Abstract
Short neuropeptide F (sNPF) is a neuropeptide that widely distributed among arthropods. This neuropeptide has been proposed to play pleiotropic roles in insects, but its physiological functions in crustaceans are poorly understood. Here, we cloned the cDNA sequences of sNPF and its putative receptor (sNPFR) from the swimming crab, Portunus trituberculatus, and determined their possible roles during ovarian development. PtsNPF encodes three sNPF mature peptides with a conserved C-terminal “RLRFG” motif. All three PtsNPF peptides concentration-dependently activated PtsNPFR expressed in HEK293T cells, with EC50 values in the nanomolar range. PtsNPF and PtsNPFR transcripts showed a broad distribution among neural and non-neural tissues. During the ovarian development, expression of PtsNPF and PtsNPFR in hepatopancreas and ovary both increased to the highest levels at the late-vitellogenic stage, a period for rapid vitellogenesis. The in vitro experiments further showed that, among the three sNPF peptides, sNPF3 treatments can induce the vitellogenin (Vg) gene and protein levels in the hepatopancreas, as well as the Vg receptor (VgR) gene and protein levels and vitellogenin (Vn) deposition in the ovary. Opposing effects were observed for the dsPtsNPFR treatments, suggesting that PtsNPFR plays a role in the PtsNPF-mediated vitellogenesis. Additionally, it was found that the PtsNPF/PtsNPFR system can activate different second messengers species in hepatopancreas and ovary, suggesting it may act via different G proteins.
1 Introduction
The short neuropeptide F (sNPF) is a small neuropeptide, characterized by an M/T/L/FRF/W/Yamide carboxyterminal motif (Bajracharya et al., 2014; Fadda et al., 2019). It typically consists of approximately 6 to 11 amino acids, in contrast to the NPF which ranges from 36 to 40 amino acids in length (Lu and Pietrantonio, 2011; Nässel and Wegener, 2011). Despite sharing carboxyterminal sequence similarities with NPF, sNPF is evolutionarily distant (Fadda et al., 2019). Unlike NPF, which is an ortholog of vertebrate neuropeptide Y (NPY), sNPF is only found in protostomian systems (Mirabeau and Joly, 2013). Similar to NPY and NPF, sNPF primarily plays a role in regulating feeding (Lee et al., 2004) and foraging (Root et al., 2011), but it also has other physiological functions, such as locomotion (Nässel and Wegener, 2011), sleep homeostasis (Juneau et al., 2019), reproduction (De Loof et al., 2001), etc. In Drosophila, sNPFs were found expressed in numerous diverse neurons, suggesting multiple distributed peptide functions (Nässel et al., 2008).
Neuropeptides mediate their biological effects mainly through specific receptors located on the cell surface (Jékely, 2013). The first insect sNPF receptor (sNPFR) was identified in Drosophila melanogaster as a G protein-coupled receptor (GPCR), which is a homologue of the vertebrate NPY type 2 receptor, and it was able to be activated by predicted sNPFs when expressed in both Xenopus oocytes and Chinese hamster ovary (CHO) cells (Mertens et al., 2002). The direct interaction of this receptor with sNPFs was subsequently confirmed by a radiolabeled sNPF-based binding assay. Since then, sNPFRs have been discovered in several other insects (Chen and Pietrantonio, 2006; Garczynski et al., 2007; Hauser et al., 2008; Liesch et al., 2013). Mutations or silencing of sNPFR were shown to result in abnormal feeding or foraging phenotypes, indicating its crucial role in the sNPF signaling system (Dillen et al., 2013; Li et al., 2022).
While the identification of the first crustacean sNPF dates back to 1998 (Sithigorngul et al., 1998), it is only in recent years that the widespread use of transcriptomics has facilitated a substantial discovery of crustacean sNPF peptides and their receptors (Christie, 2014; Christie et al., 2015; Suwansa-Ard et al., 2015; Nguyen et al., 2016; Bao et al., 2018b; Tu et al., 2021). The functional role of the sNPF system in crustaceans is primarily studied within the process of ovarian development. The administration of sNPFs was found to accelerate the ovarian development of Macrobrachium rosenbergii, as evidenced by a shorter ovarian development duration, and increasing gonadosomatic index (GSI), oocyte diameter (OD), cell proliferation and vitellogenin (Vg) concentration (Tinikul et al., 2023). This gonadal stimulating effect is consistent with the reported function of sNPF in the insect Leptinotarsa decemlineata (Cerstiaensa et al., 1999). However, it appeared that sNPFs may act as a negative regulator in vitellogenesis in the mud crab, Scylla paramamosain (Bao et al., 2018a). Although sNPF in the lepidopteran Bombyx mori is thought to exert its ovarian function via the gonadotropin juvenile hormone (JH) (Yamanaka et al., 2008; Kaneko and Hiruma, 2014), the presence of sNPFR in both insect and crustacean ovaries suggests a direct regulatory role in ovarian development (Lu and Pietrantonio, 2011; Bao et al., 2018a). Notably, crustacean sNPF is not restricted to the nervous system, but is also highly expressed in the ovary (Wang et al., 2022), suggesting its possible paracrine/autocrine functions in ovarian development.
For many economically important crustaceans, the ovarian development is not only crucial for the success of artificial breeding, but also determines their market value. The essential role of neuroendocrine regulation on crustacean ovarian development has long been a widespread consensus, but most studies were focused mainly on the CHH neuropeptide family, which has a direct or indirect inhibitory effect on vitellogenesis (Nagaraju, 2011; Webster et al., 2012). In fact, early studies have also revealed the existence of gonad-stimulating factor (GSF) in crustacean nervous systems, but its chemical nature is unclear (Nagaraju, 2011; Farhadi et al., 2021). In recent years, neuropeptides such as red pigment concentrating hormone (RPCH) and crustacean female sex hormone (CFSH) have been proposed to have gonad-stimulating functions, indicating that GSF may be a combination of multiple neuropeptides (Gong et al., 2015; Liu et al., 2018). It is still controversial whether sNPF promotes or inhibits ovarian development in crustaceans and there may be species differences (Bao et al., 2018a; Tinikul et al., 2023). Therefore, studying its regulatory function in ovarian development in more species is of great theoretical significance and can provide theoretical guidance for artificial breeding and cultivating.
In the present study, cDNAs of sNPF and its receptor were cloned from the swimming crab, Portunus trituberculatus, which is an economically crustacean species in southeastern coast of China. The P. trituberculatus sNPF (PtsNPF) prepropeptide encodes three mature sNPFs and their binding to PtsNPFR were validated in human embryonic kidney 293 (HEK293) cells using the dual-luciferase reporter system. Furthermore, the putative role of sNPF/sNPFR signaling on ovarian development were investigated using sNPF treatment and sNPFR silencing in vitro.
2 Materials and methods
2.1 Experiment animals and tissue sampling
Wild-caught female crabs (body weight, 90-300 g) were purchased from the local aquatic market in Guoju street, Ningbo City, Zhejiang Province, China. According to histological characteristics and gonadosomatic index (GSI), ovarian development was classified into four stages: pre-vitellogenin stage (GSI= 0.44 ± 0.12), early vitellogenin stage (GSI= 0.78 ± 0.16), middle vitellogenin stage (GSI= 3.28 ± 1.02), late vitellogenin stage (GSI= 7.42 ± 0.80) (Wu et al., 2007). Prior to sacrifice, all crabs were subjected to a 10-minute period of anesthesia on ice. Tissues including brain (Br), eyestalk (Es), epidermis (Ep), gill (Gi), heart (Ht), hepatopancreas (Hp), intestine (In), muscles (Ms), ovaries (Ov) and thoracic ganglion (TG) were collected for each crab and stored in RNA store (CWBIO, China) at -80°C before extraction.
2.2 RNA extraction, molecular clone and bioinformatics analysis
According to the protocol of manufacturer, total RNA was extracted from different samples using the RNA-Solv® Reagent (Omega Biotek, USA), and then dissolved in RNase-Free Water (Solarbio, China). The RNA concentrations were determined using a NanoDrop 2000 UV Spectrophotometer (Thermo Fisher Scientific, USA). Approximately 1 μg of total RNA was used for cDNA synthesis with the ABScript III RT Master Mix kit (ABclonal, China). The sequence of PtsNPF and PtsNPFR were obtained using a keyword-based screening of our RNAseq library. The open reading frames (ORF) for the two sequences were predicted using the ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), and validated using reverse-transcription PCR (RT-PCR). Gene specific primers (Supplementary Table 1) for sequence validation was designed using the Primer Premier 5.0 software and synthesized by Zhejiang Youkang Biotechnology Co., Ltd (Hangzhou, China). The RT-PCR was performed using 2×Es Taq MasterMix (Dye) (CWBIO, China). Amplifications were performed as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final elongation at 72°C for 10 min. The PCR products were verified on 1% agarose gel, purified with DiaSpin DNA Gel Extraction Kit (Sangon, China), ligated into PMD19-T vector (Takara, Japan), and then transformed into competent Escherichia coli DH5α cells. After incubation, five positive clones were selected for sequencing by Zhejiang Youkang Biotechnology Co., Ltd (Hangzhou, China).
The validated sequences were then subjected to SMART (http://smart.emblheidelberg.de/), SignalP 5.0 (https://services.healthtech.dtu.dk/services/SignalP-5.0/), and TMHMM-2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/) for prediction of conserved domains, signal peptide, and transmembrane regions, respectively. The mature peptide sequences of PtsNPF were deduced according to Veenstra (2011). For PtsNPFR, multiple sequence alignments were conducted using ClustalX software, and the resultant files were imported to the Jalview 2.11.2 software to identify conserved sequence motifs. The phylogenetic tree was constructed using the MEGA7.0.14 software with the maximum likelihood method based on the LG + G+I amino acid model, and bootstrapped with 1000 replications. The species sequence accession numbers are shown in Supplementary Table 2.
2.3 Cell culture and transient transfection
The plasmid for transient transfection was constructed by inserting the ORF of PtsNPFR into the expression vector pEGFP-N1 using restriction enzymes BamH Ⅰ and Hind III (NEB, USA). The construct was sequenced to validate the sequence and orientation. Human embryonic kidney cell lines (HEK293T) were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM, Corning, USA) supplemented with 10% fetal bovine serum (Bovogen, Australia), 100 U/mL penicillin, and 100 mg/mL streptomycin (Hyclone, USA). Cells were maintained in T25 flasks at 37°C in a humidified incubator containing 5% CO2. Cells were seeded overnight in a 60 mm culture dish and transiently co-transfected with 3 μg of the PtsNPFR/pEGFP-N1 plasmid, 3 μg of the reporter gene pCRE-luc, and 1.2 μg of internal control gene pRL-TK using 15 μL Lipofectamine 3000 transfection reagent following the manufacturer’s instructions. The empty pEGFP-N1 plasmid was used for mock transfection.
2.4 Confocal microscopy
To verify the membrane localization of PtsNPFR, the HEK293T cells transiently transfected with PtsNPFR/pEGFP-N1 plasmid were used for confocal microscope analysis. The transfected cells were fixed with 4% paraformaldehyde (PFA) for 20 min, and then stained with the cell membrane probe Dil (Beyotime, China) at 37°C for 10 min. After removing the Dil solution, the cells were washed three times with PBS and further incubated with a nuclear dye DAPI (Beyotime, China) at 37°C for 10 min. After removing the DAPI solution, the cells were washed three times with PBS, mounted in an antifade mounting medium (Solarbio, China), and imaged using a Zeiss laser scanning confocal microscope (LSM880, 294 Carl Zeiss, Oberkochen, Germany). The fluorescence detection was used green channel of 505-550 nm, blue channel of 430-490 nm and red channel of 550-585 nm with excitation of 488 nm, 405 nm and 557 nm respectively.
2.5 Dual-luciferase reporter assays
The mature PtsNPF peptides (APPSMRLRFG-NH2, SMPTLRLRFG-NH2, KDARTPALRLRFG-NH2) were synthesized by Sangon Biotech (Shanghai, China), with a purity of 98%. The transfected HEK293T was co-incubated with synthetic sNPF peptide in various concentrations (10-5-10-12 M). After incubation for 10 h, ligand-induced changes in luciferase activity were detected by the Dual-Luciferase® Reporter Assay System kit (Promega, USA). The dose-response curve was established and fitted in the Logistic equation utilizing GraphPad Prism 8.02.
2.6 In vitro assays
Female crabs at the vitellogenic stage (GSI= 4.12 ± 0.44) were selected, their hepatopancreas and ovaries were dissected, and an in vitro culture system was constructed according to the reported method (Tu et al., 2022). The in vitro experiments were divided into seven groups: Group 1 was the control group and treated with crab saline; Groups 2-4 were treated with three mature peptides, respectively; Groups 5-6 were treated with dsGFP and dssNPFR, respectively; and Group 7 was the recovery group and co-treated with dssNPFR and a mixture of sNPF peptides. The dosage for sNPF peptide treatments was set as 10-6 M, which was determined according to our pre-experiments (Supplementary Figure 3). For RNAi, cDNA fragments of the PtsNPFR (574 bp) and the green fluorescent protein (GFP, 568 bp) were cloned using specific primers with T7 promoter (Supplementary Table 1), and the dsRNA was prepared as previously described (Tu et al., 2022). The dosage for dsRNA treatments was 3 μg per well. Prior to the commencement of the experimental procedures, the tissues were incubated at 25°C for a period of 1 h. The tissues were collected at 6 and 12 h post treatments for subsequent qPCR and ELISA analysis, respectively.
2.7 Gene expression analysis
The relative mRNA expression levels were determined using quantitative real-time PCR (qPCR) with qPCR primers (Supplementary Table 1). The qPCR was carried out using the QuantStudio 3 Real-Time PCR System (Thermofisher, USA) according to the manufacturer’s instructions of the 2 × Universal SYBR Green Fast qPCR Mix (ABclonal, China). PCR conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Additional melting curve analysis was performed to confirm the product specificity, which the temperature increasing from 55 to 95°C with at a rate of 0.2°C/s. The amplification efficiencies were evaluated using the standard curve analysis by preparation of a 5-point 1:10 dilution series of cDNA. The β-actin gene was used as an internal control. For each sample, the reactions were carried out in triplicate for technical replicates. The relative mRNA expression levels were calculated using the comparative Ct (2−ΔΔCt) method (Livak and Schmittgen, 2001).
2.8 Detection of Vg, Vn, VgR, cAMP and IP3 levels
Commercial ELISA kits (Jiangsu Kete Biotechnology, China) were used to detect the contents of Vg (KT95204-A), Vn (KT300356-A), VgR (KT300343-A), and cAMP (KT0023-0A) and IP3 (KT300318-A) in the ovary and hepatopancreas explants. Briefly, 50 mg of ovary and hepatopancreas tissue fragments were homogenized with 500 μL cell lysis buffer, 10 μL protease inhibitor, 10 μL phosphatase inhibitor and 10 μL EDTA, and then centrifuged at 12000 ×g for 5 min twice at 4°C. The supernatants were collected for ELISA analysis. Furthermore, given the Vg synthesized by the hepatopancreas might be secreted into the medium, we also collected the culture medium of hepatopancreas (10 μL) for ELISA analysis.
2.9 Statistical analysis
Data were presented as the mean ± standard error of the mean (SEM), and subjected to normality testing using the Kolmogorov–Smirnov and Cochran tests prior to all statistical tests. Data that did not show a normal distribution were analyzed using the nonparametric test (Mann-Whitney). Data that showed a normal distribution were analyzed using one-way ANOVA followed by the Tukey’s test or the student’s t-test (SPSS 24.0 software). In all cases, statistical significance was accepted at P < 0.05 and noted with letters or asterisks.
3 Results
3.1 PtsNPF gene encodes three mature peptides hallmarked with conserved RLRFG motif
PtsNPF encodes 126 amino acids and consists of one signal peptide, three mature peptides, six cleavage sites and one precursor-associated peptide (Supplementary Figure 1). The predicted sNPF mature peptide sequences are APPSMRLRFG-NH2 (sNPF1), SMPTLRLRFG-NH2 (sNPF2) and KDARTPALRLRFG-NH2 (sNPF3), which are nearly identical to those identified from other crabs, and hallmarked with a conserved C-terminal RLRFG motif. In Table 1, we summarized the sNPF mature peptide sequences from a number of representative crustacean and insect species. It was found that all crustacean sNPF prepropeptides encode three mature peptides, compared to 2-4 sNPFs found in insect. Moreover, in addition to the conserved “RLRFG”, insect sNPFs have more variable C-terminal motifs such as “RLRWG” and “HLRYG”.
Table 1
| Species | Sequence | |||
|---|---|---|---|---|
| P. trituberculatus | APPSMRLRFG | SMPTLRLRFG | KDARTPALRLRFG | |
| C. arcuatus | APPSMRLRFG | SMPSLRLRFG | KDARTPALRLRFG | |
| S. paramamosain | APPSMRLRFG | SMPTLRLRFG | KDARTPALRLRFG | |
| E. sinensis | APPSMRLRFG | SMPSLRLRFG | KDARTPALRLRFG | |
| G. lateralis | AQPSMRLRFG | SMPSLRLRFG | KDARTPALRLRFG | |
| P. clarkii | APPSMRLRFG | SMPSLRLRFG | KDSRTPALRLRFG | |
| C. quadricarinatus | APPSMRLRFG | SMPSLRLRFG | KDSRIPALRLRFG | |
| N. norvegicus | GPPSLRLRFG | SMPSLRLRFG | KDTSTPRLRLRFG | |
| P. vannamei | AQPSMRLRFG | SMPSLRLRFG | KDGRTPALRLRFG | |
| P. monodon | AQPSMRLRFG | SMPSLRLRFG | KDGRTPALRLRFG | |
| P. japonicus | AQPSMRLRFG | SMPSLRLRFG | KDGRTPALRLRFG | |
| D. melanogaster | AQRSPSLRLRFG | SPSLRLRFG | KPQRLRWG | KPMRLRWG |
| A. albopictus | APQLRLRFG | SPSLRLRFG | APSQRLRWG | |
| A. gambiae | APQLRLRFG | SPSLRLRFG | APSQRLRWG | APTQRLRWG |
| M. vetustissima | KPQRLRFG | SPSLRLRFG | KPAPLRLRWG | |
| B. mori | APSMRLRFG | SPSRRLRFG | TPVRLRFG | |
| H. armigera | APSIRLRFG | SPSRRLRFG | TPVRLRFG | |
| S. gregaria | SNRSPSLRLRFG | SPSLRLRFG | ||
| A. aegypti | APQLRLRFG | SPSLRLRFG | APSQRLRWG | |
sNPF mature peptides in different crustaceans and insects.
Conservative motifs are underlined.
3.2 PtsNPFR can be activated by all three mature sNPF peptides
PtsNPFR encodes 458 amino acids and has seven transmembrane domains typical of the GPCR family. Phylogenetic analysis showed that PtsNPFR clusters with other known crustacean sNPFRs, and they are more closely to the mammalian type 2 NPYR, but relatively distant from insect sNPFRs (Supplementary Figure 2). Confocal microscopy showed that PtsNPFR/pEGFP-N1 mainly localized to the cell membrane (Figure 1A), indicating the PtsNPFR is successfully expressed in HEK293T cells. The dual-luciferase report assays showed the PtsNPFR could be activated by all three sNPF peptides, in a concentration-dependent manner. The median effective concentration (EC50) values were 3.0, 40.2, and 0.8 nM for sNPF1, sNPF2, and sNPF3, respectively, while HEK293T cells transfected with empty pEGFP-N1 showed no responses to any sNPF peptide (Figure 1B).
Figure 1
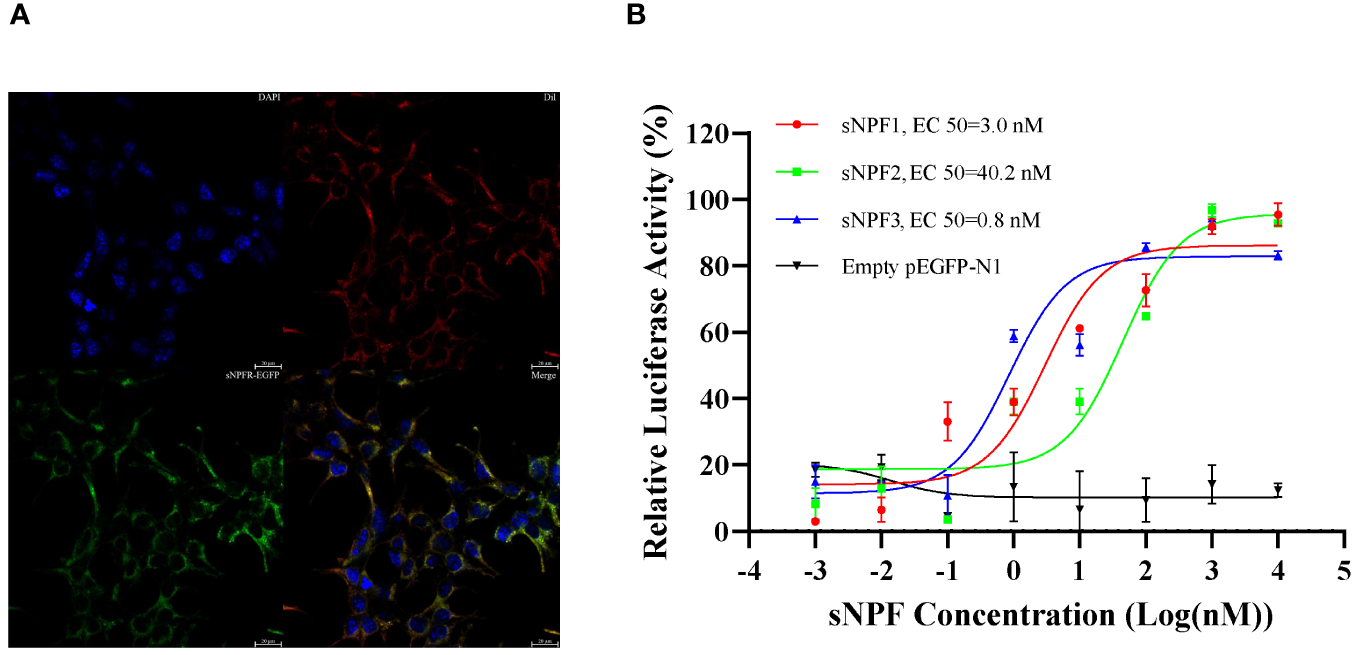
Expression of PtsNPFR in HEK293T cells and ligand-receptor activation of signaling. (A) Cells expressing PtsNPFR/pEGFP-N1 fusion protein were stained with a nuclei probe (DAPI) and a membrane plasma probe (Dil). (B) HEK293T transiently co-transfected with PtsNPFR/pEGFP-N1, pCRE-luc and pRL-TK were treated with different doses of PtsNPF1, PtsNPF2, and PtsNPF3 and the concentration of cAMP was determined. Values are plotted as mean ± SEM from three biological replicates.
3.3 PtsNPF may promote Vg synthesis and Vn deposition
qPCR analysis showed that PtsNPF and PtsNPFR have a broad tissue distribution in P. trituberculatus (Figures 2A, B). In addition to their high expression in neural tissues, their transcripts were also presented in reproductive-related tissues such as hepatopancreas and ovary. During the ovarian development, the expression of PtsNPF and PtsNPFR in hepatopancreas and ovary both increased to the highest levels at the late vitellogenic stage (Figures 2C, D), a stage for rapid vitellogenesis. We therefore investigated the in vitro effects of the three sNPF peptides on the vitellogenesis-related indexes in hepatopancreatic and ovarian explants. Among the three sNPF peptides, sNPF3 had the most notable effects, which includes the transcriptional activation of sNPFR in both tissues (Figure 3A), the elevation of Vg gene and protein levels in the hepatopancreas (Figure 3B), and the elevation of VgR gene and protein levels and Vn deposition in the ovary (Figures 3C, D). In contrast, sNPF1 and sNPF2 were only found to promote the hepatopancreas sNPFR expression and ovarian Vn deposition, while showed barely effect on other indexes.
Figure 2
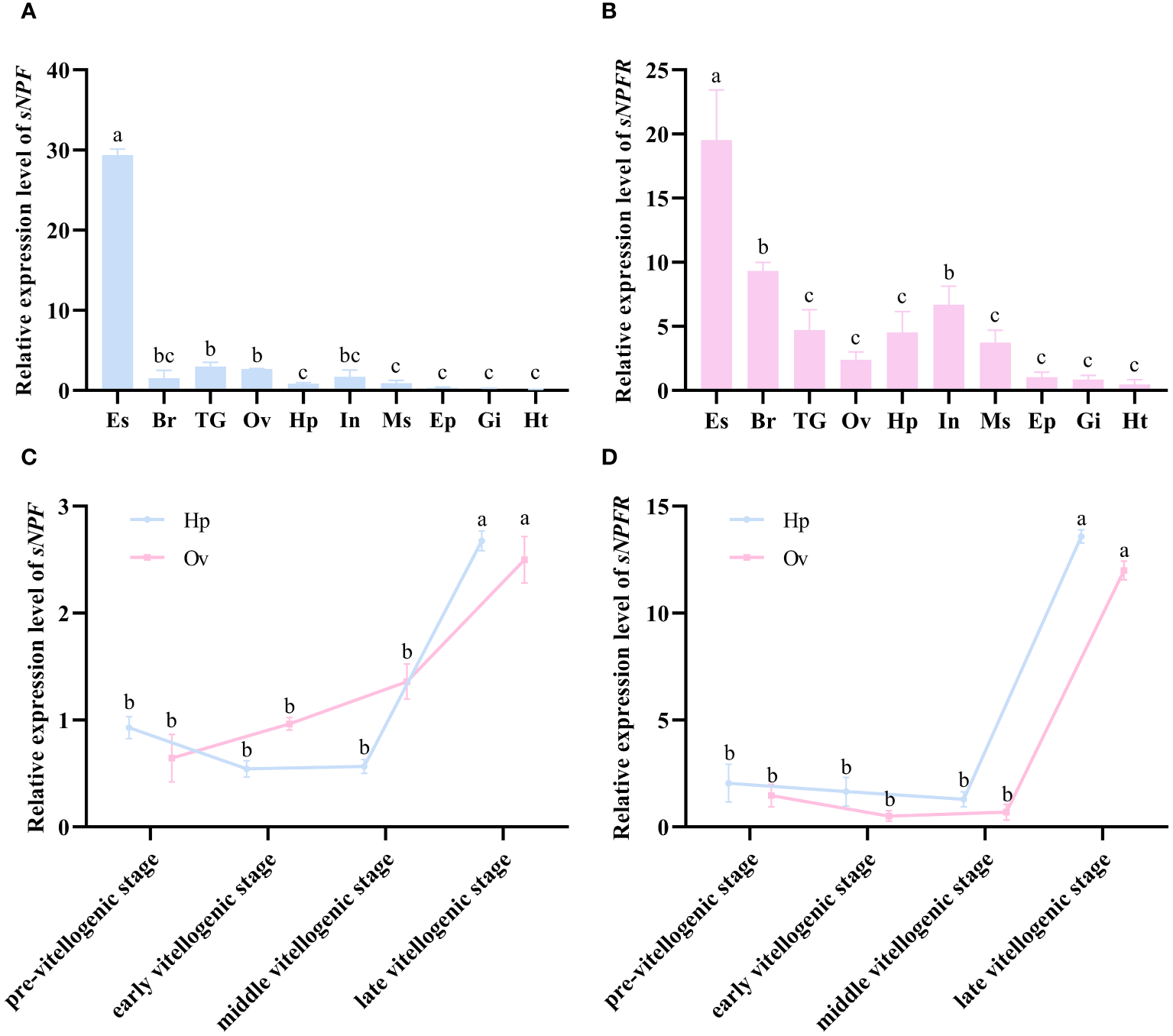
Expression profiles of PtsNPF and PtsNPFR in different tissues and during the ovarian development. The tissue distribution of PtsNPF(A) and PtsNPFR(B) in female adults. The stage-specific expression of PtsNPF(C) and PtsNPFR(D) during the ovarian development in hepatopancreas (Hp) and ovary (Ov) respectively. Tissue abbreviations are as follows: Es, eyestalk; Br, brain; TG, thoracic ganglion; Ov, ovary; Hp, hepatopancreas; In, intestine; Ms, muscle; Ep, epidermis; Gi, gill; Ht, heart. Bars represent mean ± SEM (n=3). Different letters indicate statistically significant differences (P < 0.05) within each tissue.
Figure 3
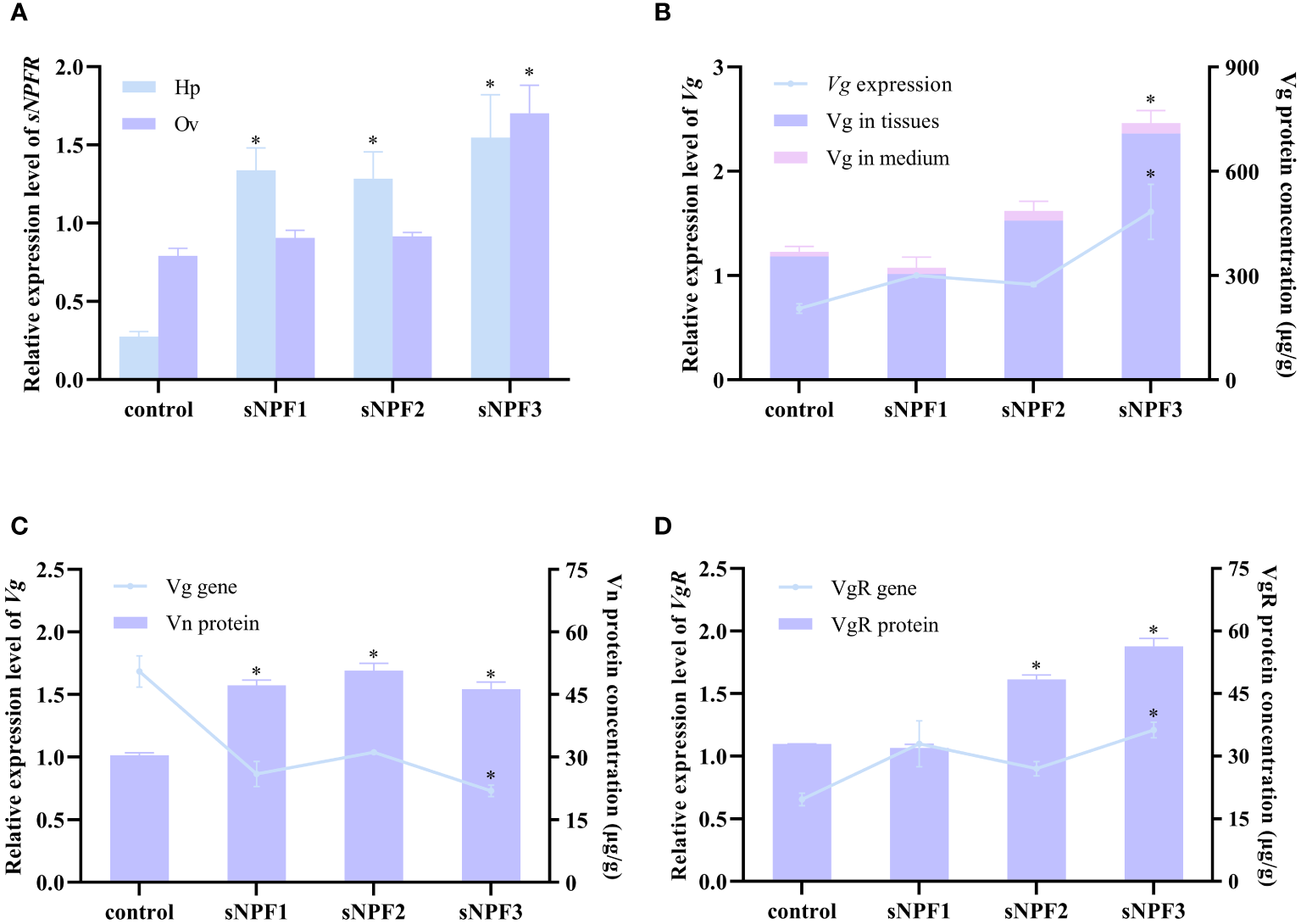
Effects of PtsNPF mature peptides on hepatopancreas and ovary explants. (A) Effects of PtsNPF peptides on PtsNPFR expression in hepatopancreas (Hp) and ovary (Ov). (B) Effects of PtsNPF peptides on Vg gene expression and protein levels in hepatopancreas explants. (C) Effects of PtsNPF peptides on Vg gene expression and Vn levels in ovary explants. (D) Effects of PtsNPF peptides on VgR gene expression and protein levels in ovary explants. Data are shown as mean ± SEM (n=3). “*” represents significant differences (P < 0.05) from the control group.
3.4 PtsNPFR is involved in the PtsNPF-mediated vitellogenesis
The role of PtsNPFR on vitellogenesis was further investigated using RNAi. In both hepatopancreas and ovary explants, dsPtsNPFR treatment showed a good RNA silencing effect. The knockdown of PtsNPFR resulted in the down-regulation of Vg gene and protein expression in the hepatopancreas, as well as the down-regulation of VgR gene and protein expression, the up-regulation of Vg gene expression and the decreased Vn levels in the ovary (Figure 4). The opposing effects of dsPtsNPFR to those observed with the sNPF3 treatment suggest that PtsNPFR plays a role in the PtsNPF-mediated vitellogenesis. However, co-treatment with dsPtsNPFR and the mixture of PtsNPF peptides demonstrated no significant effects on the aforementioned indexes when compared to the control and dsGFP groups. This indicates that the dsNPFR did not completely block the effect of PtsNPF.
Figure 4
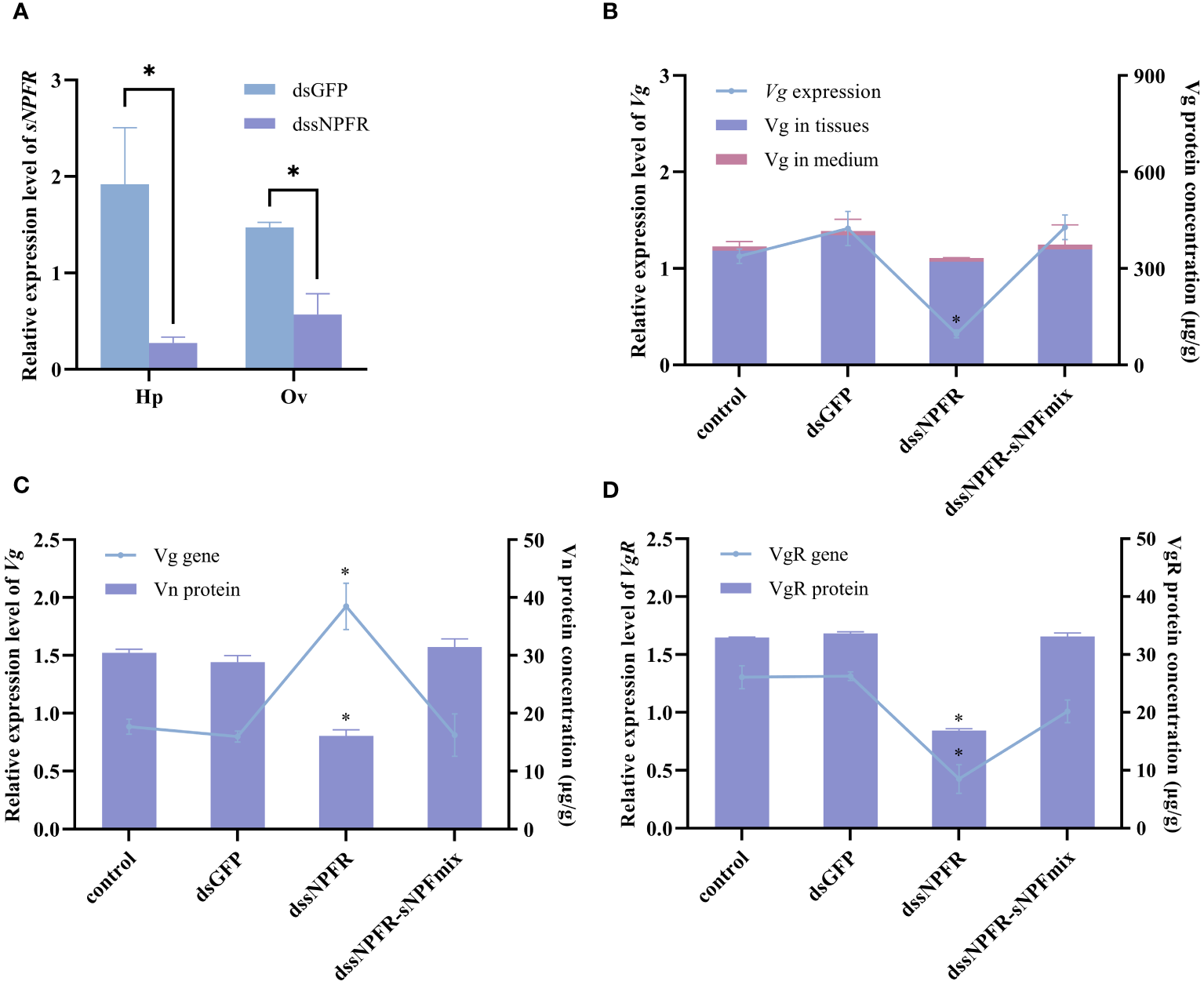
Effects of dssNPFR on hepatopancreas and ovary explants. (A) Silencing efficiency of dssNPFR in hepatopancreas (Hp) and ovary (Ov). (B) Effects of dssNPFR on Vg gene expression and protein levels in hepatopancreas explants. (C) Effects of dssNPFR on Vg gene expression and Vn levels in ovary explants. (D) Effects of dssNPFR on VgR gene expression and protein levels in ovary explants. Data are shown as mean ± SEM (n=3). “*” represents significant differences (P < 0.05) from the dsGFP group.
3.5 PtsNPF/PtsNPFR activates different second messenger species in hepatopancreas and ovary
In order to investigate the pathways of PtsNPF/PtsNPFR signaling, the levels of cAMP and IP3, two key messenger molecules in GPCR signaling, were further examined in hepatopancreas and ovary. The results demonstrated that sNPF3 was capable of elevating the cAMP level in the hepatopancreas, yet had no impact on IP3. Conversely, in the ovary, all three sNPF peptide treatments were observed to elevate the cAMP and IP3 levels. Treatment with dssNPFR was found to reduce the levels of cAMP in the hepatopancreas but had no effect on IP3, and both messenger molecule levels in the ovary were affected after treatment (Figure 5). This indicates that the PtsNPF/PtsNPFR system may have different second messenger-mediated signaling pathways in the hepatopancreas and ovary.
Figure 5
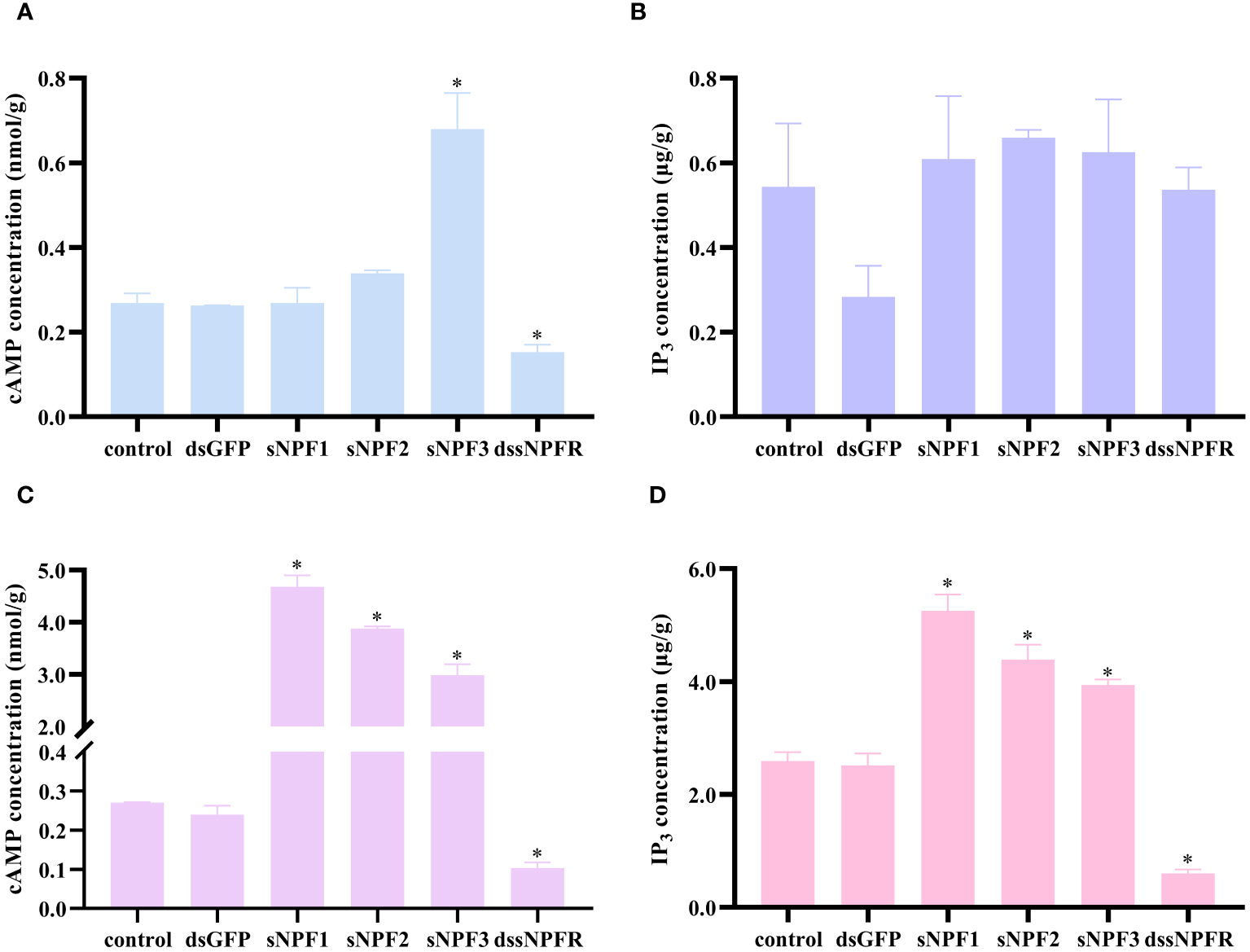
Effects of PtsNPF/PtsNPFR system on second messenger levels in hepatopancreas and ovary explants. Effects of PtsNPF peptides and dssNPFR on the levels of cAMP (A) and IP3(B) in hepatopancreas explants. Effects of PtsNPF peptides and dssNPFR on the levels of cAMP (C) and IP3(D) in ovary explants. Data are shown as mean ± SEM (n=3). “*” represents significant differences (P < 0.05) from the control group.
4 Discussion
In the present study, the cDNAs of sNPF and its putative receptor (sNPFR) were cloned and characterized in the swimming crab, P. trituberculatus. As in most other crustaceans, the PtsNPF prepropeptide can be cleaved into three mature peptides with conserved RLRFG motif. Moreover, the sequences of the three mature peptides are very similar across crustaceans, reflecting the high degree of conservation of their precursors. By contrast, the insect sNPFs are more variable, with the number of encoded sNPF mature peptides varying from 2-4 (Mertens et al., 2002; Nagata et al., 2012; Dillen et al., 2013), and some species having C-terminal sequences of RLRWG (Nässel and Wegener, 2011) or HLRYG (Bajracharya et al., 2014). Thus, the sNPF gene appears to evolve more rapidly in insects. On the other hand, although multiple alignments showed the sNPFR gene is quite conserved in arthropods (Nässel and Wegener, 2011; Bao et al., 2018a), the crustacean sNPFRs are more closely related to the vertebrate NPY type 2 receptor (NPY R2) than to the insect sNPFRs in our phylogenetic analysis. This suggests that the crustacean sNPF/sNPFR system may be evolutionarily older than that of insects.
The luciferase reporter gene system demonstrated that PtsNPFR expressed in HEK293T cells can be activated by three synthetic PtsNPF maturation peptides in a dose-dependent manner. The half-maximal effective concentration (EC50) values for PtsNPF1, PtsNPF2, and PtsNPF3 were found to be 3.0, 40.2, and 0.8 nM, respectively. This nanomolar range of sNPFR activation by its ligands is comparable to that observed in many other reports (Mertens et al., 2002; Dillen et al., 2013; Liesch et al., 2013; Bajracharya et al., 2014; Caers et al., 2016; Jiang et al., 2017). In some species, however, relatively higher concentrations of sNPF were required to activate the sNPFR, e.g., EC50 values for the three sNPF peptides were 1.2, 3.8 and 8.5 µM in the mud crab, S. paramamosain (Bao et al., 2018a), and 1.1, 2.1 and 4.1 µM in the mollusk Crassostrea gigas (Bigot et al., 2014). It is noteworthy that in Anopheles gambiae and D. melanogaster, their sNPFR had higher affinity for longer sNPFs (Feng et al., 2003; Reale et al., 2004; Garczynski et al., 2006, Garczynski et al., 2007). Similarly, among the three sNPF peptides of P. trituberculatus, sNPF3 has more amino acid residues than sNPF1 and sNPF2 and its affinity for PtsNPFR is also the highest. Our subsequent experiments also showed that sNPF3 had better effect in inducing vitellogenesis in vitro.
In addition to their presence in the nervous system, PtsNPF and PtsNPFR were also found to be widely expressed in non-neural tissues. This is consistent with the multifunctional role proposed for the sNPF/sNPFR system in insects (De Loof et al., 2001; Lee et al., 2004; Nässel et al., 2008; Nässel and Wegener, 2011; Root et al., 2011; Juneau et al., 2019). Both PtsNPF and PtsNPFR were expressed in hepatopancreas and ovary, the female reproductive tissues. Furthermore, their expression exhibited a consistent trend of change during ovarian development, which lends support to the hypothesis that sNPF plays a paracrine/autocrine role in the regulation of ovarian development (Bao et al., 2018a). The expression of PtsNPF and PtsNPFR in both tissues exhibited a significant increase during the late-vitellogenic period, which is characterized by a rapid vitellogenesis. This suggests that the sNPF/sNPFR system may promote vitellogenesis in P. trituberculatus.
Our in vitro experiments further support that the PtsNPF/PtsNPFR has a stimulatory effect on ovarian development of P. trituberculatus. As previously stated, sNPF3 exhibited a significant effect among the three peptides in hepatopancreas, which may be attributed to its higher receptor activation capacity, while three peptides all exhibited a significant effect in ovary. The effects of sNPFs treatment included elevation of Vg gene and protein levels in the hepatopancreas, elevation of VgR gene and protein levels and Vn deposition in the ovary. In contrast, interference with PtsNPFR reversed the majority of effects in sNPF3 treatment, indicating that PtsNPFR plays a pivotal role in mediating the PtsNPF functions. However, co-treated of dsPtsNPFR and sNPF peptides showed no significant difference with the crab saline group and dsGFP group in the aforementioned indexes. This could be due to two reasons: first, the treatment of dsPtsNPFR was not a complete silencing, and the increased sNPF peptide complemented the signaling strength of the PtsNPF/PtsNPFR system; second, there may be additional receptors for PtsNPF.
Our results are similar to the sNPF effects on ovarian development in giant freshwater prawn, M. rosenbergii (Tinikul et al., 2023), but discrepant to those reported in in the mud crab, S. paramamosain (Bao et al., 2018a). In the latter study, sNPF had an inhibitory effect on vitellogenesis. It is noteworthy that the regulation of ovarian development by the sNPF system is also species-specific in insects (Cerstiaensa et al., 1999; Kamruzzaman et al., 2020). However, it should be emphasized that the regulation of ovarian development by the sNPF system may involve diverse mechanisms, rather than solely its direct effects. In insects, sNPF has also been implicated in the regulation of ovarian development by modulating the synthesis of juvenile hormone (JH) (Yamanaka et al., 2008; Kaneko and Hiruma, 2014), the primary reproductive hormone in many insects. In crustaceans, it has been widely accepted that methyl farnesoate (MF), the non-epoxide form of JH III, also plays a vital role in ovarian development (Borst et al., 1987). It was found that PtsNPFR is highly expressed in the mandibular organ, the site of MF synthesis (data not shown). This suggests that the crustacean sNPF/sNPFR system may also be involved in MF synthesis. Although the potential endocrine axis of “sNPF-MF-ovarian development” was not investigated in this paper, it is reasonable to speculate that the effects of the sNPF system may differ in vitro and in vivo.
For most GPCRs, upon ligand activation, the receptor is enriched for different G proteins (Gs, Gi, Gq), which regulate the production of key effectors and second messengers (cAMP, IP3) and activate different cascades of signals (Rosenbaum et al., 2009). To investigate the possible pathways downstream PtsNPFR, we further tested the cAMP and IP3 levels in above treatments. In hepatopancreas, sNPF3 significantly elevated the level of cAMP but had no effect on the level of IP3, implying that Vg synthesis may be primally regulated by PKA and the intracellular cAMP level plays pivotal roles in regulating the expression of Vg (Chen et al., 2018). This result also explains the weak effect of sNPF1 and sNPF2 on hepatopancreas Vg synthesis. Similarly, all three sNPF peptides were able to increase the cAMP and IP3 levels in ovary, also explains their abilities in inducing ovary-related indexes. The differ in the species of second messengers changed by sNPF peptides in ovary and hepatopancreas suggests PtsNPF/PtsNPFR may make use of different signaling pathways in two tissues, interacting with Gq or Gs to activate subsequent pathways. Currently, the G protein species utilized by sNPFR remain elusive. In S. paramamosain, sNPF-sNPFR binding can result in cAMP accumulation and Ca2+ mobilization (Bao et al., 2018a), which is similar to our results. However, sNPFR appears to act via the Gi pathway in A. gambiae (Garczynski et al., 2007). Our results suggest that there may be tissue-specific utilization of G proteins by the sNPF/sNPFR system, which may have contributed to its diverse physiological functions.
In conclusion, the present study cloned the cDNA sequences of PtsNPF and its receptor PtsNPFR. PtsNPF encodes three sNPF mature peptides with conserved N-terminal “RLRFG” motifs, and all three peptides can activate the PtsNPFR expressed in HEK293T cells. Using sNPF treatments and sNPFR RNAi in vitro, we showed that PtsNPF/PtsNPFR signaling can directly promote the synthesis of vitellogenin (Vg) in hepatopancreas as well as the uptake of Vg and the deposition of vitellogenin (Vn) in ovaries. However, the indirect effects of PtsNPF on ovarian development may require further investigation. Additionally, by testing the cAMP and IP3 levels in different treatment groups, it was found that the PtsNPF/PtsNPFR might activate different G proteins in the hepatopancreas and ovary, which may explain the multifunctional roles of sNPF system.
Statements
Data availability statement
The data presented in the study are deposited in the Genbank repository, the accession numbers are PP941935 for PtsNPF and PP967214 for PtsNPFR.
Ethics statement
The animal study was approved by Committee on the Ethics of Animal Experiments of the Ningbo University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QZ: Conceptualization, Data curation, Formal Analysis, Investigation, Project administration, Validation, Writing – original draft. XL: Data curation, Software, Writing – original draft. XS: Methodology, Resources, Software, Writing – original draft. DZ: Funding acquisition, Supervision, Writing – review & editing. XX: Conceptualization, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National natural Science Foundation of China (Nos. 32373124 and U23A20248), and the K. C. Wong Magna Fund in Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1451544/full#supplementary-material
References
1
Bajracharya P. Lu H. Pietrantonio P. V. (2014). The red imported fire ant (Solenopsis invicta Buren) kept Y not F: predicted sNPY endogenous ligands deorphanize the short NPF (sNPF) receptor. PLoS One9, e109590. doi: 10.1371/journal.pone.0109590
2
Bao C. Yang Y. Huang H. Ye H. (2018a). Inhibitory role of the mud crab short neuropeptide F in vitellogenesis and oocyte maturation via autocrine/paracrine signaling. Front. Endocrinol.9. doi: 10.3389/fendo.2018.00390
3
Bao C. Yang Y. Zeng C. Huang H. Ye H. (2018b). Identifying neuropeptide GPCRs in the mud crab, Scylla paramamosain, by combinatorial bioinformatics analysis. Gen. Comp. Endocrinol.269, 122–130. doi: 10.1016/j.ygcen.2018.09.002
4
Bigot L. Beets I. Dubos M. P. Boudry P. Schoofs L. Favrel P. (2014). Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J. Exp. Biol.217, 2974–2982. doi: 10.1242/jeb.104067
5
Borst D. W. Laufer H. Landau M. Chang E. S. Hertz W. A. Baker F. C. et al . (1987). Methyl farnesoate and its role in crustacean reproduction and development. Insect Biochem.17, 1123–1127. doi: 10.1016/0020-1790(87)90133-8
6
Caers J. Peymen K. Van Hiel M. B. Van Rompay L. Van Den Abbeele J. Schoofs L. et al . (2016). Molecular characterization of a short neuropeptide F signaling system in the tsetse fly, Glossina morsitans morsitans. Gen. Comp. Endocrinol.235, 142–149. doi: 10.1016/j.ygcen.2016.06.005
7
Cerstiaensa A. Benfekihb L. Zouitenc H. Verhaerta P. De Loofa A. Schoofsa L. (1999). Led-NPF-1 stimulates ovarian development in locusts. Peptides20, 39–44. doi: 10.1016/S0196-9781(98)00152-1
8
Chen M. E. Pietrantonio P. V. (2006). The short neuropeptide F-like receptor from the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Arch. Insect Biochem.61, 195–208. doi: 10.1002/(ISSN)1520-6327
9
Chen T. Ren C. Jiang X. Zhang L. Li H. Huang W. et al . (2018). Mechanisms for type-II vitellogenesis-inhibiting hormone suppression of vitellogenin transcription in shrimp hepatopancreas: Crosstalk of GC/cGMP pathway with different MAPK-dependent cascades. PLoS One13, e0194459. doi: 10.1371/journal.pone.0194459
10
Christie A. E. (2014). Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen. Comp. Endocrinol.206, 235–254. doi: 10.1016/j.ygcen.2014.04.015
11
Christie A. E. Chi M. Lameyer T. J. Pascual M. G. Shea D. N. Stanhope M. E. et al . (2015). Neuropeptidergic signaling in the American lobster Homarus americanus: new insights from high-throughput nucleotide sequencing. PLoS One10, e0145964. doi: 10.1371/journal.pone.0145964
12
De Loof A. Baggerman G. Breuer M. Claeys I. Cerstiaens A. Clynen E. et al . (2001). Gonadotropins in insects: an overview. Arch. Insect Biochem.47, 129–138. doi: 10.1002/arch.1044
13
Dillen S. Zels S. Verlinden H. Spit J. Van Wielendaele P. Vanden Broeck J. (2013). Functional characterization of the short neuropeptide F receptor in the desert locust, Schistocerca gregaria. PLoS One8, e53604. doi: 10.1371/journal.pone.0053604
14
Fadda M. Hasakiogullari I. Temmerman L. Beets I. Zels S. Schoofs L. (2019). Regulation of feeding and metabolism by neuropeptide F and short neuropeptide F in invertebrates. Front. Endocrinol.10. doi: 10.3389/fendo.2019.00064
15
Farhadi A. Cui W. Zheng H. Li S. Zhang Y. Ikhwanuddin M. et al . (2021). The regulatory mechanism of sexual development in decapod crustaceans. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.679687
16
Feng G. Reale V. Chatwin H. Kennedy K. Venard R. Ericsson C. et al . (2003). Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Eur. J. Neurosci.18, 227–238. doi: 10.1046/j.1460-9568.2003.02719.x
17
Garczynski S. F. Brown M. R. Crim J. W. (2006). Structural studies of Drosophila short neuropeptide F: Occurrence and receptor binding activity. Peptides27, 575–582. doi: 10.1016/j.peptides.2005.06.029
18
Garczynski S. F. Crim J. W. Brown M. R. (2007). Characterization and expression of the short neuropeptide F receptor in the African malaria mosquito, Anopheles Gambiae. Peptides28, 109–118. doi: 10.1016/j.peptides.2006.09.019
19
Gong J. Ye H. Xie Y. Yang Y. Huang H. Li S. et al . (2015). Ecdysone receptor in the mud crab. J. Endocrinol.224, 273–287. doi: 10.1530/JOE-14-0526
20
Hauser F. Cazzamali G. Williamson M. Park Y. Li B. Tanaka Y. et al . (2008). A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol.29, 142–165. doi: 10.1016/j.yfrne.2007.10.003
21
Jékely G. (2013). Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci.110, 8702–8707. doi: 10.1073/pnas.1221833110
22
Jiang H. Gui S. Xu L. Pei Y. Smagghe G. Wang J. (2017). The short neuropeptide F modulates olfactory sensitivity of Bactrocera dorsalis upon starvation. J. Insect Physiol.99, 78–85. doi: 10.1016/j.jinsphys.2017.03.012
23
Juneau Z. C. Stonemetz J. M. Toma R. F. Possidente D. R. Heins R. C. Vecsey C. G. (2019). Optogenetic activation of short neuropeptide F (sNPF) neurons induces sleep in Drosophila melanogaster. Physiol. Behav.206, 143–156. doi: 10.1016/j.physbeh.2019.03.027
24
Kamruzzaman A. S. M. Mikani A. Mohamed A. A. Elgendy A. M. Takeda M. (2020). Crosstalk among Indoleamines, Neuropeptides and JH/20E in Regulation of Reproduction in the American Cockroach, Periplaneta americana. Insects11, 155. doi: 10.3390/insects11030155
25
Kaneko Y. Hiruma K. (2014). Short neuropeptide F (sNPF) is a stage-specific suppressor for juvenile hormone biosynthesis by corpora allata, and a critical factor for the initiation of insect metamorphosis. Dev. Biol.393, 312–319. doi: 10.1016/j.ydbio.2014.07.014
26
Lee K. S. You K. H. Choo J. K. Han Y. M. Yu K. (2004). Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem.279, 50781–50789. doi: 10.1074/jbc.M407842200
27
Li H. Huang X. Yang Y. Chen X. Yang Y. Wang J. et al . (2022). The short neuropeptide F receptor regulates olfaction-mediated foraging behavior in the oriental fruit fly Bactrocera dorsalis (Hendel). Insect Biochem. Mol. Biol.140, 103697. doi: 10.1016/j.ibmb.2021.103697
28
Liesch J. Bellani L. L. Vosshall L. B. (2013). Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aEgypti. PLoS Negl. Trop. Dis.7, e2486. doi: 10.1371/journal.pntd.0002486
29
Liu A. Liu J. Liu F. Huang Y. Wang G. Ye H. (2018). Crustacean female sex hormone from the mud crab Scylla paramamosain is highly expressed in prepubertal males and inhibits the development of androgenic gland. Front. Physiol.9. doi: 10.3389/fphys.2018.00924
30
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods25, 402–408. doi: 10.1006/meth.2001.1262
31
Lu H. Pietrantonio P. V. (2011). Immunolocalization of the short neuropeptide F receptor in queen brains and ovaries of the red imported fire ant (Solenopsis invicta Buren). BMC Neurosci.12, 1–15. doi: 10.1186/1471-2202-12-57
32
Mertens I. Meeusen T. Huybrechts R. De Loof A. Schoofs L. (2002). Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem. Bioph. Res. Co.297, 1140–1148. doi: 10.1016/S0006-291X(02)02351-3
33
Mirabeau O. Joly J. S. (2013). Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci.110, E2028–E2037. doi: 10.1073/pnas.1219956110
34
Nagaraju G. P. C. (2011). Reproductive regulators in decapod crustaceans: an overview. J. Exp. Biol.214, 3–16. doi: 10.1242/jeb.047183
35
Nagata S. Matsumoto S. Nakane T. Ohara A. Morooka N. Konuma T. et al . (2012). Effects of starvation on brain short neuropeptide F-1,-2, and-3 levels and short neuropeptide F receptor expression levels of the silkworm, Bombyx mori. Front. Endocrinol.3. doi: 10.3389/fendo.2012.00003
36
Nässel D. R. Enell L. E. Santos J. G. Wegener C. Johard H. A. (2008). A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci.9, 1–35. doi: 10.1186/1471-2202-9-90
37
Nässel D. R. Wegener C. (2011). A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides32, 1335–1355. doi: 10.1016/j.peptides.2011.03.013
38
Nguyen T. V. Cummins S. F. Elizur A. Ventura T. (2016). Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep.6, 38658. doi: 10.1038/srep38658
39
Reale V. Chatwin H. M. Evans P. D. (2004). The activation of G-protein gated inwardly rectifying K+ channels by a cloned Drosophila melanogaster neuropeptide F-like receptor. Eur. J. Neurosci.19, 570–576. doi: 10.1111/j.0953-816X.2003.03141.x
40
Root C. M. Ko K. I. Jafari A. Wang J. W. (2011). Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell145, 133–144. doi: 10.1016/j.cell.2011.02.008
41
Rosenbaum D. M. Rasmussen S. G. Kobilka B. K. (2009). The structure and function of G-protein-coupled receptors. Nature459, 356–363. doi: 10.1038/nature08144
42
Sithigorngul P. Saraithongkum W. Jaideechoey S. Longyant S. Sithigorngul W. (1998). Novel FMRFamide-like neuropeptides from the eyestalk of the giant freshwater prawn Macrobrachium rosenbergii. Comp. Biochem. Physiol. B. Biochem. Mol. Biol.120, 587–595. doi: 10.1016/s0196-9781(00)00382-x
43
Suwansa-Ard S. Thongbuakaew T. Wang T. Zhao M. Elizur A. Hanna P. J. et al . (2015). In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS One10, e0123848. doi: 10.1371/journal.pone.0123848
44
Tinikul Y. Tinikul R. Engsusophon A. Sobhon P. (2023). The effects of short neuropeptide F on ovarian maturation and spawning in female giant freshwater prawn, Macrobrachium rosenbergii, and associated regulatory mechanisms. Aquaculture569, 739361. doi: 10.1016/j.aquaculture.2023.739361
45
Tu S. Ge F. Han Y. Wang M. Xie X. Zhu D. (2022). Putative role of corazonin in the ovarian development of the swimming crab Portunus trituberculatus. Front. Mar. Sci.9. doi: 10.3389/fmars.2022.976754
46
Tu S. Xu R. Wang M. Xie X. Bao C. Zhu D. (2021). Identification and characterization of expression profiles of neuropeptides and their GPCRs in the swimming crab, Portunus trituberculatus. PeerJ9, e12179. doi: 10.7717/peerj.12179
47
Veenstra J. A. (2011). Neuropeptide evolution: neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen. Comp. Endocrinol.171, 160–175. doi: 10.1016/j.ygcen.2011.01.005
48
Wang M. Ye H. Miao L. Li X. (2022). Role of short neuropeptide F in regulating eyestalk neuroendocrine systems in the mud crab Scylla paramamosain. Aquaculture560, 738493. doi: 10.1016/j.aquaculture.2022.738493
49
Webster S. G. Keller R. Dircksen H. (2012). The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen. Comp. Endocrinol.175, 217–233. doi: 10.1016/j.ygcen.2011.11.035
50
Wu X. Yao G. Yang X. Cheng Y. Wang C. (2007). A study on the ovarian development of Portunus trituberculatus in East China Sea during the first reproductive cycle. Acta Oceanol. Sin.29, 120–127. doi: 10.3321/j.issn:0253-4193.2007.04.014
51
Yamanaka N. Yamamoto S. Žitňan D. Watanabe K. Kawada T. Satake H. et al . (2008). Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One3, e3048. doi: 10.1371/journal.pone.0003048
Summary
Keywords
short neuropeptide F, short neuropeptide F receptor, Portunus trituberculatus , ovarian development, RNA interference
Citation
Zhou Q, Li X, Shen X, Zhu D and Xie X (2024) Molecular characterization of a short neuropeptide F signaling system in the swimming crab, Portunus trituberculatus, and its role in ovarian development. Front. Mar. Sci. 11:1451544. doi: 10.3389/fmars.2024.1451544
Received
19 June 2024
Accepted
12 July 2024
Published
25 July 2024
Volume
11 - 2024
Edited by
Hongyu Ma, Shantou University, China
Reviewed by
Shihao Li, Chinese Academy of Sciences (CAS), China
Ting Chen, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2024 Zhou, Li, Shen, Zhu and Xie.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongfa Zhu, zhudongfa@nbu.edu.cn; Xi Xie, xiexi@nbu.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.