- 1South China Sea Environmental Monitoring Center, State Oceanic Administration, Guangzhou, China
- 2Key Laboratory of Marine Environmental Survey Technology and Application, Ministry of Natural Resources, Guangzhou, China
- 3Department of Biology, Hong Kong Baptist University, Hong Kong, Hong Kong SAR, China
- 4Key Laboratory of Tropical Marine Bioresources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Science, Guangzhou, China
Introduction: As concerns mount over the threats facing oyster reefs, awareness of the need to mitigate habitat loss and restore ecosystem services is increasing. However, challenges in identifying oysters have limited our understanding of their species- and population-level diversity, complicating efforts to establish effective marine protected areas. Therefore, this study aims to address these challenges by conducting a comprehensive survey along the coastline of Guangdong Province to assess the species composition and spatial distribution of oyster reefs across 51 intertidal sites, such as estuaries, islands, oyster farms, mudflats, and mangroves.
Methods: In total, we collected 742 oyster specimens by qualitative survey for oyster distribution and generated approximately 1,400 mitochondrial DNA sequences (N = 693 for 16S rRNA and N=706 for cytochrome c oxidase subunit I) to support genetic analysis. More than 30000 oysters sample collected by quantitative survey were applied to analyze the oyster assemblages.
Results: The analyses revealed 12 mitochondrial lineages representing three genera within the family Ostreidae. Phylogenetic analysis confirmed robust monophyletic groupings, confirming species identities and leading to the identification of two cryptic species within the genus Saccostrea. Based on DNA evidence, these two cryptic Saccostrea species were closely related to the known S. non-mordax D and H lineages. Quantitative analysis showed that Crassostrea sikamea was the most prevalent species in the study area, with an average abundance and biomass > 1,400 individuals/m2 and 4,449 grams/m2, respectively. Qualitative and quantitative assessment both revealed at least 6 species were identified at G07 (Jieyang), being the most biodiverse location of Guangdong Province.
Discussion: By mapping oyster distribution and updating the species inventory, our study provides a foundation for future research on oyster populations and informs conservation efforts aimed at preserving and restoring oyster habitats, thereby supporting marine biodiversity.
1 Introduction
Oysters (Family Ostreidae)—a prominent group of marine bivalve mollusks—play a vital role in global aquaculture, providing food and livelihoods for millions of people while significantly contributing to local and national economies. In Guangdong Province, China, cupped and rock oysters are particularly important, with an annual production of 1.15 million metric tons supporting millions of residents (Guo et al., 2018; Ma et al., 2021a). Although oysters are common, accurately identifying oyster species remains challenging owing to their pronounced phenotypic plasticity and highly variable shell morphology (Wang et al., 2010; Salvi and Mariottini, 2017; Li et al., 2018). Early taxonomic efforts of China that relied on shell characteristics often misclassified ecotypes as distinct species, leading to confusion in species identification (Harry, 1985; Guo et al., 2018).
However, by virtue of the progress in molecular identification, DNA sequence analysis has significantly advanced Ostreidae taxonomy, revealing a few ecophenotypic variants and cryptic species within cupped oysters while refining our understanding of biodiversity and phylogeny. For example, in northern China, mitochondrial DNA markers, including cytochrome c oxidase subunit I (COI) and 16S rRNA, have confirmed Crassostrea gigas as the dominant oyster species, while C. plicatula and C. talienwhanensis have been identified as ecotypes of C. gigas (Wang et al., 2008). Similarly, the designated C. rivularis in China is now recognized as the red meat oyster (C. ariakensis) or the white meat oyster (C. hongkongensis) (Wang et al., 2004). Genetic analyses have also revealed several sympatric cryptic species, leading to the identification of new species such as C. dianbaiensis (Xia et al., 2014; Sekino et al., 2015), C. zhanjiangensis (Wu et al., 2013), and Saccostrea mordoides (Cui et al., 2021). Phylogenetic studies have also provided some grounds for supporting the validity of Talonostrea talonata, the reclassification of several commercially important Crassostrea species into the genus Magallana, and the conspecific relationship between C. iredalei and C. bilineata (Salvi and Mariottini, 2016; Li et al., 2017).
On the other hand, within Saccostreinae, rock oysters were more complex led to multiple confusion and modification owing to their high morphological plasticity. Previous studies suggest a potential Saccostrea lineage that includes S. mordax (namely S. scyphophilla lineage A and B), as well as a “superspecies” of S. cuccullata which encompasses nine mitochondrial lineages (designated as lineages A–G along with S. kegaki and S. glomerata) (Lam and Morton, 2006). Based on this phylogenetic analysis, the nomenclature of S. cuccullata lineage C and S. cuccullata lineage F has been adopted and recorded in Malaysia and Singapore (Hamaguchi et al., 2014). In 2016, following the recognition that S. cuccullata is restricted to Central Western Africa and that its lineages should not represent the Indo-West Pacific Saccostrea oysters, S. cuccullata superspecies were redefined as ‘‘non-mordax’’ rock oysters (Sekino and Yamashita, 2016). As the misidentification revised, three additionally cryptic rock oyster species were confirmed and grouped into the S. non-mordax lineage, designated as lineage H, I, and J, clarifying the tentative lineage of Saccostreinae. Consequently, Saccostrea lineage F was recognized as S. malabonensis, while S. non-mordax B has been reclassified as S. echinata (Guo et al., 2018). More recently, S. non-mordax lineage J has been identified as S. spathulata (McDougall et al., 2024).
These identified cupped and rock oysters are keystone species in forming oyster reefs, which serve as vital ecosystems by providing services such as water filtration, habitat for various marine species, recreational opportunities, coastal storm protection and even potential candidates for genetic improvement (Tolley and Volety, 2005; Morris et al., 2021; Searles et al., 2021). However, 85% of the oyster reefs of the world have been lost, with most remaining reefs degraded and ecologically diminished (Jackson et al., 2001; Beck et al., 2011; Fitzsimons et al., 2019; Jiang et al., 2022). Although oyster reefs were once densely distributed in shallow marine environments of Guangdong, they have declined over the past decades owing to coastal development, overfishing, and uncontrolled nutrients and pollutants input, highlighting the need for improved conservation measures and more importantly effective restoration.
Therefore, understanding oyster species identity and their spatial distribution is essential as it informs monitoring reef health and restoration efforts. This knowledge enables more precise assessments of the oyster reef ecosystem, ultimately aiding in the conservation of oyster germplasm resources while maintaining its biodiversity (GuerraGarcía et al., 2008). Previous studies has defined the taxonomy and spatial distribution of cupped oysters (Wang et al., 2008, 2010; Guo et al., 2018), as well as population genetic diversity and phylogeography of oysters in northern and southern China (Hu et al., 2018; Liu et al., 2021; Ma et al., 2021a, b). Most of these studies have been confined to specific regions and species though. As yet, the reference and detailed reports on oyster composition and distribution of Guangdong Province are relatively rare and outdated (Cheng et al., 2022). The 2007 national coastal survey by State Oceanic Administration in China (Wu et al., 2025) showed C. ariakensis as endemic to Guangdong but found no evidence of C. hongkongensis or C. sikamea—findings that appear inconsistent with current observation. As a result, this discrepancy highlights a critical gap in our understanding of oyster biodiversity and spatial distribution within the coastal ecosystems of Guangdong.
Therefore, this study aims to address these gaps by conducting the most extensive survey of oyster reefs in Guangdong to date, covering 51 stations along the coastline of the Guangdong Province. We seek to clarify species composition, resolve taxonomic ambiguities using phylogenetic analysis, and document spatial patterns of oyster abundance and distribution via quantitative research using conventional sample plot surveys. We hope to provide a baseline for future conservation planning and contribute to the sustainable management of oyster reef ecosystems.
2 Materials and methods
2.1 Survey site
A comprehensive survey of the oyster reef ecosystem along Guangdong coastlines was conducted from July 2022 to February 2023, covering 51 sites (Supplementary Table 1; Figure 1). Herein, to encompass the oyster’s extensive natural distribution, we adopted a broad definition of oyster reefs for this study. These reefs constitute biogenic structures formed either by dense oysters’ colonies with substantial vertical reliefs —commonly termed oyster reefs or beds—or secondary structures known as oyster aggregations, where adherent oysters settle upon either hard or soft substrates (Beck et al., 2009; Baggett et al., 2014). Site selection was based on historical distribution records in intertidal areas, including mangroves, mudflats, estuaries, and even aquaculture farms (Cheng et al., 2022). Adjustments to the initially selected sites were occasionally made when necessary to account for changes in the actual oyster reef distribution. Two forms of survey, namely qualitative and quantitative survey were conducted at each site during low tide.
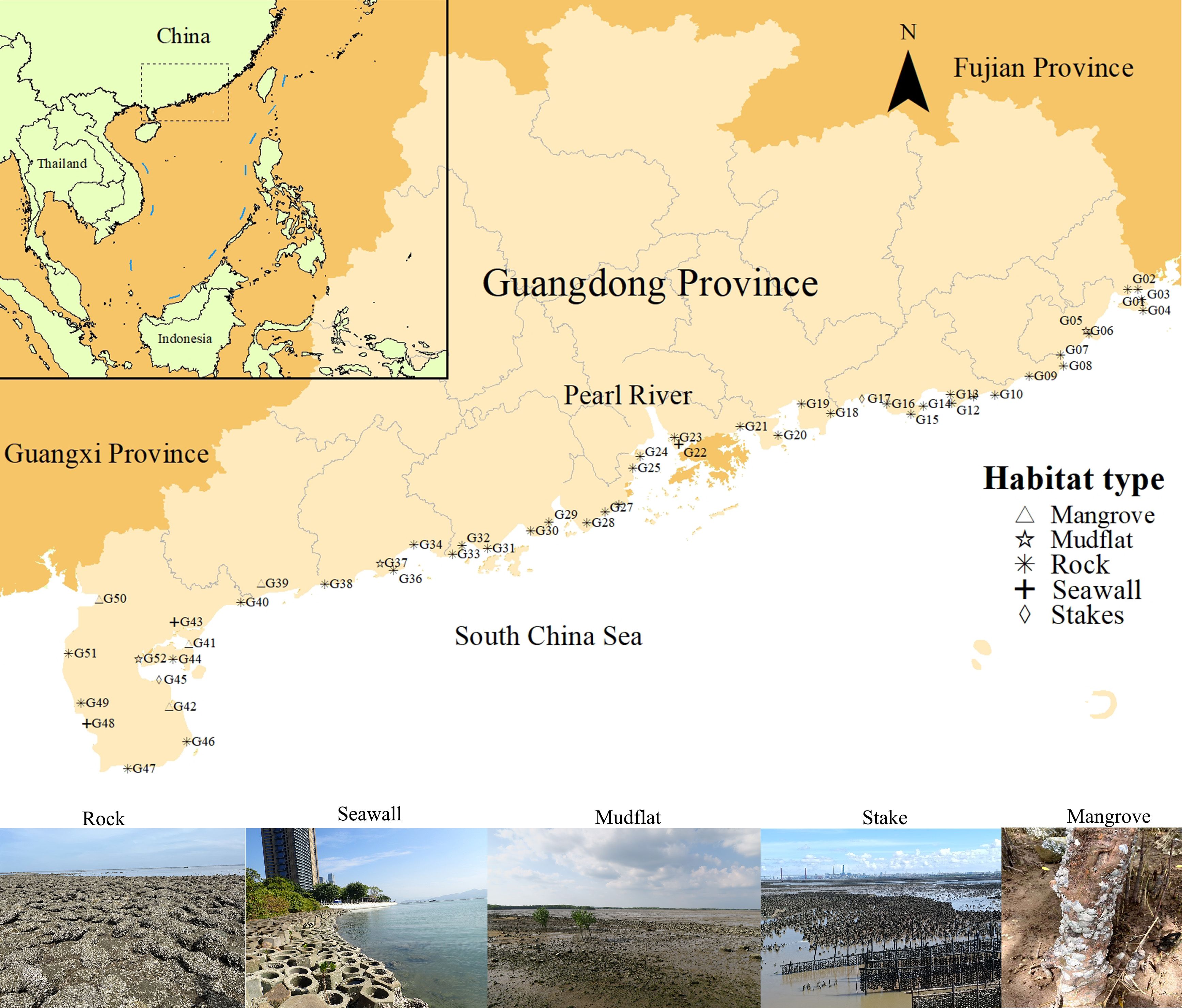
Figure 1. Survey sites of oyster reefs in Guangdong Province and their substrate types. The site numbers are numbered from east to west. Habitat distinctions are denoted by geometric markers: triangles indicating mangrove type, snowflakes indicating rock type, crosses indicating seawall type, and diamonds indicating Stakes. The frame in the upper-left corner designates the position of Guangdong Province.
2.2 Qualitative survey
At each site, GPS coordinates were recorded to map the distribution of oyster populations within a defined survey polygon. To comprehensively document diversity of oysters in the region, varying morphologies of oysters based on shell features, coloration and diagnostic pattern were collected. Totally 742 samples by qualitative survey were collected from 51 survey sites. In order to describe the original features of oysters’ specimen, specimens were photographed in situ before being removed from their substrate and individually stored in labeled plastic containers. Ultimately, the flesh was separated by sterilized scalpel and then stored at -20°C for subsequent laboratory analysis. All samples of qualitative survey were employed to molecular identification, and haplotypes separation as well as genetic diversity. Shells of all specimens were deposited in the South China Sea Monitoring Center, State Oceanic Administration.
2.3 Quantitative survey
Each oyster reef site was classified into upper, middle, and lower tidal zones based on natural tidal patterns. In the upper and lower tidal zones, three quadrats of almost 0.1 m × 0.1 m were laid out by quadrats placement methodology at random, adhering to the same guideline of intertidal zone research (Lu et al., 2024), while in the middle tidal zone, nine quadrats were collected in same method. The number of quadrats was based on the ground that nine quadrats informed by species accumulation curve was sufficient to collect all species of each site based on previous research (Supplementary Figure 1). All macrobenthic organisms, including oysters and algae within the quadrats, were removed using a brush and screwdriver and fixed in 95% ethanol. A 15 cm ruler was included in each quadrat and photographed to provide a scale reference, and each photograph was analyzed to calculate exact sample area of quadrats using ZEISS Axio Vision image processing software.
Owing to huge amounts of oysters detected by quadrats layout, tentative morphological identification was performed via the quantitative survey. Crassostrea species were identified based on established shell morphology descriptions (Huber, 2010; Wang et al., 2010). Within the genus Saccostrea, species of S. echinata, S. malabonensis, and S. scyphophilla were distinguishable. For example, S. echinata displays dense hyote (tubular) spines on its right valve and S. malabonensis exhibits a curved shell margin coupled with strong ribs forming a distinctive plicated structure, while S. scyphophilla is characterized by an orderly and jagged shell margin and purple spots on right valves. Despite this, owing to vague morphological distinction of other Saccostrea, we were unable to accurately identify them to species-level. Consequently, to ensure consistency, unresolved Saccostrea species were provisionally classified as S. non-mordax. Finally, an integrated approach was employed, wherein morphological results were guided and further validated by molecular results in the qualitative survey.
Oyster abundances were determined based on the number and weight of individuals, which were then transformed into density (ind/m2) and biomass (g/m2) metrics (Baggett et al., 2014). Biomass were measured by whole specimen with shells in wet weight after removing surface attachment and blotting excess moisture. Spatial distribution maps were generated using ArcGIS 10.7 to visualize these data.
2.4 Description of shell forms
Description of distinct shell characteristics for unresolved oysters included external and internal features. External features examined included the area of left valve attachment, the pattern of radial ribs, grooves, marginal plication, and growth lamellae, alongside the occurrence of growth lines and hyote spines (Supplementary File Figure 4). Internal features considered included the presence or absence of chalky deposits and chomata, the shape of adductor muscle scar and ligament (Littlewood, 1994; Sekino and Yamashita, 2016). The shell height was measured by vernier calipers with 0.1 mm accuracy, and the whole weight was measured by electronic scales with 0.01 g accuracy (Zhang et al., 2017). These characteristics facilitated the preliminary identification of oysters during qualitative surveys and were conducive to the discovery of new taxa within the reefs.
2.5 Molecular and morphological identification
For samples collected by qualitative survey, total genomic DNA was extracted from the adductor muscle using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s instructions. Mitochondrial DNA fragments of 16S rRNA and COI were amplified using the primer pairs ArL(F) (5’-CGCCTGTTTATCAAAAACAT-3’) and BrH(R) (5’-CCGGTCTGAACTCAGAT CACG-3’) (Palumbi et al., 1996), as well as LCO1490 (5’-GGTCAACAAATCATAA AGATATTGG-3’) and HCO2198 (5’-TAAACT TCAGGGTGACCAAAAAATCA-3’) (Folmer et al., 1994). Polymerase chain reactions were performed in a 25 µl reaction mixture containing 13.38 µl ddH2O, 2.5 µl Ex Taq Buffer (TaKaRa HS), 1.5 µl of each primer (10 mM), 2 µL template DNA, and 0.12 µl Ex Taq HS, using a BIO-Metro thermal cycler. The PCR conditions for 16S rRNA included an initial denaturation at 94°C for 3 min, followed by 40 cycles of 94°C, 45°C, and 72°C for 40, 40, and 45 s, respectively, with a final extension at 72°C for 7 min. For COI, the PCR protocol included an initial denaturation at 94°C for 3 min, followed by 5 cycles of 94°C, 45°C, and 72°C for 40, 40, and 50 s, respectively. This was followed by 35 cycles of 94°C, 51°C, and 72°C for 40, 40, and 50 s, respectively, with a final extension at 72°C for 7 min. PCR products were confirmed using 1% agarose gel electrophoresis and sequenced by Sangon Biotech Company (Guangzhou, China). The obtained sequences were initially analyzed using BioEdit 7.0. Molecular identification was conducted based on BLAST searches against the NCBI database. More importantly, the identification was further confirmed by phylogenetic tree constructed by correct and non-local reference sequence (Table 1). Haplotypes were identified using DNASP v. 5.10.01 (Librado and Rozas, 2009) and used to calculate haplotype diversity. Each identified haplotype was designated following a standardized format, combining “Hap number” with “ML+serial numbers” for qualitative surveys. The compiled sequences of S. sp.1 were deposited in NCBI under accession numbers PQ452111 and PQ459553–PQ459559 for the COI and 16S rRNA genes, respectively. Sequences of S. sp.2 were assigned under accession numbers PQ455391–PQ455392 and PQ459552 for the COI and 16S rRNA genes, respectively.
2.6 Phylogenetic analysis
DNA sequences from this study, combined with reference sequences downloaded from NCBI, were analyzed using Phylosuite v. 1.2.3 and MEGA v. 7.0.26 (Kumar et al., 2016; Zhang et al., 2020) (Table 1). Several plug-in programs were utilized, including MAFFT for sequence alignment, TrimAI for trimming redundant sequence at both ends, Gblocks for removing divergent and ambiguously aligned blocks, and ModelFinder for model selection (Gerard and Jose, 2007; Capella-Gutierrez et al., 2009; Standley, 2013). Following sequence procession, phylogenetic analysis was conducted using Bayesian inference (BI) and ML methods. A Bayesian tree was constructed with a Markov Chain Monte Carlo simulation run for 2 × 106 generations, while ML analysis was performed using IQ-Tree with bootstrap support based on 5,000 replications (Huelsenbeck, 2012). Sequence divergence among haplotype groups was calculated in MEGA v. 7.0.26 using Kimura’s two-parameter (K2P) model (Kimura, 1980).
3 Results
3.1 Species identities of oysters in Guangdong
Molecular analyses and phylogenetic assessments were conducted on representative individuals from oyster samples of distinct shell features. In total, 706 COI sequences and 693 16S rRNA sequences were generated from 742 individuals (Table 2), yielding 240 COI haplotypes and 67 16S rRNA haplotypes (Figures 2, 3).
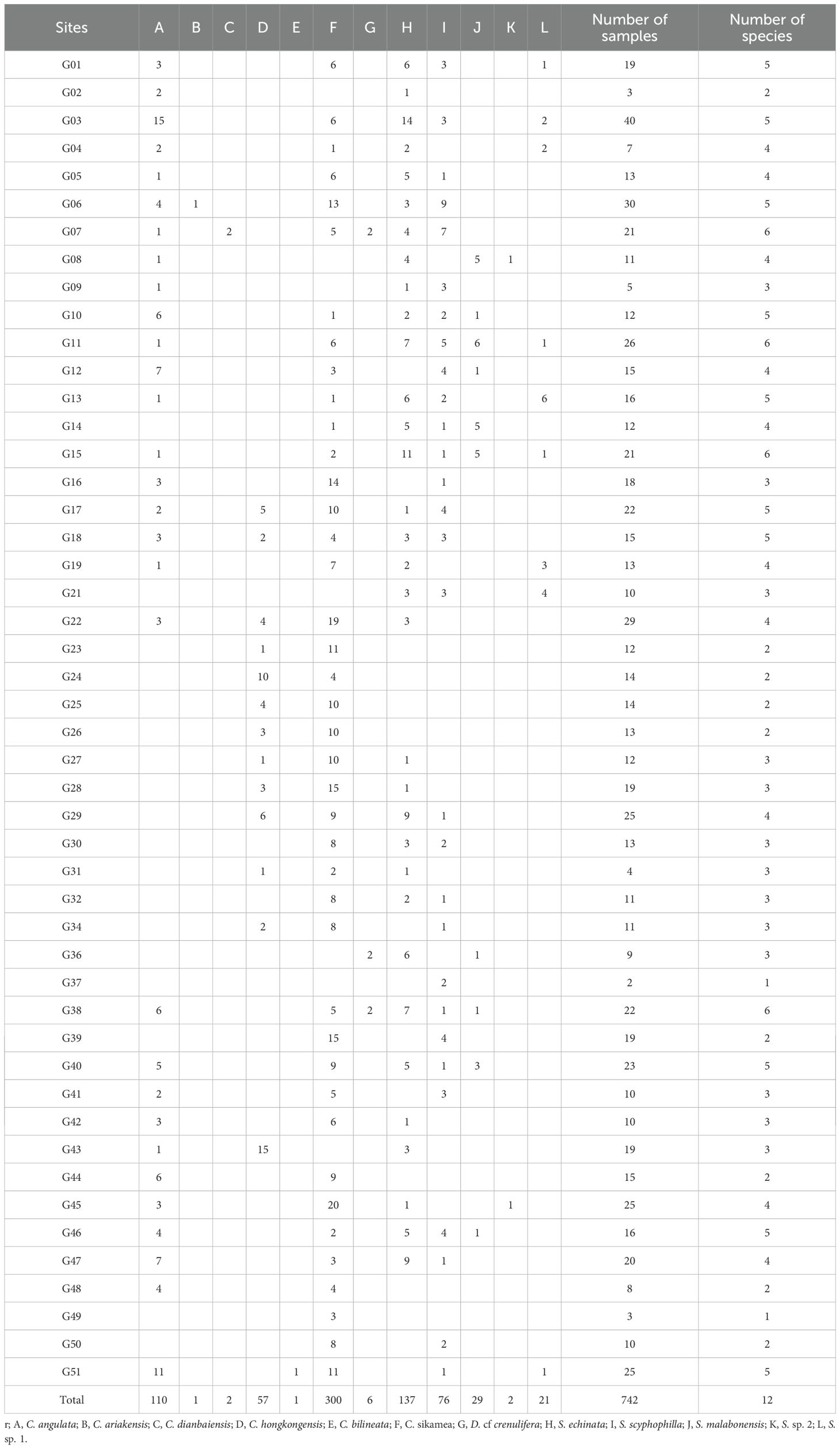
Table 2. Species identification and distribution of oysters at oyster reefs along coastline of Guangdong Province by qualitative survey.
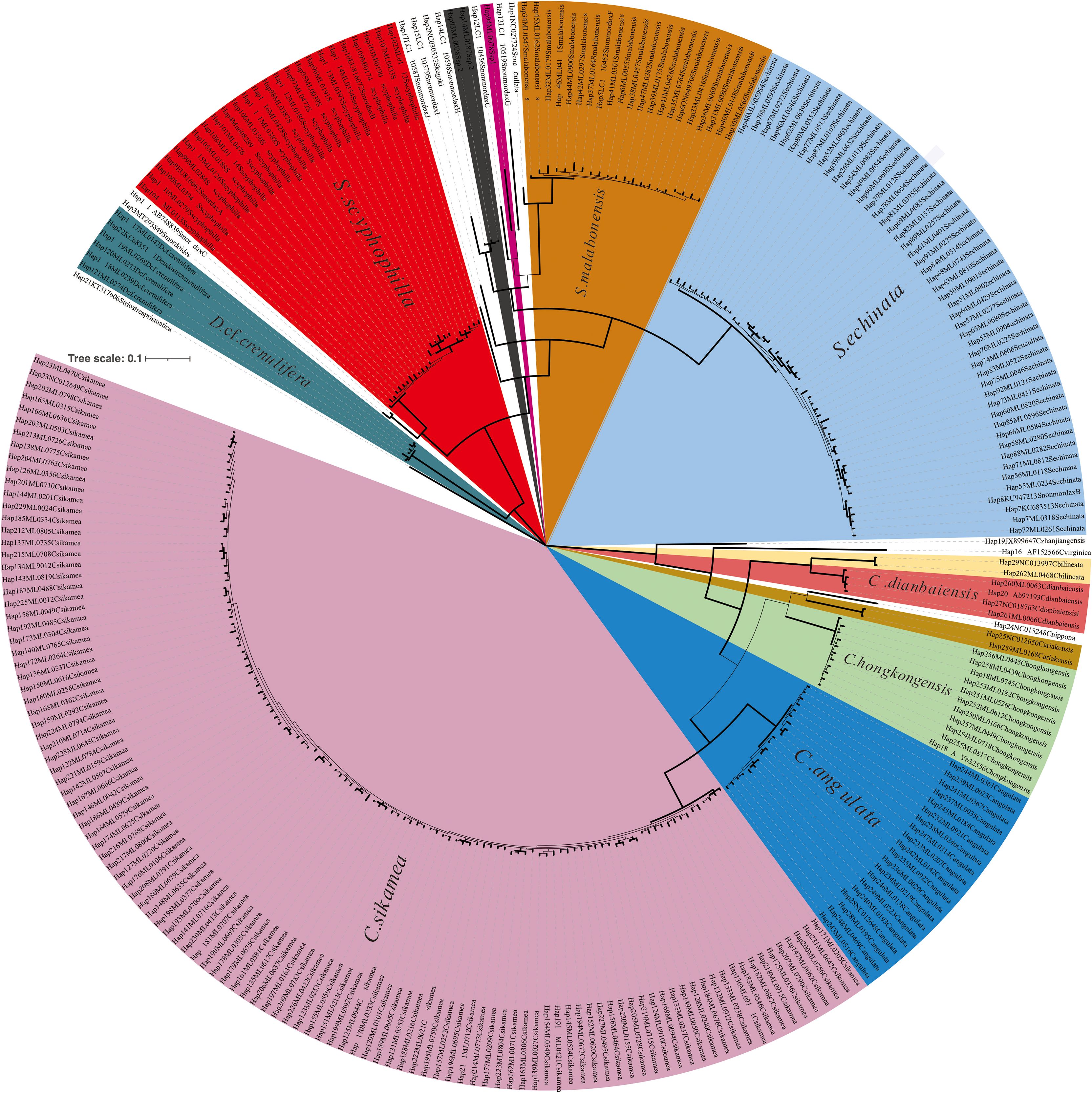
Figure 2. Bayesian phylogenetic tree of common oysters from oyster reefs along the Guangdong coastline, based on partial cytochrome oxidase I (COI) DNA sequences. Solid lines denote the posterior probability/bootstrap values greater than 60. Each taxon is labeled with its haplotype number, sequence number (or accession number), and tentative species name. The scale bar indicates the number of substitutions per site.
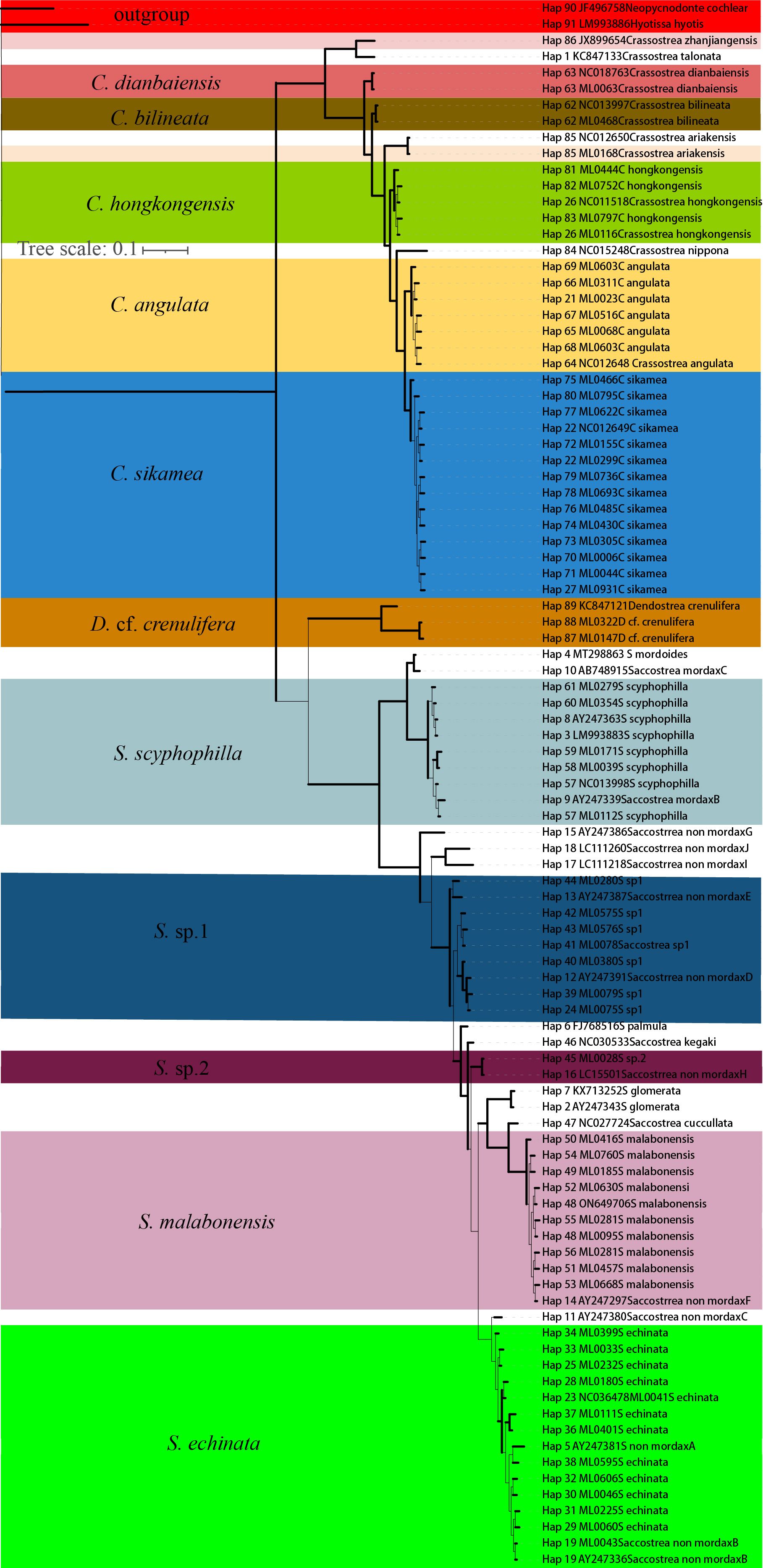
Figure 3. Bayesian phylogenetic tree of common oysters from oyster reefs along the Guangdong coastline, based on partial 16S rRNA sequences. Solid lines denote the posterior probability/bootstrap values greater than 60. See the Figure 2 legend for details.
Since the topological structures of the IQ-Tree and BI tree were generally congruent at the upper branches, with only minor difference observed at the lower branches at the interspecies level (e.g., the position of S. kegaki and S. non-mordax H) for both mitochondrial DNA, the BI tree was selected for further analysis. The corresponding IQ-Trees are provided in the Supplementary Figures 2, 3. Using Striostrea prismatica as the outgroup for BI and ML trees based on COI sequences, 12 oyster species across three genera were identified. The COI fragment used for phylogenetic analysis was 550 bp in length. The BI tree topology revealed that the representative species belonged to three subfamilies. At the species level, the following oysters formed a robust clade: C. sikamea, C. hongkongensis, C. angulata, C. ariakensis, C. dianbaiensis, C. bilineata, S. malabonensis, S. echinata, S. scyphophilla, S. sp. 1, S. sp. 2, and D. cf. crenulifera (Figures 4A–L).
Regarding BI and IQ trees based on 16S rRNA, Neopycnodonte cochlearo and Hyotissa hyotis were selected as outgroup species. In total, 12 oyster species across three genera were identified using the 16S rRNA genetic marker. The 16S rRNA fragment used for phylogenetic analysis was 495 bp in length. S. sp. 1 was closely related to S. non-mordax D, forming a sister group with S. non-mordax E, while S. sp. 2 grouped with S. non-mordax H in the 16S rRNA tree. Consequently, the 16S rRNA tree topology was largely congruent with that of the COI tree, except for variations in the interspecies relationships among S. non-mordax lineages within Saccostreinae.
3.2 Genetic structure and molecular diversity
Among the COI sequences, C. sikamea exhibited 112 haplotypes, accounting for 45.71% of the total haplotypes identified in this study. Saccostrea echinata exhibited 47 haplotypes, contributing 19.18% of the total haplotypes. Similarly, S. scyphophilla, with 23 haplotypes, exhibited the highest nucleotide and haplotype diversities with values of 0.0144 and 0.993, respectively. This pattern was consistent in genetic divergence analyses, with S. scyphophilla showing a greater pairwise distance compared to that of other rock oyster species (Tables 3, 4).
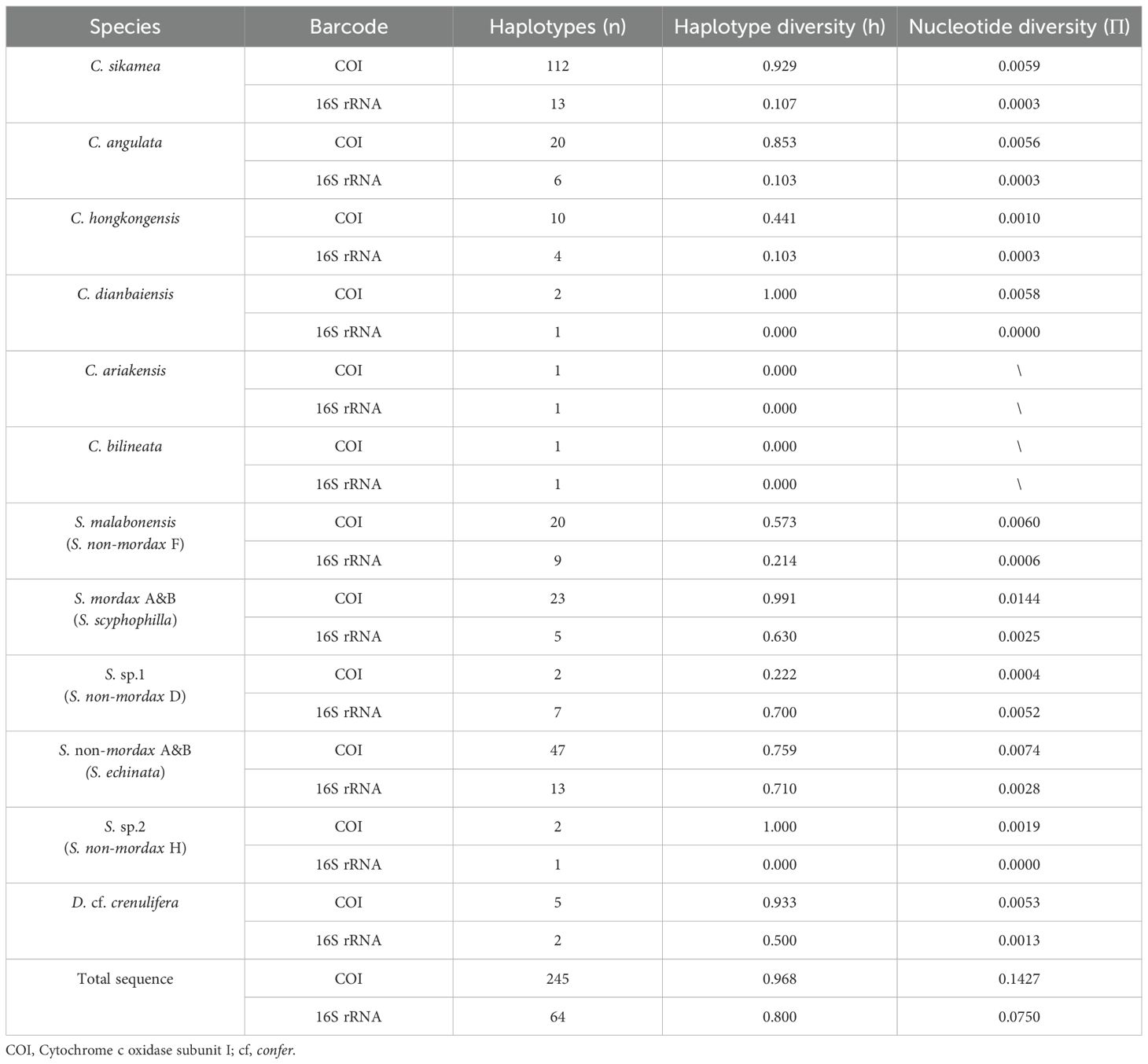
Table 3. Genetic diversity of common oysters in Guangdong Province based on cytochrome c oxidase subunit I (COI) and 16SrRNA.
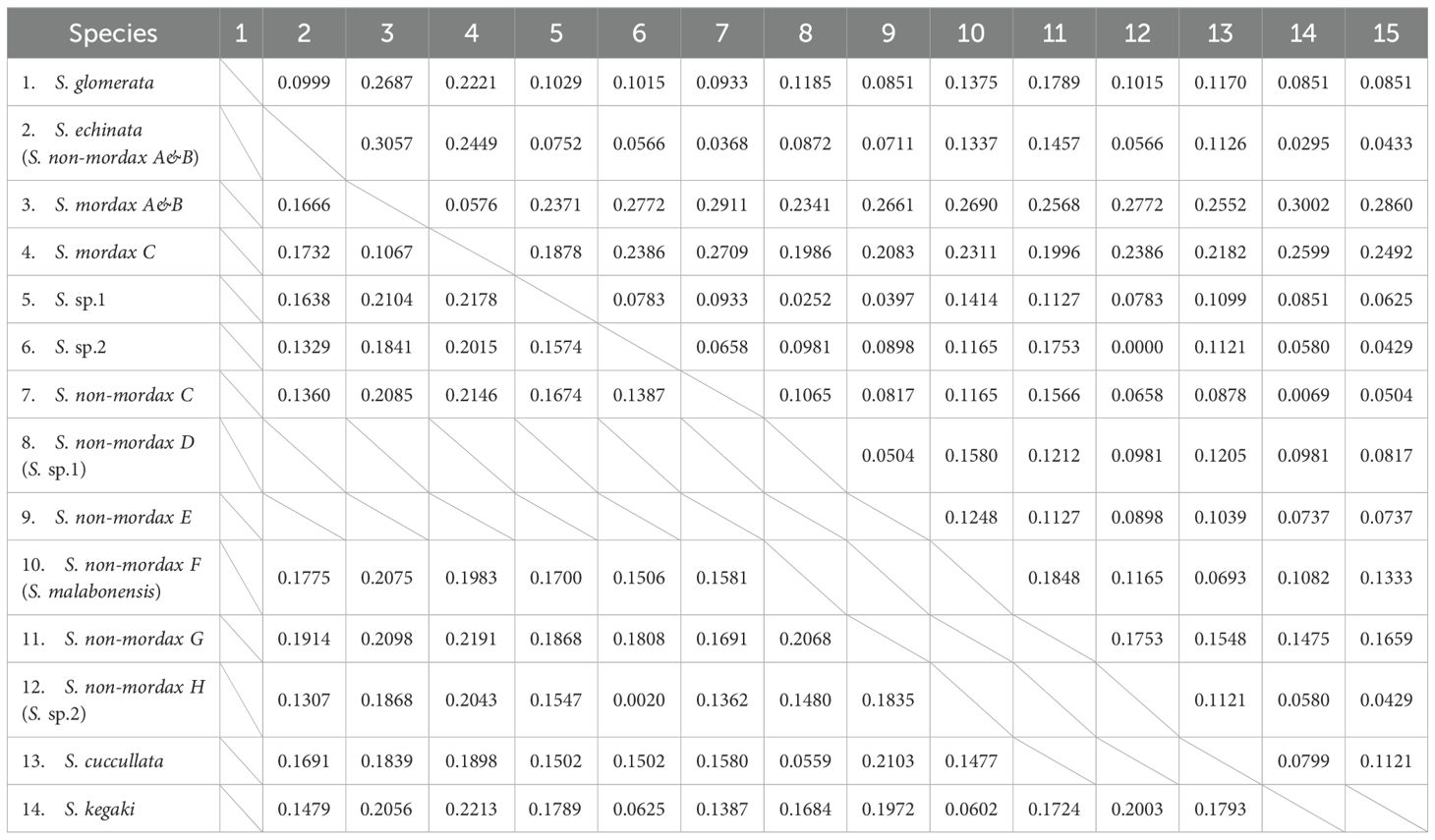
Table 4. Mean pairwise genetic divergence of the COI and 16S rRNA gene among Saccostrea oysters in Guangdong Province (upper diagonal: 16S RNA; lower diagonal: COI).
Regarding the 16S rRNA genetic locus, S. echinata and C. sikamea comprised 13 haplotypes each, representing 20.31% of the total haplotypes. S. echinata exhibited the highest nucleotide and haplotype diversities with values of 0.0028 and 0.710, respectively. A similar pattern was observed in genetic divergence analyses, with S. echinata and S. scyphophilla displaying greater pairwise distances compared to that of other rock oysters (Tables 1, 4).
3.3 Saccostrea sp. 1 and sp. 2
In this study, we described the shell morphology of the unidentified species, Saccostrea sp. 1 and sp. 2, to tentatively distinguish them from other oyster species. 9 and 23 individuals for COI sequences and 16S rRNA, respectively, were designated to Saccostrea sp. 1. These specimens were collected from rocky shores in Shantou (G3 and G4), Shanwei (G11, G13 and G15), Shenzhen (G21), Chaozhou (G01) and Xuwen (G51). Hap 41 of S. sp.1 for 16S rRNA was identified in all those regions (except Xuwen), making it the most common haplotype of Saccostrea sp. 1. Conversely, two individuals were identified as S. sp. 2, representing 2 and 1 haplotype for COI and 16S rRNA, respectively, via molecular analysis (Table 3). Specimens of S. sp. 2 were sourced from the rocky shore of Jieyang (G08) and Zhanjiang (G45).
The shell morphology of Saccostrea sp. 1 was characterized by its relatively small size compared to that of other rock oysters, with shell heights typically ranging from 20–40 mm (Figure 4G). The shell shape was usually elongated and elliptical, tapering towards the posterior end. The left valve was slightly cupped and primarily attached to the substrate. The left valve featured prominent, well-defined ribs that extended to the dorsal shell margin, forming distinct plications. The internal margin of the left valve possessed clearly visible chomata, and chalky deposits were present on the inner surface. The right valve, which featured brownish patches on its surface, was smaller compared to that of the left valve. The growth lines were prominent and concentric, forming a densely brownish growth lamella. Hyote spines were absent on the surface of the right valve. The adductor muscle scar was distinct and horseshoe-shaped, located in the posterior ventral half of the inner right valve. The hinge line was short and obliquely oriented.
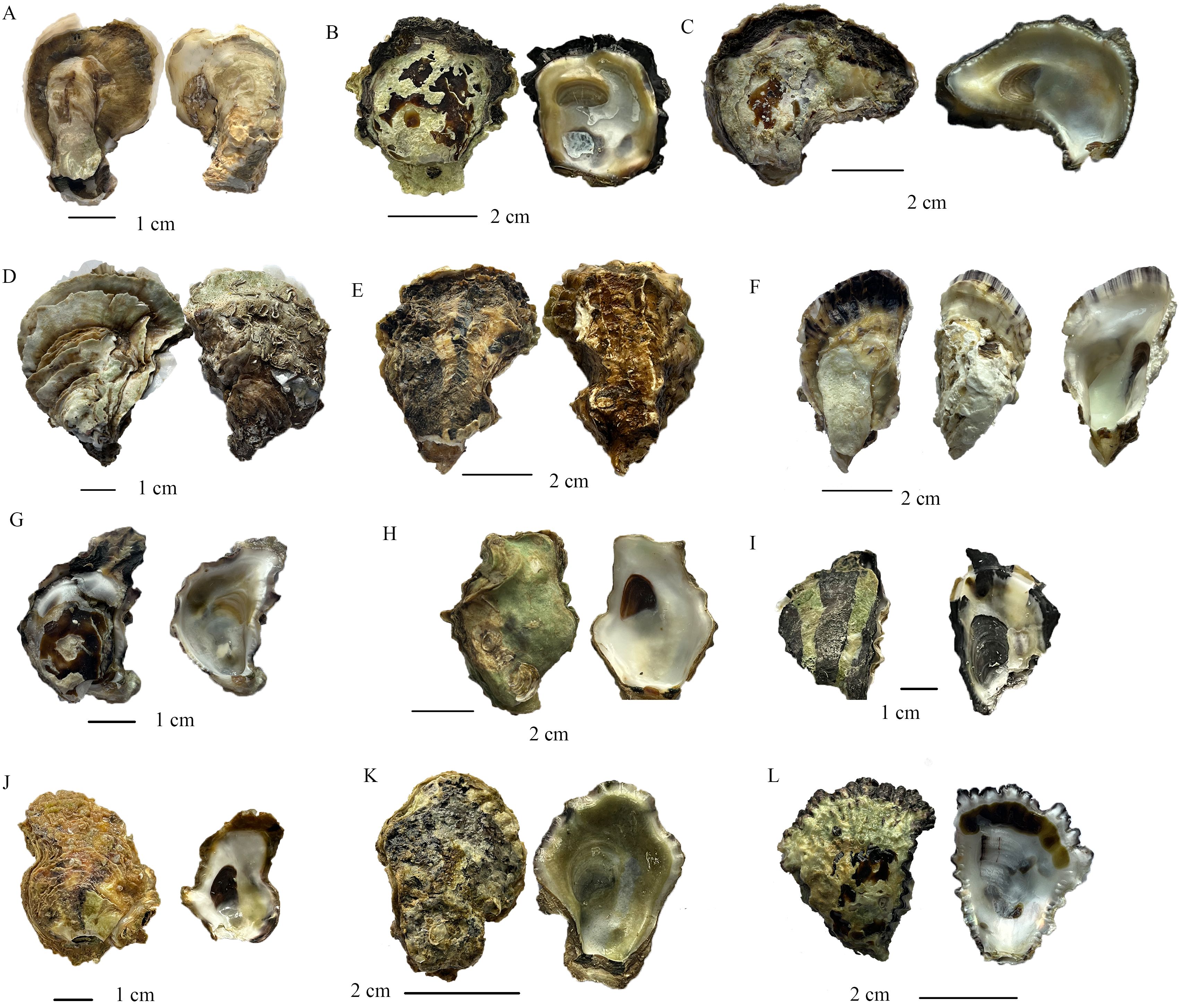
Figure 4. Shell morphology of oysters identified from oyster reefs in Guangdong Province, China. The identified oysters include the following: (A) Crassostrea dianbaiensis, (B) Saccostrea echinata, (C) S. malabonensis, (D) C. ariakensis, (E) C. angulata, (F) C. hongkongensis, (G) S. sp. 1 (S. non-mordax D in this study), (H) C. sikamea, (I) S. sp. 2 (S. non-mordax H in this study), (J) C. bilineata, (K) Dendostrea cf. crenulifera, (L) S. scyphophilla.
The detailed description of S. sp. 2 was limited owing to the scarcity of specimens. The shell shape of S. sp. 2 appeared flat and triangular (Figure 4I). There are no hyote spines observed on the outer surface of the right valve in our specimen. The growth lamellae and squamae were densely clustered along the shell margin, with distinct black deposition present. Radial ribs and the grooves between them were relatively faint but noticeable, especially along the shell margin of the left valve. Internally, marginal chomata were clearly present. The internal black and ochre pigment deposition was conspicuous, appearing as large spots. The adductor muscle scar was clearly visible and relatively large.
3.4 Geographic distribution pattern
In total, 33,549 oyster individuals for oyster morphological identification was sampled using a quantitative method to calculate density, biomass, and species dominance. The average density and biomass of C. sikamea were calculated as 1,415.38 individuals/m2 and 4,449.42 g/m2, making it the most predominated oyster species along the Guangdong shoreline (Figures 5, 6). This species was found across all survey sites, regardless of habitat type. Following C. sikamea, S. echinata had a mean density and biomass of 281.43 individuals/m2 and 1102.29 g/m2, contributing 13.11% and 13.42% to the total species richness and biomass, respectively. In this study, C. hongkongensis was the most common and dominant oyster species in the western district of the Pearl River Estuary, with an average density of 93.48 individuals/m2 and a biomass of 501.68 g/m2. Saccostrea malabonensis, although less abundant with a density and biomass of 40.45 individuals/m2 and 370.26 g/m2, respectively, was widely distributed across the study area, occurring at 40 of the 53 sites. All ambiguous Saccostrea identified in the quantitative survey (here referred to as S. non-mordax) were present in measurable quantities in our assay. The total number of species recorded at each site ranged from 1 to 6 species by qualitative survey, while G07, G11, G15, and G38 exhibited the highest richness with 6 oyster species found. At least 7 oyster species were found at G07 by quantitative method, being the most abundant area. Our study indicated G12 was the most abundant site with a density of 6779.83 ind/m2, while G08 was found to have the lowest abundance with a density of 626.16 ind/m2. G32 was the greatest area of biomass with the value reaching around 16000 g/m2. In contrast, the biomass of G46 was the lowest with only about value of 2000 g/m2 (Supplementary Table 1).
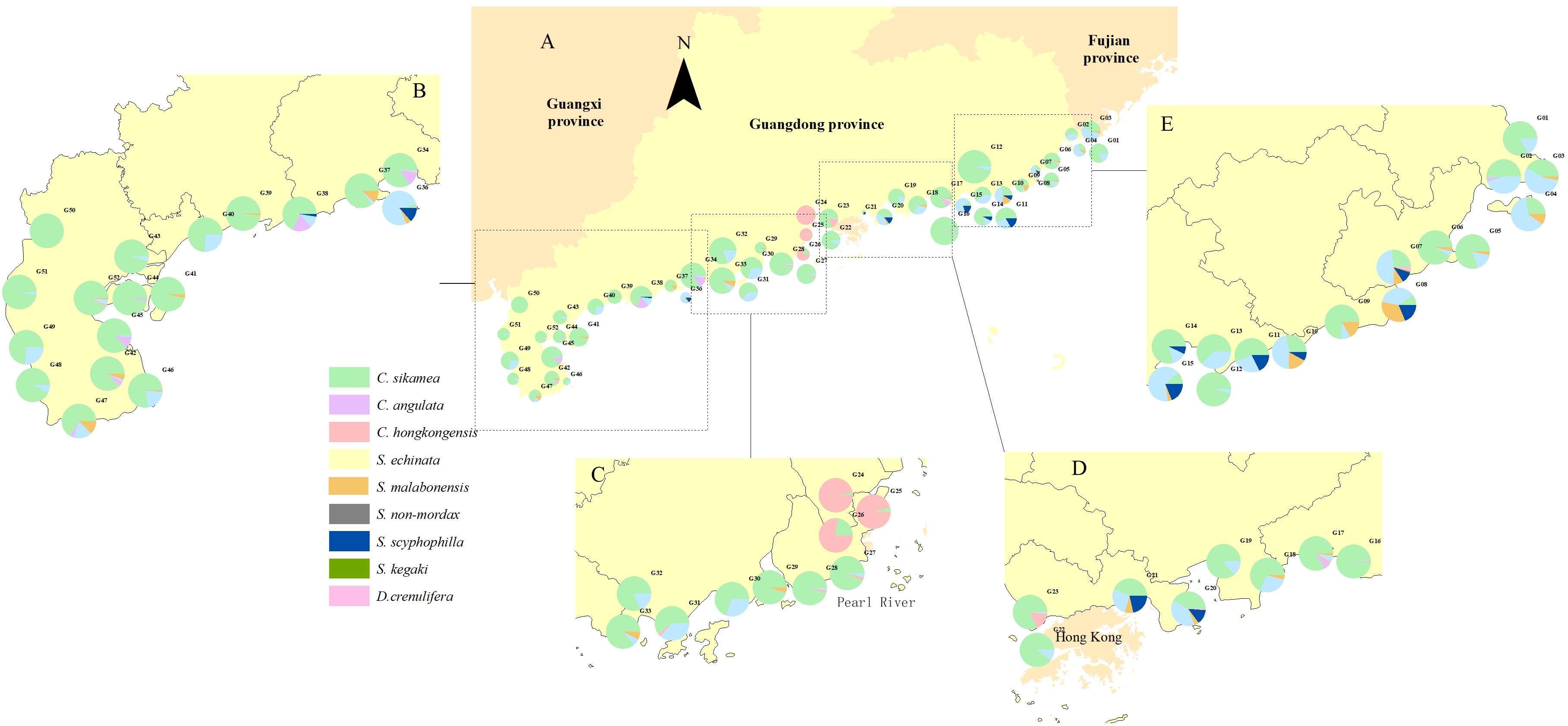
Figure 5. Spatial distribution of mean oyster density across 51 survey sites in Guangdong Province via quantitative survey. The circle size in the pie chart (A) represents the relative density of common oyster species at each site, while different colors indicate their percentage distribution within specific regions. The survey sites were divided into 4 districts: (B) the western part including Leizhou Peninsula, the central area (C, D) including Pearl River Delta, as well as the eastern part (E) of Guangdong Province.
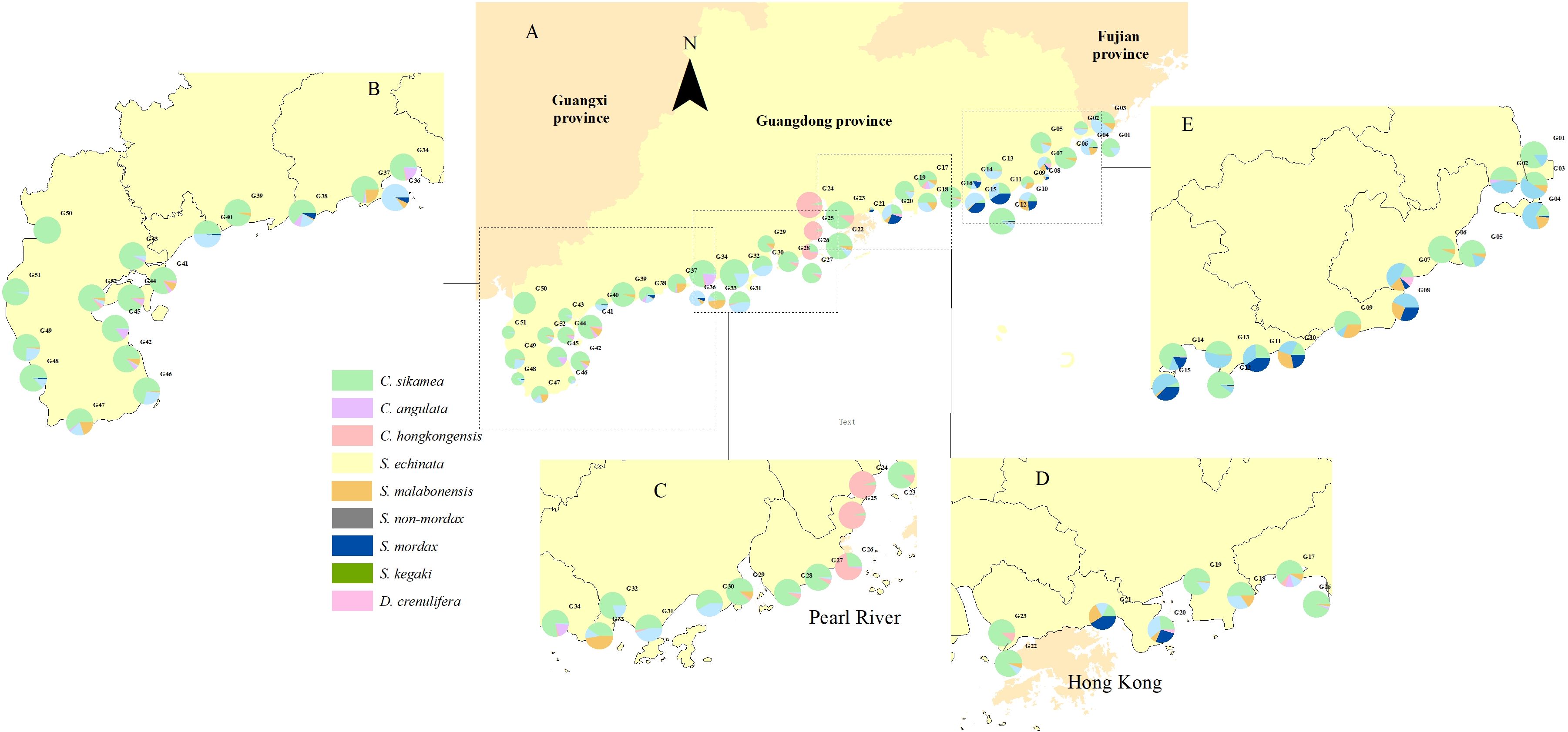
Figure 6. Spatial distribution of mean oyster biomass across 51 survey sites in Guangdong Province via quantitative survey. The circle size in the pie chart (A) represents the relative biomass of common oyster species at each site, while different colors indicate their percentage distribution within specific regions. The survey sites were divided into 5 districts: (B) the western part including Leizhou Peninsula, the central area (C, D) including Pearl River Delta, as well as the eastern part (E) of Guangdong Province.
4 Discussion
Historically, Guangdong’s estuarine and coastal systems supported some of the most extensive oyster reefs in South China, providing critical nursery habitats and contributing significantly to regional water quality. However, rapid coastal development, industrial pollution, and widespread mariculture have further compounded habitat degradation in Guangdong, threatening both wild and farmed oyster populations (Cardinale et al., 2012; Jiang et al., 2024). Given that Guangdong’s oyster reefs are influenced by complex salinity regimes driven by seasonal monsoons and freshwater discharges from the Pearl River, creating environmental heterogeneity that may promote high oyster species diversity and cryptic speciation. Therefore, a thorough investigation of oyster reefs in Guangdong Province is urgently needed.
4.1 Identification of oyster species
Currently, approximately 88 extant oyster species (Ostreidae) are recognized worldwide (Bayne et al., 2017; Li et al., 2021; Searles et al., 2021). Over 30 oyster species are found along the coast of China (Hu et al., 2019). In our analysis, population composition revealed at least 12 oyster species from three genera (Crassostrea, Saccostrea, and Dendostrea) and three subfamilies based on the phylogenetic systematics of Salvi, Raith, and Guo (Salvi et al., 2014; Raith et al., 2015; Guo et al., 2018) at the oyster reef of Guangdong. Given ongoing debates about the acceptance of new genera within Crassostreinae (e.g., Talonostrea Li and Qi, 1994, and Magallana Salvi & Mariottini, 2016), we continue to use the genus Crassostrea to represent cupped oysters in our study. Furthermore, because C. iredalei is suspected of having cryptic species and remains controversial (Guo et al., 2018), we considered it C. bilineata rather than C. iredalei. Similarly, due to challenges in phylogenetic analysis, Dendostrea was classified as D. cf. crenulifera in this study, as some D. crenulifera sequences uploaded to NCBI may be inconsistent. Further efforts are needed to clarify the relationship within Dendostrea. Our findings also suggest that S. mordax A & B lineage and S. scyphophilla are conspecific, as indicated by the placement of S. scyphophilla within the S. mordax A & B lineage in the phylogenetic tree, with minimal genetic divergence (Table 4). Therefore, it may be more accurate to refer to the entire S. mordax A& B lineage as S. scyphophilla (Raith et al., 2015; McDougall et al., 2024). S. mordax C lineage we haven’t found in our survey was referred to as S. mordoides (Cui et al., 2021).
4.2 Confusions of Saccostrinae
Genetic validation of rock oysters and phylogenetic divergence underpins conservation, sustainable utilization, and ecological restoration of these species. Saccostrinae remains insufficiently assessed by DNA analysis due to the subtle and unclear morphological distinction among Saccostrea species. Sekino and Yamashita (2016) identify 13 distinct Saccostrea lineages; however, most monophyletic clades have not been assigned valid species names. Currently, only a few of these lineages have been formally named. For example, S. non-mordax B is presumably recognized as S. echinata, while S. non-mordax F and J are assigned to S. malabonensis and S. spathulata, respectively (Cui et al., 2021; Snow et al., 2023).
S. cuccullata was once considered the sole valid Saccostrea species in the Indo-Pacific, and this superspecies was repeatedly used in earlier records of Guangdong province to represent Saccostrea (Harry, 1985; Xia, 2008). However, the geographical range of S. cuccullata is now recognized as being limited to the Atlantic, from the West Indies to the Arabian Peninsula (Huber, 2010), making this name inappropriate for oysters from the Indo-Pacific Saccostrea lineage. Our findings show that the rock oysters we collected did not cluster with S. cuccullata specimens from Madagascar (Volatiana et al., 2015), with a K2P sequence divergence of 5.6%–19.1% in COI and 2.5%–9.4% in 16S rRNA, suggesting that S. cuccullata does not occur along the Guangdong coastline. Another significant issue is the presence of S. glomerata in the northern hemisphere. Historical data have recorded the distribution of S. glomerata, referred to as “tuanju oysters,” along the Guangdong coastline (Phillips and Yim, 1981; Xu and Zhang, 2008; Wang, 2016; Cui et al., 2018). However, Lam and Morton, based on mitochondrial analysis, suggest that the common flattened oyster found in Hong Kong is not S. glomerata. They propose that native S. glomerata might only inhabit the temperate regions of the southern hemisphere (Lam and Morton, 2006). Our findings indicate that S. glomerata does not occur along the Guangdong coastline, as no taxonomic unit clustered with S. glomerata specimens from Southeastern Australia, with a K2P sequence divergence of 9.33%–26.87% in the 16S rRNA gene (Combosch et al., 2017). Considering the flattened shell form, absence of hyote spines, plication of the right valve margin, along with its relatively large size, and attachment to rocks, the oysters previously identified as S. glomerata along the southern China coastline may actually be S. malabonensis. This aligns with our findings, where S. malabonensis was identified as a common species on the rocky shores of Guangdong. Recently, the S. non-mordax lineage F was reclassified as S. malabonensis, supported by robust phylogeny, which matches our findings (Sekino and Yamashita, 2016; Cui et al., 2021). Therefore, we speculate that the occurrence and fertility of S. malabonensis in southern China may have been previously underestimated.
Our findings suggest that S. echinata may represent S. non-mordax A and B, as its tentative haplotypes are nested within lineages A and B in the 16S rRNA gene tree. S. non-mordax A and B exhibit shell morphology with hyote spines and emerge as sympatric species in our study and related research (Lam and Morton, 2006; Snow et al., 2023), with a limited K2P distance between them. This supports the idea that they may share a common lineage.
4.3 Saccostrea sp. 1 and sp. 2
In our assay, two cryptic species, Saccostrea sp. 1 and sp. 2 were found in oyster reefs along the Guangdong Province. Molecular data indicated that S. sp. 1 and S. sp. 2 were designated to known S. non-mordax lineage D and H, respectively. The average genetic distance between S. sp. 1 and other Saccostrea species ranged from 15.0%–21.7% for the COI gene and 2.5%–23.7% for the 16S rRNA gene. S. non-mordax lineage D showed the closest genetic relationship with S. sp. 1, followed by lineage E, leading us to classify S. sp 1 as lineage D rather than lineage E. This is not the first record of S. sp. 1 in Guangdong, as previous studies have documented its occurrence in Hong Kong, Hainan, and Taiwan (Lam and Morton, 2006), and its distribution has now expanded further into another district of Guangdong. S. sp. 1 appears to be exclusively distributed in adjoining areas of coastal China. The spatial distribution of Saccostrea oysters suggests that the density and biomass of S. sp. 1 may have been underestimated. However, morphological identification of S. sp. 1 remains challenging. The most distinctive shell features of the species, such as its relatively small size and flat shape tapering towards the posterior with a pointed end, are insufficient to distinguish it from other Saccostrea species, highlighting the challenge of identifying species within this genus.
Saccostrea sp. 2 formed a sister clade with known S. non-mordax H and was closely related to S. kegaki. Regarding K2P distance, S. sp. 2 showed the closest relationship to S. non-mordax H, with a genetic distance of 0% for the 16S rRNA gene and 2.00% for the COI gene. The genetic distance supports the intraspecies relationship. In contrast, S. sp. 2 and S. kegaki exhibited greater genetic divergence, with a 4.3% difference in the 16S rRNA gene and 6.3% in the COI gene, reinforcing the distinction between S. kegaki and lineage H. Considering that lineages A, B, and H both share hytote spines, Lam suggests that these lineages align with S. echinata sensu Torigoe (Torigoe and Inaba, 1981). In our phylogenetic analysis, only the IQ-Tree based on the COI gene supported this hypothesis (Supplementary Figure 2). However, another tree construction analysis indicated that lineage H did not cluster with lineages A or B but instead formed an independent lineage with S. kegaki as the basal clade (Figures 2, 3).
4.4 Transformation of oyster composition
A putative quantitative survey by morphological analysis were conducted to study oyster composition of Guangdong province. Previous studies of China’s southern coastline (Zhou and Jr., 2003; Lu et al., 2024) and the 2007 national coastal survey (Wu et al., 2025) in Guangdong Province reported substantial populations of C. ariakensis and C. angulata (Wang et al., 2010; Xia et al., 2014). In contrast, our study revealed that C. sikamea and S. echinata were the most predominant and endemic species at coastal of Guangdong Province, while historically dominant species like C. ariakensis and C. angulata were comparatively rare. Similarly, C. sikamea was also identified as the dominant oyster species along the Zhejiang coast to the north of Guangdong (Liu et al., 2021). These discrepancies may result from earlier studies relying on morphological traits or limited spatial sampling. Alternatively, climate change, widespread mariculture, and habitat alteration threatening both wild and farmed oyster populations over the past few decades could explain shifts in oyster species composition in this region. On the other hand, C. sikamea owning to its euryhaline and eurythermal characteristic possessed a specific capacity for adaption to coastline of China. A sufficient genetic variability and strength of gene flow also promote its performance of regional acclimation (Liu et al., 2021). Although historically confined to southern China, C. sikamea has recently expanded beyond its traditional range, now aggressively invading artificial shorelines in northern China (Hu and Dong, 2022). Its remarkable regional adaptation and acclimation may play a key role in its expansion. As such, our study provides an updated baseline for detecting future changes of this species and its distribution along the Guangdong coastline. From this view, the current situation underscores the need for proactive measures to safeguard and preserve oyster diversity. Additionally, C. talonata was found in mangrove of Beibu gulf and coastline in Hainan Island by previous surveys (Li and Qi, 1994; Lu et al., 2024), indicating that it was a common oyster in South China Sea. There are no specimens of C. talonata were detected in our survey though. The result indicated C. talonata was unusual species at oyster reefs in Guangdong province, which was identical to Xia’s result (Xia, 2008). The contrast might be due to its sparse distribution and its preference to subtidal area where it adhered to the surface of mesh cages or shells (Wang, 2016).
4.5 Habitat management and protection
Notably, wild-type C. hongkongensis were found between Yangjiang and Zhuhai (G24 - G37) mostly situated at Pearl River Delta in our study. However, this area suffered from habitat losses, tourist pressure, and contamination of terrestrial runoff as a result of rapid urban expansion. In consideration of the restricted area of this species and its essential position for traditional aquaculture in Guangdong Province, more efforts are needed to monitor its population dynamic process and genetic resources. Similarly, there was an ancient and naturally occurring oyster reef first documented at site G47 of Xuwen. However, the low diversity and richness of this site might suggest the poor condition recently. Establishing protected zones of this area for strengthening management may be a viable solution. In addition, our study outlined several critical regions that implied high levels of oyster diversity and abundance. At least 7 oyster species were found at site G07, which might suggest an oyster hotspot of species diversity for this area. Other sites of G38, G12 and G32 were another noteworthy area with high species richness, abundance and biomass. Overall, this study revealed some potential places for site selection of oyster habitat management, protection and restoration efforts. More importantly, it provided insights into where or which oyster species was presumably under population outbreak or population bottleneck.
5 Conclusion
In conclusion, this research enhances our understanding of oyster assemblages and the spatial distribution of oyster reefs in Guangdong Province. We collected over 30,000 oyster specimens, which were classified into 12 species across three genera within the Ostreidae family. The findings validate the monophyly of Crassostrea, Saccostrea, and Dendostrea and identify two cryptic species within Saccostrea. By combining molecular and morphological data, this study updates the species inventory of oysters along the Guangdong coastline and clarifies the existing Saccostrea identities and their systematic relationships. In the analysis of dominant species, C. sikamea and S. echinata were the most abundant across 51 survey sites. C. sikamea was more abundant than that of previous studies, while fewer C. ariakensis and C. angulata specimens were found. We also suggest several potential places including G07, G12, G32 and G38 for oyster management and protection. To conserve the environment and restore ecosystem service, implementing more effective management measures is crucial to alleviating local pressures. The findings presented here can serve as a foundation for developing corresponding management strategies.
Data availability statement
Sequences of S. sp.1 haplotype are deposited in NCBI with accession numbers PQ452111 and PQ459553-PQ459559 for COI gene and 16S rRNA gene respectively. Sequences of S. sp.2 haplotype were linked to accession number PQ455391-PQ455392 for COI gene and PQ459552 for 16S rRNA gene.
Ethics statement
The animal studies were approved by South China Sea Bureau, Ministry of Natural Resources. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. HQ: Investigation, Writing – original draft, Writing – review & editing. YC: Investigation, Writing – original draft, Writing – review & editing. J-wQ: Formal analysis, Writing – original draft, Writing – review & editing. YZ: Supervision, Validation, Writing – original draft, Writing – review & editing. JZ: Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Guangdong Science and Technology Plan Project (2021B1212110001), National Natural Science Foundation of China (42076129), National Key Research and Development Project (2022YFC3106303), and Science and Technology Development Fund Project of the South China Sea Bureau (200208007000199009).
Acknowledgments
We would like to sincerely thank Dr. Yanping Qin and Professor. Ziniu Yu for their instructive suggestion in the process of investigation. The authors would like to thank the editor and reviewers for their assistance in the manuscript revision process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1602823/full#supplementary-material
References
Baggett L. P., Powers S. P., Brumbaugh R., Coen L. D., DeAngelis B., Greene J., et al. (2014). Oyster habitat restoration monitoring and assessment handbook. Nat. Conservancy. Available online at: https://www.conservationgateway.org/ConservationPractices/Marine/Pages/oystermonitoringhandbook.aspx (Accessed August 16, 2025).
Bayne B. L., Ahrens M., Allen S. K., D’auriac M. A., and Wang H. (2017). The proposed dropping of the genus crassostrea for all pacific cupped oysters and its replacement by a new genus magallana: A dissenting view. J. Shellfish. Res. 36, 545–547. doi: 10.2983/035.036.0301
Beck M. W., Brumbaugh R. D., Airoldi L., Carranza A., Coen L. D., Crawford C., et al. (2009). Shellfish reefs at risk: a global analysis of problems and solutions. Nat. Conservancy. Available online at: https://www.conservationgateway.org/ConservationPractices/Marine/HabitatProtectionandRestoration/Pages/shellfishreefsatrisk.aspx (Accessed August 16, 2025).
Beck M. W., Brumbaugh R. D., Airoldi L., Carranza A., Coen L. D., Crawford C., et al. (2011). Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61, 107–116, 110. doi: 10.1525/bio.2011.61.2.5
Capella-Gutierrez S., Silla-Martinez J. M., and Gabaldon T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cardinale B. J., Duffy J. E., Gonzalez A., Hooper D. U., Perrings C., Venail P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 489, 59–67. doi: 10.1038/nature11148
Cheng J., Li L., Liu Q., Luo G., Quan W., Wang F., et al. (2022). Research report on the Conservation and Restoration of Oyster Reef Habitats in China (Beijing: The Nature Conservancy). Available online at: https://tnc.org.cn/content/details27_732.html (Accessed August 16, 2025).
Combosch D. J., Collins T. M., Glover E. A., Graf D. L., and Bieler R. (2017). A family-level Tree of Life for bivalves based on a Sanger-sequencing approach. Mol. Phylogenet. Evol. 107, 191–208. doi: 10.1016/j.ympev.2016.11.003
Cui Z., Hu L., Li C., Zhang Z., Guo X., and Wang H. (2021). Identification of Saccostrea mordax and a New Species Saccostrea mordoides sp. nov. (Bivalvia: Ostreidae) from China. J. Shellfish. Res. 40, 63–75. doi: 10.2983/035.040.0107
Cui Z., Huang J., Wang H., Hu L., and Liu Y. (2018). Classification and distribution of oysters in eastern coast of leizhou peninsula, China. Oceanol. Limnol. Sin. 49, 224–231. doi: 10.11693/hyhz20180300054
Fitzsimons J., Branigan S., Brumbaugh R. D., McDonald T., and zu Ermgassen P. (Eds.) (2019). Restoration guidelines for shellfish reefs (Arlington VA, USA: The Nature Conservancy).
Folmer O., Black M., Wr H., Lutz R., and Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Gerard T. and Jose C. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
GuerraGarcía J. M., Espinosa F., and GarcíaGómez J. C. (2008). Trends in taxonomy today: an overview about the main topics in taxonomy. Zool. Baetica. 19, 15–49. Available online at: https://www.ugr.es/~zool_bae/vol19/Zoo-2.pdf (Accessed August 16, 2025).
Guo X., Li C., Wang H., and Xu Z. (2018). Diversity and evolution of living oysters. J. Shellfish. Res. 37, 775–771. doi: 10.2983/035.037.0407
Hamaguchi M., Shimabukuro H., Usuki H., and Hori M. (2014). Occurrences of the Indo-West Pacific rock oyster Saccostrea cuccullata in mainland Japan. Mar. Biodivers. Records. 7, e84. doi: 10.1017/S1755267214000864
Harry H. W. (1985). Synopsis of the supraspecific classification of living oysters (bivalvia, gryphaeidae and ostreidae). Veliger 28, 121–158.
Hu L. and Dong Y. (2022). Multiple genetic sources facilitate the northward range expansion of an intertidal oyster along China’s coast. Ecol. Appl. 34, e2764. doi: 10.1002/eap.2764
Hu L., Wang H., Zhang Z., Li C., and Guo X. (2019). classification of small flat oysters of ostrea stentina species complex and a new species ostrea neostentina sp. nov. (bivalvia: ostreidae). J. Shellfish. Res. 38, 295–308. doi: 10.2983/035.038.0210
Hu L., Zhen Z., Wang H., and Tao (2018). Molecular phylogeography and population history of Crassostrea sikamea (Amemiya 1928) based on mitochondrial DNA. J. Exp. Mar. Biol. Ecol. 503, 23–30. doi: 10.1016/j.jembe.2017.11.004
Huber M. (2010). Compendium of bivalves. A full-color guide to 3,300 of the World’s Marine Bivalves. A status on Bivalvia after 250 years of research (Hackenheim: ConchBooks).
Huelsenbeck J. P. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Jackson J., Kirby M. X., Berger W. H., Bjorndal K. A., Botsford L. W., Bourque B. J., et al. (2001). Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637. doi: 10.1126/science.1059199
Jiang K., Chen C., Jiang G., Chi Y., Xu C., Kong L., et al. (2024). Genetic improvement of oysters: Current status, challenges, and prospects. Rev. Aquacult. 16, 796–817. doi: 10.1111/raq.12868
Jiang W., Shi W., Li N., Fan R., Zhang W., and Quan W. (2022). Oyster and barnacle recruitment dynamics on and near a natural reef in China: implications for oyster reef restoration. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.905373
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar S., Stecher G., and Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lam K. and Morton B. (2006). Morphological and mitochondrial-DNA analysis of the Indo-West Pacific rock oysters (Ostreidae: Saccostrea species). J. Molluscan. Stud. 72, 235–245. doi: 10.1093/mollus/eyl002
Li C., Kou Q., Zhang Z., Hu L., and Wang H. (2021). Reconstruction of the Evolutionary Biogeography Reveal the origins and diversification of Oysters (Bivalvia: Ostreidae). Mol. Phylogenet. Evol. 164, 107268. doi: 10.1016/j.ympev.2021.107268
Li L., Li A., Song K., Meng J., Guo X., Li S., et al. (2018). Divergence and plasticity shape adaptive potential of the Pacific oyster. Nat. Ecol. Evol. 2, 1751–1760. doi: 10.1038/s41559-018-0668-2
Li X. and Qi Z. (1994). Studies on the comparative anatomy, systematic classification and evolution of Chinese oysters. Stud. Mar. Sin. 35, 143–173.
Li C., Wang H., and Guo X. (2017). Classification and taxonomic revision of two oyster species from Peru: ostrea megodon (Hanley 1846) and crassostrea talonata (Li & Qi 1994). J. Shellfish. Res. 36, 359–364. doi: 10.2983/035.036.0208
Librado P. and Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Littlewood D. T. J. (1994). Molecular phylogenetics of cupped oysters based on partial 28S rRNA gene sequences. Mol. Phylogenet. Evol. 3, 221–229. doi: 10.1006/mpev.1994.1024
Liu S., Xue Q., Xu H., and Lin Z. (2021). Identification of main oyster species and comparison of their genetic diversity in zhejiang coast, south of yangtze river estuary. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.662515
Lu R., Liu Z., Chen Y., Ma P., Li C., Zhang Z., et al. (2024). Species composition and distribution of common Crassostrea and Saccostrea oysters along the coast of Hainan Island. J. Oceanol. Limnol. 42, 1–12. doi: 10.1007/s00343-024-3213-y
Ma H., Li L., Xiao S., Zhang Y., and Yu Z. (2021a). Microsatellite-based study of population genetics of Crassostrea hongkongensis in Southern China. Aquacult. Rep. 19, 100591. doi: 10.1016/j.aqrep.2021.100591
Ma H., Zhang Y., Xiang Z., Zhang Y., Qin Y., and Yu Z. (2021b). Development of tri-nucleotide microsatellite markers from Crassostrea hongkongensis using enriched genomic libraries and cross-species amplification in two closely related species. Aquacult. Rep. 19, 100592. doi: 10.1016/j.aqrep.2021.100592
McDougall C., Nenadic N., Richardson M., and Healy J. M. (2024). Molecular identification of intertidal rock oyster species in north-eastern Australia reveals new candidates for aquaculture. Aquaculture 587, 740838. doi: 10.1016/j.aquaculture.2024.740838
Morris R. L., La Peyre M. K., Webb B. M., Marshall D. A., Bilkovic D. M., Cebrian J., et al. (2021). Large-scale variation in wave attenuation of oyster reef living shorelines and the influence of inundation duration. Ecol. Appl. 31, e02382. doi: 10.1002/eap.2382
Palumbi S., Martin A., Romano S., Mcmillian W., Stice L., Grabowski G., et al. (1996). The simple fool’s guide to PCR version 2 (Honolulu: University of Hawaii Honolulu Press).
Phillips D. and Yim W. (1981). A comparative evaluation of oysters, mussels and sediments as indicators of trace metals in hong kong waters. Mar. Ecol. Prog. Ser. 6, 285–293. doi: 10.3354/meps006285
Raith M., Zacherl D. C., Pilgrim E. M., and Eernisse D. J. (2015). Phylogeny and species diversity of gulf of California oysters (Ostreidae) inferred from mitochondrial DNA*. Am. Malacol. Bull. 33, 1–21. doi: 10.4003/006.033.0206
Salvi D., Macali A., and Mariottini P. (2014). Molecular phylogenetics and systematics of the bivalve family ostreidae based on rRNA sequence-structure models and multilocus species tree. PloS One 9, e116014. doi: 10.1371/journal.pone.0108696
Salvi D. and Mariottini P. (2017). Molecular taxonomy in 2D: a novel ITS2 rRNA sequence-structure approach guides the description of the oysters’ subfamily Saccostreinae and the genus Magallana (Bivalvia: Ostreidae). Zool. J. Linn. Soc 179, 263–276. doi: 10.1111/zoj.12455
Searles A. R., Gipson E. E., Walters L. J., and Cook G. S. (2021). Oyster reef restoration facilitates the recovery of macroinvertebrate abundance, diversity, and composition in estuarine communities. Sci. Rep. 12, 8163. doi: 10.1038/s41598-022-11688-6
Sekino M., Ishikawa H., Fujiwara A., Doyola-Solis E. F. C., Lebata-Ramos M. J. H., and Yamashita H. (2015). The first record of a cupped oyster species Crassostrea dianbaiensis in the waters of Japan. Fish. Sci. 81, 267–281. doi: 10.1007/s12562-014-0838-3
Sekino M. and Yamashita H. (2016). Mitochondrial and nuclear DNA analyses of Saccostrea oysters in Japan highlight the confused taxonomy of the genus. J. Molluscan. Stud. 82, 492–506. doi: 10.1093/mollus/eyw022
Snow M., Fotedar S., Wilson N. G., and Kirkendale L. A. (2023). Clarifying the natural distribution of Saccostrea Dollfus and Dautzenberg 1920 (edible rock oyster) species in Western Australia to guide development of a fledgling aquaculture industry. Aquaculture 566, 739202. doi: 10.1016/j.aquaculture.2022.739202
Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772. doi: 10.1093/molbev/mst010
Tolley S. and Volety A. (2005). The role of oysters in habitat use of oyster reefs by resident fishes and decapod crustaceans. J. Shellfish. Res. 24, 1007–1012. doi: 10.2983/0730-8000(2005)24[1007:TROOIH]2.0.CO;2
Torigoe K. and Inaba A. (1981). On the scientific name of Japanese spiny oyster ‘Kegaki’. Venus 40, 126–134. Available online at: https://www.jstage.jst.go.jp/article/venusjjm/40/3/40_KJ00004342892/_pdf (Accessed August 16, 2025).
Volatiana J. A., Fang S., Kinaro Z. O., and Liu X. (2015). Complete mitochondrial DNA sequences of Saccostrea mordax and Saccostrea cuccullata: genome organization and phylogeny analysis. Mitochondrial. DNA Part A. DNA Mapp. Sequencing. Anal. 27, 3024. doi: 10.3109/19401736.2015.1063050
Wang H., Guo X., Zhang G., and Zhang F. (2004). Classification of jinjiang oysters Crassostrea rivularis (Gould 1861) from China, based on morphology and phylogenetic analysis. Aquaculture 242, 137–155. doi: 10.1016/j.aquaculture.2004.09.014
Wang H., Qian L., Liu X., Zhang G., and Guo X. (2010). Classification of a common cupped oyster from southern China. J. Shellfish. Res. 29, 857–866. doi: 10.2983/035.029.0420
Wang H., Zhang G., Liu X., and Guo X. (2008). Classification of common oysters from North China. J. Shellfish. Res. 27, 495–503. doi: 10.2983/0730-8000(2008)27[495:COCOFN]2.0.CO;2
Wu X., Xiao S., and Yu Z. (2013). Mitochondrial DNA and morphological identification of Crassostrea zhanjiangensis sp. nov.(Bivalvia: Ostreidae): a new species in Zhanjiang, China. Aquat. Living. Resour. 26, 273–280. doi: 10.1016/S0044-8486(03)00215-1
Wu X., Xu X., Yu Z., Wei Z., and Xia J. (2010). Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol. Phylogenet. Evol. 57, 448–454. doi: 10.1016/j.ympev.2010.05.029
Wu L., Zhang T., Zhang X., Zhang H., and Jiang C. (2025). The transformational practices of China’s coastal geological survey guided by the principles of Earth System Science over the past 40 years. Anthropocene. Coasts. 8, 11. doi: 10.1007/s44218-025-00070-4
Xia J. (2008). Identification, distribution of oysters from the South China Sea and development of microsatellite markers and application to genetic variation analysis in Crassostrea hongkongesis (Guangzhou: University of Chinese Academy of Sciences).
Xia J., Wu X., Xiao S., and Yu Z. (2014). Mitochondrial DNA and morphological identification of a new cupped oyster species Crassostrea dianbaiensis (Bivalvia: Ostreidae) in the South China Sea. Aquat. Living. Resour. 27, 41–48. doi: 10.1051/alr/2014004
Xu F. and Zhang S. (2008). An Illustrated Bivalvia Mollusca Fauna of China Seas (Beijing: Science press).
Zhang D., Gao F., Jakovli I., Zou H., and Wang G. T. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1101/489088
Zhang Y., Li J., Zhang Y., Ma H., Xiao S., Xiang Z., et al. (2017). Performance evaluation of reciprocal hybrids derived from the two brackish oysters, Crassostrea hongkongensis and Crassostrea sikamea in southern China. Aquaculture 473, 310–316. doi: 10.1016/j.aquaculture.2017.02.031
Keywords: oyster reef, species identification, biodiversity, spatial pattern, cryptic species
Citation: Zhou Z, Qiu H, Chen Y, Qiu J-w, Zhang Y and Zhang J (2025) Species identity and spatial patterns of common oysters at oyster reefs of Guangdong, China. Front. Mar. Sci. 12:1602823. doi: 10.3389/fmars.2025.1602823
Received: 30 March 2025; Accepted: 04 August 2025;
Published: 28 August 2025.
Edited by:
Anna Rita Rossi, Sapienza University of Rome, ItalyReviewed by:
Elizabeth Mansfield, Florida State University, United StatesMarc Hanke, University of Houston, United States
Copyright © 2025 Zhou, Qiu, Chen, Qiu, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghuai Zhang, emhhbmdqaW5naHVhaUBzY3MubW5yLmdvdi5jbg==
 Zihua Zhou
Zihua Zhou Hong Qiu1,2
Hong Qiu1,2 Yaohui Chen
Yaohui Chen Jian-wen Qiu
Jian-wen Qiu Yuehuan Zhang
Yuehuan Zhang