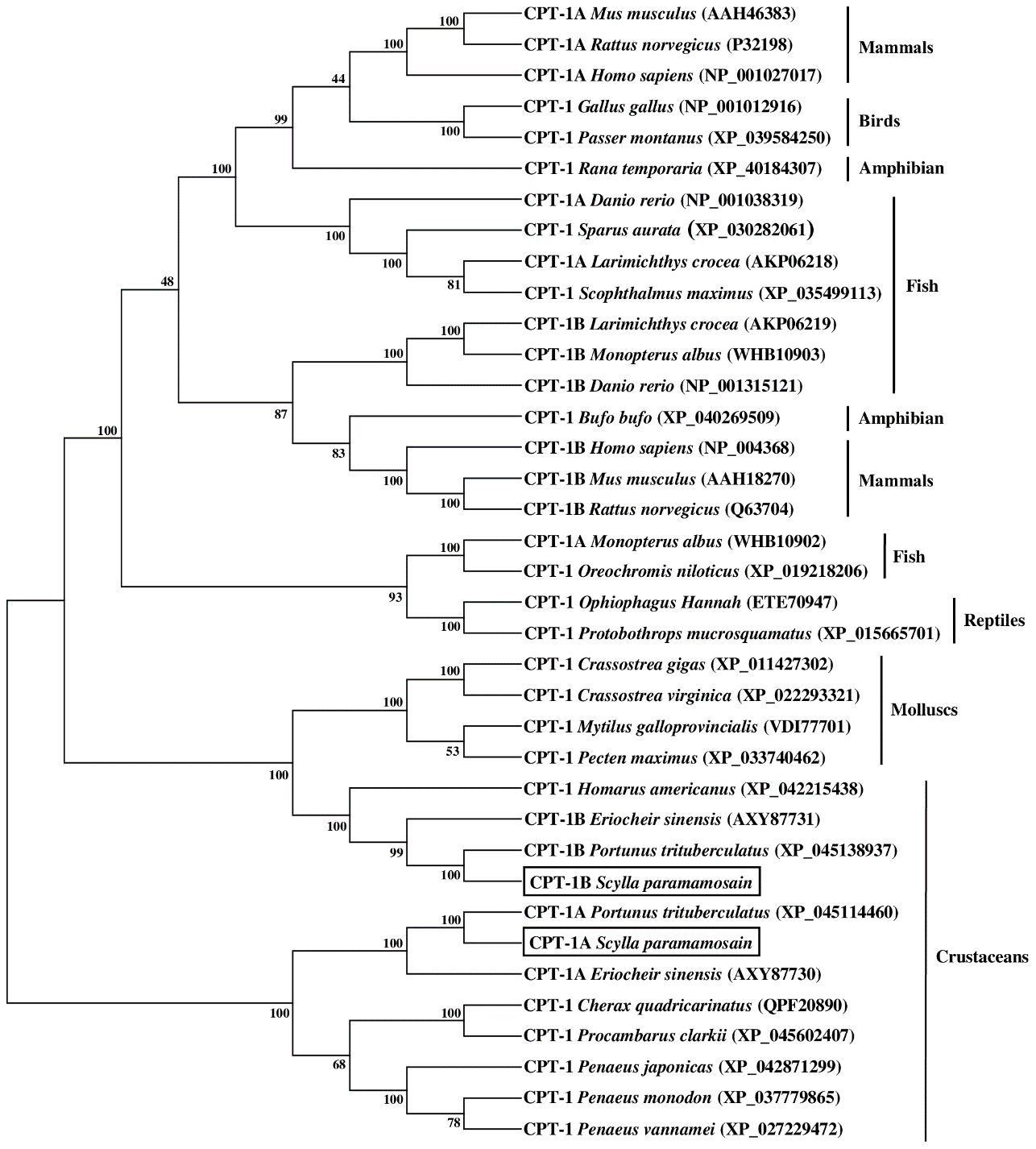
Neighbor-joining phylogenetic tree of representative vertebrate and invertebrate carnitine palmitoyltransferase 1 (CPT-1) amino acid sequences. The tree was constructed using the neighbor joining method with MEGA 7.0. The horizontal branch length is proportional to amino acid substitution rate per site. Numbers represent the frequencies with which the tree topology presented was replicated after 1000 bootstrap iterations.
Figure 4
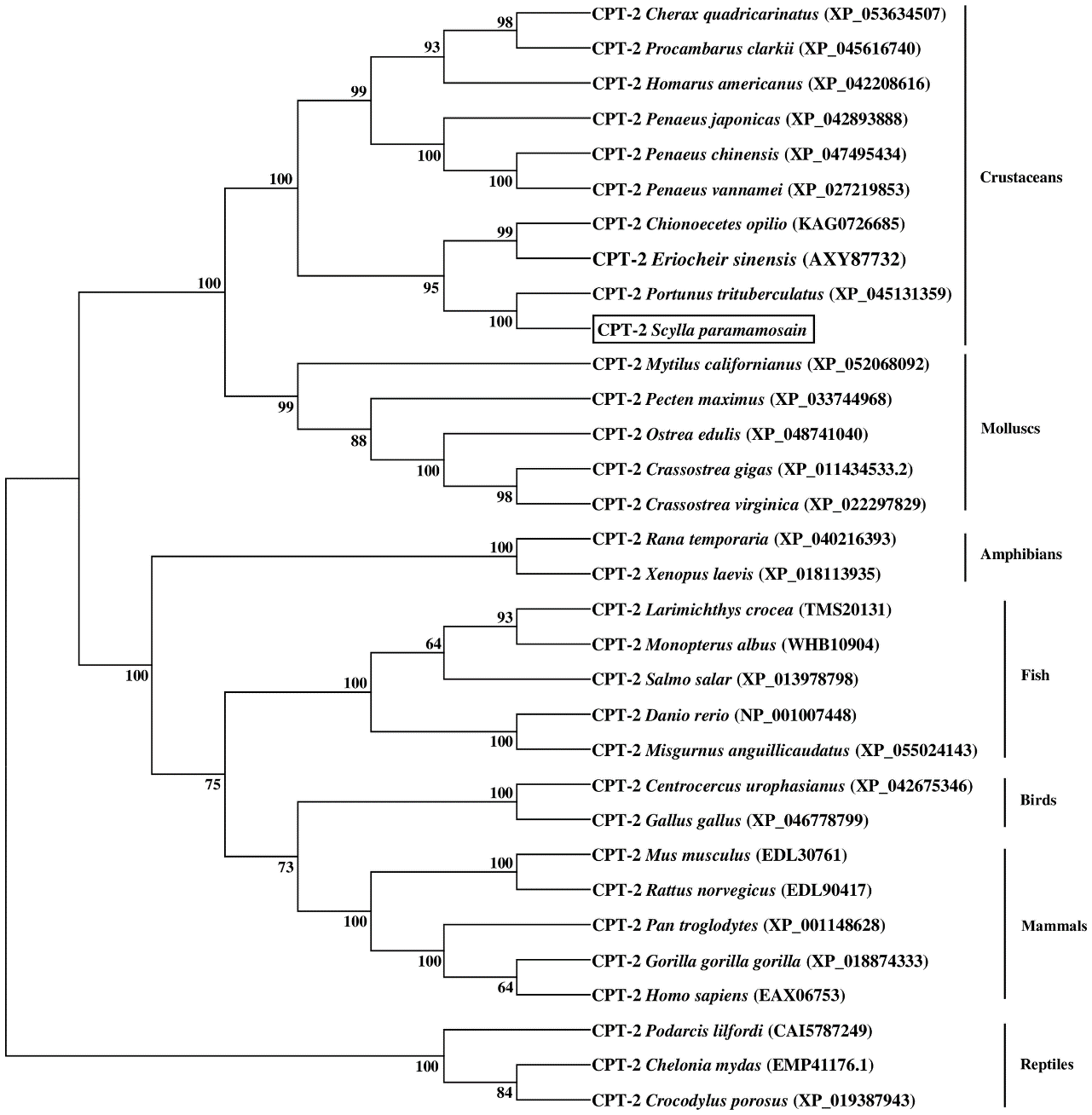
Neighbor-joining phylogenetic tree of representative vertebrate and invertebrate carnitine palmitoyltransferase 2 (CPT-2) amino acid sequences. The tree was constructed using the neighbor joining method with MEGA 7.0. The horizontal branch length is proportional to amino acid substitution rate per site. Numbers represent the frequencies with which the tree topology presented was replicated after 1000 bootstrap iterations.
3.3 Tissue distribution
The relative expression levels of cpt-1a, cpt-1b and cpt-2 in different tissues have been given in the Figure 5. The highest expression levels of cpt-1a and cpt-2 were the hepatopancreas among all the tissues (P < 0.05). The muscle, eyestalk and cranial ganglia exhibited markedly higher cpt-1a mRNA levels than the thoracic ganglia, intestine, heart and gill (P < 0.05). In addition, except for hepatopancreas, the cpt-2 was also highly expressed in the muscle when compared with the eyestalk, thoracic ganglia, intestine and gill (P < 0.05). By contrast, the muscle, eyestalk and heart were main sites of cpt-1b expression, and the muscle showed markedly higher the cpt-1b mRNA levels than the cranial ganglia, thoracic ganglia, intestine and gill (P < 0.05).
Figure 5
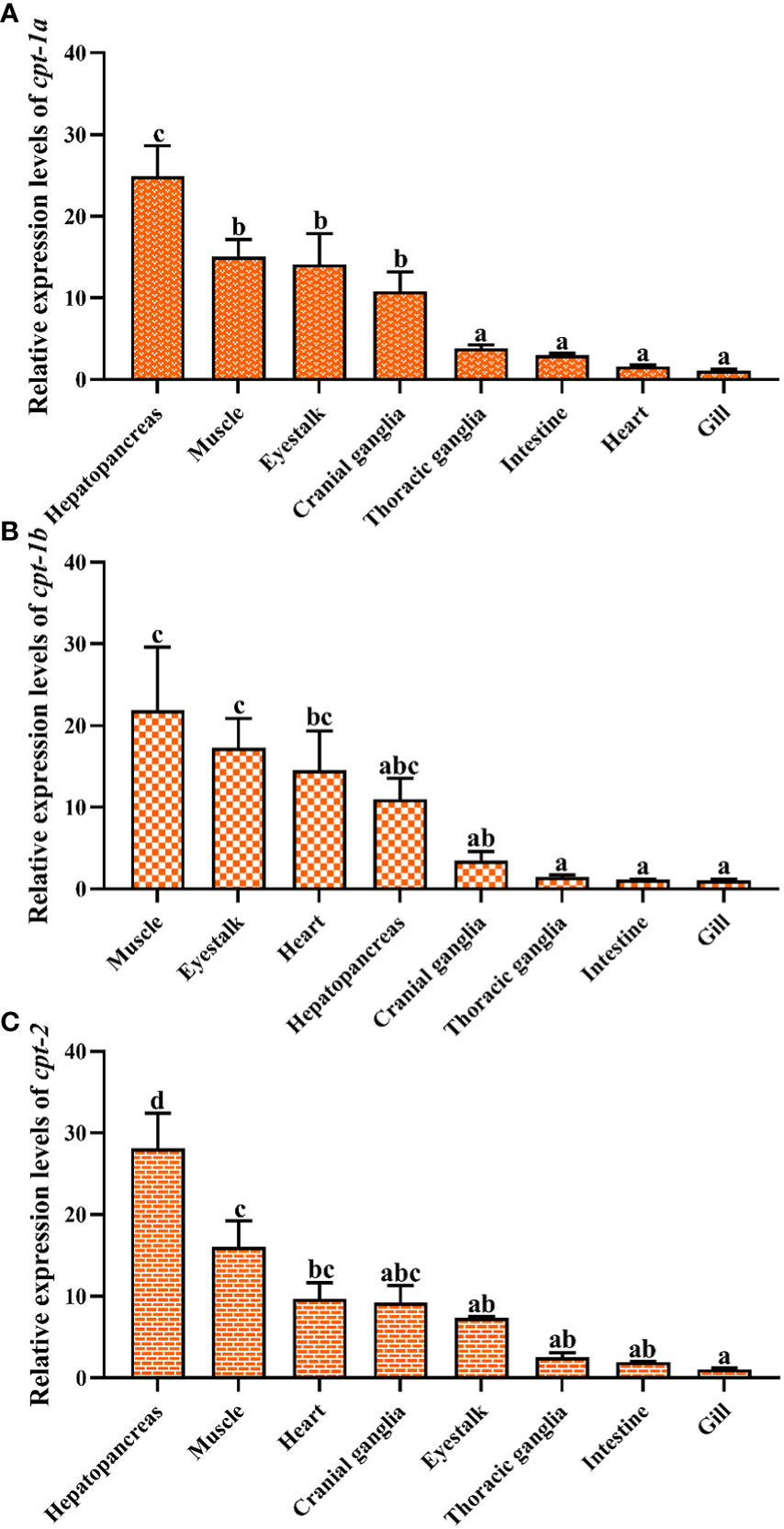
Relative expression levels of carnitine palmitoyltransferase (cpt) in different tissues of Scylla paramamosain. (A): tissue distribution of cpt-1a; (B): tissue distribution of cpt-1b; (C): tissue distribution of cpt-2. Vertical bars represented mean ± standard error of the mean (n = 6) for each tissue. Letters show significant differences (P < 0.05) among tissues as determined by one-way ANOVA followed by Duncan’s multiple comparison test.
3.4 Transcriptional levels of cpt-1a, cpt-1b and cpt-2 in response to chronic low salinity stress
The relative expression levels of cpt-1a, cpt-1b and cpt-2 in response to chronic low salinity stress have been presented in the Figure 6. The results showed that the cpt-1a transcriptional level in hepatopancreas was markedly increased in the 7‰ salinity group when compared with the 27‰, 22‰, 17‰ and 12‰ salinity groups (P < 0.05), and the 7‰ salinity group exhibited higher mRNA levels of cpt-2 in hepatopancreas than the 27‰ and 22‰ salinity groups (P < 0.05). There were no significant differences in cpt-1b expression of hepatopancreas among all groups, although the 7‰, 12‰ and 17‰ salinity groups showed higher levels than the 27‰ and 22‰ salinity groups (P > 0.05). In addition, no significant differences were observed in cpt-1a transcriptional levels of muscle among all salinity groups (P > 0.05). Compared with the 27‰ group, the cpt-1b mRNA level in muscle was significantly up-regulated in the 7‰ salinity group (P < 0.05), and the 7‰ salinity group showed markedly higher cpt-2 expression level than the 27‰, 22‰, 17‰ salinity groups (P < 0.05). Besides, the cpt-1a transcriptional levels in gill were markedly increased in the 7‰ and 12‰ salinity groups when compared with the 17‰ 22‰ and 27‰ salinity groups (P < 0.05), and 17‰ and 22‰ salinity groups also showed higher value than the 27‰ salinity group (P < 0.05). There were no significant differences in cpt-1b expression of gill among all groups (P > 0.05). The 7‰ salinity group had higher cpt-2 expression levels than the 27‰, 22‰, 17‰ and 12‰ salinity groups (P < 0.05).
Figure 6
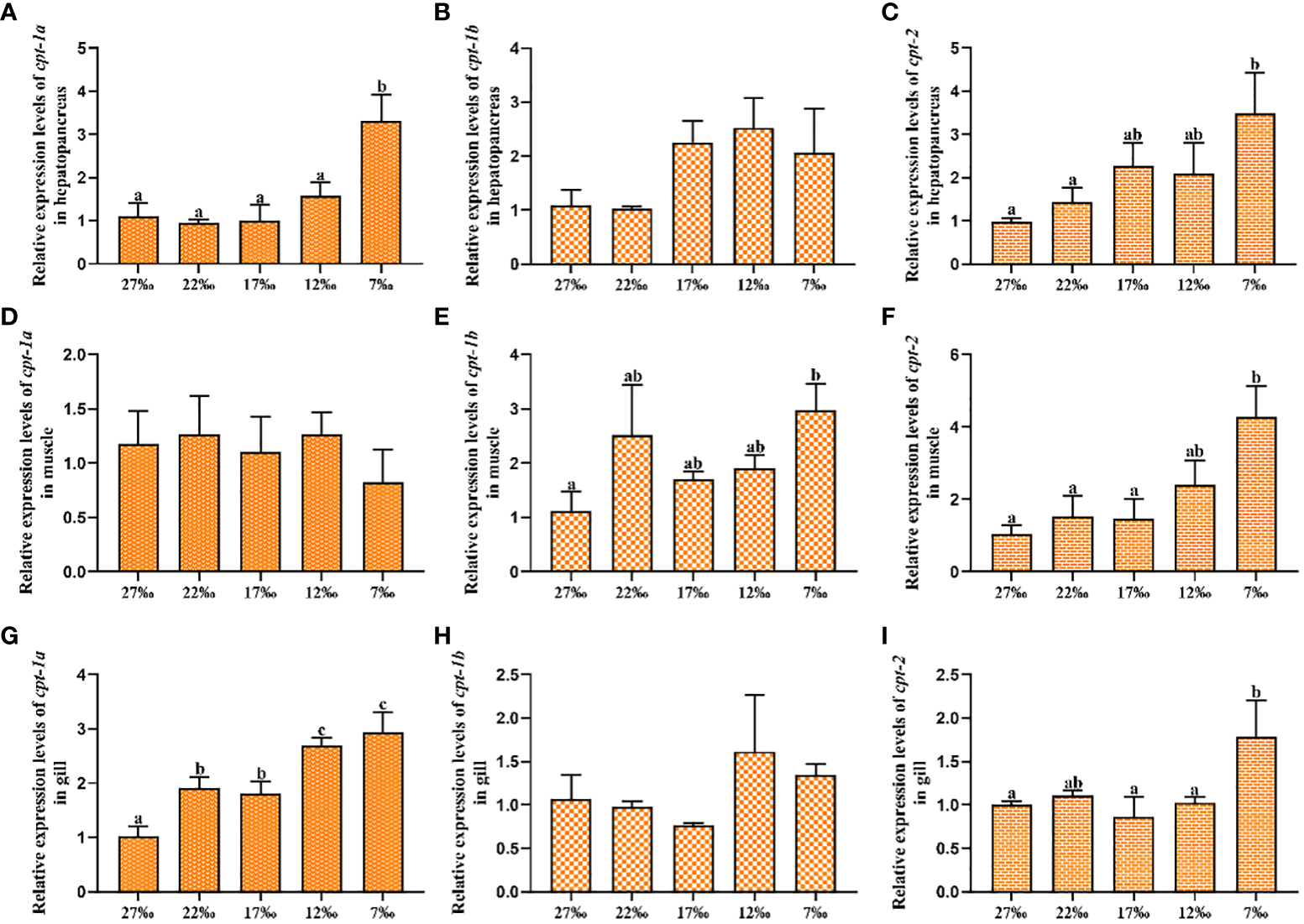
Relative expression levels of carnitine palmitoyltransferase (cpt) in hepatopancreas, muscle and gill of Scylla paramamosain in response to chronic salinity stress. (A–C): cpt-1a, cpt-1b and cpt-2 expression in hepatopancreas; (D–F): cpt-1a, cpt-1b and cpt-2 expression in muscle; (G–I): cpt-1a, cpt-1b and cpt-2 expression in gill. Bars with different superscripts are significantly different (P< 0.05, one-way ANOVA and Duncan’s multiple comparison test). Numerical values refer to seawater salinity.
3.5 Transcriptional levels of cpt-1a, cpt-1b and cpt-2 in response to acute low salinity stress
To investigate the transcriptional levels of cpt-1a, cpt-1b and cpt-2 in response to acute low salinity stress, the total RNA were isolated from the hepatopancreas and muscle at different time points. The relative expression levels of cpt-1a, cpt-1b and cpt-2 of hepatopancreas in response to acute low salinity stress have been presented in the Figure 7. Compared with the 24‰ salinity group (control group), the transcriptional levels of cpt-1a and cpt-2 in hepatopancreas were up-regulated in the 14‰ and 4‰ salinity groups at 6 h and 48 h, and the 4‰ salinity group showed markedly higher mRNA levels than the 24‰ salinity group (P < 0.05). There were no significant differences in cpt-1a and cpt-2 expression of hepatopancreas among the 24‰, 14‰ and 4‰ salinity groups at the other time points (P > 0.05). Similarly, the 14‰ and 4‰ salinity groups also exhibited higher mRNA levels of cpt-1b in hepatopancreas than the 24‰ salinity group at 48 h, while only the 4‰ and 24‰ salinity groups showed significant differences (P < 0.05). In addition, there were no significant differences in cpt-1b expression of hepatopancreas among the 24‰, 14‰ and 4‰ salinity groups at the other time points (P > 0.05), although the mRNA levels of cpt-1b in 24‰ salinity group was lower than the 14‰ and 4‰ salinity groups at 6 h.
Figure 7
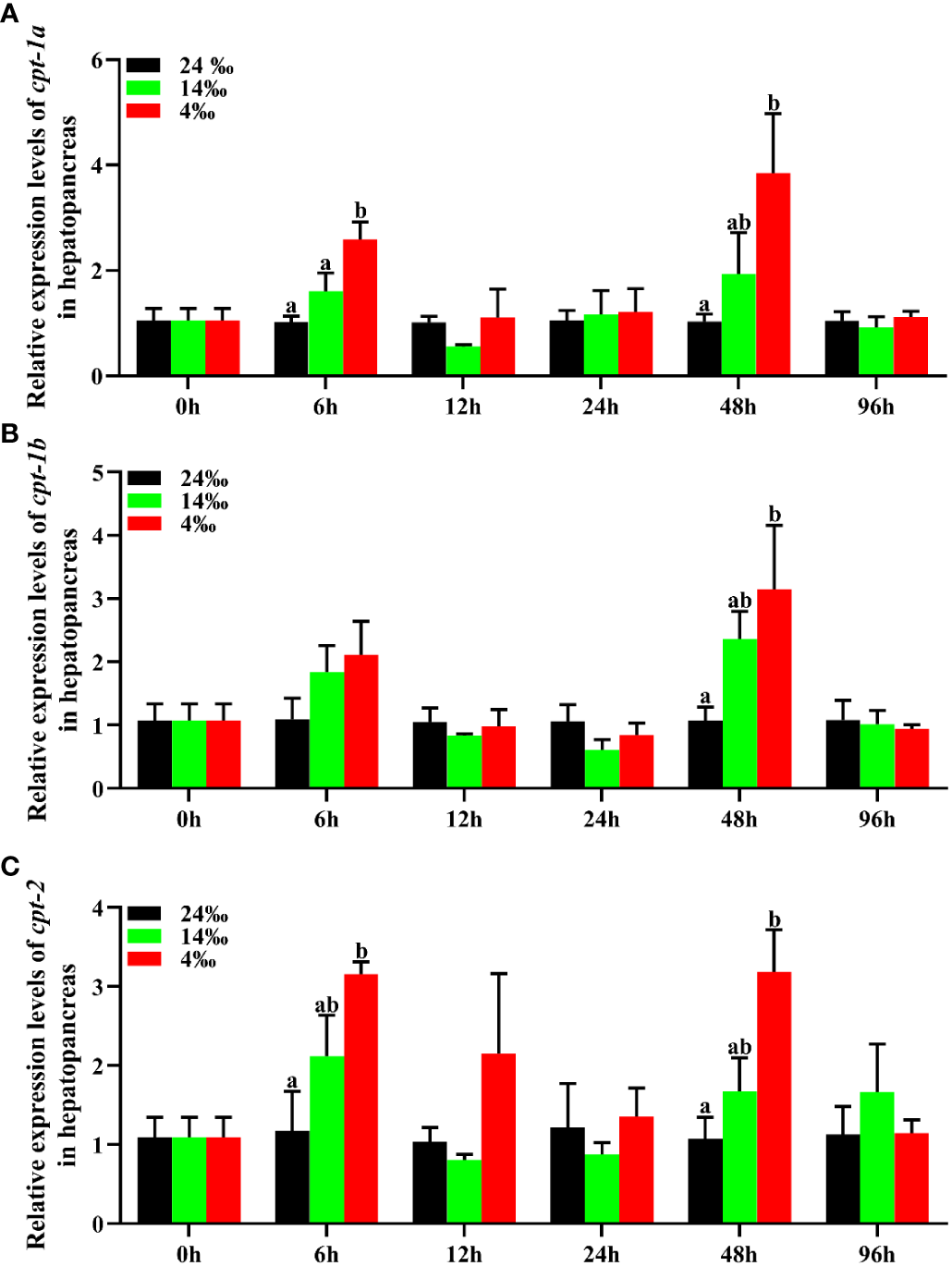
Relative expression levels of carnitine palmitoyltransferase (cpt) in hepatopancreas of Scylla paramamosain in response to acute salinity stress. (A): cpt-1a expression in hepatopancreas; (B): cpt-1b expression in hepatopancreas; (C): cpt-2 expression in hepatopancreas. Total RNA was extracted from hepatopancreas of S. paramamosain at different time points after 24‰, 14‰ and 4‰ salinity stress, respectively. Samples challenged with 24‰ salinity were adopted as control. Significance was compared between the experimental groups and the control group at the same time point. Bars with different superscripts are significantly different (P< 0.05, one-way ANOVA and Duncan’s multiple comparison test).
The relative expression levels of cpt-1a, cpt-1b and cpt-2 of muscle in response to acute low salinity stress have been presented in the Figure 8. Compared with the 24‰ salinity group, the transcriptional levels of cpt-1a and cpt-2 in muscle were up-regulated in the 14‰ and 4‰ salinity groups at 12 h and 24 h, while only the expression levels of cpt-2 in 4‰ salinity group showed significant differences with the 24‰ salinity group at 12 h (P < 0.05). Likewise, the cpt-1b expression levels of muscle in 24‰ salinity group were also lower than the 14‰ and 4‰ salinity groups at the 12 h and 24 h, and the 4‰ salinity group showed markedly higher mRNA levels than the 24‰ salinity group (P < 0.05).
Figure 8
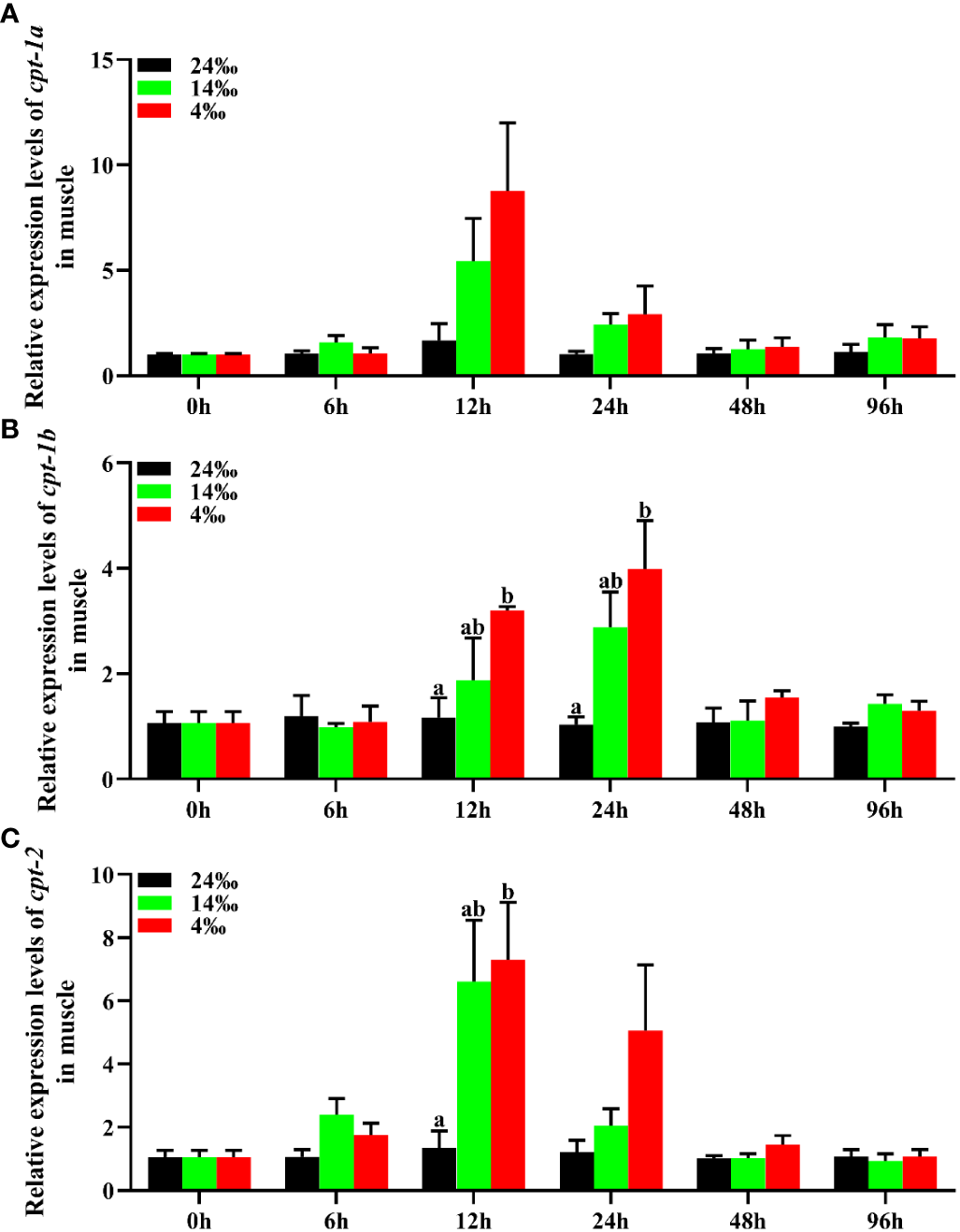
Relative expression levels of carnitine palmitoyltransferase (cpt) in muscle of Scylla paramamosain in response to acute salinity stress. (A): cpt-1a expression in muscle; (B): cpt-1b expression in muscle; (C): cpt-2 expression in muscle. Total RNA was extracted from muscle of S. paramamosain at different time points after 24‰, 14‰ and 4‰ salinity stress, respectively. Samples challenged with 24‰ salinity were adopted as control. Significance was compared between the experimental groups and the control group at the same time point. Bars with different superscripts are significantly different (P< 0.05, one-way ANOVA and Duncan’s multiple comparison test).
3.6 Transcriptional levels of cpt-1a, cpt-1b and cpt-2 in response to starvation stress
The relative expression levels of cpt-1a, cpt-1b and cpt-2 of hepatopancreas and muscle in response to starvation stress have been shown in the Figure 9. Compared with the feeding group, the transcriptional levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas were markedly up-regulated in the starvation group (P < 0.05), especially cpt-1a showing an extremely significant difference (P < 0.01). In addition, the expression levels of cpt-1a, cpt-1b and cpt-2 in muscle of starvation group were also significantly higher than those in the feeding group (P < 0.05), and the cpt-1a and cpt-1b exhibited a highly significant difference (P < 0.01).
Figure 9
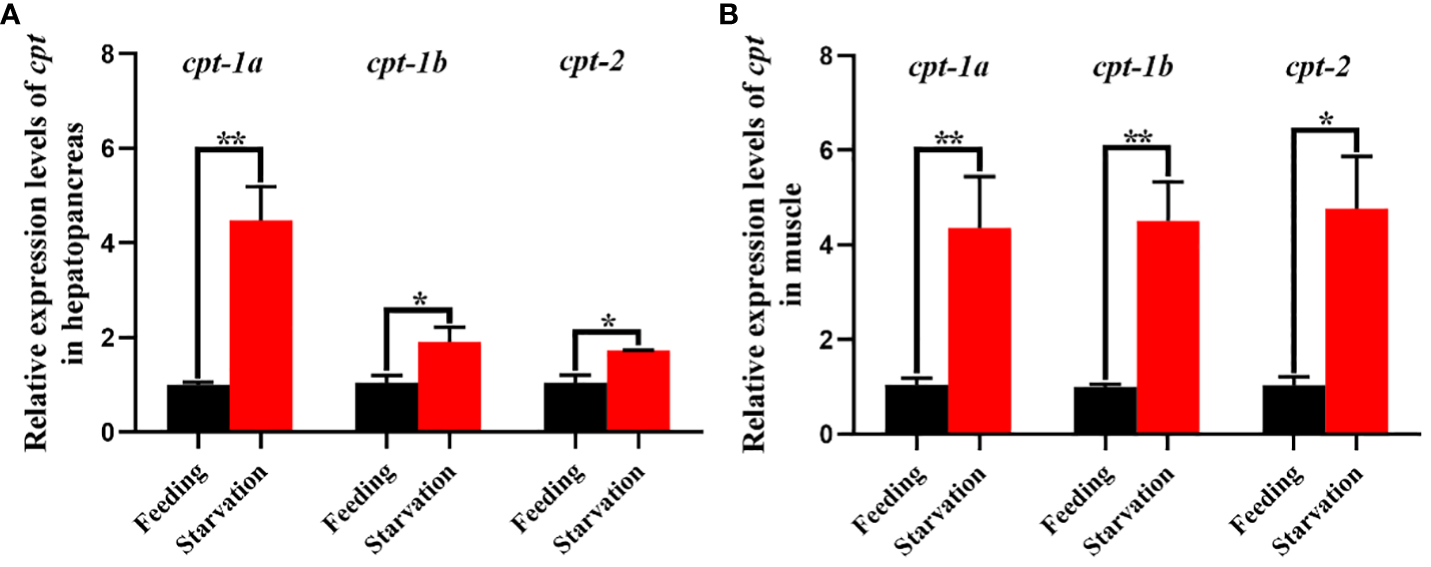
Relative expression levels of carnitine palmitoyltransferase (cpt) in hepatopancreas and muscle of Scylla paramamosain in response to starvation stress. (A): cpt-1a, cpt-1b and cpt-2 expression in hepatopancreas; (B): cpt-1a, cpt-1b and cpt-2 expression in muscle. Asterisks indicated significant differences between feeding group and starvation group (*P < 0.05 and **P < 0.01).
4 Discussion
With the rapid intensive development of aquaculture industry, increasing lipid utilization for saving dietary protein and reducing the negative effects of abnormal lipid deposition have become the needs for the healthy development of the industry (Bu et al., 2023). Therefore, the study of lipid metabolism in aquatic animals, especially lipid catabolism, is a research hotspot of nutritional physiology at present. The lipid catabolism is mainly achieved through the β-oxidation of fatty acids in peroxisome and mitochondria. Currently, the studies about β-oxidation of fatty acids in aquatic animals were more focused on fish, while few were reported in crustaceans (Lavarías et al., 2009; Liu et al., 2018).
The CPT, as rate-limiting enzymes, included CPT-1 and CPT-2 subtypes, which played critical roles in the β-oxidation of fatty acids in mitochondria (Kerner and Hoppel, 2000; Bayır et al., 2020). In the present study, the full-length cDNA sequences of three cpt isoforms were successfully cloned from the hepatopancreas of mud crab. Sequence analysis showed that the three genes were cpt-1a, cpt-1b and cpt-2 subtypes respectively. Previous studies have indicated that the cpt-1a existed the phenomenon of genome replication such as P. fulvidraco (Zheng et al., 2013), O. niloticus (Bayır et al., 2020), C. idellus (Shi et al., 2017) and Synechogobius hasta (Wu et al., 2016), while this phenomenon was not found in the present study, meanwhile, cpt-1c was also not observed. These results were in lined with the studies of E. sinensis (Liu et al., 2018) and M. albus (Chao et al., 2024), which only found three cpt isoforms named cpt-1a, cpt-1b and cpt-2. The cpt-1a, cpt-1b and cpt-2 subtypes of mud crab encoded 777, 775 and 672 amino acids respectively and these sequences of amino acids showed high similarity with mammals, fish and crustaceans, which indicated that CPT-1A, CPT-1B and CPT-2 were conserved among different species. Studies in mammals observed that the CPT-1 possessed two transmembrane structures (Bonnefont et al., 2004; Jogl et al., 2004). Similarly, Liu et al. (2018) reported that both the CPT-1A and CPT-1B of E. sinensis contained two transmembrane regions. However, the present study found that the CPT-1A of mud crab only possessed a transmembrane region and CPT-1B had no transmembrane structure, and the reason may be related to the mutation of key amino acids in genome replication. In addition, the CPT-2 of mud crab did not contain transmembrane structures possibly because it primarily played a catalytic role within the membrane. Similar results were also found in E. sinensis (Liu et al., 2018) and M. albus (Chao et al., 2024).
Studies in mammals and teleosts have indicated that the CPT-1 existed a N-terminal domain (residues 1~150), which played important roles in regulating catalytic activity and malonyl-CoA sensitivity (Cohen et al., 1998; Bonnefont et al., 2004; Jogl et al., 2004; Zheng et al., 2013; Chao et al., 2024). The present study showed that the CPT-1B of mud crab also possessed the conserved N-terminal domain, while the CPT-1A lacked the structure. Consistently, Liu et al. (2018) found that the CPT-1A of E. sinensis contained the N-terminal domain and the CPT-1B was absent. In addition, previous studies observed that the E. sinensis (Liu et al., 2018), C. idellus (Shi et al., 2017) and M. albus (Chao et al., 2024) have no N-terminal domain in CPT-2. Intriguingly, we found the CPT-2 of mud crab included a N-terminal domain in the present study, suggesting that the CPT-2 structure of mud crab was different from other species. As transporting sites of medium-chain acyl-CoA, both the CPT-1B and CPT-2 of mud crab contained an acyltransferases choActase 2 domain, while the CPT-1A was not observed. These results were consistent with the study of Liu et al. (2018), who found that both the CPT-1B and CPT-2 of E. sinensis possessed acyltransferases choActase domain, and the CPT-1A lacked. Besides, Chao et al. (2024) showed that all the CPT-1A, CPT-1B and CPT-2 of M. albus contained the acyltransferases choActase domain, and except in CPT-1B, CPT-1 peptides of C. idellus included acyltransferases choActase domain (Shi et al., 2017). The CPT-1A of mud crab above did not possess the structures of N-terminal domain and acyltransferases choActase, indicating that the CPT-1A may be not necessary for uptake of fatty acids in mitochondria of mud crab. The results of phylogenetic tree exhibited that the CPT-1A, CPT-1B and CPT-2 gathered together with their corresponding crustacean orthologues, which further supported the genes isolated in the present study belonged to the cpt family.
To determine the tissue distribution, it was of great significance for investigating physiological functions of genes. In mammals, there were three subtypes of cpt-1 including cpt-1a, cpt-1b and cpt-1c, and their expression in tissues showed tissue specificity (Price et al., 2002; Lopes-Marques et al., 2015). The cpt-1a was highly expressed in liver and kidney, the cpt-1b was mainly distributed in skeletal muscle and heart, and the cpt-1c was only detected in neurons (McGarry and Brown, 1997; Price et al., 2002). Compared with the mammals, the cpt-1 of teleosts merely possessed two subtypes named cpt-1a and cpt-1b. In addition, the cpt-1a of fish existed multiple isoforms, and different fish exhibited different expression profiles. Studies indicated that the cpt-1a of fish was mainly expressed in liver, muscle and heart (Zheng et al., 2013; Wu et al., 2016), and the cpt-1b was primarily distributed in white muscle, heart and adipose tissues (Shi et al., 2017; Chao et al., 2024). In the present study, the hepatopancreas showed markedly higher expression levels of cpt-1a than other tissues. This result was in accordance with the study of Liu et al. (2018), who reported that the cpt-1a of male and female E. sinensis was chiefly distributed in hepatopancreas compared with other tissues. The hepatopancreas of crustaceans has been regarded as center of lipid metabolism, which was akin to fat body in insects and liver in vertebrates (Wen et al., 2001). The present results may suggest that the cpt-1a play an important role in lipolysis of hepatopancreas in mud crab. Besides, the present study found that the muscle, eyestalk and heart were main sites of cpt-1b expression, which may indicate that the cpt-1b involved in β-oxidation of fatty acids in these tissues. Consistently, study in E. sinensis indicated that the cpt-1b was mainly expressed in muscle and heart (Liu et al., 2018). Similar results were also observed in M. albus (Chao et al., 2024), P. fulvidraco (Zheng et al., 2013), S. aurata (Boukouvala et al., 2010) and C. idellus (Shi et al., 2017). The results above suggested that the cpt-1a and cpt-1b of mud crab may belonged to liver and muscle isoforms, respectively. There were relatively few studies about cpt-2. The present study showed that the highest expression level of cpt-2 was the hepatopancreas among all the tissues, which was similar to cpt-1a. This result was in in agreement with the studies of E. sinensis (Liu et al., 2018), while Chao et al. (2024) and Shi et al. (2017) reported that the cpt-2 of C. idellus and M. albus was highly expressed in heart and stomach, respectively. The high expression of cpt-2 in hepatopancreas further supported that the cpt played a vital role in β-oxidation of fatty acids in hepatopancreas of mud crab.
Based on the important functions of β-oxidation of fatty acids in energy homeostasis, numerous studies have been performed to determine the regulatory mechanisms and influence factors of the β-oxidation of fatty acids in teleost (Gutières et al., 2003; Boukouvala et al., 2010; Zheng et al., 2013; Bayır et al., 2020; Li et al., 2020; Liu Q. et al., 2020; Liu Y. et al., 2020). The findings showed that the regulatory mechanisms primarily involved in the malonyl-CoA, hormone, nuclear receptor and microRNA, and the influence factors mainly included physiology, environment and diets (Bonnefont et al., 2004; Morash and McClelland, 2011; Ning et al., 2019; Li et al., 2020). By contrast, the studies about oxidation of fatty acids in crustaceans were still in infancy, which was only reported in E. sinensis, and the results found that the blend vegetable oil (soybean oil: rapeseed oil, 1: 1) instead of fish oil could markedly up-regulated the expression levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas of E. sinensis (Liu et al., 2018). Salinity stress and food shortages are two important factors that aquatic animals often need to confront, and this process is closely related to energy metabolism. Therefore, in the present study, we investigated the roles of cpt-1a, cpt-1b and cpt-2 in coping with starvation and ambient salinity stress. The results of acute low salinity stress showed that the transcriptional levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas were markedly increased in 14‰ and 4‰ groups at the 6 h and 48 h, while the mRNA levels of these three genes in muscle were markedly up-regulated in 14‰ and 4‰ groups at the 12 h and 24 h. These results indicated that the cpt-1a, cpt-1b and cpt-2 of hepatopancreas in mud crab responded to acute salinity stress earlier than muscle, and hepatopancreas and muscle showed an alternate response pattern. Similar results were also observed in the findings of Hong et al. (2019) and Zeng et al. (2004), who found that the expression of cpt was significantly increased under the condition of acute salinity stress. Consistently, the cpt-1a, cpt-1b and cpt-2 of mud crab were also significantly increased under chronic salinity conditions. The results above suggested that the cpt-1a, cpt-1b and cpt-2 of mud crab played an important role in providing energy for coping with salinity stress. As for most animals in the starvation period, especially the long-term starvation period, fatty acids are the main source of energy, and the animals meet the energy demand through regulating β-oxidation of fatty acids. Studies found that the cpt-1a expression levels of L. crocea and S. hasta were extremely up-regulated after fasting (Wang et al., 2019; Zhou et al., 2021), and Arslan et al. (2020) reported that starvation could markedly increase the transcriptional level of cpt-1b in Salmo trutta. In addition, fasting gave rise to a significant increase of CPT-1 enzymatic activity in liver of O. mykiss (Morash and McClelland, 2011). Similarly, the present study exhibited that the expression levels of cpt-1a, cpt-1b and cpt-2 in hepatopancreas and muscle were significantly up-regulated after fasting. In addition, the expression levels of cpt-1a in hepatopancreas as well as cpt-1a and cpt-1b in muscle showed more significant differences, indicating their important roles in β-oxidation of fatty acids in hepatopancreas and muscle during fasting, which also further supported the results of tissue distribution.
5 Conclusion
The full-length cDNA sequences of cpt-1a, cpt-1b and cpt-2 were isolated from the hepatopancreas of mud crab using the RACE technology in the present study, and this was first time to obtain complete sequences in crustaceans. Subsequently, we identified the homology, structural characteristics and phylogenetic tree of CPT-1A, CPT-1B and CPT-2, and compared the expression discrepancy of cpt-1a, cpt-1b and cpt-2 in different tissues. Finally, based on their functions in regulating energy homeostasis, the present study further systematically investigated the roles of cpt-1a, cpt-1b and cpt-2 in response to fasting and salinity stress. These results were conducive to increasing the knowledge of cpt phylogenetic evolution as well as their roles in energy metabolism of crustaceans.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by The Committee on the Ethics of Animal Experiments of Ningde Normal University (NDNU-LL-202308). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZL: Writing – review and editing, Writing – original draft, Formal analysis, Data curation. CH: Writing – original draft, Methodology, Investigation, Formal analysis. ZZ: Writing – original draft, Methodology, Investigation, Formal analysis. JX: Writing – original draft, Methodology, Investigation, Formal analysis. HL: Writing – original draft, Methodology, Investigation, Formal analysis. BH: Writing – original draft, Methodology, Investigation, Formal analysis. CZ: Writing – original draft, Methodology. KH: Writing – original draft, Funding acquisition, Data curation, Conceptualization. WH: Writing – original draft, Funding acquisition, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Science Foundation of Fujian Province, China (Grant number 2022J05273), Scientific Research Foundation of Ningde Normal University (Grant number 2022Y04, 2023ZX505), the United Front Special project of Good Strategies for the Construction of New Fujian (Grant number JAT22118) and the Innovation and Entrepreneurship Training Program for Undergraduate of Ningde Normal University (Grant number X202310398004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1381263/full#supplementary-material
References
1
Akieda K. Takegawa K. Ito T. Nagayama G. Yamazaki N. Nagasaki Y. et al . (2024). Unique behavior of bacterially expressed rat carnitine palmitoyltransferase 2 and its catalytic activity. Biol. Pharm. Bull.47, 23–27. doi: 10.1248/bpb.b23-00612
2
Arslan G. Bayır M. Yağanoğlu A. M. Bayır A. (2020). Changes in fatty acids, blood biochemistry and mRNA expressions of genes involved in polyunsaturated fatty acid metabolism in brown trout (Salmo trutta) during starvation and refeeding. Aquac. Res.52, 494–504. doi: 10.1111/are.14908
3
Bartlett K. Eaton S. (2004). Mitochondrial β-oxidation. Eur. J. Biochem.271, 462–469. doi: 10.1046/j.1432-1033.2003.03947.x
4
Bayır M. Arslan G. Bayır A. (2020). Identification and characterization of carnitine palmitoyltransferase 1 (cpt 1) genes in nile tilapia, oreochromis niloticus. Evol. Bioinform.16, 1–7. doi: 10.1177/117693432091325
5
Berger J. Moller D. E. (2002). The mechanisms of action of PPARs. Annu. Rev. Med.53, 409–435. doi: 10.1146/annurev.med.53.082901.104018
6
Bonnefont J. Djouadi F. Prip-Buus C. Gobin S. Munnich A. Bastin J. (2004). Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol. Aspects Med.25, 495–520. doi: 10.1016/j.mam.2004.06.004
7
Boukouvala E. Leaver M. J. Favre-Krey L. Theodoridou M. Krey G. (2010). Molecular characterization of a gilthead sea bream (Sparus aurata) muscle tissue cDNA for carnitine palmitoyltransferase 1B (CPT1B). Comp. Biochem. Phys. B.157, 189–197. doi: 10.1016/j.cbpb.2010.06.004
8
Bu X. Li Y. Lai W. Yao C. Liu Y. Wang Z. et al . (2023). Innovation and development of the aquaculture nutrition research and feed industry in China. Rev. Aquacult, 1–16. doi: 10.1111/raq.12865
9
Chao B. Chen X. Huang S. Zhou Q. Xiong L. Zhang Y. (2024). Cloning and tissue distribution analysis of swamp eel (Monopterus albus) carnitine palmyltransferase gene. Genomics Appl. Biol., 1–16.
10
China Fishery Statistical Yearbook (2023). China fishery statistical yearbook (Beijing, China: China Agriculture Press), 28.
11
Cohen I. Kohl C. McGarry J. D. Girard J. Prip-Buus C. (1998). The N-terminal domain of rat liver carnitine palmitoyltransferase 1 mediates import into the outer mitochondrial membrane and is essential for activity and malonyl-CoA sensitivity. J. Biol. Chem.273, 29896–29904. doi: 10.1074/jbc.273.45.29896
12
De Paula I. F. Santos-Araujo S. Majerowicz D. Ramos I. Gondim K. C. (2023). Knockdown of carnitine palmitoyltransferase I (CPT1) reduces fat body lipid mobilization and resistance to starvation in the insect vector Rhodnius prolixus. Front. Physiol.14. doi: 10.3389/fphys.2023.1201670
13
Dreyer C. Krey G. Keller H. Givel F. Helftenbein G. Wahli W. (1992). Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell.68, 879–887. doi: 10.1016/0092-8674(92)90031-7
14
Esau C. Davis S. Murray S. F. Yu X. X. Pandey S. K. Pear M. et al . (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab.3, 87–98. doi: 10.1016/j.cmet.2006.01.005
15
Gutières S. Damon M. Panserat S. Kaushik S. Médale F. (2003). Cloning and tissue distribution of a carnitine palmitoyltransferase I gene in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Phys. B.135, 139–151. doi: 10.1016/S1096-4959(03)00074-5
16
Hong M. Li N. Li J. Li W. Liang L. Li Q. et al . (2019). Adenosine monophosphate-activated protein kinase signaling regulates lipid metabolism in response to salinity stress in the red-eared slider turtle Trachemys scripta elegans. Front. Physiol.10. doi: 10.3389/fphys.2019.00962
17
Jogl G. Hsiao Y. S. Tong L. (2004). Structure and function of carnitine acyltransferases. Ann. New York Acad. Sci.1033, 17–29. doi: 10.1196/annals.1320.002
18
Kerner J. Hoppel C. (2000). Fatty acid import into mitochondria. BBA-Mol. Cell Biol. L.1486, 1–17. doi: 10.1016/S1388-1981(00)00044-5
19
Lavarías S. Pasquevich M. Y. Dreon M. S. Heras H. (2009). Partial characterization of a malonyl-CoA-sensitive carnitine O-palmitoyltransferase I from Macrobrachium borellii (Crustacea: Palaemonidae). Comp. Biochem. Phys. B.152, 364–369. doi: 10.1016/j.cbpb.2009.01.004
20
Li L. Li J. Ning L. Lu D. Luo Y. Ma Q. et al . (2020). Mitochondrial fatty acid β-Oxidation inhibition promotes glucose utilization and protein deposition through energy homeostasis remodeling in fish. J. Nutr.150, 2322–2335. doi: 10.1093/jn/nxaa187
21
Lin Z. Wu Z. Huang C. Lin H. Zhang M. Chen M. et al . (2023). Cloning and expression characterization of elongation of very long-chain fatty acids protein 6 (elovl6) with dietary fatty acids, ambient salinity and starvation stress in Scylla paramamosain. Front. Physiol.14. doi: 10.3389/fphys.2023.1221205
22
Liu Y. Han S. Luo Y. Li L. Chen L. Zhang M. et al . (2020). Impaired peroxisomal fat oxidation induces hepatic lipid accumulation and oxidative damage in Nile tilapia. Fish Physiol. Biochem.46, 1229–1242. doi: 10.1007/s10695-020-00785-w
23
Liu Q. Liao Y. Wu Y. Xu M. Sun Z. Ye C. (2020). Cloning and characterization of carnitine palmitoyltransferase Iα (CPT1α) from obscure puffer (Takifugu obscurus), and its gene expression in response to different lipid sources. Aquacult. Rep.18, 100424. doi: 10.1016/j.aqrep.2020.100424
24
Liu L. Long X. Deng D. Cheng Y. Wu X. (2018). Molecular characterization and tissue distribution of carnitine palmitoyltransferases in Chinese mitten crab Eriocheir sinensis and the effect of dietary fish oil replacement on their expression in the hepatopancreas. PloS One13, e0201324. doi: 10.1371/journal.pone.0201324
25
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods.25, 402–408. doi: 10.1006/meth.2001.1262
26
Lopes-Marques M. Delgado I. L. S. Ruivo R. Torres Y. Sainath S. B. Rocha E. et al . (2015). The origin and diversity of Cpt1 genes in vertebrate species. PloS One10, e0138447. doi: 10.1371/journal.pone.0138447
27
McGarry J. D. Brown N. F. (1997). The mitochondrial carnitine palmitoyltransferase system-Fom concept to molecular analysis. Eur. J. Biochem.244, 1–14. doi: 10.1111/j.1432-1033.1997.00001.x
28
Morash A. J. McClelland G. B. (2011). Regulation of carnitine palmitoyltransferase (CPT) I during fasting in rainbow trout (Oncorhynchus mykiss) promotes increased mitochondrial fatty acid oxidation. Physiol. Biochem. Zool.84, 625–633. doi: 10.1086/662552
29
Ning L. Li J. Sun S. Du Z. (2019). Fatty acid β-oxidation in fish: a review. J. Fisheries China.43, 128–142. doi: 10.11964/jfc.20181211589
30
Price N. T. van der Leij F. R. Jackson V. N. Corstorphine C. G. Thomson R. Sorensen A. et al . (2002). A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics.80, 433–442. doi: 10.1006/geno.2002.6845
31
Shi X. Sun J. Yang Z. Li X. Ji H. Li Y. et al . (2017). Molecular characterization and nutritional regulation of carnitine palmitoyltransferase (CPT) family in grass carp (Ctenopharyngodon idellus). Comp. Biochem. Phys. B.203, 11–19. doi: 10.1016/j.cbpb.2016.08.006
32
Wang C. Si L. Li W. Zheng J. (2019). A functional gene encoding carnitine palmitoyltransferase 1 and its transcriptional and kinetic regulation during fasting in large yellow croaker. Comp. Biochem. Phys. B.231, 26–33. doi: 10.1016/j.cbpb.2019.01.015
33
Wang M. Wang K. Liao X. Hu H. Chen L. Meng L. et al . (2021). Carnitine palmitoyltransferase system: a new target for anti-inflammatory and anticancer therapy? Front. Pharmacol.12. doi: 10.3389/fphar.2021.760581
34
Wen X. Chen L. Ai C. Zhou Z. Jiang H. (2001). Variation in lipid composition of Chinese mitten-handed crab, Eriocheir sinensis during ovarian maturation. Comp. Biochem. Phys. B.130, 95–104. doi: 10.1016/S1096-4959(01)00411-0
35
Wu K. Zheng J.-L. Luo Z. Chen Q.-L. Zhu Q.-L. Wei H. (2016). Carnitine palmitoyltransferase I gene in Synechogobius hasta: Cloning, mRNA expression and transcriptional regulation by insulin in vitro. Gene.576, 429–440. doi: 10.1016/j.gene.2015.10.055
36
Xu Y. Luo Z. Wu K. Fan Y. You W. Zhang L. (2017). Structure and functional analysis of promoters from two liver isoforms of CPT I in grass carp Ctenopharyngodon idella. Int. J. Mol. Sci.18, 2405. doi: 10.3390/ijms18112405
37
Zeng L. Xiong Y. Song W. Xie Z. Wang Y. (2004). The effect and mechanism of salinity stress on energy metabolism and mitochondrial autophagy in large yellow croaker. Acta Hydrobiologica Sinica., 1–10.
38
Zheng J. Luo Z. Zhu Q. Chen Q. Gong Y. (2013). Molecular characterization, tissue distribution and kinetic analysis of carnitine palmitoyltransferase I in juvenile yellow catfish Pelteobagrus fulvidraco. Genomics.101, 195–203. doi: 10.1016/j.ygeno.2012.12.002
39
Zhou R. Wu G. Qu L. Zhong X. Gao Y. Ding Z. et al . (2021). Effect of starvation on intestinal morphology, digestive enzyme activity and expression of lipid metabolism-related genes in javelin goby (Synechogobius hasta). Aquac. Res.53, 87–97. doi: 10.1111/are.15555
Summary
Keywords
β-oxidation of fatty acids, carnitine palmitoyltransferase, fasting, salinity stress, Scylla paramamosain
Citation
Lin Z, Huang C, Zhuo Z, Xie J, Lan H, Hu B, Zhang C, Han K and Huang W (2024) Molecular cloning and characterization of three carnitine palmitoyltransferase (cpt) isoforms from mud crab (Scylla paramamosain) and their roles in respond to fasting and ambient salinity stress. Front. Mar. Sci. 11:1381263. doi: 10.3389/fmars.2024.1381263
Received
03 February 2024
Accepted
22 February 2024
Published
22 March 2024
Volume
11 - 2024
Edited by
Yiming Li, Fishery Machinery and Instrument Research Institute, China
Reviewed by
Yewei Dong, Zhongkai University of Agriculture and Engineering, China
Xueshan Li, Jimei University, China
Updates
Copyright
© 2024 Lin, Huang, Zhuo, Xie, Lan, Hu, Zhang, Han and Huang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunhuang Han, hankunhuang@Foxmail.com; Weiqing Huang, 07huangweiqing163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.