Abstract
The assessment of marine microbial biodiversity is crucial for determining either the environmental status (ecological or biodiversity research) or as a first step in the biodiscovery process (biotechnological research). Regardless of the research purpose, the first step in a biodiversity assessment is sampling, which can range from ad hoc sampling expeditions to long-term monitoring campaigns. In spite of its demands for funds, infrastructure, expertise, equipment, and personnel, sampling is often not adequately planned. This results in increased likelihood for biased sampling, which can lead to misinterpretation of results, omission of valuable specimens and an unrepresentative collection of stored samples, all particularly important for the assessment of microbial biodiversity. For these reasons, we are proposing a conceptual framework to assist in better preparation of sampling, consisting of pre-sampling, sampling and post sampling steps. The manuscript guides the reader through all the necessary steps, regardless of the sampling habitat (from water column to sediment), the sampling techniques and the preservation and storage approaches including culture and biorepositories. Such a harmonized approach can be of benefit for (i) researchers in the field of ecology/biotechnology, (ii) industrial companies requiring information on the providers and availability of data, and (iii) governance structures and funders, in the light of open science principles.
1 Introduction
Marine biodiversity has continuously sparked the interest of the scientific community and the society that depends on it since the 17th and 18th centuries (Costello et al., 2010). The marine environment is a treasure trove of undiscovered life. Some estimates that out of 1 to 2 million marine species, between 75% and 90%, are still unknown (Mora et al., 2011; Rogers et al., 2023). Indeed, despite the significant advances in understanding the marine microbial diversity in the last 20 years, current estimates of biodiversity differ, sometimes by orders of magnitude, highlighting the immense potential for new scientific endeavours and the continued need for assessing marine biodiversity (Logares, 2024). The solutions for better understanding and discovering ocean life globally lie in the development of new methods, the application of consistent standards, the establishment of new working practices, and the expansion of research coverage (Rogers et al., 2023).
Field observations play a central role in assessing marine biodiversity and allow researchers to identify patterns in biodiversity that are influenced by environmental pressures. This can help address many global societal challenges (United Nations Department of Economic and Social Affairs, 2023), such as mitigating the potential impacts of climate change and pollution or contributing to an innovative society through marine (blue) biotechnology by suggesting novel products that stem from the marine environment and organisms therein.
There are major differences in sampling strategies depending on their objective, namely (i) monitoring biodiversity to determine their relationship with the environment or (ii) bioprospecting organisms for the discovery of novel compounds with biotechnological potential. When sampling is conducted to monitor biodiversity and assess the ecological status of an environment, it is usually carried out routinely at multiple times. This allows for a comparison of a before-and-after status and may be used to track changes in biodiversity patterns over time (Office of Water, 1997; Barbour, 1999; Patrício et al., 2016). Countries with transboundary marine regions typically follow international/continental monitoring programmes in order to meet legislation requirements (see a brief overview in Supplementary File S1). These activities are usually included in the national marine monitoring programmes (Barbour, 1999) of each country or result from specific research needs. On the other hand, routine sampling expeditions aimed at detecting novel bioactive compounds are usually hampered by the lack of financial resources and legal requirements needed to provide personnel, equipment, and subsequent laboratory costs. Regardless of the sampling objective (bioassessment/monitoring or discovery of bioactive compounds), traditional taxonomic methods are currently coupled with molecular approaches, including genomic sequencing (Leese et al., 2016; Compson et al., 2020). However, for biodiscovery, sampling may be followed by other indispensable steps such as bioactivity screening, culturing and preservation (Joint et al., 2010; Ingebrigtsen et al., 2017). The latter steps may be followed by further bioassay screening. In the case of microorganisms, it is important to note that culturing conditions may influence the chemical composition of extracts and consequently the bioactivity of the species (Lauritano et al., 2016).
This article focuses on techniques and approaches for effective microbial diversity assessment. The main objectives are to harmonize and justify strategies that may lead to higher (i) resource efficiency, (ii) representation of diversity units, (iii) quality of obtained data for further analyses/interpretation. We propose an operational workflow, consisting of three main parts: pre-sampling, sampling and post-sampling activities (Figure 1). Each step is discussed in sub-chapters below, along with two considerations that are critical for conducting biodiversity assessments: legislation (including ethics considerations) and the need for multidisciplinary collaboration.
Figure 1
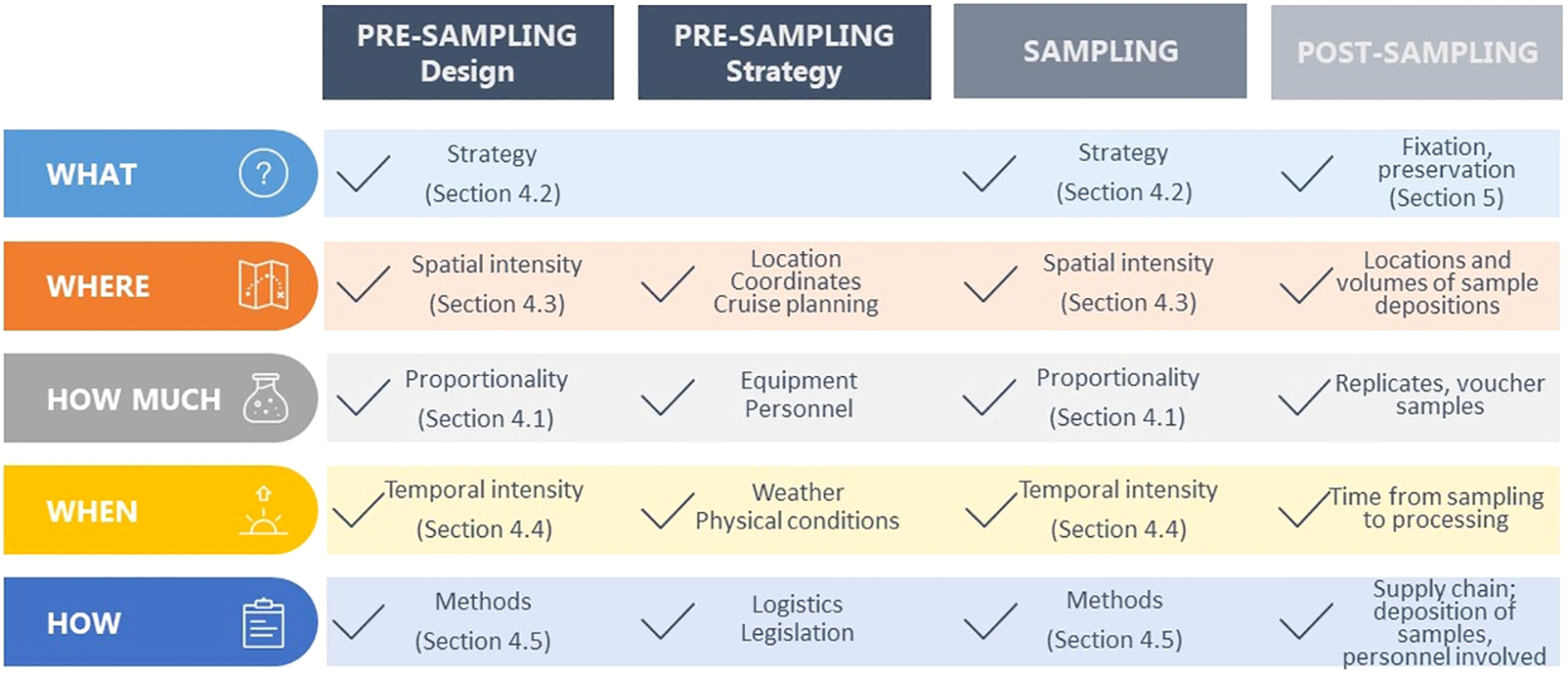
Operational flow of representative sampling strategies showing three main steps to uncover marine microbial diversity in a legal context and needed multidisciplinary approach. Pre-sampling – defining sampling design and strategy, sampling, and post-sampling that consist of fixation dependent (preservation) and independent methods (culturing, collections) and the subsequent sample management steps.
2 Targeted habitats and organisms
Microbial diversity in the oceans is exceptional and contributes to the stability and resilience of ecosystems. Microorganisms in marine ecosystems provide energy and food for higher trophic levels, while photosynthetic microorganisms produce oxygen. They play a central role in the carbon cycle, providing both organic carbon and long-term carbon storage, and recycling nutrients in marine ecosystems – all of which constitute a crucial link in food web dynamics, influence various biogeochemical processes in the water column, and have global implications for the Earth’s climate (Field et al., 1998; Fahey et al., 2017). In coastal and marine environments, most microorganisms are dispersed in the water column or attached to a substrate, e.g., sediment, forming complex microbial communities (Fazi et al., 2005; Battin et al., 2008). The abundance of microorganisms can reach up to 106 cells/mL in seawater and 109 cells/mL in the sediments of the sea floor, which therefore represents a richer source of microorganisms (Schallenberg and Kalff, 1993; Hoshino et al., 2020; Zhang et al., 2021).
2.1 Habitats
Natural or artificial environments create habitats that support living organisms and can be divided into different groups (Table 1, Figure 2). Coastal habitats are divided into (a) the nearshore terrestrial zone, (b) the intertidal zone, (c) the benthic zone, and (d) the pelagic zone (Schallenberg and Kalff, 1993). The water column in a marine pelagic habitat refers to the uppermost surface layer down to the deepest part of the ocean and includes different zones within the photic zone (epipelagic, < 200 m), the twilight zone (mesopelagic, 200 - 1,000 m), the midnight zone (bathypelagic, 1,000 - 4,000 m), the abyssal zone (abyssopelagic, 4,000 – 6,000 m) and the hadal zone (trenches, > 6,000 m) (Figure 2).
Table 1
| Habitats | Types |
|---|---|
| Coastal habitats | a) nearshore terrestrial zone b) intertidal zone c) benthic zone d) pelagic zone |
| Pelagic habitats | a) sunlight zone (epipelagic) b) twilight zone (mesopelagic) c) midnight zone (bathypelagic) d) abyssal zone (abyssopelagic) e) hadal zone (trenches) |
| Marine sediments | a) surface b) subsurface c) deep-sea sediments |
| Substrate types | a) inorganic material b) biogenic material c) artificial substrates |
Different groups of habitats and their classification.
Figure 2
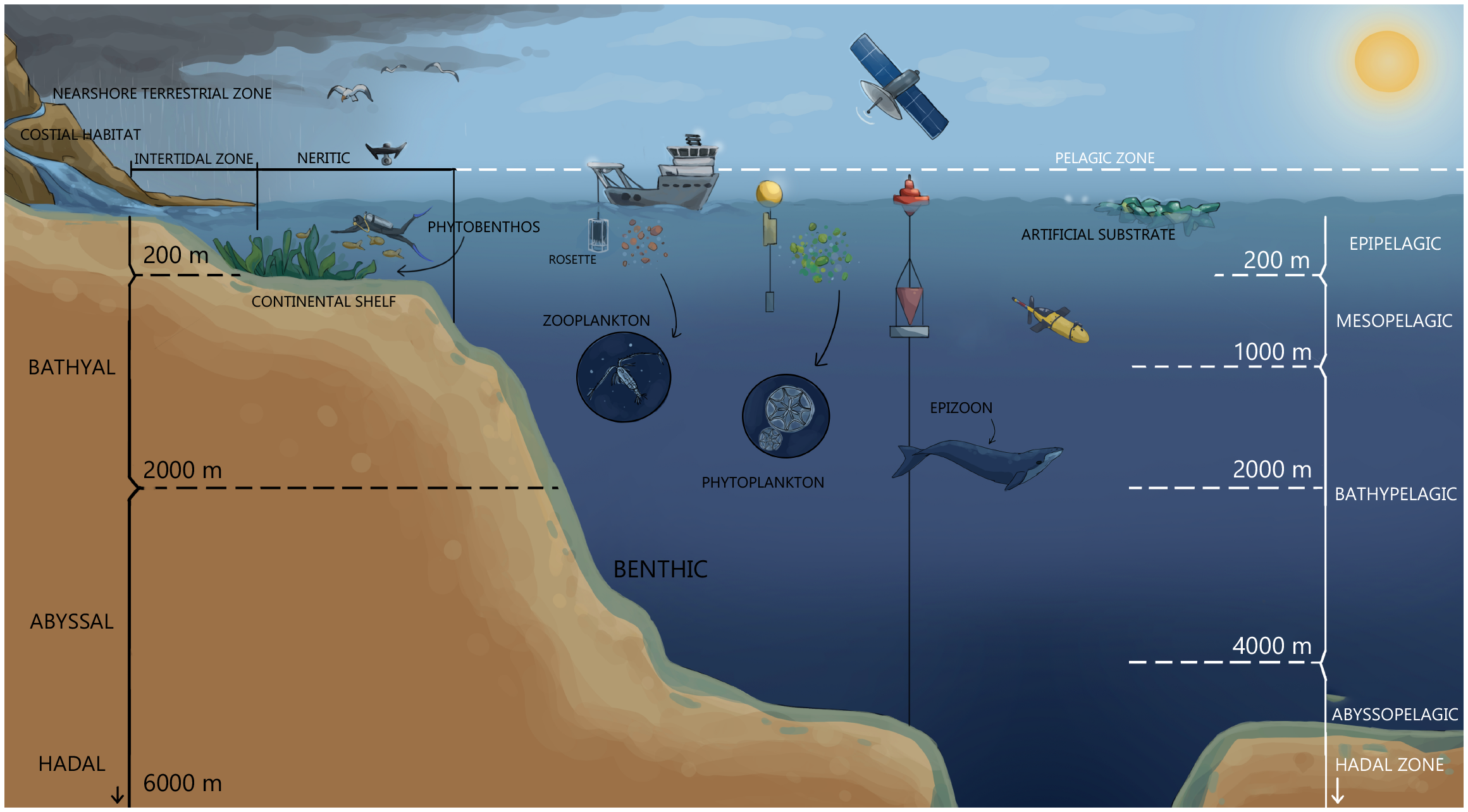
Targeted organisms and habitats – microorganisms (phytoplankton) and zooplankton in the water column, biofilm and sediments. Advanced samplers – wire-walker, sediment trap, glider, satellite, drone, and ship-based sampling during the rosette (set of niskin samplers and CTD) deployment.
Marine sediments can be classified as surface, subsurface and deep-sea sediments (Table 1). Surface sediments are found in shallow waters and nearshores. Subsurface and deep-sea sediments are often referred to as off-shore sediments (Stincone and Brandelli, 2020). Deep-sea sediments can be classified according to their depth as follows: bathyal (200–2,000 m); abyssal (2,000 – 6,000 m) and hadal (> 6,000 m) (Kamjam et al., 2017, 2018) (Figure 2).
2.2 Targeted organisms
Pelagic marine microorganisms refer to microorganisms that live in the water column of the seas and ocean (as opposed to benthic) and include plankton. Plankton are all organisms distributed in the water column that move with ocean currents (Liu et al., 2024a). From the smallest fraction, femtoplankton (< 0.2 μm), to pico- (0.2 – 2.0 μm), nano- (2 – 20 μm), micro- (20 – 200 μm), and megaplankton (> 20 cm), these include viruses, bacteria, protista, and metazoa (Figure 2). The distribution pattern of microorganisms is highly dependent on the combination of physicochemical (e.g., light availability, nutrient supply, ocean circulation, and water column stratification) and biological processes (e.g., microbial activity, zooplankton grazing pressure, viral lysis). Although circumglobal expeditions, such as Tara Ocean or Tara Pacific (De Vargas et al., 2015; Gorsky et al., 2019) have been organized for biodiversity screening focused on free-living plankton or global diversity of free-living bacteria (Pommier et al., 2007), many aquatic microorganisms colonize solid surfaces to form multi-species biofilms which offer a high pool of still uncovered marine microorganism diversity (Florio Furno et al., 2022). In marine habitats, the term substrate refers to the physical surface or material on which marine organisms live, attach, or interact with. Substrate types can vary widely in the marine environment and may significantly influence the composition of benthic (bottom-dwelling) communities. Common marine substrates include inorganic and biogenic material, as well as artificial substrates such as plastics (Figure 2, Table 1).
In coastal and marine environments, most bacteria are associated with biofilms, forming complex communities with intricate architectural organisation that contribute significantly to carbon and nutrient cycling (Fazi et al., 2005; Battin et al., 2008). Biofilm is a term used to describe structured microbial communities that occur as surface-attached communities or suspended aggregates (Coenye et al., 2022). In the marine environment, biofilms are associated with sediments, hard substrates underwater (natural and artificial), and suspended particles and aggregates. Organisms aggregate on all surfaces, as well as in the boundary layers of the water that vary in temperature, density, light, and nutrient content (e.g., thermocline, pycnocline, halocline, nitricline, etc.). Biofilm development is characterised by an increase of biomass in the early stages and changes in three-dimensional structure during maturation (Watnick and Kolter, 2000; Neu et al., 2010). Biofilms and periphyton in general are not only a novel species bank with hidden microbial diversity and functional potential in the ocean but are also increasingly recognised as a source of diverse secondary metabolites (Zhang et al., 2019; Rotter et al., 2021; Liu et al., 2024a).
Phytobenthos in coastal marine habitats grows in association with submerged substrates or colonize the first few centimetres of substrate layers (MacIntyre and Cullen, 1996; MacIntyre et al., 1996) (Figure 2). Phytobenthos communities can be distinguished according to their life traits and preferred substrates. Definitions of communities in terms of preferred substrates include “herpobenthos” (living on/in sediment) and “haptobenthos” (living on/in solid substrate). Algal communities living on/in sediment include epi-, endo-pelon (living on/in organic muddy sediment) and endopsammon (communities living on/in sandy sediment). Haptobentos represents benthic algal communities associated with various solid substrates such as rock (epilithon, endolithon), submerged macrophytes or large microalgae (periphyton, endophyton), animals (epizoon, endozoon), wood (epixylon), and sand grains (epipsammon). Organisms in benthos and water column that migrate between the two can be categorized as meroplankton, tychoplankton, or metaphyton (MacIntyre et al., 1996). There has been increasing interest in the diversity of bacteria and protists in epizoon of long-living vertebrates such as sea turtles (Kanjer et al., 2020; Trotta et al., 2021).
3 Pre-sampling
An appropriate sampling design is crucial to minimise the risk of drawing incorrect conclusions about the studied ecosystem and to minimise harming the environment/habitat (Steele, 2001). In environmental studies, sampling consistently encounters logistical bottlenecks such as time, finances, accessibility, and data integration (Albert et al., 2010). Sampling design is highly dependent on the objectives of specific projects/biodiscovery expeditions and existing prior knowledge of the system under study (Kreiner et al., 2019a, b, c). Initial sampling can be designed to develop and test metrics for application in specific environmental compartments and specific geographic regions (Stewart et al., 1986). The characteristics of organisms may differ considerably between different regions within the same ecoregion (Galitz et al., 2024). Therefore, it is often necessary to define reference conditions separately for each region (Office of Water, 2002).
Findings from previous ecosystem surveys should be integrated into the newly developed sampling strategies. This means that samples should be selected to represent a range of habitats. For example, according to the Monitoring and Sampling Manual (Water Quality and Investigation, Department of Environment and Science, 2018), water samples should be taken at a given location at approximately the same time of day or following some other natural rhythm, such as tides. Beside the daily rhythm, it is also essential to have in mind possible extreme events that can influence microbial communities like storms, heatwaves, or algal blooms. Due to the continuous and available meteorological monitoring, those events are easily visible and their possible influence on the environment needs to be addressed prior to the sampling to choose the appropriate sampling site and time. This allows for a direct comparison of survey results over time and an accurate assessment of changes. Technical guidance on appropriate sampling design is also important to maximize predictive success along environmental gradients in all areas of biology and ecology.
Representative sampling in the marine environment should ensure that the samples collected accurately reflect the habitat, relative abundance and distribution of the target organisms, and the ecological factors within the marine ecosystem. The challenges and limitations of representative sampling in the marine environment lie in the high spatial and temporal variability, limited equipment and financial constraints. It is important to integrate multidisciplinary approaches and use combinations of different methods. Five main points (described in the following subchapters) should be considered to achieve greater representativeness in sampling: proportionality, strategic sampling, sampling considering spatial and temporal variations, integration of different sampling methods (Figure 1). These should be considered already in the pre-sampling stage (Figure 1) to allow a comprehensive approach to ecosystem services assessment and an understanding of the overall ecosystem overview.
4 Sampling
4.1 Proportionally representative sampling
Proportional sampling in the marine environment is important for scientific research, environmental conservation, and natural resource management. Such sampling should ensure the representation of habitats, as well as the relative abundance and distribution of target organisms and ecological factors within the marine ecosystem. This can help address the shortcomings of previous studies, which rarely distinguished between changes in local diversity and variation in diversity across space (Keck et al., 2025). This approach is fundamental for a comprehensive understanding of the marine environment and its inhabitants. However, when designing sampling strategies that target microorganisms, proportional representation presents a significant limitation, as it depends on the availability of information on existing biodiversity. This is particularly relevant considering that individual temporal and spatial studies have typically been conducted without comparison to reference controls. Therefore, this approach requires two key considerations: (i) the need for regular monitoring campaigns to establish baseline values of microbial biodiversity in different environments, and (ii) acknowledgment of the fact that microbial diversity is vast and still largely unknown (Zinger et al., 2012; Edet et al., 2017; Santi et al., 2021; Nam et al., 2023). Therefore, microbial biodiversity assessment campaigns including both traditional (taxonomy) or new (molecular) research methods should remain a stable element in marine (micro)biology field studies, placing even stronger importance on the current global monitoring campaigns such as Tara Ocean or Tara Pacific (De Vargas et al., 2015; Kamjam et al., 2018).
4.2 Strategic sampling
Strategic sampling refers to optimized sampling that generates relevant data on abundance and distribution of species due to their high spatial and temporal variability, focusing on key areas or areas of particular importance for ecosystem services. This type of sampling strategy can allow for focused research on regions delivering specific services or protecting a particular habitat (Kreiner et al., 2019a, c, b). There are a few examples of strategic sampling (i) standardized protocols and conventions (ii) adaptive sampling strategies.
Standardized protocols are a result of long-term research of different research groups that have gone through an extensive process of intercalibration in order to generate the best possible protocol depending on the expected results. These may not refer to specific organisms, but to specific habitats or ecosystem response. An important example of a key area of interest for sampling relates to managing new species introductions, both native and non-indigenous (NIS), which are often introduced by the ballast water of vessels. Specifically, the International Maritime Organization (IMO) developed the Ballast Water Management Convention (BWMC), which provides a new international legal framework to address this threat and to also optimize Port Baseline Surveys (PBS). IMO has released guidelines that encourage port states to undertake PBS. Harmful aquatic organisms and pathogens are regarded as one of the greatest threats to the marine environment and ballast waters are one of the most frequent vectors facilitating their spread. Ports represent extremely sensitive areas for Harmful Aquatic Organisms and Pathogens (HAOP) introductions and their further proliferation. Typically, surveys of biota include sampling several different groups of organisms: hard substrate organisms, soft bottom benthos, plankton and mobile epifauna (e.g., fish). All species should be surveyed using a comprehensive sampling protocol. For example, the CRIMP protocol (Hewitt and Martin, 2001) provides guidelines for ballast water sampling (International Maritime Organization, 2004, 2007). The CRIMP protocol, which has been successfully applied to tropic and temperate marine environments, relies on SCUBA sampling and may not be applicable to all ports when alternative sampling techniques can be applied. The objectives of the PBS protocols are: to outline the steps that should be taken for a baseline survey, specify the abiotic and biotic parameters which should be analysed, quote methods and describe the report format. The distribution of sampling sites in the port area should follow a stratified sampling design (McMaugh, 2005).
Adaptive sampling strategy applies to samplings that cannot rely on standardized protocols due to the novelty of the research or specific processes. One example is sampling on fine scale, where the main question is the response of microorganisms to complex physical small-scale processes. A multidisciplinary approach employing oceanographic and meteorological models in generating a forecast of five days in advance enables guided sampling to obtain representative marine microorganisms samples. A successful example of adaptive sampling strategy was the investigation of biological response to island-trapped waves (ITWs) which may result in enhanced uplift and vertical excursion of the thermocline, upwelling of nutrients and consequently increase in net primary production. Since this physical phenomenon occurs in only a few areas in the world, in irregular time scales, the use of operational atmospheric and oceanographic models allows the prediction of intense ITW episodes and the rapid adaptation of fieldwork enabling strategic sampling in order to answer the hypothesis (Ljubešić et al., 2024).
4.3 Spatial sampling intensity
The intensity of sampling required to adequately assess microbial diversity depends on the complexity and spatial variability of microbial communities, which in turn is a function of habitat heterogeneity. In heterogeneous environments, there are several ways to reduce variability with low sampling effort. This can be achieved by (i) conducting composite sampling, i.e., combining several field samples into a single sample to account for local variation without increasing costs; (ii) selecting a single “indicator” community for sampling rather than attempting to sample all habitats, (iii) using introduced substrates (Office of Water, 2002); (iv) performing a strategy based on the vertical physicochemical profile, chlorophyll-a fluorescence and optical properties of the water column (CTD probe equipped with additional instruments, Figure 3) (Lee et al., 2005; Babić et al., 2017).
Figure 3
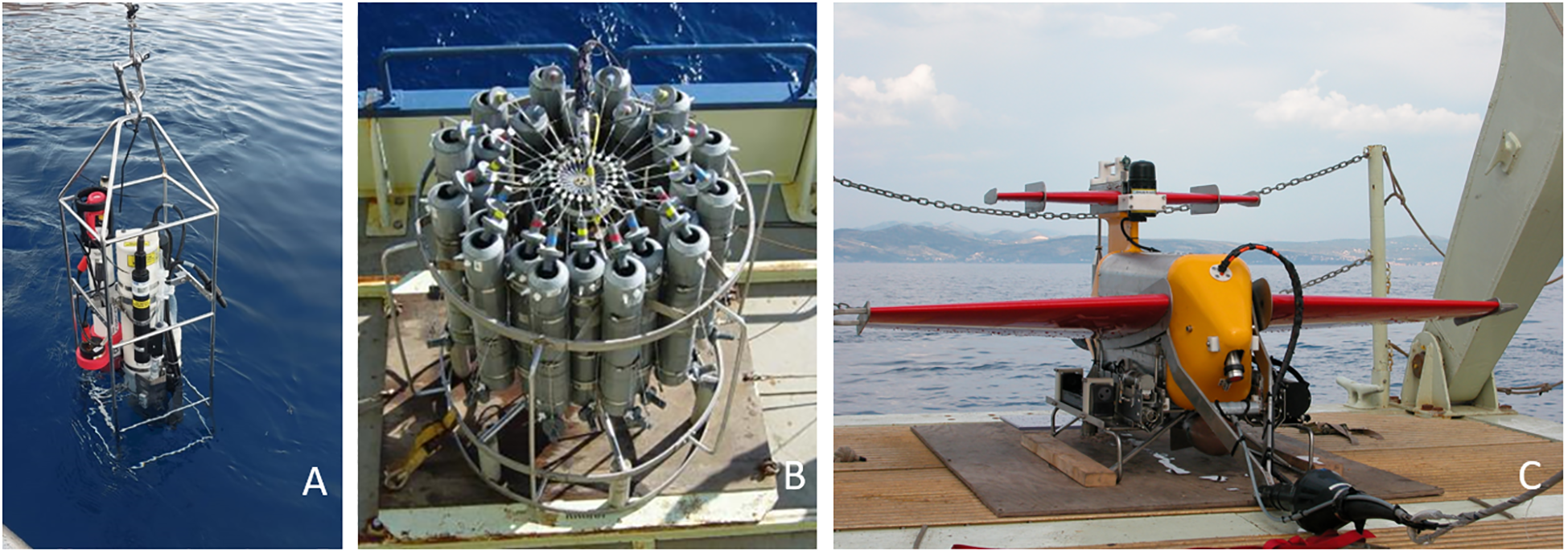
Various CTD probe set up: CTD equipped with additional instruments deployment (A); Rosette with Niskin samplers and CTD (B); CTD mounted in a vehicle that is towed by a research vessel (C).
To increase the spatial coverage, global ocean colour satellites are used (SeaWiFS, MODIS, VIIRS), which have continuously been providing global datasets of chlorophyll-a (O’Reilly et al., 1998; Vaičiūtė et al., 2021). These data not only generate insight into spatial and temporal changes in phytoplankton biomass but can also be used as an excellent tool to guide researchers in the field. Moreover, this approach allows for local (Lučić et al., 2017) or global scale (Flombaum et al., 2013; Vandermeulen et al., 2020) perspectives. Along with the continuous technological advances, another possibility in gaining global hyperspectral data is through the new PACE satellite that was launched in 2024 (NASA’s Plankton, Aerosol, Clouds, ocean, Ecosystem (PACE) mission) (Cetinić et al., 2024).
4.4 Temporal sampling intensity
Marine microorganisms typically exhibit distinct seasonal patterns in abundance and species composition, and these patterns may differ by habitat type. Some familiarity with the temporal dynamics of communities in the marine classes of interest should be gained before sampling (Office of Water, 2002). Sampling should be designed to effectively encompass the natural seasonal variability and peak community development. The required number of samples per year depends on the objectives of the survey and its specific applications. If sampling is less frequent (e.g., annually) or limited to a single assessment, the focus should be on metrics that integrate conditions over longer periods of time and are therefore not susceptible to short-term fluctuations. Nevertheless, wherever logistically or financially possible, sampling should be conducted during more than one season to allow for an integrated assessment of the biotic community in relation to their abiotic environment including nutrient conditions. When comparing habitats, they should be sampled at approximately the same time of year to minimise any disruptive influence of seasonal variation on microbial community composition (Office of Water, 2002).
4.5 Integration of different sampling methods
The choice of the sampling methods is crucial for any successful sampling campaign. The methods applied should correspond to the proportionally representative sampling, the sampling strategy, and the temporal and spatial sampling effort. Essentially, sampling methods are also influenced by existing facilities/infrastructure access and staff expertise.
4.5.1 Sediment: grab sampler, corer, sediment traps and advanced samplers
The selection of sampling methods depends on several factors such as water depth, type of samples, mass of sediments needed, characteristics of the sediments, among others (Tuit and Wait, 2020). The sampling equipment and strategies should be optimised based on the characteristics of the sediment (i.e., organic matter content, grain size, etc.). Furthermore, the target depth of the sediment layer may depend on the purpose of the study (i.e., surface sediments versus deep sediments), taking into account the depth of the oxic versus anoxic layers, and the time required to obtain samples from deep sediments.
Three types of sampling devices can be distinguished: (1) hand samplers or corers; (2) grab samplers, and (3) core samplers (Tuit and Wait, 2020) (Figure 4). Hand corers can be used for collecting surface sediment when using waders (< 1 m depth) and subsurface sediment (< 30 m depth) or driven into the sediment by divers (SCUBA). Hand sampling consists of inserting these devices into the sediments. Hand samplers include scoops, spoons, shovels, sterile plastic bags, and augers, which are used specially to collect surface sediments from shallow waters. Buckets or tube augers are other hand samplers commonly used for surface sediment sampling. However, a limitation of this method is the cross-contamination between sampling depth intervals (Schoenleber, 2005). Grab samplers, dredge samplers or mud snappers are often used for sediment collections < 30 m depth. The most common are the Birge-Ekman and Van Veen types. The Birge-Ekman sampler is mostly used to collect soft sediment in shallow waters and can be operated by handline from a boat or while wading, while the Van Veen sampler is able to collect a deeper range of sediment samples and is usually driven by a winch from a boat (Batley and Simpson, 2016). The core samplers are simple devices widely used for surface and subsurface sediment collection. These are composed of a core tube and a heavy weight to push the core tube into the sediment. These tools can be used to collect samples at different depths, are low-cost and can be manufactured from a variety of materials. They comprise gravity corers that are able to collect sediments with up to 6 m of coring length (Chen et al., 2013); piston corers, which can collect sediments up to 20 m in length in waters depths of 100 m to 5,000 m (Hollister et al., 1973); box corers, which are designed to take samples from the top of the seafloor to a maximum depth of < 1 m (Batley and Simpson, 2016; Hopkins, 2007); and vibra-corers, capable of retrieving continuous, undisturbed sediments up to 5,000 m in depth with the length of core samples up to 13 m in length (Reed et al., 2005).
Figure 4

From left to right. Van Veen grab sampler, corers used for sampling muddy sea bottom (middle photograph), hand corer (with courtesy of Eylem Atak and Daniel Bosch).
To gain high spatial and temporal resolution of sampling, automated advanced samplers are used. They have the advantage of collecting high-resolution data without the need to deploy scientists in the sea and can therefore operate in all weather conditions. Their disadvantages, however, are higher costs and maintenance requirements. Advanced core samplers have been developed to collect deep-sea sediments, including the following devices: pressure core barrel (PCB), advanced piston corer (APC), pressure core sampler (PCS), Fugro pressure corer (FPC) and many more (He et al., 2020). To collect deep-sea sediments, Remotely Operated Vehicles, ROVs (used for depths between 100 – 4,500 m), Autonomous Underwater Vehicles, AUVs (used for depths between 100 – 6,500 m) and multicorers (used for depths between 100 – 1,400 m) (Jones, 2009; Wakita et al., 2010) are often used.
Finally, sediment traps can be deployed on various platform designs, including neutrally buoyant sediment traps (Estapa et al., 2020) (Figure 5A), surface tethered sediment traps (Knauer et al., 1979) (Figure 5B) on the base of a Wirewalker profiler (Rainville and Pinkel, 2001) (Figure 5C) or simpler sediment traps that are placed at the bottom to monitor the amount of sediment collected in a given amount of time. These sediment traps collect microorganisms and particles with a high temporal and spatial resolution, in addition to collecting physico-chemical and bio-optical data by the advanced platform itself (Durkin et al., 2021, 2022) (Figure 5). Their optimisation and constant upgrade are essential in representative sampling (Omand et al., 2017; Staudinger et al., 2018; Daniel et al., 2020; Estapa et al., 2020).
Figure 5

Autonomous platforms equipped with various sensors and sediment traps: Neutrally buoyant sediment trap (A), Sediment traps (B), Wire-walker (C).
4.5.2 Water column: pumps, water samplers and nets
The biggest challenge in exploring the diversity in the water column is the patchy distribution and constant dispersion of particles, including microorganisms. Thus, the first and most important step in plankton diversity research is to effectively plan the sampling strategy, which needs to be highly adaptive, depending on the environment and the target organism group. The sampling strategy should be determined in situ to optimize discrete sample distribution based on the vertical physical-chemical profile and chlorophyll-a fluorescence (CTD probe with fluorimeter and preferably optical instruments, Figure 3A). When sampling with a scientific research vessel, the best option is a rosette of Niskin samplers equipped with a CTD profiler, where the samples are taken during the CTD cast (Figure 3B). To gain high spatial diversity, a CTD can be mounted onto an underwater vehicle – an undulator that can be autonomous and/or towed by a research vessel (Figure 3C). This enables high spatial and temporal screening of the area and subsequently adaptive sampling. Plankton can also be sampled with pumps and nets of various mesh sizes (Figure 6). The advantage of a pump is the volume of water that can be filtered, which is much greater than that collected with Niskin bottles. The seawater collected with these devices can then be (pre‐)filtered through a mesh of 200 μm and 20 μm, and additionally filtered through a 5 μm mesh to obtain a range of plankton size fractions for further analyses. The benefit of using nets with larger mesh sizes is the ability to sample greater volumes of water, however, this method also runs the risk of losing smaller organisms. For example, for ballast water sampling on board, a hand pump is used. This pump is light-weight and compact in design. It is approximately 30 cm long with a diameter of 5 cm. The water should be filtered to enable the transport of smaller volumes of water (David and Gollasch, 2015).
Figure 6
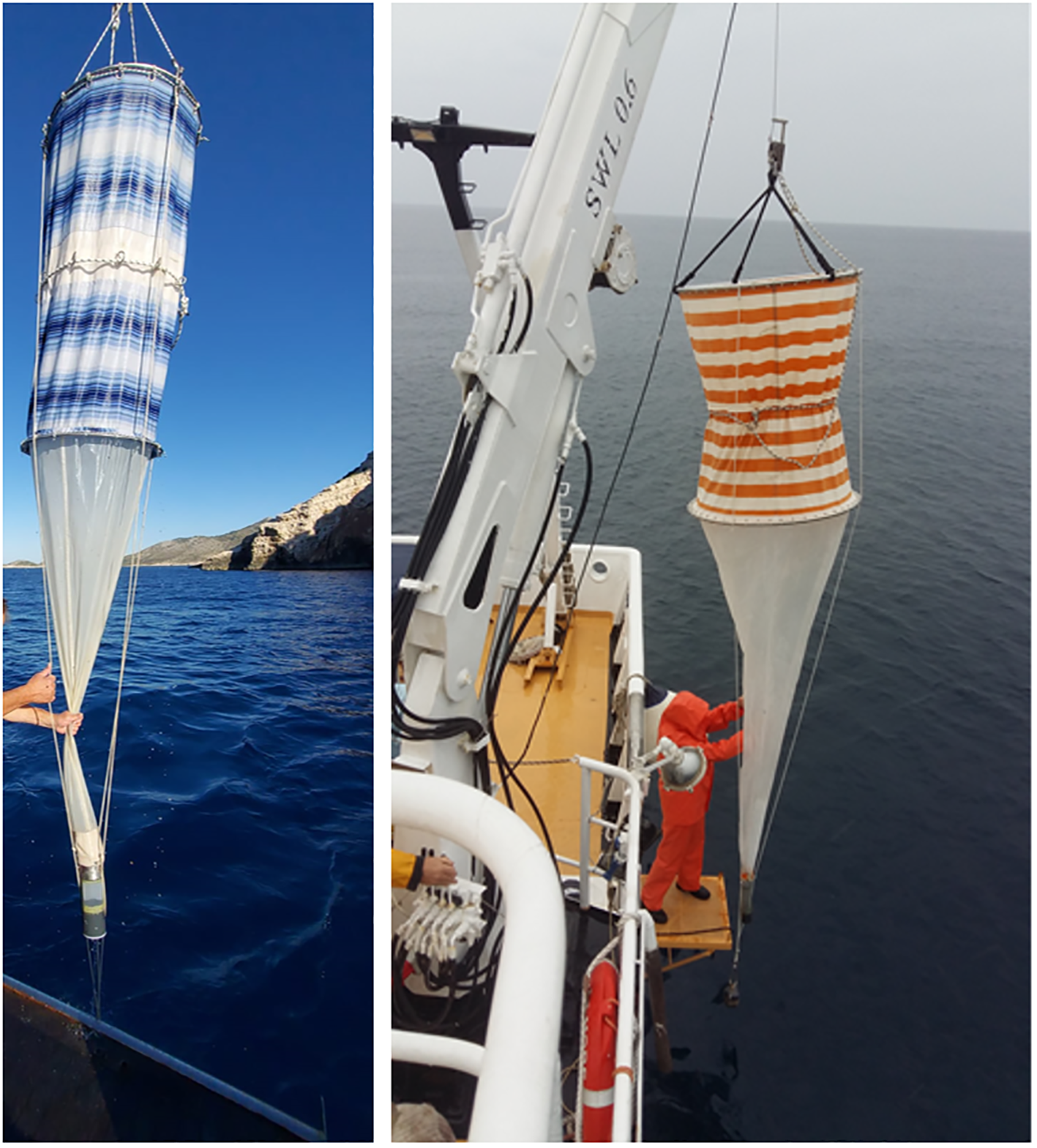
Zooplankton nets of various mesh sizes.
During the circumglobal Tara Oceans expedition (2009-2013), large volumes of sea water were collected with peristaltic pumps and plankton nets of various mesh sizes focusing on two sampling depths: subsurface mixed-layer waters and the deep chlorophyll maximum (DCM) at the top of the thermocline (De Vargas et al., 2015). The sampling design therefore enabled a wide geographical range of sampling and allowed for sampling of larger water volumes, thus elucidating much higher eukaryotic diversity of heterotrophic protists, including uncommonly sampled life forms such as parasites and photosymbiotic taxa (De Vargas et al., 2015). However, an important bottleneck has to be considered in sampling design of large expeditions, which is the lack of ship-time available for work at specific stations. To address this, the sampling approach during the following Tara Ocean expedition (2016-2018) (Gorsky et al., 2019) was optimized. The surface water was continuously pumped with a membrane pump through the hull inlet located 1.5 m under the waterline, from there, circulated through a debubbler and distributed to several flow-through instruments to optimise physical, bio-chemical, optical and genomic methods, and gain insights into the entire plankton community of the surface layer (Gorsky et al., 2019). Additionally, to collect microorganisms while sailing, the “Dolphin” sampler and “High Speed Net” (HSN) were designed to collect seawater and neustons of size < 2,000 mm and of size > 300 mm, respectively (Gorsky et al., 2019). Sampled seawater was prefiltered, and subsequently size fractionated by filtration to obtain samples for further analyses.
With regard to biotechnological applications, collection of e.g., cyanobacteria or other microalgal biomass from natural water bodies may offer a direct source of bioactive compounds (pigments, toxins, other bioactive compounds), sometimes bypassing the need for laboratory cultivation. The development of inland water crafts that can be used for the physical collection of surface blooms is gaining increasing attention and some are commercially available already. One of the oldest collection techniques has been used in Klamath Lake, USA, since 1987. The main idea of this technique is the vessel that harvests the biomass of Aphanizomenon flos-aquae (Carmichael et al., 2000) using in situ filtration. There are several pilot scale prototypes for harvesting biomass e.g., oil boom and forming fabric construction tested by Gröndahl (Gröndahl, 2009), AS-S, AS-L and AS-LAND prototypes for the collection of cyanobacteria biomass and macroalgae mats which were already patented at the State Patent Bureau of the Republic of Lithuania (Patent No. 7081) (Nature Research Centre, 2024).
4.5.3 General: diving
An excellent added value to the sediment and water column methods described above is scientific diving. The main advantage of diving is the ability to collect fine samples of benthic organisms and the water column. Also, unless the sampling tools described above are equipped with photographic or video cameras, a broader picture of the sampling area is not readily available. This can result in collecting samples which may or may not be representative. However, even if the sampling tool is equipped with a camera, the information can be overlooked due to the decreased visibility along the water column, sediment resuspension, resolution or equipment malfunction. In shallower areas, properly trained SCUBA divers are able to explore and perform direct hand sampling using various tools for sample collection, such as cores or photographic and video recording (Novosel et al., 2004; Kružić et al., 2012; Zunino et al., 2018), or deploy sensitive scientific equipment (Orlić et al., 2011) (Figure 7).
Figure 7

Underwater manual sensor mounting by diver.
5 Post sampling
5.1 Fixation-dependent approaches
Once the samples have been collected, they need to be preserved before analyses. The choice of preservation method depends on the organisms, environment, and planned methods of analyses.
Fixatives are by definition toxic, i.e., they have the ability to inhibit the growth of microorganisms. Despite this disadvantage, fixatives enable the preservation of microorganisms prior to their analysis/assessment. The main criteria for a suitable fixative are: (i) it must not inflict loss of targeted microorganisms, (ii) it must prevent microbial decomposition of the organic material during sample storage and (iii) it must ensure good preservation of morphological characteristics that allow for the identification of taxa during analysis (Institute for Standardization of Montenegro, 2014). It is therefore important to use fixatives at the recommended concentrations, consistent with laboratory protocols and guidelines, and to take safety precautions for the personnel involved. Particular attention should be paid to samples analysed by molecular methods, as fixatives can disrupt subsequent analyses.
5.1.1 Fixatives and preservatives
Various fixatives are commonly used for preservation of microorganisms from seawater, sediment, and biofilms, and the selection of an appropriate fixative or preservative depends on the purpose of the analysis.
Glutaraldehyde is a fixative known for its exceptional ability to preserve ultrastructural details. It achieves this through strong cross-linking of proteins, making it particularly suitable for electron microscopy. Its rapid penetration allows for fast fixation, and it maintains fine cellular structures with minimal distortion. Microorganisms are frequently preserved in their culture media and in a 10-50% glycerol solution at a proportion of 7:2 and stored at -80°C. When analysing prokaryotes in water samples using fluorescence microscopy or flow cytometry, samples can also be preserved with different fixatives (e.g., buffered formaldehyde solution 0.1%-1% v/v final concentration). After that, the abundance of prokaryotes can be evaluated by staining with fluorescence dyes (Porter and Feig, 1980). However, glutaraldehyde is not suitable when nucleic acid integrity is required. There is also a risk of overfixation. Additionally, glutaraldehyde is both toxic and irritating, necessitating strict safety protocols and proper laboratory ventilation.
Lugol’s solution is a mixture of iodine (I2) and potassium iodide (KI), colouring the samples in golden brown (British Standards Institute (BSI), 2011). It is a simple and an inexpensive fixative, often used for quick applications. To preserve seawater samples, 0.25 – 0.5 mL of acid Lugol’s solution per 100 mL of sample should be added immediately after sampling. For eutrophic waters, the quantity of the preservative may be increased up to 1–2 mL of Lugol’s solution per 100 mL of sample. Lugol-preserved samples should be stored in the dark and at a temperature of 4 to 8°C until analysis. Samples should be analysed within a few months. It is also unsuitable for high-resolution ultrastructural studies such as electron microscopy. The iodine component can lead to cell shrinkage and alter the refractive index of samples, and it offers limited preservation of proteins and nucleic acids.
Alcohol-based fixatives, such as ethanol, function primarily by dehydrating and precipitating cellular proteins. They are widely used because they are effective at preserving DNA and RNA, and are simple, inexpensive, and broadly accessible. Alcohols are also considered to be less hazardous than aldehydes. Ethanol is especially useful for genetic studies and is compatible with subsequent molecular techniques. On the downside, alcohols offer poor ultrastructural preservation and can cause cellular shrinkage and membrane damage. Additionally, alcohols dissolve lipids, which makes them unsuitable for lipid studies, and they may distort cellular morphology more than aldehyde-based fixatives.
Formaldehyde (CH2O), also known as formalin, is the most commonly used fixative for sample preservation. Due to the flammability of concentrated formaldehyde, the standard maximum concentration solution is 37% formaldehyde. Thus, 2% formalin is a 2% solution of the standard formaldehyde solution, not 2% formaldehyde. Formaldehyde, however, slowly transforms into formic acid and methanol (the ‘Cannizzaro reaction’), which negatively affects fixation and preservation. For this reason, formaldehyde should not be kept in stock for too long and should be buffered to pH 8 or 9. A suitable buffer is hexamethylenetetramine. If the concentration of formaldehyde exceeds 20% volume fraction, there is a risk of precipitation. It is a known carcinogen and must be handled with care. It is not ideal for the preservation of nucleic acids, as it causes fragmentation and cross-linking of DNA and RNA. Over time, formalin can polymerize into paraformaldehyde, which reduces its effectiveness.
In contrast to these classical fixatives, DNA/RNA Shield and RNAlater are not designed to preserve morphology, but rather to stabilize nucleic acids for downstream molecular applications. These agents are best described as preservatives, rather than fixatives, and they excel in preserving RNA and DNA without the need for immediate freezing.
DNA/RNA Shield is especially notable for its ability to inactivate pathogens, including viruses and bacteria, upon contact. This feature enhances biosafety during sample handling. It also allows for room-temperature storage and transport, which is particularly valuable in field conditions or settings without access to cold storage. The preservative is compatible with a wide range of sample types. Its primary limitations are its higher cost compared to traditional methods and absence of morphological preservation.
RNAlater is designed to rapidly penetrate tissues and stabilize RNA, making it a reliable choice for transcriptomic studies including qPCR, RNA sequencing, and microarray analysis. It allows short-term storage at 4°C and long-term preservation at -20°C or -80°C. RNAlater is easy to use in both laboratory and field settings and is compatible with many types of RNA extraction protocols. Like DNA/RNA Shield, RNAlater does not preserve cellular morphology.
In conclusion, the ideal choice of fixative or preservative depends on the analytical goal. When morphological preservation is critical, glutaraldehyde and formalin are the preferred options, glutaraldehyde for electron microscopy and formalin for routine analyses. When preserving nucleic acids is the priority, DNA/RNA Shield and RNAlater offer superior stabilization of RNA and DNA without requiring immediate freezing. Alcohols provide a middle ground, offering decent nucleic acid preservation with moderate morphological distortion. Lugol’s solution is best reserved for rapid, low-cost diagnostic purposes. If both structural and molecular analyses are required, the best practice is to prepare parallel samples using different preservation strategies, as no single fixative can optimally serve both purposes.
5.1.2 Cryopreservation
For microbial diversity analysis (i.e., using molecular techniques such as DNA sequencing), water is typically filtered on 0.2 µm polycarbonate filters to concentrate the microbial sample. Filters, biofilms and sediment samples are directly transferred into sterile cryovials that are RNase and DNase-free. Samples can be transported in cryogenic containers (liquid nitrogen or dry-ice) and stored at -20°C/-80°C (Bruce et al., 2021; Pawlowski et al., 2022).
When community composition is investigated by Fluorescence in situ hybridization (FISH) and Catalysed Reported FISH (CARD-FISH), samples can be fixed and subsequently cryopreserved. These hybridization techniques allow for the quantification of bacterial abundance in the indigenous marine consortium. To apply these hybridization techniques, water is sampled and triplicate aliquots (e.g., 50 mL) are immediately fixed with buffered formaldehyde solution (0.1%-1% v/v final concentration). Subsamples (1 mL, depending on cell abundance) of fixed water are filtered on polycarbonate membrane filters (pore size 0.2 µm, 47 mm diameter) by gentle vacuum pressure (< 0.2 bar) within 24 hours following fixation. Filters can be stored at -20°C until further processing (Pizzetti et al., 2016). For sediments, an efficient detachment and purification of bacterial cells associated with streambed sediment is required prior to fixation and cryopreservation (Amalfitano and Fazi, 2008). For biofilms, samples can be collected by scraping the original substrate (e.g., stones) and subsequent filtering. When working on filtered samples, however, information on the spatial distribution of specific bacterial clusters within the community structure is lost (Olapade and Leff, 2005; Fazi et al., 2007; Braun et al., 2009). To visualize specific cells, while maintaining an unaltered 3D structure of the biofilm, substantial improvements have been made by utilizing CARD FISH in combination with Confocal Laser Scanning Microscopy (see CLSMFISH in (Amann et al., 1998)). This protocol allows for the simultaneous identification and the spatial localization of cells, while maintaining an unaltered natural architecture of the biofilm. Microscope chambered glass slides (e.g., 10-well diagnostic microscope epoxy coated slides; well diameter: 6.7 mm from Thermo Scientific, Germany) can be utilized as artificial substrates for microbial growth. After collection, the slides can be fixed with buffered formaldehyde solution (0.1%-1% v/v final concentration) and stored at 4°C for less than 24 hours, or alternatively fixed with ethanol (50% final concentration) and stored at -20°C. CARD–FISH can be performed directly in the wells of the glass slide as proposed by Lupini et al. (2011). Hybridized cells are then quantified by Confocal Laser Scanning Microscopy (CLSM) and the three-dimensional reconstruction of CLSM images allows for the precise location of specific bacterial cells within the biofilm structure and details the dynamic patterns of colonization.
For long-term preservation of non-living microalgal cultures, freeze-drying (lyophilization/storage in ultra-low temperature in liquid nitrogen) is the most common technique. Lyophilization removes water from a frozen sample via sublimation and it is an effective and established technique that is used in many culture collections centres and universities (McGrath et al., 1978). There are many studies on lyophilization of microalgae (Wolkers and Oldenhof, 2021) and its effects on biomass composition, culture growth or biomass production of microalgae (Ryckebosch et al., 2011; Guldhe et al., 2014; Grossmann et al., 2018).
Cryopreservation is used to preserve live cells. Culture collections around the world use cryopreservation as long-term storage of viable cells (Frenea-Robin and Marchalot, 2022; Foo et al., 2023). For long-term preservation, cryopreservation with DMSO is the most preferred method. However, many diatom species cannot be cryopreserved using DMSO. Therefore, Stock et al. (2018) used different concentrations of DMSO and Plant Vitrification Solution 2 (PVS2) as an alternative cryoprotectant to DMSO. They have shown that the selection of cryoprotectant type and its concentration are important factors for the preservation of diatoms, with the success of preservation also species-dependent (Safarik et al., 2016).
5.2 Fixation-independent approaches
5.2.1 Immunomagnetic separation
Immunomagnetic separation (IMS) of target microorganisms is based on using magnetic nano- and microparticles with bound specific monoclonal or polyclonal antibodies (Abs) against appropriate cell surface epitopes (Liu et al., 2024b; Wang et al., 2020). When immunomagnetic particles (IMPs) are added to the cell suspension or appropriate homogenate, they selectively attach to target cells. After incubation, target cells with attached magnetic particles (including excess IMPs) are isolated using an appropriate magnetic separator. In the direct method, the appropriate Abs are coupled to the magnetic particles, which are then directly added to the cells-containing sample. In the indirect procedure, the cell suspension is first incubated with primary Abs, which bind to the target cells. Afterwards, magnetic particles with immobilized secondary Abs against the primary Ab, or protein A, or protein G are added, permitting magnetic beads to bind rapidly and firmly to the primary Abs on the target cells (Escalante-Maldonado et al., 2015; Safarik et al., 2016). Alternatively, biotin-conjugated primary Abs can be captured by streptavidin-bound magnetic particles (Aguilera et al., 2002).
5.2.2 Live samples for culture-dependent techniques
In contrast to preserved samples, live samples should be stored in the dark at a temperature of 4°C to 10°C. The storage time should not exceed 36 hours. Storage at lower temperatures slows down the physico-chemical processes and leads to a reduction in sample quality (Institute for Standardization of Montenegro, 2014). Due to the small size of the microorganisms and the lack of distinct morphological characteristics, taxonomic analyses are largely dependent on culturing. Culturing enables further taxonomic and physiological analyses of the organisms, testing of their biotechnological potential and establishing culture collections. As far as bacteria are concerned, it has been estimated that only 1% of environmental bacteria have been cultured (Hofer, 2018). Over the last few decades however, the proportion of cultured microorganisms has increased well beyond the 2% due to several innovative methods and strategies that better mimic natural conditions and target specific microbial growth requirements (Martiny, 2019). Simulated natural environment cultivation, co-culture and microdroplet encapsulation, in situ cultivation devices, targeted liquid cultivation methods, microfluidic and cell sorting technologies are methods that increased significantly the success of microbial cultivation (Stewart, 2012; Vartoukian, 2016; Kapinusova et al., 2023; Seo et al., 2023). Simulated natural environment cultivation uses devices like diffusion chambers and the ichips that allow microorganisms to be incubated in situ or under conditions closely mimicking their natural habitats. These chambers have semi-permeable membranes that permit the passage of growth factors and signalling molecules from the environment, facilitating the growth of bacteria that depend on community interactions and natural chemical cues (Vartoukian, 2016). Encapsulation of single bacterial cells in microdroplets of agarose or alginate microbeads allows bacteria to grow “together but apart,” maintaining cell-to-cell communication while preventing overgrowth by faster growers. Co-culture and microdroplet encapsulation methods are especially suitable for rare and previously uncultured lineages, such as Planctomycetales and Alphaproteobacteria (Stewart, 2012; Kapinusova et al., 2023). In situ cultivation devices use hollow-fibre membrane chambers, diffusion bioreactors, soil substrate membrane systems, and microbe domestication pods that enable cultivation directly within the natural environment or with environmental parameters maintained. These devices help provide natural growth factors and conditions that are often missing in standard laboratory media (Kapinusova et al., 2023; Seo et al., 2023). New methods like Non-Colony-Forming Liquid Cultivation (NCFLC) selectively isolate microbes that do not form colonies on agar plates by using liquid media and gentle processing of environmental samples (Seo et al., 2023). Microfluidic devices such as nanoporous microbial incubators and chemotactic droplet sorting allow high-throughput cultivation and isolation of microbes in very small volumes, facilitating the study of rare or slow-growing species (Kapinusova et al., 2023; Seo et al., 2023). Although new cultivation methods have greatly improved access to a wider range of microbes, they are not yet commonly used in most laboratories. This is mainly because they require special equipment, are complex to use, and can be expensive or not easy to use regularly, on a large scale, or in clinical labs (Chaudhary et al., 2019; Jung et al., 2021).
For growing a mixed culture expected to contain microorganisms, mainly diatoms, dinoflagellates, cryptophytes, haptophytes, and heterotrophic nanoflagellates, live samples should be inoculated in Marine Water Enrichment Solution (preferably f/2), immediately upon arrival in the laboratory. Target cells should then be isolated and filtered through Nucleopore polycarbonate membrane filters with a pore size of 3.0 µm into fresh medium using a syringe and filter holder. After filtration, isolation followed by dilution, which consists of repeatedly transferring a sub-volume of a culture (1/10 of the medium volume) into a fresh medium (9/10 of the medium volume) is used to obtain a statistically likely quantity of one cell per tube at the end of the series (Knight-Jones, 1951; Throndsen, 1978). After a desired culture is established, it should then be transferred regularly into fresh medium, at period intervals depending on the microorganism, to maintain the cells in an exponential growth phase.
In long-term culturing, storage conditions such as time, initial biomass concentration, presence or absence of light and addition of preservatives affect the biochemical composition of cultured cells. The identification of optimal conditions for long-term preservation is especially important for commercially important species. Such an example is the microalga Nannochloropsis gaditana, used commercially to produce biofuels and food additives, where optimal conditions for preservation should minimize the bacterial contamination and maximize fatty acid and pigment concentrations in cells (Camacho-Rodríguez et al., 2016).
5.2.3 Culture-independent techniques
Molecular methods, independent of microbial cultivation, are used to analyse biodiversity and community composition or to explore the biotechnologically relevant compounds produced by microorganisms in natural environments (Costa et al., 2020). They have become of increasing interest in recent years to avoid the reliance on culturing that excludes uncultured taxa from results. Currently, eDNA (environmental DNA) metabarcoding and metagenomics are mostly used to evaluate environmental biodiversity without the need to culture the samples. Both approaches offer distinct advantages and limitations. Metabarcoding, which involves amplification of short DNA fragments of selected genetic markers, is cost-effective, high-throughput, and well-suited for targeted taxonomic groups (Taberlet et al., 2012; Deiner et al., 2017). However, it is limited by PCR bias, which can preferentially amplify certain taxa, and often provides only genus-level taxonomic resolution, especially when using short reads (Clarke et al., 2014; Callahan et al., 2017). With the development of third-generation sequencing technologies, such as full-length 16S rRNA gene sequencing, species- and even sub-species-level identification has become increasingly feasible (Karst et al., 2021).
In contrast, metagenomics sequences the entire genomic DNA of a sample, avoiding primer bias and enabling the detection of both known and novel and/or uncultured taxa. It provides simultaneous taxonomic and functional profiling, offering insights into the metabolic potential of microbial communities (Quince et al., 2017; Rieder et al., 2023; Serite et al., 2023). Furthermore, metagenomic data can be used to assemble near-complete genomes (metagenome-assembled genomes), which help describe new candidate taxa and understand ecological functions (Parks et al., 2017; Serite et al., 2023).
An important distinction to highlight, which also affects the design of sampling campaigns, is between eDNA and bulk/community DNA. eDNA is generally extracted from environmental samples (e.g., water filtered in the marine environment) that capture both free DNA shed from macro and planktonic microorganisms, resulting in a mixture of biological materials. In contrast, bulk/community DNA is specifically extracted from physically collected and isolated mixtures of whole organisms or biological source materials (e.g., tissue samples, benthic organisms) obtained from the marine environment (Deiner et al., 2017). DNA metabarcoding of bulk samples is enables comparisons across studies and time points, but requires harmonization in protocols such as pre-processing, DNA extraction, PCR, and sequencing itself (Van Der Loos and Nijland, 2021). Bulk sample metabarcoding most closely resembles traditional taxonomic biodiversity biomonitoring, whereas eDNA approaches are particularly suitable for detecting ecosystem changes and identifying new, rare, invasive, or elusive species (Macher et al., 2018; Ruppert et al., 2019).
For microbial diversity analysis by metabarcoding and metagenomics, seawater from Niskin bottles is transferred in sterile bottles (1–2 L) at a given depth. Composite sampling of the entire water column can also be assessed by mixing equal amounts of different sampled depths. Samples are filtered on deck on 0.2 μm polycarbonate filters. The aliquots to be filtered depend on the abundance of free-living and particle-associated microorganisms. As a general rule of thumb, it is suggested that up to 1 L of seawater is filtered, or up to clogging of the filter (Babić et al., 2017; Mucko et al., 2018). Eukaryotic fractions of all sizes can also be filtered on 0.2 μm polycarbonate filters, but larger volumes are recommended (up to 10 L). In that case, all fractions are stored on the same filter and there is no loss of information from any fraction. Filters are stored in sterile (RNase, DNase-free) cryo vials and immediately placed in cryogenic containers (liquid nitrogen or dry-ice) until their arrival to the laboratory, where they can be stored long term at -20°C/-80°C (Matek et al., 2023).
5.2.4 Culture collections, biobanks and biorepositories
Culture collections and biobanks supply organisms for research, commercial and educational purposes. While culture collections were established with the primary goal to preserve strains, biobanks are often fulfilling more specific quality standards regarding the management of the genetic resources and their relative data (DiEuliis et al., 2016; McCluskey et al., 2017; De Vero et al., 2019). Thus, biobanks (or microbial Biological Resource centres, mBRCs) operate in a quality-controlled manner and must fulfil the quality standards required by the industry and by the scientific community (Janssens et al., 2010; Overmann, 2015). This makes mBRCs key elements in sustaining international scientific infrastructures for the development of biotechnology and the bioeconomy. The World Federation for Culture Collections (WFCC) organized a platform, data centre and information resource in World Data Centre for Microorganisms (WDCM, https://www.wdcm.org/). According to WDCM, there are 843 culture collections around the world available to both academia and bioindustry, with about 3,539,328 microbial strains (https://ccinfo.wdcm.org/). There are also specialized culture collections. As an example for algae, there are hundreds of collections worldwide, however only around 40 algal culture collections are public. These collections provide nearly 5,000 species of algae registered in the WDCM (https://brphycsoc.org/algal-culture-collections-in-the-omics-age). Regarding marine fungi, an example of a specialized collection is the Turin University Culture Collection (TUCC- https://www.tucc.unito.it/) that currently preserves more than 2,000 marine fungal strains and provides different services including the isolation, identification and characterization of marine fungi. Another microbial culture collection worth mentioning is The Microbial Culture Collection Ex within the University of Ljubljana, Slovenia (https://www.ex-genebank.com/index.php/en/about-microbial-culture-collection-ex), one of the few global collections that is specialised in the isolation and preservation of eukaryotic extremophilic fungi, but also having bacterial, archaeal and algal strains.
The main role of culture collections is to maintain and preserve strains. However, most collections do much more than that. For example, on top of maintenance and international distribution of cultures, the Culture Collection of Algae and Protozoa also provides a variety of services covering taxonomy, genomic and analytical services, patent and confidential deposits, method development, consultancy and even medium scale (> 200 L) cultivation (https://www.ccap.ac.uk). Culture collections obtain cultures from donations, exchanges and ongoing isolation. Many public culture collections have been around for decades and despite their important contribution to science and humankind, culture collections are rarely commercially independent. Public culture collections are often associated with universities and lack long-term financial stability.
The most recent and important mBRC network in Europe is MIRRI (Microbial Resource Research Infrastructure, http://www.mirri.org) which seeks to improve access to high-quality microbial resources, associated services and (meta)data by creating a pan-European, distributed infrastructure of culture collections, mBRCs and stakeholders. Within MIRRI there are many institutions working on algae, marine fungi and bacteria. Biobanks and biorepositories contribute to the preservation of marine diversity by complementing traditional in situ conservation techniques with ex situ procedures that are safe and reproducible, and guarantee a long-term storage of biological specimens. An important EU Research Infrastructure is EMBRC (European Marine Biological Resource Centre, https://www.embrc.eu/) which coordinates numerous biobanks and biorepositories specifically dedicated to provide marine biological resources to academic and industrial researchers. These networks are becoming important tools to foster research and innovation in marine biotechnology, creating interconnected pipelines by providing capacity building and services to support research innovation and development, data management and sharing, education and knowledge transfer.
5.2.5 Digitisation and data sharing
An important aspect to consider and that relates to obtaining and maintaining cultures and biorepositories is the digitisation of metadata following FAIR (findable, accessible, interoperable and reusable) principles (Wilkinson et al., 2016), which is becoming a standard requirement in modern science. It negates the need for arduously storing hard copies of data, which often require space and proper storage. There is also the possibility that physical copies of datasheets may be misplaced or damaged. Digitisation of information not only ensures the long-term storage of data but allows for such information to be easily shared with academic or industrial collaborators, stakeholders or policy makers. Biodiversity collections and data sharing have been profoundly impacted by the digital era. Indeed, the use of technology for the organisation, digitisation and sharing of vast amounts of biodiversity-related data has accelerated scientific advancements. This is evidenced within biodiversity collections, that are becoming increasingly available to a broader audience, creating more opportunities for novel scientific discoveries and knowledge creation.
However, as membership and access to information and data within several existing (public or private) biodiversity collections are often linked to paid memberships, which can be of significant costs, alternative approaches have been developed to provide open access to data and information on marine resources. An example is the Ocean4Biotech COST Action (Rotter et al., 2021), where a web database linked to local or laboratory-specific biodiversity collections was developed (http://o4b.biomarep.org/). This initiative follows on from the development of marine biorepositories as described by Reddy et al. (2021). The central aim of the database was to organize, digitise, and store vast amounts of metadata linked to specimens, including but not limited to the name of the species, sampling locations, collection dates or methods used to sample. Specimens are also associated with photographs where necessary and all collections are linked to access and benefit sharing agreements such as the Nagoya protocol as well as local and regional permits requirements. This ensures that collections follow the necessary regulations and are transparent to all end-users. The Ocean4Biotech web database has limited the need for curation of species names by directly linking to the World Register of Marine Species, WORMS (https://www.marinespecies.org/). This means that the updated species naming is done automatically on the web database, as names are updated on WORMS. The Ocean4Biotech web database is also the first of its kind to allow for the integration of multiple sources of data such as DNA, chemistry or biological activity, enabling researchers to work in a multidisciplinary way. The web database will in the future include an integrative mapping tool allowing for the visualisation of species distributions over time and their potential applications. It will also include species or strains that can be easily accessed from partner labs, allowing for much broader research scopes than is achieved when working independently. In other words, samples or strains can be added to any study to achieve a broader phylogenetic or geographic scope. This tool will be invaluable for conservationists and policy makers. It currently also includes a tracking function which allows all users to trace samples and receive live feedback as results are generated. We also ensured that all data, such as DNA and metabolites were publicly accessible on GenBank or MetaboLights.
The Ocean4Biotech web database platform is currently available to members of the recently concluded COST Action and has resulted in the development of satellites from 12 countries ranging from Ireland to South Africa (Figure 8). Each satellite web database is fully visible to its lab members and collaborators and the access of information to the public can be easily controlled. For example, some research groups may prefer to only release information following the publication of those results. This platform has provided a standard web database management system that will prevent research groups from duplicating similar efforts which is both time consuming and costly.
Figure 8
Map showing the distribution of laboratories (i.e., the satellites) currently using the Ocean4Biotech web database. For more details visit: https://biomarep.org/.
6 Inclusion of a multidisciplinary approach
Sampling is the first step when establishing environmental monitoring campaigns or driving the biodiscovery processes, both of which entail long-term and financially-consuming collaborations. For example, the time frame of discovery and marketing of novel marine-derived compounds range from a minimum of several years to several decades, with costs that can amount to over $800 million (Ferrer et al., 2019; Reddy et al., 2021). During this long timeframe, several sampling campaigns may be organized, thus generating a comprehensive understanding of a particular environment (Knight et al., 2012) and, as such, inherently demanding interdisciplinary collaborations.
Collaboration between biologists, chemists, physicists, data analysts (statisticians, bioinformaticians), and data managers is necessary to integrate and utilise the information obtained and generate knowledge (Cristini et al., 2016; Rotter et al., 2021; Sigwart et al., 2021). Collaboration between different sectors includes (i) indigenous communities that can constitute part of the scientific collaborators rather than being considered as by-standers or local stakeholders (Claudie et al., 2012). (ii) Partnerships between academia and industry that share both risks and benefits (Semple et al., 2022). (iii) The involvement of legal and economic experts to help steer biodiscovery in the right direction by considering the economic feasibility of novel products/processes while taking into account the legal framework (Rotter et al., 2021). (iv) Collaboration with non-professionals (i.e., volunteers) through their commitment, enthusiasm and acting as local stakeholders (Pocock et al., 2015). In addition, collaborative agreements can be set in place to protect indigenous intellectual property (IP), establish mutual trust, confidentiality, and a mechanism for benefit sharing if any commercialisation results from the jointly generated IP (Harden-Davies, 2017; Semple et al., 2022).
Despite their importance, limited programs have been supported to date that allow minimal direct transdisciplinary and multidisciplinary interaction (Knight et al., 2012). Nevertheless, multidisciplinary approaches remain imperative to finance by promoting and defining a legal framework supporting research and innovation (Harden-Davies, 2017).
7 Scientific innovation in the field of sampling strategies for microbial diversity in the sea and ocean
As marine environments still remain largely underexplored and undervalued (Rotter et al., 2021), advances on developing new sampling strategies, either in terms of technology, processing or harmonization of techniques, are to be expected. Seawater sampling advances have been focusing on three levels: samplers, data and sampling management.
-
Sampling devices, capable of retaining pressure, which can accurately maintain the original physical and chemical properties of the sample, have been an ongoing area of research (Wang et al., 2024). The next generation of samplers already considers modular composition, where sampling modules are coupled with sensors to record physical ocean data, providing more information for environmental monitoring (Wang et al., 2024). These can also be automated, using robotic equipment. Nevertheless, the development trends in this area continuously demand education, maintenance, update of hardware or software, technical support, manufacturing and simplification of equipment use. There are many examples of such automated platforms that collect in situ data generating continuous measurements of physical-chemical and biological data that are constantly upgraded. It is an innovative field with potential for creating niche specializations, and generating comparable sets of global data used in many circumglobal expeditions and projects offering in-depth insight in the world oceans of microbial life (Karsenti et al., 2011; Cetinić et al., 2024), carbon export (Brewin et al., 2023; Siegel et al., 2023) and biogeochemical cycles (Schlitzer et al., 2018).
-
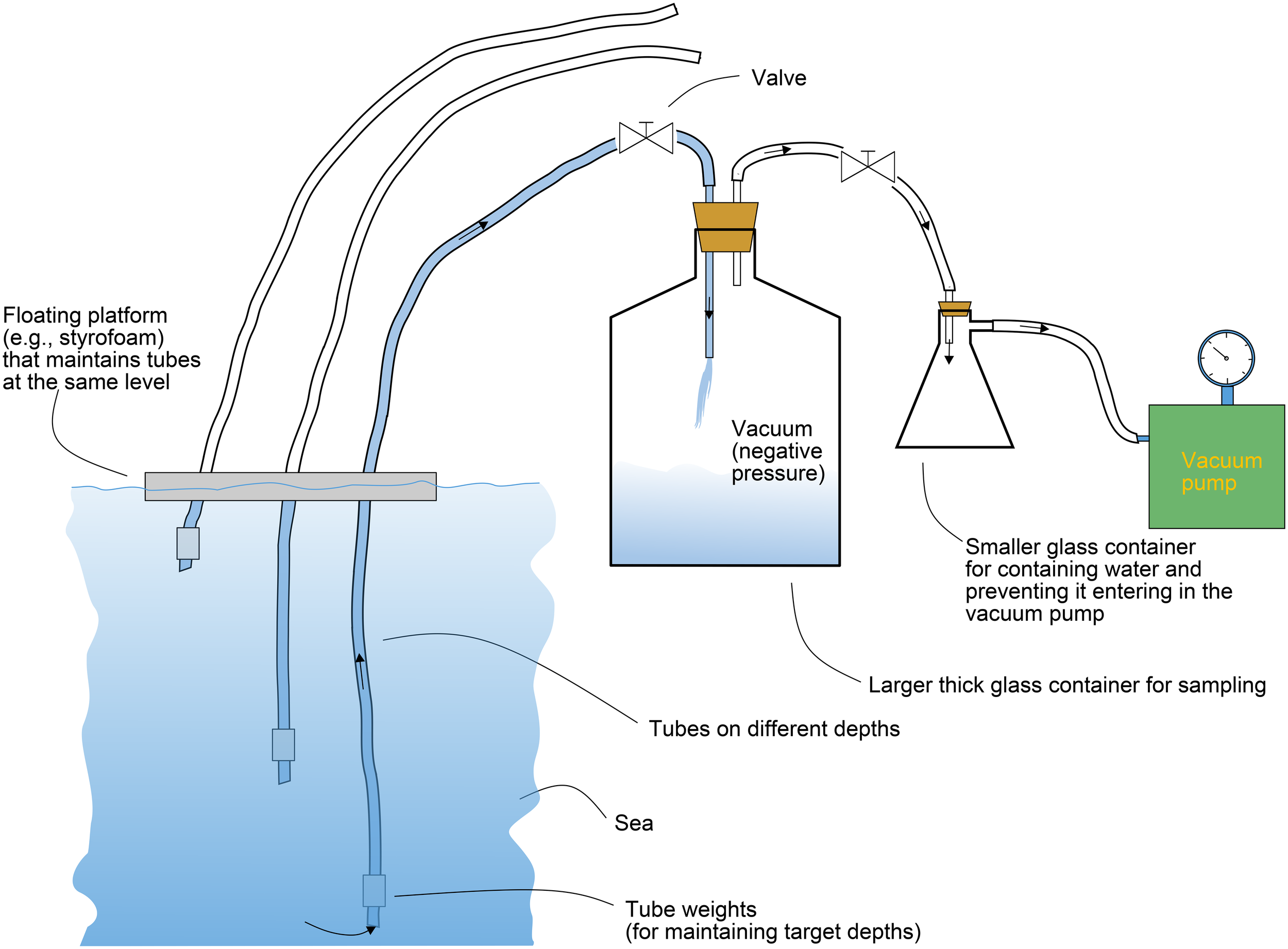
There is a clear need for adapting the samplers for representative sampling. An example that is being applied at the Marine Biology Station in Piran (National Institute of Biology), Slovenia illustrates compartmentalized sampling in water column, targeting samples in specific depths. Here, improvised vacuum samplers can be constructed. The strategy here is to insert a thin tube in the water column which will collect water samples in the first container. When the desired depth is reached, the first water container is discarded and the sampling tube is attached to the second one, thus ensuring that the sample from the desired depth alone is acquired (Figure 9). This guarantees that the sample remains intact and representative for the specific depth of interest, while protecting the pump. As seen in Figure 9, these vacuum pumps can be self-made, but they also represent an innovative potential for niche production.
-
Specific environmental conditions or emergent societal challenges demand quick adaptations to introduce novel samplers and sampling techniques. One such example is the mucilage phenomenon, i.e., gelatinous aggregates of extracellular organic matter of phytoplanktonic origin that are sporadically occurring globally under special seasonal and trophic conditions (Mecozzi et al., 2001; Ricci et al., 2014). Scientifically, these aggregates have sparked interest in discovering the causes and the predictions for their formation, but they could also present environmental challenges due to potentially representing a substrate for microbial colonization, including pathogenic forms, or causing anoxia and death of species and negative economic impacts for fisheries and tourism (Danovaro et al., 2009; Kraus and Ivošević DeNardis, 2023). The process of mucilage formation is still not fully understood (Turk et al., 2010). Therefore, when mucilage aggregates occur, they are surveyed on the surface, as well as underwater using cameras (Öztürk et al., 2021). This provides supporting data for monitoring, while still demanding effective sampling solutions. For example, one solution is using syringes and experienced SCUBA divers (Figure 10, top), as a simple solution and quick response to this phenomenon and obtaining valuable samples of this viscous material. When larger volumes need to be sampled, syringe scale up can be constructed in house (Figure 10, bottom). Another example that well illustrates the reaction to societal challenges is provided by development of samplers for microplastics on water surfaces. Here, the challenge was to develop floating samplers so only the surface layers are sampled. One such protocol using manta nets and spinnaker booms was recently developed by Kovač Viršek et al. (2016) (Figure 11). Such samples can provide information about microplastic abundance, but also potential information on the plastisphere, which harbours different microbial communities or such with different metabolic mechanisms than the surrounding water, potentially also pathogenic ones (Du et al., 2022).
-
Sampling management is being increasingly acknowledged and considerations on filtering time, sample storage, temperature, contamination prevention, storage conditions, sampling volumes, preservation conditions, costs, DNA extraction protocols, are now planned in advance (Pascoal et al., 2023; Patin and Goodwin, 2023), rather than decided ad hoc, possibly also harmonized, recorded and shared to enable a better comparison and interpretation of results from different sampling campaigns. Sampling materials also need to be taken into consideration. For example, if sampling the plastisphere, no plastic equipment or containers should be used, while on the other hand, if sampling, e.g., oil spills in search of potential microbial degradation therein and heavy metal pollution, plastic containers are preferred. Organizations that have sampling/monitoring at the core of their activities, could also design sampling routes in the plans of buildings, where one route is used for sterile equipment before sampling, while the other is used to bring and process samples post-sampling.
-
Biorepositories represent a significant advancement in biodiversity collections, representing a major innovation in itself (Reddy et al., 2021). They go beyond this by integrating modern technologies for data management, sample processing, and accessibility. In future, the automated processing of biological data, alongside cloud-based storage, will improve the efficiency and scalability of biorepositories.
Figure 9
Inhouse-made vacuum pumps and collectors with the specific use for sampling on specific water depths.
Figure 10
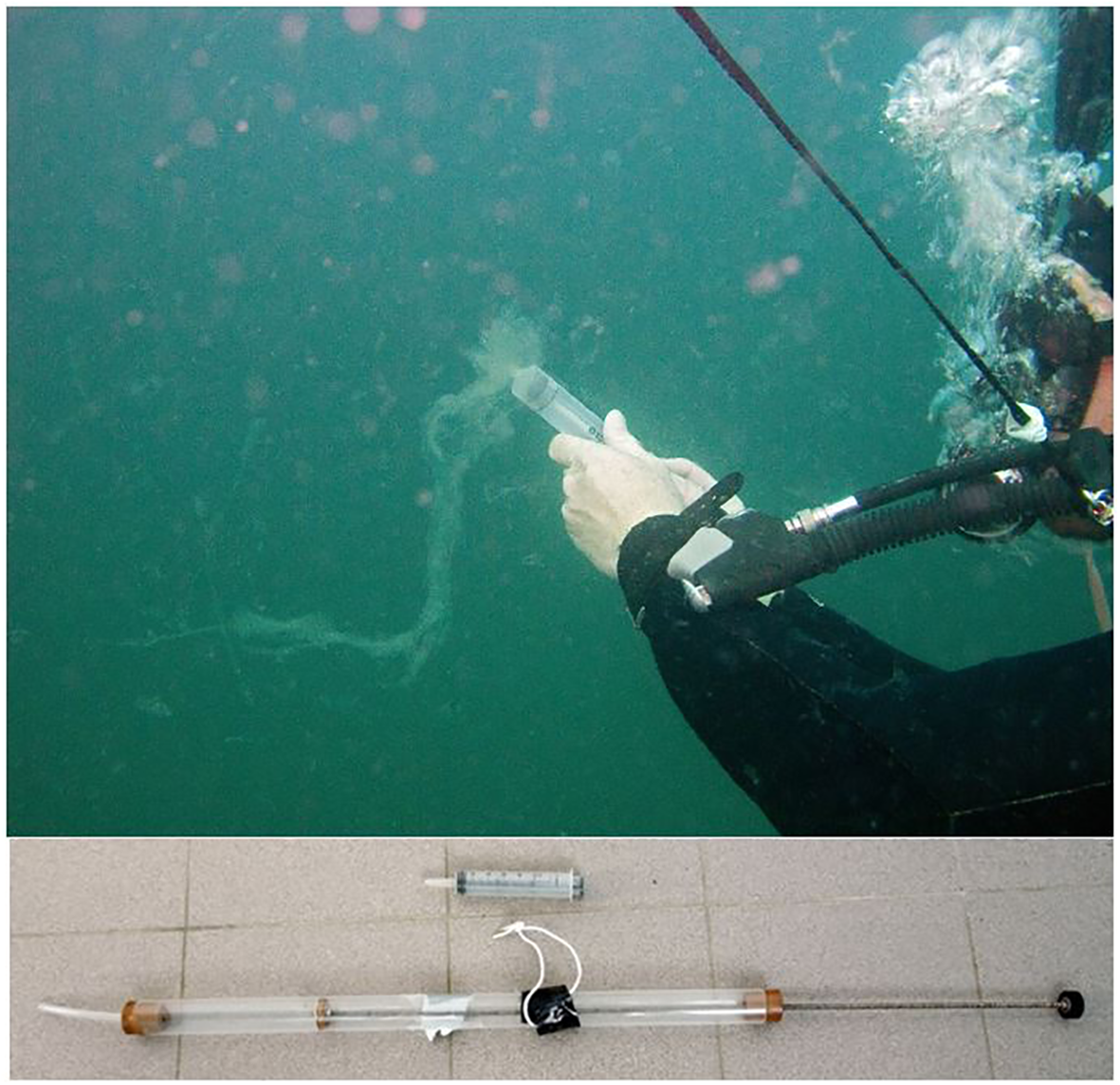
Underwater sampling of mucilage aggregates proposing a simple solution of using syringes (in smaller; top and larger; bottom) scales and experienced SCUBA divers.
Figure 11

Methodology for collecting microplastic samples from water surface, developed by Kovač Viršek, (2016). The full video is available from this reference.
Furthermore, as with many biological research facilities, there is increasing emphasis on the greening of processes and screening of samples. By reusing of equipment, wherever possible, biorepositories can reduce their environmental footprint. Additionally, research into more energy-efficient methods of cryopreservation will become increasingly important. More sustainable practices will also be explored in terms of preservatives and extraction buffers, with the goal of developing green alternatives that minimize the environmental impact while maintaining the long-term integrity fo biological samples. One exciting direction for biorepositories is decentralization, as demonstrated by initiatives such Ocean4Biotech, that was already introduced above. This approach involves distributing collections across various locations, which not only makes samples more accessible to a wider range of researchers and institutions but also improves the collaboration of biorepository networks. Decentralization encourages collaboration and ensures that valuable biodiversity data is available to a larger number of users globally (Reddy et al., 2021). To sustain and further expand these innovations, national ownership of biorepositories, coupled with funding from governments or international agencies, will be crucial. Government support can ensure that biorepositories are adequately funded and maintained over the long term, and that they remain accessible to the scientific community, industry partners and the public, facilitating the long-term preservation of biodiversity in the face of environmental and climate challenges (Reddy et al., 2021).
8 Legislation
Marine genetic resources (MGR) are currently the focus of great interest in science and biotechnology. However, for basic research and for any further knowledge exploitation, several international regulations have to be considered (Schneider et al., 2022). The Convention on Biological Diversity (CBD) is an international multilateral treaty imposing fair and equitable sharing of genetic resources. According to the Nagoya Protocol (NP), a specific CBD agreement published in 2010, any exchange of genetic materials accessed on or after October 12th, 2014 from a country of origin that is a Party to the Nagoya Protocol, must follow a strict set of rules and protocols. These include prior consent, material transfer agreements and/or Internationally Recognised Certificate of Compliance (IRCC) to ensure that genetic resources are shared fairly across different countries and their people. More information is available at the Access and Benefit-Sharing Clearing-House (ABSCH), a platform for exchanging information and a key tool for facilitating the implementation of the Nagoya Protocol (https://absch.cbd.int/).
When MGR are obtained from ex situ collections, the collection should have already acquired all the documentation for the acquisition of the resources in compliance with international regulations. When the MGR is accessed from in situ (sampling expedition), in addition to the CBD and NP, the United Nations Convention on the Law of the Sea (UNCLOS) provision has to be considered when deep sea and areas beyond national jurisdiction (ABNJ) are sampled. These regions represent the largest environments on the planet, yet are still unexplored (Rabone et al., 2019; Schneider et al., 2023).
When MGR are accessed from ex situ collections, the CBD and NP provisions apply since collections act as intermediaries in the accession, exploration and utilisation of genetic resources. Useful information can be found in the MIRRI Best Practice Manual on Access and Benefit Sharing, ABS (https://absch.cbd.int/database/A19A20/ABSCH-A19A20-SCBD-208213). This tool was designed to provide guidance for mBRCs to implement ABS institutional policies with regard to genetic resources and associated traditional knowledge, and more specifically for: (i) Managing acquisition of biological material for accession into the public collection. (ii) Transfer of material from the public collection to other mBRCs or third parties, and the delivery of other services. (iii) Access and internal use of material and benefit sharing by the mBRC. (iv) Managing documentation and data. (v) Training and awareness-raising. (vi) Managing secured collections (Microbial Resource Research Infrastructure, 2012).
Culture collections can apply for Registered Collection status giving them a legal certainty when sharing cultures. Enforcement of the NP has already been proven to slow down the number of deposits of microbial resources (Dedeurwaerdere, 2010). Similarly, an internal survey conducted by the COST Action CA18238 Ocean4Biotech also demonstrated that while researchers and innovators are determined to follow the rules and collect samples responsibly and ethically, their limited understanding of the NP, long waiting and handling times and limited contact with the National Focal Points in the country of origin have proven inhibitory to research and biodiscovery. In addition, some controversial topics, such as sharing digital material (e.g., genome sequences of collected samples) are still being discussed by the CBD, which only adds hesitation to resource sharing altogether. Additional documentation when sharing cultures across countries (e.g., phyco-certificates for algae) can be costly for culture collections operating on small budgets.
9 Conclusion
The understanding of marine habitats and the biomass demand for the responsible use of marine microbial resources and a commitment to conserve microbial biodiversity and ecosystems. The starting point for microbial biodiscovery and subsequent activities, leading to knowledge acquisition or innovative results, is the sampling of habitats. Currently, there is a limited number of harmonized publications or guidelines that are available for the international community when sampling marine habitats, either for biodiversity (ecology purposes) or biodiscovery (biotechnology purposes). To address the lack of resources, we propose a checklist that will help to effectively plan any sampling campaign targeting microbial (Figure 1). These should be categorized into pre-sampling, sampling and post-sampling activities. When careful planning is considered, the costs and effort can be optimized, allowing the already long, expensive and laborious sampling campaigns to provide effective microbial samples that can be used for immediate analyses or appropriately stored until further use. Finally, the proposed sampling operational workflow (Figure 1) allows for future integration of samples, data and results from different laboratories by minimizing sampling bias and resulting errors that can result from inadequate campaigns. This article also introduces a novel, openly available biorepository developed within Ocean4Biotech COST Action, where researchers and industrial stakeholders have a direct overview and communication with experts that are collecting and storing data from marine environments, primarily around Europe.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
ZL: Writing – original draft, Visualization, Supervision, Writing – review & editing, Conceptualization. MG: Writing – original draft, Visualization, Conceptualization, Supervision, Writing – review & editing. FA: Writing – review & editing. AB: Writing – review & editing. AD: Writing – review & editing. DD: Writing – review & editing. SF: Writing – review & editing. SG: Writing – review & editing. FG: Writing – review & editing. AK: Writing – review & editing. TM: Writing – review & editing, Visualization, Methodology. LN: Visualization, Writing – review & editing. DO: Writing – review & editing. IS: Writing – review & editing. JS: Writing – review & editing. OT: Writing – review & editing. MR: Writing – review & editing. GV: Writing – review & editing. MV: Writing – review & editing, Writing – original draft. AR: Supervision, Writing – review & editing, Writing – original draft, Funding acquisition, Visualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Croatian Science Foundation under the projects ISLAND (IP-2020-02-9524); national funds from FCT—Fundação para a Ciência e a Tecno-logia, IP, in the scope of the project UIDP/04378/2020 of the Research Unit on Applied Molecular Biosciences–UCIBIO and the project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy–i4HB and the project Laboratory Institute for Health and Bioeconomy–i4HB, and by Government of the Canary Islands – PROMOTUR Turismo de Canarias (Actuaciones de Cohesión de Destino - Canarias Ecoínsulas II), project SP3 “Islas Canarias, naturaleza marina singular: Salud y bienestar”; this publication was produced with financial assistance of the Interreg MED Programme, co-financed by the European Regional Development Fund (Project No. 8MED20_4.1_SP_001, internal ref. 8MED20_4.1_SP_001)—B-Blue project; the article was produced with the financial assistance of the Interreg Euro-MED Programme, co-financed by the European Union (Projects no. Euro-MED0300730—C4Nature - Community4Nature project and Euro-MED 0200514) –2B-BLUE). The authors acknowledge the financial support of the Slovenian Research and Innovation Agency (research core funding No. P4-0432, P1-0237, and the infrastructure program I0-0004 and the project L4-4564). This publication is based upon work from COST Action CA18238 (Ocean4Biotech) and Ocean4Biotech, the professional association joining marine biotechnologists, supported by COST (European Cooperation in Science and Technology) program.
Conflict of interest
Author LN was employed by ScotBio.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1597865/full#supplementary-material
References
1
Aguilera A. Keafer B. Rau G. Anderson D. (2002). Immunomagnetic isolation of live and preserved Alexandrium fundyense cells: species-specific physiological, chemical, and isotopic analyses. Mar. Ecol. Prog. Ser.237, 65–78. doi: 10.3354/meps237065
2
Albert C. H. Thuiller W. Yoccoz N. G. Soudant A. Boucher F. Saccone P. et al . (2010). Intraspecific functional variability: extent, structure and sources of variation. J. Ecol.98, 604–613. doi: 10.1111/j.1365-2745.2010.01651.x
3
Amalfitano S. Fazi S. (2008). Recovery and quantification of bacterial cells associated with streambed sediments. J. Microbiological Methods75, 237–243. doi: 10.1016/j.mimet.2008.06.004
4
Amann R. Lemmer H. Wagner M. (1998). Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol. Ecol.25, 205–215. doi: 10.1111/j.1574-6941.1998.tb00473.x
5
Babić I. Petrić I. Bosak S. Mihanović H. Radić I. D. Ljubešić Z. (2017). Distribution and diversity of marine picocyanobacteria community: Targeting of Prochlorococcus ecotypes in winter conditions (southern Adriatic Sea). Mar. Genomics36, 3–11. doi: 10.1016/j.margen.2017.05.014
6
Barbour M. T. (1999). Rapid bioassessment protocols for use in wadeable streams and rivers: periphyton, benthic macroinvertebrates and fish (Washington, DC, USA: US Environmental Protection Agency, Office of Water).
7
Batley G. E. Simpson S. L. (2016). “Sediment sampling, sample preparation and general analysis,” in Sediment quality assessment: a practical guide, 2nd ed (CSIRO Publishing, Clayton), 15–46.
8
Battin T. J. Kaplan L. A. Findlay S. Hopkinson C. S. Marti E. Packman A. I. et al . (2008). Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci1, 95–100. doi: 10.1038/ngeo101
9
Braun B. Richert I. Szewzyk U. (2009). Detection of iron-depositing Pedomicrobium species in native biofilms from the Odertal National Park by a new, specific FISH probe. J. Microbiological Methods79, 37–43. doi: 10.1016/j.mimet.2009.07.014
10
Brewin R. J. W. Sathyendranath S. Kulk G. Rio M.-H. Concha J. A. Bell T. G. et al . (2023). Ocean carbon from space: Current status and priorities for the next decade. Earth-Science Rev.240, 104386. doi: 10.1016/j.earscirev.2023.104386
11
British Standards Institute (BSI) (2011). “Water quality,” in Guidance on quantitative and qualitative investigations of marine phytoplankton (BS EN 15972:2011). (London, UK: British Standards Institute (BSI)).
12
Bruce K. Blackman R. Bourlat S. J. Hellström A. M. Bakker J. Bista I. et al . (2021). A practical guide to DNA-based methods for biodiversity assessment (Sofia, Bulgaria: Pensoft Publishers). doi: 10.3897/ab.e68634
13
Callahan B. J. McMurdie P. J. Holmes S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J.11, 2639–2643. doi: 10.1038/ismej.2017.119
14
Camacho-Rodríguez J. Cerón-García M. C. Macías-Sánchez M. D. Fernández-Sevilla J. M. López-Rosales L. Molina-Grima E. (2016). Long-term preservation of concentrated Nannochloropsis gaditana cultures for use in aquaculture. J. Appl. Phycol28, 299–312. doi: 10.1007/s10811-015-0572-y
15
Carmichael W. W. Drapeau C. Anderson D. M. (2000). Harvesting of Aphanizomenon flos-aquae Ralfs ex Born. & Flah. var. flos-aquae (Cyanobacteria) from Klamath Lake for human dietary use. J. Appl. phycology12, 585–595. doi: 10.1023/A:1026506713560
16
Cetinić I. Rousseaux C. S. Carroll I. T. Chase A. P. Kramer S. J. Werdell P. J. et al . (2024). Phytoplankton composition from sPACE: Requirements, opportunities, and challenges. Remote Sens. Environ.302, 113964. doi: 10.1016/j.rse.2023.113964
17
Chaudhary D. K. Khulan A. Kim J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep.9, 1–11. doi: 10.1038/s41598-019-43182-x
18
Chen J.-W. Fan W. Bingham B. Chen Y. Gu L.-Y. Li S.-L. (2013). A long gravity-piston corer developed for seafloor gas hydrate coring utilizing an in situ pressure-retained method. Energies6, 3353–3372. doi: 10.3390/en6073353
19
Clarke L. J. Soubrier J. Weyrich L. S. Cooper A. (2014). Environmental metabarcodes for insects: in silicoPCR reveals potential for taxonomic bias. Mol. Ecol. Resour.14, 1160–1170. doi: 10.1111/1755-0998.12265
20
Claudie D. J. Semple S. J. Smith N. M. Simpson B. S. (2012). “Ancient but new: developing locally driven enterprises based on traditional medicines in kuuku I’yu Northern Kaanju homelands, Cape York, Queensland, Australia,” in Indigenous People’s Innovation: Intellectual Property Pathways to Development. (Canberra, Australia: ANU E Press), 29–55.
21
Coenye T. Bové M. Bjarnsholt T. (2022). Biofilm antimicrobial susceptibility through an experimental evolutionary lens. NPJ Biofilms Microbiomes8, 82. doi: 10.1038/s41522-022-00346-4
22
Compson Z. G. McClenaghan B. Singer G. A. C. Fahner N. A. Hajibabaei M. (2020). Metabarcoding from microbes to mammals: comprehensive bioassessment on a global scale. Front. Ecol. Evol.8. doi: 10.3389/fevo.2020.581835
23
Costa O. Y. A. De Hollander M. Pijl A. Liu B. Kuramae E. E. (2020). Cultivation-independent and cultivation-dependent metagenomes reveal genetic and enzymatic potential of microbial community involved in the degradation of a complex microbial polymer. Microbiome8, 76. doi: 10.1186/s40168-020-00836-7
24
Costello M. J. Coll M. Danovaro R. Halpin P. Ojaveer H. Miloslavich P. (2010). A census of marine biodiversity knowledge, resources, and future challenges. PloS One5, e12110. doi: 10.1371/journal.pone.0012110
25
Cristini L. Lampitt R. S. Cardin V. Delory E. Haugan P. O’Neill N. et al . (2016). Cost and value of multidisciplinary fixed-point ocean observatories. Mar. Policy71, 138–146. doi: 10.1016/j.marpol.2016.05.029
26
Daniel A. Laës-Huon A. Barus C. Beaton A. D. Blandfort D. Guigues N. et al . (2020). Toward a harmonization for using in situ nutrient sensors in the marine environment. Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00773
27
Danovaro R. Fonda Umani S. Pusceddu A. (2009). Climate change and the potential spreading of marine mucilage and microbial pathogens in the Mediterranean sea. PloS One4, e7006. doi: 10.1371/journal.pone.0007006
28
David M. Gollasch S. (2015). BALMAS Ballast Water Sampling Protocol for Compliance Monitoring and Enforcement of the BWM Convention and Scientific Purposes (Korte, Slovenia, Hamburg, Germany: BALMAS project), 55 pp.
29
Dedeurwaerdere T. (2010). Global microbial commons: institutional challenges for the global exchange and distribution of microorganisms in the life sciences. Res. Microbiol.161, 414–421. doi: 10.1016/j.resmic.2010.04.012
30
Deiner K. Bik H. M. Mächler E. Seymour M. Lacoursière-Roussel A. Altermatt F. et al . (2017). Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol.26, 5872–5895. doi: 10.1111/mec.14350
31
De Vargas C. Audic S. Henry N. Decelle J. Mahé F. Logares R. et al . (2015). Eukaryotic plankton diversity in the sunlit ocean. Science348, 1261605. doi: 10.1126/science.1261605
32
De Vero L. Boniotti M. B. Budroni M. Buzzini P. Cassanelli S. Comunian R. et al . (2019). Preservation, characterization and exploitation of microbial biodiversity: the perspective of the Italian network of culture collections. Microorganisms7, 685. doi: 10.3390/microorganisms7120685
33
DiEuliis D. Johnson K. R. Morse S. S. Schindel D. E. (2016). Specimen collections should have a much bigger role in infectious disease research and response. Proc. Natl. Acad. Sci. U.S.A.113, 4–7. doi: 10.1073/pnas.1522680112
34
Du Y. Liu X. Dong X. Yin Z. (2022). A review on marine plastisphere: biodiversity, formation, and role in degradation. Comput. Struct. Biotechnol. J.20, 975–988. doi: 10.1016/j.csbj.2022.02.008
35
Durkin C. A. Buesseler K. O. Cetinić I. Estapa M. L. Kelly R. P. Omand M. (2021). A visual tour of carbon export by sinking particles. Global Biogeochemical Cycles35, e2021GB006985. doi: 10.1029/2021GB006985
36
Durkin C. A. Cetinić I. Estapa M. Ljubešić Z. Mucko M. Neeley A. et al . (2022). Tracing the path of carbon export in the ocean though DNA sequencing of individual sinking particles. ISME J.16, 1896–1906. doi: 10.1038/s41396-022-01239-2
37
Edet U. Antai S. Brooks A. Asitok A. Enya O. Japhet F. (2017). An overview of cultural, molecular and metagenomic techniques in description of microbial diversity. JAMB7, 1–19. doi: 10.9734/jamb/2017/37951
38
Escalante-Maldonado O. Kayali A. Y. Yamazaki W. Vuddhakul V. Nakaguchi Y. Nishibuchi M. (2015). Improvement of the quantitation method for the tdh+ Vibrio parahaemolyticus in molluscan shellfish based on most-probable- number, immunomagnetic separation, and loop-mediated isothermal amplification. Front. Microbiol.6. doi: 10.3389/fmicb.2015.00270
39
Estapa M. Valdes J. Tradd K. Sugar J. Omand M. Buesseler K. (2020). The neutrally buoyant sediment trap: two decades of progress. J. Atmospheric Oceanic Technol.37, 957–973. doi: 10.1175/JTECH-D-19-0118.1
40
Fahey D. W. Doherty S. J. Hibbard K. A. Romanou A. Taylor P. C. Wuebbles D. J. et al . (2017). “Ch. 2: physical drivers of climate change,” in Climate Science Special Report: Fourth National Climate Assessment, Volume I (Washington, DC, USA: U.S. Global Change Research Program). doi: 10.7930/J0513WCR
41
Fazi S. Amalfitano S. Pernthaler J. Puddu A. (2005). Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ. Microbiol.7, 1633–1640. doi: 10.1111/j.1462-2920.2005.00857.x
42
Fazi S. Amalfitano S. Pizzetti I. Pernthaler J. (2007). Efficiency of fluorescence in situ hybridization for bacterial cell identification in temporary river sediments with contrasting water content. Systematic Appl. Microbiol.30, 463–470. doi: 10.1016/j.syapm.2007.03.003
43
Ferrer M. Méndez-García C. Bargiela R. Chow J. Alonso S. García-Moyano A. et al . (2019). Decoding the ocean’s microbiological secrets for marine enzyme biodiscovery. FEMS Microbiol. Lett.366, 0378–1097. doi: 10.1093/femsle/fny285
44
Field C. B. Behrenfeld M. J. Randerson J. T. Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. science281, 237–240. doi: 10.1126/science.281.5374.237
45
Flombaum P. Gallegos J. L. Gordillo R. A. Rincón J. Zabala L. L. Jiao N. et al . (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A.110, 9824–9829. doi: 10.1073/pnas.1307701110
46
Florio Furno M. Poli A. Ferrero D. Tardelli F. Manzini C. Oliva M. et al . (2022). The culturable mycobiota of sediments and associated microplastics: from a harbor to a marine protected area, a comparative study. JoF8, 927. doi: 10.3390/jof8090927
47
Foo S. C. Mok C. Y. Ho S. Y. Khong N. M. H. (2023). Microalgal culture preservation: Progress, trends and future developments. Algal Res.71, 103007. doi: 10.1016/j.algal.2023.103007
48
Frenea-Robin M. Marchalot J. (2022). Basic principles and recent advances in magnetic cell separation. Magnetochemistry8, 11. doi: 10.3390/magnetochemistry8010011
49
Galitz A. Ekins M. Reddy M. M. Folcher E. Dumas M. Butscher J. et al . (2024). Molecular genetic biodiversity assessment of the Wallis Island sponge fauna in the Tropical Pacific. J. Mar. Biol. Ass.104, e56. doi: 10.1017/S0025315424000432
50
Gorsky G. Bourdin G. Lombard F. Pedrotti M. L. Audrain S. Bin N. et al . (2019). Expanding Tara oceans protocols for underway, ecosystemic sampling of the ocean-atmosphere interface during Tara pacific expedition, (2016–2018). Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00750
51
Gröndahl F. (2009). Removal of surface blooms of the cyanobacteria Nodularia spumigena: A pilot project conducted in the Baltic Sea. AMBIO: A J. Hum. Environ.38, 79–84. doi: 10.1579/0044-7447-38.2.79
52
Grossmann L. Ebert S. Hinrichs J. Weiss J. (2018). Effect of precipitation, lyophilization, and organic solvent extraction on preparation of protein-rich powders from the microalgae Chlorella protothecoides. Algal Res.29, 266–276. doi: 10.1016/j.algal.2017.11.019
53
Guldhe A. Singh B. Rawat I. Ramluckan K. Bux F. (2014). Efficacy of drying and cell disruption techniques on lipid recovery from microalgae for biodiesel production. Fuel128, 46–52. doi: 10.1016/j.fuel.2014.02.059
54
Harden-Davies H. (2017). Deep-sea genetic resources: New frontiers for science and stewardship in areas beyond national jurisdiction. Deep Sea Res. Part II: Topical Stud. Oceanography137, 504–513. doi: 10.1016/j.dsr2.2016.05.005
55
He S. Peng Y. Jin Y. Wan B. Liu G. (2020). Review and analysis of key techniques in marine sediment sampling. Chin. J. Mech. Eng.33, 66. doi: 10.1186/s10033-020-00480-0
56
Hewitt C. L. Martin R. B. (2001). Revised protocols for baseline port surveys for introduced marine species-survey design, sampling protocols and specimen handling. Hobart: CSIRO Marine Research.
57
Hofer U. (2018). The majority is uncultured. Nat. Rev. Microbiol.16, 716–717. doi: 10.1038/s41579-018-0097-x
58
Hollister C. D. Silva A. J. Driscoll A. (1973). A giant piston-corer. Ocean Eng.2, 159–168. doi: 10.1016/0029-8018(73)90001-2
59
Hopkins A. (2007). Recommended operating guidelines (ROG) for box coring. Available online at: https://emodnet.ec.europa.eu/sites/emodnet.ec.europa.eu/files/public/ (Accessed February 9, 2025).
60
Hoshino T. Doi H. Uramoto G.-I. Wörmer L. Adhikari R. R. Xiao N. et al . (2020). Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U.S.A.117, 27587–27597. doi: 10.1073/pnas.1919139117
61
Ingebrigtsen R. A. Hansen E. Andersen J. H. Eilertsen H. C. (2017). Field sampling marine plankton for biodiscovery. Sci. Rep.7, 15863. doi: 10.1038/s41598-017-15980-8
62
Institute for Standardization of Montenegro (2014). Water quality - Guidance standard on the enumeration of phytoplankton using inverted microscopy (Utermöhl technique) (MEST EN 15204:2014).
63
International Maritime Organization (2004). International convention for the control and management of ships’ Ballast water and sediments (BWM).
64
International Maritime Organization (2007). Guidelines for risk assessment under regulation A-4 of the BWM convention (G7) (Resolution MEPC.162(56)).
65
Janssens D. Arahal D. R. Bizet C. Garay E. (2010). The role of public biological resource centers in providing a basic infrastructure for microbial research. Res. Microbiol.161, 422–429. doi: 10.1016/j.resmic.2010.03.009
66
Joint I. Mühling M. Querellou J. (2010). Culturing marine bacteria – an essential prerequisite for biodiscovery. Microbial Biotechnol.3, 564–575. doi: 10.1111/j.1751-7915.2010.00188.x
67
Jones D. O. B. (2009). Using existing industrial remotely operated vehicles for deep-sea science. Zoologica Scripta38, 41–47. doi: 10.1111/j.1463-6409.2007.00315.x
68
Jung D. Liu L. He S. (2021). Application of in situ cultivation in marine microbial resource mining. Mar. Life Sci. Technol.3, 148–161. doi: 10.1007/s42995-020-00063-x
69
Kamjam M. Sivalingam P. Deng Z. Hong K. (2017). Deep sea actinomycetes and their secondary metabolites. Front. Microbiol.8. doi: 10.3389/fmicb.2017.00760
70
Kamjam M. Xie Q. Deng Z. Hong K. (2018). Isolation and diversity of actinomycetes from sediments of different depths between 34m and 3235m in south China sea. Chiang Mai J. Sci.45, 1595–1609.
71
Kanjer L. Majewska R. Van De Vijver B. Gračan R. Lazar B. Bosak S. (2020). Diatom diversity on the skin of frozen historic loggerhead sea turtle specimens. Diversity12, 383. doi: 10.3390/d12100383
72
Kapinusova G. Lopez Marin M. A. Uhlik O. (2023). Reaching unreachables: Obstacles and successes of microbial cultivation and their reasons. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1089630
73
Karsenti E. Acinas S. G. Bork P. Bowler C. De Vargas C. Raes J. et al . (2011). A holistic approach to marine eco-systems biology. PloS Biol.9, e1001177. doi: 10.1371/journal.pbio.1001177
74
Karst S. M. Ziels R. M. Kirkegaard R. H. Sørensen E. A. McDonald D. Zhu Q. et al . (2021). High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat. Methods18, 165–169. doi: 10.1038/s41592-020-01041-y
75
Keck F. Peller T. Alther R. Barouillet C. Blackman R. Capo E. et al . (2025). The global human impact on biodiversity. Nature641, 395–400. doi: 10.1038/s41586-025-08752-2
76
Knauer G. A. Martin J. H. Bruland K. W. (1979). Fluxes of particulate carbon, nitrogen, and phosphorus in the upper water column of the northeast Pacific. Deep Sea Res. Part A. Oceanographic Res. Papers26, 97–108. doi: 10.1016/0198-0149(79)90089-X
77
Knight R. Jansson J. Field D. Fierer N. Desai N. Fuhrman J. A. et al . (2012). Unlocking the potential of metagenomics through replicated experimental design. Nat. Biotechnol.30, 513–520. doi: 10.1038/nbt.2235
78
Knight-Jones E. W. (1951). Preliminary studies of nanoplankton and ultraplankton systematics and abundance by a quantitative culture method. ICES J. Mar. Sci.17, 140–155. doi: 10.1093/icesjms/17.2.140
79
Kraus R. Ivošević DeNardis N. (2023). Tracking the spatio-temporal distribution of organic particles to predict macroaggregation in the northern Adriatic Sea. Water15, 1665. doi: 10.3390/w15091665
80
Kreiner A. Bolton J. Branch G. Lima F. Reddy M. Serrao E. et al . (2019a). Coastal biodiversity surveys - survey 1. doi: 10.13140/RG.2.2.33130.08648
81
Kreiner A. Branch G. Ferreira L. Reddy M. Serrao E. A. Thomas M. (2019b). Coastal biodiversity surveys - survey 3. doi: 10.13140/RG.2.2.23063.75686
82
Kreiner A. Reddy M. Seabra R. Thomas M. (2019c). Coastal biodiversity surveys - survey 2. doi: 10.13140/RG.2.2.29774.64322
83
Kružić P. Sršen P. Benković L. (2012). The impact of seawater temperature on coral growth parameters of the colonial coral Cladocora caespitosa (Anthozoa, Scleractinia) in the eastern Adriatic Sea. Facies58, 477–491. doi: 10.1007/s10347-012-0306-4
84
Lauritano C. Andersen J. H. Hansen E. Albrigtsen M. Escalera L. Esposito F. et al . (2016). Bioactivity screening of microalgae for antioxidant, anti-inflammatory, anticancer, anti-diabetes, and antibacterial activities. Front. Mar. Sci.3. doi: 10.3389/fmars.2016.00068
85
Lee C. M. Askari F. Book J. Carniel S. Cushman-Roisin B. Dorman C. et al . (2005). Northern Adriatic response to a wintertime bora wind event. EoS Trans.86, 157–165. doi: 10.1029/2005EO160001
86
Leese F. Altermatt F. Bouchez A. Ekrem T. Hering D. Meissner K. et al . (2016). DNAqua-Net: Developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. RIO2, e11321. doi: 10.3897/rio.2.e11321
87
Liu X. Bian Z. Hu S. Dickinson C. F. Benjamin M. M. Jia J. et al . (2024a). The chemistry of phytoplankton. Chem. Rev.124, 13099–13177. doi: 10.1021/acs.chemrev.4c00177
88
Liu Y. Wang Z. Wang Z. Zhou J. Han J. Lu C. et al . (2024b). Rapid and simultaneous multiepitope antigen-based detection of Enterococcus by microscale thermophoresis and immunomagnetic separation. Front. Microbiol.15, 1341451. doi: 10.1016/j.heliyon.2024.e37788
89
Ljubešić Z. Mihanović H. Matek A. Mucko M. Achterberg E. P. Omand M. et al . (2024). Marine plankton community and net primary production responding to island-trapped waves in a stratified oligotrophic ecosystem. Heliyon10, e37788. doi: 10.1016/j.heliyon.2024.e37788
90
Logares R. (2024). Decoding populations in the ocean microbiome. Microbiome12, 67. doi: 10.1186/s40168-024-01778-0
91
Lučić D. Ljubešić Z. Babić I. Bosak S. Cetinić I. Vilibić I. et al . (2017). Unusual winter zooplankton bloom in the open southern Adriatic Sea. Turk J. Zool41, 1024–1035. doi: 10.3906/zoo-1702-17
92
Lupini G. Proia L. Di Maio M. Amalfitano S. Fazi S. (2011). CARD-FISH and Confocal Laser Scanner Microscopy to assess successional changes of the bacterial community in freshwater biofilms. J. Microbiol. Methods86, 248–251.
93
Macher J. Vivancos A. Piggott J. J. Centeno F. C. Matthaei C. D. Leese F. (2018). Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome c oxidase I primers. Mol. Ecol. Resour.18, 1456–1468. doi: 10.1111/1755-0998.12940
94
MacIntyre H. L. Cullen J. J. (1996). Primary production by suspended and benthic microalgae in a turbid estuary: time-scales of variability in San Antonio Bay, Texas. Mar. Ecol. Prog. Ser.145, 245–268. doi: 10.3354/meps145245
95
MacIntyre H. L. Geider R. J. Miller D. C. (1996). Microphytobenthos: the ecological role of the “Secret garden” of unvegetated, shallow-water marine habitats. I. Distribution, abundance and primary production. Estuaries19, 186. doi: 10.2307/1352224
96
Martiny A. C. (2019). High proportions of bacteria are culturable across major biomes. ISME J.13, 2125–2128. doi: 10.1038/s41396-019-0410-3
97
Matek A. Mucko M. Casotti R. Trano A. C. Achterberg E. P. Mihanović H. et al . (2023). Phytoplankton diversity and co-dependency in a stratified oligotrophic ecosystem in the south Adriatic Sea. Water15, 2299. doi: 10.3390/w15122299
98
McCluskey K. Barker K. B. Barton H. A. Boundy-Mills K. Brown D. R. Coddington J. A. et al . (2017). The U.S. Culture collection network responding to the requirements of the nagoya protocol on access and benefit sharing. mBio8, e00982–e00917. doi: 10.1128/mBio.00982-17
99
McGrath M. S. Daggett P. Dilworth S. (1978). FREEZE-DRYING OF ALGAE: CHLOROPHYTA AND CHRYSOPHYTA1. J. Phycology14, 521–525. doi: 10.1111/j.1529-8817.1978.tb02480.x
100
McMaugh T. (2005). Guidelines for surveillance for plant pests in Asia and the Pacific. ACIAR Monograph119, 192
101
Mecozzi M. Acquistucci R. Di Noto V. Pietrantonio E. Amici M. Cardarilli D. (2001). Characterization of mucilage aggregates in Adriatic and Tyrrhenian Sea: structure similarities between mucilage samples and the insoluble fractions of marine humic substance. Chemosphere44, 709–720. doi: 10.1016/S0045-6535(00)00375-1
102
Microbial Resource Research Infrastructure (2012). Best Practice Manual on Access and Benefit Sharing (Microbial Resource Research Infrastructure).
103
Mora C. Tittensor D. P. Adl S. Simpson A. G. B. Worm B. (2011). How many species are there on earth and in the ocean? PloS Biol.9, e1001127. doi: 10.1371/journal.pbio.1001127
104
Mucko M. Bosak S. Casotti R. Balestra C. Ljubešić Z. (2018). Winter picoplankton diversity in an oligotrophic marginal sea. Mar. Genomics42, 14–24. doi: 10.1016/j.margen.2018.09.002
105
Nam N. Do H. Loan Trinh K. Lee N. (2023). Metagenomics: an effective approach for exploring microbial diversity and functions. Foods12, 2140. doi: 10.3390/foods12112140
106
Nature Research Centre (2024). Final Report for LIFE project: AlgaeService for LIFE, LIFE17 ENV/LT/000407 (Nature Research Centre).
107
Neu T. R. Manz B. Volke F. Dynes J. J. Hitchcock A. P. Lawrence J. R. (2010). Advanced imaging techniques for assessment of structure, composition and function in biofilm systems. FEMS Microbiol. Ecol.72, 1–21. doi: 10.1111/j.1574-6941.2010.00837.x
108
Novosel M. Požar-Domac A. Pasarić M. (2004). Diversity and distribution of the bryozoa along underwater cliffs in the Adriatic Sea with special reference to thermal regime. Mar. Ecol.25, 155–170. doi: 10.1111/j.1439-0485.2004.00022.x
109
O’Reilly J. E. Maritorena S. Mitchell B. G. Siegel D. A. Carder K. L. Garver S. A. et al . (1998). Ocean color chlorophyll algorithms for SeaWiFS. J. Geophys. Res.103, 24937–24953. doi: 10.1029/98JC02160
110
Office of Water (1997). Monitoring guidance for determining the effectiveness of nonpoint source controls (EPA 841-B-96-004).
111
Office of Water (2002). Methods for evaluating wetland condition: Using Algae To Assess Environmental Conditions in Wetlands (EPA-822-R-02-021).
112
Olapade O. A. Leff L. G. (2005). Seasonal response of stream biofilm communities to dissolved organic matter and nutrient enrichments. Appl. Environ. Microbiol.71, 2278–2287. doi: 10.1128/AEM.71.5.2278-2287.2005
113
Omand M. Cetinić I. Lucas A. (2017). Using bio-optics to reveal phytoplankton physiology from a wirewalker autonomous platform. Oceanography30, 128–131. doi: 10.5670/oceanog.2017.233
114
Orlić M. Beg Paklar G. Dadić V. Leder N. Mihanović H. Pasarić M. et al . (2011). Diurnal upwelling resonantly driven by sea breezes around an Adriatic island. J. Geophys. Res.116, C09025. doi: 10.1029/2011JC006955
115
Overmann J. (2015). Significance and future role of microbial resource centers. Systematic Appl. Microbiol.38, 258–265. doi: 10.1016/j.syapm.2015.02.008
116
Öztürk İ.D. Mutlu S. Kaman G. Partal F. B. Demirtaş A. Çağlar S. et al . (2021). Vertical distribution of mucilage typology in the water column after a massive mucilage formation in the surface waters of the Sea of Marmara. J. Black Sea/Mediterranean Environ.27, 184–201.
117
Parks D. H. Rinke C. ChuvoChina M. Chaumeil P.-A. Woodcroft B. J. Evans P. N. et al . (2017). Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol.2, 1533–1542. doi: 10.1038/s41564-017-0012-7
118
Pascoal F. Tomasino M. P. Piredda R. Quero G. M. Torgo L. Poulain J. et al . (2023). Inter-comparison of marine microbiome sampling protocols. ISME Commun.3, 84. doi: 10.1038/s43705-023-00278-w
119
Patin N. V. Goodwin K. D. (2023). Capturing marine microbiomes and environmental DNA: A field sampling guide. Front. Microbiol.13. doi: 10.3389/fmicb.2022.1026596
120
Patrício J. Elliott M. Mazik K. Papadopoulou K.-N. Smith C. J. (2016). DPSIR—Two decades of trying to develop a unifying framework for marine environmental management? Front. Mar. Sci.3. doi: 10.3389/fmars.2016.00177
121
Pawlowski J. Bruce K. Panksep K. Aguirre F. I. Amalfitano S. Apothéloz-Perret-Gentil L. et al . (2022). Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods. Sci. Total Environ.818, 151783. doi: 10.1016/j.scitotenv.2021.151783
122
Pizzetti I. Lupini G. Bernardi Aubry F. Acri F. Fuchs B. M. Fazi S. (2016). Influence of the Po River runoff on the bacterioplankton community along trophic and salinity gradients in the Northern Adriatic Sea. Mar. Ecol.37, 1386–1397. doi: 10.1111/maec.12355
123
Pocock M. J. O. Newson S. E. Henderson I. G. Peyton J. Sutherland W. J. Noble D. G. et al . (2015). Developing and enhancing biodiversity monitoring programmes: a collaborative assessment of priorities. J. Appl. Ecol.52, 686–695. doi: 10.1111/1365-2664.12423
124
Pommier T. Canbäck B. Riemann L. Boström K. H. Simu K. Lundberg P. et al . (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol.16, 867–880. doi: 10.1111/j.1365-294X.2006.03189.x
125
Porter K. G. Feig Y. S. (1980). The use of DAPI for identifying and counting aquatic microflora1. Limnology Oceanography25, 943–948. doi: 10.4319/lo.1980.25.5.0943
126
Quince C. Walker A. W. Simpson J. T. Loman N. J. Segata N. (2017). Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol.35, 833–844. doi: 10.1038/nbt.3935
127
Rabone M. Harden-Davies H. Collins J. E. Zajderman S. Appeltans W. Droege G. et al . (2019). Access to marine genetic resources (MGR): raising awareness of best-practice through a new agreement for biodiversity beyond national jurisdiction (BBNJ). Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00520
128
Rainville L. Pinkel R. (2001). Wirewalker: an autonomous wave-powered vertical profiler. J. Atmos. Oceanic Technol.18, 1048–1051. doi: 10.1175/1520-0426(2001)018<1048:WAAWPV>2.0.CO;2
129
Reddy M. M. Jennings L. Thomas O. P. (2021). “Marine biodiscovery in a changing world,” in Progress in the Chemistry of Organic Natural Products, vol. 116 . Eds. KinghornA. D.FalkH.GibbonsS.AsakawaY.LiuJ.-K.DirschV. M. (Springer International Publishing, Cham), 1–36. doi: 10.1007/978-3-030-80560-9_1
130
Reed D. J. Finkl C. W. Khalil S. M. Maul G. A. Healy T. R. (2005). “vibracore,” in Encyclopedia of Coastal Science. Ed. SchwartzM. L. (Springer Netherlands, Dordrecht), 1026–1036. doi: 10.1007/1-4020-3880-1_335
131
Ricci F. Penna N. Capellacci S. Penna A. (2014). Potential environmental factors influencing mucilage formation in the northern Adriatic Sea. Chem. Ecol.30, 364–375. doi: 10.1080/02757540.2013.877004
132
Rieder J. Kapopoulou A. Bank C. Adrian-Kalchhauser I. (2023). Metagenomics and metabarcoding experimental choices and their impact on microbial community characterization in freshwater recirculating aquaculture systems. Environ. Microbiome18, 8. doi: 10.1186/s40793-023-00459-z
133
Rogers A. D. Appiah-Madson H. Ardron J. A. Bax N. J. Bhadury P. Brandt A. et al . (2023). Accelerating ocean species discovery and laying the foundations for the future of marine biodiversity research and monitoring. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1224471
134
Rotter A. Barbier M. Bertoni F. Bones A. M. Cancela M. L. Carlsson J. et al . (2021). The essentials of marine biotechnology. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.629629
135
Ruppert K. M. Kline R. J. Rahman M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecol. Conserv.17, e00547. doi: 10.1016/j.gecco.2019.e00547
136
Ryckebosch E. Muylaert K. Eeckhout M. Ruyssen T. Foubert I. (2011). Influence of drying and storage on lipid and carotenoid stability of the microalga Phaeodactylum tricornutum. J. Agric. Food Chem.59, 11063–11069. doi: 10.1021/jf2025456
137
Safarik I. Pospiskova K. Baldikova E. Maderova Z. Safarikova M. (2016). “Magnetic modification of cells,” in Engineering of Nanobiomaterials (Amsterdam, Netherlands: Elsevier), 145–180.
138
Santi I. Kasapidis P. Karakassis I. Pitta P. (2021). A comparison of DNA metabarcoding and microscopy methodologies for the study of aquatic microbial eukaryotes. Diversity13, 180. doi: 10.3390/d13050180
139
Schallenberg M. Kalff J. (1993). The ecology of sediment bacteria in lakes and comparisons with other aquatic ecosystems. Ecology74, 919–934. doi: 10.2307/1940816
140
Schlitzer R. Anderson R. F. Dodas E. M. Lohan M. Geibert W. Tagliabue A. et al . (2018). The GEOTRACES intermediate data product 2017. Chem. Geology493, 210–223. doi: 10.1016/j.chemgeo.2018.05.040
141
Schneider X. T. Stroil B. K. Tourapi C. Rebours C. Gaudêncio S. P. Novoveska L. et al . (2022). Responsible research and innovation framework, the Nagoya protocol and other European blue biotechnology strategies and regulations: gaps analysis and recommendations for increased knowledge in the marine biotechnology community. Mar. Drugs20, 290. doi: 10.3390/md20050290
142
Schneider X. T. Stroil B. K. Tourapi C. Rebours C. Novoveska L. Vasquez M. I. et al . (2023). Improving awareness, understanding, and enforcement of responsibilities and regulations in Blue Biotechnology. Trends Biotechnol.41, 1327–1331. doi: 10.1016/j.tibtech.2023.05.011
143
Schoenleber J. R. (2005). Field Sampling Procedures Manual (New Jersey, USA: Department of Environmental Protection).
144
Semple S. J. Staerk D. Buirchell B. J. Fowler R. M. Gericke O. Kjaerulff L. et al . (2022). Biodiscoveries within the Australian plant genus Eremophila based on international and interdisciplinary collaboration: results and perspectives on outstanding ethical dilemmas. Plant J.111, 936–953. doi: 10.1111/tpj.15866
145
Seo E.-Y. Jung D. Epstein S. S. Zhang W. Owen J. S. Baba H. et al . (2023). A targeted liquid cultivation method for previously uncultured non-colony forming microbes. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1194466
146
Serite C. P. Emami-Khoyi A. Ntshudisane O. K. James N. C. Jansen van Vuuren B. Bodill T. et al . (2023). eDNA metabarcoding vs metagenomics: an assessment of dietary competition in two estuarine pipefishes. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1116741
147
Siegel D. A. DeVries T. Cetinić I. Bisson K. M. (2023). Quantifying the ocean’s biological pump and its carbon cycle impacts on global scales. Annu. Rev. Mar. Sci.15, 329–356. doi: 10.1146/annurev-marine-040722-115226
148
Sigwart J. D. Blasiak R. Jaspars M. Jouffray J.-B. Tasdemir D. (2021). Unlocking the potential of marine biodiscovery. Nat. Prod. Rep.38, 1235–1242. doi: 10.1039/D0NP00067A
149
Staudinger C. Strobl M. Fischer J. P. Thar R. Mayr T. Aigner D. et al . (2018). A versatile optode system for oxygen, carbon dioxide, and pH measurements in seawater with integrated battery and logger. Limnology Ocean Methods16, 459–473. doi: 10.1002/lom3.10260
150
Steele B. M. (2001). “Sampling design and statistical inference for ecological assessment,” in A Guidebook for Integrated Ecological Assessments. Eds. JensenM. E.BourgeronP. S. (Springer New York, New York, NY), 79–91. doi: 10.1007/978-1-4419-8620-7_7
151
Stewart E. J. (2012). Growing unculturable bacteria. J. Bacteriol194, 4151–4160. doi: 10.1128/jb.00345-12
152
Stewart P. M. Smith E. P. Pratt J. R. McCormick P. V. Cairns J. J. (1986). Multivariate analysis of protist communities in lentic systems 1. J. protozoology33, 152–156. doi: 10.1111/j.1550-7408.1986.tb05581.x
153
Stincone P. Brandelli A. (2020). Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol.40, 306–319. doi: 10.1080/07388551.2019.1710457
154
Stock W. Pinseel E. De Decker S. Sefbom J. Blommaert L. Chepurnova O. et al . (2018). Expanding the toolbox for cryopreservation of marine and freshwater diatoms. Sci. Rep.8, 4279. doi: 10.1038/s41598-018-22460-0
155
Taberlet P. Coissac E. Pompanon F. Brochmann C. Willerslev E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol.21, 2045–2050. doi: 10.1111/j.1365-294x.2012.05470.x
156
Throndsen J. Sournia A. (1978). Phytoplankton manual. Monographs on Oceanographic Methodology A. 6, 247–248.
157
Trotta A. Cirilli M. Marinaro M. Bosak S. Diakoudi G. Ciccarelli S. et al . (2021). Detection of multi-drug resistance and AmpC β-lactamase/extended-spectrum β-lactamase genes in bacterial isolates of loggerhead sea turtles (Caretta caretta) from the Mediterranean Sea. Mar. pollut. Bull.164, 112015. doi: 10.1016/j.marpolbul.2021.112015
158
Tuit C. Wait A. (2020). A review of marine sediment sampling methods. Environ. Forensics21, 291–309. doi: 10.1080/15275922.2020.1771630
159
Turk V. Hagström Å. Kovač N. Faganeli J. (2010). Composition and function of mucilage macroaggregates in the northern Adriatic. Aquat. microbial Ecol.61, 279–289. doi: 10.3354/ame01447
160
United Nations Department of Economic and Social Affairs (2023). The Sustainable Development Goals Report 2023: Special Edition (United Nations). doi: 10.18356/9789210024914
161
Vaičiūtė D. Bučas M. Bresciani M. Dabulevičienė T. Gintauskas J. Mėžinė J. et al . (2021). Hot moments and hotspots of cyanobacteria hyperblooms in the Curonian Lagoon (SE Baltic Sea) revealed via remote sensing-based retrospective analysis. Sci. Total Environ.769, 145053. doi: 10.1016/j.scitotenv.2021.145053
162
Van Der Loos L. M. Nijland R. (2021). Biases in bulk: DNA metabarcoding of marine communities and the methodology involved. Mol. Ecol.30, 3270–3288. doi: 10.1111/mec.15592
163
Vandermeulen R. A. Mannino A. Craig S. E. Werdell P. J. (2020). 150 shades of green: Using the full spectrum of remote sensing reflectance to elucidate color shifts in the ocean. Remote Sens. Environ.247, 111900. doi: 10.1016/j.rse.2020.111900
164
Vartoukian S. R. (2016). Cultivation strategies for growth of uncultivated bacteria. J. Oral. Biosci.58, 143–149. doi: 10.1016/j.job.2016.08.001
165
Viršek M. K. Palatinus A. Koren Š. Peterlin M. Horvat P. Kržan A. (2016). Protocol for microplastics sampling on the sea surface and sample analysis. J. visualized experiments: JoVE, 55161. doi: 10.3791/55161
166
Wakita N. Hirokawa K. Ichikawa T. Yamauchi Y. (2010). Development of autonomous underwater vehicle (AUV) for exploring deep sea marine mineral resources. Mitsubishi Heavy Industries Tech. Rev.47, 73–80.
167
Wang Z. Cai R. Gao Z. Yuan Y. Yue T. (2020). Immunomagnetic separation: An effective pretreatment technology for isolation and enrichment in food microorganisms detection. Compr. Rev. Food Sci. Food Saf.19, 3802–3824. doi: 10.1111/1541-4337.12656
168
Wang Y. Chen J. Guo J. Yu Z. Lin Y. Wang Y. (2024). Advances and development in sampling techniques for marine water resources: a comprehensive review. Front. Mar. Sci.11. doi: 10.3389/fmars.2024.1365019
169
Water Quality and Investigation, Department of Environment and Science (2018). Monitoring and Sampling Manual Environmental Protection (Water) Policy 2009 (Queensland, Australia: Water Quality and Investigation, Department of Environment and Science).
170
Watnick P. Kolter R. (2000). Biofilm, city of microbes. J. Bacteriol182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
171
Wilkinson M. Dumontier M. Aalbersberg I. Appleton G. Axton M. Baak A. et al . (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data3, 160018. doi: 10.1038/sdata.2016.18
172
Wolkers W. F. Oldenhof H. (Eds.) (2021). Cryopreservation and Freeze-Drying Protocols (New York, NY: Springer US). doi: 10.1007/978-1-0716-0783-1
173
Zhang W. Ding W. Li Y.-X. Tam C. Bougouffa S. Wang R. et al . (2019). Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun.10, 517. doi: 10.1038/s41467-019-08463-z
174
Zhang Y. Yao P. Sun C. Li S. Shi X. Zhang X. et al . (2021). Vertical diversity and association pattern of total, abundant and rare microbial communities in deep-sea sediments. Mol. Ecol.30, 2800–2816. doi: 10.1111/mec.15937
175
Zinger L. Gobet A. Pommier T. (2012). Two decades of describing the unseen majority of aquatic microbial diversity. Mol. Ecol.21, 1878–1896. doi: 10.1111/j.1365-294x.2011.05362.x
176
Zunino S. Pitacco V. Mavrič B. Orlando-Bonaca M. Kružić P. Lipej L. (2018). The ecology of the Mediterranean stony coral Cladocora caespitosa (Linnaeus 1767) in the Gulf of Trieste (northern Adriatic Sea): a 30-year long story. Mar. Biol. Res.14, 307–320. doi: 10.1080/17451000.2017.1408915
Summary
Keywords
biodiscovery, marine microbial diversity, sampling protocol, sample preservation and storage, biorepositories, cultures
Citation
Ljubešić Z, Gligora Udovič M, Overlingė D, Grgurević F, Akgül F, Bacu A, Díaz-Marrero AR, Drakulović D, Fazi S, Gaudêncio SP, Kolda A, Novoveska L, Safarik I, Sousa JR, Thomas OP, Reddy MM, Varese GC, Vasquez MI, Makovec T and Rotter A (2025) Exploring marine microbial diversity: an overview of representative sampling strategies. Front. Mar. Sci. 12:1597865. doi: 10.3389/fmars.2025.1597865
Received
21 March 2025
Accepted
25 August 2025
Published
19 September 2025
Volume
12 - 2025
Edited by
Jin Zhou, Tsinghua University, China
Reviewed by
Alyse Larkin, National Oceanic and Atmospheric Administration (NOAA), United States
Francisco Pascoal, University of Porto, Portugal
Anneke Heins, Max Planck Society, Germany
Updates
Copyright
© 2025 Ljubešić, Gligora Udovič, Overlingė, Grgurević, Akgül, Bacu, Díaz-Marrero, Drakulović, Fazi, Gaudêncio, Kolda, Novoveska, Safarik, Sousa, Thomas, Reddy, Varese, Vasquez, Makovec and Rotter.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Rotter, Ana.Rotter@nib.si
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.