Abstract
Bacterivores play a key role in transferring energy through microbial food webs and are broadly classified as heterotrophic or mixotrophic based on the presence of inherent chloroplasts. Mixotrophs, especially in low-latitude regions, have recently gained attention due to their dual roles as primary producers and bacterial grazers, which enable them to thrive in nutrient-poor, stratified waters where traditional autotrophs or heterotrophs may be less competitive. This study investigated the composition of bacterivores in the coastal waters of northeastern Taiwan during the warm season (July and August 2022). A combination of LysoTracker staining, flow cytometry sorting, and 18S rRNA gene amplicon sequencing was used to rapidly identify heterotrophic and mixotrophic nanoflagellates (HNFs and MNFs, respectively) based on chloroplast autofluorescence. The dominant eukaryotic lineages included Alveolata, Stramenopiles, Opisthokonta, Chlorophyta, and Rhizaria. HNFs were primarily composed of MAST clades, choanoflagellates (Opisthokonta), Telonemia, and Radiolaria, while MNFs included cryptophytes, haptophytes, and chlorophytes such as Mamiella, Mantoniella and Tetraselmis, as well as stramenopiles like Dictyochophyceae and Chrysophyceae. Notably, several non-motile chlorophytes, including Chloropicon and members of Pycnococcaceae, were identified as potential bacterivores based on Lysotracker signals; however, their phagotrophic capability requires further confirmation in future studies. Despite the limitations of LysoTracker-based methods, this study reveals previously overlooked mixotrophic diversity and highlights the need for further validation. These findings advance our understanding of carbon flow and microbial dynamics in subtropical marine ecosystems under changing environmental conditions.
1 Introduction
Bacterivores play a crucial role in linking carbon flow from picoplankton to higher trophic levels. Based on the presence of inherent chloroplasts, they can be categorized as mixotrophic or heterotrophic. Heterotrophic eukaryotes are protists that lack plastid pigments and, in addition to consuming bacteria, may employ other trophic strategies, including osmotrophy, parasitism, saprotrophy, and symbiosis (Worden et al., 2015). Broadly defined, mixotrophy refers to organisms capable of performing photosynthesis while also assimilating dissolved organic matter through osmotrophy (Stoecker, 1998; Flynn et al., 2013). Although this strategy is widespread among phytoplankton, it is difficult to observe directly and often involves complex metabolic pathways. As a result, recent definitions of mixotrophy have more narrowly focused on phago-mixotrophs, which can be detected using experimental approaches such as fluorescently labeled bacteria and acidic vesicle staining to trace ingestion and digestion activity (Mitra et al., 2016). In addition, Mitra et al. (2016) proposed a functional classification framework for mixoplankton, dividing them into constitutive mixoplankton (CM), which possess innate photosynthetic machinery, and non-constitutive mixoplankton (NCM), which rely on plastids acquired through the ingestion of other phototrophs. NCMs can be further subdivided into generalist types, which can feed on a wide range of phototrophic prey, and plastid specialists, which tend to retain and use specific plastids for extended periods. This classification provides a mechanistic foundation for interpreting the ecological roles of diverse mixotrophs across environmental gradients. Mixotrophic eukaryotes have been shown to contribute substantially to bacterial consumption - sometimes accounting for over 50% of total bacterial ingestion in the ocean (Unrein et al., 2007; Zubkov and Tarran, 2008; Hartmann et al., 2012). However, despite this significant ecological role, the current diversity of mixotrophs is likely underestimated. While marine small eukaryotes are estimated to include more than tens of thousands of morphospecies (De Vargas et al., 2015), fewer than 500 mixotrophic species with confirmed phagotrophic capabilities are currently documented in the Mixoplankton Database (MDB) (Mitra et al., 2023). This suggests that many mixotrophs remain unidentified or experimentally unverified. Moreover, traditional laboratory-based grazing experiments using pure cultures are limited in their ability to validate mixotrophic activity in natural communities, making it difficult to fully assess in situ diversity and ecological function.
The emerging ecological importance of mixotrophy challenges the traditional dichotomy of autotrophy versus heterotrophy and highlights the need to reconsider trophic strategies in marine carbon cycling. Traditionally, marine eukaryotes have been dichotomously classified as either autotrophic or heterotrophic. However, this oversimplification fails to reflect the ecological complexity of marine food webs and may result in inaccuracies in carbon cycle models (Flynn et al., 2013). Recent studies have underscored the prevalence and ecological significance of mixotrophy in the ocean, particularly in enhancing the efficiency and nutritional quality of microbial food chains (Ward and Follows, 2016; Leles et al., 2021). Model predictions and metatranscriptomic data both indicate that mixotrophs are especially important in low-latitude regions (Edwards, 2019; Lambert et al., 2022) These mixotrophic strategies are especially relevant in subtropical marine environments, where elevated temperatures and low nutrient concentrations are common. In such oligotrophic, warm-water ecosystems, mixotrophs benefit from their metabolic flexibility, allowing them to supplement photosynthesis with phagotrophy and thus maintain growth when inorganic nutrients are scarce (Stoecker, 1998; Mitra et al., 2016). Their dual nutritional strategy provides a competitive advantage and may lead to increased dominance of mixotrophs under future climate scenarios (Stoecker et al., 2017). Experimental studies show that mixotrophic phagotrophic activity is modulated by light, nutrient availability, and prey abundance (McKie-Krisberg and Sanders, 2014; Moeller et al., 2024), while interactions such as prey selectivity and predator-derived metabolites can further shape bacterial community composition (Gerea et al., 2019). Therefore, in subtropical marine systems, mixotrophy may not only sustain microbial predator populations but also significantly influence microbial community structure and trophic interactions. Understanding the diversity and ecological roles of mixotrophic organisms in these regions is critical for improving estimates of microbial carbon fluxes, especially under changing oceanographic conditions. Although an increasing body of laboratory evidence supports bacterivory among various protists, such as Dolichomastix, Mantoniella, Nephroselmis, Pterosperma, Pyramimonas (Chlorophyta) and Dictyochophyceae and Chrysophyceae (Stramenopiles) (McKie-Krisberg et al., 2015; Bock et al., 2021; Li et al., 2022), the in situ composition and functional importance of bacterivores in natural seawater - particularly in subtropical zones - remain poorly characterized (Frias-Lopez et al., 2009; Wilken et al., 2023). The small size and dynamic trophic activity of both mixotrophic and heterotrophic bacterivores pose additional challenges for field-based studies.
Various techniques have been developed to measure eukaryotic ingestion and identify bacterivores, including the use of fluorescent beads, fluorescently labeled bacteria, and stable isotope probing such as ¹³C and 15N-labelled prey (Keller et al., 1994; McKie-Krisberg and Sanders, 2014; Orsi et al., 2018; Wilken et al., 2023). However, due to the small size and prey selectivity of bacterivores, the addition of surrogate prey may lead to underestimations of their presence and activity (Bock et al., 2021). Alternatively, the acid vesicle tracker method provides a rapid approach for identifying bacterivores, as it stains acidified vesicles indicative of phagocytic activity (Florenza et al., 2024). Despite its advantages, this method does not directly quantify ingestion rates and requires live samples, making sample preservation challenging (Beisner et al., 2019; Millette et al., 2024). Nonetheless, acid vesicle trackers have been successfully applied to detect active bacterivores in both laboratory cultures and natural environments, offering a powerful tool for investigating the bacterivory of eukaryotes (Wilken et al., 2019; Costa et al., 2022). These studies confirm that acid vesicle trackers, such as LysoTracker™ and LysoSensor™, are effective for distinguishing active bacterivores from non-phagocytic protists.
Given the ecological significance of bacterivores, understanding their prevalence and species composition is critical for refining carbon cycle estimates in marine ecosystems. In this study, we employed acid vesicle trackers to identify bacterivores and combined this approach with 18S rRNA gene amplicon sequencing to analyze their community composition in the subtropical coastal waters of northeastern Taiwan. Previous research has shown that pigmented nanoflagellates in these waters serve as important bacterivores, often exhibiting higher cyanobacterial grazing rates than non-pigmented nanoflagellates (Tsai et al., 2006, 2011). However, taxonomic information on these bacterivores remains limited, and their ecological roles are not well characterized (Chan et al., 2019). To address this gap, our study aims to identify bacterivores using LysoTracker™ Green combined with flow cytometric sorting to distinguish mixotrophic and heterotrophic bacterivores while characterizing their community composition through 18S rRNA gene amplicon sequencing for enhanced taxonomic resolution. Additionally, we assess the ecological significance of mixotrophic and heterotrophic bacterivores in a low-latitude region, considering their roles in global biogeochemical processes in the context of climate change. By integrating fluorescence-based staining with molecular sequencing, this study provides a more comprehensive understanding of the composition of the bacterivorous community.
2 Methods
2.1 Sample collection and flow cytometry cell sorting
Our study was conducted in a subtropical coastal marine ecosystem, where we sampled at seven stations on 22 July and 18 August 2022 (Figure 1A; Table 1). The distance between stations was approximately 10–12 km. Environmental variables, including temperature, salinity, and turbidity, were measured using the Ocean Seven 316Plus CTD (IDRONAUT, Italy). To study the community composition of bacterivores, 2 liters of surface seawater were collected using a 5-liter Niskin bottle and incubated on deck in a shaded area with running seawater to maintain ambient conditions. Before flow cytometry analysis, the seawater was concentrated to 15 mL using tangential flow filtration (Vivaflow® 200 50,000 MWCO PES). Aliquots of the seawater samples were stained with LysoTracker at a final concentration of 50 nM for 10 minutes under dark conditions (Sintes and Del Giorgio, 2010). The stained cells were categorized into mixotrophic nanoflagellates (MNF) and heterotrophic nanoflagellates (HNF) based on the presence of inherent chloroplasts (Figure 1B). A subsample was filtered through a 20 μm sieve prior to flow cytometry analysis, and 1,000 cells of MNF and HNF were sorted in triplicate at each station and in each month using a Beckman Coulter MoFlo XDP within 12 hours of sample collection. We filtered the remaining concentrated water onto a 0.8 μm, 47 mm diameter polycarbonate (PC) filter as a background. Then, DNA was extracted from the filters using the DNeasy® PowerWater® Kit (Qiagen) following the manufacturer’s instructions.
Figure 1
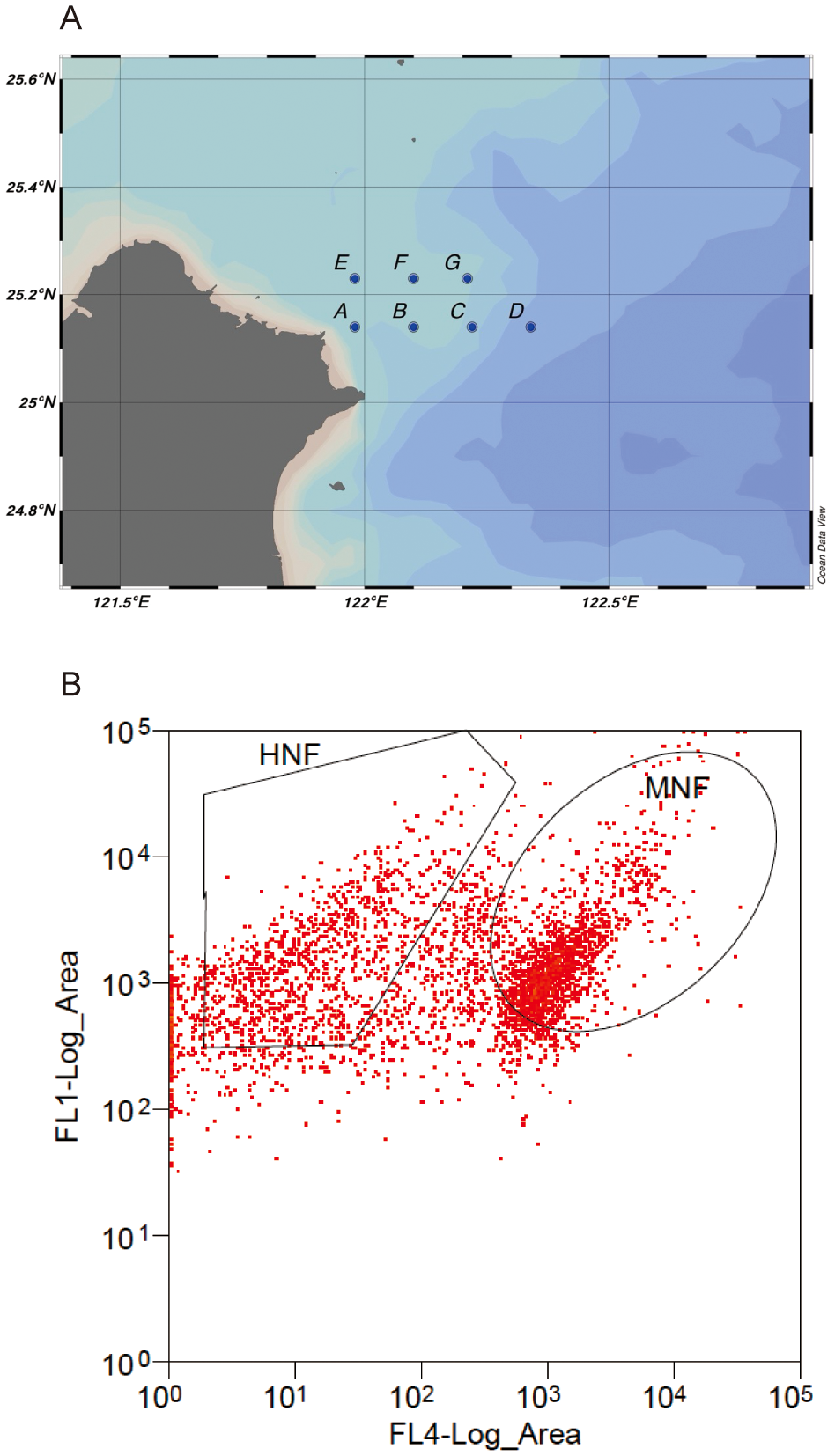
(A) The sampling locations included seven sites (A to G), with samples collected twice in July and August 2022. (B) HNF and MNF cells sorted using flow cytometry, with FL1 and FL4 representing green and red fluorescent signals, respectively, after blue excitation.
Table 1
| Month | Station | Latitude | Longitude | Temp. | Sal. | Turb. |
|---|---|---|---|---|---|---|
| Jul | A | 25.14 | 121.98 | 27.53 | 32.9 | 0.74 |
| Jul | B | 25.14 | 122.1 | 28.49 | 33.3 | 0.56 |
| Jul | C | 25.14 | 122.22 | 28.21 | 33.3 | 0.6 |
| Jul | D | 25.14 | 122.34 | 28.06 | 33.2 | 0.62 |
| Jul | E | 25.23 | 121.98 | 28.74 | 33.2 | 76 |
| Jul | F | 25.23 | 122.1 | 28.89 | 33.3 | 0.82 |
| Jul | G | 25.23 | 122.21 | 26.57 | 33.6 | 1.08 |
| Aug | A | 25.14 | 121.98 | 22.12 | 27.5 | 1.36 |
| Aug | B | 25.14 | 122.1 | 23.69 | 27.8 | 0.21 |
| Aug | C | 25.14 | 122.22 | 23.46 | 27.9 | 0.66 |
| Aug | D | 25.14 | 122.34 | 23.83 | 27.9 | 0.35 |
| Aug | E | 25.23 | 121.98 | 23.71 | 27.8 | 6.53 |
| Aug | F | 25.23 | 122.1 | 24.03 | 27.9 | -0.05 |
| Aug | G | 25.23 | 122.21 | 23.55 | 27.8 | 9.26 |
Background information for the sampling stations includes longitude (°E), latitude (°N), temperature (Temp., °C), salinity (Sal., practical salinity unit, PSU), and turbidity (Turb., formazin turbidity unit, FTU), all measured using CTD probes.
2.2 18S rRNA gene amplicon sequencing and data analysis
A total of 1,000 sorted cells were heat-lysed, and 5 µL of the lysate was subjected to genomic amplification using the REPLI-g® Mini Kit (Qiagen) with multiple displacement amplification (MDA) technology, followed by amplicon sequencing. Additionally, to assess the MDA effect on the samples, a subsample of background DNA was amplified to evaluate potential compositional variation and biases introduced during the amplification process. All DNA samples were PCR-amplified targeting the V4 region of the 18S rRNA gene using the eukaryotic universal primer set TAReuk454FWD1 (5’-[Illumina adaptor]-CCAGCASCYGCGGTAATTCC-3’) and TAReukREV3167 (5’-[Illumina adaptor]-ACTTTCGTTCTTGATYRA-3’) (Stoeck et al., 2010). A total of 112 samples were collected (1 background, 1 MDA, 3 HNF, and 3 MNF per station × 7 stations × 2 months). PCR failed in 12 samples, resulting in 100 successfully sequenced samples. Amplicon sequencing was performed on the Illumina MiSeq platform (paired-end 2 × 300 bp) at Genomics company, Taiwan. Primer sequences were removed using Cutadapt (Martin, 2011), and the reads were subsequently processed using the DADA2 pipeline (Callahan et al., 2016) with the parameter setting truncLen = c(220, 180). The resulting amplicon sequence variants (ASVs) were annotated using the classifier implemented in DADA2. Taxonomic annotation was conducted using the PR2 database version 5.0.0, assigning classifications across nine taxonomic levels: Domain, Supergroup, Division, Subdivision, Class, Order, Family, Genus, and Species, to provide a comprehensive overview of eukaryotic composition (Guillou et al., 2012) (Supplementary File 1). The raw reads from the 100 samples have been deposited in the Sequence Read Archive (SRA) of GenBank under BioProject accession number PRJNA1121045.
2.3 Data analysis
The number of sequencing reads in all samples ranged from 7,055 to 319,274. For community composition, bar plots and treemaps were generated from the original (non-resampled) dataset. Bar plots summarize composition at the division level, whereas treemaps depict genus-level composition for the HNF and MNF categories. Reads assigned to diatoms (classes Bacillariophyceae, Coscinodiscophyceae, Mediophyceae) and dinoflagellates (subdivision Dinoflagellata) were excluded as they may produce fluorescence signals unrelated to phagocytosis (see further details in the Results and Discussion section). To control for unequal sequencing depth across samples, the beta-diversity comparisons were based on resampled data in which each sample was normalized to the minimum read depth (7,055 reads) and the resampling was repeated 100× (Supplementary File 2). Bray–Curtis dissimilarities were used for non-metric multidimensional scaling (NMDS) ordinations and the accompanying ANOSIM tests, and hierarchical clustering was performed using the Ward.D2 method. All statistical analyses were conducted using R version 3.6.0.
3 Results and discussion
This study was conducted in the northeastern coastal waters of Taiwan during the warm season, specifically in July and August 2022. The average temperature across all stations was 28.07 ± 0.30°C in July (ranging from 26.57 to 28.89°C) and 23.48 ± 0.23°C in August (22.12 to 24.03°C). Similarly, average salinity was 33.25 ± 0.08 PSU in July (32.9–33.6 PSU) and 27.80 ± 0.05 PSU in August (27.5–27.9 PSU). Overall, both temperature and salinity at the sampling stations were significantly higher in July than in August (Table 1, t-test, p < 0.001 for both temperature and salinity). The 18S rRNA gene amplicon analysis revealed that the small eukaryotic community was predominantly composed of Alveolates, Stramenopiles, Opisthokonta, and Chlorophyta (Figure 2). Dinoflagellates accounted for 20% of the total reads in HNFs and 15% in MNFs, whereas diatoms accounted for 0.1% in HNFs and 68% in MNFs.
Figure 2
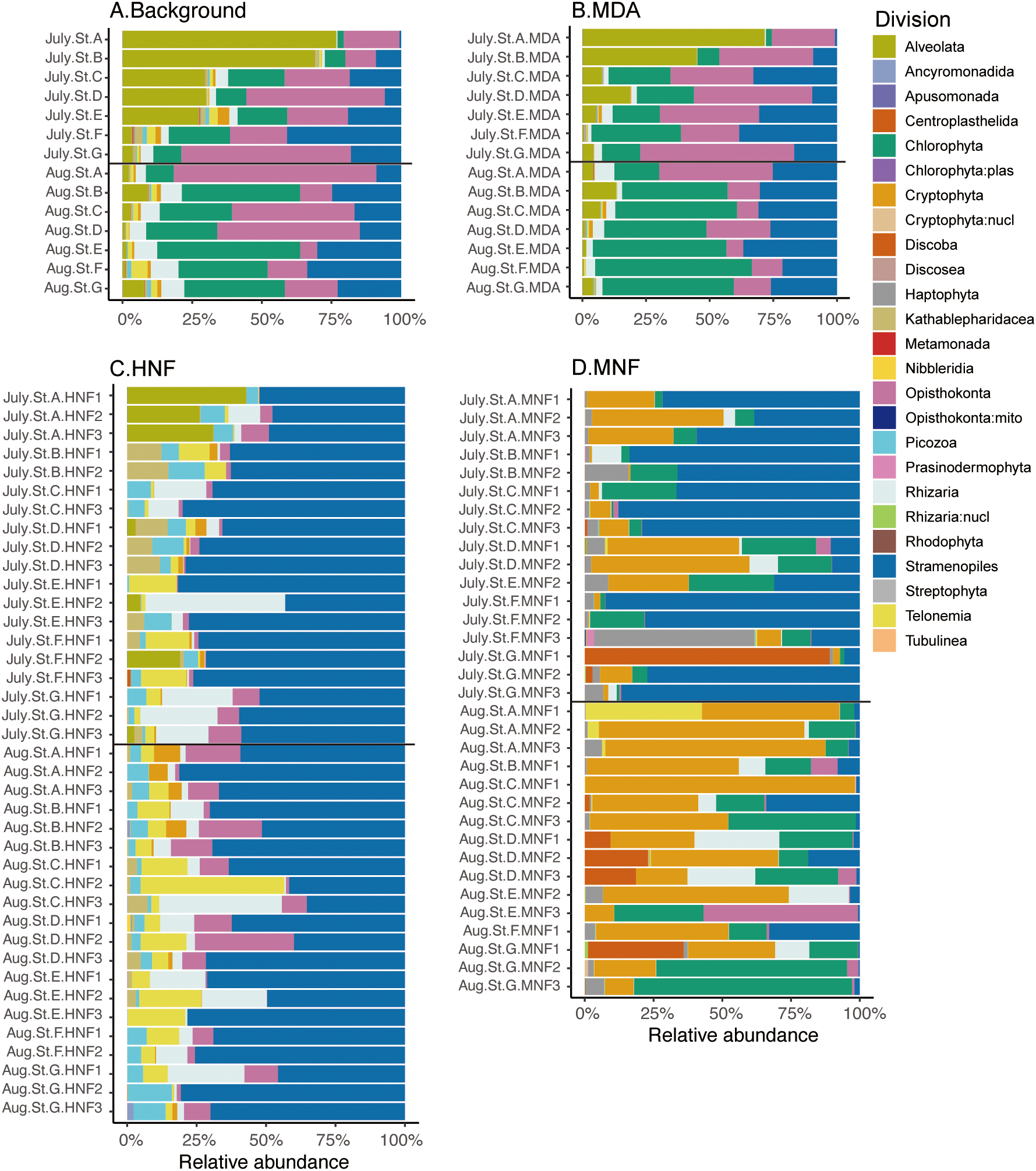
Stacked bar charts showing the taxonomic composition and relative abundance of 18S rRNA gene amplicons at the division level for (A) background samples (n = 14), (B) background samples with MDA treatment (n = 14), (C) the HNF category (n = 39), and (D) the MNF category (n = 33). Different divisions are represented by distinct colors and these plots were generated from the original dataset.
We propose that bacterivores possess acid vesicles detectable by LysoTracker staining, and the samples were subsequently categorized into heterotrophic and mixotrophic groups based on their inherent chloroplasts (Figure 1B). Non-metric multidimensional scaling (NMDS) and hierarchical cluster analyses were performed to assess the relationships among community compositions across all samples. These results revealed that background and MDA-treated samples often cluster together (Figure 3), indicating that their taxonomic compositions are not markedly different and suggesting that MDA had minimal impact on overall community composition (Figure 2). Furthermore, MNF and HNF communities generally formed distinct clusters; an exception was the July HNF samples from station A, which clustered with the background and MDA groups. By contrast, MNF and HNF communities did not cluster by sampling month (Figure 3B).
Figure 3
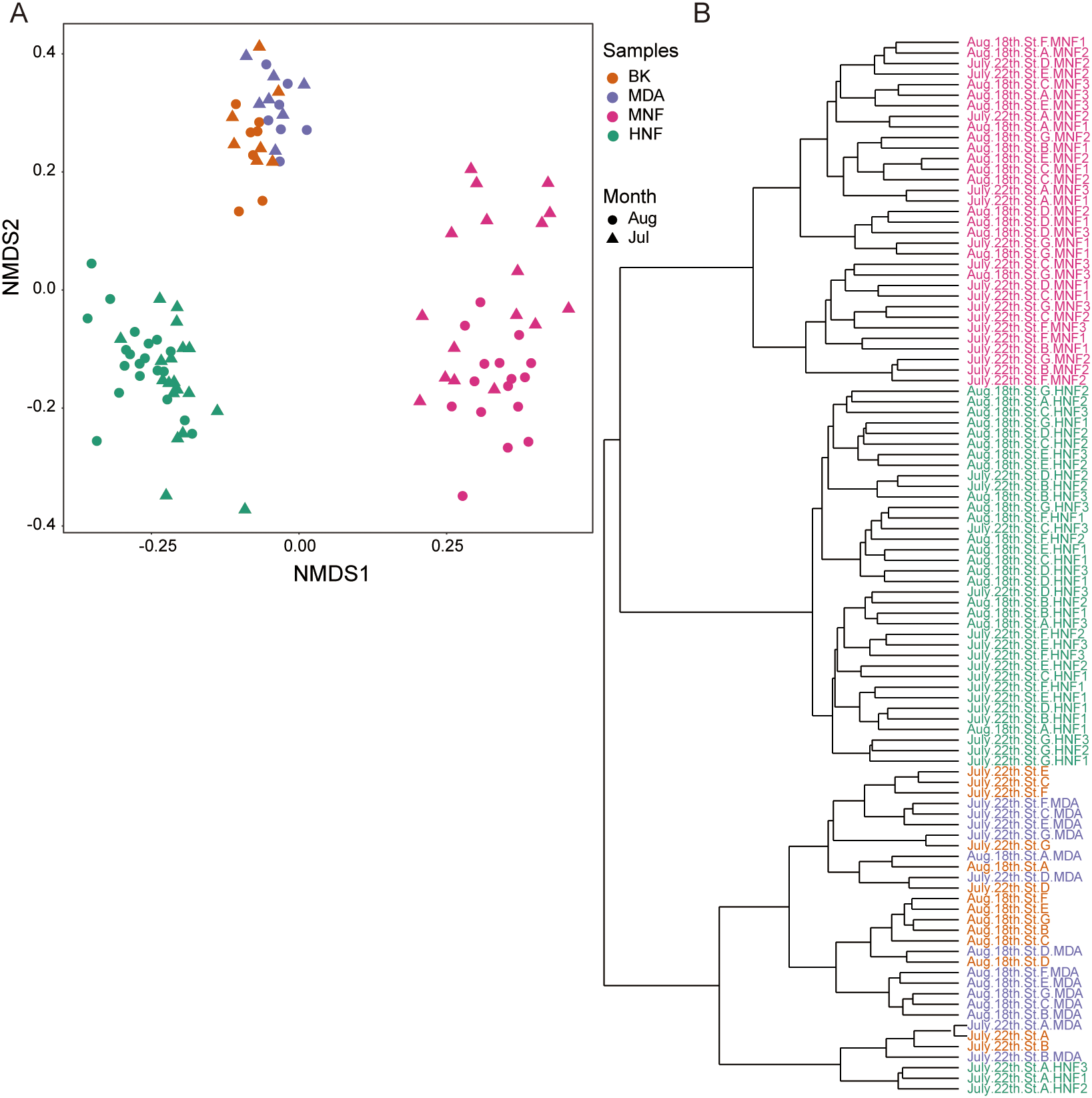
(A) Non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis dissimilarity, with colors representing sample types (Bk: background, MDA: multiple displacement amplification, MNF, HNF) and shapes indicating sampling months (July and August), (B) Hierarchical clustering dendrogram of community composition generated using the Ward.D2 method. Both dinoflagellates and diatoms were excluded from the analysis.
The 18S amplicons of HNFs were predominantly composed of the divisions Stramenopile, Rhizaria, Opisthokonta, Telonemia and Picozoa (Figures 2, 4). Among these, marine stramenopiles (MASTs) were particularly prominent, including MAST-1D, -3C, -4B, -4C, -4D, -4E, -7D, and -10 (Figure 4, Table 2). Notably, MAST-4C was abundant in the subtropical NW Pacific Ocean and has also been reported at high abundant in tropical to temperate regions (Massana et al., 2014; Lin et al., 2022b). In addition to MASTs, other stramenopiles groups such as MOCH-4, thraustochytrids, labyrinthulids, and the genus Paraphysomonas also contributed significantly to the HNF community. Within Opisthokonta, Acanthoecidae Group G and Stephanoecidae Group H (both at the family level) together accounted for more than half of the reads, and both belong to the choanoflagellates. Telonemia Group-1 and Group-2 were identified as important HNF in our dataset (Figures 4, 5). Previous studies have reported that T. antarcticum (a member of Telonemia Group-2) is capable of ingesting bacteria (Klaveness et al., 2005). However, our current understanding of the ecological roles of this group remains limited. Picozoa emerged as a notable HNF group, and their low abundance in the mixotrophic fraction suggests a potential role as algivores (Figure 4) (Yoon et al., 2011). In addition, the non-plastid cryptophyte Goniomonas sp. also contributed significantly to the HNF community (Figure 4).
Figure 4
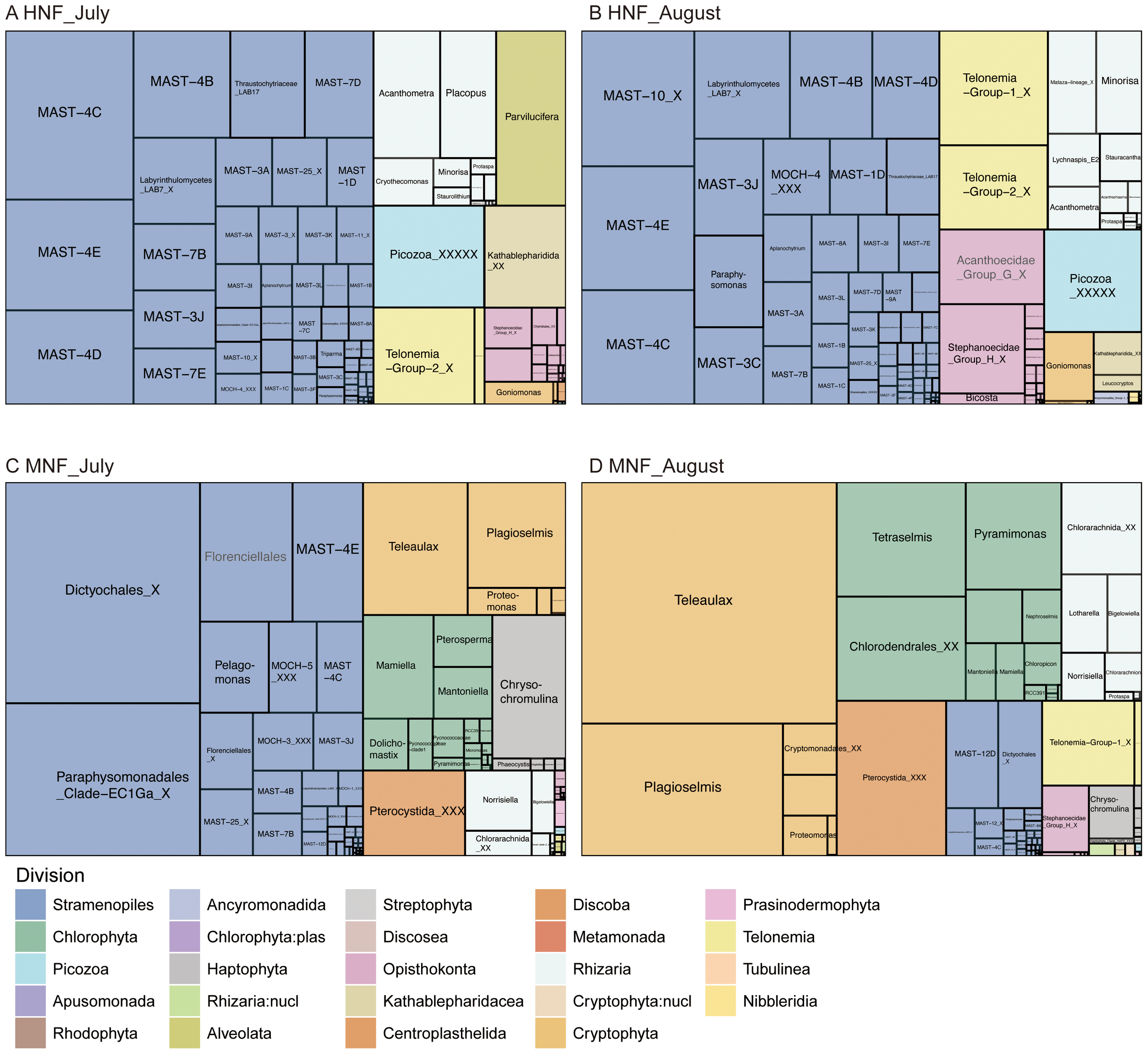
Treemaps showing the taxonomic composition and relative abundance of 18S rRNA gene amplicons at the genus level within each division for HNF in (A) July and (B) August, and for MNF in (C) July and (D) August. Divisions are indicated by distinct colors. Boxes from the same genus are merged; when genus-level assignments were unavailable (NA), higher taxonomic ranks are shown in gray text. Reads assigned to dinoflagellates and diatoms were removed. Treemaps were generated from the original dataset.
Table 2
| Month | Rank | ASV | Division | Class | Species |
|---|---|---|---|---|---|
| Jul | 1 | 8 | Stramenopiles | Sagenista | MAST-4E_sp. |
| Jul | 2 | 11 | Stramenopiles | Sagenista | MAST-4C_sp. |
| Jul | 3 | 47 | Stramenopiles | Sagenista | MAST-4D_sp. |
| Jul | 4 | 55 | Stramenopiles | Acantharea | Acanthometra_pellucida |
| Jul | 5 | 3 | Stramenopiles | Perkinsida | Parvilucifera_multicavata |
| Jul | 6 | 48 | Stramenopiles | Sagenista | MAST-4B_sp. |
| Jul | 7 | 52 | Stramenopiles | Sagenista | MAST-4C_sp. |
| Jul | 8 | 51 | Kathablepharidacea | Kathablepharidea | Kathablepharidida_XX_sp. |
| Jul | 9 | 117 | Stramenopiles | Sagenista | MAST-7D_sp. |
| Jul | 10 | 25 | Stramenopiles | Sagenista | Labyrinthulomycetes_LAB7_X_sp. |
| Aug | 1 | 8 | Stramenopiles | Sagenista | MAST-4E_sp. |
| Aug | 2 | 10 | Stramenopiles | Sagenista | MAST-10_X_sp. |
| Aug | 3 | 11 | Stramenopiles | Sagenista | MAST-4C_sp. |
| Aug | 4 | 32 | Telonemia | Telonemia_XX | Telonemia-Group-1_X_sp. |
| Aug | 5 | 25 | Stramenopiles | Sagenista | Labyrinthulomycetes_LAB7_X_sp. |
| Aug | 6 | 49 | Opisthokonta | Choanoflagellatea | Acanthoecidae_Group_G (Family level) |
| Aug | 7 | 38 | Telonemia | Telonemia_XX | Telonemia-Group-2_X_sp. |
| Aug | 8 | 41 | Stramenopiles | Sagenista | MAST-4D_sp. |
| Aug | 9 | 18 | Stramenopiles | MOCH-4 | MOCH-4_XXX_sp. |
| Aug | 10 | 48 | Stramenopiles | Sagenista | MAST-4B_sp. |
Top 10 most abundant HNF ASVs in July and August. Ranks indicate the average relative abundances across all HNF samples.
Division, Class, and Species information correspond to taxonomic classifications based on the PR2 database. Taxon names with an “X” indicate unidentified lineages that are only assigned to higher taxonomic levels in the PR2 database.
Figure 5
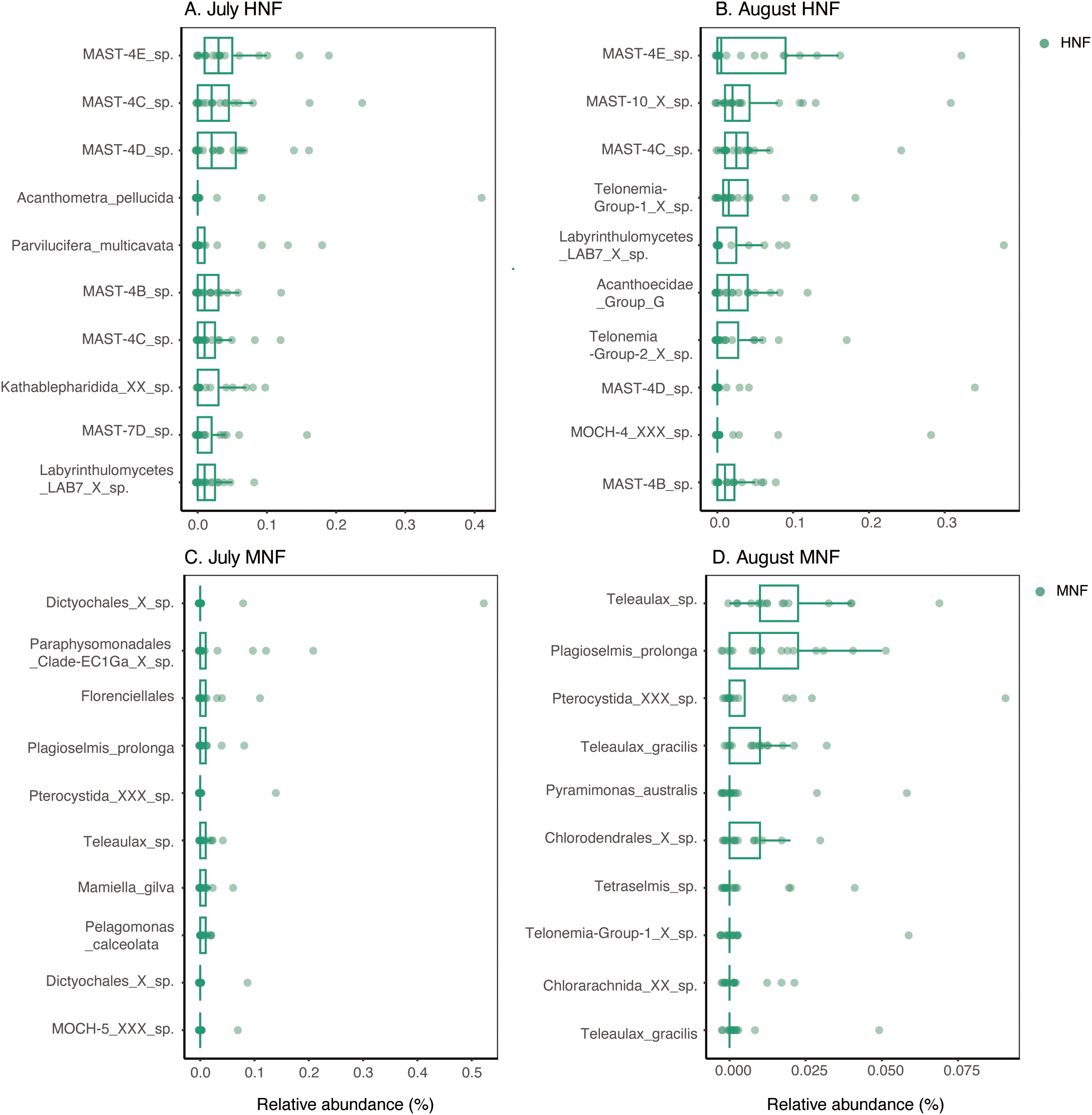
Boxplots showing the relative abundances of the top 10 ASVs in the sorted samples of (A) July HNF, (B) August HNF, (C) July MNF, and (D) August MNF. Each box represents the interquartile range of relative abundance across replicate samples, with whiskers indicating variability outside the upper and lower quartiles. Both dinoflagellates and diatoms were excluded from the analysis. Detailed taxonomic classifications of these ASVs are provided in Tables 2 and 3.
Rhizaria represents an important group of bacterivores among both HNF and MNF identified in this study (Figure 2), although the dominant genera differed between the two (Figures 4, 5). The supergroup of Rhizaria comprises Cercozoa and Radiolaria, which exhibit considerable diversity in cell size and morphology, ranging from free-living flagellates and amoeboflagellates to parasitic protists (Pawlowski and Burki, 2009). In our dataset, Cercozoa was primarily represented by Minorisa and Placopus (both classified as HNF), as well as Norrisiella, Bigelowiella, and an unassigned genus within the Chlorarachniophyceae (all classified as MNF). Radiolaria was dominated by Acanthometra, Lychnaspis_E2, and Stauracantha, all of which were categorized as HNF in this study (Figure 4). However, Acanthometra has previously been categorized as a non-constitutive mixotroph (NCM) due to its unique symbiotic relationships in the MDB (Mitra et al., 2023). The trophic modes of many Rhizaria lineages remain poorly understood, although several genera have been reported to exhibit bacterivory, such as Acanthometra, Bigelowiella, and Minorisa (Yoo and Palenik, 2021; Rodríguez-Martínez et al., 2022; Wilken et al., 2023). While most mixotrophic Rhizaria are considered NCMs (Stoecker et al., 2017), Bigelowiella and Norrisiella have been classified as constitutive mixotrophs (CMs) under the current definition of mixoplankton (Mitra et al., 2016).
Based on the 18S amplicon data obtained through flow cytometry sorting with inherent chloroplasts, we identified Stramenopiles, Cryptophyta, Chlorophyta, Centroplasthelida, Rhizaria, and Haptophyta as key mixotrophic eukaryotes in this study (Figure 4). Stramenopiles exhibit a diverse range of nutritional modes, including autotrophy, heterotrophy, osmotrophy, mixotrophy, and parasitism (Jirsová and Wideman, 2024). Within Stramenopiles, the most abundant mixotrophic reads were assigned to unknown genera within Dictyochales (classified at the family level in PR2). The second most abundant group was Paraphysomonadales Clade-EC1Ga X sp. (within Chrysophyceae) (Table 3; Figure 5), whose physiological and ecological characteristics remain poorly understood. Members of Dictyochophyceae in stramenopiles have been recognized as important mixotrophic bacterivores in the subtropical Pacific and in freshwater systems (Frias-Lopez et al., 2009; Li et al. 2021; Li et al., 2022; Florenza et al., 2024; Koppelle et al., 2024). Similarly, closely related taxa such as Florenciellales and Dictyochales were identified as mixotrophic bacterivores in this study (Table 3, Figures 4, 5). Among the mixotrophic taxa, Florenciella is a common bacterivorous genus in tropical and subtropical waters (Choi et al., 2020), where it exhibits increased grazing activity under low-nitrogen conditions (Jirsová and Wideman, 2024). The second most abundant mixotrophic division was Cryptophyta, which comprised the genera Teleaulax and Plagioselmis (Figures 2, 4C, D). Cryptophytes are recognized as important bacterivores in marine ecosystems (Unrein et al., 2014; Yoo et al., 2017). Additionally, previous studies have indicated that haptophytes play an important role as bacterivores in this region (Chan et al., 2019). In this study, we documented Chrysochromulina as the dominant genus in Haptophyta (Figures 4C, D). Although Cryptophyta and Haptophyta generally exhibited low relative abundances in the unsorted 18S survey, their sequences were enriched following sorting based on the LysoTracker signal in this study (Figure 2).
Table 3
| Month | Rank | ASV | Division | Class | Species |
|---|---|---|---|---|---|
| Jul | 1 | 26 | Stramenopiles | Dictyochophyceae | Dictyochales_X_sp. |
| Jul | 2 | 43 | Stramenopiles | Chrysophyceae | Paraphysomonadales_Clade-EC1Ga_X_sp. |
| Jul | 3 | 24 | Stramenopiles | Dictyochophyceae | Florenciellales (Family level) |
| Jul | 4 | 59 | Cryptophyta | Cryptophyceae | Plagioselmis_prolonga |
| Jul | 5 | 172 | Centroplasthelida | Pterocystida | Pterocystida_XXX_sp. |
| Jul | 6 | 60 | Cryptophyta | Cryptophyceae | Teleaulax_sp. |
| Jul | 7 | 234 | Chlorophyta | Mamiellophyceae | Mamiella_gilva |
| Jul | 8 | 94 | Stramenopiles | Pelagophyceae | Pelagomonas_calceolata |
| Jul | 9 | 197 | Stramenopiles | Dictyochophyceae | Dictyochales_X_sp. |
| Jul | 10 | 87 | Stramenopiles | MOCH-5 | MOCH-5_XXX_sp. |
| Aug | 1 | 60 | Cryptophyta | Cryptophyceae | Teleaulax_sp. |
| Aug | 2 | 59 | Cryptophyta | Cryptophyceae | Plagioselmis_prolonga |
| Aug | 3 | 172 | Centroplasthelida | Pterocystida | Pterocystida_XXX_sp. |
| Aug | 4 | 118 | Cryptophyta | Cryptophyceae | Teleaulax_gracilis |
| Aug | 5 | 138 | Chlorophyta | Pyramimonadophyceae | Pyramimonas_australis |
| Aug | 6 | 50 | Chlorophyta | Chlorodendrophyceae | Chlorodendrales_X_sp. |
| Aug | 7 | 151 | Chlorophyta | Chlorodendrophyceae | Tetraselmis_sp. |
| Aug | 8 | 126 | Telonemia | Telonemia_XX | Telonemia-Group-1_X_sp. |
| Aug | 9 | 380 | Rhizaria | Chlorarachniophyceae | Chlorarachnida_XX_sp. |
| Aug | 10 | 232 | Cryptophyta | Cryptophyceae | Teleaulax_gracilis |
Top 10 most abundant MNF ASVs in July and August.
Division, Class, and Species information correspond to taxonomic classifications based on the PR2 database. Taxon names with an “X” indicate unidentified lineages that are only assigned to higher taxonomic levels in the PR2 database. MAST ASVs were excluded from this table, as they are known to represent HNFs.
Ranks indicate the average relative abundances across all MNF samples.
In this study, we identified lots chlorophytes in the mixotrophic category, including Dolichomastix, Mamiella, Mantoniella, Pterosperma, Pyramimonas, Tetraselmis, and the Pycnococcaceae clade I (Figure 4). Early research provided limited evidence of phagotrophic capability in chlorophytes, leading to their classification as purely autotrophic (Raven, 1997). However, recent studies have demonstrated phagotrophic behavior in several chlorophyte genera, such as Micromonas, Nephroselmis, Picochlorum, Pterosperma, and Pyramimonas (McKie-Krisberg and Sanders, 2014; Bock et al., 2021; Pang et al., 2023). Although phagotrophy in Micromonas remains debated, a previous study suggested that fluorescent-labeled surrogates may adhere to its cell surface rather than being actively ingested (Jimenez et al., 2021). Picochlorum has been confirmed as a bacterivore in monoculture experiments (Pang et al., 2023); however, it appeared in relatively low read numbers in the LysoTracker staining results compared to the background samples (Figure 2). This observed inconsistency may stem from the adaptive evolution of different species or clades, as well as the dynamic trade-offs imposed by environmental factors and food availability, which constrain mixotrophic metabolisms (Edwards et al., 2023; Edward et al. 2024). These chlorophytes are categorized as CM, and although their bacterivory rates may be relatively low, such low rates may still be sufficient to meet the nutrient demands of mixoplankton in oligotrophic waters. Moreover, model predictions suggest that phagocytic activity may vary considerably within a community depending on environmental conditions (Mitra and Flynn, 2023). Therefore, phagotrophy may be limited to a small fraction of the chlorophyte population and remains difficult to quantify. In field studies, relatively few publications have employed LysoTracker staining. However, chlorophytes are rarely detected in LysoTracker-stained sorted samples from the Eastern North Pacific coastal water (Table 4), despite being abundant in marine environments. Compared to other mixotorphic groups, bacterivory in chlorophytes has been reported less frequently. A recent study in the subtropical South China Sea using a combination of fluorescently labeled bacteria (FLB) and fluorescence in situ hybridization (FISH) successfully detected phagotrophic activity in members of the Mamiellophyceae (Table 4). Their grazing rates were positively correlated with ambient bacterial abundance (Xiao et al., 2025). These findings suggest that the trophic flexibility of chlorophytes is influenced by environmental conditions, and that phagotrophy may occur only in specific marine regions.
Table 4
| Location | Methods | Dominant bacterivorous taxa | References |
|---|---|---|---|
| North Pacific Subtropical Gyre | RNA stable isotope probing technique | stramenopiles (Dictyochophyceae and Bolidomonas) | Frias-Lopez et al., 2009 |
| Gulf of Maine | Lysotracker | Stramenopiles, Alveolata, Fungi, MAST-4 chrysophytes (including heterotrophic taxa) | Martinez-Garcia et al., 2012 |
| Mediterranean Sea | FISH+FLB | Haptophyte, Cryptophyte, pigmented dinoflagellates | Unrein et al., 2014 |
| Subtropical North Pacific Gyre | 15N, 13C-labelled Micromonas pusilla | Mixotrophic dinoflagellates | Orsi et al., 2018 |
| Eastern North Pacific | Lysotracker | Cryptophytes, haptophytes and stramenopiles (chrysophytes and dictyochophytes) | Wilken et al., 2019 |
| Subtropical North Pacific Gyre | 15N, 13C-labelled Prochlorococcus | Radiolarian, chrysophytes, dictyochophytes, and specific MAST lineages (including heterotrophic taxa) | Wilken et al., 2023 |
| York River Estuary | bacteria labeled with bromodeoxyuridine (BrdU) | Dinofalgellates and Cryptophytes | Dobbertin da Costa et al., 2024 |
| Lake Långsjön | Lysosensor and FLB | Dictyochophyceae, Haptophyceae, Chrosophyceae | Florenza et al., 2024 |
| Subtropical South China Sea | FISH+FLB | Mamiellophyceae and Prymnesiophyceae | Xiao et al., 2025 |
| Subtropical Northwestern Pacific | Lysotracker | Dictyochophyceae, Chrysophyceae, Cryptophyceae, and Mamiellophyceae | This study |
Summary of major mixotrophic taxa identified in previous references.
FISH, Fluorescence in situ Hybridization; FLB, Fluorescently labeled bacteria.
So far, little is known about ecology of Mamiella gilva and Mantoniella compared to Micromonas in Mamiellophyceae. They show a preference for coastal waters in subtropical regions and some polar regions, but the dominant species differ (Yau et al., 2020; Lin et al., 2022a; Rey Redondo et al., 2024). Here, Mamiella gilva and Mantoniella sp. are identified as potential bacterivores (Table 3 and Figures 4, 5), and bacterivory by Mantoniella has been previously reported (McKie-Krisberg et al., 2015). Notably, non-motile chlorophytes such as Chloropicon and members of the Pycnococcaceae clade I, which typically lack flagella, are identified in this study as potential bacterivores based on Lysotracker signals (Figure 4). However, we cannot rule out the possibility that the observed fluorescence originated from non-phagotrophic acidic compartments, such as those involved in autophagy or other acidic compartments (Wilken et al., 2019; Koester et al., 2025). Edwards et al. (2024) did not observe clear evidence of mixotrophic growth in Chloropicon, which may reflect inter- and intra-species differences in trophic plasticity. Based on a gene-based prediction model, some Chloropicon species were predicted to be phago-mixotrophs, which is consistent with our results (Burns et al., 2015; Bock et al., 2021). However, it remains intriguing how these non-motile chlorophytes capture prey. Previous studies have indicated that chlorophytes exhibit prey selectivity, making their bacterivory difficult to detect (Bock et al., 2021). Given their dominance in picophytoplankton communities and their widespread presence in marine environments, understanding the bacterivory mechanisms of chlorophytes is essential for refining carbon flow estimates. However, their role in bacterivory has often been overlooked, potentially leading to inaccuracies in previous assessments (Guo et al., 2024). Developing improved methods to quantify bacterivory in small mixotrophic organisms at the taxonomic level remains a key research priority.
Although the use of acid vesicle staining can potentially detect phagotrophic activity in organisms that are not easily identified by traditional tracer-based ingestion assays, the method has several limitations. A major concern is non-specific staining, particularly due to background fluorescence from diatoms and dinoflagellates (Tang and Dobbs, 2007). The high proportion of diatoms within the MNF fraction, which accounted for up to half of the total reads in this study, may bias our results. Although diatoms are primarily autotrophic, vacuolar structures involved in frustule formation can be stained by LysoTracker (Desclés et al., 2008), potentially leading to their misclassification as feeding cells. In diatom-dominated systems, this could result in an overestimation of bacterivory in the MNF fraction, underscoring the need for complementary methods such as morphological validation or grazing experiments to confirm phagotrophy. Without verification by fluorescence microscopy to confirm the intracellular localization of the signal, there is a risk of false positives. For example, fluorescence originating from acidic chloroplast compartments, autophagic vacuoles, or other acidic organelles rather than from true phagosomes (Vrieling et al., 1999; Wilken et al., 2019; Koester et al., 2025). Identifying bacterivores in pico-sized algae using fluorescent surrogates remains particularly challenging, as it is often unclear whether the detected particles have been internalized or are merely attached to the cell surface (Jimenez et al., 2021). Moreover, the small size of these eukaryotes and the presence of chloroplasts can obscure ingested particles, complicating detection by fluorescence microscopy or flow cytometry. While some eukaryotes have been reported to graze on virus-like particles (González and Suttle, 1993; DeLong et al., 2023; Martínez et al., 2024), quantifying such ingestion remains difficult due to technical limitations and low signal resolution. Acid vesicle staining offers a rapid and effective means to screen for phagotrophic activity, but commercial dyes still require validation using isolated strains to ensure staining accuracy. Additionally, defining gating thresholds for green fluorescence during flow cytometric sorting is challenging due to the continuous gradient between pigment-containing (MNF) and non-pigmented (HNF) cells (Figure 1B). Despite the aforementioned limitations, acid vesicle staining provides immediate results and reduces prey-selectivity bias commonly encountered in conventional feeding experiments. A similar mixotrophic composition was observed in a freshwater system when comparing acid vesicle staining with feeding assays (Florenza et al., 2024), supporting the broader applicability of this method for the rapid screening of mixotrophic potential. Moreover, pure culture feeding experiments have shown that Lysotracker signals can indicate whether phagocytosis has occurred, but not the quantity of ingestion (Costa et al., 2022). In this study, Lysotracker staining enabled the rapid identification of potential bacterivores. However, the positive fluorescence signals detected by flow cytometry could not be definitively localized within cells, and therefore should be interpreted with caution.
Amplicon-based sequencing approaches offer high sensitivity and taxonomic resolution, enabling the rapid processing of large sample sets and the detection of hidden diversity and community dynamics. However, this approach remains qualitative, and the amplified DNA is subject to various biases, including primer mismatches, variation in rRNA gene copy numbers among taxa, and limitations of reference databases (Santoferrara et al., 2020). Due to the high cost of single-cell sorting, we sorted 1,000 cells per sample to increase DNA yield and performed multiple displacement amplification (MDA). Despite this, 12 samples failed during PCR, and MDA-treated samples may also generate erroneous sequences (Lasken and Stockwell, 2007). Owing to high variability among replicates in the sorted samples and the diatom-dominated composition of MNF libraries (>50% of reads), ranking ASVs by TMM-normalized fold change alone (Robinson et al., 2010) was not suitable. Instead, we calculated the relative abundance of each ASV to identify the most dominant taxa in the HNF and MNF categories (Figure 5).
Seasonal dynamics and water mass properties exert strong influences on both the taxonomic composition and feeding capacity of HNF and MNF. Across the two sampling months, the overall composition of HNF showed no marked changes (Figure 2), with MAST communities remaining the dominant group. In July, Acanthometra and Parvilucifera within the HNF category exhibited higher relative read proportions in the background samples under high-temperature and high-salinity conditions. In contrast, specific lineages such as MAST-10 and Telonemia Group-1 were more abundant in August (Figures 4-6), when temperature and salinity were lower. The composition of MNF, on the other hand, varied considerably between months. In July, under high-temperature and high-salinity conditions, dictyochophytes (within stramenopiles) and Pelagomonas were more abundant, whereas in August, cryptophytes and members of the class Chlorodendrophyceae (within chlorophytes) predominated (Figures 4-6). These results further suggest that temperature and salinity substantially shape both the diversity and trophic potential of HNF and MNF across seasons.
Figure 6
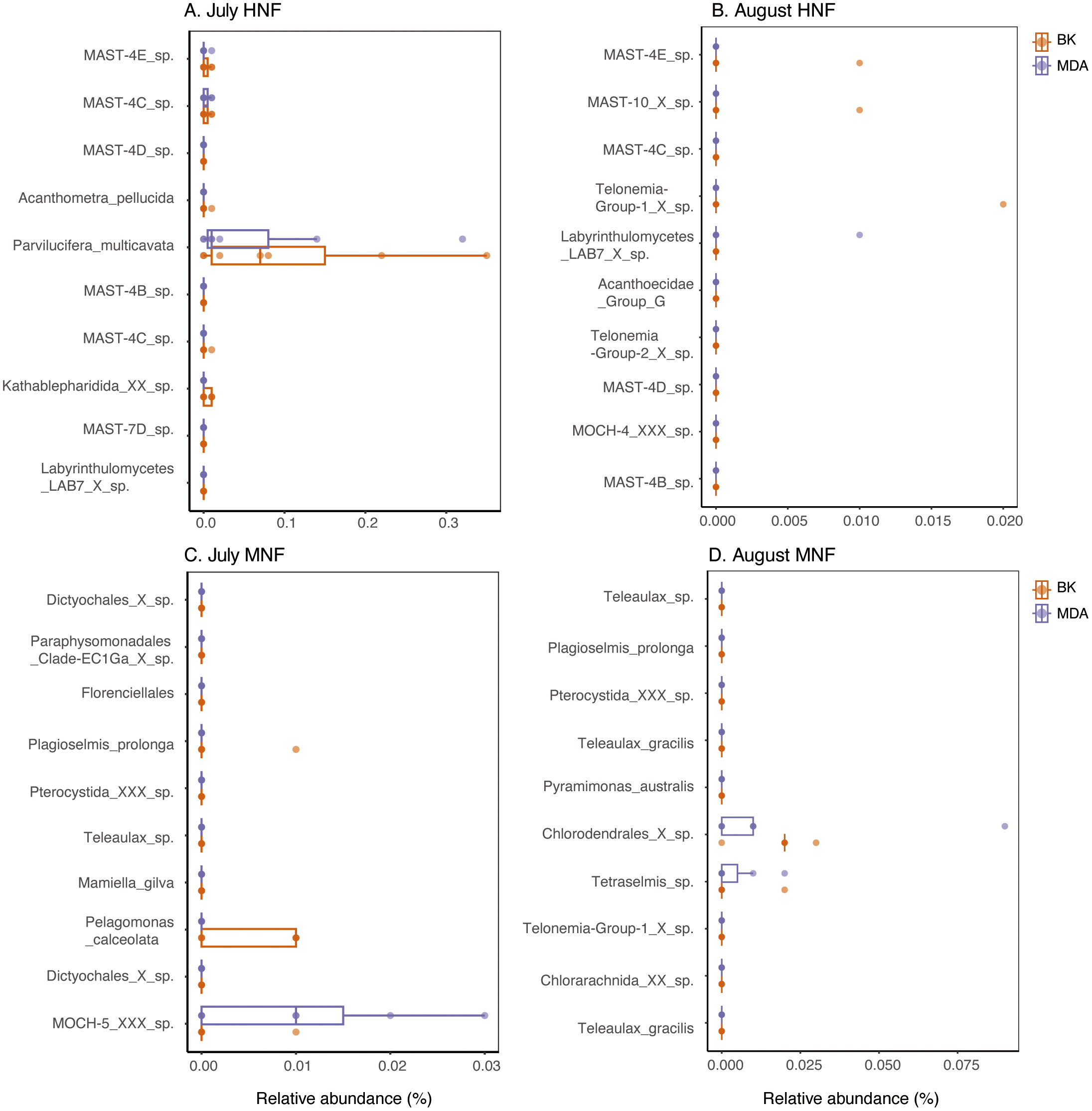
Top 10 ASVs in the sorted samples of HNF and MNF, shown as their relative abundances in the background (BK) and MDA samples for (A) July HNF, (B) August HNF, (C) July MNF, and (D) August MNF. Each point represents a sample, with orange and purple dots indicating BK and MDA samples, respectively. Both dinoflagellates and diatoms were excluded from the analysis. Detailed taxonomic classifications of these ASVs are provided in Tables 2 and 3.
This study identified potential bacterivores among the dominant oceanic genera, such as Chloropicon, Mamiella, Mantoniella, Pycnococcus, as well as stramenopiles like Dictyochophyceae and Chrysophyceae. These taxa, classified as MNFs, may possess underestimated phagotrophic capabilities that require further confirmation through direct grazing observations, quantitative assessments of grazing rates, and the identification of phagotrophy-related genes. We also documented key HNFs within the MAST lineages, choanoflagellates (Opisthokonta), Telonemia, and Rhizaria, all of which are known to play important roles in bacterivory across marine systems. Our findings are consistent with previous observations of bacterivorous groups such as choanoflagellates, chrysophytes, and MAST clades reported by Wilken et al. (2023), using ¹³C and 15N-labelled Prochlorococcus (Table 4), emphasizing the widespread ecological relevance of these taxa. Laboratory studies have shown that rising surface seawater temperatures may reduce the photosynthetic performance of mixotrophs while enhancing their heterotrophic activity (Wilken et al., 2013; Pang et al., 2023). Therefore, identifying these bacterivores across both MNFs and HNFs provides important insights into their adaptive strategies and contributes to a more comprehensive understanding of carbon and energy flow within planktonic food webs. Overall, this study expands our knowledge of the diversity, trophic strategies, and ecological roles of microbial bacterivores in the subtropical northwestern Pacific and underscores their importance in biogeochemical cycling under changing ocean conditions.
Statements
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1121045/.
Author contributions
Yun-CL: Writing – original draft, Writing – review & editing, Data curation, Methodology, Supervision. Yu-CL: Writing – original draft. K-PC: Writing – review & editing, Writing – original draft, Funding acquisition, Supervision, Resources. C-PC: Writing – review & editing, Data curation. W-CC: Methodology, Writing – review & editing, Resources, Validation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Ministry of Science and Technology, Taiwan (grant numbers MOST 109-2611-M-019-018 and MOST 110-2611-M-019-006).
Acknowledgments
The authors sincerely appreciate Ya-Han Nien and Yu-Fang Tseng for their assistance with seawater sampling and flow cytometry cell sorting, respectively, as well as Prof. Hsiao-Pei Lu for her insightful comments on the analysis. The authors also thank Sagaya John Paul Joseph for his help with English editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1602188/full#supplementary-material
Supplementary File 118S ASV table showing surface samples from the coastal water of the background (no amplification and MDA), the communities composition of HNFs and MNFs in July and August in 2023.
Supplementary File 2The 18S ASV table was subsampled to a minimum of 7,055 reads to achieve 100× coverage and reduce variation in sampling reads.
Supplementary File 3Treemaps showing the taxonomic composition and relative abundance of 18S rRNA gene amplicons at the genus level within each division in (A) background samples (n = 14), (B) background samples with MDA (n = 14), (C) the HNF category (n = 39), and (D) the MNF category (n = 33). Different divisions are represented by distinct colors. In the treemaps, data from the same genus are merged. These treemaps have not had dinoflagellate and diatom reads removed. The figures were generated using original data.
References
1
Beisner B. E. Grossart H.-P. Gasol J. M. (2019). A guide to methods for estimating phago-mixotrophy in nanophytoplankton. J. Plankton Res.41, 77–89. doi: 10.1093/plankt/fbz008
2
Bock N. A. Charvet S. Burns J. Gyaltshen Y. Rozenberg A. Duhamel S. et al . (2021). Experimental identification and in silico prediction of bacterivory in green algae. ISME J.15, 1987–2000. doi: 10.1038/s41396-021-00899-w
3
Burns J. A. Paasch A. Narechania A. Kim E. (2015). Comparative genomics of a bacterivorous green alga reveals evolutionary causalities and consequences of phago-mixotrophic mode of nutrition. Genome Biol. Evol.7, 3047–3061. doi: 10.1093/gbe/evv144
4
Callahan B. J. McMurdie P. J. Rosen M. J. Han A. W. Johnson A. J. A. Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583. doi: 10.1038/nmeth.3869
5
Chan Y.-F. Chiang K.-P. Ku Y. Gong G.-C. (2019). Abiotic and biotic factors affecting the ingestion rates of mixotrophic nanoflagellates (Haptophyta). Microb. Ecol.77, 607–615. doi: 10.1007/s00248-018-1249-2
6
Choi C. J. Jimenez V. Needham D. M. Poirier C. Bachy C. Alexander H. et al . (2020). Seasonal and geographical transitions in eukaryotic phytoplankton community structure in the Atlantic and Pacific Oceans. Front. Microbiol.11. doi: 10.3389/fmicb.2020.542372
7
Costa M. R. A. Sarmento H. Becker V. Bagatini I. L. Unrein F. (2022). Phytoplankton phagotrophy across nutrients and light gradients using different measurement techniques. J. Plankton Res.44, 507–520. doi: 10.1093/plankt/fbac035
8
DeLong J. P. Van Etten J. L. Al-Ameeli Z. Agarkova I. V. Dunigan D. D. (2023). The consumption of viruses returns energy to food chains. Proc. Natl. Acad. Sci.120, e2215000120. doi: 10.1073/pnas.2215000120
9
Desclés J. Vartanian M. El Harrak A. Quinet M. Bremond N. Sapriel G. et al . (2008). New tools for labeling silica in living diatoms. New Phytol.177, 822–829. doi: 10.1111/j.1469-8137.2007.02303.x
10
De Vargas C. Audic S. Henry N. Decelle J. Mahé F. Logares R. et al . (2015). Eukaryotic plankton diversity in the sunlit ocean. Science348, 1261605. doi: 10.1126/science.1261605
11
Dobbertin da Costa M. Gast R. J. Millette N. C. (2024). Temporal and spatial variability of constitutive mixotroph abundance and proportion. FEMS Microbiol. Ecol.100, fiae015. doi: 10.1093/femsec/fiae015
12
Edwards K. F. (2019). Mixotrophy in nanoflagellates across environmental gradients in the ocean. Proc. Natl. Acad. Sci.116, 6211–6220. doi: 10.1073/pnas.1814860116
13
Edwards K. F. Rii Y. M. Li Q. Peoples L. M. Church M. J. Steward G. F. (2024). Trophic strategies of picoeukaryotic phytoplankton vary over time and with depth in the North Pacific Subtropical Gyre. Environmental Microbiology26, e16689.
14
Edwards K. F. Li Q. McBeain K. A. Schvarcz C. R. Steward G. F. (2023). Trophic strategies explain the ocean niches of small eukaryotic phytoplankton. Proc. R. Soc B.290, 20222021. doi: 10.1098/rspb.2022.2021
15
Florenza J. Divne A. Bertilsson S. (2024). Fluorescently labeled prey surrogates in combination with fluorescence-activated cell sorting successfully discriminate actively feeding mixotrophs in a lake water sample. Limnol. Oceanogr.lno, 12545. doi: 10.1002/lno.12545
16
Flynn K. J. Stoecker D. K. Mitra A. Raven J. A. Glibert P. M. Hansen P. J. et al . (2013). Misuse of the phytoplankton–zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J. Plankton Res.35, 3–11. doi: 10.1093/plankt/fbs062
17
Frias-Lopez J. Thompson A. Waldbauer J. Chisholm S. W. (2009). Use of stable isotope-labelled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ. Microbiol.11, 512–525. doi: 10.1111/j.1462-2920.2008.01793.x
18
Gerea M. Queimaliños C. Unrein F. (2019). Grazing impact and prey selectivity of picoplanktonic cells by mixotrophic flagellates in oligotrophic lakes. Hydrobiologia831, 5–21. doi: 10.1007/s10750-018-3610-3
19
González J. Suttle C. (1993). Grazing by marine nanofiagellates on viruses and virus-sized particles: ingestion and digestion. Mar. Ecol. Prog. Ser.94, 1–10. doi: 10.3354/meps094001
20
Guillou L. Bachar D. Audic S. Bass D. Berney C. Bittner L. et al . (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res.41, D597–D604. doi: 10.1093/nar/gks1160
21
Guo X. Pang M. Zheng X. Huang L. (2024). Micromonas, a small pigmented flagellate, predominates the nanoflagellate and photosynthetic picoeukaryote communities in the northern South China Sea. Environ. Microbiol. Rep.16, e13244. doi: 10.1111/1758-2229.13244
22
Hartmann M. Grob C. Tarran G. A. Martin A. P. Burkill P. H. Scanlan D. J. et al . (2012). Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc. Natl. Acad. Sci. U.S.A.109, 5756–5760. doi: 10.1073/pnas.1118179109
23
Jimenez V. Burns J. A. Le Gall F. Not F. Vaulot D. (2021). No evidence of phago-mixotropy in Micromonas polaris (Mamiellophyceae), the dominant picophytoplankton species in the Arctic. J. Phycol.57, 435–446. doi: 10.1111/jpy.13125
24
Jirsová D. Wideman J. G. (2024). Integrated overview of stramenopile ecology, taxonomy, and heterotrophic origin. ISME J.18, wrae150. doi: 10.1093/ismejo/wrae150
25
Keller M. D. Shapiro L. P. Haugen E. M. Cucci T. L. Sherr E. B. Sherr B. F. (1994). Phagotrophy of fluorescently labeled bacteria by an oceanic phytoplankter. Microb. Ecol.28, 39–52. doi: 10.1007/BF00170246
26
Klaveness D. Shalchian-Tabrizi K. Thomsen H. A. Eikrem W. Jakobsen K. S. (2005). Telonema antarcticum sp. nov., a common marine phagotrophic flagellate. Int. J. Systematic Evolutionary Microbiol.55, 2595–2604. doi: 10.1099/ijs.0.63652-0
27
Koester J. A. Fox O. Smith E. Cox M. B. Taylor A. R. (2025). A multifunctional organelle coordinates phagocytosis and chlorophagy in a marine eukaryote phytoplankton Scyphosphaera apsteinii. New Phytol.246, 1096–1112. doi: 10.1111/nph.20388
28
Koppelle S. Ivanković M. Bengtsson M. M. Preiler C. Huisman J. Brussaard C. P. D. et al . (2024). Contrasting responses of different mixotrophic protists to light and nutrient availability. Limnol. Oceanogr.69, 1233–1246. doi: 10.1002/lno.12565
29
Lambert B. S. Groussman R. D. Schatz M. J. Coesel S. N. Durham B. P. Alverson A. J. et al . (2022). The dynamic trophic architecture of open-ocean protist communities revealed through machine-guided metatranscriptomics. Proc. Natl. Acad. Sci.119, e2100916119. doi: 10.1073/pnas.2100916119
30
Lasken R. S. Stockwell T. B. (2007). Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol.7, 19. doi: 10.1186/1472-6750-7-19
31
Leles S. G. Bruggeman J. Polimene L. Blackford J. Flynn K. J. Mitra A. (2021). Differences in physiology explain succession of mixoplankton functional types and affect carbon fluxes in temperate seas. Prog. Oceanogr.190, 102481. doi: 10.1016/j.pocean.2020.102481
32
Li Q. Edwards K. F. Schvarcz C. R. Selph K. E. Steward G. F. (2021). Plasticity in the grazing ecophysiology of Florenciella (Dichtyochophyceae), a mixotrophic nanoflagellate that consumes Prochlorococcus and other bacteria. Limnol Oceanogr66, 47–60. doi: 10.1002/lno.11585
33
Li Q. Edwards K. F. Schvarcz C. R. Steward G. F. (2022). Broad phylogenetic and functional diversity among mixotrophic consumers of Prochlorococcus. ISME J.16, 1557–1569. doi: 10.1038/s41396-022-01204-z
34
Lin Y.-C. Chin C.-P. Chen W.-T. Huang C.-T. Gong G.-C. Chiang K.-P. et al . (2022a). The spatial variation in chlorophyte community composition from coastal to offshore waters in a subtropical continental shelf system. Front. Mar. Sci.9. doi: 10.3389/fmars.2022.865081
35
Lin Y.-C. Chin C.-P. Yang J. W. Chiang K.-P. Hsieh C. Gong G.-C. et al . (2022b). How communities of marine stramenopiles varied with environmental and biological variables in the subtropical Northwestern Pacific Ocean. Microb. Ecol.83, 916–928. doi: 10.1007/s00248-021-01788-7
36
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12.
37
Martínez J. M. Talmy D. Kimbrel J. A. Weber P. K. Mayali X. (2024). Coastal bacteria and protists assimilate viral carbon and nitrogen. The ISME18, wrae231. doi: 10.1101/2024.06.14.598912
38
Martinez-Garcia M. Brazel D. Poulton N. J. Swan B. K. Gomez M. L. Masland D. et al . (2012). Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J.6, 703–707. doi: 10.1038/ismej.2011.126
39
Massana R. Del Campo J. Sieracki M. E. Audic S. Logares R. (2014). Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J.8, 854–866. doi: 10.1038/ismej.2013.204
40
McKie-Krisberg Z. M. Gast R. J. Sanders R. W. (2015). Physiological responses of three species of antarctic mixotrophic phytoflagellates to changes in light and dissolved nutrients. Microb. Ecol.70, 21–29. doi: 10.1007/s00248-014-0543-x
41
McKie-Krisberg Z. M. Sanders R. W. (2014). Phagotrophy by the picoeukaryotic green alga Micromonas pusilla: implications for Arctic Oceans. ISME J.8, 1953–1961. doi: 10.1038/ismej.2014.16
42
Millette N. C. Leles S. G. Johnson M. D. Maloney A. E. Brownlee E. F. Cohen N. R. et al . (2024). Recommendations for advancing mixoplankton research through empirical-model integration. Front. Mar. Sci.11. doi: 10.3389/fmars.2024.1392673
43
Mitra A. Caron D. A. Faure E. Flynn K. J. Gonçalves Leles S. Hansen P. J. et al . (2023). The Mixoplankton Database – diversity of photo-phago-trophic plankton in form, function and distribution across the global ocean. J. Eukaryotic Microbiol. 70, e12972. doi: 10.1111/jeu.12972
44
Mitra A. Flynn K. J. (2023). Low rates of bacterivory enhances phototrophy and competitive advantage for mixoplankton growing in oligotrophic waters. Sci. Rep.13, 6900. doi: 10.1038/s41598-023-33962-x
45
Mitra A. Flynn K. J. Tillmann U. Raven J. A. Caron D. Stoecker D. K. et al . (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist167, 106–120. doi: 10.1016/j.protis.2016.01.003
46
Moeller H. V. Archibald K. M. Leles S. G. Pfab F. (2024). Predicting optimal mixotrophic metabolic strategies in the global ocean. Sci. Adv.10, eadr0664. doi: 10.1126/sciadv.adr0664
47
Orsi W. D. Wilken S. Del Campo J. Heger T. James E. Richards T. A. et al . (2018). Identifying protist consumers of photosynthetic picoeukaryotes in the surface ocean using stable isotope probing: Identifying grazers of picoeukaryotes. Environ. Microbiol.20, 815–827. doi: 10.1111/1462-2920.14018
48
Pang M. Liu K. Gao Z. Kang C. Liu H. (2023). Temperature-light shapes the nutritional strategy of a mixotrophic green alga, Picochlorum sp. GLMF1 (Trebouxiophyceae). Mar. Ecol. Prog. Ser.713, 39–53. doi: 10.3354/meps14322
49
Pawlowski J. Burki F. (2009). Untangling the phylogeny of amoeboid potists. J. Eukaryot. Microbiol.56, 16–25. doi: 10.1111/j.1550-7408.2008.00379.x
50
Raven J. A. (1997). Phagotrophy in phototrophs. Limnol. Oceanogr.42, 198–205. doi: 10.4319/lo.1997.42.1.0198
51
Rey Redondo E. Xu Y. Yung C. C. M. (2024). Genomic characterisation and ecological distribution of:Mantoniella tinhauana: a novel Mamiellophycean green alga from the Western Pacific. Front. Microbiol.15. doi: 10.3389/fmicb.2024.1358574
52
Robinson M. D. McCarthy D. J. Smyth G. K. (2010). edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140. doi: 10.1093/bioinformatics/btp616
53
Rodríguez-Martínez R. Vaqué D. Forn I. Massana R. (2022). Dominant marine heterotrophic flagellates are adapted to natural planktonic bacterial abundances. Environ. Microbiol.24, 2421–2434. doi: 10.1111/1462-2920.15911
54
Santoferrara L. Burki F. Filker S. Logares R. Dunthorn M. McManus G. B. (2020). Perspectives from ten years of protist studies by high-throughput metabarcoding. J. Eukaryot. Microbiol.67, 612–622. doi: 10.1111/jeu.12813
55
Sintes E. Del Giorgio P. A. (2010). Community heterogeneity and single-cell digestive activity of estuarine heterotrophic nanoflagellates assessed using lysotracker and flow cytometry: Single-cell activity of flagellates. Environ. Microbiol.12, 1913–1925. doi: 10.1111/j.1462-2920.2010.02196.x
56
Stoeck T. Bass D. Nebel M. Christen R. Jones M. D. M. Breiner H. et al . (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol.19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
57
Stoecker D. K. (1998). Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications. Eur. J. Protistol.34, 281–290. doi: 10.1111/j.1365-294X.2009.04480.x
58
Stoecker D. K. Hansen P. J. Caron D. A. Mitra A. (2017). Mixotrophy in the marine plankton. Annu. Rev. Mar. Sci.9, 311–335. doi: 10.1146/annurev-marine-010816-060617
59
Tang Y. Z. Dobbs F. C. (2007). Green autofluorescence in dinoflagellates, diatoms, and other microalgae and Its implications for vital staining and morphological studies. Appl. Environ. Microbiol.73, 2306–2313. doi: 10.1128/AEM.01741-06
60
Tsai A.-Y. Chiang K.-P. Chan Y.-F. Lin Y.-C. Chang J. (2006). Pigmented nanoflagellates in the coastal western subtropical Pacific are important grazers on Synechococcus populations. J. Plankton Res.29, 71–77. doi: 10.1093/plankt/fbl058
61
Tsai A. Gong G. Sanders R. Chen W. Chao C. Chiang K. (2011). Importance of bacterivory by pigmented and heterotrophic nanoflagellates during the warm season in a subtropical western Pacific coastal ecosystem. Aquat. Microb. Ecol.63, 9–18. doi: 10.3354/ame01470
62
Unrein F. Gasol J. M. Not F. Forn I. Massana R. (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J.8, 164–176. doi: 10.1038/ismej.2013.132
63
Unrein F. Massana R. Alonso-Sáez L. Gasol J. M. (2007). Significant year-round effect of small mixotrophic flagellates on bacterioplankton in an oligotrophic coastal system. Limnol. Oceanogr.52, 456–469. doi: 10.4319/lo.2007.52.1.0456
64
Vrieling E. G. Gieskes W. W. C. Beelen T. P. M. (1999). Silicon deposition in diatoms: control by the pH inside the silicon deposition vesicle. J. Phycol.35, 548–559. doi: 10.1046/j.1529-8817.1999.3530548.x
65
Ward B. A. Follows M. J. (2016). Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc. Natl. Acad. Sci.113, 2958–2963. doi: 10.1073/pnas.1517118113
66
Wilken S. Huisman J. Naus-Wiezer S. Van Donk E. (2013). Mixotrophic organisms become more heterotrophic with rising temperature. Ecol. Lett.16, 225–233. doi: 10.1111/ele.12033
67
Wilken S. Yung C. C. M. Hamilton M. Hoadley K. Nzongo J. Eckmann C. et al . (2019). The need to account for cell biology in characterizing predatory mixotrophs in aquatic environments. Phil. Trans. R. Soc B374, 20190090. doi: 10.1098/rstb.2019.0090
68
Wilken S. Yung C. C. M. Poirier C. Massana R. Jimenez V. Worden A. Z. (2023). Choanoflagellates alongside diverse uncultured predatory protists consume the abundant open-ocean cyanobacterium Prochlorococcus. Proc. Natl. Acad. Sci. U.S.A.120, e2302388120. doi: 10.1073/pnas.2302388120
69
Worden A. Z. Follows M. J. Giovannoni S. J. Wilken S. Zimmerman A. E. Keeling P. J. (2015). Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science347, 1257594. doi: 10.1126/science.1257594
70
Xiao W. Guo J. Ma L. An L. Tong Z. Xiang M. et al . (2025). Phago-mixotrophic activity within nanophytoplankton community in a subtropical marginal sea. Limnology Oceanographylno, 70077. doi: 10.1002/lno.70077
71
Yau S. Lopes Dos Santos A. Eikrem W. Gérikas Ribeiro C. Gourvil P. Balzano S. et al . (2020). Mantoniella beaufortii and Mantoniella baffinensis sp. nov. (Mamiellales, Mamiellophyceae), two new green algal species from the high arctic. J. Phycol.56, 37–51. doi: 10.1111/jpy.12932
72
Yoo Y. D. Palenik B. (2021). Growth and grazing of the chlorarachniophyte Bigelowiella natans (Chlorarachniophyceae) on the marine cyanobacterium Synechococcus. Phycologia60, 375–383. doi: 10.1080/00318884.2021.1941567
73
Yoo Y. D. Seong K. A. Jeong H. J. Yih W. Rho J.-R. Nam S. W. et al . (2017). Mixotrophy in the marine red-tide cryptophyte Teleaulax amphioxeia and ingestion and grazing impact of cryptophytes on natural populations of bacteria in Korean coastal waters. Harmful Algae68, 105–117. doi: 10.1016/j.hal.2017.07.012
74
Yoon H. S. Price D. C. Stepanauskas R. Rajah V. D. Sieracki M. E. Wilson W. H. et al . (2011). Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science332, 714–717. doi: 10.1126/science.1203163
75
Zubkov M. V. Tarran G. A. (2008). High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature455, 224–226. doi: 10.1038/nature07236
Summary
Keywords
18S rRNA gene amplicon, flow cytometric cell sorting, heterotrophy, LysoTracker, mixotrophy
Citation
Lin Y-C, Lin Y-C, Chiang K-P, Chin C-P and Chou W-C (2025) Using an acidic vesicle tracer to identify potential bacterivores in coastal waters of the subtropical Western Pacific Ocean. Front. Mar. Sci. 12:1602188. doi: 10.3389/fmars.2025.1602188
Received
29 March 2025
Revised
30 October 2025
Accepted
05 November 2025
Published
20 November 2025
Volume
12 - 2025
Edited by
Edward Fillery, University of Toronto, Canada
Reviewed by
Qian Li, University of Hawaii at Manoa, United States
Jennifer L Beatty, University of Southern California, United States
Updates
Copyright
© 2025 Lin, Lin, Chiang, Chin and Chou.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-Chi Lin, yunchi@mail.ntou.edu.tw; Kuo-Ping Chiang, kpchiang@mail.ntou.edu.tw
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.