Abstract
The marine annelid Platynereis dumerilii, is a key model in genetics, evolution, neurobiology, ecology, and ecotoxicology. Along with its sibling species, P. cfr massiliensis, it thrives in both normal and naturally acidified environments. This makes these species ideal candidates for studying mechanisms of tolerance to acidified conditions, resembling future ocean acidification (OA) scenarios. The ATP-binding cassette (ABC) transport proteins help mitigating the adverse impacts of drugs, xenobiotics and physical stressors. There is growing evidence for their involvement to mediate tolerance towards acid-stress in bacteria and tumor cell lines. Such a function may be relevant for the ability of marine species to cope with OA and may be important to consider when predicting future OA scenarios for marine fauna. Here we addressed the question if ABC transporters of Platynereis spp. are involved in compensating adverse effects of low pH by studying ABC transporter transcript levels in marine animals exposed to various pH levels. We firstly examined P. dumerilii whole genome data (version EMBL_pdum_1.0, Genbank assembly: GCA_026936325.1) for the presence of ABC transporter genes, by homology searches, and, using the single-cell atlas database with P. dumerilii gene expression data, we then determined the presence of a potentially relevant subset of ABC transporters from the ABCB, C and G subfamilies in different organs/tissues. Finally, to assess how seawater pH affects ABC transporter expression, we conducted an in-situ reciprocal transplant experiment involving individuals of P. dumerilii/P. cfr massiliensis. Adult specimens were collected inside and outside the CO2 vents off Castello Aragonese (Ischia Island, Italy). Individuals collected from normal pH areas (8.18 ± 0.005) were transplanted to acidified conditions (7.33 ± 0.312), and vice versa, while others were placed in their original areas. We found 81 orthologs from ABC transporter subfamilies A-G, expressed in different organs/tissues including midgut, neurons, body epidermis and ectodermal cells, and somatic and visceral muscle. Following the 30 days transplant experiment, qPCR analyses were performed to examine the expression levels of seven selected genes from the ABCB, ABCC, and ABCG subfamilies (abcb_1, abcb_2, abcb_3, abcc_1, abcc_2, abcc_3, and abcg). Three of these genes were differentially expressed in specimens transplanted from normal pH to low pH areas (abcb_1 and abcg up-regulated while abcb_3 down-regulated). Based on the homology with human ABCB1 and ABCG2, which are crucial in tumor cell adaptation to acidified environments, it seems reasonable to hypothesize that abcb_1, abcb_3 and abcg play a similar role in Platynereis spp. helping in maintaining cellular homeostasis and surviving acid stress.
1 Introduction
In certain marine ecosystems, CO2 concentrations in the water are naturally elevated, which is due to gas emissions from submarine volcanism (vents) or from nutrient upwelling (González-Delgado and Hernández, 2018; Santana-Casiano et al., 2016). The pCO2 in such waters can be several folds higher than the pCO2 commonly found in oceanic surface waters (150–530 μatm) (Takahashi et al., 2014). Gradients of seawater pCO2 levels in areas with natural CO2 emissions, such as the volcanic CO2 system in the Mediterranean Sea off Castello Aragonese on Ischia Island (Bay of Naples, Italy), can range from levels predicted for oceanic surface waters by the year 2100 due to climate change (500–1370 μatm, IPCC, 2014) to extreme conditions exceeding 20,800 μatm (Hall-Spencer et al., 2008). Studies of the species inhabiting such naturally acidified marine areas can provide insights into adaptations that enable organisms to tolerate lowered pH levels. This information can be useful to predict the effects of the globally growing anthropogenic ocean acidification (OA) on marine ecosystems. OA refers to the ongoing decrease in the surface seawater pH from a global, anthropogenic increase of the CO2 concentration in seawater. This is related to the emissions of anthropogenically produced CO2 into the atmosphere from combustion of fossil fuels that sharply increased after the industrial revolution and since then remained at a high level and are even still increasing (Bindoff et al., 2019; Takahashi et al., 2014).
Although negative effects of OA to marine life have been recognized, certain marine species that inhabit naturally acidified habitats are adapted to tolerate low pH conditions and thrive in them. Understanding the molecular/cellular bases of the adaptation to hostile acidified conditions and the molecular/cellular bases of tolerance/resistance could unravel the consequences of future OA scenarios for the marine invertebrate fauna.
Various studies provided evidence that ATP-binding cassette (ABC) transporter proteins confer cellular tolerance in acid stress environments in bacteria and in tumor cells, although the mechanism how ABC transporters counteract the adverse effects of low pH to cells and tissue is not understood (Aye et al., 2009; Gao et al., 2023; Leslie et al., 2005; Matsuhashi et al., 2015; Shvarev and Maldener, 2020; Tahara et al., 2012; 2015; Uchiyama et al., 2019; Wei and Roepe, 1994; Zhu et al., 2019, 2022). Some insights on these mechanisms were obtained from studies on cyanobacteria, as Synechocystis sp (Matsuhashi et al., 2015; Tahara et al., 2012; 2015; Uchiyama et al., 2019) and Anabaena sp (Shvarev and Maldener, 2020), exposed to acidified environments (pH from 3.0 to 6.0). Other bacteria able to adapt to low pH environments, like fermented foods for Acetobacter pasteurianus (Gao et al., 2023) and Lactococcus lactis (Zhu et al., 2019; 2022), have been investigated. For all the above-mentioned species, survival in acidified environments has been attributed to the presence of ABC transporters that is likely to have a role in conferring resistance to adverse pH conditions. It was suggested that the tolerance to acid stress in bacteria is due to the ABC transport of substances crucial for the maintenance of homeostasis in cells, such as the export of lipids or negatively charged substances to regulate the extracellular structures as external barriers against H+ ions (Matsuhashi et al., 2015; Tahara et al., 2012; 2015; Uchiyama et al., 2019) or the import of ions enhancing the activity of intracellular enzymes for the equilibration of internal pH (Zhu et al., 2022). Additional important findings on the involvement of ABC transport proteins in acid stress tolerance/adaptation originated from studying Multidrug Resistance (MDR) and pH of cancer cell lines. Wei and Roepe (1994) have checked if lower extracellular pH, typical of malignant cells, contributes to the development of the MDR phenotype in colon carcinoma SW620 and HCT15 lines never exposed to chemotherapeutic drugs. Results have shown an increase in ABCB1/MDR1 protein content and gene expression in cells exposed to a CO2-enriched external environment (pH reached the value of 6.5). The adaptation of the SW620 and HCT15 lines to acid external pH has been attributed to the increased expression and functional responses of MDR related ABC transporters that are probably associated with re-equilibration of the pH of the cytosol (as shown by Keizer and Joenje, 1989), facilitating the physiological functions of the cells. These findings indicate that ABC transporters can be involved in the adaptation to low pH conditions on the cellular level; however, it is so far not clear whether ABC transporters are involved in acid-stress resistance on the multicellular organism-level.
The ABC transporters are among the largest and well conserved protein superfamilies identified to date, comprising so far ten subfamilies (ABCA to J) across all prokaryote and eukaryote taxa (Dean and Annilo, 2005; Verrier et al., 2008). There has been great attention towards the ABCB, ABCC and ABCG subfamilies because of their involvement in MDR and Multixenobiotic Resistance (MXR), denominating cellular/organismal resistance towards environmental xenobiotics (Kurelec, 1992), and their importance in bacterial antibiotic resistance (Greene et al., 2018). Originally known for their involvement in cancer drug resistance (Biedler and Riehm, 1970), they are more generally a cellular stress response as the first and last defense system widely present in several taxa including marine species and their embryos/larvae stages (Epel et al., 2008; Kurelec, 1992). Beyond drugs and xenobiotics, MDR genes are induced by other stress signals, such as heat-shock, inflammation, hypoxia, UV and X irradiation (Scotto, 2003).
Very limited information is currently available on the mechanisms of tolerance to low pH conditions of marine invertebrate species, despite their importance to understand the potential future impact of OA on marine life (Simonetti et al., 2022). The knowledge on bacterial and cancer cell resistance to acid stress provides a valuable foundation to address research on the topic. It is reasonable to hypothesize that ABC transporters may play a similar role in helping more complex species to tolerate low-pH conditions. Additionally, the mechanisms carried out by bacterial and cancer cells might also be employed by multicellular organisms to manage acidic conditions, aiding in the maintenance of homeostasis and the prevention of cell death.
The aim of the present study was to disclose the involvement of ABC proteins in adaptation to low-pH/high-pCO2 environments in the marine polychaete sibling species Platynereis dumerilii/cfr massiliensis populating naturally acidified marine coastal vents. Platynereis dumerilii (Audouin and Milne-Edwards, 1834) is a marine annelid polychaete belonging to the family Nereididae, widely distributed throughout the European seas, all along the Mediterranean coasts and the Black Sea either in normal and acidified environments (Read and Fauchald, 2023). It is considered a model species in genetics and single‐cell genomics, because its genome has been completely sequenced, which provides an important base for molecular development and evolution studies (Özpolat et al., 2021; Tessmar-Raible and Arendt, 2003) in comparative studies since its evolutionary lineage has been slow-evolving (Zantke et al., 2014), and in neurobiology, ecology (Hardege, 1999) and ecotoxicology (Bellan, 1980; Hutchinson et al., 1995). Furthermore, it has been proven to be an excellent model to address various aspects of acclimation/adaptation to OA, such as functional traits analyses (Lucey et al., 2015; Gambi et al., 2016) and evolutionary implications (Wäge et al., 2017). It is highly abundant, along with its sibling species P. crf massiliensis, in naturally acidified areas, such as in the CO2 vents off Castello Aragonese (Ischia Island, Italy) (Calosi et al., 2013; Ricevuto et al., 2012; 2014; Wäge et al., 2018), as well as in other vent systems (e.g., Vulcano vents, Italy; Vizzini et al., 2017; Wäge et al., 2017).
Firstly, the presence of ABC genes as annotated sequences from the known genome and their expression in various organs/tissues of adult specimens of P. dumerilii. crf massiliensis was investigated. Then, the contribution of 7 selected ABC transporter proteins to adaptation/tolerance to low-pH/high-pCO2 seawater conditions in two annelid sibling species was established thanks to a 30 d in-situ reciprocal transplant experiment in a naturally acidified marine area characterized by vents with a seawater pH gradient from normal pH value (8.1) down to 6.5.
2 Materials and methods
2.1 Genome-wide identification of ABC transporters in P. dumerilii
2.1.1 Identification and characterization of putative ABCs
The presence of the ABC transport proteins in the genome of P. dumerilii was investigated in tissues and organs via homology searches as detailed below. The P. dumerilii genome version EMBL_pdum_1.0 (Genbank assembly: GCA_026936325.1) was used as the reference assembly for ABC transporter identification (Mutemi et al., 2024, biorxiv).
All the available protein sequences of animal ABC transporters were downloaded from the National Center for Biotechnology Information (NCBI) database (Supplementary Table S1) and a local alignment with the P. dumerilii proteome was conducted by the BLAST tool (Basic Local Alignment Search Tool). A Pfam analysis was also carried out by the hmmer tool, version 3.3.2 (Eddy, 2011), to catch any other potential ABC transport protein sequence not identified by the BLAST analysis. The most specific domains for ABC proteins (listed in Supplementary Table S2) from https://pfam.xfam.org/ were selected and used for the analysis. Annotation and nomenclature of each P. dumerilii ABC protein were completed based on amino acid sequence similarities in terms of in silico analysis by searching for known orthologs, amino acid similarity and identity comparison with BLASTp analysis on the protein database of NCBI. Finally, annotated proteins were searched in the P. dumerilii genome to disclose the corresponding coding genes.
2.1.2 Phylogenetic analysis of ABCs
The ABC protein amino acid sequences of P. dumerilii were phylogenetically analyzed to show the evolutionary relationships among them based upon similarities and differences in their characteristics. The full-length amino acid sequences retrieved from the NCBI server were aligned using T-Coffee web server (Notredame et al., 2000) and a phylogenetic tree was constructed with iTOL v6 web server (Letunic and Bork, 2021). Furthermore, the ABC protein sequences previously retrieved from NCBI (Supplementary Table S1) were used for the phylogenetic analysis with the P. dumerilii ABC proteins to highlight evolutionary relationships between species and to support the previous ABC annotations. For this, full-length amino acid sequences were aligned using the Muscle tool, version 5.1 (Edgar, 2022) and trimmed by the ClipKIT tool, version 1.3.0 (Steenwyk et al., 2020), using the most restrictive trimming method kpic-smart-gap to retain phylogenetically informative sites and remove the others. A phylogenetic tree was constructed with a maximum likelihood method using the iq-tree tool, version 2.2.0.3 (Nguyen et al., 2014).
2.1.3 Organ/tissue distribution and expression of ABC genes
In order to understand the possible physiological function of ABC transport proteins, their distribution in the P. dumerilii body was examined through the ‘P.Dumwhole-body’ single cell atlas available at EMBL Heidelberg. The cellular atlas integrates gene expression and ultrastructure for the whole P. dumerilii body and cells are clustered in organ/tissue classes based on single-cell RNA sequencing.
A subset of 40 ABC transporter genes was selected from ABCB, C and G subfamilies and searched into the EMBL available atlas for their tissue distribution and expression pattern.
2.2 In-situ reciprocal transplant experiment
2.2.1 Study area
The transplant study was conducted at the volcanic vents off Castello Aragonese (40° 43’ 57.9’’ N, 13° 57’ 51.8 E), located on the north-eastern side of Ischia Island (Tyrrhenian Sea, Bay of Naples, Italy) (Figure 1). The naturally acidified area is characterized by thermal vents with a seawater pH gradient from normal value (8.1) down to 6.5 (0.7 x 106 L d-1 emitted gas at the north side and 1.4 x 106 L d-1 emitted gas at the south side, Hall-Spencer et al., 2008). In this area, three sites (A4, A5, A6; “A” for “Acidified”) were chosen to collect specimens of polychaetes and allocate the moorings for the in-situ reciprocal transplant experiment (Figure 1).
Figure 1
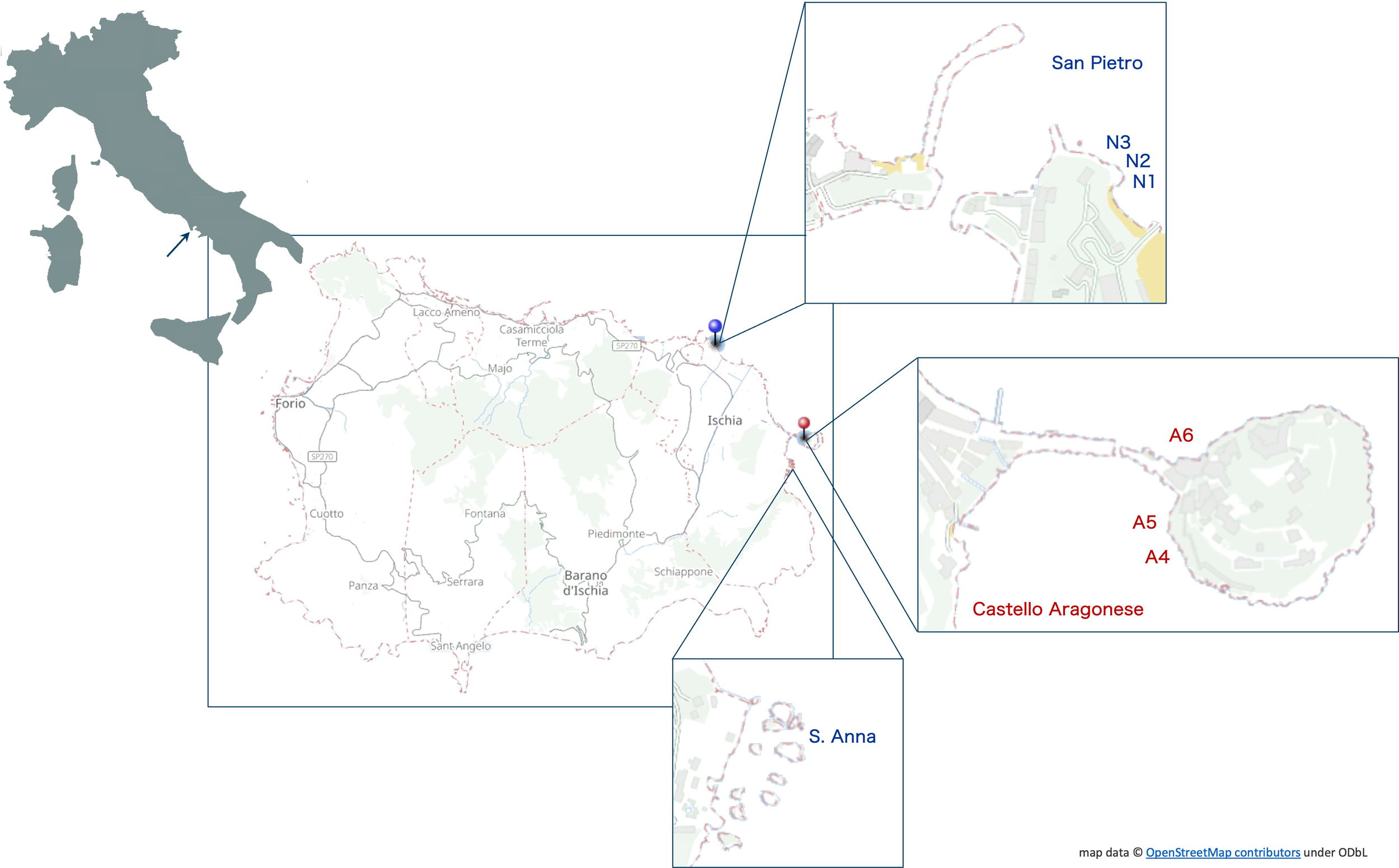
Map of the study area in the Ischia Island (Italy) indicating the three control areas where seawater was at normal pH (San Pietro, N1, N2 and N3) and the hydrothermal vents off the Castello Aragonese with acidified seawater (A4, A5, A6), where transplant experiments took place.
The three sites at normal pH (8.1) were chosen in the San Pietro area (40° 44’ 47.6’’ N; 13°56’ 40.42’’E), approximately 4 km off the vents. Indeed, the environmental characteristics of the area were very similar to those characterizing the vents in terms of exposure to light and hydrodynamic conditions, temperature, salinity, and depth, except for the pH value (Calosi et al., 2013). Specimens of polychaetes were collected nearby San Pietro, and three mooring stations (N1, N2, N3) were placed approximately 50 m from each other (Figure 1). A further site at normal pH was used for specimens’ collection due to limited abundance in the San Pietro area and named S. Anna (40° 43’ 34.51’’ N, 13° 57’ 35.7’’ E) located approximately 600 m away from the south side of the vent area of Castello, no venting activities are present in this area and the pH is in the range between 8.05 and 8.11 (Ricevuto et al., 2015a) (Figure 1).
Specimens were also collected from the acidified area of Castello Aragonese and three mooring stations (A4, A5 on the south side, and A6 on the north side) were placed again approx. 50 m from each other (Figure 1).
Seawater pH, salinity and conductivity in each site were recorded, at the beginning and at the end of the transplant experiment, as follows: A4: pH 7.25 ± 0.03, salinity 38.5 ± 0.71 ‰, conductivity 52.8 ± 0.07 µS mm-1; A5: pH 7.62 ± 0.01, salinity 39.0 ± 0.0 ‰, conductivity 53.0 ± 0.07 µS mm-1; A6: pH 6.88, salinity 38.0 ± 0.0 ‰, conductivity 52.0 ± 0.14 µS mm-1; N1-3: pH 8.18 ± 0.01, salinity 38.5 ± 0.5 ‰, conductivity 53.0 ± 0.125 µS mm-1. Long-term trend of the pH in these zones are also available in Foo et al. (2018).
2.2.2 Sample collection
Adult individuals of the two sibling species P. dumerilii/P. cfr massiliensis living in association with macroalgae, mainly the brown algae Halopteris scoparia and Dictyota spp., were collected during the first week of May 2023. Specimens of both species were collected by SCUBA divers at 1–2 m depth from the acidified area at the southern sites of Castello Aragonese and by snorkelers at 1–2 m depth from the two normal-pH sites, “San Pietro” and “S. Anna” rocks. Thalli of H. scoparia and Dictyota spp. with Platynereis individuals attached were removed and immediately transferred to containers filled with natural seawater on board and transported to the laboratory. The algae were then transferred to large plastic trays, to avoid anoxic conditions, and gently shaken to isolate Platynereis spp. specimens. After morphological identification, specimens were measured by means of millimetric paper and divided into three size groups (< 1 cm, 1–2 cm, > 2 cm length).
Organisms were maintained for 1–3 d in a temperature-controlled room at 18°C at a light regime of 12 h light/12 h dark in plastic bowls (10 individuals per bowl) supplied with approximately 200 mL of seawater taken from the sampling site to reduce the stress due to handling. pH was also checked daily and maintained constant at 8.2 for specimens collected from normal pH sites and between 7.2 and 7.6 for the acidified sites.
Additional 40 specimens from N and A areas were stored for molecular analysis of time 0 of the reciprocal transplant experiment (t0_N and t0_A groups respectively, Table 1) and then transferred into Eppendorf tubes with RNAlater solution (Sigma-Aldrich Company Ltd., Gillingham, U.K.).
Table 1
| Sample ID | Initial pH (mean) | Final pH (mean) | Transplant | Collection time |
|---|---|---|---|---|
| t0_A | 7.25-7.62 | – | – | t0 – before transplant |
| t0_N | 8.18 | – | – | t0 – before transplant |
| t30_A4 | 7.25-7.62 | 7.62 | A → A4 | t30 – end of transplant |
| t30_A5 | 7.25-7.62 | 7.25 | A → A5 | t30 – end of transplant |
| t30_A6 | 7.25-7.62 | 6.88 | A → A6 | t30 – end of transplant |
| t30_N | 8.18 | 8.18 | N → N | t30 – end of transplant |
| t30_AN | 7.25-7.62 | 8.18 | A → N | t30 – end of transplant |
| t30_NA4 | 8.18 | 7.62 | N → A4 | t30 – end of transplant |
| t30_NA5 | 8.18 | 7.25 | N → A5 | t30 – end of transplant |
| t30_NA6 | 8.18 | 6.88 | N → A6 | t30 – end of transplant |
Summary of sample IDs and experimental conditions.
2.2.3 Transplant of individuals
Individuals collected at sites with the normal seawater pH were placed in the moorings located in the acidified area and exposed for 30 d (May-June 2023) (t30_NA); other individuals were left in the moorings within the original site at normal seawater pH and left for 30 days as well (t30_N). Similarly, specimens collected from the acidified site were transferred in moorings placed in the normal-pH (t30_AN) area and some left in the acidified one and both left for 30 days (t30_A) in accordance with previous transplant studies (Ricevuto et al., 2015b) in which 30 days were selected to stimulate major pathway of response (Table 1). The experiment was designed to use two replicates of 10 individuals per each experimental plot (however, due to bad weather conditions, only one replicate was available for t30_NA4, t30_NA5, t30_NA6 treatments).
Moorings were made of cylindrical chambers hosting at least 10 individuals each and homogeneously divided according to their size. Two small cylindrical chambers were placed for each mooring. Transplantation chambers were constructed from PVC tubes (diameter 4 cm, length 11 cm) closed at both ends by a plankton net (mesh size 100 mm). The net mesh size guarantees regular water flow through the tube, allowing respiration and preventing escapes. These chambers have been used in previous experimental transplants (see Calosi et al., 2013). Small portions of H. scoparia and Dictyota spp. thalli were added to each chamber as a feeding source and a substrate to attach, to maintain polychaetes in conditions as close as possible to those they experience in their habitat. The chambers were fixed, via plastic cable ties, to a rope attached to stony moorings (Figure 2), and deployed by SCUBA divers in each exposure site, approximately at 2 m depth.
Figure 2

Stony moorings with anchored cylindrical pvc chambers where worms were allocated at the beginning (left) and 30 days later, at the end (right) of the experiment in one of the plots within the vents (A5).
After 30 days, chambers were carefully retrieved by snorkeling and rapidly transferred to the laboratory where polychaetes were checked for healthy conditions and then stored at -80°C in RNAlater for subsequent analyses.
A schematic description of the experimental plan is reported in Figure 3.
Figure 3
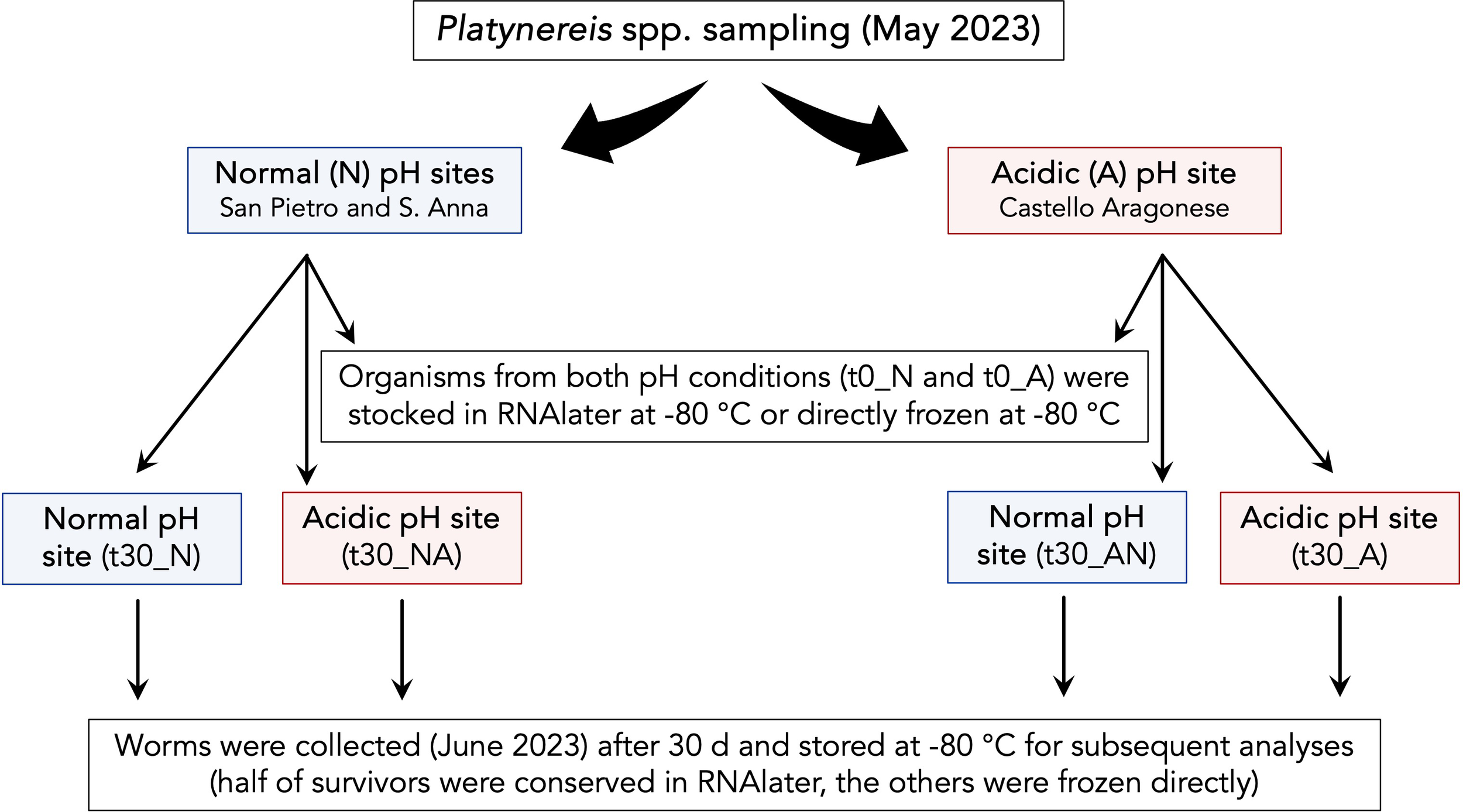
The worms’ transplant experimental plan.
2.3 Quantitative analysis of ABC transporter gene expression
2.3.1 Total RNA isolation and cDNA synthesis
Total RNA was extracted from pools of five P. dumerilii individuals (4–18 mg tissue in total) using the PureLink® RNA Mini Kit following the manufacturer’s protocol (Ambion life technologies, USA). In brief, samples stored in RNAlater at -80°C were thawed, centrifuged (6,000 rcf, 2 min, 4°C) and the supernatant was removed. The tissue was homogenized using the TissueLyser LT (Qiagen) with a run time of 3 min at 50 oscillations. The integrity of the extracted total RNA was tested on a BioPhotometer (Eppendorf 6131) and on a 1.5% agarose ‘bleach gel’ (Aranda et al., 2012, modified); the RNA concentration was quantified with a BioPhotometer (Eppendorf). First-strand cDNA was synthesized from approximately 130 ng of RNA in a final volume of 20 μL by using the qScript Ultra SuperMix® (Quantabio, USA) according to manufacturer’s instructions (8 μL RNA template, 4 μL qScript XLT cDNA SuperMix (5X), 8 μL RNase/DNase-free water).
2.3.2 Target gene selection
Based on previous studies on the involvement of ABCs in the resistance to acid conditions of bacteria species and cancer cells (Gao et al., 2023; Matsuhashi et al., 2015; Shvarev and Maldener, 2020; Tahara et al., 2012; 2015; Uchiyama et al., 2019; Wei and Roepe, 1994; Zhu et al., 2019), target genes belonging to the superfamily of ABC were selected from the P. dumerilii ABC inventory. Here the focus was on the subfamilies ABCB, ABCC, ABCG which are the largest subfamilies and in particular the abcb genes (abcb_1, abcb_2, abcb_3) were chosen for their homology to ABCB1/MDR1/P-gp and ABCB4 genes, abcc genes (abcc_1, abcc_2, abcc_3) were selected for the homology with MRPs and abcg gene is homologous to ABCG2 multidrug transporter (Supplementary Table S3). Nucleotide sequences for the selected genes were obtained from Platynereis genome (version 2.0) sequenced at EMBL. The sequences of forward and reverse primers are reported in Supplementary Table S3 and were designed through the online tool Primer3Plus (Untergasser et al., 2007).
The two housekeeping genes, 18S and a-TUB and respective primers (Supplementary Table S3) were from Wäge et al. (2018).
2.3.3 qPCR analysis of gene expression
qPCRs were performed in triplicate on an iCycleriQ5 (Bio-Rad, USA). Reaction mixes with a final volume of 20 μL contained 2 μL cDNA (diluted 1:10), 10 μL PerfeCTa® SYBR® Green SuperMix, Low ROX (Quantabio, USA) and 1 μL (10 μM) of each primer pair. A reaction without cDNA template was run in parallel to control for primer dimers and/or external contamination. qPCR conditions were: initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 15 s and extension at 72°C for 10 s. For the generation of a melt curve, 61 cycles of 10-s increasing temperature steps from 65°C to 95°C were added at the end of the qPCR run. The primer efficiency (Supplementary Table S3) was determined by a three-point cDNA dilution series (dilution factor of 5) and calculated by the iCycleriQ5. The relative quantification of gene expressions was analyzed by the formula (Livak and Schmittgen, 2001).
Where E corresponds to primer efficiency and CT represents the threshold cycle values.
2.4 Statistical analyses
For statistical analyses, data were tested for normal distribution using the Kolmogorov–Smirnov test. As normal distribution of data was not found for all cases, the significance of variations in transcription levels were evaluated using the nonparametric Kruskall-Wallis and Dunn’s post hoc tests. For all analyses, a p < 0.05 was considered statistically significant. Values were presented as means ± standard deviations (SD) and significant differences were labeled with asterisks and letters. Statistical analyses of gene expression were performed using GraphPad Prism 5 for Windows (GraphPad Software, USA).
3 Results
3.1 Genome-wide identification of ABC transporters in P. dumerilii
3.1.1 Identification, characterization and phylogenetic analysis of P. dumerilii ABC transporters
A total of 81 ABC transporters were annotated and identified in P. dumerilii proteome and genome, belonging to the 7 subfamilies from A to G, as shown in Figure 4 and listed in Supplementary Table S4.
Figure 4
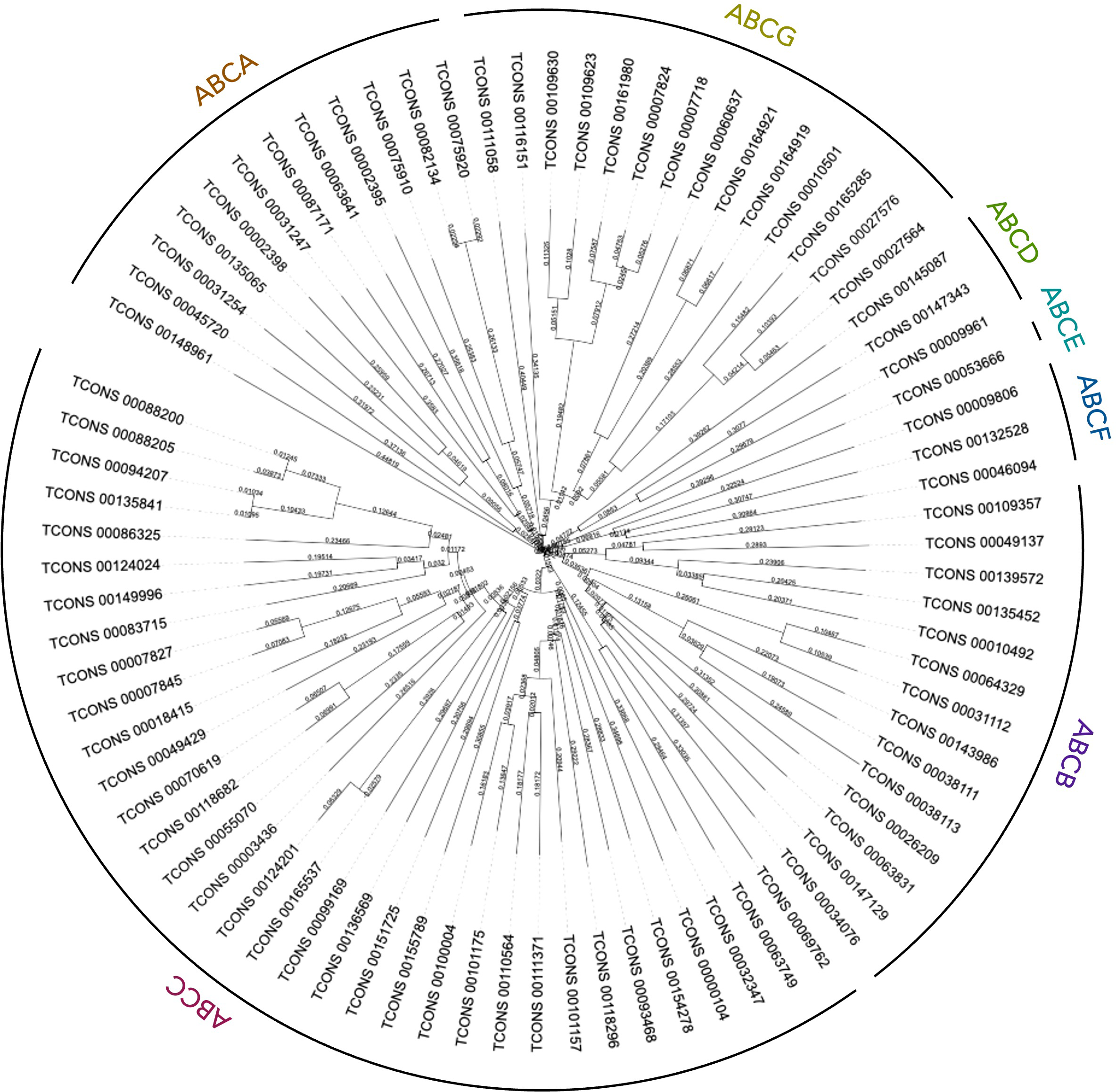
Phylogenetic tree of the amino acid sequences of P. dumerilii ABC transport proteins. The 81 ABC amino acid sequences were aligned using T-Coffee and analyzed in iTOL v6.
The largest ABC, encoded by the gene XLOC_024852, was of 2583 amino acids and the shortest ones, encoded by XLOC_028182 and XLOC_033621 of 132 (Supplementary Table S4). Subfamily C was the largest, with 34 transport proteins, followed by subfamilies B and G with 14 proteins, while 12 transport proteins belonged to the subfamily A. Subfamilies D and F included 3 proteins each and the smallest subfamily resulted the ABCE, with only 1 protein (Supplementary Table S4). No proteins from the subfamily H were found.
Among the 81 putative ABC transport proteins identified, 33 proteins were full transporters (2 nucleotide-binding domains – NBDs and 2 transmembrane domains – TMDs), belonging to the subfamilies ABCA, ABCB and ABCC, while 25 were half transporters (1 NBD and 1 TMD), from the subfamilies ABCA, ABCB, ABCC, ABCD and ABCG (Supplementary Figure S1). One ABCC transporter contained 2 NBDs and 3 TMDs; three proteins being part of the subfamilies ABCA, ABCB and ABCC contained 2 NBDs and 1 TMD and three of the subfamilies ABCB and ABCG contained 1 NBD and 2 TMDs. In addition, 16 proteins with high similarity to ABC transport proteins from other organisms missed the NBD or the TMD. Those were considered as incomplete proteins, possibly due to the completion and annotation of the P. dumerilii proteome.
Phylogenetic analysis of P. dumerilii ABC transporter proteins and all the available protein sequences of ABC transporters from NCBI database confirmed the previous annotation (Figure 5).
Figure 5
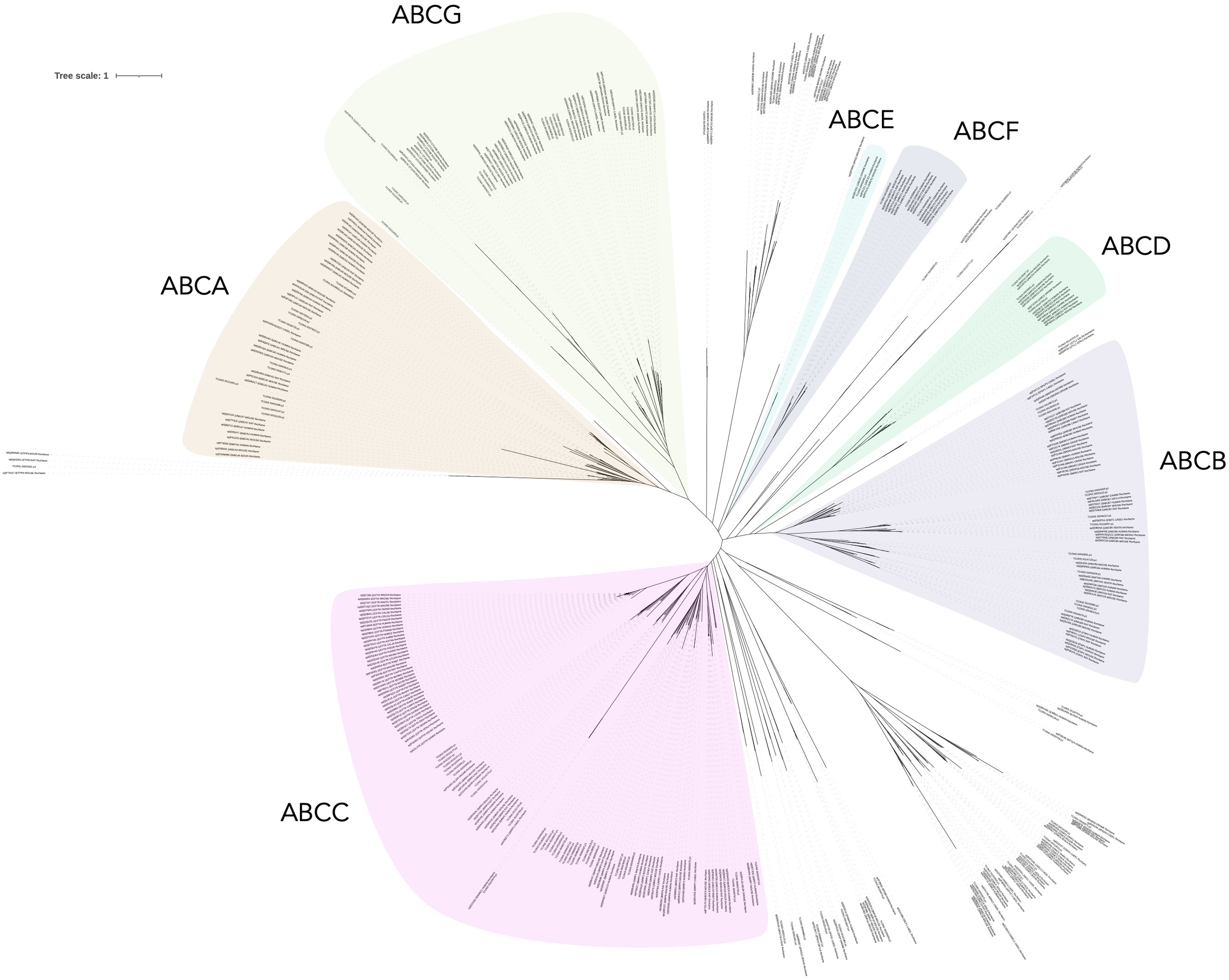
Unrooted phylogenetic tree of the amino acid sequences of P. dumerilii (Supplementary Table S4) and other animal ABC transport proteins (Supplementary Table S1). ABC amino acid sequences were aligned using the Muscle tool, version 5.1 and analyzed by ClipKIT tool, version 1.3.0.
3.1.2 Organ/tissue distribution of ABC genes expression in P. dumerilii
Thanks to the P.Dumwhole-body single cell atlas, clusters of tissue-prevalent and tissue-specific genes and genes that are differentially expressed in different tissues/organs were identified as shown in Figure 6. Most of the ABC transport proteins found in P. dumerilii are midgut-specific (8) or midgut-prevalent (9). Some showed specificity for neurons (5), body epidermis and ectodermal cells (2), and somatic (1) and visceral (1) muscle. The remaining ABCs were prevalent in unknown-tissue clades (2) or broadly expressed in all tissues (12).
Figure 6
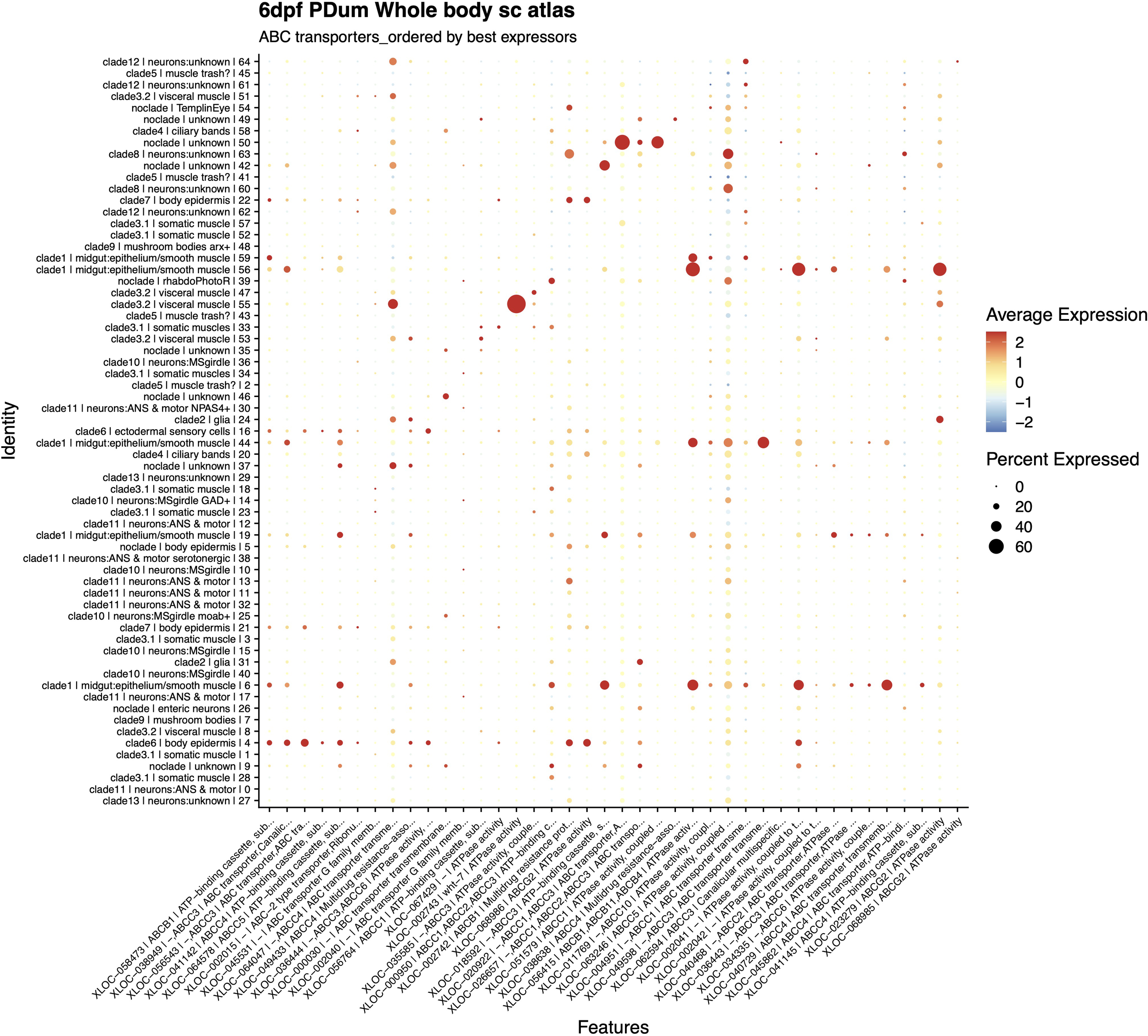
Dotplot of ABC transport protein tissue/organ distribution and expression visualizing the relative expression levels of a subset of ABC transporter genes (x-axis) across all tissue/organ clusters (y-axis). Fold gene expression levels are indicated by shades of red (increase in gene expression) and blue (decrease in gene expression), and larger dot diameter indicates that the gene expression was detected in greater proportion of cells from the cluster.
3.2 In-situ transplant experiment
At the end of the 30 days, mortality was observed only in t30_N groups (2.5%). All worms appeared in good health since they exhibited their protective mucous tubes and could actively move when taken out from chambers.
3.2.1 Differential ABC gene expression in organisms inhabiting different pH environments
As far as gene expression profile of ABC transport proteins in specimens from the transplant experiments, a comparison between specimens transplanted and those left in the original areas was performed to assess basal expression levels and modulation induced by changes in the seawater pH (Figure 7).
Figure 7
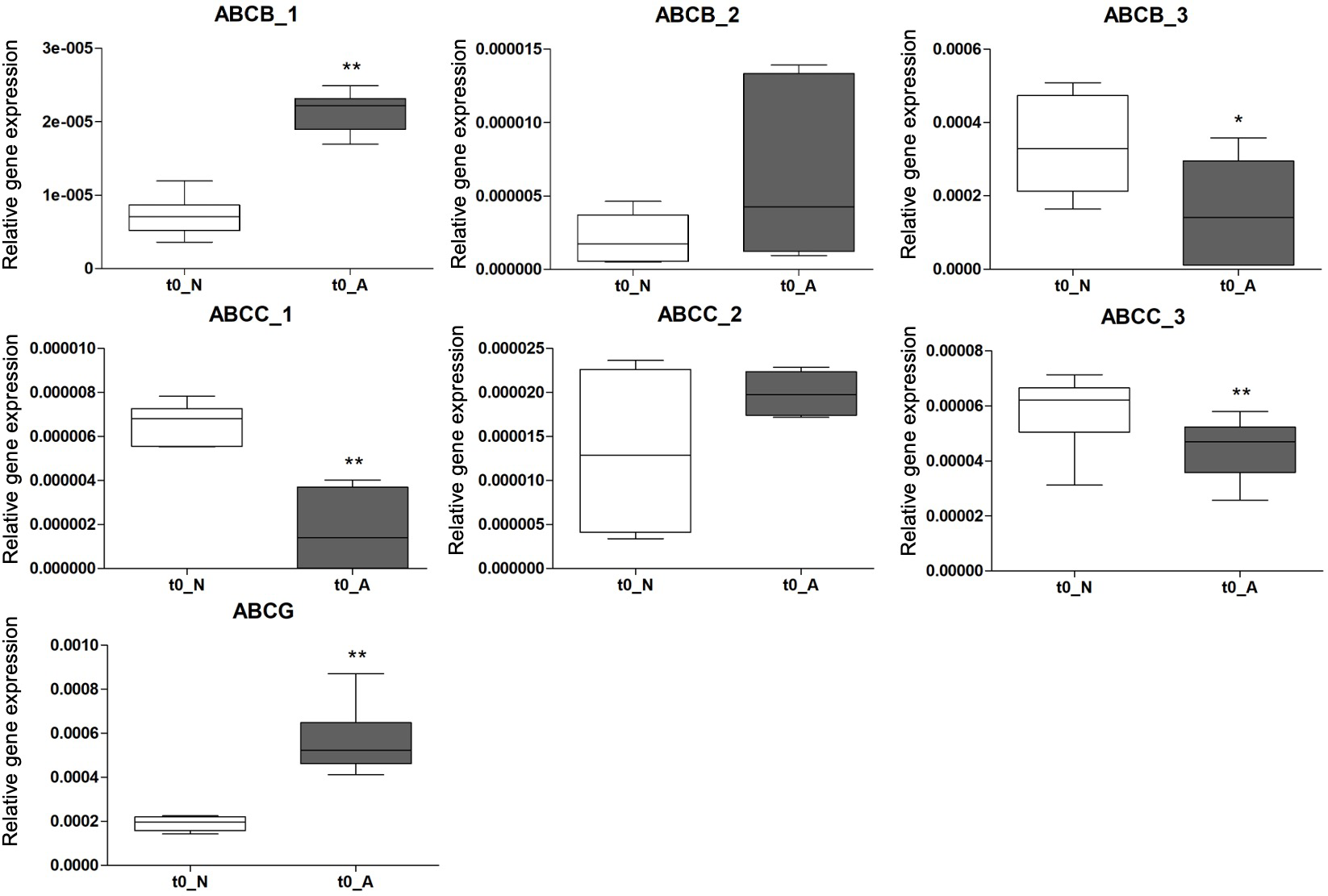
Relative gene expression of the 7 selected ABC genes in Platynereis spp. specimens collected at time 0 in normal (N) (white bars) and naturally acidified (A) (black bars) sites. Results are shown as mean ± standard deviation (SD). Asterisks indicate significant differences (*p < 0.05, **p < 0.01) between the two groups.
Overall, 5 out of the 7 ABC genes resulted to be differentially expressed at different pH conditions. In details, abcb_1 and abcg were higher expressed in worms living in low-pH sites compared to those from normal-pH areas (p < 0.01), while abcb_3, abcc_1 and abcc_3 were lower expressed (p < 0.05 for abcb_3 and p < 0.01 for abcc_1, abcc_3). No statistically significant differences were found in abcb_2 and abcc_2 gene expression by making the same comparison (Figure 7).
3.2.2 Stability gene expression of selected ABC over time
The stable expression across time (time 0–30 d) for the 7 target genes was tested to verify that ABC gene regulation during the transplant experiment was not affected by other factors except pH, so ABC gene expression was compared between t0 and after 30 days in specimens from normal pH sites and those from naturally acidified ones (t0_N vs t30_N and t0_A vs t30_A4, t30_A5, t30_A6).
abcb_3, abcc_1, abcc_3 and abcg gene expression was stable during the transplant in specimens from both sites (Figure 8).
Figure 8
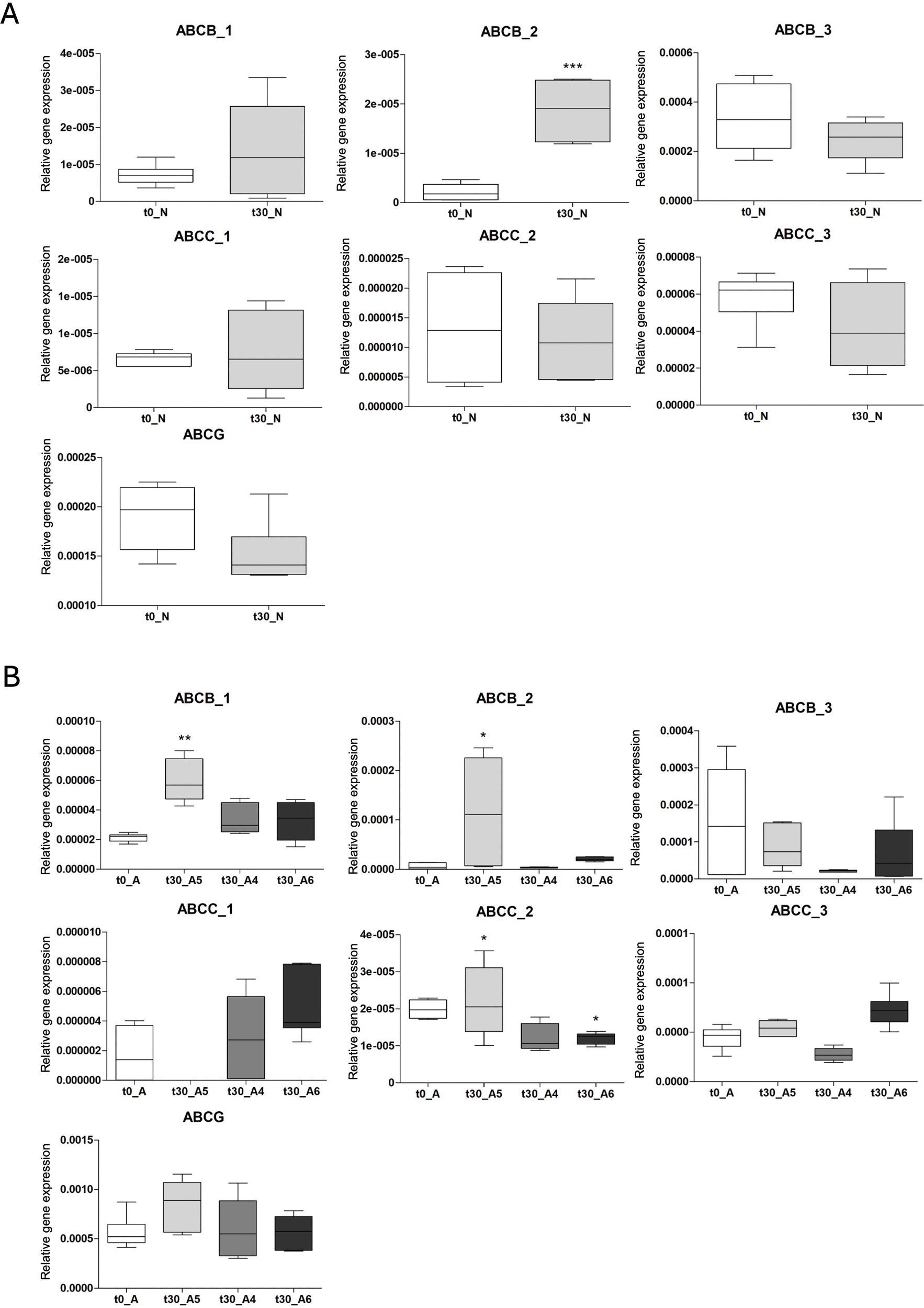
Gene expression profile of ABC genes in specimens from (A) N sites at t0 (white bars) and after 30 days (t30) (grey bars) and (B) A sites at the beginning (t0) (white bars) and at the end (t30) (grey bars) of the experiment in the three plots (A4, A5, A6). Results are shown as mean ± standard deviation (SD). Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) with the (A) t0_N group and (B) t0_A group.
No changes were observed in abcb_1 mRNA expression in normal-pH treatment (N) (Figure 8A) and it also remained stable in specimens from the lowest pH sites (6.88, t30_A6 and 7.25, t30_A4) while it was significantly higher in those from site of pH 7.62 (t30_A5 worms) compared to t0_A (p < 0.001) (Figure 8B). abcb_2 gene showed instability during the transplant experiment both in specimens from N (p < 0.001 comparing t0_N to t30_N) (Figure 8A) and A sites. In t30_A6 group the gene expression was statistically different from t0_A (p < 0.05), in t30_A5 the difference with t0A was not statistically significant but the standard deviation is high, indicating a great interindividual variability (Figure 8B). Lastly, abcc_2 gene did not change in N specimens (Figure 8A) while was downregulated in t30_A5 and t30_A6 (p < 0.05) compared to t0_A (Figure 8B).
3.2.3 Gene sensitivity to pH variation during transplant experiment
The relative gene expression for the 7 ABC genes was analyzed in Platynereis spp. specimens after translocation (N → A and A → N). ABC gene expression was firstly assessed by comparing t0_N specimens with those translocated in the 3 A sites as follows: t30_NA4, t30_NA5, t30_NA6. ABC gene expression of specimens from A sites (t0_A) was then compared to those of specimens translocated to normal-pH zones (t30_AN). Figure 9A refers to N → NA4, 5, 6 transplant and Figure 9B refers to A → AN transplant.
Figure 9
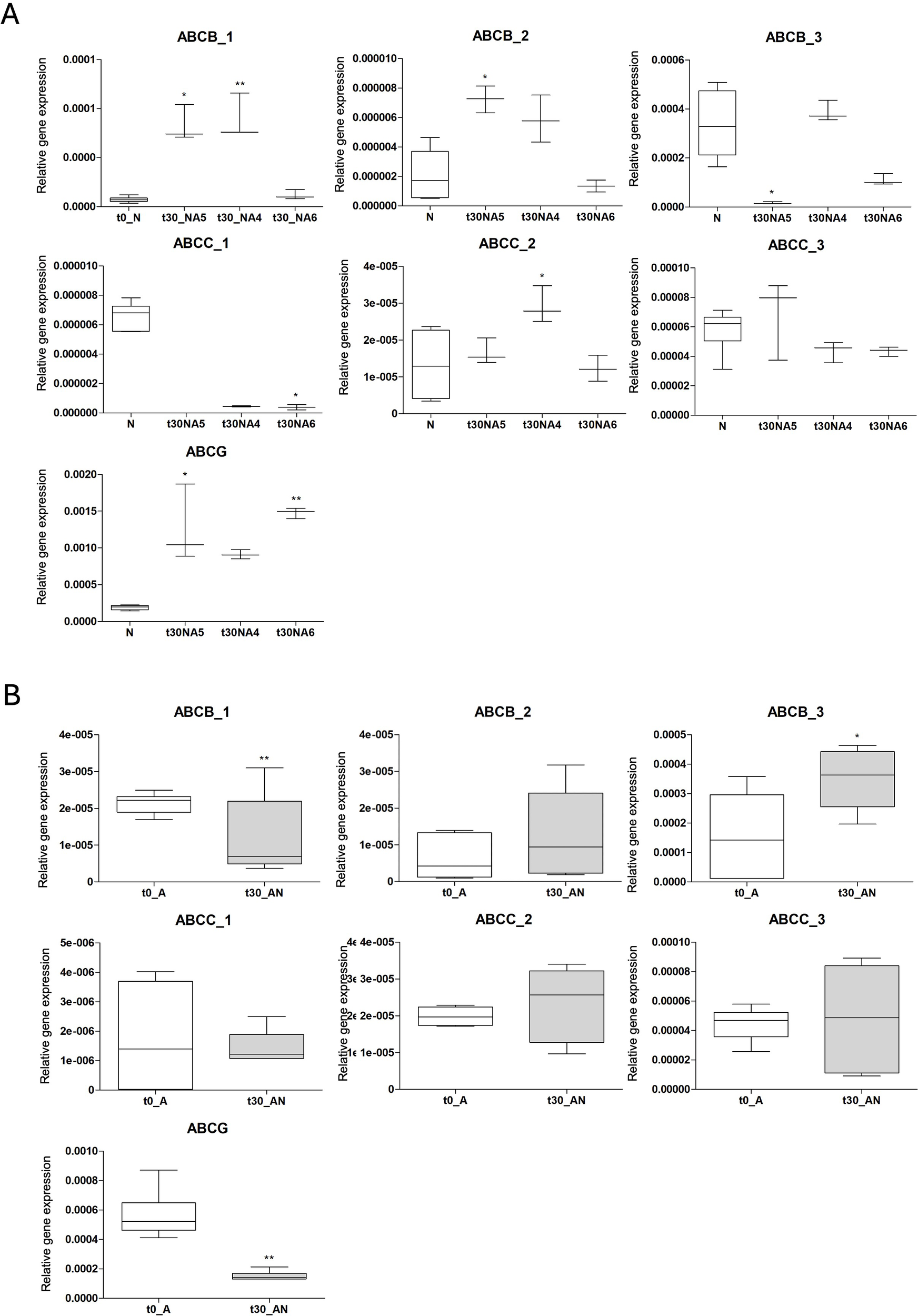
Relative gene expression levels of ABC genes in specimens transplanted from (A) N (white bars) to A4, A5, A6 (grey bars) and (B) A (white bars) to N (grey bars). Results are shown as mean ± standard deviation (SD). Asterisks indicate significant differences (*p < 0.05, **p < 0.01) with the (A) t0_N group and (B) t0_A group.
abcb_1: specimens from t30_NA4 and t30_NA5 groups showed a significant up-regulation (p < 0.01 and p < 0.05, respectively) in comparison to t0_N while t30_NA6 mRNA transcription did not vary (Figure 9A). On the contrary, abcb_1 gene expression resulted in down-regulation in specimens from A to N (p < 0.01) (Figure 9B).
abcb_2: similarly, this gene also resulted in up-regulation (p < 0.05) from t0_N group to t30_NA5 as well as t30_NA4, although not statistically significant. At lower pH (6.88, t30_NA6) no significant differences with t0_N were observed (Figure 9A). No changes in the abc gene expression were recorded in specimens translocated from A to N (Figure 9B).
abcb_3: a down-regulation was observed from t0_N to t30_NA5 (p < 0.05). Similarly, t30_NA6 specimens showed a decrease, but not significant, in abcb_3 gene expression compared to t0_N. No significant differences were detected in specimens transferred from N to A4 compared to the t0_N treatment (Figure 9A). Interestingly, the reverse transplant A to N induced an up-regulation of the abcb_3 gene (p < 0.01) (Figure 9B).
abcc_1: similarly, a down-regulation was observed from t0_N to t30_NA4 and t30_NA6 but only significant between t0_N and t30_NA6 group (p < 0.05) (Figure 9A). Unfortunately, no data was available for the t30_NA5 treatment. The individuals collected at t0_A did not show any statistically significant differences in abcc_1 gene expression with those transplanted from A to N (t30_AN) (Figure 9B).
abcc_2: no significant differences were found in t30_NA5 and t30_NA6 compared to t0_N while up-regulation of the abcc_2 was recorded in t30_NA4 specimens compared to the t0_N ones (p < 0.05) (Figure 9A). abcc_2 gene expression in treatment transplanted from A to N showed no statistically significant changes compared to t0_A (Figure 9B).
abcc_3: no significant changes in the expression of this gene were recorded (Figure 9).
abcg: an up-regulation was recorded in specimens from t0_N to t30_NA5 (p < 0.05) and between t0_N and t30_NA6 (p < 0.01) (Figure 9A). Contrarily, a down-regulation was observed in specimens translocated from A to N compared to t0_A group (p < 0.01) (Figure 9B).
4 Discussion
4.1 Genome-wide identification of ABC transporters in P. dumerilii
4.1.1 Identification, characterization and phylogenetic analysis of P. dumerilii ABC transporters
The P. dumerilii set of ABC transport proteins clearly is by far one of the largest with 81 proteins (Supplementary Table S5). From the available literature, only Tetranychus urticae and Macrostomum lignano have a higher number of ABC transporters (103 and 92 respectively) (Caña-Bozada et al., 2019; Dermauw et al., 2013), while from a minimum of 9 (Neobenedenia melleni, Caña-Bozada et al., 2019) to a maximum of 73 genes have been reported for other species (Tribolium castaneum, Broehan et al., 2013) (Supplementary Table S5). Furthermore, P. dumerilii shows a higher number of abc transporter proteins compared to marine Chordata species and Homo sapiens (Supplementary Table S5). Gene duplication has been identified as a mechanism for genomic adaptation to changing environments (Kondrashov, 2012). Therefore, the increased number of ABC transporter genes, as the one found in P. dumerilii, might be related to its ability to withstand extreme conditions, including the CO2 venting areas at Ischia Island (Calosi et al., 2013).
P. dumerilii ABC transport proteins can be divided into 7 subfamilies, labeled from A to G. This is consistent with our current understanding of ABC transporter subfamilies, as these seven subfamilies were found to be present across all taxa (Li and Nikaido, 2004; Annilo et al., 2006; Rea, 2007; Jeong et al., 2017). Notably, we observed the absence of subfamily H, which was identified in invertebrates and a single vertebrate species, the zebrafish (Danio rerio) (Dean and Annilo, 2005). Additionally, subfamily ABCI is found exclusively in plants (Verrier et al., 2008), while the subfamily ABCJ was recently proposed as a separate group, including nine proteins from the mosquito Aedes aegypti (Figueira-Mansur et al., 2020).
The ABCB, ABCC and ABCG subfamilies resulted in our study the most expanded, as observed among aquatic invertebrates (Jeong et al., 2017). Actually, these subfamilies include many highly conserved MXR-mediated related efflux transporters (abcb1/p-gp, abcc1-5, abcg2) which constitute important cellular defense mechanism towards marine pollutants in marine species (Epel, 1998; Kurelec, 1992). According to Xu et al. (2023), the number of ABCD, ABCE, and ABCF members was consistent across species, except for Platyhelminthes, which have a variable amount of such genes. As such, this confirms a high level of conservation for these transport proteins.
Phylogenetic analysis revealed that ABCE and ABCF subfamilies were more closely related to each other than to the other subfamilies (Figure 5).
Based on phylogenetic analysis including the P. dumerilii ABC transporter proteins and all protein sequences of animal ABC transporters available on the NCBI database (Figure 5) each P. dumerilii ABC transporter has homologues within the ABC transporters of H. sapiens and Mus musculus, and in many cases of Rattus norvegicus. For a more detailed characterization of P. dumerilii ABC transport proteins subfamilies see Supplementary Materials.
4.1.2 Organ/tissue distribution of ABC gene expression in P. dumerilii
ABC proteins from the subfamilies B, C and G have been discovered to be highly expressed in the midgut of different insects, including Tribolium castaneum, Conogethes punctiferali, Bactrocera dorsalis and Nilaparvata lugens. Most of them have been shown to be involved in resistance/clearance of insecticides and insecticidal proteins (Li et al., 2020; Rösner and Merzendorfer, 2020; Shwe et al., 2022; Wang et al., 2023). These findings suggest that the abundance of ABC proteins belonging to the subfamilies b, c and g found in our study in the midgut and neurons of P. dumerilii may be related to their involvement in resistance towards toxicants as well. ABC proteins of the subfamilies b and g participate in the resistance of cancer cells to low-pH environments as well. Therefore, it is reasonable to assume that ABC proteins belonging to the subfamilies B and G might have an important role in adaptation to low pH in seawater also in P. dumerilii by playing a role in physiological function maintenance and buffering the internal cell pH.
4.2 In-situ reciprocal transplant experiment
Stability of the ABC transporter gene expression was evaluated in specimens before the transplant and after 30 days of experiment in both normal- and low-pH areas as follows: t0_N vs t30_N and t0_A vs t30_A4, t30_A5, t30_A6 (Figure 8). Such assessment was needed to eventually exclude those genes that can be differentially expressed by other external factors beyond pH changes. All 7 ABC genes resulted stable except for abcb_2 and abcc_2 so these 2 were not selected for the next evaluation steps of their differential expression upon changes in pH. Hypotheses on the involvement of each abc gene in adaptation to low-pH were made based on the basal expression gene level of organisms inhabiting different pH environments (Figure 7) and verified through the ABC transcription results of the in-situ reciprocal transplant experiment (Figure 9).
abcb_1 was stably expressed across time in 2 out of 3 A sites, except in the t30_A5 vs t0_A (Figure 8B). Its expression was significantly different between the two populations inhabiting different pH habitats, with an increase in t0_A group compared to t0_N (Figure 7) so the enhancement of abcb_1 gene transcription could contribute to the resistance of Platynereis spp. to low pH. The hypothesis was confirmed thanks to the results of the transplant experiment, in which it was up-regulated in specimens translocated to low pH sites (t30_NA5 and t30_NA4) (Figure 9A) as well as down-regulated in organisms reversely translocated from A to N (t30_AN) (Figure 9B). The abcb_1 gene is homologous to the human ABCB1/MDR1 gene, found to be involved in tumor cell adaptation to acidified environments. Similarly to the Platynereis spp. abcb_1, the human MDR1 gene was detected to be up-regulated in colon carcinoma SW620 and HCT15 cell lines exposed for 5 d to pH 6.5 (Wei and Roepe, 1994). Interestingly, when the cells were shifted back to a normal extracellular pH (7.0), the gene expression decreased, in agreement with that observed in our study in specimens from A to N. Authors hypothesize that the increase in MDR protein transcription allows cells to cope with the acidic external environment by maintaining intracellular pH homeostasis (Wei and Roepe, 1994). Increased internal cell pH is correlated with the level of MDR expression by pumping acidic substances (Keizer and Joenje, 1989). Thus, it is reasonable to assume that abcb_1 in Platynereis spp. plays an important role in adaptation to low pH in seawater by playing a similar role and buffering the internal cell pH. It is worth noting that at t30_NA6 (pH 6.88) the transcription of the abcb_1 gene in transplanted Platynereis spp. specimens were at the same level as non-transplanted worms (Figure 9A). It could indicate an hormetic behavior in the abcb_1 gene expression to decreasing pH environments. The phenomenon of hormesis is defined as an adaptive dose-dependent response characterized by a biphasic pattern with a rise at lower pressure and an inhibition with increasing stress conditions (Calabrese and Baldwin, 2002). Thus, this suggests a clear initial involvement of the gene in resistance to low pH conditions based on the up-regulation of the abcb_1 gene and the ineffectiveness of this mechanism at severe pH conditions. So, different strategies are applied by the worms to survive in more severe acidified environments.
The abcb_3 gene expression did not change over time (Figure 8) and its decrease is likely to be involved in Platynereis spp. adaptation to low-pH environments, since it was down-expressed in individuals living in sites A as compared to those inhabiting normal-pH areas (Figure 7). The thesis was validated following the in-situ transplant experiment. Specimens transferred from N to A (t30_NA5 and t30_NA6) showed a down-regulation of abcb_3 gene (Figure 9A) while those transplanted from A to N sites displayed the up-regulation of the gene (Figure 9B). As abcb_1, also abcb_3 showed homology with human ABCB1/P-glycoprotein (P-gp). However, an opposite behavior of abcb_1 and abcb_3 gene expression was observed in this study supporting the hypothesis of a multifunctionality role of P-gp in aquatic invertebrates (Jeong et al., 2017). Actually, from the single cell analysis of Platynereis spp., the two abcb proteins exhibited specificity for different cell clades: abcb_1 is mostly present in the epidermis, midgut epithelium and smooth muscle cells while abcb_3 seems to occur more in neurons. Therefore, it is possible that they absolve different functions including the resistance to low pH environments.
The abcc_1 gene was stable over time (Figure 8) and by comparing t0_N and t0_A groups a significant down-expression was detected (Figure 7), confirmed also in specimens transplanted from N to A (t30_NA4, t30_NA6) (Figure 9A). However, a potential involvement in low pH resistance is not supported because of the absence of modulation in specimens translocated from A to N (Figure 8B). Similarly, although stable, abcc_3 gene did not vary in specimens from N to A (Figure 9A) and reverse (Figure 9B).
Interestingly, the abcg gene was highly expressed in specimens living in A compared to those from N (Figure 7) and up-regulated in those transplanted from N to A (Figure 9A). A down-regulation was observed in specimens transferred from A to N (Figure 9B) in agreement with the results from Cheng and To (2012), who observed an increase in the ABCG2 (homologous to Platynereis spp. abcg) gene expression in low-pH resistant human colon carcinoma HCT-116 cells after an exposure to pH 5. ABCG2 is involved in the efflux of physiological metabolites from cells (Leslie et al., 2005) and performs an anti-apoptotic function (Aye et al., 2009) so it is possible to suppose that the enhancement of abcg under acid stress might help cells to maintain their homeostasis and prevent death.
5 Conclusions
A first attempt to provide a complete inventory of the ABC transporters of the annelid P. dumerilii was carried out here; 81 ABC transporters belonging to the seven subfamilies ABCA-G were identified. These findings were crucial for the following investigation on the involvement of ABC transporter proteins in Platynereis spp. resistance/adaptation to acidified conditions.
In summary, 3 out the 7 abc transporters, namely abcb_1, abcb_3 and abcg, turned out to have a role in adaptation to low-pH/high-pCO2 environments of Platynereis spp. populating naturally acidified marine coastal areas. The homology with human ABCB1 and ABCG2, which are involved in carcinoma cell resistance to external low pH, suggests that the three here identified abc transporters play a similar role during an exposure to acidified environments participating in the maintenance of cellular homeostasis, buffering the internal pH of internal cell pH cells and preventing cell death in order to guarantee the proper functioning of cellular activities. This might be possible thanks to the trafficking of molecules and substances enrolled in these processes, such as the efflux of physiological metabolites from cells, by means of the three identified ABC transporter proteins. In future studies the assessment of function of these proteins requires more attention in order to understand why the transporters represent an adaptation mechanism to acidified conditions.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. KM: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. PR: Investigation, Writing – review & editing. TL: Validation, Writing – review & editing. VZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing. MG: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. IC: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the 2020 scholarship co-founded by the University of Siena and the Stazione Zoologica Anton Dohrn of Naples entitled ‘Molecular bases and evolutionary constraints of cellular acid tolerance in cyanobacteria and invertebrates subjected to ocean acidification and other sources of stress’ (METASTRESS) in the framework of the Doctoral Research Program in Environmental, Geological and Polar Sciences and Technologies (36th cycle) of the University of Siena. Furthermore, part of the work was performed in the framework of the Erasmus Long Life Learning Program of Dr. Simonetti S. at EMBL Heidelberg.
Acknowledgments
We are grateful to Prof. Detlev Arendt for hosting SS during her Erasmus traineeship at EMBL Heidelberg. We also acknowledge Victoria Alicia Witte and Dr. Leslie Pan from the Developmental Biology Unit at EMBL Heidelberg for their contributions to the bioinformatics analysis. Additionally, we thank Prof. Maria Cristina Fossi’s Lab (Department of Physical, Earth, and Environmental Sciences, University of Siena, Italy), particularly Dr. Cristina Panti and Dr. Giacomo Limonta, for their support with the real-time PCR instrument. We are also grateful to the Marine Protected Area staff “Regno di Nettuno” of Ischia for support at sea during the transplant experiment. At the beginning of this study, part of the PhD project of SS, MG was senior researcher at the Stazione Zoologica Anton Dohrn of Naples (Ischia Marine Center).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1573367/full#supplementary-material
References
1
Annilo T. Chen Z.-Q. Shulenin S. Costantino J. Thomas L. Lou H. et al . (2006). Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genom.88, 1–11. doi: 10.1016/j.ygeno.2006.03.001
2
Aranda P. S. LaJoie D. M. Jorcyk C. L. (2012). Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis33, 366–369. doi: 10.1002/elps.201100335
3
Aye I. L. Singh A. T. Keelan J. A. (2009). Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem. Biol. Interact.180, 327–339. doi: 10.1016/j.cbi.2009.04.012
4
Bellan G. (1980). Relationship of pollution to rocky substratum polychaetes on the French Mediterranean coast. Mar. Pollut. Bull.1, 318–321. doi: 10.1016/0025-326X(80)90048-X
5
Biedler J. L. Riehm H. (1970). Cellular resistance to actinomycin D in Chinese hamster cells in vitro: cross- resistance, radioautographic, and cytogenetic studies. Cancer. Res.30, 1174–1184.
6
Bindoff N. L. Cheung W. W. L. Kairo J. G. Arístegui J. Guinder V. A. Hallberg R. et al . (2019). “Changing ocean, marine ecosystems, and dependent communities,” in IPCC Special Report on the Ocean and Cryosphere in a Changing Climate. Eds. PörtnerH.-O.RobertsD. C.Masson- DelmotteV.ZhaiP.TignorM.PoloczanskaE.MintenbeckK.AlegríaA.NicolaiM.OkemA.PetzoldJ.RamaB.WeyerN. M. (Cambridge University Press, Cambridge, UK and New York, NY, USA), 447–587. doi: 10.1017/9781009157964.007
7
Broehan G. Kroeger T. Lorenzen M. Merzendorfer H. (2013). Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genom.14, 6. doi: 10.1186/1471-2164-14-6
8
Calabrese E. J. Baldwin L. A. (2002). Defining hormesis. Hum. Exp. Toxicol.21, 91–97. doi: 10.1191/0960327102ht217oa
9
Calosi P. Rastrick S. P. S. Lombardi C. de Guzman H. J. Davidson L. Jahnke M. et al . (2013). Adaptation and acclimatization to ocean acidification in marine ectotherms: an in situ transplant experiment with polychaetes at a shallow CO2 vent system. Phil. Trans. R. Soc Lond. B. Biol. Sci.368, 20120444. doi: 10.1098/rstb.2012.0444
10
Caña-Bozada T. V. Morales-Serna F. N. García-Gasca A. Llera-Herrerac R. Fajer-Ávila E. J. (2019). Genome-wide identification of ABC transporters in monogeneans. Mol. Biochem. Parasitol.234, 111234. doi: 10.1016/j.molbiopara.2019.111234
11
Cheng G. M. To K. K. (2012). Adverse cell culture conditions mimicking the tumor microenvironment upregulate ABCG2 to mediate multidrug resistance and a more Malignant phenotype. ISRN Oncol.2012, 746025. doi: 10.5402/2012/746025
12
Dean M. Annilo T. (2005). Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet.6, 123–142. doi: 10.1146/annurev.genom.6.080604.162122
13
Dermauw W. Osborne E. J. Clark R. M. Grbić M. Tirry L. Van Leeuwen T. (2013). A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genom.14, 317. doi: 10.1186/1471-2164-14-317
14
Eddy S. R. (2011). Accelerated profile HMM searches. PloS Comput. Biol.7, e1002195. doi: 10.1371/journal.pcbi.1002195
15
Edgar R. C. (2022). Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun.13, 6968. doi: 10.1038/s41467-022-34630-w
16
Epel D. (1998). Use of multidrug transporters as first lines of defense against toxins in aquatic organisms. Comp. Biochem. Physiol. A Mol. Integr. Physiol.120, 23–28. doi: 10.1016/S1095-6433(98)10005-3
17
Epel D. Luckenbach T. Stevenson C. N. MacManus-Spencer L. A. Hamdoun A. Smital T. (2008). Efflux transporters: newly appreciated roles in protection against pollutants. Environ. Sci. Technol.42, 3914–3920. doi: 10.1021/es087187v
18
Figueira-Mansur J. Schrago C. G. Salles T. S. Alvarenga E. S. L. Vasconcellos B. M. Melo A. C. A. et al . (2020). Phylogenetic analysis of the ATP-binding cassette proteins suggests a new ABC protein subfamily J in Aedes aEgypti (Diptera: Culicidae). BMC Genom.21, 463. doi: 10.1186/s12864-020-06873-8
19
Foo S. A. Byrne M. Ricevuto E. Gambi M. C. (2018). The carbon dioxide vents of Ischia, Italy, a natural laboratory to assess impacts of ocean acidification on marine ecosystems: an overview of research and comparisons with other vent systems. Ocean. Mar. Biol. Ann. Rev56, 237–310.
20
Gambi M. C. Musco L. Giangrande A. Badalamenti F. Micheli F. Kroeker K. J. (2016). Distribution and functional traits of polychaetes in a CO2 vent system: winners and losers among closely related species. Mar. Ecol. Prog. Ser.550, 121–134. doi: 10.3354/meps11727
21
Gao L. Shi W. Xia X. (2023). Genomic plasticity of acid-tolerant phenotypic evolution in Acetobacter pasteurianus. Appl. Biochem. Biotechnol.195, 6003–6019. doi: 10.1007/s12010-023-04353-9
22
González-Delgado S. Hernández C. (2018). The importance of natural acidified systems in the study of ocean acidification: what have we learned? Adv. Mar. Biol.80, 57–99. doi: 10.1016/bs.amb.2018.08.001
23
Greene N. P. Kaplan E. Crow A. Koronakis V. (2018). Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front. Microbiol.9. doi: 10.3389/fmicb.2018.00950
24
Hall-Spencer J. Rodolfo-Metalpa R. Martin S. Ransome E. Fine M. Turner S. M. et al . (2008). Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature454, 96–99. doi: 10.1038/nature07051
25
Hardege J. D. (1999). Nereidid polychaetes as model organisms for marine chemical ecology. Hydrobiologia402, 145–161. doi: 10.1023/A:1003740509104
26
Hutchinson T. H. Jha A. N. Dixon D. R. (1995). The polychaete Platynereis dumerilii (Audouin and Milne‐Edwards): a new species for assessing the hazardous potential of chemicals in the marine environment. Ecotoxicol. Environ. Saf.31 (3), 271–81. doi: 10.1006/eesa.1995.1074
27
IPCC . (2014). In. PachauriR. K.MeyerL. A. (Eds.), Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC, Geneva, Switzerland, 151.
28
Jeong C.-B. Kim H.-S. Kang H.-M. Lee J.-S. (2017). ATP-binding cassette (ABC) proteins in aquatic invertebrates: Evolutionary significance and application in marine ecotoxicology. Aquat. Toxicol.185, 29–39. doi: 10.1016/j.aquatox.2017.01.013
29
Keizer H. G. Joenje H. (1989). Increased cytosolic pH in multidrug-resistant human lung tumor cells: effect of verapamil. J. Natl. Cancer. Inst.81, 706–709. doi: 10.1093/jnci/81.9.706
30
Kondrashov F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc B279, 5048–5057. doi: 10.1098/rspb.2012.1108
31
Kurelec B. (1992). The multixenobiotic resistance mechanism in aquatic organisms. Crit. Rev. Toxicol.22, 23–43. doi: 10.3109/10408449209145320
32
Leslie E. M. Deeley R. G. Cole S. P. C. (2005). Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol.204, 216–237. doi: 10.1016/j.taap.2004.10.012
33
Letunic I. Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nuc. Acid. Res.49, W293–W296. doi: 10.1093/nar/gkab301
34
Li Z. Cai T. Qin Y. Zhang Y. Jin R. Mao K. et al . (2020). Transcriptional response of ATP-binding cassette (ABC) transporters to insecticide in the brown planthopper, Nilaparvata lugens (Stål). Insects11, 280. doi: 10.3390/insects11050280
35
Li X. Z. Nikaido H. (2004). Efflux-mediated drug resistance in bacteria. Drugs64, 159–204. doi: 10.2165/00003495-200464020-00004
36
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods25, 402–408. doi: 10.1006/meth.2001.1262
37
Lucey N. M. Lombardi C. De Marchi L. Schulze A. Gambi M. C. Calosi P. (2015). To brood or not to brood: are marine invertebrates that protect their offspring more resilient to ocean acidification? Sci. Rep.5, 12009. doi: 10.1038/srep12009
38
Matsuhashi A. Tahara H. Ito Y. Uchiyama J. Ogawa S. Ohta H. (2015). Slr2019, lipid A transporter homolog, is essential for acidic tolerance in Synechocystis sp. PCC6803. Photosynth. Res.125, 267–277. doi: 10.1007/s11120-015-0129-6
39
Mutemi K. N. Simakov O. Pan L. Santangeli L. Null R. Handberg-Thorsager M. et al . (2024). A genome resource for the marine annelid Platynereis dumerilii. BioRxiv, 2024.06.21.600153. doi: 10.1101/2024.06.21.600153
40
Nguyen L.-T. Schmidt H. A. von Haeseler A. Minh B. Q. (2014). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274. doi: 10.1093/molbev/msu300
41
Notredame C. Higgins D. G. Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol.302, 205–217. doi: 10.1006/jmbi.2000.4042
42
Özpolat B. D. Rande N. Williams E. A. Bezares-Calderón L. A. Andreatta G. Balavoine G. et al . (2021). The Nereid on the rise: Platynereis as a model system. EvoDevo12, 10. doi: 10.1186/s13227-021-00180-3
43
Rea P. A. (2007). Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol.58, 347–375. doi: 10.1146/annurev.arplant.57.032905.105406
44
Read G. Fauchald K. (2023). “World polychaeta database,” in Platynereis dumerilii (Audouin & Milne Edwards 1833).
45
Ricevuto E. Benedetti M. Regoli F. Spicer J. I. Gambi M. C. (2015b). Antioxidant capacity of polychaetes occurring at a natural CO2 vent system: Results of an in situ reciprocal transplant experiment. Mar. Environ. Res.112, 44–51. doi: 10.1016/j.marenvres.2015.09.005
46
Ricevuto E. Kroeker F. Ferrigno F. Micheli F. Gambi M. C. (2014). Spatio-temporal variability of polychaete colonization at volcanic CO2 vents indicates high tolerance to ocean acidification. Mar. Biol.161, 2909–2919. doi: 10.1007/s00227-014-2555-y
47
Ricevuto E. Lorenti M. Patti F. P. Scipione M. B. Gambi M. C. (2012). Temporal trends of benthic invertebrate settlement along a gradient of ocean acidification at natural CO2 vents (Tyrrhenian Sea). Biol. Mar. Mediterr.19, 49–52.
48
Ricevuto E. Vizzini S. Gambi M. C. (2015a). Ocean acidification effects on stable isotope signatures and trophic interactions of polychaete consumers and organic matter sources at a CO2 shallow vent system. J. Exp. Mar. Biol. Ecol.468, 105–117. doi: 10.1016/j.jembe.2015.03.016
49
Rösner J. Merzendorfer H. (2020). Transcriptional plasticity of different ABC transporter genes from Tribolium castaneum contributes to diflubenzuron resistance. Insect Biochem. Mol. Biol.116, 103282. doi: 10.1016/j.ibmb.2019.103282
50
Santana-Casiano J. M. Fraile-Nuez E. González-Dávila M. Baker E. T. Resing J. A. Walker S. L. (2016). Significant discharge of CO2 from hydrothermalism associated with the submarine volcano of El Hierro Island. Sci. Rep.6, 25686. doi: 10.1038/srep25686
51
Scotto K. W. (2003). Transcriptional regulation of ABC drug transporters. Oncogene22, 7496–7511. doi: 10.1038/sj.onc.1206950
52
Shvarev D. Maldener I. (2020). The HlyD-like membrane fusion protein All5304 is essential for acid stress survival of the filamentous cyanobacterium Anabaena sp. PCC 7120. FEMS Microbiol. Lett.367, fnaa108. doi: 10.1093/femsle/fnaa108
53
Shwe S. M. Prabu S. Jing D. He K. Wang Z. (2022). Synergistic interaction of Cry1Ah and Vip3Aa19 proteins combination with midgut ATP-binding cassette subfamily C receptors of Conogethes punctiferalis (Guenée) (Lepidoptera: Crambidae). Int. J. Biol. Macromol.213, 871–879. doi: 10.1016/j.ijbiomac.2022.06.019
54
Simonetti S. Zupo V. Gambi M. C. Luckenbach T. Corsi I. (2022). Unraveling cellular and molecular mechanisms of acid stress tolerance and resistance in marine species: New frontiers in the study of adaptation to ocean acidification. Mar. Pollut. Bull.185, 114365. doi: 10.1016/j.marpolbul.2022.114365
55
Steenwyk J. L. Buida T. J. III Li Y. Shen X.-X. Rokas A. (2020). ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PloS Biol.18, e3001007. doi: 10.1371/journal.pbio.3001007
56
Tahara H. Matsuhashi A. Uchiyama J. Ogawa S. Ohta H. (2015). Sll0751 and Sll1041 are involved in acid stress tolerance in Synechocystis sp. PCC 6803. Photosynth. Res.125, 233–242. doi: 10.1007/s11120-015-0153-6
57
Tahara H. Uchiyama J. Yoshihara T. Matsumoto K. Ohta H. (2012). Role of Slr1045 in environmental stress tolerance and lipid transport in the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys. Acta1817, 1360–1366. doi: 10.1016/j.bbabio.2012.02.035
58
Takahashi T. Sutherland S. C. Chipman D. W. Goddard J. G. Ho C. Newberger T. et al . (2014). Climatological distributions of pH, pCO2, total CO2, alkalinity, and CaCO3 saturation in the global surface ocean, and temporal changes at selected locations. Mar. Chem.164, 95–125. doi: 10.1016/j.marchem.2014.06.004
59
Tessmar-Raible K. Arendt D. (2003). Emerging systems: between vertebrates and arthropods, the Lophotrochozoa. Curr. Opin. Genet. Dev.13, 331–340. doi: 10.1016/S0959-437X(03)00086-8
60
Uchiyama J. Itagaki A. Ishikawa H. Tanaka Y. Kohga H. Nakahara A. et al . (2019). Characterization of ABC transporter genes, sll1180, sll1181, and slr1270, involved in acid stress tolerance of Synechocystis sp. PCC 6803. Photosynth. Res.139, 325–335. doi: 10.1007/s11120-018-0548-2
61
Untergasser A. Nijveen H. Rao X. Bisseling T. Geurts R. Leunissen J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res.35, W71–W74. doi: 10.1093/nar/gkm306
62
Verrier P. J. Bird D. Burla B. Dassa E. Forestier C. Geisler M. et al . (2008). Plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci.13, 151–159. doi: 10.1016/j.tplants.2008.02.001
63
Vizzini S. Martínez-Crego B. Andolina C. Massa-Gallucci A. Connell S. D. Gambi and M. C. (2017). Ocean acidification as a driver of community simplification via the collapse of higher-order and rise of lower-order consumers. Sci. Rep.7, 4018. doi: 10.0.4.14/s41598-017-03802-w
64
Wäge J. Rotchell J. M. Gambi M.-C. Hardege J. D. (2018). Target gene expression studies on Platynereis dumerilii and Platynereis cfr massiliensis at the shallow CO2 vents off Ischia, Italy. Estuar. Coast. Shelf Sci.207, 351–358. doi: 10.1016/j.ecss.2017.11.012
65
Wäge J. Valvassori G. Hardege J. D. Schulze and A. Gambi M. C. (2017). The sibling polychaetes Platynereis dumerilii and Platynereis massiliensis in the Mediterranean Sea: are phylogeographic patterns related to exposure to ocean acidification? Mar. Biol.164, 199. doi: 10.1007/s00227-017-3222-x
66
Wang L. He W. Lu J.-M. Sun J. Jiang S.-D. Wang J.-J. et al . (2023). Characterization and transcriptional expression of ABCG genes in Bactrocera dorsalis: Insights into their roles in fecundity and insecticidal stress response. Int. J. Biol. Macromol.253, 126836. doi: 10.1016/j.ijbiomac.2023.126836
67
Wei L.-Y. Roepe P. D. (1994). Low external pH and osmotic shock increase the expression of human MDR protein. Biochem.33, 7229–7238. doi: 10.1021/bi00189a027
68
Xu J. Zheng J. Zhang R. Wang H. Du J. Li J. et al . (2023). Identification and functional analysis of ABC transporter genes related to deltamethrin resistance in Culex pipiens pallens. Pest. Manage. Sci.79, 3642–3655. doi: 10.1002/ps.7539
69
Zantke J. Bannister S. Rajan V. B. V. Raible F. Tessmar-Raible K. (2014). Genetic and genomic tools for the marine annelid Platynereis dumerilii. Genetics197, 19–31. doi: 10.1534/genetics.112.148254
70
Zhu Z. Yang Y. Yang P. Wu Z. Zhang J. Du G. (2019). Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Microb. Cell. Factories.18, 136. doi: 10.1186/s12934-019-1188-8
71
Zhu Z. Yang P. Yang J. Zhang J. (2022). Comparative transcriptome analysis reveals the contribution of membrane transporters to acid tolerance in Lactococcus lactis. J. Biotechnol.357, 9–17. doi: 10.1016/j.jbiotec.2022.08.006
Summary
Keywords
ocean acidification, CO2 vents, Platynereis spp., ATP binding cassette (ABC) transporters, acid stress tolerance, gene expression analysis
Citation
Simonetti S, Mutemi KN, Romano P, Luckenbach T, Zupo V, Gambi MC and Corsi I (2025) Hydrothermal vents as observatories for future ocean acidification (OA) scenarios: an in-situ study to unravel the involvement of ATP binding cassette transporters in the adaptation of marine polychaetes Platynereis spp. to OA. Front. Mar. Sci. 12:1573367. doi: 10.3389/fmars.2025.1573367
Received
08 February 2025
Accepted
18 April 2025
Published
08 May 2025
Volume
12 - 2025
Edited by
Nathalie Oulhen, Brown University, United States
Reviewed by
Paul Yancey, Whitman College, United States
Wanchun Guan, Wenzhou Medical University, China
Updates
Copyright
© 2025 Simonetti, Mutemi, Romano, Luckenbach, Zupo, Gambi and Corsi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia Simonetti, silvia.simonetti2@unisi.it
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.