Abstract
Red mullet, Mullus barbatus Linnaeus, 1758, is a very important target species of high commercial value for small-scale fisheries in the Mediterranean Sea. The distribution of the species is wide encompassing the North-Eastern Atlantic (from Scandinavia to Senegal), the Mediterranean Sea and Black Sea. Climatic differences across its range could trigger specific bioenergetic demands due that ectothermic aquatic species exploit heat exchange with the external environment to regulate metabolic activities and adaptation. Mitochondrial OXPHOS (mtOXPHOS) genes are particularly involved in these processes and they have been studied in the last decades as a system that is subject to selection under determined environmental constraints. Based on the above considerations, the purpose of this work were to analyze the nucleotide sequences of the Cytochrome Oxidase I (COI) and Cytochrome b (Cytb) OXPHOS genes in seven Mediterranean populations of M. barbatus, living within a latitudinal range between the North Adriatic, the Strait of Sicily in the South, the Ionian Sea in the East and the Balearic Sea in the Western Mediterranean. The aims were to assess the genetic diversity in the studied populations and to detect the presence of positive selection on the two-target protein-encoded genes using tests of recombination and selection based on different models of evolution. The diversity indices indicated higher values of haplotype diversity in the Adriatic populations than in the remaining populations for both genes. Furthermore, a very high number of COI and Cytb private haplotypes was found in almost of populations. Signature of pervasive positive selection by FUBAR and episodic positive selection by MEME were exclusively detected in COI gene. Our results support the need to manage red mullet populations as separate sub-populations with distinct gene pools.
1 Introduction
Red mullet, Mullus barbatus Linnaeus, 1758, is an Eastern Atlantic and Mediterranean species also distributed in the Black Sea (Whitehead et al., 1984) considered an important resource for Mediterranean coastal fisheries (Reñones et al., 1995). It is a demersal species that spawns in spring, whit adults which prefer sandy and/or muddy habitats (Tserpes et al., 2002) at depths ranging from 10 to 300 meters, while larvae and juveniles are pelagic (Mamuris et al., 1998). It is one of the most valuable species in commercial landings and is then subject to high fishing pressure, which negatively affect genetic variability of populations. Furthermore, due to its wide distribution, red mullet encounters variable environmental factors throughout its range, which could trigger specific bioenergetic requirements. It is known that ectothermic aquatic species use heat exchange with the external environment to regulate metabolic activities and adaptation (Sokolova, 2018). Interestingly, a link between mitochondrial DNA variation and intraspecific behavioural diversity has been highlighted. The research aims to show how mitochondrial genetic variation, which affects energy metabolism, drives individual differences in the activity rates of organisms in different environmental contexts (Brand et al., 2024).
The efficiency of energy generation in an organism plays a crucial role in its successful metabolic performance (Lajbner et al., 2018), with mitochondria being vital for energy production and regulation. Indeed, most of the adenosine triphosphate (ATP) required by an organism is produced through oxidative phosphorylation (OXPHOS) in the mitochondria. This system is formed by five multisubunit complexes embedded in the inner mitochondrial membrane, where their activity results in ATP production (Saraste, 1999). Thirteen mitochondrial genes encode proteins in four of the five OXPHOS complexes (I, III, IV and V), together with ~72 nuclear genes that participate in all five complexes (Efremov and Sazanov, 2011). Because of this very important function, mutations in mitochondrial DNA and OXPHOS genes in particular, are generally subject to purifying selection (Jacobsen et al., 2016). However, there is evidence of directional or episodic positive selection for mitochondrial DNA (mtDNA) in response to changes in selective pressure, due to hypoxia, heat stress and cold stress, nutrient availability in several organisms (Garvin et al., 2015; Lajbner et al., 2018; Teske et al., 2019; Zhou et al., 2014). OXPHOS genes have been shown to undergo considerable adaptive evolution, and consequently the mtDNA selective neutrality hypothesis has been empirically tested and refuted in a wide range of organisms. For example, Ruiz-Pesini et al. (2004) showed that in humans living within Arctic area, three mtDNA protein-coding genes (Nicotinamide adenine dinucleotide hydrogenase 2, ND2 and 4, ND4) and ATP6) exhibit signatures of adaptive selection. Several investigation have found evidence for molecular adaptations in teleosts driven by thermal acclimation (Consuegra et al., 2015; Sebastian et al., 2020) or swimming performance (Zhang and Broughton, 2015). Recently one codon of the Nicotinamide adenine dinucleotide hydrogenase 1 (ND1) mitochondrial protein was found under positive selection in the killifish Aphanius fasciatus, an eurytherm and eurihalyne species living in transitional waters where it encounters a highly daily and seasonal variability of environmental parameters (Pappalardo et al., 2024). Positively selected sites of Cytochrome b (Cytb) were correlated with temperature adaptation on the marine mammal killer whale (Foote et al., 2011) and in the European anchovy (Silva et al., 2014); one site under positive selection was found in the Cytochrome Oxidase I (COI) of the (Pappalardo et al., 2015).
Focusing on red mullet genetic structure, conflicting results have been obtained by using different mitochondrial and nuclear molecular marker. Both a weak genetic structure among Mediterranean populations of M. barbatus and a metapopulation structure have been observed at the Mediterranean scale, indicating separate stock units (Garoia et al., 2004; Maggio et al., 2009) (Galarza et al., 2009; Matić-Skoko et al., 2018). However, for species subject to high fishing pressure, such as the overexploited red mullet, the results of genetic structure studies are crucial, as these data should be a useful tool for fisheries management of the identified subpopulations.
Based on the above considerations, in this study we explored the genetic structure of red mullet from seven Mediterranean populations through the analysis of two mtOXPHOS genes, COI and Cytb. Furthermore, we searched for signatures of positive selection on these two genes which are likely to be a target for selection due to their essential function in energy production.
2 Materials and methods
2.1 Sampling sites
Seven Mediterranean populations of M. barbatus were sampled within a latitudinal range between the North Adriatic Sea, the Strait of Sicily in the South, the Ionian Sea in the East and the Balearic Sea in the Western Mediterranean for a total of 140 individuals (Figure 1; Table 1).
Figure 1
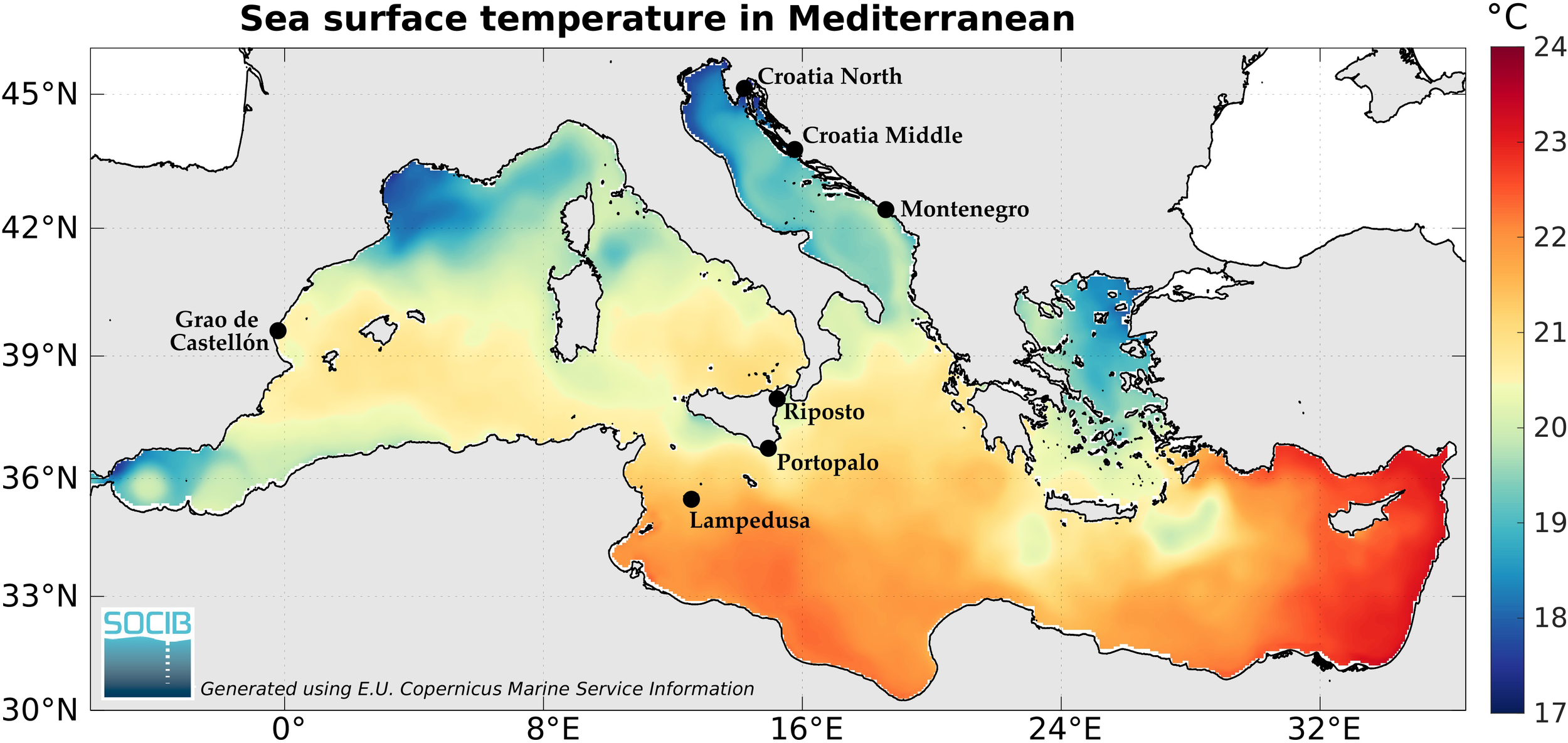
Sampling locations of Mullus barbatus on heatmap of annual mean sea surface temperature.
Table 1
| Cytochrome Oxidase I | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Collection locality | Code | n | Nh | h ± SD | π ± SD | Fu's Fs | p-value | Tajima's D | P-value |
| Croatia North | NJB | 21 | 14 | 0.948 (0.031) | 0.00497 (0.00055) | -7,328 | 0.00 | -0.554 | 0.39 |
| Croatia Middle | SJB | 21 | 12 | 0.886 (0.059) | 0.00491 (0.00353) | -4,468 | 0.01 | -1.436 | 0.06 |
| Montenegro | CGB | 21 | 14 | 0.957 (0.026) | 0.00728 (0.00119) | -4,847 | 0.01 | -0.930 | 0.17 |
| Riposto | RI | 20 | 7 | 0.837 (0.056) | 0.00365 (0.00059) | -0.629 | 0.39 | -0.628 | 0.39 |
| Portopalo | POR | 20 | 2 | 0.100 (0.088) | 0.00015 (0.00013) | -0.879 | 0.09 | -1.164 | 0.15 |
| Lampedusa | LAMP | 17 | 6 | 0.691 (0.034) | 0.00257 (0.00058) | -1.005 | 0.23 | -0.987 | 0.19 |
| Grau de Castellòn | ES | 20 | 6 | 0.811 (0.055) | 0.00279 (0.00042) | -0.483 | 0.38 | 0.962 | 0.87 |
| Cytochrome b | |||||||||
| Collection locality | Code | n | Nh | h ± SD | π ± SD | Fu's Fs | p-value | Tajima's D | P-value |
| Croatia North | NJB | 20 | 10 | 0.879 (0.052) | 0.00693 (0.00060) | -0.872 | 0.34 | 0.968 | 0.86 |
| Croatia Middle | SJB | 20 | 8 | 0.784 (0.084) | 0.00287 (0.00083) | -2,080 | 0.10 | -1.742 | 0.03 |
| Montenegro | CGB | 19 | 8 | 0.807 (0.079) | 0.00219 (0.00036) | -3,221 | 0.01 | -0.706 | 0.27 |
| Riposto | RI | 20 | 7 | 0.853 (0.043) | 0.00380 (0.00060) | -0.264 | 0.49 | -0.416 | 0.37 |
| Portopalo | POR | 18 | 6 | 0.627 (0.124) | 0.00126 (0.00032) | -2,750 | 0.01 | -1.187 | 0.10 |
| Lampedusa | LAMP | 17 | 7 | 0.838 (0.063) | 0.00488 (0.00051) | 0.082 | 0.50 | 0.689 | 0.79 |
| Grau de Castellòn | ES | 20 | 5 | 0.695 (0.081) | 0.00132 (0.00025) | -1,254 | 0.13 | -0.462 | 0.34 |
Diversity measures and neutrality tests for the collecting sites of Mullus barbatus for COI and Cytb: number of sequences (n), number of haplotypes (Nh), haplotype diversity (h), nucleotide diversity (π) and relative standard deviation (SD).
Significat values in bold.
2.2 DNA extraction, amplification and sequencing
For each sample, 25 mg of muscle tissue were dissected from the dorsal side of the specimen using sterile, disposable plastic forceps, and genomic DNA was extracted using the NucleoSpin® Tissue (Macherey-Nagel) kit according to the manufacturer’s instructions. Each DNA sample was then analyzed using the Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) in order to quantify the DNA concentration: measurements of 260/280 nm and 260/230 nm absorbance ratios were used to determine the quality and degree of purity. Finally, genomic DNA was stored at -20 C for subsequent applications. Two mitochondrial gene fragments, COI of about 650 bp and Cytb, of about 700 bp, were amplified using appropriate thermal profiles as shown in Figure 2 and specific primer pairs (VF2_t1 and FishR2_t1 for COI gene from Ivanova et al (2007) and FishcytB-F and CytBI-4R for Cytb gene from Sevilla et al. (2007). All PCR products were visualized through a 0.8% agarose gel and sequenced for both strands with the Sanger dideoxy method by EUROFINS Genomics sequencing services (https://eurofinsgenomics.eu).
Figure 2
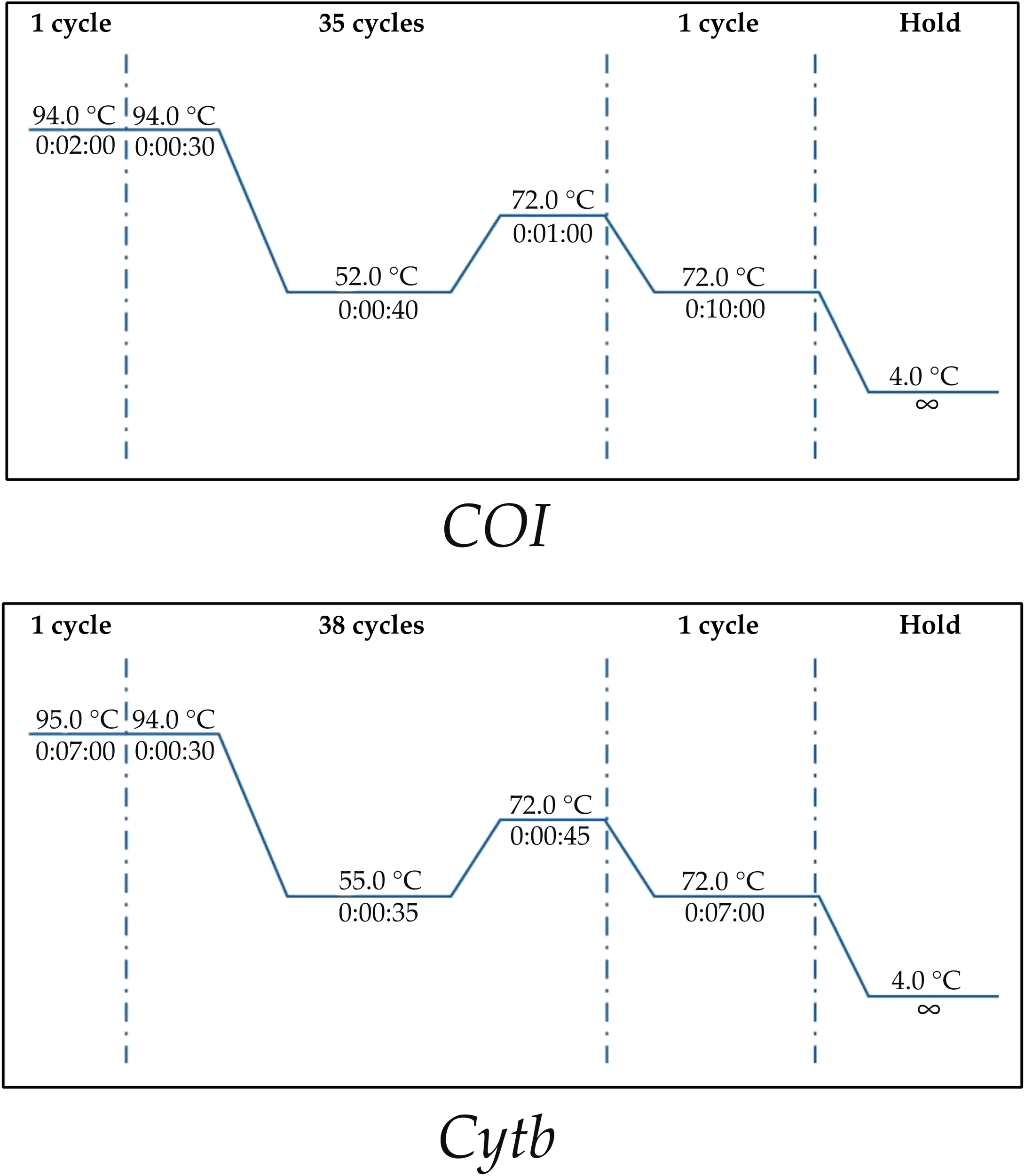
Amplification thermal profiles for each mitochondrial gene.
2.3 Bioinformatic analysis
Sequences from both genes were edited and aligned using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) (Hall, 1999) and MAFFT (Katoh et al., 2019) respectively. Mitochondrial COI and Cytb haplotypes sequences generated in this study were submitted to the GenBank database (https://support.nlm.nih.gov/kbArticle/?pn=KA-03391) (Sayers et al., 2024) (Supplementary Table S1 and Supplementary Table S2). DNA sp.6 (Rozas et al., 2017) was used to calculate haplotype (h) and nucleotide diversity (π) and number of haplotypes (N) for each gene and population (Table 1). Statistical elaboration of COI and Cytb haplotype frequencies was performed in GraphPad Prism v.8.3.0 for Windows (GraphPad Software, San Diego, CA, USA, www.graphpad.com). Relationships among the COI and Cytb haplotypes identified in each population were depicted with the median-joining (MJ) method in Network 10.2 (Fluxus-engineering.com) (Bandelt et al., 1999). Population structuring was tested with an analysis of molecular variance (AMOVA), implemented in the program ARLEQUIN version 3.5 (Excoffier and Lischer, 2010) applied to the distance matrix. The analyses of genetic variance were performed with the populations grouped into 3 distinct geographic groups: first group (Lampedusa, Portopalo, Riposto), second group (Grao de Castellon) and third group (Croatia North, Croatia Middle, Montenegro). Fst values were estimated both for population pairs and for all populations, using a “weighted” analysis of variance (Weir and Cockerham, 1984) and taking into account genetic distances between population pairs (Excoffier et al., 1992).The Tajima’s D (Tajima, 1989) and the Fu’s Fs (Fu, 1997) tests were performed to detect any possible deviation from neutrality for each population. Negative Tajima’s D values can indicate selection, but also population bottlenecks or population expansion (Tajima, 1989).
COI and Cytb haplotype datasets were then subject to phylogenetic analyses under both Bayesian Inference (BI) and Maximum Likelihood (ML) criteria to generate two unrooted trees per dataset.
Bayesian analyses were carried out in MrBayes v.3.2.7 (Ronquist et al., 2012). Nucleotide substitution models were designated in JModelTest2 under the AICc criterion (Darriba et al., 2012). For COI gene a General Time Reversible (GTR) nucleotide substitution model with gamma distributed rate variation among sites (Γ) was implemented, while standard GTR was chosen for the Cytb partition. We ran two separate phylogenetic analyses for each gene under the same MCMC specifications: four 40-million generation runs, each featuring four chains. Tree sampling occurred every 100 generations. The resulting consensus trees were summarized using the 50% majority rule. To ensure convergence was reached, we looked at two statistical criteria: the Potential Scale Reduction Factor (PSRF; Brooks and Gelman, 1998; Gelman and Rubin, 1992) and the Average Standard Deviation of Split Frequencies (ASDSF; Lakner et al., 2008). PSRF compares the variance within and between runs and should approach 1 as runs are converging, while ASDSF should approach 0 when runs are converging toward the same distribution (Ronquist et al., 2012). We ensured the PSRF was 1 and that the ASDFS of both our analyses was below a 0.001 threshold.
Maximum Likelihood analyses were carried out in RAXML-NG v1.2.2 (Kozlov et al., 2019). We applied the same models of sequence evolution used for the Bayesian analyses to both datasets. Both analyses were run with 50 starting trees computed by RAXML-NG: 25 random, and 25 obtained under the Maximum Parsimony criterion. 10000 bootstraps replicates were specified to ensure the robustness of the final trees (Felsenstein, 1985).
2.4 Recombination and selection tests
The Genetic Algorithms for Recombination Detection (GARD), part of the HyPhy package and available at www.datamonkey.org, was used to detect the presence of recombinants in the mitochondrial genes COI and Cytb. Additionally, a preliminary one-tailed Z-test in MEGA X (Kumar et al., 2018) was conducted to assess the presence of selection in each gene dataset. To identify codons under positive or purifying selection, three codon models were applied: FEL (Fixed Effects Likelihood) (Kosakovsky Pond and Frost, 2005) FUBAR (Fast, Unconstrained Bayesian Approximation for Inferring Selection) (Murrell et al., 2013), and MEME (Mixed Effects Model of Evolution) (Murrell et al., 2012). FUBAR estimates the number of non-synonymous and synonymous substitutions at each codon, given a phylogeny, and provides the posterior probability that each codon belongs to a set of classes of x (including x = 1, x < 1, or x > 1) (Murrell et al., 2013). MEME estimates the probability that a codon has undergone episodes of positive evolution, allowing the x-ratio distribution to vary across codons and branches in the phylogeny. Sites with p-values lower than 0.05 in FEL and MEME, as well as those with posterior probabilities greater than 0.9 in FUBAR, were considered to be under selection (Silva et al., 2014).
2.5 Protein structure analysis
The spatial position of sites under positive selection in a 3-dimensional space was visualized using the SWISS-MODEL server (https://swissmodel.expasy.org) with default parameters (Waterhouse et al., 2018). Spatial orientation was inferred in Orientation of Proteins in Membranes (OPM) web portal (Lomize et al., 2006; https://opm.phar.umich.edu/). PyMol was then used to visualize and edit the 3D models previously obtained in SwissModel (DeLano, 2002).
3 Results
3.1 Gene sequences and genetic variation analyses
COI and Cytb mitochondrial sequence length from red mullet sampled along a North-South thermal gradient in the Mediterranean region was 663 bp for COI and 720 bp for Cytb. The number of haplotypes ranged from 2 to 14 for COI sequences and from 5 to 10 for Cytb as shown in Supplementary Tables S1 and Supplementary Tables S2. Haplotype (h) and nucleotide (π) diversity ranged from 0.100 to 0.957 and 0.00015 to 0.00728 for COI, respectively, and 0.627 to 0.879 and 0.00126 to 0.00693 for Cytb, respectively. Neutrality tests revealed negative and significant Fu’s Fs values for COI gene of Adriatic populations of M. barbatus and for Cytb gene of the Adriatic population of Montenegro and Sicilian population of Portopalo; the Tajima D test was never significant for each gene in all populations (Table 1). The most common COI haplotype is shared between all populations (Hap3), while the second most common (Hap33) is shared between all populations with the exception of the Riposto and Portopalo populations (Figure 3). A third haplotype (Hap45) is shared only by the Spanish population of Grau de Castellon and that of Northern Croatia (Figure 3). By contrast, the percentage of private haplotypes is high in all populations with the exception of the populations of Portopalo and Lampedusa (Figure 3). The most common Cytb haplotype is the Hap6 haplotype present in all populations with the exception of Portopalo, while the second most frequent is the Hap3 haplotype shared only by specimens from the Lampedusa and Croatia North populations (Figure 3). A high percentage of private haplotypes is also present in the Cytb gene (Figure 3). The median-joining networks showing the haplotypes relationships of both genes, revealed a starlike topology with the private haplotypes originating from the most frequent and shared haplotypes (Figure 4). Note in the Cytb median-joining network the presence of two haplogroups (Hap2 and Hap35) separated by 3 mutational steps. Of the two molecular markers used to construct the ML and BI trees, only Cytb allowed the identification of well-supported haplogroups corresponding to well-defined populations (e.g. Lampedusa, Portopalo and Croatia North) (Supplementary Figures S1, S2). Results from AMOVA to test the population structuring showed the Fst values (=0.11 for COI and =0.38 for Cytb) statistically significant with respect to the null hypothesis of panmixia for both genes (Table 2). In particular, the hierarchical AMOVA organized as three groups of populations (the Sicilian populations, the Spanish population and the Adriatic populations) rejected the possible a priori hypothesis of geographic structuring (P = 0.887) for both gene (Table 2).
Figure 3
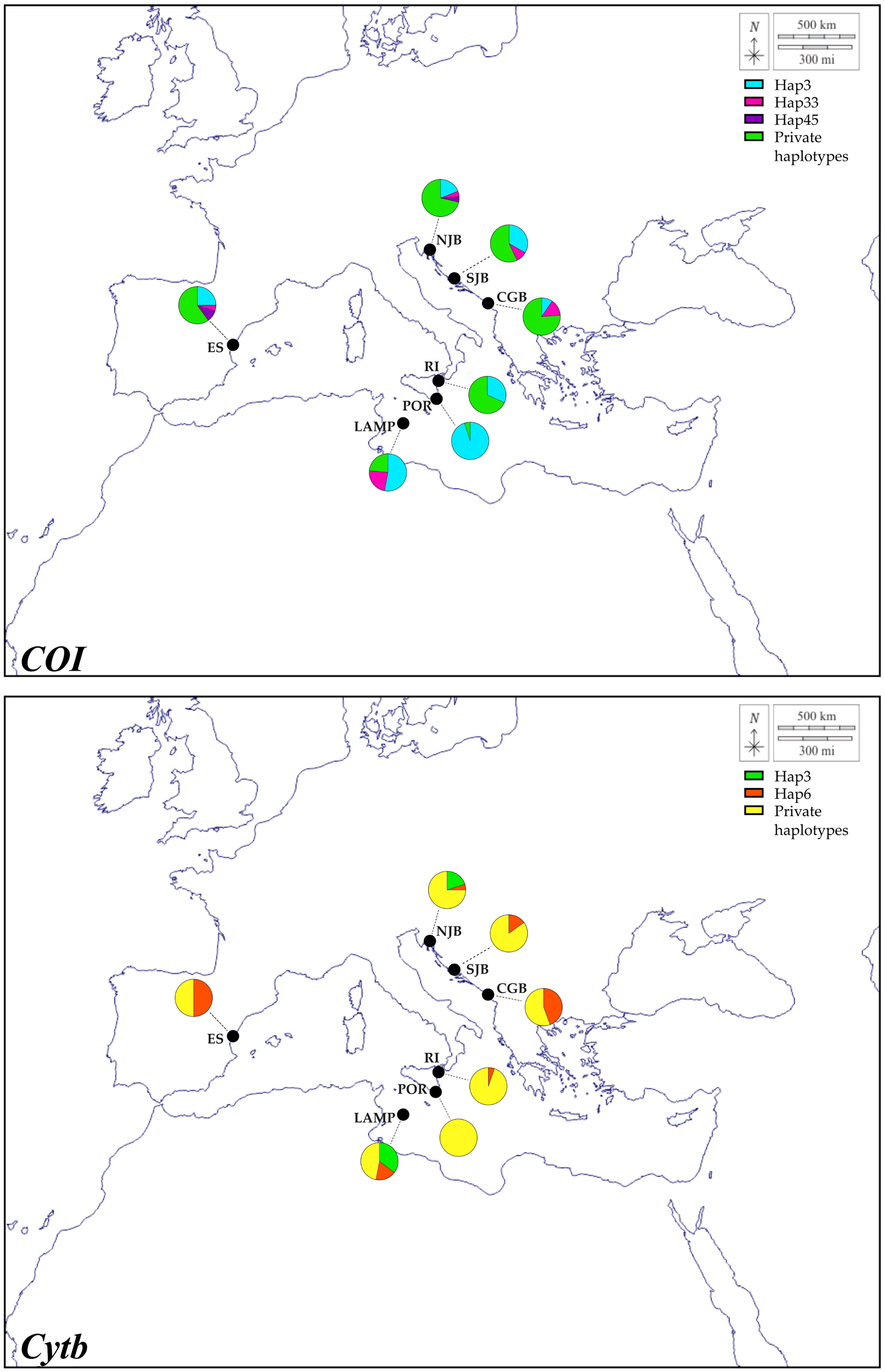
Schematic diagram of the distribution and relative abundance of mitochondrial haplotypes among Mullus barbatus population in the Mediterranean Sea.
Figure 4
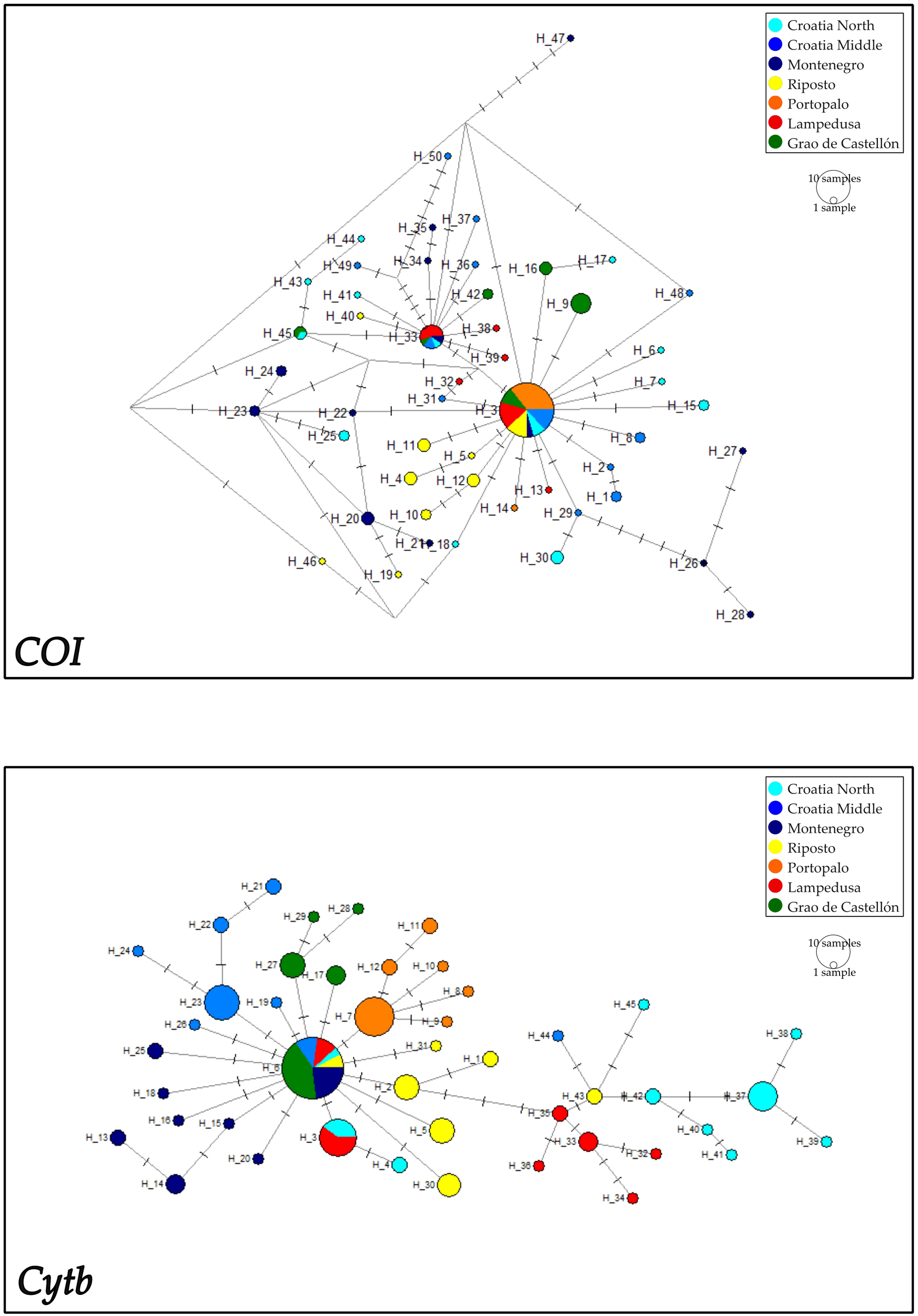
Median-joining network showing relationships among mitochondrial COI and Cytb haplotypes of Mullus barbatus.
Table 2
| COI | |||||
|---|---|---|---|---|---|
| Structured tested | Source of variation | Variance component | Percentage of variation | F statistic | P |
| All population on a unique group | Among populations | 0.15707 Va | 10.95 | Fst = 0.11 | 0.000 |
| Within populations | 1.27744 Vb | 89.05 | |||
| Three groups of populationsa | Among groups | 0.02185 Va | 1.52 | Fct = 0.01 | 0.300 |
| Among populations within groups | 0.14156 Vb | 9.82 | Fst = 0.11 | 0.000 | |
| Within popuations | 1.27744 Vc | 88.66 | Fsc = 0.10 | 0.000 | |
| Cytb | |||||
| Structured tested | Source of variation | Variance component | Percentage of variation | F statistic | P |
| All population on a unique group | Among populations | 0.73818 Va | 38.13 | Fst = 0.38 | 0.000 |
| Within populations | 1.19764 Vb | 61.87 | |||
| Three groups of populationsa | Among groups | -0.23032 Va | -12.31 | Fct = -0.12 | 0.887 |
| Among populations within groups | 0.90363 Vb | 48.30 | Fst = 0.36 | 0.000 | |
| Within popuations | 1.19764 Vb | 64.01 | Fsc = 0.43 | 0.000 | |
Results of molecular variance analysis (AMOVA) under differernt groupings in Mullus barbatus.
a First group (Lampedusa, Portopalo, Riposto), second group (Grao de Castellon), third group (Croatia North, Croatia Middle, Montenegro).
3.2 Signatures of selection in mitochondrial genes
GARD found no evidence of recombination in COI and Cytb datasets. A Z-test was used to reject the null hypothesis of neutral selection in favor of an alternative hypothesis of positive selection in both genes. Four COI codons were under purifying selection and two (4 and 207) under positive selection. The MEME method detected episodic positive selection for codon 4 and FUBAR revealed pervasive positive selection for codon 207. Two Cytb codons (32 and 79) were under purifying selection (Table 3).
Table 3
| Codon | 4 | 109 | 143 | 167 | 206 | 207 | |
|---|---|---|---|---|---|---|---|
| Selection type | positive | purifying | purifying | purifying | purifying | positive | |
| COI | FUBAR | ## | ### | ## | ## | ||
| FEL | * | *** | * | * | |||
| MEME | * | ||||||
| Codon | 32 | 79 | 143 | 167 | 206 | 207 | |
| Selection type | purifying | purifying | purifying | purifying | purifying | positive | |
| Cytb | FUBAR | ### | |||||
| FEL | * | ** | |||||
| MEME |
Positively and negatively selected sites in red mullet Mullus barbatus mitochondrial genes estimated by FUBAR (# bpp≥0.9; ## bpp≥0.95; ### bpp≥0.99) GARD, MEME and FEL models (*p < 0.05; ** p <0.01; ***p <0.001).
3.3 Spatial position of sites under positive selection
Three-dimensional mapping of COI protein showed two codons (4 and 207) under positive selection. Codon 4, under episodic positive selection by MEME, is located at the N-terminal end within the inner mitochondrial membrane (IMM). Codon 207, under pervasive positive selection by FUBAR, is located in the loop region of the intermembrane space (IMS). No evidence of positive selected codons was found in the Cytb protein (Figure 5).
Figure 5
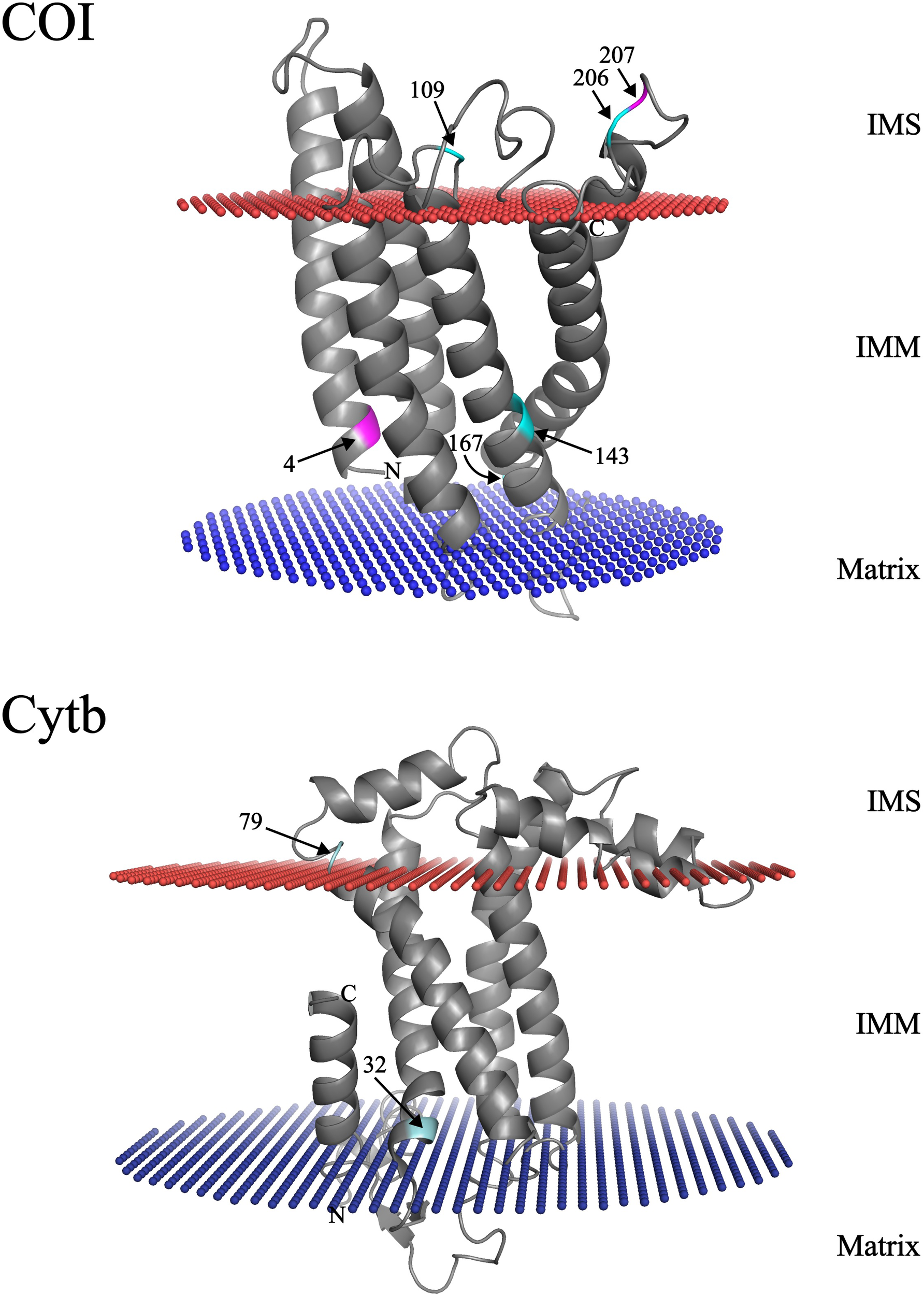
Tridimensional models depicting signs of selection in COI and Cytb proteins from Mullus barbatus. Arrows point at sites identified under different selection levels. Sites highlighted in magenta were retrieved under positive selection, while those colored in cyan are under negative selection. IMM, inner mitochondrial membrane; IMS, intermembrane space.
4 Discussion
The sequence analysis of two mitochondrial OXPHOS genes, COI and Cytb, in M. barbatus allowed to highlight: i) the genetic intraspecific variation among seven Mediterranean populations and ii) the presence of positively selected codons in the amino acidic sequence of the COI protein. The diversity indices indicated higher values of haplotype diversity in the Adriatic populations than in the remaining populations for both genes. The Sicilian population of Portopalo had the lowest diversity index values. Looking at the haplotype distribution in each population, it is evident that almost all populations showed a very high number of private COI haplotypes, with values higher than 50% in the Adriatic (ranging from 57.14% to 76.19%), in the Spanish (60%) and in the Sicilian population of Riposto (68.18%). In contrast, the other two Sicilian populations (Lampedusa and Portopalo) showed a very high prevalence of the most common COI Hap 3, with 95% in the Portopalo population and 52.94% in Lampedusa. About the Cytb, all populations also showed a high percentage of private haplotypes ranging from 50% in the Spanish population to 100% in the Sicilian population of Portopalo. Neutrality tests gave different results: Tajima’s D was not significant for all populations for each gene, while Fu’s Fs gave significant values only for the Adriatic populations with COI sequences, and only for the Adriatic population of Montenegro and the Sicilian population of Portopalo with the Cytb dataset. The statistically significant Fst values allowed to reject the null hypothesis of panmixia for both genes. By comparing these results with those obtained using either the same or other mitochondrial markers and nuclear markers, it was possible to obtain an up-to-date picture of the conservation status of this species in the Mediterranean Sea. The genetic structure of M. barbatus in the Mediterranean Sea has been assessed by allozymes (Arculeo et al., 1999) which indicated a genetic homogeneity of the populations studied due to the extensive gene flow provided by the planktonic larval stage. On the contrary, microsatellites analysis (Garoia et al., 2004; Galarza et al., 2009; Matić-Skoko et al., 2018) revealed genetic structuring in red mullet which is composed by sub-populations relatively connected to each other. In our case the statistically significant Fst values indicate the presence of moderate genetic structuring in red mullet. The negative and significant values of the Fu’s Fs test indicated recent demographic fluctuations for the Adriatic populations and the Sicilian population of Portopalo. The haplotype composition of the latter population is peculiar in that the COI sequences are almost exclusively represented by the most common Hap3 and one private haplotype, whereas the Cytb sequences are almost exclusively represented by six private haplotypes derived from the most common Hap6. Interestingly, the COI Hap3 has a higher prevalence in the most southern populations than the northern ones. In this context, it should be noted that the southern part of the Mediterranean Sea served as a refuge for marine species during glacial periods and this haplotype widely shared among all populations, can be considered the ancestral haplotype as confirmed by the median joining network. Thus, while M. barbatus populations may retain evidence of demographic changes caused by historical events, the excessive fishing pressure may have led to a decline in genetic diversity in some populations. Accordingly, Matić-Skoko et al. (2018) highlighted that high fishing pressure and other anthropic impacts together with physiological fluctuations in population size, have caused recent demographic changes in this species. The same effects of overfishing have been reported for red mullet populations in the Black Sea (Zlateva et al., 2023).
These results should also be interpreted in the light of possible future climate changes in the marine fishing grounds of M. barbatus. In this context, projections for 2035 and 2050 of the potential displacement of the main commercial demersal species, including red mullet, in the Adriatic and Ionian Seas (Panzeri et al., 2024), indicate that M. barbatus could be severely affected by climate change with a northward shift in its range. These data draw attention to the need to implement fisheries management measures for this species with reference to spatially and temporally differentiated protection plans (Panzeri et al., 2024). The search for codons under positive selection gave different results for the amino acid sequences of the two coding genes. In particular, the Cytb protein had only codons under purifying selection, whereas the COI protein had four codons under purifying selection and two codons under positive selection: codon 4 under episodic positive selection and codon 207 under pervasive positive selection. The codon 4 is located at the N-terminal end, within the inner mitochondrial membrane, where the OXPHOS protein complexes promote the translocation of protons from the mitochondrial matrix to the mitochondrial intermembrane space. In the codon 4 is present a Tyrosine (Y) which is a polar and aromatic amino acid whose aromatic side chain could interacts with other aromatic chains (stacking interactions), whereas its reactive hydroxyl group could be involved in interactions with non-carbon atoms (Betts and Russel, 2003). It is clear that the chemical properties of this amino acid, in the position it occupies, have been favored by positive episodic selection for their functional implications. In the codon 207, under pervasive positive selection, located in the loop region of the intermembrane space, there is a Proline (P), a small amino acid whose peculiar and unique structure, forming a ring, make it often present at the point where the protein chain needs to change direction (Betts and Russel, 2003), like the loop region under consideration.
The interpretation of the above results cannot disregard the crucial role played by the subunits of complex IV along the electron transmission chain (ETC) in the process of oxidative phosphorylation. The OXPHOS system produces, within the cristae of the inner mitochondrial membrane, the majority of chemical energy and the Cytochrome c Oxidase, the last complex of the ETC, plays a central role in the control of proton pumping efficiency, ATP and reactive oxygen species production (Kadenbach, 2021). Within complex IV, the COI subunit, is the core where oxygen reduction to H2O occurs, which is coupled to proton translocation across the inner membrane (Kadenbach and Hüttemann, 2015). Given this important function, it is not surprising that purifying selection is the most common form of selection acting on OXPHOS genes, and on cytochrome c oxidase in particular. However, there is also evidence that the OXPHOS genes are responsive to selective pressures exerted by variations in environmental parameters. These generally result in variable energy requirements in organisms adapting to new environmental conditions (Sebastian et al., 2020). For example, there is evidence for adaptation of mitochondrial OXPHOS genes in teleosts with different swimming performance (Zhang and Broughton, 2015) or in neotropical freshwater batoids during the transition from saltwater to freshwater environments (Nachtigall et al., 2023). The presence of codons under positive selection in the N-terminal end and in the loop region of mitochondrial intermembrane space was also detected in the killifish Aphanius fasciatus (Pappalardo et al., 2024) confirming the functional importance of the codons in this region. However, we have not considered the functional implications of the positively selected codons in the context of the OXPHOS pathway, partly due to the high complexity of interactions between OXPHOS enzymes, which is beyond the scope of this study.
5 Conclusion
Our findings, although obtained from a relatively low sample size of red mullet populations, allowed us to highlight that: i) the genetic structure of M. barbatus as revealed by sequence analysis of COI and Cytb genes, partially confirms the results of previous investigation based on microsatellites. These results support the need to manage red mullet populations as separate sub-populations with distinct gene pools; ii) the populations studied show an excess of private haplotypes, almost all of which are derived from a few highly frequent haplotypes in each gene; these results indicate that red mullet populations are overexploited and may face loss of adaptive capacity; iii) the positive selection found to act on COI of M. barbatus need to be implemented by considering the mtDNA protein-encoded of all OXPHOS complexes to explore the relationship between the site under selection and the environmental variables.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because Ethical review and approval were waived for this study due the research did not involve any kind of experimentation on fish.
Author contributions
GC: Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. MM: Methodology, Software, Writing – review & editing. TS-B: Formal Analysis, Writing – review & editing. VF: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. AP: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the University of Catania: PIACERI linea 2, 2022.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1577491/full#supplementary-material
Supplementary Figure 1Phylogenetic trees showing relationships among COI Mullus barbatus haplotypes under Maximum Likelihood (ML; top) and Bayesian Inference (BI, bottom) frameworks. Bootstrap support values for the ML analysis are displayed at nodes showing a support of 70% or higher. BI Posterior probabilities are displayed at nodes showing support ≥ 0.95.
Supplementary Figure 2Phylogenetic trees showing relationships among Cytb Mullus barbatus haplotypes under Maximum Likelihood (ML; top) and Bayesian Inference (BI, bottom) frameworks. Bootstrap support values for the ML analysis are displayed at nodes showing a support of 70% or higher. BI Posterior probabilities are displayed at nodes showing support ≥ 0.95.
References
1
Arculeo M. Brutto S. L. Cammarata M. Scalisi M. Parrinello N. (1999). Genetic variability of the Mediterranean Sea red mullet, Mullus barbatus (Pisces, Mullidae). Russ J. Genet.35, 292–296.
2
Bandelt H. J. Forster P. Rohl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol.16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
3
Betts M. J. Russel R. B. (2003). “Amino acid properties and consequences of substitutions,” in Bioinformatics for geneticists. Eds. BarnesM. R.GrayI. C. (The Atrium, Southern Gate, Chichester, West Sussex PO19 8SQ, England: John Wiley & Sons, Ltd), 289–314.
4
Brand J. A. Garcia-Gonzalez F. Dowling D. K. Wong B. B. (2024). Mitochondrial genetic variation as a potential mediator of intraspecific behavioural diversity. Tree39, 199–212. doi: 10.1016/j.tree.2023.09.009
5
Brooks S. P. Gelman A. (1998). General methods for monitoring convergence of iterative simulations. J. Comput. graph Stat.7, 434–455. doi: 10.1080/10618600.1998.10474787
6
Consuegra S. John E. Verspoor E. de Leaniz C. G. (2015). Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel Evol.47, 1–10. doi: 10.1186/s12711-015-0138-0
7
Darriba D. Taboada G. L. Doallo R. Posada D. (2012). jModelTest 2: more models, new heuristics and high-performance computing. Nat. Methods9, 772. doi: 10.1038/nmeth.2109
8
DeLano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr.40, 82–92.
9
Efremov R. G. Sazanov L. A. (2011). Structure of the membrane domain of respiratory complex I. Nature.476, 414–420. doi: 10.1038/nature10330
10
Excoffier L. Lischer H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res.10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
11
Excoffier L. Smouse P. E. Quattro J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics131.2, 479–491. doi: 10.1093/genetics/131.2.479
12
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
13
Foote A. D. Morin P. A. Durban J. W. Pitman R. L. Wade P. Willerslev E. et al . (2011). Positive selection on the killer whale mitogenome. Biol. Lett.7 (1), 7116–7118. doi: 10.1098/rsbl.2010.0638
14
Fu Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics147, 915–925. doi: 10.1093/genetics/147.2.915
15
Galarza J. A. Turner G. F. Macpherson E. Rico C. (2009). Patterns of genetic differentiation between two co-occurring demersal species: the red mullet (Mullus barbatus) and the striped red mullet (Mullus surmuletus). Can. J. Fish Aquat Sci.66, 1478–1490. doi: 10.1139/F09-098
16
Garoia F. Guarniero I. Piccinetti C. Tinti F. (2004). First microsatellite loci of red mullet (Mullus barbatus) and their application to genetic structure analysis of Adriatic shared stock. Mar biotech.6, 446–452. doi: 10.1007/s10126-004-3045-x
17
Garvin M. R. Bielawski J. P. Sazanov L. A. Gharrett A. J. (2015). Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J. Zool Syst. Evol. Res.53, 1–17. doi: 10.1111/jzs.12079
18
Gelman A. Rubin D. (1992). Inference from iterative simulation using multiple sequences. Stat. Sci.7 (4), 457–472. doi: 10.1214/ss/1177011136
19
Hall T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Ser.41, 95–98.
20
Ivanova N. V. Zemlak T. S. Hanner R. H. Hebert P. D. N. (2007). Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Resour.7, 544–548. doi: 10.1111/j.1471-8286.2007.01748.x
21
Jacobsen M. W. Da Fonseca R. R. Bernatchez L. Hansen M. M. (2016). Comparative analysis of complete mitochondrial genomes suggests that relaxed purifying selection is driving high nonsynonymous evolutionary rate of the NADH2 gene in whitefish (Coregonus ssp.). Mol. Phyl Evol.95, 161–170. doi: 10.1016/j.ympev.2015.11.008
22
Kadenbach B. (2021). The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion58, 296–302. doi: 10.1016/j.mito.2020.10.004
23
Kadenbach B. Hüttemann M. (2015). The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion24, 64–76. doi: 10.1016/j.mito.2015.07.002
24
Katoh K. Rozewicki J. Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform.20, 1160–1166. doi: 10.1093/bib/bbx108
25
Kosakovsky Pond S. L. Frost S. D. W. (2005). Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol.22, 1208–1222. doi: 10.1093/molbev/msi105
26
Kozlov A. M. Darriba D. Flouri T. Morel B. Stamatakis A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics35, 4453–4455. doi: 10.1093/bioinformatics/btz305
27
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. doi: 10.1093/molbev/msy096
28
Lajbner Z. Pnini R. Camus M. F. Miller J. Dowling D. K. (2018). Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci. Rep.8, 9500. doi: 10.1038/s41598-018-27805-3
29
Lakner C. van der Mark P. Huelsenbeck J. Larget B. Ronquist F. (2008). Efficiency of Markov chain Monte Carlo tree proposals in Bayesian phylogenetics, Syst. Biol.57, 86–103. doi: 10.1080/10635150801886156
30
Lomize M. A. Lomize A. L. Pogozheva I. D. Mosberg H. I. (2006). OPM: orientations of proteins in membranes database. Bioinformatics22, 623–625. doi: 10.1093/bioinformatics/btk023
31
Maggio T. Lo Brutto S. Garoia F. Tinti F. Arculeo M. (2009). Microsatellite analysis of red mullet Mullus barbatus (Perciformes, Mullidae) reveals the isolation of the Adriatic Basin in the Mediterranean Sea. ICES J. Mar Sci.66, 1883–1891. doi: 10.1093/icesjms/fsp160
32
Mamuris Z. Apostolidis A. P. Triantaphyllidis C. (1998). Genetic protein variation in red mullet (Mullus barbatus) and striped red mullet (M. surmuletus) populations from the Mediterranean Sea. Mar Biol.130, 353–360. doi: 10.1007/s002270050255
33
Matić-Skoko S. Šegvić-Bubić T. Mandić I. Izquierdo-Gomez D. Arneri E. Carbonara P. et al . (2018). Evidence of subtle genetic structure in the sympatric species Mullus barbatus and Mullus surmuletus (Linnaeu) in the Mediterranean Sea. Sci. Rep.8, 676. doi: 10.1038/s41598-017-18503-7
34
Murrell B. S. Mabona A. Weighill T. Sheward D. Kosakovsky Pond S. L. Scheffler K. (2013). FUBAR: A fast, unconstrained bayesian appRoximation for inferring selection. Mol. Biol. Evol.30, 1196–1205. doi: 10.1093/molbev/mst030
35
Murrell B. Wertheim J. O. Moola S. Weighill T. Scheffler K. Kosakovsky Pond S. L. (2012). Detecting individual sites subject to episodic diversifying selection. PloS Genet.8, e1002764. doi: 10.1371/journal.pgen.1002764
36
Nachtigall P. G. Loboda T. S. Pinhal D. (2023). Signatures of positive selection in the mitochondrial genome of neotropical freshwater stingrays provide clues about the transition from saltwater to freshwater environment. Mol. Genet. Genomics298, 229–241. doi: 10.1007/s00438-022-01977-0
37
Panzeri D. Reale M. Cossarini G. Salon S. Carlucci R. Spedicato M. T. et al . (2024). Future distribution of demersal species in a warming Mediterranean sub-basin. Front. Mar11. doi: 10.3389/fmars.2024.1308325
38
Pappalardo A. M. Calogero G. S. Šanda R. Giuga M. Ferrito V. (2024). Evidence for selection on mitochondrial OXPHOS genes in the mediterranean killifish aphanius fasciatus valencienne. Biology.13, 212. doi: 10.3390/biology13040212
39
Pappalardo A. M. Federico C. Sabella G. Saccone S. Ferrito V. (2015). A COI nonsynonymous mutation as diagnostic tool for intraspecific discrimination in the european anchovy engraulis encrasicolus (Linnaeus). PloS One10, e0143297. doi: 10.1371/journal.pone.0143297
40
Reñones O. Massuti E. Morales-Nin B. (1995). Life history of the red mullet Mullus surmuletus from the bottom-trawl fishery off the Island of Majorca (north-west Mediterranean). Mar Biol.123, 411–419. doi: 10.1007/BF00349219
41
Ronquist F. Teslenko M. van der Mark P. Ayres D. L. Darling A. Höhna S. et al . (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol.61, 539–542. doi: 10.1093/sysbio/sys029
42
Rozas J. Ferrer-Mata A. Sanchez-DelBarrio J. C. Guirao-Rico S. Librado P. Ramos-Onsins S. E. et al . (2017). DnaSP 6:DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol.34, 3299–3302. doi: 10.1093/molbev/msx248
43
Ruiz-Pesini E. Mishmar D. Brandon M. Procaccio V. Wallace D. C. (2004). Effects of purifying and adaptive selection on regional variation in human mtDNA. Science303, 223–226. doi: 10.1126/science.1088434
44
Saraste M. (1999). Oxidative Phosphorylation at the fin de siècle. Science283, 1488–1493. doi: 10.1126/science.283.5407.1488
45
Sayers E. W. Beck J. Bolton E. E. Brister J. R. Chan J. Comeau D. C. et al .2024Database resources of the national center for biotechnology information. Nucleic Acids Res.52, D33–D43. doi: 10.1093/nar/gkad1044
46
Sebastian W. Sukumaran S. Gopalakrishnan A. (2020). Signals of adaptive mitogenomic evolution in an indigenous Cichlid, Etroplus suratensis (Bloc) from India. Regional Stud. Marine Sci.40, 101546. doi: 10.1016/j.rsma.2020.101546
47
Sevilla R. G. Diez A. Norén M. Mouchel O. Jérome M. Verrez-Bagnis V. et al . (2007). Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol. Ecol. Notes.7, 730–734. doi: 10.1111/j.1471-8286.2007.01863.x
48
Silva G. Lima F. P. Martel P. Castilho R. (2014). Thermal adaptation and clinal mitochondrial DNA variation of european anchovy. Proc. R. Soc B.281, 20141093. doi: 10.1098/rspb.2014.1093
49
Sokolova I. (2018). Mitochondrial adaptations to variable environments and their role in animals’ Stress tolerance. Integr. Comp. Biol.58, 519–531. doi: 10.1093/icb/icy017
50
Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics123, 585–595. doi: 10.1093/genetics/123.3.585
51
Teske P. R. Sandoval-Castillo J. Golla T. R. Emami-Khoyi A. Tine M. Von Der Heyden S. et al . (2019). Thermal selection as a driver of marine ecological speciation. Proc. R. Soc. B.286, 20182023. doi: 10.1098/rspb.2018.2023
52
Tserpes G. Fiorentino F. Levi D. Cau A. Murenu M. Zamboni A. D. A. et al . (2002). Distribution of mullus barbatus and M. surmuletus (Osteichthyes: Perciformes) in the Mediterranean continental shelf: implications for management. Sci. Mar.66, 39–54. doi: 10.3989/scimar.2002.66s239
53
Waterhouse A. Bertoni M. Bienert S. Studer G. Tauriello G. Gumienny R. et al . (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res.46, W296–W303. doi: 10.1093/nar/gky427
54
Weir B. S. Cockerham C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution38 (6), 1358–1370. doi: 10.2307/2408641
55
Whitehead P. J. P. Bauchot M. L. Hureau J. C. Nielsen J. Tortonese E. (1984). Fishes of the north-eastern atlantic and the mediterranean (Bungay UK: UNESCO).
56
Zhang F. Broughton R. E. (2015). Heterogeneous natural selection on oxidative phosphorylation genes among fishes with extreme high and low aerobic performance. BMC evolutionary Biol.15, 1–15. doi: 10.1186/s12862-015-0453-7
57
Zhou T. Shen X. Irwin D. M. Shen Y. Zhang Y. (2014). Mitogenomic analyses propose positive selection in mitochondrial genes for high-altitude adaptation in galliform birds. Mitochondrion18, 70–75. doi: 10.1016/j.mito.2014.07.012
58
Zlateva I. Raykov V. Alexandrova A. Ivanova P. Chipev N. Stefanova K. et al . (2023). Effects of anthropogenic and environmental stressors on the current status of red mullet (Mullus barbatus L) populations inhabiting the Bulgarian Black Sea waters. Nat. Conserv.54, 55–79. doi: 10.3897/natureconservation.54.103758
Summary
Keywords
positive selection, OXPHOS genes, red mullet, Mediterranean, COI, Cytb
Citation
Calogero GS, Mancuso M, Segvic-Bubic T, Ferrito V and Pappalardo AM (2025) OXPHOS genes analysis in the red mullet (Mullus barbatus Linnaeus, 1758). Front. Mar. Sci. 12:1577491. doi: 10.3389/fmars.2025.1577491
Received
15 February 2025
Accepted
07 April 2025
Published
29 April 2025
Volume
12 - 2025
Edited by
Ioannis A Giantsis, Aristotle University of Thessaloniki, Greece
Reviewed by
David Osca, University of Las Palmas de Gran Canaria, Spain
Khaled Mohammed Geba, Menoufia University, Egypt
Updates
Copyright
© 2025 Calogero, Mancuso, Segvic-Bubic, Ferrito and Pappalardo.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Maria Pappalardo, pappalam@unict.it
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.