Abstract
Introduction:
Species sorting by environmental gradients is an important driver of benthic meiofaunal biodiversity in marine ecosystems, but there are few attempts to test these effects in coastal habitats.
Methods:
In this study, we evaluated the importance of habitat filtering in shaping meiofaunal communities across rocky tide pools and nearby sandy beaches in the Eastern Brazilian Marine Ecoregion, SW Atlantic. We proposed two hypotheses: (i) rocky tide pools exhibit a subset (nestedness effects) of the sandy beach meiofaunal assemblage, with lower phylogenetic diversity; and (ii) the meiofaunal assemblage composition and phylogenetic diversity vary seasonally over the year in both habitats. We used metabarcoding (V9 hypervariable region from 18S gene) from sediment samples (n = 70) to assess the meiofaunal assemblage composition and phylogenetic diversity, and tested spatial patterns of nestedness and turnover across habitats, seasons, and locations.
Results:
Compared to the neighboring sandy beaches, tide pools had higher temperatures (+ 1.8°C) and lower quality organic matter. Contrary to our hypothesis, community turnover was the main driver of meiofaunal phylogenetic diversity and composition in both tide pools and nearby sandy beaches. The tide pool assemblages showed a lower phylogenetic diversity and taxon richness than the neighboring sandy beaches.
Discussion:
Our study supports the importance of environmental drivers on benthic meiofaunal phylogenetic diversity within tide pools and sandy beaches and revealed distinct assemblages in these neighboring coastal intertidal habitats.
1 Introduction
The concept of metacommunities integrates the influence of environmental gradients and biological interactions on community ecology (Leibold et al., 2004). One key mechanism structuring marine benthic meiofaunal assemblages is species sorting, which operates through local physical effects (e.g., wave action, tidal currents) that control sediment transport and influence community turnover on sandy beaches (McLachlan and Defeo, 2018; Macher et al., 2024). Although the effects of the species-sorting paradigm in coastal benthos are well documented and support the metacommunity theory, there is limited work comparing meiofaunal beta-diversity (nestedness or turnover) in contrasting coastal habitats (Macher et al., 2024). Community turnover and nestedness are critical factors effecting the structure and functioning of metacommunities in different habitats. Turnover refers to the replacement of species between communities, while nestedness indicates a hierarchical arrangement in which species in less diverse communities are subsets of those in more diverse ones. High turnover values typically reflect strong environmental filtering or spatial heterogeneity, suggesting that different species are favored in different environmental conditions. In contrast, high nestedness implies that community composition is primarily structured by gradients in taxa richness, possibly due to differences in habitat characteristics, where poorer communities retain only the most tolerant species. Together, these patterns provide insight into the relative importance of deterministic and stochastic processes in shaping community composition (Leibold et al., 2004; Zhang et al., 2023).
Rocky tide pools offer an excellent opportunity to test ecological processes that regulate diversity in coastal ecosystems. They function as isolated natural mesocosms with distinct environmental gradients compared to those of the neighboring sandy beaches and reefs (Vinagre et al., 2015; Dias et al., 2016; Mendonça et al., 2018). Tide pools may be subjected to rapid changes in temperature, salinity and pH (Metaxas and Scheibling, 1993), and may have extreme environmental conditions with excessively high temperatures (Vinagre et al., 2018). Thus, habitat filtering of tide pool benthos may create subsets (nestedness) of nearby coastal benthic assemblages adapted to these conditions. Among the metazoan assemblages that inhabit tide pools, the benthic meiofauna is particularly diverse (Coull and Wells, 1983; Chargulaf and Tibbetts, 2015). Meiofauna metazoans include animals from at least 22 phyla that fall within the size range of 45 to 1000 μm for their entire life cycle, or for just part of their life cycle (temporary meiofauna) (Coull and Wells, 1983; McIntyre, 1969; Higgins and Thiel, 1988; Hakenkamp and Palmer, 2000; Giere, 2009). Meiofaunal assemblages can then be directly shaped by local abiotic factors, resulting in the permanence of only a subset of the regional available species pool. This abiotic-driven assemblage-shaping process is known as habitat filtering (Webb et al., 2002; Pontarp et al., 2012; Coppo et al., 2024a). This mechanism, also known as environmental filtering, has been identified as a key factor regulating the composition of fish communities and their trophic niches in tide pools (Cadotte and Tucker, 2017; Andrades et al., 2019a; Kunishima and Tachihara, 2019). Higher temperatures inside tide pools may alter meiofaunal population structure and influence top-down predation relationships (Jochum et al., 2012). As the dispersion capacity of many meiofaunal taxa is restricted, these assemblages may also be influenced by biological interactions within tide pools (Maria et al., 2012, Maria et al., 2018; Andrades et al., 2019b).
In sandy beaches, the distribution and abundance of infaunal benthos are typically associated with the swash climate, sediment grain size, and food availability (McLachlan et al., 1993; McLachlan and Brown, 2006; Blanchette et al., 2008; Griffiths et al., 2017). Over the intertidal gradient of sandy beaches, temperature and salinity are highly variable and can also influence the distribution and composition of organisms (Olafsson, 1991; Ape et al., 2018; Mitwally and Hamdan, 2021; Coppo et al., 2024b). In tropical regions, seasonal changes are less pronounced, however, meiofaunal communities have shown seasonal patterns, with higher abundance during the warmest and wettest months (Albuquerque et al., 2007). In addition to seasonal variations, temperature and salinity can fluctuate on shorter temporal scales, such as tidal cycles and diel changes. These environmental dynamics, combined with changes in the quantity and quality of organic matter, play a key role in shaping meiofaunal assemblages in sandy beach habitats (Esteves et al., 1998; Todaro and Rocha, 2004; Cisneros et al., 2011; Venturini et al., 2012; Baia and Venekey, 2019; Coppo et al., 2024b).
Understanding which factors are key to driving diversity in coastal habitats at local and regional scales is crucial to create conservation strategies to protect critical ecological processes and habitats, and to better understand temporal and spatial changes in biodiversity. Meiofaunal diversity patterns and their relation with environmental factors have been successfully assessed by eDNA metabarcoding in different coastal ecosystems (Bernardino et al., 2019; Fais et al., 2020; Bellisario et al., 2021; Castro et al., 2021; Coppo et al., 2023, 2024a; Macher et al., 2024). The use of eDNA metabarcoding not only facilitates a detailed examination of meiofaunal taxonomic composition but also enables the assessment of phylogenetic diversity, shedding light on how evolutionary relationships influence community structure (Coppo et al., 2024a; Macher et al., 2024). Understanding phylogenetic diversity is essential for revealing the impacts of habitat filtering on community composition, particularly in determining whether meiofaunal assemblages in tide pools represent a nested subset of those found in adjacent sandy beaches. Tide pools and their associated meiofaunal assemblages may serve as an experimental laboratory for future climatic conditions, and understanding changes in diversity patterns at the local scale may help to predict changes on a global scale based on predicted global temperature scenarios. Here, we aimed to assess whether the meiofaunal composition within tide pools represents a subset of meiofaunal taxa found on nearby sandy beaches, indicating habitat filtering. Additionally, we tested the seasonal influence on meiofaunal composition and diversity across both habitats. We hypothesized that: (i) the meiofaunal assemblage in rocky tide pools is a subset (nestedness) of the assemblage found on adjacent sandy beaches, exhibiting lower phylogenetic diversity; and (ii) meiofaunal composition and diversity in both habitats are shaped by seasonal variations.
2 Materials and methods
2.1 Study area and sampling
This study was carried out at the Gramuté (-19.972861, -40.138361) and Rio Preto (-20.012111, -40.154916) sandy beaches and nearby rocky shores, located within a marine protected area in the Eastern Brazilian Marine Ecoregion (Figure 1). The area is characterized by dry winters and rainy summers, with sea surface temperatures varying from 21°C to 27°C, salinity ranging from 34.6 to 36 ppt, and strong internal tidal currents with E-SE wave swells with upwelling events in spring and summer (Bernardino et al., 2015; Quintana et al., 2015; Bernardino et al., 2018; Mazzuco et al., 2019; Mazzuco et al., 2020). The Eastern Brazilian Marine Ecoregion has experienced notable warming in the last few decades (Mazzuco et al., 2020).
Figure 1
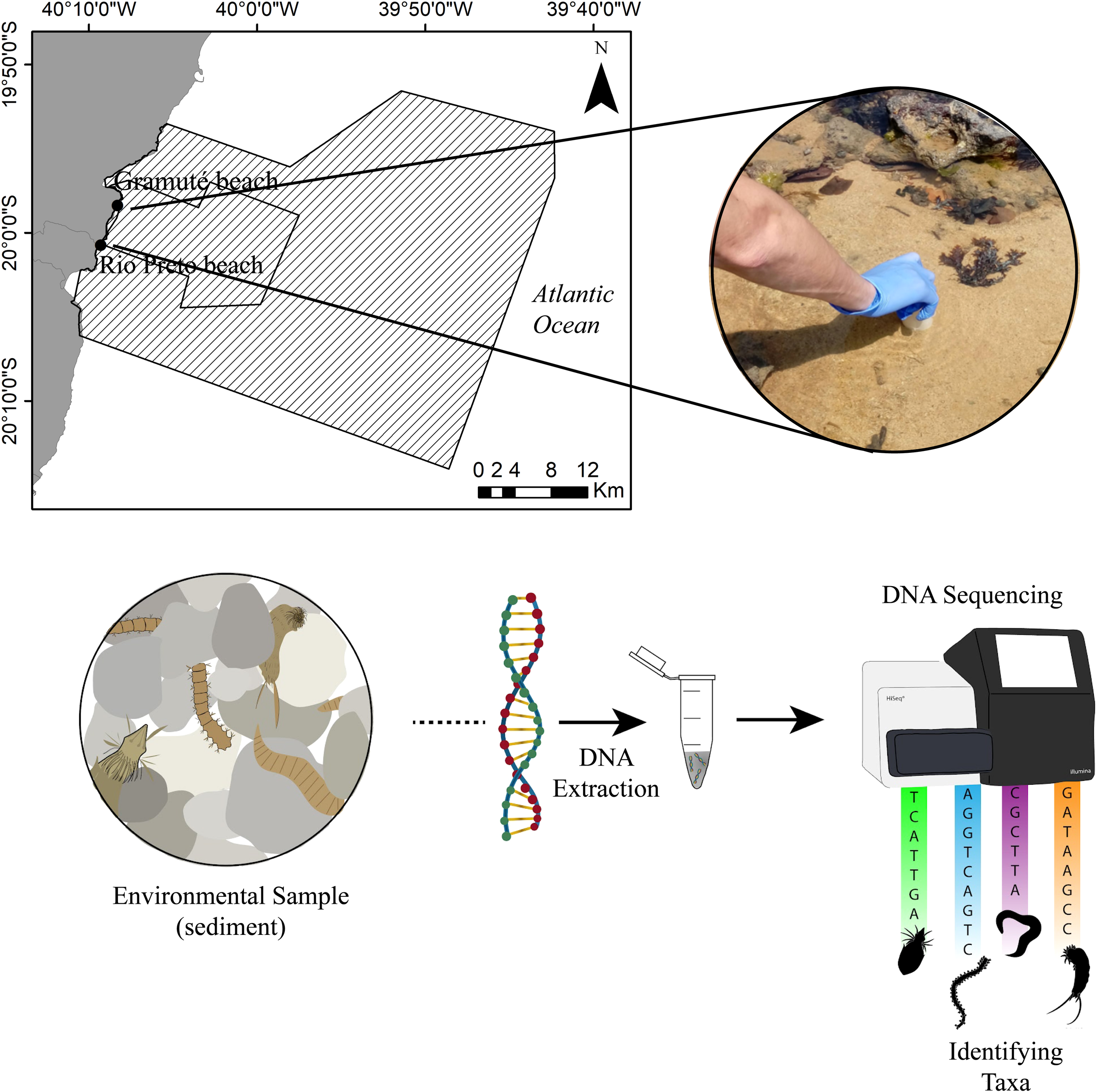
Study area (Gramuté and Rio Preto sandy beaches) within Costa das Algas Marine Protected Area (polygon area) and the workflow of sediment sampling, DNA extraction, sequencing and taxonomic identification.
During winter (June to August 2020) and spring (September to November 2020), sediment samples were collected monthly at Gramuté beach and Rio Preto beach. The sampling period was defined based on previous studies for the same region that reported a seasonal influence on meiofaunal diversity, assemblage composition and benthic recruitment (Mazzuco and Bernardino, 2022; Coppo et al., 2024b), usually associated with the seasonal presence of warm waters with high nutrient content (Mazzuco and Bernardino, 2022). Quantitative samples were collected in triplicate using sterile, DNA-free corers with 5 cm internal diameter down to 5 cm sediment depth (Figure 1). On sandy beaches, samples were collected at three stations 20 meters distant from each other in the subtidal region (approximately 0.5 meters depth; n = 9 samples per month, in each location). In the rocky tide pools, the same sampling effort was used during the low tide, with three stations 20 meters distant from each other in the intertidal region. Each rocky pool measured approximately 4.9 m² and 0.5 meters depth each (n = 9 sediment samples per month in each location). Additionally, samples were collected for sediment analysis, including grain size, total organic matter, carbonate content, protein content, carbohydrate content, lipid content, biopolymeric carbon content, protein-to-carbohydrate ratio, and carbohydrate-to-lipid ratio (n = 9 sediment samples per month, in each location). All sediment samples were transported in thermic bags with ice, and stored at -20°C until analysis. Sea surface temperature and salinity were additionally measured in situ, using a portable multiparameter (ROMERLAB RR905) and a refractometer (PCE-0100). Both parameters were measured in the surface water at each sampling location in the sandy beaches and tide pools. Field sampling was authorized by the Biodiversity Authorization and Information System of the Brazilian Institute for the Environment and Renewable Natural Resources (SISBIO-IBAMA, sampling license number 24700-1).
2.2 Sediment analysis
Sediment samples were dried for 48 hours at 60°C, then macerated and sieved using a sieve shaker through a set of meshes with openings ranging from -1.5 Φ to 4 Φ, at 1Φ intervals. To determine carbonate content, sediment samples were dried at 110°C for 4 hours, then leached with hydrochloric acid (HCl, 1:4 dilution) and dried again. The weight difference of dried sediments before and after leaching represents the carbonate content (MacCarthy, 1933). Total organic matter (TOM) was quantified by loss-on-ignition, where a known weight of dried sediment was combusted in a muffle for 3 hours at 500°C, and TOM was calculated based on the weight loss (Suguio, 1973).
Sedimentary organic biopolymers (proteins, carbohydrates, and lipids) were analyzed in triplicate (Danovaro, 2010; Neto et al., 2021). The total protein analysis (PRT) was conducted after extraction with NaOH 0.5 M and its concentration was determined following Hartree (1972) modified by Rice (1982) to compensate for phenol interference. Total carbohydrate content (CHO) was analyzed according to Gerchacov and Hatcher (1972), and total lipids (LIP) were extracted from 1 g of homogenized sediment lyophilized by ultrasonication in 10 mL of chloroform: methanol (2:0–1 v/v) (Marsh and Weinstein, 1966). Blanks were carried out for each analysis using pre-combusted sediments for 4 hours at 450 and 480°C. PRT, CHO, and LIP contents were expressed respectively as bovine serum albumin, glucose, and tripalmitin equivalents. The PRT, CHO, and LIP contents were converted to carbon equivalents using a conversion factor of 0.49, 0.40, and 0.75, respectively (Fabiano and Danovaro, 1994), and its sum was reported as biopolymeric carbon (BPC) (Fabiano et al., 1995). Additionally, protein-to-carbohydrate (PRT: CHO) and carbohydrate-to-lipid (CHO: LIP) ratios were used to assess the quality of the organic matter, represented by the state of biochemical degradation processes (Galois et al., 2000; Stelzer et al., 2021).
2.3 DNA extraction and sequencing
Before DNA extraction, sediment samples (approximately 200 g) were elutriated to extract the meiofauna from the sediment and enrich the metazoan DNA content, as suggested by Brannock and Halanych (2015). A 1 L flask was filled with 950 mL of filtered seawater. The sediment sample was then added, homogenized and allowed to settle for 30 seconds before decanting the supernatant over a 45 μm mesh sieve. The elutriation procedure was repeated ten times for each sediment sample. After that, the material retained on the sieve was rinsed into 50 mL Falcon tubes and centrifuged for 3 minutes at room temperature at 1342 X g in an Eppendorf Centrifuge 5430. After centrifugation, the sample volumes were standardized to 20 mL, then mixed and aliquoted to sterile 1mL (2 tubes per sample) and stored at -20°C (Brannock and Halanych, 2015). All glassware was cleaned using Extran® MA 02 (Merck, Darmstadt, Germany) 10% solution and autoclaved between samples to avoid cross-contamination. Sieves were sterilized by soaking for 45 minutes in 10% sodium metabisulfite solution (Creer et al., 2010; Brannock and Halanych, 2015).
The PowerSoil DNA® (Qiagen) kit was used to extract DNA from the 1mL aliquots. DNA integrity was verified on a 1% agarose gel, and the purity using a NanoDrop One spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA samples extracted from the same sediment sampling station and month were combined into a single pool, totaling three replicates per month in each habitat and location. The DNA concentration was measured using a Qubit® 4 Fluorometer (Life Technologies-Invitrogen, Carlsbad, CA, USA). Blank samples (negative controls) were carried out in triplicates for each step of the sample DNA extraction and quality checking, and run with the samples on 1% agarose gels to be sure there was no contamination. If any DNA bands were detected, the extraction was repeated.
PCR, library preparation, and sequencing were conducted by ©NGS Genomic Solutions (Piracicaba, SP, Brazil). PCR products were purified using Ampure XP beads, and Nextera XT adapters were subsequently attached. The ©NGS Genomic Solutions conducted blank samples during the PCR, checking it on 1% agarose gel, and if any DNA bands were seen, PCR was repeated. The resulting libraries were normalized to achieve a uniform concentration. After normalization, an equimolar pool was prepared with 5 µL of each library. This pool was then quantified by qPCR using the KAPA Library Quantification Kit (Roche) to estimate its concentration. Based on these data, the necessary dilutions were performed for sequencing. Metabarcoding sequencing was performed using the MiSeq Illumina platform (2 x 250 bp) with a target coverage of 100,000 paired-end reads per sample. The V9 hypervariable region from 18S SSU rRNA gene was amplified using the primers Euk_1391 forward (GTACACACCGCCCGTC) and EukBr reverse (TGATCCTTCTGCAGGTTCACCTAC) (Medlin et al., 1988; Amaral-Zettler et al., 2008; Stoeck et al., 2010). The resulting amplicons varied in size (mean 260 ± 50 bp) since the reverse primer does not have a conserved position as the forward one.
2.4 Bioinformatic pipeline
An AMD Ryzen 1950x Crucial 64 GB (16x4) DDR4–2666 MHz computer was used to run the entire bioinformatic pipeline. Demultiplexed raw paired-end reads were identified through the QIIME2 2022.8 software, after importing FastQ files as QIIME2 artifacts, denoising them via DADA2 with the denoise-paired plugin, and then by removal of low-quality bases and primer sequences (Callahan et al., 2016; Bolyen et al., 2019; Lines et al., 2023). Amplicon Sequence Variants (ASVs) were generated through the denoising process. The ASVs represent a refined taxonomic unit that allows for precise identification, reflecting true biological variants, or taxa, and were used as the main taxonomic unit in this study (Segura et al., 2024). The taxonomic assignment was generated using the DADA2 plugin (p-trim = 10, p-trunc = 160, and mean phred score = 39 ± 1; Supplementary Table S1) using the machine learning Python library scikit-learn (Pedregosa et al., 2011). The sequences were identified taxonomically using a Naïve Bayes classifier trained on Silva 138 database clustered at 99% similarity (Quast et al., 2013). Additionally, rarefaction curves were plotted to assess sampling depth per sampling site. Based on this curve, the data in samples was normalized to the minimum sequencing depth (5036 reads), allowing datasets to be analyzed and compared quantitatively using equal sequencing depth.
Further, Faith’s Phylogenetic Diversity (PD) was calculated for each sample using a diversity core-metrics-phylogenetic pipeline based on a phylogenetic tree previously generated by the align-to-tree-mafft-fasttree pipeline from the q2- phylogeny plugin in QIIME2. All raw sequence data are available online and deposited in NCBI (SRR24675047).
2.5 Statistical analysis
Here, only the main meiofaunal representatives were considered for downstream analyses, and the final dataset was composed of 9 phyla (Annelida, Arthropoda (Crustacea), Cnidaria, Echinodermata, Gastrotricha, Mollusca, Nematoda, Nemertea, and Platyhelminthes). The identified meiofaunal ASVs varied in taxonomic resolution, reaching mostly Order (22 taxa) and Family (5 taxa) levels (see Table 1). Differences in the taxon richness (number of meiofaunal ASVs) between seasons, habitats and locations were tested using Mann-Whitney U tests for non-normal data following Shapiro-Wilk normality testing, and the results were presented as Venn’s diagrams. Differences in environmental conditions (grain size, temperature, salinity, carbonate content, total organic matter, biopolymers, and biopolymeric carbon content) and meiofaunal assemblages were analyzed using a Permutational Analysis of Variance (PERMANOVA; Anderson et al., 2008). PERMANOVA was applied to test for differences across season (spring and winter), habitat (sandy beach and tide pool), location (Gramuté beach and Rio Preto beach), and their interactions. Environmental variables were analyzed based on a Euclidean similarity matrix after normalization, ensuring comparability across different scales. Normalization was performed by subtracting the mean and dividing by the standard deviation for each variable. Meiofaunal assemblage composition was analyzed using presence-absence data and based on a Jaccard dissimilarity matrix. For all comparisons, when significant main effects or interactions were detected, pairwise tests (Tukey HSD) were conducted to determine specific differences between groups.
Table 1
| Identified ASV | Season | Habitat | Location | |||
|---|---|---|---|---|---|---|
| Spring | Winter | Sandy beach | Tide pool | Gramuté | Rio Preto | |
| Annelida | 6 | 9 | 11 | 1 | 11 | 0 |
| Capitellida | 0 | 1 | 1 | 0 | 1 | 0 |
| Echiuroidea | 1 | 1 | 1 | 0 | 1 | 0 |
| Eunicida | 0 | 1 | 1 | 0 | 1 | 0 |
| Golfingiida | 0 | 1 | 1 | 0 | 1 | 0 |
| Haplotaxida | 0 | 1 | 1 | 0 | 1 | 0 |
| Phyllodocida | 1 | 1 | 1 | 0 | 1 | 0 |
| Polychaeta | 1 | 1 | 1 | 1 | 1 | 0 |
| Protodrilidae | 1 | 0 | 1 | 0 | 1 | 0 |
| Sabellida | 0 | 1 | 1 | 0 | 1 | 0 |
| Spionida | 1 | 1 | 1 | 0 | 1 | 0 |
| Terebellida | 1 | 0 | 1 | 0 | 1 | 0 |
| Arthropoda (Crustacea) | 4 | 6 | 5 | 3 | 6 | 2 |
| Calanoida | 0 | 1 | 1 | 0 | 1 | 0 |
| Harpacticoida | 1 | 1 | 1 | 0 | 1 | 0 |
| Eucarida | 1 | 1 | 1 | 1 | 1 | 1 |
| Multicrustacea | 1 | 1 | 1 | 1 | 1 | 1 |
| Cyclopoida | 0 | 1 | 0 | 1 | 1 | 0 |
| Podocopida | 1 | 1 | 1 | 0 | 1 | 0 |
| Cnidaria | 1 | 2 | 2 | 0 | 2 | 0 |
| Actiniaria | 0 | 1 | 1 | 0 | 1 | 0 |
| Zoantharia | 1 | 1 | 1 | 0 | 1 | 0 |
| Echinodermata | 1 | 2 | 2 | 0 | 2 | 0 |
| Echinoidea | 0 | 1 | 1 | 0 | 1 | 0 |
| Holothuroidea | 1 | 1 | 1 | 0 | 1 | 0 |
| Gastrotricha | 0 | 2 | 2 | 0 | 2 | 0 |
| Chaetonotida | 0 | 1 | 1 | 0 | 1 | 0 |
| Macrodasyida | 0 | 1 | 1 | 0 | 1 | 0 |
| Mollusca | 1 | 2 | 2 | 0 | 2 | 1 |
| Mytiloidea | 0 | 1 | 1 | 0 | 1 | 1 |
| Pectinida | 1 | 1 | 1 | 0 | 1 | 0 |
| Nematoda | 2 | 3 | 2 | 2 | 3 | 1 |
| Chromadorea | 1 | 1 | 1 | 0 | 1 | 0 |
| Monhysterida | 0 | 1 | 0 | 1 | 1 | 0 |
| Rhabditida | 1 | 1 | 1 | 1 | 1 | 1 |
| Nemertea | 1 | 1 | 2 | 0 | 2 | 0 |
| Monostilifera | 0 | 1 | 1 | 0 | 1 | 0 |
| Palaeonemertea | 1 | 0 | 1 | 0 | 1 | 0 |
| Platyhelminthes | 1 | 2 | 2 | 0 | 2 | 0 |
| Dalytyphloplanida | 0 | 1 | 1 | 0 | 1 | 0 |
| Rhabdocoela | 1 | 1 | 1 | 0 | 1 | 0 |
Taxa (identified Amplicon Sequence Variants) presence-absence by season (spring and winter), habitat (sandy beach and tide pool), and location (Gramuté and Rio Preto).
Presence is indicated by 1 and absence by a 0. Taxa are categorized by phyla and the total number of taxa is indicated in bold for each Phylum by column.
When only a single taxon is detected in a sample, phylogenetic diversity cannot be calculated, as it relies on branch length differences among different taxa within a phylogenetic tree. Consequently, phylogenetic diversity data are missing for Gramuté tide pool samples during winter, where only one taxon was detected. Given this, phylogenetic diversity differences were therefore also assessed using PERMANOVA due to its robustness to unbalanced designs (Anderson et al., 2008; Anderson and Walsh, 2013). To mitigate the problem of a missing level, we limited our interpretation to the main effects, excluding interaction terms from the analysis, and verified whether the significant patterns were consistent across subsets of the data. A Principal Component Analysis (PCA) was performed with meiofaunal phylogenetic diversity and environmental variables (grain size, temperature, salinity, carbonate content, total organic matter, biopolymers, and biopolymeric carbon content) by habitat (sandy beach and tide pool) to assess which environmental variables are the main drivers of meiofaunal phylogenetic diversity. Additionally, to determine which ecological process (turnover or nestedness) primarily influenced the composition of meiofaunal assemblages in both habitats (sandy beach and tide pool), total β-diversity was calculated using Jaccard´s index and decomposed it into two additive components: β-diversity resulting from turnover and β-diversity resulting from nestedness. Indices were calculated separately for each habitat by pooling all replicates across seasons and locations. This approach was chosen to provide a broad comparison of meiofaunal community structure between sandy beaches and tide pools, regardless of temporal or spatial variability. Jaccard-based turnover represents species replacement between habitats, while nestedness indicates the extent to which one assemblage is a subset of another. In this study, we used ASVs as the primary taxonomic unit to ensure high-resolution detection of community differences, minimizing potential biases associated with mixed taxonomic levels. These indices were calculated with the ‘beta.multi’ function of the betapart package (Baselga et al., 2023). Significant differences were defined when p<0.05. All graphical and analytical processes were performed in the R environment (R Core Team, 2023).
3 Results
3.1 Environmental variables at tide pools and sandy beaches
The full results of the PERMANOVAs and pairwise tests of significant interactions are given in the supplementary materials (Supplementary Tables S3-S5). Significant differences in temperature, salinity, sediment organic quality, carbonate content, and total organic matter content were observed between seasons (df = 1; Pseudo-F = 3.86; p = 0.007), habitats (df = 1; Pseudo-F = 5.29; p = 0.003), and location (df = 1; Pseudo-F = 17.6; p = 0.001), with no significant interaction effects. Higher temperatures were observed in spring compared to winter (26.8 ± 1.8 °C and 25.5 ± 1.4 °C, respectively; p = 0.031), but salinity was higher during winter months (32.5 ± 0.5 and 31.8 ± 0.7, respectively; p = 0.038). Also, water temperature was 1.8 °C higher on average (p=0.01) inside tide pools (27.0 ± 2.0 °C) when compared to sandy beaches (25.2 ± 0.8 °C), as well as salinity (32.4 ± 0.5 and 31.9 ± 0.7, respectively; p=0.001). Furthermore, temperature was higher at Gramuté (26.8 ± 1.7°C) than at Rio Preto (25.4 ± 1.0°C; p=0.015), but no significant differences were observed for salinity (32.4 ± 0.7 and 32.0 ± 0.7, respectively; p=0.158).
The sediment organic quality varied significantly across habitats (p < 0.01) with higher total lipids (LIP) and biopolymeric carbon (BPC) inside tide pools (LIP 594.5 ± 163.3 and BPC 1182.3 ± 685.9, respectively) compared to sandy beaches (LIP 317.4 ± 186.8 and BPC 792.1 ± 401.9, respectively), but we did not observe significative differences for other variables between habitats. Both locations, Gramuté beach and Rio Preto beach, showed differences in sedimentary variables. Gramuté beach had higher carbonate content (p<0.001), total organic matter content (p<0.001), carbohydrate content (p<0.001), biopolymeric carbon (p=0.026), and carbohydrate-to-lipids ratio (p<0.001). Meanwhile, Rio Preto beach showed higher lipids content (p=0.021) and protein-to-carbohydrate ratio (p<0.001). Furthermore, sediments at Gramuté beach were mainly composed of sand (95.4 ± 2.6%), with a low presence of gravel (4.6 ± 2.6%), while Rio Preto beach sediments were composed of 50.2 ± 24.3% sand and 49.8 ± 24.3% gravel. However, we did not observe significant differences in sedimentary variables among seasons.
3.2 Environmental DNA
A total of 33 meiofaunal taxa were identified. Taxon richness ranged from 18 taxa in spring to 30 taxa in winter (Figure 2), and was considerably higher in sandy beaches (31 taxa) than in tide pools (6 taxa). In terms of spatial variation, Gramuté exhibited the highest richness (33 taxa), whereas only 4 taxa were recorded at the Rio Preto location. Of the 33 taxa, 27 were exclusively found in sandy beaches, 2 were unique to tide pools, and 4 taxa occurred in both habitats (Figure 2). Meiofaunal richness was significantly higher in sandy beaches (3.50 ± 0.56) than in tide pools (1.03 ± 0.03) (U = 348.5; p < 0.001). Within sandy beaches, Gramuté showed greater taxon richness (6.00 ± 3.10) compared to the Rio Preto (0.94 ± 0.24) (U = 8.00; p < 0.001). However, no significant differences were observed between spring (2.56 ± 2.22) and winter (4.28 ± 4.01) (U = 112.5; p = 0.252). Within tide pools, no differences were found between Gramuté and Rio Preto (U = 162.00; p = 1.000) or between seasons (U = 144.5; p = 0.176).
Figure 2
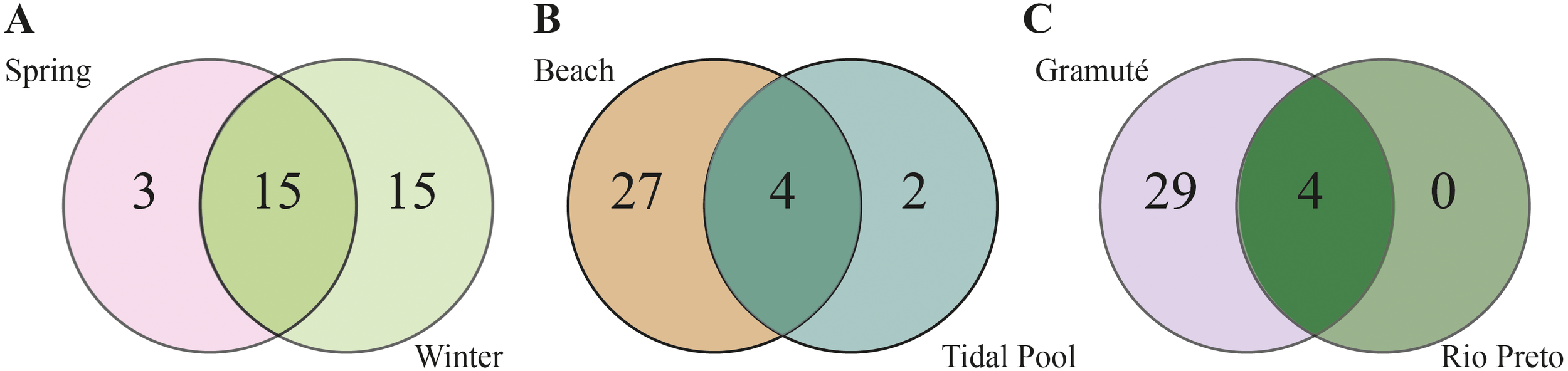
Number of meiofaunal taxa (amplicon sequence variants (ASVs)) detected by environmental DNA metabarcoding in each (A) season (spring and winter), (B) habitat (sandy beach and tide pool), and (C) location (Gramuté and Rio Preto).
Results from the PERMANOVA showed that meiofaunal assemblage composition and abundance of reads differed significantly between sandy beaches and tide pools (df = 1; Pseudo-F = 2.501; p = 0.001). Tide pools were dominated by Arthropoda (Crustacea; 3 taxa) and Nematoda (2 taxa), followed by Annelida (1 taxon) (Table 1). In contrast, sandy beach assemblages showed higher taxon richness and was primarily composed of Annelida (12 taxa), Arthropoda (Crustacea; 6 taxa), and Nematoda (2 taxa). Furthermore, Cyclopoida (Crustacea) and Monhysterida (Nematoda) were exclusively detected in tide pools, while Mollusca, Gastrotricha, Echinodermata, Cnidaria, Platyhelminthes, and Nemertea taxa were detected only in sandy beach samples.
Further differences in meiofaunal assemblage composition were also observed between the two sandy beach locations (df = 1; Pseudo-F = 3.339; p = 0.001). The Gramuté beach assemblage was composed of Arthropoda (Crustacea; 5 taxa), Nematoda (2 taxa), Annelida (12 taxa), Mollusca (2 taxa), Gastrotricha (2 taxa), Echinodermata (2 taxa), Cnidaria (2 taxa), Platyhelminthes (2 taxa), and Nemertea (2 taxa) (Table 1). At Rio Preto beach, however, only Arthropoda (Crustacea; 2 taxa), Mollusca (1 taxon), and Nematoda (1 Taxon) were detected (Table 1). Within tide pools, 4 taxa (including annelids, crustaceans, and nematodes) were exclusively detected at Gramuté and Rhabditida (Nematoda) was detected only at Rio Preto (Table 1). Seasonal differences were also detected (df = 1; Pseudo-F = 1.465; p = 0.044), with annelids and Rhabditida (Nematoda) exclusively detected during spring, whereas Cyclopoida (Crustacea), other Multicrustacea (Crustacea), Calanoida (Crustacea), Gastrotricha, and Monhysterida (Nematoda) taxa detected only during winter. We also observed significative differences in meiofauna composition in the interaction between season and location (Pseudo-F = 1.647; p = 0.026), with Rhabditida (Nematoda) only detected at Rio Preto during winter, Podocopida, Paleonemertea, Holothuroidea, Zoantharia (Cnidaria), Spionida, and Protodrilidae only detected at Gramuté during spring. Furthermore, Haplotaxida, Capitellida, Eunicida, Golfingiida, Sabellida, Calanoida (Crustacea), Actinaria (Cnidaria), Echinoidea, Gastrotricha, Monostilifera (Nemertea), and Nematoda were only detected at Gramuté during winter.
The distinct assemblage compositions between habitats were supported by high values of turnover (sandy beach = 0.939 and tide pool = 0.920) and low nestedness (sandy beach = 0.037 and tide pool = 0.017). This suggest that many taxa present in sandy beaches are not found in tide pools, and meiofaunal assemblages in tide pools are not merely a subset of the diversity found on sandy beaches, but rather represent distinct assemblages.
3.3 Phylogenetic diversity
We found significant differences on phylogenetic diversity between seasons, with higher values on winter (12.21 ± 7.66) than on spring (7.42 ± 2.54) (PERMANOVA; df = 1; Pseudo-F = 9.717; p = 0.002; Figure 3); higher in sandy beaches (8.92 ± 6.31) than in tide pools (6.55 ± 3.08) (PERMANOVA; df = 1; Pseudo-F 7.626; p = 0.005; Figure 3); and in Gramuté (10.92 ± 5.57) than in Rio Preto site (4.72 ± 1.20) (PERMANOVA; df = 1; Pseudo-F = 75.867; p = 0.001; Figure 3).
Figure 3
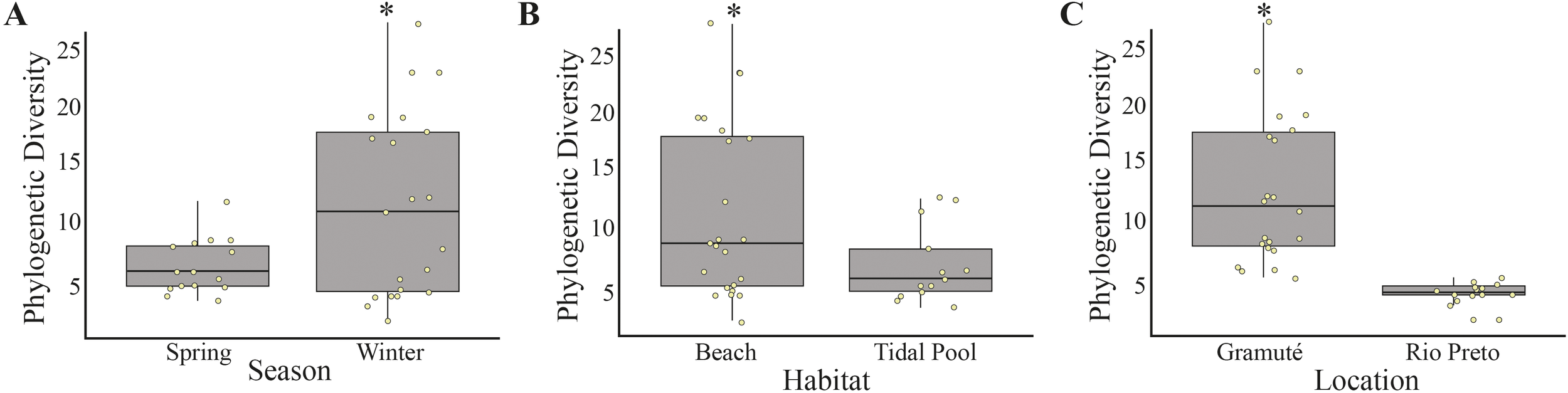
Faith’s phylogenetic diversity of meiofaunal DNA extracted from sediment samples per (A) season, (B) habitat, and (C) location. Asterisks (*) indicate significant differences.
The PCA analysis showed a clear separation between sandy beach and tide pool samples. The first axis (PCA1; 43.1%) reflected variations within habitats, primarily associated with gradients in carbonate content, TOM, CHO, %sand, and BPC content, with positive loadings, while PRT: CHO ratio showed a strong negative loading, followed by %gravel. Although PCA1 captured most of the variance, it did not clearly separate habitats. Instead, differentiation between sandy beach and tide pool samples was mainly observed along the second axis (PCA2; 19.4%), associated to LIP content, BPC content, temperature, and CHO: LIP ratio. Overall, phylogenetic diversity was positively related to CHO: LIP ratio, total organic matter (TOM) and %sand, and negatively related to %gravel, temperature, salinity, BPC, PRT, LIP content, and PRT: CHO ratio (Figure 4; Supplementary Table S6).
Figure 4
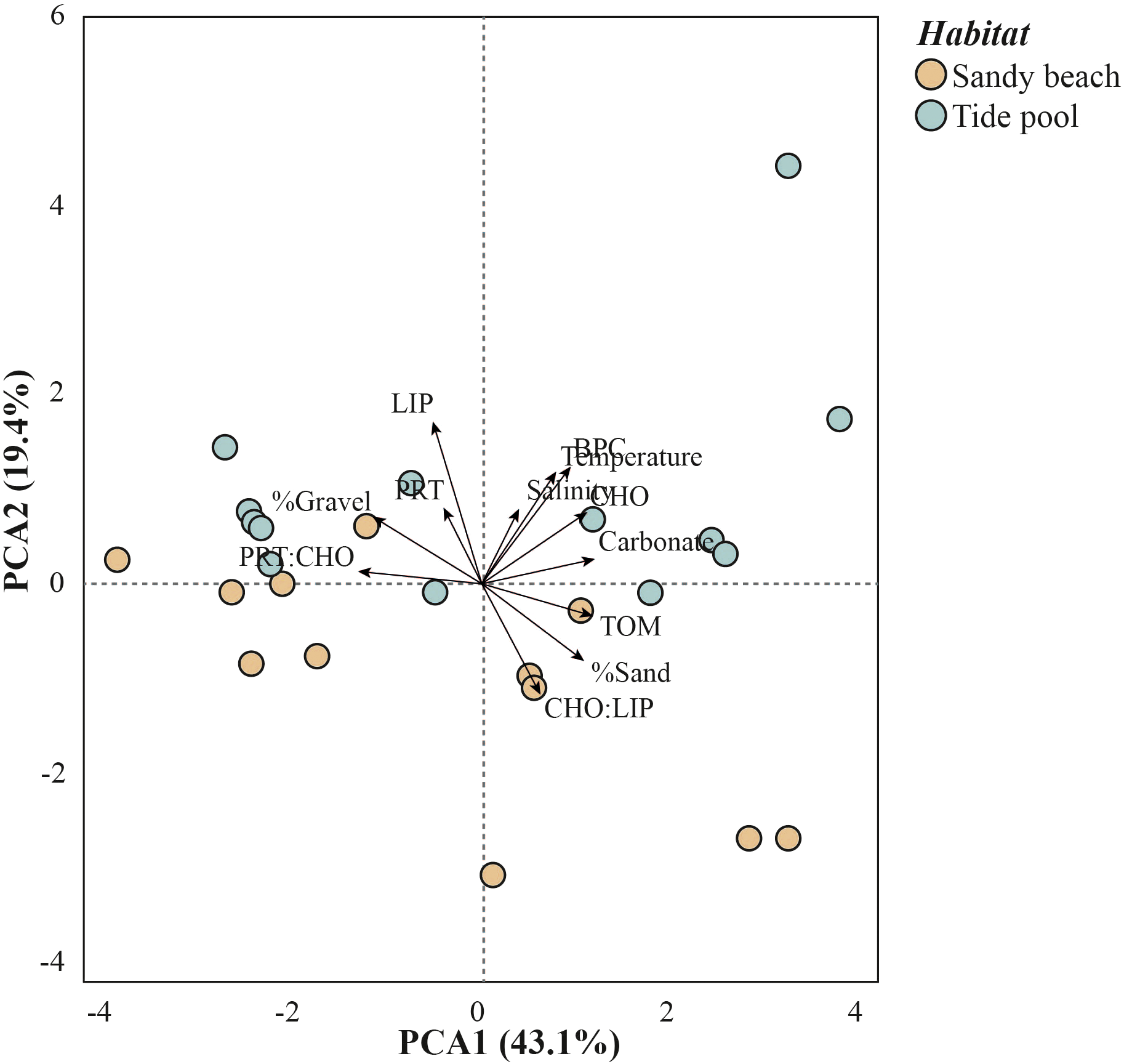
Principal Component Analysis (PCA) of meiofaunal phylogenetic diversity and environmental variables (grain size, temperature, salinity, carbonate content, total organic matter, biopolymers, and biopolymeric carbon content) in tide pools and nearby sandy beaches.
4 Discussion
The metacommunity theory provides a theoretical framework for understanding the dynamics of meiofaunal assemblages in coastal marine environments, such as sandy beaches and tide pools, proposing that biodiversity in a network of interconnected habitats results from processes of species dispersal, selection, drift, and local ecological interactions (Leibold et al., 2004). In the context of our study, the dominance of turnover over nestedness suggests that coastal neighboring habitats (sandy beaches and tide pools) may hold distinct assemblages within the metacommunity, with species adapted to specific environmental conditions and limited by dispersal barriers or competitive interactions. The different environmental conditions within tide pools, such as warmer temperatures and more labile organic matter, may act as ecological drivers shaping meiofaunal metacommunities, leading to significant differences in taxon richness (number of amplicon sequence variants (ASVs)), assemblage composition, and phylogenetic diversity between this habitat and the adjacent sandy beach. Moreover, seasonal variations observed in the studied region indicate that temporal environmental changes also play a crucial role in shaping metacommunities, highlighting the importance of considering both spatial and temporal variability in the conservation and management of these dynamic ecosystems.
Marked environmental differences were found between adjacent sandy beaches and tide pools. In general, tide pools offered warmer conditions (+ 1.8°C), higher sedimentary total lipids and biopolymeric carbon, and similar total organic matter and protein content as sandy beaches. These conditions suggest that although meiofaunal organisms may be subject to temperature stress in tide pools more frequently, the quality of organic matter available for consumers may also be an important driver shaping meiofaunal biodiversity in these coastal habitats. Previous studies have recognized the positive effects of labile sedimentary organic matter to the meiofaunal assemblages (Fabiano and Danovaro, 1994; Fabiano et al., 1995; Danovaro, 1996; Neto et al., 2021; Coppo et al., 2024b).
Meiofaunal phylogenetic diversity was positively correlated with granulometry (%Sand), sediment organic matter content (TOM) and quality (carbohydrate-to-lipid ratio, CHO: LIP), but negatively influenced by temperature, salinity, PRT, LIP and BPC content, and protein-to-carbohydrate ratio (PRT: CHO). Since phylogenetic diversity was significantly lower in tide pools, these findings suggest that meiofaunal assemblages in tide pools may be shaped by adaptations to aged organic matter (low CHO: LIP and PRT: CHO ratios) and by competitive pressure from larger meiofaunal organisms, such as crustaceans and annelids as observed in our study, which are known to be strong competitors. This aligns with previous studies that have highlighted the role of organic matter composition in shaping meiofaunal assemblages, particularly when strong competitors may limit access to fresh available organic matter (Antón et al., 2011; Neto et al., 2021). The role of interspecific competition in structuring meiofaunal assemblage aligns with the idea that phylogenetically related species cannot coexist due to overlapping ecological traits, whereas more phylogenetically distant species are less constrained by competition (Graves and Gotelli, 1993; Webb et al., 2002). Temperature was negatively associated with meiofaunal phylogenetic diversity in tide pools. This may be explained by the fact that meiofaunal communities are strongly influenced by the maximum temperature experienced in the habitat (Wieser and Schiemer, 1977; Schratzberger and Somerfield, 2020). Our findings support the idea that elevated temperatures in tide pools may reduce phylogenetic diversity by selecting for taxa with higher thermal tolerance. Additionally, we observed seasonal patterns in phylogenetic diversity and taxon richness, supporting our hypothesis. In sandy beach environments, meiofaunal phylogenetic diversity tends to show seasonal fluctuations, likely in response to significant seasonal fluctuations on characteristics of the beach system, such as temperature, salinity, rainfall, and organic matter content (Coull, 1988; Albuquerque et al., 2007; Coppo et al., 2024b). In tide pools, meiofaunal diversity and abundance can be positively influenced by the abundance of macroalgae, that may fluctuate seasonally, as well as temperature and salinity (Losi et al., 2018). However, different meiofaunal taxa may respond differently to environmental changes due to their specific adaptations of dispersion, nutrition, development, and reproduction (Curini-Galletti et al., 2012).
In sandy beaches, the meiofaunal phylogenetic diversity was positively influenced by granulometry (%Sand), temperature, and food quality and quantity (BPC content, TOM and CHO: LIP), and negatively influenced by %gravel, protein (PRT), lipids (LIP), and PRT: CHO. These results may suggest that the total organic matter content and accumulation of labile organic matter (BPC) are associated with higher meiofaunal phylogenetic diversity, even if part of it is aged or partially degraded (Lastra et al., 2006; Rodil et al., 2012; Venturini et al., 2012; Corte et al., 2022; Coppo et al., 2024b). Protein, which is a highly nutritional part of organic matter (Joseph et al., 2008) and PRT: CHO, were also negatively related to meiofaunal phylogenetic diversity in sandy beaches. This unexpected relationship may reflect strong inter-specific competition for high-quality resources, where dominant competitors may outcompete other, suppressing the overall biodiversity (Kalmykov and Kalmykov, 2012; Bianchelli et al., 2016; Coppo et al., 2024b).
Our study revealed a distinct metazoan meiofauna in coastal intertidal habitats in the SW Atlantic, demonstrating that environmental conditions of sandy beaches and tide pools influence their assemblage composition and phylogenetic diversity. Understanding the main factors that affect meiofaunal biodiversity is crucial to defining conservation strategies and priority conservation areas (Pittman et al., 2021). Therefore, our study supports existing efforts to include sandy beaches and tide pools in areas of relevant biological significance (Fanini et al., 2020; Harris and Defeo, 2022). This metabarcoding assessment is only the second molecular record of benthic animals for this region, and the first to study benthic metazoans in tide pools at this region. We recognize the importance that other eukaryotes and prokaryotes have to shape the meiobenthic community, and these taxa should be included in future assessments for this area. We understand that metabarcoding studies are influenced by different methodological limitations, such as PCR errors, primer biases, incompleteness of genetic databases with meiofaunal sequences, and the choice and number of genetic markers used. These factors can affect both the detection and accurate taxonomic assignment of meiofaunal organisms, potentially leading to underestimation of diversity or misrepresentation of community composition (Adams et al., 2019; Beng and Corlett, 2020; Steyaert et al., 2020; Castro et al., 2021). Acknowledging these limitations is essential for interpreting the results with caution and for guiding future efforts to improve molecular reference libraries and methodological protocols for meiofaunal biodiversity assessments.
Our findings also highlight the importance of temporal studies to understand how meiofaunal assemblages may vary over time in response to fluctuations in environmental conditions in tropical regions (Bernardino et al., 2016). Additionally, the reduced phylogenetic diversity and distinct taxonomic composition observed in tide pools (+ 1.8°C) highlight future risks for coastal marine habitats under climate warming. In tide pools, meiofaunal assemblages were dominated by a few taxa and phylogenetic diversity was negatively associated with higher temperature, salinity, and labile organic matter, suggesting that elevated thermal stress may lead to strong environmental filtering in these environments. Global average temperature has increased approximately 0.2°C per decade over the last 30 years and projections suggest an increase of +1.0°C to +3.0°C in the SW Atlantic by 2100 (Sánchez et al., 2015; Hoegh-Guldberg and Bruno, 2010; Schratzberger and Somerfield, 2020). Regionally, it is expected that the Eastern Brazilian Marine Ecoregion faces higher temperatures and lower rainfall, which could lead to loss of meiofaunal biodiversity and impacts on several benthic attributes and processes.
5 Conclusion
Tide pools hold different meiofaunal assemblages from those found on nearby sandy beaches, exhibiting both lower taxon richness and phylogenetic diversity. In both habitats, phylogenetic diversity varied seasonally, influenced by temperature, food availability, and organic matter quality. Although our initial hypothesis proposed that tide pool assemblages would be a nested subset of the meiofauna found on sandy beaches, our findings indicate that tide pools support unique assemblages. Additionally, our results support the second hypothesis, as significant seasonal differences were observed in meiofaunal composition and phylogenetic diversity across habitats, with higher phylogenetic diversity in winter than in spring. Furthermore, our study suggests that conditions of higher temperatures and aged organic matter may be responsible for meiofaunal assemblages with lower phylogenetic diversity. Understanding how meiofaunal assemblages respond to warming scenarios is critical in a changing ocean, underscoring the importance of long-term monitoring programs.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, sra/SRR24675047.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
GC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. SN: Formal Analysis, Investigation, Resources, Validation, Writing – review & editing. AB: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by PELD-ES grants from Fundação de Amparo à Pesquisa e Inovação do Espirito Santo (186/2021) and CNPq (441107/2020-6). GC received a scholarship from Coordenação de Aperfeiçoamento de Pessoal em Nível Superior -CAPES (88882.385194/2019-01), and a travel grant from Fundação de Amparo do Espirito Santo (393/2021). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors do not have any conflict of interest to declare. This is a PELD-HCES contribution #21
Conflict of interest
Author SN was employed by company Marítima Estudos Bênticos.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1534512/full#supplementary-material
References
1
Adams C. I. M. Knapp M. Gemmell N. J. Jeunen G.-J. Bunce M. Lamare M. D. et al . (2019). Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool? Genes10, 192. doi: 10.3390/genes10030192
2
Albuquerque E. F. Pinto A. P. B. Perez A. A. Q. Veloso V. G. (2007). Spatial and temporal changes in interstitial meiofauna on a sandy ocean beach of South America. Braz. J. Oceanogr.55, 121–131. doi: 10.1590/S1679-87592007000200005
3
Amaral-Zettler L. Peplies J. Ramette A. Fuchs B. Ludwig W. Glöckner F. O. (2008). Proceedings of the international workshop on Ribosomal RNA technology, April7–9, 2008, Bremen, Germany. Syst. Appl. Microbiol.31, 258–268. doi: 10.1016/j.syapm.2008.08.004
4
Anderson M. J. Gorley R. N. Clarke K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods (Plymouth, UK: PRIMER-E).
5
Anderson M. J. Walsh D. C. I. (2013). PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr.83, 557–574. doi: 10.1890/12-2010.1
6
Andrades R. Andrade J. M. Jesus-Junior P. S. Macieira R. M. Bernardino A. F. Giarizzo T. et al . (2019a). Multiple niche-based analyses reveal the dual life of an intertidal reef predator. Mar. Ecol. Prog. Ser.624, 131–141. doi: 10.3354/meps13027
7
Andrades R. Jackson A. L. Macieira R. M. Reis-Filho J. M. Bernardino A. F. Joyeux J.-C. et al . (2019b). Niche-related processes in island intertidal communities inferred from stable isotopes data. Ecol. Indic.104, 648–658. doi: 10.1016/j.ecolind.2019.05.039
8
Antón A. Cebrian J. Heck K. L. Duarte C. M. Sheehan K. L. Miller M.-E. C. et al . (2011). Decoupled effects (positive to negative) of nutrient enrichment on ecosystem services. Ecol. Appl.21, 991–1009. doi: 10.1890/09-0841.1
9
Ape F. Sarà G. Airoldi L. Mancuso F. P. Mirto S. (2018). Influence of environmental factors and biogenic habitats on intertidal meiofauna. Hydrobiologia807, 349–366. doi: 10.1007/s10750-017-3410-1
10
Baia E. Venekey V. (2019). Distribution patterns of meiofauna on a tropical macrotidal sandy beach, with special focus on nematodes (Caixa d’Água, Amazon Coast, Brazil). Braz. J. Oceanogr67, e19230. doi: 10.1590/s1679-87592019023006701
11
Baselga A. Orme D. Villeger S. De Bortoli J. Leprieur F. Logez M. et al . (2023). betapart: Partitioning Beta Diversity into Turnover and Nestedness Components (Santiago de Compostela, Spain: R package version 1.6). Santiago de Compostela, Spain. Available online at: https://CRAN.R-project.org/package=betapart (Accessed March 25, 2024).
12
Bellisario B. Fais M. Duarte S. Vieira P. E. Canchaya C. Costa F. O. (2021). The network structure of intertidal meiofaunal communities from environmental DNA metabarcoding surveys in Northwest Iberia. Aquat. Sci.83, 71. doi: 10.1007/s00027-021-00828-1
13
Beng K. C. Corlett R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodivers. Conserv29, 208–212. doi: 10.1007/s10531-020-01980-0
14
Bernardino A. F. Azevedo A. R. B. Pereira Filho A. C. D. Gomes L. E. O. Bissoli L. B. Barros F. C. R. (2018). “Benthic estuarine assemblages of the eastern marine Brazilian ecoregion,” in Brazilian Estuaries: A Benthic Perspective. Eds. LanaP. C.BernardinoA. F. (Springer International Publishing, Cham), 95–116.
15
Bernardino A. F. Netto S. A. Pagliosa P. R. Barros F. Christofoletti R. A. Rosa Filho J. S. et al . (2015). Predicting ecological changes on benthic estuarine assemblages through decadal climate trends along Brazilian Marine Ecoregions. Estuar. Coast. Shelf. Sci. Part A166, 74–82. doi: 10.1016/j.ecss.2015.05.021
16
Bernardino A. F. Pagliosa P. R. Christofoletti R. A. Barros F. Netto S. A. Muniz P. et al . (2016). Benthic estuarine communities in Brazil: moving forward to long term studies to assess climate change impacts. Braz. J. Oceanogr.64, 81–96. doi: 10.1590/S1679-875920160849064sp2
17
Bernardino A. F. Pais F. S. Oliveira L. S. Gabriel F. A. Ferreira T. O. Queiroz H. M. et al . (2019). Chronic trace metals effects of mine tailings on estuarine assemblages revealed by environmental DNA. PeerJ7, e8042. doi: 10.7717/peerj.8042
18
Bianchelli S. Buschi E. Danovaro R. Pusceddu A. (2016). Biodiversity loss and turnover in alternative states in the Mediterranean Sea: a case study on meiofauna. Sci. Rep.6, 34544. doi: 10.1038/srep34544
19
Blanchette C. A. Miner C. M. Raimondi P. T. Lohse D. Heady K. E. K. Broitman B. R. (2008). Biogeographical patterns of rocky intertidal communities along the Pacific coast of North America. J. Biogeogr.35, 1593–1607. doi: 10.1111/j.1365-2699.2008.01913.x
20
Bolyen E. Rideout J. R. Dillon M. R. Bokulich N. A. Abnet C. Al-Ghalith G. A. et al . (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857. doi: 10.1038/s41587-019-0209-9
21
Brannock P. M. Halanych K. M. (2015). Meiofaunal community analysis by high-throughput sequencing: comparison of extraction, quality filtering, and clustering methods. Mar. Genomics23, 67–75. doi: 10.1016/j.margen.2015.05.007
22
Cadotte M. W. Tucker C. M. (2017). Should environmental filtering be abandoned? Trends Ecol. Evol.32, 429–437. doi: 10.1016/J.TREE.2017.03.004
23
Callahan B. J. McMurdie P. J. Rosen M. J. Han A. W. Johnson A. J. A. Holmes S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583. doi: 10.1038/nmeth.3869
24
Castro L. R. Meyer R. S. Shapiro B. Shirazi S. Cutler S. Lagos A. M. et al . (2021). Metabarcoding meiofauna biodiversity assessment in four beaches of Northern Colombia: effects of sampling protocols and primer choice. Hydrobiologia848, 3407–3426. doi: 10.1007/s10750-021-04576-z
25
Chargulaf C. A. Tibbetts I. R. (2015). Spatial and temporal variation of meiofauna community structure in soft-sediment pools around Moreton Bay, Australia. Aust. J. Zool.63, 204–213. doi: 10.1071/ZO14063
26
Cisneros K. O. Smit A. J. Laudien J. Schoeman D. S. (2011). Complex, dynamic combination of physical, chemical and nutritional variables controls spatio-temporal variation of sandy beach community structure. PLoS One6, e23724. doi: 10.1371/journal.pone.0023724
27
Coppo G. C. Netto A. S. Pais F. S. Mazzuco A. C. A. Halanych K. M. Bernardino A. F. (2024a). Metabarcoding reveals meiofaunal diversity in rhodolith beds from SE Brazil. Aquat. Conserv.34, e70036. doi: 10.1002/aqc.v34.12
28
Coppo G. Pais F. S. Ferreira T. O. Halanych K. M. Donnelly K. Mazzuco A. C. et al . (2023). Transition of an estuarine benthic meiofauna assemblage 1.7 and 2.8 years after a mining disaster. PeerJ11, e14992. doi: 10.7717/peerj.14992
29
Coppo G. C. Pereira A. P. Netto A. S. Bernardino A. F. (2024b). Meiofauna at a tropical sandy beach in the SW Atlantic: the influence of seasonality on diversity. PeerJ12, e17727. doi: 10.7717/peerj.17727
30
Corte G. N. Checon H. H. Esmaeili Y. S. Defeo O. Turra A. (2022). Evaluation of the effects of urbanization and environmental features on sandy beach macrobenthos highlights the importance of submerged zones. Mar. Pollut. Bull.182, 113962. doi: 10.1016/j.marpolbul.2022.113962
31
Coull B. C. (1988). “Ecology of the marine meiofauna,” in Introduction to the Study of Meiofauna. Eds. HigginsR. P.ThielH. (Smithsonian Institution Press, Washington, D.C), 18–38.
32
Coull B. C. Wells J. B. J. (1983). Refuges from fish predation: Experiments with phytal meiofauna from the New Zealand rocky intertidal. Ecology64, 1599–1609. doi: 10.2307/1937513
33
Creer S. Fonseca V. G. Porazinska D. L. Giblin-Davis R. M. Sung W. Power D. M. et al . (2010). Ultrasequencing of the meiofaunal biosphere: practice, pitfalls, and promises. Mol. Ecol.13, 4–20. doi: 10.1111/j.1365-294X.2009.04473.x
34
Curini-Galletti M. Artois T. Delogu V. De Smet W. H. Fontaneto D. Jondelius U. et al . (2012). Patterns of diversity in soft-bodied meiofauna: dispersal ability and body size matter. PLoS One7, e33801. doi: 10.1371/journal.pone.0033801
35
Danovaro R. (1996). Detritus-Bacteria-Meiofauna interactions in a seagrass bed (Posidonia oceanica) of the NW Mediterranean. Mar. Biol.127, 1–13. doi: 10.1007/BF00993638
36
Danovaro R. (2010). “Bioavailable organic matter total and enzymatically hydrolyzable proteins, carbohydrates, and lipids,” in Methods for the Study of Deep- Sea Sediments, their Functioning and Biodiversity. Ed. DanovaroR. (CRC Press, Florida), 23–44.
37
Dias M. Roma J. Fonseca C. Pinto M. Cabral H. N. Silva A. et al . (2016). Intertidal pools as alternative nursery habitats for coastal fishes. Mar. Biol. Res.12, 331–344. doi: 10.1080/17451000.2016.1143106
38
Esteves A. M. Bloise C. Nogueira C. S. R. (1998). Variação espaço-temporal da meiofauna ao longo de um período quinzenal, em um ponto fixo da Praia Vermelha, Rio de Janeiro. Simpósio Ecossistemas Brasileiros4, 179–193.
39
Fabiano M. Danovaro R. (1994). Composition of organic matter in sediments facing a river estuary (Tyrrhenian Sea): relationships with bacteria and microphytobenthic biomass. Hydrobiologia277, 71–84. doi: 10.1007/BF00016755
40
Fabiano M. Danovaro R. Fraschetti S. (1995). A three-year time series of elemental and biochemical composition of organic matter in subtidal sandy sediments of the Ligurian Sea (northwestern Mediterranean). Continent. Shelf. Res.15, 1453–1469. doi: 10.1016/0278-4343(94)00088-5
41
Fais M. Duarte S. Vieira P. E. Sousa R. Hajibabaei M. Canchaya C. A. et al . (2020). Small-scale spatial variation of meiofaunal communities in Lima estuary (NW Portugal) assessed through metabarcoding. Estuar. Coast. Shelf. Sci.258, 106683. doi: 10.1016/j.ecss.2020.106683
42
Fanini L. Defeo O. Elliott M. (2020). Advances in sandy beach research – local and global perspectives. Estuar. Coast. Shelf. Sci.234, 106646. doi: 10.1016/j.ecss.2020.106646
43
Galois R. Blanchard G. Seguignes M. Huet V. Joassard L. (2000). Spatial distribution of sediment particulate organic matter on two estuarine intertidal mudflats: a comparison between Marennes-Oleron Bay (France) and the Humber Estuary (UK). Cont. Shelf. Res.20, 1199–1217. doi: 10.1016/S0278-4343(00)00019-4
44
Gerchacov S. M. Hatcher P. G. (1972). Improved technique for analysis of carbohydrates in the sediment. Limnol. Oceanogr.17, 938–943. doi: 10.4319/lo.1972.17.6.0938
45
Giere O. (2009). Meiobenthology: The Microscopic Motile Fauna of Aquatic Sediments (second ed.) (Berlin: Springer).
46
Graves G. R. Gotelli N. J. (1993). Assembly of avian mixed-species flocks in Amazonia. Proc. Nat. Acad. Sci.90, 1388–1391. doi: 10.1073/pnas.90.4.1388
47
Griffiths J. R. Kadin M. Nascimento F. J. A. Amelander T. Törnroos A. Bonaglia S. et al . (2017). The importance of benthic–pelagic coupling for marine ecosystem functioning in a changing world. Glob. Change. Biol.23, 2179–2196. doi: 10.1111/gcb.2017.23.issue-6
48
Hakenkamp C. C. Palmer M. A. (2000). “The ecology of hyporheic meiofauna,” in Streams and Ground Waters. Eds. JonesJ. B.MulhollandP. J. (Academic Press, Cambridge), 307–336.
49
Harris L. R. Defeo O. (2022). Sandy shore ecosystem services, ecological infrastructure, and bundles: New insights and perspectives. Ecosyst. Serv.57, 101477. doi: 10.1016/j.ecoser.2022.101477
50
Hartree E. F. (1972). Determination of proteins: a modification of the Lowry method that give a linear photometric response. Anal. Biochem.48, 422–427. doi: 10.1016/0003-2697(72)90094-2
51
Higgins R. P. Thiel H. (1988). Introduction to the Study of Meiofauna (Washington DC: Smithsonian Institution Press).
52
Hoegh-Guldberg O. Bruno J. F. (2010). The impact of climate change on the world’s marine ecosystems. Science328 (5985), 1523–1528. doi: 10.1126/science.1189930
53
Jochum M. Schneider F. D. Crowe T. P. Brose U. O’Gorman E. J. (2012). Climate-induced changes in bottom-up and top-down processes independently alter a marine ecosystem. Philos. T. R. Soc B367, 2962–2970. doi: 10.1098/rstb.2012.0237
54
Joseph M. M. Ratheesh Kumar C. S. Greesh Kumar T. R. Renjith K. R. Chandramohanakumar N. (2008). Biogeochemistry of surficial sediments in the intertidal systems of a tropical environment. Chem. Ecol.24, 247–258. doi: 10.1080/02757540802119871
55
Kalmykov L. Kalmykov V. (2012). Mechanistic mechanisms of competition and biodiversity. Nat. Prec. 7105, 1–11. doi: 10.1038/npre.2012.7105.1
56
Kunishima T. Tachihara K. (2019). What ecological role do soft-substrate tide pools play for fishes? Difference in community structures between estuarine and coastal tidal flats in subtropical Japan. Mar. Freshwater. Res.71, 737–749. doi: 10.1071/MF19019
57
Lastra M. de la Huz R. Sánchez-Mata A. G. Rodil I. F. Aerts K. Beloso S. et al . (2006). Ecology of exposed sandy beaches in northern Spain: Environmental factors controlling macrofauna communities. J. Sea. Res.55, 128–140. doi: 10.1016/j.seares.2005.09.001
58
Leibold M. A. Holyoak M. Mouquet N. Amarasekare P. Chase J. M. Hoopes M. F. et al . (2004). The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett.7, 601–613. doi: 10.1111/j.1461-0248.2004.00608.x
59
Lines R. Juggernauth M. Peverley G. Keating J. Simpson T. Mousavi-Derazmahalleh M. et al . (2023). A large scale temporal and spatial environmental DNA biodiversity survey of marine vertebrates in Brazil following the Fundao tailings dam failure. Mar. Env. Res.192, 106239. doi: 10.1016/j.marenvres.2023.106239
60
Losi V. Sbrocca C. Gatti G. Sempruci F. Rocchi M. Bianchi C. N. et al . (2018). Sessile macrobenthos (Ochrophyta) drives seasonal change of meiofaunal community structure on temperate rocky reefs. Mar. Environ. Res.142, 295–305. doi: 10.1016/j.marenvres.2018.10.016
61
MacCarthy G. R. (1933). Calcium carbonate in beach sands. J. Sediment. Res.3, 64–67. doi: 10.1306/D4268E5A-2B26-11D7-8648000102C1865D
62
Macher H.-N. Pichler M. Creer S. Martínez A. Fontaneto D. Renema W. (2024). Metacommunity theory and metabarcoding reveal the environmental, spatial, and biotic drivers of meiofaunal communities in sandy beaches. bioRxiv., 1–27. doi: 10.1101/2024.07.17.603914
63
Maria T. F. Silva Filho M. G. Souza T. P. Vanaverbeke J. Vanreusel A. Esteves A. M. (2018). Is the vertical distribution of meiofauna similar in two contrasting microhabitats? A case study of a macrotidal sandy beach. J. Exp. Mar. Bio. Ecol.502, 39–51. doi: 10.1016/j.jembe.2017.08.005
64
Maria T. F. Vanaverbeke J. Esteves A. M. De Troch M. Vanreusel A. (2012). The importance of biological interactions for the vertical distribution of nematodes in a temperate ultra-dissipative sandy beach. Estuar. Coast. Shelf. Sci.97, 114–126. doi: 10.1016/j.ecss.2011.11.030
65
Marsh J. B. Weinstein D. B. (1966). Simple charring method for determination of lipids. J. Lipid. Res.7, 574–576. doi: 10.1016/S0022-2275(20)39274-9
66
Mazzuco A. C. A. Bernardino A. F. (2022). Reef larval recruitment in response to seascape dynamics in the SW Atlantic. Sci. Rep.12, 11809. doi: 10.1038/s41598-022-11809-1
67
Mazzuco A. C. A. Stelzer P. S. Bernardino A. F. (2020). Substrate rugosity and temperature matters: patterns of benthic diversity at tropical intertidal reefs in the SW Atlantic. PeerJ8, e8289. doi: 10.7717/peerj.8289
68
Mazzuco A. C. A. Stelzer P. S. Donadia G. Bernardino J. V. Joyeux J. C. Bernardino A. F. (2019). Lower diversity of recruits in coastal reef assemblages are associated with higher sea temperatures in the tropical South Atlantic. Mar. Env. Res.148, 87–98. doi: 10.1016/j.marenvres.2019.05.008
69
McIntyre A. D. (1969). Ecology of marine meiobenthos. Biol. Rev.44, 245–288. doi: 10.1111/j.1469-185X.1969.tb00828.x
70
McLachlan A. Brown A. C. (2006). The Ecology of Sandy Shores (Burlington: Academic Press).
71
McLachlan A. Defeo O. (2018). The ecology of sandy shores (London: Academic Press).
72
McLachlan A. Jaramillo E. Donn T. E. Wessels F. (1993). Sand beach macrofauna communities: a geographical comparison. J. Coast. Res.15, 27–38.
73
Medlin L. Elwood H. J. Stickel S. Sogin M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene71, 491–499. doi: 10.1016/0378-1119(88)90066-2
74
Mendonça V. Madeira C. Dias M. Vermandele F. Archambault P. Dissanayake A. et al . (2018). What’s in a tide pool? Just as much food web network complexity as in large open ecosystems. PLoS One13, e0200066. doi: 10.1371/journal.pone.0200066
75
Metaxas A. Scheibling R. E. (1993). Community structure and organization of tidepools. Mar. Ecol. Progr.98, 187–198. doi: 10.3354/meps098187
76
Mitwally H. M. Hamdan A. M. (2021). Environmental drivers of meiofaunal natural variability, Egypt, Southeastern Mediterranean. Environ. Monit. Assess.193, 185. doi: 10.1007/s10661-021-08927-0
77
Neto J. M. Bernardino A. F. Netto S. A. (2021). Rhodolith density influences sedimentary organic matter quantity and biochemical composition, and nematode diversity. Mar. Env. Res.171, 105470. doi: 10.1016/j.marenvres.2021.105470
78
Olafsson E. (1991). Intertidal meiofauna of four sandy beaches in Iceland. Ophelia33, 55–65. doi: 10.1080/00785326.1991.10429742
79
Pedregosa F. Varoquaux G. Gramfort A. Michel V. Thirion B. Grisel O. et al . (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res.12, 2825–2830.
80
Pittman S. J. Yates K. L. Bouchet P. J. Alvarez-Berastegui D. Andréfouët S. Bell S. S. et al . (2021). Seascape ecology: identifying research priorities for an emerging ocean sustainability science. Mar. Ecol. Prog.663, 1–29. doi: 10.3354/meps13661
81
Pontarp M. Canbäck B. Tunlid A. Lundberg P. (2012). Phylogenetic analysis suggests that habitat filtering is structuring marine bacterial communities across the globe. Microb. Ecol.64, 8–17. doi: 10.1007/s00248-011-0005-7
82
Quast C. Pruesse E. Yilmaz P. Gerken J. Schweer T. Yarza P. et al . (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic. Acids Res.41, D590–D596. doi: 10.1093/nar/gks1219
83
Quintana C. O. Bernardino A. F. Moraes P. C. Valdemarsen T. Sumida P. Y. G. (2015). Effects of coastal upwelling on the structure of macrofaunal communities in SE Brazil. J. Mar. Syst.143, 120–129. doi: 10.1016/j.jmarsys.2014.11.003
84
R Core Team (2023). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
85
Rice D. L. (1982). The detritus nitrogen problem: new observations and perspectives from organic geochemistry. Mar. Ecol. Prog.9, 153–162. doi: 10.3354/meps009153
86
Rodil I. F. Compton T. J. Lastra M. (2012). Exploring macroinvertebrate species distributions at regional and local scales across a sandy beach geographic continuum. PLoS One7, e39609. doi: 10.1371/journal.pone.0039609
87
Sánchez E. Solman S. Remedio A. R. C. Berbery H. Samuelsson P. Da Rocha R. P. et al . (2015). Regional climate modelling in CLARIS-LPB: a concerted approach towards twenty first century projections of regional temperature and precipitation over South America. Clim. Dyn.45, 2193–2212. doi: 10.1007/s00382-014-2466-0
88
Schratzberger M. Somerfield P. J. (2020). Effects of widespread human disturbances in the marine environment suggest a new agenda for meiofauna research is needed. Sci. Total. Environ.728, 138435. doi: 10.1016/j.scitotenv.2020.138435
89
Segura D. Sharma D. Espin-Garcia O. (2024). Comparing subsampling strategies for metagenomic analysis in microbial studies using amplicon sequence variants versus operational taxonomic units. PLoS One19, e0315720. doi: 10.1371/journal.pone.0315720
90
Stelzer P. S. Mazzuco A. C. A. Gomes L. E. Martins J. Netto S. Bernardino A. F. (2021). Taxonomic and functional diversity of benthic macrofauna associated with rhodolith beds in SE Brazil. PeerJ9, e11903. doi: 10.7717/peerj.11903
91
Steyaert M. Priestley V. Osborne O. Herraiz A. Arnold R. Savolainen V. (2020). Advances in metabarcoding techniques bring us closer to reliable monitoring of the marine benthos. J. Appl.Ecol57, 2234–2245. doi: 10.1111/1365-2664.13729
92
Stoeck T. Bass D. Nebel M. Christen R. Jones M. D. M. Breiner H.-W. et al . (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol.19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
93
Suguio K. (1973). Introducão a Sedimentologia (São Paulo: Editora Edgard Blücher).
94
Todaro M. A. Rocha C. E. F. (2004). Diversity and distribution of marine Gastrotricha along the northern beaches of the State of São Paulo (Brazil), with description of a new species of Macrodasys (Macrodasyida, Macrodasyidae). J. Nat. Hist.38, 1605–1634. doi: 10.1080/0022293031000156169
95
Venturini N. Pita A. L. Brugnoli E. García-Rodríguez F. Burone L. Kandratavicius N. et al . (2012). Benthic trophic status of sediments in a metropolitan area (Rio de la Plata estuary): linkages with natural and human pressures. Estuar. Coast. Shelf Sci.112, 139–152. doi: 10.1016/j.ecss.2011.08.016
96
Vinagre C. Dias M. Fonseca C. Pinto M. T. Cabral H. Silva A. (2015). Use of rocky intertidal pools by shrimp species in a temperate area. Biologia70, 372–379. doi: 10.1515/biolog-2015-0046
97
Vinagre C. Mendonça V. Cereja R. Abreu-Afonso F. Dias M. Mizrahi D. et al . (2018). Ecological traps in shallow coastal waters – Potential effect of heat-waves in tropical and temperate organisms. PLoS One13, e0192700. doi: 10.1371/journal.pone.0192700
98
Webb C. O. Ackerly D. D. Kembel S. W. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst.33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
99
Wieser W. Schiemer F. (1977). The ecophysiology of some marine nematodes from Bermuda: seasonal aspects. J. Exp. Mar. Biol. Ecol.26, 97–106. doi: 10.1016/0022-0981(77)90082-X
100
Zhang K. Jiang X. Zheng P. (2023). Revealing a conservation challenge towards floodplain disconnection: decreasing turnover and increasing nestedness of mollusc metacommunities. Biodivers. Conserv.32, 2893–2908. doi: 10.1007/s10531-023-02634-7
Summary
Keywords
tide pools, sandy beach, meiofauna, benthos, metabarcoding, environmental DNA
Citation
Coppo GC, Netto SA and Bernardino AF (2025) Habitat filtering on benthic meiofauna across tide pools and nearby sandy beaches. Front. Mar. Sci. 12:1534512. doi: 10.3389/fmars.2025.1534512
Received
26 November 2024
Accepted
29 April 2025
Published
21 May 2025
Volume
12 - 2025
Edited by
Alberto Basset, University of Salento, Italy
Reviewed by
Matthew Lee, Universidad de Los Lagos, Chile
Sarah Jane Bownes, Independent Researcher, White River, South Africa
Updates
Copyright
© 2025 Coppo, Netto and Bernardino.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel Carvalho Coppo, coppogabriel@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.