Abstract
Background:
Settlement and metamorphosis are crucial developmental events in the life cycles of most marine benthic invertebrates. Neuropeptides play an important role in inducing larval settlement. However, studies on the ciliary genes and the few studies have examined the pathways regulated by neuropeptides during larval settlement.
Methods:
Here, we employed multiple molecular biology techniques to identify a ciliary gene, intraflagellar transport protein 20 (IFT20) which played a significant role in the larval settlement of Urechis unicinctus (Annelida, Echiura), and revealed the related gene pathway of IFT20 expression regulated by FILa1, a neuropeptide unique to U. unicinctus.
Results:
The IFT20 protein of U. unicinctus was highly conserved with that of other animals and localized at the ciliary base in the trochophore and the segmentation larvae of U. unicinctus. It was determined that the neuropeptide FILa1 inhibits IFT20 expression through the cAPM-PKA-CREB and Ca2+ pathways, which in turn triggers larval settlement. Knockdown the mRNA level of IFT20 resulted in a reduction in the number of vesicles at the ciliary base, the β-tubulin protein synthesis, the number and length of cilia, and the beat frequency of the circumoral cilia. Finally, the settlement rate of the larvae in the IFT20-dsRNA group increased by 3.64 times and 2.88 times compared to the control group at 48 and 72 h, respectively.
Conclusion:
Our findings provide new insights into the function of IFT20 and the signaling pathways through which neuropeptides regulate ciliary gene expression during larval settlement.
1 Introduction
Most marine benthic invertebrates, such as mollusks, annelids, and crustaceans, undergo a planktonic larval stage in their life cycle that lives in the upper layer of the water (Toonen and Pawlik, 1994). These planktic larvae eventually settle on sediments, rocks, or other surfaces and metamorphosize into benthic living juveniles (Hadfield et al., 2001; Marshall et al., 2012). The transition process of larvae from a planktic to a benthic lifestyle is a critical stage in their life cycle, essential for maintaining population numbers and community dynamics (Shikuma et al., 2014; Doll et al., 2022). During the process, cilia-mediated cessation of swimming marks the beginning of larval settlement (Maldonado and Young, 1996; Walters et al., 1999; Hadfield and Koehl, 2004). Neuropeptides are a class of biologically active molecules composed of amino acids, usually consisting of 3 to 80 amino acid residues (Williams et al., 2015). They play an important role in signal transduction in the nervous system and can affect the activity of neurons and their communication with each other (Bao et al., 2020). Neuropeptides are widely involved in the settlement process of marine benthic invertebrate larvae (Conzelmann et al., 2011, 2013; Lu et al., 2022). It has been reported that ciliary activity is regulated by neuropeptides to induce the larval settlement. For instance, in the annelid Platynereis dumerilii and the nemertean Lineus longissimus, the ciliary beat frequency is reduced by neuropeptides, thereby promoting larval settlement behavior (Conzelmann et al., 2011, 2013; Thiel et al., 2019). However, little is known about which signaling pathways and ciliary-related genes regulated by neuropeptides are involved in larval settlement and their specific roles in this process.
Intraflagellar transport (IFT) is a crucial process in the assembly and functional maintenance of cilia and flagella (Wang et al., 2021; Rosenbaum and Witman, 2002; Taschner and Lorentzen, 2016; Ishikawa and Marshall, 2017). IFT proteins consist of IFT-A and IFT-B complexes, with IFT-A comprising six proteins and IFT-B comprising sixteen proteins (Pedersen and Rosenbaum, 2008; Nakayama and Katoh, 2020). These protein complexes are essential for cilia assembly and maintenance, facilitating bidirectional intracellular transport (Jiang et al., 2023). Among these proteins, IFT20 (intraflagellar transport protein 20) has emerged as a significant player in ciliary assembly and function (Taschner et al., 2012; Su, 2020; Zhou et al., 2020; Finetti et al., 2021, 2022; Jin et al., 2022). As anchored to the Golgi apparatus by GMAP210 (Golgi microtubule-associated protein 210) (Follit et al., 2008), IFT20 facilitates the encapsulation of cilia-related materials (ciliary membrane proteins, axonemal proteins, signaling molecules, and ciliary modification proteins) from the Golgi apparatus (Keady et al., 2011; Sung and Leroux, 2013; Rezi et al., 2024), and then these vesicles encapsulating the ciliary materials are transported to the base of the cilium by IFT20 under the mediation of GM130 (Golgi matrix protein 130), aiding in the assembly and maintenance of ciliary structure and function (Roboti et al., 2015; Stoetzel et al., 2016; Lee et al., 2018). Currently, research on IFT20 mainly focuses on the cilia of internal organs or sperm flagellum in several animals, such as human (Yang et al., 2021; Li et al., 2022; Qiu et al., 2023), mouse (Yamaguchi et al., 2020; Zhang et al., 2020b; Kretschmer et al., 2023; Rezi et al., 2024), and nematode (De-Castro et al., 2021). However, studies on IFT20 related to the body surface cilia of marine animals remain a significant lack, particularly revolved in their movement and development.
The echiuran worm Urechis unicinctus, as the representative species of Echiurida (Xenopneusta, Urechidae), has a typical larval settlement process during its life cycle (Zheng et al., 2022). In our previous study, several neuropeptides (MIP, FILa, FxFa, and FRWa) have been identified to be involved in the larval settlement of U. unicinctus (Hou et al., 2020), and the expression of ciliary gene Tctex1d2 has also been shown to be regulated by the neuropeptide MIP2, thereby promoting larval settlement (Yang et al., 2024). To further elucidate the functions of neuropeptide-regulated ciliary genes in the larval settlement of U. unicinctus. Here, we identified that FILa1 significantly downregulated IFT20 expression (P < 0.05) to induce the larval settlement in U. unicinctus. Furthermore, the signaling pathways mediating FILa1 to regulate IFT20 expression were also verified. Finally, we elucidated the crucial role of IFT20 in maintaining the ciliary length and number and found that its knockdown significantly increased the larval settlement rate (3.64 times higher than the control group). These findings not only enhance our understanding of the ciliary genes involved in the larval settlement of marine benthic invertebrates but also provide broader insights into the molecular mechanisms underlying neuropeptide-mediated ciliary regulation across diverse taxa. Given the fundamental role of cilia in cell signaling and motility, these results may have implications beyond marine invertebrates, potentially contributing to research in developmental biology, evolutionary conservation of ciliary pathways.
2 Materials and methods
2.1 Animal and sample preparation
U. unicinctus adults were collected from the intertidal zone in Yantai, China. The procedures for sperm and oocyte collection, artificial fertilization, and larval rearing followed the methods outlined by Wei et al. (2022). The embryo incubation temperature was 17 ± 1°C, pH = 7.8 ± 0.06, and salinity was 28 ± 1. The larvae (early-trochophore larvae, mid-trochophore larvae, early-segmentation larvae, segmentation larvae, and worm-shaped larvae) (Supplementary Figure 1) were obtained from the tanks using a sieve, and the larvae were collected and fixed after brief centrifugation to remove water according to the methods of Bai et al. (2022). The early-segmentation larvae were frozen with liquid nitrogen, and then stored at -80°C.
2.2 Sequence analysis of U. unicinctus IFT20
The cDNA sequence of U. unicinctus IFT20 was obtained from the U. unicinctus genome and verified using the specific primers (Table 1). To validate the cDNA sequence, total RNA was extracted from early-segmentation larvae using TRIzol reagent (Yeasen, Shanghai, China), and reverse transcription to cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Yeasen, Shanghai, China). The conserved domains of IFT20 were identified using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/?term=). The three-dimensional model of IFT20 was constructed using SWISS-MODEL (http://swissmodel.expasy.org). A Neighbor-Joining phylogenetic tree was constructed using MEGA 13.4 software with bootstrap trial 1000 replicates based on the deduced amino acid sequences of IFT20 among the different species.
Table 1
| Primer name | Sequence (5’→3’) | Purpose |
|---|---|---|
| IFT20-F | ATGGCAGACGAAGCTCTC | Sequence amplification |
| IFT20-R | CTTGTTCCTCCTTGTTCAGT | |
| q-IFT20-F | AGTTAGTTGATGGTGTCTCC | qRT-PCR |
| q-IFT20-R | TGTTCTTGTTCCTCCTTGTT | |
| q-CREB-F | AAGGTGGAGGAGGATTCTAA | |
| q-CREB-R | CCGTGTCATTGGTCATAGT | |
| q-ATPS-F | GGATTGTGGAGCACTCGTTTG | |
| q-ATPS-F | CCAGCGGTTTCAGGTATTTC | |
| p-IFT20-F | CCCTCGAGGGGACTCGCCACCCAGGCGA | Plasmid construction |
| p-IFT20-R | CCAAGCTTGGTCCTTGCATTCTTCTTTTAGTTCAC | |
| ORF-CREB-F | GGTACCCGATGGTAGCATCTGTCGGGTCA | |
| ORF-CREB-R | CCTCGAGGCTAACTCTGAGCTTCCTTTTGAC | |
| d-IFT20-S | TAATACGACTCACTATAGGGAGACAATGGCAGACGAAGCTCTC | dsRNA synthesis |
| d-IFT20-A | TAATACGACTCACTATAGGGAGACACTTGTTCCTCCTTGTTCAGT | |
| d-EGFP-S | TAATACGACTCACTATAGGGAGACAAGGGCGAGGGCGATGCCACCTACGG | |
| d-EGFP-A | TAATACGACTCACTATAGGGAGACAAAGTTCACCTTGATGCCGTTC |
Primers used in this study.
The double underlines indicate restriction endonuclease sites are underlined, and the single underlines indicate T7 promoter sequences are underlined.
The IFT20 promoter sequence was obtained from U. unicinctus genome database. The genome DNA was isolated from the body wall of adult U. unicinctus using the TIANamp® Genomic DNA Kit (Tiangen Biotech, Beijing, China). PCR amplification was then performed to verify the IFT20 promoter sequence using the specific primers (Table 1). The transcription start site and upstream promoter elements were predicted using the FPROM software (http://www.softberry.com/berry.phtml?topic=fprom&group=programs&subgroup=promoter) and the Berkeley Drosophila Genome Project tool (http://www.fruitfly.org/seq_tools/promoter.html). The online tool JASPAR (http://jaspar.genereg.net/) was used to predict the binding sites of the transcription factor CREB (cAMP-response element binding protein) on IFT20 promoter.
2.3 Expression analysis of IFT20 in embryos and larvae at different development stage and adult tissues
To clarify the expression of IFT20 in U. unicinctus embryos, larvae, and various adult tissues, transcriptome data (Accession numbers: PRJNA485379, PRJNA394029, and PRJNA917787) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov). TPM (Transcripts per million) values of IFT20 were obtained using the R package and used as their expression abundance. All the data of various samples were three biological replicates.
2.4 Treatment of U. unicinctus larvae with mature peptide FILa1
The active mature peptide FILa1 was synthesized by Sangon Biotechnology (Shanghai, China) through amidation of carboxyl terminal in the last amino acid of its sequence (SLRLNDFIL). According to the method established by Lu et al. (2022), the early-segmentation larvae swimming in the upper layer water were collected and transferred to glass tubes filled with solution. The solution in treatment group was the filtered seawater (FSW) with 10 μM FILa1 mature peptide, while that of the control group was the FSW without FILa1. Each tube contained 200 larvae, and there were 3 replicates for each group. The larvae were respectively collected at 1 min, 3 min, and 5 min of the treatment, and then frozen and stored according to the methods described in section 2.1. Furthermore, early-segmentation larvae respectively treated with three concentrations of the FILa1 (5, 10, 15 μM) was performed using the above experiment system and were sampled and stored following the procedure above.
2.5 qRT-PCR
Total RNA was isolated from the treated larvae using TRIzol reagent (Yeasen, Shanghai, China) according to the manufacturer’s instructions. The first-strand cDNA was synthesized using the Hifair® AdvanceFast 1st Strand cDNA Synthesis Kit with gDNA Eraser (Yeasen, Shanghai, China). The specific primers (Table 1) were designed according to the target gene sequences, and the ATPase gene was employed as an internal reference. The qPCR-PCR was performed based on the method of Zhang et al. (2020a) using Hieff UNICON® Universal Blue SYBR Green Master Mix (Yeasen, Shanghai, China) on a qTOWER3G Real-Time PCR Thermal Cycler (Analytikjena, Jena, Germany). The relative expression level of target mRNA was calculated using the 2-ΔΔCt method.
2.6 Immunofluorescence
According to the protocol outlined by Lu et al. (2022), the larvae samples were rehydrated with a gradient methanol (100%, 75%, 50%, and 25%), treated with 3% bovine serum albumin diluted in PBT (pH 7.4). Subsequently, the samples were incubated overnight on a shaker at 4°C with the primary antibody, β-Tubulin antibody (Solarbio, Beijing, China) or IFT20 antibody (Solarbio, Beijing, China) diluted 1:200 in PBT, and then incubated for 2 h at room temperature in dark with the secondary antibody, donkey anti-rabbit Alexa Fluor 488 (Yeasen, Shanghai, China) or anti-mouse Alexa Fluor 564 (Yeasen, Shanghai, China) diluted 1:300 in PBT. Finally, the larvae were incubated in the dark with 2.5% DAPI (Solarbio, Beijing, China) for 15 min to label the nuclei. The antibody specificity was assessed using the pre-immune serum. All samples were observed using an Olympus BX43 microscope (Olympus, Tokyo, Japan). Images and final panels were prepared using ImageJ.
2.7 Dual-luciferase reporter assay
To verify the binding of transcription factor CREB to IF20 gene, the full-length CDS sequence of CREB was amplified using a pair of primers (Table 1) and cloned into the pcDNA3.1 (+) expression vector. The promoter sequence of IFT20 from U. unicinctus genome database was amplified and subcloned into the pGL4.17 reporter vector. HEK293T cells cultured in high-glucose medium (Servicebio, Wuhan, China) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NE, USA) at 37°C in 5% CO2, were used for the transfection assay. Two co-transfection groups were set: in group 1, the recombinant pGL4.17-IFT20-Luc vector (450 ng) was co-transfected with the pcDNA3.1 (+) (200 ng) and the Renilla luciferase reporter vector (20 ng); in group 2, the recombinant pGL4.17-IFT20-Luc vector (450 ng) was co-transfected with the pcDNA3.1-CREB (200 ng) and Renilla luciferase reporter vector (20 ng). The constructed vectors were transiently co-transfected into the HEK293T cells using Lipofectamine® 3000 transfection reagent (Thermo Fisher Scientific, Wilmington, NC, USA). After 48 h of the co-transfection, luciferase activity was assayed using the Dual-Luciferase® Reporter Assay System (Beyotime, Shanghai, China).
2.8 Analysis of signaling pathways mediating FILa1 to regulate IFT20 expression and trigger larval settlement
Most neuropeptides regulate down-stream gene expressions by GPCR (G protein-coupled receptors) activating the different signaling pathways (Caers et al., 2012). To clarify the signaling pathways mediating FILa1 to regulate IFT20 expression, the inhibitors/activators of cAMP-PKA and Ca2+ pathways were obtained from MedChemExpres (Princeton, USA), including the forskolin (AC activator, a diterpene compound that directly activates AC, leading to increased intracellular cAMP levels), Br-cAMP (cAMP analog, a membrane-permeable analog of cAMP that mimics endogenous cAMP signaling), H89 (PKA inhibitor, a selective inhibitor of PKA that competes with ATP for binding to the catalytic subunit), 2-APB (IP3R inhibitor, a non-specific inhibitor of IP3R that blocks intracellular calcium release from the endoplasmic reticulum), calmidazolium (calmodulin antagonist, a calmodulin (CaM) antagonist that inhibits CaM-dependent signaling by binding to its active sites), and KN-62 (CaM-II inhibitor, a specific inhibitor of calcium/CaMK-II that prevents calmodulin binding, thereby blocking its activation). The experiment was conducted in 6-well plates, and each well was added 200 early-segmentation larvae. A total of 4 groups were set, including the FILa1 treatment group, the control group, the inhibitors/activators treatment group, and the inhibitors/activators + FILa1 treatment group, and each group was set three replicated wells. In the FILa1 treated group, the larvae were treated with filtered seawater (FSW) containing the mature peptide FILa1 with a final concentration of 10 μM for 48 h. In the inhibitors/activators treated group, the larvae were treated with FSW containing the forskolin (3 μM), Br-cAMP (3 μM), H89 (0.5 μM), 2-APB (3 μM), calmidazolium (0.5 μM), or KN-62 (3 μM) for 48 h, respectively. In the inhibitors/activators + FILa1 treated group, the larvae were first treated for 1 h respectively with above inhibitors/activators, and then the mature peptide FILa1 with a final concentration of 10 μM was added to the experimental system for a further co-incubation. The control larvae were treated with only FSW without inhibitors/activators or mature peptide FILa1. The number of settled larvae was recorded after the treatment for 48 h. The settled larvae were defined as those adhering to the bottom of the plates, and unable to swim into the seawater layer. The settlement rate of larvae was calculated using the formula: (number of settled larvae/total number of larvae) × 100%. In addition, the larvae in the 4 groups after the experiment were respectively collected, and then their total RNAs were extracted, reverse transcribed according to the methods described in section 2.5. The mRNA levels of IFT20 were assessed using qRT-PCR method.
2.9 RNAi
The cDNA fragments of IFT20 and EGFP (Enhanced green fluorescent protein gene) were generated using the primers (Table 1) labeled with T7 sequences for dsRNA synthesis. The quality of the PCR products was confirmed by 1% agarose gel electrophoresis, and then the products were purified using the MolPure® PCR Purification Kit (Yeasen, Shanghai, China). The IFT20-dsRNA and EGFP-dsRNA (used as a negative control) were synthesized using the MEGAscript® RNAi Kit (Invitrogen, Waltham, MA, USA) following the manufacturer’s protocol. The quality and quantity of dsRNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1% agarose gel electrophoresis, and then the dsRNA was stored at -30°C until use.
Three groups, the IFT20-dsRNA group, EGFP-dsRNA group, and blank control group were set, and each group consisted of three replicates. The early-segmentation larvae were employed, and about 170 larvae were randomly assigned to each replicate. The larvae in the IFT20-dsRNA and EGFP-dsRNA groups were first soaked in 3 mL FSW with dsRNA (final dsRNA concentration of 50 nmol/L) for 12 h, then transferred to glass tubes containing 50 mL FSW for a further 60 h. While the larvae in the blank control group were treated only with 3 mL FSW, and all other conditions were identical to those of the dsRNA treatment groups. At the end of the treatment, the settlement rate of larvae was recorded as described in section 2.8. To assess the efficiency of gene knockdown, larvae in each group were collected at 72 h post dsRNA treatment. The procedures of sampling, RNA extraction, cDNA synthesis, and qRT-PCR were performed as described above. The primers used for qRT-PCR were shown in Table 1.
2.10 Swimming speed measurements
Videos of larval movement were recorded for both the RNAi and control groups, which were imported into ImageJ software to track the larvae’s movement trajectories using the Animal Tracker plugin. The distance traveled by the larvae in each frame was calculated using the formula S (distance) = √[(x1-x2)²+(y1-y2)²], where x and y represented the horizontal and vertical coordinates of the larvae’s position, respectively. The total distance traveled by the larvae across all frames was summed, and then divided by the swimming time to obtain the larvae’s swimming speed. Twenty-five larvae per group were used for statistical analysis.
2.11 Circumoral cilia beat frequency analysis
Following the method described by Thiel et al. (2019), single larva in the RNAi and control groups were randomly collected and transferred onto a glass slide to record the beat frequency of the circumoral cilia (a band of cilia encircling the larval body, which are the primary structures responsible for larval movement and locomotion) using a Leica DM2500 LED microscope (Leica, Weztlar, Germany). The recorded videos were imported into ImageJ software, and the Plot Z-axis Profile function was used to determine the number of frames required for one complete cycle of ciliary movement. The frequency of ciliary movement was calculated by dividing the total number of frames in the video by the number of frames required for one cycle of movement. Statistical analysis was performed using 25 larvae per group.
2.12 Sample preparation and scanning electron microscopy observation
The larvae of control and RNAi groups were fixed in 2.5% glutaraldehyde and 1% osmium tetroxide solution, respectively according to the usual method. Subsequently they were dehydrated through a series of ethanol and isopropyl acetate solution, and dried using a critical point dryer (K850, Quorum Technologies Ltd., Laughton, UK). Finally, the samples were sputter-coated with gold, and then observed and photographed using a Hitachi TM 4000 scanning electron microscope (Hitachi, Tokyo, Japan).
2.13 Sample preparation and transmission electron microscopy observation
The larvae of control and RNAi groups were fixed and dehydrated using the SEM method described above, and then transferred to pure acetone for 20 min. Next, the samples were immersed in a mixture of epoxy resin and acetone (1:1, v/v) for 1 h, followed by treatment with a mixture of epoxy resin and acetone (3:1, v/v) for 3 h, and then left in epoxy resin overnight at room temperature. The embedded samples were sectioned into 80 nm thick slices using an ultramicrotome (Leica Microsystems, Wetzlar, Germany). The sections were first stained with uranyl acetate for 30 min, and then stained with lead citrate for 15 min, and placed on copper grids. Finally, observations and imaging were performed using a Hitachi HT-7800 transmission electron microscope (Hitachi, Tokyo, Japan).
2.14 Statistical analysis
All data were presented as mean ± SEM. Significant differences among different groups were determined using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test. The independent samples t-test was used to determine significant differences between the control group and treatment group. All statistical analyses were performed using the SPSS v22 software (SPSS Inc., Chicago, IL, USA). Statistical significance was set at P < 0.05.
3 Results
3.1 Sequence characterization of U. unicinctus IFT20
The open reading frame of U. unicinctus IFT20 was 399 bp, encoding 133 amino acids (Figure 1A). Multiple sequence alignment results indicated that the IFT20 sequence was highly conserved across various species, with an alignment rate of 85.61% compared with Capitella teleta (Figure 1A). It was identified that U. unicinctus IFT20 was most closely related to that of annelids, then clustered with mollusks, and finally groups with other animals, being consistent with traditional evolutionary relationships (Figure 1B). A three-dimensional structural model of IFT20 was constructed to provide a visual representation (Figure 1C). Like other species, the deduced protein sequence of U. unicinctus IFT20 exhibited the presence of an IFT20 domain (Figure 1D).
Figure 1
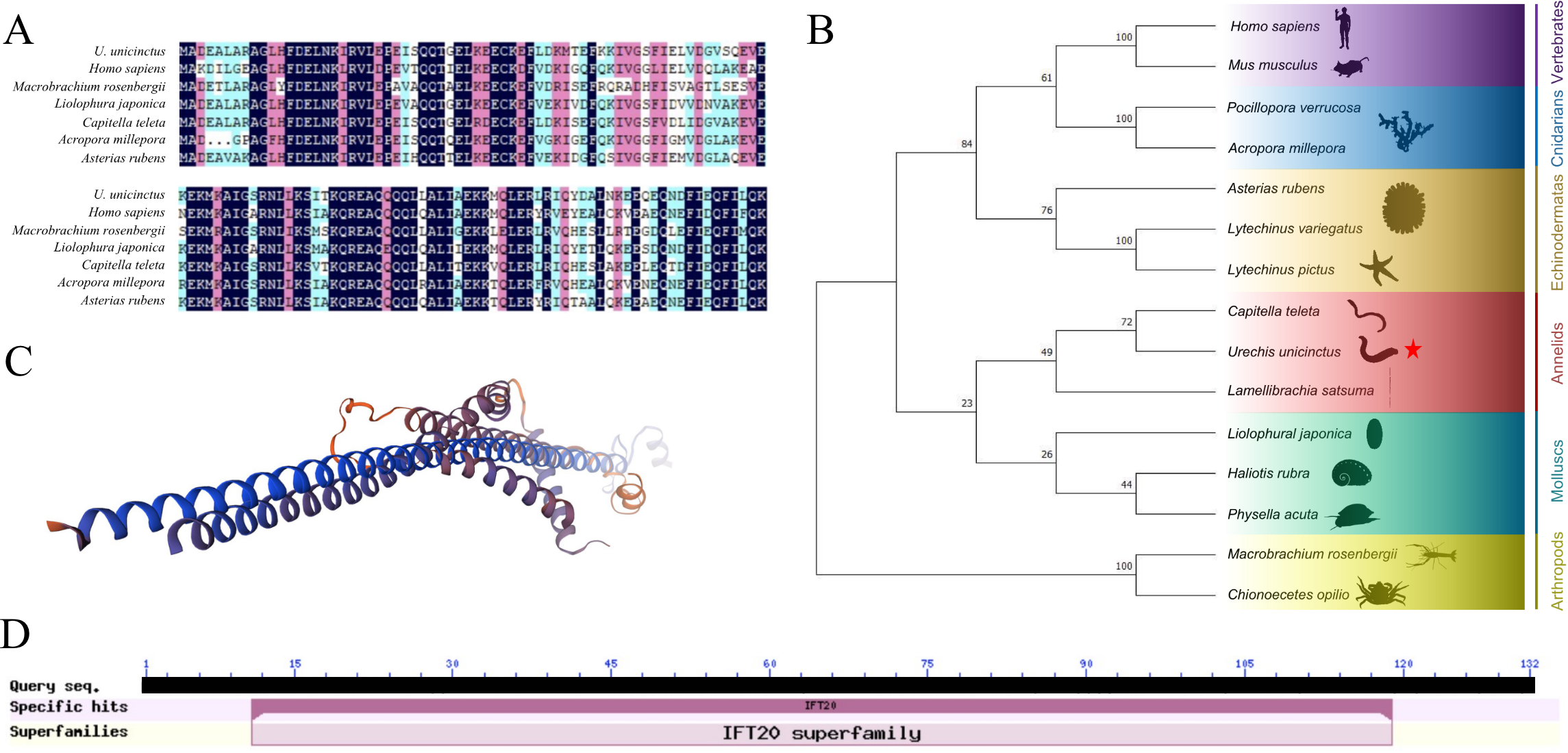
The structure and phylogenetic analysis of the IFT20. (A) Multiple alignment of the deduced amino acid sequences of IFT20 with other species; (B) Phylogenetic analysis using Neighbor-Joining method with 1000 bootstrap, the U. unicinctus IFT20 is marked with a red star on the phylogenetic tree; (C) Three-dimensional structure of U. unicicntus IFT20; (D) Conserved domain of U. unicinctusIFT20.
3.2 The mRNA expression and protein localization characteristics of the IFT20 gene
Based on the transcription data from U. unicinctus embryos and larvae, abundance of the IFT20 mRNA increased progressively during the development from 2-8 cell embryos (E) to early-trochophores (ET) which reached a peak of 7.92 in the ET, and then gradually decreased from mid-trochophores (MT) to worm-shaped larvae (WL) (Figure 2A). Based on the transcription data from the adult tissues, the IFT20 expression level was the highest in the gut, while the lowest in the coelomic fluid (Figure 2B). Immunofluorescence results showed that the localization of IFT20 was basically the same as that of β-tubulin, the microtubule marker protein, mainly localized in the circumoral cilia and the posterior cilia of the trochophore and the segmentation larvae (Figure 2C). In addition, it was also identified in the apical region and/or the posterior region of the larvae, although the localization of IFT20 in these regions varies among the larvae at various developmental stages. Furthermore, co-localization with the β-tubulin protein showed that IFT20 was localized at the base of the circumoral cilia in trochophore larvae and segmentation larvae, as well as at the base of posterior cilia in segmentation larvae (Figure 2C).
Figure 2
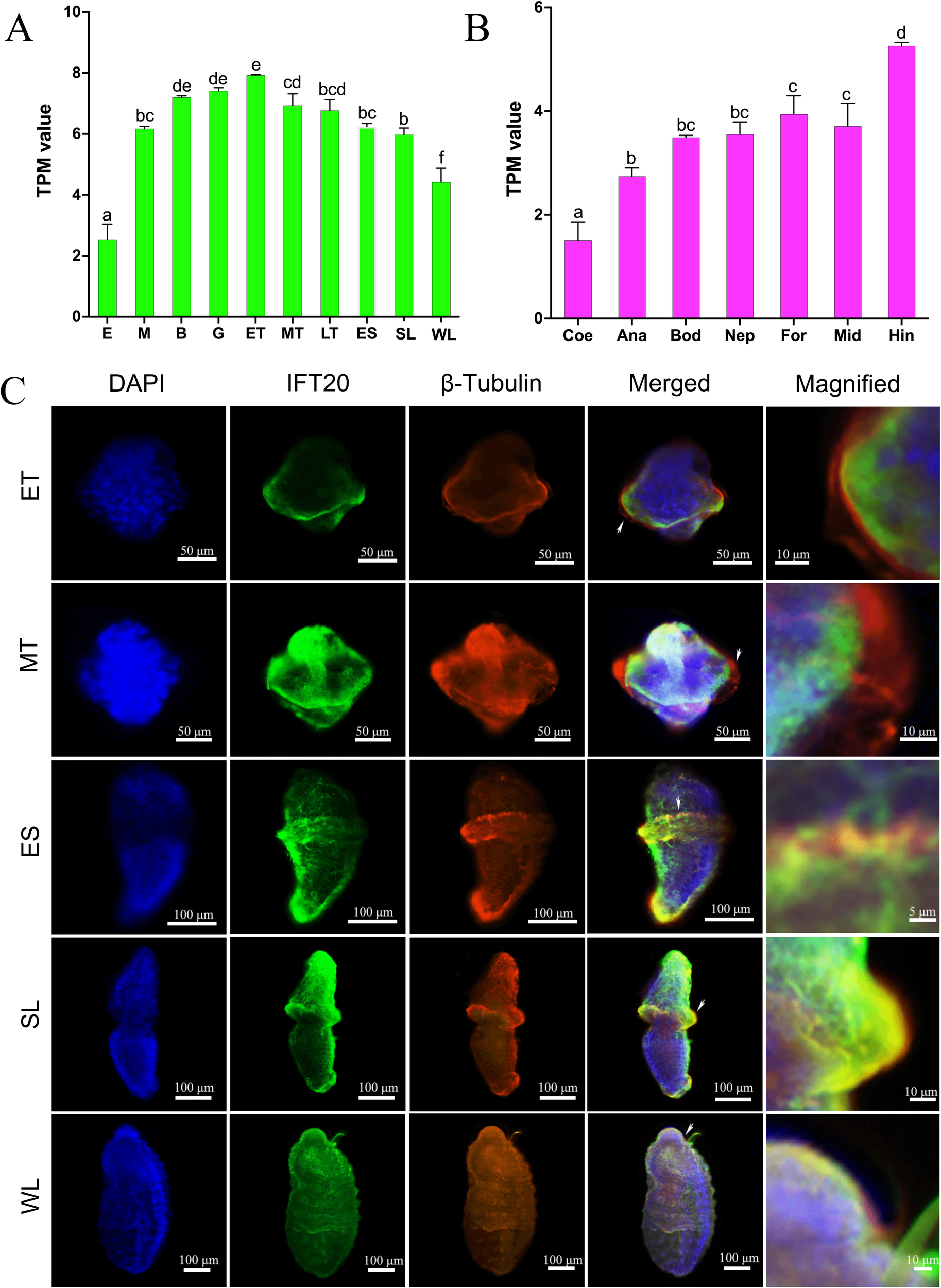
The expression and localization of the IFT20 in U. unicinctus. (A) The mRNA levels of IFT20 in U. unicinctus embryos and larvae at the different stages; (B) The mRNA levels of IFT20 in adult tissues; (C) Localizations of IFT20 and β-tubulin in U. unicinctus larvae at the different stages. E, 2-8 cell embryos; M, Multicellular embryos; B, Blastulae; G, Gastrulae; ET, Early-trochophores; MT, Mid-trochophores; LT, Late-trochophores; ES, Early-segmentation larvae; SL, Segmentation larvae; WL, Worm-shaped larvae; Bod, Body wall; Coe, Coelomic fluid; For, Fore-gut; Mid, Mid-gut; Hin, Hind-gut; Ana, Anal sac; Nep, Nephridium. Different letters indicate a significant difference at P < 0.05 in (A, B) pictures. White arrows indicate the magnified part in picture C.
3.3 The FILa1 decreased IFT20 and CREB expressions as well as IFT20 transcription regulated by CREB
The expression of U. unicinctus IFT20 in early-segmentation larvae was assessed at various time points following treated with the mature peptide FILa1. qRT-PCR results demonstrated a time-dependent downregulation of IFT20 expression in response to FILa1 induction (Figure 3A). Meanwhile, the expression levels of IFT20 decreased significantly in the larvae of the FILa1 treatment groups with the different doses than that of the control group, although no significant differences were observed between the FILa1 treatment groups (Figure 3B). Furthermore, we detected that the level of the CREB mRNA was also significantly downregulated in a time-dependent manner after the FILa1 treatment (Figure 3C). To identify the transcription factor CREB could regulate the expression of IFT20, we predicted five CREB binding sites including an 8-base palindrome sequence (5’-TGACGTCA-3’) in the promoter region of IFT20 (Figure 3D), which was recognized as a conserved CREB-binding motif. The dual-luciferase reporter assay confirmed that the transcription factor CREB can bind to the promoter sequence of IFT20 (Figure 3E). These findings suggested that the downregulation of IFT20 expression by FILa1 may be mediated by the transcription factor CREB in cAMP-PKA-CREB signaling pathway.
Figure 3
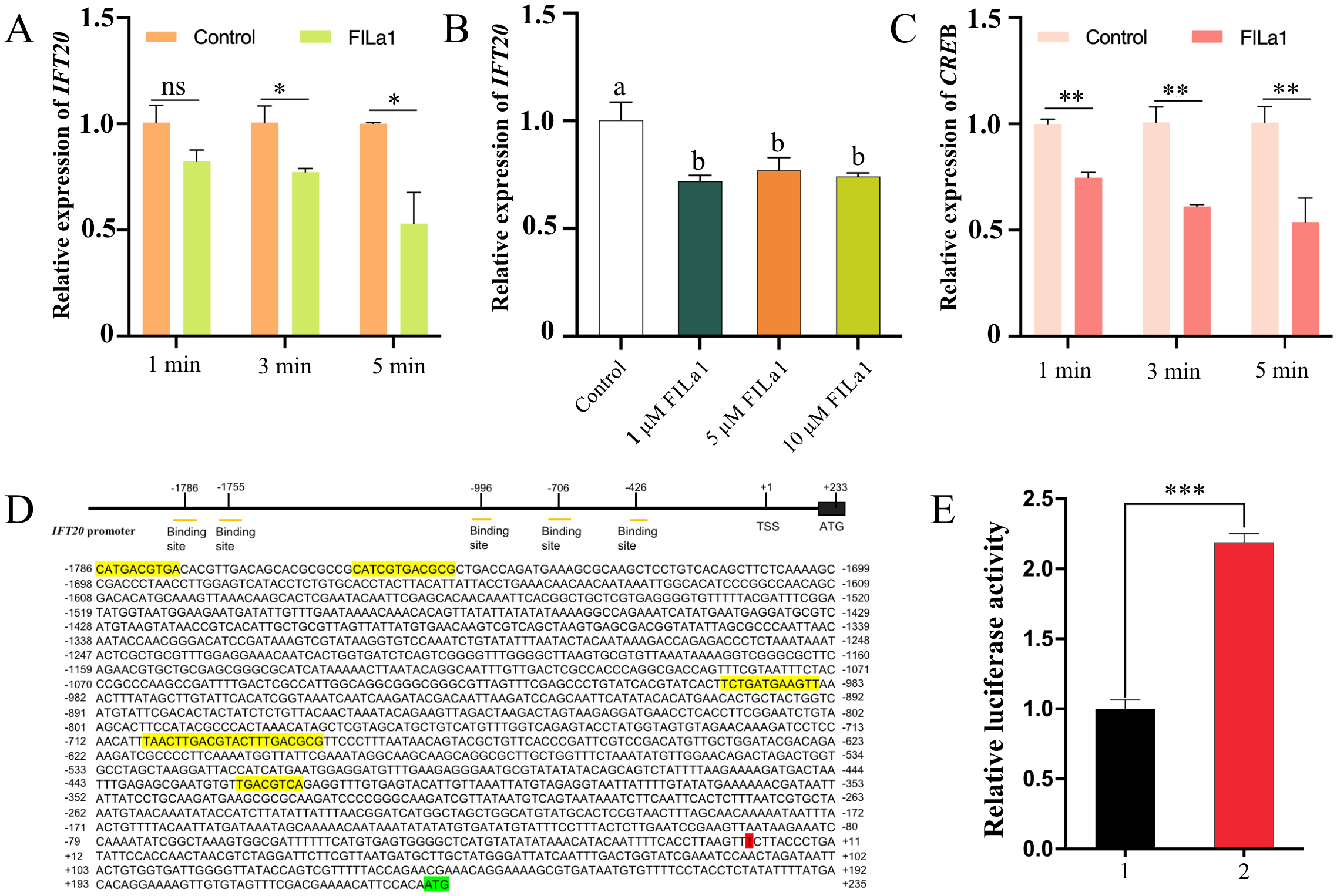
The effect of FILa1 on IFT20 and CREB mRNA levels as well as IFT20 transcriptional regulation by CREB. (A) The IFT20 expressions in the early-segmentation larvae treated by 10 µM FILa1 at the different treatment times; (B) The IFT20 expressions in the early-segmentation larvae treated by FILa1 with the different concentrations at 3 min; (C) The CREB expressions in the early-segmentation larvae treated by 10 µM FILa1 at the different times; (D) Upstream elements in the promoter region of IFT20 gene and the predicted binding sites for CREB; (E) The IFT20 transcription regulation in the HEK293T cells transfected transiently with the recombinant vectors. The expression levels of IFT20 and CREB in larvae of control groups in pictures (A–C) are set as 1.0 to calibrate the relative expression levels. The yellow shadow sequence in the picture (D) refers to the predicted IFT20 promoter sequence bound by the transcription factor CREB; the red shaded base is the transcription start site; the green shaded sequence is the start codon. 1: pcDNA3.1(+) + pGL4.17-IFT20-Luc; 2: pcDNA3.1-CREB + pGL4.17-IFT20-Luc. The significant difference is marked with * at P < 0.05, ** at P < 0.01, and *** at P < 0.001, and “ns” means no significant difference. Different letters indicate a significant difference at P < 0.05.
3.4 Identification of signal pathways mediating FILa1 to down-regulate the IFT20 expression
To further elucidate signal transduction mediating the FILa1 downregulating the expression of IFT20 to promote larval settlement, potential signaling pathways were examined. For the cAMP/PKA pathway, the expression level of IFT20 in the larvae co-treated with AC activator (forskolin) or cAMP analog (Br-cAMP) and FILa1 was significantly higher than that in the larvae treated with FILa1 alone (Figures 4A, B), and the settlement rates of the larvae in the co-treatment groups were significantly lower than that in the group treated with FILa1 alone (Figures 4A’, B’). Meanwhile, the expression level of IFT20 in the larvae co-treated with PKA inhibitor (H89) and FILa1 was not significantly different from that in the larvae treated with FILa1 alone (Figure 4C), but the settlement rate of larvae in the co-treated group was significantly higher than that in the group treated with FILa1 alone (Figure 4C’). For the Ca2+ pathway, the similar results to co-treatment of FILa1 with the AC activator (forskolin) or cAMP analog (Br-cAMP) occurred when the larvae were treated with the IP3 receptor blocker (2-APB) or the CaM antagonist (calmidazolium) (Figures 4D, E, D’, E’). The expression level of IFT20 in the larvae co-treatment with CaMK-II blocker (KN-62) and FILa1 was lower than that in the larvae treated with FILa1 alone (Figure 4F), and the settlement rate of the larvae in the co-treatment group was lower than that in the group treated with FILa1 alone (Figure 4F’).
Figure 4
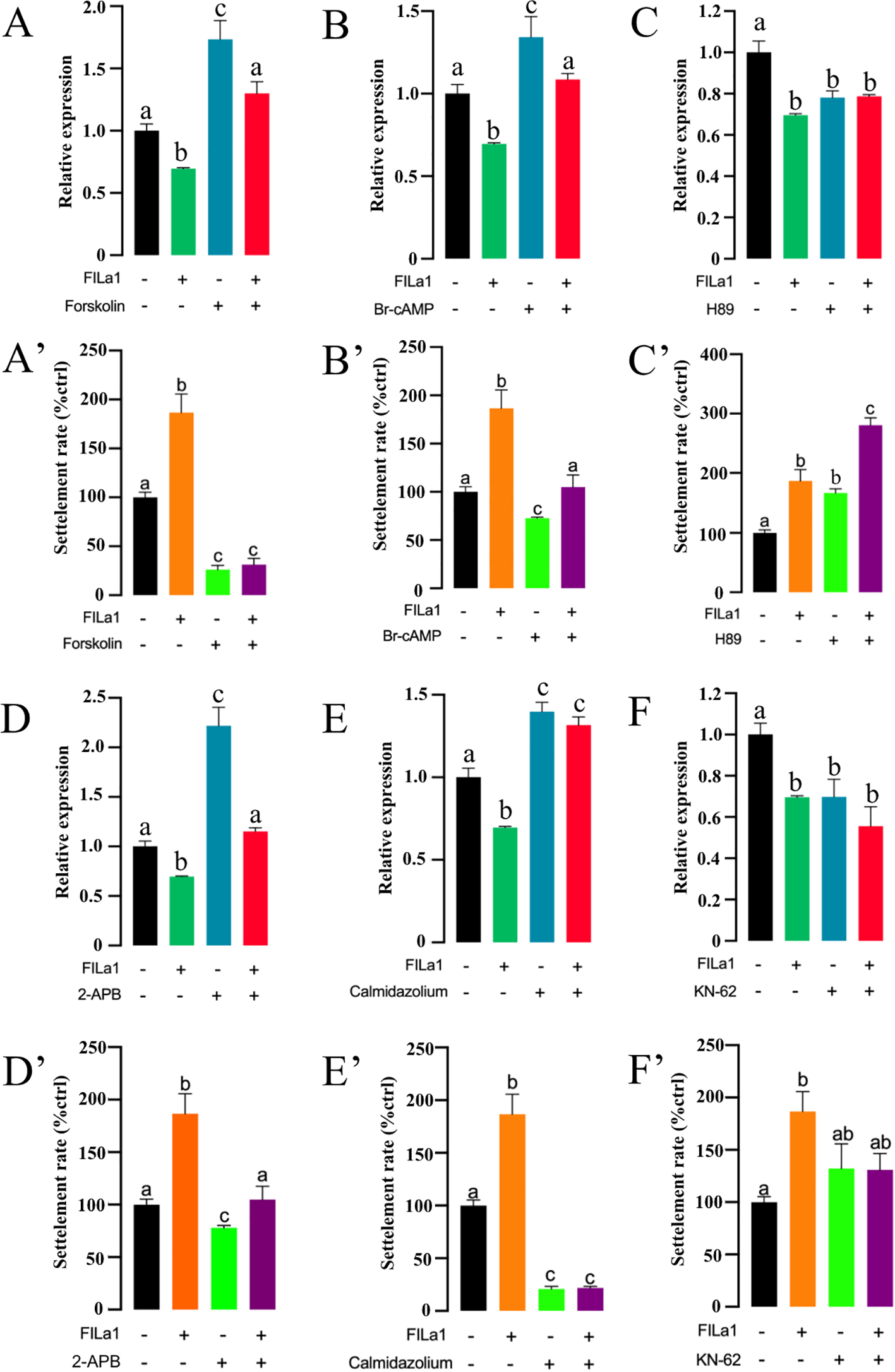
Signal transduction mediating FILa1 to regulate the IFT20 expression and the larval settlement. (A–F) The IFT20 expression in the larvae of the different groups; (A’–F’) The larval settlement rate in the different groups. Different letters indicate a significant difference at P < 0.05.
3.5 Effect of knocking down the IFT20 expression on the ciliary structure and settlement of U. unicinctus larvae
qRT-PCR detection showed that the IFT20 mRNA level in the IFT20-dsRNA group decreased significantly (P < 0.001), which was 58.37% of that in the control group at 72 h post RNAi (Figure 5A). In the IFT20-dsRNA group, a reduced swimming range of the larvae was first observed at 24 h after the RNAi (Figure 5B, Supplementary Movie S1), and the larval swimming speed significantly decreased (P < 0.001) which was 62.38% of the control group (Figure 5C). After RNAi for 72 h, the beat frequency of circumoral cilia in the larvae of IFT20-dsRNA group decreased significantly to 77.36% of that in the larvae of the control group (Figure 5D), and the ciliary length of the circumoral cilia in the larvae was also significantly reduced to 76.12% of the control group (Figures 5E, F). Moreover, the reduced positive signals of the ciliary marker protein β-tubulin were observed in IFT20-interfered larvae (Figure 5G). Furthermore, the larval settlement rates in the IFT20-dsRNA group at 48 h and 72 h were significantly higher than those of the control group, being 3.46 times (P < 0.001) and 2.88 times (P < 0.001) higher, respectively (Figure 5H).
Figure 5
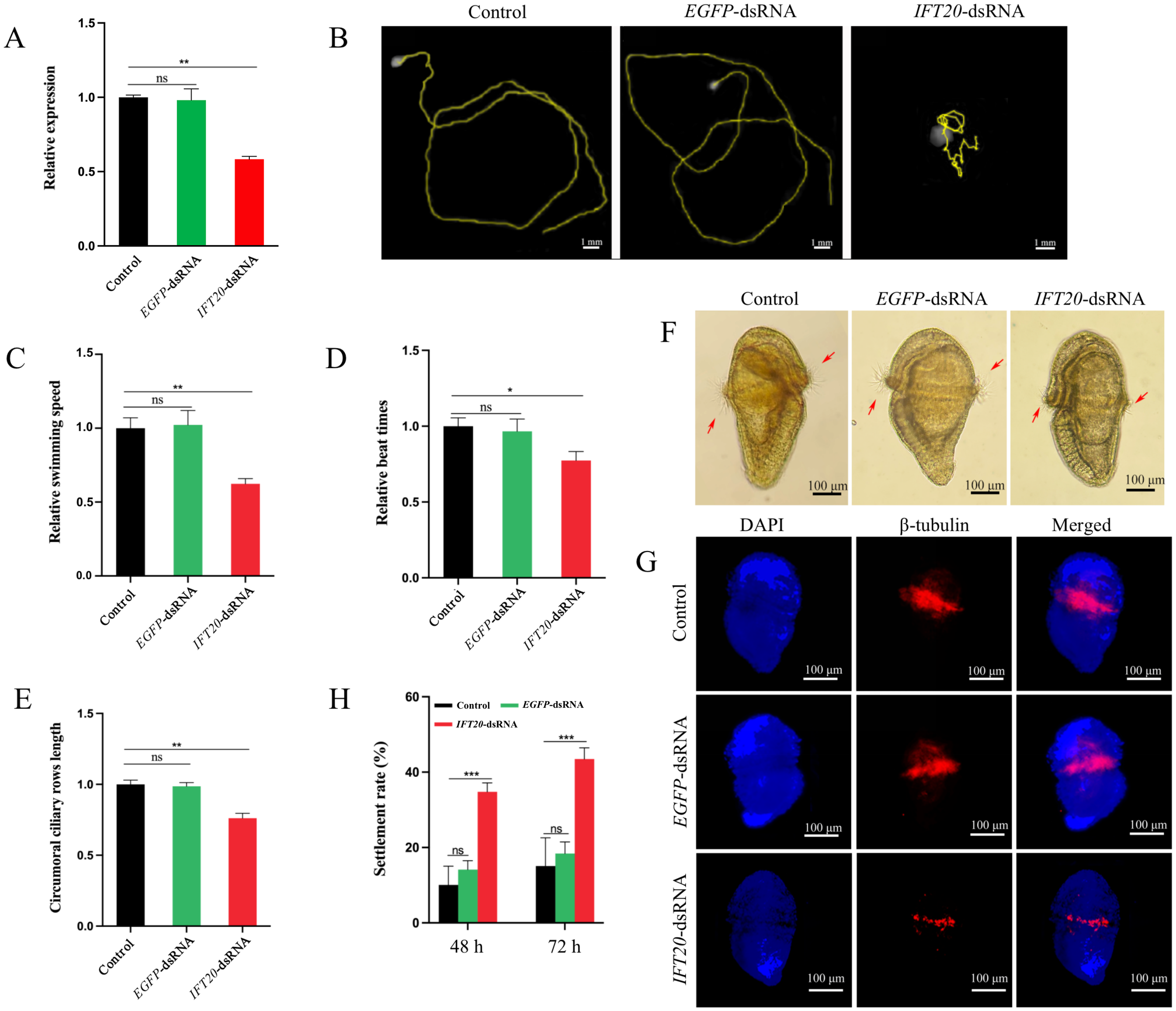
Phenotype of the larvae following the IFT20 interference. (A) Expression level of IFT20 after RNAi for 72 h; (B, C) Range and speed of the larval swimming after RNAi for 24 h; (D) Beat frequency of the circumoral cilia in the larvae after RNAi for 72 h; (E, F) Length of the circumoral cilia in the larvae after RNAi for 72 h; (G) Localizations of β-tubulin in U. unicinctus larvae after RNAi for 72 h; (H) Settlement rates of the larvae after RNAi for 48 h and 72 h, respectively. The significant difference is marked with “*” at P < 0.05, “***” at P < 0.001, and “ns” means no significant difference. Different letters indicate a significant difference at P < 0.05. The red arrows indicate the circumoral cilia in the larvae.
Furthermore, the observation was performed by scanning electron microscopy (SEM), and transmission electron microscopy (TEM) to reveal the ultrastructural changes of the cilia in the larvae after knocked down the IFT20 expression. SEM images showed that the length and number of the circumoral cilia were obviously lower in the larvae of the IFT20-dsRNA group than that of the control group (Figure 6A). TEM results showed that the number of vesicles around the base of cilia in the ciliary cells of the larvae in the IFT-dsRNA group obviously decreased, and the basal body of the cilia vacuolated (Figure 6B). In addition, significant mitochondrial damages were also indicated in the ciliary cells of the larvae in the IFT-dsRNA group, mainly manifested as the smaller size, the less prominent cristae and the decreased electron density matrix (Figure 6C).
Figure 6
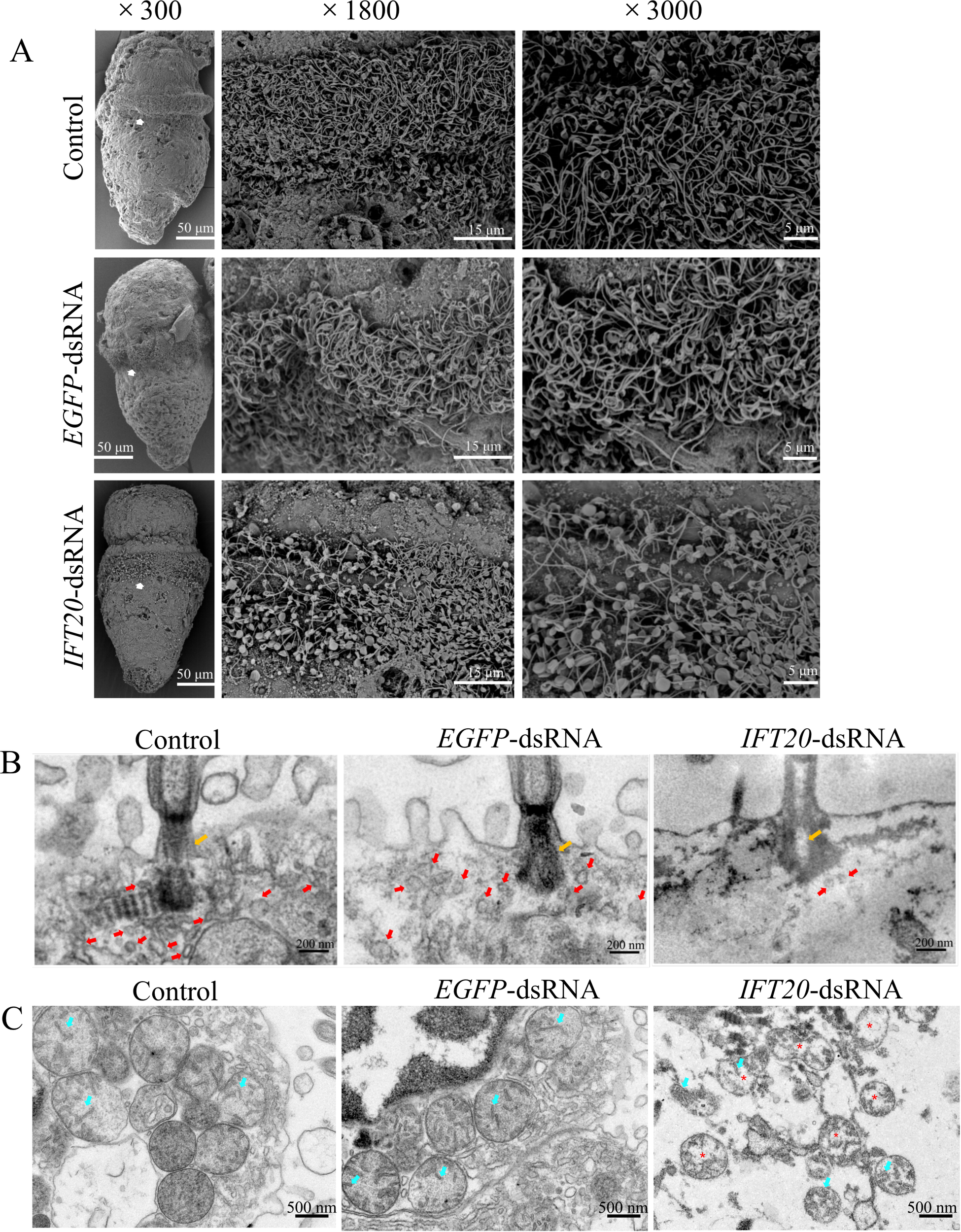
Ultrastructure observation of the cilia and the ciliary cells in the larvae after knocked down the IFT20 expression for 72 h. (A) The SEM observation; (B) The TEM observation, showing the vesicles at the base of the circumoral cilia; (C) The TEM observation, showing the organelles at the base of the circumoral cilia. The white arrows in picture (A) indicate the magnified parts; the orange arrows indicate the basal body of cilia; the red arrows indicate the vesicles; the cyan arrows indicate the ridges in mitochondria; and the red * indicates the low electron density matrix in mitochondria.
4 Discussion
Larval settlement in marine invertebrates typically involves in the cessation of swimming and the initiating substrate exploration, such as crawling or settling (Dobretsov and Rittschof, 2020). In many species, neuropeptides as a kind of signaling molecules have been reported to play a crucial role in larval settlement (Schmich et al., 1998; Grasso et al., 2011; Whalan et al., 2012; Williams, 2020; Yang et al., 2024). These neuropeptides promote the settlement behavior by inhibiting ciliary beating in response to environmental cues. In annelids, Conzelmann et al. (2013) demonstrated that neuropeptides from sensory cells can directly govern larval cilia and stop ciliary movement, causing larvae to settle. In nemerteans, the neuropeptide CCHamide can induce the settlement of Lineus longissimus larvae by reducing the frequency of cilia beating. However, some studies from transcriptome data suggested that changes in gene expression are also involved in settlement behavior of some animals, including the sponges, ascidians, gastropods, corals, shellfish, and annelids (Azumi et al., 2007; Grasso et al., 2008; Grasso et al., 2011; Heyland et al., 2011; Conaco et al., 2012; Ventura et al., 2015; Sedanza et al., 2022; Yang et al., 2024). Our previous transcriptome data has revealed that the expressions of numerous cilia genes and signaling pathways are triggered in the U. unicinctus larvae treated with neuropeptide MIP2 during settlement (Yang et al., 2024). This suggests that neuropeptides may activate a regulation cascade of ciliary gene expression to promote the settlement behavior in U. unicinctus. However, little is known about the specific roles of neuropeptide-regulated pathways and ciliary genes in larval settlement.
In this study, we first revealed that IFT20 regulated expression by the FILa1, plays a multifaceted role in the settlement behavior by impacting the ciliary structure and function. We found that the FILa1, endemic to U. unicinctus, can down-regulate the IFT20 expression in U. unicinctus larvae (Figure 3). The cAMP-PKA and Ca²+ signaling pathways are common neuropeptide-mediated pathways and have been reported to be involved in larval settlement (Conzelmann et al., 2013; Yang et al., 2024). To further identify the signaling pathways mediating FILa1 downregulating the IFT20 expression, the activators or inhibitors of the cAMP-PKA and Ca2+ signaling pathways were employed and co-treated with FILa1 to U. unicinctus larvae. We found that co-treatment with the AC activator, cAMP analog, IP3 receptor blocker (2-APB), or CaM antagonist blocked FILa1-induced IFT20 downregulation and larval settlement (Figures 4A, B, A’, B’, D, E, D’, E’). However, in larvae treated with PKA inhibitor or CaMK-II blocker together with FIL1, the effect of FILa1 on downregulating IFT20 expression was not significantly changed. A possible explanation was that the inhibitory effect of PKA inhibitor or CaMK-II blocker alone on IFT20 expression was already strong, so co-treatment of PKA inhibitor or CaMK-II blocker with FILa1 did not enhance the downregulation of IFT20. Moreover, the binding of CREB, a down-stream transcription factor in the cAMP-PKA pathway, to IFT20 gene also verified that FILa1 can regulate the expression of IFT20 through cAMP-PKA-CREB pathway (Figure 3E). In P. dumerilii, Conzelmann et al. (2013) demonstrates through in vitro experiments (CHO cells) that MIP2 induces settlement via the Ca2+ signaling pathway. Bai et al. (2022) show in vitro (HEK293T cells) that MIP1 in U. unicinctus regulates settlement behavior through cAMP signaling. However, the downstream molecules and their functions in these two neuropeptide-mediated pathways remain unclear. Normally cAMP activates PKA to regulate gene expression, and subsequently phosphorylates the transcription factor CREB to promote transcriptional activation (Moreira, 2014). In this study, we identified and elucidated the signaling pathways involved in FILa1-mediated settlement behavior (cAMP-PKA-CREB and Ca2+ pathways) through in vivo experiments and revealed for the first time that these pathways induce settlement behavior by inhibiting the expression of the ciliary gene IFT20.
IFT20 has been revealed to be involved in the transport of vesicles and regulate the related functions with cilia of human cells and several model animals. In human cells, IFT20 has been indicated to localize in the Golgi apparatus and the base of cilia for transporting vesicles from the Golgi to the ciliary base (Follit et al., 2006; Yang and Huang, 2020). The disruption of vesicle trafficking rich in IFT20 affects chondrocyte formation in humans and mice (Martin et al., 2018). In male mice, mutations of IFT20 disrupt the “9 + 2” microtubule structure of cilia, leading to a reduced sperm count (Zhang et al., 2016). In the retinal epithelium, knockout of IFT20 ablates primary cilia and leads to retinal degeneration in mice (Kretschmer et al., 2023). In addition, A few studies on IFT20 in invertebrates have also been reported. In Drosophila, IFT20 is localized in cilia (Hou et al., 2023). In C. elegans males, the lack of IFT20 leads to shorter cilia that cannot properly perform mating functions (De-Castro et al., 2021). However, localization and function of IFT20 in marine animals, especially the cilia on body surface of the larvae are unclear. In this study, we discovered that IFT20 proteins were localized at the base of the cilia in U. unicinctus larvae. This was confirmed by the co-localization of IFT20 and the ciliary protein marker β-tubulin (Figure 4). This finding is consistent with previous studies and suggests that the localization of IFT20 is a conserved characteristic across species (Rosenbaum and Witman, 2002). When IFT20 expression was knocked down, the length and number of circumoral cilia in U. unicinctus larvae were significantly reduced (Figures 5F, G, 6A). In addition, ultrastructural damages were observed by TEM, including a reduction in vesicles at the ciliary base, vacuolization at the ciliary base, and damage to the mitochondria near the ciliary base (Figures 6B, C). Therefore, we suggested that IFT20 participates in maintaining the number, length and function of the body surface cilia in U. unicinctus larvae by transporting vesicles to the ciliary base, but the FILa1 disrupts this balance, leading to settlement behavior.
5 Conclusion
In summary, we identified the IFT20 in U. unicinctus and found that its protein was located at the base of the circumoral cilia in U. unicinctus larvae. The IFT20 expression level was inhibited by mature peptide FILa1 through the cAPM-PKA-CREB and Ca2+ pathways promoting the larval settlement. In addition, our study demonstrated that the knockdown of IFT20 expression in U. unicinctus larvae reduced the number of basal body vesicles. This reduction consequently led to basal body vacuolization, a decrease in ciliary microtubule protein, a diminished beat frequency of the circumoral cilia, and a reduction in both the number and length of the circumoral cilia. Furthermore, these effects were ultimately resulting in the decreased larval swimming speed and triggered the larval settlement. The findings of this study provided new insights into the ciliary genes regulated by neuropeptides played an important role in the settlement of marine benthic invertebrates.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
LZ: Data curation, Methodology, Writing – original draft. WZ: Conceptualization, Writing – review & editing. WL: Methodology, Writing – review & editing. ZY: Data curation, Writing – review & editing. DL: Conceptualization, Writing – review & editing. ZrZ: Conceptualization, Data curation, Funding acquisition, Resources, Writing – review & editing. ZfZ: Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 42176122 and No. 32202926), the PhD Scientific Research and Innovation Foundation of Sanya Yazhou Bay Science and Technology City (HSPHDSRF-2023-02-010).
Acknowledgments
We acknowledge all funders of this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1575455/full#supplementary-material
Supplementary Figure 1Images of U. unicinctus larvae at the different stages. CC indicates the circumoral cilia.
References
1
Azumi K. Sabau S. V. Fujie M. Usami T. Koyanagi R. Kawashima T. et al . (2007). Gene expression profile during the life cycle of the urochordate Ciona intestinalis. Dev. Biol.308, 572–582. doi: 10.1016/j.ydbio.2007.05.022
2
Bai S. M. Fan S. T. Liu D. W. Zhang Z. R. Zhang Z. F. (2022). Identification and expression analysis of receptors that mediate MIP regulating larval settlement in Urechis unicinctus. Comp. Biochem. Physiol. B Biochem. Mol. Biol.260, 110732. doi: 10.1016/j.cbpb.2022.110732
3
Bao C. Liu F. Yang Y. Lin Q. Ye H. (2020). Identification of peptides and their GPCRs in the peppermint shrimp Lysmata vittata, a protandric simultaneous hermaphrodite species. Front. Endocrinol.11. doi: 10.3389/fendo.2020.00226
4
Caers J. Verlinden H. Zels S. Vandersmissen H. P. Vuerinckx K. Schoofs L. (2012). More than two decades of research on insect neuropeptide GPCRs: an overview. Front. Endocrinol.3. doi: 10.3389/fendo.2012.00151
5
Conaco C. Neveu P. Zhou H. Arcila M. L. Degnan S. M. Degnan B. M. et al . (2012). Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics13, 1–19. doi: 10.1186/1471-2164-13-209
6
Conzelmann M. Offenburger S. L. Asadulina A. Keller T. Münch T. A. Jékely G. (2011). Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl. Acad. Sci. U.S.A.108, E1174–E1183. doi: 10.1073/pnas.1109085108
7
Conzelmann M. Williams E. A. Tunaru S. Randel N. Shahidi R. Asadulina A. et al . (2013). Conserved MIP receptor - ligand pair regulates Platynereis larval settlement. Proc. Natl. Acad. Sci. U.S.A.110, 8224–8229. doi: 10.1073/pnas.1220285110
8
De-Castro A. R. G. Quintas-Gonçalves J. Silva-Ribeiro T. Rodrigues D. R. M. De-Castro M. J. G. Abreu C. M. et al . (2021). The IFT20 homolog in Caenorhabditis elegans is required for ciliogenesis and cilia-mediated behavior. MicroPubl Biol.2021. doi: 10.17912/micropub.biology.000396
9
Dobretsov S. Rittschof D. (2020). Love at first taste: induction of larval settlement by marine microbes. Int. J. Mol. Sci.21, 731. doi: 10.3390/ijms21030731
10
Doll P. C. Caballes C. F. Hoey A. S. Uthicks S. Ling S. D. Pratchett M. S. (2022). Larval settlement in echinoderms: a review of processes and patterns. Oceanogr. Mar. Biol.60, 433–494. doi: 10.1007/s10886-018-0926-4
11
Finetti F. Cassioli C. Cianfanelli V. Zevolini F. Onnis A. Gesualdo M. et al . (2021). The intraflagellar transport protein IFT20 recruits ATG16L1 to early endosomes to promote autophagosome formation in T cells. Front. Cell Dev. Biol.9. doi: 10.3389/fcell.2021.634003
12
Finetti F. Onnis A. Baldari C. T. (2022). IFT20: an eclectic regulator of cellular processes beyond intraflagellar transport. Int. J. Mol. Sci.23, 12147. doi: 10.3390/ijms232012147
13
Follit J. A. San Agustin J. T. Xu F. Jonassen J. A. Samtani R. Lo C. W. (2008). The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet.4, e1000315. doi: 10.1371/journal.pgen.1000315
14
Follit J. A. Tuft R. A. Fogarty K. E. Pazour G. J. (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell.17, 3781–3792. doi: 10.1091/mbc.e06-02-0133
15
Grasso L. C. Maindonald J. Rudd S. Hayward D. C. Saint R. Miller D. J. et al . (2008). Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics9, 1–18. doi: 10.1186/1471-2164-9-540
16
Grasso L. C. Negri A. P. Fôret S. Saint R. Hayward D. C. Miller D. J. et al . (2011). The biology of coral metamorphosis: molecular responses of larvae to inducers of settlement and metamorphosis. Dev. Biol.353, 411–419. doi: 10.1016/j.ydbio.2011.02.010
17
Hadfield M. G. Carpizo-Ituarte E. J. Del Carmen K. Nedved B. T. (2001). Metamorphic competence, a major adaptive convergence in marine invertebrate larvae. Am. Zool.41, 1123–1131. doi: 10.1093/icb/41.5.1123
18
Hadfield M. G. Koehl M. A. (2004). Rapid behavioral responses of an invertebrate larva to dissolved settlement cue. Biol. Bull.207, 28–43. doi: 10.2307/1543626
19
Heyland A. Vue Z. Voolstra C. R. Medina M. Moroz L. L. (2011). Developmental transcriptome of Aplysia californica. J. Exp. Zool B Mol. Dev. Evol.316, 113–134. doi: 10.1002/jez.b.21383
20
Hou X. T. Qin Z. K. Wei M. K. Fu Z. Liu R. Lu L. et al . (2020). Identification of the neuropeptide precursor genes potentially involved in the larval settlement in the Echiuran worm Urechis unicinctus. BMC Genomics21, 1–13. doi: 10.1186/s12864-020-07312-4
21
Hou Y. N. Zhang Y. Y. Wang Y. R. Wu Z. M. Luan Y. X. Wei Q. (2023). IFT52 plays an essential role in sensory cilia formation and neuronal sensory function in Drosophila. Insect Sci.30, 1081–1091. doi: 10.1111/1744-7917.13140
22
Ishikawa H. Marshall W. F. (2017). Intraflagellar transport and ciliary dynamics. Cold Spring Harb. Perspect. Biol.9, a021998. doi: 10.1101/cshperspect.a021998
23
Jiang M. Palicharla V. R. Miller D. Hwang S. H. Zhu H. Hixson P. et al . (2023). Human IFT-A complex structures provide molecular insights into ciliary transport. Cell Res.33, 288–298. doi: 10.1038/s41422-023-00778-3
24
Jin F. C. Zhou M. H. Chen J. J. Lin Y. Zhang Q. W. Xu Q. X. (2022). Intraflagellar transport 20 cilia-dependent and cilia-independent signaling pathways in cell development and tissue homeostasis. Int. J. Dev. Biol.66, 333–347. doi: 10.1387/ijdb.220072fj
25
Keady B. T. Le Y. Z. Pazour G. J. (2011). IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol. Biol. Cell.22, 921–930. doi: 10.1091/mbc.E10-09-0792
26
Kretschmer V. Schneider S. Matthiessen P. A. Reichert D. Hotaling N. Glasßer G. et al . (2023). Deletion of IFT20 exclusively in the RPE ablates primary cilia and leads to retinal degeneration. PLoS Biol.21, e3002402. doi: 10.1371/journal.pbio.3002402
27
Lee S. H. Joo K. Jung E. J. Hong H. Seo J. Kim J. (2018). Export of membrane proteins from the Golgi complex to the primary cilium requires the kinesin motor, KIFC1. FASEB J.32, 957–968. doi: 10.1096/fj.201700563R
28
Li L. Chen Y. Liao W. Yu Q. Lin H. Shi Y. et al . (2022). Associations of IFT20 and GM130 protein. expressions with clinicopathological features and survival of patients with lung adenocarcinoma. BMC Cancer.22, 809. doi: 10.1186/s12885-022-09905-6
29
Lu L. Zhang Z. F. Zheng Q. J. Chen Z. T. Bai S. M. Zhang Z. R. (2022). Expression characteristics and potential function of neuropeptide MIP in larval settlement of the Echiuran worm Urechis Unicinctus. J. Ocean U. China.21, 977–986. doi: 10.1007/s11802-022-4889-2
30
Maldonado M. Young C. M. (1996). Effects of physical factors on larval behavior. settlement and recruitment of four tropical demosponges. Mar. Ecol. Prog. Ser.138, 169–180. doi: 10.3354/meps138169
31
Marshall D. J. Krug P. J. Kupriyanova E. K. Byrne M. Emlet R. B. (2012). The biogeography of marine invertebrate life histories. Annu. Rev. Ecol. Evol. S.43, 97–114. doi: 10.1146/annurev-ecolsys-102710-145004
32
Martin L. Kaci N. Estibals V. Goudin N. Garfa-Traore M. Benoist-Lasselin C. (2018). Constitutively active FGFR3 disrupts primary cilium length and IFT20 trafficking in various chondrocyte models of achondroplasia. Hum. Mol. Genet.27, 1–13. doi: 10.1093/hmg/ddx374
33
Moreira I. S. (2014). Structural features of the G-protein/GPCR interactions. Biochim. Biophys. Acta Gen. Subj.1840, 16–33. doi: 10.1016/j.bbagen.2013.08.027
34
Nakayama K. Katoh Y. (2020). Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit. Rev. Biochem. Mol. Biol.55, 179–196. doi: 10.1080/10409238.2020.1768206
35
Pedersen L. B. Rosenbaum J. L. (2008). Chapter two intraflagellar transport (IFT): role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol.85, 23–61. doi: 10.1016/S0070-2153(08)00802-8
36
Qiu N. Jin H. Cui L. Zhan Y. T. Xia H. M. Jiang M. et al . (2023). IFT20 confers paclitaxel resistance by triggering β-arrestin-1 to modulate ASK1 signaling in breast cancer. Mol. Cancer Res.21, 214–227. doi: 10.1158/1541-7786.MCR-22-0289
37
Rezi C. K. Aslanyan M. G. Diwan G. D. Cheng T. Chamlali M. Junger K. et al . (2024). DLG1 functions upstream of SDCCAG3 and IFT20 to control ciliary targeting of polycystin-2. EMBO Rep.25, 3040–3063. doi: 10.1038/s44319-024-00170-1
38
Roboti P. Sato K. Lowe M. (2015). The golgin GMAP-210 is required for efficient membrane trafficking in the early secretory pathway. J. Cell Sci.128, 1595–1606. doi: 10.1242/jcs.166710
39
Rosenbaum J. L. Witman G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell bio.3, 813–825. doi: 10.1038/nrm952
40
Schmich J. Trepel S. Leitz T. (1998). The role of GLWamides in metamorphosis of Hydractinia eChinate. Dev. Genes Evol.208, 267–273. doi: 10.1007/s004270050181
41
Sedanza M. G. Alshaweesh J. Gao Y. L. Yoshida A. Kim H. J. Yamaguchi K. et al . (2022). Transcriptome dynamics of an oyster larval response to a conspecific cue-mediated settlement induction in the Pacific oyster. Crassostrea gigas. Diversity14, 559. doi: 10.3390/d14070559
42
Shikuma N. J. Pilhofer M. Weiss G. L. Hadfield M. G. Jensen G. J. Newman D. K. et al . (2014). Marine tubeworm metamorphosis induced by arrays of bacterial phage tail - like structures. Science343, 529–533. doi: 10.1126/science.1246794
43
Stoetzel C. Bär S. De Craene J. O. Scheidecker S. Etard C. Chicher J. et al . (2016). A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis-Golgi. Nat. Commun.7, 13586. doi: 10.1038/ncomms13586
44
Su S. H. (2020). Characterization of primary cilia and intraflagellar transport 20 in the epidermis (New York: Columbia University).
45
Sung C. H. Leroux M. R. (2013). The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol.15, 1387–1397. doi: 10.1038/ncb2888
46
Taschner M. Bhogaraju S. Lorentzen E. (2012). Architecture and function of IFT complex proteins in ciliogenesis. Differentiation83, S12–S22. doi: 10.1016/j.diff.2011.11.001
47
Taschner M. Lorentzen E. (2016). The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol.8, a028092. doi: 10.1101/cshperspect.a028092
48
Thiel D. Bauknecht P. Jékely G. Hejnol A. (2019). A nemertean excitatory peptide/CCHamide regulates ciliary swimming in the larvae of Lineus longissimus. Front. Zool.16, 1–14. doi: 10.1186/s12983-019-0326-9
49
Toonen R. J. Pawlik J. R. (1994). Foundations of gregariousness. Nature370, 511–512. doi: 10.1038/370511a0
50
Ventura T. Fitzgibbon Q. P. Battaglene S. C. Elizur A. (2015). Redefining metamorphosis in spiny lobsters: molecular analysis of the phyllosoma to puerulus transition in Sagmariasus verreauxi. Sci. Rep.5, 13537. doi: 10.1038/srep13537
51
Walters L. J. Miron G. Bourget E. (1999). Endoscopic observations of invertebrate larval substratum exploration and settlement. Mar. Ecol. Prog. Ser.182, 95–108. doi: 10.3354/meps182095
52
Wang W. Jack B. M. Wang H. H. Kavanaugh M. A. Maser R. L. Tran P. V. (2021). Intraflagellar transport proteins as regulators of primary cilia length. Front. Cell Dev. Biol.9. doi: 10.3389/fcell.2021.661350
53
Wei M. K. Qin Z. K. Kong D. X. Liu D. W. Zheng Q. J. Bai S. M. et al . (2022). Echiuran Hox genes provide new insights into the correspondence between Hox subcluster organization and collinearity pattern. Proc. Biol. Sci.289, 20220705. doi: 10.1098/rspb.2022.0705
54
Whalan S. Webster N. S. Negri A. P. (2012). Crustose coralline algae and a cnidarian neuropeptide. trigger larval settlement in two coral reef sponges. PLoS One7, e30386. doi: 10.1371/journal.pone.0030386
55
Williams E. A. (2020). Function and distribution of the Wamide neuropeptide superfamily in metazoans. Front. Endocrinol.11. doi: 10.3389/fendo.2020.00344
56
Williams E. A. Conzelmann M. Jékely G. (2015). Myoinhibitory peptide regulates feeding in the marine annelid Platynereis. Front. Zool.12, 1–16. doi: 10.1186/s12983-014-0093-6
57
Yamaguchi H. Terajima M. Kitami M. Wang J. He L. Saeki M. et al . (2020). IFT20 is critical for collagen biosynthesis in craniofacial bone formation. Biochem. Biophys. Res. Commun.533, 739–744. doi: 10.1016/j.bbrc.2020.09.033
58
Yang H. Huang K. (2020). Dissecting the vesicular trafficking function of IFT subunits. Front Cell. Dev. Biol.7. doi: 10.3389/fcell.2019.00352
59
Yang H. Zhang F. Long H. Lin Y. Liao J. Xia H. et al . (2021). IFT20 mediates the transport of cell migration regulators from the trans-golgi network to the plasma membrane in breast cancer cells. Front. Cell Dev. Biol.9. doi: 10.3389/fcell.2021.632198
60
Yang Z. Zhang L. Zhang W. Q. Tian X. H. Lai W. Y. Lin D. W. et al . (2024). Identification of the principal neuropeptide MIP and its action pathway in larval settlement of the echiuran worm Urechis unicinctus. BMC Genomics25, 337. doi: 10.1186/s12864-024-10228-y
61
Zhang Z. G. Li W. Zhang Y. Zhang L. Teves M. E. Liu H. et al . (2016). Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell.27, 3705–3716. doi: 10.1091/mbc.e16-05-0318
62
Zhang L. Shi R. X. Ouyang H. F. Li Y. H. (2020a). Cloning and characterization of chst11 from Procambarus clarkii involved in the host immune response of white spot syndrome virus and Aeromonas hydrophila. Fish Shellfish Immunol.102, 82–91. doi: 10.1016/j.fsi.2020.04.020
63
Zhang L. Zhen J. Huang Q. Liu H. Li W. Zhang S. et al . (2020b). Mouse spermatogenesis-associated. protein 1 (SPATA1), an IFT20 binding partner, is an acrosomal protein. Dev. Dyn.249, 543–555. doi: 10.1002/dvdy.141
64
Zheng Q. J. Wang Y. J. Chen J. Li Y. P. Zhao F. Liu D. W. et al . (2022). Effects of salinity on the growth, physiological characteristics, and intestinal microbiota of the Echiura worm (Urechis unicinctus). Front. Mar. Sci.9. doi: 10.3389/fmars.2022.912023
65
Zhou M. H. Lin Y. Zhang Z. G. (2020). Intraflagellar transport 20: New target for the treatment of Ciliopathies. Biochim. Biophys. Acta Mol. Cell Res.1867, 118641. doi: 10.1016/j.bbamcr.2019.118641
Summary
Keywords
intraflagellar transport protein 20, neuropeptide, gene pathway, larval settlement, Urechis unicinctus
Citation
Zhang L, Zhang W, Lai W, Yang Z, Lin D, Zhang Z and Zhang Z (2025) The role of IFT20 mediated by neuropeptide FILa1 in ciliary activity during larval settlement of Urechis unicinctus. Front. Mar. Sci. 12:1575455. doi: 10.3389/fmars.2025.1575455
Received
13 February 2025
Accepted
07 April 2025
Published
28 April 2025
Volume
12 - 2025
Edited by
Enric Gisbert, Institute of Agrifood Research and Technology (IRTA), Spain
Reviewed by
Sofia Priyadarsani Das, National Taiwan Ocean University, Taiwan
Satheesh Sathianeson, King Abdulaziz University, Saudi Arabia
Updates
Copyright
© 2025 Zhang, Zhang, Lai, Yang, Lin, Zhang and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengrui Zhang, zhangzhengrui@ouc.edu.cn; Zhifeng Zhang, zzfp107@ouc.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.