- 1State Key Laboratory of Marine Environmental Science, Fujian Key Laboratory of Marine Carbon Sequestration, College of Ocean and Earth Sciences, Xiamen University, Xiamen, Fujian, China
- 2Key Laboratory of Tropical Marine Ecosystem and Bioresource, Fourth Institute of Oceanography, Ministry of Natural Resources, Beihai, Guangxi, China
- 3Weihai Changqing Ocean Science Technology CO., Ltd., Weihai, Shandong, China
- 4Institute of Marine Biology and Pharmacology, Ocean College, Zhejiang University, Zhoushan, Zhejiang, China
- 5Donghai Laboratory, Zhoushan, Zhejiang, China
- 6Laboratory of Beibu Gulf Ocean Big Data Application, Fourth Institute of Oceanography, Ministry of Natural Resources, Beihai, Guangxi, China
- 7Institute of Biotechnology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
Marine microorganisms play a vital role in aquaculture by influencing water quality and the health of cultured species. Relative to conventional mono-species aquaculture, integrated multi-trophic aquaculture is more environmentally friendly and economically sustainable. However, the temporospatial dynamics of the microbial community in different aquaculture modes and their roles in the seasonal diseases of aquacultural organisms are largely unknown. Here, the seawater bacterial communities and potential pathogens were investigated over thirteen months in three different aquaculture types in Sanggou Bay, a typical maricultural area with nearly 60 years of history. Seasonality played a primary role, while aquaculture type played a subtle role in shaping the bacterioplankton community. Diverse potential pathogens were identified, and r-strategist bacteria, most of which were opportunistic pathogens and typified by Vibrio species, increased and dominated in late summer and autumn, a period of high-risk disease outbreaks in mariculture organisms. Network analyses indicated that the increase in r-strategist bacteria reduced the complexity of microbial interactions, and Vibrio halioticoli (OTU1389), a hub species with extensive positive correlations to other Vibrio OTUs, likely contributed to the increase of pathogenic vibrios due to its macroalgae degradation capability. The major potential pathogenic Vibrio OTUs were further corroborated by genome analyses and environmental simulation experiments using pathogenic Vibrio isolates, including two new species. Moreover, we found seawater dissolved oxygen, temperature, and transparency correlated significantly with the bacterioplankton temporospatial dynamics and the seasonal outbreaks of bacterial pathogens. Our results emphasize the necessity of synchronically monitoring the bacterial community, its pathogens-stimulating species, such as V. halioticoli and other macroalgae-degrading bacteria, and critical physicochemical parameters for predicting and preventing pathogen outbreaks in mariculture.
1 Introduction
Aquaculture is becoming an increasingly important food source, particularly animal proteins and micronutrients (Naylor et al., 2021). Integrated multi-trophic aquaculture (IMTA) is generally designed as a co-cultivation system of different species belonging to distinct trophic levels (Nissar et al., 2023). It has been advocated as a promising solution for sustainable aquaculture, achieving economic benefits and environmental friendliness (Chopin et al., 2001; Troell et al., 2009; Buck et al., 2018; Chang et al., 2020; Knowler et al., 2020). Numerous studies have shown the benefits of IMTA systems. For instance, Yokoyama (2013) showed that integrating oysters into sea cucumber farming significantly increased the sea cucumbers’ specific growth rates. Li et al. (2014) reported that polyculture of sea cucumbers, jellyfish, and shrimp improved total organic carbon utilization efficiency by 18.9% compared to monoculture of sea cucumbers. Nobre et al. (2010) found that IMTA increased the farm profits. Han et al. (2017) showed that oyster-seaweed co-culture turned the oyster farming seawater from a net CO2 source into a net CO2 sink. Generally, the IMTA systems enhance resource utilization and aquaculture production, improve environmental quality, reduce eutrophication, increase CO2 sequestration, and mitigate ocean acidification (Ridler et al., 2007; Han et al., 2013; Hadley et al., 2016; Mahmood et al., 2016a). IMTA has been widely developed in many countries, such as China, Canada, Chile, Australia, the USA, and Japan (Troell et al., 2003; Granada et al., 2016; Buck et al., 2018; Wartenberg et al., 2017).
However, the persistent threat of diseases remains a considerable obstacle to aquaculture (Aly and Fathi, 2024). Microbial infectious diseases cause substantial economic losses in the aquacultural industry, amounting to billions of US dollars annually (Lafferty et al., 2015; Mohamad et al., 2019; Patil et al., 2021; Vaiyapuri et al., 2021; Abd El-Hack et al., 2022; Rowley et al., 2024). Studies have reported that some bacterial pathogens are the main causes responsible for disease outbreaks, leading to massive mortality in cultivated fish, shellfish, and shrimps (Toranzo et al., 2005; Sawabe et al., 2007; Janda, 2014; Travers et al., 2015; Yang et al., 2021). Moreover, some aquaculture-originated bacteria are also human pathogens, causing foodborne or direct-contact infections (Baker-Austin et al., 2018). Besides, due to the widespread use of antibiotics in aquaculture, the appearance of antibiotic-resistant pathogens makes disease prevention and control more difficult (Cabello et al., 2016; Preena et al., 2020; Algammal et al., 2023).
The case of Sanggou Bay, with its approximately 60-year history of aquaculture (Li et al., 2018) and an annual production of over 240,000 tons of seafood from a 100 km2 aquaculture area, demonstrates the potential of IMTA (Fang et al., 2016; Hossain et al., 2022). Since adopting the IMTA model in the late 1980s, Sanggou Bay has not only increased its economic profits but also improved its ecosystem functions, including enhanced carbon sequestration through macroalgae co-cultivation (Fang et al., 2016; Li et al., 2016). Nevertheless, seasonal mortality of farmed organisms, typically peaking in late summer and early autumn, remains challenging (Mao et al., 2006). In semi-enclosed bays, tidal currents govern coastal hydrodynamics (Zeng et al., 2015). Aquaculture occupies nearly 90% of Sanggou Bay’s water surface and extends well outside the bay (Fu et al., 2013). The high aquaculture density reduces current velocity, prolongs nutrient retention, and increases phytoplankton bloom risk and disease susceptibility in cultured species (Mao et al., 2006; Wang et al., 2018a). In 2019, the Pacific abalones (Haliotis discus hannai Ino) suffered mortality rates of 30%-60%, Zhikong scallops (Chlamys farreri) 10%-20%, and Yesso scallops (Patinopecten yessoensis) 10%-30%, with the highest mortality rates observed in late summer and early autumn. Additionally, diseases in cultured and wild seaweed started in May and became the most severe in September (data from local enterprises and farmers). Therefore, predicting pathogen outbreaks is particularly important for aquaculture and human health. As conventional culturing-based methods are less time-efficient or pathogen-specific, high-throughput methods are more suitable for pathogen detection.
Microbial communities have been proposed as one of the most critical indicators for microbial disease development in aquaculture systems (Bentzon-Tilia et al, 2016). Theoretically, microbial communities could depict the health status of the aquaculture environments and ecosystems, providing valuable information about the emergence of diseases (de Bruijn et al., 2018; Liu et al., 2020). Recent studies have indicated that long-term aquaculture could change the bacterial community structure (Liang et al., 2019; Zhang et al., 2019; Liu et al., 2020), and microbes could participate in some important physiological processes of cultured species in IMTA, such as influencing the immune response of fed species and regulating the metabolic and nutritional pathways (Bentzon-Tilia et al, 2016; Deng et al., 2019; Califano et al., 2020; Langlois et al., 2021; Zhang et al., 2022). However, few studies have investigated the link between pathogen population behaviors and microbial community dynamics in aquaculture (Fu et al., 2022; Zhang et al., 2022). Disease outbreaks remain a high risk in aquaculture, highlighting the necessity for a holistic understanding of the microbial community’s roles in diseases and comprehensive pathogen detection methods.
Although IMTA’s benefits have been reported to improve water quality, stimulate probiotics, and enhance disease control (Ying et al., 2018; Lee, 2019; Zhang et al., 2022; Stabili et al., 2023), contradictory reports have also appeared (Pietrak et al., 2012; Molloy et al., 2013; Tarnecki et al., 2025). We hypothesized that the community composition and microbial interactions therein may play important roles in determining the health status of IMTA systems, in addition to their critical roles in shaping the ecological functions and stability of the ecosystems (Nawaz et al., 2022). The current study investigated the monthly changes of bacterioplankton communities in three maricultural systems (i.e., shellfish monoculture, algae monoculture, and algae-shellfish co-culture) in Sanggou Bay. It also characterized potential bacterial pathogens through a comprehensive approach, including 16S rRNA gene-based community analyses, potential bacterial pathogen screening, environmental simulation experiments, and pathogen whole-genome analyses. The value of analyzing microbial communities and interactions in mechanistically predicting bacterial pathogens’ seasonal dynamics in mariculture environments was demonstrated.
2 Materials and methods
2.1 Environmental description and sampling strategy
Sanggou Bay is a shallow, semi-enclosed coastal bay in the Yellow Sea. As one of the largest aquaculture production areas in China, the environment of Sanggou Bay is influenced by land (in particular river discharges and anthropogenic pollutions), the Yellow Sea, internal processes such as pelagic-benthic exchange, and extensive aquaculture activities (Mahmood et al., 2016b). River discharges constitute the primary source of nutrient inputs in Sanggou Bay (Mahmood et al., 2016b), and Bacillariophyta and Dinophyta usually dominate the phytoplankton communities (Han et al., 2024). Tidal currents, especially their semi-diurnal constituents, dominate the Sanggou Bay hydrodynamics, contributing to water exchange with the Yellow Sea (Zeng et al., 2015). However, the water exchange rate decreases significantly in the upper water layers due to the frictional effect caused by the densely planted seaweeds and suspended aquaculture structures (shellfish cages and nets), which occupy nearly 2/3 of the area of the entire Sanggou Bay (Zeng et al., 2015). This reduction in water exchange could impact nutrient cycling, waste transport, and, consequently, potential eutrophication and phytoplankton bloom formation (Wang et al., 2018a). In addition, elevated anthropogenic activities such as agriculture and wastewater discharges further increase the ecological risks of eutrophication and algal blooms (Mahmood et al., 2016b).
Seawater samples were collected from Sanggou Bay to investigate the monthly bacterial communities and potential pathogens in different aquaculture systems (Table 1), including shellfish monoculture (Type I, density: 38 scallop/m2 or 12 abalone/m2), algae-shellfish co-culture (Type II, density: 38 scallop/m2 or 12 abalone/m2 for shellfish and 6 kelp/m2 or 0.3–2 kg Gracilaria/m2 for algae), and algae monoculture (Type III, density: 6 kelp/m2 or 0.3–2 kg Gracilaria/m2), where Type II belonged to the IMTA mode. The stocking densities were set based on local aquaculture practices. Based on the three distinct maricultural systems, seawater from four stations along a transect (from the nearshore to the offshore aquaculture sites) was sampled monthly from February 2019 to February 2020, including S1 (Type I), SA1 (Type II), A1 (Type III), and A2 (Type III). Two additional stations, AG1 (Type II) and AG2 (Type II), were sampled from July 2019 to November 2019 to provide more samples for late summer and autumn, when diseases in cultivated marine organisms were the most severe (Figure 1A; Table 1). Sampling was conducted during mid-tide periods to minimize variability caused by tidal fluctuations. The four sampling seasons were defined as spring (March, April, and May), summer (June, July, and August), autumn (September, October, and November), and winter (December, January, and February) (Han et al., 2024).
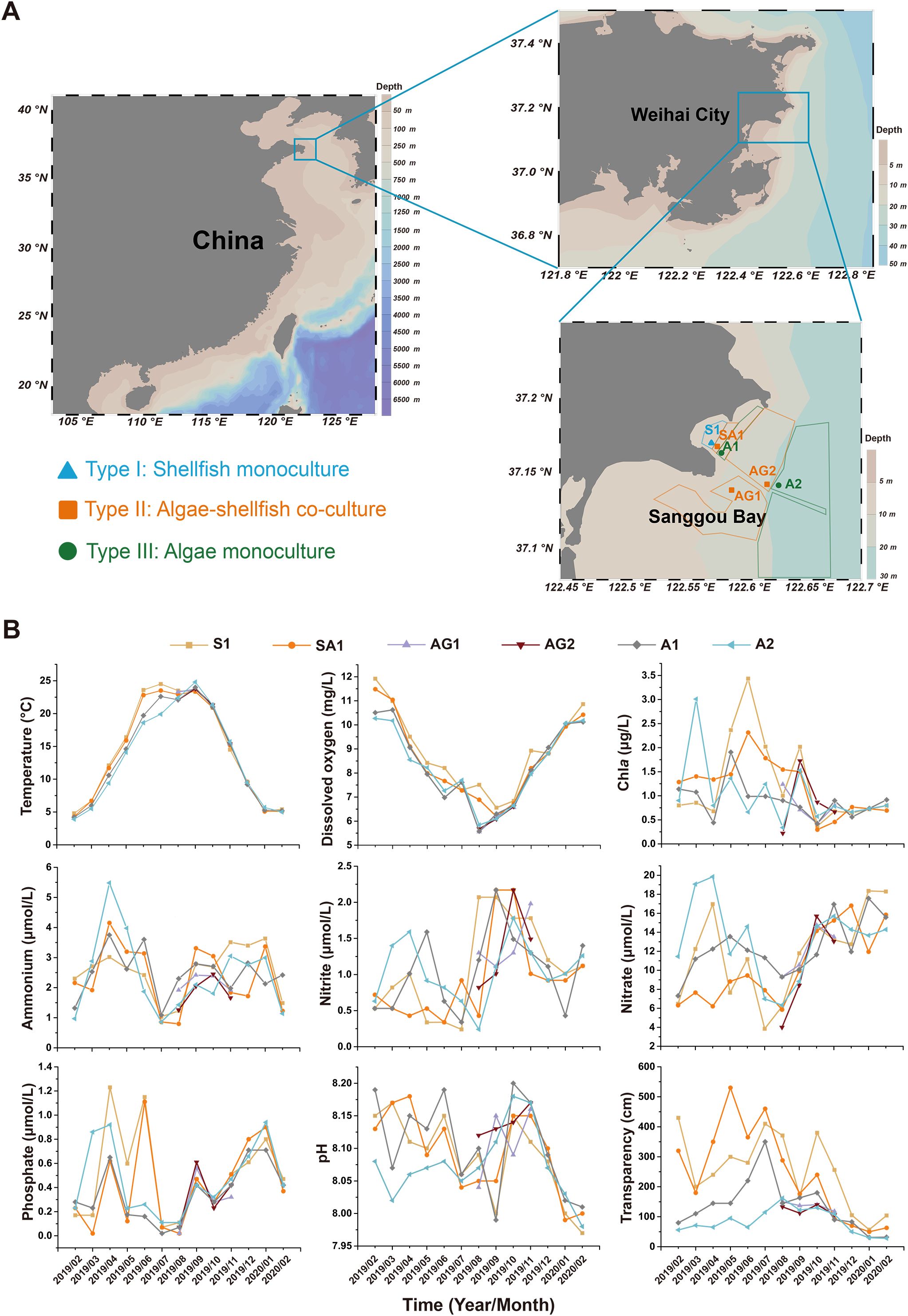
Figure 1. Locations and environmental conditions of the sampling stations in Sanggou Bay. (A) Maps showing the geographic locations of the sampling stations and cultivation areas of 3 different mariculture types. Blue: Type I (shellfish monoculture); orange: Type II (algae-shellfish co-culture); and green: Type III (algae monoculture). (B) Monthly changes in physicochemical parameters of the sampling stations from February 2019 to February 2020. Lines of different colors represent different sampling stations.
2.2 Sample collection and environmental factor measurement
Surface seawater from ~ 0.5 m depth was collected with a 2.5 L acrylic water sampler, with a total of 60 samples obtained. Measurements of in situ seawater temperature, salinity, and dissolved oxygen (DO) were taken immediately onboard using a YSI-556MPS handheld multiparameter meter (YSI Incorporated, USA). Seawater transparency was assessed with a 30 cm Secchi disk, and pH was determined in the laboratory with a pH meter (Mettler-Toledo FE28, China). Environmental factors including chlorophyll a (Chla) (Standard method: HJ897-2017), ammonium (NH4+) (Standard method: GB/T12763.4-2007), nitrite (NO2-) (Standard method: GB/T12763.4-2007), nitrate (NO3-) (Standard method: GB/T12763.4-2007), and phosphate (PO43-) (Standard method: GB/T12763.4-2007) were measured following the corresponding standard methods, respectively. For microbial community analyses, seawater samples were filtered onto 47 mm diameter polycarbonate filters with a 0.22 μm pore size (Merck Millipore Ltd., USA) under low vacuum pressure. The filters were then stored at -80°C until DNA extraction.
2.3 DNA sequencing and bacterial community taxonomic analyses
Microbial community DNA was extracted using a MagPure Stool DNA LQ Kit (Magen, China) following the manufacturer’s instructions. DNA quantity was measured with a Qubit 2.0 Fluorometer (Invitrogen, USA), and DNA quality was assessed by electrophoresis. The bacterial 16S rRNA gene V3-V4 hypervariable region was amplified using universal PCR primers 338F and 806R tagged with Illumina adapter, pad, and linker sequences (Salas-González et al., 2021; Dai et al., 2022). PCR was performed in triplicate with a 50 μL reaction mixture containing 30 ng template DNA. The PCR reaction conditions were: 94°C for 3 min, 30 cycles of 94°C for 30 s, 56°C for 45 s, 72°C for 45 s, and a final extension at 72°C for 10 min. PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter, USA), and their quality and concentration were determined using an Agilent 2010 bioanalyzer (Agilent, USA). The PCR products were sequenced on the Illumina MiSeq PE 300 platform at BGI Genomic Co., Ltd, Wuhan, China.
Paired-end raw reads were processed using FLASH (v1.2.11) to trim and merge sequences (Magoč and Salzberg, 2011). High-quality sequences were retained after removing low-quality reads based on the following criteria: (1) sliding window size = 25 nt; (2) quality score< 20; (3) reads that contained N; (4) length< 75% of the original. Operational taxonomic units (OTUs) were identified using USEARCH (v7.0.1090) at a 3% sequence dissimilarity (Edgar, 2013) and classified with the RDP Classifier (v2.2) against the Silva database (v138.1) (accessed on 11 April 2023) (Quast et al., 2012) with an 80% confidence threshold. Chimeric sequences were removed with UCHIME (v4.2.40) (Edgar, 2016; Edgar et al., 2011), and chloroplastic and non-bacterial sequences were excluded. Sample A2_2020/01 was removed due to low sequencing quality. The raw 16S rRNA gene sequences are available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1154816.
2.4 Statistical and bioinformatic analyses
Principal component analysis (PCA) and clustering analysis were performed to differentiate sampling environments using the vegan R package (v2.5-7) (Oksanen et al., 2020). Alpha-diversity indices, principal coordinate analysis (PCoA) with Bray-Curtis dissimilarities, and analysis of community similarities (ANOSIM) (Clarke, 1993) were also conducted with vegan. Canonical correspondence analysis (CCA) was performed using Canoco5 (Microcomputer Power, USA), with collinear variables removed during forward selection. Structural equation modeling (SEM) analysis was conducted via the piecewiseSEM R package to assess the direct and indirect effects of the significant environmental factors identified in the above CCA analysis on the seawater bacterial diversity and community structure (Lefcheck, 2016). Using the stats R package (v3.6.3), Spearman correlation tests assessed correlations between bacterial taxa and environmental factors, with significance at p< 0.05.
High-abundance sequences (relative abundance > 0.5% across the database) were selected to construct a bacterial 16S rRNA gene phylogenetic tree using the build function of ETE 3.1.2 (Huerta-Cepas et al., 2016), with alignment performed using MAFFT v6.861b (Katoh and Kumah, 2005) and the tree constructed with FastTree v2.1.8 (Price et al., 2010) on the GenomeNet (https://www.genome.jp/tools-bin/ete).
The iCAMP software (https://github.com/DaliangNing/iCAMP1) was used to decode community assembly mechanisms, distinguishing deterministic (homogenous and heterogeneous selection) from stochastic (dispersal limitation, homogenizing dispersal, and drift) processes based on the beta Net Relatedness Index (βNRI) and Raup-Crick metric (RC) (Zhou and Ning, 2017; Ning et al., 2020). Distance-decay relationship (DDR) analyses were performed using the Geosphere R package with Bray-Curtis similarity geographic distances (Hijmans et al., 2017).
Co-occurrence networks were constructed for the bacterial communities with the random matrix theory (RMT)-based methods using the integrated Network Analysis Pipeline (iNAP) (https://inap.denglab.org.cn/) and visualized with Gephi v.0.10.1 (Bastian et al., 2009; Feng et al., 2022). Only the bacterial species at the OTU level occurring > 50% of all samples were included in network calculations. Uniform RMT threshold parameters were applied during network construction to ensure cross-group comparability.
2.5 Potential bacterial pathogens analyses
The seawater bacterial communities were screened for potential pathogens by analyzing 16S rRNA gene sequences against three pathogen databases: the multiple bacterial pathogen detection (MBPD) database (Yang et al., 2023), the Fish Pathogen Database (FPD) (Dronen et al., 2022, Dronen et al., 2023), and the human pathogen database (Chen et al., 2016; An et al., 2020). As in previous studies (Chen et al., 2016; Li et al., 2024), BLASTn analysis was used (E-value ≤ 1 × 10-10, ≥ 99% identity).
Vibrio OTUs were further analyzed phylogenetically by comparing their sequences to the sequences of GenBank Vibrio type strains and isolates from the pathogen surveillance repository (PSR) of Weihai Changqing Ocean Science Technology Co. Ltd. This curated collection contains Vibrio strains isolated from diseased shellfish and surrounding aquaculture seawater within Sanggou Bay’s aquaculture zones from 2018 to 2022. Phylogenetic reconstruction of the 16S rRNA sequences was performed with MEGA-X (version 10.2.6) using the maximum likelihood algorithm with 1000 bootstrap replicates (Kumar et al., 2018). The top-match PSR isolates were subject to further genome analyses and environmental simulation experiments.
Accordingly, six PSR Vibrio isolates were selected and genomically sequenced. Genomic DNA was extracted using the CTAB method (Wilson, 2001). Sequencing was performed using a PacBio Sequel II and DNBSEQ platform at Beijing Genomics Institute (BGI, Shenzhen, China). Draft genomic unitigs were assembled using Canu (v1.5) (Koren et al., 2017) and refined with GATK (https://www.broadinstitute.org/gatk/) (McKenna et al., 2010). A phylogenomic tree was constructed using the genome BLAST distance phylogeny method (GBDP) on the TYGS platform (https://tygs.dsmz.de/) (accessed on 15 July 2024) (Meier-Kolthoff and Göker, 2019). Gene prediction was performed using glimmer3 (http://www.cbcb.umd.edu/software/glimmer/) with the Hidden Markov models (Delcher et al., 1999). Virulence factors and antibiotic resistance genes were identified using VFDB (Liu et al., 2022) and ARDB (Antibiotic Resistance Genes Database) (Liu and Pop, 2009), respectively. The genome sequences are deposited in NCBI GenBank (Accession numbers SAMN43439310-SAMN43439315).
Environmental simulation experiments were conducted to assess the pathogenicity of the six selected Vibrio isolates. Three species of scallops, including bay scallop (Argopecten irradians), Zhikong scallop (Chlamys farreri), and Yesso scallop (Patinopecten yessoensis), along with Pacific abalone (Haliotis discus hannai), were used as the experimental animals. They were placed at 12 ± 1°C with aerated running seawater for 7 days, with daily water changes and regular feeding (microalgae for scallops and kelp for abalones). For each species, 420 individuals (Supplementary Table S1 for body size details) were divided into seven groups, including one control group and six experiment groups (three replicates per group). Test bacteria were introduced into the rearing water of the experiment group at a final concentration of 105 individuals/mL, while the control group received an equal volume of culture medium. The feeding rate was determined 1 h after the addition of bacterial strains.
The feeding rate of scallops was determined by calculating the clearance rate (CR, L h-1 ind-1) based on the removal of Nannochloropsis particles. Optical density (OD750) was measured at the start and after 1 h. The CR (Loret et al., 2000; Vaquer et al., 2000; Wildish et al., 1992) was calculated as:
Where V is the volume of the rearing water, n is the number of experimental scallops, t is the experiment time, C0 is the concentration of Nannochloropsis at the beginning of adding, and Ct is the concentration of Nannochloropsis after 1 h.
The reduced weight of fresh kelp was used to determine the feeding rate (FR, g d-1 ind-1) of abalones following a previous study (Li X. et al., 2022):
Where W0 is the weight of kelp at the beginning of adding, Wt is the weight of kelp after 1 h, n is the number of abalones, and t is the experiment time.
3 Results
3.1 Temporospatial dynamics of environmental condition in Sanggou Bay
The environmental condition of Sanggou Bay showed apparent seasonal variation, with seawater temperature and DO showing greater consistency across sampling stations (Figure 1B). Higher water temperatures and lower DO concentrations were observed from June to September, with the highest temperature appearing at station A2 (24.8°C) in August and the lowest DO appearing at sampling station AG2 (5.56 mg/L) in September. Concentrations of Chla and inorganic nutrients, including ammonium, nitrite, nitrate, and phosphate, varied across stations with less pronounced seasonal patterns. Low concentrations of seawater ammonium, nitrite, nitrate, and phosphate were usually observed in July and August and Chla in August. PCA and clustering analysis showed that seasonal variation outweighed spatial differences in shaping the sampling environment (Supplementary Figures S1, S2). Therefore, subsequent analyses mainly focused on seasonality-related bacterial communities and pathogens.
3.2 Bacterial community composition and diversity
After quality filtering, 1,987,548 valid bacterial 16S rRNA gene sequences were identified, representing 4069 OTUs across 59 samples. A total of 50 phyla, 107 classes, 225 orders, 314 families, and 571 genera were taxonomically assigned. Thirteen dominant bacterial classes (relative abundance of each > 0.5%) were identified, with Alphaproteobacteria (33.89%), Gammaproteobacteria (33.01%), and Bacteroidia (16.32%) being the most abundant (Figure 2A). Twenty-five dominant bacterial families were identified, with Rhodobacteraceae (28.78%), Flavobacteriaceae (11.63%), and Moraxellaceae (5.14%) being the most abundant (Figure 2A). The bacterial community in Sanggou Bay showed seasonal and aquaculture-type variations (Supplementary Figure S3).
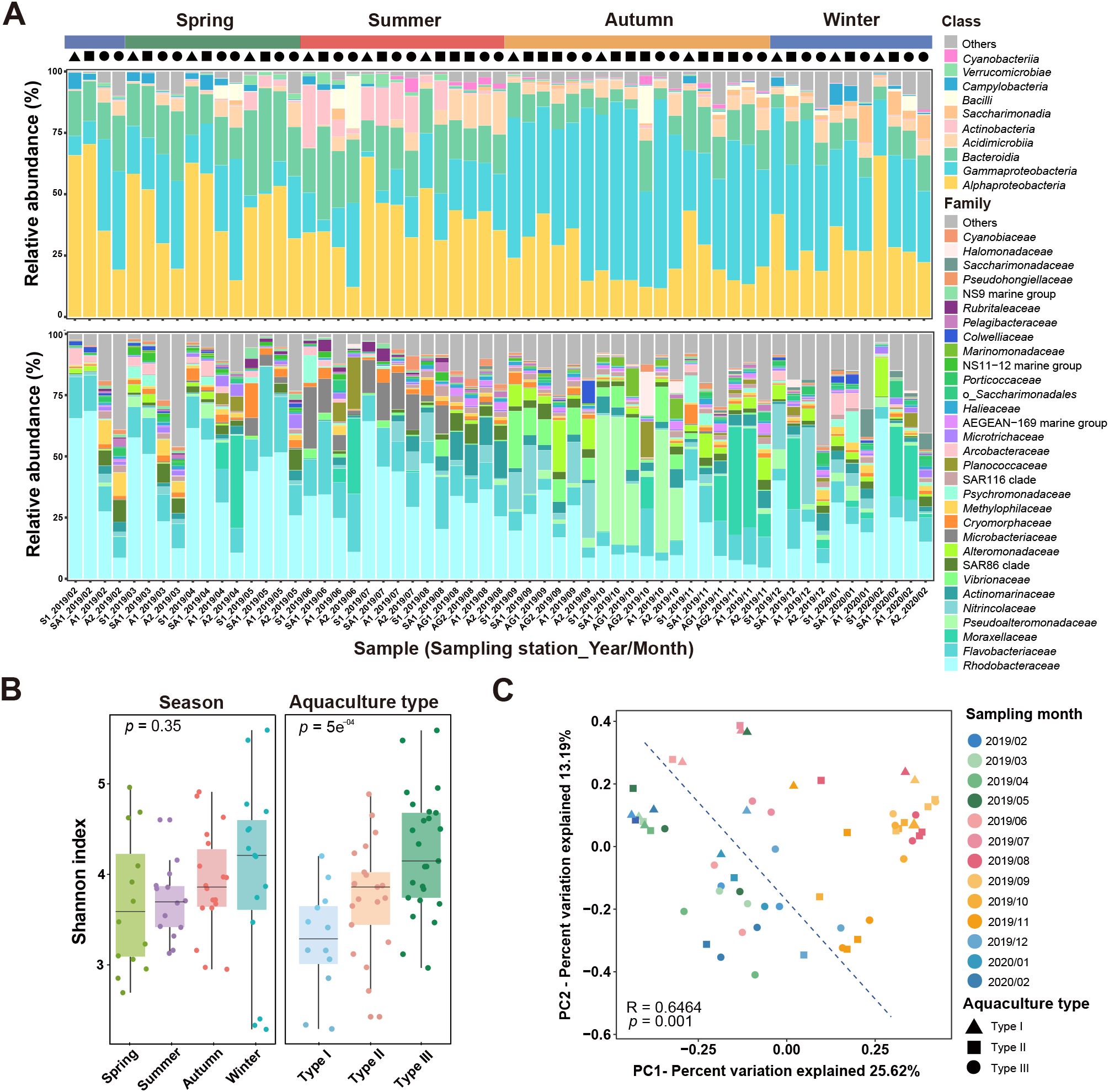
Figure 2. Community composition and diversity of seawater bacteria in Sanggou Bay. (A) Bacterial community compositions at class level and family level, respectively. Classes or families representing< 0.5% of the total community composition are classified as “others.” (B) Comparisons of community Shannon biodiversity indices in different seasons and aquaculture types. (C) Principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarity illustrating the variation in bacterial community structure. Colors represent months, and shapes represent aquaculture types.
The Shannon biodiversity of the seawater bacterial communities varied with season and aquaculture type (Figure 2B). However, only the aquaculture type showed a significant influence (ANOVA, p< 0.05) on the bacterial diversity, with the highest diversity being observed in Type III and the lowest diversity being observed in Type I (Figure 2B). These suggest that cultivating algae might have increased the diversity of seawater bacteria in Sanggou Bay, likely due to the enhanced effect of photosynthesis-driven seawater quality enhancement. In contrast, the shellfish monoculture might decrease the seawater bacterial diversity, and including algae cultivation in shellfish aquaculture (i.e., the IMTA mode) might enhance seawater quality and thus enhance bacterioplankton community diversity.
The PCoA result showed that seasonality might be a major factor driving the shift of seawater bacterial community structure, with similar bacterial communities being found in winter and spring and in summer and autumn, respectively (Figure 2C). ANOSIM statistical analyses showed that the bacterial community difference among different seasons or aquaculture types was highly significant (p< 0.01), and those between each pair of seasons or aquaculture types were generally significant (p< 0.05), as well, except for the pair between the algae-shellfish co-culture system (Type II) and the shellfish monoculture system (Type I) (Table 2).
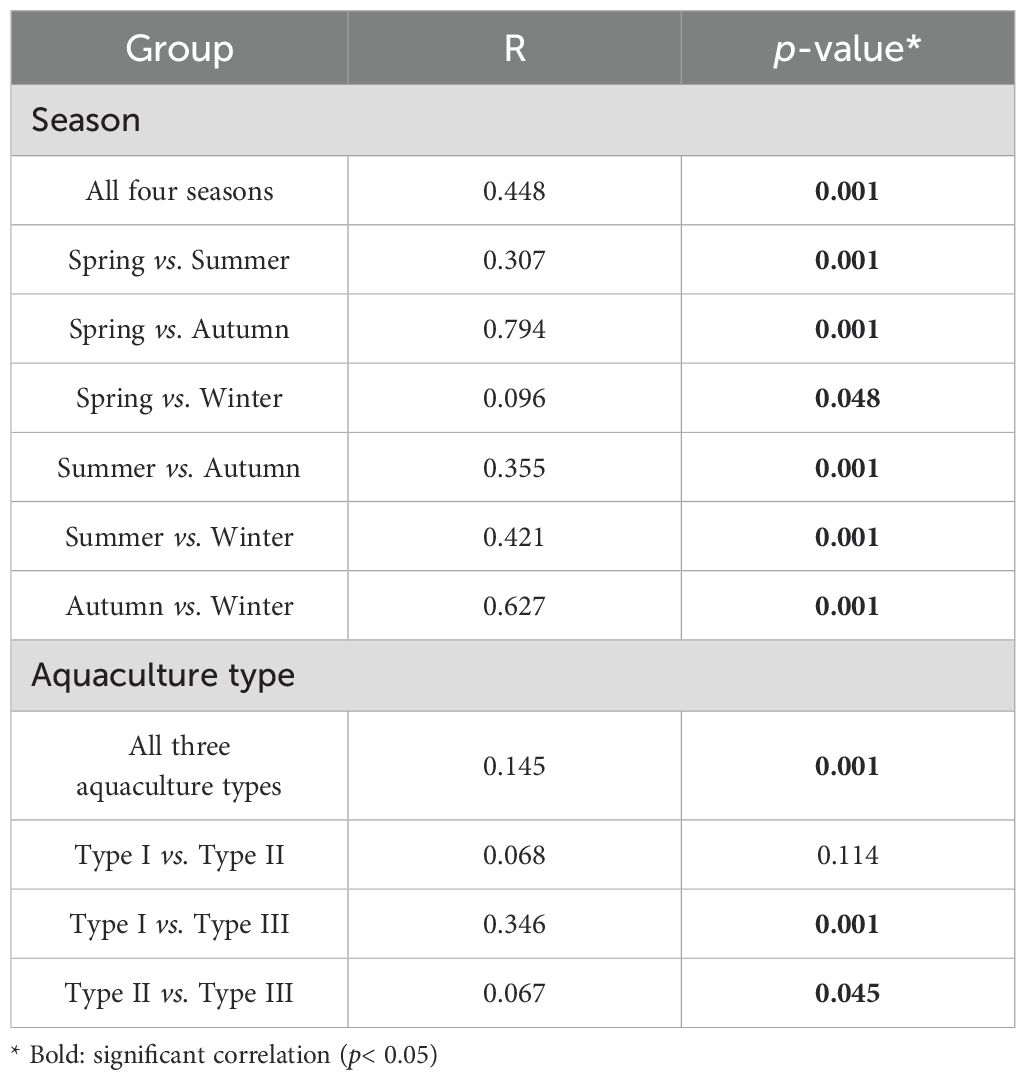
Table 2. Two-way crossed analysis of similarity (ANOSIM) of bacterial community structure based on Bray-Curtis dissimilarity across different seasons and aquaculture types (999 permutations).
3.3 Dynamics of bacterial taxa in response to seasonality and aquaculture type
A total of 41 OTUs were identified as abundant OTUs (relative abundance > 0.5%), exhibiting varying seasonal patterns (Figure 3A). The five most dominant OTUs, ranked by abundance, were Otu1 (8.46%), Otu6 (5.31%), Otu16 (4.31%), Otu4 (3.71%), and Otu3 (3.08%). Of these, the relative abundance of Otu1 (Sulfitobacter sp.) was higher in winter and spring, Otu6 (Rhodobacteraceae HIMB11) in summer, Otu16 (Planktomarina sp.) in spring and summer, Otu4 (Psychrobacter sp.) in autumn and winter, and Otu3 (Pseudoalteromonas sp.) in autumn (Figures 3A,B). In addition to seasonality, the bacterial community composition of the abundant OTUs also varied in response to the different aquaculture types (Figure 3B). For the five most abundant OTUs, Otu1 had the highest relative abundance in Type I, Otu6 in Type II, Otu16 in Type I and Type III, Otu4 in Type II and Type III, and Otu3 in Type I and Type II (Figure 3B).
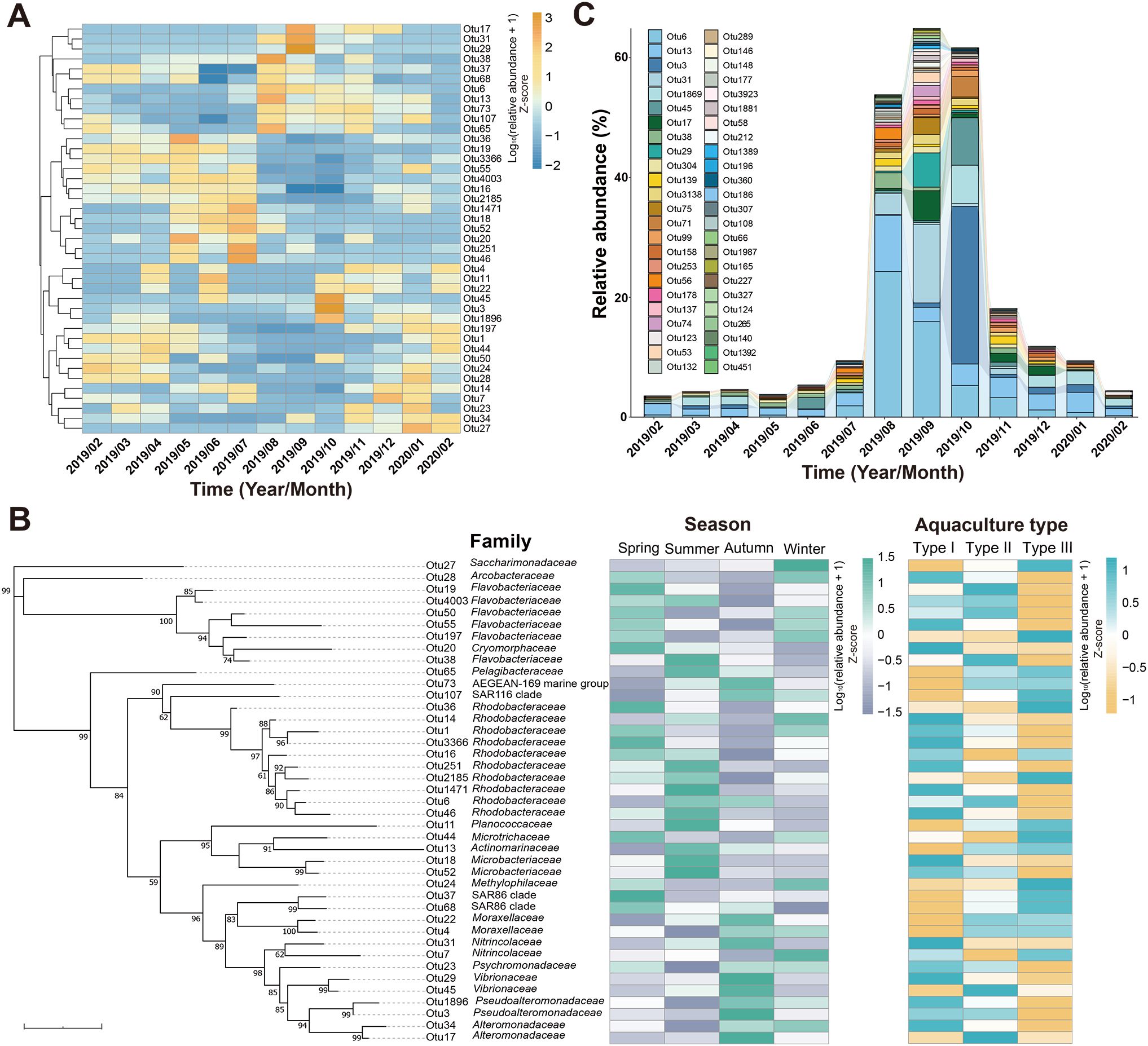
Figure 3. Distribution patterns of the abundant OTUs and OTUs with an increased relative abundance in late summer and early autumn. (A) Heatmap of abundant OTUs showing their monthly variations. The relative abundances were shown in the z-score scale of the log-transformed values for easy visibility of the temporal variation patterns. (B) Phylogenetic tree of the 16S rRNA gene sequences showing the taxonomies of the abundant OTUs and their distributional variations related to seasons and aquaculture types. Bootstrap values greater than 50% were shown. The heatmaps on the right show the relative abundance of each OTU in different seasons and aquaculture types. The relative abundances were shown in the z-score scale of the log-transformed values for easy visibility of the variation patterns. (C) Monthly dynamics of the OTUs that showed increased relative abundance in later summer and early autumn.
Forty-eight OTUs exhibited increased relative abundance during late summer and early autumn, a period of high disease risks, including common marine bacteria, such as those affiliated with Rhodobacteraceae, Pseudoalteromonadaceae, Actinomarinaceae, Vibrionaceae, Flavobacteriaceae, and Alteromonadaceae (Figure 3C).
3.4 Drivers of bacterial composition and community structure
Spearman non-parametric correlation analysis revealed significant correlations (p< 0.05) between several dominant bacterial OTUs and seawater parameters, including DO, temperature, transparency, NO3-, NO2-, TN, PO4-, and Chla (Figure 4A). In addition, differences in the interrelationships between aquaculture types and high-abundance bacterial taxa were found. Shellfish monoculture had significant positive correlations (p< 0.05) with the abundances of certain OTUs affiliated with Rhodobacteraceae and Flavobacteriaceae (2.97% of the total community) and significant negative correlations (p< 0.05) with certain OTUs affiliated with Actinomarinaceae, Microtrichaceae, Methylophilaceae, Nitrincolaceae, Rhodobacteraceae, SAR86 clade, Pelagibactereae, SAR116 clade, and Planococcaceae (12.24%). Algae monoculture had significant positive correlations with the abundances of certain OTUs affiliated with SAR86 clade, SAR116 clade, Pelagibacterceae, Actinomarinaceae, Methylophilaceae, Planococcaceae, and AEGEAN-169 marine group (9.48%) and significant negative correlations with certain OTUs affiliated with Rhodobacteraceae, Arcobacteraceae, and Flavobacteriaceae (5.92%). Algae-shellfish co-culture only negatively correlated with the abundance of one OTU affiliated with Microtrichaceae (0.53%).
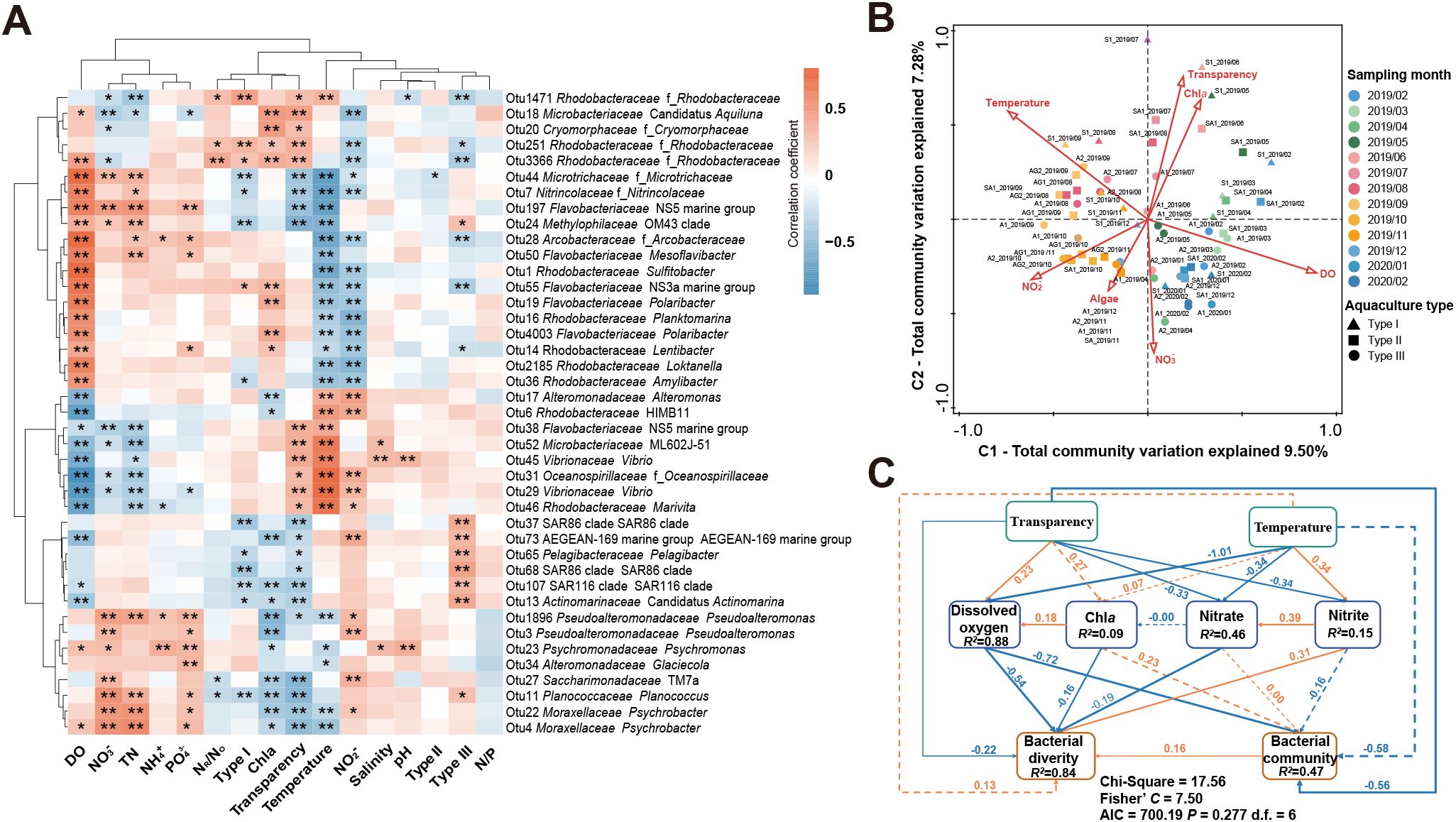
Figure 4. Relationships between environmental conditions and the seawater bacterial communities. (A) Heatmap plot showing the relationship between the abundant OTUs and environmental factors. The plot was produced at OTU, family, and genus taxonomic ranks. The color bar indicates the r values. * indicates 0.01 ≤ p< 0.05. ** indicates p< 0.01. (B) Canonical correspondence analysis (CCA) ordination plot for the first two dimensions of the relationships between the environmental factors and the seawater bacterial communities. (C) Structural equation modeling (SEM) result showing the effects of environmental factors on bacterial Shannon diversity and community structure. Orange and blue arrows indicate positive and negative relationships, respectively. Solid and dashed lines indicate significant (p< 0.05) and nonsignificant relationships. Numbers near pathway arrows indicate the standard path coefficients. R2 represents the proportion of variance explained for each dependent variable.
The CCA result identified seven significant environmental factors (p< 0.05) mainly contributing to the variation of the bacterial communities, including seawater DO, transparency, temperature, NO2-, Chla, NO3-, and algae cultivation (Figure 4B). Seawater DO made the highest contribution (8.5%) among these factors. The SEM analysis further confirmed seawater DO, along with seawater transparency, as the most significant environmental factor (p< 0.05) directly affecting (negatively) the seawater bacterial community variation (Figure 4C). In contrast, seawater temperature exerted an indirect effect, through the effect of seawater DO, on the bacterial community variation (Figure 4C). The SEM result also revealed that the seawater bacterial diversity was directly affected by seawater DO, transparency, Chla, NO3-, and NO2- (p< 0.05) and indirectly affected by seawater temperature through its effects on seawater DO, NO3-, and NO2- (p< 0.05) (Figure 4C). The SEM result provides a more insightful explanation than the CCA result by revealing additional information about the interactions among the key environmental factors in driving the variation of the seawater bacterial communities.
The CCA results provided a limited explanation (< 20%) for the total variation of the bacterial communities, indicating the presence of other contributing factors. Indeed, the iCAMP analysis results showed that the stochastic processes, especially dispersal limitation and drift, were the primary bacterial community assembly mechanisms (Figure 5A). Among the deterministic processes, homogenous selection contributed the most to the bacterial community assembly (Figure 5A). Results of the DDR analyses statistically showed a significant but weak geographic distance decay (p< 0.001, r2 = 0.002) of the bacterial community similarity, further confirming the role of dispersal limitation in the assembly of the seawater bacterial communities in Sanggou Bay (Figure 5B).
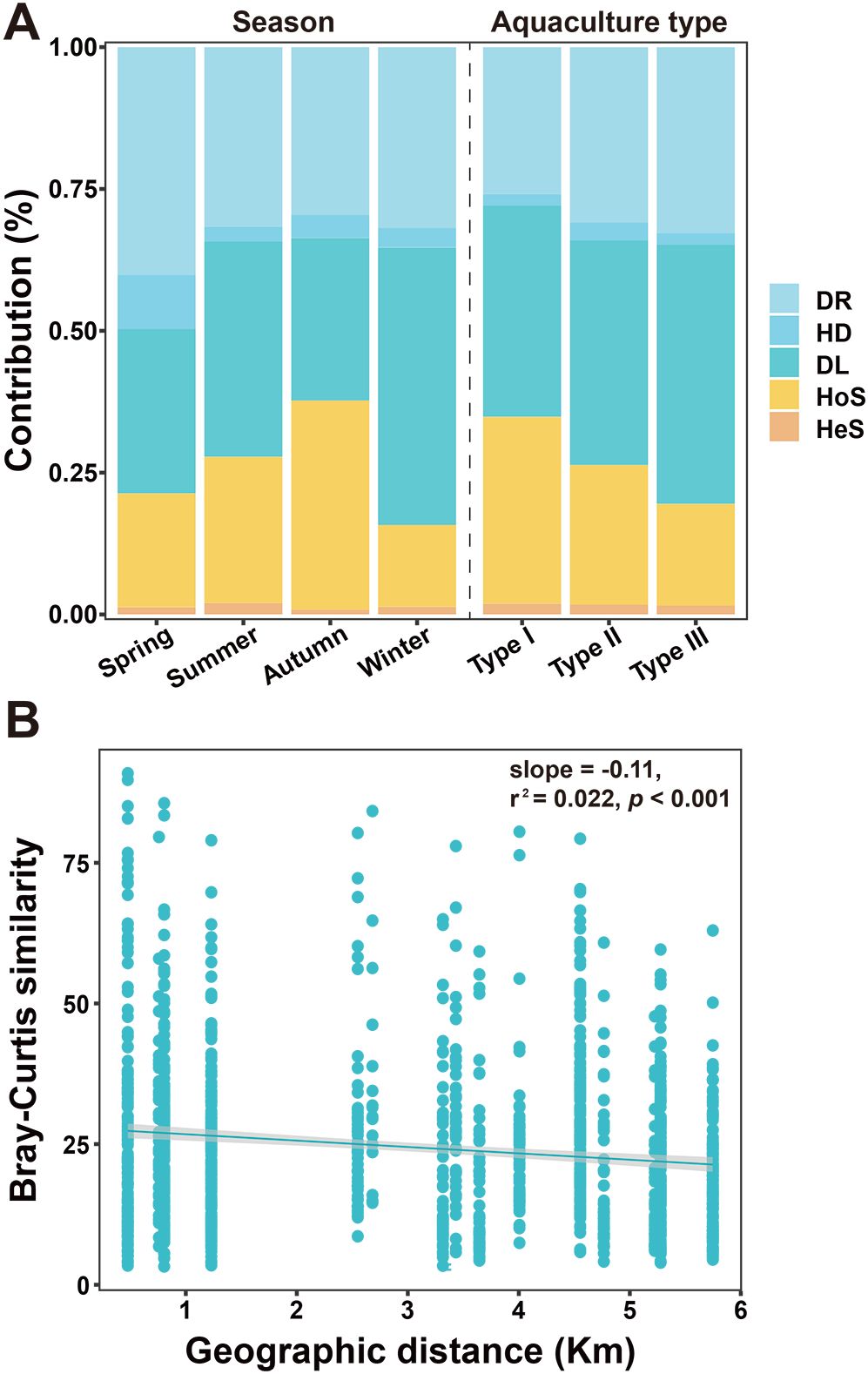
Figure 5. (A) The contributions of different mechanisms to the assembly of the seawater bacterial communities in Sanggou Bay analyzed using iCAMP. Percentage values indicate the specific contributions of deterministic and stochastic processes in different seasons and aquaculture types. Deterministic processes include homogenous (HoS) and heterogeneous (HeS) selection, and stochastic processes include dispersal limitation (DL), homogenizing dispersal (HD), and drift (DR). (B) Distance-decay relationship (DDR) analysis result showing the bacterial community Bray-Curtis similarity against geographic distances in Sanggou Bay.
The iCAMP analysis results also showed the seasonal variation in the different bacterial community assembly mechanisms, with autumn exhibiting the highest contribution by homogenous selection and the lowest contribution by dispersal limitation (Figure 5A). A further analysis of the bacterial community assembly mechanisms was conducted across different aquaculture types. The stochastic processes exhibited the highest contribution in Type III (80.51%), followed by Type II (73.66%) and Type I (65.18%). Conversely, the deterministic processes, predominantly homogenous selection, exhibited a higher contribution in Type I (32.92%) than in the other two aquaculture types (Type II: 24.61%, Type III: 17.92%) (Figure 5A).
3.5 Dynamics of putative interactions in the bacterial communities
Ecological network analyses were conducted to examine potential microbial interactions within the bacterial communities in different seasons and aquaculture types, respectively (Figure 6). The topological properties of each network are listed in Supplementary Table S2. Positive interactions predominated over negative interactions in all networks (Supplementary Table S2). Seasonal comparisons revealed a substantial network structural variation. The autumn network exhibited markedly reduced complexity, containing merely 81 nodes (OTUs) and 97 links, approximately 2.28 to 4.06-fold fewer nodes, and 4.51 to 14.25-fold fewer edges than the spring network (329 nodes, 1382 links), the summer network (185 nodes, 437 links), and the winter network (240 nodes, 687 links) (Figures 6A, B; Supplementary Table S2). Consistent with this pattern, network connectivity metrics showed autumn had the lowest average degrees (avgK = 2.395), much lower than spring (8.401), summer (4.724), and winter (5.725) (Supplementary Table S2). The nodes with the maximum degrees also varied seasonally, with Otu138 (affiliated with SAR116 clade) in spring, Otu139 (Flavobacteriaceae) in summer, Otu1389 (Vibrionaceae) and Otu148 (Nitrincolaceae) in autumn, and Otu154 (Woeseiaceae) in winter (Supplementary Table S2). These findings indicate the seasonality of the microbial interactions and network complexity, presenting autumn as a particularly unique situation.
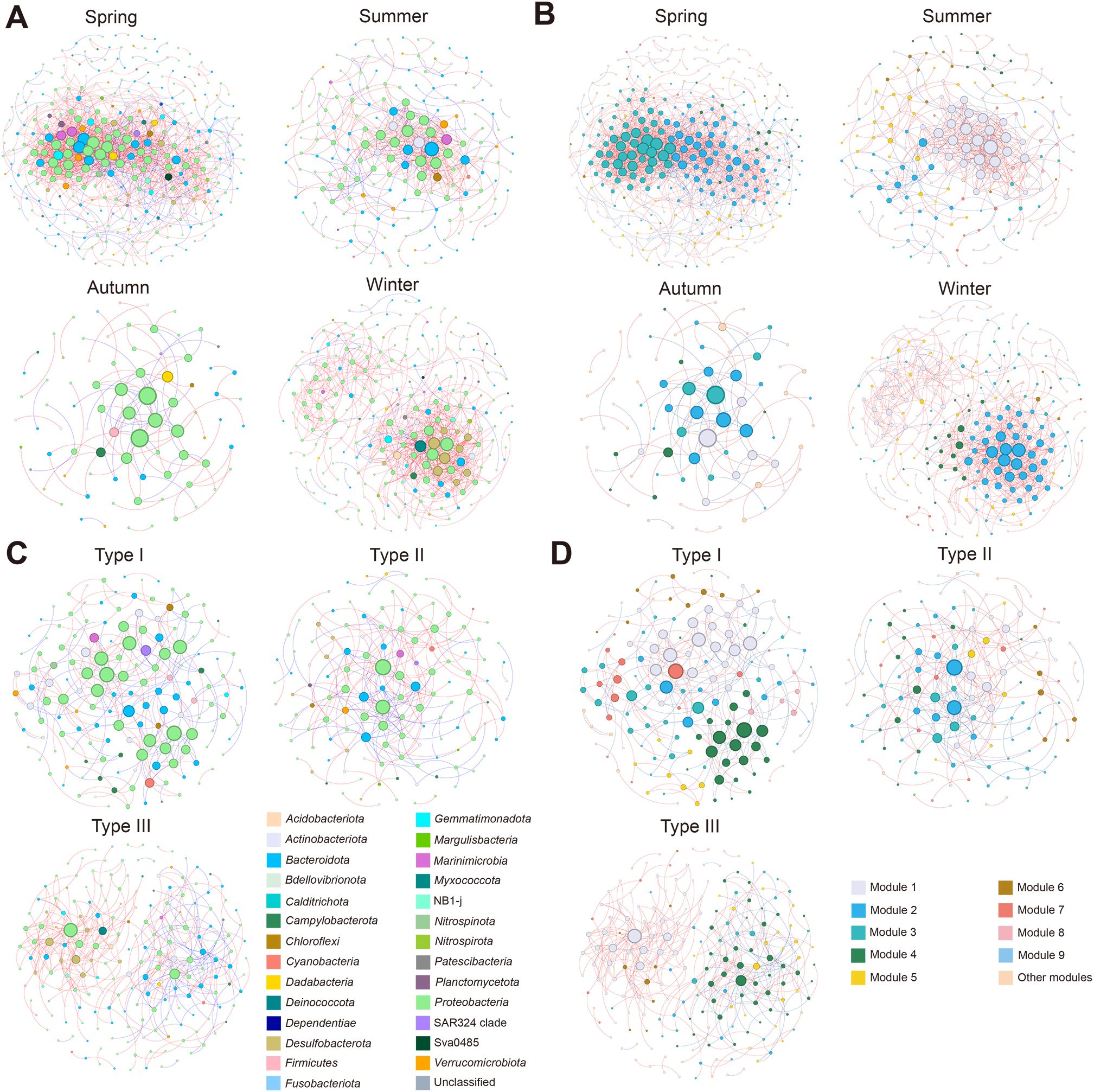
Figure 6. Results of ecological interaction network analyses of the seawater bacterial communities. (A) Ecological networks constructed for different seasons, with bacterial taxa distinguished by colors according to their affiliated phyla. (B) Ecological networks constructed for different seasons, with bacterial taxa distinguished by colors according to network modules. (C) Ecological networks constructed for different aquaculture types, with bacterial taxa distinguished by colors according to their affiliated phyla. (D) Ecological networks constructed for different aquaculture types, with bacterial taxa distinguished by colors according to network modules. Each node represents an OTU (i.e., a species), and its size represents the degree level, with larger nodes indicating greater degrees. The edge between each two nodes represents a positive (red) or negative (blue) interaction. “Other modules” include the network modules with less than five nodes.
The network topological properties also varied significantly among different aquaculture types, with the highest number of nodes (190) and links (368) in Type III, followed by Type I (152 nodes, 274 links) and Type II (132 nodes, 221 links) (Figures 6C, D; Supplementary Table S2). In addition, the highest network degrees were observed in Type III (3.874), followed by Type I (3.605) and Type II (3.348) (Supplementary Table S2). The nodes with the maximum degrees were Otu1655 (Rhodobacteraceae) and Otu356 (Psychromonadaceae) in Type I, Otu6 (Rhodobacteraceae) in Type II, and Otu154 (Woeseiaceae) in Type III, respectively (Supplementary Table S2).
3.6 Seasonal dynamics of potential pathogenic bacteria
Forty-two potential pathogenic OTUs spanning six phyla and 35 genera were identified, including 24 genera of animal pathogens, eight genera of zoonotic pathogens, two genera of plant pathogens, and one genus of fungus pathogens (Supplementary Table S3). Plant and fungus pathogens were excluded in subsequent analyses due to their limited ecological relevance to the Sanggou Bay aquaculture species. Bacteria in the Vibrio genus emerged as the predominant potential pathogens, accounting for 1.57% of total bacterial 16S rRNA gene sequences, followed by Mycobacterium (0.02%), Arthrobacter (0.01%), and Prevotella (0.01%). All other potential pathogenic genera exhibited relative abundances< 0.01%. Monthly distribution showed that the abundances of putative Vibrio pathogens progressively increased from May, peaking in October (Figure 7A; Supplementary Figure S4). Sampling-station-specific distribution showed that the Vibrio abundances were high across all sampling stations in September and October, with Otu45, Otu75, and Otu685 representing putative pathogens (Figure 7B).
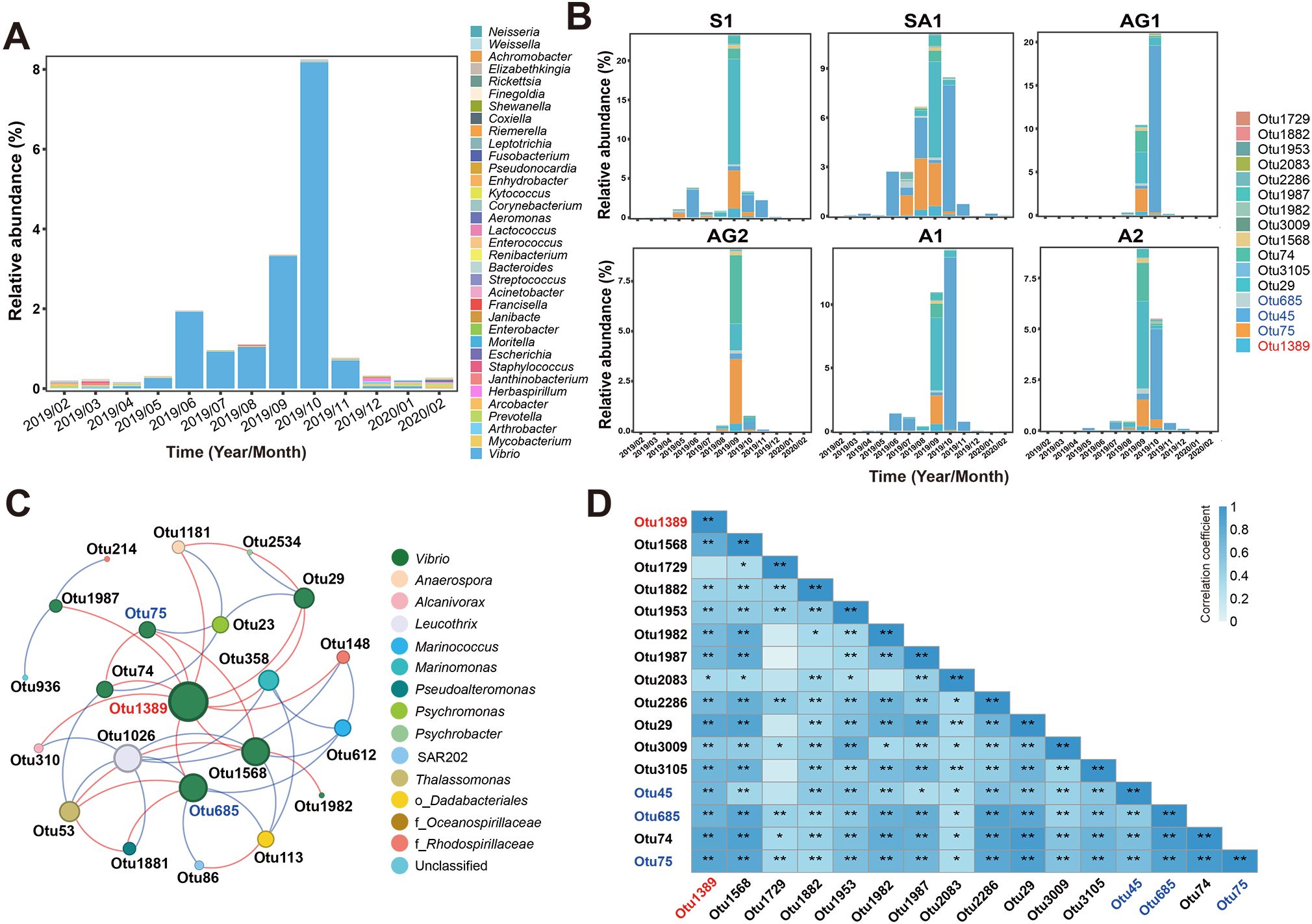
Figure 7. Analytical results of potential bacterial pathogens in the seawater bacterial communities of Sanggou Bay. (A) Monthly variation of potential pathogenic bacteria at the genus level. (B) Monthly variations of OTUs annotated as Vibrio in each sampling station. OTUs identified as pathogenic Vibrio are labeled in blue. (C) A partial ecological network of the autumn bacterial communities specifically showing the Vibrio OTUs’ interactions with each other and with other bacteria. The node colors indicate different bacterial genera or families if the OTUs cannot be annotated to the genus level. Each node represents an OTU (i.e., a species), and its size represents the degree level, with larger nodes indicating greater degrees. The edge between each two nodes represents a positive (red) or negative (blue) interaction. Otu1389 is labeled in red, and putative pathogenic Vibrio OTUs are labeled in blue. (D) A heatmap of Spearman correlation between OTUs assigned to Vibrio. Otu1389 is labeled in red. OTUs identified as pathogenic Vibrio are labeled in blue. * indicates 0.01 ≤ p< 0.05. ** indicates p< 0.01.
A subnetwork was extracted from the autumn network (Figures 6A, B) to explore the microbial interactions, focusing on the Vibrio OTUs (Figure 7C). Notably, Otu1389 (annotated as Vibrio halioticoli) showed positive correlations with six other Vibrio OTUs, including two putative pathogens (Otu75 and Otu685). Otu1389 was detected in 39 of 59 samples and exhibited a high relative abundance in autumn, especially in September (0.56% of the bacterial community). Spearman correlation analysis further found significant and positive correlations between Otu1389 and other 14 Vibrio OTUs (Figure 7D). Its hub positioning in the network and extensive associations with other Vibrio OTUs suggested a pivotal role of the Otu1389-represented bacterium in supporting the growth of other Vibrio species in autumn (see the Discussion section for further reasoning).
Through 16S rRNA gene sequence phylogenetic analysis, three putative pathogenic Vibrio OTUs identified in the seawater bacterial communities were found to match pathogenic Vibrio strains from the previously established PSR (Supplementary Figure S5), providing further supporting evidence that these OTUs may represent authentic Vibrio pathogens. Precisely, Otu45 (0.32%-7.92%, 2.90% on average from August to October) and Otu75 (0.24%-2.81%, 1.21% on average from August to October), two of the most dominant Vibrio OTUs, particularly in high-disease-risk late summer and autumn, matched pathogenic Vibrio strains ScY-2108 and ScY-2109, and ScY-2106 and WaG-2229, respectively, while less-abundant Otu685 matched pathogenic Vibrio strains AbY-1905 and AbY-2104. These six pathogenic strains were thus selected for further whole genome sequencing and environmental simulation experiments. Phylogenomic analysis showed that strains ScY-2106 and WaG-2229 belonged to V. alginolyticus, strains AbY-1905 and AbY-2104 belonged to V. mediterranei, and strains ScY-2108 and ScY-2109 might represent two new species of Vibrio, tentatively named V. sanggoullaii and V. sanggoullbii, respectively (Supplementary Figure S6). The six pathogenic Vibrio strains harbored diverse virulence factors, such as those involved in adherence (OmpU, tcpI, and Type IV pili), antimicrobial activity/competitive advantage (AcrAB), biofilm formation (PlcR-PapR quorum sensing and lateral flagella), effector delivery (HIS, P. syringae TTSS effectors, and T3SS1), exoenzyme production (NanH/VCNA, HAP, and streptococcal enolase), exotoxin production (haemolysin, cytolysin, phytotoxin phaseolotoxin, TLH, Ymt, and PlaB), immune modulation (Capsular polysaccharide, LOS, and LPS), motility (flagella), invasion (K1 capsule), nutrient uptake (iron uptake, siderophore biosynthesis, vibriobactin/vulnibactin, and vibrioferrin), post-translational modification (Mip), regulation (Fur), and stress survival (urease and ClpP) (Supplementary Tables S4–S9). Antibiotic resistance genes were found in the genomes of all six strains (Supplementary Table S10). Further antibiotic susceptibility tests showed all strains were resistant to 60.0%-83.3% of the tested antibiotics (Supplementary Tables S11–S16).
The survival and feeding/clearance rates were measured in the environmental simulation experiments to assess the impact of these six Vibrio pathogens on farmed shellfish (Figure 8). Although no significant mortality was observed (The survival rates ranged from 87.50% to 100%, details not shown), all Vibrio strains reduced the feeding/clearance rates (0.05%-98.72%, 24.15% on average), with Argopecten irradians and Chlamys farreri showing to be the most pronouncedly affected (Figure 8). Argopecten irradians demonstrated a significant decrease in clearance rate on the first day following the challenge with all six pathogenic Vibrio strains. This rate remained notably lower than that of the control group throughout the five-day experiments. A similar pattern was observed for Chlamys farreri, except for strains AbY-1905 and ScY-2108 on the first day. The tested pathogens had less effect on the clearance/feeding rate of Patinopecten yessoensis and Haliotic discus hannai (Figure 8). Patinopecten yessoensis exhibited a decrease in clearance rate on the third day following bacterial challenge. In contrast, Haliotic discus hannai exhibited heightened sensitivity to strains AbY-2104, WaG-2229, and ScY-2109, resulting in a conspicuous decline in feeding rate on the first day of pathogen challenging, with sustained reduction in comparison to the control groups throughout the experimental duration. These findings highlight the potential negative impacts of Vibrio pathogens on the shellfish’s physiology.
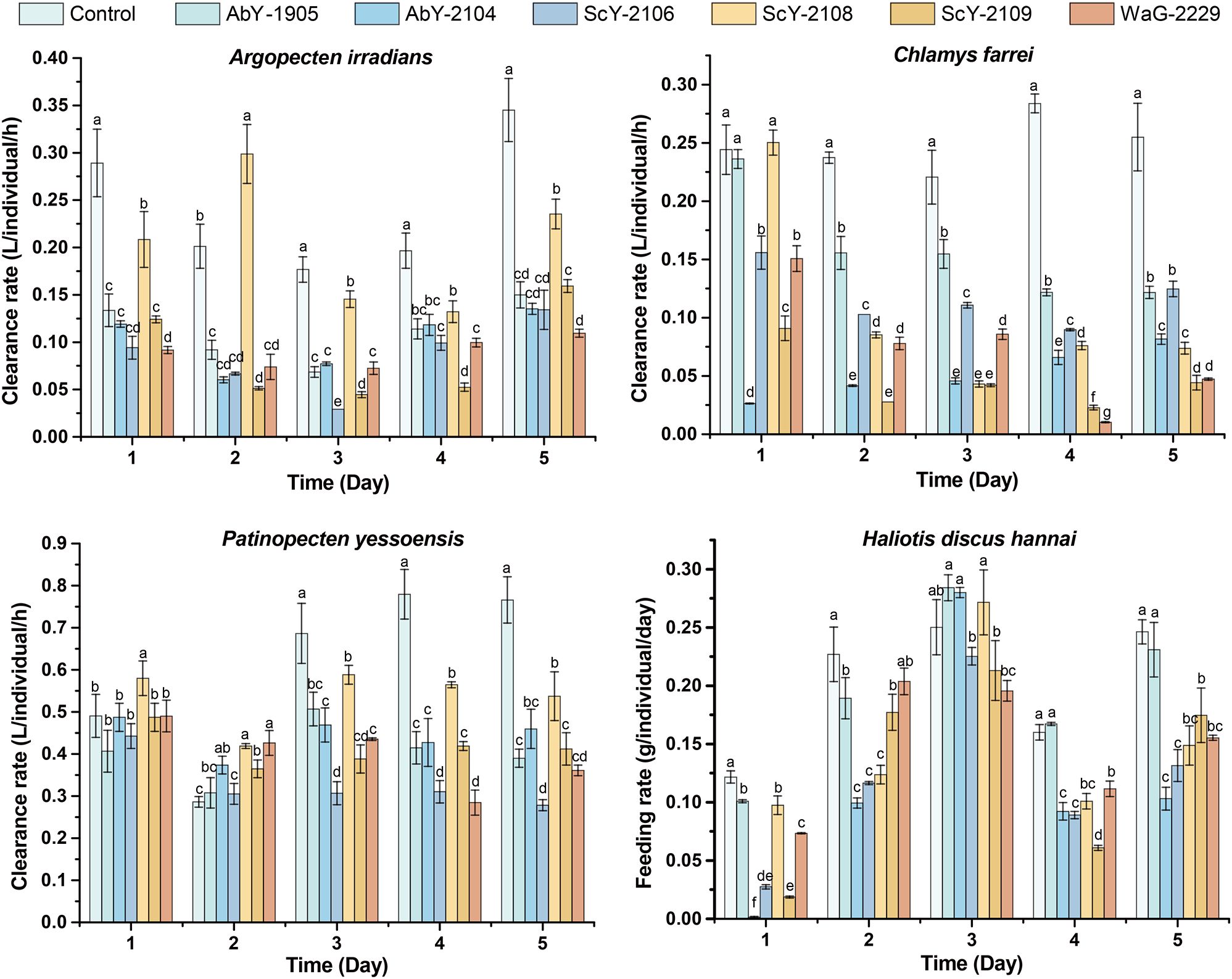
Figure 8. Feeding/clearance rate dynamics of farmed shellfish. Significant differences (ANOVA test, p< 0.05) among different groups at each time point are presented by different letters. Error bars represent standard deviation.
4 Discussion
4.1 Seasonal dynamics of bacterial communities in mariculture environments
Certain microbial taxa maintained stable across seasonal and aquaculture-type gradients. The surface seawater bacterial communities exhibited high diversities yet maintained persistent dominance of three classes: Alphaproteobacteria (mainly Rhodobacteraceae), Gammaproteobacteria (taxa varied among different samples), and Bacteroidia (mainly Flavobacteriaceae) (Figure 2A; Supplementary Figure S3). These results are consistent with published studies in other aquaculture environments, such as the Laoshan Bay (Fang et al., 2021), shrimp culture ponds (Hu et al., 2022), Yantai intensive mariculture systems (Wang et al., 2018b), and Dongying intensive mariculture systems (Zhao et al., 2017). Rhodobacteraceae bacteria act as metabolic generalists through organoheterotrophy, photoheterotrophy, and non-obligate chemolithotrophy (Luo and Moran, 2014), enabling the utilization of various labile and recalcitrant organic substrates (Dang and Lovell, 2016). In addition, Rhodobacteraceae are usually associated with algae, fecal pellets, other marine particles, and submerged surfaces (Wagner-Döbler and Biebl, 2006; Mayali et al., 2008; Dang and Lovell, 2016). Moreover, specific Rhodobacteraceae bacteria could inhibit and kill bacterial pathogens (Bruhn et al., 2005; Zhao et al., 2019), correlating with maintaining the health of farmed animals at higher relative abundances (Xue et al., 2017; Hu et al., 2022). Flavobacteriaceae specialize in macromolecule degradation (Dang and Lovell, 2016; Enke et al., 2019; Gavriilidou et al., 2020). Studies showed that Flavobacteriaceae were often associated with the surface of algae, marine invertebrates, fish, and detrital particles (Dang and Lovell, 2016; Gavriilidou et al., 2020). Bacteria in the Rhodobacteraceae and Flavobacteriaceae families may play an essential role in particulate matter degradation, nutrient recycling, organic waste scavenging, and farmed animals’ health.
Bacteria can be classified into r-strategists (fast-growing, small body size, opportunistic strategy) or K-strategists (slow-growing, large body size, conservative strategy) (Reznick et al., 2002). The r/K selection theory explains seasonal pathogen proliferation. In the current study, specific bacterial taxa exhibited an obvious seasonal pattern, among which the relative abundance of Vibrionaceae, Pseudoalteromonadaceae, and Alteromonadaceae increased significantly in late summer and autumn. These bacteria are classified as r-strategists, while Rhodobacteraceae and Flavobacteriaceae that were consistently dominant in aquaculture seawater in our study, are classified as K-strategists (Lauro et al., 2009; Vadstein et al., 2018; Pettersen et al., 2021; Wietz et al., 2022). A recent study suggests that the rise in r-strategist bacteria increases homogeneous selection pressure (Liu and Salles, 2024), which aligns with our results. Homogeneous selection contributed the most to the bacterial community assembly in autumn, accompanied by increased r-strategist bacteria in Sanggou Bay mariculture seawater (Figure 5A).
The rise of r-strategist bacteria may reduce microbial interactions. Bacterial pathogens are usually r-strategists, with Vibrio species serving as typical representatives (Andrews and Harris, 1986; Vine et al., 2004). It has been shown that some health-indicator bacteria, such as those affiliated with Psychromonadaceae and Thiotrichaceae, appear significantly reduced in marine organisms’ intestines or surrounding seawater during pathogenic bacteria infection (Baliga et al., 2021; Reyes et al., 2024). Network analyses in our current study (Figure 7C) identified negative correlations between pathogenic Vibrionaceae (Otu75, Otu685) and health-indicator bacteria (Psychromonadaceae, Otu23; Thiotrichaceae, Otu1026). Ecological network analyses also revealed a significant reduction in bacterial interactions and network complexity in autumn (Figures 6A, B). These results suggest that pathogenic Vibrio species may inhibit the growth of health-indicator bacteria. Mechanistically, Vibrio bacteria often employ type VI secretion systems and/or toxin-antitoxin systems for competition (Kostiuk et al., 2017; Song et al., 2022). Therefore, elevated r-strategist pathogens may reduce the bacterial diversity and interaction complexity in the mariculture ecosystems. In contrast, K-strategist bacteria could protect farmed species from pathogens, improving survival, growth, and stress robustness (D’Alvise et al., 2013; Vadstein et al., 2018; Pettersen et al., 2021). Therefore, maintaining a high abundance of K-strategist bacteria and monitoring the dynamics of r-strategist bacteria could help prevent bacterial disease outbreaks in mariculture.
4.2 Bacterial diversity and community structure in different aquaculture types
Macroalgae-based aquaculture harbors the highest bacterial diversity. The bacterial Shannon diversity was the highest in type III (algae monoculture) and the lowest in type I (shellfish monoculture) (Figure 2B). Previous studies have shown that intensive monoculture of marine animals reduces bacterial diversity in coastal seawaters compared to non-culture ecosystems (Wang et al., 2018b), whereas kelp cultivation can enhance bacterial biodiversity (Wang et al., 2020; Sun et al., 2023). Macroalgae cultivation plays an increasingly important role in modern aquaculture, which is not only seen as the potential to improve seawater quality by assimilating excess inorganic nutrients but also as a possible “carbon sink” for CO2 sequestration, let alone providing an important source of food and natural products (Gross, 2003; Xiao et al., 2017; Duarte et al., 2022; Yong et al., 2022). Moreover, algae could secrete diverse and abundant dissolved organic matter, leading to complex algae-bacteria interactions and increased microbial community diversity and functionality (Singh and Reddy, 2014; Li H. et al., 2022; Weigel et al., 2022; Sun et al., 2023).
The dominant bacterial taxa exhibited ecosystem-specific functional traits. ANOSIM revealed a significant structural divergence among bacterial communities (Table 2), with each aquaculture type harboring unique OTUs (61 in type I, 151 in type II, and 828 in type III). In addition, although many (273) bacterial families were common among different aquaculture types, the relative abundances of these bacterial families varied in distinct aquaculture types. For example, the bacterial families Rhodobacteraceae, Microbacteriaceae, and Cryomorphaceae were more abundant in type I, whereas Methylophilaceae, Microtrichaceae, and Woeseiaceae dominated type III (Supplementary Figure S3). Rhodobacteraceae, known for their versatile carbon, nitrogen, and sulfur metabolic capabilities, may help detoxify metabolic wastes from farmed marine animals (Hu et al., 2022; Ahmed and Campbell, 2025). Microbacteriaceae play a crucial role in degrading and remineralizing polycyclic aromatic hydrocarbon compounds, which is important for detoxifying chemical contaminants and farmed organisms’ health (Jacques et al., 2008; Tonon et al., 2014; Wang et al., 2019; Hu et al., 2022). Methylophilaceae are reported to produce plant hormones such as cytokinins to promote the growth of terrestrial plants (Ivanova et al., 2000). Although the effect of Methylophilaceae on macroalgae is unclear, high abundances of these bacteria have often been found in algae cultures and aquatic angiosperm studies (Crump and Koch, 2008; Zhang et al., 2022). Microtrichaceae were reported to be one of the core phycosphere families in the macroalgae microbiomes (Lu et al., 2023). A recent study on coastal microbial populations showed that Woeseiaceae carried polysaccharide utilization loci predicted to target laminarin, alginate, and α-glucan (Viver et al., 2024). In our current study, Woeseiaceae (Otu154) was found to be the module hub with the maximum degrees in the type III (algae monoculture) bacterial network (Supplementary Table S2), which may be due to its ability to degrade algal polysaccharides. Farmed organisms may alter seawater’s chemical and nutritional conditions, selecting specific bacteria that adapt to the changed environment and contribute to inorganic nutrient transformation and organic waste scavenging (Dang and Lovell, 2016; Enke et al., 2019; Gavriilidou et al., 2020). These data suggest that bacterial communities play an essential role in the health status of the aquaculture ecosystems (Deng et al., 2021; Ebeling et al., 2006; Hu et al., 2022).
4.3 Environmental drivers of the mariculture bacterial communities
Seawater bacterial communities’ temporospatial dynamics were closely related to certain environmental factors, including seawater DO, transparency, temperature, NO2-, NO3-, Chla, and algae culture (Figure 4B). Previous studies also indicated that DO, temperature, and Chla were closely correlated with bacterial communities in mariculture systems, especially for surface waters (Alfiansah et al., 2018; Guan et al., 2020; Wang et al., 2021). Seawater DO is the most significant environmental factor affecting the bacterioplankton communities in the present study (Figure 4B). The highest DO concentrations were observed in late winter (February) (10.27-11.92 mg/L), while the lowest values were observed in late summer or early autumn (August or September) (5.56-7.51 mg/L) (Figure 1B). Our results are consistent with previous studies on surface waters of certain coastal seas and mariculture ecosystems (Li et al., 2022; Mahmood et al., 2016b; Zhou et al., 2020). Possible reasons for the observed seasonal DO dynamics include higher metabolic rates (particularly respiration) of marine organisms (including both macro- and microorganisms) and lower oxygen solubility in warmer seasons than in colder seasons in temperate marine waters (Dang, 2020; Dang and Jiao, 2014). Oxygen is essential for aerobic bacterial respiration and growth. Seawater deoxygenation could lead to a decreased oxygen utilization rate of bacteria (Lee et al., 2002). Our current study observed a relatively higher Shannon diversity in winter (Figure 2B). Long-term analyses showed that elevated DO could increase microbial diversity (Riding et al., 2019). Moreover, a reduction in DO concentration may increase facultative anaerobes, including pathogens like vibrios (Johnson et al., 2012; Guo et al., 2022), which partially explains the increased abundance of vibrios in autumn in Sanggou Bay (Figure 7A). The finding highlights the potential deoxygenation risk of mariculture for both farmed marine animals and coastal ecosystems, particularly in warmer seasons.
In the current study, a significant difference in seawater temperature was observed among different seasons, with the high temperatures observed from July to September (23.1 ± 1.6°C) and the low temperatures observed from January to March (5.3 ± 0.8°C) (Figure 1B). Temperature changes strongly influence bacterial community structure and the population dynamics of many OTUs (Figures 4A, B), consistent with previous studies (Pomeroy and Wiebe, 2001). The temperature effect is usually based on its influence on bacterial growth and production rates (White et al., 1991). In addition, elevated temperatures reduce the microbial interactions (Rajeev et al., 2021). A study by Zhou et al. (2021) also found that seawater warming could reduce prokaryotic network complexity, which aligns with our finding that microbial network nodes and links were significantly lower in summer (nodes: 185, links: 437) and autumn (nodes: 81, links: 97), compared to winter (nodes: 240, links: 687) and spring (nodes: 329, links: 1382) (Figures 6A, B; Supplementary Table S2). Moreover, the SEM result further revealed that the effects of temperature on bacterial community structure and diversity were indirect (Figure 4C), emphasizing the multiplex importance of temperature and its complex interactions with other environmental factors in driving the dynamics of bacterial communities.
Seawater transparency is another important environmental factor in aquaculture, particularly for shellfish production. The growth of phytoplankton, which is the natural feed for bivalves, is affected by seawater transparency (Tilzer et al., 1994). The current study identified a significant relationship (Spearman correlation, p< 0.05) between seawater transparency and Chla. In addition, CCA and SEM analyses both identified seawater transparency as a key environmental factor influencing the bacterial communities (Figures 4B,C). Low transparency is typically linked to high seawater turbidity and elevated particulate material concentrations, which increase (micro)niche diversity and complexity in the otherwise seemingly homogenous seawater environment (Dang and Lovell, 2016). This provides a rational explanation for the observed importance of seawater transparency in driving the variation of bacterioplankton community structure and diversity in the studied mariculture ecosystems.
4.4 Seasonal dynamics and putative driving factors of pathogenic bacteria
Seasonal changes in bacterial communities are key to understanding disease outbreaks. Disease outbreaks of aquatic organisms are usually associated with an increased abundance of pathogens (de Bruijn et al., 2018). Diseases and mortality in high-temperature seasons are common in cultivated shellfish, including abalones, scallops, and oysters (Xiao et al., 2005; Ashton et al., 2020; Bansemer et al., 2023). Pathogenic diseases also occur in some cultivated macroalgae in these seasons (Eggert et al., 2010), as high temperatures promote bacterial pathogen proliferation and pathogenicity (Colwell, 1996; Yang et al., 2021; Wang et al., 2024). In September, the highest seawater temperature was observed, coinciding with the peak mortality of the farmed animals in Sanggou Bay in 2019, which increased from late August and peaked in September, reaching an estimated ~30%-60% mortality (survey results from Weihai Changqing Ocean Science Technology Co., Ltd., data not shown). This mortality pattern largely corresponded to the monthly pattern of potential bacterial pathogens, with the bacterial genus Vibrio as a typical and the most dominant member (Figures 7A, B).
Six pathogenic Vibrio strains were found to match certain Vibrio OTUs in the seawater bacterial communities (Supplementary Figure S5), and the genomes of these strains carried a variety of virulence factors, which could lead to the massive mortality of marine organisms (Frans et al., 2011; Darshanee Ruwandeepika et al., 2012) (Supplementary Tables S4-Supplementary Tables S9). Eight exotoxin-encoding genes were identified, including hemolysin III (hlyIII) and thermolabile hemolysin (TLH). Among the various hemolysins, hlyIII was first found in V. vulnificus (Chen et al., 2004), and TLH was found in V. parahaemolyticus (Taniguchi et al., 1985). As the most widely distributed bacterial toxins in vibrios, hemolysins exert diverse effects during infection (Shinoda, 1999). Hemolysins can act on erythrocyte membranes and cause rupture of blood cells in infected animals (Zhang and Austin, 2005). Hemolysins can also cause tissue necrosis, leading to massive mortality in cultivated marine organisms (Jia et al., 2010; Wong et al., 2012; Paria et al., 2021). One-component proteins and secreted effectors of Type III secretion system (T3SS), widely implicated in Vibrio pathogenesis, were also predicted in the genomes of ScY-2106 and WaG-2229. T3SS is usually regarded as a pathogenic island that can inject a variety of effector proteins into host cells. T3SS effectors exert their virulence by manipulating the modification of host protein functions or signaling pathways (Calder et al., 2014; Portaliou et al., 2016; Li et al., 2019). The genomes of Vibrio isolates provide supporting evidence for the potential pathogen screening results of the bacterial communities. Further evidence was provided by the environmental simulation experiment, which showed that all six pathogenic Vibrio isolates could reduce the feeding/clearance rate of the tested shellfish (Figure 8), demonstrating that bacterial pathogens at near-natural densities (105 individuals/mL) in seawater could reduce the physiological activities of shellfish.
Vibrio bacteria pose a grave threat to public health as they naturally inhabit estuarine and coastal environments, with some species being human pathogens closely associated with seafood animals (Sepala Dahanayake et al., 2023; Brauge et al., 2024). Unraveling the temporospatial dynamics of the mariculture microbiomes and the pathogenic members therein, especially in warm seasons, is paramount in human and farmed animals’ health. Vibrio typically increases with rising water temperatures (Maugeri et al., 2004; Turner et al., 2009), and a similar trend was observed in the present study (Figure 7B; Supplementary Figure S4). This trend is further exacerbated by anthropogenic stressors such as global warming and intensified aquacultural activities, leading to a surge in the abundance of pathogenic and harmful (e.g., antibiotic-resistant) vibrios and the incidence of Vibrio diseases (Vezzulli et al., 2013; Ina-Salwany et al., 2019; Sanches-Fernandes et al., 2022).
Along with high temperatures, seawater deoxygenation also increases pathogen abundance, infection, and marine animal mortality (Mikulski et al., 2000; Phippen and Oliver, 2017). In Sanggou Bay, the highest abundances of bacterial pathogens, such as Vibrio species, coincided with the lowest seawater DO levels (Figure 4A), showing that seasonal deoxygenation stimulated the rise of pathogenic bacteria, especially Vibrio (Guo et al., 2022). Moreover, low-oxygen conditions can upregulate bacterial virulence gene expression in marine animals’ intestines and hypoxic seawater (Liu et al., 2011; El-Malah et al., 2014; Fan et al., 2014), mechanistically explaining the observed ecological phenomenon. In addition, hypoxia adversely impacts the survival, growth, metabolism, behavior, and immunity of marine animals (McCormick and Levin, 2017; Song et al., 2024; Waller et al., 2024), amplifying the deoxygenation effects on the losses of mariculture industries, fisheries, and environmental health. Future seawater oxygen reduction, driven by global warming and anthropogenic eutrophication, could further exacerbate the pathogenic threat (Guo et al., 2022). Our results indicate the importance of monitoring seawater DO in predicting the health status of the farmed shellfish and maricultural ecosystems.
The proliferation of algal degraders may act as a stimulator for pathogen outbreaks in shellfish mariculture. This study identified V. halioticoli (Otu1389) (Supplementary Figure S5) as a key biotic factor driving seasonal Vibrio dynamics in Sanggou Bay. This OTU emerged as a hub node with the highest degrees in the autumn bacterial network (Figures 6A, B, 7C) and showed significant positive correlations with almost all the other Vibrio OTUs (including all the pathogenic Vibrio species identified) (Figure 7D). V. halioticoli is commonly found in aquaculture seawater and the guts of herbivorous invertebrates, particularly abalones, known for its alginate lyase activities (Sawabe et al., 1998; Sugimura et al., 2000). This bacterium can form biofilms and has been detected on seaweed surfaces (Tanaka et al., 2002; Dash et al., 2009). The degradation of macroalgal alginate by V. halioticoli releases simple sugars and short-chain organic acids (Sawabe et al., 2003), promoting the growth of other heterotrophic bacteria, including pathogens. Analyses of the genomes of two V. halioticoli strains ASM49669 (GenBank accession number: GCA_000496695.1) and IAM14596 (GCA_003568965.1) through CAZy database annotation (https://www.cazy.org/Home.html) (Drula et al., 2022) revealed an extensive algal polysaccharide-degrading repertoire, including numerous algal polysaccharides-related lyases and hydrolases (Supplementary Figure 9). Moreover, genomic virulence factor analyses via the Virulence Factor Database (VFDB) identified numerous virulence determinants in Vibrio halioticoli, likely involved in Vibrio-algae interactions (Supplementary Table 17). Alginate degradation may cause the disruption of macroalgal cell walls, resulting in algal rotting and death (Zhu et al., 2016; Costa et al., 2021), consistent with the observation that diseases in cultivated Gracilaria and wild kelp became the most severe in September (data from Weihai Changqing Ocean Science Technology Co., Ltd.). The degraded macroalgae release cellular contents in the form of dissolved and particulate organic matter into the surrounding seawater, further stimulating the growth of opportunistic bacteria (i.e., the r-strategist bacteria, including opportunistic pathogens). This hypothesized scenario is supported by the observed low seawater DO and transparency in autumn (Figure 1B). The degraded macroalgal materials stimulated microbial community respiration and increased seawater turbidity. These findings suggest that the bacterial degradation of macroalgae in the IMTA systems may be an important biotic factor leading to the outbreak of shellfish pathogenic bacteria. Simultaneously monitoring algae-degrading bacteria and the health status of mass-cultivated macroalgae should be a prerequisite for preventing pathogenic diseases in mariculture.
5 Conclusions
In the present study, temporospatial bacterial community diversities and structures were investigated, and potential bacterial pathogens and their dynamics were also explored in three distinct aquaculture systems in Sanggou Bay. Seawater bacterial communities showed obvious season- and aquaculture-type-related dynamics. Environmental factors, such as seawater DO, transparency, temperature, NO2-, NO3-, Chla, and algae culture, may significantly influence the bacterial communities in Sanggou Bay. By performing a combined analysis of bacterial communities and pathogenic strains as well as environmental simulation experiments, it was found that the outbreaks of pathogenic bacteria are closely related to the seasonal changes in bacterial communities and seawater DO, temperature, and transparency. Furthermore, macroalgae-degrading bacteria, such as V. halioticoli, were found for the first time as a putative biotic factor stimulating the growth of pathogenic bacteria in high-temperature seasons via their active degradation and transformation of macroalgal biomass, thus providing enriched carbon and energy sources to the pathogens. For future aquaculture practices, some optimizing strategies are proposed: (1) synchronically monitoring the bacterial communities and critical physicochemical parameters, including seawater dissolved oxygen, temperature, and transparency to provide valuable information for the prediction of pathogenic bacteria in the marine aquaculture systems; (2) monitoring the health status of cultivated macroalgae (e.g., relative growth rate and tissue necrosis index) and the temporospatial dynamics of macroalgae-degrading bacteria, especially during high-temperature seasons; and (3) optimizing the balance between shellfish and macroalgae cultivations and carefully managing harvest timing. These practical strategies may help mitigate the outbreaks of seasonal pathogenic diseases for farmed marine animals in the IMTA systems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LL: Conceptualization, Investigation, Project administration, Validation, Writing – original draft, Funding acquisition. LN: Formal Analysis, Writing – review & editing. CA: Writing – review & editing. DZ: Funding acquisition, Writing – review & editing. PZ: Formal Analysis, Writing – review & editing. XJL: Data curation, Formal Analysis, Investigation, Software, Visualization, Writing – original draft, Writing – review & editing. XL: Formal Analysis, Funding acquisition, Methodology, Resources, Writing – review & editing. HD: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the National Key Research and Development Program of China (2020YFA0608302), the National Natural Science Foundation of China (42076111, 42141003, 42188102, and 42376084), the Natural Science Foundation of Shandong Province (ZR2021QC220), the Beijing Natural Science Foundation (8222047), and the Zhejiang Provincial Natural Science Foundation of China (LZ24D060003).
Conflict of interest
Author LL was employed by the company Weihai Changqing Ocean Science Technology CO., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1581190/full#supplementary-material
References
Abd El-Hack M. E., El-Saadony M. T., Ellakany H. F., Elbestawy A. R., Abaza S. S., Geneedy A. M., et al. (2022). Inhibition of microbial pathogens in farmed fish. Mar. Pollut. Bull. 183, 114003. doi: 10.1016/j.marpolbul.2022.114003
Ahmed M. A. and Campbell B. J. (2025). Genome-resolved adaptation strategies of Rhodobacterales to changing conditions in the Chesapeake and Delaware Bays. Appl. Environ. Microb. 91, e02357–e02324. doi: 10.1128/aem.02357-24
Alfiansah Y. R., Hassenrück C., Kunzmann A., Taslihan A., Harder J., and Gärdes A. (2018). Bacterial abundance and community composition in pond water from shrimp aquaculture systems with different stocking densities. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02457
Algammal A., Hetta H. F., Mabrok M., and Behzadi P. (2023). Emerging multidrug-resistant bacterial pathogens “superbugs”: A rising public health threat. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1135614
Aly S. M. and Fathi M. (2024). Advancing aquaculture biosecurity: a scientometric analysis and future outlook for disease prevention and environmental sustainability. Aquacult. Int. 32, 8763–8789. doi: 10.1007/s10499-024-01589-y
An X. L., Wang J. Y., Pu Q., Li H., Pan T., Li H. Q., et al. (2020). High-throughput diagnosis of human pathogens and fecal contamination in marine recreational water. Environ. Res. 190, 109982. doi: 10.1016/j.envres.2020.109982
Andrews J. H. and Harris R. F. (1986). r-and K-selection and microbial ecology. Adv. Microb. Ecol. 9, 99–147. doi: 10.1007/978-1-4757-0611-6_3
Ashton E. C., Guist S., Roberts D., and Sigwart J. D. (2020). Effects of environmental factors and husbandry practices on summer mortality events in the cultivated Pacific oyster Crassostrea gigas in the North of Ireland. J. Shellfish Res. 39, 13–20. doi: 10.2983/035.039.0102
Baker-Austin C., Oliver J. D., Alam M., Ali A., Waldor M. K., Qadri F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers 4, 1–19. doi: 10.1038/s41572-018-0005-8
Baliga P., Goolappa P. T., Shekar M., Girisha S., Ramesh K., Udyavara V., et al. (2021). Bacterial community changes in Penaeus vannamei Boone 1931 surface and rearing water during Enterocytozoon hepatopenaei infection. Asian Fish. Sci. 34, 168–180. doi: 10.33997/j.afs.2021.34.2.006
Bansemer M. S., Buss J. J., Macaulay G., Bradley T., Knowles G., Dang C., et al. (2023). Risk factors for summer mortality in greenlip abalone (Haliotis laevigata) and hybrid abalone (H. laevigata × Haliotis rubra): A case-control study. Aquaculture 577, 739928. doi: 10.1016/j.aquaculture.2023.739928
Bastian M., Heymann S., and Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. ICWSM Conf. 3, 361–362. doi: 10.1609/icwsm.v3i1.13937
Bentzon-Tilia M., Sonnenschein E. C., and Gram L. (2016). Monitoring and managing microbes in aquaculture–Towards a sustainable industry. Microb. Biotechnol. 9, 576–584. doi: 10.1111/1751-7915.12392
Brauge T., Mougin J., Ells T., and Midelet G. (2024). Sources and contamination routes of seafood with human pathogenic Vibrio spp.: A Farm-to-Fork approach. Compr. Rev. Food Sci. F. 23, e13283. doi: 10.1111/1541-4337.13283
Bruhn J. B., Nielsen K. F., Hjelm M., Hansen M., Bresciani J., Schulz S., et al. (2005). Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl. Environ. Microb. 71, 7263–7270. doi: 10.1128/AEM.71.11.7263-7270.2005
Buck B. H., Troell M. F., Krause G., Angel D. L., Grote B., and Chopin T. (2018). State of the art and challenges for offshore integrated multi-trophic aquaculture (IMTA). Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00165
Cabello F. C., Godfrey H. P., Buschmann A. H., and Dölz H. J. (2016). Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 16, e127–e133. doi: 10.1016/S1473-3099(16)00100-6
Calder T., de Souza Santos M., Attah V., Klimko J., Fernandez J., Salomon D., et al. (2014). Structural and regulatory mutations in Vibrio parahaemolyticus type III secretion systems display variable effects on virulence. FEMS Microbiol. Lett. 361, 107–114. doi: 10.1111/1574-6968.12619
Califano G., Kwantes M., Abreu M. H., Costa R., and Wichard T. (2020). Cultivating the macroalgal holobiont: effects of integrated multi-trophic aquaculture on the microbiome of Ulva rigida (Chlorophyta). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00052
Chang Z. Q., Neori A., He Y. Y., Li J. T., Qiao L., Preston S. I., et al. (2020). Development and current state of seawater shrimp farming, with an emphasis on integrated multi-trophic pond aquaculture farms, in China–a review. Rev. Aquacult. 12, 2544–2558. doi: 10.1111/raq.12457
Chen Q. L., An X. L., Li H., Su J. Q., Ma Y. B., and Zhu Y.-G. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92, 1–10. doi: 10.1016/j.envint.2016.03.026
Chen Y. C., Chang M. C., Chuang Y. C., and Jeang C. L. (2004). Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr. Microbiol. 49, 175–179. doi: 10.1007/s00284-004-4288-5
Chopin T., Buschmann A. H., Halling C., Troell M., Kautsky N., Neori A., et al. (2001). Integrating seaweeds into marine aquaculture systems: a key toward sustainability. J. Phycol. 37, 975–986. doi: 10.1046/j.1529-8817.2001.01137.x
Clarke K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Colwell R. R. (1996). Global climate and infectious disease: the cholera paradigm. Science 274, 2025–2031. doi: 10.1126/science.274.5295.2025
Costa M., Pio L., Bule P., Cardoso V., Alfaia C. M., Coelho D., et al. (2021). An individual alginate lyase is effective in the disruption of Laminaria digitata recalcitrant cell wall. Sci. Rep. 11, 9706. doi: 10.1038/s41598-021-89278-1
Crump B. C. and Koch E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. App. Environ. Microb. 74, 5948–5957. doi: 10.1128/AEM.00952-08
D’Alvise P. W., Lillebø S., Wergeland H. I., Gram L., and Bergh Ø. (2013). Protection of cod larvae from vibriosis by Phaeobacter spp.: a comparison of strains and introduction times. Aquaculture 384, 82–86. doi: 10.1016/j.aquaculture.2012.12.013
Dai T. J., Wen D. H., Bates C. T., Wu L. W., Guo X., Liu S., et al. (2022). Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 13, 175. doi: 10.1038/s41467-021-27857-6
Dang H. Y. (2020). Grand challenges in microbe-driven marine carbon cycling research. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01039
Dang H. Y. and Jiao N. Z. (2014). Perspectives on the microbial carbon pump with special reference to microbial respiration and ecosystem efficiency in large estuarine systems. Biogeosciences 11, 3887–3898. doi: 10.5194/bg-11-3887-2014
Dang H. Y. and Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. R 80, 91–138. doi: 10.1128/mmbr.00037-15
Darshanee Ruwandeepika H. A., Sanjeewa Prasad Jayaweera T., Paban Bhowmick P., Karunasagar I., Bossier P., and Defoirdt T. (2012). Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the Harveyi clade. Rev. Aquacult. 4, 59–74. doi: 10.1111/j.1753-5131.2012.01061.x
Dash S., Jin C., Lee O. O., Xu Y., and Qian P.-Y. (2009). Antibacterial and antilarval-settlement potential and metabolite profiles of novel sponge-associated marine bacteria. J. Ind. Microbiol. Biotechnol. 36, 1047–1056. doi: 10.1007/s10295-009-0588-x
de Bruijn I., Liu Y., Wiegertjes G. F., and Raaijmakers J. M. (2018). Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 94, fix161. doi: 10.1093/femsec/fix161
Delcher A. L., Harmon D., Kasif S., White O., and Salzberg S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641. doi: 10.1093/nar/27.23.4636
Deng M., Zhao X. L., Senbati Y., Song K., and He X. G. (2021). Nitrogen removal by heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas sp. DM02: Removal performance, mechanism and immobilized application for real aquaculture wastewater treatment. Bioresource Technol. 322, 124555. doi: 10.1016/j.biortech.2020.124555
Deng Y., Zhou F., Ruan Y. J., Ma B., Ding X. Y., Yue X. M., et al. (2019). Feed types driven differentiation of microbial community and functionality in marine integrated multitrophic aquaculture system. Water 12, 95. doi: 10.3390/w12010095
Dronen K., Roalkvam I., Nilsen H., Olsen A. B., Dahle H., and Wergeland H. (2022). Presence and habitats of bacterial fish pathogen relatives in a marine salmon post-smolt RAS. Aquacult. Rep. 26, 101312. doi: 10.1016/j.aqrep.2022.101312
Dronen K., Roalkvam I., Tungland K., Dahle H., Nilsen H., Olsen A., et al. (2023). How to define fish pathogen relatives from a 16S rRNA sequence library and Pearson correlation analysis between defined OTUs from the library: Supplementary data to the research article “Presence and habitats of bacterial fish pathogen relatives in a marine salmon post-smolt RAS. Data Brief 46, 108846. doi: 10.1016/j.dib.2022.108846
Drula E., Garron M. L., Dogan S., Lombard V., Henrissat B., and Terrapon N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Duarte C. M., Bruhn A., and Krause-Jensen D. (2022). A seaweed aquaculture imperative to meet global sustainability targets. Nat. Sustain. 5, 185–193. doi: 10.1038/s41893-021-00773-9
Ebeling J. M., Timmons M. B., and Bisogni J. (2006). Engineering analysis of the stoichiometry of photoautotrophic, autotrophic, and heterotrophic removal of ammonia-nitrogen in aquaculture systems. Aquaculture 257, 346–358. doi: 10.1016/j.aquaculture.2006.03.019
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C., Haas B. J., Clemente J. C., Quince C., and Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eggert A., Peters A. F., and Küpper F. C. (2010). “The potential impact of climate change on endophyte infections in kelp sporophytes,” in Seaweeds and their role in globally changing environments. Eds. Seckbach J., Einav R., and Israel A. (Springer Press, Dordrecht), 139–154. doi: 10.1007/978-90-481-8569-6_9
El-Malah S. S., Yang Z. Q., Hu M. Z., Li Q. C., Pan Z. M., and Jiao X. A. (2014). Vibrio parahaemolyticus strengthens their virulence through modulation of cellular reactive oxygen species in vitro. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00168
Enke T. N., Datta M. S., Schwartzman J., Cermak N., Schmitz D., Barrere J., et al. (2019). Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, 1528–1535.E6. doi: 10.1016/j.cub.2019.03.047
Fan F. X., Liu Z., Jabeen N., Birdwell L. D., Zhu J., and Kan B. (2014). Enhanced interaction of Vibrio cholerae virulence regulators TcpP and ToxR under oxygen-limiting conditions. Infect. Immun. 82, 1676–1682. doi: 10.1128/iai.01377-13
Fang G. J., Yu H. L., Sheng H. X., Tang Y. L., and Liang Z. L. (2021). Comparative analysis of microbial communities between water and sediment in Laoshan Bay marine ranching with varied aquaculture activities. Mar. Pollut. Bull. 173, 112990. doi: 10.1016/j.marpolbul.2021.112990
Fang J. G., Zhang J., Xiao T., Huang D. J., and Liu S. M. (2016). Integrated multi-trophic aquaculture (IMTA) in Sanggou Bay, China. Aquacult. Env. Interact. 8, 201–205. doi: 10.3354/aei00179
Feng K., Peng X., Zhang Z., Gu S. S., He Q., Shen W. L., et al. (2022). iNAP: an integrated network analysis pipeline for microbiome studies. iMeta 1, e13. doi: 10.1002/imt2.13
Frans I., Michiels C. W., Bossier P., Willems K. A., Lievens B., and Rediers H. (2011). Vibrio Anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. fish Dis. 34, 643–661. doi: 10.1111/j.1365-2761.2011.01279.x
Fu M. Z., Pu X. M., Wang Z. L., and Liu X. J. (2013). Integrated assessment of mariculture ecosystem health in Sanggou Bay. Acta Ecol. Sin. 33, 238-248. doi: 10.5846/stxb201106270960 (in Chinese with English abstract)
Fu S. Z., Wang Q. Y., Wang R., Zhang Y. X., Lan R. T., He F. L., et al. (2022). Horizontal transfer of antibiotic resistance genes within the bacterial communities in aquacultural environment. Sci. Total Environ. 820, 153286. doi: 10.1016/j.scitotenv.2022.153286
Gavriilidou A., Gutleben J., Versluis D., Forgiarini F., van Passel M. W. J., Ingham C. J., et al. (2020). Comparative genomic analysis of Flavobacteriaceae: insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genomics 21, 569. doi: 10.1186/s12864-020-06971-7
Granada L., Sousa N., Lopes S., and Lemos M. F. (2016). Is integrated multitrophic aquaculture the solution to the sectors’ major challenges?–a review. Rev. Aquacult. 8, 283–300. doi: 10.1111/raq.12093
Gross E. M. (2003). Allelopathy of aquatic autotrophs. Crit. Rev. Plant Sci. 22, 313–339. doi: 10.1080/713610859
Guan X. Y., Wang B., Duan P., Tian J. S., Dong Y., Jiang J. W., et al. (2020). The dynamics of bacterial community in a polyculture aquaculture system of Penaeus chinensis, Rhopilema esculenta and Sinonovacula constricta. Aquac. Res. 51, 1789–1800. doi: 10.1111/are.14528
Guo Y. Y., Wu C., and Sun J. (2022). Pathogenic bacteria significantly increased under oxygen depletion in coastal waters: A continuous observation in the central Bohai Sea. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1035904
Hadley S., Wild-Allen K., Johnson C., and Macleod C. (2016). Quantification of the impacts of finfish aquaculture and bioremediation capacity of integrated multi-trophic aquaculture using a 3D estuary model. J. Appl. Phycol. 28, 1875–1889. doi: 10.1007/s10811-015-0714-2
Han T. T., Jiang Z. J., Fang J. G., Zhang J. H., Mao Y. Z., Zou J., et al. (2013). Carbon dioxide fixation by the seaweed Gracilaria lemaneiformis in integrated multi-trophic aquaculture with the scallop Chlamys farreri in Sanggou Bay, China. Aquacult. Int. 21, 1035–1043. doi: 10.1007/s10499-012-9610-9
Han J. C., Liu X., Pan K. H., Liu J. J., Sun Y., Jin G. Y., et al. (2024). Impacts of integrated multi-trophic Aquaculture on phytoplankton in Sanggou Bay. J. Ocean U. China 23, 835–843. doi: 10.1007/s11802-024-5665-2
Han T. T., Shi R. J., Qi Z. H., Huang H. H., Liang Q. Y., and Liu H. X. (2017). Interactive effects of oyster and seaweed on seawater dissolved inorganic carbon systems: implications for integrated multi-trophic aquaculture. Aquacult. Environ. Interact. 9, 469–478. doi: 10.3354/aei00246
Hijmans R. J., Williams E., Vennes C., and Hijmans M. R. J. (2017). Package ‘geosphere’. Spherical trigonometry 1, 1–45. Available at: https://github.com/rspatial/geosphere (Accessed November 15, 2024).
Hossain A., Senff P., and Glaser M. (2022). Lessons for coastal applications of IMTA as a way towards sustainable development: A review. Appl. Sci. 12, 11920. doi: 10.3390/app122311920
Hu D., Wang L. P., Zhao R., Zeng J., and Shao Z. Z. (2022). Core microbiome involved in nitrite removal in shrimp culture ponds. Aquac. Res. 53, 1663–1675. doi: 10.1111/are.15698
Huerta-Cepas J., Serra F., and Bork P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Ina-Salwany M., Al-saari N., Mohamad A., Mursidi F. A., Mohd-Aris A., Amal M., et al. (2019). Vibriosis in fish: A review on disease development and prevention. J. Aquat. Anim. Health 31, 3–22. doi: 10.1002/aah.10045
Ivanova E., Doronina N., Shepelyakovskaya A., Laman A., Brovko F., and Trotsenko Y. A. (2000). Facultative and obligate aerobic methylobacteria synthesize cytokinins. Microbiology 69, 646–651. doi: 10.1023/A:1026693805653
Jacques R. J., Okeke B. C., Bento F. M., Teixeira A. S., Peralba M. C., and Camargo F. A. (2008). Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresource Technol. 99, 2637–2643. doi: 10.1016/j.biortech.2007.04.047
Janda J. M. (2014). Shewanella: a marine pathogen as an emerging cause of human disease. Clin. Microbiol. News. 36, 25–29. doi: 10.1016/j.clinmicnews.2014.01.006
Jia A. R., Woo N. Y. S., and Zhang X. H. (2010). Expression, purification, and characterization of thermolabile hemolysin (TLH) from Vibrio alginolyticus. Dis. Aquat. Organ. 90, 121–127. doi: 10.3354/dao02225
Johnson C. N., Bowers J. C., Griffitt K. J., Molina V., Clostio R. W., Pei S., et al. (2012). Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microb. 78, 7249–7257. doi: 10.1128/AEM.01296-12
Katoh K. and Kumah K. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Knowler D., Chopin T., Martínez-Espiñeira R., Neori A., Nobre A., Noce A., et al. (2020). The economics of integrated multi-trophic aquaculture: where are we now and where do we need to go? Rev. Aquacult. 12, 1579–1594. doi: 10.1111/raq.12399
Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., and Phillippy A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kostiuk B., Unterweger D., Provenzano D., and Pukatzki S. (2017). T6SS intraspecific competition orchestrates Vibrio cholerae genotypic diversity. Int. Microbiol. 20, 130–137. doi: 10.2436/20.1501.01.294
Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lafferty K. D., Harvell C. D., Conrad J. M., Friedman C. S., Kent M. L., Kuris A. M., et al. (2015). Infectious diseases affect marine fisheries and aquaculture economics. Ann. Rev. Mar. Sci. 7, 471–496. doi: 10.1146/annurev-marine-010814-015646
Langlois L., Akhtar N., Tam K. C., Dixon B., and Reid G. (2021). Fishing for the right probiotic: host-microbe interactions at the interface of effective aquaculture strategies. FEMS Microbiol. Rev. 45, fuab030. doi: 10.1093/femsre/fuab030
Lauro F. M., McDougald D., Thomas T., and Cavicchioli R. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533. doi: 10.1073/pnas.0903507106
Lee C. S. (2019). Challenges and opportunities of IMTA in Hawaii and beyond. Bull. Japan Fisheries Res. Educ. Agency 49, 129–134.
Lee C. W., Kudo I., Yokokawa T., Yanada M., and Maita Y. (2002). Dynamics of bacterial respiration and related growth efficiency, dissolved nutrients and dissolved oxygen concentration in a subarctic coastal embayment. Mar. Freshwater Res. 53, 1–7. doi: 10.1071/MF01003
Lefcheck J. S. (2016). piecewiseSEM: Piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573–579. doi: 10.1111/2041-210X.12512
Li J. W., Dong S. L., Gao Q. F., Wang F., Tian X. L., and Zhang S. S. (2014). Total organic carbon budget of integrated aquaculture system of sea cucumber Apostichopus japonicus, jellyfish Rhopilema esculenta and shrimp Fenneropenaeus chinensis. Aquac. Res. 45, 1825–1831. doi: 10.1111/are.12131
Li X. J., Liang Y. T., Wang Z. H., Yao Y. Y., Chen X. L., Shao A. R., et al. (2022). Isolation and characterization of a novel Vibrio natriegens-infecting phage and its potential therapeutic application in abalone aquaculture. Biology 11, 1670. doi: 10.3390/biology11111670
Li C., Liu J. H., Chen X., Ren H. W., Su B., Ma K., et al. (2022). Determinism governs the succession of disturbed bacterioplankton communities in a coastal maricultural ecosystem. Sci. Total Environ. 828, 154457. doi: 10.1016/j.scitotenv.2022.154457
Li R. H., Liu S. M., Zhang J., Jiang Z. J., and Fang J. G. (2016). Sources and export of nutrients associated with integrated multi-trophic aquaculture in Sanggou Bay, China. Aquacult. Env. Interac. 8, 285–309. doi: 10.3354/aei00177
Li L. Z., Meng H. M., Gu D., Li Y., and Jia M. D. (2019). Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 222, 43–51. doi: 10.1016/j.micres.2019.03.003
Li L. J., Xu F., Xu J. X., Yan Y., Su J. Q., Zhu Y. G., et al. (2024). Spatiotemporal changes of antibiotic resistance, potential pathogens, and health risk in kindergarten dust. Environ. Sci. Technol. 58, 3919–3930. doi: 10.1021/acs.est.3c07935
Li H. M., Zhang Y. Y., Liang Y. T., Chen J., Zhu Y. C., Zhao Y. T., et al. (2018). Impacts of maricultural activities on characteristics of dissolved organic carbon and nutrients in a typical raft-culture area of the Yellow Sea, North China. Mar. Pollut. Bull. 137, 456–464. doi: 10.1016/j.marpolbul.2018.10.048
Li H. M., Zhang Z. H., Xiong T. Q., Tang K. X., He C., Shi Q., et al. (2022). Carbon sequestration in the form of recalcitrant dissolved organic carbon in a seaweed (kelp) farming environment. Environ. Sci. Technol. 56, 9112–9122. doi: 10.1021/acs.est.2c01535
Liang Y. T., Zhang Y. Y., Zhou C., Li H. M., Kang X. M., Wang L., et al. (2019). Cumulative impact of long-term intensive mariculture on total and active bacterial communities in the core sediments of the Ailian Bay, North China. Sci. Total Environ. 691, 1212–1224. doi: 10.1016/j.scitotenv.2019.07.200
Liu Z. G., Iqbal M., Zeng Z. B., Lian Y. X., Zheng A. F., Zhao M. M., et al. (2020). Comparative analysis of microbial community structure in the ponds with different aquaculture model and fish by high-throughput sequencing. Microb. Pathogenesis 142, 104101. doi: 10.1016/j.micpath.2020.104101
Liu B. and Pop M. (2009). ARDB–antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
Liu X. P. and Salles J. F. (2024). Bridging ecological assembly process and community stability upon bacterial invasions. ISME J. 18, wrae066. doi: 10.1093/ismejo/wrae066
Liu Z., Yang M. H., Peterfreund G. L., Tsou A. M., Selamoglu N., Daldal F., et al. (2011). Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc. Natl. Acad. Sci. U.S.A. 108, 810–815. doi: 10.1073/pnas.1014640108
Liu B., Zheng D. D., Zhou S. Y., Chen L. H., and Yang J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkn656
Loret P., Le Gall S., Dupuy C., Blanchot J., Pastoureaud A., Delesalle B., et al. (2000). Heterotrophic protists as a trophic link between picocyanobacteria and the pearl oyster Pinctada margaritifera in the Takapoto lagoon (Tuamotu Archipelago, French Polynesia). Aquat. Microb. Ecol. 22, 215–226. doi: 10.3354/ame022215
Lu D. C., Wang F. Q., Amann R. I., Teeling H., and Du Z. J. (2023). Epiphytic common core bacteria in the microbiomes of co-located green (Ulva), brown (Saccharina) and red (Grateloupia, Gelidium) macroalgae. Microbiome 11, 126. doi: 10.1186/s40168-023-01559-1
Luo H. W. and Moran M. A. (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. R. 78, 573–587. doi: 10.3354/ame022215
Magoč T. and Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mahmood T., Fang J. G., Jiang Z. J., and Zhang J. (2016a). Carbon and nitrogen flow, and trophic relationships, among the cultured species in an integrated multi-trophic aquaculture (IMTA) bay. Aquacult. Env. Interac. 8, 207–219. doi: 10.3354/aei00152
Mahmood T., Fang J. G., Jiang Z. J., and Zhang J. (2016b). Seasonal nutrient chemistry in an integrated multi-trophic aquaculture region: case study of Sanggou Bay from North China. Chem. Ecol. 32, 149–168. doi: 10.1080/02757540.2015.1121246
Mao Y. Z., Zhou Y., Yang H. S., and Wang R. C. (2006). Seasonal variation in metabolism of cultured Pacific oyster, Crassostrea gigas in Sanggou bay, China. Aquaculture 253, 322–333. doi: 10.1016/j.aquaculture.2005.05.033
Maugeri T., Carbone M., Fera M., Irrera G., and Gugliandolo C. (2004). Distribution of potentially pathogenic bacteria as free living and plankton associated in a marine coastal zone. J. Appl. Microbiol. 97, 354–361. doi: 10.1111/j.1365-2672.2004.02303.x
Mayali X., Franks P. J., and Azam F. (2008). Cultivation and ecosystem role of a marine Roseobacter clade-affiliated cluster bacterium. Appl. Environ. Microb. 74, 2595–2603. doi: 10.1128/AEM.02191-07
McCormick L. R. and Levin L. A. (2017). Physiological and ecological implications of ocean deoxygenation for vision in marine organisms. Philos. T. R. Soc A 375, 20160322. doi: 10.1098/rsta.2016.0322
McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Meier-Kolthoff J. P. and Göker M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182. doi: 10.1038/s41467-019-10210-3
Mikulski C. M., Burnett L. E., and Burnett K. G. (2000). The effects of hypercapnic hypoxia on the survival of shrimp challenged with Vibrio parahaemolyticus. J. Shellfish Res. 19, 301–311. doi: 10.1016/j.jenvp.2020.103556
Mohamad N., Amal M. N. A., Yasin I. S. M., Saad M. Z., Nasruddin N. S., Al-saari N., et al. (2019). Vibriosis in cultured marine fishes: a review. Aquaculture 512, 734289. doi: 10.1016/j.aquaculture.2019.734289
Molloy S. D., Pietrak M. R., Bricknell I., and Bouchard D. A. (2013). Experimental transmission of infectious pancreatic necrosis virus from the blue mussel, Mytilus edulis, to cohabitating Atlantic Salmon (Salmo salar) smolts. Appl. Environ. Microbiol. 79, 5882–5890. doi: 10.1128/AEM.01142-13
Nawaz M. Z., Subin Sasidharan R., Alghamdi H. A., and Dang H. Y. (2022). Understanding interaction patterns within deep-sea microbial communities and their potential applications. Mar. Drugs 20, 108. doi: 10.3390/md20020108
Naylor R. L., Hardy R. W., Buschmann A. H., Bush S. R., Cao L., Klinger D. H., et al. (2021). A 20-year retrospective review of global aquaculture. Nature 591, 551–563. doi: 10.1038/s41586-021-03308-6
Ning D. L., Yuan M. T., Wu L. W., Zhang Y., Guo X., Zhou X. S., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11, 4717. doi: 10.1038/s41467-020-18560-z
Nissar S., Bakhtiyar Y., Arafat M. Y., Andrabi S., Mir Z. A., Khan N. A., et al. (2023). The evolution of integrated multi-trophic aquaculture in context of its design and components paving way to valorization via optimization and diversification. Aquaculture 565, 739074. doi: 10.1016/j.aquaculture.2022.739074
Nobre A. M., Robertson-Andersson D., Neori A., and Sankar K. (2010). Ecological–economic assessment of aquaculture options: comparison between abalone monoculture and integrated multi-trophic aquaculture of abalone and seaweeds. Aquaculture 306, 116–126. doi: 10.1016/j.aquaculture.2010.06.002
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2020). R package for community ecologists: popular ordination methods, ecological null models & diversity analysis, vegan 2.5-6.1. Available online at: https://CRAN.R-project.org/package=vegan (Accessed March 21, 2024).
Paria P., Behera B. K., Mohapatra P. K. D., and Parida P. K. (2021). Virulence factor genes and comparative pathogenicity study of tdh, trh and tlh positive Vibrio parahaemolyticus strains isolated from Whiteleg shrimp, Litopenaeus vannamei (Boone 1931) in India. Infect. Genet. Evol. 95, 105083. doi: 10.1016/j.meegid.2021.105083
Patil P. K., Geetha R., Ravisankar T., Avunje S., Solanki H. G., Abraham T. J., et al. (2021). Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV). Aquaculture 533, 736231. doi: 10.1016/j.aquaculture.2020.736231
Pettersen J. P., Gundersen M. S., and Almaas E. (2021). Robust bacterial co-occurence community structures are independent of r- and K-selection history. Sci. Rep. 11, 23497. doi: 10.1038/s41598-021-03018-z
Phippen B. L. and Oliver J. D. (2017). Impact of hypoxia on gene expression patterns by the human pathogen, Vibrio vulnificus, and bacterial community composition in a North Carolina estuary. GeoHealth 1, 37–50. doi: 10.1002/2016GH000024
Pietrak M. R., Molloy S. D., Bouchard D. A., Singer J. T., and Bricknell I. (2012). Potential role of Mytilus edulis in modulating the infectious pressure of Vibrio Anguillarum 02β on an integrated multi-trophic aquaculture farm. Aquaculture 326, 36–39. doi: 10.1002/2016GH000024
Pomeroy L. R. and Wiebe W. J. (2001). Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23, 187–204. doi: 10.3354/ame023187
Portaliou A. G., Tsolis K. C., Loos M. S., Zorzini V., and Economou A. (2016). Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189. doi: 10.1016/j.tibs.2015.09.005
Preena P. G., Swaminathan T. R., Kumar V. J. R., and Singh I. S. B. (2020). Antimicrobial resistance in aquaculture: A crisis for concern. Biologia 75, 1497–1517. doi: 10.2478/s11756-020-00456-4
Price M. N., Dehal P. S., and Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5, e9490. doi: 10.1371/journal.pone.0009490
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rajeev M., Sushmitha T., Aravindraja C., Toleti S. R., and Pandian S. K. (2021). Thermal discharge-induced seawater warming alters richness, community composition and interactions of bacterioplankton assemblages in a coastal ecosystem. Sci. Rep. 11, 17341. doi: 10.1038/s41598-021-96969-2
Reyes G., Andrade B., Betancourt I., Panchana F., Preciado C., and Bayot B. (2024). Bacterial communities and signatures in the stomach and intestine of juvenile Penaeus (litopenaeus) vannamei shrimp affected by acute hepatopancreatic necrosis disease. Heliyon 10, e33034. doi: 10.1016/j.heliyon.2024.e33034
Reznick D., Bryant M. J., and Bashey F. (2002). r- and K-selection revisited: the role of population regulation in life–history evolution. Ecology 83, 1509–1520. doi: 10.1890/0012-9658(2002)083[1509:RAKSRT]2.0.CO;2
Riding R., Liang L., Lee J. H., and Virgone A. (2019). Influence of dissolved oxygen on secular patterns of marine microbial carbonate abundance during the past 490 Myr. Palaeogeogr. Palaeocl. 514, 135–143. doi: 10.1016/j.palaeo.2018.10.006
Ridler N., Wowchuk M., Robinson B., Barrington K., Chopin T., Robinson S., et al. (2007). Integrated multi – trophic aquaculture (IMTA): a potential strategic choice for farmers. Aquacult. Econ. Manage. 11, 99–110. doi: 10.1080/13657300701202767
Rowley A. F., Baker-Austin C., Boerlage A. S., Caillon C., Davies C. E., Duperret L., et al. (2024). Diseases of marine fish and shellfish in an age of rapid climate change. iScience 27, 110838. doi: 10.1016/j.isci.2024.110838
Salas-González I., Reyt G., Flis P., Custódio V., Gopaulchan D., Bakhoum N., et al. (2021). Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science 371, eabd0695. doi: 10.1126/science.abd0695
Sanches-Fernandes G. M., Sá-Correia I., and Costa R. (2022). Vibriosis outbreaks in aquaculture: addressing environmental and public health concerns and preventive therapies using gilthead seabream farming as a model system. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.904815
Sawabe T., Inoue S., Fukui Y., Yoshie K., Nishihara Y., and Miura H. (2007). Mass mortality of Japanese abalone Haliotis discus hannai caused by Vibrio harveyi infection. Microbes Environ. 22, 300–308. doi: 10.1264/jsme2.22.300
Sawabe T., Setoguchi N., Inoue S., Tanaka R., Ootsubo M., Yoshimizu M., et al. (2003). Acetic acid production of Vibrio halioticoli from alginate: a possible role for establishment of abalone–V. halioticoli association. Aquaculture 219, 671–679. doi: 10.1016/S0044-8486(02)00618-X
Sawabe T., Sugimura I., Ohtsuka M., Nakano K., Tajima K., Ezura Y., et al. (1998). Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. Int. J. Syst. Evol. Micr. 48, 573–580. doi: 10.1099/00207713-48-2-573
Sepala Dahanayake P., Majeed S., Kumarage P. M., and Heo G.-J. (2023). Molluscan shellfish: a potential source of pathogenic and multidrug-resistant Vibrio spp.J. Consum. Prot. Food S. 18, 227–242. doi: 10.1007/s00003-023-01445-w
Shinoda S. (1999). Protein toxins produced by pathogenic vibrios. J. Nat. Toxins 8, 259–269. Available at: https://europepmc.org/article/med/10410336impact (Accessed December 13, 2023).
Singh R. P. and Reddy C. (2014). Seaweed-microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 88, 213–230. doi: 10.1111/1574-6941.12297
Song J. J., Farhadi A., Tan K., Lim L., and Tan K. (2024). Impact of anthropogenic global hypoxia on the physiological response of bivalves. Sci. Total Environ. 926, 172056. doi: 10.1016/j.scitotenv.2024.172056
Song X. J., Lin Z., and Yuan W. S. (2022). Toxin-antitoxin systems in pathogenic Vibrio species: a mini review from a structure perspective. 3 Biotech. 12, 125. doi: 10.1016/j.scitotenv.2024.172056
Stabili L., Giangrande A., Arduini D., Borghese J., Petrocelli A., Alabiso G., et al. (2023). Environmental quality improvement of a mariculture plant after its conversion into a multi-trophic system. Sci. Total Environ. 884, 163846. doi: 10.1016/j.scitotenv.2023.163846
Sugimura I., Sawabe T., and Ezura Y. (2000). Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar. Biotechnol. 2, 65–73. doi: 10.1007/s101269900010
Sun Y., Li H. J., Wang X. C., Li H. B., and Deng Y. (2023). Kelp culture enhances coastal biogeochemical cycles by maintaining bacterioplankton richness and regulating its interactions. Msystems 8, e00002–e00023. doi: 10.1128/msystems.00002-23
Tanaka R., Sawabe T., Yoshimizu M., and Ezura Y. (2002). Distribution of Vibrio halioticoli around an abalone-farming center in Japan. Microbes Environ. 17, 6–9. doi: 10.1264/jsme2.2002.6
Taniguchi H., Ohta H., Ogawa M., and Mizuguchi Y. (1985). Cloning and expression in Escherichia coli of Vibrio parahaemolyticus thermostab le direct hemolysin and thermolabile hemolysin genes. J. Bacteriol. 162, 510–515. doi: 10.1128/jb.162.2.510-515.1985
Tarnecki A. M., Brennan N. P., Guttman L., and Main K. L. (2025). Variability in prokaryotic and eukaryotic periphyton communities in marine recirculating integrated multi-trophic aquaculture systems. Aquaculture 600, 742210. doi: 10.1016/j.aquaculture.2025.742210
Tilzer M., Gieskes W., Heusel R., and Fenton N. (1994). The impact of phytoplankton on spectral water transparency in the Southern Ocean: implications for primary productivity. Polar Biol. 14, 127–136. doi: 10.1007/BF00234975
Tonon L. A. C., Moreira A., and Thompson F. (2014). “The family erythrobacteraceae,” in The prokaryotes: alphaproteobacteria and betaproteobacteria. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., and Thomopson F. (Springer Press, Berlin, Heidelberg), 213–235.
Toranzo A. E., Magariños B., and Romalde J. L. (2005). A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246, 37–61. doi: 10.1016/j.aquaculture.2005.01.002
Travers M.-A., Miller K. B., Roque A., and Friedman C. S. (2015). Bacterial diseases in marine bivalves. J. Invertebr. Pathol. 131, 11–31. doi: 10.1016/j.jip.2015.07.010
Troell M., Halling C., Neori A., Chopin T., Buschmann A. H., Kautsky N., et al. (2003). Integrated mariculture: asking the right questions. Aquaculture 226, 69–90. doi: 10.1016/S0044-8486(03)00469-1
Troell M., Joyce A., Chopin T., Neori A., Buschmann A. H., and Fang J. G. (2009). Ecological engineering in aquaculture–potential for integrated multi-trophic aquaculture (IMTA) in marine offshore systems. Aquaculture 297, 1–9. doi: 10.1016/j.aquaculture.2009.09.010
Turner J. W., Good B., Cole D., and Lipp E. K. (2009). Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J. 3, 1082–1092. doi: 10.1038/ismej.2009.50
Vadstein O., Attramadal K. J., Bakke I., and Olsen Y. (2018). K-selection as microbial community management strategy: a method for improved viability of larvae in aquaculture. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02730
Vaiyapuri M., Pailla S., Badireddy M. R., Pillai D., Nagarajarao R. C., and Mothadaka M. P. (2021). Antimicrobial resistance in Vibrios of shrimp aquaculture: Incidence, identification schemes, drivers and mitigation measures. Aquacult. Res. 52, 2923–2941. doi: 10.1111/are.15142
Vaquer C. D. A., Lam-Höai T., Rougier C., Mazouni N., Lautier J., Collos Y., et al. (2000). Feeding rate of the oyster Crassostrea gigas in a natural planktonic community of the Mediterranean Thau Lagoon. Mar. Ecol. Prog. Ser. 205, 171–184. doi: 10.3354/meps205171
Vezzulli L., Colwell R. R., and Pruzzo C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65, 817–825. doi: 10.1007/s00248-012-0163-2
Vine N. G., Leukes W. D., and Kaiser H. (2004). In vitro growth characteristics of five candidate aquaculture probiotics and two fish pathogens grown in fish intestinal mucus. FEMS Microbiol. Lett. 231, 145–152. doi: 10.1016/S0378-1097(03)00954-6
Viver T., Knittel K., Amann R., and Orellana L. H. (2024). Deep long-read metagenomic sequencing reveals niche differentiation in carbon cycling potential between benthic and planktonic microbial populations (Preprint). BioRxiv 2024-06, 597336. doi: 10.1101/2024.06.04.597336
Wagner-Döbler I. and Biebl H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Waller M. J., Humphries N. E., Womersley F. C., Loveridge A., Jeffries A. L., Watanabe Y., et al. (2024). The vulnerability of sharks, skates, and rays to ocean deoxygenation: Physiological mechanisms, behavioral responses, and ecological impacts. J. Fish Biol. 105, 482–511. doi: 10.1111/jfb.15830
Wang B., Cao L., Micheli F., Naylor R. L., and Fringer O. B. (2018a). The effects of intensive aquaculture on nutrient residence time and transport in a coastal embayment. Environ. Fluid. Mech. 18, 1321–1349. doi: 10.1007/s10652-018-959
Wang J. H., Lu J., Zhang Y. X., Wu J., Zhang C., Yu X., et al. (2018b). High-throughput sequencing analysis of the microbial community in coastal intensive mariculture systems. Aquacult. Eng. 83, 93–102. doi: 10.1016/j.aquaeng.2018.10.001
Wang W., Tang K., and Wang X. (2024). High temperatures increase the virulence of Vibrio bacteria towards their coral host and competing bacteria via type VI secretion systems. PloS Biol. 22, e3002788. doi: 10.1371/journal.pbio.3002788
Wang C. X., Wang Y. B., Liu P. Y., Sun Y. Y., Song Z. L., and Hu X. K. (2021). Characteristics of bacterial community structure and function associated with nutrients and heavy metals in coastal aquaculture area. Environ. Pollut. 275, 116639. doi: 10.1016/j.envpol.2021.116639
Wang W. L., Wu L., Xu K., Xu Y., Ji D. H., Chen C. S., et al. (2020). The cultivation of Pyropia haitanensis has important impacts on the seawater microbial community. J. Appl. Phycol. 32, 2561–2573. doi: 10.1007/s10811-020-02068-6
Wang M., Yi M. M., Lu M. X., Zhu X., Chen G. L., Gao F. Y., et al. (2019). Effect of in situ sediment remediation combining oyster shells and bottom microporous aeration on nitrogen removal and microbiota. Aquac. Res. 50, 331–341. doi: 10.1111/are.13907
Wartenberg R., Feng L., Wu J. J., Mak Y. L., Chan L. L., Telfer T. C., et al. (2017). The impacts of suspended mariculture on coastal zones in China and the scope for integrated multi-trophic aquaculture. Ecosyst. Health Sust. 3, 1340268. doi: 10.1080/20964129.2017.1340268
Weigel B. L., Miranda K. K., Fogarty E. C., Watson A. R., and Pfister C. A. (2022). Functional insights into the kelp microbiome from metagenome-assembled genomes. Msystems 7, e01422–e01421. doi: 10.1128/msystems.01422-21
White P. A., Kalff J., Rasmussen J. B., and Gasol J. M. (1991). The effect of temperature and algal biomass on bacterial production and specific growth rate in freshwater and marine habitats. Microb. Ecol. 21, 99–118. doi: 10.1007/BF02539147
Wietz M., López-Pérez M., Sher D., Biller S. J., and Rodriguez-Valera F. (2022). Microbe Profile: Alteromonas macleodii – a widespread, fast-responding, ‘interactive’marine bacterium. Microbiology 168, 1236. doi: 10.1099/mic.0.001236
Wildish D., Kristmanson D., and Saulnier A. (1992). Interactive effect of velocity and seston concentration on giant scallop feeding inhibition. J. Exp. Mar. Biol. Ecol. 155, 161–168. doi: 10.1016/0022-0981(92)90060-N
Wilson K. (2001). Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 56, 2.4. 1–2.4. 5. doi: 10.1002/0471142727.mb0204s56
Wong S. K., Zhang X. H., and Woo N. Y. (2012). Vibrio alginolyticus thermolabile hemolysin (TLH) induces apoptosis, membrane vesiculation and necrosis in sea bream erythrocytes. Aquaculture 330, 29–36. doi: 10.1016/j.aquaculture.2011.12.012
Xiao X., Agusti S., Lin F., Li K., Pan Y. R., Yu Y., et al. (2017). Nutrient removal from Chinese coastal waters by large-scale seaweed aquaculture. Sci. Rep. 7, 46613. doi: 10.1038/srep46613
Xiao J., Ford S. E., Yang H. S., Zhang G. F., Zhang F. S., and Guo X. M. (2005). Studies on mass summer mortality of cultured zhikong scallops (Chlamys farreri Jones et Preston) in China. Aquaculture 250, 602–615. doi: 10.1016/j.aquaculture.2005.05.002
Xue S. X., Xu W., Wei J. L., and Sun J. S. (2017). Impact of environmental bacterial communities on fish health in marine recirculating aquaculture systems. Vet. Microbiol. 203, 34–39. doi: 10.1016/j.vetmic.2017.01.034
Yang X. R., Jiang G. F., Zhang Y. Z., Wang N. Q., Zhang Y. L., Wang X. F., et al. (2023). MBPD: A multiple bacterial pathogen detection pipeline for One Health practices. iMeta 2, e82. doi: 10.1002/imt2.82
Yang B., Zhai S. Y., Li X., Tian J., Li Q., Shan H. W., et al. (2021). Identification of Vibrio alginolyticus as a causative pathogen associated with mass summer mortality of the Pacific Oyster (Crassostrea gigas) in China. Aquaculture 535, 736363. doi: 10.1016/j.aquaculture.2021.736363
Ying C., Chang M. J., Hu C. H., Chang Y. T., Chao W. L., Yeh S. L., et al. (2018). The effects of marine farm-scale sequentially integrated multi-trophic aquaculture systems on microbial community composition, prevalence of sulfonamide-resistant bacteria and sulfonamide resistance gene sul1. Sci. Total Environ. 643, 681–691. doi: 10.1016/j.scitotenv.2018.06.204
Yokoyama H. (2013). Suspended culture of the sea cucumber Apostichopus japonicus below a Pacific oyster raft – potential for integrated multi-trophic aquaculture. Aquacult. Res. 46, 825–832. doi: 10.1111/are.12234
Yong W. T. L., Thien V. Y., Rupert R., and Rodrigues K. F. (2022). Seaweed: a potential climate change solution. Renew. Sust. Energ. Rev. 159, 112222. doi: 10.1016/j.rser.2022.112222
Zeng D. Y., Huang D. J., Qiao X. D., He Y. Q., and Zhang T. (2015). Effect of suspended kelp culture on water exchange as estimated by in situ current measurement in Sanggou Bay, China. J. Marine Syst. 149, 14–24. doi: 10.1016/j.jmarsys.2015.04.002
Zhang X. H. and Austin B. (2005). Haemolysins in vibrio species. J. Appl. Microbiol. 98, 1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x
Zhang M. Y., Pan L. Q., Huang F., Gao S., Su C., Zhang M. Z., et al. (2019). Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 506, 280–293. doi: 10.1016/j.aquaculture.2019.03.038
Zhang M. Q., Yang J. L., Lai X. X., Li W., Zhan M. J., Zhang C. P., et al. (2022). Effects of integrated multi-trophic aquaculture on microbial communities, antibiotic resistance genes, and cultured species: A case study of four mariculture systems. Aquaculture 557, 738322. doi: 10.1016/j.aquaculture.2022.738322
Zhao Z. L., Wang J., Han Y., Chen J. W., Liu G. F., Lu H., et al. (2017). Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 220, 909–918. doi: 10.1016/j.envpol.2016.10.075
Zhao W. J., Yuan T., Piva C., Spinard E. J., Schuttert C. W., Rowley D. C., et al. (2019). The probiotic bacterium Phaeobacter inhibens downregulates virulence factor transcription in the shellfish pathogen Vibrio coralliilyticus by N-acyl homoserine lactone production. Appl. Environ. Microb. 85, e01545–e01518. doi: 10.1128/AEM.01545-18
Zhou F. X., Lu X., Chen F. J., Zhu Q. M., Meng Y. F., Chen C. Q., et al. (2020). Spatial-monthly variations and influencing factors of dissolved oxygen in surface water of Zhanjiang Bay, China. J. Mar. Sci. Eng. 8, 403. doi: 10.3390/jmse8060403
Zhou Y. Q., Sun B. Y., Xie B. H., Feng K., Zhang Z. J., Zhang Z., et al. (2021). Warming reshaped the microbial hierarchical interactions. Global Change Biol. 27, 6331–6347. doi: 10.1111/gcb.15891
Keywords: seawater bacterial community, monthly dynamics, bacterial pathogens, seasonal diseases, Vibrio, marine deoxygenation, integrated multi-trophic aquaculture
Citation: Lu L, Ni L, Ai C, Zhang D, Zheng P, Li X, Liu X and Dang H (2025) Seasonal dynamics of microbial communities link to summer-autumn aquaculture disease outbreaks in Sanggou Bay. Front. Mar. Sci. 12:1581190. doi: 10.3389/fmars.2025.1581190
Received: 21 February 2025; Accepted: 28 April 2025;
Published: 21 May 2025.
Edited by:
Sun Cuici, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Fulin Sun, Chinese Academy of Sciences (CAS), ChinaZhihua Li, Shandong University, China
Copyright © 2025 Lu, Ni, Ai, Zhang, Zheng, Li, Liu and Dang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuejing Li, bGl4dWVqaW5nMTk4OUB4bXUuZWR1LmNu; Xin Liu, WGluTGl1QGJhYnJjLmFjLmNu; Hongyue Dang, RGFuZ0hZQHhtdS5lZHUuY24=
 Longfei Lu1,2,3
Longfei Lu1,2,3 Chunxiang Ai
Chunxiang Ai Dongdong Zhang
Dongdong Zhang Pengfei Zheng
Pengfei Zheng Xuejing Li
Xuejing Li Hongyue Dang
Hongyue Dang