Abstract
The ongoing rise in atmospheric CO2 levels and the consequent global warming make it increasingly difficult to maintain the global temperature within the 1.5 - 2°C target set by the Paris Agreement. Therefore, strategies to remove carbon dioxide from the atmosphere are being developed, with ocean alkalinity enhancement (OAE) gaining most attention. Within OAE, ocean liming- the addition of quicklime (CaO) or hydrated lime (Ca(OH)2)- can not only remove CO2 from the atmosphere but potentially counteract the effects of ocean acidification. Although quite attractive, these technologies have yet to be tested regarding ecological safety and efficacy. Here we report the impacts of ocean liming on the abundance, composition and extracellular enzymatic activity (EEA) rates of a North Atlantic planktonic community. The results demonstrate that OAE led to a decreased phytoplankton development, mainly diatoms. The bacterial response to OAE was community-specific, with a consistent increase in the relative abundance of the order Oceanospirillales. OAE also led to increased EEA rates, especially within the bacterial community. These findings suggest that while initial effects on phytoplankton may be limited, the specific impacts on bacterial groups suggest that OAE could influence the remineralization of organic matter. If our results apply to other communities, OAE might initially affect marine microbial dynamics, but further studies are needed to determine if these effects are long-term.
1 Introduction
The continued increase of atmospheric CO2 and the consequent warming of planet Earth (Calvin et al., 2023) further distances the goal of keeping the global temperature increase between 1.5 - 2°C (United Nations Framework Convention on Climate Change, 2015). Relying only on the reduction of emissions has been deemed insufficient to achieve the Paris Agreement terms, highlighting the need for carbon capture and negative emission technologies (NETs) (Gattuso et al., 2021; Keller et al., 2014; Rau et al., 2012).
One of the most promising technologies proposed within NETs is Ocean Alkalinity Enhancement (OAE). This approach mimics natural weathering processes by adding alkaline compounds or solutions to the ocean, which enhances CO2 sequestration and storage in the form of bicarbonate (HCO3-) and carbonate ions (CO3²-), thereby increasing ocean alkalinity (Hartmann et al., 2013; Kheshgi, 1995; Renforth and Henderson, 2017). The addition of alkalinity can be previously CO2-equilibrated, initially increasing only total alkalinity (TA) when added, or non-CO2-equilibrated, leading also to a decrease in pCO2 and increase in pH (Hartmann et al., 2023).
Among the various OAE strategies under investigation, ocean liming — the dispersal of quicklime (CaO) or hydrated lime (Ca(OH)2) on the ocean surface — has recently been reconsidered as a potential OAE solution (Foteinis et al., 2022; Renforth et al., 2013; Renforth and Henderson, 2017). When it is not pre-equilibrated with atmospheric CO2, this approach causes a rapid increase in pH upon addition, enhancing CO2 uptake and helping to counteract ocean acidification (Feng et al., 2016; Gattuso et al., 2018; Ilyina et al., 2013).
Several in-depth studies using Earth System Models have evaluated the feasibility of this strategy focusing on its carbon sequestration capacity, potential biochemical impacts and estimated costs (Burt et al., 2021; González and Ilyina, 2016; Keller et al., 2014; Lenton et al., 2018; Paquay and Zeebe, 2013). One major concern with OAE’s ecological impact is the immediate effects of its deployment and whether the affected ecosystems can recover from the added alkalinity, since it could lead to long-term changes that could reshape the ecosystem (Bach et al., 2019; Renforth and Henderson, 2017). To address these challenges and determine safe limits for OAE application, the scientific community is actively working to provide timely insights to policy makers (Oschlies et al., 2023).
Currently, studies investigating the effects of OAE on the marine ecosystem are being conducted. Although studies are progressing rapidly with initial results now being published, they primarily focus on phytoplankton population dynamics and single species (e.g. diatoms, coccolithophores) (Ferderer et al., 2022, 2024; Guo et al., 2022; Hutchins et al., 2023; Marín-Samper et al., 2024) which play a critical role in carbon cycling and are the foundation of the marine food web. While the importance of phytoplankton is undeniable, marine heterotrophic bacteria, which constitute a major portion of the microbial community, have received comparatively less attention. These bacteria not only form intricate relationships with phytoplankton—ranging from mutualistic to antagonistic (Seymour et al., 2017)—but also play a pivotal role in carbon cycling through respiration and the remineralization of organic matter produced by phytoplankton (Azam and Malfatti, 2007; Caron, 1994; Hoppe, 1991). This degradation process is mediated by hydrolytic enzymes, which are sensitive to shifts in ocean chemistry (Yamada and Suzumura, 2010). Consequently, it is vital to assess how OAE might impact bacterial extracellular enzymatic activity and the subsequent effects on the balance between surface ocean remineralization and the sinking of organic matter. Such research is particularly urgent in oligotrophic systems, where nutrient availability is limited, and bacterial processes are critical to ecosystem functioning.
This study aimed to assess the potential effects of ocean alkalinity enhancement through ocean liming on a marine microbial community, specifically phytoplankton abundance and distribution, marine bacteria community dynamics through time and potential effects on the microbial food web.
2 Materials and methods
2.1 Experimental setup
The effects of OAE were investigated on a natural marine microbial community from the North Atlantic, specifically offshore Terceira Island, Azores (38°39'20"N 27°16'54"W). On May 24, 2022, surface seawater was manually collected by filling a 40 L polypropylene bottle approximately 40 m from the shoreline at Negrito (Angra do Heroísmo), resulting in an estimated 40 L of water.
The collected seawater was immediately filtered through a 200 μm mesh, to remove larger zooplankton. As zooplankton are typically sparse and unevenly distributed in the area, their removal helped ensure consistency across treatments. In-situ measurements of temperature, salinity, and total pH (pHt) were taken to characterize the seawater and to calibrate climate chamber conditions.
The filtered community (< 200 μm), hereafter referred to as the Total Community (TC), was gently and continuously mixed in the laboratory to maintain homogeneity and prevent sedimentation. No nutrients were added to the samples, as naturally occurring concentrations of nitrate (8 μM), phosphate (0.2 μM), and silica (≈46 μM) were already present in Negrito Bay at the time of collection.
To isolate the effects of OAE on heterotrophic bacteria, a Bacterioplankton Community (BC) was prepared by filtering the TC through a 0.7 μm glass microfiber filter (GF/F), thereby removing most eukaryotic microbes. The filtrate, primarily containing bacteria, was collected for bacterioplankton treatments.
Both TC and BC were each split into two treatments: control (no alkalinity alteration) and TA addition, where alkalinity was increased by 250 μmol kg-¹ using CaO (see next section for details). Each treatment was transferred into experimental bottles and randomly arranged in a climate chamber (Aralab) set to 18°C (matching in-situ conditions), under a 14/10 h light/dark cycle and a light intensity of 150 ± 10 μmol m-² s-¹. Bottles were gently mixed twice daily and repositioned daily to reduce positional bias.
Samples were taken on t3 and t6 to capture both the exponential growth and stationary phases of phytoplankton, during which bacterial dominance increases. This 2 × 2 factorial design included two community types (TC and BC) and two alkalinity conditions (control and TA addition), resulting in four experimental treatments (see Table 1).
Table 1
| Community type | Alkalinity | Condition |
|---|---|---|
| Total (< 200 μm) | Control | TC-C |
| Total | TA addition (T) | TC-T |
| Bacterioplankton (< 0.7μm) | Control | BC-C |
| Bacterioplankton | TA addition | BC-T |
Treatments tested in the experiment.
TC, Total Community; BC, Bacterioplankton community; C, Control; T, TA addition.
2.2 TA addition
CaO solubility was previously tested by comparing “expected” versus “real” TA enhancement. The results of the optimization protocol showed a 70% efficiency of this alkalinization method, which led to the correction of the addition to assure the achievement of wanted final TA concentration (Δ 250 μmol kg-1).
According to Equation 1, theoretically, one mole of CaO (Calcidrata, purity above 75%), increases the alkalinity by two moles via HCO3-.
Seawater collected on site was filtered (0.22 µm) using a peristaltic pump to reduce microbial content and was used to prepare an intermediate, highly alkaline solution referred to as the “mother solution”. This TA-enriched solution (~4000 μmol kg-¹ increase) was prepared by rapidly dissolving 0.0896 g of CaO in ultrapure water through vigorous shaking in a 15 mL Falcon tube. The resulting slurry was then transferred to a 1 L Duran bottle containing a magnetic stirrer, and filtered seawater was added until the final volume reached 1 L. Subsequently, 60–70 mL of the “mother solution” were added to each TA addition treatment bottle (1 L final volume) to increase alkalinity by 250 μmol kg-¹.
2.3 Carbonate chemistry
TA samples were filtered through a polyethersulfone (PES) 0.2 µm syringe filter and measured in a Metrohm Titrino Plus 848 coupled with an 869 Compact Sample Changer, with correction being carried out by the certified reference material supplied by Dickson et al. (2003). Total pH (pHt) was measured using a glass electrode (WTW,pH 340i) and corrected using TRIS buffer supplied by Dickson et al. (2003).The remaining parameters of the carbonate system were calculated through the CO2SYS script for excel Lewis et al. (1998), using as input salinity, temperature, total phosphorus and silica, TA, and pH, and using the equilibrium constants defined by Lueker et al. (2000).
2.4 Inorganic nutrients
Samples for dissolved inorganic nutrients were filtered through a 0.2 µm syringe filter on t0, t3 and t6 and frozen until processing. Nitrate, nitrite, phosphate and silicate concentrations were determined spectrophotometrically (Varian Cary 50) according to Hansen and Koroleff (1999).
2.5 Composition and abundance of microphytoplankton and coccolithophores
Microphytoplankton composition and abundance of functional groups were assessed through light microscopy (Nikon TS100) at 200 X magnification on t0, corresponding to the initial community, and then on t3 and t6 of the experiment. Briefly, at each time point, between 100–200 mL were collected, preserved with 2% Lugol, and kept in the dark at room temperature until further analysis. The samples were then processed after two months, where they were placed in an Utermöhl chamber (HYDRO-BIOS KIEL), set to sediment, and the cells counted.
To deepen the information on the phytoplankton community, calcifying nanoplankton, represented here by coccolithophores due to their dominance in the modern ocean alongside diatoms and dinoflagellates (Falkowski et al., 2004) and its relevance in the Azores region (Narciso et al., 2016) were studied. Coccolithophore abundance was determined at the beginning of the experiment (t0) and at the end (t6). Samples were filtered through cellulose nitrate membrane filters and then rinsed with low mineral water to eliminate salt artifacts. The filters were then air dried and stored in petri dishes. A sector of approximately 30° was cut out of each filter and placed on a glass slide, where the mounting medium (Entellam) was previously added, and a coverslip was placed on top. The mounted sample was then heated for about 30 seconds to degas the sample and facilitate visualization and left overnight to finish the mounting process. Lastly, the coccolithophores were identified and counted on a polarization microscope (Leica DM 2700 P).
2.6 Heterotrophic bacteria community composition
The bacterial community was determined by filtering approximately 90–150 mL through a 0.22 µm polycarbonate filter (47 mm diameter) using a vacuum pump. The filters were carefully removed, immersed in ethanol absolute and stored at -20°C. Afterwards, DNA was extracted from the filters using the FastDNA™ Spin Kit for Soil (MP Biomedicals) following the manufacturer’s instructions, with an additional step regarding the processing of the polycarbonate filters. In brief, the filters were cut into smaller pieces under sterile conditions before being placed in the Lysing Matrix E tube for further processing. After obtaining the DNA, the samples were sent to STABVIDA (Lisbon) for metagenomic paired-end sequencing of the 16S rRNA gene on Illumina MiSeq Platform, using the 515F (5’-3’): GTGCCAGCMGCCGCGGTAA and 806R (5’-3’): GGACTACHVGGGTWTCTAAT primers that target the V4 region of the subunit (Caporaso et al., 2011). Algorithms in QIIME2 (version 2020.8) were used to transform the amplicon libraries into an amplicon sequence variant (ASV) abundance table. Taxonomy was assigned to ASVs using the SILVA database of the 16S reference sequences at 99% similarity (version 138, released December 2019).
2.7 Extracellular enzymatic activity
The extracellular enzymatic activity of leucine aminopeptidase (LAP) and alkaline phosphatase (ALP) was determined based on the protocol developed by Hoppe (1983) with some minor modifications. In brief, the fluorogenic substrate analogs solutions, L-leucine-7-amido-4-methylcoumarine hydrochloride (Leu-MCA) and 4-methylumbelliferyl phosphate (MUF-Phos) were added to the sterile 96-well microplates at the final concentration of 32 μM. For alkaline phosphatase samples, 30 μL of enhancement solution (MUF) was added before measurements. All measurements were performed on a FLUOstar Omega equipment with the following wavelengths, λ excitation= 360 nm; λ emission= 450 nm. The fluorescence values obtained for LAP and ALP were then used to estimate the concentration by means of the calibration standards L-Leucine 7-amido-4-methylcoumarin hydrochloride and 4-Methylumbelliferyl, respectively.
2.8 Data analysis and bioinformatics
All statistical analyses were performed on RStudio version 2023.06.1 + 524. Multifactorial ANOVA was performed using as factors the alkalinity (control vs TA addition) treatments for carbonate chemistry parameters, microphytoplankton abundance and enzymatic activity along Tukey multiple pairwise-comparisons test with 95% confidence interval. For the microphytoplankton community, a non-metric multidimensional scaling (NMDS) ordination based on Bray-Curtis dissimilarities of Hellinger-transformed abundance data was performed to visualize differences in community composition across treatments. Additionally, a permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis dissimilarities was conducted to statistically assess the effect of OAE on community structure (vegan package, R). For the bacterial community, alpha diversity indexes (Shannon’s H, Peilou’s J and Fisher alpha) were calculated using vegan package. Beta diversity was visualized through principal coordinates analysis (PCoA) based on Bray-Curti’s dissimilarities (phyloseq package) while the significant effects of OAE were assessed through PERMANOVA. A taxon-specific analysis for all relevant orders (relative abundance > 2.5%) was performed through a multifactorial ANOVA applied to general linearized model (GLM) (vegan package). Heatmap was generated using Euclidian distance matrix of 50 most abundant ASV as input.
3 Results
3.1 Carbonate chemistry
The manipulation resulted in an increase of total alkalinity (TA) by approximately 170 μmol kg-¹, whereas the intended increase was 250 μmol kg-¹, yielding an alkalinization efficiency of approximately 70%. And consequently, the pHt also increased (average value 8.28) in TA addition treatments (Figure 1). While the TA concentration remained constant throughout the whole experiment, pHt values decreased in the bacterioplankton and increased in the total community by the end of the experiment (Figure 1). Concomitantly, CO2 levels, which initially showed an opposite trend to pHt by starting lower in the TA addition samples (2-fold) compared to the control, in both communities (Figure 1), increased in the bacterioplankton community but decreased in the total community by the end of the experiment.
Figure 1
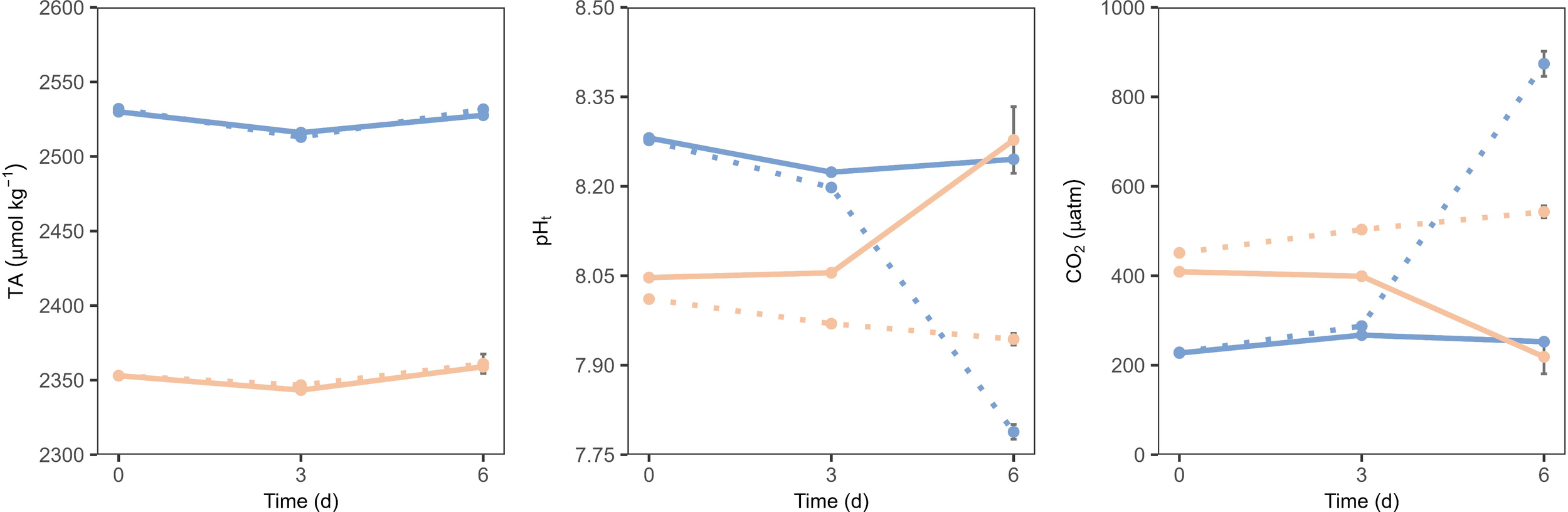
Carbonate chemistry throughout the experiment: total pH; TA and CO2. Peach lines represent the control, and blue lines represent TA addition. Solid lines represent the total community, and dashed lines represent the bacterioplankton community. Bars represent the standard error.
3.2 Effects of OAE on phytoplankton
The total abundance of phytoplankton (including both microphytoplankton and coccolithophores) was estimated visually using light microscopy and showed a marked increase over the course of the experiment, rising approximately 50- to 100-fold by t6 (Figure 2A). Nevertheless, phytoplankton development was decreased under the TA addition treatment by 3-fold (P = 0.0214) as evidenced by lower increase compared to the control and lower uptake of inorganic nutrients (Supplementary Table S2).
Figure 2
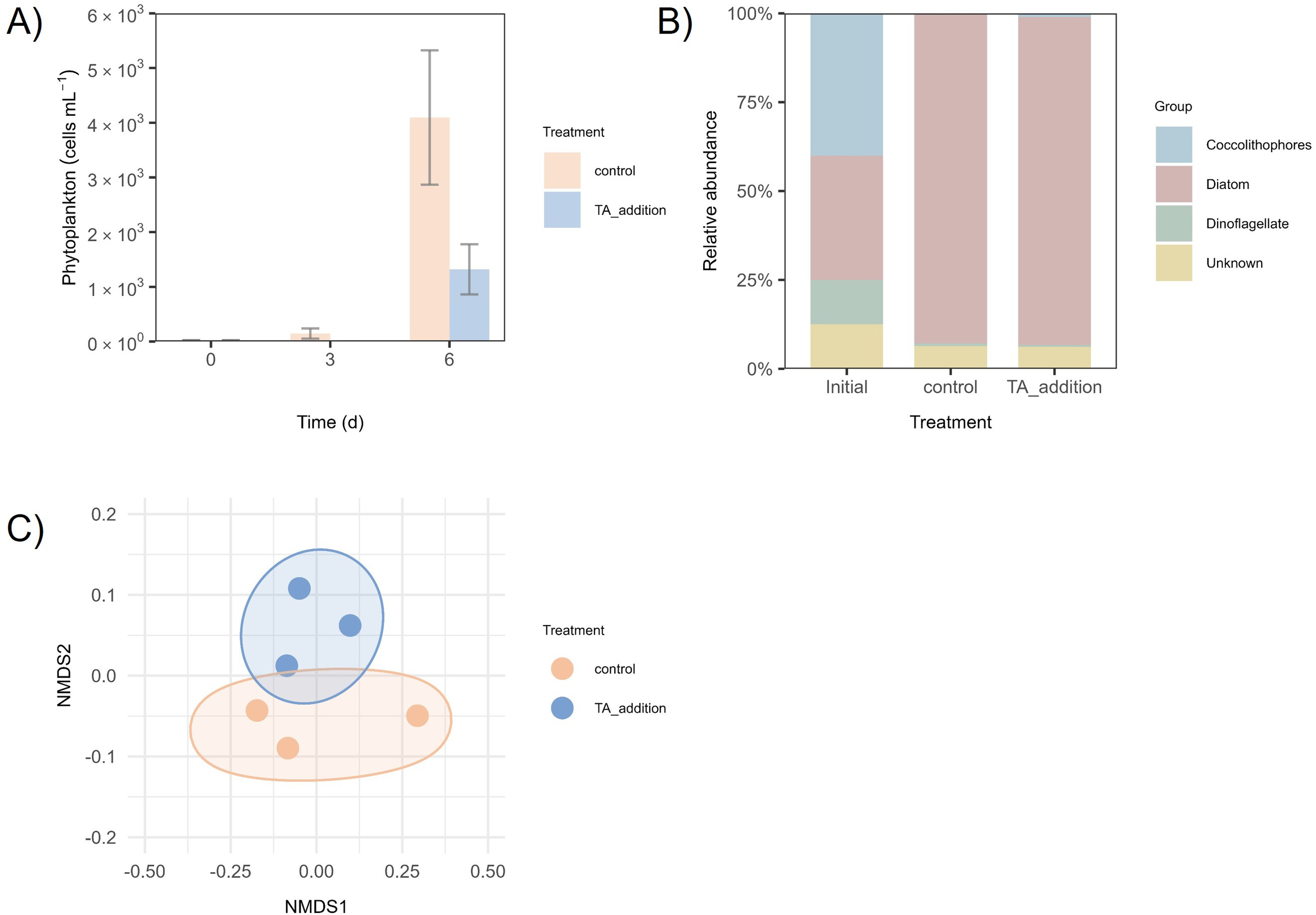
(A) Phytoplankton abundance throughout the experiment. Peach lines represent the control, and blue lines represent TA addition; (B) Relative abundance of functional groups of phytoplankton of the total community on t6. C, Control; T, TA addition; (C) NMDS plot based on Bray-Curtis dissimilarities.
The increase in abundance was mainly driven by diatoms in all treatments, shifting from an initial community, composed of 40% coccolithophores, 35% diatoms, 12% dinoflagellates and 13% unidentified organisms (Figure 2B) to diatoms (92-98%), followed by dinoflagellates (1-9%) in all treatments (Figure 2B). This was corroborated by the decrease in silica on t6 (Supplementary Table S2). Meanwhile, coccolithophores, which initially made up a large portion of the community, were almost completely absent by the end of the experiment, detected only at 1% on the TA addition treatment. Still, the PERMANOVA model revealed no significant OAE influence (P = 0.1) on microphytoplankton community composition as displayed also by the NMDS ordination (Figure 2C).
3.3 Effects of OAE on bacteria
The bacterial communities were predominantly composed of ASVs classified within the phyla Proteobacteria and Bacteroidota, specifically the orders Alteromonadales, Oceanospirillales and Rhodobacterales, and Flavobacteriales, respectively (Figure 3A).
Figure 3
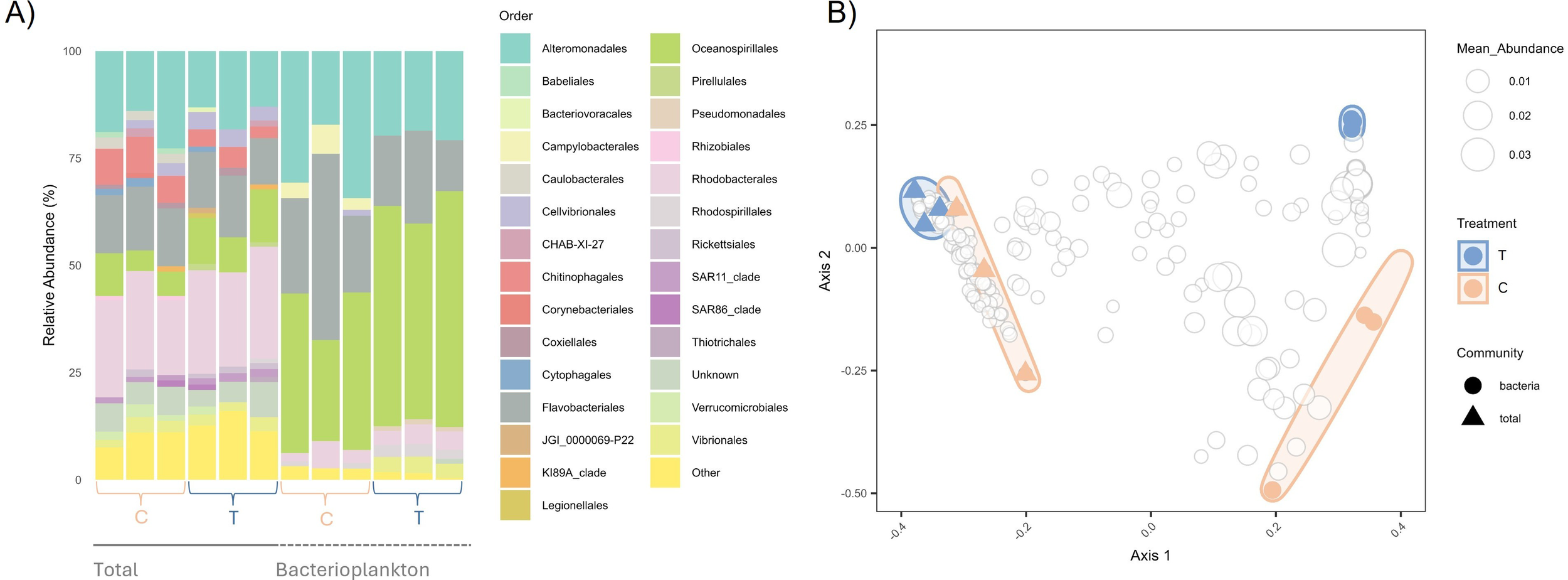
(A) Relative abundance of the 20 most abundant orders in the overall dataset for Bacterioplankton and Total communities. C, Control; T, TA addition; (B) Principal coordinates analysis (PCoA) based on Bray-Curtis dissimilarities.
The bacterioplankton community (BC), derived from the total community (TC) by filtration, showed a distinct composition with lower taxonomic richness, as indicated by Shannon H, Pielou J, and Fisher alpha indices (Figures 3A; Supplementary Figure S1). Furthermore, there is a clear separation between bacterioplankton and total communities, evidenced in the heatmap dendrogram (Figure 4) and confirmed in the PCoA (Figure 3B), except for replicate 2 from the bacterioplankton community that is grouped with the total community (Figure 4). Still, the filtration retained important taxonomic similarities, namely the presence of ASVs classified to order Alteromonadales, Flavobacteriales, Oceanospirillales and Rhodobacterales (Figure 3).
Figure 4
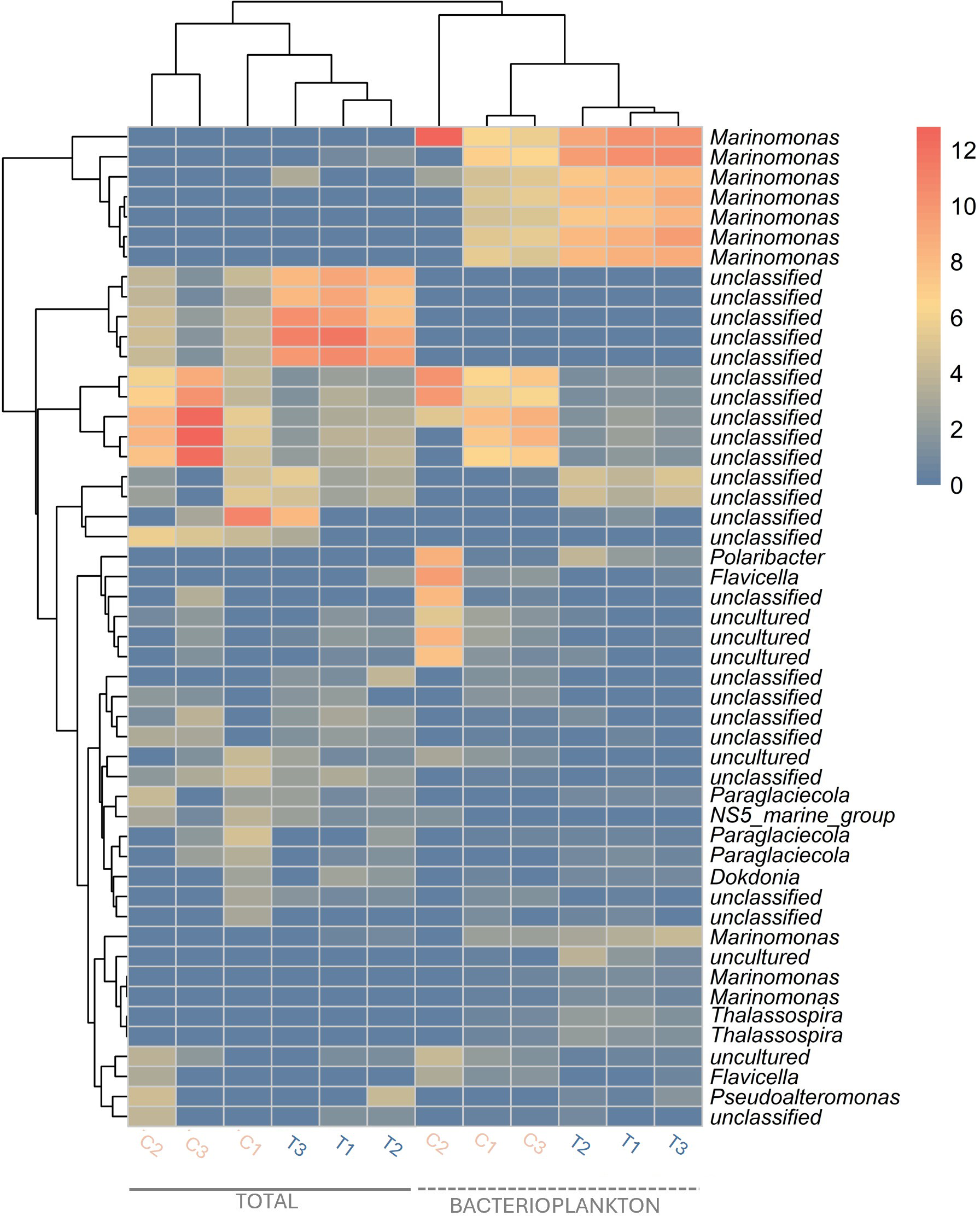
Heatmap of the 50 most abundant ASVs in each sample. C, Control; T, TA addition.
Although the PERMANOVA revealed no overall effect (P = 0.1) of TA addition on both TC and BC community composition, an in-depth look into the TC revealed that some orders were affected by TA addition, specifically orders Chitinophagales, Cellvibrionales and Oceanospirillales (Figure 5; Supplementary Table S1). In fact, the relative abundance of Chitinophagales, mainly composed of ASVs from the Saprospiraceae family, was reduced under TA addition (P = 0.008). In contrast, Cellvibrionales represented by Halieaceae, Porticoccaceae, Spongiibacteraceae and Cellvibrionaceae families responded positively to TA addition increasing their abundance (P = 0.019), alongside Oceanospirillales (mostly Marinomonas sp.) to a lesser extent (P = 0.064).
Figure 5
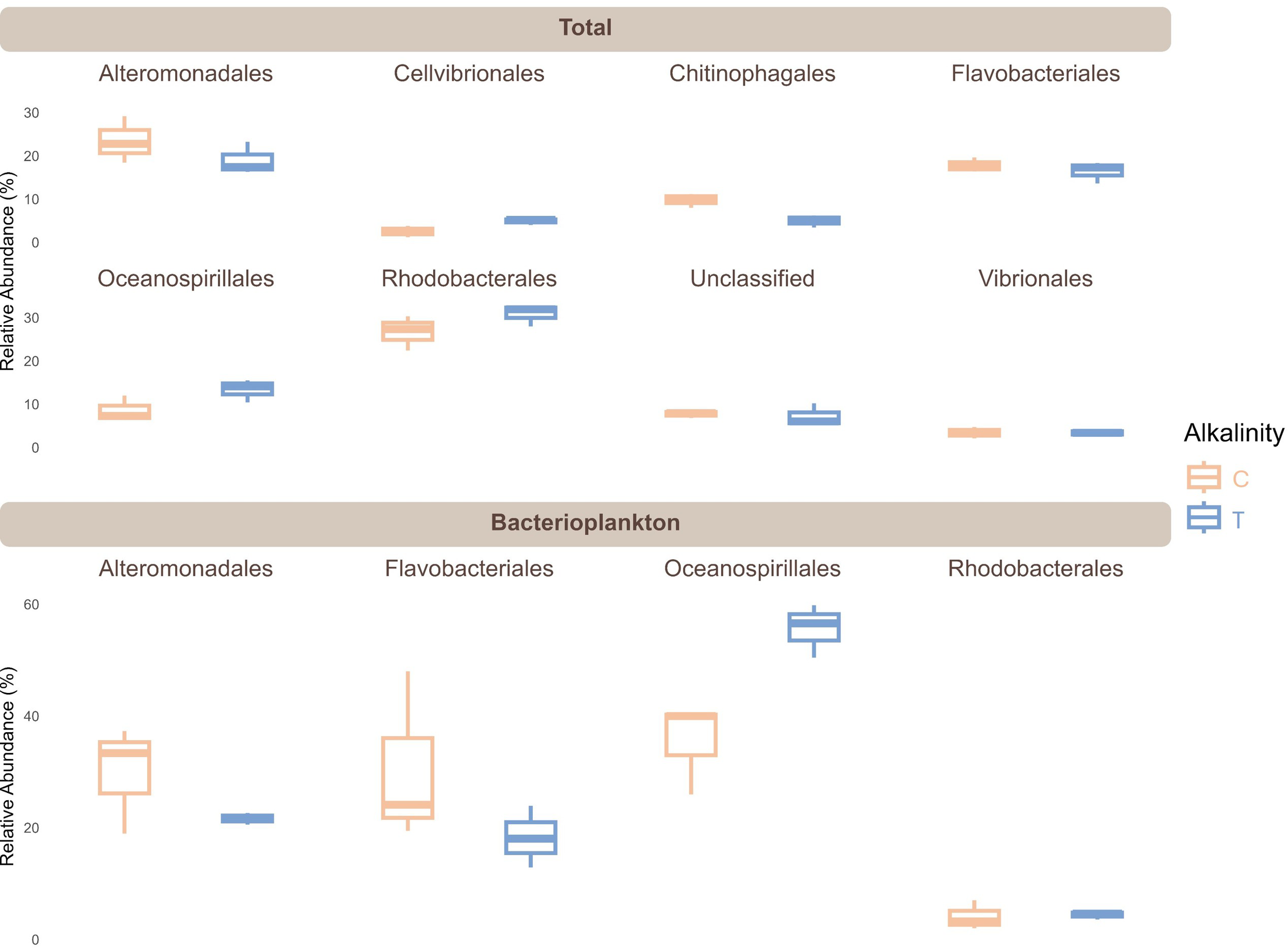
Boxplots of abundance of most relevant bacterial orders with mean relative abundance > 2.5% in the multifactorial ANOVA within the total and 769 bacterioplankton communities. C, Control; T, TA addition.
Furthermore, an analysis of the BC community (Figure 5; Supplementary Table S1) revealed that only the order Oceanospirillales, composed almost entirely of the Marinomonas genus, was significantly affected by the alkalinity treatment, exhibiting a positive response to TA addition (P = 0.044) (Figure 5; Supplementary Table S1).
3.4 OAE effects on extracellular enzymatic activity
LAP activity increased in both communities until t3 under all treatments, with no evident effect of OAE (Figure 6). From t3 to t6, responses diverged: in the total community, LAP activity decreased regardless of treatment, while in the bacterioplankton community, activity continued to rise under TA addition but remained stable in the control. By t6, LAP activity was higher in the bacterioplankton community than in the total community. However, the difference between treatments was marginal (P = 0.065; Figure 6). Overall, OAE did not affect LAP in the total community, but led to an increase in the bacterioplankton community under TA addition by the end of the experiment.
Figure 6
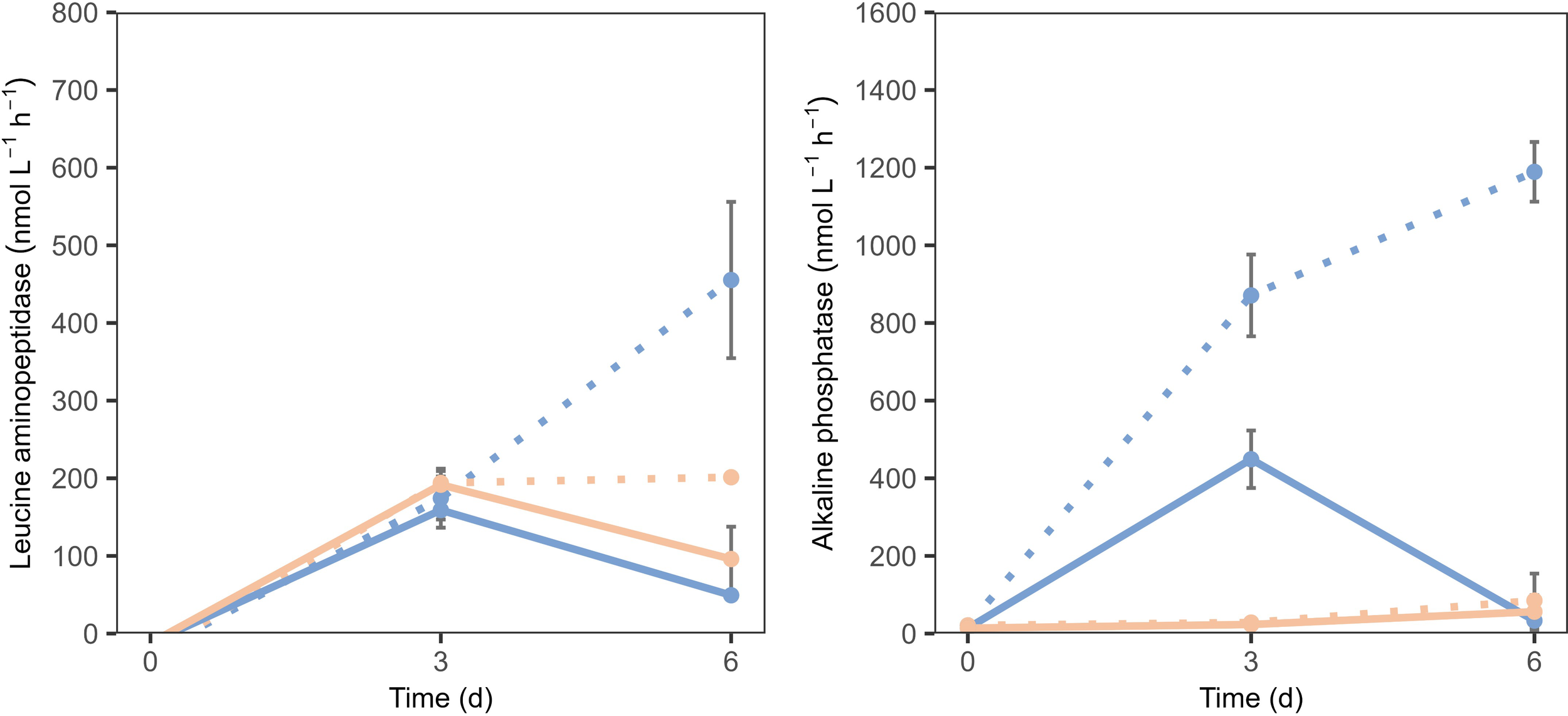
Extracellular enzymatic activity of LAP and ALP. Peach lines represent the control, and blue lines represent TA addition. Solid lines represent the total community, and dashed lines represent the bacterioplankton community. Bars represent the standard error.
Regarding ALP activity, rates were significantly higher under TA addition treatment compared to the control, increasing 41-fold in the bacterioplankton community and 32-fold in the total community (Figure 6). However, by the end of the experiment (t6), this trend persisted in the bacterioplankton community, whereas ALP activity in the total community declined sharply (Figure 6).
4 Discussion
4.1 Ecological and physiological impacts of OAE on phytoplankton
This study showed a lower phytoplankton development, mostly composed of diatoms, under OAE but no changes in community composition could be inferred. Given the initially high inorganic nutrient concentrations, diatom dominance was anticipated due to their rapid nutrient uptake (Margalef, 1978; Smetacek, 1999). Although the non-equilibrated OAE strategy employed in this study influenced both pCO2 and pH, it is unlikely that the observed decreased in diatom bloom development was driven by pH changes. In the TA addition treatment, the initial pH increase reached 8.3, which remains well below the threshold considered “safe” in previous studies, where pH levels approached 9 (Chen and Durbin, 1994; Pedersen and Hansen, 2003).
Instead, the bloom decrease might be related to decreased pCO2 concentration (≈ 228 μatm) under TA addition compared the communities from the control (pCO2 ≈ 400 μatm) a known limiting factor in phytoplankton growth (Riebesell et al., 1993). Lab experiments with the diatom Thalassiosira pseudonana reported decreased growth rates on day 8, but only under CO2 non-equilibrated OAE conditions, where DIC levels had declined. No such decrease was observed under CO2-equilibrated conditions (Oberlander et al., 2025). CO2 ingassing is required to counterbalance the drawdown resulting from initial alkalinization. In a real-world scenario, this CO2 depletion would be naturally compensated, meaning the effects reported here likely represent an overestimation.
Previous studies on the effects of OAE on diatoms have yielded contrasting results. A decrease in biogenic silica (BSi) buildup was reported in one study (Ferderer et al., 2022), whereas a separate experiment using a pure culture of the cosmopolitan marine diatom Chaetoceros sp. under CO2-equilibrated OAE found no significant differences in BSi production (Gately et al., 2023). Regarding studies conducted in the North Atlantic, research simulating CO2-equilibrated OAE reported no significant changes in primary production, abundance, or composition of nano- and microphytoplankton under alkalinization conditions similar to those presented here (Marín-Samper et al., 2024; Xin et al., 2024). However, the authors suggest that low nutrient availability, particularly the absence of detectable nitrate and nitrite (NOx) until phase II, may have masked potential OAE effects. In contrast, a study conducted in the North Atlantic Gyre observed a decline in net primary production under OAE conditions (Subhas et al., 2022). Given that inorganic nutrient concentrations are key determinants of phytoplankton community responses to changes in the carbonate system (Bach et al., 2019), it is expected that the findings of this study may differ from those reported in the oligotrophic conditions of the North Atlantic.
Regarding potential byproducts released during OAE via liming that could have contributed to the bloom delay, studies using steel slag for OAE have reported no effects on microphytoplankton (Guo et al., 2024). Given that steel slag contains significantly more impurities than CaO, the notion that OAE via CaO could introduce contaminants appears unfounded. Moreover, some authors suggest that steel slag may have even released trace metals, potentially exerting a fertilizing effect. However, their experimental conditions were not directly comparable to ours, as their initial pH was 7.8 with an fCO2 of 600 µatm, substantially higher than the pCO2 of 400 µatm in our study.
4.2 Ecological and physiological impacts of OAE on bacteria
OAE deployment did not have an overall effect on marine bacteria composition, evidenced by the PERMANOVA results, nevertheless, some bacterial orders were significantly affected. In the total community, where bacteria coexisted with phytoplankton, which provided essential resources to support its growth and development (Amin et al., 2012; Eigemann et al., 2022; Kieft et al., 2021; Seymour et al., 2017), the abundance of Saprospiraceae, was lower under OAE treatment, correlating with the phytoplankton trend. This was expected since the Saprospiraceae family exhibit an epiphytic lifestyle, living attached to particles and other organisms such as algae and filamentous bacteria (Korlević et al., 2021; Miranda et al., 2013; Xia et al., 2008). However, orders Cellvibrionales and Oceanospirillales were present at higher number in the TA addition treatments, hinting at a possible benefit from OAE. Cellvibrionales is a bacterial order commonly found in marine environments (Spring et al., 2015), responsible for degrading organic material from phytoplankton (Delgadillo-Nuño et al., 2023). During a phytoplankton bloom, the bacterial community composition shifts as the bloom progresses. Cellvibrionales have been reported to be more abundant in the early stages of the bloom (Pontiller et al., 2022). In the TA addition treatment, the decrease bloom, evidenced by lower microphytoplankton biomass and reduced nutrient drawdown, likely explains the higher abundance of Cellvibrionales.
Finally, the effect of OAE on Oceanospirillales was more evident in the Bacterioplankton community, where the order was overrepresented, in comparison to the total community. Also, while in the total community, the majority of the ASVs belonged to the Marinomonas genus, other genera were also present like Pseudohongiella sp. and to a lesser extent Neptuniibacter sp, in the bacterioplankton community, Oceanospirillales were almost exclusively Marinomonas sp. strains. In this order, OAE application led to an increase in abundance across both communities, a potential benefit due to the release of co-factors such as trace metals released alongside CaO deployment (Hartmann et al., 2013). Also, while in the total community Oceanospirillales account for approximately 10% of the relative abundance, in the bacterioplankton community under the TA addition treatment, Marinomonas sp. represented about 50% of the ASVs, making the effects of the alkalinization more noticeable. The Marinomonas domination observed in both communities under OAE might have been due to its metabolic versatility like the potential for assimilating nitrate (Maeda et al., 2019).
4.3 Effects of OAE on EEA and remineralization
The effects of OAE on the bacterial remineralization of organic matter were assessed through the activity of extracellular enzymes, namely LAP, involved in the breakdown of naturally occurring peptides (Caruso and Zaccone, 2000) and ALP, responsible for hydrolyzing organic phosphorus compounds into inorganic phosphate (Hoppe, 2003). Our data showed no evidence of the effect of OAE on bacterial LAP activity in the total community. However, there was an increase in LAP activity, although not significant, on the bacterioplankton community. Notably, LAP activity was generally higher in the bacterioplankton community compared to the total community, which is unexpected given that LAP is typically induced by the presence of peptides in seawater (Shindoh et al., 2021) and is often correlated with organic matter produced by phytoplankton (Shi et al., 2019). One explanation for the increase in LAP activity could be attributed to changes in the bacterial composition (Spilling et al., 2023). Furthermore, ALP activity was also stimulated by OAE in both communities, but only persisted on the bacterioplankton community, having dissipated in the total community by the end of the experiment. Although ALP activity is typically triggered under PO4 depletion, no increase in ALP rates were observed in the control treatments. One possible explanation for the observed increase of activity under OAE is the increase in calcium (Ca2+) levels, which acts as a co-factor in the production of the marine alkaline phosphatase PhoX (Kathuria and Martiny, 2011; Sebastian and Ammerman, 2009). ALP activity in bacteria can also be stimulated by carbon depletion (Hoppe, 2003; Hoppe and Ullrich, 1999), which can explain why the activity remained high in the bacterioplankton but dropped in the total community.
5 Conclusion
The results presented in this study report an immediate response of a marine microbial community to the deployment of OAE strategy themed ocean liming. The phytoplankton community, dominated by diatoms, showed reduced development in response to the OAE but no changes in community composition were reported. In contrast, the effects of ocean liming on the bacterial composition were dependent on the type of community, with order Oceanospirillales, mainly the Marinomonas genus increasing in abundance as a response to OAE. Furthermore, OAE led to an increase in the bacterial rates of alkaline phosphatase on both communities. According to our findings OAE might initially affect marine microbial dynamics, highlighting the need for more studies to assess long-term effects of OAE deployment.
Statements
Data availability statement
The original contributions presented in the study are publicly available in the European Nucleotide Archive (ENA) under project accession number PRJEB90456 and study accession number ERP173458.
Author contributions
IC: Methodology, Data curation, Investigation, Conceptualization, Formal Analysis, Writing – review & editing, Writing – original draft. SR: Writing – review & editing, Methodology. AL: Methodology, Investigation, Writing – review & editing. NG: Writing – review & editing, Resources. MC: Methodology, Resources, Writing – review & editing. EBA: Writing – review & editing, Funding acquisition. JBR: Conceptualization, Investigation, Methodology, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the FCT project of IITAA with the reference UIDB/00153/2020, by the ARM Program supported by DOE, in the framework of the ENA project through the agreement between LANL and the University of the Azores, IC is financed by National Funds through the Portuguese funding agency, FCT -Fundação para a Ciência e a Tecnologia with the grant UI/BD/150890/2021, SCR is supported by FCT grant UIDP/00153/2020, and JBR is supported by FCT, within Contrato-Programa, Apoio Institucional.
Acknowledgments
We would like to thank Dominika Jurášková for her extra hands in sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1602158/full#supplementary-material
References
1
Amin S. A. Parker M. S. Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev.76, 667–684. doi: 10.1128/MMBR.00007-12
2
Azam F. Malfatti F. (2007). Microbial structuring of marine ecosystems. Nature Reviews Microbiology5 (10), 782–791. doi 10.1038/nrmicro1747
3
Bach L. T. Gill S. J. Rickaby R. E. M. Gore S. Renforth P. (2019a). CO2 removal with enhanced weathering and ocean alkalinity enhancement: potential risks and co-benefits for marine pelagic ecosystems. Front. Climate1. doi: 10.3389/fclim.2019.00007
4
Bach L. T. Hernández-Hernández N. Taucher J. Spisla C. Sforna C. Riebesell U. et al . (2019b). Effects of elevated CO 2 on a natural diatom community in the subtropical NE Atlantic. Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00075
5
Burt D. J. Fröb F. Ilyina T. (2021). The sensitivity of the marine carbonate system to regional ocean alkalinity enhancement. Front. Climate3. doi: 10.3389/fclim.2021.624075
6
Calvin K. Dasgupta D. Krinner G. Mukherji A. Thorne P. W. Trisos C. et al . (2023). IPCC 2023: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Core Writing Team. Eds. LeeH.RomeroJ. (Geneva, Switzerland: IPCC). doi: 10.59327/IPCC/AR6-9789291691647
7
Caporaso J. G. Lauber C. L. Walters W. A. Berg-Lyons D. Lozupone C. A. Turnbaugh P. J. et al . (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci.108, 4516–4522. doi: 10.1073/pnas.1000080107
8
Caron D. A. (1994). Inorganic nutrients, bacteria, and the microbial loop. Microbial Ecology28 (2), 295–298. doi 10.1007/BF00166820
9
Caruso G. Zaccone R. (2000). Estimates of leucine aminopeptidase activity in different marine and brackish environments. J. Appl. Microbiol.89, 951–959. doi: 10.1046/j.1365-2672.2000.01198.x
10
Chen C. Y. Durbin E. G. (1994). Effects of pH on the growth and carbon uptake of marine phytoplankton. Marine Ecology Progress Series, 109, 83–94. doi: 10.3354/meps109083
11
Delgadillo-Nuño E. Teira E. Pontiller B. Lundin D. Joglar V. Pedrós-Alió C. et al . (2023). Coastal upwelling systems as dynamic mosaics of bacterioplankton functional specialization. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1259783
12
Dickson A. G. Afghan J. D. Anderson G. C. (2003). Reference materials for oceanic CO2 analysis: a method for the certification of total alkalinity. Mar. Chem.80, 185–197. doi: 10.1016/S0304-4203(02)00133-0
13
Eigemann F. Rahav E. Grossart H. Aharonovich D. Sher D. Vogts A. et al . (2022). Phytoplankton exudates provide full nutrition to a subset of accompanying heterotrophic bacteria via carbon, nitrogen and phosphorus allocation. Environ. Microbiol.24, 2467–2483. doi: 10.1111/1462-2920.15933
14
Falkowski P. G. Katz M. E. Knoll A. H. Quigg A. Raven J. A. Schofield O. et al . (2004). The evolution of modern eukaryotic phytoplankton. Science305, 354–360). doi: 10.1126/science.1095964
15
Feng E. Y. Keller D. P. Koeve W. Oschlies A. (2016). Could artificial ocean alkalinization protect tropical coral ecosystems from ocean acidification? Environ. Res. Lett.11, 74008. doi: 10.1088/1748-9326/11/7/074008
16
Ferderer A. Chase Z. Kennedy F. Schulz K. G. Bach L. T. (2022). Assessing the influence of ocean alkalinity enhancement on a coastal phytoplankton community. Biogeosciences19, 5375–5399. doi: 10.5194/bg-19-5375-2022
17
Ferderer A. Schulz K. G. Riebesell U. Baker K. G. Chase Z. Bach L. T. (2024). Investigating the effect of silicate- and calcium-based ocean alkalinity enhancement on diatom silicification. Biogeosciences21 (11), 2777–2794. doi 10.5194/bg-21-2777-2024
18
Foteinis S. Andresen J. Campo F. Caserini S. Renforth P. (2022). Life cycle assessment of ocean liming for carbon dioxide removal from the atmosphere. J. Cleaner Production370, 133309. doi: 10.1016/j.jclepro.2022.133309
19
Gately J. A. Kim S. M. Jin B. Brzezinski M. A. Iglesias-Rodriguez M. D. (2023). Coccolithophores and diatoms resilient to ocean alkalinity enhancement: A glimpse of hope?. Science Advances, 9 (24). doi: 10.1126/sciadv.adg6066
20
Gattuso J. P. Magnan A. K. Bopp L. Cheung W. W. L. Duarte C. M. Hinkel J. et al . (2018). Ocean solutions to address climate change and its effects on marine ecosystems. Front. Mar. Sci.5. doi: 10.3389/fmars.2018.00337
21
Gattuso J.-P. Williamson P. Duarte C. M. Magnan A. K. (2021). The potential for ocean-based climate action: negative emissions technologies and beyond. Front. Climate2. doi: 10.3389/fclim.2020.575716
22
González M. F. Ilyina T. (2016). Impacts of artificial ocean alkalinization on the carbon cycle and climate in Earth system simulations. Geophysical Res. Lett.43, 6493–6502. doi: 10.1002/2016GL068576
23
Guo J. A. Strzepek R. Willis A. Ferderer A. Bach L. T. (2022). Investigating the effect of nickel concentration on phytoplankton growth to assess potential side-effects of ocean alkalinity enhancement. Biogeosciences19 (15), 3683–3697. doi 10.5194/bg-19-3683-2022
24
Guo J. A. Strzepek R. F. Swadling K. M. Townsend A. T. Bach L. T. (2024). Influence of ocean alkalinity enhancement with olivine or steel slag on a coastal plankton community in Tasmania. Biogeosciences21, 2335–2354. doi: 10.5194/bg-21-2335-2024
25
Hansen H. Koroleff F. (1999). Determination of nutrients. Methods of Seawater Analysis. Wiley, pp. 159–228. doi: 10.1002/9783527613984.ch10
26
Hartmann J. Suitner N. Lim C. Schneider J. Marín-Samper L. Arístegui J. et al . (2023). Stability of alkalinity in ocean alkalinity enhancement (OAE) approaches - consequences for durability of CO2storage. Biogeosciences20, 781–802. doi: 10.5194/bg-20-781-2023
27
Hartmann J. West A. J. Renforth P. Köhler P. de la Rocha C. L. Wolf-Gladrow D. A. et al . (2013). Enhanced chemical weathering as a geoengineering strategy to reduce atmospheric carbon dioxide, supply nutrients, and mitigate ocean acidification. Rev. Geophysics51, 113–149. doi: 10.1002/rog.20004
28
Hoppe H.-G. (1983). Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl-substrates. Mar. Ecol. Prog. Ser.11, 299–308. doi: 10.3354/meps011299
29
Hoppe H. G. (1991). Microbial Extracellular Enzyme Activity: A New Key Parameter in Aquatic Ecology (pp. 60–83). doi 10.1007/978-1-4612-3090-8_4
30
Hoppe H.-G. (2003). Phosphatase activity in the sea. Hydrobiologia493, 187–200. doi: 10.1023/A:1025453918247
31
Hoppe L. H.-G. Ullrich S. (1999). Profiles of ectoenzymes in the Indian Ocean: phenomena of phosphatase activity in the mesopelagic zone. Aquatic Microbial Ecology, vol. 19, 139–148. doi: 10.3354/ame019139
32
Hutchins D. A. Fu F.-X. Yang S.-C. John S. G. Romaniello S. J. Andrews M. G. et al . (2023). Responses of globally important phytoplankton species to olivine dissolution products and implications for carbon dioxide removal via ocean alkalinity enhancement. Biogeosciences20 (22), 4669–4682. doi 10.5194/bg-20-4669-2023
33
Ilyina T. Wolf-Gladrow D. Munhoven G. Heinze C. (2013). Assessing the potential of calcium-based artificial ocean alkalinization to mitigate rising atmospheric CO2 and ocean acidification. Geophysical Res. Lett.40, 5909–5914. doi: 10.1002/2013GL057981
34
Kathuria S. Martiny A. C. (2011). Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol.13, 74–83. doi: 10.1111/j.1462-2920.2010.02310.x
35
Keller D. P. Feng E. Y. Oschlies A. (2014). Potential climate engineering effectiveness and side effects during a high carbon dioxide-emission scenario. Nat. Commun.5 (1), 3304. doi: 10.1038/ncomms4304
36
Kheshgi H. S. (1995). Sequestering atmospheric carbon dioxide by increasing ocean alkalinityEnergy, 20 (9), 915–922. doi: 10.1016/0360-5442(95)00035-F
37
Kieft B. Li Z. Bryson S. Hettich R. L. Pan C. Mayali X. et al . (2021). Phytoplankton exudates and lysates support distinct microbial consortia with specialized metabolic and ecophysiological traits. Proc. Natl. Acad. Sci.118 (41), e2101178118. doi: 10.1073/pnas.2101178118
38
Korlević M. Markovski M. Zhao Z. Herndl G. J. Najdek M. (2021). Seasonal dynamics of epiphytic microbial communities on marine macrophyte surfaces. Front. Microbiol.12. doi: 10.3389/fmicb.2021.671342
39
Lenton A. Matear R. J. Keller D. P. Scott V. Vaughan N. E. (2018). Assessing carbon dioxide removal through global and regional ocean alkalinization under high and low emission pathways. Earth System Dynamics9, 339–357. doi: 10.5194/esd-9-339-2018
40
Lewis E. Wallace D. Allison L. J. (1998). Program developed for CO₂ system calculations (ORNL/CDIAC-105)Oak Ridge National Laboratory. doi: 10.2172/639712
41
Lueker T. J. Dickson A. G. Keeling C. D. (2000). Ocean pCO calculated from dissolved inorganic carbon, 2 alkalinity, and equations for K and K : validation based on 1–2 laboratory measurements of CO in gas and seawater at 2 equilibrium. Mar. Chem.70, 105–119. doi: 10.1016/S0304-4203(00)00022-0
42
Maeda S. I. Aoba R. Nishino Y. Omata T. (2019). A novel bacterial nitrate transporter composed of small transmembrane proteins. Plant Cell Physiol.60, 2180–2192. doi: 10.1093/pcp/pcz112
43
Margalef R. (1978). Life forms of phytoplankton as survival alternatives in an unstable environment. Oceanology Acta1, 493–509.
44
Marín-Samper L. Arístegui J. Hernández-Hernández N. Ortiz J. Archer S. D. Ludwig A. et al . (2024). Assessing the impact of CO2-equilibrated ocean alkalinity enhancement on microbial metabolic rates in an oligotrophic system. Biogeosciences21, 2859–2876. doi: 10.5194/bg-21-2859-2024
45
Miranda L. N. Hutchison K. Grossman A. R. Brawley S. H. (2013). Diversity and abundance of the bacterial community of the red macroalga porphyra umbilicalis: did bacterial farmers produce macroalgae? PloS One8, e58269. doi: 10.1371/journal.pone.0058269
46
Narciso Á. Gallo F. Valente A. Cachão M. Cros L. Azevedo E. B. et al . (2016). Seasonal and interannual variations in coccolithophore abundance off Terceira Island, Azores (Central North Atlantic). Continental Shelf Res.117, 43–56. doi: 10.1016/j.csr.2016.01.019
47
Oberlander J. L. Burke M. E. London C. A. Macintyre H. L. (2025). Assessing the impacts of simulated ocean alkalinity enhancement on viability and growth of nearshore species of phytoplankton. Biogeosciences22, 499–512. doi: 10.5194/bg-22-499-2025
48
Oschlies A. Stevenson A. Bach L. T. Fennel K. Rickaby R. E. M. Satterfield T. et al . (2023). Guide to Best Practices in Ocean Alkalinity Enhancement Research. doi: 10.5194/sp-2-oae2023
49
Paquay F. S. Zeebe R. E. (2013). Assessing possible consequences of ocean liming on ocean pH, atmospheric CO2 concentration and associated costs. Int. J. Greenhouse Gas Control17, 183–188. doi: 10.1016/j.ijggc.2013.05.005
50
Pedersen M. F. Hansen P. J. (2003). Effects of high pH on a natural marine planktonic community. Mar. Ecol. Prog. Ser.260, 19–31. doi: 10.3354/meps260019
51
Pontiller B. Martínez-García S. Joglar V. Amnebrink D. Pérez-Martínez C. González J. M. et al . (2022). Rapid bacterioplankton transcription cascades regulate organic matter utilization during phytoplankton bloom progression in a coastal upwelling system. ISME J.16, 2360–2372. doi: 10.1038/s41396-022-01273-0
52
Rau G. H. McLeod E. L. Hoegh-Guldberg O. (2012). The need for new ocean conservation strategies in a high-carbon dioxide world. Nat. Climate Change2, 720–724. doi: 10.1038/nclimate1555
53
Renforth P. Henderson G. (2017). Assessing ocean alkalinity for carbon sequestration. Rev. Geophysics55, 636–674. doi: 10.1002/2016RG000533
54
Renforth P. Jenkins B. G. Kruger T. (2013). Engineering challenges of ocean liming. Energy60, 442–452. doi: 10.1016/j.energy.2013.08.006
55
Riebesell U. Wolf-Gladrow D. Smetacek V. (1993). Carbon dioxide limitation of marine phytoplankton growth rates. Nature361, 249–251. doi: 10.1038/361249a0
56
Sebastian M. Ammerman J. W. (2009). The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J.3, 563–572. doi: 10.1038/ismej.2009.10
57
Seymour J. R. Amin S. A. Raina J.-B. Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol.2, 17065. doi: 10.1038/nmicrobiol.2017.65
58
Shi Z. Xu J. Li X. Li R. Li Q. (2019). Links of extracellular enzyme activities, microbial metabolism, and community composition in the river-impacted coastal waters. J. Geophysical Research: Biogeosciences124, 3507–3520. doi: 10.1029/2019JG005095
59
Shindoh S. Obayashi Y. Suzuki S. (2021). Induction of extracellular aminopeptidase production by peptides in some marine bacterial species. Microbes Environments36 (1), ME20150. doi: 10.1264/jsme2.ME20150
60
Smetacek V. (1999). Diatoms and the ocean carbon cycle. Protist150, 25–32. doi: 10.1016/S1434-4610(99)70006-4
61
Spilling K. Piiparinen J. Achterberg E. P. Arístegui J. Bach L. T. Camarena-Gómez M. T. et al . (2023). Extracellular enzyme activity in the coastal upwelling system off Peru: A mesocosm experiment. Biogeosciences20, 1605–1619. doi: 10.5194/bg-20-1605-2023
62
Spring S. Scheuner C. Göker M. Klenk H.-P. (2015). A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front. Microbiol.6. doi: 10.3389/fmicb.2015.00281
63
Subhas A. V. Marx L. Reynolds S. Flohr A. Mawji E. W. Brown P. J. et al . (2022). Microbial ecosystem responses to alkalinity enhancement in the North Atlantic Subtropical Gyre. Front. Climate4. doi: 10.3389/fclim.2022.784997
64
United Nations Framework Convention on Climate Change (2015). Paris Agreement to the United Nations Framework Convention on Climate Change, 12 December 2015 (T.I.A.S. No. 16-1104). Available online at: https://unfccc.int/resource/docs/2015/cop21/eng/10a01.pdf.
65
Xia Y. Kong Y. Thomsen T. R. Halkjær Nielsen P. (2008). Identification and ecophysiological characterization of epiphytic protein-hydrolyzing saprospiraceae (“ Candidatus epiflobacter” spp.) in Activated Sludge. Appl. Environ. Microbiol.74, 2229–2238. doi: 10.1128/AEM.02502-07
66
Xin X. Goldenberg S. U. Taucher J. Stuhr A. Arístegui J. Riebesell U. (2024). Resilience of phytoplankton and microzooplankton communities under ocean alkalinity enhancement in the oligotrophic ocean. Environ. Sci. Technol. 58 (47), 20918–20930. doi: 10.1021/acs.est.4c09838
67
Yamada N. Suzumura M. (2010). Effects of seawater acidification on hydrolytic enzyme activities. Journal of Oceanography66 (2), 233–241. doi 10.1007/s10872-010-0021-0
Summary
Keywords
OAE, phytoplankton, marine heterotrophic bacteria, extracellular enzymatic activity, 16S, global change, microbial loop, carbon dioxide removal
Citation
de Castro I, Ribeiro SC, Louvado A, Gomes NCM, Cachão M, Brito de Azevedo E and Barcelos e Ramos J (2025) Ocean liming effect on a North Atlantic microbial community: changes in composition and rates. Front. Mar. Sci. 12:1602158. doi: 10.3389/fmars.2025.1602158
Received
28 March 2025
Accepted
30 June 2025
Published
31 July 2025
Volume
12 - 2025
Edited by
Yuri Artioli, Plymouth Marine Laboratory, United Kingdom
Reviewed by
Gerhard Josef Herndl, University of Vienna, Austria
Paraskevi Pitta, Hellenic Centre for Marine Research (HCMR), Greece
Updates
Copyright
© 2025 de Castro, Ribeiro, Louvado, Gomes, Cachão, Brito de Azevedo and Barcelos e Ramos.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inês de Castro, inesmcastro@ua.pt
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.