- 1Department of Fisheries, University of Papua, Manokwari, Indonesia
- 2Research and Community Outreach Division, University of Papua, Manokwari, Indonesia
- 3Research Center for Conservation of Marine and Inland Waters Resources, National Research and Innovation Agency, Bogor, Indonesia
- 4Department of Environmental Conservation, University of Massachusetts, Amherst, MA, United States
- 5Marine Mammal and Turtle Division, Southwest Fisheries Science Center, National Marine Fisheries Service, National Oceanic and Atmospheric Administration, La Jolla, CA, United States
Leatherback (Dermochelys coriacea) populations are endangered globally and there is a need to better understand their genetic diversity and structure in order to inform conservation efforts. Most nesting in the western Pacific is concentrated in the Bird’s Head Seascape (BHS) region of West Papua, Indonesia. Previous genetic assessment based on limited mtDNA sequences inadequately represented the demographic complexity that is now evident for West Papua leatherbacks. In this study, we quantified the genetic diversity, connectivity and structure for leatherback populations in the BHS by integrating nuclear and mitochondrial (mt) DNA data. We compared 763-bp sequences of the mtDNA control region and data from 17 microsatellite loci at two beaches, Jeen Yessa (JY) and Jeen Syuab (JS) that represent temporally separated nesting populations. We then leveraged reduced-representation (RAD-capture) and whole genome resequencing approaches to generate genome-wide SNPs. We detected low genetic diversity for all datatypes. A total of 11 mtDNA haplotypes were identified, including two new haplotypes and three previously reported from Atlantic populations. Pairwise tests of haplotype and genotype (microsatellite) frequencies found no evidence of structure between the JY (boreal summer) and JS (boreal winter) populations. Furthermore, admixture and principal components analyses of genomic SNP datasets did not identify any clear genetic structure. These results suggest that the BH leatherbacks represent a single genetic stock based on current criteria for defining population Management Units for sea turtles. This study provides the basis for further population structure assessment that includes other nesting sites in Indonesia and the broader western Pacific.
Introduction
Leatherback turtles (Dermochelys coriacea) are highly migratory marine reptiles threatened with extinction in the Pacific (Wallace et al., 2013). In the eastern Pacific, female turtles lay their eggs on beaches along the coast of Mexico and Central America, and these nesting populations have collapsed (Sarti et al., 2007). In the Indo and Western Pacific, rookeries/nesting populations are extinct or severely depleted, with one last relatively large nesting population in West Papua, Indonesia that has been declining at an alarming rate (Dutton et al., 2007; Tapilatu et al., 2013; Lontoh et al., unpublished1). One of the interesting features of the Bird’s Head leatherback population (Figure 1) is that nesting occurs year-round, unlike most other populations where nesting is seasonal. Nesting activity is bi-modal, with a peak during the boreal summer at the Jeen Yessa (formerly called Jamursba Medi) beaches and a second peak that primarily occurs at the second beach, Jeen Syuab (formerly called Wermon), during the boreal winter. However, it is unclear whether these temporal and spatial nesting patterns represent two demographically distinct populations (Hitipeuw et al., 2007; Tapilatu et al., 2013). Previous studies based on limited sampling and short (496-bp) mtDNA sequences found that the eastern and western Pacific nesting populations were distinct but failed to detect any sub-structuring among the nesting within these two regions (Dutton et al., 1999, 2007). These findings led to the general belief that small leatherback nesting aggregations scattered throughout Melanesia combined with the dense focal point on the northwest coast of Papua-Indonesia, all belonged to a single western Pacific genetic stock (Dutton et al., 2007) that was distinct from the Indo-Pacific (Malaysia) stock, and a single homogeneous eastern Pacific genetic stock (Dutton et al., 1999; Wallace et al., 2010, 2023). However, mtDNA, particularly with shorter sequences, may not provide sufficient fine-scale resolution when the amount of haplotype frequency overlap between nearby nesting populations becomes more widespread (Velez-Zuazo et al., 2008; LeRoux et al., 2012; Komoroske et al., 2017). This is particularly the case for leatherbacks, which are characterized by a low level of mtDNA variation globally (Dutton et al., 1999). The authors of these earlier studies themselves speculated that the apparent homogeneity was likely due to the lack of informative genetic markers rather than the actual absence of stock structure (Dutton et al., 1999, 2007). This was resolved by Dutton et al. (2013), who were able to detect fine-scale structuring in Atlantic leatherbacks using a combination of longer (763-bp) mtDNA sequences and nuclear (microsatellite) markers, however inadequate sampling of key nesting sites has prevented accurate characterization of fine-scale stock structure for leatherbacks within the Pacific.
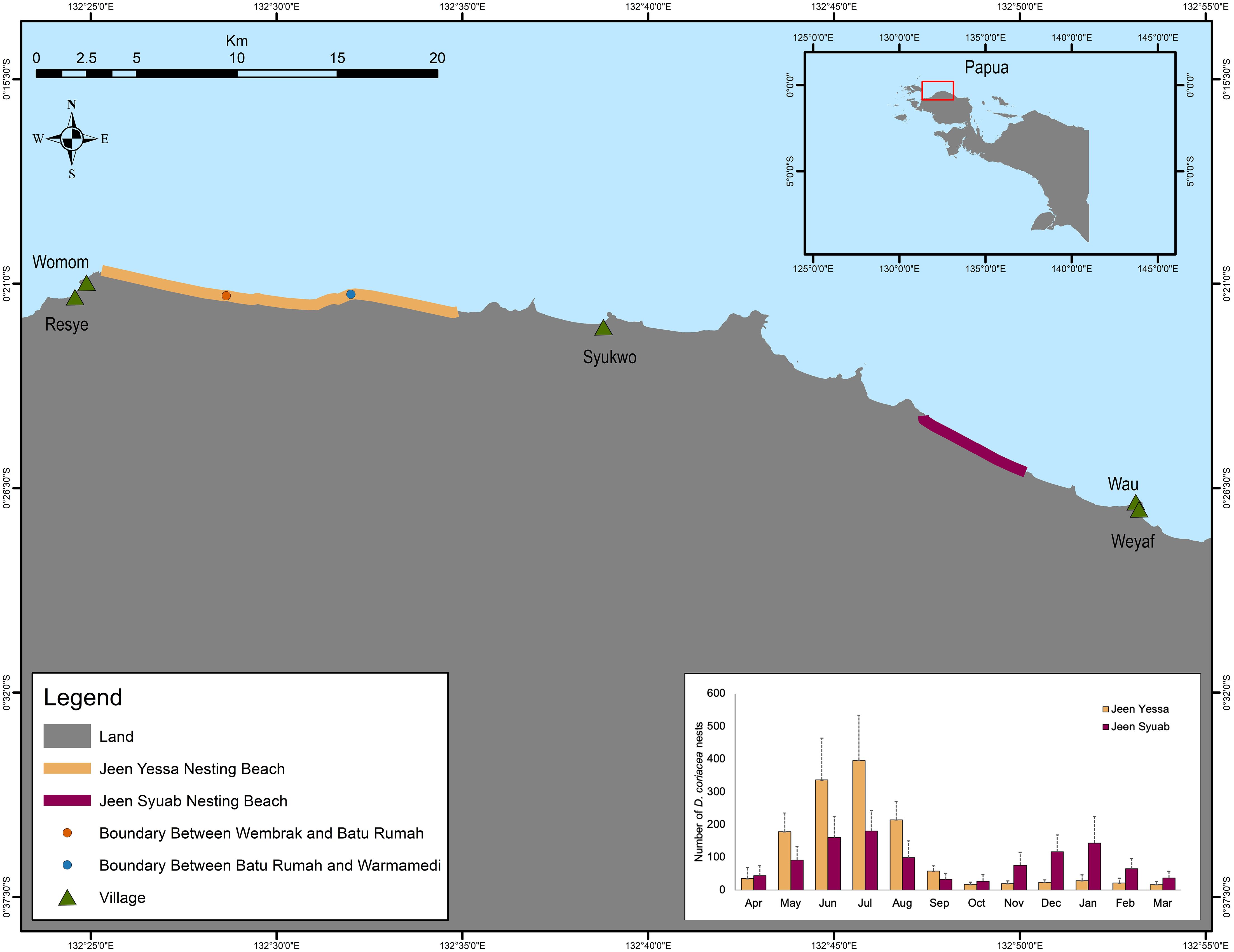
Figure 1. Nesting sites on the northwest coast of the Bird’s Head region of Papua and leatherback monthly nesting activity distribution at each site.
Recently, the application of high-throughput sequencing (HTS) technologies, including Rapture (RAD-Capture; Ali et al., 2016) and whole genome resequencing (WGR) provide further opportunities to dramatically expand the number of loci available for stock structure analysis. Komoroske et al. (2019) developed a flexible Rapture platform using restriction-site associated DNA (RAD) and capture baits to identify thousands of genome-wide Single Nucleotide Polymorphisms (SNPs) for six sea turtle species, including leatherbacks. The availability of a high quality reference genome for leatherback turtles (Bentley et al., 2023), now also allows WGR for optimizing the ability to detect genomic variation in leatherbacks.
Here, we conduct genetic analysis of a comprehensive set of samples from leatherbacks nesting across the Bird’s Head Peninsula using mtDNA sequencing and an array of 17 polymorphic microsatellite loci (Dutton et al., 2013) as well as genome-wide nuclear SNPs to characterize genetic diversity and test for fine-scale population structure between the boreal summer and winter nesting populations at Jeen Syuab and Jeen Yessa. This study aimed to 1) quantify the genetic diversity of leatherbacks nesting in the Jeen Yessa and Jeen Syuab areas; 2) describe the genetic relationship among the Bird’s Head Seascape (BHS) leatherback nesting populations; and 3) identify the management units (MU) for leatherback turtle populations in Papua-Indonesia.
Materials and methods
Sample collection
We collected blood or skin samples from leatherback turtles from the two monitored index beaches, Jeen Yessa and Jeen Syuab at Bird’s Head, Papua-Indonesia (Figure 1). Samples were collected between 2000 and 2011 from nesting females using protocols described in Dutton et al. (2013).
Laboratory procedures
Total genomic DNA was extracted from tissues obtained in different years using one of the following methods: phenol/chloroform (modified from Sambrook et al., 1989), sodium chloride extraction (modified from Miller et al., 1988), a modified DNEasy® Qiagen extraction kit (Qiagen, Valencia, CA, USA), following standard laboratory protocols (Dutton et al., 2013).
Mitochondrial DNA
Primers LCM15382 (5’ GCTTAACCCTAAAGCATTGG 3’) and H950g (5’GTCTCGGATTTAGGGGTTT 3’) (Abreu-Grobois et al., 2006) were used to amplify an 832-base-pair (bp) fragment at the 5’ end of the mitochondrial control region (mtCR) as described in Dutton et al. (2013). We assigned haplotypes by comparing aligned sequences against a local reference library of published and unpublished leatherback haplotype sequences using Geneious Pro 5.6.3 (http://www.geneious.com). Sequences of both the forward and reverse strands from each individual were compared to confirm variable positions. We standardized nomenclature of haplotypes based on 763-bp alignments for leatherbacks, as described in Dutton et al. (2013). We also aligned sequences to the published 496-bp reference data for haplotypes from Pacific nesting populations in Dutton et al. (1999, 2007) to allow comparisons.
Nuclear DNA
Microsatellites
We genotyped genomic DNA using multiplexed PCR reactions for 17 microsatellite loci using the published reaction schemes for the 17 microsatellite primers as follows: LB99, 14-5, LB110, LB128, LB141, LB142, LB145, LB143, LB133, LB123, LB125, LB157, LB158 (Roden and Dutton, 2011); D1 and C102 (Dutton and Frey, 2009); N32 (Dutton, 1995); and D107 (Dutton et al., 2013). All PCR products were checked for amplification using 2% agarose gels with ethidium bromide staining. Microsatellite alleles were separated by capillary electrophoresis on Applied Biosystems Genetic Analyzers (ABI 3100, ABI 3130 or ABI Prism 3730) using ROX 500 fluorescent size standard and fragments were scored using Genescan 3.1, Genotyper 2.0, or GeneMapper 4.0 software (Applied Biosystems, Foster City, CA, USA). We ran each PCR reaction and genotyping plate with positive and negative controls to ensure high genotyping quality and contamination-free reactions.
Single-nucleotide polymorphisms
We generated genome-wide SNP data using reduced representation (RAD-capture or Rapture; Ali et al., 2016) and whole genome resequencing (WGR; Therkildsen and Palumbi, 2017) approaches. We generated Rapture SNP data for a total of 206 individuals using the sea turtle platform as described in Komoroske et al. (2019), and WGR libraries for 58 individuals using an Illumina Nextera PCR-based protocol adjusted for one tenth reaction sizes, as described in Bentley et al. (2023), adapted from Therkildsen and Palumbi (2017). We used a Fragment Analyzer (Agilent, Santa Clara, CA, USA) to assess DNA quality and quantity (prior to library preparation) and to assess library quality and quantity. Libraries were then pooled based on normalized concentrations and sequenced either at UC Davis Genomics Core Facility (Rapture libraries; Davis, CA, USA; Illumina HiSeq 3000) or Novogene (WGR libraries; Sacramento, CA, USA; NovaSeq 6000) using 150-bp PE sequencing chemistry. Raw sequence files for each dataset were then processed as described in Komoroske et al. (2019) and Bentley et al. (2023) to generate. bam files for downstream analyses. In brief, raw sequence files were demultiplexed and checked for quality using FastQC (Andrews et al., 2012), followed by quality trimming using Trimmomatic v 0.39 (Bolger et al., 2014) with default parameters. Reads were then aligned to the leatherback turtle (D. coriacea) reference genome (Bentley et al., 2023) with BWA-MEM v0.7.17 (Li and Durbin, 2009; Li, 2013), followed by marking PCR duplicates and adding read group headers using Picard-Tools v2.23.2 using the MarkDuplicates and AddOrReplaceReadGroups functions, respectively (http://broadinstitute.github.io/picard).
Data analysis
Data were grouped into two populations for further analysis to represent the boreal summer and winter nesting population: the summer population included nesters sampled primarily from Jeen Yessa from May through September, but included some turtles that nested in Jeen Syuab (from June-August), which are part of the broader Jeen Yessa summer nesting population (Figure 1). Samples from Jeen Syuab were collected primarily during the boreal “winter” nesting season (December – February). For the purposes of this study the Jeen Yessa population refers to data for the boreal summer nesting population.
For the mtDNA data we calculated haplotype (h) and nucleotide (π) diversity for each population using Arlequin v 3.5.1.2 (Excoffier and Lischer, 2010). For the microsatellite data set, we used strataG (Archer et al., 2017) to estimate allelic richness (AR), observed heterozygosity (Ho), expected heterozygosity (He), and the number of alleles per locus (na). Microsatellite loci were screened for linkage disequilibrium and null alleles according to Roden and Dutton (2011). We tested for deviations from Hardy–Weinberg (HW) equilibrium via Markov chain permutation (Guo and Thompson, 1992) using Genepop 4.1 (Raymond and Rousset, 1995).
For the mtDNA data, we tested for differentiation between Jeen Yessa and Jeen Syuab (summer vs winter nesting populations) by conducting an analysis of molecular variance (AMOVA) (Excoffier et al., 1992), pairwise FST comparisons, and pairwise exact tests of population differentiation with Arlequin v 3.5.1.2 (Excoffier and Lischer, 2010). Significance values for AMOVA were obtained from 10,000 permutations. Exact tests of population differentiation were conducted with 100,000 permutations and 10,000 dememorization steps (Raymond and Rousset, 1995). We compared the Bird’s Head sequences with published data for other Pacific nesting sites by aligning our sequences to the 496-bp reference sequences from Dutton et al. (2007) and tested for stock structure based on the truncated haplotypes. In order to estimate differentiation between Jeen Syuab and Jeen Yessa with the microsatellite data set we used StrataG (Archer et al., 2017) to calculate pairwise FST (Weir and Cockerham, 1984) and FST analogs, including F’ST (Meirmans and Hedrick, 2011), G’ST, Jost’s D and χ2 (Winter, 2012). In order to represent the relationships between the haplotypes found within our BHS-Papua Indonesia study area, we generated a network using PopART using a Median-joining method (Bandelt et al., 1999) using default settings in the software Network 4 (http://www.fluxus-engineering.com). We also generated a network to represent relationships between a representative set of published haplotypes globally.
Following pre-processing steps as outlined above, Rapture and WGR datasets were analyzed in parallel for genetic diversity and population structure using ANGSD (Korneliussen et al., 2014) and associated programs that estimate genotype likelihoods (GL) for use in downstream analyses to incorporate uncertainty in individual genotypes at low and/or variable sequencing coverage (Lou et al., 2021). We first estimated heterozygosity for each dataset using GL estimates at all sites and then assessed population structure using GL estimates at variant sites (i.e. SNPs) only via PCAngsd (Meisner and Albrechtsen, 2018). Briefly, PCAngsd uses genotype likelihood estimations at variant sites in principal component analysis to estimate individual allele frequencies and subsequent admixture proportions. We estimated admixture over multiple values of K (1-3) corresponding to ancestry clusters and evaluated most supported values of K using log likelihoods and the amount of variance explained in the first two components of the PCA.
Results
Mitochondrial DNA
We obtained mtCR sequence data from 426 specimens of D. coriacea at two sampling localities in Papua-Indonesia, Jeen Yessa and Jeen Syuab. We identified eleven variable sites within the 763-bp fragment, all of which consisted of transitions (Supplementary Table S1). All variable sites (total number of mutations) are 6 singleton variable sites and 5 parsimony informative sites. Site positions of singleton variable sites (two variants) are 134, 234, 267, 358, 414, and 674; and site positions of parsimony informative sites (two variants) are 199, 212, 292, 606, and 616 (Supplementary Table S1).
Jeen Yessa (JY/JM), with 299 sequences, had 11 haplotypes, while Jeen Syuab (JS/WER), with 127 sequences, had 8 haplotypes, with Dc9.1 accounting for over 81% at both locations. Three rare haplotypes (Dc6.1, and two new haplotypes Dc1.6 and Dc20.1) were only found at Jeen Yessa (Table 1). Five (496bp) haplotypes previously reported in Pacific populations (Dc1, Dc4, Dc5, Dc8, Dc9) were found, while three haplotypes (Dc7, Dc11, and Dc12) observed in East Pacific populations were not detected (Dutton et al., 1999, 2007). Haplotypes Dc1.1, Dc1.4, and Dc4.1 were also found, previously reported only in Atlantic populations (Dutton et al., 2013). The parsimony network for haplotypes within the Pacific shows a star-shaped phylogroup of six closely related haplotypes clustered around one common widespread haplotype (Dc4; Figures 2A, B). A second phylogroup consists of five haplotypes, with Dc9 widespread in the Western Pacific and separated by one step from Dc8 (Figure 2B). Nine of the twelve Pacific haplotypes are present in Indonesia. The global haplotype network indicates a third haplogroup anchored by Dc1.1, common and widespread in the Atlantic (and South Africa in the Indian Ocean) (Figure 2B).
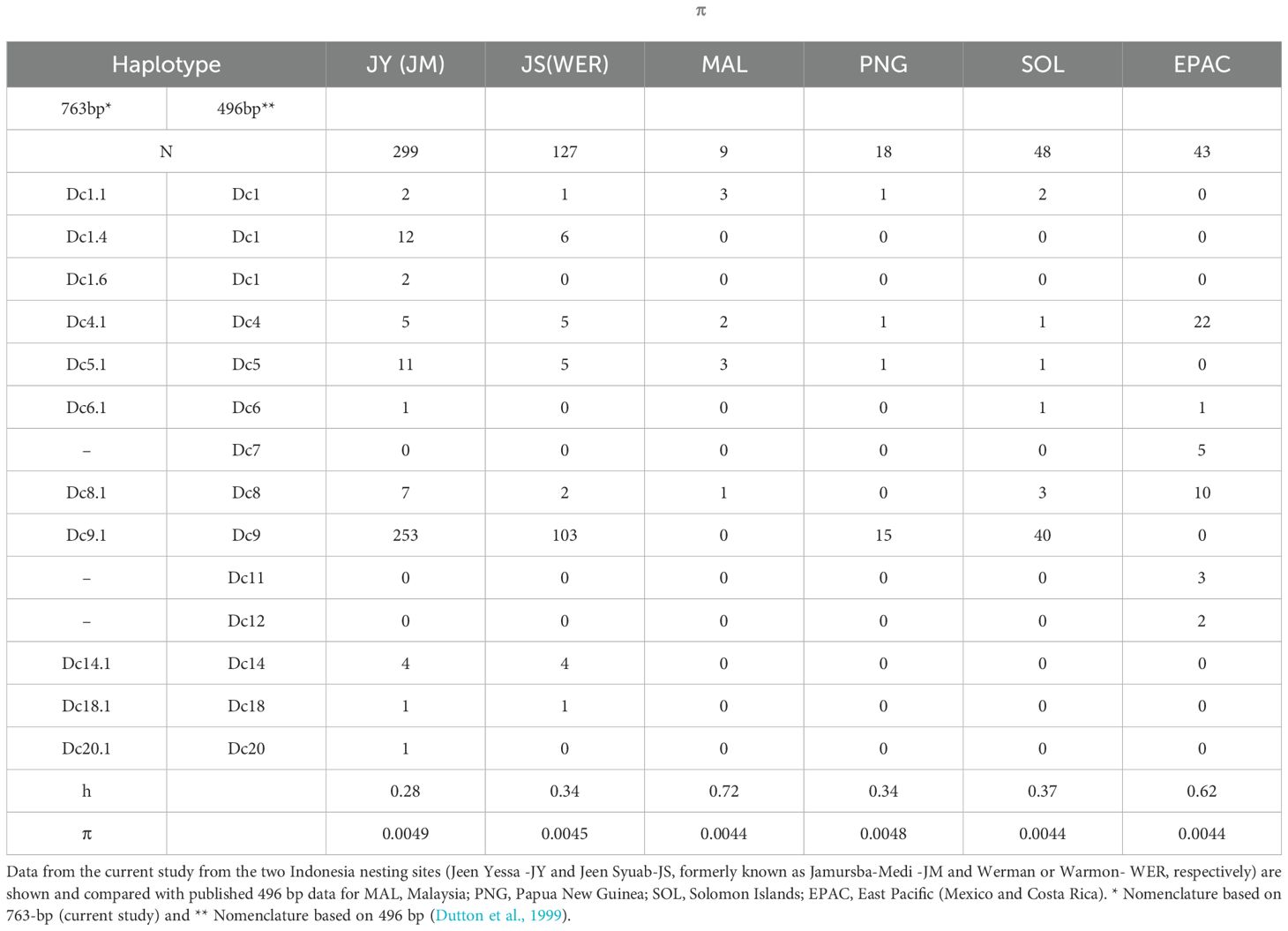
Table 1. MtDNA haplotype frequencies, haplotype (h) and nucleotide diversities (π) for West, East and Indo-Pacific nesting sites.
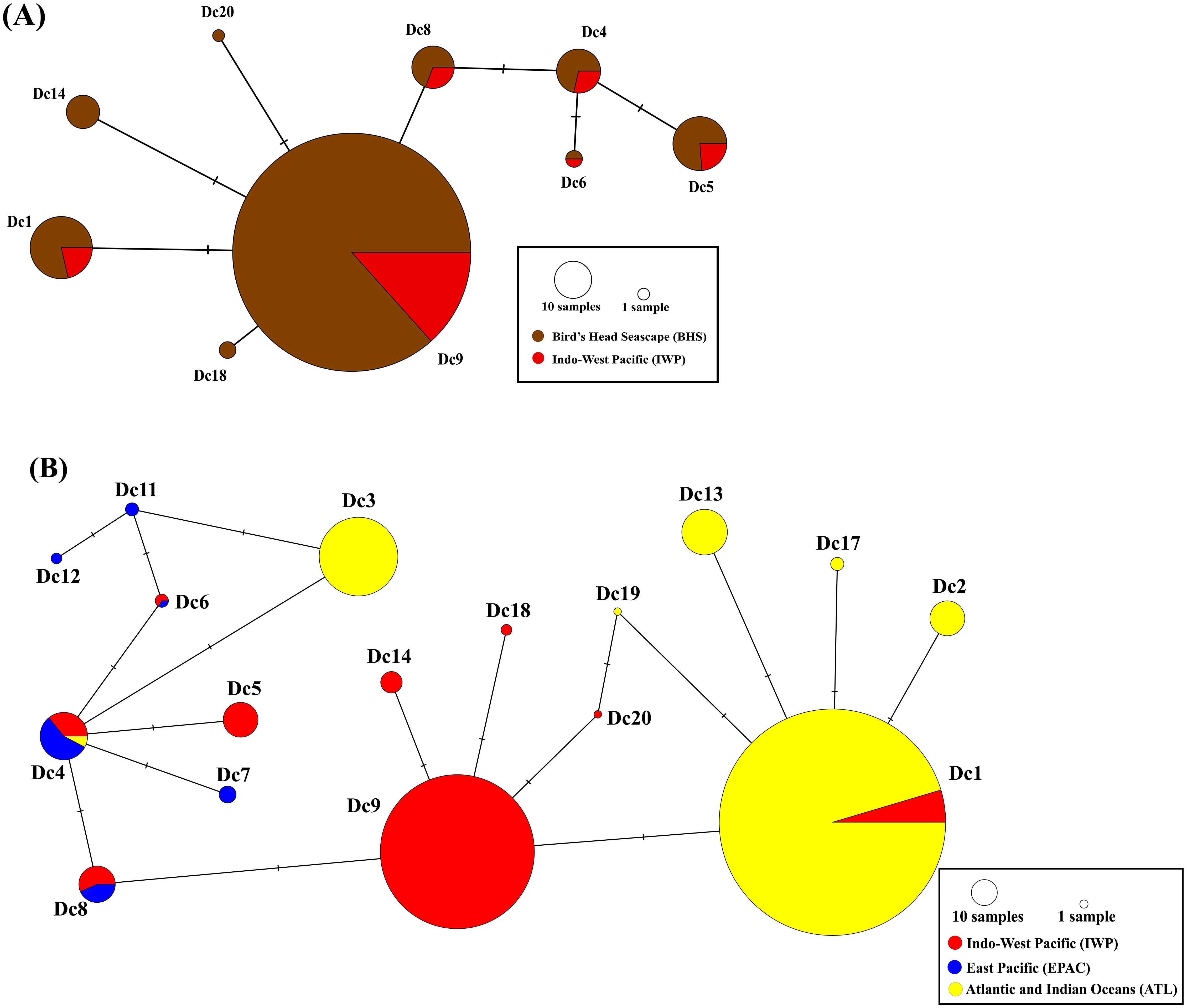
Figure 2. The most parsimonious median joining network of the 496bp of the mtDNA control region for leatherback haplotypes in (A) The Indo-West Pacific (IWP) (BHS, Indonesia (brown), IWP (PNG, Malaysia, Solomon Islands) red and (B) Global rookeries (IWP-red; East Pacific EPAC (Mexico, Costa Rica-blue; Atlantic and Indian Ocean ATL-yellow). The number of mutations between haplotypes is illustrated by dashes in connecting lines. The node size is proportional to the haplotype frequency in the overall sample set (Table 1, Dutton et al., 2013). Colors denote the regions where individual haplotypes were detected and the proportions of shared haplotypes that were distributed among rookeries in different regions (brown=BHS, red= IWP, blue= East Pacific, yellow = Atlantic and Indian Oceans).
Both nesting sites had a haplotype diversity (h) of approximately 0.3, with nucleotide diversities ranging from π = 0.00435 to 0.00488 (Table 1). There was no significant differentiation between Jeen Yessa and Jeen Syuab based on the 763bp results of the pairwise (and exact) tests from this study (FST = 0.002, p>0.05). The AMOVA results based on the 496bp dataset indicated significant population sub-structuring across the Indo, West and East Pacific (Supplementary Table S2). Pairwise FST and exact tests further indicated that the East Pacific was highly differentiated from all the Indo and West Pacific populations (FST =0.15-0.61, p<0.0001), and that only Malaysia was significantly differentiated from the other West Pacific populations (FST =0.46-0.60, p<0.0001; Supplementary Table S3).
Nuclear DNA
We analyzed microsatellite genotypes from 407 samples (291 from JY, 116 from JS; Supplementary Table S4). None of the loci deviated significantly (p < 0.05) from Hardy-Weinberg equilibrium and we found no evidence of linkage disequilibrium. The Jeen Yessa and Jeen Syuab allele frequencies were not significantly different (p>0.3 for FST, F’ST, G’ST, and Jost’s D; p>0.1 for χ2; Supplementary Table S5). Admixture analyses from both datasets of genome-wide SNPs (Rapture = 30,625 SNPs n=202, WGR =3,237,629 SNPs n=58) also indicated no clear genetic structure between Jeen Syuab and Jeen Yessa (Figures 3A, C). In the WGR dataset (available at https://www.ncbi.nlm.nih.gov/bioproject/1254794), we found a value of K=1 to have the most support, though log-likelihood values were similar for values from K=1-3 (K=1: -7707055.87, K=2: -7591677.81, K=3: -7587428.34). Our PCA supported this lack of clustering (Figure 3B), and only 6.02% of variance was explained with the first two PCs. The results in the Rapture dataset were similar (Figure 3D) (K=1: -3694032.20, K=2: -3644746.35, K=3: -3644714.36; 25.64% variance explained with PC1-2). Heterozygosity was estimated to be 0.093 and 0.019 for Jeen Syab, and 0.106 and 0.033 for Jeen Yessa for WGR and Rapture SNPs, respectively. Mean Heterozygosity (for JS and JY combined) was 0.524 based on the microsatellite data (Supplementary Table S6).
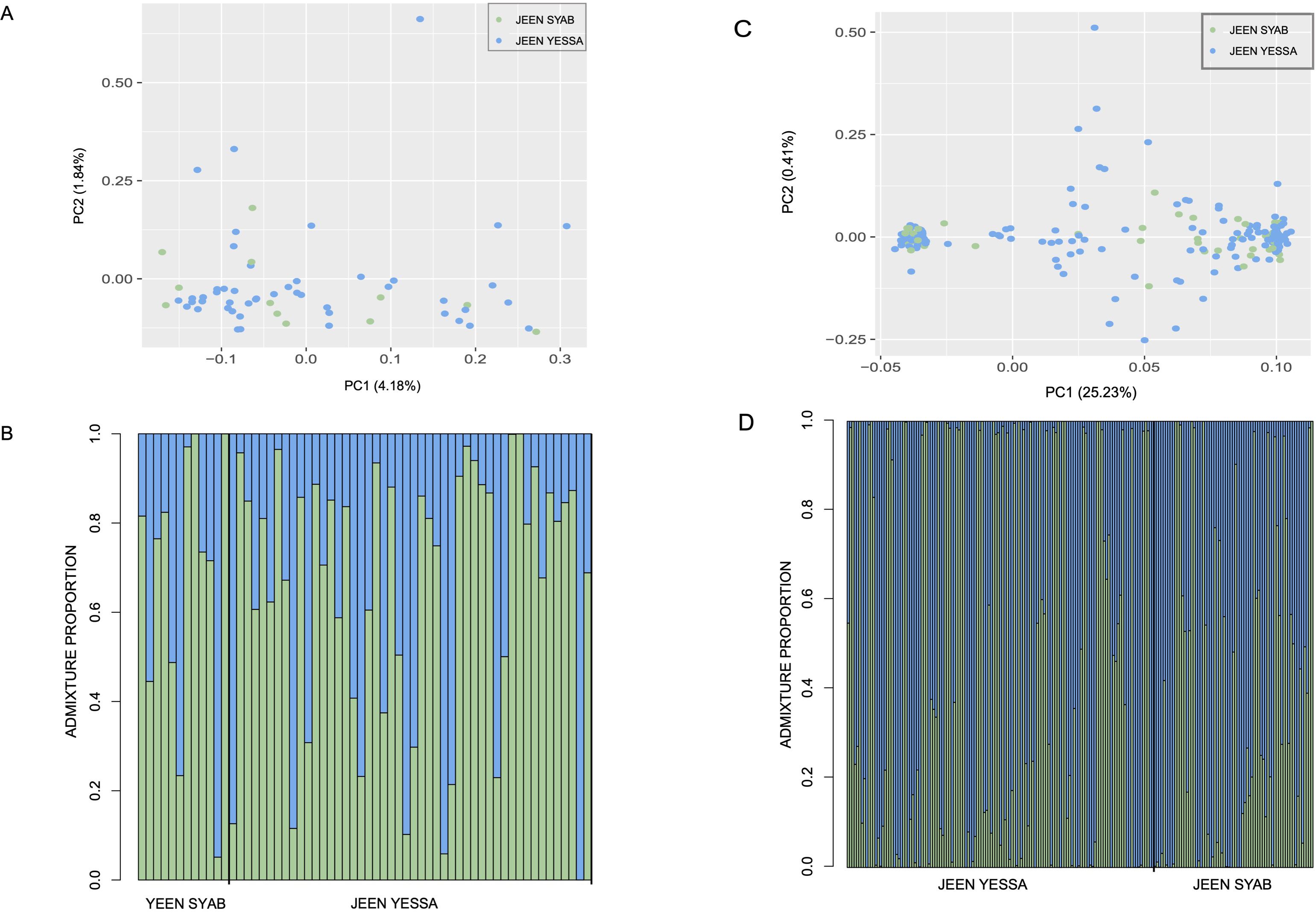
Figure 3. (A) Admixture plot for WGR SNP data and K=2 where each color represents the proportion of genetic variance belonging to each group, and black line separates Jeen Yessa individuals from Jeen Syuab individuals. Each bar represents the genetic variance partitioned to each group for one individual. (B) PCA for WGR SNP data where each point represents one individual from Jeen Yessa or Jeen Syuab. The amount of variance explained by each PC is denoted in parentheses. (C, D) are analogous plots for the Rapture SNP data.
Discussion
This study provides a comprehensive integration of nuclear DNA (nDNA) and mtCR sequence data to address genetic diversity, connectivity, and structure for leatherback turtle populations in the Bird’s Head Seascape, Papua-Indonesia. MtCR is used to elucidate deep divergences between populations, and nDNA microsatellites and SNPs can detect recent structuring (Silver-Gorges et al., 2020). The lack of significant differentiation between Jeen Yessa and Jeen Syuab with either mitochondrial or nuclear DNA markers together provide convincing evidence that the BHS-Papua leatherbacks all comprise one genetic stock (or MU) following the criteria defined by Moritz (1994).
Genetic diversity and population structure
Leatherback turtles in the BHS were generally characterized by low mtDNA nucleotide diversities. These results are consistent with prior studies reporting slower microevolutionary rates in turtles relative to other vertebrates (Avise et al., 1992; Bowen et al., 1993), and leatherbacks having gone through a bottleneck relatively recently (~100,000 years ago), resulting in closely related haplotypes and an evolutionary history of global recolonization (Dutton et al., 1999). The new data we report here for Indonesia further advances this scenario, reflected by the relatively large number of closely related rare haplotypes, clustered around a dominant common haplotype (Dc9.1) that is closely related to the globally distributed ancestral haplotype Dc1.1. Our results for BHS further show that Dc1 is predominantly (87.5%) represented by the 763-bp variants Dc1.4 and Dc1.6 as opposed to Dc1.1 exclusively in the Northwest Atlantic, and predominantly (>80%) in the Southeast Atlantic and Indian Ocean (Dutton et al., 2013). Although the longer sequences did not identify further variation among Dc9, the most common BHS haplotype, our findings illustrate the importance of sequencing the full 763-bp mtCR in order to accurately identify baseline mtDNA haplotype variation. A more comprehensive phylogeographic analysis will be possible when additional data are available for all the other Pacific populations (see Wongfu et al., 2022; Piboon et al., 2025). Furthermore, future work to identify additional variation in the mitogenome for leatherbacks (beyond the mtCR) is warranted, particularly with regard to Dc9.1, in order to improve the power to detect regional stock structure within the West and Indo Pacific. Mitogenomic variation that permits subdividing common mtCR haplotypes into population-informative mitogenomic haplotypes has been found in Pacific green turtles (Chelonia mydas; Frey et al., 2025), Atlantic green turtles (Shamblin et al., 2012), Mediterranean loggerheads (Caretta caretta; Tolve et al., 2024), and Kemp’s ridley lineages (Lepidochelys kempii; Frandsen et al., 2020). The WGR sequence data we generated for the BHS leatherbacks can be further analyzed to recover and contribute mitogenome sequences for future studies when analogous mitogenomic data is available for other populations for comparison.
This low genetic diversity is consistent with those previously reported for D. coriacea globally (Dutton et al., 1999), in the Atlantic (Dutton et al., 2013), and in the Pacific including Papua-Indonesia (Dutton et al., 2007). Preliminary data reported recently from the relatively small numbers of nesting leatherbacks along the coast of West Sumatra (Western Indonesia) found four haplotypes based on notably higher nucleotide site variability (22 mtCR variable sites) compared to other regions (Maslim et al., 2016); however, the haplotype sequence quality could not be verified and the nucleotide variability, and hence number of reported Dc4 haplotype variants is likely an artifact (Maslim, pers. comm). West Sumatra is one of several areas in Indonesia where additional leatherback genetic sampling is warranted to further characterize the regional population diversity (Maslim et al., in prep2). Atlantic nesting populations had haplotype diversities ranging from 0.112 to 0.533 (Vargas et al., 2019) or 0.112 to 0.498 (Dutton et al., 2013), so the haplotype diversity for the BHS (0.28-0.34) that we found is within this range for comparable studies based on longer sequence haplotypes. Interestingly, the Malaysian population that is now extinct, has the highest haplotype diversity (even based on shorter sequence data) based on samples collected just before extinction, over twice as much as our results for BHS.
Our results taken together suggest that the BHS leatherback population is connected locally both by contemporary gene flow as well as more broadly, regionally, by historic patterns of migration and colonization. It is possible that the demographic isolation between the summer and winter nesters is not yet discernable (genomically) because recent (and still unfolding) separation combined with slow microevolutionary rates of turtles (Avise et al., 1992) dampen genetic signals of their isolation. The datasets here provide a basis for further study to explore the role this region has played in shaping the genetic diversity for leatherbacks, and the potential for providing a source for maintaining diversity for leatherbacks across the Indo, West and East Pacific into the future, given that it represents one of the last remaining sizeable populations in this broad region. The geographic and oceanographic features of the BHS are somewhat unique given its location at the northeastern entrance of the ‘Indonesian Throughflow’ which transports water from the Pacific to the Indian Ocean and is spatially and temporally dynamic (Vranes and Gordon, 2005). The inter-annual variation in the IT is associated with the ENSO and Asian monsoons. During the southeast monsoon, the South Equatorial Current (SEC) travels west across the northern coast of the BHS, merging with the Halmahera Eddy and joining the Northern Equatorial Counter Current (NECC) flowing east. The SEC reverses direction during the northwest monsoon (Mangubhai et al., 2012). Strong tidal currents likely promote good larval connectivity among reefs (DeBoer et al., 2008) and genetic connectivity of whale sharks (Toha et al., 2016, 2020) in the Indian and Pacific Oceans including in the BHS. However, given the complex life-history of sea turtles, it is unclear how these regional current patterns influence the genetic connectivity across the region, or how much seasonal reversals of local currents drive hatchling dispersal patterns and promote genetic connectivity between the Jeen Syuab and Jeen Yessa populations (see Gaspar et al., 2012).
Implications for conservation
The IUCN has classified the leatherback turtle (D. coriacea) as a vulnerable species (Wallace et al., 2013). Over recent decades, the species has experienced a significant decline (Benson et al., 2011), with a reported annual nesting decrease of 5.9% at Jeen Yessa and Jeen Syuab beaches since 1984 (Tapilatu et al., 2013). Therefore, protecting and conserving the D. coriacea population in Papua is a conservation priority. Sea turtles have had legal protections in Indonesia since 1999 (Government Regulation (PP) number 7, regarding the Preservation of Plant and Wildlife) that prohibit trade in sea turtle products and by-products and there have been efforts since the 1970’s to study, monitor and protect the BHS leatherbacks. The efforts have recently scaled up and become more consistent and organized under a holistic and science-based management plan (Pakiding et al., 2020) that our results can further inform.
According to Parra et al. (2018) understanding the genetic population structure and demographic history is essential for biota conservation. Genetic diversity has become highly important in the conservation of species, especially in population studies within the last decade. It indicates the population health, resilience, and capacity to adapt to environmental change (Hughes et al., 2008). Due to its role in tracking and preventing harm to populations, genetics play an important role in conservation (Schwartz et al., 2007; Parra et al., 2018). The results in this study indicate that Indonesian leatherbacks have low genetic diversity that is on par with other leatherback populations. This is generally lower than many other vertebrate species, including other sea turtles, however recent studies have found that this is a consequence of longer-term low sustained population sizes, rather than rapid loss of genetic diversity with recent population declines (Bentley et al., 2023). Nonetheless, having lower genetic diversity may limit adaptive capacity for future environmental change. The WGR dataset here for Indonesian leatherbacks represents the first of its kind published for any population of sea turtles and provides a basis to further explore functional genomics, identify genes under selection and begin to assess the adaptive potential of leatherbacks as a species in future studies that include other populations around the world.
The variation in migration and foraging strategies exhibited by the BHS leatherbacks (Bailey et al., 2012) underscore the importance of and the need for ecosystem‐based management and coordinated Pacific‐wide conservation efforts. Thus, it is important to be able to identify oceanic and coastal areas that are critical for population dynamics and those that are of high conservation priority at the population level (Wallace et al., 2018). Our results now provide a baseline for conducting stock ID of fisheries bycatch, strandings and foraging turtles, essential for assessing threats and re-drawing RMU boundaries (see Wallace et al., 2023). Jeen Yessa had three unique mtDNA haplotypes (Dc1.6, Dc6.1, and Dc20.1), with Dc1.6 and Dc20.1 being newly identified and potentially useful for tracking the BHS stock in foraging areas and fisheries by-catch. Recent studies have used mixed-marker approaches combining mtDNA and microsatellites to improve the accuracy and precision of individual stock assignments for Atlantic leatherbacks (Stewart et al., 2013; Roden et al., 2017) and Pacific green turtles (Horne et al., 2023), and ongoing projects attempting to assess origin of the leatherback bycatch in the Pacific can now apply these approaches in a more meaningful way (Roden et al., 2025; Zárate et al., 2025).
Data availability statement
The datasets presented in this study can be found in online repositories. The Genbank IDs of the mtDNA haplotype sequences can be found in the Supplementary Material. The WGS data are available at https://www.ncbi.nlm.nih.gov/bioproject/1254794.
Ethics statement
Ethical review and approval was not required for the animal study because all research in this study complied with all applicable animal welfare laws. Samples are archived in the US National Marine Fisheries Service (NMFS) Marine Turtle Molecular Research Sample Collection at the Southwest Fisheries Science Center and were collected under the national and local authorizations and CITES permit conditions and imported under CITES permit (https://www.fisheries.noaa.gov/west-coast/science-data/marine-mammal-and-sea-turtle-research-tissue-collection).
Author contributions
AT: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. DL: Conceptualization, Data curation, Resources, Validation, Writing – review & editing. FP: Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing. AP: Formal analysis, Methodology, Software, Writing – review & editing. LK: Data curation, Formal analysis, Methodology, Resources, Writing – original draft, Writing – review & editing. PD: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funding provided by National Oceanic and Atmospheric Administration’s National Marine Fisheries Service. Partial funding for fieldwork and sample collection was provided by the U.S. Fish and Wildlife Service, Balai Besar Konservasi Sumber Daya Alam Papua Barat, WWF Indonesia, and the Papua Barat Provincial Government.
Acknowledgments
We would like to acknowledge UNIPA, WWF-Indonesia, NOAA-Fisheries and Regency governments of West Papua Province. We also thank the beach patrollers at Jeen Yessa and Jeen Syuab beaches. We thank Erin LaCasella, Suzanne Roden, Amy Frey (NOAA-Fisheries Southwest Fisheries Science Center), Jamie Adkins and Blair Bentley (University of Massachusetts at Amherst) for technical advice and help with laboratory and data analysis. We thank Kelly Stewart and Suzanne Roden for edits on an earlier draft of the manuscript. Samples were legally and ethically collected under national and international permits (CITES permits- USA: 02US844694/9, 06US844694/9, 07US844694/9, 10US844694/9, 11US844694/9; Indonesia: 11111/IV/SATS-LN/2003, 16825/IV/SATS-LN/2006, 02222/IV/SATS-LN/2008, 16642/IV/SATS-LN/2010, 19568/IV/SATS-LN/2010, 14520/IV/SATS-LN/2011).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1569466/full#supplementary-material
Footnotes
- ^ Lontoh, D. N., Swabra, Y., Batubara, P. P., Mau, J., Leleran, A., Wanaputra, A. A., et al. (unpublished). Overview of marine turtle nesting trends at the Jeen Womom Coastal Park, Tambrauw Regency, Papua Barat Daya Province, Indonesia. Biodiversitas.
- ^ Maslim, A., Nijland, R., Bista, I., Dutton, P. H., and Becking, L. E. (under review). Assessing applicability of eDNA-based sampling for population monitoring of leatherback turtles in the Northeast Indian Ocean.
References
Abreu-Grobois F. A., Horrocks J. A., Formia A., Dutton P. H., LeRoux R. A., Velez-Zuazo X., et al. (2006). “New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis,” in Proceedings of the 26th annual symposium on sea turtle biology. Eds. Frick M., Panagopoulous A., Rees A. F., and Williams K. (NOAA, Myrtle Beach), 179.
Ali O., O’Rourke S., Amish S., Meek M., Luikart G., Jeffres C., et al. (2016). RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 202, 389–400. doi: 10.1534/genetics.115.183665
Andrews S., Krueger F., Segonds-Pichon A., Biggins L., Krueger C., and Wingett S. (2012). FastQC: A quality control tool for high throughput sequence data (Babraham, UK). Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Archer F. I., Adams P. E., and Schneiders B. B. (2017). StrataG: An r package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 17, 5–11. doi: 10.1111/1755-0998.12559
Avise J. C., Bowen B. W., Lamb T., Meylan A. B., and Bermingham E. (1992). Mitochondrial DNA evolution at a turtle’s pace: evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Mol. Biol. Evol. 9, 457–473. doi: 10.1093/oxfordjournals.molbev.a040735
Bailey H., Benson S. R., Shillinger G. L., Bograd S. J., Dutton P. H., Eckert S. A., et al. (2012). Identification of distinct movement patterns in Pacific leatherback turtle populations influenced by ocean conditions. Ecol. Appl. 22, 735–747. doi: 10.1890/11-0633
Bandelt H.-J., Forster P., and Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Benson S. R., Eguchi T., Foley D. G., Forney K. A., Bailey H., Hitipeuw C., et al. (2011). Large-scale movements and high-use areas of western Pacific leatherback turtles. Dermochelys coriacea. Ecosphere 2, 1–27. doi: 10.1890/ES11-00053.1
Bentley B. P., Carrasco-Valenzuela T., Ramos E. K. S., Pawar H., Arantes L. S., Alexander A., et al. (2023). Divergent sensory and immune gene evolution in sea turtles with contrasting demographic and life histories. Proc. Natl. Acad. Sci. 120, e2201076120. doi: 10.1073/pnas.220107612
Bolger A. M., Lohse M., and Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114 ± 2120. doi: 10.1093/bioinformatics/btu170
Bowen B. W., Nelson W. S., and Avise J. C. (1993). A molecular phylogeny for marine turtles: trait mapping, rate assessment, and conservation relevance. Proc. Natl. Acad. Sci. U.S.A. 90, 5574–5577. doi: 10.1073/pnas.90.12.5574
DeBoer T. S., Subia M. D., Erdmann M. V., Kovitvongsa K., and Barber P. H. (2008). Phylogeography and limited genetic connectivity in the endangered boring giant clam across the Coral Triangle. Conserv. Biol. 22, 1255–1266. doi: 10.1111/j.1523-1739.2008.00983.x
Dutton P. H. (1995). Molecular evolution of the sea turtles with special reference to the leatherback, Dermochelys coriacea (College Station: Ph.D. dissertation, Texas A&M University).
Dutton P. H., Bowen B. W., Owens D. W., Barragan A. R., and Davis S. K. (1999). Global phylogeography of the leatherback turtle (Dermochelys coriacea). J. Zool 248, 397–409. doi: 10.1111/j.1469-7998.1999.tb01038.x
Dutton P. H. and Frey A. (2009). Characterization of polymorphic microsatellite markers for the green turtle (Chelonia mydas). Mol. Ecol. Resour. 9, 354–356. doi: 10.1111/j.1755-0998.2008.02443.x
Dutton P. H., Hitipeuw C., Zein M., Benson S. R., and Al-Ghais S. M. (2007). Status and genetic structure of nesting populations of leatherback turtles (Dermochelys coriacea) in the Western Pacific. Chelonian Conserv. Biol. 6, 47–53. doi: 10.2744/1071-8443
Dutton P. H., Roden S. E., Stewart K. R., LaCasella E., Tiwari M., Formia A., et al. (2013). Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv. Genet. 14, 625–636. doi: 10.1007/s10592-013-0456-0
Excoffier L. and Lischer H. E. L. (2010). Arlequin suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier L., Smouse P. E., and Quattro J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. doi: 10.1093/genetics/131.2.479
Frandsen H., Figueroa D. F., and George J. A. (2020). Mitochondrial genomes and genetic structure of the Kemp’s ridley sea turtle (Lepidochelys kempii). Ecol. Evol. 10, 249–262. doi: 10.1002/ece3.5891
Frey A., LaCasella E. L., Jensen M. P., and Dutton P. H. (2025). Whole mitochondrial DNA sequencing improves resolution of population structure for Pacific green turtles (Chelonia mydas). 12. doi: 10.3389/fmars.2025.1581306
Gaspar P., Benson S. R., Dutton P. H., Réveillère A., Jacob G., Meetoo C., et al. (2012). Oceanic dispersal of juvenile leatherback turtles: going beyond passive drift modeling. Mar. Ecol. Prog. Ser. 457, 265–284. doi: 10.3354/meps09689
Guo S. W. and Thompson E. A. (1992). Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48, 361–372. doi: 10.2307/2532296
Hitipeuw C., Dutton P. H., Benson S., Thebu J., and Bakarbessy J. (2007). Population status and internesting movement of leatherback turtles, Dermochelys coriacea, nesting on the Northwest coast of Papua, Indonesia. Chelonian Conserv. Biol. 6, 28–36. doi: 10.2744/1071-8443(2007)6[28:PSAIMO]2.0.CO;2
Horne J. B., Roden S. E., LaCasella E. L., Frey A., Martin S. L., Jones T. T., et al. (2023). Origins of green turtle fishery bycatch in the central pacific revealed by mixed genetic markers. Front. Mar. Sci. 10, 1112842. doi: 10.3389/fmars.2023.1112842
Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., and Vellend M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. doi: 10.1111/j.1461-0248.2008.01179.x
Komoroske L. M., Jensen M. P., Stewart K. R., Shamblin B. M., and Dutton P. H. (2017). Advances in the application of genetics in marine turtle biology and conservation. Front. Mar. Sci. 4, 156. doi: 10.3389/fmars.2017.00156
Komoroske L. M., Miller M. R., O’Rourke S. M., Stewart K. R., Jensen M. P., and Dutton P. H. (2019). A versatile rapture (RAD-capture) platform for genotyping marine turtles. Mol. Ecol. Resour. 19, 497–511. doi: 10.1111/1755-0998.12980
Korneliussen T. S., Albrechtsen A., and Nielsen R. (2014). ANGSD: analysis of next generation sequencing data. BMC Bioinf. 15, 1–13. doi: 10.1186/s12859-014-0356-4
LeRoux R. A., Dutton P. H., Abreu-Grobois F. A., Lagueux C. J., Campbell C. L., Delcroix E., et al. (2012). Re-examination of population structure and phylogeography of hawksbill turtles in the Wider Caribbean using longer mtDNA sequences. J. Hered. 103, 806–820. doi: 10.1093/jhered/ess055
Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. Available at: http://arxiv.org/abs/1303.3997.
Li H. and Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lou R. N., Jacobs A., Wilder A. P., and Therkildsen N. O. (2021). A beginner’s guide to low-coverage whole genome sequencing for population genomics. Mol. Ecol. 30, 5966–5993. doi: 10.1111/mec.16077
Mangubhai S., Erdmann M. V., Wilson J. R., Huffard C. L., Ballamu F., Hidayat N. I., et al. (2012). Papuan Bird’s Head Seascape: emerging threats and challenges in the global center of marine biodiversity. Mar. Pol. Bull. 64, 2279–2295. doi: 10.1016/j.marpolbul.2012.07.024
Maslim M., Farajallah A., and Zamani N. P. (2016). Leatherback turtle (Dermochelys coriacea) populations in Sumatra: genetic diversity and connectivity pattern. AACL Bioflux 9, 276–283. https://bioflux.com.ro/docs/2016.276-283.pdf.
Meirmans P. G. and Hedrick P. W. (2011). Assessing population structure: Fst and related measures. Mol. Ecol. Resour. 11, 5–18. doi: 10.1111/j.1755-0998.2010.02927.x
Meisner J. and Albrechtsen A. (2018). Inferring Population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731. doi: 10.1534/genetics.118.301336
Miller S., Dykes D., and Polesky H. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucl. Acids Res. 16, 11:1215. doi: 10.1093/nar/16.3.1215
Moritz C. (1994). Defining “Evolutionarily significant units” for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Pakiding F., Zohar K., Allo A. Y. T., Keroman S., Lontoh D., Dutton P. H., et al. (2020). Community engagement: an integral component of a multifaceted conservation approach for the transboundary western Pacific leatherback. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.549570
Parra G. J., Cagnazzi D., Jedensjo M., Ackermann C., Frere C., Seddon J., et al. (2018). Low genetic diversity, limited gene flow and widespread genetic bottleneck effects in a threatened dolphin species, the Australian humpback dolphin. Biol. Conserv. 220, 192–200. doi: 10.1016/j.biocon.2017.12.028
Piboon P., Brown J., Kaewmong K., Kittiwattanawong K., and Nganvongpanit K. (2025). Biology, nesting behavior, genetic diversity, and conservation of leatherback sea turtles: Insights from Thailand and global perspectives. Ecol. Evol. 15, e71014 1–15. doi: 10.1002/ece3.71014
Raymond M. and Rousset F. (1995). An exact test for population differentiation. Evolution 49, 1280–1283. doi: 10.2307/2410454
Roden S. E. and Dutton P. H. (2011). Isolation and characterization of 14 polymorphic microsatellite loci in the leatherback turtle (Dermochelys coriacea) and cross-species amplification. Conserv. Genet. Resour. 3, 49–52. doi: 10.1007/s12686-010-9284-4
Roden S. E., LaCasella E. L. L., Alfaro-Shigueto J., Balazs G. H., de Paz Campos N., Donoso M., et al. (2025). “Genetic stock identification of fisheries bycatch provides insights into differences in broad-scale distribution patterns of leatherbacks in the North and Southeast Pacific,” in Proceedings of the Forty-Second Annual Symposium on Sea Turtle Biology and Conservation- Pattaya, Thailand. NOAA Technical Memorandum NOAA NMFS-SEFSC-785: 290 p. doi: 10.25923/b53w-tq22
Roden S. E., Stewart K. R., James M. C., Dodge K. L., Dell’Amico F., and Dutton P. H. (2017). Genetic fingerprinting reveals natal origins of male leatherback turtles encountered in the Atlantic Ocean and Mediterranean Sea. Mar. Biol. 164, 1–9. doi: 10.1007/s00227-017-3211-0
Sambrook J., Fritsch E. F., and Maniatis T. (1989). Molecular Cloning: A Laboratory Manual (New York, New York: Cold Spring Harbour Laboratory Press).
Sarti L., Barragan A. R., Garcia Munoz D., Garcia N., Huerta P., and Vargas F. (2007). Conservation and biology of the leatherback turtle in the Mexican Pacific. Chel. Conserv. Biol. 6, 70–78. doi: 10.2744/1071-8443(2007)6[70:CABOTL]2.0.CO;2
Schwartz M. K., Luikart G., and Waples R. S. (2007). Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22, 25–33. doi: 10.1016/j.tree.2006.08.009
Shamblin B. M., Bjorndal K. A., Bolten A. B., Hillis-Starr Z. M., Lundgren I., Naro-Maciel E., et al. (2012). Mitogenomic sequences better resolve stock structure of southern Greater Caribbean green turtle rookeries. Mol. Ecol. 21, 2330–2340. doi: 10.1111/j.1365-294X.2012.05530.x
Silver-Gorges I., Koval J., Rodriguez-Zarate J. C., Paladino F. V., and Jordan M. (2020). Large-scale connectivity, cryptic population structure, and relatedness in Eastern Pacific Olive ridley sea turtles (Lepidochelys olivacea). Ecol. Evol. 10, 8688–8704. doi: 10.1002/ece3.6564
Stewart K. R., James M. C., Roden S., and Dutton P. H. (2013). Assignment tests, telemetry and tag-recapture data converge to identify natal origins of leatherback turtles foraging in Atlantic Canadian waters. J. Anim. Ecol. 82, 791–803. doi: 10.1111/jane.2013.82.issue-4
Tapilatu R. F., Dutton P. H., Tiwari M., Wibbels T., Ferdinandus H. V., Iwanggin W. G., et al. (2013). Long-term decline of the western Pacific leatherback, Dermochelys coriacea: a globally important sea turtle population. Ecosphere 4, 25. doi: 10.1890/ES12-00348.1
Therkildsen N. O. and Palumbi S. R. (2017). Practical low-coverage genomewide sequencing of hundreds of individually barcoded samples for population and evolutionary genomics in nonmodel species. Mol. Ecol. Resour. 17, 194–208. doi: 10.1111/1755-0998.12593
Toha A. H. A., Dailami M., Anwar S., Setiawan J. B., Jentewo Y., Lapadi I., et al. (2020). The genetic relationships and Indo-Pacific connectivity of whale sharks (Rhincodon typus) with particular reference to mitochondrial COI gene sequences from Cendrawasih Bay, Papua, Indonesia. Biodiversitas 21, 2159–2171. doi: 10.13057/biodiv/d210544
Toha A. H. A., Widodo N., Subhan B., Himawan M. R., Tania C., Noor B. A., et al. (2016). Close genetic relatedness of whale sharks, Rhincodon typus in the Indo-Pacific region. AACL Bioflux 9, 458–465. doi: 10.5339/qproc.2016.iwsc4.31
Tolve L., Iannucci A., Garofalo L., Ninni A., Dondona A. C., Ceciarini I., et al. (2024). Whole mitochondrial genome sequencing provides new insights into the phylogeography of loggerhead turtles (Caretta caretta) in the Mediterranean Sea. Mar. Biol. 171, 19. doi: 10.1007/s00227-023-04325-x
Vargas S. M., Lins L. S. F., Molfetti E., Ho S. Y. W., Monteiro D., Barreto J., et al. (2019). Revisiting the genetic diversity and population structure of the critically endangered leatherback turtles in the South-west Atlantic Ocean: insights for species conservation. J. Mar. Biol. Assoc. UK. 99, 31–41. doi: 10.1017/S002531541700193X
Velez-Zuazo X., Ramos W. D., Van Dam R. P., Diez C. E., Abreu-Grobois A., and Mcmillan W. O. (2008). Dispersal, recruitment and migratory behaviour in a hawksbill sea turtle aggregation. Mol. Ecol. 17, 839–853. doi: 10.1111/j.1365-294X.2007.03635.x
Vranes K. and Gordon A. L. (2005). Comparison of Indonesian Throughflow transport observations, Makassar Strait to eastern Indian Ocean. Geophysical Res. Lett. 32, 1–5. doi: 10.1029/2004GL022158
Wallace B. P., DiMatteo A. D., Hurley B. J., Finkbeiner E. M., Bolten A. B., Chaloupka M. Y., et al. (2010). Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PloS One 5, e15465. doi: 10.1371/journal.pone.0015465
Wallace B. P., Posnik Z. A., Hurley B. J., DiMatteo A. D., Bandimere A., Rodriguez I., et al. (2023). Marine turtle regional management units 2.0: an updated framework for conservation and research of wide-ranging megafauna species. Endang. Species Res. 52, 209–223. doi: 10.3354/esr01243
Wallace B. P., Tiwari M., and Girondot M. (2013). “Dermochelys coriacea,” in The IUCN red list of threatened species 2013, e.T6494A43526147. 2013. doi: 10.2305/IUCN.UK.2013-2.RLTS.T6494A43526147.en (Accessed April 18, 2025).
Wallace B. P., Zolkewitz M., and James M. C. (2018). Discrete, high-latitude foraging areas are important to energy budgets and population dynamics of migratory leatherback turtles. Sci. Rep. 8, 11017. doi: 10.1038/s41598-018-29106-1
Weir B. S. and Cockerham C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
Winter D. J. (2012). MMOD: an R library for the calculation of population differentiation statistics. Mol. Ecol. Resour. 12, 1158–1160. doi: 10.1111/j.1755-0998.2012.03174.x
Wongfu C., Prasitwiset W., Poommouang A., Buddhachat K., Brown J. L., Chomdej S., et al. (2022). Genetic diversity in leatherback turtles (Dermochelys coriacea) along the Andaman Sea of Thailand. Diversity 14, 764. doi: 10.3390/d14090764
Keywords: population genetics, sea turtles, mtDNA, microsatellites, RAD-capture, whole genome resequencing
Citation: Toha AHA, Lontoh D, Pakiding F, Prasetyo AP, Komoroske LM and Dutton PH (2025) Population structure and genetic diversity of leatherback turtles (Dermochelys coriacea) in Bird’s Head Seascape, Papua-Indonesia. Front. Mar. Sci. 12:1569466. doi: 10.3389/fmars.2025.1569466
Received: 31 January 2025; Accepted: 07 April 2025;
Published: 21 May 2025.
Edited by:
Molly E. Lutcavage, University of Massachusetts, United StatesReviewed by:
Khaled Mohammed Geba, Menoufia University, EgyptKarima Fadhlaoui-Zid, Tunis El Manar University, Tunisia
Copyright © 2025 Toha, Lontoh, Pakiding, Prasetyo, Komoroske and Dutton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter H. Dutton, UGV0ZXIuRHV0dG9uQG5vYWEuZ292
 Abdul Hamid A. Toha
Abdul Hamid A. Toha Deasy Lontoh
Deasy Lontoh Fitryanti Pakiding
Fitryanti Pakiding Andhika P. Prasetyo
Andhika P. Prasetyo Lisa M. Komoroske
Lisa M. Komoroske Peter H. Dutton
Peter H. Dutton