- 1Department of Marine Biology, Faculty of Marine Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of Microbiology, Kaduna State University, Kaduna, Nigeria
- 3Department of Marine Biology and Fisheries, Faculty of Marine Science and Environments, Hodeidah University, Hodeidah, Yemen
- 4Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia
- 5Department of Animal Production, Faculty of Agriculture, Sana’a University, Sanaa, Yemen
Genome sequences of sponge bacterial endophytes will be useful for understanding the bioactive compound synthetic potential and molecular mechanisms of sponge-bacteria interactions. In this study, the complete genome of the bacterium E9 isolated from the Red Sea sponge species was sequenced and its antibiofilm activity was assessed through laboratory assay. Experiments indicated the strong antibacterial and antibiofilm activity of the extracts of bacterium E9. Complete genome sequencing reveals that genome assembly generated a single chromosome of 2123451 base pairs with a guanine-cytosine (GC) content of 32.9% with 2420 protein coding sequences and a gene/genome ratio of 83.7%. The bacterial strain was identified as Staphylococcus epidermidis based on phylogenetic analysis. A total of 9 biosynthetic gene clusters were identified in the genome using the open-source platform AntiSMASH. The observed antibacterial and antibiofilm activity of the strain E9 may be due the presence of gene clusters such as nonribosomal peptides, lasso peptides and terpenes. Overall, the whole genome analysis indicated the bacterium’s capability to adapt in diverse environments including invertebrate hosts, and bioactive compound synthesis.
1 Introduction
The marine environment, particularly the Red Sea, has emerged as a promising source of novel bioactive compounds with potential therapeutic applications (El-Hossary et al., 2020). Of particular interest are the symbiotic bacteria associated with marine invertebrates, such as sponges, which have evolved to produce a diverse array of secondary metabolites (Liang et al., 2023). These compounds often serve as chemical defenses for their hosts and represent a vast, largely untapped reservoir of potential agents with therapeutic properties including antifouling and antibiofilm (Pawlik, 1993; Zhang et al., 2022). Marine microorganisms are potential future sources of effective antifouling and antibiofilm agents (Satheesh et al., 2016; Sukmarini et al., 2024). These compounds can impede microbial attachment, growth, cell communication, and the subsequent formation of biofilms (Rendueles and Ghigo, 2012). Additionally, they can disrupt bacterial signaling by inactivating quorum sensing mechanisms and inhibiting the production of enzymes responsible for degrading bacterial signals and polymers (Vimala, 2016). Notably, some of these compounds exhibit multiple bioactivities, including antimicrobial, anti-algal, and antilarval properties, and have been successfully isolated from heterotrophic marine bacteria associated with sponges (Freeman et al., 2021).
The bioactive compounds produced by these microorganisms are the result of secondary metabolism, which occurs through enzymatic pathways encoded by specific biosynthetic genes within their genomes. This biosynthetic capability is often associated with specialized cells, and intriguingly, the resulting secondary metabolites may not confer any direct benefit to the producing organism (Luckner, 2013). Within the genomes, the genes encoding these enzymes or regulatory proteins are typically clustered together in what is referred to as a biosynthetic gene cluster (BGC) (Naughton et al., 2017). Medema et al. (2015) define a BGC as a “physically clustered group of two or more genes in a particular genome that collectively encodes a biosynthetic pathway for the production of a specialized metabolite, including its chemical variants”.
Recent advancements in genomic technologies have revolutionized our approach to discovering bioactive compounds. The advent of high-throughput sequencing and improved computational tools has made it possible to sequence entire bacterial genomes quickly and accurately (Viju et al., 2021). This has opened new avenues for research and innovation in biotechnology and pharmacology. One of the most significant developments is genome mining, a powerful bioinformatic strategy that allows researchers to identify biosynthetic gene clusters (BGCs) within bacterial genomes. These BGCs are groups of co-located genes that work together to produce secondary metabolites, which are compounds not directly involved in the normal growth, development, or reproduction of the organism (Naughton et al., 2017; Adnani et al., 2017; Sandoval-Powers et al., 2021). The secondary metabolites identified from the marine bacteria based on biosynthetic origin include polyketide derivatives, terpenoids, omega-3 polyunsaturated fatty acids and amino acid derivatives (Giordano et al., 2015; Wibowo et al., 2023). Many of these secondary metabolites hold great promise for pharmaceutical development due to their bioactive properties such as antibacterial, antifungal, antiviral, antibiofilm, anti-inflammatory and anticancer activities (Wibowo et al., 2023).
By analyzing complete bacterial genomes, researchers can systematically search for and characterize BGCs (Naughton et al., 2017). This process involves comparing genetic sequences to known BGCs and predicting the structures and functions of the resulting compounds (Adnani et al., 2017). Genome mining has already led to the discovery of numerous novel antibiotics, anticancer agents, and other therapeutically valuable compounds (Viju et al., 2021). The complete genome analysis of bacteria, combined with genome mining techniques, provides an efficient and effective way to uncover and harness the vast potential of bacterial secondary metabolites.
The Red Sea, known for its unique environmental conditions, harbors a rich diversity of marine life, including numerous sponge species and their associated microbiota (DiBattista et al., 2016). Bacteria from the Red Sea found in association with sponges were reported to show particular promise and possess an unusually high number of BGCs compared to their counterparts from other environments (Othoum et al., 2018). This study focuses on an endophytic bacterial strain E9 isolated from a Red Sea sponge identified as Hyrtios erectus. The bacteria were subjected to antibacterial and antibiofilm activity against fouling bacteria. By employing whole-genome sequencing and subsequent bioinformatic analysis to detect BGCs, we aim to characterize the genome of this strain, identify and analyze the BGCs present within its genome and to assess the potential of these BGCs to produce bioactive compounds.
2 Materials and methods
2.1 Collection of sponge samples
The sampling for this study was conducted along the Jeddah Red Sea coast at Obhur, a creek located in the North of Jeddah region of Saudi Arabia (N21°42.562′ E39°05.764′). Approvals were obtained from the relevant authorities to collect sponge samples at appropriate depths. Sites with potential for high biodiversity were selected and documented through photography for future reference. The collected sponge sample was identified as previously described by Sánchez-Rodríguez et al. (2018).
2.2 Isolation of bacteria from the sponge
Bacterial isolates were obtained from the surfaces of sponges using culture-based techniques as outlined by Anand et al. (2006) and Satheesh et al. (2012). In brief, the surfaces of the sponge samples were cleaned to eliminate loosely attached bacteria by washing them multiple times with filtered seawater (FSW) that had been sterilized through autoclaving. Following this, FSW was added to the sponges and agitated for one hour. The resulting solution was then used for bacterial isolation through serial dilution. An aliquot of 1 ml of the solution was transferred into sterilized tubes containing 9 ml of FSW, creating serial dilutions ranging from 10–1 to 10-6. Each dilution was inoculated onto Zobell Marine Agar (ZMA) plates using the spread plate method, where 0.1 ml of the dilution was transferred to the plates and evenly spread with a sterilized spreader in duplicate. The plates were aerobically incubated at 30°C for 72 hours, with observations recorded every 24 hours. Pure cultures of individual bacterial colonies were obtained by subculturing onto fresh ZMA plates. This was followed by morphological evaluation of the colonies and microscopic observation following Gram staining.
2.3 Extraction of bioactive crude extracts
Bioactive components were extracted from the bacterium isolated from the sponge using ethyl acetate as a solvent, following the procedure established by Sánchez-Rodríguez et al. (2018). Briefly, the bacterial strain was cultured overnight in Zobell marine broth (ZMB) at 30°C and 5 ml of overnight culture was transferred to a 500 ml Erlenmeyer flask containing ZMB filled to half capacity (250 mL). These flasks were incubated with shaking at 25°C for 5 days. After incubation, the cultures were centrifuged at 9,000 rpm for 20 minutes at 4°C to separate the supernatant and cell residue. Equal volume of ethyl acetate (selected for its low toxicity and medium polarity suitable for extracting both polar and nonpolar bioactive compounds) was added to the culture supernatant (CS) in an Erlenmeyer flask and shaken at room temperature for 24 hours. The mixture was then separated using a separating funnel into solvent and liquid phases. This extraction process was repeated three times with the liquid phase before discarding it. The collected solvent phase was filtered through Whatman No. 1 filter paper. the solvents were removed to concentrate the extracts in a rotary evaporator (Buchi, Switzerland) at 50°C, 240 mbar pressure. The crude extract was dissolved in 10% Dimethyl sulphoxide (DMSO) at a concentration of 25 mg ml-1 and then stored in a refrigerator until required for further use.
2.4 Antibacterial and antibiofilm activity
The antibacterial and antibiofilm activities were assessed against five selected biofilm-forming strains identified as microfouling bacteria which were previously reported (Abdulrahman et al., 2021). DMSO (100%) was used as the negative control while the extract concentration was maintained at 1mg/ml. These evaluations were conducted using the agar well diffusion method and the microtiter assay method respectively as previously described (Teanpaisan et al., 2017). The tested biofilm-forming bacteria strains included Pseudoalteromonas sp. IMB1 (ON003955), Halomonas sp. IMB2 (ON415519), Vibrio alginolyticus IMB11 (ON003958), Pseudoalteromonas gelatinilytica IMB14 (ON003961), and Pseudoalteromonas gelatinilytica IMB15 (ON003962). All evaluations were done in triplicates. The biofilm formation inhibition assay was conducted by adding 100 μL of bacterial suspensions (adjusted to McFarland standard) and 100 μL of the test extracts (from the stock solution) into each well of a microtiter plate. The plates were then incubated at 28°C for 48 hours. After incubation, the absorbance was measured at a wavelength of 570 nm. The percentage of biofilm inhibition (BI) was calculated using the following formula:
Where:
● OD Control represents the optical density of the control (untreated) wells.
● OD Sample represents the optical density of the wells treated with the test extracts.
2.5 Genome extraction, sequencing, assembly and annotation
2.5.1 Sample preparation and sequencing
Genomic DNA was extracted from the samples using the E.Z.N.A® DNA extraction kit (OMEGA) following the manufacturer’s instructions. Library preparation was performed using the Illumina TruSeq™ Nano DNA Sample Prep Kit according to the manufacturer’s protocols. Briefly, the extracted genomic DNA was fragmented to an average size of 300–500 bp using a Covaris M220 ultrasonic device. The fragmented DNA underwent end-repair, followed by the addition of an A base to the 3’ ends and ligation of index adapters. The library was then enriched through 8 cycles of PCR amplification. To recover the target size fragments, the amplified library was purified using 2% agarose gel electrophoresis. The concentration of the purified library was quantified using the TBS380 Picogreen assay (Invitrogen). Sequencing was performed on the Illumina Hiseq 4000 platform using a 2 x 150 bp paired-end sequencing strategy.
2.5.2 Quality control and data processing
Raw sequencing data underwent quality control using Trimmomatic-0.39 (Bolger et al., 2014). Sequences containing adapter contamination were removed, and bases with a quality score below Q20 were trimmed from the ends of the reads. Reads with more than 10% ambiguous bases (N) or shorter than 75 bp were discarded. The quality of the cleaned data was assessed, including metrics such as GC content and the percentage of bases with quality scores Q20 and Q30.
2.5.3 Bioinformatics analysis
The cleaned sequencing data was analyzed using a comprehensive bioinformatics pipeline. First, we used unicycler (https://github.com/rrwick/Unicycler) to perform genome assembly with default parameters and received the optimal results of the assembly. GC depth and genome size information were calculated by custom Perl scripts to judge whether the DNA sample was contaminated or not.
2.5.4 Genome assembly and quality control
Raw paired-end Illumina reads were trimmed and quality-controlled using Trimmomatic (version 0.36) (Bolger et al., 2014) with parameters SLIDINGWINDOW:4:15 and MINLEN:75. These clean, quality-controlled reads were used for subsequent analyses. Raw PacBio reads were converted to FASTA format using Samtools Fasta (Li et al., 2009). The Illumina data was utilized to evaluate genome complexity and to correct the long reads. The strain genome was then circularized using Circulator (Hunt et al., 2015). This whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JBLZIG000000000. The version described in this paper is JBLZIG010000000. The details of BioProject (PRJNA1228063) and BioSample (SAMN47005962) were also available in the same database.
2.5.5 Phylogenetic analysis
A phylogenetic tree based on the 16S rRNA gene sequence of strain E9 was constructed using MEGA11 software (Tamura et al., 2021). Additional 16S rRNA gene sequences from other species were obtained from the NCBI database for comparison. Sequence alignment was performed using the MUSCLE algorithm (Edgar, 2004). The phylogenetic tree was then generated using the Neighbor-Joining method with 1000 bootstrap replications (Saitou and Nei, 1987).
2.5.6 Functional annotation
Functional annotation of the assembled genome was conducted using various tools and databases, including Clusters of Orthologous Groups (COG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2023), to identify functional genes and metabolic pathways. The predicted protein-coding sequences were functionally annotated through comparison against several known protein databases using BLASTP with an E-value threshold of ≤ 1e-5. To ensure biological significance, only the optimal alignment result was retained as the gene’s database alignment information when multiple alignments were found. All predicted gene models were aligned against these databases to provide comprehensive functional annotations. The results from these multiple annotation sources were integrated to provide a holistic understanding of each gene’s potential function and its role in biological pathways.
2.5.7 Biosynthesis gene cluster analysis
The gene mining open-source platform AntiSMASH (Antibiotics and Secondary Metabolite Analysis Shell, bacterial version 5.0: http://antismash.secondarymetabolites.org) was used as the tool for BGC analysis. In brief, the complete sequence (gbk file) of the bacterium was uploaded to the AntiSMASH platform and submitted for analysis. After 10 minutes of processing, the results displayed were analyzed and compared with the BGCs submitted in the BGC database Mi-BIG (Minimum Information about a Biosynthetic Gene cluster).
3 Results
3.1 Antibacterial and antibiofilm activity
The antibacterial activity of the crude extract from the marine bacterium E9 was assessed against various biofilm-forming strains. The extract exhibited moderate antibacterial effects, with inhibition zones ranging from 11 mm against Halomonas sp. IMB2 to 16.1 mm against Pseudoalteromonas gelatinilytica IMB 14. DMSO controls didn’t show antibacterial activity in this assay. These results are summarized in Table 1. The antibiofilm activity of the extract varied among the tested microfouling bacterial strains. The extract exhibited highest biofilm inhibition by achieving approximately 55% inhibition against strain IMB14. This was closely followed by the antibiofilm activity against strain IMB11 with 50% inhibition. The extract also showed strong antibiofilm activity of about 40% inhibition against strain IMB1. In contrast, strains IMB15 and IMB2 displayed lower inhibition levels, at approximately 30% and 20%, respectively after treated with the extract of strain E9. These findings are illustrated in Figure 1.
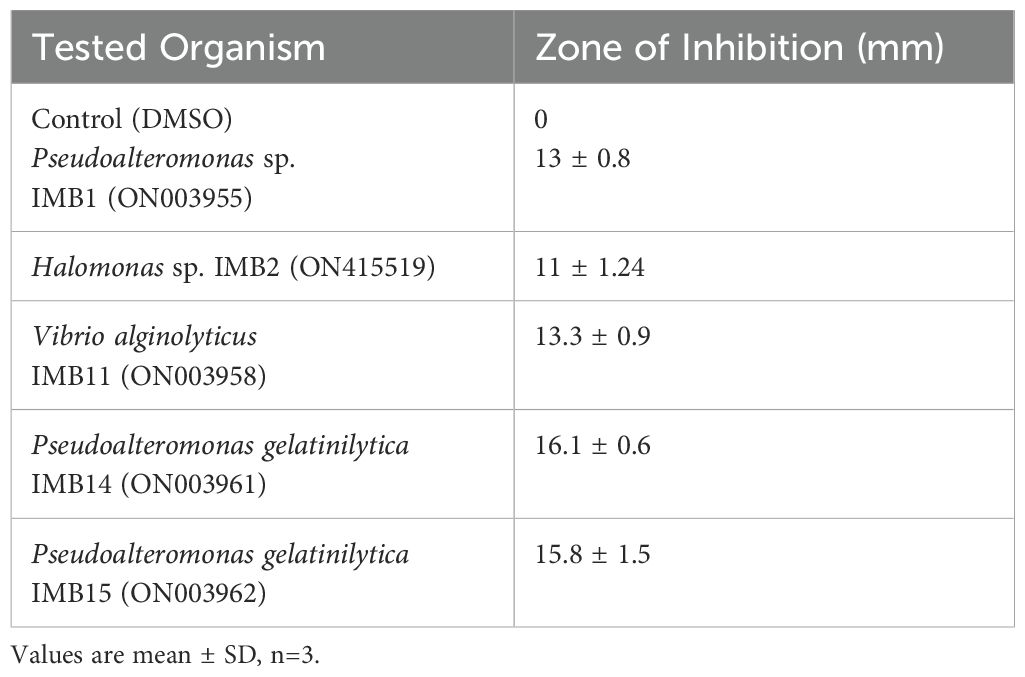
Table 1. Antibacterial activity of crude extract (1000 µg/ml) of Staphylococcus epidermidis E9 against some microfouling organisms.
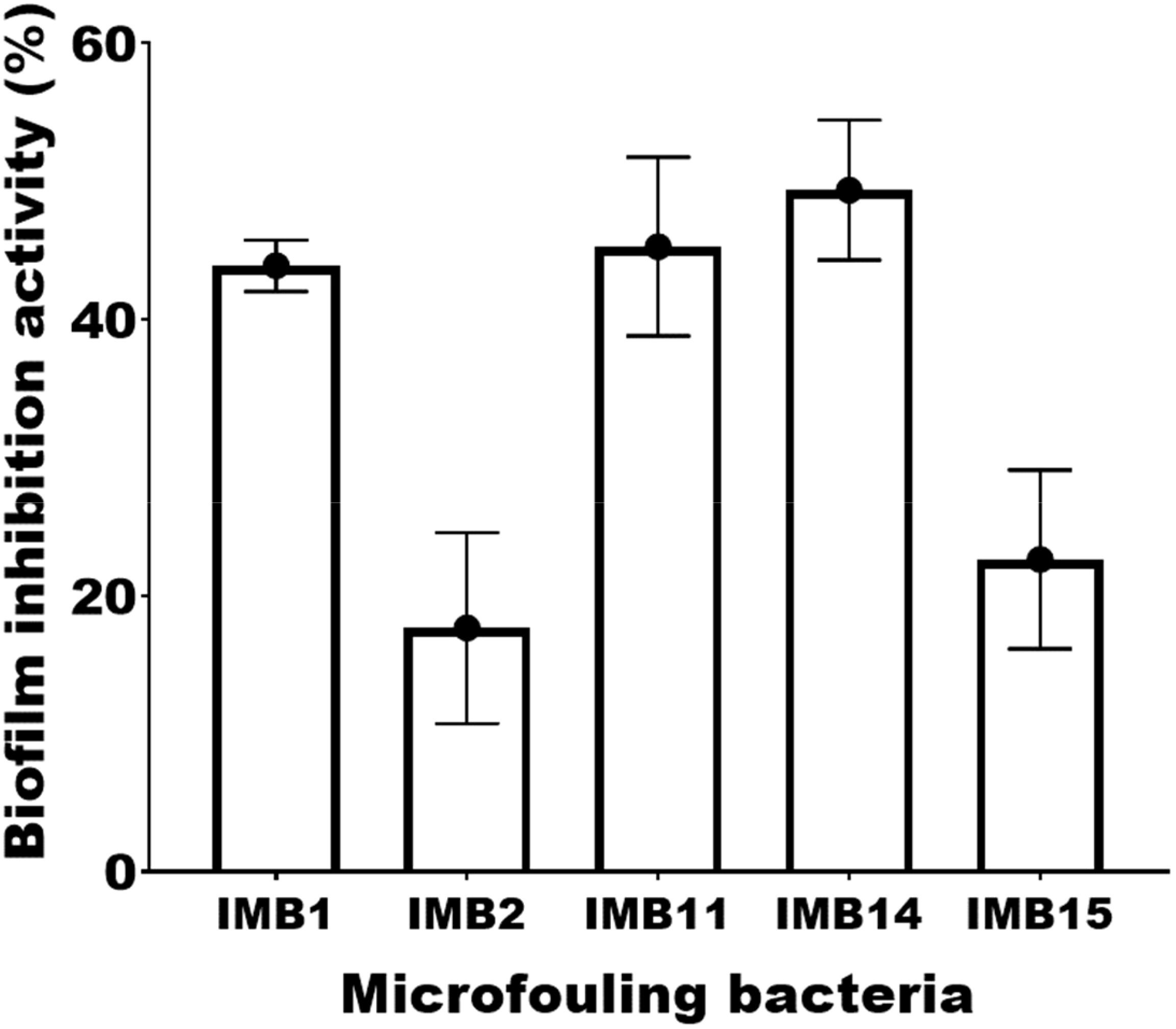
Figure 1. Antibiofilm activity of the extract of sponge endophytic bacterium E9 against bacteria isolated from the substrates. Pseudoalteromonas sp. (IMB1), Halomonas sp. (IMB2), Vibrio alginolyticus (IMB11). Pseudoalteromonas gelatinilytica (IMB14), Pseudoalteromonas gelatinilytica (IMB15).
3.2 Identification and whole genome analysis of the bacterium E9
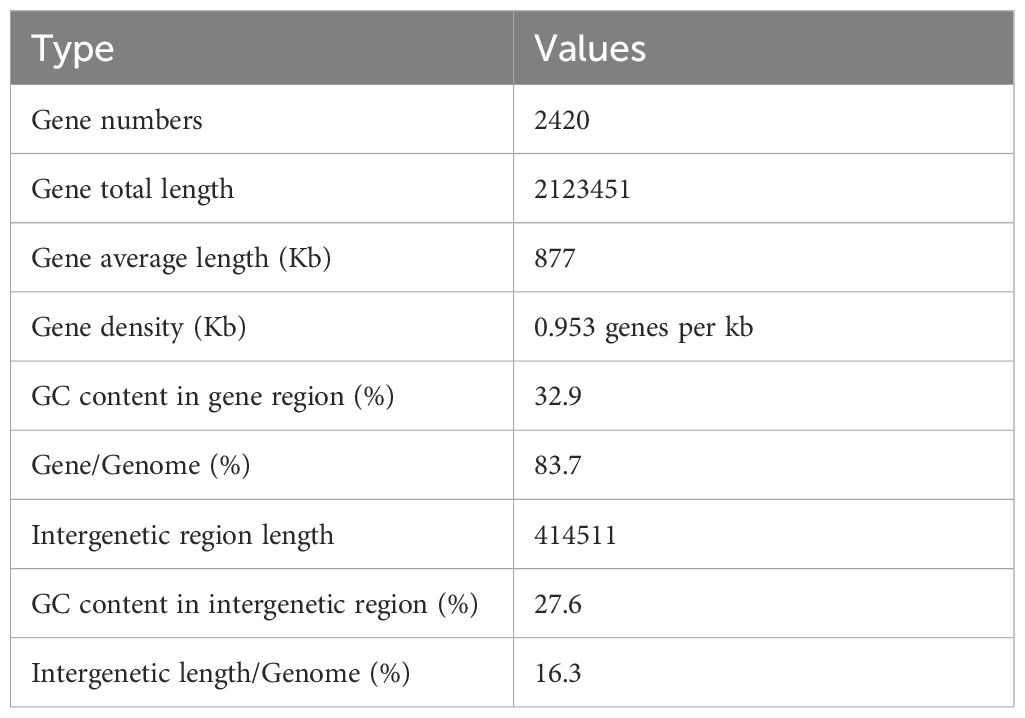
The organism shows large, rounded colonies that are transparent but not mucoid on ZMA plates and appear as Gram-positive cocci following Gram staining and microscopy. The sequencing results revealed the organism as Staphylococcus epidermidis after using the NCBI BLAST which recorded similarity in the range of 98% with other previously reported organisms (Figure 2). The genome circle map of strain E9 is presented in Figure 3. The important characteristics of the genome such as the distribution of genes on positive and antisense chains, GC content, homologous genes, genome islands and COG functional genes are displayed in the genome circle map. The study and analysis of the genome of bacteria reveal several important features of the genome structure and gene distribution. According to the data in Table 2, the bacterium E9 has 2420 gene numbers with an average gene length of about 877 Kb, which results in a gene density of 0.953 genes per kb.

Figure 2. Phylogenetic tree of the bacterial strain E9. The strain was identified as Staphylococcus epidermidis based on phylogenetic analysis.

Figure 3. A genome circle map of strain E9. The circles represent genome-size markers of the functional classification.
3.3 Functional classification of genes
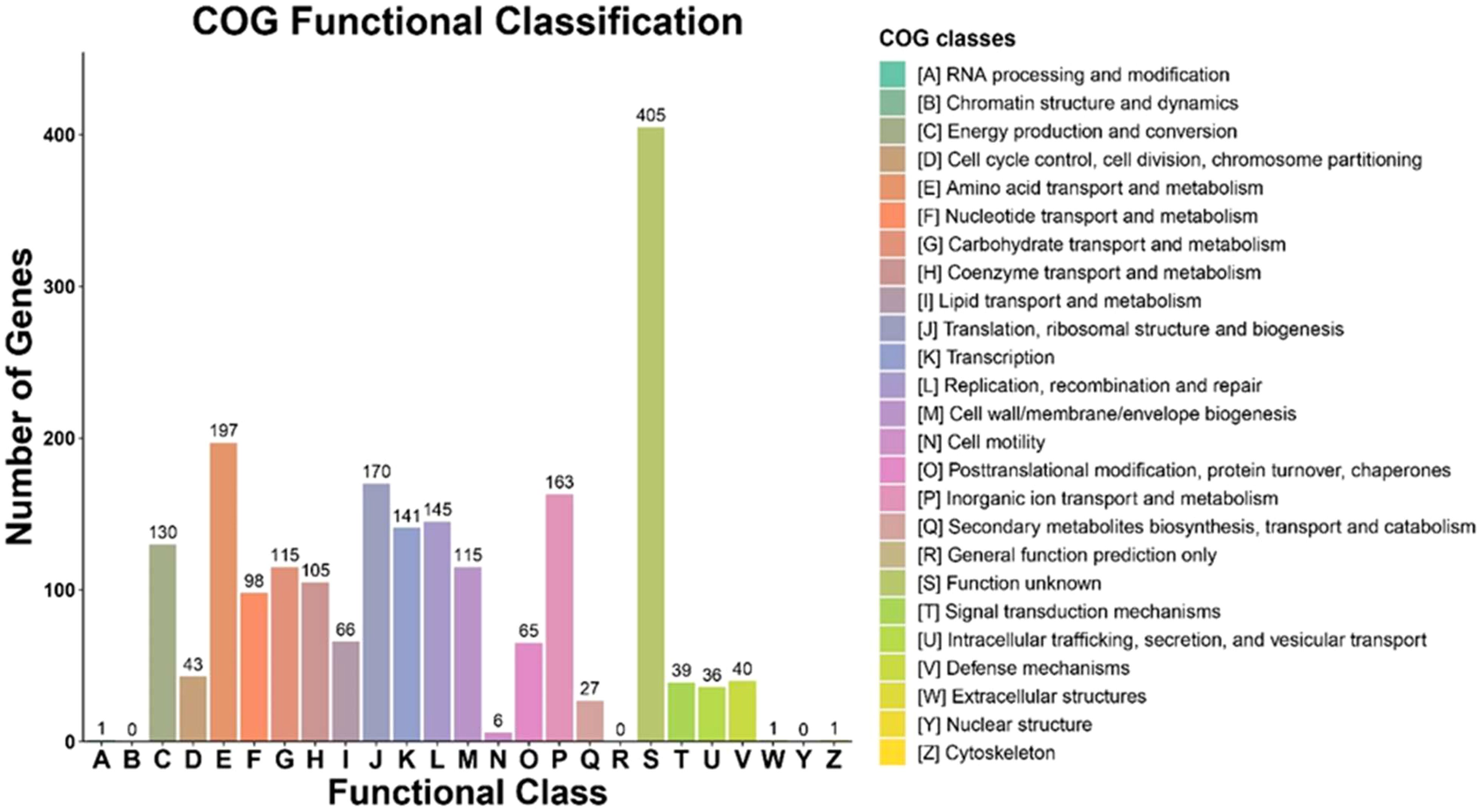
COG functional classification analysis revealed that the majority of genes of the bacterium E9 were involved in metabolic activities (Figure 4). COG analysis revealed that the largest role was played by unknown functions with 405 genes, followed by amino acid transport and metabolism with 197 genes, and then translation, ribosomal structure and biogenesis and inorganic ion transport and metabolism with 170 and 163 genes, respectively. The genes encoding secondary metabolites synthesis and defense mechanisms were 27 and 40 respectively. The gene ontology (GO) analysis also revealed that a larger percentage of genes encoding the metabolic and cellular functions were present in the genome of the strain E9 (Figure 5). Regarding molecular function genes, those encoding catalytic activity topped the list followed by the genes encoding binding activity (Figure 5).

Figure 5. Gene ontology (GO) function analysis showing cellular component, molecular function and biological process in the genome of bacterial strain E9.
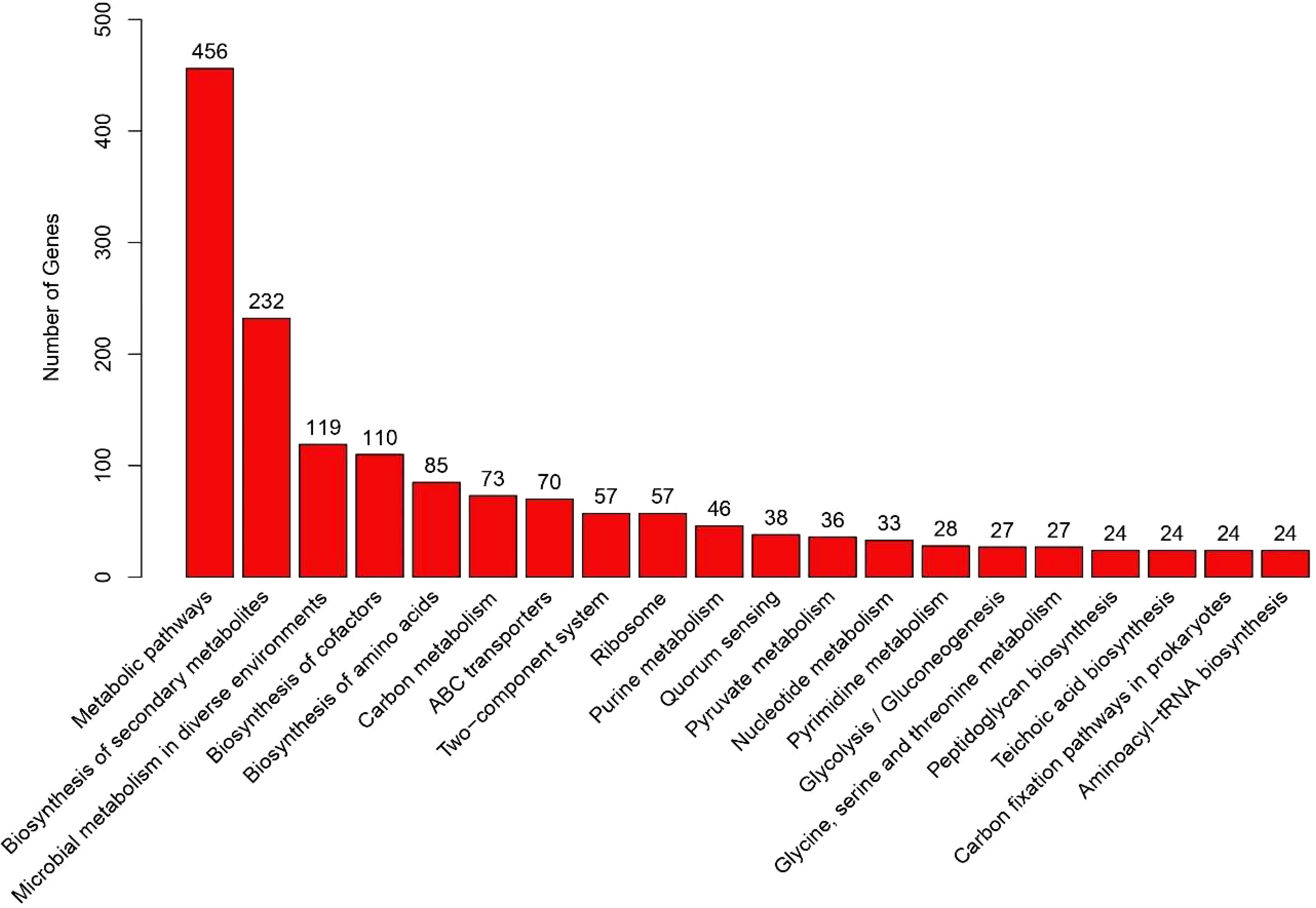
KEGG ORTHOLOGY (KO) of the genome showed the presence of a higher number of genes encoding metabolic pathways (Figure 6). Following the metabolic pathway genes, those encoding the biosynthesis of secondary metabolites were also predicted in large numbers (232 genes). KO analysis also revealed the presence of genes (38 genes) encoding quorum sensing, an essential function of the bacteria living on surfaces or associated with other organisms.
3.4 Biosynthetic gene cluster analysis
The BGC finder AntiSMASH identified a total of 9 BGCs from the genome of the bacterium E9 (Table 3). The identified BGCs were belonging to NRPS, terpene, siderophores, lasso peptide, opine-like-metallophore and cyclic-lactone-autoinducer. Whereas the predicted products include staphylopine, aureusimine, staphyloferrin, Belactosin, carnobacteriocin, bicereucin, Mutanocyclin and listeriocytocin.
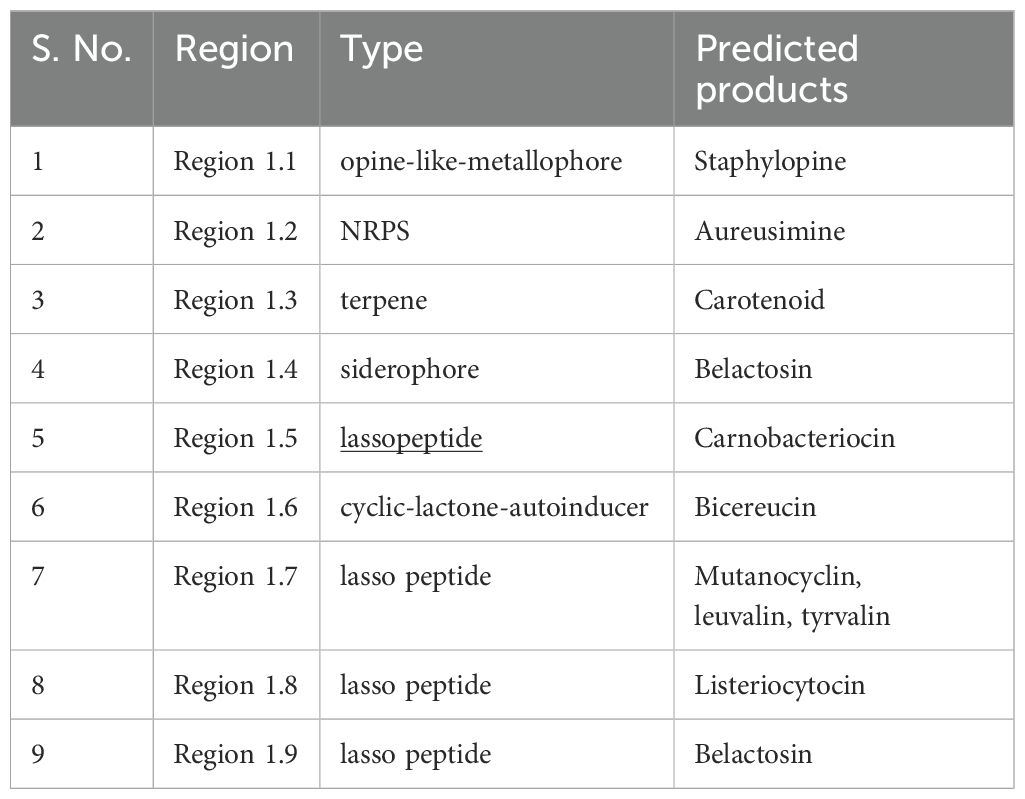
Table 3. Biosynthetic gene clusters and the predicted secondary metabolites from the genome of bacterial strain E9.
4 Discussion
Bacteria associated with sponges and other invertebrates are considered a prolific source of bioactive molecules with antibiofilm and antifouling activities (Satheesh et al., 2016). The findings from this study on the antibacterial and antibiofilm activity of the crude extract from the sponge-associated marine bacterium E9 provide valuable insights into the potential applications of marine-derived compounds in combating biofouling and biofilm formation on submerged surfaces. The moderate antibacterial activity observed against biofilm-forming strains, with inhibition zones ranging from 11 mm to 16.1 mm, aligns with previous studies that have reported varying degrees of antibacterial efficacy from marine bacteria. For instance, a study by Al-Daghistani et al. (2021) demonstrated that extracts from marine bacteria exhibited significant inhibition zones against pathogenic bacteria, with some strains showing maximum inhibition zones as high as 25 mm against some bacteria.
The antibiofilm activity of the E9 extract revealed that strain IMB14 had the highest inhibition rate at approximately 55%, which is comparable to findings from other marine-derived extracts. For example, extracts from sponge-associated Alcanivorax sp. demonstrated biofilm inhibition ranging from 46% to 71% against various biofilm-forming bacteria (Jamal and Satheesh, 2022). The varying levels of biofilm inhibition observed in this study highlight the complexity of biofilm formation and the need for targeted approaches when developing antibiofilm agents. The strong inhibition rates noted for strains IMB11 and IMB1 at around 50% and 40%, respectively, further emphasize the potential of marine bacteria as sources of bioactive compounds. The lower inhibition levels observed for strains IMB15 and IMB2 (30% and 20% respectively) suggest that not all strains respond equally to the crude extract, which is consistent with research indicating that different bacterial species exhibit varying susceptibilities to antibiofilm agents (Balasubramanian et al., 2018).
The bacterium E9 isolated from the sponge was identified as Staphylococcus epidermidis based on whole genome sequencing and the phylogenetic tree construction. S. epidermidis is a well-known human pathogenic strain, that usually inhabits in human skin and mucus membranes (Severn and Horswill, 2023). However, previous studies confirmed the presence of S. epidermidis in marine environments that expanded their ecological role (Gunn and Colwell, 1983). Previously Zhang et al. (2016) reported the antibacterial activity of S. epidermidis strain isolated from marine fish. Further, Zaghloul et al. (2021) isolated S. epidermidis strain from the marine waters which produce biocement. The association of S. epidermidis strain with marine sponges was also recorded previously (Paul et al., 2021).
Antibiofilm activities of S. epidermidis strains were reported previously against pathogenic bacterium such as S. aureus (Iwase et al., 2010). Protease enzymes produced by S. epidermidis strains are reported to be the reason for the biofilm inhibitory activity (Glatthardt et al., 2020; Vandecandelaere et al., 2014; Iwase et al., 2010; Sugimoto et al., 2013). Glatthardt et al. (2020) confirmed the production of small molecules by commensal S. epidermidis strain that disrupts the biofilms. Further, another study by Nakatsuji et al. (2017) reported that S. epidermidis commensal strains produce antimicrobial compounds to inhibit S. aureus. This indicates that commensal S. epidermidis strains produce metabolites that inhibit the attachment or growth of other bacteria.
Complete genome sequencing data of many pathogenic S. epidermidis strains are available in genomic databases (Chaudhry and Patil, 2016). However, the whole genome of this strain associated with marine organisms is very limited and most of the sequences available in public databases are based on 16S rRNA sequencing. Whole genome analysis is important to understand the ecological role of this bacterium specifically with the sponge host. Whole genome analysis of the S. epidermidis strain in this study showed a gene density of 0.953 genes per kb indicating a relatively compact genome structure that minimizes intergenic regions (Ng, 2022). The GC content in gene regions recorded a moderate level of GC-rich sequences of about 32.9%, which in turn affects the stability of genes and their expression patterns (Kato et al., 2003; Ng, 2022). The Intergenetic region length indicated 414511 base pairs with lower GC content values of about 27.6% indicating less evolutionary pressures compared to gene regions (Ng, 2022; Hou and Lin, 2009). Intergenetic length constituted about 16.3% of the genome, which may play a role in many regulatory functions and control of gene expression (Rayburn, 1993).
The functional analysis indicated the presence of genes responsible for the secondary metabolites and defense mechanisms in the genome of S. epidermidis. The secondary metabolite production is important for the bacteria as these molecules play significant role in interaction with other organisms, signaling and against other competitors (Petersen et al., 2020; Giordano et al., 2015). The secondary metabolite producing capability of this strain was further confirmed by KEGG Orthology analysis which predicted 232 genes related to the biosynthesis of secondary metabolites. Also, BGC analysis revealed that the genome of the bacterium contained some gene clusters for the production of bioactive metabolites such as lasso peptides, NRPS and terpenes The compounds predicted through BGC analysis participate in different biological functions that include antimicrobial activity, signaling and metal chelation. These functions are essential for the microorganisms to live in diverse environments including invertebrate hosts (Whalen et al., 2019; Wang et al., 2022). Further, the presence of BGCs related to terpene biosynthesis and NRPS revealed the biotechnological potentials of this strain particularly for drug discovery (Calisto et al., 2025; Yi et al., 2024). Among the potential bioactive compounds predicted through BGCs analysis, previous studies reported the production of carotenoids by many Staphylococcus species (Solouki Nezhad et al., 2019; Leidy et al., 2020; Manrique-Moreno et al., 2022). Further, presence of lasso peptide (bacteriocin) cluster genes in Staphylococcus species was previously reported by Carson et al. (2017). Additionally, Staphylococcal bacteriocins exhibited strong antimicrobial activities including antibiofilm activity against Staphylococcus aureus (Newstead et al., 2020).
The genome of the S. epidermidis strain also contains 38 genes relevant for quorum sensing, which is essential for communication, biofilm formation and interaction with the host organisms (Wu and Luo, 2021; Rutherford and Bassler, 2012). The GO analysis of the genome indicated the prevalence of genes encoding catalytic activities. Enzyme production is one of the important functions of bacteria living in stressful conditions such as marine environments, which indicates the capability of the bacteria to carry out biochemical transformations for acquiring nutrients or molecular interactions with other organisms (Gu et al., 2024; Hobbs et al., 2018). Due to the opportunistic pathogenic nature of S. epidermidis (Burke et al., 2024), further analysis will provide more insights into the potential role of this endophyte on the sponge host.
In conclusion, whole genome analysis of S. epidermidis strain isolated from the marine sponge revealed the presence of genes responsible for diverse functions. The predicted secondary metabolites indicated the bacterial strain’s antimicrobial activity, ecological adaptability and competitiveness in different environmental conditions. The antibiofilm activity exhibited by the extracts of S. epidermidis showed the potential applications of this bacterium in medicine, biotechnology and environmental areas. The interaction of this bacterium with marine invertebrates may be a symbiotic relationship or as pathogens due to it’s opportunistic pathogenic behavior. Hence further studies on understating this relationship will be useful for potential applications in bioactive compound synthesis. Further, the presence of large numbers of genes with unknown functions would be of interest for deeper analysis of novel metabolic pathways that may provide the basis for the synthesis of bioactive molecules.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, JBLZIG000000000.
Author contributions
MJ: Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – review & editing. IA: Formal analysis, Investigation, Visualization, Writing – original draft. RH: Formal analysis, Investigation, Writing – original draft. MA-M: Investigation, Methodology, Resources, Writing – review & editing. SS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research work was funded by Institutional Fund Projects under grant no. (IFPIP-569-150-1443). The authors gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdulrahman I., Jamal M. T., Alshaery M., Al-Maaqar S. M., Satheesh S. (2021). Isolation and identification of biofilm bacteria from microfouling assemblage developed on artificial materials submerged in the red sea. JKAU: Mar. Sci. 31, 45–54. doi: 10.4197/Mar.31-2.4
Adnani N., Rajski S. R., Bugni T. S. (2017). Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod Rep. 34, 784–814. doi: 10.1039/c7np00009j
Al-Daghistani H. I., Abu-Niaaj L. F., Bustanji Y., Al-Hamaideh K. D., Al-Salamat H., Nassar M. N., et al. (2021). Antibacterial and cytotoxicity evaluation of arum hygrophilum bioss. Eur. Rev. Med. Pharmacol. Sci. 25, 7306–7316. doi: 10.26355/eurrev_202112_27424
Anand T. P., Bhat A. W., Shouche Y. S., Roy U., Siddharth J., Sarma S. P. (2006). Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of south east India. Microbiological Res. 161, 252–262. doi: 10.1016/j.micres.2005.09.002
Balasubramanian S., Skaf J., Holzgrabe U., Bharti R., Förstner K. U., Ziebuhr W., et al. (2018). A new bioactive compound from the marine sponge-derived streptomyces sp. Sbt348 inhibits staphylococcal growth and biofilm formation. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01473
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burke Ó., Zeden M. S., O’Gara J. P. (2024). The pathogenicity and virulence of the opportunistic pathogen staphylococcus epidermidis. Virulence 15, 2359483. doi: 10.1080/21505594.2024.2359483
Calisto R., Godinho O., Devos D. P., Lage O. M. (2025). Genome-based in silico assessment of biosynthetic gene clusters in planctomycetota: Evidences of its wide divergent nature. Genomics 117, 110965. doi: 10.1016/j.ygeno.2024.110965
Carson D. A., Barkema H. W., Naushad S., De Buck J. (2017). Bacteriocins of non-aureus staphylococci isolated from bovine milk. Appl. Environ. Microbiol. 83, e01015–e01017. doi: 10.1128/AEM.01015-17
Chaudhry V., Patil P. B. (2016). Genomic investigation reveals evolution and lifestyle adaptation of endophytic staphylococcus epidermidis. Sci. Rep. 6, 19263. doi: 10.1038/srep19263
DiBattista J. D., Roberts M. B., Bouwmeester J., Bowen B. W., Coker D. J., Lozano-Cortés D. F., et al. (2016). A review of contemporary patterns of endemism for shallow water reef fauna in the red sea. J. Biogeography 43, 423–439. doi: 10.1111/jbi.12649
Edgar R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
El-Hossary E. M., Abdel-Halim M., Ibrahim E. S., Pimentel-Elardo S. M., Nodwell J. R., Handoussa H., et al. (2020). Natural products repertoire of the red sea. Marine Drugs 18, 457. doi: 10.3390/md18090457
Freeman C. J., Easson C. G., Fiore C. L., Thacker R. W. (2021). Sponge–microbe interactions on coral reefs: Multiple evolutionary solutions to a complex environment. Front. Marine Sci. 8. doi: 10.3389/fmars.2021.705053
Giordano D., Coppola D., Russo R., Denaro R., Giuliano L., Lauro F. M., et al. (2015). “Chapter four - marine microbial secondary metabolites: Pathways, evolution and physiological roles,” in Advances in microbial physiology, vol. 66 . Ed. Poole R. K. (Oxford: Academic Press), 357–428.
Glatthardt T., Campos J. C. M., Chamon R. C., de Sá Coimbra T. F., Rocha G. A., de Melo M. A. F., et al. (2020). Small molecules produced by commensal staphylococcus epidermidis disrupt formation of biofilms by staphylococcus aureus. Appl. Environ. Microbiol 86, e02539–19. doi: 10.1128/aem.02539-19
Gu J., Qiu Q., Yu Y., Sun X., Tian K., Chang M., et al. (2024). Bacterial transformation of lignin: Key enzymes and high-value products. Biotechnol. Biofuels Bioproducts 17, 2. doi: 10.1186/s13068-023-02447-4
Gunn B. A., Colwell R. R. (1983). Numerical taxonomy of staphylococci isolated from the marine environment. Int. J. Systematic Evolutionary Microbiol. 33, 751–759. doi: 10.1099/00207713-33-4-751
Hobbs J. K., Pluvinage B., Boraston A. B. (2018). Glycan-metabolizing enzymes in microbe–host interactions: The streptococcus pneumoniae paradigm. FEBS Lett. 592, 3865–3897. doi: 10.1002/1873-3468.13045
Hou Y., Lin S. (2009). Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PloS One 4, e6978. doi: 10.1371/journal.pone.0006978
Hunt M., Silva N. D., Otto T. D., Parkhill J., Keane J. A., Harris S. R. (2015). Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16, 294. doi: 10.1186/s13059-015-0849-0
Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., et al. (2010). Staphylococcus epidermidis esp inhibits staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. doi: 10.1038/nature09074
Jamal M. T., Satheesh S. (2022). Antibiofilm activity of secondary metabolites of sponge-associated bacterium Alcanivorax sp. From the red sea. Front. Marine Sci. 9. doi: 10.3389/fmars.2022.980418
Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. (2023). Kegg for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592. doi: 10.1093/nar/gkac963
Kato T., Asamizu E., Nakamura Y., Tabata S. (2003). Genome analysis of a flowering plant, arabidopsis thaliana. In: Nagata T., Tabata S. (eds) Brassicas and Legumes From Genome Structure to Breeding. Biotechnology in Agriculture and Forestry, vol 52. Springer, Berlin, Heidelberg. doi: 10.1007/978-3-662-05036-1_2
Leidy C., Perez M. I., Reina R. M. M., Trier S., Herrfurth C., Lopez G. D., et al. (2020). Carotenoid content and composition in exponential, stationary and biofilm states of staphylococcus aureus and their influence on membrane biophysical properties. Biophys. J. 118, 321a. doi: 10.1016/j.bpj.2019.11.1806
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang J., She J., Fu J., Wang J., Ye Y., Yang B., et al. (2023). Advances in natural products from the marine-sponge-associated microorganisms with antimicrobial activity in the last decade. Marine Drugs 21, 236. doi: 10.3390/md21040236
Luckner M. (2013). Secondary metabolism in microorganisms, plants and animals (Berlin Heidelberg Gmbh: Springer Science and Business Media).
Manrique-Moreno M., Jemioła-Rzemińska M., Múnera-Jaramillo J., López G. D., Suesca E., Leidy C., et al. (2022). Staphylococcus aureus carotenoids modulate the thermotropic phase behavior of model systems that mimic its membrane composition. Membranes 12, 945. doi: 10.3390/membranes12100945
Medema M. H., Kottmann R., Yilmaz P., Cummings M., Biggins J. B., Blin K., et al. (2015). Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625. doi: 10.1038/nchembio.1890
Nakatsuji T., Chen T. H., Narala S., Chun K. A., Two A. M., Yun T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680. doi: 10.1126/scitranslmed.aah4680
Naughton L. M., Romano S., O’Gara F., Dobson A. D. W. (2017). Identification of secondary metabolite gene clusters in the pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01494
Newstead L. L., Varjonen K., Nuttall T., Paterson G. K. (2020). Staphylococcal-produced bacteriocins and antimicrobial peptides: their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics 9, 40. doi: 10.3390/antibiotics9020040
Ng W. (2022). Statistical analysis of number of genes and chromosome lengths of different microbial species. bioRxiv. doi: 10.1101/2022.08.13.503871
Othoum G., Bougouffa S., Razali R., Bokhari A., Alamoudi S., Antunes A., et al. (2018). In silico exploration of red sea bacillus genomes for natural product biosynthetic gene clusters. BMC Genomics 19, 382. doi: 10.1186/s12864-018-4796-5
Paul S. I., Rahman M. M., Salam M. A., Khan M. A. R., Islam M. T. (2021). Identification of marine sponge-associated bacteria of the Saint Martin’s island of the bay of bengal emphasizing on the prevention of motile aeromonas septicemia in labeo rohita. Aquaculture 545, 737156. doi: 10.1016/j.aquaculture.2021.737156
Pawlik J. R. (1993). Marine invertebrate chemical defenses. Chem. Rev. 93, 1911–1922. doi: 10.1021/cr00021a012
Petersen L.-E., Kellermann M. Y., Schupp P. J. (2020). “Secondary metabolites of marine microbes: From natural products chemistry to chemical ecology, Youmares 9 - the oceans: Our research, our future,” in Proceedings of the 2018 conference for young marine researcher in oldenburg, Germany, 159–180 (Cham: Springer International Publishing). doi: 10.1007/978-3-030-20389-4_8
Rayburn A. L. (1993). “Comparative studies of genome content,” in Methods in enzymology, vol. 224. (San Diego, CA: Elsevier), 204–212.
Rendueles O., Ghigo J.-M. (2012). Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 36, 972–989. doi: 10.1111/j.1574-6976.2012.00328.x
Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect. Med. 2, a012427. doi: 10.1101/cshperspect.a012427
Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sánchez-Rodríguez D. E., Ortiz-Aguirre I., Aguila-Ramírez R. N., Rico-Virgen E. G., González-Acosta B., Hellio C. (2018). Marine bacteria from the gulf of california with antimicrofouling activity against colonizing bacteria and microalgae. Rev. Biología Trop. 66, 1649–1663. doi: 10.15517/rbt.v66i4.31963
Sandoval-Powers M., Králová S., Nguyen G.-S., Fawwal D. V., Degnes K., Lewin A. S., et al. (2021). Streptomyces poriferorum sp. Nov., a novel marine sponge-derived actinobacteria species expressing anti-mrsa activity. Systematic Appl. Microbiol. 44, 126244. doi: 10.1016/j.syapm.2021.126244
Satheesh S., Ba-akdah M. A., Al-Sofyani A. A. (2016). Natural antifouling compound production by microbes associated with marine macroorganisms: A review. Electronic J. Biotechnol. 19, 26–35. doi: 10.1016/j.ejbt.02.002
Satheesh S., Soniamby A. R., Sunjaiy Shankar C. V., Mary Josephine Punitha S. (2012). Antifouling activities of marine bacteria associated with sponge (sigmadocia sp.). J. Ocean Univ. China 11, 354–360. doi: 10.1007/s11802-012-1927-5
Severn M. M., Horswill A. R. (2023). Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat. Rev. Microbiol. 21, 97–111. doi: 10.1038/s41579-022-00780-3
Solouki Nezhad R. S., Asaadi H., Eshaghi Milasi Y., Yazdansetad S. (2019). Isolation and molecular identification of carotenoid-producing bacteria. Nova Biologica Reperta 6, 61–69. doi: 10.29252/nbr.6.1.61
Sugimoto S., Iwamoto T., Takada K., Okuda K., Tajima A., Iwase T., et al. (2013). Staphylococcus epidermidis esp degrades specific proteins associated with staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol 195, 1645–1655. doi: 10.1128/jb.01672-12
Sukmarini L., Atikana A., Hertiani T. V. (2014). Antibiofilm activity of marine microbial natural products: potential peptide- and polyketide-derived molecules from marine microbes toward targeting biofilm-forming pathogens. J Nat Med 7, 1–20. doi: 10.1007/s11418-023-01754-2
Tamura K., Stecher G., Kumar S. (2021). Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Teanpaisan R., Kawsud P., Pahumunto N., Puripattanavong J. (2017). Screening for antibacterial and antibiofilm activity in thai medicinal plant extracts against oral microorganisms. J. Traditional Complementary Med. 7, 172–177. doi: 10.1016/j.jtcme.2016.06.007
Vandecandelaere I., Depuydt P., Nelis H. J., Coenye T. (2014). Protease production by staphylococcus epidermidis and its effect on staphylococcus aureus biofilms. Pathog Dis. 70, 321–331. doi: 10.1111/2049-632x.12133
Viju N., Punitha S. M. J., Satheesh S. (2021). An analysis of biosynthesis gene clusters and bioactivity of marine bacterial symbionts. Curr. Microbiol 78, 2522–2533. doi: 10.1007/s00284-021-02535-4
Vimala R. (2016). Marine organisms: A potential source of natural antifouling metabolites. Int. J. ChemTech Res. 9, 208–217.
Wang K.-L., Dou Z.-R., Gong G.-F., Li H.-F., Jiang B., Xu Y. (2022). Anti-larval and anti-algal natural products from marine microorganisms as sources of anti-biofilm agents. Marine Drugs 20, 90. doi: 10.3390/md20020090
Whalen K. E., Becker J. W., Schrecengost A. M., Gao Y., Giannetti N., Harvey E. L. (2019). Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7, 93. doi: 10.1186/s40168-019-0711-9
Wibowo J. T., Bayu A., Aryati W. D., Fernandes C., Yanuar A., Kijjoa A., et al. (2023). Secondary metabolites from marine-derived bacteria with antibiotic and antibiofilm activities against drug-resistant pathogens. Marine Drugs 21, 50. doi: 10.3390/md21010050
Wu L., Luo Y. (2021). Bacterial quorum-sensing systems and their role in intestinal bacteria-host crosstalk. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.611413
Yi Y., Liang L., de Jong A., Kuipers O. P. (2024). A systematic comparison of natural product potential, with an emphasis on ripps, by mining of bacteria of three large ecosystems. Genomics 116, 110880. doi: 10.1016/j.ygeno.2024.110880
Zaghloul E. H., Ibrahim H. A. H., El-Badan D. E. S. (2021). Production of biocement with marine bacteria; staphylococcus epidermidis edh to enhance clay water retention capacity. Egyptian J. Aquat. Res. 47, 53–59. doi: 10.1016/j.ejar.2020.08.005
Zhang J., Jiang H., Jiang J., Huang G. (2016). Isolation and identification of staphylococcus epidermidis s14 screening extracellular antimicrobial metabolites. Am. J. Biochem. Biotechnol. 12, 56–63. doi: 10.3844/ajbbsp.2016.56.63
Keywords: sponge-associated bacteria, bioactive metabolites, antibacterial compounds, complete genome, biofilms, biosynthetic gene cluster
Citation: Jamal MT, Abdulrahman I, Amran RH, Al-Matary MA and Satheesh S (2025) Complete genome sequencing and antibiofilm activity of an endophytic bacterium associated with marine sponge Hyrtios erectus collected from the Red Sea. Front. Mar. Sci. 12:1588772. doi: 10.3389/fmars.2025.1588772
Received: 06 March 2025; Accepted: 21 April 2025;
Published: 14 May 2025.
Edited by:
Santhiyagu Prakash, Tamil Nadu Fisheries University, IndiaReviewed by:
Ramasamy Ramasubburayan, Saveetha University, IndiaManikandan Gurusamy, Tshwane University of Technology, South Africa
Copyright © 2025 Jamal, Abdulrahman, Amran, Al-Matary and Satheesh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sathianeson Satheesh, c3NhdGhpYW5lc29uQGthdS5lZHUuc2E=; c2F0aGVlc2hfczIwMDVAeWFob28uY28uaW4=
 Mamdoh T. Jamal
Mamdoh T. Jamal Idris Abdulrahman1,2
Idris Abdulrahman1,2 Ramzi H. Amran
Ramzi H. Amran Mohammed A. Al-Matary
Mohammed A. Al-Matary Sathianeson Satheesh
Sathianeson Satheesh