Abstract
The endosymbiosis between cnidarians and photosynthetic dinoflagellates of the Symbiodiniaceae family forms the foundation of coral reef ecosystems. Prolonged environmental shifts can disrupt the cnidarian–Symbiodiniaceae partnership, triggering dysbiosis and coral bleaching and ultimately resulting in coral starvation, mortality, and the collapse of reef ecosystems. Despite its significance, critical gaps remain in our understanding of the cellular mechanisms governing symbiosis and dysbiosis. Innate immune genes and pathways are highly conserved across the Metazoa, including in cnidarians. Among these is NADPH oxidase (NOX), a key enzyme responsible for generating reactive oxygen species (ROS), primarily for microbial degradation within phagolysosomes. In this study, we hypothesize that NOX plays a role in the regulation of cnidarian–Symbiodiniaceae symbiosis and the host phagosomal maturation process. We investigated NOX function in relation to symbiotic state and heat stress in the sea anemone Exaiptasia diaphana (commonly called aiptasia), a model for cnidarian–Symbiodiniaceae symbiosis and dysbiosis. Our findings show that NOX gene and protein expression is suppressed in the symbiotic state, supporting the hypothesis that symbionts modulate host innate immunity. However, upon heat treatment, we observed increased NOX expression and activity along with NOX localization around algal symbionts, suggesting that host phagosomal maturation processes are engaged during bleaching. We propose a model where the phagocytic NOX complex becomes activated during symbiosis breakdown and bleaching. Our findings support the hypothesis that in situ degradation, facilitated by ROS generated by NOX, plays a key role in the process of dysbiosis. This work contributes to our understanding of cnidarian innate immunity, highlighting critical steps in dysbiosis and phagosomal maturation processes within cnidarian–Symbiodiniaceae symbiosis.
1 Introduction
The endosymbiosis between cnidarians and photosynthetic dinoflagellates, from the family Symbiodiniaceae, forms the foundation of coral reef ecosystems (Fisher et al., 2015; LaJeunesse et al., 2018). The cnidarian–Symbiodiniaceae partnership is sensitive to environmental shifts that can trigger symbiosis dysfunction and algal loss, known as dysbiosis. Prolonged dysbiosis, commonly called bleaching due to the paling of coral hosts from loss of algae, results in coral starvation, mortality, reduced fitness, and the subsequent devastation of coral reef ecosystems, with severe ecological, cultural, and economic consequences worldwide (Eddy et al., 2021; Ranjan et al., 2023). Our ability to address this ecosystem-level crisis is limited by substantial gaps in our understanding of the cellular mechanisms governing symbiosis and dysbiosis (Weis, 2019).
The establishment of endosymbiosis between animals and their intracellular symbionts involves core cellular processes such as interpartner signaling, vesicular trafficking, metabolic integration, nutrient exchange, phagocytosis, and immune modulation. In most symbiotic cnidarians, the symbionts reside within a host-derived vacuolar membrane, called the symbiosome, within host gastrodermal cells (Wakefield and Kempf, 2001). Bidirectional metabolic exchange occurs across the symbiosome, with each partner providing for the metabolic needs of the other. The symbiont produces photosynthetically fixed carbon that fulfills most of the host’s nutritional requirements, while the symbiont receives inorganic nutrients, such as inorganic carbon and nitrogen, from the host (Muscatine et al., 1981; Whitehead and Douglas, 2003; Yellowlees et al., 2008). This intimate partnership requires the cnidarian immune system to manage the uptake and survival of beneficial Symbiodiniaceae while also targeting and removing foreign invaders (Jacobovitz et al., 2021).
Cnidarians, such as corals and sea anemones, share much of their innate immune repertoire with that found in vertebrates. During the initiation of cnidarian–Symbiodiniaceae symbiosis, a variety of microbe-associated molecular patterns (MAMPs) on Symbiodiniaceae bind to host pattern recognition receptors (PRRs), triggering signaling cascades that can lead to microbial destruction or uptake, depending on the microbe being detected (Detournay and Weis, 2011; Dunn and Weis, 2009; Maruyama et al., 2022; Neubauer et al., 2016; Parkinson et al., 2018; Tivey et al., 2020). Downstream immune pathways, including the complement system, sphingosine rheostat, transforming growth factor beta (TGFβ), and nuclear factor-kappa B (NF-κB), engage with Symbiodiniaceae, playing essential roles in establishing the partnership (Detournay et al., 2012; Detournay and Weis, 2011; Kitchen and Weis, 2017; Mansfield et al., 2017; Neubauer et al., 2017; Poole et al., 2016; Poole and Weis, 2014; Williams et al., 2018). Both the initiation and maintenance of cnidarian–Symbiodiniaceae symbiosis are linked to immune suppression in the host, potentially serving as a survival strategy for the symbiont to evade recognition and phagolysosomal digestion by inhibiting the maturation of phagosomes (Lehnert et al., 2014; Mansfield et al., 2017; Wolfowicz et al., 2016). Characterization of the symbiosome environment reveals its lysosome-like qualities, including a low pH (~4) and the presence of several lysosomal and late phagolysosomal markers, including lysosomal-associated membrane protein 1 (LAMP1), Niemann–Pick type C2 (NPC2), and vacuolar-type ATPase (V-ATPase) (Barott and Tresguerres, 2015; Dani et al., 2017; Jacobovitz et al., 2021). During dysbiosis, many immune-related components and pathways are upregulated (Traylor-Knowles et al., 2017; Van De Water et al., 2018; Mansfield et al., 2017; Richier et al., 2008; DeSalvo et al., 2010), suggesting a re-engagement of the phagosomal machinery in response to dysbiosis. However, our understanding of phagosomal maturation in relation to symbiosis remains limited.
Numerous studies investigating cnidarians in various symbiotic states, including bleaching and dysbiosis, have documented significant alterations in host gene expression that reflect stress and dysregulation (Cleves et al., 2020; Cui et al., 2023; Cziesielski et al., 2018; Kirk and Weis, 2016; Lehnert et al., 2014; Sunagawa et al., 2009). These studies have identified a number of symbiosis-associated genes that are promising candidates for functional analysis (Mohamed et al., 2016). One such gene encodes nicotinamide adenine dinucleotide phosphate oxidase, (NADPH oxidase or NOX) (Cui et al., 2023; Lehnert et al., 2014; Meyer and Weis, 2012), which has increased expression at high seawater temperatures and bleaching corals and anemones (summarized in Supplementary Table S1) (Cziesielski et al., 2018; DeSalvo et al., 2010). Metazoan NOXs are oxidoreductases involved in the controlled production of superoxide anions (O2• –) and other reactive oxygen species (ROS) (Vermot et al., 2021). Their primary function is to generate highly reactive superoxide anions (O2• –) for microbial degradation in microbicidal phagosomes (Segal, 2005; Vermot et al., 2021). In addition, NOX plays critical roles in maintaining redox homeostasis, regulating cell growth, differentiation, and apoptosis through a variety of signaling pathways (Vermot et al., 2021). The phagocytic NOX complex (Figure 1) is present across the Metazoa as a multiunit transmembrane protein comprised of cytosolic (p47phox and p67phox) and membrane-bound subunits (NOX2 and p22phox) (Kawahara and Lambeth, 2007; Sumimoto, 2008). In vertebrates, the core NOX complex is formed by NOX2 and p22phox, which facilitate electron transfer across the membrane, while the cytosolic subunits, p47phox and p67phox, assemble and translocate to the membrane to activate the NOX complex (Sumimoto, 2008). Upon activation, the NOX complex transfers electrons across the membrane, leading to the production of superoxide, which is involved in microbial killing and in signaling pathways.
Figure 1
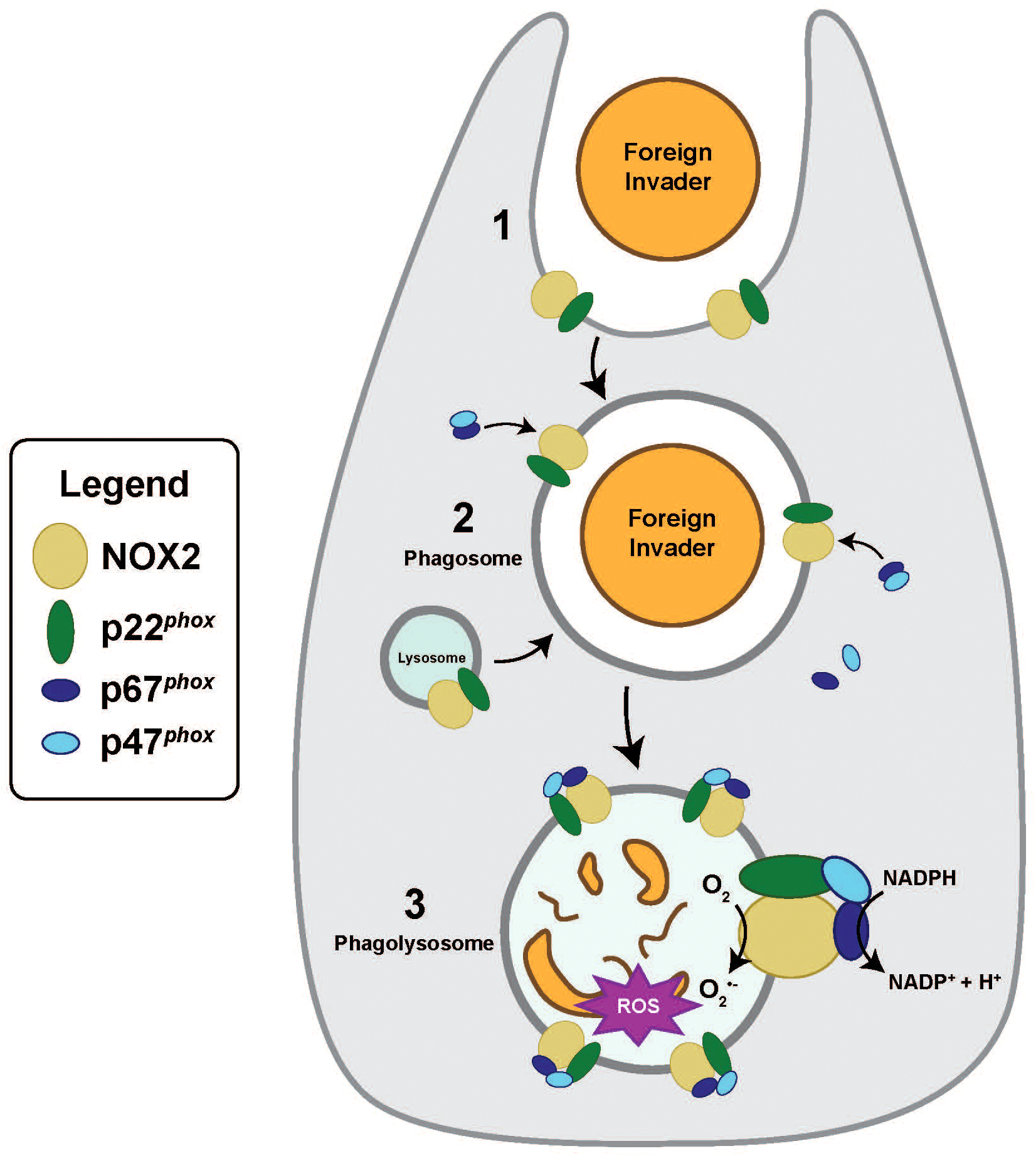
Activation of the canonical phagocytic NOX complex. (1) Following the internalization and detection of foreign invaders, a nascent phagosome is formed. (2) During the phagosomal maturation process, lysosomes will fuse with phagosome, and the NOX complex becomes activated. Activated cytosolic factors p47phox and p67phox assemble and then translocate from the cytosol to membrane-bound NOX2 and p22phox. (3) Phagolysosomes are microbicidal, producing reactive oxygen species (ROS) to degrade foreign invaders. Phagolysosomal membranes possess activated NOX that catalyzes the transfer of one electron from NADPH to O2, reducing molecular oxygen to produce superoxide anions (O2•–) and downstream ROS.
In corals and sea anemones, NOX inhibitors have been shown to reduce the production of extracellular superoxide, suggesting that NOX contributes to immunity by generating ROS for microbicidal degradation (Hutton and Smith, 1996; Saragosti et al., 2010). Furthermore, tissue-specific transcriptomes of the symbiotic sea anemone, Exaiptasia diaphana (commonly called aiptasia), shows distinct gastrodermal-versus-epidermal expression profiles that shift in response to symbiotic state (Supplementary Table S1) (Cui et al., 2023). Despite the presence of NOX and its demonstrated activity, a functional analysis of its role in cnidarian–algal symbiosis has not yet been performed. Consequently, our understanding of the role of NOX in cnidarian immunity and symbiosis remains limited.
In this study, we used aiptasia as a model system to investigate the role of NOX in a cnidarian–Symbiodiniaceae symbiosis, with a focus on its function across different symbiotic states and during heat stress. Aiptasia serves as a tractable model system for exploring cellular mechanisms governing cnidarian symbiosis, benefiting from a variety of genomic resources that facilitate cellular and molecular investigations (Baumgarten et al., 2015; Jacobovitz et al., 2023). Based on our findings, we develop a model where the phagocytic NOX complex becomes activated on the symbiosome during heat stress, suggesting that the host phagosomal maturation process is engaged in symbiosis breakdown and bleaching.
2 Materials and methods
2.1 Animal maintenance
A clonal culture of aiptasia (culture ID: H2 originally collected from Hawaii) (Xiang et al., 2013), harboring the symbiont Breviolum minutum (LaJeunesse et al., 2018; Xiang et al., 2013), was maintained in 300 mL of artificial seawater (ASW) at 25°C ( ± 0.1°C) in an incubator (Percival AL41L4) with coral lights (Zoo Med Ocean Sun F25T8/10,000k) at 25 µmol photons/m2/s with a 12-h:12-h light/dark cycle. Aposymbiotic anemones were initially generated by menthol bleaching (Matthews et al., 2016) and maintained in the dark at 25°C. The anemones were fed three times a week ad libitum with freshly hatched Artemia nauplii enriched with S.presso (Brine Shrimp Direct) and given fresh ASW changes 4–8 h post-feeding.
2.2 Experimental design for NOX expression, localization, and activity as a function of symbiotic state and heat stress
All experiments exploring NOX function—gene expression, protein expression, localization, and activity—in aiptasia followed the same experimental design unless otherwise noted. Prior to each experiment, three aposymbiotic and 12 symbiotic anemones were moved to six-well plates containing FSW, starved for 5 days, and kept in the conditions described under the subsection “Animal maintenance”. Aposymbiotic anemones were screened for symbiont contamination by looking for symbiont chlorophyll auto-fluorescence using fluorescence microscopy (green excitation at 510–540 nm by using a Zeiss Stemi 305 stereoscope). Only fully aposymbiotic animals were chosen for the experiment.
NOX gene or protein expression, localization, or activity was first compared in symbiotic vs. aposymbiotic animals. Symbiotic and aposymbiotic anemones were sampled at day 0 at an ambient temperature of 25°C ( ± 0.1°C) (N = 3 animals per treatment), flash-frozen in liquid nitrogen, and stored for later processing. Subsequently, the remaining nine symbiotic anemones were subjected to heat stress of 32°C ( ± 0.1°C) by placement in an incubator (Percival AL30L2) for 5 days, with the initial target temperature reaching 32°C within 4 h. Heat-treated symbiotic anemones were sampled on days 1, 3, and 5 (N = 3 animals per treatment), flash-frozen in liquid nitrogen, and stored for later processing. Elevated temperature conditions were maintained in the incubator (Percival AL30L2) and kept in light conditions as described under the subsection “Animal maintenance”. During the heat treatment, the anemones were not fed, and pre-heated FSW was used for daily water changes.
2.3 Identification of NOX genes
NCBI Basic Local Alignment Search Tool (BLAST) was used to search for NOX sequences in aiptasia genome database (Aip-v1 GCF 001417965.1) using human NOX gene sequences (NCBI accession nos.: CYBB (NOX2), NM_000397; NCF2 (p67phox), NM_000433; CYBA (p22phox), NM_000101; NCF1 (p47phox), NM_000265) as the query. Full-length NOX sequences were subsequently identified by aligning several aiptasia adult and larvae transcriptomes (for accuracy) from the NCBI SRA database (adult accession nos.: aposymbiotic, SRX757525; partially populated, SRX757526; symbiotic, SRX757528 and larvae accession nos.: aposymbiotic, SRX757531; symbiotic, SRX757532) and referencing the aiptasia genome from the NCBI database (Aip-v1 GCF 001417965.1) in Geneious (v10.2.6) (Baumgarten et al., 2015; Drummond et al., 2010; Hughes et al., 2018; Lehnert et al., 2014).
2.4 Development of qPCR primers
Conserved regions of NOX genes were targeted for designing qPCR primers in Geneious (v10.2.6) (Drummond et al., 2010). To test the NOX primer pairs, a standard PCR was performed, followed by qPCR with the addition of a melt curve. Primer products were confirmed to be a single product of target size, and amplification efficiency was confirmed to be at least 90% by analyzing the slope of standard curves generated from serial dilutions of template cDNA (Supplementary Table S2). Nuclear ribosomal large subunit 10 (L10) was selected as the reference gene for elevated temperature-treated anemones and amplification specificity of A. pallida DNA (Poole et al., 2016).
2.5 Quantification of NOX gene expression via qPCR
The gene expression of NOX genes NOX2 1-3, p67phox, and p22phox were examined in symbiotic and aposymbiotic anemones (~0.5-cm pedal disc diameter) at day 0 and in symbiotic animals during heat treatment on days 1, 3, and 5. Each time point had three biological replicates, and each qPCR reaction was run in triplicate. RNA was extracted from each anemone sample using a hybrid Trizol/chloroform RNeasy protocol (RNeasy Mini Kit by Qiagen) as described in Poole et al. (2016), followed by a DNase step (DNase I by Sigma Aldrich) and PCR inhibitor removal (OneStep PCR Inhibitor Removal Kit by ZYMO), following the manufacturer’s instructions. cDNA was synthesized with oligo d(T)23 VN primers following the manufacturer’s instructions (ProtoScript II First Strand cDNA Synthesis Kit by NEB). After confirming the cDNA and primer quality, qPCR was performed to quantify the NOX expression levels of each sample using 25 ng of cDNA template and 20 µL reactions (SsoAdvanced Universal SYBR Green Supermix by BIO-RAD). A BioRad CFX96 Real-Time System was used to carry out PCR reactions using a two-step amplification phase (95°C/10 min, followed by 40 cycles of 95°C/10 s and 58°C/30 s) with the addition of a melt curve. No-reverse transcription, no-primer, and no-template controls were included as negative controls. L10, a reference gene for heat stress, was used to normalize the expression levels of target genes (Poole et al., 2016).
2.6 Gene expression data analysis
Gene expression data were analyzed following the Livak method, commonly known as ΔΔCt (Livak and Schmittgen, 2001). For each sample and target gene, ΔCt values were calculated by subtracting the Ct value of the target gene from the Ct value of the reference gene (L10). The ΔCt value was then subtracted from the mean ΔCt value of corresponding genes from reference samples to calculate the relative expression ΔΔCt. Fold change gene expression was then calculated as 2-ΔΔCt for each gene from each sample. For the comparison between symbiotic states, aposymbiotic samples served as the reference, and for the heat treatment comparison, day 0 symbiotic samples served as the reference. After confirming normality with the Shapiro–Wilk test and homogeneity of variance with Levene’s test, unpaired two-sided t-tests were performed, followed by a false discovery rate (FDR) correction to account for multiple comparisons for each gene in RStudio (R Core Team, 2015). Differences in gene expression were considered statistically significant at p <0.05.
2.7 Symbiont quantification in hosts
Symbiotic anemones followed the same experimental design as described under the subsection “Experimental design for NOX expression, localization, and activity as a function of symbiotic state and heat stress”. Whole anemones were homogenized in RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) with protease inhibitors (mini cOmplete tablets by Sigma-Aldrich). Algal cell densities were quantified from two technical replicates of 10 µL homogenate using an automated cell counter and cell counting chamber slides (Countess II by Life Technologies) and normalized to total anemone protein in the supernatant (Qubit 4 Fluorometer and Qubit Protein Assay kit by ThermoFisher Scientific). Data were plotted using “ggplot2” package in RStudio (R Core Team, 2015). To determine changes in algal density as a function of the heat treatment, normality and homogeneity of variance were evaluated with the Shapiro–Wilk and Levene’s tests, and then unpaired two-sided T-tests were performed between each heat treatment day compared to day 0, followed by a false discovery rate (FDR) to account for multiple comparisons in RStudio (R Core Team, 2015). Differences in algal cell densities were considered statistically significant at p <0.05.
2.8 Generation of polyclonal purified antibodies for NOX subunits
Two NOX subunits were targeted: p67phox, due to its established role in activating NOX through translocation from the cytosol to the NOX complex, and NOX2-3 (AIPGENE 22656), which is upregulated in the gastrodermis of symbiotic anemones relative to aposymbiotic anemones (Cui et al., 2023; Sumimoto, 2008). Custom-made rabbit polyclonal anti-p67phox and anti-NOX2–3 antibodies were developed by GenScript USA Inc. using three synthetic peptide antigens each for aiptasia p67phox and NOX2-3 (Supplementary Table S3). Geneious, Phyre2 (version 2.0), EzMol (v2.1), and NCBI BLAST were used to identify peptides based on a variety of factors, such as antigenicity, 3-D structure, and an absence of overlapping homology with the symbiont genome. For each synthesized peptide, GenScript immunized two rabbits per antibody and subsequently provided purified antibody, peptide antigen, and a single combined pre-immune serum from the two rabbits in equal proportions. Immunoblots were conducted to examine individual antibody specificity, size estimates of targets, and match staining among each set of antibodies. A detailed description of the testing of each anti-p67phox and anti-NOX2–3 is provided in Supplementary Methods and Supplementary Figure S1. Based on this testing, one anti-p67phox and anti-NOX2–3 were selected to quantify protein expression and for localization. Pre-immune serum (1:1,000 dilution in RIPA buffer) and peptide-blocked antibodies were used as negative controls for the selected anti-p67phox and anti-NOX2–3 antibodies to rule out any cross-reactivity and non-specific binding of the antibody, respectively.
2.9 Quantification of NOX protein expression via immunoblots
The protein expression of putative p67phox and NOX2–3 was quantified in symbiotic and aposymbiotic anemones (~0.5-cm pedal disc diameter) at day 0 and in heat-treated symbiotic anemones at days 1, 3, and 5 (N = 3 anemones per treatment) using anti-p67phox and anti-NOX2–3 immunoblots. Whole anemones were homogenized in RIPA lysis buffer with protease inhibitors (mini cOmplete tablets by Sigma-Aldrich). The homogenates were centrifuged at 12,000 × g for 10 min at 4°C, and the supernatants were placed in a new tube. Protein concentration was determined using a Qubit 4 Fluorometer and Qubit Protein Assay kit (ThermoFisher Scientific). Furthermore, 30 µg of homogenate with 5% beta-mercaptoethanol was boiled for 10 minutes and resolved by SDS-PAGE in a 4%–20% gradient gel initially at 50 V for 10 min, followed by 175 V for 60 min at room temperature (Mini Protean TGX gels by BIO-RAD). The proteins were electrophoretically transferred onto a nitrocellulose membrane at 75 V for 100 min at 4°C. The membrane was blocked in TBST/5% non-fat dry milk (by Carnation) for 2 h at RT with gentle shaking (60 rpm) and then incubated with anti-NOX2–3 primary antibody at a 1:2,000 concentration overnight at 4°C. Following five washes with TBST, the membrane was incubated with a 1:5,000 concentration of a goat anti-rabbit IgG-HRP secondary antibody (Santa Cruz Biotechnology) for 1 h at RT.
Antibody labeling was visualized by chemiluminescence (BioRad ECL substrate kit) using a myECL imager (Thermo Scientific). After imaging for anti-NOX2, the membrane was stripped (0.2 M glycine, 0.1% SDS, 1% Tween 20; pH 2.2) and probed with a 1:2,000 concentration of the anti-p67phox primary antibody, followed by another secondary antibody incubation. All primary and secondary antibodies were diluted in TBST/5% non-fat dry milk. Protein sizes were determined using a colorimetric protein molecular weight ladder (Bio-Rad Precision Plus Protein Dual Color Standard). Protein expression was quantified using FIJI ImageJ and calculated as corrected total fluorescence = [integrated density − (area × mean gray value of background)] (Schindelin et al., 2012). For the symbiotic state comparison, aposymbiotic samples served as the reference, and for the heat treatment comparison, day 0 symbiotic samples served as the reference. The mean ± SD for each sampling time was calculated for the three biological replicates and compared to reference samples. After confirming normality with the Shapiro–Wilk test and homogeneity of variance with Levene’s test, we performed unpaired two-sided t-tests in RStudio (R Core Team, 2015). To account for multiple comparisons between heat treatment day and day 0, a false discovery rate (FDR) correction was applied. Differences in protein expression were considered statistically significant if p <0.05.
2.10 Localization of NOX via immunofluorescence microscopy
2.10.1 Whole mounts
The localization of anti-p67phox and anti-NOX2–3 was visualized from small symbiotic and aposymbiotic anemones (eight- to 12-tentacle stage and a pedal disc diameter of 1 to 2 mm) at day 0 and in symbiotic animals during heat treatment on days 1, 3, and 5 (N = 3 for all time points and treatments). The anemones were relaxed in FSW with 3.5% w/v MgCl2 for 5–10 min and then fixed in 3.7% v/v formaldehyde/0.2% glutaraldehyde in PBS (pH 7.4) at 4°C for 8 h. After several washes in PBS with 0.2% Triton X-100 (PBST), the samples were blocked in PBST with 5% BSA and 5% normal goat serum at 4°C for 12 h. The samples were then incubated in primary antibodies (1:100 for anti-p67phox and anti-NOX2-3) at 4°C for 12 h, followed by several washes with PBST. Following this, the samples were stained with 10 µg/mL Hoechst 33342 (Invitrogen) and a secondary (1:2,000; goat anti-rabbit Alexa Fluor 488 by Santa Cruz Biotechnology) antibody at 4°C for 12 h. All antibodies were diluted in PBS with 5% goat serum, 5% BSA, and 0.2% Triton X-100. Stained anemones were washed with PBST and then PBS several times. The samples were then incubated in 70% glycerol diluted in PBS at 4°C for 2 to 3 h and followed by 87% glycerol diluted in PBS at 4°C for 2 to 3 h. The stained anemones were mounted in 87% glycerol on glass slides with no. 1.5 coverslips and stored at 4°C before imaging.
2.10.2 Cryosections
To generate anemone cryosections, symbiotic anemones (0.5-cm pedal disc diameter) were starved for 5 days, relaxed in 3.5% w/v MgCl2 for 5–10 min, and fixed in 4% paraformaldehyde in FSW at 4°C overnight. The fixed anemones were fully submerged in 10%, 20%, and 30% sucrose sequentially as a cryoprotectant at 4°C, embedded with OCT medium, and rapidly frozen in isopentane cooled by liquid nitrogen. The tentacles were sectioned (20 μm) at −20°C with a cryostat (Microm500, Zeiss, Inc., Germany), cold-mounted onto positively charged glass slides, and stored at -80°C until staining. The sections were thawed and rehydrated in PBS for 10 min and then blocked in 5% goat serum in 1× PBS for 30 min at 37°C. The slides were incubated in either 1:100 anti-p67phox or anti-NOX2–3 for 60 min at 37°C, followed by several washes in PBST. The samples were then incubated with 10 µg/mL Hoechst 33342 (Invitrogen) and secondary (1:2,000; goat anti-rabbit Alexa Fluor 488 by Santa Cruz Biotechnology) antibodies for 60 min at 37°C, followed by several washes. The slides were mounted with 87% glycerol diluted in PBS on glass slides with no. 1.5 coverslips and stored for a short term at 4°C before imaging.
2.10.3 Confocal imaging
Z-stacks were acquired on an LSM 780 NLO Confocal Microscope (ZEISS). Separate channels for Hoechst, Alexa Fluor 488, and chlorophyll autofluorescence were captured with a 405-nm diode (410–483 nm), an argon laser (491–554 nm), and a 633-nm HeNe laser (647–722 nm), respectively. Simultaneously, using laser illumination, a transmitted light detector (T-PMT) acquired transmitted light images in bright field. Confocal z-stack images were merged to create a maximum intensity projection using ZEN Black and ZEN lite (Carl Zeiss AG).
2.11 Measurement of NOX activity
NOX activity was measured by quantifying the respiratory burst of superoxide and other downstream ROS in symbiotic anemones (0.5-cm pedal disc diameter) at days 0 and 1 of heat treatment (N = 3 for each treatment, including the controls and inhibitors described below). The Cell-ROX Green Reagent (Life Technologies) specifically detects ROS, particularly superoxide anion (O2• –), through fluorescence upon oxidation. Live anemones were incubated in the dark with 5 mM Cell-ROX Green Reagent for 2 h and then homogenized, following the manufacturer’s protocol. ROS was quantified emission from supernatant using a fluorescent plate reader in nanometers (SpectraMax M3 fluorescent plate reader by Molecular Devices) at 490-nm excitation and 525-nm, and fluorescence was expressed as relative fluorescence units (RFU). The assay involved incubating anemones with reagents to assess ROS production and the effects of specific inhibitors and controls. Diphenyleneiodonium chloride (DPI) was applied at a concentration of 25 µM for 30 min to selectively inhibit NOX-derived ROS production by blocking NOX activation (Lambeth, 2004). DPI treatment assessed NOX’s contribution to overall ROS levels. As a positive control, menadione was used at 12.5 µM for 30 min to induce ROS release, confirming the assay system’s responsiveness to oxidative stress. N-Acetylcysteine (NAC) at 25 µM for 5 min was used as a negative antioxidant control to sequester and neutralize ROS. All fluorescence values were normalized to total host protein per sample using a Qubit 4 Fluorometer and Qubit Protein Assay kit, and the results were reported as RFU per microgram of protein. After confirming normality with the Shapiro–Wilk test and homogeneity of variance with Levene’s test, we compared the ambient and heat-stressed groups across the four conditions using unpaired two-sided t-tests, applying a FDR correction for multiple comparisons in RStudio (R Core Team, 2015). Differences in NOX activity were considered statistically significant at p <0.05.
3 Results
3.1 NOX gene expression was downregulated in symbiotic compared to aposymbiotic anemones and moderately upregulated in symbiotic anemones subjected to elevated temperature
Five NOX genes were identified in aiptasia, including three NOX2-like genes (termed NOX2 1–3), p22phox, and p67phox (Supplementary Table S2). No p47phox ortholog was detected, suggesting that this sub-unit is either absent from aiptasia or so highly divergent that it lacks recognizable conserved domains. Of the five NOX genes in aiptasia, NOX2-3, p22phox, and p67phox were significantly downregulated (p < 0.01) in symbiotic compared to aposymbiotic anemones with mean fold changes of -2.13, -3.26, and -1.70, respectively (Figure 2A). While NOX2–1 and NOX2–2 showed an initial increase in expression on day 1 of heat stress, followed by a downward trend on days 3 and 5, these changes were not statistically significant (Figure 2B).
Figure 2
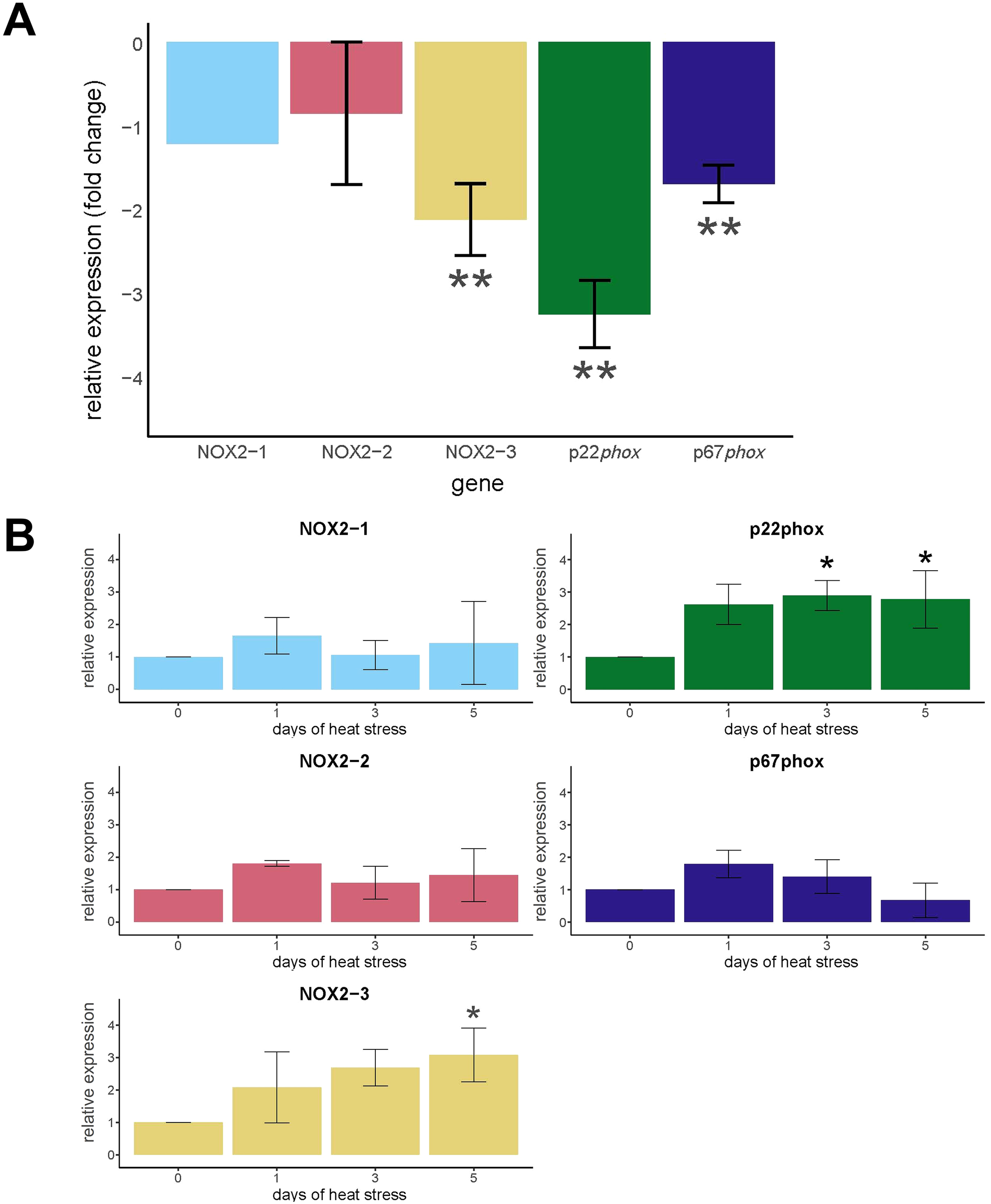
Expression of NOX genes of aiptasia as a function of symbiotic state and heat treatment. (A) Relative amounts of NOX expression were determined by qRT-PCR of cDNA from aposymbiotic and symbiotic anemones at ambient temperature (N = 3). The values are expression in symbiotic anemones relative (fold change) to aposymbiotic anemones set at 0. (B) Relative amounts of NOX expression were determined by qRT-PCR of cDNA from symbiotic anemones during days 1, 3, and 5 of heat treatment and at day 0 (N = 3). The values are expression in symbiotic anemones relative (fold change) to symbiotic anemones at day 0 set at 0. (A, B) The values are means ± SD. The asterisks indicate statistically significant differences compared to (A) aposymbiotic anemones and (B) symbiotic anemones at day 0 calculated on ΔΔCt values as determined by unpaired T-tests with FDR correction. *p < 0.05, **p < 0.01.
During the heat treatment of aiptasia, NOX2–3 and p22phox exhibited increases in transcript levels compared to day 0. NOX2–3 expression was significantly higher (p < 0.05) at day 5, with a 2.50-fold change compared to day 0 levels (Figure 2B). In addition, p22phox expression was significantly elevated (p < 0.05) at days 3 and 5 of heat treatment with a 1.89- and 2.24-fold change, respectively, compared to day 0 levels (Figure 2B). The increase in NOX2–3 and p22phox expression coincided with a significant decrease in algal density in host tissues at day 5 (Supplementary Figure S2). NOX2-1, NOX2-2, and p67phox gene expression levels did not differ from day 0 levels during the heat treatment as determined by unpaired two-sided T-tests (Figure 2B).
3.2 NOX protein expression was downregulated in symbiotic compared to aposymbiotic anemones and NOX2 expression increased in symbiotic anemones subjected to prolonged elevated temperatures
Both putative p67phox and NOX2–3 protein expression was significantly lower in the symbiotic compared to aposymbiotic anemone homogenates (p < 0.01; Figures 3A, B). Overall, putative p67phox and NOX2–3 protein expression was variable during the heat treatment of aiptasia. Putative p67phox protein levels during heat treatment did not differ from day 0 levels (Figure 3C). However, putative NOX2–3 protein levels were significantly higher (p < 0.05) at day 3 of heat treatment than at day 0 (Figure 3D).
Figure 3
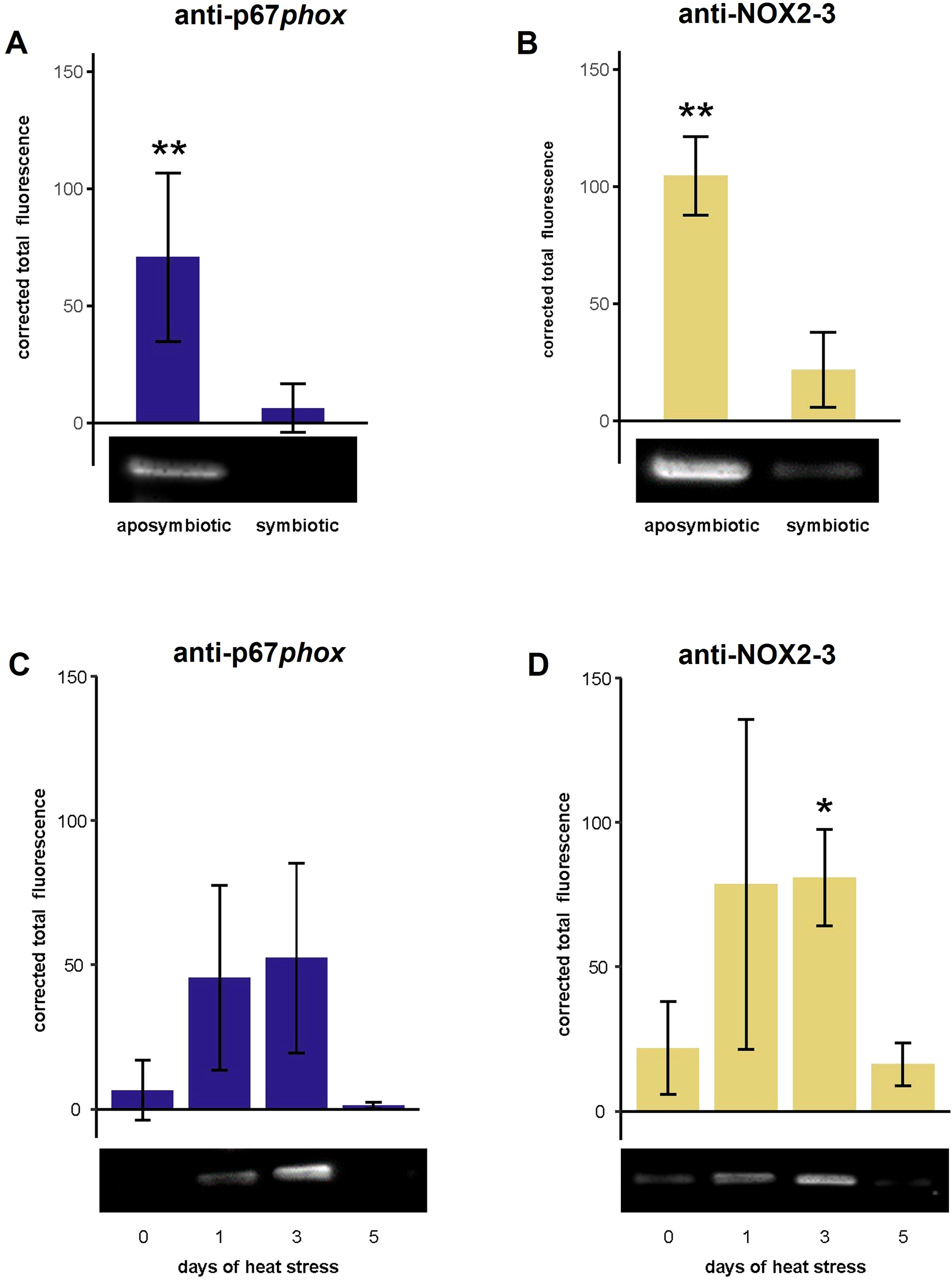
The protein expression of NOX subunits, p67phox and NOX2-3, of aiptasia as a function of symbiotic state and heat treatment. Western blots were performed to quantify protein expression using anti-p67phox and anti-NOX2–3 and then quantifying the corrected total fluorescence. (A, B) Relative amounts of putative NOX protein were determined from aposymbiotic and symbiotic anemones at ambient temperature (N = 3). The values are expression in symbiotic anemones relative (fold change) to aposymbiotic anemones set to 0. (C, D) Relative amounts of NOX protein were determined from symbiotic anemones during heat treatment (days 1, 3, and 5) and at day 0 (N = 3). The values are expression in symbiotic anemones relative (fold change) to symbiotic anemones at day 0. The values are means ± SD. The asterisks indicate statistically significant differences in protein expression between (A, B) aposymbiotic anemones and symbiotic anemones as determined by unpaired T-tests and (C, D) symbiotic anemones during 1, 3, and 5 days of heat treatment compared to day 0 as determined by unpaired T-tests with FDR correction. *p < 0.05, **p < 0.01.
3.3 NOX localized to both gastrodermis and epidermis in symbiotic and aposymbiotic anemones but accumulated around symbionts in symbiotic anemones subjected to elevated temperatures
Immunostaining with anti-p67phox and anti-NOX2–3 revealed diffuse localization in the gastrodermis and epidermis in both symbiotic and aposymbiotic anemone tentacles at ambient conditions on day 0 (Figure 4A). Staining intensities were greater in symbiotic compared to aposymbiotic anemones. In addition, both anti-p67phox and anti-NOX2–3 exhibited faint staining near the algal symbionts in the gastrodermis.
Figure 4
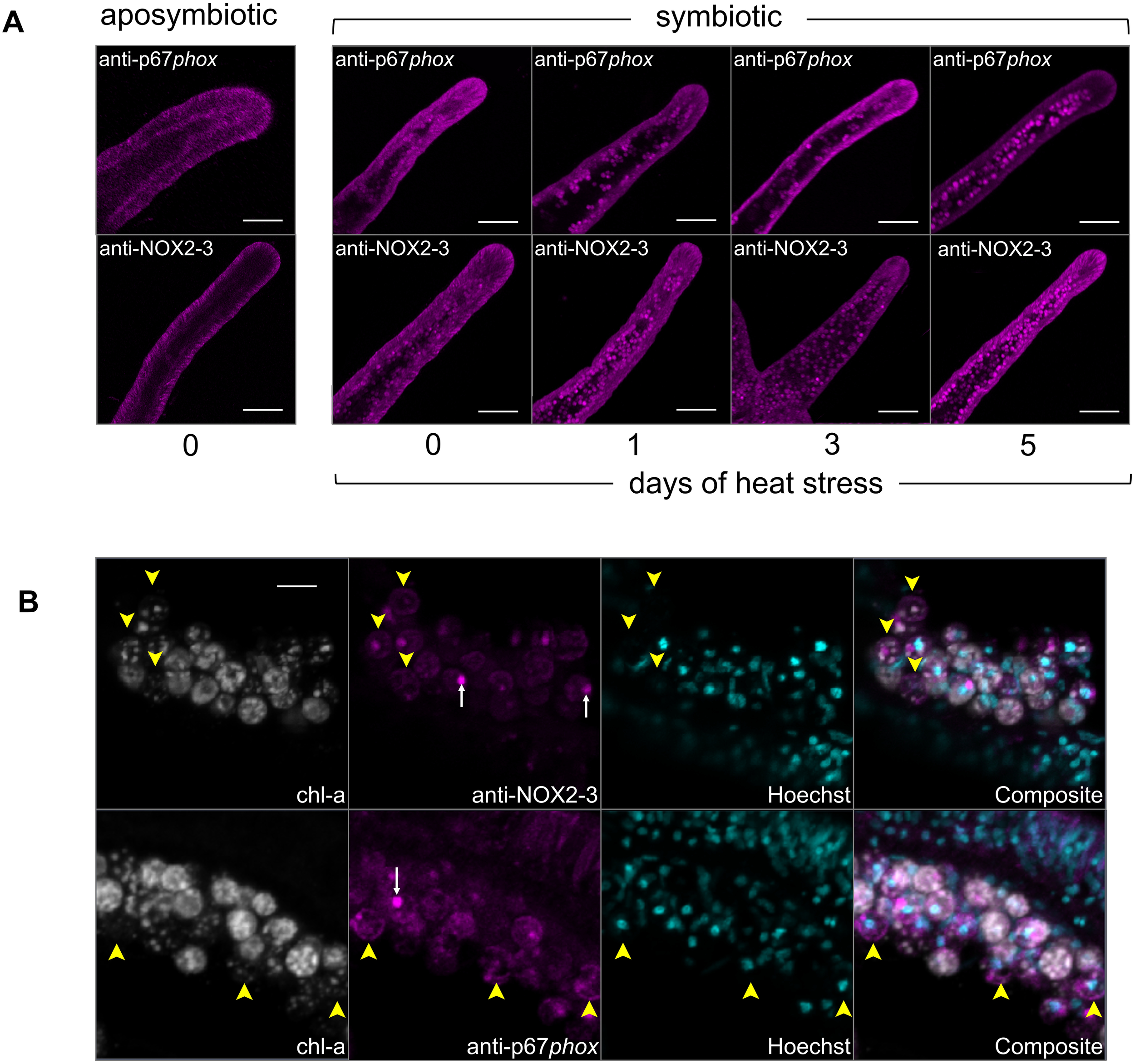
The immunofluorescence localization of NOX subunits, p67phox and NOX2-3, of aiptasia as a function of symbiotic state and heat treatment using confocal microscopy. (A) Images of small aposymbiotic and symbiotic aiptasia anemone tentacles at day 0 and during 1, 3, and 5 days of heat treatment. Magenta: anti-p67phox (top row) and anti-NOX2-3 (bottom row) detected with argon and emission at 491–554 nm. (B) Images of aiptasia anemone tentacle cryosections, separate channels, and then composite, showing anti-p67phox and anti-NOX2–3 localized near degraded algal symbionts during heat treatment (day 3). White: algal chlorophyll detected with excitation at 633 nm laser and emission at 647–722 nm. Magenta: anti-NOX2–3 detected with argon and emission at 491–554 nm. Cyan: Hoechst detected with 633-nm HeNe laser and emission at 647–722 nm. Labels: Yellow arrowheads: anti-p67phox and anti-NOX2–3 localized at degraded algal symbiont. White arrow: algal pyrenoids. Scale bars: (A) 100 µm and (B) 10 µm.
Through the heat treatment time course, the initially diffuse signal of both anti-p67phox and anti-NOX2–3 within gastrodermal cells was gradually replaced by label concentrated near the algal symbionts or possibly symbiosomes (the images do not have sufficient resolution to distinguish between the two) in both antibody treatments (Figure 4A). This was especially pronounced on day 5 of the heat treatment, which coincided with significantly decreased algal densities in host tissues (Supplementary Figure S2). Furthermore, anti-p67phox and anti-NOX2–3 exhibited staining in a ring-like pattern around algae that were pigment-depleted, suggesting that these algae were unhealthy, degrading, or dead (Figure 4B). Both anti-p67phox and anti-NOX2–3 exhibited staining that co-localized with the algal symbionts (Supplementary Figure S3); however, these patterns did not correspond with algal chlorophyll fluorescence, aside from the autofluorescent pyrenoids, an organelle within algae. This indicates that there was no spectral overlap of antibody staining and the algal autofluorescence (Figure 4B).
3.4 Elevated temperature increased NOX activity, measured by superoxide production, in symbiotic anemones
Superoxide production increased on day 1 of heat treatment compared to ambient conditions on day 0, as measured by the CellROX assay (Figure 5). Negative controls reduced the superoxide levels, while positive controls increased the superoxide production, verifying the assay’s sensitivity to ROS (Figure 5). Co-incubation of anemones with the NOX inhibitor (DPI) reduced the superoxide production by approximately 50% for the heat treatment, linking the observed superoxide production to NOX activity (Figure 5).
Figure 5
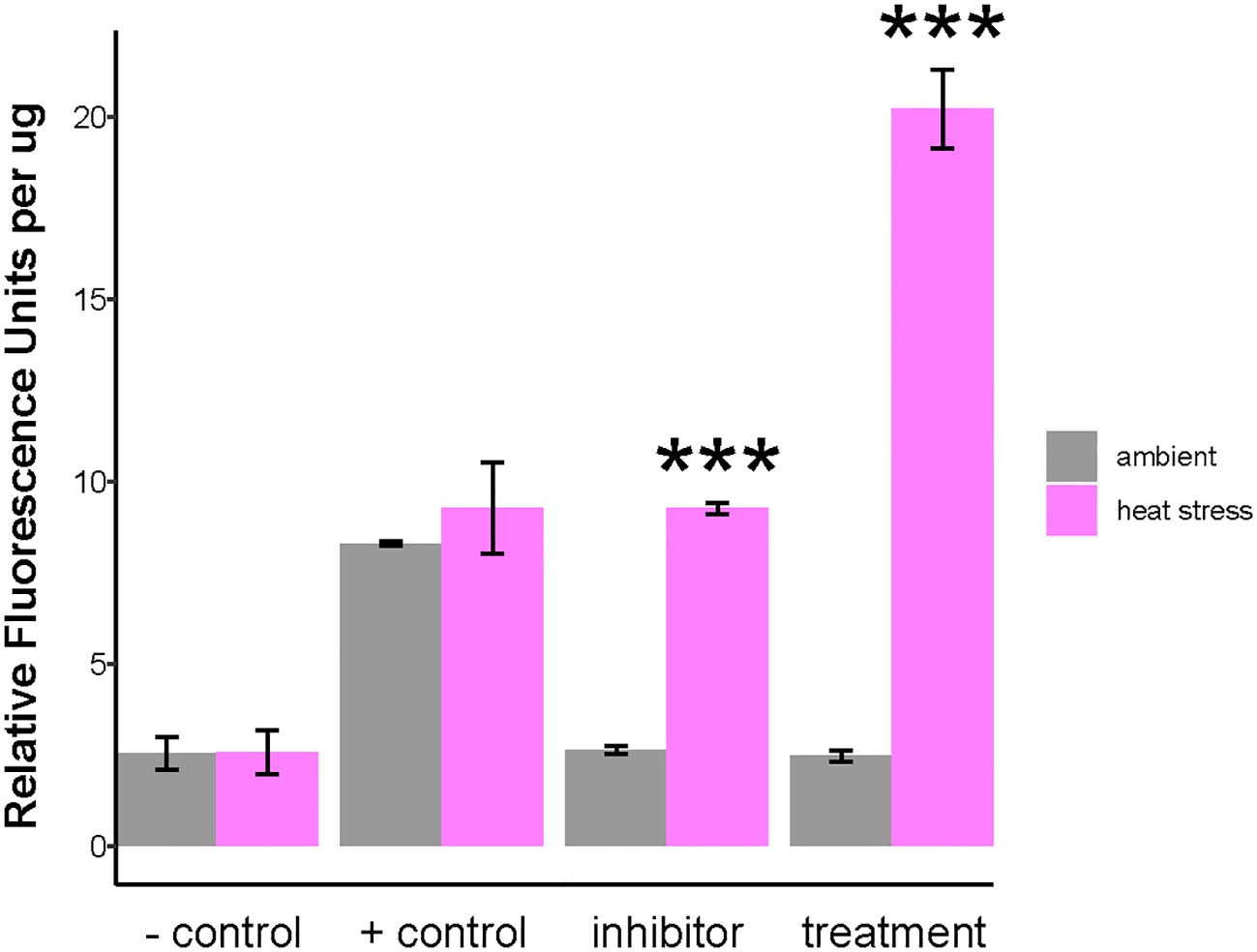
Quantification of NOX activity as measured by superoxide production in symbiotic anemones as a function of heat stress. NOX activity in Aiptasia was investigated using the superoxide marker, CellROX Green. CellROX Green fluorescence was quantified from symbiotic anemone homogenates at ambient temperature (gray) and after 24-h of heat stress (magenta). Negative control: N-acetylcysteine (NAC). Positive control: menadione. Exp: experimental anemones. Inhibitor: diphenyleneiodonium chloride (DPI). Bars are means ± SD. Asterisks indicate statistically significant differences in Relative Fluorescence Units per µg between heat stress (day 1) relative to ambient as determined by unpaired T-tests with FDR correction. *** p < 0.001.
4 Discussion
4.1 NOX is suppressed in the symbiotic state
Our results show decreased expression of the NOX complex in the symbiotic state, suggesting that algal symbionts actively modulate the host innate immune response during symbiosis (Figures 2A, B, 3A). Immunosuppression is a well-documented feature of cnidarian–Symbiodiniaceae symbiosis (Mansfield and Gilmore, 2019; Matthews et al., 2017). Our findings are consistent with the observed twofold change in NOX2 expression in symbiotic anemones compared to aposymbiotic anemones (potentially misannotated as DUOX2) (Lehnert et al., 2014). Furthermore, the absence of immunolabel around the algae and NOX activity in symbiotic host tissues at ambient temperatures suggests an absence of an active NOX complex, an indication that NOX may not play a role in the homeostatic regulation of native algal symbiont populations in a healthy association (Figures 4A, 5). Tissue-specific transcriptomes corroborate our findings, where p67phox and NOX2 are downregulated in the symbiotic gastrodermis (Supplementary Table S1) (Cui et al., 2023).
Our observation of reduced host NOX gene and protein expression in the presence of algae suggests that the algae are actively modulating NOX activation to avoid destruction by the host (Figures 2A, B, 3A). Microbes, whether parasitic or mutualistic, have evolved numerous strategies to persist within their hosts, including strategies to avoid phagosomal destruction—for instance, the bacterial pathogen Salmonella spp. and endoparasites Plasmodium falciparum and Leishmania spp. prevent the assembly of the NOX complex in their hosts using a variety of regulatory mechanisms and thus prevent an oxidative burst that would destroy them in the host phagosome (Amer, 2002; Flannagan et al., 2009; Sacks and Sher, 2002). Precisely how Symbiodiniaceae may evade the oxidative burst in their cnidarian host phagosomes, however, remains a compelling question. The NOX complex activator p67phox typically requires phosphorylation and subsequent translocation to the phagosomal membrane to form the active NOX complex (Belambri et al., 2018). One hypothesis that would explain our observed differential NOX expression and activity is that the algal symbionts modulate upstream signaling pathways that regulate NOX gene and protein expression, ultimately suppressing NOX function. Both the master immune regulators NF-κB and activator protein-1 (AP-1) are NOX regulators in other systems (Manea et al., 2008, 2012) and have been shown to be downregulated in the symbiotic state in aiptasia (Oakley et al., 2016). These two transcription factors are possible targets for immune modulation by the symbionts.
NOX2–1 and NOX2–2 expression did not vary with symbiotic state, suggesting a function unrelated to algal symbiosis (Figure 2A). NOX inhibitors have been shown to reduce the production of extracellular superoxide (O2•−) in a coral and non-symbiotic anemone, indicating that an active NOX complex is present in the epidermis (Hutton and Smith, 1996; Saragosti et al., 2010). Consequently, NOX2–1 and NOX2–2 may be expressed in tissues, such as the epidermis, that frequently encounter microbes. This possibility is consistent with NOX diversity in other metazoans which commonly have multiple isoforms NOX2, each differing in its tissue distribution, biochemistry, and specific function (Nazari et al., 2023). Two homologs of ancestral NOX have been identified in the non-symbiotic anemone Nematostella vectenesis and a coral, Stylophora pistillata, indicating that gene duplications are a common feature for cnidarian NOXs (Massari et al., 2022). A comprehensive phylogenetic analysis of NOX genes in cnidarians is warranted to fully characterize the evolutionary history of the NOX family. This information would improve our understanding of the functions of NOX isoforms, especially those that maintain redox homeostasis and influence cell proliferation and differentiation (Sumimoto, 2008; Vermot et al., 2021).
Our results suggest that NOX is not active in the ongoing maintenance of the healthy symbiosis; however, a role for NOX in inter-partner recognition during the initial onset of symbiosis remains unexplored—for example, is the NOX complex activated when encountering inappropriate non-native or free-living Symbiodiniaceae during initial colonization? Recent work has shown that non-symbiotic microalgae are expelled via vomocytosis, a specialized immune mechanism, rather than being digested internally. This suggests that NOX may function exclusively with phagocytosed native symbionts; however, additional research is needed to test this. Furthermore, does NOX contribute to symbiont shuffling in response to changing environmental conditions (Baker, 2003)? Does it play a role in modulating pathogenic infection to fight off infectious diseases? These questions are ripe for future research.
4.2 Phagosomal maturation and the NOX complex are engaged during bleaching
The observed increase in NOX2–3 gene and protein expression during heat stress, coupled with the increased immunolabeling of NOX in a ring around algal symbionts during heat treatments, suggests the activation of the phagolysosomal machinery during elevated temperature stress (Figures 2B, 3B, C, 4B). Although a faint signal is visible within some algal cells, a future colloidal-gold immuno-TEM study would provide the precise positioning of each NOX component relative to the symbiosome membrane. This is consistent with NOX being a marker of late phagolysosomes in vertebrates (Lee et al., 2020) and suggests that phagosomal maturation is engaged during cnidarian dysbiosis and bleaching. In addition, the presence of immunolabel surrounding apparently degraded symbionts and measurement of increased NOX activity during incubation in elevated temperature indicate the presence of active phagolysosomes. Because transcriptomic responses to heat peak on days 3 and 5, extending NOX activity assays to these later time points would provide a more complete picture of the enzyme’s activity. Symbiont degradation during thermal stress and bleaching has been observed in corals and anemones (Ainsworth and Hoegh-Guldberg, 2008; Brown et al., 1995; Dani et al., 2016; Dunn et al., 2004; Franklin et al., 2004; Strychar et al., 2004). Therefore, our results point to in situ degradation as a bleaching mechanism, facilitated by ROS generated by NOX.
While the mechanisms regulating the expression and activity of the NOX complex are not yet understood in cnidarians, many innate immune pathways known to influence NOX activity in vertebrates are also upregulated in symbiotic cnidarians undergoing dysbiosis, including Toll-like receptors (Van De Water et al., 2018), AP-1 (Jacobovitz et al., 2021; Traylor-Knowles et al., 2017), Rab GTPases (Barshis et al., 2013; Kaniewska et al., 2012), NF-κB (DeSalvo et al., 2010; Mansfield et al., 2017; Traylor-Knowles et al., 2017), myeloid differentiation primary response 88 (Myd88) (Van De Water et al., 2018), and tumor necrosis factor receptor-associated factor (TRAF) (Barshis et al., 2013; DeSalvo et al., 2010; Kaniewska et al., 2012; Seneca and Palumbi, 2015; Van De Water et al., 2018). Our results, together with these extensive findings, strongly suggest that NOX is playing an active role during cnidarian dysbiosis.
4.3 Proposed model for a phagocytic NOX complex in aiptasia and future directions
Currently, the intracellular niche of symbiotic dinoflagellates in the cnidarian host is best described as late phagolysosomes/lysosomes. While a variety of studies have characterized the features of symbiosome, it has remained uncertain how the algal symbionts avoided digestion while residing inside of a lysosomal-like organelle (Barott and Tresguerres, 2015; Dani et al., 2017; Jacobovitz et al., 2021). Based on our results, we propose a model of the healthy symbiotic state where algal symbionts modulate the host immune response by downregulating NOX genes and/or suppressing NOX activity to avoid digestion by the host (Figure 6). Our model further suggests that during symbiosis breakdown and bleaching, a phagocytic NOX complex becomes activated on the symbiosome membrane and participates in the destruction of the symbionts via in situ degradation. Upon activation, p67phox translocates from the cytosol of the symbiotic gastrodermal host cell to the symbiosome, where it associates with the NOX2/p22phox heterodimer, thereby forming an active NOX complex. This, in turn, generates an oxidative burst to degrade algae symbionts (Bedard and Krause, 2007; Hordijk, 2006).
Figure 6
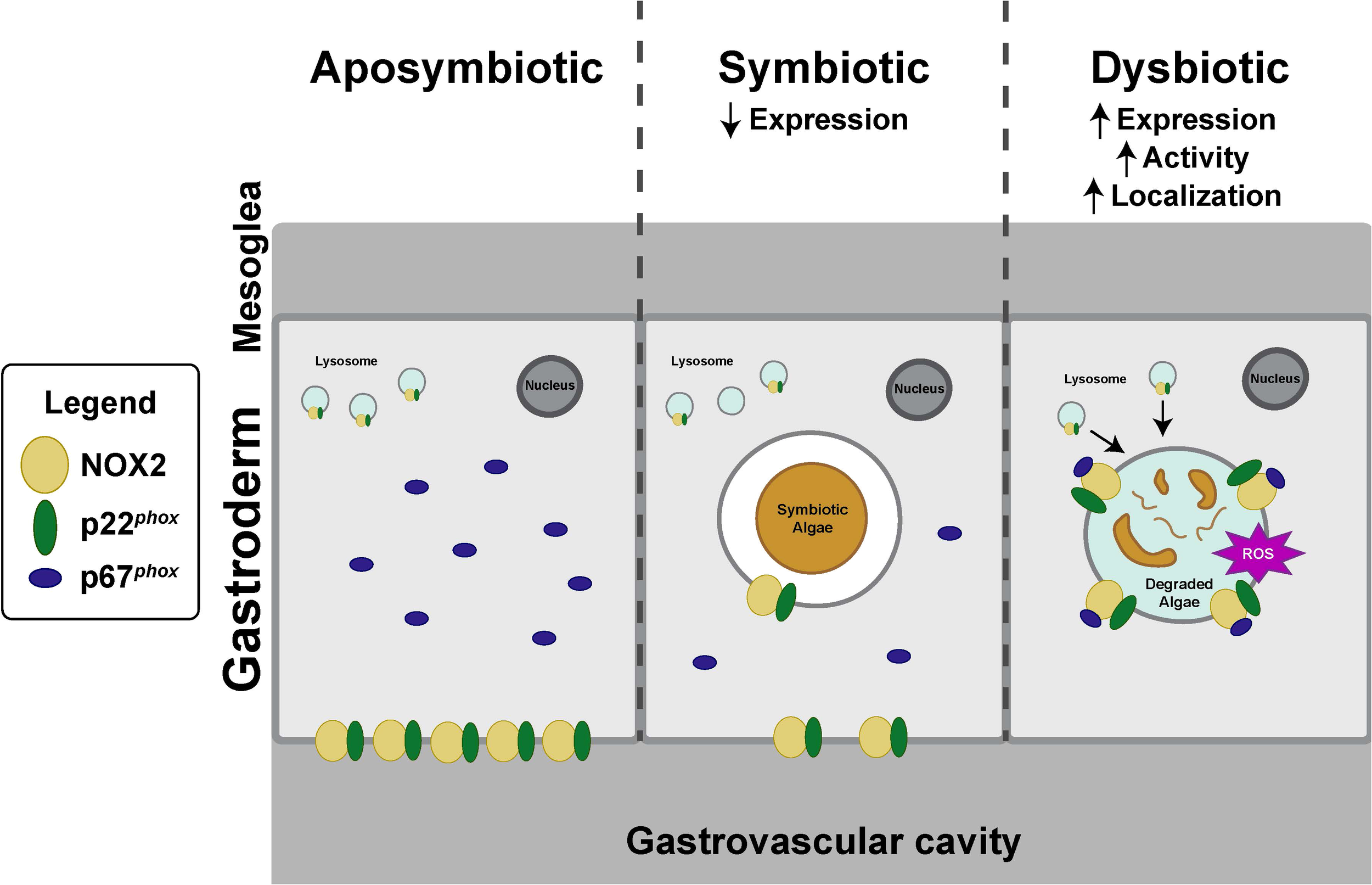
Model for NOX activity on symbiosomes in aposymbiotic, symbiotic, and dysbiotic states. (Aposymbiotic) NOX is expressed and localized in both epidermal and gastrodermal cells; however, the NOX complex remains unassembled and inactive. (Symbiotic) NOX localizes to the epidermis, gastrodermis, and the symbiosome, but NOX expression is decreased relative to the aposymbiotic state. (Dysbiotic) During dysbiosis, NOX activity and expression are increased relative to the symbiotic state. To activate the NOX complex, p67phox migrates from the host cytosol and attaches to membrane-bound heterodimers, NOX2–3 and p22phox, located within the symbiosome of a host gastrodermal cell. The activated NOX ultimately produces ROS to degrade the algal symbiont, resulting in algal loss.
Our expression data indicate that p22phox is responsive to both symbiosis and heat stress, pointing to its significant role in this model. Future work could incorporate p22phox protein assays to verify its inferred role in NOX assembly and to refine the model proposed here. Given the recent methodological advances, such as closing the aiptasia life cycle in the laboratory and CRISP–Cas9 gene editing in coral, loss-of-function approaches should now be feasible, allowing the targeted knockout of p22phox and other NOX components to verify how they function in symbiont persistence and symbiosis breakdown and bleaching (Cleves et al., 2018; Maegele et al., 2023). In addition, comparative work would help determine the level of conservation of the NOX-mediated mechanism across Cnidaria. Intriguingly, two ancestral copies of the NOX1/2/3 lineage have been reported in the non-symbiotic anemone Nematostella vectensis and in the symbiotic coral Stylophora pistallata, showing that gene duplication is a recurring theme in cnidarian NOX evolution (Massari et al., 2022). A comprehensive phylogenetic survey of cnidarian NOX genes—coupled with expression and functional assays in representative anthozoans, scyphozoans, and hydrozoans—will be essential to map the evolutionary history of the family and to clarify the distinct physiological roles of each NOX isoform.
Our study provides insight into the role of NOX in early-diverging metazoans, particularly in the context of cnidarian–Symbiodiniaceae symbiosis, and its implications for comparative immunology. We show that NOX homologs are suppressed in the symbiotic state, but these form an active phagocytic NOX complex during heat-induced symbiosis breakdown and bleaching. This activation of the host phagosomal maturation points to in situ degradation as a bleaching mechanism. Further research into NOX pathways, their role in symbiosis, and their impact on bleaching will be crucial to understand coral health and resilience in the face of elevated seawater temperatures.
Supplementary methods
Selection of polyclonal purified antibodies for NOX subunits
Anti-p67phox
The staining patterns among anti-p67phox (a, b, c) were highly variable. Anti-p67phox b demonstrated remarkable specificity, binding exclusively to a single band at approximately 30 kDa (Supplementary Figure S1). Conversely, anti-p67phox a and c displayed reduced specificity and more background staining (Supplementary Figure S1). Anti-p67phox a and c recognized shared target at 67 and 42 kDa (Supplementary Figure S1). As a control, peptide antigens were incubated with their corresponding antibody before the Western blot, which led to the absence of any bands in this control, supporting the specificity of the anti-p67phox a, b, and c (data not shown). For anti-p67phox a, b, and c, no target bands were labeled in a single combined pre-immune serum which was taken from two rabbits in equal proportions, ruling out any cross-reactivity and non-specific binding of the antibodies (Supplementary Figure S1; data not shown for anti-p67phox a and c). Due to high specificity, anti-p67phox b was selected to examine putative p67phox expression and localization during heat stress.
The target mass of anti-p67phox b is lower than the predicted value, although down-shifts are well documented for this protein. The vertebrate p67phox in neutrophils undergoes post-translational trimming, and the N-terminus leads to bands below the predicted size (Han et al., 1998). In addition, p67phox is unusually proline- and acid-rich, which reduces SDS binding and can accelerate migration by up to 15 kDa (Guan et al., 2015). Commercial antibodies (e.g., Abcam ab31098, MyBioSource MBS7104129) consistently report vertebrate p67phox antibody targets below their predicted size due to natural N-terminal truncation or alternative starts.
Anti-NOX2-3
The staining patterns differed among each anti-NOX2-3 (x, y, z). Anti-NOX2–3 x and y showed a common band stained at 40 kDa. Anti-NOX2–3 x selectively bound to a single band at 40 kDa and showed a heavy smear, indicative of substantial glycosylation (Supplementary Figure S1). Anti-NOX2–3 y displayed binding to three targets at 35, 40, and 50 kDa, and anti-NOX2–3 z revealed a faint band to a single target with a molecular weight greater than 245 kDa (Supplementary Figure S1). As a control, peptide antigens were incubated with their corresponding antibody before the Western blot, which led to the absence of any bands in this control, supporting the specificity of anti-NOX2 x, y, and z (data not shown). For anti-NOX2 x, y, and z, no target bands were labeled in a single combined pre-immune serum which was taken from two rabbits in equal proportions, ruling out any cross-reactivity and non-specific binding of the antibodies (Supplementary Figure S1; data not shown for anti-NOX y and z).
Anti-NOX2–3 x was selected to examine putative NOX2–3 expression and localization due to its high specificity and characteristic glycosylation (Kleinberg et al., 1989; Paclet et al., 2004). While the anti-NOX2–3 x target band is lower than predicted, integral membrane proteins typically migrate 10%–30% faster on SDS-PAGE due to incomplete SDS binding and residual secondary structures (Paclet et al., 2004; Rath et al., 2009).
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
VS: Writing – original draft, Software, Writing – review & editing, Funding acquisition, Investigation, Resources, Project administration, Formal analysis, Conceptualization, Visualization, Validation, Methodology, Data curation. NT: Writing – review & editing, Validation, Formal analysis, Conceptualization, Resources. SM: Validation, Resources, Conceptualization, Writing – review & editing. MR: Formal analysis, Writing – review & editing, Resources, Validation, Methodology, Conceptualization, Investigation, Data curation, Software. CV: Investigation, Resources, Validation, Writing – review & editing, Conceptualization, Formal analysis. LH: Validation, Investigation, Conceptualization, Writing – review & editing, Resources. AP: Methodology, Conceptualization, Supervision, Writing – review & editing. VW: Methodology, Supervision, Funding acquisition, Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Science Foundation (IOS: 1645164 and 2124119 to VW), the American Microscopical Society (AMS Student Research Fellowship to VS), and OSU Integrative Biology Department (Zoology Department Research Fund (ZoRF) to VS).
Acknowledgments
We are grateful to members (Jason Presnell, Trevor Tivey, Jack Koch, Sam Bedgood, Holland Elder, Erick White, Jun Cai, and Olivia Burleigh), affiliated lab members (Nate Kirk, Keith Garrison, Lucia Pita, Keyla Plichon, Rowan McLachlan, and Yasmin Gabay), undergraduate researchers (Tara Wirsching, Fernanda Rodriguez, Bryan Acevedo-Adames, Meghan Bray, Bismah Mirza, and Danielle Timm), and anemone husbandry staff of the Weis lab for feedback during the execution of our experiment. We are also grateful to Oregon State University (OSU) Confocal Microscopy Facility of the Center for Quantitative Life Sciences (CQLS) (NSF DBI: 1337774 to OSU CQLS), Magnusson Lab in the OSU Department of Biomedical Sciences, and Barretto, Burke, and Phillips Labs in the OSU Department of Integrative Biology for support with imaging and molecular biological experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1596098/full#supplementary-material
Supplementary Figure 1Western blot analysis of anti-p67phox and anti-NOX2–3 with symbiotic aiptasia protein. Three polyclonal anti-p67phox (a, b, c) and three anti-NOX2-3 (x, y, z) primary antibodies were each generated against a unique peptide. All six antibodies were tested with symbiotic anemone protein (N = 1) at 1:2,000 dilution and 1:2,000 dilution with their corresponding peptide (peptide block) to confirm antibody specificity. Corresponding pre-immune rabbit serum (1:1,000 dilution) is shown for anti-p67phox b and anti-NOX2–3 x. The ladder is labeled in kilodalton.
Supplementary Figure 2Algal cell densities from symbiotic anemones during heat treatment. Parallel heat stress experiments were performed on symbiotic anemones, corresponding with gene and protein expression experiment. Day 0 represents ambient temperature (25°C) and 1, 3, and 5 represent subsequent days of heat treatment (32°C). The dots represent the number of algal cells within an individual symbiotic anemone. The line represents the median value for each time. The asterisks indicate statistically significant differences in algal abundance between heat treatment (days 1, 3, and 5) relative to day 0 as determined by unpaired T-tests with FDR correction. *p < 0.05.
Supplementary Figure 3Controls for immunofluorescence localization of NOX subunits, p67phox and NOX2-3, of aiptasia during heat treatment and with respect to symbiotic state. Confocal images (separate channels) of small (12+ tentacles) aposymbiotic and symbiotic aiptasia tentacles at day 0 and during heat treatment (days 1, 3, and 5). Magenta: (A) anti-p67phox, (B) anti-NOX2-3, and (C) control with no immunostaining (day 0) detected with argon and emission at 491–554 nm. White: B. minutum chlorophyll detected with excitation at 633-nm laser and emission at 647–722 nm. Cyan: Hoechst detected with 633-nm HeNe laser and emission at 647–722 nm. Composite: Merging of all fluorescent channels. T-PMT: bright field-like image. Scale bar: (A, B) 100 µm.
Supplementary Table 1Published reports of NADPH oxidase (NOX) complex genes in corals and anemones.
Supplementary Table 2Primer sequences and their primer efficiencies used in qPCR experiments of NOX subunits.
Supplementary Table 3Peptide sequences used to generate anti-p67phox and anti-NOX2.
References
1
Abcam . (n.d.). Anti-NOXA2/p67phox antibody (ab31098). Retrieved June 12, 2025, from Abcam product datasheet.
2
Ainsworth T. D. Hoegh-Guldberg O. (2008). Cellular processes of bleaching in the Mediterranean coral Oculina patagonica. Coral Reefs27(3), 593–597. doi: 10.1007/s00338-008-0355-x
3
Amer A. (2002). A phagosome of one’s own: A microbial guide to life in the macrophage. Curr. Opin. Microbiol.5, 56–61. doi: 10.1016/S1369-5274(02)00286-2
4
Baker A. C. (2003). Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecology Evolution Systematics34(1), 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
5
Barott K. Tresguerres M. (2015). Immunolocalization of proteins in corals: The V-type H+-ATPase proton pump. BIO-PROTOCOL5(17), e1573. doi: 10.21769/BioProtoc.1573
6
Barshis D. J. Ladner J. T. Oliver T. A. Seneca F. O. Traylor-Knowles N. Palumbi S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A110, 1387–1392. doi: 10.1073/pnas.1210224110
7
Baumgarten S. Simakov O. Esherick L. Y. Liew Y. J. Lehnert E. M. Michell C. T. et al . (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci.112, 11893–11898. doi: 10.1073/pnas.1513318112
8
Bedard K. Krause K. H. (2007). The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev.87, 245–313. doi: 10.1152/physrev.00044.2005
9
Belambri S. A. Rolas L. Raad H. Hurtado-Nedelec M. Dang P. M. El-Benna J. (2018). NADPH oxidase activation in neutrophils: Role of the phosphorylation of its subunits. Eur. J. Clin. Invest.48, e12951. doi: 10.1111/eci.12951
10
Brown B. E. Le Tissier M. D. A. Bythell J. C. (1995). Mechanisms of bleaching deduced from histological studies of reef corals sampled during a natural bleaching event. Mar. Biol.122, 655–663. doi: 10.1007/BF00350687
11
Cleves P. A. Krediet C. J. Lehnert E. M. Onishi M. Pringle J. R. (2020). Insights into coral bleaching under heat stress from analysis of gene expression in a sea anemone model system. Proc. Natl. Acad. Sci.117. doi: 10.1073/pnas.2015737117
12
Cleves P. A. Strader M. E. Bay L. K. Pringle J. R. Matz M. V. (2018). CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc. Natl. Acad. Sci.115, 5235–5240. doi: 10.1073/pnas.1722151115
13
Cui G. Konciute M. K. Ling L. Esau L. Raina J.-B. Han B. et al . (2023). Molecular insights into the Darwin paradox of coral reefs from the sea anemone Aiptasia. Sci. Adv.9, eadf7108. doi: 10.1126/sciadv.adf7108
14
Cziesielski M. J. Liew Y. J. Cui G. Schmidt-Roach S. Campana S. Marondedze C. et al . (2018). Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proc. R. Soc. B: Biol. Sci.285. doi: 10.1098/rspb.2017.2654
15
Dani V. Priouzeau F. Mertz M. Mondin M. Pagnotta S. Lacas-Gervais S. et al . (2017). Expression patterns of sterol transporters NPC1 and NPC2 in the cnidarian–dinoflagellate symbiosis. Cell. Microbiol.19. doi: 10.1111/cmi.12753
16
Dani V. Priouzeau F. Pagnotta S. Carette D. Laugier J.-P. Sabourault C. (2016). Thermal and menthol stress induce different cellular events during sea anemone bleaching. Symbiosis69, 175–192. doi: 10.1007/s13199-016-0406-y
17
DeSalvo M. Sunagawa S. Voolstra C. Medina M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser.402, 97–113. doi: 10.3354/meps08372
18
Detournay O. Schnitzler C. E. Poole A. Z. Weis V. M. (2012). Regulation of cnidarian–dinoflagellate mutualisms: Evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev. Comp. Immunol.38. doi: 10.1016/J.DCI.2012.08.008
19
Detournay O. Weis V. M. (2011). Role of the sphingosine rheostat in the regulation of cnidarian-dinoflagellate symbioses. Biol. Bull.221, 261–269. doi: 10.1086/BBLv221n3p261
20
Drummond A. J. Ashton B. Buxton S. Cheung M. Cooper A. Duran C. et al . (2010). Geneious v.10.2.6. (Auckland, New Zealand: Biomatters Ltd).
21
Dunn S. R. Thomason J. C. Le Tissier M. D. A. Bythell J. C. (2004). Heat stress induces different forms of cell death in sea anemones and their endosymbiotic algae depending on temperature and duration. Cell Death Differentiation11, 1213–1222. doi: 10.1038/sj.cdd.4401484
22
Dunn S. R. Weis V. M. (2009). Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ. Microbiol.11 (1), 268–276. doi: 10.1111/j.1462-2920.2008.01774.x
23
Eddy T. D. Lam V. W. Y. Reygondeau G. Cisneros-Montemayor A. M. Greer K. Palomares M. L. D. et al . (2021). Global decline in capacity of coral reefs to provide ecosystem services. One Earth4, 1278–1285. doi: 10.1016/j.oneear.2021.08.016
24
Fisher R. O’Leary R. A. Low-Choy S. Mengersen K. Knowlton N. Brainard R. E. et al . (2015). Species richness on coral reefs and the pursuit of convergent global estimates. Curr. Biol.25, 500–505. doi: 10.1016/j.cub.2014.12.022
25
Flannagan R. S. Cosío G. Grinstein S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol.7, 355–366. doi: 10.1038/nrmicro2128
26
Franklin D. J. Hoegh-Guldberg O. Jones R. J. B. J. (2004). Cell death and degeneration in the symbiotic dinoflagellates of the coral Stylophora pistillata during bleaching. Mar. Ecol. Prog. Ser.272, 117–130. doi: 10.3354/meps272117
27
Guan Y. Zhu Q. Huang D. Zhao S. Lo L. J. Peng J. (2015). An equation to estimate the difference between theoretically predicted and SDS-PAGE–displayed molecular weights for an acidic peptide. Scientific Reports.5, 13370. doi: 10.1038/srep13370
28
Han C. H. Freeman J. L. R. Lee T. Motalebi S. A. Lambeth J. D. (1998). Regulation of the Neutrophil Respiratory Burst Oxidase. J. Biol. Chem.273 (27), 16663–16668. doi: 10.1074/jbc.273.27.16663
29
Hordijk P. L. (2006). Regulation of NADPH oxidases: The role of Rac proteins. Circ. Res.98, 453–462. doi: 10.1161/01.RES.0000204727.46710.5e
30
Hughes T. P. Anderson K. D. Connolly S. R. Heron S. F. Kerry J. T. Lough J. M. et al . (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science359. doi: 10.1126/science.aan8048
31
Hutton D. M. C. Smith V. J. (1996). Antibacterial properties of isolated amoebocytes from the sea anemone Actinia equina. Biol. Bull.191, 441–451. doi: 10.2307/1543017
32
Jacobovitz M. R. Hambleton E. A. Guse A. (2023). Unlocking the complex cell biology of coral–dinoflagellate symbiosis: A model systems approach. Annu. Rev. Genet.57, 411–434. doi: 10.1146/annurev-genet-072320-125436
33
Jacobovitz M. R. Rupp S. Voss P. A. Maegele I. Gornik S. G. Guse A. (2021). Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. Nat. Microbiol.6, 769–782. doi: 10.1038/s41564-021-00897-w
34
Kaniewska P. Campbell P. R. Kline D. I. Rodriguez-Lanetty M. Miller D. J. Dove S. et al . (2012). Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS One7. doi: 10.1371/journal.pone.0034659
35
Kawahara T. Lambeth J. D. (2007). Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evolutionary Biol.7, 1–29. doi: 10.1186/1471-2148-7-178
36
Kirk N. L. Weis V. M. (2016). “Animal–Symbiodinium symbioses: Foundations of coral reef ecosystems,” in The Mechanistic Benefits of Microbial Symbionts. Ed. HurstC. J. (Springer International Publishing), 269–294. doi: 10.1007/978-3-319-28068-4_10
37
Kitchen S. A. Weis V. M. (2017). The sphingosine rheostat is involved in the cnidarian heat stress response but not necessarily in bleaching. J. Exp. Biol.220, 269–294. doi: 10.1242/jeb.153858
38
Kleinberg M. E. Rotrosen D. Malech H. L. (1989). Asparagine-linked glycosylation of cytochrome b558 large subunit varies in different human phagocytic cells. J. Immunol.143, 4152–4157. doi: 10.4049/jimmunol.143.12.4152
39
LaJeunesse T. C. Parkinson J. E. Gabrielson P. W. Jeong H. J. Reimer J. D. Voolstra C. R. et al . (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol.28. doi: 10.1016/j.cub.2018.07.008
40
Lambeth J. D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol.4(3), 181–189. doi: 10.1038/nri1312
41
Lee H.-J. Woo Y. Hahn T.-W. Jung Y. M. Jung Y.-J. (2020). Formation and maturation of the phagosome: A key mechanism in innate immunity against intracellular bacterial infection. Microorganisms8(9), 1298. doi: 10.3390/microorganisms8091298
42
Lehnert E. M. Mouchka M. E. Burriesci M. S. Gallo N. D. Schwarz J. A. Pringle J. R. (2014). Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. Genes|Genomes|Genetics4. doi: 10.1534/g3.113.009084
43
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods25(4), 402–408. doi: 10.1006/meth.2001.1262
44
Maegele I. Rupp S. Özbek S. Guse A. Hambleton E. A. Holstein T. W. (2023). A predatory gastrula leads to symbiosis-independent settlement in Aiptasia. Proc. Natl. Acad. Sci.120. doi: 10.1073/pnas.2311872120
45
Manea A. Manea S. A. Florea I. C. Luca C. M. Raicu M. (2012). Positive regulation of NADPH oxidase 5 by proinflammatory-related mechanisms in human aortic smooth muscle cells. Free Radical Biol. Med.52, 1497–1507. doi: 10.1016/j.freeradbiomed.2012.02.018
46
Manea A. Manea S. A. Gafencu A. V. Raicu M. Simionescu M. (2008). AP-1–dependent transcriptional regulation of NADPH oxidase in human aortic smooth muscle cells. Arteriosclerosis Thrombosis Vasc. Biol.28, 878–885. doi: 10.1161/ATVBAHA.108.163592
47
Mansfield K. M. Carter N. M. Nguyen L. Cleves P. A. Alshanbayeva A. Williams L. M. et al . (2017). Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci. Rep.7, 16025. doi: 10.1038/s41598-017-16168-w
48
Mansfield K. M. Gilmore T. D. (2019). Innate immunity and cnidarian-Symbiodiniaceae mutualism. Dev. Comp. Immunol.90, 199–209. doi: 10.1016/j.dci.2018.09.020
49
Maruyama S. Mandelare-Ruiz P. E. McCauley M. Peng W. Cho B. G. Wang J. et al . (2022). Heat stress of algal partner hinders colonization success and alters the algal cell surface glycome in a cnidarian-algal symbiosis. Microbiol. Spectr.10, e0156722. doi: 10.1128/spectrum.01567-22
50
Massari M. Nicoll C. R. Marchese S. Mattevi A. Mascotti M. L. (2022). Evolutionary and structural analyses of the NADPH oxidase family in eukaryotes reveal an initial calcium dependency. Redox Biol.56, 102436. doi: 10.1016/j.redox.2022.102436
51
Matthews J. L. Crowder C. M. Oakley C. A. Lutz A. Roessner U. Meyer E. et al . (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc. Natl. Acad. Sci.114, 13194–13199. doi: 10.1073/pnas.1710733114
52
Matthews J. L. Sproles A. E. Oakley C. A. Grossman A. R. Weis V. M. Davy S. K. (2016). Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J. Exp. Biol.219. doi: 10.1242/jeb.128934
53
Meyer E. Weis V. M. (2012). Study of cnidarian-algal symbiosis in the “omics” age. Biol. Bull.223, 44–65. doi: 10.1086/BBLv223n1p44
54
Mohamed A. R. Cumbo V. Harii S. Shinzato C. Chan C. X. Ragan M. A. et al . (2016). The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: The symbiosome as an arrested early phagosome. Mol. Ecol.25, 3127–3141. doi: 10.1111/mec.13659
55
Muscatine L. McCloskey L. R. Marian R. E. (1981). Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnology Oceanography26(4), 601–611. doi: 10.4319/lo.1981.26.4.0601
56
Nazari B. Jaquet V. Krause K.-H. (2023). NOX family NADPH oxidases in mammals: Evolutionary conservation and isoform-defining sequences. Redox Biol.66, 102851. doi: 10.1016/j.redox.2023.102851
57
Neubauer E. F. Poole A. Z. Detournay O. Weis V. M. Davy S. K. (2016). The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ4, e2692. doi: 10.7717/peerj.2692
58
Neubauer E.-F. Poole A. Z. Neubauer P. Detournay O. Tan K. Davy S. K. et al . (2017). A diverse host thrombospondin-type-1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian-dinoflagellate symbiosis. eLife6. doi: 10.7554/eLife.24494
59
Oakley C. A. Ameismeier M. F. Peng L. Weis V. M. Grossman A. R. Davy S. K. (2016). Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia: Symbiosis and the Aiptasia proteome. Cell. Microbiol.18. doi: 10.1111/cmi.12564
60
Paclet M.-H. Henderson L. M. Campion Y. Morel F. Dagher M.-C. (2004). Localization of Nox2 N-terminus using polyclonal antipeptide antibodies. Biochem. J.382(Pt 3), 981–986. doi: 10.1042/BJ20040954
61
Parkinson J. E. Tivey T. R. Mandelare P. E. Adpressa D. A. Loesgen S. Weis V. M. (2018). Subtle differences in symbiont cell surface glycan profiles do not explain species-specific colonization rates in a model cnidarian-agal symbiosis. Front. Microbiol.9. doi: 10.3389/fmicb.2018.00842
62
Poole A. Z. Kitchen S. A. Weis V. M. (2016). The role of complement in cnidarian-dinoflagellate symbiosis and immune challenge in the sea anemone Aiptasia pallida. Front. Microbiol.7. doi: 10.3389/fmicb.2016.00519
63
Poole A. Z. Weis V. M. (2014). TIR-domain-containing protein repertoire of nine anthozoan species reveals coral–specific expansions and uncharacterized proteins. Dev. Comp. Immunol.46. doi: 10.1016/j.dci.2014.06.002
64
Ranjan D. Chandravanshi S. Verma P. Singh M. B. Verma D. K. Maurya P. et al . (2023). Effects of coral reef destruction on humans and the environment. Int. J. Environ. Climate Change13. doi: 10.9734/ijecc/2023/v13i102708
65
Rath A. Glibowicka M. Nadeau V. G. Chen G. Deber C. M. (2009). Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A106, 1760–1765. doi: 10.1073/pnas.0813167106
66
R Core Team . (2015). RStudio: Integrated Development for R (RStudio, Inc). Available online at: http://www.rstudio.com/ (Accessed February 18, 2023).
67
Richier S. Rodriguez-Lanetty M. Schnitzler C. E. Weis V. M. (2008). Response of the symbiotic cnidarian Anthopleura elegantissima transcriptome to temperature and UV increase. Comp. Biochem. Physiol. - Part D: Genomics Proteomics3. doi: 10.1016/j.cbd.2008.08.001
68
Sacks D. Sher A. (2002). Evasion of innate immunity by parasitic protozoa. Nat. Immunol.3, 1041–1047. doi: 10.1038/ni1102-1041
69
Saragosti E. Tchernov D. Katsir A. Shaked Y. (2010). Extracellular production and degradation of superoxide in the coral Stylophora pistillata and cultured Symbiodinium. PLoS One5(9), e12508. doi: 10.1371/journal.pone.0012508
70
Schindelin J. Arganda-Carreras I. Frise E. Kaynig V. Longair M. Pietzsch T. et al . (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods9, 676–682. doi: 10.1038/nmeth.2019
71
Segal A. W. (2005). How neutrophils kill microbes. Annu. Rev. Immunol.23(1), 197–223. doi: 10.1146/annurev.immunol.23.021704.115653
72
Seneca F. O. Palumbi S. R. (2015). The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol.24, 1467–1484. doi: 10.1111/mec.13125
73
Strychar K. B. Coates M. Sammarco P. W. (2004). Loss of Symbiodinium from bleached Australian scleractinian corals (Acropora hyacinthus, Favites complanata and Porites solida). Mar. Freshw. Res.55, 135–144. doi: 10.1071/MF03080
74
Sumimoto H. (2008). Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J.275, 3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x
75
Sunagawa S. Wilson E. C. Thaler M. Smith M. L. Caruso C. Pringle J. R. et al . (2009). Generation and analysis of transcriptomic resources for a model system on the rise: The sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics10. doi: 10.1186/1471-2164-10-258
76
Tivey T. R. Parkinson J. E. Mandelare P. E. Adpressa D. A. Peng W. Dong X. et al . (2020). N-linked surface glycan biosynthesis, composition, inhibition, and function in cnidarian-dinoflagellate symbiosis. Microbial Ecol. doi: 10.1007/s00248-020-01487-9
77
Traylor-Knowles N. Rose N. H. Palumbi S. R. (2017). The cell specificity of gene expression in the response to heat stress in corals. J. Exp. Biol.220(10), 1837–1845. doi: 10.1242/jeb.155275
78
Van De Water J. A. J. M. Chaib De Mares M. Dixon G. B. Raina J. Willis B. L. Bourne D. G. et al . (2018). Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef-building coral. Mol. Ecol.27, 1065–1080. doi: 10.1111/mec.14489
79
Vermot A. Petit-Härtlein I. Smith S. M. E. Fieschi F. (2021). NADPH oxidases (NOX): An overview from discovery, molecular mechanisms to physiology and pathology. Antioxidants10. doi: 10.3390/antiox10060890
80
Wakefield T. S. Kempf S. C. (2001). Development of host- and symbiont-specific monoclonal antibodies and confirmation of the origin of the symbiosome membrane in a cnidarian–dinoflagellate symbiosis. Biol. Bull.200. doi: 10.2307/1543306
81
Weis V. M. (2019). Cell biology of coral symbiosis: Foundational study can inform solutions to the coral reef crisis. Integr. Comp. Biol.59(4), 845–855. doi: 10.1093/icb/icz067
82
Whitehead L. F. Douglas A. E. (2003). Metabolite comparisons and the identity of nutrients translocated from symbiotic algae to an animal host. J. Exp. Biol.206(18), 3149–3157. doi: 10.1242/jeb.00539
83
Williams L. M. Fuess L. E. Brennan J. J. Mansfield K. M. Salas-Rodriguez E. Welsh J. et al . (2018). A conserved Toll-like receptor-to-NF-κB signaling pathway in the endangered coral Orbicella faveolata. Dev. Comp. Immunol.79, 128–136. doi: 10.1016/j.dci.2017.10.016
84
Wolfowicz I. Baumgarten S. Voss P. A. Hambleton E. A. Voolstra C. R. Hatta M. et al . (2016). Aiptasia sp. Larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci. Rep.6. doi: 10.1038/srep32366
85
Xiang T. Hambleton E. A. DeNofrio J. C. Pringle J. R. Grossman A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J. Phycology49. doi: 10.1111/jpy.12055
86
Yellowlees D. Rees T. A. V. Leggat W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ.31(5), 679–694. doi: 10.1111/j.1365-3040.2008.01802.x
Summary
Keywords
cnidarian-dinoflagellate symbiosis, immunity, host-microbe interaction, dysbiosis, coral
Citation
Sawiccy V, Tjandra NW, Maruyama S, Ruggeri M, Vo C, Harmon LA, Poole AZ and Weis VM (2025) Regulatory role of NADPH oxidases in symbiosis and dysbiosis in the sea anemone Aiptasia. Front. Mar. Sci. 12:1596098. doi: 10.3389/fmars.2025.1596098
Received
19 March 2025
Accepted
29 July 2025
Published
01 September 2025
Volume
12 - 2025
Edited by
Sebastian Fraune, Heinrich Heine University of Düsseldorf, Germany
Reviewed by
Angel Llamas, University of Cordoba, Spain
Joachim Surm, Ludwig Maximilian University of Munich, Germany
Updates
Copyright
© 2025 Sawiccy, Tjandra, Maruyama, Ruggeri, Vo, Harmon, Poole and Weis.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valeri Sawiccy, vsawiccy@uoregon.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.