In the published article, there was an error. Due to an oversight, the newly isolated phage M1 was incorrectly named “vB_PsaP_M1” in the original article. To more accurately reflect its morphology and host specificity, the correct designation should be “vB_PaeM_M1”. All the instances of the incorrect name “vB_PsaP_M1” have been corrected with “vB_PaeM_M1” in the article title, in the legends for Figures 1–7 (12 instances), in Supplementary Tables 1, 3–5 and in Supplementary Image 1, in Abstract (4 instances), Introduction (3 instances), Materials and methods (9 instances), Results and discussion (36 instances), and Conclusion (2 instances). Please note that this naming correction does not affect any of the study’s results or conclusions, only the phage’s designation was incorrect. We sincerely apologize for this error.
Figure 1
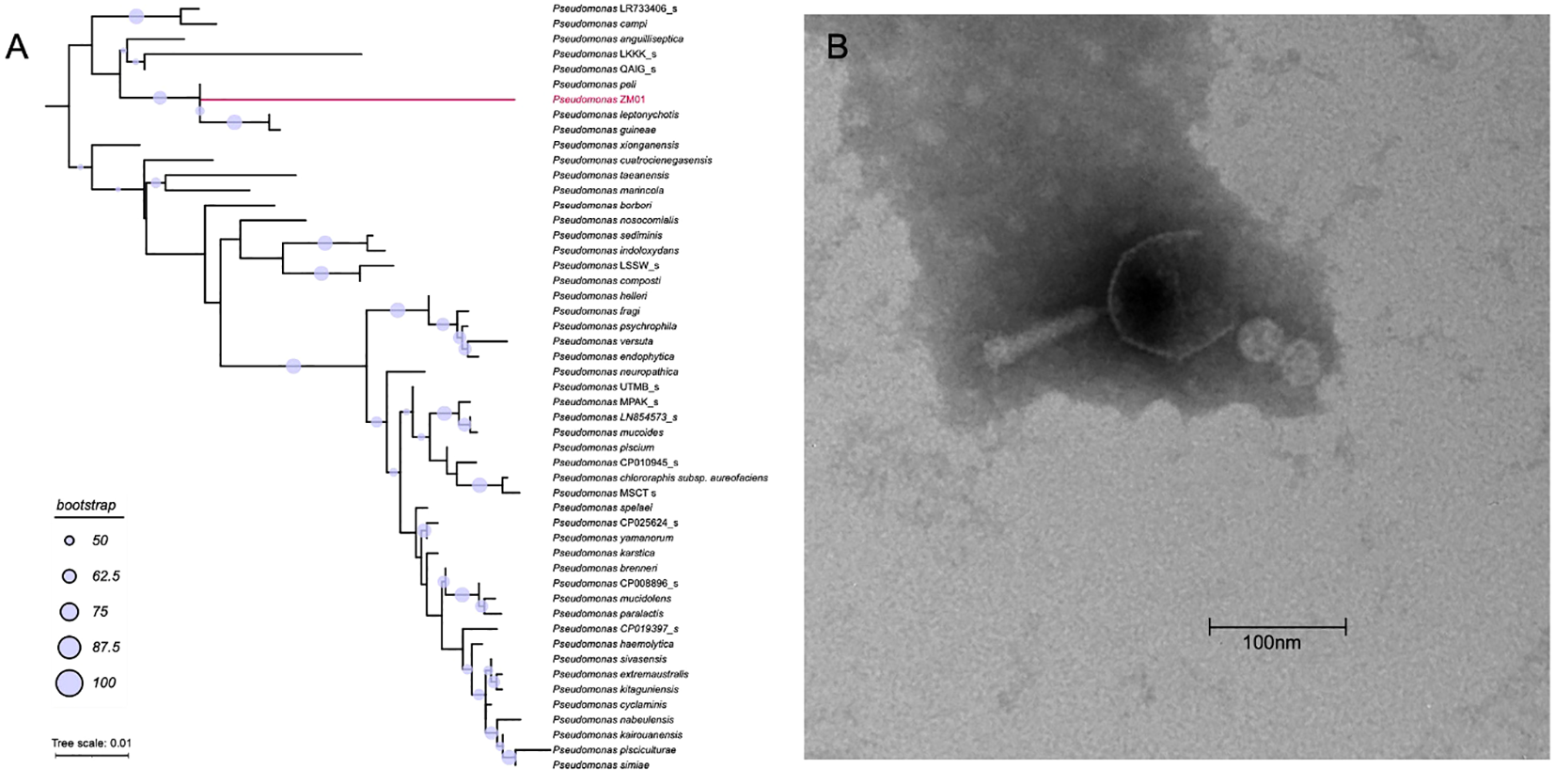
(A) The phylogenic tree based on the 16S rRNA gene of Pseudomonas ZM01 and other 120 reference 16S rRNA gene sequences of Pseudomonas ZM01. (B) Transmission electron micrograph of vB_PaeM_M1.
Figure 2
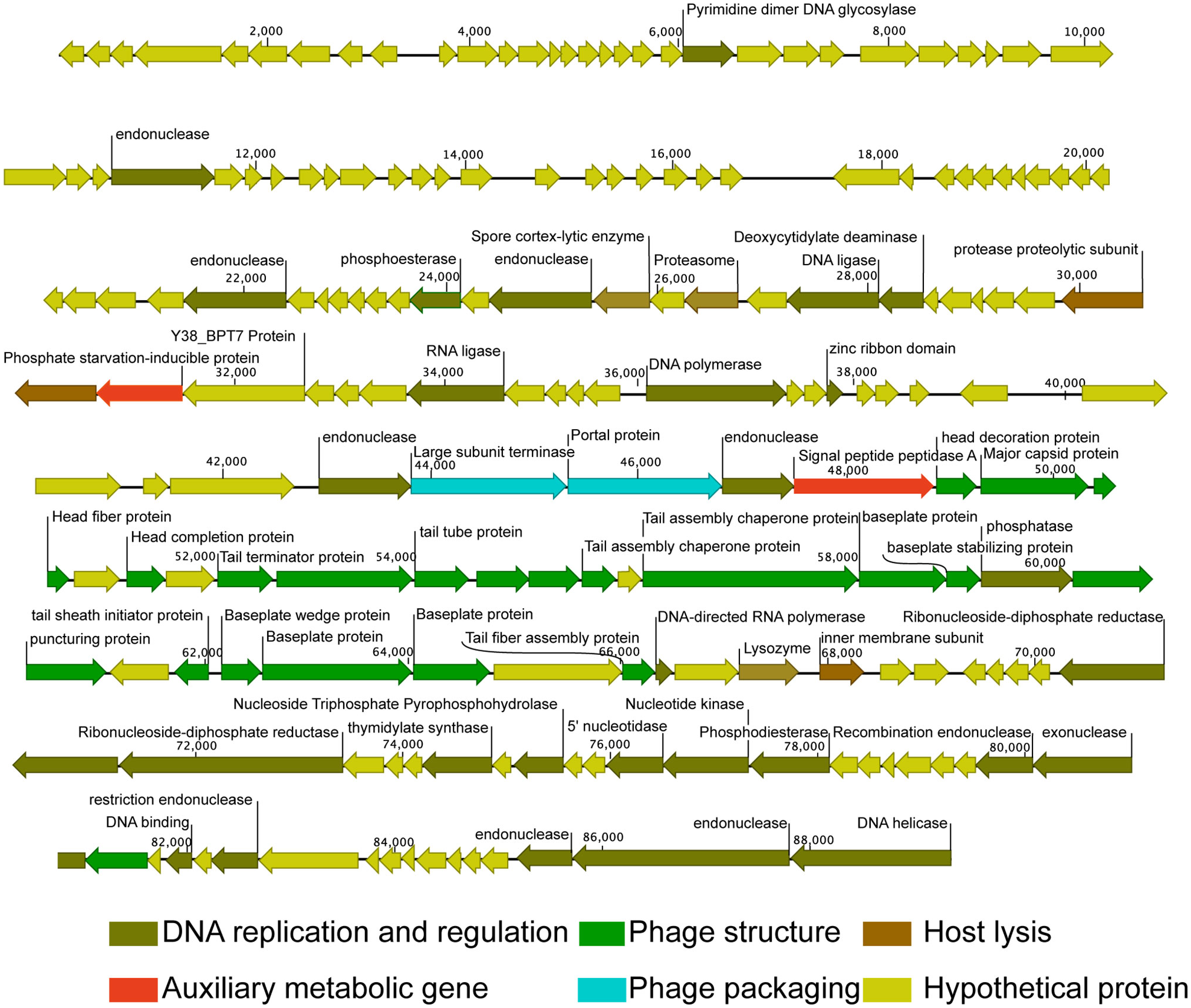
Genome map of Pseudomonas phage vB_PaeM_M1. Putative functional categories were defined according to annotation and are represented by different colors. The length of each arrow represents the length of each gene.
Figure 3
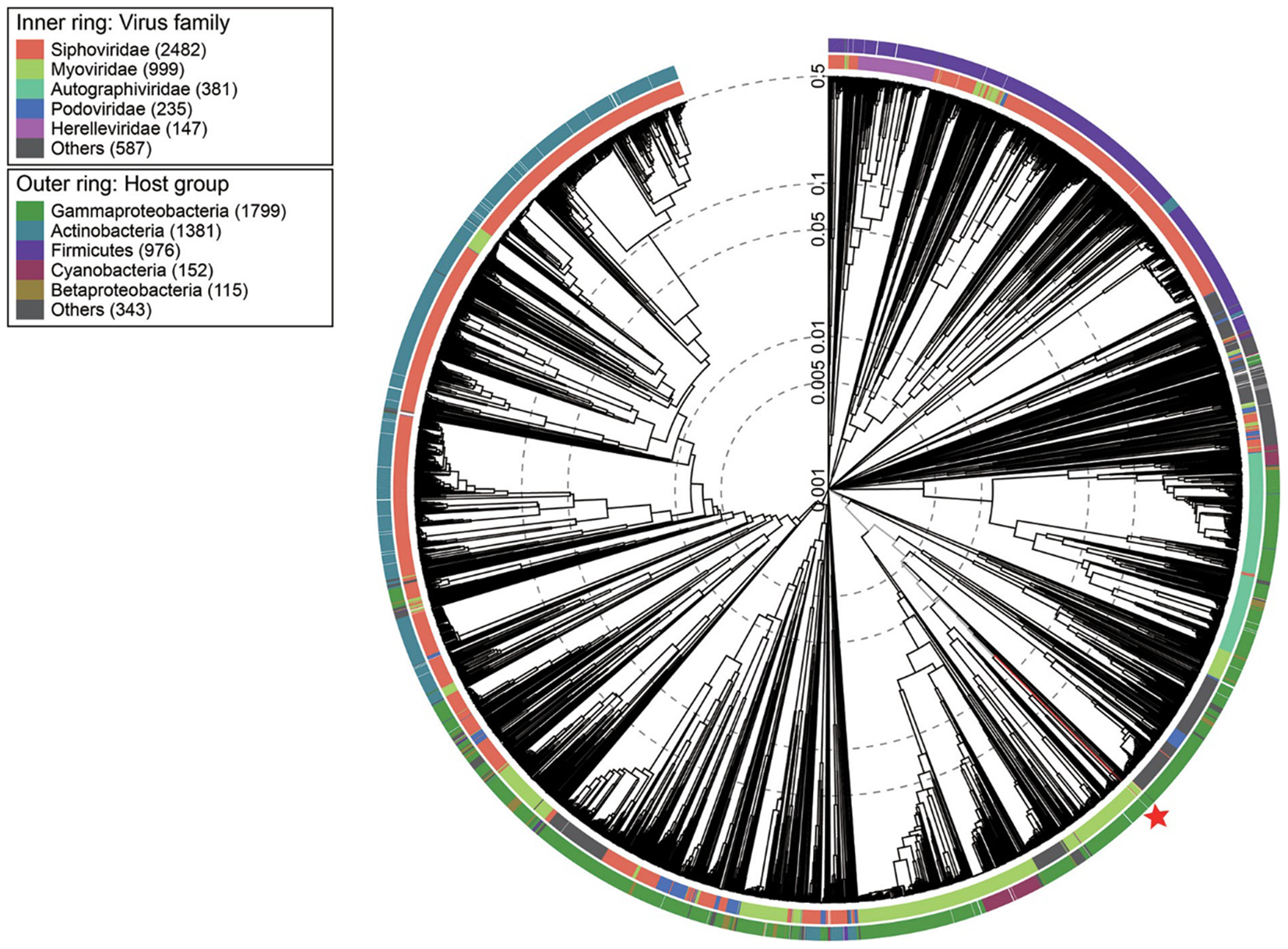
Viral phylogenetic tree of whole phage vB_PaeM_M1 genomes represented in the circular view. The colored rings represent the host groups (outer ring) and virus families (inner ring). These trees are calculated by BIONJ according to the genome distance matrix and take the midpoint as the root.
Figure 4
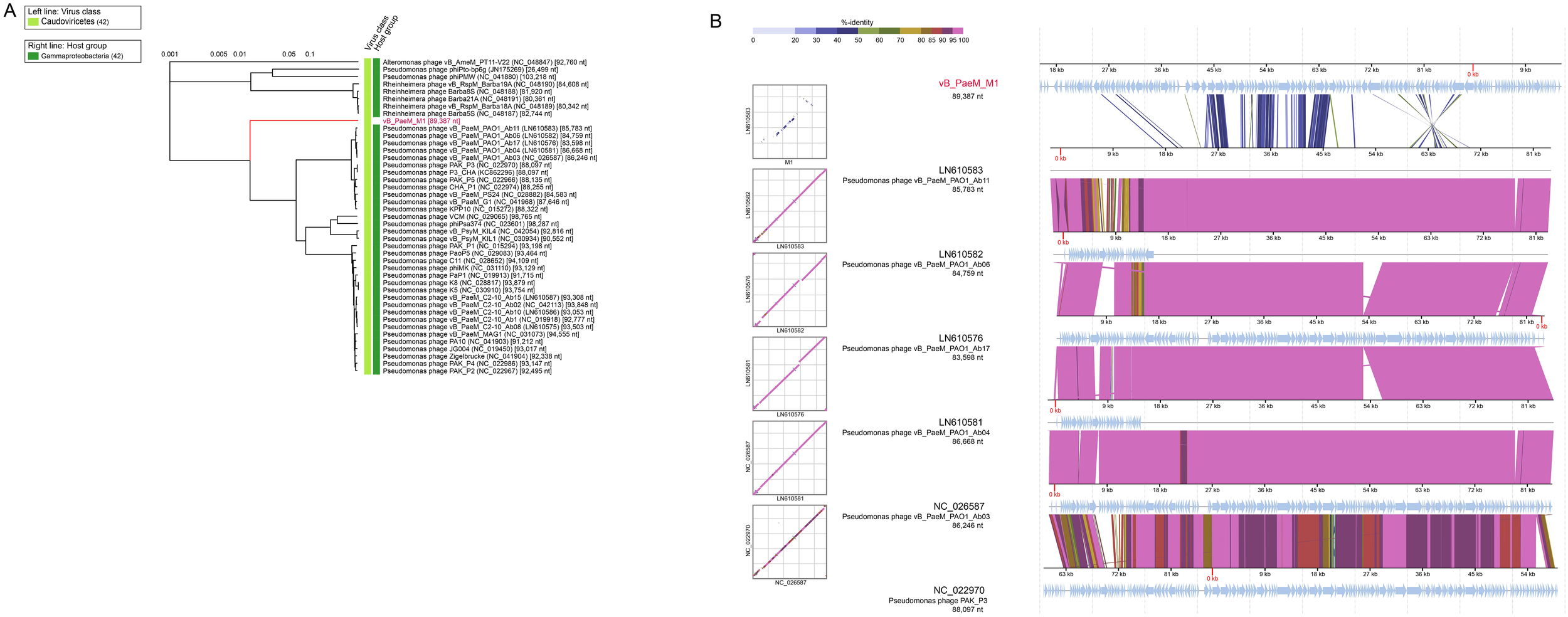
Phylogenetic and comparative genomic analyses of Pseudomonas phage vB_PaeM_M1. (A) Phylogenetic tree of Pseudomonas phage vB_PaeM_M1 and the 36 closest virus genomes. (B) Genomic comparisons between Pseudomonas phage vB_PaeM_M1 and vB_PaeM_M1-like viruses. The shading below each genome indicates sequence similarities between the genomes, with different colors representing the levels of similarity.
Figure 5
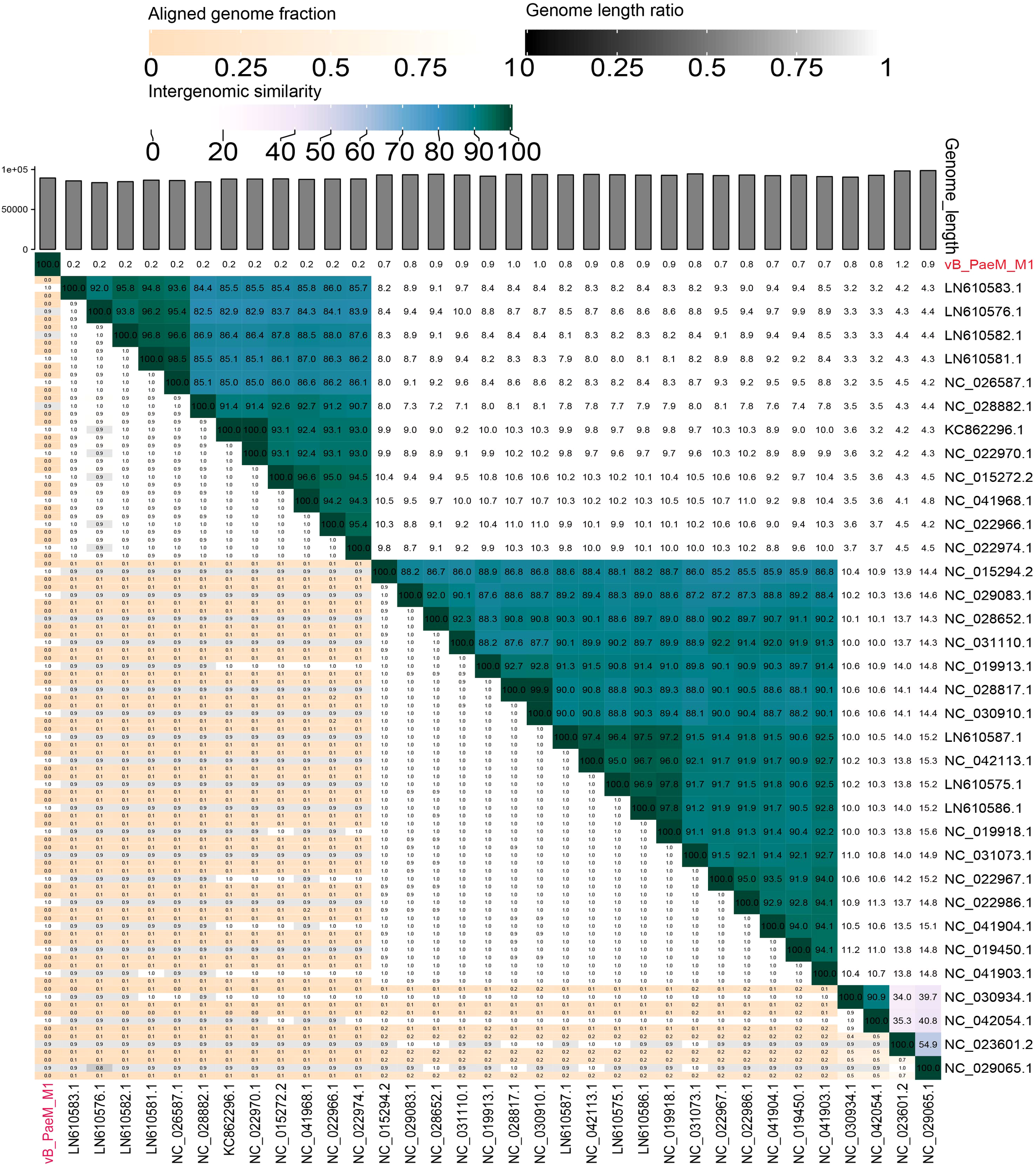
Shared gene heat map of vB_PaeM_M1 and vB_PaeM_M1 like virus in rectangular phylogenetic tree. In the right half, color coding allows rapid visualization of phage genome clustering based on intergenic similarity: the closer the relationship between genomes, the darker the color. These numbers represent the similarity values of each genome pair. In the left half, the three indicator values of each genome pair are expressed in order from top to bottom: aligned partial genome 1 (for the genome found in this row), genome length ratio (for two genomes in this pair), and aligned partial genome 2 (for the genome found in this column). Darker colors emphasize lower values, indicating that there is only a small number of genome aligned genome pairs (orange to white gradient), or there is a large difference in the length of the two genomes (black to white gradient). As the distance between phages increases, the aligned genomic fragments are expected to decrease.
Figure 6
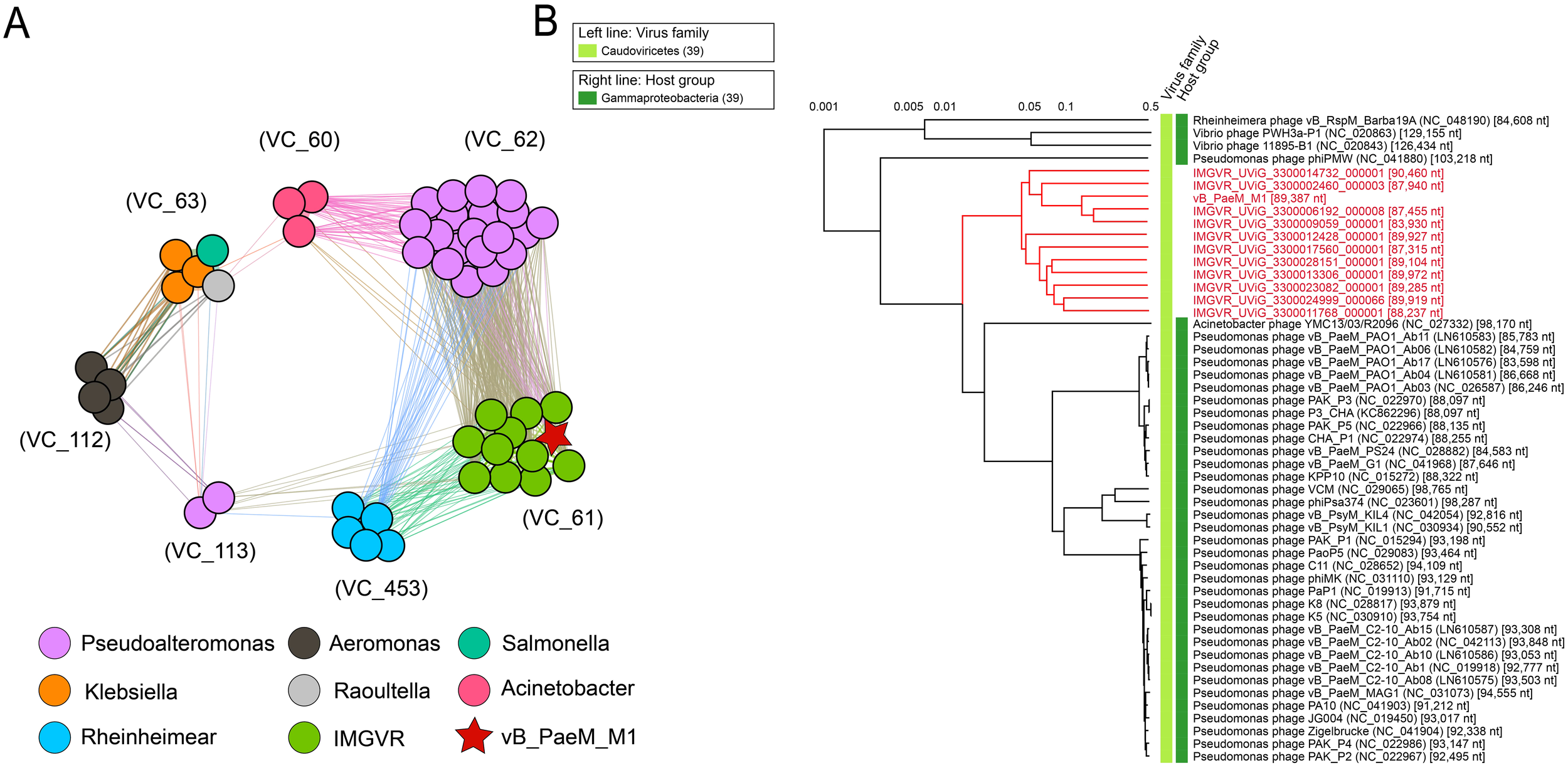
(A) vB_PaeM_M1, virus from NCBI RefSeq database, and related environmental viral sequences from Integrated Microbial Genomes/Virus (IMG/VR). Different colors of nodes represent different kinds of viruses. Edges represent similarity scores between genomes. VCs are generated by vcontact2, and the network is visualized by gephi version 0.9.2. (B) Whole-genome-based phylogenetic tree constructed by vB_PaeM_M1, metagenomic assembled uncultured viral genomes (UViGs) and isolated viruses. The members in VC_61 are shown in red.
Figure 7
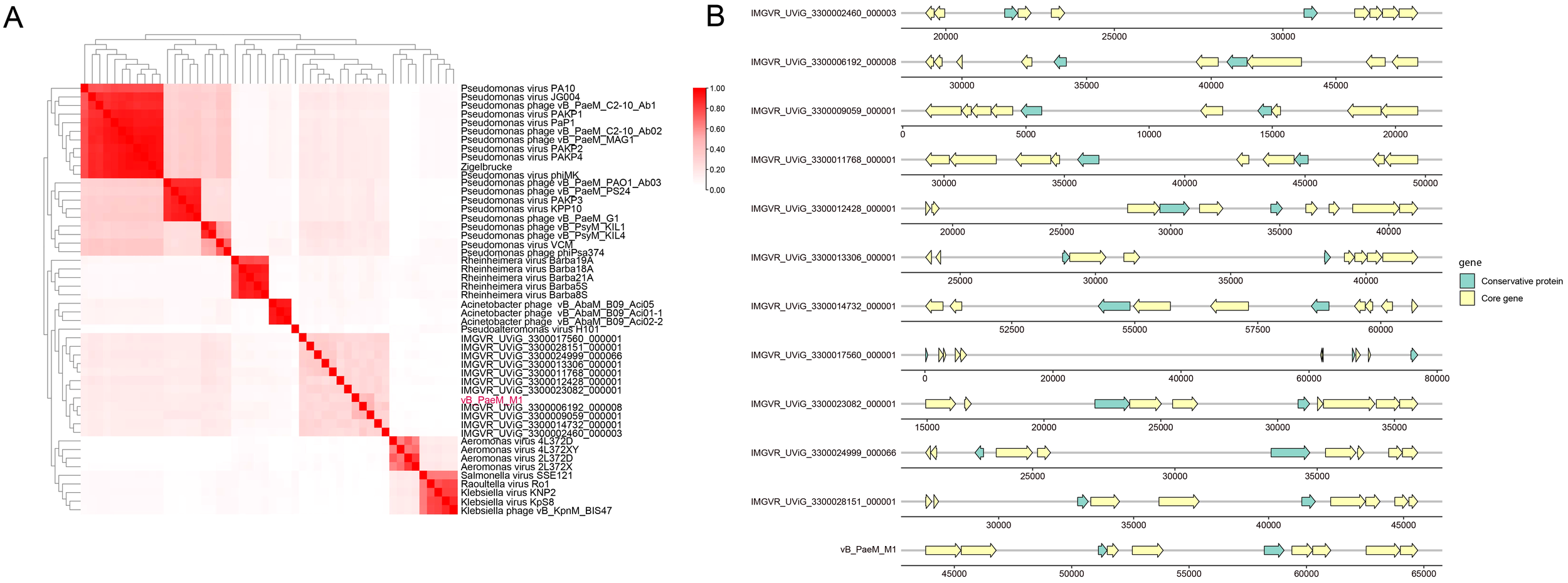
(A) Heat map of gene sharing rate between vB_PaeM_M1 and other phages. The sharing rate is displayed by color depth. (B) The gene map of VC_61. Core genomes are represented by yellow arrows and the conservative protein are represented by green arrows.
In the published article, there was an error in Figures 4–7 as published. “vB_PsaP_M1” should be changed with “vB_PaeM_M1”. The corrected Figures 4–7 and their captions appear below.
A correction has been made to Data availability statement. This sentence previously stated:
“The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.”
The corrected Data availability statement appears below:
“The complete genome sequence of Pseudomonas phage vB_PaeM_M1 is available in the GenBank database under accession number OP714164. In addition, the 16S rRNA gene sequence of Pseudomonas ZM01 has also been deposited in NCBI under accession number OP788125.”
The authors apologize for the errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Statements
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Summary
Keywords
bacteriophage, Pseudomonas , genomic and comparative genomic analysis, phylogenetic analysis, Psaeviridae
Citation
Ren L, Liu Y, Liang Y, Liu B, McMinn A, Zheng K, Wang Z, Wang H, Shao H, Sung YY, Mok WJ, Wong LL and Wang M (2025) Corrigendum: Characterization and genomic analysis of a novel Pseudomonas phage vB_PaeM_M1, representing a new viral family, Psaeviridae. Front. Mar. Sci. 12:1580141. doi: 10.3389/fmars.2025.1580141
Received
20 February 2025
Accepted
19 March 2025
Published
24 April 2025
Volume
12 - 2025
Edited and reviewed by
Hao Chen, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2025 Ren, Liu, Liang, Liu, McMinn, Zheng, Wang, Wang, Shao, Sung, Mok, Wong and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yantao Liang, liangyantao@ouc.edu.cn; Baohong Liu, gsyylbh@163.com; Min Wang, mingwang@ouc.edu.cn
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.