- Shanghai Engineering Research Center of Hadal Science and Technology, College of Oceanography and Ecological Science, Shanghai Ocean University, Shanghai, China
Hadal amphipods have developed unique survival strategies to cope with extreme conditions, such as low temperatures and high hydrostatic pressure, with their gut microbes playing a crucial role in this adaptation. Nevertheless, the specific mechanisms and functional contributions of these microbial communities remain poorly understood. In this study, a gut symbiotic fungus, Chaetomium madrasense HM411, which has cellulose degrading ability, was isolated in the gut of Hirondellea gigas from the Mariana Trench. Comparison of enzyme production between the terrestrial and deep-sea strains of C. madrasense (HM412 and HM411, respectively) showed that the deep-sea strain exhibited significantly higher endoglucanase activity at 15°C, representing a 1.2-fold increase. Furthermore, growth rate analysis indicated that C. madrasense HM411 maintained significantly higher proliferation rates at 15°C, suggesting psychrotolerant traits in the deep-sea derived strain. Transcriptomic analysis revealed distinct low-temperature enzyme production advantages in C. madrasense HM411 compared to strain HM412, particularly in the regulation of Carbohydrate-Active Enzymes, extracellular enzyme secretion, stress protein production, and cell wall/membrane modifications. These findings suggest that C. madrasense HM411 may have evolved unique enzymatic and genomic adaptations to thrive in hadal environments, potentially playing important roles in recalcitrant substance degradation under extreme conditions.
1 Introduction
The biological definition of the hadal zone is where water depths exceed 6,000 m (Wolff, 1970). It represents only 1% to 2% of the seafloor area, but encompasses the deepest 45% of the vertical depth gradient (Wolff, 1959). Its unique geography creates an environment characterized by high hydrostatic pressure, limited food, and low temperatures (Bartlett, 1992). Despite the extreme environmental conditions in the trenches, there is still an active and diverse biological community. Amphipods have been found in high abundance within many of the major hadal trenches, such as the Peru-Chile Trench, Kermadec Trench, and the Mariana Trench (Kilgallen, 2015; Lacey et al., 2016; Wei et al., 2023). This phenomenon is primarily attributed to the vertical sinking flux of particulate organic matter (POM) from surface waters (Oguri et al., 2013). Hirondellea gigas is one of the dominant species in the Mariana Trench. It feeds on decaying animals and plants and occupies an important position in the hadal ecosystem, which is nutrient poor but relatively rich in complex carbohydrates (Shi et al., 2018). To survive in this nutrient-limited environment, H. gigas relies on the enzymatic breakdown of complex carbohydrates derived from sinking detritus (Kobayashi et al., 2019). Hadal symbiotic microorganisms play a crucial role in host adaptation to extreme environmental conditions through multiple mechanisms. As demonstrated by the previous study (Sun et al., 2022), gut-associated microbial communities enhanced host metabolic capabilities through the production of specialized enzymes that facilitate the degradation of complex organic compounds and carbohydrates.
As a result of coevolution with their specific hosts, microbial symbionts have developed a range of unique biochemical traits, rendering them highly efficient producers of biologically active natural products (Xiao et al., 2023). Seven novel cytoglobosins (Cytoglobosins A–G) were isolated from Chaetomium globosum QEN-14, a fungal symbiont of the marine green alga Ulva pertusa. These compounds exhibited cytotoxic activity against a human lung adenocarcinoma cell line (A549) (Cui et al., 2010). In addition to producing bioactive compounds, symbiotic microbes contribute to host survival by degrading complex substrates. For instance, macrogenomic analysis revealed a remarkably high abundance of carbohydrate metabolism genes (particularly cellulases) within the H. gigas gut microbial community. Functional annotation further demonstrated the involvement of these genes in cellulose and hemicellulose catabolic pathways (Chan et al., 2021). Additionally, three fungi (Cadophora sp. TS2, Emericellopsis sp. TS11, and Pseudogymnoascus sp. TS12) were isolated from deep-sea sponges, which exhibited high CMCase and xylanase activities (Batista-García et al., 2017). The complexity of the abyssal environment involving high pressures, low temperatures, and limited light availability may contribute to the significant differences between the enzymes generated by marine microorganisms and homologous enzymes from terrestrial microorganisms (Zhang and Kim, 2010). For instance, a hyperthermophilic protease isolated from the deep-sea methanogenic archaeon Methanococcus jannaschii exhibited 3.4-fold higher activity and 2.7-fold greater thermal stability under high hydrostatic pressure (50.7 MPa), while its terrestrial homologue showed pressure-induced inactivation at atmospheric pressure (0.1 MPa) (Michels and Clark, 1997). Indeed, enzymes of marine symbiotic microbial origin exhibit novel biocatalytic properties, such as hyperthermal stability, high salt tolerance, cold adaptation, and hydrophilicity (Barzkar and Sohail, 2020).
In marine ecosystems, microbial cellulases serve as essential catalysts for organic matter decomposition and carbon cycling, mediating the transformation of recalcitrant substrates into bioavailable energy sources within benthic and pelagic food webs (John J et al., 2022). Although marine bacteria are involved in cellulose metabolism, their enzyme activity is limited by the low temperatures of trenches, a challenge that may be alleviated by marine fungi. For example, marine bacteria maintain metabolic homeostasis through transcriptome reprogramming at low temperatures (0°C), but cellulase activity remains lower than in mesophilic conditions (Riccardi et al., 2023). In contrast, marine fungi adapt to low temperatures by accumulating osmoregulatory compounds and secreting antifreeze proteins (Robinson, 2001). Furthermore, they maintain enzymatic stability under cold conditions through structural modifications of key proteins (Duarte et al., 2018). Despite this potential, no fungal cellulase has been characterized from hadal amphipods, and fewer than 5% of published studies on hadal microbial communities focus on eukaryotes (Fan et al., 2022). The exploration of these symbiotic strains possessing cellulose degradation potential will not only unveil the survival strategies of abyssal amphipods in extreme environments but also elucidate how they leverage limited resources to sustain their life processes.
To address the critical gap in understanding fungal contributions to hadal amphipods, we characterized Chaetomium madrasense HM411, a cellulolytic fungus isolated from H. gigas in the Mariana Trench. Its low-temperature tolerance and cellulase activity were evaluated via in vitro biochemical assays, and comparative transcriptome analyses were employed to disclose the molecular mechanism underlying cellulase production in C. madrasense HM411 under low-temperature conditions. Our study provides functional and molecular evidence for the presence of a fungal symbiont with cellulose-degrading capacity in the gut of H. gigas. Furthermore, the low-temperature-adapted fungi identified here (e.g., C. madrasense HM411) have biotechnological potential for cellulose degradation, providing a template for engineering robust industrial biocatalysts.
2 Materials and methods
2.1 Sample collection and fungal isolation
H. gigas samples were collected from the Mariana Trench during a research expedition in September 2021 aboard the research vessel “Explore I”, using a macrobiological pressure-retaining sampler. The sampling sites were located at 11°19.6098′ N, 142°11.283′ E (10,895 m depth) and 11°22.584′ N, 142°35.3102′ E (10,910 m depth), where the ambient temperature typically ranges between 1-4°C (Yasuhara and Danovaro, 2016). All samples (n=6) were collected from the above two sites. Immediately after collection, H. gigas specimens were stored at −80°C. To ensure biological representativeness, gut contents from three selected individuals per site were processed independently. Before dissection, the samples were individually thawed at 4°C and rinsed with distilled water. Subsequently, the amphipods were dissected using a sterile scalpel to isolate intestinal tissues, which were then transferred into sterile centrifuge tubes for fungal isolation experiments. Fungi were isolated following the method described in our previous study (Cui et al., 2024). Briefly, 200 µL of diluted gut contents were spread onto potato dextrose agar (PDA), tryptone soy agar (TSA), and malt extract agar (MEA) plates and incubated at 20°C for 2–3 days. Visible colonies were transferred to fresh medium for purification, and single colonies were obtained through three rounds of subculturing.
2.2 DNA extraction, phylogenetic analysis, and morphological characterization
Fungal identification was conducted based on a combination of morphological characteristics and conserved sequence analysis. Pure fungal cultures were grown on PDA at 28°C for 3 days. Fungal hyphae were collected from petri dishes and strain DNA was extracted using the TIAN combi DNA Lyse & Det PCR kit [Tiangen Biotech (Beijing) Co. Ltd.], following the manufacturer’s instructions. PCR amplification and sequencing were performed using the following primers: ITS1 and ITS4 for the internal transcribed spacer (ITS) region, and RPB2AM-1bf and RPB2AM-7R (Miller and Huhndorf, 2005) for the second largest subunit of the DNA-directed RNA polymerase II (RPB2) gene region (all primers used in this study were listed in Supplementary Table S1).
PCR amplification was performed under the following conditions: initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min; final extension at 72°C for 5 min; and storage at 4°C (Deng et al., 2023). The amplified ITS and RPB2 sequences were subjected to a sequence similarity search using BLASTn against the NCBI nucleotide database. Highly similar sequences were aligned and comparatively analyzed using MEGA X. All sequences were uniformly trimmed at both 5’ and 3’ ends to ensure positional homology in the alignment. Phylogenetic trees were performed using the neighbor-joining algorithm in MEGA X, with nodal support evaluated through 1,000 bootstrap replications. Microscopic characterization of the target fungus was performed using an OLYMPUS BX53 microscope (Olympus Corporation, Tokyo, Japan) to observe the ascomycete fruiting bodies.
Screening for cellulase-producing fungi was conducted using a plate assay method. C. madrasense strains were initially screened for cellulase activity by culturing on agar medium supplemented with 1% carboxymethyl cellulose (CMC) as the sole carbon source. The plates were incubated at 28°C for 5 days. After incubation, the plates were stained with 1% Congo red staining solution for 15 min, followed by destaining with 1 M NaCl buffer solution for 15 min (n=3 biological replicates). The formation of a clear zone around the fungal colony was considered indicative of cellulase activity. Cellulolytic fungi were identified based on their ability to hydrolyze cellulose, as evidenced by the presence of a clearance zone surrounding the colony (Khokhar et al., 2012).
2.3 Comparative analysis of growth characteristics of two fungal strains from different origins
Fresh mycelial plugs (2 mm²) obtained from the edges of fungal colonies were inoculated onto PDA (0 M NaCl, 0.5 M NaCl) and 0 M NaCl microcrystalline cellulose agar (MCC) (Supplementary Table S2), with 3 biological replicates set up for each condition. All plates were incubated at 28°C for 3 days, and colony morphology was observed. Additionally, mycelial fragments (2 mm2) were excised from the edges of actively growing colonies of each isolate and inoculated into the center of 0 M NaCl MCC and the plates were incubated at 15°C, 20°C, and 28°C (n=3 replicates per temperature). Colony diameters were measured daily for 3 days using a measuring scale, with three perpendicular measurements per colony to minimize observer bias.
2.4 Assay of endoglucanase activity of C. madrasense
Mycelial discs of uniform size were inoculated into an enrichment medium and incubated for 2 days. Subsequently, 1% of the enriched culture was transferred to MCC medium and further incubated at 180 rpm in a rotary shaker (n=3 biological replicates). Endoglucanase activity was quantified using the 3,5-dinitrosalicylic acid reagent (DNS) assay to measure the concentration of reducing sugars released by cellulase hydrolysis. The methods for measuring cellulase activity were performed in accordance with those described by Wei (2007). A 0.5 mL enzyme solution was mixed with 1 mL of 1% CMC-Na buffer solution and incubated at 50°C for 30 min. After terminating the reaction by adding 1.5 mL DNS and heating at 100°C for 10 min, the samples were immediately cooled on ice to minimize nonspecific color development. The absorbance at 540 nm was then measured (n=3 biological replicates). Substitute the absorbance into the glucose standard curve and calculate the glucose concentration (Supplementary Figure S2). One unit of cellulase activity (U) was defined as the amount of enzyme required to catalyze the release of 1 μmol of glucose from 1% CMC-Na under specific reaction conditions (pH 4.8, 50°C, 30 min). The enzyme unit (symbol U), also known as the international unit (IU), is a measure of an enzyme’s catalytic activity (Helal et al., 2021).
2.5 Transcriptome assembly and transcript quantification
To perform RNA sequencing and transcriptomics analysis, C. madrasense was incubated at 180 rpm at 15°C on MCC medium for 6 days, after which biomass was harvested. The mycelia were fully ground using liquid nitrogen and preserved in RNAiso Plus (Takara, Japan) at −80°C. The RNA extraction, transcriptomic sequencing, and bioinformatics analysis were accomplished by Novogene Bioinformatics Technology (Beijing, China). RNA integrity was assessed using an Agilent 2100 Bioanalyzer. Following standard protocols, high-quality RNA was used for mRNA enrichment, fragmentation, reverse transcription, end repair, amplification, and circularization to construct the sequencing library. After passing quality control, the library was loaded onto the Illumina sequencing platform for high-throughput sequencing. To minimize batch effects, all samples were harvested simultaneously, processed in the same RNA extraction batch, and sequenced in a single Illumina run. Gene expression levels were quantified as FPKM (fragments per kilobase of transcript per million mapped reads) using RSEM (v1.3.1). Analysis of differentially expressed genes (DEGs) was performed using DESeq2 (1.20.0). Q value ≤ 0.05 and |log2 Fold-Change| ≥ 1.0 were set as the threshold for significant differential expression.
2.6 RNA extraction, RT-qPCR, and transcriptome validation
To validate the reliability of the RNA-Seq results, selected differentially expressed genes were used to design primers for quantitative reverse transcription PCR (RT-qPCR) analysis. Total RNA was obtained from Novozymes and synthesized according to the instructions from the manufacturer of the PrimeScript RT Reagent Kit (Takara, Japan). Gene expression levels were quantified using the 2-ΔΔCt method, and the fold changes obtained from RNA-Seq were compared with those from RT-qPCR. The correlation coefficient (R²) was calculated to assess the consistency between the two methods. β-actin was used as the internal reference gene, and the primer sequences are provided in Supplementary Table S1.
2.7 Statistical analysis
All experiments were performed at least three times, with three biological replicates per sample. All data were analyzed using SPSS Statistics 27 for Windows (SPSS Inc.). After testing for normality using the Shapiro-Wilk test and Levene’s test for all data, one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test were used to assess differences between groups. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, while non-significant differences were denoted as n.s (P > 0.05). Results are presented as the mean ± standard error (SD) of three replicates. For multiple comparisons, we applied the Benjamini-Hochberg false discovery rate (FDR) correction with Q < 0.05 as the significance threshold. The line and bar graphs were drawn using Origin 2024b. The heatmap was drawn using TBtools software. Multi-group difference scatterplots were plotted using Omicshare tool.
3 Results
3.1 Isolation and characterization of the intestinal commensal fungus H. gigas
An abyssal fungal strain was successfully isolated from the gut microbiota of amphipods inhabiting the Mariana Trench. Phylogenetic characterization based on ITS and RPB2 of conserved sequences confirmed the taxonomic classification of strain HM411 as C. madrasense (Figure 1A).
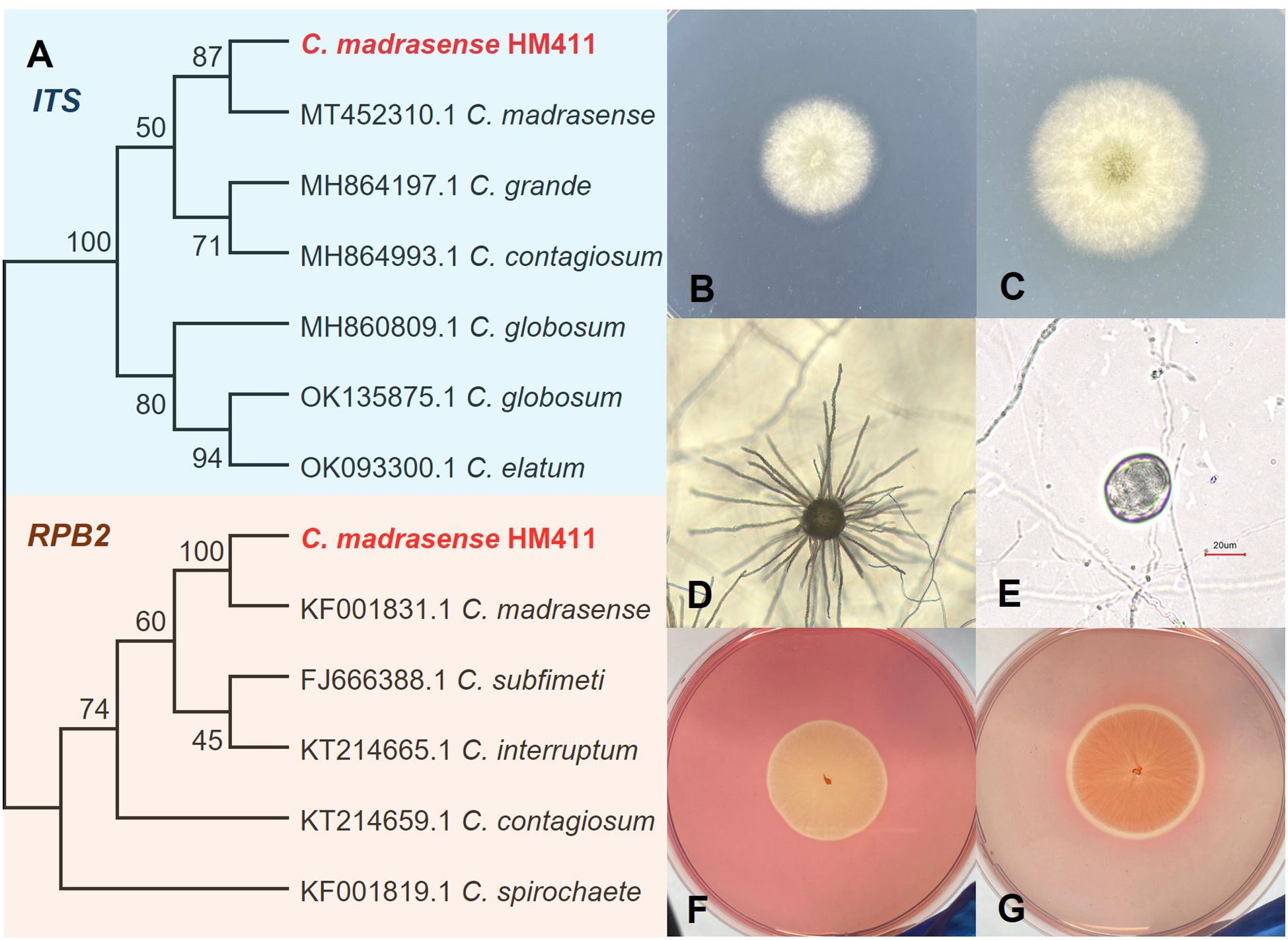
Figure 1. Identification of gut microbial fungus C. madrasense HM411. (A) Phylogenetic tree of C. madrasense based on ITS, RPB2 sequences. C. madrasense HM411 owned the most closet relative to C. madrasense CBS (MT 452310) under ITS amplification, with 100% homology in RPB2. Bootstrap analysis was performed with 1,000 replicates. (B, C) Plate morphology of C. madrasense on 0 M NaCl PDA after 3–4 days of incubation at 28°C. (D) Ascoma. (E) Ascospores. (F, G) C. madrasense HM411 showed a cellulose degradation activity index of 1.11, forming a hydrolysis zone on CMC-containing agar medium.
The morphological characteristics of C. madrasense HM411 were systematically investigated. When cultured on 0.5 M NaCl PDA, the strain exhibited white aerial hyphae (Figure 1B) and produced distinctive yellow exudates in the central region during prolonged incubation (Figure 1C). The colony demonstrated robust growth, reaching 6.50 ± 0.12 cm (n=3 biological replicates) in diameter following 7 days of incubation at 28°C. Microscopic examination revealed the presence of typical ascomata and ascospores (Figures 1D, E), with epidermal, spherical structures delineated by the filaments. The mature ascospores displayed an olivaceous-brown pigmentation and exhibited ellipsoidal to subglobose morphology.
As members of the genus Chaetomium are widely recognized for their cellulolytic capabilities in both cellulosic waste degradation and single cell protein (SCP) production (Darwish and Abdel-Azeem, 2020), we evaluated the cellulolytic potential of C. madrasense HM411. The strain demonstrated sustained growth on CMC agar and formed distinct cellulose-hydrolyzed hyaline rings upon Congo red staining, thereby confirming its cellulose-degrading capacity. These findings substantiate the cellulolytic potential of this abyssal fungal strain (Figures 1F, G).
3.2 C. madrasense HM411 exhibits superior low-temperature tolerance compared to C. madrasense HM412
To evaluate the cellulose-degrading specificity of hadal-derived C. madrasense, a comparative terrestrial strain (C. madrasense CGMCC 3.17112), originally isolated from cotton, was obtained from the China General Microbiological Culture Collection Center (CGMCC) and designated as C. madrasense HM412 (Supplementary Figure S1). In the same medium, both strains exhibited similar colony morphology. Visually, the growth diameter of C. madrasense in 0 M NaCl MCC appeared larger than that in (0 M, 0.5 M NaCl) PDA under the same incubation conditions (Figure 2A), suggesting that cellulose may be a more favorable carbon source for C. madrasense than PDA components (n=3 biological replicates).
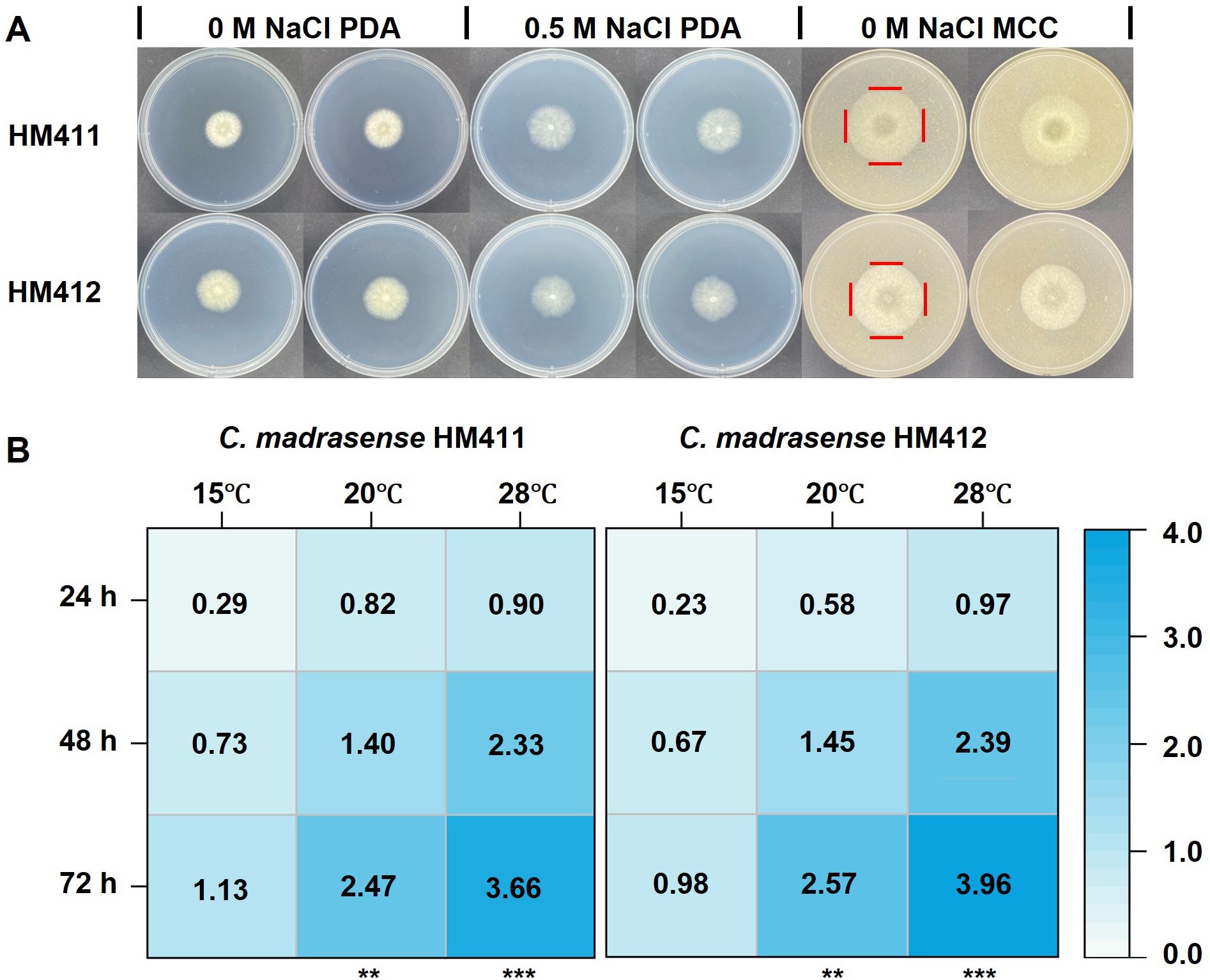
Figure 2. Comparative analysis of colony morphology and growth characteristics of C. madrasense HM411 and HM412. (A) Colony morphology of C. madrasense HM411 and HM412 cultured on 0 M NaCl PDA, 0.5 M NaCl PDA and 0 M NaCl MCC, following 3 days incubation at 28°C. (B) Heat maps showed the growth diameter of two C. madrasense inoculated in cellulose-producing medium at different temperatures. **P < 0.01, ***P < 0.001.
To assess the impact of temperature on the growth of the strains, we inoculated fungus discs of the same size on 0 M NaCl MCC, and measured the growth diameters of the two strains at different temperatures, with three biological replicates set up for each strain. The mean colony diameters of C. madrasense from marine and terrestrial sources, calculated from three independent replicates, were 3.66 ± 0.03 cm and 3.96 ± 0.03 cm, respectively, after 72 hours of incubation at 28°C. Whereas, the diameter of C. madrasense colonies of marine and terrestrial origin was 1.13 ± 0.02 cm and 0.98 ± 0.02 cm (n=6 measurements from 3 biological replicates). after 72 hours of incubation at 15°C, respectively (Figure 2B). The colony diameter of C. madrasense HM412 was larger than that of C. madrasense HM411 (p < 0.0001) under the incubation condition at 28°C, whereas at 15°C, the larger colony diameter was that of C. madrasense HM411 (p < 0.001), which might indicate that C. madrasense HM411 showed greater low-temperature tolerance than terrestrial source strains.
3.3 Comparison of the enzymatic productivity of C. madrasense HM411 and HM412 under different temperature conditions
Temperature represents a critical environmental factor influencing the viability and enzymatic productivity of cellulase-producing microbial strains (Sohail et al., 2009; Rosyida et al., 2015). To evaluate the thermal adaptability of C. madrasense HM411, endoglucanase production was assessed across a temperature gradient (10, 15, 20, 25, and 28°C). The experimental results demonstrated that the optimal temperature range for endoglucanase production by C. madrasense HM411 was 25-28°C, with enzyme synthesis reaching a stabilization phase after 5 days of incubation. Notably, the strain exhibited remarkable psychrotolerant characteristics, maintaining metabolic activity and enzymatic production at 15°C, albeit with an extended stabilization phase observed at 16 days. These findings highlight the strain’s adaptive capacity to suboptimal thermal conditions while maintaining cellulolytic functionality. While, until the end of the experiment (i.e., 18 days), no production of endoglucanase was detected under 10°C culture conditions (Figure 3A).
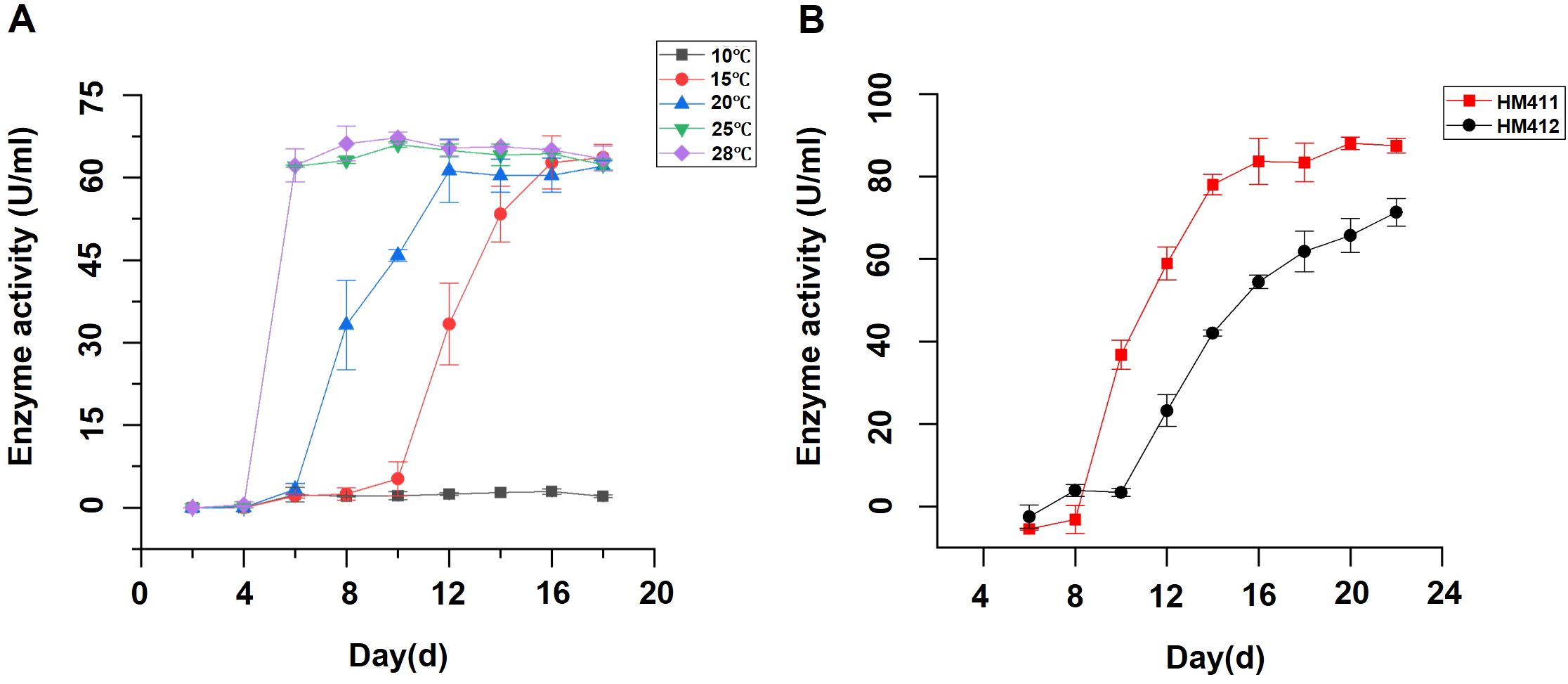
Figure 3. Temperature-dependent endoglucanase production profiles of C. madrasense HM411 and comparative analysis of enzymatic production with C. madrasense HM412. (A) Endoglucanase production profiles of marine-derived C. madrasense HM411 at different cultivation temperatures. (B) Comparative analysis of endoglucanase production between marine-derived C. madrasense HM411 and terrestrial-derived C. madrasense HM412 at 15°C.
To investigate the cellulolytic potential of two C. madrasense strains from different origins under low-temperature conditions, we evaluated their endoglucanase production capabilities at 15°C. Comparative analysis revealed that C. madrasense HM411 showed significantly higher enzymatic productivity than C. madrasense HM412 under identical conditions (p < 0.01). Specifically, C. madrasense HM412 reached the enzyme production stabilization phase at 22 days, demonstrating endoglucanase activity levels of 71.3 ± 3.4 U/mL (n=3 biological replicates), while C. madrasense HM411 achieved significantly higher activity levels of 87.5 ± 1.8 U/mL (n=3 biological replicates) during the same period (Figure 3B).
3.4 Transcriptome overview, differentially expressed genes, and enrichment analysis
C. madrasense HM411 demonstrated superior endoglucanase activity and a significantly reduced fermentation period at 15°C compared to C. madrasense HM412. To elucidate the molecular mechanisms underlying the enhanced cellulase production and enzymatic activity of C. madrasense HM411 under low-temperature conditions, we conducted a comparative transcriptomic analysis of the two C. madrasense strains from distinct origins.
The reliability of the RNA-Seq data was supported by the raw read information (Supplementary Table S4) and the high correlation and consistency observed among the three biological replicates (Supplementary Figure S3A). The RT-qPCR analysis results reflected that the trend of transcript level changes of these genes was consistent with those obtained from the comparative transcriptome analysis (Supplementary Figure S4).
Comparative transcriptome analysis showed that there were 1,788 DEGs between C. madrasense HM411 and C. madrasense HM412 at 6 days of cellulose induction in 15°C (|log2FC| ≥ 1 and adjusted padj ≤ 0.05), of which 1,089 genes were up-regulated, and 699 genes were down-regulated (Supplementary Figure S3B).
We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis on DEGs and selected the top 10 items for further enrichment. These DEGs are mainly involved in protein synthesis, processing and degradation, primary metabolism, small molecule metabolism such as galactose, sucrose, fructose, aminosugar, and nucleotide sugar metabolism. In addition, the enrichment of differentially expressed genes in amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, and fructose and mannose metabolism may reflect enhanced metabolic flexibility in C. madrasense HM411, enabling better adaptation to low-temperature conditions through coordinated regulation of multiple metabolic pathways. This regulation of sugar metabolism may also enhance its metabolic efficiency and adaptive capacity (Figure 4A). GO enrichment analysis showed that genes involved in the regulation of intracellular metabolic activities, degradation of macromolecular substances were significantly enriched, in addition to genes related to transcription, regulation, protein synthesis, metabolism and transport using DNA as a template (Figure 4B). Energy metabolism is important for protein synthesis (Huang et al., 2017), and we hypothesize that up-regulation of related genes may benefit C. madrasense HM411 to resist low-temperature stress and produce cellulase. Next, TBtools (v1.120) was used to draw a heatmap containing the main marker genes associated with strains that resist low temperatures and produce enzymes. These include marker genes encoding enzymes associated with strain secretion. Notably, genes encoding secreted enzymes (e.g., endoglucanase CBH1, chitinase E3.2.1.14) were upregulated in C. madrasnese HM411. Additionally, genes associated with glycerol and choline synthesis, such as alcohol dehydrogenase (adh) and acetylcholinesterase (ACHE), were identified. These genes have been attributed to extracellular protein secretion and intracellular metabolism, including cell wall components, and cell membranes (Figure 4C), which may be related to cellulase production by C. madrasense HM411 at low temperatures.
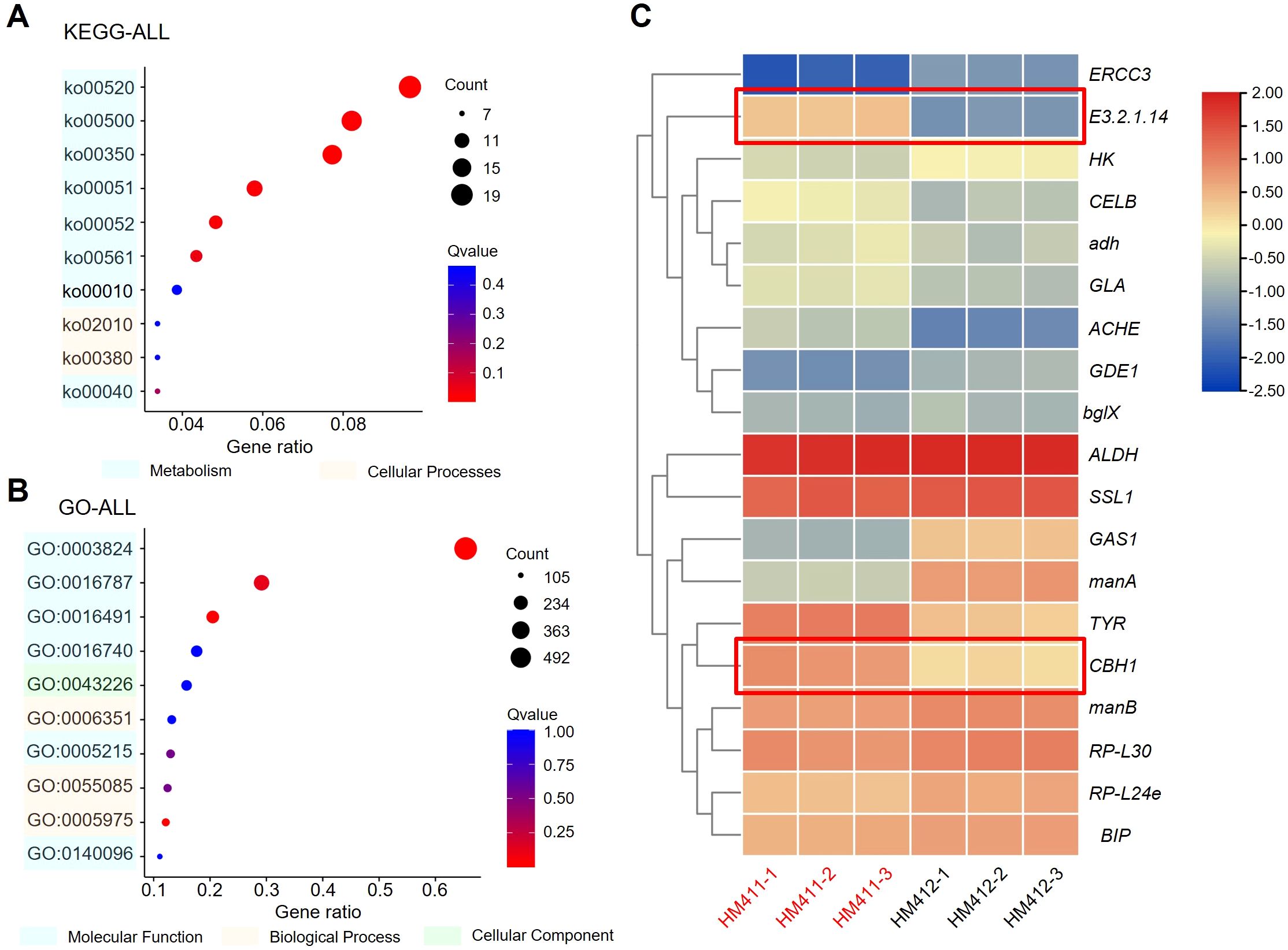
Figure 4. DEG overview of C. madrasense HM411 and C. madrasense HM12 cultured under 15°C for 6 days. (A, B) Top 10 terms in KEGG (A) and GO (B) enriched from DEGs of the C. madrasense HM411 groups (log2 fold-change > 1.5 or < −1.5). Among of them, amino sugar and nucleotide sugar metabolism (ko00520; 20 candidate genes in HM411) was occupied mostly in KEGG enrichments, including starch and sucrose metabolism (ko00500), tyrosine metabolism (ko00350), and Fructose and mannose metabolism (ko00051). In the top terms of GO enrichments, there were three main functional categories: biological process, cellular component and molecular function. The catalytic activity (GO:0003824) occupied the majority with 619 candidate genes in C. madrasense HM411. (C) Normalized FPKM average value of marker genes analyzed in this study.
3.5 Analysis of differential expression of CAZyme genes and cellulase synthesis under low temperature stress
CAZymes are key enzymes for nutrient conversion and utilization in fungi, mainly including 6 enzyme families: auxiliary activities (AAs), glycosyl hydrolases (GHs), carbohydrate binding modules (CBMs), carbohydrate esterases (CEs), glycosyl transferases (GTs), and polysaccharide lyases (PLs) (Hao et al., 2019; Li et al., 2020). We identified 378 genes as CAZyme genes by comparison with the CAZy database, of which 268 were up-regulated and 110 were down-regulated in expression (Figure 5A), including 37 secreted proteins. In C. madrasense HM411, the differential genes were mainly concentrated in the AA and GH families, in which the expression of glycoside hydrolase genes, GH5, GH6, GH7, GH11, GH16, GH18, and GH43 families, was significantly up-regulated (log2FC > 1, padj < 0.05) (Supplementary Table S5), and the major cellulase genes were distributed in the GH5, GH6, GH7, GH9 families.
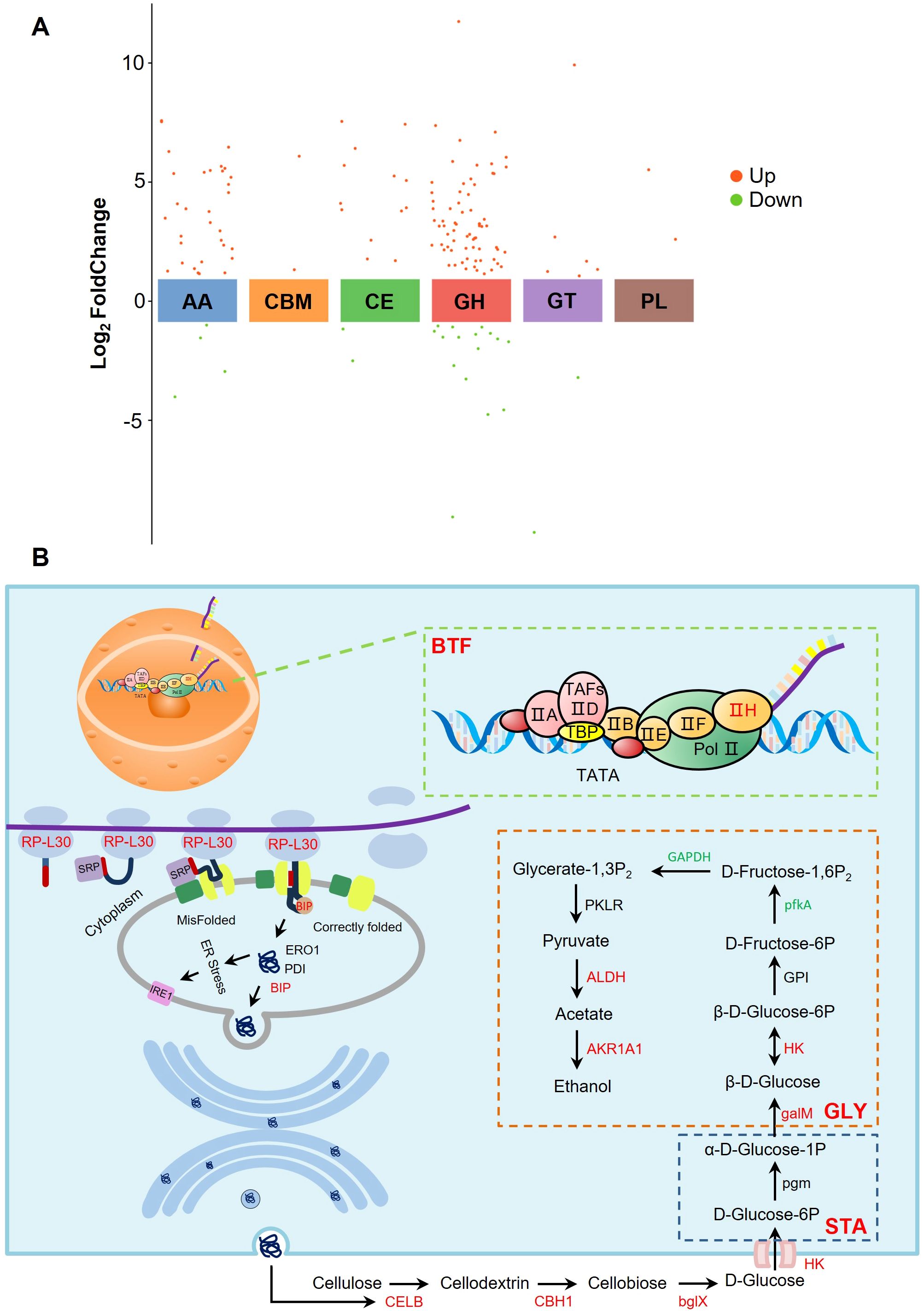
Figure 5. Analysis of differential expression of CAZyme gene and cellulase synthesis under low temperature stress. (A) Expression profile of different CAZyme families. (B) Extracellular protein synthesis pathway and cellulose metabolism pathway. BTF, basal transcription factors; GLY, Glycolysis; STA, starch and sucrose metabolism; Genes labeled in red, green, and black represent up-regulated, down-regulated, and non-significantly expressed genes, respectively.
Cellulose degradation is carried out by three classes of enzymes, mainly endoglucanase (EG) (EC 3.2.1.4), cellobiose hydrolase (CBH) (EC 3.2.1.91), and β-glucosidase (BG) (EC 3.2.1.21) (Henriksson et al., 2000; Yuan et al., 2001; Amer et al., 2017). In addition, oxidoreductases such as cellobiose dehydrogenase (CDH) (EC 1.1.99.18) may act synergistically with classical GH to accelerate the enzymatic conversion of polysaccharides (Cameron and Aust, 2001). Significant up-regulation of CELB (log2FC = 5.69, padj < 0.05), CBH1 (log2FC = 3.11, padj < 0.05), and bglX (log2FC = 2.13, padj < 0.05) involved in the cellulose metabolism pathway was detected in RNA-Seq, corresponding to EG, CBH, and BG, respectively (Figure 5B), in addition to CDH (log2FC = 3.46, padj < 0.05), a cellobiose dehydrogenase that accelerates the enzymatic conversion of polysaccharides (Supplementary Table S6). The degradation product of cellulose, glucose, is specifically recognized and bound by glucose transport proteins and enters the cell by changing its own conformation to participate in glycolysis and ethanol fermentation pathways. The up-regulation of HK (log2FC = 1.08, padj < 0.05) and galM (log2FC = 4.76, padj < 0.05) genes within the starch and sucrose metabolic pathways, along with the enrichment of GAPDH, ALDH, and AKR1A1 genes in the glycolytic pathway, was observed. These findings suggest that C. madrasense HM411 cells may possess enhanced capabilities for cellulose degradation and ethanol synthesis, thereby providing efficient energy metabolism for cellular functions.
Cellulase secretion begins with the transcription of DNA in the nucleus, and in the RNA-Seq we detected up-regulation of the TFIIH2 (log2FC = 1.22, padj < 0.05) and XPB (log2FC = 1.34, padj < 0.05) genes. TFIIH2 assists RNA polymerase II to bind to the promoter region of the gene, promotes the formation of the transcription initiation complex, and stabilizes the structure of the transcription initiation complex (Schilbach et al., 2017). Up-regulation of RP-L30 (log2FC = 1.18, padj < 0.05) and RP-L24 (log2FC = 1.09, padj < 0.05) genes involved in encoding the 60S large subunit of the ribosome was detected, and the L30 protein helps to maintain the normal function and stability of the ribosome and ensures that the translation process proceeds efficiently (White et al., 2004). The L24 protein helps maintain ribosome activity and accuracy, ensuring proper decoding of mRNA and accurate amino acid incorporation (Dresios et al., 2000). The up-regulation of TFIIH2, RP-L30, and RP-L24 genes may indicate that C. madrasense HM411 has a better DNA transcription capacity than C. madrasense HM412 under the same low temperature conditions.
Peptides entering the ER lumen need to be folded with the assistance of the binding protein BiP1, a homolog of Kar2 in Streptococcus that plays a role in the folding of ER proteins (Seppä and Makarow, 2005). Thus, up-regulation of the BIP (log2FC = 1.28, padj < 0.05) gene may help C. madrasense HM411 maintain intracellular protein homeostasis. Unfortunately, endoplasmic reticulum stress is induced when misfolded proteins accumulate in the endoplasmic reticulum. IRE1 is able to sense this stress signal and, through its own nucleic acid endonuclease activity, splices the X-box-binding protein 1 (XBP1) mRNA to convert it into the active XBP1 protein, a potent transcription factor that enters the nucleus to activate a series of factors involved in ER protein folding, mass control, lipid synthesis and transport, among other processes (Xu et al., 2016). Therefore, we hypothesized that C. madrasense HM411 attempted to restore protein folding homeostasis by enhancing the molecular chaperone-assisted folding function through the up-regulated BIP gene. And there were no significant changes in PDI, ERO1 and IRE1 genes, indicating that oxidative protein folding and IRE1-mediated UPR signaling pathway in C. madrasense HM411 cells are relatively stable at the current stage and have not yet been significantly activated, and that the cells still have a certain ability to cope with protein folding under the existing mechanism. Folded proteins undergo complex assembly and modification in preparation for vesicle transport, which begins with vesicle growth.
3.6 Cell wall, cell membrane differential genes
In our results, low-temperature stress induced changes in the transcript levels of DEGs associated with cell wall components (Figure 6A). We detected the activated expression of the genes related to chitinase synthesis, which attack the crystal structure of chitin by disrupting glycosidic glycoconjugates, which further leads to a decrease in chitin content within the cell wall. The β-mannosidase-encoding gene (manB) showed up-regulated expression (log2FC = 1.12, padj < 0.05). Such an enzyme is indispensable for the complete degradation of mannans. Moreover, the decline in mannans will result in a reduction in mannoprotein molecules. Mannan is a macromolecular polysaccharide that is composed of mannose and glucose via β-(1-2) glycosidic linkages and β-(1-3) glycosidic linkages. The expression of genes related to mannose metabolizing enzymes will decrease the mannose content, thereby influencing the formation of mannan. In addition, key genes responsible for prolonging β-1,3-glucan were down-regulated. Taken together, we hypothesize that reduced cell wall stability may favor improved tolerance to low temperatures. Furthermore, the key genes accountable for elongating β-1,3-glucan were down-regulated. All things considered, we put forward the hypothesis that the decreased stability of the cell wall might contribute to enhanced tolerance to low temperatures.
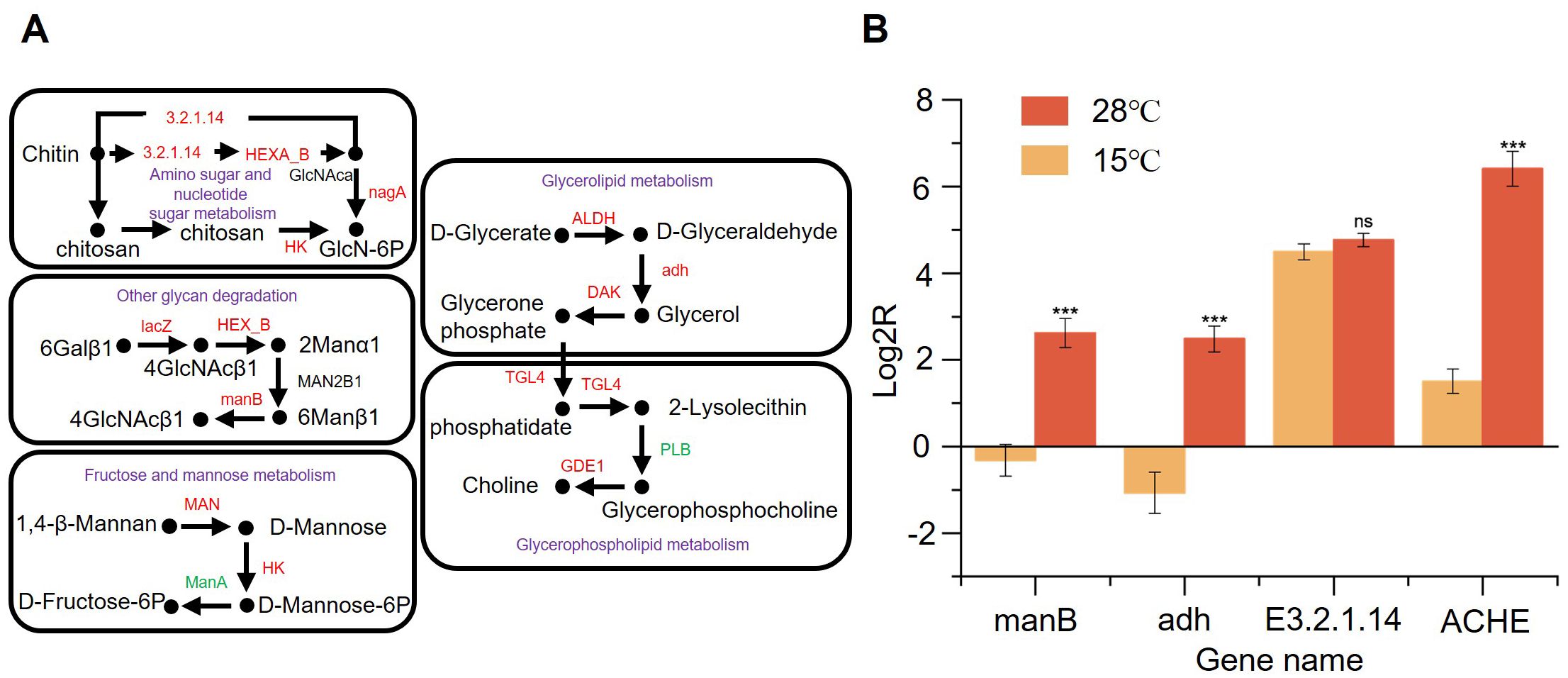
Figure 6. Comparative analysis of endoglucanase secretion efficiency between C. madrasense HM411 and HM412 at 15°C through differential gene expression profiling of cell wall and cell membrane-related pathways. (A) Main cell wall component biodegradation pathways and main cell membrane component biosynthesis pathways. (B) Relative mRNA expression level of 4 genes under different temperature. Error bars represent the standard deviations from three independent experiments. n.s, not significant (P > 0.05), ***P < 0.001.
Low temperature or heat stress is capable of disrupting the stability and fluidity of the membrane. Meanwhile, the stability and fluidity of the membrane dictate the cell’s tolerance towards stress conditions and directly influence a multitude of membrane associated physiological processes (Santos et al., 2019). KEGG analysis of DEGs showed enrichment of glycerolipid and glycerophospholipid metabolic pathways involved in the response to low temperature stress. Enrichment of genes associated with the glycerol ester and glycerophospholipid metabolic pathways was observed. The upregulation of ALDH (log2FC = 1.17, padj < 0.05), adh (log2FC = 2.82, padj < 0.05), and DAK (log2FC = 4.86, padj < 0.05) expression in the glyceride metabolic pathway led to increased intracellular glycerophospholipid levels. Glycerophospholipid acyl ketone participates in pyruvate synthesis during glycolysis, which subsequently promotes the tricarboxylic acid cycle (TCA) to generate substantial ATP through substrate phosphorylation, thereby supplying energy for cellular activities in fungal cells. In the glycerophospholipid metabolic pathway, the upregulation of GDE1 enhances the hydrolysis of glycerophosphodiester and accelerates glycerophospholipid metabolism. This may result in alterations in intracellular glycerophospholipid levels, potentially influencing the composition and fluidity of cell membranes.
To verify the above analysis, we incubated C. madrasense HM411 and HM412 at 15°C and 28°C for 6 days (Figure 6B). Subsequently, RNA was extracted, and the expression of genes involved in the synthesis of E3.2.1.14, manB, adh, and ACHE were determined using β-actin as an internal reference. It should be noted that the relative expression of manB (7.6 ± 2.5-fold, ΔΔCt = -2.62 ± 0.33, n=3 biological replicates), adh (11.4 ± 4.6-fold, ΔΔCt = -2.48 ± 0.30, n=3 biological replicates), E3.2.1.14 (1.2 ± 0.2-fold, ΔΔCt = -4.77 ± 0.15, n=3 biological replicates) and ACHE (30.3 ± 9.8-fold, ΔΔCt = -6.41 ± 0.40, n=3 biological replicates) genes in HM411 at 15°C was higher than that at 28°C. The increased relative expression of manB and E3.2.1.14 may directly influence intracellular chitin content. This could lead to reduced cell wall stability in C. madrasense HM411, and decreased obstruction of extracellular substances. Consequently, the uptake of nutrients, such as glucose and amino acids, into the cell may be facilitated. Glucose, amino acids and other nutrients enter the cell smoothly, and enhance intracellular material metabolic activities. The increased expression of adh and ACHE genes may have enhanced cell membrane fluidity, facilitating the movement of various transporter proteins and signal receptors. This improved membrane dynamics enables cells to more rapidly and accurately perceive external low-temperature signals, which are subsequently transduced intracellularly to activate the expression of relevant genes and initiate the cellulase synthesis pathway.
4 Discussion
The intestinal tract of H. gigas harbors a diverse repertoire of polysaccharide-hydrolyzing enzymes that facilitate the metabolism of debris-derived sugars, representing a critical adaptive strategy for survival in extreme environments while contributing to ecosystem energy flux (Kobayashi et al., 2019). However, key questions remain unresolved, particularly regarding both culturable and unculturable fractions of its gut microbiota, their environmental adaptation mechanisms, as well as their collective roles in driving carbon cycling and energy flow, require further investigation. In this study, we successfully isolated a symbiotic fungal strain, C. madrasense HM411, which has potential for cellulose degradation capacity. Notably, this represents the first report of cellulose-degrading potential in an abyssal-origin strain of C. madrasense, whereas previous studies on terrestrial and marine isolates of this species have primarily focused on their bioactive natural product potential (Tian and Li, 2022). The strain exhibited robust extracellular endoglucanase activity, reaching 63.4 ± 2.3 U/mL at 28°C. Comparative analysis revealed that the deep-sea derived C. madrasense HM411 showed 22.7% greater endoglucanase activity at 15°C than its terrestrial counterpart. This may indicate that C. madrasense HM411 is more adapted to the low-temperature environment. These findings not only advance our understanding of microbial extremophile physiology but also underscore the ecological and biotechnological potential of hadal fungi in carbon turnover and low-temperature enzyme discovery. Future research should focus on elucidating the metabolic regulation mechanisms of these strains in extreme environments and exploring their potential applications in biotechnology and ecological restoration. While this study provides insights into the cellulolytic potential of hadal-adapted fungal symbionts, we acknowledge that our culture-dependent approach may introduce bias relative to the total fungal diversity. The observed culturable fungi likely represent only a subset of the in situ fungal community, as many oligotrophic or slow-growing taxa may resist laboratory cultivation (Tedersoo et al., 2014).
The cellulase production of the two strains was determined at 15°C, and it was found that C. madrasense HM411 entered the enzyme producing stable phase earlier than strain C. madrasense HM412. The analysis of the transcriptome data revealed that the CAZymes were dominated by GHs, AAs, and CEs, which are essential for the degradation of polysaccharides as well as the oxidative degradation of cellulose and chitin (Lombard et al., 2014). The expression of GHs, AAs and CEs-related genes was higher in C. madrasense HM411 compared to CBMs and GTs. Among these genes, cellulase (AA9) is the most up-regulated CAZyme, followed by xylanase (GH43), which is the most up-regulated of the family of GHs with the highest number of differentiated genes. These enzymes are essential for the degradation of cellulose and serve as an important source of carbon and nitrogen for marine organisms. Additionally, in gut microbiota, the GH43 family of enzymes (e.g., xynB, log2FC = 1.59, padj < 0.05) helps to break down complex carbohydrates and affect gut microbiota metabolism and host health (Flint et al., 2012; Xu et al., 2019).
The genes encoding cellulase, a key enzyme for cellulose hydrolysis, were significantly upregulated. This upregulation enhances the strain’s ability to efficiently degrade cellulose-based polysaccharides in the environment, converting them into small-molecule sugars that can be directly absorbed and utilized by cells, thereby providing essential carbon and energy for cellular growth and metabolism (Hegazy et al., 2018; Mattam et al., 2022). Meanwhile, the up-regulation of genes related to the secretory protein synthesis pathway further enhanced the strain’s ability to exchange substances and interact with the external environment (Baeza et al., 2022). These secreted proteins may include hydrolytic enzymes, transporter proteins, and signaling molecules, which facilitate nutrient uptake (e.g., polysaccharides, oligosaccharides, and amino acids) and enhance the strain’s ability to cope with low-temperature stress (Wouters et al., 2000). We support the hypothesis that C. madrasense HM411 strain, by virtue of its gene regulatory mechanisms and strong metabolic capacity, is able to better utilize the rich and diverse polysaccharide resources in the environment compared to terrestrial sources C. madrasense HM412, under 15°C culture conditions. This hypothesis can be analyzed by comparative transcriptomic analysis of C. madrasense HM411 and reference strains (e.g., C. madrasense HM412) grown on polysaccharide as medium at 15°C. This not only facilitates the survival and reproduction of the strain in the host intestine, but also at the macro level of marine ecosystems, C. madrasense HM411 may play an important role in marine carbon cycling and nutrient dynamics. Scientific studies have demonstrated that extracellular enzymes produced by marine microorganisms enhance carbon sequestration by degrading algal and zooplankton-derived detritus, thereby accelerating the vertical flux of organic carbon from surface waters to the deep ocean (Arnosti, 2011). While these enzymatic properties may facilitate carbon cycling, direct quantification of their ecological impact requires future in situ stable isotope probing (SIP) experiments. A limitation of this study is the use of β-actin as the sole reference gene for RT-qPCR normalization. While β-actin showed stable expression here, future studies should validate additional reference genes (e.g., GAPDH). Additionally, while three biological replicates per group align with field standards, larger sample sizes would enhance statistical power. Future work should incorporate power analyses to optimize experimental designs.
Fungi in abyssal environments face many extremes, including environmental conditions such as low temperature, high pressure, and darkness. Fungi are critical for carbon and nutrient cycling in cold ecosystems, and saprophytic fungi actively decompose organic matter by secreting various hydrolytic enzymes (Wang et al., 2017). Under temperature stress conditions, the tertiary structures of molecules such as enzymes and other functional proteins may be damaged and they will not function properly. Fungi can produce heat or cold marker molecules, such as heat shock proteins and chaperones to help repair functional structures (Bai et al., 2015; Tiwari et al., 2015), extracellular hydrolytic enzymes such as chitinolytic enzymes (Fenice, 2016), as well as increased production of heat shock proteins to minimize protein misfolding (Kroll et al., 2014; Miteva-Staleva et al., 2017). In our study, a series of gene expression changes closely related to the cold acclimatization mechanism of fungi were identified. The genes involved in encoding heat shock proteins showed a clear trend of up-regulation, suggesting that the fungus actively increases the synthesis of heat shock proteins in response to low temperatures in order to enhance its own tolerance to low temperature stress (Hao et al., 2022). The up-regulation of the gene encoding chitinase could mean that the fungus is focusing more on obtaining nutrients such as glucose and amino acids from the environment as sustenance to further optimize its survival strategy (Langner and Göhre, 2016). Based on these results, it can be suggested that C. madrasense HM411 strains may have acquired more unique and effective adaptive strategies during the evolutionary process than strains of terrestrial origin, making them more adaptable to the extreme environmental conditions of low temperatures in the hadal zone. However, comparisons with single terrestrial isolates may not fully represent the ecological plasticity of this species. Future studies should include a broader diversity of marine and terrestrial strains, particularly those from extreme environments (e.g., polar regions or hot springs), to better elucidate potential niche-specific adaptations.
Our work emphasizes the potential importance of H. gigas symbiotic fungi, in particular C. madrasense HM411. Its cellulase synthesis potential was studied by enzyme producing primary and secondary screens. Our study verifies the presence of cellulase-producing symbiotic fungi in the gut of the H. gigas from the Mariana Trench, which may be helpful in the digestion of difficult to biodegrade substances by H. gigas. Furthermore, our study provides valuable insights into the adaptation mechanisms of symbiotic fungi to extreme environments, particularly their capacity to produce functional enzymes under low-temperature conditions. These findings not only enhance our understanding of microbial symbiosis in deep-sea ecosystems but also hold potential for biotechnological applications.
Data availability statement
The datasets presented in this study can be found in online repositories. Raw sequencing reads for the transcript information in this work were submitted to the NCBI Sequence Read Archive under sequential accession numbers from SRR31859304 to SRR31859309.
Author contributions
PJ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. YC: Writing – review & editing, Investigation, Visualization. YX: Investigation, Writing – review & editing. SW: Investigation, Writing – review & editing. JF: Writing – review & editing, Resources, Supervision. XY: Resources, Supervision, Writing – review & editing, Conceptualization, Funding acquisition, Investigation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Natural Science Foundation of China 42476091, and Shanghai Municipal Education Commission (2023ZKZD53).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1602692/full#supplementary-material
References
Amer A., Nasim F. u.-H., Kashfa B., and Bibi A. (2017). Microbial β-Glucosidase: sources, production and applications. Available online at: https://iris.uniroma1.it/handle/11573/1227144 (Accessed December 22, 2024).
Arnosti C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Baeza M., Zúñiga S., Peragallo V., Gutierrez F., Barahona S., Alcaino J., et al. (2022). Response to cold: A comparative transcriptomic analysis in eight cold-adapted yeasts. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.828536
Bai Y., Wang S., Zhong H., Yang Q., Zhang F., Zhuang Z., et al. (2015). Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature. Sci. Rep. 5, 14582. doi: 10.1038/srep14582
Barzkar N. and Sohail M. (2020). An overview on marine cellulolytic enzymes and their potential applications. Appl. Microbiol. Biotechnol. 104, 6873–6892. doi: 10.1007/s00253-020-10692-y
Batista-García R. A., Sutton T., Jackson S. A., Tovar-Herrera O. E., Balcázar-López E., Sánchez-Carbente M., et al. (2017). Characterization of lignocellulolytic activities from fungi isolated from the deep-sea sponge Stelletta normani. PloS One 12, e0173750. doi: 10.1371/journal.pone.0173750
Cameron M. D. and Aust S. D. (2001). Cellobiose dehydrogenase–an extracellular fungal flavocytochrome. Enzyme Microb. Technol. 28, 129–138. doi: 10.1016/S0141-0229(00)00307-0
Chan J., Geng D., Pan B., Zhang Q., and Xu Q. (2021). Metagenomic insights into the structure and function of intestinal microbiota of the hadal amphipods. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.668989
Cui C.-M., Li X.-M., Li C.-S., Proksch P., and Wang B.-G. (2010). Cytoglobosins A–G, cytochalasans from a marine-derived endophytic fungus, chaetomium globosum QEN-14. J. Nat. Prod. 73, 729–733. doi: 10.1021/np900569t
Cui Y., Xiao Y., Wang Z., Ji P., Zhang C., Li Y., et al. (2024). Microbial community structure and functional traits involved in the adaptation of culturable bacteria within the gut of amphipods from the deepest ocean. Microbiol. Spectr. 0, e00723–e00724. doi: 10.1128/spectrum.00723-24
Darwish A. M. G. and Abdel-Azeem A. M. (2020). “Chaetomium enzymes and their applications,” in Recent developments on genus chaetomium. Ed. Abdel-Azeem A. M. (Springer International Publishing, Cham), 241–249. doi: 10.1007/978-3-030-31612-9_9
Deng L., Zhong M., Li Y., Hu G., Zhang C., Peng Q., et al. (2023). High hydrostatic pressure harnesses the biosynthesis of secondary metabolites via the regulation of polyketide synthesis genes of hadal sediment-derived fungi. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1207252
Dresios J., Derkatch I. L., Liebman S. W., and Synetos D. (2000). Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39, 7236–7244. doi: 10.1021/bi9925266
Duarte A. W. F., Santos d., Aparecida J., Vitti V. M., Freitas V. J. M., Hugo M. V., et al. (2018). Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 38, 600–619. doi: 10.1080/07388551.2017.1379468
Fan S., Wang M., Ding W., Li Y.-X., Zhang Y.-Z., and Zhang W. (2022). Scientific and technological progress in the microbial exploration of the hadal zone. Mar. Life Sci. Technol. 4, 127–137. doi: 10.1007/s42995-021-00110-1
Fenice M. (2016). The psychrotolerant antarctic fungus lecanicillium muscarium CCFEE 5003: A powerful producer of cold-tolerant chitinolytic enzymes. Molecules 21, 447. doi: 10.3390/molecules21040447
Flint H. J., Scott K. P., Duncan S. H., Louis P., and Forano E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Hao H., Zhang J., Wang H., Wang Q., Chen M., Juan J., et al. (2019). Comparative transcriptome analysis reveals potential fruiting body formation mechanisms in Morchella importuna. AMB Express 9, 103. doi: 10.1186/s13568-019-0831-4
Hao H., Zhang J., Wu S., Bai J., Zhuo X., Zhang J., et al. (2022). Transcriptomic analysis of Stropharia rugosoannulata reveals carbohydrate metabolism and cold resistance mechanisms under low-temperature stress. AMB Express 12, 56. doi: 10.1186/s13568-022-01400-2
Hegazy W. K., Abdel-Salam M. S., Hussain A. A., Abo-Ghalia H. H., and Hafez S. S. (2018). Improvement of cellulose degradation by cloning of endo-β-1, 3-1, 4 glucanase (bgls) gene from Bacillus subtilis BTN7A strain. J. Genet. Eng. Biotechnol. 16, 281–285. doi: 10.1016/j.jgeb.2018.06.005
Helal G. A., Khalil R. R., Galal Y. G., Soliman S. M., and Abd Elkader R. S. (2021). Studies on cellulases of some cellulose-degrading soil fungi. Arch. Microbiol. 204, 65. doi: 10.1007/s00203-021-02705-9
Henriksson G., Johansson G., and Pettersson G. (2000). A critical review of cellobiose dehydrogenases. J. Biotechnol. 78, 93–113. doi: 10.1016/S0168-1656(00)00206-6
Huang M., Bao J., Hallström B. M., Petranovic D., and Nielsen J. (2017). Efficient protein production by yeast requires global tuning of metabolism. Nat. Commun. 8, 1131. doi: 10.1038/s41467-017-00999-2
John J A., Samuel M. S., Govarthanan M., and Selvarajan E. (2022). A comprehensive review on strategic study of cellulase producing marine actinobacteria for biofuel applications. Environ. Res. 214, 114018. doi: 10.1016/j.envres.2022.114018
Khokhar I., Haider M. S., Mushtaq S., and Mukhtar I. (2012). Isolation and screening of highly cellulolytic filamentous fungi. J. Appl. Sci. Environ. Manage. 16, 223–226. Available at: https://www.ajol.info/index.php/jasem/article/view/90862 (Accessed December 29, 2024).
Kilgallen N. M. (2015). Three new species of Hirondellea (Crustacea, Amphipoda, Hirondelleidae) from hadal depths of the Peru-Chile Trench. Mar. Biol. Res. 11, 34–48. doi: 10.1080/17451000.2014.889309
Kobayashi H., Shimoshige H., Nakajima Y., Arai W., and Takami H. (2019). An aluminum shield enables the amphipod Hirondellea gigas to inhabit deep-sea environments. PloS One 14, e0206710. doi: 10.1371/journal.pone.0206710
Kroll K., Pähtz V., and Kniemeyer O. (2014). Elucidating the fungal stress response by proteomics. J. Proteomics 97, 151–163. doi: 10.1016/j.jprot.2013.06.001
Lacey N. C., Rowden A. A., Clark M. R., Kilgallen N. M., Linley T., Mayor D. J., et al. (2016). Community structure and diversity of scavenging amphipods from bathyal to hadal depths in three South Pacific Trenches. Deep Sea Res. Part Oceanogr. Res. Pap . 111, 121–137. doi: 10.1016/j.dsr.2016.02.014
Langner T. and Göhre V. (2016). Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr. Genet. 62, 243–254. doi: 10.1007/s00294-015-0530-x
Li X., Han C., Li W., Chen G., and Wang L. (2020). Insights into the cellulose degradation mechanism of the thermophilic fungus Chaetomium thermophilum based on integrated functional omics. Biotechnol. Biofuels 13, 143. doi: 10.1186/s13068-020-01783-z
Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Mattam A. J., Chaudhari Y. B., and Velankar H. R. (2022). Factors regulating cellulolytic gene expression in filamentous fungi: an overview. Microb. Cell Factories 21, 44. doi: 10.1186/s12934-022-01764-x
Michels P. C. and Clark D. S. (1997). Pressure-enhanced activity and stability of a hyperthermophilic protease from a deep-sea methanogen. Appl. Environ. Microbiol. 63, 3985–3991. doi: 10.1128/aem.63.10.3985-3991.1997
Miller A. N. and Huhndorf S. M. (2005). Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Mol. Phylogenet. Evol. 35, 60–75. doi: 10.1016/j.ympev.2005.01.007
Miteva-Staleva J. G., Krumova E. T., Vassilev S. V., and Angelova M. B. (2017). Cold-stress response during the stationary-growth phase of Antarctic and temperate-climate Penicillium strains. Microbiology 163, 1042–1051. doi: 10.1099/mic.0.000486
Oguri K., Kawamura K., Sakaguchi A., Toyofuku T., Kasaya T., Murayama M., et al. (2013). Hadal disturbance in the Japan Trench induced by the 2011 Tohoku–Oki Earthquake. Sci. Rep. 3, 1915. doi: 10.1038/srep01915
Riccardi C., Calvanese M., Ghini V., Alonso-Vásquez T., Perrin E., Turano P., et al. (2023). Metabolic robustness to growth temperature of a cold- adapted marine bacterium. mSystems 8, e01124–e01122. doi: 10.1128/msystems.01124-22
Robinson C. H. (2001). Cold adaptation in Arctic and Antarctic fungi. New Phytol. 151, 341–353. doi: 10.1046/j.1469-8137.2001.00177.x
Rosyida V. T., Indrianingsih A. W., Maryana R., and Wahono S. K. (2015). Effect of temperature and fermentation time of crude cellulase production by trichoderma reesei on straw substrate. Energy Proc. 65, 368–371. doi: 10.1016/j.egypro.2015.01.065
Santos T., Viala D., Chambon C., Esbelin J., and Hébraud M. (2019). Listeria monocytogenes biofilm adaptation to different temperatures seen through shotgun proteomics. Front. Nutr. 6. doi: 10.3389/fnut.2019.00089
Schilbach S., Hantsche M., Tegunov D., Dienemann C., Wigge C., Urlaub H., et al. (2017). Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209. doi: 10.1038/nature24282
Seppä L. and Makarow M. (2005). Regulation and Recovery of Functions of Saccharomyces cerevisiae Chaperone BiP/Kar2p after Thermal Insult. Eukaryot. Cell 4, 2008–2016. doi: 10.1128/ec.4.12.2008-2016.2005
Shi L., Xiao W., Liu Z., Pan B., and Xu Y. (2018). Diet change of hadal amphipods revealed by fatty acid profile: A close relationship with surface ocean. Mar. Environ. Res. 142, 250–256. doi: 10.1016/j.marenvres.2018.10.012
Sohail M., Siddiqi R., Ahmad A., and Khan S. A. (2009). Cellulase production from Aspergillus Niger MS82: effect of temperature and pH. New Biotechnol. 25, 437–441. doi: 10.1016/j.nbt.2009.02.002
Sun Q., Vega N. M., Cervantes B., Mancuso C. P., Mao N., Taylor M. N., et al. (2022). Enhancing nutritional niche and host defenses by modifying the gut microbiome. Mol. Syst. Biol. 18, e9933. doi: 10.15252/msb.20209933
Tedersoo L., Bahram M., Põlme S., Kõljalg U., Yorou N. S., Wijesundera R., et al. (2014). Global diversity and geography of soil fungi. Science 346, 1256688. doi: 10.1126/science.1256688
Tian Y. and Li Y. (2022). A review on bioactive compounds from marine-derived chaetomium species. J. Microbiol. Biotechnol. 32, 541–550. doi: 10.4014/jmb.2201.01007
Tiwari S., Thakur R., and Shankar J. (2015). Role of heat-shock proteins in cellular function and in the biology of fungi. Biotechnol. Res. Int. 2015, 132635. doi: 10.1155/2015/132635
Wang M., Tian J., Xiang M., and Liu X. (2017). Living strategy of cold-adapted fungi with the reference to several representative species. Mycology 8, 178–188. doi: 10.1080/21501203.2017.1370429
Wei (2007). Screening of high-yield cellulase-producing strains (Lanzhou, China: Lanzhou University).
Wei T., Liao Y., Wang Y., Li J., and He L. (2023). Comparably characterizing the gut microbial communities of amphipods from littoral to hadal zones. J. Mar. Sci. Eng. 11, 2197. doi: 10.3390/jmse11112197
White S. A., Hoeger M., Schweppe J. J., Shillingford A., Shipilov V., and Zarutskie J. (2004). Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA 10, 369–377. doi: 10.1261/rna.2159504
Wolff T. (1959). The hadal community, an introduction. Deep Sea Res. 6, 95–124. doi: 10.1016/0146-6313(59)90063-2
Wolff T. (1970). The concept of the hadal or ultra-abyssal fauna. Deep Sea Res. Oceanogr. Abstr. 17, 983–1003. doi: 10.1016/0011-7471(70)90049-5
Wouters J. A., Rombouts F. M., Kuipers O. P., de Vos W. M., and Abee T. (2000). The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23, 165–173. doi: 10.1016/S0723-2020(00)80001-6
Xiao Y., Yan F., Cui Y., Du J., Hu G., Zhai W., et al. (2023). A symbiotic bacterium of Antarctic fish reveals environmental adaptability mechanisms and biosynthetic potential towards antibacterial and cytotoxic activities. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1085063
Xu B., Dai L., Zhang W., Yang Y., Wu Q., Li J., et al. (2019). Characterization of a novel salt-, xylose- and alkali-tolerant GH43 bifunctional β-xylosidase/α-l-arabinofuranosidase from the gut bacterial genome. J. Biosci. Bioeng. 128, 429–437. doi: 10.1016/j.jbiosc.2019.03.018
Xu Z., Chikka M. R., Xia H., and Ready D. F. (2016). Ire1 supports normal ER differentiation in developing Drosophila photoreceptors. J. Cell Sci. 129, 921–929. doi: 10.1242/jcs.180406
Yasuhara M. and Danovaro R. (2016). Temperature impacts on deep-sea biodiversity. Biol. Rev. 91, 275–287. doi: 10.1111/brv.12169
Yuan S., Wu Y., and Cosgrove D. J. (2001). A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 127, 324–333. doi: 10.1104/pp.127.1.324
Keywords: gut microbes, amphipods, the Mariana Trench, psychrotolerant adaptation, cellulase, transcriptomic analyses
Citation: Ji P, Cui Y, Xiao Y, Wan S, Fang J and Yu X (2025) Low-temperature adaptation of Chaetomium madrasense, a symbiotic gut fungus of amphipods in the Mariana Trench: cellulase activity and transcriptome analysis. Front. Mar. Sci. 12:1602692. doi: 10.3389/fmars.2025.1602692
Received: 04 April 2025; Accepted: 10 June 2025;
Published: 26 June 2025.
Edited by:
Valerio Mazzella, Anton Dohrn Zoological Station Naples, ItalyReviewed by:
Lucia Bongiorni, National Research Council (CNR), ItalyGiulio Barone, National Research Council (CNR), Italy
Copyright © 2025 Ji, Cui, Xiao, Wan, Fang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Yu, eHl1QHNob3UuZWR1LmNu
 Paiyao Ji
Paiyao Ji Yu Xiao
Yu Xiao Xi Yu
Xi Yu