- 1State Key Laboratory of Submarine Geoscience, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
- 2Key Laboratory of Marine Ecosystem Dynamics, Ministry of Natural Resources and Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
- 3School of Oceanography, Shanghai Jiao Tong University, Shanghai, China
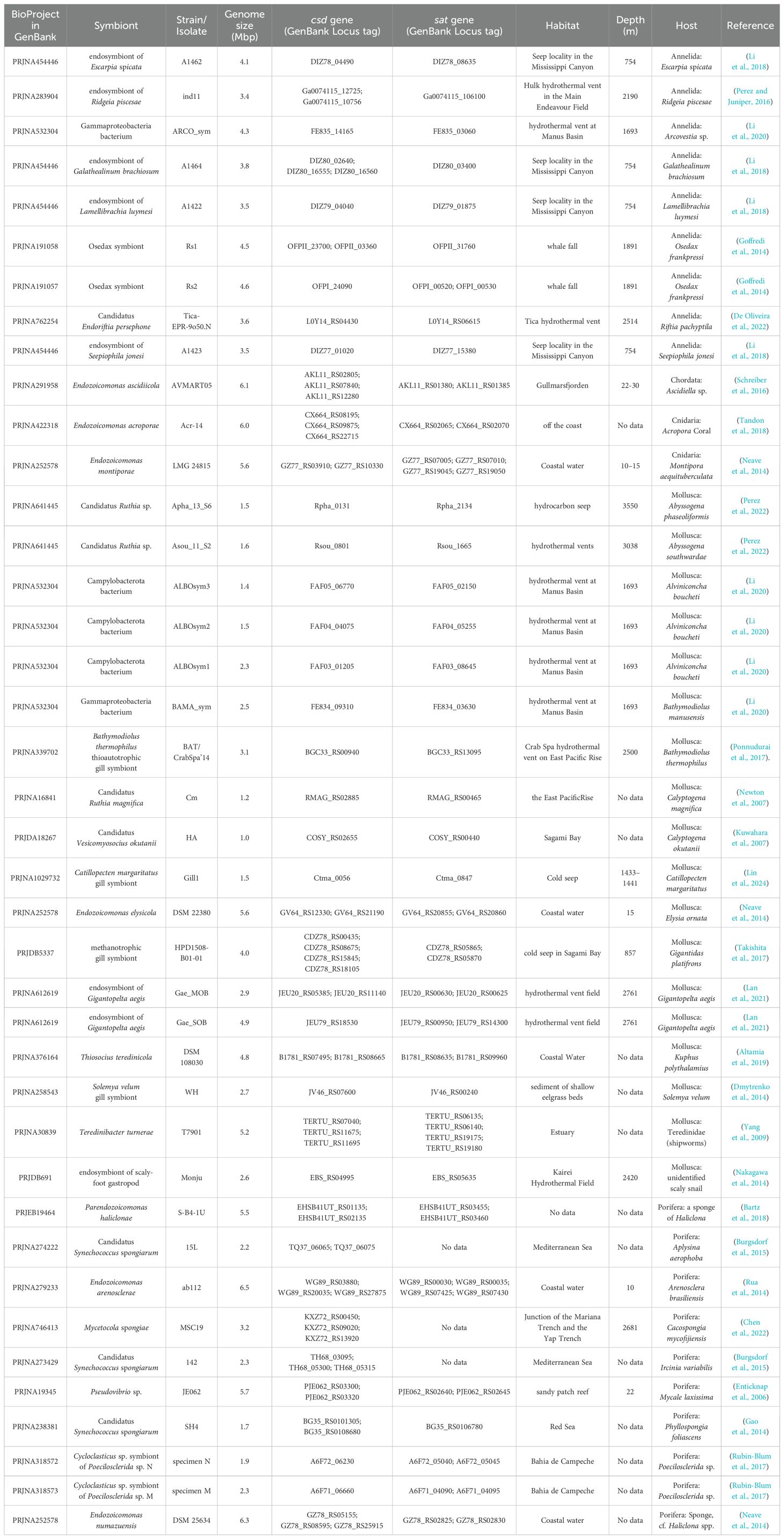
Various invertebrates, with microorganisms as their symbionts, inhabit diverse and dynamically changing environments such as hydrothermal vents (HVs) and cold seeps (CSs). The ongoing \dispersal of these symbionts is crucial for their biogeographic distribution and connectivity, which in turn facilitates the persistence of mutualistic relationships. To gain insights into the mechanisms underlying the adaptation of symbionts in response to environmental changes, this perspective analyzed two genes related to sulfur metabolism in the symbionts, based on their genome annotations. Our findings revealed that the gene encoding cysteine desulfurase (CSD) is ubiquitous among these symbionts, regardless of their geographic locations, hosts, or genome sizes. This suggests that these symbionts possess the ability to utilize sulfur from cysteine. Similarly, genes encoding sulfate adenylyltransferase (SAT), which is essential for sulfate assimilation, are also widely present in the genomes of the symbionts, with notable exceptions being some isolates from sponges. Notably, most of the investigated symbionts possess both sat and csd genes, hinting at their capability to utilize both organic and inorganic sulfur resources. The presence of both sat and csd genes may confer an advantage to the symbionts while cessation of hydrothermal and cold seep activity or during their dispersal among isolated locales. Further comparative genomic studies, particularly those focusing on the versatile adaptation strategies of symbionts across different life stages, can enhance our understanding of their ecological fitness and broaden our knowledge about how these symbiotic microorganisms successfully dwell in the dynamic marine environments.
Introduction
Globally, the emissions of fluids and gases from the seafloor, such as those from widespread hydrothermal vents (HVs) and cold seeps (CSs), serve as resources to chemoautotrophic microorganisms. These microorganisms harness energy from oxidization of reducing chemicals (e.g., H2S, S0, H2, CH4) to fix inorganic carbon into biomass (Dick, 2019). A variety of invertebrate species host chemoautotrophic microorganisms as symbionts, often within gills or trophosomes, forming close nutritional relationship with them (Dubilier et al., 2008). Since the discovery of chemosynthetic symbioses between bacteria and invertebrates at marine hydrothermal vents on the Galapagos Rift, it has been realized that chemosynthetic symbioses occur worldwide in a diverse range of habitats. These include cold seeps, whale and wood falls, and shallow-water coastal sediments (Dubilier et al., 2008). To gain a deeper understanding of these featured mutualistic associations, numerous genomes of marine chemoautotrophic symbiotic microorganisms have been sequenced. The genomic data reveal the genetic blueprint of their metabolic capabilities, and the symbionts’ metabolic roles within the symbiosis and their adaptations to intracellular conditions have been studied (Kuwahara et al., 2007; Newton et al., 2007). For instance, in the genome of the symbiont Candidatus Vesicomyosocius okutanii, which is approximately 1.0 million base pairs (Mbps) in size (Table 1), genes that are unnecessary for an intracellular life stage, as well as some essential genes, appear to have been lost (Kuwahara et al., 2007). Another example is the genome of an endosymbiont associated with armored snails (approximately 2.6 Mbps in size, Table 1). Compared to three other free-living relatives, this genome is smaller and displays features that are consistent with ongoing genome reduction (Nakagawa et al., 2014). Moreover, a recent genomic analysis of the ectosymbiont of Catillopecten margaritatus revealed that the symbiont genome is significantly smaller than its free-living relatives and has lost cellular components required for free-living (Lin et al., 2024). The accumulating genome sequences of symbionts provide an opportunity for studying the mechanisms of the adaptation by genomic comparisons. Recently, extensive genomic analyses have unveiled a widespread conservation of metabolic pathways for sulfur oxidation across sulfur-oxidizing symbionts derived from diverse host species and habitats (Sudo et al., 2024). It is hypothesized that the expansion and diversification of the SoxY gene family, which encodes a pivotal sulfur anion carrier protein integral to the sulfur-oxidizing multi-enzyme complex, may enhance the metabolic versatility of sulfur-oxidizing symbionts (Sudo et al., 2024).
Environmental adaptations
Numerous genome-based studies have explored the partnerships between the hosts and symbionts and their adaptations to the reducing marine environments. However, there are few analyses conducted regarding their adaptation to the chemical changes while cessation of hydrothermal and cold seep activity or dispersal. Since symbionts play critical roles in supporting their hosts, the transmission of these symbionts between generations of the hosts is of paramount importance. Previous studies have provided evidences for the horizontal transmissions of microbial symbionts and its importance in maintaining mutualisms (Cary et al., 1993; Kádár et al., 2005; Nussbaumer et al., 2006; Wentrup et al., 2014; Breusing et al., 2022; Hauer et al., 2023).
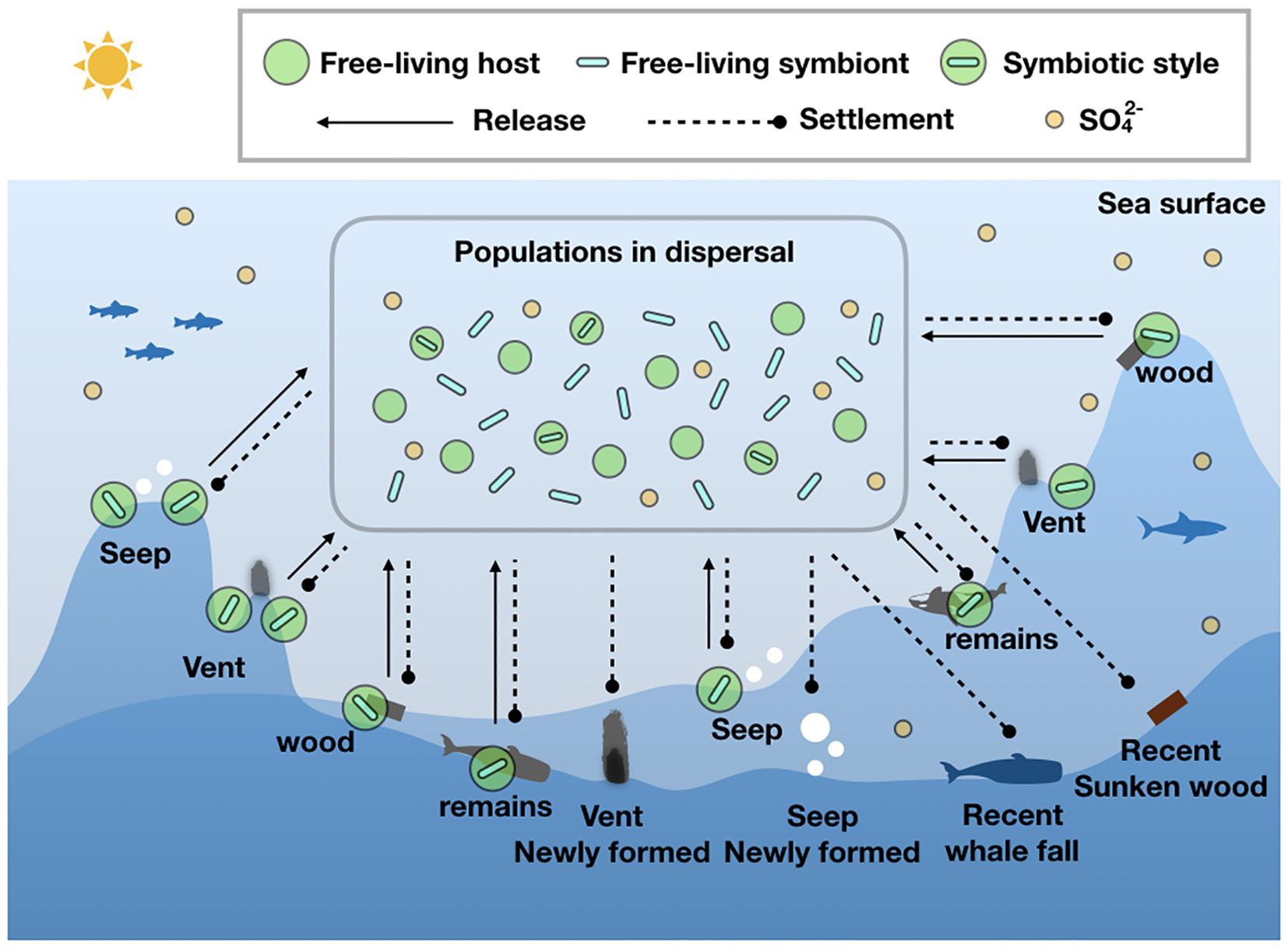
Building upon observations of faunal overlap across diverse chemosynthetic communities, Smith et al. postulated that whale carcasses might function as stepping stones for fauna dispersal, potentially facilitating the colonization of new habitats situated hundreds of kilometers apart, such as hydrothermal vents (Smith et al., 1989; Sumida et al., 2016). As illustrated in Figure 1, the dispersal of marine fauna serves as a crucial link in the chain of symbiont transmission. The discontinuous distribution of hydrothermal vents and cold seeps suggest they themselves can serve as ‘stepping stones’ for faunal dispersion. Considering that sporadic volcanic and tectonic events destroy existing vent fields and create new ones, HVs are dynamically changing habitats accompanied by extirpation (local extinction) and novel colonization of species (Vrijenhoek, 2010). The organic falls, such as whale falls and sunken wood, are also isolated habitats on the seafloor that undergo dynamic changes. Therefore, ongoing dispersal of symbionts, whether in host-associated or free-living lifestyle, may facilitate persistence of mutualistic relationship and enable the spread and colonize a wide range of habitable sites (Figure 1). However, the symbiotic microorganisms from reducing marine environments (e.g. HVs and CSs), may have to encounter situations with significant chemical changes, such as cessation of hydrothermal and cold seep activity. Moreover, during dispersal in seawater, leaving HVs and CSs, the symbionts face crucial chemical changes, such as the form of sulfur changing from reducing sulfur compounds to sulfate. The intriguing question that remains is how these symbionts adapt to these environmental changes. Given that sulfur is a vital element for cells (Zhou et al., 2024), to adapt to the changes, symbionts may modulate their intracellular sulfur sources, e.g. cysteine (an organic sulfur source), or sulfate from the environment. Therefore, it is highly meaningful to study the plasticity of sulfur metabolism. The genes csd (encoding cysteine desulfurase) and sat (encoding sulfate adenylyltransferase) are involved in the initiation of pathways utilizing cysteine and sulfate as substrates, respectively. From this perspective, the presence of these two genes is examined in the sequenced genomes of symbionts (Table 1) to study their potential for sulfur metabolic plasticity. This perspective aims to promote further research into the adaptive mechanisms employed by symbionts.
Presence of genes encoding cysteine desulfurase in symbiont genomes
The sulfur-containing amino acid cysteine, regardless of whether derived from the hosts or the symbionts themselves, serves as a sulfur source for symbiotic microorganisms. Cysteine desulfurase (CSD) catalyzes the conversion of L-cysteine to L-alanine and sulfane sulfur via the formation of a protein-bound cysteine persulfide intermediate on a conserved cysteine residue (Hidese et al., 2011). The persulfide sulfur atoms could be utilized in various biosynthetic pathways to produce sulfur-containing biofactors, such as iron–sulfur clusters, molybdopterin, transfer RNA thionucleosides, biotin, thiamin, and lipoic acid. The biofactors play pivotal roles in numerous essential and diverse cellular processes, including DNA repair, respiration, intermediary metabolism, gene regulation, and redox sensing (Das et al., 2021). Given the essential function of CSDs in the biosynthesis of these sulfur-containing biofactors, we investigated the presence of the gene encoding CSD in the genomes of symbiotic microorganisms.
Despite thriving in markedly different geochemical conditions, all investigated symbiotic microorganisms exhibited the presence of the csd gene in their genome annotations (Gene Locus tag shown in Table 1). The investigation covered symbionts from a variety of habitat types, including hydrothermal vents, cold seeps, whale remains, sunken wood, and others, as listed in Table 1. The habitats are distributed across a wide range of geographic locations and depths, spanning from shallow to deep sea environments (Table 1). The seep located at Aleutian Trench in Pacific Ocean, reaching a depth of 3550, is inhibited by Candidatus Ruthia sp. Apha_13_S6 and its host Abyssogena phaseoliformis (Perez et al., 2022). As shown in Table 1, the genomes of two dominant endosymbionts Rs1 and Rs2 living in deep-sea worm Osedax frankpressi, collected from a whale fall, contained genes belonging to the CSD family. Additionally, the chemolithoautotrophic sulfur-oxidizing endosymbiotic bacterium strain Thiosocius teredinicola DSM 108030T, isolated from the giant shipworm Kuphus polythalamius in sunken wood (Distel et al., 2017), also possesses the csd gene (Table 1).
The csd genes were present in microorganisms belonging to distinct clades. For example, the mussel B. manusensis and tubeworm Arcovestia ivanovi, both sampled from PACManus hydrothermal area, are colonized by Gammaproteobacteria from different clades, specifically isolate BAMA_sym and ARCO_sym, respectively (Li et al., 2020). The csd genes were detected in the genome annotations of both isolates (see Table 1). Additionally, the csd genes were also present in genome annotations of all three Epsilonproteobacteria symbionts harbored in vent-mouth-dwelling snail Alviniconcha boucheti, suggesting that CSD is crucial to their survival. Furthermore, these csd genes are not limited to sulfur-oxidizing endosymbionts but are also found in methane-oxidizing endosymbiont. For instance, the genes were detected in both the genome of sulfur-oxidizing endosymbiont isolate Gae_SOB and methane-oxidizing endosymbiont isolate Gae_MOB in the deep-sea snail Gigantopelta aegis (Table 1). Notably, regardless of how small the genome is, this gene is always present (Table 1). Larger genomes frequently harbor multiple csd genes, with many genomes possessing three, and in the case of the symbiont isolate HPD1508-B01-01, the genome even contains up to four (Table 1).
As shown in the Table 1, the gene encoding CSD is present in the annotations of all symbiont genomes, regardless of whether the host belongs to Annelida, Mollusca, Porifera, Cnidaria, or Chordata. In summary, all symbiotic genomes investigated in this perspective possess genes annotated as encoding CSD, highlighting the significance of cysteine and CSD to the symbionts.
Presence of genes encoding sulfate adenylyltransferase in symbiont genomes
In addition to organic sulfur sources, sulfate is abundant in modern ocean, with concentrations reaching approximately 28 mM, making it the second most prevalent anion in seawater (Fakhraee et al., 2024). Consequently, seawater serves as a potential sulfur resource for cells. While assimilatory sulfate reduction is increasingly being documented in marine microorganisms (e.g., Methanothermococcus thermolithotrophicus DSM 2095 and Phototrophicus methaneseepsis ZRK33) (Jespersen and Wagner, 2023; Zheng et al., 2024), the potential for symbionts to assimilate sulfate as a sulfur source is also worth further investigation. Bacterial sulfate assimilation pathways involve the activation of inorganic sulfur through intermediates such as adenosine 5′-phosphosulfate (APS) or 3′-phosphoadenosine 5′-phosphosulfate (PAPS) (Williams et al., 2002). PAPS has been recognized as a universal sulfuryl donor in cells, such as the substrate for producing sulfolipids. The biosynthetic pathway of PAPS in bacteria initiates with the formation of adenosine 5´-phosphosulfate from ATP and inorganic sulfate, a reaction catalyzed by sulfate adenylyltransferase (SAT) (Duffel, 2023). Therefore, we also investigated the presence of the sat gene encoding SAT in the genomes of symbiotic microorganisms.
Similarly, all investigated symbiotic microorganisms exhibit the presence of genes encoding SAT in their genome annotations, except for some isolates from sponges, such as Mycetocola spongiae MSC19T and Candidatus Synechococcus spongiarum isolates 15L and 142 (Table 1). M. spongiae MSC19T, belonging to the Actinobacteria phylum, was isolated from the deep-sea sponge Cacospongia mycofijiensis. Its genome comprises a single circular chromosome of 3.2 Mbps (Chen et al., 2022). However, no sat gene was annotated in its genome. Given that the genome coverage has reached 800x (see its Assembly in BioProject), it is unlikely that the absence of the sat gene is due to insufficient sequencing depth. Intriguingly, gene annotation revealed that M. spongiae MSC19T possesses three genes encoding CSD family proteins (Table 1), suggesting that cysteine could serve as an important sulfur source. Isolates 15L and 142 of the Candidatus S. spongiarum were symbionts of the sponge Aplysina aerophoba and Ircinia variabilis, respectively (Burgsdorf et al., 2015). Neither sat gene was annotated in their genomes (Table 1). Considering that sponges can reproduce vegetatively by fission or budding (Fields and Levin, 2020), one of the possible explanations for this phenomenon is that the symbionts, such as M. spongiae MSC19T, may stick to its host through the vegetative reproduction and utilize organic sulfur instead of acquiring the sulfate from environment, which could be result of adaptative evolution, but this requires further in-depth research. As uptake of environmental symbionts bears a risk of infection to the host by cheaters (Douglas, 2008), maintaining the host and symbiont together (without separation) may help prevent infection and maintain the persistence of mutualistic relationship. Notably, not all symbionts in sponges lack the gene for SAT. For instance, Pseudovibrio sp. strain JE062, isolated from sponge Mycale laxissima, possesses sat gene (Table 1). Previous study has provided evidence for vertical transmission of bacterium Pseudovibrio sp. strain JE062 via the larvae of sponge Mycale laxissima (Enticknap et al., 2006). Further research in connection with other genes in the sulfate assimilation pathway is required.
Discussion
Cysteine serves as a common sulfur source in cells. The csd gene is ubiquitous among symbionts, regardless of their geographic location, hosts, or genome sizes. The conserved presence of genes encoding CSD in symbiotic microorganisms suggest their ability to utilize sulfur from cysteine. Cysteine may originate from the host or from the microorganism. This capability may assist symbionts in harnessing cysteine as a sulfur source during dispersion. Similarly, genes encoding SAT, which is crucial for the assimilation of inorganic sulfur, have also been broadly detected in the genomes of symbiotic microorganisms. Most of the symbiotic microorganisms investigated in this context possess both genes encoding SAT and CSD. The presence of sat and csd gene may benefit the dispersal of symbionts among isolated locales. There are variations in copy number of genes from the CSD and SAT families per genome, which may be shaped by interaction with hosts and local environmental conditions, conferring ecological advantages. In contrast, symbionts from sponges harboring only csd gene may be the result of evolution due to the formation of intimate symbiosis with their hosts.
Notably, even the symbiotic microorganisms with small genomes possess gene from both the SAT and CSD families (Table 1), highlighting their importance for the survival of these microorganisms. For example, the genome of Candidatus V. okutanii HA is approximately 1.0 Mbps in size, and the genes that are unnecessary for an intracellular lifestyle, as well as some essential genes (e.g., ftsZ for cytokinesis), appear to be absent (Kuwahara et al., 2007). Reductive evolution of the genome might be ongoing in the vertically transmitted Calyptogena symbionts (Kuwahara et al., 2007). Despite this reduction, the sat gene still exists in this small symbiont genome. Similarly, the sat gene is also present in the small genome of Candidatus R. magnifica Cm, which is 1.2 Mbps in size (see Table 1). The sat gene is present in the genome of gill symbiont isolate Gill1 from Catillopecten margaritatus as well (Table 1). Genomic analysis of this symbiont reveals that its genome is substantially smaller than those of its free-living relatives and has lost cellular components required for free-living (Lin et al., 2024). The presence of sat genes in the compact genomes of these symbionts indicates that sat may play a pivotal role in the functioning of these symbionts, despite the overall reduction in their genome size. It is crucial to acknowledge that these analyses are based solely on gene annotation, and their metabolic functions require further experimental validation.
Perspective
With advances in large-scale, high-throughput sequencing and assembly technologies, the high-quality genome sequences generated from projects like the Aquatic Symbiosis Genomics Project, which covers a wide range of aquatic host organisms and their microbial symbionts (McKenna et al., 2024), will help us better understand how these organisms interact with each other and their environment. In this perspective, we attempt to attract broader attention by linking genomic traits of symbionts to their adaptation. By a preliminary genomic analysis, this perspective highlights the conservation of the two genes, csd and sat, across diverse marine symbiotic microorganisms from varied hosts and reducing marine habitats, indicating their importance. Further studies on the metabolic flexibility in utilizing both organic and inorganic sulfur sources are needed, such as functional demonstration and ecological relevance. Besides, HV and CS ecosystems are chemically complex and exhibit distinct chemical profiles relative to seawater. Moreover, the marine symbiotic microorganisms experience various environmental transitions, such as changes in temperature, hydrostatic pressure, pH, nutrient availability, and osmotic stress, and have evolved traits to overcome many of these stressors (Apprill, 2020). Starting from the studies on how these symbionts might acquire and utilize sulfur during the dispersal, further explorations on metabolic plasticity in response to environmental changes will advance our understanding of the complex adaptation strategies. Dispersal of symbionts and their hosts is crucial for their biogeographic distribution and connectivity. Metabolic plasticity may prolong the survival of symbionts under adverse conditions. The dispersal capacity of symbionts may aid the mutualism persistence across habitats in the oceans. Understanding the dispersal patterns of the symbionts has implications for spatio-temporal dynamics and biodiversity conservation. The studies on metabolic plasticity will also improve our interpretation of their ecological roles in connection with biogeochemical conditions. The genomic traits that symbionts evolve to adapt to these conditions may enhance their tolerance and adaptations to environmental stressors. Therefore, this perspective concludes with a call for further research on metabolic plasticity to deepen our understanding of the connections between the genomic traits of symbionts from reducing habitats and environmental adaptation in the oceans. By combining comparative genomic analysis with consideration of lifestyles (such as symbiotic and free-living life stages) and animal behaviors (e.g. reproductive modes), we can better comprehend their versatile adaptation strategies and ecological fitness. Specifically, studying conserved genes and their functions in the core genome will aid in understanding the shared mechanisms of adaptation. Additionally, by analyzing the accessory genomes with considerations of geographical differences and animal behaviors, we can gain insights into their specificity, such as unique adaptive features. Furthermore, differences in gene expression, translation or post-translational modifications may play roles in adapting to free-living or host-associated lifestyles. Comparisons of transcriptomes and proteomes between free-living and host-associated symbiont populations may yield additional clues about how symbiont adapt to different lifestyles. An integrative investigation would help us comprehensively elucidate the adaptability of these symbionts to their respective hosts and free-living lifestyle.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
PZ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. X-QH: Writing – original draft, Writing – review & editing. PX: Funding acquisition, Resources, Writing – review & editing. D-SZ: Funding acquisition, Resources, Writing – review & editing. C-SW: Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFC2811401), the National Natural Science Foundation of China (42376133), and the Oceanic Interdisciplinary Program of Shanghai Jiao Tong University (No. SL2022ZD108).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altamia M. A., Shipway J. R., Concepcion G. P., Haygood M. G., and Distel D. L. (2019). Thiosocius teredinicola gen. nov., sp. nov., a sulfur-oxidizing chemolithoautotrophic endosymbiont cultivated from the gills of the giant shipworm, Kuphus polythalamius. Int. J. Systematic Evolutionary Microbiol. 69, 638–644. doi: 10.1099/ijsem.0.003143
Apprill A. (2020). The role of symbioses in the adaptation and stress responses of marine organisms. Annu. Rev. Mar. Sci. 12, 291–314. doi: 10.1146/annurev-marine-010419-010641
Bartz J.-O., Blom J., Busse H.-J., Mvie J. B., Hardt M., Schubert P., et al. (2018). Parendozoicomonas haliclonae gen. nov. sp. nov. isolated from a marine sponge of the genus Haliclona and description of the family Endozoicomonadaceae fam. nov. comprising the genera Endozoicomonas, Parendozoicomonas, and Kistimonas. Systematic Appl. Microbiol. 41, 73–84. doi: 10.1016/j.syapm.2017.11.004
Breusing C., Genetti M., Russell S. L., Corbett-Detig R. B., and Beinart R. A. (2022). Horizontal transmission enables flexible associations with locally adapted symbiont strains in deep-sea hydrothermal vent symbioses. Proc. Natl. Acad. Sci. U.S.A. 119, e2115608119. doi: 10.1073/pnas.2115608119
Burgsdorf I., Slaby B. M., Handley K. M., Haber M., Blom J., Marshall C. W., et al. (2015). Lifestyle evolution in cyanobacterial symbionts of sponges. mBio 6, e00391–e00315. doi: 10.1128/mBio.00391-15
Cary S. C., Warren W., Anderson E., and Giovannoni S. J. (1993). Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol. Mar. Biol. Biotechnol. 2, 51–62.
Chen Y., Pan T., Chai G., and Li Z. (2022). Complete genome of Mycetocola spongiae MSC19T isolated from deep-sea sponge Cacospongia mycoFijiensis indicates the adaptation to deep-sea environment and sponge-microbe symbioses. Mar. Genomics 63, 100955. doi: 10.1016/j.margen.2022.100955
Das M., Dewan A., Shee S., and Singh A. (2021). The multifaceted bacterial cysteine desulfurases: from metabolism to pathogenesis. Antioxidants 10, 997. doi: 10.3390/antiox10070997
De Oliveira A. L., Srivastava A., Espada-Hinojosa S., and Bright M. (2022). The complete and closed genome of the facultative generalist Candidatus Endoriftia persephone from deep-sea hydrothermal vents. Mol. Ecol. Resour. 22, 3106–3123. doi: 10.1111/1755-0998.13668
Dick G. J. (2019). The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat. Rev. Microbiol. 17, 271–283. doi: 10.1038/s41579-019-0160-2
Distel D. L., Altamia M. A., Lin Z., Shipway J. R., Han A., Forteza I., et al. (2017). Discovery of chemoautotrophic symbiosis in the giant shipworm Kuphus polythalamia (Bivalvia: Teredinidae) extends wooden-steps theory. Proc. Natl. Acad. Sci. U.S.A. 114, E3652-E3658. doi: 10.1073/pnas.1620470114
Dmytrenko O., Russell S. L., Loo W. T., Fontanez K. M., Liao L., Roeselers G., et al. (2014). The genome of the intracellular bacterium of the coastal bivalve, Solemya velum: a blueprint for thriving in and out of symbiosis. BMC Genomics 15, 924. doi: 10.1186/1471-2164-15-924
Douglas A. E. (2008). Conflict, cheats and the persistence of symbioses. New Phytol. 177, 849–858. doi: 10.1111/j.1469-8137.2007.02326.x
Dubilier N., Bergin C., and Lott C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. doi: 10.1038/nrmicro1992
Duffel M. W. (2023). “Sulfotransferases,” in Reference Module in Biomedical Sciences (Elsevier), B978032395488400005X. doi: 10.1016/B978-0-323-95488-4.00005-X
Enticknap J. J., Kelly M., Peraud O., and Hill R. T. (2006). Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72, 3724–3732. doi: 10.1128/AEM.72.5.3724-3732.2006
Fakhraee M., Crockford P. W., Bauer K. W., Pasquier V., Sugiyama I., Katsev S., et al. (2024). The history of Earth’s sulfur cycle. Nat. Rev. Earth Environ. 6, 106–125. doi: 10.1038/s43017-024-00615-0
Fields C. and Levin M. (2020). Why isn’t sex optional? Stem-cell competition, loss of regenerative capacity, and cancer in metazoan evolution. Communicative Integr. Biol. 13, 170–183. doi: 10.1080/19420889.2020.1838809
Gao Z.-M., Wang Y., Tian R.-M., Wong Y. H., Batang Z. B., Al-Suwailem A. M., et al. (2014). Symbiotic adaptation drives genome streamlining of the cyanobacterial sponge symbiont “ Candidatus synechococcus spongiarum. mBio 5, e00079–e00014. doi: 10.1128/mBio.00079-14
Goffredi S. K., Yi H., Zhang Q., Klann J. E., Struve I. A., Vrijenhoek R. C., et al. (2014). Genomic versatility and functional variation between two dominant heterotrophic symbionts of deep-sea Osedax worms. ISME J. 8, 908–924. doi: 10.1038/ismej.2013.201
Hauer M. A., Breusing C., Trembath-Reichert E., Huber J. A., and Beinart R. A. (2023). Geography, not lifestyle, explains the population structure of free-living and host-associated deep-sea hydrothermal vent snail symbionts. Microbiome 11, 106. doi: 10.1186/s40168-023-01493-2
Hidese R., Mihara H., and Esaki N. (2011). Bacterial cysteine desulfurases: versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 91, 47–61. doi: 10.1007/s00253-011-3336-x
Jespersen M. and Wagner T. (2023). Assimilatory sulfate reduction in the marine methanogen Methanothermococcus thermolithotrophicus. Nat. Microbiol. 8, 1227–1239. doi: 10.1038/s41564-023-01398-8
Kádár E., Bettencourt R., Costa V., Santos R. S., Lobo-da-Cunha A., and Dando P. (2005). Experimentally induced endosymbiont loss and re-acquirement in the hydrothermal vent bivalve Bathymodiolus azoricus. J. Exp. Mar. Biol. Ecol. 318, 99–110. doi: 10.1016/j.jembe.2004.12.025
Kuwahara H., Yoshida T., Takaki Y., Shimamura S., Nishi S., Harada M., et al. (2007). Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, calyptogena okutanii. Curr. Biol. 17, 881–886. doi: 10.1016/j.cub.2007.04.039
Lan Y., Sun J., Chen C., Sun Y., Zhou Y., Yang Y., et al. (2021). Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis. Nat. Commun. 12, 1165. doi: 10.1038/s41467-021-21450-7
Li Y., Liles M. R., and Halanych K. M. (2018). Endosymbiont genomes yield clues of tubeworm success. ISME J. 12, 2785–2795. doi: 10.1038/s41396-018-0220-z
Li L., Wang M., Li L., Du Z., Sun Y., Wang X., et al. (2020). Endosymbionts of metazoans dwelling in the PACManus hydrothermal vent: diversity and potential adaptive features revealed by genome analysis. Appl. Environ. Microbiol. 86, e00815–e00820. doi: 10.1128/AEM.00815-20
Lin Y.-T., Ip J. C.-H., He X., Gao Z.-M., Perez M., Xu T., et al. (2024). Scallop-bacteria symbiosis from the deep sea reveals strong genomic coupling in the absence of cellular integration. ISME J. 18, wrae048. doi: 10.1093/ismejo/wrae048
McKenna V., Archibald J. M., Beinart R., Dawson M. N., Hentschel U., Keeling P. J., et al. (2024). The Aquatic Symbiosis Genomics Project: probing the evolution of symbiosis across the Tree of Life. Wellcome Open Res. 6, 254. doi: 10.12688/wellcomeopenres.17222.2
Nakagawa S., Shimamura S., Takaki Y., Suzuki Y., Murakami S., Watanabe T., et al. (2014). Allying with armored snails: the complete genome of gammaproteobacterial endosymbiont. ISME J. 8, 40–51. doi: 10.1038/ismej.2013.131
Neave M. J., Michell C. T., Apprill A., and Voolstra C. R. (2014). Whole-genome sequences of three symbiotic endozoicomonas strains. Genome Announc 2, e00802–e00814. doi: 10.1128/genomeA.00802-14
Newton I. L. G., Woyke T., Auchtung T. A., Dilly G. F., Dutton R. J., Fisher M. C., et al. (2007). The calyptogena magnifica chemoautotrophic symbiont genome. Science 315, 998–1000. doi: 10.1126/science.1138438
Nussbaumer A. D., Fisher C. R., and Bright M. (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345–348. doi: 10.1038/nature04793
Perez M., Breusing C., Angers B., Beinart R. A., Won Y.-J., and Young C. R. (2022). Divergent paths in the evolutionary history of maternally transmitted clam symbionts. Proc. R. Soc B. 289, 20212137. doi: 10.1098/rspb.2021.2137
Perez M. and Juniper S. K. (2016). Insights into Symbiont Population Structure among Three Vestimentiferan Tubeworm Host Species at Eastern Pacific Spreading Centers. Appl. Environ. Microbiol. 82, 5197–5205. doi: 10.1128/AEM.00953-16
Ponnudurai R., Sayavedra L., Kleiner M., Heiden S. E., Thürmer A., Felbeck H., et al. (2017). Genome sequence of the sulfur-oxidizing Bathymodiolus thermophilus gill endosymbiont. Stand Genomic Sci. 12, 50. doi: 10.1186/s40793-017-0266-y
Rua C. P. J., Trindade-Silva A. E., Appolinario L. R., Venas T. M., Garcia G. D., Carvalho L. S., et al. (2014). Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis. PeerJ 2, e419. doi: 10.7717/peerj.419
Rubin-Blum M., Antony C. P., Borowski C., Sayavedra L., Pape T., Sahling H., et al. (2017). Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat. Microbiol. 2, 17093. doi: 10.1038/nmicrobiol.2017.93
Schreiber L., Kjeldsen K. U., Obst M., Funch P., and Schramm A. (2016). Description of Endozoicomonas ascidiicola sp. nov., isolated from Scandinavian ascidians. Systematic Appl. Microbiol. 39, 313–318. doi: 10.1016/j.syapm.2016.05.008
Smith C. R., Kukert H., Wheatcroft R. A., Jumars P. A., and Deming J. W. (1989). Vent fauna on whale remains. Nature 341, 27–28. doi: 10.1038/341027a0
Sudo M., Osvatic J., Taylor J. D., Dufour S. C., Prathep A., Wilkins L. G. E., et al. (2024). SoxY gene family expansion underpins adaptation to diverse hosts and environments in symbiotic sulfide oxidizers. mSystems 9, e01135–e01123. doi: 10.1128/msystems.01135-23
Sumida P. Y. G., Alfaro-Lucas J. M., Shimabukuro M., Kitazato H., Perez J. A. A., Soares-Gomes A., et al. (2016). Deep-sea whale fall fauna from the Atlantic resembles that of the Pacific Ocean. Sci. Rep. 6, 22139. doi: 10.1038/srep22139
Takishita K., Takaki Y., Chikaraishi Y., Ikuta T., Ozawa G., Yoshida T., et al. (2017). Genomic evidence that methanotrophic endosymbionts likely provide deep-sea bathymodiolus mussels with a sterol intermediate in cholesterol biosynthesis. Genome Biol. Evol. 9, 1148–1160. doi: 10.1093/gbe/evx082
Tandon K., Chiang P.-W., Chen W.-M., and Tang S.-L. (2018). Draft genome sequence of endozoicomonas acroporae strain acr-14T, isolated from acropora coral. Genome Announc 6, e01576–e01517. doi: 10.1128/genomeA.01576-17
Vrijenhoek R. C. (2010). Genetic diversity and connectivity of deep-sea hydrothermal vent metapopulations: HYDROTHERMAL VENT METAPOPULATIONS. Mol. Ecol. 19, 4391–4411. doi: 10.1111/j.1365-294X.2010.04789.x
Wentrup C., Wendeberg A., Schimak M., Borowski C., and Dubilier N. (2014). Forever competent: deep-sea bivalves are colonized by their chemosynthetic symbionts throughout their lifetime. Environ. Microbiol. 16, 3699–3713. doi: 10.1111/1462-2920.12597
Williams S. J., Senaratne R. H., Mougous J. D., Riley L. W., and Bertozzi C. R. (2002). 5′-adenosinephosphosulfate lies at a metabolic branch point in mycobacteria. J. Biol. Chem. 277, 32606–32615. doi: 10.1074/jbc.M204613200
Yang J. C., Madupu R., Durkin A. S., Ekborg N. A., Pedamallu C. S., Hostetler J. B., et al. (2009). The complete genome of teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (Shipworms). PloS One 4, e6085. doi: 10.1371/journal.pone.0006085
Zheng R., Wang C., and Sun C. (2024). Deep-sea in situ and laboratory multi-omics provide insights into the sulfur assimilation of a deep-sea Chloroflexota bacterium. mBio 15, e00004–e00024. doi: 10.1128/mbio.00004-24
Keywords: hydrothermal vent, cold seep, symbiont, dispersal, cysteine desulfurase, sulfate adenylyltransferase
Citation: Zhou P, He X-Q, Xu P, Zhang D-S and Wang C-S (2025) Exploring environmental adaptation mechanisms of symbiotic microorganisms in marine reducing ecosystems: harnessing genomic comparison to unveil the underlying mechanisms. Front. Mar. Sci. 12:1571722. doi: 10.3389/fmars.2025.1571722
Received: 07 February 2025; Accepted: 07 July 2025;
Published: 30 July 2025.
Edited by:
Heng-Lin Cui, Jiangsu University, ChinaReviewed by:
Qing-lei Sun, Chinese Academy of Sciences (CAS), ChinaZengfeng Du, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Zhou, He, Xu, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Xu, eHVwZW5nQHNpby5vcmcuY24=; Chun-Sheng Wang, d2FuZ3Npb0BzaW8ub3JnLmNu
 Peng Zhou
Peng Zhou Xue-Qing He1,2
Xue-Qing He1,2 Peng Xu
Peng Xu Dong-Sheng Zhang
Dong-Sheng Zhang Chun-Sheng Wang
Chun-Sheng Wang